- 1Department of Genetics and Biotechnology, Faculty of Biology, National and Kapodistrian University of Athens, Athens, Greece
- 2Department of Environmental Engineering, University of Patras, Agrinio, Greece
Methicillin-resistant Staphylococcus aureus (MRSA) strains are important human pathogens and a significant health hazard for hospitals and the food industry. They are resistant to β-lactam antibiotics including methicillin and extremely difficult to treat. In this study, we show that the Staphylococcus aureus COL (MRSA) strain, with a known complete genome, can easily survive and grow under acidic and alkaline conditions (pH5 and pH9, respectively), both planktonically and as a biofilm. A microarray-based analysis of both planktonic and biofilm cells was performed under acidic and alkaline conditions showing that several genes are up- or down-regulated under different environmental conditions and growth modes. These genes were coding for transcription regulators, ion transporters, cell wall biosynthetic enzymes, autolytic enzymes, adhesion proteins and antibiotic resistance factors, most of which are associated with biofilm formation. These results will facilitate a better understanding of the physiological adjustments occurring in biofilm-associated S. aureus COL cells growing in acidic or alkaline environments, which will enable the development of new efficient treatment or disinfection strategies.
Introduction
Staphylococcus aureus is a Gram-positive cluster-forming aerobic coccus that is commonly found on the skin and the respiratory tract of humans and animals. It is recognized as a cause of serious nosocomial infection and especially methicillin-resistant S. aureus (MRSA) strains are considered a major public health hazard. MRSA is prevalent in hospitals, prisons, and nursing homes, where people with open wounds, invasive devices such as catheters, and weakened immune systems are at greater risk of nosocomial infection. S. aureus is known to efficiently colonize the biomaterials that are used for medical implants and devices. In the event of a biomaterial-associated infection, the device must be substituted, something that seriously burdens the patient, while relapsing infections remain possible. In cases where the device cannot be substituted, the patient faces a higher mortality risk (Arciola et al., 2012). In addition, MRSA strains have been found present in retailed meat products (O’Brien et al., 2012), dairy products (Normanno et al., 2007), seafood (Kumar et al., 2016), green leafs of pre-cut salads (Doulgeraki et al., 2017), the hands of food industrial workers (Kamal et al., 2013) and the equipment and surfaces related to food preparation (Gibson et al., 1999), therefore explaining the alarmingly increasing reports on food-borne acquired MRSA outbreaks (Jones et al., 2002; Harris et al., 2010; Centers for Disease Control and Prevention, 2018). Only in the United States, 72,444 cases of MRSA infections were reported in 2014, while the morbidity rate reached 11.8% (Hassoun et al., 2017).
Staphylococcus aureus strains have the ability to form biofilms (BFs), multicellular communities covered by a thick polysaccharide layer, which contribute significantly to antibiotic and detergent resistance (Christensen et al., 1994; Götz, 2002; de Souza et al., 2014). In general, bacterial BFs are multi-layered complex communities which in their mature form contain specific three-dimensional structures that are separated by fluid channels. Depending on the position of cells, they are allowed to differentially express proteins throughout the BF. The formation of BFs is generally regarded as a four-step process that includes: (a) an initial attachment of cells to the surface through ionic or hydrophobic interactions, (b) the accumulation in multiple bacterial layers, mediated by microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), (c) BF maturation with the production of extracellular capsular exopolysaccharide (PNAG) and several exoproteins which mediate the attachment of S. aureus cells on surfaces and eDNA, rendering encapsulated cells resistance to phagocytosis and antibiotics, and (d) detachment of BF cells and dispersal in a planktonic state form to initiate a new cycle of BF formation elsewhere, guided by numerous environmental signals, signal transduction pathways and effectors (Patti et al., 1994; Stoodley et al., 2002; Bischoff et al., 2004; Archer et al., 2011; Arciola et al., 2012; Atwood et al., 2015).
The effect of environmental pH on BF formation can influence several important biological processes. For example, wound pH is known to gradually decrease while the wound is healing, due to lactic acid production and other factors. Bacterial BFs can lead to serious infection, if they are tolerant to low pH or antiseptics (Percival et al., 2014; Jones et al., 2015). Moreover, acidic and alkaline detergents are frequently used to decontaminate clinical surfaces and surgical instruments (Lemmer et al., 2004), as well as food-processing surfaces and equipment (Sharma and Beuchat, 2004; Akbas and Cag, 2016). Acidic or alkaline sanitizers are also used to disinfect fruit and vegetables (Park et al., 2011) and orthopedic hardware (Moussa et al., 1996), conditions that can easily allow the survival of tolerant BF-forming bacteria and cause infections.
Staphylococcus aureus BF cells exhibit a different phenotype with respect to bacterial physiology, metabolism and gene transcription compared to planktonic cells (Donlan and Costerton, 2002). The ability of S. aureus to form BF and its morphology were strongly influenced by significant pH changes (Jones et al., 2015). When weakly acidic and alkaline detergents were used against S. aureus BFs on stainless steel surfaces, BF-associated cell numbers were reduced, but the BFs were not completely removed (Ueda and Kuwabara, 2007). Lastly, alkaline and acidic pHs were shown to inhibit S. aureus BF formation and reduced its amount and thickness (Nostro et al., 2012).
A number of transcriptomic studies using planktonic S. aureus cells that grew in liquid media with acidic or alkaline pH have been published (Weinrick et al., 2004; Bore et al., 2007; Anderson et al., 2010; Rode et al., 2010). These reports have identified few genes whose expression is affected by pH changes, but have not clearly defined specific functional or regulatory mechanisms yet, neither have they contributed to the transcriptomic adjustments occurring in BF-associated MRSA cells growing under acidic and alkaline conditions. Thus, the aim of this study was to detect genes that are differentially expressed in S. aureus COL BF cells, under acidic and alkaline conditions (pH5 and pH9, respectively) with the use of DNA microarrays. Two modes of growth were studied: BF-associated cells on a porous nitrocellulose membrane placed on solid media and planktonic cells in liquid medium. Gene expression levels at environments of different pH and growth modes were measured and compared, in order to gain better knowledge about the molecular mechanisms connecting pH-related stress response with BF-formation and pathogenicity in this important human pathogen. This study will help understanding how the pathogen survives and responds under acidic and alkaline conditions, which will lead to the design of better treatment or disinfection strategies.
Materials and Methods
Bacterial Strains, Media, and Cultures
Tryptone Soya Broth (TSB) and Tryptone Soya Agar (TSA) were used for growing S. aureus COL (MRSA) in this study. 100 μL of an overnight pre-culture were used to inoculate 10 mL of the same medium in sterile glass shake flasks. The flasks were incubated for 5 h at 37°C (150 rpm). HCl and NaOH solutions (1 M) were used to adjust the pH. Colony forming units per mL of liquid culture were determined by serial dilutions and colony enumeration (at least three biological replicates in each case). Biomass from these cultures was harvested for RNA isolation, immediately dissolved in RNAlater® reagent (Ambion, United States), as advised by the manufacturer, and stored at −80°C for further use.
Biofilm Formation
Four 100 μL drops of a 5-h pre-culture were pipetted on a nitrocellulose membrane (pore size 0.45 μm; Sartorius, United Kingdom), which was placed on TSA with different pH (5, 7, and 9) and allowed to grow statically for 24 h at 37°C. Determination of colony forming units per nitrocellulose disk (at least three biological replicates in each case) and biomass harvestation with RNAlater® reagent were performed as described above.
Total RNA Extraction and First-Strand cDNA Synthesis
Biomass pellets treated with RNAlater® reagent were dissolved in an aqueous solution of lysostaphin (0.2 U/μL) and incubated at 37°C for 30 min. The samples were transferred into new Eppendorf tubes containing 0.2 g of glass beads (0.6 mm diameter), 750 μL of RA1 lysis buffer (Nucleospin® RNA II kit; Cat. No. 740955.50; Macherey-Nagel, Germany) and 1% β-mercaptoethanol (Sigma-Aldrich, United Kingdom). The samples were vortexed thrice for 30 sec and total RNA was then isolated as suggested by the Nucleospin® RNA II kit instructions. Two elution steps were performed at the end of the procedure. The quality of the extracted RNA was determined by spectrophotometry and gel electrophoresis in a 1.4% agarose gel with DEPC-treated distilled water. First-strand cDNA synthesis was performed by using the PrimeScriptTM 1st strand cDNA Synthesis Kit (Cat. No. 6110A; Takara, Japan).
DNA Microarrays
1.5 μg of synthesized first-strand cDNA were hybridized on a GeneChip® S. aureus Genome Array (Cat. No. 900514; Affymetrix, United States), following the procedure suggested by the GeneChip® Expression Analysis Technical Manual (Affymetrix; P/N 702232 Rev. 3). Biological duplicates were used (n = 2).
Microarray Data Analysis
The raw microarray data were first normalized by using the statistical language R (TM4 protocol at: https://github.com/dfci-cccb/www.tm4.org/blob/master/normalizing.html). Data filtering was performed by using MeV (MultiExperiment Viewer Quickstart Guide v. 4.2). The selected variance filter value was 50. Finally, the filtered data were exported to Excel and the Log2 ratios of the average gene expression values of the two compared conditions were calculated [Log2(Expr1/Expr2)]. A two-tailed paired t-test was also performed in Excel, using the gene expression values of each gene for the two compared conditions. Gene expression differences were considered to be significant only if the p-value was <0.05. Databases KEGG and Aureowiki were used for confirming gene annotation and function. For gene annotation, the files provided by Affymetrix for this specific microarray product were used1.
Results
Staphylococcus aureus can either grow planktonically in the bloodstream or colonize body surfaces, such as the nasopharyngeal mucosa. During the second mode of growth, the pathogen is using cell surface proteins to attach on specific mucosal ligands and then produces a polysaccharide layer that stabilizes and protects the bacterial colony. It was shown by preliminary results that S. aureus COL can grow well and form BF even at extreme pH environments (pH 4-10; Efthimiou et al., 2015). Therefore, we chose to study gene expression at pH5 and pH9, as the BF levels on polystyrene surfaces were comparable, although slightly lower, with these at pH7.
In this study, S. aureus COL (genome sequence PRJNA238) grew well in both liquid and solid media, under acidic, neutral and alkaline conditions. Total planktonic growth reached 109 in acidic and 1010 CFU/10 mL in neutral and alkaline TSB and TSA, respectively (p-value = 0.0009) (Figure 1). The same was observed for biofilm growth. This indicates that the pathogen is possibly more tolerant to highly alkaline than acidic environments.
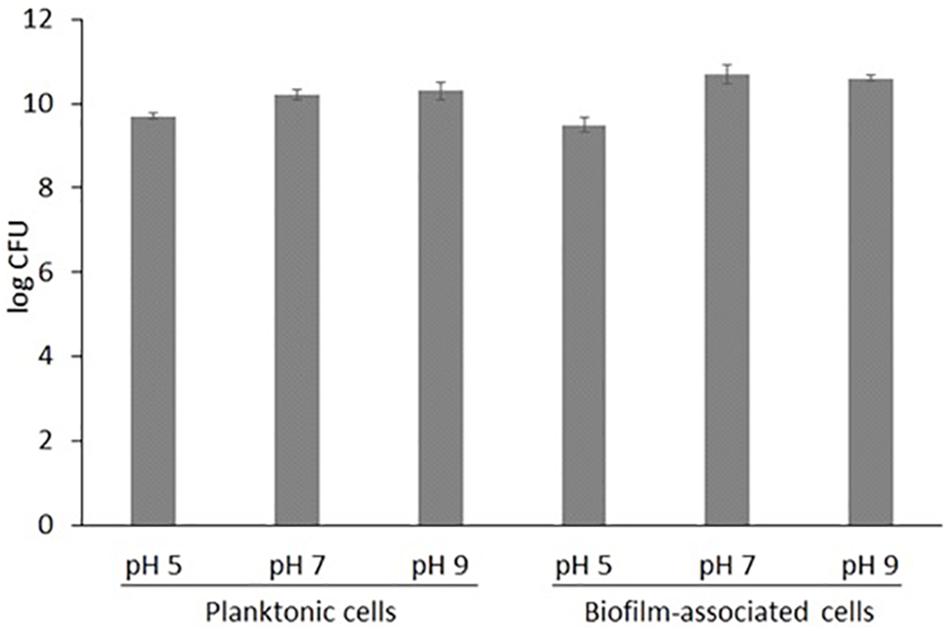
Figure 1. Total growth of S. aureus COL in liquid TSB medium (log CFU in 10 mL) and nitrocellulose filters placed on solid TSA medium (log CFU per disk) (n = 3, where n the number of biological replicates; ± STDEV error bars are also shown).
Out of the over 3,300 open reading frame genes imprinted on the array (Cat No. 900154) eight were found to be over-expressed in BF-forming S. aureus COL cells growing under alkaline conditions. They all had a log2 fold-change ratio above 3.16 and a p-value < 0.0061 (Table 1A). These genes encoded three transcriptional regulators (CodY, MecA and CtsR), one capsular biosynthesis enzyme (CapC) and four other proteins (two cell wall proteins IsdC or SirD and IsaB; the von Willebrand matrix secreted surface protein, VWbp; and a hypothetical protein similar to SceD, which is involved in autolysis). Under acidic conditions, eleven genes were over-expressed in BFS. aureus COL cells and all of them had a log2 fold-change ratio above 3.12 and a p-value < 0.0316 (Table 2A). These encoded three genes coding for cell surface proteins (MapW, Efb/FnbA and the secreted VWbp), two cell wall-associated proteins (CapC and EbsB), an enzyme with possible phosphomannomutase activity (PgcA) and an ABC transporter (CymR). Additionally, genes coding for a terminase (SACOL 0366), a methionine import protein (MetN2), a hypothetical protein (SACOL2556) and a probable type I site-specific deoxyribonuclease LldI chain (HsdM) were up-regulated.
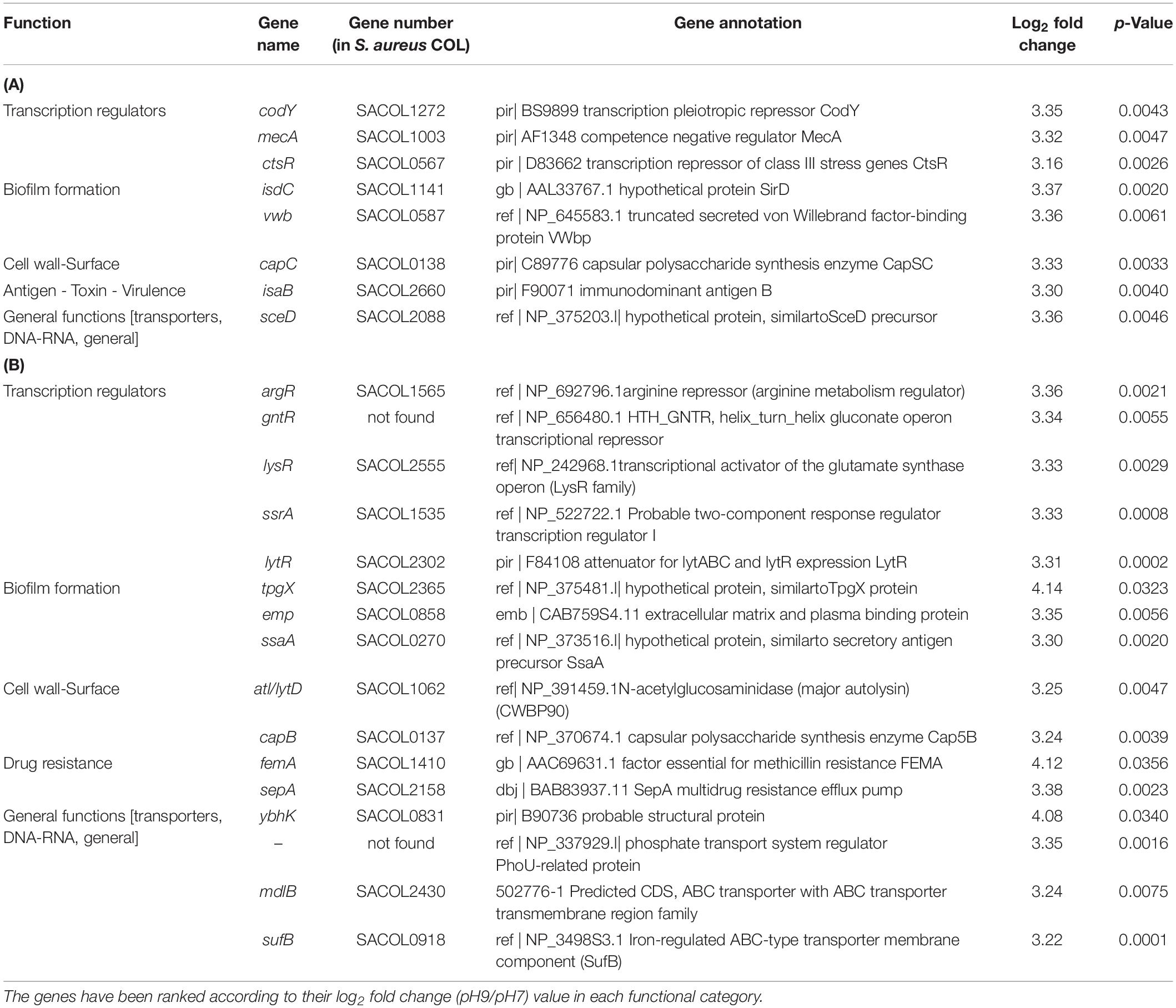
Table 1. Significantly upregulated genes at pH 9 in S. aureus COL biofilm-associated (A) and planktonic cells (B) (p < 0.05).
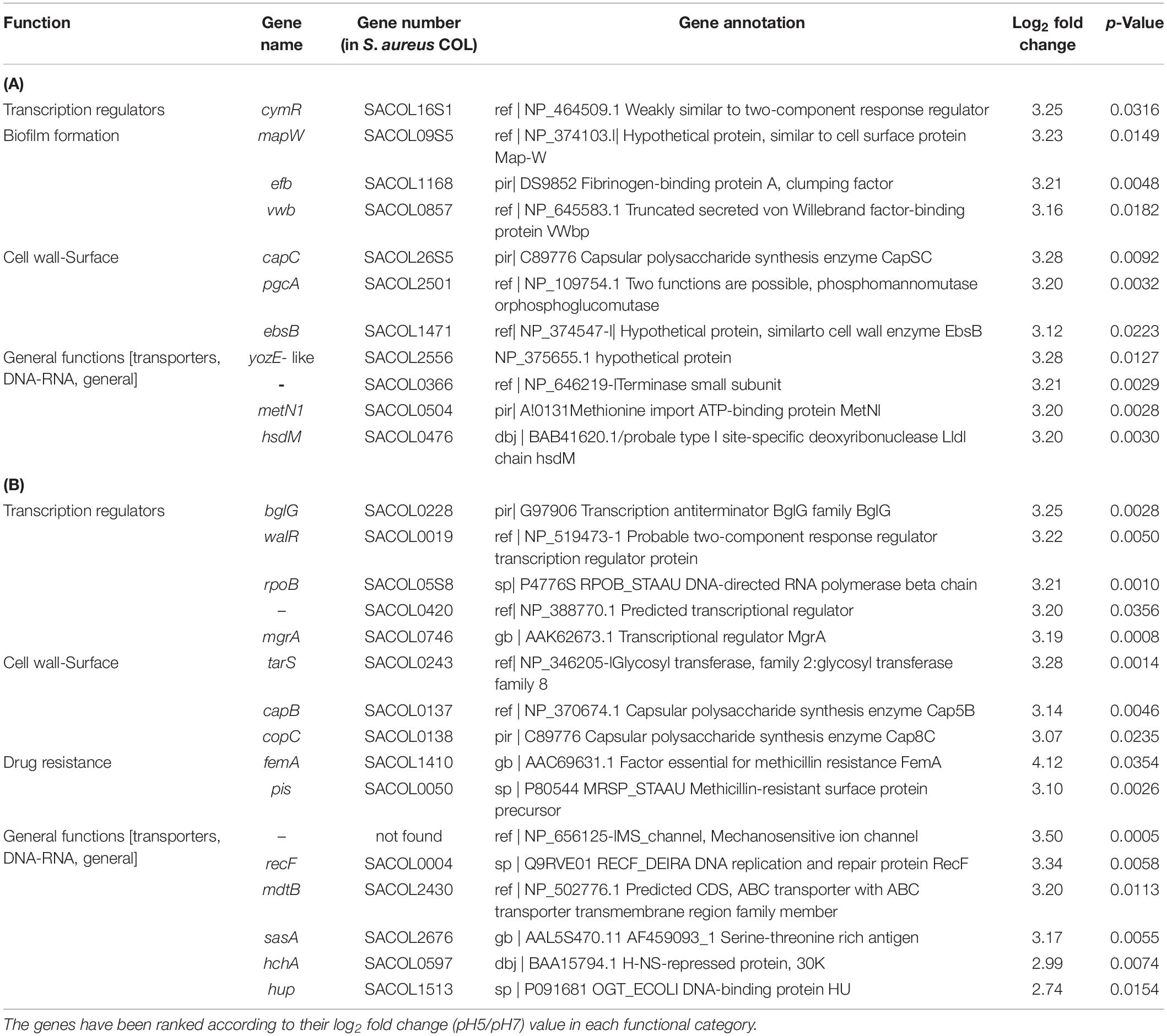
Table 2. Significantly upregulated genes at pH 5 in S. aureus COL biofilm-associated (A) and planktonic cells (B) (p < 0.05).
In planktonic cells growing at pH 9, sixteen genes were over-expressed, with log2 fold-change ratios between 3.22–4.14 and a p-value < 0.0356 (Table 1B). They included genes encoding five transcription regulators, one protein associated with drug resistance (the essential factor for methicillin resistance, FEMA) and nine other proteins with various functions. The transcription factors included an attenuator for lytABC and LytR expression (LytR), a two-component response regulator (SrrA), the repressors for arginine (ArgR) and gluconate biosynthesis (GntR), and an activator of glutamate synthase (LysR/CidR). The other up-regulated genes coded for two ABC transporters with transmembrane functions (MdlB and an iron regulating, SufB), a phosphate transport system regulator (PhoU-related protein), a major autolysin (Atl/LytD), a hypothetical; protein similar to secretory antigen precursor SsaA (LysM), the capsule biosynthetic enzyme CapB, a cell wall-related enzyme SepA, an extracellular matrix and plasma binding protein (Emp), and two probable proteins (similar to TpgX protein and a structural protein, similar to YbhK).
A similar number of genes (16) were also over-expressed in planktonic cells growing at pH5, with log2 fold-change ratios between 2.74–4.12 and a p-value < 0.0354 (Table 2B). They included three genes associated with DNA or RNA functions (the replication and repair protein RecF, the DNA-binding protein Hup, and the DNA-directed RNA polymerase beta chain, RpoB), four gene products involved in regulation of transcription (the transcription regulators MgrA, WalR, SACOL0420, and the antiterminator BglG), two capsular biosynthesis enzymes (CapB and CapC), a serine-threonine rich antigen (SasA), two genes associated with methicillin resistance (the essential factor for methicillin resistance, FEMA, and the methicillin-resistance surface protein precursor, Pls), an ABC transporter (MdlB), a glycosyl transferase involved in colanic acid biosynthesis (TarS), and two factors with ill-defined function (a mechanosensitive ion channel protein and an H-NS-repressed protein HchA).
In BF cells growing at pH9, the down-regulated genes coded for two transcriptional regulators (AirR and the two-component response-regulatorNreB), a phosphate transport system regulator (PhoU-related protein) and a cell surface protein (MapW). All had log2 fold change ratios between −3.24 and −3.34 and p-values < 0.0042 (Table 3A). At acidic conditions (pH5) 18 genes were down-regulated in BF-forming cells. Two of these were as at pH9 (AirR and SACOL0420), three coded for transcriptional regulators (HutR, SACOL2517, and BglG), two were associated with toxin production (Ssl1 and Ssl11) and another three with cell surface (EpiG, MapW and elastin binding protein EbsS). Down-regulated were also the signal peptide precursor AgrD, the cell division FtsK, the RNA polymerase beta subunit RpoB, the translation elongation factor TufA, the probable translational initiation factor (InfB), a stress response DNA-binding protein Dps, a peptidoglycan hydrolase (LytM) and interestingly enough the plasmid mobilizing protein Mob. All had log2 fold change ratios between −2.37 and −4.05 and p-values < 0.0365 (Table 4A).
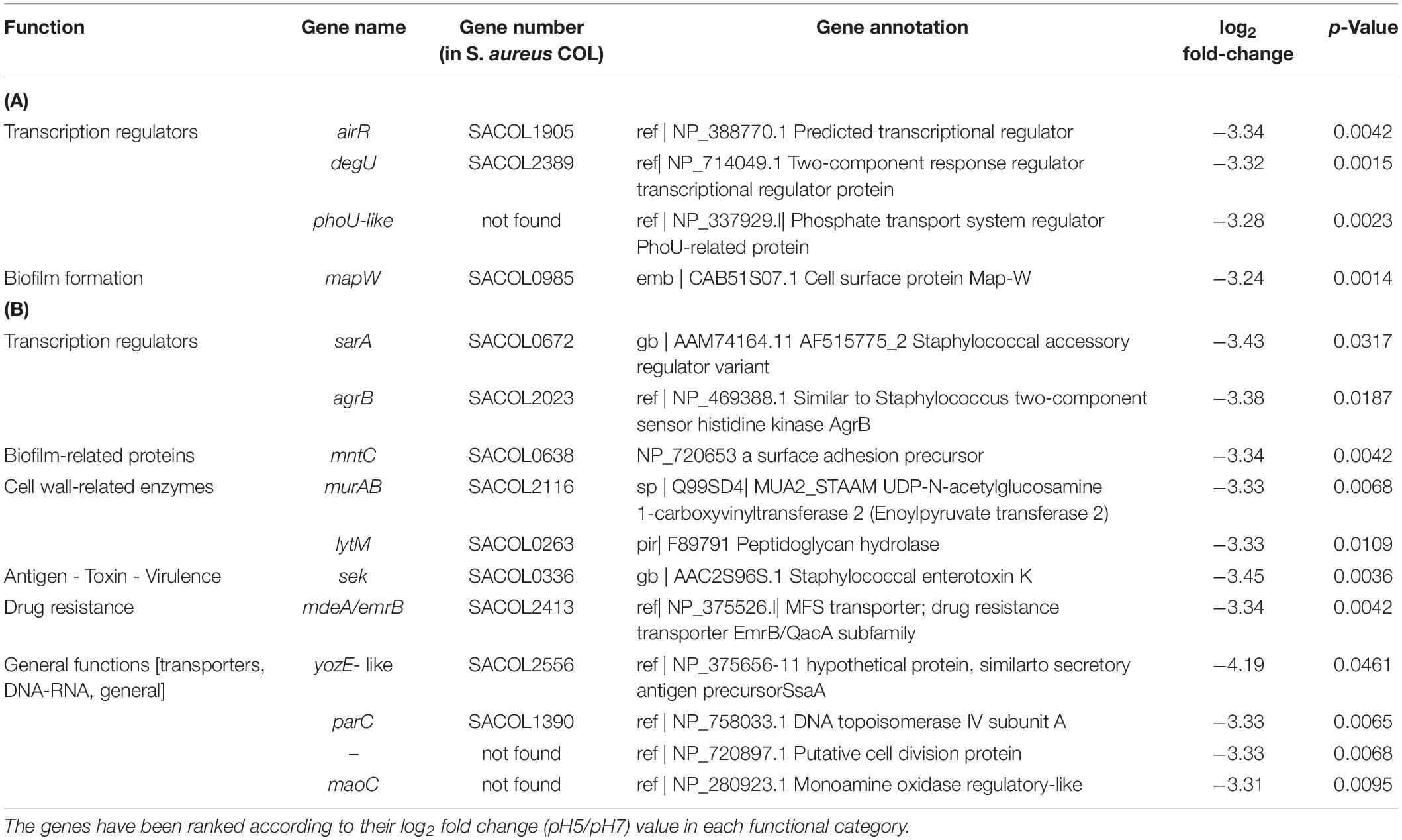
Table 3. Significantly down-regulated genes at pH 9 in S. aureus COL biofilm-associated (A) and planktonic cells (B) (p < 0.05).
Table 4. Significantly down-regulated genes at pH 5 in S. aureus COL biofilm-associated (A) and planktonic cells (B) (p < 0.05).
For the planktonic cells under alkaline conditions, 11 genes were clearly down-regulated (log2 fold change ratios between −3.31 and −3.38 and all p-values < 0.0317, Table 3B). These included the staphylococcal accessory regulator SarA, the two-component sensor of histidine kinase AgrB and the staphylococcal enterotoxin K (Sek). Also, a monoamine oxidase (MaoC1), a DNA topoisomerase IV subunit A (ParC), a surface adhesin precursor (MntC), a peptidoglycan hydrolase (LytM), a putative UDP-N-acetylglucosamine 1-carboxyvinyltransferase 2 (MurAB), a hypothetical multidrug resistance protein (MdeA/EmrB), a putative cell division protein (NP_720653.1) and a multispecies conserved protein (YozE-like, SACOL2556).
Similarly, for the planktonic cells in acidic conditions 12 genes were down-regulated (log2 fold change ratios between −3.05 and −3.53 and p-values < 0.0308, Table 4B). Genes coding for MaoC1, ParC and MntC were down-regulated as under alkaline conditions, and in addition the amino acid ABC transporter (MetN2), the host factor protein (Hfq), the staphylococcal accessory regulator SarA, exotoxin 3 (Ssl14), fibrinogen-binding protein (Scc), immunodominant antigen B (IsaB), cell surface protein (MapW), a parvulin-like PPIase precursor (PrsA) and a putative permease (NP_437451.1).
Figure 2 summarizes the gene expression changes of genes associated with BF formation that were observed in this study, under different conditions.
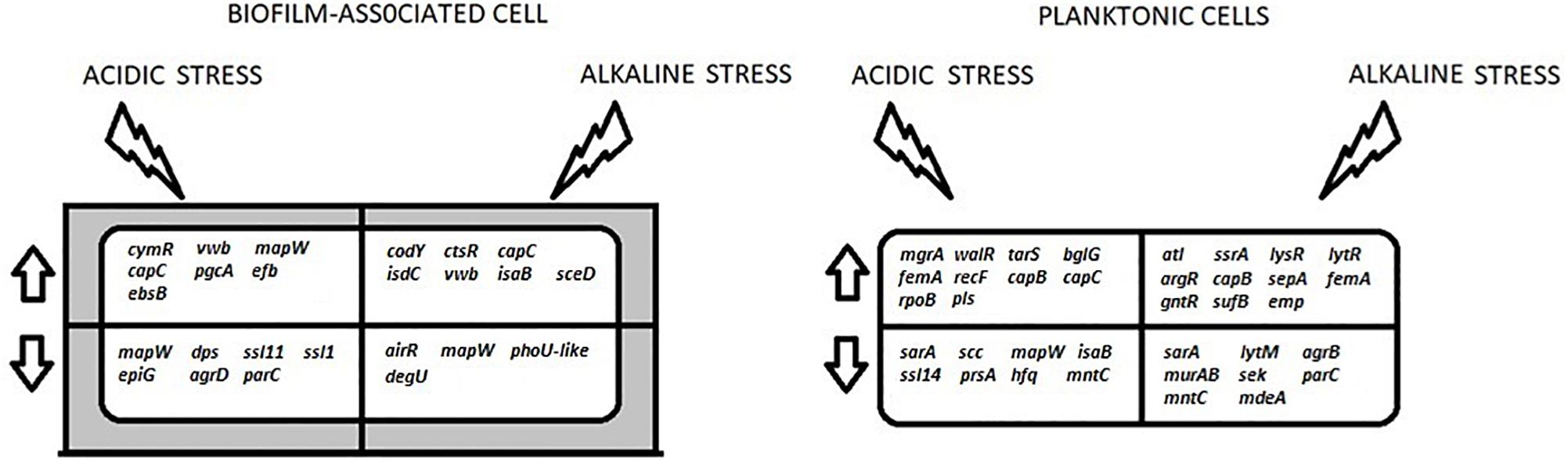
Figure 2. A summary of the key differentially-expressed genes that are discussed in this study. These genes are involved in the processes of transcription regulation (ctsR, argR, lysR, ssrA, lytR, cymR, bglG, walR, rpoB, mgrA, airR, degU, sarA, agrD, phoU-like, agrB, dps, hfq), cell wall biosynthesis (capB, capC, tarS, pgcA, ebsB), BF formation (mapW, efb, isdC, vwb, trgX, emp, ssaA, mntC, epiG, scc), autolysis (atl, murAB, lytM), virulence (isaB, ssl11, ssll1, ssl14, sek) and antibiotic resistance (femA, sepA, pls, mdeA). The arrows indicate up- and down-regulation. The shaded area around the BF-associated cell represents the extracellular matrix.
Discussion
Staphylococci are commensal bacteria and the most prevalent on human skin and mucous. Due to their ability to freely form BFs and, as such, to persist on indwelling medical devices, they are the most frequent culprits of nosocomial infections and cause severe problems to patients who have undergone surgical operations (Trindade et al., 2017). The discovery that in BF formed by several pathogenic bacteria, including S. aureus, the BF-associated bacteria are up to 1,000-fold more tolerant to antibiotics than their genetically identical planktonic cells, attracted the interest of many scientists. As a result, research on the molecular regulatory mechanisms that influence BF formation in S. aureus has intensified during the past two decades (Pratten et al., 2001; Beenken et al., 2004; Resch et al., 2005; Archer et al., 2011; Periasamy et al., 2012; Guilhen et al., 2017; Otto, 2018). The first two microarray studies on S. aureus BF formation were performed in a murine model of catheter-based BF (Beenken et al., 2004) and dialysis membranes laid on agar plates (Resch et al., 2005). In both cases over-expression of large numbers of genes coding for cell wall-associated proteins, transport proteins, secreted proteins, enzymes and transcription regulators was observed in BF cells. These findings were later verified by a proteomic analysis of BF cells and correlation with the transcriptomic profiles of S. aureus (Resch et al., 2006). Similarly, transcriptomic studies that focused on S. aureus in liquid cultures have identified over-expression of a variety of genes, when the cells grew under acidic or alkaline conditions. For example, under acidic conditions cap5B, cap8C, other capsule biosynthesis genes, isaA, ssaA, an autolysin gene and fnbA, ctsR, phoP were up-regulated (Weinrick et al., 2004; Bore et al., 2007), whereas fnbB, Na+/H+ antiporters, and lytR were down-regulated (Anderson et al., 2010; Rode et al., 2010). Similarly, Na+/H+ antiporter and Cap5 enzyme genes were over-expressed under alkaline conditions (Anderson et al., 2010). The fact that most of these genes were also identified as differentially expressed in our study confirms that they are indeed important when the cells are exposed to pH-associated stress. Overall, under the same pH conditions, more genes were up- or down-regulated in planktonic cells than in BF cells (Tables 1–4).
Bacteria can survive and even thrive under harsh environmental conditions often due to their capacity to form BFs. Abundance or depletion of nutrients, carbon and nitrogen sources, presence or absence of oxygen, electron acceptors, acidic or alkaline conditions, etc., proved to be major environmental stress factors that induce or prevent BF formation in S. aureus (Weinrick et al., 2004; Boles and Horswill, 2008; Anderson et al., 2010; Rode et al., 2010; Mashruwala et al., 2017). The ability of bacteria to sense chemical and physical characteristics of their surroundings and adjust gene expression require mechanisms that take decisions in response to environmental condition changes, and these are mainly based on protein–DNA interactions defined by transcriptional factors (TFs) and their targets around promoters. In S. aureus 135 TFs and sigma factors of various family groups have been identified with only half of them experimentally characterized to date (Ibarra et al., 2013). Our results show that the best characterized global transcription regulators like Agr, SarA, AirR, CodY, CtsR, MgrA, LysR, WalR, SrrA, along with several other less studied TFs, like CidR, BglG, ArgR, LytR, CymR, HutR, Hup, InfB, NreB, Dps, as well as some of putative function like the GntR family SACOL2516, the Xre family SACOL0420, the Mer family SACOL2517, all of which are associated with BF formation, were differentially expressed under acidic or alkaline conditions. Of the above TFs, pivotal role play those that are also members of two-component systems (TCS), signal transduction mechanisms utilized by most bacteria to monitor and respond to environmental stimuli. TCSs are composed by a membrane protein sensor (histidine kinase) and a response regulator which receives the information from the kinase and brings about the relevant response. Out of the 16 known TCSs in S. aureus only WalKR is essential (Dubrac et al., 2007) and as our results show, apart from WalKR and AgrBDCA five more TCSs were differentially expressed, i.e., three associated with oxygen availability in the media (SrrAB, AirRS and NreAB), one involved in autolysis (LytRS), and one sensing K+ limitation or salt stress (KdpDE).
In association with its involvement in BF formation, the agr Quorum-Sensing (QS) system of S. aureus, is the best studied in the bacterium. AgrB and AgrD act to generate the auto-inducing peptide AIP (QS molecule) which, after reaching an extracellular threshold concentration, stimulates activation of the TCS regulatory system AgrC (sensor) and AgrA (regulator). Under normal growth conditions, during BF formation, the Agr QS system is repressed to stop the expression of S. aureus colonization factors (Novick, 2003) and it gets activated mostly in the bacteria of the outer BF layers leading to the dispersal (Thoendel et al., 2011). In this respect, the down-regulation of AgrD, the peptide precursor of AIP, in BF cells at pH5 is explained by the effort of cells to promote BF formation as a defense against the acidic conditions. Similarly, the down-regulation of AgrB, the membrane protein which secrets the AIP product, is also expected in planktonic cells at pH9, in the same way as shown in S. aureus strain UAMS-1 where agrB expression was up-regulated in early growth stages and completely shut off at later stages (Grande et al., 2014). The accessory gene regulator (agrA) and the staphylococcal accessory regulator (sarA) have opposing roles in S. aureus BF formation and exert pleiotropic effects on the expression of molecules responsible for binding to different surfaces, controlling large numbers of target genes involved in virulence, autolysis, stress responses and metabolic processes (Pratten et al., 2001; Bischoff et al., 2004). The primary regulatory role of SarA is to repress the production of extracellular nucleolytic and proteolytic enzymes in early BF formation and once BF have developed and matured, agr expression leads to up-regulation of a number of virulence factors (Luong and Lee, 2002). Therefore, its down-regulation in planktonic cells at pH9 should have been accompanied by the induction of virulence factors, which is not the case (Tables 1, 3). However, although our results contradict with those obtained from BF cells grown under normal growth conditions, they are in full agreement with studies showing that agr expression in BF development strongly depends on environmental conditions and clearly underline the influence of the alkaline environment (Yarwood et al., 2004). Bearing also in mind that LytM belongs to the staphylin-type peptidase family and SsaA is a staphyloxanthin biosynthesis protein, their down-regulation in planktonic cells at pH9 along with SarA and AgrB is easily understood. MgrA, an important member of the SarA family, was also up-regulated in planktonic cells at pH5, along with WalR and TarS. MgrA is a global regulator that modulates the expression of 5–10% of the S. aureus genome, controls autolysis, virulence, a number of large surface proteins and most importantly, activates the agr system, thus repressing BF formation in S. aureus (Luong et al., 2006; Crosby et al., 2016). Known to form in combination with the TCS ArlRS a regulatory cascade and regulate BF formation (Crosby et al., 2016), mgrA’s over-expression was more or less expected in the absence of ArlRS activity. However, this is not the case for WalR, the response regulator of the two-component system (TCS) WalKR (also known as YycGF), which positively controls BF formation in S. aureus and many genes involved in cell wall degradation, like atlA, lytM, isaA, sceD, ssaA, and four ssaA-related genes (Dubrac et al., 2007). This TCS positively regulates major virulence genes involved in host matrix interactions (like efb, emp, fnbA and fnbB), oxidative stress resistance and vancomycin resistance (Delaune et al., 2012). Since none of the above-mentioned genes was up-regulated under acidic conditions, the most likely explanation is the loss of proper WalR function due to conformational changes that take place in its functional domains, i.e., the DNA-binding domain and the phosphorylation reception domain, under these pH conditions. This may happen by an alteration of the phosphorylation state of the conserved histidine residue in the cytoplasmic kinase, therefore preventing the response regulator to bind specifically on its DNA targets.
Two TCSs associated with oxygen availability in the media, AirRS and NreBC, were down-regulated in BF cells at pH9, along with a putative phosphate transport system regulator, PhoU-like, which encodes a probable transcriptional regulatory protein homologous to PhoU proteins involved in the down-regulation of phosphate uptake. AirRis the DNA-binding response regulator of the TCS AirSR that senses oxygen and redox changes, and also regulates pathways for nitrate respiration and lactose catabolism, as well as virulence factors (Sun et al., 2012). In a concerted action with SrrAB they reduce agr expression under conditions of low oxygen availability and are required for BF formation (Yarwood et al., 2001; Ulrich et al., 2007). AirR was also down-regulated at pH5, underlining the adverse effects of pH stress on its function and verifying its importance for BF formation. Knowing that AirR’s activity state is determined by oxidation of an Fe-S cluster present in AirS (Sun et al., 2012), it is feasible to assume that the prolonged exposure of cells to alkaline or acidic conditions could have destroyed this cluster in AirS, -which is essential for its kinase activity-, and thus hamper DNA binding activity of AirR and allow BF formation. A similar dissociation of Fe-S at pH9 would have also been the explanation for the down-regulation of NreB, the sensor histidine kinase protein of the TCS, a cytoplasmic protein containing four conserved cysteine residues that together comprise a Fe-S cluster (Kamps et al., 2004). In regard to the putative PhoU-like phosphate transport regulator it should be pointed out that recently the function of two phoU genes has been experimentally determined in S. epidermidis proving that only one of these is an important regulator of BF formation and drug tolerance (Wang et al., 2017). Although sequence similarity of the PhoU-like COL gene with either of these genes is low, its up-regulation in planktonic cells grown at alkaline conditions signifies the important role of this putative protein in response to pH stress.
In BF cells growing at acidic conditions, we have recorded a moderate up-regulation of KdpD, regulator of the kdpDE TCS (Supplementary Table S1). This TCS responds to K+ limitation and salt concentrations and it is up-regulated by agr through repressing rot translation (Xue et al., 2011). Its up-regulation in both BF and planktonic cells at pH5 is regarded as an indication of no K+ limitation in the media and also confirms its dependence on the agr system. Under the same growth conditions CymR, MetN2, and HsdM were also up-regulated in BF cells. CymR is the master regulator of cysteine metabolism in S. aureus known to control host sulfur source utilization and also play a role in BF formation (Soutourina et al., 2009). MetN2, is an ABC transporter ATP-binding protein (member of the three-gene operon metN2 > SACOL0505 > 0506), involved in metabolism. Their up-regulation is in full agreement with results which show that CymR under stress requires more cysteine and therefore up-regulates several target genes including metNPQ (Chang et al., 2006). HsdM, is the site specific restriction-modification enzyme used in the bacterial defense system against foreign DNA. Therefore, its up-regulation in BF cells may be due to the bacterial sensing of excess extracellular DNA produced by the lysis of outer layer cells and the strengthening of matrix that takes place in the formed BF.
The staphylococcal respiratory response TCS, SrrAB, is critical for anaerobic growth of S. aureus as the membrane component SrrB senses oxygen limitation and signals the cytoplasmic SrrA to repress transcription of the accessory gene regulator agr (Yarwood et al., 2001). Under normal conditions SrrA binds to its own promoter (autoregulation) as well as to numerous other promoters as its complex regulatory role has been revealed by a recent microarray analysis which shows that a ΔsrrA mutation affects the transcription of 230 genes in normal growth conditions, and 51 under decreased oxygen (Wu et al., 2015). Impaired respiration leads to increased cell lysis via increased expression of atlA, resulting also to the release of DNA, cytosolic proteins and BF formation (Mashruwala et al., 2017). This is in full agreement with our observation that in planktonic cells grown at pH9, LytR, the transcriptional regulator of the S. aureus TCS system LytRS and affector of murein hydrolase activity, and AtlA, a murein hydrolase, were up-regulated. Due to their lytic activities both proteins are linked with cell wall synthesis, autolysis and release of genomic DNA that eventually becomes an important part of the BF matrix thus positively affecting BF formation (Lehman et al., 2015). The concerted up-regulation of the above genes along with genes involved in general metabolism like phoU, argR (arginine metabolism repressor), lysS (transcriptional activator of the glutamate synthase operon), gntR (gluconate operon transcriptional regulator), ybhK (a putative phospho-L-lactate transferase-like protein) and sufB (an ABC-type transporter membrane component), strongly indicate that they contribute to cell lysis and release of genomic DNA. In particular the strong influence of arginine on polysaccharide intercellular adhesin synthesis and BF formation that has been recorded in S. aureus (Zhu et al., 2007) the up-regulation of ArgR is expected to withhold BF formation in planktonic cells. The glutamate metabolism activator (LysR, also known as CidR in other strains) could also be involved in stress response as poly-γ-DL-glutamic acid is a virulence factor that protects S. epidermidis against high salt concentrations and additionally mediates resistance to antimicrobial peptides and phagocytosis (Fey and Olson, 2010). Similarly, as the iron-regulated transporter SufB is associated with resistance to oxidative stress it can act as a protection mechanism due to its non-specific DNA binding ability (Mashruwala et al., 2015). Proteins SufB and SufU synthesize inorganic cofactors called iron-sulfur (Fe-S) clusters, which are required for functional Fe-S proteins. Mutant S. aureus cells that are unable to transfer iron-sulfur clusters have impaired pathogenicity and are more sensitive to stress due to endogenous reactive oxygen species (ROS) which leads to DNA damage, and exogenously supplied ROS and reactive nitrogen species. Thus, as the planktonic cells grown under alkaline conditions are in late exponential phase, the over-expression of lytic enzymes can be explained as the preparation of the cells to form BF as soon as the conditions become favorable (i.e., as soon as they reach post-exponential phase and there is a clear nutrient depravation or when the pH drops to pH7, which happens after 2–3 days). Interestingly enough, none of the known for S. aureus lytic enzymes was up-regulated in planktonic cells under acidic conditions, underlining the strong repression of the corresponding genes.
The major TRs, CodY and CtsR were over-expressed in BFs under alkaline conditions. The importance of CodY as a global regulator of S. aureus has been revealed by a genome-wide analysis using DNaseI foot-printing assays which has shown that it has more than 200 direct gene targets (Majerczyk et al., 2010). As a DNA-binding protein it interacts directly with chromosomal DNA containing a conserved sequence stem-loop motif and affects BF formation both positively and negatively depending on the strain (Brinsmade, 2017). It has been suggested that under normal growth conditions it suppresses BF formation in methicillin-resistant strains (such as the MRSA strain used in this study) and promotes BF formation in methicillin-susceptible strains (Atwood et al., 2015). This contradiction with our findings can be attributed either to the different strains used or to the adverse effect of alkaline conditions on the bacterium. Nevertheless, the known repression of virulence gene expression by CodY (Waters et al., 2016) is also recorded in our results. CtsR is a global transcriptional regulator of protein quality control which under normal conditions is active as a repressor binding to its cognate DNA operator sequences. In S. aureus it is the first of a four-gene operon (ctsR, SACL0568, SACL0569, clpC) and its protein acts as the negative regulator of the Class III family of heat shock genes clpC, clpB and clpP; the latter acting as a global regulator on regulons involved in virulence, oxidative stress response, autolysis and DNA repair (Michel et al., 2006). Under exposure to stress ctsR losses its ability to bind DNA because of conformational changes and that leads to an un-induced transcription of target genes (Derre et al., 1999). Thus, the over-expression of ctsR in BF cells grown under alkaline conditions indicates that the gene exerts its negative control on clp shock genes, including the ClpC protease. This comes to agreement with the over-expression of the negative regulator of competence MecA that we observed under the same conditions, a protein which binds to ClpC and prevents proteolysis (Tian et al., 2013). The need for autolysis in order to maintain the BF and enhance the rigidity of extracellular matrix by the release of eDNA is compensated at alkaline conditions by the over-expression of the lytic transglycosylase SceD, which is known to promote BF formation and is essential for nasal colonization in cotton rats (Stapleton et al., 2007), and the secreted immunodominant surface protein IsaB, which has the ability to bind eDNA and stabilize the extracellular matrix (Gibert et al., 2014).
Under acidic conditions along with AirR and AgrD several TFs, namely HutR, BglG, InfB, Dps, SACOL0420 and SACOL2517 were also down-regulation in BF cells together with proteins RpoB, FtsK and TufA, of general cell maintenance functions. HutR is a transcriptional regulator of the LysR family, which is the most common type of bacterial DNA-binding proteins, acting as either activators or repressors of gene expression and considered as a putative repressor of the histidine utilization operon (Ibarra et al., 2013). The BglG family transcriptional anti-terminators are DNA-binding proteins that regulate the expression of bacterial genes and operons, whose products are required for utilization of phosphoenolpyruvate:sugar phosphotransferase system carbohydrates (Fux et al., 2004). In S. aureus COL the bglG gene that was down-regulated was SACOL0228, the mannitol operon transcriptional antiterminator. It is noted that there are three more bglG loci in S. aureus COL genome with highly similar protein sequences (SACOL0403 and SACOL2147, mannitol operon transcriptional anti-terminators; and SACOL2662, activator of the mannose operon). SACOL2517 is a putative transcriptional regulator of the MerR family, contains a HTH domain and shows extended similarity with the gluconate operon transcription regulator GntR. InfB is the translation initiation factor IF-2, one of the essential components for the initiation of protein synthesis. It protects formyl-methionyl-tRNA from hydrolysis and promotes its binding to the 30S ribosomal subunits. Dps is a DNA-binding stress response protein for which very little is known about its exact function in S. aureus. In E. coli it protects DNA in a non-specific way from acid-induced damage (Jeong et al., 2008). However, the same study shows that even in the presence of Dps, protein RecA is needed for the repair of acid-induced DNA damage. In addition to the above, rpoB, the DNA directed RNA polymerase βchain coding gene, ftsK, the gene coding for a protein required for cell division and chromosome partition and the translational elongation factor tufA involved in peptide chain formation were also down-regulated in BF cells under acidic conditions. Taking into consideration the functions of all the above down-regulated TFs and proteins, we may assume that in order to maintain the BF, cells at pH5 drastically reduce metabolic activities, thus preventing cell lysis and dispersal which would require the protection of DNA. In support of this hypothesis may also be considered the down-regulation of mob, which codes for a protein causing the mobilization of S. aureus COL plasmid pT181, hence it prevents the potential horizontal gene transfer of plasmid carried antibiotic resistance genes. Finally, the down-regulation of SACOL0420 in BF cells in contrast with its up-regulation in planktonic cells at pH5, indicates that this putative transcriptional regulator of the Xre family, may play an important role in BF formation. Its sequence analysis shows that it contains a signal transduction peptide, REC and HTH domains, and it is the first gene of an operon 0420 > 0421 > 0422 > 0424, which is putatively regulated by NreC. Therefore, since its true function is still unknown it certainly merits more attention in the future.
In the early stages of BF formation S. aureus cells are passively adsorbed on the material surface through electrostatic and hydrophobic interactions. Following the initial cell adhesion and formation of a monolayer, a cell to cell aggregation and accumulation in bacterial multilayered architecture is mediated by MSCRAMMs, proteins with differential binding specifications for host matrix components and all containing an LPXTG motif that allows them to anchor on surfaces (Patti et al., 1994; Stoodley et al., 2002). All these MSCRAMMs are covalently attached to cell wall peptidoglycan by the membrane-associated enzyme sortase that recognizes the LPXTG motif and their corresponding genes are controlled and up-regulated by the sigma B (Bischoff et al., 2004). Several transcriptomic studies with pathogenic isolates of S. aureus have shown that under normal pH conditions all these genes are up-regulated in BF and planktonic cells (Dunman et al., 2001; Resch et al., 2005; Lindsay et al., 2006; Wang et al., 2012). It is, therefore, interesting to note that in our study only two of these proteins were differentially expressed in BF cells under the pH stress conditions, i.e., the secreted VWF-binding protein (Vwb), which was up-regulated under both pH regimes, and EbsS, an elastin binding protein, the only adhesive trans-membrane MCRAMM that contains the pentapeptide motif NPQTN instead of LPXTG and was the only down-regulated MSCRAMM family protein at pH5. The two large surface associated glycoproteins, VWF and SasA, both mediating platelet adhesion at sites of endothelial damage were up-regulated in planktonic cells grown at pH5 (Tables 1, 2). The Von Willebrand factor (VWF) is a large, multimeric glycoprotein mediating platelet adhesion at sites of endothelial damage and Vwb interacts with VWF and the surface protein clumping factor A (ClfA), thus anchoring S. aureus to vascular endothelium under shear stress, enhancing its ability to cause tissue damage or systemic disease (Claes et al., 2017). It appears therefore, that in BF-associated cells Vwb not only can adhere on polystyrene surfaces under both alkaline and acidic conditions, but obviously plays a pivotal role in cell to cell aggregation and accumulation under these stress conditions. SasA, also known as Srap (serine-rich adhesin for platelets) mediates the direct binding of S. aureus to platelets and contributes to infective endocarditis is a less studied MSCRAMM protein which contains the LPXTG motif (Siboo et al., 2005). SasA has been found responsible for binding to gp340 -a factor that in the oral cavity induces salivary aggregation with bacteria and promotes S. aureus adhesion to tissues such as the teeth and mucosa- via the N-acetyl-neuraminic acid moiety (Kukita et al., 2013). Thus, its up-regulation is considered as an additional tool for binding on abiotic surface. The down-regulated at pH5 protein EbsS, due to its structure, is under different regulatory control as it is known that the constitutively expressed sortase SrtA is responsible for anchoring all LPXTG-containing surface proteins, whereas SrtB is specialized to carry out the specific iron-regulated cell wall sorting of a NPQTN signal proteins like EbsS and IsdC (Mazmanian et al., 2002).
Biofilm maturation starts when the intrinsic regulatory program of BF formation begins to produce the matrix, consisting of extracellular capsular polysaccharides, proteins and eDNA. Altogether these molecules organize cells in three-dimensional structures, separated by fluid channels which are vital in delivering nutrients into BF deeper layers, as well as to deliver auto-inducing peptides that sense population densities (QS) and are subsequently used to trigger dispersal of cells and virulence factors (Boles et al., 2010; Thoendel et al., 2011; Otto, 2013). Concerning cell wall enzymes, in accordance to what has been observed under normal pH conditions (Resch et al., 2006; Beenken et al., 2012) it is interesting to note that the capsular polysaccharide biosynthesis enzymes Cap8C and Cap5B were up-regulated at both pH regimes, the former in BF cells –as well as in planktonic cells at pH5- and the latter in planktonic cells (Tables 1, 2). Thus, apart from their established role in BF development, the up-regulation of Cap5B and Cap8C is most likely due to the effort of cells to repair their capsule and cell wall after damage caused by the alkaline or acidic environment. Knowing that in a previous study under mild acid conditions (pH5.5) both Cap8C and Cap5B were significantly up-regulated in BF cells (Weinrick et al., 2004), the lack of induction of Cap5B at slightly lower pH in our work possibly underlines the importance of small pH differences on gene expression in BFs. Facilitated by LytR-A-Psr family enzymes, Cap5B is known to covalently attach to the glycan strands of peptidoglycan (Chan et al., 2014), and this seems to be the case in planktonic cells growing at pH9 that showed up-regulation of both lytR and cap5B (Table 1). It is pointed out that the over-production of type 8 capsular polysaccharides was previously found to augment S. aureus virulence, leading to longer persistence in the bloodstream, the liver, and the spleen of experimental mice (Luong and Lee, 2002), however, without influencing the pathogen’s susceptibility to vancomycin (Jansen et al., 2013). EbsB, a putative cell wall enzyme and PgcA, a phosphomannomutase/phosphoglucomutase family protein were also up-regulated in BF cells at pH5. EbsB contains a nucleic acid-binding motif, which may have an additional role either as a regulator or in association with eDNA. PgcA is involved in the biosynthesis of UDP-N-glucosamnine and a pgcA-transposon inactivated gene in S. aureus was shown to have drastically reduced methicillin resistance, although its femA gene remained intact (Wu et al., 1996). In addition, under the same pH conditions, SACOL0366 a gene encoding for phage terminase small subunit was also up-regulated together with yozM (a prophage-derived-like uncharacterized gene in Bacillus subtilis) and several other moderately up-regulated (1.55–1.82 times, see Supplementary Table S1) S. aureus phage associated proteins, like a hydrolase and a head protein of phage phi-11, a protein of phage phi-13, a phage anti repressor protein, a conserved phage associated protein, a hypothetical pathogenicity island, and a couple of phage associated proteins similar to those of other bacteria (Supplementary Table S1). Since the small terminase subunit forms a nucleoprotein structure that helpsto position the terminase large subunit at the packaging initiation site that interacts with the double-stranded phage DNA, its over-expression in combination with that of all the above mentioned phage-associated proteins BF cells under acidic conditions is somehow alarming and should be taken into consideration for appropriate use of disinfectants and sanitizers against S. aureus. Equally important seems to be the over-expression of tarS in planktonic cells at pH5 because TarS is a glycosyl transferase (member of the operon ispD > 0241 > 0242 > tarS) which glycosylates cell wall teichoic acid polymers, a process that is specifically responsible for methicillin resistance in MRSA (Sobhanifar et al., 2016). The over-expression in the same cells of three genes associated with stress responses (hchA, recF and hup), is understood under the view of their functions. HchA, a chaperone nucleoid-associated protein H-NS, is known to repress transcription by forming extended DNA-H-NS complexes and capturing early unfolding intermediates under prolonged conditions of severe stress, finally releasing them when cells return to physiological conditions (Mujacic et al., 2004). RecF is a DNA replication and repair protein that can be used by the cell for repairing acidic stress-induced DNA damage, and in combination with the histone-like DNA-binding protein Hup which is capable of wrapping DNA to stabilize it, prevent its denaturation under extreme environmental conditions (Castro et al., 2011). Hup has been also found to repress the E. coli bgl operon (Dole et al., 2004), and according to our results a similar association between the two loci seems to exist in S. aureus. Under alkaline conditions in planktonic cells, two more cell wall associated genes, murAB and lytM, were down-regulated, along with parC, yozE and NP_72897. MurAB, is the UDP-N-acetylglucosamine 1-carboxyvinyltransferase 2, which is involved in glycan synthesis and is known to be over-expressed in Streptococcus suis BFs growing under normal conditions (Wang et al., 2012), and LytM - also down-regulated in BF cells at pH5- is the peptidoglycan hydrolase previously considered as the only autolysin of S. aureus. Its role as an autolysin has recently been disputed as it was proved to be an early exponential phase protein whose expression was down-regulated by Agr, but still indicates that LytM plays an important role in BF development (Singh et al., 2010). YozE, a hypothetical YozE_SAM_like protein, belongs to a family of proteins with a four-helix motif similar to sterile alpha motif (SAM) domains and is likely to involve binding to DNA (Swapna et al., 2012). NP_72897 is a putative cell division protein, and ParC, is the DNA topoisomerase IV subunit A, responsible for relaxing supercoiled DNA and very important in bacteria replication, where the circular chromosome becomes catenated or linked. In addition, ParC is known to bind to the ica cluster, which is involved in the extracellular matrix production (Jefferson et al., 2004). Therefore, it becomes evident that under alkaline stress the planktonic cells are slowing down chromosome replication and cell division in an effort to focus on functions of defense mechanisms against cell damage.
During BF maturation and at the exponential growth phase S. aureus normally produces several surface binding proteins, which are subsequently secreted. The surface binding proteins IsdC, MntC, SsaA, SasA, the substrate binding proteins MapW, Efb, Emp, Scc, EpiG, Pls and the envelope and membrane associated proteins TpgX, NP_437451,NP_406103 were all differentially expressed. In BF cells grown at pH9, apart from Vwb and SceD, the cell wall surface anchor protein involved in heme uptake, IsdC, was also up-regulated. IsdC induces BF formation in S. lugdunensis grown under iron limitation (Missineo et al., 2014), yet under these conditions isdC and a variety of virulence factors are repressed in S. aureus (Hammer and Skaar, 2011). Thus, the absence of induction in the expression of any of the known virulence genes under alkaline conditions is most likely due to the simultaneous up-regulation of the global regulator CodY, which is known to negatively regulate virulence gene expression (Majerczyk et al., 2008). The only other cell surface associated protein that was down-regulated in BF cells grown at pH9 was MapW. The same protein was down-regulated under acidic conditions in both BF and planktonic cells. MapW, is a cell surface protein with no LPXTG sequence, which is not recognized and linked to the peptidoglycan by a sortase, but it is released by digestion with lysostaphin. It does not bind to soluble extracellular matrix proteins but functions as an endogenous adhesion substrate in the attachment to plastic surfaces and eukaryotic cells via interaction with staphylococcal surface adhesions (Kreikemeyer et al., 2002). Its down-regulation in both BF cells and planktonic cells strongly suggests that this protein can only be involved in cell attachment to surfaces at neutral pH environments. Together with EbsB, a hypothetical Map-like protein, the fibrinogen-binding protein Efb, and a putative protein (NP_406103) were over-expressed in BF cells at pH5. The hypothetical Map-like protein appears unique as it has no significant similarity with any of the known Map proteins of S. aureus. However, as map-like proteins contain up to six MAP domains, each containing a 31-residue sub-domain sharing striking sequence homology with a segment present in the peptide binding groove of the beta chain of the MHC class II proteins from different mammalian species, it is expected to have a different binding ability than MapW. Efb is a secreted virulence factor that helps the pathogen to evade human neutrophils, impairs wound healing and inhibits the formation of platelet-leukocyte complexes (Posner et al., 2016). The NP_406103 putative membrane protein shows some similarity with gntR gene of other S. aureus strains, a gene coding for glucokinase. On the contrary, EpiG/BsaG, another extracellular surface associated protein, the epidermin immunity protein F was down-regulated in BF cells. Thus, this differential expression of extracellular surface proteins under the same pH stress is thought to reflect to the influence of these conditions on the ability/inability of cells to adhere on the polystyrene surface. Since EpiG and Scc were found to interact with biotic and abiotic surfaces in a zinc-dependent way (Geoghegan et al., 2013; Nakakido et al., 2014), reducing the availability of zinc ions could lead to the development of novel therapeutic or disinfection strategies for controlling S. aureus infections.
In planktonic cells MntC, a conserved manganese-binding surface protein and an ABC (ATP-binding cassette) transporter system component was down-regulated under both pH regimes. Using ELISA tests it was recently demonstrated that MntC is also an ion-scavenging factor with a marked ability to bind to several extracellular matrix and coagulation cascade components, including laminin, collagen type IV, cellular and plasma fibronectin, plasminogen and fibrinogen, hence a potential virulence factor (Salazar et al., 2014). Therefore, its down-regulation at both pH conditions is likely due to iron repletion, as in the case of S. epidermidis which was found to withstand higher variations in iron availability when grown planktonically (Oliveira et al., 2017). In planktonic cells grown under alkaline conditions, TpgX, Emp and SsaA were over-expressed. TpgX is a hypothetical protein which following a proteomic profiling of S. aureus was shown to be cell envelope-associated (Resch et al., 2006). Emp, a secretory extracellular matrix and plasma binding protein, is an adhesin that displays a broad binding specificity to the host cell extracellular matrix proteins fibronectin, fibrinogen, collagen, and vitronectin (McGavin et al., 1993). Evidently, the up-regulation of these two proteins mediates the adherence on the abiotic surface in spite of the alkaline environment. SsaA, under normal pH conditions is expressed at slightly higher levels in BF cells than in planktonic cells (Resch et al., 2005). Therefore, its up-regulation at pH9 in planktonic cells, should be taken seriously into consideration because as an antigen associated with S. aureus surface and staphyloxanthin biosynthesis is considered as a virulence factor. Of equal interest is the up-regulation in planktonic cell growing at pH5 of two genes involved in S. aureus virulence, pls and sasA. As mentioned before, SasA mediates the direct binding of S. aureus to platelets and contributes to infective endocarditis. The plasmin-sensitive protein gene pls which was found to be a virulence factor in mouse septic arthritis model is encoded by the staphylococcal cassette chromosome mec type I in MRSA that also encodes the methicillin-conferring mecA and further genes and has been found to stimulate BF formation (Bleiziffer et al., 2017). Finally, five more proteins associated with cell surface IsaB, Scc, NP_437451 and chaperones PrsaA, Hfq, were down-regulated in planktonic cells grown under acidic conditions. NP_437451 is a putative membrane transport protein, associated with cell surface. IsaB is an extracellular nucleic acid binding protein with no sequence specificity, which elicits an immune response during septicemia and is generally classified as a virulence factor. However, its role in virulence has not been defined yet (Mackey-Lawrence and Jefferson, 2013). Scc is a fibrinogen-binding protein which facilitates attachment to fibrinogen during colonization of biotic surfaces which is over-expressed during BF formation in vitro and is crucial for the colonization of medical devices by healthcare S. aureus strains (Geoghegan et al., 2013). PrsA is a post-translocational chaperone lipoprotein that is involved in both glycopeptide and oxacillin resistance in S. aureus. More specifically, disruption of prsA leads to notable alterations in the sensitivity to glycopeptides and dramatically decreases the resistance of S. aureus COL (MRSA) to oxacillin (Jousselin et al., 2012). Hfq, an RNA chaperone that binds small regulatory RNAs and mRNAs, negatively regulates translation in response to envelope stress, environmental stress and changes in metabolite concentrations and upon over-expression decreases persister cell formation (Guisbert et al., 2007). The down-regulation of all these genes involved in virulence and antibiotic resistance in planktonic cells, is rather promising because it confirms that alkaline conditions can be safely used as disinfectants to prevent BF formation on food processing and/or indwelling medical devices. Finally, the down-regulation of the putative monoamine oxidase regulator gene maoC1, at both pH9 and pH5, is rather interesting in view of its putative function. Bacterial cells respond to monoamine compounds, such as tyramine, dopamine, octopamine, or norepinephrine, and induce the syntheses of tyramine oxidase encoded by tynA and maoA. The monoamine oxidase regulator gene moaR of several bacteria was found to play a central role in the positive regulation of the expression of the monoamine regulon (moa) including the atsBA, maoCA, moaEF and tyn operons (Murooka et al., 1996). Thus, a similar role may be envisaged for the S. aureus maoC1 and its importance may be associated with the enzymatic activity of FabI in the essential fatty acid biosynthesis pathway, as studies with antisense RNA have shown (Ji et al., 2004).
Staphylococcal enterotoxins are important causative agents in staphylococcal toxic shock syndrome and food poisoning (Orwin et al., 2001). Staphylococcal superantigen-like (SSL) proteins are encoded by a cluster of fourteen ssl genes and contribute to the S. aureus virulence. Despite their structural similarity to superantigens, SSLs do not bind to T-cell receptors or major histocompatibility complex class II molecules but they target components of innate immunity and myeloid cells (Hermans et al., 2012). In this context, it is important to underline that no exotoxin or enterotoxin genes were over-expressed under highly alkaline or acidic conditions in both BF and planktonic cells (Tables 3, 4). This is in sharp contrast with results from other researchers working with cells growing at pH7 who have found genes that can lead to toxic shock and sepsis as the exotoxins 6 (ssl1) and 15 (ssl11) over-expressed in both BF and planktonic cells, and in addition, in planktonic cells the genes encoding exotoxin 3(ssl14) and enterotoxin K (sek) also up-regulated (Aguilar et al., 2017). This indicates that the pathogen is not launching a damaging offensive against host tissue when defending against a highly alkaline or acidic environment, a finding that could have direct clinical importance in the case of vaginal S. aureus infections, since the pH of this organ is low (between 3.8 and 4.5) due to its acidic secretions. Also, S. aureus is known to invade and survive for a short amount of time in the lysosomal compartment of non-phagocytic cells (pH 4.5 to 5.5), before escaping into the cytosol (Anderson et al., 2010). Although with a hypothetical function, the up-regulation of mdlB in planktonic cells at both pHs, merits particular attention as it codes for a putative ABC transporter permease and ATP–binding protein which may be exporting toxin(s).
As for genes involved directly or indirectly in antibiotic resistance of MRSAs we recorded four genes that were differentially expressed only in planktonic cells. Up-regulated were the genes femA, at both pHs, sepA at pH9, pls at pH5, and down-regulated was mdeA at pH9. FemA, codes fo FEMA, an aminoacyl-transferase which catalyzes the formation of the pentaglycine interpeptide bridge in S. aureus peptidoglycan and is considered as a factor influencing the level of methicillin resistance. It also strengthens the cell wall and is involved in dormancy (Savijoki et al., 2016). When femA was inactivated, mutant cells had a reduced peptidoglycan glycine content, reduced cell wall turnover, reduced whole-cell autolysis, and increased sensitivity toward β-lactam antibiotics (Maidhof et al., 1991). This is in agreement with the up-regulation of the capsule biosynthesis and autolytic enzymes observed in our study. Interestingly, acidic pH was found to restore susceptibility of methicillin-resistant Staphylococcus aureus to β-lactam antibiotics (Lemaire et al., 2008). Gene sepA encodes a drug efflux protein with four predicted transmembrane segments, which proved to be a multi-drug resistance gene when cloned from S. aureus to E. coli conferring the reduction of susceptibility to acriflavine and the acceleration of ethidium bromide efflux from the E. coli cells (Narui et al., 2002). As mentioned before, Pls is a methicillin-resistant surface protein precursor which could be involved in methicillin resistance by S. aureus (Bleiziffer et al., 2017), and a similar role may be attributed to the down-regulated at pH5 hypothetical mdeA/emrB gene which codes for a multi-drug resistance-related transporter.
Conclusion
Our results show that when S. aureus COL grows under highly acidic or alkaline conditions it attempts to respond to the resulting stress, repair its cell wall, protect itself by forming BF when an appropriate surface is provided, strengthen the BF by release of extracellular DNA and boost its resistance to antibiotics. In the meantime, it reduces its virulence (e.g., toxin production), as it has entered a defensive mode.
Interestingly, although the exact role in S. aureus BF formation of many of the transcription factors, stress response systems and adhesion proteins that were described above are not fully demonstrated yet, their involvement in BF formation is rather apparent. By examining the effect of alkaline and acidic pH on the gene expression of MRSA BF cells for first time, we have contributed important data to the understanding of cellular adjustments that might influence colonization, virulence and antibiotic resistance in this defensive growth mode. Overall, our results showed that S. aureus COL can easily grow at highly alkaline and acidic environments and led to the identification of several genes that were differentially expressed under these conditions and could be involved in stress response, virulence and antibiotic resistance pathways in this important pathogen. Understanding how the pathogen survives and responds under these conditions will certainly influence the design of better treatment or disinfection strategies in the future.
Data Availability Statement
The datasets generated and analyzed for this study can be found in the GEO repository (GSE138075) at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138075.
Author Contributions
GE, MT, and KP conceived and designed the experiments. GE performed the experiments. GE, GT, MT, and KP analyzed the data. GE, MT, and KP wrote the manuscript.
Funding
This work has been funded by the Network of Excellence «THALES – ‘BIOFILMS’», Pr. Code MIS 380229, to which we are grateful.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank to Prof. Sophia Kathariou (North Carolina State University, United States) for providing the bacterial strain and useful advice.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02393/full#supplementary-material
TABLE S1 | Diffentially-expressed genes at different conditions (alkaline or acidic pH; biofilm or planktonic growth), with a log2 ratio between 1.5 and 2 or –1.5 and –2.
Footnotes
References
Aguilar, J. L., Varshney, A. K., Pechuan, X., Dutta, K., Nosanchuk, J. D., and Fries, B. C. (2017). Monoclonal antibodies protect from Staphylococcal Enterotoxin K (SEK) induced toxic shock and sepsis by USA300 Staphylococcus aureus. Virulence 8, 741–750. doi: 10.1080/21505594.2016.1231295
Akbas, M. Y., and Cag, S. (2016). Use of organic acids for prevention and removal of Bacillus subtilis biofilms on food contact surfaces. Food. Sci. Technol. Int. 22, 587–597. doi: 10.1177/1082013216633545
Anderson, K. L., Roux, C. M., Olson, M. W., Luong, T. T., Lee, C. Y., Olson, R., et al. (2010). Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 60, 208–250. doi: 10.1111/j.1574-695X.2010.00736.x
Archer, N. K., Mazaitis, M. J., Costerton, J. W., Leid, J. G., Powers, M. E., and Shirtliff, M. E. (2011). Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2, 445–459. doi: 10.4161/viru.2.5.17724
Arciola, C. R., Campoccia, D., Speziale, P., Montanaro, L., and Costerton, J. W. (2012). Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 33, 5967–5982. doi: 10.1016/j.biomaterials.2012.05.031
Atwood, D. N., Loughran, A. J., Courtney, A. P., Anthony, A. C., Meeker, D. G., Spencer, H. J., et al. (2015). Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiol. Open 4, 436–451. doi: 10.1002/mbo3.250
Beenken, K. E., Dunman, P. M., McAleese, F., Macapagal, D., Murphy, E., Projan, S. J., et al. (2004). Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186, 4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004
Beenken, K. E., Spencer, H., Griffin, L. M., and Smeltzer, M. S. (2012). Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect. Immun. 80, 1634–1638. doi: 10.1128/IAI.06134-11
Bischoff, M., Dunman, P., Kormanec, J., Macapagal, D., Murphy, E., Mounts, W., et al. (2004). Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186, 4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004
Bleiziffer, I., Eikmeier, J., Pohlentz, G., McAulay, K., Xia, G., Hussain, M., et al. (2017). The plasmin-sensitive protein Pls in methicillin-resistant Staphylococcus aureus (MRSA) is a glycoprotein. PLoS Pathog. 13:e1006110. doi: 10.1371/journal.ppat.1006110
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PloS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Boles, B. R., Thoendel, M., Roth, A. J., and Horswill, A. R. (2010). Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146
Bore, E., Langsrud, S., Langsrud, Ø., Rode, T. M., and Holck, A. (2007). Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153, 2289–2303. doi: 10.1099/mic.0.2007/005942-0
Brinsmade, S. R. (2017). CodY, a master integrator of metabolism and virulence in Gram positive bacteria. Curr. Genet. 63, 417–425. doi: 10.1007/s00294-016-0656-5
Castro, S. L., Nelman-Gonzalez, M., Nickerson, C. A., and Ott, C. M. (2011). Induction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl. Environ. Microbiol. 77, 6368–6378. doi: 10.1128/AEM.00175-11
Centers for Disease Control and Prevention (2018). Methicillin-Resistant Staphylococcus aureus (MRSA). Available at: www.cdc.gov/mrsa (accessed January 02, 2018).
Chan, Y. G., Kim, H. K., Schneewind, O., and Missiakas, D. (2014). The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J. Biol. Chem. 289, 15680–15690. doi: 10.1074/jbc.M114.567669
Chang, W., Small, D. A., Toghrol, F., and Bentley, W. E. (2006). Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188, 1648–1659. doi: 10.1128/JB.188.4.1648-1659.2006
Christensen, G. D., Baldassarri, L., and Simpson, W. A. (1994). “Colonization of medical devices by coagulase-negative staphylococci,” in Infections associated with indwelling medical devices, eds A. L. Bisno, and F. A. Waldvogel (Washington DC: American Society for Microbiology), 45–78.
Claes, J., Liesenborghs, L., Peetermans, M., Veloso, T. R., Missiakas, D., Schneewind, O., et al. (2017). Clumping factor A, von Willebrand factor-binding protein and von Willebrand factor anchor Staphylococcus aureus to the vessel wall. J. Thromb. Haemost. 15, 1009–1019. doi: 10.1111/jth.13653
Crosby, H. A., Schlievert, P. M., Merriman, J. A., King, J. M., Salgado-Pabón, W., and Horswill, A. R. (2016). The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 12:e1005604. doi: 10.1371/journal.ppat.1005604
de Souza, E. L., Meira, Q. G., de Medeiros Barbosa, I., Athayde, A. J., da Conceição, M. L., and de Siqueira Júnior, J. P. (2014). Biofilm formation by Staphylococcus aureus from food contact surfaces in a meat-based broth and sensitivity to sanitizers. Braz. J. Microbiol. 45, 67–75. doi: 10.1590/s1517-83822014000100010
Delaune, A., Dubrac, S., Blanchet, C., Poupel, O., Mader, U., Hiron, A., et al. (2012). The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 80, 3438–3453. doi: 10.1128/IAI.00195-12
Derre, I., Rapoport, G., and Msadek, T. (1999). CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol. 31, 117–131. doi: 10.1046/j.1365-2958.1999.01152.x
Dole, S., Nagarajavel, V., and Schnetz, K. (2004). The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52, 589–600. doi: 10.1111/j.1365-2958.2004.04001.x
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/cmr.15.2.167-193.2002
Doulgeraki, A. I., Efthimiou, G., Paramithiotis, S., Pappas, K. M., Typas, M. A., and Nychas, G.-J. (2017). Effect of rocket (Eruca sativa) extract on MRSA growth and proteome: metabolic adjustments in plant-based media. Front. Microbiol. 8:782. doi: 10.3389/fmicb.2017.00782
Dubrac, S., Boneca, I. G., Poupel, O., and Msadek, T. (2007). New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269. doi: 10.1128/JB.00645-07
Dunman, P. M., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S., et al. (2001). Transcriptional profiling-based identification of Staphylococcus aureus genes regulated by agr and/or sarA loci. J. Bacteriol. 183, 7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001
Efthimiou, G., Pappas, K. M., and Typas, M. A. (2015). “Study of biofilm formation by the hospital-acquired and recently considered as foodborne pathogen Staphylococcus aureus subsp. aureus COL (MRSA),” in Proceedings of the 6th Mikrobiokosmos Conference, (Athens), 100–101.
Fey, P. D., and Olson, M. E. (2010). Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5, 917–933. doi: 10.2217/fmb.10.56
Fux, L., Nussbaum-Shochat, A., Lopian, L., and Amster-Choder, O. (2004). Modulation of monomer conformation of the BglG transcriptional antiterminator from Escherichia coli. J. Bacteriol. 186, 6775–6781. doi: 10.1128/JB.186.20.6775-6781.2004
Geoghegan, J. A., Monk, I. R., O’Gara, J. P., and Foster, T. J. (2013). Subdomains N2N3 of fibronectin binding protein A mediate Staphylococcus aureus biofilm formation and adherence to fibrinogen using distinct mechanisms. J Bacteriol. 195, 2675–2683. doi: 10.1128/JB.02128-12
Gibert, L., Didi, J., Marlinghaus, L., Lesouhaitier, O., Legris, S., Szabados, F., et al. (2014). The major autolysin of Staphylococcus lugdunensis, AtlL, is involved in cell separation, stress-induced autolysis and contributes to bacterial pathogenesis. FEMS Microbiol. Lett. 352, 78–86. doi: 10.1111/1574-6968.12374
Gibson, H., Taylor, J. H., Hall, K. E., and Holah, J. T. (1999). Effectiveness of cleaning techniques used in the food industry in terms of the removal of bacterial biofilms. J. Appl. Microbiol. 87, 41–48. doi: 10.1046/j.1365-2672.1999.00790.x
Götz, F. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x
Grande, R., Nistico, L., Sambanthamoorthy, K., Longwell, M., Iannitelli, A., Cellini, L., et al. (2014). Temporal expression of agrB, cidA, and alsS in the early development of Staphylococcus aureus UAMS-1 biofilm formation and the structural role of extracellular DNA and carbohydrates. Pathog. Dis. 70, 414–422. doi: 10.1111/2049-632X.12158
Guilhen, C., Forestier, C., and Balestrino, D. (2017). Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol. Microbiol. 105, 188–200. doi: 10.1111/mmi.13698
Guisbert, E., Rhodius, V. A., Ahuja, N., Witkin, E., and Gross, C. (2007). Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 189, 1963–1973. doi: 10.1128/JB.01243-06
Hammer, N. D., and Skaar, E. P. (2011). Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 65, 129–147. doi: 10.1146/annurev-micro-090110-102851
Harris, S. R., Feil, E. J., Holden, M. T., Quail, M. A., Nickerson, E. K., Chantratita, N., et al. (2010). Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474. doi: 10.1126/science.1182395
Hassoun, A., Linden, P. K., and Friedman, B. (2017). Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. Crit. Care 21:211. doi: 10.1186/s13054-017-1801-3
Hermans, S. J., Baker, H. M., Sequeira, R. P., Langley, R. J., Baker, E. N., and Fraser, J. D. (2012). Structural and functional properties of staphylococcal superantigen-like protein 4. Infect. Immun. 80, 4004–4013. doi: 10.1128/IAI.00764-12
Ibarra, J. A., Pérez-Rueda, E., Ronan, K., Carroll, R. K., and Shaw, L. N. (2013). Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14:126. doi: 10.1186/1471-2164-14-126
Jansen, A., Szekat, C., Schröder, W., Wolz, C., Goerke, C., Lee, J. C., et al. (2013). Production of capsular polysaccharide does not influence Staphylococcus aureus vancomycin susceptibility. BMC Microbiol. 13:65. doi: 10.1186/1471-2180-13-65
Jefferson, K. K., Pier, D. B., Goldmann, D. A., and Pier, G. B. (2004). The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186, 2449–2456. doi: 10.1128/jb.186.8.2449-2456.2004
Jeong, K. C., Hung, K. F., Baumler, D. J., Byrd, J. J., and Kaspar, C. W. (2008). Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 8:181. doi: 10.1186/1471-2180-8-181
Ji, Y., Yin, D., Fox, B., Holmes, D. J., Payne, D., and Rosenberg, M. (2004). Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231, 177–184. doi: 10.1016/S0378-1097(03)00931-5
Jones, E. M., Cochrane, C. A., and Percival, S. L. (2015). The effect of pH on the extracellular matrix and biofilms. Adv. Wound Care 4, 431–439. doi: 10.1089/wound.2014.0538
Jones, T. F., Kellum, M. E., Porter, S. S., Bell, M., and Schaffner, W. (2002). An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 8, 82–84. doi: 10.3201/eid0801.010174
Jousselin, A., Renzoni, A., Andrey, D. O., Monod, A., Lew, D. P., and Kelley, W. L. (2012). The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 3629–3640. doi: 10.1128/AAC.06264-11
Kamal, R. M., Bayoumi, M. A., and Abd El Aal, S. F. A. (2013). MRSA detection in raw milk, some dairy products and hands of dairy workers in Egypt, a mini-survey. Food Control 33, 49–53. doi: 10.1016/j.foodcont.2013.02.017
Kamps, A., Achebach, S., Fedtke, I., Unden, G., and Götz, F. (2004). Staphylococcal NreB: an O2-sensing histidine protein kinase with an O2-labile iron–sulphur cluster of the FNR type. Mol. Microbiol. 52, 713–723. doi: 10.1111/j.1365-2958.2004.04024.x
Kreikemeyer, B., McDevitt, D., and Podbielski, A. (2002). The role of the Map protein in Staphylococcus aureus matrix protein and eukaryotic cell adherence. Int. J. Med. Microbiol. 292, 283–295. doi: 10.1078/1438-4221-00212
Kukita, K., Kawada-Matsuo, M., Oho, T., Nagatomo, M., Oogai, Y., Hashimoto, M., et al. (2013). Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect. Immun. 81, 1870–1879. doi: 10.1128/IAI.00011-13
Kumar, L. R. G., Kasim, A. K., Lekshmi, M., Nayak, B. B., and Kumar, S. (2016). Incidence of methicillin-resistant staphylococci in fresh seafood. Adv. Microbiol. 6, 399–406. doi: 10.4236/aim.2016.66039
Lehman, M. K., Bose, J. L., Sharma-Kuinkel, B. K., Moormeier, D. E., Endres, J. L., Sadykov, M. R., et al. (2015). Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Mol. Microbiol. 95, 723–737. doi: 10.1111/mmi.12902
Lemaire, S., Fuda, C., Van Bambeke, F., Tulkens, P. M., and Mobashery, S. (2008). Restoration of susceptibility of methicillin-resistant Staphylococcus aureus to beta-lactam antibiotics by acidic pH: role of penicillin-binding protein PBP 2a. J. Biol. Chem. 283, 12769–12776. doi: 10.1074/jbc.M800079200
Lemmer, K., Mielke, M., Pauli, G., and Beekes, M. (2004). Decontamination of surgical instruments from prion proteins: in vitro studies on the detachment, destabilization and degradation of PrPSc bound to steel surfaces. J. Gen. Virol. 85, 3805–3816. doi: 10.1099/vir.0.80346-0
Lindsay, J. A., Moore, C. E., Day, N. P., Peacock, S. J., Witney, A. A., Stabler, R. A., et al. (2006). Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188, 669–676. doi: 10.1128/JB.188.2.669-676.2006
Luong, T. T., Dunman, P. M., Murphy, E., Projan, S. J., and Lee, C. Y. (2006). Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188, 1899–1910. doi: 10.1128/JB.188.5.1899-1910.2006
Luong, T. T., and Lee, C. Y. (2002). Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence. Infect. Immun. 70, 3389–3395. doi: 10.1128/iai.70.7.3389-3395.2002
Mackey-Lawrence, N. M., and Jefferson, K. K. (2013). Regulation of Staphylococcus aureus immunodominant antigen B is a cell-surface associated nucleic acid binding protein. Microbiol. Res. 168, 113–118. doi: 10.1186/1471-2180-9-61
Maidhof, H., Reinicke, B., Blumel, P., Berger-Bachi, B., and Labischinski, H. (1991). FemA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol. 173, 3507–3513. doi: 10.1128/jb.173.11.3507-3513.1991
Majerczyk, C. D., Dunmann, P. M., Luong, T. T., Lee, C. Y., Sadykov, M. R., Somerville, G. A., et al. (2010). Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877. doi: 10.1128/JB.00220-10
Majerczyk, C. D., Sadykov, M. R., Luong, T. T., Lee, C. Y., Somerville, G. A., and Sonenshein, A. L. (2008). Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265. doi: 10.1128/JB.01545-07
Mashruwala, A. A., Guchte, A. V., and Boyd, J. M. (2017). Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. eLife 6:e23845. doi: 10.7554/eLife.23845
Mashruwala, A. A., Pang, Y. Y., Rosario-Cruz, Z., Chahal, H. K., Benson, M. A., Mike, L. A., et al. (2015). Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol. Microbiol. 95, 383–409. doi: 10.1111/mmi.12860
Mazmanian, S. K., Ton-That, H., Su, K., and Schneewind, O. (2002). An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 2293–2298. doi: 10.1073/pnas.032523999
McGavin, M. H., Krajewska-Pietrasik, D., Rydén, C., and Höök, M. (1993). Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect. Immun. 61, 2479–2485.
Michel, A., Agerer, F., Hauck, C. R., Herrmann, M., Ullrich, J., Hacker, J., et al. (2006). Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796. doi: 10.1128/JB.00074-06
Missineo, A., Di Poto, A., Geoghegan, J. A., Rindi, S., Heilbronner, S., Gianotti, V., et al. (2014). IsdC from Staphylococcus lugdunensis induces biofilm formation under low-iron growth conditions. Infect. Immun. 82, 2448–2459. doi: 10.1128/IAI.01542-14
Moussa, F. W., Gainor, B. J., Anglen, J. O., Christensen, G., and Simpson, W. A. (1996). Disinfecting agents for removing adherent bacteria from orthopaedic hardware. Clin. Orthop. Relat. Res. 329, 255–262. doi: 10.1097/00003086-199608000-00032
Mujacic, M., Bader, M. W., and Baneyx, F. (2004). Escherichia coli Hsp31 functions as a holding chaperone that cooperates with the DnaK-DnaJ-GrpE system in the management of protein misfolding under severe stress conditions. Mol. Microbiol. 51, 849–859. doi: 10.1046/j.1365-2958.2003.03871.x
Murooka, Y., Azakami, H., and Yamashita, M. (1996). The monoamine regulon including syntheses of arylsulfatase and monoamine oxidase in bacteria. Biosci. Biotechnol. Biochem. 60, 935–941. doi: 10.1271/bbb.60.935
Nakakido, M., Aikawa, C., Nakagawa, I., and Tsumoto, K. (2014). The staphylococcal elastin-binding protein regulates zinc-dependent growth/biofilm formation. J. Biochem. 156, 155–162. doi: 10.1093/jb/mvu027
Narui, K., Noguchi, N., Wakasugi, K., and Sasatsu, M. (2002). Cloning and characterization of a novel chromosomal drug efflux gene in Staphylococcus aureus. Biol. Pharm. Bull. 25, 1533–1536. doi: 10.1248/bpb.25.1533
Normanno, G., Corrente, M., La Salandra, G., Dambrosio, A., Quaglia, N. C., Parisi, A., et al. (2007). Methicillin-resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food. Microbiol. 117, 219–222. doi: 10.1016/j.ijfoodmicro.2007.04.006
Nostro, A., Cellini, L., Di Giulio, M., D’Arrigo, M., Marino, A., Blanco, A. R., et al. (2012). Effect of alkaline pH on staphylococcal biofilm formation. APMIS 120, 733–742. doi: 10.1111/j.1600-0463.2012.02900.x
Novick, R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x
O’Brien, A. M., Hanson, B. M., Farina, S. A., Wu, J. Y., Simmering, J. E., Wardyn, S. E., et al. (2012). MRSA in conventional and alternative retail pork products. PLoS One 7:e30092. doi: 10.1371/journal.pone.0030092
Oliveira, F., França, A., and Cerca, N. (2017). Staphylococcus epidermidis is largely dependent on iron availability to form biofilms. Int. J. Med. Microbiol. 307, 552–563. doi: 10.1016/j.ijmm.2017.08.009
Orwin, P. M., Leung, D. Y., Donahue, H. L., Novick, R. P., and Schlievert, P. M. (2001). Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69, 360–366. doi: 10.1128/IAI.69.1.360-366.2001
Otto, M. (2013). Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Ann. Rev. Med. 64, 175–188. doi: 10.1146/annurev-med-042711-140023
Otto, M. (2018). Staphylococcal biofilms. Microbiol. Spectrum 6:GPP3-0023-2018. doi: 10.1128/microbiolspec.GPP3-0023-2018
Park, S. H., Choi, M. R., Park, J. W., Park, K. H., Chung, M. S., Ryu, S., et al. (2011). Use of organic acids to inactivate Escherichia coli O157:H7, Salmonella Typhimurium, and Listeria monocytogenes on organic fresh apples and lettuce. J. Food. Sci. 76, M293–M298. doi: 10.1111/j.1750-3841.2011.02205.x
Patti, J. M., Allen, B. L., McGavin, M. J., and Höök, M. (1994). MSCRAMM-mediated adherence of microorganisms to host tissues. Ann. Rev. Microbiol. 48, 585–617. doi: 10.1146/annurev.mi.48.100194.003101
Percival, S. L., McCarty, S., Hunt, J. A., and Woods, E. J. (2014). The effects of pH on wound healing, biofilms, and antimicrobial efficacy. Wound Repair Regen. 22, 74–86. doi: 10.1111/wrr.12125
Periasamy, S., Joo, H.-S., Duong, A. C., Bach, T.-H. L., Tan, V. Y., Chatterjee, S. S., et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl.Acad. Sci. U.S.A. 109, 1281–1286. doi: 10.1073/pnas.1115006109
Posner, M. G., Upadhyay, A., Abubaker, A. A., Fortunato, T. M., Vara, D., Canobbio, I., et al. (2016). Extracellular fibrinogen-binding protein (Efb) from Staphylococcus aureus inhibits the formation of platelet-leukocyte complexes. J. Biol. Chem. 291, 2764–2776. doi: 10.1074/jbc.M115.678359
Pratten, J., Foster, S. J., Chan, P. F., Wilson, M., and Nair, S. P. (2001). Staphylococcus aureus accessory regulators: expression within biofilms and effect on adhesion. Microbes Infect. 3, 633–637. doi: 10.1016/S1286-4579(01)01418-6
Resch, A., Leicht, S., Saric, M., Pasztor, L., Jakob, A., Gotz, F., et al. (2006). Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6, 1867–1877. doi: 10.1002/pmic.200500531
Resch, A., Rosenstein, R., Nerz, C., and Götz, F. (2005). Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71, 2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005
Rode, T. M., Møretrø, T., Langsrud, S., Langsrud, O., Vogt, G., and Holck, A. (2010). Responses of Staphylococcus aureus exposed to HCl and organic acid stress. Can. J. Microbiol. 56, 777–792. doi: 10.1139/w10-057
Salazar, N., Castiblanco-Valencia, M. M., Silva, L. B. D., Castro, Í. A. D., Monaris, D., Masuda, H. P., et al. (2014). Staphylococcus aureus manganese transport protein C (MntC) is an extracellular matrix- and plasminogen-binding protein. PLoS One 9:e112730. doi: 10.1371/journal.pone.0112730
Savijoki, K., Skogman, M., Fallarero, A., Nyman, T. A., Sukura, A., Vuorela, P., et al. (2016). Penicillin G increases the synthesis of a suicidal marker (CidC) and virulence (HlgBC) proteins in Staphylococcus aureus biofilm cells. Int. J. Med. Microbiol. 306, 69–74. doi: 10.1016/j.ijmm.2015.11.006
Sharma, M., and Beuchat, L. R. (2004). Sensitivity of Escherichia coli O157:H7 to commercially available alkaline cleaners and subsequent resistance to heat and sanitizers. Appl. Environ. Microbiol. 70, 1795–1803. doi: 10.1128/aem.70.3.1795-1803.2004
Siboo, I. R., Chambers, H. F., and Sullam, P. M. (2005). Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 73, 2273–2280. doi: 10.1128/IAI.73.4.2273-2280.2005
Singh, V. K., Carlos, M. R., and Singh, K. (2010). Physiological significance of the peptidoglycan hydrolase, LytM, in Staphylococcus aureus. FEMS Microbiol. Lett. 311, 167–175. doi: 10.1111/j.1574-6968.2010.02087.x
Sobhanifar, S., Worrall, L. J., King, D. T., Wasney, G. A., Baumann, L., Gale, R. T., et al. (2016). Structure and mechanism of Staphylococcus aureus TarS, the wall teichoic acid β-glycosyltransferase involved in methicillin resistance. PLoS Pathog. 12:e1006067. doi: 10.1371/journal.ppat.1006067
Soutourina, O., Poupel, O., Coppée, J.-Y., Danchin, A., Msadek, T., and Martin-Verstraete, I. (2009). CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol. 73, 194–211. doi: 10.1111/j.1365-2958.2009.06760.x
Stapleton, M. R., Horsburgh, M. J., Hayhurst, E. J., Wright, L., Jonsson, I. M., Tarkowski, A., et al. (2007). Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 189, 7316–7325. doi: 10.1128/JB.00734-07
Stoodley, P., Sauer, K., Davies, D. G., and Costerton, J. W. (2002). Biofilms as complex differentiated communities. Ann. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Sun, F., Ji, Q., Jones, M. B., Deng, X., Liang, H., Frank, B., et al. (2012). AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J. Am. Chem. Soc. 134, 305–314. doi: 10.1021/ja2071835
Swapna, G. V., Rossi, P., Montelione, A. F., Benach, J., Yu, B., Abashidze, M., et al. (2012). Three structural representatives of the PF06855 protein domain family from Staphyloccocus aureus and Bacillus subtilis have SAM domain-like folds and different functions. J. Struct. Funct. Genomics 13, 163–170. doi: 10.1007/s10969-012-9134-6
Thoendel, M., Kavanaugh, J. S., Flack, C. E., and Horswill, A. R. (2011). Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151. doi: 10.1021/cr100370n
Tian, X. L., Dong, G., Liu, T., Gomez, Z. A., Wahl, A., Hols, P., et al. (2013). MecA protein acts as a negative regulator of genetic competence in Streptococcus mutans. J. Bacteriol. 195, 5196–5206. doi: 10.1128/JB.00821-13
Trindade, M., Vieira, C., Ferreira, J., Sousa, J., Haddad, J., Farias, L., et al. (2017). Dynamic of biofilm formation on surgical clamps by Staphylococcus aureus and Acinetobacter baumannii strains. Br. J. Med. Med. Res. 19, 1–9. doi: 10.9734/BJMMR/2017/31167
Ueda, S., and Kuwabara, Y. (2007). Susceptibility of biofilm Escherichia coli, Salmonella enteritidis and Staphylococcus aureus to detergents and sanitizers. Biocontrol Sci. 12, 149–153. doi: 10.4265/bio.12.149
Ulrich, M., Bastian, M., Cramton, S. E., Ziegler, K., Pragman, A. A., Bragonzi, A., et al. (2007). The staphylococcal respiratory response regulator SrrAB induces icagene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 65, 1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x
Wang, X., Han, H., Lv, Z., Lin, Z., Shang, Y., Xu, T., et al. (2017). PhoU2 but not PhoU1 as an important regulator of biofilm formation and tolerance to multiple stresses by participating in various fundamental metabolic processes in Staphylococcus epidermidis. J. Bacteriol. 199:e00219-17. doi: 10.1128/JB.00219-17
Wang, Y., Yi, L., Wu, Z., Shao, J., Liu, G., Fan, H., et al. (2012). Comparative proteomic analysis of Streptococcus suis biofilms and planktonic cells that identified biofilm infection-related immunogenic proteins. PLoS One 7:e33371. doi: 10.1371/journal.pone.0033371
Waters, N. R., Samuels, D. J., Behera, R. K., Livny, J., Rhee, K. Y., Sadykov, M. R., et al. (2016). A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 101, 495–514. doi: 10.1111/mmi.13404
Weinrick, B., Dunman, P. M., McAleese, F., Murphy, E., Projan, S. J., Fang, Y., et al. (2004). Effect of mild acid on gene expression in Staphylococcus aureus. J Bacteriol. 186, 8407–8423. doi: 10.1128/JB.186.24.8407-8423.2004
Wu, S., de Lencastre, H., Sali, A., and Tomasz, A. (1996). A phoshoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb. Drug Resist. 2, 277–286. doi: 10.1089/mdr.1996.2.277
Wu, Y., Wu, Y., Zhu, T., Han, H., Liu, H., Xu, T., et al. (2015). Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J. Bacteriol. 197, 459–476. doi: 10.1128/JB.02231-14
Xue, T., You, Y., Hong, D., Sun, H., and Sun, B. (2011). The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect. Immun. 79, 2154–2167. doi: 10.1128/IAI.01180-10
Yarwood, J. M., Bartels, D. J., Volper, E. M., and Greenberg, E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850. doi: 10.1128/jb.186.6.1838-1850.2004
Yarwood, J. M., McCormick, J. K., and Schlievert, P. M. (2001). Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183, 1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001
Zhu, Y., Weiss, E. C., Otto, M., Fey, P. D., Smeltzer, M. S., and Somerville, G. A. (2007). Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 75, 4219–4226. doi: 10.1128/IAI.00509-07
Keywords: MRSA, biofilm, alkaline, acidic, microarray, transcription factors
Citation: Efthimiou G, Tsiamis G, Typas MA and Pappas KM (2019) Transcriptomic Adjustments of Staphylococcus aureus COL (MRSA) Forming Biofilms Under Acidic and Alkaline Conditions. Front. Microbiol. 10:2393. doi: 10.3389/fmicb.2019.02393
Received: 18 July 2019; Accepted: 02 October 2019;
Published: 18 October 2019.
Edited by:
Spyridon Ntougias, Democritus University of Thrace, GreeceReviewed by:
Marina Papadelli, University of Peloponnese, GreeceMichalis D. Omirou, Agricultural Research Institute, Cyprus
Copyright © 2019 Efthimiou, Tsiamis, Typas and Pappas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georgios Efthimiou, Zy5lZnRoaW1pb3VAaHVsbC5hYy51aw==
†Present address: Georgios Efthimiou, Biomedical Sciences, University of Hull, Kingston upon Hull, United Kingdom
 Georgios Efthimiou
Georgios Efthimiou George Tsiamis
George Tsiamis Milton A. Typas
Milton A. Typas Katherine M. Pappas
Katherine M. Pappas