- 1School of Biological Sciences, Seoul National University, Seoul, South Korea
- 2Institute of Molecular Biology and Genetics, Seoul National University, Seoul, South Korea
- 3Department of Systems Biotechnology, Chung-Ang University, Anseong, South Korea
- 4School of Natural and Environmental Sciences, Newcastle University, Newcastle upon Tyne, United Kingdom
The present study was designed to clarify the taxonomic status of two species classified as Bacillus cereus sensu lato, namely B. cereus sensu stricto and Bacillus thuringiensis. To this end, nearly 900 whole genome sequences of strains assigned to these taxa were the subject of comparative genomic and phylogenomic analyses. A phylogenomic tree based on core gene sequences showed that the type strains of B. cereus and B. thuringiensis formed a well-supported monophyletic clade that was clearly separated from corresponding clades composed of the remaining validly published species classified as B. cereus sensu lato. However, since average nucleotide identity and digital DNA–DNA hybridization similarities between the two types of Bacillus were slightly higher than the thresholds used to distinguish between closely related species we conclude that B. cereus and B. thuringiensis should continue to be recognized as validly published species. The B. thuringiensis strains were assigned to two genomically distinct groups, we propose that these taxa be recognized as genomovars, that is, as B. thuringiensis gv. thuringiensis and B. thuringiensis gv. cytolyticus. The extensive comparative genomic data clearly show that the distribution of pesticidal genes is irregular as strains identified as B. thuringiensis were assigned to several polyphyletic groups/subclades within the B. cereus–B. thuringiensis clade. Consequently, we recommend that genomic or equivalent molecular systematic features should be used to identify B. thuringiensis strains as the presence of pesticidal genes cannot be used as a diagnostic marker for this species. Comparative taxonomic studies are needed to find phenotypic properties that can be used to distinguish between the B. thuringiensis genomovars and between them and B. cereus.
Introduction
Bacillus cereus sensu lato, also known as the B. cereus group, is a phylogenetically defined taxon within the genus Bacillus (Cohn, 1872) which encompasses an array of Gram-stain-positive, rod-shaped, facultatively anaerobic, endospore-forming bacteria that are common in natural habitats (Guinebretiere et al., 2013; Liu et al., 2017b; Patino-Navarrete and Sanchis, 2017). The group currently contains 21 validly published species (Liu et al., 2017a) which include Bacillus anthracis (Cohn, 1872), the causal agent of anthrax (Ezzell and Welkos, 1999; Moayeri et al., 2015); B. cereus (Frankland and Frankland, 1887), an opportunistic pathogen that causes food poisoning (Kotiranta et al., 2000; Bottone, 2010); Bacillus thuringiensis (Berliner, 1915), which produces insecticidal toxins widely used as biological control agents (Bravo et al., 2013; Raymond and Federici, 2017); and Bacillus toyonensis (Jimenez et al., 2013), which is used as a probiotic in animal nutrition. Members of these and related species assigned to the B. cereus group have been extensively studied given their economic and medical importance (Bottone, 2010; Bravo et al., 2013; Lacey et al., 2015; Hong et al., 2016; Kumari and Sarkar, 2016; Soni et al., 2016; Raymond and Federici, 2017).
Bacillus cereus sensu stricto is a common soil organism that is better known as a source of toxins associated with two forms of food poisoning, emesis and diarrhea. Emesis is caused by the toxin peptide cereulide that is encoded by ces genes located on a mega-virulence plasmid related to the B. anthracis toxin plasmid XO1 (Ehling-Schulz et al., 2004, 2005, 2015). Cereulide-producing B. cereus strains, in contrast to their diarrheal counterparts, form a single evolutionary lineage of closely related strains (Ehling-Schulz et al., 2006). Diarrheal food poisoning is caused by the single or combined action of heat-labile enterotoxins (Ehling-Schulz et al., 2004). In particular, three enterotoxins expressed by chromosomal genes (Fagerlund et al., 2010) are linked to this condition: the protein complexes hemolysin BL (Hbl), its non-hemolytic counterpart (Nhe), and the single protein cytotoxin K (CytK) (Stenfors Arnesen et al., 2008; Ceuppens et al., 2011). However, cytK and corresponding genes on the hbl operon are also evident in species of B. cereus sensu lato as a consequence of extensive lateral gene transfer events (Bohm et al., 2015); food poisoning toxicity can also be affected by transcription and unknown environmental factors (Jessberger et al., 2015). In turn, B. thuringiensis strains and associated parasporal crystal proteins are widely used as biological agents (Bt toxins) to control insect pests (Palma et al., 2014); the ability to synthesize crystal and cytotoxic enterotoxins are encoded by plasmid-borne cry and cyt genes, respectively (Schnepf et al., 1998; Palma et al., 2014). Many Bt toxins have been reported and classified based on amino acid sequences1 (Table 1; Crickmore et al., 1998; Berry and Crickmore, 2017).
Phenotypic and genotypic approaches have been used to characterize species assigned to the B. cereus group. Phenotypic markers include biochemical (Logan and Berkeley, 1984), colonial (Flugge, 1886), and plasmid-encoded features (Rasko et al., 2005) and genotypic methods by various procedures, such as DNA–DNA hybridization (Hill et al., 2004), multilocus enzyme electrophoresis (Carlson et al., 1994; Vilas-Boas et al., 2002), pulsed-field gel electrophoresis (Zhong et al., 2007), and single (Ko et al., 2004; La Duc et al., 2004) and multilocus sequence analyses of conserved housekeeping genes (Helgason et al., 2004; Priest et al., 2004). Recently combinations of these two approaches have been used for classifying members of the B. cereus group (Jimenez et al., 2013; Liu et al., 2017a). Such studies have clarified relationships among species classified as B. cereus sensu lato (Liu et al., 2017b) even though problems remain, notably in distinguishing between B. anthracis, B. cereus, and B. thuringiensis (Patino-Navarrete and Sanchis, 2017). The historical distinction between these taxa based on plasmid expressed features is not reliable; the loss of cry genes from B. thuringiensis strains, for instance, makes them indistinguishable from B. cereus sensu stricto (Lin et al., 1998). Further, members of these taxa are very difficult to differentiate using genotypic criteria, as witnessed by the low genetic diversity seen from analyses of 16S rRNA (Daffonchio et al., 2006) and protein-coding gene sequences (Priest et al., 2004) while DNA–DNA hybridization values between representatives of these species have been reported to be above the 70% cut-off point used to circumscribe prokaryotic species (Liu et al., 2017a). Genotypic properties such as these have led to suggestions that these taxa be recognized as a single species (Helgason et al., 2000, 2004; Rasko et al., 2005) or as subspecies of B. cereus sensu stricto (Ash et al., 1991). However, in the meantime, B. anthracis has been distinguished from B. cereus and B. thuringiensis using several taxonomic procedures (Radnedge et al., 2003; Zhong et al., 2007; Patino-Navarrete and Sanchis, 2017).
Classifications based on whole-genome sequences and associated bioinformatic tools are not only clarifying relationships between closely related bacteria that proved difficult to resolve using conventional taxonomic procedures (Carro et al., 2018; Gupta et al., 2018; Nouioui et al., 2018; Sangal et al., 2018) but are also providing improved metrics for recognizing species boundaries (Chun and Rainey, 2014; Chun et al., 2018), such as those based on average nucleotide identity (ANI; Goris et al., 2007; Arahal, 2014) and digital DNA–DNA hybridization (dDDH; Auch et al., 2010; Meier-Kolthoff et al., 2013). Similarly, dDDH values of 79–80% have been set for the recognition of sub-species (Meier-Kolthoff et al., 2014; Nouioui et al., 2018). B. cereus sensu lato species fall below the ANI and dDDH thresholds for separating species (95–96 and 70%, respectively), apart from B. cereus sensu stricto and B. thuringiensis (96.8 and 71.2%) and Bacillus mycoides and Bacillus weihenstephanensis (97.6 and 78.2% (Liu et al., 2017a). The latter is now considered to be a later heterotypic synonym of B. mycoides (Liu et al., 2018), but the relationship between B. cereus and B. thuringiensis has still to be resolved. However, it is now clear that the presence or absence of plasmid-bearing genes cannot be used to separate these taxa (Liu et al., 2015), a result in agreement to those of earlier studies (Kolsto et al., 2009; Zwick et al., 2012).
The present study was designed to determine the taxonomic relationship between B. cereus and B. thuringiensis based on comparisons of high-quality whole-genome sequences of nearly 900 strains. By linking phylogenomic relationships and the distribution of genes encoding toxin and other taxonomic markers, we propose that the bona fide members of B. cereus and B. thuringiensis be classified into three genomically coherent groups, B. cereus, B. thuringiensis genomovar thuringiensis, and B. thuringiensis genomovar cytolyticus; emended descriptions are given of B. cereus and B. thuringiensis.
Materials and Methods
Genome Data Acquisition and Filtering Out Low-Quality Genomes
Genome sequences and corresponding annotated protein sequences of the 973 B. cereus and B. thuringiensis strains were downloaded from the EzBioCloud database (Yoon et al., 2017a; Supplementary Table S1). Genomes with either >500 contigs or an N50 size under 20,000 bp were excluded from the dataset as they were considered to be poorly assembled, as were genomes found by CheckM to be either incomplete (<95%) or had a contamination rate above 5% (Parks et al., 2015). The completeness of the genomes was also checked using bacterial core genes extracted by UBCG software (v.3.02; Na et al., 2018); when >5% of the 92 core gene set was absent the genomes were excluded from further analysis. Outlier strains with long branches were removed by TreeShrink (Mai and Mirarab, 2018) from the FastTree (Price et al., 2010)-driven phylogenomic tree. The resultant 898 strains were the subject of further studies.
Phylogenomic Tree Reconstruction of the Bacillus cereus–Bacillus thuringiensis Clade
Based on the core genes extracted using the UBCG software, a maximum-likelihood phylogenomic tree of the filtered B. cereus and B. thuringiensis strains was constructed by IQ-TREE version 1.6.7 (Nguyen et al., 2015) with 1000 bootstrap replications (Minh et al., 2013; Hoang et al., 2018). The model used for the phylogenetic estimations was automatically detected as the GTR + F + R10 model by using the ModelFinder algorithm implemented in IQ-TREE (Kalyaanamoorthy et al., 2017). UBCG software executed with the RAxML (v. 8.2.8) option was used to generate a more rigorous maximum-likelihood phylogenomic tree which included genomes of the type species belonging to the B. cereus group and related taxa (Stamatakis, 2014; Na et al., 2018). The iTOL v3 webserver3 (Letunic and Bork, 2016) was used to display various gene contents with the phylogenetic trees.
Calculation of the Overall Genome Sequence Relatedness Among the Strains of B. cereus and B. thuringiensis
Pairwise relatedness values based on whole-genome sequences were calculated using OrthoANIu software for ANI (Yoon et al., 2017b) and GGDC v. 2.1 for dDDH (Meier-Kolthoff et al., 2013).
Detection of Toxin-Related Genes in B. cereus and B. thuringiensis Genomes
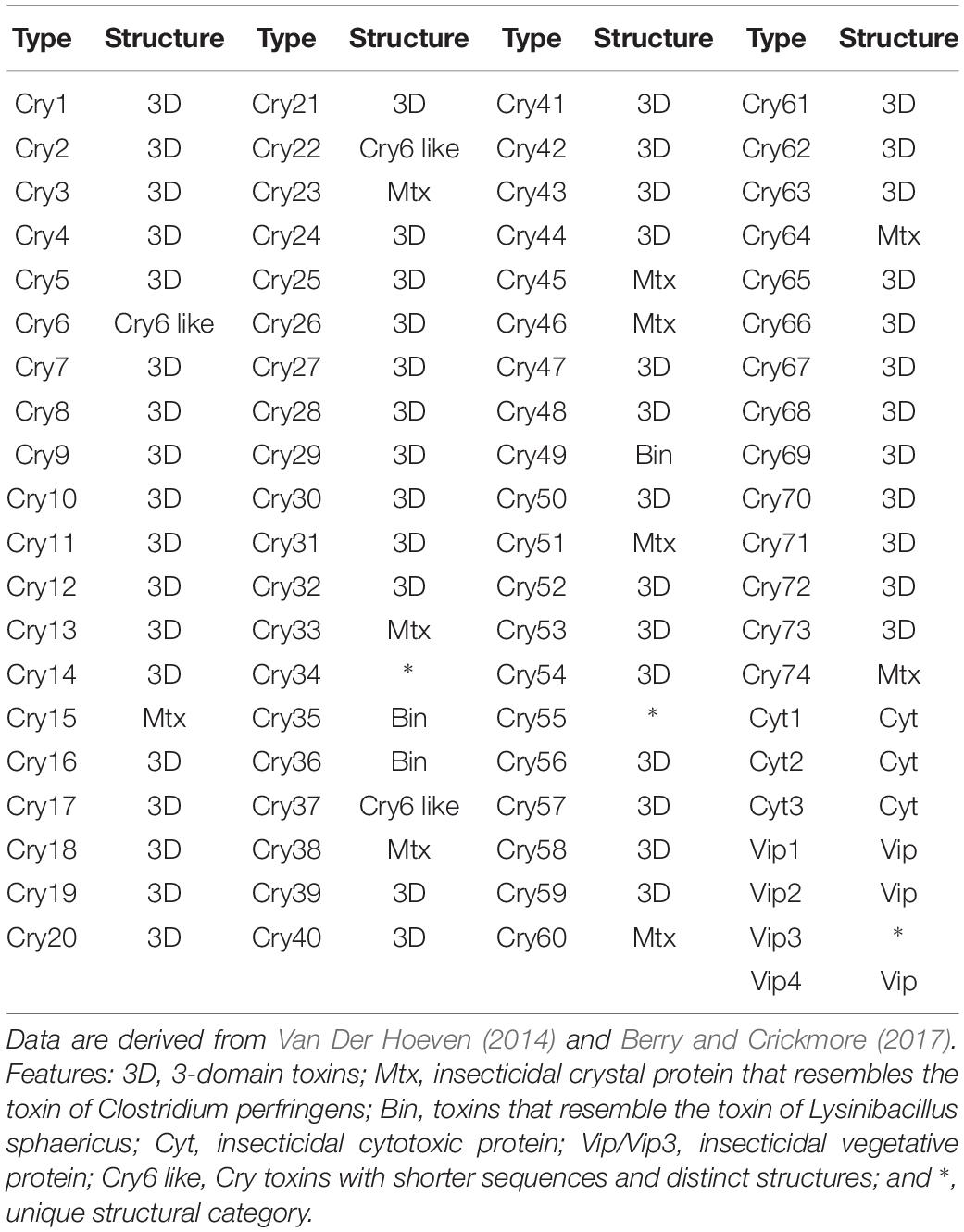
Bt toxin genes used as references for the genomic analyses were downloaded from the Bt toxin nomenclature database4 (Crickmore et al., 1998); the structural classifications of the Bt toxins were taken from an earlier study (Van Der Hoeven, 2014). Other reference pathogenic genes detected in the B. cereus group were used as described previously (Kovac et al., 2016). The gene sequences were downloaded from the UniProt database (Apweiler et al., 2004) and additional information drawn from SwissProt (Boutet et al., 2016). The presence or absence of genes homologous to the reference toxins was determined by the tblastn search from the standalone BLAST package (Camacho et al., 2009). Parameters were set as default options except for the e-value cutoff 1e−5. Hits with ≥70% sequence identity and ≥70% alignment length were used as a cutoff to recognize homologs (Yoon et al., 2017a). In addition, a 78% sequence identity cutoff was applied to pesticidal genes for annotating secondary subgroups (Crickmore et al., 1998).
Results
Genomic Characteristics and Relatedness of the B. cereus and B. thuringiensis Strains
In general, most of the genomes identified as either B. cereus or B. thuringiensis were found to have genome sizes within the range 5.0–7.9 Mbp while the corresponding G + C content ranged from 33.8 to 35.4 mol% (Supplementary Table S1). It is evident from the phylogenomic tree that the B. cereus and B. thuringiensis strains are closely related and separated from the type strains of other species belonging to B. cereus sensu lato (Figure 1). The ANI and dDDH values between type strains of B. cereus and B. thuringiensis were 96.71 and 71.20%, respectively (Table 2), that is greater than the species boundary cutoffs (95–96 and 70%, respectively). Within the clade encompassing the B. cereus and B. thuringiensis strains, three subclades were recognized at the 96% ANI cutoff; these were considered to correspond to B. cereus, B. thuringiensis genomovar cytolyticus, and B. thuringiensis genomovar thuringiensis (Figure 2A).
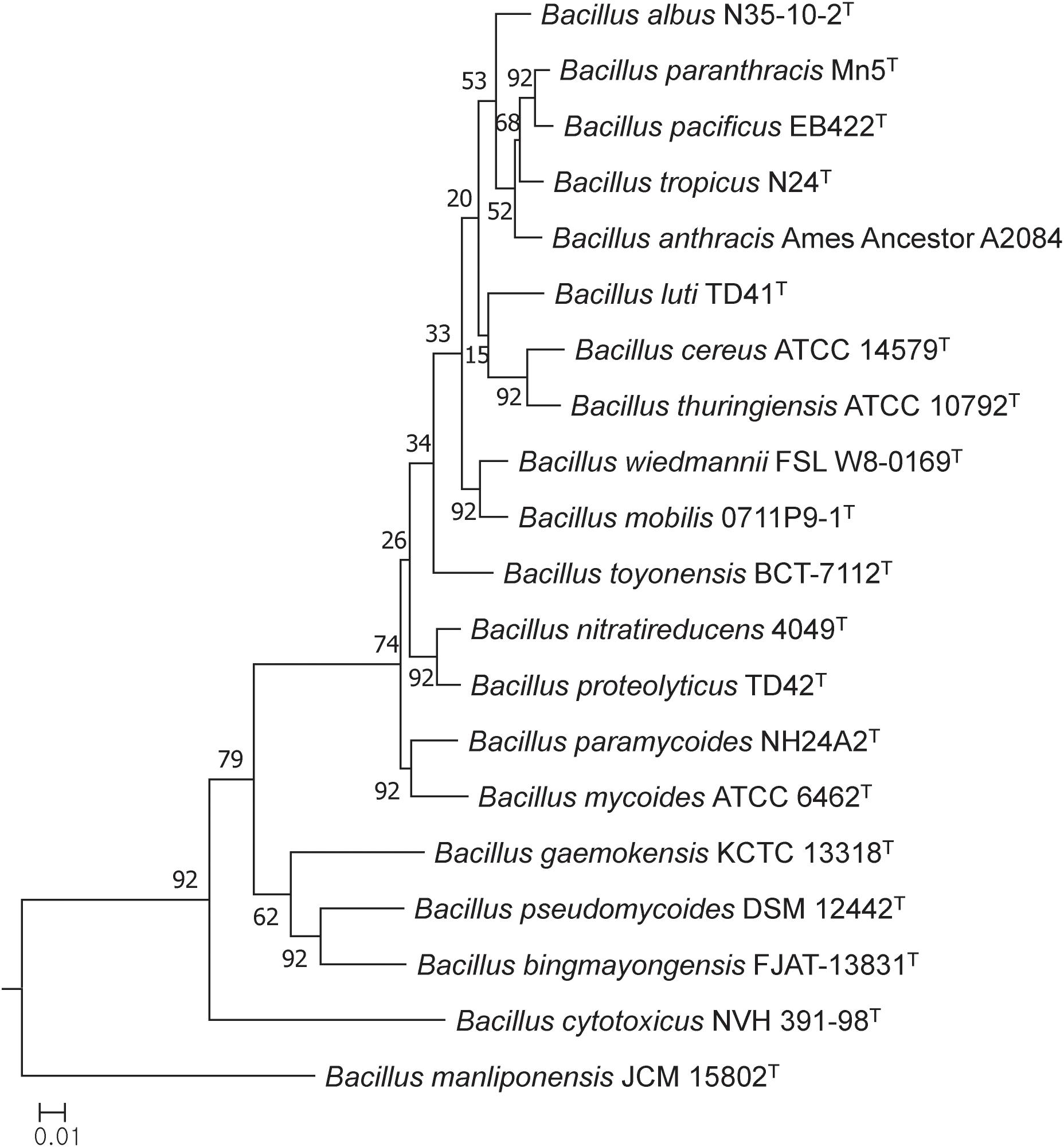
Figure 1. Maximum-likelihood tree based on 92 single copy core gene sequences showing relationships between the type trains of B. cereus and B. thuringiensis and between them and corresponding strains of other B. cereus sensu lato species. The numbers at the nodes are bootstrap values based on 1000 replicates. The bar stands for the number of substitutions per nucleotide.
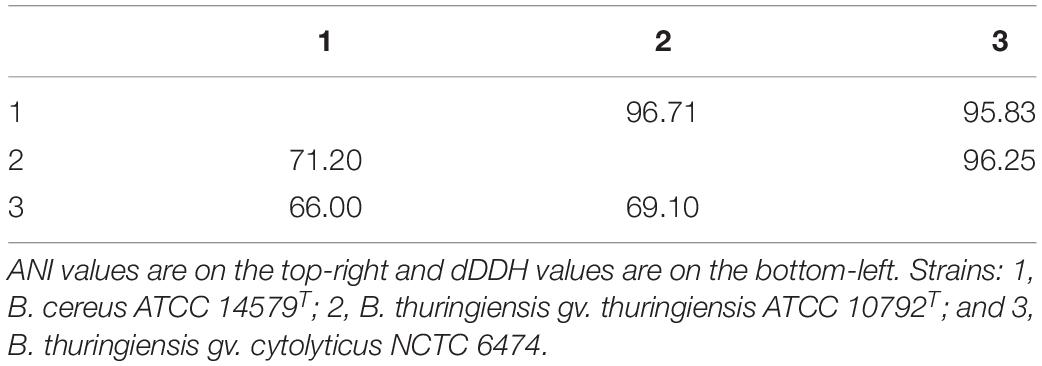
Table 2. Pairwise ANI (Yoon et al., 2017b) and dDDH (Meier-Kolthoff et al., 2013) values between the type/representative strains of three subclades of the B. cereus–B. thuringiensis clade.
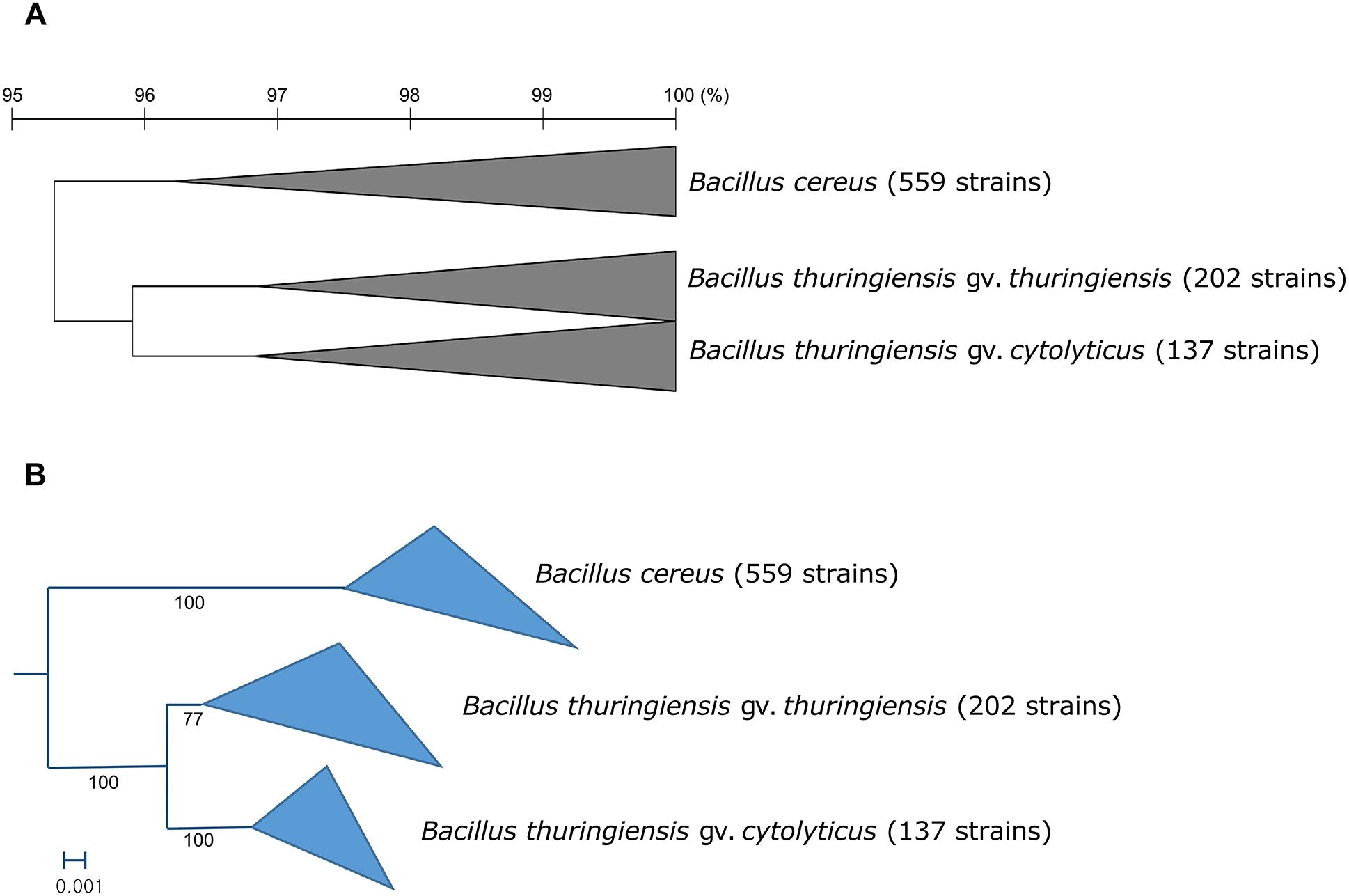
Figure 2. (A) Collapsed OrthoANIu dendrogram and (B) collapsed phylogenomic tree based on 92 universal bacterial genes showing relationships between the three subclades of the B. cereus–B. thuringiensis clade. Bacillus toyonensis BCT-7112T was used as the outgroup (not shown). The scale bar on the OrthoANIu dendrogram stands for OrthoANIu values (%). Bar on the phylogenomic tree indicates the number of substitutions per nucleotide.
Phylogenomic Overview of B. cereus and B. thuringiensis
The topology of the genome-based phylogenetic tree based on universal bacterial core genes showed that the analyzed strains could be assigned to three subclades (Figure 2B). This result indicates that the current taxonomic annotation in public databases is incorrect because strains annotated as either B. cereus or B. thuringiensis are irregularly positioned across the phylogenetic tree.
The Absence of Five Genes Involved in Emesis
It can be seen from the genomic mining data that five genes (cesA, cesB, cesC, cesD, and cesE) involved in producing the vomiting-induced endotoxin cereulide (Ezzell and Welkos, 1999) were absent from all of the analyzed genomes. In contrast, two transcriptional regulators involved in activating emesis, namely abrB and codY (Lucking et al., 2015), were universally present (Supplementary Table S2).
Distribution of B. cereus Toxin Genes
Multiple types of diarrheal causative toxins are formed by members of the B. cereus group, such as those expressed by operons (nhe and hbl) and genes (cytK, entA, and entFM); those present in the nhe operon were found in all but three of the genomes, namely strains GeD10, BGSC 4BW1, and SJ-S28. Similarly, four genes (hblA, hblB, hblC, and hblD) belonging to the hbl operon were completely absent from 29 out of the 898 strains belonging to the B. cereus–B. thuringiensis clade. The genomes from most of the strains carried a single gene associated with enterotoxin production; 898 entA genes, 843 cytK genes, and 896 entFM genes. In addition, only four genomes lacked cerA, one cerB, and four clo (Supplementary Table S2). The distributions of B. cereus toxin genes among the strains of B. cereus–B. thuringiensis clade were visualized in the phylogenomic tree (Figure 3).
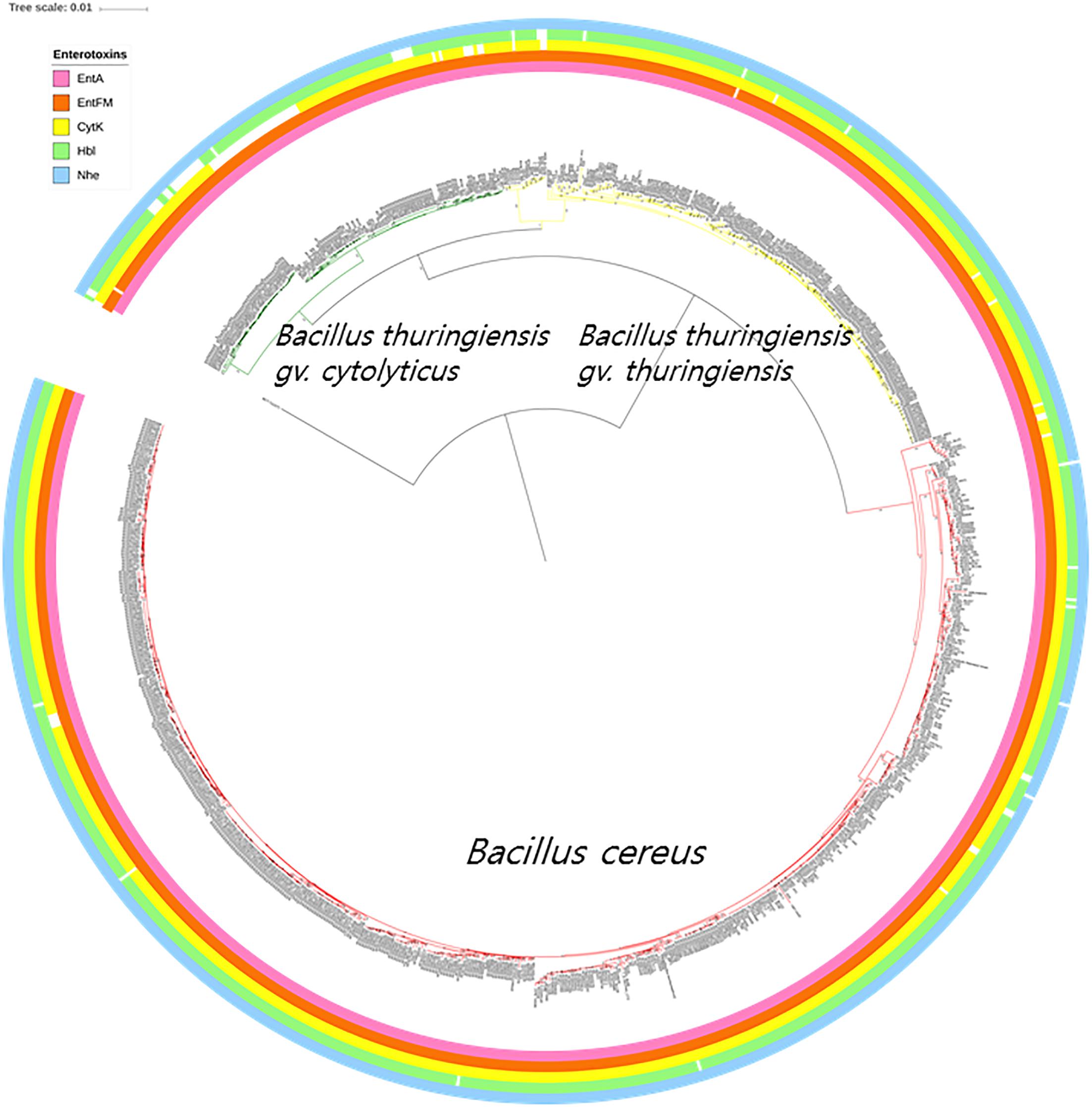
Figure 3. The bacterial core gene-based phylogenomic tree of Bacillus cereus and Bacillus thuringiensis strains together with labeled enterotoxin genes. The numbers at each branching point are bootstrap values. Bar indicates the nucleotide substitution rate per site. Bacillus toyonensis BCT-7112T was used as the outgroup. Taxonomic groups are indicated in different colors: red, B. cereus; yellow, B. thuringiensis gv. thuringiensis; and green, B. thuringiensis gv. cytolyticus. Genes are displayed on the outer circles: EntA, enterotoxin A; EntFM, enterotoxin FM; CytK, cytotoxin K; Hbl, hemolysin BL; Nhe, non-hemolytic enterotoxin.
The Absence of Anthrax-Related Genes
Genes associated with the production of anthrax toxin (atxA, cya, lef, and pagA) were not detected in the genomes of the B. cereus and B. thuringiensis strains even though some of them possessed two capsule forming genes (Supplementary Table S2). Also, the analyzed strains do not have the capsular synthesis regulator genes acpA and acpB (Drysdale et al., 2004). This suggests that the capsule formation genes may not be expressed unless an unknown transcription factor reacts with them (Supplementary Table S2).
The Intermittent Distribution of Bt Toxins in the Phylogenomic Tree
It is apparent from the genomic data that Bt toxin genes are present in strains distributed across the three subclades though unlike the diarrheal toxin operons (nhe and hbl) they do not predominant in these subclades (Figure 4). Cry genes with three domains (Cry-3D) were prevalent compared with other types of Cry genes. It can also be concluded from the phylogenetic analysis that Cry-3D genes have frequently been transferred among members of the B. cereus group, especially toxins belonging to the Cry1 and Cry2 groups (Supplementary Table S2). Some of the Cry toxins detected in the genomes were found to have distinct structures. Cry toxins homologous to the Mtx toxin of Clostridium perfringens were detected in 15 strains while seven strains were shown to contain a distinct structure and shorter sequences of the Cry toxin (Cry6, Cry22, and Cry37; named as “Cry6-like toxins”). Only strain 62 contained a Cry toxin homologous with the toxin of Lysinibacillus sphaericus (Cry-Bin). Further, some strains contained a pair of Cry proteins that acted as conjugative toxins. Two neighboring strains were found to have the Cry23–Cry37 pair (BGSC 4AA1 and BGSC 4BR1) and strain 62 possesses a Cry34–Cry35 pair. It is particularly interesting that Cyt, Vip, and other types of pesticidal toxins were associated with the genomes of strains spread across the phylogenetic tree (Figure 4).
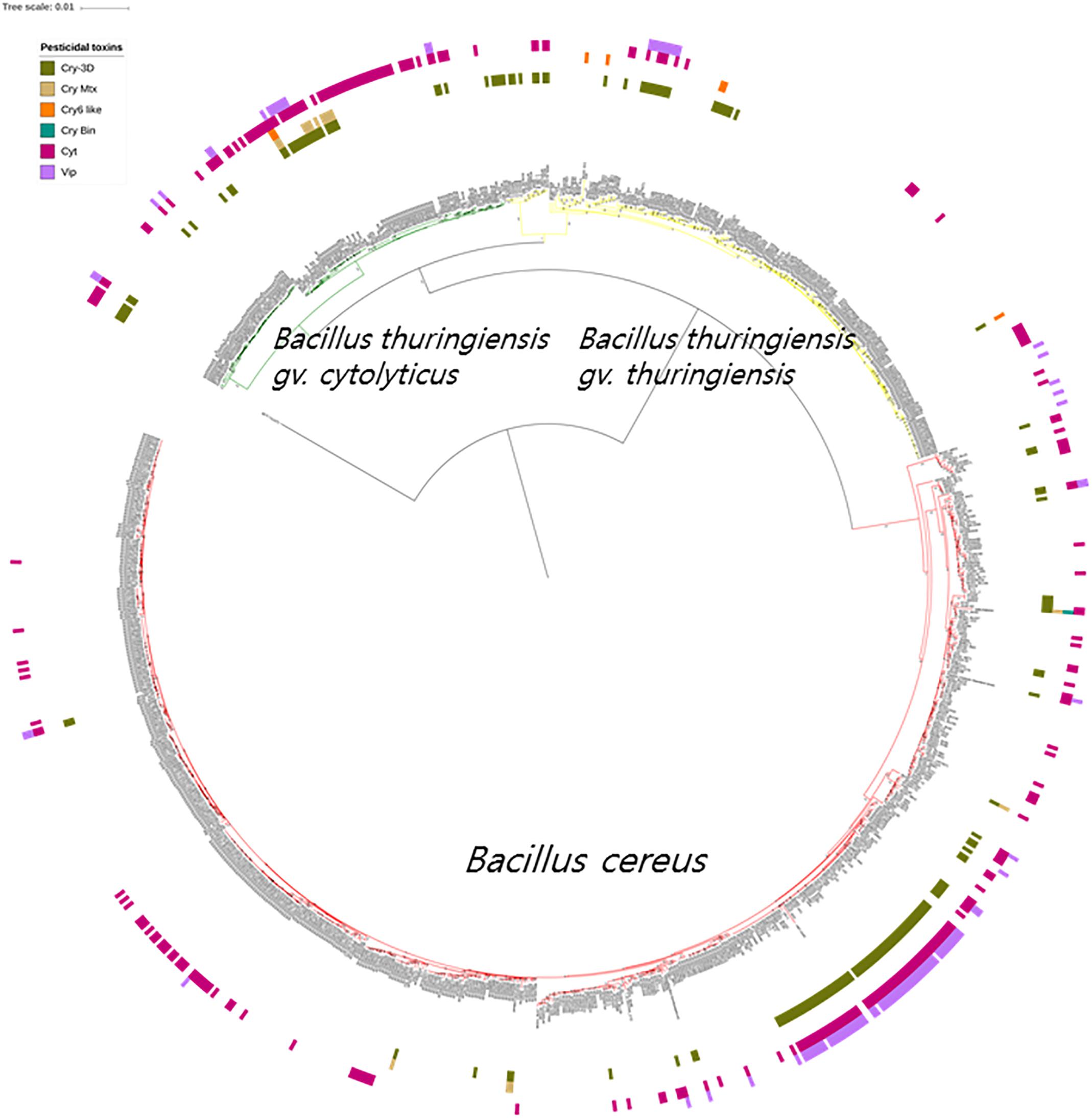
Figure 4. The bacterial core gene-based phylogenomic tree of Bacillus cereus and Bacillus thuringiensis strains together with labeled pesticidal genes. The numbers at each branching points are bootstrap values. Bar indicates the nucleotide substitution rate per site. Bacillus toyonensis BCT-7112T was used as the outgroup. Taxonomic groups are indicated in different colors: red, B. cereus; yellow, B. thuringiensis gv. thuringiensis; and green, B. thuringiensis gv. cytolyticus. Genes are displayed on the outer circles: Cry 3D, insecticidal crystal protein with three protein domains; Cry Mtx, insecticidal crystal protein that sequentially resembles the toxin of Clostridium perfringens; Cry Bin, a conjugated insecticidal crystal protein that sequentially resembles the toxin of Lysinibacillus sphaericus; Cry6 like, Cry toxins with shorter sequence and distinct structures; Cyt, insecticidal cytotoxic protein; Vip, insecticidal vegetative protein.
Discussion
This study was undertaken to try and resolve the confusion over the taxonomic status of B. cereus (Frankland and Frankland, 1887) and B. thuringiensis (Berliner, 1915), taxa of importance to agriculture, industry, and medicine (Bottone, 2010; Bravo et al., 2013; Lacey et al., 2015; Hong et al., 2016; Kumari and Sarkar, 2016; Soni et al., 2016; Raymond and Federici, 2017). The outcome of the study together with those of previous phylogenomic analyses of species classified as B. cereus sensu lato (Liu et al., 2015, 2017b, 2018) is a timely reminder that advances in the classification, nomenclature, and identification of prokaryotes reflect the introduction of new metrics for the recognition of species and genera (Chun and Rainey, 2014; Chun et al., 2018) which are based on better quality taxonomic data drawn from the application of new technologies (Goodfellow et al., 2014). This study also provides further evidence that analyses of high-quality genome sequences provide a framework for the clarification of taxonomically complex groups of prokaryotes (Carro et al., 2018; Sangal et al., 2018). Further, genome-based taxonomic investigations have proved clarifying the status of taxa of biotechnological, ecological, and medical importance, as exemplified by work on the phylum Actinobacteria (Nouioui et al., 2018) and more specifically the genera Amycolatopsis (Sangal et al., 2018) and Mycobacterium (Gupta et al., 2018).
The phylogenetic tree based on core genome sequences shows that the type strains of B. cereus and B. thuringiensis form a well-supported clade that can be distinguished from corresponding lineages composed of strains of other validly published species classified as B. cereus sensu lato. In general, better resolution was found between the type strains in the core gene tree than in the corresponding 16S rRNA gene tree generated by Liu et al. (2017a).
The genomic screening data underpin and extend those from many previous studies which found that diarrheal and Bt toxin genes cannot be used to distinguish between B. cereus and B. thuringiensis (Carlson et al., 1996; Helgason et al., 2000; Priest et al., 2004; Rasko et al., 2007; Stenfors Arnesen et al., 2008; Liu et al., 2015; Miller et al., 2018). In the present study, multiple diarrheal toxin genes (e.g., cytK, entA, entFM, hbl, and nhe) were found in nearly all members of the B. cereus–B. thuringiensis clade whereas strains with Bt toxin genes were shown to be polyphyletic in the phylogenomic tree (Figures 3, 4). Emetic toxin genes involved in the production of cereulide (Ezzell and Welkos, 1999) were not present in any of the B. cereus and B. thuringiensis strains thereby providing further evidence that diarrheal and emetic B. cereus belong to different evolutionary lineages (Ehling-Schulz et al., 2005; Carlin et al., 2006). In this respect, it is interesting that the reference strain of emetic B. cereus, namely AH 187 (=F4810/72) (Rasko et al., 2007) has been shown to be a member of Bacillus paranthracis, a species assigned to B. cereus sensu lato by Liu et al. (2017a)5. In turn, none of the B. cereus and B. thuringiensis strains contained genes (atxA, cya, lef, and pagA) related to those implicated in the production of anthrax toxins (Kovac et al., 2016).
Within the monophyletic clade encompassing the B. cereus and B. thuringiensis strains, three subclades were consistently recovered by ANI-based hierarchical clustering (Figure 2A) and bacterial core gene-based phylogenetic analysis (Figure 2B), respectively. The subclade that includes B. cereus type strain represents the B. cereus whereas the two other subclades were designated as two genomovars, namely thuringiensis and cytolyticus of B. thuringiensis. We propose strain NCTC 6474 (NCBI genome accession GCA_900445335.1) as the representative strain of B. thuringiensis gv. cytolyticus. The two genomovars proposed in this study may be equated with the rank of subspecies and formally described as such when sufficient supporting phenotypic data are acquired. It is noteworthy that genomovars represent genomically coherent taxa at the intra-species level that is not covered by the International Code of Nomenclature of Prokaryotes (Parker et al., 2019), as spelled out by Rule 14a and in Appendix 10.
One of the advantages of genome-based classification is the use of objective criteria in the definition of species. In this study, the ANI and dDDH values found between the type strains of B. cereus and B. thuringiensis were slightly higher than the generally accepted species boundary cutoffs. Despite this, we decided against combining the two species into single species as the name B. thuringiensis has been extensively used in various microbiological disciplines, especially in agriculture and biotechnology. Instead, we have provided a genome-based taxonomic framework where B. cereus and B. thuringiensis isolates can be identified not by the unreliable biomarkers (e.g., toxin genes) but by objective molecular methods.
Conclusion
In conclusion, on the basis of large-scale genomic analyses, we propose that B. thuringiensis be divided into two genomovars and the isolates of B. cereus and B. thuringiensis be identified by genome-based methods, but not by phenotypic or genotypic characterization involving insecticidal genes.
Emended Description of Bacillus cereus Frankland and Frankland, 1887
Data are taken from the present study and from the description of B. cereus as given by De Vos and Logan (2011).
Gram-stain-positive, facultatively anaerobic, usually motile rods (1.0–1.2 × 3.0–5.0 μm) that occur singly and in pairs and chains; and form ellipsoidal, sometimes cylindrical, subterminal, sometimes paraientral spores in unswollen sporangia; spores may lie obliquely in the sporangia. The sporangia of some strains carry parasporal bodies adjacent to spores. These crystalline protein inclusions, which vary in shape, are found outside the exosporium and are readily separated from liberated spores. The crystalline proteins and cytolytic proteins are prototoxins which may be toxic for certain insects and other invertebrates, including flatworms, mites, nematodes, and protozoa. The ability to synthesize parasporal bodies is plasmid-borne and may be lost on subculture. Cells grown on glucose agar produce large amounts of storage material giving them a vacuolated to foaming appearance. Cells are characteristically large (2–7 μm in diameter) and vary in shape from circular to irregular with entire to undulate, crenate, or fimbriate edges; they usually have matt or granular textures, but smooth and moist colonies may occur. The minimum temperature for growth is usually 10−20°C, and the maximum 40−50°C. Catalase positive and oxidase negative. Voges–Proskauer positive. Citrate is used as a sole carbon source. Endospores are widespread in soil and many other environments. The diarrheal enterotoxins are widely present, but emetic enterotoxins are absent. Genome sizes range from 5.1 to 7.9 Mbp and corresponding DNA G + C values are within the range 33.8–35.4%, based on 559 genome sequences.
Type strain is ATCC 14579T (=DSM 31T=JCM 2152T=LMG 6923T=NCIMB 9373T=NRRL B-3711T=IAM 12605T).
Emended Description of Bacillus thuringiensis Berliner, 1915
Data are taken from the present study and from the description of B. thuringiensis as given by De Vos and Logan (2011).
Gram-stain-positive, facultatively anaerobic, usually motile rods (1.0–1.2 × 3.0–5.0 μm) that occur singly and in pairs and chains; and form ellipsoidal, sometimes cylindrical, subterminal, sometimes paraientral spores in unswollen sporangia; spores may lie obliquely in the sporangia. The sporangia of some strains carry parasporal bodies adjacent to the spores. These crystalline protein inclusions, which vary in shape, are found outside the exosporium and are readily separated from liberated spores. The crystalline proteins and cytolytic proteins are prototoxins which may be toxic for certain insects and other invertebrates, including flatworms, mites, nematodes, and protozoa. The ability to synthesize parasporal bodies is plasmid-borne and may be lost on subculture. Cells grown on glucose agar produce large amounts of storage material giving them a vacuolated to foaming appearance. Cells are characteristically large (2–7 μm in diameter) and vary in shape from circular to irregular with entire to undulate, crenate, or fimbriate edges; they usually have matt or granular textures, but smooth and moist colonies may occur. The minimum temperature for growth is usually 10–15°C, and the maximum 40–50°C. Catalase positive and oxidase negative. Voges–Proskauer positive. Citrate is used as a sole carbon source. Endospores are widespread in soil and many other environments. The diarrheal enterotoxin is present in most strains. Strains can be assigned to two genomovars based on genomic relatedness; B. thuringiensis genomovar cytolyticus and B. thuringiensis genomovar thuringiensis. The majority of strains belonging to genomovar cytolyticus produce cytolytic toxins whereas few of those in genomovar thuringiensis do so. Genome sizes range from 5.0 to 7.1 Mbp and corresponding DNA G + C values are within the range 33.8–35.4%, based on 339 genome strains.
Type strain is ATCC 10792T (=CCUG 7429T=CIP 53.137T=DSM 2046T=HAMBI 478T=JCM 20386T=LMG 7138T=NBRC 101235T=NCAIM B.01292T=NCCB 70008T=NRRL HD-735T=VKM B-1544T).
Data Availability
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ezbiocloud.net/.
Author Contributions
IB, MG, and JC derived the idea for the whole project. IB performed the data collection and phylogenomic analysis, and IB, KL, MG, and JC wrote the manuscript. All authors approved the manuscript.
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) of Korea (918013-04-2-SB010), and also by the National Research Foundation of Korea (NRF-2014M3C9A3063541) funded by the Ministry of Science and ICT, South Korea.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01978/full#supplementary-material
Footnotes
- ^ http://www.btnomenclature.info
- ^ https://www.ezbiocloud.net/tools/ubcg
- ^ https://itol.embl.de/
- ^ http://www.btnomenclature.info/
- ^ https://www.ezbiocloud.net/genome/explore?puid=696
References
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., et al. (2004). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119.
Arahal, D. R. (2014). “Whole-genome analyses: average nucleotide identity,” in Methods in Microbiology, eds. M. Goodfellow, I. C. Sutcliffe, and J. Chun (Amsterdam: Elsevier), 103–122.
Ash, C., Farrow, J. A., Dorsch, M., Stackebrandt, E., and Collins, M. D. (1991). Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41, 343–346. doi: 10.1099/00207713-41-3-343
Auch, A. F., Von Jan, M., Klenk, H. P., and Goker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Berliner, E. (1915). Über die schlaffsucht der mehlmottenraupe (Ephestia kühniella Zell) und ihren erreger Bacillus thuringiensis n. sp. Zeitschrift fur angewandte Entomologie Berlin 2, 29–56. doi: 10.1111/j.1439-0418.1915.tb00334.x
Berry, C., and Crickmore, N. (2017). Structural classification of insecticidal proteins - towards an in silico characterisation of novel toxins. J. Invertebr. Pathol. 142, 16–22. doi: 10.1016/j.jip.2016.07.015
Bohm, M. E., Huptas, C., Krey, V. M., and Scherer, S. (2015). Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol. Biol. 15:246. doi: 10.1186/s12862-015-0529-4
Bottone, E. J. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23, 382–398. doi: 10.1128/CMR.00073-09
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/swiss-prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Bravo, A., Gomez, I., Porta, H., Garcia-Gomez, B. I., Rodriguez-Almazan, C., Pardo, L., et al. (2013). Evolution of Bacillus thuringiensis cry toxins insecticidal activity. Microb Biotechnol. 6, 17–26. doi: 10.1111/j.1751-7915.2012.00342.x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carlin, F., Fricker, M., Pielaat, A., Heisterkamp, S., Shaheen, R., Salonen, M. S., et al. (2006). Emetic toxin-producing strains of Bacillus cereus show distinct characteristics within the Bacillus cereus group. Int. J. Food Microbiol. 109, 132–138. doi: 10.1016/j.ijfoodmicro.2006.01.022
Carlson, C. R., Caugant, D. A., and Kolsto, A. B. (1994). Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 60, 1719–1725.
Carlson, C. R., Johansen, T., and Kolsto, A. B. (1996). The chromosome map of Bacillus thuringiensis subsp. canadensis HD224 is highly similar to that of the Bacillus cereus type strain ATCC 14579. FEMS Microbiol. Lett. 141, 163–167. doi: 10.1016/0378-1097(96)00216-9
Carro, L., Nouioui, I., Sangal, V., Meier-Kolthoff, J. P., Trujillo, M. E., Montero-Calasanz, M. D. C., et al. (2018). Genome-based classification of Micromonosporae with a focus on their biotechnological and ecological potential. Sci. Rep. 8:525. doi: 10.1038/s41598-017-17392-0
Ceuppens, S., Van De Wiele, T., Boon, N., and Uyttendaele, M. (2011). Gastrointestinal passage of Bacillus cereus. Commun. Agric. Appl. Biol. Sci. 76, 3–6.
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., Da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Chun, J., and Rainey, F. A. (2014). Integrating genomics into the taxonomy and systematics of the bacteria and archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324. doi: 10.1099/ijs.0.054171-0
Crickmore, N., Zeigler, D. R., Feitelson, J., Schnepf, E., Van Rie, J., Lereclus, D., et al. (1998). Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 807–813.
Daffonchio, D., Raddadi, N., Merabishvili, M., Cherif, A., Carmagnola, L., Brusetti, L., et al. (2006). Strategy for identification of Bacillus cereus and Bacillus thuringiensis strains closely related to Bacillus anthracis. Appl. Environ. Microbiol. 72, 1295–1301. doi: 10.1128/aem.72.2.1295-1301.2006
De Vos, P., and Logan, N. A. (2011). Bergey’s Manual of Systematic Bacteriology. New York, NY: Springer.
Drysdale, M., Bourgogne, A., Hilsenbeck, S. G., and Koehler, T. M. (2004). atxA controls Bacillus anthracis capsule synthesis via acpA and a newly discovered regulator, acpB. J. Bacteriol. 186, 307–315. doi: 10.1128/jb.186.2.307-315.2004
Ehling-Schulz, M., Frenzel, E., and Gohar, M. (2015). Food-bacteria interplay: pathometabolism of emetic Bacillus cereus. Front. Microbiol. 6:704. doi: 10.3389/fmicb.2015.00704
Ehling-Schulz, M., Fricker, M., Grallert, H., Rieck, P., Wagner, M., and Scherer, S. (2006). Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6:20. doi: 10.1186/1471-2180-6-20
Ehling-Schulz, M., Fricker, M., and Scherer, S. (2004). Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 48, 479–487. doi: 10.1002/mnfr.200400055
Ehling-Schulz, M., Svensson, B., Guinebretiere, M. H., Lindback, T., Andersson, M., Schulz, A., et al. (2005). Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151, 183–197. doi: 10.1099/mic.0.27607-0
Ezzell, J. W., and Welkos, S. L. (1999). The capsule of Bacillus anthracis, a review. J. Appl. Microbiol. 87:250. doi: 10.1046/j.1365-2672.1999.00881.x
Fagerlund, A., Lindback, T., and Granum, P. E. (2010). Bacillus cereus cytotoxins Hbl, Nhe and CytK are secreted via the sec translocation pathway. BMC Microbiol. 10:304. doi: 10.1186/1471-2180-10-304
Frankland, G. C., and Frankland, P. F. (1887). Studies on some new microorganisms obtained from air. R. Soc. Lond. Philos. Trans. B Biol. Sci. 178, 257–287. doi: 10.1098/rstb.1887.0011
Goodfellow, M., Sutcliffe, I., and Chun, J. (2014). Methods in microbiology preface. New Approaches Prokaryotic Systematics 41:348.
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Guinebretiere, M. H., Auger, S., Galleron, N., Contzen, M., De Sarrau, B., De Buyser, M. L., et al. (2013). Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int. J. Syst. Evol. Microbiol. 63, 31–40. doi: 10.1099/ijs.0.030627-0
Gupta, R. S., Lo, B., and Son, J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Helgason, E., Okstad, O. A., Caugant, D. A., Johansen, H. A., Fouet, A., Mock, M., et al. (2000). Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis–one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66, 2627–2630. doi: 10.1128/aem.66.6.2627-2630.2000
Helgason, E., Tourasse, N. J., Meisal, R., Caugant, D. A., and Kolsto, A. B. (2004). Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70, 191–201. doi: 10.1128/aem.70.1.191-201.2004
Hill, K. K., Ticknor, L. O., Okinaka, R. T., Asay, M., Blair, H., Bliss, K. A., et al. (2004). Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 70, 1068–1080. doi: 10.1128/aem.70.2.1068-1080.2004
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hong, M., Wang, Q., Tang, Z., Wang, Y., Gu, Y., Lou, Y., et al. (2016). Association of genotyping of Bacillus cereus with clinical features of post-traumatic endophthalmitis. PLoS One 11:e0147878. doi: 10.1371/journal.pone.0147878
Jessberger, N., Krey, V. M., Rademacher, C., Bohm, M. E., Mohr, A. K., Ehling-Schulz, M., et al. (2015). From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front. Microbiol. 6:560. doi: 10.3389/fmicb.2015.00560
Jimenez, G., Urdiain, M., Cifuentes, A., Lopez-Lopez, A., Blanch, A. R., Tamames, J., et al. (2013). Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst. Appl. Microbiol. 36, 383–391. doi: 10.1016/j.syapm.2013.04.008
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Ko, K. S., Kim, J. W., Kim, J. M., Kim, W., Chung, S. I., Kim, I. J., et al. (2004). Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR gene. Infect. Immun. 72, 5253–5261. doi: 10.1128/iai.72.9.5253-5261.2004
Kolsto, A. B., Tourasse, N. J., and Okstad, O. A. (2009). What sets Bacillus anthracis apart from other Bacillus species? Annu. Rev. Microbiol. 63, 451–476. doi: 10.1146/annurev.micro.091208.073255
Kotiranta, A., Lounatmaa, K., and Haapasalo, M. (2000). Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2, 189–198. doi: 10.1016/s1286-4579(00)00269-0
Kovac, J., Miller, R. A., Carroll, L. M., Kent, D. J., Jian, J., Beno, S. M., et al. (2016). Production of hemolysin BL by Bacillus cereus group isolates of dairy origin is associated with whole-genome phylogenetic clade. BMC Genomics 17:581. doi: 10.1186/s12864-016-2883-z
Kumari, S., and Sarkar, P. K. (2016). Bacillus cereus hazard and control in industrial dairy processing environment. Food Control 69, 20–29. doi: 10.1016/j.foodcont.2016.04.012
La Duc, M. T., Satomi, M., and Venkateswaran, K. (2004). Bacillus odysseyi sp. nov., a round-spore-forming Bacillus isolated from the mars odyssey spacecraft. Int. J. Syst. Evol. Microbiol. 54, 195–201. doi: 10.1099/ijs.0.02747-0
Lacey, L. A., Grzywacz, D., Shapiro-Ilan, D. I., Frutos, R., Brownbridge, M., and Goettel, M. S. (2015). Insect pathogens as biological control agents: back to the future. J. Invertebr. Pathol. 132, 1–41. doi: 10.1016/j.jip.2015.07.009
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Lin, S. F., Schraft, H., and Griffiths, M. W. (1998). Identification of Bacillus cereus by fourier transform infrared spectroscopy (FTIR). J. Food Prot. 61, 921–923. doi: 10.4315/0362-028x-61.7.921
Liu, Y., Du, J., Lai, Q., Zeng, R., Ye, D., Xu, J., et al. (2017a). Proposal of nine novel species of the Bacillus cereus group. Int. J. Syst. Evol. Microbiol. 67, 2499–2508. doi: 10.1099/ijsem.0.001821
Liu, Y., Lai, Q., Du, J., and Shao, Z. (2017b). Genetic diversity and population structure of the Bacillus cereus group bacteria from diverse marine environments. Sci. Rep. 7:689. doi: 10.1038/s41598-017-00817-1
Liu, Y., Lai, Q., Goker, M., Meier-Kolthoff, J. P., Wang, M., Sun, Y., et al. (2015). Genomic insights into the taxonomic status of the Bacillus cereus group. Sci. Rep. 5:14082. doi: 10.1038/srep14082
Liu, Y., Lai, Q., and Shao, Z. (2018). Genome analysis-based reclassification of Bacillus weihenstephanensis as a later heterotypic synonym of Bacillus mycoides. Int. J. Syst. Evol. Microbiol. 68, 106–112. doi: 10.1099/ijsem.0.002466
Logan, N. A., and Berkeley, R. C. (1984). Identification of Bacillus strains using the API system. J. Gen. Microbiol. 130, 1871–1882. doi: 10.1099/00221287-130-7-1871
Lucking, G., Frenzel, E., Rutschle, A., Marxen, S., Stark, T. D., Hofmann, T., et al. (2015). Ces locus embedded proteins control the non-ribosomal synthesis of the cereulide toxin in emetic Bacillus cereus on multiple levels. Front. Microbiol. 6:1101. doi: 10.3389/fmicb.2015.01101
Mai, U., and Mirarab, S. (2018). TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genomics 19:272. doi: 10.1186/s12864-018-4620-2
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Hahnke, R. L., Petersen, J., Scheuner, C., Michael, V., Fiebig, A., et al. (2014). Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 9:2. doi: 10.1186/1944-3277-9-2
Miller, R. A., Jian, J. H., Beno, S. M., Wiedmann, M., and Kovac, J. (2018). Intraclade Variability in toxin production and cytotoxicity of Bacillus cereus group type strains and dairy-associated isolates. Appl. Environ. Microbiol. 84:e02479-17. doi: 10.1128/AEM.02479-17
Minh, B. Q., Nguyen, M. A., and Von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Moayeri, M., Leppla, S. H., Vrentas, C., Pomerantsev, A. P., and Liu, S. H. (2015). Anthrax pathogenesis. Annu. Rev. Microbiol. 69, 185–208.
Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I., and Chun, J. (2018). UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285. doi: 10.1007/s12275-018-8014-6
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nouioui, I., Carro, L., Garcia-Lopez, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-based taxonomic classification of the phylum actinobacteria. Front. Microbiol. 9:2007. doi: 10.3389/fmicb.2018.02007
Palma, L., Munoz, D., Berry, C., Murillo, J., and Caballero, P. (2014). Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins 6, 3296–3325. doi: 10.3390/toxins6123296
Parker, C. T., Tindall, B. J., and Garrity, G. M. (2019). International code of nomenclature of prokaryotes. Int. J. Syst. Evol. Microbiol. 69, S1–S111. doi: 10.1099/ijsem.0.000778
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Patino-Navarrete, R., and Sanchis, V. (2017). Evolutionary processes and environmental factors underlying the genetic diversity and lifestyles of Bacillus cereus group bacteria. Res. Microbiol. 168, 309–318. doi: 10.1016/j.resmic.2016.07.002
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Priest, F. G., Barker, M., Baillie, L. W., Holmes, E. C., and Maiden, M. C. (2004). Population structure and evolution of the Bacillus cereus group. J. Bacteriol. 186, 7959–7970. doi: 10.1128/jb.186.23.7959-7970.2004
Radnedge, L., Agron, P. G., Hill, K. K., Jackson, P. J., Ticknor, L. O., Keim, P., et al. (2003). Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69, 2755–2764. doi: 10.1128/aem.69.5.2755-2764.2003
Rasko, D. A., Altherr, M. R., Han, C. S., and Ravel, J. (2005). Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29, 303–329.
Rasko, D. A., Rosovitz, M. J., Okstad, O. A., Fouts, D. E., Jiang, L., Cer, R. Z., et al. (2007). Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 189, 52–64. doi: 10.1128/jb.01313-06
Raymond, B., and Federici, B. A. (2017). In defense of Bacillus thuringiensis, the safest and most successful microbial insecticide available to humanity - a response to EFSA. FEMS Microbiol. Ecol. doi: 10.1093/femsec/fix084 [Epub ahead of print].
Sangal, V., Goodfellow, M., Blom, J., Tan, G. Y. A., Klenk, H. P., and Sutcliffe, I. C. (2018). Revisiting the taxonomic status of the biomedically and industrially important genus Amycolatopsis, using a phylogenomic approach. Front. Microbiol. 9:2281. doi: 10.3389/fmicb.2018.02281
Schnepf, E., Crickmore, N., Van Rie, J., Lereclus, D., Baum, J., Feitelson, J., et al. (1998). Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 775–806.
Soni, A., Oey, I., Silcock, P., and Bremer, P. (2016). Bacillus spores in the food industry: a review on resistance and response to novel inactivation technologies. Compr. Rev. Food Sci. Food Saf. 15, 1139–1148. doi: 10.1111/1541-4337.12231
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stenfors Arnesen, L. P., Fagerlund, A., and Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606. doi: 10.1111/j.1574-6976.2008.00112.x
Van Der Hoeven, N. (2014). Bacillus Thuringiensis Toxins: their Mode of Action and the Potential Interaction Between them Statistical Consultancy in Ecology, Ecotoxicology and Agricultural Research. Report 14/001 Hauppauge, NY: Commissie Genetische Modificatie.
Vilas-Boas, G., Sanchis, V., Lereclus, D., Lemos, M. V., and Bourguet, D. (2002). Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 68, 1414–1424. doi: 10.1128/aem.68.3.1414-1424.2002
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017a). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017b). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zhong, W., Shou, Y., Yoshida, T. M., and Marrone, B. L. (2007). Differentiation of Bacillus anthracis, B. Cereus, and B. thuringiensis by using pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 73, 3446–3449. doi: 10.1128/aem.02478-06
Keywords: Bacillus cereus, Bacillus thuringiensis, Bt toxin, Cry toxin, phylogenomic analysis
Citation: Baek I, Lee K, Goodfellow M and Chun J (2019) Comparative Genomic and Phylogenomic Analyses Clarify Relationships Within and Between Bacillus cereus and Bacillus thuringiensis: Proposal for the Recognition of Two Bacillus thuringiensis Genomovars. Front. Microbiol. 10:1978. doi: 10.3389/fmicb.2019.01978
Received: 16 April 2019; Accepted: 12 August 2019;
Published: 23 August 2019.
Edited by:
David W. Ussery, University of Arkansas for Medical Sciences, United StatesReviewed by:
Mohammad Ali Amoozegar, University of Tehran, IranChristopher Dunlap, Agricultural Research Service, U.S. Department of Agriculture, United States
David W. Dyer, The University of Oklahoma Health Sciences Center, United States
Copyright © 2019 Baek, Lee, Goodfellow and Chun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Goodfellow, bS5nb29kZmVsbG93QG5jbC5hYy51aw==; bWljaGFlbC5nb29kZmVsbG93QG5jbC5hYy51aw==; Jongsik Chun, amNodW5Ac251LmFjLmty
 Inwoo Baek
Inwoo Baek Kihyun Lee
Kihyun Lee Michael Goodfellow4*
Michael Goodfellow4* Jongsik Chun
Jongsik Chun