- Bacterial Epidemiology and Antimicrobial Resistance Research Unit, U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, Athens, GA, United States
Salmonella enterica is a common foodborne illness in the United States and globally. An increasing number of Salmonella infections are resistant to antibiotics, and many of the genes responsible for those resistances are carried by plasmids. Plasmids are important mediators of horizontal gene exchange, which could potentially increase the spread of antibiotic resistance (AR) genes. Twenty-eight different incompatibility groups of plasmids have been described in Enterobacteriaceae. Incompatibility groups differ in their accessory gene content, replication mechanisms, and their associations with Salmonella serotypes and animal sources. Plasmids also differ in their ability to conjugate or be mobilized, essential genes, and conditions required for transfer. It is important to understand the differences in gene content and transfer mechanisms to accurately determine the impact of plasmids on the dissemination and persistence of antibiotic resistance genes. This review will cover the most common plasmid incompatibility groups present in S. enterica with a focus on the transfer mechanisms and associated antibiotic resistance genes.
Introduction
Non-typhoidal Salmonella enterica (NTS) is a leading cause of bacterial foodborne illness, causing over one million infections per year in the United States and more than 93 million globally (Majowicz et al., 2010; Scallan et al., 2011). Salmonellosis generally presents as mild to severe gastroenteritis within 72 h of ingestion of contaminated food or water. Most illnesses are self-limiting, and otherwise healthy individuals usually recover within seven days of symptom onset without antibiotics (Kurtz et al., 2017). However, immunocompromised patients and invasive infections may require antibiotic treatment (Crump et al., 2015). Infections are usually treated with sulfamethoxazole/trimethoprim, ciprofloxacin, or cephalosporins, but resistance to these and other antibiotics has been increasing in Salmonella since the 1980s (Theilman and Guerrant, 2004; Crump et al., 2015). The Centers for Disease Control and Prevention (CDC) predicted that 16% of NTS infections in the United States between 2015 and 2018 were resistant to one antibiotic or more (CDC, 2019).
One factor influencing the spread of antibiotic resistance (AR) is horizontal gene transfer (HGT). Mobile genetic elements, including plasmids, phage, and transposons, can facilitate HGT via conjugation, transduction, and transformation, respectively (Mazel and Davies, 1999; Frost et al., 2005). Plasmids specifically have featured prominently as agents of HGT associated with AR in Enterobacteriaceae, including Salmonella (Carattoli, 2003; Gillings, 2014; Brown-Jaque et al., 2015). In the United States, AR Salmonella infections are increasing, highlighting the need for tracking plasmids containing AR genes (CDC, 2019; Tack et al., 2020).
Conjugative plasmids are self-transmissible, giving them the potential to increase the spread of AR genes. The essential components for conjugation are: the origin of transfer (oriT), MOB genes, and the mate-pair formation (MPF) genes (Smillie et al., 2010; Banuelos-Vazquez et al., 2017). The MOB genes process the DNA being replicated and transferred to the new cell. The MPF complex forms a channel between the two cells for the DNA to travel through. This protein complex is a subfamily of Type-4 secretion system. However, the genes encoding those proteins vary by plasmid type (Smillie et al., 2010; Wallden et al., 2010; Li et al., 2019). Combined, the MOB and mate-pair genes are sometimes called the tra genes because individual genes are often named tra.
In order for conjugation to occur, the relaxasome, a protein complex responsible for processing plasmid DNA to prepare it for transfer, must form at the oriT (Lawley et al., 2003b; Wong et al., 2012; Waksman, 2019). In the presence of the mate-pair structures, the relaxase then nicks the plasmid DNA and guides it through the translocation channel encoded by the MPF genes (Alvarez-Martinez and Christie, 2009; Ilangovan et al., 2015). However, not all mobile plasmids are conjugative. Non-conjugative plasmids must contain an oriT in order for the plasmid to be mobilized, but require a “helper plasmid” that possesses all the necessary MPF genes, which mobilizable plasmids utilize to complete the conjugation process (Loftie-Eaton and Rawlings, 2012). Mobilizable plasmids generally also contain a relaxase. There are eight families of relaxase genes: MOBB, MOBC, MOBF, MOBH, MOBP, MOBQ, MOBT, and MOBV (Francia et al., 2004; Garcillan-Barcia et al., 2009; Guglielmini et al., 2011).
Plasmids are typed based on incompatibility with other plasmids, which are defined as the inability of two plasmids to be maintained together in the same cell line (Novick et al., 1976). Replication genes can differ by incompatibility group. These genes can be detected with PCR for plasmid typing (Carattoli et al., 2005). Relaxase genes can also be used for typing by a similar process (Compain et al., 2014). Twenty-eight incompatibility groups have been isolated in Enterobacteriaceae (Carattoli, 2009; Rozwandowicz et al., 2018). Plasmids of different incompatibility groups differ in their host range, size, and transfer mechanism (Rozwandowicz et al., 2018). They can also differ by the AR genes they carry.
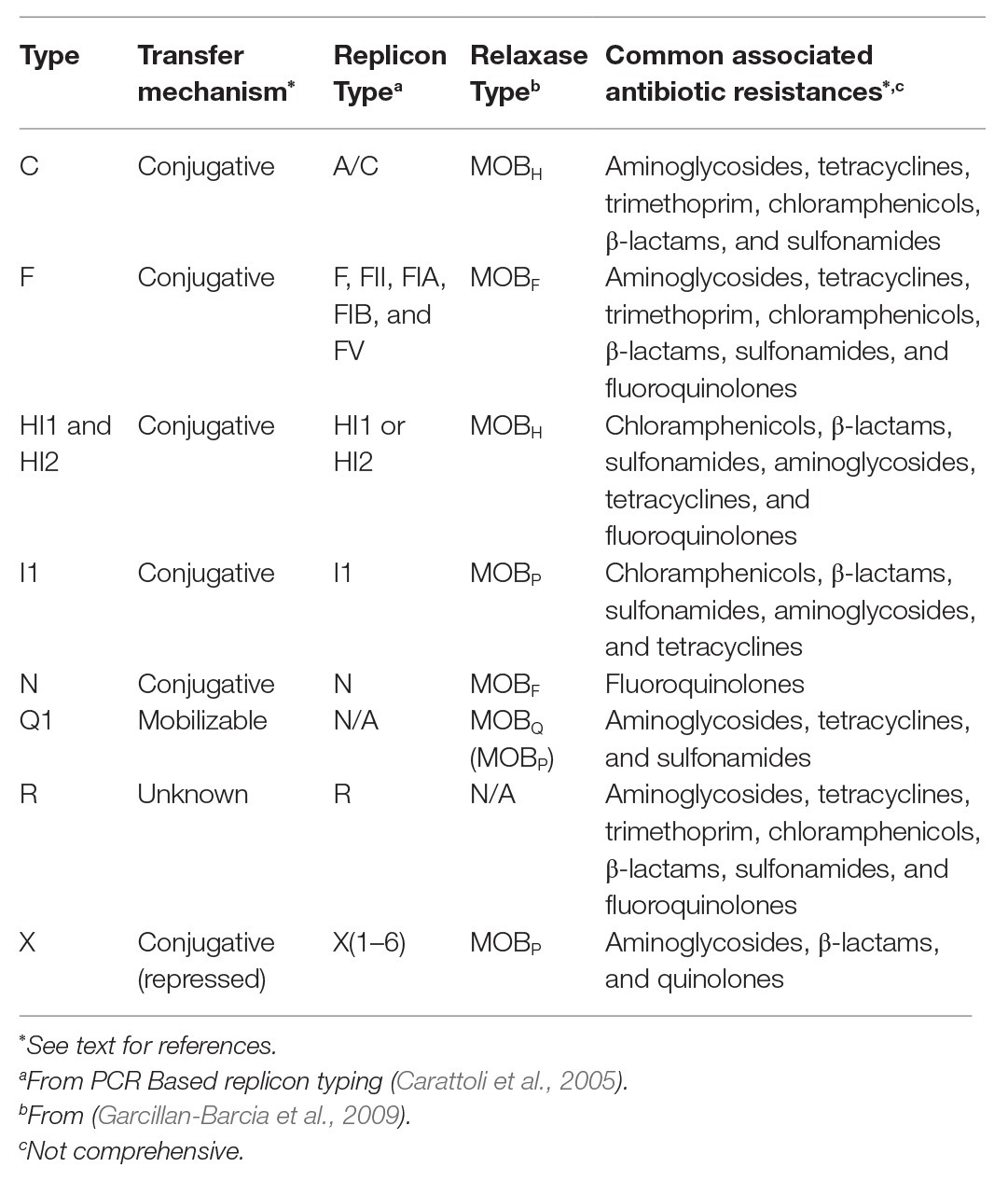
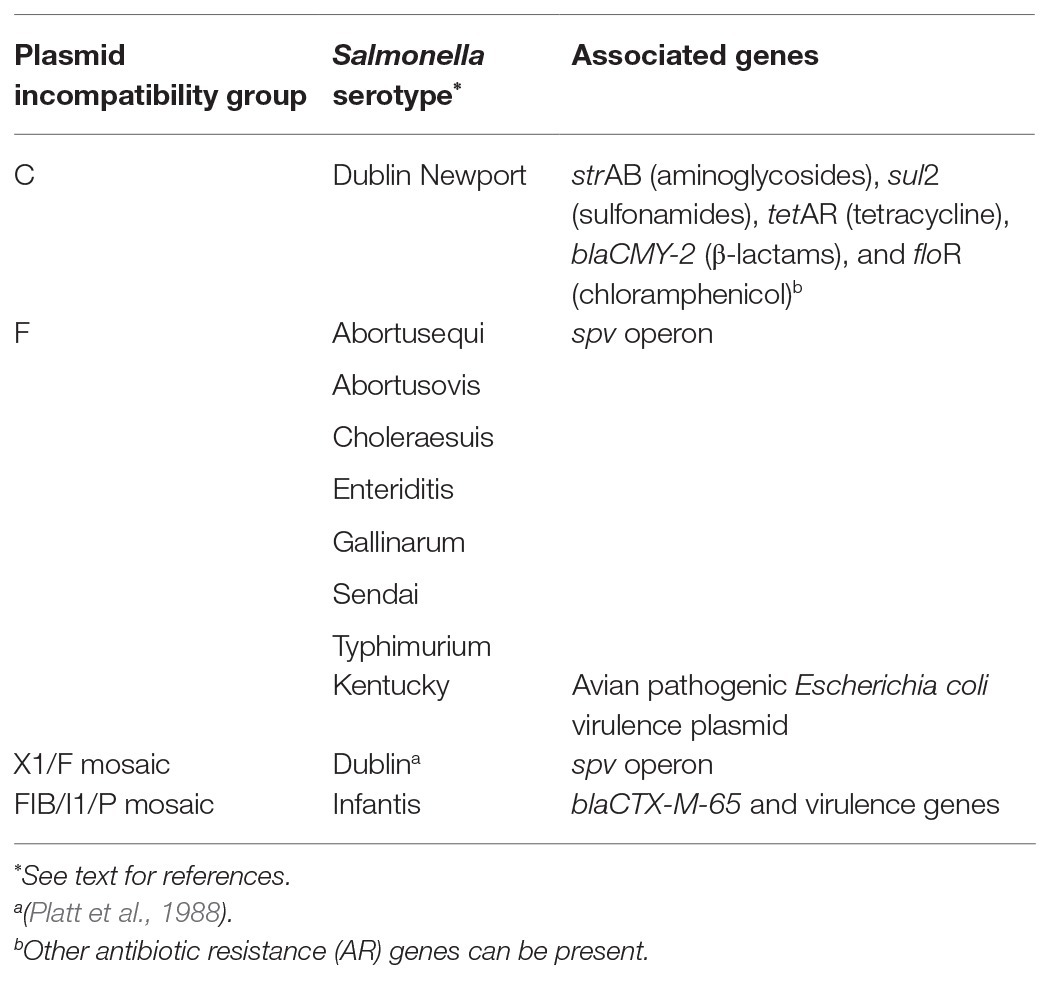
Despite genetic similarities, other Enterobacteriaceae species like Escherichia coli, are not good predictors of AR in Salmonella, even from the same sample (Nyirbahizi et al., 2020). Additionally, distribution of plasmid types and accessory genes these plasmids carry can differ in Salmonella compared to other Enterobacteriaceae, although estimates of plasmid frequencies can be biased by the tendency to sequence clinically relevant strains from humans and animals (Williams et al., 2013). Further, even though Salmonella contain plasmids that can be found in many organisms, serotypes of Salmonella differ in the frequencies of plasmids they contain, and new plasmids can emerge within any serotype. While some serotypes are more likely to contain certain plasmids than others, these plasmids can exist in any serotype, and no incompatibility group is confined to a single serotype. As Salmonella is an important human pathogen known to develop AR and potentially transfer it between animals and humans, it is recognized globally as a sentinel organism for surveillance programs like the National Antimicrobial Resistance Monitoring System (NARMS). Salmonella also contains unique plasmids; therefore, this review will focus on characteristics of the major incompatibility groups found in Salmonella, which are summarized in Table 1. Presented examples of serotypes that can contain certain plasmids more frequently than others are summarized in Table 2.
IncC
One of the most well-studied incompatibility groups found in Salmonella is IncA/C. Initial studies indicated that the IncA and IncC groups were closely related and exhibited entry exclusion toward each other so the groups were combined (Hedges, 1974). However, it was confirmed that IncA and IncC plasmids are compatible, and the use of the IncA/C term should be discontinued (Ambrose et al., 2018a). Since IncA/C has been grouped together historically, plasmids that have not been reclassified will be designated as IncA/C because PCR is not a strong enough tool to differentiate them (Ambrose et al., 2018a).
IncC plasmids are large, low copy number, broad host range, and frequently contain AR genes (Harmer and Hall, 2015). In contrast, only five IncA plasmids have been isolated (none from Salmonella; Ambrose et al., 2018a). Most plasmids are conjugative, but co-carriage of another large plasmid may increase the transfer rate (Han et al., 2018). IncC plasmids encode a MOBH relaxase (Garcillan-Barcia et al., 2009). Essentials for IncC plasmid transfer are the AcaC and AcaD proteins, which serve as master activators for the tra genes and at least two chromosomally encoded genomic islands: Salmonella genomic island 1 (SGI1) and a mobilizable chromosomal element in Vibrio mimicus (Carraro et al., 2014a). The genes traN, traH, and traG have also been determined to be essential for IncC transfer and can facilitate the mobilization of SGI1 (Carraro et al., 2017). IncC plasmids contain other tra genes, but their functions have not been confirmed experimentally (Harmer and Hall, 2015; Ambrose et al., 2018b). The relaxase-like gene mobI is also required for IncC transfer (Carraro et al., 2014b). While the biochemical function of the gene is unknown, it is located immediately downstream of the oriT and essential for plasmid transfer, but unrelated to SGI1 mobilization (Hegyi et al., 2017). In IncC plasmids, the gene eexC facilitates entry exclusion toward IncA plasmids as well as undefined incompatibility groups by recognizing traG (Humbert et al., 2019).
IncA/C plasmids have been isolated in Salmonella that contain up to 10 AR genes for more than five classes of antibiotics. The most common AR genes carried by IncA/C are strAB (aminoglycosides), sul2 (sulfonamides), tetAR (tetracycline), blaCMY-2 (β-lactams), and floR (chloramphenicols). Other genes for resistance to aminoglycosides, tetracyclines, trimethoprim, chloramphenicols, and cephalosporins have also been identified (Welch et al., 2007; Hoffmann et al., 2017; Cao et al., 2018). In Salmonella isolated in the United States, IncA/C plasmids have been found in several different serotypes and animal commodities, especially cattle (Lindsey et al., 2009; Folster et al., 2017; Mollenkopf et al., 2017). Notably, IncA/C plasmids have been found in many Salmonella Newport isolates (Table 2; Cao et al., 2015). Salmonella Newport has been isolated from cattle containing IncC plasmids with AR genes for at least five classes of antibiotics (Cao et al., 2018). Isolates in a 2017 study of human and dairy cattle associated Salmonella of serotypes Dublin, Newport, and Typhimurium showed that all Dublin and Newport isolates tested contained sul2, strA, strB, tetA, and blaCMY-2. More than 75% of all isolates tested also contained floR, and one-third of the isolates contained an IncA/C plasmid (Carroll et al., 2017). However, more investigation is needed to explain why these serotypes are more likely to contain IncC plasmids.
IncF
IncF plasmids are large (>80 kb), low-copy number, and host restricted to Enterobacteriaceae. They are important because many virulence-associated plasmids, those that allow a host bacterium to cause a more virulent infection, of Salmonella are IncF (Silva et al., 2017; Rozwandowicz et al., 2018). An example of virulence genes in Salmonella plasmids is the spv genes (Boyd and Hartl, 1998). The spv operon increases the invasive nature of Salmonella and host cytotoxicity (Guiney and Fierer, 2011). Serotypes Typhimurium, Dublin, Enteritidis, Choleraesuis, Abortusovis, Abortusequi, Gallinarum, and Sendai usually contain a virulence plasmid (Table 2; Boyd and Hartl, 1998; Uzzau et al., 2000; Anzai et al., 2005; Guiney and Fierer, 2011). Salmonella Kentucky isolates have been identified containing an IncF virulence plasmid acquired from an avian pathogenic E. coli (APEC) strain that may confer an advantage in an avian host (Table 2; Johnson et al., 2010).
The conjugation systems of IncF plasmids are diverse. They can be categorized into five classes based on the genes regulating the expression of the transfer genes; only two of the groups, one and three, have been seen in Salmonella (Fernandez-Lopez et al., 2016). The transfer pilus is one of the defining features of IncF plasmids as they are similar across all IncF plasmids but distinct from other incompatibility groups (Lawley et al., 2003b; Fernandez-Lopez et al., 2016). Relaxase genes carried by IncF plasmids are classified as MOBF (subtype MOBF12; Garcillan-Barcia et al., 2009). In contrast to other incompatibility groups, IncF plasmids can carry multiple types of replicon associated genes, such as FIA, FII, or FIB (Villa et al., 2010).
IncF plasmids have been isolated that contain AR genes. IncF plasmids in Salmonella isolated in China carried fluoroquinolone resistance genes (Chen et al., 2018). IncF plasmids isolated from Salmonella I 4,[5],12:i:-, the monophasic variant of Typhimurium, carried blaTEM-1, cmlA (chloramphenicols), and an integron containing drfA (trimethoprim), aadA1 and aadA2 (aminoglycosides), and sul3 (Garcia et al., 2014). In Salmonella isolated in the United States, IncF plasmids have been associated with strAB, tetA, tetC, tetD, aphA (aminoglycosides), and sul2 (Han et al., 2012; McMillan et al., 2019).
IncHI
First described in Salmonella Typhi in 1972, IncHI plasmids are classified into three groups: HI1, HI2, and HI3 (Anderson and Smith, 1972; Rozwandowicz et al., 2018). IncHI plasmids are generally conjugative and very large, containing up to 300,000 base pairs (Gilmour et al., 2004). HI1 and HI2 plasmids have a well-conserved backbone structure with regions of variation (Holt et al., 2007). However, HI1 plasmids have two distinct lineages based on the presence and absence of accessory gene regions (Cain and Hall, 2013). HI1 and HI2 plasmids are often isolated in Salmonella, but not host-restricted (Maher and Taylor, 1993; Holt et al., 2007; Chen et al., 2016).
Conjugation in IncHI plasmids is temperature-dependent. Conjugation is optimized at 27°C, but repressed at 37°C, the normal body temperature for warm blooded hosts of Salmonella (Maher et al., 1993). Relaxase genes of both HI1 and HI2 plasmids are classified as MOBH subtype MOBH11 (Garcillan-Barcia et al., 2009). In HI1 and HI2 plasmids, the transfer gene region is split into two locations, Tra1 and Tra2 (Taylor et al., 1984). Tra1 contains 14 genes, including five MPF genes and five non-essential genes (Lawley et al., 2002). Tra2 contains 28 genes, including four involved in partitioning the plasmid between two cells, 11 MPF genes, 10 non-essential genes, and two that regulate transfer frequency (Lawley et al., 2003a). A novel Ig-containing protein (contains an imunoglobulin-like motif), RSP, is also required for conjugation (Huttener et al., 2019).
IncHI plasmids have been isolated that contain heavy metal resistance genes as well as the tetB gene of the transposon Tn10 (Gilmour et al., 2004; Cain and Hall, 2012a,b). In addition to tetB, plasmids are also associated with resistance genes for streptomycin and sulfonamides (Rozwandowicz et al., 2018). In the globally circulating MDR Salmonella Typhi strain, HI1 plasmids carry cat (chloramphenicol), strAB, tetAR, sul2, and blaTEM-1 (Phan et al., 2009). IncHI plasmids have also been associated with qnr genes (fluroquinolones) and ESBL genes (Chen et al., 2016). Genes conferring colistin resistance have been found on HI2 plasmids in Salmonella in several countries, including Canada and Portugal (Figueiredo et al., 2016; Mulvey et al., 2018; Lima et al., 2019).
IncI1
IncI1 plasmids are large, conjugative, and host-limited to Enterobacteriaceae (Carattoli et al., 2018). The prototypical plasmid, R64, was isolated from Salmonella Typhimurium during a study examining the differences in plasmids before they were classified by incompatibility (Meynell and Datta, 1966; Sampei et al., 2010). IncI1 genetic structure is well-conserved, except for the accessory gene region, which varies by individual plasmid (Carattoli et al., 2018). The transfer region is approximately 54 kb and contains at least 49 genes, up to 35 of which are essential, depending on the plasmid (Komano et al., 1990, 2000). IncI1 contain a MOBP relaxase, generally subtype MOBP12 (Garcillan-Barcia et al., 2009). Cefotaxime (126 mg/L, half the minimum inhibitory concentration) can cause an upregulation of transfer genes in IncI1 plasmids that contain cefotaxime resistance genes (Moller et al., 2017).
While genetically similar to the transfer region of IncF plasmids, IncI1 plasmids exhibit distinct differences: two types of pili and a unique shufflon system. Both pili, one thick and one thin, are required for conjugation on a surface, while only the thin pilus is required for conjugation in broth (Bradley, 1983; Komano et al., 2000). The shufflon system allows for several variants of the pilV gene, which encodes adhesins that recognize lipopolysaccharide targets on the cell surface during conjugation (Komano et al., 1986; Sampei et al., 2010). This variation allows IncI1 plasmids to transfer to a broader range of hosts. For example, E. coli and Salmonella require a different pilV variant for transfer (Ishiwa and Komano, 2003; Sekizuka et al., 2017).
IncI1 plasmids are subdivided into hundreds of sequence types with a pMLST scheme based on the pilL (pilus biosynthesis), sogS (primase), ardA (restriction-modification enzyme), repI1 (RNAI), and a region between the trbA and pndC genes (Garcia-Fernandez et al., 2008). They are also divided into clonal complexes that contain specific AR genes (Carattoli et al., 2018). IncI1 plasmids play a major role in the dissemination and persistence of β-lactamase genes (Carattoli et al., 2018). The blaCMY-2 gene is especially prevalent among IncI1 plasmids (Folster et al., 2011). IncI1 plasmids are also frequently implicated as carriers of AR genes in Salmonella associated with poultry (Folster et al., 2014). In Salmonella Heidelberg, IncI1 plasmids have been associated with an integron containing aadA, aac(3')-IId (aminoglycosides), and sul1, blaTEM-1, and tetA; some isolates were responsible for an outbreak associated with turkey in the United States in 2011 (Folster et al., 2012; Han et al., 2012).
Other Incompatibility Groups
Other incompatibility groups have been found carrying AR genes in Salmonella. IncN plasmids are a small (30–70 kb) broad host range plasmid categorized by a pMLST scheme based on the repN, korA, and traJ genes (Garcia-Fernandez et al., 2011; Rozwandowicz et al., 2018). They contain a MOBF relaxase (Garcillan-Barcia et al., 2009). The transfer region of IncN plasmids is split into three discontinuous segments (Winans and Walker, 1985). In Salmonella, IncN plasmids have been associated with qnr genes (Garcia-Fernandez et al., 2009; Kim et al., 2013).
IncX plasmids range from 30 to 50 kb in size and are divided into six subgroups: X1–X6 (Compain et al., 2014; Rozwandowicz et al., 2018). Although most common in Salmonella and E. coli, transfer is possible to Pseudomonas aeruginosa (Tardif and Grant, 1983; Johnson et al., 2012). Conjugation in IncX plasmids is naturally repressed, but can be derepressed by suspending cells in an ammonium acetate solution (Bradley, 1980). IncX plasmids contain a MOBP relaxase, but the subtype differs by individual plasmid (Garcillan-Barcia et al., 2009). In Salmonella isolated in the United States, IncX plasmids have been associated with resistance to β-lactams and aminoglycosides (McMillan et al., 2019). IncX plasmids carrying ESBL and quinolone resistance genes have also been isolated (Rozwandowicz et al., 2018).
Mobilizable Plasmids
Mobilizable plasmids in Salmonella also contain AR genes. IncQ1 plasmids are small (10–12 kb), have a well-conserved structure, and broad host range. They are generally associated with strAB, tetAR, and sul2, although other AR genes have been identified (Poirel et al., 2010; Oliva et al., 2017). IncQ1 plasmids replicate via strand-displacement and contain a relaxase gene, mobA, fused to a repB primase. The relaxase genes of IncQ1 plasmids are diverse, sometimes only exhibiting 84% homology between different plasmids (Loftie-Eaton and Rawlings, 2012). This could explain why IncQ1 relaxases can be classified as either MOBQ or MOBP (Garcillan-Barcia et al., 2009). IncQ1 plasmids can be mobilized by large plasmids including types F, I1, N, P, W, and X (Willetts and Crowther, 1981; Francia et al., 2004).
IncR plasmids were first described in 2009 in isolates of Klebsiella and Salmonella Montevideo (Garcia-Fernandez et al., 2009). They are not thought to be conjugative since they do not contain any tra genes or a relaxase, but could be mobilizable (Chen et al., 2006; Bielak et al., 2011; Rozwandowicz et al., 2018). Although IncR plasmids are rarely reported, an outbreak strain of Salmonella Newport in the United States reported between 2018 and 2019 contained an IncR plasmid carrying genes for resistance to up to five different classes of antibiotics: trimethoprim/sulfamethoxazole, tetracycline, chloramphenicol, β-lactams, and aminoglycosides (Plumb et al., 2019). IncR plasmids have also been associated with resistance to quinolones (Rozwandowicz et al., 2018).
Discussion
Understanding how plasmids transfer in Salmonella is of the utmost importance in determining the risk of the dissemination of AR genes. If a plasmid carrying AR genes is conjugative, genes may have a higher risk of dissemination than those carried by non-conjugative plasmids. Furthermore, if a plasmid is mobilizable, it is important to know if the cell carries a helper plasmid and of what type; conjugative plasmids mobilize non-conjugative plasmids at different rates (Cabezon et al., 1997). However, it is important to remember that presence of conjugation genes does not guarantee conjugation. External conditions influence transfer efficiency; for example, inflammation has been shown to increase rates of transfer for IncI1 plasmids from Salmonella to E. coli (Stecher et al., 2012).
Plasmids are common in Salmonella and differ in their gene content and transfer genes. Tracking these plasmids is important because some can be associated with certain AR genes, like β-lactamases with I1 plasmids (Carattoli et al., 2018). However, as AR genes are frequently associated with transposons, genes are not fixed to specific plasmids, and AR genes can emerge on new plasmids at any time. Additionally, plasmids can contain multiple replicons. One study found that 66% of Salmonella plasmids investigated were mosaic, meaning that they contained sequences from multiple different plasmids as a result of recombination; many were mosaics of plasmids from different incompatibility groups (Boyd et al., 1996; Pesesky et al., 2019). Recently, a Salmonella Infantis strain containing the mosaic pESI-like plasmid (plasmid for emerging Salmonella infantis) carrying blaCTX-M-65 has emerged in poultry and caused human infections around the world (Table 2; Aviv et al., 2014; Tate et al., 2017; Brown et al., 2018). Although this gene has been isolated on other plasmids in E. coli, the gene is only found on the pESI plasmid in Salmonella Infantis. This plasmid has contributed to the rise of Infantis as one of the dominant serotypes in poultry in Europe and in the United States, where the prevalence of the serotype increased nearly 70% in the past few years (Tack et al., 2020). In addition to blaCTX-M-65, this plasmid carries other AR genes and genes that convey advantages over other Salmonella within a poultry host.
Plasmids in Salmonella may acquire AR genes seemingly at random, but their proliferation in a population is not random. The pESI plasmid is a good example of how a strain with a plasmid containing AR genes can rapidly spread by co-selection. Plasmids in Salmonella often carry more than one AR gene, or genes for resistance to heavy metals and/or biocides. In these cases, even if an antibiotic is withdrawn from the environment, other selective pressures encouraging plasmid carriage may remain. Further, plasmids may contain other factors, like fimbriae or nutrient acquisition systems, that give the host bacterium an advantage that further selects for the plasmid. Even without another selecting factor, plasmids may not carry a high fitness cost to the host bacterium. In some cases, AR genes carried by plasmid can have a lower cost of fitness than those carried chromosomally (Vogwill and MacLean, 2014).
Co-selection could also play a part in the associations between serotypes and certain plasmid types, but further investigation is needed to fully understand these correlations. For example, it is unknown why IncC plasmids are more common in serotypes Newport and Dublin. In the United States, these serotypes are also more frequently isolated from cattle sources, but whether the relationship of the plasmid is related to the serotype, the source, or a combination of factors remains unknown. Co-selection could also be responsible for the association of the pESI plasmid with serotype Infantis in poultry but does not explain why the plasmid has not been isolated in any other poultry-related serotypes. In this case, another factor such as a barrier to conjugation must be influencing the serotype association.
Surveillance is necessary, not just of Salmonella and other important human and animal pathogens, but of the plasmids they carry. Tracking plasmids and the genes they carry would allow for a better understanding of co-selection of AR genes and the associations of plasmids with Salmonella serotypes. Plasmid surveillance will not prevent the spread of AR, but it would provide information for designing mitigation strategies that account for these factors. Further study is needed to assess the contribution of plasmids to the spread of AR in Salmonella, both genotypically and phenotypically.
Author’s Note
The mention of trade names or commercial products in this manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author Contributions
EM, CJ, and JF all wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
EM, CJ, and JF were supported by U.S. Department of Agriculture (USDA) project plans 6040-32000-006-00 and 6040-32000-009-00.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Elizabeth Ottesen, Anna Karls, Jan Mrazek, and Fred Quinn for their feedback on early versions of this review.
References
Alvarez-Martinez, C. E., and Christie, P. J. (2009). Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808. doi: 10.1128/MMBR.00023-09
Ambrose, S. J., Harmer, C. J., and Hall, R. M. (2018a). Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 96–97, 7–12. doi: 10.1016/j.plasmid.2018.02.002
Ambrose, S. J., Harmer, C. J., and Hall, R. M. (2018b). Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid 99, 40–55. doi: 10.1016/j.plasmid.2018.08.001
Anderson, E. S., and Smith, H. R. (1972). Chloramphenicol resistance in the typhoid bacillus. Br. Med. J. 3, 329–331. doi: 10.1136/bmj.3.5825.525
Anzai, T., Kuwamoto, Y., Hobo, S., Niwa, H., Katayama, Y., Ode, H., et al. (2005). The importance of a 95-kb virulence plasmid in the pathogenicity of Salmonella abortusequi in horses. J. Equine Sci. 16, 111–116. doi: 10.1294/jes.16.111
Aviv, G., Tsyba, K., Steck, N., Salmon-Divon, M., Cornelius, A., Rahav, G., et al. (2014). A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ. Microbiol. 16, 977–994. doi: 10.1111/1462-2920.12351
Banuelos-Vazquez, L. A., Torres Tejerizo, G., and Brom, S. (2017). Regulation of conjugative transfer of plasmids and integrative conjugative elements. Plasmid 91, 82–89. doi: 10.1016/j.plasmid.2017.04.002
Bielak, E., Bergenholtz, R. D., Jorgensen, M. S., Sorensen, S. J., Hansen, L. H., and Hasman, H. (2011). Investigation of diversity of plasmids carrying the blaTEM-52 gene. J. Antimicrob. Chemother. 66, 2465–2474. doi: 10.1093/jac/dkr331
Boyd, E. F., and Hartl, D. L. (1998). Salmonella virulence plasmid: modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics 149, 1183–1190.
Boyd, E. F., Hill, C. W., Rich, S. M., and Hartl, D. L. (1996). Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics 143, 1091–1100.
Bradley, D. E. (1980). Determination of pili by conjugative bacterial drug resistance plasmids of incompatibility groups B, C, H, J, K, M, V, and X. J. Bacteriol. 141, 828–837. doi: 10.1128/JB.141.2.828-837.1980
Bradley, D. E. (1983). Derepressed plasmids of incompatibility group I1 determine two different morphological forms of pilus. Plasmid 9, 331–334. doi: 10.1016/0147-619x(83)90011-2
Brown, A. C., Chen, J. C., Watkins, L. K. F., Campbell, D., Folster, J. P., Tate, H., et al. (2018). CTX-M-65 extended-spectrum beta-lactamase-producing Salmonella enterica serotype infantis, United States1. Emerg. Infect. Dis. 24, 2284–2291. doi: 10.3201/eid2412.180500
Brown-Jaque, M., Calero-Caceres, W., and Muniesa, M. (2015). Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7. doi: 10.1016/j.plasmid.2015.01.001
Cabezon, E., Sastre, J. I., and de la Cruz, F. (1997). Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254, 400–406. doi: 10.1007/s004380050432
Cain, A. K., and Hall, R. M. (2012a). Evolution of a multiple antibiotic resistance region in IncHI1 plasmids: reshaping resistance regions in situ. J. Antimicrob. Chemother. 67, 2848–2853. doi: 10.1093/jac/dks317
Cain, A. K., and Hall, R. M. (2012b). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67, 1121–1127. doi: 10.1093/jac/dks004
Cain, A. K., and Hall, R. M. (2013). Evolution of IncHI1 plasmids: two distinct lineages. Plasmid 70, 201–208. doi: 10.1016/j.plasmid.2013.03.005
Cao, G., Allard, M. W., Hoffmann, M., Monday, S. R., Muruvanda, T., Luo, Y., et al. (2015). Complete sequences of six IncA/C plasmids of multidrug-resistant Salmonella enterica subsp. enterica serotype Newport. Genome Announc. 3, e00027–e00115. doi: 10.1128/genomeA.00027-15
Cao, G., Allard, M., Hoffmann, M., Muruvanda, T., Luo, Y., Payne, J., et al. (2018). Sequence analysis of IncA/C and IncI1 plasmids isolated from multidrug-resistant Salmonella Newport using single-molecule real-time sequencing. Foodborne Pathog. Dis. 15, 361–371. doi: 10.1089/fpd.2017.2385
Carattoli, A. (2003). Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 5, 113–122. doi: 10.21775/cimb.005.113
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli, A., Villa, L., Fortini, D., and Garcia-Fernandez, A. (2018). Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid. 102392. doi: 10.1016/j.plasmid.2018.12.001 (in press).
Carraro, N., Durand, R., Rivard, N., Anquetil, C., Barrette, C., Humbert, M., et al. (2017). Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet. 13:e1006705. doi: 10.1371/journal.pgen.1006705
Carraro, N., Matteau, D., Luo, P., Rodrigue, S., and Burrus, V. (2014a). The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 10:e1004714. doi: 10.1371/journal.pgen.1004714
Carraro, N., Sauve, M., Matteau, D., Lauzon, G., Rodrigue, S., and Burrus, V. (2014b). Development of pVCR94DeltaX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front. Microbiol. 5:44. doi: 10.3389/fmicb.2014.00044
Carroll, L. M., Wiedmann, M., den Bakker, H., Siler, J., Warchocki, S., Kent, D., et al. (2017). Whole-genome sequencing of drug-resistant Salmonella enterica isolates from dairy cattle and humans in New York and Washington states reveals source and geographic associations. Appl. Environ. Microbiol. 83, e00140–e00217. doi: 10.1128/AEM.00140-17
Chen, K., Dong, N., Zhao, S., Liu, L., Li, R., Xie, M., et al. (2018). Identification and characterization of conjugative plasmids that encode ciprofloxacin resistance in Salmonella. Antimicrob. Agents Chemother. 62, e00575–e00618. doi: 10.1128/AAC.00575-18
Chen, W., Fang, T., Zhou, X., Zhang, D., Shi, X., and Shi, C. (2016). IncHI2 plasmids are predominant in antibiotic-resistant Salmonella isolates. Front. Microbiol. 7:1566. doi: 10.3389/fmicb.2016.01566
Chen, Y. -T., Shu, H. -Y., Li, L. -H., Liao, T. -L., Wu, K. -M., Shiau, Y. -R., et al. (2006). Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 50, 3861–3866. doi: 10.1128/aac.00456-06
Compain, F., Poisson, A., Le Hello, S., Branger, C., Weill, F. X., Arlet, G., et al. (2014). Targeting relaxase genes for classification of the predominant plasmids in Enterobacteriaceae. Int. J. Med. Microbiol. 304, 236–242. doi: 10.1016/j.ijmm.2013.09.009
Crump, J. A., Sjolund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Fernandez-Lopez, R., de Toro, M., Moncalian, G., Garcillan-Barcia, M. P., and de la Cruz, F. (2016). Comparative genomics of the conjugation region of f-like plasmids: five shades of F. Front. Mol. Biosci. 3:71. doi: 10.3389/fmolb.2016.00071
Figueiredo, R., Card, R. M., Nunez, J., Pomba, C., Mendonca, N., Anjum, M. F., et al. (2016). Detection of an mcr-1-encoding plasmid mediating colistin resistance in Salmonella enterica from retail meat in Portugal. J. Antimicrob. Chemother. 71, 2338–2340. doi: 10.1093/jac/dkw240
Folster, J. P., Grass, J. E., Bicknese, A., Taylor, J., Friedman, C. R., and Whichard, J. M. (2017). Characterization of resistance genes and plasmids from outbreaks and illness clusters caused by Salmonella resistant to ceftriaxone in the United States, 2011–2012. Microb. Drug Resist. 23, 188–193. doi: 10.1089/mdr.2016.0080
Folster, J. P., Pecic, G., McCullough, A., Rickert, R., and Whichard, J. M. (2011). Characterization of bla(CMY)-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog. Dis. 8, 1289–1294. doi: 10.1089/fpd.2011.0944
Folster, J. P., Pecic, G., Rickert, R., Taylor, J., Zhao, S., Fedorka-Cray, P. J., et al. (2012). Characterization of multidrug-resistant Salmonella enterica serovar Heidelberg from a ground turkey-associated outbreak in the United States in 2011. Antimicrob. Agents Chemother. 56, 3465–3466. doi: 10.1128/AAC.00201-12
Folster, J. P., Tolar, B., Pecic, G., Sheehan, D., Rickert, R., Hise, K., et al. (2014). Characterization of blaCMY plasmids and their possible role in source attribution of Salmonella enterica serotype Typhimurium infections. Foodborne Pathog. Dis. 11, 301–306. doi: 10.1089/fpd.2013.1670
Francia, M. V., Varsaki, A., Garcillan-Barcia, M. P., Latorre, A., Drainas, C., and de la Cruz, F. (2004). A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28, 79–100. doi: 10.1016/j.femsre.2003.09.001
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Garcia, P., Hopkins, K. L., Garcia, V., Beutlich, J., Mendoza, M. C., Threlfall, J., et al. (2014). Diversity of plasmids encoding virulence and resistance functions in Salmonella enterica subsp. enterica serovar typhimurium monophasic variant 4,[5], 12:i:- strains circulating in Europe. PLoS One 9:e89635. doi: 10.1371/journal.pone.0089635
Garcia-Fernandez, A., Chiaretto, G., Bertini, A., Villa, L., Fortini, D., Ricci, A., et al. (2008). Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 61, 1229–1233. doi: 10.1093/jac/dkn131
Garcia-Fernandez, A., Fortini, D., Veldman, K., Mevius, D., and Carattoli, A. (2009). Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63, 274–281. doi: 10.1093/jac/dkn470
Garcia-Fernandez, A., Villa, L., Moodley, A., Hasman, H., Miriagou, V., Guardabassi, L., et al. (2011). Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 66, 1987–1991. doi: 10.1093/jac/dkr225
Garcillan-Barcia, M. P., Francia, M. V., and de la Cruz, F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–687. doi: 10.1111/j.1574-6976.2009.00168.x
Gillings, M. R. (2014). Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78, 257–277. doi: 10.1128/MMBR.00056-13
Gilmour, M. W., Thomson, N. R., Sanders, M., Parkhill, J., and Taylor, D. E. (2004). The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52, 182–202. doi: 10.1016/j.plasmid.2004.06.006
Guglielmini, J., Quintais, L., Garcillan-Barcia, M. P., de la Cruz, F., and Rocha, E. P. (2011). The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222. doi: 10.1371/journal.pgen.1002222
Guiney, D. G., and Fierer, J. (2011). The role of the spv genes in Salmonella pathogenesis. Front. Microbiol. 2:129. doi: 10.3389/fmicb.2011.00129
Han, J., Lynne, A. M., David, D. E., Tang, H., Xu, J., Nayak, R., et al. (2012). DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS One 7:e51160. doi: 10.1371/journal.pone.0051160
Han, J., Pendleton, S. J., Deck, J., Singh, R., Gilbert, J., Johnson, T. J., et al. (2018). Impact of co-carriage of IncA/C plasmids with additional plasmids on the transfer of antimicrobial resistance in Salmonella enterica isolates. Int. J. Food Microbiol. 271, 77–84. doi: 10.1016/j.ijfoodmicro.2018.01.018
Harmer, C. J., and Hall, R. M. (2015). The A to Z of A/C plasmids. Plasmid 80, 63–82. doi: 10.1016/j.plasmid.2015.04.003
Hedges, R. W. (1974). R factors from providence. J. Gen. Microbiol. 81, 171–181. doi: 10.1099/00221287-81-1-171
Hegyi, A., Szabo, M., Olasz, F., and Kiss, J. (2017). Identification of oriT and a recombination hot spot in the IncA/C plasmid backbone. Sci. Rep. 7:10595. doi: 10.1038/s41598-017-11097-0
Hoffmann, M., Pettengill, J. B., Gonzalez-Escalona, N., Miller, J., Ayers, S. L., Zhao, S., et al. (2017). Comparative sequence analysis of multidrug-resistant IncA/C plasmids from Salmonella enterica. Front. Microbiol. 8:1459. doi: 10.3389/fmicb.2017.01459
Holt, K. E., Thomson, N. R., Wain, J., Phan, M. D., Nair, S., Hasan, R., et al. (2007). Multidrug-resistant Salmonella enterica serovar paratyphi A harbors IncHI1 plasmids similar to those found in serovar typhi. J. Bacteriol. 189, 4257–4264. doi: 10.1128/JB.00232-07
Humbert, M., Huguet, K. T., Coulombe, F., and Burrus, V. (2019). Entry exclusion of conjugative plasmids of the IncA, IncC, and related untyped incompatibiliy groups. J. Bacteriol. 201, e00731–e00818. doi: 10.1128/JB.00731-18
Huttener, M., Prieto, A., Aznar, S., Bernabeu, M., Glaria, E., Valledor, A. F., et al. (2019). Expression of a novel class of bacterial Ig-like proteins is required for IncHI plasmid conjugation. PLoS Genet. 15:e1008399. doi: 10.1371/journal.pgen.1008399
Ilangovan, A., Connery, S., and Waksman, G. (2015). Structural biology of the gram-negative bacterial conjugation systems. Trends Microbiol. 23, 301–310. doi: 10.1016/j.tim.2015.02.012
Ishiwa, A., and Komano, T. (2003). Thin pilus PilV adhesins of plasmid R64 recognize specific structures of the lipopolysaccharide molecules of recipient cells. J. Bacteriol. 185, 5192–5199. doi: 10.1128/jb.185.17.5192-5199.2003
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Johnson, T. J., Thorsness, J. L., Anderson, C. P., Lynne, A. M., Foley, S. L., Han, J., et al. (2010). Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS One 5:e15524. doi: 10.1371/journal.pone.0015524
Kim, J. H., Cho, J. K., and Kim, K. S. (2013). Prevalence and characterization of plasmid-mediated quinolone resistance genes in Salmonella isolated from poultry in Korea. Avian Pathol. 42, 221–229. doi: 10.1080/03079457.2013.779636
Komano, T., Funayama, N., Kim, S. A., and Nisioka, T. (1990). Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J. Bacteriol. 172, 2230–2235. doi: 10.1128/jb.172.5.2230-2235.1990
Komano, T., Kubo, A., Kayanuma, T., Furuichi, T., and Nisioka, T. (1986). Highly mobile DNA segment of IncI alpha plasmid R64: a clustered inversion region. J. Bacteriol. 165, 94–100. doi: 10.1128/jb.165.1.94-100.1986
Komano, T., Yoshida, T., Narahara, K., and Furuya, N. (2000). The transfer region of IncI1 plasmid R64: similarities between R64 tra and legionella icm/dot genes. Mol. Microbiol. 35, 1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x
Kurtz, J. R., Goggins, J. A., and McLachlan, J. B. (2017). Salmonella infection: interplay between the bacteria and host immune system. Immunol. Lett. 190, 42–50. doi: 10.1016/j.imlet.2017.07.006
Lawley, T. D., Gilmour, M. W., Gunton, J. E., Standeven, L. J., and Taylor, D. E. (2002). Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184, 2173–2180. doi: 10.1128/jb.184.8.2173-2180.2002
Lawley, T. D., Gilmour, M. W., Gunton, J. E., Tracz, D. M., and Taylor, D. E. (2003a). Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 185, 581–591. doi: 10.1128/jb.185.2.581-591.2003
Lawley, T. D., Klimke, W. A., Gubbins, M. J., and Frost, L. S. (2003b). F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224, 1–15. doi: 10.1016/s0378-1097(03)00430-0
Li, Y. G., Hu, B., and Christie, P. J. (2019). Biological and structural diversity of type IV secretion systems. Microbiol. Spectr. 7:PSIB-0012-2018. doi: 10.1128/microbiolspec.PSIB-0012-2018
Lima, T., Domingues, S., and Da Silva, G. J. (2019). Plasmid-mediated colistin resistance in Salmonella enterica: a review. Microorganisms 7:55. doi: 10.3390/microorganisms7020055
Lindsey, R. L., Fedorka-Cray, P. J., Frye, J. G., and Meinersmann, R. J. (2009). Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75, 1908–1915. doi: 10.1128/AEM.02228-08
Loftie-Eaton, W., and Rawlings, D. E. (2012). Diversity, biology and evolution of IncQ-family plasmids. Plasmid 67, 15–34. doi: 10.1016/j.plasmid.2011.10.001
Maher, D., Sherburne, R., and Taylor, D. E. (1993). H-Pilus assembly kinetics determined by electron microscopy. J. Bacteriol. 175, 2175–2183. doi: 10.1128/jb.175.8.2175-2183.1993
Maher, D., and Taylor, D. E. (1993). Host range and transfer efficiency of incompatibility group HI plasmids. Can. J. Microbiol. 39, 581–587. doi: 10.1139/m93-084
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Mazel, D., and Davies, J. (1999). Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56, 742–754. doi: 10.1007/s000180050021
McMillan, E. A., Gupta, S. K., Williams, L. E., Jové, T., Hiott, L. M., Woodley, T. A., et al. (2019). Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 10:832. doi: 10.3389/fmicb.2019.00832
Meynell, E., and Datta, N. (1966). The relation of resistance transfer factors to the F-factor (sex-factor) of Escherichia coli K12. Genet. Res. 7, 134–140. doi: 10.1017/s0016672300009538
Mollenkopf, D. F., Mathys, D. A., Dargatz, D. A., Erdman, M. M., Habing, G. G., Daniels, J. B., et al. (2017). Genotypic and epidemiologic characterization of extended-spectrum cephalosporin resistant Salmonella enterica from US beef feedlots. Prev. Vet. Med. 146, 143–149. doi: 10.1016/j.prevetmed.2017.08.006
Moller, T. S. B., Liu, G., Boysen, A., Thomsen, L. E., Luthje, F. L., Mortensen, S., et al. (2017). Treatment with cefotaxime affects expression of conjugation associated proteins and conjugation transfer frequency of an IncI1 plasmid in Escherichia coli. Front. Microbiol. 8:2365. doi: 10.3389/fmicb.2017.02365
Mulvey, M. R., Bharat, A., Boyd, D. A., Irwin, R. J., and Wylie, J. (2018). Characterization of a colistin-resistant Salmonella enterica 4,[5],12:i:- harbouring mcr-3.2 on a variant IncHI-2 plasmid identified in Canada. J. Med. Microbiol. 67, 1673–1675. doi: 10.1099/jmm.0.000854
Novick, R. P., Clowes, R. C., Cohen, S. N., Curtiss, R. III, Datta, N., and Falkow, S. (1976). Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol. Rev. 40, 168–189. doi: 10.1128/mmbr.40.1.168-189.1976
Nyirbahizi, E., Tyson, G. H., Dessai, U., Zhao, S., Kebara, C., Crarey, E., et al. (2020). Evaluation of Escherichia coli as an indicator for antimicrobial resistance in Salmonella recovered from the same food or animal ceca samples. Food Control 115:107280. doi: 10.1016/j.foodcont.2020.107280
Oliva, M., Monno, R., D’Addabbo, P., Pesole, G., Dionisi, A. M., Scrascia, M., et al. (2017). A novel group of IncQ1 plasmids conferring multidrug resistance. Plasmid 89, 22–26. doi: 10.1016/j.plasmid.2016.11.005
Pesesky, M. W., Tilley, R., and Beck, D. A. C. (2019). Mosaic plasmids are abundant and unevenly distributed across prokaryotic taxa. Plasmid 102, 10–18. doi: 10.1016/j.plasmid.2019.02.003
Phan, M. D., Kidgell, C., Nair, S., Holt, K. E., Turner, A. K., Hinds, J., et al. (2009). Variation in Salmonella enterica serovar typhi IncHI1 plasmids during the global spread of resistant typhoid fever. Antimicrob. Agents Chemother. 53, 716–727. doi: 10.1128/AAC.00645-08
Platt, D. J., Taggart, J., and Heraghty, K. A. (1988). Molecular divergence of the serotype-specific plasmid (pSLT) among strains of Salmonella typhimurium of human and veterinary origin and comparison of pSLT with the serotype specific plasmids of S. enteridits and S. dublin. J. Med. Microbiol. 27, 277–284. doi: 10.1099/00222615-27-4-277
Plumb, I. D., Schwensohn, C. A., Gieraltowski, L., Tecle, S., Schneider, Z. D., Frieman, J., et al. (2019). Outbreak of Salmonella Newport infections with decreased susceptibility to azithromycin linked to beef obtained in the United States and soft cheese obtained in Mexico-United States, 2018–2019. MMWR Morb. Mortal. Wkly Rep. 68, 713–717. doi: 10.15585/mmwr.mm6833a1
Poirel, L., Carattoli, A., Bernabeu, S., Bruderer, T., Frei, R., and Nordmann, P. (2010). A novel IncQ plasmid type harbouring a class 3 integron from Escherichia coli. J. Antimicrob. Chemother. 65, 1594–1598. doi: 10.1093/jac/dkq166
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Sampei, G., Furuya, N., Tachibana, K., Saitou, Y., Suzuki, T., Mizobuchi, K., et al. (2010). Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 64, 92–103. doi: 10.1016/j.plasmid.2010.05.005
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Sekizuka, T., Kawanishi, M., Ohnishi, M., Shima, A., Kato, K., Yamashita, A., et al. (2017). Elucidation of quantitative structural diversity of remarkable rearrangement regions, shufflons, in IncI2 plasmids. Sci. Rep. 7:928. doi: 10.1038/s41598-017-01082-y
Silva, C., Puente, J. L., and Calva, E. (2017). Salmonella virulence plasmid: pathogenesis and ecology. Pathog. Dis. doi: 10.1093/femspd/ftx070 [Epub ahead of print]
Smillie, C., Garcillan-Barcia, M. P., Francia, M. V., Rocha, E. P., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Stecher, B., Denzler, R., Maier, L., Bernat, F., Sanders, M. J., Pickard, D. J., et al. (2012). Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterbacteriaceae. Proc. Natl. Acad. Sci. U. S. A. 109, 1269–1274. doi: 10.1073/pnas.1113246109
Tack, D. M., Ray, L., Griffin, P. M., Cieslak, P. R., Dunn, J., Rissman, T., et al. (2020). Preliminary incidence and trends of infections with pathogenes transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 2016–2019. MMWR Morb. Mortal. Wkly Rep. 69, 509–514. doi: 10.15585/mmwr.mm6917a1
Tardif, G., and Grant, R. B. (1983). Transfer of plasmids from Escherichia coli to Pseudomonas aeruginosa: characterization of a Pseudomonas aeruginosa mutant with enhanced recipient ability for enterobacterial plasmids. Antimicrob. Agents Chemother. 24, 201–208. doi: 10.1128/AAC.24.2.201
Tate, H., Folster, J. P., Hsu, C. -H., Chen, J., Hoffmann, M., Li, C., et al. (2017). Comparitive analysis of extended-spectrum-β-lactamase CTX-M-65 producing Salmonella enterica serovar infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother. 61, e00488–e00517. doi: 10.1128/AAC.00488-17
Taylor, D. E., Brose, E. C., Kwan, S., and Yan, W. (1984). Mapping of transfer regions within incompatibility group HI plasmid R27. J. Bacteriol. 162, 1221–1226. doi: 10.1128/JB.162.3.1221-1226.1985
Theilman, N. M., and Guerrant, R. L. (2004). Acute infectious diarrhea. N. Engl. J. Med. 350, 38–47. doi: 10.1056/nejmcp031534
Uzzau, S., Gulig, P. A., Paglietti, B., Leori, G., Stocker, B. A., and Rubino, S. (2000). Role of the Salmonella abortusovis virulence plasmid in the infection of BALB/c mice. FEMS Microbiol. Lett. 188, 15–18. doi: 10.1111/j.1574-6968.2000.tb09161.x
Villa, L., Garcia-Fernandez, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Vogwill, T., and MacLean, R. C. (2014). The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol. Appl. 8, 284–295. doi: 10.1111/eva.12202
Waksman, G. (2019). From conjugation to T4S systems in gram-negative bacteria: a mechanistic biology perspective. EMBO Rep. 20:e47012. doi: 10.15252/embr.201847012
Wallden, K., Rivera-Calzada, A., and Waksman, G. (2010). Microreview: type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12, 1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x
Welch, T. J., Fricke, W. F., McDermott, P. F., White, D. G., Rosso, M. L., Rasko, D. A., et al. (2007). Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. doi: 10.1371/journal.pone.0000309
Willetts, N., and Crowther, C. (1981). Mobilization of the non-conjugative IncQ plasmid RSF1010. Genet. Res. 37, 311–316. doi: 10.1017/s0016672300020310
Williams, L. E., Wireman, J., Hilliard, V. C., and Summers, A. O. (2013). Large plasmids of Escherichia coli and Salmonella encode highly diverse arrays of accessory genes on common replicon families. Plasmid 69, 36–48. doi: 10.1016/j.plasmid.2012.08.002
Winans, S. C., and Walker, G. C. (1985). Conjugal transfer system of the IncN plasmid pKM101. J. Bacteriol. 161, 402–410. doi: 10.1128/JB.161.1.402-410.1985
Keywords: Salmonella, plasmids, conjugation, antibiotic resistance, horizontal gene transfer
Citation: McMillan EA, Jackson CR and Frye JG (2020) Transferable Plasmids of Salmonella enterica Associated With Antibiotic Resistance Genes. Front. Microbiol. 11:562181. doi: 10.3389/fmicb.2020.562181
Edited by:
Hemda Garelick, Middlesex University, United KingdomReviewed by:
Christopher Morton Thomas, University of Birmingham, United KingdomAntonio Juárez, University of Barcelona, Spain
Copyright © 2020 McMillan, Jackson and Frye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth A. McMillan, ZWxpemFiZXRoLm1jbWlsbGFuQHVzZGEuZ292
 Elizabeth A. McMillan
Elizabeth A. McMillan Charlene R. Jackson
Charlene R. Jackson Jonathan G. Frye
Jonathan G. Frye