- 1Microbial Biogeochemistry, Research Area Landscape Functioning, Leibniz Centre for Agricultural Landscape Research (ZALF), Müncheberg, Germany
- 2Institute of Forest Genetics, Johann Heinrich von Thünen Institute, Waldsieversdorf, Germany
- 3Integrative Infection Biology Crops-Livestock, University of Hohenheim, Stuttgart, Germany
The invasive ascomycete Hymenoscyphus fraxineus has been threatening Fraxinus excelsior populations throughout Europe for over two decades. Since the infection and first colonization by the pathogen occurs in leaves, leaf-colonizing microorganisms have been discussed as a barrier and as possible biocontrol agents against the disease. To identify fungal groups with health-supporting potential, we compared the fungal microbiota of compound leaves from susceptible and tolerant ash trees in four ash stands with high H. fraxineus exposure. The fungal communities were analyzed both culture-independently by ITS2 amplicon sequencing and by the taxonomic classification of 1,704 isolates using matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) or sequencing of the entire ITS region. The fungal community structure did not show significant differences depending on the health status. However, for several OTUs and a MALDI group, a significantly higher abundance was found in tolerant ash trees. Thus, the yeast Papiliotrema flavescens was significantly increased and accounted for 12.3% of the mycobiome of tolerant ashes (OTU0003), and it had also a distinctly higher abundance among the isolates. The filamentous ascomycete Sarocladium strictum was increased 24-fold among the isolates of tolerant trees, but its abundance was comparably low. An in vitro screening for the growth inhibition of the pathogen via cocultivation resulted in 28 yeast-like isolates and 79 filamentous fungi with antagonistic activity. A statistical cocultivation test on two H. fraxineus strains confirmed six of the yeast-like isolates that suppressed H. fraxineus significantly, from 39–50%, two of them through a fungicidal effect. The highest inhibition rates among the yeasts were found for three isolates belonging to Aureobasidium pullulans and P. flavescens. The cocultivation test of the filamentous isolates revealed higher effects compared to the yeasts. Four isolates showed significant inhibition of both H. fraxineus strains with a rate of 72–100%, and five further isolates inhibited only one H. fraxineus strain significantly. The most effective isolates were members of the genus Cladosporium. During the next step, in planta tests will be necessary to verify the efficacy of the antagonistic isolates and to assess their suitability as biocontrol agents.
Introduction
In recent decades, ash dieback has spread from north-eastern Poland to all over Europe, and it threatens the future of the common ash (Fraxinus excelsior) because of its associated severe damage and high mortality rates (Kowalski and Holdenrieder, 2009; Timmermann et al., 2011; McKinney et al., 2014; Skovsgaard et al., 2017). This disease is caused by the invasive ascomycete Hymenoscyphus fraxineus (Kowalski, 2006; Queloz et al., 2011; Baral et al., 2014). The fungus was introduced from East Asia, where it lives in asymptomatic association with the Manchurian ash (Fraxinus mandshurica) (Zhao et al., 2013; Gross et al., 2014; Han et al., 2014; Cleary et al., 2016). H. fraxineus infections come from airborne ascospores that germinate on leaves and penetrate the cuticula and the epidermis. Starting from the infected leaves, the pathogen is able to colonize the petioles and subsequently the twigs and shoots. This leads to blockages of the xylem vessels as well as lesions of the cambium and finally the crown starts dying back (Gross et al., 2014). Infection and colonization by the pathogen can be prevented or reduced by genetically mediated defense mechanisms. Thus, several ash species, such as F. mandshurica and Fraxinus ornus, the native flowering ash, tolerate this pathogen based on weak susceptibility (Gross et al., 2014; Kirisits and Schwanda, 2015; Drenkhan et al., 2017). A hereditary resilience seems to also be present within the common ash, which is reflected in the occurrence of individual healthy-looking trees in infested stands (Stener, 2013; Lobo et al., 2014; Wohlmuth et al., 2018). Resistance is considered as a polygenic trait, the specific function of which is still poorly understood (Wohlmuth et al., 2018; Stocks et al., 2019). However, it is assumed that the resilience of the host is significantly influenced by its microbiome (Berg et al., 2017; Compant et al., 2019; Liu et al., 2020). Bacteria and fungi that colonize the surface and the endosphere of the plant can suppress invaders directly by resource competition, parasitism and antibiosis or indirectly by activating the plant’s own defense system (Latz et al., 2018; Fadiji and Babalola, 2020). In addition, some endophytic microorganisms are able to promote plant growth and development, resulting in an increased resilience in response to stress and pathogens (Glick, 2012; Hossain et al., 2017). These different mechanisms, which can occur simultaneously, are activated in interaction with the plant’s metabolism and the environmental conditions (Berg and Smalla, 2009; Berg et al., 2015).
To assess the role of fungal endophytes in ash dieback tolerance, the structure of leaf- and twig-associated fungal communities of tolerant and diseased ashes was investigated over a series of studies (Cross et al., 2017; Hanackova et al., 2017a; Schlegel et al., 2018; Kosawang et al., 2019). Different approaches showed significant variations in the community structure depending on the plant location, season and plant genotype. In particular, the composition of twig-associated fungal communities differed significantly between ash species, i.e., Fraxinus angustifolia, Fraxinus latifolia, F. mandshurica, and F. ornus but also between genotypes of F. excelsior. However, a direct and distinctive relation to ash dieback susceptibility could not be detected (Kosawang et al., 2019; Griffiths et al., 2020). Griffiths et al. (2020) observed that higher H. fraxineus infection levels were associated with higher fungal diversity and the presence of specific fungal genera.
The complex interactions between the resistance, plant genotype and associated microorganisms make it challenging to identify specific microbial groups with disease-suppressing potential. It is assumed that health-supporting taxa should primarily be present in tolerant hosts (Schulz et al., 2019). However, only a few species have been identified so far, which are clearly associated with H. fraxineus tolerance. Cleary et al. (2016) analyzed the phyllosphere fungal communities of the dieback-tolerant ash F. mandshurica and identified Mycosphaerella as the dominant fungal genus in these trees. Mycosphaerella was found in all the F. mandshurica samples collected in eastern Russia and was limited to this ash species. A study on the leaf petioles of common ashes showed increased frequencies of Aureobasidium pullulans in healthy trees, while in diseased trees, Alternaria and Cytospora species were also frequently represented (Davydenko et al., 2013). Other investigations focused on the in vitro testing of ash leaf endophytes for antagonism against H. fraxineus. A row of fungal isolates obtained from tolerant ash species as well as F. excelsior indicated antagonistic activity in cocultivation with the pathogen (Hanackova et al., 2017a; Kosawang et al., 2018; Halecker et al., 2020). However, cocultivation experiments also showed that both partners, H. fraxineus and the endophytic isolate, are often inhibited by mutual antagonisms (Schulz et al., 2015; Halecker et al., 2020). Therefore, comprehensive tests are required to identify effective antagonistic candidates whose colonization and activities are not or only weakly suppressed by H. fraxineus.
In this study, we combined a high-throughput sequencing approach and cultivation methods to analyze the fungal communities of susceptible and tolerant common ashes. The comparative analyses were aimed at detecting the fungal groups that are specific to tolerant trees and might therefore be involved in disease suppression. In parallel, the isolates were subjected to an extensive screening for H. fraxineus growth inhibition. Based on the primary screening, the best isolates were evaluated in a statistical cocultivation test for their antagonism against the pathogen.
Materials and Methods
Study Site and Sampling
Compound ash leaf material was obtained by sampling of ash trees from four forest districts in Northeast Germany in July of 2017. Observations taken during long-term monitoring projects revealed a very low occurrence of single trees with low susceptibility in stands that were severely affected by H. fraxineus (Sündermann and Jütte, 2014). In each of the districts (A–D), four pairs of adjacent trees consisting of an affected (K) and a tolerant (P) tree were chosen for sampling. Details of the sampling campaign and sample preparation are described in Ulrich et al. (2020). Accordingly, the same leaf samples were used in this study to analyze the fungal community.
Amplicon Sequencing and Analysis of the ITS2 Region
For high comparability among analyses, the total DNA used to investigate of the bacterial community (Ulrich et al., 2020) was also applied to study the fungal community. For this purpose, the ITS2 region was chosen because it has a lower length variability compared to the ITS1 region combined with higher genetic diversity. It also reduces the pitfalls caused by introns in the flanking rDNA of the ITS1 region of some species (Tedersoo et al., 2015). The ITS2 region was amplified with the primers fITS7 (5′-GTGARTCATCGAATCTTTG-3′) (Ihrmark et al., 2012) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990), preventing the amplification of the host plant DNA. PCR amplification was conducted as described by Ihrmark et al. (2012). The primers were extended with a heterogeneity spacer and a barcode sequence. The amplicons were checked with gel electrophoresis, purified with an MSB Spin PCRapace kit (Invitek, Essen, Germany) and mixed to equimolar DNA concentrations. The library preparation and Illumina MiSeq 300-bp paired-end sequencing were performed at LGC Genomics Berlin.
The raw sequence data were analyzed with Mothur v. 1.39.1 (Schloss et al., 2009; RRID:SCR_011947) in accordance with the MiSeq SOP (Kozich et al., 2013). In addition, the ITS2 region was extracted with ITSx 1.1.2 (Bengtsson-Palme et al., 2013) by removing the flanking 5.8S and 28S rRNA gene fragments. Chimeras were removed using the Uchime algorithm. For phylogenetic identification, the sequences were compared to the dynamic UNITE database (version 8.2) using a confidence threshold of 80%. The operational taxonomic units (OTUs) were generated with the VSEARCH algorithm without using a multiple sequence alignment (Edgar, 2010) at a similarity of 97%. Singletons were discarded as suggested in the UPARSE pipeline (Edgar, 2013). The phylogenetic assignment of the OTUs was checked manually using BLAST searches of the UNITE database as well as the NCBI taxonomy browser. All the samples were subsampled to the minimum number of sequences among all the samples. The paired sequence reads were deposited in the public Sequence Read Archive (SRA, RRID:SCR_004891) repository under the accession number PRJNA611938.
Statistical analyses were performed using the phyloseq, vegan and ape packages in R 3.6.0, as well as MicrobiomeAnalyst (Chong et al., 2020; RRID:SCR_015022). The alpha diversity indices Chao 1, Shannon, and Simpson were calculated. Significant differences between the fungal communities were tested using an Analysis of Similarity (ANOSIM). The phylogenetic composition of the ash microbiome was visualized with a Krona chart (Ondov et al., 2011). The differential abundance at different taxonomic levels was analyzed with the metagenomeSeq tool using a false discovery rate (FDR) for multiple test correction (Paulson et al., 2013). An indicator species analysis was performed with PC-Ord v. 7.02 (McCune and Mefford, 2015) by applying the procedure by Dufrêne and Legendre (1997).
Cultivation of Fungal Epi/Endophytes
Ground material of compound ash leaves were serially diluted in 0.8% NaCl, plated on PDA (Merck, Darmstadt) and incubated for seven to ten days at room temperature. To determine the population densities, colony forming units (CFU) were counted and expressed as CFU per gram of fresh weight. Approximately 50 isolates per sample were randomly selected for analysis. From the filamentous fungi, the mycelium was subcultivated on PDA and pure cultures were scraped from the Petri dishes using a sterile scalpel and transferred to agar slants (PDA) for storage and further processing. Yeast-like fungi were purified by streak plate method and stored as cryo-cultures (−80°C) in nutrient broth containing 40% glycerol until analysis.
Dereplication and Classification of Fungal Isolates
Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) was applied as a reliable method for the dereplication and classification of yeasts in environmental analyses (Chudejova et al., 2017; Huschek and Witzel, 2019). Prior to this measurement, the yeast-like isolates were freshly inoculated on CASO agar (Fluka, Buchs, Switzerland) and cultivated for 24 h at 25°C. Mass spectra were obtained based on the whole-cell measurement protocol as described by Ulrich et al. (2020). The analysis was performed on a microflexTM LT/SH MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) using Flex Control 3.4 software. Isolates showing spectra with a score value >2.3 (highly probable species identification) were considered as identified by the Bruker database. Isolates with a lower score were compared with each other and grouped on the basis of a score value >2.3.
Representative strains of the unidentified groups were taxonomically classified by sequencing of the complete fungal ITS rRNA region. Total DNA was extracted from single colonies by resuspending in 20 μl of 25 mM NaOH/0.25% SDS followed by incubation for 15 min at 95°C. The amplification was performed using the universal primer pair ITS1F (Gardes and Bruns, 1993) and ITS4 (White et al., 1990). PCRs were performed using PCR Master Mix 2× (Thermo Fisher Scientific, Darmstadt, Germany) containing 0.2 μM of each primer. The thermal cycling conditions were as follows: an initial denaturation step of 95°C for 5 min; 10 cycles of 95°C for 30 s, 60°C for 30 s with a touchdown of −1°C by cycle and 72°C for 1 min, 30 cycles of 95°C for 30 s, 50°C for 30 s and 72°C for 1 min. The PCR products were checked on a 1% agarose gel and the fragments of approximately 600–700 bp were sequenced using the primer ITS1F. The fungal isolates were taxonomically assigned using the UNITE fungal ITS database version 8.0 (Kõljalg et al., 2013) and the species names were approved by the NCBI taxonomy browser. After supplementing the MALDI BiotyperTM database with the spectra of the reference strains, a reliable taxonomic identification of all the fungal isolates was achieved. All the MALDI groups were assigned at the species level.
For the classification of filamentous fungi, small pieces of mycelia (ca. 0.5 cm2) were scraped from agar cultures using a sterile scalpel. To extract the DNA, a modified CTAB protocol was used according to Doyle (1990). In brief, for the first lysis step, the mycelium was frozen at −80°C in 2 ml tubes containing 8–10 ceramic beads (1.4 mm; Omni International, Kennesaw, United States) and subsequently ground for 25 s with a speed of 7.45 m/s using the Bead Ruptor 24 (Omni International). After the addition of 750 μL of CTAB buffer and sample incubation of the samples for 30 min at 65°C, the procedure followed the regular protocol. Filamentous fungal isolates were identified by sequencing the entire ITS region of ribosomal DNA as described above and assigned at the species level as well.
A community analysis of culturable fungi was performed by combining all the isolates classified by either MALDI-TOF MS or direct sequencing. The B2P sample was removed from the analysis as an outlier due to the small number of only seven isolates. For the statistical analysis, the number of isolates per sample was normalized using the CSS method (Paulson et al., 2013). Significant differences in the community composition between tolerant and susceptible trees were tested at all taxonomic levels using the MetagenomeSeq tool (MicrobiomeAnalyst, PFDR < 0.05).
In vitro Screening and Statistical Test for Antagonistic Activity Against H. fraxineus
All the fungal isolates were screened for antagonistic activity against H. fraxineus P3. To simulate the conditions of their natural habitat, cultivation was performed on PDA medium enriched with ash shoot extract (30 g ash leaves/L PDA) at 22°C (Hanackova et al., 2017a). Due to its slow growth, H. fraxineus P3 was incubated for 5 to 6 days before the isolates for testing were struck out. Depending on the growth type of the isolates (yeasts or filamentous fungi), two different approaches were used:
The antagonistic potential of yeast-like fungi was estimated using an assay according to Pane and Zaccardelli (2015). An agar plug (ø 5 mm) containing fresh H. fraxineus mycelium was placed in the center of the Petri dish (ø 8 cm), whereas four different yeast counterparts were spread around the four edges of the plate. Plates, solely carrying H. fraxineus served as control. The colony radius (r) of H. fraxineus was measured after seven and 14 days until the control reached the edge of the plate. The inhibitory effect was estimated using the formula percent growth inhibition = 100 × [(rcontrol – rcocultivation)/rcontrol]. Isolates with inhibition rates >30% were chosen for a subsequent statistical test including three replicates and two H. fraxineus strains (P3, HF23). In this approach, only one isolate per plate was spread out along the edges of the plate, and the inhibition rates were calculated by measuring the diameter of the H. fraxineus mycelium.
The filamentous fungi were tested in dual cultures with agar plugs (ø 5 mm) of the fungal isolates and H. fraxineus mycelium at a distance of 4 cm. The test included a self-inhibition control by pairing two H. fraxineus colonies of the same strain and a negative control by pairing H. fraxineus with an agar plug without mycelium. Similar to Hanackova et al. (2017a), the growth of H. fraxineus was measured as the colony radius on a connective line between the two colonies as well as on a line measuring 45° up and down from this line after seven and 14 days of dual culture. The inhibition of H. fraxineus was estimated after averaging the three measured values and using the formula mentioned above. Isolates that indicated higher inhibition rates compared to the self-inhibition were evaluated again in a statistical test, as outlined above.
In addition, the statistical tests on the yeasts and filamentous fungi were ultimately used to evaluate the vitality of the residual H. fraxineus mycelium after confrontation with the fungal isolates. For this purpose, agar plugs with the remaining H. fraxineus mycelium (P3, HF23) were picked from the three replicates and incubated separately on PDA with ash shoot extract. The growth of the mycelium was assessed after 2 weeks in comparison to the untreated control.
To verify the pathogenicity of the H. fraxineus isolates P3 and HF23, 2-year-old ash seedlings were inoculated with either isolate using an approach modified according to Przybyl (2003). The typical symptoms of ash dieback (Gross et al., 2014) were monitored over the course of 8 weeks, and the virulence of both isolates was confirmed.
Results
Fungal Community Structure of Susceptible and Tolerant Ashes
ITS2-Based Community Profiling
The mycobiomes of 16 tolerant and 16 susceptible trees were studied by amplicon sequencing of the ITS2 rRNA region. Although only visibly healthy leaves and petioles were sampled from both the tolerant and the diseased trees, an extremely high proportion of H. fraxineus (86%) was found in the fungal community of one sample (C2K). Due to this contamination, this sample was excluded from further analysis. All the other samples contained a low proportion of the pathogen H. fraxineus with an average of 0.03% of the fungal community. In total, the sequences could be assigned to 1012 operational taxonomic units (OTUs) with an average of 210 OTUs per sample. For the statistical analyses, the sequences were rarefied to 10,707 reads per sample.
The comparison of fungal communities from tolerant and susceptible trees showed no significant differences in the genetic diversity (e.g., with a mean Shannon index of 2.72 ± 0.23 vs. 2.65 ± 0.44). An analysis of the composition of the mycobiome revealed small but significant differences in grouping by sample location (forest districts) and health status (ANOSIM R = 0.1898, P < 0.001). Figure 1 displays the differences in the fungal community structure. The second axis indicated some deviations between tolerant and susceptible ashes for forest districts A and C. However, the pairwise comparisons of the health status did not show significant differences for any of the districts.
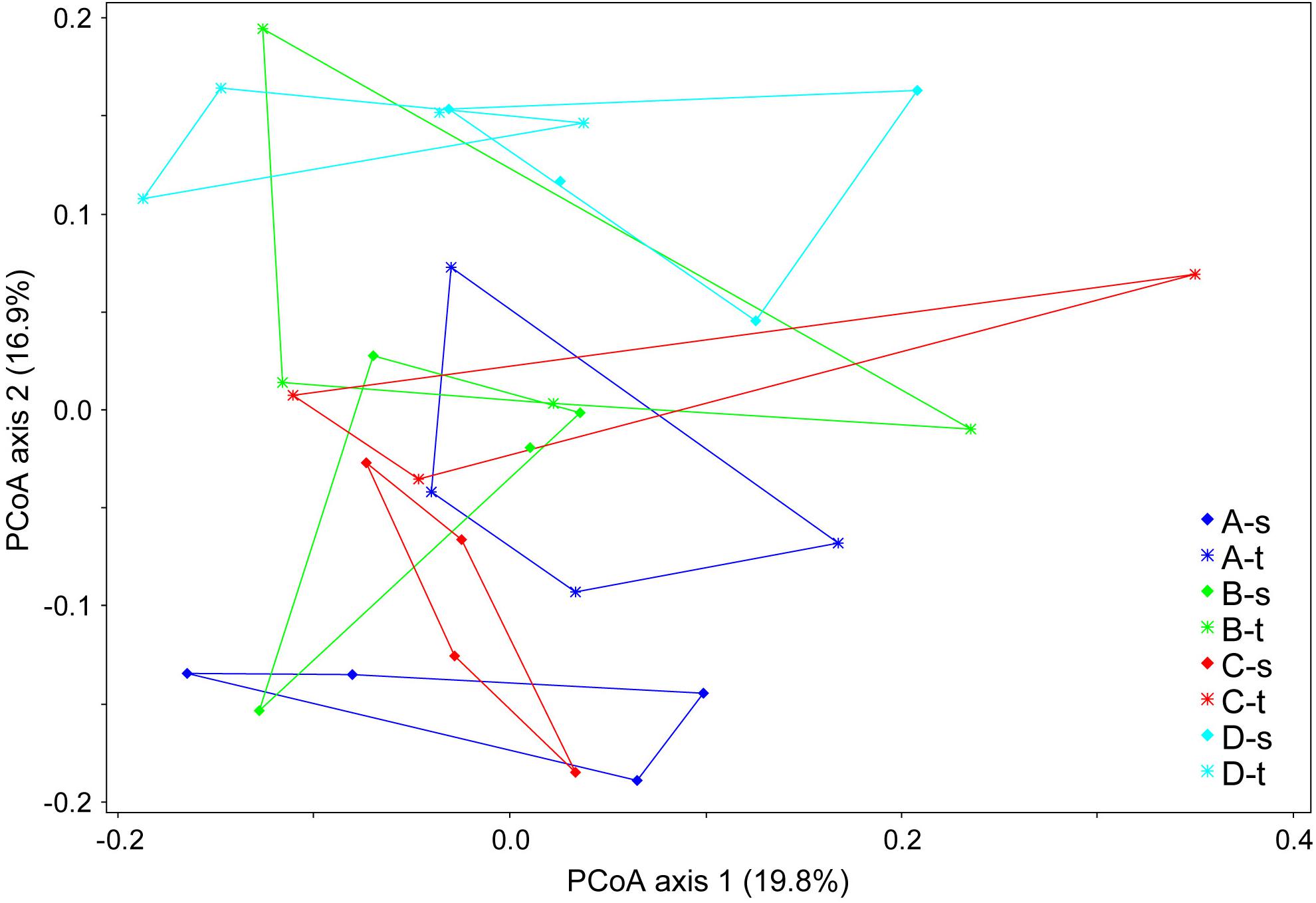
Figure 1. Community clustering of the mycobiome of tolerant (t) and susceptible (s) ashes from the four forest districts (A–D). A principle coordinate analysis (PCoA) was applied based on a Bray-Curtis distance matrix.
In total, 61% of the sequences were assigned to the phylum Ascomycota and 38% to the phylum Basidiomycota, and only 0.46% of the fungal sequences could not be classified (Figure 2). At the class level, Dothideomycetes dominated the fungal community with 54%, followed by Tremellomycetes with 28%. At the species level, A. pullulans showed the highest abundance with 32%, followed by Cladosporium spp. (11%), Papiliotrema flavescens (10%) and Vishniacozyma carnescens (9%). Thus, four species represented more than 60% of the fungal community.
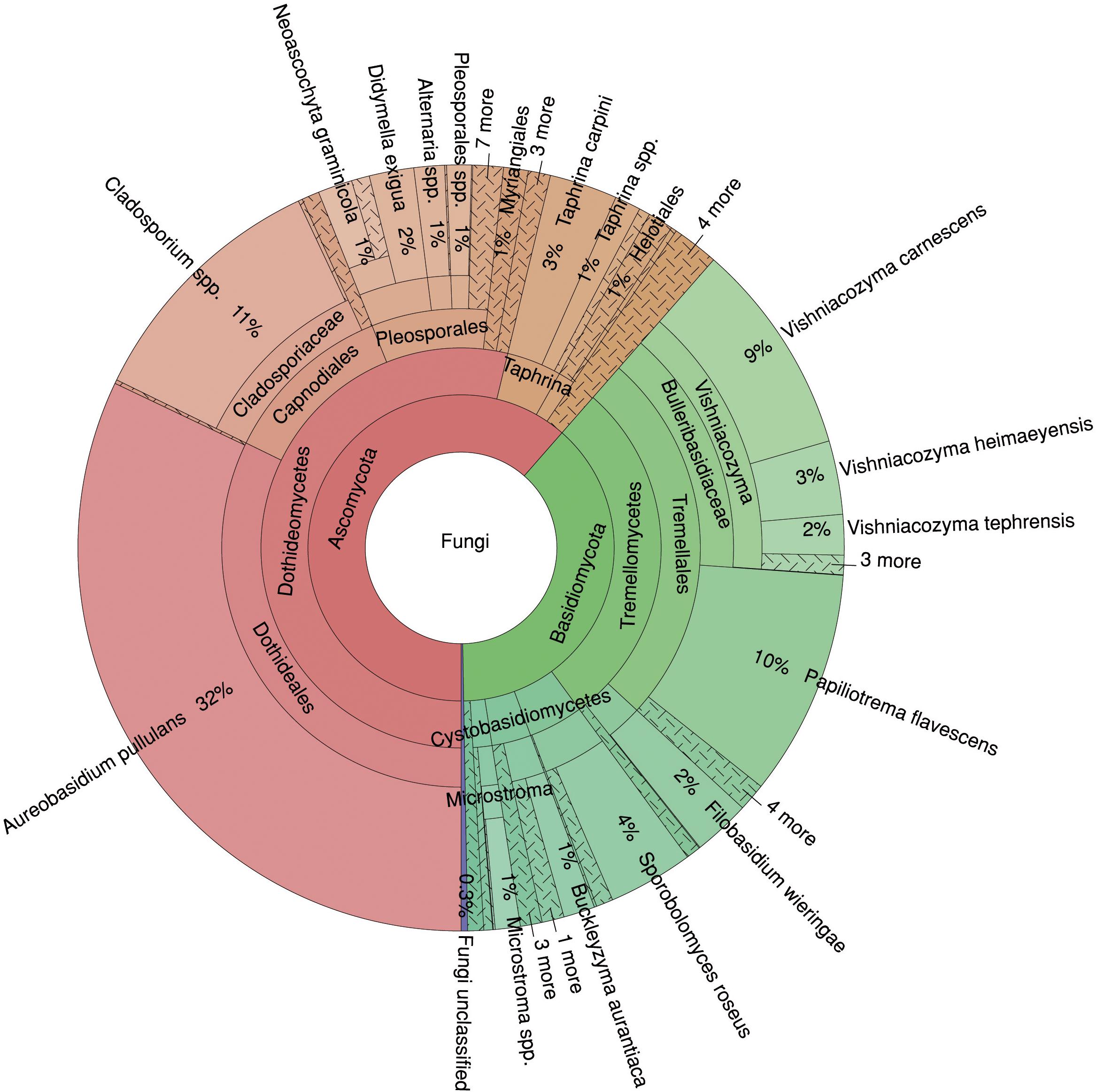
Figure 2. Relative proportions of taxa in the mycobiome of Fraxinus excelsior. The Krona diagram shows six taxonomic levels from phylum to species.
When comparing the mycobiome at the species level, only the yeast P. flavescens was significantly increased in tolerant trees (approximately 2-fold). At the OTU level, the differential abundance analysis revealed 18 OTUs with a proportion of more than 0.02% that were significantly increased in tolerant trees (Figure 3). Remarkably, a group identified as P. flavescens (OTU0003) accounted for 12.3% of the fungal community in tolerant ashes. Additional OTUs with higher amounts in tolerant plants (e.g., OTU0014, 0026, and 0036) belonged to Microstroma, uncl. Dothidiomycetes and Neosetophoma. However, the abundances of these taxa were comparably low.
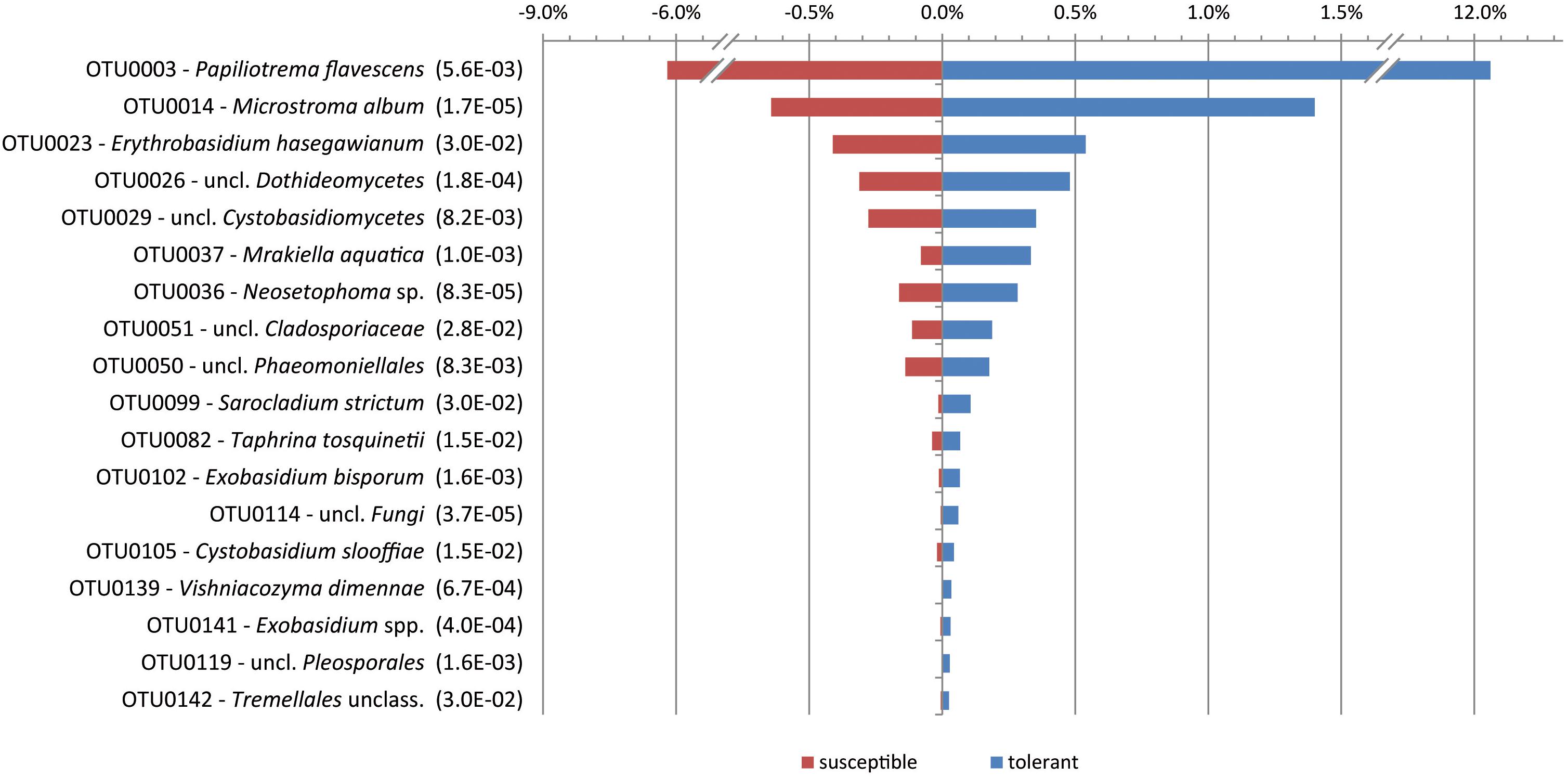
Figure 3. Significantly increased OTUs in tolerant ashes. The OTUs are sorted according to their relative abundance (>0.02), and the level of significance (FDR) is given in brackets.
An indicator species analysis at the OTU level revealed four groups for the tolerant ash trees. It again concerns P. flavescens (OTU0003), but Microstroma album (OTU0014), Vishniacozyma dimennae (OTU0139) and Exobasidium spp. (OTU0141) could also be proven (p < 0.05).
Culturable Fungal Communities
In addition to the marker gene analysis, the culturable leaf-associated fungal community was compared between tolerant and susceptible ash trees. The population densities of the fungi ranged between 1 × 104 and 4 × 105 CFU/g fresh weight with no significant differences depending on the health status. Previous studies showed that the leaf fungal density is very changeable with yeasts ranging between 10 and 1010 and filamentous fungi between 102 and 108 CFU/g (Thompson et al., 1993; Inacio et al., 2002). In total, 931 filamentous isolates were classified via ITS rRNA sequencing and 773 yeast-like isolates via MALDI-TOF MS. The 1,704 isolates were grouped at the species level into 49 phylotypes belonging to 42 genera.
The majority of fungal isolates was assigned to the phylum Ascomycota (72%) followed by Basidiomycota with 28%. At the class level, Dothideomycetes (63%), and Tremellomycetes (28%) dominated the community. At the species level, a clear dominance of the saprophytic yeasts A. pullulans (50.5%), P. flavescens (14.1%), and V. carnescens (9%) was observed. The most abundant species A. pullulans, which is known for its switch between yeast-like and filamentous growth mode, accounted for 28% of the yeasts and 69% of the filamentous fungi. Thus, the filamentous fungi were clearly dominated by A. pullulans, whereas P. flavescens was the predominant yeast (31%).
When comparing trees with different health statuses, P. flavescens occurred in higher abundance in tolerant trees (Figure 4). However, this difference could not be statistically ensured. Sarocladium strictum isolates made up a lower proportion but showed a significant difference with a 24-fold increase in tolerant trees.
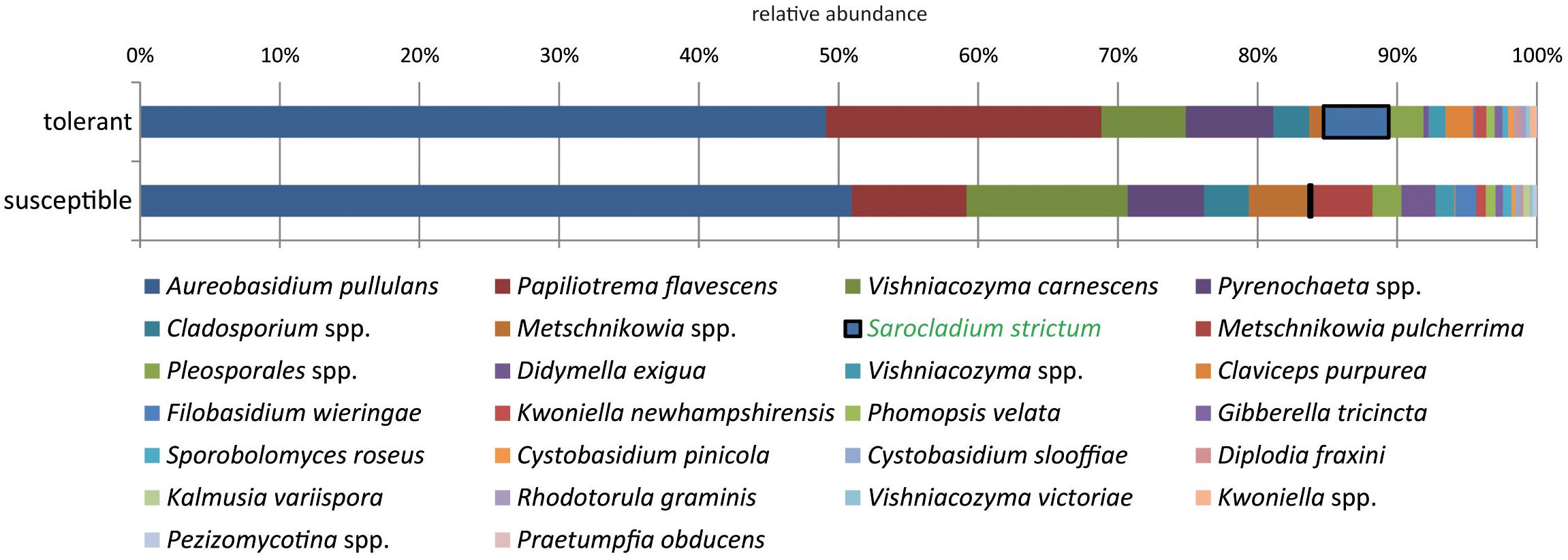
Figure 4. Comparison of the culturable fungal community from tolerant and susceptible ashes at the species level. The species significantly increased in resistant trees is highlighted.
In vitro Antagonistic Activity of Fungal Isolates
Yeast-Like Fungi
To detect potential antagonists, the yeast isolates were screened for growth inhibition of H. fraxineus P3. Overall, the screening revealed a high proportion of isolates with slight inhibitory effects. One-fifth of the isolates inhibited H. fraxineus by 31–50%. The potential antagonists were obtained in a comparable ratio from susceptible and tolerant trees. They were dominated by A. pullulans (56%), followed by the Basidiomycota members P. flavescens (23%) and Vishniacozyma spp. (13%). Other taxa such as Metschnikowia sp., Cystobasidium pinicola and Kwoniella newhampshirensis were represented in frequencies between 5% and less than 1%. The highest inhibition rates were achieved by A. pullulans isolates with 37% on average, while the mean inhibition of the other taxa was slightly lower with 32–34%.
Based on the primary screening, 28 isolates with an inhibition rate above 30% were subjected to the statistical test on antagonism against two H. fraxineus isolates. The isolates were derived from all the locations and from both susceptible and tolerant trees. An overview on the antagonistic potential of the yeasts is given in Table 1. In total, six of the 28 isolates significantly inhibited the growth of H. fraxineus HF23 at 39–50%. The H. fraxineus strain P3 was inhibited by these yeasts over a similar range, but this finding could not be statistically ensured. After cocultivation, the vitality of the remaining H. fraxineus mycelium was also examined. Ten out of the 28 isolates were able to completely kill H. fraxineus P3, whereas the vitality of HF23 was only slightly injured. Remarkably, four of the nine isolates of P. flavescens, a species that was found to have increased in tolerant trees, were able to devitalize H. fraxineus P3. When summarizing all the antagonistic features, the best isolates were A3P046, A3K040, and A3P071. D3K042 and C4K010 showed a strong fungistatic effect during the cocultivation test indicated by the ability of the pathogen to regrow. The antagonistic activity is exemplarily shown for isolates A3K040 and A3P071 in Figure 5. In addition to the growth inhibition, a lightening of the mycelium was visible, indicating an ongoing lysis that started approximately 10 days after cocultivation. This effect was differently pronounced in both isolates.
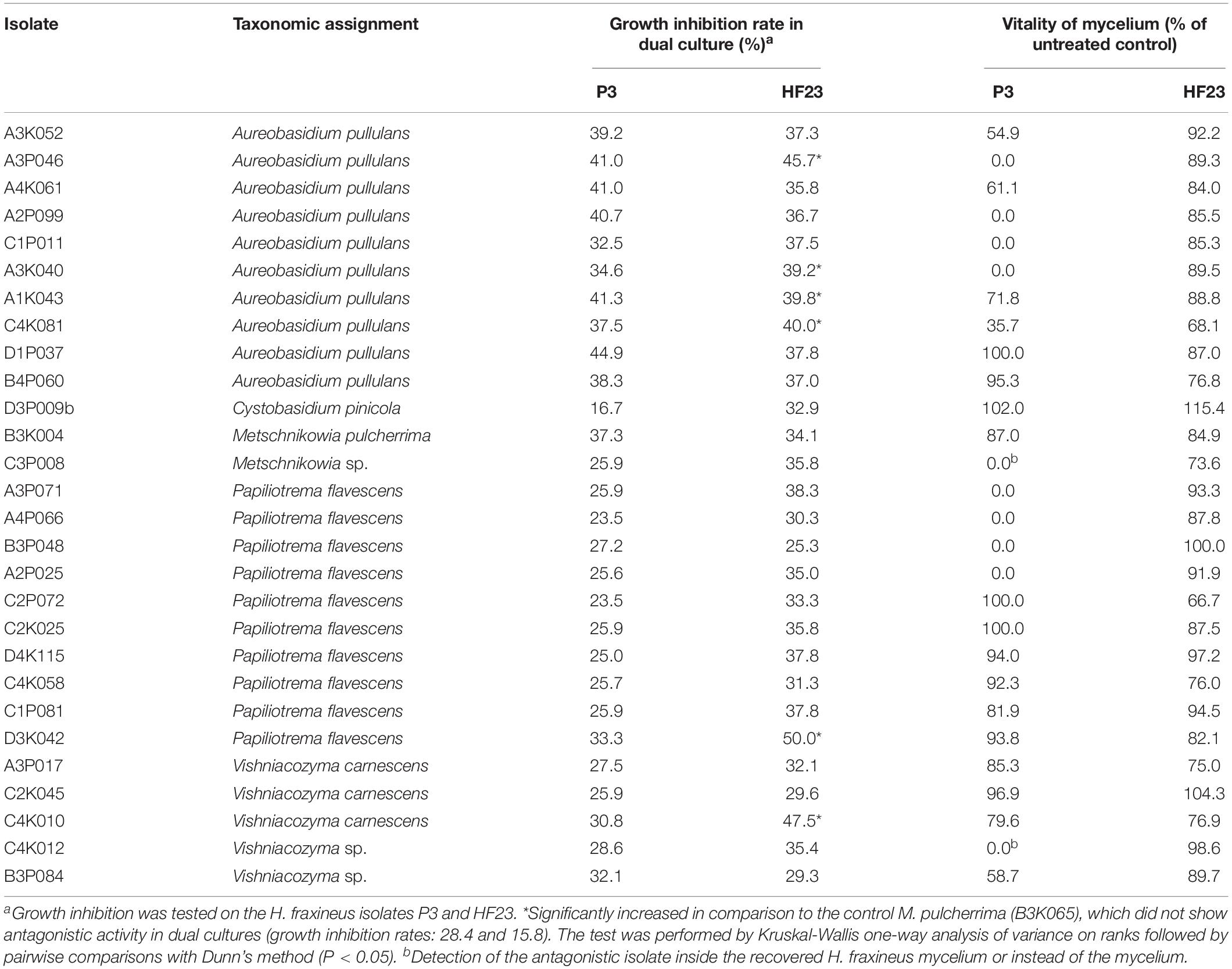
Table 1. Antagonistic activity of yeast-like epi/endophytic fungi as assessed by the growth inhibition of H. fraxineus and the vitality of the remaining pathogen mycelium.
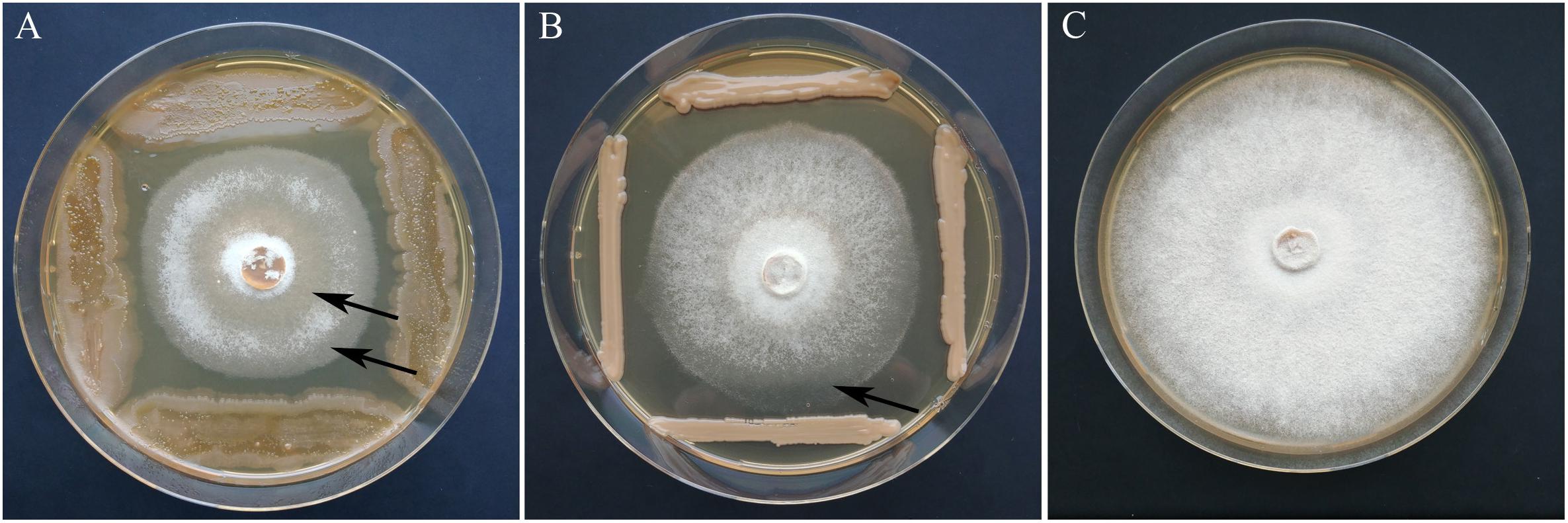
Figure 5. Growth inhibition of H. fraxineus P3 after cocultivation with A. pullulans A3K040 (A) and P. flavescens A3P071 (B) in comparison to the control (C). The clearing of the H. fraxineus mycelium which indicates an ongoing lysis of the pathogen is marked by arrows.
Filamentous Fungi
Due to the high proportion of A. pullulans among the filamentous fungi, more than half of the fungi tested here belonged to this species. To avoid testing clones of the same strain, only a selection of A. pullulans strains was tested per sample. Accordingly, the screening of filamentous fungi included 695 isolates. As a result, 79 isolates (11.4% of the total isolates) inhibited the pathogen growth by at least 43%, i.e., higher than the mean self-inhibition rate of H. fraxineus P3. The highest values were determined for the Cladosporium isolates C1K002 and A3K053 with inhibition rates of 70 and 60%. The positively screened isolates obtained in comparable proportions from tolerant and susceptible trees represented members of Aureobasidium (40%), Pyrenochaeta (16%), Cladosporium (10%), Gibberella (10%), and Pleosporales (8%) as well as Diplodia, Didymella, Sarocladium, and Phomopsis with lower occurrence. The inhibition rates per group varied between 41 and 55%.
Most likely due to the different test procedures, the filamentous isolates inhibited H. fraxineus P3 by 48%, whereas the mean inhibition rate of yeast-like isolates was 37%. Otherwise, not only the procedure but also the morphological status of the fungal isolate could be of importance for the antagonistic properties of fungi.
Out of the 79 antagonistic isolates, 37 were derived from different samples and taxonomic groups. To exclude clones from the subsequent statistical test, only these isolates were selected. As shown in Table 2, four isolates suppressed either H. fraxineus strain significantly with inhibition rates from 72 to 100%. The inhibitory effects were distinctly above the level of self-inhibition for H. fraxineus. Five further isolates inhibited one of the H. fraxineus strains significantly with rates between 57 and 93%.
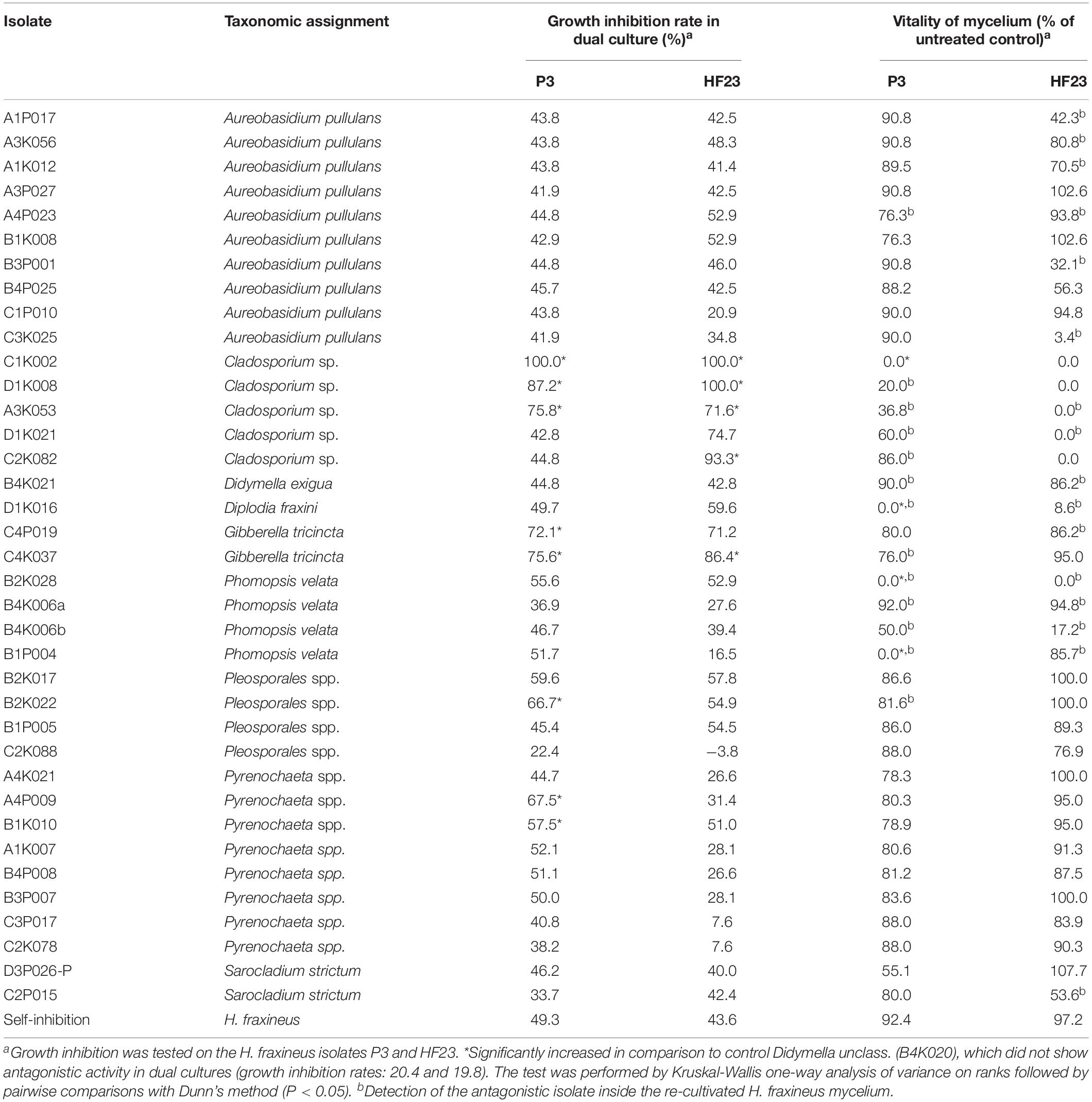
Table 2. Antagonistic activity of filamentous epi/endophytic fungi as assessed by the growth inhibition of H. fraxineus and a vitality test of the remaining pathogen mycelium.
The most effective isolates C1K002 and D1K008, which inhibited the pathogen almost completely, were members of the genus Cladosporium. Both isolates showed a rapid growth by strong sporulation and were already able to suppress the H. fraxineus strains during the first week of cocultivation. Accordingly, the growth of H. fraxineus was affected even in its initial phase. After 14 days, no or very few mycelia from H. fraxineus had grown (Figure 6 and Table 2). A third Cladosporium isolate (A3K053) showed an almost comparable antagonistic activity, with inhibition rates of 72 to 76%. Additionally, B2K028 and D1K016 were able to kill the pathogen more or less completely. This killing was achieved by the ingrowing of the antagonistic fungus into the H. fraxineus colony. Consequently, the antagonistic isolate was predominantly detected in the re-cultivation approach. The fourth isolate with clear significant effects, C4K037, was identified as Gibberella tricincta. Like other strains of this species, this isolate also tended to invade the H. fraxineus colony, which partly led to its occurrence in the re-cultivated mycelium of the pathogen (Table 2). In general, fungicidal effects that became visible during the re-cultivation approach have been observed within several taxonomic groups. These effects particularly concern isolates of fast-growing species: for example, Diplodia fraxini, Cladosporium sp., Phomopsis velata, and to a lesser extent, A. pullulans, and S. strictum.
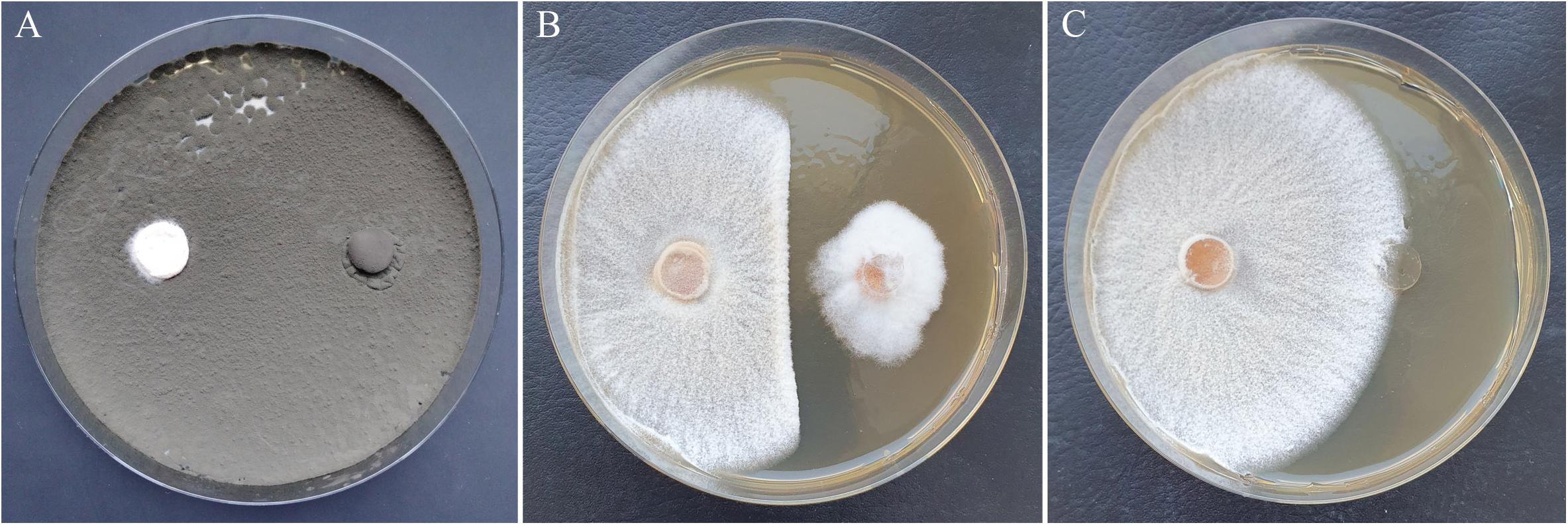
Figure 6. Growth inhibition of H. fraxineus HF23 after cocultivation with Cladosporium sp. C1K002 (A) and Sarocladium strictum D3P026-P (B) in comparison to the H. fraxineus control (C). Left plate side: H. fraxineus, right plate side: agar plug with antagonist (A, B) or without mycelium (C).
Discussion
To identify fungal taxa associated with tolerance to H. fraxineus, we analyzed the fungal communities of ash leaves by ITS2 amplicon sequencing and culturing. Leaf-colonizing fungi are considered as a barrier to invaders such as H. fraxineus that spread starting from leaf infections (Cross et al., 2017; Hanackova et al., 2017b). The indigenous fungi fight against the invading pathogen directly by resource and space competition, partly supported by antibiosis, parasitism or indirectly by the induction of plant resistance (Busby et al., 2016; Jia et al., 2020).
Our study was based on the assumption that both epi- and endophytic fungi interact with the pathogen in a similar way. During and directly after the invasion and penetration of epidermal tissue, the pathogenic fungus is in contact with both habitats, the phyllosphere and endosphere of the ash leaves. Aside from that, several indigenous microorganisms can switch between the two habitats (Hardoim et al., 2015; van Overbeek and Saikkonen, 2016). Accordingly, both habitats were sampled in a combined approach.
Culture-independent as well as culturing approaches showed a strong presence of saprophytic yeasts, especially within the phyllosphere communities (Slavikova et al., 2007; Davydenko et al., 2013; Hanackova et al., 2017a; Kemler et al., 2017). Yeasts thrive on plant exudates, whereas plants can benefit from the secondary metabolites produced by yeast, e.g., auxins, chelates, and glycolipids (Fonseca and Inacio, 2006; Kemler et al., 2017). In addition, several yeasts possess antifungal activities based on resource competition, antibiosis, biofilm formation or the production of volatile organic compounds (Liu et al., 2013; Freimoser et al., 2019). Therefore, an intensive colonization of leaves with yeasts can contribute to higher plant fitness and pathogen resistance. While yeasts are very common in the phyllosphere, the endophytic community is dominated by filamentous fungi (Prior et al., 2017). In our study, the fungal community was clearly dominated by the yeast-like fungus A. pullulans. This species is a very abundant colonizer of plant surfaces but was also often isolated as an endophyte from trees (Albrectsen et al., 2010; Martin-Garcia et al., 2012), including common ash (Slavikova et al., 2007; Davydenko et al., 2013; Hanackova et al., 2017a). As a pleiomorphic fungus, it is known to switch between yeast and mycelial forms in response to its environmental conditions (Ramos and García Acha, 1975; Campbell et al., 2004). Among the next-most abundant fungi, P. flavescens and V. carnescens belong to yeasts, while Pyrenochaeta spp. and Cladosporium spp. have filamentous growth habits.
The fungal communities of tolerant and susceptible F. excelsior trees showed a similar genetic diversity and community structure. Nevertheless, the differential abundance analysis and the indicator species analysis revealed several OTUs as well as an isolate group with significant relations to the health status of the trees. In addition to the predominant P. flavescens OTU0003, several less abundant OTUs and the isolate group S. strictum could be detected. Strains from the genus Papiliotrema are known for their protective effect against fungal disease-related pre- and postharvest losses of different crops. Especially isolates of P. flavescens (formerly Cryptococcus flavescens) are widely used as biocontrol agents against head blight on cereals caused by Fusarium graminearum (Dunlap and Schisler, 2010; McSpadden Gardener et al., 2014; Schisler et al., 2014). Similarly, the postharvest infestation of different fruits with common pathogens such as Botrytis cinerea, Alternaria alternata, and Penicillium expansum were shown to be reduced effectively by P. flavescens and P. laurentii strains (Qin and Tian, 2004; Zhang et al., 2007; Kheireddine et al., 2018). Various Sarocladium strains are discussed as potential antagonists due to the formation of antifungal substances such as antibiotics and hydrolytic enzymes (Wicklow et al., 2005; Guimarães et al., 2017). In addition, S. strictum has been shown to have disease-reducing effects through mycoparasitism, e.g., on Botrytis cinerea and Helminthosporium solani (Rivera et al., 2007; Choi et al., 2008). However, the use of S. strictum as a biocontrol agent might be limited due to the putative human pathogenic potential of this species (Schell and Perfect, 1996; Sharma et al., 2013). Consequently, the comprehensive analysis by applying a marker gene study and a culturing approach resulted in two distinct fungal groups which are specific to tolerant ash trees and might be associated with the ability of the trees to resist the pathogen.
As a result of the cocultivation test on antagonism, several yeast isolates were found to possess an antagonistic capacity by a direct growth reduction of H. fraxineus (of up to 50%) and/or an ongoing mycelium lysis, resulting in the complete killing of one of the pathogen strains. Cell-lytic effects from the secretion of exoenzymes have been described for several antagonistic yeasts (Bar-Shimon et al., 2004; Zhang et al., 2011, 2012). The lysis of fungal cell walls usually involves various synergistically acting enzymes, including glucanases, chitinases and proteases (Salazar and Asenjo, 2007). In particular, numerous strains of A. pullulans harbor a broad spectrum of lytic enzymes. This enzymatic activity as well as the production of other antimicrobial compounds make A. pullulans a versatile biocontrol agent (Federici, 1982; Prasongsuk et al., 2018). Similar to P. flavescens, A. pullulans but also Metschnikowia and Vishniacozyma strains can effectively mitigate pre- and postharvest infections (Mari et al., 2012; Gramisci et al., 2018; Pawlikowska et al., 2019).
Compared to the yeasts, the filamentous fungi usually showed stronger antagonistic effects, even if the differences of the applied tests were taken into account. Three Cladosporium isolates (C1K002, D1K008, and A3K053) suppressed both H. fraxineus strains significantly with inhibition rates of 72–100%. The strong effects were also indicated by a clear reduction in the pathogen vitality. Fungicidal effects were also observed for Diplodia fraxini D1K016 and Phomopsis velata B2K028. All these isolates were representatives of fast-growing species that affected and displaced the pathogen in dual culture by strong sporulation or by overgrowing. Hanackova et al. (2017a) tested a number of endophytic fungi of common ashes in a similar approach and found comparable high inhibition rates. The best results were also achieved for fast-growing species, e.g., Botrytis cinerea and Phoma macrostoma. However, Diplodia fraxini, species of Phomopsis (syn. Diaporthe) and Botrytis cinerea are also known as plant pathogens (Elena et al., 2018; Hua et al., 2018; Sessa et al., 2018). Moreover, a promising antagonist (Hypoxylon rubiginosum) was also found through the detection of the antifungal metabolite phomopsidin, although the in vitro inhibitory effect was less pronounced (Halecker et al., 2020). Remarkably, studying endophytic fungi from dieback-tolerant ash species including F. mandshurica and F. ornus resulted in somewhat lower antagonistic activity against H. fraxineus, with inhibition rates of 40–64% (Kosawang et al., 2018).
Three of the isolates with the highest antagonistic activity were members of the genus Cladosporium. This genus was found to be the second-most abundant taxon among the leaf fungal communities of the dieback-tolerant Manchurian ash (Cleary et al., 2016). Furthermore, Cladosporium is closely related to the genus Mycosphaerella (Braun et al., 2003), which represented the predominant fungal group of the Manchurian ash (Cleary et al., 2016). Both genera include numerous widespread species varying in their ecological roles, including plant pathogens, mutualistic endophytes, saprophytes or hyperparasites on other fungi (Crous, 2009; Bensch et al., 2012). The ability of Cladosporium species to parasitize other fungi, in particular those causing rust diseases of agricultural and forestry crops (i.e., Puccinia sp.) has been demonstrated for decades in numerous studies (Tsuneda and Hiratsuka, 1979; Moricca et al., 2005; Zhan et al., 2014; Torres et al., 2017). One Cladosporium species (C. delicatulum) was also found to be a mycoparasite on Taphrina pruni, the plum gall pathogen of Prunus species (Amanelah Baharvandi and Zafari, 2015). Thus, recent data indicate that Cladosporium species have potential in plant diseases management. However, further studies are needed to clarify the biology of the antagonism. Since some Cladosporium strains can also be pathogenic to vertebrate hosts, including humans (Sandoval-Denis et al., 2016), the biosafety of the respective strains must be guaranteed on a case-specific basis (Torres et al., 2017).
The two H. fraxineus strains used in the dual culture experiment showed a different sensitivity to a range of antagonists. The H. fraxineus strain P3 was more damaged by some yeast isolates, while HF23 showed a higher sensitivity to individual filamentous fungi, including isolates of Cladosporium. H. fraxineus is known for its intrinsic diversity, which was indicated by the differing growth rates, enzymatic activity and virulence of various isolates (Junker, 2013; Junker et al., 2017). Against the background that the ash dieback invasion of Europe started with only two strains, a further increase in genetic diversity can be expected (McMullan et al., 2018). Accordingly, subsequent in planta tests of antagonistic activity should be performed with infected petioles representing the natural H. fraxineus populations from a typical ash forest. It remains unclear to what extent our results from the in vitro studies can be confirmed using this approach. Another critical aspect of in vitro testing is the lack of interactions of the antagonists with the plant holobiont. The complex plant-microbiota interaction can lead to both a reduction and an enhancement of the desired effect (Knudsen et al., 1997; Schulz et al., 2015; Latz et al., 2018).
In conclusion, the comparative analysis of the leaf-inhabiting fungi from susceptible and tolerant ashes revealed a high similarity in the community structure. Only a few fungal groups primarily belonging to P. flavescens and S. strictum, showed a distinctly higher abundance in tolerant ashes. These specific groups are suggested to improve pathogen tolerance, which could be mediated by competition, by inducing a systemic resistance or just by altering the microbial community structure (Pieterse et al., 2014; Romera et al., 2019). Additionally, in vitro tests of antagonism against H. fraxineus indicated clear inhibitory effects of various yeasts and particularly of several fast growing filamentous fungi. Both, the yeasts with the relatively mild inhibitory effects and the filamentous species with stronger activities, particularly Cladosporium sp. C1K002 and D1K008, could be involved in disease suppression. Based on these results, for the next step, in planta tests are necessary to evaluate these promising groups and isolates with respect to their efficacy and stability in inhibiting H. fraxineus. In this context, possible shifts in the microbial community should be studied and might be correlated with the plant health as well.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA611938.
Author Contributions
RB and KU performed the experiments, analyzed the data, and prepared the manuscript. UB performed the MALDI-TOF MS analysis. KU, MK, and AU conducted the microbiome analysis. All authors listed here substantially contributed to and approved the manuscript. AU and MK supervised the entire study.
Funding
This study was supported by Grant 22006116 from the Fachagentur Nachwachsende Rohstoffe (FNR), Germany.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank our colleagues from the forest district of Lendershagen, as well as Ben Bubner, Volker Schneck and Falk Schäfer for monitoring the ash plots and for their assistance with the sampling campaign. We are grateful to Ulrike Klauß and Heidrun Mattauch for providing excellent technical assistance.
References
Albrectsen, B. R., Bjorken, L., Varad, A., Hagner, A., Wedin, M., Karlsson, J., et al. (2010). Endophytic fungi in European aspen (Populus tremula) leaves-diversity, detection, and a suggested correlation with herbivory resistance. Fungal Divers. 41, 17–28. doi: 10.1007/s13225-009-0011-y
Amanelah Baharvandi, H., and Zafari, D. (2015). Identification of Cladosporium delicatulum as a mycoparasite of Taphrina pruni. Arch. Phytopathol. 48, 688–697. doi: 10.1080/03235408.2015.1099886
Baral, H.-O., Queloz, V., and Hosoya, T. (2014). Hymenoscyphus fraxineus, the correct scientific name for the fungus causing ash dieback in Europe. IMA Fungus 5, 79–80. doi: 10.5598/imafungus.2014.05.01.09
Bar-Shimon, M., Yehuda, H., Cohen, L., Weiss, B., Kobeshnikov, A., Daus, A., et al. (2004). Characterization of extracellular lytic enzymes produced by the yeast biocontrol agent Candida oleophila. Curr. Genet. 45, 140–148. doi: 10.1007/s00294-003-0471-7
Bengtsson-Palme, J., Ryberg, M., Hartmann, M., Branco, S., Wang, Z., Godhe, A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919. doi: 10.1111/2041-210x.12073
Bensch, K., Braun, U., Groenewald, J. Z., and Crous, P. W. (2012). The genus Cladosporium. Stud. Mycol. 72, 1–379. doi: 10.3114/sim0003
Berg, G., Koberl, M., Rybakova, D., Muller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. doi: 10.1093/femsec/fix050
Berg, G., Rybakova, D., Grube, M., and Köberl, M. (2015). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13.
Braun, U., Crous, P., Dugan, F., Groenewald, J. Z., and Hoog, S. (2003). Phylogeny and taxonomy of Cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycol. Prog. 2, 3–18. doi: 10.1007/s11557-006-0039-2
Busby, P. E., Ridout, M., and Newcombe, G. (2016). Fungal endophytes: modifiers of plant disease. Plant Mol. Biol. 90, 645–655. doi: 10.1007/s11103-015-0412-0
Campbell, B., Siddique, A.-B., McDougall, B., and Seviour, R. (2004). Which morphological forms of the fungus Aureobasidium pullulans are responsible for pullulan production? FEMS Microbiol. Lett. 232, 225–228. doi: 10.1016/S0378-1097(04)00076-X
Choi, G., Kim, J.-C., Jang, K., Cho, K., and Kim, H. (2008). Mycoparasitism of Acremonium strictum BCP on Botrytis cinerea, the gray mold pathogen. J. Microbiol. Biotechnol. 18, 167–170.
Chong, J., Liu, P., Zhou, G., and Xia, J. (2020). Using microbiomeanalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799–821. doi: 10.1038/s41596-019-0264-1
Chudejova, K., Bohac, M., Skalova, A., Rotova, V., Papagiannitsis, C. C., Hanzlickova, J., et al. (2017). Validation of a novel automatic deposition of bacteria and yeasts on MALDI target for MALDI-TOF MS-based identification using MALDI Colonyst robot. PLoS One 12:e0190038. doi: 10.1371/journal.pone.0190038
Cleary, M., Nguyen, D., Marciulyniene, D., Berlin, A., Vasaitis, R., and Stenlid, J. (2016). Friend or foe? Biological and ecological traits of the European ash dieback pathogen Hymenoscyphus fraxineus in its native environment. Sci. Rep. 6:21895. doi: 10.1038/srep21895
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Cross, H., Sønstebø, J. H., Nagy, N. E., Timmermann, V., Solheim, H., Børja, I., et al. (2017). Fungal diversity and seasonal succession in ash leaves infected by the invasive ascomycete Hymenoscyphus fraxineus. New Phytol. 213, 1405–1417. doi: 10.1111/nph.14204
Crous, P. (2009). Taxonomy and phylogeny of the genus Mycosphaerella and its anamorphs. Fungal Divers. 38, 1–24.
Davydenko, K., Vasaitis, R., Stenlid, J., and Menkis, A. (2013). Fungi in foliage and shoots of Fraxinus excelsior in eastern Ukraine: a first report on Hymenoscyphus pseudoalbidus. Forest Pathol. 43, 462–467. doi: 10.1111/efp.12055
Drenkhan, R., Solheim, H., Bogacheva, A., Riit, T., Adamson, K., Drenkhan, T., et al. (2017). Hymenoscyphus fraxineus is a leaf pathogen of local Fraxinus species in the Russian far east. Plant Pathol. 66, 490–500. doi: 10.1111/ppa.12588
Dufrêne, M., and Legendre, P. (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366.
Dunlap, C. A., and Schisler, D. A. (2010). Fluidized-bed drying and storage stability of Cryptococcus flavescens OH 182.9, a biocontrol agent of Fusarium head blight. Biocontrol Sci. Technol. 20, 465–474. doi: 10.1080/09583150903572377
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Elena, G., León, M., Abad-Campos, P., Armengol, J., Mateu-Andrés, I., and Güemes-Heras, J. (2018). First report of Diplodia fraxini causing dieback of Fraxinus angustifolia in Spain. Plant Dis. 102, 2645–2645. doi: 10.1094/pdis-05-18-0792-pdn
Fadiji, A. E., and Babalola, O. O. (2020). Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioeng. Biotechnol. 8:467. doi: 10.3389/fbioe.2020.00467
Federici, F. (1982). Extracellular enzymatic activities in Aureobasidium pullulans. Mycologia 74, 738–743. doi: 10.2307/3792859
Fonseca, A., and Inacio, J. P. (2006). “Phylloplane yeasts,” in Biodiversity and Ecophysiology of Yeasts, eds G. Peter and C. Rosa (Berlin: Springer), 263–301.
Freimoser, F. M., Rueda-Mejia, M. P., Tilocca, B., and Migheli, Q. (2019). Biocontrol yeasts: mechanisms and applications. World J. Microbiol. Biotechnol. 35:154. doi: 10.1007/s11274-019-2728-4
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for Basidiomycetes – application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:15. doi: 10.6064/2012/963401
Gramisci, B. R., Lutz, M. C., Lopes, C. A., and Sangorrín, M. P. (2018). Enhancing the efficacy of yeast biocontrol agents against postharvest pathogens through nutrient profiling and the use of other additives. Biol. Control 121, 151–158. doi: 10.1016/j.biocontrol.2018.03.001
Griffiths, S. M., Galambao, M., Rowntree, J., Goodhead, I., Hall, J., O’Brien, D., et al. (2020). Complex associations between cross−kingdom microbial endophytes and host genotype in ash dieback disease dynamics. J. Ecol. 108, 291–309. doi: 10.1111/1365-2745.13302
Gross, A., Holdenrieder, O., Pautasso, M., Queloz, V., and Sieber, T. N. (2014). Hymenoscyphus pseudoalbidus, the causal agent of European ash dieback. Mol. Plant Pathol. 15, 5–21. doi: 10.1111/mpp.12073
Guimarães, R. A., da Silva, Lobo, V. L., Côrtes, M. V. C. B., Corsi de Filippi, M. C., and Prabhu, A. S. (2017). Characterization of Sarocladium oryzae and its reduction potential of rice leaf blast. Pesq. Agropec. Trop. Goiânia 47, 41–52.
Halecker, S., Wennrich, J.-P., Rodrigo, S., Andrée, N., Rabsch, L., Baschien, C., et al. (2020). Fungal endophytes for biocontrol of ash dieback: the antagonistic potential of Hypoxylon rubiginosum. Fungal Ecol. 45:100918. doi: 10.1016/j.funeco.2020.100918
Han, J.-G., Shrestha, B., Hosoya, T., Lee, K.-H., Sung, G.-H., and Shin, H.-D. (2014). First report of the ash dieback pathogen Hymenoscyphus fraxineus in Korea. Mycobiology 42, 391–396. doi: 10.5941/MYCO.2014.42.4.391
Hanackova, Z., Havrdova, L., Cerny, K., Zahradnik, D., and Koukol, O. (2017a). Fungal endophytes in ash shoots – diversity and inhibition of Hymenoscyphus fraxineus. Balt. For. 23, 89–106.
Hanackova, Z., Koukol, O., Cmokova, A., Zahradnik, D., and Havrdova, L. (2017b). Direct evidence of Hymenoscyphus fraxineus infection pathway through the petiole-shoot junction. Forest Pathol. 47:12370. doi: 10.1111/efp.12370
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/mmbr.00050-14
Hossain, M. M., Sultana, F., and Islam, S. (2017). “Plant growth-promoting fungi (PGPF): phytostimulation and induced systemic resistance,” in Plant-Microbe Interactions in Agro-Ecological Perspectives, eds D. Singh, H. Singh, and R. Prabha (Singapore: Springer).
Hua, L., Yong, C., Zhanquan, Z., Boqiang, L., Guozheng, Q., and Shiping, T. (2018). Pathogenic mechanisms and control strategies of Botrytis cinerea causing post-harvest decay in fruits and vegetables. Food Qual. Saf. 2, 111–119. doi: 10.1093/fqsafe/fyy016
Huschek, D., and Witzel, K. (2019). Rapid dereplication of microbial isolates using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a mini-review. J. Adv. Res. 19, 99–104. doi: 10.1016/j.jare.2019.03.007
Ihrmark, K., Bödeker, I. T. M., Cruz-Martinez, K., Friberg, H., Kubartova, A., Schenck, J., et al. (2012). New primers to amplify the fungal ITS2 region – evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677.
Inacio, J., Pereira, P., de Carvalho, M., Fonseca, A., Amaral-Collaco, M. T., and Spencer-Martins, I. (2002). Estimation and diversity of phylloplane mycobiota on selected plants in a Mediterranean-type ecosystem in Portugal. Microb. Ecol. 44, 344–353. doi: 10.1007/s00248-002-2022-z
Jia, Q., Qu, J., Mu, H., Sun, H., and Wu, C. (2020). Foliar endophytic fungi: diversity in species and functions in forest ecosystems. Symbiosis 80, 103–132. doi: 10.1007/s13199-019-00663-x
Junker, C. (2013). Pathogenese und Ansätze zur Kontrolle von Hymenoscyphus Pseudoalbidus – Erreger der Eschentriebsterbens: Variabilität von Virulenz, Morphologie, Biochemie und Sekundärstoffwechsel. Ph.D. thesis, Technische Universität Braunschweig, Brunswick.
Junker, C., de Vries, J., Eickhorst, C., and Schulz, B. (2017). Each isolate of Hymenoscyphus fraxineus is unique as shown by exoenzyme and growth rate profiles. Balt. For. 23, 25–40.
Kemler, M., Witfeld, F., Begerow, D., and Yurkov, A. (2017). “Phylloplane yeasts in temperate climates,” in Yeasts in Natural Ecosystems: Diversity, eds P. Buzzini, M.-A. Lachance, and A. Yurkov (Cham: Springer International Publishing), 171–197.
Kheireddine, A., Essghaier, B., Hedi, A., Dhieb, C., and Sadfi-Zouaoui, N. (2018). New epiphytic yeasts able to reduce grey mold disease on apples. Plant Prot. Sci. 54, 248–257. doi: 10.17221/103/2017-pps
Kirisits, T., and Schwanda, K. (2015). First definite report of natural infection of Fraxinus ornus by Hymenoscyphus fraxineus. Forest Pathol. 45, 430–432.
Knudsen, I. M. B., Hockenhull, J., Jensen, D. F., Gerhardson, B., Hökeberg, M., Tahvonen, R., et al. (1997). Selection of biological control agents for controlling soil and seed-borne diseases in the field. Eur. J. Plant Pathol. 103, 775–784. doi: 10.1023/A:1008662313042
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Kosawang, C., Amby, D. B., Bussaban, B., McKinney, L. V., Xu, J., Kjaer, E. D., et al. (2018). Fungal communities associated with species of Fraxinus tolerant to ash dieback, and their potential for biological control. Fungal Biol. 122, 110–120. doi: 10.1016/j.funbio.2017.11.002
Kosawang, C., Sorensen, H., Kjaer, E. D., Dilokpimol, A., McKinney, L. V., Collinge, D. B., et al. (2019). Defining the twig fungal communities of Fraxinus species and Fraxinus excelsior genotypes with differences in susceptibility to ash dieback. Fungal Ecol. 42:100859. doi: 10.1016/j.funeco.2019.08.003
Kowalski, T. (2006). Chalara fraxinea sp nov associated with dieback of ash (Fraxinus excelsior) in Poland. Forest Pathol. 36, 264–270. doi: 10.1111/j.1439-0329.2006.00453.x
Kowalski, T., and Holdenrieder, O. (2009). Pathogenicity of Chalara fraxinea. Forest Pathol. 39, 1–7. doi: 10.1111/j.1439-0329.2008.00565.x
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/Aem.01043-13
Latz, M. A. C., Jensen, B., Collinge, D. B., and Jorgensen, H. J. L. (2018). Endophytic fungi as biocontrol agents: elucidating mechanisms in disease suppression. Plant Ecol. Divers. 11, 555–567. doi: 10.1080/17550874.2018.1534146
Liu, H., Brettell, L. E., Qiu, Z., and Singh, B. K. (2020). Microbiome-mediated stress resistance in plants. Trends Plant Sci. 25, 733–743. doi: 10.1016/j.tplants.2020.03.014
Liu, J., Sui, Y., Wisniewski, M., Droby, S., and Liu, Y. (2013). Review: utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int. J. Food Microbiol. 167, 153–160. doi: 10.1016/j.ijfoodmicro.2013.09.004
Lobo, A., Hansen, J. K., McKinney, L. V., Nielsen, L. R., and Kjær, E. D. (2014). Genetic variation in dieback resistance: growth and survival of Fraxinus excelsior under the influence of Hymenoscyphus pseudoalbidus. Scand. J. For. Res. 29, 519–526. doi: 10.1080/02827581.2014.950603
Mari, M., Martini, C., Spadoni, A., Rouissi, W., and Bertolini, P. (2012). Biocontrol of apple postharvest decay by Aureobasidium pullulans. Postharvest Biol. Technol. 73, 56–62. doi: 10.1016/j.postharvbio.2012.05.014
Martin-Garcia, J., Muller, M. M., and Diez, J. J. (2012). ITS-based comparison of endophytic mycota in twigs of native Populus nigra and cultivated P. x euramericana (cv. I-214) stands in Northern Spain. Ann. For. Sci. 69, 49–57. doi: 10.1007/s13595-011-0129-4
McCune, B., and Mefford, M. J. (2015). PC-ORD. Multivariate Analysis of Ecological Data, 7.0 Edn. Gleneden Beach, OR: MjM Software.
McKinney, L. V., Nielsen, L. R., Collinge, D. B., Thomsen, I. M., Hansen, J. K., and Kjaer, E. D. (2014). The ash dieback crisis: genetic variation in resistance can prove a long-term solution. Plant Pathol. 63, 485–499. doi: 10.1111/ppa.12196
McMullan, M., Rafiqi, M., Kaithakottil, G., Clavijo, B. J., Bilham, L., Orton, E., et al. (2018). The ash dieback invasion of Europe was founded by two genetically divergent individuals. Nat. Ecol. Evol. 2, 1000–1008. doi: 10.1038/s41559-018-0548-9
McSpadden Gardener, B. B., Anderson Paul, P., Boehm, M. J., Rong, X., and Schisler, D. (2014). Methods for using Crytococcus flavescens Strains for Biological Control of Fusarium Head Blight. Papers in Plant Pathology – Patent Application Publication US Google Patent No. 2014/0271560A1.
Moricca, S., Ragazzi, A., and Assante, G. (2005). “Biocontrol of rust fungi by Cladosporium tenuissimum,” in Rust Diseases of Willow and Poplar, eds M. H. Pei and A. R. McCracken (Wallingford: CAB International), 213–219.
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12:385. doi: 10.1186/1471-2105-12-385
Pane, C., and Zaccardelli, M. (2015). Evaluation of Bacillus strains isolated from solanaceous phylloplane for biocontrol of Alternaria early blight of tomato. Biol. Control 84, 11–18. doi: 10.1016/j.biocontrol.2015.01.005
Paulson, J. N., Stine, O. C., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1205. doi: 10.1038/Nmeth.2658
Pawlikowska, E., James, S. A., Breierova, E., Antolak, H., and Kregiel, D. (2019). Biocontrol capability of local Metschnikowia sp. isolates. Anton. Leeuw. Int. J. G. 112, 1425–1445. doi: 10.1007/s10482-019-01272-w
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Wees, S. C. M. V., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Prasongsuk, S., Lotrakul, P., Ali, I., Bankeeree, W., and Punnapayak, H. (2018). The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol. 63, 129–140. doi: 10.1007/s12223-017-0561-4
Prior, R., Feige, A., and Begerow, D. (2017). Antagonistic activity of the phyllosphere fungal community. Sydowia 69, 183–198. doi: 10.12905/0380.sydowia69-2017-0483
Przybyl, K. (2003). Effect of Pseudomonas spp. on inoculation of young plants of Fraxinus excelsior stem with Diplodia mutila. Dendrobiology 50, 29–32.
Qin, G., and Tian, S. (2004). Biocontrol of postharvest diseases of jujube fruit by Cryptococcus laurentii combined with a low dosage of fungicides under different storage conditions. Plant Dis. 88, 497–501. doi: 10.1094/PDIS.2004.88.5.497
Queloz, V., Grunig, C., Berndt, R., Kowalski, T., Sieber, T., and Holdenrieder, O. (2011). Cryptic speciation in Hymenoscyphus albidus. Forest Pathol. 41, 133–142. doi: 10.1111/j.1439-0329.2010.00645.x
Ramos, S., and García Acha, I. (1975). A vegetative cycle of Pullularia pullulans. Trans. Br. Mycol. Soc. 64, 129–135, IN8–IN9. doi: 10.1016/S0007-1536(75)80083-0
Rivera, V., Freeman, T., Gudmestad, N., and Secor, G. (2007). Mycoparasitism of Helminthosporium solani by Acremonium strictum. Phytopathology 97, 1331–1337. doi: 10.1094/PHYTO-97-10-1331
Romera, F. J., Garcia, M. J., Lucena, C., Martinez-Medina, A., Aparicio, M. A., Ramos, J., et al. (2019). Induced systemic resistance (ISR) and Fe deficiency responses in dicot plants. Front. Plant Sci. 10:287. doi: 10.3389/fpls.2019.00287
Salazar, O., and Asenjo, J. A. (2007). Enzymatic lysis of microbial cells. Biotechnol. Lett. 29, 985–994. doi: 10.1007/s10529-007-9345-2
Sandoval-Denis, M., Gene, J., Sutton, D. A., Wiederhold, N. P., Cano-Lira, J. F., and Guarro, J. (2016). New species of Cladosporium associated with human and animal infections. Persoonia 36, 281–298. doi: 10.3767/003158516X691951
Schell, W. A., and Perfect, J. R. (1996). Fatal, disseminated Acremonium strictum infection in a neutropenic host. J. Clin. Microbiol. 34, 1333–1336.
Schisler, D. A., Core, A. B., Boehm, M. J., Horst, L., Krause, C., Dunlap, C. A., et al. (2014). Population dynamics of the Fusarium head blight biocontrol agent Cryptococcus flavescens OH 182.9 on wheat anthers and heads. Biol. Control 70, 17–27. doi: 10.1016/j.biocontrol.2013.11.011
Schlegel, M., Queloz, V., and Sieber, T. N. (2018). The endophytic mycobiome of european ash and sycamore maple leaves – geographic patterns, host specificity and influence of ash dieback. Front. Microbiol. 9:2345. doi: 10.3389/fmicb.2018.02345
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/Aem.01541-09
Schulz, B., Haas, S., Junker, C., Andree, N., and Schobert, M. (2015). Fungal endophytes are involved in multiple balanced antagonisms. Curr. Sci. 109, 39–45.
Schulz, B., Rabsch, L., and Junker, C. (2019). “Chemical warfare in the plant microbiome leads to a balance of antagonisms and a healthy plant,” in Seed Endophytes: Biology and Biotechnology, eds S. K. Verma and J. F. J. White (Berlin: Springer).
Sessa, L., Abreo, E., Bettucci, L., and Lupo, S. (2018). Diversity and virulence of Diaporthe species associated with wood disease symptoms in deciduous fruit trees in Uruguay. Phytopathol. Mediterr. 56, 431–444.
Sharma, A., Hazarika, N. K., Barua, P., Shivaprakash, M. R., and Chakrabarti, A. (2013). Acremonium strictum: report of a rare emerging agent of cutaneous hyalohyphomycosis with review of literatures. Mycopathologia 176, 435–441. doi: 10.1007/s11046-013-9709-1
Skovsgaard, J. P., Wilhelm, G. J., Thomsen, I. M., Metzler, B., Kirisits, T., Havrdova, L., et al. (2017). Silvicultural strategies for Fraxinus excelsior in response to dieback caused by Hymenoscyphus fraxineus. Forestry 90, 455–472. doi: 10.1093/forestry/cpx012
Slavikova, E., Vadkertiova, R., and Vranova, D. (2007). Yeasts colonizing the leaf surfaces. J. Basic Microbiol. 47, 344–350. doi: 10.1002/jobm.200710310
Stener, L.-G. (2013). Clonal differences in susceptibility to the dieback of Fraxinus excelsior in southern Sweden. Scand. J. Forest Res. 28, 205–216. doi: 10.1080/02827581.2012.735699
Stocks, J. J., Metheringham, C. L., Plumb, W. J., Lee, S. J., Kelly, L. J., Nichols, R. A., et al. (2019). Genomic basis of European ash tree resistance to ash dieback fungus. Nat. Ecol. Evol. 3, 1686–1696. doi: 10.1038/s41559-019-1036-6
Sündermann, J., and Jütte, K. (2014). Eschentriebsterben – Forst- und holzwirtschaftliche Strategien zum Umgang mit dem neuartigen Eschentriebsterben; Teilvorhaben 1: Nachhaltige Eschenholzproduktion sowie Erprobung effizienter und bodenschonender Holzernteverfahren auf sensiblen Nassstandorten. Schlussbericht FNR-Verbundvorhaben No. 22007410. Available online at: https://www.fnr.de/index.php?id=11150&fkz=22007410 (accessed August 3, 2020).
Tedersoo, L., Anslan, S., Bahram, M., Polme, S., Riit, T., Liiv, I., et al. (2015). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. Mycokeys 10, 1–43. doi: 10.3897/mycokeys.10.4852
Thompson, I. P., Bailey, M. J., Fenlon, J. S., Fermor, T. R., Lilley, A. K., Lynch, J. M., et al. (1993). Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil 150, 177–191. doi: 10.1007/BF00013015
Timmermann, V., Børja, I., Hietala, A., Kirisits, T., and Solheim, H. (2011). Ash dieback: pathogen spread and diurnal patterns of ascospore dispersal, with special emphasis on Norway. EPPO Bull. 41, 14–20. doi: 10.1111/j.1365-2338.2010.02429.x
Torres, D. E., Rojas-Martínez, R. I., Zavaleta-Mejía, E., Guevara-Fefer, P., Márquez-Guzmán, G. J., and Pérez-Martínez, C. (2017). Cladosporium cladosporioides and Cladosporium pseudocladosporioides as potential new fungal antagonists of Puccinia horiana Henn., the causal agent of chrysanthemum white rust. PLoS One 12:e0170782. doi: 10.1371/journal.pone.0170782
Tsuneda, A., and Hiratsuka, Y. (1979). Mode of parasitism of a mycoparasite, Cladosporium gallicola, on western gall rust, Endocronartium harknessii. Can. J. Plant Pathol. 1, 31–36. doi: 10.1080/07060667909501480
Ulrich, K., Becker, R., Behrendt, U., Kube, M., and Ulrich, A. (2020). A comparative analysis of ash leaf-colonizing bacterial communities identifies putative antagonists of Hymenoscyphus fraxineus. Front. Microbiol. 11:966. doi: 10.3389/fmicb.2020.00966
van Overbeek, L. S., and Saikkonen, K. (2016). Impact of bacterial–fungal interactions on the colonization of the endosphere. Trends Plant Sci. 21, 230–242. doi: 10.1016/j.tplants.2016.01.003
White, T. J., Burns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal genes for phylogenetics,” in PCR Protocols. A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Wicklow, D. T., Roth, S., Deyrup, S. T., and Gloer, J. B. (2005). A protective endophyte of maize: Acremonium zeae antibiotics inhibitory to Aspergillus flavus and Fusarium verticillioides. Mycol. Res. 109, 610–618. doi: 10.1017/S0953756205002820
Wohlmuth, A., Essl, F., and Heinze, B. (2018). Genetic analysis of inherited reduced susceptibility of Fraxinus excelsior L. seedlings in Austria to ash dieback. Forestry 91, 514–525. doi: 10.1093/forestry/cpy012
Zhan, G., Tian, Y., Wang, F., Chen, X., Guo, J., Jiao, M., et al. (2014). A novel fungal hyperparasite of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS One 9:e111484. doi: 10.1371/journal.pone.0111484
Zhang, D., Spadaro, D., Valente, S., Garibaldi, A., and Gullino, M. L. (2011). Cloning, characterization and expression of an exo-1,3-ß-glucanase gene from the antagonistic yeast, Pichia guilliermondii strain M8 against grey mold on apples. Biol. Control 59, 284–293.
Zhang, D., Spadaro, D., Valente, S., Garibaldi, A., and Gullino, M. L. (2012). Cloning, characterization, expression and antifungal activity of an alkaline serine protease of Aureobasidium pullulans PL5 involved in the biological control of postharvest pathogens. Int. J. Food Microbiol. 153, 453–464. doi: 10.1016/j.ijfoodmicro.2011.12.016
Zhang, H., Zheng, X., and Yu, T. (2007). Biology control of postharvest diseases of peach with Cryptococcus laurentii. Food Control 18, 287–291. doi: 10.1016/j.foodcont.2005.10.007
Keywords: ash dieback, antagonism, Fraxinus excelsior, mycobiome, cocultivation, phyllosphere
Citation: Becker R, Ulrich K, Behrendt U, Kube M and Ulrich A (2020) Analyzing Ash Leaf-Colonizing Fungal Communities for Their Biological Control of Hymenoscyphus fraxineus. Front. Microbiol. 11:590944. doi: 10.3389/fmicb.2020.590944
Received: 03 August 2020; Accepted: 02 October 2020;
Published: 22 October 2020.
Edited by:
Yong Wang, Guizhou University, ChinaReviewed by:
Qian Guoliang, Nanjing Agricultural University, ChinaJia Liu, Chongqing University of Arts and Sciences, China
Copyright © 2020 Becker, Ulrich, Behrendt, Kube and Ulrich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Ulrich, YXVscmljaEB6YWxmLmRl
 Regina Becker
Regina Becker Kristina Ulrich2
Kristina Ulrich2 Undine Behrendt
Undine Behrendt Michael Kube
Michael Kube Andreas Ulrich
Andreas Ulrich