- 1Faculty of Engineering, Tottori University, Tottori, Japan
- 2Center for Research on Green Sustainable Chemistry, Tottori University, Tottori, Japan
- 3Department of Engineering, Graduate School of Sustainability Science, Tottori University, Tottori, Japan
- 4Department of Chemistry and Biotechnology, Graduate School of Engineering, Tottori University, Tottori, Japan
Geobacillus kaustophilus HTA426 is a thermophilic bacterium whose genome harbors numerous insertion sequences (IS). This study was initially conducted to generate mutant genes for thermostable T7 RNA polymerase in G. kaustophilus; however, relevant experiments unexpectedly identified that the organism transposed multiple IS elements and produced derivative cells that expressed a silent gene via transposition. The transposed elements were diverse and included members of the IS4, IS701, IS1634, and ISLre2 families. The transposition was relatively active at elevated temperatures and generated 4–9 bp of direct repeats at insertion sites. Transposition was more frequent in proliferative cells than in stationary cells but was comparable between both cells when sigX, which encodes an extra-cytoplasmic function sigma factor, was forcibly expressed. Southern blot analysis indicated that IS transposition occurred under growth inhibitory conditions by diverse stressors; however, IS transposition was not detected in cells that were cultured under growth non-inhibitory conditions. These observations suggest that G. kaustophilus enhances IS transposition via sigX-dependent stress responses when proliferative cells were prevented from active propagation. Considering Geobacillus spp. are highly adaptive bacteria that are remarkably distributed in diverse niches, it is possible that these organisms employ IS transposition for environmental adaptation via genetic diversification. Thus, this study provides new insights into adaptation strategies of Geobacillus spp. along with implications for strong codependence between mobile genetic elements and highly adaptive bacteria for stable persistence and evolutionary diversification, respectively. This is also the first report to reveal active IS elements at elevated temperatures in thermophiles and to suggest a sigma factor that governs IS transposition.
Introduction
Insertion sequences (IS) are a simple class of mobile genetic elements that propagate themselves or change the position in the host’s genetic material via replicative or non-replicative transposition, respectively (Vandecraen et al., 2018). An IS element is flanked by short inverted repeats and encodes a transposase that catalyzes transposition. In addition, the IS element may carry regulatory genes essential for transposition but does not carry accessory genes. A total of 32 IS families are classified, and most encode DDE-type (named for a conserved amino acid triad) transposases (Siguier et al., 2015). Transposition potentially causes deleterious mutations; therefore, IS elements were initially considered parasitic and selfish factors that multiply without conferring a survival advantage to the host organism (Schrader and Schmitz, 2019). However, it is now known that IS transposition can provide evolutionary adaptation for their hosts via gene inactivation and/or modulated expression of the neighboring genes (Vandecraen et al., 2018).
Insertion sequences that employ DDE-type transposases can achieve either replicative transposition or non-replicative transposition (Bouuaert and Chalmers, 2010). Non-replicative transposition results from cut-and-paste mechanisms where the transposase expressed from an IS element excises the IS from the original site and integrates it into another site. Replicative transposition is performed by either copy-and-paste (donor-primed replication) or copy-in (target-primed replication) mechanisms. In the former mechanism, the transposase releases a single-stranded and circular IS element from the original site. The IS element undergoes replication and eventually integrates into another site. The latter mechanism uses transposases to nick IS termini and directly ligate them with another site. This results in formation of a Shapiro intermediate followed by a cointegrate molecule via DNA replication. Subsequently, the intermediate undergoes recombination to generate the final products. All these mechanisms integrate IS elements into target sites while generating gaps. The gaps are then filled by the DNA repair systems of the host organisms; therefore, IS elements are generally flanked by direct repeats (DR).
Geobacillus spp. are gram-positive thermophiles that can form endospores. The species rapidly grow at >55°C but are unusually distributed in diverse habitats including cool or ambient environments (Suzuki, 2018). This is partially explained by the distribution of their endospores (Zeigler, 2014). However, it is equally possible that Geobacillus spp. grow as proliferative cells in numerous environments because they often exhibit unique capacities to reproduce in the respective habitats (Suzuki, 2018). Pan-genomic analysis suggests that Geobacillus spp. have remarkably diversified their genomes (Bezuidt et al., 2016). Geobacillus spp. can also rapidly generate mutant genes for thermostable variants from thermolabile protein genes (Liao et al., 1986; Suzuki et al., 2015; Kobayashi et al.,2015a,b; Wada et al., 2016). In the species, mutagenesis is apparently induced when proliferative cells are faced with growth inhibition by antibiotics (Suzuki et al., 2018). These observations suggest that Geobacillus spp. are active in environmental adaptation via genetic diversification.
We have studied generation and selection of thermostable proteins in Geobacillus kaustophilus HTA426 (Suzuki et al., 2015; Kobayashi et al.,2015a,b; Wada et al., 2016). This strain originates from a mud sample of the Mariana Trench and can grow between 42°C and 74°C (Takami et al., 2004a). In related studies, we unexpectedly discovered that multiple IS elements could perform transposition in this organism. Mobile genetic elements in thermophiles are insufficiently characterized; therefore, we analyzed this phenomenon in detail. Here, we report that G. kaustophilus performs frequent and genome-wide IS transposition potentially via sigX-dependent stress responses when proliferative cells were prevented from active propagation.
Materials and Methods
Genetic Tools
Plasmid pGAM46 was previously constructed for marker-free gene integration into the amyA gene in G. kaustophilus (Suzuki et al., 2012). Plasmid pGKE75 (Kobayashi et al., 2015a) was used for forcible gene expression under the control of the gk704 promoter (Pgk704) of G. kaustophilus (Suzuki et al., 2013b). Plasmid pGKE74 was constructed from pGKE75 via elimination of the Pgk704 region. Plasmids were introduced into G. kaustophilus using conjugative plasmid transfer from Escherichia coli (Suzuki et al., 2013a). Chromosomal gene replacement was performed using the procedure previously described (Suzuki et al., 2012). The primer sequences are summarized in the Supplementary Table 1.
Construction of Plasmids
Table 1 summarizes relevant plasmids. T7 promoter (PT7) was amplified from pET-16b (Merck KGaA, Darmstadt, Germany) using the primers t7–250F and t70R. The pyrF gene encoding for orotidine 5′-phosphate decarboxylase of G. kaustophilus was amplified using the primers pyrF0F and pyrFTR. The PT7 and pyrF fragments were cloned between the HindIII and SphI sites and the SphI and BamHI sites, respectively, of pGAM46 to give pGAM46PT7-pyrF carrying the PT7-pyrF cassette. The gene for T7 RNA polymerase (T7RP) was amplified from E. coli BL21(DE3) using the primers t7RP0F and t7RPTR and cloned between the SphI and BamHI sites of pGKE75 to give pGKE75-T7RP, which carried the Pgk704-T7RP cassette. To construct the Pgk704-sigB cassette, Pgk704 was amplified from pGKE75 using the primers gk704–250F and sigB0R; sigB encoding for sigma factor B (SigB) of G. kaustophilus was amplified using the primers sigB0F and sigBTR. These fragments were combined using fusion PCR and cloned between the SphI and BamHI sites to give pGKE74Pgk704-sigB. To construct the Pgk704-rsbV cassette, Pgk704 was amplified using the primers gk704–250F and rsbV0R; rsbV encoding for anti-SigB antagonist (RsbV) of G. kaustophilus was amplified using the primers rsbV0F and rsbVTR. These fragments were combined and cloned between the HindIII and BamHI sites of pGKE74 to give pGKE74Pgk704-rsbV. To construct the Pgk704-sigX cassette, Pgk704 was amplified using the primers gk704–250F and sigX0R; sigX encoding for sigma factor X (SigX) of G. kaustophilus was amplified using the primers sigX0F and sigXTR. These fragments were combined and cloned between the HindIII and BamHI sites to give pGKE74Pgk704-sigX.
Bacterial Strains
Table 1 summarizes thermophilic strains used in this study. G. kaustophilus strains MK242 and MK480 were previously constructed from G. kaustophilus HTA426 (Suzuki et al., 2015). The Pgk704-bgaB cassette in MK242 and MK480 was replaced with the PT7-pyrF cassette using pGAM46PT7-pyrF to give G. kaustophilus strains MK536 and MK534, respectively. G. kaustophilus MK534 was transformed with pGKE75-T7RP to provide G. kaustophilus MK534T7RP. G. kaustophilus MK536 was transformed with pGKE74, pGKE74Pgk704-rsbV, pGKE74Pgk704-sigB, and pGKE74Pgk704-sigX to generate strains MK536p74, MK536rsbV, MK536sigB, and MK536sigX, respectively. G. kaustophilus MK536up1 was derived from G. kaustophilus MK536 via intrinsic IS transposition.
Culture Conditions
Geobacillus kaustophilus was cultured in Luria–Bertani (LB; Nacalai Tesque, Kyoto, Japan) or semisynthetic (MM, MU, MC, or MN) media. The MM medium contained inorganic salts (K2SO4, 0.3 g/L; Na2HPO4⋅12H2O, 2.5 g/L; NH4Cl, 1 g/L; MgSO4, 0.4 g/L; MnCl2⋅4H2O, 3 mg/L; CaCl2⋅2H2O, 5 mg/L; and FeCl3⋅6H2O, 7 mg/L), 0.1% trace element solution (Amartey et al., 1991), Tris–HCl (10 mM, pH 7.5), uracil (10 mg/L), casamino acids (1 g/L), and D-glucose (10 g/L). The other semisynthetic media were based on MM medium; however, MU medium lacked uracil. The MC medium lacked both casamino acids and D-glucose, and MN lacked casamino acids and NH4Cl. Solid media contained agar (20 g/L). Kanamycin (5 mg/L) was added when necessary. The optical density at 600 nm (OD600) was monitored using an infrared-dependent detector (OD-Monitor A; Taitec, Saitama, Japan).
Generation Assay of Uracil Prototrophs From G. kaustophilus MK536
Geobacillus kaustophilus MK536 was precultured overnight at 60°C in LB medium (5 mL). The cells were collected by centrifugation (14,000 × g, 10 s) and suspended in sterile water (1 mL) to remove medium elements. Cells were collected again by centrifugation and resuspended in sterile water (0.15 mL). The suspension was used to prepare a dilution series in sterile water, which was plated on MU media and then incubated at 65°C for 72 h to obtain uracil prototrophs. The dilution series was also incubated on MM plates for 24 h to determine viable cell concentrations. The generation frequency of uracil prototrophs was defined as the ratio of generated uracil prototrophs to incubated viable cells (105–106 cfu).
Isolation of Genomic DNA
Mixtures were vigorously agitated during each addition of reagents. G. kaustophilus was cultured at 60°C in LB medium (30 mL). Cells were collected by centrifugation (4,400 × g, 5 min) and suspended in TEG buffer (3 mL) that contained Tris–HCl (25 mM, pH 8.0), ethylenediaminetetraacetic acid (10 mM), D-glucose (50 mM), lysozyme (1 mg/mL), and ribonuclease A (1 μg/mL). Following incubation at 37°C for 30 min, the suspension was mixed with sodium dodecyl sulfate (10%, 0.3 mL) and proteinase K (4 μg) and then incubated at 60°C for 30 min. The homogenate was supplemented with NaCl (5 M, 0.3 mL), cetyltrimethylammonium bromide (5%, 0.3 mL), and phenol/chloroform/isoamyl alcohol (25:24:1, 0.3 mL). After centrifugation (4,400 × g, 10 min), the aqueous supernatant was transferred to a conical tube and mixed with an equal volume of ethanol. The tube was repeatedly inverted to precipitate genomic DNA, which was washed twice with ethanol (70%, 1 mL) and dissolved in TE buffer (1 mL) that contained Tris–HCl (10 mM, pH 7.5) and ethylenediaminetetraacetic acid (1 mM). For next-generation sequencing, genomic DNA was further purified using a NucleoSpin gDNA Clean-up (Takara Bio, Otsu, Japan).
Sequencing Analysis
The pyrF upstream region in uracil prototrophs was amplified using the primers pyrF200R and amyA1300R. The amplicons were purified using the GenElute Agarose Spin Columns (Sigma Aldrich, St. Louis, MO, United States) and sequenced with Applied Biosystems 3730xl DNA Analyzer (Thermo Fisher Scientific, Waltham, MA, United States). Cycle sequencing reactions were performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) with the primers amyA200F, amyA400F, amyA400R, amyA800R, amyA1300R, and/or pyrF20R. The library for next-generation sequencing was constructed using an NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA, United States) and validated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States). Sequences were obtained as 150 bp pair-end reads on a NovaSeq 6000 system (Illumina, San Diego, CA, United States). The assembly was first performed using Velvet (Zerbino and Birney, 2008) with default parameters. Based on the assembly, sequencing reads were aligned and assembled into contigs using SSPACE (Boetzer et al., 2011) and GapFiller (Boetzer and Pirovano, 2012). The draft sequence was compared with the complete sequence of G. kaustophilus HTA426 (Takami et al., 2004b) using BLAST1 to identify IS elements that had transposed. The read sequences were also mapped to the genome sequence of G. kaustophilus HTA426 using Burrows–Wheeler Aligner (Li and Durbin, 2010) and analyzed using Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013) to confirm the IS transposition. The IS elements transposed in G. kaustophilus MK536up1 were amplified using the following primers: is25F and is25R (at GK3299 locus); is28F and is28R (between GK1097 and GK1098 loci); is72F and is72R (at GK0885 locus); and is87F and is87R (between GK0301 and GK0302 loci). Amplicons were sequenced to determine the intact sequences.
Transcription Analysis
Geobacillus kaustophilus strains MK536 and MK536up1 were cultured in MM medium at 60°C. Cells were collected at OD600 = 1, and RNA was purified using an RNAprotect Bacteria Reagent and RNeasy Mini Kit (Qiagen, Venlo, Netherlands) with gDNA Eraser (Takara Bio). The pyrF transcript was detected using endpoint reverse transcription-polymerase chain reaction (RT-PCR). The RT reaction was performed using a PrimeScript RT reagent Kit (Takara Bio) with the PyrFTR primer, whereas PCR was performed using Quick Taq HS DyeMix (Toyobo, Osaka, Japan) with two sets of primers: pyrF0F and pyrF600R (primer A) and is701250F and pyrF200R (primer B). Thermal cycles comprised 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 2 min. The reaction without reverse transcriptase was used as negative control. The transcription of rpoB, which encodes for RNA polymerase β subunit, was detected as positive control using the primers rpoB2800F and rpoB3800R.
Southern Blot Analysis of ISGka1/ISGka2 Transposition
Geobacillus kaustophilus MK536 was precultured in LB medium (5 mL) at 60°C. The cells were washed with sterile water (see above), and an aliquot (105–106 cfu) was incubated at 65°C on MU plates for 72 h. Generated colonies were purified on MU plates and used as uracil prototrophs, whereas background cells (without colony formation) were purified on LB plates and used as uracil auxotrophs. Washed cells (105–106 cfu) were also incubated at 65°C in liquid MU (20 mL) for 48 h. Cells were recovered on LB plates and screened using MM and MU plates to distinguish between uracil prototrophs and auxotrophs. In addition, washed cells were incubated at 65°C for as long as possible using media that prevent active cell growth: MC and MN media and LB medium supplemented with kanamycin (5 mg/L) or chloramphenicol (10 mg/L). Cells were recovered on LB plates from the resultant cultures. For successive culture under growth non-inhibitory conditions, MK536 was inoculated in LB medium (100 mL) and incubated at 65°C for 24 h. An aliquot of the culture (1 mL) was inoculated in fresh medium and further incubated under the same conditions. This process was repeated an additional five times. Subsequently, cells were colonized on LB plates. Respective clones were analyzed by Southern blotting to detect IS elements of G. kaustophilus (ISGka1 and ISGka2). Genomic DNA (25 μg) was digested with DraI and MunI. The products were separated on an agarose gel (0.9%) by electrophoresis and transferred onto a nylon membrane to hybridize with a digoxigenin-labeled DNA probe. The probe was synthesized using a PCR DIG Probe Synthesis Kit (Roche, Basel, Switzerland) with the primers is701250F and is701800R. Hybridized DNA was detected using a DIG Nucleic Acid Detection Kit (Roche).
Generation Assay of Uracil Prototrophs and Rifampicin-Resistant Mutants From MK536 Derivatives
Geobacillus kaustophilus strains MK536p74, MK536rsbV, MK536sigB, and MK536sigX were precultured at 60°C in LB medium (20 mL). After the culture had reached proliferative phase (OD600 = 1) and stationary phase (plus four additional hours of incubation), the cells were collected and washed with sterile water (see above). An aliquot (105–106 cfu) was incubated at 65°C for 96 h on MU plates to obtain uracil prototrophs. The aliquot was also incubated on MM plates for 24 h to determine viable cell concentrations. The generation frequency of uracil prototrophs was defined as the ratio of generated uracil prototrophs to incubated viable cells (105–106 cfu). In addition, proliferative and stationary cells were spread on LB plates supplemented with rifampicin (10 mg/L) and incubated at 65°C for 24 h to obtain rifampicin-resistant mutants. The aliquots were also incubated on LB plates for 24 h to determine viable cell concentrations. The generation frequency of rifampicin-resistant mutants was defined as the ratio of generated rifampicin-resistant mutants to incubated viable cells (105–108 cfu).
Statistical and Bioinformatic Analyses
Statistical significance was analyzed using unpaired Student’s t-tests (one-tailed) with Microsoft Excel 2016. The sequence comparison was performed using ClustalW2, and IS elements were predicted using ISsaga (Varani et al., 2011).
Results
Unexpected Generation of Uracil Prototrophs From G. kaustophilus MK536
Geobacillus kaustophilus MK536 lacks the pyrF gene essential for pyrimidine biosynthesis. Although the strain harbors intact pyrF under the PT7 control (Figure 1), the gene is theoretically silent in the absence of T7RP that functions at elevated temperatures. Therefore, G. kaustophilus MK536 is auxotrophic for uracil; however, this strain unexpectedly produced uracil prototrophs when incubated on minimum medium without uracil (MU) at 65°C. The prototrophs appeared following incubation for >48 h (Figure 2A). When a prototroph (termed MK536up1) was randomly selected and again incubated on MU plates, the majority of cells (>95%) formed colonies within 24 h as similar to those formed by the wild-type strain (HTA426). The prototrophy was stable and remained so throughout subculturing. The generation of uracil prototrophs was more rapid at 70°C but was most frequent at 65°C with prolonged incubation (Figure 2B). The optimal growth temperature of G. kaustophilus is 65°C (Suzuki, 2017); thus, uracil prototrophs were actively generated at temperatures where G. kaustophilus efficiently propagates. Prototrophs were not generated from a control strain lacking the PT7-pyrF cassette (MK242).
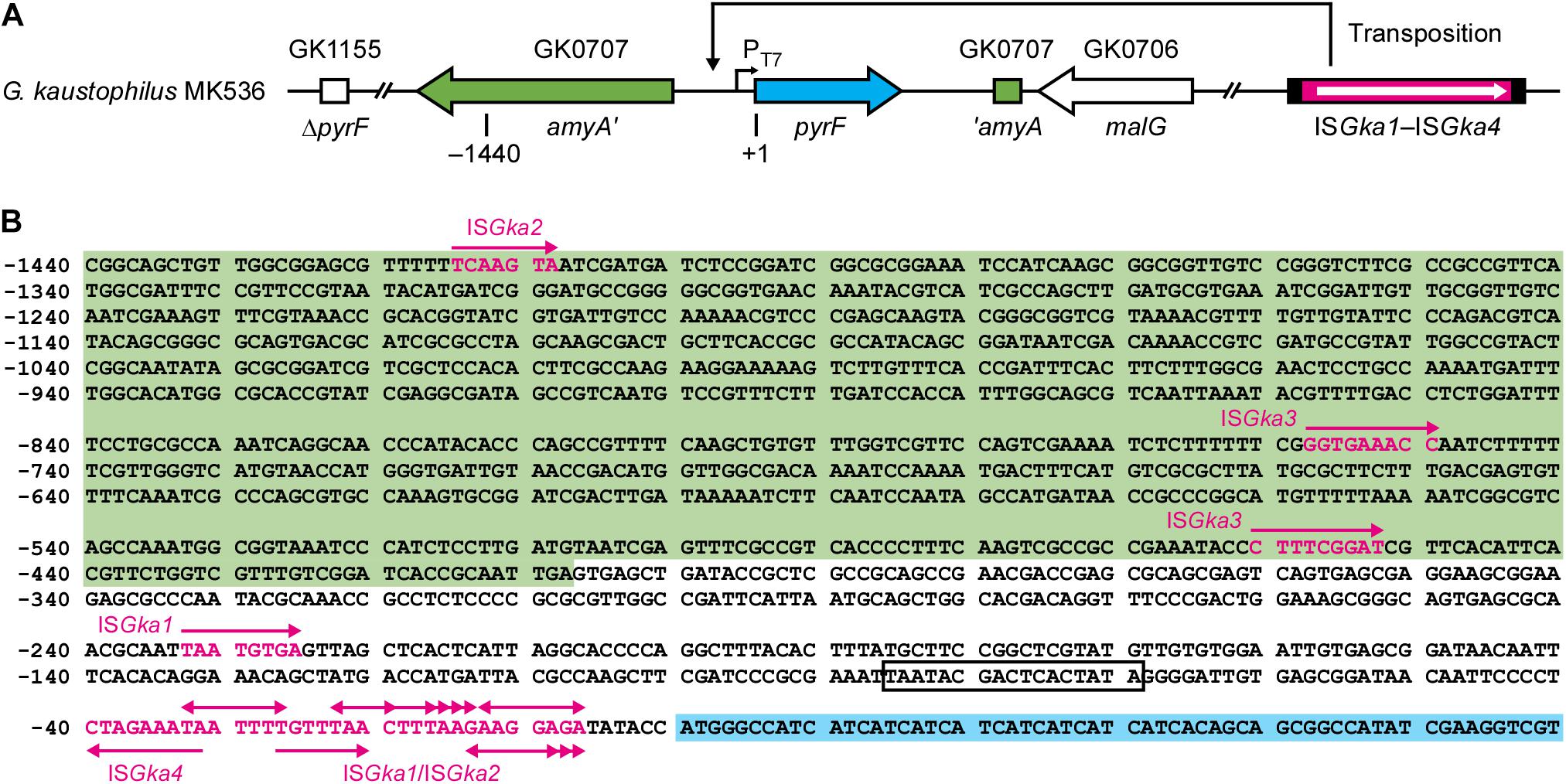
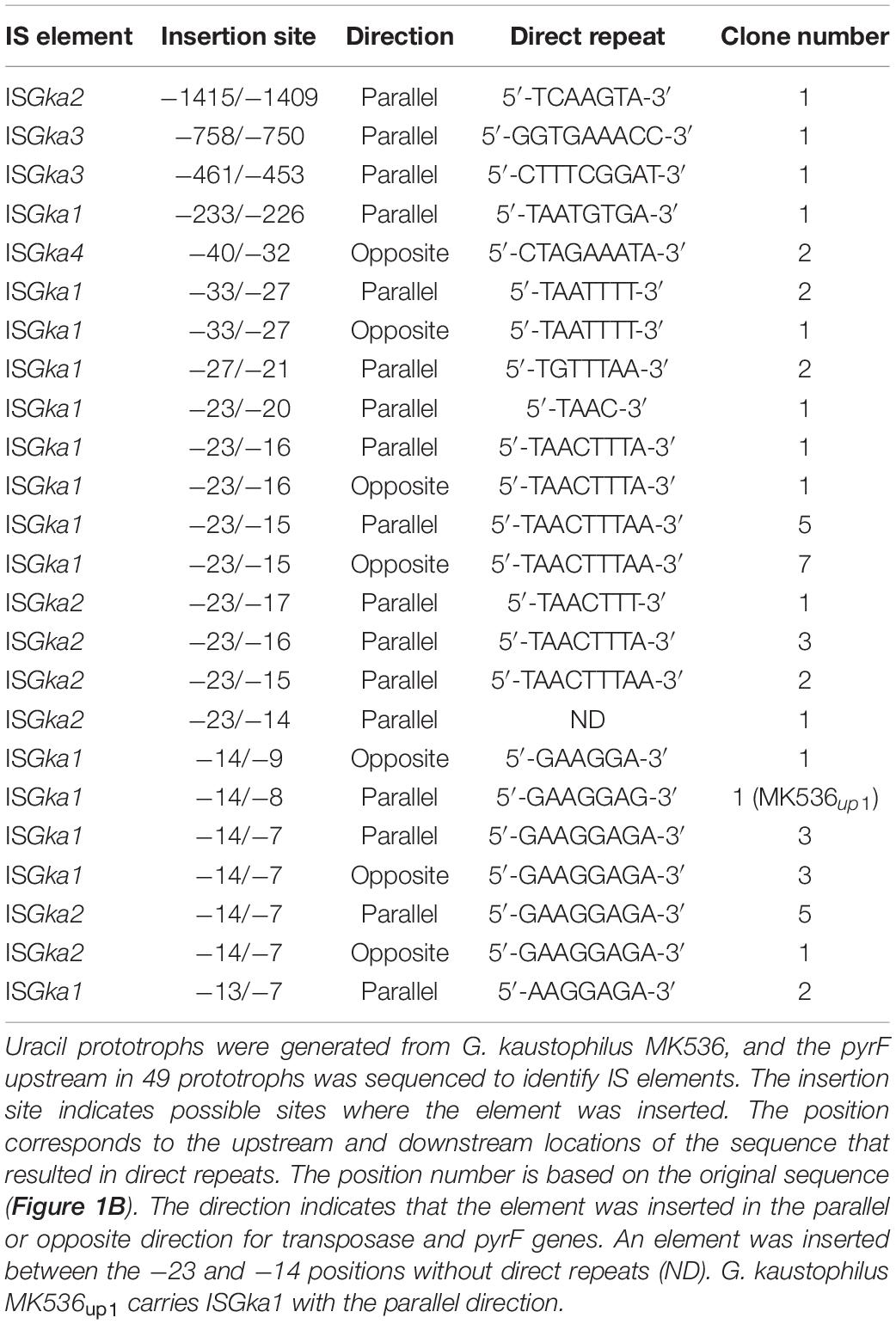
Figure 1. Organization of relevant genes in Geobacillus kaustophilus MK536. (A) G. kaustophilus MK536 lacks pyrF at GK1155 locus (ΔpyrF) but separately integrates pyrF (cyan) at GK0707 locus along with amyA disruption (green). The pyrF gene is under the control of T7 promoter (PT7), which is inactive in the absence of T7 RNA polymerase; therefore, G. kaustophilus MK536 is essentially auxotrophic for uracil. The chromosome contains insertion sequences (IS) termed ISGka1, ISGka2, ISGka3, and ISGka4 (magenta) at three (GK0302, GK2085, and GK2942), two (GK0390 and GK1725), one (GK0169), and three loci (GK0015, GK2451, and GK3431), respectively. The numbers indicate positions where the initiation codon of pyrF is defined as +1. (B) Nucleotide sequence between the positions −1440 and +60. The amyA and pyrF regions are shown with green and cyan backgrounds, respectively, and PT7 is boxed. Uracil prototrophs were identified to carry IS elements in this region. Magenta arrows indicate the position and insertion direction of the IS elements, whereas magenta letters indicate sequences that resulted in direct repeats. Table 2 summarizes detailed information on the elements.
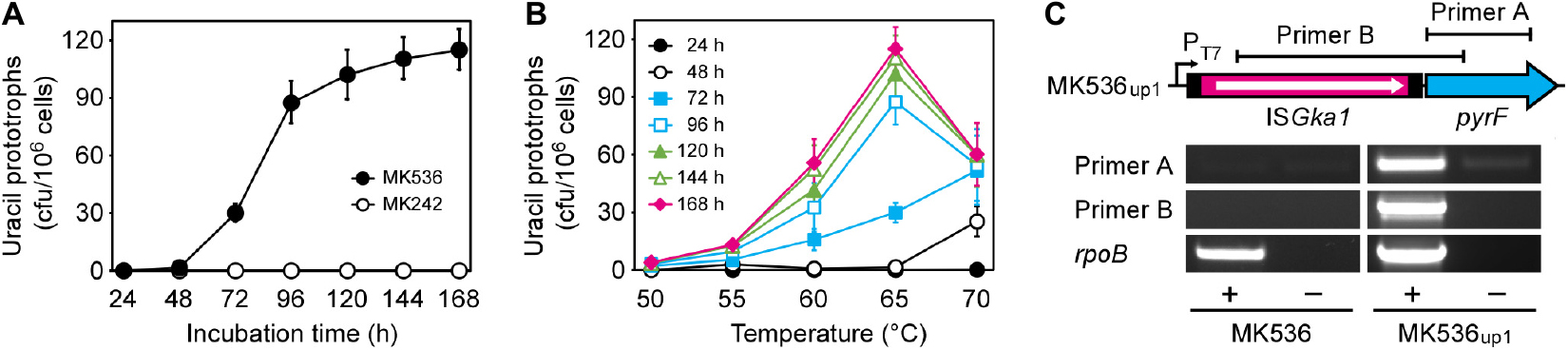
Figure 2. Generation of uracil prototrophs from G. kaustophilus MK536. (A) The time course of prototroph generation. G. kaustophilus strains MK536 (solid) and MK242 (hollow) were incubated at 65°C on MU plates to determine the generation frequencies of uracil prototrophs every 24 h. Data are presented as the mean ± standard error of cumulative numbers (n = 4–8). (B) The temperature profiles for prototroph generation. G. kaustophilus MK536 was incubated at 50–70°C on MU plates to determine the generation frequencies of uracil prototrophs every 24 h. Data are presented as the mean ± standard error of cumulative numbers (n = 4–8). (C) Transcription analysis of pyrF in G. kaustophilus strains MK536 and MK536up1. Total RNA was purified from cells that were cultured in MM medium and underwent reverse transcription (+). The products were used as templates for PCR to amplify the pyrF (0.6 kb) and ISGka1-pyrF (1.4 kb) regions using primers A and B, respectively. The reaction without reverse transcriptase was used as negative control (–). A partial region (1.0 kb) of rpoB transcripts was detected as positive control.
Uracil Prototrophs Carry IS Elements in the pyrF Upstream Region
For uracil prototrophy, it was hypothesized that PT7 mutations were responsible for T7RP-independent pyrF expression because uracil prototrophs were not generated from G. kaustophilus MK242; thus, we sequenced upstream of pyrF in 49 prototrophs including G. kaustophilus MK536up1. All the sequences lacked mutations but did contain different IS elements that encoded for DDE-type transposases (Figure 1). The IS elements were precisely flanked by DR that originated from the insertion site; therefore, it was probable that the event resulted from transposition but not from heterologous recombination. Among the 49 elements, 31 and 14 were completely identical with the IS elements at GK0302, GK2085, and GK2942 loci (termed ISGka1; Supplementary Figure 1) and those at GK0390 and GK1725 loci (ISGka2; Supplementary Figure 1), respectively. The ISGka2 sequence was highly homologous to the element at GK0778 locus but was distinguishable via a mutation. The other four sequences were completely identical to the IS element at GK0169 locus (ISGka3; Supplementary Figure 2) or those at GK0015, GK2451, and GK3431 loci (ISGka4; Supplementary Figure 3). The homologs of ISGka3 and ISGka4 were further identified at three (GK1006, GK1161, and GK1712) and five (GK0785, GK0875, GK1016, GK1720, and GKP33) loci, respectively. Based on sequence similarities, ISGka1 and ISGka2 were together classified to the IS701 family, whereas ISGka3 and ISGka4 were classified to the IS4 and ISLre2 families, respectively.
Insertion Features of the IS Elements
Table 2 summarizes the classification, position, direction, and DR sequence of IS elements identified at the pyrF upstream site. The ISGka3 sequence was inserted in the parallel direction with the transposase and pyrF genes, whereas ISGka4 was inserted in the opposing direction. In agreement with observations for IS4 and ISLre2 families (Siguier et al., 2015), ISGka3 and ISGka4 generated 9 bp of DR. The ISGka1 and ISGka2 sequences were inserted in both directions and occasionally at distant locations from the pyrF gene. Their transposition generated 4–9 bp of DR in disagreement with the observations of another set of IS701 members, which are known to generate 4 or 5 bp of DR (Siguier et al., 2015). Hot spots were observed immediately upstream of the pyrF gene and were favored by ISGka1 and ISGka2. Consensus sequences were not identified around insertion sites, although hot spots were abundant in adenine and thymine positions (Figure 1B).
Transcription of pyrF in G. kaustophilus MK536up1
Endpoint RT-PCR analysis showed that the pyrF gene was positively transcribed in MK536up1 but not in MK536; there was a continuous transcript from ISGka1 upstream to pyrF (Figure 2C). Although faint bands were detected for MK536up1 samples without RT via genomic DNA contamination, band signals were stronger for samples with RT. These observations suggested that G. kaustophilus MK536up1 became prototrophic for uracil by pyrF expression via leaky and read-through transcription of the transposase gene and/or via active transcription from another promoter in ISGka1.
Genome-Wide IS Transposition in G. kaustophilus MK536up1
Next-generation sequencing of G. kaustophilus MK536up1 provided 8 × 106 reads and 169 contigs with 318 depth, which were compared with the complete genome sequence of G. kaustophilus HTA426 (Takami et al., 2004b). In addition to ISGka1 that transposed to the pyrF upstream region, ISGka2 and an IS element of the IS1634 family were inserted at two and one loci, respectively, (Figure 3). The ISGka2 sequence was flanked by 7 or 8 bp of DR. The IS1634 member was identical to the IS elements at GK0145 and GK3302 loci (termed ISGka5; Supplementary Figure 4) and generated 6 bp of DR. The PCR assays confirmed that these elements were absent at the respective loci in G. kaustophilus MK536. Although another IS1634 member was also identified at GK3299 locus, the element already existed in G. kaustophilus MK536; thus, it was likely that this transposed during construction of G. kaustophilus MK536 from G. kaustophilus HTA426. IS deletion was not identified in the genome sequence, which suggested that these elements achieved replicative transposition. Many mutations (134 single nucleotide variants, 50 deletions, and 81 insertions) were identified; however, it was unclear whether these mutations occurred during generation of MK536up1 from MK536 or during construction of MK536 from HTA426.
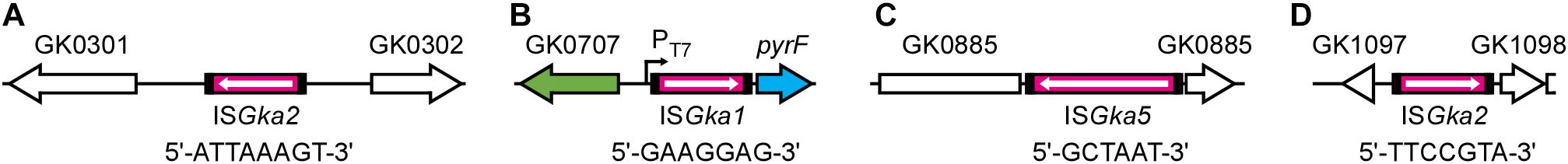
Figure 3. IS transposition in G. kaustophilus MK536up1. Genomic DNA was purified from G. kaustophilus MK536up1 and sequenced to identify genome-wide IS transposition. Magenta boxes indicate IS elements identified to achieve transposition in G. kaustophilus MK536up1 where arrows indicate the direction of transposase genes. Short sequences indicate direct repeats. ISGka2 was identified between transposase genes at GK0301 and GK0302 loci (A), whereas ISGka1 was identified at the pyrF upstream site (B). ISGka5 was identified in a hypothetical gene at GK0885 locus (C). ISGka2 was also identified between hypothetical and RNA methyltransferase genes at GK1097 and GK1098 loci, respectively, (D).
Southern Blot Analysis of ISGka1/ISGka2 Transposition
Geobacillus kaustophilus MK536 was incubated at 65°C for 72 h on MU plates to isolate uracil prototrophs, and eight clones were analyzed by Southern blotting that collectively detects ISGka1 and ISGka2 (Figure 4A). Six bands were present in the MK536 samples. The band lengths corresponded to the theoretical ones predicted from the sequences around the loci GK0778 (7.5 kb), GK2085 (2.8 kb), GK0302 (2.7 kb), GK0390 (2.4 kb), GK1725 (2.2 kb), and GK2942 (2.0 kb). The original bands remained present in the seven prototrophs; thus, ISGka1 and ISGka2 generally achieved replicative transposition that provided new information on the IS701 family. One prototroph lost the band at GK0302 locus. Because an IS element uses either replicative or non-replicative mechanism, the loss seemed to result from a band shift via IS insertion around the locus, as observed for G. kaustophilus MK536up1 (Figure 3A). A band was further shared at 4.2 kb, which was attributable to the pyrF upstream region that carries ISGka1 or ISGka2 on the basis of the theoretical length. Additional bands were also detected in four prototrophs. This supported IS transposition occurring in a genome-wide manner. In addition to uracil prototrophs, background cells that remained uracil auxotrophs were recovered from MU plates and analyzed by Southern blotting to show that the six clones increased the band signals at diverse lengths but not at 4.2 kb (Figure 4B). Notably, successive cultures under non-inhibitory growth conditions in LB medium resulted in undetectable ISGka1/ISGka2 transposition (Figure 4C). These observations implied that ISGka1/ISGka2 transposition extensively occurred in cells (>75%) on MU plates and that cells became uracil prototrophs when an IS element fortuitously transposed to the pyrF upstream region.
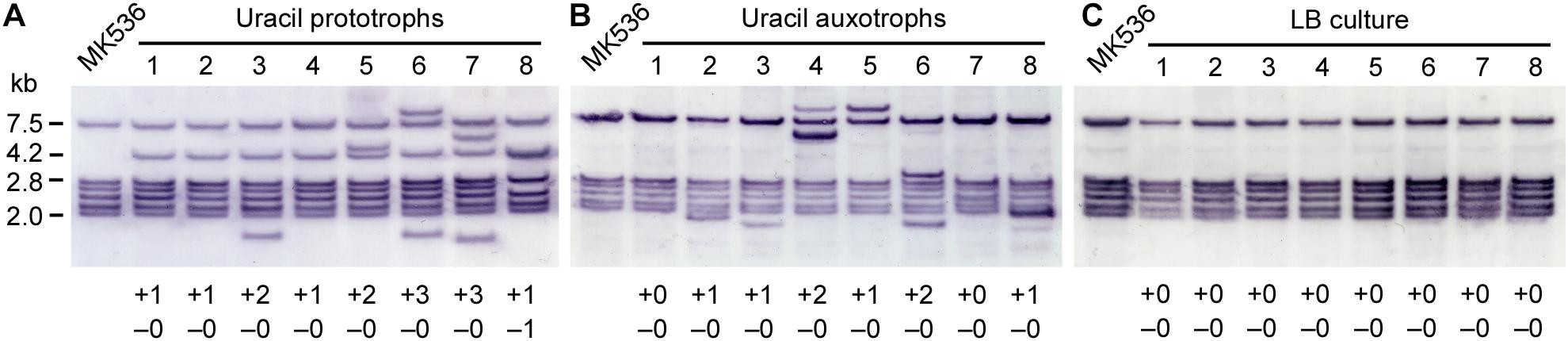
Figure 4. Southern blot analysis of ISGka1/ISGka2 transposition. (A,B) G. kaustophilus MK536 (105–106 cfu) was incubated at 65°C on MU plates for 72 h. Grown colonies were used as uracil prototrophs (A), whereas background cells were used as uracil auxotrophs (B). Eight clones were randomly selected and analyzed by Southern blotting. The probe was designed to detect both ISGka1 and ISGka2. The six bands for MK536 were attributable to fragments from the loci GK0778 (7.5 kb), GK2085 (2.8 kb), GK0302 (2.7 kb), GK0390 (2.4 kb), GK1725 (2.2 kb), and GK2942 (2.0 kb). At 4.2 kb, the pyrF upstream was detected carrying an IS element. The numbers at the bottom indicate an increase (+) and decrease (–) in bands. (C) G. kaustophilus MK536 was successively (seven times) cultured at 65°C in LB medium. Cells were purified on LB plates, and eight clones were analyzed by Southern blotting.
Culture Conditions for ISGka1/ISGka2 Transposition
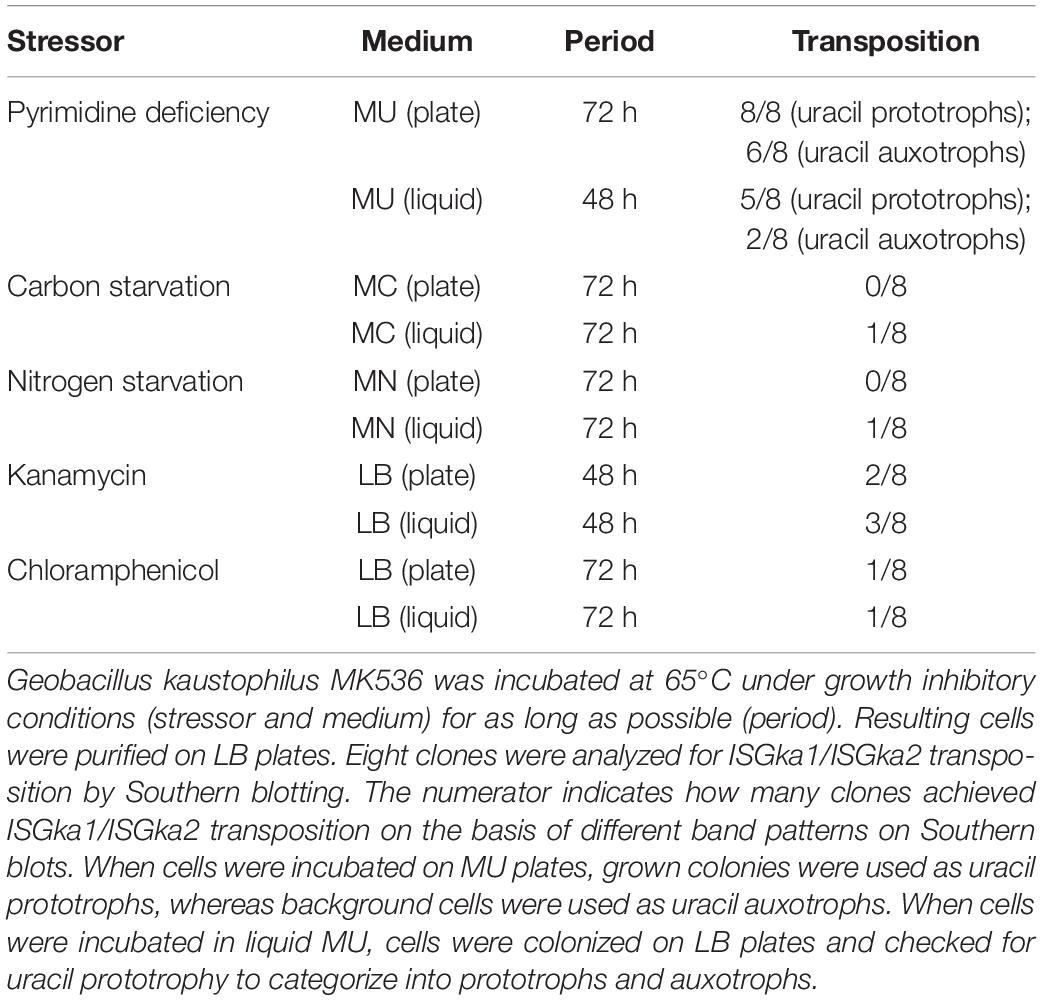
Geobacillus kaustophilus MK536 was incubated in liquid MU at 65°C for 48 h and then grown on LB plates. Any subsequent colonies were classified as uracil prototrophs or auxotrophs, and eight clones were analyzed by Southern blotting to detect ISGka1 and ISGka2. The signals were changed in five prototrophs and two auxotrophs (Table 3), which suggested that ISGka1/ISGka2 transposition was enhanced not only on MU plates but also in liquid MU. The 4.2 kb signal was not detected in the prototrophs; however, PCR analysis confirmed that the pyrF upstream region had lengthened, suggesting that these clones might carry an IS element other than ISGka1 or ISGka2 at the pyrF upstream site. Similarly, MK536 cells were incubated in diverse media to prevent the cells from active propagation and then eight clones were recovered on LB plates to analyze ISGka1/ISGka2 transposition. Incubation was performed for 72 h; however, the incubation time was shortened to 48 h when cells were not recovered. Table 3 summarizes the incubation conditions and the number of clones that achieved ISGka1/ISGka2 transposition. Transposition was detected for cells that underwent growth inhibition by kanamycin, chloramphenicol, or carbon or nitrogen starvation. These observations suggested that IS transposition was enhanced under growth inhibitory conditions and may be regulated via a stress response pathway.
Expression of sigX Enhances Generation of Uracil Prototrophs From G. kaustophilus MK536
To see whether stress response regulators govern IS transposition in G. kaustophilus, we constructed MK536 derivatives that forcibly expressed rsbV (MK536rsvV), sigB (MK536sigB), and sigX (MK536sigX). The cells were precultured until they reached proliferative and stationary growth phases and then incubated at 65°C on MU plates to determine the generation frequency of uracil prototrophs (Figure 5A). As with G. kaustophilus MK536, the control strain (MK536p74) generated prototrophs following incubation for >48 h. Similar observations were also made for MK536rsbV and MK536sigB. In these strains, the generation frequency was higher in proliferative cells than in stationary cells. Although the stationary cells of MK536rsbV and MK536sigB exhibited lower frequency than did those of MK536p74, the difference was not substantial. In contrast, MK536sigX more rapidly and frequently generated uracil prototrophs than did the other strains. Moreover, the generation frequency was comparable between proliferative and stationary cells at >48 h. These observations suggested that IS transposition was enhanced via sigX-dependent stress responses and that the response was stronger in proliferative cells.
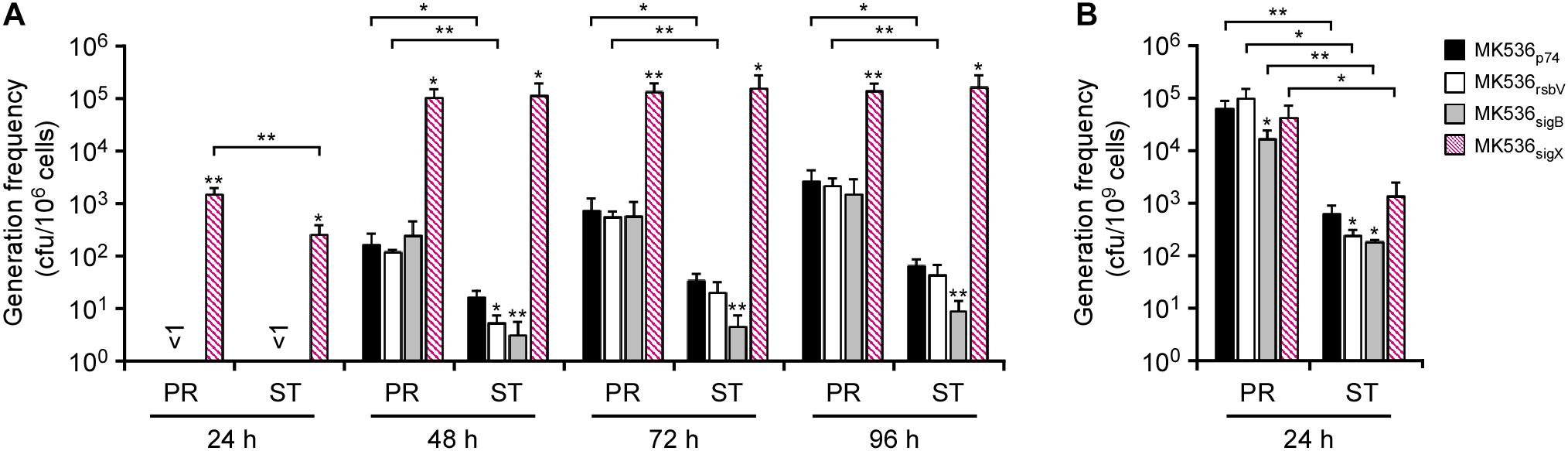
Figure 5. The effects of rsbV, sigB, and sigX expression on the generations of uracil prototrophs (A) and rifampicin-resistant mutants (B). (A) G. kaustophilus strains MK536p74 (solid), MK536rsbV (hollow), MK536sigB (gray), and MK536sigX (diagonal) were precultured until the proliferative (PR) and stationary (ST) phases were achieved and then incubated at 65°C on MU plates for 24–96 h to determine the generation frequencies of uracil prototrophs. (B) Proliferative and stationary cells were incubated at 65°C on LB plates supplemented with rifampicin (10 mg/L) for 24 h to determine the generation frequencies of rifampicin-resistant mutants. Data are presented as the mean ± standard error of cumulative numbers (n = 4). Statistical significance is shown between respective strains and MK536p74 (asterisks directly above the bars) and between proliferative and stationary cells (asterisks above the connection lines). Single and double asterisks indicate P < 0.15 and P < 0.05, respectively.
Expression of sigX Has Negligible Effects on Rifampicin-Resistant Mutations
Geobacillus spp. apparently induce mutagenesis when proliferative cells are exposed to rifampicin and efficiently generate rifampicin-resistant cells via rpoB mutations (Suzuki et al., 2018). To see whether the mutagenesis depends on sigX-dependent stress responses, MK536 derivatives (MK536p74, MK536rsbV, MK536sigB, and MK536sigX) were assessed by generation frequency assay of rifampicin-resistant mutants. Cells were precultured until they reached proliferative and stationary phases and incubated for 24 h at 65°C on LB plates supplemented with rifampicin. Grown colonies were counted to determine that the generation frequency of rifampicin-resistant mutants was higher in proliferative cells than in stationary cells for all of the strains, but substantial differences were not observed between the respective strains (Figure 5B). This suggested that the rifampicin-resistant mutations were independent from sigX-dependent stress responses.
Discussion
Isothermal and transcription-based amplification of nucleic acids can be performed using T7RP, which is responsible for strong transcription from PT7 (Niemz et al., 2011). Because amplification performance is potentially improved when conducted at higher temperatures, thermostable T7RP variants are of biotechnological importance (Boulain et al., 2013). This study was originally designed to generate mutant genes for thermostable T7RP variants in G. kaustophilus MK534T7RP. The strain is auxotrophic for uracil by the ΔpyrF genotype but carries pyrF under control of PT7 (PT7-pyrF); therefore, it could become prototrophic with functional expression of T7RP from pGKE75-T7RP. In addition, this strain lacks genes for DNA repair (mutS, mutL, mutY, ung, and mfd) and thereby could serve as an error-prone strain. We expected that G. kaustophilus MK534T7RP would generate mutant genes for thermostable T7RP variants via culture and that such genes could be found in clones prototrophic for uracil at elevated temperatures. As expected, MK534T7RP generated uracil prototrophs at 65°C; however, similar prototrophs were intrinsically generated from a control strain that lacked pGKE75-T7RP (MK536). Uracil prototrophs were not generated from another control strain without PT7-pyrF (MK242); therefore, we assumed PT7 mutations in uracil prototrophs and unexpectedly identified diverse IS elements at the pyrF upstream region (Figure 1). IS elements are known to cause neighboring gene expression from internal promoters or via formation of hybrid promoters (Zhang and Saier, 2016; Vandecraen et al., 2018). In fact, pyrF transcription was detected in MK536up1 in contrast to MK536 (Figure 2C). In G. kaustophilus MK536up1, a promoter upstream of the transposase gene apparently contributes to pyrF transcription. However, considering several prototrophs carried ISGka1/ISGka2 or ISGka4 at the pyrF upstream region in the opposite direction (Table 2), they seem to harbor promoters with the opposite direction. Possible opposite promoters identified in ISGka1/ISGka2 and ISGka4 are shown in Supplementary Figures 1 and 3, respectively.
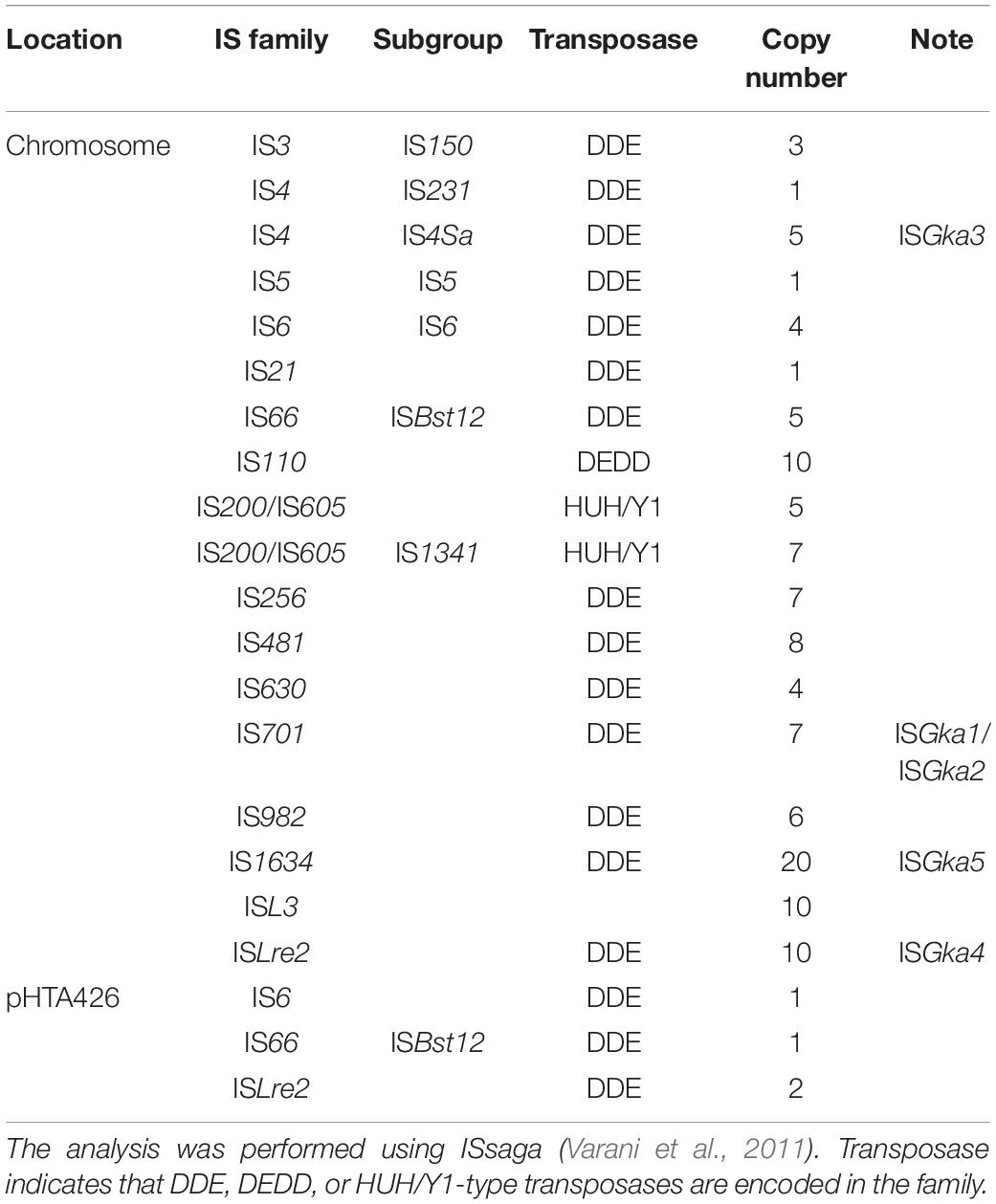
Geobacillus kaustophilus HTA426 harbors a circular chromosome of 3.5 Mb and a large plasmid of 48 kb (Takami et al., 2004b), which have been predicted to carry 118 and 4 copies of possible IS elements, respectively (Table 4). The elements that achieved transposition include members of the families IS4 (ISGka3), IS701 (ISGka1 and ISGka2), IS1634 (ISGka5), and ISLre2 (ISGka4). The copy number of these elements is 11, which suggest that >9% of the total elements (122 copies) are functional in G. kaustophilus. Among the elements, ISGka1 and ISGka2 were most frequently identified in the pyrF upstream region (Table 2). Their transposition was also more frequent in the MK536up1 genome (Figure 3). Possible reasons for this include a higher copy number, replicative transposition, and abundant sequences to which ISGka1 and ISGka2 preferentially transpose. All the elements (ISGka1–ISGka5) transposed at elevated temperatures, indicating that these elements are thermophilic, which is in agreement with their distribution in thermophilic species of the family Bacillaceae (e.g., Aeribacillus, Anoxybacillus, Bacillus, and Geobacillus genera). Although several reports describe IS transposition in thermophiles (Sen and Oriel, 1990; Natarajan and Oriel, 1991; Xu et al., 1993; Schleper et al., 1994; Gregory and Dahlberg, 2008) or genetic modification using mesophilic mobile elements (Carr et al., 2015), no IS element has been hitherto demonstrated to preferentially function in thermophiles at elevated temperatures; thus, this study is the first to identify thermophilic IS elements. Mobile elements can be used for gene discovery and gene delivery (Picardeau, 2010; Narayanavari et al., 2017). Therefore, ISGka1–ISGka5 have the potential to expand the genetic tools for thermophiles.
Southern blot analysis suggested that ISGka1/ISGka2 transposition frequently occurred in a genome-wide manner. Genome-wide transposition has been supported by the MK536up1 genome, which carries additional IS elements at four loci (Figure 3). ISGka1/ISGka2 transposition was detected not only in cells that became uracil prototrophs but also in background cells that remained uracil auxotrophs; however, transposition was not detected when cells were cultured under growth non-inhibitory conditions (Figure 4). These observations suggest that G. kaustophilus enhances IS transposition in response to pyrimidine deficiency and generated numerous mutants, including uracil prototrophs, where an element was fortunately transposed into the pyrF upstream region. The idea is consistent with growth of uracil prototrophs on MU plates after >48 h incubation. Given that prototrophs had randomly arisen during preculture, the colonies should have appeared within 24 h, as observed for G. kaustophilus strains HTA426 and MK536up1. Uracil prototrophs identified in liquid MU supposedly carried another type of IS elements in the pyrF upstream region, which seems to perform transposition earlier than ISGka1 or ISGka2 under this condition because prototrophs generated earlier in the liquid culture rapidly grow and could become dominant throughout the subsequent phases. This observation also supports the IS transposition enhanced under pyrimidine deficient conditions.
Stress-induced transposition has been reported for several organisms by respective stressors (Zhang and Saier, 2016; Vandecraen et al., 2018; Lee et al., 2019, 2020). In G. stearothermophilus CU21, IS4 and IS21 members have been reported to achieve transposition during growth inhibition by chloramphenicol exposure (Xu et al., 1993). Notably, ISGka1/ISGka2 transposition was detected in eight clones that underwent growth inhibition by antibiotic exposure or starvation; therefore, IS transposition may be enhanced by growth inhibition regardless of stressors. Although transposition was not observed under similar starvation conditions on solid media, this was attributed to nutrient contaminants in the agar enabling minimal growth under conditions of incomplete starvation. We note that transposition enhanced by growth inhibition helps organisms achieve genetic diversification that results in environmental adaptation exclusively during the period of growth inhibition; in parallel, transposition permitted by hosts is advantageous for IS elements in terms of their propagation. Geobacillus spp. harbor numerous IS elements in their genomes (Suzuki, 2018), which may reflect codependence relationships between Geobacillus spp. and IS elements for evolutionary diversification and stable persistence, respectively.
Bacillus subtilis 168 is a model bacterium that is phylogenetically related to G. kaustophilus (Suzuki, 2018). In B. subtilis, various stress responses are positively regulated by SigB where the function is repressed by RsbW (anti-SigB) via complex formation, whereas RsbW is released by RsbV (anti-SigB antagonist); therefore, SigB can be activated by RsbV (van Schaik and Abee, 2005). In G. kaustophilus, homologous genes for RsbV, RsbW, and SigB have been identified at the GK3422, GK3423, and GK3424 loci, respectively, (Takami et al., 2004b). Extra-cytoplasmic function (ECF) sigma factors are also known to positively regulate stress responses (van Schaik and Abee, 2005). Although B. subtilis employs multiple ECF sigma factors (e.g., SigM, SigV, SigW, SigX, SigY, and SigZ), only the homologs for sigW and sigX at their respective loci GK0150 and GK2254 have been identified in G. kaustophilus (Takami et al., 2004b).
Because IS transposition was apparently enhanced by growth inhibition, we focused on RsbV, SigB, SigW, and SigX as potential regulators that govern IS transposition in G. kaustophilus and constructed rsbV (MK536rsbV), sigB (MK536sigB), and sigX (MK536sigX) expressers. Despite repetitive trials, neither a sigW expressor nor most deletion mutants (ΔsigW, ΔsigX, ΔrsbV, or ΔrsbW) could be constructed. Notably, uracil prototrophs were rapidly and substantially generated from MK536sigX in comparison with the other constructs (Figure 5A); therefore, it is possible that G. kaustophilus enhances IS transposition via sigX-dependent stress responses. This is the first observation suggesting that a sigma factor regulates IS transposition. In B. subtilis, SigX-dependent promoters share tgtAACnww and CGwCww consensus sequences at −35 and −10 regions, respectively, (Huang and Helmann, 1998). However, similar regions were not found upstream of transposase genes in ISGka1–ISGka5 (Supplementary Figures 1–4), and therefore SigX seems to indirectly regulate IS transposition rather than to directly bind to IS elements. It is also noteworthy that SigX is involved in controlling several processes related to cell envelope modification in B. subtilis (Souza et al., 2014). Conceivably, G. kaustophilus may enhance IS transposition by sensing cell surface damage driven by growth inhibition via sigX-dependent stress responses.
Uracil prototrophs were equally generated from proliferative and stationary cells of MK536sigX in contrast to other strains (Figure 5A). This profile suggests that G. kaustophilus enhances IS transposition under SigX regulation when proliferative cells are prevented from active propagation, thus potentially enabling immediate adaptation via genetic diversification. Geobacillus spp. can form robust endospores in the stationary phase (Suzuki, 2018); therefore, IS transposition may be enhanced as an adaptation strategy specific to the proliferative phase. We previously observed that exposure of G. kaustophilus to rifampicin apparently induced mutagenesis to produce rifampicin-resistant mutants and the induction was stronger in proliferative cells than in stationary cells (Suzuki et al., 2018). Although this manner implies that mutagenesis may be also governed by SigX in parallel to IS transposition, rifampicin-resistant mutants were comparably generated between MK536p74 and MK536sigX (Figure 5B); therefore, mutagenesis is not under the SigX control. Multiple mechanisms appear to be employed to generate genetic diversification in G. kaustophilus and potentially other Geobacillus spp. This characteristic may be a primary reason why Geobacillus spp. are highly adaptive organisms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRA database PRJNA699136.
Author Contributions
HS had conceived the experiment plan, supervised the experiment process, and wrote the original manuscript. JK examined T7RP mutations and identified IS transposition. MT and MO analyzed the transposition frequency and insertion sites of IS elements. TT performed the Southern blot and mutation assay. MO performed the transcriptome and genome analyses. TO had supervised the experiment process. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the following organizations: the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, Bio-oriented Technology Research Advancement Institution, Japan; the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry, Japan; the Institute for Fermentation, Osaka, Japan; and the Nagase Science and Technology Foundation; and JSPS KAKENHI (17K06925).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.650461/full#supplementary-material
Footnotes
References
Amartey, S. A., Leak, D. J., and Hartley, B. S. (1991). Development and optimization of a defined medium for aerobic growth of Bacillus stearothermophilus LLD-15. Biotechnol. Lett. 13, 621–626. doi: 10.1007/bf01086315
Bezuidt, O. K., Pierneef, R., Gomri, A. M., Adesioye, F., Makhalanyane, T. P., Kharroub, K., et al. (2016). The Geobacillus pan-genome: implications for the evolution of the genus. Front. Microbiol. 7:723. doi: 10.3389/fmicb.2016.00723
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Boulain, J. C., Dassa, J., Mesta, L., Savatier, A., Costa, N., Muller, B. H., et al. (2013). Mutants with higher stability and specific activity from a single thermosensitive variant of T7 RNA polymerase. Protein Eng. Des. Sel. 26, 725–734. doi: 10.1093/protein/gzt040
Bouuaert, C. C., and Chalmers, R. M. (2010). Gene therapy vectors: the prospects and potentials of the cut-and-paste transposons. Genetica 138, 473–484. doi: 10.1007/s10709-009-9391-x
Carr, J. F., Gregory, S. T., and Dahlberg, A. E. (2015). Transposon mutagenesis of the extremely thermophilic bacterium Thermus thermophilus HB27. Extremophiles 19, 221–228. doi: 10.1007/s00792-014-0663-8
Gregory, S. T., and Dahlberg, A. E. (2008). Transposition of an insertion sequence, ISTth7, in the genome of the extreme thermophile Thermus thermophilus HB8. FEMS Microbiol. Lett. 289, 187–192. doi: 10.1111/j.1574-6968.2008.01389.x
Huang, X., and Helmann, J. D. (1998). Identification of target promoters for the Bacillus subtilis σX factor using a consensus-directed search. J. Mol. Biol. 279, 165–173. doi: 10.1006/jmbi.1998.1765
Kobayashi, J., Furukawa, M., Ohshiro, T., and Suzuki, H. (2015a). Thermoadaptation-directed evolution of chloramphenicol acetyltransferase in an error-prone thermophile using improved procedures. Appl. Microbiol. Biotechnol. 99, 5563–5572. doi: 10.1007/s00253-015-6522-4
Kobayashi, J., Tanabiki, M., Doi, S., Kondo, A., Ohshiro, T., and Suzuki, H. (2015b). Unique plasmids generated via pUC replicon mutagenesis in an error-prone thermophile derived from Geobacillus kaustophilus HTA426. Appl. Environ. Microbiol. 81, 7625–7632. doi: 10.1128/aem.01574-15
Lee, C., Choi, N., Bae, M. K., Choo, K., and Lee, S. (2019). Transposition of insertion sequences was triggered by oxidative stress in radiation-resistant bacterium Deinococcus geothermalis. Microorganisms 7:446. doi: 10.3390/microorganisms7100446
Lee, C., Choo, K., and Lee, S. (2020). Active transposition of insertion sequences by oxidative stress in Deinococcus geothermalis. Front. Microbiol. 11:558747. doi: 10.3389/fmicb.2020.558747
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Liao, H., McKenzie, T., and Hageman, R. (1986). Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc. Natl. Acad. Sci. USA 83, 576–580. doi: 10.1073/pnas.83.3.576
Narayanavari, S. A., Chilkunda, S. S., Ivics, Z., and Izsvák, Z. (2017). Sleeping beauty transposition: from biology to applications. Crit. Rev. Biochem. Mol. Biol. 52, 18–44. doi: 10.1080/10409238.2016.1237935
Natarajan, M. R., and Oriel, P. (1991). Conjugal transfer of recombinant transposon Tn916 from Escherichia coli to Bacillus stearothermophilus. Plasmid 26, 67–73. doi: 10.1016/0147-619x(91)90037-w
Niemz, A., Ferguson, T. M., and Boyle, D. S. (2011). Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 29, 240–250. doi: 10.1016/j.tibtech.2011.01.007
Picardeau, M. (2010). Transposition of fly mariner elements into bacteria as a genetic tool for mutagenesis. Genetica 138, 551–558. doi: 10.1007/s10709-009-9408-5
Schleper, C., Roder, R., Singer, T., and Zillig, W. (1994). An insertion element of the extremely thermophilic archaeon Sulfolobus solfataricus transposes into the endogenous beta-galactosidase gene. Mol. Gen. Genet. 243, 91–96. doi: 10.1007/bf00283880
Schrader, L., and Schmitz, J. (2019). The impact of transposable elements in adaptive evolution. Mol. Ecol. 28, 1537–1549. doi: 10.1111/mec.14794
Sen, S., and Oriel, P. (1990). Transfer of transposon Tn916 from Bacillus subtilis to Thermus aquaticus. FEMS. Microbiol. Lett. 67, 131–134. doi: 10.1111/j.1574-6968.1990.tb13849.x
Siguier, P., Gourbeyre, E., Varani, A., Bao, T., and Chandler, M. (2015). Everyman’s guide to bacterial insertion sequences. Microbiol. Spectr. 3:MDNA3-0030-2014. doi: 10.1128/microbiolspec.MDNA3-0030-2014
Souza, B. M., Castro, T. L., Carvalho, R. D., Seyffert, N., Silva, A., Miyoshi, A., et al. (2014). σECF factors of gram-positive bacteria: a focus on Bacillus subtilis and the CMNR group. Virulence 5, 587–600. doi: 10.4161/viru.29514
Suzuki, H. (2017). “Geobacillus kaustophilus HTA426: a model organism for moderate thermophiles,” in Advances in Medicine and Biology, ed. L. V. Berhardt (New York, USA: Nova Science Publishers), 75–108.
Suzuki, H. (2018). Peculiarities and biotechnological potential of environmental adaptation by Geobacillus species. Appl. Microbiol. Biotechnol. 102, 10425–10437. doi: 10.1007/s00253-018-9422-6
Suzuki, H., Kobayashi, J., Wada, K., Furukawa, M., and Doi, K. (2015). Thermoadaptation-directed enzyme evolution in an error-prone thermophile derived from Geobacillus kaustophilus HTA426. Appl. Environ. Microbiol. 81, 149–158. doi: 10.1128/aem.02577-14
Suzuki, H., Murakami, A., and Yoshida, K. (2012). Counterselection system for Geobacillus kaustophilus HTA426 through disruption of pyrF and pyrR. Appl. Environ. Microbiol. 78, 7376–7383. doi: 10.1128/aem.01669-12
Suzuki, H., Taketani, T., Kobayashi, J., and Ohshiro, T. (2018). Antibiotic resistance mutations induced in growing cells of Bacillus-related thermophiles. J. Antibiot. 71, 382–389. doi: 10.1038/s41429-017-0003-1
Suzuki, H., Wada, K., Furukawa, M., Doi, K., and Ohshima, T. (2013a). A ternary conjugation system for the construction of DNA libraries for Geobacillus kaustophilus HTA426. Biosci. Biotechnol. Biochem. 77, 2316–2318. doi: 10.1271/bbb.130492
Suzuki, H., Yoshida, K., and Ohshima, T. (2013b). Polysaccharide-degrading thermophiles generated by heterologous gene expression in Geobacillus kaustophilus HTA426. Appl. Environ. Microbiol. 79, 5151–5158. doi: 10.1128/aem.01506-13
Takami, H., Nishi, S., Lu, J., Shimamura, S., and Takaki, Y. (2004a). Genomic characterization of thermophilic Geobacillus species isolated from the deepest sea mud of the Mariana Trench. Extremophiles 8, 351–356. doi: 10.1007/s00792-004-0394-3
Takami, H., Takaki, Y., Chee, G., Nishi, S., Shimamura, S., Suzuki, H., et al. (2004b). Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 32, 6292–6303. doi: 10.1093/nar/gkh970
Thorvaldsdóttir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. doi: 10.1006/plas.1993.1001
van Schaik, W., and Abee, T. (2005). The role of σB in the stress response of Gram-positive bacteria – targets for food preservation and safety. Curr. Opin. Biotechnol. 16, 218–224. doi: 10.1016/j.copbio.2005.01.008
Vandecraen, J., Chandler, M., Aertsen, A., and Van Houdt, R. (2018). The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 43, 709–730. doi: 10.1080/1040841x.2017.1303661
Varani, A. M., Siguier, P., Gourbeyre, E., Charneau, V., and Chandler, M. (2011). ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12:R30. doi: 10.1186/gb-2011-12-3-r30
Wada, K., Kobayashi, J., Furukawa, M., Doi, K., Ohshiro, T., and Suzuki, H. (2016). A thiostrepton resistance gene and its mutants serve as selectable markers in Geobacillus kaustophilus HTA426. Biosci. Biotechnol. Biochem. 80, 368–375. doi: 10.1080/09168451.2015.1079478
Xu, K., He, Z., Mao, Y., Sheng, R., and Sheng, Z. (1993). On two transposable elements from Bacillus stearothermophilus. Plasmid 29, 178–182.
Zeigler, D. R. (2014). The Geobacillus paradox: why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology 160, 1–11. doi: 10.1099/mic.0.071696-0
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Keywords: extra-cytoplasmic function sigma factor, IS4, IS701, IS1634, ISLre2, stress-induced transposition, thermophile, transposable element
Citation: Suzuki H, Taketani T, Tanabiki M, Ohara M, Kobayashi J and Ohshiro T (2021) Frequent Transposition of Multiple Insertion Sequences in Geobacillus kaustophilus HTA426. Front. Microbiol. 12:650461. doi: 10.3389/fmicb.2021.650461
Received: 07 January 2021; Accepted: 03 March 2021;
Published: 24 March 2021.
Edited by:
Richard Allen White III, University of North Carolina at Charlotte, United StatesReviewed by:
Fernando Rodriguez, Marine Biological Laboratory (MBL), United StatesSusan Wessler, University of California, Riverside, United States
Rajesh K. Sani, South Dakota School of Mines and Technology, United States
Copyright © 2021 Suzuki, Taketani, Tanabiki, Ohara, Kobayashi and Ohshiro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirokazu Suzuki, aGlyb2thcEB4cG9zdC5wbGFsYS5vci5qcA==; aGlyb2thenVzdXp1a2lAdG90dG9yaS11LmFjLmpw
†Present address: Jyumpei Kobayashi, Research Institute of Innovative Technology for the Earth, Kyoto, Japan
 Hirokazu Suzuki
Hirokazu Suzuki Tatsunari Taketani3
Tatsunari Taketani3 Misaki Ohara
Misaki Ohara Jyumpei Kobayashi
Jyumpei Kobayashi