- State Key Laboratory of Agricultural Microbiology, Key Laboratory of Horticultural Plant Biology (MOE), College of Plant Science and Technology, Institute of Urban and Horticultural Entomology, Huazhong Agricultural University, Wuhan, China
The Asian citrus psyllid (Diaphorina citri) is a major pest of citrus trees as it transmits Candidatus Liberibacter asiaticus (CLas). The composition of a host’s microbiota can affect the evolution and ecological distribution of the host. This study monitored the compositional shifts in the citrus psyllid microbiota through all the life stages (egg, nymph 1–5 stages, and adult) by next-generation sequencing (NGS) and quantitative real-time PCR. There were clear differences in both α- and β-diversity of microbiota through the psyllid life stages. Microbiota diversity was markedly higher in the nymph 2–5 stages than in the adult, egg, and nymph 1 stages. Proteobacteria were dominant in all the life stages of D. citri, representing >97.5% of the total bacterial community, and Candidatus Profftella armature was the dominant genus in all the life stages. Data from the qPCR analysis showed an exponential increase in the populations of three D. citri endosymbionts: Candidatus Profftella armature, Candidatus Carsonella ruddii, and Wolbachia. The gut bacterium Pantoea was present in all the life stages, but it was markedly higher in the nymph 2–5 stages. The microbiota composition substantially differed among the egg–nymph 1, nymphs 2–5, and adult stages. Therefore, we successfully characterized the microbiota dynamics and thus identified a microbiota shift during the life cycle of D. citri by 16S rRNA gene sequencing and quantitative PCR. Moreover, 16S rRNA gene sequencing suggested that D. citri acquired the ability to bear CLas in the nymph 1 stage. This study enhances our understanding of microbial establishment in the developing D. citri and provides a reference resource for the identification of potential biocontrol approaches against this pest.
Introduction
Recently, microbiotas of insects have attracted great interest, mainly because of the ecological and economical importance of insects (Engel and Moran, 2013; Sugio et al., 2014). In many cases, microbes allow their insect hosts to survive on nutrient-deficient food resources, such as plant sap, conferring a competitive advantage on them (Beck et al., 2018). In addition, arthropod-associated microbes play numerous roles during the life cycle of the host arthropod, including development, reproduction, speciation, immunity, and defense against predators (Oliver et al., 2014; Macke et al., 2017).
The sap-sucking Asian citrus psyllid, Diaphorina citri Kuwayama (Hemiptera: Liviidae), is one of the most serious agricultural pests in the citrus-growing regions around the world. It is the vector of Candidatus Liberibacter asiaticus (CLas). This bacterial species causes the highly destructive huanglongbing (HLB) disease, also called citrus greening, in citrus trees (Grafton-Cardwell et al., 2013). Like many other plant-feeding insects, they rely on bacterial endosymbionts to acquire the amino acids and other nutrients missing in their diet. The obligate bacterial endosymbionts, Candidatus Carsonella ruddii, Candidatus Profftella armatura, and Wolbachia have cooperative and close associations with D. citri (Thao et al., 2000; Nachappa et al., 2011; Nakabachi et al., 2013b). The roles of Carsonella and Profftella in D. citri’s life cycle have been deduced by genome sequencing, but Wolbachia’s role is still relatively elusive (Saha et al., 2012). Carsonella acts as a nutritional symbiont (Tamames et al., 2007; Nakabachi et al., 2013a), and Profftella has been predicted to confer a defense advantage because it produces diaphorin, a mildly cytotoxic polyketide (Nakabachi et al., 2013b; Ramsey et al., 2015). Many studies have attempted to explore the endosymbionts, the internal microbiota, of D. citri. PCR-RFLP and DGGE methods have been performed to evaluate presence of bacteria in D. citri (Subandiyah et al., 2000; Yin et al., 2011). Additionally, the diversity of the cultivable microbiota of D. citri has recently been investigated by culture-dependent methods and 16S rDNA clone library (Kolora et al., 2015). Quantitative real-time PCR (qPCR) analyses showed that the Carsonella, Profftella, and Wolbachia levels were positively correlated with each other (Dossi et al., 2014; Chu et al., 2016). Previous studies have provided valuable information regarding the general bacterial diversity and identity of common internal bacteria and potential endosymbionts. However, cloning is a laborious and costly technique with a limited outcome for the detection of scarce bacteria (Zolnik et al., 2016), and many of the early studies were descriptive and confined to adults. There is limited information about the lifelong dynamics and trans-generational transfer of D. citri’s microbiota. We used next-generation sequencing (NGS) in our study, given that this technique has been powerful for in-depth compositional and functional analysis of microbiotas and characterization of the relationships between the microbial communities (Hovatter et al., 2011; Costello et al., 2012).
Wild populations of D. citri were used in this study. The bacterial community associated to different life stages of D. citri (egg, nymph 1–5 stages, and adults) has been analyzed through Illumina MiSeq sequencing. Insects exhibit complex symbiotic interactions with microbes, which have a strong impact on insect physiology and provide resources for developing species-specific pest management tactics (Douglas, 2007; Hamby and Becher, 2016). Understanding microbiota composition and dynamics during D. citri’s life cycle will enhance the pest management strategies in the future. From a fundamental perspective, this knowledge is also important to elucidate the role of bacteria in D. citri’s life.
Materials and Methods
Sample Collection, Processing, and DNA Extraction
Diaphorina citri at seven different life stages were collected from a navel orange orchard in Ganzhou city, Jiangxi Province (32°13′ N, 114°08′ E), China. The psyllid eggs were harvested with a sterilized blade, and then pooled together. The adults and nymphs were individually collected with a camel hairbrush. Sexing was not performed during any of the life stages. The psyllid samples were cleaned with successive washes of 3% hydrogen peroxide (1 min vortex), 70% ethanol (twice for 30 s each), and deionized H2O (for 2 min) to remove environmental contaminants before DNA extraction. Samples of each life stage (egg, nymph 1–5 stages, and adult) were divided into three pools, with 300 eggs, 200 nymphs 1–3, 100 nymphs 4–5, and 80 adults per pool. Total nucleic acid was extracted using the Qiagen DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA, United States), according to the manufacturer’s instructions.
Microbiota Sample Preparation and Sequencing
Separate amplicon libraries were prepared for each psyllid sample according to the Illumina MiSeq 16S Metagenomic Sequencing Library Preparation protocol (Klindworth et al., 2013). Amplicon libraries targeting the V3–V4 hypervariable regions of the 16S rRNA gene were generated (Illumina, San Diego, CA, United States). PCR amplification was performed with primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GACTACHVGGGTWTCTAAT-3′) under the following thermal cycling conditions: 95°C for 3 min, followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. All psyllid DNA samples were amplified in triplicates and subsequently pooled to minimize PCR bias.
The amplicons were purified using solid phase reversible immobilization (SPRI) beads to remove DNA < 60 base pairs (bp) long (primers and primer dimers), and DNA > 700 bp (Off-target PCR products). The resulting amplicon was approximately 550 bp long. Sequencing was performed with an Illumina MiSeq using the MISEQ REAGENT KIT (version 2; Illumina, Inc., San Diego, CA, United States).
Statistical Analyses of D. citri’s Microbiota From Different Psyllid Life Stages
Illumina FASTQ files were demultiplexed and quality-filtered (q20) with quantitative insights in microbial ecology (QIIME 1.9.1) (Caporaso et al., 2010). Paired-end reads were aligned using FLASH and Trimmomatic, and then assigned to operational taxonomic units (OTUs) using a 97% pairwise identity threshold. The Greengenes taxonomic database was used with an open reference OTU picking strategy for taxonomic assignment1.
For the analysis of the psyllid life stages, the sequence data were rarefied to a depth of 12,141 sequences. The results were plotted on a rarefaction curve, and analysis of variance (ANOVA)-Tukey’s test was performed to assess the differences between the psyllid life stages. The microbial alpha diversity indices were estimated to uncover the bacterial diversity (Simpson and Shannon) and species richness (Chao1 and ACE). The microbial beta diversity was estimated using weighted and unweighted UniFrac analyses to compare different samples. The adonis function, that performs a permutational multivariate analysis of variance (PERMANOVA) with 999 permutations was used to determine the percent variation explained by developmental stages, as well as the significance and effect size for both weighted and unweighted UniFrac distance matrices. Principal coordinate analysis (PCoA) was used to compare differences among psyllid samples based on both UniFrac distance matrices.
Quantitative Real-Time PCR (qPCR)
Both the forward and reverse primers (338F and 806R) used for the 16S rRNA gene sequencing contained multiple mismatches to Carsonella genome, biasing the amplification against Carsonella’s 16S rRNA gene (Morrow et al., 2017). To quantify Carsonella and determine the density of the other three known symbionts (Profftella, Wolbachia, and CLas) at different stages of D. citri, we conducted an absolute qPCR analysis as described previously (Chu et al., 2016).
Both TaqMan (wingless, CLas) and SYBR green methods (Carsonella, Profftella, and Wolbachia) were used in this study (Table 1). The wingless host gene established by Li et al. (2008) was selected as endogenous control. Standard curves were generated to ensure that quantification of the same target gene on different qPCR plates was comparable. The qPCR assays were conducted in triplicates with a CFX384 Touch Real-time PCR cycler (Bio-Rad, Hercules, CA, United States). Each reaction mixture was composed of PCR Master Mix (Roche), 0.4 μL of each primer (10 μmol/L), and 1 μL of DNA template (15 ng) in a final volume of 10 μL. The bacterial concentration in each sample was calculated relative to the Ct values in the standard curves, which were generated by using 10-fold serial dilutions of specific DNA fragments.
For each sample, the copy numbers of the target genes in the template DNA were calculated as described by Whelan et al. (2003). Subsequently, the symbiont copy number was divided by the wingless gene copy number in the same sample. The CLas density of the samples was compared by dividing infected D. citri into high- and low-infection groups using a density threshold of 0.02 copies of CLas 16S rDNA per copy of wingless gene (Chu et al., 2016). Statistical analyses (chi-square test of homogeneity and ANOVA) were conducted on the symbiont densities. To satisfy the condition of normality for ANOVA, symbiont densities were transformed for statistical analysis using log (1 + x) (Wolbachia) or log (Carsonella and Profftella) as previously described (Chu et al., 2016).
Results
A total of 21 samples, with three pools for each category (egg, nymph 1–5 stages, and adult) generated 943,384 raw reads in one Illumina MiSeq flow cell. After filtering, 820,407 sequences were assigned taxonomy. This resulted in 31,174–44,938 high-quality merged sequences per sample (Table 2). On average, the conditional uncovered probability of the samples indicated a high likelihood that over 99% of the bacterial taxa in the samples were covered. Rarefaction curves from a depth of 1,000–100,000 sequences indicated enough sequencing coverage as demonstrated by the observation that the OTU accumulation curves reached a plateau (Supplementary Figure S1).
16S rRNA Gene Sequencing Data
In total, 451 distinct OTUs were clustered across the seven life stages. The alpha diversity indices were estimated to determine the bacterial diversity (Simpson and Shannon indices) and species richness (Chao1 and ACE indices) (Table 3). Across the various life stages, there were significant differences in the bacterial diversity estimated by Simpson and Shannon indices (ANOVA, F = 11.078, df = 4, 20, p < 0.001; Table 3), and as D. citri matured from nymph to adult, the bacterial diversity showed a declining trend. The bacterial diversity of nymphs 2–5 was significantly higher than that of adult (p < 0.001), egg (p < 0.01), and nymph 1 (p < 0.01), but the bacterial diversity did not significantly differ among the egg, nymph 1, or adult stages (p > 0.15). Analysis of species richness estimated by Chao1 and ACE indices indicated differences among the life stages, but this difference did not reach a significant level (ANOVA, F = 2.025, df = 4, 20, p = 0.13; Table 3). The average species richness of nymphs 2–5 was higher than that of adult (p > 0.10), egg (p > 0.10), and nymph 1 (p > 0.10).
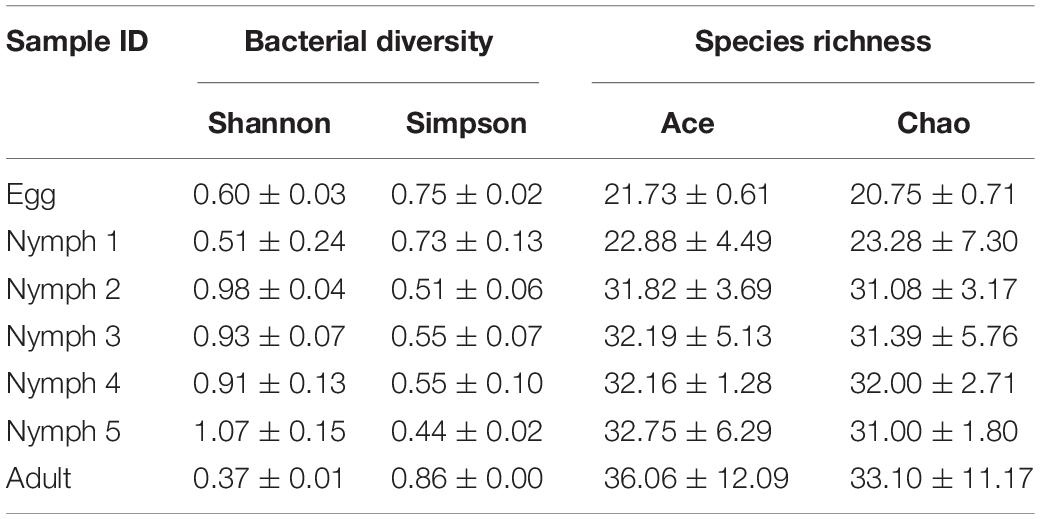
Table 3. Estimation of species richness and diversity of D. citri microbiota during psyllid life stages.
Community Characterization
The phylum with the highest relative abundance (97.5% of all sequences) across all the developmental stages was found to be the Proteobacteria. The remaining sequences were classified into Cyanobacteria, Actinobacteria, Firmicutes, Bacteroidetes, and Acidobacteria. Among the Proteobacteria, the dominant class was β-Proteobacteria (73.0% of all Proteobacteria), followed by γ-Proteobacteria (18.7%), and α-Proteobacteria (5.83%). Cyanobacteria were only present during the egg, nymph 1 and nymph 2 stages with a decreasing abundance as D. citri developed (from 11.8 to 0.03%). Profftella was clearly the dominant genus (mean = 58.1–84%) during all the stages and seven genera (along with one family-level taxon and one Norank-c-Cyanobacteria) showed a relative abundance of 1% or greater during at least one stage (Figure 1). The adults had the highest proportion of Profftella (84%), and thus had fewer genera with a relative abundance ≥1%. Wolbachia was the second most abundant genus in the adults (13.6%) and was detected in the samples from all other life stages (>1.1%). Pantoea was widely present during all the stages of D. citri but was more abundant during the nymph 2–5 stages (>17%). Thiopseudomonas and Dechlorobacter were only found in egg and nymph 1 samples. Rosenbergiella was only detected in the samples from nymph 2–5 stages (Figure 1).
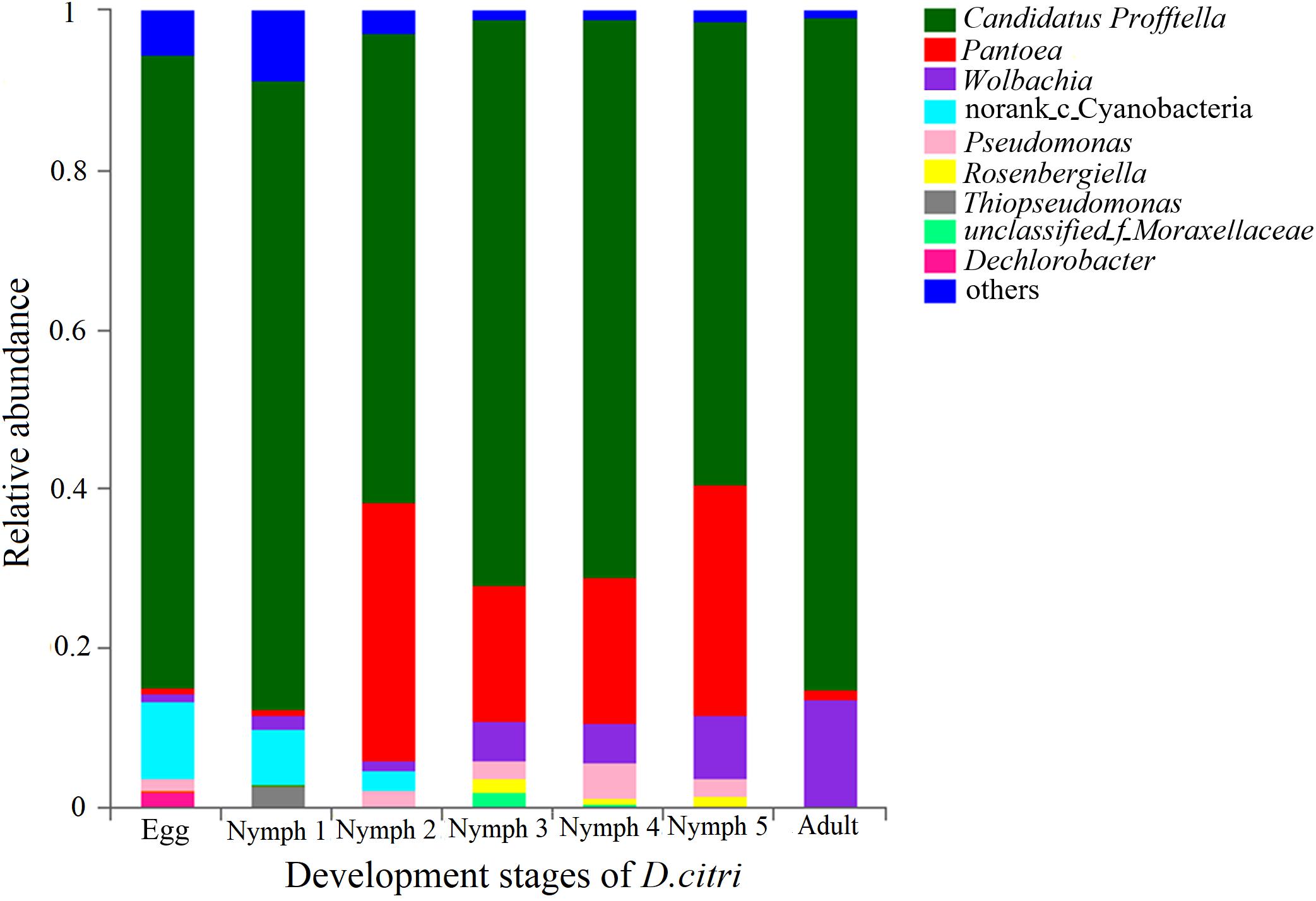
Figure 1. Relative abundances of the bacterial genera in the pooled psyllid samples from the seven developmental stages. Only taxa with a relative abundance ≥1% in at least one sample were analyzed.
The average relative abundance of CLas across the seven life stages was 0‱ (egg), 0.25‱ (nymph 1), 1.25‱ (nymph 2), 3.22‱ (nymph 3), 3.78‱ (nymph 4), 7.96‱ (nymph 5), and 25.51‱ (adult), increasing with age (Figure 2).
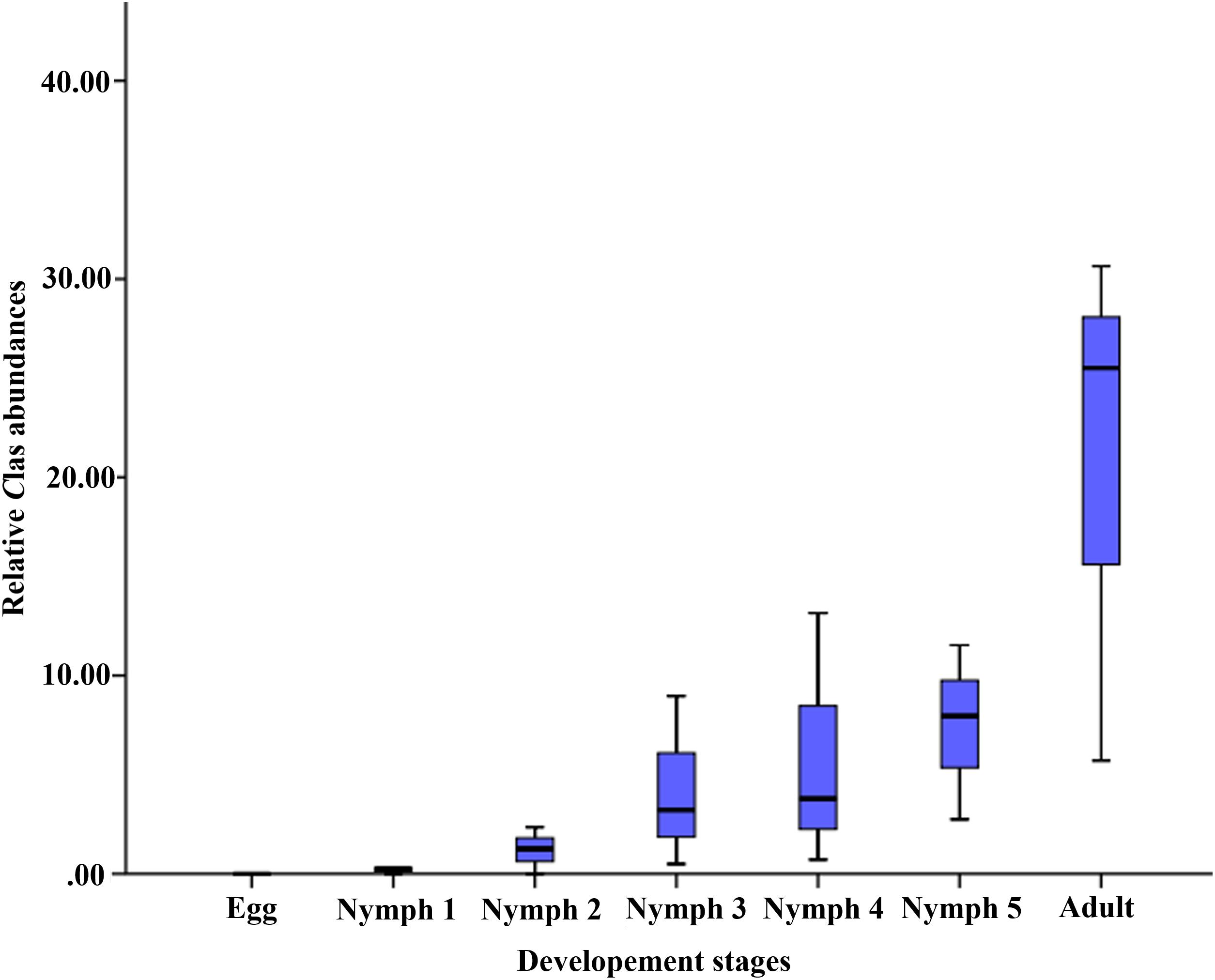
Figure 2. Relative CLas abundances (on the y-axis as CLas ‱ of all 16S rRNA sequences in samples of the corresponding life stage).
Taxonomic Composition of D. citri’s Microbiota as Identified by 16S rRNA Gene Sequencing
Bacterial communities across the seven life stages were significantly different according to a PERMANOVA test (unweighted F = 1.47, R2 = 0.42, p = 0.006; weighted F = 5.36, R2 = 0.73, p = 0.001). Pairwise PERMANOVA comparisons (Table 4) revealed differences among egg, nymphs and adults’ samples except for comparisons of egg and nymph1 (p = 0.6012). Figure 3 illustrated the ordination by PCoA of the Illumina sequencing profiles. The unweighted UniFrac (which does not account for abundance data) PCoA explained 18.4% (axis 1) and 13.11% (axis 2) of the variation across the life stages, with the egg and nymph 1 samples clustering together (p = 0.6012) (Figure 3A). On the other hand, the weighted UniFrac PCoA explained 68.18% (axis 1) and 13.51% (axis 2) of the variation, showing that samples from the adults did not cluster with those of earlier stages (p < 0.05) (Figure 3B).
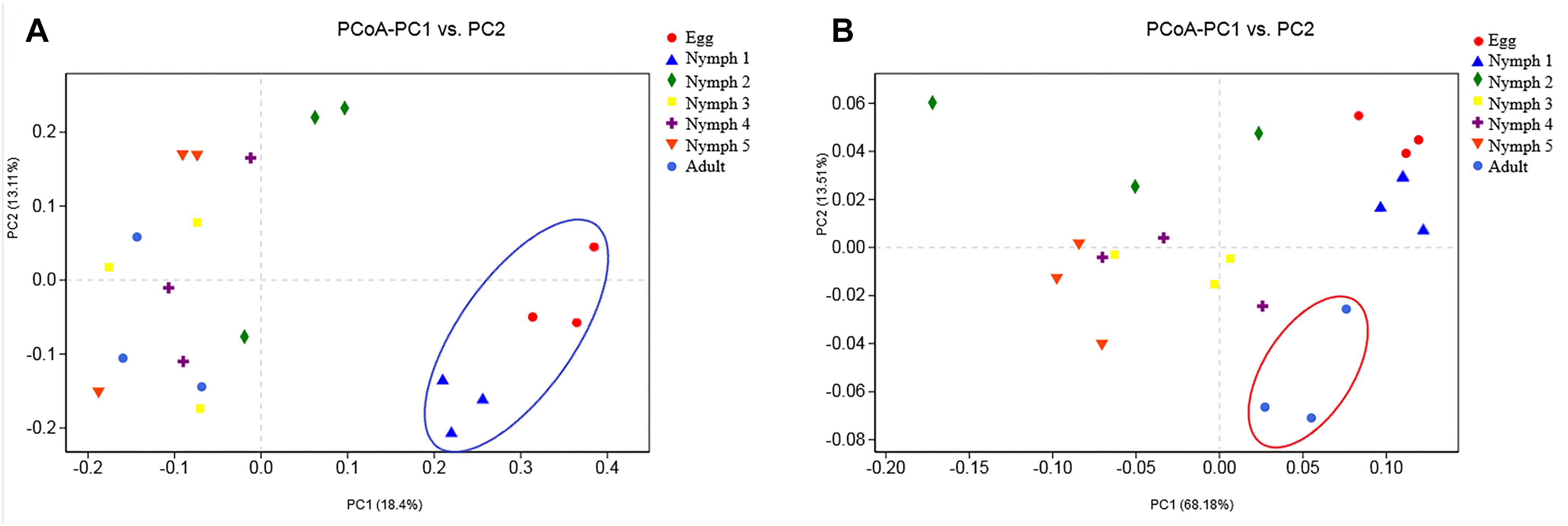
Figure 3. Principal coordinate analysis of the β-diversity values during the seven developmental stages. (A) Abundance was ignored in the unweighted UniFrac distances; (B) but considered in the weighted UniFrac distances. Confidence ellipsoids around the samples indicate the degree of variation.
Population Shift in the Abundant Bacteria Species Across the Life Stages as Shown by qPCR
By 16S rRNA gene sequencing, Carsonella sequence reads in D. citri individuals ranged between 0 and 0.2%. However, in the qPCR experiment, the presence of Carsonella was confirmed in all individuals, and the content was high (Figure 4B). All the three investigated endosymbionts of D. citri showed exponential increases from egg to adult stage (Figure 4). Profftella, the secondary endosymbiont, was the dominant genus during D. citri development (Figure 4A). Even though Carsonella and Wolbachia showed trends like Profftella, these two species were not as abundant (Figures 4B,C).
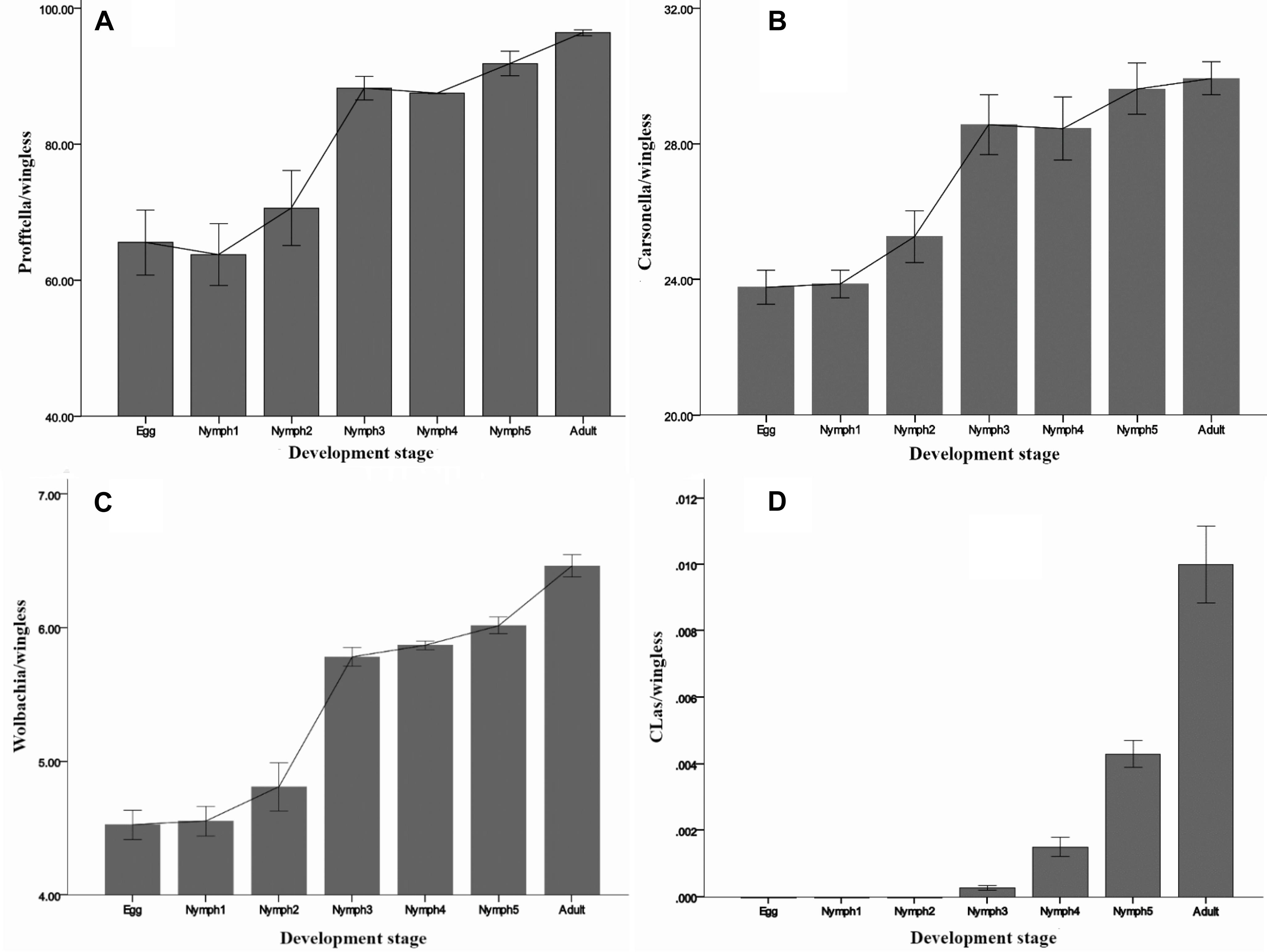
Figure 4. Densities of (A) Profftella, (B) Carsonella, (C) Wolbachia, and (D) CLas in D. citri across the seven life stages. The error bars indicate the standard errors.
CLas was detected during the nymph 3–5 and adult stages, but at a low level (<0.02 copies). Inconsistent with the MiSeq sequencing results, CLas was not detected in the egg and nymph 1–2 samples by qPCR (Figure 4D). Taken together, these results suggest that in this case qPCR has limitations even though it is a highly sensitive technique. For some low-abundance templates, the adequate amplification for signal detection above the fluorescent background noise may not occur, resulting in false-negatives. NGS due to its high-throughput can detect large amounts of information about the total and active members of bacterial communities at the gene sequence level and can quantify low-abundance species (0–5%) (Webster et al., 2010; van Elsas and Boersma, 2011). Quigley et al. (2014) evaluated the ability of NGS to quantitatively analyze samples with known densities of Symbiodinium types and demonstrated that NGS is more sensitive and quantitative than qPCR. Likely this also happened in our case, but further investigations are needed to elucidate the level of CLas detection.
Discussion
A complex microbiota is the basis for the normal survival and development of insects from fertilized eggs to adults (Dillon and Dillon, 2004; Duguma et al., 2013). Here we reported the patterns in microbiota diversity across all the developmental stages of D. citri, the primary vector of HLB. RFLP analysis (Subandiyah et al., 2000), qPCR (Dossi et al., 2014; Hussain et al., 2017; Mann et al., 2018), and the Roche 454 pyrosequencing technology (Kolora et al., 2015) have been used in previous studies on psylla microbiotas. Previous studies have provided valuable information regarding the general bacterial diversity and identity of common internal bacteria and potential endosymbionts. However, many of the early studies were descriptive and confined to adults. There is limited information about the lifelong dynamics of D. citri’s microbiota.
As in many other insects, D. citri microbiota was mainly dominated by species of the Proteobacteria phylum. Diversity estimation revealed that the bacterial communities of D. citri significantly differed across the psyllid life stages. The diversity indices of the bacterial communities (Shannon index: 0.37–0.98, Simpson index: 0.44–0.86) were significantly lower than those reported in drosophila flies (Shannon index: 1.99, Simpson index: 0.70) (Wong et al., 2013). Among the herbivore insects examined to date, sap-feeders, such as psyllids, aphids, and whiteflies show the lowest microbial diversity (Shannon’s index: 0.40–1.46, Simpson’s index: 0.15–0.84). Phloem sap’s low bacterial content and simple biological processes may contribute to the low bacterial diversity in sap-feeders (Jing et al., 2014).
Microbiomes display temporal and spatial variability across host developmental stages (Shade and Handelsman, 2012). The beta diversity revealed by PERMANOVA test indicates that the microbiota of D. citri differed across the life stages (egg, nymph 1–5 stages, and adult). The microbiotas of egg and nymph 1 were highly different from those of subsequent stages, with a much lower alpha diversity and unique microbial composition dominated by Profftella and Norank-c-Cyanobacteria. The microbiota of nymph 1 more closely resembles that of the egg, possibly because psyllids in the nymph 1 stage have just hatched from their eggs and have little contact with outside microbes. Some of these bacteria species, such as members of the genus Profftella, are vertically acquired through transovarial transmission in D. citri. The photosynthetic bacteria, Cyanobacteria, are not found in the adults, and thus may be acquired from breeding sites (Thiery et al., 1991). One bacterial group that appears to be particularly important in the nymph 2–5 stages is Pantoea, and its abundance is significantly higher than that in other stages. Pantoea are ubiquitous plant pathogens but they have also been described as part of the gut microbiota in D. citri and some aphid species (Engel and Moran, 2013). Some taxa of Pantoea have been known to degrade and utilize different types of plant materials (Morales-Jiménez et al., 2009). The recruitment of Pantoea in nymphs 2–5 may play a key role in the initiation of digesting external nutrient sources. However, its dominance in D. citri was transient. In the adult stage, Profftella and Wolbachia are the most abundant genera in D. citri. Wolbachia has been shown to spread throughout the insect’s somatic tissues and germline (Dobson et al., 1999). Increased abundance of Wolbachia is thought to be associated with organ development (Dobson et al., 1999; Ramsey et al., 2017). The effects of developmental stages on the microbiota of other herbivore insects were also reported. In the sweet potato whitefly (Bemisia tabaci), the bacteria diversity was the highest in the nymph stage and then decreased at maturity stages (Indiragandhi et al., 2010), while in the desert locusts (Schistocerca gregaria), there was often a successional increase in diversity with age (Dillon et al., 2010).
Besides providing a general insight into the microbiota of D. citri, our study also analyzed the dynamics of the known endosymbionts. Psyllids have a specialized organ called bacteriome, where coexisting microbes have developed intimate interactions (Spaulding and von Dohlen, 1998). D. citri’s bacteriome harbors bacterial endosymbionts Carsonella, Profftella, and Wolbachia. The primary endosymbiont Carsonella resides in psyllid bacteriocytes in the external cell layer of the bacteriome, whereas Profftella is found in the syncytial cytoplasm within the interior of the bacteriome (Nakabachi et al., 2013b; Hosseinzadeh et al., 2019). Both bacteria are vertically transmitted from mother to offspring (Hosseinzadeh et al., 2019). Wolbachia is also localized to the outside layer of the bacteriome, as is the case with Carsonella (Hosseinzadeh et al., 2019). Unlike the bacteriocyte-limited endosymbionts Profftella and Carsonella, Wolbachia is scattered throughout the D. citri body and has the highest density in the Malpighian tubules (Hosseinzadeh et al., 2019). Wolbachia is predominantly transmitted maternally, but also occasionally horizontally between species (Werren et al., 2008; Zug et al., 2012). We identified that the infection titers of the three endosymbionts tended to increase with successive psyllid life stages. The exponential growth of the three endosymbionts across the stages of D. citri is synchronized with the organogenesis of the bacteriome as previously observed under microscopy (Dossi, 2013). Simonet et al. (2016) suggested that the rapid increase in the numbers of both endosymbionts and bacteriocytes during insect development was linked to the insect’s high nutritional demands given that the nutritional needs of the insect increase as it develops. The metabolic specialization between symbionts and D. citri indicates mutually indispensable associations, suggesting that these symbionts may be potential targets for vector control. Furthermore, the adaptative changes in the host’s immune response against the endosymbionts may also explain the changes in symbiotic density during host development (Nishikori et al., 2009; Douglas et al., 2010). Continuous multiplication of the symbionts may be due to their tolerance to or escape from the host immune responses (Welchman et al., 2009). Nevertheless, further research is needed to elucidate the growth dynamics of the endosymbionts in detail (Login and Heddi, 2013; Ramsey et al., 2017).
CLas has previously been shown to colonize D. citri starting from the nymph 2 stage (Hung et al., 2004). These results were obtained by PCR and qPCR assays. qPCR is a powerful culture-independent method used to quantity microbial fractions or organisms, but limitations exist in terms of resolution. Very low DNA concentrations as well as the presence of inhibitors and contaminants may result in false negative results (Gryndler et al., 2013; Uyaguari-Diaz et al., 2016). Here we used MiSeq sequencing method and found that D. citri could also acquire CLas in the nymph 1 stage. In our study, D. citri eggs were CLas-negative, which was in line with previous reports (Grafton-Cardwell et al., 2013; Grafton-Cardwell and Daugherty, 2013).
Overall, in this study we showed that bacterial colonization of D. citri is an active and continuous process and the composition of the bacterial community varies throughout the psyllid life cycle. Our findings demonstrated the plasticity of symbiont density in response to host development. Additionally, we showed that CLas-acquisition ability of D. citri could start as early as nymph 1 stage. Further studies are required to determine whether these strong differences in microbiota diversity and composition across the life stages of D. citri may affect its CLas transmission. The major limitation of our study is that a single psyllid population from a single citrus field was used. Recent studies have shown that differences in the genetic backgrounds of D. citri populations can affect CLas and endosymbiont densities (Chu et al., 2016; Ammar et al., 2018). It remains to be elusive whether such factors also influence microbiota dynamics, but worth further investigation.
Data Availability
All the bacterial 16S rRNA amplicon and metagenomic sequencing data have been deposited in NCBI’s Short Read Archive under the accession number SRP178960.
Author Contributions
LM and HZ designed the study, conducted the data analysis, and wrote the manuscript. LM performed the fieldwork and the laboratory and data analyses. XL and XC assisted in collecting the research data. All authors contributed to the manuscript revision, and read and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (No. 2017YFD0202000), National Natural Science Foundation of China (Nos. 31872931 and 31572008), China Agricultural Research System (No. CARS-26), and the Fundamental Research Funds for the Central Universities (No. 2662019PY055).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01948/full#supplementary-material
FIGURE S1 | Rarefaction curves of observed OTUs (97% threshold) versus the number of the sampled reads in D. citri samples.
Footnotes
References
Ammar, E. D., Hall, D. G., Hosseinzadeh, S., and Heck, M. (2018). The quest for a non-vector psyllid: natural variation in acquisition and transmission of the huanglongbing pathogen ‘Candidatus liberibacter asiaticus’ by Asian citrus psyllid isofemale lines. PLoS One 13:e0195804. doi: 10.1371/journal.pone.0195804
Beck, J. J., Alborn, H. T., Block, A. K., Christensen, S. A., Hunter, C. T., Rering, C. C., et al. (2018). Interactions among plants, insects, and microbes: elucidation of inter-organismal chemical communications in agricultural ecology. J. Agric. Food. Chem. 66, 6663–6674. doi: 10.1021/acs.jafc.8b01763
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chu, C. C., Gill, T. A., Hoffmann, M., and Pelz-Stelinski, K. S. (2016). Inter-population variability of endosymbiont densities in the asian citrus psyllid (Diaphorina citri kuwayama). Microb. Ecol. 71, 999–1007. doi: 10.1007/s00248-016-0733-9
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J., and Relman, D. A. (2012). The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. doi: 10.1126/science.1224203
Coy, M. R., Hoffmann, M., Gibbard, H. K., Kuhns, E. H., Pelz-Stelinski, K. S., and Stelinski, L. L. (2014). Nested-quantitative PCR approach with improved sensitivity for the detection of low titer levels of Candidatus liberibacter asiaticus in the Asian citrus psyllid, Diaphorina citri kuwayama. J. Microbiol. Methods 102, 15–22. doi: 10.1016/j.mimet.2014.04.007
Dillon, R. J., and Dillon, V. M. (2004). The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92. doi: 10.1146/annurev.ento.49.061802.123416
Dillon, R. J., Webster, G., Weightman, A. J., and Charnley, A. K. (2010). Diversity of gut microbiota increases with aging and starvation in the desert locust. Antonie Van Leeuwenhoek 97, 69–77. doi: 10.1007/s10482-009-9389-5
Dobson, S. L., Bourtzis, K., Braig, H. R., Jones, B. F., Zhou, W., Rousset, F., et al. (1999). Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem. Mol. Bio. 29, 153–160. doi: 10.1016/s0965-1748(98)00119-2
Dossi, F. C. A. (2013). Morfogênese Do Bacterioma e Multiplicação De Simbiontes Ao Longo Do Desenvolvimento De Diaphorina Citri (Hemiptera: Liviidae) E Sua Resposta Ao Estresse Térmico. Ph.D. thesis, Universidade de São Paulo, São Paulo.
Dossi, F. C. A., da Silva, E. P., and Cônsoli, F. L. (2014). Population dynamics and growth rates of endosymbionts during Diaphorina citri (Hemiptera, Liviidae) ontogeny. Microb. Ecol. 68, 881–889. doi: 10.1007/s00248-014-0463-9
Douglas, A. E. (2007). Symbiotic microorganisms: untapped resources for insect pest control. Trends Biotechnol. 25, 338–342. doi: 10.1016/j.tibtech.2007.06.003
Douglas, A. E., Bouvaine, S., and Russell, R. R. (2010). How the insect immune system interacts with an obligate symbiotic bacterium. Proc. R. Soc. B Biol. Sci. 278, 333–338. doi: 10.1098/rspb.2010.1563
Duguma, D., Rugman-Jones, P., Kaufman, M. G., Hall, M. W., Neufeld, J. D., Stouthamer, R., et al. (2013). Bacterial communities associated with Culex mosquito larvae and two emergent aquatic plants of bioremediation importance. PLoS One 8:e72522. doi: 10.1371/journal.pone.0072522
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Grafton-Cardwell, E. E., and Daugherty, M. P. (2013). Asian Citrus Psyllid and Huanglongbing Disease. Pest Notes Publication 74155. St Davis, CA: University of California Agriculture and Natural Resources.
Grafton-Cardwell, E. E., Stelinski, L. L., and Stansly, P. A. (2013). Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 58, 413–432. doi: 10.1146/annurev-ento-120811-153542
Gryndler, M., Trilèová, J., Hršelová, H., Streiblová, E., Gryndlerová, H., and Jansa, J. (2013). Tuber aestivum vittad. mycelium quantified: advantages and limitations of a qPCR approach. Mycorrhiza 23, 341–348. doi: 10.1007/s00572-012-0475-6
Hamby, K. A., and Becher, P. G. (2016). Current knowledge of interactions between Drosophila suzukii and microbes, and their potential utility for pest management. J. Pest. Sci. 89, 621–630. doi: 10.1007/s10340-016-0768-1
Hosseinzadeh, S., Shams-Bakhsh, M., Mann, M., Fattah-Hosseini, S., Bagheri, A., Mehrabadi, M., et al. (2019). Distribution and variation of bacterial endosymbiont and “Candidatus liberibacter asiaticus” titer in the huanglongbing insect vector, Diaphorina citri kuwayama. Microb. Ecol. 78, 206–222. doi: 10.1007/s00248-018-1290-1
Hovatter, S. R., Dejelo, C., Case, A. L., and Blackwood, C. B. (2011). Metacommunity organization of soil microorganisms depends on habitat defined by presence of Lobelia siphilitica plants. Ecology 92, 57–65. doi: 10.1890/10-0332.1
Hung, T. H., Hung, S. C., Chen, C. N., Hsu, M. H., and Su, H. J. (2004). Detection by PCR of Candidatus liberibacter asiaticus, the bacterium causing citrus huanglongbing in vector psyllids: application to the study of vector–pathogen relationships. Plant Pathol. 53, 96–102. doi: 10.1111/j.1365-3059.2004.00948.x
Hussain, M., Akutse, K. S., Ravindran, K., Lin, Y., Bamisile, B. S., Qasim, M., et al. (2017). Effects of different temperature regimes on survival of Diaphorina citri and its endosymbiotic bacterial communities. Environ. Microbiol. 19, 3439–3449. doi: 10.1111/1462-2920.13821
Indiragandhi, P., Yoon, C., Yang, J. O., Cho, S., Sa, T. M., and Kim, G. H. (2010). Microbial communities in the developmental stages of B and Q biotypes of sweet potato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). J. Korean Soc. Appl. Biol. Chem 53, 605–617. doi: 10.3839/jksabc.2010.093
Jing, X., Wong, A. C. N., Chaston, J. M., Colvin, J., McKenzie, C. L., and Douglas, A. E. (2014). The bacterial communities in plant phloem-sap-feeding insects. Mol. Ecol. 23, 1433–1444. doi: 10.1111/mec.12637
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Kolora, L. D., Powell, C. M., Hunter, W., Bextine, B., and Lauzon, C. R. (2015). Internal extracellular bacteria of Diaphorina citri kuwayama (Hemiptera: Psyllidae), the asian citrus psyllid. Curr. Microbiol. 70, 710–715. doi: 10.1007/s00284-015-0774-1
Li, W., Duan, Y., Brlansky, R. H., Twieg, E., and Levy, L. (2008). “Incidence and population of ‘Candidatus liberibacter asiaticus’ in Asian citrus psyllids (Diaphorina citri) on citrus plants affected by huanglongbing in florida,” in Proceedings of the 2008 International Research Conference Huanglongbing, Orlando, FL.
Li, W., Hartung, J. S., and Levy, L. (2006). Quantitative real-time PCR for detection and identification of Candidatus liberibacter species associated with citrus huanglongbing. J. Microbiol. Meth. 66, 104–115. doi: 10.1016/j.mimet.2005.10.018
Login, F. H., and Heddi, A. (2013). Insect immune system maintains long-term resident bacteria through a local response. J. Insect Physiol. 59, 232–239. doi: 10.1016/j.jinsphys.2012.06.015
Macke, E., Tasiemski, A., Massol, F., Callens, M., and Decaestecker, E. (2017). Life history and eco-evolutionary dynamics in light of the gut microbiota. Oikos 126, 508–531. doi: 10.1111/oik.03900
Mann, M., Fattah-Hosseini, S., Ammar, E. D., Stange, R., Warrick, E., Sturgeon, K., et al. (2018). Diaphorina citri nymphs are resistant to morphological changes induced by Candidatus liberibacter asiaticus in midgut epithelial cells. Infect. Immun. 86:e00889-17. doi: 10.1128/IAI.00889-17
Morales-Jiménez, J., Zúñiga, G., Villa-Tanaca, L., and Hernández-Rodríguez, C. (2009). Bacterial community and nitrogen fixation in the red turpentine beetle, dendroctonus valens leconte (coleoptera: curculionidae: scolytinae). Microb. Ecol. 58, 879–891. doi: 10.1007/s00248-009-9548-2
Morrow, J. L., Hall, A. A., and Riegler, M. (2017). Symbionts in waiting: the dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome 5:58. doi: 10.1186/s40168-017-0276-4
Nachappa, P., Levy, J., Pierson, E., and Tamborindeguy, C. (2011). Diversity of endosymbionts in the potato psyllid, Bactericera cockerelli (Hemiptera: Triozidae), vector of zebra chip disease of potato. Curr. Microbiol. 62, 1510–1520. doi: 10.1007/s00284-011-9885-5
Nakabachi, A., Ueoka, R., Oshima, K., Teta, R., Mangoni, A., Gurgui, M., et al. (2013b). Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 23, 1478–1484. doi: 10.1016/j.cub.2013.06.027
Nakabachi, A., Nikoh, N., Oshima, K., Inoue, H., Ohkuma, M., Hongoh, Y., et al. (2013a). The 160-kilobase genome of the bacterial endosymbiont carsonella. Science 314, 267–267. doi: 10.1126/science.1134196
Nishikori, K., Morioka, K., Kubo, T., and Morioka, M. (2009). Age-and morph-dependent activation of the lysosomal system and Buchnera degradation in aphid endosymbiosis. J. Insect Physiol. 55, 351–357. doi: 10.1016/j.jinsphys.2009.01.001
Oliver, K. M., Smith, A. H., and Russell, J. A. (2014). Defensive symbiosis in the real world–advancing ecological studies of heritable, protective bacteria in aphids and beyond. Func. Ecol. 28, 341–355. doi: 10.1111/1365-2435.12133
Quigley, K. M., Davies, S. W., Kenkel, C. D., Willis, B. L., Matz, M. V., and Bay, L. K. (2014). Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS One 9:e94297. doi: 10.1371/journal.pone.0094297
Ramsey, J. S., Chavez, J. D., Johnson, R., Hosseinzadeh, S., Mahoney, J. E., Mohr, J. P., et al. (2017). Protein interaction networks at the host–microbe interface in Diaphorina citri, the insect vector of the citrus greening pathogen. R. Soc. Open. Sci. 4:160545. doi: 10.1098/rsos.160545
Ramsey, J. S., Johnson, R. S., Hoki, J. S., Kruse, A., Mahoney, J., Cilia, M., et al. (2015). Metabolic interplay between the asian citrus psyllid and its profftella symbiont: an achilles’ heel of the citrus greening insect vector. PLoS One 10:e0140826. doi: 10.1371/journal.pone.0140826
Saha, S., Hunter, W. B., Reese, J., Morgan, J. K., Marutani-Hert, M., Huang, H., et al. (2012). Survey of endosymbionts in the Diaphorina citri metagenome and assembly of a Wolbachia wDi draft genome. PLoS One 7:e50067. doi: 10.1371/journal.pone.0050067
Shade, A., and Handelsman, J. (2012). Beyond the venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Simonet, P., Duport, G., Gaget, K., Weiss-Gayet, M., Colella, S., Febvay, G., et al. (2016). Direct flow cytometry measurements reveal a fine-tuning of symbiotic cell dynamics according to the host developmental needs in aphid symbiosis. Sci. Rep. 6:19967. doi: 10.1038/srep19967
Spaulding, A. W., and von Dohlen, C. D. (1998). Phylogenetic characterization and molecular evolution of bacterial endosymbionts in psyllids (Hemiptera: Sternorrhyncha). Mol. Biol. Evol. 15, 1506–1513. doi: 10.1093/oxfordjournals.molbev.a025878
Subandiyah, S., Nikoh, N., Tsuyumu, S., Somowiyarjo, S., and Fukatsu, T. (2000). Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloidea). Zool. Sci. 17, 983–989. doi: 10.2108/zsj.17.983
Sugio, A., Dubreuil, G., Giron, D., and Simon, J. C. (2014). Plant–insect interactions under bacterial influence: ecological implications and underlying mechanisms. J. Exp. Bot. 66, 467–478. doi: 10.1093/jxb/eru435
Tamames, J., Gil, R., Latorre, A., Peretó, J., Silva, F. J., and Moya, A. (2007). The frontier between cell and organelle: genome analysis of Candidatus carsonella ruddii. BMC Evol. Biol. 7:181. doi: 10.1186/1471-2148-7-181
Thao, M. L., Moran, N. A., Abbot, P., Brennan, E. B., Burckhardt, D. H., and Baumann, P. (2000). Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl. Environ. Microbiol. 66, 2898–2905. doi: 10.1094/PHYTO.1998.88.9.950
Thiery, I., Nicolas, L., Rippka, R., and Marsac, N. T. D. (1991). Selection of cyanobacteria isolated from mosquito breeding sites as a potential food source for mosquito larvae. Appl. Environ. Microbiol. 57, 1354–1359. doi: 10.1016/0167-7799(91)90065-P
Uyaguari-Diaz, M. I., Chan, M., Chaban, B. L., Croxen, M. A., Finke, J. F., Hill, J. E., et al. (2016). A comprehensive method for amplicon-based and metagenomic characterization of viruses, bacteria, and eukaryotes in freshwater samples. Microbiome 4:20. doi: 10.1186/s40168-016-0166-1
van Elsas, J. D., and Boersma, F. G. H. (2011). A review of molecular methods to study the microbiota of soil and the mycosphere. Eur. J. Soil Biol. 47, 77–87. doi: 10.1016/j.ejsobi.2010.11.010
Webster, N. S., Taylor, M. W., Behnam, F., Lucker, S., Rattei, T., Whalan, S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12, 2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x
Welchman, D. P., Aksoy, S., Jiggins, F., and Lemaitre, B. (2009). Insect immunity: from pattern recognition to symbiont-mediated host defense. Cell Host Microbe 6, 107–114. doi: 10.1016/j.chom.2009.07.008
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Whelan, J. A., Russell, N. B., and Whelan, M. A. (2003). A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 278, 261–269. doi: 10.1016/S0022-1759(03)00223-0
Wong, A. C., Chaston, J. M., and Douglas, A. E. (2013). The inconstant gut microbiota of drosophila species revealed by 16S rRNA gene analysis. ISME J. 7, 1922–1932. doi: 10.1038/ismej.2013.86
Yin, Y., Liu, T., Tian, S., Hu, X., Wu, D., and Wang, Z. (2011). Microbial diversity in Diaphorina citri (Homoptera: Psyllidae) estimated by 16S rDNA analysis using DGGE and RFLP. Acta. Entomol. Sin. 54, 664–674. doi: 10.1017/S0022112010006427
Zolnik, C. P., Prill, R. J., Falco, R. C., Daniels, T. J., and Kolokotronis, S. O. (2016). Microbiome changes through ontogeny of a tick pathogen vector. Mol. Ecol. 25, 4963–4977. doi: 10.1111/mec.13832
Keywords: Illumina MiSeq sequencing, quantitative real-time PCR, bacterial community, Diaphorina citri, developmental stages
Citation: Meng L, Li X, Cheng X and Zhang H (2019) 16S rRNA Gene Sequencing Reveals a Shift in the Microbiota of Diaphorina citri During the Psyllid Life Cycle. Front. Microbiol. 10:1948. doi: 10.3389/fmicb.2019.01948
Received: 22 February 2019; Accepted: 08 August 2019;
Published: 23 August 2019.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Antonios Alekos Augustinos, Hellenic Agricultural Organisation, GreeceElena Crotti, University of Milan, Italy
Copyright © 2019 Meng, Li, Cheng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Zhang, aG9uZ3l1LnpoYW5nQG1haWwuaHphdS5lZHUuY24=
 Lixue Meng
Lixue Meng Xiaoyang Li
Xiaoyang Li Xiaoqin Cheng
Xiaoqin Cheng Hongyu Zhang
Hongyu Zhang