Abstract
Nicotine is a major N-heterocyclic aromatic alkaloid produced in tobacco plants and the main toxic chemical in tobacco waste. Due to its complex physiological effects and toxicity, it has become a concern both in terms of public health and the environment. A number of bacteria belonging to the genera Arthrobacter and Pseudomonas can degrade nicotine via the pyridine and pyrrollidine pathways. Recently, a novel hybrid of the pyridine and pyrrolidine pathways (also known as the VPP pathway) was found in the Rhizobiale group bacteria Agrobacterium tumefaciens S33, Shinella sp. HZN7 and Ochrobactrum sp. SJY1 as well as in other group bacteria. The special mosaic pathway has attracted much attention from microbiologists in terms of the study of their molecular and biochemical mechanisms. This will benefit the development of new biotechnologies in terms of the use of nicotine, the enzymes involved in its catabolism, and the microorganisms capable of degrading the alkaloid. In this pathway, some metabolites are hydroxylated in the pyridine ring or modified in the side chain with active groups, which can be used as precursors for the synthesis of some important compounds in the pharmaceutical and agricultural industries. Moreover, some enzymes may be used for industrial biocatalysis to transform pyridine derivatives into desired chemicals. Here, we review the molecular and biochemical basis of the hybrid nicotine-degrading pathway and discuss the electron transport in its oxidative degradation for energy conservation and bacterial growth.
Introduction
Nicotine is the main alkaloid produced in tobacco (Bush et al., 1999). In addition to the central nervous system, it has complex physiological effects on humans, causing people to get addicted to tobacco and develop various smoking-related diseases (Hecht, 1999; Campain, 2004; Benowitz, 2010). Due to its toxicity, it is also an environmental concern when considering the smoke and the waste accumulation during tobacco consumption and manufacturing (Novotny and Zhao, 1999). Its production and consumption are thus regulated according to the World Health Organization Framework Convention on Tobacco Control. For centuries, however, tobacco has been widely planted as an important economic nonfood crop around the world. It is therefore imperative to develop alternative technologies to utilize tobacco and dispose of waste. Nicotine has attracted much interest as a substrate to synthesize or biotransform into renewable functionalized pyridines, such as drugs and insecticides, as well as a precursor for synthesis of these molecules (Wang et al., 2012). For example, some nicotine-degrading bacteria can specifically hydroxylate the pyridine ring of nicotine at the 6-position or 2- and 5-positions, which is a difficult procedure when using chemical methods (Schmid et al., 2001; Wang et al., 2005; Ondachi and Comins, 2010). These bacteria or their enzymes involved in nicotine catabolism, could consequently be used as biocatalysts in the processes.
Many microorganisms can degrade nicotine via various biochemical pathways, including the demethylation pathway in Aspergillus oryzae (Meng et al., 2010), the pyridine pathway in Arthrobacter (Brandsch, 2006; Figure 1A), and the pyrrolidine pathway in Pseudomonas (Wang et al., 2007; Tang et al., 2012, 2013; Figure 1C). The molecular mechanisms underlying the pyridine and pyrrolidine pathways in bacteria have been well investigated. Recently, a novel hybrid of the pyridine and pyrrolidine pathways (also called the VPP pathway) was discovered in the Rhizobiale group bacterium Agrobacterium tumefaciens S33 (Wang et al., 2012; Figure 1B) and was later also found in Shinella sp. HZN7 (Ma et al., 2014), Ochrobactrum sp. SJY1 (Yu et al., 2015), and other group bacteria, such as Sphingomonas melonis TY (Wang et al., 2017) and Pseudomonas geniculata N1 (Wang et al., 2019b). Here, we summarize the progress in molecular and biochemical studies on the hybrid pathway, considering its unique mosaic feature and valuable functionalized pyridine intermediates. Although most core genes involved in the pathway have been identified in these bacteria, the enzymatic properties and kinetics are mainly determined for those from the three Rhizobiale group bacteria, which are the main focus of this mini review; for further information, the reader is referred to a recent review paper (Mu et al., 2020).
Figure 1
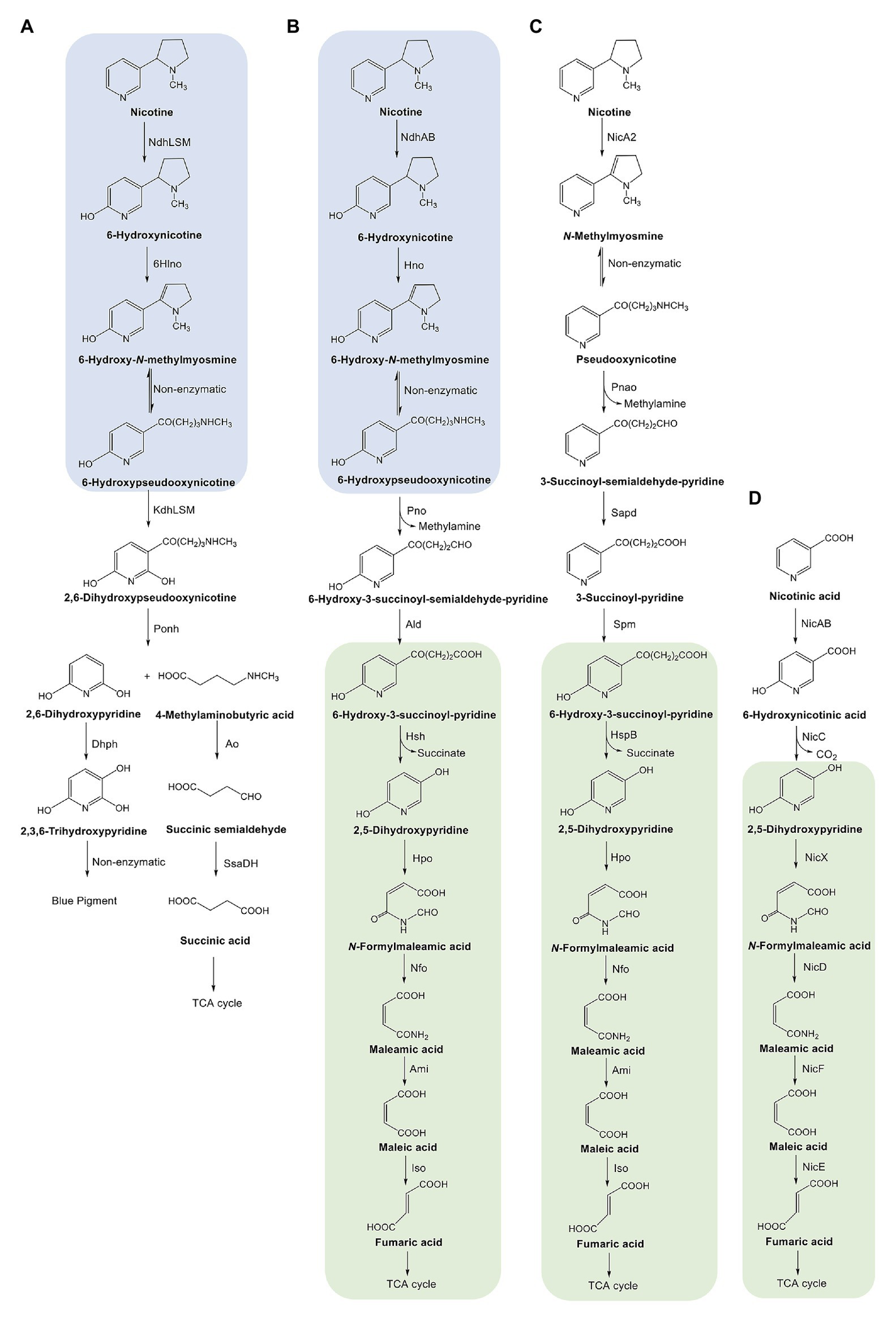
Scheme depicting the pathways for degrading nicotine and nicotinate in bacteria. (A) Pyridine pathway of nicotine degradation in Arthrobacter. Ndh, nicotine dehydrogenase; 6Hlno, 6-hydroxy-L-nicotine oxidase; Kdh, ketone dehydrogenase; Ponh, 2,6-dihydroxypseudooxynicotine hydrolase; Dhph, 2,6-dihydroxypyridine 3-hydroxylase. (B) A hybrid of pyridine and pyrrolidine pathways for nicotine degradation discussed in this review. See Figure 2 legend for enzyme abbreviations. (C) Pyrrolidine pathway of nicotine degradation in Pseudomonas. NicA, nicotine oxidoreductase; Pnao, pseudooxynicotine amine oxidase; Sapd, 3-succinoylsemialdehyde-pyridne dehydrogenase; SpmABC, 3-succinoylpyridine monooxygenase; HspB, 6-hydroxy-3-succinoylpyridine hydroxylase; Hpo, 2,5-dihydroxypyridine dioxygenase; Nfo, N-formylmaleamate deformylase; Ami, maleamate amidohydrolase; Iso, maleate cis/trans-isomerase. (D) Nicotinate-degrading pathway in Pseudomonas. NicAB, nicotinate hydroxylase; NicC, 6-hydroxynicotinate monooxygenase; NicX, 2,5-dihydroxypyridine dioxygenase; NicD, N-formylmaleamate deformylase; NicF, maleamate amidohydrolase; NicE, maleate cis/trans-isomerase. The parts shaded in blue indicate the same steps in the pyridine and hybrid pathways; those shaded in green indicate the same steps in the hybrid, pyrrolidine, and nicotinate-degrading pathways. The figure is modified from Huang et al., 2017, permitted by a Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).
A Hybrid of the Pyridine and Pyrrolidine Pathways for Nicotine Degradation in Bacteria
The bacterial catabolism of nicotine includes two crucial actions: hydroxylation of the pyridine ring and dehydrogenation of the pyrrolidine ring of nicotine. Different pathways follow varying sequences. The pyridine pathway (Figure 1A) starts by attacking the pyridine ring to hydroxylate nicotine into 6-hydroxynicotine. The product is then dehydrogenated in the pyrrolidine ring to form 6-hydroxy-N-methylmyosmine, which is then spontaneously hydrolyzed to break the pyrrolidine ring and produce 6-hydroxy-pseudooxynicotine. In contrast, the pyrrolidine pathway (Figure 1C) starts by dehydrogenating the pyrrolidine ring into N-methylmyosmine, which is then spontaneously hydrolyzed to pseudooxynicotine. After oxidation of pseudooxynicotine to 3-succinoyl-semialdehyde-pyridine and further dehydrogenation to 3-succinoyl-pyridine, the pyridine ring is hydroxylated to produce 6-hydroxy-3-succinoyl-pyridine. The hybrid pathway (Figure 1B) conforms with the pyridine pathway in the steps from nicotine to 6-hydroxy-pseudooxynicotine. However, the oxidation of 6-hydroxy-pseudooxynicotine to 6-hydroxy-3-succinoyl-pyridine via its semialdehyde intermediate in the hybrid is similar to that of the non-hydroxylated intermediates in the pyrrolidine pathway. The first common intermediate of the hybrid and the pyrrolidine pathways is 6-hydroxy-3-succinoyl-pyridine. The next steps for the breakdown of the side chain of 6-hydroxy-3-succinoyl-pyridine and decomposition of the pyridine ring in the hybrid are identical to those in the pyrrolidine pathway. Interestingly, the steps for 2,5-dihydroxypyridine degradation are also identical to those in the nicotinate-degrading pathway (Figure 1D).
Among the intermediates of the hybrid pathway, several are hydroxylated in the pyridine ring or modified in the side chain with active groups, which can be used as precursors for the synthesis of some compounds of pharmaceutical and agricultural importance combined with chemical methods (Roduit et al., 1997; Nakano et al., 1999; Wang et al., 2005). For example, 6-hydroxynicotine and 6-hydroxy-3-succinoylpyridine can be chemically transformed into 2,5- or 3,5-disubstituted pyridines, such as the insecticide Imidacloprid, the anti-Parkinson agent SIB-1508Y, and the analgesic epibatidine. The disubstituted pyridine 2,5-dihydroxypyridine can be chemically catalyzed into 5-aminolevulinic acid, which is applied as a universal precursor for porphyrins, herbicides, and plant growth hormones as well as cancer therapy. This provides opportunities to develop new technologies for utilizing tobacco and nicotine.
The Molecular Basis for the Hybrid Pathway
Most of the genes responsible for the hybrid pathway in A. tumefaciens S33, Shinella sp. HZN7, and Ochrobactrum sp. SJY1 have been identified by genomic, transcriptomic, and biochemical analyses and gene disruption experiments (Yu et al., 2015, 2016; Qiu et al., 2016; Huang et al., 2017). They form a large cluster located on a genomic island in the circular chromosome of strain S33 (Figure 2A), which also harbors a second linear chromosome (Huang et al., 2017). In contrast, the nicotine-degrading gene cluster in Shinella sp. HZN7 is on 155-kb plasmid pShin-05, which harbors one circular chromosome and 12 plasmids. For Ochrobactrum sp. SJY1, the location of the nicotine-degrading gene cluster is not clear because only the whole genome shotgun sequence is available. Despite these differences, the arrangement of the gene clusters in the three strains is highly similar (Figure 2A). They are spaced by several genes annotated to encode transposase, group II intron reverse transcriptase/maturase, and DDE endonuclease. In addition, the upstream and/or downstream regions of the gene cluster in S33 and HZN7 contain several gene modules encoding mobile genetic elements for transposition (for example, intergrase and tranposases), conjugal transfer (TraGDCFBHR and TrbIHGFLKJEDCB), and plasmid partition and replication initiation (RepABC), indicating that the nicotine-degrading genes evolved via complicated recombination and integration through transposition and conjunction. However, there is no gene for conjugal transfer in strain SJY1. Moreover, transcriptional analyses indicate that these genes are nicotine-inducible, and some genes are co-transcribed and can form single contiguous transcripts (Yu et al., 2015; Huang et al., 2017).
Figure 2
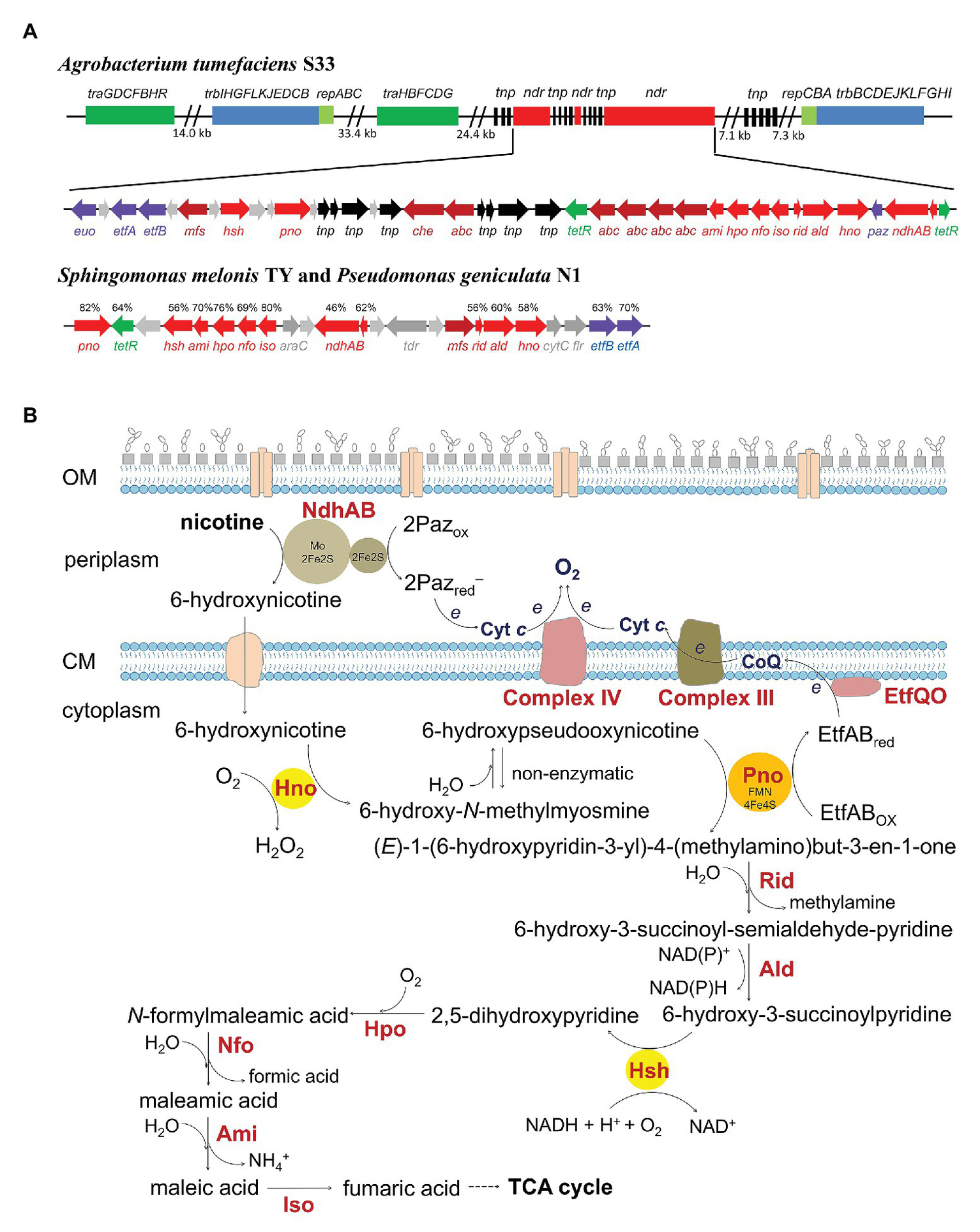
The genetic organization of the gene cluster involved in the hybrid nicotine-degrading pathway in A. tumefaciens S33, S. melonis TY, and P. geniculata N1 (A) and the proposed biochemical process of the hybrid for nicotine catabolism (B). traGDCFBHR and trbIHGFLKJEDCB, conjugal transfer proteins; repABC, plasmid partitioning and replication initiation proteins; ndr, nicotine-degrading gene cluster; tnp, transposase; euo, electron transfer flavoprotein:ubiquinone oxidoreductase (EtfQO); etfAB, electron transfer flavoprotein; mfs, major facilitator superfamily transporter; hsh, 6-hydroxy-3-succinoyl-pyridine hydroxylase; pno, 6-hydroxypseudooxynicotine dehydrogenase; che, chemotaxis protein; abc, ABC transporter; tetR, TetR family transcriptional regulator; ami, maleamate amidohydrolase; hpo, 2,5-dihydroxypyridine dioxygenase; nfo, N-formylmaleamate deformylase; iso, maleate cis/trans-isomerase; rid, Rid family protein; ald, aldehyde dehydrogenase; hno, 6-hydroxynicotine oxidase; paz, pseudoazurin; ndhAB, nicotine dehydrogenase; araC, helix-turn-helix (HTH) transcriptional regulator containing an AraC family HTH domain; tdr, TonB-dependent receptor; cytC, cytochrome c; flr, flavin reductase. The protein sequence identities compared with those from S33 are shown above the genes. The genetic organization in strain S33 in (A) is modified from Huang et al., 2017, permitted by a Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).
The core nicotine-degrading genes are highly conserved in the three strains, and their encoding proteins are almost identical. Most functional genes and their encoding enzymes have been characterized (see next section). There are still some genes whose functions are only predicted by annotation, including the genes for one major facilitator superfamily transporter (MFS), one chemotaxis protein (Che), five putrescine/spermidine ABC transporters (ABC), and two TetR family transcriptional regulators (TetR). They are predicted to function in substrate sensing and transport as well as transcriptional regulation in nicotine catabolism.
For the cases of S. melonis TY and P. geniculata N1 (Figure 2A), the organization, arrangement, and sequence of their nicotine-degrading genes is highly identical, which is different from the case of the three Rhizobiale species (Wang et al., 2017, 2019b). They are colocalized and tightly clustered without interruption by any mobile genetic element. The nicotine-degrading enzymes show 56–75% and 46–82% identity in DNA sequence and protein sequence, respectively, to those of Rhizobiale species. In addition, there are two genes for TetR transcriptional regulator and helix-turn-helix transcriptional regulator, which contains an AraC family HTH domain, in the gene cluster, and there are no genes for the ABC transporter, Che, Euo (electron transfer flavoprotein:ubiquinone oxidoreductase), or Paz (pseudoazurin). The MFS shows no significant similarity to the proteins of Rhizobiale species.
Alignment of the protein sequences of the enzymes with the corresponding enzymes involved in the pyridine and pyrrolidine pathways shows that the first three enzymes in the hybrid exhibit low identity to those in the pyridine pathway though they catalyze the same reactions (see the next section; Huang et al., 2017). In contrast, the enzymes for the late steps from 6-hydroxy-3-succinoylpyridine to fumaric acid in the hybrid are highly similar to those of the pyrrolidine pathway and the nicotinate-degrading pathway. This indicates that the enzymes for the starting steps of the hybrid evolve independently from the pyridine pathway, while others for the late steps are closely related to the enzymes in the pyrrolidine pathway and the nicotinate-degrading pathway in molecular evolution. The evolution of the hybrid pathway is therefore not a simple fusion of the genes involved in the pyridine and pyrrolidine pathways but a result of multiple complicated later gene transfers.
The Biochemical Basis for the Hybrid Pathway
In the hybrid pathway, nicotine is catabolized into fumaric acid via 10 steps, which then enters the TCA cycle. Nine core enzymes are responsible for the catalytic process, and the third step is a non-enzymatic reaction (Figure 2B). Among them, six are oxidoreductase, two are hydrolase, and one is an isomerase. In A. tumefaciens S33, nicotine dehydrogenase NdhAB, which catalyzes the initial step of hydroxylation of nicotine at the 6-position carbon atom of the pyridine ring, is located in the periplasm of cells, although it was co-purified with 6-hydroxy-pseudooxynicotine dehydrogenase (Pno) at the beginning because of their strong nonspecific interactions (Li et al., 2016; Yu et al., 2017a). Other enzymes in the pathway are found in the cytoplasm. There is a typical twin-arginine translocation system (Tat) signal peptide at the N-terminus of NdhA, which transports NdhAB into the periplasm after synthesis in the cytoplasm. The two-component enzyme harbors a molybdopterin cofactor on NdhA (82.4 kDa) and two [2Fe2S] clusters on NdhB (17.1 kDa), which is similar to the corresponding subunits of the classical three-component molybdopterin-containing hydroxylases belonging to the xanthine oxidase family such as nicotine dehydrogenase NdhLMS from A. nicotinovorans. However, NdhAB lacks a FAD-binding subunit, the middle size subunit of the classical hydroxylase. These cofactors of hydroxylase play important roles in transporting electrons from the reducing substrate to the electron acceptor. The typical hydroxylase usually uses NAD(P)+ or O2 as an electron acceptor, which reacts at the FAD site, and the electrons transfer between molybdopterin and FAD via the intervening [2Fe2S] clusters. Due to the lack of FAD, NdhAB cannot use NAD(P)+ or O2 but utilizes pseudoazurin (Paz) as its physiological electron acceptor. Paz is a 17-kDa blue copper protein and is also transported into the periplasm with the guidance of its special signal peptide. This is consistent with the fact that the gene paz is adjacent to ndhAB and that its transcriptional level is significantly upregulated when strain S33 grew on nicotine, which is similar to other nicotine-degrading genes. In the presence of Paz, NdhAB presents an activity of 110.5 U/mg at pH 7.0 and 30°C. The apparent Km values for nicotine and Paz are 1.6 and 3.6 μM, respectively. The 18O-labeled experiments indicate that the oxygen atom in the hydroxyl group of the product 6-hydroxynicotine is derived from H2O.
The oxidation of 6-hydroxynicotine is catalyzed by 6-hydroxynicotine oxidase (Hno) in the cytoplasm of cells (Yu et al., 2017a,b). The substrate 6-hydroxynicotine must be transported into the cytoplasm before oxidation. The predicted chemotaxis protein (Che), ABC transporter (Abc), and major facilitator superfamily transporter (Mfs) may be responsible for its sensing and transport. Their genes are clustered with the core nicotine-degrading genes and have been verified to be involved in nicotine degradation by transcriptomic analysis (Huang et al., 2017). Hno (48.7 kDa) binds an FAD with a Rossmann-like domain, and converts 6-hydroxynicotine into 6-hydroxy-N-methylmyosmine in the presence of O2 with the formation of H2O2 like other flavin-containing amine oxidases. Hno has a 38.9% identity to nicotine oxidase Nox from Pseudomonas sp. HZN6, 38.4% to nicotine oxidoreductase NicA2 from P. putida S16, and 24.8% to 6-hydroxy-L-nicotine oxidase from A. nicotinovorans. It presents a specific activity of 26.4 U/mg at 37°C and pH 9.2 with an apparent Km of 0.067 mM for 6-hydroxynicotine. The product 6-hydroxy-N-methylmyosmine is spontaneously hydrolyzed into 6-hydroxy-pseudooxynicotine. The pyrrolidine ring of nicotine is consequently opened.
The intermediate 6-hydroxy-pseudooxynicotine is further dehydrogenated into 6-hydroxy-3-succinoyl-semialdehyde-pyridine by 6-hydroxy-pseudooxynicotine dehydrogenase (Pno), which was formerly thought to be an oxidase since it catalyzes a reaction similar to that by pseudooxynicotine amine oxidase (Pao or Pnao) from Pseudomonas sp. HZN6 and P. putida S16 (Li et al., 2016; Wang et al., 2019a). By coupling with the reaction of 6-hydroxynicotine oxidation by Hno, the activity of Pno at 32.3 U/mg and an apparent Km of 0.37 mM for 6-hydroxy-pseudooxynicotine were estimated at pH 8.5 and 30°C. Because 6-hydroxy-pseudooxynicotine is not commercially available, its analog pseudooxynicotine was used as the substrate to characterize the enzyme. It presents a similar activity (52.8 U/mg) and apparent Km (0.42 mM) for pseudooxynicotine. In the assays, DCPIP serves as an artificial electron acceptor since NAD(P)+, O2, or ferredoxin does not function. Pno is a 73-kDa protein harboring an FMN, a [4Fe4S]-cluster, and an ADP. It shows almost no identity to Pao/Pano (less than 12%), but exhibits high identity to histamine dehydrogenase (48%) from Pimelobacter simplex, trimethylamine dehydrogenase (40%) from Methylophilus methylotrophus W3A1, and dimethylamine dehydrogenase (39%) from Hyphomicrobium sp. strain X. Recently, the crystal structure of the enzyme from P. geniculata N1 (82% identity to Pno from strain S33) was determined (Liu et al., 2020). It shows that FMN is not covalently bound to the protein and the isoalloxazine ring is planar, which is distinct from the case of trimethylamine dehydrogenase and histamine dehydrogenase. Differently, the enzyme from P. geniculata N1 is found to catalyze 6-hydroxy-pseudooxynicotine to 6-hydroxy-3-succinoyl-semialdehyde-pyridine and 6-hydroxy-3-succinoyl-pyridine with a very low activity (Wang et al., 2019b). Pno from strain S33 utilizes electron transfer flavoprotein (EtfAB) as the physiological electron acceptor, which is similar to trimethylamine dehydrogenase and dimethylamine dehydrogenase. The reduced EtfAB can be reoxidized by electron transfer flavoprotein:ubiquinone oxidoreductase (Euo) with CoQ as the electron acceptor. The etfAB and euo genes are located in the nicotine-degrading gene cluster, and their transcriptional levels are also up-regulated in the presence of nicotine. The 18O-labeling experiments showed that the oxygen atom in the aldehyde group of the product 3-succinoyl-semialdehyde-pyridine was derived not from O2 but from H2O. This evidence indicates that Pno is a new member of EC 1.5.8., which is a group of dehydrogenases acting on the CH-NH group of electron donors with a flavin (EtfAB) as an electron acceptor. Considering the role of Pno in vivo, it is assumed that 6-hydroxy-pseudooxynicotine is dehydrogenated to an enamine intermediate [(E)-1-(6-hydroxypyridin-3-yl)-4-(methylamino)but-3-en-1-one] by Pno, which is further hydrolyzed to form the deamination product 6-hydroxy-3-succinoyl-semialdehyde-pyridine and methylamine.
It is known that enamine is a highly reactive and unstable compound, which can be isomerized to an imine (Borchert et al., 2019). The two intermediates usually can be spontaneously hydrolyzed into a harmless ketoacid, but this reaction is very slow because of the shortage of free water molecules in vivo, which causes their accumulation in cells. This will lead to metabolic damage to the cells because they can covalently bind to pyridoxal 5′-phosphate (PLP) to inactivate PLP-dependent enzymes. RidA protein has been found to preempt the damage of enamine compounds by hydrolyzing them via its deaminase activity (Lambrecht et al., 2013). In the nicotine-degrading gene cluster of strain S33, an ORF is annotated to encode a Rid6 subfamily protein, whose transcriptional level is also up-regulated in the presence of nicotine. We predict that Rid6 is required to remove the toxic enamine/imine intermediate produced in the Pno reaction in vivo.
In the next step of the reaction, 6-hydroxy-3-succinoyl-semialdehyde-pyridine is oxidized to 6-hydroxy-3-succinoyl-pyridine by a predicted NAD(P)-dependent aldehyde dehydrogenase (Ald), which is encoded by an ORF adjacent to the hno gene in the nicotine-degrading gene cluster (Huang et al., 2017). Like other genes in the cluster, its transcriptional level presents a similar up-regulated pattern with nicotine. The protein (49.9 kDa) has 35% identity to the NADP-dependent 3-succinoyl-semialdehyde-pyridine dehydrogenase (Sapd) from Pseudomonas sp. HZN6 (Qiu et al., 2012). Its detailed biochemical properties require further characterization.
The sixth step is oxidative decarboxylation of 6-hydroxy-3-succinoyl-pyridine to 2,5-dihydroxypyridine and succinic acid by 6-hydroxy-3-succinoyl-pyridine hydroxylase (Hsh; Li et al., 2014). Hsh is a 90 kDa homodimeric FAD-containing monooxygenase, which exhibits 62.8% identity to with HSP hydroxylase (HspB) in the pyrrolidine pathway of P. putida S16. The enzyme also presents a high identity to p-nitrophenol monooxygenase from Pseudomonas sp. WBC-3 (38%) and Pseudomonas sp. NyZ402 (37%), but low identification (approximately 17%) with 6-hydroxynicotinate monooxygenase from P. putida KT2440. The protein has two conserved Rossmann fold motifs, which each bind FAD and NADH. However, FAD is easily disassociated from the enzyme during purification. The purified Hsh demonstrates a specific activity of 18.8 U/mg, an apparent Km of 0.15 mM for 6-hydroxy-3-succinoyl-pyridine, and an apparent Km of 0.1 mM for NADH at pH 8.0 and 30°C. The enzyme can also use NADPH to substitute NADH in the reaction with a specific activity of 12.9 U/mg and an apparent Km of 0.35 mM. 18O-labeling experiments for HspB from P. putida S16 showed that the oxygen atom incorporated into the product 2,5-dihydroxypyrdine is derived from O2, while the oxygen introduced into the product succinic acid is from H2O (Tang et al., 2011; Yu et al., 2014). Since the second oxygen atom from O2 is finally reduced to H2O, H2O is both a substrate and product in the reaction. Rapid reaction kinetic experiments have shown that HspB is first reduced on its flavin by NADH, then oxidized by O2 to form a C(4a)-hydroperoxyflavin intermediate, which further reacts with the substrate 6-hydroxy-3-succinoyl-pyridine to form 2,5-dihydroxypyrdine and succinic acid (Yu et al., 2014).
The four enzymes responsible for the last steps of conversion from 2,5-dihydroxypyrdine to fumaric acid in the hybrid pathway are very close to those in the pyrrolidine pathway in P. putida S16 and the nicotinate-degrading pathway in P. putida KT2440 (Jiménez et al., 2008; Tang et al., 2012; Yao et al., 2013; Huang et al., 2017). The oxidation of 2,5-dihydroxypyridine to break the pyridine ring into N-formylmaleamic acid is catalyzed by 2,5-dihydroxypyridine dioxygenase (Hpo, 37.6 kDa for monomer), which exhibits 81% identity to the enzyme from P. putida S16 and 42.9% to NicX from P. putida KT2440. The enzyme from strain S16 is an Fe(II)-dependent homotrimer and presents a specific activity of 20.5 U/mg and an apparent Km of 0.14 uM for 2,5-dihydroxypyridine in the presence of Fe2+ at pH 7.5 and 20°C (Tang et al., 2012; Yao et al., 2013). N-formylmaleamate deformylase (Nfo, 28.1 kDa) catalyzes the hydrolysis of N-formylmaleamic acid into maleamic and formic acids. The enzyme exhibits an identity of 62.3% to Nfo from strain S16 and 58.2% identity to NicD from strain KT2440. Nfo from strain S16 shows a specific activity of 1.1 U/mg and an apparent Km of 0.94 mM for N-formylmaleamic acid at pH 6.7 and 20°C. Next the hydrolysis of maleamic acid into maleic acid and ammonia is catalyzed by maleamate amidohydrolase (Ami, 22.5 kDa), which has 64.8% identity to Ami from strain S16 and 36.7% to NicF from strain KT2440. Finally, maleate cis/trans-isomerase (Iso, 26.8 kDa) catalyzes the isomerization of maleic acid into fumaric acid, which exhibits an identity of 78% to Iso from strain S16 and 71.6% to NicE from strain KT2440. Iso from strain S16 presents a specific activity of 9.4 U/mg and an apparent Km of 0.69 mM for maleic acid at pH 8.4 and 30°C. The crystal structures of Nfo, Ami, and Iso from strain S16 have been reported, providing a comprehensive understanding of the catalytic mechanisms of these enzymes (Chen et al., 2013; Wu et al., 2014). Finally, the end product fumaric acid enters the classic TCA cycle for further catabolism.
A. tumefaciens S33, Shinella sp. HZN7, and Ochrobactrum sp. SJY1 can grow aerobically with nicotine as the sole source of carbon and nitrogen. They catabolize the alkaloid with the nicotine-degrading enzymes to support their life. Meanwhile, they conserve energy from its oxidation. In connection with the TCA cycle, oxidation of nicotine provides metabolites, reducing power (electrons), and energy (ATP) for anabolism. Among the six oxidoreductive reactions in the hybrid pathway, three are catalyzed by oxidases which transfer electrons from their substrates directly to O2 (Figure 2B). The other three reactions deliver electrons to an electron carrier. NdhAB utilizes Paz as its electron acceptor, which can further deliver the electrons to cytochrome c and then to the final electron acceptor O2 catalyzed by cytochrome c oxidase, or directly to O2 considering the pseudospecificity between Paz and cytochrome c (Yu et al., 2017a). Pno transfers the electron to EtfAB, and then to CoQ catalyzed by Euo, which enters the classic electron transport chain (ETC; Wang et al., 2019a). Ald uses NAD(P) as an electron acceptor, and the reduced equivalent can be reoxidized when it takes part in the reaction of Hsh. Alternately, the reduced equivalent also can be reoxidized by NADH dehydrogenase complex, by which the electrons enter the classic ETC. The routes for electron transport involved in nicotine oxidation via the hybrid pathway are thus constructed, and ATP can be further synthesized via chemiosmosis.
Perspectives
The genomic, transcriptomic, and biochemical analyses have revealed the mysterious mechanism of the novel hybrid pathway. To completely understand the molecular mechanism, further studies on the molecular evolution, transcription regulation, substrate sensing and transportation, and detoxification of reactive intermediates are still needed. In addition, it is crucial to resolve the structures of some key enzymes, transcriptional regulators, and chemotaxis proteins to discover their catalytic mechanism and substrate sensing/interaction. This will benefit the development of new technologies, for example, transforming nicotine into valuable chemicals or constructing bio-sensors for detecting nicotine and its metabolites with wild-type or engineered whole cells and enzymes. Recently, whole cells of genetically engineered strain S33 have been used as catalysts to transform nicotine from tobacco wastes to 6-hydroxynicotine (Yu et al., 2017b). An enzymatic route has also been constructed to produce 2,5-dihydroxypyridine (Li et al., 2014). A more extensive investigation of the enzymes will consequently pave the way for their application within industrial biocatalysis.
Statements
Author contributions
HH and SW designed the minireview. All authors wrote, revised, and approved the manuscript.
Funding
This work was supported by the grant from the National Natural Science Foundation of China (grant no. 31970057) and the 111 Project (no. B16030).
Acknowledgments
We thank Professor Ping Xu from Shanghai Jiao Tong University for his valuable support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Benowitz N. L. (2010). Nicotine addiction. N. Engl. J. Med.362, 2295–2303. doi: 10.1056/NEJMra0809890
2
Borchert A. J. Ernst D. C. Downs D. M. (2019). Reactive enamines and imines in vivo: lessons from the RidA paradigm. Trends Biochem. Sci.44, 849–860. doi: 10.1016/j.tibs.2019.04.011
3
Brandsch R. (2006). Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol.69, 493–498. doi: 10.1007/s00253-005-0226-0
4
Bush L. Hempfling W. P. Burton H. (1999). “Biosynthesis of nicotine and related compounds” in Analytical determination of nicotine and related compounds and their metabolites. eds. GorrodJ. W.JacobP. I. (Amsterdam: Elsevier).
5
Campain J. A. (2004). Nicotine: potentially a multifunctional carcinogen?Toxicol. Sci.79, 1–3. doi: 10.1093/toxsci/kfh106
6
Chen D. D. Tang H. Z. Lv Y. Zhang Z. Y. Shen K. L. Lin K. et al . (2013). Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside. Mol. Microbiol.87, 1237–1244. doi: 10.1111/mmi.12163
7
Hecht S. S. (1999). Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst.91, 1194–1210. doi: 10.1093/jnci/91.14.1194
8
Huang H. Yu W. Wang R. Li H. Xie H. Wang S. (2017). Genomic and transcriptomic analyses of Agrobacterium tumefaciens S33 reveal the molecular mechanism of a novel hybrid nicotine-degrading pathway. Sci. Rep.7:4813. doi: 10.1038/s41598-017-05320-1
9
Jiménez J. I. Canales A. Jimenez-Barbero J. Ginalski K. Rychlewski L. Garcia J. L. et al . (2008). Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. U. S. A.105, 11329–11334. doi: 10.1073/pnas.0802273105
10
Lambrecht J. A. Schmitz G. E. Downs D. M. (2013). RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. MBio4, e00033–e00013. doi: 10.1128/mBio.00033-13
11
Li H. Xie K. Huang H. Wang S. (2014). 6-Hydroxy-3-succinoylpyridine hydroxylase catalyzes a central step of nicotine degradation in Agrobacterium tumefaciens S33. PLoS One9:e103324. doi: 10.1371/journal.pone.0103324
12
Li H. Xie K. Yu W. Hu L. Huang H. Xie H. et al . (2016). Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in Agrobacterium tumefaciens S33. Appl. Environ. Microbiol.82, 1745–1755. doi: 10.1128/AEM.03909-15
13
Liu G. Wang W. He F. Zhang P. Xu P. Tang H. (2020). Structural insights into 6-hydroxypseudooxynicotine amine oxidase from Pseudomonas geniculata N1: the key enzyme involved in nicotine degradation. Appl. Environ. Microbiol. doi: 10.1128/AEM.01559-20 [Epub ahead of print]
14
Ma Y. Wei Y. Qiu J. Wen R. Hong J. Liu W. (2014). Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl. Microbiol. Biotechnol.98, 2625–2636. doi: 10.1007/s00253-013-5207-0
15
Meng X. J. Lu L. L. Gu G. F. Xiao M. (2010). A novel pathway for nicotine degradation by Aspergillus oryzae 112822 isolated from tobacco leaves. Res. Microbiol.161, 626–633. doi: 10.1016/j.resmic.2010.05.017
16
Mu Y. Chen Q. Parales R. E. Lu Z. M. Hong Q. He J. et al . (2020). Bacterial catabolism of nicotine: catabolic strains, pathways and modules. Environ. Res.183:109258. doi: 10.1016/j.envres.2020.109258
17
Nakano H. Wieser M. Hurh B. Kawai T. Yoshida T. Yamane T. et al . (1999). Purification, characterization and gene cloning of 6-hydroxynicotinate 3-monooxygenase from Pseudomonas fluorescens TN5. Eur. J. Biochem.260, 120–126. doi: 10.1046/j.1432-1327.1999.00124.x
18
Novotny T. E. Zhao F. (1999). Consumption and production waste: another externality of tobacco use. Tob. Control.8, 75–80. doi: 10.1136/tc.8.1.75
19
Ondachi P. W. Comins D. L. (2010). Synthesis of fused-ring nicotine derivatives from (S)-nicotine. J. Organomet. Chem.75, 1706–1716. doi: 10.1021/jo9026929
20
Qiu J. Ma Y. Wen Y. Chen L. Wu L. Liu W. (2012). Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl. Environ. Microbiol.78, 2154–2160. doi: 10.1128/AEM.07025-11
21
Qiu J. Yang Y. Zhang J. Wang H. Ma Y. He J. et al . (2016). The complete genome sequence of the nicotine-degrading bacterium Shinella sp. HZN7. Front. Microbiol.7:1348. doi: 10.3389/fmicb.2016.01348
22
Roduit J. P. Wellig A. Kiener A. (1997). Renewable functionalized pyridines derived from microbial metabolites of the alkaloid (S)-nicotine. Heterocycles45, 1687–1702. doi: 10.3987/COM-97-7827
23
Schmid A. Dordick J. S. Hauer B. Kiener A. Wubbolts M. Witholt B. (2001). Industrial biocatalysis today and tomorrow. Nature409, 258–268. doi: 10.1038/35051736
24
Tang H. Wang L. Wang W. Yu H. Zhang K. Yao Y. et al . (2013). Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet.9:e1003923. doi: 10.1371/journal.pgen.1003923
25
Tang H. Yao Y. Wang L. Yu H. Ren Y. Wu G. et al . (2012). Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci. Rep.2:377. doi: 10.1038/srep00377
26
Tang H. Yao Y. Zhang D. Meng X. Wang L. Yu H. et al . (2011). A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J. Biol. Chem.286, 39179–39187. doi: 10.1074/jbc.M111.283929
27
Wang S. Huang H. Xie K. Xu P. (2012). Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl. Microbiol. Biotechnol.95, 1567–1578. doi: 10.1007/s00253-012-4007-2
28
Wang S. N. Liu Z. Tang H. Z. Meng J. Xu P. (2007). Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype a strain S16. Microbiology153, 1556–1565. doi: 10.1099/mic.0.2006/005223-0
29
Wang S. N. Xu P. Tang H. Z. Meng J. Liu X. L. Ma C. Q. (2005). “Green” route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a Pseudomonas sp. Environ. Sci. Technol.39, 6877–6880. doi: 10.1021/es0500759
30
Wang R. Yi J. Shang J. Yu W. Li Z. Huang H. et al . (2019a). 6-Hydroxypseudooxynicotine dehydrogenase delivers electrons to electron transfer flavoprotein during nicotine degradation by Agrobacterium tumefaciens S33. Appl. Environ. Microbiol.85, e00454–e00419. doi: 10.1128/AEM.00454-19
31
Wang H. X. Zhi X. Y. Qiu J. G. Shi L. X. Lu Z. M. (2017). Characterization of a novel nicotine degradation gene cluster ndp in Sphingomonas melonis TY and its evolutionary analysis. Front. Microbiol.8:337. doi: 10.3389/fmicb.2017.00337
32
Wang W. Zhu X. Liu X. Wu W. Xu P. Tang H. (2019b). Cloning and characterization the nicotine degradation enzymes 6-hydroxypseudooxynicotine amine oxidase and 6-hydroxy-3-succinoylpyridine hydroxylase in Pseudomonas geniculata N1. Int. Biodeterior. Biodegrad.142, 83–90. doi: 10.1016/j.ibiod.2019.05.003
33
Wu G. Chen D. D. Tang H. Z. Ren Y. L. Chen Q. H. Lv Y. et al . (2014). Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases. Mol. Microbiol.91, 1009–1021. doi: 10.1111/mmi.12511
34
Yao Y. X. Tang H. Z. Ren H. X. Yu H. Wang L. J. Zhang W. et al . (2013). Iron(II)-dependent dioxygenase and N-formylamide deformylase catalyze the reactions from 5-hydroxy-2-pyridone to maleamate. Sci. Rep.3:3235. doi: 10.1038/srep03235
35
Yu H. Hausinger R. P. Tang H. Z. Xu P. (2014). Mechanism of the 6-hydroxy-3-succinoyl-pyridine 3-monooxygenase flavoprotein from Pseudomonas putida S16. J. Biol. Chem.289, 29158–29170. doi: 10.1074/jbc.M114.558049
36
Yu W. Li H. Xie K. Huang H. Xie H. Wang S. (2016). Genome sequence of the nicotine-degrading Agrobacterium tumefaciens S33. J. Biotechnol.228, 1–2. doi: 10.1016/j.jbiotec.2016.04.029
37
Yu H. Tang H. Zhu X. Li Y. Xu P. (2015). Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1. Appl. Environ. Microbiol.81, 272–281. doi: 10.1128/AEM.02265-14
38
Yu W. Wang R. Huang H. Xie H. Wang S. (2017a). Periplasmic nicotine dehydrogenase NdhAB utilizes pseudoazurin as its physiological electron acceptor in Agrobacterium tumefaciens S33. Appl. Environ. Microbiol.83, e01050–e01017. doi: 10.1128/AEM.01050-17
39
Yu W. Wang R. Li H. Liang J. Wang Y. Huang H. et al . (2017b). Green route to synthesis of valuable chemical 6-hydroxynicotine from nicotine in tobacco wastes using genetically engineered Agrobacterium tumefaciens S33. Biotechnol. Biofuels10:288. doi: 10.1186/s13068-017-0976-9
Summary
Keywords
nicotine, degrading pathway, enzymes, gene cluster, catabolism, functionalized pyridines
Citation
Huang H, Shang J and Wang S (2020) Physiology of a Hybrid Pathway for Nicotine Catabolism in Bacteria. Front. Microbiol. 11:598207. doi: 10.3389/fmicb.2020.598207
Received
24 August 2020
Accepted
07 October 2020
Published
12 November 2020
Volume
11 - 2020
Edited by
Kylie Allen, Virginia Tech, United States
Reviewed by
Jiguo Qiu, Nanjing Agricultural University, China; Qing Chen, Zaozhuang University, China
Updates
Copyright
© 2020 Huang, Shang and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuning Wang, shuningwang@sdu.edu.cn
This article was submitted to Microbial Physiology and Metabolism, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.