- 1Department of Biology, University of Florence, Sesto Fiorentino, Italy
- 2G.I.D.A. SpA, Prato, Italy
Improvement of industrial productions through more environment-friendly processes is a hot topic. In particular, land and marine environment pollution is a main concern, considering that recalcitrant compounds can be spread and persist for a long time. In this context, an efficient and cost-effective treatment of wastewater derived from industrial applications is crucial. Phytodepuration has been considered as a possible solution and it is based on the use of plants and their associated microorganisms to remove and/or transform pollutants. In this work we investigated the culturable microbiota of Phragmites australis roots, sampled from the constructed wetlands (CWs) pilot plant in the G.I.D.A. SpA wastewater treatment plant (WWTP) of Calice (Prato, Tuscany, Italy) before and after the CW activation in order to check how the influx of wastewater might affect the resident bacterial community. P. australis specimens were sampled and a panel of 294 culturable bacteria were isolated and characterized. This allowed to identify the dynamics of the microbiota composition triggered by the presence of wastewater. 27 out of 37 bacterial genera detected were exclusively associated to wastewater, and Pseudomonas was constantly the most represented genus. Moreover, isolates were assayed for their resistance against eight different antibiotics and synthetic wastewater (SWW). Data obtained revealed the presence of resistant phenotypes, including multi-drug resistant bacteria, and a general trend regarding the temporal evolution of resistance patterns: indeed, a direct correlation linking the appearance of antibiotic- and SWW-resistance with the time of exposure to wastewater was observed. In particular, nine isolates showed an interesting behavior since their growth was positively affected by the highest concentrations of SWW. Noteworthy, this study is among the few investigating the P. australis microbiota prior to the plant activation.
Introduction
Plants and microorganisms have been living in association for a very long time. In fact, arbuscular mycorrhizal mutualism is believed to have had a key importance in the terrestrialization process and in the evolution and diversification of plant phototrophs (Selosse and Le Tacon, 1998; Heckman et al., 2001). Different microorganisms (bacteria and fungi) can establish (more or less) deep associations with plants; some of them exhibit an endophytic lifestyle, in that they colonize plant tissues internally, although a more specific definition of endophytes states that they are organisms which, at some moment of their life cycle, colonize the internal plant tissues without causing any type of harm to the host (Patriquin and Döbereiner, 1978). Potential endophytes often inhabit the surrounding soil, especially rhizosphere, from where they can enter plant tissues switching to an endophytic lifestyle. They may thus enter plant tissues through wounds, germinating radicles, emergence points of lateral roots or root elongation and differentiation zones (Reinhold-Hurek et al., 2006; Sturz et al., 2010). Once inside, bacteria adapt to different environmental conditions (e.g., pH, osmotic pressure, carbon source, and availability of oxygen) and overcome plant defense responses (Zeidler et al., 2004).
The plant host and the bacterial endophytes create a mutualistic interaction, with bacteria gaining nutrients and a niche to colonize (Sturz et al., 2010). Even though the exact role of endophytes within plant tissues has not been fully understood yet, it is well-established that in many cases endophytes are beneficial to plants (Schlaeppi and Bulgarelli, 2015; Wani et al., 2015). The most common functions observed for bacterial endophytes are (i) uptake of nutrients (e.g., N, P, S, Mg, Fe, and Ca; Duijff et al., 1999; Çakmakçi et al., 2006), (ii) biosynthesis of phytohormones promoting plant growth (Spaepen et al., 2007), (iii) 1-aminocyclopropane-1-carboxylate deaminase activity (ACC; Glick et al., 2007), (iv) nitrogen fixation (Doty et al., 2009), (v) prevention of pathogenic infections (Weller, 2007; Pérez-García et al., 2011), (vi) acceleration of seedling emergence (Hardoim et al., 2008), and (vii) tolerance to pollution and stresses (Ryan et al., 2008; Lugtenberg and Kamilova, 2014).
In the context of the present work, particularly important is the ability of plant-associated bacteria to increase tolerance to pollution and/or increase the ability of plants to detoxify polluted environments. Environmental pollution, especially water pollution, represents a concern of considerable prominence in the current society. In this regard, phytodepuration is the over-arching term for a group of technologies that utilizes plants and the associated rhizospheric microorganisms to remove and/or transform contaminants leached from soils/sediments and from used water streams (He et al., 2017; Saxena et al., 2020). It represents an environmental-friendly and a valuable solution for environmental cleanup, in particular for wastewater treatment, and it is popular because of its cost effectiveness, aesthetic advantages, and long-term applicability (Puvanakrishnan et al., 2019). In the present manuscript the term “phytodepuration” has been used to indicate specifically the remediation process regarding water and wastewater, rather than “phytoremediation,” which has a more general meaning, encompassing applications regarding, for example, soil remediation.
The constructed wetlands (CW) are engineered systems designed to mimic the self-purification processes of natural wetlands. For decades, CW have been successfully used for treating wastewater of different origins and have been identified as a sustainable wastewater management option worldwide (Wang et al., 2017), demonstrating their ability to eliminate diffuse pollutants from urban, rural, and industrial emissions. In literature, the effectiveness of the use of CW in the treatment of sewage containing heavy metals and high salinity is reported (Vymazal, 2011). This process is due to the interaction between plants, microorganisms, soil, and polluting substances (Zhou et al., 2013).
In CW, the rhizosphere is the mainly involved plant compartment, where multiple different physiochemical and biological processes occur (Stottmeister et al., 2003). The common reed Phragmites australis is one of the most employed plant species, because of its ability to flourish in marshy areas and swamps and the high detoxification and phytodepuration potential. Moreover, it is widely used to treat industrial wastewater containing heavy metals (Zhang et al., 2017). One peculiar characteristic of P. australis is that its internal environment is characterized by a relatively constant osmotic gradient determined by the downward transportation of Na+ from stems to roots (Vasquez et al., 2006). For this reason, P. australis is also well-adapted to salty ecosystems. In CW, vegetation is responsible for only a small amount of pollutant removal (0.02%; Zhang et al., 2017), while its main function is to provide additional oxygen and organic matter for microbial growth (Zhou et al., 2013). Indeed, microorganisms have been described as the main actors of pollutant removal in CW (Zhang et al., 2017). Phytodepuration has proved to effectively remove or neutralize hazardous environmental contaminants and it is predicted to have a growing application in the next years. However, this process presents some limitations, such as the toxic effects of pollutants on the growth and health of the plants (Glick, 2003). In fact, plant biomass is critical for phytodepuration (Germaine et al., 2010) and even hyperaccumulator plants, which can accumulate concentrations of toxic elements up to 100-fold higher than other plant species, usually exhibit a reduced growth. Also, phytodepuration may determine the accumulation of contaminants in plant tissues, which, in turn, is responsible for ecological and airborne exposure issues (Ho et al., 2012). In this scenario, rhizobacteria and endophytic bacteria can aid plants by supporting their growth (Tesar et al., 2002; Shaw and Burns, 2004; Chaudhry et al., 2005), reducing phytotoxicity effects, increasing pollutant uptake and removal (Glick and Stearns, 2011), reducing the release of toxic compounds into the atmosphere (Barac et al., 2004), removing contaminants and/or accumulating heavy metals (Germaine et al., 2010; Ho et al., 2012).
The experimental plant of Calice (Prato, Italy), managed by G.I.D.A. SpA, has therefore set itself as a goal to verify the action of this association in tertiary treatment of landfill leachate (LFL; Coppini et al., 2019).
The aim of this work was to characterize the cultivable bacterial communities associated to the roots of P. australis plants in Calice CW and to analyze their temporal dynamics before and after the activation of the plant for 22 months. This allowed the assessment of wastewater influx effect in shaping the composition of pre-existing bacterial communities. Moreover, bacteria isolated from roots were tested for their ability to grow in the presence of synthetic wastewater (SWW), along with their resistance against a panel of antibiotics commonly used to treat infections in humans. To the best of our knowledge, this work is among the few taking in consideration the bacterial composition of endophytes before the activation of CW, and likely the first regarding this issue in P. australis.
Materials and Methods
Site Description
P. australis plants were obtained from the CWs pilot plant managed by G.I.D.A. SpA and located at Calice Wastewater Treatment Plant (WWTP) in Prato, Italy. The CW of Calice was designed for the tertiary treatment of LFL. This CW is located downstream of a membrane bio-reactor (MBR) designed to pretreat a mixture of LFLs before their discharging in the main line of a full-scale WWTP, which treats both urban and industrial wastewater.
Constructed wetlands medium, used as substrate for the growth of P. australis, consists of four layers of gravel and sand; proceeding from the top to the bottom they are (thickness of layers and diameter range of particles are reported in brackets, respectively): gravel (20 cm; 5–10 mm) – sand (60 cm; 0.1–0.4 mm and 0.02–0.1 mm) – gravel (10 cm; 5–10 mm) – gravel (10 cm; 40–70 mm). CW implant was designed with two parallel lines, named “Line A” and “Line B,” respectively, with a total surface area of 1,620 m2. Each line is a two-stage subsurface flow system (SFS), consisting of a vertical system (SFS-v) followed by a horizontal one (SFS-h). The SFS-v of Line A is subdivided into four parallel separated tanks (SFS-v1, SFS-v2, SFS-v3 e SFS-v4), while the SFS-v of Line B is composed by two tanks (SFS-v5 e SFS-v6). Furthermore, both SFS-h lines are composed by three tanks, each one receiving the same hydraulic load. The maximum hydraulic load supplied to the entire system was 95 m3/day corresponding to a 1.9-day Hydraulic Retention Time for the horizontal stage (Coppini et al., 2019).
Sampling and Isolation of Bacteria
Samples from the roots of P. australis were collected using sterile plastic bags and immediately transported to the laboratory for the subsequent processing. All procedures described hereinafter were carried out under sterile conditions to avoid external contaminations. Samples of three different specimens of P. australis growing in three different tanks were grouped and pooled before starting any procedure. Two pools were obtained from both SFS-v and SFS-h for each sampling campaign. 1 g of fresh tissue from each pool was surface-sterilized with 1% v/v HClO solution at room temperature to remove epiphytic bacteria and then washed three times with sterile water. Aliquots of 100 μL of water from the last wash were plated in triplicate as sterility controls. Subsequently, samples were homogenously pottered in a sterile mortar with the addition of 2 mL of 0.9% w/v NaCl sterile solution. Serial dilutions of tissue extracts were plated in triplicate on trypticase soy agar (TSA) medium (Biolife) and incubated at 30°C for 48 h. The total number of aerobic heterotrophic fast-growing bacteria of each sample was expressed as colony forming units per gram of roots (CFU/g), and it was determined as an average of three replicates. Isolated bacteria were name-coded according to the portion of the CW from whom they were isolated (V for the SFS-v and H for the SFS-h, respectively) and the pool of origin collected during the five samplings (1–2, 3–4, 5–6, 7–8, and 9–10 for the first, second, third, fourth, and fifth sampling, respectively).
Amplification and Sequencing of 16S rRNA Gene
Polymerase chain reaction (PCR) were performed to amplify the 16S rRNA coding gene. 2 μL of colony thermal lysate were used as template for a PCR in 1X DreamTaq Buffer (Thermo Scientific) containing 200 μM of each dNTPs, 0.2 μM of primers P0 (5′-GAGAGTTTGATCCTGGCTCAG-3′) and P6 (5′-CTACGGCTACCTTGTTACGA-3′; Di Cello and Fani, 1996), 2 U of DreamTaq DNA Polymerase (Thermo Scientific) in a final volume of 25 μl. The PCR cycling for 16S rRNA gene amplification was 95°C for 3 min followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, then a final extension at 72°C for 10 min. A Bio-Rad T100 thermal cycler was used. Sequencing of 16S rRNA gene was performed by IGA Technology Services Srl (Udine, Italy).
Taxonomic and Phylogenetic Analyses
Taxonomic affiliation of isolates was determined through the alignment of sequences to those of type strains downloaded from the ribosomal database project (RDP; Cole et al., 2014) using BioEdit (Hall, 1999). The obtained alignment was then used to build a phylogenetic tree through MEGA7 (Kumar et al., 2016), applying the Neighbor-Joining algorithm with a 1000-bootstrap resampling.
Antibiotic Resistance Assay
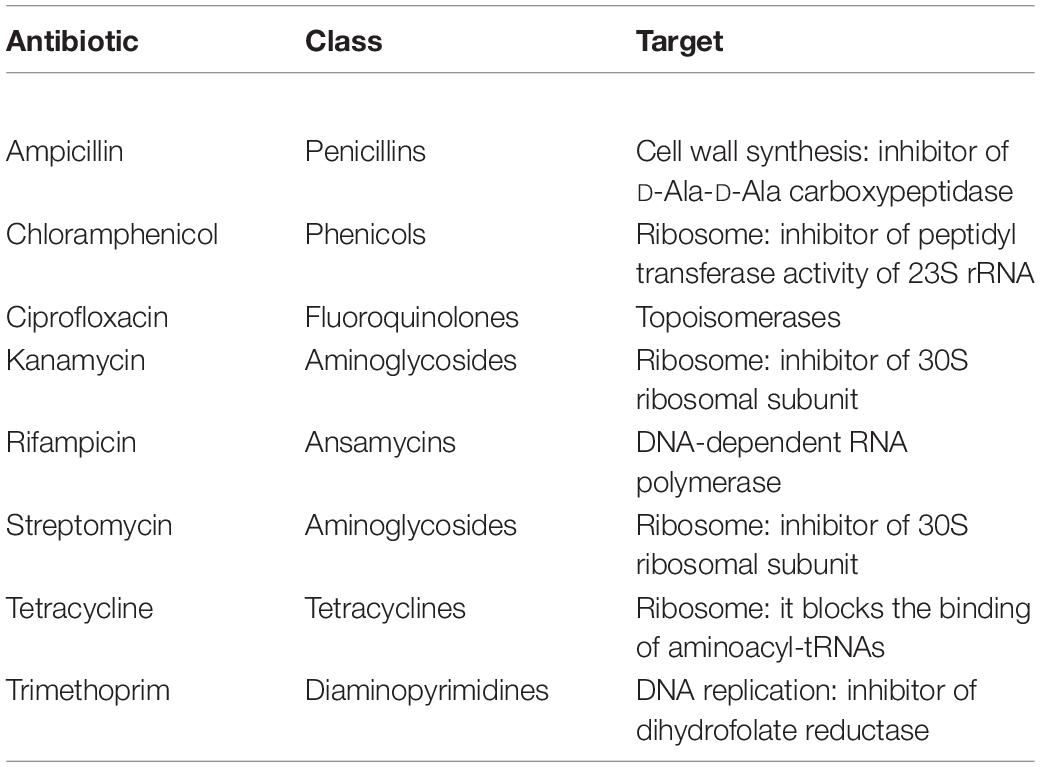
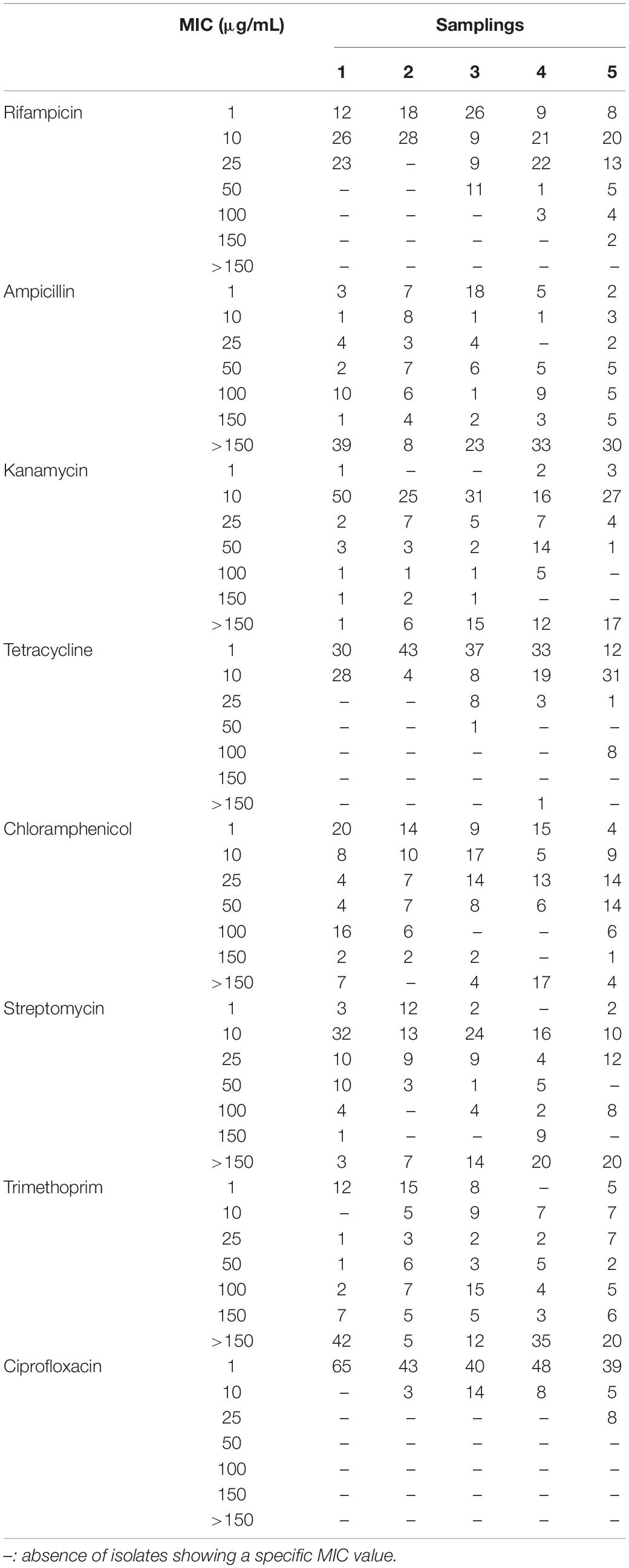
Isolates were tested for their resistance against eight antibiotics (i.e., rifampicin, ampicillin, kanamycin, tetracycline, chloramphenicol, streptomycin, trimethoprim and ciprofloxacin) at six different concentrations (i.e., 1 – 10 – 25 – 50 – 100 – 150 μg/mL; Table 1). Bacteria were firstly grown overnight on TSA (Biolife) at 30°C, then a single colony was resuspended in 100 μL of 0.9 w/v NaCl sterile solution. The obtained suspensions were streaked on Mueller–Hinton II Agar (Biolife) plates supplemented with the tested antibiotics. Bacteria were also cultivated on the same medium in the absence of antibiotics, using these cultures as control to evaluate the presence of growth inhibition in presence of antibiotics. All plates were incubated at 30°C and growth performances were evaluated after 48 h. The minimal inhibitory concentration (MIC) value for each antibiotic was considered as the lowest concentration of the compound that inhibited visible growth of the tested isolate.
Growth in Presence of Synthetic Wastewater
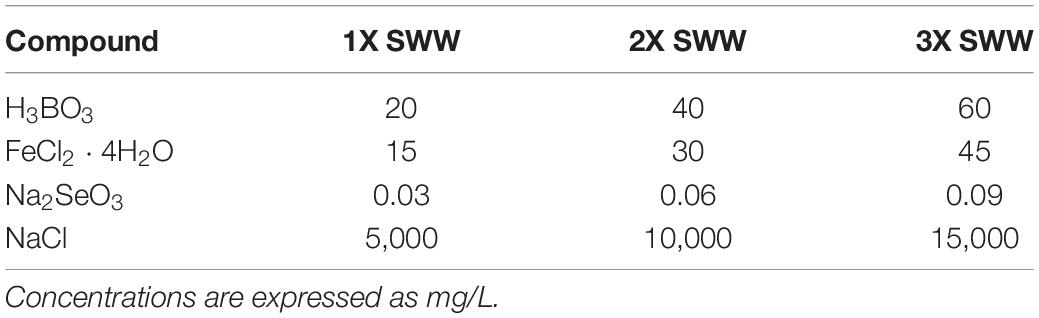
Growth of strains isolated from roots of P. australis in presence of SWW was assayed through the broth microdilution methods (Wiegand et al., 2008) using trypticase soy broth (TSB) medium (Biolife). The bacterial inoculum for the experiment was prepared by dissolving an isolated bacterial colony in 10 ml of TSB medium after 24 h-growth at 30°C on TSA. The inoculum was incubated overnight at 30°C under shaking. Upon incubation, absorbance at 600 nm was measured and adjusted to 0.1. The experiment was performed using 96-well plates. Each well contained 10 μL of bacterial inoculum, 80 μL of TSB medium and 10 μL of 10X, 20X, and 30X SWW, to reach the final concentration of 1X, 2X, and 3X, respectively. The composition of SWWs used for this assay is shown in Table 2. Growth performances in presence of SWW were evaluated calculating the ratio between the OD600 of cultures in presence of SWW (herein after indicated as OD600SWW) and OD600 of controls (i.e., bacteria grown in TSB lacking SWW). Bacterial isolates were considered sensitive to SWW when this parameter assumed values <0.7, while they were evaluated as resistant when it was >1.3.
Results
Bacterial Counts
P. australis plants were sampled from the CW in Calice during a period of 22 months, spanning from March 2017 to December 2018; 5 samplings were conducted, with the first one (i.e., March 2017) performed before the activation of the CW (Table 3). This experimental strategy allowed us to compare the composition of the cultivable bacterial community associated to the P. australis roots before and after the beginning of wastewater influx. The titer of viable bacteria associated to roots was determined as described in section “Materials and Methods”. Data obtained revealed that there were no great differences between the CFU counts in SFS-v and SFS-h, exception for the third and fourth samplings in which CFU values were higher in SFS-h (Table 3).
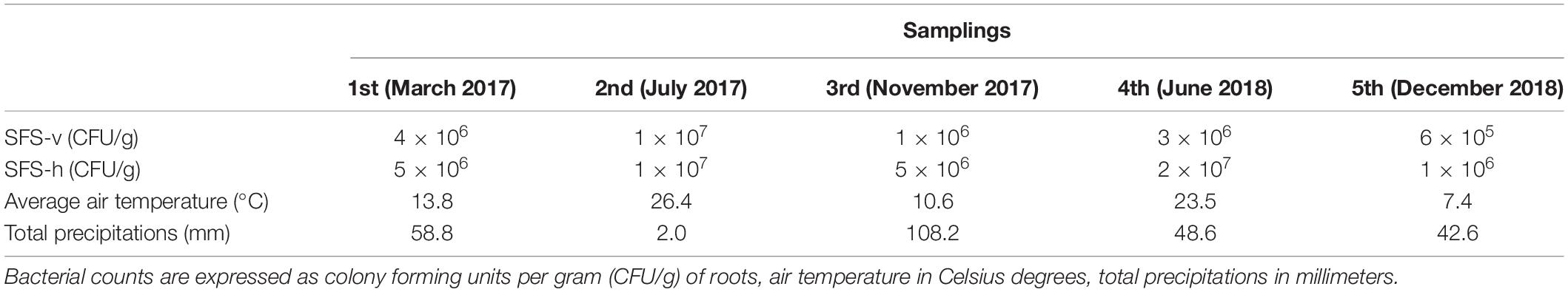
Table 3. Bacterial counts in roots of Phragmites australis collected during the five samplings and meteorological conditions registered monthly.
In general, bacterial load was quite constant during the experiment and fluctuations might be related to different factors, such as wastewater composition, frequency of raining, and/or seasonal variations. Indeed, it is likely that the weather exerted a main effect on bacterial growth since the highest bacterial loads were observed during summer (i.e., second and fourth samplings, respectively), when the higher temperatures probably facilitated bacterial growth and the poor precipitations probably caused a higher concentration of wastewater.
Taxonomic Affiliation of Cultivable Bacteria
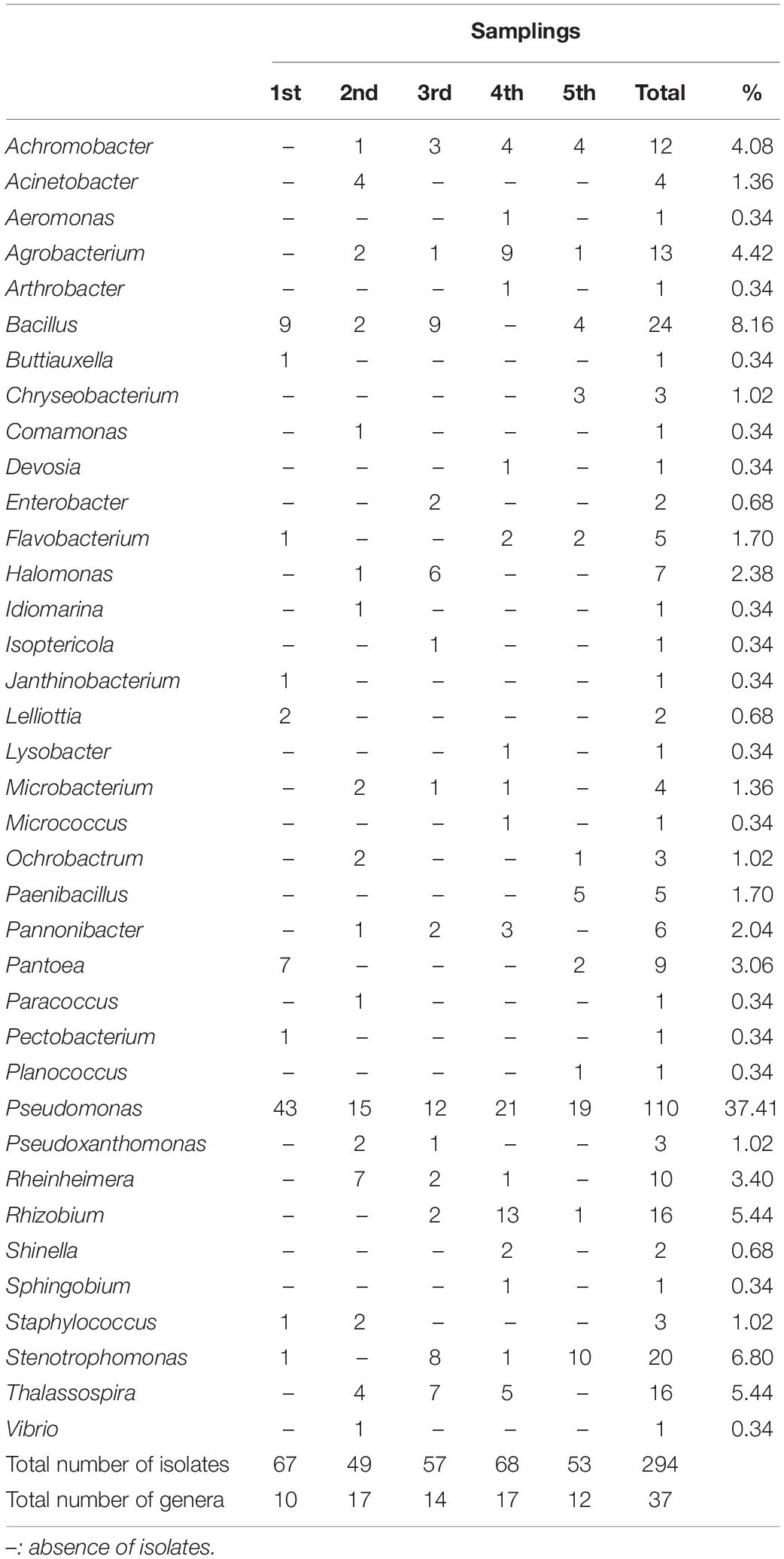
A total of 294 isolates (67, 49, 57, 68, and 53 from the first, second, third, fourth, and fifth sampling, respectively) were isolated from the P. australis roots. The attention was focused on bacteria isolated from this plant compartment because it has been reported that it is primarily involved in the depuration process (Riva et al., 2020). Each of the 294 isolates underwent a taxonomic characterization; to this purpose the amplification, sequencing, and analysis of 16S rRNA coding gene(s) were performed as described in section “Materials and Methods.”
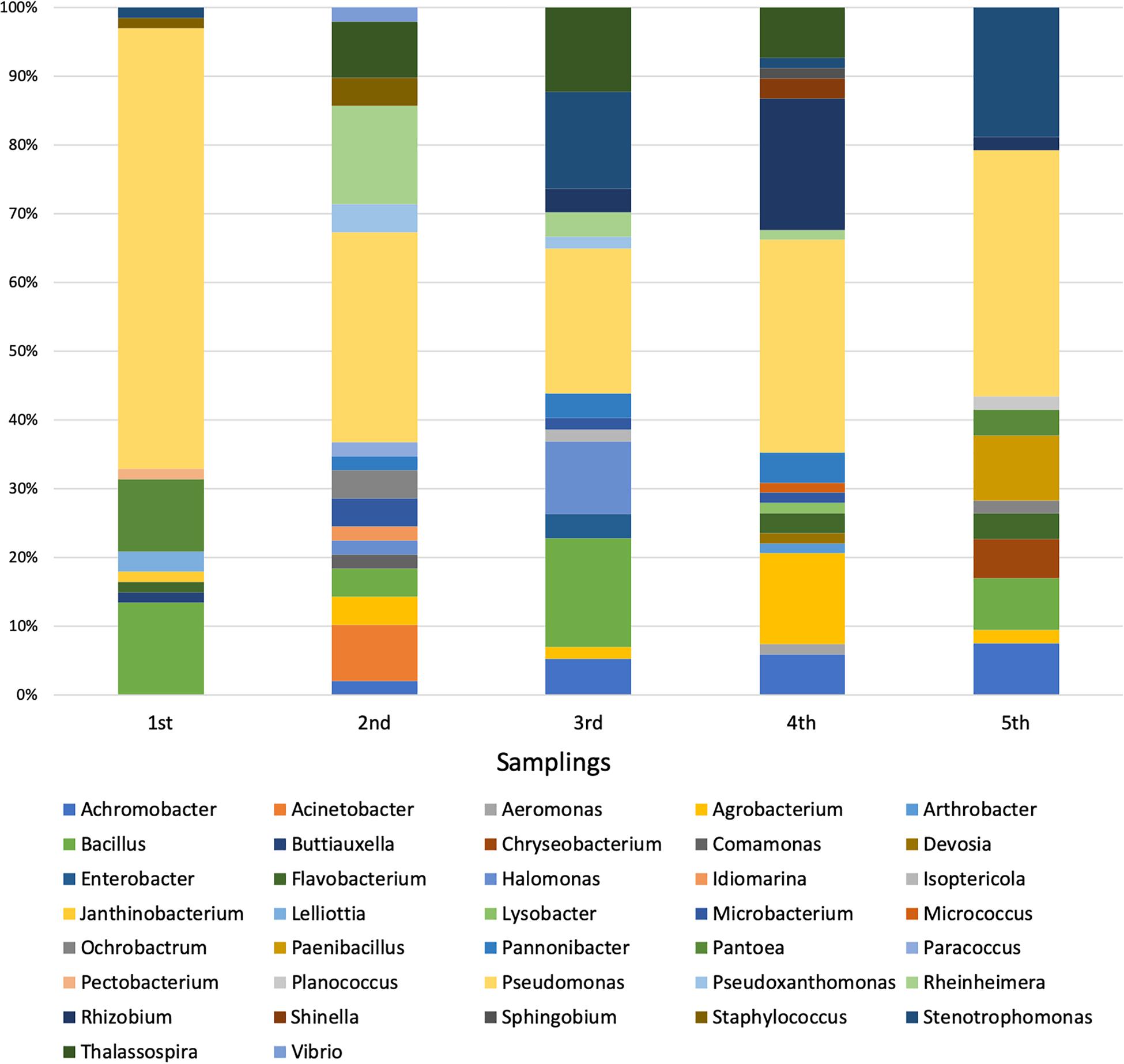
Each sequence was submitted to Genbank and was assigned the accession number reported in Supplementary Table S1. The comparative analysis of each sequence with those available in databases allowed to split the 294 isolates into 37 different genera (Supplementary Figures S1–S30). The analysis revealed that 254 isolates were Gram-negative while 40 were Gram-positive bacteria. Moreover, a total of four different phyla were represented, with 246 belonging to Proteobacteria (59 Alphaproteobacteria, 14 Betaproteobacteria, and 173 Gammaproteobacteria), 8 to Bacteroidetes (all belonging to Flavobacteriia class), 33 to Firmicutes (all belonging to Bacilli class), and 7 to Actinobacteria (all belonging to Actinobacteria class). The most represented genus was Pseudomonas, whose members accounted for 37% of all isolates, as shown in Table 4. The abundance of Pseudomonas was not directly related to the activation of the CW, because it was the most represented genus even before the influx of wastewater (Figure 1).
Among the 37 genera, only 4 were exclusively present during the first sampling (i.e., Buttiauxella, Janthinobacterium, Lelliottia, and Pectobacterium), while 27 started being present from the second one on (i.e., Achromobacter, Acinetobacter, Aeromonas, Agrobacterium, Arthrobacter, Chryseobacterium, Comamonas, Devosia, Enterobacter, Halomonas, Idiomarina, Isoptericola, Lysobacter, Microbacterium, Micrococcus, Ochrobactrum, Paenibacillus, Pannonibacter, Paracoccus, Planococcus, Pseudoxanthomonas, Rheinheimera, Rhizobium, Shinella, Sphingobium, Thalassospira, and Vibrio). Hence, it is possible that the bacteria belonging to these 27 genera might derive from the wastewater, although it cannot be established whether they were present in wastewater with either urban or industrial origin. In addition to this, we cannot a priori exclude the possibility that they were already present in the pre-existing community (even though in low percentage) and that the presence of the wastewater might have exerted a selective pressure favoring their reproduction. In most cases, the phylogenetic trees showed a narrow taxonomic distribution of isolates, which clustered together in the same branch. For example, all Acinetobacter strains were phylogenetically close to A. haemolyticus (Supplementary Figure S1), all Achromobacter isolates formed a distinct cluster and were close to A. spanius (Supplementary Figure S5), all Chryseobacterium were related to C. indoltheticum (Supplementary Figure S7), all Paenibacillus belonged to the same cluster and were close to P. tundrae (Supplementary Figure S19), all Pannonibacter were affiliated to P. phragmitetus (Supplementary Figure S24), and all Thalassospira isolates formed a separate branch in the phylogenetic tree (Supplementary Figure S29). Moreover, Rheinhemera isolates distributed in close branches which included R. aquimaris, R. pacifica, and R. nanhaiensis (Supplementary Figure S22). On the contrary, a higher phylogenetic diversity was observed in the case of Bacillus (Supplementary Figure S4), Enterobacteriales (Supplementary Figure S10), Pseudomonas (Supplementary Figure S20), Rhizobiales (Supplementary Figure S23), and Stenotrophomonas (Supplementary Figure S28). However, with the exception of the bacteria belonging to the Enterobacteriales order, in the case of these genera the formation of distinct clusters was observed.
Resistance Against Antibiotics
All 294 isolates were tested for their resistance against a panel of eight antibiotics used to treat human infections as described in section “Materials and Methods.” These compounds were chosen because they are representatives of diverse antibiotic classes and they are directed toward different cellular targets (Table 1). Data obtained are shown in Table 5 and Supplementary Table S2 and revealed that, overall, the most effective antibiotics were rifampicin, tetracycline and, above all, ciprofloxacin. On the contrary, the more tolerated antibiotic was ampicillin, especially in the case of Pseudomonas and Stenotrophomonas (Supplementary Table S2). Although resistant bacteria were isolated since the first sampling, a correlation between the time of exposure to the wastewater (i.e., earlier vs later samplings) and the increase of MIC values was observed for almost all antibiotics (Table 5 and Supplementary Table S2).
On one hand, tests with rifampicin, ciprofloxacin and tetracycline showed a progressive increase in the number of isolates having the highest MIC values going from the first to the fifth sampling. On the other one, although MIC values were not determined in the assayed conditions because isolates were able to grow even in presence of the highest concentration of antibiotic, in the case of kanamycin and streptomycin the number of isolates with MIC > 150 μg/mL increased during time. Finally, clear trends were not detected using ampicillin, chloramphenicol and trimethoprim: indeed, there were bacteria able to survive in the presence of the highest concentration since the first sampling and, also, the frequency of resistant isolates was not subjected to temporal variations.
According to the MIC breakpoints provided by the European Committee on Antimicrobial Susceptibility Testing (EUCAST – Breakpoint tables for interpretation of MICs and zone diameters; Version 10.0, 20201), relatively to the antibiotics assayed in this work and limiting to the species reported by EUCAST, six isolates could be defined as multi-drug resistant strains, since they were resistant against at least three different antibiotics (Supplementary Table S2). In detail, two Lelliottia (V2R14 and H1R21) and two Enterobacter (H5R6 and H5R7) isolates were resistant to ampicillin, chloramphenicol, and ciprofloxacin; lastly, the two Pantoea isolates H9R2 and H9R15 that were resistant to ampicillin, chloramphenicol, trimethoprim, and ciprofloxacin.
Growth in the Presence of SWW
The 294 isolated P. australis root-associated endophytes were also tested for their ability to grow in the presence of SWW containing B, Fe, and Se since these elements are critical for the WWTP studied in this work. Selection of bacteria able to grow efficiently in the presence of these compounds is of relevant interest because CW might be enriched with these more tolerant microorganisms, which, in turn, might increase the pollutant removal efficiency in wastewater. All bacterial isolates were assayed for their growth in TSB medium supplemented with three different concentration of SWW: 1X (i.e., a mix of H3BO3, FeCl2 ⋅ 4H2O, Na2SeO3, and NaCl at the maximum concentrations allowed by law for sewer emission), and 2X and 3X in which TSB medium was supplemented with two- and threefold higher concentrations of 1X SWW, respectively.
In general, the presence of 1X SWW did not alter the growth of isolates, indicating that these endophytes can tolerate the presence of the tested compounds (Figure 2 and Supplementary Table S3). The analysis of data shown in Supplementary Table S3 and Figure 2 also revealed that 211 bacterial isolates were able to grow efficiently also in the presence of either 2X SWW or 3X SWW. Interestingly, eight isolates (i.e., V2R8, H3R17, H4R18, V6R1, V5R1, V8R24, H9R1, and H10R8), belonging to the genera Bacillus, Planococcus, Pseudomonas, and Rheinheimera, showed a positive correlation between growth and concentration of SWW: indeed, the higher the SWW concentration, the higher the growth of these bacteria. This finding suggests that these bacteria could represent good candidates for future applications and for improvements of phytodepuration efficiency and pollutant removal. Moreover, the isolate H9R16 deserves further investigations, since it showed the highest growth increase (about 350%) in presence of SWW. The analysis of the 16S rDNA phylogenetic tree revealed that it joined bacteria belonging to the Bacillus gibsonii species (Supplementary Figure S4), alkaliphilic bacteria exploited for production of alkaline proteases (Martinez et al., 2013; Deng et al., 2014).
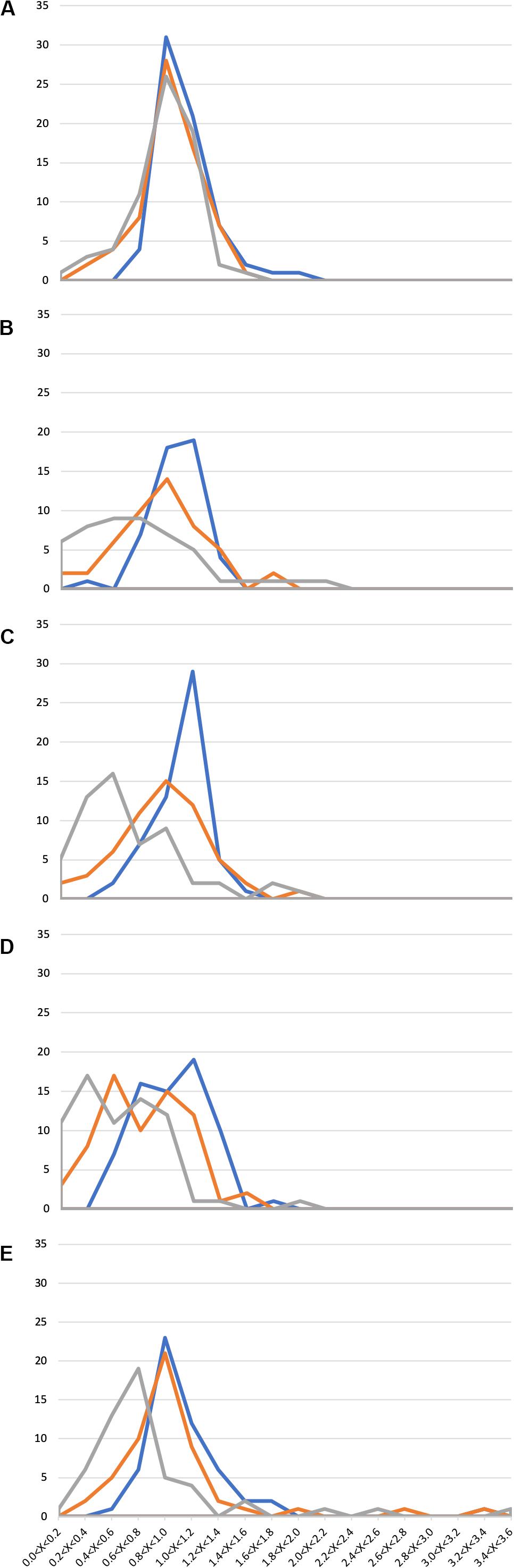
Figure 2. Growth of 294 Phragmites australis root-associated bacteria in the presence of synthetic wastewater (SWW). Ranges of OD600SWW/OD600 values are reported in the x axis, while the number of isolates is reported in the y axis. (A) 1st sampling; (B) 2nd sampling; (C) 3rd sampling; (D) 4th sampling; and (E) 5th sampling. Blue: 1X SWW; Orange: 2X SWW; Gray: 3X SWW.
Similarly to the case of antibiotic resistance, also in this assay a correlation between the time of exposure to wastewater in the CW and the appearance of more resistant isolates was highlighted. Indeed, a progressive increase of the number of isolates showing an augmented growth (measured as OD600SWW/OD600 ratio as described in section “Materials and Methods”) was observed from the first to the last sampling. For instance, several isolates with an OD600SWW/OD600 > 2.4 (Figure 2E) were detected only during the last sampling.
Changings of resistance profiles along time, considering those against either antibiotics or SWW, were particularly clear in the case of isolates belonging to the genus Pseudomonas. As shown in Figure 3 by the mean of principal component analysis (PCA) performed with the PAST4 software (Hammer et al., 2001), the formation of two different main clusters was observed with the main part of isolates from the first sampling clustering independently from those isolated from all the other four samplings.
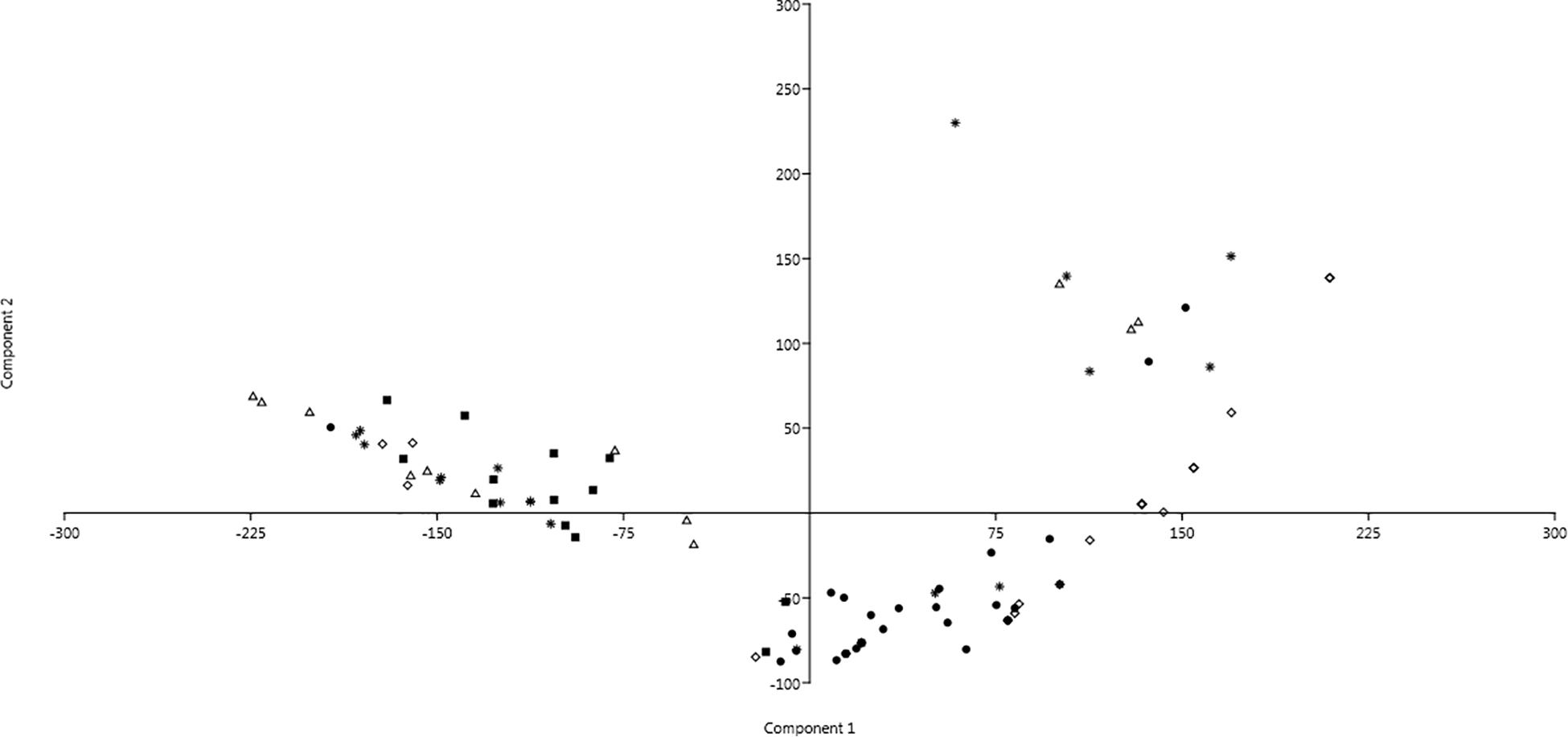
Figure 3. Principal component analysis (PCA) showing profiles of resistance against antibiotics and SWW of isolates belonging to the genus Pseudomonas. Dot: 1st sampling; Filled square: 2nd sampling; Triangle: 3rd sampling; Diamond: 4th sampling; and Star: 5th sampling.
Discussion
The aim of this work was the analysis of the composition, the phenotypic characterization, and the temporal dynamics of the cultivable microbiota associated to the roots of P. australis grown in the CW of Calice (Prato, Italy) before and after the activation of the CW.
The composition of root cultivable microbiota was determined through five samplings spanning from March 2017 to December 2018. We focused on cultivable bacterial communities because their isolation and characterization might permit the identification of strains particularly resistant to the antibiotics and/or to the compounds present in the wastewater. Hence, these strains could be used to construct a “synthetic” consortium that, in turn, might be exploited in pilot-experiments with the goal of increasing the phytodepuration efficiency of the plant. The taxonomic analysis was performed on 294 cultivable bacteria through the analysis of the 16S rRNA genes. Even though we are completely aware that the number of isolates could not be representative of the entire community, the analysis performed gave useful hints on the effect of the activation of CW on bacterial community composition. Data obtained revealed that the wastewater income exerted a shaping effect on the bacterial composition. Overall, 37 bacterial genera were disclosed and six of them were detected both before and after the activation of CW. Moreover, bacteria belonging to 27 different genera were detected only after the activation of the CW, while, on the contrary, 4 genera that were present at the beginning were not found in the following samplings. The most represented genus in all five samplings was Pseudomonas, which accounted for the 37% of all isolates.
As it might be expected, the effect of the wastewater income was exerted not only at the taxonomic level, but also at the phenotypic one. Each of the 294 bacterial isolates was assayed for its resistance against a panel of eight antibiotics belonging to different chemical classes and acting toward different cellular targets. This analysis was performed since it is known that WWTP are reservoirs of antibiotic resistance genes and/or resistant bacteria. Data obtained revealed the presence of several resistant (or multi-resistant) strains and, mostly important, that the number of antibiotic resistant isolates and the degree of antibiotic resistance increased over time, from the first to the last sampling. This strongly suggests that the wastewater income might generate a selective pressure favoring the growth of those isolates intrinsically resistant to antibiotics, even though it cannot be a priori excluded the possibility of horizontal gene transfer (HGT) events and/or the acquisition of resistance through mutations in chromosomal genes.
All the 294 isolates were assayed also for their ability to grow in SWW containing three different concentrations of B, Se, Fe, and NaCl. Analogously to the antibiotic resistance pattern, we detected a similar correlation between SWW exposure and the ability to grow in its presence. Interestingly, among isolates able to grow in the presence of these compounds, nine of them showed an increasing growth at the highest concentrations of SWW: these isolates will deserve a specific focus to identify the molecular mechanisms behind this intriguing behavior. Moreover, a PCA carried out on Pseudomonas strains, isolated from all the five samplings, furtherly suggested that the main event changing the resistance patterns against antibiotics and SWW was the activation of the plant (i.e., when conveyance of the permeate into the tanks occurred). The characterization of heavy metal resistant strains may be crucial to better understand the diffusion of antibiotic resistance genes in the environment. As a matter of fact, it has been reported that the occurrence of multiple heavy metal resistance markers is associated with the onset of antibiotic resistance (Wales and Davies, 2015; Wu et al., 2018; Zhu et al., 2019). This might be due to the co-localization of resistance genes against antibiotics and heavy metals in the same mobile genetic element(s) and, as a consequence, the accumulation of heavy metals in the environment can cause the selection of antibiotic resistant species. So, the dissemination of these heavy metal resistance genes represents an issue that should not be underestimated. Moreover, monitoring the presence of bacteria resistant to antibiotics and/or heavy metals specifically in WWTPs should be considered as a priority to contrast the spreading of multi-drug resistant (MDR) pathogens. Indeed, WWTPs represent hotspots for HGT events, because of the mixing of bacteria from diverse sources (e.g., households, hospitals, industries, etc.), the high bacterial densities, stressful conditions triggering SOS responses and presence of antibiotics at sublethal concentrations (Karkman et al., 2018). It must be also considered that although HGT occurring in WWTPs might not directly regard human pathogens, these could acquire resistance markers from harmless bacteria which act as vectors as soon as effluent is released in the environment (Manaia, 2017).
To deeply characterize this phenomenon, future work could take advantage of emulsion, paired isolation and concatenation PCR (epicPCR) as previously reported (Spencer et al., 2016; Hultman et al., 2018). Indeed, this kind of analysis could allow the “tagging” of resistance genes with phylogenetic markers, such as 16S rRNA gene, helping to compare these pairs in wastewater entering the WWTP and in effluents. However, this would be limited to target resistance markers with known sequence and for whom it is thereof possible to design specific primers.
Conclusion
The experimental approach used in this work revealed that the cultivable bacterial community existing prior to the plant activation underwent fluctuations in terms of both taxonomy and resistance to antibiotics and SWW compounds. As it might be expected, the influx of wastewater exerted a selective pressure on the resident bacterial community, selecting and/or bringing bacterial strains progressively more resistant to SWW and/or antibiotics. We are completely aware that the analysis of the entire community (both cultivable and uncultivable) might give more detailed insights into the composition of the total community. In spite of this, only the selection of particular cultivable strains, i.e., more resistant to SWW and antibiotics, can permit the formulation of a synthetic bacterial community to improve the phytodepuration properties of P. australis.
Data Availability Statement
The datasets generated in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MK110895, MK110946, MK110896, MK110920, MK110921, MK110947, MK110948, MK110922, MK110897, MK110949, MK110898, MK110950, MK110899, MK110959, MK110923, MK110925, MK110924, MK110960, MK110926, MK110945, MK110957, MK110927, MK110928, MK110929, MK110930, MK110931, MK110932, MK110900, MK110901, MK110902, MK110933, MK110934, MK110935, MK110958, MK110936, MK110937, MK110938, MK110939, MK110940, MK110941, MK110942, MK110943, MK110903, MK110904, MK110905, MK110906, MK110907, MK110908, MK110909, MK110910, MK110911, MK110912, MK110951, MK110913, MK110914, MK110915, MK110952, MK110916, MK110953, MK110917, MK110954, MK110918, MK110944, MK110961, MK110919, MK110955, MK110956, MK134509, MK134489, MK134488, MK134487, MK134486, MK134554, MK134508, MK134547, MK134496, MK138850, MK134502, MK134511, MK134551, MK134555, MK134559, MK134558, MK134549, MK138851, MK134553, MK134542, MK134557, MK134546, MK134544, MK134543, MK134541, MK134540, MK134510, MK134497, MK134490, MK134495, MK134494, MK134493, MK134499, MK134505, MK134552, MK134556, MK134545, MK134548, MK134500, MT165525, MK134507, MK134504, MK134503, MK134550, MK134501, MK134498, MK134506, MK134492, MK134491, MK134518, MK130934, MK130935, MK134524, MK134539, MK134538, MK134515, MK138852, MK134526, MK130907, MK130906, MK130937, MK134534, MK134533, MK138853, MK130915, MK130913, MK130910, MK130921, MK130917, MK134514, MK130914, MK134516, MK130911, MK134532, MK134531, MK134530, MK134528, MK134485, MK138854, MK130912, MK130908, MK130920, MK130932, MK130933, MK130936, MK134513, MK130922, MK130919, MK134521, MK134512, MK134537, MK134536, MK134535, MK134522, MK134519, MK134517, MK134529, MK134525, MK134523, MK134520, MK134527, MK130916, MK130931, MK130909, MK130923, MK130918, MK130945, MK130957, MK130901, MK130905, MK133358, MK138868, MK138869, MK138870, MK138872, MK138874, MK138875, MK130924, MK130928, MK138881, MK138862, MK130903, MK138867, MK138878, MK138861, MK130939, MK138863, MK130949, MK130953, MK130904, MK138876, MK138879, MK130926, MK130927, MK130929, MK138880, MK130930, MK138882, MK138883, MK138884, MK138885, MK138886, MK138887, MK130940, MK130941, MK130943, MK138889, MK130944, MK130902, MK130900, MK130946, MK138877, MK138855, MK138856, MK138857, MK138858, MK138859, MK138860, MK138864, MK138865, MK138866, MK138871, MK138873, MK130925, MK130942, MK130948, MK130950, MK130955, MK138888, MK130951, MK130952, MK130954, MK130956, MK130947, MT165526, MT165527, MT165547, MT165551, MT165553, MT165552, MT165528, MT165529, MT165530, MT165531, MT165532, MT165533, MT165534, MT165557, MT165554, MT165535, MT165536, MT165555, MT165558, MT165559, MT165561, MT165548, MT165562, MT165560, MT165549, MT165563, MT165564, MT165537, MT165538, MT165565, MT165539, MT165540, MT165566, MT165570, MT165569, MT165567, MT165571, MT165568, MT165578, MT165550, MT165541, MT165542, MT165543, MT165572, MT165544, MT165545, MT165573, MT165546, MT165574, MT165556, MT165575, MT165576, and MT165577.
Author Contributions
RF, EC, and DF conceived the project. AV, EM, CF, and RF designed the experiments. AV, EM, CF, SV, SD, LC, and SC performed the experiments. RF supervised the experiments. AV, EM, and RF analyzed the results. AV wrote the original draft of the manuscript. AV, SD, LC, SC, CF, EM, EC, and RF reviewed and edited the manuscript. All authors read and approved the manuscript.
Conflict of Interest
EC and DF were employed by G.I.D.A. SpA.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01652/full#supplementary-material
FIGURE S1–S30 | Phylogenetic trees.
TABLE S1 | List of bacteria isolated from roots of Phragmites australis and used in this work. Accession numbers of 16S rRNA gene partial sequences are reported.
TABLE S2 | MIC (μg/ml) values of bacterial isolates described in this work.
TABLE S3 | Growth of 294 bacterial isolates associated to Phragmites australis roots in TSB in presence of three different concentrations of synthetic wastewater (SWW).
Footnotes
References
Barac, T., Taghavi, S., Borremans, B., Provoost, A., Oeyen, L., Colpaert, J. V., et al. (2004). Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 22, 583–588. doi: 10.1038/nbt960
Çakmakçi, R., Dönmez, F., Aydın, A., and Şahin, F. (2006). Growth promotion of plants by plant growth-promoting rhizobacteria under greenhouse and two different field soil conditions. Soil Biol. Biochem. 38, 1482–1487. doi: 10.1016/j.soilbio.2005.09.019
Chaudhry, Q., Blom-Zandstra, M., Gupta, S., and Joner, E. J. (2005). Utilising the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ. Sci. Pollut. Res. Int. 12, 34–48. doi: 10.1065/espr2004.08.213
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Coppini, E., Palli, L., Antal, A., Del Bubba, M., Miceli, E., Fani, R., et al. (2019). Design and start-up of a constructed wetland as tertiary treatment for landfill leachates. Water Sci. Technol. 79, 145–155. doi: 10.2166/wst.2019.030
Deng, A., Zhang, G., Shi, N., Wu, J., Lu, F., and Wen, T. (2014). Secretory expression, functional characterization, and molecular genetic analysis of novel halo-solvent-tolerant protease from Bacillus gibsonii. J. Microbiol. Biotechnol. 24, 197–208. doi: 10.4014/jmb.1308.08094
Di Cello, F., and Fani, R. (1996). A molecular strategy for the study of natural bacterial communities by PCR-based techniques. Minerva Biotecnol. 8, 126–134.
Doty, S. L., Oakley, B., Xin, G., Kang, J. W., Singleton, G., Khan, Z., et al. (2009). Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 47, 23–33. doi: 10.1007/BF03179967
Duijff, B. J., Recorbet, G., Bakker, P. A., Loper, J. E., and Lemanceau, P. (1999). Microbial antagonism at the root level is involved in the suppression of fusarium wilt by the combination of nonpathogenic Fusarium oxysporum Fo47 and Pseudomonas putida WCS358. Phytopathology 89, 1073–1079. doi: 10.1094/PHYTO.1999.89.11.1073
Germaine, K. J., Liu, X., Cabellos, G. G., Hogan, J. P., Ryan, D., and Dowling, D. N. (2010). Bacterial endophyte-enhanced phytoremediation of the organochlorine herbicide 2,4-dichlorophenoxyacetic acid. FEMS Microbiol. Ecol. 57, 302–310. doi: 10.1111/j.1574-6941.2006.00121.x
Glick, B. R. (2003). Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnol. Adv. 21, 383–393. doi: 10.1016/s0734-9750(03)00055-7
Glick, B. R., and Stearns, J. C. (2011). Making phytoremediation work better: maximizing a plant’s growth potential in the midst of adversity. Int. J. Phytoremediation 13(Suppl. 1), 4–16. doi: 10.1080/15226514.2011.568533
Glick, B. R., Todorovic, B., Czarny, J., Cheng, Z., and Mcconkey, B. (2007). Promotion of plant growth by bacterial ACC deaminase. CRC Crit. Rev. Plant Sci. 26, 227–242. doi: 10.1080/07352680701572966
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.12691/ajmr-2-6-8
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Paleontol. Electron. 4, 1–9.
Hardoim, P. R., van Overbeek, L. S., and Elsas, J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. doi: 10.1016/j.tim.2008.07.008
He, Y., Langenhoff, A. A. M., Sutton, N. B., Rijnaarts, H. H. M., Blokland, M. H., Chen, F., et al. (2017). Metabolism of ibuprofen by Phragmites australis: uptake and phytodegradation. Environ. Sci. Technol. 51, 4576–4584. doi: 10.1021/acs.est.7b00458
Heckman, D. S., Geiser, D. M., Eidell, B. R., Stauffer, R. L., Kardos, N. L., and Hedges, S. B. (2001). Molecular evidence for the early colonization of land by fungi and plants. Science 293, 1129–1133. doi: 10.1126/science.1061457
Ho, Y. N., Mathew, D. C., Hsiao, S. C., Shih, C. H., Chien, M. F., Chiang, H. M., et al. (2012). Selection and application of endophytic bacterium Achromobacter xylosoxidans Strain F3B for improving phytoremediation of phenolic pollutants. J. Hazard. Mater. 21, 43–49. doi: 10.1016/j.jhazmat.2012.03.035
Hultman, J., Tamminen, M., Pärnänen, K., Cairns, J., Karkman, A., and Virta, M. (2018). Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 94:fiy038. doi: 10.1093/femsec/fiy038
Karkman, A., Do, T. T., Walsh, F., and Virta, M. P. J. (2018). Antibiotic-resistance genes in waste water. Trends Microbiol. 26, 220–228. doi: 10.1016/j.tim.2017.09.005
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lugtenberg, B., and Kamilova, F. (2014). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Manaia, C. M. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 25, 173–181. doi: 10.1016/j.tim.2016.11.014
Martinez, R., Jakob, F., Tu, R., Siegert, P., Maurer, K. H., and Schwaneberg, U. (2013). Increasing activity and thermal resistance of Bacillus gibsonii alkaline protease (BgAP) by directed evolution. Biotechnol. Bioeng. 10, 711–720. doi: 10.1002/bit.24766
Patriquin, D. G., and Döbereiner, J. (1978). Light microscopy observations of tetrazolium-reducing bacteria in the endorhizosphere of maize and other grasses in brazil. Can. J. Microbiol. 24, 734–742. doi: 10.1139/m78-122
Pérez-García, A., Romero, D., and de Vicente, A. (2011). Plant protection and growth stimulation by microorganisms: biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotechnol. 22, 187–193. doi: 10.1016/j.copbio.2010.12.003
Puvanakrishnan, R., Sivasubramanian, S., and Hemalatha, T. (2019). Microbes and Enzymes: Basics and Applied. Chennai: MJP Publisher.
Reinhold-Hurek, B., Maes, T., Gemmer, S., Van Montagu, M., and Hurek, T. (2006). An endoglucanase is involved in infection of rice roots by the not-cellulose-metabolizing Endophyte azoarcus sp. strain BH72. Mol. Plant Microbe Interact. 19, 181–188. doi: 10.1094/MPMI-19-0181
Riva, V., Riva, F., Vergani, L., Crotti, E., Borin, S., and Mapelli, F. (2020). Microbial assisted phytodepuration for water reclamation: environmental benefits and threats. Chemosphere 241:124843. doi: 10.1016/j.chemosphere.2019.124843
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Saxena, G., Purchase, D., Mulla, S. I., Saratale, G. D., and Bharagava, R. N. (2020). Phytoremediation of heavy metal-contaminated sites: eco-environmental concerns, field studies, sustainability issues, and future prospects. Rev. Environ. Contam. Toxicol. 249, 71–131. doi: 10.1007/398_2019_24
Schlaeppi, K., and Bulgarelli, D. (2015). The plant microbiome at work. Mol. Plant Microbe Interact. 28, 212–217. doi: 10.1094/MPMI-10-14-0334-FI
Selosse, M. A., and Le Tacon, F. (1998). The land flora: a phototroph–fungus partnership? Trends Ecol. Evol. 13, 15–20. doi: 10.1016/S0169-5347(97)01230-5
Shaw, L. J., and Burns, R. G. (2004). Enhanced mineralization of [U-14C] 2,4-dichlorophenoxyacetic acid in soil from the rhizosphere of trifolium pratense. Appl. Environ. Microbiol. 70, 4766–4774. doi: 10.1128/AEM.70.8.4766-4774.2004
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Spencer, S. J., Tamminen, M. V., Preheim, S. P., Guo, M. T., Briggs, A. W., Brito, I. L., et al. (2016). Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 10, 427–436. doi: 10.1038/ismej.2015.124
Stottmeister, U., Wiessner, A., Kuschk, P., Kappelmeyer, U., Kästner, M., Bederski, O., et al. (2003). Effects of plants and microorganisms in constructed wetlands for wastewater treatment. Biotechnol. Adv. 22, 93–117. doi: 10.1016/j.biotechadv.2003.08.010
Sturz, A. V., Christie, B. R., and Nowak, J. (2010). Bacterial endophytes: potential role in developing sustainable systems of crop production. CRC Crit. Rev. Plant Sci. 19, 1–30. doi: 10.1080/07352680091139169
Tesar, M., Reichenauer, T. G., and Sessitsch, A. (2002). Bacterial rhizosphere populations of black poplar and herbal plants to be used for phytoremediation of diesel fuel. Soil Biol. Biochem. 34, 1883–1892. doi: 10.1016/S0038-0717(02)00202-X
Vasquez, E. A., Glenn, E. P., Guntenspergen, G. R., Brown, J. J., and Nelson, S. G. (2006). Salt tolerance and osmotic adjustment of Spartina alterniflora (Poaceae) and the invasive M Haplotype of Phragmites australis (Poaceae) along a salinity gradient. Am. J. Bot. 93, 1784–1790. doi: 10.3732/ajb.93.12.1784
Vymazal, J. (2011). Constructed wetlands for wastewater treatment: five decades of experience. Environ. Sci. Technol. 45, 61–69. doi: 10.1021/es101403q
Wales, A. D., and Davies, R. H. (2015). Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604. doi: 10.3390/antibiotics4040567
Wang, M., Zhang, D. Q., Dong, J. W., and Tan, S. K. (2017). Constructed wetlands for wastewater treatment in cold climate – A review. J. Environ. Sci. 57, 293–311. doi: 10.1016/j.jes.2016.12.019
Wani, Z. A., Ashraf, N., Mohiuddin, T., and Riyaz-Ul-Hassan, S. (2015). Plant-endophyte symbiosis, an ecological perspective. Appl. Microbiol. Biotechnol. 99, 2955–2965. doi: 10.1007/s00253-015-6487-3
Weller, D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256. doi: 10.1094/PHYTO-97-2-0250
Wiegand, I., Hilpert, K., and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Wu, C., Lin, C., Zhu, X., Liu, H., Zhou, W., Lu, J., et al. (2018). The β-Lactamase Gene Profile And A Plasmid-Carrying Multiple Heavy Metal Resistance Genes of Enterobacter cloacae. Int. J. Genomics 2018:4989602. doi: 10.1155/2018/4989602
Zeidler, D., Zähringer, U., Gerber, I., Dubery, I., Hartung, T., Bors, W., et al. (2004). Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. U.S.A. 101, 15811–15816. doi: 10.1073/pnas.0404536101
Zhang, C., Wang, B., Dai, X., Li, S., Lu, G., and Zhou, Y. (2017). Structure and function of the bacterial communities during rhizoremediation of hexachlorobenzene in Constructed Wetlands. Environ. Sci. Pollut. Res. Int. 24, 11483–11492. doi: 10.1007/s11356-017-8463-1
Zhou, Y., Tigane, T., Li, X., Truu, M., Truu, J., and Mander, U. (2013). Hexachlorobenzene dechlorination in constructed wetland mesocosms. Water Res. 47, 102–110. doi: 10.1016/j.watres.2012.09.030
Keywords: Phragmites australis, phytodepuration, wastewater, endophytes, antibiotic resistance, metal resistance
Citation: Vassallo A, Miceli E, Fagorzi C, Castronovo LM, Del Duca S, Chioccioli S, Venditto S, Coppini E, Fibbi D and Fani R (2020) Temporal Evolution of Bacterial Endophytes Associated to the Roots of Phragmites australis Exploited in Phytodepuration of Wastewater. Front. Microbiol. 11:1652. doi: 10.3389/fmicb.2020.01652
Received: 27 March 2020; Accepted: 25 June 2020;
Published: 17 July 2020.
Edited by:
Margherita Sosio, Naicons Srl, ItalyReviewed by:
Gaurav Saxena, Jawaharlal Nehru University, IndiaShaohua Chen, Institute of Urban Environment (CAS), China
Copyright © 2020 Vassallo, Miceli, Fagorzi, Castronovo, Del Duca, Chioccioli, Venditto, Coppini, Fibbi and Fani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renato Fani, cmVuYXRvLmZhbmlAdW5pZmkuaXQ=
 Alberto Vassallo
Alberto Vassallo Elisangela Miceli1
Elisangela Miceli1 Camilla Fagorzi
Camilla Fagorzi Lara Mitia Castronovo
Lara Mitia Castronovo Sara Del Duca
Sara Del Duca Renato Fani
Renato Fani