- 1Pfizer Vaccine Research and Development, Pearl River, NY, United States
- 2Pfizer Vaccines Medical Development, Scientific and Clinical Affairs, Collegeville, PA, United States
- 3Pfizer Medical Development, Scientific and Clinical Affairs, Tadworth, United Kingdom
by da Silva, R. A. G., Karlyshev, A. V., Oldfield, N. J., Wooldridge, K. G., Bayliss, C. D., Ryan, A., et al. (2019). Front. Microbiol. 10:2847. doi: 10.3389/fmicb.2019.02847
Introduction
Meningitis and/or septicemia caused by Neisseria meningitidis remains a serious public health concern. The factor H binding protein (FHbp) is a surface lipoprotein and an antigen component of the two vaccines that are licensed in the US for the prevention of N. meningitidis serogroup B (MenB) disease (TRUMENBA®, 2014; BEXSERO®, 2015). Both vaccines were licensed based on the demonstration that they induce antibody responses that kill MenB isolates in serum bactericidal activity assays using human complement (hSBA). hSBA titers of >1:4 are the established correlate of protection against meningococcal disease (Goldschneider et al., 1969).
Conclusions From da Silva et al. (2019)
Four classes of FHbp signal peptide (SP) sequences (SP1-SP4) were identified. After screening 18 MenB strains, the authors provide data illustrating that FHbp variants coding for classes SP2-SP4 are not exported to the cell surface via the traditional lipoprotein pathway. These FHbp variants could still be detected on the cell surface (albeit at levels lower than SP1 FHbp variants) and were able to bind the cognate ligand, factor H. FHbp-specific monoclonal antibodies were used to conclude that strains expressing SP2-4 FHbp variants were less susceptible in hSBAs than strains expressing SP1 FHbp variants (which utilize the traditional lipoprotein transport mechanism). The authors note that SP1 FHbp variants are representative of <12% of invasive MenB isolates collected in the UK during 2009-2017 and caution that statements regarding the breadth of coverage for Trumenba® need to consider the SP sequence of FHbp variants expressed by target strains used to demonstrate the immune response to vaccination.
Rebuttal
da Silva et al. (2019) suggest that Trumenba® coverage (and to a lesser extent Bexsero®) may be overstated considering their findings. The immune response to Trumenba® was defined during clinical development using strains expressing FHbp variants that: (i) were heterologous to vaccine antigens, (ii) were representative of prevalent variants, (iii) express antigen at representative levels, and (iv) were selected in an unbiased manner (Donald et al., 2017; Harris et al., 2017). A considerable body of data was used to support vaccine licensure by national regulatory authorities. In the European Summary of Product Characteristics, the data for Trumenba® highlight the potential to kill over 91% of isolates (https://www.ema.europa.eu/en/documents/product-information/trumenba-epar-product-information_en.pdf) and 78% for Bexsero® (https://www.ema.europa.eu/en/documents/product-information/bexsero-epar-product-information_en.pdf). Licensure in both instances was supported by assays that determined the level of FHbp expression using thousands of MenB strains collected systematically from different geographical regions so that the breadth of coverage could be ascertained. These data were generated prior to the recognition of alternative FHbp maturation pathways that are experimentally detailed in the da Silva et al. (2019) publication. In the US, Trumenba® licensure was based on demonstrating the ability of vaccine induced antibodies to kill a broad range of MenB strains with diverse FHbp variants and as such is the only vaccine approved for broad coverage against diverse MenB strains. The authors suggest that their findings translate to the potential for ineffective coverage elicited by Trumenba®. We are concerned that this misleading statement will impact discussions between medical professionals and their patients regarding decisions to vaccinate. For this reason, we are providing details of the MenB strains that were used to demonstrate the breadth of coverage of Trumenba®.
Assessments of the breadth of coverage to vaccination with Trumenba® in phase 3 clinical trials used four primary hSBA strains and ten additional test strains that met the requirements described earlier. Characteristics of these strains are listed in Table 1. SP sequences corresponding to each of the four classes of SP allelic variants are represented multiple times. Therefore, regardless of the mechanism of biochemical processing, these clinically relevant hSBA strains each express surface exposed FHbp and are susceptible to Trumenba® immune sera (Harris et al., 2017; Ostergaard et al., 2017; Taha et al., 2017). Thus, the concerns raised in da Silva et al. (2019) regarding the potential coverage of Trumenba® can now be shown to be unfounded. Likewise, the proposed algorithm using SP sequences to predict whether an isolate expresses FHbp at levels sufficient to be susceptible to the bactericidal activity of Trumenba® immune sera would also not be relevant based on the results we present here.
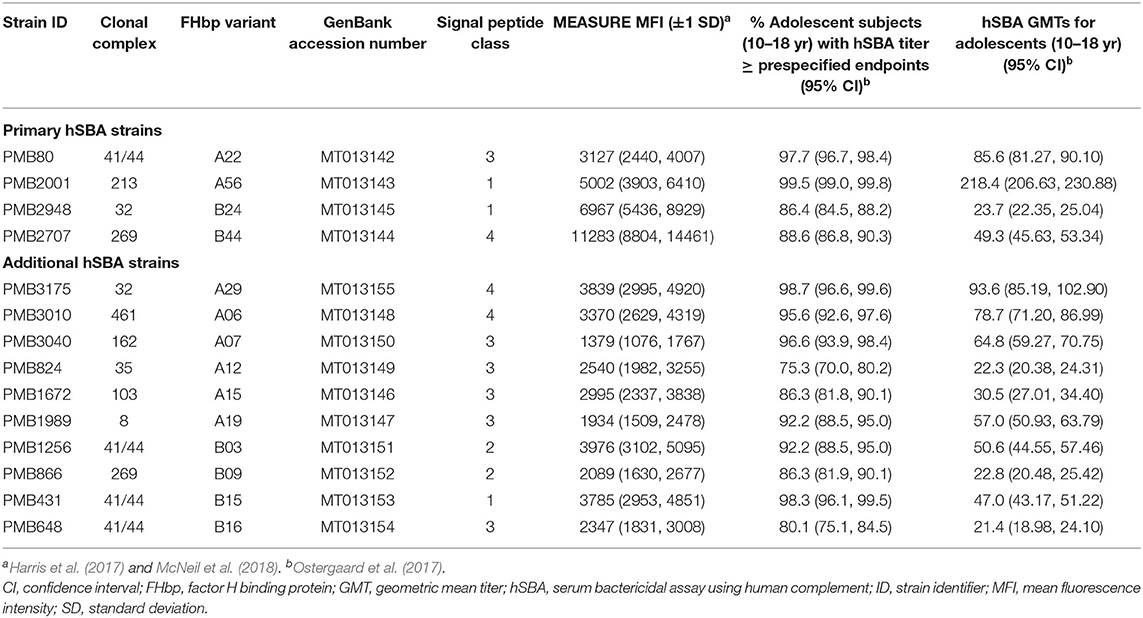
Table 1. Characteristics of primary and additional hSBA strains used to determine the immune response to Trumenba®.
The da Silva et al. (2019) manuscript also cites a report which concluded that FHbp expressed at levels of at least 757 molecules per cell (~135 pg FHbp/μg of cellular extract) is required for killing by FHbp-specific antibodies derived from mice immunized with Bexsero® (Biagini et al., 2016). Pfizer has demonstrated that strains expressing lower levels of FHbp (<1,000 mean fluorescence intensity, or the equivalent <30 pg FHbp/μg of cellular extract) can be killed in hSBAs using Trumenba® clinical immune sera (McNeil et al., 2018). The difference in the FHbp threshold level sufficient for killing may be attributed to differences in the bactericidal responses elicited by the vaccines or the reagents and methods used, including the source of immune sera (mouse for Bexsero®, human for Trumenba®) or the source of complement (rabbit and rSBAs for Bexsero®, human and hSBAs for Trumenba®). Nonetheless, both sets of results should be considered to provide a balanced hypothesis on the level of expression required for FHbp-specific antibody-mediated killing. Indeed, the basis for many of the conclusions in da Silva et al. (2019) draw upon non-clinical data obtained for Bexsero®.
Conclusion
The experimental findings reported by da Silva et al. (2019) clearly demonstrate that post-translational processing and surface localization of FHbp are complex, challenging traditional mechanisms of lipoprotein maturation in bacteria. Though the results reported are mechanistically interesting, the proposed impact of these findings on the breadth of coverage of Trumenba®, a vaccine licensed to prevent MenB infections, has not taken into account the hSBA data that was used to support licensure of Trumenba®. The results presented herein illustrate that MenB strains that code for each of the four SP alleles express FHbp at the bacterial surface and are susceptible to complement-mediated killing by antibodies elicited by Trumenba®. The data summarized in this Commentary address the concerns raised in da Silva et al. (2019) and highlight the potential for Trumenba® to prevent meningococcal disease caused by diverse MenB strains.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
PL, RD, PB, JF, and AA are Pfizer employees.
Acknowledgments
This commentary was funded by Pfizer Inc. Editorial and writing assistance was provided by Nataliya Kushnir, Pfizer Inc.
References
BEXSERO® (2015). (Meningococcal Group B Vaccine) [Package Insert]. Sovicille: GlaxoSmithKline. Available online at: https://www.gsksource.com/pharma/content/dam/GlaxoSmithKline/US/en/Prescribing_Information/Bexsero/pdf/BEXSERO.PDF (accessed January 25, 2020).
Biagini, M., Spinsanti, M., De Angelis, G., Tomei, S., Ferlenghi, I., Scarselli, M., et al. (2016). Expression of factor H binding protein in meningococcal strains can vary at least 15-fold and is genetically determined. Proc. Natl. Acad. Sci. U.S.A. 113, 2714–2719. doi: 10.1073/pnas.1521142113
da Silva, R. A. G., Karlyshev, A. V., Oldfield, N. J., Wooldridge, K. G., Bayliss, C. D., Ryan, A., et al. (2019). Variant signal peptides of vaccine antigen, FHbp, impair processing affecting surface localization and antibody-mediated killing in most meningococcal isolates. Front. Microbiol. 10:2847. doi: 10.3389/fmicb.2019.02847
Donald, R. G., Hawkins, J. C., Hao, L., Liberator, P., Jones, T. R., Harris, S. L., et al. (2017). Meningococcal serogroup B vaccines: estimating breadth of coverage. Hum. Vaccin. Immunother. 13, 255–265. doi: 10.1080/21645515.2017.1264750
Goldschneider, I., Gotschlich, E. C., and Artenstein, M. S. (1969). Human immunity to the meningococcus. I. The role of humoral antibodies. J. Exp. Med. 129, 1307–1326.
Harris, S. L., Donald, R. G., Hawkins, J. C., Tan, C., O'Neill, R., McNeil, L. K., et al. (2017). Neisseria meningitidis serogroup B vaccine, bivalent rLP2086, induces broad serum bactericidal activity against diverse invasive disease strains including outbreak strains. Pediatr. Infect. Dis. J. 36, 216–223. doi: 10.1097/INF.0000000000001399
McNeil, L. K., Donald, R. G. K., Gribenko, A., French, R., Lambert, N., Harris, S. L., et al. (2018). Predicting the susceptibility of meningococcal serogroup B isolates to bactericidal antibodies elicited by bivalent rLP2086, a novel prophylactic vaccine. mBio 9:e00036–18. doi: 10.1128/mBio.00036-18
Ostergaard, L., Vesikari, T., Absalon, J., Beeslaar, J., Ward, B. J., Senders, S., et al. (2017). A bivalent meningococcal B vaccine in adolescents and young adults. N. Engl. J. Med. 377, 2349–2362. doi: 10.1056/NEJMoa1614474
Taha, M. K., Hawkins, J. C., Liberator, P., Deghmane, A. E., Andrew, L., Hao, L., et al. (2017). Bactericidal activity of sera from adolescents vaccinated with bivalent rLP2086 against meningococcal serogroup B outbreak strains from France. Vaccine 35, 1530–1537. doi: 10.1016/j.vaccine.2017.01.066
TRUMENBA® (2014). (Meningococcal Group B Vaccine) [Package Insert]. Philadelphia, PA: Wyeth Pharmaceuticals Inc., a subsidiary of Pfizer Inc. Available online at: http://labeling.pfizer.com/ShowLabeling.aspx?id=1796 (accessed January 25, 2020).
Keywords: signal peptide, vaccine, FHbp, serogroup B, Neisseria meningitidis, Trumenba®, Bexsero®, antibody-mediated killing
Citation: Liberator P, Donald RGK, Balmer P, Findlow J and Anderson AS (2020) Commentary: Variant Signal Peptides of Vaccine Antigen, FHbp, Impair Processing Affecting Surface Localization and Antibody-Mediated Killing in Most Meningococcal Isolates. Front. Microbiol. 11:538209. doi: 10.3389/fmicb.2020.538209
Received: 04 March 2020; Accepted: 28 September 2020;
Published: 06 November 2020.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Rino Rappuoli, GlaxoSmithKline, ItalySeyed Davar Siadat, Pasteur Institute of Iran (PII), Iran
Copyright © 2020 Liberator, Donald, Balmer, Findlow and Anderson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Annaliesa S. Anderson, YW5uYWxpZXNhLmFuZGVyc29uQHBmaXplci5jb20=
 Paul Liberator1
Paul Liberator1 Annaliesa S. Anderson
Annaliesa S. Anderson