- 1Department of Plant Biotechnology, School of Biotechnology, Madurai Kamaraj University, Madurai, India
- 2Department of Biological Sciences, Texas Tech University, Lubbock, TX, United States
Geminiviruses are plant DNA viruses that infect a wide range of plant species and cause significant losses to economically important food and fiber crops. The single-stranded geminiviral genome encodes a small number of proteins which act in an orchestrated manner to infect the host. The fewer proteins encoded by the virus are multifunctional, a mechanism uniquely evolved by the viruses to balance the genome-constraint. The host-mediated resistance against incoming virus includes post-transcriptional gene silencing, transcriptional gene silencing, and expression of defense responsive genes and other cellular regulatory genes. The pathogenicity property of a geminiviral protein is linked to its ability to suppress the host-mediated defense mechanism. This review discusses what is currently known about the targets and mechanism of the viral suppressor AC2/AL2/transcriptional activator protein (TrAP) and explore the biotechnological applications of AC2.
Introduction
Geminiviruses are single-stranded (ss) DNA viruses that cause major losses to a number of economically important crops throughout the world (Scholthof et al., 2011; Rojas et al., 2018). Geminiviridae constitutes the largest family of plant viruses with nine genera and 485 species (Zerbini et al., 2017). Geminiviruses are characterized by their small, circular, ssDNA genomes encapsidated in twinned-icosahedral particles. They are vector-transmissible and infect both monocotyledonous and dicotyledonous plants (Zerbini et al., 2017). The viral genomes are either monopartite or bipartite with circular ssDNA molecules of 2.5 to 5.5 × 103 nucleotides. Bipartite geminiviruses, with DNA A and DNA B components, possess a highly conserved common region (CR) of ~200 nucleotides. An inverted repeat within the CR forms a hairpin loop, and within the loop is the conserved 9-nt sequence 5'-TAATATT↓AC-3'.
Geminiviruses are classified into nine genera namely Becurtovirus, Begomovirus, Capulavirus, Curtovirus, Eragrovirus, Grablovirus, Mastrevirus, Topocuvirus, and Turncurtovirus, based on the genome organization, host-range, and the type of insect vector which transmits the virus (Zerbini et al., 2017). Viruses in the genus Begomovirus have mono- or bipartite genomes while those of all other genera possess monopartite genome organization. Geminiviruses exhibit bidirectional transcription and encode 5–7 proteins that exploit and reprogram host machineries to establish infection (Hanley-Bowdoin et al., 2013; Aguilar et al., 2020). Two systems of gene nomenclature are currently in use. Both designate genes and gene products by numbers. One nomenclature denotes genes as virion-sense (V) or complementary-sense (C), whereas the other indicates genes as oriented in the rightward (R; virion-sense; clockwise) or leftward (L; complementary-sense; counter clockwise) direction of the genome map. We have used in this review, the nomenclature based on the virion-sense (V) and complementary-sense (C) strands.
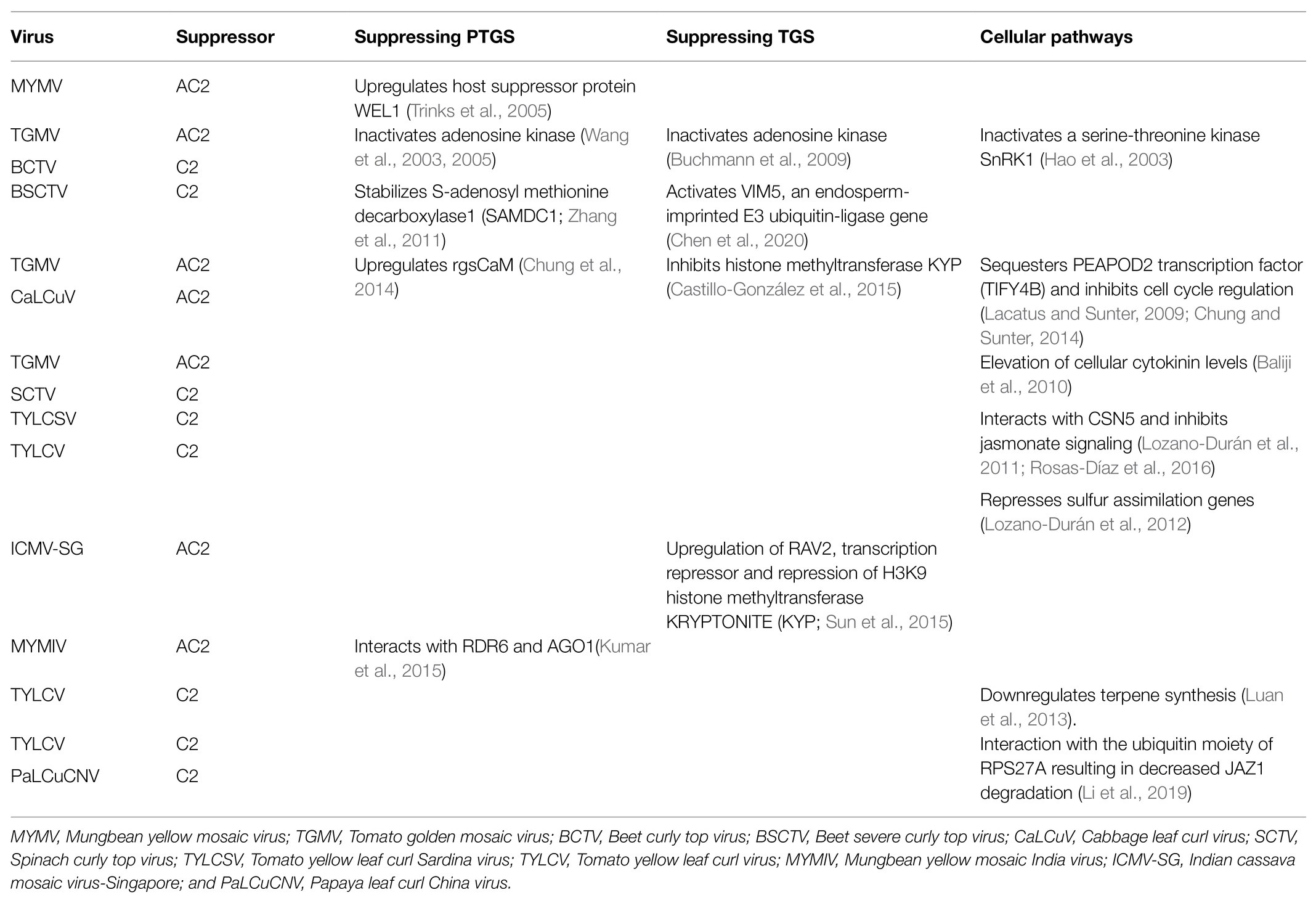
Begomovirus represents the largest and the best-studied geminivirus genus. The bipartite genomes of begomoviruses are designated as DNA A and DNA B (Figure 1A). DNA A encodes five or six open reading frames (ORFs) and DNA B encodes two ORFs. The virion-sense ORFs AV1 and AV2 of DNA A encode coat protein (CP) and pre-coat protein, respectively (Padidam et al., 1996). The complementary-sense strand ORFs of DNA A encode the replication-associated protein (AC1/Rep), the transcriptional activator protein (AC2/TrAP), the replication enhancer protein (AC3/REn), and AC4 (Hanley-Bowdoin et al., 2000). DNA B ORFs encode the nuclear-shuttle protein (BV1/NSP) and the movement protein (BC1/MP; Schaffer et al., 1995).
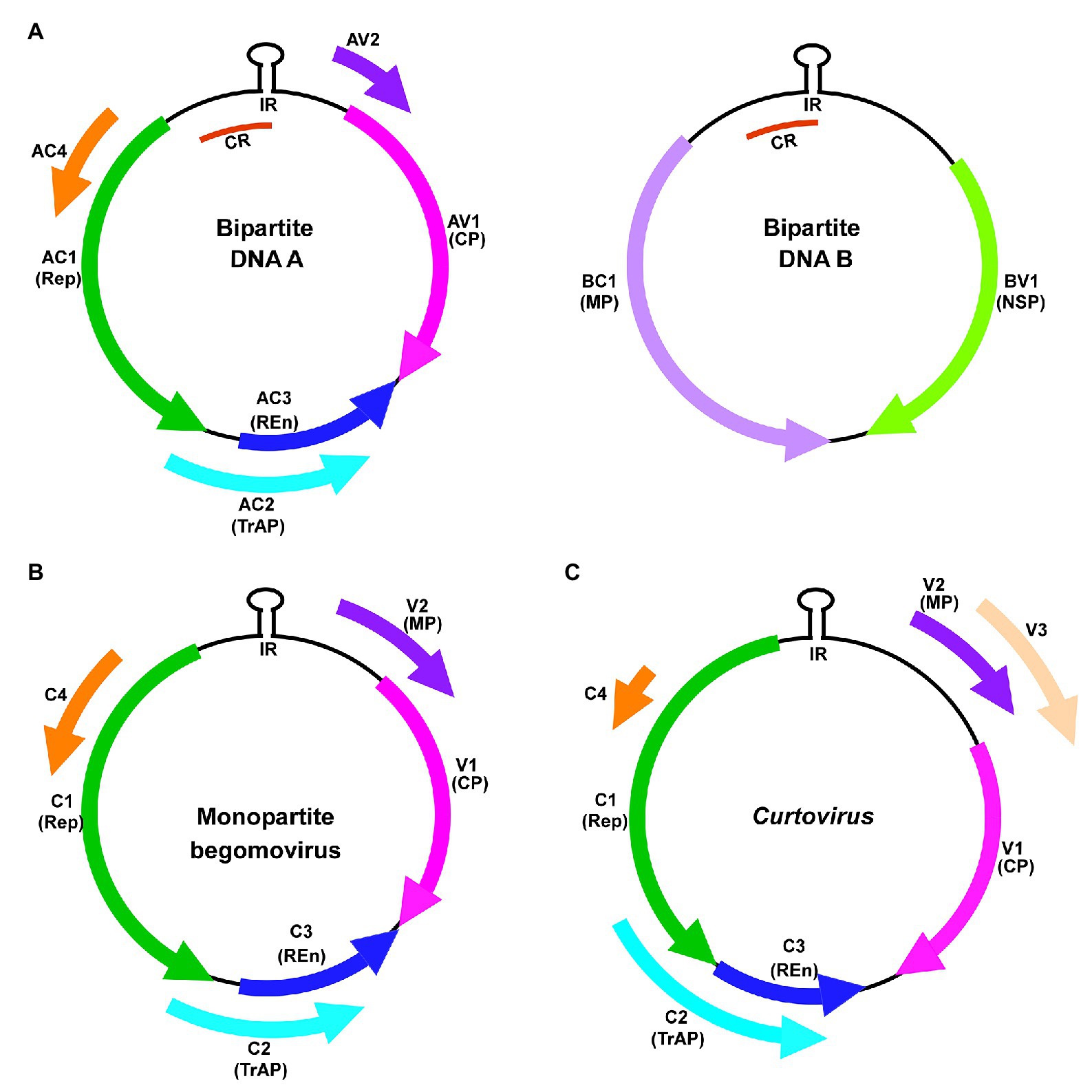
Figure 1. Genomic organization of (A) bipartite begomovirus, (B) monopartite begomovirus, and (C) curtovirus. Virion-sense (V) or complementary-sense (C) strand open reading frames (ORFs) and corresponding protein products are coded by color. CR, common region; CP, coat protein; IR, intergenic region; MP, movement protein; NSP, nuclear shuttle protein; REn, replication enhancer protein; Rep, replication-associated protein; and TrAP, transcriptional activator protein.
Monopartite begomoviruses lack the DNA B component (Kheyr-Pour et al., 1991; Navot et al., 1991). The single, circular ssDNA genome encodes six ORFs, two in the virion-sense (V1 and V2) and four in the complementary-sense (C1, C2, C3, and C4) strands (Figure 1B). Some monopartite begomoviruses do not cause typical disease symptoms in infected plants when acting alone. They require an ssDNA satellite molecule (betasatellite/alphasatellite/deltasatellites/defective satellites) to cause symptomatic disease and viral DNA accumulation (Saunders et al., 2000; Fiallo-Olivé et al., 2012, 2016).
Curtovirus genome organization is like that of monopartite begomoviruses (Figure 1C). The virion-sense strand of curtoviruses is more complex with three ORFS V1, V2, and V3. V1 encodes the coat protein and V3 encodes the movement protein. V2/Reg is a unique curtovirus protein that regulates relative ssDNA and dsDNA levels and hence the name “Reg” (Hormuzdi and Bisaro, 1993). The complementary-sense strand encodes four genes namely Rep/C1, C2, REn/C3, and C4 (Stanley and Latham, 1992; Stanley et al., 1992). Rep/C1 (Luna and Lozano-Durán, 2020) and Ren/C3 are highly conserved between curtovirus and begomovirus. Curtovirus REn was shown to functionally complement the REn mutation in a bipartite begomovirus (Hormuzdi and Bisaro, 1995). However, curtovirus C2, and the positional homolog of begomovirus AC2, display only partial sequence and functional homology between them (Luna and Lozano-Durán, 2020).
Although the position of AC2/C2 ORFs is conserved in most geminivirus genera, the functions of AC2/C2 are known only in curtovirus and begomovirus. AC2, a 15 kDa multifunctional protein, is also known as C2, L2, AL2, or transcriptional activator protein (TrAP). AC2 is a delayed early gene product which transactivates late viral genes, CP, and NSP (Sunter and Bisaro, 1991, 1992; Haley et al., 1992; Shivaprasad et al., 2005), suppresses host defense mechanism (Bisaro, 2006; Raja et al., 2010), and acts as a symptom determinant (Hao et al., 2003; Rajeswaran et al., 2007; Siddiqui et al., 2008). C2, a positional homolog of AC2 in monopartite begomoviruses, is highly similar to AC2 in sequence and function (Figure 2A; Noris et al., 1996; van Wezel et al., 2001, 2002, 2003; Dong et al., 2003; Gopal et al., 2007). The C2 protein which is encoded by the curtoviruses Beet curly top virus (BCTV), Beet severe curly top virus (BSCTV), and Spinach curly top virus (SCTV) exhibits very little sequence similarity when compared to begomovirus AC2/C2 except for the conserved Cys-His residues in the middle (Figures 2A,D; Luna and Lozano-Durán, 2020). AC2 has three highly conserved functional domains: (1) N-terminal basic domain with a bipartite nuclear localization signal (NLS), which consists of four consecutive arginine residues (Figures 2A–C; Dong et al., 2003; Trinks et al., 2005). (2) A conserved zinc finger-like domain comprising the conserved cysteine and histidine residues (CCHC) which is present in the middle. (3) The C-terminal which possesses an acidic transactivation domain (Figures 2A,B; Trinks et al., 2005). This review focuses on the host targets and versatile mechanisms deployed by the geminiviral silencing suppressor protein AC2/C2 to counter the plant defense.
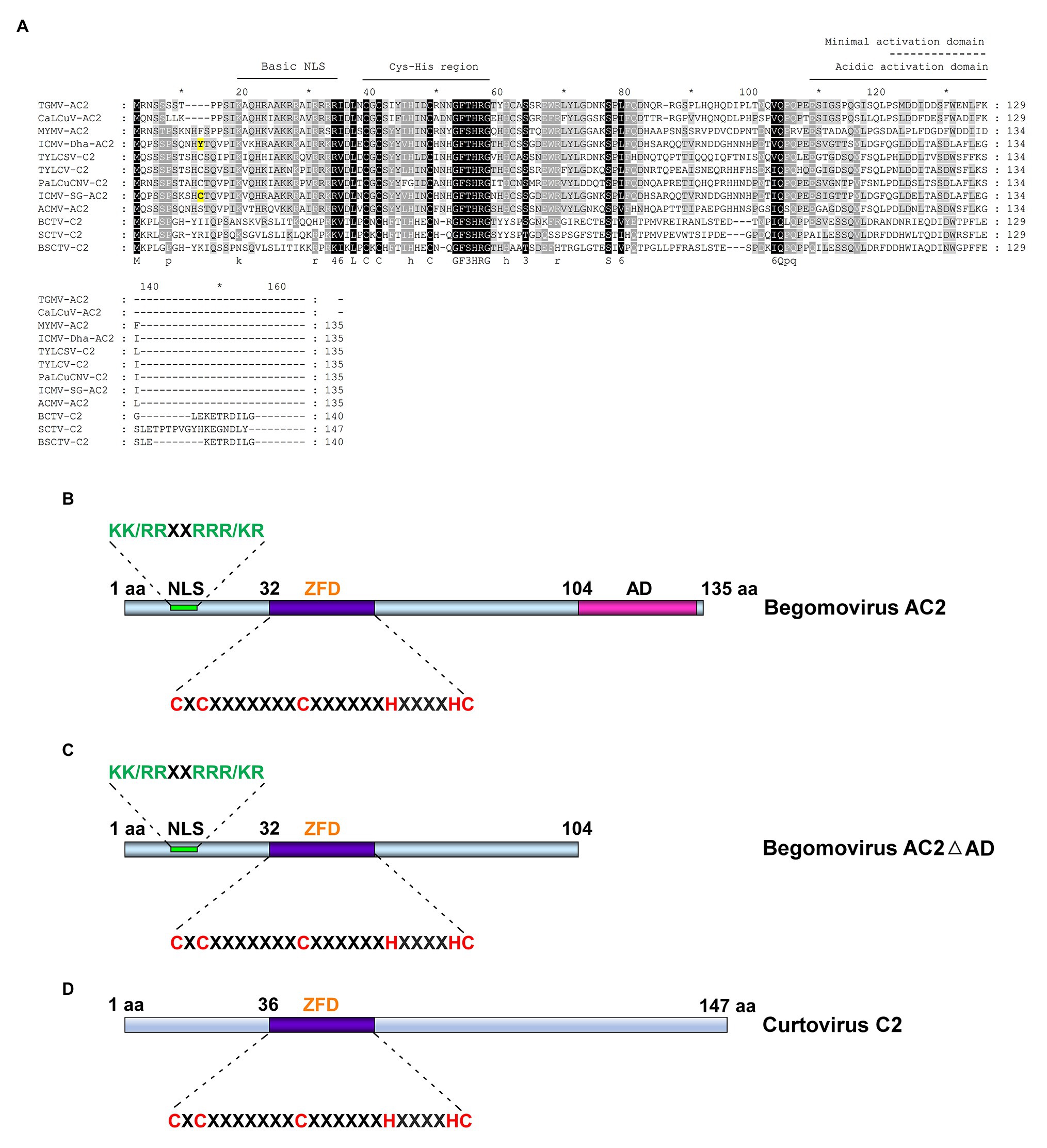
Figure 2. Alignment of AC2 of bipartite, C2 proteins of monopartite begomovirus, and C2 of curtovirus. (A) Clustal alignment of AC2/C2 amino sequences of Tomato golden mosaic virus (TGMV; acc. no. NC_001507); Mungbean yellow mosaic virus (MYMV; acc. no. AJ132575); African cassava mosaic virus (ACMV; acc. no. NC_001467); Indian cassava mosaic virus-Singapore (ICMV-SG; acc. no. JX518289); Indian cassava mosaic virus-Dharwad (ICMV-Dha; acc. no. GQ924760); Cabbage leaf curl virus (CaLCuV; acc. no. NC_00386); Tomato yellow leaf curl Sardinia virus (TYLCSV; acc. no. L27708); Tomato yellow leaf curl virus (TYLCV; acc. no. AM282874); Papaya leaf curl China virus (PaLCuCNV; acc. no. FN256260); Beet curly top virus (BCTV; acc. no. AF379637); Beet severe curly top virus (BSCTV; acc. no. U02311); and Spinach curly top virus (SCTV; Acc. No. AY548948). The conserved N-terminal basic domain, the Cys-His residues in the middle and the C-terminal acidic activation domain are marked. The Tyr residue in ICMV-Dha strain and Cys residue in pathogenic ICMV-SG strain are highlighted in yellow. (B) Schematic representation of begomovirus AC2 involved in activation domain-dependent silencing suppression. (C) Schematic representation of begomovirus AC2 with deletion in activation domain (AC2ΔAD) involved in activation domain-independent silencing suppression. (D) Schematic representation of curtovirus C2 involved in activation domain-independent silencing suppression. NLS, nuclear localization signal; ZFD, zinc-finger like domain; and AD, activation domain.
Plant Defense and Viral Counter-Defense Mechanisms
RNA silencing is a very effective antiviral defense mechanism. RNA silencing has evolved as the first line of defense against invading nucleic acids including viruses, transposons, transgenes, and repetitive sequences (Baulcombe, 2004; Voinnet, 2005; Shabalina and Koonin, 2008). In addition to structured ssRNA viral genomes, DNA viral transcripts which are structured and overlapping transcripts also act as precursors for viral siRNA pathway (Blevins et al., 2011; Aregger et al., 2012). The dsRNA is processed into 21 nt siRNAs by DICER-LIKE4/DCL4 protein (Figure 3A; Akbergenov et al., 2006; Blevins et al., 2006). DCL2 expression stimulates transitivity and secondary siRNA production and increases silencing efficiency in the absence of DCL4 (Parent et al., 2015). When DCL2 and DCL4 are present together, abundant RNAs from viruses and transgenes are processed hierarchically first by DCL4 which has high affinity and processivity to restrict off-target silencing caused by the secondary siRNAs generated by the transitivity-prone DCL2 (Parent et al., 2015). DCL1-mediated processing of geminiviral dsRNA into 21 nt siRNA is inefficient (Blevins et al., 2006); however, it acts as a positive regulator by making viral dsRNAs available to other DCLs to be processed into siRNAs (Blevins et al., 2006; Csorba et al., 2015). DCL1 also acts as a negative regulator of viral silencing by downregulating DCL4 and DCL3 (Qu et al., 2008). The 21 nt siRNA generated by DCL4 cleaves the target viral transcript in association with ARGONAUTE (AGO) proteins. AGO2 acts as a second layer of defense when AGO1 is suppressed in Arabidopsis (Harvey et al., 2011). AGO7 acts in coordination with AGO1 for viral clearance (Qu et al., 2008) but preferentially targets less structured viral RNA. The role of AGO7 in geminiviral defense is not clear.
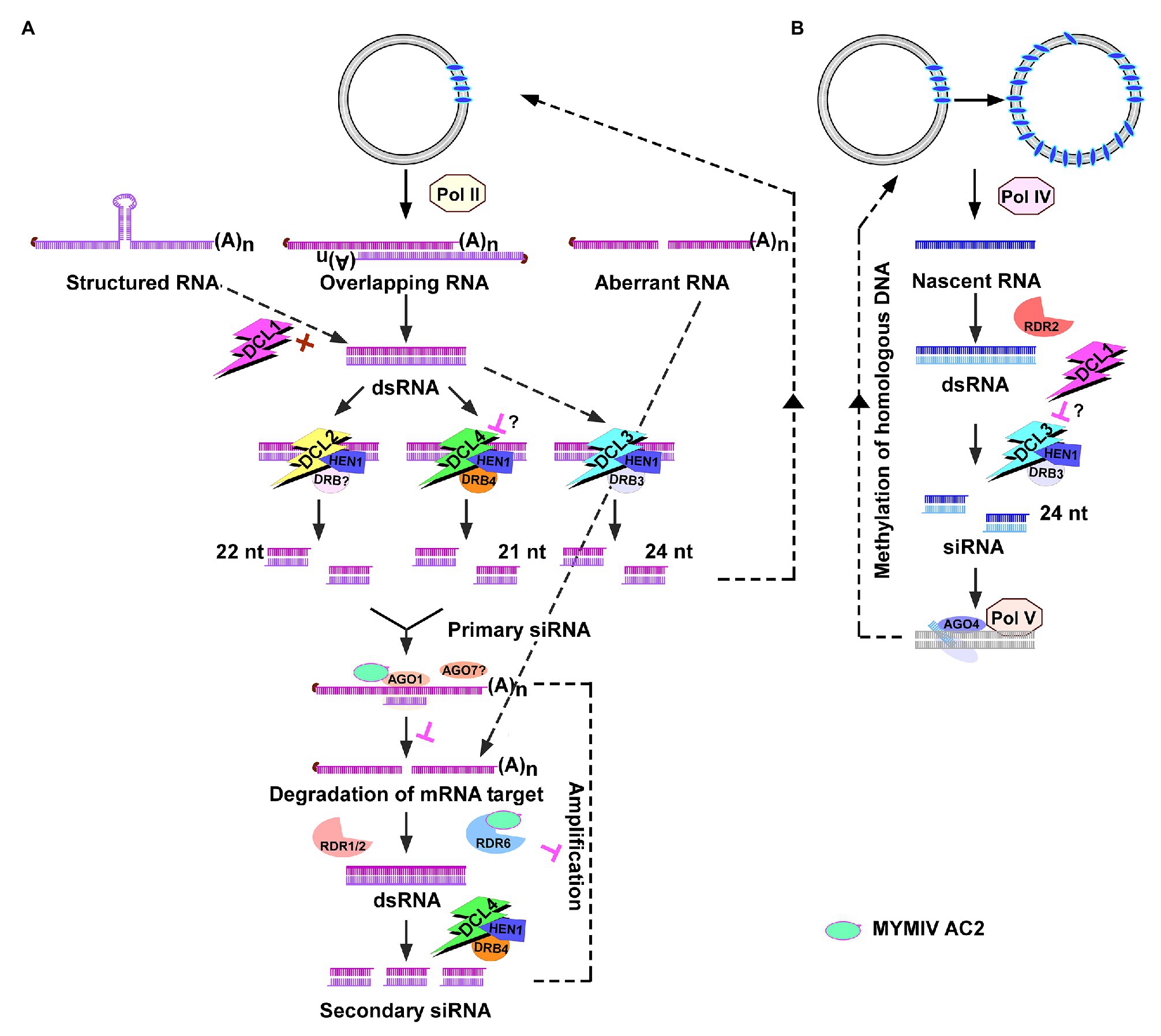
Figure 3. Geminiviral siRNA pathways. (A) Post-transcriptional gene silencing (PTGS) pathway. The host RNA polymerase transcribes the replicative form of virus into structured/overlapping/aberrant transcripts which are processed into 21 nt siRNA by DICER-LIKE4/DCL4. DCL2 generates 22 nt siRNAs in the absence of DCL4. DCL1 acts as a positive regulator (+) by facilitating dsRNA access to other DCLs. The role of DCL1 as a negative regulator (┴) of DCL4 and DCL3 in geminiviral siRNA pathway is not clear (?). The 21 nt siRNAs-Argonaute1 (AGO1) target viral transcripts for slicing by the endo-ribonuclease activity of AGO1. The role of AGO7 in geminiviral defense is not clear (?). The sliced mRNA transcript serves as a template for RNA-DEPENDENT RNA POLYMERASE1/2/6 (RDR1, -2, -6) to convert it into dsRNA, which thereafter generates secondary siRNAs to amplify the host defense response. Suppression of PTGS by binding AGO1 and RDR6 by Mungbean yellow mosaic India virus (MYMIV) AC2 is denoted. (B) Transcriptional gene silencing (TGS) pathway. The Pol II transcribed dsRNA is processed by DCL3 (when DCL2 and DCL4 are saturated) into 24 nt siRNAs which cause de novo methylation of viral genome. The methylated viral genome is transcribed by Pol IV to produce a nascent transcript, which is converted to dsRNA by RNA-DEPENDENT RNA POLYMERASE2/RDR2. dsRNA is processed by DCL3 into 24 nt siRNAs. The complex of 24 nt siRNA-AGO4 targets the Pol V-generated scaffold transcript to mediate RNA-directed DNA methylation. DRB, double-stranded RNA binding proteins and HEN1, HUA ENHANCER1 methyltransferase.
The AGO-siRNA sliced target transcript serves as a template for RNA-DEPENDENT RNA POLYMERASE1/RDR1, RDR2, and RDR6 to convert it into dsRNA, which thereafter generates secondary siRNAs in RNA viruses (Garcia-Ruiz et al., 2010). Interestingly, geminiviral mRNAs appear to be poor templates for RDR-dependent secondary siRNA biogenesis. Aregger et al. (2012) showed that the viral siRNAs which accumulate in Cabbage leaf curl virus (CaLCuV)-infected Arabidopsis are RDR1/2/6-independent primary siRNAs. Interestingly, RDR6 was shown as a target of Mungbean yellow mosaic India virus (MYMIV) AC2 (Kumar et al., 2015). Although DCL4 has been implicated in the generation of 21 nt siRNAs, DCL2 and DCL3 appear to work in concert to generate the antiviral response (Figures 3A,B). Abundance of 21, 22, and 24 nt siRNAs is observed in cassava and Nicotiana benthamiana infected with African cassava mosaic virus (ACMV; Akbergenov et al., 2006). Prevalence of 21–22 nt siRNAs was observed in tomato infected with Tomato yellow leaf curl Sardinia virus (TYLCSV; Miozzi et al., 2013) and preferential accumulation of 22 nt siRNAs was observed in tomato and N. benthamiana infected with Tomato yellow leaf curl China virus (TYLCCNV; Yang et al., 2011). Interestingly, 24 nt siRNAs generated by DCL3 (Figure 3B) are the most abundant siRNAs in Arabidopsis infected with CaLCuV (Blevins et al., 2006).
RNA-directed DNA methylation (RdDM) of cytosine residues and histone H3 lysine 9 dimethylation (H3K9me2) of chromatin are hallmarks of epigenetic defense mechanism evolved by the plants against invading DNAs including geminiviruses and transposons. The equilibrium between repressed and active viral chromatin determines the outcome of infection and symptom remission. Host recovery is tightly associated with the equilibrium favoring repressed state (Ceniceros-Ojeda et al., 2016; Coursey et al., 2018). The canonical RdDM machinery includes RNA DEPENDENT RDR2, DCL3, AGO4, and two plant-specific RNA polymerases, Pol IV and Pol V (Zhang et al., 2018). Pol IV transcribes heterochromatic regions to produce a nascent transcript (Herr et al., 2005; Daxinger et al., 2009), which is recruited to the cajal bodies in the nucleolus, where they are converted into dsRNA by RDR2 (Li et al., 2006). The resultant dsRNA is processed by DCL3 into 24 nt siRNAs, which are methylated by HEN1 and used by AGO4 to base pair with the Pol V-generated scaffold transcript (Wierzbicki et al., 2008) to mediate RNA-directed DNA methylation (Figure 3B). Scaffold transcripts which originate from intergenic non-coding sequences are required for silencing adjacent siRNA-generating loci (Wierzbicki et al., 2009). AGO4 recruits chromatin modifying methyltransferase (MTase) and also slices the scaffold transcript which serves as a template for RDR2-mediated dsRNA production and amplification of 24 nt siRNA. Methylation of the invading geminiviral genome is mediated by 24 nt siRNAs (Raja et al., 2008; Buchmann et al., 2009). Interestingly, Pol IV and Pol V were shown to be not essential for de novo methylation of geminiviral genome (Jackel et al., 2016). Arabidopsis mutants pol IV and pol V reduced accumulation of all classes of virus-derived siRNAs suggesting that they are not so essential in viral siRNA biogenesis including 24 nt siRNA biogenesis. While Pol IV and Pol V were not essential in establishing cytosine methylation in the viral genome, they were found to be critical for maintenance and amplification of methylation. Incidentally, the mechanism involving Pol II-RDR6-mediated methylation of retrotransposon (Marí-Ordóñez et al., 2013) was evoked to explain the de novo methylation of geminiviral genome (Jackel et al., 2016; Figure 3B). A study by Marí-Ordóñez et al. (2013) showed that EVADE (EVD), a retrotransposon, generates high levels of Pol II-RDR6 dependent dsRNA upon proliferation which could eventually quench the DCL2 and DCL4 dicers involved in 21 and 22 nt siRNA biogenesis. Thus, the Pol II-RDR6 dependent dsRNA are processed by DCL3 into 24 nt siRNAs from transcribed regions of the retrotransposon. The key role of Pol IV and Pol V in establishing chromatin methylation and in enabling recovery emphasized the importance of these enzymes in mounting antiviral defense through methylation.
The presence of a robust viral counter defense mechanism is underscored by the ubiquitous presence of one or more silencing suppressor proteins in the armor of a virus. The arms race between silencing and silencing suppression results in resistance or susceptibility to the pathogen. Geminiviruses encode several proteins namely AC2/C2, AC4/C4, AV2/V2, βC1, and Rep that suppress RNA silencing by targeting various components of the post-transcriptional gene silencing (PTGS) machinery, transcriptional gene silencing (TGS) machinery, and cellular regulatory genes (reviewed in Hanley-Bowdoin et al., 2013; Kumar, 2019; Rishishwar and Dasgupta, 2019; Yang et al., 2019; Guerrero et al., 2020). Of all the known geminiviral suppressor proteins, AC2 is the most well-studied and is known to target multiple plant genes and proteins. In this review, we have taken a comprehensive approach to document all known targets of the geminiviral suppressor protein AC2 and the interconnecting and/or unique mechanisms evolved by the viruses to counter the plant defense mechanism.
AC2-Mediated Suppression of Host Defense Mechanism
AC2 of begomoviruses and C2 of curtoviruses act as suppressors of silencing through two broadly classified mechanisms: (i) activation domain-dependent silencing suppression- begomovirus AC2 with a C-terminal acidic activation domain (Figure 2B) is involved in this mechanism and (ii) activation domain-independent silencing suppression, which is manifested by curtovirus C2 which lacks the activation domain (Figure 2D) and certain begomovirus AC2 proteins with deletions of the activation domain (Figure 2C). AC2/C2 also interacts with and inactivates many cellular regulatory proteins to circumvent the innate defense mechanism which is independent of siRNA-mediated silencing (Guerrero et al., 2020).
Activation Domain-Dependent Silencing Suppression
Suppression activity of AC2 was first demonstrated by Voinnet et al. (1999) in the N. benthamiana line 16c, in which the previously established silencing of gfp was reverted by the expression of the ACMV AC2 in a potato virus X (PVX) vector. Similar studies with C2 of Tomato yellow leaf curl virus-China (TYLCV-C), a monopartite begomovirus, revealed that the suppression of silencing was associated with the presence of an intact zinc finger-like motif (van Wezel et al., 2002) and an NLS (Dong et al., 2003). East African cassava mosaic Cameroon virus (EACMCV) and Indian cassava mosaic virus (ICMV) are known to suppress silencing (Vanitharani et al., 2004). Although the mechanism of suppression is not clear, transient AC2 expression enabled ~8-fold increase in synergistic mixed infection with recovery type viruses ACMV and Sri Lankan cassava mosaic virus (SLCMV), which exhibit recovery from symptoms 2 to 3 weeks after infection (Vanitharani et al., 2004). The activation domain-dependent silencing suppression is also known as “transcription-dependent silencing suppression” (Bisaro, 2006). Trinks et al. (2005) demonstrated the requirement of the intact activation domain of Mungbean yellow mosaic virus (MYMV) AC2 in addition to the NLS and Zn-finger motifs to suppress silencing. The need for NLS, Zn-finger motif, and activation domain of AC2 for efficient silencing suppression suggested that MYMV and ACMV AC2 and TYLCV-C C2 regulate silencing suppression in the host cell nucleus and are dependent on DNA interaction and transcriptional activation.
Suppression of PTGS by Transactivation of Host Suppressor WEL1
Transient expression of MYMV and ACMV AC2 in Arabidopsis protoplasts upregulated the expression of 30 plant genes. One such gene was Werner’s exonuclease-like 1 (WEL1), a homolog of Werner Syndrome-like exonuclease (WEX). WEX, an RNase D exonuclease-like protein acts as a positive regulator of post-transcriptional gene silencing (PTGS; Glazov et al., 2003). WEL1 does not have the complete “DEDDY” signature conserved in WEX. Hence, AC2-mediated upregulation of WEL1 is likely to exert a dominant negative effect on WEX function (Figure 4A; Trinks et al., 2005). Thereby, a novel mechanism of AC2-mediated induction of host silencing suppressors was proposed by Trinks et al. (2005). The authors showed induction of WEL1 and five other genes by MYMV and ACMV AC2 in Arabidopsis and Nicotiana plumbaginifolia protoplasts. However, upregulation of WEL1 upon ACMV or MYMV viral infection or an increase in viral load upon transgenic over-expression of WEL1 has not been demonstrated to date; thus, the role of WEL1 as a host silencing suppressor remains to be confirmed. Similarly, the authors did not evaluate whether silencing of WEL1 abolished AC2-mediated suppression in N. benthamiana 16c line. Interestingly, Trinks et al. (2005) demonstrated that mutations of all three functional domains (NLS1-, ZF-, and AD-) abolished transactivation as well as silencing suppression property of AC2. MYMV AC2 being a small, multifunctional protein, the loss of silencing suppression of AC2 mutants by impaired folding or loss of interaction with host factors cannot be discounted.
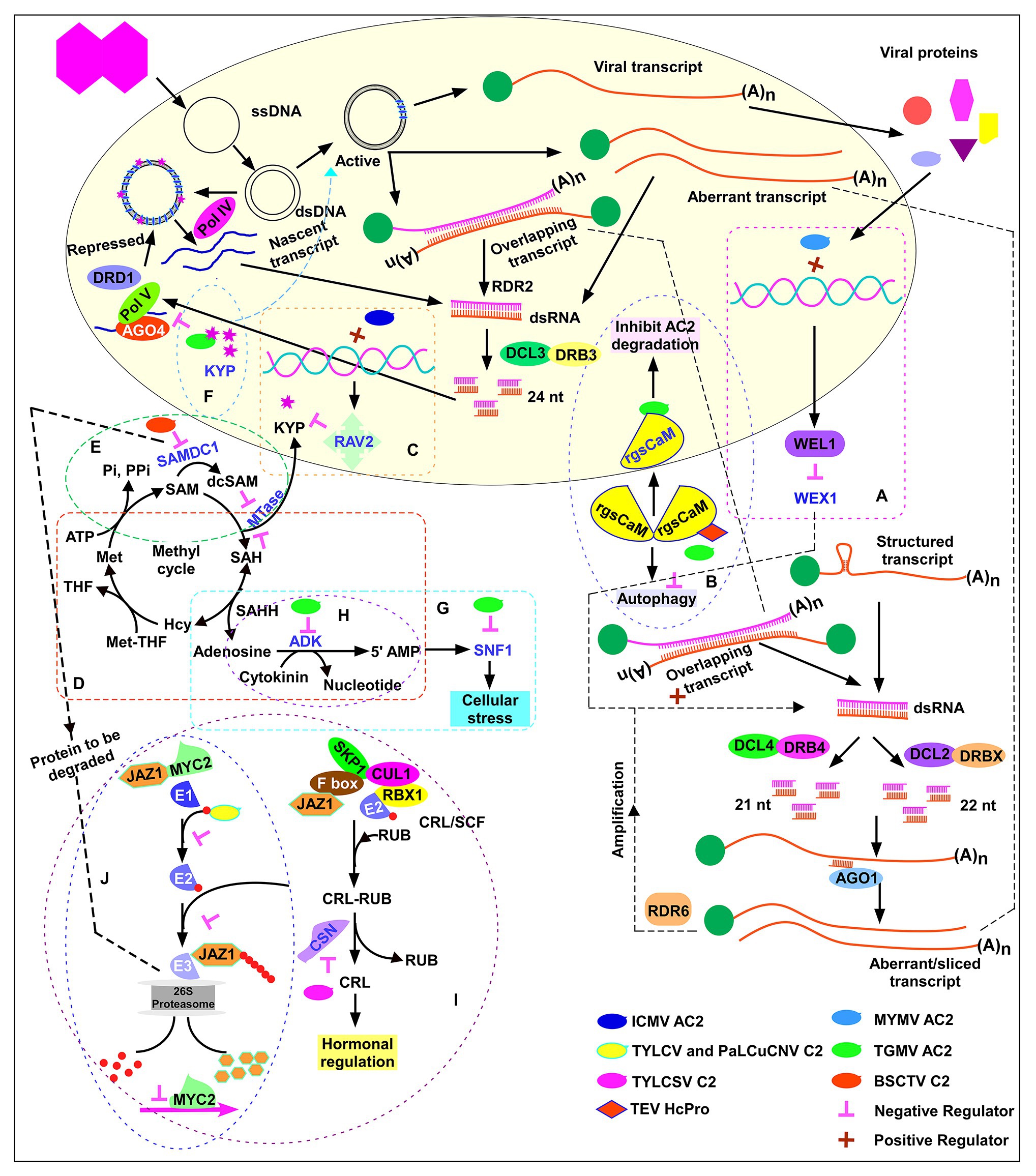
Figure 4. AC2-mediated suppression of host defense mechanism. Activation domain-dependent mechanism: suppression of PTGS by activating the host silencing suppressors (A) Werner’s exonuclease-like 1 (WEL1) by MYMV AC2, (B) calmodulin-like protein by TGMV AC2, and (C) RELATED TO ABI3 and VP1 (RAV2), transcription repression and downregulation of H3K9 histone methyltransferase KRYPTONITE (KYP) by Indian cassava mosaic virus (ICMV) AC2. Activation domain-independent mechanism: (D) The methyl cycle and suppression of PTGS/TGS by inactivating adenosine kinase by TGMV AC2 and BCTV C2. (E) Suppression of PTGS by stabilizing S-adenosyl methionine decarboxylase1 by BSCTV C2. (F) Suppression of TGS by inhibiting KYP enzymatic activity by TGMV and CaLCuV AC2 binding. (G) Suppression mediated by inactivation of SNF1 kinase by TGMV AC2 and BCTV C2. (H) Suppression mediated by elevation of cellular cytokinin levels by TGMV AC2 and C2 of SCTV. (I) Suppression mediated by inhibition of jasmonate signaling pathway by C2 of TYLCSV. (J) Suppression mediated by competitive binding of C2 of TYLCV and PaLCuCNV to ubiquitin.
Suppression of PTGS by Transactivation of Calmodulin-Like Protein
Tomato golden mosaic virus (TGMV) AC2 induces a calmodulin-like protein Nb-rgsCaM (Chung et al., 2014). rgsCaM over-expression leads to an increase in viral DNA load. rgsCaM was shown to be induced by HcPro, a silencing suppressor of Tobacco etch virus (TEV; Anandalakshmi et al., 1998) and over-expression of rgsCaM was shown to reverse PTGS (Anandalakshmi et al., 2000), suggesting the role of rgsCaM as an endogenous silencing suppressor. rgsCaM prevents TEV HcPro and Cucumber mosaic virus suppressor 2b from binding to dsRNAs/siRNAs and reduce the suppressor protein stability by autophagy, resulting in a more potent RNAi defense against viral infection. rgsCaM over-expressing lines were less susceptible to the virus (Nakahara et al., 2012). In contrast, rgsCaM over-expression resulted in increased susceptibility to TGMV and CaLCuV (Chung et al., 2014) likely because TGMV and CaLCuV are DNA viruses, whereas the viruses studied by Nakahara et al. (2012) were RNA viruses. TGMV AC2 does not bind dsRNAs/siRNAs unlike RNA viral suppressors HcPro, P19, and 2b. rgsCaM self-interaction was observed in cytoplasm while interaction with TGMV AC2 sequestered rgsCaM to the nucleus (Figure 4B). It was speculated that AC2-mediated localization of rgsCaM to the nucleus is the likely mechanism evolved by TGMV to evade degradation of AC2 by autophagy and thereby effectively suppress the plant defense mechanism. The cajal bodies in the nucleolus are the sites of 24 nt siRNA biogenesis (Pontes and Pikaard, 2008). Chung et al. (2014) speculated that nuclear localization of rgsCaM by TGMV AC2 might interfere with the overall host siRNA biogenesis and make the plants more susceptible to the virus. More studies are needed in future to confirm this hypothesis. TYLCCNV βC1 was shown to act as suppressor of PTGS by upregulating rgsCaM (Li et al., 2014). A recent study showed that TYLCCNV βC1-upregulated rgsCaM interacts with Suppressor of Gene Silencing 3 (SGS3), a cofactor of RDR6 and induces autophagic degradation of it and thereby suppresses PTGS (Li et al., 2017).
Suppression of TGS by Transactivation of Host Repressor
Indian cassava mosaic virus-Singapore (ICMV-SG) displayed higher pathogenicity in comparison to ICMV-Dharwad (ICMV-Dha). A single point mutation that changes Tyr to Cys in ICMV-Dha AC2 (ICMVDhaY11C) significantly increased pathogenicity (Figure 2A; Sun et al., 2015). Increased ICMV-SG infection was associated with increased repression of H3K9 histone MTase KRYPTONITE (NbKYP), a key enzyme for maintenance of chromatin methylation. Downregulation of KYP was directly correlated with an increase in RELATED TO ABI3 and VP1 (RAV2), a transcription repressor. RAV2 is known to regulate RNA silencing and to get upregulated by suppressor proteins of potyvirus HcPro and cucomovirus P38 in Arabidopsis (Endres et al., 2010). Transient expression of ICMV-SG, ICMV-Dharwad (ICMV-Dha), and the ICMV-DhaY11CAC2 mutant in N. benthamiana 16c-TGS reversed the TGS of the 16c line as evidenced by reactivation of green fluorescence (Sun et al., 2015). Silencing of NbRAV2 significantly reduced the viral titer, thus indicating a novel mechanism of silencing suppression. Activation of the putative transcription repressor NbRAV2 by ICMV-AC2 interferes with TGS by suppressing the expression of KYP (Figure 4C). It is not known whether the RAV2 repressor protein directly binds to the KYP promoter sequence to downregulate its expression. A RAV2-dependent upregulation of Arabidopsis FIERY1 (FRY1) and CML38 (rgsCaM homolog) was observed in TuMV HC-Pro transgenic lines (Endres et al., 2010). It will be useful to study whether ICMV-SG induced RAV2 protein can induce rgsCaM in N. benthamiana.
Activation Domain-Independent Silencing Suppression
Reports on AC2-mediated silencing suppression in TGMV suggest that not all viruses require C-terminal activation domain in AC2 for mediating silencing suppression. The activation domain-independent silencing suppression observed in TGMV is also referred as “transcription-independent silencing suppression” (Bisaro, 2006). The unique mechanism evolved by the viruses encoding activation domain-independent silencing suppressors is by interfering with the methyl cycle. S-adenosyl methionine (SAM) is the methyl donor for most transmethylation reactions and is an essential MTase co-factor. An increase in the accumulation of SAM analogs would competitively inhibit MTase and prevent methylation and associated silencing. Activation domain-independent silencing suppressors interfere with the methyl cycle and increase the cellular levels of SAM analogs which compete with SAM. In addition to methyl cycle interference, TGMV and CaLCuV AC2 directly target and inhibit the H3K9me2 histone MTase Su(var)3-9 homolog 4/Kryptonite (SUVH4/KYP), an enzyme critical for histone methylation.
Suppression of PTGS by Inactivating Adenosine Kinase
TGMV AC2 with activation domain deletion (AC2-∆AD; Figure 2C) and BCTV C2 (Figure 2D) interact with adenosine kinase (ADK) and inhibit the synthesis of 5'-AMP from adenosine and ATP (Figure 4D; Wang et al., 2003). C2 of BCTV and C2 of SCTV, both curtoviruses, lack the transcriptional activation domain which is present in AC2/C2 of begomoviruses (Sunter et al., 1994; Baliji et al., 2007). Although AC2 self-interacts and moves into the nucleus for transcriptional activation, AC2:ADK and C2:ADK complexes form in the cytoplasm (Yang et al., 2007). This emphasizes the dispensability of the activation domain for ADK interaction. AC2/C2 expression inhibited ADK activity in Escherichia coli, yeast, and transgenic plants (Wang et al., 2003). ADK plays a key role in the methyl cycle and in SAM-dependent MTase activity. MTase catalyzes methyl group transfer from SAM to a methyl acceptor converting SAM to S-adenosyl-homocysteine (SAH). S-adenosyl-homocysteine hydrolase (SAHH) hydrolyzes SAH to homocysteine (Hcy) and adenosine (Figure 4D). SAHH catalyzed reaction is reversible, and the removal of adenosine is essential to tilt the equilibrium of reaction toward hydrolysis of SAH, which otherwise lies strongly toward synthesis of SAH. SAH is also a competitive inhibitor of MTase. Phosphorylation of adenosine to 5'-AMP by ADK prevents resynthesis of SAH and promotes flux through the methyl cycle to regenerate SAM. Removal of adenosine by phosphorylation facilitates methionine (Met) synthesis, wherein methionine synthase catalyzes the transfer of a methyl group from methylated folic acid [methyltetrahydrofolate (MTHF)] to homocysteine. Addition of adenosine to the sulfur group of methionine regenerates SAM. Thus, the removal of SAH is critical as it can strongly compete with SAM and inhibit MTase (Figure 4D). ADK-deficient plants displayed defects in silencing, thus implying an indirect role for ADK in regulating methyl cycle and silencing.
Wild-type AC2 and AC2-∆AD of TGMV and C2 of BCTV suppressed PTGS which was directed against gfp in the N. benthamiana 16c line in an activation domain-independent manner (Wang et al., 2005). The local suppression caused by AC2 could be mimicked by using an invert repeat of ADK or by addition of an adenosine homolog (A-134974) that inhibited ADK activity (Wang et al., 2005). Methylation of the coding region of a gene is a hallmark of PTGS (Ingelbrecht et al., 1994; English et al., 1996). Thus, AC2-mediated inactivation of ADK is likely to have caused suppression of PTGS by its interference in the methylation of coding region (Raja et al., 2010).
Suppression of PTGS by Stabilizing S-Adenosyl Methionine Decarboxylase1
BSCTV C2 interaction with S-adenosyl methionine decarboxylase1 (SAMDC1) and resultant suppression of de novo DNA methylation were observed in Arabidopsis (Zhang et al., 2011). SAMDC1 is a key enzyme in the conversion of SAM to decarboxylate-S-adenosyl methionine (dcSAM). dcSAM acts as an aminopropyl donor for the biosynthesis of spermidine and spermine. dcSAM also competes with SAM and acts as a competitive inhibitor of MTase (Figure 4E). Hence, SAM/dcSAM balance is a key determinant of transmethylation. SAMDC1 possesses a conserved PEST (proline, glutamine, serine, and threonine) sequence that is associated with proteins with rapid turnover rates. BSCTV C2 interacts with SAMDC1 in the PEST region and stabilizes the enzyme by attenuating 26S proteasome-mediated degradation (Figure 4E). The resultant increase in dcSAM/SAM ratio affects the host de novo methylation. Infection of wild-type plants with BSCTV C2− mutant strain (engineered by introducing a stop codon in the C2 ORF) or infection of SAMDC1 mutant plant with wild-type BSCTV strain resulted in enhanced DNA methylation, reduced viral titer and reduced susceptibility confirming the key role of C2 and SAMDC1 in regulating host DNA methylation. Agroinfiltration of BSCTV C2 and SAMDC1 genes reverted gfp silencing in N. benthamiana 16c (Zhang et al., 2011).
Suppression of TGS by Inactivating Adenosine Kinase and Stabilizing SAMDC1
Adenosine kinase phosphorylation of adenosine is a prerequisite for sustaining cellular SAM levels. Thus, AC2-mediated interaction and inactivation of ADK invoked the possibility of methylation-mediated repression of viral genome as a possible host defense mechanism against the virus. The Arabidopsis mutants adk, cmt3, drm1/2, drb3, clsy1, pol IV, and pol V (mutants of RNA-directed DNA methylation components) were hypersensitive to viral infection. Cytosine methylation level of the viral genome was significantly reduced in the hypersusceptible mutant plants (Raja et al., 2008, 2014; Jackel et al., 2016). Infection of Arabidopsis with a BCTV mutant lacking C2 (BCTV C2−) resulted in a host recovery phenotype. The viral DNA in the recovered tissue was hypermethylated, suggesting methylation of the viral genome to be the cause of host recovery. Interestingly, ago4, dcl3, drb3, and pol V mutant Arabidopsis plants did not recover when infected with BCTV C2− (Raja et al., 2008, 2014; Jackel et al., 2016), whereas pol IV and clsy1 mutants displayed a delayed recovery phenotype (Jackel et al., 2016). This finding underlined the requirement of AGO4, DCL3, DRB3, Pol IV, and Pol V for methylation of viral genome and resultant host recovery. These results confirmed that methylation of viral genome has been evolved as an epigenetic defense mechanism against geminiviruses (Raja et al., 2008, 2014; Jackel et al., 2016; Coursey et al., 2018).
The AC2/C2-mediated suppression of methylation was studied in the N. benthamiana (16-TGS) line, in which the 35S promoter-driven gfp transgene is transcriptionally silenced. The silencing of the 16-TGS plant was suppressed by wild-type AC2 and AC2-∆AD of TGMV and CaLCuV and also by BCTV C2 when expressed from PVX vectors (Buchmann et al., 2009). Knocking down of SAHH and ADK expression using a Tobacco rattle virus VIGS vector reversed gfp silencing in 16-TGS. These observations further implied that reversal of silencing mediated by inhibition of methyl cycle is one of the prominent mechanisms evolved by viruses to suppress silencing (Buchmann et al., 2009). Infection of N. benthamiana 16-TGS with TGMV, CaLCuV, and BCTV restored GFP fluorescence in a manner consistent with the tissue tropism exhibited by the virus. TGMV and CaLCuV infection restored GFP expression in symptomatic vascular and mesophyll cells (Buchmann et al., 2009; Raja et al., 2010). Interestingly, BCTV-mediated suppression of gfp silencing was confined to the vascular tissue, which corroborated well with the vascular specificity of the virus. Transgenic expression of AC2 and AC2-∆AD of TGMV, BCTV C2, and dsADK under a dexamethasone (dex)-inducible promoter reversed the methylation of transcriptionally silenced loci in Arabidopsis. The reversal of methylation by AC2 and AC2-∆AD of TGMV, and BCTV C2 was found to be locus non-specific. Four independent regions, including one gene-coding region, two transposable elements, and one repetitive DNA region were used as markers to study the effect of AC2 and AC2-∆AD of TGMV, and BCTV C2 on cytosine methylation. Reversal of cytosine methylation resulted in ectopic expression of TGS-silenced loci namely, a putative F-box gene, a retrotransposon AtSN1 (SINE element), and Athila (LTR element) in all analyzed transgenic plants. The ability of AC2 and AC2-∆AD of TGMV and BCTV C2 to suppress TGS indicates that the suppression is activation domain-independent. TGS in the CACTA-like transposon was reversed only upon wild-type AC2 induction. This invoked the necessity of transcriptional activation domain for mediating suppression of the CACTA TGS-loci. Thus, AC2 transgenic plants reduced cytosine methylation of a wide spectrum of genomic regions and reversed methylation of the TGS loci in Arabidopsis by non-specifically inhibiting cellular transmethylation reactions (Buchmann et al., 2009). Although, methylation extension assays confirmed a decrease in 5-methylcytosine methylation of a wide range of genomic regions in AC2 and C2 transgenic lines (Buchmann et al., 2009), a whole-genome bisulfite sequencing would provide data on AC2 or C2-mediated reversal of genome-wide cytosine methylation at single-nucleotide resolution.
The attenuation of SAMDC1 degradation by BSCTV C2 may also result in suppression of TGS. SAMDC1 stabilization resulted in an increase in dcSAM/SAM ratio, which in turn reduced the host de novo methylation. BSCTV-C2 mutant infection resulted in enhanced DNA methylation of the viral genome (Zhang et al., 2011). In an interesting assay, an FWA genomic fragment containing tandem repeats in its promoter region was used to study the role of BSCTV C2 on de novo methylation. Methylation of the FWA promoter and consequent silencing of the FWA gene is essential for normal flowering. The absence of de novo methylation of FWA transgene in a MTase double mutant Arabidopsis plant (drm1/drm2) caused a delay in flowering (Cao and Jacobsen, 2002). Similar late flowering phenotype was observed when the FWA gene was transformed into C2-expressing transgenic lines (Zhang et al., 2011). This suggested a role for BSCTV-C2 in interfering with the de novo methylation of the FWA transgene.
Developmental Stage-Specific Silencing Suppression
CaLCuV AC2 with the transcription activation domain deletion, and BCTV C2 reversed PTGS and TGS in the vegetative phase of plants indicating a transcription-independent mechanism of silencing suppression. Interestingly, only CaLCuV AC2 but not BCTV C2 could reverse PTGS and TGS upon onset of flowering, indicating that a transcription-dependent activity is required during the reproductive transition. A third suppression mechanism was demonstrated in which CaLCuV AC21–114 lacking the transcription activation domain but not BCTV C2 effectively reversed TGS in reproductive plants (Jackel et al., 2015). Although TGS reversal was observed only in the vegetative phase of plants (plants with prolonged vegetative growth), reduced ADK activity was observed in both vegetative and reproductive plants upon BCTV infection or by silencing of ADK suggesting that TGS and ADK inhibition are uncoupled in reproductive N. benthamiana plants. In contrast, SAHH silencing resulted in reversal of TGS in both vegetative and reproductive plants. Thus, a new mechanism of TGS reversal that is independent of both transcription activation and ADK inactivation was observed in reproductive N. benthamiana plants expressing AC21–114.
Suppression of TGS by Inhibiting KYP
Interestingly, TGMV and CaLCuV AC2 were shown to inhibit the enzymatic activity of KYP by binding to the catalytic domain and thereby decreasing CHH methylation in gene-rich regions (Castillo-González et al., 2015). Over-expression of KYP enriched H3K9me2 mark of viral chromatin, leading to formation of viral heterochromatin. As a counter-defense strategy, inhibition of KYP activity by TGMV and CaLCuV AC2 protein restored the euchromatic status of the minichromosome which allowed active replication and transcription of viral genes and suppression of the host defense mechanism (Figure 4F). While kyp mutant was hypersusceptible to CaLCuV infection and accumulated significantly higher viral titers when compared to wild-type, over-expression of the KYP transgene in the kyp mutant reduced the disease severity significantly.
The necessity of functional AC2 for ssDNA accumulation (Hayes and Buck, 1989) and the requirement of AC2-mediated transactivation of the CP and NSP genes for systemic infection (Sunter and Bisaro, 1992) is well-known. CaLCuV AC2− strain was engineered by introducing a premature stop codon and was used to infect wild-type and kyp mutant plants. While CaLCuV lacking functional AC2 did not show any symptom or systemic accumulation of the mutant virus in wild-type plants, the kyp mutants showed low level of sustained systemic infection of the mutant CaLCuV. Although, the mutant viral titer was very low in kyp mutants, this exciting study confirmed the role of KYP in inhibiting viral replication. Whole genome bisulfite sequencing revealed that AC2 inhibited KYP-dependent CHH methylation. Interaction of AC2 with KYP blocks its methylation activity and relaxes the viral chromatin and facilitates viral replication (Figure 4F; Ré and Manavella, 2015).
Suppression of TGS by Ectopic Expression of VIM5
A recent work by Chen et al. (2020) revealed a unique mechanism in which BSCTV-encoded C2 recruits a host imprinted gene VIM5 to evade host silencing. While over-expressing BSCTV Rep transgene that also contained the C2-C3 promoter sequence and C2 N-terminal sequence (C2N), the authors serendipitously observed accumulation of the C2N transcript in addition to the Rep transcript in one of the transgenic Arabidopsis lines. Additional transcription from the C2-C3 promoter was correlated with hypomethylation of the C2-C3 promoter. Ectopic vegetative transcription of the endosperm-imprinted E3 ubiquitin ligase-encoding gene VARIANT IN METHYLATION5 (VIM5) was observed only in the C2N transcript-expressing transgenic line, when compared to wild-type Arabidopsis. The role of VIM5 in hypomethylation was substantiated by over-expressing VIM5 in another Rep transgenic line that did not originally accumulate the C2N transcript; Over-expression of VIM5 triggered accumulation of the C2N transcript which correlated with decreased DNA methylation at the transgenic C2-C3 promoter. Infection with BSCTV resulted in transient expression of the host endosperm-imprinted E3 ubiquitin ligase-encoding gene VIM5 in rosette leaf tissues. BSCTV infection of Arabidopsis vim5 mutants resulted in delayed accumulation of viral early gene transcripts C2 and C3, thus confirming the role of VIM5 in contributing to the early expression of C2 and C3 from the viral genome. VIM5 was shown to interact with host CG MTase MET1 and the CHG MTase CMT3 and promote 26S proteasomal degradation of MET1 and CMT3. MTase mutant plants met1 and cmt3 displayed reduced viral methylation at the C2-C3 promoter. Similarly, infection of a modified BSCTV with substitutions at CG and CHG sites in the C2-C3 promoter showed higher viral accumulation in vim5 mutant when compared with the unmodified BSCTV. Thus, early expression of BSCTV silencing suppressor C2 and replication enhancer protein C3 immediately after the expression of Rep, by activating an imprinted E3 ubiquitin-ligase gene, is a strategy evolved by the virus to inhibit viral DNA methylation and establish disease (Chen et al., 2020).
Suppression of PTGS by Inhibiting RDR6 and AGO1
Silencing suppression activity of MYMIV AC2 was demonstrated in gfp silenced Nicotiana xanthi plants (Karjee et al., 2008; Rahman et al., 2012). The activation domain mutant of MYMIV-AC2 possessed the suppression activity. A two-pronged mechanism involving physical interaction of AC2 with RDR6 and AGO1 was shown as the basis of PTGS suppression of MYMIV AC2 (Figure 3A). AC2-mediated inhibition of RNA-dependent RNA polymerase activity and slicing activity of AGO1 protein resulted in reduced siRNA accumulation (Figure 3A; Kumar et al., 2015). An in planta assay using invert repeat of RDR6 to reverse gfp silencing only partially mimicked the suppression activity of AC2. The possibility of RDR6 paralogs partially compensating for the absence of RDR6 cannot be discounted. Future studies involving rdr6 and ago1 mutant plants and testing them for enhanced susceptibility would reveal any redundant function of paralogs.
Suppression of Defense Mechanism by Inactivating Cellular Regulatory Genes
Inactivation of SNF1 Kinase
AC2 of TGMV and C2 of BCTV have been shown to interact with SNF1-related kinase (SnRK1), a serine-threonine kinase, and inactivate it (Hao et al., 2003). SnRK1 has been implicated as a key regulator of cellular stress response including innate defense mechanism. AC2-SnRK1 interaction alters the cellular stress metabolism and causes a novel enhanced susceptibility (Hao et al., 2003). Although SNF1 inactivation does not involve suppression of silencing, interaction of AC2 with ADK reduces the cellular AMP levels (Figure 4D; Wang et al., 2003). AMP acts as an activator of SNF1 (Figure 4G), suggesting a dual strategy evolved by the virus to attenuate the cellular metabolism by inactivating SNF1 by direct interaction and indirectly by reducing cellular AMP levels (Figure 4G). Arabidopsis SnRK1 was shown to phosphorylate the serine residue at 109 (S109) of CaLCuV AC2 protein. A phosphomimic mutation of the S109 reduced viral DNA accumulation and delayed symptom appearance in Arabidopsis, thus revealing phosphorylation of viral protein as a host defense mechanism against an invading virus (Shen et al., 2014). Guerrero et al. (2020) report that AC2 of all old world begomoviruses and cutoviruses C2 lack the S109 residue. While majority of the new world begomovirus AC2s possess the conserved SnRK1 phosphorylation site, some new world begomoviruses including TGMV AC2 lack the S109 residue and hence are not phosphorylated. Instead, TGMV AC2 is shown to inhibit the kinase activity of SnRK1 (Hao et al., 2003).
Elevation of Cellular Cytokinin Levels
Baliji et al. (2010) demonstrated a novel consequence of ADK inhibition by AC2 of TGMV and C2 of SCTV. ADK has a role in maintaining the cellular cytokinin level. ADK-mediated phosphorylation of cytokinin converts the bioactive form of cytokinin to a less bioactive form (Figure 4H). Silencing of ADK in Arabidopsis increased the cellular cytokinin levels, substantiated by increased activity of a cytokinin-responsive promoter. Over-expression of CaLCuV AC2 and SCTV C2 in Arabidopsis resulted in increased expression of endogenous cytokinin-responsive promoters. Thus, geminivirus AC2, by inactivating ADK, increased the cellular cytokinin levels. An enhanced cytokinin level is a prerequisite for cell cycle progression and to maintain an active state of replication. Increased cytokinin resulted in an enhanced susceptibility phenotype. Interestingly, cytokinin is known to negatively regulate the expression of SULTR1;2, a high affinity sulfate transporter in Arabidopsis roots (Maruyama-Nakashita et al., 2004). Cytokinin binding to the receptor cytokinin response 1/wooden leg/Arabidopsis histidine kinase 4 (CRE1/WOL/AHK) is the cue for negative regulation of sulfur assimilation, while cre1 mutant is insensitive to cytokinin. In sultr1;2 mutant, SULTR1;1 was downregulated by cytokinin. Downregulation of sulfate transporters by cytokinin correlated with the decrease in sulfate uptake. Another study by Ohkama et al. (2002) showed that exogenous application of cytokinins upregulated sulfur responsive genes APS reductase 1 (APR1) and SULTR1;2 through a pathway independent of sulfur starvation; likely through increasing sucrose concentration which is known to upregulate APR1 gene (Kopriva et al., 1999). Exogenous application of cysteine and glutathione (GSH) resulted in downregulation of ATP sulfurylase, APS reductase, and sulfate transporter (Vauclare et al., 2002).
TYLCSV-C2 transgenic plants repressed the expression of three genes involved in sulfur assimilation namely ATP sulfurylase 3 (APS3), APS reductase 1 (APR1), and APS reductase 3 (APR3) and also accumulated reduced sulfur (Lozano-Durán et al., 2012). Adequate sulfate supply in tobacco plant resulted in a suppressed and delayed Tobacco mosaic virus (TMV) symptom development through a phenomenon named sulfur-induced resistance (SIR) or sulfur-enhanced defense (SED; Höller et al., 2010), thus revealing a role for sulfur in plant defense. Exogenous treatment of the TYLCSV-C2 plants with methyl jasmonate (MeJA) reversed the repression of sulfur assimilation genes (Lozano-Durán et al., 2012). TYLCSV-C2 transgenic plants over-accumulated cysteine and glutathione. The authors could not explain the reason behind the over-accumulation of cysteine and glutathione when the sulfur assimilation genes were repressed. Lozano-Durán et al. (2012) proposed that the C2 protein might suppress SIR/SED by suppressing jasmonate signaling pathway. However, it would be interesting to study in future if TYLCSV-C2 plants had increased cellular cytokinin levels similar to TGMV-AC2, CaLCuV-AC2, and SCTV-C2 plants. Also, it is relevant to study if upregulation of sulfur assimilation genes by cytokinin is initially through sucrose-dependent pathway. It is also important to study if the reduction observed in the expression of sulfur assimilation genes is because of the accumulation of cysteine and glutathione in TYLCSV-C2 transgenic plants. It would be informative to understand if the sulfate reduction in C2 transgenic plants might be because of the negative regulation of sulfate transport pathway by cytokinin binding to CRE1 receptor. Further studies are necessary to understand cross-talk between cytokinin, jasmonate, and sulfur assimilation pathways.
Inhibition of Jasmonate Signaling Pathway
COP9 signalosome (CSN), a highly conserved protein complex with eight subunits that resembles the 19S lid of the 26S proteasome (Dohmann et al., 2008), regulates the activity of E3 ligases. The CSN complex comprises eight subunits, named CSN1–CSN8, where CSN5 is the only catalytic subunit. TYLCSV C2 interacts with CSN5 and interferes with the cellular ubiquitination machinery (Lozano-Durán et al., 2011). Ubiquitination occurs through a cascade of enzymatic reactions namely ubiquitin activation by E1, conjugation by E2, and ligation by E3. E3 ligase comprises the multisubunit Cullin RING Ligases (CRLs). Among CRLs, cullin 1-based group or SCF (for Skp1/Cullin1/F-box), is comprised of four proteins, cullin 1 (CUL1), S-phase kinase-associated protein (SKP1/ASK), the RING subunit RBX1 (RING box 1) and an F-box substrate binding protein. The CRL activity is regulated by covalent attachment and removal of ubiquitin-like protein RUB (related to ubiquitin; Figure 4I). CSN5 is associated with the derubylation activity (Gusmaroli et al., 2007). CSN is a multisubunit isopeptidase which removes the RUB moiety from CRLs to function in vivo. C2 of TYLCSV interacts with CSN5 and inhibits the derubylation of CRL (Figure 4I). Rubylated CUL1 alters several SCF-dependent hormonal processes and also suppresses jasmonate responses in C2 transgenic plants. Jasmonate signaling has been implicated in defense response and suppression of jasmonate response resulted in enhanced susceptibility phenotype, which was reverted upon exogenous treatment of methyl jasmonate (MeJA). TYLCV C2 and BCTV C2 were also shown to interact with CSN5 suggesting that CSN5-C2 interaction is a conserved function in geminiviruses (Lozano-Durán et al., 2011).
Jasmonate receptor SCF complex is also the receptor for the bacterial toxin coronatine, which is secreted by the plant pathogenic Pseudomonas syringae pv. tomato DC3000 (Pto DC3000). Coronatine application on plants facilitates stomatal opening and increases infection (Melotto et al., 2006; Geng et al., 2014). The hindrance of SCF complex by TYLCV C2 (Lozano-Durán et al., 2011) makes TYLCV/TYLCSV C2 transgenic plants less sensitive to coronatine, as evidenced by reduced Pto DC3000 bacterial growth (Rosas-Díaz et al., 2016). The mutant COR− strain and wild-type strain had similar bacterial growth in C2 transgenic plants (Rosas-Díaz et al., 2016) confirming the role of C2 in altering SCF function (Lozano-Durán et al., 2011).
Jasmonic acid, which is involved in biotic and abiotic stress responses, is also known to induce the production of secondary metabolites including alkaloids, anthocyanins, and terpenoid compounds (Devoto et al., 2005). Whitefly-infested plants showed increased terpenoid production which was decreased in virus-infected plants (Luan et al., 2013). TYLCV-infected tobacco plants manifested increased survival and fecundity of whiteflies (Li et al., 2019). Terpene synthesis genes were downregulated in TYLCV and TYLCV-whitefly co-infected plants compared to whitefly-infested plants. TYLCV C2 over-expression lines had decreased expression of terpene synthesis genes including MYC2, a transcription factor. MYC2 is under the regulatory control of JAZ1 protein and ubiquitination-mediated degradation of JAZ1 protein is the switch of MYC2 activation (Niu et al., 2011). TYLCV and Papaya leaf curl China virus (PaLCuCNV) C2 were shown to interact with RPS27A, a fusion protein consisting of ubiquitin at the N terminus and ribosomal protein S27a at the C terminus. C2 interaction was with the ubiquitin moiety of RPS27A. Li et al. (2019) demonstrated a novel suppression mechanism in which TYLCV C2 competitively bound to ubiquitin, which resulted in decreased JAZ1 protein ubiquitination (Figure 4J). Consequently, the MYC2 bound to JAZ1 is stabilized and interferes with the ability of MYC2 to induce the expression of downstream defense genes.
Suppression by Inhibiting Cell Cycle Regulator
Arabidopsis PEAPOD2 transcription factor (TIFY4B) was shown to interact with AC2 and promoter sequences of TGMV and CaLCuV CP (Lacatus and Sunter, 2009) and TGMV NSP (Berger and Sunter, 2013). TIFY4B is known to limit cell proliferation in leaf epidermis and vascular tissues (White, 2006), which is substantiated by increased TIFY4B expression in callus tissue and during inflorescence emergence (Vanholme et al., 2007). TIFY4B has three conserved domains: PPD, TIFY, and CCT_2. While TIFY4B was shown to localize to nucleus (Lacatus and Sunter, 2009), mutant versions of TIFY4B including an 84–150-amino acid version were localized in the cytoplasm (Chung and Sunter, 2014). TGMV and CaLCuV AC2 interaction with the mutant TIFY4B altered the localization to nucleus. Increased TIFY4B expression was observed upon viral infection while the geminivirus infection is expected to downregulate a repressor of cell cycle progression. Over-expression of TIFY4B resulted in increased mean latent period and reduced CP expression suggesting a role of TIFYB in antiviral defense, wherein TIFYB inhibits cell proliferation, and therefore, viral replication. Chung and Sunter (2014) proposed a suppression mechanism in which AC2 sequesters TIFY4B and inhibits its role in cell cycle regulation, thereby creating a conducive environment for viral replication.
Summary
This review highlights the different targets and mechanisms evolved by geminivirus AC2/C2 to counter PTGS and TGS. Recent studies on cellular proteins that are targets of AC2/C2 have opened a third defense mechanism which does not involve PTGS and TGS (Table 1). Many studies show how a single suppressor protein like TGMV AC2 could deploy multiple targets. TGMV AC2 targets rgsCaM and ADK to suppress PTGS, targets ADK, and KYP to suppress TGS and inactivates SnRK1 and TIFY4B to inhibit the cellular defense response (Table 1). It is likely that silencing suppression is not limited to targeting of one viral protein and a host protein but rather involves a concerted effect on multiple host proteins as manifested in TGMV AC2. It would be interesting to see if other viral suppressor proteins also act on multiple targets. Most of our current knowledge regarding suppressor proteins and their targets is from curtoviruses and begomoviruses. Positional homologs of AC2/C2 are observed in other geminiviruses namely Eragovirus, Topocuvirus, and Turncurtovirus; however, they are not known to encode functional AC2/C2. Advancing our knowledge of suppressor proteins in other genera is essential. For example, Grapevine red blotch virus (GRBV), the type member of Grablovirus, is known to be a serious threat to the Californian wine industry. Understanding the GRBV suppressor proteins and their plant targets can present cogent strategies for mitigating this threat to a multibillion-dollar industry. Most of our current knowledge on silencing and silencing suppression is from dicot plants Arabidopsis and N. benthamiana. The genus Mastrevirus are monocot-infecting geminiviruses with 41 species. The only silencing suppressor protein identified till date in Mastrevirus is the Rep protein from Wheat dwarf virus which binds to ss- and ds-siRNA (Wang et al., 2014). It would be interesting to see if DCL5 and AGO18, the unique silencing machinery components of monocots, are targets of Mastrevirus.
Future Directions
Future research in AC2 suppressor protein should be focused on addressing the question of how the small AC2/AL2 protein (15 kDa) has evolved an ability to interact with so many different targets and make an impact on viral pathogenesis and plant metabolism. It would be interesting to know which of the studied target/targets is relevant in viral pathogenesis. Editing of the target genes in plants and study of viral pathogenesis and RNA silencing in the mutant plants infected with viruses will help in a more thorough evaluation of the AC2-interacting plant proteins. Abutilon mosaic virus (AbMV) AC2 was shown to act as a brake in geminivirus replication and was reported to enhance PTGS rather than suppress it (Krenz et al., 2015). It would be interesting to study if any other geminiviral AC2/C2 function as a facilitator of silencing. Besides the uniqueness of AbMV AC2 acting as a PTGS enhancer, a greater challenge is to understand how AbMV AC2 evolved as an enhancer. Does this confer any advantage to the virus or did the host evolve to inhibit viral replication or symptom development?
One major focus of future study should be in exploiting AC2 silencing suppressor function for biotechnological applications. The toxic effect of AC2, when expressed as a transgene, has been reported by several groups. This has limited the use of geminivirus AC2 in transgenic technology. Mutations of functional domains have been shown to alleviate the toxic effect (Rajeswaran et al., 2007). A study to precisely identify the amino acid residue/residues that contribute to the toxic effect of the 135 aa AC2 protein is highly desirable. DNA viral vectors containing only the replication origin and with Rep gene provided in cis or in trans have found application in biopharming (Rybicki and Martin, 2014). Bean yellow dwarf virus (BeYDV), a mild dicot infecting Mastrevirus, has been deconstructed and used widely as a replicon vector (Regnard et al., 2010). Huang et al. (2010) demonstrated high level expression of Ebola virus glycoprotein GP1 (6D8) recombinant monoclonal antibodies in N. benthamiana leaves by co-expressing the transgene in BeYDV replicon along with Rep and Tomato bushy stunt virus (TBSV) p19, a ds siRNA binding suppressor protein. Earlier work by Lacombe et al. (2018) has demonstrated that a combination of P19, P0, and P1 suppressors, that act at distinct steps of the RNA silencing pathway, allowed the highest ectopic protein expression. If the toxic effect of geminiviral AC2 can be negated without compromising on the silencing suppression function, AC2, which is known to target multiple steps of gene silencing, could find extensive use for over-expressing recombinant proteins in plants and plant cell cultures.
Both RNA and DNA viruses are widely used in Virus-Induced Gene Silencing (VIGS). The ease with which DNA viruses can be manipulated, the fact that geminiviral CP can be replaced with the gene of interest without affecting the systemic movement of virus (only in certain viruses), the recombinant vector derived from replacing CP is not vector transmissible and the broad host range of DNA viral vectors makes DNA viruses a good choice for VIGS over RNA viruses (Robertson, 2004). While several studies have shown the application of geminivirus as VIGS vector (Yang et al., 2017), the presence of suppressor proteins including AC2/C2 was shown to antagonize the silencing effect caused by the VIGS vector. The silencing efficiency is depleted as the suppressor protein accumulates in the plants. A null mutant in MYMIV-AC2, when used as VIGS vector, increased the silencing efficiency (Pandey et al., 2009). However, the mutation in the AC2 ORF reduced MYMIV replication efficiency by about 25%. A null mutation in CaLCuV AC2 abolished the viral replication (Castillo-González et al., 2015). Hence, future studies should investigate the possibility of mutating the AC2/C2 to mute the suppression function alone without compromising on the replication efficiency of the VIGS vector. Geminivirus VIGS vectors will help in performing functional genomics in a wide range of plants.
Baltes et al. (2014) demonstrated genome engineering using the BeYDV-based replicon system. Desired DNA changes were made when TALENs and CRISPR/Cas9 system were delivered using the BeYDV-based replicon system. A recent study by Mao et al. (2018) showed improved gene-editing efficiency by silencing AGO1 or by co-expressing TBSV p19 protein as part of CRISPR/Cas9 cassette. The challenge of gene editing technology is in identifying the edited events. Mao et al. (2018) established a suave means to phenotypically identify the edited events in the T1 generation and T-DNA segregated events in T2 generation. By co-expressing p19 as part of the CRISPR/Cas9 cassette, the authors could group the T1 plants based on the severity of p19-induced leaf phenotype alteration. T1 plants displaying severe phenotype associated with p19 over-expression were selected as edited lines. T2 seedlings from the gene edited T1 events were again grouped based on the leaf phenotype. Segregated plants with wild-type leaf phenotype were then identified as gene-edited plants from which the T-DNA was segregated out. The geminiviral suppressor protein AC2 can be co-expressed to increase the genome editing efficiency. Interestingly, AC2 is more attractive than TBSV p19 for the following reasons: (1) AC2 over-expression is known to cause pronounced phenotype in leaf (Siddiqui et al., 2008; Castillo-González et al., 2015). (2) TGMV AC2 over-expression resulted in early flowering (Castillo-González et al., 2015). (3) AC2 over-expression is likely to result in genome-wide hypomethylation (Castillo-González et al., 2015), and hence the tightly regulated genes involved in meiotic recombination are also likely to be upregulated. (4) MYMV AC2 and ACMV AC2, when transiently expressed in Arabidopsis protoplasts, upregulated the expression of the meiotic recombination protein AtDMC1 (Trinks et al., 2005; Da Ines et al., 2013). Thus, future studies would pave way to the evolution of AC2 from a core viral protein to a potent molecular tool with myriad applications.
Author Contributions
SS: conceptualization and writing-original draft preparation. KV: conceptualization, reviewing, and editing. Both authors contributed to the article and approved the submitted version.
Funding
The costs of publication are supported by California Department of Food and Agriculture Pierce’s Disease Board, agreement number 20-0248-000-SA to SS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Christopher Rock, Texas Tech University for critical reading of the manuscript.
References
Aguilar, E., Garnelo Gomez, B., and Lozano-Duran, R. (2020). Recent advances on the plant manipulation by geminiviruses. Curr. Opin. Plant Biol. 56, 56–64. doi: 10.1016/j.pbi.2020.03.009
Akbergenov, R., Si-Ammour, A., Blevins, T., Amin, I., Kutter, C., Vanderschuren, H., et al. (2006). Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 34, 462–471. doi: 10.1093/nar/gkj447
Anandalakshmi, R., Marathe, R., Ge, X., Herr, J. M., Mau, C., Mallory, A., et al. (2000). A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290, 142–144. doi: 10.1126/science.290.5489.142
Anandalakshmi, R., Pruss, G. J., Ge, X., Marathe, R., Mallory, A. C., Smith, T. H., et al. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. U. S. A. 95, 13079–13084. doi: 10.1073/pnas.95.22.13079
Aregger, M., Borah, B. K., Seguin, J., Rajeswaran, R., Gubaeva, E. G., Zvereva, A. S., et al. (2012). Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 8:e1002941. doi: 10.1371/journal.ppat.1002941
Baliji, S., Lacatus, G., and Sunter, G. (2010). The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 402, 238–247. doi: 10.1016/j.virol.2010.03.023
Baliji, S., Sunter, J., and Sunter, G. (2007). Transcriptional analysis of complementary sense genes in spinach curly top virus and functional role of C2 in pathogenesis. Mol. Plant-Microbe Interact. 20, 194–206. doi: 10.1094/MPMI-20-2-0194
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A., and Voytas, D. F. (2014). DNA replicons for plant genome engineering. Plant Cell 26, 151–163. doi: 10.1105/tpc.113.119792
Berger, M. R., and Sunter, G. (2013). Identification of sequences required for AL2-mediated activation of the tomato golden mosaic virus-yellow vein BR1 promoter. J. Gen. Virol. 94, 1398–1406. doi: 10.1099/vir.0.050161-0
Bisaro, D. M. (2006). Silencing suppression by geminivirus proteins. Virology 344, 158–168. doi: 10.1016/j.virol.2005.09.041
Blevins, T., Rajeswaran, R., Aregger, M., Borah, B. K., Schepetilnikov, M., Baerlocher, L., et al. (2011). Massive production of small RNAs from a non-coding region of cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 39, 5003–5014. doi: 10.1093/nar/gkr119
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H. S., et al. (2006). Four plant dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. doi: 10.1093/nar/gkl886
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G., and Bisaro, D. M. (2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. doi: 10.1128/JVI.01771-08
Cao, X., and Jacobsen, S. E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12, 1138–1144. doi: 10.1016/S0960-9822(02)00925-9
Castillo-González, C., Liu, X., Huang, C., Zhao, C., Ma, Z., Hu, T., et al. (2015). Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. elife 4:e06671. doi: 10.7554/eLife.06671
Ceniceros-Ojeda, E. A., Rodríguez-Negrete, E. A., and Rivera-Bustamante, R. F. (2016). Two populations of viral minichromosomes are present in a geminivirus-infected plant showing symptom remission (recovery). J. Virol. 90, 3828–3838. doi: 10.1128/JVI.02385-15
Chen, Z. Q., Zhao, J. H., Chen, Q., Zhang, Z. H., Li, J., Guo, Z. X., et al. (2020). DNA geminivirus infection induces an imprinted E3 ligase gene to epigenetically activate viral gene transcription. Plant Cell 32, 3256–3272. doi: 10.1105/tpc.20.00249
Chung, H. Y., Lacatus, G., and Sunter, G. (2014). Geminivirus AL2 protein induces expression of, and interacts with, a calmodulin-like gene, an endogenous regulator of gene silencing. Virology 460–461, 108–118. doi: 10.1016/j.virol.2014.04.034
Chung, H. Y., and Sunter, G. (2014). Interaction between the transcription factor AtTIFY4B and begomovirus AL2 protein impacts pathogenicity. Plant Mol. Biol. 86, 185–200. doi: 10.1007/s11103-014-0222-9
Coursey, T., Regedanz, E., and Bisaro, D. M. (2018). Arabidopsis RNA polymerase V mediates enhanced compaction and silencing of geminivirus and transposon chromatin during host recovery from infection. J. Virol. 92, e1320–e1417. doi: 10.1128/JVI.01320-17
Csorba, T., Kontra, L., and Burgyán, J. (2015). Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479–480, 85–103. doi: 10.1016/j.virol.2015.02.028
Da Ines, O., Degroote, F., Goubely, C., Amiard, S., Gallego, M. E., and White, C. I. (2013). Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 9:e1003787. doi: 10.1371/journal.pgen.1003787
Daxinger, L., Kanno, T., Bucher, E., van der Winden, J., Naumann, U., Matzke, A. J., et al. (2009). A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 28, 48–57. doi: 10.1038/emboj.2008.260
Devoto, A., Ellis, C., Magusin, A., Chang, H. S., Chilcott, C., Zhu, T., et al. (2005). Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 58, 497–513. doi: 10.1007/s11103-005-7306-5
Dohmann, E. M., Levesque, M. P., De Veylder, L., Reichardt, I., Jürgens, G., Schmid, M., et al. (2008). The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135, 2013–2022. doi: 10.1242/dev.020743
Dong, X., van Wezel, R., Stanley, J., and Hong, Y. (2003). Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77, 7026–7033. doi: 10.1128/JVI.77.12.7026-7033.2003
Endres, M. W., Gregory, B. D., Gao, Z., Foreman, A. W., Mlotshwa, S., Ge, X., et al. (2010). Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog. 6:e1000729. doi: 10.1371/journal.ppat.1000729
English, J. J., Mueller, E., and Baulcombe, D. C. (1996). Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8, 179–188. doi: 10.2307/3870263
Fiallo-Olivé, E., Martínez-Zubiaur, Y., Moriones, E., and Navas-Castillo, J. (2012). A novel class of DNA satellites associated with New World begomoviruses. Virology 426, 1–6. doi: 10.1016/j.virol.2012.01.024
Fiallo-Olivé, E., Tovar, R., and Navas-Castillo, J. (2016). Deciphering the biology of deltasatellites from the New World: maintenance by New World begomoviruses and whitefly transmission. New Phytol. 212, 680–692. doi: 10.1111/nph.14071
Garcia-Ruiz, H., Takeda, A., Chapman, E. J., Sullivan, C. M., Fahlgren, N., Brempelis, K. J., et al. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 22, 481–496. doi: 10.1105/tpc.109.073056
Geng, X., Jin, L., Shimada, M., Kim, M. G., and Mackey, D. (2014). The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta 240, 1149–1165. doi: 10.1007/s00425-014-2151-x
Glazov, E., Phillips, K., Budziszewski, G. J., Schöb, H., Meins, F., and Levin, J. Z. (2003). A gene encoding an RNase D exonuclease-like protein is required for post-transcriptional silencing in Arabidopsis. Plant J. 35, 342–349. doi: 10.1046/j.1365-313X.2003.01810.x
Gopal, P., Pravin Kumar, P., Sinilal, B., Jose, J., Kasin Yadunandam, A., and Usha, R. (2007). Differential roles of C4 and betaC1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 123, 9–18. doi: 10.1016/j.virusres.2006.07.014
Guerrero, J., Regedanz, E., Lu, L., Ruan, J., Bisaro, D. M., and Sunter, G. (2020). Manipulation of the plant host by the geminivirus AC2/C2 protein, a central player in the infection cycle. Front. Plant Sci. 11:591. doi: 10.3389/fpls.2020.00591
Gusmaroli, G., Figueroa, P., Serino, G., and Deng, X. W. (2007). Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis cullin3-based E3 ligases. Plant Cell 19, 564–581. doi: 10.1105/tpc.106.047571
Haley, A., Zhan, X., Richardson, K., Head, K., and Morris, B. (1992). Regulation of the activities of African cassava mosaic virus promoters by the AC1, AC2, and AC3 gene products. Virology 188, 905–909. doi: 10.1016/0042-6822(92)90551-Y
Hanley-Bowdoin, L., Bejarano, E. R., Robertson, D., and Mansoor, S. (2013). Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. doi: 10.1038/nrmicro3117
Hanley-Bowdoin, L., Settlage, S. B., Orozco, B. M., Nagar, S., and Robertson, D. (2000). Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35, 105–140.
Hao, L., Wang, H., Sunter, G., and Bisaro, D. M. (2003). Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15, 1034–1048. doi: 10.1105/tpc.009530
Harvey, J. J., Lewsey, M. G., Patel, K., Westwood, J., Heimstädt, S., Carr, J. P., et al. (2011). An antiviral defense role of AGO2 in plants. PLoS One 6:e14639. doi: 10.1371/journal.pone.0014639
Hayes, R. J., and Buck, K. W. (1989). Replication of tomato golden mosaic virus DNA B in transgenic plants expressing open reading frames (ORFs) of DNA A: requirement of ORF AL2 for production of single-stranded DNA. Nucleic Acids Res. 17, 10213–10222. doi: 10.1093/nar/17.24.10213
Herr, A. J., Jensen, M. B., Dalmay, T., and Baulcombe, D. C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120. doi: 10.1126/science.1106910
Höller, K., Király, L., Künstler, A., Müller, M., Gullner, G., Fattinger, M., et al. (2010). Enhanced glutathione metabolism is correlated with sulfur-induced resistance in tobacco mosaic virus-infected genetically susceptible Nicotiana tabacum plants. Mol. Plant-Microbe Interact. 23, 1448–1459. doi: 10.1094/MPMI-05-10-0117
Hormuzdi, S. G., and Bisaro, D. M. (1993). Genetic analysis of beet curly top virus: evidence for three virion sense genes involved in movement and regulation of single- and double-stranded DNA levels. Virology 193, 900–909. doi: 10.1006/viro.1993.1199
Hormuzdi, S. G., and Bisaro, D. M. (1995). Genetic analysis of beet curly top virus: examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206, 1044–1054. doi: 10.1006/viro.1995.1027
Huang, Z., Phoolcharoen, W., Lai, H., Piensook, K., Cardineau, G., Zeitlin, L., et al. (2010). High-level rapid production of full-size monoclonal antibodies in plants by a single-vector DNA replicon system. Biotechnol. Bioeng. 106, 9–17. doi: 10.1002/bit.22652
Ingelbrecht, I., Van Houdt, H., Van Montagu, M., and Depicker, A. (1994). Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc. Natl. Acad. Sci. U. S. A. 91, 10502–10506. doi: 10.1073/pnas.91.22.10502
Jackel, J. N., Buchmann, R. C., Singhal, U., and Bisaro, D. M. (2015). Analysis of geminivirus AL2 and L2 proteins reveals a novel AL2 silencing suppressor activity. J. Virol. 89, 3176–3187. doi: 10.1128/JVI.02625-14
Jackel, J. N., Storer, J. M., Coursey, T., and Bisaro, D. M. (2016). Arabidopsis RNA polymerases IV and V are required to establish H3K9 methylation, but not cytosine methylation, on geminivirus chromatin. J. Virol. 90, 7529–7540. doi: 10.1128/JVI.00656-16
Karjee, S., Islam, M. N., and Mukherjee, S. K. (2008). Screening and identification of virus-encoded RNA silencing suppressors. Methods Mol. Biol. 442, 187–203. doi: 10.1007/978-1-59745-191-8_14
Kheyr-Pour, A., Bendahmane, M., Matzeit, V., Accotto, G. P., Crespi, S., and Gronenborn, B. (1991). Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res. 19, 6763–6769. doi: 10.1093/nar/19.24.6763
Kopriva, S., Muheim, R., Koprivova, A., Trachsel, N., Catalano, C., Suter, M., et al. (1999). Light regulation of assimilatory sulphate reduction in Arabidopsis thaliana. Plant J. 20, 37–44. doi: 10.1046/j.1365-313X.1999.00573.x
Krenz, B., Deuschle, K., Deigner, T., Unseld, S., Kepp, G., Wege, C., et al. (2015). Early function of the Abutilon mosaic virus AC2 gene as a replication brake. J. Virol. 89, 3683–3699. doi: 10.1128/JVI.03491-14
Kumar, R. V. (2019). Plant antiviral immunity against geminiviruses and viral counter-defense for survival. Front. Microbiol. 10:1460. doi: 10.3389/fmicb.2019.01460
Kumar, V., Mishra, S. K., Rahman, J., Taneja, J., Sundaresan, G., Mishra, N. S., et al. (2015). Mungbean yellow mosaic Indian virus encoded AC2 protein suppresses RNA silencing by inhibiting Arabidopsis RDR6 and AGO1 activities. Virology 486, 158–172. doi: 10.1016/j.virol.2015.08.015
Lacatus, G., and Sunter, G. (2009). The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virology 392, 196–202. doi: 10.1016/j.virol.2009.07.004
Lacombe, S., Bangratz, M., Brizard, J. P., Petitdidier, E., Pagniez, J., Sérémé, D., et al. (2018). Optimized transitory ectopic expression of promastigote surface antigen protein in Nicotiana benthamiana, a potential anti-leishmaniasis vaccine candidate. J. Biosci. Bioeng. 125, 116–123. doi: 10.1016/j.jbiosc.2017.07.008
Li, F., Huang, C., Li, Z., and Zhou, X. (2014). Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 10:e1003921. doi: 10.1371/journal.ppat.1003921
Li, P., Liu, C., Deng, W. H., Yao, D. M., Pan, L. L., Li, Y. Q., et al. (2019). Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 15:e1007607. doi: 10.1371/journal.ppat.1007607
Li, C. F., Pontes, O., El-Shami, M., Henderson, I. R., Bernatavichute, Y. V., Chan, S. W., et al. (2006). An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126, 93–106. doi: 10.1016/j.cell.2006.05.032
Li, F., Zhao, N., Li, Z., Xu, X., Wang, Y., Yang, X., et al. (2017). A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 13:e1006213. doi: 10.1371/journal.ppat.1006213
Lozano-Durán, R., García, I., Huguet, S., Balzergue, S., Romero, L. C., and Bejarano, E. R. (2012). Geminivirus C2 protein represses genes involved in Sulphur assimilation and this effect can be counteracted by jasmonate treatment. Eur. J. Plant Pathol. 134, 49–59. doi: 10.1007/s10658-012-0021-6
Lozano-Durán, R., Rosas-Díaz, T., Gusmaroli, G., Luna, A. P., Taconnat, L., Deng, X. W., et al. (2011). Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23, 1014–1032. doi: 10.1105/tpc.110.080267
Luan, J. B., Yao, D. M., Zhang, T., Walling, L. L., Yang, M., Wang, Y. J., et al. (2013). Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol. Lett. 16, 390–398. doi: 10.1111/ele.12055
Luna, A. P., and Lozano-Durán, R. (2020). Geminivirus-encoded proteins: not all positional homologs are made equal. Front. Microbiol. 11:878. doi: 10.3389/fmicb.2020.00878
Mao, Y., Yang, X., Zhou, Y., Zhang, Z., Botella, J. R., and Zhu, J. K. (2018). Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol. 19:149. doi: 10.1186/s13059-018-1529-7
Marí-Ordóñez, A., Marchais, A., Etcheverry, M., Martin, A., Colot, V., and Voinnet, O. (2013). Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45, 1029–1039. doi: 10.1038/ng.2703
Maruyama-Nakashita, A., Nakamura, Y., Yamaya, T., and Takahashi, H. (2004). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38, 779–789. doi: 10.1111/j.1365-313X.2004.02079.x
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. doi: 10.1016/j.cell.2006.06.054
Miozzi, L., Pantaleo, V., Burgyán, J., Accotto, G. P., and Noris, E. (2013). Analysis of small RNAs derived from tomato yellow leaf curl Sardinia virus reveals a cross reaction between the major viral hotspot and the plant host genome. Virus Res. 178, 287–296. doi: 10.1016/j.virusres.2013.09.029
Nakahara, K. S., Masuta, C., Yamada, S., Shimura, H., Kashihara, Y., Wada, T. S., et al. (2012). Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. U. S. A. 109, 10113–10118. doi: 10.1073/pnas.1201628109
Navot, N., Pichersky, E., Zeidan, M., Zamir, D., and Czosnek, H. (1991). Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology 185, 151–161. doi: 10.1016/0042-6822(91)90763-2
Niu, Y., Figueroa, P., and Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154. doi: 10.1093/jxb/erq408
Noris, E., Jupin, I., Accotto, G. P., and Gronenborn, B. (1996). DNA-binding activity of the C2 protein of tomato yellow leaf curl geminivirus. Virology 217, 607–612. doi: 10.1006/viro.1996.0157
Ohkama, N., Takei, K., Sakakibara, H., Hayashi, H., Yoneyama, T., and Fujiwara, T. (2002). Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol. 43, 1493–1501. doi: 10.1093/pcp/pcf183
Padidam, M., Beachy, R. N., and Fauquet, C. M. (1996). The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology 224, 390–404. doi: 10.1006/viro.1996.0546
Pandey, P., Choudhury, N. R., and Mukherjee, S. K. (2009). A geminiviral amplicon (VA) derived from tomato leaf curl virus (ToLCV) can replicate in a wide variety of plant species and also acts as a VIGS vector. Virol. J. 6:152. doi: 10.1186/1743-422X-6-152
Parent, J. S., Bouteiller, N., Elmayan, T., and Vaucheret, H. (2015). Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 81, 223–232. doi: 10.1111/tpj.12720
Pontes, O., and Pikaard, C. S. (2008). siRNA and miRNA processing: new functions for Cajal bodies. Curr. Opin. Genet. Dev. 18, 197–203. doi: 10.1016/j.gde.2008.01.008
Qu, F., Ye, X., and Morris, T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105, 14732–14737. doi: 10.1073/pnas.0805760105
Rahman, J., Karjee, S., and Mukherjee, S. K. (2012). MYMIV-AC2, a geminiviral RNAi suppressor protein, has potential to increase the transgene expression. Appl. Biochem. Biotechnol. 167, 758–775. doi: 10.1007/s12010-012-9702-z
Raja, P., Jackel, J. N., Li, S., Heard, I. M., and Bisaro, D. M. (2014). Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 88, 2611–2622. doi: 10.1128/JVI.02305-13
Raja, P., Sanville, B. C., Buchmann, R. C., and Bisaro, D. M. (2008). Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82, 8997–9007. doi: 10.1128/JVI.00719-08
Raja, P., Wolf, J. N., and Bisaro, D. M. (2010). RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochim. Biophys. Acta 1799, 337–351. doi: 10.1016/j.bbagrm.2010.01.004
Rajeswaran, R., Sunitha, S., Shivaprasad, P. V., Pooggin, M. M., Hohn, T., and Veluthambi, K. (2007). The mungbean yellow mosaic begomovirus transcriptional activator protein transactivates the viral promoter-driven transgene and causes toxicity in transgenic tobacco plants. Mol. Plant-Microbe Interact. 20, 1545–1554. doi: 10.1094/MPMI-20-12-1545
Regnard, G. L., Halley-Stott, R. P., Tanzer, F. L., Hitzeroth, I. I., and Rybicki, E. P. (2010). High level protein expression in plants through the use of a novel autonomously replicating geminivirus shuttle vector. Plant Biotechnol. J. 8, 38–46. doi: 10.1111/j.1467-7652.2009.00462.x
Rishishwar, R., and Dasgupta, I. (2019). Suppressors of RNA silencing encoded by geminiviruses and associated DNA satellites. Virusdisease 30, 58–65. doi: 10.1007/s13337-018-0418-8
Robertson, D. (2004). VIGS vectors for gene silencing: many targets, many tools. Annu. Rev. Plant Biol. 55, 495–519. doi: 10.1146/annurev.arplant.55.031903.141803
Rojas, M. R., Macedo, M. A., Maliano, M. R., Soto-Aguilar, M., Souza, J. O., Briddon, R. W., et al. (2018). World management of geminiviruses. Annu. Rev. Phytopathol. 56, 637–677. doi: 10.1146/annurev-phyto-080615-100327
Rosas-Díaz, T., Macho, A. P., Beuzón, C. R., Lozano-Durán, R., and Bejarano, E. R. (2016). The C2 protein from the geminivirus tomato yellow leaf curl Sardinia virus decreases sensitivity to jasmonates and suppresses jasmonate-mediated defences. Plan. Theory 5:8. doi: 10.3390/plants5010008
Rybicki, E. P., and Martin, D. P. (2014). Virus-derived ssDNA vectors for the expression of foreign proteins in plants. Curr. Top. Microbiol. Immunol. 375, 19–45. doi: 10.1007/82_2011_185
Saunders, K., Bedford, I. D., Briddon, R. W., Markham, P. G., Wong, S. M., and Stanley, J. (2000). A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. U. S. A. 97, 6890–6895. doi: 10.1073/pnas.97.12.6890
Schaffer, R. L., Miller, C. G., and Petty, I. T. (1995). Virus and host-specific adaptations in the BL1 and BR1 genes of bipartite geminiviruses. Virology 214, 330–338. doi: 10.1006/viro.1995.0042
Scholthof, K. B., Adkins, S., Czosnek, H., Palukaitis, P., Jacquot, E., Hohn, T., et al. (2011). Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. doi: 10.1111/j.1364-3703.2011.00752.x
Shabalina, S. A., and Koonin, E. V. (2008). Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587. doi: 10.1016/j.tree.2008.06.005
Shen, W., Dallas, M. B., Goshe, M. B., and Hanley-Bowdoin, L. (2014). SnRK1 phosphorylation of AL2 delays cabbage leaf curl virus infection in Arabidopsis. J. Virol. 88, 10598–10612. doi: 10.1128/JVI.00761-14
Shivaprasad, P. V., Akbergenov, R., Trinks, D., Rajeswaran, R., Veluthambi, K., Hohn, T., et al. (2005). Promoters, transcripts, and regulatory proteins of mungbean yellow mosaic geminivirus. J. Virol. 79, 8149–8163. doi: 10.1128/JVI.79.13.8149-8163.2005
Siddiqui, S. A., Sarmiento, C., Truve, E., Lehto, H., and Lehto, K. (2008). Phenotypes and functional effects caused by various viral RNA silencing suppressors in transgenic Nicotiana benthamiana and N. tabacum. Mol. Plant-Microbe Interact. 21, 178–187. doi: 10.1094/MPMI-21-2-0178
Stanley, J., and Latham, J. R. (1992). A symptom variant of beet curly top geminivirus produced by mutation of open reading frame C4. Virology 190, 506–509. doi: 10.1016/0042-6822(92)91243-N
Stanley, J., Latham, J. R., Pinner, M. S., Bedford, I., and Markham, P. G. (1992). Mutational analysis of the monopartite geminivirus beet curly top virus. Virology 191, 396–405. doi: 10.1016/0042-6822(92)90201-Y
Sun, Y. W., Tee, C. S., Ma, Y. H., Wang, G., Yao, X. M., and Ye, J. (2015). Attenuation of histone methyltransferase KRYPTONITE-mediated transcriptional gene silencing by geminivirus. Sci. Rep. 5:16476. doi: 10.1038/srep16476
Sunter, G., and Bisaro, D. M. (1991). Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180, 416–419. doi: 10.1016/0042-6822(91)90049-H
Sunter, G., and Bisaro, D. M. (1992). Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4, 1321–1331. doi: 10.1105/tpc.4.10.1321
Sunter, G., Stenger, D. C., and Bisaro, D. M. (1994). Heterologous complementation by geminivirus AL2 and AL3 genes. Virology 203, 203–210. doi: 10.1006/viro.1994.1477
Trinks, D., Rajeswaran, R., Shivaprasad, P. V., Akbergenov, R., Oakeley, E. J., Veluthambi, K., et al. (2005). Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79, 2517–2527. doi: 10.1128/JVI.79.4.2517-2527.2005
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Vanitharani, R., Chellappan, P., Pita, J. S., and Fauquet, C. M. (2004). Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78, 9487–9498. doi: 10.1128/JVI.78.17.9487-9498.2004
van Wezel, R., Dong, X., Liu, H., Tien, P., Stanley, J., and Hong, Y. (2002). Mutation of three cysteine residues in tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol. Plant-Microbe Interact. 15, 203–208. doi: 10.1094/MPMI.2002.15.3.203
van Wezel, R., Liu, H., Tien, P., Stanley, J., and Hong, Y. (2001). Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localized in the nucleus. Mol. Plant-Microbe Interact. 14, 1125–1128. doi: 10.1094/MPMI.2001.14.9.1125
van Wezel, R., Liu, H., Wu, Z., Stanley, J., and Hong, Y. (2003). Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J. Virol. 77, 696–700. doi: 10.1128/JVI.77.1.696-700.2003
Vauclare, P., Kopriva, S., Fell, D., Suter, M., Sticher, L., von Ballmoos, P., et al. (2002). Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5'-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 31, 729–740. doi: 10.1046/j.1365-313X.2002.01391.x
Voinnet, O. (2005). Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220. doi: 10.1038/nrg1555
Voinnet, O., Pinto, Y. M., and Baulcombe, D. C. (1999). Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. U. S. A. 96, 14147–14152. doi: 10.1073/pnas.96.24.14147
Wang, H., Buckley, K. J., Yang, X., Buchmann, R. C., and Bisaro, D. M. (2005). Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79, 7410–7418. doi: 10.1128/JVI.79.12.7410-7418.2005
Wang, Y., Dang, M., Hou, H., Mei, Y., Qian, Y., and Zhou, X. (2014). Identification of an RNA silencing suppressor encoded by a mastrevirus. J. Gen. Virol. 95, 2082–2088. doi: 10.1099/vir.0.064246-0
Wang, H., Hao, L., Shung, C. Y., Sunter, G., and Bisaro, D. M. (2003). Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15, 3020–3032. doi: 10.1105/tpc.015180
White, D. W. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 103, 13238–13243. doi: 10.1073/pnas.0604349103
Wierzbicki, A. T., Haag, J. R., and Pikaard, C. S. (2008). Noncoding transcription by RNA polymerase pol IVb/pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. doi: 10.1016/j.cell.2008.09.035
Wierzbicki, A. T., Ream, T. S., Haag, J. R., and Pikaard, C. S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41, 630–634. doi: 10.1038/ng.365
Yang, X., Baliji, S., Buchmann, R. C., Wang, H., Lindbo, J. A., Sunter, G., et al. (2007). Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J. Virol. 81, 11972–11981. doi: 10.1128/JVI.00617-07
Yang, Q.-Y., Ding, B., and Zhou, X.-P. (2017). Geminiviruses and their application in biotechnology. J. Integr. Agric. 16, 2761–2771. doi: 10.1016/S2095-3119(17)61702-7
Yang, X., Guo, W., Li, F., Sunter, G., and Zhou, X. (2019). Geminivirus-associated betasatellites: exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 24, 519–529. doi: 10.1016/j.tplants.2019.03.010
Yang, X., Wang, Y., Guo, W., Xie, Y., Xie, Q., Fan, L., et al. (2011). Characterization of small interfering RNAs derived from the geminivirus/betasatellite complex using deep sequencing. PLoS One 6:e16928. doi: 10.1371/journal.pone.0016928
Zerbini, F. M., Briddon, R. W., Idris, A., Martin, D. P., Moriones, E., Navas-Castillo, J., et al. (2017). ICTV virus taxonomy profile: geminiviridae. J. Gen. Virol. 98, 131–133. doi: 10.1099/jgv.0.000738
Zhang, Z., Chen, H., Huang, X., Xia, R., Zhao, Q., Lai, J., et al. (2011). BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23, 273–288. doi: 10.1105/tpc.110.081695
Keywords: geminivirus, AC2/C2, transcriptional activator, suppressor protein, silencing suppressor, biopharming, genome-editing
Citation: Veluthambi K and Sunitha S (2021) Targets and Mechanisms of Geminivirus Silencing Suppressor Protein AC2. Front. Microbiol. 12:645419. doi: 10.3389/fmicb.2021.645419
Edited by:
Rajarshi Kumar Gaur, Deen Dayal Upadhyay Gorakhpur University, IndiaReviewed by:
Claudia Castillo-González, Texas A&M University, United StatesManoj Prasad, National Institute of Plant Genome Research (NIPGR), India
Copyright © 2021 Veluthambi and Sunitha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sukumaran Sunitha, c3VuaXRoYS5zdWt1bWFyYW5AdHR1LmVkdQ==
 Karuppannan Veluthambi1
Karuppannan Veluthambi1 Sukumaran Sunitha
Sukumaran Sunitha