- 1Key Laboratory of Special Pathogens and Biosafety, Center for Biosafety Mega-Science, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China
- 2University of Chinese Academy of Sciences, Beijing, China
The RNA-dependent RNA polymerases (RdRPs) encoded by the RNA viruses are a unique class of nucleic acid polymerases. Each viral RdRP contains a 500–600 residue catalytic module with palm, fingers, and thumb domains forming an encircled human right hand architecture. Seven polymerase catalytic motifs are located in the RdRP palm and fingers domains, comprising the most conserved parts of the RdRP and are responsible for the RNA-only specificity in catalysis. Functional regions are often found fused to the RdRP catalytic module, resulting in a high level of diversity in RdRP global structure and regulatory mechanism. In this review, we surveyed all 46 RdRP-sequence available virus families of the positive-strand RNA viruses listed in the 2018b collection of the International Committee on Virus Taxonomy (ICTV) and chose a total of 49 RdRPs as representatives. By locating hallmark residues in RdRP catalytic motifs and by referencing structural and functional information in the literature, we were able to estimate the N- and C-terminal boundaries of the catalytic module in these RdRPs, which in turn serve as reference points to predict additional functional regions beyond the catalytic module. Interestingly, a large number of virus families may have additional regions fused to the RdRP N-terminus, while only a few of them have such regions on the C-terminal side of the RdRP. The current knowledge on these additional regions, either in three-dimensional (3D) structure or in function, is quite limited. In the five RdRP-structure available virus families in the positive-strand RNA viruses, only the Flaviviridae family has the 3D structural information resolved for such regions. Hence, future efforts to solve full-length RdRP structures containing these regions and to dissect the functional contribution of them are necessary to improve the overall understanding of the RdRP proteins as an evolutionarily integrated group, and our analyses here may serve as a guideline for selecting representative RdRP systems in these studies.
Introduction
First identified in the 1950s in the mengovirus and poliovirus (PV) related studies (Reich et al., 1961, 1962), the RNA-dependent RNA polymerases (RdRPs) encoded by the RNA viruses catalyze the RNA synthesis from RNA templates, and are responsible for the viral genome replication and transcription processes. As the essential and the most conserved protein from the RNA viruses, the RdRPs are attractive systems both for understanding the fundamentals of nucleic acid synthesis and for developing antiviral strategies. Each RdRP contains a catalytic module (or catalytic core) with an overall architecture resembling an encircled human right hand that is composed of the palm, fingers, and thumb domains (Hansen et al., 1997; Ago et al., 1999; Bressanelli et al., 1999; Lesburg et al., 1999). The size of the catalytic module is typically about 50 and 70 kilo-Dalton (kD) for primer-dependent and de novo RdRPs, respectively, regarding the RdRP initiation mode (Lesburg et al., 1999; Thompson and Peersen, 2004). However, the size of the RdRP protein could reach 240–450 kD (Kinsella et al., 2004; Liang et al., 2015; Gerlach et al., 2015), often due to the requirement of incorporating other functional modules or as a result of coevolution with different host species. To some extent, the conservation of the catalytic module and the diversity of the full-length RdRP protein are two important aspects in understanding this unique class of polymerases. In this review, we surveyed all 46 RdRP-sequence available virus families listed in the International Committee on Virus Taxonomy (ICTV)1 2018b collection from the positive-strand RNA virus category [ssRNA(+) in the ICTV genome composition assignment] and tend to provide an RdRP reference map based on known information of RdRP structure and function. The main purpose of this work is to place the current knowledge of these RdRPs in a broader context and to facilitate future studies in RdRPs with representative primary structure and/or with distinct functions beyond the catalytic module, thereby help improve the overall understanding of RdRP structure, function, and evolution. Based on sequence availability in the United States National Center for Biotechnology Information (NCBI) database2, one representative RdRP amino acid sequence was chosen for each virus family by giving the priority to the ICTV-suggested type species (Table 1). As an exception, one representative sequence was chosen for each virus genus for the Flaviviridae family, since very different RdRPs primary structure has been identified in this virus family. For simplicity, we used residues in the PV RdRP (also known as the 3Dpol protein) to define conserved sites.
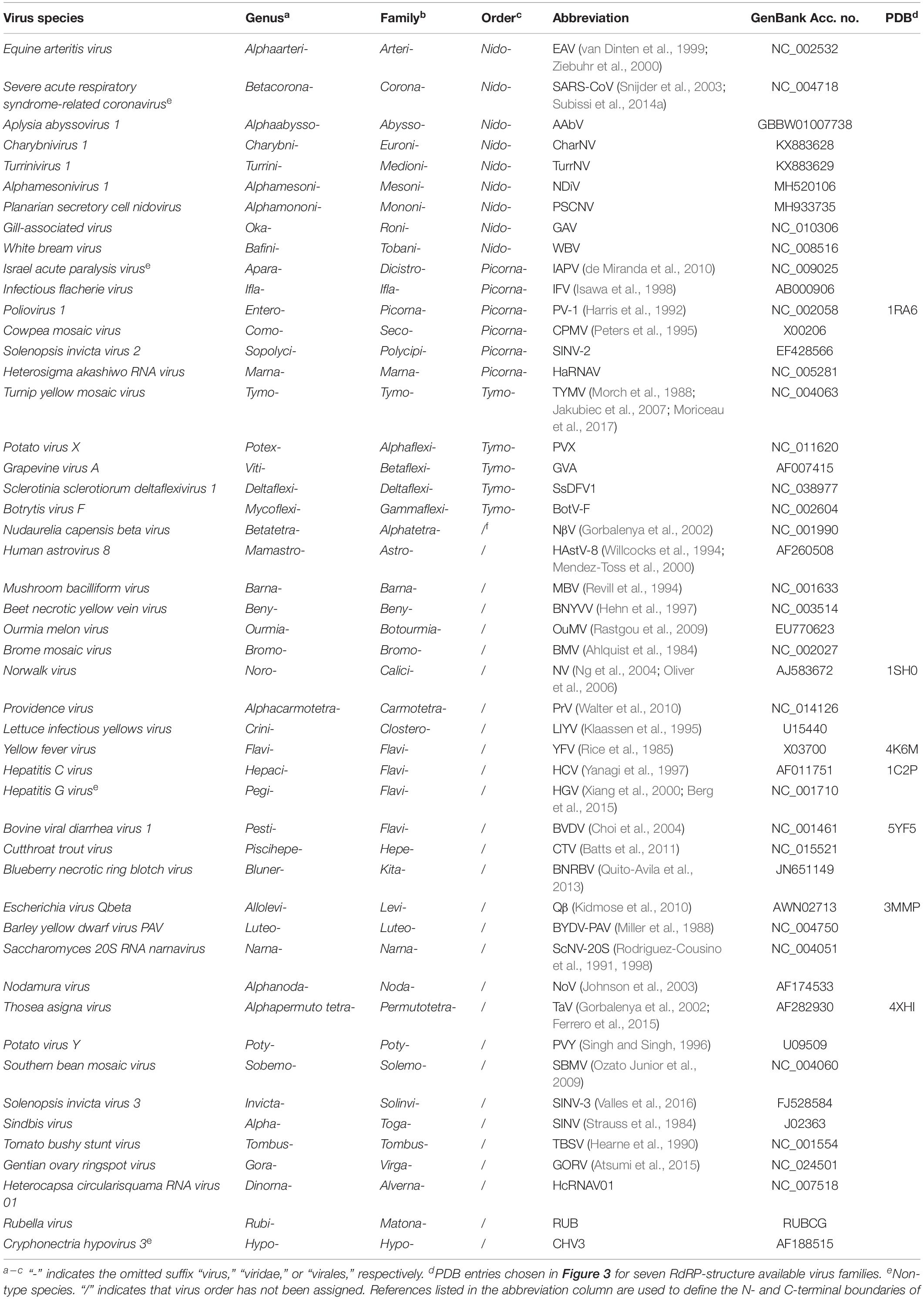
Table 1. Virus taxonomy assignments, virus name abbreviations, GenBank accession numbers, and related Protein Data Bank (PDB) entries of the representative positive-strand RNA virus RdRPs chosen for analyses in this study.
Conserved Catalytic Motifs and Residues as Reference Points for Defining the Boundaries of the RdRP Catalytic Module
A large number of RdRPs from the positive-strand RNA viruses are proteolytic products of viral polyproteins (Palmenberg, 1990; Wimmer and Nomoto, 1993; Reed and Rice, 2000; Bartenschlager et al., 2010; Pietila et al., 2017). Since not all related proteolytic cleavage sites have been reported for some of the virus families, we were only able to define N- and C-terminal boundaries for 33 RdRPs among the 49 representatives (Figure 1 and Table 1). For the rest of the RdRPs, 7 of them only have a defined C-terminus, and 9 of them have both termini undefined based on our best knowledge. Hence, functional studies to identify polyprotein proteolytic sites are necessary to improve the global picture RdRP primary structure diversity, and our analyses are based on incomplete boundary assignments. The overall size of the RdRPs with clear boundaries ranges from ∼460 to ∼1930 residues, indicating that the primary structure of these RdRPs are quite diverse and potential functional regions are likely integrated into some of these RdRP proteins.
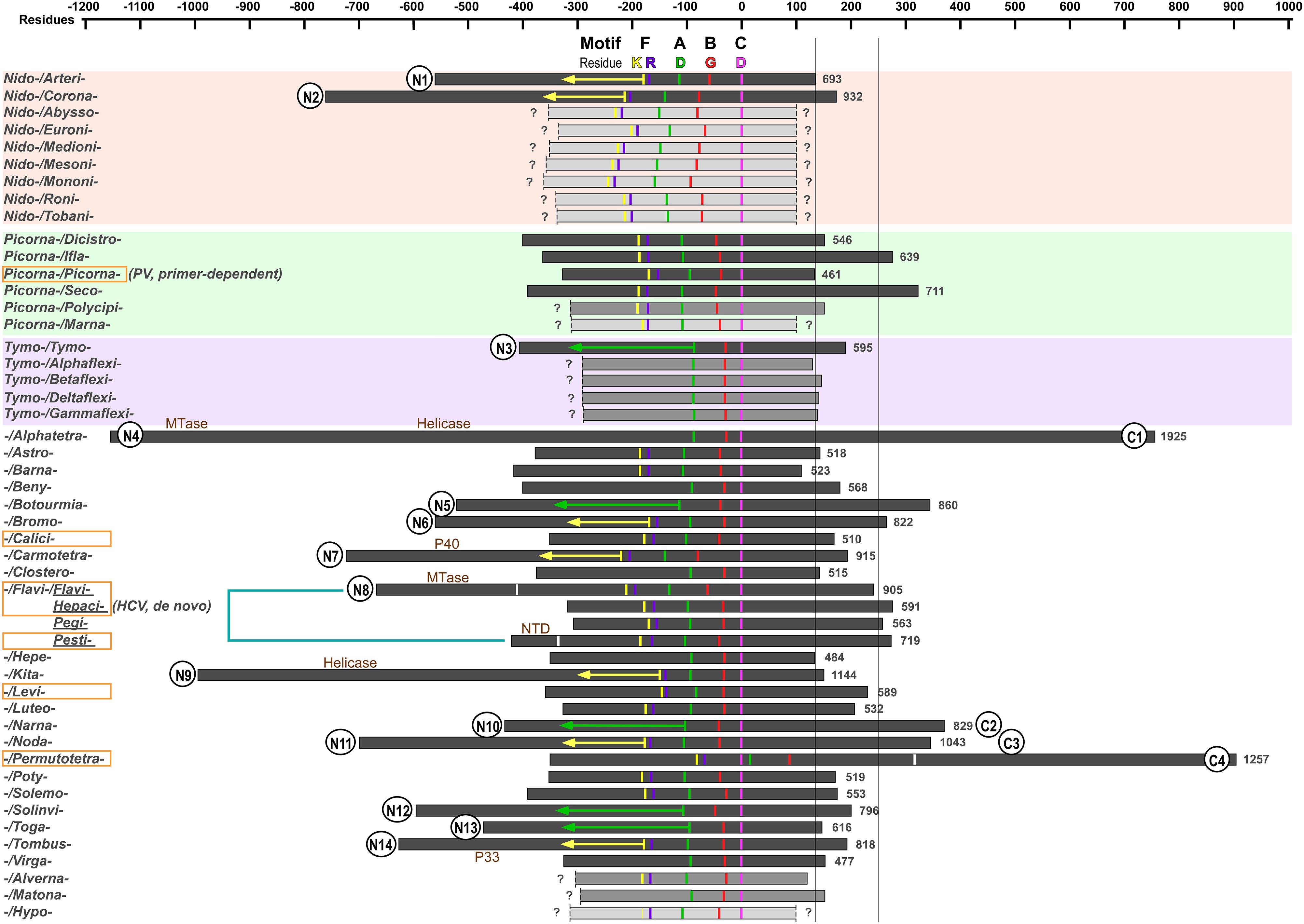
Figure 1. The primary structure comparison of RNA-dependent RNA polymerases (RdRPs) from representative positive-strand RNA viruses. Orders, families, genera, and species assignments based on the ICTV 2018b collection are listed in Table 1. The virus species names are listed in alphabetical order giving the priority to virus order, then to virus family, and then to virus genus. The conserved motif C aspartic acid (magenta, corresponding to PV RdRP D328) is used as the origin in the scale bar. Conserved residues in motifs A, B, and F are also labeled: motif F lysine (corresponding to PV RdRP K159) in yellow; motif F arginine (corresponding to PV RdRP R174) in purple; motif A aspartic acid (corresponding to PV RdRP D233) in green; motif B glycine (corresponding to PV RdRP G289) in red. The orange rectangle indicates that 3D structures are available in that virus family (or virus genus in case of the Flaviviridae). The boundaries of the RdRP catalytic module defined by reported 3D structures are indicated by the white bars for the Flaviviridae and Permutotetraviridae RdRPs. Numbers on the right side of individual RdRP indicate the amino acids numbers for the full-length RdRPs. The question mark (?) indicates undefined boundaries of the RdRP proteins. The yellow and green arrows (150 and 230 residues in length, respectively) are used to estimate the N-terminal boundary of the RdRPs. The two long vertical bars (130 and 250 residues to the origin) indicate the C-terminal boundary of the RdRP catalytic module of the primer-dependent PV 3Dpol and the de novo HCV NS5B and are used to help predict additional functional regions. Wherever available in literature, the name of additional functional regions are labeled. Circles placed at the RdRP termini indicate predicted additional regions. The numbers following the “N” and “C” simply refer to the number of families possibly having additional regions at the RdRP N- and C-termini, respectively.
In order to assign or identify possible functional regions beyond the RdRP catalytic module, we use the RdRP catalytic motifs and highly conserved residues to help estimate the boundaries of the catalytic module. The RdRP active site is surrounded by the palm, fingers, and thumb domains with seven catalytic motifs (motifs A–G) distributed within the palm (motifs A–E) and fingers (motifs F–G) (Poch et al., 1989; Gorbalenya et al., 2002; Bruenn, 2003; te Velthuis, 2014; Wu et al., 2015) (see an alignment of motif A–C of the 49 representative RdRP sequences in Figure 2). RdRPs share motifs A/C/D with DNA-dependent polymerases and A–F with the reverse transcriptases (RTs, RNA-dependent DNA polymerases) (Poch et al., 1989; Delarue et al., 1990; Gong and Peersen, 2010), while motif G is an RdRP hallmark motif that may participate in RNA template binding and post-catalysis RdRP translocation on the template (Gorbalenya et al., 2002; Shu and Gong, 2016). Although each catalytic motifs may be well conserved at the levels of virus genus and family, highly conserved residues across different virus families can only be identified in motifs A/B/C/F, and only three residues are absolutely conserved (Figure 2). Among these three residues, two aspartic acid residues in motifs A and C (corresponding to the PV RdRP residues D233 and D328) participate in the coordination interactions with the two divalent metal ions essential for the phosphoryl transfer reaction, and are also required for other classes of polymerases (Beese and Steitz, 1991; Huang et al., 1998; Li et al., 1998; Yin and Steitz, 2004; Zamyatkin et al., 2008; Gong and Peersen, 2010; Appleby et al., 2015). The third absolutely conserved residue is a glycine (corresponding to the PV RdRP residue G289) in motif B. This residue is typically adjacent to an serine and this SG dipeptide plays essential roles in recognizing the 2′-hydroxyl group of the nucleotide triphosphate (NTP) substrate, while the corresponding peptide bond flip accompanies a subtle conformational change of the NTP-induced RdRP active site closure identified by crystallography (Gong and Peersen, 2010; Appleby et al., 2015; Shu and Gong, 2016). It has also been suggested that this glycine residue may be essential for a 3-Å tip movement of the motif B loop (corresponding to the PV RdRP residues 288–292) that could participate in the aforementioned post-catalysis RdRP translocation during each nucleotide addition cycle (NAC) (Sholders and Peersen, 2014). Under either situation, the backbone flexibility of this glycine residue likely explains its requirement at this position. In some cases, the serine residue is replaced by a threonine or even rarely by other residues, and the threonine substitution can also be found in nucleoside analog drug-resistant virus stains (Figure 2; Dutartre et al., 2006; Lam et al., 2012; Flint et al., 2014), suggesting that the side-chain hydroxyl group is the core conservative part of this residue. Motif F typically contains several basic residues and is known to interact with the triphosphate and base moieties of the NTP substrate. Among these residues, one lysine and one arginine (corresponding to the PV RdRP residues K159 and R174) have the highest conservation level (Bruenn, 2003). Hence, we use the absolutely conserved motif C aspartic acid (the first D in the signature sequence XGDD) as the reference point to align all RdRP sequences and use the aforementioned conserved residues to label motifs A/B/C/F (Figure 1). In this way, the relative spacing of these key motifs can be compared in all representative sequences. Typically, the seven motifs appear in the order of G-F-A-B-C-D-E and follow the same protein folding topology. Very interestingly, the RdRPs from the Permutotetraviridae family has a different motif order of G-F-C-A-B-D-E. While its spatial organization of the motifs is consistent with that of other RdRPs, the folding topology is permutated (Gorbalenya et al., 2002; Ferrero et al., 2015). A similar situation was found in RdRPs from the Birnaviridae family in the double-stranded (ds) RNA virus category (Gorbalenya et al., 2002; Pan et al., 2007). These exceptions suggest that the swapping of the motifs could occur during protein evolution, while the catalytic function could remain largely unaffected. Besides the similarity in the order of RdRP catalytic motifs between the Birnaviridae and the Permutotetraviridae, the Birnaviridae viruses also use a VPg (viral protein genome linked)-mediated initiation mechanism for genome replication and a polyprotein coding strategy that are often found in the positive-strand RNA viruses (Lee et al., 1977; Pan et al., 2007). These observations suggest that the evolutionary boundary between the positive-strand and ds RNA viruses are not definite.
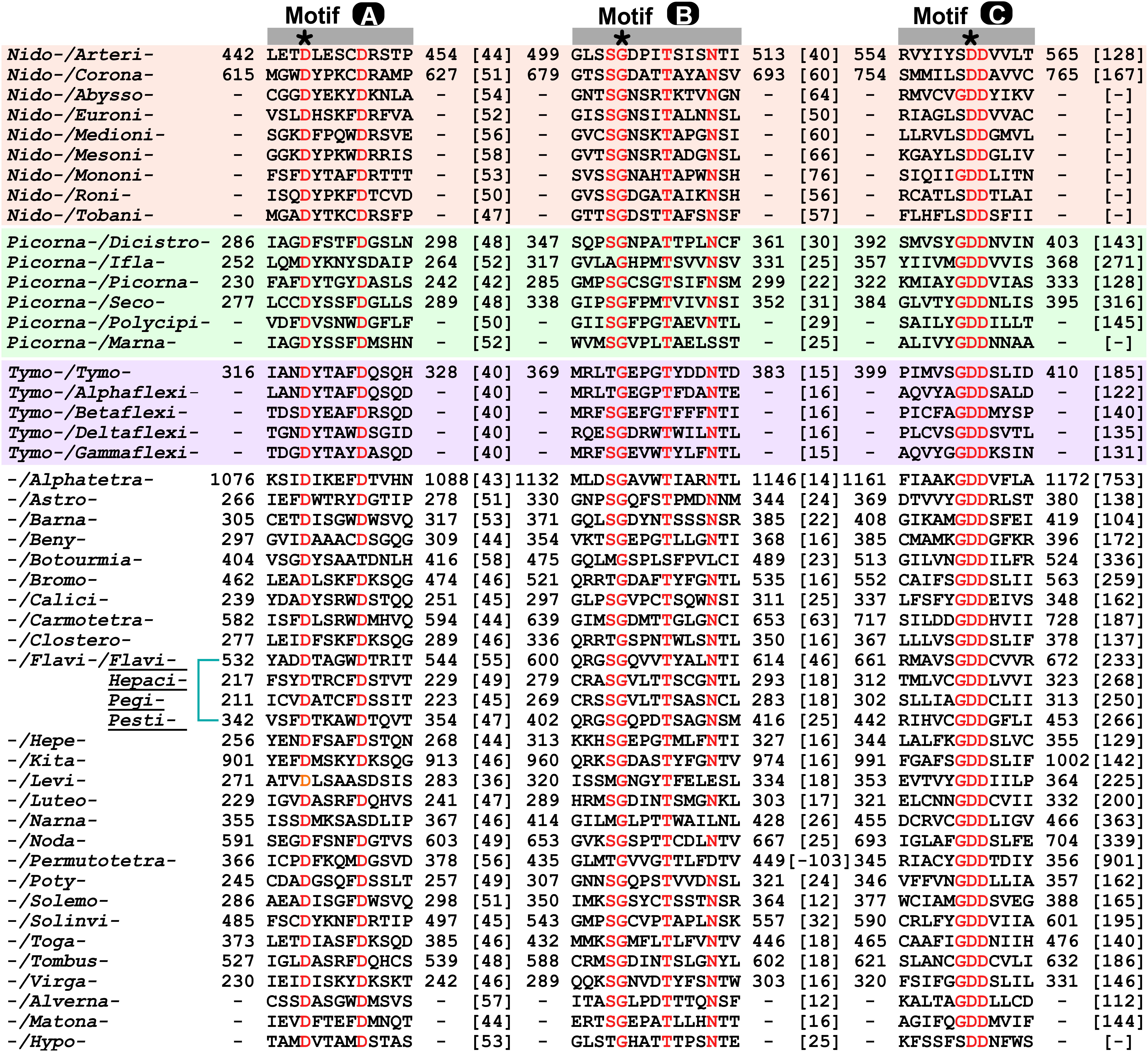
Figure 2. Sequence alignment of motifs A-C of RdRPs from representative positive-strand RNA viruses. The order of motif sequences from top to bottom are consistent with that in Figure 1 and Table 1. Highly conserved residues, including three absolutely conserved residues (labeled by asterisks), are shown in red. Numbers within the brackets indicate the number of residues not shown.
Next, we use two representative RdRPs, the PV 3Dpol and the hepatitis C virus (HCV) NS5B to help estimate the boundaries of the catalytic module using conserved residues in motifs A/B/C/F as the reference points (Figure 1). The first reason for choosing these two representatives is that these two proteins are known not to contain functional regions beyond the catalytic module except that the NS5B protein has a 21-residue membrane anchor at its C-terminus (Schmidt-Mende et al., 2001). The second reason is that 3Dpol and NS5B represent RdRPs that utilize primer-dependent and de novo mechanisms to initiate the RNA synthesis, respectively (Wimmer and Nomoto, 1993; Zhong et al., 2000). Both of these proteins have its N-terminus ∼150 or ∼230 residues away from the conserved motif F lysine or motif A aspartic acid (corresponding to the yellow and green bars in Figure 1), respectively, while the residue distances between C-terminal boundary of the catalytic module and the conserved motif C aspartic acid are different (∼130 residues for 3Dpol vs. ∼250 residues for NS5B). The thumb domain usually starts from 50–60 residues after the XGDD sequence and ends at the C-terminal boundary of the catalytic module. The primer-dependent 3Dpol contains four helices in the thumb, while the de novo NS5B contains seven. If compared to 3Dpol, NS5B has one insertion between the third and fourth helices, three extra helices after the fourth helix, and a C-terminal extension (Hansen et al., 1997; Lesburg et al., 1999). It has been suggested that the insertion and the extension together form a priming platform, interacting with the 3′-end of the template and the initiating NTPs to facilitate the de novo initiation (Luo et al., 2000; Appleby et al., 2015). In subsequent analyses, we use 150 or 230 residues from motif F lysine or motif A aspartic acid to estimate the N-terminal boundary (corresponding to the yellow and green arrows in Figure 1) and 250 residues from the motif C aspartic acid to estimate the C-terminal boundary (corresponding to the vertical bar on the right hand side in Figure 1) for RdRPs without three-dimensional (3D) structure reported.
Representative 3D Structures of RdRPs From the Positive-Strand RNA Viruses
Three-dimensional RdRPs structures have been reported for about 20 positive-strand RNA virus species (Hansen et al., 1997; Lesburg et al., 1999; Ng et al., 2002, 2004; Choi et al., 2004; Ferrer-Orta et al., 2004; Love et al., 2004; Fullerton et al., 2007; Malet et al., 2007; Yap et al., 2007; Campagnola et al., 2008; Takeshita and Tomita, 2010; Wu et al., 2010; Lu and Gong, 2013; Vives-Adrian et al., 2014; Ferrero et al., 2015; Bi et al., 2017; Upadhyay et al., 2017; Wang et al., 2017; Liu et al., 2018). However, these species only cover five virus families (Picornaviridae, Caliciviridae, Flaviviridae, Leviviridae, and Permutotetraviridae) (Figures 1, 3). Among structure-available RdRPs in each virus family, only the RdRPs from the Flaviviridae exhibit apparent global structure diversity and have three distinct structural forms. Therefore, a total of seven RdRP structures, including three from the Flaviviridae, were chosen as representatives for a schematic illustration of RdRP global structure diversity in positive-strand RNA viruses (Figure 3). Among these seven structures, five of them do not contain functional regions beyond the polymerase catalytic module (Lesburg et al., 1999; Ng et al., 2002; Thompson and Peersen, 2004; Takeshita and Tomita, 2010; Ferrero et al., 2015), although the full-length Thosea asigna virus (TaV) RdRP does contain a large C-terminal region (discussed below). While the structural details are quite different, all these structures are composed of the palm, fingers, and thumb domains and share similar global architecture. The flavivirus NS5 and the pestivirus NS5B, both from the Flaviviridae family, are the only RdRP structures contain additional functional regions (Lu and Gong, 2013; Liu et al., 2018). The N-terminal ∼260 residues of the flavivirus NS5 is a methyltransferase (MTase) that participates in the 5′-capping process of the virus RNA genome (Egloff et al., 2002; Koonin, 1993). Based on full-length NS5 crystal structures solved in Japanese encephalitis virus (JEV), dengue virus (DENV), and Zika virus (ZIKV), the MTase adopts the Rossmann fold and interacts with the RdRP fingers domain intra-molecularly in two different modes, one represented by the JEV and ZIKV structures and the other represented by the DENV structures (Lu and Gong, 2013; Upadhyay et al., 2017; Zhao et al., 2015). The N-terminal ∼90 residues of the pestivirus NS5B folds into a small α/β globular domain (namely NTD). The NTD forms intra-molecular interactions with the RdRP palm domain (Li et al., 2018; Liu et al., 2018). Collectively, only a couple of representative RdRP structural forms contain functional regions beyond the RdRP catalytic module. However, the following primary structure analysis suggest that numerous representative RdRPs may have functional regions fused to the catalytic module, in particular to the N-terminus.
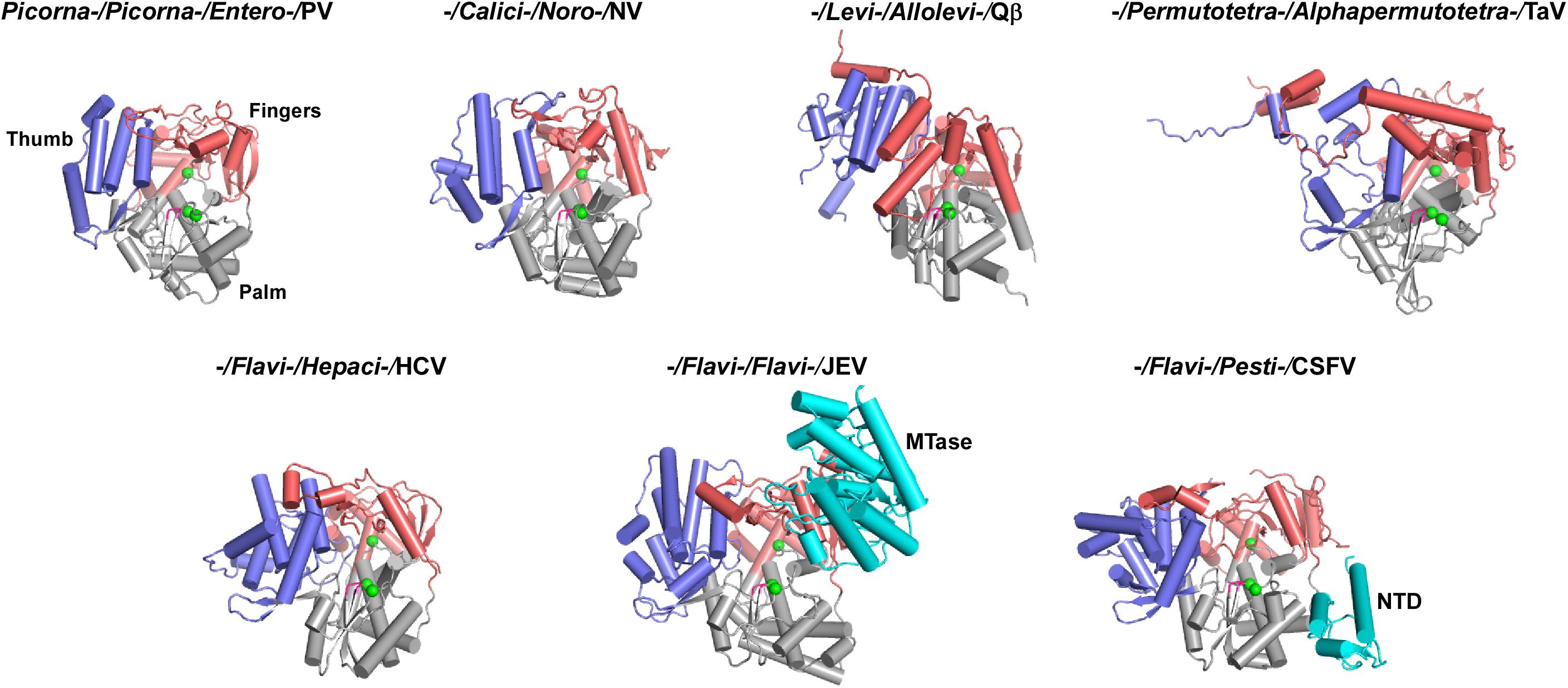
Figure 3. Global views of seven representative RdRPs 3D structures. RdRP structures are shown in cartoon representations. If available, order/family/genus/species assignments are shown on top of each structure. PV, poliovirus, PDB entry 1RA6 (chain A); NV, norovirus, PDB entry 1SH0 (chain A); Qβ, bacteriophage Qβ, PDB entry 3MMP (chain G); TaV, Thosea asigna virus, PDB entry 4XHI (chain A); HCV, hepatitis C virus, PDB entry 1C2P (chain A); JEV, Japanese encephalitis virus, PDB entry 4K6M (chain A); CSFV, classical swine fever virus, PDB entry 5YF5 (chain A). Coloring scheme: RdRP palm in gray, thumb in blue, fingers in pink, and signature sequence XGDD in magenta. The α-carbon atom of the three absolutely conserved amino acid residues (labeled by asterisks in Figure 2) are shown as green spheres. The N-terminal additional regions, if present, are shown in cyan.
The Diversity of the RdRP Primary Structure in the Positive-Strand RNA Viruses
With the assignment of the key motifs and the size estimation between these motifs and boundaries of the catalytic module, we are able to predict whether functional regions exist at both N- and C-terminal sides of the catalytic module and the approximate size of these regions using the aforementioned criterion, in particular, for those RdRPs with defined boundaries. Interestingly, most of these additional functional regions with an estimated size of 100 residues or larger were found at the N-terminal side of the catalytic module (14 out of 30 families with defined N-terminus), while much fewer showed up at the C-terminal side (4 out of 37 families with defined C-terminus) (Figure 1). The preference of “recruiting” N-terminal regions may be related to two factors. Firstly, among all 46 virus families surveyed, 22 families have the RdRP coding region located at the 3′-end of a polyprotein open reading frame (ORF), while 16 families have the RdRP coding region in the middle of an ORF and 8 families have the RdRP coding region as an independent ORF. Secondly, the de novo RdRPs tend to have important initiation elements located at its C-terminus (e.g., the aforementioned priming platform components in HCV NS5B). Both of these factors could reduce the opportunity for RdRPs to recruit additional regions to their C-termini during their evolution. Among these RdRPs containing additional functional regions, some of them have long drawn attentions in the related field but without much advances in structure and related functional characterization of the additional regions. The Coronaviridae nsp12 and Arteriviridae nsp9 have a ∼200–400 residue N-terminal region with both the structure and function remaining elusive (Gorbalenya et al., 1989; Xu et al., 2003; Beerens et al., 2007; te Velthuis et al., 2010). The Coronaviridae nsp8, which can form a supercomplex with nsp7 (each protein contributes eight copies) (Zhai et al., 2005), may have RNA-dependent primase activities (Imbert et al., 2006), and were shown to facilitate the nsp12 RdRP activities along with nsp7 (Subissi et al., 2014b). However, whether and how the N-terminal region of nsp12 might participate in RdRP catalysis or interactions with nsp7/nsp8 remain unclarified. The Permutotetraviridae RdRP that contains a ∼600-residue C-terminal region only has the catalytic module structure solved (Ferrero et al., 2015). The nearly 2000-residue Alphatetraviridae RdRP has an MTase and a helicase in its N-terminal region and a ∼500-residue C-terminal region with unknown function (Gorbalenya et al., 2002). The Kitaviridae RdRP has a helicase in its N-terminal region (Quito-Avila et al., 2013). The Togaviridae nsP4 protein has a ∼150-residue N-terminal region that may interact with other viral replication proteins (Lemm et al., 1990; Tomar et al., 2006). The N-terminal ∼300–350 residues of the Tombusviridae and Carmotetraviridae RdRPs can be produced as proteins (named P33 and P40, respectively) due to a UAG stop codon within the RdRP ORF (Kidmose et al., 2010; Walter et al., 2010). However, the function of these N-terminal regions either as individual proteins or as portions of the RdRP proteins is unknown. Solving the 3D structures of these RdRPs, in particular in their full-length form, is essential to the understanding of these RdRPs. Aside from these RdRPs with relatively large additional regions, some RdRPs with small additional regions may have evolved important functions as well. In the Flaviviridae family RdRPs, the hepacivirus and pestivirus NS5B proteins have a 21–24 residue hydrophobic membrane anchor at their C-termini, facilitating its involvement in the replication complex that is located in membranous vesicles derived from endoplasmic reticulum (ER) (Lai et al., 1999; Schmidt-Mende et al., 2001; Appel et al., 2006; Romero-Brey and Bartenschlager, 2014). The ∼90-residue NTD of the pestivirus NS5B modulates the fidelity of RNA synthesis through its intra-molecular interactions with the RdRP palm domain (Liu et al., 2018). Therefore, it will also be quite interesting to dissect the mechanisms involving small but potentially functional regions that may not be readily predicted in our boundary analysis.
Discussion
Viral RdRPs represent a unique nucleic acid polymerase class of and is the only class that does not involve DNA in any stages of the synthesis. To preserve their RNA template and ribonucleotide triphosphate (rNTP) substrate specificity, the seven catalytic motifs are the central segments to preserve during virus evolution. While we have mainly focused on the diversity and variations beyond the catalytic modules, attentions may also be drawn to the variations within the catalytic module but excluding the catalytic motifs. For the representative RdRPs that we surveyed in this study, the spacing between certain catalytic motifs could vary to a great extent. For example, the residue distance between the conserved motif B serine and the motif C aspartic acid in these RdRPs ranges from 30 to 94, corresponding to a motif spacing of 12–76 residues (Figures 1, 2). We hypothesize that such regions, if located at or near the RdRP protein surface, may have been utilized by the positive-strand RNA viruses as evolutionary “hot spot,” in particular in their host adaptation processes. Further investigations are needed to test this hypothesis.
Although we tried to survey the positive-strand RNA virus RdRPs in a comprehensive manner, there are still several limitations in our analyses. Firstly, the virus species that have not been assigned at the virus family level are not included in our analyses. Secondly, some virus families contain a large number of virus genera (e.g., 47 in Picornaviridae and 14 in Tombusviridae), suggesting high-level of genome and RdRP diversity within individual families. Moreover, RdRP primary structure diversity and genome-level diversity may not be consistent. The Flaviviridae is such an example with only four genera but three drastically different primary RdRP structures. Therefore, choosing one representative RdRP for each family (with the Flaviviridae as the only exception) may not be sufficient and ideal. Thirdly, the boundary estimation of the catalytic module is only a crude assessment. For example, the distance between the N-terminal boundary of the TaV RdRP catalytic module to the conserved motif F lysine is at least 100 residues longer than estimated distance using the 150-residue criterion used in our analyses (Ferrero et al., 2015).
In summary, we collected representative RdRPs encoded by positive-strand RNA viruses mainly at the level of virus family. By locating highly conserved residues within catalytic motifs within the RdRP ORF and by referencing structural and functional information of RdRPs in the literature, we tried to estimate the boundaries of the RdRP catalytic module in the full-length RdRP. Numerous regions beyond the RdRP catalytic module exist and many of them have either structure, or function, or both to be determined. Collectively, the global structure and regulatory functions related to regions beyond the catalytic module of the positive-strand RNA virus polymerases are quite diverse, and the current knowledge of these proteins is limited to only a few virus families. One purpose of our analyses is to provide a general guideline for researchers interested in these RdRP proteins and related viral systems to selectively or systematically investigate RdRPs with representative features. The global view of the positive-strand RNA virus RdRPs will continue to evolve with new virus species assigned, new structures determined, new functional regions identified, and new mechanisms dissected.
Author Contributions
Both authors surveyed the literature, analyzed the data, and wrote the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFA0507200, 2016YFC1200400, and 2018YFD0500100); the National Natural Science Foundation of China (31670154 and 31802147); the Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory, Chinese Academy of Sciences, China (2018ACCP-MS06); the “One-Three-Five” Strategic Programs, Wuhan Institute of Virology, Chinese Academy of Sciences, China (Y605191SA1).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Ago, H., Adachi, T., Yoshida, A., Yamamoto, M., Habuka, N., Yatsunami, K., et al. (1999). Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7, 1417–1426.
Ahlquist, P., Dasgupta, R., and Kaesberg, P. (1984). Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J. Mol. Biol. 172, 369–383. doi: 10.1016/s0022-2836(84)80012-1
Appel, N., Schaller, T., Penin, F., and Bartenschlager, R. (2006). From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 281, 9833–9836. doi: 10.1074/jbc.r500026200
Appleby, T. C., Perry, J. K., Murakami, E., Barauskas, O., Feng, J., Cho, A., et al. (2015). Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775. doi: 10.1126/science.1259210
Atsumi, G., Tomita, R., Yamashita, T., and Sekine, K. T. (2015). A novel virus transmitted through pollination causes ring-spot disease on gentian (Gentiana triflora) ovaries. J. Gen. Virol. 96, 431–439. doi: 10.1099/vir.0.071498-0
Bartenschlager, R., Cosset, F. L., and Lohmann, V. (2010). Hepatitis C virus replication cycle. J. Hepatol. 53, 583–585. doi: 10.1016/j.jhep.2010.04.015
Batts, W., Yun, S., Hedrick, R., and Winton, J. (2011). A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res. 158, 116–123. doi: 10.1016/j.virusres.2011.03.019
Beerens, N., Selisko, B., Ricagno, S., Imbert, I., van der Zanden, L., Snijder, E. J., et al. (2007). De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J. Virol. 81, 8384–8395. doi: 10.1128/jvi.00564-07
Beese, L. S., and Steitz, T. A. (1991). Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 10, 25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x
Berg, M. G., Lee, D., Coller, K., Frankel, M., Aronsohn, A., Cheng, K., et al. (2015). Discovery of a novel human pegivirus in blood associated with Hepatitis C Virus co-infection. PLoS Pathog. 11:e1005325. doi: 10.1371/journal.ppat.1005325
Bi, P., Shu, B., and Gong, P. (2017). Crystal structure of the coxsackievirus A16 RNA-dependent RNA polymerase elongation complex reveals novel features in motif A dynamics. Virol. Sin. 32, 548–552. doi: 10.1007/s12250-017-4066-8
Bressanelli, S., Tomei, L., Roussel, A., Incitti, I., Vitale, R. L., Mathieu, M., et al. (1999). Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 96, 13034–13039.
Bruenn, J. A. (2003). A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 31, 1821–1829. doi: 10.1093/nar/gkg277
Campagnola, G., Weygandt, M., Scoggin, K., and Peersen, O. (2008). Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J. Virol. 82, 9458–9464. doi: 10.1128/JVI.00647-08
Choi, K. H., Groarke, J. M., Young, D. C., Kuhn, R. J., Smith, J. L., Pevear, D. C., et al. (2004). The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. U.S.A. 101, 4425–4430. doi: 10.1073/pnas.0400660101
de Miranda, J. R., Cordoni, G., and Budge, G. (2010). The acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J. Invertebr. Pathol. 103(Suppl. 1), S30–S47. doi: 10.1016/j.jip.2009.06.014
Delarue, M., Poch, O., Tordo, N., Moras, D., and Argos, P. (1990). An attempt to unify the structure of polymerases. Protein Eng. 3, 461–467. doi: 10.1093/protein/3.6.461
Dutartre, H., Bussetta, C., Boretto, J., and Canard, B. (2006). General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob. Agents Chemother. 50, 4161–4169. doi: 10.1128/aac.00433-06
Egloff, M. P., Benarroch, D., Selisko, B., Romette, J. L., and Canard, B. (2002). An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21, 2757–2768. doi: 10.1093/emboj/21.11.2757
Ferrero, D. S., Buxaderas, M., Rodriguez, J. F., and Verdaguer, N. (2015). The structure of the RNA-dependent RNA polymerase of a permutotetravirus suggests a link between primer-dependent and primer-independent polymerases. PLoS Pathog. 11:e1005265. doi: 10.1371/journal.ppat.1005265
Ferrer-Orta, C., Arias, A., Perez-Luque, R., Escarmis, C., Domingo, E., and Verdaguer, N. (2004). Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279, 47212–47221. doi: 10.1074/jbc.m405465200
Flint, M., McMullan, L. K., Dodd, K. A., Bird, B. H., Khristova, M. L., Nichol, S. T., et al. (2014). Inhibitors of the tick-borne, hemorrhagic fever-associated flaviviruses. Antimicrob. Agents Chemother. 58, 3206–3216. doi: 10.1128/AAC.02393-14
Fullerton, S. W., Blaschke, M., Coutard, B., Gebhardt, J., Gorbalenya, A., Canard, B., et al. (2007). Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 81, 1858–1871. doi: 10.1128/jvi.01462-06
Gerlach, P., Malet, H., Cusack, S., and Reguera, J. (2015). Structural insights into Bunyavirus replication and its regulation by the vRNA promoter. Cell 161, 1267–1279. doi: 10.1016/j.cell.2015.05.006
Gong, P., and Peersen, O. B. (2010). Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510. doi: 10.1073/pnas.1007626107
Gorbalenya, A. E., Koonin, E. V., Donchenko, A. P., and Blinov, V. M. (1989). Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17, 4847–4861. doi: 10.1093/nar/17.12.4847
Gorbalenya, A. E., Pringle, F. M., Zeddam, J. L., Luke, B. T., Cameron, C. E., Kalmakoff, J., et al. (2002). The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 324, 47–62. doi: 10.1016/s0022-2836(02)01033-1
Hansen, J. L., Long, A. M., and Schultz, S. C. (1997). Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5, 1109–1122. doi: 10.1016/s0969-2126(97)00261-x
Harris, K. S., Reddigari, S. R., Nicklin, M. J., Hammerle, T., and Wimmer, E. (1992). Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J. Virol. 66, 7481–7489.
Hearne, P. Q., Knorr, D. A., Hillman, B. I., and Morris, T. J. (1990). The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177, 141–151. doi: 10.1016/0042-6822(90)90468-7
Hehn, A., Fritsch, C., Richards, K. E., Guilley, H., and Jonard, G. (1997). Evidence for in vitro and in vivo autocatalytic processing of the primary translation product of beet necrotic yellow vein virus RNA 1 by a papain-like proteinase. Arch. Virol. 142, 1051–1058. doi: 10.1007/s007050050141
Huang, H., Chopra, R., Verdine, G. L., and Harrison, S. C. (1998). Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282, 1669–1675. doi: 10.1126/science.282.5394.1669
Imbert, I., Guillemot, J. C., Bourhis, J. M., Bussetta, C., Coutard, B., Egloff, M. P., et al. (2006). A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25, 4933–4942. doi: 10.1038/sj.emboj.7601368
Isawa, H., Asano, S., Sahara, K., Iizuka, T., and Bando, H. (1998). Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: evidence for evolutionary relationships among insect, mammalian and plant picorna(-like) viruses. Arch. Virol. 143, 127–143. doi: 10.1007/s007050050273
Jakubiec, A., Drugeon, G., Camborde, L., and Jupin, I. (2007). Proteolytic processing of turnip yellow mosaic virus replication proteins and functional impact on infectivity. J. Virol. 81, 11402–11412. doi: 10.1128/jvi.01428-07
Johnson, K. L., Price, B. D., and Ball, L. A. (2003). Recovery of infectivity from cDNA clones of nodamura virus and identification of small nonstructural proteins. Virology 305, 436–451. doi: 10.1006/viro.2002.1769
Kidmose, R. T., Vasiliev, N. N., Chetverin, A. B., Andersen, G. R., and Knudsen, C. R. (2010). Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 10884–10889. doi: 10.1073/pnas.1003015107
Kinsella, E., Martin, S. G., Grolla, A., Czub, M., Feldmann, H., and Flick, R. (2004). Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology 321, 23–28. doi: 10.1016/j.virol.2003.09.046
Klaassen, V. A., Boeshore, M. L., Koonin, E. V., Tian, T., and Falk, B. W. (1995). Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly-transmitted, bipartite closterovirus. Virology 208, 99–110. doi: 10.1006/viro.1995.1133
Koonin, E. V. (1993). Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74(Pt 4), 733–740. doi: 10.1099/0022-1317-74-4-733
Lai, V. C., Kao, C. C., Ferrari, E., Park, J., Uss, A. S., Wright-Minogue, J., et al. (1999). Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73, 10129–10136.
Lam, A. M., Espiritu, C., Bansal, S., Micolochick Steuer, H. M., Niu, C., Zennou, V., et al. (2012). Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob. Agents Chemother. 56, 3359–3368. doi: 10.1128/AAC.00054-12
Lee, Y. F., Nomoto, A., Detjen, B. M., and Wimmer, E. (1977). A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. U.S.A. 74, 59–63. doi: 10.1073/pnas.74.1.59
Lemm, J. A., Durbin, R. K., Stollar, V., and Rice, C. M. (1990). Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64, 3001–3011.
Lesburg, C. A., Cable, M. B., Ferrari, E., Hong, Z., Mannarino, A. F., and Weber, P. C. (1999). Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6, 937–943.
Li, W., Wu, B., Soca, W. A., and An, L. (2018). Crystal structure of classical swine fever virus NS5B reveals a novel N-Terminal domain. J. Virol. 92:e00324-18. doi: 10.1128/JVI.00324-18
Li, Y., Korolev, S., and Waksman, G. (1998). Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17, 7514–7525. doi: 10.1093/emboj/17.24.7514
Liang, B., Li, Z., Jenni, S., Rahmeh, A. A., Morin, B. M., Grant, T., et al. (2015). Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327. doi: 10.1016/j.cell.2015.06.018
Liu, W., Shi, X., and Gong, P. (2018). A unique intra-molecular fidelity-modulating mechanism identified in a viral RNA-dependent RNA polymerase. Nucleic Acids Res. 46, 10840–10854. doi: 10.1093/nar/gky848
Love, R. A., Maegley, K. A., Yu, X., Ferre, R. A., Lingardo, L. K., Diehl, W., et al. (2004). The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure 12, 1533–1544.
Lu, G., and Gong, P. (2013). Crystal structure of the full-length Japanese Encephalitis Virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 9:e1003549. doi: 10.1371/journal.ppat.1003549
Luo, G., Hamatake, R. K., Mathis, D. M., Racela, J., Rigat, K. L., Lemm, J., et al. (2000). De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74, 851–863. doi: 10.1128/jvi.74.2.851-863.2000
Malet, H., Egloff, M. P., Selisko, B., Butcher, R. E., Wright, P. J., Roberts, M., et al. (2007). Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 282, 10678–10689.
Mendez-Toss, M., Romero-Guido, P., Munguia, M. E., Mendez, E., and Arias, C. F. (2000). Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81, 2891–2897. doi: 10.1099/0022-1317-81-12-2891
Miller, W. A., Waterhouse, P. M., and Gerlach, W. L. (1988). Sequence and organization of barley yellow dwarf virus genomic RNA. Nucleic Acids Res. 16, 6097–6111. doi: 10.1093/nar/16.13.6097
Morch, M. D., Boyer, J. C., and Haenni, A. L. (1988). Overlapping open reading frames revealed by complete nucleotide sequencing of turnip yellow mosaic virus genomic RNA. Nucleic Acids Res. 16, 6157–6173. doi: 10.1093/nar/16.13.6157
Moriceau, L., Jomat, L., Bressanelli, S., Alcaide-Loridan, C., and Jupin, I. (2017). Identification and molecular characterization of the chloroplast targeting domain of Turnip yellow mosaic virus replication proteins. Front. Plant Sci. 8:2138. doi: 10.3389/fpls.2017.02138
Ng, K. K., Cherney, M. M., Vazquez, A. L., Machin, A., Alonso, J. M., Parra, F., et al. (2002). Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277, 1381–1387. doi: 10.1074/jbc.m109261200
Ng, K. K., Pendas-Franco, N., Rojo, J., Boga, J. A., Machin, A., Alonso, J. M., et al. (2004). Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279, 16638–16645. doi: 10.1074/jbc.m400584200
Oliver, S. L., Asobayire, E., Dastjerdi, A. M., and Bridger, J. C. (2006). Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology 350, 240–250. doi: 10.1016/j.virol.2006.02.027
Ozato Junior, T., Gaspar, J. O., and Belintani, P. (2009). Completion of the nucleotide sequence of a Brazilian isolate of Southern bean mosaic virus. J. Phytopathol. 157, 573–575. doi: 10.1111/j.1439-0434.2008.01516.x
Palmenberg, A. C. (1990). Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44, 603–623. doi: 10.1146/annurev.micro.44.1.603
Pan, J., Vakharia, V. N., and Tao, Y. J. (2007). The structure of a birnavirus polymerase reveals a distinct active site topology. Proc. Natl. Acad. Sci. U.S.A. 104, 7385–7390. doi: 10.1073/pnas.0611599104
Peters, S. A., Mesnard, J. M., Kooter, I. M., Verver, J., Wellink, J., and van Kammen, A. (1995). The cowpea mosaic virus RNA 1-encoded 112 kDa protein may function as a VPg precursor in vivo. J. Gen. Virol. 76(Pt 7), 1807–1813. doi: 10.1099/0022-1317-76-7-1807
Pietila, M. K., Hellstrom, K., and Ahola, T. (2017). Alphavirus polymerase and RNA replication. Virus Res. 234, 44–57. doi: 10.1016/j.virusres.2017.01.007
Poch, O., Sauvaget, I., Delarue, M., and Tordo, N. (1989). Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x
Quito-Avila, D. F., Brannen, P. M., Cline, W. O., Harmon, P. F., and Martin, R. R. (2013). Genetic characterization of Blueberry necrotic ring blotch virus, a novel RNA virus with unique genetic features. J. Gen. Virol. 94, 1426–1434. doi: 10.1099/vir.0.050393-0
Rastgou, M., Habibi, M. K., Izadpanah, K., Masenga, V., Milne, R. G., Wolf, Y. I., et al. (2009). Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 90, 2525–2535. doi: 10.1099/vir.0.013086-0
Reed, K. E., and Rice, C. M. (2000). Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242, 55–84. doi: 10.1007/978-3-642-59605-6_4
Reich, E., Franklin, R. M., Shatkin, A. J., and Tatum, E. L. (1961). Effect of actinomycin D on cellular nucleic acid synthesis and virus production. Science 134, 556–557. doi: 10.1126/science.134.3478.556
Reich, E., Franklin, R. M., Shatkin, A. J., and Tatumel, E. L. (1962). Action of actinomycin D on animal cells and viruses. Proc. Natl. Acad. Sci. U.S.A. 48, 1238–1245.
Revill, P. A., Davidson, A. D., and Wright, P. J. (1994). The nucleotide sequence and genome organization of mushroom bacilliform virus: a single-stranded RNA virus of Agaricus bisporus (Lange) Imbach. Virology 202, 904–911. doi: 10.1006/viro.1994.1412
Rice, C. M., Lenches, E. M., Eddy, S. R., Shin, S. J., Sheets, R. L., and Strauss, J. H. (1985). Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229, 726–733. doi: 10.1126/science.4023707
Rodriguez-Cousino, N., Esteban, L. M., and Esteban, R. (1991). Molecular cloning and characterization of W double-stranded RNA, a linear molecule present in Saccharomyces cerevisiae. Identification of its single-stranded RNA form as 20 S RNA. J. Biol. Chem. 266, 12772–12778.
Rodriguez-Cousino, N., Solorzano, A., Fujimura, T., and Esteban, R. (1998). Yeast positive-stranded virus-like RNA replicons. 20 S and 23 S RNA terminal nucleotide sequences and 3′ end secondary structures resemble those of RNA coliphages. J. Biol. Chem. 273, 20363–20371. doi: 10.1074/jbc.273.32.20363
Romero-Brey, I., and Bartenschlager, R. (2014). Membranous replication factories induced by plus-strand RNA viruses. Viruses 6, 2826–2857. doi: 10.3390/v6072826
Schmidt-Mende, J., Bieck, E., Hugle, T., Penin, F., Rice, C. M., Blum, H. E., et al. (2001). Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276, 44052–44063.
Sholders, A. J., and Peersen, O. B. (2014). Distinct conformations of a putative translocation element in poliovirus polymerase. J. Mol. Biol. 426, 1407–1419. doi: 10.1016/j.jmb.2013.12.031
Shu, B., and Gong, P. (2016). Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. U.S.A 113, E4005–E4014. doi: 10.1073/pnas.1602591113
Singh, M., and Singh, P. (1996). Nucleotide sequence and genome organization of a Canadian isolate of the common strain of potato virus Y (PVY). Can. J. Plant Pathol. 18, 209–224. doi: 10.1080/07060669609500615
Snijder, E. J., Bredenbeek, P. J., Dobbe, J. C., Thiel, V., Ziebuhr, J., Poon, L. L., et al. (2003). Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004. doi: 10.1016/s0022-2836(03)00865-9
Strauss, E. G., Rice, C. M., and Strauss, J. H. (1984). Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133, 92–110. doi: 10.1016/0042-6822(84)90428-8
Subissi, L., Imbert, I., Ferron, F., Collet, A., Coutard, B., Decroly, E., et al. (2014a). SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antiviral Res. 101, 122–130. doi: 10.1016/j.antiviral.2013.11.006
Subissi, L., Posthuma, C. C., Collet, A., Zevenhoven-Dobbe, J. C., Gorbalenya, A. E., Decroly, E., et al. (2014b). One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 111, E3900–E3909. doi: 10.1073/pnas.1323705111
Takeshita, D., and Tomita, K. (2010). Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl. Acad. Sci. U.S.A. 107, 15733–15738. doi: 10.1073/pnas.1006559107
te Velthuis, A. J. (2014). Common and unique features of viral RNA-dependent polymerases. Cell Mol. Life Sci. 71, 4403–4420. doi: 10.1007/s00018-014-1695-z
te Velthuis, A. J., Arnold, J. J., Cameron, C. E., van den Worm, S. H., and Snijder, E. J. (2010). The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 38, 203–214. doi: 10.1093/nar/gkp904
Thompson, A. A., and Peersen, O. B. (2004). Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23, 3462–3471. doi: 10.1038/sj.emboj.7600357
Tomar, S., Hardy, R. W., Smith, J. L., and Kuhn, R. J. (2006). Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 80, 9962–9969. doi: 10.1128/jvi.01067-06
Upadhyay, A. K., Cyr, M., Longenecker, K., Tripathi, R., Sun, C., and Kempf, D. J. (2017). Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr. F Struct. Biol. Commun. 73, 116–122. doi: 10.1107/S2053230X17001601
Valles, S. M., Oi, D. H., Becnel, J. J., Wetterer, J. K., LaPolla, J. S., and Firth, A. E. (2016). Isolation and characterization of Nylanderia fulva virus 1, a positive-sense, single-stranded RNA virus infecting the tawny crazy ant, Nylanderia fulva. Virology 496, 244–254. doi: 10.1016/j.virol.2016.06.014
van Dinten, L. C., Rensen, S., Gorbalenya, A. E., and Snijder, E. J. (1999). Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J. Virol. 73, 2027–2037.
Vives-Adrian, L., Lujan, C., Oliva, B., van der Linden, L., Selisko, B., Coutard, B., et al. (2014). The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site. J. Virol. 88, 5595–5607. doi: 10.1128/JVI.03502-13
Walter, C. T., Pringle, F. M., Nakayinga, R., de Felipe, P., Ryan, M. D., Ball, L. A., et al. (2010). Genome organization and translation products of Providence virus: insight into a unique tetravirus. J. Gen. Virol. 91, 2826–2835. doi: 10.1099/vir.0.023796-0
Wang, C., Wang, C., Li, Q., Wang, Z., and Xie, W. (2017). Crystal structure and thermostability characterization of enterovirus D68 3D(pol). J. Virol. 91:e00876-17.
Willcocks, M. M., Brown, T. D., Madeley, C. R., and Carter, M. J. (1994). The complete sequence of a human astrovirus. J. Gen. Virol. 75(Pt 7), 1785–1788. doi: 10.1099/0022-1317-75-7-1785
Wimmer, E., and Nomoto, A. (1993). Molecular biology and cell-free synthesis of poliovirus. Biologicals 21, 349–356. doi: 10.1006/biol.1993.1095
Wu, J., Liu, W., and Gong, P. (2015). A structural overview of RNA-dependent RNA polymerases from the Flaviviridae family. Int. J. Mol. Sci. 16, 12943–12957. doi: 10.3390/ijms160612943
Wu, Y., Lou, Z., Miao, Y., Yu, Y., Dong, H., Peng, W., et al. (2010). Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China. Protein Cell 1, 491–500. doi: 10.1007/s13238-010-0061-7
Xiang, J., Wunschmann, S., Schmidt, W., Shao, J., and Stapleton, J. T. (2000). Full-length GB virus C (Hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J. Virol. 74, 9125–9133. doi: 10.1128/jvi.74.19.9125-9133.2000
Xu, X., Liu, Y., Weiss, S., Arnold, E., Sarafianos, S. G., and Ding, J. (2003). Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 31, 7117–7130. doi: 10.1093/nar/gkg916
Yanagi, M., Purcell, R. H., Emerson, S. U., and Bukh, J. (1997). Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. U.S.A. 94, 8738–8743. doi: 10.1073/pnas.94.16.8738
Yap, T. L., Xu, T., Chen, Y. L., Malet, H., Egloff, M. P., Canard, B., et al. (2007). Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81, 4753–4765. doi: 10.1128/jvi.02283-06
Yin, Y. W., and Steitz, T. A. (2004). The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116, 393–404. doi: 10.1016/s0092-8674(04)00120-5
Zamyatkin, D. F., Parra, F., Alonso, J. M., Harki, D. A., Peterson, B. R., Grochulski, P., et al. (2008). Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283, 7705–7712. doi: 10.1074/jbc.M709563200
Zhai, Y., Sun, F., Li, X., Pang, H., Xu, X., Bartlam, M., et al. (2005). Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12, 980–986. doi: 10.1038/nsmb999
Zhao, Y., Soh, T. S., Zheng, J., Chan, K. W., Phoo, W. W., Lee, C. C., et al. (2015). A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 11:e1004682. doi: 10.1371/journal.ppat.1004682
Zhong, W., Ferrari, E., Lesburg, C. A., Maag, D., Ghosh, S. K., Cameron, C. E., et al. (2000). Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74, 9134–9143. doi: 10.1128/jvi.74.19.9134-9143.2000
Keywords: positive-strand RNA virus, RNA-dependent RNA polymerase, genome replication, structure, catalytic motif
Citation: Jia H and Gong P (2019) A Structure-Function Diversity Survey of the RNA-Dependent RNA Polymerases From the Positive-Strand RNA Viruses. Front. Microbiol. 10:1945. doi: 10.3389/fmicb.2019.01945
Received: 13 May 2019; Accepted: 07 August 2019;
Published: 22 August 2019.
Edited by:
Aartjan Te Velthuis, University of Cambridge, United KingdomReviewed by:
Carmen Hernandez, Polytechnic University of Valencia, SpainJulien Lescar, Nanyang Technological University, Singapore
Copyright © 2019 Jia and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Gong, Z29uZ3BlbmdAd2guaW92LmNu
 Hengxia Jia
Hengxia Jia Peng Gong
Peng Gong