- 1Centre for Integrative Ecology, School of Life and Environmental Sciences, Deakin University, Geelong, VIC, Australia
- 2Deakin Genomics Centre, Deakin University, Geelong, VIC, Australia
- 3School of Science, Monash University Malaysia, Petaling Jaya, Malaysia
by Moriuchi, R., Dohra, H., Kanesaki, Y., and Ogawa, N. (2019). Front. Microbiol. 10:133. doi: 10.3389/fmicb.2019.00133
Moriuchi et al. reported a comprehensive reclassification of bacterial strains from the genera Cupriavidus and Ralstonia based on percentage of conserved proteins (POCP), average nucleotide identity (ANI), multilocus sequence analysis and 16S rRNA gene sequence. In the study, conflicting results were repeatedly observed for the taxonomic classification of strain PBA that was initially identified as Ralstonia sp. PBA based on 16S rRNA gene sequence (Gan et al., 2011b; Moriuchi et al., 2019).
Strain PBA was isolated as a co-culture with Hydrogenophaga intermedia PBC from textile wastewater a decade ago. The co-culture could grow on 4-aminobenzenesulfonate (4-ABS), a recalcitrant dye intermediate (Wagner and Reid, 1931), as the sole nitrogen, carbon, and sulfur source to a relatively high cell density (Gan et al., 2011b). In this syntrophic relationship, strain PBA is the sole provider of p-aminobenzoate, an essential vitamin required for the growth of H. intermedia PBC, the main 4-ABS degrader (Gan et al., 2011a, 2017). In light of new genomic resources, the initial taxonomic assignment of strain PBA has also been previously questioned by Kim and Gan (2017) given its closer phylogenetic affiliation to the genus Cupriavidus than to the genus Ralstonia. Unfortunately, both recent genome-based taxonomic classifications of strain PBA (Kim and Gan, 2017; Moriuchi et al., 2019) suffered from incomplete and biased taxon sampling (restricted mostly to members from the genus Ralstonia and Cupriavidus) that can result in the misinterpretation of evolutionary relationships (Heath et al., 2008). The taxonomic affiliation of strain PBA should be inferred from a comprehensive phylogenomic analysis that includes all genera with genome availability from the family Burkholderiaceae.
A total of 428 Burkholderiaceae (including strain PBA) and 15 non-Burkholderiaceae genome assemblies were obtained from the NCBI RefSeq database (accessed on 30th May 2019). The genomes were processed using two microbial phylogenomic analysis pipelines e.g., GToTree v1.2.1 (Lee, 2019) and PhylophlAN v0.99 (Segata et al., 2013) that identify single copy bacterial genes (GToTree: n = 203, Betaproteobacteria HMM set; PhylophIAN: n = 400) and produce concatenated protein alignment. Maximum likelihood tree construction from the protein alignments used IQTree v.1.6.8 with 1,000 ultrafast bootstrap replicates (Nguyen et al., 2014). In both phylogenomic trees, the Ralstonia and Cupriavidus clusters received maximal support and are sister taxa to the exclusion of strain PBA (Figures 1A,B). The updated phylogenomic placement of strain PBA in light of extensive taxon sampling precludes its genus assignment to the genus Ralstonia or Cupriavidus and suggests that it is a member of a hitherto undescribed genus within the family Burkholderiaceae. Within the Genome Taxonomy Database (Parks et al., 2018) that inferred standardized bacteria taxonomy from conserved proteins present in 143,512 bacterial genomes (GTDB release R04-RS89), strain PBA was still assigned to its own genus (g__AKCV01) despite an even more extensive taxon sampling of 4,378 genomes from the family Burkholderiaceae (https://gtdb.ecogenomic.org/tree?r=g__AKCV01 accessed on 1st August 2019).
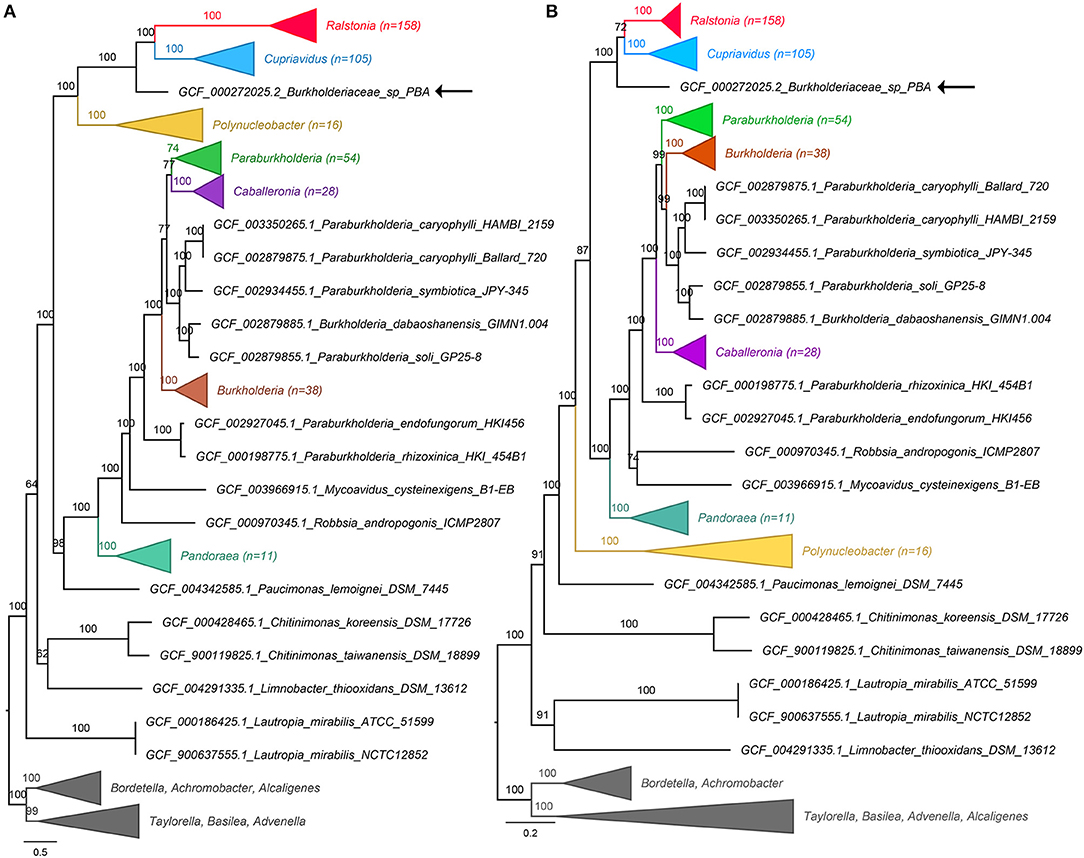
Figure 1. Genome-based phylogeny of Burkholderiaceae. Construction of the phylogenomic trees used protein alignments generated from (A) PhyloPhlAN and (B) GToTree. Numbers in brackets indicate number of taxa in the collapsed clades. GCF codes preceding the species names are RefSeq Assembly numbers. The positions of strain PBA in both trees are shown by black arrows. Branch lengths and labels indicate the number of substitutions per site and IQTree ultrafast bootstrap support values, respectively. The uncollapsed trees are available in the Zenodo database (http://doi.org/10.5281/zenodo.3258920).
Given the concordance observed from these independent analyses, the taxonomic assignment of strain PBA has been updated from Ralstonia sp. PBA to Burkholderiaceae sp. PBA in the NCBI database (Bioproject: PRJNA78957; BioSample: SAMN02471424) (Gan et al., 2012) pending future genus description. To facilitate future strain description and comparison, strain PBA has been deposited in the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ) under the accession number DSM 106616. Furthermore, the concatenated alignments, uncollapsed phylogenomic trees and genome information are also made available in the Zenodo database (http://doi.org/10.5281/zenodo.3258920).
Author Contributions
HG performed data analysis and wrote the manuscript.
Funding
This research was supported by the Deakin Centre of Integrative Ecology.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author is grateful to the Deakin Centre of Integrative Ecology for financial support.
References
Gan, H. M., Chew, T. H., Tay, Y.-L., Lye, S. F., and Yahya, A. (2012). Genome sequence of Ralstonia sp. strain PBA, a bacterium involved in the biodegradation of 4-aminobenzenesulfonate. Am. Soc. Microbiol. 194, 5139–5140. doi: 10.1128/JB.01165-12
Gan, H. M., Ibrahim, Z., Shahir, S., and Yahya, A. (2011a). Identification of genes involved in the 4-aminobenzenesulfonate degradation pathway of Hydrogenophaga sp. PBC via transposon mutagenesis. FEMS Microbiol. Lett. 318, 108–114. doi: 10.1111/j.1574-6968.2011.02245.x
Gan, H. M., Lee, Y. P., and Austin, C. M. (2017). Nanopore long-read guided complete genome assembly of Hydrogenophaga intermedia, and genomic insights into 4-aminobenzenesulfonate, p-aminobenzoic acid and hydrogen metabolism in the genus Hydrogenophaga. Front. Microbiol. 8:1880. doi: 10.3389/fmicb.2017.01880
Gan, H. M., Shahir, S., Ibrahim, Z., and Yahya, A. (2011b). Biodegradation of 4-aminobenzenesulfonate by Ralstonia sp. PBA and Hydrogenophaga sp. PBC isolated from textile wastewater treatment plant. Chemosphere 82, 507–513. doi: 10.1016/j.chemosphere.2010.10.094
Heath, T. A., Zwickl, D. J., Kim, J., and Hillis, D. M. (2008). Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Syst. Biol. 57, 160–166. doi: 10.1080/10635150701884640
Kim, K., and Gan, H. M. (2017). A glimpse into the genetic basis of symbiosis between Hydrogenophaga and their helper strains in the biodegradation of 4-aminobenzenesulfonate. J. Genomics 5, 77–82. doi: 10.7150/jgen.20216
Lee, M. D. (2019). GToTree: a user-friendly workflow for phylogenomics. Bioinformatics. btz188. doi: 10.1093/bioinformatics/btz188
Moriuchi, R., Dohra, H., Kanesaki, Y., and Ogawa, N. (2019). Complete genome sequence of 3-chlorobenzoate-degrading bacterium Cupriavidus necator NH9 and reclassification of the strains of the genera Cupriavidus and Ralstonia based on phylogenetic and whole-genome sequence analyses. Front. Microbiol. 10:133. doi: 10.3389/fmicb.2019.00133
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evolu. 32, 268–274. doi: 10.1093/molbev/msu300
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P.-A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Segata, N., Börnigen, D., Morgan, X. C., and Huttenhower, C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4:2304. doi: 10.1038/ncomms3304
Keywords: Ralstonia, Burkholderiaceae, Cupriavidus, phylogenomics, taxonomy
Citation: Gan HM (2019) Commentary: Complete Genome Sequence of 3-Chlorobenzoate-Degrading Bacterium Cupriavidus necator NH9 and Reclassification of the Strains of the Genera Cupriavidus and Ralstonia Based on Phylogenetic and Whole-Genome Sequence Analyses. Front. Microbiol. 10:2011. doi: 10.3389/fmicb.2019.02011
Received: 28 June 2019; Accepted: 16 August 2019;
Published: 28 August 2019.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Roberta Fulthorpe, University of Toronto Scarborough, CanadaZhang Yong, Southwest University, China
Jorgen Johannes Leisner, University of Copenhagen, Denmark
Copyright © 2019 Gan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Ming Gan, aGFuLmdhbkBkZWFraW4uZWR1LmF1
 Han Ming Gan
Han Ming Gan