- College of Food Science and Engineering, Gansu Agricultural University, Lanzhou, China
Doushen, a traditional Chinese fermented soybean product, may be spiced or spicy depending on whether pepper powder is added. While numerous studies have investigated the microbial communities of other fermented foods, little is known on the fungal diversity of Doushen. Therefore, in this study, we investigated the fungal community and diversity in both spiced and spicy Doushen. Our results revealed that fungal species richness significantly differed between the samples with different flavors. A total of nine phyla and 188 fungal genera were identified, and Ascomycota and Aspergillus were predominant in all samples. Based on linear discriminant analysis, a total of 57 OTUs were significantly different between the two samples. Results of non-metric multidimensional scaling and unweighted pair-group analysis suggested that the presence of pepper powder affects the microbial community in Doushen. Network analysis showed that microbial interactions between fungal communities in Doushen with different flavors were significantly different. The results on the enumeration and identification of fungi were consistent with the composition of the dominant genera in the samples with different flavors. This study provides a theoretical basis for future research on food ecology in Doushen.
Introduction
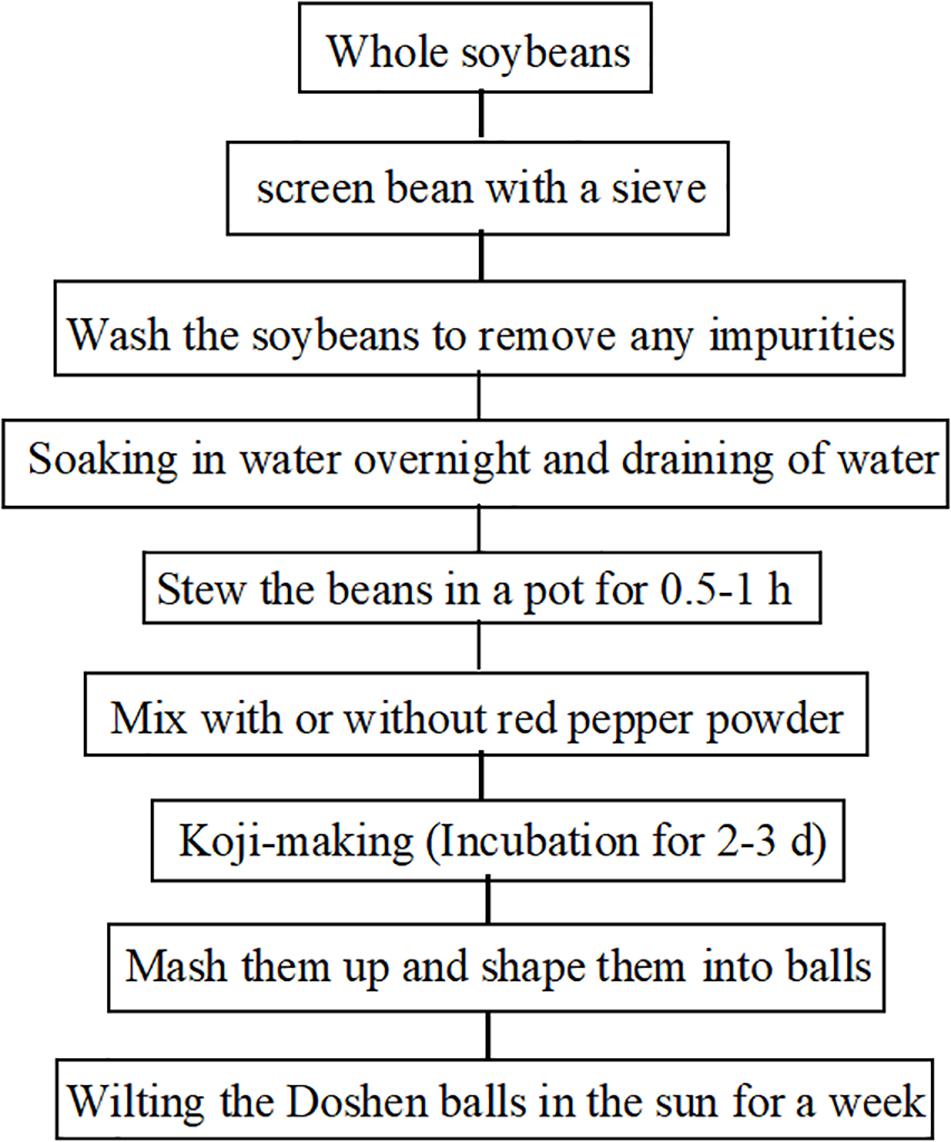
Traditional fermented soybean-based foods, including natto (Liu et al., 2018), tempeh (Noriyuki, 1990), douchi (Zhang et al., 2018), sufu (Qiu et al., 2018), miso (Jayachandran and Xu, 2019), soy sauce (Li et al., 2018), and soybean paste (Inoue et al., 2016) have been consumed in Asia for centuries. Currently, these foods are becoming recognized and accepted by Western countries (Wang et al., 2007; Zhang et al., 2007; Wang et al., 2010). Doushen, a douchi-like product, has been produced and consumed widely as seasoning in Henan Province for thousands of years. Due to its delicious taste and unique flavor, Doushen is being favored by local people in Zhoukou, Xuchang, Shangqiu, and Kaifeng. The traditional preparation process of Doushen consists of screening, cleaning, soaking, cooking, mixing, koji-making, mashing, and wilting in the sun (Figure 1). The end products are processed into Doushen balls of approximately 4–6 cm in diameter. During the production of Doushen, raw soybeans are heat-treated sufficiently to kill most of the microorganisms except for the heat-resistant ones. However, cooked soybeans are fermented under exposed conditions; hence, a variety of endogenous and exogenous microorganisms may be involved in the fermentation process. After fermentation, soybeans used in Doushen are usually ground and sun-dried. Environmental factors might shape the microbial communities in Doushen. Therefore, abundant and diverse microbial populations may exist in this specific food ecosystem.
A variety of fungi, including Aspergillus, Mucor, Rhizopus, Saccharomyces, Pichia, and Lichtheimia, play a pinnacle role in the production of traditional fermented soybean products (Chen et al., 2011; Yang et al., 2016; Zhang et al., 2018). During the process of fermentation, fungi produce various enzymes including glutaminase, cellulase, proteases, lipases, amylases, and fibrinolytic enzymes. Under the action of these enzymes, proteins, sugars, lipids, and other compounds in soybeans are degraded into ketones, esters, and acids, thereby contributing to taste and aroma (Kim et al., 2010; Yebra and Monedero, 2010; Chen et al., 2012; Chen et al., 2014). Over the last four decades, several studies have analyzed the fungal community of traditional fermented soybean products using culture-dependent or culture-independent methods (Kim et al., 2010; Chen et al., 2011; Chen et al., 2012; Nam et al., 2012; Chen et al., 2014; Ji et al., 2014; Yang et al., 2016; Zhang et al., 2018). To date however, no studies have evaluated the microbial community in Doushen.
Traditional culture-dependent and culture-independent approaches have been used to investigate the microbial community of fermented soybean products. These methods provide little information on microbial community composition and structure (Kim et al., 2010; Chen et al., 2011; Chen et al., 2012, 2014). Recently, high-throughput sequencing has been used in ecosystem exploration. This technology circumvents the limitations associated with culture-dependent methods and, consequently, has been widely applied to analyze the microbial diversity in various natural environments such as soil, air, water, plants, and various human microbiota (Arumugam et al., 2011; Fu et al., 2015; Helena et al., 2016; Qin et al., 2016; Kozyaeva et al., 2017). Recently, numerous traditional fermented foods such as douchi (Yang et al., 2016; Zhang et al., 2018), soybean paste (Nam et al., 2012), cheese (Ahram et al., 2018), yogurt (Lisa et al., 2013), Qula (Zhu et al., 2018), and Pu-er tea (Zhang et al., 2016) have been analyzed using high-throughput sequencing In this study, we used high-throughput sequencing to evaluate the fungal community diversity in Doushen with different flavors obtained from Henan province. Our findings may assist in understanding the complex fungal community structure in Doushen, thereby providing a solid theoretical basis to select appropriate strains suitable for high-quality Doushen.
Methods
Sample Collection
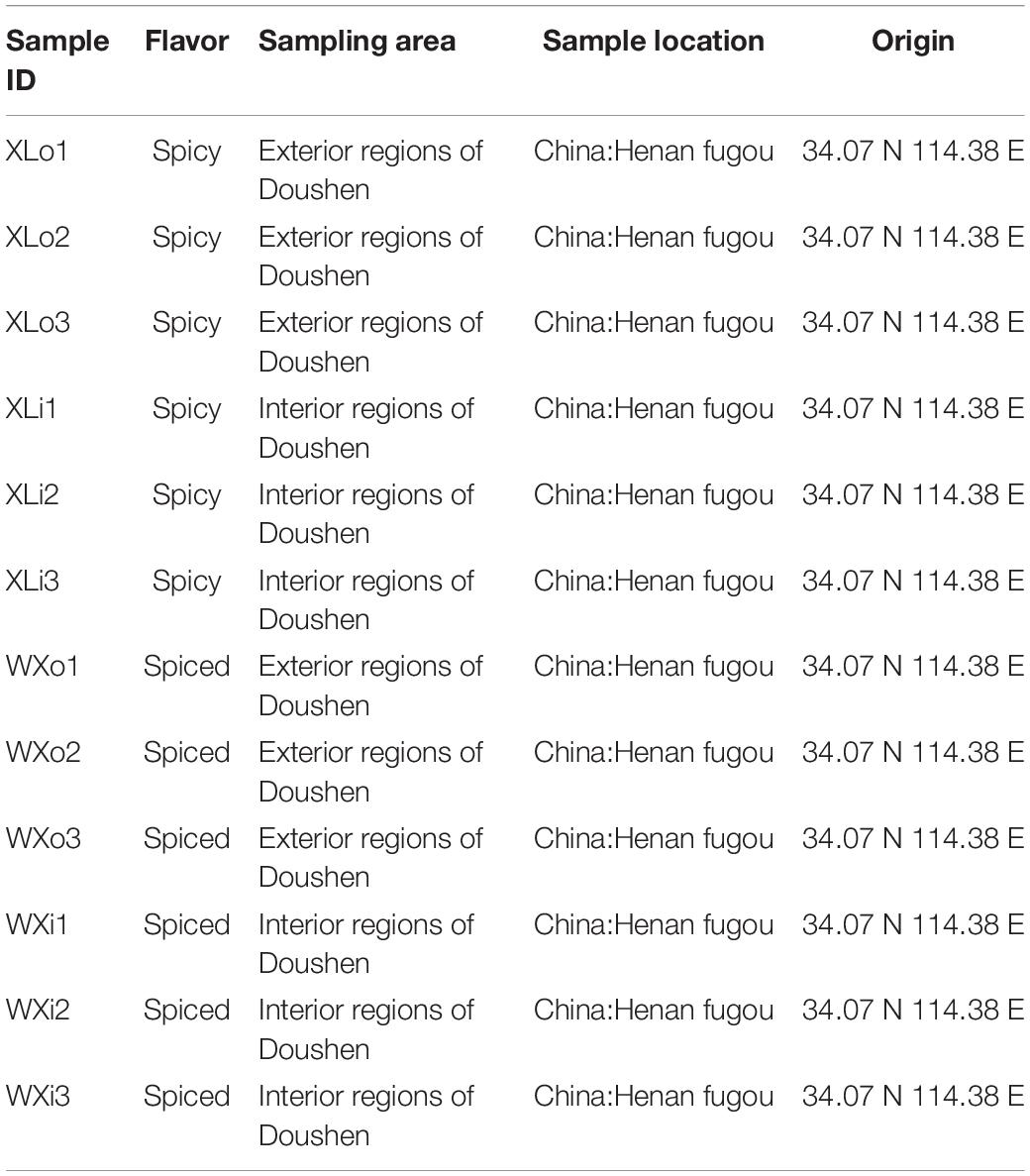
Homemade Doushen samples with different flavor (spicy and spiced) were obtained from local producers in Fugou County (Henan province, China). Five Doushen balls of each flavor were randomly collected from one producer. A total of 30 samples were placed into an ice box for transportation to the lab. Each Doushen ball was divided into exterior and interior regions at a ∼2.5-cm depth from the surface using a saw. Two grams of each exterior or interior sample were collected from one ball. Subsequently, the five samples obtained from the same producer were pooled into a composite sample, and stored at −80°C. All the samples were divided into four groups based on sampling location and flavor. The first group includes the sample from the exterior of spicy Doushen (XLo), the second group includes the sample from the interior of spicy Doushen (XLi), the third group includes the sample from the exterior of spiced Doushen (WXo), and the fourth group includes the sample from the interior of spiced Doushen (WXi) (Table 1).
Physiochemical Characterization of Doushen
The moisture, lipid, and protein contents of samples were determined by AOAC methods (AOAC, 2000). Amino-type nitrogen and total acid contents were determined by the titration method described by Judoamidjojo (1986).
Plate Count of Fungi
The total number of viable fungal cells in Doushen was determined using a standard viable cell counting method. Briefly, 1 g of Doushen samples was diluted to 100 mL normal saline (0.9% NaCl). The diluted samples were spread on potato-dextrose agar (PDA, BD, United States) for the enumeration of fungi. The agar plates were incubated at 28°C for 5-day, and the number of fungi in Doushen was calculated as colony forming units (CFU) per gram. The colonies on the plates were randomly selected and cultivated to extract total DNA. The internal transcribed spacer (ITS) region of nuclear ribosomal DNA was amplified using specific primers (ITS1 and ITS4). The PCR amplification products were purified and sequenced at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). Sequences were blasted in NCBI web site for identification of fungi.
DNA Extraction, PCR Amplification, and Sequencing
DNA of all Doushen samples was extracted with a Soil DNA Kit D5625-01 (Power Soil DNA Isolation kit, MOBIO Laboratories, Inc., United States) according to the manufacturer’s instructions. DNA concentration and quality was determined by using an ultraviolet visible spectrophotometer (Thermo Scientific, NC 2000, United States).
The universal primer pairs ITS1-F (CTTGGTCATTTAG AGGAAGTAA) and ITS2 (GCTGCGTTCTTCATCGA TGC) were used to amplify the fungal ITS rDNA gene targeting the ITS1-ITS2 region (Wu et al., 2018). The PCR thermal cycle profile was as follows: initial denaturation for 2 min at 98°C; followed by 25 cycles of denaturation at 98°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; then a final extension at 72°C for 5 min, after which the samples were cooled to 10°C for later use.
All PCR products were sequenced on an Illumina Miseq PE300 Sequencing platform (Illumina, Inc., San Diego, CA, United States) at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). Raw sequences were uploaded into the Sequence Read Archive at NCBI under the accession number PRJNA493155.
Processing of High-Throughput Sequencing Data
Raw Illumina fastq files generated from the high-throughput sequencing were demultiplexed, quality filtered, and analyzed following pipelines of Mothur (V.1.31.2) and QIIME (V1.7.0) (Schlosset et al., 2009; Caporaso et al., 2010). Quality sequences were classified into operational taxonomic units (OTUs) with a cutoff of 97% identity using the de novo OTU selection strategy. Taxonomies were assigned by the RDP classifier (Version 2.2) (Cole et al., 2003) against the UNITE databases (Koljalg et al., 2013) with 80% confidence threshold.
Data Analysis
Alpha diversity indices of Chao1, Shannon, and Good’s coverage were calculated using QIIME following the tutorial1 for each sample. The indices were subsequently tested for significant differences between the different groups using the Duncan’s test.
Non-metric multidimensional scaling (NMDS) and a Heat map were constructed using the R package2 (v2.15.3) to quantify differences in community composition. The difference of fungal community structures among Doushen samples was analyzed by Adonis permutational multivariate analysis (Adonis/PERMANOVA) and analysis of similarities (ANOSIM) (Mcardle and Anderson, 2001; Lozupone et al., 2007).
Unweighted pair-group method with arithmetic means (UPGMA) clustering was conducted using QIIME following the guidance3.
To identify differentially abundant taxa between different groups in Doushen samples, LDA effect size (LEfSe) was performed using the online Galaxy work flow framework4 (LDA score ≥2.0 and p ≤ 0.05; Segata et al., 2011).
The SparCC algorithm was used to analyze the correlation of all fungal genera in the abundance and variety of the Doushen samples, including positive correlation and negative correlation (Friedman and Alm, 2012). Statistical analysis was also performed on the screening of the correlation score >0.6 with a significance level less than 0.05. Then correlation networks were made using the biocloud tools, a free online platform for data analysis.5
Results
Physiochemical Characterization of Doushen
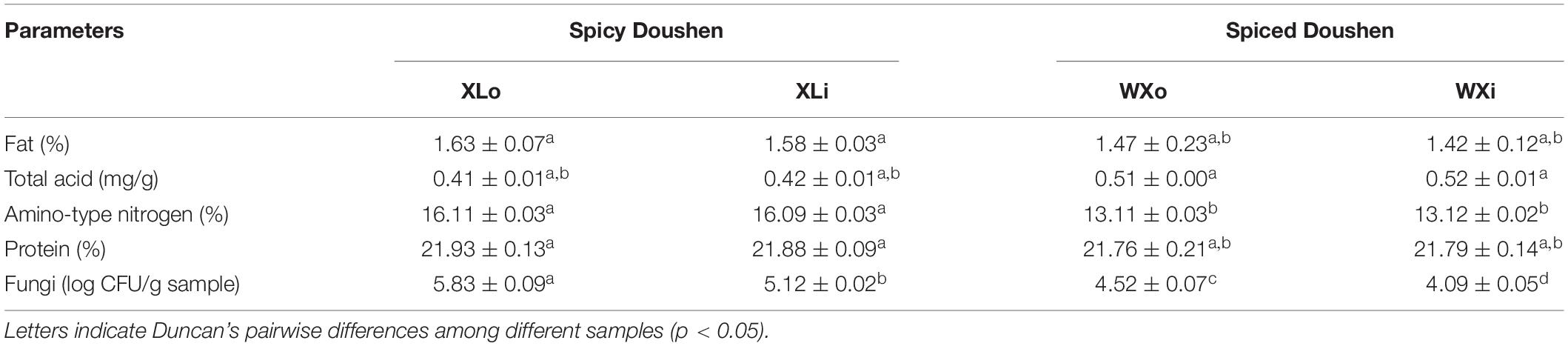
There was no significant difference in protein content between the spicy and spiced Doushen samples (p > 0.05; Table 2). However, there were differences in the amino-type nitrogen content (p < 0.05). In addition, no significant differences were obtained in fat and total acid content between the two types of Doushen samples (p < 0.05).
Sequencing and Classification
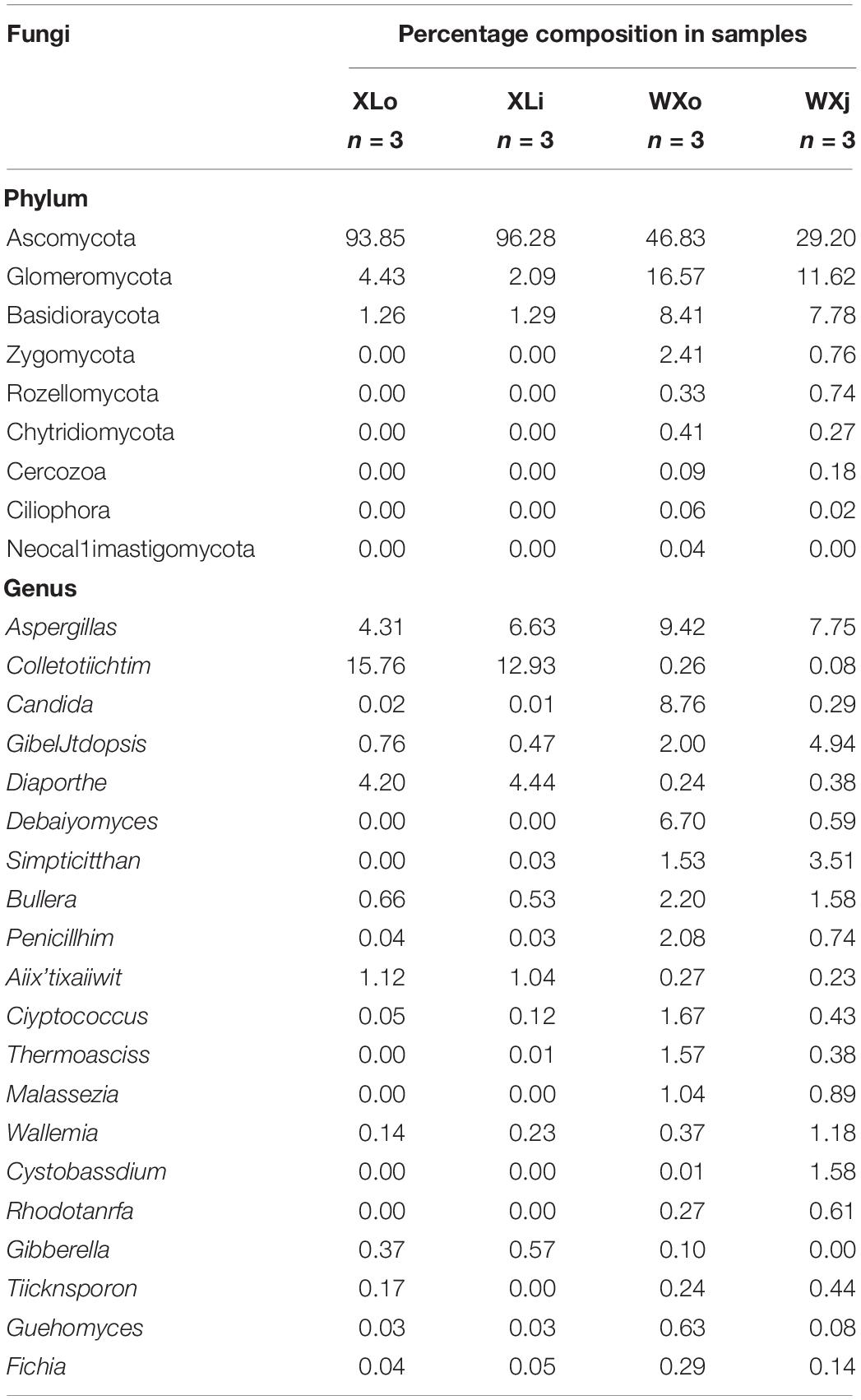
Total DNA was extracted from the Doushen samples, and the ITS rDNA gene was amplified by PCR. PCR products were sequenced using Illumina MiSeq platform at Shanghai Personal Biotechnology Co. Ltd. (Shanghai, China). After removing low quality and chimeric sequences, 774,761 high-quality sequencing reads were generated from 12 Doushen samples, with an average of 64,563 reads per sample (Table 3). A total of 9,051 OTUs were generated from the high-quality sequences, with an average of 754 OTUs per sample. OTUs were significantly higher in spicy Doushen samples (1,027 OTUs for XLo and 1,002 OTUs for XLi) than in spiced Doushen samples (615 OTUs for WXo and 373 OTUs for WXi). In addition, the samples from the exterior area (XLo and WXo) contained more OTUs than the samples from the interior area (Xli and WXi).
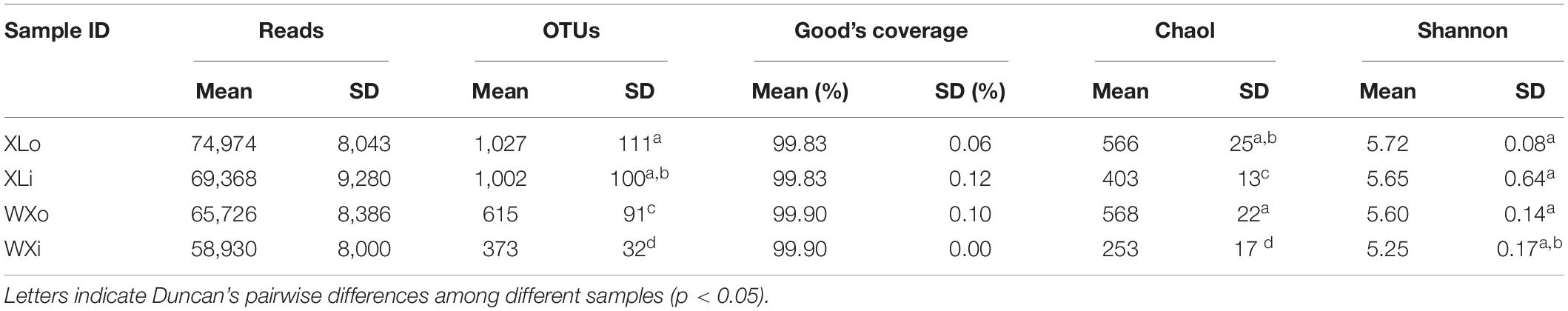
Table 3. Reads, OTUs, Good’s Coverage, Chaol, and Shannon’s indices for ITS rRNA sequencing of the Doushen samples.
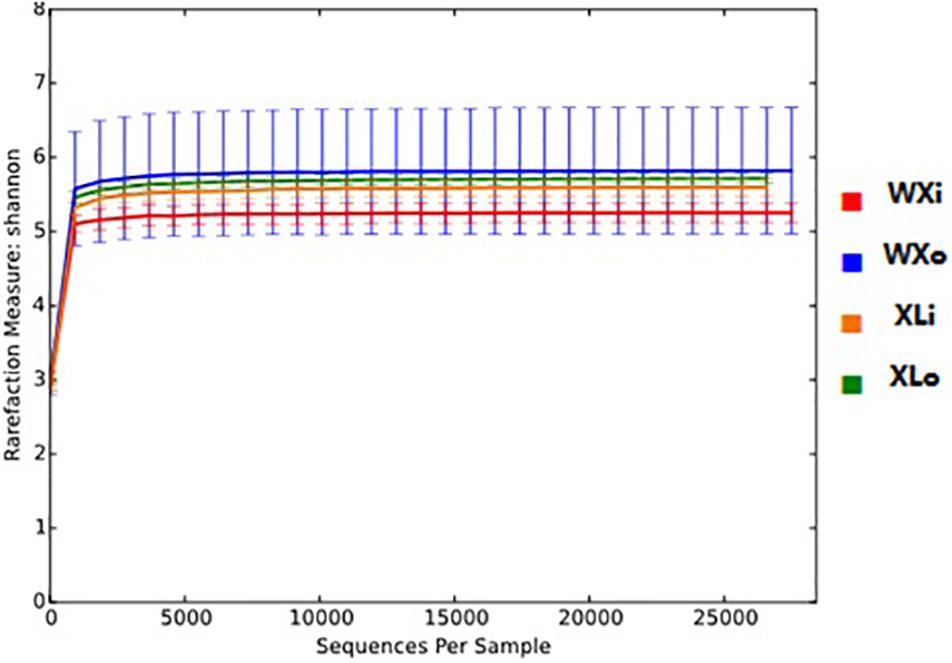
In the study, the Good’s coverage of all the samples was >98% (Table 3), indicating that the sequencing depth was sufficient to saturate the fungal diversity. The rarefaction curve for Shannon diversity indices was nearly parallel with the x-axis (Figure 2), suggesting that the majority of fungi present in Doushen had been captured.
Analysis of Alpha Diversity
Shannon and Chao1 indexes were used to assess alpha diversity (Table 3). There were no significant differences in Shannon indexes among the samples (p > 0.05), implying that the fungal diversity difference of the Doushen samples was not significant. However, the results showed that fungal species richness was different (p < 0.05) between Doushen samples with different flavor. In addition, the results showed that the exterior area of Doushen samples had higher fungal diversity than the interior area, consistent with the differences in fungal counts and OTUs.
Comparison of Fungal Communities
To investigate the microbial community and diversity in Doushen, the fungal ITS rRNA gene sequences were classified at both phylum and genus levels. At the phylum level, there were nine phyla detected in Doushen samples. Ascomycota were predominant in WXi, WXo, Xli, and XLo, with a relative abundance >20% (Table 4). The relative abundance of Ascomycota in spicy Doushen was particularly high (>90%); its abundance was significantly different among XLo, WXo, and XLi (p < 0.05; Figure 3). Glomeromycota and Basidiomycota, which were detected in all Doushen samples, were particularly more abundant in spiced Doushen (p < 0.05; Figure 3). The abundance of Zygomycota was higher in spiced Doushen than in spicy Doushen (p < 0.05; Figure 3).
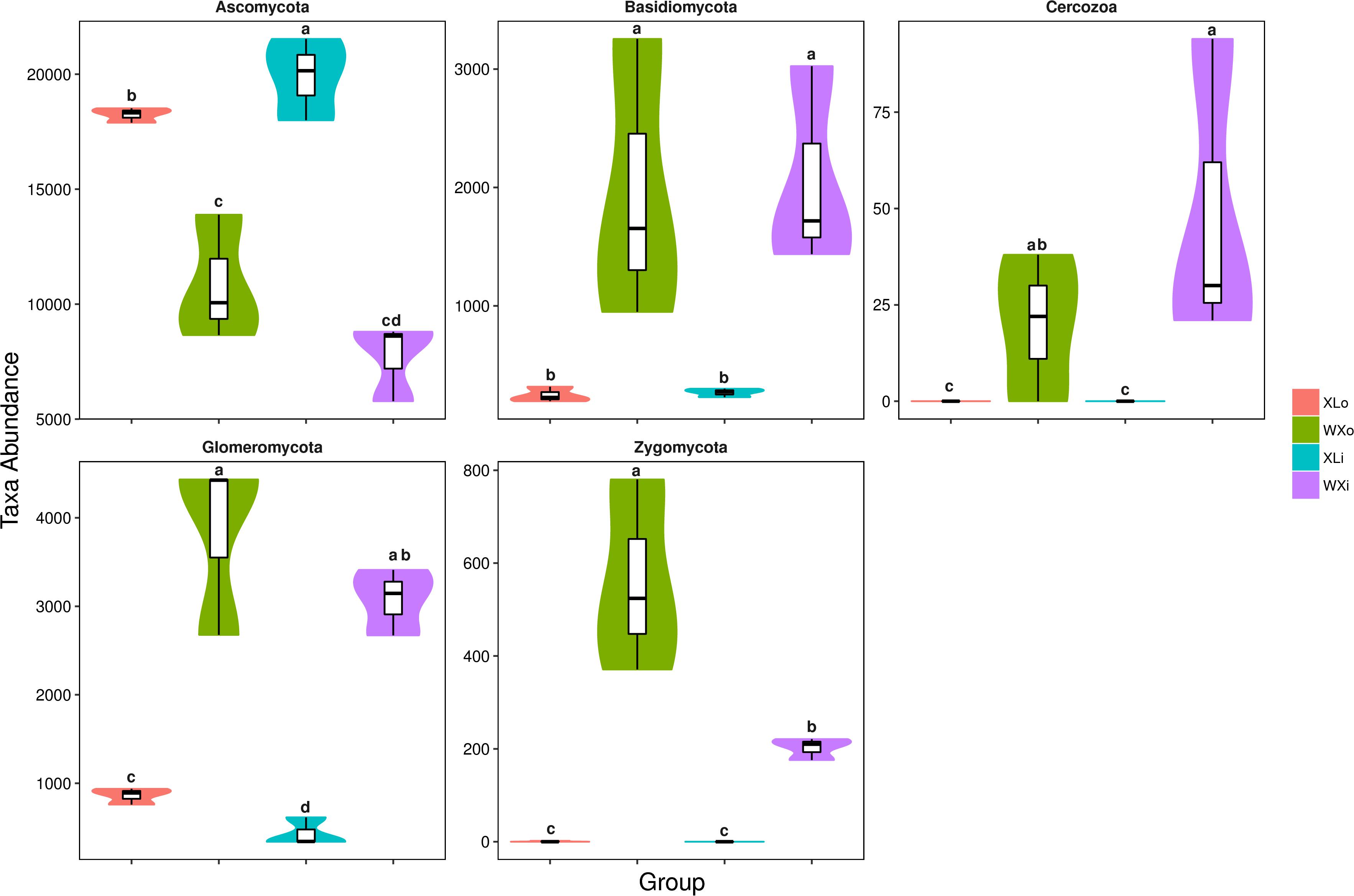
Figure 3. Taxonomic profiles of statistically significant fungi at the phylum level. Different letters indicate significance at p < 0.05.
At the genus level, there were 188 genera tested in the Doushen samples. The relative abundance of the top 20 fungal genera is shown in Table 4. A total of 15 dominant genera were obtained in all samples, with a relative abundance >1%. Aspergillus was the only dominant genus present in all samples, which had a higher relative abundance in spiced Doushen (XLo and XLi). Three genera (Colletotrichum, Diaporthe, and Acremonium) were only dominant in spicy Doushen. Among them, Colletotrichum was the most-dominant genus, with a relative abundance >10%, followed by Diaporthe (>4%). Ten genera (Candida, Gibellulopsis, Debaryomyces, Simplicillium, Bullera, Penicillium, Cryptococcus, Thermoascus, Malassezia, Wallemia, and Cystobasidium) were only dominant in spiced Doushen. Among these, three dominant genera (Gibellulopsis, Simplicillium, and Bullera) were present in all spiced Doushen samples. Six genera (Candida, Debaryomyces, Penicillium, Cryptococcus, Thermoascus, and Malassezia) were only dominant in WXo samples (the exterior area of spiced Doushen). The other two genera (Wallemia and Cystobasidium) were only dominant in WXi samples (the interior area of spiced Doushen). Additionally, a total of 173 non-dominant genera (relative abundance <1%) were identified in the samples.
To identify the specific taxa within each group, LEfSe analysis was applied to compare the microbiota between different groups (Figure 4). In the XLi group, 14 taxa were enriched including one phylum (Ascomycota), two classes (Eurotiomycetes and Dothideomycetes), three orders (Diaporthales, Hypocreales, and Pleosporales), four families (Diaporthaceae, Botryosphaeriaceae, Mycosphaerellaceae, and Nectriaceae), and four genera (Diaporthe, Hannaella, Gibberella, and Sphaerulina). The fungal groups enriched in the XLo group were one class (Sordariomycetes), one order (Ascosphaerales), two families (Inocybaceae and Glomerellaceae), and five genera (Ascosphaera, Inocybe, Colletotrichum, Acremonium, and Monographella). In the WXi group, 15 taxa were enriched including one phylum (Zygomycota), two classes (Tremellomycetes and Saccharomycetes), four orders (Chaetothyriales, Mucorales, Saccharomycetales, and Tremellales), two families (Davidiellaceae and Lichtheimiaceae), and six genera (Cryptococcus, Penicillium, Cladosporium, Staphylotrichum, Candida, and Lichtheimia). The fungal groups enriched in the WXo group were two phyla (Basidiomycota and Glomeromycota), three classes (Microbotryomycetes, Glomeromycetes, and Agaricomycetes), five orders (Archaeosporales, Sporidiobolales, Trichocomaceae, Malasseziales, and Sordariales), three families (Plectosphaerellaceae, Cordycipitaceae, and Ambisporaceae), and four genera (Gibellulopsis, Simplicillium, Rhodotorula, and Bullera). In total, 57 OTUs were significantly different between samples with different flavors.
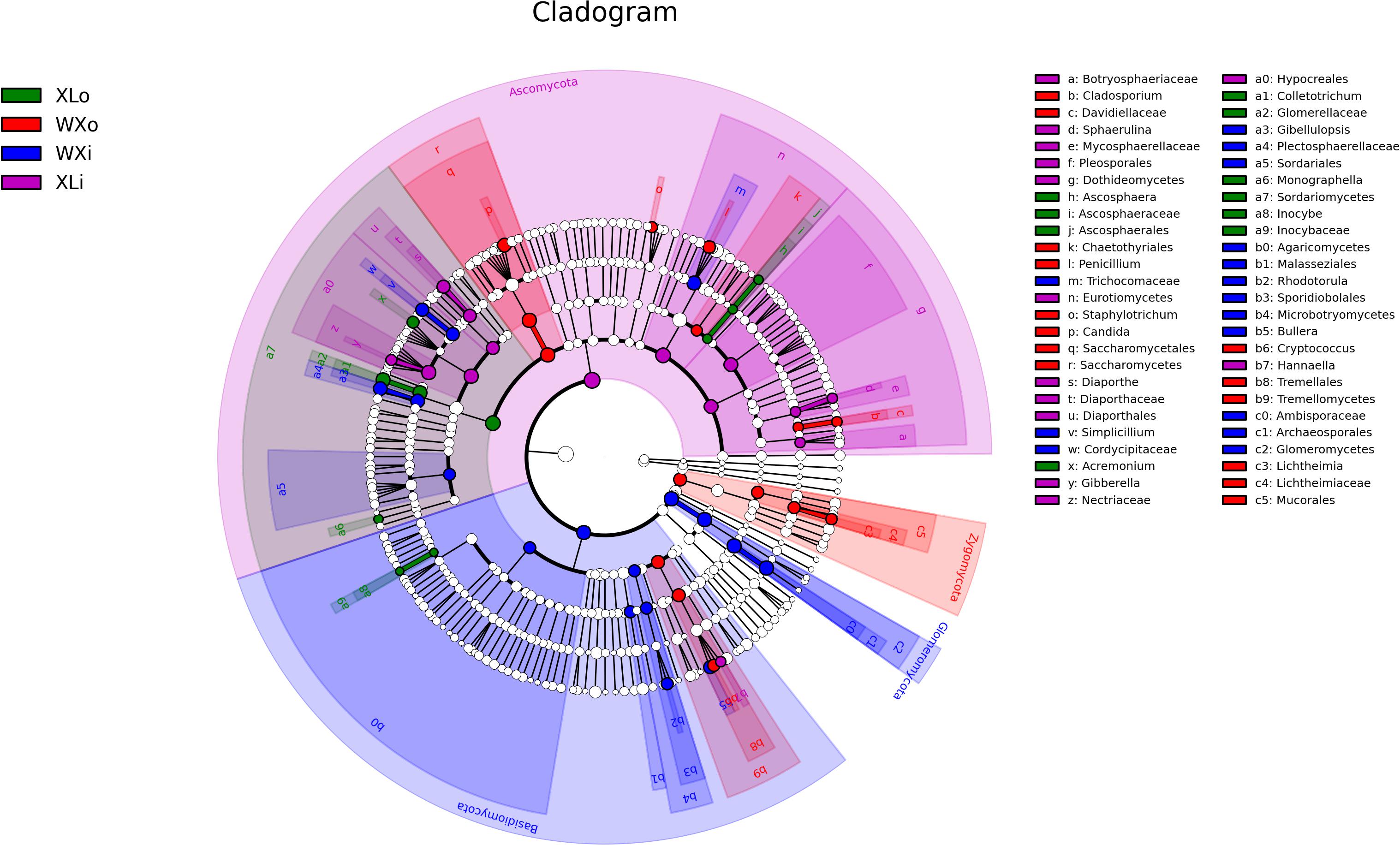
Figure 4. Linear discriminant analysis in microbial community compositions in Doushen samples. The node size represents the difference in relative abundance.
Analysis of Beta Diversity
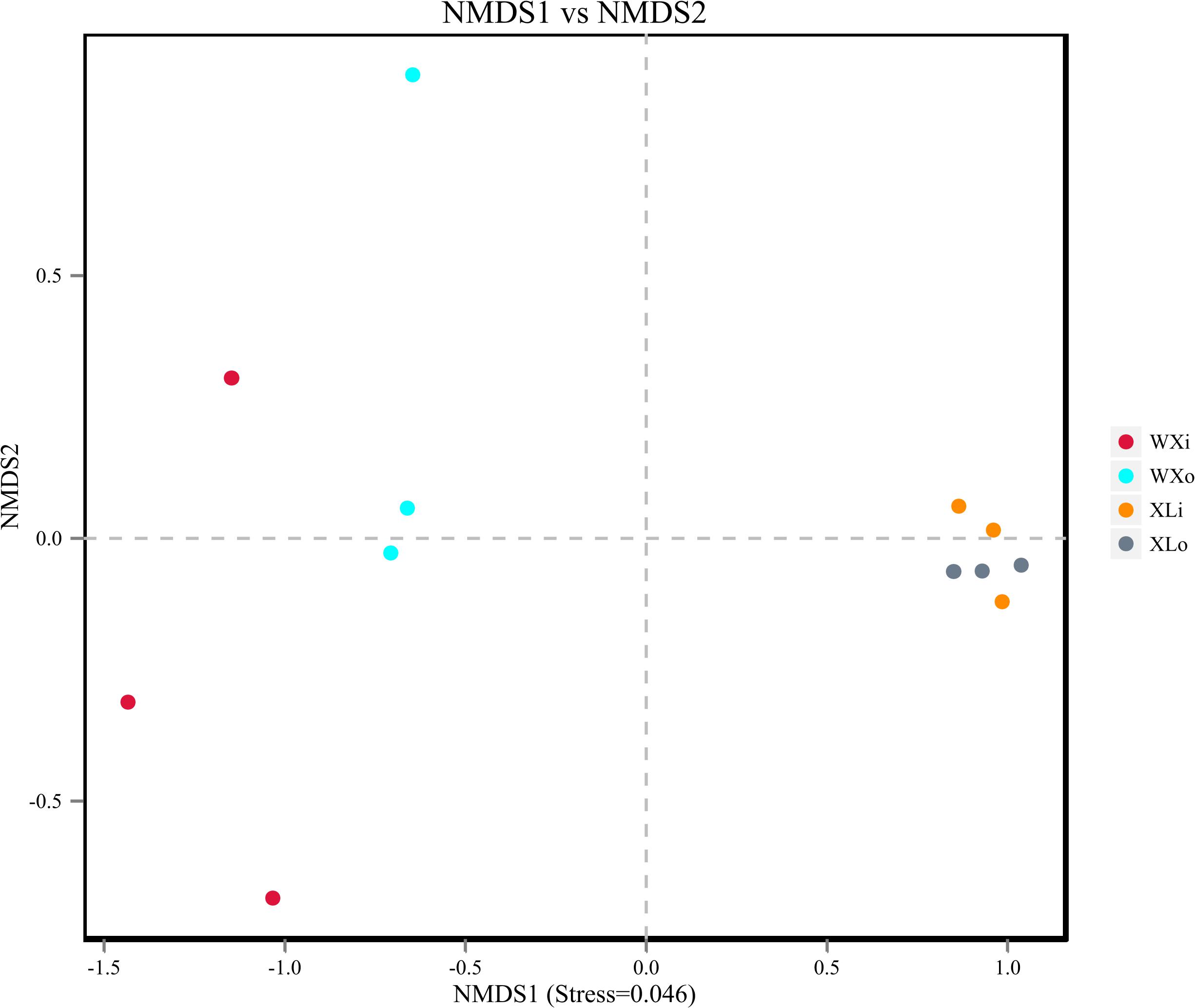
Beta diversity results of NMDS based on unweighted uniFrac indicated the data distribution of Doushen samples and is shown in Figure 5. Samples of the same flavor grouped together tightly indicating that the fungal community structure of XLo and XLi has a high degree of similarity and small differences. This situation also applies to the fungal community structure of the WXo and WXi. In addition, the XL samples are more tightly clustered than WX samples in the present study. The results were much similar when comparing the samples with different flavors by using unweighted pair-group analysis (Figure 6). Adonis/PERMANOVA analysis was performed out on the sample, the results showed that p = 0.001 (p < 0.05). Results of ANOSIM’s statistical analysis also indicated that flavor (the addition of pepper powder) was a crucial factor influencing the fungal composition of the different samples (R = 0.6235, P = 0.002).
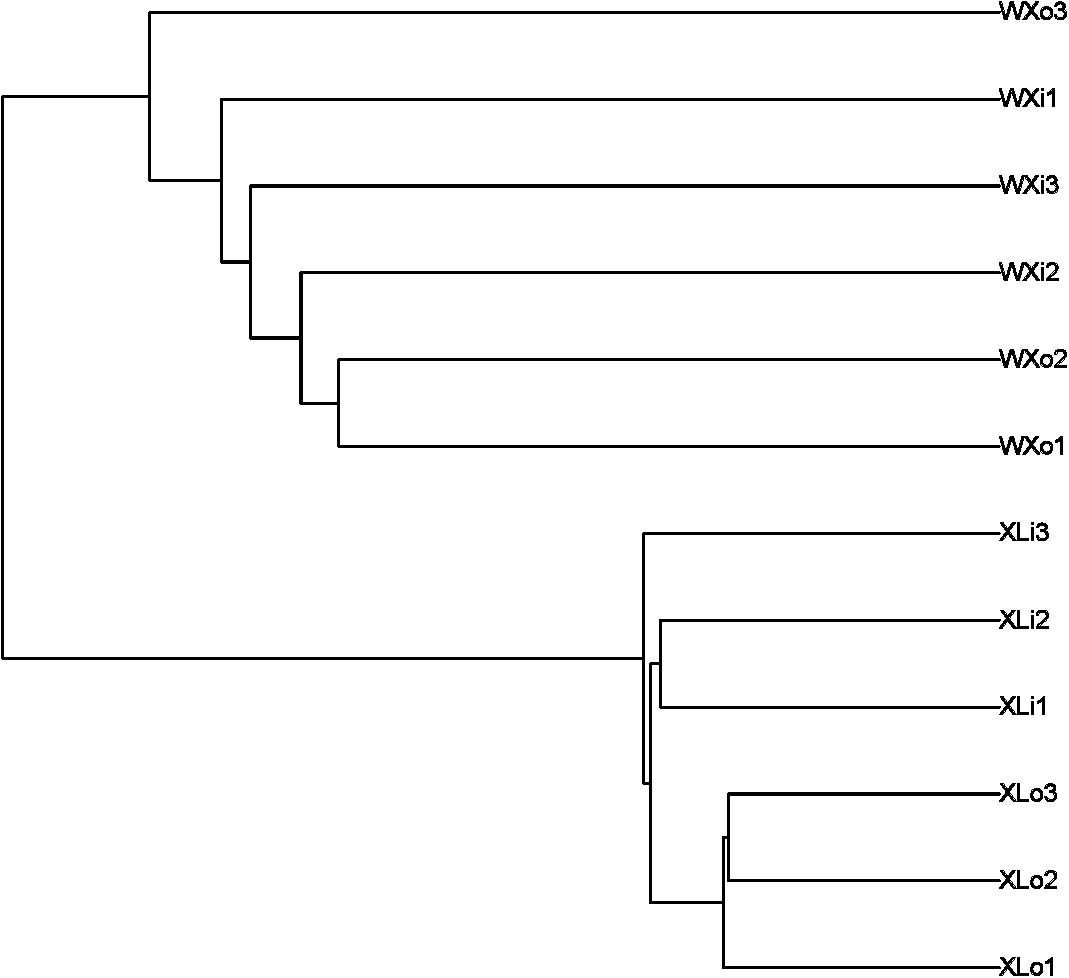
Figure 6. The unweighted pair-group analysis (UPGMA) based on UniFrac distance for fungal communities of Doushen samples.
Interactions of Fungi in Doushen Samples
In this study, SparCC algorithm was used to calculate the relationship between genera identified in Doushen and it was visualized as the network (v 3.7.1) (Figures 7, 8). The network for fungal communities in spicy Doushen consisted of 50 nodes and 34 edges (Figure 7). Results suggest the network is cooperative, and the ratio of cooperative and non-cooperative interaction is 20:14. Trichothecium had a negative relationship with Serendipita, but had a positive relationship with three other genera: Lentinula, Arthrocatena, and Rhizomucor. All effects related to Massarina were positive, including three other genera: Peroneutypa, Microascus, and Slimplicillium.
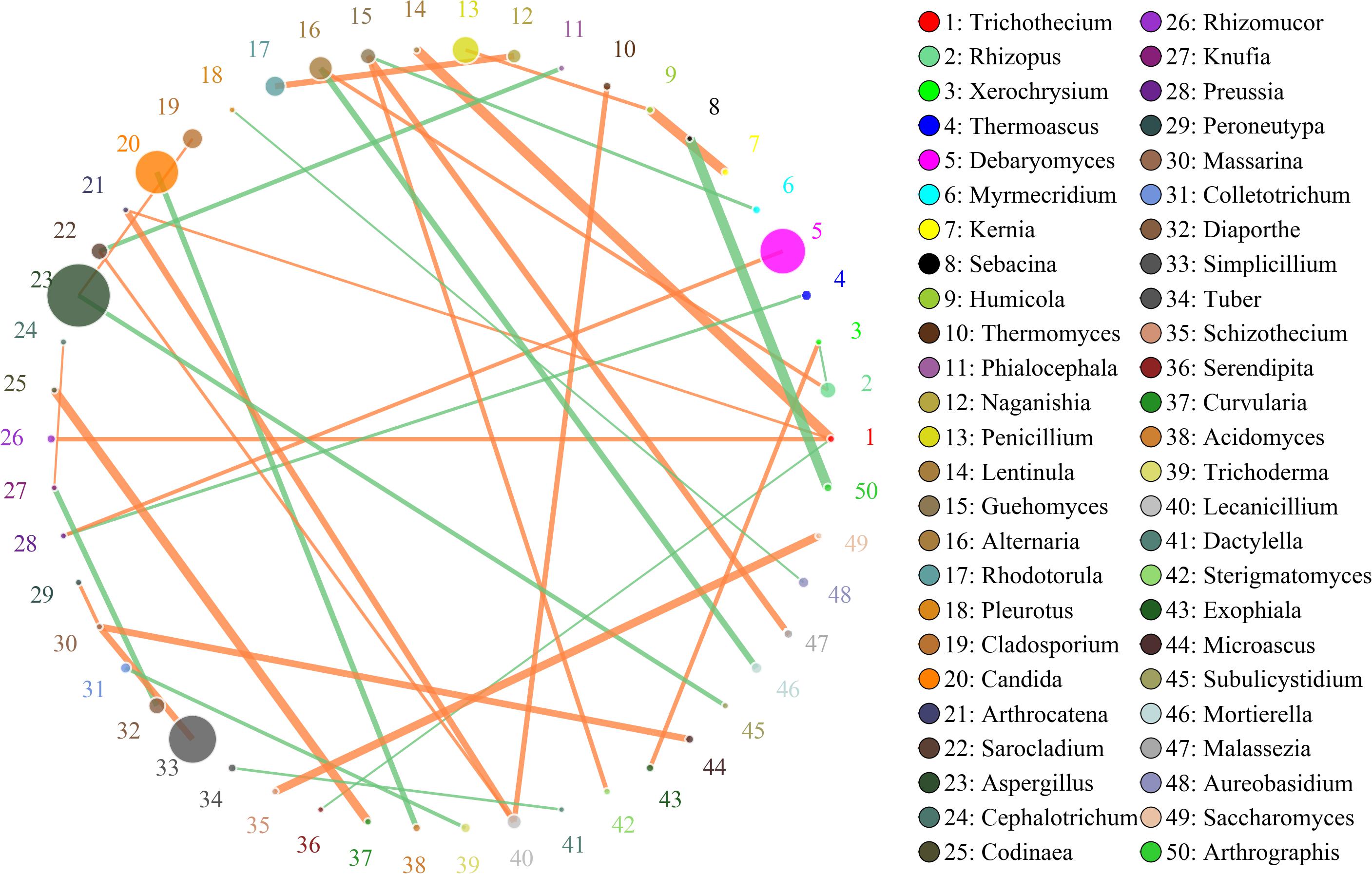
Figure 7. Networks of microbial interaction in the spicy doushen samples. Circle is on behalf of the species, size of the circle represents the abundance. Lines represent the correlation between the two species, line thickness to represent the strength of the correlation, the color of the line: orange represents the positive correlation, green represents the negative correlation.
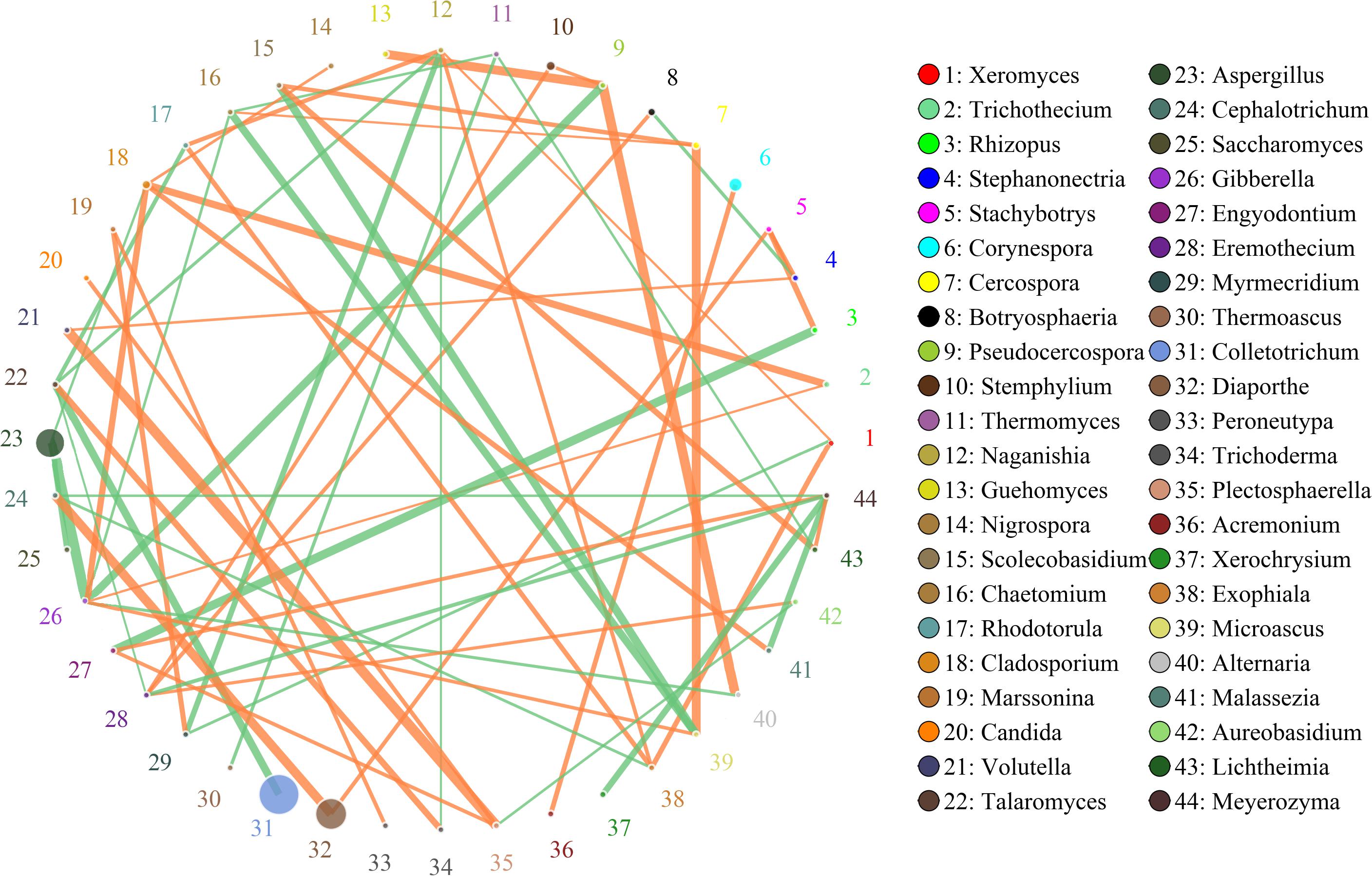
Figure 8. Networks of microbial interaction in the spiced doushen samples. Circle is on behalf of the species, size of the circle represents the abundance. Lines represent the correlation between the two species, line thickness to represent the strength of the correlation, the color of the line: orange represents the positive correlation, green represents the negative correlation.
The network for microbial community in spiced Doushen consists of 44 nodes and 61 edges (Figure 8). There were three fungal genera were found to be hub genera (≥6 edges per node) in the network, including Naganishia, Gibberella, and Meyerozyma. Results suggest the network is cooperative, and the ratio of cooperative and non-cooperative interaction is 35:26. Aspergillus had a negative relationship with three other genera: Cladosporium, Saccharomyces, and Gibberella. All of the correlation coefficients associated with Marssonina were positively correlated. These included Myrmecridium and Peroneutypa. Gibberella was found to have a negative relationship with three genera: Alternaria, Chaetomium, and Pseudocercospora, while have a positive relationship with three other genera: Cladosporium, Microascus, and Trichothecium. Cladosporium had a positive relationship with three other genera: Malassezia, Nigrospora, and Trichothecium.
Enumeration and Identification of Fungi
The number of fungal colonies in the WXo and WXi groups was 4.52 log CFU/g sample and 4.09 log CFU/g sample, respectively. The fungal counts in spicy Doushen were 5.83 log CFU/g sample in XLo and 5.12 log CFU/g sample in XLi. The enumeration results showed that the fungal population was lower in spiced Doushen than in spicy Doushen (p < 0.05). In addition, the results revealed that the number of fungi was lower in the interior area than in the exterior area of Doushen balls (Table 2).
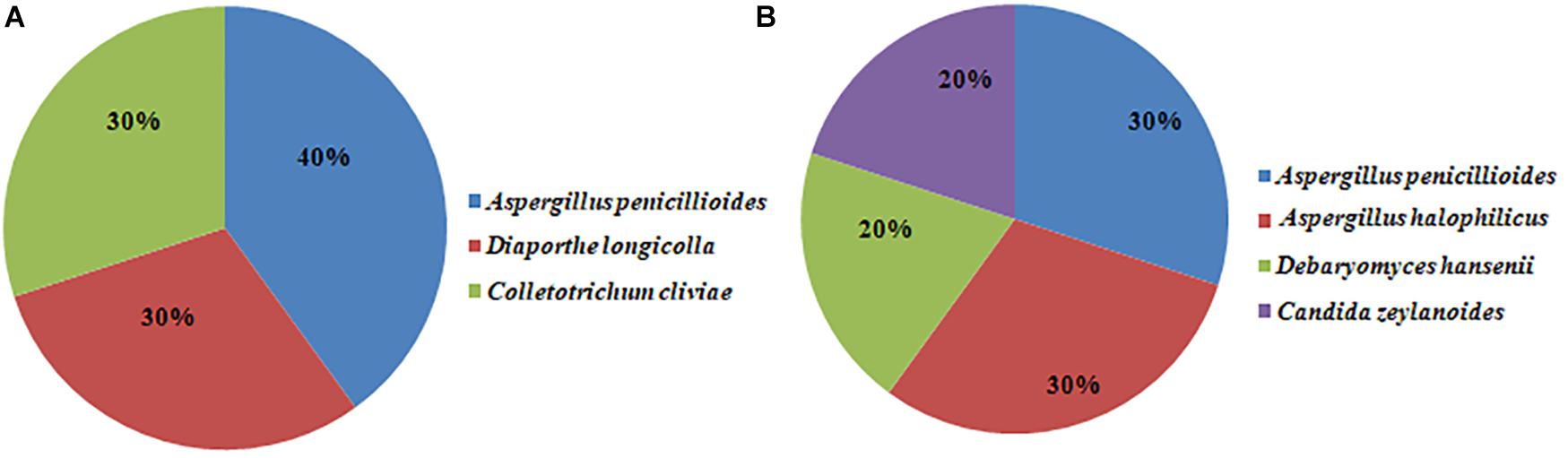
Twenty fungal strains were randomly selected from the Doushen samples and identified by ITS rRNA sequencing. Aspergillus penicillioides, Diaporthe longicolla, and Colletotrichum cliviae were detected in spicy Doushen and accounted for 40, 30, and 30%, respectively (Figure 9 and Table 5). A. halophilicus, A. penicillioides, Debaryomyces hansenii, and Candida zeylanoides were identified in spiced Doushen, accounting for 30, 30, 20, and 20%, respectively (Figure 9 and Table 5). These results were consistent with the composition of the dominant genera in the samples with different flavors.
Discussion
Different types of traditional fermented soybean products with special flavor have been produced and consumed around the world for centuries (Noriyuki, 1990; Wang et al., 2010; Li et al., 2018; Liu et al., 2018; Qiu et al., 2018; Zhang et al., 2018). Doushen, a douchi-like fermented soybean product, has been produced in Henan Province for a long time. The differences between Doushen and other traditional fermented soybean products is that the soybeans used for Doushen need to be minced. The study of microbial mechanisms is gaining considerable attention within the scientific community. In this study, the fungal diversity in two types of homemade traditional Doushen was evaluated by high-throughput sequencing. A total of nine fungal phyla and 188 fungal genera were detected in the Doushen samples. At the phylum level, Ascomycota, Basidiomycota, Rozellomycota, Chytridiomycota, Glomeromycota, and Zygomycota were previously detected in Douchi. However, Cercozoa and Ciliophora have not been previously detected in Douchi (Chen et al., 2011; Zhang et al., 2018). At the genus level, Aspergillus, Diaporthe, Colletotrichum, Gibellulopsis, Simplicillium, Bullera, Penicillium, Acremonium, Cryptococcus, Wallemia, Pichia, Yarrowia, and Candida were also detected in Douchi (Chen et al., 2011; Ji et al., 2014; Zhang et al., 2018). Among them, the relative abundance of Pichia, Candida, and Yarrowia was comparatively lower (Zhang et al., 2018). Aspergillus, Diaporthe, Colletotrichum, Gibellulopsis, and Simplicillium have a higher relative abundance in Doushen than in douchi (Zhang et al., 2018). However, Debaryomyces and Cystobasidium were not detected in a previous study (Zhang et al., 2018). The findings revealed a plethora of different types of fungi in the samples. Fermented soybean products are usually prepared with cooked soybeans and contain both endogenous and exogenous microorganisms involved in the fermentation process. Therefore, there is a variety of microorganisms present in the fermented soybean products (Chen et al., 2011; Yang et al., 2016).
Our study also demonstrated that the microorganisms in Doushen samples with different flavors exhibit a large degree of biodiversity. At the phylum level, the relative abundance of Basidiomycota, Cercozoa, and Zygomycotawas significantly different between spicy Doushen and spiced Doushen (Figure 3 and Table 4). The same applies to the relative abundance of Colletotrichum and Candida at the genus level (Table 4). The present study also showed that specific members of the fungal genera were dominant in the Doushen samples with different flavors. Aspergillus, Colletotrichum, Diaporthe, and Acremonium were the predominant fungi in spicy Doushen, whereas Aspergillus, Candida, Debaryomyces, Gibellulopsis, Simplicillium, Bullera, Penicillium, Cryptococcus, Thermoascus, and Malassezia were predominant in spiced Doushen. Given that Doushen is naturally fermented from steamed soybeans and do not require a starter, these fungi may emerge from the environment or other raw materials. There have been some reports that soybeans can be the main source of microorganisms in soybean fermentation (Mulyowidarso et al., 1989; Mulyowidarso et al., 1990; Ji et al., 2014). The present study showed that fungal diversity in different parts of Doushen samples was significantly different. These results imply that aeration and water content may be another important determinant, as previously reported in doenjang-meju (Ji et al., 2014). Chili Pepper, a key ingredient of several fermented soybean products, is present in Doushen products. Doushen with different flavors (spicy and spiced) were prepared with and without red pepper powder. Previous studies have revealed that chili pepper affects the microbial distribution in Kimchi and intestinal flora (Jeong et al., 2013; Cao et al., 2019). In this study, besides the significant difference in fungal composition between two types of Doushen samples, we found that the XL samples were more tightly clustered than WX samples (Figure 5). The result indicated that pepper powder exerted a very strong selection on fungal community structure of Doushen samples. Due to the absence of pepper powder, the fungal communities in the WX samples were more random than that in the XL samples. Previous studies have indicated that both deterministic and stochastic processes play an important role in microbial community assembly in natural environments (Chen et al., 2019; Hou et al., 2019; Gao et al., 2020). Future studies should use the ecological null models to decipher the relative importance of deterministic and stochastic processes in the microbial communities of Doushen.
Additionally, we obtained significant differences in microbial interactions between Doushen samples with different flavors, indicating that the Doushen microbial ecosystem may depend on different interactive relationships. Unique co-occurrence patterns were observed for Doushen samples with different flavors, which is likely to reflect adaptation to different environmental and nutritional conditions. Different types of fungi have different roles in soybean fermentation and could result in a complex relationship between species diversity and ecosystem function. Aspergillus, the most common genus of mold found in the environment, includes approximately 185 species, 20 species of which are harmful to humans and animals (Moy et al., 2012). However, Aspergillus was the predominant fungus in Douchi during koji-making and has been detected in meju, soy sauce, sofu, bean paste, and other traditional fermented soybean products (Lee et al., 2010; Moy et al., 2012; Li et al., 2017; He et al., 2019). Within the Aspergillus genus, the most common species is A.oryzae, which secretes enzymes that digest raw materials during soybean fermentation (He et al., 2019). Candida, a genus of yeast-like fungi, comprises pathogenic species that cause oral thrush. However, some species of Candida play important roles in enhancing the taste and quality of fermented soybean products, such as Douchi and soy sauce (Suezawa et al., 2006; Cao et al., 2010; Ying et al., 2010; Feng et al., 2012). Debaryomyces is an ascomycetous yeast species present in traditional fermented soybean foods such as Sufu, soybean pastes, doenjang, and kanjang (Azizul et al., 2015; Sun et al., 2018). The best-known species of this genus, D. hansenii, can grow under high salt concentrations and can secret various peptidases and proteases, which contribute to the aroma and flavor of fermented foods (Azizul et al., 2015). Cryptococcus is a genus of encapsulated yeast, which is found in fermented cereals, traditional llama meat sausages, traditional fermented soybean foods, alcoholic beverages, cheese, fermented pepper, and soy sauce (Corsetti et al., 2001; Volkmar et al., 2009; Lucía et al., 2014; Azizul et al., 2015; Zhao et al., 2016). Several species of Cryptococcus can produce extracellular enzymes such as lipase, pectinase, cellulase, xylanase, and laccase (Gomes et al., 2000; Kamini et al., 2000; Nahoko et al., 2012). The genus Thermoascus includes some species that can grow at high temperatures (Hosoya et al., 2014). These fungi have been identified in tea, olive cake, fruit juice, and other agricultural products and can produce amylase, xylanase, and cellulase (Adams, 1992; Wareing, 1997; Ping et al., 2018). Bullera, a genus belonging to Ballistosporous, can produce conidia and are present in diverse plants (phyllosphere), dairy products, and various environments (Nakase, 2000; Mushtaq et al., 2013; Silva-Bedoya et al., 2014). Some species of Bullera are producers of β-galactosidase, toxins, and coenzyme Q10 (Golubev and Nakase, 1998; Cho et al., 2003). Other genera, such as Gibellulopsis, Simplicillium, Penicillium, Colletotrichum, and Diaporthe, mainly include a number of fungal pathogens (Araceli et al., 2010; Udayanga et al., 2012). Numerous species of these genera produce extracellular enzymes such as glucoamylase, lipase, pectinase, and cellulase, which may affect product quality (Bahkali, 1992; Araceli et al., 2010; Prajapati et al., 2013). Some species of Penicillium, Colletotrichum, and Diaporthe produce fungal toxins under certain conditions (Kung et al., 1976; Gunnar et al., 2012; Zhang et al., 2012). In the present study, the exact function of the microorganisms has not been determined for lack of information on the species. Therefore, to assess their role on Doushen quality, future studies should isolate and grow these fungi.
Conclusion
High-throughput sequencing is a powerful method for exploring a large diversity of microorganisms in an array of environments. Until recently, few studies have focused on food microbiota. In this study, fungal diversity and communities in Doushen samples with different flavors were studied using high-throughput sequencing. This is the first study that applies this technology to study food ecology in Doushen. The results provided detailed information on the fungal diversity of Doushen, which is important for the production of Doushen.
Data Availability Statement
The datasets generated for this study can be found in the PRJNA493155.
Author Contributions
QL, ZZ, and WZ contributed conception and design of the study. YZ is co-first author. ZZ and YZ performed the statistical analysis. QL and YC wrote the first draft of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This work was supported by the Special funds for discipline construction of GAU (GAU-XKJS-2018-247), the National Natural Science Fund of China (31760466, 31960486, and 21864002), and the Enterprise research transformation and industrialization project (No. 2018-SF-C29) for financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ http://qiime.org/scripts/alpha_diversity.html
- ^ https://www.r-project.org/
- ^ http://qiime.org/1.4.0/scripts/jackknifed_beta_diversity.html
- ^ http://huttenhower.sph.harvard.edu/galaxy/
- ^ https://international.biocloud.net/zh/software/tools/
References
Adams, P. R. (1992). Growth and amylase production of Thermoascus aurantiacus Miehe. Biotechnol. Appl. Biochem. 15, 311–313.
Ahram, O., Eric, B. M., and Deog, H. (2018). Screening for potential probiotic bacteria from Korean fermented soybean paste: in vitro and Caenorhabditis elegans model testing. LWT Food Sci. Technol. 88, 132–138. doi: 10.1016/j.lwt.2017.10.007
AOAC (2000). “Official methods of analysis,” Association of Official Analytical Chemists (Gait Hersburg, Maryland).
Araceli, M. R., Marcela, G., María, C. G., and Laura, L. (2010). Pectinolytic enzyme production by Colletotrichum truncatum, causal agent of soybean anthracnose. Rev. Iberoam. Micol. 27, 186–190. doi: 10.1016/j.riam.2010.06.002
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Azizul, H., Weon, T. S., Chung, E. H., Hee, Y. L., Min, J. A., and Kim, M. C. (2015). Culture-independent analysis of yeast diversity in Korean traditional fermented soybean foods (doenjang and kanjang) based on 26SrRNA sequence. J. Korean Soc. Appl. Biol. Chem. 58, 377–385. doi: 10.1016/S0955-0674(00)00238-6
Bahkali, A. H. (1992). In vitro production of pectolytic and cellulolytic enzymes by Colletotrichum lindemuthianum isolated from soybean grown in Saudi Arabia. World J. Microbiol. Biotechnol. 8, 55–59. doi: 10.1007/BF01200685
Cao, S. Y., Zhao, C. N., Xu, X. Y., Tang, G. Y., Corke, H., Gan, R. Y., et al. (2019). Dietary plants, gut microbiota, and obesity: Effects and mechanisms. Trends Food Sci. Technol. 92, 194–204. doi: 10.1016/j.tifs.2019.08.004
Cao, X., Hou, L., Lu, M., and Wang, C. (2010). Improvement of soy-sauce flavour by genome shuffling in Candida versatilis to improve salt stress resistance. Int. J. Food Sci. Technol. 45, 17–22. doi: 10.1111/j.1365-2621.2009.02085.x
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, T. T., Jiang, S. Y., Xiong, S. Q., Wang, M. J., Zhu, D. C., and Wei, H. (2012). Application of denaturing gradient gel electrophoresis to microbial diversity analysis in Chinese Douchi. J. Sci. Food Agric. 92, 2171–2176. doi: 10.1002/jsfa.5604
Chen, T. T., Wang, M. J., Jiang, S. Y., Xiong, S. Q., Zhu, D. C., and Wei, H. (2011). Investigation of the microbial changes during koji-making process of Douchi by culture-dependent techniques and PCR-DGGE. Int. J. Food Sci. Technol. 46, 1878–1883. doi: 10.1111/j.1365-2621.2011.02696.x
Chen, T. T., Wang, M. J., Li, S. J., Wang, M. J., Wu, Q. L., and Wei, H. (2014). Molecular identification of microbial community in surface and undersurface Douchi during post-fermentation. Food Sci. Biotechnol. 20, 1633–1638. doi: 10.1111/1750-3841.12417
Chen, W., Ren, K., Isabwe, A., Chen, H., Liu, M., and Yang, J. (2019). Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome 7:138. doi: 10.1186/s40168-019-0749-8
Cho, Y. J., Shin, H. J., and Bucke, C. (2003). Purification and biochemical properties of a galactooligosaccharide producing β-galactosidase from Bullera singularis. Biotechnol. Lett. 25, 2107–2111. doi: 10.1023/B:BILE.0000007077.58019.bb
Cole, J. R., Chai, B., Marsh, T. L., Farris, R. J., Wang, Q., Kulam, S. A., et al. (2003). The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31, 442–443. doi: 10.1007/BF02860455
Corsetti, A., Rossi, J., and Gobbetti, M. (2001). Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69, 1–10. doi: 10.1016/s0168-1605(01)00567-0
Feng, J., Zhan, X. B., Wang, D., Zhang, L. M., and Lin, C. C. (2012). Identification andanalysis of the metabolic functions of a high-salt-tolerant halophilic aromatic yeast Candida etchellsii for soy sauce production. World J. Microbiol. Biotechnol. 28, 1451–1458. doi: 10.1007/s11274-011-0945-6
Friedman, J., and Alm, E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. doi: 10.1371/journal.pcbi.1002687
Fu, Y. J., Li, X., Zheng, S. H., Du, J., and Liang, A. H. (2015). Classification and identification of bacteria in the soil treated by AcMNPV using high-throughput sequencing technique. Biotechnol. Bioproc. E 20, 931–936. doi: 10.1007/s12257-015-0228-4
Gao, C., Montoya, L., Xu, L., Madera, M., Hollingsworth, J., Purdom, E., et al. (2020). Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nat. Commun. 11:34. doi: 10.1038/s41467-019-13913-9
Golubev, V. I., and Nakase, T. (1998). Mycocinogeny in the genus Bullera: killer activity of Bulleraunica and intrageneric killer-sensitive relationships. Microbiology 67, 184–188.
Gomes, J., Gomes, I., and Steiner, W. (2000). Thermolabile xylanase of the Antarctic yeast Cryptococcus adeliae: production and properties. Extremophiles 4, 227–235. doi: 10.1007/s007920070024
Gunnar, S. E., Thomas, R., Mattis, B. W., Frode, F., Aksel, B., and Angel, M. (2012). Poisoning by penicillium toxins—Clinical and brain receptor effects. Toxicol. Lett. 211, 58–59. doi: 10.1016/j.toxlet.2012.03.231
He, B., Li, H., Hu, Z., Zhang, Y., Sun, M., Qiu, S., et al. (2019). Difference in microbial community and taste compounds between Mucor-type and Aspergillus-type Douchi during koji-making. Food Res. Int. 121, 136–143. doi: 10.1016/j.foodres.2019.03.031
Helena, K., Maria, P., and Tea, H. (2016). Effective detection of indoor fungi by metabarcoding. Ann. Microbiol. 66, 495–498. doi: 10.1007/s13213-015-1118-x
Hosoya, K., Nakayama, M., Tomiyama, D., Matsuzawa, T., Imanishi, Y., Ueda, S., et al. (2014). Risk analysis and rapid detection of the genus Thermoascus, food spoilage fungi. Food Control 41, 7–12. doi: 10.1016/j.foodcont.2013.12.021
Hou, L., Mulla, S. I., Niño-Garcia, J. P., Ning, D., Rashid, A., Hu, A., et al. (2019). Deterministic and stochastic processes driving the shift in the prokaryotic community composition in wastewater treatment plants of a coastal Chinese city. Appl. Microbiol. Biotechnol. 103, 9155–9168. doi: 10.1007/s00253-019-10177-7
Inoue, Y., Kato, S., Saikusa, M., Suzuki, C., Otsubo, Y., Tanaka, Y., et al. (2016). Analysis of the cooked aroma and odorants that contribute to umami aftertaste of soy miso (Japanese soybean paste). Food Chem. 213, 521–528. doi: 10.1016/j.foodchem.2016.06.106
Jayachandran, M., and Xu, B. J. (2019). An insight into the health benefits of fermented soy products. Food Chem. 271, 362–371. doi: 10.1016/j.foodchem.2018.07.158
Jeong, S. H., Lee, H. J., Jung, J. Y., Lee, S. H., Seo, H. Y., Park, W. S., et al. (2013). Effects of red pepper powder on microbial communities and metabolites during kimchi fermentation. Int. J. Food Microbiol. 160, 252–259. doi: 10.1016/j.ijfoodmicro.2012.10.015
Ji, Y. J., Se, H. L., and Che, O. J. (2014). Microbial community dynamics during fermentation of doenjang-meju, traditional Korean fermented soybean. Int. J. Food Microbiol. 185, 112–120. doi: 10.1016/j.ijfoodmicro.2014.06.003
Judoamidjojo, M. (1986). The Studies on Kecap-Indigenous Seasoning of Indonesia. Ph.D. thesis, The Tokyo University of Agriculture, Tokyo.
Kamini, N., Fujii, T., Kurosu, T., and Iefuji, H. (2000). Production, purification and characterization of an extracellular lipase from the yeast, Cryptococcus sp. S-2. Process Biochem. 36, 317–324. doi: 10.1016/S0032-9592(00)00228-4
Kim, T., Lee, J. H., Park, M. H., and Kim, H. Y. (2010). Analysis of bacterial and fungal communities in Japanese and Chinese-fermented soybean pastes using nested PCR-DGGE. Curr. Microbiol. 60, 315–320. doi: 10.1007/s00284-009-9542-4
Koljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Kozyaeva, V. V., Grouzdev, D. S., Dziuba, M. V., Kolganova, T. V., and Kuznetsov, B. B. (2017). Diversity of magnetotactic bacteria of the Moskva River. Microbiology 86, 106–112. doi: 10.1134/S0026261717010088
Kung, H. C., Chipley, J. R., Latshaw, J. D., and Kerr, K. M. (1976). Mycotoxicosis in chicks produced by toxins from Phomopsis sp. or Diaporthephaseolorum var. sojae. Avian Dis. 20, 504–522. doi: 10.2307/1589383
Lee, J. H., Kim, T. W., Lee, H., Chang, H. C., and Kim, H. Y. (2010). Determination of microbial diversity in meju, fermented cooked soya beans, using nested PCR-denaturing gradient gel electrophoresis. Lett. Appl. Microbiol. 51, 388–394. doi: 10.1111/j.1472-765X.2010.02906.x
Li, H., Lin, L., Feng, Y., Zhao, M., Li, X., Zhu, Q., et al. (2018). Enrichment of antioxidants from soy sauce using macroporous resin and identification of 4-ethylguaiacol, catechol, daidzein, and 4-ethylphenol as key small molecule antioxidants in soy sauce. Food Chem. 240, 885–892. doi: 10.1016/j.foodchem.2017.08.001
Li, X., Zhao, C., Zheng, C., Liu, J., Vu, V. H., Wang, X., et al. (2017). Characteristics of microbial community and aroma compounds in traditional fermentation of Pixian broad bean paste as compared to industrial fermentation. Int. J. Food Prop. 20, 2520–2531. doi: 10.1080/10942912.2017.1373358
Lisa, S., Tikam, C. D., Paolo, G., and Paolo, G. (2013). Next-generation sequencing and its potential impact on food microbial genomics. Ann. Microbiol. 1, 21–37. doi: 10.1007/s13213-012-0478-8
Liu, Y., Su, H., and Song, H. L. (2018). Comparison of four extraction methods, SPME, DHS, SAFE, Versus SDE, for the analysis of flavor compounds in Natto. Food Anal. Method 2, 343–354. doi: 10.1007/s12161-017-1005-0
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β-diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/aem.01996-06
Lucía, M. M., Beatriz, P., Carmela, B., and Graciela, V. (2014). Diversity and enzymatic profile of yeasts isolated from traditional llama meat sausages from north-western Andean region of Argentina. Food Res. Int. 62, 572–579. doi: 10.1016/j.foodres.2014.04.008
Mcardle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.2307/2680104
Moy, Y. S., Lu, T. J., and Chou, C. C. (2012). Volatile components of the enzyme- ripened sufu, a Chinese traditional fermented product of soy bean. J. Biosci. Bioeng. 113, 196–201. doi: 10.1016/j.jbiosc.2011.09.021
Mulyowidarso, R. K., Fleet, G. H., and Buckle, K. A. (1989). The microbial ecology of soybean soaking for tempeh production. Int. J. Food Microbiol. 8, 35–46. doi: 10.1016/0168-1605(89)90078-0
Mulyowidarso, R. K., Fleet, G. H., and Buckle, K. A. (1990). Association of bacteria with the fungal fermentation of soybean tempeh. J. Appl. Bacteriol. 68, 43–47. doi: 10.1111/j.1365-2672.1990.tb02546.x
Mushtaq, M., Sharfun, N., and Hashmi, M. H. (2013). Screening of killer-sensitive pattern (ksp) for biotyping yeast strains isolated from dairy products. Pak. J. Bot. 45, 1039–1044.
Nahoko, N., Kazuo, M., Hiroaki, T., Tsutomu, F., and Haruyuki, I. (2012). Comparison of laccase production levels in Pichiapastoris and Cryptococcus sp. S-2. J. Biosci. Bioeng. 115, 394–399. doi: 10.1016/j.jbiosc.2012.10.025
Nakase, T. (2000). Expanding world of ballistosporous yeasts. Distribution in the phyllosphere, systematics and phylogeny. J. Gen. Appl. Microbiol. 46, 189–216. doi: 10.2323/jgam.46.189
Nam, Y. D., Lee, S. Y., and Lim, S. I. (2012). Microbial community analysis of Korean soybean pastes by next-generation sequencing. Int. J. Food Microbiol. 155, 36–42. doi: 10.1016/j.ijfoodmicro.2012.01.013
Ping, L., Wang, M., Yuan, X., Cui, F., Huang, D., Sun, W., et al. (2018). Production and characterization of a novel acidophilic and thermostable xylanase from Thermoascus aurantiacu. Int. J. Biol. Macromol. 109, 1270–1279. doi: 10.1016/j.ijbiomac.2017.11.130
Prajapati, V. S., Trivedi, U. B., and Patel, K. C. (2013). Kinetic and thermodynamic characterization of glucoamylase from Colletotrichum sp. KCP1. Indian J. Microbiol. 54, 87–93. doi: 10.1007/s12088-013-0413-0
Qin, Y. C., Fu, Y. M., Dong, C., Jia, N. N., and Liu, H. (2016). Shifts of microbial communities of wheat (Triticumaestivum L.) cultivation in a closed artificial ecosystem. Appl. Microbiol. Biotechnol. 100, 4085–4095. doi: 10.1007/s00253-016-7317-y
Qiu, S., Wang, Y., Cheng, Y. Q., Liu, Y., Yadav, M. P., and Yin, L. J. (2018). Reduction of biogenic amines in sufu by ethanol addition during ripening stage. Food Chem. 239, 1244–1252. doi: 10.1016/j.foodchem.2017.07.056
Schlosset, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: Opensource, platform-independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Silva-Bedoya, L. M., Ramírez-Castrillón, M., and Osorio-Cadavid, E. (2014). Yeast diversity associated to sediments and water from two Colombian artificial lakes. Braz. J. Microbiol. 45, 135–142. doi: 10.1590/S1517-83822014005000035
Suezawa, Y., Kimura, I., Inoue, M., Gohda, N., and Suzuki, M. (2006). Identification andtyping of miso and soy sauce fermentation yeasts, Candida etchellsii and C. versatilis, based on sequence analyses of the D1D2 domain of the 26S ribosomal RNA gene, andthe region of internal transcribed spacer 1, 5.8S ribosomal RNA gene. J. Agric. Chem. Soc. Jpn. 70, 348–354. doi: 10.1271/bbb.70.348
Sun, X., Lu, G., Luan, Y., Zhao, Z., Yang, H., and Su, D. (2018). Analyses of microbial community of naturally homemade soybean pastes in Liaoning Province of China by Illumina Miseq Sequencing. Food Res. Int. 111, 50–57. doi: 10.1016/j.foodres.2018.05.006
Udayanga, D., Liu, X., Crous, P. W., McKenzie, E. C., Chukeatirote, E., and Hyde, K. D. (2012). A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers. 56, 157–171. doi: 10.1007/s13225-012-0190-9
Volkmar, P., Ann-Christin, A., Matilda, O., Bart, T., Teun, B., and Johan, S. (2009). Cryptococcus cerealis sp. nov. a psychrophilic yeast species isolated from fermented cereals. Antonie Van Leeuwenhoek 96, 635–643. doi: 10.1007/s10482-009-9379-7
Wang, L. J., Cheng, Y. Q., Yin, L. J., Bhandari, B., Saito, M., and Li, L. T. (2010). Changes during processing and sodium chloride supplement on the physical and chemical properties of Douchi. Int. J. Food Prop. 13, 131–141. doi: 10.1080/10942910802256149
Wang, L. J., Li, D., Zou, L., Chen, X. D., Cheng, Y. Q., Yamaki, K., et al. (2007). Antioxidative activity of Douchi (A Chinese Traditional Salt-Fermented Soybean Food) extracts during Its Processing. Int. J. Food Prop. 10, 385–396. doi: 10.1080/10942910601052715
Wareing, P. W. (1997). Incidence and detection of thermotolerant and thermophilic fungi from maize with particular reference to Thermoascus species. Int. J. Food Microbiol. 35, 137–145. doi: 10.1016/s0168-1605(96)01212-3
Wu, Y., Chi, X., Zhang, Q., Chen, F., and Deng, X. (2018). Characterization of the salivary microbiome in people with obesity. PeerJ 6:e4458. doi: 10.7717/peerj.4458
Yang, L., Yang, H. L., Tu, Z. C., and Wang, X. L. (2016). High-throughput sequencing of microbial community diversity and dynamics during douchi fermentation. PLoS One 11:e0168166. doi: 10.1371/journal.pone.0168166
Yebra, M. J., and Monedero, V. (2010). Perspectives of engineering lactic acid bacteria for biotechnological polyol production. Appl. Microbiol. Biotechnol. 4, 1003–1015. doi: 10.1007/s00253-010-2494-6
Ying, L., Zhao, H., Zhao, M., and Cui, C. (2010). Relationships between antioxidant activity and quality indices of soy sauce: an application of multivariate analysis. Int. J. Food Sci. Technol. 45, 133–139. doi: 10.1111/j.1365-2621.2009.02112.x
Zhang, G. M., Fang, B. H., Chen, H., and Li, X. L. (2012). Characteristics of the Toxin extracted from liquid culture of Colletotrichumcapsici F. nicotianae. Appl. Biochem. Biotechnol. 167, 52–61. doi: 10.1007/s12010-012-9656-1
Zhang, J. H., Eizo, T., Fan, J. F., and Li, L. T. (2007). Chemical components of Aspergillus-type Douchi, a Chinese traditional fermented soybean product, change during the fermentation process. Int. J. Food Sci. Technol. 3, 263–268. doi: 10.1111/j.1365-2621.2005.01150.x
Zhang, W. B., Luo, Q. Q., Zhu, Y., Ma, J., Cao, L., Yang, M., et al. (2018). Microbial diversity in two traditional bacterial douchi from Gansu province in northwest China using Illumina sequencing. PLoS One 13:e0197527. doi: 10.1371/journal.pone.0194876
Zhang, Y., Skaar, I., Sulyok, M., Liu, X., Rao, M., and Taylor, J. W. (2016). The microbiome and metabolites in fermented Pu-erh tea as revealed by high-throughput sequencing and quantitative multiplex metabolite analysis. PLoS One 11:e0157847. doi: 10.1371/journal.pone.0157847
Zhao, L., Li, Y., Jiang, L., and Deng, F. (2016). Determination of fungal community diversity in fresh and traditional Chinese fermented pepper by pyrosequencing. FEMS Microbiol. Lett. 363:fnw273. doi: 10.1093/femsle/fnw273
Keywords: Doushen, high-throughput sequencing, flavor, fungal community, diversity
Citation:: Luo Q, Zhu Y, Zhang Z, Cao Y and Zhang W (2020) Variations in Fungal Community and Diversity in Doushen With Different Flavors. Front. Microbiol. 11:447. doi: 10.3389/fmicb.2020.00447
Received: 08 January 2020; Accepted: 02 March 2020;
Published: 20 March 2020.
Edited by:
Laurent Dufossé, Université de la Réunion, FranceReviewed by:
Cheng Gao, University of California, Berkeley, United StatesSanjay K. Singh, Agharkar Research Institute, India
Copyright © 2020 Luo, Zhu, Zhang, Cao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weibing Zhang, NDUzMzAzMDFAcXEuY29t
 Qiaoqiao Luo
Qiaoqiao Luo Yan Zhu
Yan Zhu Zhongming Zhang
Zhongming Zhang Yingying Cao
Yingying Cao Weibing Zhang
Weibing Zhang