- 1Infectious Diseases and Immunology Division, CSIR-Indian Institute of Chemical Biology, Kolkata, India
- 2Molecular Genetics Laboratory, Infection and Immunology Division, Translational Health Science and Technology Institute, NCR Biotech Science Cluster, Haryana, India
The stringent response, an adaptive response to nutrient limitation and exposure to xenobiotics in bacteria, is mediated by two intracellular signaling molecules, pppGpp and ppGpp, together called (p)ppGpp. The cellular level of (p)ppGpp in bacterial cells is controlled by the Rel/Spo family of proteins. In the cholera pathogen, Vibrio cholerae, (p)ppGpp metabolism is regulated by the products of at least three genes relA, spoT, and relV. In this study, we identify and characterize the function of the guanosine-5′-triphosphate 3′-diphosphate pyrophosphatase A (GppA) encoding gene gppA of V. cholerae. Genomic analysis indicates that the gppA locus is conserved in vibrios and organized as a bicistronic operon along with the rhlB gene. We engineered the genome of V. cholerae to develop different mutants devoid of GppA and/or other phosphate metabolic enzymes. Our findings indicate that in V. cholerae, GppA plays an important role in the conversion of pppGpp to ppGpp during amino acid deprivation but not during glucose starvation. Quantitative analyses of the gppA transcript level reveal its differential expression pattern at different growth phases and starvation conditions. It has been observed that the GppA deficiency during amino acid starvation condition could be complemented by overexpressing the exopolyphosphatase coding gene ppx of V. cholerae. By deletion analysis, we further demonstrate that the amino and carboxy terminal sequences flanking the Ppx-GppA motif of the GppA protein of V. cholerae are also important for its enzymatic function.
Introduction
The enteric pathogen Vibrio cholerae faces various physicochemical stresses while living within or outside of the human intestine. Among multiple environmental stresses, nutritional stress is important for survival since it determines the growth and multiplication of the pathogen under such conditions. Like other bacterial pathogens, V. cholerae has evolved with complex gene regulatory networks to cope with various environmental stresses, among which the stringent response (SR) is important. The SR is typically characterized by strong repression of transcription of stable RNAs like transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), etc. (Cashel et al., 1996; Potrykus and Cashel, 2008; Gaca et al., 2015), upregulation of transcription of genes coding for the enzymes involved in amino acid biosynthesis (Stephens et al., 1975; Choy, 2000), and inhibition of replication (Wang et al., 2007). Regulations of all the above stated cellular processes under nutritional stress conditions are crucial for the survival of bacteria. The SR is essentially managed by two intracellular small signaling molecules, guanosine 3’-diphosphate 5’-triphosphate (pppGpp) and guanosine 3’,5’-bis(diphosphate) (ppGpp), together called (p)ppGpp (Cashel et al., 1996; Potrykus and Cashel, 2008). While in Escherichia coli, intracellular (p)ppGpp metabolism is controlled by two multidomain containing proteins RelA and SpoT, in V. cholerae, apart from these two enzymes, a small alarmone synthetase, called RelV, is also involved in (p)ppGpp metabolism as shown in Figure 1 (Haralalka et al., 2003; Das and Bhadra, 2008; Das et al., 2009; Dasgupta et al., 2014). RelA, the product of the relA gene, is a ribosome-associated protein and responsible for (p)ppGpp synthesis under amino acid starvation (Cashel et al., 1996). RelA is able to synthesize pppGpp and ppGpp by using guanosine triphosphate (GTP) and guanosine diphosphate (GDP) as substrates, respectively (Mechold et al., 2002; Sajish et al., 2009; Kudrin et al., 2018). On the other hand, SpoT, the product of the spoT gene, has a strong (p)ppGpp hydrolase activity and weak (p)ppGpp synthetase activity under different stress conditions (Xiao et al., 1991; Seyfzadeh et al., 1993; Das et al., 2009). Interestingly, in V. cholerae, the RelV enzyme, which is unique among Gram-negative bacteria, also contributes in (p)ppGpp synthesis under glucose and fatty acid starvations (Das et al., 2009; Dasgupta et al., 2014). Furthermore, unlike RelA and SpoT, RelV is a small single-domain-containing protein (Das et al., 2009; Dasgupta et al., 2014). In E. coli, apart from RelA and SpoT, another well-characterized enzyme, called guanosine pentaphosphate 5′-phosphohydrolase A (GppA), is associated with the (p)ppGpp metabolic cycle (Somerville and Ahmed, 1979; Hara and Sy, 1983; Keasling et al., 1993; Mechold et al., 2013). The main function of GppA is to convert pppGpp to ppGpp by removing the terminal γ-phosphate from 5’-position of pppGpp. Initial mutational analysis by Somerville and Ahmed (1979) indicated the presence of the GppA enzyme in E. coli for the conversion of pppGpp to ppGpp. However, Hara and Sy (1983) were the first to purify, characterize, and show the substrate specificity of GppA enzyme of E. coli. Recently, Mechold et al. (2013) have successfully manipulated the expression of GppA to demonstrate differential accumulation of ppGpp and pppGpp in E. coli cells, which allowed them to conclude that pppGpp is less potent than ppGpp with respect to regulation of growth rate, RNA/DNA ratios, ribosomal RNA P1 promoter transcription inhibition, threonine operon promoter activation, RpoS induction, etc. After the discovery of GppA, another phosphatase, called exopolyphosphatase (Ppx), was identified in E. coli (Akiyama et al., 1993). Ppx has been shown to hydrolyze the inorganic polyphosphate (polyP) to Pi (Akiyama et al., 1993). PolyP, a polymer of hundreds of phosphate residues, accumulates in E. coli and in other microbes in response to various stresses, including nutritional starvation (Kuroda et al., 2001). PolyP in E. coli and in other bacteria is synthesized by the enzyme polyphosphate kinase (Ppk), and the genes encoding Ppk and Ppx are usually physically linked (Akiyama et al., 1993; Kuroda et al., 1997, 2001; Ogawa et al., 2000). Extensive in silico analysis further indicated that GppA and Ppx are homologous proteins, and both of them belong to the sugar kinase/actin/hsp70 superfamily (Reizer et al., 1993; Koonin, 1994). It has been shown that like Ppx, the GppA enzyme of E. coli also has exopolyphosphatase activity, and thus, it is a bifunctional enzyme (Keasling et al., 1993). Similarly, a modest pppGpp hydrolase activity of E. coli Ppx has previously been demonstrated (Kuroda et al., 1997). It may be mentioned here that the members of Actinobacteria, like mycobacteria, corneybacteria, actinomycetes, etc., as well as certain other Gram-negative pathogens may encode homologous “Ppx-GppA” motif containing enzymes instead of separate Ppx and GppA proteins. The “Ppx-GppA” family of proteins share homology with both Ppx and GppA, and thus, they may have the ability to hydrolyze both polyP and pppGpp as substrates (Kristensen et al., 2004, 2008; Choi et al., 2012; Malde et al., 2014; Song et al., 2019).
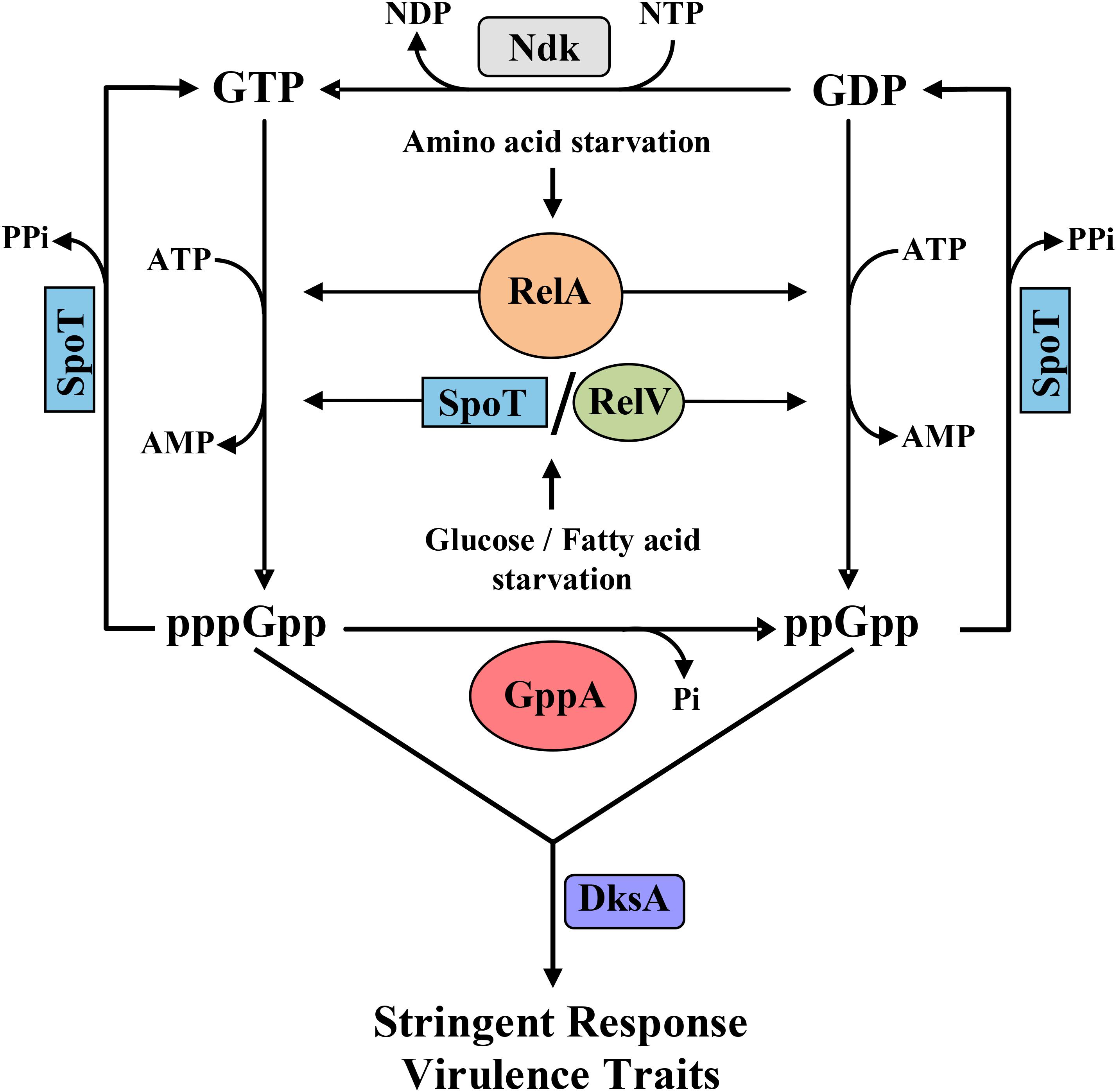
Figure 1. Schematic diagram showing the stringent response (SR) metabolic cycle involving different enzymes of V. cholerae. As shown, the RelA enzyme is activated during amino acid starvation, while glucose or fatty acid starvation induces SpoT and RelV followed by synthesis of pppGpp and ppGpp using guanosine triphosphate (GTP) and guanosine diphosphate (GDP) as substrates, respectively. pppGpp is converted to ppGpp by the enzyme GppA (this study). SpoT, the bifunctional enzyme, has a strong hydrolase activity and thus hydrolyzes pppGpp and ppGpp to GTP and GDP, respectively. GDP is subsequently converted to GTP by the nucleotide diphosphate kinase (Ndk) enzyme. Synthesized pppGpp/ppGpp molecules with the help of the regulatory protein DksA controls the SR and several critical virulence traits of V. cholerae like cholera toxin production, biofilm formation, motility, and protease expression.
Evolution of GppA supports that ppGpp is most likely the crucial molecule for the execution of SR in bacteria. Over the years, our group has characterized and established the entire SR regulatory genetic circuits including the discovery of the unique (p)ppGpp synthetase gene relV in V. cholerae as shown in Figure 1 (Haralalka et al., 2003; Das and Bhadra, 2008; Das et al., 2009; Pal et al., 2011, 2012; Dasgupta et al., 2014). In this (p)ppGpp metabolic cycle, conversion of GDP to GTP is carried out by the enzyme nucleoside diphosphate kinase, called Ndk, which has been characterized by the other workers (Dar et al., 2011). We have shown that the SR regulatory circuit is also linked with the modulation of certain critical virulence-related traits of V. cholerae (Haralalka et al., 2003; Pal et al., 2012; Basu et al., 2017; Basu and Bhadra, 2019). However, at present, no information is available about the functional aspect of the gppA gene in the cholera pathogen, the product of which converts pppGpp to ppGpp. Furthermore, very little information is available about the level of pppGpp and ppGpp under amino acid or glucose starved condition. Like in E. coli, V. cholerae genome also carries the physically linked ppk-ppx genes, which are involved in polyP metabolism (Heidelberg et al., 2000; Ogawa et al., 2000). In this report, we have carried out extensive genetic analysis of the gppA gene and show that the product of the gene is linked with the (p)ppGpp metabolism in V. cholerae (Figure 1). Like in E. coli, Ppx is also homologous to GppA in V. cholerae; therefore, mutational approaches were adopted to analyze the function of ppx in the presence or absence of the gppA gene. It was found that the overexpression of V. cholerae Ppx can complement the GppA function.
(Part of this work has been presented in the 54th United States–Japan Joint Panel Conference on Cholera and Other Bacterial Enteric Diseases held on 10–13 December 2019 at Osaka University, Osaka, Japan).
Materials and Methods
Bacterial Strains, Plasmids, Media, and Growth Conditions
Bacterial strains and plasmids used in this study are described in Table 1. For cloning purpose, the plasmid pBluescript II KS(+) and E. coli DH5α strain were used unless stated otherwise. Both E. coli and V. cholerae cells were routinely grown in Luria broth (LB; Difco, United States) at 37°C with aeration, and for plate culture, Luria agar (LA; Difco) was used, which contained 1.5% (w/v) Bacto agar (Difco) as described earlier (Haralalka et al., 2003). Antibiotics (Sigma-Aldrich, United States) were used in the following concentrations: ampicillin (Ap), 100 μg ml–1; kanamycin (Km), 40 μg ml–1; streptomycin (Sm), 100 μg ml–1; chloramphenicol (Cm), 3 μg ml–1 for V. cholerae; and 30 μg ml–1 for E. coli. Bacterial strains were stored at −70°C in LB containing 20% (v/v) sterile glycerol (Verma et al., 2019) and were always taken out before doing experiments to avoid the development of any uncharacterized mutation. The growth of bacterial cultures was monitored by measuring optical density at 600 nm (OD600) using a spectrophotometer (Hitachi, Model U-5100).
Molecular Biological Methods
For chromosomal and plasmid DNA preparations, restriction enzyme digestion, DNA ligation, bacterial transformation, conjugation, agarose gel electrophoresis, etc., standard molecular biological methods (Ausubel et al., 1989) were followed. Restriction and nucleic-acid-modifying enzymes were purchased from New England Biolabs, Inc., United States and were used essentially as instructed by the manufacturer. Electrocompetent V. cholerae cells were made as described previously from this laboratory (Das and Bhadra, 2008), and transformants were selected on LA plates containing appropriate antibiotics.
Construction of Plasmids and Mutants
Throughout the study, the expression vector pBADcam (Table 1), a derivative of pBAD24 (∼4.5 kb; Apr; Table 1) carrying the Cm gene (cam) cassette, was used. To construct pBADcam, the plasmid pPROTet.E (2.2 kb; Cmr; Table 1) was digested with EcoRV, and ∼0.8 kb digested product carrying the cam cassette was ligated to ScaI-digested pBAD24; the ligation mixture was transformed into E. coli DH5α. Among multiple clones obtained, one was selected for use and named pBADcam (∼5.3 kb; Cmr; Table 1). Details of the primers used in this study are described in Supplementary Table S1. The recombinant plasmid pGppA (Table 1) was constructed by amplifying the gppA Open Reading Frame (ORF) (∼1.6 kb) using the primers gppAorf-F/gppAorf-R (Supplementary Table S1) and the genomic DNA of V. cholerae O1 El Tor strain N16961 [Table 1; henceforth will be called wild type (Wt)], followed by double digestion with the restriction enzymes EcoRI/PstI and cloning in similarly digested expression vector pBADcam. Similarly, the ppx ORF (∼1.6 kb) carrying plasmid pPpx (Table 1) was constructed using the primers ppxORF-F/ppxORF-R (Supplementary Table S1) and genomic DNA of V. cholerae Wt strain. The PCR amplified ppx ORF containing fragment was double digested with XbaI/HindIII and cloned in similarly digested vector pBADcam. For the construction of deletion mutants, we always used the suicide vector pKAS32 (Apr; Table 1), which was maintained in E. coli SM10λpir (Kmr; Table 1). Details of the recombinant suicide plasmids, pKΔgAK and pKPXUDCam, constructed in this study are given in Table 1. Apart from cam gene, the kan gene cassette from the plasmid pUC4K (Table 1) was used for creation of a deletion allele of a desired gene of V. cholerae. To construct any deletion mutant of V. cholerae, the corresponding recombinant suicide plasmid carrying the mutant allele was conjugally transferred from E. coli SM10λpir to V. cholerae cells, and the desired deletion mutant was selected by double crossover event using appropriate antibiotics as described previously (Haralalka et al., 2003; Das et al., 2009). Details of the deletion mutants of V. cholerae constructed in this study are given in Table 1.
For construction of plasmids carrying amino (N)- and/or carboxy (C)-terminal coding region deleted gppA gene, each of such desired fragments was PCR amplified using specific set of primers (Supplementary Table S1) and genomic DNA of V. cholerae Wt strain as templates followed by digestion of the desired fragment with EcoRI/PstI and cloning of that DNA fragment in similarly digested vector DNA pBADcam. For amplification of 5′-end deleted fragment of gppA ORF each of the forward primers (Supplementary Table S1) always carried an artificially inserted ATG start codon. Similarly, for the amplification of 3′-end deleted fragment of the gppA ORF, each of the reverse primers (Supplementary Table S1) always carried an artificially inserted TAA stop codon. Restriction digestion and DNA sequencing further confirmed the recombinant plasmids.
Detection of Intracellular (p)ppGpp Accumulation
Intracellular accumulation of (p)ppGpp in V. cholerae cells under various starvation conditions was determined by radiolabeling of bacterial cells with [32P]-H3PO4 (BRIT, Mumbai, India) using 3-(N-morpholino) propanesulfonic acid (MOPS) medium with different supplements followed by lysis of cells and thin layer chromatography (TLC) essentially as described previously (Haralalka et al., 2003; Das and Bhadra, 2008; Das et al., 2009). For (p)ppGpp labeling under no starvation conditions, the same MOPS medium mentioned above was used, but it contained all amino acids plus glucose. Bacterial cells were labeled as mentioned earlier (Haralalka et al., 2003; Das and Bhadra, 2008; Das et al., 2009) followed by autoradiography. Densitometric analysis of (p)ppGpp spots on autoradiogram was done using the ImageJ software1. Abundance of pppGpp in each lane relative to the total (pppGpp + ppGpp) was calculated by dividing the spot intensity of pppGpp with that of total intensity value of pppGpp + ppGpp.
Reverse Transcriptase PCR Assay
For reverse transcriptase PCR (RT-PCR) assay, V. cholerae cells were grown in LB at an OD600 value of ∼1.0 followed by extraction of total cellular RNA using TRI Reagent (Sigma-Aldrich, United States) essentially as described previously (Pal et al., 2012; Basu et al., 2017; Basu and Bhadra, 2019). Purity and quantitation of prepared RNA were done spectrophotometrically. RT-PCR assay was carried out using the Qiagen One Step RT-PCR kit as directed by the manufacturer (Qiagen, Germany). The PCR-amplified product was checked by agarose gel electrophoresis using appropriate molecular size markers. To confirm the absence of any contaminating DNA in prepared RNA samples, PCR assay of each sample was also done with Taq DNA polymerase (Invitrogen). Lack of amplification in the absence of RT confirmed that the desired PCR product was generated only from complementary DNA (cDNA). For quantitative RT-PCR (qRT-PCR) assay, total cellular RNA was prepared from bacterial cells grown in LB medium to an OD600 value of ∼0.5, 1.0, or 1.5 as mentioned above. Reactions were done using the One Step SYBR®PrimeScriptTM RT-PCR Kit II essentially as described by the manufacturer (Takara Bio Inc., Japan). qRT-PCR assay was done using the CFX96TM Real-Time System (Bio-Rad, United States). The primer set gppArtm-F/gppArtm-R (Supplementary Table S1) was used for qRT-PCR analysis. Relative expression values (R) were calculated using the equation R = 2–(Δ CTtarget – CTreference), where CT is the fractional threshold cycle. In each experiment, as an internal control, the recA-specific primers recA-F/recA-R (Supplementary Table S1) were used. Each assay was repeated at least thrice. To check the expression status of gppA during amino acid starvation, glucose starvation, and in the absence of any starvation, total cellular RNA was extracted from V. cholerae following the same experimental conditions as done for (p)ppGpp labeling experiment. The level of gppA transcripts was measured by qRT-PCR analysis. As an internal control, the gyrA-specific primers, gyrArtm-F/gyrArtm-R (Supplementary Table S1), were used as recommended earlier (He et al., 2012). As positive controls, we used the V. cholerae biofilm related regulatory genes, vpsR and vpsT, which have been shown to be upregulated during amino acid starvation (He et al., 2012). Each RT-PCR experiment was repeated at least thrice.
DNA Sequencing and Computational Analyses
For confirmation of plasmid constructs, DNA sequencing was done using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc., United States) essentially as instructed by the manufacturer. The samples were run on an ABI3130 Genetic Analyzer using the pop-7 polymer (Applied Biosystems Inc.). DNA sequence data were compiled and analyzed by using the Chromas 1.452. DNA sequences were obtained from J. Craig Venter Institute (JCVI)3, and protein domain information was obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG)4. BLASTN and BLASTP programs were used to search for homologous nucleotide or protein sequences, respectively, in the database5. Clustal Omega software6 was used for the alignment of protein sequences and further modified through GeneDoc software version 2.7.0007. For designing primers, Primer3 software8 was used. For designing qRT-PCR primers, Primer Express 3.0 software (Applied Biosystems, United States) was used.
Statistical Analysis
Where needed, pairwise comparison of data for each sample was analyzed for statistical significance using Student’s t-test.
Results
In silico Analysis of the gppA Gene of V. cholerae
Since no information is currently available about the gppA gene of V. cholerae, BLASTN9 analysis of the whole-genome-sequenced V. cholerae Wt strain N16961 (Table 1) was carried out using the E. coli gppA gene sequence as a query. Such analysis indicated that the large chromosome of V. cholerae indeed carries a 1.5-kb long gppA gene (JCVI annotation no. VC0304). Comparison of the gppA gene sequence of V. cholerae indicated 63% nucleotide sequence identity with that of E. coli by using the BLAST Global Alignment Tool. While the V. cholerae gppA gene codes for a 497 amino acid long protein, the E. coli GppA is composed of 494 amino acids (see footnote 3). BLASTP analysis of V. cholerae GppA protein shows 57% identity (279 out of 491 amino acids) and 72% similarity with that of E. coli GppA. Genetic organization of the gppA gene in the genomes of V. cholerae, other Vibrio spp., and E. coli indicated that the gppA locus is highly conserved in vibrios (Figure 2A). BioCyc analysis10 suggested that the gppA gene along with its upstream region is physically linked with the rhlB gene (JCVI annotation no. VC0305), and they transcribe as a bicistronic operon. KEGG analysis of the 497 amino acid long GppA protein of V. cholerae revealed that it carries a highly conserved Ppx-GppA motif of 281 amino acids long (position 23 to 303), which is similar to the Ppx-GppA motif of E. coli GppA enzyme (Figure 2B). We also did BLASTN analysis for the V. cholerae ppx gene (JCVI annotation no. VC0722) and found that it encodes 523 amino acids long exopolyphosphatase. BLASTP analysis indicated that the V. cholerae Ppx has 40.5% identity with that of GppA (Figure 2B). Furthermore, amino acid sequence alignment of the GppA and Ppx proteins of V. cholerae, other Vibrio spp., E. coli, and thermophilic Aquifex aeolicus indicated substantial conservation of the residues including those needed for the pppGpp hydrolysis function (see Figure 2B). Analyses of the crystal structures of the E. coli Ppx and GppA proteins indicated the presence of four distinct domains, I–IV (Rangarajan et al., 2006; Song et al., 2019). It is evident that the V. cholerae Ppx and GppA proteins also carry similar domains as shown in Figure 2B.
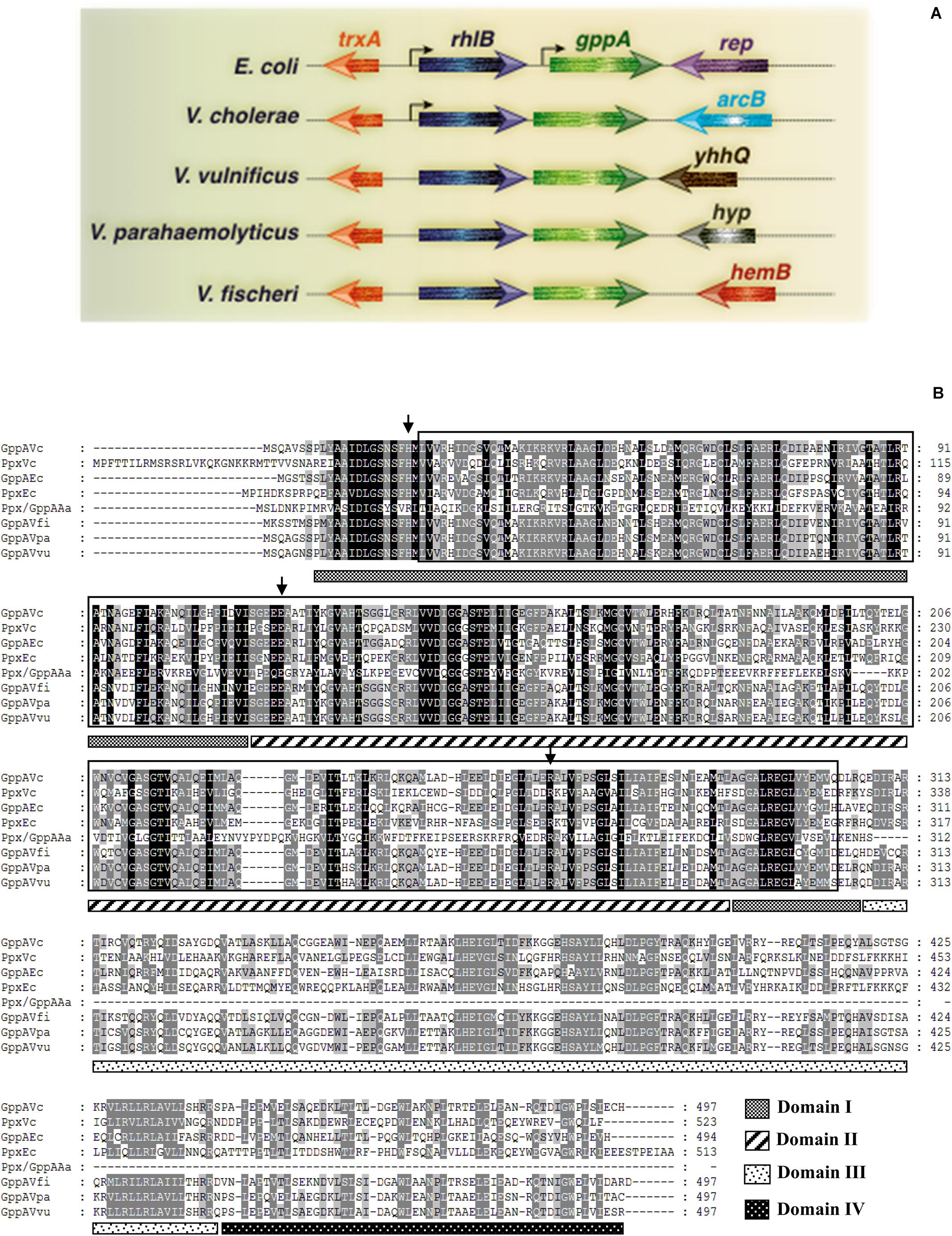
Figure 2. (A) Schematic diagram (not drawn to scale) showing genomic arrangement of the gppA gene in V. cholerae, E. coli, and other Vibrio spp. Names of different organisms are given in the left margin. Physically linked genes of gppA and their homologs are as indicated. Arrowheads indicate direction of transcription of genes. Thin gray line represents intergenic or chromosomal DNA. Bend arrows indicate putative monocistronic and bicistronic transcription of the gppA-rhlB genes in E. coli and V. cholerae, respectively. (B) Comparison of the amino acid sequences of Ppx-GppA motif containing proteins. Alignment was done using the Clustal Omega software (https://www.ebi.ac.uk/Tools/msa/clustalo/) and further modification by GeneDoc software version 2.7.000. Aligned proteins include GppA (GppAVc) and Ppx (PpxVc) of V. cholerae with those of E. coli GppA (GppAEc), E. coli Ppx (PpxEc), A. aeolicus Ppx/GppA (Ppx/GppAAa), V. fischeri GppA (GppAVfi), V. parahaemolyticus GppA (GppAVpa), and V. vulnificus GppA (GppAVvu). Dark and light shades indicate identical and similar amino acid, respectively. Boxed amino acid sequence denotes highly conserved Ppx-GppA motif (usually about 280 amino acids long). Small vertical arrows indicate conserved critical amino acid residues needed for phosphohydrolase activity. Four probable domains (domains I–IV) are indicated as different patterned bars. The domains are marked considering the PpxEc protein as a reference. Numbers in the right margin indicate amino acids length of each protein.
Transcriptional Organization of the gppA Operon of V. cholerae
Comparison of the gppA genetic loci of V. cholerae and E. coli revealed distinct difference in organization of different ORFs (Figure 2A). Bioinformatic analysis indicated that the rhlB-gppA genes of V. cholerae are arranged in a bicistronic operon since there is only a 7-bp gap between these two genes. The transcription orientations of the flanking ORFs of the rhlB-gppA operon are in reverse orientations (Figure 2A). In sharp contrast, the E. coli gppA gene appears to be organized as a monocistronic transcription unit since there is a 135-bp long intergenic region with predicted promoter sequence between the rhlB and gppA genes (Figure 2A). While the downstream region of gppA of V. cholerae contains the arcB gene encoding a sensor histidine kinase, in E. coli, the rep gene encoding an ATP-dependent DNA helicase is present in the same position (Figure 2A). In this study, we predicted that the V. cholerae gppA gene (JCVI annotation no. VC304) cotranscribes with the rhlB gene (JCVI annotation no. VC0305), and they are arranged in a bicistronic operon. To confirm this, V. cholerae cells were grown till late exponential phase (OD600 = 1.0), and total cellular RNAs were prepared for RT-PCR analysis. When the primer pair gppASeq-F (VC0305 ORF specific) and gppArtm-R (Supplementary Table S1) was used, as expected, a desired cDNA fragment of ∼0.33 kb in size was obtained (Figure 3). Similarly, when gppA or VC0305 ORF specific internal primers (gppArtm-F/gppA-R or rhlB-F/rhlB-R; Supplementary Table S1) were used, in each case, the desired cDNA fragment of size ∼0.56 or 0.37 kb, respectively, was obtained (Figure 3). The RT-PCR results confirmed that the gppA and VC0305 (rhlB) genes are indeed arranged in an operon, and they are cotranscribed. Next, we wanted to ensure that in the gppA-deleted V. cholerae strain NΔgAK (Table 1; henceforth will be called ΔgppA), the transcription of the neighboring gene rhlB (VC0305) was not affected. It is to be noted that during construction of the ΔgppA mutant, the closest rhlB ORF was kept intact; however, transcription of the arcB gene was not tested. For this purpose, the transcript status of rhlB in ΔgppA strain was examined by RT-PCR using the same set of primer rhlB-F/rhlB-R (Supplementary Table S1). While no cDNA was detectable for the gppA gene, rhlB ORF-specific primer set was indeed able to amplify the desired cDNA fragment of ∼0.37 kb using the ΔgppA mutant (Figure 3).
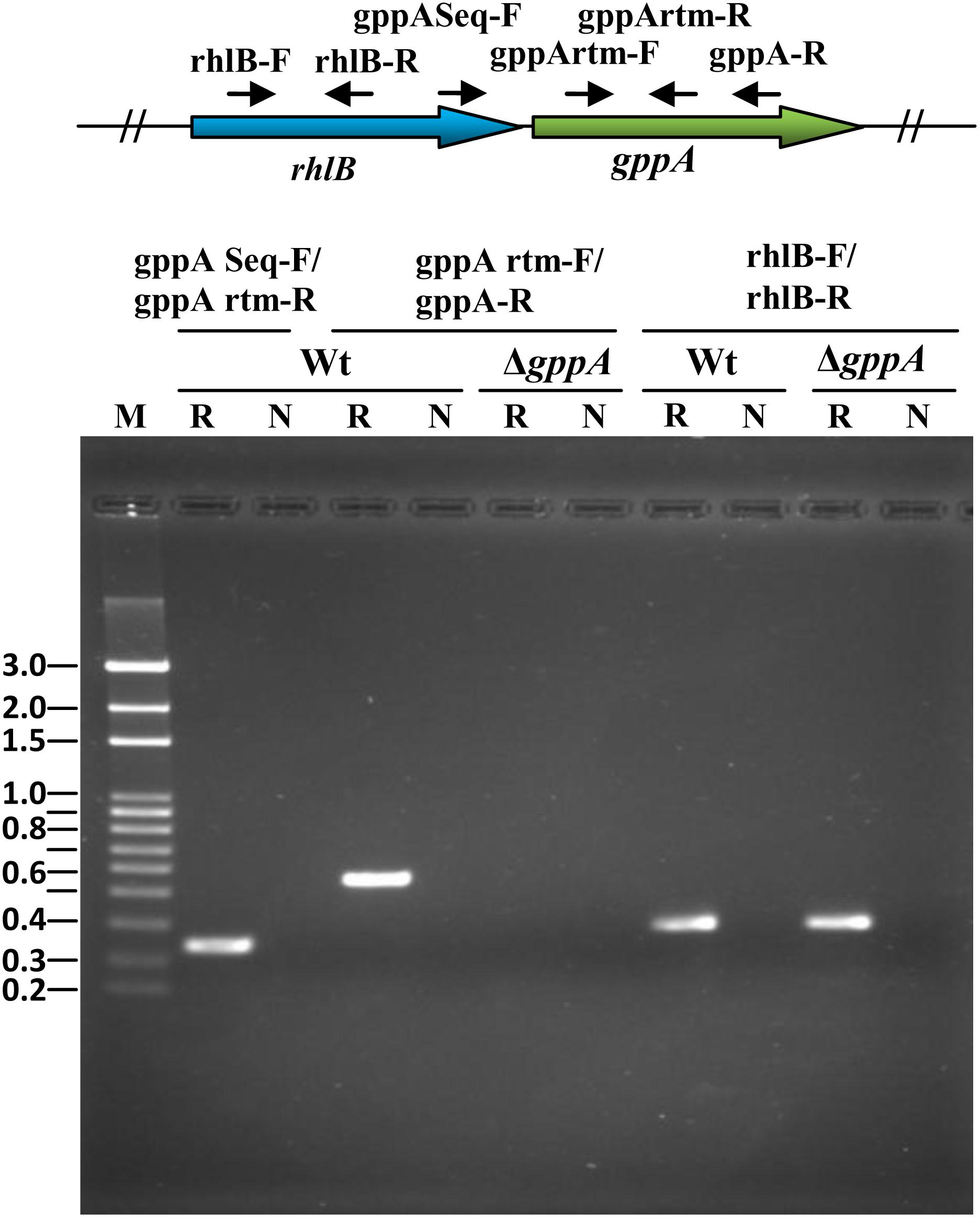
Figure 3. Reverse transcription PCR (RT-PCR) analysis to determine bicistronic arrangement of V. cholerae rhlB-gppA genes. Primers were designed exclusively from the rhlB-gppA locus, and small solid arrows indicate positions and directions of these primers. Primer sets used for RT-PCR analysis are indicated above each lane. Wt and ΔgppA V. cholerae strains are indicated above the lanes. “R” indicates RT with Taq polymerase enzyme, and “N” denotes use of only Taq DNA polymerase as a negative control. “M” denotes 100-bp DNA ladder used as molecular size markers, and their sizes (in kb) are given in the left margin.
GppA Modulates (p)ppGpp Homeostasis During Amino Acid Starvation
In order to understand the functions of GppA in regulating the intracellular pppGpp and ppGpp levels in V. cholerae under various starved conditions, intracellular (p)ppGpp labeling assay was performed using the ΔgppA mutant (Table 1) and its parental Wt strain. Before performing different experiments with the ΔgppA mutant, its growth kinetics in LB and M9 minimal medium were compared with that of Wt cells. However, in both the media, ΔgppA mutant showed no defect in growth when compared with the Wt (Supplementary Figures S1A,B). We have shown previously that the RelA of V. cholerae exclusively produces ppGpp under amino acid starved condition (Haralalka et al., 2003; Das and Bhadra, 2008). It may be argued here that under amino acid starvation, V. cholerae RelA may use GDP as a major substrate to synthesize ppGpp or it could use GTP as a substrate to produce pppGpp, which is rapidly converted to ppGpp by GppA. Interestingly, when ΔgppA cells were starved for amino acids along with its isogenic Wt strain and labeled with 32P followed by autoradiography, a significant increase (∼10-fold, see Figure 4) in the intracellular concentration of pppGpp in ΔgppA mutant was observed compared to the Wt (Figure 4A). The result suggests that, under amino acid starvation, GTP is also used as a substrate by the V. cholerae RelA to produce pppGpp followed by conversion to ppGpp by GppA. But in the case of ΔgppA mutant, which is devoid of GppA, this conversion is not possible, and thus, the mutant cells showed accumulation of substantial amount of pppGpp. This observation is also supported by densitometric analysis of the (p)ppGpp spots using the ImageJ software as shown in Figure 4B. This is further supported by the complementation assay using the ΔgppA cells carrying the V. cholerae gppA gene expressing plasmid pGppA (Table 1). As expected, such an assay and (p)ppGpp spot quantification clearly showed significant accumulation of ppGpp in ΔgppA(pGppA) cells like that of the parental Wt strain (Figures 4D,E). It is important to mention here that functional complementation by GppA was also observed in the case of ΔgppA(pGppA) in the absence of arabinose (Figure 4D), probably due to the leaky expression of gppA from the plasmid pGppA. By comparison of the profiles of pppGpp and ppGpp under amino acid starvation in V. cholerae relA+ gppA+ (Wt) and relA+ gppA– (ΔgppA) genetic backgrounds, it appears that the GDP is a more preferred substrate for RelA than GTP (Figure 4A). In support of this conclusion, it may be mentioned here that the E. coli RelA is able to synthesize pppGpp and ppGpp using GTP and GDP as substrates, respectively (Sajish et al., 2009; Kudrin et al., 2018). It has been shown that pppGpp allosterically activates RelA leading to its increased efficiency in the synthesis of ppGpp from GDP (Kudrin et al., 2018). Thus, through all these genetic analyses, we first time established the function of GppA in V. cholerae.
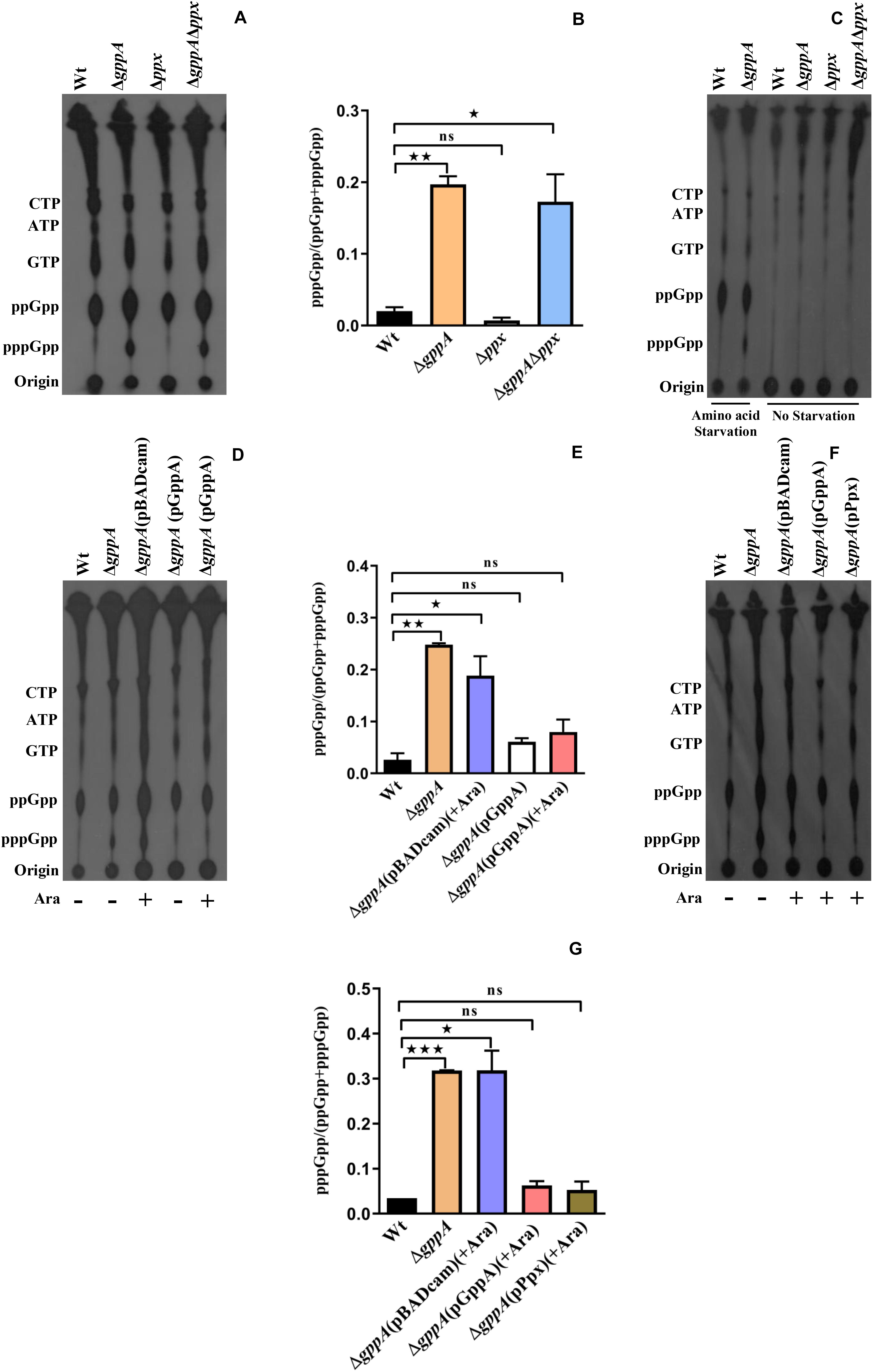
Figure 4. (A) Accumulation of (p)ppGpp in V. cholerae cells under amino-acid-starved condition. Bacterial cells were labeled with 32P-orthophosphoric acid, extracted, resolved by TLC followed by autoradiography. Strains used are Wt, ΔgppA, Δppx, and ΔgppAΔppx. Distinct accumulation of pppGpp in ΔgppA is evident. The presence of other labeled nucleotides including pppGpp/ppGpp is indicated in the left margin. (B) Densitometry of the (p)ppGpp spots detected in the autoradiogram was carried out by ImageJ software, and the ratio of pppGpp/(pppGpp + ppGpp) was plotted. Values represent the average of two independent experiments (n = 2), and one representative autoradiogram is shown in (A). Error bars indicate standard deviation. (*P < 0.05; **P < 0.01; ns, non-significant). (C) Accumulation of (p)ppGpp in V. cholerae cells without any starvation (see details in text). Strains used are Wt, ΔgppA, Δppx, and ΔgppAΔppx. (p)ppGpp accumulation in Wt and ΔgppA under amino acid starvation condition served as controls. The presence of 32P-labeled nucleotides is indicated in the left margin. (D) Complementation analysis of the ΔgppA mutant of V. cholerae. Autoradiogram shows poor or no accumulation of pppGpp in ΔgppA(pGppA) strain in which gppA was expressed through the plasmid pGppA using 0.2% arabinose (Ara) as an inducer. Wt, ΔgppA, and ΔgppA carrying the empty plasmid [ΔgppA(pBADcam)] strains were used as controls. The presence of labeled nucleotides including pppGpp/ppGpp as indicated in the left margin. (E) Densitometric analysis of the (p)ppGpp spots was done as described in (B). Values represent the average of two independent experiments (n = 2), and one representative autoradiogram is shown in (D). Error bars indicate standard deviation. (*P < 0.05; **P < 0.01; ns, non-significant). (F) Autoradiogram showing (p)ppGpp accumulation in ΔgppA mutant by overexpressing ppx. Induction of both gppA and ppx expression through the plasmids pGppA and pPpx, respectively, was done using 0.2% arabinose as an inducer. Controls used are as given in (D). The presence of labeled nucleotides including pppGpp/ppGpp as indicated in the left margin. (G) Densitometric analysis of the (p)ppGpp spots was done as described in (B). Values represent the average of two independent experiments (n = 2), and one representative autoradiogram is shown in (F). Error bars indicate standard deviation. (*P < 0.05; ***P < 0.001; ns, non-significant).
Overexpression of Ppx Complements ΔgppA Mutant During Amino Acid Starvation
Since the ppx gene of V. cholerae showed high homology with the gppA gene (Figure 2B), we also constructed V. cholerae ppx and gppA ppx deletion mutants to test the pppGpp level during amino acid starvation in the presence and absence of functional ppx gene. V. cholerae ppx and gppA ppx mutants constructed for this purpose were designated as NΔPPX (hereafter will be called Δppx) and NΔgPx (henceforth will be designated ΔgppAΔppx), respectively (Table 1). While Δppx behaved like Wt, the ΔgppAΔppx double mutant accumulated similar amount of pppGpp like in ΔgppA cells (Figure 4A). This was further verified by densitometric quantification of the (p)ppGpp spots (Figure 4B). To confirm that the spots shown in Figure 4A are indeed of (p)ppGpp, as a control experiment, the same set of bacterial strains was used to label (p)ppGpp using 32P without inducing any starvation. As expected, there was no accumulation of pppGpp and ppGpp in nutrient supplemented strains compared to the amino-acid-starved Wt and ΔgppA cells (Figure 4C). Apparently, the result suggests that the Ppx enzyme is probably unable to convert pppGpp to ppGpp, which could be due to its low expression/activity under the experimental condition used. To test this hypothesis, we have constructed a Ppx-overexpressing plasmid pPpx (Table 1). When Ppx was overexpressed in ΔgppA cells through the recombinant plasmid pPpx, it was able to complement the GppA function (Figure 4F). Densitometric analysis of the (p)ppGpp spots also supported this result as shown in Figure 4G. The result suggests that, under natural physiological condition, the basal level of Ppx probably has no or very weak pppGpp to ppGpp conversion activity. However, high abundance of the Ppx protein achieved by overexpression in ΔgppA cells most probably helped in overcoming this defect and converted pppGpp to ppGpp. The result warrants further studies to elucidate the exact role of Ppx in the metabolism of (p)ppGpp in V. cholerae under different nutritional stress conditions.
GppA Mediated Conversion of pppGpp to ppGpp Is Not Observed During Glucose Starvation Condition
We further examined the status of pppGpp and ppGpp in V. cholerae ΔgppA mutant under glucose starvation. Interestingly, there was no difference in intracellular pppGpp and ppGpp levels between ΔgppA and isogenic Wt cells (Figure 5A). We extended our analysis to test the pppGpp and ppGpp levels in V. cholerae ΔgppA, Δppx, and ΔppxΔgppA mutants. As expected, no difference was observed in the intracellular pppGpp and ppGpp levels among Wt, ΔgppA, Δppx, and ΔgppAΔppx mutants either in the autoradiogram or by densitometric analysis (Figures 5A,B). At present, the mechanism(s) underscoring this contrasting function of V. cholerae GppA under amino-acid- and glucose-starved conditions is not clear. The possibility of the presence of interacting partner(s) that modulate GppA activity during glucose starvation or differential expression pattern of gppA during glucose and amino acid starvations cannot be ruled out and needs further investigation.
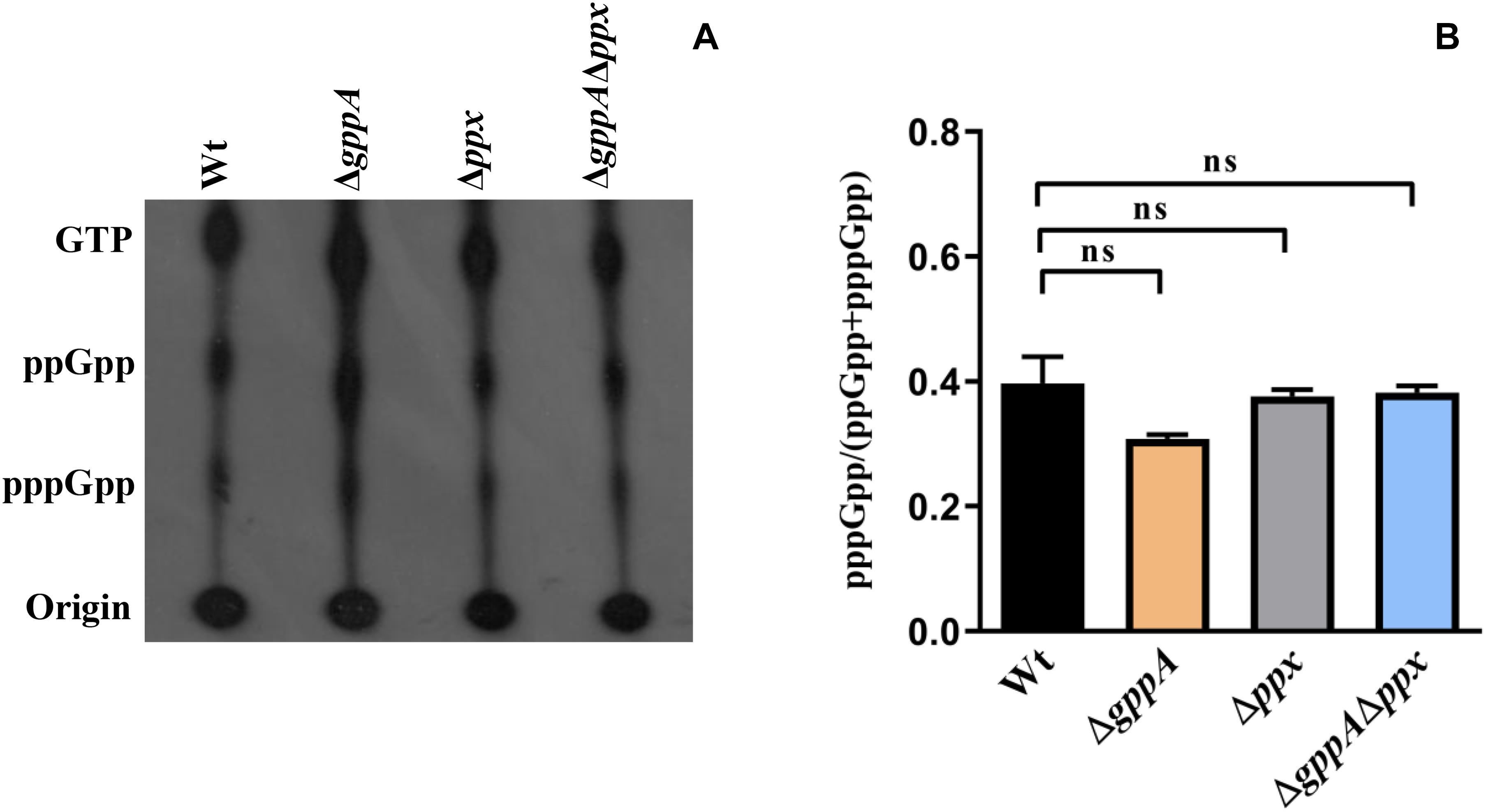
Figure 5. (A) Accumulation of (p)ppGpp in V. cholerae cells under glucose starvation. Strains used are: Wt, ΔgppA, Δppx, and ΔgppAΔppx. The presence of labeled nucleotides including pppGpp/ppGpp is indicated in the left margin. (B) Densitometry of the (p)ppGpp spots detected in the autoradiogram was carried out by ImageJ software and the ratio of pppGpp/(pppGpp + ppGpp) was plotted. Values represent the average of two independent experiments (n = 2) and one representative autoradiogram is shown in Panel A. Error bars indicate standard deviation. (ns = non-significant).
Expression of the gppA Gene Increases During Stationary Growth Phase and Amino Acid Starvation Condition
After establishing the function of gppA gene in amino-acid-starved cells, we attempted to test its regulation, if any, during various growth phases and starvation conditions. To this end, we performed quantitative estimation of the gppA gene transcripts under various growth phases of V. cholerae cells in nutrient-rich LB medium using qRT-PCR assay. As shown inFigure 6A, there was significant increase (∼2.5-fold) in gppA transcripts level in stationary phase cells of V. cholerae Wt strain compared to that of early or mid-log phase cells. We further extended our analysis to explore the expression level of the gppA gene in V. cholerae under both amino acid as well as glucose-starved conditions with respect to non-starved condition in MOPS medium using qRT-PCR assay (see details in section “Materials and Methods”). Interestingly, qRT-PCR analysis indicated a significant downregulation (∼1.8-fold) of gppA expression in glucose-starved V. cholerae cells compared to non-starved bacteria (Figure 6B). On the other hand, it has been observed that gppA expression goes up (∼2.5-fold) during amino acid starvation compared to non-starved cells (Figure 6B). When we compared the gppA expression between glucose and amino acid starved cells, a significant (∼4.5-fold) upregulation was observed in the latter condition (Figure 6B). This differential expression pattern of gppA during amino acid and glucose starvations may be responsible for the lower level of pppGpp accumulation in Wt V. cholerae cells under amino acid starvation. To support this observation, as positive controls, we measured the expression levels of two biofilm regulatory genes vpsR and vpsT of V. cholerae. It has previously been reported that the vpsR and vpsT genes are upregulated during amino acid starvation conditions (He et al., 2012). Therefore, we compared the expression of gppA, vpsT, and vpsR in amino-acid-starved V. cholerae cells. As expected, the expression of both vpsR and vpsT genes are indeed upregulated (∼5-fold) during amino-acid-starved condition compared to non-starved V. cholerae cells (Figure 6B).
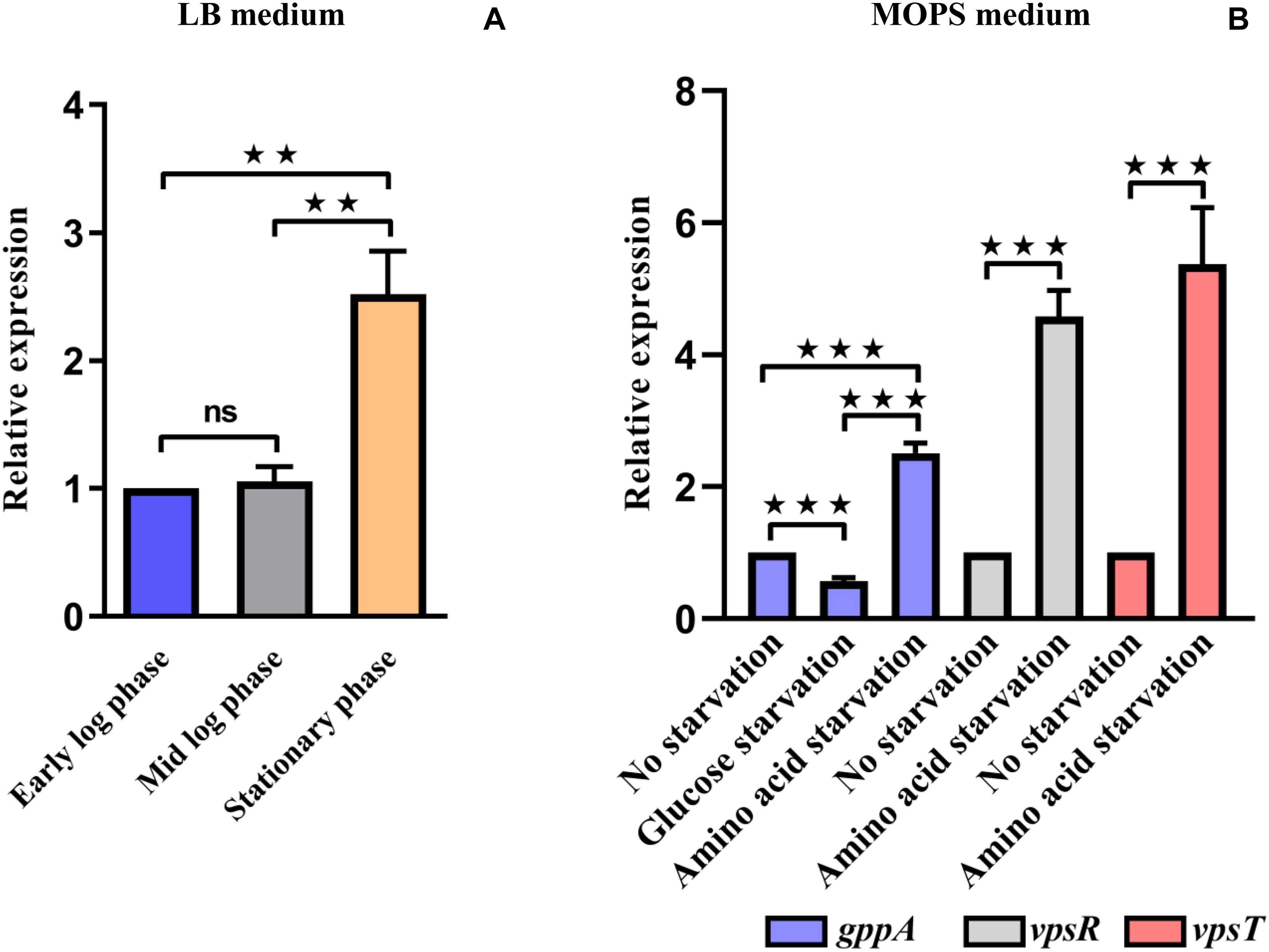
Figure 6. (A) Quantitative reverse transcription PCR (qRT-PCR) analysis of the expression of V. cholerae gppA gene in Luria broth (LB) medium under different growth phases as indicated (see details in text). Values represent the average of three independent experiments (n = 3). Error bars indicate standard deviation. (**P < 0.01; ns, non-significant). (B) qRT-PCR analysis of the expression of V. cholerae gppA gene under different starvation conditions in 3-(N-morpholino) propanesulfonic acid (MOPS) medium as indicated. As positive controls, the biofilm regulatory genes vpsR and vpsT were used (see details in text). Values represent the average of three independent experiments (n = 3). Error bars indicate standard deviation. (***P < 0.001).
Flanking Sequences Surrounding the Ppx-GppA Motif of GppA Are Important for Its Function
To determine the importance of the N- and C-terminal regions as well as the conserved domains (I–IV) of the V. cholerae GppA protein in pppGpp to ppGpp conversion function, several progressive deletion constructs (Figure 7A) were made by cloning each of the truncated gppA ORF under the PBAD promoter of the plasmid pBADcam (Table 1), and each recombinant clone was used for complementation in V. cholerae ΔgppA strain by assaying the ability of the expressed mutant alleles in intracellular conversion of pppGpp to ppGpp during amino acid starvation. While the ΔgppA mutant strains carrying the plasmid pGppA1-313 [ΔgppA(pGppA1-313)] or pGppA1-310 [ΔgppA(pGppA1-310)], expressing truncated GppA where 184 or 187 amino acids were deleted, respectively, were able to complement the GppA function, the strain ΔgppA(pGppA1-272), ΔgppA(pGppA1-303), or ΔgppA(pBG1-308), where truncated gppA expressed 225, 194, or 189 C-terminally amino acids deleted proteins, respectively, failed to complement (Figure 7B). On the other hand, the ΔgppA mutant strains carrying the plasmid pGppA13-497 [ΔgppA(pGppA13-497)] or pGppA18-497 [ΔgppA(pGppA18-497)], expressing N-terminally 12 or 17 amino acids deleted proteins, were able to complement the GppA function but not by the strain ΔgppA(pGppA23-497) or ΔgppA(pGppA20-497), expressing N-terminally 22 or 19 amino acids deleted GppA, respectively (Figure 7C). Finally, based on the above results, a recombinant plasmid pGppA18-310 (Table 1) containing the truncated gppA gene was constructed (Figure 7A), which should express N-terminally (17 amino acids) and C-terminally (187 amino acids) deleted GppA protein. As expected, when the plasmid pGppA18-310 was introduced into the ΔgppA mutant, it was able to complement the GppA function (Figure 7D). Thus, it appears that the functional N- and C-terminal boundary of GppA resides between the amino acid positions 18 and 310. In fact, the conserved Ppx-GppA motif actually falls within this minimal functional region of the protein (Figure 2B). To our knowledge this is the first report about functional boundary determination of a bacterial GppA protein.
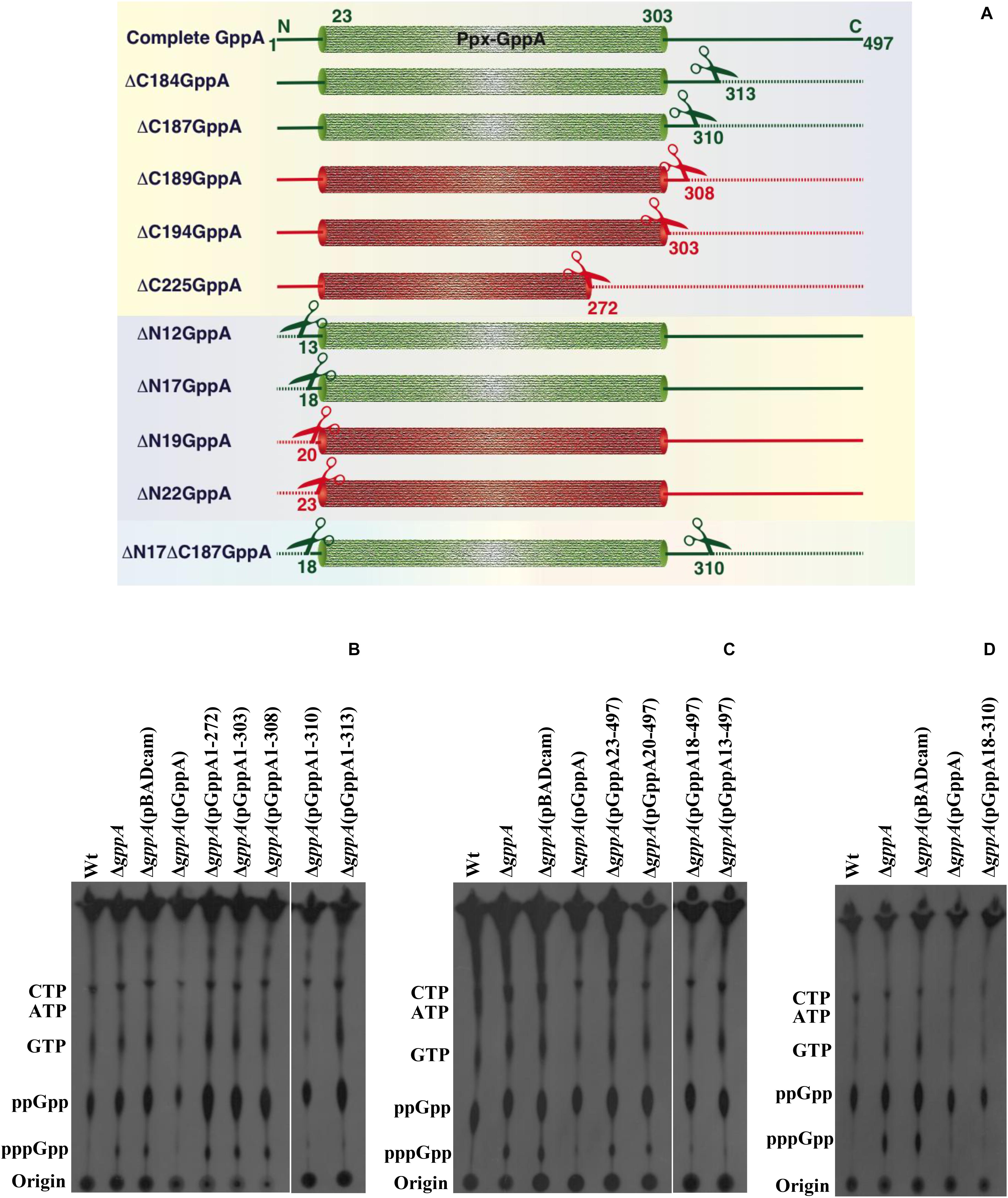
Figure 7. Functional analysis of the N- or/and C-terminal coding region deleted mutant alleles of the gppA gene of V. cholerae. (A) Schematic diagram (not drawn to scale) showing N- or/and C-terminal deleted regions of the V. cholerae GppA protein. Cylindrical-shaped portion of the GppA protein represents the putative Ppx-GppA motif, which spans from amino acid positions 23 to 303 as indicated. Scissors indicate position of each deletion, and dashed line represents deleted amino acid sequence from N- or/and C-terminal regions of GppA. Green and red colors denote functionally active and inactive GppA protein, respectively. N- and C-terminally deleted amino acid sequence of the GppA protein is symbolized by ΔN and ΔC, respectively, followed by the number of amino acids deleted as shown in the left margin. Intact GppA protein is 497 amino acids long, and the stretch of amino acid sequences present after deletion from N- or C-terminal of the GppA protein are shown at the corresponding ends. (B) Complementation analysis using C-terminal encoding sequence deleted alleles. Each C-terminally truncated GppA protein was expressed using 0.2% arabinose as an inducer in the ΔgppA mutant through the respective recombinant plasmid as shown above each lane. For the detection of pppGpp/ppGpp, thin layer chromatography (TLC) analysis of each strain was done followed by autoradiography. The presence of labeled nucleotides including pppGpp/ppGpp is indicated in the left margin. Autoradiogram shows complementation in ΔgppA(pGppA1-310) and ΔgppA(pGppA1-313) strains but not in ΔgppA(pGppA1-272), ΔgppA(pGppA1-303), and ΔgppA(pGppA1-308) strains. Wt, ΔgppA, ΔgppA carrying the empty vector pBADcam [ΔgppA(pBADcam)] and ΔgppA carrying the full-length GppA expressing clone pGppA [ΔgppA(pGppA)] were used as controls. (C) Complementation analysis using N-terminal encoding sequence deleted alleles. Each N-terminally truncated GppA protein was expressed using 0.2% arabinose as an inducer in the ΔgppA mutant through the respective recombinant plasmid as shown above each lane. TLC analysis of each strain was done followed by autoradiography. The presence of labeled nucleotides including pppGpp/ppGpp is indicated in the left margin. Autoradiogram shows complementation in strains ΔgppA(pGppA18-497) and ΔgppA(pGppA13-497) but not in strains ΔgppA(pGppA23-497) and ΔgppA(pGppA20-497). Control strains used as indicated in (B). (D) Determination of minimal functional region of the GppA protein. Here, both N- and C-terminal coding regions deleted gppA clone pGppA18-310 was used for complementation in ΔgppA mutant. For the expression of the truncated GppA protein, 0.2% arabinose was used. TLC analysis was done as described in (B). Strains used are Wt, ΔgppA, ΔgppA(pBADcam), ΔgppA(pGppA), and ΔgppA(pGppA18-310). The presence of labeled nucleotides including pppGpp and ppGpp is indicated in the left margin.
Effect of Deletion of the gppA Gene on Virulence Phenotypes of V. cholerae
We have also examined the effect of deletion of the gppA gene on certain virulence phenotypes of V. cholerae like cholera toxin production, biofilm formation, and motility. However, no defect in any of these virulence phenotypes was observed in the ΔgppA mutant (Supplementary Figure S2).
Discussion
The V. cholerae SR pathway gene gppA, the product of which converts pppGpp to ppGpp (Figure 1), has not been characterized previously. Therefore, the main objective of this study was to characterize the gppA gene function by genetic and mutational approaches. We found that unlike in E. coli, the gppA gene in the genome of V. cholerae is arranged in an operon along with rhlB (Figure 3). In E. coli, the rhlB gene codes for an ATP-dependent RNA helicase, which is a component of the RNA degradasome complex (Khemici et al., 2005). Although at present no information is available about the function of RhlB of V. cholerae, the arrangement of the rhlB-gppA genes in an operon has raised a possibility that RhlB could be involved in the regulation of the SR in this pathogen, which needs investigation.
We found that the accumulation of pppGpp in ΔgppA cells is high (∼10-fold) compared to the isogenic Wt strain during amino acid starvation but not under glucose deficient condition (Figures 4, 5). We have shown earlier that under amino acid starvation, V. cholerae RelA synthesizes (p)ppGpp (Haralalka et al., 2003), while during glucose starvation, SpoT and RelV enzymes are responsible for (p)ppGpp synthesis (Das and Bhadra, 2008; Das et al., 2009; Dasgupta et al., 2014). Therefore, it may be argued here that Wt cells produce both pppGpp and ppGpp using GTP and GDP as substrates, respectively, during amino acid starvation, but GppA most likely converts most of the pppGpp molecules to ppGpp (Figure 4). In contrast, under glucose starvation, there was no difference in pppGpp/ppGpp accumulation in Wt and ΔgppA cells suggesting that the result is independent of GppA. It may be possible that the affinity of GppA to the pppGpp substrate is altered under glucose starvation stress, or may be some unknown factor is needed to activate the GppA enzyme under this condition, which needs further work. In this context, it may be noted that in E. coli, significant amounts of pppGpp accumulate in ΔgppA mutant compared to an isogenic Wt strain under both amino acid and glucose starvation conditions (Somerville and Ahmed, 1979). However, our result on glucose starvation of V. cholerae cells does not support this finding in E. coli, which could be organism specific.
In this study, we have also characterized the ppx gene of V. cholerae to determine whether it has any gppA-like function. This is because both Ppx and GppA contain conserved Ppx-GppA motif, which appears to be needed for their similar enzymatic functions (Kristensen et al., 2008). Recently, it has been reported that deletion of the ppx1 gene of the enteropathogen Campylobacter jejuni leads to increase in intracellular accumulation of pppGpp as well as polyP (Malde et al., 2014). We found that the overexpression of Ppx can complement the GppA function in amino acid starved ΔgppA V. cholerae cells (Figure 4). Similarly, Kuroda et al. (1997) have reported complementation of the GppA function in an E. coli ΔgppAΔppkx mutant strain (GppA-Ppk_Ppx-) by overexpressing the Ppx enzyme. Very recently, the structure of the Ppx/GppA protein of the Gram-negative bacterium Helicobacter pylori has been solved (Song et al., 2019). It was found that the H. pylori Ppx/GppA enzyme is ∼27-fold less efficient in hydrolyzing pppGpp compared to its polyP hydrolase activity. Analysis of the crystal structures of Ppx and GppA also supports that the active site of Ppx can efficiently bind the narrower polyP but not the wider pppGpp molecule. Similarly, the structure of the active site of GppA allows pppGpp to bind efficiently and hydrolyze it to ppGpp (Song et al., 2019). Thus, it could be plausible that overexpression of V. cholerae Ppx in ΔgppA cells may hydrolyze pppGpp and complement the gppA mutant phenotype due its probable weak pppGppase activity. However, further studies are needed to understand the exact role of Ppx in the complex metabolic network of (p)ppGpp in V. cholerae.
Currently, the expression profile of the gppA gene in different growth phases is not available for any Gram-negative bacteria. Such analysis indicated that in V. cholerae, the gppA gene expression is constitutive, although there is modest increase in transcripts level during the stationary growth phase (Figure 6A). This is likely since during stationary phase, bacterial cells face exhaustion of nutrients, which may trigger SR-related gene expression including gppA. Since gppA expression during amino acid starvation has been observed to be high compared to that of glucose-starved cells (Figure 6B), which could be one of the reasons for no accumulation of pppGpp in amino-acid-starved Wt cells.
After establishing the function of the gppA gene, we determined the core functional region of the GppA protein of V. cholerae by progressive deletion analysis of the N- and C-terminal regions carrying the conserved domains (I–IV) and Ppx-GppA motif (Figure 2B). However, extensive complementation analysis of N- or/and C-terminal coding deletion alleles allowed us to validate that the Ppx-GppA motif of V. cholerae GppA actually spans from amino acid positions 18 to 310 residues, and this is the first report where we have determined the potential minimal functional boundaries of the GppA protein (Figure 7). It is important to mention here that the GppA protein of A. aeolicus, the crystal structure of which has been reported, is only 312 amino acids long. On the other hand, in Gram-negative bacteria including V. cholerae, GppA is composed of about 500 amino acids (Figure 2B). Our deletion analysis indicated that the Ppx-GppA motif containing the core functional region of V. cholerae GppA is 293 amino acids long, which is very close to that of A. aeolicus GppA complete protein. As mentioned, the presence of four distinct domains (I–IV) in E. coli Ppx and GppA have been reported (Rangarajan et al., 2006; Song et al., 2019). While domains I and II form the active sites of the Ppx and GppA proteins, domain III has similarity with the hydrolase domain of the SpoT enzyme and is probably responsible for the dimer formation of Ppx in E. coli (Rangarajan et al., 2006). On the other hand, domain IV showed structural similarities with the cold shock-associated RNA-binding protein (Rangarajan et al., 2006; Song et al., 2019). Our deletion analysis highlights particularly the importance of the conserved N-terminal domains I and II of the GppA protein of V. cholerae because disruption of these domains but not the C-terminal domains III and IV leads to functional loss of GppA (Figure 7). Thus, it seems that similar functions are being carried out by the domains I and II of V. cholerae GppA protein.
In conclusion, the present study shows that the deletion of V. cholerae gppA gene leads to accumulation of substantial amounts of pppGpp molecules during amino-acid-starved condition. At present, however, it is not clearly known which of the alarmones, ppGpp or pppGpp, is most potent in regulating the different phenotypes of SR in bacteria including V. cholerae. Since ppGpp is synthesized directly from GDP and almost in all bacteria, the level of intracellular ppGpp under nutritional stress conditions is higher than pppGpp, it is likely that ppGpp is the key player of SR in bacteria (Cashel et al., 1996; Potrykus and Cashel, 2008; Sajish et al., 2009; Mechold et al., 2013). In addition, the presence of functional GppA in Gram-negative bacteria further supports the hypothesis that ppGpp is the critical molecule for controlling SR in bacteria. It also appears from this study that the activation of the RelA and GppA enzymes are probably linked with amino acid starvation condition, which may help in maintaining the lower pppGpp level in V. cholerae. Further studies on other Gram-negative bacteria may help in establishing this unique link between RelA and GppA.
Data Availability Statement
Data presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
RKB and BD designed the study. DR and SD performed the experiments. DR, SD, BD, and RKB analyzed the data. DR, BD, and RKB wrote the manuscript. All authors have contributed to the article and approved the submitted version.
Funding
The work was supported by the research grant (MLP118) of Council of Scientific and Industrial Research (CSIR) and also in part by Dept. of Biotechnology (DBT), Govt. of India (Grant No. BT/MB/THSTI/HMC-SFC/2011) to BD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Pratap Koyal and Shibprasad Sharma for their excellent technical assistance in this work. DR is grateful for the research fellowship from the University Grant Commission (UGC), Government of India. SD is grateful for the research fellowship from CSIR.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.564644/full#supplementary-material
Footnotes
- ^ https://imagej.nih.gov
- ^ http://www.technelysium.com.au/chromas.html
- ^ www.jcvi.org
- ^ www.genome.jp/kegg/
- ^ www.ncbi.nlm.nih.gov
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ www.psc.edu/biomed/genedoc
- ^ http://frodo.wi.mit.edu/
- ^ https://blast.ncbi.nlm.nih.gov/blast.cgi
- ^ http://biocyc.org/VCHO/
References
Akiyama, M., Crooke, E., and Kornberg, A. (1993). An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268, 633–639.
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., et al. (1989). Current Protocols in Molecular Biology. New York, NY: John Wiley and Sons.
Basu, P., and Bhadra, R. K. (2019). Post-transcriptional regulation of cholera toxin production in Vibrio cholerae by the stringent response regulator DksA. Microbiology 165, 102–112. doi: 10.1099/mic.0.000743
Basu, P., Pal, R. R., Dasgupta, S., and Bhadra, R. K. (2017). DksA-HapR-RpoS axis regulates haemagglutinin protease production in Vibrio cholerae. Microbiology 163, 900–910. doi: 10.1099/mic.0.000469
Cashel, M., Gentry, D. R., Hernandes, V. J., and Vinella, D. (1996). “The stringent respons,” in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, 2nd Edn, Vol. 2, ed. F. C. Neidhardt (Washington, DC: ASM Press), 1458–1496.
Choi, M. Y., Wang, Y., Wong, L. L., Lu, B. T., Chen, W. Y., Huang, J. D., et al. (2012). The two PPX-GppA homologues from Mycobacterium tuberculosis have distinct biochemical activities. PLoS One 7:e42561. doi: 10.1371/journal.pone.0042561
Choy, H. E. (2000). The study of guanosine 5’-diphosphate 3’-diphosphate-mediated transcription regulation in vitro using a coupled transcription-translation system. J. Biol. Chem. 275, 6783–6789. doi: 10.1074/jbc.275.10.6783
Dar, H. H., Prasad, D., Varshney, G. C., and Chakraborti, P. K. (2011). Secretory nucleoside diphosphate kinases from both intra- and extracellular pathogenic bacteria are functionally indistinguishable. Microbiology 15, 3024–3035. doi: 10.1099/mic.0.049221-0
Das, B., and Bhadra, R. K. (2008). Molecular characterization of Vibriocholerae ΔrelA ΔspoT double mutants. Arch. Microbiol. 189, 227–238.
Das, B., Pal, R. R., Bag, S., and Bhadra, R. K. (2009). Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p) ppGpp synthetase gene. Mol. Microbiol. 72, 380–398. doi: 10.1111/j.1365-2958.2009.06653.x
Dasgupta, S., Basu, P., Pal, R. R., Bag, S., and Bhadra, R. K. (2014). Genetic and mutational characterization of the small alarmone synthetase gene relV of Vibrio cholerae. Microbiology 160, 1855–1866. doi: 10.1099/mic.0.079319-0
Gaca, A. O., Colomer-Winter, C., and Lemos, J. A. (2015). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156. doi: 10.1128/jb.02577-14
Hara, A., and Sy, J. (1983). Guanosine 5’-triphosphate, 3’-diphosphate 5’-phosphohydrolase: purification and substrate specificity. J. Biol. Chem. 258, 1678–1683.
Haralalka, S., Nandi, S., and Bhadra, R. K. (2003). Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J. Bacteriol. 185, 4672–4682. doi: 10.1128/jb.185.16.4672-4682.2003
He, H., Cooper, J. N., Mishra, A., and Raskin, D. M. (2012). Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol. 194, 2962–2972. doi: 10.1128/jb.00014-12
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483.
Keasling, J. D., Bertsch, L., and Kornberg, A. (1993). Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. U.S.A. 90, 7029–7033. doi: 10.1073/pnas.90.15.7029
Khemici, V., Poljak, L., Toesca, I., and Carpousis, A. J. (2005). Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNaseE. Proc. Natl. Acad. Sci. U.S.A. 102, 6913–6918. doi: 10.1073/pnas.0501129102
Koonin, E. V. (1994). Yeast protein controlling inter-organelle communication is related to bacterial phosphatases containing the Hsp 70-type ATP-binding domain. Trends Biochem Sci. 19, 156–157. doi: 10.1016/0968-0004(94)90275-5
Kristensen, O., Laurberg, M., Liljas, A., Kastrup, J. S., and Gajhede, M. (2004). Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family. Biochemistry 43, 8894–8900. doi: 10.1021/bi049083c
Kristensen, O., Ross, B., and Gajhede, M. (2008). Structure of the PPX/GPPA phosphatase from Aquifex aeolicus in complex with the alarmone ppGpp. J. Mol. Biol. 375, 1469–1476. doi: 10.1016/j.jmb.2007.11.073
Kudrin, P., Dzhygyr, I., Ishiguro, K., Beljantseva, J., Maksimova, E., Oliveira, S. R. A., et al. (2018). The ribosomal A-site finger is crucial for binding and activation of the stringent factor RelA. Nucleic Acids Res. 46, 1973–1983. doi: 10.1093/nar/gky023
Kuroda, A., Murphy, H., Cashel, M., and Kornberg, A. (1997). Guanosine tetra-and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272, 21240–21243. doi: 10.1074/jbc.272.34.21240
Kuroda, A., Nomura, K., Ohtomo, R., Kato, J., Ikeda, T., Takiguchi, N., et al. (2001). Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293, 705–708. doi: 10.1126/science.1061315
Malde, A., Gangaiah, D., Chandrashekhar, K., Pina-Mimbela, R., Torrelles, J. B., and Rajashekara, G. (2014). Functional characterization of exopolyphosphatase/guanosine pentaphosphate phosphohydrolase (PPX/GPPA) of Campylobacter jejuni. Virulence 5, 521–533. doi: 10.4161/viru.28311
Mechold, U., Murphy, H., Brown, L., and Cashel, M. (2002). Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184, 2878–2888. doi: 10.1128/jb.184.11.2878-2888.2002
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S., and Cashel, M. (2013). Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189. doi: 10.1093/nar/gkt302
Ogawa, N., Tzeng, C. M., Fraley, C. D., and Kornberg, A. (2000). Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J. Bacteriol. 182, 6687–6693. doi: 10.1128/jb.182.23.6687-6693.2000
Pal, R. R., Bag, S., Dasgupta, S., Das, B., and Bhadra, R. K. (2012). Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J. Bacteriol. 194, 5638–5648. doi: 10.1128/jb.00518-12
Pal, R. R., Das, B., Dasgupta, S., and Bhadra, R. K. (2011). Genetic components of stringent response in Vibrio cholerae. Indian J. Med. Res. 133, 212–217.
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Rangarajan, E. S., Nadeau, G., Li, Y., Wagner, J., Hung, M. N., Schrag, J. D., et al. (2006). The structure of the exopolyphosphatase (PPX) from Escherichia coli O157:H7 suggests a binding mode for long polyphosphate chains. J. Mol. Biol. 359, 1249–1260. doi: 10.1016/j.jmb.2006.04.031
Reizer, J., Reizer, A., Saier, M. H. Jr., Bork, P., and Sander, C. (1993). Exopolyphosphate phosphatase and guanosine pentaphosphate phosphatase belong to the sugar kinase/actin/hsp 70 superfamily. Trends Biochem. Sci. 18, 247–248. doi: 10.1016/0968-0004(93)90172-j
Sajish, M., Kalayil, S., Verma, S. K., Nandicoori, V. K., and Prakash, B. (2009). The significance of EXDD and RXKD motif conservation in Rel proteins. J. Biol. Chem. 284, 9115–9123. doi: 10.1074/jbc.m807187200
Seyfzadeh, M., Keener, J., and Nomura, M. (1993). spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 11004–11008. doi: 10.1073/pnas.90.23.11004
Somerville, C. R., and Ahmed, A. (1979). Mutants of Escherichia coli defective in the degradation of guanosine 5’-triphosphate, 3’-diphosphate (pppGpp). Mol. Gen. Genet. 169, 315–323. doi: 10.1007/bf00382277
Song, H., Dharmasena, M. N., Wang, C., Shaw, G. X., Cherry, S., Tropea, J. E., et al. (2019). Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori. FEBS J. 287, 1865–1885. doi: 10.1111/febs.15120
Stephens, J. C., Artz, S. W., and Ames, B. N. (1975). Guanosine 5’-diphosphate 3’-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino-acid deficiency. Proc. Natl. Acad. Sci. U.S.A. 72, 4389–4393. doi: 10.1073/pnas.72.11.4389
Verma, J., Bag, S., Saha, B., Kumar, P., Ghosh, T. S., Dayal, M., et al. (2019). Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 116, 6226–6231.
Wang, J. D., Sanders, G. M., and Grossman, A. D. (2007). Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128, 865–875. doi: 10.1016/j.cell.2006.12.043
Keywords: Vibrio cholerae, stringent response, (p)ppGpp, GppA, Ppx, amino acid starvation, glucose starvation
Citation: Rakshit D, Dasgupta S, Das B and Bhadra RK (2020) Functional Insights Into the Role of gppA in (p)ppGpp Metabolism of Vibrio cholerae. Front. Microbiol. 11:564644. doi: 10.3389/fmicb.2020.564644
Received: 22 May 2020; Accepted: 08 September 2020;
Published: 29 September 2020.
Edited by:
Ulrike Kappler, The University of Queensland, AustraliaReviewed by:
Mohammad Roghanian, Umeå University, SwedenCheng-Cai Zhang, Chinese Academy of Sciences (CAS), China
Copyright © 2020 Rakshit, Dasgupta, Das and Bhadra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rupak K. Bhadra, cnVwYWtiaGFkcmFAaWljYi5yZXMuaW4=; Bhabatosh Das, YmhhYmF0b3NoQHRoc3RpLnJlcy5pbg==
†Present address: Shreya Dasgupta, Clinical Medicine Division, ICMR-National Institute of Cholera and Enteric Diseases, Kolkata, India
 Dipayan Rakshit
Dipayan Rakshit Shreya Dasgupta
Shreya Dasgupta Bhabatosh Das2*
Bhabatosh Das2*