- 1National Research Center for Wildlife Borne Diseases, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 2College of Life Sciences, University of the Chinese Academy of Sciences, Beijing, China
- 3Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public, Laboratory of Wild Animal Conservation and Utilization, Guangdong Institute of Applied Biological Resources, Guangzhou, China
- 4Beijing Wildlife Rescue Center, Beijing Municipal Bureau of Landscape and Forestry, Beijing, China
- 5Division of Biological Sciences, Science, Technology, Engineering and Mathematics, University of Washington, Bothell, WA, United States
Campylobacter jejuni (C. jejuni) is considered as an opportunistic zoonotic pathogen that may cause gastroenteritis in humans and other animals. Wild birds may be as potential vectors of C. jejuni around urban and suburban areas. Here, 520 samples were collected from 33 wild bird species in urban and suburban areas, Beijing. In total 57 C. jejuni were isolated from seven species. It was found that Nineteen (33.33%, 19/57) isolates were resistant to at least one of 11 antibiotics, especially streptomycin (36.84%) and four isolates resistant to all. Nineteen (33.33%, 19/57) isolates were multi-drug resistance. Multilocus sequence typing (MLST) analysis of the isolates showed that 36 different sequence types (STs) belonged to four Clonal complexes and unassigned. Twenty STs (55.56%) and six alleles among them were first detected. Virulence genes including flaA, cadF, and the cytolethal distending toxin (CDT) gene cluster, were detected in all isolates, but truncated cdt gene clusters only detected in the isolates from the crow, daurian jackdaw and silver pheasant. In conclusion, it was the first detection of C. jejuni involved truncated cdt gene clusters from the silver pheasant. These wild birds around urban and suburban areas may pose potential public health problems as reservoir vectors of C. jejuni.
Introduction
Campylobacter jejuni is a gram-negative spiral rod bacterium that causes gastroenteritis in humans and other animals. In 2016, a total of 246,307 confirmed cases of human campylobacteriosis were reported in the European Union (EU), representing almost 70% of all the reported human cases of zoonoses (European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC), 2017). There was a significantly increasing trend over the period 2008–2016. Compared with European countries, reports on campylobacteriosis in Asian countries, including China are limited, and the campylobacteriosis prevalence in humans is generally low (Wang et al., 2015).
The clinical presentation of campylobacteriosis includes watery or bloody diarrhea lasting for a median duration of 6 days, with 80% of patients having cramps and fever (Friedman et al., 2004). In some cases, the infection can lead to extra-intestinal complications and severe autoimmune disorders, such as pancreatitis, cholecystitis, obstructive hepatitis, Guillain-Barré syndrome (GBS), and Miller Fisher syndrome. Generally, the patients with campylobacteriosis are self-limited and disappear after 1 week without any specific treatment. However, in some relatively severe cases, antimicrobial chemotherapy is required. Macrolides and quinolones are commonly used as first-line therapies, and tetracycline, doxycycline, and chloramphenicol are alternative drugs (Ma et al., 2017).
Resistant strains and multi-drug resistance (MDR) strains are increasingly reported in humans and animals which may be induced by increasing use of antibiotics in humans, domestic animals and poultry (Franiczek et al., 2012; Karaiskos and Giamarellou, 2014; Alsan et al., 2015). C. jejuni has a high capacity to transfer genetic elements that lead to the combination of different strains. This characteristic may allow of C. jejuni transfer antibiotic resistance genes easily (de Boer et al., 2002; Wilson et al., 2003; Avrain et al., 2004), The abuse of antibiotics could increase the selection pressure and decrease the effectiveness of antibiotics further (Klena et al., 2004; Laxminarayan et al., 2013).
In Campylobacter, mutations in the 23S rRNA genes shown to contribute to macrolide resistance (Payot et al., 2006). C. jejuni resistance to tetracycline is usually associated with the tet(O) gene, which is carried on transmissible plasmids or located chromosomally. Ribosomal protein – Tet(O), which encoded by tet(O) gene can confer the resistance by displacing tetracycline from its primary binding site on the ribosome (Connell et al., 2003; Gibreel et al., 2004b). C. jejuni resistance to fluoroquinolones is mainly associated with mutations in the DNA gyrase gene (gyrA) (Smith and Fratamico, 2010). Accordingly, it is necessary to monitor the antibiotic resistance and research the antibiotic resistance mechanism of C. jejuni in wildlife.
The process of infection involved in adhesion, colonization, invasion and toxin production, especially, flaA genes, cadF genes, and cdt gene clusters are necessary for cell pathology and virulence in humans (Wei et al., 2018). These three virulence genes are frequently researched in isolates from various hosts, especially humans and poultry. However, little is known about these virulence genes in wild bird isolates (Wei et al., 2018).
The consumption of feces-contaminated raw or undercooked poultry has been identified as an important transmission vehicle for human campylobacteriosis (Wei et al., 2018). However, there is evidence that non-food-borne exposure of C. jejuni may contribute to the burden of illness as well. Thus contamination of the environment by domestic and wild birds feces may constitute an additional risk for human infection via environmental water or direct contact with them (French et al., 2009). Some studies suggested that C. jejuni is a commensal microorganism in the intestine of many wild and domestic animals, particularly avian species, and they can be natural reservoirs of C. jejuni (Oravcova et al., 2014). Furthermore, migratory birds may play a direct or indirect role in the zoonotic transmission of Campylobacter through the foods with fecal contamination (Kapperud et al., 2003; Gardner et al., 2012). C. jejuni has been isolated from wild birds such as pigeons, crows, geese, ducks, gulls, and cranes (Broman et al., 2002; Chen et al., 2011). Isolates from black-headed gulls (Chroicocephalus ridibundus), Sandhill cranes (Grus canadensis), and European starlings (Sturnus vulgaris) have been implicated in human disease (Palmer et al., 1983; Broman et al., 2002, 2004; Waldenstrom et al., 2002; Pearson et al., 2007; Karaiskos and Giamarellou, 2014).
Although C. jejuni is the main cause of bacterial diarrhea, there are still limited data on C. jejuni in China, especially for a wide range of free-living and migrating birds in various parts of cities, and the threat to public health cannot be ignored. Here, we investigated the prevalence, genetic diversity (Multilocus sequence typing), antimicrobial resistance patterns and virulence genes (flaA genes, cadF genes, and cdt gene cluster) of Campylobacter in wild birds in Beijing, China.
Results
The Prevalence of Campylobacter jejuni
In total, 520 samples were collected from 33 species and 12 sites during the 4 months in Beijing, China (Figure 1). C. jejuni was isolated from 57 samples (10.96%, 57/520) including seven species. In the positive C. jejuni samples, 24.19% (15/62) were from crow (Corvus sp.), 51.67% (31/60) were from daurian jackdaw (Corvus dauurica), 14.29% (1/7) were from silver pheasant (Lophura nycthemera), 8.57% (6/70) were from mallard (Anas platyrhynchos), 6.25% (1/16) were from mandarin duck (Aix galericulata), 12.5% (1/8) were from black swan (Cygnus atratus) and 7.69% (2/26) were from rock pigeon (Columba livia). Crows (Corvus sp.) and Daurian jackdaws (Corvus dauurica) had significantly higher positive rates than other species (P < 0.007). Mallard (Anas platyrhynchos), mandarin duck (Aix galericulata) and black swan (Cygnus atratus) showed a relatively low positive rate between 6.25 and 12.5%. Silver Pheasant (Lophura nycthemera), family Phasianidae showed a similar positive rate as chicken, which is one of the most important reservoirs of C. jejuni. Rock pigeon (Columba livia), had relatively lower positive rate than others. This is the first time that C. jejuni has been isolated from black swan (Cygnus atratus) (Table 1).
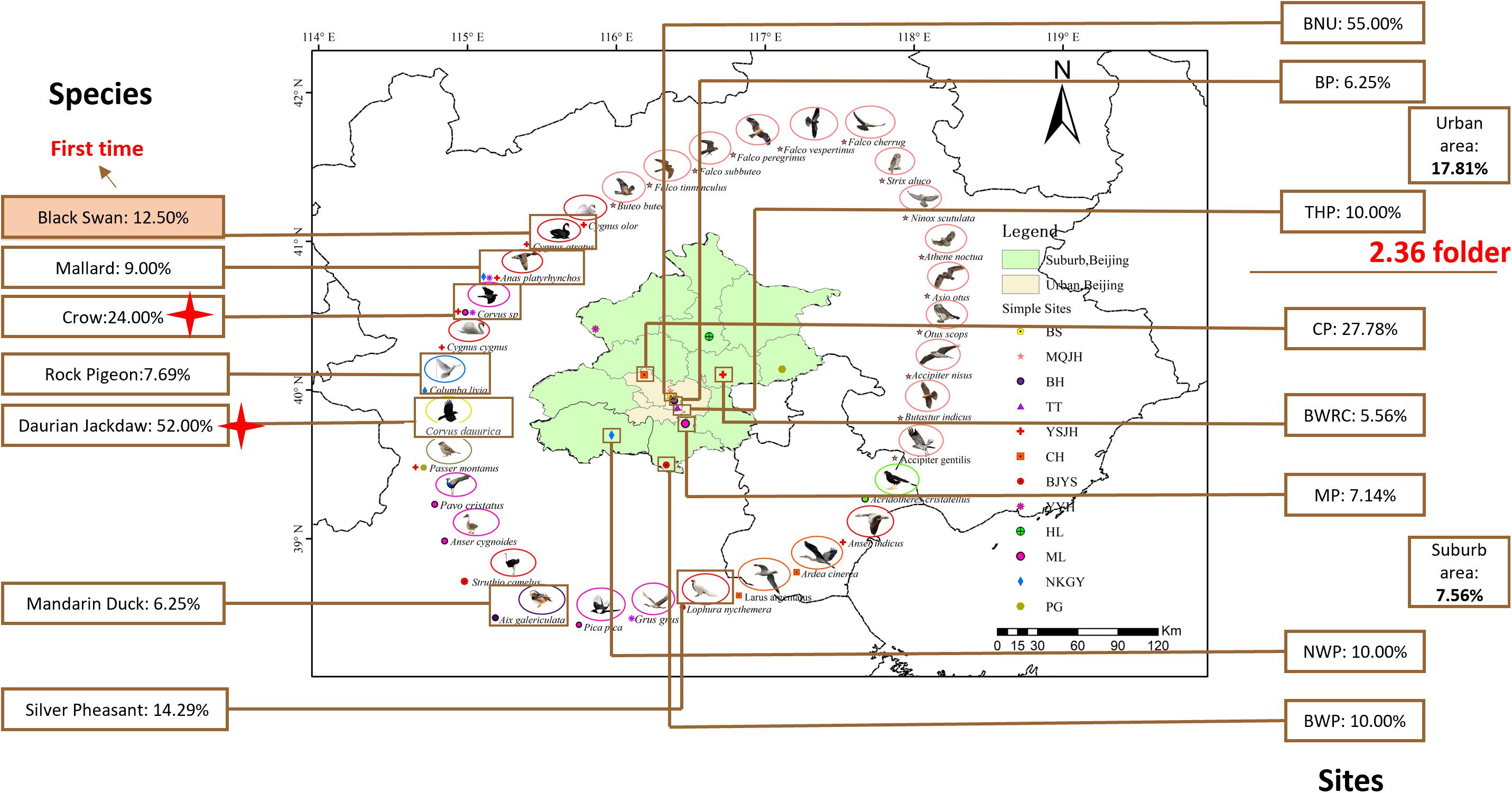
Figure 1. Samples collect sites and species in Beijing, China. Green represent Suburb area, Beijing; fleshcolor represent Urban area, Beijing. Images around the map of Beijing represent 33 species and the small icons represent 11 different sites. The data in the box represent the rate of the positive C. jejuni samples from different species and sites. Red tetragon represent crows and daurian jackdaws of family Corvids have significantly high positive rate than other species (P < 0.007). BNU, Beijing Normal University; BP, Beihai Park; THP, Temple of Heaven Park; CP, Cuihu Park; BWRC, Beijng Wildlife rescue center; BWP, Beijing Wildlife Park; MP, Milu Park; NWP, Niukouyu Wetland Park; BRRC, Beijing Raptor rescue center; WDL, Wild Duck Lake; HL, Hongluo Lake; XR, Xiyu Reservoir.
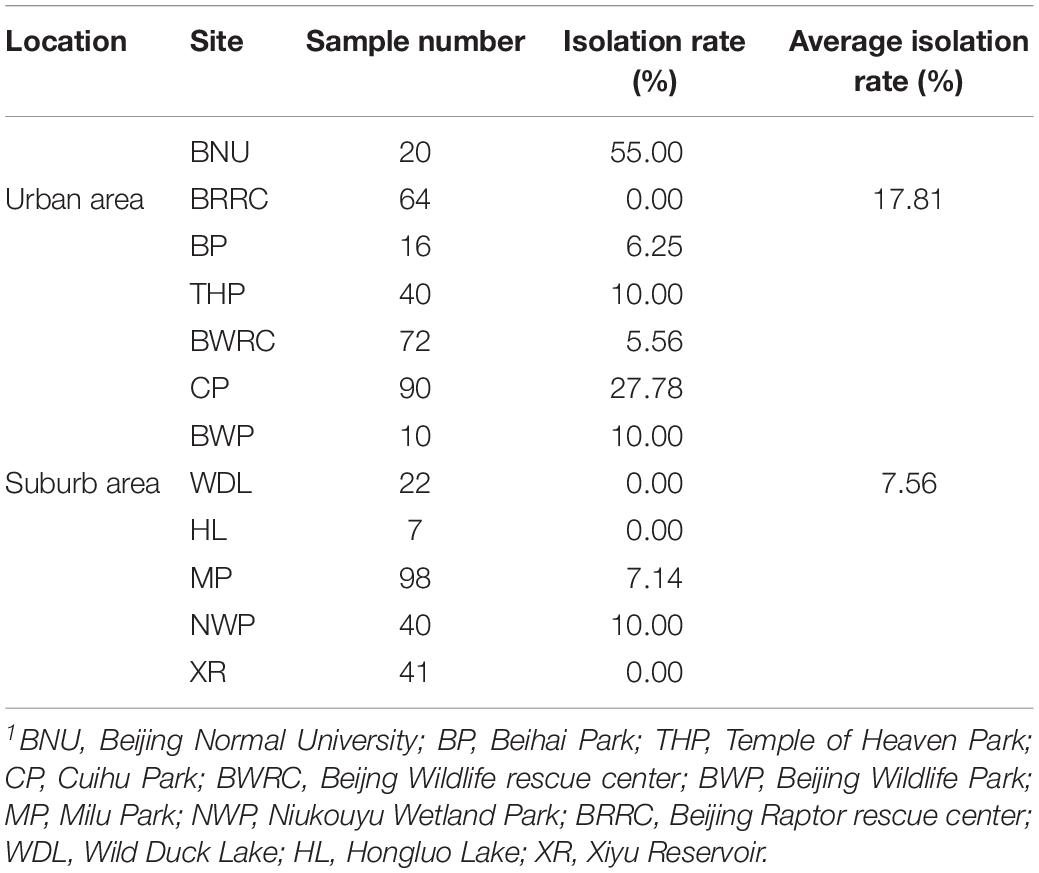
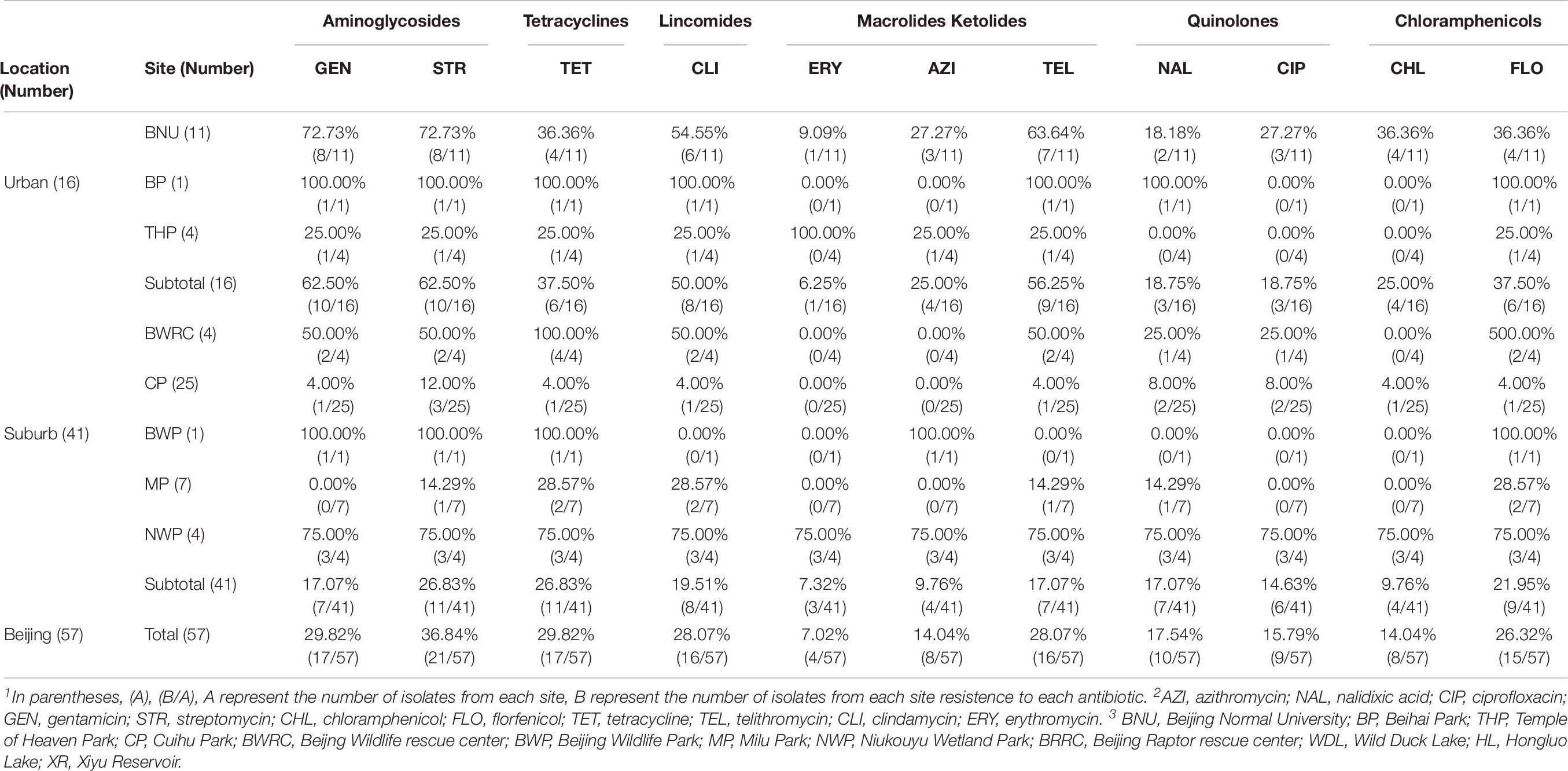
The 12 collection sites were divided into two groups, including urban area and suburban area. Totally, all the positive samples of C. jejuni were from 8 sites, including Beijing Normal University (BNU) (55.00%), Beihai Park (BP) (6.25%), Temple of Heaven Park (THP) (10.00%), Cuihu Park (CP) (27.78%), Beijing Wildlife Rescue Center (BWRC) (5.56%), Beijing Wildlife Park (BWP) (10.00%), Milu Park (MP) (7.14%), and Niukouyu Wetland Park (NWP) (10.00%). The highest was BNU (55%) which located in an urban area, and the lowest was BWRC (5.46%) which located in the suburb area. The average positive rate in urban areas (17.81%) was 2.36 times higher than in suburb areas (7.66%) (Table 2).
Multilocus Sequence Typing for Campylobacter jejuni
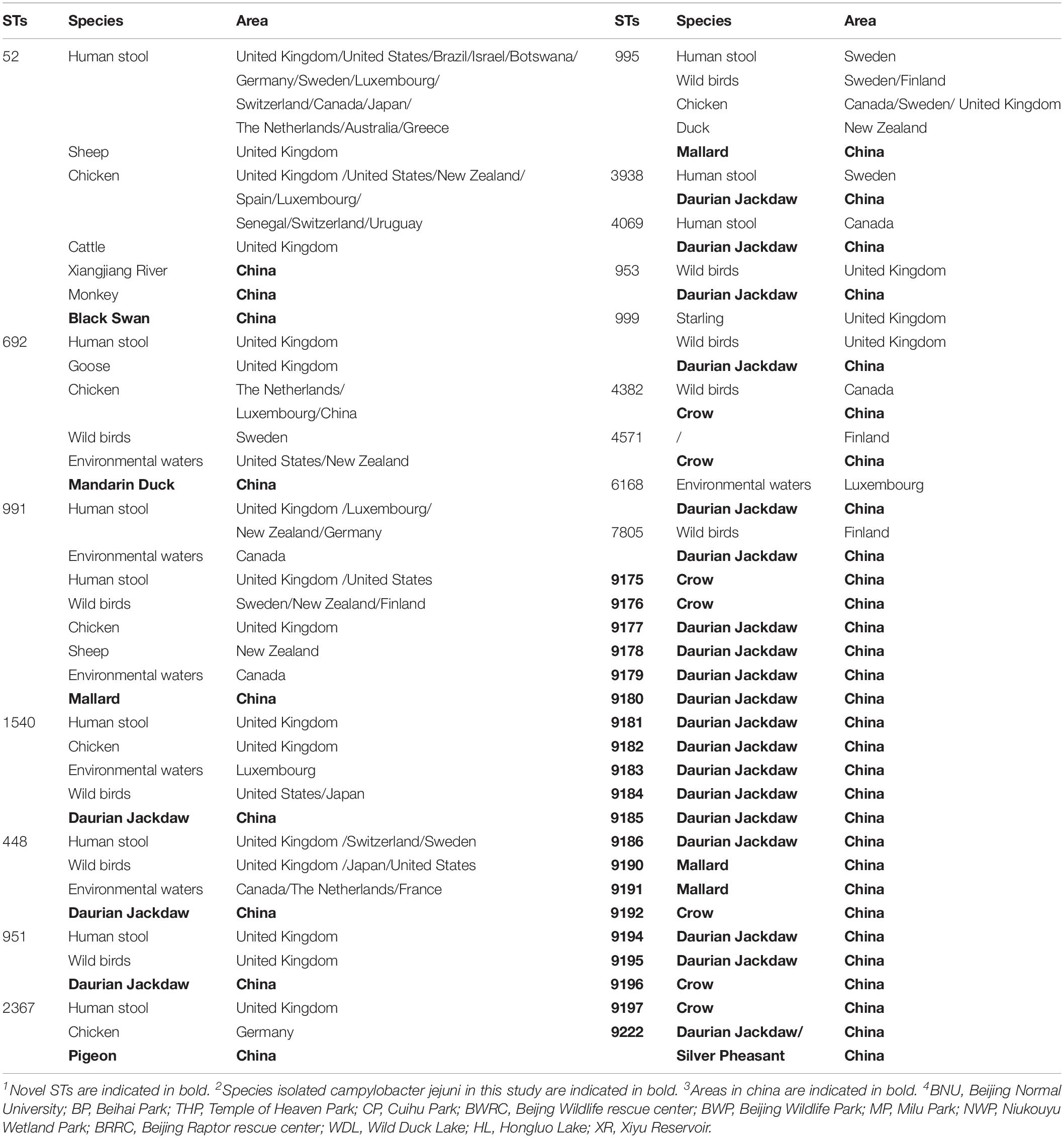
To discover the genetic diversity of C. jejuni in wild birds and explore the role of wild birds in disease transmission, the genotype of C. jejuni isolates from wild birds were tested by multilocus sequence typing (MLST). MLST, which uses seven genes to build a classification system, is a common way to reveal genetic diversity. Fifty-seven isolates were divided into 36 different sequence types (STs) that clustered into four clonal complexes (CCs) and unassigned.
The same bird species could carry a variety of STs from different individuals (Table 3). Twenty-three STs were identified from 31 C. jejuni isolates in daurian jackdaws, and 7 STs from 15 isolates in crows. Overall, 52.6% of novel STs were first discovered in the present study (Table 3), especially more than 10 novel STs were from one or more new allelic genes, and six novel alleles were found (aspA484, aspA485, glnA673, glyA754, tkt718, tkt719) (Supplementary Table S1). Moreover, 84.5% (49 strains) of the STs did not belong to any clonal complex (CC). Three STs (ST-692, ST-991, ST-9191) belonged to ST-692 clonal complex and ST-52, ST-952, and ST-1275 complex comprised three STs, respectively (Table 3).
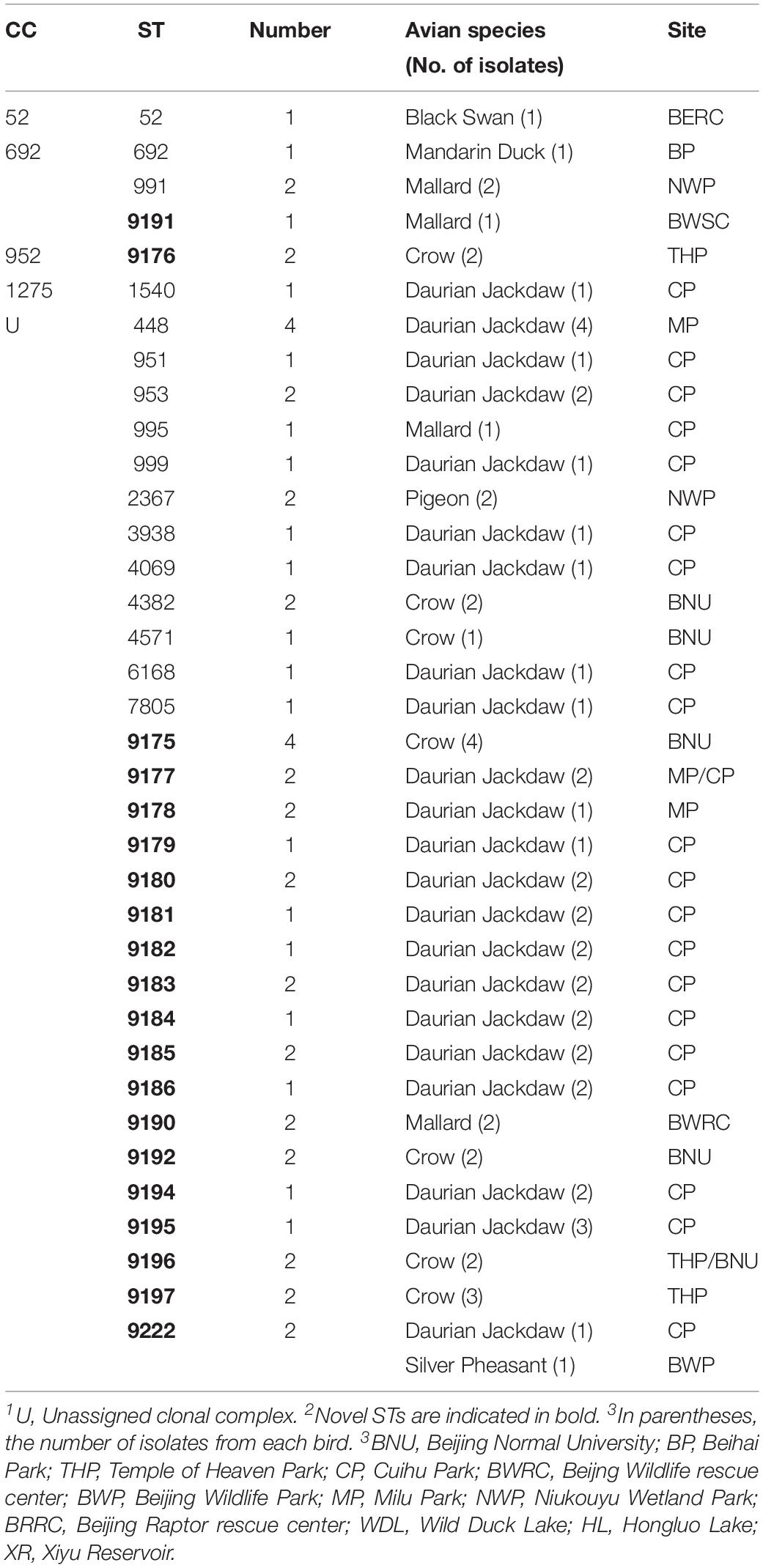
Table 3. Clonal complex (CC) and sequence type (ST) distribution of Campylobacter in wild bird species.
Phylogenetic Analysis of Campylobacter jejuni Strains
The minimum spanning tree for 57 C. jejuni isolates and other reference isolates from the PubMLST database was constructed (Supplementary Figure S1). The genetic diversity of the C. jejuni isolates showed that nine STs (ST-995, ST-991, ST-2367, ST-3938, ST692, ST-52, ST1540, ST448, ST-951, ST4069) were found in wild birds and human, 5 STs (ST-995, ST-991, ST-2367, ST-52, ST692, ST1540) in wild birds, chicken and humans, 2 STs (ST-448 and ST-995) in monkeys, wild birds and humans, and 2 STs (ST-52 and ST-995) in 4 hosts (wild birds, humans, chicken, dogs/monkeys). Researches previously reported showed that humans and other animals have the same predominant STs, which was suggested that these animals are important reservoirs of human domestically acquired infections (Olkkola et al., 2016). In this study, the isolates from black swan, mandarin duck, mallard, daurian jackdaw, and rock pigeon were found in humans (Table 4). Therefore these results above indicated that the C. jejuni could transmit between different species (Tables 3, 4 and Supplementary Figure S1).
Antibiotic Resistance of Campylobacter jejuni
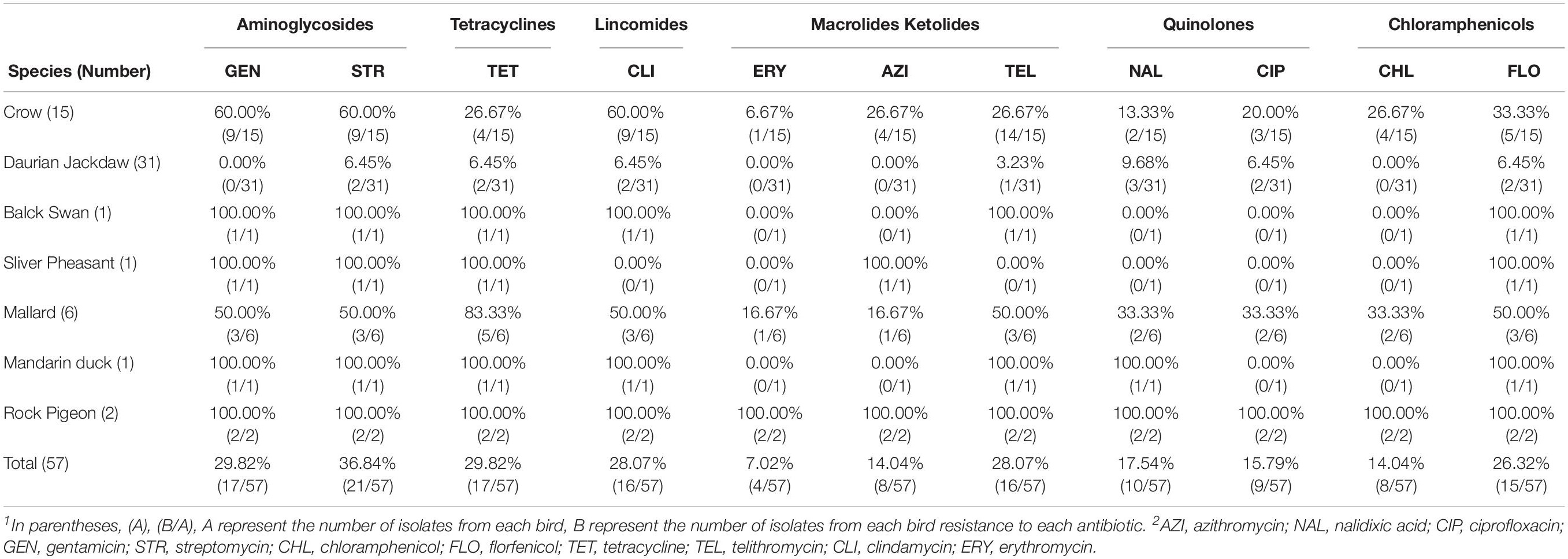
The antibiotic resistance profile of C. jejuni isolates was evaluated by 11 antibiotics according to the recommendations of the Clinical Laboratory Standards Institute (CLSI, 2015). Antibacterial resistance revealed that streptomycin (36.84%) was most common, followed by tetracycline (29.82%), gentamicin (29.82%), clindamycin (28.07%), telithromycin (28.07%), florfenicol (26.32%), nalidixic acid (17.54%), and ciprofloxacin (15.79%), azithromycin (14.04%), chloramphenicol (14.04%), erythromycin (7.02%) (Figure 2 and Table 5).
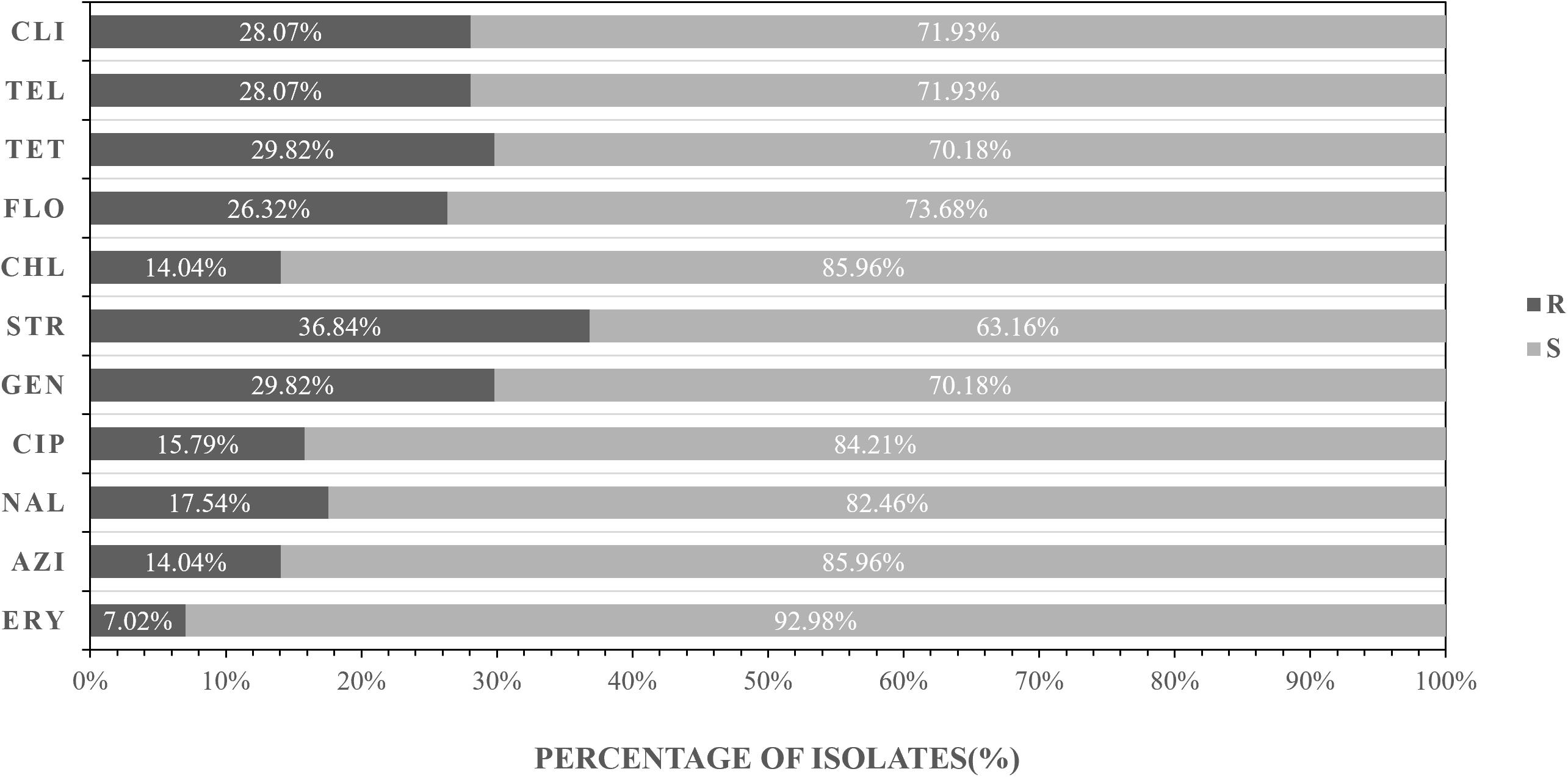
Figure 2. Frequency of resistance to 11 antibiotics among the 57 Campylobacter jejuni isolates. R represents resistant, I represents intermediate, S represents sensitive. AZI, azithromycin; NAL, nalidixic acid; CIP, ciprofloxacin; GEN, gentamicin, STR, streptomycin; CHL, streptomycin, FLO, florfenicol; TET, tetracycline, TEL, telithromycin, CLI, clindamycin; ERY, erythromycin.
The isolates from different locations and sites showed different antimicrobial profile. In the urban area, the antimicrobial efficiency of three antibiotics commonly used in humans and animals are as follows: streptomycin (62.50%), gentamicin (62.50%), and telithromycin (50.00%) (Table 6). In the suburb area, the rate of antibiotic resistance was low, ranging from 7.32 to 26.83% (Table 6).
Multi-drug resistance of bacteria is also common in this study. Nineteen (33.33%, 19/57) isolates were MDR. In detail, The main antibiotics producing drug resistance were streptomycin, gentamicin, and clindamycin (26.32%) (Figure 3 and Supplementary Table S2).
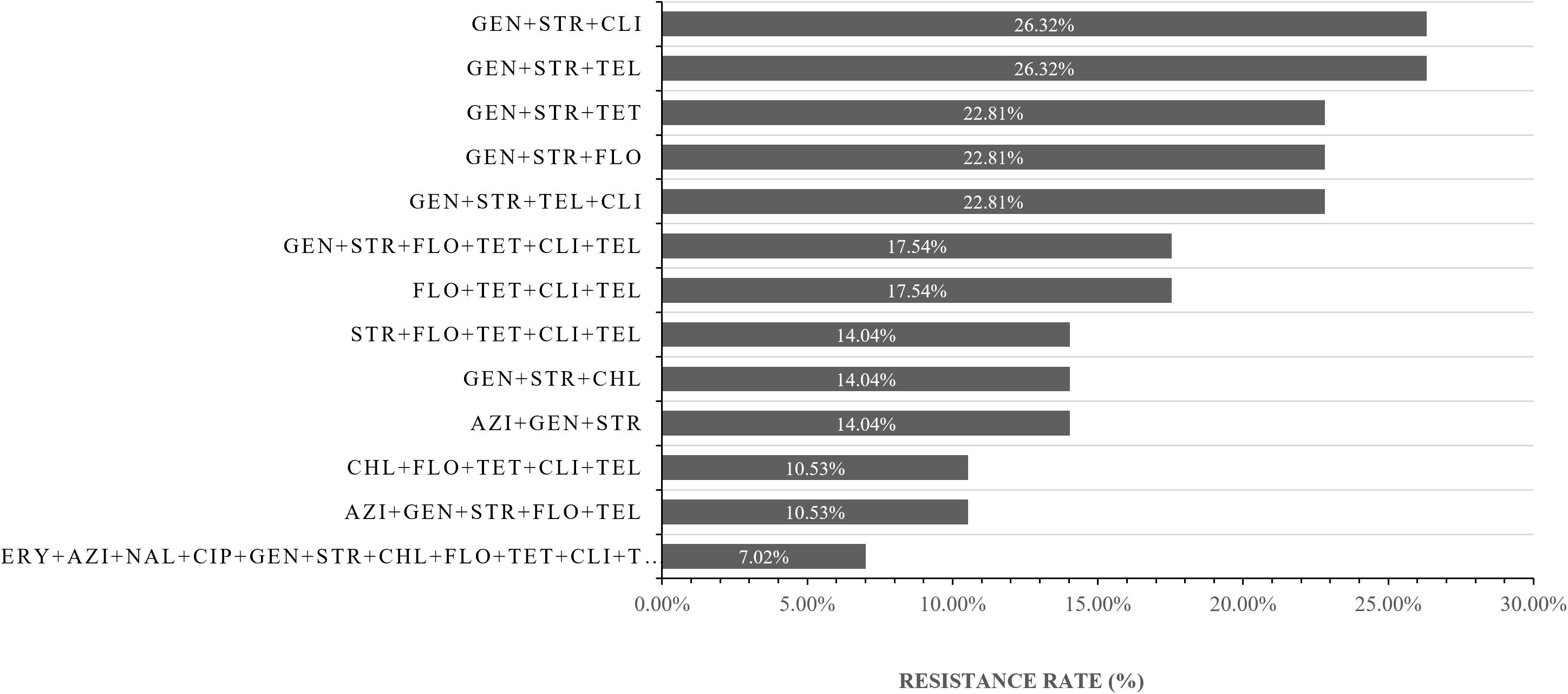
Figure 3. The resistance spectrum of strains of Campylobacter jejuni to various antibiotic combinations. The X-axis represents the resistance rate of Campylobacter jejuni, The Y-axis represents a series of combination of antibiotics. Thirty-five (61.4%, 35/57) isolates were multi-drug resistance (resistant to more than two antibiotics respectively). AZI, azithromycin; NAL, nalidixic acid; CIP, ciprofloxacin; GEN, gentamicin; STR, streptomycin; CHL, chloramphenicol; FLO, florfenicol; TET, tetracycline; TEL, telithromycin; CLI, clindamycin; ERY, erythromycin.
Antibiotic Resistance Mechanism of Campylobacter jejuni
The characteristics of macrolide resistance associated with the genes were analyzed in 4 resistant isolates and 53 susceptible isolates, and this helps investigate the molecular mechanisms of the macrolide-resistant isolates. The A2075G mutation in the 23S rRNA gene, which is responsible for high-level resistance to macrolide, was not detected in all of the resistant strains and susceptible isolates (Supplementary Figure S2A).
All Campylobacter isolates were also investigated for the presence of the tet(O) gene associated with the resistance to tetracycline. The tet(O) marker was detected in all but three resistant strains(3/17) (Supplementary Figure S2B).
To investigate the molecular mechanisms of the fluoroquinolones resistant isolates, Multiplex PCR was designed, which uses three primers in a single reaction. Only the isolates with the gene mutation generated a product with the reverse primer with mutation and the conserved forward primer, whereas all 57 strains generated the gyrA PCR product with reverse and forward conserved primers. 87.50%(7/8) C. jejuni resistant isolates generated the specific product in MAMA-PCR that indicated the mutation Thr-86-to-Ile (ACA→ATA for C. jejuni), while none of the susceptible strains gave positive results (Supplementary Figure S2C).
All Campylobacter isolates were also investigated for the presence of the aphA-3 gene associated with the resistance to Aminoglycosides. The aphA-3 marker was detected in all but one resistant strains(1/17) (Supplementary Figure S2B).
Determination of the Presence of Virulence Genes
To determine whether virulence differences exist among isolates from different wild birds, we tested the virulence genes including the cdt gene cluster, flaA gene and cadF gene, the gene that encodes an adhesin that involves in colonization for all C. jejuni isolates.
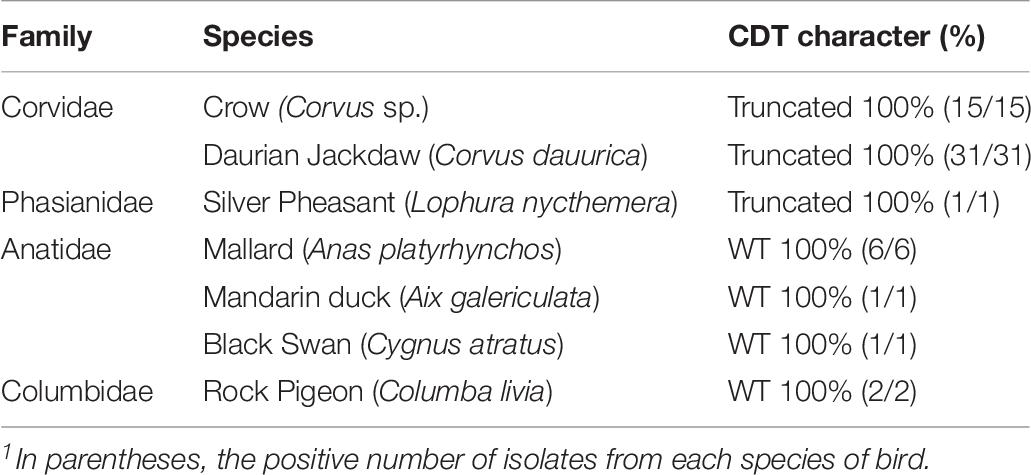
The results showed that all isolates contained flaA genes, cadF genes, and cdt gene cluster. Further analysis indicated that the truncated cdt gene clusters (approximately 1400 bp) only existed in the isolates from crows, daurian jackdaws and silver pheasants. In particular, the truncated cdt gene clusters were first detected in C. jejuni isolates from silver pheasant. C. jejuni isolates from Corvidae are more likely to carry truncated cdt gene clusters, other species belong to family Corvids and Phasianidae may also have deletions within the cdt Gene Cluster and still need a further survey. These results above indicated that bacteria in the family of Anatidae and Columbidae might have complete cdt gene clusters, but further research is still needed for two families species (Table 7).
Discussion
Camphylobacter jejuni carried by wild birds has been identified to be potentially pathogenic to humans and other animals (Kapperud et al., 2003; Gardner et al., 2012). The majority of studies have focused on food sources that may cause human campylobacterosis, while studies on wild birds are rare, especially in China. In order to get a better understanding of the distribution and transmission of C. jejuni in wild birds, we analyzed the prevalence, genetic diversity, antimicrobial resistance and virulence genes of C. jejuni in Beijing, China.
In the present study, we found that 10.96% of fecal samples collected from 33 species carried with C. jejuni. Previous studies demonstrated that the prevalence of C. jejuni among different countries and wild birds species varied from 1.4 to 72.7% (Ramonaite et al., 2015). Among these species we found that the highest prevalence of C. jejuni was daurian jackdaw 51.67%, followed by crow 24.19%, silver pheasant 14.29%, black swan 12.5%, mallard 8.57%, rock pigeon 7.69%, and mandarin duck 6.25%. C. jejuni isolated from different wild birds with different prevalence which in agreement with other studies that the prevalence of Campylobacter spp. in different taxonomic families wild birds were diverse (Waldenstrom et al., 2007; French et al., 2009).
It is believed that the variation of C.jejuni prevalence in wild bird species is due to ecological factors including feeding habits, habitat preferences, and migration patterns (Waldenstrom et al., 2002; Colles et al., 2009; Waldenstrom et al., 2010; Colles et al., 2011). Located in urban, BNU had the highest prevalence of C. jejuni. Although the proportion of human disease attributable to environmental sources is relatively low, we could not ignore the effect on humans (Wilson et al., 2008; Sheppard et al., 2009; Colles et al., 2011). Most importantly, to our knowledge, it is the first detection of C. jejuni carried by black swans in China. These data strengthen the hypothesis that the high prevalence of C. jejuni in wild birds might provide evidence of wild birds being a natural reservoir of C. jejuni (Oravcova et al., 2014).
Multilocus sequence typing was the golden standard to use for comparison between isolates from different sources because of its high reproducibility and accessible to comparability amongst laboratories worldwide (Dingle et al., 2001). Previous studies have shown that human patients and other animals have the same predominant STs suggesting that these animals are important reservoirs of human domestically acquired infections (Olkkola et al., 2016). From our results, 36 different STs belonging to 4 CCs and unassigned, among which 20 were novel. Of the 20 novel STs, 25% were from one or more new allelic sequences, and a total of 6 novel alleles were found (aspA484, aspA485, glnA673, glyA754, tkt718, tkt719). No new allele sequences were found for another 75% new STs, and these STs resulted from novel combinations of alleles already existed in the PubMLST database. These results indicated that mutation frequency in the MLST alleles is substantially lower than the recombination frequency, which in agreement with previous research (Schouls et al., 2003). ST448, ST951, and ST52 these isolates from daurian jackdaw and black swan, however other researches indicated that all of these three strains isolated from other animals, poultry, and humans (Griekspoor et al., 2010; Olkkola et al., 2016). So wild birds as a potential source of known and novel multilocus sequence types of C. jejuni may have the potential to transmit to other animals, poultry, and humans.
The antibiotic resistance profile of C. jejuni isolates from these animals was determined using 11 antibiotics. The results of antimicrobial susceptibility testing in this study indicated that the isolates were in general resistant to the tested antibiotics at rates ranging from 7.02 to 36.84%. The high rate of resistance to streptomycin and gentamicin was seen among C. jejuni isolates from these birds. All Campylobacter isolates were also investigated for the presence of the aphA-3 gene associated with the resistance to Aminoglycosides. The most common form of resistance to Aminoglycosides related antibiotics involves the synthesis of 3′-aminoglycoside phosphotransferases [APH(3′)](Gibreel et al., 2004a). The aphA-3 marker was detected in all but one resistant strains(1/17), which is consistent with previous research results.
From the different locations and sites, the antibiotic resistance profile performed differently. In urban areas, the isolates from wild birds have high antibiotic resistance, which might be due to contaminated environment water. The reasonable interpretation for this difference may be human activities, such as antibiotic abuse, are more active in urban areas than in the suburbs, resulting in birds in different habitats getting different resistant bacteria from the environment. However, the relationship between high antibiotic resistance and contaminated environment water needs further study. As previous study indicated that C. jejuni isolates detected in crows and pigeons as potential infection sources to humans (Ramonaite et al., 2015), it was worth noting that four strains from wild birds (crow, mallard, and rock pigeon) are resistant to all 11 antibiotics which may be an important indicator of public health safety.
All wild birds isolates had flaA genes, cadF genes, and cdt gene clusters, which in agreement with the previous study (Sen et al., 2018). The results suggest that these three genes are conserved amongst different sources. As before, C. jejuni isolates from crows had a truncated gene cluster of about 1400 bp (Sen et al., 2018), C. jejuni isolates from daurian jackdaw also had a truncated gene cluster (Kovanen et al., 2019). However, the most unexpected results were isolates from silver pheasant which also have a truncated gene cluster of about 1400 bp. To our knowledge, it was the first detection of the isolates from silver pheasant also have a truncated gene cluster of about 1400 bp. The CDT toxin is a tripartite protein formed by the expression of three tandem genes, cdtA, cdtB, and cdtC where cdtB encodes the active component of the toxin, while cdtA and cdtC are responsible for binding and internalization of the toxin (Pickett et al., 1996). Some research confirmed that C. jejuni 81-176 cdtBs– strains were significantly attenuated in HeLa cytotoxicity assays, while still holding some toxigenicity. However, C. jejuni NCTC 11168 cdtB– strains produced no detectable cytotoxicity in HeLa cell (Purdy et al., 2000). So, if these isolates with a truncated cdt gene cluster still retaining the toxigenicity required further verification.
Conclusion
Wild birds as a reservoir of potentially pathogenic C. jejuni strains and can be a vector of disease transmission. However, further studies are needed to link the high occurrence of Campylobacter in wild birds to human campylobacteriosis cases and transmission to other animals.
Experimental Procedures
Samples Collection and Campylobacter Isolation
Five hundred and twenty fecal samples were collected from 33 species in 12 sites, Beijing, China, between January 2018 and April 2018 (Figure 1 and Table 1). Samples were collected in sterile tubes and stored at 4 to 7°C for 2 to 6 h before culturing in Lab (Sen et al., 2018). Fecal samples were inoculated onto Modified Charcoal Cefoperazone Deoxycholate Agar (mCCDA) containing Cefoperazone, Rifampicin, and Amphotericin B (Qingdao Hope Bio-Technology, Co., Ltd.), with incubation at 37°C under microaerophilic conditions (CampyGen; Oxoid Limited, Hampshire, United Kingdom) for 48 to 168 h. Then picked white to translucent colonies subculture onto mCCDA for further characterization.
Campylobacter Identification
All positive samples were used to extract DNA by adding 100 μl 0.25% SDS and boiling in a heater block at 95°C for 10 min, followed by centrifugation at 12,000 g for 5 min. Template DNA was stored at −20°C until used for PCR and at least 1 year without any degradation (Sen et al., 2018). Then Campylobacter spp. identified by a qPCR method based on the 16S rRNA gene and primers used as previously described (Lund and Madsen, 2006) (Table 8), The qPCR conditions as follows: 1 cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s, with a final cycle of 72°C for 5 min. For determining the presence of Campylobacter jejuni, Campylobacter coli, Campylobacter lari, and Campylobacter upsaliensis, a multiplex PCR method was performed with the isolates using the lipid A gene (lpxA) as previously described (Klena et al., 2004), PCR conditions were as follows: 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, extension at 72°C for 1min and a final extension step at 72°C for 5 min. The presence of flaA, cadF and CDT gene cluster in 57 isolates were determined by PCR using the primer sets described in Table 8 (Nachamkin et al., 1993; Konkel et al., 1999; Bang et al., 2003). The thermocycling conditions which can be found in the respective references in Table 8. The PCR products were detected on 1% Agarose gels and verified by sequencing.
Multilocus Sequence Typing for Campylobacter jejuni
Multilocus sequence typing was performed by amplifying and sequencing seven housekeeping genes loci, aspA, glnA, gltA, glyA, pgm, tkt, and uncA, and primers used for C. jejuni are listed in Table 9 (Dingle et al., 2001). The PCR reaction conditions were as follows: initial denaturation at 94°C for 5 min; followed by 35 cycles of 94°C for 2 min, 50°C for 1 min and 72°C for 1 min; last denaturation at 72°C for 7 min. The PCR products were detected on 1% Agarose gels and verified by sequencing. Allele numbers and STs were assigned using the Campylobacter MLST database1.
Antimicrobial Susceptibility Testing for Campylobacter jejuni
Antimicrobial resistance analysis was performed on 57 Campylobacter isolates. All isolates were cultured overnight before testing. The C. jejuni strains were tested against phenotypic resistance to 11 antimicrobial agents (erythromycin, azithromycin, nalidixic acid, ciprofloxacin, gentamicin, streptomycin, chloramphenicol, florfenicol, tetracycline, telithromycin, and clindamycin) (Zhongchuan biology technology Company, Qingdao, China) by the agar dilution method according the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2015). Mueller-Hinton agar (Oxoid) with dilutions ranging from 64 to 0.5 μg/mL for erythromycin, azithromycin, nalidixic acid, ciprofloxacin, gentamicin, streptomycin, chloramphenicol, florfenicol, tetracycline, telithromycin, and clindamycin was prepared. Subcultured colonies were harvested and suspended in sterile water to a standardized cell density (0.5 McFarland), and 2 μL of approximately 104 CFU of bacteria was pipetted into each well (after diluting in PBS). The plates were incubated under a microaerophilic atmosphere at 42°C for 24 h. The MIC values were defined as the lowest concentration that produces complete inhibition of C. jejuni growth. For quality control, the reference strain C. jejuni ATCC 33560 was included. The C. jejuni isolates were considered resistant to chloramphenicol (CHL), erythromycin (ERY), ciprofloxacin (CIP), nalidixic acid (NAL) tetracycline (TET), streptomycin (STR), and telithromycin (TEL) at MICs of ≥32, ≥ 32, ≥ 4, ≥ 64, ≥ 16, ≥ 16, and ≥ 16 μg/ml, respectively. For gentamicin (GEN), florfenicol (FLO), clindamycin (CLI) and azithromycin (AZI), the isolates with MICs ≥ 8 μg/ml were considered resistant.
Determination of Mechanisms of Antimicrobial Resistance
All strains tested for the antibiotic resistance were examined for the presence of molecular background of the appearing resistance. For the determination of macrolide resistance, the 23S rRNA genes mutations were detected by the use of PCR for C. jejuni (Zhou et al., 2016). For the determination of fluoroquinolone resistance, the gyrA mutations were detected by the use of the Mismatch Amplification Mutation Assay – PCR (MAMA-PCR) suitable for C. jejuni (Wieczorek, 2011). The presence of tet(O) gene associated with the resistance to tetracyclines was also detected (Wieczorek, 2011). The presence of aphA-3 gene associated with the resistance to aminoglycosides was also detected (Gibreel et al., 2004a) primers used for determination of Mechanisms of Antimicrobial Resistance are listed in Table 10.
The 23S rRNA genes PCR reaction conditions were as follows: initial denaturation at 94°C for 2 min; followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; last denaturation at 72°C for 7 min. The tet(O) gene PCR reaction conditions were as follows: initial denaturation at 94°C for 2 min; followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; last denaturation at 72°C for 7 min. The gyrA gene PCR reaction conditions were as follows: initial denaturation at 94°C for 2 min; followed by 35 cycles of 94°C for 30 s, 54°C for 30 s and 72°C for 30 s; last denaturation at 72°C for 7 min. The tet(O) gene PCR reaction conditions were as follows: initial denaturation at 94°C for 2 min; followed by 35 cycles of 94°C for 30 s, 52°C for 30 s and 72°C for 30 s; last denaturation at 72°C for 7 min. The primers used for these three genes are listed in Table 10. The PCR products were detected on 1% Agarose gels and verified by sequencing, and the sequences were analyzed to identify mutations using the BLAST program of the GenBank sequence database.
Phylogenetic Analysis
The MLST profiles were clustered with the Bionumerics software, version 7.6 by using a categorical coefficient and a graphing method called the minimum spanning tree as described before (Schouls et al., 2004). The minimum spanning tree showing the relatedness of 57 C. jejuni strains and other C. jejuni strains from the PubMLST database, which was based on the STs. Each ST is represented by a circle that is proportional to the number of isolate species comprising that ST. Circles (STs) are linked by lines indicating allelic variation. The different color of each ST indicates the animal host from which each isolate was recovered (red-human, green -wild bird, blue-chicken, yellow-dog, sky blue-monkey). Thick and short lines connect single-locus variants, thin and longer lines connect double-locus variants and dashed lines represent three or more allele differences. For MLST, a maximum neighbor difference of 2 was used to create complexes. Background shading highlights clonal complexes.
Accession Number(s)
Multilocus sequence typing sequences of the bird isolates that represented novel STs were deposited in the PubMLST database (see footnote 1) to assign new sequence types and allelic profiles.
Statistical Analysis
Statistical analysis of the prevalence of positive rate in the C. jejuni isolates was performed using the chi-squared test with SPSS version 16.0. A value of p < 0.05 was considered to be statistically significant.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
JD, HH, JL, and CW designed the project. JD and JH performed the main experiments. JD, JL, CW, and ML wrote and revised the manuscript. BJW, BW, HC, JJ, and KS conducted part of the experiments.
Funding
This work was supported by China Agriculture Research System Poultry-related Science and Technology Innovation Team of Peking (BAIC04-2018), Special Fund Project for the Introduced Leading Talents of Guangdong Academy of Sciences (2016GDASRC-0205), the External Cooperation Program of BIC, Chinese Academy of Sciences (152111KYSB20150023), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA19050204), the Wildlife-Borne Disease Surveillance Program from the State Forestry and Grassland Administration of the People’s Republic of China, and Beijing Wildlife Rescue & Rehabilitation Center, China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02433/full#supplementary-material
Footnotes
References
Alsan, M., Schoemaker, L., Eggleston, K., Kammili, N., Kolli, P., and Bhattacharya, J. (2015). Out-of-pocket health expenditures and antimicrobial resistance in low-income and middle-income countries: an economic analysis. Lancet Infect. Dis. 15, 1203–1210. doi: 10.1016/S1473-3099(15)00149-8
Avrain, L., Vernozy-Rozand, C., and Kempf, I. (2004). Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J. Appl. Microbiol. 97, 134–140. doi: 10.1111/j.1365-2672.2004.02306.x
Bang, D., Nielsen, E., Scheutz, F., Pedersen, K., Handberg, K., and Madsen, M. (2003). PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J. Appl. Microbiol. 94, 1003–1014. doi: 10.1046/j.1365-2672.2003.01926.x
Broman, T., Palmgren, H., Bergstrom, S., Sellin, M., Waldenstrom, J., Danielsson-Tham, M. L., et al. (2002). Campylobacter jejuni in black-headed gulls (Larus ridibundus): prevalence, genotypes, and influence on C. jejuni epidemiology. J. Clin. Microbiol. 40, 4594–4602. doi: 10.1128/jcm.40.12.4594-4602.2002
Broman, T., Waldenstrom, J., Dahlgren, D., Carlsson, I., Eliasson, I., and Olsen, B. (2004). Diversities and similarities in PFGE profiles of Campylobacter jejuni isolated from migrating birds and humans. J. Appl. Microbiol. 96, 834–843. doi: 10.1111/j.1365-2672.2004.02232.x
Chen, J., Sun, X.-T., Zeng, Z., and Yu, Y.-Y. (2011). Campylobacter enteritis in adult patients with acute diarrhea from 2005 to 2009 in Beijing, China. Chin. Med. J. 124, 1508–1512.
CLSI (2015). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard, 10th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Colles, F. M., Ali, J. S., Sheppard, S. K., McCarthy, N. D., and Maiden, M. C. J. (2011). Campylobacter populations in wild and domesticated Mallard ducks (Anas platyrhynchos). Environ. Microbiol. Rep. 3, 574–580. doi: 10.1111/j.1758-2229.2011.00265.x
Colles, F. M., McCarthy, N. D., Howe, J. C., Devereux, C. L., Gosler, A. G., and Maiden, M. C. J. (2009). Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European Starling). Environ. Microbiol. 11, 258–267. doi: 10.1111/j.1462-2920.2008.01773.x
Connell, S. R., Trieber, C. A., Dinos, G. P., Einfeldt, E., Taylor, D. E., and Nierhaus, K. H. (2003). Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22, 945–953. doi: 10.1093/emboj/cdg093
de Boer, P., Wagenaar, J., Achterberg, R., van Putten, J., Schouls, L., and Duim, B. (2002). Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44, 351–359. doi: 10.1046/j.1365-2958.2002.02930.x
Dingle, K., Colles, F., Wareing, D., Ure, R., Fox, A., Bolton, F., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39, 14–23.
European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC) (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:e05077. doi: 10.2903/j.efsa.2017.5077
Franiczek, R., Sobieszczańska, B., Turniak, M., Kasprzykowska, U., Krzyzanowska, B., Jermakow, K., et al. (2012). ESBL-producing Escherichia coli isolated from children with acute diarrhea - antimicrobial susceptibility, adherence patterns and phylogenetic background. Adv. Clin. Exp. Med. 21, 187–192.
French, N., Midwinter, A., Holland, B., Collins-Emerson, J., Pattison, R., Colles, F., et al. (2009). Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children’s playgrounds. Appl. Environ. Microbiol. 75, 779–783. doi: 10.1128/AEM.01979-08
Friedman, C., Hoekstra, R., Samuel, M., Marcus, R., Bender, J., Shiferaw, B., et al. (2004). Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3), S285–S296.
Gardner, T. J., Fitzgerald, C., and Xavier, C. (2012). Outbreak of Campylobacteriosis associated with consumption of raw peas (vol 53, pg 26, 2011). Clin. Infect. Dis. 54, 1040–1040. doi: 10.1093/cid/cir249
Gibreel, A., Skold, O., and Taylor, D. E. (2004a). Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb. Drug Resist. 10, 98–105. doi: 10.1089/1076629041310127
Gibreel, A., Tracz, D. M., Nonaka, L., Ngo, T. M., Connell, S. R., and Taylor, D. E. (2004b). Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 48, 3442–3450. doi: 10.1128/aac.48.9.3442-3450.2004
Griekspoor, P., Engvall, E. O., Olsen, B., and Waldenstrom, J. (2010). Multilocus sequence typing of Campylobacter jejuni from broilers. Vet. Microbiol. 140, 180–185. doi: 10.1016/j.vetmic.2009.07.022
Kapperud, G., Espeland, G., Wahl, E., Walde, A., Herikstad, H., Gustavsen, S., et al. (2003). Factors associated with increased and decreased risk of Campylobacter infection: a prospective case-control study in Norway. Am. J. Epidemiol. 158, 234–242. doi: 10.1093/aje/kwg139
Karaiskos, I., and Giamarellou, H. (2014). Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin. Pharmacother. 15, 1351–1370. doi: 10.1517/14656566.2014.914172
Klena, J., Parker, C., Knibb, K., Ibbitt, J., Devane, P., Horn, S., et al. (2004). Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J. Clin. Microbiol. 42, 5549–5557. doi: 10.1128/jcm.42.12.5549-5557.2004
Konkel, M., Gray, S., Kim, B., Garvis, S., and Yoon, J. (1999). Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J. Clin. Microbiol. 37, 510–517.
Kovanen, S., Rossi, M., Pohja-Mykra, M., Nieminen, T., Raunio-Saarnisto, M., Sauvala, M., et al. (2019). Population genetics and characterization of Campylobacter jejuni isolates from western jackdaws and game birds in Finland. Appl. Environ. Microbiol. 85:e02365-18. doi: 10.1128/AEM.02365-18
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A., Wertheim, H., Sumpradit, N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/S1473-3099(13)70318-9
Lund, M., and Madsen, M. (2006). Strategies for the inclusion of an internal amplification control in conventional and real time PCR detection of Campylobacter spp. in chicken fecal samples. Mol. Cell. Probes 20, 92–99. doi: 10.1016/j.mcp.2005.10.002
Ma, H., Su, Y., Ma, L., Ma, L., Li, P., Du, X., et al. (2017). Prevalence and characterization of Campylobacter jejuni isolated from retail chicken in Tianjin, China. J. Food Prot. 80, 1032–1040. doi: 10.4315/0362-028X.JFP-16-561
Nachamkin, I., Bohachick, K., and Patton, C. M. (1993). Flagellin gene typing of campylobacter-jejuni by restriction-fragment-length-polymorphism analysis. J. Clin. Microbiol. 31, 1531–1536.
Olkkola, S., Nykasenoja, S., Raulo, S., Llarena, A. K., Kovanen, S., Kivisto, R., et al. (2016). Antimicrobial resistance and multilocus sequence types of Finnish Campylobacter jejuni isolates from multiple sources. Zoonoses Public Health 63, 10–19. doi: 10.1111/zph.12198
Oravcova, V., Zurek, L., Townsend, A., Clark, A., Ellis, J., Cizek, A., et al. (2014). American crows as carriers of vancomycin-resistant enterococci with vanA gene. Environ. Microbiol. 16, 939–949. doi: 10.1111/1462-2920.12213
Palmer, S. R., Gully, P. R., White, J. M., Pearson, A. D., Suckling, W. G., Jones, D. M., et al. (1983). Water-Borne outbreak of Campylobacter gastroenteritis. Lancet 1, 287–290. doi: 10.1016/s0140-6736(83)91698-7
Payot, S., Bolla, J. M., Corcoran, D., Fanning, S., Megraud, F., and Zhang, Q. J. (2006). Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 8, 1967–1971. doi: 10.1016/j.micinf.2005.12.032
Pearson, B. M., Gaskin, D. J. H., Segers, R., Wells, J. M., Nuijten, P. J. A., and van Vliet, A. H. M. (2007). The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J. Bacteriol. 189, 8402–8403. doi: 10.1128/jb.01404-07
Pickett, C. L., Pesci, E. C., Cottle, D. L., Russell, G., Erdem, A. N., and Zeytin, H. (1996). Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp cdtB genes. Infect. Immun. 64, 2070–2078.
Purdy, D., Buswell, C. M., Hodgson, A. E., McAlpine, K., Henderson, I., and Leach, S. A. (2000). Characterisation of cytolethal distending toxin (CDT) mutants of Campylobacter jejuni. J. Med. Microbiol. 49, 473–479. doi: 10.1099/0022-1317-49-5-473
Ramonaite, S., Novoslavskij, A., Zakariene, G., Aksomaitiene, J., and Malakauskas, M. (2015). High prevalence and genetic diversity of Campylobacter jejuni in wild crows and pigeons. Curr. Microbiol. 71, 559–565. doi: 10.1007/s00284-015-0881-z
Schouls, L. M., Reulen, S., Duim, B., Wagenaar, J. A., Willems, R. J. L., Dingle, K. E., et al. (2003). Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41, 15–26. doi: 10.1128/jcm.41.1.15-26.2003
Schouls, L. M., van der Heide, H. G. J., Vauterin, L., Vauterin, P., and Mooi, F. R. (2004). Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186, 5496–5505. doi: 10.1128/jb.186.16.5496-5505.2004
Sen, K., Lu, J., Mukherjee, P., Berglund, T., Varughese, E., and Mukhopadhyay, A. K. (2018). Campylobacter jejuni colonization in the crow gut involves high deletion within the cytolethal distending toxin gene cluster. Appl. Environ. Microbiol. 84:e1893-17.
Sheppard, S. K., Dallas, J. F., Strachan, N. J. C., MacRae, M., McCarthy, N. D., Wilson, D. J., et al. (2009). Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis 48, 1072–1078. doi: 10.1086/597402
Smith, J. L., and Fratamico, P. M. (2010). Fluoroquinolone resistance in Campylobacter. J. Food Prot. 73, 1141–1152. doi: 10.4315/0362-028x-73.6.1141
Waldenstrom, J., Axelsson-Olsson, D., Olsen, B., Hasselquist, D., Griekspoor, P., Jansson, L., et al. (2010). Campylobacter jejuni colonization in wild birds: results from an infection experiment. PLoS One 5:8. doi: 10.1371/journal.pone.0009082
Waldenstrom, J., Broman, T., Carlsson, I., Hasselquist, D., Achterberg, R. P., Wagenaar, J. A., et al. (2002). Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68, 5911–5917. doi: 10.1128/aem.68.12.5911-5917.2002
Waldenstrom, J., On, S. L. W., Ottvall, R., Hasselquist, D., and Olsen, B. (2007). Species diversity of Campylobacteria in a wild bird community in Sweden. J. Appl. Microbiol. 102, 424–432.
Wang, X., Wang, J., Sun, H., Xia, S., Duan, R., Liang, J., et al. (2015). Etiology of childhood infectious diarrhea in a developed region of china: compared to childhood diarrhea in a developing region and adult diarrhea in a developed region. PLoS One 10:e0142136. doi: 10.1371/journal.pone.0142136
Wei, B., Kang, M., and Jang, H.-K. (2018). Genetic characterization and epidemiological implications of Campylobacter isolates from wild birds in South Korea. Transbound. Emerg. Dis. 66, 56–65. doi: 10.1111/tbed.12931
Wieczorek, K. (2011). Resistance to quinolones and tetracycline and its molecular background among campylobacter strains isolated in Poland. Bull. Vet. Inst. Pulawy 55, 613–618.
Wilson, D., Bell, J., Young, V., Wilder, S., Mansfield, L., and Linz, J. (2003). Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149, 3603–3615. doi: 10.1099/mic.0.26531-0
Wilson, D. J., Gabriel, E., Leatherbarrow, A. J. H., Cheesbrough, J., Gee, S., Bolton, E., et al. (2008). Tracing the source of Campylobacteriosis. PLoS Genet. 4:9.
Zhou, J. Y., Zhang, M. J., Yang, W. N., Fang, Y. Q., Wang, G. Q., and Hou, F. Q. (2016). A seventeen-year observation of the antimicrobial susceptibility of clinical Campylobacter jejuni and the molecular mechanisms of erythromycin-resistant isolates in Beijing, China. Int. J. Infect. Dis. 42, 28–33. doi: 10.1016/j.ijid.2015.11.005
Keywords: emergence, Campylobacter jejuni, wild birds, MLST, multi-resistance, CDT gene cluster
Citation: Du J, Luo J, Huang J, Wang C, Li M, Wang B, Wang B, Chang H, Ji J, Sen K and He H (2019) Emergence of Genetic Diversity and Multi-Drug Resistant Campylobacter jejuni From Wild Birds in Beijing, China. Front. Microbiol. 10:2433. doi: 10.3389/fmicb.2019.02433
Received: 26 February 2019; Accepted: 09 October 2019;
Published: 29 October 2019.
Edited by:
Ziad Daoud, University of Balamand, LebanonReviewed by:
Heriberto Fernandez, Austral University of Chile, ChilePhilippe Lehours, Université Bordeaux Segalen, France
Zhangqi Shen, China Agricultural University (CAU), China
Marja-Liisa Hänninen, University of Helsinki, Finland
Copyright © 2019 Du, Luo, Huang, Wang, Li, Wang, Wang, Chang, Ji, Sen and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxuan He, aGVoeEBpb3ouYWMuY24=
†These authors have contributed equally to this work
 Juan Du1,2†
Juan Du1,2† Chengmin Wang
Chengmin Wang Meng Li
Meng Li Keya Sen
Keya Sen Hongxuan He
Hongxuan He