Abstract
The relationship between microbial community and host has profound effects on the health of animals. A balanced gastrointestinal (GI) microbial population provides nutritional and metabolic benefits to its host, regulates the immune system and various signaling molecules, protects the intestine from pathogen invasion, and promotes a healthy intestinal structure and an optimal intestinal function. With the fast development of next-generation sequencing, molecular techniques have become standard tools for microbiota research, having been used to demonstrate the complex intestinal ecosystem. Similarly to other mammals, the vast majority of GI microbiota in cats (over 99%) is composed of the predominant bacterial phyla Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. Many nutritional and clinical studies have shown that cats’ microbiota can be affected by several different factors including body condition, age, diet, and inflammatory diseases. All these factors have different size effects, and some of these may be very minor, and it is currently unknown how important these are. Further research is needed to determine the functional variations in the microbiome in disease states and in response to environmental and/or dietary modulations. Additionally, further studies are also needed to explain the intricate relationship between GI microbiota and the genetics and immunity of its host. This review summarizes past and present knowledge of the feline GI microbiota and looks into the future possibilities and challenges of the field.
Introduction
The intestinal microbiome is a complex collection of microorganisms (i.e., bacteria, archaea, viruses, fungi, and protozoa) (Frank et al., 2007). Based on the study of the small ribosomal subunit RNA (16S rRNA), current phylogenetic research has demonstrated that the mammalian gastrointestinal (GI) tract harbors hundreds to thousands of microbial phylotypes (Suchodolski et al., 2009). According to recent reports, approximately 1010 to 1014 microbes are present in the mammals’ GI tract (Frank et al., 2007), which is around ten times the total amount of host cells (Hoffmann et al., 2016). This complex system is composed of the mutual interaction between host cells and resident microorganisms and is known as the gastrointestinal microbiome (Suchodolski, 2011a).
Domestic cats (Felis catus) are an obligate carnivore which depend on high intakes of animal tissue to meet its nutrition requirements. This has led to a metabolic adaptation to a low-glucose and high-protein metabolism (Macdonald et al., 1984; Verbrugghe et al., 2012). Compared to humans or other mammals, cats are less dependent on the intestinal microbiota for energy acquisition through microbial fermentation. Nevertheless, a stable and balanced microbiota remains critical for the maintenance of intestinal health (Deng and Swanson, 2015). Similarly to other mammals, the dominant bacterial phyla in the feline GI tract are Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Figure 1). However, according to the literature (Suchodolski, 2011a; Minamoto et al., 2012; Barko et al., 2018; Tizard and Jones, 2018), the percentages of these bacterial groups often differ among species and individuals. These variations may be caused by the animals’ living environment or by the different experimental methods used (Deng and Swanson, 2015).
FIGURE 1
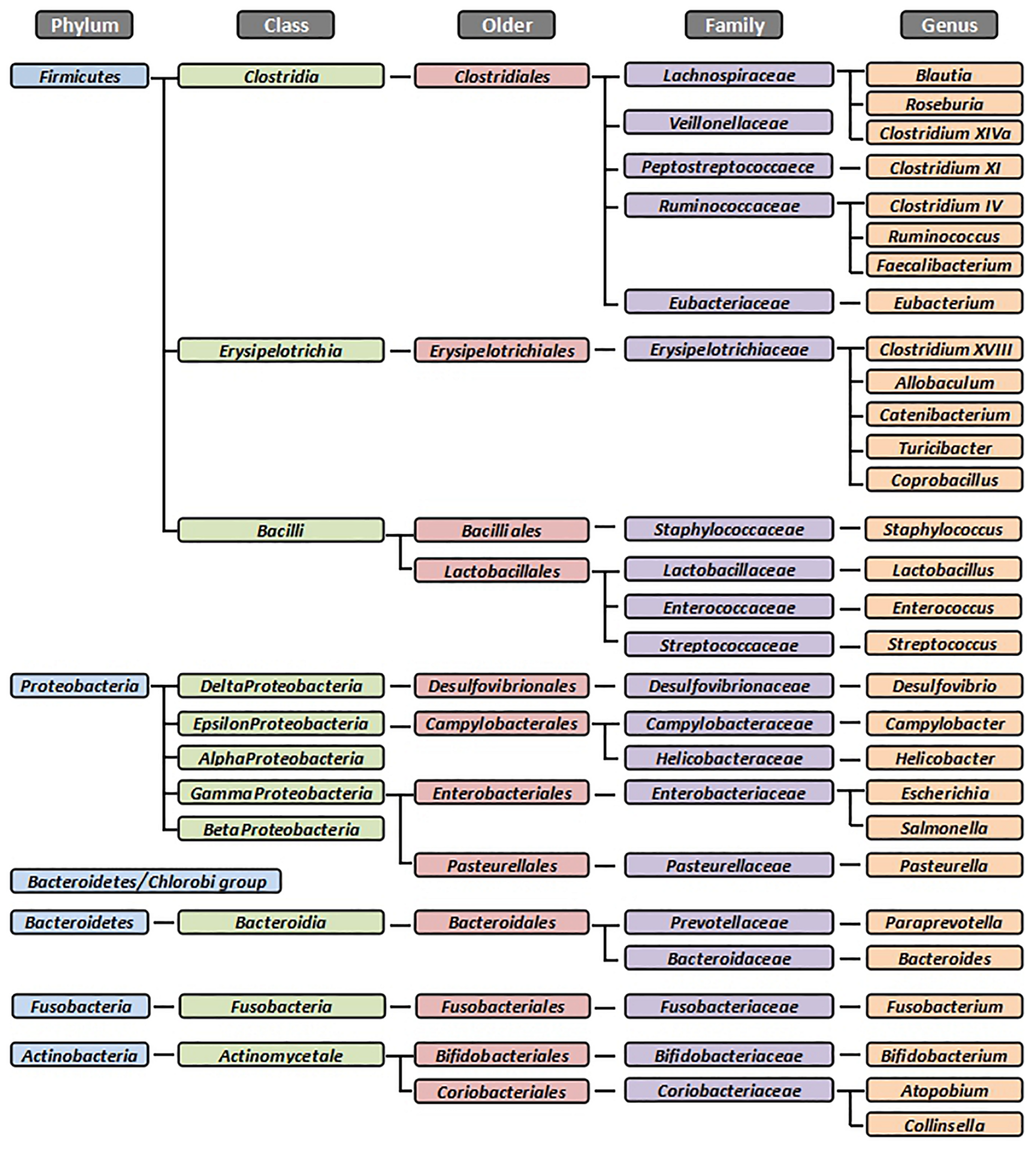
The dominant bacterial groups in the feline gastrointestinal tract. Summarized from Osbaldiston and Stowe (1971), Johnston et al. (1993, 2000, 2001), Inness et al. (2007), Desai et al. (2008), Janeczko et al. (2008), Ritchie et al. (2008), Abecia et al. (2010), Barry et al. (2010, 2012), Garcia-Mazcorro et al. (2011), Jia et al. (2011a, b), Sparkes et al. (1998b), and Tun et al. (2012). All bacteria were identified from fecal samples or intestinal biopsies by the molecular method.
Research on the GI microbiota of cats is a field in permanent expansion. Recently developed molecular techniques have improved the knowledge on the composition, alterations, and function of the feline gastrointestinal ecosystem (Desai et al., 2008; Ritchie et al., 2008; Swanson et al., 2011). Several exogenous factors (e.g., diet) have been demonstrated to influence the microbiota composition to some extent (Desai et al., 2008; Ritchie et al., 2008; Barry et al., 2010; Middelbos et al., 2010; Swanson et al., 2011). Nevertheless, the microbiota is resilient to most environmental influences, rapidly returning to its pretreatment state (Middelbos et al., 2010). Several studies on humans and dogs have reported that the administration of antibiotics could cause more profound shifts of the GI microbiota composition, and some bacterial groups could remain depressed for several weeks (Dethlefsen et al., 2008; Suchodolski et al., 2009; Torres-Henderson et al., 2017; Whittemore et al., 2018, 2019). Recent evidence both in humans and in animals including cats has already shown implicating alterations in the composition of the GI microbiota to chronic enteropathies (Inness et al., 2007; Janeczko et al., 2008). Moreover, extraintestinal disorders, such as atopic dermatitis, have been associated with GI dysbiosis due to the mutual interaction between GI microbiome and host immunity (Penders et al., 2007). These results highlight the importance of maintaining the balance of the gastrointestinal ecosystem.
This review focuses on the feline GI microbiota and summarizes the past and present knowledge on the GI microbiota in cats, including characterization techniques, composition, roles in health and disease, effects to different treatments, and future directions. All citations in this review were obtained from the online open database Google Scholar,1 the National Centre for Biotechnology Information (NCBI),2 and ScienceDirect,3 using search terms “microbiota/microbiome in cats” or “feline microbiota/microbiome,” within the time frame from 1990 to the present.
Composition
Result From Traditional Cultivation Methods
At the early stages of microbial research, traditional cultivation techniques were the most common method to characterize intestinal microbiota. Many pioneer researchers explored this area using culture-based methods and observed the bacterial composition of intestinal and fecal samples in cats. The most abundant cultivable groups found were Bacteroides, Clostridium, Enterococcus, Streptococcus, Fusobacteria, and Eubacteria (Osbaldiston and Stowe, 1971; Terada et al., 1993; Sparkes et al., 1998a, b; Johnston et al., 2000, 2001). Similarly to humans and other species, the abundance of microbiota in the cat’s intestine increases along the gut (Handl et al., 2011; Deng and Swanson, 2015). The proximal intestine contains a practically equal distribution of aerobic and anaerobic bacteria, while distal portions are predominantly colonized by anaerobic bacterial groups (Minamoto et al., 2012). Interestingly, some culture-based studies have suggested that, compared to bacterial counts in the small intestine of dogs and humans, cats display a relatively higher number of bacteria, especially anaerobic (Johnston et al., 1993; Greetham et al., 2002). Nevertheless, phenotypical and biochemical characterization systems have often failed to precisely identify many microbes present in the GI tract (Suchodolski, 2011a; Deng and Swanson, 2015). Hence, DNA sequencing of cultured isolations was often required, which have given a vigorous boost to the development of today’s molecular techniques (Suchodolski, 2011b).
Results From Molecular Techniques
The variable region of the 16S rRNA gene of bacteria contains the signature of a phylogenetic group and even species, based on the revelation of this knowledge; many new tools for analysis of microbial community became available (Tun et al., 2012). In the last 20 years, several molecular tools have facilitated a more in-depth characterization of the complex intestinal microbiota (Tannock, 2005). These tools have now largely replaced traditional bacterial culture methods, becoming the standard approach to study the microbial ecology (Suchodolski et al., 2008). Current molecular techniques include fluorescence in situ hybridization (FISH), polymerase chain reaction (PCR, i.e., PCR/DGGE, qPCR), and sequencing (i.e., 454-pyrosequencing, shotgun sequencing) (Tannock, 2005; Suchodolski et al., 2009, 2015; Swanson et al., 2011); details are summarized in Table 1. Molecular techniques allow the characterization of unidentified gastrointestinal microorganisms in the past (Swanson et al., 2011). Combining them with metagenomics tools, these techniques can also provide description of the functional potentials of microbiota (Tun et al., 2012).
TABLE 1
| Techniques | Purpose | Summary |
| FISH | Detection and quantification of bacterial cells | Fluorescent dye-labeled oligonucleotide probe hybridizes to ribosomal RNA sequence in cells fixed on slides with wells. Enumeration by epifluorescence microscopy |
| PCR/DGGE | Profiling the composition of bacterial communities for comparative analysis | Separation of 16S rDNA fragments from different bacterial types is based on differences in chemical stability, through a linearly increasing gradient of chemical denaturants. The profile of DNA fragments represents the genetic fingerprint of the community. |
| Qpcr | Quantification of bacteria | PCR primers and a labeled probe (often incorporating a reporter dye and a quencher molecule) are used to measure the real-time accumulation of a specific target sequence |
| 454-Pyrosequencing | Detecting the nucleotide incorporated | A single strand of DNA is used as a template to synthesize the sequence of its complementary strand, which is determined by a chain of reactions resulting in light being emitted when a specific nucleotide or length of nucleotides are added to the complementary sequence |
| Shotgun sequencing | Determining the sequence of entire chromosomes and genomes | Based on producing random fragments of DNA that are then assembled by computers that order fragments by finding overlapping ends |
Frequently used methods in feline microbiota studies.
FISH, fluorescence in situ hybridization; PCR/DGGE, polymerase chain reaction combined with denaturing gradient gel electrophoresis; qPCR, real-time quantitative polymerase chain reaction. Summarized from Tannock (2005), Suchodolski et al. (2009, 2015), Suchodolski (2011b), and Swanson et al. (2011).
Fish
Studies using FISH have indicated that the total count of bacteria present in the cat’s intestine is approximately 10.5 log10 cells/g feces (Minamoto et al., 2012). The most abundant populations in intestines of young (1–3 years old) and senior (8–14 years old) cats belong to the Atopobium group (probe Ato291) – including Coriobacteriaceae, Clostridium cluster XIVa – and lactic acid bacteria, including Bifidobacteria (Abecia et al., 2010; Jia et al., 2011a, b). Studies on fecal samples from adult cats have shown similar results, as well as feces of cats with inflammatory bowel diseases (IBD) (Inness et al., 2007; Abecia et al., 2010). However, the total count of Bifidobacteria in healthy adult cats varied between two studies (approximately 11% vs. 30%). FISH targets 16S rRNA, diametrically quantifying bacteria using fluorescent-labeled probes. Nevertheless, this tool is too labor intensive, and the probes need to be designed specifically for a particular bacterial group (Tal Gavriel, 2018). Thereby, FISH is generally not appropriated for studies with a large sample size.
Sequencing Techniques
Sequencing techniques, either based on the construction of 16S rRNA gene clone libraries or recent high-throughput methods such as 454-pyrosequencing or Illumina sequencing, have allowed the identification of previously uncharacterized bacterial groups.
Five different bacterial phyla were identified in the stomach and intestines of healthy cats using traditional Sanger sequencing, with sequences predominantly classified in phylum Firmicutes (68%), followed by Proteobacteria (14%), Bacteroidetes (10%), Fusobacteria (5%), and Actinobacteria (4%) (Ritchie et al., 2008; Minamoto et al., 2012). Most clones belong to order Clostridiales (54%), followed by Lactobacillales in jejunum and Bacteroidales in ileum and colon. However, different percentages of abundance were found in another study that used the 60-kDa chaperonin (cpn60) gene as target. In this study, the most abundant phylum found was Firmicutes, followed by Actinobacteria, Bacteroidetes, and Proteobacteria (Desai et al., 2008). Although the Sanger technique was one of the first methods used in human and animal research, this method is laborious and has limited throughput, as amplicons must be cloned into bacteria, with individual bacterial colonies sequenced (Sanger et al., 1977; Tal Gavriel, 2018).
High-throughput sequencing techniques, such as 454-pyrosequencing or Illumina sequencing, are capable of sequencing thousands to millions of base pairs in a short amount of time, allowing for in-depth study of the microbiota and relative quantification of amplicons. Firmicutes (92%) and Actinobacteria (7.3%) were the most abundant phylum reported in the fecal sample of cats using these techniques (Handl et al., 2011). The phylum Firmicutes consisted the predominant class Clostridia (65%), Erysipelotrichi (13%), and Bacilli (9%); the class Clostridia was dominated by Clostridium XIVa and XI and Ruminococcus; the class Bacilli mostly consisted of the order Lactobacillales, which was dominated by genera Enterococcus and Lactobacillus; the class Erysipelotrichia only consisted of the order Erysipelotrichales, which mainly comprised genera Turicibacter, Catenibacterium, and Coprobacillus (Handl et al., 2011). Some feline studies reported similar distributions of fecal bacterial groups (Desai et al., 2008; Garcia-Mazcorro et al., 2011). Although high-throughput sequencing techniques have been only used in the last decade, these techniques have their disadvantages as well, since the use of universal bacterial primers may underestimate specific bacteria. Moreover, because of the semi-quantitative abundances, detected dynamics by sequencing methods could not precisely present the actual taxon densities (Props et al., 2017). Additionally, due to its cost, only small numbers of samples were analyzed in most of the studies. Therefore, available information obtained from these studies should be considered with caution.
Next-generation sequencing platforms allow metagenomics approaches (i.e., shotgun genomic sequencing). These approaches allow identifying the genes of host and microbes and thereby are able to assess the functional aspect of the microbiome (Gill et al., 2006). Using 454-pyrosequencing, one study revealed that the predominant phylum of fecal microbiota in cats were Bacteroidetes/Chlorobi group (68%), Firmicutes (13%), Proteobacteria (6%), Actinobacteria (1.2%), and Fusobacteria (0.7%) (Tun et al., 2012). Another study with the metagenomics analysis platform MG-RAST, reported Bacteroidetes/Chlorobi group (36.1%), Firmicutes (36.3%), Proteobacteria (12.4%), and Actinobacteria (7.7%) were predominant phyla (Barry et al., 2010). Afterward, using shotgun 454-pyrosequencing, research showed that the dominant bacterial phyla included Firmicutes (36–50%), Bacteroidetes (24–36%), and Proteobacteria (11–12%) (Barry et al., 2012). Metagenomics approaches can also characterize the expression of microbial genes; the major functional metabolic categories are carbohydrate, protein DNA, and amino acid, respectively accounting for 13, 9, 8, 7, and 6% of the feline metagenome (Tun et al., 2012). Apparently, more research needs to be established to investigate in depth the feline metagenome.
Discrepancy During the Characterization
Currently, there are several tools to describe the gastrointestinal microbiota. However, discrepancies of microbial abundance are often observed when using different techniques (Abecia et al., 2010; Minamoto et al., 2012). These differences can be partially attributed to different sensitivities and specificities among the different methods. For instance, different sampling sites and methodologies (DNA extraction protocols or PCR primers) have been used in different sequencing studies (Baker et al., 2003; Zoetendal et al., 2004). The sample size may affect the statistical analyses due to limited power (Tal Gavriel, 2018). Collection and processing methods and storage conditions of samples may also result in alterations in the quality of those samples (Ott et al., 2004; Kieler et al., 2016; Tal et al., 2017). Moreover, the specific characteristics of cat populations need to be considered. Variations in the microbiota composition between conventional and specific-pathogen-free cats have been reported, as well as a variation between indoor and outdoor cats (Desai et al., 2008; Ritchie et al., 2008). Furthermore, the composition of the intestinal microbiota could vary between anatomical sites and between the luminal and mucosa-adherent tissues (Ott et al., 2004). Considering these limitations, researchers should exert caution when interpreting results of different experiments and techniques.
Due to the high diversity of the microbial community, less abundant bacterial groups may escape identification, even when using high-throughput sequencing techniques with broad-range primers (Hoffmann et al., 2016). The additional applications of PCR analysis is needed to detect specific groups with low proportion. Significantly, no optimal DNA extraction protocol or PCR-based identification method exists for accurate characterization of all microorganisms, and therefore, the various methods available should be used complementarily.
Non-bacterial Composition
Aside from bacteria, the mammalian gut harbors many other microbes including archaea, fungi, viruses, and parasites. The intricate relationships between these organisms, the host, and the bacteria are unclear. Current research using pan fungal primers has revealed several fungal components of the cats’ microbiota. Eukaryote (1%) and fungi (0.02%) have been found in feces of cats (Suchodolski et al., 2008; Handl et al., 2011). A study based on 454-pyrosequencing of 18S rRNA gene identified four fungal phyla in feces of cats; Ascomycota (>90%) and Neocallimastigomycota (>5%) were the predominant phylum. Ascomycota was the only and most abundant fungal phylum, dominated by genera Saccharomyces and Aspergillus (58.31 and 11%) (Suchodolski, 2011a). Research on viral components in the feline GI tract remains lacking. Shotgun sequencing of viral dsDNA found only one order of Bacteriophages, Caudovirales (Barry et al., 2010). Di Sabatino (2019) reported that viruses accounted for approximately 0.07% of all sequences, and most of them belonged to the Caudovirales order and an additional unclassified order. This accounted for a total of 18 families and 42 genera. In addition, Archaea was reported for the first time in this study. This domain accounted for 0.77% of sequences, consisting of five phyla (Crenarchaeota, Euryarchaeota, Korarchaeota, Nanoarchaeota, and Thaumarchaeota) and twelve classes.
Role of GI Microbiota
Nutrition and Metabolism
In addition to the production of energy by using nutrients, the intestinal microbiota produces many metabolites that may have an effect on the host’s health. For example, carbohydrate fermentation leads to the production of short-chain fatty acids (SCFA) (Ramakrishna and Roediger, 1990), further promoting the process of intestinal gluconeogenesis and supporting the formation of lipids; protein fermentation leads to the production of SCFA, ammonia, and branched chain fatty acids (BCFAs), and some studies have revealed that protein fermentation increases phenolic metabolites in humans, which could affect the host’s health (Windey et al., 2012). However, as strict carnivores, cats have several-fold higher intakes of protein compared to other mammals, which seem to have no issues with the carcinogenic effects of protein fermentation (Rissetto et al., 2011). Additionally, certain bacterial groups in the GI tract also play different specific roles. For instance, Escherichia coli (E. coli) and Bacteroides spp. produce vitamin K2 (Ramotar et al., 1984), Enterococcus spp. synthesize folate (Camilo et al., 1996), Lactobacillus and Bifidobacterium spp. contribute to the salvage of bile acids (Ridlon et al., 2006), and Enterococcus casseliflavus and Eubacterium ramulus promote transformation of polyphenols (Schneider et al., 1999), a group of compounds that may confer a health benefit to the host due to their high antioxidant activities (Lambert et al., 2007).
Promoting Intestinal Structure and Function
Comparative studies between germ-free and specific-pathogen-free animals indicated the essential role of intestinal microbiota on the development of the GI structure and function (Cario et al., 2007; Lutgendorff et al., 2008; Al-Asmakh and Zadjali, 2015). A damaged intestinal epithelial integrity, a decreased intestinal osmolarity, and fatty acid concentrations were found in germ-free mice in a study by Stappenbeck et al. (2002). This evidence highlighted the importance of the microbiota in the development of the intestinal structure. Despite the lack of studies in cats, it is broadly accepted that the main role of the microbiota might be similar among different mammal species. For instance, an ex vivo study in cats demonstrated that SCFAs stimulate contraction of colon longitudinal muscles in cats, implying that bacteria-produced SCFAs could possibly promote colonic motility; this result is similar to the findings in humans (Rondeau et al., 2003).
Barrier and Protection
Gastrointestinal microbiota promotes colonization resistance, providing a microbial barrier against potential pathogens by competitive exclusion (Brosey et al., 2000; Lawley and Walker, 2013). It also stimulates the host to produce various antimicrobial compounds such as antimicrobial peptides (AMP) (Othman et al., 2008; Hooper, 2009). Microbial metabolites can also induce AMP expression; for example, SCFAs and lithocholic acid were reported to induce expression of LL-37 cathelicidin by different pathways, which serves a critical role in mammalian innate immune defense against invasive bacterial infection (Schauber et al., 2003; Kida et al., 2006; Termen et al., 2008). Research on this field has been traditionally carried out by human medicine groups using rodent models. Whereas the authors are not aware of any publications on this topic regarding cats, similar effects could be assumed.
Immunomodulation
Gastrointestinal microbiota contributes to gut immunomodulation together with both innate and adaptive immune systems (Suzuki et al., 2010; Geuking et al., 2011; Chung et al., 2012). Studies have shown that probiotic administration can have immunomodulatory effects. For example, the supplementation of Lactobacillus acidophilus DSM13241 (2 × 108 CFU/d for 4.5 weeks) in healthy adult cats increased the phagocytic capacity in the peripheral granulocytes and decreased the concentration of endotoxins in plasma (Marshall-Jones et al., 2006).
Regulation Outside the Gut
A healthy microbial ecosystem is not only important for the GI tract itself. There is growing evidence on the close interaction between the gut microbiota and the body’s major neuroendocrine system, the hypothalamic–pituitary–adrenal (HPA) axis, which controls various body processes in response to stress (Sudo et al., 2004; Sudo, 2006). The role of microbiota on the development of neural processes is being currently studied and termed the “brain-gut microbial axis” (Forsythe et al., 2009; Rhee et al., 2009). This area has not been investigated in cats yet.
Relationship With Disease
A dysregulated GI mucosal homeostasis has been associated with several diseases (Honda and Takeda, 2009). For instance, IBD often presents itself together with an altered abundance of groups of the feline microbiota, particularly Enterobacteriaceae and Desulfovibrio (Inness et al., 2007; Janeczko et al., 2008). The GI microbiota plays a vital role on the pathologies that affect the gut, but the relationship remains unclear. For instance, several enteric pathogens, such as Salmonella, Campylobacter jejuni, Clostridium perfringens (C. perfringens), and E. coli, can also be found in healthy cats (Marks et al., 2011). The microbiota is a contributor to intestinal homeostasis; it is consequently associated with the progress of diseases.
Factors Influencing Microbiota in Healthy Cats
Age, Gender, and Neutering
Aging has been associated with a number of changes in the gut of animals. These changes may lead to an increasing incidence of several chronic diseases (Bermingham et al., 2012). A study on the impacts of neutering, neutering age, and gender on the GI microbiota of cats during growth revealed that, when diets and environments are controlled, the only significant association found was age (Deusch et al., 2015). Masuoka et al. (2017) used cultivation as well as real-time PCR to assess feline fecal microbiota in five different age groups (12.6 ± 0.5 days; 7.5 ± 0.5 weeks; 2.5 ± 0.5 years; 11.6 ± 1.6 years; 17.5 ± 1.2 years), finding that the composition of feline fecal microbiota changed with age. The age-related shifts however differed from results from humans and dogs, which involved Bifidobacteria, C. perfringens, Lactobacillus, Enterobacteriaceae, and Enterococcus. Nevertheless, because this study did not use the sequencing method, the results may not be comparable. In addition, the species composition of Lactobacillus was only found in preweaning and young groups but not in aged and senile cats, suggesting that Lactobacillus species may be not as important for cat health as in the case of humans and dogs. A similar conclusion was found in another longitudinal study, which did not identify Bifidobacteria in young cats but demonstrated similar Lactobacilli abundance in cats at different ages (8–260 weeks) using the 16S rRNA gene-based method (Bermingham et al., 2018). In this study, the most dominant group in young cats that were fed kibbled diets was an unclassified Peptostreptococcaceae (22.5%), whereas in older cats fed with the same diet, the most dominant group was Prevotella (29.7%). The abundance of Lactobacillus in older cats was significantly lower compared to that found in young cats.
Although several studies have investigated the impacts of age on the GI microbiota of cats, these studies are often confounded by differences in diet, by individual variation, and by the different methodologies used to analyze the microbiome. Moreover, the results of these studies are often inconsistent, and the impact of gender and neutering remains unclear as well. Nevertheless, these studies have shown that the significance of single component bacteria (e.g., Lactobacillus and Bifidobacteria) in the GI microbiota in cats may be different from that of dogs or humans. This is an interesting finding that could lead to further research.
Diet
The nutritional composition of food has been proved to influence the intestinal function, microbial composition, and metabolism (GrzesKowiak et al., 2015). Pet food is highly variable regarding the format (i.e., dry vs. wet, conventional vs. alternative), macronutrient sources and composition, and content of functional ingredients such as prebiotics and probiotics. Several diet-related factors affecting the GI microbiota have been reported by nutritional studies focused on the GI microbiota (summary in Table 2).
TABLE 2
| References | Diet | Method | Alterations of microbiota |
| Bermingham et al. (2013a) | Preweaning and postweaning diet (n = 5 per group) | 454-Pyrosequencing | Postweaning diet: Fusobacteria↓ Firmicutes, Actinobacteria ↑ |
| Kerr et al. (2014) | Extruded diets and whole chicks (n = 4) | 454-Pyrosequencing | Extruded diets: Faecalibacterium, Succinivibrio ↑ |
| Chicks: Lachnospiraceae, Peptococcus, Pseudobutyrivibrio ↑ | |||
| Young et al. (2016) | Kibbled and canned diet (n = 5) | Shotgun sequencing | Kibbled diets: Lactobacillus, Bifidobacterium, Collinsella ↑ |
| Lubbs et al. (2009) | Different protein concentration (n = 4) | qPCR DGGE | High protein: Bifidobacterium↓ C. perfringens ↑ |
| Hooda et al. (2013) | Protein: carbohydrate ratio (n = 7) | 454-Pyrosequencing | Moderate protein and moderate carbohydrate vs. high protein and low carbohydrate: Actinobacteria ↑, Fusobacteria↓ |
| Barry et al. (2012) | Add 3 prebiotic substances (n = 4) | Shotgun sequencing | Fructooligosaccharides: Actinobacteria ↑ Pectin: Firmicutes, total bacteria ↑ |
| Barry et al. (2014) | Add 2 prebiotic substances (n = 6) | qPCR DGGE | Oligofructose + insulin: Bifidobacteria spp., E. coli↓ |
| Pinna (2014) | Add 6 prebiotic substances and 2 levels protein (n = 4) | FISH | Lactitol and pectins: Enterobacteriaceae↓ |
| High protein: C. perfringens ↑ Lactobacillus spp., Enterococci↓ | |||
| Santos et al. (2018) | Add spray-dried yeast cell wall (n = 4) | qPCR | Bifidobacterium spp., Lactobacillus spp. ↑ C. perfringens, E. coli↓ |
| Deb-Choudhury et al. (2018) | Add wool hydrolysate, insulin and cellulose (n = 8) | qPCR | Wool hydrolysate and cellulose: Prevotella, Bulleidia, Faecalibacterium, Ruminococcaceae↓; Fusobacterium ↑ |
Summary of available research on the dietary effects on feline intestinal microbiota.
Type of Diets
A 454-pyrosequencing-based feline study found that cats that were fed a dry diet [32.91% crude protein, 11.05% crude fat, and 1.88% crude fiber; dry matter (DM)] displayed a higher abundance of Actinobacteria and lower abundance of Fusobacteria and Proteobacteria than those fed with wet food (41.87% crude protein, 42.39% crude fat, and 1.62% crude fiber; DM). However, changes on the microbiota composition cannot be attributed to any specific nutrients (Bermingham et al., 2013b). Kerr et al. (2014) compared the fecal bacterial population in cats fed chicken-based extruded diets (38.9% crude protein and 14.4% crude fat, DM) with those fed commercial 1–3-day-old chicks (71.4% crude protein and 20.0% crude fat, DM). Cats fed chicks showed a higher abundance of genera: Peptococcus, Pseudobutyrivibrio, and unclassified Lachnospiraceae, while those fed extruded diets displayed a higher abundance of Faecalibacterium and Succinivibrio. Furthermore, Lactobacillus and Bifidobacterium were only detected in cats fed with extruded diets (Kerr et al., 2014). A recent study investigated the impacts of preweaning and postweaning diets (kibbled or canned) in cats using metagenomic sequencing and MG-RAST (Young et al., 2016). In this study, cats fed with kibbled diets (35.3% crude protein, 20.2% crude fat, and 1.8% crude fiber; DM) had higher proportions of Lactobacillus, Bifidobacterium, and Collinsella than those fed with canned diets (45.3% crude protein, 37.6% crude fat, and 1.5% crude fiber; DM). Several metagenomic differences in cats fed with canned diet revealed the related metabolic pathways, associated with vitamin biosynthesis, metabolism, and transport (Young et al., 2016).
Conventional commercial cat foods generally fall into two broad categories: dry (approximately 30–40% crude protein and 30–40% crude fat) or wet (approximately 40–50% crude protein and 40–50% crude fat) (Villaverde and Fascetti, 2014). Furthermore, the consumption of “alternative” pet food, for instance fresh/refrigerated, raw/frozen, and dehydrated varieties of pet food, is becoming a new customer trend (Berschneider, 2002). Studies demonstrated that short-term dietary exposure to diet leads to large shifts in the fecal bacterial population that have the potential to affect the cat’s ability to process macronutrients in the diet, therefore impacting the functional capacities of the microbiota and its interaction with the host. In addition, metagenomic research has suggested that the modulation of the microbiome function through diet may be an important method for improving the nutrition of companion animals. Further research is needed to determine the impact of these shifts on the long-term health of domestic cats and the impact of the newly developed dietary types.
Energy Source Composition
Traditional commercial cat food often contains 30–40% of protein (Hooda et al., 2013). However, cats are obligate carnivores, having evolved on diets rich in protein and fat (Plantinga et al., 2011; Depauw et al., 2013); therefore, the content of dietary protein has always been a hot topic in feline research. In a 2009 study, Lubbs et al. (2009) identified several variations in the GI microbiota composition in adult cats that were fed diets with different proportions of proteins. Cats fed moderate protein (MP, approximately 30% crude protein) had a bacterial similarity index (Dice’s similarity coefficient) of 66.7% while those fed high protein (HP, approximately 50% crude protein) had 40.6%. Similarly as in dogs and humans, a higher dietary protein concentration decreased Bifidobacterium and increased C. perfringens populations (Zentek et al., 2003; Singh et al., 2017). In addition, Bermingham et al. (2013a) showed significant differences in the GI microbiota of pregnant and lactating queens that were fed either a moderate protein:fat:carbohydrate kibbled diet (35:20:28%, DM), a preweaning diet, or a high protein:fat:carbohydrate canned (45:37:2%, DM) postweaning diet. However, the preweaning diet had a low impact on the fecal microbial population of weaned kittens, whereas the postweaning diet changed considerably the microbial community profile. Moreover, kittens fed with the postweaning diet had a lower abundance of Firmicutes and Actinobacteria and a higher abundance of Fusobacteria when compared with those fed the preweaning diet (Bermingham et al., 2013a). Hooda et al. (2013) also showed an increased Actinobacter abundance and decreased Fusobacteria abundance at the phylum level by moderate protein diet and moderate carbohydrate (34% protein and 19% fat; DM) diet compared with those fed a high-protein, low-carbohydrate diet (53% protein and 24% fat; DM) using 454-pyrosequencing in 8–16-week-old kittens. Using the same samples from Hooda’s study, Deusch et al. (2014) investigated the microbiome function by using shotgun sequencing, revealing a great dietary influence on the pathways associated with amino acid biosynthesis and metabolism.
Many studies have been conducted to investigate the impact of dietary macronutrient concentrations on the GI microbiota of humans and dogs. However, only few studies have studied the effect of dietary carbohydrates/protein/fat concentration in cats. Moreover, it is complicated to reach any accurate conclusions about the types of nutrients that could cause the observed changes in the microbial population, as the macronutrient contents and ingredient composition of diets differed largely in these studies. Furthermore, cats, as obligate carnivores, consume little plant materials under natural conditions; therefore, data collected from adapted carnivores and omnivores, such as dogs and humans, in other nutritional studies cannot be simply extrapolated to cats (Verbrugghe et al., 2012).
Supplementation of Fibers
In 2010, Barry et al. (2010) evaluated the effects of dietary supplementation of 4% cellulose, fructooligosaccharides, or pectin on the fecal microbial population in cats. In this study, an increased proportion of fecal Bifidobacterium and a decreased proportion of E. coli were observed due to the supplementation of fructooligosaccharides. Dietary supplementation of pectin also showed a marked increase in the fecal proportions of C. perfringens, E. coli, and Lactobacillus. The same authors had also performed a similar study using a metagenomic approach and indicated that a 4% fructooligosaccharide supplementation leads to a greater abundance of Actinobacteria, while pectin supplementation leads to a greater abundance of Firmicutes and total bacteria (Barry et al., 2012). Furthermore, KEGG pathway analysis displayed a strong association between carbohydrates, clustering-based subsystems, protein metabolism, and amino acids and derivatives. In addition, a functional prediction by COG suggested a strong association between amino acid metabolism and transport, and carbohydrate transport and metabolism (Barry et al., 2012). In a different study from the same research group, the effects of fructan supplementation in senior cats were explored. On this occasion, oligofructose or oligofructose + inulin was individually supplemented to the diets at 1% (DM), causing a decrease in Bifidobacteria spp. and E. coli concentrations (Barry et al., 2014). The results of these studies indicate that, despite the cat being an obligate carnivore, the phylogeny and gene content of its gut microbiome are similar to those of omnivores.
Some studies have attempted to search new functional ingredients for cats, but the alterations of microbiota in current reports are not obvious. An in vitro study in 2014 evaluated the influence of different prebiotics and diets with two levels of protein on the fecal microbiota of cats. Supplementation of 2 g/L lactitol and pectin from citrus fruit reduced the count of Enterobacteriaceae, and high-protein diets with no supplementation increased the count of C. perfringens and decreased the counts of Lactobacillus and Enterococci (Pinna, 2014). Santos et al. (2018) investigated the impacts of increasing the concentration of the spray-dried yeast’s cell wall (0, 0.2, 0.4, 0.6%) in diets on the fecal bacterial composition of healthy adult cats showing a decrease in C. perfringens and E. coli and an increase in Bifidobacterium and Lactobacillus with increasing yeast cell wall. In addition, a dietary supplementation of 2% wool hydrolysate has been recently reported to influence the composition of the fecal microbiota in a similar way as cellulose supplementation (Deb-Choudhury et al., 2018).
The supplementation of fibers has been frequently applied in animal nutrition to improve the quality of the food or the performance and/or health of the animals (Jones and Jew, 2007). Because of the obvious effects modulating the microbial environment, some recent studies have also investigated its potential benefits for cats. Despite the short colon and the lack of a functional cecum as part of evolutionary adaptations to a strict carnivorous diet, considerable microbial fermentation occurs in the hindgut of domestic cats (Verbrugghe et al., 2012). The microbiota of domestic cats has been showed to be capable of fermenting a broad range of dietary fibers, including prebiotics. As a consequence, many attempts have been established to evaluate the effects of fibers in cats. Nevertheless, the shifts of the microbiota in most studies are so small that researchers usually cannot draw accurate conclusions.
Future Direction
Numerous studies on GI microbiota have revealed that diets alter the population and functionality of the community of microorganisms in the gut of humans and animals (David et al., 2014). The research on nutritional intervention in the last decade has demonstrated the extent to which the intestinal microbial community could be affected by diet changes (Sonnenburg et al., 2004). This can be summarized as three major aspects: (i) the microbiome rapidly responds to short-term macronutrient changes, (ii) long-term dietary habits are a dominant force in determining the composition of an individual’s gut microbiota, and (iii) a change in the diet has significantly variable effects in different individuals. Regarding the first theme, plenty of attempts to study this response using different designs and multiple techniques have been performed, as listed in the previous sections. However, there are few long-term studies investigating the effect of diet on the feline microbiota; the study on individual variability remains lacking. In addition, different results can be often found within similar designed studies, partially due to the different techniques being employed. These limitations have pointed out a very broad direction for future research. With the current development of metagenomics, it may be necessary to repeat some existing studies that showed marked shifts in the microbial population in order to obtain detailed information about the functional alterations on the microbiota.
Diseases
Similar to the findings in humans, an unbalanced GI microbiota in cats could lead to gastrointestinal disorders, caused not only by the proliferation of enteropathogens in the GI tract but also by the various metabolic processes in which the GI microbiota participates (Deng and Swanson, 2015). The brief introduction of research progress on the alteration of feline microbiota caused by GI disease, metabolic disease and others is presented in Table 3.
TABLE 3
| References | Sample | Disease | Method | Alterations of microbiota |
| Suchodolski et al. (2015) | Feces | Acute (n = 19) or chronic diarrhea (n = 29) | Sequencing and qPCR | Burkholderiales, Enterobacteriaceae, Streptococcus, Collinsella↑ Campylobacterales, Bacteroidaceae, Megamonas, Helicobacter, Roseburia↓ |
| Inness et al. (2007) | Feces | IBD (n = 11) | FISH | IBD: Total bacteria, Bifidobacterium spp., Bacteroides↓ Desulfovibrio↑ |
| Janeczko et al. (2008) | Intestine Biopsies | IBD (n = 13) | FISH | Enterobacteriaceae↑ |
| Fischer et al. (2017) | Feces | Obesity (n = 8) | 16S rRNA sequencing | Firmicutes, Peptostreptococcaceae↑ Bacteroidetes↓ |
| Pallotto et al. (2018) | Feces | Obesity (n = 4) | Illumina sequencing | Restriction of diet: Actinobacteria↑; Bacteroidetes↓ |
| Ghosh et al. (2013) | Ileum Biopsies | Severe systemic ill (n = 50) | FISH | Enterococcus faecalis↑ |
| Weese et al. (2015) | Rectum | Immunodeficiency (n = 16) | Illumina sequencing | Bifidobacteriales, Lactobacillales, Aeromonadales↑ |
| Rossi et al. (2017) | Feces | Megacolon (n = 3) and constipation (n = 7) | qPCR | Probiotic (SLAB51TM): Lactobacillus spp., Bacteroidetes↑ |
| Schmid et al. (2018) | Feces | Severe GI diseases (n = 6) | Illumina sequencing | Omeprazole: Bifidobacterium spp.↑ Streptococcus, Lactobacillus, Clostridium, Faecalibacterium spp.↓ |
| Marsilio et al. (2019) | Feces | IBD (n = 13) and SCL (n = 14) | Illumina sequencing | Enterobacteriaceae, Streptococcaceae↑ Ruminococcaceae, Turicibacteraceae, Bifidobacterium, Bacteroidetes↓ |
| Kieler et al. (2019) | Feces | Diabetes mellitus (n = 23) | Illumina sequencing | Anaerotruncus, Dialister, Ruminococcaceae↓ |
Summary of available research on the intestinal microbiota in cats with diseases.
Gastrointestinal Disease
Janeczko et al. (2008) suggested an increase in the number of Enterobacteriaceae in the duodenum of cats with IBD, showing a positive correlation between increased microbial counts and the severity of histological inflammation. Cats with IBD also displayed lower counts of total bacteria, Bacteroides, and Bifidobacterium and a higher count of Desulfovibrio compared to healthy cats (Inness et al., 2007). Desulfovibrio spp. are sulfate-reducing bacteria that are capable of producing hydrogen sulfides, which might be implicated in the IBD pathogenesis in cats (Janeczko et al., 2008). Nevertheless, in a different study with a similar experimental design, no statistical variation of FISH counts was observed in cats with IBD compared with healthy cats (Abecia et al., 2010). Ramadan et al. (2014) described the alterations of fecal bacterial composition in cats with chronic diarrhea and the effects of diet intervention, finding strong correlations between fecal score and Coriobacteriaceae, the Enterobacteriaceae family, and an unclassified genus from the order Clostridiales. Fecal microbiota in cats with acute and chronic diarrhea showed that, compared to healthy cats, diarrheal cats had an increased fecal microbial abundance of order Burkholderiales, family Enterobacteriaceae, and genera Streptococcus and Collinsella. Lower abundance of order Campylobacterales, family Bacteroidaceae, and genera Megamonas, Helicobacter, and Roseburia was also described (Suchodolski et al., 2015). Moreover, chronic diarrheal cats (>21 days duration) had a higher abundance of class Erysipelotrichia and genus Lactobacillus, while acute diarrheal cats had a higher abundance of phylum Bacteroidetes (Suchodolski et al., 2015). The impact of a multi-strain probiotic (SLAB51TM) was investigated in constipated and megacolon cats. No differences were found in the microbial population of healthy control cats and constipated cats. However, the administration of probiotics markedly increased the presence of Lactobacillus and Bacteroidetes in constipated cats (Rossi et al., 2017). Using Illumina sequencing, Marsilio et al. (2019) revealed a decreased alpha diversity in cats with IBD and small cell lymphoma (SCL) and a significant difference in bacterial communities with a small effect size. Cats with IBD and SCL both displayed a higher abundance of Enterobacteriaceae and Streptococcaceae and a lower abundance of Ruminococcaceae, Turicibacteraceae, Bifidobacterium, and Bacteroidetes (Marsilio et al., 2019).
Microbiota imbalance, often defined as “dysbiosis,” refers to any disturbance of the normal microbial content that can disrupt the symbiotic relationships between microorganisms and the host, potentially leading to the onset of pathologies (Carding et al., 2015). Recent studies have demonstrated alterations in the GI microbiome due to dietary modifications in cats; only a few studies have evaluated alterations in the intestinal bacterial communities in cats with GI disease. In addition, many of these studies focused exclusively on a particular microbial group using FISH. Furthermore, some of these studies attempted to predict the relationship by correlation analysis, which cannot demonstrate causation.
Metabolic Disease
Fecal microbial communities of cats suffering from type 2 diabetes have been indicated to differ from those from healthy cats (Kieler et al., 2019). This new information has raised the interest on the role of environmental factors as the central link between metabolic diseases and the intestinal microbial population (Sonnenburg et al., 2004). Several studies in obese dogs have found a higher abundance of the phyla Actinobacteria and Proteobacteria and genus Roseburia, Bifidobacteriaceae, and Eubacterium, when compared to fecal microbiota in healthy or lean dogs (Handl et al., 2013; Forster et al., 2018). There are also a few studies that have explored this field in cats.
Bell et al. (2014) attempted to compare the fecal bacterial populations of insulin-treated diabetic and non-diabetic cats, however, no apparent shift was observed in the fecal microbiota composition between diabetic and non-diabetic cats. Using 16S rRNA gene amplicon metabarcoding, Kieler et al. (2019) found that diabetic cats have a decreased gut microbial diversity and a lack of butyrate-producing bacteria genera Anaerotruncus, Dialister, and unknown Ruminococcaceae. Significant differences in fecal bacterial groups were detected between lean cats and overweight or obese cats, but the variations could not be ascribed to the shifts of any particular bacteria (Kieler et al., 2016). Fischer et al. (2017) revealed associations between the shifts in GI microbiota composition and obesity, energy restriction, and neutering. In this study, lean neutered cats displayed a higher abundance of Firmicutes and lower abundance of Bacteroidetes than the cats in other groups, similarly to previous findings in obese rodents and humans. Additionally, increased Firmicutes in lean neutered cats was assigned to the increase in Peptostreptococcaceae; decreased fat mass in obese cats by energy restriction was linked to the decreased abundance of genus Sarcina and the alterations of a few low-abundance bacterial genera (Fischer et al., 2017). Feeding a moderate-protein and high-fiber diet caused weight loss in overweight cats, resulting in a reduction in Bacteroidetes abundance and an increase in Actinobacteria abundance (Pallotto et al., 2018). Contrarily, Tal et al. (2020) did not observe significant differences between the fecal microbiome in obese cats during weight loss plan. However, some significantly enriched taxa, mainly belonging to Firmicutes, were noted in the linear discriminant analysis effect size test in obese cats before weight loss compared to lean cats (Tal et al., 2020).
Others
Ghosh et al. (2013) identified significant differences in the abundance of Enterococcal groups in the ileum mucosa between healthy and terminally ill kittens: E. hirae was the predominant species of Enterococci found in healthy kittens. This species generally lacks virulence traits. Contrarily, E. faecalis commonly has several virulence traits and many antimicrobial resistances, while its abundance was found to be higher in the terminally ill kittens. Moreover, a great number of E. coli was present in the ileum mucosa of kittens with terminal illness and not detected in all kittens with adherent E. hirae (Ghosh et al., 2013). Weese et al. (2015) evaluated the rectal microbial composition of cats infected with feline immunodeficiency virus (FIV); they found that the relative abundance of Bifidobacteriales, Lactobacillales, and Aeromonadales was significantly increased. Schmid et al. (2018) evaluated the impact of prolonged omeprazole administration on the fecal microbiota of healthy cats to identify targets for analysis in a larger subset of cats with GI disease. There was no significant shift of the microbial community and species richness in cats with omeprazole administration. Nevertheless, transient shifts were observed in cats subjected to omeprazole administration, which were associated with a decreased proportion of Bifidobacterium and increased proportion of Streptococcus, Lactobacillus, Clostridium, and Faecalibacterium (Schmid et al., 2018).
Future Direction
A balanced intestinal microbial ecosystem is essential for feline GI health. Profound shifts of GI microbiota have been not only demonstrated in chronic and acute GI diseases but also suggested that those shifts may have a potential role in some extraintestinal diseases (Carding et al., 2015). Nevertheless, most studies have only investigated GI and metabolic diseases and there is little research involving extra-alimentary diseases. Furthermore, the vast majority of recent research on diseases has focused on certain pathogens rather than focusing on the interaction between microbiota and gut. Moreover, many studies have attempted to analyze the relationship between microbiota and disease by using correlations, which cannot demonstrate causation. It would be preferable to design studies aimed at demonstrating a cause–effect relationship or at a minimum acknowledge that in the cases of co-occurring disease and altered microbial status is not possible to identify neither the trigger nor the result. In addition, despite several bacteria being significantly altered, discussion about the size effect is lacking in many papers. The presentation of quantitative effect sizes in addition to p-values will improve the ability to perform meta-analysis and to evaluate potentially relevant biological effects. Finally, future research on disease needs to consider how alterations in the microbiota can result in a range of local and systemic diseases. A combination of multiple-omics technologies, such as metagenomics, transcriptomics, and metabolomics, can fill the gap of knowledge in this field. Fortunately, researchers are currently at the initial steps to relate the phylogenetic shift to functional alteration and broaden the knowledge about the biomarker and therapeutic agents of diseases. Future progress on microbiota and diseases is, therefore, to be expected.
Future Prospect
Research on microbiota in cats benefits of not only the health of the cats themselves but also the health of their owners, since companion animals have the same living environment, similar dietary pattern, and microbial communities as humans (Song et al., 2013). Furthermore, as obligate carnivores, cats have evolved to thrive on a high-protein, high-fat diet; this type of diet is detrimental to humans or other omnivores. Thereby, this unique metabolic pattern associated with microbial activity in cats could serve as a valuable comparative model for the research on the interactions between intestinal microbiota and host metabolism (Eisert, 2011). Moreover, there is some evidence revealed that cats can be a good research model for some human pathologies such as cancer, type 2 diabetes mellitus, and human immunodeficiency virus (Mcniel, 2001; Henson and O’Brien, 2006; Bailey et al., 2010; Bienzle, 2014), all of which have a close connection with the intestinal microbiota. Unfortunately, progress in the study of the feline microbiota has been small compared with the number of studies in humans or other animals, including dogs. This fact could be explained by the lack of funding and the relatively small number of feline research groups. As usual, industry sponsors and specific research foundations will be extremely crucial to the advance and development of this field.
Great progress has been made in feline research to study the phylogenetic information of GI microbial community, however, the knowledge of microbiota in cats is still at the preliminary stage which can hardly be utilized in practice (Figure 2). In addition, there are some limitations regarding the techniques used in the study of the GI microbiota. For instance, existing sequencing methods often find difficulties identifying the small amounts of microbial DNA present in samples, particularly when the sample sizes are small (Minamoto et al., 2012). Moreover, discrepancies among different analysis tools are warranted to be decreased in the future, and the combined use of multiple techniques and comparisons among different experimental designs can promote progress in this direction. Furthermore, studies in humans and other animals are moving on to combinatorial approaches, such as metagenomics and metatranscriptomics, which could help link microbiome to host health (Quince et al., 2017; Ishii et al., 2018). These new techniques are eagerly anticipated in the future feline studies.
FIGURE 2
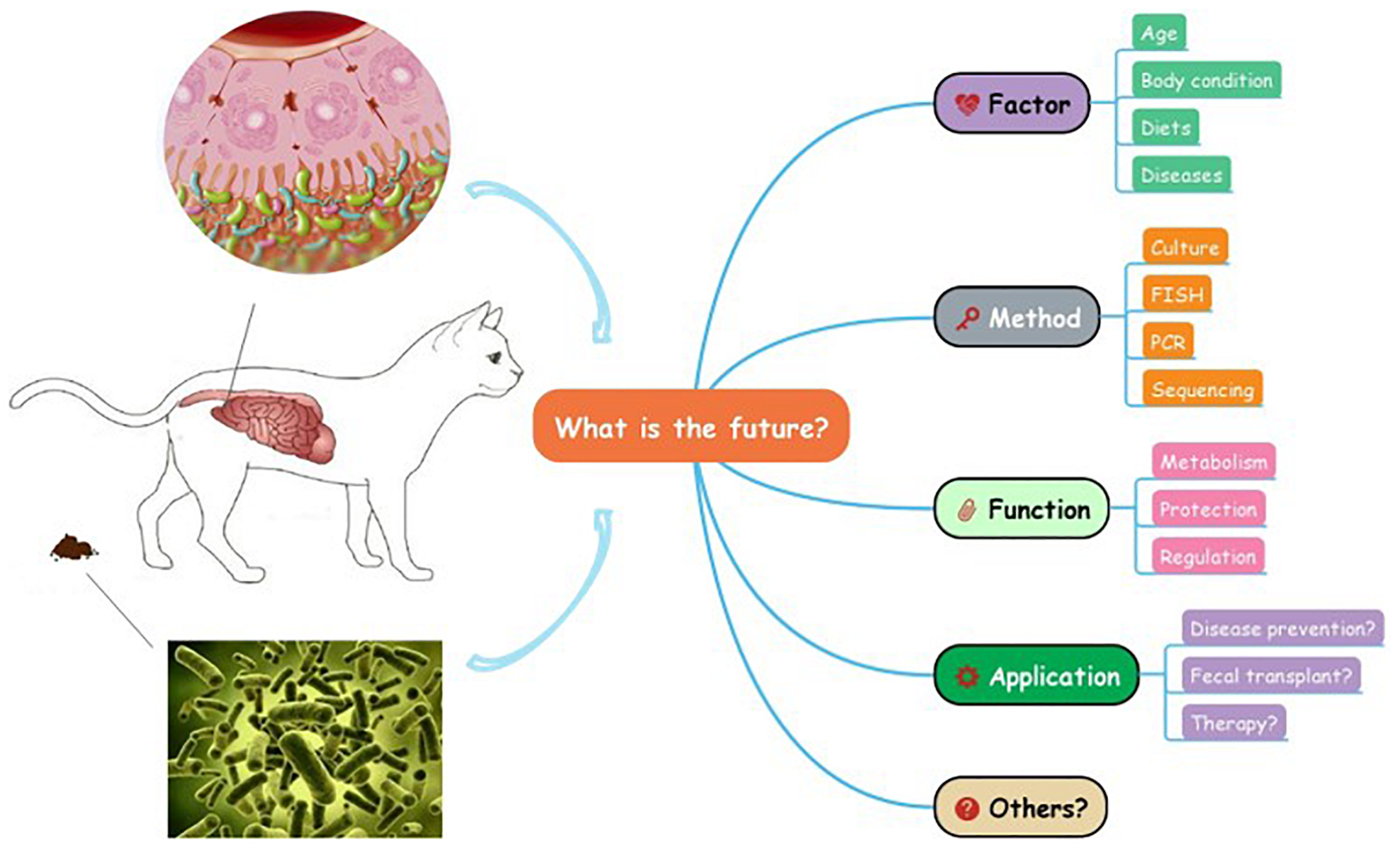
What is the future of the gastrointestinal microbiota in cats?
The study of nutrition is one of the most important subjects in life science; however, the vast majority of information on feline nutrition is usually extrapolated from humans. Dietary and metabolic interspecies differences need to be considered; therefore, a database of feline microbiomes, metagenomes, and metabolomes needs to be established. Most of the available research regarding pathologies affecting cats focuses mostly on certain pathogens associated with intestinal conditions rather than the cause/effect relationships between the GI microbiome and the GI health (Marks et al., 2011). To better understand the host–microbe interactions, it is important to study the alterations on the bacterial metabolic function and to investigate the host’s response with the help of metabolomics and transcriptomics. It is also crucial to demonstrate how alterations in the microbiota can result in a range of local and systemic pathologies. The microbiota of cats affected with different diseases, such as diabetes mellitus and feline immunodeficiency virus, should be further researched. Additionally, future applications of microbiota are still expanding in cats. Fecal transplant, for instance, has successfully cured Clostridioides difficile infection and IBD in humans and dogs (Gianotti and Moss, 2017; Niina et al., 2019) and ulcerative colitis in cats (Furmanski and Mor, 2017). Finally, all this new information could be applied to design an environmental and dietary strategy and to develop next-generation prebiotics, probiotics, or drugs, which could modulate both the composition and the function of the GI microbial populations and benefit intestinal and host health.
Statements
Author contributions
YL and CS worked on the original draft preparation. AM worked on the language revision. AV, TV, and MH reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AbeciaL.HoylesL.KhooC.FrantzN.McCartneyA. L. (2010). Effects of a novel galactooligosaccharide on the faecal microbiota of healthy and inflammatory bowel disease cats during a randomized, double-blind, cross-over feeding study.Int. J. Probiotics Prebiotics561–68.
2
Al-AsmakhM.ZadjaliF. (2015). Use of germ-free animal models in microbiota- related research.J. Microbiol. Biotechnol.251583–1588. 10.4014/jmb.1501.01039
3
BaileyM. T.DowdS. E.ParryN. M.GalleyJ. D.SchauerD. B.LyteM. (2010). Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium.Infec. Imm.781509–1519. 10.1128/iai.00862-09
4
BakerG.SmithJ.CowanD. (2003). Review and re-analysis of domain-specific 16S primers.J. Microbiol. Meth.55541–555. 10.1016/j.mimet.2003.08.009
5
BarkoP. C.McmichaelM. A.SwansonK. S.WilliamsD. A. (2018). The gastrointestinal microbiome: a review.J. Vet. Intern. Med.329–25.
6
BarryK. A. (2010). Indices of Gut Health and Intestinal Microbial Ecology of the Cat as Affected by Ingestion of Select Carbohydrates Varying in Fermentative Capacity.PhD Thesis, University of Illinois, Urbana, IL.
7
BarryK. A.HernotD. C.Van LooJ.FaheyG. C.De GodoyM. R. (2014). Fructan supplementation of senior cats affects stool metabolite concentrations and faecal microbiota concentrations, but not nitrogen partitioning in excreta.J. Anim. Sci.924964–4971. 10.2527/jas.2013-7510
8
BarryK. A.MiddelbosI. S.Vester BolerB. M.VesterB. M.SwansonK. S.FaheyG. C. (2012). Effects of dietary fiber on the feline gastrointestinal metagenome.J. Proteome Res.115924–5933. 10.1021/pr3006809
9
BarryK. A.WojcickiB. J.MiddelbosI. S.VesterB. M.SwansonK. S.FaheyG. C. (2010). Dietary cellulose, fructooligosaccharides, and pectin modify faecal protein catabolites and microbial populations in adult cats.J. Anim. Sci.882978–2987. 10.2527/jas.2009-2464
10
BellE. T.SuchodolskiJ. S.AnithaI.FleemanL. M.CookA. K.SteinerJ. M.et al (2014). Faecal microbiota of cats with insulin-treated diabetes mellitus.PLoS One9:e108729. 10.1371/journal.pone.0108729
11
BerminghamE. N.SandraK.WayneY.KerrK. R.SwansonK. S.RoyN. C.et al (2013a). Post-weaning diet affects faecal microbial composition but not selected adipose gene expression in the cat (Felis catus).PLoS One8:e80992. 10.1371/journal.pone.0080992
12
BerminghamE. N.WayneY.ButowskiC. F.MoonC. D.MacleanP. H.RosendaleD.et al (2018). The faecal microbiota in the domestic cat (Felis catus) is influenced by interactions between age and diet, a five year longitudinal study.Front. Microbiol.9:1231. 10.3389/fmicb.2018.01231
13
BerminghamE. N.WeidgraafK.HekmanM.RoyN. C.TavendaleM. H.ThomasD. G. (2012). Seasonal and age effects on energy requirements in domestic short-hair cats (Felis catus) in a temperate environment.J. Anim. Physiol. Anim. Nutr.97522–530. 10.1111/j.1439-0396.2012.01293.x
14
BerminghamE. N.YoungW.KittelmannS.KerrK. R.SwansonK. S.RoyN. C.et al (2013b). Dietary format alters faecal bacterial populations in the domestic cat (Felis catus).Microbiol. Open2173–181. 10.1002/mbo3.60
15
BerschneiderH. M. (2002). Alternative diets.Clin. Tech. Small Anim. Pract.171–5.
16
BienzleD. (2014). Fiv in cats - a useful model of hiv in people?Vet. Immunol. Immunopathol.159171–179. 10.1016/j.vetimm.2014.02.014
17
BroseyB. P.HillR. C.ScottK. C. (2000). Gastrointestinal volatile fatty acid concentrations and pH in cats.Am. J. Vet. Res.61359–361. 10.2460/ajvr.2000.61.359
18
CamiloE.ZimmermanJ.MasonJ. B.GolnerB.RussellR.SelhubJ.et al (1996). Folate synthesized by bacteria in the human upper small intestine is assimilated by the host.Gastroenterology110991–998. 10.1053/gast.1996.v110.pm8613033
19
CardingS.VerbekeK.VipondD. T.CorfeB. M.OwenL. J. (2015). Dysbiosis of the gut microbiota in disease.Microb. Ecol. Health Dis.26:26191.
20
CarioE.GerkenG.PodolskyD. K. (2007). Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function.Gastroenterology1321359–1374. 10.1053/j.gastro.2007.02.056
21
ChungH.PampS. J.HillJ. A.SuranaN. K.EdelmanS. M.TroyE. B.et al (2012). Gut immune maturation depends on colonization with a host-specific microbiota.Cell1491578–1593. 10.1016/j.cell.2012.04.037
22
DavidL. A.MauriceC. F.CarmodyR. N.GootenbergD. B.ButtonJ. E.WolfeB. E.et al (2014). Diet rapidly and reproducibly alters the human gut microbiome.Nature505559. 10.1038/nature12820
23
Deb-ChoudhuryS.BerminghamE. N.YoungW.BarnettM. P. G.KnowlesS. O.HarlandD.et al (2018). The effects of a wool hydrolysate on short-chain fatty acid production and faecal microbial composition in the domestic cat (Felis catus).Food Funct.94107–4121. 10.1039/c7fo02004j
24
DengP.SwansonK. S. (2015). GI microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges.Br. J. Nutr.113S6–S17.
25
DepauwS.HestaM.Whitehouse-TeddK.VanhaeckeL.VerbruggheA.JanssensG. P. (2013). Animal fibre: the forgotten nutrient in strict carnivores? First insights in the cheetah.J. Anim. Physiol. Anim. Nutr.97146–154. 10.1111/j.1439-0396.2011.01252.x
26
DesaiA. R.MusilK. M.CarrA. P.HillJ. E. (2008). Characterization and quantification of feline faecal microbiota using cpn60 sequence-based methods and investigation of animal-to-animal variation in microbial population structure.Vet. Microbiol.137120–128. 10.1016/j.vetmic.2008.12.019
27
DethlefsenL.HuseS.SoginM. L.RelmanD. A. (2008). The pervasive effects of an antibiotic on the human GI microbiota, as revealed by deep 16S rRNA sequencing.PLoS Biol.6:e280. 10.1371/journal.pbio.0060280
28
DeuschO.O’FlynnC.ColyerA.MorrisP.AllawayD.JonesP. G.et al (2014). Deep Illumina based shotgun sequencing reveals dietary effects on the structure and function of the faecal microbiome of growing kittens.PLoS One9:e101021. 10.1371/journal.pone.0101021
29
DeuschO.O’FlynnC.ColyerA.SwansonK. S.AllawayD.MorrisP. (2015). A longitudinal study of the feline faecal microbiome identifies changes into early adulthood irrespective of sexual development.PLoS One10:e0144881. 10.1371/journal.pone.0144881
30
Di SabatinoB. (2019). Dietary Management of Obesity Minimally Effects Variation of the Feline Faecal Microbiota’s Metabolic Function.Master’s thesis, University of Guelph, Guelph, ON.
31
EisertR. (2011). Hypercarnivory and the brain: protein requirements of cats reconsidered.J. Comp. Physiol. B1811–17. 10.1007/s00360-010-0528-0
32
FischerM. M.KesslerA. M.KiefferD. A.KnottsT. A.KimK.WeiA.et al (2017). Effects of obesity, energy restriction and neutering on the faecal microbiota of cats.Br. J. Nutr.118513–524. 10.1017/s0007114517002379
33
ForsterG. M.StockmanJ.NoyesN.HeubergerA. L.BroecklingC. D.BantleC. M.et al (2018). A comparative study of serum biochemistry, metabolome and microbiome parameters of clinically healthy, normal weight, overweight, and obese companion dogs.Top. Companion Anim. M.33126–135. 10.1053/j.tcam.2018.08.003
34
ForsytheP.SudoN.DinanT.TaylorV. H.BienenstockJ. (2009). Mood and gut feelings.Brain Behav. Imm.24316–323.
35
FrankD. N.AmandA. L. S.FeldmanR. A.BoedekerE.HarpazN.PaceN. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases.Proc. Natl. Acad. Sci. U.S.A.10413780–13785. 10.1073/pnas.0706625104
36
FurmanskiS.MorT. (2017). First case report of fecal microbiota transplantation in a cat in Israel.Isr. J. Vet. Med.1235–41.
37
Garcia-MazcorroJ. F.LanerieD. J.DowdS. E.PaddockC. G.GrutznerN.SteinerJ. M.et al (2011). Effect of a multi-species synbiotic formulation on faecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing.FEMS Microbiol. Ecol.78542–554. 10.1111/j.1574-6941.2011.01185.x
38
GeukingM. B.CahenzliJ.LawsonM. A.NgD. C.SlackE.HapfelmeierS.et al (2011). Intestinal bacterial colonizati on induces mutualistic regulatory T cell responses.Immunity34794–806. 10.1016/j.immuni.2011.03.021
39
GhoshA.BorstL.StaufferS. H.SuyemotoM.MoisanP.ZurekL.et al (2013). Mortality in kittens is associated with a shift in ileum mucosa-associated enterococci from Enterococcus hirae to biofilm-forming Enterococcus faecalis and adherent Escherichia coli.J. Clin. Microbiol.513567–3578. 10.1128/jcm.00481-13
40
GianottiR. J.MossA. C. (2017). Fecal microbiota transplantation: from clostridium difficile to inflammatory bowel disease.Gastroenterol. Hepatol.13209–213.
41
GillS. R.PopM.DeboyR. T.EckburgP. B.TurnbaughP. J.SamuelB. S.et al (2006). Metagenomic analysis of the human distal gut microbiome.Science3121355–1359.
42
GreethamH. L.GiffardC.HutsonR. A.CollinsM. D.GibsonG. R. (2002). Bacteriology of the Labrador dog gut: a cultural and genotypic approach.J. Appl. Microbiol.93640–646. 10.1046/j.1365-2672.2002.01724.x
43
GrzesKowiakL.EndoA.BeasleyS.SalminenS. (2015). Microbiota and probiotics in canine and feline welfare.Anaerobe3414–23. 10.1016/j.anaerobe.2015.04.002
44
HandlS.DowdS. E.Garcia-MazcorroJ. F.SteinerJ. M.SuchodolskiJ. S. (2011). Massive parallel 16s rrna gene pyrosequencing reveals highly diverse faecal bacterial and fungal communities in healthy dogs and cats.FEMS Microbiol. Ecol.76301–310. 10.1111/j.1574-6941.2011.01058.x
45
HandlS.GermanA. J.HoldenS. L.DowdS. E.SteinerJ. M.HeilmannR. M.et al (2013). Faecal microbiota in lean and obese dogs.FEMS Microbiol. Ecol.84332–343. 10.1111/1574-6941.12067
46
HensonM. S.O’BrienT. D. (2006). Feline models of type 2 diabetes mellitus.ILAR J.47234–242. 10.1093/ilar.47.3.234
47
HoffmannA. R.ProctorL. M.SuretteM. G.SuchodolskiJ. S. (2016). The microbiome the trillions of microorganisms that maintain health and cause disease in humans and companion animals.Vet. Pathol.5310–21. 10.1177/0300985815595517
48
HondaK.TakedaK. (2009). Regulatory mechanisms of immune responses to intestinal bacteria.Mucosal Immunol.2187–196. 10.1038/mi.2009.8
49
HoodaS.BolerB. M. V.KerrK. R.DowdS. E.SwansonK. S. (2013). The gut microbiome of kittens is affected by dietary protein:carbohydrate ratio and associated with blood metabolite and hormone concentrations.Br. J. Nutr.1091637–1646. 10.1017/s0007114512003479
50
HooperL. V. (2009). Do symbiotic bacteria subvert host immunity?Nat. Rev. Microbiol.7367–374. 10.1038/nrmicro2114
51
InnessV. L.McCartneyA. L.KhooC.GrossK. L.GibsonG. R. (2007). Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp.J. Anim. Physiol. An. Nutr.918–53.
52
IshiiS. I.SuzukiS.TenneyA.NealsonK. H.BretschgerO. (2018). Comparative metatranscriptomics reveals extracellular electron transfer pathways conferring microbial adaptivity to surface redox potential changes.ISME J.122844–2863. 10.1038/s41396-018-0238-2
53
JaneczkoS.AtwaterD.BogelE.Greiter-WilkeA.GeroldA.BaumgartM.et al (2008). The relationship of mucosal bacteria to duodenal histopathology, cytokine mRNA, and clinical disease activity in cats with inflammatory bowel disease.Vet. Microbiol.128178–193. 10.1016/j.vetmic.2007.10.014
54
JiaJ.FrantzN.KhooC.GibsonG. R.RastallR. A.McCartneyA. L. (2011a). Investigation of the faecal microbiota of geriatric cats.Lett. Appl. Microbiol.53288–293. 10.1111/j.1472-765x.2011.03105.x
55
JiaJ.FrantzN.KhooC.GibsonG. R.RastallR. A.McCartneyA. L. (2011b). Investigation of the faecal microbiota of kittens: monitoring bacterial succession and effect of diet.FEMS Microbiol. Ecol.78395–404. 10.1111/j.1574-6941.2011.01172.x
56
JohnstonK.LamportA.BattR. M. (1993). An unexpected bacterial flora in the proximal small intestine of normal cats.Vet. Rec.132362–363. 10.1136/vr.132.14.362
57
JohnstonK. L.LamportA. I.BallevreO. P.BattR. M. (2000). Effects of oral administration of metronidazole on small intestinal bacteria and nutrients of cats.Am. J. Vet. Res.611106–1112. 10.2460/ajvr.2000.61.1106
58
JohnstonK. L.SwiftN. C.Van HijfteM. F.RutgersH. C.LamportA.BallevreO.et al (2001). Comparison of the bacterial flora of the duodenum in healthy cats and cats with signs of gastrointestinal tract disease.J Am. Vet. Med. A21848–51. 10.2460/javma.2001.218.48
59
JonesP. J.JewS. (2007). Functional food development: concept to reality.Trend Food Sci. Technol.18387–390. 10.1016/j.tifs.2007.03.008
60
KerrK. R.DowdS. E.SwansonK. S. (2014). Faecal microbiota of domestic cats fed raw whole chicks v. an extruded chicken-based diet.J. Nutr. Sci.31–5.
61
KidaY.ShimizuT.KuwanoK. (2006). Sodium butyrate up-regulates cathelicidin gene expression via activator protein-1 and histone acetylation at the promoter region in a human lung epithelial cell line, EBC-1.Mol. Immunol.431972–1981. 10.1016/j.molimm.2005.11.014
62
KielerI. N.MoLbakL.HansenL. L.Hermann-BankM. L.BjornvadC. R. (2016). Overweight and the feline gut microbiome - a pilot study.J. Anim. Physiol. Anim. Nutr.100478–484. 10.1111/jpn.12409
63
KielerI. N.OstoM.HugentoblerL.PuetzL.GilbertM. T. P.HansenT.et al (2019). Diabetic cats have decreased gut microbial diversity and a lack of butyrate producing bacteria.Sci. Rep.91–13.
64
LambertJ. D.SangS.YangC. S. (2007). Possible controversy over dietary polyphenols: benefits vs risks.Chem. Res. Toxicol.20583–585. 10.1021/tx7000515
65
LawleyT. D.WalkerA. W. (2013). Intestinal colonization resistance.Immunology1381–11. 10.1111/j.1365-2567.2012.03616.x
66
LubbsD. C.VesterB. M.FastingerN. D.SwansonK. S. (2009). Dietary protein concentration affects intestinal microbiota of adult cats: a study using DGGE and qPCR to evaluate differences in microbial populations in the feline gastrointestinal tract.J. Anim. Physiol. Anim. Nutr.93113–121. 10.1111/j.1439-0396.2007.00788.x
67
LutgendorffF.AkkermansL. M.SöderholmJ. D. (2008). The role of microbiota and probiotics in stress-induced gastro-intestinal damage.Curr. Mol. Med.8282–298. 10.2174/156652408784533779
68
MacdonaldM. L.RogersQ. R.MorrisJ. G. (1984). Nutrition of the domestic cat, a mammalian carnivore.Annu. Rev. Nutr.4521–562. 10.1146/annurev.nu.04.070184.002513
69
MarksS. L.RankinS. C.ByrneB. A.WeeseJ. S. (2011). Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control.J. Vet. Intern. Med.251195–1208. 10.1111/j.1939-1676.2011.00821.x
70
Marshall-JonesZ. V.BaillonM. L. A.CroftJ. M.ButterwickR. F. (2006). Effects of Lactobacillus acidophilus DSM13241 as a probiotic in healthy adult cats.Am. J. Vet. Res.671005–1012.
71
MarsilioS.PillaR.SarawichitrB.ChowB.HillS. L.AckermannM. R.et al (2019). Characterization of the fecal microbiome in cats with inflammatory bowel disease or alimentary small cell lymphoma.Sci. Rep.91–11.
72
MasuokaH.ShimadaK.Kiyosue-YasudaT.KiyosueM.OishiY.KimuraS.et al (2017). Transition of the intestinal microbiota of cats with age.PLoS One12:e0181739. 10.1371/journal.pone.0181739
73
McnielE. A. (2001). Vaccine-associated sarcomas in cats: a unique cancer model.Clin. Orthop. Relat. Res.38221–27. 10.1097/00003086-200101000-00005
74
MiddelbosI. S.BolerB. M. V.QuA.WhiteB. A.SwansonK. S.FaheyG. C. (2010). Phylogenetic characterization of faecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing.PLoS One5:e9768. 10.1371/journal.pone.0009768
75
MinamotoY.HoodaS.SeemaH.SwansonK. S.SuchodolskiJ. S. (2012). Feline gastrointestinal microbiota.Anim. Health Res. Rev.1364–77. 10.1017/s1466252312000060
76
NiinaA.KibeR.SuzukiR.YuchiY.TeshimaT.MatsumotoH.et al (2019). Improvement in clinical symptoms and fecal microbiome after fecal microbiota transplantation in a dog with inflammatory bowel disease.Vet. Med.10197–201. 10.2147/vmrr.s230862
77
OsbaldistonG. W.StoweE. C. (1971). Microflora of alimentary tract of cats.Am. J. Vet. Res.321399–1405.
78
OthmanM.AgueroR.LinH. C. (2008). Alterations in intestinal microbial flora and human disease.Curr. Opin. Gastroen.2411–16. 10.1097/mog.0b013e3282f2b0d7
79
OttS. J.MusfeldtM.TimmisK. N.HampeJ.WenderothD. F.SchreiberS. (2004). In vitro alterations of intestinal bacterial microbiota in faecal samples during storage.Diagn. Micr. Infec. Dis.50237–245. 10.1016/j.diagmicrobio.2004.08.012
80
PallottoM. R.De GodoyM. R. C.HolscherH. D.BuffP. R.SwansonK. S. (2018). Effects of weight loss with a moderate-protein, high-fiber diet on body composition, voluntary physical activity, and faecal microbiota of obese cats.Am. J. Vet. Res.79181–190. 10.2460/ajvr.79.2.181
81
PendersJ.ThijsC.Van de BrandtP. A.KummelingI.SnijdersB.StelmaF.et al (2007). GI microbiota composition and development of atopic manifestations in infancy: the koala birth cohort study.Gut56661–667. 10.1136/gut.2006.100164
82
PinnaC. (2014). In vitro effect of dietary protein level and nondigestible oligosaccharides on feline faecal microbiota.J. Anim. Sci.925593–5602. 10.2527/jas.2013-7459
83
PlantingaE. A.BoschG.HendriksW. H. (2011). Estimation of the dietary nutrient profile of free-roaming feral cats: possible implications for nutrition of domestic cats.Br. J. Nutr.106S35–S48.
84
PropsR.KerckhofF. M.RubbensP.De VriezeJ.SanabriaE. H.WaegemanW.et al (2017). Absolute quantification of microbial taxon abundances.ISME J.11584–587. 10.1038/ismej.2016.117
85
QuinceC.WalkerA. W.SimpsonJ. T.LomanN. J.SegataN. (2017). Shotgun metagenomics, from sampling to analysis.Nat. Biotechnol.35833–844. 10.1038/nbt.3935
86
RamadanZ.XuH.LaflammeD.Czarnecki-MauldenG.LiQ. J.LabudaJ.et al (2014). Faecal microbiota of cats with naturally occurring chronic diarrhea assessed using 16S rRNA gene 454-pyrosequencing before and after dietary treatment.J. Vet. Intern. Med.2859–65. 10.1111/jvim.12261
87
RamakrishnaB. S.RoedigerW. E. (1990). Bacterial short chain fatty acids: their role in gastrointestinal disease.Digest. Dis.8337–345. 10.1159/000171266
88
RamotarK.ConlyJ. M.ChubbH.LouieT. J. (1984). Production of menaquinones by intestinal anaerobes.J. Infect. Dis.150213–218. 10.1093/infdis/150.2.213
89
RheeS. H.PothoulakisC.MayerE. A. (2009). Principles and clinical implications of the brain-gut-enteric microbiota axis.Nat. Rev. Gastroenterol. Hepatol.6306–314. 10.1038/nrgastro.2009.35
90
RidlonJ. M.KangD. J.HylemonP. B. (2006). Bile salt biotransformations by human intestinal bacteria.J. Lipid Res.47241–259. 10.1194/jlr.r500013-jlr200
91
RissettoK.VillamilJ. A.SeltingK. A.TylerJ.HenryC. J. (2011). Recent trends in feline intestinal neoplasia: an epidemiologic study of 1,129 cases in the veterinary medical database from 1964 to 2004.J. Am. Anim. Hosp. Assoc.4728–36. 10.5326/jaaha-ms-5554
92
RitchieL. E.SteinerJ. M.SuchodolskiJ. S. (2008). Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis.FEMS Microbiol. Ecol.66590–598. 10.1111/j.1574-6941.2008.00609.x
93
RondeauM. P.MeltzerK.MichelK. E.McManusC. M.WashabauR. J. (2003). Short chain fatty acids stimulate feline colonic smooth muscle contraction.J. Feline Med. Surg.5167–173. 10.1016/s1098-612x(03)00002-0
94
RossiG.JergensA.CerquetellaM.BerardiS.Di CiccoE.BassottiG.et al (2017). Effects of a probiotic (slab51TM) on clinical and histologic variables and microbiota of cats with chronic constipation/megacolon: a pilot study.Benef. Microbes9101–110. 10.3920/bm2017.0023
95
SangerF.NicklenS.CoulsonA. R. (1977). DNA sequencing with chain-terminating inhibitors.Proc. Natl. Acad. Sci. U.S.A.745463–5467. 10.1073/pnas.74.12.5463
96
SantosJ. P. F.AquinoA. A.GlóriaM. B. A.Avila-CamposM. J.ObaP. M.SantosK. M.et al (2018). Effects of dietary yeast cell wall on faecal bacteria and fermentation products in adult cats.J. Anim. Physiol. Anim. Nutr.1021091–1101. 10.1111/jpn.12918
97
SchauberJ.SvanholmC.TermenS.IfflandK.MenzelT.ScheppachW.et al (2003). Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways.Gut52735–741. 10.1136/gut.52.5.735
98
SchmidS. M.SuchodolskiJ. S.PriceJ. M.TolbertM. K. (2018). Omeprazole minimally alters the faecal microbial community in six cats: a pilot study.Front. Vet. Sci.5:79. 10.3389/fvets.2018.00079
99
SchneiderH.SchwiertzA.CollinsM. D.BlautM. (1999). Anaerobic transformation of quercetin-3-glucoside by bacteria from the human intestinal tract.Arch. Microbiol.17181–91. 10.1007/s002030050682
100
SinghR. K.ChangH. W.YanD.LeeK. M.UcmakD.WongK.et al (2017). Influence of diet on the gut microbiome and implications for human health.J. Transl. Med.15:73.
101
SongS. J.LauberC.CostelloE. K.LozuponeC. A.HumphreyG.Berg-LyonsD.et al (2013). Cohabiting family members share microbiota with one another and with their dogs.eLife2:e00458.
102
SonnenburgJ. L.AngenentL. T.GordonJ. I. (2004). Getting a grip on things: how do communities of bacterial symbionts become established in our intestine?Nat. Immunol.5:569. 10.1038/ni1079
103
SparkesA. H.PapasouliotisK.SunvoldG.WerrettG.Gruffydd-JonesG.EganE. A.et al (1998a). Effect of dietary supplementation with fructoolig osaccharides on faecal flora of healthy cats.Am. J. Vet. Res.59436–440.
104
SparkesA. H.PapasouliotisK.SunvoldG.WerrettG.Gruffydd-JonesT. J.ReinhartG. (1998b). Bacterial flora in the duodenum of healthy cats, and effect of dietary supplementation with fructo-oligosaccharides.Am. J. Vet. Res.59431–435.
105
StappenbeckT. S.HooperL. V.GordonJ. I. (2002). Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells.Proc. Natl. Acad. Sci. U.S.A.9915451–15455. 10.1073/pnas.202604299
106
SuchodolskiJ. S. (2011a). Companion animals symposium: microbes and gastrointestinal health of dogs and cats.J. Anim. Sci.891520–1530. 10.2527/jas.2010-3377
107
SuchodolskiJ. S. (2011b). Intestinal microbiota of dogs and cats: a bigger world than we thought.Vet. Clin. N. Am. Small Anim. Pract.41261–272. 10.1016/j.cvsm.2010.12.006
108
SuchodolskiJ. S.CamachoJ.SteinerJ. M. (2008). Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis.FEMS Microbiol. Ecol.66567–578. 10.1111/j.1574-6941.2008.00521.x
109
SuchodolskiJ. S.DowdS. E.WestermarckE.SteinerM.WolcottR. D.SpillmannT.et al (2009). The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rDNA sequencing.BMC Microbiol.10:210. 10.1186/1471-2180-9-210
110
SuchodolskiJ. S.FosterM. L.SohailM. U.LeuteneggerC.QueenE. V.SteinerJ. M.et al (2015). The faecal microbiome in cats with diarrhoea.PLoS One10:e0127378. 10.1371/journal.pone.0127378
111
SudoN. (2006). Stress and GI microbiota: does postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response?Int. Congr. Ser.1287350–354. 10.1016/j.ics.2005.12.019
112
SudoN.ChidaY.AibaY.SonodaJ.OyamaN.YuX. N.et al (2004). Postnatal microbial colonization programs the hypothalamic- pituitary-adrenal system for stress response in mice.J. Physiol.558263–275. 10.1113/jphysiol.2004.063388
113
SuzukiK.MaruyaM.KawamotoS.SitnikK.KitamuraH.AgaceW. W.et al (2010). The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut.Immunity3371–83. 10.1016/j.immuni.2010.07.003
114
SwansonK. S.DowdS. E.SuchodolskiJ. S.MiddelbosI. S.VesterB. M.BarryK. A.et al (2011). Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice.ISME J.5639–649. 10.1038/ismej.2010.162
115
TalM.VerbruggheA.GomezD. E.ChauC.WeeseJ. S. (2017). The effect of storage at ambient temperature on the feline faecal microbiota.BMC Vet. Res.13:256. 10.1186/s12917-017-1188-z
116
TalM.WeeseJ. S.GomezD. E.HestaM.SteinerJ. M.VerbruggheA. (2020). Bacterial fecal microbiota is only minimally affected by a standardized weight loss plan in obese cats.BMC Vet. Res.16:112. 10.1186/s12917-020-02318-2
117
Tal GavrielM. (2018). The Effect of Obesity and Weight Loss on the Feline Faecal Microbiota, Inflammation and Intestinal Health Markers.Ph.D. thesis, University of Guelph, Guelph, ON.
118
TannockG. W. (2005). New perceptions of the GI microbiota: Implications for future research.Gastroenterol. Clin. North Am.34361–382. 10.1016/j.gtc.2005.05.006
119
TeradaA.HaraH.KatoS.KimuraT.FujimoriI.HaraK.et al (1993). Effect of lactosucrose (4G-beta-D-galactosylsucrose) on faecal flora and faecal putrefactive products of cats.J. Vet. Med. Sci.55291–295. 10.1292/jvms.55.291
120
TermenS.TollinM.RodriguezE.SveinsdóttirS. H.JóhannessonB.CederlundA.et al (2008). PU1 and bacterial metabolites regulate the human gene CAMP encoding antimicrobial peptide LL-37 in colon epithelial cells.Mol. Immunol.453947–3955. 10.1016/j.molimm.2008.06.020
121
TizardI. R.JonesS. W. (2018). The microbiota regulates immunity and immunologic diseases in dogs and cats.Vet. Clin. N. Am. Small Anim. Pract.48307–322. 10.1016/j.cvsm.2017.10.008
122
Torres-HendersonC.SummersS.SuchodolskiJ.LappinM. R. (2017). Effect of Enterococcus faecium strain SF68 on gastrointestinal signs and fecal microbiome in cats administered amoxicillin-clavulanate.Top. Companion Anim. Med.32104–108. 10.1053/j.tcam.2017.11.002
123
TunH. M.BrarM. S.KhinN.JunL.HuiR. K.DowdS. E.et al (2012). Gene-centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing.J. Microbiol. Meth.88369–376. 10.1016/j.mimet.2012.01.001
124
VerbruggheA.HestaM.DaminetS.JanssensG. P. (2012). Peculiarities of one-carbon metabolism in the strict carnivorous cat and the role in feline hepatic lipidosis.Crit. Rev. Food Sci. Nutr.52172–182.
125
VillaverdeC.FascettiA. J. (2014). Macronutrients in feline health.Vet. Clin. Small Anim.44699–717. 10.1016/j.cvsm.2014.03.007
126
WeeseJ. S.NicholsJ.JalaliM.LitsterA. (2015). The rectal microbiota of cats infected with feline immunodeficiency virus infection and uninfected controls.Vet. Microbiol.18096–102. 10.1016/j.vetmic.2015.08.012
127
WhittemoreJ. C.StokesJ. E.LaiaN. L.PriceJ. M.SuchodolskiJ. S. (2018). Short and long-term effects of a synbiotic on clinical signs, the fecal microbiome, and metabolomic profiles in healthy research cats receiving clindamycin: a randomized, controlled trial.PeerJ6:e5130. 10.7717/peerj.5130
128
WhittemoreJ. C.StokesJ. E.PriceJ. M.SuchodolskiJ. S. (2019). Effects of a synbiotic on the fecal microbiome and metabolomic profiles of healthy research cats administered clindamycin: a randomized, controlled trial.Gut Microbes10521–539. 10.1080/19490976.2018.1560754
129
WindeyK.De PreterV.VerbekeK. (2012). Relevance of protein fermentation to gut health.Mol. Nutr. Food Res.56184–196. 10.1002/mnfr.201100542
130
YoungW.MoonC. D.ThomasD. G.CaveN. J.BerminghamE. N. (2016). Pre- and postweaning diet alters the faecal metagenome in the cat with differences in vitamin and carbohydrate metabolism gene abundances.Sci. Rep.7:41198.
131
ZentekJ.MarquartB.PietrzakBallevreT. O.RochatF. (2003). Dietary effects on bifidobacteria and Clostridium perfringens in the canine intestinal tract.J. Anim. Physiol. Anim. Nutr.87397–407. 10.1046/j.0931-2439.2003.00451.x
132
ZoetendalE. G.CollierC. T.KoikeS.MackieR. I.GaskinsH. R. (2004). Molecular ecological analysis of the gastrointestinal microbiota: a review.J. Nutr.134465–472. 10.1093/jn/134.2.465
Summary
Keywords
microbiome, gastrointestinal tract, molecular techniques, nutrition and diseases, feline
Citation
Lyu Y, Su C, Verbrugghe A, Van de Wiele T, Martos Martinez-Caja A and Hesta M (2020) Past, Present, and Future of Gastrointestinal Microbiota Research in Cats. Front. Microbiol. 11:1661. doi: 10.3389/fmicb.2020.01661
Received
28 April 2020
Accepted
25 June 2020
Published
24 July 2020
Volume
11 - 2020
Edited by
Amparo Latorre, University of Valencia, Spain
Reviewed by
Jan S Suchodolski, Texas A&M University, United States; Katie Lynn Summers, United States Department of Agriculture (USDA), United States
Updates
Copyright
© 2020 Lyu, Su, Verbrugghe, Van de Wiele, Martos Martinez-Caja and Hesta.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Myriam Hesta, myriam.hesta@ugent.be
This article was submitted to Microbial Symbioses, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.