- 1Laboratory of Applied Microbiology and Biotechnology, Department of Bioscience Engineering, University of Antwerp, Antwerp, Belgium
- 2Department of Neurobiology, The University of Chicago, Chicago, IL, United States
- 3Division of Immunology, Department of Pathology, Institute of Infectious Disease & Molecular Medicine, University of Cape Town, Cape Town, South Africa
- 4APC Microbiome Ireland, University College Cork, Cork, Ireland
- 5Teagasc Food Research Centre, Fermoy, Ireland
- 6Institute for Biomedical Sciences, Shinshu University, Nagano, Japan
- 7Department of Zoology, University of Oxford, Oxford, United Kingdom
- 8Department of Biochemistry, University of Oxford, Oxford, United Kingdom
- 9Centre for Human Microbiome and Probiotic Research, Lawson Health Research Institute, London, ON, Canada
- 10Department of Microbiology and Immunology, The University of Western Ontario, London, ON, Canada
- 11Department of Surgery, The University of Western Ontario, London, ON, Canada
The opportunities in the fields of probiotics and prebiotics to a great degree stem from what we can learn about how they influence the microbiota and interact with the host. We discuss recent insights, cutting-edge technologies and controversial results from the perspective of early career researchers innovating in these areas. This perspective emerged from the 2019 meeting of the International Scientific Association for Probiotics and Prebiotics - Student and Fellows Association (ISAPP-SFA). Probiotic and prebiotic research is being driven by genetic characterization and modification of strains, state-of-the-art in vitro, in vivo, and in silico techniques designed to uncover the effects of probiotics and prebiotics on their targets, and metabolomic tools to identify key molecules that mediate benefits on the host. These research tools offer unprecedented insights into the functionality of probiotics and prebiotics in the host ecosystem. Young scientists need to acquire these diverse toolsets, or form inter-connected teams to perform comprehensive experiments and systematic analysis of data. This will be critical to identify microbial structure and co-dependencies at body sites and determine how administered probiotic strains and prebiotic substances influence the host. This and other strategies proposed in this review will pave the way for translating the health benefits observed during research into real-life outcomes. Probiotic strains and prebiotic products can contribute greatly to the amelioration of global issues threatening society. The intent of this article is to provide an early career researcher’s perspective on where the biggest opportunities lie to advance science and impact human health.
Introduction
Trillions of microbes inhabit the human body, collectively forming the human microbiota. These microbes create complex, organ-specific and adaptive ecosystems, which continually impact the host’s physiology. The microbiota and its overall genetic material (the microbiome) consist of bacteria (bacteriome), archaea (archeome), fungi (mycobiome), viruses (virome), and parasites (parasitome). Together they play a pivotal part in human and animal physiology through influencing digestion, immune development, vitamin production, and likely behavior and mental wellbeing (Belkaid and Hand, 2014; Biesalski, 2016; Borre et al., 2014; Leung et al., 2018). Beneficial effects of certain microorganisms are reflected in the concept of probiotics, defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). Also, certain microbial substrates, called prebiotics, can be selectively utilized by host microorganisms, thereby conferring a health benefit (Gibson et al., 2017). Thus, by supplementing with appropriate probiotics and/or prebiotics, it is possible to provide added benefits to human health.
The fields of probiotics, prebiotics, and related microbiome research have seen remarkable advances in the last decades (Sanders et al., 2019). Novel tools offer alternatives to traditional in vitro and in vivo models (Table 1), allowing more efficient generation of convincing data on the probiotic and prebiotic mode of action, their effect on the microbiome, and the resulting clinical health outcomes. Numerous clinical trials provide evidence of strain-specific probiotic and prebiotic benefits for a range of health conditions, including diarrhea (Guandalini, 2011) vaginal dysbiosis (Reid et al., 2003), respiratory infections (Hatakka et al., 2001), bowel function (Vandeputte et al., 2017) body weight (Nicolucci et al., 2017) and bone mineralization (Abrams et al., 2005). These findings have sparked an interest from the press and general public, emphasizing the need for correct and accessible scientific communication around these topics. Taken together, this calls for a discussion on the research possibilities and implementation of probiotics and prebiotics and their interaction with the host microbiome (Figure 1).
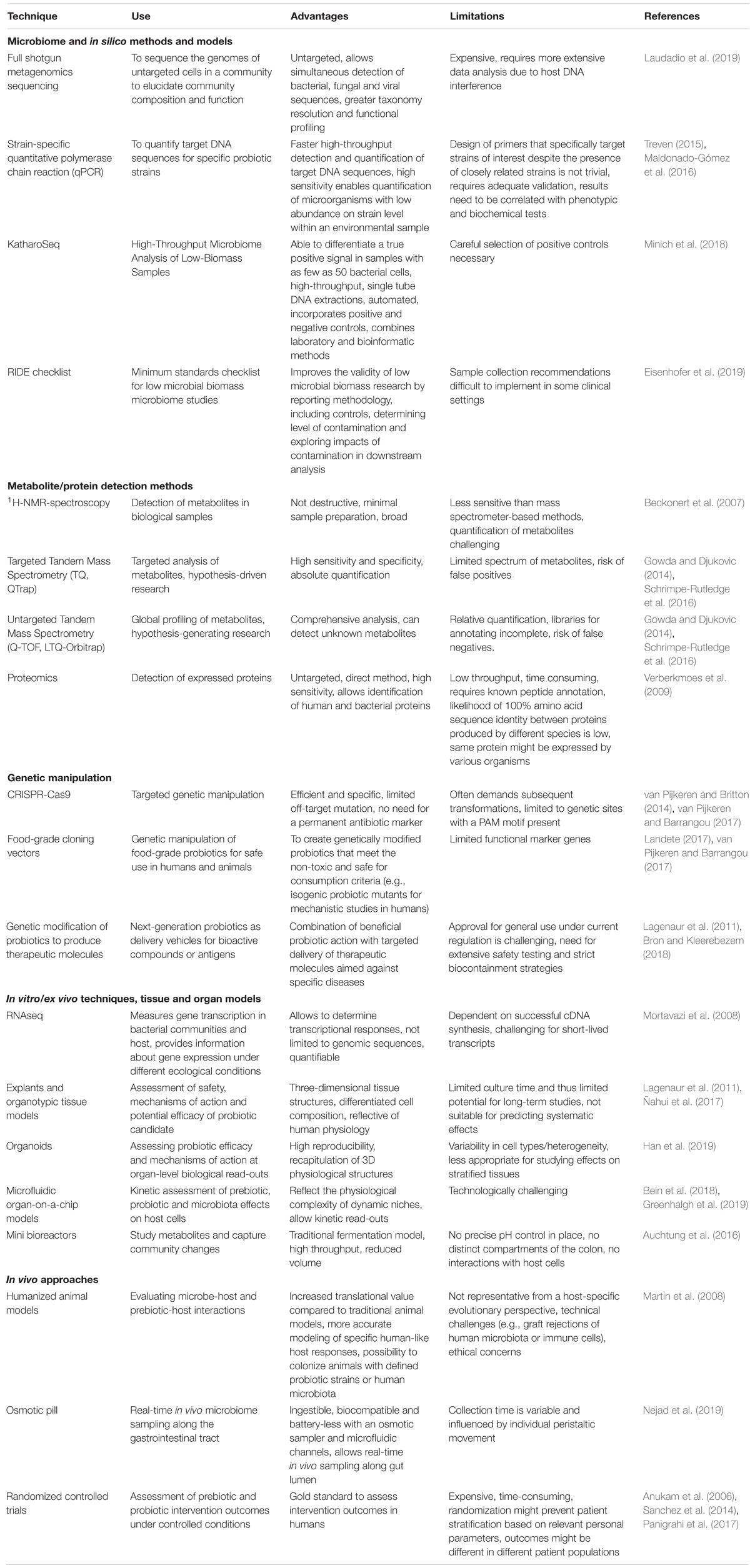
Table 1. Promising in vitro, in vivo, and in silico techniques for probiotics and prebiotics research.
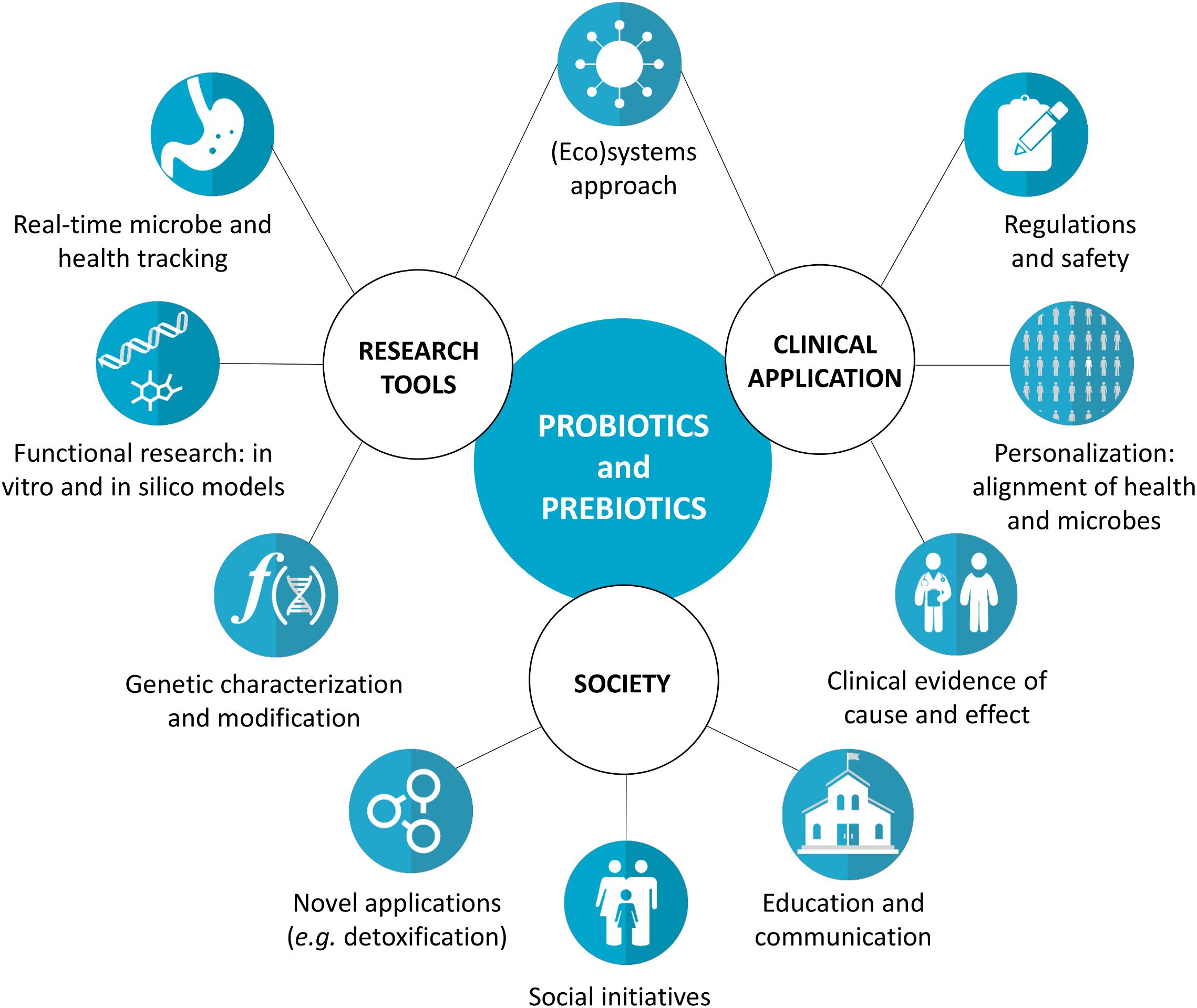
Figure 1. Key concepts for the development of the probiotics and prebiotics fields. We propose a systems approach linking research tools and clinical application to translate the potential health benefits observed during research into real-life outcomes. For example, not only the probiotics and prebiotics themselves should be considered, but also the real-time interactions with the host, microbial functionality, individual factors, and the regulation and safety aspects. Furthermore, various environmental and social needs can and should be addressed by probiotics and prebiotics.
Developments in Probiotics and Prebiotics Research
Considerations for Probiotics Research
Genetic Characterization and Modification of Probiotics
Bioinformatics and in silico approaches contribute to a more detailed understanding of beneficial microorganisms, thereby allowing their targeted usage and safety assessment. As whole genome sequencing (WGS) is available at a reasonable cost, we advise that qualitative WGS and rigorous annotation should become the standard practice prior to marketing new probiotic strains. Newly sequenced genomes should be deposited and made publicly available via standard central databases (e.g., GenBank1, DDBJ2, ENA3). A rigorous sequence quality control and annotation should be carried out (Smits, 2019) identifying the mobile and other genetic elements (e.g., CRISPR arrays) and predicting their functional properties, thereby estimating the safety of probiotic candidates regarding virulence factors and possible antibiotic resistance gene transfer. The European Food Safety Authority (EFSA) and recent publications recommend WGS to improve the monitoring of foodborne antimicrobial resistance (Collineau et al., 2019; Efsa et al., 2019) and workflows to assess risk-related gene traits based on WGS are available (Salvetti et al., 2016). WGS is also useful to assess genetic instability and ensure the retention of regions linked to the strain’s health benefits, as demonstrated for the Lacticaseibacillus rhamnosus GG variants with and without spaCBA pili genes (Sybesma et al., 2013).
Genetic manipulation is an important tool to study probiotic mechanisms of action [e.g., by using isogenic mutant strains (Lebeer et al., 2011)] and to potentially create improved strains (Lebeer et al., 2018; Petrova et al., 2018). However, a lack of sufficient genetic tools available for some probiotic species, especially food-grade systems for bifidobacteria, and a legal framework for the use of genetically manipulated/enhanced organisms limits the research progress (Allain et al., 2015; Asto et al., 2019). In addition to ethical and legal questions, it is not yet common practice to send genetic constructs to biological repositories. The considerable effort it takes to make these constructs and regulations within the research institutions are probably the main reasons to keep this genetic material in-house. We therefore advocate for a better sharing of genetic constructs by providing the material to the scientific community through existing repositories (e.g., AddGene, European Nucleotide Archive). This can save a substantial investment of time and research funds and will serve to enlarge the genetic tools box for probiotics. In particular, the development of safe vectors (e.g., food-grade vectors) is a necessary step in genetically tweaking probiotic strains for industrial and pharmaceutical applications (Landete, 2017), including specialized probiotics designed to deliver bioactive compounds to more effectively target specific diseases (Table 1). Both fundamental and applied probiotics research would benefit from a vast investment in genetic elements, and a better understanding of their mechanism of action and specificity (Allain et al., 2015). Genetic and bioinformatic training and experience in these interdisciplinary areas will thus be key for researchers making progress in the probiotics field.
In vitro Models in Probiotics Research
Animal models are not strictly necessary for preclinical assessment of probiotics. While humanized animal models can be implemented (Table 1), it is highly challenging to develop ones that simulate microbe-host interaction in humans for niches demonstrating unique physiological features such as the lactobacilli-dominated vaginal niche characterized by a low pH (Miller et al., 2016). Human-based in vitro and ex vivo models followed by small studies with healthy volunteers and larger clinical intervention studies are invariably required to draw more precise and relevant conclusions on probiotic safety, action, and health benefits.
Recent cutting-edge in vitro and ex vivo approaches based on human cells and tissues pave the way beyond in vitro cell lines and animal models (Table 1). Reproducible human organoids have been used to recapitulate irritable bowel syndrome manifestations and the restorative effects of L. rhamnosus GG (Han et al., 2019). Sophisticated organs-on-chips combine advances in human cell culturing with microelectronics and microfluidics to discover the anti-cancer potential of probiotic and synbiotic formulations, however, they are limited due to lack of exposure to host defenses (Bein et al., 2018; Greenhalgh et al., 2019). Cervico-vaginal tissue explants and organotypic tissue models circumvent this by combining human epithelial and immune cells and have previously allowed to identify anti-HIV-1 effects of wild type and genetically modified lactobacilli (Lagenaur et al., 2011; Ñahui Palomino et al., 2017).
It is notable that on September 10, 2019, the United States Environmental Protection Agency released a memorandum stating that studies on mammals will be eliminated by 2035. This has implications for early career scientists developing their future line of research, opening opportunities for implementation of alternative in vitro models.
Considerations for Prebiotics Research
In vitro Methods to Study Prebiotics and Their Targets
Early prebiotic research was focused on the ability of compounds to stimulate bifidobacterial growth in the gut. Now, research has expanded to prebiotics targeting other species in the gut, but also in the vagina, lung, and skin (Gibson et al., 2017; Collins et al., 2018). This offers opportunities for prebiotics beyond dietary non-digestible carbohydrates targeting colonic microbiota. Studies are warranted to identify novel prebiotics, especially by experts in chemistry and food science, and engineers designing delivery systems. In addition, synbiotics can exert a synergistic effect on host health with promising clinical effects against neonatal sepsis (Panigrahi et al., 2017) and insulin resistance (Eslamparast et al., 2014).
A diverse array of in vitro, in vivo, and in silico-based techniques has been applied to understand the mechanisms of action and efficacy of prebiotics (Table 1). The traditional method has been a bottom-up approach, where candidate prebiotics are screened by in vitro techniques, such as single strain culturing and chemostat fermentation of stool samples, before employing them in animal models and human intervention studies.
A novel top-down systems biology-based approach has recently been developed through in silico methods, which involve computer modeling and analysis of biological processes and interactions. For this, existing metagenomics and genomic data from human studies are mined to identify potential novel beneficial species able to prosper with the compound being tested. The prebiotics for these species are then selected by metabolic models based on microbial genomic data (Shoaie et al., 2015; Tramontano et al., 2018). Attempts have been made to create metabolic models of microbial inhabitants of the gut, to unravel the complex cross-feeding that occurs within the gut microbiota (Shoaie et al., 2013). We argue that in silico techniques are useful complements to classical techniques, and their implementation has the potential to improve the design of human studies.
Importance of Metabolomics for Prebiotics
The recent expansion of the prebiotic concept to include other types of compounds in addition to non-digestible carbohydrates, such as polyphenols and certain fatty acids (e.g., polyunsaturated fatty acids), has resulted in greater emphasis on untargeted metabolome analysis (Bindels et al., 2015). Microbial biotransformation of dietary phytochemicals (e.g., polyphenols) leads to a diverse array of metabolites, more bioavailable than the parent compound. These could enhance health effects (Duenas et al., 2015). Evidence suggesting bacteria contribute to approximately 70% of fecal and 15% of serum metabolites highlights the importance of understanding metabolite origin (Visconti et al., 2019). An in silico approach including annotating metabolite data to host, diet or bacterial derived origins would contribute to this understanding (Shaffer et al., 2019). Proving the origin of a metabolite can be achieved in vitro and in vivo by a dual isotope/radio labeling of novel prebiotics to build databases for annotating and mapping metabolic networks through elucidating the pharmacokinetics of the prebiotics and the mechanism and site of action of the metabolites (Birkemeyer et al., 2005).
For those pursuing prebiotic research, this multi-dimensional approach should uncover how these compounds function within the diverse gut microbiome or at other sites.
Discussion on Untangling the In Vivo Effects of Probiotics and Prebiotics
Translating Effects of Probiotics and Prebiotics Into Real-Life Outcomes
Two recent publications questioned the in vivo benefits and safety of probiotics (Rao et al., 2018; Suez et al., 2018), but failed to provide sufficient, substantiated evidence on actual harm or lack of efficacy. Sweeping generalizations on probiotics as a whole should be avoided by properly documenting the strains used, and emphasizing their strain-specificity and mechanisms of action (Lebeer et al., 2018; Sanders et al., 2019). Probiotic strains with a safe history of use should be tested in small pilot trials in humans with detailed sampling and safety assessment, especially in high risk populations and regarding long-term effects (Langille et al., 2013; Sanders et al., 2019). When strains pose a safety concern, postbiotics, or “bioactive compounds produced by food-grade micro-organisms during a fermentation process,” might be an efficient alternative (Wegh et al., 2019). Subsequently, large well-designed and properly controlled trials in the target host are needed to obtain evidence of benefit, optimal dose, and intended clinical uses (Sanders et al., 2019).
Understanding the influence of interpersonal differences on clinical outcomes would greatly contribute to the efficacy of probiotic and prebiotic interventions (Mullish et al., 2020; Rodriguez et al., 2020). However, this is not a trivial task, and it is often not yet clear why a strain or compound is more effective in some individuals than others (Sanchez et al., 2014). Future research should focus on stratification of clinical trials based on individual characteristics of the participants, including sex, ethnicity, diet, and the functional characteristics of their microbiome. Already, the need for microbiome research to emphasize function rather than microbiota composition has been raised (Reid et al., 2019). This requires metabolomic tools and small molecule identification aligned with clinical evidence of cause and effect.
We advocate for adopting a systems approach that integrates readouts of functional microbial and health data in a range of samples (including stool, urine, blood, mucosa and saliva) from clinical trials. The readouts should focus on functional interactions with the host and how they are influenced by additional factors (e.g., drugs, pollutants, nutrients). This can be reasonably well achieved with a combination of in silico, in vitro, and in vivo approaches (Table 1). Precise metabolic modeling may predict which probiotic or prebiotic would be most likely to help an individual realize an effective personalized nutrition strategy for treating disease and promoting health (Tebani and Bekri, 2019). Response profiles to probiotic interventions can and should be evaluated in humans, even if it requires complicated tools like assessing the transcriptome. Such studies reveal that different probiotic Lactobacillus/Lacticaseibacillus strains induce different gene-regulatory pathways in the small intestinal mucosa of healthy volunteers (van Baarlen et al., 2011).
Approaches like this provide avenues for understanding the individual-specific effects of different probiotic strains and facilitate their rationally designed clinical application. An exciting future goal would be to develop personalized probiotics and prebiotics. However, given there is no single healthy microbiome (Lloyd-Price et al., 2016), the personalized concept may not require individual remedies, but rather sufficient options to manipulate functions shared by many.
Probiotics, Prebiotics and the Microbiome: An Ecological Perspective
When exploring probiotic and prebiotic effects at distant sites, it is critical to consider the complex microbial ecosystems of those niches. The administered probiotic should ideally promote homeostasis. Longitudinal and body niche-specific microbiome sampling (Gajer et al., 2012) is important to assess the dynamic changes resulting from probiotic and prebiotic interventions. This is helped by non-invasive devices, such as an osmotic pill for real-time in vivo microbiome sampling of difficult to access niches in the gut (Nejad et al., 2019) (Table 1). This pill is ingestible, biocompatible and does not require a battery. It has an integrated osmotic sampler and microfluidic channels, allowing for real-time sampling of the gut lumen and its microbiome without the need for colonoscopy. This will help us understand the microbiome’s response to probiotic or prebiotic administration and pave the way for lab-on-a-pill devices.
The manner in which probiotic strains adjust to the conditions of the target site are largely unknown. Global gene expression profiling and studying isogenic mutants showed that exopolysaccharide production affects lactobacilli survival in the human (Marco et al., 2010) and murine (Lebeer et al., 2011) intestine. Interplay with environmental factors (e.g., medication, pollutants, nutrients) must also be investigated to truly appreciate factors that can influence probiotic strain activity and health of the host. Ultimately, an integrative approach would not simply establish dose-response relationships to treatment, but rather attempt to align what enters our system, how it is processed, and what microbes can be delivered to improve the net effect. It is thus inevitable that systematic approaches should be implemented to elucidate how an administered probiotic or prebiotic interacts with the host at various levels (e.g., the immune stimulator/modulator effect, microbiome, medication interactions).
The ecologic perspective will not be complete without identifying how changes in the relative abundance of one species impacts the relative abundances of others. Interdependency of sequencing data overlooks the real population dynamics. An ecosystem includes different interactions: co-operation, competition, exploitation, mutualism, co-dependency, commensalism, and amensalism, and these interactions can play a role in microbiome stability (Coyte et al., 2015). Importantly, the microbiome datasets obtained with high throughput sequencing are compositional, as they consist of proportions with a constant sum, therefore compositionally appropriate tools should be applied for their analysis (Gloor et al., 2017). To assess these complex interactions, evaluation of relative microbial community composition (e.g., by 16S rRNA gene or shotgun DNA sequencing) should be combined with species-specific qPCR for abundance, whole-transcriptome shotgun sequencing and proteomics to determine function (Quince et al., 2017) (Table 1). These approaches will allow to identify functional changes, adaptability and ecological interactions of probiotics and prebiotics with indigenous host microorganism communities. Large datasets generated using these multi “omics” could facilitate the hypothesis-driven studies at different levels of evidence.
Impact Beyond the Bench
Probiotics, and to a lesser extent prebiotics, are now widely available to people in developed countries. Nevertheless, continual education is needed as too often the media or companies misrepresent what these are and what they can do (Reid et al., 2019). On a global front, people in developed and especially developing countries could benefit from probiotic products due to effects against infectious diseases, but the lack of affordable and well-documented strains is a hindrance (Kort et al., 2015). Various probiotic initiatives to better influence developing countries, including Westernheadseast.ca, Yoba4Life.org, and Yogurito (Argentina), have proven that populations in low-income regions derive benefits beyond probiotic-mediated health, as seen by facilitating economic development and fighting malnutrition with local resources (Reid et al., 2018, 2020). Every country should take action to implement programs allowing the poorest of their society access to fermented foods and probiotics that reduce the risk of key diseases: diabetes, malnutrition, infections.
We could also harness the power of beneficial microbes to ameliorate the impact of worldwide problems. A study showed that the probiotic Lacticaseibacillus rhamnosus GR-1 can sequester the heavy metals lead and cadmium, effectively reducing their translocation across the intestinal epithelium in vitro (Daisley et al., 2019). Thus, administration of certain probiotic strains may offer a simple and effective option to reduce the amount of heavy metals absorbed from foods in contaminated regions of the world. Novel marine probiotics can also help prevent ecological damage, for example by increasing coral resistance to bleaching (Rosado et al., 2019). When such applications become feasible, many opportunities will arise for early career researchers in the field of probiotics and prebiotics.
Concluding Remarks and Future Perspectives
Current technological and methodological developments offer exciting possibilities for probiotics and prebiotics research and applications. New tools allowing real-time studies in humans and following a microbe as it integrates into an existing microbiota, as well as systems that can quantify levels of health, will drive this field forward. Read-outs on what microbes are present, their interaction with the host and the influence of environmental factors (e.g., drugs, nutrients) will become standard when going for a physical examination in the future.
Novel sampling systems will elucidate how an applied probiotic or prebiotic interacts with the host at various levels, including the immune system, metabolism and all components of the microbiome. Ultimately, an integrative approach will support a form of personalized medicine to establish dose-response relationships for treatment, but moreso attempt to align what enters our system, how it is processed, and which probiotics or prebiotics deliver the best desired effects. Mechanistic insights into effector molecules will pave the way for emerging concepts, such as postbiotics.
As early career scientists, we want to be part of a society that uses beneficial microbes to help solve global problems, such as reducing the risk and impact of disease (including viruses and pandemics) and removing drugs and toxins from our food and environment. These will be exciting times with many career paths open for probiotics and prebiotics research in the sciences and applied to many other disciplines.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This publication was made possible with financial support from the International Scientific Association for Probiotics and Prebiotics (ISAPP). IS was funded by IOF POC ReLACT (University of Antwerp) and also supported by IWT SBO ProCure during her postdoctoral work. HD was supported by BrightFocus Foundation postdoctoral fellowship (Grant Number A2019032F) and Alzheimer’s Association Research Fellowship (AARF) program. DV was a JSPS International Research Fellow (Grant Number FY2018 P18782). A-UH was receiving a departmental scholarship at the University of Cape Town. CS was funded by Science Foundation Ireland Research Grant SFI/12/RC/227.
Conflict of Interest
GR acknowledged providing advice to several companies that sell probiotic products.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The input of Dr. Mary Ellen Sanders is greatly appreciated. We gratefully acknowledge the guidance and financial support of travel and meeting expenses provided by the International Scientific Association for Probiotics and Prebiotics (ISAPP). We also thank all the members of the ISAPP Students and Fellows Association (ISAPP-SFA) for their enthusiasm and participation in ongoing scientific discussions.
Footnotes
References
Abrams, S. A., Griffin, I. J., Hawthorne, K. M., Liang, L., Gunn, S. K., Darlington, G., et al. (2005). A combination of prebiotic short- and long-chain inulin-type fructans enhances calcium absorption and bone mineralization in young adolescents. Am. J. Clin. Nutr. 82, 471–476. doi: 10.1093/ajcn/82.2.471
Allain, T., Aubry, C., Natividad, J. M., Chatel, J.-M., Langella, P., and Bermúdez-Humarán, L. G. (2015). “Chapter 11: engineering lactic acid bacteria and bifidobacteria for mucosal delivery of health molecules,” in Biotechnology of Lactic Acid Bacteria: Novel Applications, 2nd Edn, eds F. Mozzi, R. R. Raya, and G. M. Vignolo (Hoboken, NJ: Wiley Blackwell), 2016.
Anukam, K. C., Osazuwa, E., Osemene, G. I., Ehigiagbe, F., Bruce, A. W., and Reid, G. (2006). Clinical study comparing probiotic Lactobacillus GR-1 and RC-14 with metronidazole vaginal gel to treat symptomatic bacterial vaginosis. Microb. Infect. 8, 2772–2776. doi: 10.1016/j.micinf.2006.08.008
Asto, E., Méndez, I., Rodríguez-Prado, M., Cuñe, J., Espadaler, J., and Farran-Codina, A. (2019). Effect of the degree of polymerization of fructans on ex vivo fermented human gut microbiome. Nutrients 11:1293. doi: 10.3390/nu11061293
Auchtung, J. M., Robinson, C. D., Farrell, K., and Britton, R. A. (2016). “MiniBioReactor arrays (MBRAs) as a tool for studying C. difficile physiology in the presence of a complex community,” in Clostridium Difficile: Methods in Molecular Biology, eds A. Roberts and P. Mullany (New York, NY: Humana Press).
Beckonert, O., Keun, H., Ebbels, T., Bundy, J., Holmes, E., Lindon, J. C., et al. (2007). Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2, 2692–2703. doi: 10.1038/nprot.2007.376
Bein, A., Shin, W., Jalili-Firoozinezhad, S., Park, M. H., Sontheimer-Phelps, A., Tovaglieri, A., et al. (2018). Microfluidic organ-on-a-chip models of human intestine. Cell. Mol. Gastroenterol. Hepatol. 5, 659–668. doi: 10.1016/j.jcmgh.2017.12.010
Belkaid, Y., and Hand, T. W. (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. doi: 10.1016/j.cell.2014.03.011
Biesalski, H. K. (2016). Nutrition meets the microbiome: micronutrients and the microbiota. Ann. N. Y. Acad. Sci. 1372, 53–64. doi: 10.1111/nyas.13145
Bindels, L. B., Delzenne, N. M., Cani, P. D., and Walter, J. (2015). Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 12, 303–310. doi: 10.1038/nrgastro.2015.47
Birkemeyer, C., Luedemann, A., Wagner, C., Erban, A., and Kopka, J. (2005). Metabolome analysis: the potential of in vivo labeling with stable isotopes for metabolite profiling. Trends Biotechnol. 23, 28–33. doi: 10.1016/j.tibtech.2004.12.001
Borre, Y. E., Moloney, R. D., Clarke, G., Dinan, T. G., and Cryan, J. F. (2014). “The impact of microbiota on brain and behavior: mechanisms & therapeutic potential,” in Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease. Advances in Experimental Medicine and Biology, eds M. Lyte and J. Cryan (New York, NY: Springer).
Bron, P. A., and Kleerebezem, M. (2018). Lactic acid bacteria for delivery of endogenous or engineered therapeutic molecules. Front. Microbiol. 9:1821. doi: 10.3389/fmicb.2018.01821
Collineau, L., Boerlin, P., Carson, C. A., Chapman, B., Fazil, A., Hetman, B., et al. (2019). Integrating whole-genome sequencing data into quantitative risk assessment of foodborne antimicrobial resistance: a review of opportunities and challenges. Front. Microbiol. 10:1107. doi: 10.3389/fmicb.2018.1107
Collins, S. L., McMillan, A., Seney, S., van der Veer, C., Kort, R., Sumarah, M. W., et al. (2018). Promising prebiotic candidate established by evaluation of lactitol, lactulose, raffinose, and oligofructose for maintenance of a Lactobacillus-dominated vaginal microbiota. Appl. Environ. Microbiol. 84:e02200-17.
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Daisley, B. A., Monachese, M., Trinder, M., Bisanz, J. E., Chmiel, J. A., Burton, J. P., et al. (2019). Immobilization of cadmium and lead by Lactobacillus rhamnosus GR-1 mitigates apical-to-basolateral heavy metal translocation in a Caco-2 model of the intestinal epithelium. Gut Microb. 10, 321–333. doi: 10.1080/19490976.2018.1526581
Duenas, M., Muñoz-González, I., Cueva, C., Jiménez-Girón, A. M., Sánchez-Patán, F., Santos-Buelga, C., et al. (2015). A survey of modulation of gut microbiota by dietary polyphenols. Biomed. Res. Intern. 2015:850902.
Efsa, E., Aerts, M., Battisti, A., Hendriksen, R., Kempf, I., Teale, C., et al. (2019). Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J. 17:e05709.
Eisenhofer, R., Minich, J. J., Marotz, C., Cooper, A., Knight, R., and Weyrich, L. S. (2019). Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 27, 105–117. doi: 10.1016/j.tim.2018.11.003
Eslamparast, T., Zamani, F., Hekmatdoost, A., Sharafkhah, M., Eghtesad, S., Malekzadeh, R., et al. (2014). Effects of synbiotic supplementation on insulin resistance in subjects with the metabolic syndrome: a randomised, double-blind, placebo-controlled pilot study. Br. J. Nutr. 112, 438–445. doi: 10.1017/s0007114514000919
Gajer, P., Brotman, R. M., Bai, G., Sakamoto, J., Schütte, U. M. E., Zhong, X., et al. (2012). Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 4:132ra52. doi: 10.1126/scitranslmed.3003605
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V., and Egozcue, J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2018.2224
Gowda, G. A. N., and Djukovic, D. (2014). Overview of mass spectrometry-based metabolomics: opportunities and challenges. Methods Mol. Biol. 1198, 3–12. doi: 10.1007/978-1-4939-1258-2_1
Greenhalgh, K., Ramiro-Garcia, J., Heinken, A., Ullmann, P., Bintener, T., Pacheco, M. P., et al. (2019). Integrated in vitro and in silico modeling delineates the molecular effects of a synbiotic regimen on colorectal-cancer-derived cells. Cell Rep. 27, 1621–1632.
Guandalini, S. (2011). Probiotics for prevention and treatment of diarrhea. J. Clin. Gastroenterol. 45, S149–S153.
Han, X., Lee, A., Huang, S., Gao, J., Spence, J. R., and Owyang, C. (2019). Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut Microb. 10, 59–76. doi: 10.1080/19490976.2018.1479625
Hatakka, K., Savilahti, E., Pönkä, A., Meurman, J. H., Poussa, T., Näse, L., et al. (2001). Effect of long term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised trial. Br. Med. J. 322:1327. doi: 10.1136/bmj.322.7298.1327
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Kort, R., Westerik, N., Mariela Serrano, L., Douillard, F. P., Gottstein, W., Mukisa, I. M., et al. (2015). A novel consortium of Lactobacillus rhamnosus and Streptococcus thermophilus for increased access to functional fermented foods. Microb. Cell Factor. 14:195.
Lagenaur, L. A., Sanders-Beer, B. E., Brichacek, B., Pal, R., Liu, X., Liu, Y., et al. (2011). Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunol. 4, 648–657. doi: 10.1038/mi.2011.30
Landete, J. M. (2017). A review of food-grade vectors in lactic acid bacteria: from the laboratory to their application. Crit. Rev. Biotechnol. 37, 296–308. doi: 10.3109/07388551.2016.1144044
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Laudadio, I., Fulci, V., Stronati, L., and Carissimi, C. (2019). Next-generation metagenomics: methodological challenges and opportunities. OMICS 23, 327–333. doi: 10.1089/omi.2019.0073
Lebeer, S., Bron, P. A., Marco, M. L., van Pijkeren, J.-P., Motherway, M., Hill, C., et al. (2018). Identification of probiotic effector molecules: present state and future perspectives. Curr. Opin. Biotechnol. 49, 217–223. doi: 10.1016/j.copbio.2017.10.007
Lebeer, S., Claes, I. J., Verhoeven, T. L., Vanderleyden, J., and De Keersmaecker, S. C. (2011). Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb. Biotechnol. 4, 368–374. doi: 10.1111/j.1751-7915.2010.00199.x
Leung, J. M., Graham, A. L., and Knowles, S. L. C. (2018). Parasite-microbiota interactions with the vertebrate gut: synthesis through an ecological lens. Front. Microbiol. 9:843. doi: 10.3389/fmicb.2018.0843
Lloyd-Price, J., Abu-Ali, G., and Huttenhower, C. (2016). The healthy human microbiome. Genome Med. 8:51. doi: 10.1186/s13073-016-0307-y
Maldonado-Gómez, M. X., Martínez, I., Bottacini, F., O’Callaghan, A., Ventura, M., van Sinderen, D., et al. (2016). Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microb. 20, 515–526. doi: 10.1016/j.chom.2016.09.001
Marco, M. L., de Vries, M. C., Wels, M., Molenaar, D., Mangell, P., Ahrne, S., et al. (2010). Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 4, 1481–1484. doi: 10.1038/ismej.2010.61
Martin, F. P., Wang, Y., Sprenger, N., Yap, I. K., Lundstedt, T., Lek, P., et al. (2008). Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 4:157. doi: 10.1038/msb4100190
Miller, E. A., Beasley, D. E., Dunn, R. R., and Archie, E. A. (2016). Lactobacilli dominance and vaginal pH: why is the human vaginal microbiome unique? Front. Microbiol. 7:1936. doi: 10.3389/fmicb.2018.01936
Minich, J. J., Zhu, Q., Janssen, S., Hendrickson, R., Amir, A., Vetter, R., et al. (2018). KatharoSeq enables high-throughput microbiome analysis from low-biomass samples. mSystems 3:e0218-17.
Mortavazi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mullish, B. H., Michael, D. R., McDonald, J. A. K., Masetti, G., Plummer, S. F., and Marchesi, J. R. (2020). Identifying the factors influencing outcome in probiotic studies in overweight and obese patients: host or microbiome? Gut doi: 10.1136/gutjnl-2020-321110 [Epub ahed of print].
Ñahui Palomino, R. A., Zicari, S., Vanpouille, C., Vitali, B., and Margolis, L. (2017). Vaginal Lactobacillus inhibits HIV-1 replication in human tissues ex vivo. Front. Microbiol. 8:906.
Nejad, H. R., Oliveira, B. C. M., Sadeqi, A., Dehkharghani, A., Kondova, I., Langermans, J. A. M., et al. (2019). Ingestible osmotic pill for in vivo sampling of gut microbiome. Adv. Intellig. Syst. 1:1900053. doi: 10.1101/690982
Nicolucci, A. C., Hume, M. P., Martínez, I., Mayengbam, S., Walter, J., and Reimer, R. A. (2017). Prebiotics reduce body fat and alter intestinal microbiota in children who are overweight or with obesity. Gastroenterology 153, 711–722. doi: 10.1053/j.gastro.2017.05.055
Panigrahi, P., Parida, S., Nanda, N. C., Satpathy, R., Pradhan, L., Chandel, D. S., et al. (2017). A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412. doi: 10.1038/nature23480
Petrova, M. I., van den Broek, M. F. L., Spacova, I., Verhoeven, T. L. A., Balzarini, J., Vanderleyden, J., et al. (2018). Engineering Lactobacillus rhamnosus GG and GR-1 to express HIV-inhibiting griffithsin. Int. J. Antimicrob. Agents 52, 599–607. doi: 10.1016/j.ijantimicag.2018.07.013
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J., and Segata, N. (2017). Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844. doi: 10.1038/nbt.3935
Rao, S., Rehman, A., Yu, S., and Andino, N. M. (2018). Brain fogginess, gas and bloating: a link between SIBO, probiotics and metabolic acidosis. Clin. Transl. Gastroenterol. 9:162.
Reid, G., Charbonneau, D., Erb, J., Kochanowski, B., Beuerman, D., Poehner, R., et al. (2003). Oral use of Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 significantly alters vaginal flora: randomized, placebo-controlled trial in 64 healthy women. FEMS Immunol. Med. Microbiol. 35, 131–134. doi: 10.1016/s0928-8244(02)00465-0
Reid, G., Gadir, A. A., and Dhir, R. (2019). Probiotics: reiterating what they are and what they are not. Front. Microbiol. 10:424. doi: 10.3389/fmicb.2018.0424
Reid, G., Kort, R., Alvarez, S., Bourdet-Sicard, R., Benoit, V., Cunningham, M., et al. (2018). Expanding the reach of probiotics through social enterprises. Benef. Microb. 9, 707–715. doi: 10.3920/bm2018.0015
Reid, G., Sybesma, W., Matovu, W., Onyango, A., Westerik, N., Musika, I., et al. (2020). Empowering women through probiotic fermented food in East Africa. J. Glob. Health 10:010330. doi: 10.7189/jogh.10.010330
Rodriguez, J., Hiel, S., Neyrinck, A. M., Le Roy, T., Pötgens, S. A., Leyrolle, Q., et al. (2020). Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut doi: 10.1136/gutjnl-2019-319726
Rosado, P. M., Leite, D. C. A., Duarte, G. A. S., Chaloub, R. M., Jospin, G., Nunes da Rocha, U., et al. (2019). Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Salvetti, E., Orrù, L., Capozzi, V., Martina, A., Lamontanara, A., Keller, D., et al. (2016). Integrate genome-based assessment of safety for probiotic strains: Bacillus coagulans GBI-30, 6086 as a case study. Appl. Microbiol. Biotechnol. 100, 4595–4605. doi: 10.1007/s00253-016-7416-9
Sanchez, M., Darimont, C., Drapeau, V., Emady-Azar, S., Lepage, M., Rezzonico, E., et al. (2014). Effect of Lactobacillus rhamnosus CGMCC1.3724 supplementation on weight loss and maintenance in obese men and women. Br. J. Nutr. 111, 1507–1519. doi: 10.1017/s0007114513003875
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R., and Rastall, R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616. doi: 10.1038/s41575-019-0173-3
Schrimpe-Rutledge, A. C., Codreanu, S. G., Sherrod, S. D., and McLean, J. A. (2016). Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 27, 1897–1905. doi: 10.1007/s13361-016-1469-y
Shaffer, M., Thurimella, K., Quinn, K., Doenges, K., Zhang, X., Bokatzian, S., et al. (2019). AMON: annotation of metabolite origins via networks to better integrate microbiome and metabolome data. BMC Bioinform. 20:614. doi: 10.1186/s12864-019-6014-614
Shoaie, S., Ghaffari, P., Kovatcheva-Datchary, P., Mardinoglu, A., Sen, P., Pujos-Guillot, E., et al. (2015). Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 22, 320–331. doi: 10.1016/j.cmet.2015.07.001
Shoaie, S., Karlsson, F., Mardinoglu, A., Nookaew, I., Bordel, S., and Nielsen, J. (2013). Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci. Rep. 3:2532.
Smits, T. H. M. (2019). The importance of genome sequence quality to microbial comparative genomics. BMC Genomics 20:662. doi: 10.1186/s12864-019-6014-5
Suez, J., Zmora, N., Zilberman-Schapira, G., Mor, U., Dori-Bachash, M., Bashiardes, S., et al. (2018). Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.
Sybesma, W., Molenaar, D., van IJcken, W., Venema, K., and Kort, R. (2013). Genome instability in Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 79, 2233–2239. doi: 10.1128/aem.03566-12
Tebani, A., and Bekri, S. (2019). Paving the way to precision nutrition through metabolomics. Front. Nutr. 6:41. doi: 10.3389/fmicb.2018.041
Tramontano, M., Andrejev, S., Pruteanu, M., Klünemann, M., Kuhn, M., Galardini, M., et al. (2018). Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 3, 514–522. doi: 10.1038/s41564-018-0123-9
Treven, P. (2015). Strategies to develop strain-specific PCR based assays for probiotics. Benef. Microb. 6, 887–898. doi: 10.3920/bm2015.0009
van Baarlen, P., Troost, F., van der Meer, C., Hooiveld, G., Boekschoten, M., Brummer, R. J., et al. (2011). Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4562–4569. doi: 10.1073/pnas.1000079107
van Pijkeren, J.-P., and Barrangou, R. (2017). Genome editing of food-grade lactobacilli to develop therapeutic probiotics. Microbiol. Spectr. 5:13. doi: 10.1128/microbiolspec.BAD-0013-2016
van Pijkeren, J. P., and Britton, R. A. (2014). Precision genome engineering in lactic acid bacteria. Microb. Cell Factor. 13(Suppl. 1), S10–S10.
Vandeputte, D., Falony, G., Vieira-Silva, S., Wang, J., Sailer, M., Theis, S., et al. (2017). Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 66, 1968–1974. doi: 10.1136/gutjnl-2016-313271
Verberkmoes, N., Russell, A., Shah, M., Godzik, A., Rosenquist, M., Halfvarson, J., et al. (2009). Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3, 179–189. doi: 10.1038/ismej.2008.108
Visconti, A., Le Roy, C. I., Rosa, F., Rossi, N., Martin, T. C., Mohney, R. P., et al. (2019). Interplay between the human gut microbiome and host metabolism. Nat. Commun. 10:4505.
Keywords: probiotics, prebiotics, review, young scientist, ISAPP, SFA
Citation: Spacova I, Dodiya HB, Happel A-U, Strain C, Vandenheuvel D, Wang X and Reid G (2020) Future of Probiotics and Prebiotics and the Implications for Early Career Researchers. Front. Microbiol. 11:1400. doi: 10.3389/fmicb.2020.01400
Received: 08 April 2020; Accepted: 29 May 2020;
Published: 24 June 2020.
Edited by:
Michael Gänzle, University of Alberta, CanadaReviewed by:
Angelica Thomaz Vieira, Federal University of Minas Gerais, BrazilManoj Kumar, ICMR-National Institute for Research in Environmental Health, India
Laure B. Bindels, Catholic University of Louvain, Belgium
Copyright © 2020 Spacova, Dodiya, Happel, Strain, Vandenheuvel, Wang and Reid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irina Spacova, aXJpbmEuc3BhY292YUB1YW50d2VycGVuLmJl
†These authors have contributed equally to this work and share second authorship
 Irina Spacova
Irina Spacova Hemraj B. Dodiya
Hemraj B. Dodiya Anna-Ursula Happel
Anna-Ursula Happel Conall Strain
Conall Strain Dieter Vandenheuvel1,6†
Dieter Vandenheuvel1,6† Xuedan Wang
Xuedan Wang Gregor Reid
Gregor Reid