- Guangdong Provincial Engineering Laboratory of Biomass High Value Utilization, Institute of Bioengineering, Guangdong Academy of Sciences, Guangzhou, China
Keratinases belong to a class of proteases that are able to degrade keratins into amino acids. Microbial keratinases play important roles in turning keratin-containing wastes into value-added products by participating in the degradation of keratin. Keratin is found in human and animal hard tissues, and its complicated structures make it resistant to degradation by common proteases. Although breaking disulfide bonds are involved in keratin degradation, keratinase is responsible for the cleavage of peptides, making it attractive in pharmaceutical and feather industries. Keratinase can serve as an important tool to convert keratin-rich wastes such as feathers from poultry industry into diverse products applicable to many fields. Despite of some progress made in isolating keratinase-producing microorganisms, structural studies of keratinases, and biochemical characterization of these enzymes, effort is still required to expand the biotechnological application of keratinase in diverse fields by identifying more keratinases, understanding the mechanism of action and constructing more active enzymes through molecular biology and protein engineering. Herein, this review covers structures, applications, biochemistry of microbial keratinases, and strategies to improve its efficiency in keratin degradation.
Introduction
Keratin is an important structural protein in some hard tissues in which it plays a protective role by forming a barrier between the organ and its environment. Keratin is a fibrous protein that is insoluble in water and other solvents. Due to the structure of keratin stabilized by disulfide bonds and hydrogen bonds, keratin is resistant to degradation by common proteases such as trypsin and pepsin. Keratin is one of the ubiquitous proteins in nature and found in many organs such as feather of birds, hair, wools, and nails of mammals (McKittrick et al., 2012; Chilakamarry et al., 2021). Keratin is among the most abundant renewable organic polymers in nature after cellulose, lignin, hemicellulose, pectin, and chitin (Lange et al., 2016; Bealer et al., 2020). Keratin-containing wastes such as feathers from poultry industry represent an attractive resource for carbon, sulfur, and nitrogen that can be converted into other products (Bhari et al., 2021).
Keratinous wastes are rich in amino acids (Qiu et al., 2020) and could affect the atmosphere, water sources, and soil if they are not treated properly (Hassan et al., 2020). On the other hand, this type of wastes serves as a low-cost resource for amino acids or can be converted into animal feeds and fertilizers (Pettett and Ipek, 2004; Gurav and Jadhav, 2013). Compared with other natural polymers such as cellulose, starch, and collagen, extraction of keratin is a challenging process. Quite a few strategies such as physical, chemical, and biological methods are applied in keratin extraction. Although chemical and physical treatments are efficient strategies to treat keratinous wastes, a large amount of energy is needed and amino acids were destroyed during treatment. As keratin does not accumulate in nature, microorganisms are playing the major role in its degradation and recycling. Therefore, keratinous wastes threatening the environment can be converted into value-added products by using microbial treatment (de Menezes et al., 2021; Nnolim and Nwodo, 2021). Extensive studies have been carried out to search suitable microorganisms and obtain optimized processes to make full use of keratinous wastes (Gradišar et al., 2000; Sangali and Brandelli, 2000; Kim et al., 2001; Rai and Mukherjee, 2011). It has been shown that wastes such as feathers can be degraded by bacteria and fungi to produce other important products such as amino acids or proteins with added values (Callegaro et al., 2018; Shanmugasundaram et al., 2018; Bohacz, 2019; Tamreihao et al., 2019; Chaudhary et al., 2021). Therefore, conversion of the wastes using microorganisms is the most environmentally friendly method while more studies are still needed to improve the degradation efficiency of keratins. As the amount of keratin-containing wastes is increasing rapidly due to various reasons, keratin derived from the wastes should be fully utilized by serving as a source of proteins, amino acids, and a low-cost resource for producing other products.
The structure of keratin explains their relatively stable existence and resistance to chemicals. Keratin can be classified as α-keratin and β-keratin according to the composition of amino acids and the secondary structure of polypeptide chains (Fraser and Parry, 2008, 2011; Lange et al., 2016). It is shown that α-keratin is mainly present in mammals and β-keratin is in avian and reptilian tissues. The polypeptide chains are packed into the final structure through disulfide bonds formed by cysteine residues, hydrogen bonds, and hydrophobic interactions (Vidmar and Vodovnik, 2018). Cysteine residues play a key role in the structural stability of keratin by forming intra- or intermolecular disulfide bonds (Barone et al., 2005). Keratin is also classified as hard keratin and soft keratin based on the content of cysteine (Jin et al., 2017). As basic units of keratin are polypeptides, keratinases play a major role in keratin degradation by breaking the disulfide bonds and peptidic bonds (Fraser and Parry, 2008; Hassan et al., 2020; de Menezes et al., 2021). One of the features of keratinases is that they are able to cleave a sequence with hydrophobic residues at the P1 position (Brandelli et al., 2010). Most keratinases were reported to degrade keratins in the presence of disulfide reducers or reducing agents while such reducing environment should not be required for a true keratinase (Qiu et al., 2020). Currently, the identified keratinases produced by microorganisms can be classified into at least 14 protease families (Qiu et al., 2020). In addition to journal publications, quite a few patent literatures reported preparation, extraction, and recombinant production of keratinases. The related patents can be obtained from different resources (Yahaya et al., 2021). Over 20,000 records can be obtained in google patent when keratinase was used as a searching keyword1. In this review, the structure and function of keratinases from bacteria and fungi are discussed. With accumulated knowledge in understanding microbial degradation of keratin, keratin-rich wastes are considered as a valuable and low-cost resource that can be converted into diverse products such as feed and fertilizers.
Mechanism of Action for Keratinase
Keratinases can be understood as a class of proteases that are able to degrade keratin by cleaving the peptide bonds (Gupta and Ramnani, 2006; Sahni et al., 2015). Although the identified keratinases are serine and metalloproteases that are able to break the peptide bond in peptide chains, they recognize hydrophobic substrates and affect the disulfide bonds. Most keratinases require other enzymes to break the disulfide, and two steps, namely, keratin peptide releasing and peptide degradation are included in keratin degradation. Reduction reaction can be catalyzed by disulfide reductases or reducing agents (Sangali and Brandelli, 2000; Rahayu et al., 2012; Lange et al., 2016). Microbial keratinases are usually secreted into the medium when the microorganism was cultivated in a keratin-containing medium (Friedrich et al., 1999; Monod, 2008; Jayalakshmi et al., 2010). This is not surprising as keratins are not soluble and not able to be transported into cells. Studies have shown that diverse keratinases with different molecular weights, optimal pH values, and optimal temperatures are produced by microorganisms. Most microbial keratinases are secreted into the extracellular matrix in the presence of keratin or keratin-containing substrates (Vidmar and Vodovnik, 2018; Nnolim and Nwodo, 2021). Some microorganisms are able to produce extracellular and intracellular keratinases simultaneously. Cell-bound keratinases are also identified, and this type of enzymes might be of great interest for industrial application as well, which is due to the fact that they are immobilized on the cell surface and can be easily used in waste treatment. With the development of molecular biology and accumulation of genome sequences of microorganisms, microbial keratinases and their mechanism of keratin degradation can be predicted through bioinformatics (Li et al., 2018; Song et al., 2018; Pinski et al., 2020; Zolfaghari Emameh et al., 2021). A recent report showing a careful and detail classification of current keratinases provides a clear view to understand their mechanism of action and explains the requirement of multiple enzymes to achieve complete degradation of keratin or keratinous wastes (Qiu et al., 2020).
Keratinase-Producing Microorganisms
Keratinase-producing microorganisms are widely distributed in nature, and they can be readily isolated from the environment (Vidmar and Vodovnik, 2018). Bacteria, fungi, and actinobacteria are able to produce keratinases and use keratin as the carbon and nitrogen sources in the minimal medium. To isolate a keratinase-producing microorganism, keratins or keratin-containing wastes such as feathers are usually present in the cultural medium, implying that microbial keratinase production is an inducible process (Brandelli et al., 2010). To isolate keratinase-producing microorganisms, the following steps including sample collection, assay development, strain identification, and characterization are usually applied (Figure 1). First, samples need to be collected from the environment. The samples can be soils, keratin-containing wastes, or water that was contaminated with wastes (Jeevana Lakshmi et al., 2013; Gegeckas et al., 2014). Feathers are one of the most commonly used substrates in screening, and degradation of feathers can be readily monitored by observing the changes in shapes and releasing of proteins into the solution. It has been noted that several factors such as location, water content, keratin composition, and weather of the local environment need to be considered to make sure that a collection of microorganisms can be obtained. Second, a medium for the microorganism growth needs to be set up. As the media for enriching bacteria or fungi are different, a suitable medium is important in the screening (De Azeredo et al., 2006; Fakhfakh et al., 2011; Mazotto et al., 2013; Parrado et al., 2014; Arokiyaraj et al., 2019). In addition, other parameters such as cultural time and temperature should be considered based on the experimental objectives. For example, if a team plans to isolate a thermally stable keratinase, a higher temperature in screening might enhance the rate of success (Wu et al., 2017). In the case of isolating microbial consortia, the enrichment step might not be important as it might reduce the content of certain microorganisms. Third, an assay for measuring protease activity should be set up for ranking keratin degradation efficiencies caused by different keratinases (Iglesias et al., 2017). It will be useful to monitor both changes in the shape of keratin substrate and the composition of the proteins secreted by the microorganisms. Last, a reliable method for microorganism identification is needed (Herzog et al., 2016). In addition to identifying the amino acid sequence of the enzyme through analyzing the MS data carefully, knowing the genome of the screened microorganisms will provide more information to understand mechanism of action for the identified keratinase and a strategy to express certain types of enzymes (Sittipol et al., 2021).
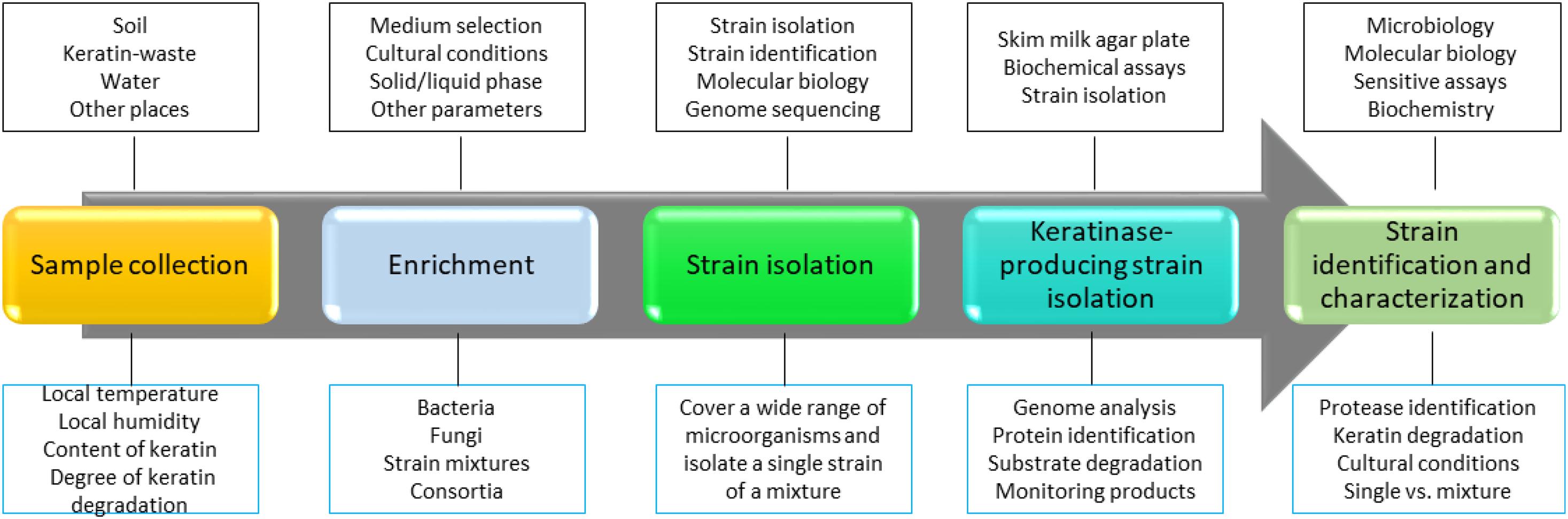
Figure 1. A strategy to screen keratinase-producing microorganisms. In this strategy, a method to identify keratinase-producing strain is critical. Molecular biology, bioinformatics, biochemistry, and sensitive analytical methods such as MS are critical in the screening. In addition, as keratin degradation is a complicated step, sample isolation and enriching steps are important to make sure that the desired strains are sustained. Many keratinase-producing strains have been isolated and identified from the environment (Nnolim and Nwodo, 2021).
Bacillus stains are the predominate bacteria that are able to produce keratinases. The species include Bacillus subtilis, Bacillus pumilus, Bacillus lichenifomis, and Bacillus cereus (Zaraî Jaouadi et al., 2015; Bhari et al., 2018; Gegeckas et al., 2018). Some other bacteria are able to degrade feathers with a high efficiency (Kim et al., 2005; Zaraî Jaouadi et al., 2015; Abdel-Naby et al., 2017; Arokiyaraj et al., 2019; Hamiche et al., 2019). Keratin-degrading fungi can be isolated from some tissues of human and animals as the produced keratinase might be important for the fungal infection (Friedrich et al., 1999; Bohacz and Korniłłowicz-Kowalska, 2019; Zhang et al., 2019). It was shown that fungi such as Chrysosporium (Bohacz, 2016; Gurung et al., 2018) and Trichophyton (Zhang et al., 2019) were able to degrade keratins through their produced keratinases (Shadzi et al., 2002; Hassan et al., 2020). Due to the function of keratins, keratinases in some pathogenic fungi might be essential for the invasion by breaking the barrier between the tissue and the environment. Streptomyces is the predominant actinobacterium that is able to produce keratinases (Li et al., 2020). Quite a few reports have shown that keratinolytic actinobacteria can be isolated from different environment. Some actinobacteria are able to produce thermally stable keratinases which have great potential to be widely used in industry (Nnolim and Nwodo, 2021). The detail introduction of keratinase-producing microorganisms have been described in several reviews (Papadopoulos, 1989; Onifade et al., 1998; Korniłłowicz-Kowalska and Bohacz, 2011; McKittrick et al., 2012; Sahni et al., 2015; Kanoksilapatham and Intagun, 2017; Pahua-Ramos et al., 2017; Verma et al., 2017; Kowalczyk et al., 2018; Li et al., 2018; Vidmar and Vodovnik, 2018; Li, 2019; Hassan et al., 2020; Kumar, 2020; Qiu et al., 2020; Yahaya et al., 2021).
Although many keratinase producers have been isolated and identified (Cavello et al., 2020; Jagadeesan et al., 2020; Moridshahi et al., 2020; Nnolim et al., 2020b; Reis et al., 2020), the isolation and characterization of keratinase-producing microorganisms are still an important task. The keratin degradation efficiency can be improved when more keratinases are applied (Peng et al., 2019). Therefore, a mixture of microorganism-microbial consortia might have great potential in converting keratin-rich waste into valuable products (Kang et al., 2020; Nasipuri et al., 2020). It is challenging to have a microbial consortium because the amount of the organism in the system will be affected under different conditions. It is also possible to set up a microbial consortium to improve keratin degradation by mixing several microorganisms which have been well characterized. This is a feasible method in industrial applications.
Keratin Degradation by Keratinases
Several mechanisms have been proposed based on the accumulated studies (Korniłłowicz-Kowalska and Bohacz, 2011; Li et al., 2020). The challenge in keratin degradation is due to the presence of high content of disulfide bonds (Korniłłowicz-Kowalska and Bohacz, 2011). As most keratinases are proteases responsible for breaking peptide bonds, other enzymes or chemicals are needed to affect the disulfide bonds and reduce the forces for keratin packing to make proteins accessible to the proteases (Kasperova et al., 2013). Therefore, at least two steps including disulfide bond breakage and proteolysis are involved in keratin degradation (Figure 2). It has been noted that fungi and bacteria use similar ways to degrade keratins with a slight difference as mechanical destruction is also playing a role in degradation by fungi (Korniłłowicz-Kowalska and Bohacz, 2011). It was shown that mechanical destruction, production of inorganic sulfite, and involvement of disulfide reductase can contribute to the step of disulfide bond breakage (Korniłłowicz-Kowalska and Bohacz, 2011). In the protein degradation step, keratinases are able to break polypeptides into amino acids. There are several protease families responsible for keratin degradation (Qiu et al., 2020). To convert keratin into amino acids completely, multiple keratinases are required as different enzymes prefer to different cleavage sites. The diversity and classification of keratinase based on the amino acid sequence and conserved domains in MEROPS have been described in detail in a recent review (Qiu et al., 2020). Some disulfide reductases involved in keratin degradation were shown (Kasperova et al., 2013; Navone and Speight, 2018; Table 1). The roles of these enzymes in keratin degradation have been described in other literatures, which will not be described here (Kasperova et al., 2013; Lange et al., 2016; Mercer and Stewart, 2018; Flückger et al., 2020).
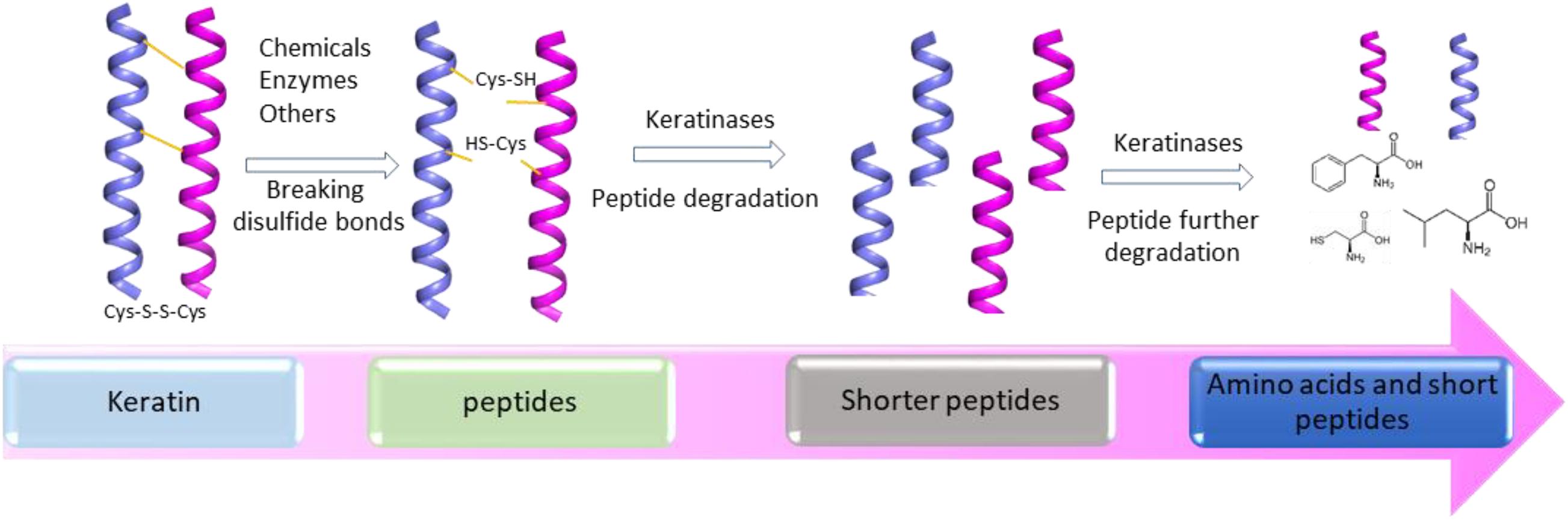
Figure 2. A simplified diagram showing the degradation of keratin by proteases. Two-step disulfide bond breakage and polypeptide degradation are usually included in keratin degradation. Keratins are simplified as helices. The degradation includes the steps such as releasing of keratin and degradation of keratin by multiple enzymes, which has been described in several reviews (Korniłłowicz-Kowalska and Bohacz, 2011; Sharma and Devi, 2018; Vidmar and Vodovnik, 2018; Qiu et al., 2020).
Accumulated studies have shown that the crude microbial culture exhibited higher keratin degradation efficiency than the purified enzymes. A keratinase-degrading system can be developed by carefully analyzing the components or enzymes that are critical for keratin degradation. The crude culture of a microorganism is a mixture of enzymes which can be used in keratin treatment. Therefore, a mixture of enzymes can be readily obtained by exploring the effects of cultural conditions on keratin degradation. Two important elements are important in this strategy. One is to have a good strain to work with and the other is to have an optimized fermentation condition to produce an enzymatic system for keratin degradation.
Structure of Keratinase
Keratinases are serine and metalloproteases, and their active sites are formed by several conserved residues. Crystal structures of several keratinases demonstrate the structural basis for their activity and provide insights into designing more stable and efficient enzymes for industrial applications (Betzel et al., 2001; Gupta et al., 2017). These structures can also be utilized as a template to obtain homology models of other keratinases (Kluskens et al., 2002).
The crystal structure of a serine protease from family S8 produced by Tritirachium album was obtained (Figure 3; Betzel et al., 2001). Crystal structures of other two members, namely, Fervidolysin from Fervidobacterium pennivorans (Kim et al., 2004) and rMtaKer from Meiothermus taiwanensis WR-220 (Wu et al., 2017) in S8 family were also determined (Figure 3). Proteases in this family contain both α and β structures. Despite their difference in overall folding, these proteases contain a conserved catalytic triad formed by Asp, His, and Ser residues which are critical for the cleavage of peptide bonds (Figure 3). In addition to determining the folding of the active site in this type of proteases, the structure of rMtaKer provides the molecular basis for substrate and protease interactions (Wu et al., 2017). The molecular interaction between P1–P4 residues and the protease was well resolved, which paved a way to design a suitable substrate for enzymatic assays (Wu et al., 2017; Figure 4). In addition, the structures of these proteases also provide insights into the structural basis for the thermal stability of the enzyme, which is useful for improving the stability of keratinases through site-directed mutagenesis.
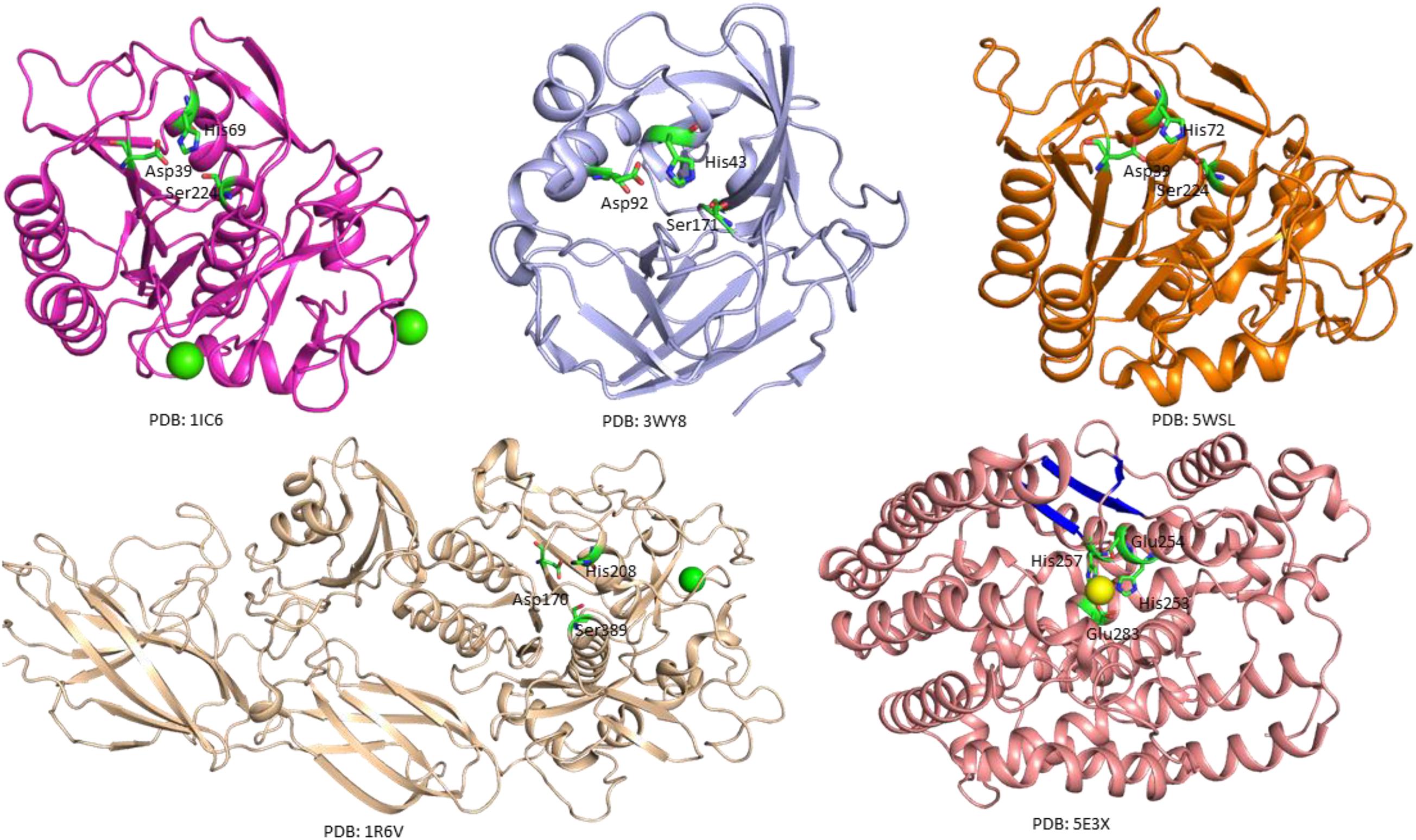
Figure 3. Crystal structures of microbial keratinases. The crystal structures of five keratinases were shown to understand their mechanism of action. The PDB access codes of the structures are indicated. The amino acids in active sites are highlighted in sticks and labeled with sequence numbers. Ca2+ and Co2+ atoms in the structures are shown as yellow and green spheres, respectively. In the crystal structure of fervidolyis (PDB:1R6V) (Kim et al., 2004), His208 was mutated into Ala. Ala 208 was labeled as His208 in this figure to show the active site. The β-sheet structures in FisCP (PDB: 5E3X) are highlighted in blue (Lee et al., 2015). All the figures were made using PyMOL (www.pymol.org). The details of the structures can be found in the reports (Betzel et al., 2001; Kim et al., 2004; Teruo et al., 2015; Wu et al., 2017).
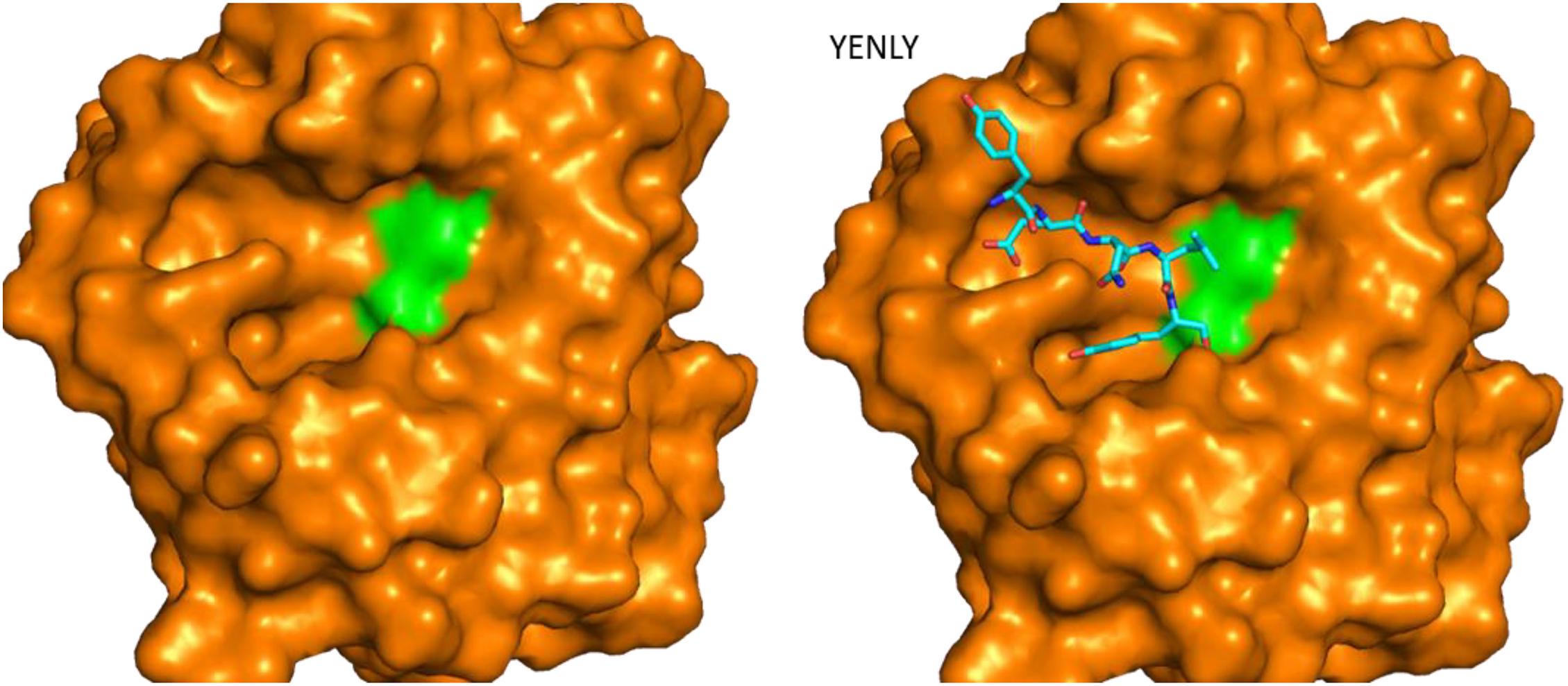
Figure 4. Structure of rMtaKer and its insights into protease and substrate interactions. Surface presentation of one keratinase in the absence and presence of a peptide sequence binding to the active site. The crystal structure of the protease (PDB ID 5WSL) is shown (Wu et al., 2017). The orientation of the figure is similar to those in Figure 3. The residues forming the catalytic triad are shown in green, and the peptide from the adjacent molecule in the crystal structure is shown in sticks. The peptide sequence is shown as sticks in the figure.
A crystal structure of keratin-degrading enzyme from Paenarthrobacter nicotinovorans was determined (Figure 3; Teruo et al., 2015). This protease consists of mainly β-sheets to form two β-barrels. The active site with the catalytic triad formed by His43, Asp92, and Ser171 is located between these two barrels (Teruo et al., 2015). The catalytic mechanism of these serine proteases has been well described. The following steps are critical for substrate degradation. The substrate needs to bind to the active site, followed by the peptide cleavage and release of the shorter segments. Proteases from M32 family have different secondary structures. A crystal structure of FisCP from Fervidobacterium islandicum AW-1 showed that the protease contained main helical structures with a short β-sheet close to the active site. The active site contains several amino acids (His253, Glue254, His257, and Glue283) and a Co2+ atom which is important for coordination of the substrate (Figure 3).
Biochemistry of Keratinases
Based on the identified enzymes and the sequences deposited in databases, the molecular weights of keratinase range from 20 to 130 kDa (Kanoksilapatham and Intagun, 2017; Sharma and Devi, 2018; Pinski et al., 2020; Qiu et al., 2020). The optimal conditions for enzymatic activity are diverse as different enzymes prefer different pH values, temperatures, and substrates (Tatineni et al., 2007; Tiwary and Gupta, 2010b; Rai and Mukherjee, 2011; Khodayari and Kafilzadeh, 2018; Arokiyaraj et al., 2019; Nnolim et al., 2020a). The classification of keratinases based on the amino acid sequence provides a unique and clear view toward the function and mechanism of keratinases (Qiu et al., 2020), which suggests that obtaining the amino acid sequence of the newly identified keratinases is an important task. Although the effect of keratinase producers on breaking disulfide bonds should be measured (Jaouadi et al., 2010), most studies explored the capabilities of breaking peptide bonds by keratinases (Benkiar et al., 2013; Jaouadi et al., 2013; Ben Elhoul et al., 2016; Omrane Benmrad et al., 2019). To validate the capability to degrade keratin, several substrates were adopted in the enzymatic assay (Böckle et al., 1995; Brandelli et al., 2010; Table 2).
Two types of the substrate were frequently used in the assays (Table 2). One type is the natural keratin such as feathers, wool, and pig bristles or the substrate derived from keratin-rich materials (Laba and Rodziewicz, 2014; Jin et al., 2017). As the solubility of the natural keratin is low, it is challenging to compare the enzymatic activities of keratinases obtained from different studies. Azo-keratin is frequently used as a substrate of keratinase and can be prepared using keratin derived from different sources (Herzog et al., 2016; Gonzalo et al., 2020). Azo-keratin is not a commercial product and is prepared by coupling keratins with a diazotized aryl amide to result in a deep red-orange compound (Riffel et al., 2003). The activity of keratinase is obtained by measuring changes of absorbance at 450 nm. Azo-keratin derived from different keratin-containing materials will be different and might not be an ideal substrate to compare enzymatic activities obtained from different laboratories (Riffel et al., 2003; Qiu et al., 2020). Keratin azure is another frequently used substrate in keratinase assay (Lin et al., 1992; Korkmaz et al., 2004; Tork et al., 2016). The enzymatic activity was defined according to the changes of the absorbance at 450 nm after mixing substrate with enzyme for a certain time (Jaouadi et al., 2010; Tork et al., 2016). Keratin azure is commercially available and very suitable for characterization of keratinases with a preference to degrade α-keratins. Keratin-containing materials such as feathers are also frequently utilized in biochemical assays in which feather degradation can be detected through observing the release of amino acids, the shape changes of feathers, and reduction of feather mass (Kim et al., 2005; Anbu et al., 2008; Cãlin et al., 2017; Wu et al., 2017; Bohacz and Korniłłowicz-Kowalska, 2019; Peng et al., 2019). The advantage of using natural keratin is that the keratin-degrading capability of keratinases can be evaluated directly (Gurung et al., 2018; Thankaswamy et al., 2018). The disadvantage is that the substrate degradation requires collaborative effort of different types of enzymes. The native substrate might not reflect the biochemical parameters of a single enzyme. A recent study applied recombinant feather protein in the protease assay. The recombinant protein was soluble in solution, giving rise to accurate analysis of the keratinase (Jin et al., 2017). Therefore, purified enzymes and modified substrates are critical for characterizing the enzymes.
The other type of substrates soluble in solution was applied in the enzymatic assay (Brandelli et al., 2010). Azocasein was utilized in several studies to determine whether a protein harbors protease activity (Tork et al., 2013, 2016). It is a non-specific protease substrate and its hydrolysis results in releases of the azo dye which can be detected by monitoring the absorbance at 366 nm (Tork et al., 2013, 2016). Synthetic peptides were also useful in the protease assays. As the sequence of the peptidic substrates can be modified easily, this type of assay can be utilized to explore the most suitable substrate for a keratinase (Böckle et al., 1995). The cleavage of peptides are able to be monitored using reversed-phase chromatography while this method is not a robust assay (Mäkinen et al., 1994). Peptidic substrates containing p-nitroanilide (pNA) were used to study the preference for P1 and P2 residues (Macedo et al., 2008; Benkiar et al., 2013; Zaraî Jaouadi et al., 2015; Bouacem et al., 2016). As the release of pNA can be detected by measuring the absorbance at 405 nm, pNA derivatives are very suitable to explore the effect of environmental conditions and other chemicals on the activity of keratinases (Rozs et al., 2001). Several keratinases produced by Bacillus sp. have been characterized using this type of substrates (Macedo et al., 2008). Fluorogenic substrates containing amino-4-methylcoumarin (AMC) have been widely utilized in the enzymatic assays of serine proteases (Bakhtiar et al., 2005; Silveira et al., 2009). The substrate cleavage can be confirmed with fluorescence spectroscopy. This method is very sensitive and provides an accurate way to characterize proteases. Effect of ions on keratinase activity was explored using AMC-derived substrates (Bakhtiar et al., 2005). Based on accumulated studies, it is a good strategy to adopt multiple protease assays to characterize keratinases obtained from different microorganisms, which will provide an insightful view to these enzymes. The biochemical features of keratinases can be obtained from several reviews (Onifade et al., 1998; Brandelli et al., 2010; Sahni et al., 2015; Kanoksilapatham and Intagun, 2017; Pahua-Ramos et al., 2017; Sharma and Devi, 2018; Zhang et al., 2019; Hassan et al., 2020; Qiu et al., 2020). The crystal structure of rMtaKer provides useful information for the molecular interaction between the enzyme and a peptide sequence from the C-terminal of this enzyme (Figure 4; Wu et al., 2017). This structure will be useful for designing a suitable substrate for enzymatic assays.
In summary, a sensitive biochemical assay needs to be set up as different keratinases may prefer different substrates. In addition, effects of different additives such as reducing reagents on keratin degradation could provide useful insights into understanding keratin degradation and expanding the application of these enzymes.
Microbial Production of Keratinases
Application of keratinases in industry requires a large amount of enzymes. Therefore, fermentation is essential to produce these enzymes in a large scale to meet the demands from industry (Zaghloul et al., 2011). Fermentation parameters such as carbon source, nitrogen source, temperature, and others need to be optimized as these parameters have an impact on the production of the keratinase. Wastes from industries such as feathers can be added into the cultural medium (Deniz et al., 2021). In addition, other wastes such as wheat and soy beans can be used as a substrate for producing keratinases (Syed et al., 2009; Prakash et al., 2010a). Interestingly, a study showed that glucose and ammonium nitrate are not good sources for growing Stenotrophomonas maltophilia to degrade feathers (Qu et al., 2018). Accumulated studies suggest that different microorganisms require various fermentation conditions for large-scale production, indicating that careful exploratory studies are necessary prior to the large-scale production of the enzymes (Jagadeesan et al., 2020; de Menezes et al., 2021; Deniz et al., 2021; Sharma and Kango, 2021). It was demonstrated that the solid-state fermentation increased keratinase production compared with the commonly used submerged fermentation (Inácio et al., 2018). In addition to feathers and human hair used in solid-state fermentation, other low-cost resources from agriculture can be considered in fermentation (Awad et al., 2011).
Recombinant techniques are applied to the production of keratinases (Descamps et al., 2003; Liu et al., 2013b; Fang et al., 2014; Yong et al., 2020; Yahaya et al., 2021). This method is particularly meaningful for keratinases that are produced by pathogenic microorganisms (Muhammed et al., 2021) and the mutants with an improved enzymatic activity and stability (Zhang et al., 2020). The recombinant production of keratinases does not require the application of keratin as the carbon and nitrogen sources. It is possible to purify the recombinant enzymes in a fast way when an affinity purification tag is introduced. Several studies demonstrate that it is feasible to produce recombinant keratinases. Keratinases from bacteria can be produced in Escherichia coli (Tiwary and Gupta, 2010a). It has been shown that the gene kerA encoding a keratinase from Bacillus licheniformis was expressed in Escherichia coli and Bacillus subtilis while the yield was lower than that of the wild type. An improved yield was observed by integration of multiple copies of kerA into the chromosome (Wang et al., 2004). Therefore, producing keratinases using recombinant techniques is of great interest while extensive studies are still needed to obtain the recombinant keratinase with an improved activity.
The purification of keratinases is important for enzymatic characterization and other application (Brandelli et al., 2015). In waste treatment, the enzyme purification is not needed for reducing cost and improving efficiency. To obtain a keratinase with a high purity, several strategies can be utilized. Ammonium sulfate precipitation, gel filtration chromatography, and ion-exchange chromatography are commonly used methods in the purification (Brandelli et al., 2015). For recombinant proteins, the affinity chromatography can be used in purification based on the affinity tag that is incorporated into the target protein. Keratinases from different bacteria have been purified for biochemical characterization. Techniques such as the aqueous two-phase system are applicable to obtain a large amount of enzymes (Bach et al., 2012; Sala et al., 2014). A carefully experimental design is vital when a large quantity of pure enzymes is needed as the purification could be an expensive step.
It has been noted that recombinant techniques are still needed for producing keratinases with a high purity, keratinases with mutations, and keratinases originated from a pathogenic microorganism (Liu et al., 2014). Recombinant protein expression systems and host and gene cloning strategies need to be explored (Gong et al., 2020). As the recombinant protein is critical for exploring the function of keratinases, it is useful in studying the function and activity of the enzymes.
Application of Keratinases
Keratin exists widely in nature and is a valuable source of carbon, nitrogen, and sulfur which can be converted into diverse products (McKittrick et al., 2012; Wang et al., 2016). Keratinases have extensive industrial and biotechnological applications due to their ability to degrade keratins (Onifade et al., 1998; Gupta and Ramnani, 2006; Rai et al., 2009; Syed et al., 2009; Brandelli et al., 2010; Sahni et al., 2015; Kanoksilapatham and Intagun, 2017; Gegeckas et al., 2018; Vidmar and Vodovnik, 2018). Their applications are summarized (Figure 5) and have been described in several literatures (Gupta and Ramnani, 2006; Brandelli et al., 2010; Vidmar and Vodovnik, 2018; Hassan et al., 2020; Nnolim and Nwodo, 2021).
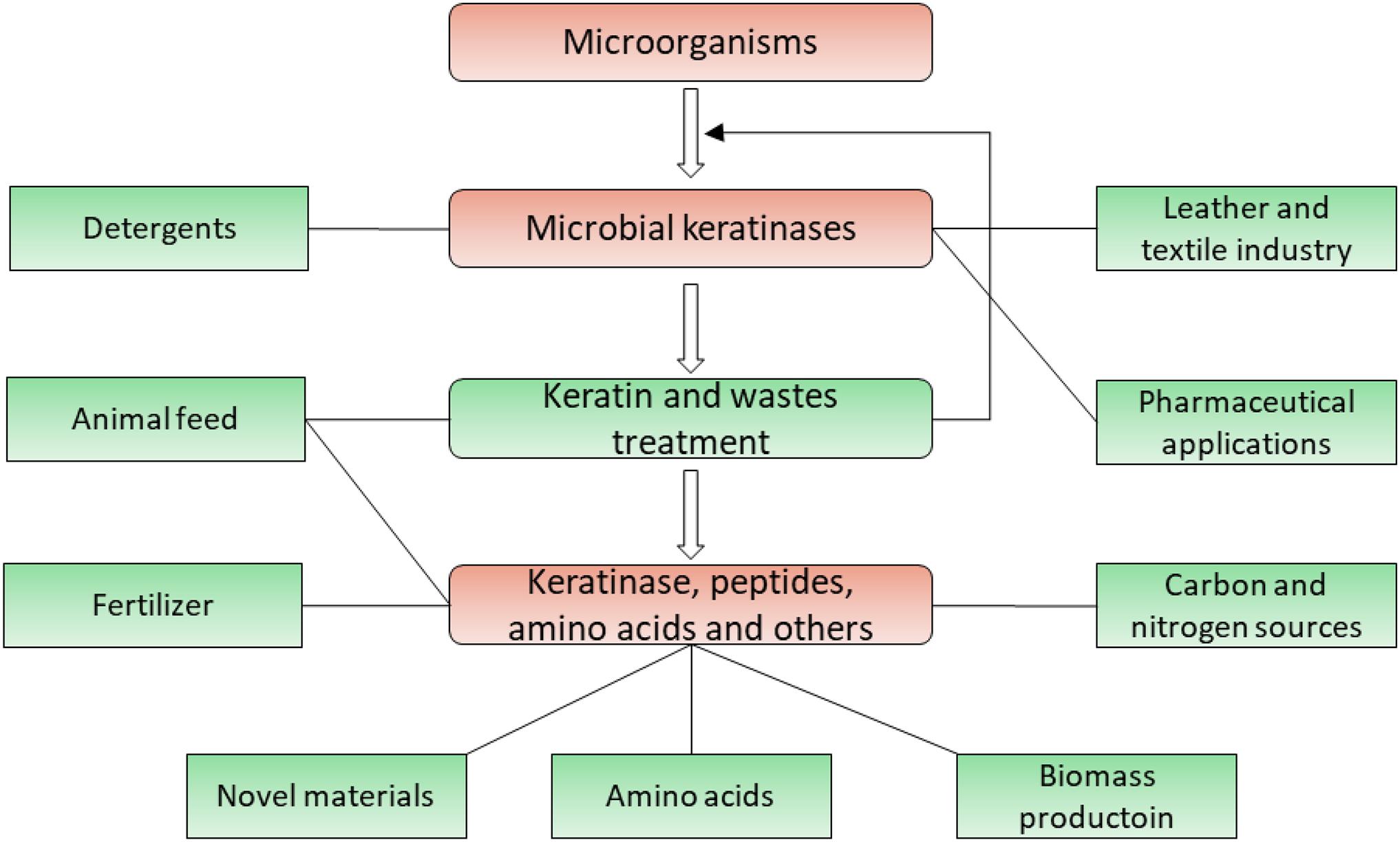
Figure 5. Application of microbial keratinases in different fields. Applications of keratinases and their products are highlighted in green, which has been introduced in multiple reports (Onifade et al., 1998; Gupta and Ramnani, 2006; Brandelli et al., 2010; Chaturvedi et al., 2014; Sahni et al., 2015; Sharma and Gupta, 2016; Vidmar and Vodovnik, 2018).
Improvement of Keratinases
To improve the activity and thermal stability of keratinases, mutagenesis was applied (Fang et al., 2010; Wang et al., 2015; de Paiva et al., 2018; Su et al., 2019). The mutagenesis studies were carried out by treating whole cells with reagents and modifying genes using molecular biology. In a study, ethyl methanesulfonate (EMS) was used to treat the whole cells of Bacillus subtilis LFB-FIOCRUZ 1266, and the resulting mutant strains exhibited higher feather hydrolysis compared with the wild-type strain (de Paiva et al., 2018). Treatment of feather-degrading Deinococcus ficus with UV resulted in mutants with an improved and decreased keratinolytic activity (Zeng et al., 2011). Other chemical reagents such as ethidium bromide (EtBr) and N-methy-N’-nitro-N-nitroso-guanidine (MNG) were also able to cause mutations in the genes, which affected the keratinase activity (Cai et al., 2008; Vidmar and Vodovnik, 2018).
Protein engineering was also applied to cause an augmentation of the keratinase activity (Fang et al., 2019). When the amino acid sequence and structure of a keratinase are available, the rational protein design can play a role in improving the activity and thermal stability of a keratinase. When amino acids essential for the protease activity, metal binding, and thermal stability are identified, computer-based methods will enable researchers to design proteins with improved enzymatic activities and thermal stabilities. This strategy has been successfully applied to the keratinase of Bacillus licheniformis BBE11 (Liu et al., 2013a). Four amino acid substitutions (N122Y, N217S, A193P, N160C) were designed, and the corresponding genes were expressed in Bacillus subtilis WB60. A mutant keratinase with the N122Y substitution exhibited an approximately 5.6-fold increase in catalytic efficiency, suggesting that this is an efficient strategy in improving activity and stability (Liu et al., 2013a). Other methods such as PCR-based methods and direct evolution will play a role in obtaining more potent keratinases (Vidmar and Vodovnik, 2018). When the regulatory mechanism of a keratinase is understood, the modification on other regions of the keratinase can also improve its activity and stability (Fang et al., 2016b; Peng et al., 2021). In a study, the N- and C-terminal regions of KerSMD were replaced with those regions of a homogenous keratinase. Replacing the N-terminal region resulted in a mutant exhibiting more than a twofold catalytic activity toward casein catalytic efficiency. Replacing the C-terminal region improved keratinases activity using succinyl-Ala-Ala-Pro-Phe-p-nitroanilide as a substrate in a biochemical assay. Replacing both N- and C-terminal regions resulted in a mutant with an improved thermal stability (Fang et al., 2016b).
It is important to improve the enzymatic characteristics of the keratinase while caution has to be taken when the whole-cell-based mutagenesis is used. All the mutant strains should meet the safety requirement from certain authorities. Compared with random mutagenesis, structure-guided protein engineering is of great interest as the mutation is well managed. To carry out such studies efficiently, the amino acid sequence and structures need to be known. Recombinant protein production is therefore a strategy to play an important role in this process. Researchers have to make sure that the strains with modified genes are acceptable in industrial applications.
Conclusion
Keratin is a rich resource in nature, and the amount of keratin-rich wastes is increasing annually. Keratinases play important roles in keratin recycle and have diverse applications in different fields. Studies need to be carried out to obtain active enzymes and enlarge their applications. Microbiology, molecular biology, structural biology, computation and biochemistry will play important roles in the research field of keratinases.
Author Contributions
QL drafted and revised the manuscript.
Funding
This research was supported by funds from the “Hundred-Talent Program” (Grant Nos. 2020GDASYL-20200102010 and 2020GDASYL-20200102009), Guangdong Academy of Sciences, China.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
QL appreciates the support from Institute of Bioengineering, Guangdong Academy of Sciences, China.
Footnotes
References
Abdel-Naby, M. A., El-Refai, H. A., and Ibrahim, M. H. A. (2017). Structural characterization, catalytic, kinetic and thermodynamic properties of Keratinase from Bacillus pumilus FH9. Int. J. Biol. Macromol. 105, 973–980. doi: 10.1016/j.ijbiomac.2017.07.118
Anbu, P., Hilda, A., Sur, H.-W., Hur, B.-K., and Jayanthi, S. (2008). Extracellular keratinase from Trichophyton sp. HA-2 isolated from feather dumping soil. Int. Biodeterior. Biodegradation 62, 287–292. doi: 10.1016/j.ibiod.2007.07.017
Arokiyaraj, S., Varghese, R., Ali Ahmed, B., Duraipandiyan, V., and Al-Dhabi, N. A. (2019). Optimizing the fermentation conditions and enhanced production of keratinase from Bacillus cereus isolated from halophilic environment. Saudi J. Biol. Sci. 26, 378–381. doi: 10.1016/j.sjbs.2018.10.011
Awad, G. E. A., Esawy, M. A., Salam, W. A., Salama, B. M., Abdelkader, A. F., and El-diwany, A. (2011). Keratinase production by Bacillus pumilus GHD in solid-state fermentation using sugar cane bagasse: optimisation of culture conditions using a Box-Behnken experimental design. Ann. Microbiol. 61, 663–672. doi: 10.1007/s13213-010-0187-0
Bach, E., Sant’Anna, V., Daroit, D. J., Corrêa, A. P. F., Segalin, J., and Brandelli, A. (2012). Production, one-step purification, and characterization of a keratinolytic protease from Serratia marcescens P3. Process Biochem. 47, 2455–2462. doi: 10.1016/j.procbio.2012.10.007
Bakhtiar, S., Estiveira, R. J., and Hatti-Kaul, R. (2005). Substrate specificity of alkaline protease from alkaliphilic feather-degrading Nesterenkonia sp. AL20. Enzyme Microb. Technol. 37, 534–540. doi: 10.1016/j.enzmictec.2005.04.003
Barone, J. R., Schmidt, W. F., and Liebner, C. F. E. (2005). Thermally processed keratin films. J. Appl. Polymer Sci. 97, 1644–1651. doi: 10.1002/app.21901
Bealer, E. J., Onissema-Karimu, S., Rivera-Galletti, A., Francis, M., Wilkowski, J., Salas-de la Cruz, D., et al. (2020). Protein–Polysaccharide composite materials: fabrication and applications. Polymers 12:464.
Ben Elhoul, M., Zaraî Jaouadi, N., Rekik, H., Omrane Benmrad, M., Mechri, S., Moujehed, E., et al. (2016). Biochemical and molecular characterization of new keratinoytic protease from Actinomadura viridilutea DZ50. Int. J. Biol. Macromol. 92, 299–315. doi: 10.1016/j.ijbiomac.2016.07.009
Benkiar, A., Nadia, Z. J., Badis, A., Rebzani, F., Soraya, B. T., Rekik, H., et al. (2013). Biochemical and molecular characterization of a thermo- and detergent-stable alkaline serine keratinolytic protease from Bacillus circulans strain DZ100 for detergent formulations and feather-biodegradation process. Int. Biodeterior. Biodegradation 83, 129–138. doi: 10.1016/j.ibiod.2013.05.014
Betzel, C., Gourinath, S., Kumar, P., Kaur, P., Perbandt, M., Eschenburg, S., et al. (2001). Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 Å resolution. Biochemistry 40, 3080–3088. doi: 10.1021/bi002538n
Bhari, R., Kaur, M., and Sarup Singh, R. (2021). Chicken feather waste hydrolysate as a superior biofertilizer in agroindustry. Curr. Microbiol. 78, 2212–2230. doi: 10.1007/s00284-021-02491-z
Bhari, R., Kaur, M., Singh, R. S., Pandey, A., and Larroche, C. (2018). Bioconversion of chicken feathers by Bacillus aerius NSMk2: a potential approach in poultry waste management. Bioresour. Technol. Rep. 3, 224–230. doi: 10.1016/j.biteb.2018.07.015
Böckle, B., Galunsky, B., and Müller, R. (1995). Characterization of a keratinolytic serine proteinase from Streptomyces pactum DSM 40530. Appl. Environ. Microbiol. 61, 3705–3710.
Bockle, B., and Muller, R. (1997). Reduction of disulfide bonds by Streptomyces pactum during growth on chicken feathers. Appl. Environ. Microbiol. 63, 790–792.
Bohacz, J. (2016). Biodegradation of feather waste keratin by a keratinolytic soil fungus of the genus Chrysosporium and statistical optimization of feather mass loss. World J. Microbiol. Biotechnol. 33:13. doi: 10.1007/s11274-016-2177-2
Bohacz, J. (2019). Changes in mineral forms of nitrogen and sulfur and enzymatic activities during composting of lignocellulosic waste and chicken feathers. Environ. Sci. Pollut. Res. 26, 10333–10342. doi: 10.1007/s11356-019-04453-2
Bohacz, J., and Korniłłowicz-Kowalska, T. (2019). Fungal diversity and keratinolytic activity of fungi from lignocellulosic composts with chicken feathers. Process Biochem. 80, 119–128. doi: 10.1016/j.procbio.2019.02.012
Bouacem, K., Bouanane-Darenfed, A., Zaraî Jaouadi, N., Joseph, M., Hacene, H., Ollivier, B., et al. (2016). Novel serine keratinase from Caldicoprobacter algeriensis exhibiting outstanding hide dehairing abilities. Int. J. Biol. Macromol. 86, 321–328. doi: 10.1016/j.ijbiomac.2016.01.074
Brandelli, A., Daroit, D. J., and Riffel, A. (2010). Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 85, 1735–1750. doi: 10.1007/s00253-009-2398-5
Brandelli, A., Sala, L., and Kalil, S. J. (2015). Microbial enzymes for bioconversion of poultry waste into added-value products. Food Res. Int. 73, 3–12. doi: 10.1016/j.foodres.2015.01.015
Busk, P. K., and Lange, L. (2015). Classification of fungal and bacterial lytic polysaccharide monooxygenases. BMC Genomics 16:368. doi: 10.1186/s12864-015-1601-6
Cai, C.-G., Lou, B.-G., and Zheng, X.-D. (2008). Keratinase production and keratin degradation by a mutant strain of Bacillus subtilis. J. Zhejiang Univ. Sci. B 9, 60–67. doi: 10.1631/jzus.B061620
Cãlin, M., Constantinescu-Aruxandei, D., Alexandrescu, E., Rãut, I., Doni, M. B., Arsene, M.-L., et al. (2017). Degradation of keratin substrates by keratinolytic fungi. Electron. J. Biotechnol. 28, 101–112. doi: 10.1016/j.ejbt.2017.05.007
Callegaro, K., Welter, N., and Daroit, D. J. (2018). Feathers as bioresource: microbial conversion into bioactive protein hydrolysates. Process Biochem. 75, 1–9. doi: 10.1016/j.procbio.2018.09.002
Cavello, I., Urbieta, M. S., Cavalitto, S., and Donati, E. (2020). Bacillus cytotoxicus isolated from a pristine natural geothermal area reveals high keratinolytic activity. Microorganisms 8:796.
Chaturvedi, V., Bhange, K., Bhatt, R., and Verma, P. (2014). Production of kertinases using chicken feathers as substrate by a novel multifunctional strain of Pseudomonas stutzeri and its dehairing application. Biocatal. Agric. Biotechnol. 3, 167–174. doi: 10.1016/j.bcab.2013.08.005
Chaudhary, L., Siddiqui, M. H., Vimal, A., and Bhargava, P. (2021). Biological degradation of keratin by microbial keratinase for effective waste management and potent industrial applications. Curr. Protein Pept. Sci. 22:CPPS-EPUB-114223. doi: 10.2174/1389203722666210215151952
Chilakamarry, C. R., Mahmood, S., Saffe, S. N. B. M., Arifin, M. A. B., Gupta, A., Sikkandar, M. Y., et al. (2021). Extraction and application of keratin from natural resources: a review. 3 Biotech 11:220. doi: 10.1007/s13205-021-02734-7
De Azeredo, L. A. I., De Lima, M. B., Coelho, R. R. R., and Freire, D. M. G. (2006). Thermophilic protease production by Streptomyces sp. 594 in submerged and solid-state fermentations using feather meal. J. Appl. Microbiol. 100, 641–647. doi: 10.1111/j.1365-2672.2005.02791.x
de Menezes, C. L. A., Santos, R. D. C., Santos, M. V., Boscolo, M., da Silva, R., Gomes, E., et al. (2021). Industrial sustainability of microbial keratinases: production and potential applications. World J. Microbiol. Biotechnol. 37:86. doi: 10.1007/s11274-021-03052-z
de Paiva, D. P., de Oliveira, S. S. A., Mazotto, A. M., Vermelho, A. B., and de Oliveira, S. S. (2018). Keratinolytic activity of Bacillus subtilis LFB-FIOCRUZ 1266 enhanced by whole-cell mutagenesis. 3 Biotech 9:2. doi: 10.1007/s13205-018-1527-1
Demirkan, E., Kut, D., Sevgi, T., Dogan, M., and Baygin, E. (2020). Investigation of effects of protease enzyme produced by Bacillus subtilis 168 E6-5 and commercial enzyme on physical properties of woolen fabric. J. Textile Inst. 111, 26–35. doi: 10.1080/00405000.2019.1624069
Deniz, I., Demir, T., Oncel, S. S., Hames, E. E., and Vardar-Sukan, F. (2021). Effect of agitation and aeration on keratinase production in bioreactors using bioprocess engineering aspects. Protein J. doi: 10.1007/s10930-021-09978-5
Descamps, F., Brouta, F., Vermout, S., Monod, M., Losson, B., and Mignon, B. (2003). Recombinant expression and antigenic properties of a 31.5-kDa keratinolytic subtilisin-like serine protease from Microsporum canis. FEMS Immunol. Med. Microbiol. 38, 29–34. doi: 10.1016/s0928-8244(03)00101-9
Dozie, I. N. S., Okeke, C. N., and Unaeze, N. C. (1994). A thermostable, alkaline-active, keratinolytic proteinase from chrysosporium keratinophilum. World J. Microbiol. Biotechnol. 10, 563–567. doi: 10.1007/BF00367668
Fakhfakh, N., Ktari, N., Haddar, A., Mnif, I. H., Dahmen, I., and Nasri, M. (2011). Total solubilisation of the chicken feathers by fermentation with a keratinolytic bacterium, Bacillus pumilus A1, and the production of protein hydrolysate with high antioxidative activity. Process Biochem. 46, 1731–1737. doi: 10.1016/j.procbio.2011.05.023
Fang, N., Zhong, C.-Q., Liang, X., Tang, X.-F., and Tang, B. (2010). Improvement of extracellular production of a thermophilic subtilase expressed in Escherichia coli by random mutagenesis of its N-terminal propeptide. Appl. Microbiol. Biotechnol. 85, 1473–1481. doi: 10.1007/s00253-009-2183-5
Fang, Z., Sha, C., Peng, Z., Zhang, J., and Du, G. (2019). Protein engineering to enhance keratinolytic protease activity and excretion in Escherichia coli and its scale-up fermentation for high extracellular yield. Enzyme Microb. Technol. 121, 37–44. doi: 10.1016/j.enzmictec.2018.11.003
Fang, Z., Zhang, J., Du, G., and Chen, J. (2016a). Improved catalytic efficiency, thermophilicity, anti-salt and detergent tolerance of keratinase KerSMD by partially truncation of PPC domain. Sci. Rep. 6:27953. doi: 10.1038/srep27953
Fang, Z., Zhang, J., Liu, B., Du, G., and Chen, J. (2013). Biochemical characterization of three keratinolytic enzymes from Stenotrophomonas maltophilia BBE11-1 for biodegrading keratin wastes. Int. Biodeterior. Biodegradation 82, 166–172. doi: 10.1016/j.ibiod.2013.03.008
Fang, Z., Zhang, J., Liu, B., Du, G., and Chen, J. (2016b). Enhancement of the catalytic efficiency and thermostability of Stenotrophomonas sp. keratinase KerSMD by domain exchange with KerSMF. Microb. Biotechnol. 9, 35–46. doi: 10.1111/1751-7915.12300
Fang, Z., Zhang, J., Liu, B., Jiang, L., Du, G., and Chen, J. (2014). Cloning, heterologous expression and characterization of two keratinases from Stenotrophomonas maltophilia BBE11-1. Process Biochem. 49, 647–654. doi: 10.1016/j.procbio.2014.01.009
Flückger, S., Igareta, N. V., and Seebeck, F. P. (2020). Convergent evolution of fungal cysteine dioxygenases. ChemBioChem 21, 3082–3086. doi: 10.1002/cbic.202000317
Fraser, R. D. B., and Parry, D. A. D. (2008). Molecular packing in the feather keratin filament. J. Struct. Biol. 162, 1–13. doi: 10.1016/j.jsb.2008.01.011
Fraser, R. D. B., and Parry, D. A. D. (2011). The structural basis of the filament-matrix texture in the avian/reptilian group of hard β-keratins. J. Struct. Biol. 173, 391–405. doi: 10.1016/j.jsb.2010.09.020
Friedrich, J., Gradišar, H., Mandin, D., and Chaumont, J. P. (1999). Screening fungi for synthesis of keratinolytic enzymes. Lett. Appl. Microbiol. 28, 127–130. doi: 10.1046/j.1365-2672.1999.00485.x
Gegeckas, A., Gudiukaitë, R., and Citavicius, D. (2014). Keratinolytic proteinase from Bacillus thuringiensis AD-12. Int. J. Biol. Macromol. 69, 46–51. doi: 10.1016/j.ijbiomac.2014.05.024
Gegeckas, A., Šimkutë, A., Gudiukaitë, R., and Čitavičius, D. J. (2018). Characterization and application of keratinolytic paptidases from Bacillus spp. Int. J. Biol. Macromol. 113, 1206–1213. doi: 10.1016/j.ijbiomac.2018.03.046
Gong, J.-S., Ye, J.-P., Tao, L.-Y., Su, C., Qin, J., Zhang, Y.-Y., et al. (2020). Efficient keratinase expression via promoter engineering strategies for degradation of feather wastes. Enzyme Microb. Technol. 137:109550. doi: 10.1016/j.enzmictec.2020.109550
Gonzalo, M., Espersen, R., Al-Soud, W. A., Cristiano Falco, F., Hägglund, P., Sørensen, S. J., et al. (2020). Azo dying of α-keratin material improves microbial keratinase screening and standardization. Microb. Biotechnol. 13, 984–996. doi: 10.1111/1751-7915.13541
Gradišar, H., Kern, S., and Friedrich, J. (2000). Keratinase of Doratomyces microsporus. Appl. Microbiol. Biotechnol. 53, 196–200. doi: 10.1007/s002530050008
Grumbt, M., Monod, M., Yamada, T., Hertweck, C., Kunert, J., and Staib, P. (2013). Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J. Invest. Dermatol. 133, 1550–1555. doi: 10.1038/jid.2013.41
Gupta, R., and Ramnani, P. (2006). Microbial keratinases and their prospective applications: an overview. Appl. Microbiol. Biotechnol. 70:21. doi: 10.1007/s00253-005-0239-8
Gupta, S., Tewatia, P., Misri, J., and Singh, R. (2017). Molecular modeling of cloned Bacillus subtilis keratinase and its insinuation in psoriasis treatment using docking studies. Indian J. Microbiol. 57, 485–491. doi: 10.1007/s12088-017-0677-x
Gurav, R. G., and Jadhav, J. P. (2013). A novel source of biofertilizer from feather biomass for banana cultivation. Environ. Sci. Pollut. Res. 20, 4532–4539. doi: 10.1007/s11356-012-1405-z
Gurung, S. K., Adhikari, M., Kim, S. W., Bazie, S., Kim, H. S., Lee, H. G., et al. (2018). Discovery of two Chrysosporium species with keratinolytic activity from field soil in Korea. Mycobiology 46, 260–268. doi: 10.1080/12298093.2018.1514732
Hamiche, S., Mechri, S., Khelouia, L., Annane, R., El Hattab, M., Badis, A., et al. (2019). Purification and biochemical characterization of two keratinases from Bacillus amyloliquefaciens S13 isolated from marine brown alga Zonaria tournefortii with potential keratin-biodegradation and hide-unhairing activities. Int. J. Biol. Macromol. 122, 758–769. doi: 10.1016/j.ijbiomac.2018.10.174
Hassan, M. A., Abol-Fotouh, D., Omer, A. M., Tamer, T. M., and Abbas, E. (2020). Comprehensive insights into microbial keratinases and their implication in various biotechnological and industrial sectors: a review. Int. J. Biol. Macromol. 154, 567–583. doi: 10.1016/j.ijbiomac.2020.03.116
Herzog, B., Overy, D. P., Haltli, B., and Kerr, R. G. (2016). Discovery of keratinases using bacteria isolated from marine environments. Syst. Appl. Microbiol. 39, 49–57. doi: 10.1016/j.syapm.2015.10.004
Iglesias, M. S., Sequeiros, C., García, S., and Olivera, N. L. (2017). Newly isolated Bacillus sp. G51 from Patagonian wool produces an enzyme combination suitable for felt-resist treatments of organic wool. Bioprocess Biosyst. Eng. 40, 833–842. doi: 10.1007/s00449-017-1748-4
Inácio, F. D., Martins, A. F., Contato, A. G., Brugnari, T., Peralta, R. M., and Souza, C. G. M. D. (2018). Biodegradation of human keratin by protease from the basidiomycete Pleurotus pulmonarius. Int. Biodeterior. Biodegradation 127, 124–129. doi: 10.1016/j.ibiod.2017.11.010
Jagadeesan, Y., Meenakshisundaram, S., Saravanan, V., and Balaiah, A. (2020). Sustainable production, biochemical and molecular characterization of thermo-and-solvent stable alkaline serine keratinase from novel Bacillus pumilus AR57 for promising poultry solid waste management. Int. J. Biol. Macromol. 163, 135–146. doi: 10.1016/j.ijbiomac.2020.06.219
Jaouadi, B., Abdelmalek, B., Fodil, D., Ferradji, F. Z., Rekik, H., Zaraî, N., et al. (2010). Purification and characterization of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour. Technol. 101, 8361–8369. doi: 10.1016/j.biortech.2010.05.066
Jaouadi, N. Z., Rekik, H., Badis, A., Trabelsi, S., Belhoul, M., Yahiaoui, A. B., et al. (2013). Biochemical and molecular characterization of a serine keratinase from Brevibacillus brevis US575 with promising keratin-biodegradation and hide-dehairing activities. PLoS One 8:e76722. doi: 10.1371/journal.pone.0076722
Jayalakshmi, T., Krishnamoorthy, P. K. P., Kumar, G. R., and Sivamani, P. S. P. (2010). Isolation and screening of a feather-degrading keratinolytic actinomycetes from Actinomyces sp. J. Am. Sci. 6, 45–48.
Jeevana Lakshmi, P., Kumari Chitturi, C. M., and Lakshmi, V. V. (2013). Efficient degradation of feather by keratinase producing Bacillus sp. Int. J. Microbiol. 2013:7. doi: 10.1155/2013/608321
Jin, H.-S., Park, S. Y., Kim, K., Lee, Y.-J., Nam, G.-W., Kang, N. J., et al. (2017). Development of a keratinase activity assay using recombinant chicken feather keratin substrates. PLoS One 12:e0172712. doi: 10.1371/journal.pone.0172712
Kang, D., Jacquiod, S., Herschend, J., Wei, S., Nesme, J., and Sørensen, S. J. (2020). Construction of simplified microbial consortia to degrade recalcitrant materials based on enrichment and dilution-to-extinction cultures. Front. Microbiol. 10:3010. doi: 10.3389/fmicb.2019.03010
Kanoksilapatham, W., and Intagun, W. (2017). A review: biodegradation and applications of keratin degrading microorganisms and keratinolytic enzymes, focusing on thermophiles and thermostable serine proteases. Am. J. Appl. Sci. 14, 1016–1023. doi: 10.3844/ajassp.2017.1016.1023
Kasperova, A., Cahlikova, R., Kunert, J., Sebela, M., Novak, Z., and Raska, M. (2014). Exposition of dermatophyte Trichophyton mentagrophytes to L-cystine induces expression and activation of cysteine dioxygenase. Mycoses 57, 672–678. doi: 10.1111/myc.12220
Kasperova, A., Kunert, J., Horynova, M., Weigl, E., Sebela, M., Lenobel, R., et al. (2011). Isolation of recombinant cysteine dioxygenase protein from Trichophyton mentagrophytes. Mycoses 54, e456–e462. doi: 10.1111/j.1439-0507.2010.01948.x
Kasperova, A., Kunert, J., and Raska, M. (2013). The possible role of dermatophyte cysteine dioxygenase in keratin degradation. Med. Mycol. 51, 449–454. doi: 10.3109/13693786.2013.794310
Khodayari, S., and Kafilzadeh, F. (2018). Separating keratinase producer bacteria from the soil of poultry farms and optimization of the conditions for maximum enzyme production. Eur. J. Exp. Biol. 8:35. doi: 10.21767/2248-9215.100076
Kim, J. M., Lim, W. J., and Suh, H. J. (2001). Feather-degrading Bacillus species from poultry waste. Process Biochem. 37, 287–291. doi: 10.1016/S0032-9592(01)00206-0
Kim, J. M. K. U., Choi, Y. M. S. C., and Suh, H. J. K. U. (2005). Preparation of feather digests as fertilizer with Bacillus pumilis KHS-1. J. Microbiol. Biotechnol. 15, 427–476.
Kim, J.-S., Kluskens, L. D., de Vos, W. M., Huber, R., and van der Oost, J. (2004). Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin. J. Mol. Biol. 335, 787–797. doi: 10.1016/j.jmb.2003.11.006
Kluskens, L. D., Voorhorst, W. G., Siezen, R. J., Schwerdtfeger, R. M., Antranikian, G., van der Oost, J., et al. (2002). Molecular characterization of fervidolysin, a subtilisin-like serine protease from the thermophilic bacterium Fervidobacterium pennivorans. Extremophiles 6, 185–194. doi: 10.1007/s007920100239
Korkmaz, H., Hur, H., and Dincer, S. (2004). Characterization of alkaline keratinase of Bacillus licheniformis strain HK-1 from poultry waste. Ann. Microbiol. 54, 201–211.
Korniłłowicz-Kowalska, T., and Bohacz, J. (2011). Biodegradation of keratin waste: theory and practical aspects. Waste Manag. 31, 1689–1701. doi: 10.1016/j.wasman.2011.03.024
Kowalczyk, P., Mahdi-Oraibi, S., Misiewicz, A., Gabzdyl, N., Miskiewicz, A., and Szparecki, G. (2018). Feather-Degrading bacteria: their biochemical and genetic characteristics. Arab. J. Sci. Eng. 43, 33–41. doi: 10.1007/s13369-017-2700-2
Kumar, A. (2020). Aspergillus nidulans: a potential resource of the production of the native and heterologous enzymes for industrial applications. Int. J. Microbiol. 2020:8894215. doi: 10.1155/2020/8894215
Laba, W., and Rodziewicz, A. (2014). Biodegradation of hard keratins by two Bacillus Strains. Jundishapur. J. Microbiol. 7:e8896. doi: 10.5812/jjm.8896
Lange, L., Huang, Y., and Busk, P. K. (2016). Microbial decomposition of keratin in nature-a new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 100, 2083–2096. doi: 10.1007/s00253-015-7262-1
Lee, Y.-J., Dhanasingh, I., Ahn, J.-S., Jin, H.-S., Choi, J. M., Lee, S. H., et al. (2015). Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1. Biochem. Biophys. Res. Commun. 468, 927–933. doi: 10.1016/j.bbrc.2015.11.058
Li, F., Wang, Y., Li, C., Marquez-Lago, T. T., Leier, A., Rawlings, N. D., et al. (2018). Twenty years of bioinformatics research for protease-specific substrate and cleavage site prediction: a comprehensive revisit and benchmarking of existing methods. Brief. Bioinform. 20, 2150–2166. doi: 10.1093/bib/bby077
Li, Q. (2019). Progress in microbial degradation of feather waste. Front. Microbiol. 10:2717. doi: 10.3389/fmicb.2019.02717
Li, Z.-W., Liang, S., Ke, Y., Deng, J.-J., Zhang, M.-S., Lu, D.-L., et al. (2020). The feather degradation mechanisms of a new Streptomyces sp. isolate SCUT-3. Commun. Biol. 3:191. doi: 10.1038/s42003-020-0918-0
Lin, X., Lee, C.-G., Casale, E. S., and Shih, J. C. H. (1992). Purification and characterization of a keratinase from a feather-degrading Bacillus licheniformis Strain. Appl. Environ. Microbiol. 58, 3271–3275.
Liu, B., Zhang, J., Fang, Z., Gu, L., Liao, X., Du, G., et al. (2013a). Enhanced thermostability of keratinase by computational design and empirical mutation. J. Ind. Microbiol. Biotechnol. 40, 697–704. doi: 10.1007/s10295-013-1268-4
Liu, B., Zhang, J., Gu, L., Du, G., Chen, J., and Liao, X. (2014). Comparative analysis of bacterial expression systems for keratinase production. Appl. Biochem. Biotechnol. 173, 1222–1235. doi: 10.1007/s12010-014-0925-z
Liu, B., Zhang, J., Li, B., Liao, X., Du, G., and Chen, J. (2013b). Expression and characterization of extreme alkaline, oxidation-resistant keratinase from Bacillus licheniformis in recombinant Bacillus subtilis WB600 expression system and its application in wool fiber processing. World J. Microbiol. Biotechnol. 29, 825–832. doi: 10.1007/s11274-012-1237-5
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with the folin phenol reagent. J. Biol. Chem. 193, 265–275.
Macedo, A. J. J., Silva, W. O. D. S. O. B., and Termignoni, C. (2008). Properties of a non collagen-degrading Bacillus subtilis keratinase. Can. J. Microbiol. 54, 180–188.
Mäkinen, P. L., Mäkinen, K. K., and Syed, S. A. (1994). An endo-acting proline-specific oligopeptidase from Treponema denticola ATCC 35405: evidence of hydrolysis of human bioactive peptides. Infect. Immun. 62, 4938–4947.
Mazotto, A. M., Couri, S., Damaso, M. C. T., and Vermelho, A. B. (2013). Degradation of feather waste by Aspergillus niger keratinases: comparison of submerged and solid-state fermentation. Int. Biodeterior. Biodegradation 85, 189–195. doi: 10.1016/j.ibiod.2013.07.003
McKittrick, J., Chen, P. Y., Bodde, S. G., Yang, W., Novitskaya, E. E., and Meyers, M. A. (2012). The structure, functions, and mechanical properties of keratin. JOM 64, 449–468. doi: 10.1007/s11837-012-0302-8
Mercer, D. K., and Stewart, C. S. (2018). Keratin hydrolysis by dermatophytes. Med. Mycol. 57, 13–22. doi: 10.1093/mmy/myx160
Mitsuiki, S., Ichikawa, M., Oka, T., Sakai, M., Moriyama, Y., Sameshima, Y., et al. (2004). Molecular characterization of a keratinolytic enzyme from an alkaliphilic Nocardiopsis sp. TOA-1. Enzyme Microb. Technol. 34, 482–489. doi: 10.1016/j.enzmictec.2003.12.011
Monod, M. (2008). Secreted proteases from dermatophytes. Mycopathologia 166:285. doi: 10.1007/s11046-008-9105-4
Monod, M., Léchenne, B., Jousson, O., Grand, D., Zaugg, C., Stöcklin, R., et al. (2005). Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology 151, 145–155. doi: 10.1099/mic.0.27484-0
Moridshahi, R., Bahreini, M., Sharifmoghaddam, M., and Asoodeh, A. (2020). Biochemical characterization of an alkaline surfactant-stable keratinase from a new keratinase producer, Bacillus zhangzhouensis. Extremophiles 24, 693–704. doi: 10.1007/s00792-020-01187-9
Muhammed, N. S., Hussin, N., Lim, A. S., Jonet, M. A., Mohamad, S. E., and Jamaluddin, H. (2021). Recombinant production and characterization of an extracellular subtilisin-like serine protease from Acinetobacter baumannii of fermented food origin. Protein J. 1–17. doi: 10.1007/s10930-021-09986-5
Nasipuri, P., Herschend, J., Brejnrod, A. D., Madsen, J. S., Espersen, R., Svensson, B., et al. (2020). Community-intrinsic properties enhance keratin degradation from bacterial consortia. PLoS One 15:e0228108. doi: 10.1371/journal.pone.0228108
Navone, L., and Speight, R. (2018). Understanding the dynamics of keratin weakening and hydrolysis by proteases. PLoS One 13:e0202608. doi: 10.1371/journal.pone.0202608
Nnolim, N. E., Mpaka, L., Okoh, A. I., and Nwodo, U. U. (2020a). Biochemical and molecular characterization of a thermostable alkaline metallo-keratinase from Bacillus sp. Nnolim-K1. Microorganisms 8:1304.
Nnolim, N. E., and Nwodo, U. U. (2021). Microbial keratinase and the bio-economy: a three-decade meta-analysis of research exploit. AMB Express 11:12. doi: 10.1186/s13568-020-01155-8
Nnolim, N. E., Okoh, A. I., and Nwodo, U. U. (2020b). Proteolytic bacteria isolated from agro-waste dumpsites produced keratinolytic enzymes. Biotechnol. Rep. 27:e00483. doi: 10.1016/j.btre.2020.e00483
Omrane Benmrad, M., Mechri, S., Zaraî Jaouadi, N., Ben Elhoul, M., Rekik, H., Sayadi, S., et al. (2019). Purification and biochemical characterization of a novel thermostable protease from the oyster mushroom Pleurotus sajor-caju strain CTM10057 with industrial interest. BMC Biotechnol. 19:43. doi: 10.1186/s12896-019-0536-4
Onifade, A. A., Al-Sane, N. A., Al-Musallam, A. A., and Al-Zarban, S. (1998). A review: potentials for biotechnological applications of keratin-degrading microorganisms and their enzymes for nutritional improvement of feathers and other keratins as livestock feed resources. Bioresour. Technol. 66, 1–11. doi: 10.1016/S0960-8524(98)00033-9
Pahua-Ramos, M. E., Hernández-Melchor, D. J., Camacho-Pérez, B., and Quezada-Cruz, M. (2017). Degradation of chicken feathers: a review. Biotechnol. Ind. J. 13, 658–666.
Papadopoulos, M. C. (1989). Effect of processing on high-protein feedstuffs: a review. Biol. Wastes 29, 123–138. doi: 10.1016/0269-7483(89)90092-X
Parrado, J., Rodriguez-Morgado, B., Tejada, M., Hernandez, T., and Garcia, C. (2014). Proteomic analysis of enzyme production by Bacillus licheniformis using different feather wastes as the sole fermentation media. Enzyme Microb. Technol. 57, 1–7. doi: 10.1016/j.enzmictec.2014.01.001
Peng, Z., Mao, X., Zhang, J., Du, G., and Chen, J. (2019). Effective biodegradation of chicken feather waste by co-cultivation of keratinase producing strains. Microb. Cell Fact. 18:84. doi: 10.1186/s12934-019-1134-9
Peng, Z., Zhang, J., Song, Y., Guo, R., Du, G., and Chen, J. (2021). Engineered pro-peptide enhances the catalytic activity of keratinase to improve the conversion ability of feather waste. Biotechnol. Bioeng. doi: 10.1002/bit.27771
Pettett, L. M., and Ipek, K. (2004). Development of an environmentally friendly biofertilizer with keratin degrading and antibiotic producing actinomycetes. Actinomycetologica 18, 34–42. doi: 10.3209/saj.18_34
Pinski, A., Zur, J., Hasterok, R., and Hupert-Kocurek, K. (2020). Comparative genomics of Stenotrophomonas maltophilia and Stenotrophomonas rhizophila revealed characteristic features of both species. Int. J. Mol. Sci. 21:4922.
Prakash, P., Jayalakshmi, S. K., and Sreeramulu, K. (2010a). Production of keratinase by free and immobilized cells of Bacillus halodurans strain PPKS-2: partial characterization and its application in feather degradation and dehairing of the goat skin. Appl. Biochem. Biotechnol. 160, 1909–1920. doi: 10.1007/s12010-009-8702-0
Prakash, P., Jayalakshmi, S. K., and Sreeramulu, K. (2010b). Purification and characterization of extreme alkaline, thermostable keratinase, and keratin disulfide reductase produced by Bacillus halodurans PPKS-2. Appl. Microbiol. Biotechnol. 87, 625–633. doi: 10.1007/s00253-010-2499-1
Qiu, J., Wilkens, C., Barrett, K., and Meyer, A. S. (2020). Microbial enzymes catalyzing keratin degradation: classification, structure, function. Biotechnol. Adv. 44:107607. doi: 10.1016/j.biotechadv.2020.107607
Qu, F., Chen, Q., Ding, Y., Liu, Z., Zhao, Y., Zhang, X., et al. (2018). Isolation of a feather-degrading strain of bacterium from spider gut and the purification and identification of its three key enzymes. Mol. Biol. Rep. 45, 1681–1689. doi: 10.1007/s11033-018-4311-8
Rahayu, S., Syah, D., and Thenawidjaja Suhartono, M. (2012). Degradation of keratin by keratinase and disulfide reductase from Bacillus sp. MTS of Indonesian origin. Biocatal. Agric. Biotechnol. 1, 152–158. doi: 10.1016/j.bcab.2012.02.001
Rai, S. K., Konwarh, R., and Mukherjee, A. K. (2009). Purification, characterization and biotechnological application of an alkaline β-keratinase produced by Bacillus subtilis RM-01 in solid-state fermentation using chicken-feather as substrate. Biochem. Eng. J. 45, 218–225. doi: 10.1016/j.bej.2009.04.001
Rai, S. K., and Mukherjee, A. K. (2011). Optimization of production of an oxidant and detergent-stable alkaline β-keratinase from Brevibacillus sp. strain AS-S10-II: application of enzyme in laundry detergent formulations and in leather industry. Biochem. Eng. J. 54, 47–56. doi: 10.1016/j.bej.2011.01.007
Ramnani, P., Singh, R., and Gupta, R. (2005). Keratinolytic potential of Bacillus licheniformis RG1: structural and biochemical mechanism of feather degradation. Can. J. Microbiol. 51, 191–196. doi: 10.1139/w04-123
Reis, S. V. D., Beys-da-Silva, W. O., Tirloni, L., Santi, L., Seixas, A., Termignoni, C., et al. (2020). The extremophile Anoxybacillus sp. PC2 isolated from Brazilian semiarid region (Caatinga) produces a thermostable keratinase. J. Basic Microbiol. 60, 809–815. doi: 10.1002/jobm.202000186
Riffel, A., Lucas, F., Heeb, P., and Brandelli, A. (2003). Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch. Microbiol. 179, 258–265. doi: 10.1007/s00203-003-0525-8
Rozs, M., Manczinger, L., Vágvölgyi, C., and Kevei, F. (2001). Secretion of a trypsin-like thiol protease by a new keratinolytic strain of Bacillus licheniformis. FEMS Microbiol. Lett. 205, 221–224. doi: 10.1111/j.1574-6968.2001.tb10951.x
Sahni, N., Sahota, P. P., and Gupta, P. U. (2015). Bacterial keratinases and their prospective applications: a review. Int. J. Curr. Microbiol. Appl. Sci. 4, 768–783.
Sala, L., Gautério, G. V., Younan, F. F., Brandelli, A., Moraes, C. C., and Kalil, S. J. (2014). Integration of ultrafiltration into an aqueous two-phase system in the keratinase purification. Process Biochem. 49, 2016–2024. doi: 10.1016/j.procbio.2014.07.013
Sangali, S., and Brandelli, A. (2000). Isolation and characterization of a novel feather-degrading bacterial strain. Appl. Biochem. Biotechnol. 87, 17–24.
Shadzi, S., Chadeganipour, M., and Alimoradi, M. (2002). Isolation of keratinophilic fungi from elementary schools and public parks in Isfahan, Iran. Mycoses 45, 496–499. doi: 10.1046/j.1439-0507.2002.00798.x
Shanmugasundaram, O. L., Syed Zameer Ahmed, K., Sujatha, K., Ponnmurugan, P., Srivastava, A., Ramesh, R., et al. (2018). Fabrication and characterization of chicken feather keratin/polysaccharides blended polymer coated nonwoven dressing materials for wound healing applications. Mat. Sci. Eng. C 92, 26–33. doi: 10.1016/j.msec.2018.06.020
Sharma, I., and Kango, N. (2021). Production and characterization of keratinase by Ochrobactrum intermedium for feather keratin utilization. Int. J. Biol. Macromol. 166, 1046–1056. doi: 10.1016/j.ijbiomac.2020.10.260
Sharma, R., and Devi, S. (2018). Versatility and commercial status of microbial keratinases: a review. Rev. Environ. Sci. Bio Technol. 17, 19–45. doi: 10.1007/s11157-017-9454-x
Sharma, S., and Gupta, A. (2016). Sustainable management of keratin waste biomass: applications and future perspectives. Braz. Arch. Biol. Technol. 59, 1–14.
Silveira, S. T., Jaeger, M. K., and Brandelli, A. (2009). Kinetic data and substrate specificity of a keratinase from Chryseobacterium sp. strain kr6. J. Chem. Technol. Biotechnol. 84, 361–366. doi: 10.1002/jctb.2048
Sittipol, D., Rodpan, S., Ajingi, Y. S., Lohnoo, T., Lerksuthirat, T., Kumsang, Y., et al. (2021). Identification, overexpression, purification, and biochemical characterization of a novel hyperthermostable keratinase from Geoglobus acetivorans. 3 Biotech 11:2. doi: 10.1007/s13205-020-02538-1
Song, J., Wang, Y., Li, F., Akutsu, T., Rawlings, N. D., Webb, G. I., et al. (2018). iProt-Sub: a comprehensive package for accurately mapping and predicting protease-specific substrates and cleavage sites. Brief. Bioinform. 20, 638–658. doi: 10.1093/bib/bby028
Su, C., Gong, J.-S., Sun, Y.-X., Qin, J., Zhai, S., Li, H., et al. (2019). Combining pro-peptide engineering and multisite saturation mutagenesis to improve the catalytic potential of keratinase. ACS Synth. Biol. 8, 425–433. doi: 10.1021/acssynbio.8b00442 x1
Syed, D. G., Lee, J. C., Li, W.-J., Kim, C.-J., and Agasar, D. (2009). Production, characterization and application of keratinase from Streptomyces gulbargensis. Bioresour. Technol. 100, 1868–1871. doi: 10.1016/j.biortech.2008.09.047
Tamreihao, K., Mukherjee, S., Khunjamayum, R., Devi, L. J., Asem, R. S., and Ningthoujam, D. S. (2019). Feather degradation by keratinolytic bacteria and biofertilizing potential for sustainable agricultural production. J. Basic Microbiol. 59, 4–13. doi: 10.1002/jobm.201800434
Tatineni, R., Doddapaneni, K. K., Potumarthi, R. C., and Mangamoori, L. N. (2007). Optimization of keratinase production and enzyme activity using response surface methodology with streptomyces sp7. Appl. Biochem. Biotechnol. 141:187. doi: 10.1007/bf02729061
Teruo, S., Yumiko, H., Aki, K., Toyoyuki, O., Megumi, T., Ayumi, A., et al. (2015). Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease. Protein Pept. Lett. 22, 63–72. doi: 10.2174/0929866521666140919100851
Thankaswamy, S. R., Sundaramoorthy, S., Palanivel, S., and Ramudu, K. N. (2018). Improved microbial degradation of animal hair waste from leather industry using Brevibacterium luteolum (MTCC 5982). J. Clean. Prod. 189, 701–708. doi: 10.1016/j.jclepro.2018.04.095
Tiwary, E., and Gupta, R. (2010a). Extracellular expression of keratinase from Bacillus licheniformis ER-15 in Escherichia coli. J. Agric. Food Chem. 58, 8380–8385. doi: 10.1021/jf100803g
Tiwary, E., and Gupta, R. (2010b). Medium optimization for a novel 58kDa dimeric keratinase from Bacillus licheniformis ER-15: biochemical characterization and application in feather degradation and dehairing of hides. Bioresour. Technol. 101, 6103–6110. doi: 10.1016/j.biortech.2010.02.090
Tork, S. E., Shahein, Y. E., El-Hakim, A. E., Abdel-Aty, A. M., and Aly, M. M. (2013). Production and characterization of thermostable metallo-keratinase from newly isolated Bacillus subtilis NRC 3. Int. J. Biol. Macromol. 55, 169–175. doi: 10.1016/j.ijbiomac.2013.01.002
Tork, S. E., Shahein, Y. E., El-Hakim, A. E., Abdel-Aty, A. M., and Aly, M. M. (2016). Purification and partial characterization of serine-metallokeratinase from a newly isolated Bacillus pumilus NRC21. Int. J. Biol. Macromol. 86, 189–196. doi: 10.1016/j.ijbiomac.2016.01.060
Verma, A., Singh, H., Anwar, S., Chattopadhyay, A., Tiwari, K. K., Kaur, S., et al. (2017). Microbial keratinases: industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 37, 476–491. doi: 10.1080/07388551.2016.1185388
Vidmar, B., and Vodovnik, M. (2018). Microbial keratinases: enzymes with promising biotechnological applications. Food Technol. Biotechnol. 56, 312–328. doi: 10.17113/ftb.56.03.18.5658
Wang, B., Yang, W., McKittrick, J., and Meyers, M. A. (2016). Keratin: structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Progress Mater. Sci. 76, 229–318. doi: 10.1016/j.pmatsci.2015.06.001
Wang, J.-J., Rojanatavorn, K., and Shih, J. C. H. (2004). Increased production of Bacillus keratinase by chromosomal integration of multiple copies of the kerA gene. Biotechnol. Bioeng. 87, 459–464. doi: 10.1002/bit.20145
Wang, X. C., Zhao, H. Y., Liu, G., Cheng, X. J., and Feng, H. (2015). Improving production of extracellular proteases by random mutagenesis and biochemical characterization of a serine protease in Bacillus subtilis S1-4. Genet. Mol. Res. 15:gmr.15027831. doi: 10.4238/gmr.15027831
Wu, W.-L., Chen, M.-Y., Tu, I. F., Lin, Y.-C., EswarKumar, N., Chen, M.-Y., et al. (2017). The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles. Sci. Rep. 7:4658. doi: 10.1038/s41598-017-04723-4
Yahaya, R. S. R., Normi, Y. M., Phang, L. Y., Ahmad, S. A., Abdullah, J. O., and Sabri, S. (2021). Molecular strategies to increase keratinase production in heterologous expression systems for industrial applications. Appl. Microbiol. Biotechnol. 105, 3955–3969. doi: 10.1007/s00253-021-11321-y
Yamamura, S., Morita, Y., Hasan, Q., Yokoyama, K., and Tamiya, E. (2002). Keratin degradation: a cooperative action of two enzymes from Stenotrophomonas sp. Biochem. Biophys. Res. Commun. 294, 1138–1143. doi: 10.1016/S0006-291X(02)00580-6
Yong, B., Fei, X., Shao, H., Xu, P., Hu, Y., Ni, W., et al. (2020). Recombinant expression and biochemical characterization of a novel keratinase BsKER71 from feather degrading bacterium Bacillus subtilis S1-4. AMB Express 10:9. doi: 10.1186/s13568-019-0939-6
Zaghloul, T. I., Embaby, A. M., and Elmahdy, A. R. (2011). Biodegradation of chicken feathers waste directed by Bacillus subtilis recombinant cells: scaling up in a laboratory scale fermentor. Bioresour. Technol. 102, 2387–2393. doi: 10.1016/j.biortech.2010.10.106
Zaraî Jaouadi, N., Rekik, H., Ben Elhoul, M., Zohra Rahem, F., Gorgi Hila, C., Slimene Ben Aicha, H., et al. (2015). A novel keratinase from Bacillus tequilensis strain Q7 with promising potential for the leather bating process. Int. J. Biol. Macromol. 79, 952–964. doi: 10.1016/j.ijbiomac.2015.05.038
Zeng, Y.-H., Shen, F.-T., Tan, C.-C., Huang, C.-C., and Young, C.-C. (2011). The flexibility of UV-inducible mutation in Deinococcus ficus as evidenced by the existence of the imuB–dnaE2 gene cassette and generation of superior feather degrading bacteria. Microbiol. Res. 167, 40–47. doi: 10.1016/j.micres.2011.02.008
Zhang, M., Jiang, L., Li, F., Xu, Y., Lv, S., and Wang, B. (2019). Simultaneous dermatophytosis and keratomycosis caused by Trichophyton interdigitale infection: a case report and literature review. BMC Infect. Dis. 19:983. doi: 10.1186/s12879-019-4612-0
Zhang, R.-X., Gong, J.-S., Su, C., Qin, J., Li, H., Li, H., et al. (2020). Recombinant expression and molecular engineering of the keratinase from Brevibacillus parabrevis for dehairing performance. J. Biotechnol. 320, 57–65. doi: 10.1016/j.jbiotec.2020.06.016
Keywords: keratinase, waste treatment, keratin, microorganisms, structure, protease
Citation: Li Q (2021) Structure, Application, and Biochemistry of Microbial Keratinases. Front. Microbiol. 12:674345. doi: 10.3389/fmicb.2021.674345
Received: 01 March 2021; Accepted: 17 May 2021;
Published: 23 June 2021.
Edited by:
Kian Mau Goh, University of Technology Malaysia, MalaysiaReviewed by:
Arun Gupta, Universiti Malaysia Pahang, MalaysiaBassem Jaouadi, Centre of Biotechnology of Sfax, Tunisia
Suman Kumar Halder, Vidyasagar University, India
Copyright © 2021 Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingxin Li, cWluZ3hpbl9saUBvdXRsb29rLmNvbQ==
 Qingxin Li
Qingxin Li