- 1Key Laboratory of Fermentation Engineering (Ministry of Education), National “111” Center for Cellular Regulation and Molecular Pharmaceutics, Hubei Provincial Cooperative Innovation Center of Industrial Fermentation, College of Bioengineering, Hubei University of Technology, Wuhan, China
- 2ABI Group, College of Marine Science and Technology, Zhejiang Ocean University, Zhoushan, China
- 3State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, China
- 4Tobacco Research Institute of Hubei Province, Wuhan, China
2-Phenylethanol (2-PE) is an important flavouring ingredient with a persistent rose-like odour, and it has been widely utilized in food, perfume, beverages, and medicine. Due to the potential existence of toxic byproducts in 2-PE resulting from chemical synthesis, the demand for “natural” 2-PE through biotransformation is increasing. L-Phenylalanine (L-Phe) is used as the precursor for the biosynthesis of 2-PE through the Ehrlich pathway by Saccharomyces cerevisiae. The regulation of L-Phe metabolism in S. cerevisiae is complicated and elaborate. We reviewed current progress on the signal transduction pathways of L-Phe sensing, uptake of extracellular L-Phe and 2-PE synthesis from L-Phe through the Ehrlich pathway. Moreover, the anticipated bottlenecks and future research directions for S. cerevisiae biosynthesis of 2-PE are discussed.
Introduction
2-Phenylethanol (2-PE) is a higher alcohol with a rose-like odour. 2-PE contributes significantly to the flavour and aroma of beer, bread, cocoa, cheese, soy sauce, and other fermented foods and has been widely used in the perfume, cosmetics, and food industries (Chung et al., 2000; Stark et al., 2002). 2-PE is also the precursor for the production of 2-phenylethyl acetate (2-PEA), which is an important flavouring agent with floral and rose-like odours (Carlquist et al., 2015). Moreover, 2-PE is utilized in sanitation and hygiene products, which mainly rely on its antifungal and antibacterial characteristics (Etschmann et al., 2002). Currently, the Flavour and Extract Manufacturers Association (FEMA), the Food and Drug Administration (FDA), the Joint Expert Committee on Food Additives (JECFA), the Council of Europe (COE), and other international organizations have approved the use of 2-PE as a flavouring agent in food (Scognamiglio et al., 2012).
At present, the global market demand for 2-PE is increasing every year, with an annual global demand of 1,000 tons in 2011, equal to a market value of $700 million (Hua and Xu, 2011). Currently, most 2-PE is synthesized by chemical methods, including the Friedel-Craft reaction of ethylene oxide with benzene, catalytic reduction of styrene oxide, and oxidation of propylene with 2-phenylethyl hydroperoxide (Martínez-Avila et al., 2018). This process involves high temperatures (>300°C) and toxic chemicals (benzene and styrene) and leads to the restricted availability of 2-PE (Chreptowicz et al., 2016; Martínez-Avila et al., 2018). Concerning environmental issues and health hazards, 2-PE from chemical synthesis is less preferred or restricted in the food and cosmetics industries. Although 2-PE is found naturally in some plants, such as rose, hyacinths, and jasmine, and natural 2-PE can be extracted from essential oils of flowers, the volume of natural 2-PE extracted from flowers is limited, and the market price of 2-PE is as high as $1,000/kg (Hua and Xu, 2011).
The United States Food and Drug Administration and European legislation have determined that 2-PE from microbial synthesis is considered “natural” (Hua and Xu, 2011). Therefore, the biotransformation of natural 2-PE has received increasing attention and may be the most effective alternative (Lukito et al., 2019). Previous studies have demonstrated that Saccharomyces cerevisiae, Kluyveromyces marxianus, Kluyveromyces lactis, Pichia fermentans, Pichia anomala, Schizosaccharomyces pombe, Yarrowia lipolytica, Zygosaccharomyces rouxii, and Hansenula anomala can synthesize 2-PE from L-phenylalanine (L-Phe) or glucose through the Ehrlich pathway and phenylpyruvate (PPA) pathway (Romagnoli et al., 2015; Martínez-Avila et al., 2018; Cordente et al., 2019; Hassing et al., 2019).
Although microorganisms possess the ability to synthesize 2-PE from glucose through the PPA pathway, the process is very complex, with many metabolic branches competing for carbon flow. In addition, 2-PE itself is toxic to microbial cells, and 2-PE biosynthesis is strongly feedback-inhibited by L-Phe. Therefore, the efficiency of the phenylpyruvate pathway is very low (Hassing et al., 2019).
2-Phenylethanol can also be efficiently synthesized from L-Phe through the Ehrlich pathway, which consists of three steps, transamination, decarboxylation, and reduction (Hazelwood et al., 2008). To improve 2-PE production, various methods have been employed, including strain mutagenesis and selection, medium composition and culture condition optimization, and in situ product removal techniques (Hua and Xu, 2011; Martínez-Avila et al., 2018; Qian et al., 2019; Wang et al., 2019). In this review, we focus on the regulation of the sensing, transportation, and catabolism of L-Phe to produce 2-PE.
Signal Transduction of L-Phe
Yeast cells can use various amino acids for growth. To discriminate amino acids, S. cerevisiae has evolved to have a complete extracellular amino acid-sensing system [Ssy1-Ptr3-Ssy5 signalling sensor system or Ssy1-Ptr3-Ssy5 (SPS) sensor system] and intracellular amino acid-sensing system [target of rapamycin (TOR) pathway], which are crucial for sensing extracellular and intracellular amino acids, respectively (Klasson et al., 1999; Forsberg and Ljungdahl, 2001; Schneper et al., 2004; Zhang et al., 2018).
External L-Phe Sensing by the SPS Sensor System
Extracellular amino acids can be detected via the SPS sensor pathway in S. cerevisiae, and some amino acid permeases are regulated by the SPS sensor pathway. The sensing of extracellular amino acids is mostly controlled by the signal transduction pathway, which in turn, regulates the dynamic interactions between transcription factors and specific promoter binding sites (Figure 1). The SPS sensor system is a plasma membrane (PM)-localized complex consisting of three core components, Ssy1, Ptr3, and Ssy5 (Forsberg and Ljungdahl, 2001). Ssy1 is an important component of the PM that exhibits high sequence similarity with amino acid permease families (Didion et al., 1998; Gaber et al., 2003). However, Ssy1 differs from amino acid permeases because of its long cytoplasmically oriented N-terminal domain and inability to transport amino acids. In addition, Ssy1 serves as an extracellular amino acid receptor and a scaffold that concatenates Ptr3 and Ssy5, as well as other membrane proteins, through its long cytoplasmically oriented N-terminal domain (Iraqui et al., 1999b; Klasson et al., 1999).
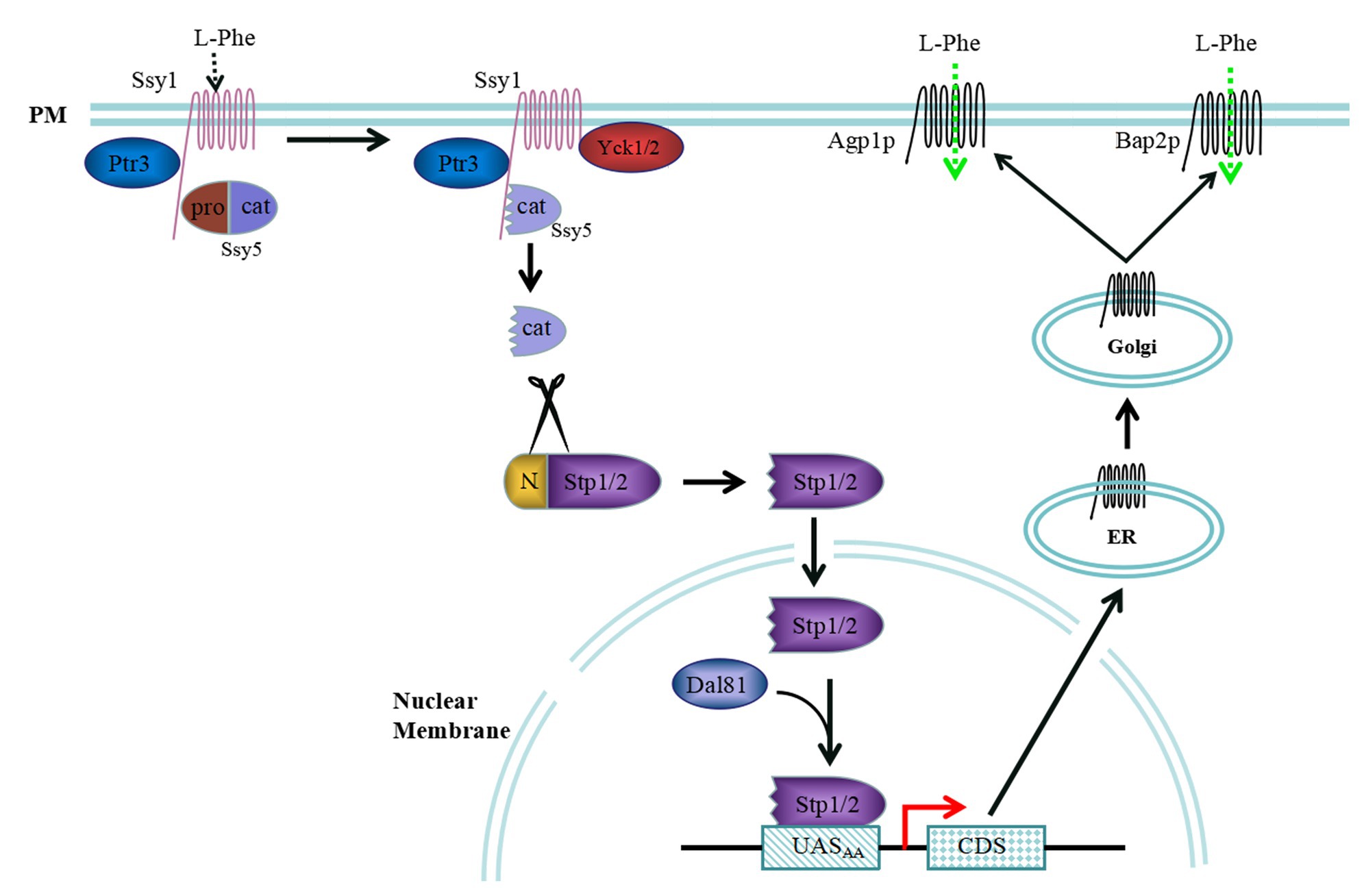
Figure 1. Extracellular L-Phenylalanine (L-Phe) signalling pathway mediated by the Ssy1-Ptr3-Ssy5 (SPS) sensor system. When L-Phe is the sole nitrogen source, Ssy1 may react to it and recruit casein kinase Yck1/2 to phosphorylate the N-terminus of Ssy5. The N-terminus of Ssy5 is ubiquitinated and degraded to free the Cat domain, which cleaves the N-terminal regulatory domains of Stp1 and Stp2. The shorter forms of Stp1 and Stp2 target the nucleus and bind the UASAA elements AGP1 and BAP2, activating expression of these genes. Then, Agp1p and Bap2p are secreted into the membrane of the endoplasmic reticulum, where they are processed, modified, and transferred to the Golgi apparatus for further processing and packaging. Finally, Agp1p and Bap2p are localized to the cell membrane and transport extracellular L-Phe into cells.
Ssy5, a core component of the SPS, is a serine protease expressed as an inactive zymogen that contains a regulatory N-terminal pro-domain and noncovalently bound C-terminal catalytic-domain (Martins et al., 2018). The endoprotease activity of Ssy5 is inhibited by its own regulatory N-terminal pro-domain. In response to extracellular amino acids, Ssy5 undergoes an autocatalytic event upon proteasomal degradation of its regulatory N-terminal pro-domain, leading to the activation of its endo-protease activity (Martins et al., 2019). Once an extracellular amino acid signal is received, the Ssy1 conformation is altered, which recruits the casein kinase Yck1/2. The N-terminal pro-domain of Ssy5 is phosphorylated by Yck1/2, as facilitated by Ptr3, which is then modified by the ubiquitin ligase complex. Finally, the pro-domain is degraded by the 26S proteasome, freeing the Cat domain (Abdel-Sater et al., 2011; Omnus and Ljungdahl, 2013).
The transcription factors Stp1 and Stp2 are cleaved by the freed Cat domain, and the shortened Stp1 and Stp2 peptides are translocated into the nucleus, where they bind to the promoter region of the SPS regulatory gene to induce transcription (Tumusiime et al., 2011; Omnus and Ljungdahl, 2014). Stp1 and Stp2 are initially produced with N-terminal regulatory domains, preventing them from entering the nucleus. Stp1 and Stp2 are homologous 10-kDa zinc finger transcription factors that serve as downstream effectors of the SPS sensor system, and their N-terminal domains are crucial for their activity (Andréasson and Ljungdahl, 2002). Stp1 and Stp2 possess two regulatory motifs, I (RI) and II (RII), and RII has an endoprotease-processing site that is required for the cleavage of Stp1 between cysteine 85 and serine 86 by the Ssy5 Cat-domain (Andréasson and Ljungdahl, 2004; Omnus et al., 2016). Upon amino acid induction, the Stp1/2 N-terminal regulatory domains are degraded by the SPS sensor controller Ssy5 signalling protease, and the shorter Stp1/2 are then localized to the nucleus, where they bind to the specific upstream activation element UASAA of the targeted genes, namely, AGP1, BAP2, BAP3, GNP1, DIP5, and MUP1. However, complete Stp1 and Stp2 are widely distributed in the cytoplasm and cannot enter the nucleus in the absence of amino acids.
When L-Phe is used as the sole nitrogen source to produce 2-PE, Ssy1 located in the plasma membrane may react with it and recruit casein kinase Yck1/2 to phosphorylate the Ssy5 N-terminus. The Ssy5 N-terminus is then ubiquitinated and degraded to free the Cat domain, which cleaves the N-terminal regulatory domains of Stp1 and Stp2. The shorter forms of Stp1 and Stp2 pass through the nuclear membrane and bind to the UASAA elements AGP1, BAP2, and BAP3, inducing the expression of these three genes. Agp1p, Bap2p, and Bap3p are secreted into the endoplasmic reticulum membrane adjacent to the nuclear membrane, where they are processed and modified and then transferred to the Golgi apparatus for further processing and packaging. Finally, Agp1p, Bap2p, and Bap3p are localized to the cell membrane and transport extracellular L-Phe into cells (Figure 1).
In addition, a small amount of complete Stp1 and Stp2 can leak into the nucleus, where they can bind to the upstream activation sequence of a target gene to facilitate Dal81/Uga35 function. However, the nuclear membrane protein Asi1-3 localized to the nuclear membrane plays important roles in the cytoplasmic retention of Stp1 and Stp2 and can prevent the complete Stp1 and Stp2 proteins from entering the nucleus (Zargari et al., 2007).
Intracellular Amino Acid Sensing by the TOR Pathway
In eukaryotic cells, the TOR signalling pathway controls cell growth and proliferation. Yeast cells recognize intracellular amino acid conditions through the TOR-sensing pathway, which responds to the availability of amino acids (Schneper et al., 2004). The TOR signalling cascade includes the EGOC complex, an upstream regulatory element, TOR complex 1 (TORC1), and downstream effectors (Tap42-PPase and Sch9). TORC1 is inhibited by rapamycin and is structurally and functionally conserved. As the core component of the TOR signalling pathway, TORC1 is composed of the Tor1, Kog1, Tco89, and Lst8 proteins (Reinke et al., 2004), and TORC1 primarily recognizes the amino acid/nitrogen conditions in yeast cells. Upon nitrogen starvation/reduction or rapamycin treatment, TORC1 activity is restrained. An increase in nitrogen or cycloheximide treatment results in the activation of TORC1 (Binda et al., 2009). The intracellular amino acid signal is transmitted to the related protein of TORC1 through the upstream component EGOC to activate or inhibit the activity of TORC1 (Figure 2).
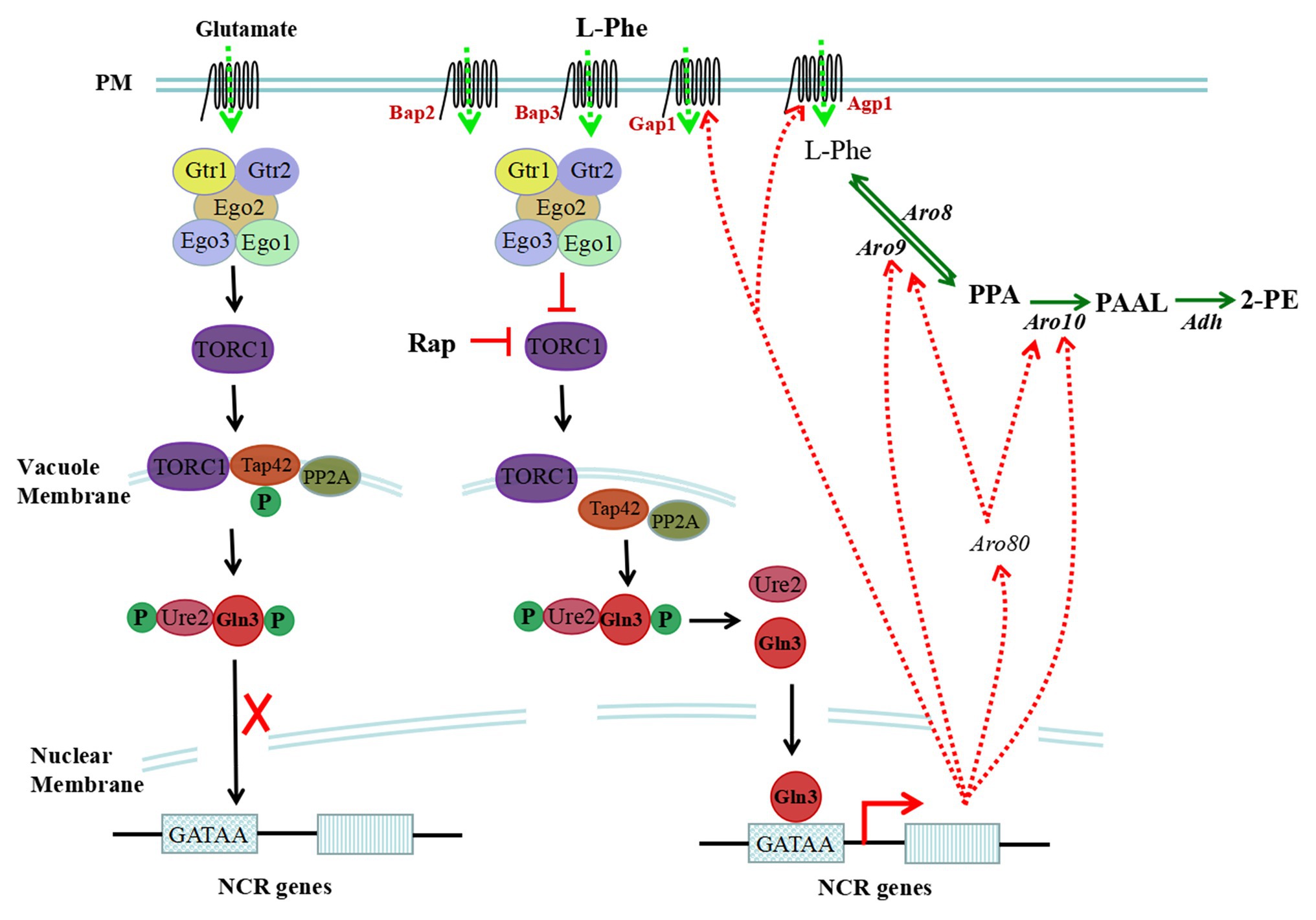
Figure 2. Sensing and regulation of intracellular L-Phe levels through the target of rapamycin (TOR) pathway. The TOR signalling cascade includes the EGOC complex, TOR complex 1 (TORC1), and downstream effectors (Tap42-PPase and Sch9). Intracellular L-Phe is sensed by the EGOC complex, and TORC1 can be inhibited. However, upon rapamycin treatment or in the presence of a poor nitrogen source, the activity of TORC1 can be restrained, which results in the dephosphorylation of Tap42, freeing it from the vacuole membrane. Gln3 is dephosphorylated by freed Tap42 and is freed from Ure2; then, it targets the nucleus and binds to the GATAA/G motif of nitrogen catabolite repression (NCR)-sensitive genes, activating the transcription of NCR-sensitive genes. In the presence of glutamate, TORC1 is activated and Gln3 cannot be dephosphorylated and resides in the cytoplasm, which represses the expression of NCR-sensitive genes.
EGOC includes structural subunits and regulatory subunits in S. cerevisiae, and the structural subunits are EGO/Rag complexes consisting of Ego1, Ego2, and Ego3 (Dubouloz et al., 2005; Powis et al., 2015). EGOC is bound to a vacuole membrane by Ego1. The regulatory subunit consists of Gtr1 and Gtr2, and the nucleotide-binding status of Gtr1 and Gtr2 regulates the activity of TORC1 (Binda et al., 2009; Zhang et al., 2018). Upon binding to GTP, Gtr1 interacts with the Tco89 and Kog1 proteins, and TORC1 is activated to block the utilization of poor nitrogen sources, which is hindered by leucine starvation (Kim et al., 2008; Broach, 2012; Powis et al., 2015). The nucleotide binding state of Gtr1 is regulated by several regulatory factors, including guanine nucleotide exchange factor (GEF; Sancak et al., 2008; Binda et al., 2009), GTPase activating protein (GAP; Powis and De Virgilio, 2016), and the Seh1-associated complex proteins Npr2 and Npr3 (Neklesa and Davis, 2009). These regulatory factors are sensitive to the levels of intracellular nitrogen and amino acids and activate or inhibit the activity of TORC1 to modulate intracellular nitrogen and amino acid metabolism through the downstream regulatory factors Tap42 and Sch9. Intracellular glutamate, sensed by the EGOC complex, activates TORC1 and represses the expression of nitrogen catabolite repression (NCR)-sensitive genes in S. cerevisiae. However, L-Phe is a nonpreferred nitrogen source for S. cerevisiae and is sensed by EGOC, which may restrain the activity of TORC1 and derepress the expression of NCR-sensitive genes (Figure 2).
Regulation of NCR via the TOR Signalling Pathway
Saccharomyces cerevisiae exhibits a hierarchical preference for nitrogen sources, which are usually classified as preferred nitrogen sources and poor nitrogen sources. Saccharomyces cerevisiae cultured in the presence of nitrogen sources with different qualities presents sequential utilization of preferred, intermediate, and poor nitrogen sources, which is controlled by NCR (Henschke and Jiranek, 1993; Boer et al., 2007). The preferred nitrogen sources include ammonium salts, glutamate, glutamine, asparagine, and other nitrogen sources. The poor nitrogen sources include methionine, proline, allantoin, γ-aminobutyric acid, urea, and other nitrogen sources (Magasanik and Kaiser, 2002; Godard et al., 2007). However, leucine and phenylalanine are considered to be “intermediate” nitrogen sources (Boer et al., 2007). The rough classification of nitrogen sources is generally based on the following two criteria: the extent to which an individual nitrogen source supports growth when it is the sole nitrogen source and the extent to which a nitrogen source prevents the utilization of poorer nitrogen sources (Magasanik and Kaiser, 2002).
The classification of nitrogen sources and the priority of nitrogen source assimilation vary. Currently, the order of L-Phe assimilation has not been clearly established and differs between different yeast strains and environmental conditions. In a previous study, when different brewing and wine yeast strains were cultured in anaerobic synthetic medium that mimicked grape must supplied with various nitrogen compounds, L-Phe was consumed early along with aspartate, threoine, glutamate, histidine, methionine, serine, and glutamine. Ammonium and tryptophan were consumed late (Crépin et al., 2012).
However, based on transcriptomic analysis, L-Phe is considered a nonpreferred nitrogen source. When S. cerevisiae ∑1278b was grown in aerobic minimal buffer medium with glucose and 21 different nitrogen sources as the sole nitrogen source, L-Phe supported slower growth and exerted a weaker NCR effect. Saccharomyces cerevisiae CEN. PK113-7D was grown in aerobic glucose-limited chemostat cultures with various amino acids as nitrogen sources, and L-Phe exerted an “intermediate” NCR response. There was no direct correlation between the growth rate of each nitrogen source and the degree of NCR (Boer et al., 2007; Godard et al., 2007).
NCR is modulated by four GATA family transcription factors, including the transcriptional activators Gln3/Gat1 and transcriptional repressors Dal80/Gzf3. In the presence of preferred nitrogen sources, Gln3 and Ure2 form complexes in the cytoplasm, which repress the transcription of NCR-sensitive genes. However, in the presence of nonpreferred nitrogen sources, limited nitrogen or added rapamycin, Gln3 is dephosphorylated and freed. Then, it is targeted to the nucleus and binds to the GATAA/G motifs of NCR-sensitive gene promoters, activating gene transcription (Beck and Hall, 1999; Cox et al., 2000; Kulkarni et al., 2001; Tate et al., 2018).
Tap42, an important downstream effector of TORC1, is necessary for the dephosphorylation of Gln3. The extent of Gln3 phosphorylation is synergistically affected by Tap42 and protein phosphatase 2A (PP2A; Beck and Hall, 1999). In the presence of rich nitrogen sources, the activation of TORC1 results in the phosphorylation of Tap42, which combines with PP2A to form a complex located in the vacuole membrane. When Gln3 cannot be dephosphorylated and resides in the cytoplasm, NCR-sensitive genes cannot be expressed (Shamji et al., 2000; Duvel et al., 2003). In the presence of nonpreferred nitrogen sources, limited nitrogen or added rapamycin, TORC1 is inhibited, resulting in the dephosphorylation of Tap42, freeing it from the vacuole membrane. When Gln3 is dephosphorylated by freed Tap42, it is localized to the nucleus and binds to the GATAA/G motifs of NCR-sensitive genes, activating their transcription. In addition to the Tap42-PP2A complex, Ure2 also affects the subcellular localization of Gln3. Ure2 acts as the anchor of Gln3 to maintain its residence in the cytoplasm (Broach, 2012). When Ure2 is inactivated, Gln3 constitutively targets the nucleus (Salmon and Barre, 1998).
The genes related to L-Phe metabolism and regulated by NCR include the permeases GAP1 and AGP1, aromatic aminotransferase ARO9, phenylpyruvate decarboxylase ARO10, aromatic amino acid transcription factor ARO80, and NAD-dependent glutamate dehydrogenase GDH2. With L-Phe as the sole nitrogen source or added rapamycin, the expression levels of these genes can be upregulated.
Uptake of Extracellular L-Phe
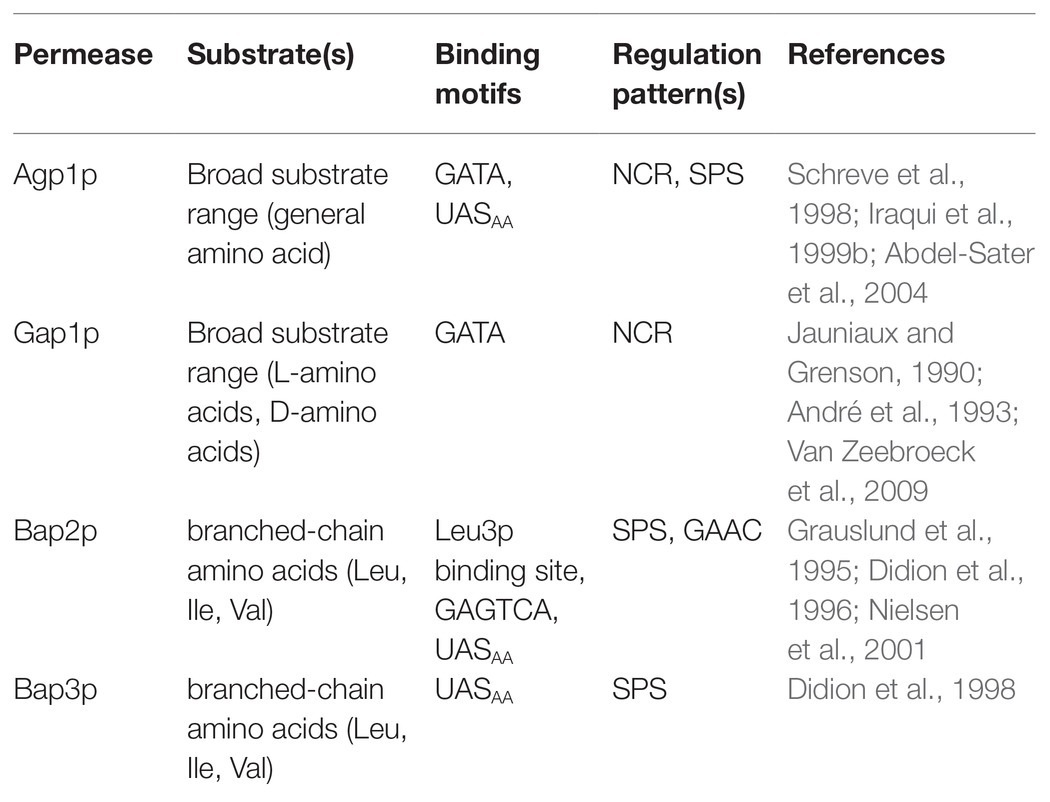
Amino acids are important nitrogen-containing compounds and play central roles in growth and metabolism. Amino acid transporter (AAP) families with conserved sequences and architectural characteristics are critical for the transportation of amino acids (Cain and Kaiser, 2011; Wong et al., 2012). Amino acid permeases driven by proton gradients constitute the largest nitrogen source transport system in S. cerevisiae and play central roles in nitrogen metabolism and protein synthesis (Horák, 1997; Zhang et al., 2019). To date, 24 AAPs have been reported in S. cerevisiae, and each of them consists of approximately 600 amino acids (Grauslund et al., 1995; Zhu et al., 1996). In addition, these transporters share a similar conformation comprising 12 transmembrane domains and cytoplasmically oriented N-terminal and C-terminal domains (Grauslund et al., 1995). These transporters are critical for transporting amino acids and other amines. Permeases that have been reported to transport L-Phe through the PM include Agp1p, Bap2p, Bap3p, and Gap1p (Table 1; Sáenz et al., 2014; Zhang et al., 2019).
Agp1p encoded by the AGP1 gene is a general amino acid permease and has a broad substrate range and low substrate affinity. Earlier research found that asparagine and glutamine are the major substrates of Agp1p in S. cerevisiae YCC5, with Km < 1.0 mM. Moreover, Agp1p can transport L-Phe and other uncharged amino acids when these amino acids are present in millimolar concentrations (Schreve et al., 1998). Iraqui found that L-Phe can be effectively transported by Agp1p and that SSY1 is required for the transcriptional induction of the AGP1 gene (Iraqui et al., 1999b). Agp1p may be regulated according to the L-Phe titre and/or that of other nitrogen sources, and Agp1p is regulated simultaneously by the SPS-sensing pathway and NCR. The AGP1 promoter region has a cis-sequence called UASAA and several 5'-GATA-3' motifs, which can bind to Stp1 and GATA family transcription factors separately, activating transcription of the AGP1 gene (Schreve et al., 1998; Iraqui et al., 1999b; Abdel-Sater et al., 2004; Tate et al., 2010). The UASAA element consists of two inversely repetitive 5'-CGGC-3' motifs separated by six nucleotides.
When S. cerevisiae is grown with L-Phe as the sole nitrogen source to produce 2-PE, the cells recognize extracellular L-Phe through the SPS-sensing pathway. The signal is transmitted to the downstream effector factor Stp1, which then passes through the nuclear membrane and binds the UASAA elements of AGP1, inducing the expression of AGP1 genes (Figure 1). Extracellular L-Phe can be transported into the cytoplasm and serves as a non-preferred nitrogen source to exert a weak NCR effect sensed by the TOR pathway. In addition, upon rapamycin treatment or in the presence of a poor nitrogen source, the activity of TORC1 is restrained. Gln3 is dephosphorylated by the downstream effector factor of TOR and targets the nucleus, where it binds to 5'-GATA-3' motifs, activating transcription of the AGP1 gene (Figure 2).
Gap1p is also a broad-range nitrogen source transporter that can transport all natural L-amino acids, such as L-Phe, some D-amino acids, γ-aminobutyric acid, and polyamines. Transcription of the GAP1 gene in S. cerevisiae is mainly regulated by the NCR pathway and the general amino acid control pathway (GAAC pathway) and can be inhibited in the presence of ammonium salts (Jauniaux and Grenson, 1990; André et al., 1993; Van Zeebroeck et al., 2009).
Bap2p is a branched-chain amino acid permease that mainly transports leucine, isoleucine, and valine with high efficiency and high affinity (Grauslund et al., 1995). Bap2p can also transport L-Phe. Moreover, when S. cerevisiae Y294 was grown with ammonium as the sole nitrogen source and the cells were harvested and transferred to a buffer system containing L-Phe and leucine, Bap2p was the major L-Phe transporter in this specific environment, where extracellular leucine is an important trigger for the induction of BAP2 gene transcription (Didion et al., 1996; Sáenz et al., 2014). The BAP2 promoter contains a Leu3p-binding site, one or two Gcn4p-binding sites (GAGTCA motif) and a UASAA motif, and transcription of the BAP2 gene is regulated by Leu, the SPS sensor system and the GAAC pathway (Grauslund et al., 1995; Nielsen et al., 2001). In addition to the three amino acids mentioned above, Bap3p can also transport L-Phe. Bap3p is a branched-chain amino acid permease that is very similar to Bap2p and has high affinity for branched-chain amino acids. However, the regulation of BAP3 gene transcription is controlled by the SPS sensor system (Didion et al., 1998).
2-PE Synthesis Via The Ehrlich Pathway
Major Enzymes of the Ehrlich Pathway
Two pathways in S. cerevisiae lead to the synthesis of 2-PE, the phenylpyruvate pathway and the Ehrlich pathway (Figure 3). When L-Phe is the precursor, 2-PE is synthesized by the Ehrlich pathway, which consists of three steps: conversion of L-Phe to phenylpyruvate by aromatic aminotransferases, decarboxylation of phenylpyruvate to phenylacetaldehyde (PAAL) by phenylpyruvate decarboxylase, and finally, reduction of PAAL to 2-PE by alcohol dehydrogenase (Hazelwood et al., 2008; Qian et al., 2019). Two isoenzymes are involved in the first step, aminotransferases I and II, which are encoded by ARO8 and ARO9, respectively. ARO8 is constitutively expressed and regulated by the general control of the amino acid biosynthesis pathway (Iraqui et al., 1998).
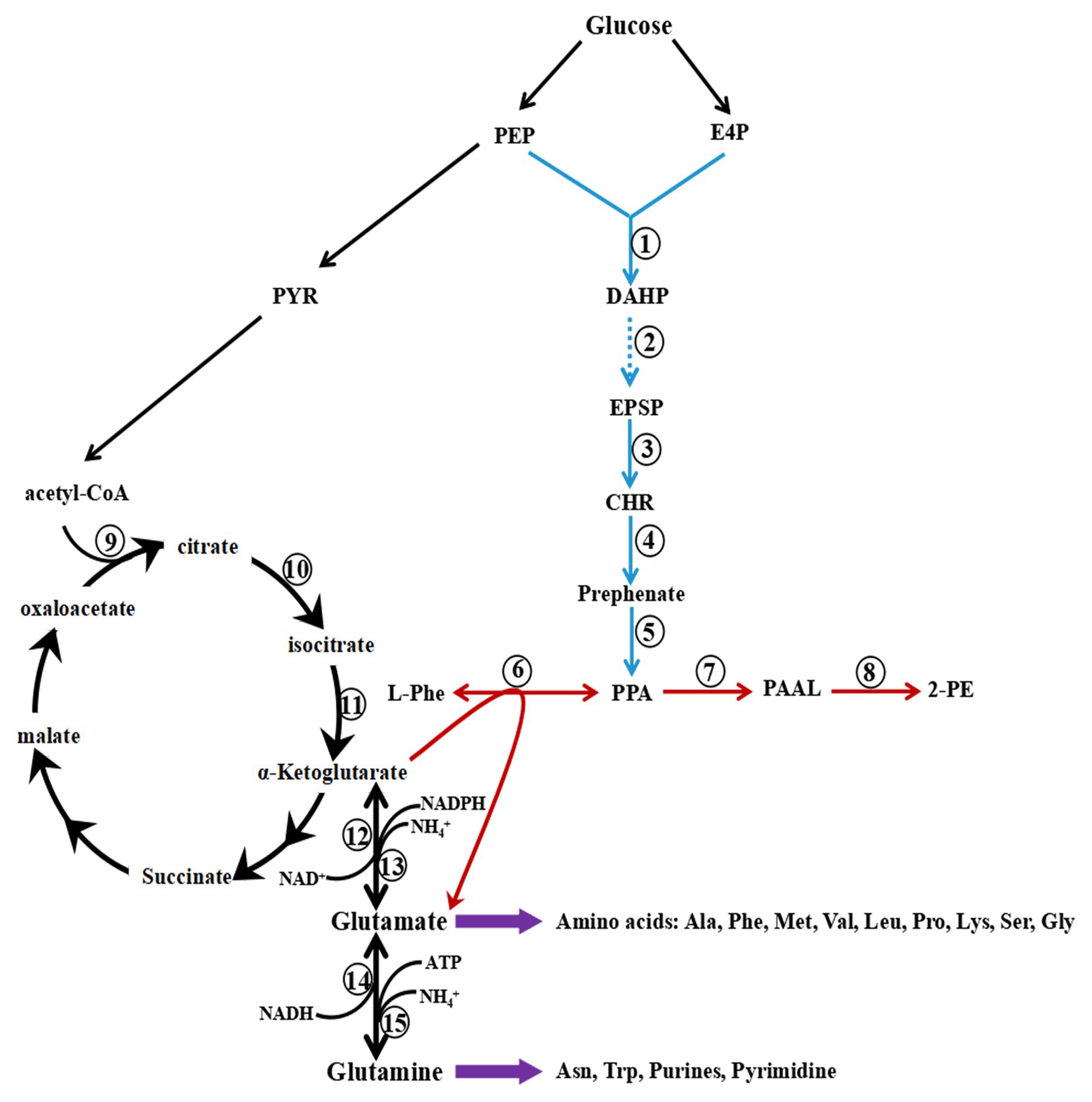
Figure 3. Metabolic pathway of 2-Phenylethanol (2-PE) production in Saccharomyces cerevisiae. The Ehrlich pathway (red) and phenylpyruvate pathway (the combination of red and blue) produce 2-PE. PEP, phosphoenolpyruvate; PYR, pyruvate; E4P, erythrose-4-phosphate; DAHP, 3-deoxy-D-arabinoheptulosonate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SHK, shikimate; S3P, shikimate-3-phosphate; EPSP, 5-enolpyruvyshikimate-3-phosphate; CHR, chorismite; L-Phe, L-phenylalanine; PPA, phenylpyruvate; PAAL, phenylacetaldehyde; and 2-PE, 2-phenylethanol. 1, phospho-2-dehydro-3-deoxyheptonate aldolase ARO3/4; 2, pentafunctional AROM polypeptide ARO1; 3, chorismate synthase ARO2; 4, chorismate mutase ARO7; 5, prephenate dehydratase PHA2; 6, aminotransferase ARO8/9; 7, decarboxylase ARO10; 8, dehydrogenase ADH; 9, citrate T-cell target antigen CTT1/2; 10, aconitase ACO1; 11, isocitrate dehydrogenase IDH1/2; 12, NAD-dependent glutamate dehydrogenase 2 GDH2; 13, NADP-dependent glutamate dehydrogenase 3 GDH3; 14, NADH-dependent glutamate synthase 1 GLT1; and 15, glutamate-ammonia ligase GLN1.
The expression level of ARO9 is upregulated in the presence of aromatic amino acids (L-Phe, tryptophan, or tyrosine), poor nitrogen sources (urea or proline) or the addition of rapamycin. When S. cerevisiae is cultured in the presence of preferred nitrogen sources, the transcription of ARO9 is low or negligible (Iraqui et al., 1999a; Godard et al., 2007). The enzymes that catalyse the second step are thiamine diphosphate-dependent decarboxylases, including Aro10p, Pdc5p, Pdc6p, Pdc1p, and Thi3p. When L-Phe is the sole nitrogen source, Aro10p is the primary decarboxylase catalyst for the decarboxylation of phenylpyruvate to produce phenylacetaldehyde (Vuralhan et al., 2005). Similar to the expression of ARO9, the transcription of ARO10 is low or negligible in the presence of preferred nitrogen sources. ARO10 is induced by aromatic amino acids (L-Phe, tryptophan, or tyrosine), poor nitrogen sources (urea or proline) or the addition of rapamycin (Eden et al., 2007).
In the Ehrlich pathway, the flux from L-Phe to phenylacetaldehyde limits the efficiency of 2-PE biosynthesis. Therefore, the production of 2-PE can be effectively elevated by overexpression of the crucial genes of the Ehrlich pathway, ARO8, ARO9, and ARO10. It has been demonstrated that overexpression of the ARO9, ARO10, and ARO80 genes in S. cerevisiae W303-1B results in an increased 2-PE titre. Coexpression of the ARO9, ARO10, and ARO80 genes and disruption of ALD3 results in a yield of 2-PE of 4.8 g/L (Kim et al., 2014). Subsequently, in S. cerevisiae S288c, overexpression of ARO8 and ARO10 increases the yield of 2-PE by 9.3 and 16.3%, respectively, and coexpression of ARO8 and ARO10 leads to an increase in the yield of 2-PE by 36.8% (Yin et al., 2015).
The last step of the Ehrlich pathway involves the reduction of phenylacetaldehyde to 2-PE by alcohol dehydrogenases. The main genes encoding alcohol dehydrogenases are ADH1, ADH2, ADH3, ADH4, and ADH5. The final reaction can be catalysed by any of these alcohol dehydrogenases (Dickinson et al., 2003). However, although the overexpression of different alcohol dehydrogenase genes did not affect the efficiency of 2-PE synthesis in S. cerevisiae YPH499, coexpression of ADH and ARO10 increased the concentration of 2-PE by 6.5-fold (Shen et al., 2016). In addition, phenylacetaldehyde is competitively oxidized to phenylacetic acid. The ratio of acid to alcohol depends on the cellular redox status and cultivation conditions. Under anaerobic conditions, S. cerevisiae generates excess NADH, resulting in the final reduction step of the Ehrlich pathway, favouring the synthesis of 2-PE (Vuralhan et al., 2003). 2-PE synthesis efficiency can also be improved by eliminating the competitive pathway or increasing the supply of the cofactor NADH (Kim et al., 2014). In addition, deficiency of NADH and/or L-Phe may limit the efficiency of 2-PE synthesis in the Ehrlich pathway. It has been demonstrated that when GDH2, GAP1, ARO8, ARO10, and ADH2 were coexpressed in S. cerevisiae YS58, the intracellular level of L-Phe was increased and NADH was regenerated, leading to the concentration of 2-PE increasing to 6.3 g/L (Wang et al., 2018).
Cis/Trans-Acting Regulatory Factors of the Ehrlich Pathway
In the Ehrlich pathway, ARO9 and ARO10 encode the crucial enzymes of L-Phe metabolism, and their expression levels affect 2-PE production. The expression of ARO9/10 is synergistically regulated by GATA factors and Aro80, but the details of the regulatory mechanism are unclear. The promoters of ARO9, ARO10, and ARO80 contain GATA motifs, which are the binding sites for Gln3 and Gat1 (Figure 4; Eden et al., 2007). In the presence of a poor nitrogen source or upon rapamycin treatment, Gln3 is freed from the Ure2 protein and targets the nucleus, binding to the GATAA/G motifs of the ARO9, ARO10, and ARO80 promoter genes to activate transcription. In addition to the binding site for Gln3, the ARO9, ARO10, and ARO80 gene promoters have Aro80 binding sites consisting of four CCG repeats separated by 7 bp. Aro80 constitutively binds to a pair of adjacent CCG motifs with different orientations and spacings, and the binding state is not affected by intracellular aromatic amino acids (Macpherson et al., 2006). Aro80, a member of the Zn2Cys6 family of proteins, can activate the expression of ARO9 and ARO10 in the presence of aromatic amino acids. The expression of ARO9 and ARO10 is synergistically regulated by Aro80 and Gln3/Gat1, respectively. Gln3/Gat1 indirectly affects the activity of Aro80, which is required for the binding of Gln3/Cat1 to the ARO9 and ARO10 promoter genes (Lee and Hahn, 2013).
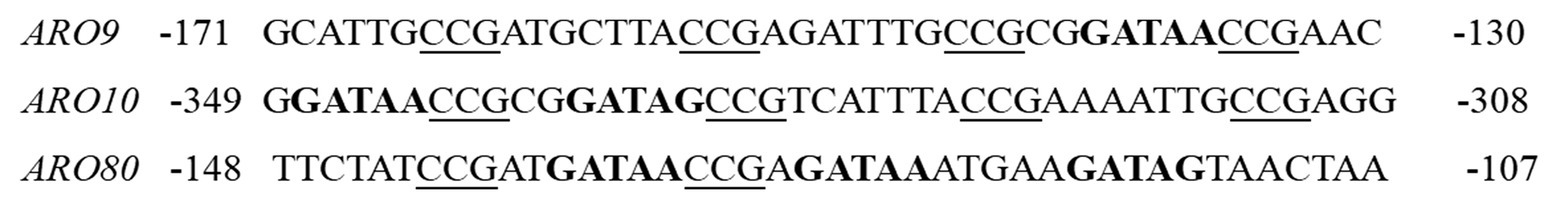
Figure 4. Promoter sequences of Aro80 target genes. CCG triplets, the binding sites of Aro80, are underlined, and potential GATA factor binding sites (GATAA/G) are indicated in bold.
Compared with modifications of the core genes of the Ehrlich pathway, modifications of regulatory factors are simple and efficient alternatives that can activate or inhibit the expression of multiple genes simultaneously. Aro80 and GATA can regulate transcription of the crucial genes ARO9 and ARO10. It has been demonstrated that overexpression of ARO80 can upregulate the transcription levels of ARO9 and ARO10, resulting in a significant increase in the 2-PE titre (Kim et al., 2014). Similarly, modification of the GATA factor can also effectively improve the efficiency of 2-PE synthesis; for example, overexpression of GLN3 and GAT1 in S. cerevisiae YS58 led to an increase in 2-PE production (Wang et al., 2018).
Additionally, recent studies have shown that mepanipyrim and tetraconazole residues could affect the biosynthesis of volatile aromatic compounds during the winemaking process (Sieiro-Sampedro et al., 2019a,b). In particular, tetraconazole seems to accelerate the Ehrlich pathway, and several genes of the Ehrlich pathway (BAT1, PDC1, PDC5, ADH1, and SFA1) are upregulated in tetraconazole-enriched medium. Therefore, the activity of aminotransferase, decarboxylase, and dehydrogenase may be enhanced by tetraconazole (Sieiro-Sampedro et al., 2020).
The Effect of L-Phe Catabolism in the Ehrlich Pathway
In the Ehrlich pathway, L-Phe is transaminated to phenylpyruvate, and α-ketoglutarate is the acceptor of the amino group and converted into glutamate, followed by the conversion of phenylpyruvate into 2-PE. Glutamate, a crucial intermediate, is converted into glutamine by glutamate synthase or glutamate-ammonia ligase, and then, purine and pyrimidine are synthesized to maintain cell growth (Figure 3). In addition, glutamate is used for the biosynthesis of other amino acids, such as alanine, methionine, leucine, phenylalanine, serine, and proline. Glutamate and glutamine are the hubs of nitrogen metabolism; 85% of cellular nitrogen is derived from the amino nitrogen of glutamate, and the remaining 15% is derived from the amide nitrogen of glutamine (Ljungdahl and Daignan-Fornier, 2012). As a preferred nitrogen source, glutamate may inhibit the transport and catabolism of L-Phe when it accumulates in yeast cells. However, glutamate is the byproduct of L-Phe catabolism via the Ehrlich pathway, which may lead to the accumulation of glutamate.
To avoid the accumulation of glutamate and alleviate the inhibition of L-Phe utilization, glutamate is deaminated to produce α-ketoglutarate and NH4+ by NAD-dependent glutamate dehydrogenase or by NADP-dependent glutamate dehydrogenase encoded by GDH2 and GDH3, respectively. This conjecture is consistent with the results of a previous study: the expression levels of GDH2 and GDH3 are upregulated in S. cerevisiae CEN.PK113-7D with L-Phe as the sole nitrogen source (Boer et al., 2007). In addition, the reaction resupplies α-ketoglutarate and NADH for L-Phe catabolism via the Ehrlich pathway. Therefore, when S. cerevisiae is cultured with L-Phe as the sole nitrogen source, intracellular glutamate undergoes rapid synthesis and catabolism to support cell growth and biosynthesis.
α-Ketoglutarate is another important substrate of the Ehrlich pathway, and its intracellular concentration also affects the efficiency of 2-PE synthesis from L-Phe. In addition, α-ketoglutarate is a crucial intermediate of the tricarboxylic acid cycle (TCA). Therefore, α-ketoglutarate is the converging point of L-Phe catabolism and glucose metabolism. α-Ketoglutarate is modulated by the retrograde regulation (RTG) pathway, which mainly regulates the expression of CTT1/2, IDH1/2, and ACO1 (Giannattasio et al., 2005). In the presence of a poor nitrogen source, the genes related to the synthesis of α-ketoglutarate are upregulated by the RTG pathway to meet the demand of α-ketoglutarate (Broach, 2012). Therefore, with L-Phe as the sole nitrogen source, the expression of CTT1/2, IDH1/2, and ACO1 might also be upregulated to satisfy the needs of the Ehrlich pathway.
Conclusion and Perspectives
Saccharomyces cerevisiae is generally recognized as safe (GRAS) and is typically used in food and industrial production. Saccharomyces cerevisiae can also serve as one of most promising microorganisms for the biosynthesis of natural 2-PE. Saccharomyces cerevisiae recognizes extracellular L-Phe through the SPS-sensing pathway, regulates the expression of genes that are critical for the transport of L-Phe, and then recognizes intracellular L-Phe through the TOR-sensing pathway. Finally, L-Phe is converted into 2-PE via the Ehrlich pathway. The currently known transporters involved in the transport of L-Phe are Agp1p, Bap2p, Bap3p, and Gap1p.
However, the four permeases contributing to L-Phe transmembrane transport could be finely tuned based on different nitrogen sources. When S. cerevisiae Y294 is grown with ammonium salt as the sole nitrogen source, the harvested cells are transferred to the buffer system containing L-Phe and leucine, in which Bap2p is the principal L-Phe transporter. Agp1p is the major transporter of L-Phe when S. cerevisiae Y294 gap1∆ is cultured in synthetic medium. The transporters of L-Phe are regulated synergistically by different pathways. For example, Bap2p is regulated simultaneously by the SPS-sensing pathway and GAAC pathway, and Agp1p is regulated simultaneously by the SPS-sensing pathway and NCR. The details of the regulatory mechanism of L-Phe transport remain unclear, and further study is necessary to determine how the regulatory pathways synergistically regulate the transport of L-Phe.
Although, we have gained a certain understanding of the catabolism and regulation of L-Phe, the highest production of 2-PE by the Ehrlich pathway reached 6.3 g/L through reconstruction of the metabolic module. However, 2-PE induces high levels of toxicity in yeast cells, which is the biggest bottleneck to the process for improving 2-PE production (Jin et al., 2018; Dai et al., 2020). In situ product removal, two-phase extraction and in situ product adsorption are used to alleviate the toxicity of 2-PE, which has been shown to be effective but uneconomical. Many efforts have been made to screen robust strains (Lu et al., 2016; Dai et al., 2020; Zhan et al., 2020). However, the molecular mechanism of 2-PE tolerance remains unclear. Recently, Hap5 was discovered to be a necessary regulator of 2-PE resistance in Candida glycerinogenes (Wang et al., 2020). Therefore, further study on the toxicity of 2-PE in yeast cells would lead to the mechanism of 2-PE tolerance, which would provide more theoretical guidance for further increasing the production of 2-PE.
L-Phenylalanine has been proven to be a nonpreferred nitrogen source in S. cerevisiae, and the crucial genes of the Ehrlich pathway are modulated by NCR in the presence of preferred nitrogen sources, blocking synthesis of 2-PE on the industrial scale. The details of the regulatory mechanism should be further studied to avoid repression to augment the efficient synthesis of 2-PE.
Although good results were achieved with L-Phe as the precursor (Wang et al., 2019), the high price of L-Phe might be a good economic barrier for the scaled-up application of this bioprocess. Saccharomyces cerevisiae possesses the native ability to synthesize 2-PE from glucose via the phenylpyruvate pathway, which is very complex, contains many branches competing for carbon flow and is strongly feedback-inhibited by L-Phe. Therefore, the efficiency of the phenylpyruvate pathway is very low (Hassing et al., 2019; Wang et al., 2019). To reduce the cost, the development of cheaper substrates or robust strains might be the best alternative to accomplish industrial production. Many efforts have been made to improve the de novo synthesis of nonconventional yeast or develop robust strains. It has been demonstrated that engineered Pichia pastoris can produce 2-PE from simple sugars, and the concentration of 2-PE increased to 1,169 mg/L through genetic engineering strategies (Kong et al., 2020). Moreover, 2-PE can be synthesized by solid-state fermentation using low-cost raw materials by Pichia kudriavzevii, and the maximum 2-PE titre was 27.2 mg per gram of dry substrate (Martinez-Avila et al., 2020). Interestingly, hydrolysed corn stover or molasses can be used as a carbon source to produce 2-PE by nonconventional yeast or Bacillus licheniformis, with higher 2-PE concentrations of 3.67 g/L and 4.41 g/L, respectively (Mierzejewska et al., 2019; Zhan et al., 2020). To reduce the cost, in current studies, Escherichia coli has been developed for the biotransformation of L-Phe, glucose, or glycerol to 2-PE with resting cells, leading to a higher 2-PE concentration (9.1 g/L) from glucose or glycerol (Liu et al., 2018; Lukito et al., 2019; Sekar et al., 2019).
Understanding the meticulous process of L-Phe metabolism and current research will facilitate the ultimate industrial production of 2-PE, 2-PEA, and other valuable derivatives of 2-PE. Moreover, branched-chain amino acids (leucine, valine, and isoleucine), aromatic amino acids (tyrosine, and tryptophan), and sulfur-containing amino acids (methionine) are assimilated by the Ehrlich pathway to produce other higher alcohols, such as 2-methylbutanol, propanol, and 3-methylbutanol. This research could play a guiding role in the production of other higher alcohols.
Author Contributions
XC and CY conceived the review. JD and HX wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundations of China (grant nos. 31871789 and 41876114), Open Funding Project of the State Key Laboratory of Biocatalysis and Enzyme Engineering (no. SKLBEE2018013), the Natural Science Foundation of Hubei Provincial Department of Education (no. B2016046), the Natural Science Foundation of Zhejiang Province (no. LY18D060007), Major special projects of China Tobacco Corporation (110202001039(XJ-01)), and Key technology projects of Hubei tobacco company (027Y2019-003).
Conflict of Interest
The authors declare that this study received funding from China Tobacco Corporation and Hubei tobacco company. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
References
Abdel-Sater, F., Iraqui, I., Urrestarazu, A., and André, B. (2004). The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae. Genetics 166, 1727–1739. doi: 10.1534/genetics.166.4.1727
Abdel-Sater, F., Jean, C., Merhi, A., Vissers, S., and Andre, B. (2011). Amino acid signaling in yeast: activation of Ssy5 protease is associated with its phosphorylation-induced ubiquitylation. J. Biol. Chem. 286, 12006–12015. doi: 10.1074/jbc.M110.200592
André, B., Hein, C., Grenson, M., and Jauniaux, J. C. (1993). Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol. Gen. Genet. 237, 17–25. doi: 10.1007/BF00282779
Andréasson, C., and Ljungdahl, P. O. (2002). Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16, 3158–3172. doi: 10.1101/gad.239202
Andréasson, C., and Ljungdahl, P. O. (2004). The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol. Cell. Biol. 24, 7503–7513. doi: 10.1128/MCB.24.17.7503-7513.2004
Beck, T., and Hall, M. N. (1999). The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692. doi: 10.1038/45287
Binda, M., Péli-Gulli, M. P., Bonfils, G., Panchaud, N., Urban, J., Sturgill, T. W., et al. (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573. doi: 10.1016/j.molcel.2009.06.033
Boer, V. M., Tai, S. L., Vuralhan, Z., Arifin, Y., Walsh, M. C., Piper, M. D., et al. (2007). Transcriptional responses of Saccharomyces cerevisiae to preferred and nonpreferred nitrogen sources in glucose-limited chemostat cultures. FEMS Yeast Res. 7, 604–620. doi: 10.1111/j.1567-1364.2007.00220.x
Broach, J. R. (2012). Nutritional control of growth and development in yeast. Genetics 192, 73–105. doi: 10.1534/genetics.111.135731
Cain, N. E., and Kaiser, C. A. (2011). Transport activity-dependent intracellular sorting of the yeast general amino acid permease. Mol. Biol. Cell 22, 1919–1929. doi: 10.1091/mbc.E10-10-0800
Carlquist, M., Gibson, B., Karagul Yuceer, Y., Paraskevopoulou, A., Sandell, M., Angelov, A. I., et al. (2015). Process engineering for bioflavour production with metabolically active yeasts - a mini-review. Yeast 32, 123–143. doi: 10.1002/yea.3058
Chreptowicz, K., Wielechowska, M., Glowczyk-Zubek, J., Rybak, E., and Mierzejewska, J. (2016). Production of natural 2-phenylethanol: from biotransformation to purified product. Food Bioprod. Process. 100, 275–281. doi: 10.1016/j.fbp.2016.07.011
Chung, H. Jr., Lee, S. L., and Chou, C. C. (2000). Production and molar yield of 2-phenylethanol by Pichia fermentans L-5 as affected by some medium components. J. Biosci. Bioeng. 90, 142–147. doi: 10.1016/S1389-1723(00)80101-2
Cordente, A. G., Schmidt, S., Beltran, G., Torija, M. J., and Curtin, C. D. (2019). Harnessing yeast metabolism of aromatic amino acids for fermented beverage bioflavouring and bioproduction. Appl. Microbiol. Biotechnol. 103, 4325–4336. doi: 10.1007/s00253-019-09840-w
Cox, K. H., Rai, R., Distler, M., Daugherty, J. R., Coffman, J. A., and Cooper, T. G. (2000). Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J. Biol. Chem. 275, 17611–17618. doi: 10.1074/jbc.M001648200
Crépin, L., Nidelet, T., Sanchez, I., Dequin, S., and Camarasa, C. (2012). Sequential use of nitrogen compounds by Saccharomyces cerevisiae during wine fermentation: a model based on kinetic and regulation characteristics of nitrogen permeases. Appl. Environ. Microbiol. 78, 8102–8111. doi: 10.1128/AEM.02294-12
Dai, J., Li, K., Song, N., Yao, W., Xia, H., Yang, Q., et al. (2020). Zygosaccharomyces rouxii, an aromatic yeast isolated from chili sauce, is able to biosynthesize 2-phenylethanol via the Shikimate or Ehrlich pathways. Front. Microbiol. 11:597454. doi: 10.3389/fmicb.2020.597454
Dickinson, J. R., Salgado, L. E., and Hewlins, M. J. (2003). The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278, 8028–8034. doi: 10.1074/jbc.M211914200
Didion, T., Grauslund, M., Kielland-Brandt, M. C., and Andersen, H. A. (1996). Amino acids induce expression of BAP2, a branched-chain amino acid Permease gene in Saccharomyces cerevisiae. J. Bacteriol. 178, 2025–2029. doi: 10.1128/JB.178.7.2025-2029.1996
Didion, T., Regenberg, B., Jorgensen, M. U., Kielland-Brandt, M. C., and Andersen, H. A. (1998). The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27, 643–650. doi: 10.1046/j.1365-2958.1998.00714.x
Dubouloz, F., Deloche, O., Wanke, V., Cameroni, E., and De Virgilio, C. (2005). The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19, 15–26. doi: 10.1016/j.molcel.2005.05.020
Duvel, K., Santhanam, A., Garrett, S., Schneper, L., and Broach, J. R. (2003). Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11, 1467–1478. doi: 10.1016/S1097-2765(03)00228-4
Eden, E., Lipson, D., Yogev, S., and Yakhini, Z. (2007). Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 3:e39. doi: 10.1371/journal.pcbi.0030039
Etschmann, M. M., Bluemke, W., Sell, D., and Schrader, J. (2002). Biotechnological production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 59, 1–8. doi: 10.1007/s00253-002-0992-x
Forsberg, H., and Ljungdahl, P. O. (2001). Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21, 814–826. doi: 10.1128/MCB.21.3.814-826.2001
Gaber, R. F., Ottow, K., Andersen, H. A., and Kielland-Brandt, M. C. (2003). Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot. Cell 2, 922–929. doi: 10.1128/EC.2.5.922-929.2003
Giannattasio, S., Liu, Z., Thornton, J., and Butow, R. A. (2005). Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J. Biol. Chem. 280, 42528–42535. doi: 10.1074/jbc.M509187200
Godard, P., Urrestarazu, A., Vissers, S., Kontos, K., Bontempi, G., van Helden, J., and Andre, B. (2007). Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 27, 3065–3086. doi: 10.1128/MCB.01084-06
Grauslund, M., Didion, T., Kielland-Brandt, M. C., and Andersen, H. A. (1995). BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269, 275–280. doi: 10.1016/0167-4889(95)00138-8
Hassing, E. J., De Groot, P. A., Marquenie, V. R., Pronk, J. T., and Daran, J. M. G. (2019). Connecting central carbon and aromatic amino acid metabolisms to improve de novo 2-phenylethanol production in Saccharomyces cerevisiae. Metab. Eng. 56, 165–180. doi: 10.1016/j.ymben.2019.09.011
Hazelwood, L. A., Daran, J. M., van Maris, A. J. A., Pronk, J. T., and Dickinson, J. R. (2008). The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266. doi: 10.1128/AEM.02625-07
Henschke, P. A., and Jiranek, V. (1993). “Metabolism of nitrogen compounds” in Wine microbiology and biotechnology. ed. Fleet, G. H. (Chur, Switzerland: Harwood Academic Publishers), 77–164.
Horák, J. (1997). Yeast nutrient transporters. Biochim. Biophys. Acta 1331, 41–79. doi: 10.1016/s0304-4157(96)00015-9
Hua, D., and Xu, P. (2011). Recent advances in biotechnological production of 2-phenylethanol. Biotechnol. Adv. 29, 654–660. doi: 10.1016/j.biotechadv.2011.05.001
Iraqui, I., Vissers, S., Andre, B., and Urrestarazu, A. (1999a). Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 3360–3371. doi: 10.1128/mcb.19.5.3360
Iraqui, I., Vissers, S., Bernard, F., De Craene, J. O., Boles, E., Urrestarazu, A., et al. (1999b). Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19, 989–1001. doi: 10.1128/mcb.19.2.989
Iraqui, I., Vissers, S., Cartiaux, M., and Urrestarazu, A. (1998). Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257, 238–248. doi: 10.1007/s004380050644
Jauniaux, J. C., and Grenson, M. (1990). GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190, 39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x
Jin, D. F., Gu, B. T., Xiong, D. W., Huang, G. C., Huang, X. P., Liu, L., et al. (2018). A Transcriptomic analysis of Saccharomyces cerevisiae under the stress of 2-phenylethanol. Curr. Microbiol. 75, 1068–1076. doi: 10.1007/s00284-018-1488-y
Kim, B., Cho, B. R., and Hahn, J. S. (2014). Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol. Bioeng. 111, 115–124. doi: 10.1002/bit.24993
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P., and Guan, K. L. (2008). Regulation of TORC1 by rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945. doi: 10.1038/ncb1753
Klasson, H., Fink, G. R., and Ljungdahl, P. O. (1999). Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19, 5405–5416. doi: 10.1128/MCB.19.8.5405
Kong, S., Pan, H., Liu, X., Li, X., and Guo, D. (2020). De novo biosynthesis of 2-phenylethanol in engineered Pichia pastoris. Enzym. Microb. Technol. 133:109459. doi: 10.1016/j.enzmictec.2019.109459
Kulkarni, A. A., Abul-Hamd, A. T., Rai, R., El Berry, H., and Cooper, T. G. (2001). Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J. Biol. Chem. 276, 32136–32144. doi: 10.1074/jbc.M104580200
Lee, K., and Hahn, J. S. (2013). Interplay of Aro80 and GATA activators in regulation of genes for catabolism of aromatic amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 88, 1120–1134. doi: 10.1111/mmi.12246
Liu, C., Zhang, K., Cao, W., Zhang, G., Chen, G., Yang, H., et al. (2018). Genome mining of 2-phenylethanol biosynthetic genes from Enterobacter sp. CGMCC 5087 and heterologous overproduction in Escherichia coli. Biotechnol. Biofuels 11:305. doi: 10.1186/s13068-018-1297-3
Ljungdahl, P. O., and Daignan-Fornier, B. (2012). Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190, 885–929. doi: 10.1534/genetics.111.133306
Lu, X. Y., Wang, Y. Q., Zong, H., Ji, H., Zhuge, B., and Dong, Z. L. (2016). Bioconversion of L-phenylalanine to 2-phenylethanol by the novel stress-tolerant yeast Candida glycerinogenes WL2002-5. Bioengineered 7, 418–423. doi: 10.1080/21655979.2016.1171437
Lukito, B. R., Wu, S. K., Saw, H. J. J., and Li, Z. (2019). One-pot production of natural 2-phenylethanol from L-phenylalanine via cascade biotransformations. ChemCatChem 11, 831–840. doi: 10.1002/cctc.201801613
Macpherson, S., Larochelle, M., and Turcotte, B. (2006). A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70, 583–604. doi: 10.1128/MMBR.00015-06
Magasanik, B., and Kaiser, C. A. (2002). Nitrogen regulation in Saccharomyces cerevisiae. Gene 290, 1–18. doi: 10.1016/S0378-1119(02)00558-9
Martínez-Avila, O., Sánchez, A., Font, X., and Barrena, R. (2018). Bioprocesses for 2-phenylethanol and 2-phenylethyl acetate production: current state and perspectives. Appl. Microbiol. Biotechnol. 102, 9991–10004. doi: 10.1007/s00253-018-9384-8
Martinez-Avila, O., Sanchez, A., Font, X., and Barrena, R. (2020). 2-phenylethanol (rose aroma) production potential of an isolated pichia kudriavzevii through solid-state fermentation. Process Biochem. 93, 94–103. doi: 10.1016/j.procbio.2020.03.023
Martins, A., Pfirrmann, T., Heessen, S., Sundqvist, G., Bulone, V., Andreasson, C., et al. (2018). Ssy5 is a signaling serine protease that exhibits atypical biogenesis and marked S1 specificity. J. Biol. Chem. 293, 8362–8378. doi: 10.1074/jbc.RA118.002457
Martins, A., Ring, A., Omnus, D. J., Heessen, S., Pfirrmann, T., and Ljungdahl, P. O. (2019). Spatial and temporal regulation of the endoproteolytic activity of the SPS-sensor-controlled Ssy5 signaling protease. Mol. Biol. Cell 30, 2709–2720. doi: 10.1091/mbc.E19-02-0096
Mierzejewska, J., Dabkowska, K., Chreptowicz, K., and Sokolowska, A. (2019). Hydrolyzed corn stover as a promising feedstock for 2-phenylethanol production by nonconventional yeast. J. Chem. Technol. Biotechnol. 94, 777–784. doi: 10.1002/jctb.5823
Neklesa, T. K., and Davis, R. W. (2009). A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 5:e1000515. doi: 10.1371/journal.pgen.1000515
Nielsen, P. S., van den Hazel, B., Didion, T., de Boer, M., Jørgensen, M., Planta, R. J., et al. (2001). Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264, 613–622. doi: 10.1007/s004380000347
Omnus, D. J., and Ljungdahl, P. O. (2013). Rts1-protein phosphatase 2A antagonizes Ptr3-mediated activation of the signaling protease Ssy5 by casein kinase I. Mol. Biol. Cell 24, 1480–1492. doi: 10.1091/mbc.E13-01-0019
Omnus, D. J., and Ljungdahl, P. O. (2014). Latency of transcription factor Stp1 depends on a modular regulatory motif that functions as cytoplasmic retention determinant and nuclear degron. Mol. Biol. Cell 25, 3823–3833. doi: 10.1091/mbc.E14-06-1140
Omnus, D. J., Manford, A. G., Bader, J. M., Emr, S. D., and Stefan, C. J. (2016). Phosphoinositide kinase signaling controls ER-PM cross-talk. Mol. Biol. Cell 27, 1170–1180. doi: 10.1091/mbc.E16-01-0002
Powis, K., and De Virgilio, C. (2016). Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov 2:15049. doi: 10.1038/celldisc.2015.49
Powis, K., Zhang, T., Panchaud, N., Wang, R., De Virgilio, C., and Ding, J. (2015). Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting rag GTPase-dependent TORC1 signaling. Cell Res. 25, 1043–1059. doi: 10.1038/cr.2015.86
Qian, X. J., Yan, W., Zhang, W. M., Dong, W. L., Ma, J. F., Ochsenreither, K., et al. (2019). Current status and perspectives of 2-phenylethanol production through biological processes. Crit. Rev. Biotechnol. 39, 235–248. doi: 10.1080/07388551.2018.1530634
Reinke, A., Anderson, S., Mccaffery, J. M., Yates, J. 3rd., Aronova, S., Chu, S., et al. (2004). TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752–14762. doi: 10.1074/jbc.M313062200
Romagnoli, G., Knijnenburg, T. A., Liti, G., Louis, E. J., Pronk, J. T., and Daran, J. M. (2015). Deletion of the Saccharomyces cerevisiae ARO8 gene, encoding an aromatic amino acid transaminase, enhances phenylethanol production from glucose. Yeast 32, 29–45. doi: 10.1002/yea.3015
Sáenz, D. A., Chianelli, M. S., and Stella, C. A. (2014). L-phenylalanine transport in Saccharomyces cerevisiae: participation of GAP1, BAP2, and AGP1. J. Amino Acids 2014:283962. doi: 10.1155/2014/283962
Salmon, J. M., and Barre, P. (1998). Improvement of nitrogen assimilation and fermentation kinetics under enological conditions by derepression of alternative nitrogen-assimilatory pathways in an industrial Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 64, 3831–3837. doi: 10.1128/AEM.64.10.3831-3837.1998
Sancak, Y., Peterson, T. R., Shaul, Y. D., Lindquist, R. A., Thoreen, C. C., Bar-Peled, L., et al. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. doi: 10.1126/science.1157535
Schneper, L., Düvel, K., and Broach, J. R. (2004). Sense and sensibility: nutritional response and signal integration in yeast. Curr. Opin. Microbiol. 7, 624–630. doi: 10.1016/j.mib.2004.10.002
Schreve, J. L., Sin, J. K., and Garrett, J. M. (1998). The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J. Bacteriol. 180, 2556–2559. doi: 10.1128/JB.180.9.2556-2559.1998
Scognamiglio, J., Jones, L., Letizia, C. S., and Api, A. M. (2012). Fragrance material review on phenylethyl alcohol. Food Chem. Toxicol. 50(Suppl. 2), S224–S239. doi: 10.1016/j.fct.2011.10.028
Sekar, B. S., Lukito, B. R., and Li, Z. (2019). Production of natural 2-phenylethanol from glucose or glycerol with coupled Escherichia coli strains expressing L-phenylalanine biosynthesis pathway and artificial biocascades. ACS Sustain. Chem. Eng. 7, 12231–12239. doi: 10.1021/acssuschemeng.9b01569
Shamji, A. F., Kuruvilla, F. G., and Schreiber, S. L. (2000). Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10, 1574–1581. doi: 10.1016/S0960-9822(00)00866-6
Shen, L., Nishimura, Y., Matsuda, F., Ishii, J., and Kondo, A. (2016). Overexpressing enzymes of the Ehrlich pathway and deleting genes of the competing pathway in Saccharomyces cerevisiae for increasing 2-phenylethanol production from glucose. J. Biosci. Bioeng. 122, 34–39. doi: 10.1016/j.jbiosc.2015.12.022
Sieiro-Sampedro, T., Alonso-del-Real, J., Briz-Cid, N., Rial-Otero, R., Querol, A., and Simal-Gandara, J. (2020). The effect of two antifungal commercial formulations on the metabolism of a commercial Saccharomyces cerevisiae strain and their repercussion on fermentation evolution and phenylalanine catabolism. Food Microbiol. 92:103554. doi: 10.1016/j.fm.2020.103554
Sieiro-Sampedro, T., Figueiredo-Gonzalez, M., Gonzalez-Barreiro, C., Simal-Gandara, J., Cancho-Grande, B., and Rial-Otero, R. (2019a). Impact of mepanipyrim and tetraconazole in Mencia wines on the biosynthesis of volatile compounds during the winemaking process. Food Chem. 300:125223. doi: 10.1016/j.foodchem.2019.125223
Sieiro-Sampedro, T., Pose-Juan, E., Briz-Cid, N., Figueiredo-Gonzalez, M., Torrado-Agrasar, A., Gonzalez-Barreiro, C., et al. (2019b). Mepanipyrim residues on pasteurized red must influence the volatile derived compounds from Saccharomyces cerevisiae metabolism. Food Res. Int. 126:108566. doi: 10.1016/j.foodres.2019.108566
Stark, D., Munch, T., Sonnleitner, B., Marison, I. W., and Von Stockar, U. (2002). Extractive bioconversion of 2-phenylethanol from L-phenylalanine by Saccharomyces cerevisiae. Biotechnol. Prog. 18, 514–523. doi: 10.1021/bp020006n
Tate, J. J., Georis, I., Dubois, E., and Cooper, T. G. (2010). Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J. Biol. Chem. 285, 17880–17895. doi: 10.1074/jbc.M109.085712
Tate, J. J., Rai, R., and Cooper, T. G. (2018). More than one way in: three Gln3 sequences required to relieve negative Ure2 regulation and support nuclear Gln3 import in Saccharomyces cerevisiae. Genetics 208, 207–227. doi: 10.1534/genetics.117.300457
Tumusiime, S., Zhang, C., Overstreet, M. S., and Liu, Z. (2011). Differential regulation of transcription factors Stp1 and Stp2 in the Ssy1-Ptr3-Ssy5 amino acid sensing pathway. J. Biol. Chem. 286, 4620–4631. doi: 10.1074/jbc.M110.195313
Van Zeebroeck, G., Bonini, B. M., Versele, M., and Thevelein, J. M. (2009). Transport and signaling via the amino acid binding site of the yeast Gap1 amino acid transceptor. Nat. Chem. Biol. 5, 45–52. doi: 10.1038/nchembio.132
Vuralhan, Z., Luttik, M. a. H., Tai, S. L., Boer, V. M., Morais, M. A., Schipper, D., et al. (2005). Physiological characterization of the ARO10-dependent, broad-substrate-specificity 2-Oxo acid decarboxylase activity of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 3276–3284. doi: 10.1128/AEM.71.6.3276-3284.2005
Vuralhan, Z., Morais, M. A., Tai, S. L., Piper, M. D., and Pronk, J. T. (2003). Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69, 4534–4541. doi: 10.1128/AEM.69.8.4534-4541.2003
Wang, Y., Zhang, H., Lu, X., Zong, H., and Zhuge, B. (2019). Advances in 2-phenylethanol production from engineered microorganisms. Biotechnol. Adv. 37, 403–409. doi: 10.1016/j.biotechadv.2019.02.005
Wang, Y., Zhang, Z., Lu, X., Zong, H., and Zhuge, B. (2020). Transcription factor Hap5 induces gsh2 expression to enhance 2-phenylethanol tolerance and production in an industrial yeast Candida glycerinogenes. Appl. Microbiol. Biotechnol. 104, 4093–4107. doi: 10.1007/s00253-020-10509-y
Wang, Z., Jiang, M., Guo, X., Liu, Z., and He, X. (2018). Reconstruction of metabolic module with improved promoter strength increases the productivity of 2-phenylethanol in Saccharomyces cerevisiae. Microb. Cell Fact. 17:60. doi: 10.1186/s12934-018-0907-x
Wong, F. H., Chen, J. S., Reddy, V., Day, J. L., Shlykov, M. A., Wakabayashi, S. T., et al. (2012). The amino acid-polyamine-organocation superfamily. J. Mol. Microbiol. Biotechnol. 22, 105–113. doi: 10.1159/000338542
Yin, S., Zhou, H., Xiao, X., Lang, T., Liang, J., and Wang, C. (2015). Improving 2-phenylethanol production via Ehrlich pathway using genetic engineered Saccharomyces cerevisiae strains. Curr. Microbiol. 70, 762–767. doi: 10.1007/s00284-015-0785-y
Zargari, A., Boban, M., Heessen, S., Andréasson, C., Thyberg, J., and Ljungdahl, P. O. (2007). Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J. Biol. Chem. 282, 594–605. doi: 10.1074/jbc.M609201200
Zhan, Y., Zhou, M., Wang, H., Chen, L., Li, Z., Cai, D., et al. (2020). Efficient synthesis of 2-phenylethanol from L-phenylalanine by engineered Bacillus licheniformis using molasses as carbon source. Appl. Microbiol. Biotechnol. 104, 7507–7520. doi: 10.1007/s00253-020-10740-7
Zhang, P., Chen, Q., Fu, G., Xia, L., and Hu, X. (2019). Regulation and metabolic engineering strategies for permeases of Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 35:112. doi: 10.1007/s11274-019-2684-z
Zhang, W., Du, G., Zhou, J., and Chen, J. (2018). Regulation of sensing, transportation, and catabolism of nitrogen sources in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 82:e00040. doi: 10.1128/mmbr.00040-17
Keywords: 2-phenylethanol, sensing of L-phenylalanine, uptake of L-phenylalanine, Ehrlich pathway, Saccharomyces cerevisiae
Citation: Dai J, Xia H, Yang C and Chen X (2021) Sensing, Uptake and Catabolism of L-Phenylalanine During 2-Phenylethanol Biosynthesis via the Ehrlich Pathway in Saccharomyces cerevisiae. Front. Microbiol. 12:601963. doi: 10.3389/fmicb.2021.601963
Edited by:
Nuno Pereira Mira, University of Lisbon, PortugalReviewed by:
Benoit Divol, Stellenbosch University, South AfricaOscar Martínez Avila, Universitat de Vic - Universitat Central de Catalunya, Spain
Copyright © 2021 Dai, Xia, Yang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiong Chen, Y3gxNjNfcXhAMTYzLmNvbQ==; Chunlei Yang, eWNsMTkzNzM3QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Jun Dai
Jun Dai Huili Xia
Huili Xia Chunlei Yang4*
Chunlei Yang4* Xiong Chen
Xiong Chen