- 1School of Life Sciences, University of Essex, Colchester, United Kingdom
- 2School of Water, Energy and Environment, Cranfield University, Cranfield, United Kingdom
- 3Institute for Analytics and Data Science, University of Essex, Wivenhoe Park, Essex, United Kingdom
- 4Archipelagos Institute of Marine Conservation, Samos, Greece
In September 2017 the Agia Zoni II sank in the Saronic Gulf, Greece, releasing approximately 500 tonnes of heavy fuel oil, contaminating the Salamina and Athens coastlines. Effects of the spill, and remediation efforts, on sediment microbial communities were quantified over the following 7 months. Five days post-spill, the concentration of measured hydrocarbons within surface sediments of contaminated beaches was 1,093–3,773 μg g–1 dry sediment (91% alkanes and 9% polycyclic aromatic hydrocarbons), but measured hydrocarbons decreased rapidly after extensive clean-up operations. Bacterial genera known to contain oil-degrading species increased in abundance, including Alcanivorax, Cycloclasticus, Oleibacter, Oleiphilus, and Thalassolituus, and the species Marinobacter hydrocarbonoclasticus from approximately 0.02 to >32% (collectively) of the total bacterial community. Abundance of genera with known hydrocarbon-degraders then decreased 1 month after clean-up. However, a legacy effect was observed within the bacterial community, whereby Alcanivorax and Cycloclasticus persisted for several months after the oil spill in formerly contaminated sites. This study is the first to evaluate the effect of the Agia Zoni II oil-spill on microbial communities in an oligotrophic sea, where in situ oil-spill studies are rare. The results aid the advancement of post-spill monitoring models, which can predict the capability of environments to naturally attenuate oil.
Introduction
On the 10th of September 2017, the Agia Zoni II tanker sank in the inner Saronic Gulf, Greece, releasing approximately 500 metric tonnes of heavy fuel oil into the waters and contaminating the surrounding coastlines (IOPC, 2017). The Hellenic Coast Guard deployed ∼600 m of oil boom out to sea within 8 h, which increased to ∼9 km of booms and absorbents in the following weeks. Despite this response, the oil had spread far from the spill site and impacted over 4 km of the Salamina coastline and over 25 km of the Athens Riviera including Glyfada and the port of Piraeus, due to a change in the wind direction (Figure 1). An extensive clean-up response was undertaken on all contaminated beaches, including manual removal of tar balls, flushing and trenching, high-powered washing, removal of sediments for either washing/replacement or disposal at landfill, and the use of absorbents (see Figure 1 and Supplementary Table S1 for sample location and specific treatment/action undertaken at each site). On the 30th November, 2017 the shipwreck was removed, and the clean-up operations ceased in February 2018. On the 28th April, 2018 the Greek government lifted maritime restrictions.
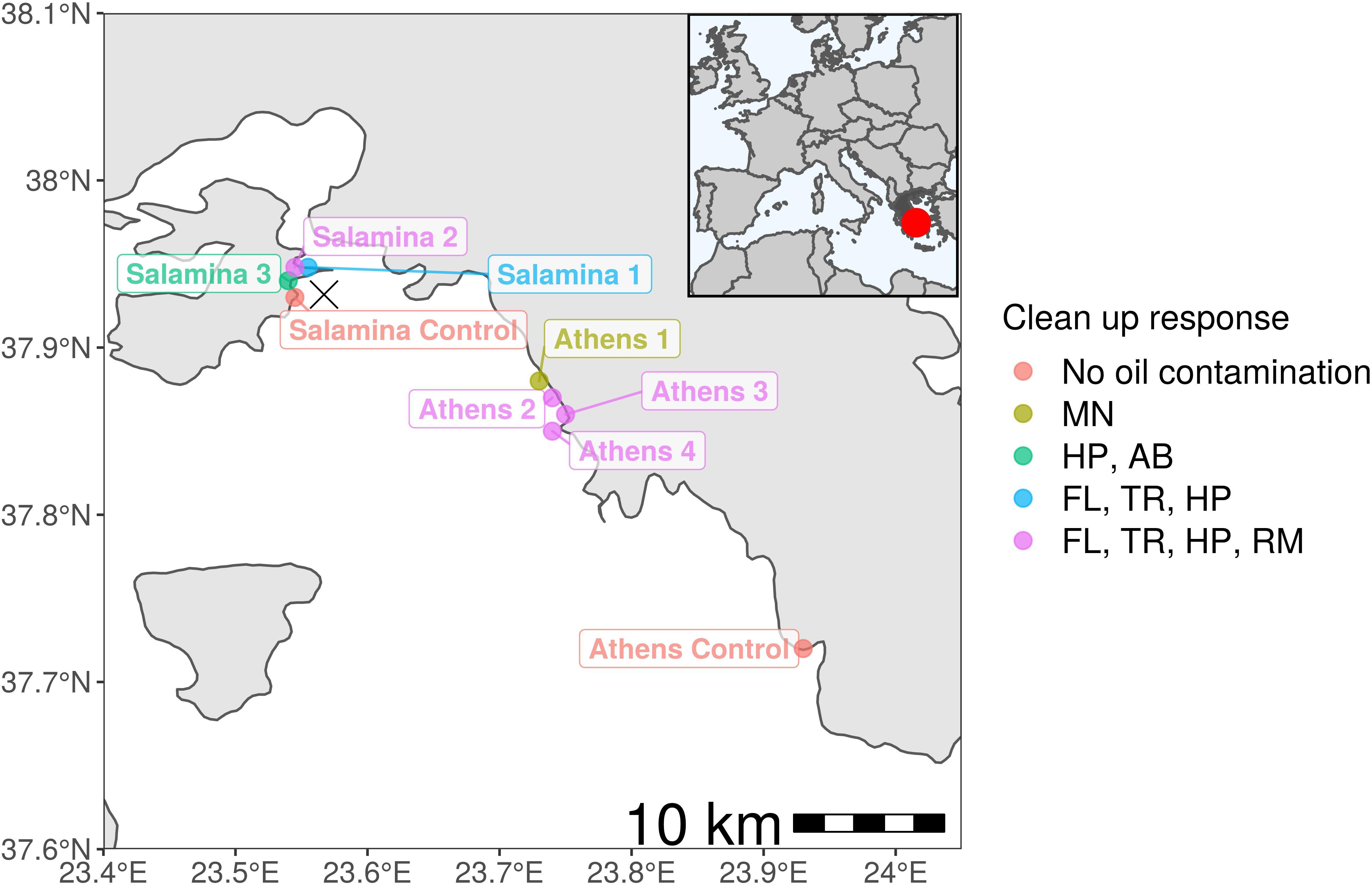
Figure 1. Sampling sites from the Agia Zoni II oil-spill (marked as an X where tanker sank) in Greece (inlet; dark gray; red dot highlights spill-site) and affected Athens Riviera and Salamina coastline. Abbreviations for clean-up response applied at oil-contaminated sample sites (Athens 1–4 and Salamina 1–3): MN (manual removal of tar balls), HP (high-powered washing, to remove oil from hard surfaces), AB (use of absorbents to collect floating oil from the water surface), FL (flushing of sediment with medium pressure water), TR (trenching, used in conjunction with flushing to collect oil), and RM (removal of coastal sediment, either washed and replaced or disposed in landfill).
Oil spills dramatically alter marine microbial community composition, resulting in a decrease in species richness and diversity, in conjunction with selection for oil-degrading bacteria (Head et al., 2006; McGenity et al., 2012). Certain microbes can degrade a range of hydrocarbons found in crude oil and its derivatives, including obligate hydrocarbonoclastic bacteria (OHCB), which use hydrocarbons as an almost exclusive source of carbon and energy (Yakimov et al., 2007). While OHCB have been demonstrated to degrade other compounds in pure culture (Radwan et al., 2019; Zadjelovic et al., 2020), there is still limited evidence that the OHCB are competitive for non-hydrocarbon substrates in the environment. This is evidenced by the fact OHCB are typically present in extremely low numbers in uncontaminated environments but rapidly increase in abundance following oil-spills (Yakimov et al., 2004a; Atlas and Hazen, 2011; Acosta-González et al., 2015; Nogales and Bosch, 2019). Therefore, while it is evident that OHCB use a few other carbon and energy sources, for clarity we refer to them hereafter as “OHCB” to distinguish between these highly adapted and competitive hydrocarbon degraders and those more metabolically diverse bacteria, which can also degrade hydrocarbons. Since the deepwater horizon well blowout, there has been increased emphasis on understanding oil-spill microbial ecology, with much of the focus on deep sea oil plumes (Hazen et al., 2010; Mason et al., 2012; Redmond and Valentine, 2012), salt marsh sediments (Beazley et al., 2012), or benthic sediments (Mason et al., 2014). Though some studies have focused on coastal sediments (Kostka et al., 2011; Lamendella et al., 2014; Huettel et al., 2018) most were conducted either ex situ (Röling et al., 2002; Da Silva et al., 2009; Almeida et al., 2013) or retrospectively (De La Huz et al., 2005; Short et al., 2007; Boufadel et al., 2010), and represented a bias toward a single oil-spill event, deepwater horizon. Therefore, the in situ establishment and succession of specialized oil-degrading microbial taxa, in the Mediterranean immediately following an oil spill, are less well understood. This is especially true for coastal sediments in the Mediterranean (see Supplementary Table S2). Understanding how such ecosystems, and particularly oil-degrading microbial communities, respond to such events will allow for better post-spill monitoring (Kirby et al., 2018). Exposure of environments to oil can also lead to a long-lasting adaptation within the microbial community. This phenomenon of prior exposure can be important in determining the rate at which any subsequent hydrocarbon inputs may be biodegraded (Leahy and Colwell, 1990). Such knowledge will also assist in designing or improving models such as the Ecological Index of Hydrocarbon Exposure (EIHE) (Lozada et al., 2014) and inform future oil-spill response, such as indicating the thoroughness of clean-up efforts and identifying when tourist/fishing activities can recommence.
The Mediterranean is an oligotrophic sea with extremely low levels of phosphorus (Thingstad et al., 2005) and is virtually an enclosed basin with limited oceanic exchange. Phosphorous is a macronutrient that is vital for microbial growth and especially for hydrocarbon degradation (Seah and McFerran, 2016); and therefore, the availability of these nutrients in the presence of hydrocarbons is vital (Ron and Rosenberg, 2014). From 1977 to 2018 the Mediterranean has been subject to 989 recorded oil-pollution incidents (REMPEC, 2018). Additionally, the Mediterranean has been the location of three of the world’s top 20 largest tanker oil spills; including the tankers Haven (1991, 144,000 tonnes, Italy), Irenes Serenade (1980, 100,000 tonnes, Greece), and Independenta (1979, 95,000 tonnes, Turkey) (ITOPF, 2019). The majority of accidents resulting in oil-pollution occur in the Eastern Mediterranean, especially around Greece (Girin and Carpenter, 2019). The Mediterranean currently contains two refineries, two oil terminals, and three oil ports (REMPEC, 2018), with an additional 480 active shipping ports, 50% of which are in located around Greece and Italy (Xue et al., 2004). The Mediterranean is highly socio-politically complex with 21 countries bordering the sea, and future predictions indicate increased levels of oil and gas drilling in the region (Union et al., 2016). Oil pollution in the Mediterranean therefore represents a significant threat to the environment. Despite this, there remains a paucity of information on how oil pollution affects Mediterranean sediment microbial communities.
Our early sampling (5 days post spill) of the Agia Zoni II incident allowed a very rare opportunity to investigate the immediate impact on the microbial community and identify the earliest key colonizing oil-degrading bacteria from an oil spill that covered a large area of coastal sediment, in a region where fishing and tourism play a pivotal socio-economic role. We quantified hydrocarbon concentrations across seven contaminated and two uncontaminated sites 5 days post-spill and after remediation. Additionally, we determined oil-spill-induced changes in gene abundance, diversity, and composition of the sediment microbial communities using qPCR and high throughput sequencing of the 16S rRNA gene.
Results
Sediment Hydrocarbon Concentrations
Five days after the oil spill, the surface sediments at three sample sites on the Athens Riviera were visibly contaminated by hydrocarbons (Figure 2). The concentrations of the aliphatic hydrocarbons fraction including n-alkanes from undecane (C11) to dotriacontane (C32) and the branched alkanes (pristane and phytane) ranged from 1,536 ± 557 to 2,803 ± 549 μg g–1 dry sediment (Figure 2). The concentrations of the aromatic fraction, containing the 2–5 fused-ring polycyclic aromatic hydrocarbons (PAHs), including methylnaphthalenes, dimethylnapthalenes, methylphenanthrenes, and methylanthracenes ranged from 88.20 ± 13.50 to 332 ± 99 μg g–1 dry sediment. One-month post-spill, the concentration of total n-alkanes, branched alkanes, and PAHs reduced to almost undetectable levels at both the Athens Riviera and Salamina sites (Figure 3). Additionally, we found no significant differences in phosphate, nitrate, and ammonium concentrations between contaminated sites and uncontaminated control sites when compared to coastal water samples (Supplementary Figure S1).
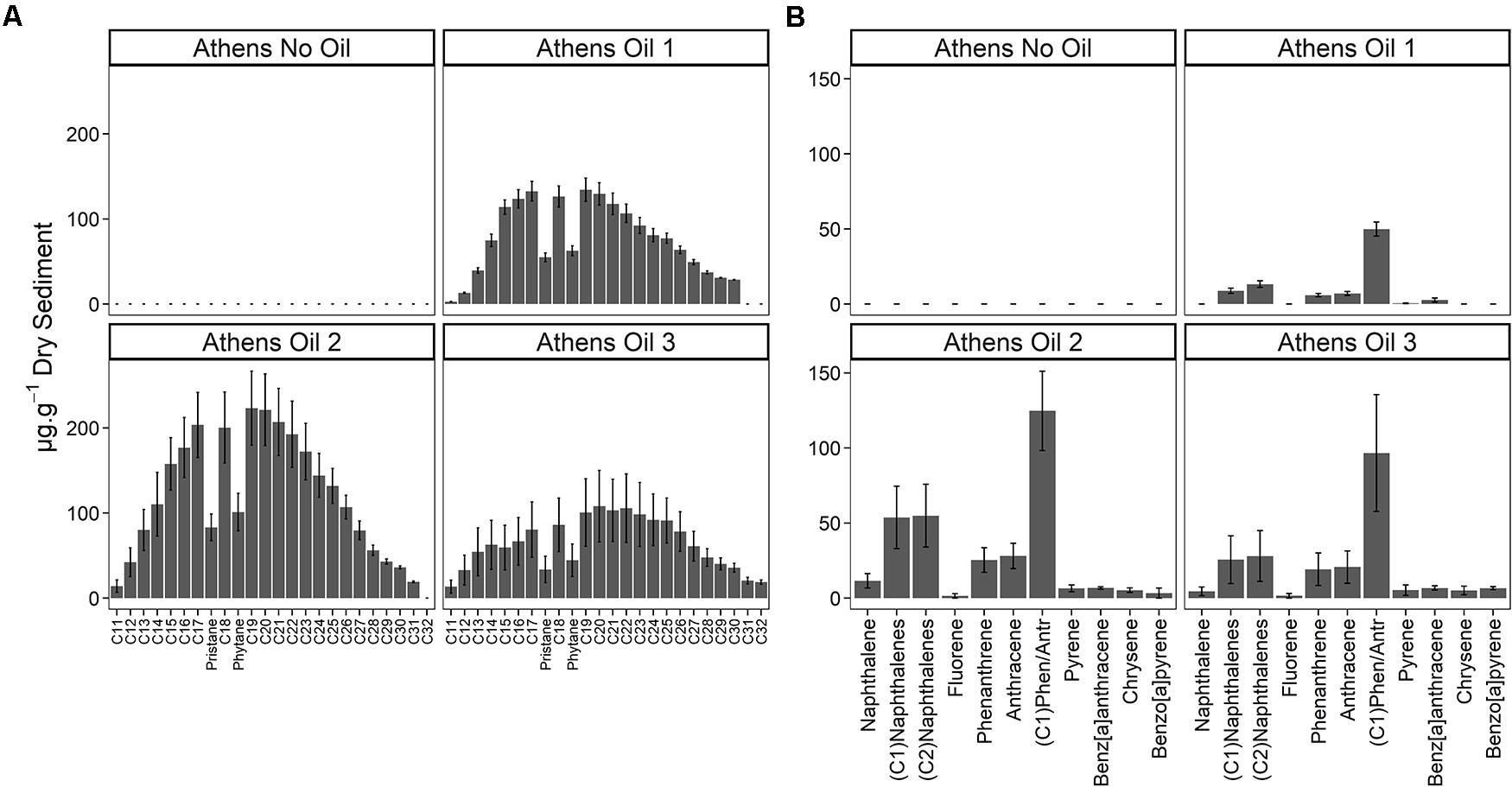
Figure 2. Hydrocarbon concentrations (mean ± SE, n = 3) of the three contaminated sites (Athens Oil 1-3) and the uncontaminated control site (Athens No Oil), September 2017. (A) n-alkanes from C11 to C32 and the branched alkanes pristane and phytane. (B) PAHs from 2-ring naphthalene to 5-ring benzo[a]pyrene. No samples were taken for Salamina in September 2017.
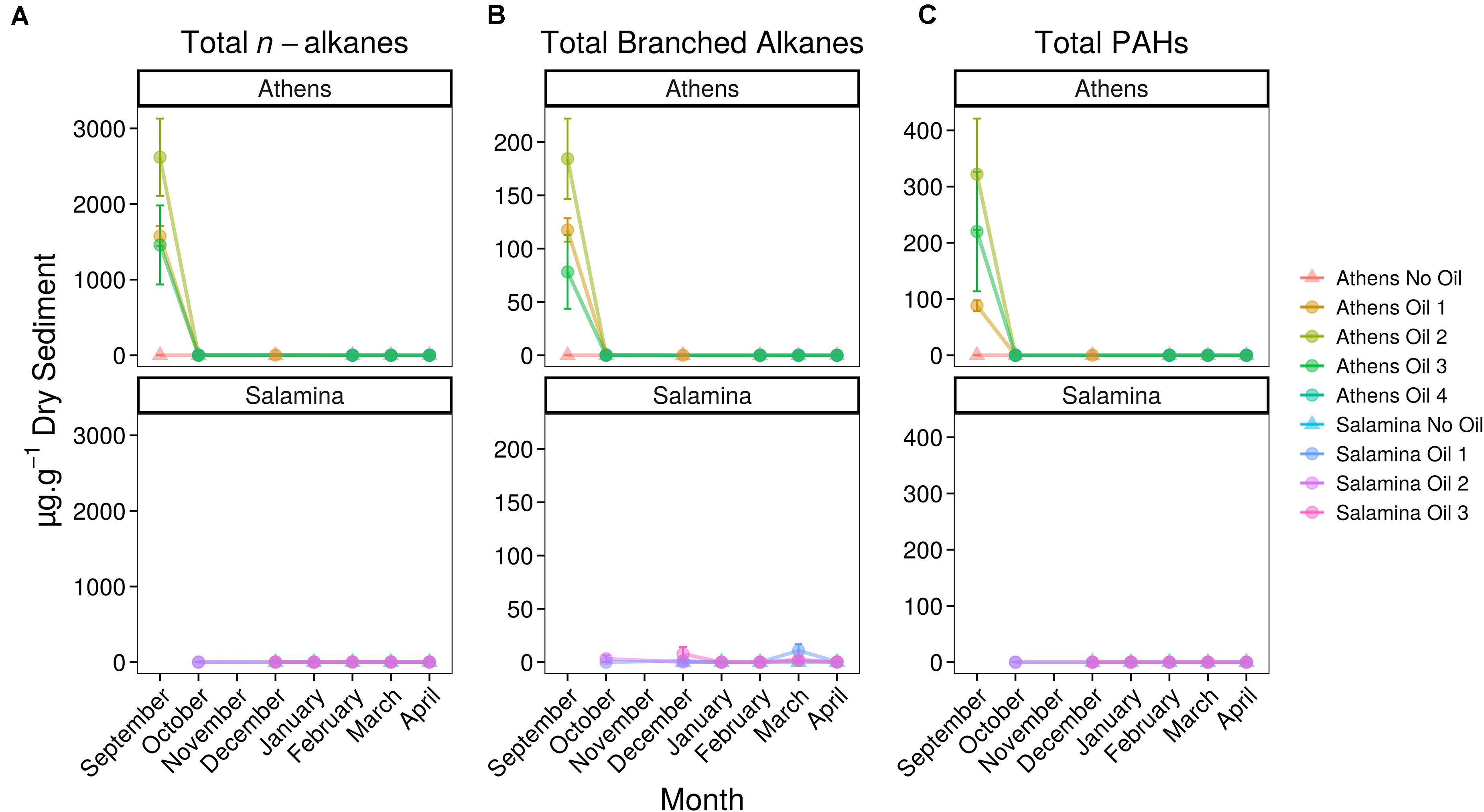
Figure 3. Concentrations (mean ± SE, n = 3) of hydrocarbons in control (Suffix “No Oil”) and contaminated (Suffix “Oil”) sediments along the Athens Riviera and Salamina coastline, sampled from September 2017 to April 2018. (A) Total n-alkanes, (B) total branched alkanes (pristane and phytane), and (C) total PAHs. No samples were collected for all Salamina sites in September 2017, “Salamina No Oil” in October 2017, or all Salamina and Athens sites in November 2017.
Effects of the Oil-Spill on Sediment Microbial Community Composition and Abundance
In order to evaluate any effect that the oil-spill had on microbial community composition or abundance, 16S rRNA bacterial and archaeal genes were analyzed by qPCR and amplicon library sequencing [see operational taxonomic units (OTU) tables in Supplementary Tables S3, S4]. High-throughput 16S rRNA gene sequencing resulted in an average of 55,591 (range 8,093–209,105) and 49,983 (range 3,822–196,697) sequence reads for bacteria and archaea, respectively. Five days post-spill, the mean bacterial 16S rRNA gene abundance (1.6 × 108 copies g–1 dry sediment) in Athens contaminated sediments was ∼2.5 fold greater, but not statistically significant, than in the uncontaminated control sediments (Supplementary Figure S2A). Archaeal 16S rRNA gene abundance was typically two orders of magnitude lower than the Bacteria (Supplementary Figure S2B) and no significant changes were observed in archaeal 16S rRNA gene abundance in response to the oil spill. Simpson Diversity Index analysis of archaeal and bacterial 16S rRNA gene amplicon libraries revealed no significant difference between contaminated (average across all samples, archaea 0.35 and bacteria 0.99) and uncontaminated (archaea 0.26 and bacteria 0.99) sites, at any month. Archaeal community analysis revealed a high relative abundance of Candidatus Giganthauma and Nitrosopumilus, though no significant differences were observed between contaminated and uncontaminated sediment samples. This suggests archaea did not play a significant role in biodegradation of, and were unaffected by, the oil (Supplementary Figure S3).
Operational taxonomic units affiliated to numerous OHCB in contaminated sites 5 days post-spill were significantly more relatively abundant in comparison to uncontaminated control sites. These changes included higher relative abundance of members of Marinobacter, closely related to M. hydrocarbonoclasticus, Alcanivorax spp., Oleibacter spp., Oleiphilus spp., and Thalassolituus spp. (Figure 4) in comparison to uncontaminated sites. In October and December higher relative abundance of Cycloclasticus, a known PAH-degrading genus (Dyksterhouse et al., 1995) was also observed in the contaminated site “Salamina Oil 1.” There was also significantly higher relative abundance of sequences affiliated to more catabolically versatile genera or species (i.e., those that use a wider range of substrates than the OHCB whose substrate range in the environment is typically restricted to hydrocarbons) in comparison to uncontaminated sites. These genera have been shown to contain representatives that can degrade hydrocarbons; including Marinobacter spp., Idiomarina spp., Alteromonas spp., Vibrio spp., Erythrobacter spp., and Roseovarius spp. (Figure 5; see Supplementary Table S2). Dissimilarities in bacterial community composition were evident between uncontaminated and contaminated sites in September 2017 immediately after the oil-spill. However, from October 2017 onward the bacterial community composition at the oiled sites become increasingly similar to as those found at the uncontaminated sites (Supplementary Figure S4).
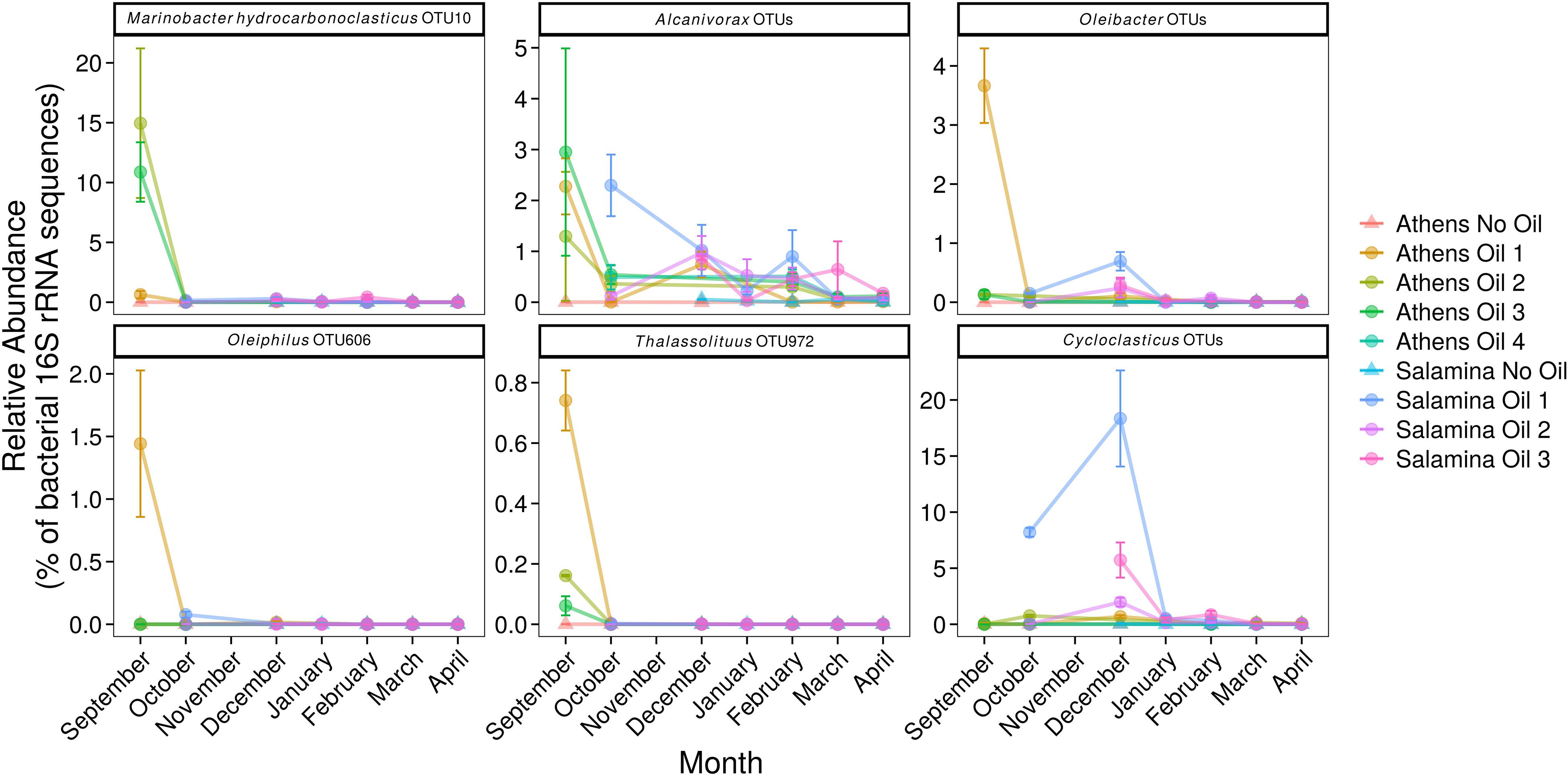
Figure 4. Relative abundance (% of the bacterial community; mean ± SE, n = 3) of 16S rRNA gene OTUs assigned to putative obligate hydrocarbonoclastic bacteria (including OTU-10 which has a 100% identity match to M. hydrocarbonoclasticus) in sediments from control (Suffix “No Oil”) and contaminated (Suffix “Oil”) sites along the Athens Riviera and Salamina coastline, from September 2017 to April 2018. No samples were collected for all Salamina sites in September 2017, “Salamina No Oil” in October 2017, or all Salamina and Athens sites in November 2017. Bacteria and Archaea OTU tables are available in Supplementary Tables S3, S4.
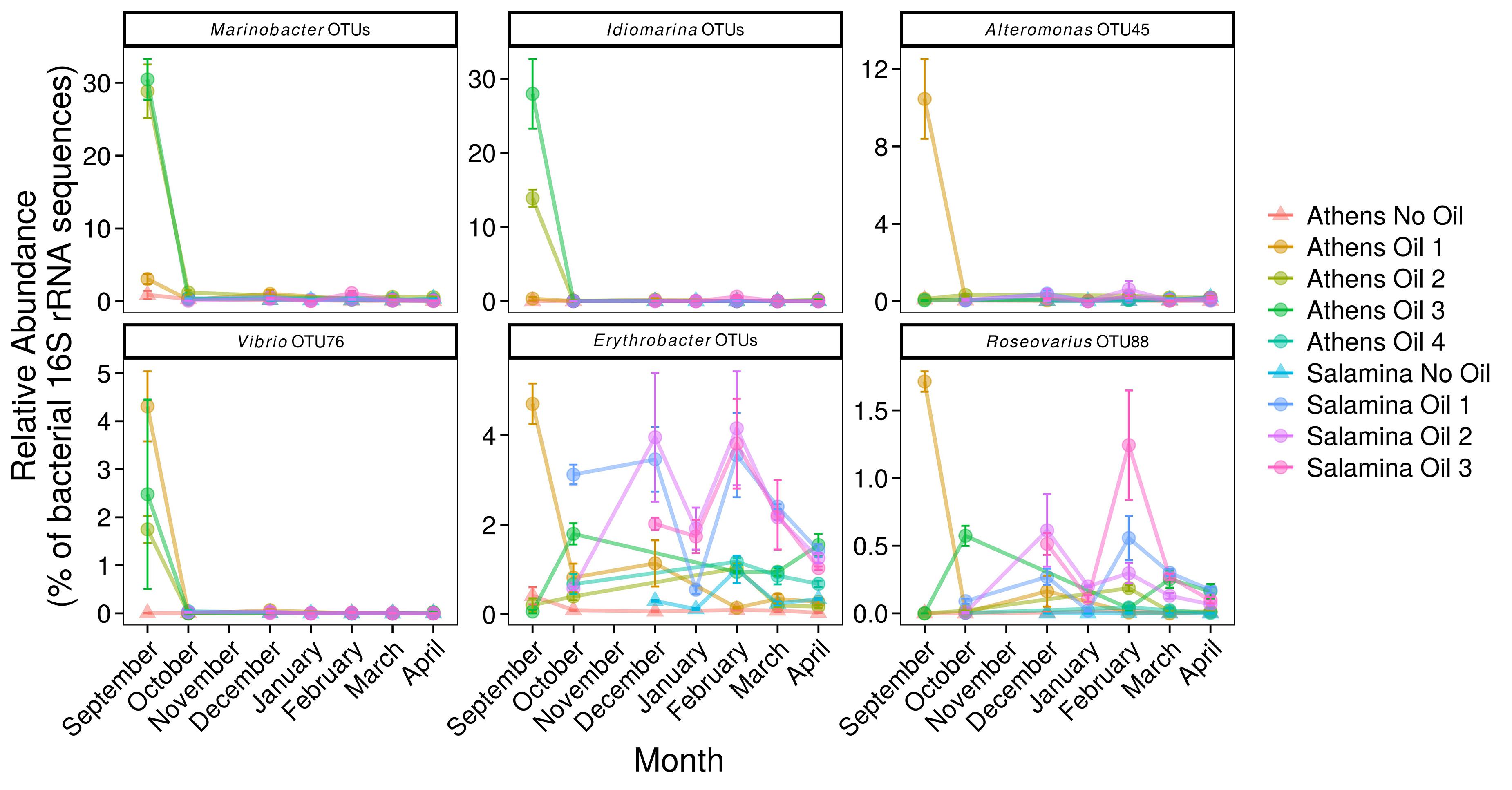
Figure 5. Relative abundance (% of the bacterial community; mean ± SE, n = 3) of 16S rRNA gene OTUs assigned to Bacteria associated with oil-degradation, in sediments from control (Suffix “No Oil”) and contaminated (Suffix “Oil”) sites along the Athens Riviera and Salamina coastline, from September 2017 to April 2018. No samples were collected for all Salamina sites in September 2017, “Salamina No Oil” in October 2017, or all Salamina and Athens sites in November 2017. Bacteria and archaea OTU tables are available in Supplementary Tables S3, S4.
Several of the OTUs that were significantly more relatively abundant at contaminated versus uncontaminated sites were affiliated to OHCB and specifically to known alkane degraders. For example, OTU-10 (100% 16S rRNA sequence identity to Marinobacter hydrocarbonoclasticus; Figures 4, 6), had significantly higher relative abundance at some contaminated sites (coef. = 10.88, z = 18.13, p < 0.001) compared to uncontaminated sites. Five days post-spill, OTU-10, represented 15 and 11% at “Athens Oil 2 and 3,” respectively, but sharply decreased in relative abundance to virtually undetectable levels from October onward. The increase in the relative abundance of M. hydrocarbonoclasticus represented an increase of 2.14 × 107 16S rRNA gene copies assigned to M. hydrocarbonoclasticus compared to uncontaminated Athens sediments. Five OTUs assigned to the OHCB genus Alcanivorax (Figures 4, 6) were significantly more relatively abundant in all contaminated sites (September 2.19% ± 1.52%; coef. = 2.28, z = 6.40, p < 0.001) compared to uncontaminated sites where it was generally undetectable. From October the relative abundance of Alcanivorax OTUs steadily decreased in the contaminated sites, though always remained at least 5- to 297-fold greater than the uncontaminated control sites, where they were often undetectable. Similarly, OTUs from the genus Oleibacter (OTU-200 and OTU-265) and OTU-606 from Oleiphilus (Figures 4, 6) were significantly more abundant in “Athens Oil 1” immediately after the oil-spill in September (coef. = 1.44, z = 13.35, p < 0.001) from uncontaminated control levels (3.67 ± 0.89% and 1.44 ± 0.83%, respectively). However, in April the relative abundance of both genera decreased to levels below 0.01%. Finally, OTU-972 assigned to Thalassolituus (Figures 4, 6), a genus containing species that can degrade a wide range of alkanes (Yakimov et al., 2004b; Gregson et al., 2018), was significantly more relatively abundant in sediments from contaminated sites in September (0.74%; coef. = 0.16, z = 7.52, p < 0.001) compared to uncontaminated control sediments. The relative abundance of Thalassolituus decreased in the months thereafter to undetectable levels. The increase in the relative abundance of Thalassolituus represented an increase of 1.22 × 106 16S rRNA gene copies compared to uncontaminated Athens sediments, even though Thalassolituus was the least abundant OCHB genus.
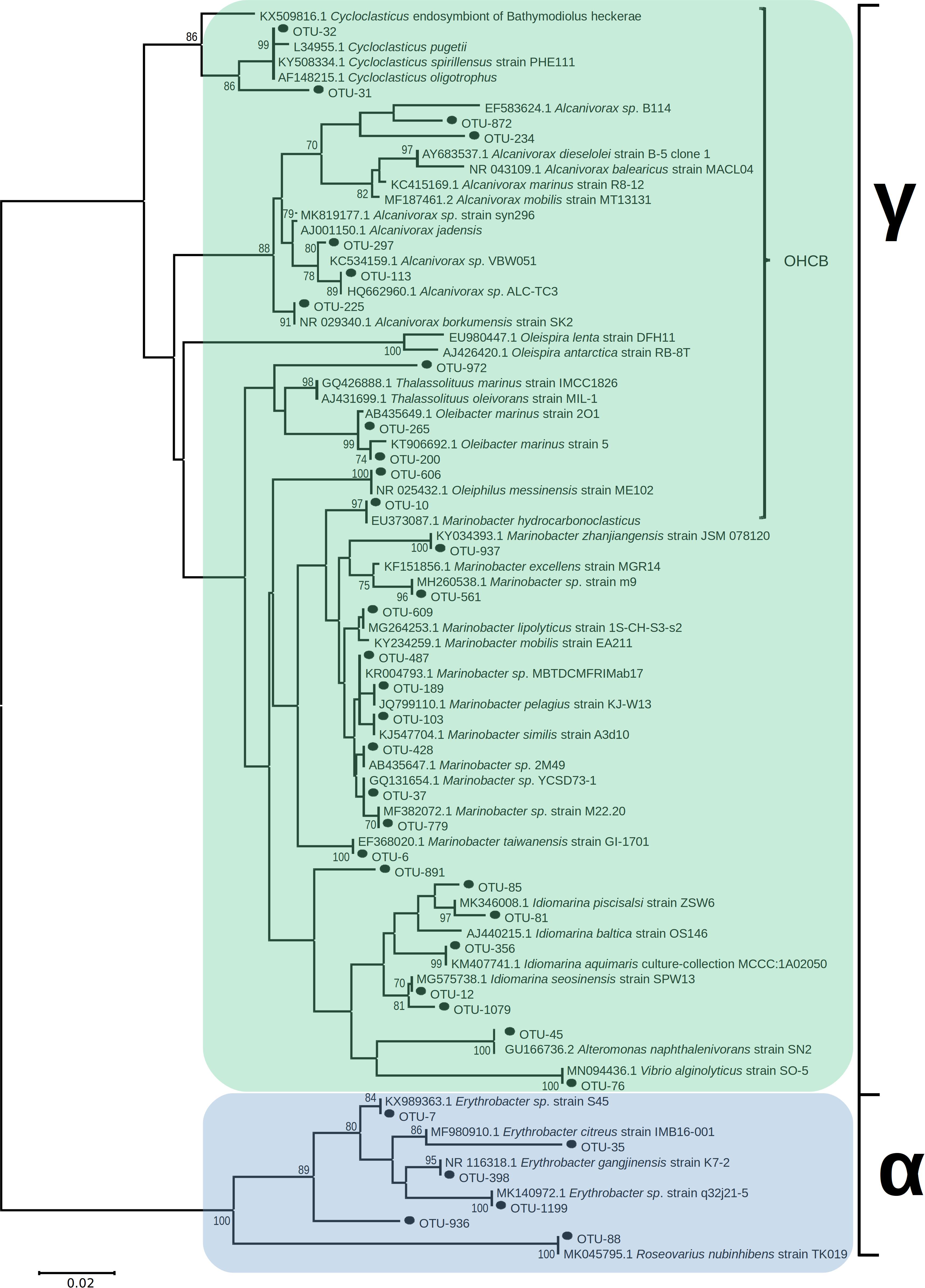
Figure 6. Unrooted neighbor-joining phylogeny of 16S rRNA bacterial OTUs, in sediments from contaminated sites along the Athens Riviera and Salamina coastline, from September 2017 to April 2018, aligned with known hydrocarbon-degrading bacteria and closest relatives; bootstrap values >70 displayed (1,000 iterations). Evolutionary distances computed by Maximum Composite Likelihood protocol (using the Tamura-Nei model, Tamura and Nei, 1993), sum of branch length = 1.15. Analysis involved 82 nucleotide sequences (36 OTUs) with a total of 258 positions in the final dataset. Evolutionary analyses were conducted in MEGA7. Bacteria and archaea OTU tables are available in Supplementary Tables S3, S4.
No known PAH-degrading OHCB were detected five days post-spill when high PAH concentrations were measured. However, OTUs from more metabolically diverse genera (which have been demonstrated to degrade PAHs; see Supplementary Table S2) were observed in significantly higher relative abundance (coef. = 1.75, z = 6.06, p < 0.001) compared to uncontaminated levels, including Alteromonas spp. (10.46 ± 2.91% at “Athens Oil 1”), Idiomarina spp. (13.96 ± 11.75% at “Athens Oil 2 and 3”), and Vibrio spp. (2.09 ± 2.03% across “Athens Oil 1, 2, and 3”). OTU-31 and OTU-32, assigned to the PAH-degrading genus Cycloclasticus (Figures 4, 6), had significantly greater relative abundance in some contaminated locations compared with uncontaminated control sites. For example, in October and December they contributed up to 19% of relative abundance at “Salamina Oil 1” (coef. = 18.31, z = 19.21, p < 0.001), sharply reducing thereafter.
Ten OTUs belonging to the genus Marinobacter (excluding sequences assigned to the OHCB species M. hydrocarbonoclasticus) (Figures 5, 6), were significantly more relatively abundant in sediments taken in September from “Athens Oil 2 and 3” (30%; coef. = 27.94, z = 43.21, p < 0.001) relative to the uncontaminated sites, decreasing sharply from October. Similarly, five OTUs assigned to the genus Idiomarina (Figures 5, 6) were also significantly greater in relative abundance (coef. = 13.88, z = 23.40, p < 0.001), at 14 and 28% from sediments in “Athens Oil 2 and 3,” respectively, decreasing from October onward. OTU-45 assigned to the genus Alteromonas and OTU-76 assigned to the genus Vibrio (Figures 5, 6) significantly increased in relative abundance (coef. = 10.37, z = 26.11, p < 0.001), from uncontaminated control levels, particularly at contaminated “Athens Oil 1” to approximately 10 and 3% respectively, before sharply decreasing from October onward.
Two genera from the class Alphaproteobacteria also increased following oil contamination, which included five OTUs assigned the genus Erythrobacter and OTU-88 assigned to the species Roseovarius spp. (Figures 5, 6), which were significantly higher in relative abundance at contaminated sites from September to April, relative to levels at the uncontaminated sites.
Five days after the oil spill in September there was a strong positive correlation (R2 = 0.80, p < 0.01) between the relative abundance of 16S rRNA sequences assigned to hydrocarbon-degrading bacteria (all OTUs displayed in Figure 6) and the concentration of measured hydrocarbons. In addition, the EIHE, which quantifies the proportion of the bacterial community with hydrocarbon bioremediation potential on a scale of 0–1, whereby 1 represents 100% (Lozada et al., 2014), was calculated at 0.52 ± 0.14 in September. This EIHE result was significantly (p < 0.05) higher than the control site at 0.30 ± 0.11. At the genus level, some correlative relationships between bacterial genera containing hydrocarbon-degraders were observed. OTUs from the genera Marinobacter and Idiomarina typically co-occurred and were significantly correlated (R2 = 0.80, p < 0.05). A difference in the bacterial community composition occurred at different contaminated sites, despite the presence of similar hydrocarbons. For example, at “Athens Oil 2 and 3,” when Marinobacter and Idiomarina were in high relative abundance (collectively 46%) Alteromonas, Oleibacter, Oleiphilus, and Thalassolituus had low relative abundance of approximately 0.64%, collectively. In contrast, when Marinobacter and Idiomarina were in low relative abundance (collectively 4% at “Athens Oil 1” in September) Alteromonas, Oleibacter, Oleiphilus, and Thalassolituus had a relative abundance of approximately 16%, collectively, at the same site in September. The full correlation analysis showing the co-occurrence of taxa is provided in Supplementary Figure S5.
Discussion
Sediment Hydrocarbon Concentrations and Clean-Up Operations
Five days after the oil spill, GC-MS analysis revealed extensive oil contamination in the surface sediments at multiple beaches along the Athens Riviera. This contamination occurred due to rapid transfer of the oil from the water onto the coastal sediments within the first few days of the spill, due to sustained prevailing winds (Parinos et al., 2019). The level of total measured hydrocarbons, 2,158 ± 800 μg g–1 dry sediment, was similar to that observed from surface sediments in the Exxon Valdez oil-spill, 4,636 ± 1,628 μg g–1 sediment (Bragg et al., 1994). However, Agia Zoni II hydrocarbon sediment concentrations were much lower than that observed in buried sediments in Pensacola Beach, Florida (11,000 μg g–1 sediment total petroleum hydrocarbons) which was impacted by the Macondo oil spill (Huettel et al., 2018). Unbranched alkanes and PAHs from sediments after the Agia Zoni II oil spill were undetected from October 2017 onward; though some branched alkanes persisted at low concentrations (primarily at contaminated Salamina sites, Oct–Apr 7.31 ± 7.04 μg g–1 sediment).
The near complete reduction of hydrocarbons to undetectable levels, from October 2017 onward, is in line with measurements taken by the Hellenic Centre for Marine Research (HCMR) (Parinos et al., 2019) and is indicative of efficient physical removal and other clean-up operations, which occurred immediately after the oil-spill incident and continued through to February 2018. The preferential removal of the more readily degradable linear n-alkanes, as indicated by the presence of branched alkanes and no measurable n-alkanes or PAHs from October 2017 onward, would suggest biodegradation was occurring. The extent to which the removal of hydrocarbons can be attributed to either biodegradation, natural processes, or remediation efforts cannot be determined from our data. However, hydrocarbon data from Parinos et al. (2019) suggested the early onset of biodegradation, which was sustained for 85 days post-oil-spill, especially amongst phenanthrenes.
While absorbents, flushing, and sediment washing were used, a large proportion of the clean-up operations at the study sites consisted of direct sediment removal (whether contaminated or not) (International Oil Pollution Compensation Funds, 2018). Though effective in the removal of oil (Dave and Ghaly, 2011), the bulk removal of coastal sediment can be environmentally damaging (Owens, 1972; Gundlach and Hayes, 1978; Petersen et al., 2002), and, has economic and environmental costs associated with disposal, often to landfill sites. In the case of Agia Zoni II, some sediment washing and replacement occurred, but these responses were not adopted to the same degree as removal, likely due to the need for a rapid clean up to minimize the economic impact from loss of tourism.
Ecology of Obligate Hydrocarbonoclastic Bacteria
An oil-spill perturbation is indicative of a discrete short term disturbance event (Shade et al., 2012), whereby changes in microbial community are largely trait driven, by the ability of individual species to occupy available niches due to hydrocarbon-degradation processes and pressures. This often manifests as a large increase in the absolute abundance of Bacteria, due to selection for hydrocarbon-degrading bacteria (Head et al., 2006), which was observed here with a ∼2.5-fold increase, but not significant, in the absolute abundance of the bacterial 16S rRNA gene immediately following the oil-spill. In particular, there was significantly greater relative abundance of genera with known hydrocarbon-degrading bacteria at contaminated sites in comparison to uncontaminated sites. These notable increases, along with a history of hydrocarbon pollution in the Saronic Gulf, indicates an indigenous community of hydrocarbon-degrading bacteria, albeit at mostly very low or undetectable levels, allowing for rapid colonization of oiled sediments and growth. Hydrocarbons within sediments five days after the oil spill supported the growth of OHCB, which rapidly became dominant, and were almost undetectable in uncontaminated sediments.
Alcanivorax is an obligate degrader of branched- and n-alkanes (Yakimov et al., 1998), although it has been shown to enhance degradation of PAHs (McKew et al., 2007). Alcanivorax is ubiquitous in the marine environment and is observed in a variety of hydrocarbon polluted environments (see Supplementary Table S2). In agreement with the results presented here, Alcanivorax is often observed to become abundant early on in coastal beach sediments and decrease in abundance as n-alkanes are removed from the environment (Head et al., 2006; Kostka et al., 2011; Rodriguez-R et al., 2015). Alcanivorax persisted for many months after hydrocarbons (unbranched alkanes and PAHs) were undetected within the environment. This legacy effect could be caused by several processes, including the use of wax esters (Rontani, 2010) or polyhydroxyalkanoates (Możejko-Ciesielska and Kiewisz, 2016) as a carbon and energy store for use when nutrient availability (e.g., N & P) is more favorable for cellular growth. Alternatively, this persistence could be due to the low levels of branched alkanes, such as pristane and phytane, observed within the sediments in the months following September. Unlike most OHCB, certain species of Alcanivorax can use branched alkanes, for example, A. borkumensis SK2T (Yakimov et al., 1998; Gregson et al., 2019), A. dieselolei B-5 (Liu et al., 2011), and A. hongdengensis A-11-3 (Wang and Shao, 2012).
Other alkane-degrading OHCB genera detected included Oleibacter (Teramoto et al., 2011), Oleiphilus (Golyshin et al., 2002), and Thalassolituus (Yakimov et al., 2004b; Gregson et al., 2018); though their abundance was primarily restricted to “Athens Oil 1” sediments. Oleibacter, Oleiphilus, and Thalassolituus have all been observed to increase in abundance in oil-contaminated marine environments, though until now evidence is lacking for detection in coastal sandy sediments (see Supplementary Table S2). Previous studies of oil-contaminated sediment (Coulon et al., 2012) and seawater (Teramoto et al., 2013) observed Oleibacter and Alcanivorax in similar abundances, in line with our observations. However, in seawater-based laboratory experiments Thalassolituus has been observed to outcompete other alkane-degraders such as Alcanivorax (McKew et al., 2007), through its use of medium- and long-chained alkanes (Gregson et al., 2019); this was not observed in the sediment samples collected from the Agia Zoni II oil-spill, whereby Alcanivorax was in greater abundance.
Cycloclasticus is a PAH-degrading genus (Dyksterhouse et al., 1995), apart from one lineage that degrades very short-chained alkanes (symbiont of Bathymodiolus, undetected in this study; Rubin-Blum et al., 2017). Cycloclasticus, which has been observed to grow in many marine environments including sandy coastal sediments (see Supplementary Table S2), was mostly undetected in sediments from Athens, but was observed in high abundance from October through December in Salamina sediments (note that no Salamina samples were taken in September). As found with Alcanivorax, Cycloclasticus demonstrated an oil-legacy effect, persisting in the sediments for several months after PAH contamination was undetectable. Once again this could be due to carbon and energy storage, or the persistence of trace amounts of high molecular weight PAHs. Alternatively, hydrocarbons can be undetected through analytical methods such as GC-MS (McKenna et al., 2013) when weathered hydrocarbons from an oil spill undergo transformation into oxygenated hydrocarbons (though currently there is no evidence that Cycloclasticus is able to degrade oxygenated hydrocarbons) (Kiruri et al., 2013), which can persist for years within oil/sand aggregates (Aeppli et al., 2018).
Ecology of Metabolically Versatile Oil-Degrading Bacteria and Microbial Interactions
The specialist PAH degrader, Cycloclasticus, was mostly undetected in sediments collected in September from Athens, possibly because it was outcompeted by more metabolically versatile bacteria that can also degrade PAHs. Potential candidates for this PAH degradation are the genera Idiomarina [including OTUs most closely matched to PAH degraders I. seosinensis (Yuan et al., 2015; Gomes et al., 2018) (100% similarity) and I. piscisalsi (Nzila et al., 2018) (98.83% similarity)] and Alteromonas [including an OTU most closely matched to PAH degrading A. naphthalenivorans (Jin et al., 2011) (100% similarity)]. Idiomarina and Alteromonas can use a range of substrates for growth, including PAHs, and in this region (Athens) may have outcompeted Cycloclasticus, in contrast to the latter months in Salamina where Cycloclasticus dominated. Given the dominance of Cycloclasticus in October/December in Salamina, it could be speculated that this genus was also dominant in September, in contrast to Athens where more metabolically versatile bacteria became established. Alternatively, differences in the clean-up response may partly account for the observed differences in Cycloclasticus relative abundance, as, for example, the remediation process of sediment removal did not occur at “Salamina Oil 1 and 3” (see Figure 1 and Supplementary Table S1), where an increased relative abundance of Cycloclasticus occurred. This is also the case for sediments from “Athens Oil 1” where the presence of Cycloclasticus was observed, but not in “Athens Oil 2 and 3” sediments.
Idiomarina and Alteromonas have been found in a range of oil-contaminated environments, including sandy coastal sediments (see Supplementary Table S2). In this study it was observed that while both these genera increased in abundance in contaminated sites compared to uncontaminated sites, they did not co-occur. Idiomarina did, however, positively correlate with the genus Marinobacter; including OTU-10 most closely related to M. hydrocarbonoclasticus, which has been shown to degrade a range of alkanes (Gauthier et al., 1992). The co-occurrence of Idiomarina and Marinobacter was most notable between sediments from “Athens Oil 1” and “Athens Oil 2 and 3” and was to the competitive exclusion of other genera with known oil-degrading bacteria, including: Alteromonas, Erythrobacter, Oleibacter, Oleiphilus, and Roseovarius, which co-occurred. Alteromonas and Oleibacter have been recorded previously to co-occur (Neethu et al., 2019). The dominance of Idiomarina and Marinobacter, at “Athens Oil 2 and 3,” could potentially be due to indigenous species having a priority effect, providing a competitive advantage, or potentially due to source point pollution events. Alternatively, the absence of M. hydrocarbonoclasticus and Idiomarina from “Athens Oil 1” sediments could be due to the different remediation efforts at this site, where visible oil contamination was not as evident and thus only manual removal of tar balls occurred (see Figure 1 and Supplementary Table S1). The positive relationship between Marinobacter and Idiomarina has been observed before, including in oil-sludge samples (Albokari et al., 2015) and in sandy coastal sediments from the Macondo oil-spill where these two genera dominated (Joye et al., 2014). Additionally, Joye et al. (2014) observed that Alcanivorax was able to co-occur with Marinobacter and Idiomarina, as also observed in sediments from this study.
This study also demonstrates the ability of Vibrio (which is a genus that contains species that can degrade alkanes (Al-Awadhi et al., 2012; Imron and Titah, 2018) and PAHs (Melcher et al., 2002) to grow in several oil-contaminated environments; see Supplementary Table S2) to also avoid competitive-exclusion by Marinobacter and Idiomarina. This finding could indicate the ability of certain Vibrio to degrade a wide range of hydrocarbons, potentially including branched alkanes, like Alcanivorax, which may indicate why these genera are unaffected by the prevailing dominance of Marinobacter and Idiomarina. Indeed, certain species of Vibrio are known to attach to algal cells (Kumazawa et al., 1991; Hood and Winter, 1997) and many algae are known to produce branched alkanes (Binark et al., 2000) so perhaps there is a phototrophic–heterotroph interaction (McGenity et al., 2012) through the utilization of branched alkanes by Vibrio. However, increased relative abundance in Vibrio spp. was not matched with increased branched alkane concentration in Salamina, and therefore this remains unverified. This co-occurrence could also occur due to Vibrio using PAHs.
Lastly, species within the genera Erythrobacter and Roseovarius can degrade a range of hydrocarbons and grow in oil-contaminated marine environments, including sandy coastal sediments (see Supplementary Table S2). Erythrobacter and Roseovarius maintained consistently higher abundances at the previously oiled sites in comparison to uncontaminated sites. Suggesting that once the genera have established a presence within the community, through the utilization of hydrocarbons, they are able to maintain this even in the absence of such carbon and energy sources.
Are Archaea Affected by Oil-Contamination Within Coastal Sediments?
In agreement with many studies, archaeal abundance and diversity was mostly unaffected in the sediment samples taken from after the Agia Zoni II oil-spill (Redmond and Valentine, 2012; Urakawa et al., 2012; King et al., 2015; Sanni et al., 2015). Archaea are not commonly considered as hydrocarbon-degraders, apart from certain Halobacteria (McGenity, 2010; Oren, 2017) which typically do not prevail in coastal seawater (exception herein being the genera Haladaptatus and Halogranum). There is growing evidence that growth of nitrifying-archaea, especially from the genus Nitrosopumilus, is inhibited by crude oil (Urakawa et al., 2012, 2019), though some studies do not show any significant inhibition by oil on archaeal populations (Newell et al., 2014; Bernhard et al., 2016). In this study, while there were some between-site differences in the relative abundance of OTUs assigned to Nitrosopumilus these were not significantly different. By quantifying archaeal species which are sensitive to oil-spill disturbances, post-oil-spill monitoring models can be made more efficient by including them as sentinel species (Kirby et al., 2018).
Post-Oil-Spill Monitoring of Microbial Communities
The ability of those responding to an oil-spill (oil-spill responders, Government bodies, local authorities, oil-industries, NGOs) to evaluate the efficiency of clean-up operations, and thus provide guidance on future risk-based scenarios, is a vital endeavor to protect the environment and reduce socioeconomic impact. This has been recognized across the globe, and in the United Kingdom the cross-government initiative “PREMIAM” was published to this accord (Kirby et al., 2018). Understanding how microbes respond to such perturbations can assist to this regard and has the potential to be incorporated into tools such as the Spill Impact Mitigation Assessment (IPIECA and IOGP, 2017). Combining the data from both hydrocarbon analysis, microbial ecology, and other environmental sources, a more detailed and holistic understanding can be revealed. Microbial community analysis can highlight whether biodegradation may be taking place and the likelihood of the environment to naturally attenuate the oil-spill. Nutrient concentration analysis would reveal whether the environment is nutrient limited and should therefore be supplemented with fertilizer. Hydrocarbon analysis can detect the proportion of light and heavy molecules that could indicate the need for additional remediation operations.
Using microbial ecology as a post-oil-spill monitoring tool is still in its infancy, however, models and indices to assist in this effort are available. Lozada et al. (2014) published an “EIHE,” whereby hydrocarbon contamination was assumed based on the relative abundance of genera known to contain hydrocarbon-degrading bacteria. The simplicity of this model is both a strength and a weakness. It is simple enough that it can easily be adapted as oil-spill microbial knowledge progresses, but it is too simple to fully establish hydrocarbon exposure. For example, in this study our uncontaminated control sites had an EIHE index of 0.30 ± 0.11 (on a scale of 0–1), despite no hydrocarbons being detected. This is due to the model including all species within functionally diverse bacterial genera such as Pseudomonas which includes >200 different species, of which only a small minority are known to degrade hydrocarbons. The EIHE could be improved further by analyzing microbial community composition at a finer detail, perhaps at a species level, though this would require a tool to phylogenetically match OTUs to known oil-degrading species. Additionally, the inclusion of sentinel microbes, those that are adversely affected by oil-contamination (i.e., ammonia-oxidizing archaea; Urakawa et al., 2012, 2019), would also add comprehension to the index. By advancing the knowledge of oil-spill microbial ecology, understanding the genetics and metabolic capabilities of all hydrocarbon-degrading bacteria and their microbial community interactions, and then combining this with hydrocarbon and environmental data, models such as the EIHE can be improved.
Experimental Procedures
Sampling Locations and Schedule
Sediments were surveyed at both the surface (0–5 cm) and deeper depths (down to approximately 60 cm) on initial and subsequent site visits. However, only surface sediments were observed to be oil contaminated, so all analyses were focused on this upper layer. Samples were collected, in triplicate, in sterile 50 ml sterile polypropylene containers from control (uncontaminated) and contaminated (oil contaminated) sites along the Athens Riviera and Salamina Island coast, Greece (see Figure 1 and Supplementary Table S1). Sediments were randomly sampled (n = 3) across the distance of each beach. Additionally, coastal seawater samples were collected (we were unable to obtain seawater samples from Athens or Salamina for September 2017) for nutrient analysis [ammonium (NH4+), phosphate (PO43–), silicate (SiO2–), nitrate (NO3–), and nitrite (NO2–)] using a SEAL Analytical AA3 HR AutoAnalyzer tandem JASCO FP-2020 Plus fluorescence detector. Samples were collected in September 2017 5 days after the oil-spill, when oiling of sediment was visible, at the Athens sites. However, we were unable to obtain sediment samples from all Salamina site for September 2017. Thereafter, samples were collected over a 7-month period (except for October when no samples for “Salamina No Oil” were collected and November when no samples for all site were collected). Samples were immediately frozen on collection and stored at −20°C.
Hydrocarbon Degradation (GC-MS)
Hydrocarbons were extracted from thawed sediments. Samples (2 g) were dried with 2 g of anhydrous sodium (Na2SO4). Dried samples were extracted with 12 ml of hexane:dichloromethane (50:50) in 20 ml vials with Teflon-lined screw caps by horizontal shaking at 150 oscillations per min over 16 h and finally sonicated for 30 min at 20°C. After centrifugation 12,000 × g for 5 min, extracts were cleaned on Florisil§ columns by elution with hexane. Extracts were transferred in conical tubes and evaporated to 0.7 ml over an ice bath to minimize loss of light PAHs. Deuterated alkanes (nonadecane C19d40 and triacontane C30d62 at 10 μg ml–1) and PAH (naphthalene-d8 and anthrancene-d10 at 10 μg ml–1) internal standards were added to each sample and quantification was performed on an Agilent 7890A Gas Chromatography system coupled with a Turbomass Gold Mass Spectrometer with Triple-Axis detector, operating at 70 eV in positive ion mode, using conditions as previously described by Coulon et al. (2007). External multilevel calibrations were carried out using alkanes [Standard Solution (C8–C40); Sigma], methylated-PAHs (1-methylnapthalene, 2-methylanthracene, and 9,10-dimethylanthracene; Sigma), and PAH (QTM PAH Mix; Sigma) standards, the concentrations of which ranged from 1.125 to 18 μg ml–1. For quality control, a calibration standard (10 μg ml–1) and a blank were analyed every 10 samples. We quantified alkanes between C10 and C36 including pristane and phytane and the following PAHs: naphthalene; all isomers of methyl-, dimethyl- and trimethyl-naphthalenes; acenaphthylene; acenaphthene; fluorene; phenanthrene; all isomers of methyl- and dimethyl-phenanthrenes/anthracenes; fluoranthene; pyrene; all isomers of methyl- and dimethyl-pyrene; chrysene; all isomers of methyl- and dimethyl-chrysene. Only those hydrocarbons detected are shown in Figure 3.
qPCR Analysis of Bacterial and Archaeal 16S rRNA Genes
DNA was extracted from 0.25 g of thawed sediment samples with a DNeasy PowerSoil Kit (Qiagen), according to the manufacturer’s instructions. The primers used for quantification of archaeal 16S rRNA gene were 344f – ACGGGGYGCAGCAGGCGCGA (Raskin et al., 1994) and 915r – GTGCTCCCCCGCCAATTCCT (Stahl and Amann, 1991), and for bacterial 16S rRNA gene, 341f – CCTACGGGNGGCWGCAG, and 785r – GACTACHVGGGTATCTAATCC (Klindworth et al., 2013) were used. These primers have been successfully used to quantify archaeal (Huby et al., 2020) and bacterial (Clark et al., 2020) 16S rRNA gene abundances in environmental samples previously. Furthermore, inspection of standard curves showed that all assays produced satisfactory efficiency (85%) and R2 values (>0.99). All qPCR reactions were performed using a CFX384TM Real-Time PCR Detection System (BioRad) using reagents, cycle conditions, and standards as previously described (McKew and Smith, 2015; Tatti et al., 2016).
Amplicon Sequencing and Bioinformatics
Amplicon libraries were prepared, as per Illumina instructions, by a 25-cycle (bacteria) and 31-cycle (archaea) PCR. PCR primers were the same as those used for qPCR but flanked with Illumina Nextera overhang sequences. A unique combination of Nextera XT Indices (Illumina) were added to PCR products from each sample, via an eight-cycle PCR. PCR products were quantified using PicoGreen and pooled in equimolar concentrations. Quantification of the amplicon libraries was determined via NEBNext® Library Quant Kit for Illumina (New England BioLabs Inc.), prior to sequencing on the Illumina MiSeq® platform, using a MiSeq® 600 cycle v3 reagent kit and 20% PhiX sequencing control standard. Raw sequence data have been submitted to the European Nucleotide Archive database under accession number PRJEB33987. Sequence output from the Illumina MiSeq platform were analyzed within BioLinux (Field et al., 2006), using a bioinformatics pipeline as described by Dumbrell et al. (2016). Forward sequence reads were quality trimmed using Sickle (Joshi and Fass, 2011) prior to error correction within Spades (Nurk et al., 2013) using the BayesHammer algorithm (Nikolenko et al., 2013). The quality filter and error corrected sequence reads were dereplicated, sorted by abundance, and clustered into OTUs at the 97% sequence identity level via VSEARCH (Rognes et al., 2016). Singleton OTUs were discarded, as well as chimeras using reference based chimera checking with UCHIME (Edgar et al., 2011). Lastly, taxonomic assignment was conducted with the Ribosomal Database Project (RDP) classifier (Wang et al., 2007). Non-locus-specific, or artifactual, OTUs were discarded prior to statistical analyses, along with any OTUs that had <70% identity with any sequence in the RDP database.
Phylogenetic Analysis
The neighbor-joining protocol (Nei and Saitou, 1987) was used to infer the evolutionary history of partial 16S rRNA gene sequence Bacterial OTUs, aligned with known hydrocarbon-degrading bacteria and closest neighboring accessions using MUSCLE (Edgar, 2004). Bootstrapping analysis (1,000 iterations) was conducted to determine the percentage of time associated taxa clustered together in replicate trees (Felsenstein, 1985); only bootstrap values >70% are shown. Evolutionary distances, units in the number of base substitutions per site, were calculated with the use of Maximum Composite Likelihood protocol (Hanson and McElroy, 2015). Phylogenetic analyses were conducted in MEGA7.
Statistical Analysis
Prior to community analysis, sequence data were rarefied to the lowest library sequence value (8,093). Data were first tested for normality (Shapiro–Wilks test), those data which were normally distributed were tested for significance with ANOVAs or appropriate linear models. Non-normally distributed data were analyzed using appropriate GLMs (Generalized Linear Models) as follows. The relative abundance of OTUs or genera in relation to oil exposure, site, and sample month were modeled using multivariate negative binomial GLMs (Wang et al., 2010). Here, the number of sequences in each library was accounted for using an offset term, as described previously (Alzarhani et al., 2019). The abundance of archaeal and bacterial 16S rRNA gene copies was also modeled using negative binomial GLMs (Venables and Ripley, 2002). The significance of model terms was assessed via likelihood ratio tests. The EIHE (Lozada et al., 2014) was calculated using the script available at the ecolFudge GitHub page1 (Clark, 2019) and EIHE values modeled using Poisson GLMs. Correlations were performed using the Pearson’s correlation with an alpha of 0.05. All statistical analyses were carried out in R3.6.1 (R Development Core Team, 2011) using a variety of packages available through the references (Venables and Ripley, 2002; Becker et al., 2016; Auguie, 2017; Lenth, 2020). All plots were constructed using the (Wickham, 2010) and “patchwork” (Pedersen, 2019) R packages.
Data Availability Statement
The datasets generated for this study can be found in the European Nucleotide Archive under accession number PRJEB33987.
Author Contributions
GT, CW, TC, and BM carried out the sampling campaign. AM provided the country logistics and sampling support. GT and BG performed the molecular work. GT, PC, and FC performed the analytical chemistry. GT, BM, and DC performed the data and statistical analysis. GT wrote the manuscript, supplementary information, and produced all figures and tables. All authors reviewed the manuscript, supplementary information, figures, and tables. BM, TM, CW, TC, LH, and AM secured the funding. All authors contributed to the article and approved the submitted version.
Funding
We would like to acknowledge and thank the National Environmental Research Council (NERC) for primary funding of this work (NE/R016569/1). Additionally, we thank NERC EnvEast (NE/L002582/1) for funding of this work. DC was supported by NERC funding while contributing to this work (NE/P011624/1). BM also acknowledges funding from the Eastern Academic Research Consortium.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Thodoris Tsimpidis and volunteers at Archipelagos Institute of Marine Conservation (Samos, Greece), for their support during sample collection. We acknowledge ITOPF for providing technical information regarding clean-up operations. Additionally, we acknowledge John Green, Tania Cresswell-Maynard, and Ben Archer from the University of Essex for providing technical and sampling support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01706/full#supplementary-material
Footnotes
References
Acosta-González, A., Martirani-von Abercron, S. M., Rosselló-Móra, R., Wittich, R. M., and Marqués, S. (2015). The effect of oil spills on the bacterial diversity and catabolic function in coastal sediments: a case study on the Prestige oil spill. Environ. Sci. Pollut. Res. 22, 15200–15214. doi: 10.1007/s11356-015-4458-y
Aeppli, C., Swarthout, R. F., O’Neil, G. W., Katz, S. D., Nabi, D., Ward, C. P., et al. (2018). How persistent and bioavailable are oxygenated deepwater horizon oil transformation products? Environ. Sci. Technol. 52, 7250–7258. doi: 10.1021/acs.est.8b01001
Al-Awadhi, H., Dashti, N., Kansour, M., Sorkhoh, N., and Radwan, S. (2012). Hydrocarbon-utilizing bacteria associated with biofouling materials from offshore waters of the Arabian Gulf. Int. Biodeterior. Biodegrad. 69, 10–16. doi: 10.1016/j.ibiod.2011.12.008
Albokari, M., Mashhour, I., Alshehri, M., Boothman, C., and Al-Enezi, M. (2015). Characterization of microbial communities in heavy crude oil from Saudi Arabia. Ann. Microbiol. 65, 95–104. doi: 10.1007/s13213-014-0840-0
Almeida, C. M. R., Reis, I., Couto, M. N., Bordalo, A. A., and Mucha, A. P. (2013). Potential of the microbial community present in an unimpacted beach sediment to remediate petroleum hydrocarbons. Environ. Sci. Pollut. Res. 20, 3176–3184. doi: 10.1007/s11356-012-1240-2
Alzarhani, A. K., Clark, D. R., Underwood, G. J. C., Ford, H., Cotton, T. E. A., and Dumbrell, A. J. (2019). Are drivers of root-associated fungal community structure context specific? ISME J. 13, 1330–1344. doi: 10.1038/s41396-019-0350-y
Atlas, R. M., and Hazen, T. C. (2011). Oil biodegradation and bioremediation: a tale of the two worst spills in U.S. history. Environ. Sci. Technol. 45, 6709–6715. doi: 10.1021/es2013227
Auguie, B. (2017). gridExtra: Functions in Grid Graphics. R Package Version 2.3. Available online at: https://cran.r-project.org/web/packages/gridExtra/index.html (accessed September 9, 2017).
Beazley, M. J., Martinez, R. J., Rajan, S., Powell, J., Piceno, Y. M., Tom, L. M., et al. (2012). Microbial community analysis of a coastal salt marsh affected by the Deepwater Horizon oil spill. PLoS One 7:e41305. doi: 10.1371/journal.pone.0041305
Becker, R. A., Wilks, A. R., Brownrigg, R., Minka, T. P., and Deckmyn, A. (2016). Package “maps”: Draw Geographical Maps. R Packag. version 2.3-6. Available online at: https://cran.r-project.org/web/packages/maps/maps.pdf (accessed April 3, 2018).
Bernhard, A. E., Sheffer, R., Giblin, A. E., Marton, J. M., and Roberts, B. J. (2016). Population dynamics and community composition of ammonia oxidizers in salt marshes after the Deepwater Horizon oil spill. Front. Microbiol. 7:854. doi: 10.3389/fmicb.2016.00854
Binark, N., Güven, K. C., Gezgin, T., and Ünlü, S. (2000). Oil pollution of marine algae. Bull. Environ. Contam. Toxicol. 64, 866–872. doi: 10.1007/s0012800083
Boufadel, M. C., Sharifi, Y., Van Aken, B., Wrenn, B. A., and Lee, K. (2010). Nutrient and oxygen concentrations within the sediments of an Alaskan beach polluted with the Exxon Valdez oil spill. Environ. Sci. Technol. 44, 7418–7424. doi: 10.1021/es102046n
Bragg, J. R., Prince, R. C., Harner, E. J., and Atlas, R. M. (1994). Effectiveness of bioremediation for the Exxon Valdez oil spill. Nature 368, 413–418. doi: 10.1038/368413a0
Clark, D. R. (2019). ecolFudge. Available online at: https://github.com/Dave-Clark/ecolFudge (accessed October 1, 2019).
Clark, D. R., McKew, B. A., Dong, L. F., Leung, G., Dumbrell, A. J., Stott, A., et al. (2020). Mineralization and nitrification: archaea dominate ammonia-oxidising communities in grassland soils. Soil Biol. Biochem. 143, 107725. doi: 10.1016/j.soilbio.2020.107725
Coulon, F., Chronopoulou, P. M., Fahy, A., Païssé, S., Goñi-Urriza, M., Peperzak, L., et al. (2012). Central role of dynamic tidal biofilms dominated by aerobic hydrocarbonoclastic bacteria and diatoms in the biodegradation of hydrocarbons in coastal mudflats. Appl. Environ. Microbiol. 78, 3638–3648. doi: 10.1128/AEM.00072-12
Coulon, F., McKew, B. A., Osborn, A. M., McGenity, T. J., and Timmis, K. N. (2007). Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ. Microbiol. 9, 177–186. doi: 10.1111/j.1462-2920.2006.01126.x
Da Silva, A. C., De Oliveira, F. J. S., Bernardes, D. S., and De França, F. P. (2009). Bioremediation of marine sediments impacted by petroleum. Appl. Biochem. Biotechnol. 153, 58–66. doi: 10.1007/s12010-008-8457-z
Dave, D., and Ghaly, A. E. (2011). Remediation technologies for marine oil spills: a critical review and comparative analysis. Am. J. Environ. Sci. 7, 424–440. doi: 10.3844/ajessp.2011.424.440
De La Huz, R., Lastra, M., Junoy, J., Castellanos, C., and Viéitez, J. M. (2005). Biological impacts of oil pollution and cleaning in the intertidal zone of exposed sandy beaches: preliminary study of the “prestige” oil spill. Estuar. Coast. Shelf Sci. 65, 19–29. doi: 10.1016/j.ecss.2005.03.024
Dumbrell, A. J., Ferguson, R. M. W., and Clark, D. R. (2016). “Microbial community analysis by single-amplicon high-throughput next generation sequencing: data analysis – from raw output to ecology,” in Hydrocarbon and Lipid Microbiology Protocols, eds T. J. McGenity, K. N. Timmis, and B. Nogales, (Berlin: Springer Protocols Handbooks), 155–206. doi: 10.1007/8623_2016_228
Dyksterhouse, S. E., Gray, J. P., Herwig, R. P., Lara, J. C., and Staley, J. T. (1995). Cycloclasticus pugetti gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45, 116–123. doi: 10.1099/00207713-45-1-116
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Field, D., Houten, S., Thurston, M., Swan, D., Tiwari, B., Booth, T., et al. (2006). Open software for biologists: from famine to feast. Nat. Biotechnol. 24, 801–803. doi: 10.1038/nbt0706-801
Gauthier, M. J., Lafay, B., Christen, R., Fernandez, L., Acquaviva, M., Bonin, P., et al. (1992). Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a New, Extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42, 568–576. doi: 10.1099/00207713-42-4-568
Girin, M., and Carpenter, A. (2019). “Shipping and oil transportation in the mediterranean sea,” in Pollution in the Mediterranean Sea: Part I - The International Context, eds A. Carpenter and A. G. Kostianoy, (Berlin: Springer International Publishing). doi: 10.1007/698_2017_6
Golyshin, P. N., Chernikova, T. N., Abraham, W. R., Lünsdorf, H., Timmis, K. N., and Yakimov, M. M. (2002). Oleiphilaceae fam. nov., to include Oleiphilus messinensis gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int. J. Syst. Evol. Microbiol. 52, 901–911. doi: 10.1099/ijs.0.01890-0
Gomes, M. B., Gonzales-Limache, E. E., Sousa, S. T. P., Dellagnezze, B. M., Sartoratto, A., Silva, L. C. F., et al. (2018). Exploring the potential of halophilic bacteria from oil terminal environments for biosurfactant production and hydrocarbon degradation under high-salinity conditions. Int. Biodeterior. Biodegrad. 126, 231–242. doi: 10.1016/j.ibiod.2016.08.014
Gregson, B. H., Metodieva, G., Metodiev, M. V., Golyshin, P. N., and McKew, B. A. (2018). Differential protein expression during growth on medium versus long-chain alkanes in the obligate marine hydrocarbon-degrading bacterium Thalassolituus oleivorans MIL-1. Front. Microbiol. 9:3130. doi: 10.3389/fmicb.2018.03130
Gregson, B. H., Metodieva, G., Metodiev, M. V., and McKew, B. A. (2019). Differential protein expression during growth on linear versus branched alkanes in the obligate marine hydrocarbon-degrading bacterium Alcanivorax borkumensis SK2T. Environ. Microbiol. 21, 2347–2359. doi: 10.1111/1462-2920.14620
Gundlach, E. R., and Hayes, M. O. (1978). Vulnerability of coastal environments to oil spill impacts. Mar. Technol. Soc. J. 12, 18–27.
Hanson, K. M., and McElroy, E. J. (2015). Anthropogenic impacts and long-term changes in herpetofaunal diversity and community composition on a barrier island in the Southeastern United States. Herpetol. Conserv. Biol. 10, 765–780. doi: 10.1073/pnas.0404206101
Hazen, T. C., Dubinsky, E. A., DeSantis, T. Z., Andersen, G. L., Piceno, Y. M., Singh, N., et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330, 204–208. doi: 10.1126/science.1195979
Head, I. M., Jones, D. M., and Röling, W. F. M. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4, 173–182. doi: 10.1038/nrmicro1348
Hood, M. A., and Winter, P. A. (1997). Attachment of Vibrio cholerae under various environmental conditions and to selected substrates. FEMS Microbiol. Ecol. 22, 215–223. doi: 10.1016/S0168-6496(96)00092-X
Huby, T. J. C., Clark, D. R., McKew, B. A., and McGenity, T. J. (2020). Extremely halophilic archaeal communities are resilient to short-term entombment in halite. Environ. Microbiol. doi: 10.1111/1462-2920.14913
Huettel, M., Overholt, W. A., Kostka, J. E., Hagan, C., Kaba, J., Wells, W. B., et al. (2018). Degradation of Deepwater Horizon oil buried in a Florida beach influenced by tidal pumping. Mar. Pollut. Bull. 126, 488–500. doi: 10.1016/j.marpolbul.2017.10.061
Imron, M. F., and Titah, H. S. (2018). Optimization of diesel biodegradation by vibrio alginolyticus using Box-Behnken design. Environ. Eng. Res. 23, 374–382. doi: 10.4491/eer.2018.015
International Oil Pollution Compensation Funds (2018). Agia Zoni II. Available online at: https://www.iopcfunds.org/incidents/incident-map#1740-09-September-2017 (accessed September 9, 2017).
IOPC (2017). Incident Report: Agia Zoni II. 1–5. Available online at: https://documentservices.iopcfunds.org/wp-content/uploads/sites/2/2019/10/IOPC-OCT19-3-11_e.pdf (accessed September 17, 2018).
IPIECA, and IOGP (2017). Guidelines on Implementing Spill Impact Mitigation Assessment, SIMA. IPIECA/IOGP Oil Spill Response JIP. Available online at: http://www.ipieca.org/resources/ (accessed May 28, 2020).
Jin, H. M., Jeong, H., Moon, E. J., Math, R. K., Lee, K., Kim, H. J., et al. (2011). Complete genome sequence of the polycyclic aromatic hydrocarbon-degrading bacterium Alteromonas sp. strain SN2. J. Bacteriol. 193, 4292–4293. doi: 10.1128/JB.05252-11
Joshi, N. A., and Fass, J. N. (2011). Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ files (Version 1.33). Available online at: https://github.com/najoshi/sickle.
Joye, S. B., Teske, A. P., and Kostka, J. E. (2014). Microbial dynamics following the macondo oil well blowout across gulf of Mexico environments. Bioscience 64, 766–777. doi: 10.1093/biosci/biu121
King, G. M., Kostka, J. E., Hazen, T. C., and Sobecky, P. A. (2015). Microbial responses to the deepwater horizon oil spill: from coastal wetlands to the deep sea. Ann. Rev. Mar. Sci. 7, 377–401. doi: 10.1146/annurev-marine-010814-015543
Kirby, M. F., Brant, J., Moore, J., and Lincoln, S. (2018). PREMIAM – Pollution Response in Emergencies – Marine Impact Assessment and Monitoring: Post-incident monitoring guidelines. Secone Edition. Science Series Technical Report. Lowestoft. Available online at: https://www.cefas.co.uk/premiam/guidelines/ (accessed May 28, 2020).
Kiruri, L. W., Dellinger, B., and Lomnicki, S. (2013). Tar balls from deep water horizon oil spill: environmentally persistent free radicals (EPFR) formation during crude weathering. Environ. Sci. Technol. 47, 4220–4226. doi: 10.1021/es305157w
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, 1–11. doi: 10.1093/nar/gks808
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 77, 7962–7974. doi: 10.1128/AEM.05402-11
Kumazawa, N. H., Fukuma, N., and Komoda, Y. (1991). Attachment of Vibrio parahaemolyticus strains to Estuarine Algae. J. Vet. Med. Sci. 53, 201–205. doi: 10.1292/jvms.53.201
Lamendella, R., Strutt, S., Borglin, S., Chakraborty, R., Tas, N., Mason, O. U., et al. (2014). Assessment of the deepwater horizon oil spill impact on gulf coast microbial communities. Front. Microbiol. 5:130. doi: 10.3389/fmicb.2014.00130
Leahy, J. G., and Colwell, R. R. (1990). Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54, 305–315. doi: 10.1128/mmbr.54.3.305-315.1990
Lenth, R. (2020) emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.4.7. Available online at: https://CRANR-project.org/package=emmeans
Liu, C., Wang, W., Wu, Y., Zhou, Z., Lai, Q., and Shao, Z. (2011). Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5. Environ. Microbiol. 13, 1168–1178. doi: 10.1111/j.1462-2920.2010.02416.x
Lozada, M., Marcos, M. S., Commendatore, M. G., Gil, M. N., and Dionisi, H. M. (2014). The bacterial community structure of hydrocarbon-polluted marine environments as the basis for the definition of an ecological index of hydrocarbon exposure. Microbes Environ. 29, 269–276. doi: 10.1264/jsme2.me14028
Mason, O. U., Hazen, T. C., Borglin, S., Chain, P. S. G., Dubinsky, E. A., Fortney, J. L., et al. (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 6, 1715–1727. doi: 10.1038/ismej.2012.59
Mason, O. U., Scott, N. M., Gonzalez, A., Robbins-Pianka, A., Bælum, J., Kimbrel, J., et al. (2014). Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 8, 1464–1475. doi: 10.1038/ismej.2013.254
McGenity, T. J. (2010). “Halophilic hydrocarbon degraders,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. N. Timmis, (Berlin: Springer). doi: 10.1007/978-3-540-77587-4_142
McGenity, T. J., Folwell, B. D., McKew, B. A., and Sanni, G. O. (2012). Marine crude-oil biodegradation: a central role for interspecies interactions. Aquat. Biosyst. 8, 1–19. doi: 10.1186/2046-9063-8-10
McKenna, A. M., Nelson, R. K., Reddy, C. M., Savory, J. J., Kaiser, N. K., Fitzsimmons, J. E., et al. (2013). Expansion of the analytical window for oil spill characterization by ultrahigh resolution mass spectrometry: beyond gas chromatography. Environ. Sci. Technol. 47, 7530–7539. doi: 10.1021/es305284t
McKew, B. A., Coulon, F., Yakimov, M. M., Denaro, R., Genovese, M., Smith, C. J., et al. (2007). Efficacy of intervention strategies for bioremediation of crude oil in marine systems and effects on indigenous hydrocarbonoclastic bacteria. Environ. Microbiol. 9, 1562–1571. doi: 10.1111/j.1462-2920.2007.01277.x
McKew, B. A., and Smith, C. J. (2015). “Real-time PCR approaches for analysis of hydrocarbon-degrading bacterial communities,” in Hydrocarbon and Lipid Microbiology Protocols, eds T. J. McGenity, K. N. Timmis, and B. N. Fernandez, (Berlin: Springer Protocols Handbooks). doi: 10.1007/8623_2015_64
Melcher, R. J., Apitz, S. E., and Hemmingsen, B. B. (2002). Impact of irradiation and polycyclic aromatic hydrocarbon spiking on microbial populations in marine sediment for future aging and biodegradability studies. Appl. Environ. Microbiol. 68, 2858–2868. doi: 10.1128/AEM.68.6.2858-2868.2002
Możejko-Ciesielska, J., and Kiewisz, R. (2016). Bacterial polyhydroxyalkanoates: still fabulous? Microbiol. Res. 192, 271–282. doi: 10.1016/j.micres.2016.07.010
Neethu, C. S., Saravanakumar, C., Purvaja, R., Robin, R. S., and Ramesh, R. (2019). Oil-spill triggered shift in indigenous microbial structure and functional dynamics in different marine environmental matrices. Sci. Rep. 9:1354. doi: 10.1038/s41598-018-37903-x
Nei, M., and Saitou, N. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Newell, S. E., Eveillard, D., Mccarthy, M. J., Gardner, W. S., Liu, Z., and Ward, B. B. (2014). A shift in the archaeal nitrifier community in response to natural and anthropogenic disturbances in the northern Gulf of Mexico. Environ. Microbiol. Rep. 6, 106–112. doi: 10.1111/1758-2229.12114
Nikolenko, S. I., Korobeynikov, A. I., and Alekseyev, M. A. (2013). BayesHammer: bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14:S7. doi: 10.1186/1471-2164-14-S1-S7
Nogales, B., and Bosch, R. (2019). “Microbial communities in hydrocarbon-polluted harbors and marinas,” in Microbial Communities Utilizing Hydrocarbons and Lipids: Members, Metagenomics and Ecophysiology, ed. T. McGenity, (Cham: Springer). doi: 10.1007/978-3-319-60063-5_18-1
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A., Korobeynikov, A., Lapidus, A., et al. (2013). “Assembling genomes and mini-metagenomes from highly chimeric reads,” in Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), eds M. Deng, R. Jiang, F. Sun, and X. Zhang, (Berlin: Springer), 158–170. doi: 10.1007/978-3-642-37195-0_13
Nzila, A., Jung, B. K., Kim, M. C., Ibal, J. C., Budiyanto, F., Musa, M. M., et al. (2018). Complete genome sequence of the polycyclic aromatic hydrocarbons biodegrading bacterium Idiomarina piscisalsi strain 10PY1A isolated from oil-contaminated soil. Korean J. Microbiol. 54, 289–292. doi: 10.7845/kjm.2018.8034
Oren, A. (2017). “Aerobic hydrocarbon-degrading Archaea,” in Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes. Handbook of Hydrocarbon and Lipid Microbiology, ed. T. J. McGenity, (Cham: Springer), 1–12. doi: 10.1007/978-3-319-60053-6_5-1
Owens, E. H. (1972). “The cleaning of gravel beaches polluted by oil,” in Proceedings of the 13th International Conference on Coastal Engineering, Vancouver. doi: 10.9753/icce.v13.143
Parinos, C., Hatzianestis, I., Chourdaki, S., Plakidi, E., and Gogou, A. (2019). Imprint and short-term fate of the Agia Zoni II tanker oil spill on the marine ecosystem of Saronikos Gulf. Sci. Total Environ. 693, 133568. doi: 10.1016/j.scitotenv.2019.07.374
Pedersen, T. L. (2019). patchwork: The Composer of Plots. Cran. Available online at: https://cran.r-project.org/package=patchwork (accessed June 22, 2020).
Petersen, J., Michel, J., Zengel, S., White, M., Lord, C., and Plank, C. (2002). Environmental Sensitivity Index Guidelines. Version 3.0. NOAA Technical Memorandum NOS OR&R 11. Seattle, WA: NOAA.
R Development Core Team (2011). R: A Language and Environment for Statistical Computing. Vienna: R Core Team.
Radwan, S. S., Khanafer, M. M., and Al-Awadhi, H. A. (2019). Ability of the so-called obligate hydrocarbonoclastic bacteria to utilize nonhydrocarbon substrates thus enhancing their activities despite their misleading name. BMC Microbiol. 19:41. doi: 10.1186/s12866-019-1406-x
Raskin, L., Stromley, J. M., Rittmann, B. E., and Stahl, D. A. (1994). Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60, 1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994
Redmond, M. C., and Valentine, D. L. (2012). Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. U.S.A. 109, 20292–20297. doi: 10.1073/pnas.1108756108
REMPEC (2018). Mediterranean Integrated GIS on Marine Pollution Risk Assessment and Response. Valletta: REMPEC.
Rodriguez-R, L. M., Overholt, W. A., Hagan, C., Huettel, M., Kostka, J. E., and Konstantinidis, K. T. (2015). Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 9, 1928–1940. doi: 10.1038/ismej.2015.5
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Röling, W. F. M., Milner, M. G., Jones, D. M., Lee, K., Daniel, F., Swannell, R. J. P., et al. (2002). Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl. Environ. Microbiol. 68, 5537–5548. doi: 10.1128/AEM.68.11.5537-5548.2002
Ron, E. Z., and Rosenberg, E. (2014). Enhanced bioremediation of oil spills in the sea. Curr. Opin. Biotechnol. 27, 191–194. doi: 10.1016/j.copbio.2014.02.004
Rontani, J.-F. (2010). “Production of wax esters by bacteria,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. Timmis, (Berlin: Springer), 459–470. doi: 10.1007/978-3-540-77587-4_34
Rubin-Blum, M., Antony, C. P., Borowski, C., Sayavedra, L., Pape, T., Sahling, H., et al. (2017). Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat. Microbiol. 2:17093. doi: 10.1038/nmicrobiol.2017.93
Sanni, G. O., Coulon, F., and McGenity, T. J. (2015). Dynamics and distribution of bacterial and archaeal communities in oil-contaminated temperate coastal mudflat mesocosms. Environ. Sci. Pollut. Res. 22, 15230–15247. doi: 10.1007/s11356-015-4313-1
Seah, C. H., and McFerran, K. S. (2016). The transition to practice experience of five music therapy graduates. Nord. J. Music Ther. 25, 352–371. doi: 10.1080/08098131.2015.1080288
Searle, S. R., Speed, F. M., and Milliken, G. A. (1980). Population marginal means in the linear model: an alternative to least squares means. Am. Stat. 34, 216–221. doi: 10.1080/00031305.1980.10483031
Shade, A., Peter, H., Allison, S. D., Baho, D. L., Berga, M., Bürgmann, H., et al. (2012). Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3:417. doi: 10.3389/fmicb.2012.00417
Short, J. W., Irvine, G. V., Mann, D. H., Maselko, J. M., Pella, J. J., Lindeberg, M. R., et al. (2007). Slightly weathered Exxon Valdez oil persists in Gulf of Alaska beach sediments after 16 years. Environ. Sci. Technol. 41, 1245–1250. doi: 10.1021/es0620033
Stahl, D. A., and Amann, R. (1991). “Development and application of nucleic acid probes in bacterial systematics,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow, (Chichester: John Wiley & Sons Ltd), 205–248.
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Tatti, E., McKew, B. A., Whitby, C., and Smith, C. J. (2016). Simultaneous dna-rna extraction from coastal sediments and quantification of 16S rRNA genes and transcripts by real-time PCR. J. Vis. Exp. 2016:e54067. doi: 10.3791/54067
Teramoto, M., Ohuchi, M., Hatmanti, A., Darmayati, Y., Widyastuti, Y., Harayama, S., et al. (2011). Oleibacter marinus gen. nov., sp. nov., a bacterium that degrades petroleum aliphatic hydrocarbons in a tropical marine environment. Int. J. Syst. Evol. Microbiol. 61, 375–380. doi: 10.1099/ijs.0.018671-0
Teramoto, M., Queck, S. Y., and Ohnishi, K. (2013). Specialized Hydrocarbonoclastic bacteria prevailing in seawater around a port in the strait of malacca. PLoS One 8:e66594. doi: 10.1371/journal.pone.0066594
Thingstad, T. F., Krom, M. D., Mantoura, R. F. C., Flaten, C. A. F., Groom, S., Herut, B., et al. (2005). Nature of phosphorus limitation in the ultraoligotrophic eastern Mediterranean. Science 309, 1068–1071. doi: 10.1126/science.1112632
Union, E. U. E., and Ttf, D. International Association of Oil & Gas (2016). Europe Exploration and Production Trends 2016. London: International Association of Oil & Gas.
Urakawa, H., Garcia, J. C., Barreto, P. D., Molina, G. A., and Barreto, J. C. (2012). A sensitive crude oil bioassay indicates that oil spills potentially induce a change of major nitrifying prokaryotes from the Archaea to the Bacteria. Environ. Pollut. 164, 42–45. doi: 10.1016/j.envpol.2012.01.009
Urakawa, H., Rajan, S., Feeney, M. E., Sobecky, P. A., and Mortazavi, B. (2019). Ecological response of nitrification to oil spills and its impact on the nitrogen cycle. Environ. Microbiol. 21, 18–33. doi: 10.1111/1462-2920.14391
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S (Fourth Edition). New York, NY: Springer-Verlag.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, W., and Shao, Z. (2012). Genes involved in alkane degradation in the alcanivorax hongdengensis strain A-11-3. Appl. Microbiol. Biotechnol. 94, 437–448. doi: 10.1007/s00253-011-3818-x
Wang, W., Wang, L., and Shao, Z. (2010). Diversity and abundance of oil-degrading bacteria and alkane hydroxylase (alkB) genes in the subtropical seawater of Xiamen island. Microb. Ecol. 60, 429–439. doi: 10.1007/s00248-010-9724-4
Wilke, C. O. (2015). Cowplot: Streamlined Plot Theme and Plot Annotations for ggplot2. R Packag. Version 0.5.0. Available online at: https://cran.r-project.org/web/packages/cowplot/index.html (accessed July 11, 2019).
Xue, X., Hong, H., and Charles, A. T. (2004). Cumulative environmental impacts and integrated coastal management: the case of Xiamen. China. J. Environ. Manage. 71, 271–283. doi: 10.1016/j.jenvman.2004.03.006
Yakimov, M. M., Gentile, G., Bruni, V., Cappello, S., D’Auria, G., Golyshin, P. N., et al. (2004a). Crude oil-induced structural shift of coastal bacterial communities of rod bay (Terra Nova Bay, Ross Sea, Antarctica) and characterization of cultured cold-adapted hydrocarbonoclastic bacteria. FEMS Microbiol. Ecol. 49, 419–432. doi: 10.1016/j.femsec.2004.04.018
Yakimov, M. M., Giuliano, L., Denaro, R., Crisafi, E., Chernikova, T. N., Abraham, W. R., et al. (2004b). Thalassolituus oleivorans gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int. J. Syst. Evol. Microbiol. 54, 141–148. doi: 10.1099/ijs.0.02424-0
Yakimov, M. M., Golyshin, P. N., Lang, S., Moore, E. R. B., Abraham, W. R., Lünsdorf, H., et al. (1998). Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon- degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48, 339–348. doi: 10.1099/00207713-48-2-339
Yakimov, M. M., Timmis, K. N., and Golyshin, P. N. (2007). Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18, 257–266. doi: 10.1016/j.copbio.2007.04.006
Yuan, J., Lai, Q., Sun, F., Zheng, T., and Shao, Z. (2015). The diversity of PAH-degrading bacteria in a deep-sea water column above the southwest Indian ridge. Front. Microbiol. 6:853. doi: 10.3389/fmicb.2015.00853
Keywords: Mediterranean, hydrocarbons, Agia Zoni II, oil spill, Alcanivorax, Cycloclasticus, Idiomarina, Greece
Citation: Thomas GE, Cameron TC, Campo P, Clark DR, Coulon F, Gregson BH, Hepburn LJ, McGenity TJ, Miliou A, Whitby C and McKew BA (2020) Bacterial Community Legacy Effects Following the Agia Zoni II Oil-Spill, Greece. Front. Microbiol. 11:1706. doi: 10.3389/fmicb.2020.01706
Received: 12 March 2020; Accepted: 29 June 2020;
Published: 17 July 2020.
Edited by:
Tony Gutierrez, Heriot-Watt University, United KingdomReviewed by:
Joaquim Vila, University of Barcelona, SpainCamilla Lothe Nesbø, University of Alberta, Canada
Casey R. J. Hubert, University of Calgary, Canada
Copyright © 2020 Thomas, Cameron, Campo, Clark, Coulon, Gregson, Hepburn, McGenity, Miliou, Whitby and McKew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gareth E. Thomas, Z3Rob21hYkBlc3NleC5hYy51aw==; Boyd A. McKew, Ym95ZC5tY2tld0Blc3NleC5hYy51aw==
 Gareth E. Thomas
Gareth E. Thomas Tom C. Cameron1
Tom C. Cameron1 Pablo Campo
Pablo Campo Dave R. Clark
Dave R. Clark Frederic Coulon
Frederic Coulon Benjamin H. Gregson
Benjamin H. Gregson Terry J. McGenity
Terry J. McGenity Corinne Whitby
Corinne Whitby Boyd A. McKew
Boyd A. McKew