- 1Department of Respiratory Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 2Department of Clinical Laboratory Medicine, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
- 3Shanghai Key Laboratory of Tuberculosis, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
- 4Jiangxi Provincial Key Laboratory of Medicine, Clinical Laboratory of the Second Affiliated Hospital of Nanchang University, Nanchang, China
- 5Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
Many antimicrobial resistance genes usually located on transferable plasmids are responsible for multiple antimicrobial resistance among multidrug-resistant (MDR) Gram-negative bacteria. The aim of this study is to characterize a carbapenemase-producing Enterobacter hormaechei 1575 isolate from the blood sample in a tertiary hospital in Wuhan, Hubei Province, China. Antimicrobial susceptibility test showed that 1575 was an MDR isolate. The whole genome sequencing (WGS) and comparative genomics were used to deeply analyze the molecular information of the 1575 and to explore the location and structure of antibiotic resistance genes. The three key resistance genes (blaSFO–1, blaNDM–1, and mcr-9) were verified by PCR, and the amplicons were subsequently sequenced. Moreover, the conjugation assay was also performed to determine the transferability of those resistance genes. Plasmid files were determined by the S1 nuclease pulsed-field gel electrophoresis (S1-PFGE). WGS revealed that p1575-1 plasmid was a conjugative plasmid that possessed the rare coexistence of blaSFO–1, blaNDM–1, and mcr-9 genes and complete conjugative systems. And p1575-1 belonged to the plasmid incompatibility group IncHI2 and multilocus sequence typing ST102. Meanwhile, the pMLST type of p1575-1 was IncHI2-ST1. Conjugation assay proved that the MDR p1575-1 plasmid could be transferred to other recipients. S1-PFGE confirmed the location of plasmid with molecular weight of 342,447 bp. All these three resistant genes were flanked by various mobile elements, indicating that the blaSFO–1, blaNDM–1, and mcr-9 could be transferred not only by the p1575-1 plasmid but also by these mobile elements. Taken together, we report for the first time the coexistence of blaSFO–1, blaNDM–1, and mcr-9 on a transferable plasmid in a MDR clinical isolate E. hormaechei, which indicates the possibility of horizontal transfer of antibiotic resistance genes.
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) has recently emerged as a serious threat to modern healthcare, challenging our present antibiotic treatment strategy (Chen et al., 2014). Moreover, the carbapenem-resistant and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae are also classified as the “priority pathogens” by the World Health Organization in 2017 (WHO, 2017; Tacconelli et al., 2018). Among all these Enterobacteriaceae isolates, Enterobacter hormaechei is a notorious nosocomial pathogen contributing to various infections, such as bacteremia, endocarditis, and lower respiratory, urinary tract, and intra-abdominal infections (Xu et al., 2015).
Recently, reports about the coexistence of a rare ESBL gene blaSFO–1 and carbapenemase genes were increased (Zhou et al., 2020). New Delhi metallo-lactamase (NDM-1), a β-lactam hydrolase, constitutes a critical and growingly important medical issue, since its resistance trait compromises the efficacy of almost all lactams (except aztreonam), including carbapenems (Dortet et al., 2014). Compared with other broad-spectrum β-lactamase, the blaSFO–1 gene is a low-incidence antimicrobial resistance gene and usually not subject to systematic monitoring, which puts it at risk of being missed (Zhao et al., 2015; Zhou et al., 2020). With the increase of infections caused by carbapenemase-producing bacteria and the lack of novel antibiotics (Gurjar, 2015), polymyxins have become the last-resort therapies in the treatment of infections caused by this kind of multidrug-resistant (MDR) bacterial (Olaitan et al., 2014; Yilmaz et al., 2016). Thus, once the strains are resistant to both carbapenems and polymyxins, the treatment will be very tough. The first plasmid-mediated colistin resistance gene mcr-1 was identified in China from the plasmid of Escherichia coli and Klebsiella pneumoniae in IncI2 (Liu et al., 2016). mcr-1 remains the main plasmid-mediated myxobacteria resistance gene, but mcr-2 to mcr-8 has been identified in different species in humans and animals (Wang et al., 2018; Nang et al., 2019). mcr-9 has also been identified in Swedish ESBL isolates, including Enterobacter cloacae, E. coli, Klebsiella acidophilus, and Citrobacter freundii (Börjesson et al., 2020). Of particular concern is the spread of mcr genes into CRE, which would create strains that are potentially pan-drug resistant (PDR). So mobile colistin-resistant genes (mcr) have become an increasing public health concern.
It is common for the coexistence of mcr-9 with carbapenemases, such as blaNDM–1, blaVIM–4, and blaIMP–4 (Chavda et al., 2019; Chen et al., 2020). However, in this study, we found a clinical isolate of carbapenem-resistant E. hormaechei, which possessed the rare coexistence of blaSFO–1, blaNDM–1, and mcr-9 genes. And we also explored the molecular basis for antibiotic resistance of this strain.
Materials and Methods
Bacterial Isolation and Identification
The E. hormaechei 1575 was isolated from the blood sample in a tertiary hospital in Wuhan, Hubei Province, China. The cultured bacteria were stored in glycerol broth at 80°C. And then samples were cultured on Colombian blood Agar plate and identified by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) according to the manufacturer’s instructions and also by whole genome sequencing (WGS) (discussed below). Escherichia coli American Type Culture Collection (ATCC) 25922 was used as control strains for the identification of the species.
Antimicrobial Susceptibility Testing
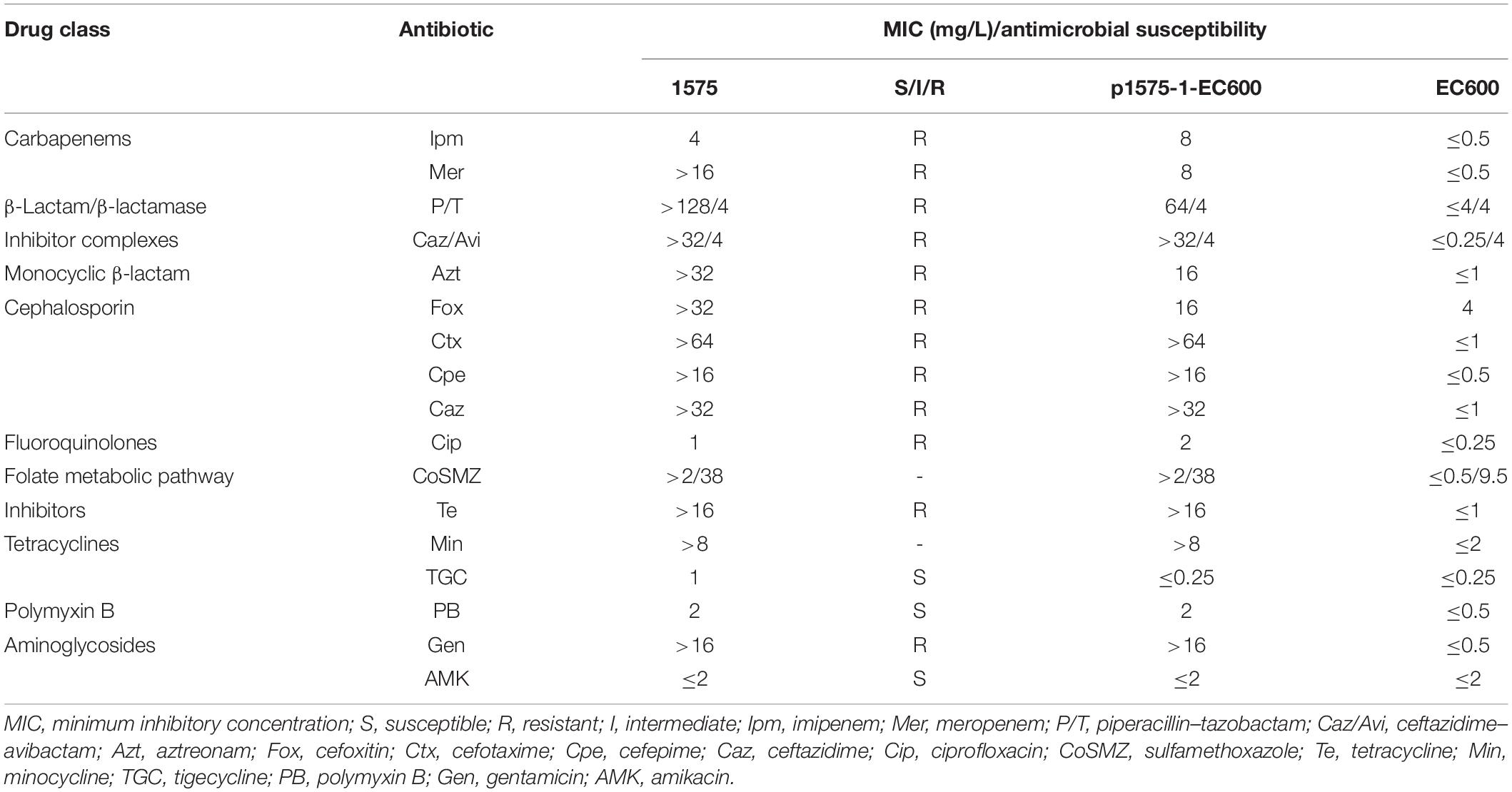
A total of 17 antimicrobial agents were tested, including imipenem (Ipm), meropenem (Mer), piperacillin–tazobactam (P/T), ceftazidime–avibactam (Caz/Avi), aztreonam (Azt), cefoxitin (Fox), cefotaxime (Ctx), cefepime (Cpe), ceftazidime (Caz), gentamicin (Gen), amikacin (AMK), ciprofloxacin (Cip), sulfamethoxazole (CoSMZ), tetracycline (Te), minocycline (Min), tigecycline (TGC), and polymyxin B (PB). The minimum inhibitory concentrations (MICs) of antimicrobial agents for the bacteria tested were determined using the broth microdilution method, and the susceptibility breakpoints were interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI) guideline (Clinical and Laboratory Standards Institute (CLSI), 2020), except for tigecycline and colistin, for which we used the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (Clinical and Laboratory Standards Institute (CLSI), 2020; EUCAST, 2020). AST was repeated three times in our study. E. coli ATCC 25922 was used as a control strain for the AST.
Carbapenemase Phenotype Confirmation Testing
The modified carbapenem inactivation test (mCIM) was performed, according to CLSI 2020 standards [Clinical and Laboratory Standards Institute (CLSI), 2020], to verify carbapenemase production by the isolate. The tested strain 1575 was incubated with a meropenem disk (10 μg, OXOID, United Kingdom) immersed in the 2 ml of TSB suspension at 37°C for 4 h. E. coli ATCC 25922 was used as an indicator and with its 0.5 McFarland suspension uniformly coated on the Mueller Hinton Agar (MHA) plate. After the plate was dried for 3–10 min, the meropenem disk was removed from the suspension, and the excess medium was squeezed out. It was then placed on the MHA plate and incubated at 37°C for 18–24 h.
Whole Genome Sequencing and Bioinformatics Analysis
Bacterial genomic DNA was isolated using the UltraClean Microbial Kit (Qiagen, NW, Germany) and sequenced from a sheared DNA library with average size of 15 kb (ranged from 10 to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, United States), as well as a paired-end library with an average insert size of 350 bp (ranged from 150 to 600 kb) on a HiSeq sequencer (Illumina, CA, United States). Sequencing libraries were constructed using the NEBNext® UltraTM II DNA Library Prep Kit for Illumina® (second-generation sequencing) and the SMRTbell® Express Template Prep Kit 2.0 kit (third-generation sequencing) and then loaded onto NovaSeq S4 flowcell and SMRT Cell 8 M DNA sequencing chip, respectively. The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread (Hackl et al., 2014), and then the corrected PacBio reads were assembled de novo utilizing SMARTdenovo1. Antimicrobial resistance genes were identified by ResFinder 3.2 available at Center for Genomic Epidemiology2. The plasmid incompatibility groups, pMLST, and multilocus sequence typing (MLST) were identified by PlasmidFinder 2.13, pMLST 2.04, and MLST 2.0 software5, respectively. To verify whether the plasmid was also a conjugative plasmid, we used the OriT Finder website6 to conduct a detailed analysis of the conjugation module. The IS elements can be directly determined from the known website7. We used blast8 to determine similar plasmids by comparing their coverages and identities. The circular representation of p1575 was generated with CGview9. The plasmid linear graph was analyzed by Easyfig software10.
PCR Amplifications and Sequencing
The isolate was verified for the presence of blaSFO–1-positive strains using PCR with the primers blaSFO–1-forward and blaSFO–1-reverse. Meanwhile, the other carbapenemase genes responsible for carbapenem resistance (blaKPC, blaVIM, blaGES, blaIMP, blaSPM, blaOXA–23, blaOXA–48, blaSME, blaSIM, and blaNDM) (Queenan and Bush, 2007; Nordmann et al., 2011) and the colistin resistance gene mcr-9 were also detected by PCR. The DNA fragments were analyzed using gel electrophoresis on 1% agarose gels, and the amplicons were subsequently sequenced on both strands by TSINGKE sequencing (Table 1).
Conjugation Experiment
The horizontal transferability of blaSFO–1, blaNDM–1, and mcr-9 was examined using conjugation assay. The E. hormaechei 1575 was used as donor strain, and the E. coli EC600 (rifampicin-resistant) was used as the recipient strain. The donors and recipients were cultured to the logarithmic phase (OD600 = 0.4–0.6), mixed in a 1:1 ratio, centrifuged at 8,000 g for 1 min, and resuspended them in 20 μl of Luria Bertani (LB) broth. The resuspension was spotted on the LB plates and incubated overnight at 37°C. The spots were then transferred to 15-ml centrifuge tubes and washed with 3 ml of LB broth. Subsequently, the serial dilutions were plated onto MH agar plates containing cefotaxime (8 μg/ml) and rifampicin (200 μg/ml). The donor cells and recipient cells were used separately as controls to ensure the effectiveness of the screening plate antibiotics. All transconjugants were confirmed by PCR for the presence of blaSFO–1, blaNDM–1, and mcr-9 genes. Transconjugants were subjected to susceptibility assays. The conjugation frequency was calculated as the number of transconjugants per donor cell.
S1 Pulsed-Field Gel Electrophoresis
S1 pulsed-field gel electrophoresis (S1-PFGE) was performed to obtain plasmid profiles in donor strains, recipient strains, and transconjugants, as described previously (Chen et al., 2011). Briefly, the isolates were embedded in 10 g/L of Seakem Gold gel, digested with endonuclease S1 nuclease (Takara, Dalian, China), and subjected to pulsed-field gel electrophoresis (parameters: 14°C, voltage 6 V/cm, electric field angle 120°, conversion time 4.0–40 s, and electrophoresis 19 h). The genomic DNA of Salmonella enterica serovar Braenderup H9812 strain cut with XbaI was used as a control standard strain and a molecular size marker.
Nucleotide Accession Number
The complete nucleotide sequences of the chromosome of 1575, p1575-1, and p1575-2 were deposited as GenBank accession numbers CP068287, CP068288, and CP068289, respectively.
Results
Enterobacter hormaechei 1575 Was a Multidrug-Resistant Strain and Produced Carbapenemase
To clarify the antibiotic-resistant phenotype of E. hormaechei 1575, we tested the susceptibility of 17 antibiotics in this strain. As the results showed (Table 2), E. hormaechei 1575 was resistant to all β-lactam antibiotics (cephalosporins, carbapenems, penicillins, and monocyclic β-lactams), aminoglycosides, quinolones, and tetracycline. We found that 1575 was only susceptible to tigecycline, amikacin, and polymyxin B. Notably, for the ceftazidime–avibactam, a novel carbapenemase inhibitor, this isolate also exhibited high-level resistance.
Since E. hormaechei 1575 was resistant to both carbapenems and ceftazidime–avibactam, we used the mCIM assay to test preliminary whether this isolate produces carbapenemases. The result showed that E. hormaechei 1575 was positive for the mCIM assay, indicating that the isolate produced carbapenemases. Combining this strain with resistance to ceftazidime–avibactam, we speculated that E. hormaechei 1575 produced metallo-carbapenemase.
Enterobacter hormaechei 1575 Co-harboring blaSFO–1, blaNDM–1, and mcr-9 Resistance Genes
Through the resistance phenotype assays, we evaluated the clinical treatment challenges brought by this strain. Here, we continued to explore the associated molecular mechanism that contributed to such phenotype.
We used WGS to deeply mine the genomic information of the MDR bacteria. We found two plasmids in this isolate (named p1575-1 and p1575-2); and p1575-1 (CP068288) was larger with approximately 342,447 bp and sheltered multiple antibiotic resistance genes, especially including β-lactam resistance genes blaSFO–1, blaNDM–1, and colistin resistance gene mcr-9 (Table 3). Besides, consistent with its multidrug resistance phenotype, p1575-1 also had multiple genes mediating resistance to quinolone (qnrS1), aminoglycosides [aac(3)-IId, aph(3′)-Ia, and aph(6)-Id], β-lactams (blaTEM–1B and blaLAP–2), bleomycin bleMBL, trimethoprims (dfrA14 and dfrA19), and MLS—macrolide [mph(A)] and tetracycline [tet(D)]. p1575-2 was a small plasmid of approximately 1,699 bp, with no resistance genes located on. We found the antibiotic-resistant plasmid by second-generation sequencing and further analyzed it by third-generation sequencing. Then we applied the PCR assay to verify these resistance genes. In addition, MLST analysis showed that E. hormaechei 1575 belonged to clone group ST102.
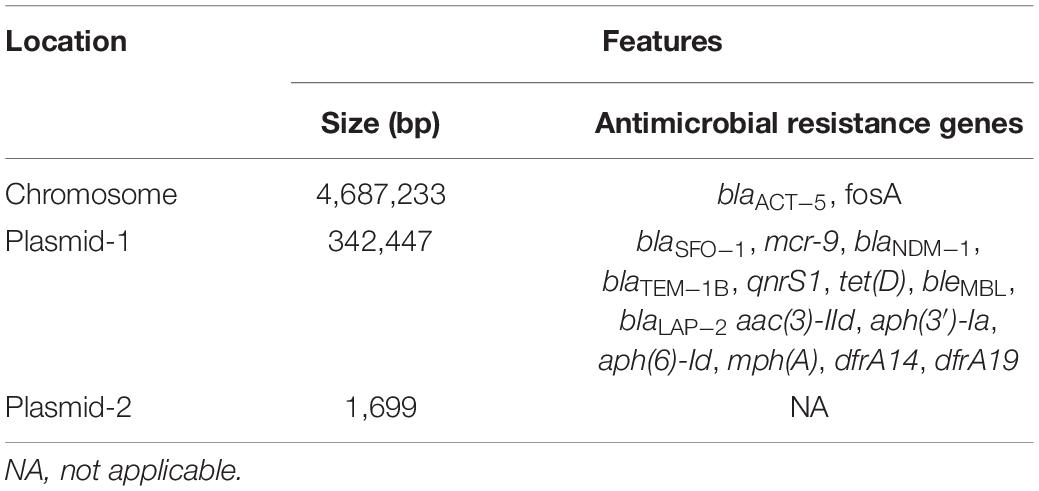
Table 3. General features, antimicrobial resistance genes, and mobile genetic elements of plasmids p1575-1 and p1575-2.
Comparative Genomics of the Plasmid p1575-1 Carrying blaSFO–1, blaNDM–1, and mcr-9
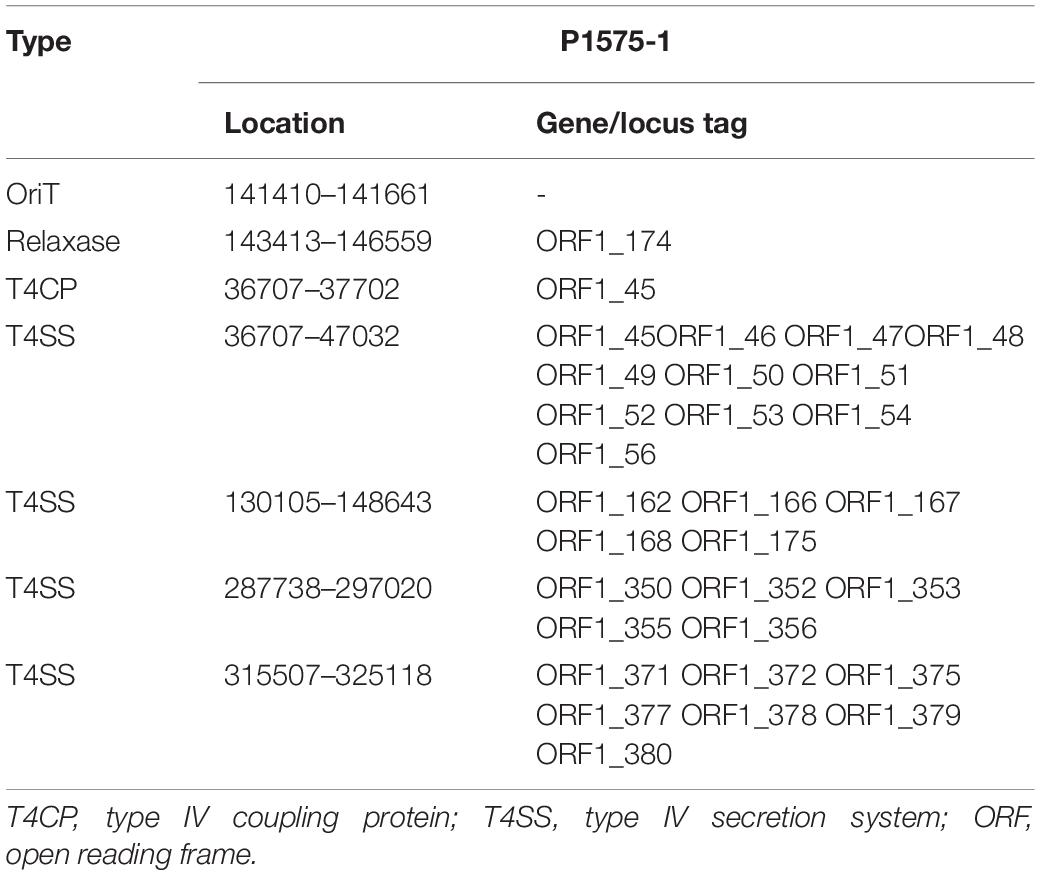
We have known that the plasmid p1575-1 was the key plasmid for the contribution of the MDR phenotype; thus, exploring the characteristics of p1575-1 is the key to elucidating the spread of such bacteria and the mechanism of antibiotic resistance. p1575-1 is a 342,447-bp circular molecule with an average G + C content of 47.93% and was predicted to encode a total of 386 coding sequences (CDSs). Through the PlasmidFinder (see text footnote 3) and pMLST website (see text footnote 4), p1575-1 was typed as IncHI2 with Double Locus Sequence Type DLST1. To verify whether the p1575-1 plasmid was also a conjugative plasmid, we used the OriT Finder website (see text footnote 6) to conduct a detailed analysis of the conjugation module. Through the analysis, we identified the complete conjugative modules on the plasmid p1575-1, including the origin of transfer site (oriT), relaxase gene, gene encoding type IV coupling protein (T4CP), and gene cluster for bacterial type IV secretion system (T4SS) (Table 4). These results strongly suggested that p1575-1 is an MDR plasmid that can be transferred autonomously (Figure 1).
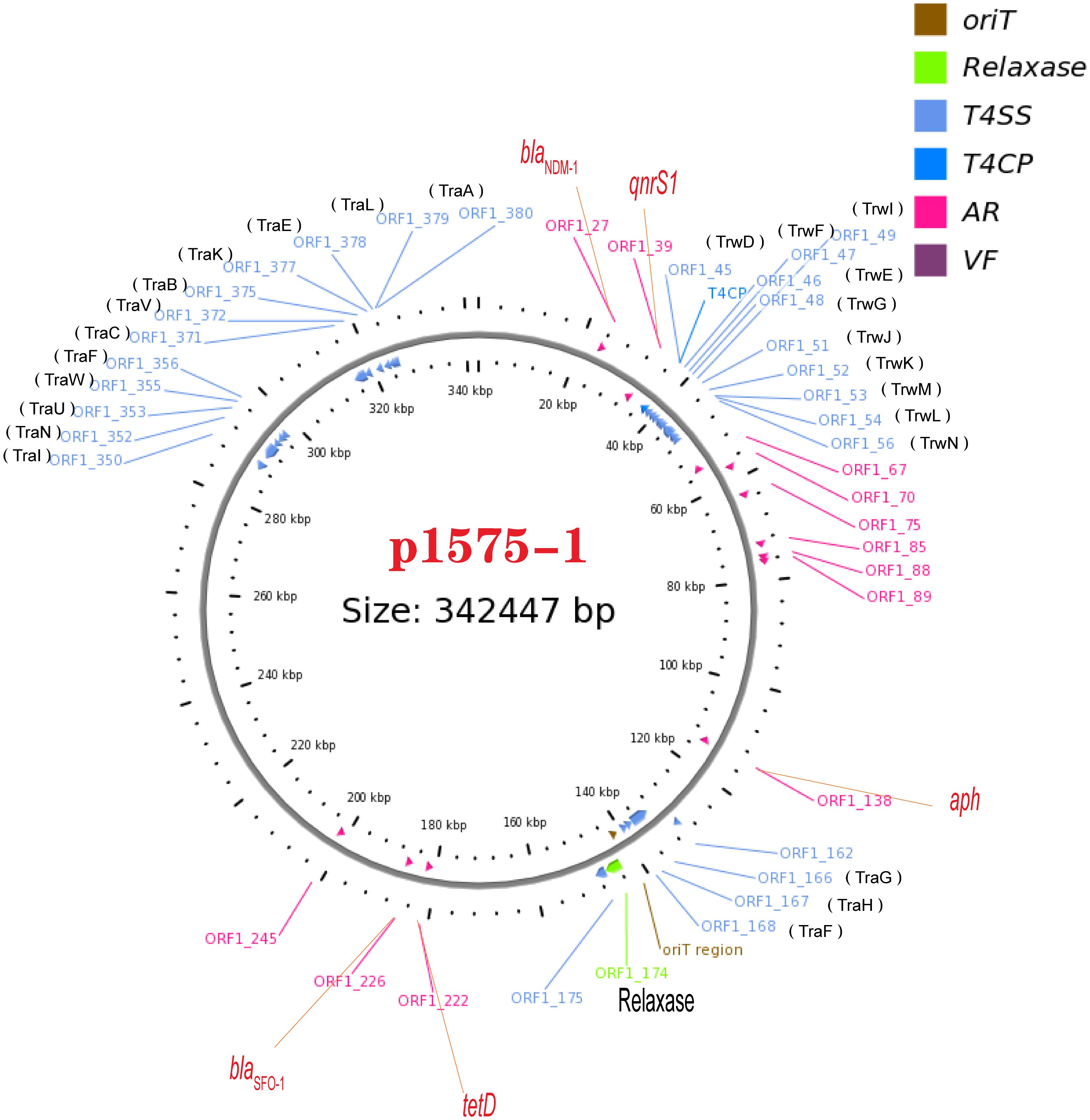
Figure 1. A conjugative plasmid p1575-1. AR (ARGs), acquired antibiotic resistance determinant genes; VF, virulence factors; ORF1-27, blaNDM–1; ORF1-39, qnrS1; ORF1-174, Relaxase; ORF1-222, tetD; ORF1-226, blaSFO–1.
Moreover, we obtained three plasmids from the National Center for Biotechnology Information (NCBI) GenBank database for comparative analysis with our target plasmids. We found that p1575-1 had high homology with pNIHE14-1904-mcr9 (GenBank accession no. LC570845.1) from E. hormaechei, with 91% query coverage and 99.97% sequence similarity. The other two plasmids held only 89% query coverage (pECL-90-2, CP061746.1) and 88% query coverage (pIHI2-323, CP049189.1) (Figure 2A). These results also suggested that these plasmids might have evolved from a single ancestor, or one might have evolved from the other. Finally, the pNIHE14-1904-mcr9 was chosen as the reference plasmid for genome analysis, because of the high query coverage and sequence similarity (Figure 2B). The conjugative system of p1575-1 shared greater than 99% identity to that of pNIHE14-1904-mcr9, which in turn confirmed that the p1575-1 plasmid was a conjugative IncHI2 plasmid. Moreover, two IS26 units were found on p1575-1. The first was the IS26–blaSFO–1–IS26 transposable unit containing the SFO-1 ESBL gene (blaSFO–1) (Figure 3A). The other was the IS26–blaLAP–2–qnrS1–IS26 module (Figure 3D) with two resistance genes included (blaLAP–2 and qnrS1). Remarkably, only p1575-1 plasmid harbored the second MDR gene IS26 unit compared with other three IncHI2 plasmids.
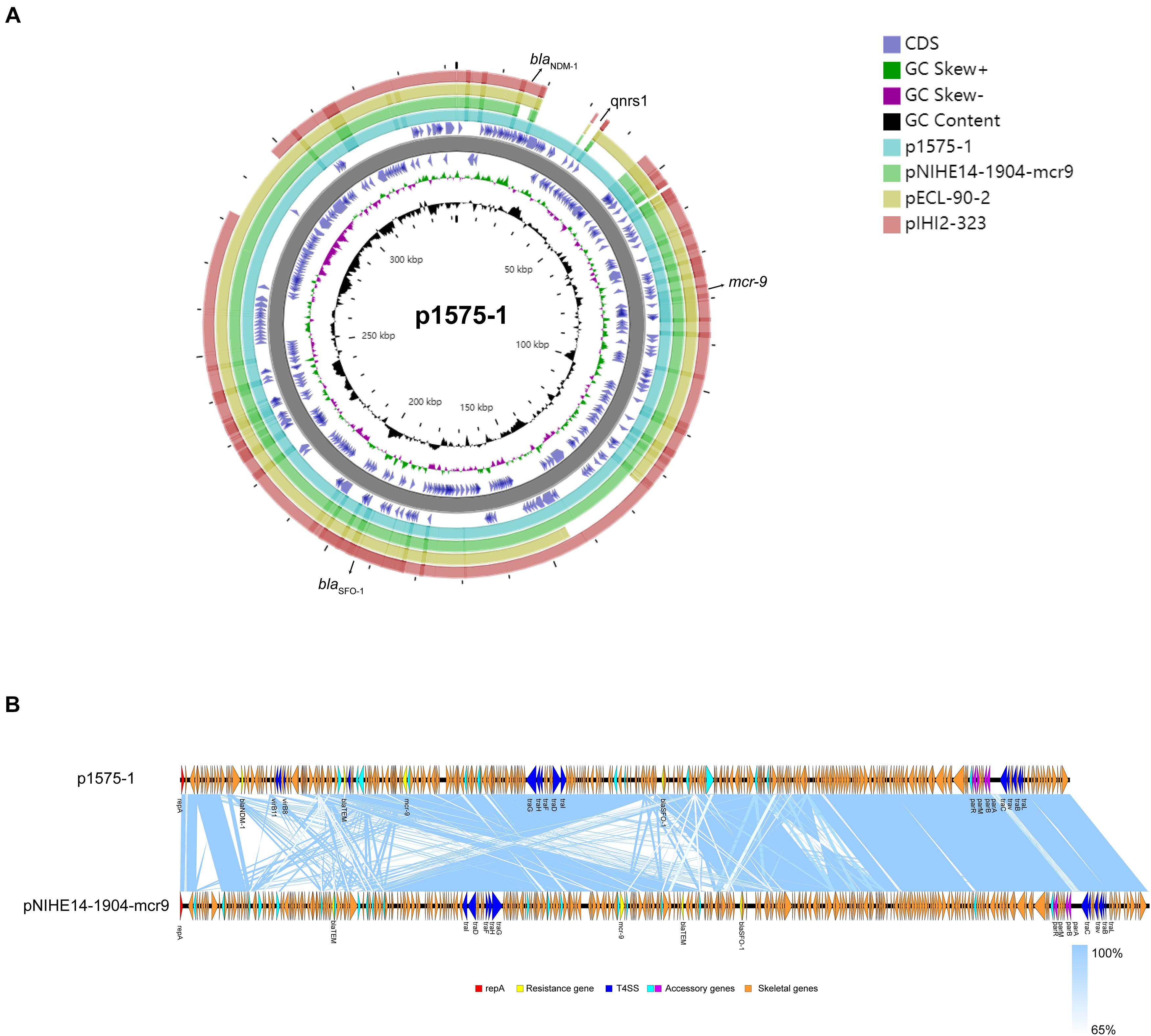
Figure 2. (A) Ring diagram representation of plasmid p1575-1. From the inside to the outside, the first circle represents the scale; the second circle represents GC content; the third circle represents the GC skew; the fourth and sixth circles represent the COG to which each CDS belongs; the fifth circle represents the backbone; the seventh to 10th circles represent p1575-1, pNIHE14-1904-mcr9, pECL-90-2, and pIHI2-323, respectively. GC, guanine + cytosine; blaSFO–1, extended-spectrum β-lactamases (ESBLs); blaNDM–1, New Delhi metallo-β-lactamase-1 gene; mcr-9, colistin resistance gene; qnrs1, fluoroquinolones gene. (B) Comparative analysis of the mcr-9-harboring plasmid characterized in this study with closely related plasmid pNIHE14-1904-mcr9. Open reading frames (ORFs) are portrayed by arrows and are depicted in different colors based on their predicted gene functions. The genes associated with the T4SS are indicated by dark blue arrows, while the genes involved in replication are indicated by red arrows. Resistance genes are indicated by yellow arrows, and accessory genes are indicated by light blue and purple arrows. Orange arrows represent the skeletal gene of the plasmid, and blue shading denotes shared regions of homology among different plasmids.
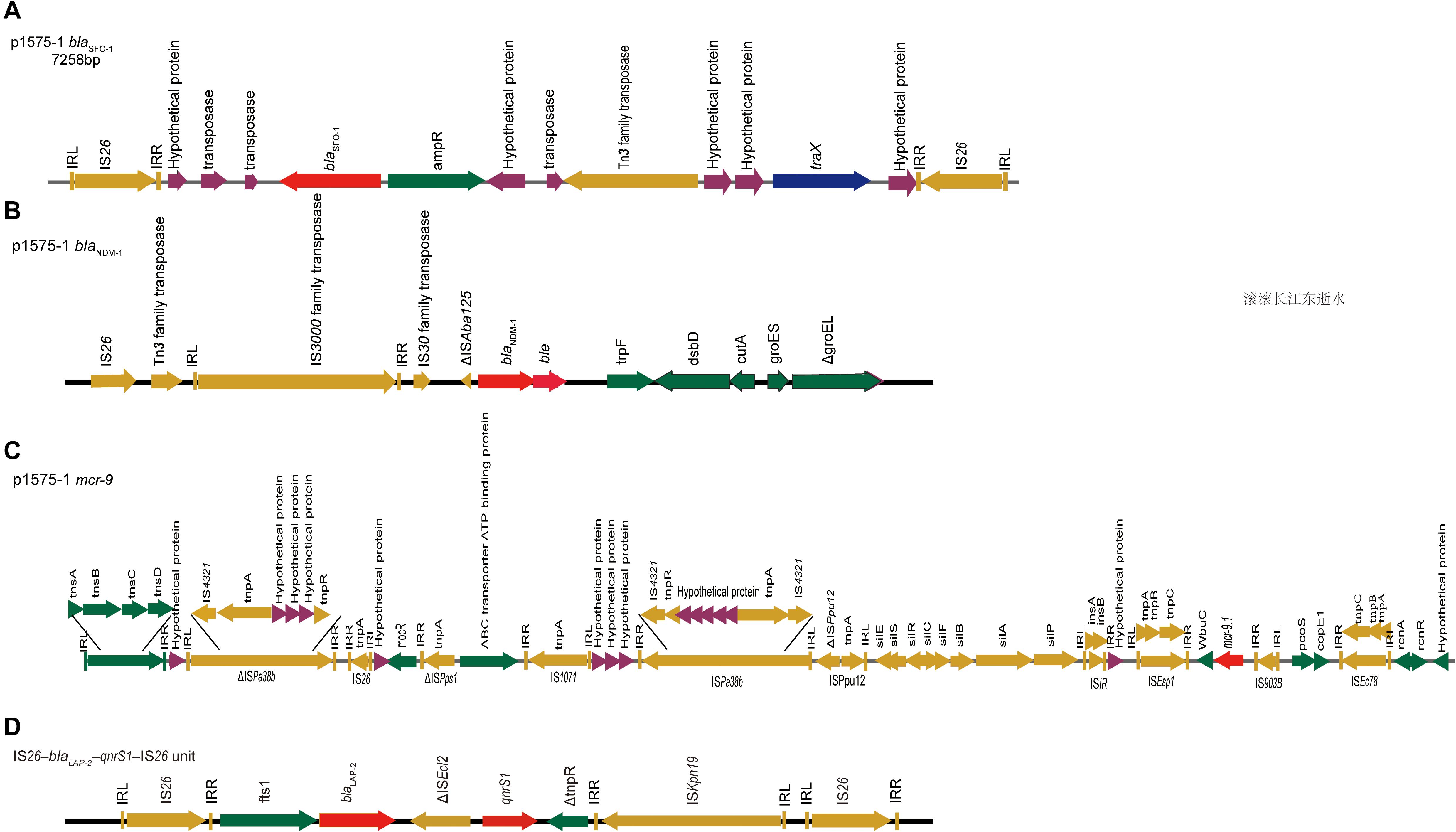
Figure 3. blaSFO–1, blaNDM–1, and mcr-9 gene contigs. Genetic environments surrounding the blaNDM–1, blaSFO–1 and mcr-9 genes in plasmid p1575-1. (A) The DNA fragments flanking the blaSFO–1 gene in plasmid p1575-1. (B) The DNA fragments flanking the blaNDM–1 gene in plasmid p1575-1. (C) The DNA fragments flanking the mcr-9 gene in plasmid p1575-1. (D) The IS26–blaLAP–2–qnrS1–IS26 module. Colored arrows indicate open reading frames, with dark green, dark yellow, dark blue, purple, and red arrows representing other genes, mobile and accessory elements, the individual conjugation-related genes, hypothetical proteins and transposases, and antibiotic resistance genes, respectively.
Overall, these findings revealed that the p1575-1 plasmid was an MDR conjugative plasmid, which carried three key resistance genes (blaSFO–1, blaNDM–1, and mcr-9) and complete conjugative systems.
p1575-1 Plasmid Could Transfer blaSFO–1, blaNDM–1, and mcr-9 Genes
We found that the p1575-1 plasmid carried complete conjugative systems. Hence, we applied the conjugation assay to prove whether the MDR p1575-1 plasmid could infect other strains autonomously by conjugation. We identified that p1575-1 was able to be transferred to the rifampicin-resistant Escherichia coli EC600 via conjugation, p1575-1-EC600; and the conjugation frequency was estimated at (0.5–2) × 10–6 per donor cell. Then S1-PFGE revealed that E. hormaechei 1575 and p1575-1-EC600 contained the large plasmid (p1575-1) (336.5–398.4 kb) (Figure 4), consistent with the result of WGS. Besides, another plasmid (named p1575-2) was also found by WGS, a small plasmid approximately 1,699 bp, with no resistance genes on and could not be detected by S1-PFGE. Transconjugants were subjected to susceptibility assays. The antimicrobial susceptibility patterns are shown in Table 2. The transconjugants showed similar antibiotic susceptibility profile to the donor strain E. hormaechei 1575. The MICs of transconjugants were decreased compared with those of 1575, but they were both sensitive to tigecycline.
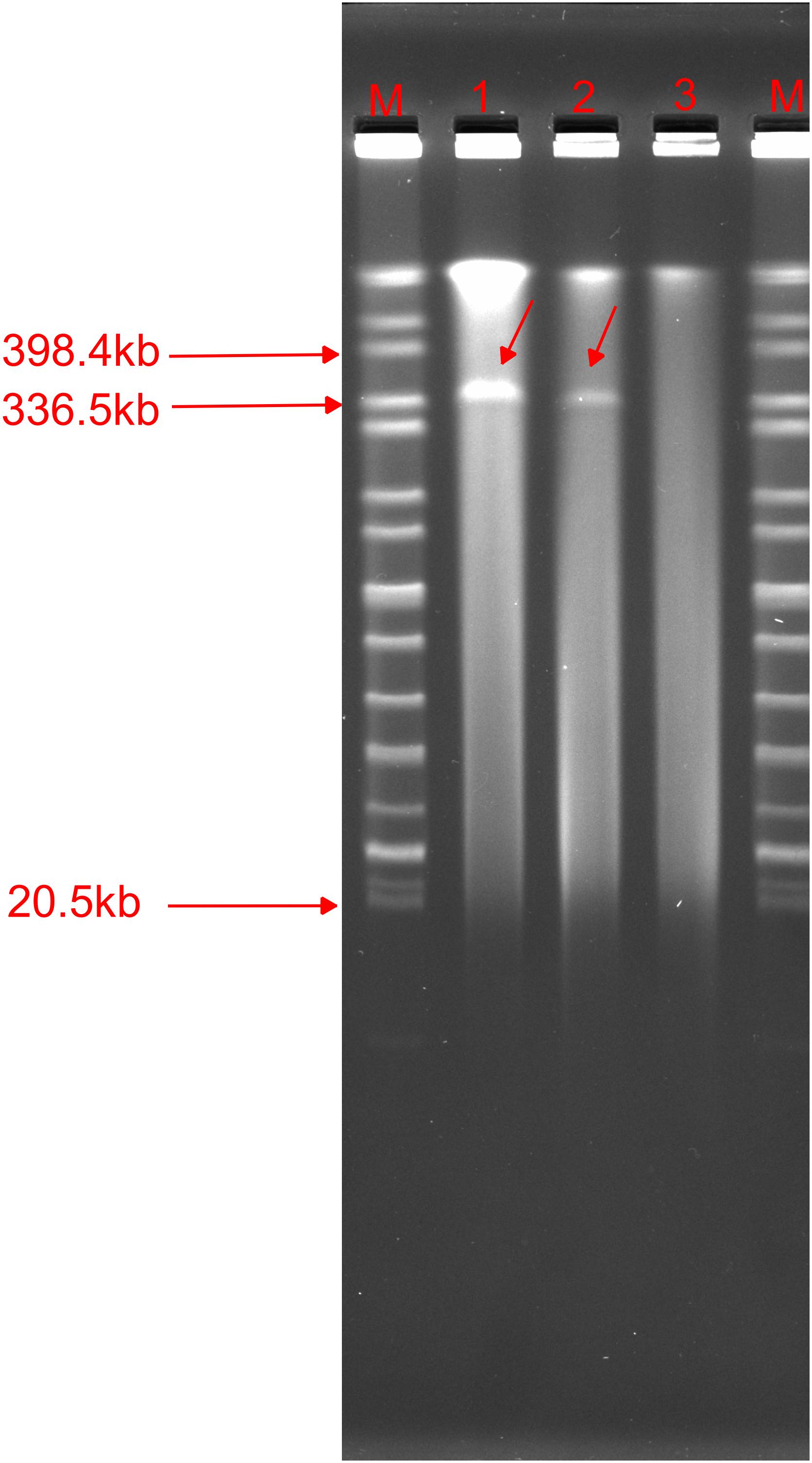
Figure 4. S1-nuclease pulsed-field gel electrophoresis profiles. M, Salmonella enterica serotype Braenderup strain H9812; 1,1575; 2, p1575-1-EC600; 3, EC600.
Mobile Genetic Elements Associated With blaSFO–1, blaNDM–1, and mcr-9
Besides an in-depth analysis of the characteristics of MDR plasmids, we also analyzed the mobile elements flanking the resistant genes.
Our results showed that blaSFO–1 was located on a 7,258-bp IS26 unit (IS26–traX-ampR-blaSFO–1-IS26) (Figure 3A). Genetic mapping of blaSFO–1 revealed that IS26 and ampR were upstream and downstream of blaSFO–1, respectively. Tn3 family, Tn3 transposase DDE domain protein, and IncF plasmid conjugative transfer pilin acetylase, traX, were located downstream of ampR. Genetic mapping of blaNDM–1 revealed that the insertion sequence IS3000 was interrupted by the insertion of a truncated ΔISAba125 element. A bleomycin resistance gene, bleMBL, and dsbD, encoded oxidoreductase superfamily protein, were downstream of blaNDM–1 (Figure 3B). For blaNDM–1, a high similar genetic pattern was also observed in other plasmids, pNDM-BTR (McGann et al., 2015) (GenBank accession number KF534788, IncN1) and pNDM1-CBG (accession number CP046118, unpublished). In plasmid 1575-1, IS5 family transposase (IS903B) was located upstream of mcr-9.1, whereas wbuC, IS1R, sil, mocR, IS26, and tnsDCBA were located downstream (Figure 3C). Besides, there were many other insertion sequences on the backbone where mcr-9 was located. However, qseB and qseC regulatory genes were not found in association with the mcr-9 gene.
Discussion
The spread of blaNDM-1 among bacterial pathogens is of concern not only because of resistance to carbapenems but also because such pathogens typically are resistant to multiple antimicrobial drug classes, which leaves few treatment choices available (Kumarasamy et al., 2010; Moellering, 2010; Walsh, 2010). Not like the blaNDM-1, which receives widespread attention, the blaSFO–1 gene is not included in the routine surveillance, but it could be an effective weapon that various gram-negative bacteria could use to resist β-lactams (Matsumoto and Inoue, 1999); therefore, the prevalence of the coexistence of the blaSFO–1 gene and carbapenemase genes might be underestimated. Some studies reported the coexistence of blaSFO–1 and blaNDM–1 β-lactamase genes and fosfomycin resistance gene fosA3 in Escherichia coli clinical isolate (Zhao et al., 2015) and the co-occurrence of mcr-9 and blaNDM–1 in Enterobacter cloacae (Yuan et al., 2019; Faccone et al., 2020; Lin et al., 2020). However, in this study, we not only found the coexistence of mcr-9 and blaNDM–1, but also a rare gene blaSFO–1 was detected on the same transferable plasmid. The presence of blaSFO–1 might confer resistance to more antibiotics. mcr is a family of genes found to promote colistin resistance in bacteria. As we all know, polymyxin antibiotic would be a good choice for blaNDM–1-positive strains, but we found mcr-9 (Börjesson et al., 2020) in E. hormaechei 1575, which could reduce the sensitivity of the strain to polymyxin and increase its clinical menacing. Notably, the novel antibiotic ceftazidime–avibactam was also ineffective against 1575. Tigecycline is a last-resort antibiotic that is used to treat severe infections caused by extensively drug-resistant bacteria (Tasina et al., 2011) and may be used as a therapeutic drug for 1575. All these results indicated the E. hormaechei 1575 was MDR isolates and could only choose limited antibiotics. The presence of drug resistance genes strongly correlated with resistant phenotypes. The E. hormaechei 1575 was confirmed to produce carbapenemase. At the same time, two cases of MDR E. cloacae isolates had been reported to be ST102 in China (Cao et al., 2017), and this kind of strain was also found in our study. High attention should be given to its subsequent epidemic.
Previous studies showed that multiple resistance transfer of plasmids could result from rare gene capture events mediated by different mobile genetic elements, clustering, and combinatorial evolution of resistance genes and related mobile elements (Partridge and Tsafnat, 2018). Through the WGS and comparative genomics, we clarified that the key to mediating the antibiotic resistance of this strain was the p1575-1 resistant plasmid. The p1575-1 identified in this study was an IncHI2 conjugative plasmid, representing one of the most frequently encountered plasmid types in Enterobacteriaceae (Carattoli, 2009). Notably, IncHI2 plasmids are also broad-host-range, large (>250 kb) conjugative plasmids that mobilize metal and drug resistance genes within gram-negative pathogens (Bertrand et al., 2006; Novais et al., 2006; Roy Chowdhury et al., 2019). Meanwhile, IncHI2-ST1 plasmids always contributed to the dissemination of carbapenemase-encoding genes and are also reported frequently to play a critical role in the evolution of complex resistance phenotypes within disease-causing strains of Enterobacteriaceae (Roy Chowdhury et al., 2019). Moreover, IncHI2 plasmids contain the conjugal transfer gene regions tra1 and tra2, likely contributing to the spread of resistance in the environment (Sherburne et al., 2000). In this study, we analyzed the conjugative modules of the p1575-1 plasmid and evaluated its mobility with conjugation assay. Like the classical IncHI2 plasmids, the p1575-1 plasmid held a complete conjugative system, and the conjugation frequencies ranged from 0.5 × 10–6 to 2 × 10–6 per donor cell. The IncHI2-type conjugative plasmids harboring mcr-9 were also discovered previously, and the conjugation frequencies of those plasmid were 10–4 (Lin et al., 2020) or 2.03 × 10–7 (−5.42 × 10–8) (Cha et al., 2020), which were similar to our findings. Through the analysis, we identified the complete conjugative modules on the plasmid p1575-1, strongly suggesting that p1575-1 could be transferred autonomously. In addition to the conjugative plasmids, the capture, accumulation, and dissemination of resistance genes are largely due to the actions of mobile genetic elements, including insertion sequences, transposons, gene cassettes, and integrons. In this study, we found that all these three resistance genes were flanked by several mobile elements. The blaSFO–1 was located in an IS26 composite transposon. IS6 family elements IS26 have played a pivotal role in the dissemination of resistance determinants in Gram-negative bacteria; thus, blaSFO–1 held the potential to transfer to other strains. AmpR, a class of DNA-binding regulatory protein, belongs to the LysR family of transcriptional regulators (Henikoff et al., 1988; Bartowsky and Normark, 1993). AmpR is confirmed to be a transcriptional activator in the presence of certain β-lactam antibiotics in the culture medium and a repressor in their absence (Lindberg et al., 1988). The presence of ampR seems to be a disadvantage for the host strain because E. cloacae become highly resistant to β-lactams (Matsumoto and Inoue, 1999). The movement of IS26 is originally demonstrated to occur by replicative transposition. Moreover, the blaSFO-1 genes in previous identifications were located on non-conjugative plasmids (Guo et al., 2012). In our study, the conjugative blaSFO–1-blaNDM–1-mcr-9-bearing plasmid belonged to IncHI2, which is a kind of broad-host-range mobile plasmid and might greatly accelerate the dissemination of the blaSFO–1 genes. Previous reports showed that the blaNDM–1 genes in Enterobacteriaceae were usually on 50- to 200-kb plasmids belonging to IncL/M, IncHI1, IncFIIs, IncF, or untypable (Ahmad et al., 2018). ISAba125 and Tn125 are always associated with the blaNDM–1 gene. Upstream of the blaNDM–1 gene, a truncated insertion sequence, ISAba125, was identified, which provides a promoter for the expression of blaNDM–1 (Carattoli et al., 2012), and the presence of ble andΔtnpA genes suggests a possible hypothesis that blaNDM–1 originates from Acinetobacter baumannii (Poirel et al., 2012; Toleman et al., 2012). Besides, phosphoribosylanthranilate isomerase gene trpF was identified in the downstream sequences of the bleMBL gene (Liu et al., 2013). In addition, qnrS1 in IS26–blaLAP–2–qnrS1–IS26 unit (3D) was also found, consistent with our AST results. In the IncHI2 plasmid, the mcr-9 allele always inserted an IS903B element and an ISEsp1, encoding a cupin fold metalloprotein, wbuC family (Yuan et al., 2019; Börjesson et al., 2020), which was consistent with our results. Because mcr-9.1 was located between IS903B and IS26, these flanking sequences can also be potentially transferred to other bacteria along with mcr-9.1. All results indicated that the resistant plasmid carried by E. hormaechei 1575 can be spontaneously transmitted to other strains through conjugation, which had great potential to cause clinical epidemics. qseB and qseC regulatory genes were found in association with the mcr-9 gene and played an important role in mediating polymyxin resistance (Chavda et al., 2019; Kieffer et al., 2019). The lack of two key regulators (qseB and qseC) may explain why E. hormaechei 1575 carrying mcr-9 did not exhibit a high resistance level to colistin (MIC, 2 μg/ml). Serious importance needs to be taken on this phenomenon.
In this study, all the resistant genes located on the p1575-1 plasmid were found to be chimeric with multiple IS sequences and various mobile elements, indicating that blaSFO–1, blaNDM–1, and mcr-9 could be transferred not only by the p1575-1 plasmid but also by these mobile genes.
Conclusion
In this study, we report the coexistence of blaSFO–1, blaNDM–1, and mcr-9 encoding one transferable IncHI2 plasmid in an E. hormaechei isolate. The co-occurrence of blaSFO–1, blaNDM–1, and mcr-9 (as well as many associated resistance genes) caused E. hormaechei 1575 to be highly resistant not only to carbapenems but also to novel antibiotic ceftazidime–avibactam. At the same time, enough attention should be given to the dissemination of colistin resistance genes mcr-9, as polymyxin has been considered to be the “last-resort” antibiotic to treat human infections caused by CRE. Yet more worryingly, these genes are associated with various mobile elements and conjugative plasmids. The presence of multiple mobile elements indicates that horizontal gene transfer events play a key role in the acquisition of antibiotic resistance and the evolution of plasmids. Future studies are necessary to evaluate the prevalence of these plasmids among clinical isolates in China and other countries.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
As the E. hormaechei clinical isolate in this study was part of the routine hospital laboratory procedure, the Ethics Committee of the Shanghai Pulmonary Hospital of Tongji University School of Medicine approved our study.
Author Contributions
All authors contributed to data analysis and drafting or revising the article, gave the final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Funding
This study was supported by grants from the Jiangxi Provincial Department of Science and Technology, China (20181BBG70030) covering the each section of this study, including the design of the study and collection, analysis, and interpretation of the data and manuscript preparation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the excellent technical assistance provided by Fangzhou Chen.
Footnotes
- ^ https://github.com/ruanjue/smartdenovo
- ^ https://cge.cbs.dtu.dk/services/ResFinder/
- ^ https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^ https://cge.cbs.dtu.dk/services/pMLST/
- ^ https://cge.cbs.dtu.dk/services/MLST/
- ^ https://bioinfo-mml.sjtu.edu.cn/oriTfinder/
- ^ https://www-is.biotoul.fr/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://stothard.afns.ualberta.ca/cgview_server/
- ^ http://mjsull.github.io/Easyfig/
References
Ahmad, N., Khalid, S., Ali, S., and Khan, A. (2018). blaOccurrence of Variants Among Enterobacteriaceae from a neonatal intensive care unit in a Northern India hospital. Front. Microbiol. 9:407. doi: 10.3389/fmicb.2018.00407
Bartowsky, E., and Normark, S. (1993). Interactions of wild-type and mutant AmpR of Citrobacter freundii with target DNA. Mol. Microbiol. 10, 555–565. doi: 10.1111/j.1365-2958.1993.tb00927.x
Bertrand, S., Weill, F., Cloeckaert, A., Vrints, M., Mairiaux, E., Praud, K., et al. (2006). Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 44, 2897–2903. doi: 10.1128/jcm.02549-05
Börjesson, S., Greko, C., Myrenas, M., Landén, A., Nilsson, O., and Pedersen, K. (2020). A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying bla from horses in Sweden. J. Glob. Antimicrob. Resist. 20, 285–289. doi: 10.1016/j.jgar.2019.08.007
Cao, X., Cheng, L., Zhang, Z., Ning, M., Zhou, W., Zhang, K., et al. (2017). Survey of clinical extended-spectrum beta-lactamase-producing enterobacter cloacae isolates in a Chinese tertiary hospital, 2012-2014. Microb. Drug. Resist. (Larchmont, N.Y.) 23, 83–89. doi: 10.1089/mdr.2015.0128
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/aac.01707-08
Carattoli, A., Villa, L., Poirel, L., Bonnin, R., and Nordmann, P. (2012). Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 56, 783–786. doi: 10.1128/aac.05116-11
Cha, M. H., Woo, G. J., Lee, W., Kim, S. H., Woo, J. H., Kim, J., et al. (2020). Emergence of transferable mcr-9 gene-carrying colistin-resistant Salmonella enterica Dessau ST14 isolated from retail chicken meat in Korea. Foodborne Pathog Dis. 17, 720–727. doi: 10.1089/fpd.2020.2810
Chavda, K. D., Westblade, L. F., Satlin, M. J., Hemmert, A. C., Castanheira, M., Jenkins, S. G., et al. (2019). First report of bla (VIM-4)- and mcr-9-Coharboring Enterobacter species isolated from a pediatric patient. mSphere 4:e00629-19. doi: 10.1128/mSphere.00629-19
Chen, L., Mathema, B., Chavda, K., DeLeo, F., Bonomo, R., and Kreiswirth, B. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
Chen, W., Hu, Z., Wang, S., Huang, D., Wang, W., Cao, X., et al. (2020). Characterization of a clinical Enterobacter hormaechei strain belonging to epidemic clone ST418 co-carrying blaNDM-1, blaIMP-4 and mcr-9.1. bioRxiv [preprint] 2020.2009.2025.314500. doi: 10.1101/2020.09.25.314500
Chen, Y., Zhou, Z., Jiang, Y., and Yu, Y. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemothe. 66, 1255–1259. doi: 10.1093/jac/dkr082
Clinical and Laboratory Standards Institute (CLSI) (2020). Performance Standards for Antimicrobial Susceptibility Testing. CLSI supplement M100, 30th Edn. Wayne: Clinical and Laboratory Standards Institute.
Dortet, L., Poirel, L., and Nordmann, P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed. Res. Int. 2014:249856. doi: 10.1155/2014/249856
EUCAST (2020). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. Available online at: http://www.eucast.org (accessed January 20, 2020).
Faccone, D., Martino, F., Albornoz, E., Gomez, S., Corso, A., and Petroni, A. (2020). Plasmid carrying mcr-9 from an extensively drug-resistant NDM-1-producing Klebsiella quasipneumoniae subsp. quasipneumoniae clinical isolate. Infect. Genet. Evol. : J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 81:104273. doi: 10.1016/j.meegid.2020.104273
Guo, Q., Wang, P., Ma, Y., Yang, Y., Ye, X., and Wang, M. (2012). Co-production of SFO-1 and DHA-1 β-lactamases and 16S rRNA methylase ArmA in clinical isolates of Klebsiella pneumoniae. J. Antimicrob. Chemother. 67, 2361–2366. doi: 10.1093/jac/dks244
Gurjar, M. (2015). Colistin for lung infection: an update. J. Intens. Care 3:3. doi: 10.1186/s40560-015-0072-9
Hackl, T., Hedrich, R., Schultz, J., and Förster, F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics (Oxford, England) 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Henikoff, S., Haughn, G., Calvo, J., and Wallace, J. (1988). A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. U.S.A. 85, 6602–6606. doi: 10.1073/pnas.85.18.6602
Kieffer, N., Royer, G., Decousser, J. W., Bourrel, A. S., Palmieri, M., Ortiz De La Rosa, J. M., et al. (2019). mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63, e965–e919. doi: 10.1128/aac.00965-19
Kumarasamy, K., Toleman, M., Walsh, T., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/s1473-3099(10)70143-2
Lin, M., Yang, Y., Yang, Y., Chen, G., He, R., Wu, Y., et al. (2020). mcr-9Co-occurrence of and in isolated from a patient with bloodstream infection. Infect. Drug Resist. 13, 1397–1402. doi: 10.2147/idr.S248342
Lindberg, F., Lindquist, S., and Normark, S. (1988). Genetic basis of induction and overproduction of chromosomal class I beta-lactamase in nonfastidious gram-negative bacilli. Rev. Infect. Dis. 10, 782–785. doi: 10.1093/clinids/10.4.782
Liu, Y., Wang, Y., Walsh, T., Yi, L., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Liu, Z., Li, W., Wang, J., Pan, J., Sun, S., Yu, Y., et al. (2013). Identification and characterization of the first Escherichia coli strain carrying NDM-1 gene in China. PLoS ONE 8:e66666. doi: 10.1371/journal.pone.0066666
Matsumoto, Y., and Inoue, M. (1999). Characterization of SFO-1, a plasmid-mediated inducible class A beta-lactamase from Enterobacter cloacae. Antimicrob. Agents Chemother. 43, 307–313. doi: 10.1128/aac.43.2.307
McGann, P., Snesrud, E., Ong, A., Appalla, L., Koren, M., Kwak, Y., et al. (2015). War wound treatment complications due to transfer of an IncN plasmid harboring bla(OXA-181) from Morganella morganii to CTX-M-27-producing sequence type 131 Escherichia coli. Antimicrob. Agents Chemother. 59, 3556–3562. doi: 10.1128/aac.04442-14
Moellering, R. (2010). NDM-1–a cause for worldwide concern. New England J. Med. 363, 2377–2379. doi: 10.1056/NEJMp1011715
Nang, S., Li, J., and Velkov, T. (2019). mcrThe rise and spread of plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841x.2018.1492902
Nordmann, P., Poirel, L., Carrër, A., Toleman, M., and Walsh, T. (2011). How to detect NDM-1 producers. J. Clin. Microbiol. 49, 718–721. doi: 10.1128/jcm.01773-10
Novais, A., Cantón, R., Valverde, A., Machado, E., Galán, J., Peixe, L., et al. (2006). Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob. Agents Chemother. 50, 2741–2750. doi: 10.1128/aac.00274-06
Olaitan, A., Morand, S., and Rolain, J. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Partridge, S., and Tsafnat, G. (2018). Automated annotation of mobile antibiotic resistance in Gram-negative bacteria: the Multiple Antibiotic Resistance Annotator (MARA) and database. J. Antimicrob. Chemother. 73, 883–890. doi: 10.1093/jac/dkx513
Poirel, L., Bonnin, R., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/aac.05620-11
Queenan, A., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/cmr.00001-07 table of contents,
Roy Chowdhury, P., Fourment, M., DeMaere, M., Monahan, L., Merlino, J., Gottlieb, T., et al. (2019). Identification of a novel lineage of plasmids within phylogenetically diverse subclades of IncHI2-ST1 plasmids. Plasmid 102, 56–61. doi: 10.1016/j.plasmid.2019.03.002
Sherburne, C., Lawley, T., Gilmour, M., Blattner, F., Burland, V., Grotbeck, E., et al. (2000). The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28, 2177–2186. doi: 10.1093/nar/28.10.2177
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/s1473-3099(17)30753-3
Tasina, E., Haidich, A. B., Kokkali, S., and Arvanitidou, M. (2011). Efficacy and safety of tigecycline for the treatment of infectious diseases: a meta-analysis. Lancet Infect. Dis. 11, 834–844. doi: 10.1016/s1473-3099(11)70177-3
Toleman, M., Spencer, J., Jones, L., and Walsh, T. (2012). blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 2773–2776. doi: 10.1128/aac.06297-11
Walsh, T. (2010). Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents, 36S3, S8–S14. doi: 10.1016/s0924-8579(10)70004-2
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
WHO. (2017). WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Geneva: World Health Organization.
Xu, Y., Liu, Y., Liu, Y., Pei, J., Yao, S., and Cheng, C. (2015). Bacteriophage therapy against Enterobacteriaceae. Virol. Sin. 30, 11–18. doi: 10.1007/s12250-014-3543-6
Yilmaz, G., Dizbay, M., Guven, T., Pullukcu, H., Tasbakan, M., Guzel, O., et al. (2016). Risk factors for infection with colistin-resistant gram-negative microorganisms: a multicenter study. Ann. Saudi Med. 36, 216–222. doi: 10.5144/0256-4947.2016.216
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J., et al. (2019). Enterobacter hormaecheiCoproduction Of MCR-9 And NDM-1 By colistin-resistant isolated from bloodstream infection. Infect. Drug Resist. 12, 2979–2985. doi: 10.2147/idr.S217168
Zhao, J., Zhu, Y., Li, Y., Mu, X., You, L., Xu, C., et al. (2015). Coexistence of SFO-1 and NDM-1 β-lactamase genes and fosfomycin resistance gene fosA3 in an Escherichia coli clinical isolate. FEMS Microbiol. Lett. 362, 1–7. doi: 10.1093/femsle/fnu018
Zhou, K., Zhou, Y., Zhang, C., Song, J., Cao, X., Yu, X., et al. (2020). Dissemination of a ‘rare’ extended-spectrum β-lactamase gene bla(SFO-1) mediated by epidemic clones of carbapenemase-producing Enterobacter hormaechei in China. Int J Antimicrob. Agents 56:106079. doi: 10.1016/j.ijantimicag.2020.106079
Keywords: Enterobacter hormaechei, plasmid, blaSFO–1, blaNDM–1, mcr-9, IncHI2, WGS, mobile elements
Citation: Ai W, Zhou Y, Wang B, Zhan Q, Hu L, Xu Y, Guo Y, Wang L, Yu F and Li X (2021) First Report of Coexistence of blaSFO–1 and blaNDM–1 β-Lactamase Genes as Well as Colistin Resistance Gene mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter hormaechei. Front. Microbiol. 12:676113. doi: 10.3389/fmicb.2021.676113
Received: 04 March 2021; Accepted: 27 April 2021;
Published: 18 June 2021.
Edited by:
Che-Hsin Lee, National Sun Yat-sen University, TaiwanReviewed by:
Linda Falgenhauer, University of Giessen, GermanyCheng-Yen Kao, National Yangming University, Taiwan
Copyright © 2021 Ai, Zhou, Wang, Zhan, Hu, Xu, Guo, Wang, Yu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangxing Wang, Mzg4MDVAMTYzLmNvbQ==; Fangyou Yu, d3pqeHlmeUAxNjMuY29t; Xiaolong Li, bGl4aWFvbG9uZ2NxQDE2My5jb20=
†These authors have contributed equally to this work
 Wenxiu Ai
Wenxiu Ai Ying Zhou
Ying Zhou Bingjie Wang
Bingjie Wang Qing Zhan2,3
Qing Zhan2,3 Fangyou Yu
Fangyou Yu