- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2MARA-CABI Joint Laboratory for Bio-Safety, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Bacterial symbionts are omnipresent in insects, particularly aphids, and often exert important effects on the host ecology; however, examples of symbionts that mediate herbivore-plant interactions remain limited. Here, three clones with identical genetic backgrounds were established: a Hamiltonella defensa-free clone, H. defensa-infected clone and H. defensa-cured clone. H. defensa infection was found to increase the fitness of Sitobion miscanthi by increasing the total number of offspring and decreasing the age of first reproduction. Furthermore, gene expression studies and phytohormone measurement showed that feeding by the Hamiltonella-infected clone suppressed the salicylic acid (SA)- and jasmonic acid (JA)-related defense pathways and SA/JA accumulation in wheat plants relative to feeding by the other two clones. Additionally, after feeding by the Hamiltonella-infected clone, the activity levels of the defense-related enzymes polyphenol oxidase (PPO) and peroxidase (POD) in wheat plants were significantly decreased compared with the levels observed after feeding by the other two clones. Taken together, these data reveal for the first time the potential role of H. defensa of S. miscanthi in mediating the anti-plant defense responses of aphids.
Introduction
Virtually all plants in nature protect themselves against a variety of insect species by using different strategies, including constitutive defense and induced defense mechanisms (Hall, 1999). Constitutive defenses, which are effective against generalist herbivores, are based on secondary metabolites of diverse chemical origins (Schoonhoven et al., 2005). In addition to constitutive defenses, which are always present in the plant, the induction of defenses by herbivorous insect attack is mediated mainly by phytohormones such as jasmonic acid (JA) and salicylic acid (SA). The current theory posits that the JA pathway is frequently induced by chewing-biting herbivores and necrotrophic pathogens (Kawazu et al., 2012) and that the SA pathway is primarily induced by piercing-sucking herbivores and biotrophic pathogens (Glazebrook, 2005).
As one of the largest groups of phloem-feeding insects, aphids (Hemiptera: Aphidoidea) are economically important pests that cause enormous agricultural losses worldwide. The induction of SA-related defense pathways by aphid feeding has been demonstrated in many aphid-plant interactions, for example, for the green peach aphid (Myzus persicae) in Arabidopsis (Moran and Thompson, 2001), the greenbug aphid (Schizaphis graminum) in sorghum (Zhu-Salzman et al., 2004) and the potato aphid (Macrosiphum euphorbiae) in tomato (Martinez et al., 2003). Moreover, several genes, such as lipoxygenase (LOX), involved in the JA-related defense pathway are induced by the feeding of the cabbage aphid (Brevicoryne brassicae) on wild cabbage (Li et al., 2016) and the feeding of the soybean aphid (Aphis glycines) on soybean (Selig et al., 2016).
As a result of the long-term coevolution of plants and herbivores, some herbivores have evolved innovative methods of avoiding detection or manipulating plant defenses (Walling, 2010). This evolution has been demonstrated by findings indicating that some herbivores produce effector molecules to suppress plant defenses in response to phloem feeding (Zhang et al., 2013), oral secretions (Consales et al., 2012; Chung et al., 2013) and egg deposition (Bruessow et al., 2010). Recently, insect-associated symbionts, which usually play hidden roles in insect-plant interactions (Frago et al., 2012), have been recognized as potential agents through which insects mediate plant defense responses (Oliver et al., 2014).
Many insects harbor various types of maternally inherited microbial symbionts (Oliver et al., 2014) that provide essential nutrients and/or have important effects on host insect ecology and physiology (Chen et al., 2000; Montllor et al., 2002; Russell and Moran, 2005; Sakurai et al., 2005; Oliver et al., 2010, 2014; Guidolin and Cônsoli, 2017). Additionally, emerging evidence has revealed that insect-associated symbionts may suppress plant defense responses (Barr et al., 2010; Su et al., 2016) and detoxify plant secondary metabolites and even chemical pesticides (Oliver et al., 2010; Berasategui et al., 2016; Cheng et al., 2017). Therefore, these symbionts may have potential functions in manipulating the “arms race” between insects and host plants. The strategies through which herbivore-associated microbial symbionts manipulate the antagonistic crosstalk between insects and plant defense responses require greater attention.
The grain aphid, Sitobion miscanthi Takahashi, is one of the most widespread wheat aphids in China and frequently harbors several secondary symbionts (S-symbionts). Hamiltonella defensa, a well-studied S-symbiont in Acyrthosiphon pisum that confers conditional adaptive advantages to its host by protecting the insect host against natural enemies (Oliver et al., 2003, 2005; Ferrari et al., 2004), was also detected in S. miscanthi in China (Li et al., 2014). Sporadic reports have noted that insect symbionts mediate plant defense responses (Barr et al., 2010; Chung et al., 2013; Su et al., 2016), but there is still relatively little information available on a clear role for H. defensa in regulating aphid-plant interactions, especially in the wheat aphid S. miscanthi.
In our previous study, Hamiltonella-free and Hamiltonella-infected aphid clones were established by microinjection, but these clones also carried co-infections with two additional secondary symbionts, Regella insecticola and Spiroplasma (Li et al., 2018). To rule out the effects of other symbionts on follow-up experiments, an additional antibiotic treatment was performed to construct an aphid clone without S-symbionts other than H. defensa. Subsequently, natural Hamiltonella-free, Hamiltonella-infected and Hamiltonella-cured aphid clones without any other S-symbionts were also established through an improved antibiotic treatment. To identify the distribution of H. defensa in aphids, we examined the position of H. defensa and Buchnera aphidicola in the embryo of S. miscanthi using fluorescence in situ hybridization (FISH). Ecological fitness indices were compared among the Hamiltonella-free, Hamiltonella-infected and Hamiltonella-cured clones. Finally, the effects of H. defensa on the expression of JA- and SA-related defense pathway genes, SA and JA accumulation and the activity of defense-related enzymes in plants were investigated. Our results show that H. defensa mediated the anti-plant defense responses by suppressing the expression of SA and JA-related defense pathway genes and SA and JA production in plants and decreasing defense-related enzyme activity in wheat plants, all of which resulted in improved aphid fitness.
Materials and Methods
Experimental Aphid Clones
The following clones of S. miscanthi established by a single female were used in this study: a Hamiltonella-free clone (DZ) was collected from a Dezhou wheat field in Shandong Province, China, and a natural Hamiltonella-infected clone (YX) was collected from a Yuxi wheat field in Yunnan Province, China, according to our previous studies (Li et al., 2018).
H. defensa Artificial Infection and Antibiotic Elimination
The injection of H. defensa and antibiotic elimination was performed according to our previous work (Li et al., 2018). Then, DZ and DZ-HT contained no known secondary symbionts and DZ-H contained only H. defensa were established after an additional antibiotic treatment experiment (Supplementary Method “Details About Microinjection and Antibiotic Treatment”). The infected and cured clones were not used in any experiment until at least 10 generations had passed to eliminate any negative effects associated with mechanical damage and the antibiotic treatment (Koga et al., 2003), and aphids from the infected and cured clones were retested by PCR before the next step in the experiment assay.
Gene Amplification, Sequencing, and Phylogenetic Analysis
To ensure that the newly infected and cured aphid clones were produced by microinjection and antibiotic treatment rather than contamination and to verify whether the aphid genotype of the new H. defensa-acquired S. miscanthi DZ-H clone was contaminated, the mitochondrial cytochrome oxidase I (COI) sequences of the S. miscanthi native Hamiltonella-infected YX clone, Hamiltonella-free DZ clone, artificial Hamiltonella-infected DZ-H clone and Hamiltonella-cured DZ-HT clone were amplified and used to build a maximum likelihood tree with Kimura’s two-parameter distance and 1000 bootstrap resampling iterations with MEGA software, version 7.0.26 (Kumar et al., 2001). The nucleotide sequence accession numbers of the COI genes from the S. miscanthi YX, DZ, DZ-H, and DZ-HT clones were MH805861, MH805862, MH805863, and MH805864, respectively.
To verify the consistent identity of the H. defensa strain during artificial infection, we adapted the multilocus approach developed in a previous study (Henry et al., 2013). Two H. defensa housekeeping genes (dnaA and recJ) and one APSE (a bacteriophage produced by H. defensa) locus, P3, were selected and analyzed in the YX and DZ-H clones. A phylogenetic tree was constructed by using the maximum likelihood method in MEGA. The nucleotide sequences of the dnaA, recJ, and P3 genes from the H. defensa donor S. miscanthi YX clone described in this paper have been deposited in GenBank under the accession numbers MH884762, MH823746, and MH910617. The primer information is listed in Supplementary Table S1. The cycling conditions were 94°C for 4 min; followed by 35 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 1 min; and 4°C for the final elongation. The reaction products were analyzed with a model 3500 ABI PRISM DNA sequencer (Perkin-Elmer, New York, NY, United States).
Relative Changes in B. aphidicola Abundance by Quantitative PCR
After the clones were reared for 10 generations, DNA was extracted from 30 mixed first-instar nymphs of the DZ, DZ-H, and DZ-HT clones, and the B. aphidicola relative abundance was quantified by real-time PCR. All data were compared with the DZ clone as a control. The primers used in this study are based on our previous work (Li et al., 2018). Three biological replicates were performed.
Fluorescence in situ Hybridization (FISH)
Fluorescence in situ hybridization (FISH) was performed according to a previous study (Sakurai et al., 2005) with some modifications [Supplementary Method “Details About Fluorescence in situ Hybridization (FISH)”]. The probe ApisBuch-Cy5 (5′-Cy5-CCTCTTTTGGGTAGATCC-3′) targeted the 16S rRNA of Buchnera spp. (Kumar et al., 2001), and the probe ApisHami-Cy3 (5′-Cy3-CCAGAT TCCCAGACTTTACTCA-3′) targeted the 16S rRNA of H. defensa. To confirm the specificity of the detection, a series of control experiments were conducted as previously described (Qian et al., 2018): Hamiltonella-free aphid embryos and Hamiltonella-cured aphid embryos were probed with the H. defensa 16S rRNA probe to confirm the result of H. defensa hybridization.
Fitness Measurements
Thirty nymphs from the DZ, DZ-H, and DZ-HT clones were selected at random and individually placed in petri dishes containing wheat seedlings whose roots were inserted into water in 1.5 ml tubes and kept at 20°C under a long-day (16 h) light cycle. Fitness indices, including total number of offspring and age of first reproduction, were monitored daily until all nymphs had completed their whole life cycle. We measured the offspring of five newly emerged adults collected from each clone and performed 6 replications.
Aphid Infestation Treatments
At the two-leaf stage, 30 newly molted adults of the DZ, DZ-H, and DZ-HT clones were transferred to the first leaf (the oldest leaf) of wheat, and the movement of the aphids was restricted by a plastic cage (2.7 cm × 2.7 cm × 2.7 cm) clipped onto the leaf to prevent their escape. Empty cages were clipped onto uninfested plants. Each pot contained one wheat plant and was kept in the culture room at 20 ± 1°C with 75% relative humidity and a light: dark photoperiod of 16:8 (L: D) hours. After 30 min, all aphids had begun settling and feeding, and this time was recorded as 0 h. A 100-mg quantity of plant tissue from each clip-caged leaf was harvested after 24, 48, or 72 h of aphid feeding for RNA extraction and assessed for the induction of genes associated with plant defense, as described below. In addition, plant tissues not subjected to aphid feeding were harvested at the different time points as controls. All treatments had three biological replicates.
Wheat RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Wheat leaf tissues were ground in liquid nitrogen. Total RNA was extracted with an RNeasy Plus kit following the manufacturer’s protocol. RNA concentration was evaluated using a NanoDrop 1000 spectrophotometer, and then 1 μg of RNA was used to synthesize first-stand cDNA using the EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) according to the manufacturer’s protocol. Each cDNA sample was generated in triplicate from each biological replicate. The genes PR-1, PAL and β-1,3-GA, which encode pathogenesis-related protein 1, phenylalanine ammonia lyase, and beta-1,3-glucanase, respectively, were selected as SA-marker genes, and the genes AOS, LOX and FAD, which encode allene oxide synthase, lipoxygenase and Ω-3 fatty acid desaturase, respectively, were selected as JA marker genes because they are strongly induced in response to insects and involved in the synthesis of these two plant hormones (Zhang et al., 2013; Züst and Agrawal, 2016). To quantify the PR-1, PAL, β-1,3-GA, AOS, LOX, and FAD transcript levels, qRT-PCR was performed. All treatments had three biological replicates, and each replicate consisted of three technical replicates. The primers for the SA and JA marker genes are shown in Supplementary Table S1. The abundances of the SA and JA marker genes were normalized to that of the plant housekeeping gene β-actin to obtain the relative abundances. All data were compared with the control to obtain the relative value.
Quantification of SA and JA
Wheat leaves were subjected to feeding by Hamiltonella-free, Hamiltonella-infected and Hamiltonella-cured aphids as described above. Ten leaves were used for each treatment, and leaf tissues were harvested 24, 48, and 72 h after placing aphids from three individual plants were pooled as one biological replicate per clone and time point. Three biological replicates were performed. SA and JA were extracted and measured using GC/MS as described previously (Tooker et al., 2008).
Enzyme Activity Assays
The methods used to determine the wheat plant defense-related enzyme activities were largely the same as previously reported methods (Lang et al., 2017). The activities of POD and PPO were evaluated using their respective diagnostic kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). POD activity was determined by a spectrophotometer (UV-2000, UNICO, Shanghai, China) following the change in absorption at 420 nm due to guaiacol oxidation (Maehly and Chance, 1995). The PPO activity was assayed according to the methods of a previous study (Cai et al., 2010). Each test had three biological replicates.
Statistical Analysis
Treatment effects were assessed with an analysis of variance (ANOVA) or Student’s t-test using IBM SPSS statistics version 21 (ver. 21, SPSS, Inc., Chicago, IL, United States). For assays in which two or more treatments were compared, Tukey’s honestly significant difference (HSD) multiple comparison test (P < 0.05) was used to determine whether the treatments were significantly different.
Results
Establishment of Aphid Clones
Our previous work revealed that the YX clone was infected with three S-symbionts—R. insecticola, H. defensa and Spiroplasma sp.—and that the DZ clone was infected with R. insecticola and Spiroplasma sp. With the improvement of previous manipulation methods, a Hamiltonella-infected clone (DZ-H) and a new Hamiltonella-cured clone (DZ-HT), which had an identical genetic background to the DZ clone, were constructed. To rule out the effects of other S-symbionts on the following experiments, an additional antibiotic treatment was performed to eliminate S-symbionts other than H. defensa in the DZ, DZ-H, and DZ-HT clones. PCR detection revealed that H. defensa was completely cured in the DZ-HT clone after antibiotic treatment (Supplementary Figure S1). To rule out contamination of the new Hamiltonella-infected DZ-H clone and Hamiltonella-cured DZ-HT by the native Hamiltonella-infected YX clone and Hamiltonella-free DZ clone, the mitochondrial cytochrome oxidase I (COI) sequences of the YX, DZ, DZ-H, and DZ-HT clones were aligned with MEGA software, and a maximum likelihood tree was built (Figure 1A). The results showed that the sequence of the COI mitochondrial gene was strictly identical among the DZ, DZ-H, and DZ-HT clones but distinct in the YX clone, indicating that the newly infected and cured H. defensa aphid clones were derived from microinjection and antibiotic treatment.
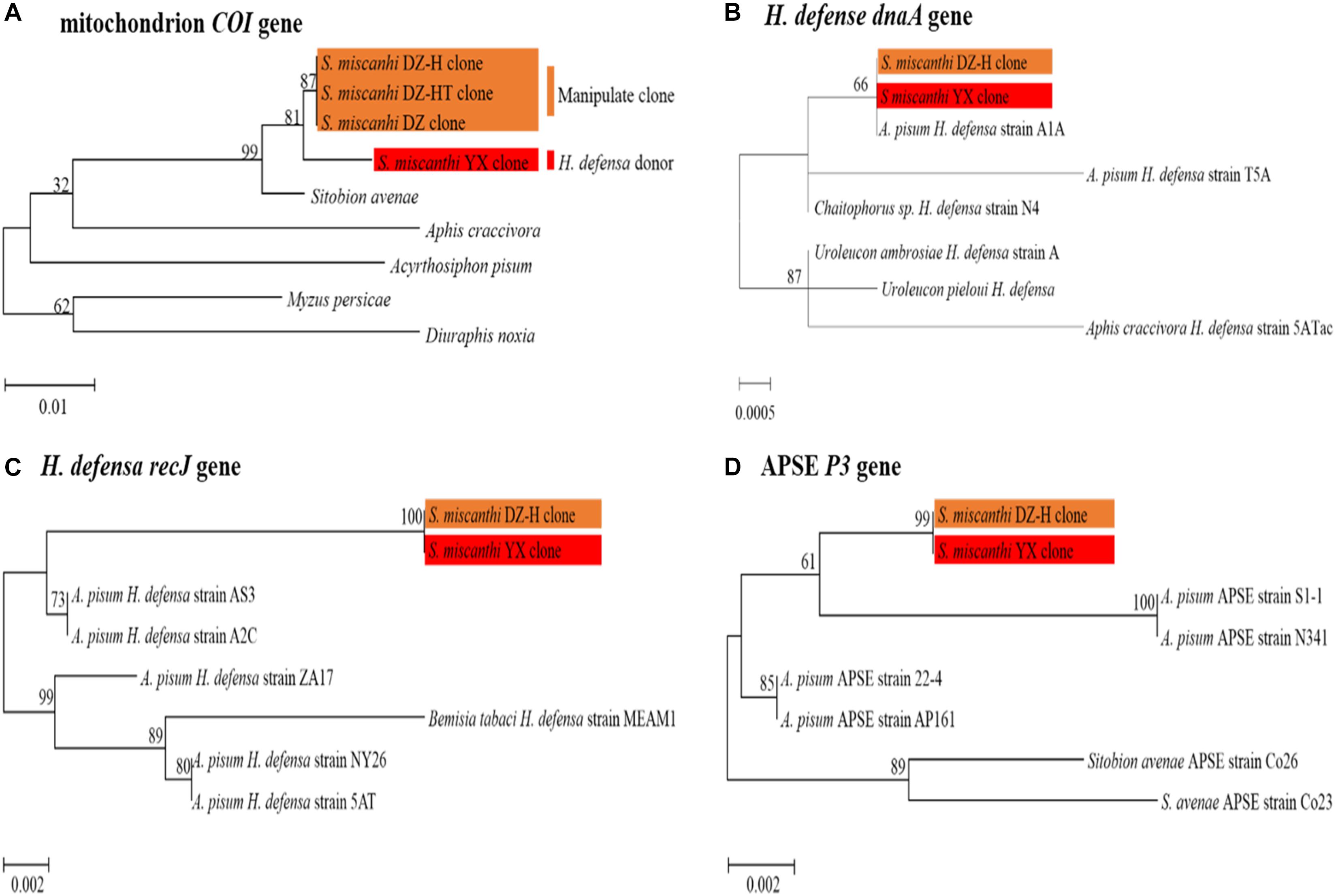
Figure 1. Phylogenetic trees were constructed with molecular evidence. (A) Phylogenetic analysis of different clones based on the COI gene. (B–D) The maximum likelihood (ML) tree was constructed based on H. defensa and APSE multilocus gene sequences. The orange frame indicates the manipulated clones, and the red frame indicates the H. defensa donor Sitobion miscanthi YX clone. The bar indicates the estimated number of substitutions per site.
To verify the consistent identity of the H. defensa strain during microinjection, a phylogenetic tree was constructed by using two H. defensa housekeeping genes (dnaA and recJ) and one APSE (a bacteriophage secreted from H. defensa) gene (P3). The results showed that all of the orthologous genes were completely identical in the native Hamiltonella-infected S. miscanthi YX clone and artificial Hamiltonella-infected DZ-H clone (Figures 1B–D). These findings indicated that the H. defensa strain remained consistent throughout the microinjection process.
Relative Abundance of B. aphidicola in the DZ, DZ-H, and DZ-HT Clones
Quantitative PCR results showed that at the first instar stage, the relative abundance of B. aphidicola in DZ, DZ-H, and DZ-HT were not significantly different after the antibiotic treatment (Figure 2), however, the abundance of H. defensa in the DZ-HT clone could not be detected after antibiotic treatment, as was the case in the DZ clone. Moreover, the other S-symbionts were also not detected after an additional antibiotic treatment in all clones (data not shown). These results show that microinjection and a moderate concentration of antibiotics could specifically cure targeted symbionts without affecting other symbionts.
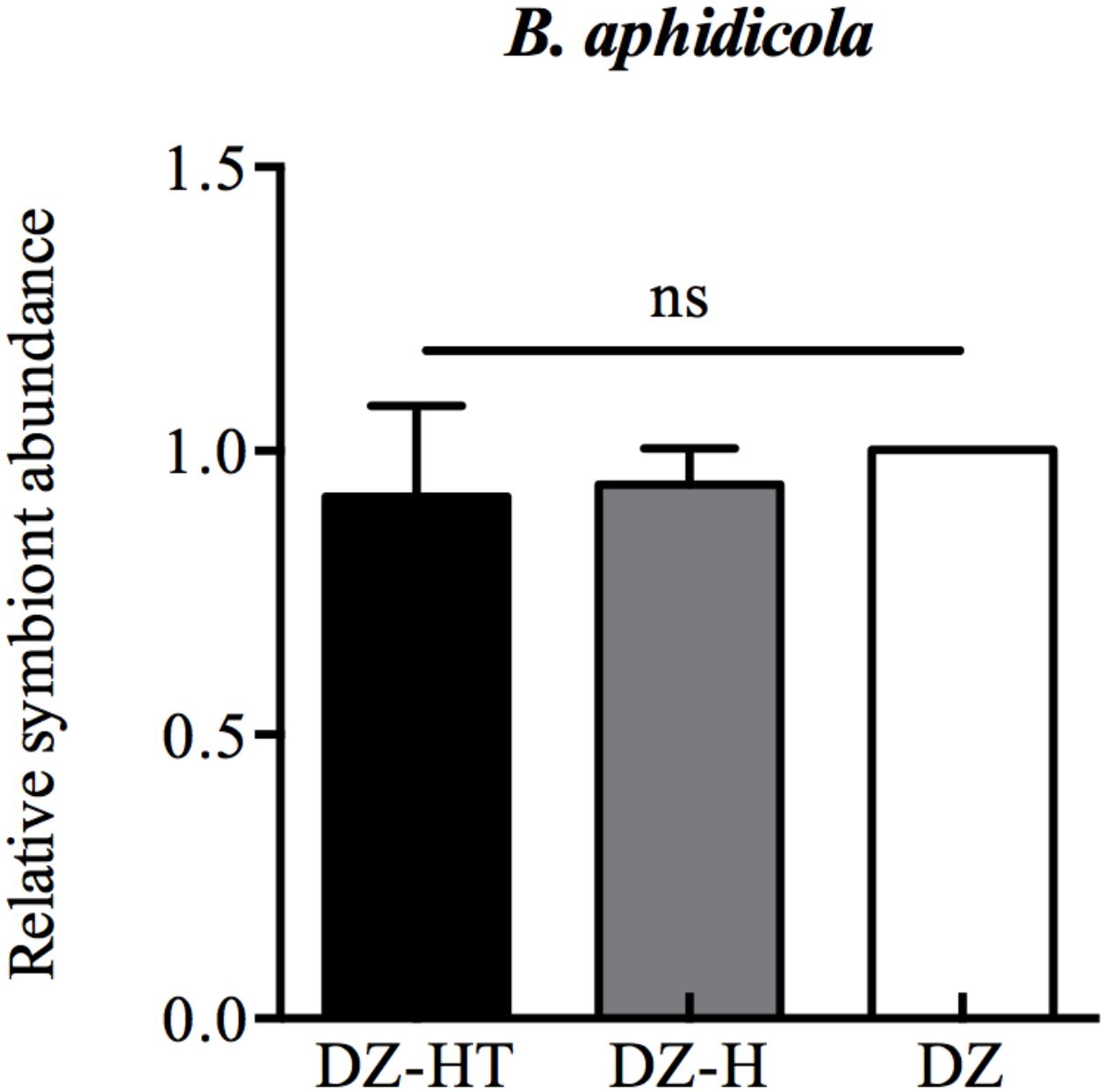
Figure 2. Relative changes in B. aphidicola abundance after microinjection and antibiotic treatment designed to specifically infect and eliminate H. defensa. qPCR analysis of DNA extracted from microinjection-treated aphids (DZ-H clone), antibiotic-treated aphids (DZ-HT clone), and Hamiltonella-free aphids (DZ clone). Microinjection and antibiotic treatments did not have an effect on the abundance of B. aphidicola. Bars represent the standard errors of the means, different letters indicate significant differences based on one-way ANOVA followed by Tukey’s HSD multiple comparison test (P < 0.01), and ns indicates no significant difference.
In situ Hybridization of H. defensa and B. aphidicola
Whole-mount FISH revealed the cellular localizations of H. defensa and B. aphidicola (Figure 3). In aphid embryos of the Hamiltonella-infected DZ-H clone, strong signals of the Hamiltonella-specific probe (red) were found in sheath cells, secondary mycetocytes and hemolymph (Figures 3A–E). Interestingly, the sheath cells and secondary bacteriocytes were located on the periphery of bacteriocytes, where B. aphidicola (green) was harbored. Signals of B. aphidicola, but not H. defensa, were detected in the Hamiltonella-free clone DZ and the Hamiltonella-cured clone DZ-HT (Figures 3F,G), and the no-probe control confirmed the specificity of the detected signals (data not shown).
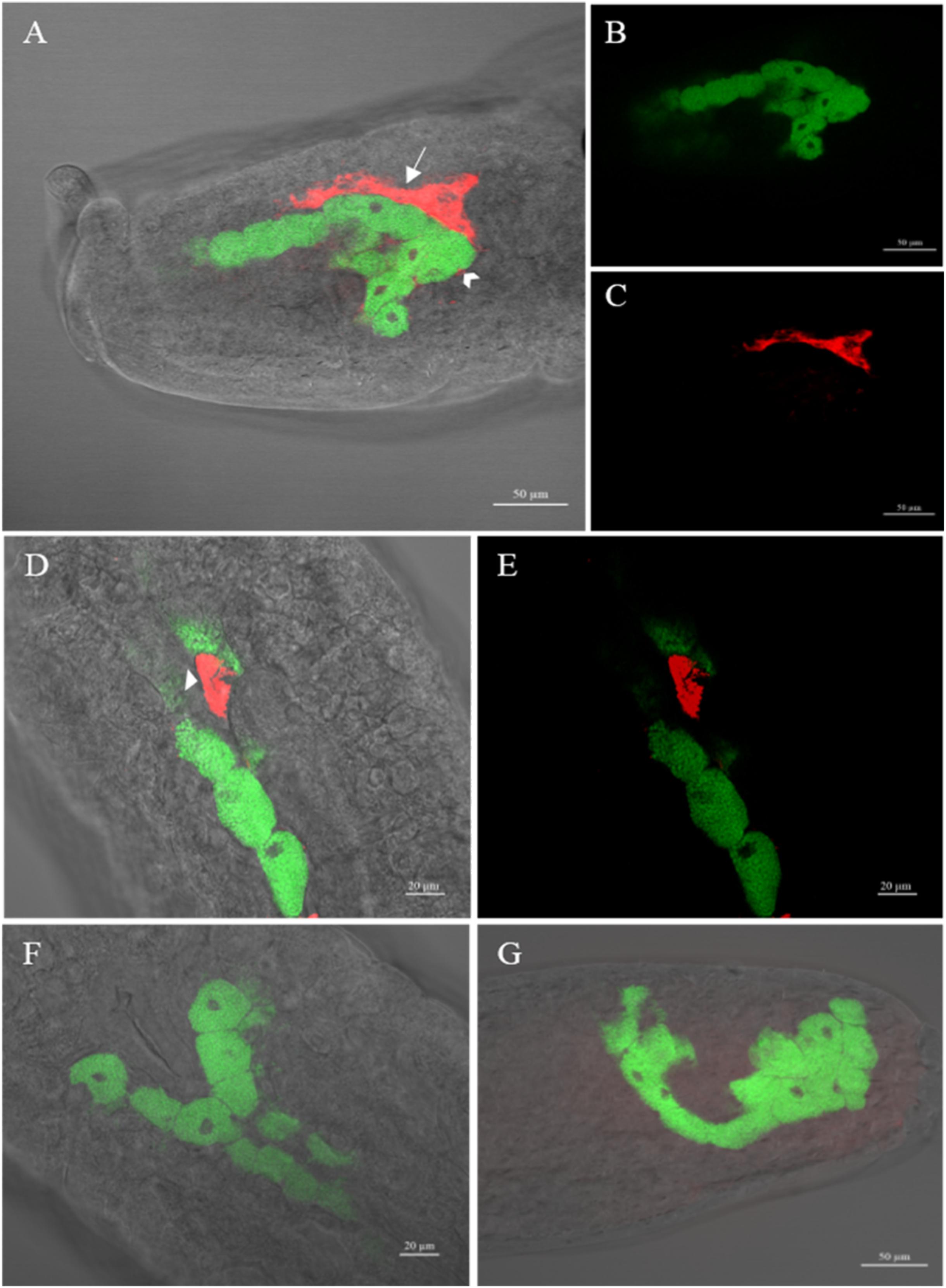
Figure 3. Whole-mount in situ hybridization of aphid embryos targeting H. defensa (red) and B. aphidicola (green). (A–E) Different embryos of the artificial Hamiltonella-infected aphid clone DZ-H in which sheath cells, secondary bacteriocytes and hemolymph harboring H. defensa are seen in addition to a number of bacteriocytes harboring B. aphidicola. (F,G) An embryo of the Hamiltonella-free aphid clone DZ and Hamiltonella-cured aphid clone DZ-HT in which H. defensa was not detected but B. aphidicola was detected. Swallowtail, sheath cell; arrowhead, secondary bacteriocyte; arrow, hemolymph.
Fitness Measurements
Two aphid demographic parameters—the total number of offspring and the age of first reproduction—were compared among the Hamiltonella-free clone (DZ), Hamiltonella-infected clone (DZ-H) and Hamiltonella-cured clone (DZ-HT) with identical genetic backgrounds (Figure 4). The total number of offspring differed significantly among the three clones (F2,15 = 11.816, P < 0.001). With an average number of 112.3 offspring it was significantly higher in the DZ-H clone than in DZ (85.3) and in DZ-HT (82.2) (Figure 4A). Meanwhile, the aphid age of first reproduction also exhibited significant variation among the three clones (F2,87 = 13.101, P < 0.001). With an average age of first reproduction of 7.6 days it was significantly lower in DZ-H clone than in DZ (8.3 days) and DZ-HT (8.6 days) (Figure 4B).
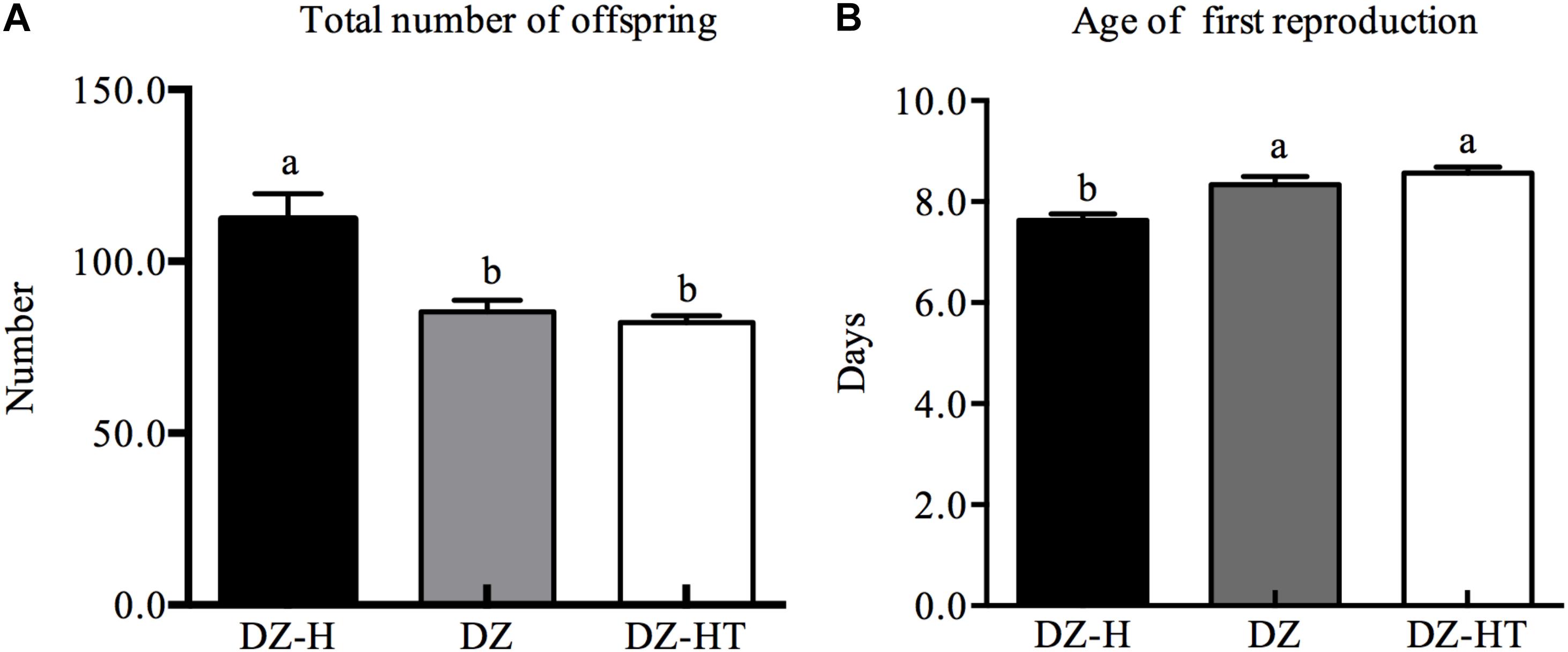
Figure 4. Comparison of aphid fitness measurements among Hamiltonella-infected (DZ-H), Hamiltonella-free (DZ) and Hamiltonella-cured (DZ-HT) S. miscanthi clones. (A) Total number of offspring. (B) Age of first reproduction. Bars represent the standard errors of the means, and different letters above the bars indicate significant differences based on one-way ANOVA followed by Tukey’s HSD multiple comparison test (P < 0.05).
Effect of H. defensa on the Plant SA and JA Defense Pathways
To investigate whether H. defensa suppresses plant defenses, we measured SA/JA defense marker gene expression in wheat plants at different time points after feeding by the DZ, DZ-H, and DZ-HT clones. Some of the key genes involved in the SA and JA defense pathways were found to be differentially expressed in wheat leaves (Figure 5). Compared with the levels observed after feeding by the DZ and DZ-HT clones, the relative levels of expression of the SA-related defense genes PR-1, PAL, and β-1,3-GA in wheat after feeding by the DZ-H clone were not significantly different at 24 h (PR-1: F2,6 = 3.237, P = 0.111; PAL: F2,6 = 1.810, P = 0.243; β-1,3-GA: F2,6 = 0.026, P = 0.975) but were exhibited significant variation at other time points. The results in Figures 5A–C show that the SA-defense-related gene levels were significantly lower in the leaves feeding by DZ-H clone than DZ and DZ-HT at 48 and 72 h (PR-1: F2,6 = 67.541, P < 0.001, and F2,6 = 34.385, P = 0.001, for 48 and 72 h, respectively; PAL: F2,6 = 14.608, P = 0.005, and F2,6 = 15.519, P = 0.004, for 48 and 72 h, respectively; β-1,3-GA: F2,6 = 37.899, P < 0.001, and F2,6 = 51.814, P < 0.001, for 48 and 72 h, respectively).
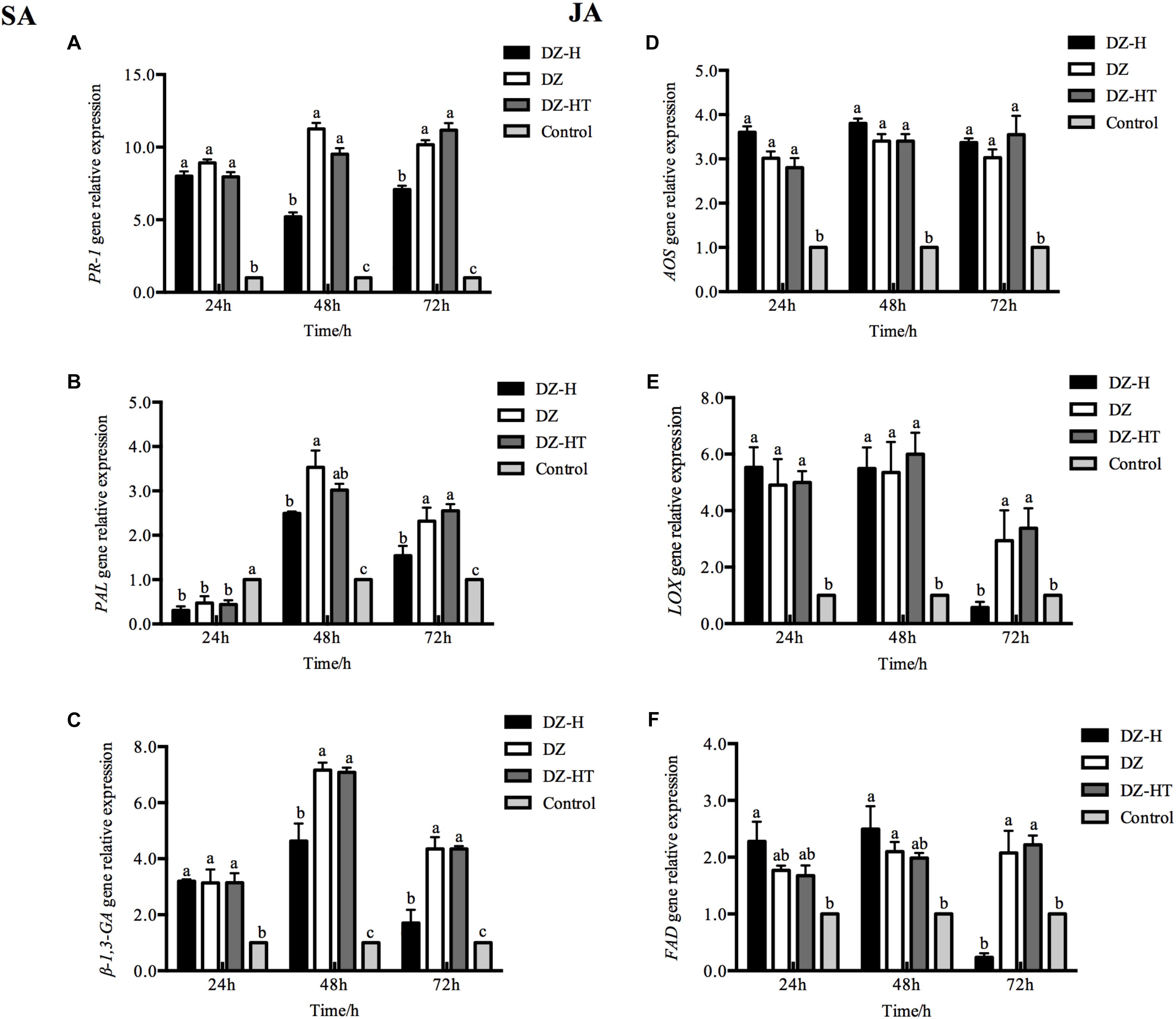
Figure 5. Effect of H. defensa infection on the induced plant SA and JA defense pathways. (A–C) Expression levels of SA-regulated genes in plants infested by aphids with H. defensa infection. (D–F) Expression levels of JA-regulated genes in plants infested by aphids with H. defensa infection. Bars represent the standard errors of the means, and different letters above the bars indicate significant differences based on one-way ANOVA followed by Tukey’s HSD multiple comparison test (P < 0.05).
The relative levels of expression of some JA-related defense genes differed from those of SA-related defense genes. Compared with the levels observed after feeding by the DZ and DZ-HT clones, the relative gene expression levels of LOX and FAD were not significantly different at 24 and 48 h (LOX: F2,6 = 0.680, P = 0.542, and F2,6 = 0.449, P = 0.658, for 24 and 48 h, respectively; FAD: F2,6 = 1.980, P = 0.219, and F2,6 = 1.064, P = 0.402, for 24 and 48 h, respectively) but differed significantly at 72 h after feeding by the DZ-H clone. The results in Figures 5D–F show that the JA-defense-related gene levels were significantly lower in the leaves feeding by DZ-H clone than DZ and DZ-HT at 72 h (LOX: F2,6 = 12.174, P = 0.008; FAD: F2,6 = 19.762, P = 0.002). However, the relative gene expression level of AOS in wheat plants was not exhibit significant variation at any of the monitored times after feeding by the DZ, DZ-H, and DZ-HT clones. Both the plant SA- and JA-related defense genes were upregulated after feeding by the DZ, DZ-H, and DZ-HT clones; however, the upregulated expression levels of the relative genes were significantly lower after feeding by the Hamiltonella-infected clone than by the Hamiltonella-free clone and Hamiltonella-cured clone. Therefore, these results indicate that the presence of H. defensa in aphids may play a role in suppressing plant defense responses.
Effect of H. defensa on Wheat SA and JA Accumulation
To determine if an antagonistic interaction between the SA and JA signaling pathways is mediated by H. defensa, we measured endogenous SA and JA levels in wheat plants at different time points after infestation and feeding by the DZ, DZ-H, and DZ-HT clones. Compared to the DZ and DZ-HT clones, the SA concentrations in wheat subjected to feeding by the DZ-H clone were not significantly different at 24 h (F2,6 = 1.070, P = 0.401) but differed significantly at 48 and 72 h. With an average concentration of 91.1 and 105.3 ng/g it was significantly lower in DZ-H clone than in DZ (155.7 and 153.4 ng/g) and DZ-HT (149.8 and 158.3 ng/g) (Figure 6A) (F2,6 = 56.653, P < 0.001 and F2,6 = 61.863, P < 0.001, for 48 and 72 h, respectively) (Figure 6A). However, the JA concentrations in wheat fed on by the DZ-H clone for 72 h was 105.3 ng/g, which is significantly lower than the wheat after being fed on by clones DZ (153.4 ng/g) or DZ-HT (158.3 ng/g) (F2,6 = 22.617, P = 0.002) (Figure 6B).
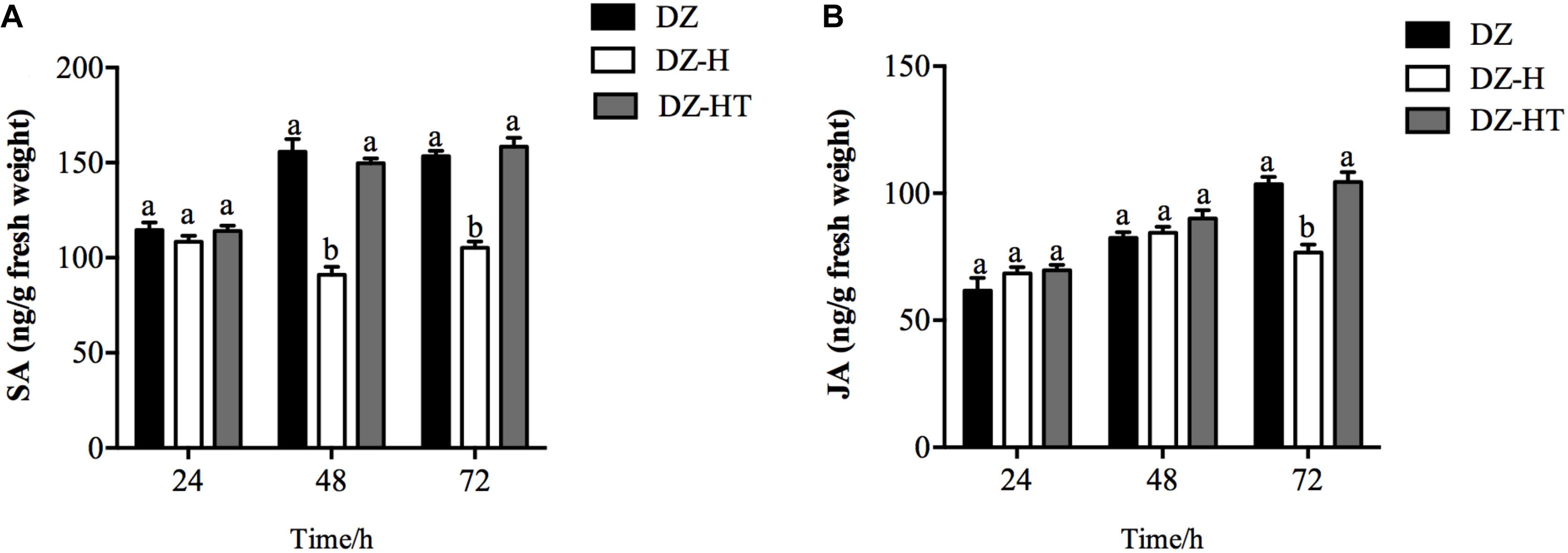
Figure 6. Salicylic acid and JA accumulation in plants infested by aphids with or without H. defensa infection. SA level (A) and JA level (B) were assessed in wheat plants after different periods of infestation with the DZ, DZ-H, and DZ-HT clones. Bars represent standard errors of the means, and different letters above the bars indicate significant differences based on a one-way ANOVA followed by Tukey’s HSD multiple comparison test (P < 0.05).
Effect of H. defensa on Wheat Defense-Related Enzyme Activity
To investigate the mechanism by which H. defensa improves the fitness of S. miscanthi for wheat plants, we measured defense-related enzyme (PPO and POD) activity in plants (Figure 7). After the plants were fed on by the DZ-H clone for 24 and 48 h, the activity of PPO in the plants was 12.7 and 23.1 U/mg, respectively, which were significantly lower than the activity after being fed on by the clones DZ (60.7 and 57.7 U/mg) or DZ-HT (47.3 and 54.7 U/mg) (F2,6 = 44.723, P < 0.001, and F2,6 = 19.093, P = 0.003, for 24 and 48 h, respectively). The activity of POD in plants fed on by the DZ-H clone for 48 h was 185.5 U/mg, which is significantly lower than the activity after being fed on by clones DZ (286.2 U/mg) or DZ-HT (277.5 U/mg) (F2,6 = 33.394, P = 0.001), but there was no significant difference in POD activity among the plants fed on by different clones after 24 h of feeding (F2,6 = 0.594, P = 0.582) (Figures 7A,B).
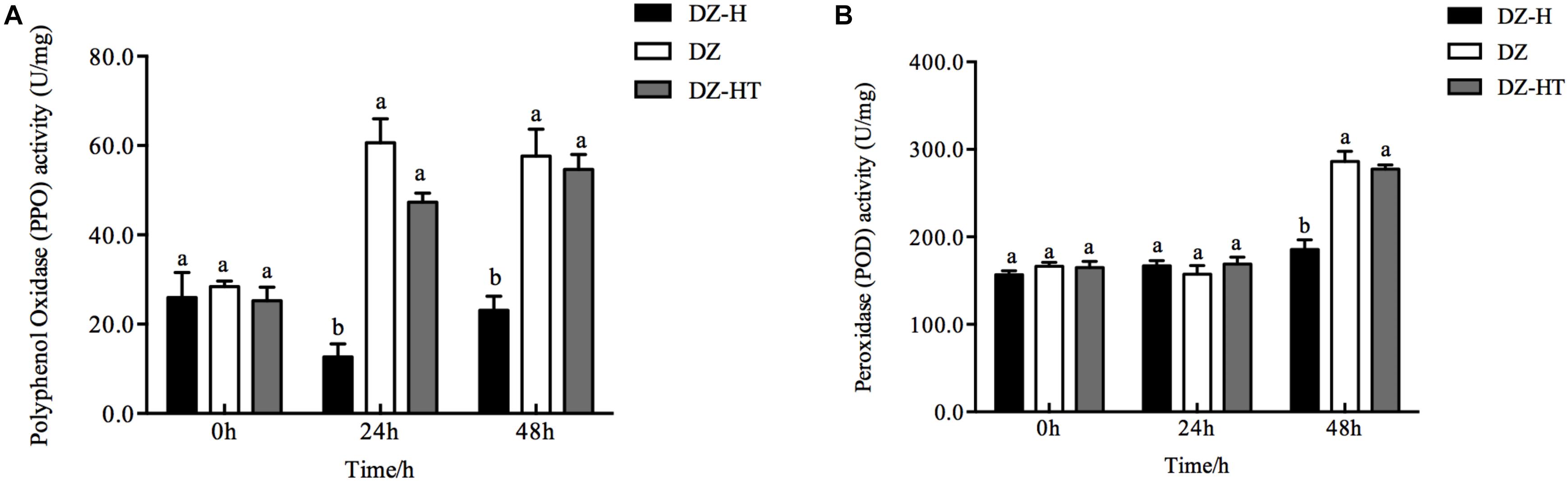
Figure 7. Effect of H. defensa on wheat defense-related enzyme activity. Activities of PPO (A) and POD (B) were assessed in wheat plants after different periods of infestation with the DZ, DZ-H, and DZ-HT clones. Bars represent standard errors of the means, and different letters above the bars indicate significant differences based on a one-way ANOVA followed by Tukey’s HSD multiple comparison test (P < 0.05).
Discussion
Insect symbionts are usually hidden players in insect-plant interactions (Frago et al., 2012). Numerous studies have shown that microbial symbionts play important roles in insect survival and confer conditional adaptive advantages, including providing nutrition, protecting the host insect against natural enemies (Oliver et al., 2003, 2005, 2010), and detoxifying toxins (Cheng et al., 2017). In certain insects, symbiotic viruses suppress the host insect defense reaction (Asgari and Johnson, 2000). However, very little is known about how the endosymbionts of herbivores may mediate host plant defenses. Here, we illustrated strategies mediated by aphid symbionts against plant defense responses.
In our previous work, we established a Hamiltonella-infected clone and a Hamiltonella-reduced clone (Li et al., 2018) and to rule out the effects of remnant H. defensa and other S-symbionts, an improved method of antibiotic treatment was performed to completely eliminate H. defensa and other S-symbionts without affecting the P-symbiont B. aphidicola. In this study, the Hamiltonella-infected clone was used, and a new Hamiltonella-cured clone that presented a consistent relative abundance of B. aphidicola was established. Then, H. defensa and B. aphidicola were identified in the aphid embryos by FISH. Notably, strong signals of H. defensa in the Hamiltonella-infected clone DZ-H were found in sheath cells, secondary bacteriocytes and hemolymph, which were located close to the bacteriocytes that contained B. aphidicola. This observation in artificial Hamiltonella-infected DZ-H clones was similar to our previous study that used the natural Hamiltonella-infected YX clone (Qian et al., 2018), indicating the diverse distribution of H. defensa in aphids and the lack of connection between infection methods and distribution. The subsequent fitness measurement results showed that infection by H. defensa significantly increased the fitness of S. miscanthi, as evidenced by the greater total number of offspring and lower age of first reproduction with H. defensa infection. This result was consistent with our previous work (Li et al., 2018), and complete elimination of H. defensa caused a significant decrease in the total number of offspring, even more so than clones with decreased relative abundance of H. defensa. A prior report indicated that a close relationship with symbionts can be costly for the host insect, negatively affecting insect development (Oliver et al., 2006; Vorburger and Gouskov, 2011; Polin et al., 2014). Interestingly, in our study, the fitness of S. miscanthi increased after infecting with H. defensa. Therefore, we presume that these direct costs may be caused by a trade-off between allocating resources to symbiosis and improving the adaptive ability of the host insect. These results indicate that the change in fitness caused by H. defensa infection may be due to the specific strains of H. defensa and their host aphid species.
Plants initiate phytohormone biosynthesis and antiherbivore defense responses when attacked by herbivores. Therefore, herbivorous insects have evolved strategies to circumvent plant defense responses; one such strategy is to inhibit the core gene expression of plant defense-related pathways and phytohormone production. Although it has been reported that herbivorous insects overcome plant defenses (Musser et al., 2010), only recently emerging evidence has indicated that the symbionts harbored in host insects mediate plant defense pathways. In maize roots, larvae of the western corn rootworm Diabrotica virgifera virgifera influence the downregulation of JA defense pathway genes via infection with Wolbachia sp. (Barr et al., 2010). In tomato, larvae of the potato beetle Leptinotarsa decemlineata inhibit JA-responsive gene expression and JA accumulation, but this inhibition was not observed when the insects were treated with antibiotics (Chung et al., 2013). Interestingly, a recent study has shown that an indirect suppression response mediated by H. defensa occurs in pea aphid through attenuating the yield of host plant volatiles (Frago et al., 2017). Moreover, in whitefly, infection with H. defensa resulted in suppression of the plant JA defense response (Su et al., 2016); however, in the present study, we found that the presence of H. defensa in S. miscanthi significantly decreased the expression of the SA-responsive antiherbivore genes from 48 to 72 h after feeding but suppressed only some JA-responsive genes at 72 h (Figure 5). Meanwhile, SA and JA accumulation also displayed similar tendencies to the results of gene expression measurement. Therefore, in our study, SA is important in establishing resistance early in the infestation process, and JA is important in later facilitation of resistance. It has been well-documented that the two hormones SA and JA are natural antagonists, most likely as part of a plant’s strategy to fine-tune its defenses (Thaler et al., 2012). If one pathway is genetically manipulated, hormonal “crosstalk” can be measured in the other (Züst and Agrawal, 2016). It has been reported that the different hormone signaling pathways are to some extent activated during green peach aphid (Myzus persicae) infestation, and the SA signaling pathway is the predominant one (Rodriguez et al., 2014), while JA-responsive genes are repressed (Kerchev et al., 2013). However, the application of JA and SA induction treatments in tomato resulted in increased aphid resistance, suggesting a role for both pathways in the activation of defenses against aphids (Cooper et al., 2004). This result also suggests that the activation of SA signaling by aphid feeding does not necessarily suppress JA-related defenses. It is likely that, in plant-aphid interactions, the JA and SA defense pathways may function antagonistically or synergistically, depending on the timing, level and interaction between the host plant and aphid species (Pieterse et al., 2009). Therefore, in our study, this antagonism and synergism may be the reason that the inhibition of JA-responsive gene expression and JA accumulation by H. defensa infection was less pronounced and slower than that of SA-responsive gene expression and accumulation. Plant attack by herbivores is associated with oxidative damage at the cellular level through the accumulation of reactive oxygen species (ROS) (Kariola et al., 2005). Several protective enzymes, such as POD and PPO, are involved in ROS detoxification. Therefore, enhancement of the activities of these protective enzymes is one of the most essential elements of plant defense responses (Bednarski et al., 2013). Moreover, the induction of plant defense responses, such as the SA/JA defense pathway, can be influenced by ROS accumulation (Baker et al., 1997). In our study, the activity of the defense-related enzymes PPO and POD in plants was decreased after feeding by the Hamiltonella-infected clone, indicating that H. defensa can help aphids overcome the host plant defense response in some ways. The result was similar to that of a previous study in photo beetle larvae (Chung et al., 2013).
Conclusion
Insect symbionts are newly identified participants that mediate plant-insect interactions during their long-term coevolution and play important roles in the defense and antidefense responses of plants and insects, respectively. Although we did not investigate in this study whether this defense suppression was induced by H. defensa or proteins and other molecules originating from H. defensa invasion in aphid oral secretions, our previous work has indicated that H. defensa can be detected in the aphid stylet and horizontally transmitted into wheat plants (Qian et al., 2018). Therefore, in combination with our previous work, this study has revealed antidefense strategies in aphids through H. defensa infection in response to the plant defense response. Notably, in our study, we verified the function of a specific H. defensa genotype in inhibiting plant defense responses using different S. miscanthi clones with identical genetic backgrounds, whereas in nature, aphids exist with additional strains of H. defensa (Chevignon et al., 2018) or with different genetic backgrounds. Thus, future studies are required to establish whether this difference is a general effect or a specific attribute of this particular combination of aphid and symbiont genotypes. Symbionts exist in many pests of important crops; thus, understanding the symbiont-mediated mechanism responsible for the suppression of plant defenses may provide important guidance to field arrangements of wheat varieties and the application of reasonable insecticide doses to reduce the potential threat of aphid outbreaks caused by symbiont-mediated aphid fitness enhancement. In addition, these results help generate a strategy for meeting the increased demand for novel insect pest management created by a growing human population and global climate change.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
QL, JF, JC, and MH conceived and designed the experiments. QL, JS, and YZ performed the experiments. QL analyzed the data. QL and JC wrote the manuscript. All the authors read and approved the final version of the manuscript.
Funding
This study was financially supported by the National Key R&D Plan of China (2016YFD0300700 and 2017YFD0201700) and the Opening Fund of the State Key Laboratory for the Biology of Plant Diseases and Insect Pests (SKLOF201601).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Yanxia Liu for rearing the aphids.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02419/full#supplementary-material
References
Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S. P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733.
Barr, K. L., Hearne, L. B., Briesacher, S., Clark, C. L., and Davis, G. E. (2010). Microbial symbionts in insects influence down-regulation of defense genes in maize. PLoS One 5:e11339. doi: 10.1371/journal.pone.0011339
Bednarski, W., Borowiak-Sobkowiak, B., Wilkaniec, B., Samardakiewicz, S., and Morkunas, I. (2013). Oxidative stress in pea seedling leaves in response to Acyrthosiphon pisum infestation. Phytochemistry 93, 49–62. doi: 10.1016/j.phytochem.2013.02.011
Berasategui, A., Axelsson, K., Schmidt, A., Borg-Karlson, A. K., Gershenzon, J., Terenius, O., et al. (2016). The gut microbiota of the pine weevil (Hylobius abietis, Coleoptera: Curculionidae) is similar across Europe and resembles that of other confer-feeding beetles. Mol. Ecol. 24, 4014–4031. doi: 10.1111/mec.13702
Bruessow, F., Gouhierdarimont, C., Buchala, A., Metraux, J. P., and Reymond, P. (2010). Insect eggs suppress plant defence against chewing herbivores. Plant J. 62, 876–885. doi: 10.1111/j.1365-313X.2010.04200.x
Cai, K., Gao, D., Luo, S., Zeng, R., Yang, J., and Zhu, X. (2010). Physiological and cytological mechanisms of silicon-induced resistance in rice against blast disease. Physiol. Plant 134, 324–333. doi: 10.1111/j.1399-3054.2008.01140.x
Chen, D. Q., Montllor, C. B., and Purcell, A. H. (2000). Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 95, 315–323. doi: 10.1111/jeb.12260
Cheng, D., Guo, Z., Riegler, M., Xi, Z., Liang, G., and Xu, Y. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (hendel). Microbiome 5:13. doi: 10.1186/s40168-017-0236-z
Chevignon, G., Boyd, B. M., Brandt, J. W., Oliver, K. M., and Strand, M. R. (2018). Culture-facilitated comparative genomics of the facultative symbiont Hamiltonella defensa. Gen. Biol. Evol. 10, 786–802. doi: 10.1093/gbe/evy036
Chung, S. H., Rosa, C., Scully, E. D., Peiffer, M., Tooker, J. F., Hoover, K., et al. (2013). Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Nati. Acad. Sci. U.S.A. 39, 15728–15733. doi: 10.1073/pnas.1308867110
Consales, F., Schweizer, F., Erb, M., Gouhierdarimont, C., Bodenhausen, N., Bruessow, F., et al. (2012). Insect oral secretions suppress wound-induced responses in Arabidopsis. J. Exp. Bot. 63, 727–737. doi: 10.1093/jxb/err308
Cooper, W. C., Jia, L., and Goggin, F. L. (2004). Acquired and R-gene mediated resistance against the potato aphid in tomato. J. Chem. Ecol. 30, 2527–2542. doi: 10.1007/s10886-004-7948-9
Ferrari, J., Darby, A. C., Daniell, T. J., Godfray, H. C. J., and Douglas, A. E. (2004). Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29, 60–65. doi: 10.1111/j.1365-2311.2004.00574.x
Frago, E., Dicke, M., and Godfray, H. C. (2012). Insect symbionts as hidden players in insect-plant interactions. Trends Ecol. Evol. 27, 705–711. doi: 10.1016/j.tree.2012.08.013
Frago, E., Mala, M., Weldegergis, B. T., Yang, C., Mclean, A., Godfray, H. C. J., et al. (2017). Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat. Commun. 8:1860. doi: 10.1038/s41467-017-01935-0
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Guidolin, A. S., and Cônsoli, F. L. (2017). Symbiont diversity of aphis (Toxoptera) citricidus, (Hemiptera: Aphididae) as influenced by host plants. Microl. Ecol. 73, 201–210. doi: 10.1007/s00248-016-0892-8
Hall, A. (1999). Induced responses to herbivory. Plant Pathol. 48, 122–123. doi: 10.1046/j.1365-2311.1999.00178.x
Henry, L. M., Peccoud, J., Simon, J. C., Hadfield, J. D., Maiden, M. J. C., Ferrari, J., et al. (2013). Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 23, 1713–1717. doi: 10.1016/j.cub.2013.07.029
Kariola, T., Brader, G., Li, J., and Palva, E. T. (2005). Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17, 282–294. doi: 10.1105/tpc.104.025817
Kawazu, K., Mochizuki, A., Sato, Y., Sugeno, W., Murata, M., Seo, S., et al. (2012). Different expression profiles of jasmonic acid and salicylic acid inducible genes in the tomato plant against herbivores with various feeding modes. Arthropod Plant Interact. 6, 221–230. doi: 10.1007/s11829-011-9174-z
Kerchev, P. I., Karpinska, B., Morris, J. A., Hussain, A., Verrall, S. R., Hedley, P. E., et al. (2013). Vitamin C and the abscisic acid-insensitive 4 (ABI4) transcription factor are important determinants of aphid resistance in Arabidopsis. Antioxid. Redox Signal. 18, 2091–2105. doi: 10.1089/ars.2012.5097
Koga, R., Tsuchida, T., and Fukatsu, T. (2003). Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. Lond. B 270, 2543–2550. doi: 10.1098/rspb.2003.2537
Kumar, S., Tamura, K., Jakobsen, I. B., and Nei, M. (2001). MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. doi: 10.1093/bioinformatics/17.12.1244
Lang, Y., Han, Y., Pei, L., Fei, L., Ali, S., and Hou, M. (2017). Silicon amendment is involved in the induction of plant defense responses to a phloem feeder. Sci. Rep. 7:4232. doi: 10.1038/s41598-017-04571-2
Li, Q., Fan, J., Sun, J. X., Wang, M. Q., and Chen, J. L. (2018). Effect of the secondary symbiont Hamiltonella defensa on fitness and relative abundance of Buchnera aphidicola of wheat aphid, Sitobion miscanthi. Front. Microbiol. 9:582. doi: 10.3389/fmicb.2018.00582
Li, T., Xiao, J. H., Wu, Y. Q., and Huang, D. W. (2014). Diversity of bacterial symbionts in populations of Sitobion miscanthi (Hemiptera: Aphididae) in China. Environ. Entomol. 43, 605–611. doi: 10.1603/EN13229
Li, Y., Dicke, M., Kroes, A., Liu, W., and Gols, R. (2016). Interactive effects of cabbage aphid and caterpillar herbivory on transcription of plant genes associated with phytohormonal signalling in wild cabbage. J. Chem. Ecol. 42, 793–805. doi: 10.1007/s10886-016-0738-3
Maehly, A. C., and Chance, B. (1995). The assay of catalases and peroxidases. Methods Biochem. Anal. 1, 357–424. doi: 10.1002/9780470110171.ch14
Martinez, O. D. I., Xie, Q., and Kaloshian, I. (2003). Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol. Plant Microbe Interact. 16, 699–708. doi: 10.1094/mpmi.2003.16.8.699
Montllor, C. B., Maxmen, A., and Purcell, A. H. (2002). Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27, 189–195. doi: 10.1046/j.1365-2311.2002.00393.x
Moran, P. J., and Thompson, G. A. (2001). Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 125, 1074–1085. doi: 10.1104/pp.125.2.1074
Musser, R. O., Hummusse, S. M., Eichenseer, H., Peiffer, M., Ervin, G., Murphy, J. B., et al. (2010). Herbivory: caterpillar saliva beats plant defences. Nature 416, 599–600. doi: 10.1038/416599a
Oliver, K. M., Degnan, P. H., Burke, G. R., and Moran, N. A. (2010). Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55, 247–266. doi: 10.1146/annurev-ento-112408-085305
Oliver, K. M., and Martinez, A. J. (2014). How resident microbes modulate ecologically important traits of insects. Curr. Opin. Ins. Sci. 4, 1–7. doi: 10.1016/j.cois.2014.08.001
Oliver, K. M., Moran, N. A., and Hunter, M. S. (2005). Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl. Acad. Sci. U.S.A. 102, 12795–12800. doi: 10.1073/pnas.0506131102
Oliver, K. M., Moran, N. A., and Hunter, M. S. (2006). Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. B 273, 1273–1280. doi: 10.1098/rspb.2005.3436
Oliver, K. M., Russell, J. A., Moran, N. A., and Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U.S.A. 100, 1803–1807. doi: 10.1073/pnas.0335320100
Oliver, K. M., Smith, A. H., and Russell, J. A. (2014). Defensive symbiosis in the real world-advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 28, 341–355. doi: 10.1111/1365-2435.12133
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Polin, S., Simon, J. C., and Outreman, Y. (2014). An ecological cost associated with protective symbionts of aphids. Ecol. Evol. 4, 836–840. doi: 10.1002/ece3.991
Qian, L., Jia, F., Jingxuan, S., Man-Qun, W., and Julian, C. (2018). Plant-mediated horizontal transmission of Hamiltonella defensa in the wheat aphid Sitobion miscanthi. J. Agricul. Food Chem. 66, 13367–13377. doi: 10.1021/acs.jafc.8b04828
Rodriguez, P. A., Stam, R., Warbroek, T., and Bos, J. I. (2014). Mp10 and mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol Plant Microbe Interact. 27, 30–39. doi: 10.1094/MPMI-05-13-0156-R
Russell, J. A., and Moran, N. A. (2005). Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl. Environ. Microbiol. 71, 7987–7994. doi: 10.1128/aem.71.12.7987-7994.2005
Sakurai, M., Koga, R., Tsuchida, T., Meng, X. Y., and Fukatsu, T. (2005). Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbl. 71, 4069–4075. doi: 10.1128/aem.71.7.4069-4075.2005
Schoonhoven, L. M., Loon, J. J. A. V., and Dicke, M. (2005). Insect-Plant Biology. New York, NY: Oxford University Press.
Selig, P., Keough, S., Nalam, V. J., and Nachappa, P. (2016). Jasmonate-dependent plant defenses mediate soybean thrips and soybean aphid performance on soybean. Arthropod Plant Interact. 10, 273–282. doi: 10.1007/s11829-016-9437-9
Su, Q., Oliver, K. M., Xie, W., Wu, Q., Wang, S., and Zhang, Y. (2016). The whitefly−associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Funct. Ecol. 29, 1007–1018. doi: 10.1111/1365-2435.12405
Thaler, J. S., Humphrey, P. T., and Whiteman, N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. doi: 10.1016/j.tplants.2012.02.010
Tooker, J. F., Rohr, J. R., Abrahamson, W. G., and Moraes, C. M. (2008). Gall insects can avoid and alter indirect plant defenses. New Phytol. 178, 657–671. doi: 10.1111/j.1469-8137.2008.02392.x
Vorburger, C., and Gouskov, A. (2011). Only helpful when required: a longevity cost of harbouring defensive symbionts. J. Evol. Biol. 24, 1611–1617. doi: 10.1111/j.1420-9101.2011.02292.x
Zhang, P. J., Li, W. D., Huang, F., Zhang, J. M., Xu, F. C., and Lu, Y. B. (2013). Feeding by whiteflies suppresses downstream jasmonic acid signaling by eliciting salicylic acid signaling. J. Chem. Ecol. 39, 612–619. doi: 10.1007/s10886-013-0283-2
Zhu-Salzman, K., Salzman, R. A., Ahn, J. E., and Koiwa, H. (2004). Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol. 134, 420–431. doi: 10.1104/pp.103.028324
Keywords: Hamiltonella defensa, Sitobion miscanthi, anti-plant defense, defense pathway, enzyme
Citation: Li Q, Fan J, Sun J, Zhang Y, Hou M and Chen J (2019) Anti-plant Defense Response Strategies Mediated by the Secondary Symbiont Hamiltonella defensa in the Wheat Aphid Sitobion miscanthi. Front. Microbiol. 10:2419. doi: 10.3389/fmicb.2019.02419
Received: 16 August 2019; Accepted: 07 October 2019;
Published: 25 October 2019.
Edited by:
Amparo Latorre, University of Valencia, SpainReviewed by:
Christoph Vorburger, Swiss Federal Institute of Aquatic Science and Technology, SwitzerlandTong Li, Henan Academy of Agricultural Sciences, China
Copyright © 2019 Li, Fan, Sun, Zhang, Hou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: JuLian Chen, Y2hlbmp1bGlhbkBjYWFzLmNu
 Qian Li
Qian Li Jia Fan
Jia Fan JingXuan Sun1
JingXuan Sun1 Yong Zhang
Yong Zhang