- 1Bacterial Infection Disease Control of Institute, Shandong Center for Disease Control and Prevention, Jinan, China
- 2Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 3Department of Environment and Health, School of Public Health, Shandong University, Jinan, China
- 4Department of Supervise Sampling, Shandong Institute for Food and Drug Control, Jinan, China
- 5Shandong Academy of Clinical Medicine, Shandong Provincial Hospital, Jinan, China
Raoultella ornithinolytica is an opportunistic pathogen of the Enterobacteriaceae family and has been implicated in nosocomial infections in recent years. The aim of this study was to characterize a carbapenemase-producing R. ornithinolytica isolate and three extended-spectrum β-lactamase (ESBL)-producing R. ornithinolytica isolates from stool samples of adults in a rural area of Shandong Province, China. The species were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and 16S rDNA sequence analysis. Antimicrobial susceptibility test showed that all four isolates were multidrug-resistant (MDR). The whole genome sequence (WGS) of these isolates was determined using an Illumina HiSeq platform, which revealed MDR-related genes. The S1 nuclease-pulsed-field gel electrophoresis (S1-PFGE) was used to characterize the plasmids carried by the R. ornithinolytica isolates. The blaNDM-1 and blaCTX-M-3 genes were probed using Southern blotting, which confirmed the location of both genes on the same plasmid with molecular weight of 336.5–398.4 kb. The transferability of blaNDM-1 and blaCTX-M was also confirmed by conjugation assays. Finally, BLAST analysis of both genes showed that mobile genetic elements were associated with the spread of drug resistance genes. Taken together, we report the presence of conjugative blaNDM-1 and blaCTX-M plasmids in R. ornithinolytica isolates from healthy humans, which indicate the possibility of inter-species transfer of drug resistance genes. To the best of our knowledge, this is the first study to isolate and characterize carbapenemase-producing R. ornithinolytica and ESBL-producing R. ornithinolytica isolates from healthy human hosts.
Introduction
Raoultella is a genus of encapsulated Gram-negative aerobic bacilli of the Enterobacteriaceae family (Luo et al., 2017) that was initially part of the genus Klebsiella, but later reclassified based on the 16S rDNA sequence and the rpoB, gyrA, and gyrB genes (Drancourt et al., 2001). Raoultella ornithinolytica is one of the three species of the genus Raoultella (Singh et al., 2017) and naturally exists in the soil, water, and plants (Ayoade et al., 2018). R. ornithinolytica can cause pneumonia, biliary or urinary tract infections and bacteremia and such cases are being increasingly reported (Sekowska et al., 2015; Van Cleve et al., 2018). Both organ-specific and systemic R. ornithinolytica infections can be life-threatening, especially in cancer patients after abdominal surgery (Hajjar et al., 2018). Therefore, it is vital to understand the pathogenic potential of R. ornithinolytica isolates in humans (De Petris and Ruffini, 2018). Cephalosporins, quinolones, and carbapenems are routinely used against this pathogen, and multi-drug resistant (MDR) R. ornithinolytica isolates have been reported in recent years (Wang et al., 2019).
Widespread use of third-generation cephalosporins and other β-lactam antibiotics in the past decades have led to the emergence of third-generation cephalosporin-resistant bacteria that produce extended spectrum β-lactamases (ESBLs) and AmpC β-lactamases that are, respectively, encoded by plasmids and chromosomes (Kola et al., 2012; Dohmen et al., 2015). Consistent with this, infections caused by the ESBL-producing Gram-negative bacilli (GNB) have become increasingly prevalent worldwide, both in the healthcare and community settings, and pose significant therapeutic challenges (Wang et al., 2015). The class A serine CTX-M-type is the most common plasmid-encoded ESBLs that are produced by drug-resistant pathogens (He et al., 2016). The genetic elements encoding these CTX-M enzymes constantly evolve via random mutations and recombination between different resistance genes (Canton et al., 2012). A genetic epidemiological study on ESBLs found that blaCTX-M has replaced blaSHV and blaTEM as the most common ESBL-encoding genes (Xia et al., 2014). While CTX-M-15 and CTX-M-14 are the most prevalent ESBLs worldwide (He et al., 2016), blaCTX-M-14 has been identified as the most prevalent ESBL gene in China, and epidemiological surveillance in Asia, Latin America, and Europe has revealed a dramatic increase in cephalosporin-resistant Escherichia coli and Klebsiella spp. strains due to spread of the CTX-M ESBLs (Zhang et al., 2016).
Carbapenem-resistant Enterobacteriaceae (CRE) is a serious public health concern worldwide because of its rapid spread and limited therapeutic drugs (Zheng et al., 2018). Metallo-β-lactamases (MBLs) are produced by many species of Gram-negative bacteria and confer resistance to carbapenems, cephalosporins, and penicillins except monobactams (Tada et al., 2019). New Delhi Metallo-beta-Lactamase 1 (NDM-1) is a plasmid-associated Ambler class B β-lactamase/carbapenemase that was first reported in clinical E. coli and Klebsiella pneumoniae isolates from an Indian patient in Sweden in 2008 (Yong et al., 2009). Subsequent cases of carbapenemase-producing isolates have since been reported in Britain, Australia, India, Russia, etc. (Yong et al., 2009; Kumarasamy et al., 2010; Bocanegra-Ibarias et al., 2017), and clinical isolates of R. ornithinolytica from urethral effluent, fester, and rectum samples have recently been found to produce this enzyme (Li et al., 2012; Khajuria et al., 2013; Zhou et al., 2013; Zheng et al., 2015; Paskova et al., 2018).
R. ornithinolytica infections are largely nosocomial and have rarely been reported in a healthy community (Seng et al., 2016). Nevertheless, the high rates of antimicrobial resistance in R. ornithinolytica isolates should be characterized in order to provide a basis for treating infections. To this end, we conducted a cross-sectoral study as part of the Sino-Swedish Integrated Multisectoral Partnership for Antibiotic Resistance Containment (IMPACT) in the Shandong Province in China using a One Health approach. The aim of this project is to study the relationship between the development of drug resistance in human (symbiotic and clinical), zoonotic, food, and environmental isolates of R. ornithinolytica. We identified NDM-1 and ESBL-producing R. ornithinolytica strains from healthy subjects and analyzed the drug resistance phenotypes and underlying mechanisms. To the best of our knowledge, this is the first report detailing the presence of an NDM-producing R. ornithinolytica strain in the human gut microbiota.
Methods
Bacterial Isolation and Identification
A total of 1,380 fecal samples were collected from healthy people in rural communities in July 2017 according to a previously described sampling procedure (Sun et al., 2018). Briefly, the samples were collected into ESwab tubes (Copan, Brescia, Italy) and stored at −80°C until cultivation. After thawing, the fecal samples were cultured on ChromID CARBA agar and ChromID ESBL agar plates (bioMérieux, Marcy l’Etoile, France) for 18 h at 37°C to respectively screen for the carbapenemase- and ESBL-producing R. ornithinolytica strains. The suspected R. ornithinolytica colonies identified based on color and morphology were picked and sub-cultured on CHROMagar Orientation agar (CHROMagar Company, Paris, France) overnight at 37°C. The resulting isolates were identified using MALDI-TOF MS and then confirmed by 16S rDNA sequence analysis against the bacterial 16S rDNA gene sequence in GenBank. The genomic average nucleotide identity (ANI) was calculated as described previously (Jiang et al., 2018).
DNA Extraction and PCR
DNA was extracted from pure cultures of R. ornithinolytica using a Gentra Puregene Yeast/Bact. Kit (QIAGEN, Hilden, Germany), and the 16S rDNA sequences were amplified by PCR with primers designed using the Primer 5.0 software (3′-AGAGTTTGATCCTGGCTCAG-5′/3′-GGTTACCTTGTTAGGACTT-5′). The optimized cycling conditions were: initial denaturation at 94°C for 5 min followed by 30 cycles of amplification each with 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min and final extension at 72°C for 5 min. The PCR product was detected by capillary electrophoresis as described previously (Zhang et al., 2019). The blaNDM-1, blaCTX-M-3, and blaCTX-M-14 genes were amplified by PCR using primers published previously (Shimizu et al., 2017; Chetri et al., 2019), and the products were confirmed by capillary electrophoresis followed by Sanger sequencing.
Antimicrobial Resistance Test
To verify carbapenemase production by the isolates, resistance against imipenem, ertapenem, and/or meropenem were determined using the modified Hodge test. ESBL-production was verified using the double disk diffusion method with cefotaxime, ceftazidime, and/or clavulanic acid according to Clinical and Laboratory Standards Institute (CLSI, 2017).
Antimicrobial Susceptibility Test
A total of 17 antibiotics belonging to 13 antimicrobial classes were tested, including cephalosporins (cefotaxime and ceftazidime), cephamycins (cefoxitin), β-lactam/β-lactamase inhibitor complexes (amoxicillin-clavulanate and piperacillin-tazobactam), carbapenems (imipenem and meropenem), penicillins (ampicillin), aminoglycosides (gentamicin and amikacin), fluoroquinolones (ciprofloxacin), folate metabolic pathway inhibitors (trimethoprim-sulfamethoxazole), tetracycline, chloramphenicols (florfenicol), colistin, furantoin, and tigecycline. The minimal inhibitory concentrations (MICs) of colistin and tigecycline were determined by the broth microdilution method and of other antibiotics using the agar dilution method. The results were interpreted according to CLSI (2017) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) (version 8.1, 2018) guidelines. E. coli ATCC25922 was used as the quality control strain. Since clinical breakpoints of florfenicol are not available for Enterobacteriaceae in EUCAST or CLSI, the resistance breakpoint of >16 mg/L was selected based on the epidemiological cut-off values for the closely related E. coli and Salmonella spp. (Chi et al., 2019).
Whole-Genome Sequencing
Whole-genome sequencing was performed on the extracted DNA by Sangon Biotech (Shanghai, China) using the Illumina HiSeq sequencing platform. The quality of the high-throughput sequence data was assessed by FastQC1. SPAdes 3.11.0 was used for assembling raw sequences (Hu et al., 2011). All draft genomes were deposited in the NCBI database under accession number VJYE00000000-VJYH00000000. Acquired antimicrobial resistance genes and plasmid replicons were identified using ResFinder 2.1 and PlasmidFinder 1.32, respectively. The gene sequences surrounding blaNDM-1 and blaCTX-M were annotated using RAST3 and Easyfig 2.2.3 (Chi et al., 2019). To find the core genes of the R. ornithinolytica genomes, Roary was used for SNPs analysis (Page et al., 2015). Maximum likelihood-based phylogenetic reconstruction was performed with RAxML version 8.2.10 (Stamatakis, 2014) and visualized with FastTree (Price et al., 2009).
S1 Nuclease-Pulsed-Field Gel Electrophoresis and Southern Blotting
The location of blaNDM-1, blaCTX-M-3, and blaCTX-M-14 on the plasmids was validated by S1-PFGE and southern blotting. Briefly, the isolates were embedded in 10 g/L Seakem Gold gel, digested with endonuclease S1 nuclease (TakaRa, Dalian, China), and subjected to pulsed-field gel electrophoresis (Parameters: 14°C, voltage 6 V/cm, electric field angle 120°, conversion time 2.16–63.8 s, and electrophoresis 16 h). The DNA fragments were transferred horizontally to a nylon membrane (Millipore, USA), and hybridized with three digoxin-labeled probes obtained by PCR amplification (Yu et al., 2002) and the Dig High Prime DNA Labeling and Detection Starter Kit (Roche Diagnostics). The genomic DNA of Salmonella enterica serovar Braenderup H9812 strain cut with XbaI was used as the DNA marker.
Conjugation Assay
The horizontal transferability of blaNDM-1, blaCTX-M-3, and blaCTX-M-14 were examined using conjugation assay using E. coli J53 (azide-resistant) or E. coli EC600 (rifampicin-resistant) as the recipient strains. The recipient bacteria and the target single colony were inoculated into 2 ml LB liquid medium and cultured at 37°C with constant shaking for about 6.5 h till the logarithmic growth phase. The recipient and donor bacteria culture broths were mixed at 1:2 v/v (200 μl and 400 μl), inoculated in 2 ml LB liquid medium, and incubated at 37°C for 12 h. The following day, 100 μl of the culture broth was uniformly spread on sodium azide-resistant agar containing cefotaxime (imipenem) and rifampicin and cultured at 37°C for 18 h. The single colonies were picked and purified overnight. Conjugants were identified by the presence of antibiotic resistance genes that were detected using PCR as described.
Results
Multidrug-Resistant Raoultella ornithinolytica Strains Were Isolated From the Human Fecal Samples
One carbapenemase-producing (ROF058) and three ESBL-producing (ROE007, ROE058, and ROI014) R. ornithinolytica strains were isolated from 1,380 fecal samples (Table 1) and confirmed by MALDI-TOF MS, 16S rDNA sequencing, and ANI analysis (Figure 1A). Phylogenetic analysis revealed that isolates ROE007 and ROE058 are clonally related although they were recovered from different villagers in the same natural village (Figure 1B). The blaNDM-1, blaCTX-M-3, and blaCTX-M-14 genes were subsequently identified in all the strains, and multidrug resistance was confirmed by antimicrobial susceptibility tests. ROF058 was resistant to 12 antibiotics other than carbapenem, but sensitive to amikacin, tigecycline, furantoin, colistin, and trimethoprim-sulfamethoxazole. The resistance profiles of the three ESBL-producing isolates were similar, and all were resistant to gentamicin, tetracycline, cefotaxime, trimethoprim-sulfamethoxazole, and ampicillin. Furthermore, all trans-conjugants exhibited MDR phenotypes similar to the donor strain (Table 1).
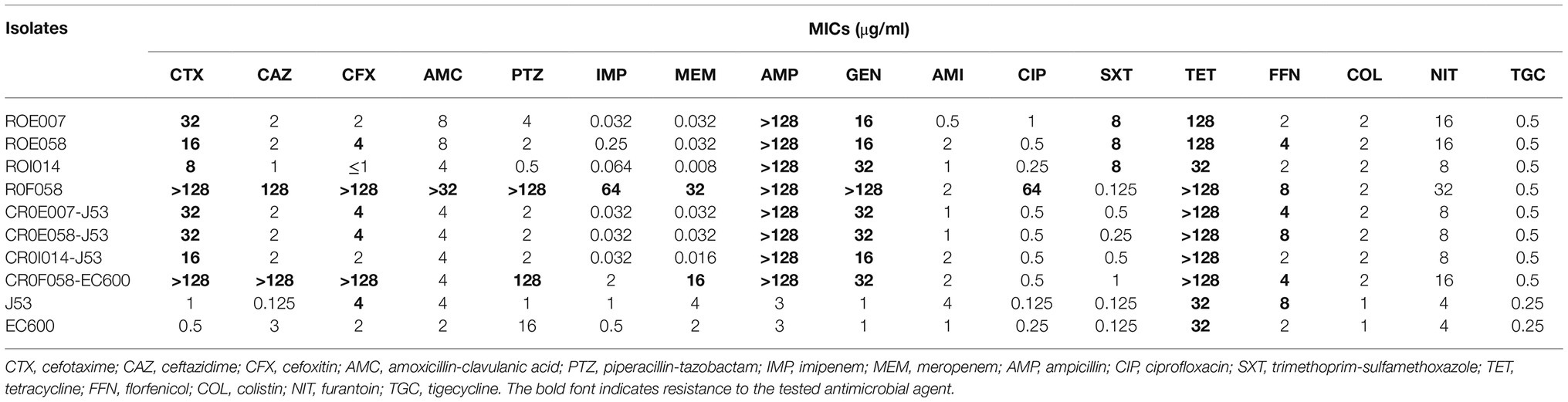
Table 1. The minimum inhibitory concentrations of tested antimicrobial agents against the R. ornithinolytica isolates and the respective conjugants.
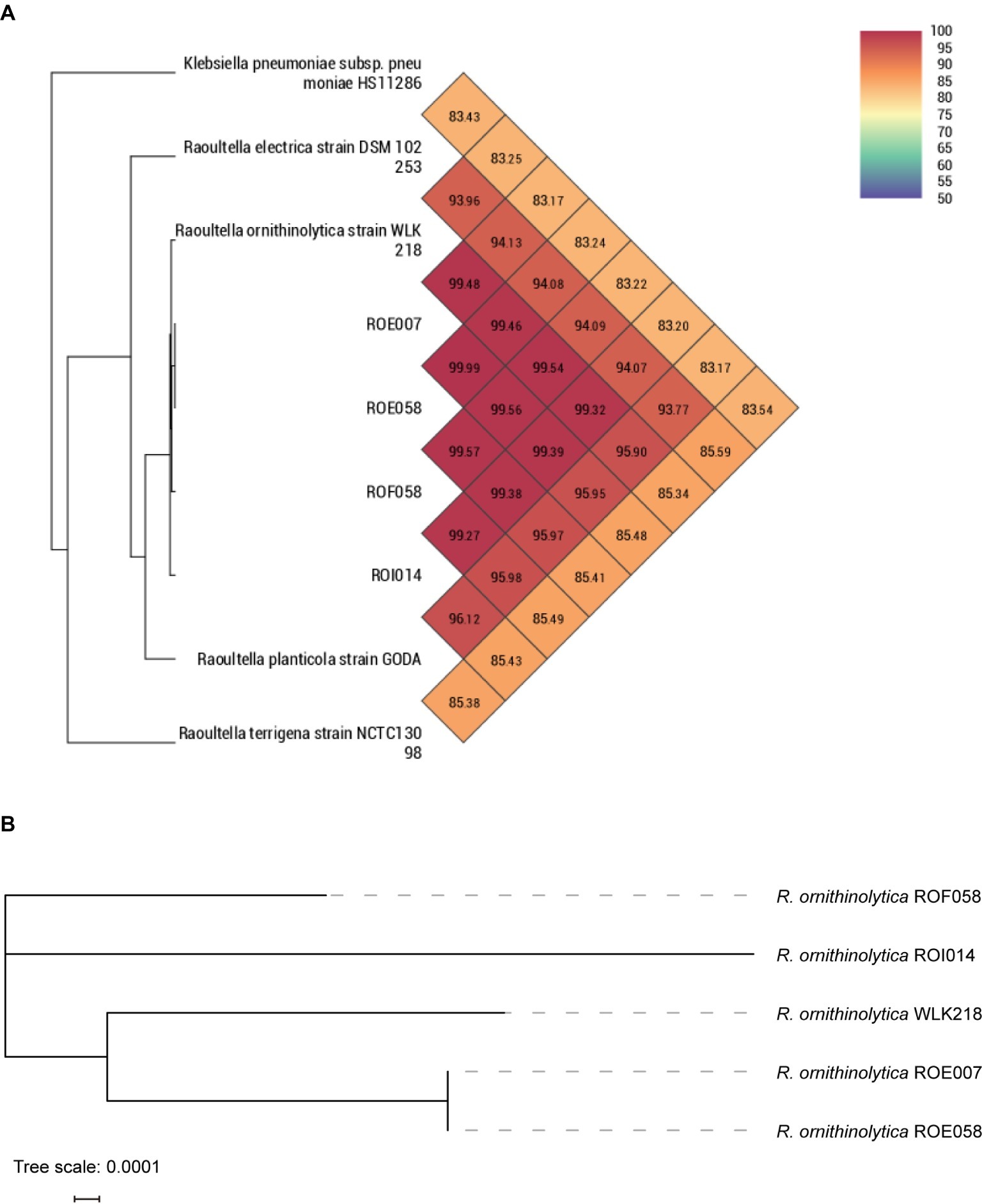
Figure 1. Phylogenetic comparison depicting the relationship of R. ornithinolytica strain isolated in this study with other Raoultella species. (A) ANI analysis of R. ornithinolytica isolates with other Raoultella species. (B) Core genome phylogeny for the R. ornithinolytica isolates.
The Raoultella ornithinolytica Strains Harbor Multidrug-Resistant Genes
MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, and XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (Magiorakos et al., 2012). Whole genome sequencing showed that ROF058 is an extensively drug-resistant (XDR) strain, while ROE007, ROE058, and ROI014 are MDR strains (Table 1). In addition to the carbapenemase-encoding blaNDM-1, ROF058 also carries genes encoding for other ESBLs (blaCTX-M-3, blaOXA-1, blaTEM-1B, and blaDHA-1), as well as resistance factors against, aminoglycosides [aac(3)-IId], rifampicins (arr-3), chloramphenicols (catB3), tetracyclines [tet(D)], quinolones [qnrB4 and aac(6′)-Ib-cr], fosfomycin (fosA), and sulfonamides (sul1) (Figure 2). The remaining isolates harbored genes encoding the ESBLs (blaCTX-M-14), quinolones (qnrS1), aminoglycosides [aac(3)-IId], sulfonamides (sul1), tetracyclines [tet(A)], sulfanilamides (dfrA1), and fosfomycins (fosA) resistance genes.
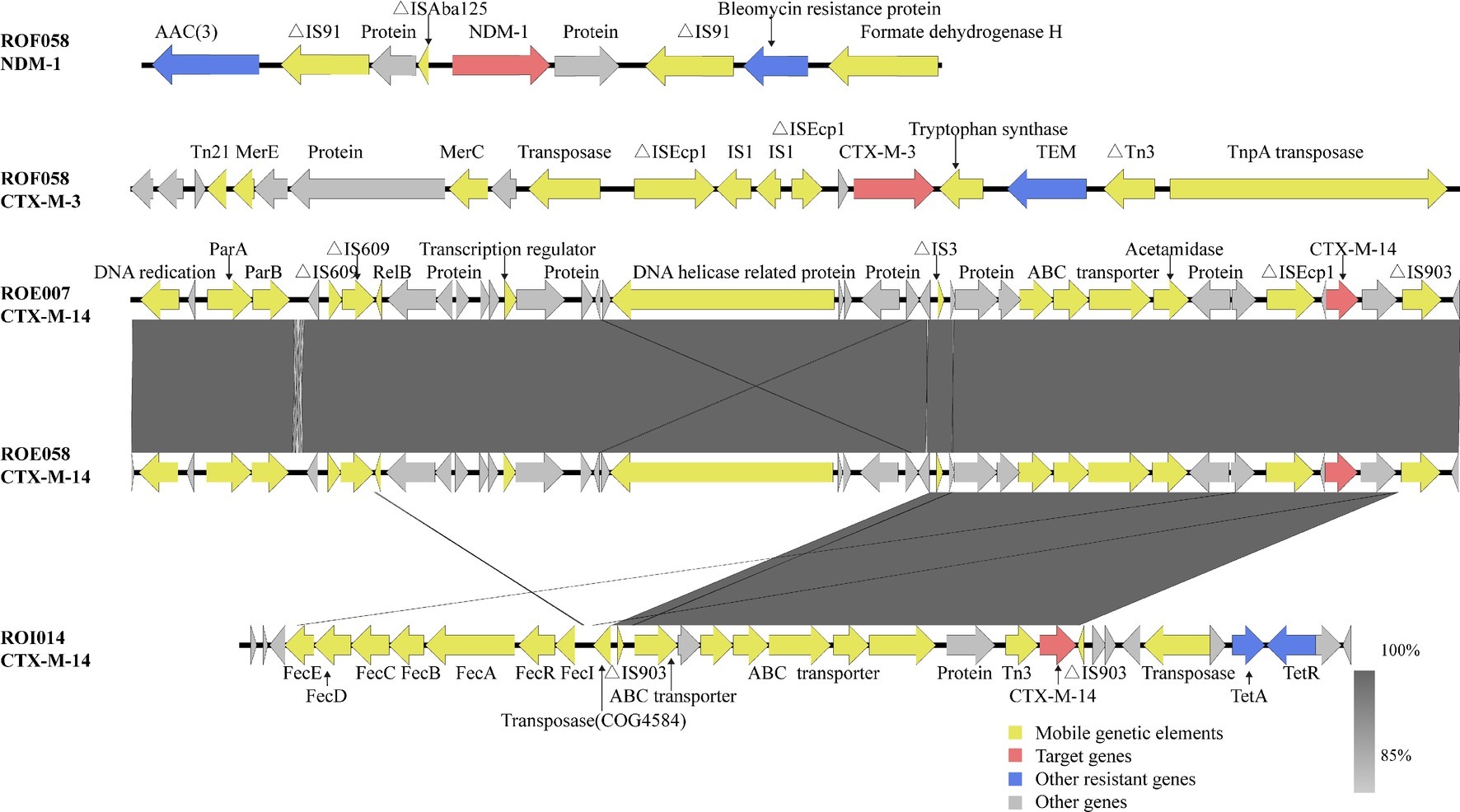
Figure 2. Genetic analysis of the blaNDM-1 and blaCTX-M genes. Arrows represent direction of transcription. Red open reading frames (ORFs) indicate NDM-1/CTX-M, blue ORFs drug resistance-associated proteins, yellow ORFs mobile genetic elements, and gray ORFs other proteins or proteins of unknown function.
The Multidrug-Resistant Genes Are Located on Mobile Genetic Elements
Whole genome sequencing of strain ROF058 generated 5,747,150 clean reads, which were then assembled to 209 contigs with a GC content of 51.38%; WGS of strain R0E007 generated 5,214,213 clean reads, which were then assembled to 203 contigs with a GC content of 49.36%; WGS of strain ROE058 generated 5,235,762 clean reads, which were then assembled to 199 contigs with a GC content of 50.28%; WGS of strain ROI014 generated 5,510,178 clean reads, which were then assembled to 215 contigs with a GC content of 54.66%. The plasmids in the different isolates were identified using the Center for Genomic Epidemiology program (Luo et al., 2017), which showed that ROF058 harbored the IncFIB(K)-type plasmid, ROE007 and ROE058 contained IncFIB(K) and IncFII-type plasmids, and ROI014 carried IncFIB(K) and IncFII plasmids. In addition, S1-PFGE and southern blot hybridization revealed three different plasmids in the R. ornithinolytica isolates ranging from 104.5 to 398.4 kb (Figure 3). ROFO58 contained the larger plasmid (336.5–398.4 kb) harboring the blaNDM-1 and blaCTX-M-3 genes. R0E007 and ROE058 had identical plasmid profiles, and the blaCTX-M-14 gene was located on a 138.9 kb plasmid in both isolates. The blaCTX-M-14 gene in ROI014 was located on a 216.9–244.4 kb plasmid. Furthermore, the blaNDM-1 and blaCTX-M-3 genes could be transferred from each of the isolates into recipient E. coli strains via conjugation (Table 1), which was confirmed by PCR (data not shown).
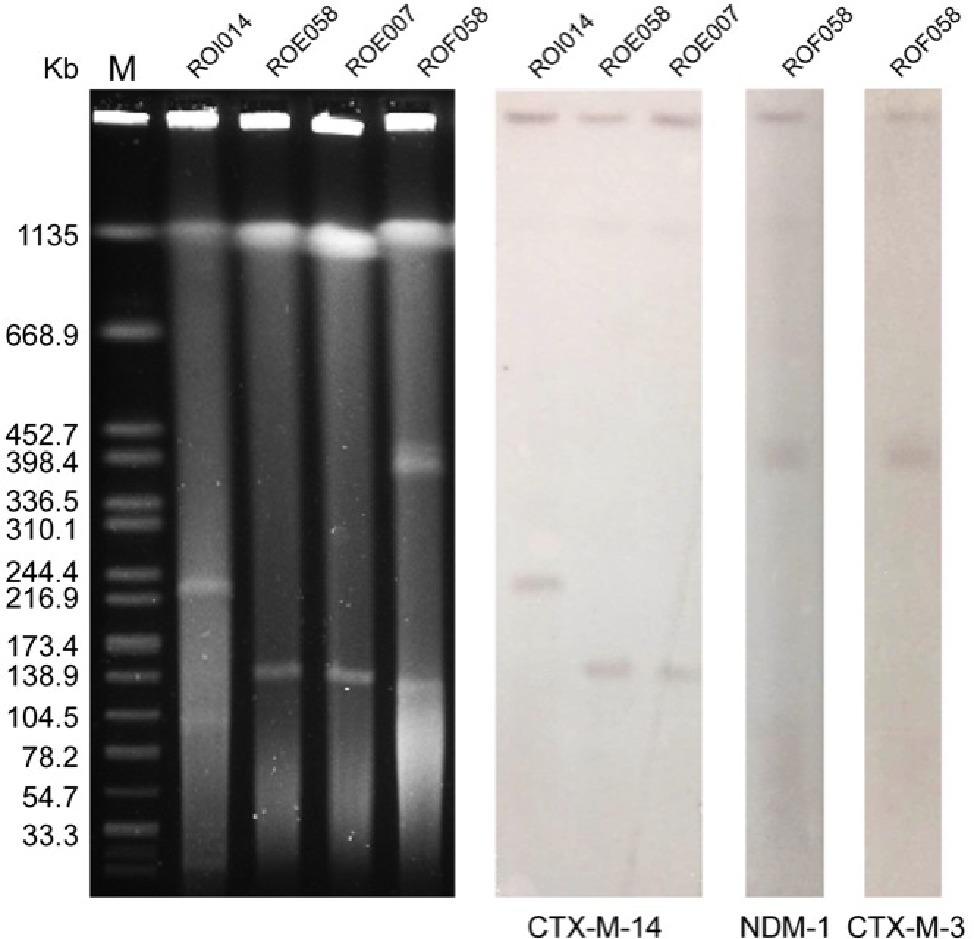
Figure 3. S1-PFGE analysis of the R. ornithinolytica isolates and southern hybridization. Lane M, molecular weight marker Salmonella Braenderup H9812; Line 1, strain ROI014; Line 2, ROEO58; Line 3, strain ROE007; Line 4, strain ROF058; Line 5, CTX-M-14 hybridization for ROI014; Line 6, CTX-M-14 hybridization for ROE058; Line 7, CTX-M-14 hybridization for ROE007; Line 8, NDM-1 hybridization for ROFO58; Line 9, CTX-M-3 hybridization for ROFO58.
Discussion
Intermittent but lethal nosocomial infections by R. ornithinolytica have been increasingly reported in the past decade (De Petris and Ruffini, 2018), and the advent of techniques like MALDI-TOF MS has enabled identification of new isolates (De Jong et al., 2013; Seng et al., 2016). Notable reports of R. ornithinolytica infections include sepsis in a patient with mitochondrial disease in Spain (Sanchez-Codez et al., 2019), urinary tract infection in a pediatric patient in Turkey (Buyukcam et al., 2019), and ventilator associated pneumonia in two immunocompetent trauma patients in USA (Van Cleve et al., 2018). R. ornithinolytica has also been implicated in joint infections and appendicitis in the clinical setting (Beye et al., 2018; Hajjar et al., 2018). In addition, MDR strains of this pathogen have emerged in recent years, primarily due to the spread of the resistance genes. The first NDM-1-producing R. ornithinolytica strain was isolated from the pus of hospitalized patients in India in 2013 (Khajuria et al., 2013) and was followed by reports of resistant R. ornithinolytica in USA, China, India, Spain, Korea, Bangladesh, and Brazil. In addition, some resistant strains have also been isolated from animals, vegetables, wastewater, and river water. The major drug resistance genes identified in these isolates include blaNDM-1, blaKPC-2, mcr-1, blaCTX-M-15, blaIMP-4, and blaOXA-1 (Zhou et al., 2014; Hasan et al., 2015; Zheng et al., 2015; Hernandez-Garcia et al., 2018; Miao et al., 2018; Yoon et al., 2018; Carramaschi et al., 2019; Wang et al., 2019). However, little is known regarding the carbapenemase- and ESBL-producing R. ornithinolytica strains. To the best of our knowledge, this is the first report on resistant R. ornithinolytica isolated from human gut microbiota.
Studies show that bacterial strains producing NDM-1 are resistant to most of the antibiotics used clinically and are only sensitive to a few such as colistin and tigecycline (Khan et al., 2017). However, Wang et al. (2018) described a strain of K. pneumoniae isolated from both humans and animals that was positive for the NDM-1 gene and showed resistance to polymyxin and colistin (Wang et al., 2018). Consistent with this, our drug susceptibility test showed that ROFO58 was resistant to cefotaxime, ceftazidime, gentamicin, tetracycline, and ciprofloxacin. R. ornithinolytica is naturally resistant to aminopenicillin due to the presence of the chromosomal class A β-lactamase gene (Vasaikar et al., 2017), and all isolates did exhibit resistance to ampicillin. The presence of drug resistance genes strongly correlated with resistant phenotypes. Therefore, the WGS approach can rapidly detect antibiotic resistance genes that have been annotated and predict drug-resistance in a particular isolate. Although all trans-conjugants showed an MDR phenotype similar to that of the donor strain in our study, the NDM-1-producing isolate had lower resistance to gentamicin and were sensitive to imipenem, ciprofloxacin, and amoxicillin-clavulanic acid. This could be due to factors other than the resistance genes. For example, studies have shown that the same resistance gene confers different susceptibility patterns when under the control of different promoters (Zhou et al., 2019).
A previous report showed that the blaNDM-1 gene in Enterobacteriaceae is located on a rapidly transferable 50–200 kb plasmid belonging to several incompatibility groups such as IncL/M, IncHI1, IncFIIs, IncF, or untypable (Ahmad et al., 2018). In this study, the plasmid carrying blaNDM-1 was larger than 300 kb and of type IncFIB. Furthermore, this plasmid could be transferred from R0F058 to E. coli EC600 but not to E. coli J53. We surmised therefore that the plasmid was rare and selectively transferred. Epidemiological and genetic studies have shown that plasmid ligation and transposition of mobile genetic elements play an important role in the horizontal transmission of blaNDM-1 (Khajuria et al., 2013). BLAST analysis showed the presence of the aminoglycoside acetyltransferase gene [aac(3)] and bleomycin resistance gene upstream and downstream of blaNDM-1, respectively. There were two insertion sequences (ΔIS91) flanking both resistance genes, indicating its important role in the spread of blaNDM-1 and other resistant genes. In addition, a remnant of ΔISAba125 was located upstream of blaNDM-1 (Figure 2). A previous study has shown that partial ISAba125 is a promoter of the blaNDM-1 gene (Poirel et al., 2011). The blaNDM-1 and blaCTX-M-3 genes were detected in the ROF058 trans-conjugant, indicating that blaNDM-1 can be co-transferred with blaCTX-M-3 and across species, and thereby disseminate drug resistance. Although clinically important bacteria such as Enterobacter cloacae and Klebsiella pneumoniae produce both NDM-1 and CTX-M-3 (Rubin et al., 2014; Hu et al., 2016), we have shown the co-existence of blaNDM-1 and blaCTX-M-3 in one plasmid for the first time.
The isolates in our study carried the blaCTX-M-3 and blaCTX-M-14 genes, which is similar to the CTX-M types of ESBL-producing K. pneumoniae isolated from environmental samples (Chi et al., 2019). This likely indicates spread of resistant bacteria between human hosts and the environment. The blaCTX-M genes are usually present on the IncF, IncI, IncN, IncHI2, IncL/M, and IncK plasmid types (Zhang et al., 2016). WGS analysis showed that three ESBL-producing R. ornithinolytica isolates in our study carry the IncFIC plasmid, along with a ΔISEcp1 sequence upstream of blaCTX-M-3 in ROF058, and blaCTX-M-14 in ROE007 and ROE058. A study showed that 97.6% of the blaCTX-M genes have an upstream ΔISEcp1 sequence (Adamski et al., 2015), which plays an important role in the transposition of blaCTX-M and other ESBL genes (Decano et al., 2019). R0E007 and ROE058 isolated from the same village showed an identical sequence flanking blaCTX–M–14, i.e., ΔISEcp1-blaCTX-M-14-ΔIS903, which could be the result of either the clonal expansion of the original host bacteria or lateral transfer of the genetic elements. A linear fecE-fecD-fecC-fecB-fecA-fecR-fecI ferric citrate genes were detected upstream of blaCTX-M-14 in ROI014, which is very similar to that reported in an environmental K. pneumoniae isolate (accession no. KF914891) from the same area (Chi et al., 2019). These findings strongly indicate the spread of drug-resistance genes between K. pneumoniae and R. ornithinolytica.
To summarize, we characterized R. ornithinolytica isolates from healthy human feces in terms of their antibiotic susceptibility, drug resistance genes, and the transfer mechanism of the mobile genetic elements. The strains co-producing NDM-1 and CTX-M can confer higher levels of resistance to multiple antibiotics and can transfer the genes to other strains via plasmids by conjugation. Therefore, the patterns of antibiotic resistance and transmission should be closely monitored for R. ornithinolytica, especially in healthy individuals.
Data Availability Statement
The datasets generated for this study can be found in the National Center for Biotechnology Information, VJYE00000000-VJYH00000000, https://submit.ncbi.nlm.nih.gov/.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee for Shandong Center for Disease Control and Prevention; Shandong Center for Disease Control and Prevention. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SW and LX contributed to perform the experiments, data analysis, and manuscript writing. XC and YL contributed to perform the experiments. XC, ZK, PH, HX, ZB, and BZ contributed to the sample collection and data analysis. BZ, ZB, and HX contributed to conceive and design the experiments and reviewed the article.
Funding
This study was funded by the National Natural Science Foundation of China (Nos 81741098 and 71073098), the Mega Project on Infectious Disease Control and Prevention (2018ZX10714002-003-004), and Shandong key research and development plan project (2017GSF218062).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the participants and administrators for their support during the study. The authors would like to thank the faculty members and graduate students from Zhejiang University (Hangzhou, Zhejiang, China) for their warm-hearted help in performing the experiments.
Footnotes
References
Adamski, C. J., Cardenas, A. M., Brown, N. G., Horton, L. B., Sankaran, B., Prasad, B. V., et al. (2015). Molecular basis for the catalytic specificity of the CTX-M extended-spectrum beta-lactamases. Biochemistry 54, 447–457. doi: 10.1021/bi501195g
Ahmad, N., Khalid, S., Ali, S. M., and Khan, A. U. (2018). Occurrence of blaNDM variants among Enterobacteriaceae from a neonatal intensive care unit in a Northern India hospital. Front. Microbiol. 9:407. doi: 10.3389/fmicb.2018.00407
Ayoade, F., Mada, P. K., and Alam, M. (2018). Fat necrosis and polymicrobial wound infection caused partly by Raoultella ornithinolytica after reduction mammoplasty. BMJ Case Rep. pii: bcr-2018-224234. doi: 10.1136/bcr-2018-224234
Beye, M., Hasni, I., Seng, P., Michelle, C., La Scola, B., Raoult, D., et al. (2018). Genomic analysis of a Raoultella ornithinolytica strain causing prosthetic joint infection in an immunocompetent patient. Sci. Rep. 8:9462. doi: 10.1038/s41598-018-27833-z
Bocanegra-Ibarias, P., Garza-Gonzalez, E., Morfin-Otero, R., Barrios, H., Villarreal-Trevino, L., Rodriguez-Noriega, E., et al. (2017). Molecular and microbiological report of a hospital outbreak of NDM-1-carrying Enterobacteriaceae in Mexico. PLoS One 12:e0179651. doi: 10.1371/journal.pone.0179651
Buyukcam, A., Liste, U., Bicakcigil, A., Kara, A., and Sancak, B. (2019). A case of Raoultella ornithinolytica urinary tract infection in a pediatric patient. J. Infect. Chemother. 25, 467–469. doi: 10.1016/j.jiac.2018.12.011
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carramaschi, I. N., Castro, E. A. R., Leite, J. A., Queiroz, M. M. C., Boas, M., Rangel, K., et al. (2019). First report of Raoultella ornithinolytica carrying blaKPC-2 isolated from a dipteran muscoid collected in a garbage from a public hospital in Rio de Janeiro, Brazil. Rev. Inst. Med. Trop. Sao Paulo 61:e32. doi: 10.1590/S1678-9946201961032
Chetri, S., Bhowmik, D., Dhar, D., Chakravarty, A., and Bhattacharjee, A. (2019). Effect of concentration gradient carbapenem exposure on expression of blaNDM-1 and acrA in carbapenem resistant Escherichia coli. Infect. Genet. Evol. 73, 332–336. doi: 10.1016/j.meegid.2019.05.024
Chi, X., Berglund, B., Zou, H., Zheng, B., Borjesson, S., Ji, X., et al. (2019). Characterization of clinically relevant strains of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae occurring in environmental sources in a rural area of China by using whole-genome sequencing. Front. Microbiol. 10:211. doi: 10.3389/fmicb.2019.00211
CLSI (2017). Performance standards for antimicrobial susceptibility testing. 27th Edn. CLSI supplement M100. Wayne, PA, USA: Clinical and Laboratory Standards Institute.
De Jong, E., De Jong, A. S., Smidts-Van Den Berg, N., and Rentenaar, R. J. (2013). Differentiation of Raoultella ornithinolytica/planticola and Klebsiella oxytoca clinical isolates by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Diagn. Microbiol. Infect. Dis. 75, 431–433. doi: 10.1016/j.diagmicrobio.2012.12.009
De Petris, L., and Ruffini, E. (2018). Roultella ornithinolytica infection in infancy: a case of febrile urinary tract infection. CEN Case Rep. 7, 234–236. doi: 10.1007/s13730-018-0333-2
Decano, A. G., Ludden, C., Feltwell, T., Judge, K., Parkhill, J., and Downing, T. (2019). Complete assembly of Escherichia coli sequence type 131 genomes using long reads demonstrates antibiotic resistance gene variation within diverse plasmid and chromosomal contexts. mSphere 4, pii: e00130-19. doi: 10.1128/mSphere.00130-19
Dohmen, W., Bonten, M. J., Bos, M. E., Van Marm, S., Scharringa, J., Wagenaar, J. A., et al. (2015). Carriage of extended-spectrum beta-lactamases in pig farmers is associated with occurrence in pigs. Clin. Microbiol. Infect. 21, 917–923. doi: 10.1016/j.cmi.2015.05.032
Drancourt, M., Bollet, C., Carta, A., and Rousselier, P. (2001). Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int. J. Syst. Evol. Microbiol. 51, 925–932. doi: 10.1099/00207713-51-3-925
Hajjar, R., Schwenter, F., Su, S. H., Gasse, M. C., and Sebajang, H. (2018). Community-acquired infection to Raoultella ornithinolytica presenting as appendicitis and shock in a healthy individual. J. Surg. Case Rep. 2018:rjy097. doi: 10.1093/jscr/rjy097
Hasan, B., Olsen, B., Alam, A., Akter, L., and Melhus, A. (2015). Dissemination of the multidrug-resistant extended-spectrum beta-lactamase-producing Escherichia coli O25b-ST131 clone and the role of house crow (Corvus splendens) foraging on hospital waste in Bangladesh. Clin. Microbiol. Infect. 21, e1001–e1004. doi: 10.1016/j.cmi.2015.06.016
He, D., Chiou, J., Zeng, Z., Chan, E. W., Liu, J. H., and Chen, S. (2016). Comparative characterization of CTX-M-64 and CTX-M-14 provides insights into the structure and catalytic activity of the CTX-M class of enzymes. Antimicrob. Agents Chemother. 60, 6084–6090. doi: 10.1128/AAC.00917-16
Hernandez-Garcia, M., Leon-Sampedro, R., Perez-Viso, B., Morosini, M. I., Lopez-Fresnena, N., Diaz-Agero, C., et al. (2018). First report of an OXA-48- and CTX-M-213-producing Kluyvera species clone recovered from patients admitted in a University Hospital in Madrid, Spain. Antimicrob. Agents Chemother. 62, pii: e01238-18. doi: 10.1128/AAC.01238-18
Hu, L., Liu, Y., Deng, L., Zhong, Q., Hang, Y., Wang, Z., et al. (2016). Outbreak by ventilator-associated ST11 K. pneumoniae with co-production of CTX-M-24 and KPC-2 in a SICU of a tertiary teaching hospital in Central China. Front. Microbiol. 7:1190. doi: 10.3389/fmicb.2016.01190
Hu, Y., Zhang, W., Liang, H., Liu, L., Peng, G., Pan, Y., et al. (2011). Whole-genome sequence of a multidrug-resistant clinical isolate of Acinetobacter lwoffii. J. Bacteriol. 193, 5549–5550. doi: 10.1128/JB.05617-11
Jiang, X., Liu, W., and Zheng, B. (2018). Complete genome sequencing of Comamonas kerstersii 8943, a causative agent for peritonitis. Sci. Data 5:180222. doi: 10.1038/sdata.2018.222
Khajuria, A., Praharaj, A. K., Grover, N., and Kumar, M. (2013). First report of blaNDM-1 in Raoultella ornithinolytica. Antimicrob. Agents Chemother. 57, 1092–1093. doi: 10.1128/AAC.02147-12
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, genetics and worldwide spread of New Delhi metallo-beta-lactamase (NDM): a threat to public health. BMC Microbiol. 17:101. doi: 10.1186/s12866-017-1012-8
Kola, A., Kohler, C., Pfeifer, Y., Schwab, F., Kuhn, K., Schulz, K., et al. (2012). High prevalence of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J. Antimicrob. Chemother. 67, 2631–2634. doi: 10.1093/jac/dks295
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Li, J., Liu, C., Li, B., Yuan, H., Yang, J., and Zheng, B. (2012). Identification and molecular characterization of a novel DyP-type peroxidase from Pseudomonas aeruginosa PKE117. Appl. Biochem. Biotechnol. 166, 774–785. doi: 10.1007/s12010-011-9466-x
Luo, J., Yao, X., Lv, L., Doi, Y., Huang, X., Huang, S., et al. (2017). Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob. Agents Chemother. 61, pii: e01139-17. doi: 10.1128/AAC.01139-17
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Miao, M., Wen, H., Xu, P., Niu, S., Lv, J., Xie, X., et al. (2018). Genetic diversity of carbapenem-resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in Eastern China. Front. Microbiol. 9:3341. doi: 10.3389/fmicb.2018.03341
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paskova, V., Medvecky, M., Skalova, A., Chudejova, K., Bitar, I., Jakubu, V., et al. (2018). Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front. Microbiol. 9:1549. doi: 10.3389/fmicb.2018.01549
Poirel, L., Dortet, L., Bernabeu, S., and Nordmann, P. (2011). Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55, 5403–5407. doi: 10.1128/AAC.00585-11
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Rubin, J. E., Peirano, G., Peer, A. K., Govind, C. N., and Pitout, J. D. (2014). NDM-1-producing Enterobacteriaceae from South Africa: moving towards endemicity? Diagn. Microbiol. Infect. Dis. 79, 378–380. doi: 10.1016/j.diagmicrobio.2014.04.003
Sanchez-Codez, M., Lubian-Gutierrez, M., Blanca-Garcia, J. A., and Perez Aragon, C. (2019). Leclercia adecarboxylata and Raoultella ornithinolytica catheter-related infection in a child with mitocondrial disease. Arch. Argent. Pediatr. 117, e147–e149. doi: 10.5546/aap.2019.e147
Sekowska, A., Dylewska, K., Gospodarek, E., and Bogiel, T. (2015). Catheter-related blood stream infection caused by Raoultella ornithinolytica. Folia Microbiol. 60, 493–495. doi: 10.1007/s12223-015-0390-2
Seng, P., Boushab, B. M., Romain, F., Gouriet, F., Bruder, N., Martin, C., et al. (2016). Emerging role of Raoultella ornithinolytica in human infections: a series of cases and review of the literature. Int. J. Infect. Dis. 45, 65–71. doi: 10.1016/j.ijid.2016.02.014
Shimizu, T., Harada, K., Tsuyuki, Y., Kimura, Y., Miyamoto, T., Hatoya, S., et al. (2017). In vitro efficacy of 16 antimicrobial drugs against a large collection of beta-lactamase-producing isolates of extraintestinal pathogenic Escherichia coli from dogs and cats. J. Med. Microbiol. 66, 1085–1091. doi: 10.1099/jmm.0.000535
Singh, M., Kaur, I., Mundi, D. K., and Kaur, A. (2017). ENT infection caused by Raoultella ornithinolytica. Niger. J. Clin. Pract. 20, 914–917. doi: 10.4103/njcp.njcp_337_16
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sun, Q., Wang, Y., Hulth, A., Xiao, Y., Nilsson, L. E., Li, X., et al. (2018). Study protocol for one health data collections, analyses and intervention of the Sino-Swedish integrated multisectoral partnership for antibiotic resistance containment (IMPACT). BMJ Open 8:e017832. doi: 10.1136/bmjopen-2017-017832
Tada, T., Sekiguchi, J. I., Watanabe, S., Kuwahara-Arai, K., Mizutani, N., Yanagisawa, I., et al. (2019). Assessment of a newly developed immunochromatographic assay for NDM-type metallo-beta-lactamase producing Gram-negative pathogens in Myanmar. BMC Infect. Dis. 19:565. doi: 10.1186/s12879-019-4147-4
Van Cleve, J. R., Boucher, B. A., Smith, D. V., and Croce, M. A. (2018). Ventilator associated pneumonia caused by Raoultella ornithinolytica in two immunocompetent trauma patients. Respir. Med. Case Rep. 24, 135–137. doi: 10.1016/j.rmcr.2018.05.006
Vasaikar, S., Obi, L., Morobe, I., and Bisi-Johnson, M. (2017). Molecular characteristics and antibiotic resistance profiles of Klebsiella isolates in Mthatha, Eastern Cape Province, South Africa. Int. J. Microbiol. 2017:8486742. doi: 10.1155/2017/8486742
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front. Microbiol. 10:228. doi: 10.3389/fmicb.2019.00228
Wang, J. T., Wu, U. I., Lauderdale, T. L., Chen, M. C., Li, S. Y., Hsu, L. Y., et al. (2015). Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS One 10:e0121668. doi: 10.1371/journal.pone.0146050
Xia, S., Fan, X., Huang, Z., Xia, L., Xiao, M., Chen, R., et al. (2014). Dominance of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS One 9:e100707. doi: 10.1371/journal.pone.0100707
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Yoon, E. J., Kang, D. Y., Yang, J. W., Kim, D., Lee, H., Lee, K. J., et al. (2018). New Delhi metallo-beta-lactamase-producing Enterobacteriaceae in South Korea between 2010 and 2015. Front. Microbiol. 9:571. doi: 10.3389/fmicb.2018.00571
Yu, W. L., Winokur, P. L., Von Stein, D. L., Pfaller, M. A., Wang, J. H., and Jones, R. N. (2002). First description of Klebsiella pneumoniae harboring CTX-M beta-lactamases (CTX-M-14 and CTX-M-3) in Taiwan. Antimicrob. Agents Chemother. 46, 1098–1100. doi: 10.1128/AAC.46.4.1098-1100.2002
Zhang, X., Hao, X., Huo, S., Lin, W., Xia, X., Liu, K., et al. (2019). Isolation and identification of the Raoultella ornithinolytica-ZK4 degrading pyrethroid pesticides within soil sediment from an abandoned pesticide plant. Arch. Microbiol. 201, 1207–1217. doi: 10.1007/s00203-019-01686-0
Zhang, J., Zhou, K., Zheng, B., Zhao, L., Shen, P., Ji, J., et al. (2016). High prevalence of ESBL-producing Klebsiella pneumoniae causing community-onset infections in China. Front. Microbiol. 7:1830. doi: 10.3389/fmicb.2016.01830
Zheng, B., Lv, T., Xu, H., Yu, X., Chen, Y., Li, J., et al. (2018). Discovery and characterisation of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int. J. Antimicrob. Agents 51, 273–275. doi: 10.1016/j.ijantimicag.2017.09.005
Zheng, B., Zhang, J., Ji, J., Fang, Y., Shen, P., Ying, C., et al. (2015). Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob. Agents Chemother. 59, 7086–7089. doi: 10.1128/AAC.01363-15
Zhou, G., Guo, S., Luo, Y., Ye, L., Song, Y., Sun, G., et al. (2013). NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerg. Infect. Dis. 20. doi: 10.3201/eid2002.121263
Zhou, G., Guo, S., Luo, Y., Ye, L., Song, Y., Sun, G., et al. (2014). NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerg. Infect. Dis. 20, 340–342. doi: 10.3201/eid2002.121263
Keywords: Raoultella ornithinolytica, carbapenemase, extended-spectrum β-lactamase, blaNDM-1, blaCTX-M, whole-genome sequencing
Citation: Wang S, Xu L, Chi X, Li Y, Kou Z, Hou P, Xie H, Bi Z and Zheng B (2019) Emergence of NDM-1- and CTX-M-3-Producing Raoultella ornithinolytica in Human Gut Microbiota. Front. Microbiol. 10:2678. doi: 10.3389/fmicb.2019.02678
Edited by:
Charlene Renee Jackson, United States Department of Agriculture (USDA), United StatesReviewed by:
Ignasi Roca Subirà, Instituto Salud Global Barcelona (ISGlobal), SpainJorge Aníbal Reyes, Universidad San Francisco de Quito, Ecuador
Viviane Zahner, Oswaldo Cruz Foundation (Fiocruz), Brazil
Copyright © 2019 Wang, Xu, Chi, Li, Kou, Hou, Xie, Bi and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hengjie Xie, eGhqQG1haWwuc2R1LmVkdS5jbg==; Zhenwang Bi, Ynp3am5AMTYzLmNvbQ==; Beiwen Zheng, emhlbmdid0B6anUuZWR1LmNu
†These authors have contributed equally to this work
 Shuang Wang
Shuang Wang Liuchen Xu1†
Liuchen Xu1† Xiaohui Chi
Xiaohui Chi Beiwen Zheng
Beiwen Zheng