- 1Centre for Antimicrobial Resistance, Alberta Health Services/Alberta Precision Laboratories/University of Calgary, Calgary, AB, Canada
- 2Department of Pathology and Laboratory Medicine, University of Calgary, Calgary, AB, Canada
- 3Department of Microbiology, Immunology and Infectious Diseases, University of Calgary, Calgary, AB, Canada
- 4Department of Medicine, University of Calgary, Calgary, AB, Canada
- 5The Calvin, Phoebe and Joan Snyder Institute for Chronic Diseases, University of Calgary, Calgary, AB, Canada
- 6Alberta Health Services, Calgary, AB, Canada
The timely detection of Methicillin-resistant Staphylococcus aureus (MRSA) is crucial for antimicrobial therapy and a key factor to limit the hospital spread of MRSA. Currently available commercial MRSA detection assays target the 3′ end of the orfX gene and the right extremity of Staphylococcal Cassette Chromosome mec (SCCmec). These assays suffer from both false positive due to SCC-like elements that lack mecA and false negative results due to the inability to detect new or variant SCCmec cassettes with the existing primers. We developed a novel MRSA detection scheme, designed to circumvent issues present in the existing commercial assays. Our assay demonstrated specificity and accuracy, capable of detecting prototypic strains of SCCmec types I-XIII [C(t) values ranged 8.58–26.29]. Previous false positive isolates (N = 19) by Xpert MRSA nasal assay were accurately classified with our assay. Further validation with 218 randomly selected clinical isolates (73 MRSA, 75 MSSA, 43 MR-CoNS, and 27 MS-CoNS) confirmed its feasibility and practicality. Testing assay performance with 88 direct clinical swabs from 33 patients showed that the assay was 96.6% in agreement with clinical culture results. Our novel MRSA detection assay targets both the S. aureus specific sequence and the mecA/mecC genes simultaneously to overcome the false positive and false negative deficits of currently available commercial assays. The results validate our assay and confirmed its feasibility and practicality. The assay is not affected by SCCmec types and only needs modification if new mec homologs emerge and establishes a new platform for other emerging SCCmec types.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) strains emerged shortly after clinical methicillin use (Barber, 1961; Jevons, 1961) through acquisition of the methicillin resistance gene mecA, which is carried on a mobile genetic element, staphylococcal cassette chromosome mec (SCCmec) (International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements [IWG-SCC], 2009). The SCCmec element is characterized by terminal repeat regions, two main genetic components (the ccr and the mec gene complexes), and three joining regions (J1–J3) which are located between the ccr and the mec gene complexes and repeat regions (Ito et al., 2001). Currently 13 SCCmec types have been described (I-XIII), based on the nature of the ccr and the mec gene complexes that they carry, and further classified into subtypes according to differences in their J region DNA (Lakhundi and Zhang, 2018). The J regions can be shared between SCCmec types, or can be unique to a specific type. Strains of MRSA have spread and become established as major nosocomial pathogens worldwide (Crossley et al., 1979; Panlilio et al., 1992; Voss et al., 1994; Ayliffe, 1997; Fluit et al., 2001), and more recently have emerged as a major cause of community-acquired infections (Vandenesch et al., 2003; Lindsay and Holdenm, 2004). Although 1st generation cephalosporins (e.g., cefazolin and cephalexin) and isoxazolyl penicillins (oxacillin, cloxacillin) are still the agents of choice for treatment of methicillin-susceptible S. aureus (MSSA) infection, MRSA is resistant to most β-lactam agents, including cephalosporins (with the exception of ceftibiprole and ceftaroline) and carbapenems. The timely detection of MRSA is crucial for optimal management of this pathogen and for limiting the nosocomial spread of this organism.
Early detection is complicated by the fact that in clinical samples S. aureus (SA) is often mixed with less pathogenic coagulase-negative staphylococci (CoNS), both of which can harbor mecA. Conventional methods for the detection of MRSA in clinical microbiology laboratories depend on growth of the organism with selective media, which is time consuming and requires 2–3 days. Nucleic acid amplification methods are also used to discriminate MRSA from MSSA and CoNS, however, traditional genetic identification methods require PCR amplification of a pure bacterial culture.
Currently, two FDA-approved commercially available PCR based assays are widely used to detect MRSA directly from clinical samples, including IDI-MRSA/GeneOhm MRSA (BD Diagnostics, Franklin Lakes, NJ, United States) and GeneXpert MRSA (Cepheid, Sunnyvale, CA, United States). Both assays are similar, using quantitative real-time PCR-based methods targeting the 3′ end of the orfX gene in SA, in conjunction with the J3 region at the right extremity of staphylococcal cassette chromosome mec (SCCmec), but not targeting mecA directly. However, both assays are problematic in that they produce false negative and false positive results. False negatives arise from the inability to detect new, variant and non-typeable SCCmec cassettes with the existing primers, and false positives from the presence of SCC-like elements that do not contain mecA, which are incorrectly amplified (Desjardins et al., 2006; Malhotra-Kumar et al., 2008; Kelley et al., 2009; Arbefeville et al., 2011; Stamper et al., 2011). Moreover, any emerging variant SCCmec new types will not be detected. In recent years these assays have been updated to improve sensitivities and specificities, and to include the mecA/C genes, such as with the BD Max MRSA XT, Xpert gen3, and BD Max StaphSR assays (Ellem et al., 2015; Lepainteur et al., 2015; Silbert et al., 2015, 2017; Becker et al., 2016; Mendes et al., 2016). They still, however, rely on detection at the SCCmec-orfX junction and could suffer from the lack of ability to detect emerging SCCmec variants.
Here we present a novel scheme for the direct detection of MRSA from clinical samples, which is designed to circumvent the false positive and negative issues of the currently available commercial assays. This novel 2-step assay uses a small number of primers that simultaneously target both the mecA/C and SA specific orfX genes and are able to accurately detect all MRSA tested to date, including SCCmec control types I-XIII.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains were grown overnight on Tryptic soy agar plates at 37°C. Control strains, including SCCmec I (NCTC10442), type II (N315), type IIIHg (85/2082), type IIIA (JCSC290), type IVa (CA05), type IVb (8/6-3P), type IVc (MR108), type IVd (JCSC4469), IVg (JCSC6673), IVh (JCSC6674), IVi (JCSC 6668), IVj (JCSC6670), type V (WIS [WBG8318]-JCSC3624), VII (JCSC6082), IX (JCSC6943), X (JCSC6945), and XI (LGA251) were obtained from K. Hiramatsu and T. Ito at the Juntendo University in Tokyo, Japan. SCCmec IIA (AR14/0298), IIB (AR05/0.1345), IIC (AR14/2188), IID (AR13/3635.2), IIE (AR13/3330.2), IVE (AR43/3330.1), and IVF (AR43/3636.1) were obtained from D. Coleman at the University of Dublin, Ireland. SCCmec VI (HDE288) was obtained from H. de Lencastre at The Rockefeller University, New York, United States. SCCmec XIII was obtained from M. Stegger at the Statens Serum Institut in Copenhagen, Denmark. SCCmec IIb (05MS-150) and VIII (C10682) were recovered from patients in our local hospitals or clinics. Strains Iowa 1-20 were obtained from S. Richter, University of Iowa Health Care, Iowa, United States (Arbefeville et al., 2011).
DNA Extraction
DNA was extracted by rapid boiling method as previously described (Zhang et al., 2004, 2008). Alternatively, purified DNA was extracted using the QIAamp DNA mini kit (Qiagen Inc., Toronto, ON, Canada) as per the recommended protocol, with elution in 50 μl of sterile distilled water.
Primer Design
Primers and probes for the MRSA detection real-time PCR assay were designed following comprehensive analysis of the orfX and mecA genes, in both CoNS and SA. Primers and their sequences are listed in Table 1.
Long Range PCR Conditions
Round one LR-PCR used primers SA-3e, Umec-F and Umec-FX (Table 1). DNA template was derived from either crude boiled extracts, or from Qiagen kit extraction as described. One μl of template DNA was added to 24 μl of reaction mixture containing 0.2 mM of each primer, 1X LA PCR buffer II (TaKaRa Bio inc), 0.4 mM of each dNTP (TaKaRa Bio inc), and 1 unit of LA Taq HS (TaKaRa Bio inc). Amplification was performed in a 2720 thermal cycler (Applied Biosystems, Foster City, CA, United States) using 15 cycles of 98°C for 10 s, 58°C for 15 s, and 68°C for 20 min, followed by holding at 4°C.
Magnetic Bead Capture and PCR Product Washing
Ten μl of 10 mg/ml streptavidin coated magnetic beads (Dynabeads M-270 streptavidin; Invitrogen) was used for each LR-PCR reaction. Beads were pooled in a 1.5 ml microcentrifuge tube and washed twice in 1 ml of 1X binding and washing buffer containing 5 mM Tris-HCL, pH7.5, 0.5 mM EDTA, and 1 M NaCl, with separation from the buffer accomplished with a DynaMag-2 (Invitrogen, Oslo, Norway). Following the washes, beads were re-suspended in 20 μl of 2X binding and washing buffer (10 mM Tris-HCL, pH7.5, 1 mM EDTA, 2 M NaCl) per reaction, and 20 μl of suspension added to each LR-PCR tube. Tubes were incubated at room temperature for 45 min, with gentle mixing by inversion every 5 min. Following binding, magnetic beads were washed two times with 100 μl of 1X binding and washing buffer, with mixing and separation accomplished using a DynaMag-96 Side (Invitrogen, Oslo, Norway). The samples were re-suspended in 100 μl of 10 mM Tris buffer, then 50 μl transferred to each of 2 real-time PCR tubes. Using the magnet, buffer was removed from the tubes, leaving the DNA bound beads as template for the second round real-time PCR.
Round 2 Real-Time PCR Conditions
Round 2 real-time PCR was set up as 2 reactions for each sample. Detection of the SA specific orfX gene was accomplished with the first reaction, using primers Arb3 and SA-4. The 5′ PrimeTime qPCR 5′ nuclease probe, SA-HEX-1, was used for PCR product detection (Integrated DNA Technologies, Skokie, IL, United States) (Table 1). Detection of the mecA/C gene was accomplished with the second reaction, using primers Arb2 along with primers Umec-R and Umec-RX. The 5′ PrimeTime qPCR 5′ nuclease probes mec-FAM-2, mec-FAM-3 and mec-FAM-4 were used for PCR product detection in this reaction (Integrated DNA technologies, Skokie, IL, United States) (Table 1). The reactions were made using 1 mM of each of the appropriate primers, along with 0.25 mM of each corresponding probe and 1X PrimeTime gene expression master mix (Integrated DNA technologies, Skokie, Illinois), in a final volume of 10 μl. Ten μl of the orfX mix was added to one of the tubes containing washed beads, while 10 μl of the mecA mix was added to the second tube of washed beads. Both reactions were run simultaneously in the same real-time PCR thermocycler with the following conditions: an initial incubation at 95°C for 2 min was followed by 41 cycles of 95°C for 10 s and 65°C for 20 s, with at HEX read for the tubes containing the orfX mix, or a FAM read for tubes containing the mecA mix. Reactions were considered positive when the C(t) value was between 1-38, and negative if there was no C(t) value, or if it was greater than 38.
Assay Sensitivity, Specificity and Validation
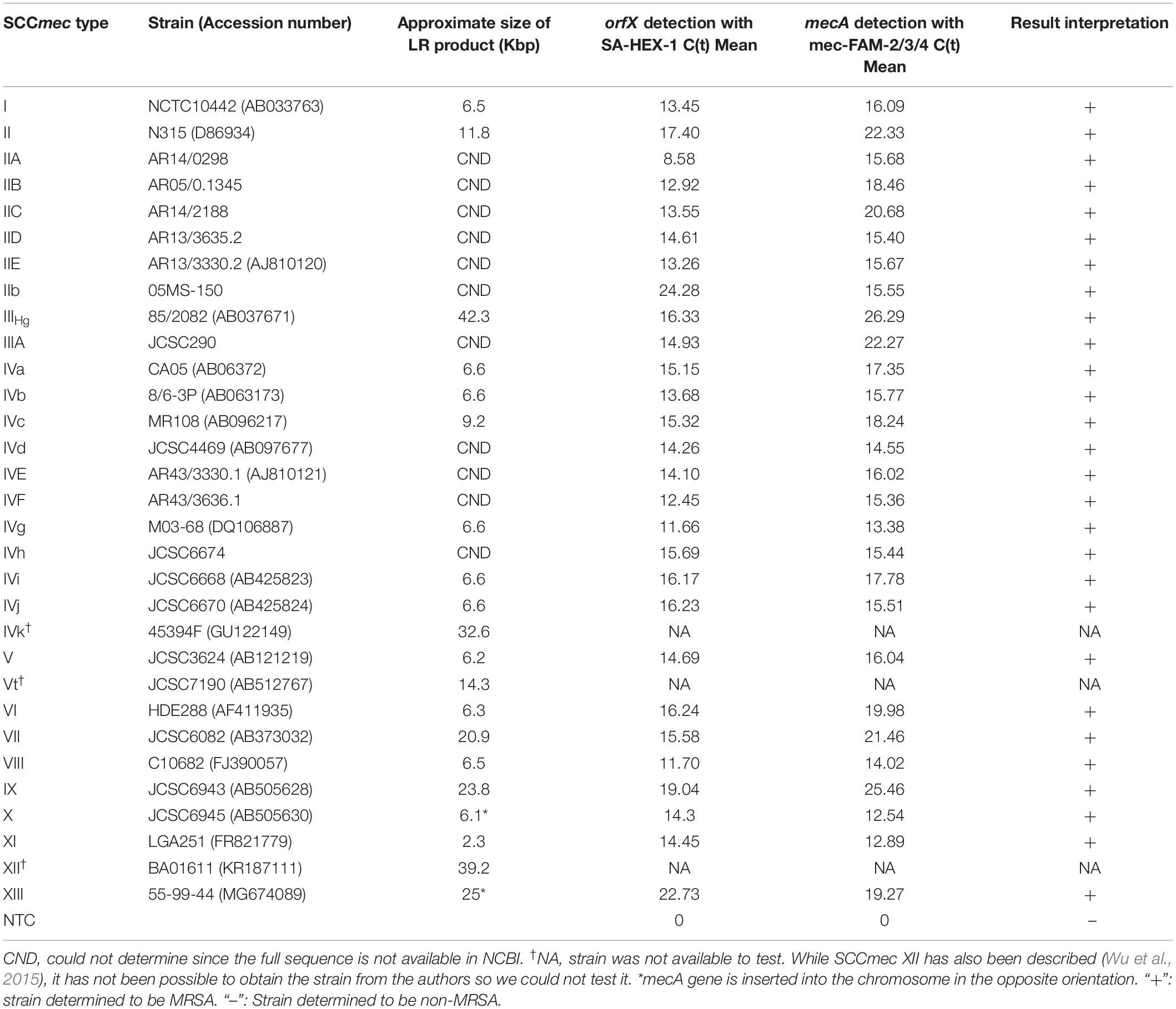
The ability of our MRSA detection assay to detect a large variety of SCCmec types was assessed using SCCmec control strains, including types I, II, IIA, IIB, IIC, IID, IIE, IIb, IIIHg, IIIa, IVa, IVb, IVc, IVd, IVE, IVF, IVg, IVh, IVi, IVj, V, VI, VII, VIII, IX, X, XI, and XIII (Table 2). Assay cross-reactivity was determined using a collection of 40 non-MRSA, including 30 non-staphylococcal strains (Table 3). One μl of Qiagen purified DNA was used as template.
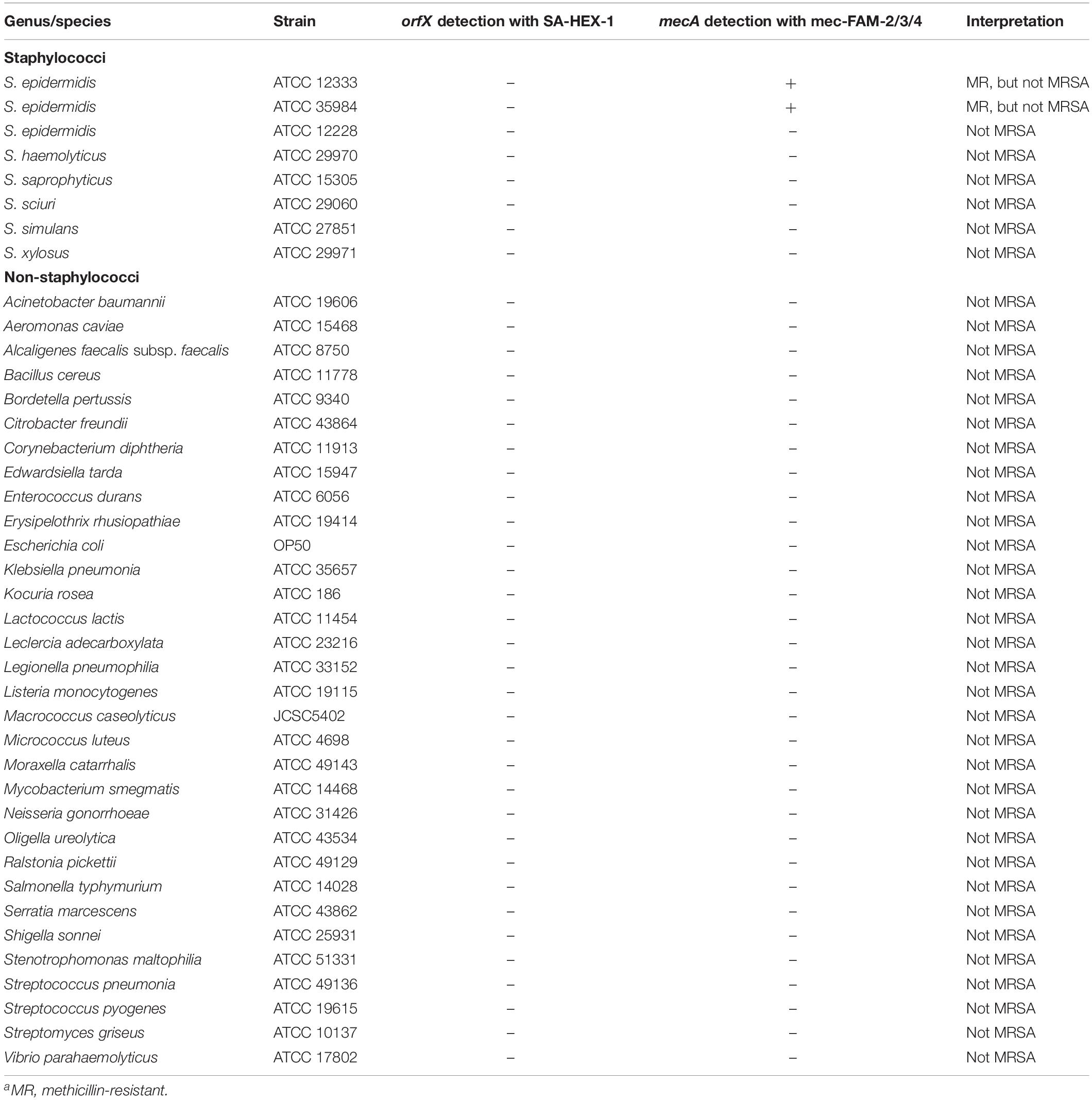
Table 3. Assay specificity in various strains of coagulase-negative staphylococci and non-staphylococcal bacteria.
To determine assay sensitivity, the correlation of 1.0 McFarland standard being equal to 3 × 108CFU/ml was initially used. Control strains carrying SCCmec II, III, IVa, IX, X, and XI were suspended in saline to a concentration of 1.0 Mcfarland standard. Exact colony counts were further confirmed by plating dilutions of the suspension on TSA plates, and growing overnight at 37°C. One ml of suspension was pelleted and the DNA extracted following standard procedures using the QIAamp DNA mini kit, and eluted with 50 μl of sdH2O. DNA concentration of the extract was determined with a Qubit 3 fluorometer using the Qubit dsDNA BR kit (Invitrogen). Ten fold serial dilution of the DNA were subsequently used to assess assay sensitivity.
The ability of the assay to correctly classify isolates falsely identified as MRSA by the Xpert MRSA nasal assay (Arbefeville et al., 2011) was determined. One μl of boiled DNA from 20 MSSA strains (Iowa 1–20) was tested. The assay was subsequently validated using a collection of strains that were obtained from random clinical samples that had been extensively characterized molecularly (Zhang et al., 2004, 2008). DNA was extracted using the boiling method and 1 μl used in the assay.
Clinical Applicability Using Direct Patient Swabs
The ability of the assay to detect MRSA directly from patient samples was assessed with 88 swabs collected from hospitalized patients previously known to be MRSA positive or from a convenience sample of patients attending the sexually transmitted infection clinic in our health region. Duplicate swabs were collected from multiple sites on each patient appropriate to the clinical setting [nasal (N), throat (T), perianal-perineal (K), groin (G), Z-swab (Z), wound (W), axilla (A), or vaginal (V) swab]. One swab was subjected to routine clinical culture and identification, followed by genetic characterization of each isolate with a PCR multiplex assay capable of distinguishing CoNS from SA and methicillin-resistant vs. methicillin-susceptible isolates (Zhang et al., 2008). Isolates were also subjected to SCCmec typing (Zhang et al., 2012) as previously described. DNA was isolated from the second swab using the QIAmp DNA minikit (Qiagen Inc., Toronto, ON, Canada) following protocol D for isolation of genomic DNA from gram-positive bacteria, using 200 μg/ml of lysostaphin. The entire swab was deposited into the microcentrifuge tube for extraction purposes, and DNA was eluted from the column with 50 μl of sterile water. One μl of the eluent was used as the template in real-time PCR assay. When PCR was found to be positive and clinical culture negative, the swab used for clinical culture was placed into 30 ml of Tryptic soy broth and grown overnight at 37°C. Fifty μl of the overnight culture was transferred to a blood plate and subjected to clinical identification and molecular typing, as described above.
Results
A Novel Scheme for Direct MRSA Detection Designed to Overcome the Deficits of Commercially Available Assays
Currently available commercial MRSA detection assays rely on quantitative real-time PCR-based methods targeting the 3′ end of the orfX gene along with the right extremity of SCCmec, but not directly targeting the mecA gene. Both, however, suffer from the detection of false positives (due to SCC-like elements that lack mecA) and false negatives (due to the inability to detect new, variant or non-typeable SCCmec cassettes with the existing primers). We developed a novel MRSA detection assay scheme, designed to overcome issues present in the existing commercial assays.
Our novel assay is comprised of 2 PCR steps (Figure 1), including an initial long range (LR-) PCR reaction (Round 1), the product of which acts as the template for the second round real-time PCR reactions (Round 2). Forward and reverse primers for LR-PCR are located in the orfX and mecA/C genes, creating a PCR product that ranges in size from 2.3 to 42.3 kb (Tables 1, 2). By simultaneously targeting both of these regions, the assay was designed to eliminate templates that do not contain the mecA gene, as well as anything that is not SA. Primer Umec-F targets a conserved sequence in the mecA/mecC gene, present in all SCCmec types except types X and XIII. Since the orientation of mecA in SCCmec X and XIII is reversed compared to the other types, primer Umec-FX was included to effectively detect those types, as well as any future SCCmec types with the reversed mecA orientation. Primer SA-3e targets a region that is specific to the SA orfX gene and is universally found in all SA with available sequences. The target sequence differs significantly enough from that found in CoNS, thereby preventing amplification from CoNS orfX genes. Primer SA-3e also carries a 5′ biotin label, allowing the LR-PCR products to be captured with streptavidin coated magnetic beads, thereby concentrating them and purifying them of residual LR-PCR reaction components. Magnetic beads containing captured LR-PCR product from Round 1 were added directly to the round 2 real-time PCR reaction mixtures as template. All 3 LR-PCR primers were also designed with a 20 or 21 bp tail sequence on the 5′ end that do not match any bacterial genomes published thus far (Table 1, underlined sequence). These right (orfX side) and left (mecA side) tail sequences act as templates for round 2 PCR primers, eliminating the possibility of amplification directly from any contaminating chromosomal DNA in the Round 2 reactions.
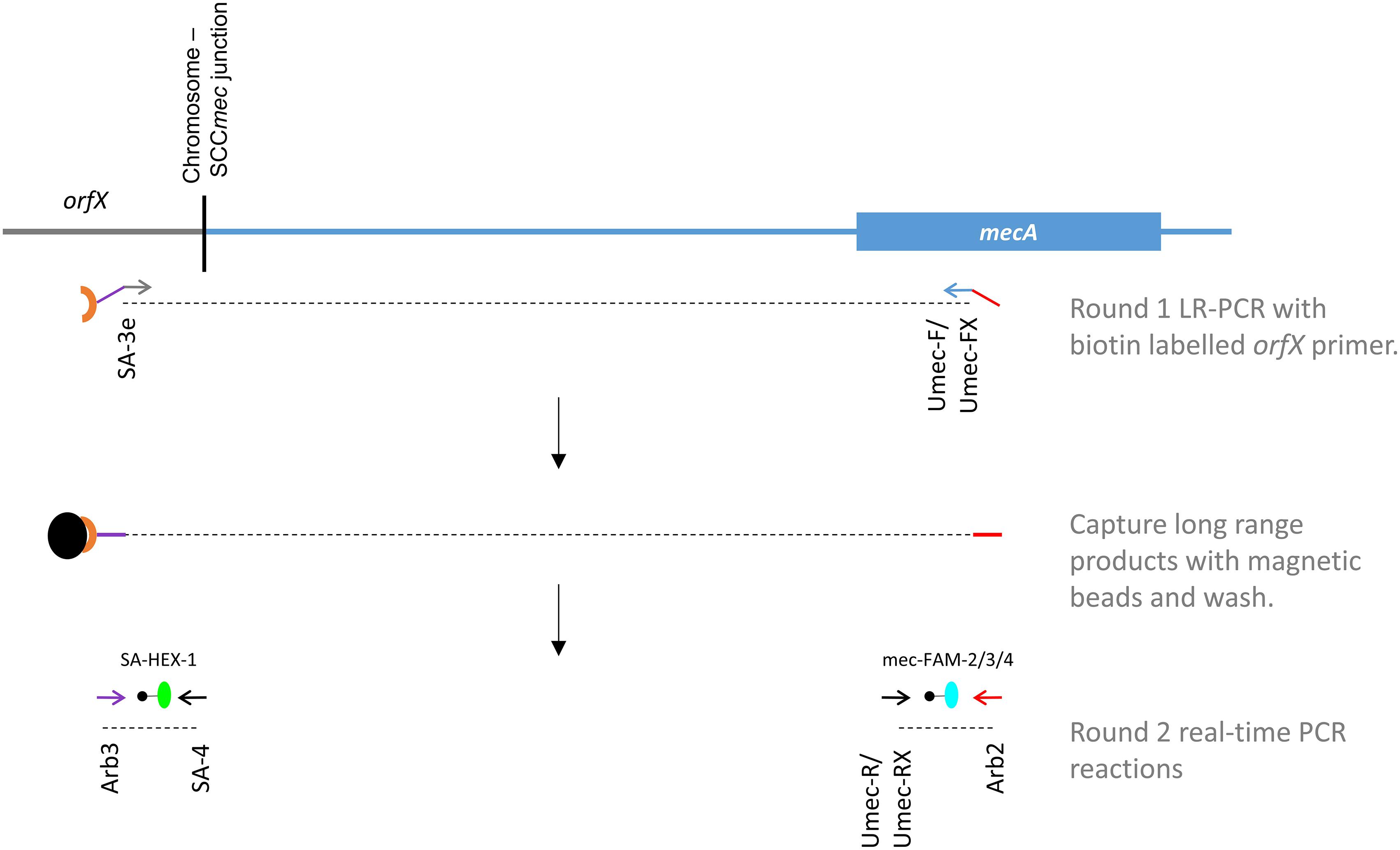
Figure 1. A new scheme for the MRSA directly clinical sample detection, designed to circumvent deficits in commercially available assays. Primers for the initial long range (LR) PCR are located in the orfX and mecA genes (green and blue arrows respectively) and contain tail sequences (indicated by purple and red lines) that are unique, and function as templates for Round 2 primers (indicated by purple and red arrows). Primer SA-3e is biotin labeled (brown crescent), allowing the LR-PCR product to be captured by streptavidin coated magnetic beads (black orb), and concentrated/purified. The Round 2 real-time PCR reactions are detected with probes specific to SA’s orfX (green orb) and mecA (blue orb).
The second round real-time PCR was designed as 2 separate reactions; one to detect the orfX gene, and the other to detect the mecA gene. For the orfX reaction, amplification primer SA-4 is specific to a region in the SA orfX gene, while primer Arb3 is specific to the right side- tail of primer SA-3e. Probe SA-HEX-1, also specific to a region in the SA orfX gene, was used to detect the PCR product specific for SA. For the mecA reaction, primer Arb2 is specific to the left side tail of UmecF/UmecFX, while primer UmecR is specific to the mecA gene in the typical orientation, and Umec-RX is specific to the mecA gene in the reversed orientation. Because of slight sequence variations in the mecA gene sequence, probes mec-FAM-2 and mec-FAM-3 were both needed to detect the PCR products, and mec-FAM-4 needed for detecting the product from SCCmec X and XIII.
Many layers of specificity were built into the assay by careful selection of the target sequences for the long-range primers, round 2 primers and probes. However, due to the erroneous nature of long-range PCR, whereby short PCR products can be generated from one correctly annealing and one mis-priming primer, we determined that it was essential to detect from both the mecA and orfX sides. While these mis-primed products are too few to detect on a traditional gel, they become amplified in the Round 2 reactions and create signals in one reaction or the other. In MSSA we noted that there was a positive signal from the orfX reaction, while in MR-CoNS we noted a positive signal from the mecA reaction. In each case, however, the signal from the other reaction was negative, allowing us to discriminate between these strains and true MRSA, which are the only ones positive for both the mecA and orfX reactions.
A diagram of the assay steps and approximate timelines with the detailed explanation is outlined in Figure 2.

Figure 2. New MRSA detection assay timeline. The assay steps and approximate timelines are diagramed with the detailed explanation in the right side notes. Automation in a closed system could decrease assay time, minimize the numbers of manual procedures required, and reduce or eliminate the possibility of contamination.
New Assay Detecting SCCmec Types I-XIII
The ability of the assay to detect a wide variety of SCCmec types was assessed with 28 types or subtypes, including SCCmec I-XIII. When the set of SCCmec types was tested, all 28 types and subtypes were found to be positive for both orfX and mecA reactions and, consequently, positively identified as MRSA with our assay (Table 2). Corresponding real-time PCR curves for each of the round 2 amplicons are shown in Figures 3A,B. C(t) values for the orfX reaction ranged from 8.58 (type IIA) to 24.28 (type IIb), while C(t) values for the mecA reaction ranged from 12.54 (type X) to 26.29 (type IIIHg). During initial assay development, LR-PCR products from representative SCCmec types I-XI were run on a 0.7% agarose gel to visualize them (Figure 4A). Anticipated bands were not seen for any SCCmec type, however, with the small number of cycles being used, this result was not unexpected. Multiple bands were seen for some types (such as in type I), while in other types a smear was seen (such as type II and VII–XI). As with the LR-PCR products, second round real-time PCR reaction products were also run on a 2% agarose gel during assay development (Figure 4B). With the orfX reaction, the anticipated band was seen strongly at 196 bp for all representative SCCmec types, along with weaker bands that were larger in size. For the mecA reaction, multiple bands were seen for most SCCmec types, with the expected one at 213 bp being weak (or equivalent to the other ones) (data not shown). The notable exception was in SCCmec X, where the expected band at 179 bp was the only one visible. Despite the non-ideal results seen with these gels, the small number of full-length amplicons generated during LR-PCR were sufficient to act to act as templates for the second round reactions, with the product correctly detected by virtue of the probe specificity (Figure 3).
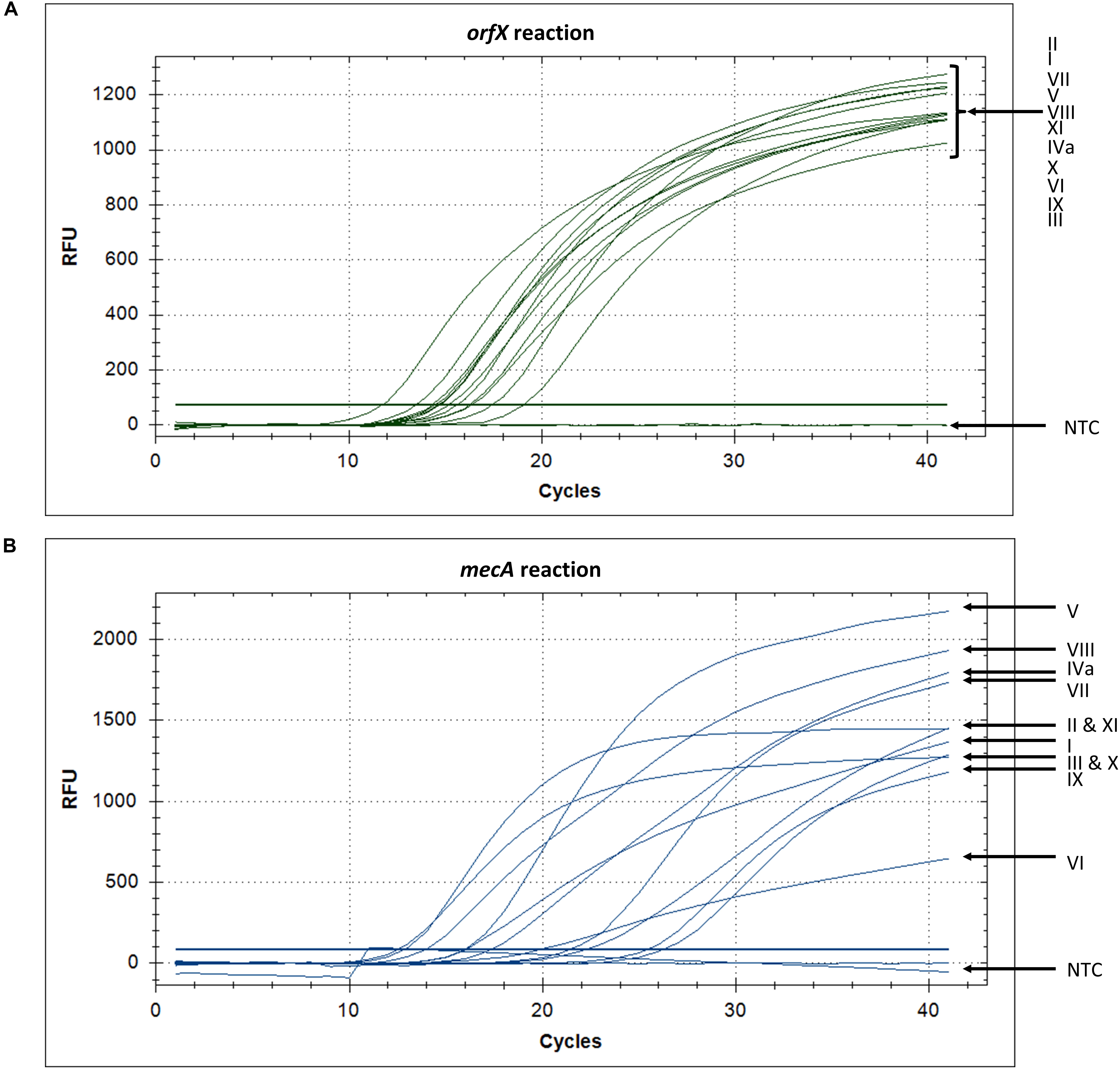
Figure 3. orfX and mecA reaction amplification curves show that SCCmec types I–XI are detected. (A) Detection of SCCmec types with the orfX reaction, using fluorophore HEX, shows that all types pass the threshold and have C(t) values between cycles 11 and 19, while the no template control (NTC) remains negative below the threshold. (B) Detection of SCCmec types with the mecA reaction, using fluorophore FAM, shows that all types pass the threshold and have C(t) values between cycles 13 and 27, while the no template control (NTC) remains negative below the threshold.
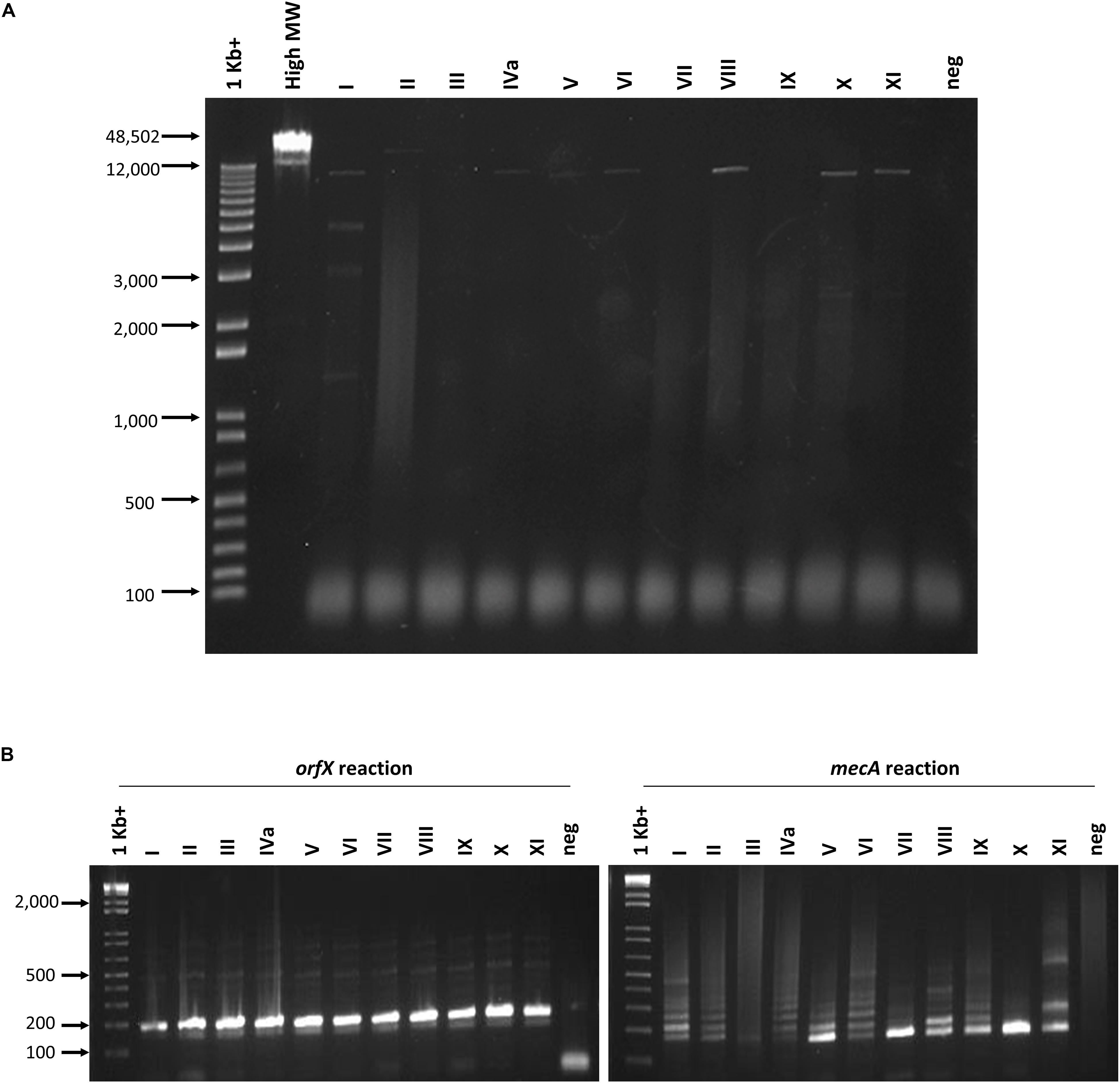
Figure 4. Agarose gel electrophoresis of round 1 and round 2 PCR reactions. (A) No PCR products corresponding to the correct sized product were seen following 10 cycles of long range amplification. Lanes from left to right correspond to the following samples: 1 Kb + marker (Invitrogen), High molecular weight marker (Invitrogen), SCCmec I–XI, no template negative control. (B) Round 2 orfX and mecA reactions show the correct sized amplicon in the orfX reaction, while most types produce weak and multiple bands in the mecA reaction. Lanes from left to right in each gel correspond to the following samples: 1 Kb + marker (Invitrogen), SCCmec I-XI, no template negative control.
Specificity of the Assay
Assay specificity was assessed using a panel of 40 non-MRSA, including 32 non-Staphylococci strains. In total there were 6 CoNS species (including 3 strains of S. epidermidis), and 31 strains from other diverse genera (including 2 species of Streptococcus) (Table 3). All of the isolates, including the 7 CoNS species, were negative with the orfX reaction, and all but 2 of the isolates were negative with the mecA reaction. This indicates that the assay does not cross react with other species and is very specific to MRSA. The noted exceptions were ATCC29971 (S. xylosus) and ATCC35984 (S. epidermidis), both of which are MR-CoNS and, consequently, positive for the mecA target.
Sensitivity of the Assay
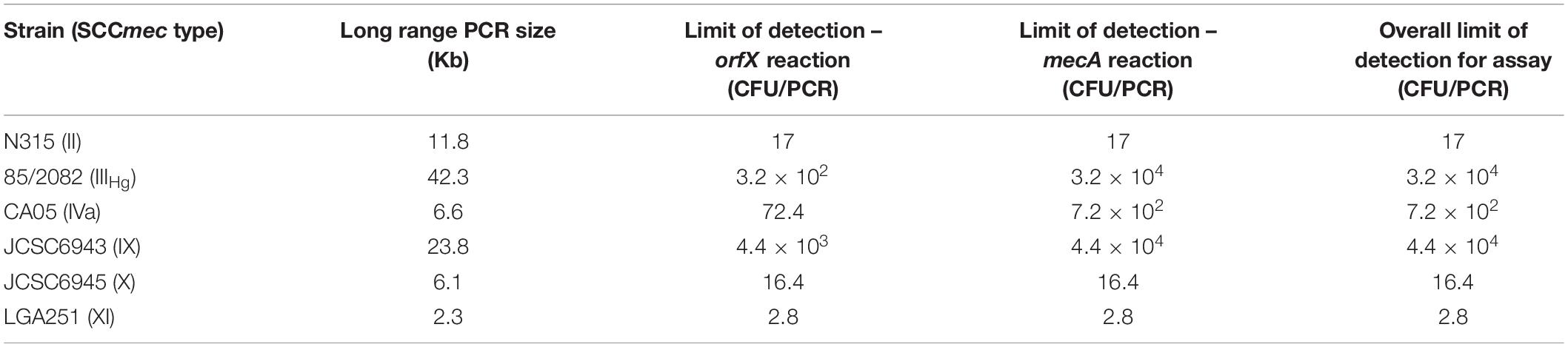
MRSA assay sensitivity will be impacted by the length of the long range PCR product (with longer products amplified with lower efficiency) as well as the efficiency of interaction of the specific primer pair in relation to the template DNA. As such, assay sensitivity was examined with 6 representative SCCmec types, covering a range of orfX-mecA interval sizes, both mecA and mecC genes, as well as the reversed mecA of type X. Under standard conditions, the limit of detection for the orfX reaction was 2.8–4.4 × 103 CFU/PCR, and the limit of detection for the mecA reaction was 2.8–4.4 × 104 CFU/PCR. As both targets need to be positive for a sample to be considered MRSA positive, the overall limit of detection for the assay is limited by the least sensitive reaction (the mecA reaction in most cases) (Table 4).
Previous False Positive Isolates Were Accurately Classified With Our MRSA Detection Assay
Strains Iowa 1–20, previously identified as false MRSA positives with the Xpert MRSA assay (Arbefeville et al., 2011), were tested with our MRSA detection assay and all 20 were found to be positive for the orfX reaction, while all but one were negative for the mecA reaction. Iowa-16 was positive for both the orfX and mecA reactions and classified as MRSA, which is in agreement with the Arbefeville study (Arbefeville et al., 2011).
Assay Validation With Random Clinical Isolates
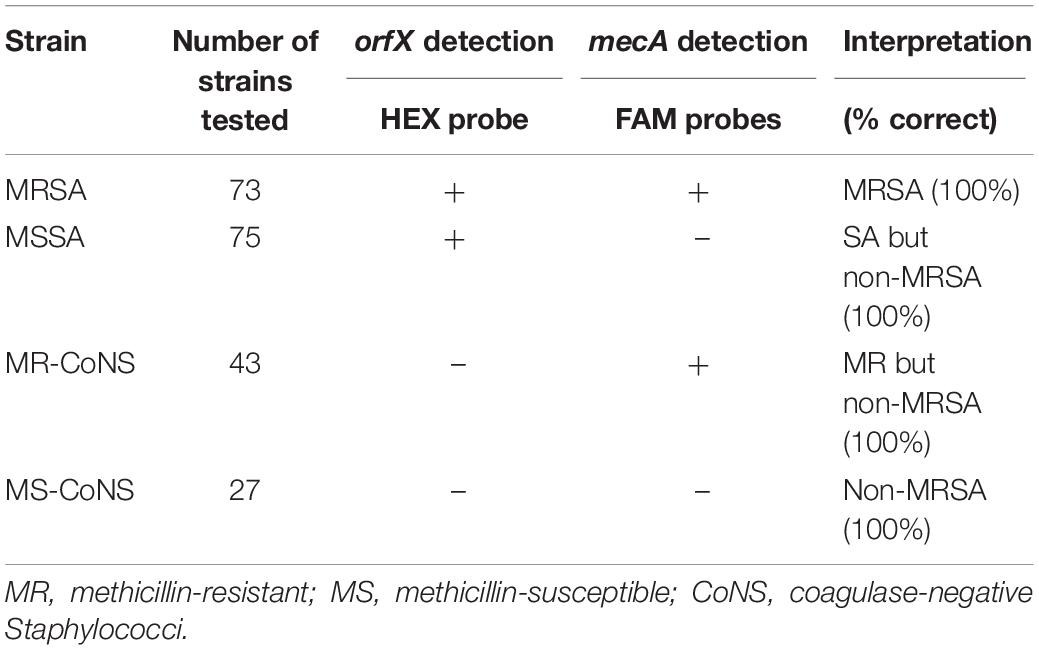
Validation of our assay was done using a collection of 218 randomly selected clinical isolates that had previously undergone extensive genetic characterization. Among them were 73 MRSA, 75 MSSA, 43 MR-CoNS, and 27 MS-CoNS isolates (Table 5). Of the 73 MRSA, 41 strains were previously determined to be SCCmec II, 10 were type IIIHg, 3 were type IVa, 1 was type IVb and 18 carried an unknown SCCmec type. All 73 MRSA were positive in both the mecA and orfX reactions, with the orfX reaction having a lower C(t) than the mecA reaction [approximately 10 C(t) values lower (data not shown)]. All 75 MSSA had positive C(t) value for the orfX reaction but remained negative for the mecA reaction. Similarly, all 43 MR-CoNS had a positive C(t) value for the mecA reaction but remained negative for the orfX reaction. All 27 MS-CoNS were negative for both the orfX and mecA reactions. Table 5 summarizes results for the 218 clinical isolates, showing that the assay is capable of correctly identifying MRSA with 100% accuracy.
Assay Applicability Determined With Direct Clinical Samples
A total of 42 samples were obtained from patients attending the sexually transmitted clinic, representing 14 patients with sampling from 3 different body sites for each patient. As seen in Table 6, none of the samples from the clinic were positive for MRSA using the real-time PCR assay, and no MRSA was isolated by routine clinical culture, meaning there was 100% agreement between the two methods. An additional 46 samples were obtained from hospital inpatients previously known to be MRSA positive, representing 19 patients with 2–3 body sites sampled for each patient. As shown in Table 7, there was a high degree of concordance between the real-time PCR assay results and clinical culture results. By clinical culture and PCR testing 23 and 25 patients were found to be MRSA positive, respectively, with 95.5% (84/88) agreement from the swabs. The PCR assay was able to detect low and high levels of MRSA on the swabs, ranging from 2 to several 100 colonies present on the plate. The results for a total of four swabs differed between the two assays. One swab was PCR negative, but 5–10 MRSA colonies were detected on the culture plate. One swab was PCR positive but plate culture negative, however, following overnight incubation of the swab in Tryptic soy broth (TSB), MRSA was detected in culture. Finally, two swabs were PCR positive but culture negative, and no MRSA was detected even after overnight incubation of the swabs in TSB broth. SCCmec typing of the MRSA that were isolated by clinical culture revealed that the majority of them (20/27) belonged to SCCmec IVa, 1/27 to SCCmec IVc, 2/27 to SCCmec II and 2/27 to SCCmec V. Two could not be determined because they were not isolated on plate culture, leaving no strain to type.
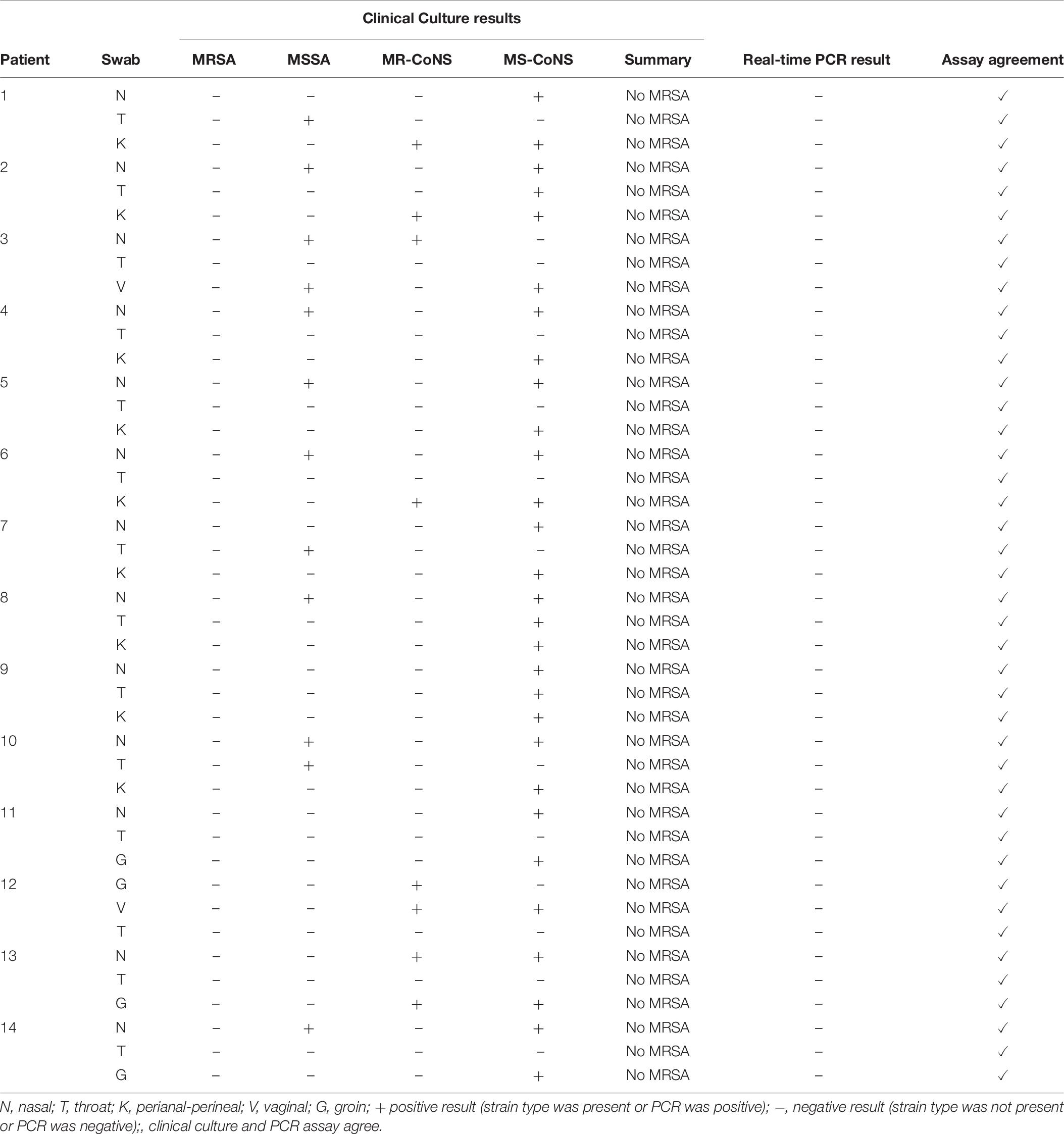
Table 6. Assay applicability assessed with swabs from random patients attending an outpatient clinic.
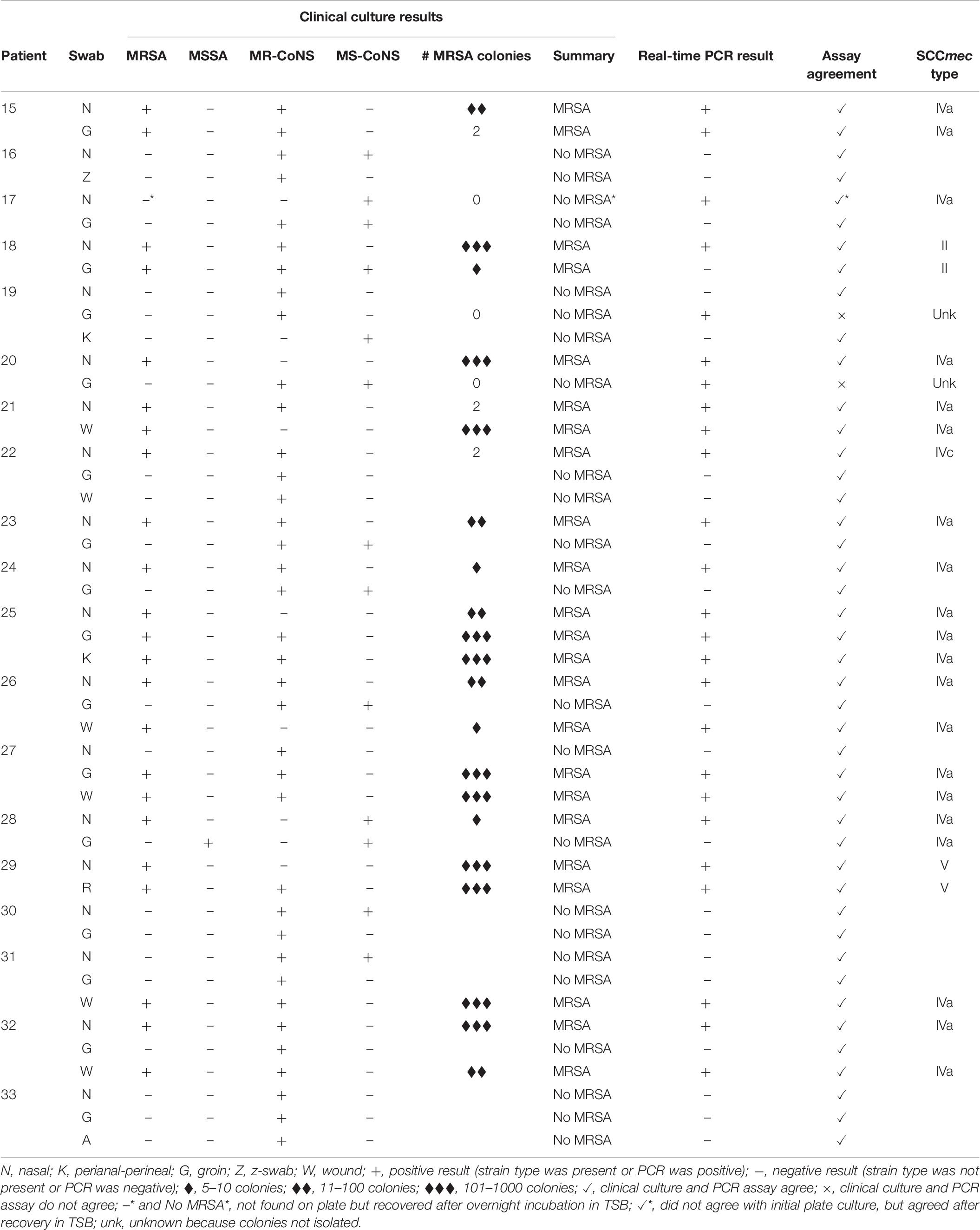
Table 7. Assay applicability assessed with swabs from hospital inpatients previously known to be MRSA positive.
Discussion
Molecular detection methods have been developed but are hampered by the fact that clinical samples can be mixed, containing combinations of MSSA, MRSA, and MS- or MR-CoNS. Detection of the mecA gene does not necessarily indicate the presence of MRSA, as MR-CoNS could also be the source of the gene.
Two FDA-approved commercially available PCR based assays that have been described and are widely used to detect MRSA directly from clinical samples, include the IDI-MRSA/GeneOhm MRSA (BD Diagnostics) and GeneXpert MRSA (Cepheid). In the IDI-MRSA assay, there is a SA specific primer (Xsau325) in the orfX gene, immediately upstream of attB, along with a SA specific probes which gives species specificity. The other five SCCmec specific primers (mecii574, meciii519, meciv511, mecv492, mecvii512) are in the J3 region of SCCmec, and are used along with Xsau325 to amplify the junction region of MRSA (Huletsky et al., 2004). The GeneXpert MRSA assay is similarly designed, with primers spanning the chromosome orfX-SCCmec junction, with amplification and detection occurring in separate chambers of single-use disposable cartridge. Multiple studies, however, have found that both assays are problematic in that they produce false negatives from their inability to detect variant or new SCCmec types, and false positives from their detection of SCC-like elements that do not contain mecA primers (Desjardins et al., 2006; Malhotra-Kumar et al., 2008; Kelley et al., 2009; Arbefeville et al., 2011; Stamper et al., 2011). A study by Stamper et al. found that the GeneOhm MRSA assay had a sensitivity and specificity of 89 and 91.7% respectively, with false positives resulting from retained segments of the right extremity-junction sequences in non-mecA containing SCC-like elements (Stamper et al., 2011). Likewise, a study by Arbefeville et al. (2011) found that the rate of false positives for the Xpert MRSA assay was 7.7%, due to mecA dropout strains with remnants of SCCmec cassettes. Both assays are also unable to detect the newly emerging variant SCCmec types.
In 2015, the Xpert MRSA was updated to include targets for the mecA and mecC genes, in conjuction with the existing SCCmec-orfX junction (Becker et al., 2016). The new assay, GeneXpert MRSA Gen 3, was also updated to detect SCCmec VI–XI, on top of the I–V that it previously detected. A study by Lepainteur et al. (2015) detecting MRSA directly from nasal swabs showed that the assay had a sensitivity of 95.7% and a specificity of 100%. The BD Max MRSA XT (extended detection technology) kit was also introduced to detect mecC as well as mecA and, in the same study by Lepainteur, was shown to have a sensitivity and specificity of 87.5 and 97.1% respectively (Lepainteur et al., 2015). BD also released the BD Max StaphSR assay, which is a multiplex assay targeting the SCCmec-orfX junction, nuc and mecA/C genes, with a sensitivity of 99.1–100% and specificity of 100% when detecting from blood culture (Ellem et al., 2015). Further evaluation of the BD Max Staph SR and Max MRSA XT assays showed sensitivities of 94.3–99.1% and specificities of 97.7–99.8% (Silbert et al., 2015, 2017; Mendes et al., 2016). These assays, however, all target the SCCmec-orfX junction, and could be limited in their ability to detect newly emerging SCCmec types and variants.
Another FDA-approved assay, the cobas® vivoDX MRSA test was announced in December 2019. The assay is a bioluminescence-based method that uses bacteriophage technology to detect MRSA in nasal swabs in as little as 5 h. While limited information is currently available about the assay (no publications available at present), they do report in media reports that the assay has a false negative rate of approximately 10%, and a false positive rate of 1.4%.
With the goal of improving point of care detection, we developed a new real-time PCR assay for the detection of MRSA in clinical samples that circumvents the major problems encountered with commercially available assays. Our assay simultaneously targets both the chromosomal orfX region and the mecA and has the advantage of only needing a small number of primers to detect a large range of SCCmec types. Because of the highly homologous nature of both the orfX and mecA genes, these limited primers are capable of detecting all SCCmec types tested to date, including SCCmec XI, which carries the new mecC homolog as well as SCCmec X and XIII, with mecA in the reversed orientation.
With the assay, we were able to correctly detect 28 SCCmec types and subtypes, ranging from SCCmec I to XIII. The high degree of assay specificity was further demonstrated by testing 40 non-MRSA isolates including 32 non-Staphylococcal species, and 218 clinical isolates that had previously undergone extensive genetic testing.
Applicability of the assay was assessed using clinical swabs from inpatients previously known to be MRSA positive, as well as from random patient samples from a clinic in our health region. Results indicate that the assay has a high level of sensitivity and accuracy. The one culture positive-PCR negative result could represent a false negative because the MRSA strain carried SCCmec II, which has a larger orfX-mecA size. However, the type II control strain had a low experimental limit of detection of 17 cells/reaction. It’s also possible that the discrepancy and false negative resulted from sampling error. The finding of 3 PCR positive and culture negative swabs suggests that the sensitivity of the real-time PCR assay is superior to that of culture, although as mentioned, sampling of the duplicate swabs could potentially account for the discrepancies, particularly if low levels of bacteria were present, illustrated by the one broth enriched positive culture. The differences could also be due to the presence of non-viable MRSA on the swabs. All three patients were on antibiotic therapy prior to the samples being collected. Assuming that the three swabs were in fact positive for MRSA, the real-time PCR assay correctly identified 87/88 (98.9%) of the swabs, while clinical culture correctly identified 85/88 (96.6%). An interesting observation was that the real-time PCR assay was able to detect MRSA with higher sensitivity than the experimental limits of detection suggested. Fourteen of the 25 swabs that were positive for MRSA had ≤100 colonies on the culture plates, with 9 of those having ≤ 10.
This novel MRSA detection assay has successfully overcome the issue of false negatives from new or novel SCCmec types, and false positives from non-mecA containing SCC-like elements (Desjardins et al., 2006; Malhotra-Kumar et al., 2008; Kelley et al., 2009; Arbefeville et al., 2011; Stamper et al., 2011). False negatives are eliminated due to the universal nature of the targets and primers chosen for the long-range PCR. By amplifying from these universal regions, rather than targeting sequences specific to each SCCmec type, this assay has been shown to detect the majority of SCCmec types and subtypes described to date (types IVk and XII were not available) and should, in theory, be effective in detecting newly discovered types. In fact, of the 73 MRSA clinical samples that were used to validate the assay, 18 of them were untypeable using previously described SCCmec typing assays (McClure et al., 2010; Zhang et al., 2012). Despite carrying an unknown SCCmec type, the assay was able to accurately classify them as MRSA. The second deficit of previously described MRSA detection assays was the issue of false positives, but with our design, and the simultaneous targeting of both mecA and orfX during the long-range PCR, these false positives have been eliminated. While the Arbefeville et al. (2011) study did not have a clear explanation for the false reactivity in the Xpert MRSA assay, they postulated that it could be due to the presence of SCCmec-orfX right extremity components. Screening these isolates with our assay correctly classified 19 of the strains as MSSA, and one as MRSA. SCC-like elements present in CoNS are also negative for both targets with our assay, eliminating this potential source of false positives. Of the 27 MS-CoNS clinical isolates that were tested, 3 of them were determined to have a non-mecA SCC element in previous studies (data not shown), and all remained negative for both targets with this assay. ATCC12228 is also described as carrying a non-mecA SCC-like element, known as SCCpbp–4 (Mongkolrattanothai et al., 2004), and was negative for both targets with our assay.
In its current form, our novel assay does have some limitations. At present, 15 cycles of long-range PCR are used in the first round of amplification, which require 5.5 h to run. When factoring in sample preparation, handling time, magnetic bead capture/washing and the second round of PCR, the overall assay requires approximately 8–9 h (Figure 2). While this time frame is longer than the rapid turn-around times available with the 2 FDA approved assays, it is significantly shorter than the time required for traditional microbiology culture methods. Automation is an option for reducing assay time, as is decreasing the number of cycles and/or extension time of the long-range PCR. Initial attempts have been made to reduce the extension time to 10 min instead of 20 min and have shown promising results in both control strains and in clinical samples, which could potentially reduce the overall time from originally 8–9 h to about 6 h (Figure 2). Another limitation of the assay is the number of manual steps required, each of which increases the chances of contamination. Once again, automation in a closed system is a viable option for mitigating this problem. Currently, the assay clinical applicability was validated using the direct patient swab examples collected from different body sites including nasal, throat, perianal-perineal, groin, Z-swab, wound, axilla, and vaginal swabs. The possibility of application of the assay in other direct examples such as pus, wound, sputum and etc., should be assessed in future.
Conclusion
Our MRSA detection real-time PCR assay has proven to be an effective method of identifying MRSA directly in clinical samples. Due to its design, in which both the orfX and mecA/C genes are simultaneously targeted, false positive and false negatives observed with other clinically approved point of care detection assays are eliminated. The assay can detect all SCCmec types tested to date and, in theory, should be effective at detecting most future ones that arise.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics Statement
The protocol for collection of the clinical swab samples from patients in this study was approved by the Conjoint Health Research Ethics Board, University of Calgary under Ethics ID: REB13-0219_MOD3 and REB16-0623_MOD1, on October 22, 2018 and April 9, 2019, respectively.
Author Contributions
KZ conceived, designed, and supervised the work. J-AM, OO, and LW performed the experiments and analyzed the data. JC, AU-T, and TL collected and provided the clinical isolates. J-AM and KZ structured and drafted the manuscript. J-AM, JC, and KZ edited the manuscript. All authors reviewed and approved the manuscript.
Funding
This work was supported in part by the operating grants (FRN: ARE-147623 and ARF-151557) from the Canadian Institutes of Health Research (CIHR), Canada, and in part by an operating fund from the Centre for Antimicrobial Resistance (CAR), Alberta Health Services, Alberta, Canada.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. Hiramatsu and T. Ito (Juntendo University, Japan) for generously providing us with SCCmec types I, II, IIIHg, IIIA, IVa, IVb, IVc, IVd, IVg, IVh, IVi, IVj, V, VII, IX, X, and XI prototypic strains, D. Coleman (University of Dublin, Ireland) for SCCmec IIA, IIB, IIC, IID, IIE, IVE, and IVF prototypic strains, H. de Lencastre (The Rockefeller University, United States) for SCCmec VI strain, M. Stegger (Statens Serum Institut, Denmark) for SCCmec XIII strain, and S. Richter (University of Iowa Health Care, United States) for Iowa 1–20 isolates.
References
Arbefeville, S. S., Zhang, K., Kroeger, J. S., Howard, W. J., Diekema, D. J., and Richter, S. S. (2011). Prevalence and genetic relatedness of methicillin-susceptible Staphylococcus aureus isolates detected by the Xpert MRSA nasal assay. J. Clin. Microbiol. 49, 2996–2999. doi: 10.1128/JCM.00046-11
Ayliffe, G. A. (1997). The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24(Suppl. 1), S74–S79. doi: 10.1093/clinids/24.supplement_1.s74
Barber, M. (1961). Methicillin-resistant staphylococci. J. Clin. Pathol. 14, 385–393. doi: 10.1136/jcp.14.4.385
Becker, K., Denis, O., Roisin, S., Mellmann, A., Idelevich, E. A., and Knaack, D. (2016). Detection of mecA- and mecC-positive methicillin-resistant Staphylococcus aureus (MRSA) isolates by the new Xpert MRSA Gen 3 PCR assay. J. Clin. Microbiol. 54, 180–184. doi: 10.1128/JCM.02081-15
Caetano-Anolles, G. (1993). Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3, 85–94. doi: 10.1101/gr.3.2.85
Crossley, K., Landesman, B., and Zaske, D. (1979). An outbreak of infections caused by strains of Staphylococcus aureus resistant to methicillin and aminoglycosides. II. Epidemiologic studies. J. Infect. Dis. 139, 280–287. doi: 10.1093/infdis/139.3.280
Desjardins, M., Guibord, C., Lalonde, B., Toye, B., and Ramotar, K. (2006). Evaluation of the IDI-MRSA assay for detection of methicillin-resistant Staphylococcus aureus from nasal and rectal specimens pooled in a selective broth. J. Clin. Microbiol. 44, 1219–1223. doi: 10.1128/JCM.44.4.1219-1223.2006
Ellem, J. A., Olma, T., and O’Sullivan, M. V. (2015). Rapid detection of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus directly from positive blood cultures by use of the BD Max StaphSR assay. J. Clin. Microbiol. 53, 3900–3904. doi: 10.1128/JCM.02155-15
Fluit, A. C., Verhoef, J., and Schmitz, F. J. (2001). Frequency of isolation and antimicrobial resistance of gram-negative and gram-positive bacteria from patients in intensive care units of 25 European university hospitals participating in the European arm of the SENTRY Antimicrobial Surveillance Program 1997-1998. Eur. J. Clin. Microbiol. Infect. Dis. 20, 617–625. doi: 10.1007/s100960100564
Huletsky, A., Giroux, R., Rossbach, V., Gagnon, M., Vaillancourt, M., Bernier, M., et al. (2004). New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42, 1875–1884. doi: 10.1128/jcm.42.5.1875-1884.2004
International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements [IWG-SCC] (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother 53, 4961–4967. doi: 10.1128/AAC.00579-09
Ito, T., Katayama, Y., Asada, K., Mori, N., Tsutsumimoto, K., Tiensasitorn, C., et al. (2001). Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45, 1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001
Jevons, M. P. (1961). “Celbenin” - resistant Staphylococci. Br. Med. J. 1, 124–125. doi: 10.1136/bmj.1.5219.124-a
Kelley, P. G., Grabsch, E. A., Howden, B. P., Gao, W., and Grayson, M. L. (2009). Comparison of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay, BD GeneOhm MRSA assay, and culture for detection of nasal and cutaneous groin colonization by MRSA. J. Clin. Microbiol. 47, 3769–3772. doi: 10.1128/JCM.00303-09
Lakhundi, S., and Zhang, K. (2018). Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 3:e00020-18. doi: 10.1128/CMR.00020-18
Lepainteur, M., Delattre, S., Cozza, S., Lawrence, C., Roux, A. L., and Rottman, M. (2015). Comparative evaluation of Two PCR-based methods for detection of methicillin-resistant Staphylococcus aureus (MRSA): Xpert MRSA Gen 3 and BD-Max MRSA XT. J. Clin. Microbiol. 53, 1955–1958. doi: 10.1128/jcm.03679-14
Lindsay, J. A., and Holdenm, M. T. (2004). Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12, 378–385. doi: 10.1016/j.tim.2004.06.004
Malhotra-Kumar, S., Haccuria, K., Michiels, M., Ieven, M., Poyart, C., Hryniewicz, W., et al. (2008). Current trends in rapid diagnostics for methicillin-resistant Staphylococcus aureus and glycopeptide-resistant Enterococcus species. J. Clin. Microbiol. 46, 1577–1587. doi: 10.1128/JCM.00326-08
McClure, J. A., Conly, J. M., Elsayed, S., and Zhang, K. (2010). Multiplex PCR assay to facilitate identification of the recently described Staphylococcal cassette chromosome mec type VIII. Mol. Cell Probes. 24, 229–232. doi: 10.1016/j.mcp.2010.01.001
Mendes, R. E., Watters, A. A., Rhomberg, P. R., Farrell, D. J., and Jones, R. N. (2016). Performance of BD Max StaphSR for Screening of Methicillin-Resistant Staphylococcus aureus Isolates among a contemporary and diverse collection from 146 Institutions Located in Nine U.S. census regions: prevalence of mecA dropout mutants. J. Clin. Microbiol. 54, 204–207. doi: 10.1128/JCM.02047-15
Mongkolrattanothai, K., Boyle, S., Murphy, T. V., and Daum, R. S. (2004). Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 48, 1823–1836. doi: 10.1128/aac.48.5.1823-1836.2004
Panlilio, A. L., Culver, D. H., Gaynes, R. P., Banerjee, S., Henderson, T. S., Tolson, J. S., et al. (1992). Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect. Control Hosp. Epidemiol. 13, 582–586. doi: 10.1086/646432
Silbert, S., Gostnell, A., Kubasek, C., and Widen, R. (2017). Evaluation of the BD max StaphSR assay for detecting methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) in ESwab-collected wound samples. J. Clin. Microbiol. 55, 2865–2867. doi: 10.1128/JCM.00641-17
Silbert, S., Kubasek, C., Galambo, F., Vendrone, E., and Widen, R. (2015). Evaluation of BD Max StaphSR and BD Max MRSAXT Assays Using ESwab-Collected Specimens. J. Clin. Microbiol. 53, 2525–2529. doi: 10.1128/JCM.00970-15
Stamper, P. D., Louie, L., Wong, H., Simor, A. E., Farley, J. E., and Carroll, K. C. (2011). Genotypic and phenotypic characterization of methicillin-susceptible Staphylococcus aureus isolates misidentified as methicillin-resistant Staphylococcus aureus by the BD GeneOhm MRSA assay. J. Clin. Microbiol. 49, 1240–1244. doi: 10.1128/JCM.02220-10
Vandenesch, F., Naimi, T., Enright, M. C., Lina, G., Nimmo, G. R., Heffernan, H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9, 978–984. doi: 10.3201/eid0908.030089
Voss, A., Milatovic, D., Wallrauch-Schwarz, C., Rosdahl, V. T., and Braveny, I. (1994). Methicillin-resistant Staphylococcus aureus in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 13, 50–55. doi: 10.1007/bf02026127
Wu, Z., Li, F., Liu, D., Xue, H., and Zhao, X. (2015). Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase. CcrC2. Antimicrob. Agents Chemother. 59, 7597–7601. doi: 10.1128/AAC.01692-15
Zhang, K., McClure, J. A., and Conly, J. M. (2012). Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Mo.l Cell. Probes. 26, 218–221. doi: 10.1016/j.mcp.2012.04.002
Zhang, K., McClure, J. A., Elsayed, S., Louie, T., and Conly, J. M. (2008). Novel multiplex PCR assay for simultaneous identification of community-associated methicillin-resistant Staphylococcus aureus strains USA300 and USA400 and detection of mecA and panton-valentine leukocidin genes, with discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 46, 1118–1122. doi: 10.1128/JCM.01309-07
Zhang, K., Sparling, J., Chow, B. L., Elsayed, S., Hussain, Z., Church, D. L., et al. (2004). New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42, 4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004
Keywords: Staphylococcus aureus, methicillin-resistant Staphylococcus aureus (MRSA), staphylococcal cassette chromosome mec (SCCmec), coagulase negative staphylococci (CoNS), MRSA rapid detection assay, molecular diagnosis
Citation: McClure J-A, Conly JM, Obasuyi O, Ward L, Ugarte-Torres A, Louie T and Zhang K (2020) A Novel Assay for Detection of Methicillin-Resistant Staphylococcus aureus Directly From Clinical Samples. Front. Microbiol. 11:1295. doi: 10.3389/fmicb.2020.01295
Received: 25 March 2020; Accepted: 20 May 2020;
Published: 18 June 2020.
Edited by:
Santi M. Mandal, Indian Institute of Technology Kharagpur, IndiaReviewed by:
Suvankar Ghorai, SRM Institute of Science and Technology, IndiaJiun-Ling Wang, National Cheng Kung University, Taiwan
Tsai-Ling Lauderdale, National Health Research Institutes, Taiwan
Copyright © 2020 McClure, Conly, Obasuyi, Ward, Ugarte-Torres, Louie and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kunyan Zhang, a3poYW5nQHVjYWxnYXJ5LmNh
 Jo-Ann McClure1
Jo-Ann McClure1 Kunyan Zhang
Kunyan Zhang