- 1Carlsberg Research Laboratory, Group Research, Copenhagen, Denmark
- 2National Institute for Food, Technical University of Denmark, Kongens Lyngby, Denmark
- 3MS-Omics ApS, Vedbaek, Denmark
Brettanomyces yeasts have gained popularity in many sectors of the biotechnological industry, specifically in the field of beer production, but also in wine and ethanol production. Their unique properties enable Brettanomyces to outcompete conventional brewer’s yeast in industrially relevant traits such as production of ethanol and pleasant flavors. Recent advances in next-generation sequencing (NGS) and high-throughput screening techniques have facilitated large population studies allowing the selection of appropriate yeast strains with improved traits. In order to get a better understanding of Brettanomyces species and its potential for beer production, we sequenced the whole genome of 84 strains, which we make available to the scientific community and carried out several in vitro assays for brewing-relevant properties. The collection includes isolates from different substrates and geographical origin. Additionally, we have included two of the oldest Carlsberg Research Laboratory isolates. In this study, we reveal the phylogenetic pattern of Brettanomyces species by comparing the predicted proteomes of each strain. Furthermore, we show that the Brettanomyces collection is well described using similarity in genomic organization, and that there is a direct correlation between genomic background and phenotypic characteristics. Particularly, genomic patterns affecting flavor production, maltose assimilation, beta-glucosidase activity, and phenolic off-flavor (POF) production are reported. This knowledge yields new insights into Brettanomyces population survival strategies, artificial selection pressure, and loss of carbon assimilation traits. On a species-specific level, we have identified for the first time a POF negative Brettanomyces anomalus strain, without the main spoilage character of Brettanomyces species. This strain (CRL-90) has lost DaPAD1, making it incapable of converting ferulic acid to 4-ethylguaiacol (4-EG) and 4-ethylphenol (4-EP). This loss of function makes CRL-90 a good candidate for the production of characteristic Brettanomyces flavors in beverages, without the contaminant increase in POF. Overall, this study displays the potential of exploring Brettanomyces yeast species biodiversity to find strains with relevant properties applicable to the brewing industry.
Introduction
In 1904, the Danish scientist Niels Hjelte Claussen isolated and identified a new yeast species at the Carlsberg Research Laboratory (CRL—formerly Carlsberg Laboratory). The yeast was named “Brettanomyces” meaning “British fungus,” after being found in an English stock ale beer. Curiously, Brettanomyces was then the first microorganism in the history being patented (Claussen, 1904, 1906). In continuity to Claussen’s work, Holger Schiønning characterized more Brettanomyces isolates who he named “Torula” between the years 1905 and 1908 (Schiønning, 1908; Andersion, 2012). From then, Brettanomyces species have been isolated in wineries and breweries all over the world, as well as other substrates like sodas, olives, kombucha, and bioethanol production plants (Schifferdecker et al., 2014; Smith and Divol, 2016). In its sexual form, it is also referred to as Dekkera, a genus comprising the most frequently found species Dekkera/Brettanomyces bruxellensis and Dekkera/Brettanomyces anomalus (for review, see Smith, 2011). In addition, other asexual species of Brettanomyces have been described, such as Brettanomyces naardenensis, Brettanomyces custersianus, and Brettanomyces nanus (Kurtzman et al., 2011; Tiukova et al., 2019; Roach and Borneman, 2020). The potential of Brettanomyces species for brewing is controversial, as it is usually recognized as a spoilage yeast, being the cause of major economic losses in many production facilities (Gilliland, 1961; Lebleux et al., 2020). Its main spoilage feature is the production of phenolic off-flavor (POF), more specifically 4-ethylguaiacol (4-EG) and 4-ethylphenol (4-EP), imparting displeasing characters to the final product, described as horsy, barnyard, leather, band-aid, or medicine (Heresztyn, 1986; Chatonnet et al., 1992; Callemien and Collin, 2010). Nevertheless, Brettanomyces species are playing an important role in spontaneously fermented traditional Belgian beers, such as Lambics or Gueuzes, and are also contributing with “funky” characters to farmhouse ales (Spitaels et al., 2014; Crauwels et al., 2015a; Steensels et al., 2015). Over the last decade, the craft beer sector has constantly demanded novel attractive flavors, and there has been a rising interest in understanding Brettanomyces species, exploiting its potential in beer fermentation and ethanol production (for review, see Colomer et al., 2019).
Understanding the relationship between genotype and phenotype is essential for strain selection and optimization of brewing. However, the lack of genomic tools to perform gene deletions in Brettanomyces strains is still a bottleneck to establish convincing genotype-phenotype correlations (Miklenić et al., 2015; Varela et al., 2018). Moreover, while yeast breeding via sporulation and mating is a common practice for strain improvement in brewer’s yeast (Sanchez et al., 2012; Steensels et al., 2014; Gibson et al., 2017), it is still unclear if there is such a tool available for Brettanomyces species. To date, several assemblies of B. bruxellensis species have been published (Curtin et al., 2012; Piškur et al., 2012; Borneman et al., 2014; Crauwels et al., 2014; Olsen et al., 2015; Fournier et al., 2017) displaying a wide intra-specific variability of B. bruxellensis species (Woolfit et al., 2007; Hellborg and Piškur, 2009; Borneman et al., 2014). There are frequent variations at genomic level, including re-organizations and gene duplications, with the number of chromosomes varying from 4 to 9 (Woolfit et al., 2007; Gounot et al., 2019). While most of the population is diploid (2n) or triploid (3n) (Borneman et al., 2014; Avramova et al., 2018a), up to five alleles for certain locus have been reported (Avramova et al., 2018a). In addition, there is an increasing number of studies focusing on the human influence on the domestication of conventional brewer’s yeast suggesting that yeast populations have been pushed toward a concrete phenotype: POF negative, high flocculation, maltose assimilation (Gallone et al., 2016; Gonçalves et al., 2016; Preiss et al., 2017; Peter et al., 2018). Artificial selection pressure has also been investigated for Brettanomyces species, mainly with the usage of SO2 as a wine preservative. Recent studies have found a correlation between a triploid state and SO2 tolerance (Curtin and Pretorius, 2014; Avramova et al., 2018b). Interestingly, another recent study, using microsatellite genotyping of 1488 different B. bruxellensis isolates concluded that ploidy explains much of the population variance and can be correlated to SO2 tolerance (Avramova et al., 2018a).
The biochemical pathways involved in beer fermentation and aroma formation in brewer’s yeasts have been extensively studied. Fewer studies have been done in Brettanomyces yeasts, although genetic similarities with brewer’s yeasts have been reported (Curtin and Pretorius, 2014). One of them is the use of promoter rewiring for survival strategies, based on producing, accumulating, and consuming ethanol (Ihmels et al., 2005; Rozpedowska et al., 2011). A crucial trait for an efficient beer fermentation is maltose utilization. The MAL locus is commonly duplicated as an adaptive response to the substrate (Needleman et al., 1984; Vanoni et al., 1989; Gallone et al., 2016). While complex regulatory pathways involved in maltose utilization have been characterized in brewer’s yeast (Jespersen et al., 1999; Horák, 2013), maltose metabolism in Brettanomyces is still unclear. Different sugar consumption patterns have been reported, with variations in maltose and maltotriose utilization (Crauwels et al., 2017; Smith and Divol, 2018). Ethanol production is closely linked to sugar utilization, and Brettanomyces has been widely described as a potential bioethanol producer (Blomqvist et al., 2010; Aguilar-Uscanga et al., 2011; Schifferdecker et al., 2016). A unique property of Brettanomyces yeasts is their high β-glucosidase activity, conferring the capability to break beta linked substrates (Daenen et al., 2004, 2008b). This activity allows the release of aromatic monoterpene alcohols from hops, enhancing “flowery” and “citrus” characters in beer. Additionally, it facilitates Brettanomyces long-lasting survival in wooden barrels by enhancing the break-down of cellobiose, the main disaccharide present in wood (for review, see Steensels et al., 2015). Two ORFs coding for β-glucosidases have been identified, and other non-specific exo-glucanases (EXG) have been suggested (Crauwels et al., 2014; Kuo et al., 2018).
Quantification of volatile compounds (VOCs) is a common practice to predict the flavor and aroma of a beverage. VOCs are secondary metabolites produced by the yeast during fermentation, generally consisting of higher alcohols, esters, ketones, and phenols. Higher alcohols are commonly related to solvent-like aromas and are mainly products of the catabolism of aromatic amino acids (leucine, isoleucine, valine) through the Ehrlich pathway (Hazelwood et al., 2008). Esters are associated with pleasant fruity flavors and comprise two types: acetate esters and ethyl esters. Acetate esters are normally synthesized with acetyl-CoA reacting with ethanol or a higher alcohol, the most frequent being isoamyl acetate (banana aroma), ethyl acetate (lipstick aroma), and 2-phenylacetate (honey aroma) (Bisson and Karpel, 2010). These reactions are catalyzed by either Atf1 or Atf2, and can be reversed by the action of Iah1 esterase (Fujii et al., 1994; Fukuda et al., 2000). As a result, the ratio between higher alcohols and acetate esters remains dependent on the balance of Atf1/Iah1 activities (Fukuda et al., 1998). Ethyl esters are described as pineapple, grape, and tropical fruit aromas and are formed in a reaction of an activated fatty acid with ethanol by the activity of ethanol acyl transferases: Eeb1 and Eht1 (Saerens et al., 2006). In addition, formation of the “buttery” off-flavor diacetyl during alcoholic fermentation presents a challenge for brewers. Diacetyl is formed by a non-enzymatic reaction from acetolactate leaking out of the cell. Synthesis of acetolactate is mediated by the ILV2 and ILV6 genes among others (Gjermansen et al., 1988; Duong et al., 2011). As acetolactate is an intermediate in the branched-chain amino acids (BCAA) biosynthetic pathways, it can be feedback regulated by valine (Krogerus and Gibson, 2013). Furthermore, POF production by brewer’s yeasts has been well characterized. 4-Vinylguaiacol (4-VG) production is a result of a dimeric interaction of Pad1 and Fdc1 proteins decarboxylating ferulic acid from the media (Mukai et al., 2010). In Brettanomyces species, POF production consists of a two-step conversion pathway, with an additional reduction step of 4-VG to 4-EG (Heresztyn, 1986; Godoy et al., 2009). A phenolic acid decarboxylase (BbPad1) has been identified as being responsible for the decarboxylation of ferulic acid (Godoy et al., 2014), and a superoxide dismutase (BbSod) has been suggested for the second reduction step (4-VG to 4-EG) (Romano et al., 2017). The same pathway is used to produce 4-vinylphenol (4-VP) and 4-EP, using coumaric acid as a substrate. The proportion of 4-EG/4-EP in Brettanomyces fermented beer is 3:1, while in wine it is 1:1 or lower (for review, see Crauwels et al., 2015a). As 4-EP is generally more unpleasant than 4-EG, such ratio could play a significant role in explaining why Brettanomyces species are more undesired in wine (Crauwels et al., 2015a; Lentz and Harris, 2015; Cibrario et al., 2020).
In order to gain a better understating of the genetic diversity and the brewing capabilities of Brettanomyces, we sequenced the whole genome of 84 strains including the species B. bruxellensis, B. anomalus, B. custersianus, and B. naardenensis, derived from different sources worldwide, and make them available to the scientific community. The genomes were annotated, the function of each gene was predicted, and the strains were compared with each other. Strains were grouped according to the genomic organization, absence or presence of brewing-relevant genes was investigated. Essential brewing traits were investigated, including wort fermentation and flavor analysis along with in vitro assays to predict specific enzymatic activity. Finally, the genotype–phenotype relationship has been investigated, revealing new population trends and highlighting new application areas of Brettanomyces.
Materials and Methods
Brettanomyces Strain Collection and Culture Conditions
Brettanomyces strains were selected from CRL yeast strain collection. Strains used in this study are mainly isolated from beer. Isolates from wine, kombucha, sodas, olives, and bioethanol production plants, among others, are also included. The geographical origin of the strains is very diverse, comprising isolates from Europe, South America, Africa, and Australia (Figure 1 and Table 1). We have included two of the oldest known Brettanomyces isolates in the world, obtained at the CRL in Denmark between the years 1904–1908 (CRL-49, CRL-50) (Schiønning, 1908).
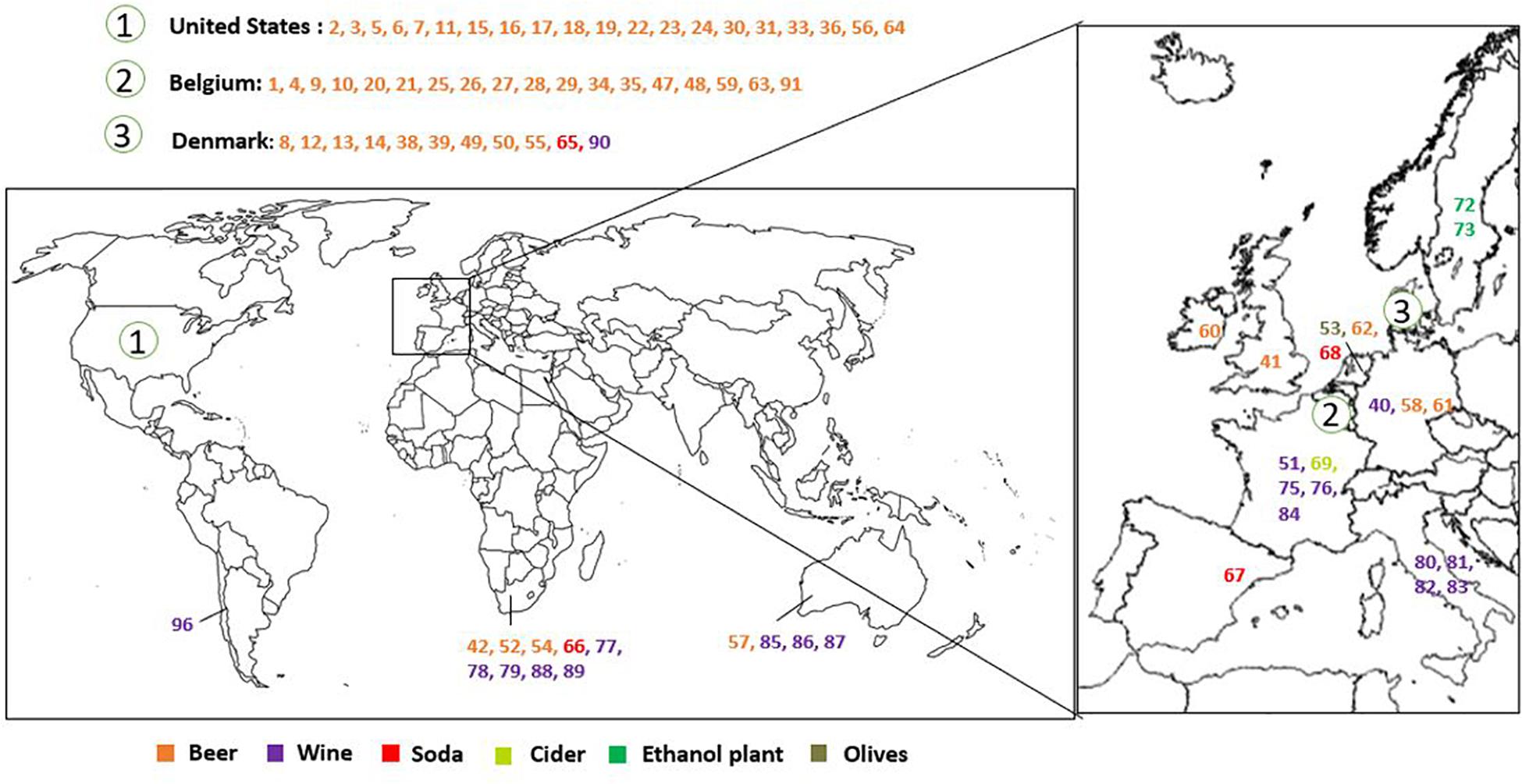
Figure 1. Geographical distribution and substrate of origin of the Brettanomyces strains included in the study.
For strain isolation from beverages, Wallerstein Laboratory Nutrient (WLN) agar containing bromocresol green supplemented with cycloheximide (4 μg/mL) was used as a selection media. Brettanomyces strains were differentiated based on color and morphology when metabolizing bromocresol green. Preliminary species differentiation was performed by polymerase chain reaction (PCR) amplification of the internal transcriber spacer (ITS) region followed by gel electrophoresis (Fujita et al., 2001). In the current study, 84 isolates were used, comprising 64 D. bruxellensis, 14 D. anomalus, three B. naardenensis, and three B. custersianus. For general cultivation, strains were grown in yeast peptone dextrose (YPD) at 25°C with agitation (100 r/min, IKA KS 501 digital). For storage purposes, YPD supplemented with 20% glycerol was used to freeze the cultures at −80°C. To perform high-throughput phenotyping, the strain collection was arranged in triplicates in Greiner Flat-Bottom Microtiter Plate (MTP, 96-well format). Biomek FXP Automated Workstation (Beckman–Coulter) was employed for general liquid handling and for in vitro assays.
Yeast Whole-Genome Sequencing
For genomic DNA isolation, single colonies were inoculated in liquid YPD and grown for 1 week in 50 mL shake flasks. Biomass was collected by centrifugation (4000 g, 10 min, 4°C), washed with sterile water, and stored at −4°C in 70% ethanol. Pellets were delivered for DNA extraction, short insert size library preparation (<800 bp), 150 bp paired-end sequencing on Illumina HiSeq4000 whole-genome sequencing (performed by BGI Europe®). In order to get high-quality genome assemblies, representatives for each species, were selected for PACBio RSII technology long-read sequencing (CRL-49 as B. anomalus, CRL-53 as B. custersianus, and CRL-65 as B. naardenensis) (performed by BGI Europe®). For the B. bruxellensis group, the publicly available genome of the isolate UMY321 was obtained from NCBI (Fournier et al., 2017).
Bioinformatics Analysis: Genome Assembly, Gene Calling, Prediction of Function, and Phylogenetic Comparison
De novo assembly for all the 84 yeast strains was performed using ABySS (Assembly By Short Sequences), a parallelized sequence assembler. Strain species were identified by blasting against the internal transcribed spacer (ITS) region from fungi type and reference material database of NCBI.
The PacBio raw sequencing reads were assembled with Canu software (Koren et al., 2017) using the default parameters. Each Canu assembly was used as a reference to map the corresponding short reads using fast and versatile mapper (Minimap2) (Li, 2018). The resulting SAM files were transformed into sorted and indexed BAM files using Samtools (Li et al., 2009). Finally, we used a tool for polishing our Canu assemblies with the Illumina short reads to produce an improved draft assembly for each of the PacBio sequenced yeast.
We next divided the whole pool into four groups (according to its species) and for each one of them defined the reference. For the B. anomalus species, the “polished” sequence of CRL-49 was used as a reference; the CRL-53 was used as a reference for the group B. custersianus, while the CRL-65 served as a reference for the B. naardenensis group. The genome of the D. bruxellensis UMY321 strain served as a reference for the B/D. bruxellensis group (Fournier et al., 2017). It is publicly available from NCBI and the authors obtained the most complete and contiguous de novo assembly of D. bruxellensis using a combination of Nanopore long-read and Illumina short-read sequencing data (Fournier et al., 2017). For all the four groups, we mapped the short Illumina reads to the reference using BWA-MEM algorithm and set a filter on MAPQ (mapping quality) 30, in order to get only uniquely mapped reads. We then generated a consensus sequence for each genome which was used for the downstream analyses.
For gene prediction, we used Augustus with the consensus sequence generated on the mapping step (Stanke et al., 2006). The Saccharomyces cerevisiae S288C was used as the closest related species. For each yeast, the amino acid sequence and the general feature file (GFF) were saved for the later steps. To predict the function of each open reading frame (ORF), a query against the Protein Database of NCBI with keywords “brettanomyces, saccharomyces, saccharomycopsis” was done, resulting in 1,318,613 hits, which were then used to build a BLAST DB (functional_db). Each of the proteins of the three references (CRL-49, CRL-53, CRL-65) were searched against the functional_db using an e-value cut-off of 1e-4 and in the case of success the function was then transferred from the hit to the query.
To compare the 85 yeast genomes, OrthoFinder, a platform for comparative genomics was used (Emms and Kelly, 2015, 2019). Orthogroups, gene orthologs, and complete set of gene tree were computed from raw amino acid sequences with OrthoFinder. The species tree was inferred from the set of unrooted orthogroup gene trees, and a consensus tree was taken of all individual estimates using STAG algorithm (Emms and Kelly, 2018), and was rooted using STRIDE (Emms and Kelly, 2017). The metadata consisting of “Source,” “Species,” and “Continent” was added in the dataset, and the phylogenetic tree was imported on CLC Genomics Workbench 11 software (QIAGEN Bioinformatics®). The tree visualization was elaborated under the Circular cladogram and radial display (QIAGEN, 2016). CLC Genomics Software was used for general genome analysis and data handling. BLAST function on CLC was used to predict regions of loss, and hits scoring > 95% of the total BLAST score were considered as valid for gene presence.
Phenotypic Characterization and in vitro Assays
Small-Scale Wort Fermentation
Yeast strains were propagated in triplicates in YPD media in 96-well plate format (Greiner Bio-one®) at room temperature with agitation at 100 r/min for 5 days. Cultures were diluted 1:20 in water prior to inoculation. 10 μL of the yeast dilution was inoculated into standard pilsner wort (Viking malt®) for a final volume of 270 μL/well; 96 well-plates (CR1496c—Enzyscreen®) were sealed with anaerobic lids and kept at 25°C with agitation at 200 r/min in a GrowthProfiler 960 (Enzyscreen®), with pictures automatically taken every 30 min to monitor cell growth. To obtain a relative growth value, pictures were converted to G-Value using the image analysis software (Enzyscreen). Fermentations were harvested after 7 days, cells were removed by centrifugation (3000 g, 4°C, 5 min), and supernatant was kept for further analysis.
Determination of Ethanol and Acetic Acid in Beers
Quantification of ethanol and acetic acid was performed in 96-well format following indicated microplate assay procedure. In order to quantify ethanol content, samples were diluted 1:200 in water and determined using the Ethanol Assay Kit K-ETOH® from Megazyme. In order to quantify acetic acid content, samples were diluted 1:100 in water and determined using Acetic Acid (Acetate Kinase Manual Format) K-ACETRM® from Megazyme. Both kits are based on stoichiometric reactions using NADH as a cofactor and measurements in the decrease/increase of absorbance at 340 nm (Megazyme®). A calibration curve of known concentrations was performed for the accurate determination of each compound. Optical density was measured in a Spark®Multimode Plate Reader (TECAN).
Determination of Volatile Compounds in Beer With Gas Chromatography/Mass Spectrometry
The main flavor-active compounds produced by the Brettanomyces collection were analyzed using gas chromatography/mass spectrometry analysis. The beer sample size was 200 μL. Analytes were extracted using liquid–liquid extraction with methyl-tert-butyl ether directly in the vial. The analysis was performed using a mid-polarity column (ZebronTM ZB-1701, GC Cap. Column 30 m × 0.25 mm × 0.25 μm) installed in a GC (7890B, Agilent) coupled with a quadrupole detector (59977B, Agilent). The system was controlled by ChemStation (Agilent). The GC-method was set up as described by Pinu and Villas-Boas (2017). Raw data were converted to netCDF format using Chemstation (Agilent), before the data were imported and processed in Matlab R2014b (Mathworks, Inc.). Quantifications were performed using external calibration lines.
Assay for Utilization of Glucose and Maltose With Biolog
Cells were grown in 96-well plates in YPD until saturation. Cultures were diluted in sterile water 1:104 before inoculation in the appropriate media. Media consisted of Yeast Nitrogen Base with amino acids supplemented with 1% glucose and maltose, respectively, as a carbon source. Strains were inoculated in triplicates into Biolog® 96-well plates and incubated at 25°C without agitation. Growth kinetics was monitored with OmniLog®Biolog. The quantification was coupled with formation of the purple formazan due to reduction of the tetrazolium dye by metabolically active cells. The quantification of the purple formazan formation was performed by Omnilog®Biolog software and its output is given in B values. Data handling was performed using OmniLog_FileManagement Software.
Assay for Growth on Cellobiose and Extracellular β-Glucosidase Activity
Brettanomyces strains were inoculated in triplicates into 96-well plates containing Yeast Peptone Cellobiose (2% Cellobiose) and incubated for 1 week at 25°C. Absorbance was measured at 600 nm to monitor growth. Cultures were spun down and supernatant was collected to measure extracellular β-glucosidase activity. The enzymatic beta-glucosidase test was performed with MAK129® β-glucosidase assay kit from Sigma–Aldrich. P-nitrophenyl-β-D-glucopyranoside (β-NPG) was used as a substrate and variation of optical density was measured at 405 nm after 20 min incubation at 37°C. Absorbance on 96-well plate format was measured on Spark®Multimode Plate Reader (TECAN). The assay results are given in units/L, one unit is the amount of enzyme that catalyzes the hydrolysis of 1.0 μmole substrate per minute at pH = 7.
Screening for POF and Quantification of Volatile Phenols With UPLC
Phenolic off-flavor production was screened using an absorbance-based method based on the uptake of ferulic acid (Steensels, 2017). Yeast cells were grown in triplicates in 96-well plates with YPD media for 7 days. Cultures were diluted 1:100 in water before inoculation in a new 96-well plate with YPD supplemented with ferulic acid at 0.1 mg/mL. After 1 week cultivation at 25°C with agitation (100 r/min, IKA KS 501 digital), plates were centrifuged (4000 g, 5 min, 4°C), 100 μL of supernatant was collected, and absorbance was measured at 325 nm using a Spark®Multimode Plate Reader (TECAN).
During wort fermentations, phenolic compounds (ferulic acid, coumaric acid, 4-EG, 4-EP) were quantified by ultra performance liquid chromatography (UPLC) (Waters) with PDA detection (280 nm). The separation was achieved using the BEH Phenyl Ultra Column (2.1 × 100 mm, 1.7 μm) and a flow of 0.5 mL/min. The injection volume was 1 μL. The mobile phase was 99.9% A, 0.1% B between 0 and 3 min. followed by a gradient up to 45% B for 5 min. Eluent A was contained 3% formic acid, 10% methanol in water and eluent B was 100% methanol. Calibration standards were prepared in methanol in the range 0.1–10 mg/L by dilution of a 10 mg/L standard mix. Beer samples were filtered on a 0.2 μm filter, diluted 1.5× with eluent A and vortexed for 5 s. Compounds were identified by retention time and ID was confirmed by spiking with standard solution.
Intermediate Fermentations
Strains were propagated in pilsner wort in 50 mL Erlenmeyer shake flasks. A pitching rate of 100,000 cells/mL was determined using a Cellometer X2 (Nexelom Bioscience) to count the cells. Fermentations were performed in duplicates in 250 mL Duran bottles containing 200 mL of standard pilsner wort (Viking Malt). Cumulative pressure was monitored with ANKOM RF Gas Production System® (ANKOM). Fermentations were stopped after 7 days, cells were removed by centrifugation (4000 g, 10 min, 4°C) and the supernatant was used for analysis.
Principal Component Analysis and Statistical Analysis
Principal component analysis (PCA) was performed on all 84 individuals, described by 12 variables. PCA was performed using the R software (version 3.5.2). Missing values were addressed by imputing them using the imputePCA function. PCA was performed using first five principal components, which explained 75% of the variance. A biplot displaying PC scores of different strains (points) and loadings for each variable (cos2-vector) was produced. Variables are colored by cos2 (squared coordinates) high cos2 indicates a good representation, a low-bad. The colors are: red = good, green = bad. Observations are shown as balls, which are colored according the grouping following the Genetic.Cluster. For each group, there is a bigger ball representing the mean of the group (centroid). Correlation and PCA scores were extracted and included. Statistical analysis was performed on phenotypical data and in vitro analysis, with one-way ANOVA followed by Tukey post hoc test (p < 0.05) using R software (version 3.5.2). The detailed output of the statistical analysis can be found at Supplementary Material (Statistical tests).
Results
Genetic Clustering of Brettanomyces Species
In order to study the population structure of Brettanomyces genus, we sequenced the whole genome of 84 Brettanomyces strains, comprising the species B. bruxellensis (64), B. anomalus (14), B. custersianus (3), and B. naardenensis (3). Through comparison of protein sequences, a phylogenetic tree comparing all 84 isolates was constructed (Figure 2A). The different Brettanomyces species cluster together as shown by the branch color (Figure 2A). The metadata layer 1 displaying the substrate of isolation infers that the genomic set-up of Brettanomyces population is strongly influenced by the media they survive in. Interestingly, a major clade comprising strains isolated from beer is formed. This clade contains mainly commercial strains applied for craft brewing referred to as “Farmhouse” and another clade where the majority of strains are isolated from spontaneously fermented Belgian Lambic beers, which is labeled as “Lambic.” Moreover, a sub-clade within the “Lambic” group could be identified, where the strains come from various natural origins; ethanol plants, barrel-aged beers, and matured wines. Therefore, this sub-clade is labeled as “Wood/Wild.” The old Carlsberg B. bruxellensis strain CRL-50 is located at the edge of the “Farmhouse” cluster, indicating that after deposition this strain could have been applied for brewing purposes. With regard to the wine spoilage isolates, the genetic divergence is much higher as displayed by branch lengths in Figure 2B. Genetic similarities among beer strains are the first hint of artificial selection pressure, in which strains with desired phenotypes have been recently applied. It is observed that the geographical distribution of the beer isolates seems to be more homogenous than wine isolates. In contrast, the artificial selection of wine-spoilage Brettanomyces has occurred in a reversed way. The results indicate that strains have evolved in parallel in several wine regions in the world, developing resistance to wine preservatives, and resulting in higher genetic divergences (Curtin and Pretorius, 2014; Avramova et al., 2018a). In regard to wine isolates, two main clades are found. As previously described (Borneman et al., 2014; Avramova et al., 2018b), population structure of B. bruxellensis displays a variable ploidy of 2n/3n. The cluster “Wine 2n” comprises mainly diploid strains [also described as (CBS2499-like) in Avramova et al., 2018a], and the reference genome UMY321, while the cluster “Wine 3n” comprises mainly triploid strains (AWRI1499-like) in Avramova et al. (2018a). Moreover, there are some isolates highly divergent from the rest of strains, CRL-63, -52 labeled as (“Tequila”-like) in Avramova et al. (2018a) and showing potential signs to be the common ancestors in B. bruxellensis species.
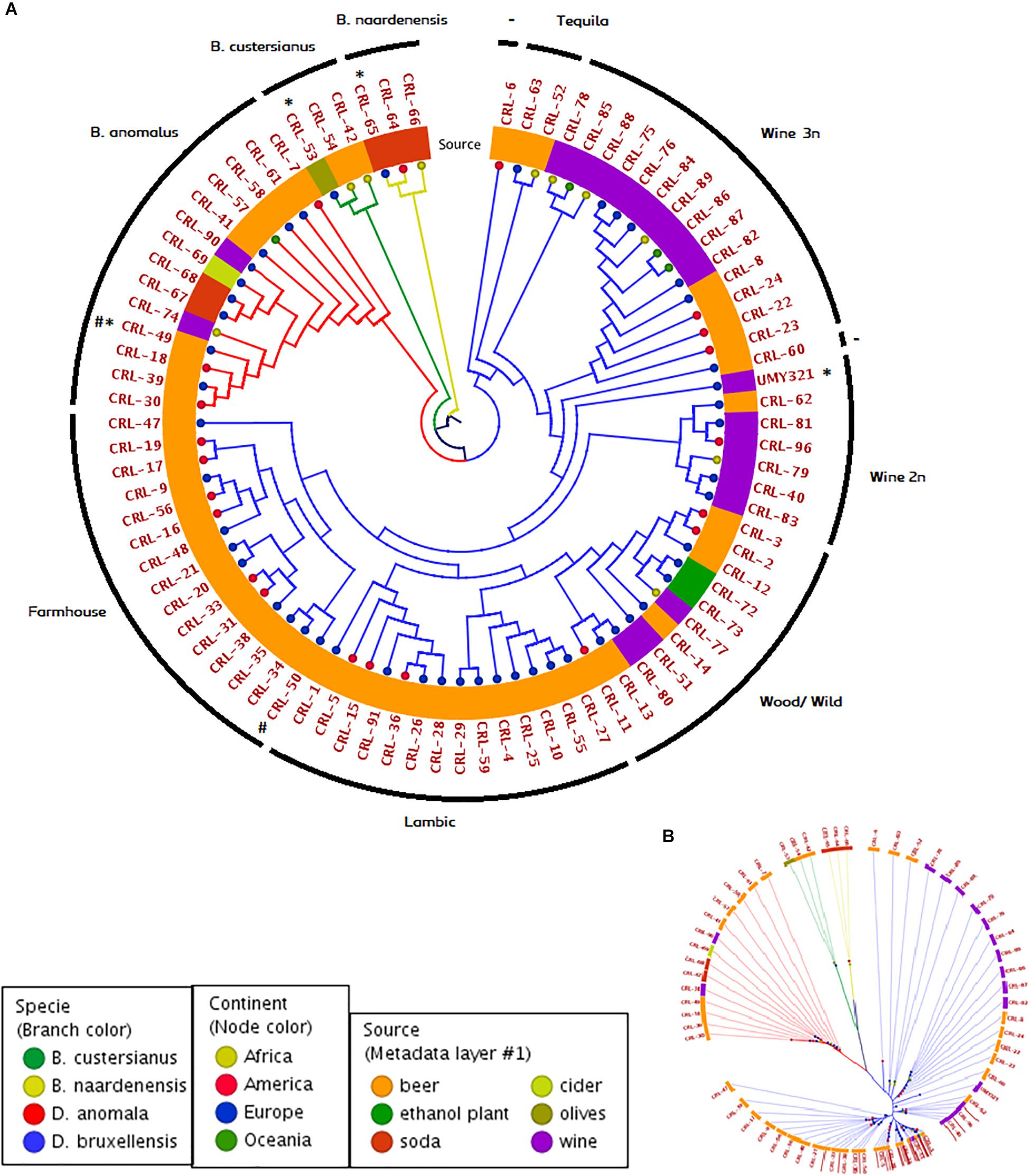
Figure 2. Genomic phylogeny of Brettanomyces population. (A) Circular claudogram of 84 Brettanomyces species included in this study. Phylogenetic tree was produced by comparison of the predicted proteins in the whole genome of each strain. Strains used as reference genome are marked with (∗) Old CRL isolates are marked with (#). Branches are colored according to species, tree nodes according to continent of origin, and metadata layer is colored according to substrate of isolation. Yeasts have been grouped in different genetic clusters according to its location in the tree and genetic cluster names are indicated in the external circle. (B) Same phylogenetic tree in radial form. Branch length and separation displays genomic divergence between Brettanomyces isolates.
Chromosomal Organization of the Reference B. bruxellensis UMY321 Strain: Distribution of Functional Genes
The high-quality reference genome UMY321 was downloaded from NCBI (Fournier et al., 2017). The genome is organized in eight scaffolds, which could be considered as a chromosome level structure. All the ORFs of the genome were identified, and the function of each gene was predicted. The most relevant genes for brewing were pinpointed, and we produced a chromosome-like map with an overview of the genome distribution (Figure 3). The list of predicted gene function and its relevance is explained in Supplementary Table S1. Despite the genome of Brettanomyces being highly dynamic and karyotypes strongly variable (Hellborg and Piškur, 2009; Curtin and Pretorius, 2014), we could display an overview of the polymorphic subtelomeric regions where more variation is expected (Brown et al., 2010). The centromeric loci CEN1, CEN2, responsible for chromosome rearrangements and change of ploidy (Ishchuk et al., 2016) is located in scaffold I. Scaffold I also contains a maltose assimilation cluster (MAL31, MAL11, IMA1), similar to the one already described in brewer’s yeast in which is essential for maltose degradation (Needleman et al., 1984; Horák, 2013; Needleman and Michels, 2015). In this case, no transcriptional activator such as ScMAL13 is present, but just an α-glucosidase surrounded by maltose permeases and transporters. The nitrate assimilation cluster is spotted in the subtelomeric region of scaffold IV, and both β-glucosidases (BGL1, 2) are found on the edges of scaffolds V and II, respectively. BbPAD1, gene responsible for POF phenotype, is located at the shortest scaffold (VIII). As a general overview, most diacetyl, ester, and ethanol production genes are found at conserved regions, therefore low variability in these phenotypes could be expected. Furthermore, several events of gene duplication are present, especially abundant in scaffold VI. This phenomenon is commonly seen as a mechanism of adaptation by yeast (Hittinger and Carroll, 2007; Ames et al., 2010; Dunn et al., 2012; Gorter de Vries et al., 2017; Harari et al., 2018). For example, three repetitions of the major isomaltase IMA1 are present, displaying copy number repetitions to enhance utilization of carbon sources (Duval et al., 2010; Naumoff and Naumov, 2010; Teste et al., 2010). These numerous repetitions of IMA1 are also common in brewing yeasts, with sequences sharing high similarities one to another (Libkind et al., 2011; Deng et al., 2014). ADH6 responsible for aldehyde reduction (Larroy et al., 2002) is also present in triplicate along the genome and could have a potential role in environmental adaptation and Brettanomyces survival. Duplication of ADH6 as a response to cell damage has been described in brewing yeast, as protein abundance increase when DNA-replication-stress is induced (Tkach et al., 2012). Moreover, three copies of isoamyl acetate esterase IAH1 are noticed, which are presumably involved in acetate ester degradation (Fukuda et al., 2000; Colomer et al., 2019). The FLO gene family is often duplicated at the subtelomeric regions, a phenomenon already well-described in brewing yeast (Teunissen and Steensma, 1995). This gene family often show intrinsic tandem repeats and occasionally are transcriptionally silent (Van Mulders et al., 2009).
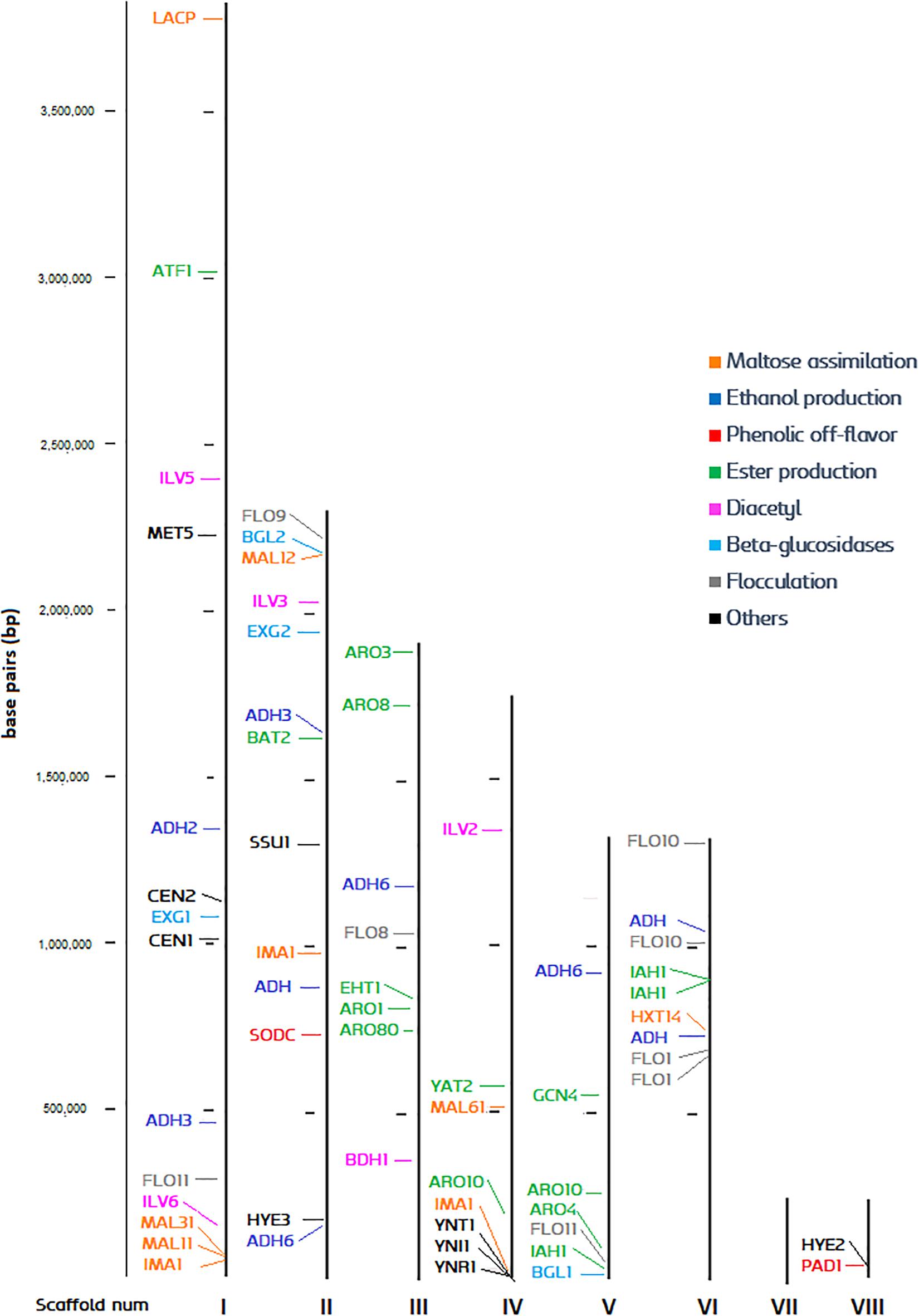
Figure 3. Distribution of brewing-relevant genes across the B. bruxellensis UMY321 reference genome.
Genomic Variability in Brettanomyces Populations
A phylogenetic tree based on predicted functional ORFs was created with the aim of assessing genomic clustering among isolates (Figure 2). The identified genes from Figure 3 were used to perform a multi-BLAST search for each one of them, and a heatmap was created to assess major divergence among the previously classified genomic clusters (Figure 4). As the reference B. bruxellensis strain UMY321 is a diploid wine isolate strain, most of the “Wine 2n” cluster display a full genome coverage. Several strains of the “Wine 3n” cluster are missing an outer region of scaffold V, where BGL1 is located among others. This is also the case in the “Wild/Wood” group. With regard to the beer isolates “Lambic” and “Farmhouse,” most isolates are missing the subtelomeric segments of scaffold I (where a sugar-transporter LACP is located) and scaffold II (BGL2-region). The lack of this β-glucosidase in beer-isolated strains has been observed before (Crauwels et al., 2015b). All strains in the “Lambic” sub-group are missing large internal region of the scaffold II and correspondingly lacking a maltase (MAL12). A high number of strains in the “Lambic” cluster are also missing the nitrogen assimilation cluster in the subtelomeric region of scaffold IV.
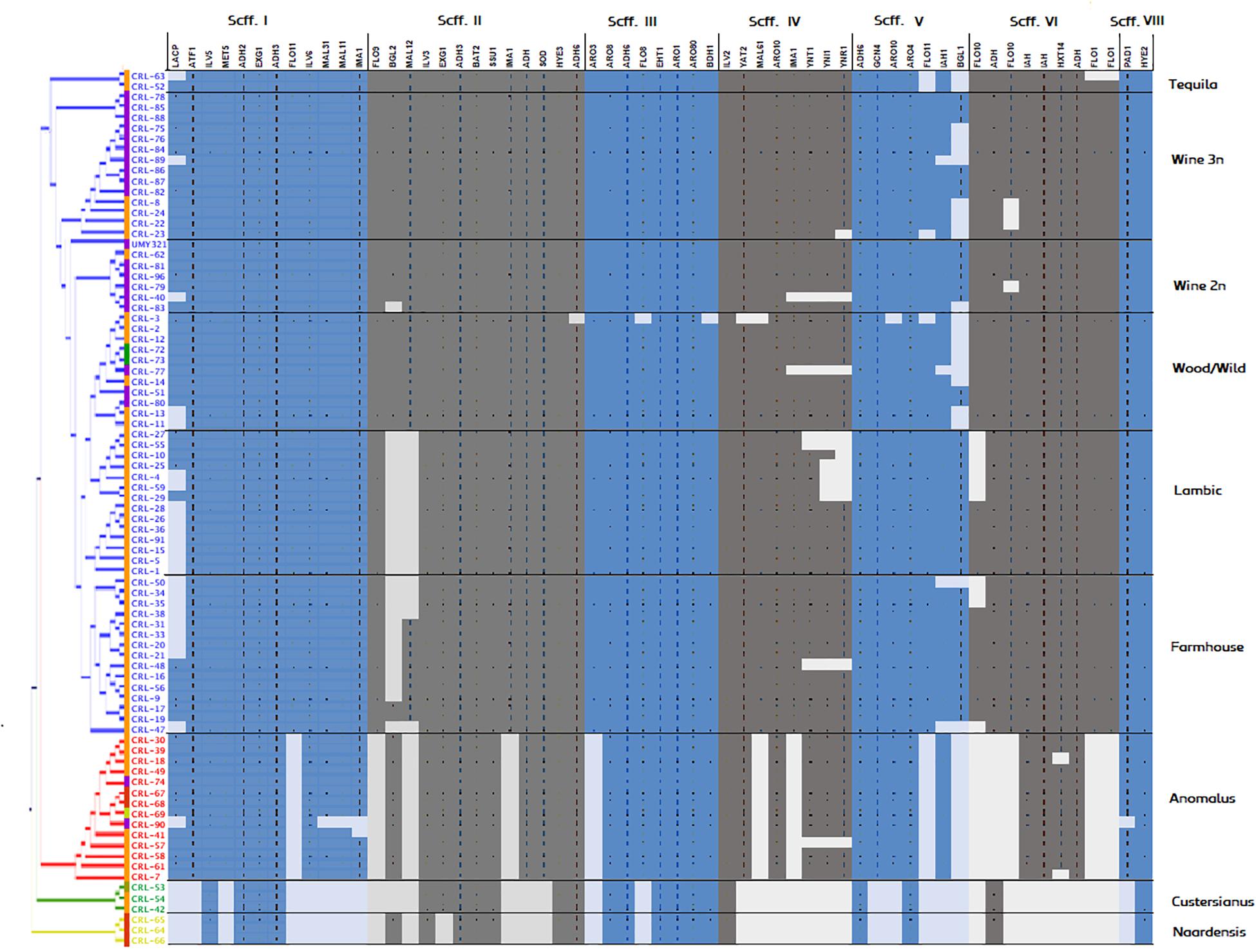
Figure 4. Genomic coverage of Brettanomyces strains included in the study. The reference genes spotted in Figure 3 were subtracted and a BLAST search was performed for each gene, respectively. To predict regions of loss, hits scoring > 95% of the total BLAST score were considered as valid for gene presence. Coloring in the genomic heatmap indicates absence/presence of the respective gene. Two strains showed a low read-mapping coverage and were removed from the figure (CRL-6, CRL-60).
The “Anomalus,” “Custersianus,” and “Naardenensis” groups were mapped to a different reference genome. Therefore, the genomic coverage is lower and unevenly distributed among the scaffolds. Nevertheless, while most of the essential brewing genes have a corresponding homologous in the “Anomalus” group, similarities in “Naardenensis” and “Custersianus” group are hardly found, inferring the high divergence of this species and the reference group.
Fermentation Performance and Production of Flavor Compounds in Brettanomyces Population
In order to assess the brewing potential of the Brettanomyces population, strains were inoculated into Pilsner wort, and fermentation was monitored in a 96-well plate in triplicates. Growth was quantified with a Growth-profiler® Enzyscreen and growth kinetics curves were created (Figure 5A). Fermentations were stopped after 7 days when most of the strains had reached the stationary phase. Plates were centrifuged and the supernatant was collected for analysis. Acetic acid content was quantified varying from 5 to 20 g/L (Figure 5B and Supplementary Table S3). Also, ethanol content was measured from 0.5 to 2.5% v/v (Figure 5C and Supplementary Table S3). It should also be noted that for certain strains, the standard deviations are large among replicates (Supplementary Table S3). Despite plates being sealed with anaerobic lids, such high acetic acid/ethanol ratios indicate that oxygen was present during the wort fermentation (Aguilar Uscanga et al., 2003), and carbon utilization could have been poor in certain isolates. VOCs were quantified and a violin plot was produced to get an overview of the flavor distribution among genetic clusters (Figure 5D and Supplementary Table S2). In regard to fusel alcohols, the production of Isobutanol, 2-phenylethanol and isoamyl alcohol is generally higher in strains belonging to the genomic cluster (“Lambic,” “Farmhouse”), also “Anomalus.” Consistently, production of acetate esters is above average in such beer-related clusters. Certain strains such as CRL-21 and CRL-25 produce an outstanding amount of acetate esters, reaching levels above 26 mg/L of isoamyl acetate and 170 mg/L of ethyl acetate. Ethyl caprylate could only be measured in four groups and in small amounts. Diacetyl was detected in all samples except in “Naardenensis” group, most of it above sensory threshold of detection (0.05 mg/L).
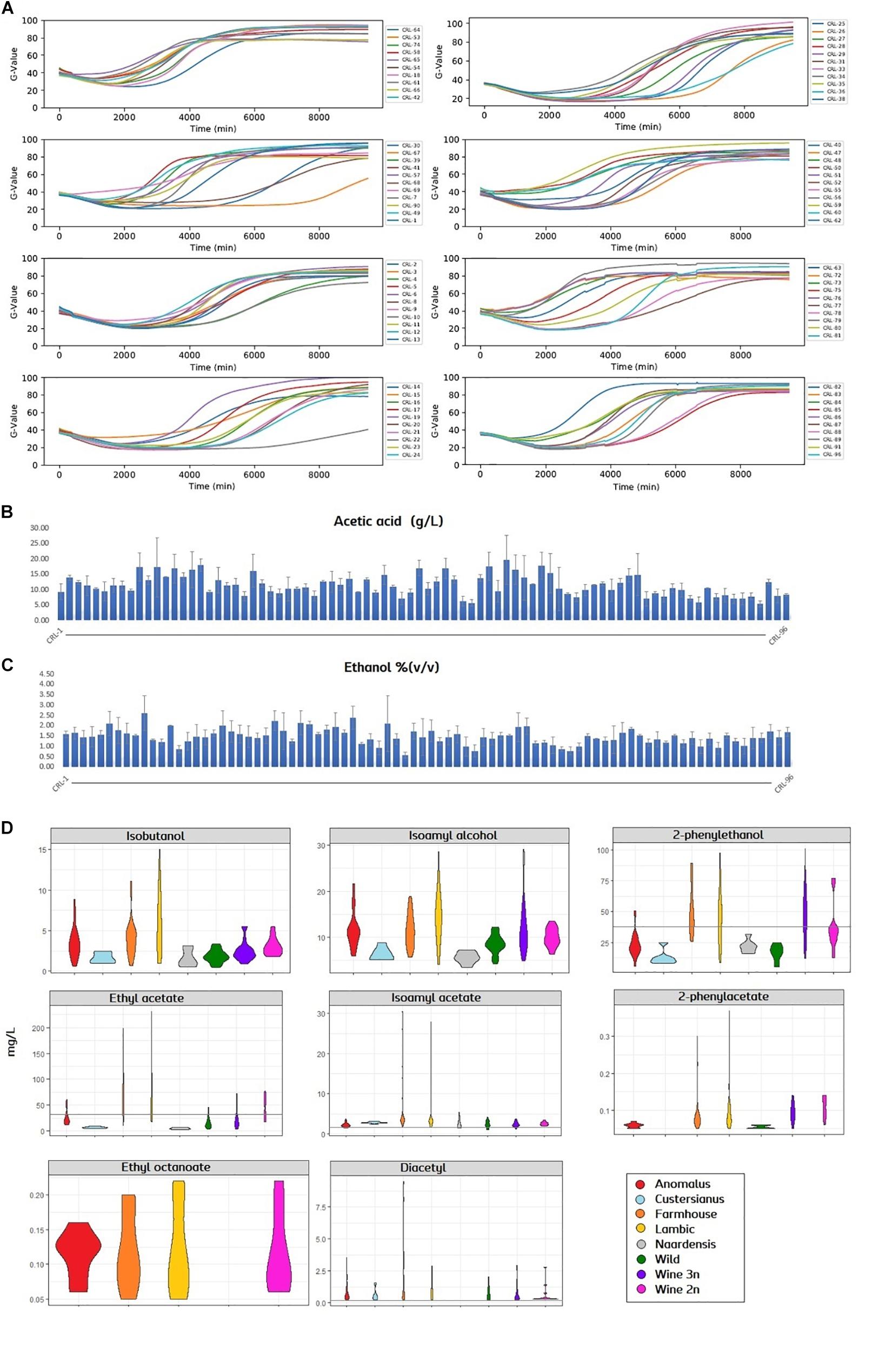
Figure 5. (A) Growth curves of Brettanomyces strain collection in pilsner wort, shown by means (n = 3). Growth-related values (G-value, y-axis) were extracted with the image analysis software (Enzyscreen). Fermentation time is indicated in minutes (x-axis). (B,C) Acetic acid and ethanol content of beers produced. Error bars show standard deviation (n = 3). (D) Violin plot for the main volatiles measured in beers. Strains are grouped according to its genetic cluster. Sensory detection threshold is indicated by a continuous line. When the line is not present, measured values are below detection threshold. Plots were obtained using ggplot2 package in R. Quantified values and statistical test can be found in Supplementary Table S2 and Statistical Test 1, respectively.
Brettanomyces Population Patterns in α- and β-Glucosidase Activity
Minimal media with a sole carbon source (glucose or maltose) was used in order to get an overview of α-glucosidase activity among the Brettanomyces collection. Growth was monitored by quantifying the colorimetric change in relation to NAD+ utilization, using Omnilog® (BIOLOG). Kinetics of glucose and maltose utilization of the Brettanomyces collection is displayed in Figure 6. All the strains efficiently grow in the presence of glucose, confirming that glucose is the preferred substrate by Brettanomyces species (Blomqvist et al., 2010). However, when maltose is applied as a sole carbon source, a wide variability of responses is observed. Maltose is usually taken at lower rates than glucose, and several strains are not able to assimilate the sugar at all. These results indicate that alpha-glucosidase activity is strongly variable in between Brettanomyces species, and is not functional in some strains. Generally, there is a longer lag-phase when maltose is present, indicating that a regulatory mechanism is involved which can require adaptation time.
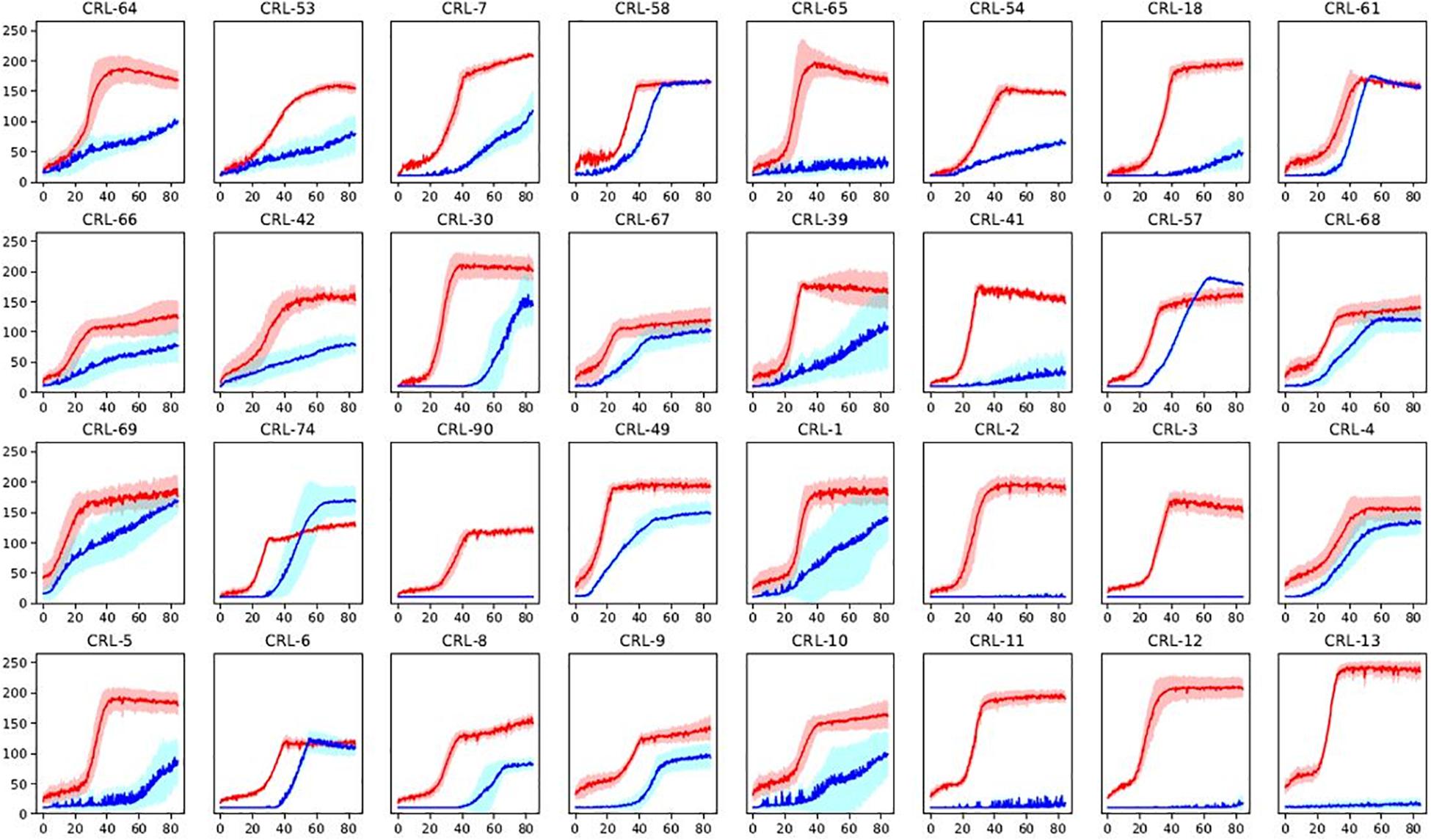
Figure 6. Sugar utilization of a representative part of Brettanomyces collection. Strains grown in media with glucose (red) or maltose (blue) as sole carbon source. Shaded curves are representing standard deviations between triplicates. Y-axis represents yeast growth quantified by Bvalue in Biolog. X-axis represents time in hours. Graphs of the full-collection can be found at Supplementary Figure S1. Statistical tests of the current data on maltose assimilation can be found in Supplementary Material.
To test β-glucosidase activity, strains were grown in YPC, using cellobiose as a sole carbon source, and OD600 was measured after 7 days. After that, yeast cells were removed by centrifugation, supernatant was collected, and conversion of beta substrates on the remaining media was measured. A Scatter plot comparing both activities was produced with the Unscrambler®CAMO (Figure 7). Strains are color-grouped according to the possession of genes. As expected, growth in cellobiose is strongly correlated with the beta-glucosidase activity in the extracellular fraction (Figure 7). Two β-glucosidases can be found in B. bruxellensis, and strains can have either none of them, just one, or both ORFs. Strains possessing no ORF or single BbBGL1 do not grow efficiently on cellobiose, and consequently no β-glucosidase activity is observed. In contrast, strains possessing BbBGL2 or both ORF display abundant growth on cellobiose and extracellular beta-glucosidase activity (Figure 7). These results clearly indicate that BbBGL2 plays a major role in break-down beta glucosides, while the role of BbBGL1 is minimum. As previously reported, the remaining Brettanomyces species (anomalus, custersianus, naardenensis) can grow efficiently on cellobiose and showed high β-glucosidase activity (Daenen et al., 2008a; Wu et al., 2014; Vervoort et al., 2016). β-glucosidase activity in the cellobiose-active strains is diverse as shown by x-axis in Figure 7. B. bruxellensis strain CRL-52 scored the highest conversion of β-glucosides measured in 2,56 units/L.
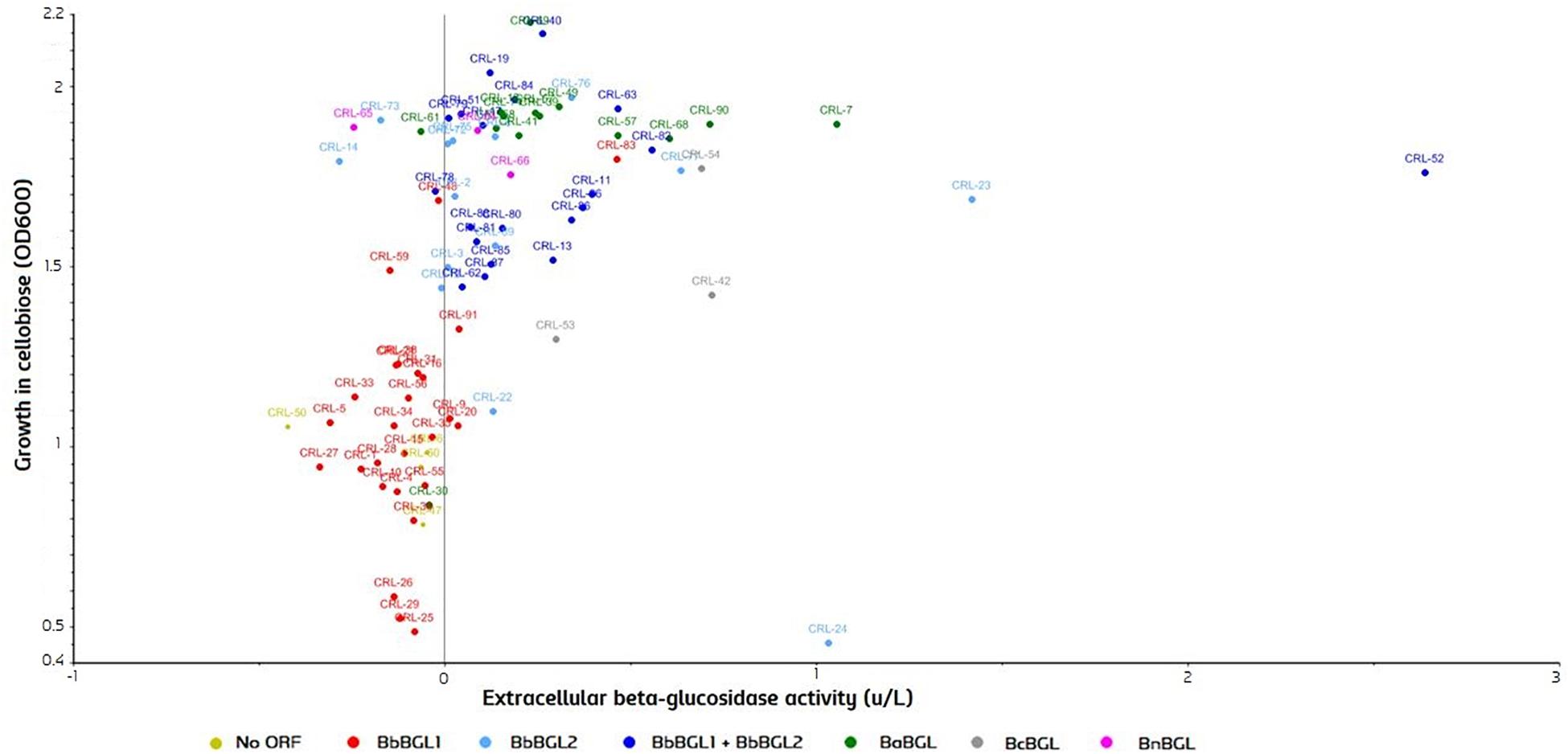
Figure 7. β-glucosidase activity in cultures grown on cellobiose as sole carbon source (OD600) and extracellular activity measured (units/L). Strains are grouped according to the identified ORF in the genome. Statistical tests of the current experimental data can be found in Supplementary Material.
Phenolic Off-Flavor Phenotype of Brettanomyces Population and Detection of POF-Brettanomyces anomalus CRL-90
The POF phenotype of the Brettanomyces population was investigated by measuring the uptake of the precursor ferulic acid. Strains were grown in YPD + 100 mg/L ferulic acid in triplicates in 96-deep well MTP format. After 7 days, the residual ferulic acid was detected by OD measurement at 325 nm. On one hand, results shown in Figure 8A indicate that both B. custersianus and B. naardenensis species are not able to assimilate ferulic acid. Only one B. naardenensis strain (CRL-65) seems to convert ferulic acid and be POF+. On the other hand, all strains of B. anomalus and B. bruxellensis are able to metabolize ferulic acid. Interestingly, one strain belonging to B. anomalus species (CRL-90) with a minimum conversion of ferulic acid was identified. To test CRL-90 POF phenotype further, wort fermentations in 250 mL glass bottles were performed, and the content of volatile phenols in the resulting beer was quantified (Figures 8B,C). While both 4-EP and 4-EG are detected in the control stain CRL-2, 4-EP is absent and a minimum amount of 4-EG is present after fermentation with CRL-90. The phenolic pathway intermediates 4-VP and 4-VG are below threshold in all fermentations. These results confirm the inability of CRL-90 to convert phenolic acids and therefore the first Brettanomyces/Dekkera species without POF up to date is reported. Such results are consistent with the genotypic data, as CRL-90 strain is missing BaPAD1 gene responsible for decarboxylation of ferulic acid to 4-VG. When the corresponding scaffold is compared to CRL-49 reference genome, strain CRL-90 is missing the side region where BaPAD1 is located (Figure 8D). Additionally, as loss-of-function mutations (such as frameshifts or premature stop codons) in ScPAD1 and ScFDC1 are commonly found (Gallone et al., 2016; Gonçalves et al., 2016; Preiss et al., 2017), we investigated their presence in BbPAD1. Nevertheless, amino acid sequences of BbPAD1 resulted highly similar and none of the POF inactivation mechanisms previously mentioned were found.
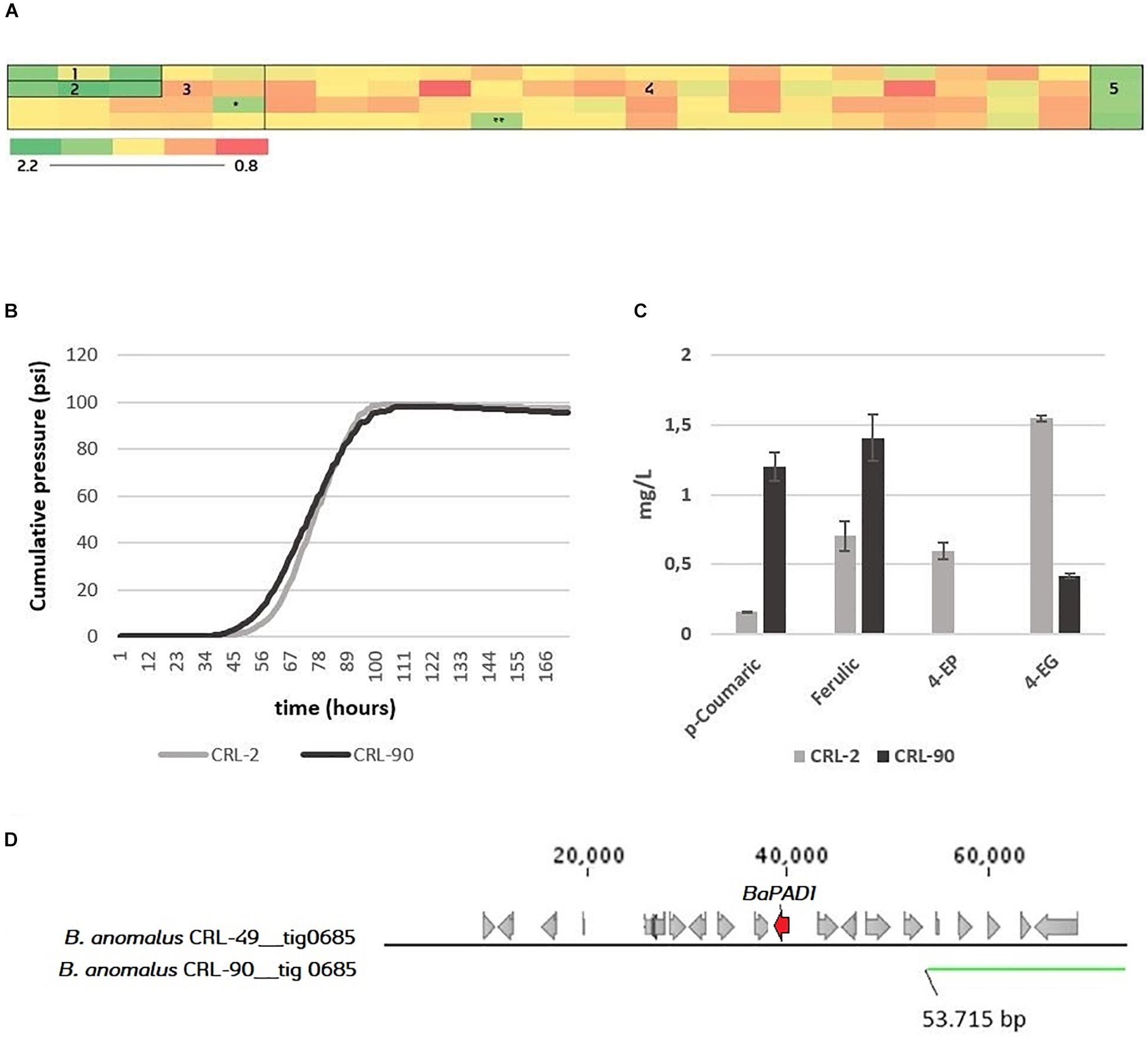
Figure 8. (A) Screening of the 84 Brettanomyces strains for uptake of ferulic acid. Heatmap displays averaged values of OD325 (n = 3). Standard deviations can be found at Supplementary Table S2. 1 = B. naardenensis; 2 = B. custersianus; 3 = B. anomalus; 4 = B. bruxellensis; 5 = Blanc; ∗ = CRL-90. ∗∗ = Low growth. For ferulic concentration, see Supplementary Table S3. (B) Pilsner wort fermentation of CRL-2 and CRL-90. Average values (n = 2). (C) Phenolic acids (p-coumaric, ferulic) and volatile phenols (4-ethylphenol, 4-ethylguaiacol) in mg/L at the end of fermentation. (D) Genomic setup of CRL-90 compared to the reference CRL-49. The figure has been produced with BLAST tool, comparing the contig nr. 685 where DaPAD1 is present. In Supplementary Table S4, information about the presence of neighboring ORFs with its respective function is given.
Genotype/Phenotype Correlation and Principal Component Analysis Based on Experimental Data
The genotype–phenotype correlation of the Brettanomyces population was investigated by grouping the strains according to its genomic cluster and plotting the phenotypical data generated (Figure 9). To summarize, B. naardenensis and B. custersianus confirm to be far distinct species, with specific properties as shown by low acetic acid and ethanol production along with ferulic acid consumption. With regard to alpha and beta-glucosidase activity, clear population trends are observed. “Anomalus” group displays powerful carbon assimilation, scoring high values in both cellobiose and maltose utilization. In regard to B. bruxellensis species, strains belonging to beer group (“Farmhouse,” “Lambic”) display efficient maltose utilization coupled to poor cellobiose assimilation. The opposite effect was observed in “Wine” and “Wild” isolates, as slow growth in maltose is linked to efficient growth in cellobiose. Our observations suggest that B. bruxellensis strains rely either on its α- or β-glucosidase activity to survive, and unnecessary traits for survival are lost over time. Furthermore, the population trends were visualized using a PCA of all previously quantified phenotypical parameters. A bi-plot was produced, with strains colored according to its genetic cluster (Figure 10). The weight of the variables into each PC is displayed by cos2 function with arrow length. Production of VOCs (as shown by concentrations) imposed a strong impact across PC1, explaining 36.5% of the variance. Variable loadings and correlation plot can be found in Supplementary Figures S2, S3. The Brettanomyces population distribution across the matrix in Figure 10 suggests that strain phenotypical properties can be predicted according to the genetic cluster they belong to.
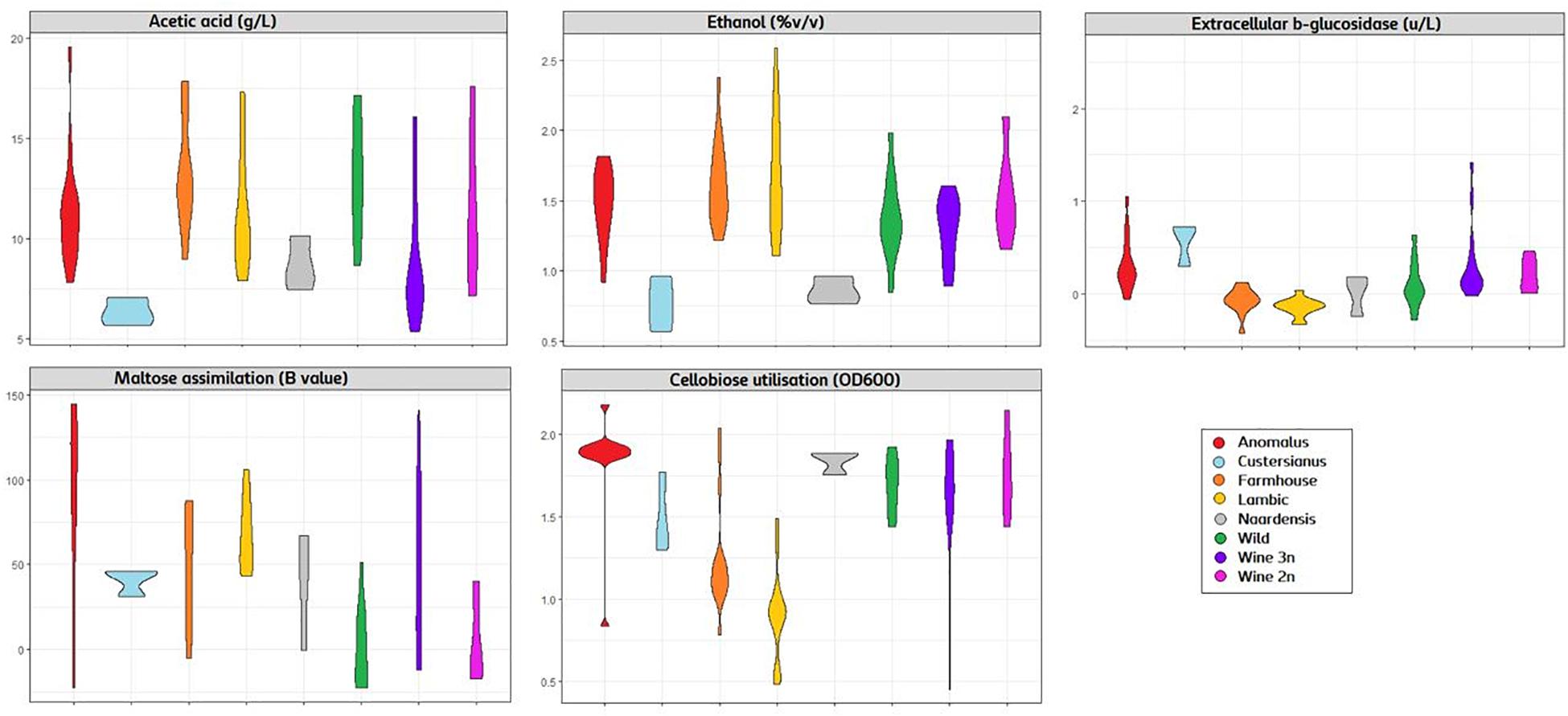
Figure 9. Violin plot with phenotypical measurements of Brettanomyces population. Strains are grouped according to the genomic cluster assigned in Figure 2.
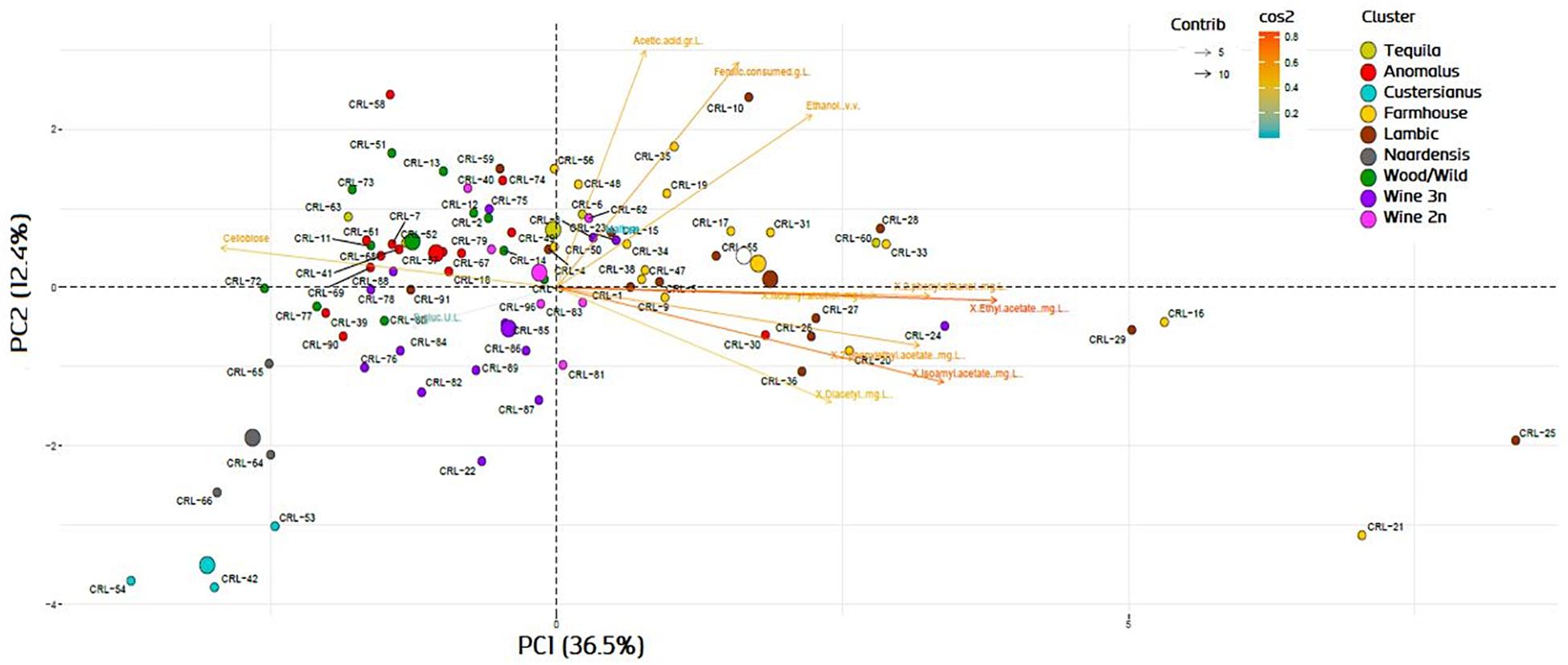
Figure 10. Bi-plot with principal component analysis variable loadings of Brettanomyces strains included in the study. Variable loadings are measured by cos2 function.
Discussion
In this study, we have aimed at providing a base to identify potential applications for Brettanomyces in brewing. A strain collection of Brettanomyces has been genotypically and phenotypically analyzed. Yeast strains have been clustered according to its predicted protein content, revealing specific genomic setups that enable survival in certain niches such as wine, beer, and wooden barrels. A large amount of functional genes have been identified, and the population has been sorted according to their presence or absence. The strains have also been tested for their brewing potential and a correlation with the genomic cluster has been carried out.
Throughout the study, several signs of artificial selection pressure have been observed. The short genetic divergence of beer isolates indicates that strains with similar characteristics have been selected for brewing. Such selection could have happened either in a selected manner, in Farmhouse brewing or in a spontaneous way in Lambic brewing (Spitaels et al., 2014; Colomer et al., 2019). This has led to a set of strains with a high degree of sugar attenuation and high ester production. In contrast, strains isolated from wine are largely divergent, whereas Brettanomyces species are considered spoilage yeasts. The pressure for natural adaptation is likely the cause of the genomic diversity observed. One example is the development of a triploid state for SO2 tolerance, pushing Brettanomyces wine-related population toward this characteristic (Borneman et al., 2014; Avramova et al., 2018a). Moreover, a singular cluster of strains has been characterized, containing strains isolated from bioethanol production plants and wooden barrels. This set of strains contain special properties, such as the inability of utilizing maltose as a substrate, balanced by the efficient degradation of cellobiose. In addition, B. custersianus and B. naardenensis species demonstrate interesting traits for brewing applications, like POF—phenotype and high β-glucosidase activity. In addition, B. custersianus displays a highly flocculent phenotype which is desired to ease brewing downstream processes. However, the low maltose assimilation and the production of several off-flavors such as H2S prevent these two species from being applied in brewing, at least in the short term. The potential application of B. custersianus and B. naardenensis for low alcoholic beers and secondary fermentation strains could be explored further.
Furthermore, B. anomalus has proven its potential for brewing, with an efficient metabolism of maltose, high β-glucosidase activity, and remarkable production of flavors (Figures 5D, 9). Additionally, one B. anomalus strain (CRL-90) was found to have a POF negative phenotype, related to the loss of the subtelomeric region containing BaPAD1. This confirms that BaPad1 protein is essential for the decarboxylation of ferulic acid (Godoy et al., 2014). CRL-90 is also the first strain within the Brettanomyces genus where such loss of POF function is described. Interestingly, the loss of the subtelomeric region containing PAD1-FDC1 genetic cluster has been recently reported as a mechanism of POF inactivation in lager yeasts of the Saaz-type (Gallone et al., 2019; Langdon et al., 2019). Curiously, CRL-90 was also the only Brettanomyces/Dekkera strain with the inability of assimilating maltose, lacking also the maltose assimilation cluster (MAL31, MAL11, IMA1). This observation points toward a possible evolutionary correlation between the traits of maltose assimilation and POF production in Brettanomyces species (Shen et al., 2018).
In Farmhouse and Lambic brewing, Brettanomyces strains have been generally applied as “primary” fermentation strains, selecting toward an efficient maltose metabolism. Curiously, most of the B. bruxellensis beer isolates “Farmhouse” and “Lambic” score low on β-glucosidase activity and cannot metabolize cellobiose. This is due to the lack of BbBGL2 gene which has shown to be essential on breaking-down beta-substrates (Figure 7). When possessing BbBGL2 B. bruxellensis can consume cellobiose, and have more probabilities to survive long term in wooden barrels or grape-peels (Crauwels et al., 2015a; Steensels et al., 2015). Therefore, BbBGL2 could be considered as a potential gene target in production plants where Brettanomyces is not desired. As the concentration of α-glucosides in beer is substantially higher than the β-glucosides, most beer isolates have lost this non-essential ORF. In contrast, the absence of BbBGL1 does not restrict the digestion of beta substrates, as shown by “Wine 3n” and “Wild” population. In terms of extracellular β-glucosidase activity, CRL-52 is outstanding from the rest of the population (Figure 7). Despite being a beer isolate, CRL-52 is genomically distinct from the main B. bruxellensis beer isolates, and could also be considered a potential common ancestor (Figure 2). CRL-52 was isolated in a South African “Bantu” brewery, where malted millet is used as raw material, requiring a different adaptation to the substrate composition for its survival. Moreover, CRL-52 is among the few strains capable to metabolize both maltose and cellobiose efficiently, confirming that unnecessary traits for survival (either α- or β-glucosidase activity) could have been eliminated in certain Brettanomyces populations over time.
Interestingly, the yeasts in the “Lambic” genetic cluster are missing nitrate assimilation ORFs, pointing toward nitrate uptake not being an essential requirement for survival in beer (Figure 4). Beer wort is widely known as an amino acid-rich media, therefore other nitrogen sources are available to use by this Brettanomyces population (Parente et al., 2018). Maltose assimilation in the studied B. bruxellensis isolates still remains ambiguous. Despite the maltose assimilation cluster (MAL31, MAL11, IMA1) is found in all the isolates, mostly beer and some wine isolates are capable of assimilating maltose, while “wild” strains are not. A gene coding for a maltase, MAL12, was initially thought to contribute to the population variance aiding the break-down of α-linked sugars (Crauwels et al., 2015b). Unexpectedly, most beer strains are missing this ORF and are still able to metabolize maltose, which was also seen before (Crauwels et al., 2017). While maltose assimilation has been extensively elucidated in brewer’s yeast, with complex regulatory pathways containing several repressors and regulators (Novak et al., 2004; Brickwedde et al., 2018), further research is needed to uncover such sugar metabolism in Brettanomyces. Despite population trends in regard to maltose assimilation phenotype are observed, the role of specific proteins is still uncertain. As in S. cerevisiae, maltose metabolism is probably determined by gene duplication and increase in copy number (Teste et al., 2010; Needleman and Michels, 2015).
With regard to flavor production, certain strains with high production of acetate esters have been identified (CRL-21 and CRL-25). Such phenotype could be due to a higher copy number of ATF1 gene, as unique nucleotide sequences in any of these strains could not be identified. Production of fusel alcohols and acetate esters is generally higher in beer isolates, most likely due to efficient catabolism of amino acid precursors (valine, leucine, isoleucine, phenylalanine) (Hazelwood et al., 2008; Celińska et al., 2019). Furthermore, the off-flavor diacetyl was measured at extensive ranges, and no correlation could be defined. Most of the diacetyl-related ILV genes are located in centromeric-like regions and conserved in B. bruxellensis strains. As this off-flavor is linked to the fermentation–maturation stage of the beer, and its flavor detection threshold is remarkably low, it can become challenging to rush into certain conclusions for production and prevention of this metabolite (Dasari and Kölling, 2011; Pires et al., 2015).
Conclusion
The present study reveals an extensive evaluation of the Brettanomyces population diversity worldwide, while also connects phenotypical traits to genotypes. A deep understanding of the huge range of properties Brettanomyces species are capable of offers a novel perspective into the phenotypic variants of this species. This opens opportunities for its industrial application, particularly in the field of beer brewing. This knowledge could be applied to satisfy the high demand for development of craft beers with novel flavors and beers with low- to non-alcohol content.
Data Availability Statement
The genome sequence datasets generated in this study are available in NCBI BioProject number PRJNA592329.
Author Contributions
MC conducted the experiments and wrote the manuscript. AC and RF supported bioinformatics analysis and figure creation. KO and LJ performed the quantification and data handling of volatile compounds. MC, NS, and JF designed the experiments. NS and JF contributed to supervision support. The manuscript was critically reviewed by all the authors and approved.
Funding
This work was supported by Innovation Fund Denmark. MC was a recipient of an Industrial Ph.D. fellowship (project 5189-00057) granted by Innovation Fund Denmark.
Conflict of Interest
A patent application to protect some findings of this work has been filed by Carlsberg Breweries. AC, RF, KO, NS, and JF were employed by the company Carlsberg AS. LJ was employed by the company MS-Omics.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Judita Gartner for kindly donating a large set of Brettanomyces strains to CRL. The authors also thank the Brewing Science and Technology department at CRL for providing beer wort, and Group Development in Carlsberg Group for analytics support. The authors thank Michael Katz, Jesper Harholt, and Christoph Dockter for a critical review of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00637/full#supplementary-material
References
Aguilar Uscanga, M. G., Délia, M. L., and Strehaiano, P. (2003). Brettanomyces bruxellensis: effect of oxygen on growth and acetic acid production. Appl. Microbiol. Biotechnol. 61, 157–162. doi: 10.1007/s00253-002-1197-z
Aguilar-Uscanga, M. G., Garcia-Alvarado, Y., Gomez-Rodriguez, J., Phister, T., Delia, M. L., and Strehaiano, P. (2011). Modelling the growth and ethanol production of Brettanomyces bruxellensis at different glucose concentrations. Lett. Appl. Microbiol. 53, 141–149. doi: 10.1111/j.1472-765X.2011.03081.x
Ames, R. M., Rash, B. M., Hentges, K. E., Robertson, D. L., Delneri, D., and Lovell, S. C. (2010). Gene duplication and environmental adaptation within yeast populations. Genome Biol. Evol. 2, 591–601. doi: 10.1093/gbe/evq043
Andersion, R. A. Y. (2012). Brewery One Yeast or Two? Pure Yeast and Top Fermentation. Brew. Hist. 149, 30–38.
Avramova, M., Cibrario, A., Peltier, E., Coton, M., Coton, E., Schacherer, J., et al. (2018a). Brettanomyces bruxellensis population survey reveals a diploid-triploid complex structured according to substrate of isolation and geographical distribution. Sci. Rep. 8:4136. doi: 10.1038/s41598-018-22580-7
Avramova, M., Vallet-Courbin, A., Maupeu, J., Masneuf-Pomarède, I., and Albertin, W. (2018b). Molecular diagnosis of Brettanomyces bruxellensis’ sulfur dioxide sensitivity through genotype specific method. Front. Microbiol. 9:1260. doi: 10.3389/fmicb.2018.01260
Bisson, L. F., and Karpel, J. E. (2010). Genetics of Yeast Impacting Wine Quality. Annu. Rev. Food Sci. Technol. 1, 139–162. doi: 10.1146/annurev.food.080708.100734
Blomqvist, J., Eberhard, T., Schnürer, J., and Passoth, V. (2010). Fermentation characteristics of Dekkera bruxellensis strains. Appl. Microbiol. Biotechnol. 87, 1487–1497. doi: 10.1007/s00253-010-2619-y
Borneman, A. R., Zeppel, R., Chambers, P. J., and Curtin, C. D. (2014). Insights into the Dekkera bruxellensis genomic landscape: comparative genomics reveals variations in ploidy and nutrient utilisation potential amongst wine isolates. PLoS Genet. 10:e1004161. doi: 10.1371/journal.pgen.1004161
Brickwedde, A., Brouwers, N., van den Broek, M., Gallego Murillo, J. S., Fraiture, J. L., Pronk, J. T., et al. (2018). Structural, physiological and regulatory analysis of maltose transporter genes in Saccharomyces eubayanus CBS 12357T. Front. Microbiol. 9:1786. doi: 10.3389/fmicb.2018.01786
Brown, C. A., Murray, A. W., and Verstrepen, K. J. (2010). Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 20, 895–903. doi: 10.1016/j.cub.2010.04.027
Callemien, D., and Collin, S. (2010). Structure, organoleptic properties, quantification methods, and stability of phenolic compounds in beer-A review. Food Rev. Int. 26, 1–84. doi: 10.1080/87559120903157954
Celińska, E., Borkowska, M., Białas, W., Kubiak, M., Korpys, P., Archacka, M., et al. (2019). Genetic engineering of Ehrlich pathway modulates production of higher alcohols in engineered Yarrowia lipolytica. FEMS Yeast Res. 19:foy122. doi: 10.1093/femsyr/foy122
Chatonnet, P., Dubourdie, D., Boidron, J.-N., and Pons, M. (1992). The origin of ethylphenols in wines. J. Sci. Food Agric. 60, 165–178. doi: 10.1002/jsfa.2740600205
Cibrario, A., Miot-sertier, C., Paulin, M., Bullier, B., Riquier, L., Perello, M. C., et al. (2020). Brettanomyces bruxellensis phenotypic diversity, tolerance to wine stress and wine spoilage ability. Food Microbiol. 87:103379. doi: 10.1016/j.fm.2019.103379
Claussen, N. H. (1904). On a method for the application of Hansen’s pure yeast system in the manufacturing of well-conditioned english stock beers. J. Inst. Brew. 10, 308–331. doi: 10.1002/j.2050-0416.1904.tb04656.x
Claussen, N. H. (1906). Manufacture of English beers or malt liquors. U.S. Patent No 1904208464A. Washington, DC: U.S. Patent and Trademark Office. doi: 10.1002/j.2050-0416.1904.tb04656.x
Colomer, M. S., Funch, B., and Forster, J. (2019). The raise of Brettanomyces yeast species for beer production. Curr. Opin. Biotechnol. 56, 30–35. doi: 10.1016/j.copbio.2018.07.009
Crauwels, S., Steensels, J., Aerts, G., Willems, K. A., Verstrepen, K. J., and Lievens, B. (2015a). Brettanomyces bruxellensis, essential contributor in spontaneous beer fermentations providing novel opportunities for the brewing industry. Brew. Sci. 68, 110–121.
Crauwels, S., Van Assche, A., de Jonge, R., Borneman, A. R., Verreth, C., Troels, P., et al. (2015b). Comparative phenomics and targeted use of genomics reveals variation in carbon and nitrogen assimilation among different Brettanomyces bruxellensis strains. Appl. Microbiol. Biotechnol. 99, 9123–9134. doi: 10.1007/s00253-015-6769-9
Crauwels, S., Van Opstaele, F., Jaskula-Goiris, B., Steensels, J., Verreth, C., Bosmans, L., et al. (2017). Fermentation assays reveal differences in sugar and (off-) flavor metabolism across different Brettanomyces bruxellensis strains. FEMS Yeast Res. 17:fow105. doi: 10.1093/femsyr/fow105
Crauwels, S., Zhu, B., Steensels, J., Busschaert, P., De Samblanx, G., Marchal, K., et al. (2014). Assessing genetic diversity among Brettanomyces yeasts by DNA fingerprinting and whole-genome sequencing. Appl. Environ. Microbiol. 80, 4398–4413. doi: 10.1128/AEM.00601-14
Curtin, C. D., Borneman, A. R., Chambers, P. J., and Pretorius, I. S. (2012). De-novo assembly and analysis of the heterozygous triploid genome of the wine spoilage yeast Dekkera bruxellensis AWRI1499. PLoS One 7:e33840. doi: 10.1371/journal.pone.0033840
Curtin, C. D., and Pretorius, I. S. (2014). Genomic insights into the evolution of industrial yeast species Brettanomyces bruxellensis. FEMS Yeast Res. 14, 997–1005. doi: 10.1111/1567-1364.12198
Daenen, L., Saison, D., Sterckx, F., Delvaux, F. R., Verachtert, H., and Derdelinckx, G. (2008a). Screening and evaluation of the glucoside hydrolase activity in Saccharomyces and Brettanomyces brewing yeasts. J. Appl. Microbiol. 104, 478–488. doi: 10.1111/j.1365-2672.2007.03566.x
Daenen, L., Sterckx, F., Delvaux, F. R., Verachtert, H., and Derdelinckx, G. (2008b). Evaluation of the glycoside hydrolase activity of a Brettanomyces strain on glycosides from sour cherry (Prunus cerasus L.) used in the production of special fruit beers. FEMS Yeast Res. 8, 1103–1114. doi: 10.1111/j.1567-1364.2008.00421.x
Daenen, L., Vanderhaegen, B., Verachtert, H., and Derdelinckx, G. (2004). Flavour enhancement in beer by yeast beta-glucosidase activity. Commun. Agric. Appl. Biol. Sci. 69, 73–76.
Dasari, S., and Kölling, R. (2011). Cytosolic localization of acetohydroxyacid synthase Ilv2 and its impact on diacetyl formation during beer fermentation. Appl. Environ. Microbiol. 77, 727–731. doi: 10.1128/AEM.01579-10
Deng, X., Petitjean, M., Teste, M. A., Kooli, W., Tranier, S., François, J. M., et al. (2014). Similarities and differences in the biochemical and enzymological properties of the four isomaltases from Saccharomyces cerevisiae. FEBS Open Bio 4, 200–212. doi: 10.1016/j.fob.2014.02.004
Dunn, B., Richter, C., Kvitek, D. J., Pugh, T., and Sherlock, G. (2012). Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 22, 908–924. doi: 10.1101/gr.130310.111
Duong, C. T., Strack, L., Futschik, M., Katou, Y., Nakao, Y., Fujimura, T., et al. (2011). Identification of Sc-type ILV6 as a target to reduce diacetyl formation in lager brewers’ yeast. Metab. Eng. 13, 638–647. doi: 10.1016/j.ymben.2011.07.005
Duval, E. H., Alves, S. L., Dunn, B., Sherlock, G., and Stambuk, B. U. (2010). Microarray karyotyping of maltose-fermenting Saccharomyces yeasts with differing maltotriose utilization profiles reveals copy number variation in genes involved in maltose and maltotriose utilization. J. Appl. Microbiol. 109, 248–259. doi: 10.1111/j.1365-2672.2009.04656.x
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Emms, D. M., and Kelly, S. (2017). STRIDE: species tree root inference from gene duplication events. Mol. Biol. Evol. 34, 3267–3278. doi: 10.1093/molbev/msx259
Emms, D. M., and Kelly, S. (2018). STAG: Species Tree Inference from All Genes. bioRxiv [Preprint]. doi: 10.1101/267914
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. doi: 10.1186/s13059-019-1832-y
Fournier, T., Gounot, J.-S., Freel, K., Cruaud, C., Lemainque, A., Aury, J.-M., et al. (2017). High-quality de novo genome assembly of the Dekkera bruxellensis yeast isolate using Nanopore MinION sequencing. G3 7, 3243–3250. doi: 10.1534/g3.117.300128
Fujii, T., Nagasawa, N., Iwamatsu, A., Bogaki, T., Tamai, Y., and Hamachi, M. (1994). Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 60, 2786–2792. doi: 10.1128/aem.60.8.2786-2792.1994
Fujita, S. I., Senda, Y., Nakaguchi, S., and Hashimoto, T. (2001). Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J. Clin. Microbiol. 39, 3617–3622. doi: 10.1128/JCM.39.10.3617-3622.2001
Fukuda, K., Kiyokawa, Y., Yanagiuchi, T., Wakai, Y., Kitamoto, K., Inoue, Y., et al. (2000). Purification and characterization of isoamyl acetate-hydrolyzing esterase encoded by the IAH1 gene of Saccharomyces cerevisiae from a recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 53, 596–600. doi: 10.1007/s002530051662
Fukuda, K., Yamamoto, N., Kiyokawa, Y., Yanagiuchi, T., Wakai, Y., Kitamoto, K., et al. (1998). Balance of activities of alcohol acetyltransferase and esterase in Saccharomyces cerevisiae is important for production of isoamyl acetate. Appl. Environ. Microbiol. 64, 4076–4078. doi: 10.1128/aem.64.10.4076-4078.1998
Gallone, B., Steensels, J., Mertens, S., Dzialo, M. C., Gordon, J. L., Wauters, R., et al. (2019). Interspecific hybridization facilitates niche adaptation in beer yeast. Nat. Ecol. Evol. 3, 1562–1575. doi: 10.1038/s41559-019-0997-9
Gallone, B., Steensels, J., Prahl, T., Soriaga, L., Saels, V., Herrera-Malaver, B., et al. (2016). Domestication and Divergence of Saccharomyces cerevisiae Beer Yeasts. Cell 166, 1397–1410.e16. doi: 10.1016/j.cell.2016.08.020
Gibson, B., Geertman, J. M. A., Hittinger, C. T., Krogerus, K., Libkind, D., Louis, E. J., et al. (2017). New yeasts-new brews: modern approaches to brewing yeast design and development. FEMS Yeast Res. 17:fox038. doi: 10.1093/femsyr/fox038
Gilliland, R. (1961). Brettanomyces: occurrence, characteristics and effects on beer flavor. J. Inst. Brew. 67:257. doi: 10.1002/j.2050-0416.1961.tb01791.x
Gjermansen, C., Nilson, T., and Holmnberg, S. (1988). Towards diacetyl-less brewer’s yeast. Influence of ILV2 and ILV5 mutations. J. Basic Micribiol. 28, 175–183. doi: 10.1002/jobm.3620280304
Godoy, L., García, V., Peña, R., Martínez, C., and Ganga, M. A. (2014). Identification of the Dekkera bruxellensis phenolic acid decarboxylase (PAD) gene responsible for wine spoilage. Food Control 45, 81–86. doi: 10.1016/j.foodcont.2014.03.041
Godoy, L., Garrido, D., Martínez, C., Saavedra, J., Combina, M., and Ganga, M. A. (2009). Study of the coumarate decarboxylase and vinylphenol reductase activities of Dekkera bruxellensis (anamorph Brettanomyces bruxellensis) isolates. Lett. Appl. Microbiol. 48, 452–457. doi: 10.1111/j.1472-765X.2009.02556.x
Gonçalves, M., Pontes, A., Almeida, P., Barbosa, R., Serra, M., Libkind, D., et al. (2016). Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 26, 2750–2761. doi: 10.1016/j.cub.2016.08.040
Gorter de Vries, A. R., Pronk, J. T., and Daran, J.-M. G. (2017). Industrial relevance of chromosomal copy number variation in Saccharomyces yeasts. Appl. Environ. Microbiol. 83, e3206–e3216. doi: 10.1128/aem.03206-16
Gounot, J.-S., Neuveglise, C., Freel, K. C., Devillers, H., Piskur, J., Friedrich, A., et al. (2019). High complexity and degree of genetic variation in Brettanomyces bruxellensis population. bioRxiv [Preprint]. doi: 10.1101/826990
Harari, Y., Ram, Y., and Kupiec, M. (2018). Frequent ploidy changes in growing yeast cultures. Curr. Genet. 64, 1001–1004. doi: 10.1007/s00294-018-0823-y
Hazelwood, L. A., Daran, J. M., Van Maris, A. J. A., Pronk, J. T., and Dickinson, J. R. (2008). The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266. doi: 10.1128/AEM.02625-07
Hellborg, L., and Piškur, J. (2009). Complex nature of the genome in a wine spoilage yeast, Dekkera bruxellensis. Eukaryot. Cell 8, 1739–1749. doi: 10.1128/ec.00115-09
Heresztyn, T. (1986). Metabolism of volatile phenolic compounds from hydroxycinnamic acids by Brettanomyces yeast. Arch. Microbiol. 146, 96–98. doi: 10.1007/BF00690165
Hittinger, C. T., and Carroll, S. B. (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681. doi: 10.1038/nature06151
Horák, J. (2013). Regulations of sugar transporters: insights from yeast. Curr. Genet. 59, 1–31. doi: 10.1007/s00294-013-0388-8
Ihmels, J., Bergmann, S., Gerami-Nejad, M., Yanai, I., McClellan, M., Berman, J., et al. (2005). Molecular biology: rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309, 938–940. doi: 10.1126/science.1113833
Ishchuk, O. P., Zeljko, T. V., Schifferdecker, A. J., Wisén, S. M., Hagström, Å. K., Rozpȩdowska, E., et al. (2016). Novel centromeric loci of the wine and beer yeast Dekkera bruxellensis CEN1 and CEN2. PLoS One 11:e0161741. doi: 10.1371/journal.pone.0161741
Jespersen, L., Cesar, L. B., Meaden, P. G., and Jakobsen, M. (1999). Multiple α-glucoside transporter genes in brewer’s yeast. Appl. Environ. Microbiol. 65, 450–456. doi: 10.1128/aem.65.2.450-456.1999
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: Scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Krogerus, K., and Gibson, B. R. (2013). Influence of valine and other amino acids on total diacetyl and 2,3-pentanedione levels during fermentation of brewer’s wort. Appl. Microbiol. Biotechnol. 97, 6919–6930. doi: 10.1007/s00253-013-4955-1
Kuo, H. P., Wang, R., Huang, C. Y., Lai, J. T., Lo, Y. C., and Huang, S. T. (2018). Characterization of an extracellular β-glucosidase from Dekkera bruxellensis for resveratrol production. J. Food Drug Anal. 26, 163–171. doi: 10.1016/j.jfda.2016.12.016
Kurtzman, C. P., Fell, J. W., and Boekhout, T. (eds) (2011). “The yeasts: a taxonomic study,” in The Yeasts, 5th Edn. (Amsterdam: Elsevier), 1–3. doi: 10.1017/CBO9781107415324.004
Langdon, Q. K., Peris, D., Baker, E. C. P., Opulente, D. A., Nguyen, H. V., Bond, U., et al. (2019). Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 3, 1576–1586. doi: 10.1038/s41559-019-0998-8
Larroy, C., Fernández, M. R., González, E., Parés, X., and Biosca, J. A. (2002). Characterization of the Saccharomyces cerevisiae YMR318C (ADH6) gene product as a broad specificity NADPH-dependent alcohol dehydrogenase: relevance in aldehyde reduction. Biochem. J. 361(Pt 1), 163–172. doi: 10.1042/bj3610163
Lebleux, M., Abdo, H., Coelho, C., Basmaciyan, L., Albertin, W., Maupeu, J., et al. (2020). New advances on the Brettanomyces bruxellensis biofilm mode of life. Int. J. Food Microbiol. 318:108464. doi: 10.1016/j.ijfoodmicro.2019.108464
Lentz, M., and Harris, C. (2015). Analysis of growth inhibition and metabolism of hydroxycinnamic acids by brewing and spoilage strains of Brettanomyces yeast. Foods 4, 581–593. doi: 10.3390/foods4040581
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Libkind, D., Hittinger, C. T., Valerio, E., Goncalves, C., Dover, J., Johnston, M., et al. (2011). Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 14539–14544. doi: 10.1073/pnas.1105430108
Miklenić, M., Žunar, B., Štafa, A., and Svetec, I. K. (2015). Improved electroporation procedure for genetic transformation of dekkera/brettanomyces bruxellensis. FEMS Yeast Res. 15:fov096. doi: 10.1093/femsyr/fov096
Mukai, N., Masaki, K., Fujii, T., Kawamukai, M., and Iefuji, H. (2010). PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J. Biosci. Bioeng. 109, 564–569. doi: 10.1016/j.jbiosc.2009.11.011
Naumoff, D. G., and Naumov, G. I. (2010). Discovery of a novel family of α-glucosidase IMA genes in yeast Saccharomyces cerevisiae. Biochem. Biophys. 432, 114–116. doi: 10.1134/s1607672910030051
Needleman, R. B., Kaback, D. B., Dubin, R. A., Perkins, E. L., Rosenberg, N. G., Sutherland, K. A., et al. (1984). MAL6 of Saccharomyces: a complex genetic locus containing three genes required for maltose fermentation. Proc. Natl. Acad. Sci. U.S.A. 81, 2811–2815. doi: 10.1073/pnas.81.9.2811
Needleman, R. B., and Michels, C. (2015). Repeated family of genes controlling maltose fermentation in Saccharomyces carlsbergensis. Mol. Cell. Biol. 3, 796–802. doi: 10.1128/mcb.3.5.796
Novak, S., Zechner-Krpan, V., and Marić, V. (2004). Regulation of maltose transport and metabolism in Saccharomyces cerevisiae. Food Technol. Biotechnol. 42, 213–218.
Olsen, R. A., Bunikis, I., Tiukova, I., Holmberg, K., Lötstedt, B., Pettersson, O. V., et al. (2015). De novo assembly of Dekkera bruxellensis: a multi technology approach using short and long-read sequencing and optical mapping. GigaScience 4:56. doi: 10.1186/s13742-015-0094-1
Parente, D. C., Cajueiro, D. B. B., Moreno, I. C. P., Leite, F. C. B., De Barros Pita, W., and De Morais, M. A. (2018). On the catabolism of amino acids in the yeast Dekkera bruxellensis and the implications for industrial fermentation processes. Yeast 35, 299–309. doi: 10.1002/yea.3290
Peter, J., De Chiara, M., Friedrich, A., Yue, J.-X., Pflieger, D., Bergström, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Pinu, F. R., and Villas-Boas, S. G. (2017). Rapid quantification of major volatile metabolites in fermented food and beverages using gas chromatography-mass spectrometry. Metabolites 7:37. doi: 10.3390/metabo7030037
Pires, E. J., Teixeira, J. A., Brányik, T., Brandão, T., and Vicente, A. A. (2015). Continuous beer fermentation - diacetyl as a villain. J. Inst. Brew. 121, 55–61. doi: 10.1002/jib.205
Piškur, J., Ling, Z., Marcet-Houben, M., Ishchuk, O. P., Aerts, A., LaButti, K., et al. (2012). The genome of wine yeast Dekkera bruxellensis provides a tool to explore its food-related properties. Int. J. Food Microbiol. 157, 202–209. doi: 10.1016/j.ijfoodmicro.2012.05.008
Preiss, R., Tyrawa, C., Van Der Merwe, G., Biology, C., Laboratories, E., Author, C., et al. (2017). Traditional norwegian Kveik Yeasts: underexplored domesticated Saccharomyces cerevisiae Yeasts. BioRxiv [Preprint]. doi: 10.1101/194969
Roach, M. J., and Borneman, A. R. (2020). New genome assemblies reveal patterns of domestication and adaptation across Brettanomyces (Dekkera) species. BMC Genomics 21:194. doi: 10.1186/s12864-020-6595-z
Romano, D., Valdetara, F., Zambelli, P., Galafassi, S., De Vitis, V., Molinari, F., et al. (2017). Cloning the putative gene of vinyl phenol reductase of Dekkera bruxellensis in Saccharomyces cerevisiae. Food Microbiol. 63, 92–100. doi: 10.1016/j.fm.2016.11.003
Rozpedowska, E., Hellborg, L., Ishchuk, O. P., Orhan, F., Galafassi, S., Merico, A., et al. (2011). Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts. Nat. Commun. 2:302. doi: 10.1038/ncomms1305
Saerens, S. M. G., Verstrepen, K. J., Van Laere, S. D. M., Voet, A. R. D., Van Dijck, P., Delvaux, F. R., et al. (2006). The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 281, 4446–4456. doi: 10.1074/jbc.M512028200
Sanchez, R. G., Solodovnikova, N., and Wendland, J. (2012). Breeding of lager yeast with Saccharomyces cerevisiae improves stress resistance and fermentation performance. Yeast 29, 343–355. doi: 10.1002/yea.2914
Schifferdecker, A. J., Dashko, S., Ishchuk, O. P., and Piškur, J. (2014). The wine and beer yeast Dekkera bruxellensis. Yeast 31, 323–332. doi: 10.1002/yea.3023
Schifferdecker, A. J., Siurkus, J., Andersen, M. R., Joerck-Ramberg, D., Ling, Z., Zhou, N., et al. (2016). Alcohol dehydrogenase gene ADH3 activates glucose alcoholic fermentation in genetically engineered Dekkera bruxellensis yeast. Appl. Microbiol. Biotechnol. 100, 3219–3231. doi: 10.1007/s00253-015-7266-x
Schiønning, H. (1908). On Torula in English Beer Manufacture. Journal of the Institute of Brewing 15, 2–35. doi: 10.1002/j.2050-0416.1909.tb02237.x
Shen, X., Opulente, D. A., Kominek, J., Kurtzman, C. P., Hittinger, C. T., Rokas, A., et al. (2018). Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175, 1533–1545.e20. doi: 10.1016/j.cell.2018.10.023
Smith, B. D., and Divol, B. (2016). Brettanomyces bruxellensis, a survivalist prepared for the wine apocalypse and other beverages. Food Microbiol. 59, 161–175. doi: 10.1016/j.fm.2016.06.008
Smith, B. D., and Divol, B. (2018). The carbon consumption pattern of the spoilage yeast Brettanomyces bruxellensis in synthetic wine-like medium. Food Microbiol. 73, 39–48. doi: 10.1016/j.fm.2017.12.011
Smith, M. T. (2011). “Dekkera van der Walt (1964),” in The Yeasts, Vol. 2, eds C. P. Kurtzman and J. W. Fell, (Amsterdam: Elsevier), 373–377. doi: 10.1016/B978-0-444-52149-1.00025-2
Spitaels, F., Wieme, A. D., Janssens, M., Aerts, M., Daniel, H. M., Van Landschoot, A., et al. (2014). The microbial diversity of traditional spontaneously fermented lambic beer. PLoS One 9:e95384. doi: 10.1371/journal.pone.0095384
Stanke, M., Keller, O., Gunduz, I., Hayes, A., Waack, S., and Morgenstern, B. (2006). AUGUSTUS: A b initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439. doi: 10.1093/nar/gkl200
Steensels, J. (2017). Rapid Screening Method for Phenolic Off-Flavor (POF) Production in Yeast. J. Am. Soc. Brew. Chem. 75, 318–323. doi: 10.1094/ASBCJ-2017-4142-01
Steensels, J., Daenen, L., Malcorps, P., Derdelinckx, G., Verachtert, H., and Verstrepen, K. J. (2015). Brettanomyces yeasts - From spoilage organisms to valuable contributors to industrial fermentations. Int. J. Food Microbiol. 206, 24–38. doi: 10.1016/j.ijfoodmicro.2015.04.005
Steensels, J., Meersman, E., Snoek, T., Saels, V., and Verstrepen, K. J. (2014). Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 80, 6965–6975. doi: 10.1128/aem.02235-14
Teste, M. A., Marie François, J., and Parrou, J. L. (2010). Characterization of a new multigene family encoding isomaltases in the yeast Saccharomyces cerevisiae, the IMA family. J. Biol. Chem. 285, 26815–26824. doi: 10.1074/jbc.M110.145946
Teunissen, A. W. R. H., and Steensma, H. Y. (1995). The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11, 1001–1013. doi: 10.1002/yea.320111102
Tiukova, I. A., Jiang, H., Dainat, J., Hoeppner, M. P., Lantz, H., Piskur, J., et al. (2019). Assembly and analysis of the genome sequence of the yeast Brettanomyces naardenensis CBS 7540. Microorganisms 7:489. doi: 10.3390/microorganisms7110489
Tkach, J. M., Yimit, A., Lee, A. Y., Riffle, M., Costanzo, M., Jaschob, D., et al. (2012). Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14, 966–976. doi: 10.1038/ncb2549
Van Mulders, S. E., Christianen, E., Saerens, S. M. G., Daenen, L., Verbelen, P. J., Willaert, R., et al. (2009). Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 9, 178–190. doi: 10.1111/j.1567-1364.2008.00462.x
Vanoni, M., Sollitti, P., Goldenthal, M., and Marmur, J. (1989). Structure and regulation of the multigene family controlling maltose fermentation in budding yeast. Prog. Nucleic Acid Res. Mol. Biol. 37, 281–322. doi: 10.1016/S0079-6603(08)60701-1
Varela, C., Lleixà, J., Curtin, C., and Borneman, A. (2018). Development of a genetic transformation toolkit for Brettanomyces bruxellensis. FEMS Yeast Res. 18:foy070. doi: 10.1093/femsyr/foy070
Vervoort, Y., Herrera-Malaver, B., Mertens, S., Guadalupe Medina, V., Duitama, J., Michiels, L., et al. (2016). Characterization of the recombinant Brettanomyces anomalus β-glucosidase and its potential for bioflavouring. J. Appl. Microbiol. 121, 721–733. doi: 10.1111/jam.13200
Woolfit, M., Rozpȩdowska, E., Piškur, J., and Wolfe, K. H. (2007). Genome survey sequencing of the wine spoilage yeast Dekkera (Brettanomyces) bruxellensis. Eukaryot. Cell 6, 721–733. doi: 10.1128/EC.00338-06
Keywords: genomics, high-throughput screening, brewing fermentation, phenolic off-flavor, 4-ethylguaiacol, maltose assimilation, beta-glucosidase, Dekkera bruxellensis
Citation: Colomer MS, Chailyan A, Fennessy RT, Olsson KF, Johnsen L, Solodovnikova N and Forster J (2020) Assessing Population Diversity of Brettanomyces Yeast Species and Identification of Strains for Brewing Applications. Front. Microbiol. 11:637. doi: 10.3389/fmicb.2020.00637
Received: 18 September 2019; Accepted: 20 March 2020;
Published: 09 April 2020.
Edited by:
Vittorio Capozzi, Institute of Food Production Sciences (CNR), ItalyReviewed by:
Marta Avramova, INRA Institut National de la Recherche Agronomique Centre Montpellier, FranceDiego Libkind, CONICET Instituto Andino Patagónico en Tecnologías Biológicas y Geoambientales (IPATEC), Argentina
Copyright © 2020 Colomer, Chailyan, Fennessy, Olsson, Johnsen, Solodovnikova and Forster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jochen Forster, Sm9jaGVuLmZvcnN0ZXJAY2FybHNiZXJnLmNvbQ==
 Marc Serra Colomer
Marc Serra Colomer Anna Chailyan
Anna Chailyan Ross T. Fennessy
Ross T. Fennessy Kim Friis Olsson
Kim Friis Olsson Lea Johnsen
Lea Johnsen Natalia Solodovnikova
Natalia Solodovnikova Jochen Forster
Jochen Forster