Abstract
Probiotic strain Eurotium cristatum was isolated from Chinese Fuzhuan brick-tea and tested for its in vitro activity against aflatoxigenic Aspergillus flavus. Results indicated that E. cristatum can inhibit the radial growth of A. flavus. Furthermore, this inhibition might be caused by E. cristatum secondary metabolites. The ability of culture filtrate of strain E. cristatum against growth and aflatoxin B1 production by toxigenic A. flavus was evaluated in vitro. Meanwhile, the influence of filtrate on spore morphology of A. flavus was analyzed by scanning electron microscopy (SEM). Results demonstrated that both radial growth of A. flavus and aflatoxin B1 production were significantly weakened following increases in the E. cristatum culture filtrate concentration. In addition, SEM showed that the culture filtrate seriously damaged hyphae morphology. Gas chromatography mass spectrometry (GC/MS) analysis of the E. cristatum culture supernatant revealed the presence of multiple antifungal compounds. Real-time quantitative polymerase chain reaction (RT-qPCR) analysis showed that the expression of aflatoxin biosynthesis-related genes (aflD, aflQ, and aflS) were down-regulated. Importantly, this latter occurrence resulted in a reduction of the AflS/AflR ratio. Interestingly, cell-free supernatants of E. cristatum facilitated the effective degradation of aflatoxin B1. In addition, two degradation products of aflatoxin B1 lacking the toxic and carcinogenic lactone ring were identified. A toxicity study on the HepG2 cells showed that the degradation compounds were less toxic when compared with AFB1.
Introduction
Aflatoxins, toxic derivatives of difuran coumarin, are predominantly synthesized by Aspergillus species such as Aspergillus flavus and Aspergillus parasiticus (Karabulut et al., 2014). Several forms of aflatoxin exist with aflatoxin B1 (AFB1) being the most toxic (Adebo et al., 2017; Gong et al., 2019). Aflatoxin B1 poses serious problems to human health and is known to cause acute or chronic toxicity aflatoxicosis resulting in mutagenic, carcinogenic, teratogenic, and immunosuppressive effects in humans (Sherif et al., 2009; Bbosa et al., 2013; Santini and Ritieni, 2013). It is estimated that aflatoxins may play a causative role in 4.6–28.2% of all global hepatocellular carcinoma cases (Liu and Wu, 2010). AFB1 has been classified as Group I carcinogen to humans by the International Agency for Research on Cancer (IARC, 1993). In addition to causing serious health issues, aflatoxins pollution in food and feed results in significant economic losses (Wu, 2015). Thus, multifarious physical (such as artificial removal, physisorption, temperature, and humidity control) and chemical (such as alkali and oxidation treatments, chemical agents) methods have been developed to alleviate associated problems in recent decades (Torres et al., 2014; Ismail et al., 2018). However, these methods not only consume large amounts of energy, but also significantly reduce the nutritional efficiency of food while adversely impacting available natural resources (Wang et al., 2015; Peng et al., 2018). Moreover, with the excessive application of synthetic fungicides as well as the potential for extended exposure, numerous fungi have developed resistance (Pham et al., 2014; Hawkins et al., 2019). For the afore-mentioned reasons, an effective alternative is urgently required.
Bio-control agents represent an attractive alternative in relation to the reduction or elimination of aflatoxin contamination (Tsitsigiannis et al., 2012; Medina-Cordova et al., 2016; Nguyen et al., 2017). These strategies are beneficial as they offer milder means of removing or degrading toxic materials; they can also ensure that significant losses in aesthetic and nutritional value are ameliorated. Previous studies have reported that some bacterial or fungal strains including Bacillus subtilis, Nocardia asteroides, Rhodococcus erythropolis, and Aspergillus niger can inhibit aflatoxins (Chaudhary et al., 2001; Wu et al., 2009; Cserhati et al., 2013; Zhang et al., 2017). And the most promising method for reducing contamination levels of aflatoxins is introduction of non-aflatoxin producing strains to soil where they compete with wild-type aflatoxigenic populations (Jaime-Garcia and Cotty, 2007; Wu and Khlangwiset, 2010; Alshannaq et al., 2018). A non-toxigenic A. flavus Afla-Guard (NRRL 21882) isolated by Dorner and Lamb (2006) have been used commercially on the peanuts in the southeastern United States. In addition, the atoxigenic A. flavus K49 (NRRL 30797, isolated from Maize) and AF36 (NRRL 18543, isolated from Cottonseed) also have been registered as a biopesticide for the management of aflatoxin-producing fungi during the crop production (Abbas et al., 2011). Although these strains could reduce aflatoxin contamination effectively, they still face significant challenge (Ehrlich, 2014). One of the major problems is a potential risk of introducing a heavy dose of A. flavus strains. The diversity of A. flavus populations may trigger other toxic secondary metabolites even imposed an additional burden on food safety and food quality especially with global warming (Alshannaq et al., 2018). Thus, it is imperative that identify other effective strains for the treatment of aflatoxin contamination.
Eurotium cristatum, which has been commonly known as “golden flower,” is used as a non-toxic and safe fungus for food fermentation (such as dark tea, okara, kudzu root) (Yazdani et al., 2011; Zhang et al., 2018; Chan et al., 2019; Gu et al., 2019). And it is the main probiotic fungus traditionally used in Fuzhuan brick dark tea in China (Peng et al., 2011; Shi et al., 2019). Actually, some studies suggesting that E. cristatum was useful in regulating the blood/lipid balance and cholesterol metabolism, enhancing immunity, alleviating obesity, and modulating gut microbiota (Liu et al., 2016; Chen et al., 2018; Du et al., 2019; Kang et al., 2019). Moreover, previous studies have reported that filtrate of E. cristatum exhibits significant inhibitory activity on some bacterial or fungal strains (such as Staphylococcus aureus, Escherichia coli, and Magnaporthe grisea) (Xu et al., 2015; Du et al., 2017). Therefore, in this study, a probiotic strain of E. cristatum isolated from Fuzhuan tea was used for the first time to control A. flavus.
Here, we evaluated the ability of E. cristatum isolated from Fuzhuan tea to inhibit the in vitro production of AFB1 by toxigenic A. flavus. The results of this study will provide scientists with a better understanding of the mechanisms underpinning aflatoxin inhibition by E. cristatum.
Materials and Methods
Chemicals and Fungal Strains
Aflatoxin B1 standard was purchased from Shanghai Yuanye Bio-Technology Co., Ltd., China. And other chemicals were purchased from Sigma-Aldrich, United States.
A single E. cristatum strain HNYYWX.21 was isolated from Fuzhuan brick-tea in our laboratory (Wang et al., 2019). The AFB1-producing A. flavus strain used in this study was originally isolated from natural spices and kept in our laboratory. These strains were grown on potato dextrose agar (PDA) medium (potato 200 g/L, glucose 20 g/L, agar 15 g/L) for 7 days at 28 ± 2°C until good sporulation was observed. Conidial suspensions (1 × 107 conidia/mL) were prepared from sporulated cultures with sterile 0.01% (vol/vol) Tween 80.
Growth Curves of A. flavus and E. cristatum
Ten microliters of spore suspension (1 × 106 spores/mL) of A. flavus and 10 μL with 1 × 107 spores/mL of E. cristatum were inoculated on PDA medium for 5 days at 28 ± 2°C, respectively. The growth curve was obtained by measuring the colony diameter every 24 h. All treatments were repeated three times.
Preparation of the E. cristatum Culture Filtrate
The E. cristatum culture filtrate was prepared according to a method described by Xing et al. (2017) with minor modifications. Fifty milliliters of potato dextrose broth (PDB) was inoculated [2% (v/v) inoculum] with a conidial suspension (107 spores/mL) at 28 ± 2°C in a rotary shaker incubator (120 r/min) for 10 days. The culture filtrate was obtained following centrifugation at 10,000 g for 10 min at 4°C and subsequent passage through a 0.45-μm pore size filter (Millipore, United States); the filtrate was stored at −20°C until further required.
In vitro Antifungal Assay
Competition Assay on PDA Plates
An antagonism experiment was performed using relative growth of A. flavus. Briefly, 10 μL of an A. flavus fungal suspension at a concentration of 106 spores/mL were inoculated onto the center of a PDA plate, and 10 μL of the E. cristatum (107 spores/mL) suspension and sterile water (used as control) were independently inoculated on 5-mm diameter sterile filter paper discs that were positioned 25 mm from the center of the plate. The plates were incubated at 28 ± 2°C and the radial growth of each fungal colony was measured by a digital caliper every 24 h until the control group was overgrown with agar plate. All experiments were repeated for three times.
The inhibition ratio (%) = (r−r′)/r × 100%, where r (mm) represents the radius of the A. flavus colony (from the center to the control group) and r′ (mm) represents the growth of the A. flavus colony from the center toward the E. cristatum colony.
Effect of the E. cristatum Culture Filtrate on A. flavus Growth and AFB1 Production
The influence of the E. cristatum culture filtrate on the radial growth of the A. flavus mycelium was assayed according to a method described by Xu et al. (2013) with some minor modifications. The E. cristatum culture filtrate was adjusted to concentrations of 10, 20. and 40% (v/v) with PDA, respectively. Ten microliters of the A. flavus conidial suspension (at a concentration of 106 spores/mL) was inoculated onto each plate; the plates were incubated in the darkness at 28 ± 2°C for 5 days. Equal volume of water replaced culture filtrate served as a control. The diameter of the A. flavus colony was assayed every 24 h and the level of AFB1 in the PDA medium was analyzed after the culture period. All treatments were repeated three times.
Effect of the Culture Filtrate on the Ultrastructure of A. flavus
Potato dextrose broth (10 mL) containing culture filtrate [40% (v/v)] was inoculated with A. flavus conidia [106 spores/mL, 2% (vol/vol)] and incubated at 28 ± 2°C in a rotary shaker (120 r/min). PDB with water[(40% (v/v)] was used as a control. All treatments were repeated three times. The germination of A. flavus spores was investigated by scanning electron microscopy (SEM) after 24 h according to a method published by Sangmanee and Hongpattarakere (2014). After incubation, fungal mycelia were washed four times (10 min per wash) with sterile water. Next, the fungal mycelia were fixed overnight at 4°C with 2.5% glutaraldehyde. The samples were subsequently washed with sterile water to remove free glutaraldehyde; this was followed by dehydration in a graded series of ethanol (10, 30, 50, 70, 85, 95, 100, and 100%) for 10 min. The samples were then dried in a critical point drier (CPD) with liquid carbon dioxide, coated with gold in a polaron sputter coater and examined using a Nova Nano SEM-450 (FEI, United States).
Effect of the Culture Filtrate on the Expression of Genes Associated With AFB1 Production
Ten microliters of the A. flavus conidial suspension (at a concentration of 106 spores/mL) was inoculated into 50 mL of PDB containing 30% (v/v) of the culture filtrate. PDB with 30% (v/v) of water was used as a control group. They were incubated in the dark at 28 ± 2°C on a rotary shaker at 120 r/min in triplicate. After 3 days, the expression of genes associated with AFB1 production were assayed by real-time quantitative polymerase chain reaction (RT-qPCR).
Total RNA Extraction
RNA was isolated from fungal hyphae after 3 days. Approximately 100 mg of ground mycelium in liquid N2 was treated with 1 mL of Trizol (Sigma-Aldrich) according to the manufacturer’s instructions. RNA samples were treated with RNase-Free DNase (QIAGEN, Germany) to remove genomic DNA. The concentration and purity (A260/A280 ratio) of RNA were determined by a NanoDrop spectrophotometer (IMPLEN, Germany). RNA samples with A260/A280 values ranging from 1.97 (control group) to 2.05 (E. cristatum culture filtrate group) were used in this study. RNA was detected by 1% agarose gel electrophoresis, until the appearances of 5S band, clear 28S and clear 18S. Then the purified RNA was stored at −80°C until further analysis.
cDNA Synthesis
Reverse transcription was performed using the Fastking RT Kit (with gDNase) (QIAGEN, Germany). First-strand cDNA was obtained from 1 μg of total RNA in a 20-μL reaction mixture. Each reaction mixture was incubated at 42°C for 15 min followed by 95°C for 3 min and the product was stored at −20°C until further required.
Real-Time PCR
In this study, RT-qPCR was performed in a LightCycler96 detection system (Roche, Switzerland). Primers for genes associated with AFB1 such as aflR, aflS, aflD, aflQ, VeA, LaeA, and β-tubulin (as an internal control) used in this study were listed in Table 1, and they were used to understand the relationship between aflatoxin biosynthesis and the active compound of E. cristatum culture filtrate. Three replicates of both template free-negative control and endogenous control were used for each run. Each RT-qPCR reaction consisted of 10 μL of SuperReal PreMix Color (SYBR Green) (QIAGEN, Germany), different concentrations of each primer, and 100 ng of cDNA template in a final volume of 20 μL. Cycling conditions were as follows: 95°C for 15 min, 95°C for 10 s, 58°C (56°C for aflQ) for 30 s, 72°C for 32 s (40 cycles) in a 20-μL reaction mixture. A melt curve was generated at the end of every run to ensure PCR product uniformity by heating at 95°C for 10 s, 65°C for 60 s and 97°C for 1 s. All samples were analyzed in triplicate and qPCR reactions were repeated three times.
TABLE 1
| Gene | Nucleotide sequence (5′-3′) | References | Concentration (μM) | Melting temperature (°C) | Product size (bp) | Accession number |
| β-tubulin | F: TCTTCATGGTTGGCTTCGCT | Peromingo et al., 2017 | 6 | 88.39 | 98 | FR775333.1 |
| R: CTTGGGGTCGAACATCTGCT | 6 | |||||
| aflR | F: GATCTGGCTGGTCAGGAGCA | Chen et al., 2019 | 6 | 89.57 | / | / |
| R: CGCCTGAAACGGTGGTAGTG | 6 | |||||
| aflS | F: TGGTGCGACCATATTTACA | Peromingo et al., 2017 | 6 | 85.81 | 94 | AF441435.2 |
| R: GGTTGGGTCACGAACTGTTT | 6 | |||||
| aflD | F: ATGCTCCCGTCCTACTGTTT | Liang et al., 2015 | 6 | 88.32 | 108 | XM_002379908 |
| R: ATGTTGGTGATGGTGCTGAT | 6 | |||||
| aflQ | F: TTAAGGCAGCGGAATACAAG | Moon et al., 2016 | 8 | 90.32 | / | / |
| R: GACGCCCAAAGCCGAACACAAA | 8 | |||||
| VeA | F: TTGTCGTGTGCGGATTCG | Cary et al., 2007 | 6 | 86.51 | / | / |
| R: CTCATCGTAGTCGTAGTCATCG | 6 | |||||
| LaeA | F: AAAGGTTGCTCGCTGGTACA | Liu et al., 2017 | 6 | 84.65 | 121 | / |
| R: GACTTCTGACGAAATGCGCC | 6 |
Nucleotide sequences of primers for RT-qPCR assays.
The symbol “/” indicates that it is not mentioned in the reference.
Relative Gene Expression
Relative quantification of the expressions of the aflR, aflS, aflD, aflQ, VeA, and LaeA genes were done compared to the housekeeping gene β-tubulin. Data analysis was performed using the 2–ΔΔCt analysis method (Livak and Schmittgen, 2001). The PCR efficiency of each genes were practically equal since the amplification curves of the target and reference genes were parallel to each other in the exponential amplification interval. The specificity of the reactions was checked by analyzing the melt curves, which displayed a single sharp peak (Luo et al., 2005).
AFB1 Degradation by the E. cristatum Culture Filtrate
The effect of the culture filtrate of E. cristatum on AFB1 was studied according to a method published by Rao et al. (2017). One microliter of AFB1 standard was added to 1 mL of the culture filtrate until a final concentration of 1000 ng/mL was achieved; this mixture was subsequently incubated at 28 ± 2°C in the dark in a shaker incubator at 200 r/min for 0, 3, and 5 days. One milliliter of the PDB (with 1000 ng/mL AFB1) and the E. cristatum culture filtrate as the internal control were incubated under same conditions, respectively. All treatments were repeated three times. The remnant AFB1 and associated metabolites in the liquid medium were analyzed by high performance liquid chromatography (HPLC) and liquid chromatography quadrupole time of flight-mass spectrometry (LC-qTOF/MS).
Analysis of AFB1 by HPLC
AFB1 determination was performed using a Shimadzu LC-20A equipped with a fluorescence detector (RF-20A). AFB1 accumulated in the culture media was extracted according to a method by Zhou et al. (2017) with some modifications. Briefly, 2 g of PDA medium (1 mL of liquid medium) was placed in a 4-mL Eppendorf tube and 1 mL of chloroform was added. The mixture was blended at 3200 r/min for 90 s and sonicated (100 watts) for 20 min; the sample was subsequently centrifuged for 10 min at 8000 r/min. This procedure was repeated three times and the organic layer containing AFB1 was combined and then evaporated to dryness under an N2 stream at 50°C. The derivatizing reaction was performed by adding 200 μL of n-hexane and 100 μL of trifluoroacetic acid (TFA) for 1 h at 45°C in the darkness. After evaporating to dryness once more and redissolution in a 1000-μL mixture of 2/8 (v/v) acetonitrile/water, the mixture was violently shaken for 2 min and sonicated for 20 min at 100 watts of power. Next, each solution was filtered through a sterile 0.22-μm pore size filter (Millipore, United States) before injection into the HPLC. A series of reference compounds (10, 20, 50, 100, and 1000 ng/mL) at different concentrations was used to quantify the AFB1 content.
High performance liquid chromatography analysis of AFB1 was performed as follows: a XDB-C18 column (4.6 × 250 mm, 5 μm, Agilent, United States) was operated at 40°C with a flow rate of 1 mL/min with acetonitrile-water as mobile phase. A starting mobile phase of 20% acetonitrile was orderly escalated to 30% within 13 min, then the gradient elution was switched to 40% over 5 min and this was subsequently maintained for 1 min. Next, the eluted ratio returned to 20% once more over 2 min, and this ratio was maintained for 7 min. The injection volume was 20 μL. The excitation and detection wavelengths were 360 and 440 nm, respectively. The calibration curves for AFB1 (10–1000 ng/mL) by HPLC revealed a good linear relationship (R2 ≥ 0.99) between the detector response and the amounts of the AFB1 standards. The limit of detection (LOD) obtained in this study were 0.27 ng/mL.
Analysis of AFB1 Degradation Products by LC-qTOF/MS
Samples were extracted three times using equal volumes of chloroform. The samples were subsequently dried with N2 and re-dissolved in acetonitrile/water (2/8, v/v). LC/MS was performed on an LC-30A + SelexION + TripleTOF5600 + system (AB SCIEX, United States) equipped with a Agilent Plus C18 column (2.1 × 150 mm, 5 μm). The mobile phase for elution was composed of 70% acetonitrile (0.1% formic acid in water) with a flow rate of 0.4 mL/min. The total run time was 32 min and the injection volume was 25 μL. MS was performed using the following conditions (Iram et al., 2016): positive-ion mode, the capillary voltage and temperature were 3.5 kV and 300°C, respectively. Nitrogen was used as the collision gas. LC/MS analysis was performed to identify any potential degraded products in a full-scan mode within the range of m/z 100–2000.
Evaluation of the E. cristatum Culture Filtrate by GC/MS
Prior to extraction, the culture filtrate of E. cristatum to which 40 ng/mL 4-methyl-1-pentanol (internal standard) was added was equilibrated at 40°C for 30 min. PDB as the control group was incubated under same conditions, and all experiments were carried out three times independently. Gas chromatography mass spectrometry (GC/MS) was performed on a GC/MS-QP2010 Ultra system (Shimadzu, Japan). Chromatographic separation was performed on a DB-1MS column (60 m × 0.25 mm ID, 0.25 μm film thickness). Helium was used as the carrier gas with a flow rate of 1 mL/min. An inlet temperature of 40°C with splitless injection was employed. The temperature program was as follows: 40°C for 3 min; 4°C/min ramp to 120°C; 6°C/min ramp to 240°C and subsequent hold for 2 min. The total run time was 55 min. MS was performed using the following conditions: ion source temperature 230°C, transfer line temperature 230°C, scan mass range 35–500 amu, solvent delay time 3 min. The assay was subsequently performed according to the internal standard method.
Cytotoxicity Studies
HepG2 Cell Culture and Preparation of Samples
The human hepatocyte carcinoma HepG2 cell line was cultured in Dulbecco’s modified Eagle’s medium (10% fetal bovine serum, 1% non-essential amino acids, 100 U/mL penicillin, 100 μg/mL streptomycin) (D5796, Sigma-Aldrich) at 37°C in a 5% CO2 humidified atmosphere. The AFB1 degradation products (20 mL) and AFB1 stock solution (20 mL of 1 μg/mL of AFB1) were dried with N2 and re-dissolved in 0.1% (v/v) dimethyl sulfoxide (DMSO). Untreated AFB1 stock solution was used as the control group. And HepG2 cells grew under normal conditions and treated with DMSO [0.1% (v/v)] as the nulling group (internal control) and blank group, respectively. All experiments were repeated six times.
MTT Assay
MTT assay, a way to detect cell proliferation, was performed as described by Adebo et al. (2016) with some modifications. Briefly, 100 μL of HepG2 culture at a density of 1 × 103 cells/mL was pipetted into 96-well plates. After overnight stabilization, cells were treated in 100 μL of fresh culture medium with appropriate concentrations of the tested compounds and then incubated for 48 h. Next, 10 μL of MTT [50 mg/mL; 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] were added to the culture medium and the plates were incubated for a further 4 h. All the culture conditions were at 37°C in a 5% CO2 humidified atmosphere. After removing the culture medium, 150 μL of DMSO was added to the plates. The plates were subsequently incubated for 10 min in a rotary shaker incubator (120 r/min) in the darkness and the absorbance was read in a victor X3 (Perkin Elmer, United States) at a wavelength of 490 nm.
Cell viability = (ODe−ODb)/(ODn−ODb) × 100%, where ODe is the absorbance of the experimental group (cells with AFB1 stock solution and AFB1 degradation products); ODn is the absorbance of the nulling group; ODb is the absorbance of the blank group.
Statistical Analysis
Data analysis was performed by Student’s t-test and ANOVA using IBM SPSS V20 (SPSS/IBM, Chicago, IL, United States). The means for each treatment were separated by Tukey’s test using a level of significance of 0.05.
Results and Discussion
Antagonistic Assay of E. cristatum Against A. flavus
Aspergillus flavus and E. cristatum were co-cultured on PDA plates for 5 days. E. cristatum exhibited mycelial growth inhibition against A. flavus (Figure 1B) with an inhibition ratio of 63.74%. The inhibition of fungal growth might be attributed to competition for nutrients and/or space or the synthesis of inhibitory metabolites. However, the average growth rate of E. cristatum (4.39 mm/day) was obviously lower than that of A. flavus (15.64 mm/day) under the same conditions (p < 0.05, Figure 1A). Shi et al. (2019) have reported that metabolites produced by E. cristatum inhibit the growth of B. subtilis. Similarly, Du et al. (2014) observed that metabolites of E. cristatum such as cristatumin A, cristatumin D, and 3-O-(a-D-ribofuranosyl) might be active against some bacterias and fungi (such as B. subtilis, S. aureus, and M. grisea). Thus, we speculate that the inhibition of E. cristatum on A. flavus also might be caused by the synthesis of secondary metabolites by E. cristatum. Hence, further studies were required to determine the effects of E. cristatum culture supernatant.
FIGURE 1
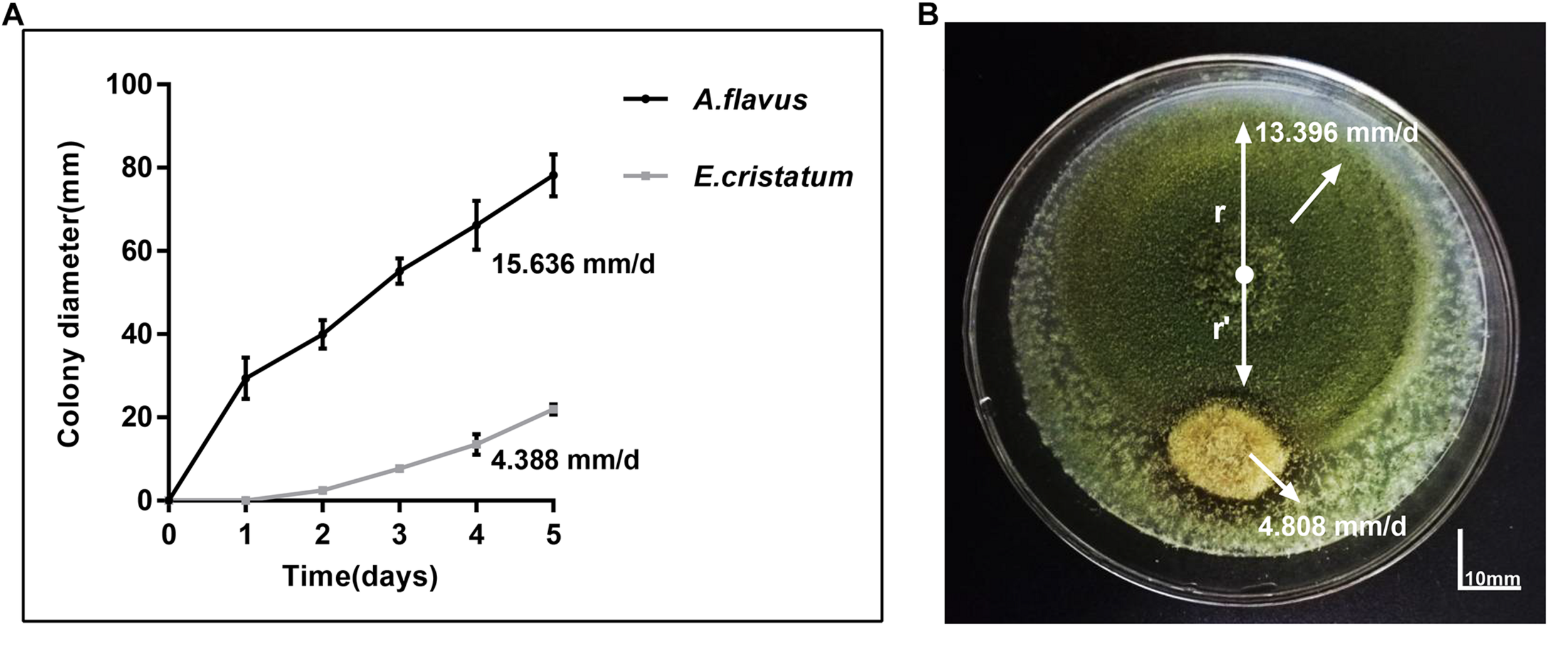
Confronting incubation assay and growth curve for A. flavus and E. cristatum.(A) Growth curves of A. flavus and E. cristatum in PDA at 28 ± 2°C for 5 days. Data spots represent mean values of three determinations ± standard deviation. (B)A. flavus was inoculated at the center of the PDA plate and E. cristatum and sterile water (control) were inoculated at 25 mm from the center of the plate.
Effect of the E. cristatum Culture Filtrate on Growth and Aflatoxin Accumulation in A. flavus
To test whether E. cristatum secretes unknown compound(s) into medium that confer A. flavus inhibition, we filter-sterilized (0.45 μm filter) the 10-day-old culture of E. cristatum grown in PDB, and combined the cell-free culture with A. flavus and 40 mL of fresh PDA. As a control, a set of water were mixed with the PDA. A. flavus grown on PDA plates with different concentrations of the E. cristatum culture filtrate and water were observed every 24 h. As shown in Figure 2A, the radial growth of A. flavus was significantly inhibited by the E. cristatum culture filtrate (p < 0.05). And the ratios of inhibition were ranged from 5.90 to 44.17%. Xu et al. (2013) reported that a 2% (v/v) of 10-fold concentrated the culture filtrate of strain A. niger (FS10) inhibits the growth of A. flavus (49.6%) while impeding AFB1 production (94.5%). The latter study also revealed that there is a direct correlation between biological growth and AFB1 production. Thus, in this study, the effect of the E. cristatum culture filtrate on aflatoxin production in PDA medium was also determined on the seventh day. According to the results shown in Figure 2B, the AFB1 production was significantly (p < 0.01) reduced from 139.04 to 40.41 ng/mL when the PDA medium contained only 10% culture filtrate. The AFB1 enormously decreased from 84.87 to 23.07 ng/mL, and resulting in a 72.52% inhibition when the culture filtrate concentration was 40%. similarly, Alshannaq et al. (2018) reported that the culture filtrate of strain Aspergillus oryzae M2040 in concentrations of 10 and 25% (V:V in water or PDB) could significantly inhibit A. flavus NRRL 3357 spore recovery and AFB1 production (p < 0.01). Although Shi et al. (2019) and Zhang et al. (2019) have demonstrated that E. cristatum exhibits antimicrobial activity, this study is the first to report on the inhibition of aflatoxigenic A. flavus.
FIGURE 2
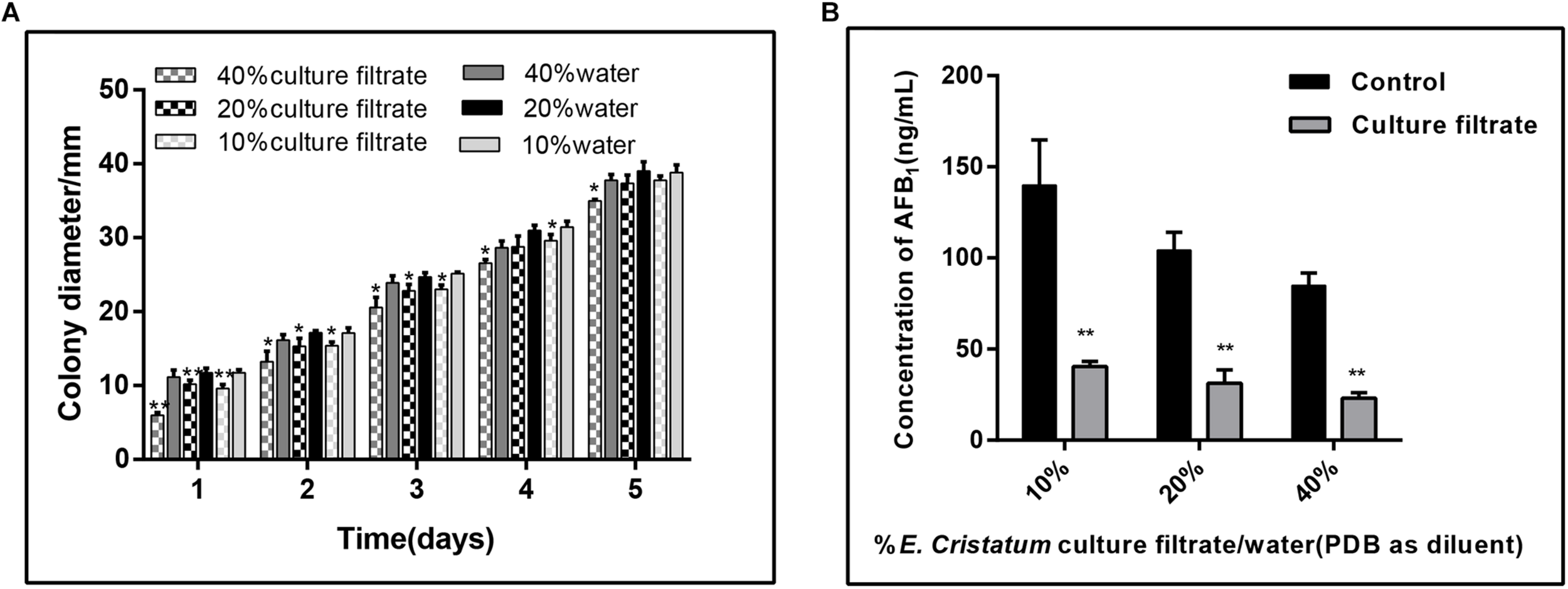
Effects of the E. cristatum culture filtrate on growth and AFB1 production of A. flavus. (A) The colony diameter of A. flavus in PDA medium with 10, 20, and 40% E. cristatum culture filtrate or water at 28 ± 2°C for the first 5 days. (B) AFB1 content in PDA medium with different concentrations of E. cristatum culture filtrate and water at 28 ± 2°C for 5 days. Values in each column followed by one (p < 0.05) or two (p < 0.01) asterisks are significantly different from the control according to the Student’s t-test.
Fungal Morphology Following Scanning Electron Microscopy
The effects of the E. cristatum culture filtrate on spore germination and morphology of A. flavus were studied by SEM after 24 h. Typical SEM images of treated and untreated (PDB with water) spore suspensions are depicted in Figure 3. Importantly, the E. cristatum culture filtrate did not affect spore germination, while fungal morphology was affected. This result is in accordance with a study published by Sangmanee and Hongpattarakere (2014). Complete mycelial structures often exhibit increased resistance to antimicrobial treatment (Liang et al., 2019). Sun et al. (2016) concluded that the antifungal effect of some the associated compounds may be partly ascribed to damage caused to the hyphal cell structure. As illustrated in Figure 3A, untreated A. flavus hyphae retained an elongated structure with a smooth appearance. Conversely, as shown in Figure 3B, the hyphae of the culture filtrate treatment were wrinkled and folded. Similar phenomena have been observed when A. flavus were treated with volatile organic compounds from Streptomyces alboflavus TD-1 (Yang et al., 2019) and Streptomyces yanglinensis 3–10 (Shakeel et al., 2018). This shriveled morphology might be attributed to interactions between components of the culture filtrate of E. cristatum and A. flavus and the cell walls; it is possible that associated interactions lead to visible membrane invagination.
FIGURE 3
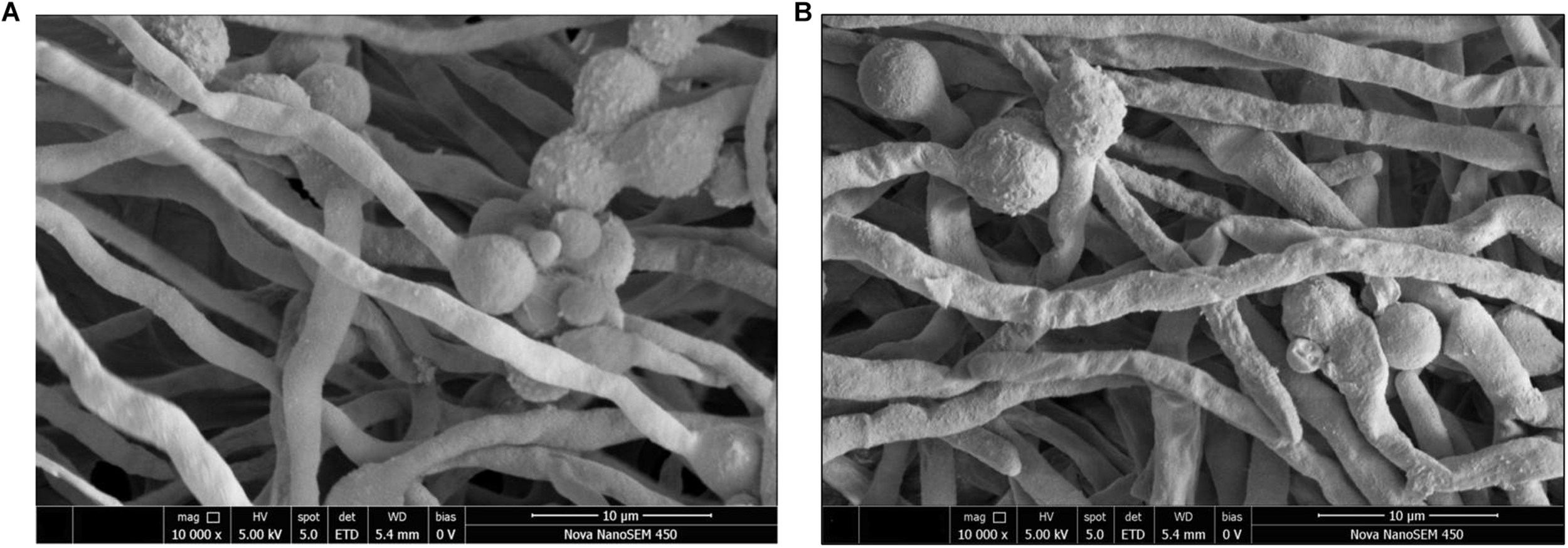
Morphology of A. flavus spores inoculated in PDB medium with the E. cristatum culture filtrate (B) and water (A).
Gene Expression Studies
In order to analyze the effect of the E. cristatum culture filtrate on aflatoxin gene activation, A. flavus was grown in PDB containing 30% (v/v) E. cristatum culture filtrate for 3 days. Six genes (aflR, aflS, aflD, aflQ, VeA, and LaeA) were investigated relatively to reference gene (β-tubulin). As shown in Figure 4, the E. cristatum culture filtrate repressed the expressions of aflD (2.94-fold), aflQ (8.33-fold), and aflS (4.16-fold). LaeA, veA, and aflR gene expressions were not significantly impacted by the culture filtrate. The aflR gene is known to encode a major transcriptional regulator of aflatoxin biosynthesis genes, while aflS might be involved in the regulation of aflatoxin biosynthesis through the regulation of other genes (Tominaga et al., 2006; Wang B. et al., 2017). The aflD gene encodes the first stable intermediate norsolorinic acid during aflatoxin formation (Abdel-Hadi et al., 2010), while aflQ encodes an oxidoreductase, which is required for the final steps associated with the conversion of sterigmatocystin to AFB1 (Cary et al., 2012). In this study, the transcript level of aflR did not obviously change, while that of aflS was significantly downregulated (p < 0.05), and the ratio aflR:aflS was above 1. Upon normal expression of aflR and aflS, sufficient quantities of AflS protein combine with AflR protein to form the AflS (4)-AflR (1) complex; the formation of this complex results in the synthesis of natural levels of toxin (Kong et al., 2014). Here, the aflR/aflS balance was upset was following down-regulation of aflS. Although aflR:aflS ratio above one would lead to an activation of AFB1 biosynthesis (Schmidt-Heydt et al., 2009). In our study, a ratio above one was not correlated with high AFB1 accumulation. Similar results were also obtained by Verheecke et al. (2015). Additionally, expression of the two structural genes, aflD and aflQ, was also significantly reduced (p < 0.01). Quantitative PCR showed that lack of aflS transcript led to a reduction of aflD expression (Meyers et al., 1998). In addition, two- to four-folds reductions in aflD, aflQ, and aflS were observed when A. flavus had a reduction in AFB1 (Moon et al., 2018). Thus, the repressions of aflS, aflD, and aflQ expressions could be the main reasons for a reduction in the formation of AFB1 in our experimental conditions.
FIGURE 4
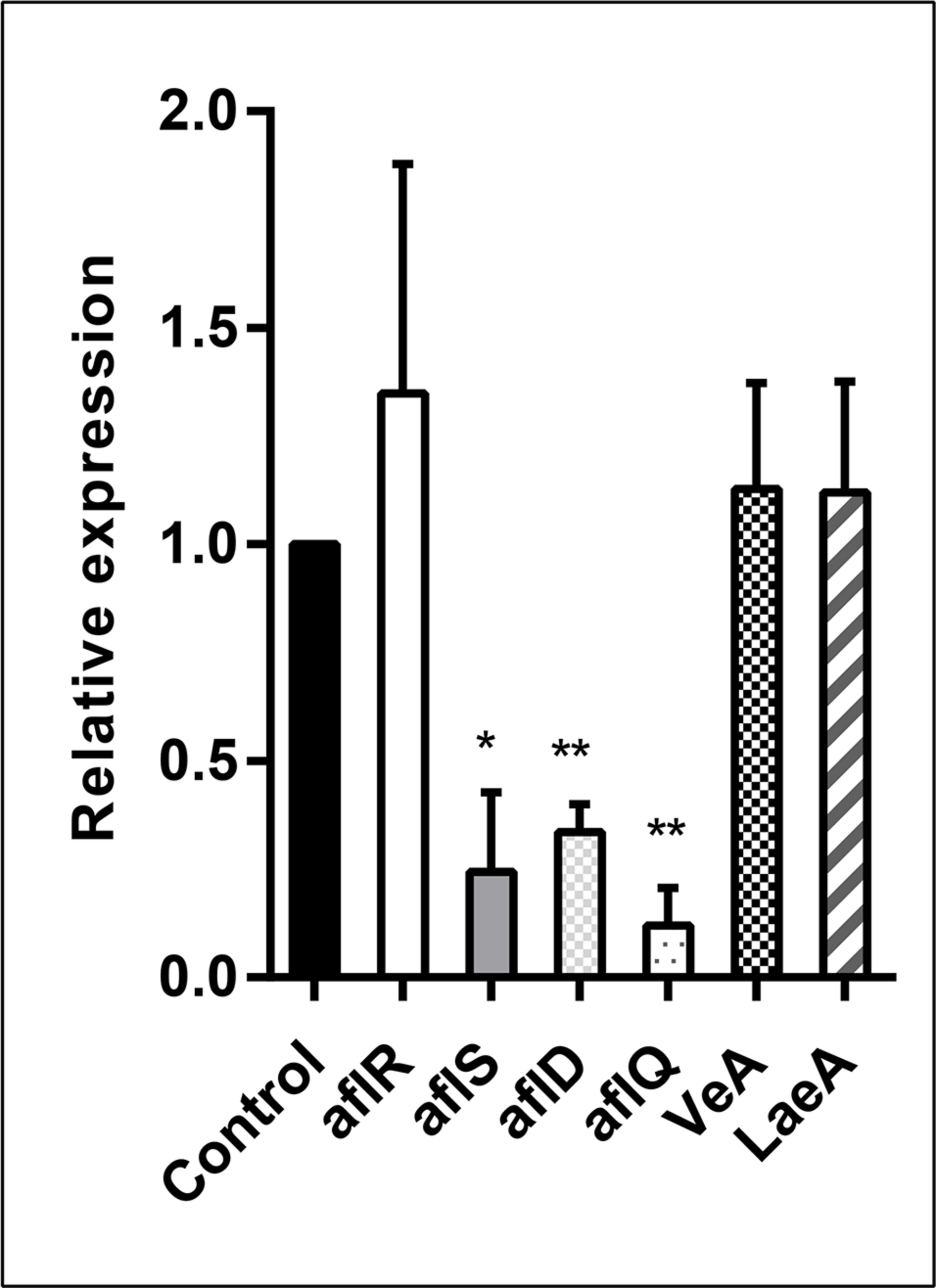
Relative expression values of the aflR, aflS, aflD, aflQ, VeA, and LaeA genes in A. flavus at 28 ± 2°C after 3 days. The β-tubulin gene was used as an internal control to normalize the expression data. The error bars represent the standard deviations from three independent experiments with three replicates each. Values in each column followed by one (p < 0.05) or two (p < 0.01) asterisks are significantly different from the control according to the Student’s t-test.
Degradation of AFB1 in the E. cristatum Culture Filtrate at Different Exposure Times
In this study, the AFB1 degradation ability of the E. cristatum culture filtrate was confirmed by HPLC. As shown in Figure 5A, the concentration of AFB1 in the control were stable during the incubation time. And the total ion chromatograms of aflatoxin B1 of control group was showed in Figure 5B. However, the concentration of the AFB1 was reduced from 942.24 to 517.47 ng/mL when disposed the E. cristatum culture filtrate after 3 days. And the percentage of AFB1 degradation was approximately 47.42%. The microbial culture supernatants as the degrading matrix have been widely studied for their AFB1 degradation potential (Verheecke et al., 2016). The culture supernatants of A. niger D15-Lcc2#3, Pleurotus ostreatus St2-3, Phanerochaete chrysosporium ME-446, and Bjerkandera adusta SCC0169 also had the ability to degrade AFB1. AFB1 degradation rates by these culture filtrates were respectively, 55.0, 35.90, 13.77, and 28.19% after 3 days at 30°C (Alberts et al., 2009). In comparison, our 47.42% degradation rate was relatively high. Moreover, following increased incubation times, our result showed that the AFB1 degradation rate was 58.85%, and only 353.45 ng/mL of AFB1 was detected after 5 days (p < 0.01). This latter result indicates that the residual quantity of AFB1 was positively correlated with exposure times; this is in accordance with studies performed by Sangare et al. (2014) and Petchkongkaew et al. (2008). However, the supernatant of Stenotrophomonas maltophilia 35-3 was able to reduce 84.80% of AFB1 (initial concentration 2 mg/L) at 37°C when exposed a more longer time (90 h) (Guan et al., 2008). Therefore, further optimizations of degradation conditions (such as temperature, pH, metal ions) are necessary.
FIGURE 5
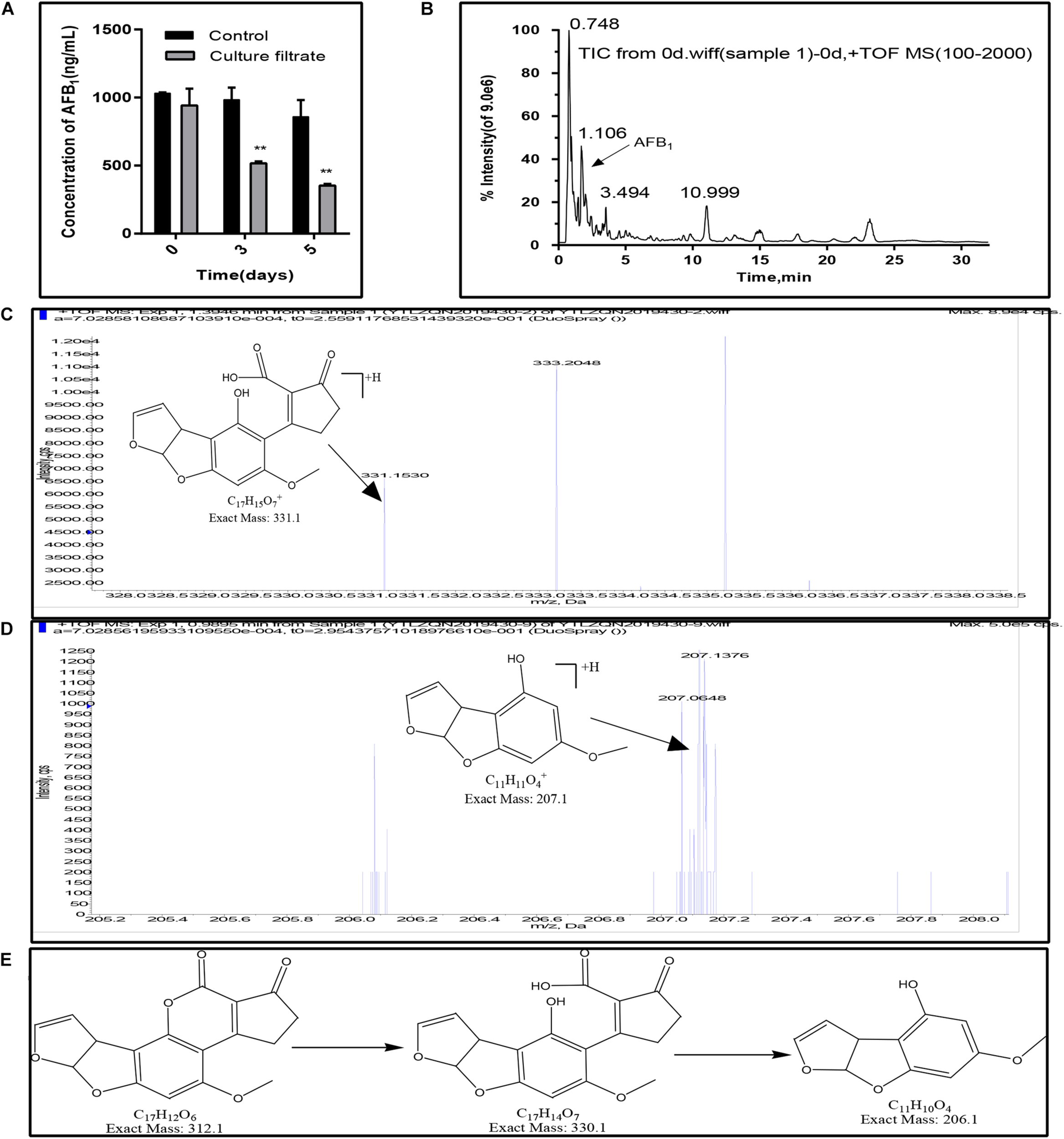
AFB1 degradation and fragmentation of biodegradation products of AFB1. (A) Residual aflatoxin B1 (ng/mL) in the E. cristatum culture filtrate and PDB over different time-periods at 28 ± 2°C. Error bars represent standard deviations of the means following three replicates each. (B) Electrospray ionization (ESI) total ion chromatogram (TIC) scan of control group (1000 ng/mL AFB1 in PDB broth, after incubation in the dark at 28 ± 2°C). (C) Putative degraded Product 1. (D) Putative degraded Product 2. (E) The deduced biodegradation pathway of AFB1.
Furthermore, AFB1 degradation products have been observed and identified using LC-qTOF/MS. Compared with the control (AFB1 with PDB and just only the E. cristatum culture filtrate), two degraded products with suggested molecular formulae of C17H14O7 (product 1) and C11H10O4 (product 2) with [M + H]+ ion peaks at m/z 331.15 (Figure 5C) and 207.06 (Figure 5D) were detected in the treated samples following Aanalyst TF1.7.1 analysis. Generally, aflatoxicol, aflatoxin B2a (AFB2a), and aflatoxin D1 (AFD1) were the most reported AFB1 degradation products. Nakazato et al. (1990) found that Eurotium herbariorum could convert AFB1 to aflatoxicol-A (AFL-A) by reducing the cyclopentenone carbonyl of AFB1, then AFL-A was converted to aflatoxicol-B (AFL-B) by the actions of medium components or organic acids produced from the fungi, this study also found that A. niger could convert AFB1 to AFB2a by producing organic acids to lowering pH of the medium. Eshelli et al. (2015) studied the metabolomics of the AFB1 degradation by the R. erythropolis culture filtrate. The author stated that AFB1 degradation may involve the formation of the β-keto acid structure, followed by hydrolysis of the lactone ring resulting in a metabolite with 330 amu (product 1 in this study). The hydrolysis was followed by decarboxylation of the open lactone ring yielding to AFD1, this process was also involved the formation of AFD2. In this study, as shown in Figure 5E, AFB1 degradation may involve the hydrolysis of the lactone ring resulting in product 1 with 330 m/z, followed by decarboxylation of the open lactone ring yielding a product known as AFD2 with 206 m/z (product 2), where the lactone carbonyl and cyclopentenone ring disappeared. Although the products still retained the 8,9-dihydrofuran double bond, the associated toxicities were much less than that of AFB1 (Eshelli et al., 2015).
Composition of the E. cristatum Culture Filtrate
Putative metabolites (Table 2) of the E. cristatum culture filtrate were analyzed using GC/MS (PDB medium was used as the control). Numerous substances, which have been shown to elicit significant antimicrobial activities in previous studies, were observed in the present study. Of the volatile components detected, alcohols, including 1-hexanol and 1-octen-3-ol, were the predominant substances detected. Due to the fact that these materials non-selectively adsorbed and predominantly accumulated in the cell membrane, thereby inhibiting membrane function, it was deduced that these substances exhibit antimicrobial activity (Ingram and Buttke, 1984). 1-octen-3-ol at 4.87 ng/L was the most abundant of these substances observed in this study. (Zunino et al., 2015) found that 1-hexanol (4.24 mM) can effectively inhibit fumonisin production by Fusarium verticillioides. Haidar et al. (2016) reported that 1-octen-3-ol can strongly inhibit the growth of Phaeomoniella chlamydospora. Similarly, Xiong et al. (2017) revealed that 1-octen-3-ol can inhibit fungal growth and spore germination, while also suggesting that this compound can change the permeability of the cell membrane.
TABLE 2
| Number | RT | RI | Compounds | Concentrations (ng/L) |
| 1 | 4.542 | 408 | Acetaldehyde | 0.10 ± 0.02 |
| 2 | 11.071 | 715 | (E)-2-pentenal | 0.66 ± 0.35 |
| 3 | 11.211 | 790 | Acetylacetone | 3.12 ± 2.96 |
| 4 | 15.19 | 860 | 1-hexanol | 1.25 ± 0.30 |
| 5 | 20.233 | 969 | 1-octen-3-ol | 4.87 ± 0.91 |
| 6 | 21.029 | 1005 | Octanal | 0.46 ± 0.00 |
| 7 | 22.926 | 1018 | D-limonene | 0.29 ± 0.02 |
| 8 | 23.448 | 1029 | Acetophenone | 0.30 ± 0.08 |
| 9 | 26.554 | 1059 | 1,2-dimethoxy-benzene | 0.18 ± 0.06 |
| 10 | 27.093 | 1124 | 3,5,5-trimethyl-hexanoic acid | 3.33 ± 1.94 |
| 11 | 31.849 | 1262 | Thymol | 0.93 ± 0.76 |
| 12 | 32.456 | 1293 | (R)-(+)-Citronellic acid | 0.39 ± 0.01 |
| 13 | 34.526 | 1331 | Propanoic acid,2-methy, l,3-hydroxy-2,2,4-trimethylpentyl ester | 1.15 ± 0.72 |
| 14 | 40.041 | 1605 | Pentanoic-acid,2,2,4-trimethyl-3-carboxyisopropyl, isobutyl ester | 1.32 ± 1.43 |
Metabolites in the E. cristatum culture filtrate identified by GC/MS.
Aldehydes have stronger antifungal activity than alcohols (Ando et al., 2012). (E)-2-pentenal (0.66 ng/L), acetaldehyde (0.10 ng/L), and octanal (0.46 ng/L) were the predominant aldehydes analyzed. Previous studies have shown that acetaldehyde can inhibit the growth of Rhizopus stolonifer, Botrytis cinerea, Alternaria alternata, and Penicillium digitatum (Zhou et al., 2018). Wright et al. (2000) indicated that octanal (100 μL/L) had potent fungitoxicity against A. parasiticus with an inhibition ratio of 76%; this study also revealed that octanal also inhibited AFB1 production. In addition, thymol (0.93 ng/L), acetophenone (0.30 ng/L) and d-limonene (0.29 ng/L) also exhibited remarkable antimicrobial activities. A study by Lambert et al. (2001) suggested that thymol inhibits the anthracnose pathogen and is positively correlated with damage to the membrane; these occurrences affect pH homeostasis and the equilibrium of inorganic ions. Boukaew and Prasertsan (2018) previously reported on the inhibitory activities of acetophenone (100 μL/L) in relation to the growth and sporulation of both A. flavus TISTR 3041 and A. parasiticus TISTR 3276 on PDA plates. Our results reveal that the E. cristatum culture filtrate is bioactive and volatile-rich, and individual quantitative analysis of the inhibitory activities of these compounds will be required in future studies.
Cell Viability Assay
In this study, the cytotoxicity of the AFB1 degraded products was assessed using human liver cancer cells via the MTT assay. Percentage cell viability is presented in Figure 6. Results indicated that the percentage cell viability was reduced to 77.10 and 94.17%, respectively, when cells were exposed to AFB1 stock solution and the AFB1 degradation (3 days) products. Conversely, when cells were treated with longer degradation periods (5 days) AFB1 extracts, evident growth-promoting effects (104.95%) were observed (p < 0.05). AFB1 is a well-documented hepatocarcinogen, and it can cause DNA damage by forming DNA-adducts, also can affect RNA translation and induce oxidative stress (Madalena et al., 2018). Since AFB1 is predominantly metabolized in the liver, the human hepatoma HepG2 cell model is considered to be the most suitable system for testing its in vitro toxicity (Domijan et al., 2019). Previous studies have showed that AFB1 could decrease HepG2 survival (Chan et al., 2003; Wang C. et al., 2017; Madalena et al., 2018). But in this study, AFB1 degradation extracts (5 days) stimulated the growth of HepG2 cells. Specifically, AFB1 has a two-phase effect on cells: low (0.5–1 μg/mL) doses promote the proliferation of HepG2 cells, while high doses inhibit proliferation (Wang C. et al., 2017). Therefore, our results suggest that AFB1 was degraded to relatively low levels by the E. cristatum culture filtrate. This occurrence may lead to the synthesis of other possibly less or non-toxic compounds. Furthermore, the cell-free culture extracts did not exhibit cytotoxicity to HepG2 cells and may even have stimulated the growth of HepG2 cells. Therefore, the degraded (5 days) AFB1 extracts exhibit increased stimulatory effects compared with AFB1 alone. Similar results were also observed by Ma et al. (2012).
FIGURE 6
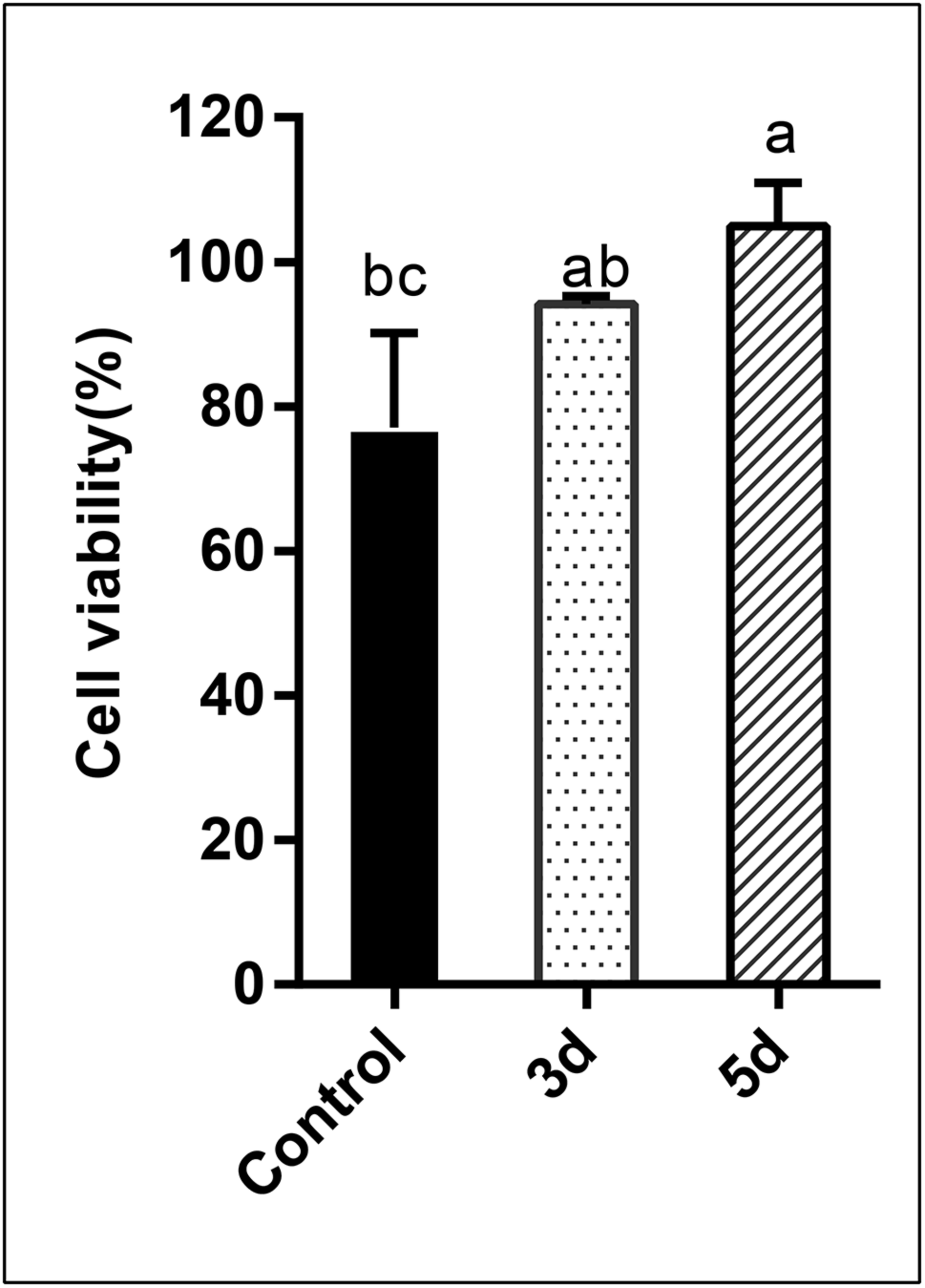
Analysis of the cytotoxicity of AFB1 degradation by cell-free E. cristatum culture filtrate using MTT assay. The error bars represent the standard deviations from three independent experiments with six replicates each. The same letters in each column indicate no significant differences among the data at the 5% level following the Tukey test.
Conclusion
Our study showed that E. cristatum exhibited potential biocontrol activity against aflatoxigenic A. flavus. Thus, E. cristatum may represent an antagonist due to its capacity to reduce both fungal growth and AFB1 biosynthesis levels. GC/MS analysis indicated that there are many antifungal substances present in the E. cristatum culture filtrate. qRT-PCR analysis revealed a significant reduction in the AflS/AflR ratio. In addition, our results confirmed that atoxigenic E. cristatum could effectively degrade AFB1. Furthermore, two degradation products, where the lactone ring was destroyed, were identified by LC-qTOF/MS. In addition, cytotoxicity test showed that the degradation compounds were less toxic than AFB1. Further studies are required to investigate E. cristatum culture filtrate.
Statements
Data availability statement
GenBank accession numbers for our nucleotide sequences: A. flavus: MN759628, HNYYWX.21: MN759629.
Author contributions
QZ, YQ, TY, and YY conceived and designed the experiments. QZ performed the experiments and analyzed the data. QZ and TY wrote the manuscript. XW, YG, YZ, and YW contributed reagents, materials, and analysis tools. All authors read and approved the final manuscript.
Funding
This research was supported by the Shaanxi Special Project of China (2018ZDXM-NY-084).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AbbasH. K.ZablotowiczR. M.HornB. W.PhillipsN. A.JohnsonB. J.JinX.et al (2011). Comparison of major biocontrol strains of non-aflatoxigenic Aspergillus flavus for the reduction of aflatoxins and cyclopiazonic acid in maize.Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess.28198–208. 10.1080/19440049.2010.544680
2
Abdel-HadiA.CarterD.MaganN. (2010). Temporal monitoring of the nor-1 (aflD) gene of Aspergillus flavus in relation to aflatoxin B1 production during storage of peanuts under different water activity levels.J. Appl. Microbiol.1091914–1922. 10.1111/j.1365-2672.2010.04820.x
3
AdeboO. A.NjobehP. B.GbashiS. (2017). Review on microbial degradation of aflatoxins.Crit. Rev. Food Sci. Nutr.573208–3217. 10.1080/10408398.2015.1106440
4
AdeboO. A.NjobehP. B.SiduS.TlouM. G.MavumengwanaV. (2016). Aflatoxin B-1 degradation by liquid cultures and lysates of three bacterial strains.Int. J. Food Microbiol.23311–19. 10.1016/j.ijfoodmicro.2016.06.007
5
AlbertsJ. F.GelderblomW. C. A.BothaA.van ZylW. H. (2009). Degradation of aflatoxin B-1 by fungal laccase enzymes.Int. J. Food Microbiol.13547–52. 10.1016/j.ijfoodmicro.2009.07.022
6
AlshannaqA. F.GibbonsJ. G.LeeM.-K.HanK.-H.HongS.-B.YuJ.-H. (2018). Controlling aflatoxin contamination and propagation of Aspergillus flavus by a soy-fermenting Aspergillus oryzae strain.Sci. Rep.8:16871. 10.1038/s41598-018-35246-1
7
AndoH.HatanakaK.OhataI.Yamashita-KitaguchiY.KurataA.KishimotoN. (2012). Antifungal activities of volatile substances generated by yeast isolated from Iranian commercial cheese.Food Control26472–478. 10.1016/j.foodcont.2012.02.017
8
BbosaG. S.KityaD.LubegaA.Ogwal-OkengJ.AnokbonggoW. W.KyegombeD. B. (2013). “Review of the biological and health effects of aflatoxins on body organs and body systems,” in Aflatoxins - Recent Advances and Future Prospects, ed.Razzaghi-AbyanehM. (London: InTech).
9
BoukaewS.PrasertsanP. (2018). Inhibitory effects of acetophenone or phenylethyl alcohol as fumigant to protect soybean seeds against two aflatoxin-producing fungi.J. Food Sci. Technol. Mysore555123–5132. 10.1007/s13197-018-3458-6
10
CaryJ. W.Harris-CowardP. Y.EhrlichK. C.MooreG. G.WeiQ. J.BhatnagarD. (2012). Functional and phylogenetic analysis of the Aspergillus ochraceoroseus aflQ (ordA) gene ortholog.Mycologia104857–864. 10.3852/11-328
11
CaryJ. W.ObrianG. R.NielsenD. M.NiermanW.Harris-CowardP.BhatnagarJ. Y. D.et al (2007). Elucidation of veA-dependent genes associated with aflatoxin and sclerotial production in Aspergillus flavus by functional genomics.Appl. Microbiol. Biotechnol.761107–1118. 10.1007/s00253-007-1081-y
12
ChanH. T.ChanC.HoJ. W. (2003). Inhibition of glycyrrhizic acid on aflatoxin B-1-induced cytotoxicity in hepatoma cells.Toxicology188211–217. 10.1016/s0300-483x(03)00087-8
13
ChanL.TakahashiM.LimP. J.AoyamaS.MakinoS.FerdinandusF.et al (2019). Eurotium cristatum fermented okara as a potential food ingredient to combat diabetes.Sci. Rep.9:17536. 10.1038/s41598-019-54021-4
14
ChaudharyA.MorS.SinghK. (2001). Biodegradation of aflatoxins by Aspergillus niger.Indian J. Anim. Sci.71877–880.
15
ChenG.XieM.DaiZ.WanP.YeH.ZengX.et al (2018). Kudingcha and fuzhuan brick tea prevent obesity and modulate gut microbiota in high-fat diet fed mice.Mol. Nutr. Food Res.62:e1700485. 10.1002/mnfr.201700485
16
ChenY.KongQ.LiangY. (2019). Three newly identified peptides from Bacillus megaterium strongly inhibit the growth and aflatoxin B-1 production of Aspergillus flavus.Food Control9541–49. 10.1016/j.foodcont.2018.07.040
17
CserhatiM.KrisztB.KrifatonC.SzoboszlayS.HahnJ.TothS.et al (2013). Mycotoxin-degradation profile of Rhodococcus strains.Int. J. Food Microbiol.166176–185. 10.1016/j.ijfoodmicro.2013.06.002
18
DomijanA.-M.CermakA. M. M.VulicA.BujakI. T.PavicicI.PleadinJ.et al (2019). Cytotoxicity of gamma irradiated aflatoxin B-1 and ochratoxin A.J. Environ. Sci. Health B54155–162. 10.1080/03601234.2018.1536578
19
DornerJ. W.LambM. C. (2006). Development and commercial use of afla-Guard, an aflatoxin biocontrol agent.Mycotoxin Res.2233–38. 10.1007/bf02954555
20
DuF.-Y.LiX.LiX.-M.ZhuL.-W.WangB.-G. (2017). Indolediketopiperazine alkaloids from Eurotium cristatum EN-220, an endophytic fungus isolated from the marine alga Sargassum thunbergii.Mar. Drugs15:24. 10.3390/md15020024
21
DuF.-Y.LiX.-M.SongJ.-Y.LiC.-S.WangB.-G. (2014). Anthraquinone derivatives and an orsellinic acid ester from the marine alga-derived endophytic fungus Eurotium cristatum EN-220.Helv. Chim. Acta97973–978. 10.1002/hlca.201300358
22
DuH.WangQ.YangX. (2019). Fu brick tea alleviates chronic kidney disease of rats with high fat diet consumption through attenuating insulin resistance in skeletal muscle.J. Agric. Food Chem.672839–2847. 10.1021/acs.jafc.8b06927
23
EhrlichK. C. (2014). Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: advantages and limitations.Front. Microbiol.5:50. 10.3389/fmicb.2014.00050
24
EshelliM.HarveyL.Edrada-EbelR.McNeilB. (2015). Metabolomics of the bio-degradation process of aflatoxin B1 by actinomycetes at an initial pH of 6.0.Toxins7439–456. 10.3390/toxins7020439
25
GongA.-D.DongF.-Y.HuM.-J.KongX.-W.WeiF.-F.GongS.-J.et al (2019). Antifungal activity of volatile emitted from Enterobacter asburiae Vt-7 against Aspergillus flavus and aflatoxins in peanuts during storage.Food Control106:106718. 10.1016/j.foodcont.2019.106718
26
GuQ.DuanG.YuX. (2019). Bioconversion of flavonoid glycosides from Hippophae rhamnoides leaves into flavonoid aglycones by Eurotium amstelodami.Microorganisms7:122. 10.3390/microorganisms7050122
27
GuanS.JiC.ZhouT.LiJ.MaQ.NiuT. (2008). Aflatoxin B-1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium.Int. J. Mol. Sci.91489–1503. 10.3390/ijms9081489
28
HaidarR.RoudetJ.BonnardO.DufourM. C.Corio-CostetM. F.FertM.et al (2016). Screening and modes of action of antagonistic bacteria to control the fungal pathogen Phaeomoniella chlamydospora involved in grapevine trunk diseases.Microbiol. Res.192172–184. 10.1016/j.micres.2016.07.003
29
HawkinsN. J.BassC.DixonA.NeveP. (2019). The evolutionary origins of pesticide resistance.Biol. Rev.94135–155. 10.1111/brv.12440
30
IARC (1993). OCHRATOXIN-A IARC Monographs on the Evaluation Of Carcinogenic Risks To Humans Some Naturally Occurring Substances: Food Items And Constituents, Heterocyclic Aromatic Amines And Mycotoxins, Vol. 56. Lyon: IARC, 489–540.
31
IngramL. O.ButtkeT. M. (1984). Effects of alcohols on micro-organisms.Adv. Microb. Physiol.25253–300. 10.1016/s0065-2911(08)60294-5
32
IramW.AnjumT.IqbalM.GhaffarA.AbbasM. (2016). Structural elucidation and toxicity assessment of degraded products of aflatoxin B1 and B2 by aqueous extracts of Trachyspermum ammi.Front. Microbiol.7:346. 10.3389/fmicb.2016.00346
33
IsmailA.GonçalvesB. L.de NeeffD. V.PonzilacquaB.CoppaC. F. S. C.HintzscheH.et al (2018). Aflatoxin in foodstuffs: occurrence and recent advances in decontamination.Food Res. Int.11374–85. 10.1016/j.foodres.2018.06.067
34
Jaime-GarciaR.CottyP. J. (2007). Influences of application timing on sporulation of the biocontrol product Aspergillus flavus AF36 in cotton fields of Arizona.Phytopathology97S168–S168.
35
KangD.SuM.DuanY.HuangY. (2019). Eurotium cristatum, a potential probiotic fungus from Fuzhuan brick tea, alleviated obesity in mice by modulating gut microbiota.Food Funct.105032–5045. 10.1039/c9fo00604d
36
KarabulutS.PaytakovG.LeszczynskiJ. (2014). Reduction of aflatoxin B1 to aflatoxicol: a comprehensive DFT study provides clues to its toxicity.J. Sci. Food Agric.943134–3140. 10.1002/jsfa.6663
37
KongQ.ChiC.YuJ.ShanS.LiQ.LiQ.et al (2014). The inhibitory effect of Bacillus megaterium on aflatoxin and cyclopiazonic acid biosynthetic pathway gene expression in Aspergillus flavus.Appl. Microbiol. Biotechnol.985161–5172. 10.1007/s00253-014-5632-8
38
LambertR. J. W.SkandamisP. N.CooteP. J.NychasG. J. E. (2001). A study of the minimum inhibitory concentration and mode of action of oregano essential oil, thymol and carvacrol.J. Appl. Microbiol.91453–462. 10.1046/j.1365-2672.2001.01428.x
39
LiangD.XingF.SelvarajJ. N.LiuX.WangL.HuaH.et al (2015). Inhibitory effect of cinnamaldehyde, citral, and eugenol on aflatoxin biosynthetic gene expression and aflatoxin B-1 biosynthesis in Aspergillus flavus.J. Food Sci.80M2917–M2924. 10.1111/1750-3841.13144
40
LiangY.KongQ.YaoY.XuS.XieX. (2019). Fusion expression and anti-Aspergillus flavus activity of a novel inhibitory protein DN-AflR.Int. J. Food Microbiol.290184–192. 10.1016/j.ijfoodmicro.2018.10.015
41
LiuT.ChenR.ChenD.YingJ.MengQ.HaoB.et al (2016). Effect of CGMCC No. 8730 Eurotium cristatum fermented dark tea extract on body weight and blood lipid in two dyslipidemia rat models.Wei Sheng Yan Jiu45637–642.
42
LiuX.GuanX.XingF.LvC.DaiX.LiuY. (2017). Effect of water activity and temperature on the growth of Aspergillus flavus, the expression of aflatoxin biosynthetic genes and aflatoxin production in shelled peanuts.Food Control82325–332. 10.1016/j.foodcont.2017.07.012
43
LiuY.WuF. (2010). Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment.Environ. Health Perspect.118818–824. 10.1289/ehp.0901388
44
LivakK. J.SchmittgenT. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method.Methods25402–408. 10.1006/meth.2001.1262
45
LuoM.LiangX. Q.DangP.HolbrookC. C.BausherM. G.LeeR. D.et al (2005). Microarray-based screening of differentially expressed genes in peanut in response to Aspergillus parasiticus infection and drought stress.Plant Sci.169695–703. 10.1016/j.plantsci.2005.05.020
46
MaY.KongQ.HuaH.LuoT.JiangY. (2012). Aflatoxin B1 up-regulates insulin receptor substrate 2 and stimulates hepatoma cell migration.PLoS One7:e47961. 10.1371/journal.pone.0047961
47
MadalenaM.SobralC.FariaM. A.CunhaS. C.FerreiraI. M. P. L. V. O. (2018). Toxicological interactions between mycotoxins from ubiquitous fungi: impact on hepatic and intestinal human epithelial cells.Chemosphere202538–548. 10.1016/j.chemosphere.2018.03.122
48
Medina-CordovaN.Lopez-AguilarR.AscencioF.CastellanosT.Campa-CordovaA. I.AnguloC. (2016). Biocontrol activity of the marine yeast Debaryomyces hansenii against phytopathogenic fungi and its ability to inhibit mycotoxins production in maize grain (Zea mays L.).Biol. Control9770–79. 10.1016/j.biocontrol.2016.03.006
49
MeyersD. M.ObrianG.DuW. L.BhatnagarD.PayneG. A. (1998). Characterization of aflJ, a gene required for conversion of pathway intermediates to aflatoxin.Appl. Environ. Microbiol.643713–3717.
50
MoonY.-S.ChoiW.-S.ParkE.-S.BaeI. K.ChoiS.-D.PaekO.et al (2016). Antifungal and antiaflatoxigenic methylenedioxy-containing compounds and piperine-like synthetic compounds.Toxins8:240. 10.3390/toxins8080240
51
MoonY.-S.KimH.-M.ChunH. S.LeeS.-E. (2018). Organic acids suppress aflatoxin production via lowering expression of aflatoxin biosynthesis-related genes in Aspergillus flavus.Food Control88207–216. 10.1016/j.foodcont.2018.01.017
52
NakazatoM.MorozumiS.SaitoK.FujinumaK.NishimaT.KasaiN. (1990). Interconversion of aflatoxin B1 and aflatoxicol by several fungi.Appl. Environ. Microbiol.561465–1470. 10.1128/aem.56.5.1465-1470.1990
53
NguyenP.-A.StrubC.FontanaA.Schorr-GalindoS. (2017). Crop molds and mycotoxins: alternative management using biocontrol.Biol. Control10410–27. 10.1016/j.biocontrol.2016.10.004
54
PengX.-Y.ZhangW.-M.LiuS.-Y.LeiC.-X.ZhouX.-M.LiuS.-Q.et al (2011). Isolation and identification of “golden-flower” fungus in fuzhuan tea from Hunan area.J. Fungal Res.9157–161.
55
PengZ.ChenL.ZhuY.HuangY.HuX.WuQ.et al (2018). Current major degradation methods for aflatoxins: a review.Trends Food Sci. Technol.80155–166.
56
PeromingoB.RodriguezM.DelgadoJ.AndradeM. J.RodriguezA. (2017). Gene expression as a good indicator of aflatoxin contamination in dry-cured ham.Food Microbiol.6731–40. 10.1016/j.fm.2017.05.008
57
PetchkongkaewA.TaillandierP.GasaluckP.LebrihiA. (2008). Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): screening for aflatoxin B1 and ochratoxin A detoxification.J. Appl. Microbiol.1041495–1502. 10.1111/j.1365-2672.2007.03700.x
58
PhamC. D.ReissE.HagenF.MeisJ. F.LockhartS. R. (2014). Passive surveillance for azole-resistant Aspergillus fumigatus, United States, 2011-2013.Emerg. Infect. Dis.201498–1503. 10.3201/eid2009.140142
59
RaoK. R.VipinA. V.HariprasadP.AppaiahK. A. A.VenkateswaranG. (2017). Biological detoxification of aflatoxin B-1 by Bacillus licheniformis CFR1.Food Control71234–241. 10.1016/j.foodcont.2016.06.040
60
SangareL.ZhaoY.FollyY. M. E.ChangJ.LiJ.SelvarajJ. N.et al (2014). Aflatoxin B1 degradation by a Pseudomonas strain.Toxins63028–3040. 10.3390/toxins6103028
61
SangmaneeP.HongpattarakereT. (2014). Inhibitory of multiple antifungal components produced by Lactobacillus plantarum K35 on growth, aflatoxin production and ultrastructure alterations of Aspergillus flavus and Aspergillus parasiticus.Food Control40224–233. 10.1016/j.foodcont.2013.12.005
62
SantiniA.RitieniA. (2013). Aflatoxins: Risk, Exposure and Remediation.London: IntechOpen.
63
Schmidt-HeydtM.Abdel-HadiA.MaganN.GeisenR. (2009). Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature.Int. J. Food Microbiol.135231–237. 10.1016/j.ijfoodmicro.2009.07.026
64
ShakeelQ.LyuA.ZhangJ.WuM.LiG.HsiangT.et al (2018). Biocontrol of Aspergillus flavus on peanut kernels using Streptomyces yansingensis 3-10.Front. Microbiol.9:1049. 10.3389/fmicb.2018.01049
65
SherifS. O.SalamaE. E.Abdel-WahhabM. A. (2009). Mycotoxins and child health: the need for health risk assessment.Int. J. Hyg. Environ. Health212347–368. 10.1016/j.ijheh.2008.08.002
66
ShiJ.LiuJ.KangD.HuangY.KongW.XiangY.et al (2019). Isolation and characterization of benzaldehyde derivatives with anti-inflammatory activities from Eurotium cristatum, the dominant fungi species in fuzhuan brick tea.ACS Omega46630–6636. 10.1021/acsomega.9b00593
67
SunQ.ShangB.WangL.LuZ. S.LiuY. (2016). Cinnamaldehyde inhibits fungal growth and aflatoxin B-1 biosynthesis by modulating the oxidative stress response of Aspergillus flavus.Appl. Microbiol. Biotechnol.1001355–1364. 10.1007/s00253-015-7159-z
68
TominagaM.LeeY. H.HayashiR.SuzukiY.YamadaO.SakamotoK.et al (2006). Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains.Appl. Environ. Microbiol.72484–490. 10.1128/aem.72.1.484-490.2006
69
TorresA. M.BarrosG. G.PalaciosS. A.ChulzeS. N.BattilaniP. (2014). Review on pre- and post-harvest management of peanuts to minimize aflatoxin contamination.Food Res. Int.6211–19. 10.1016/j.foodres.2014.02.023
70
TsitsigiannisD. I.DimakopoulouM.AntoniouP. P.TjamosE. C. (2012). Biological control strategies of mycotoxigenic fungi and associated mycotoxins in Mediterranean basin crops.Phytopathol. Mediterr.51158–174.
71
VerheeckeC.LibozT.AnsonP.DiazR.MathieuF. (2015). Reduction of aflatoxin production by Aspergillus flavus and Aspergillus parasiticus in interaction with Streptomyces.Microbiology161967–972. 10.1099/mic.0.000070
72
VerheeckeC.LibozT.MathieuF. (2016). Microbial degradation of aflatoxin B1: current status and future advances.Int. J. Food Microbiol.2371–9. 10.1016/j.ijfoodmicro.2016.07.028
73
WangB.HanX.BaiY.LinZ.QiuM.NieX.et al (2017). Effects of nitrogen metabolism on growth and aflatoxin biosynthesis in Aspergillus flavus.J. Hazard. Mater.324691–700. 10.1016/j.jhazmat.2016.11.043
74
WangS.-Q.HuangG.-Q.LiY.-P.XiaoJ.-X.ZhangY.JiangW.-L. (2015). Degradation of aflatoxin B1 by low-temperature radio frequency plasma and degradation product elucidation.Eur. Food Res. Technol.241103–113. 10.1007/s00217-015-2439-5
75
WangC.LiZ.WangH.QiuH.ZhangM.LiS.et al (2017). Rapid biodegradation of aflatoxin B1 by metabolites of Fusarium sp WCQ3361 with broad working temperature range and excellent thermostability.J. Sci. Food Agric.971342–1348. 10.1002/jsfa.7872
76
WangX.ZhangY.RenT.ZhaoQ.YueT.YuanY. (2019). Isolation and identification of Eurotium cristatum from Fuzhuan tea and its application in liquid-state fermentation.Food Sci.40172–178. 10.7506/spkx1002-6630-20180820-207
77
WrightM. S.Greene-McDowelleD. M.ZeringueH. J.BhatnagarD.ClevelandT. E. (2000). Effects of volatile aldehydes from Aspergillus-resistant varieties of corn on Aspergillus parasiticus growth and aflatoxin biosynthesis.Toxicon381215–1223. 10.1016/s0041-0101(99)00221-4
78
WuF. (2015). Global impacts of aflatoxin in maize: trade and human health.World Mycotoxin J.8137–142. 10.3920/wmj2014.1737
79
WuF.KhlangwisetP. (2010). Evaluating the technical feasibility of aflatoxin risk reduction strategies in Africa.Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess.27658–676. 10.1080/19440041003639582
80
WuQ.JezkovaA.YuanZ.PavlikovaL.DohnalV.KucaK. (2009). Biological degradation of aflatoxins.Drug Metab. Rev.411–7. 10.1080/03602530802563850
81
XingF.WangL.LiuX.SelvarajJ. N.WangY.ZhaoY.et al (2017). Aflatoxin B-1 inhibition in Aspergillus flavus by Aspergillus niger through down-regulating expression of major biosynthetic genes and AFB(1) degradation by atoxigenic A. flavus.Int. J. Food Microbiol.2561–10. 10.1016/j.ijfoodmicro.2017.05.013
82
XiongC.LiQ.LiS. H.ChenC.ChenZ. Q.HuangW. L. (2017). In vitro antimicrobial activities and mechanism of 1-octen-3-ol against food-related bacteria and pathogenic fungi.J. Oleo Sci.661041–1049. 10.5650/jos.ess16196
83
XuD.WangH.ZhangY.YangZ.SunX. (2013). Inhibition of non-toxigenic Aspergillus niger FS10 isolated from Chinese fermented soybean on growth and aflatoxin B1 production by Aspergillus flavus.Food Control32359–365. 10.1016/j.foodcont.2012.12.013
84
XuL.MengW.CaoC.WangJ.ShanW.WangQ. (2015). Antibacterial and antifungal compounds from marine fungi.Mar. Drugs133479–3513. 10.3390/md13063479
85
YangM.LuL.PangJ.HuY.GuoQ.LiZ.et al (2019). Biocontrol activity of volatile organic compounds from Streptomyces alboflavus TD-1 against Aspergillus flavus growth and aflatoxin production.J. Microbiol.57396–404. 10.1007/s12275-019-8517-9
86
YazdaniD.Zainal AbidinM. A.TanY. H.KamaruzamanS. (2011). Molecular identification of Aspergillus and Eurotium species isolated from rice and their toxin-producing ability.Mikrobiologiia80707–713.
87
ZhangB.LuZ.XuX.DongM. (2018). Solid-state fermentation with Eurotium cristatum HC-18 o improve antioxidant activity of kudzu (Pueraria lobata) root.J. Food Nutr. Res.57384–395.
88
ZhangL.MaQ.MaS.ZhangJ.JiaR.JiC.et al (2017). Ameliorating effects of Bacillus subtilis ANSB060 on growth performance, antioxidant functions, and aflatoxin residues in ducks fed diets contaminated with aflatoxins.Toxins9:1. 10.3390/toxins9010001
89
ZhangP.JiaC.DengY.ChenS.ChenB.YanS.et al (2019). Anti-inflammatory prenylbenzaldehyde derivatives isolated from Eurotium cristatum.Phytochemistry158120–125. 10.1016/j.phytochem.2018.11.017
90
ZhouG.ChenY.KongQ.MaY.LiuY. (2017). Detoxification of aflatoxin B1 by Zygosaccharomyces rouxii with solid state fermentation in peanut meal.Toxins9:42. 10.3390/toxins9010042
91
ZhouT.WangX. H.YeB. S.ShiL.BaiX. L.LaiT. F. (2018). Effects of essential oil decanal on growth and transcriptome of the postharvest fungal pathogen Penicillium expansum.Postharvest Biol. Technol.145203–212. 10.1016/j.postharvbio.2018.07.015
92
ZuninoM. P.HerreraJ. M.PizzolittoR. P.RubinsteinH. R.ZygadloJ. A.DambolenaJ. S. (2015). Effect of selected volatiles on two stored pests: the fungus Fusarium verticillioides and the maize weevil Sitophilus zeamais.J. Agric. Food Chem.637743–7749. 10.1021/acs.jafc.5b02315
Summary
Keywords
Eurotium cristatum, aflatoxin B1, Aspergillus flavus, gene expression, degradation products
Citation
Zhao Q, Qiu Y, Wang X, Gu Y, Zhao Y, Wang Y, Yue T and Yuan Y (2020) Inhibitory Effects of Eurotium cristatum on Growth and Aflatoxin B1 Biosynthesis in Aspergillus flavus. Front. Microbiol. 11:921. doi: 10.3389/fmicb.2020.00921
Received
09 December 2019
Accepted
17 April 2020
Published
15 May 2020
Volume
11 - 2020
Edited by
Jae-Hyuk Yu, University of Wisconsin–Madison, United States
Reviewed by
John G. Gibbons, University of Massachusetts Amherst, United States; Belén Patiño, Complutense University of Madrid, Spain
Updates
Copyright
© 2020 Zhao, Qiu, Wang, Gu, Zhao, Wang, Yue and Yuan.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianli Yue, Yuetl@nwafu.edu.cnYahong Yuan, yyh324@126.com
This article was submitted to Fungi and Their Interactions, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.