- 1Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China, Ministry of Education, Southwest Forestry University, Kunming, China
- 2State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 3College of Life Science, University of Chinese Academy of Sciences, Beijing, China
- 4College of Plant Protection, Jilin Agricultural University, Changchun, China
- 5College of Biodiversity Conservation, Southwest Forestry University, Kunming, China
Four new species within the genus Absidia, A. globospora, A. medulla, A. turgida, and A. zonata, are proposed based on a combination of morphological traits, physiological features, and molecular evidences. A. globospora is characterized by globose sporangiospores, a 1.0- to 3.5-μm-long papillary projection on columellae, and sympodial sporangiophores. A. medulla is characterized by cylindrical to oval sporangiospores, a 1.0- to 4.5-μm-long bacilliform projection on columellae, and spine-like rhizoids. A. turgida is characterized by variable sporangiospores, up to 9.5-μm-long clavate projections on columellae, and swollen top of the projection and inflated hyphae. A. zonata is characterized by cylindrical to oval sporangiospores, a 2.0- to 3.5-μm-long spinous projection on columellae, and as many as eight whorled sporangiophores. Phylogenetic analyses based on sequences of internal transcribed spacer rDNA and D1–D2 domains of LSU rDNA support the novelty of these four species within the Absidia. All new species are illustrated, and an identification key to all the known species of Absidia in China is included.
Introduction
The genus Absidia Tiegh. (Cunnighamellaceae, Mucorales, Mucoromycetes, Mucoromycota) was proposed by van Tieghem (1876). Absidia members are ubiquitous in soil and also often associated with warm decaying plant matter, such as compost heaps. Some Absidia can be used to produce chitin, chitosan, and chitooligosaccharides (Kaczmarek et al., 2019) and hydrocortisone (Chen et al., 2020). Absidia species typically have sporangiophores arising from stolons, rhizoids never opposite the sporangiophores, pyriform sporangia and their deliquescent wall, obvious apophyses, a septum beneath the sporangium, and zygospores surrounded by appendages from the suspensors (Hoffmann et al., 2007; Hoffmann, 2010).
The classification and circumscription of the Absidia have been debated since it was described. According to zygospore morphology, Hesseltine and Ellis (1964) divided Absidia into two subgenera, the subgenus Absidia and the subgenus Mycocladus (Beauverie) Hesselt. & J.J. Ellis. Different from the former, the subgenus Mycocladus does not form any appendages from the suspensors of zygospores. This classification framework was followed by Schipper (1990), who further divided the subgenus Absidia into six groups. However, this kind of delimitation are not accepted nowadays, and six genera are synonymized with the genus Absidia instead (Hawksworth et al., 1995). They are Tieghemella Berl. & De Toni 1888, Mycocladus Beauverie 1900, Lichtheimia Vuill 1903, Proabsidia Vuill 1903, Pseudoabsidia Bainier 1903, and Protoabsidia Naumov 1935. Among these synonyms, Lichtheimia, Mycocladus, Pseudoabsidia, and Protoabsidia lack appendages.
Recently, a combined study of molecular phylogenetics, morphology, and physiology has provided a more reliable delimitation among Absidia species (Hoffmann et al., 2007), where Absidia was classified into three groups: (1) the thermotolerant species with an optimal growth temperature of 37–45°C, which were then transferred into the genus Lichtheimia (Hoffmann et al., 2009a); (2) the mesophilic species with an optimal growth temperature of 25–34°C, which have been accepted up to now as Absidia sensu stricto; and (3) the mycoparasitic species, potential to parasitize other mucoralean hosts with optimal growth temperatures below 30°C, which were then transferred into the genus Lentamyces Kerst. Hoffm. & K. Voigt (Hoffmann and Voigt, 2008). Currently, 37 species have been reported worldwide in Absidia (Hesseltine and Ellis, 1961, 1964, 1966; Ellis and Hesseltine, 1965, 1966; Index Fungorum1). Among these species, 13 were reported in the last decade using the strategy of combing morphology, physiology, and phylogeny: Absidia caatinguensis D.X. Lima and A.L. Santiago, Absidia cornuta D.X. Lima, C.A. de Souza, H.B. Lee, and A.L. Santiago; Absidia jindoensis Hyang B. Lee, and T.T.T. Nguyen; Absidia koreana Hyang B. Lee, Hye W. Lee, and T.T. Nguyen; Absidia multispora T.R.L. Cordeiro, D.X. Lima, Hyang B. Lee, and A.L. Santiago; Absidia panacisoli T. Yuan Zhang, Ying Yu, He Zhu, S.Z. Yang, T.M. Yang, Meng Y. Zhang, and Yi X. Zhang; Absidia pararepens Jurjeviæ, M. Kolaøík, and Hubka; Absidia pernambucoensis D.X. Lima, C.M. Souza-Motta, and A.L. Santiago; Absidia saloaensis T.R.L. Cordeiro, D.X. Lima, Hyang B. Lee, and A.L. Santiago; Absidia stercoraria Hyang B. Lee, H.S. Lee, and T.T.T. Nguyen; Absidia terrestris Rosas de Paz, Dania García, Guarro, Cano, and Stchigel; Absidia bonitoensis C.L. Lima, D.X. Lima, Hyang B. Lee, and A.L. Santiago; and Absidia ovalispora H. Zhao and X.Y. Liu (Ariyawansa et al., 2015; Li et al., 2016; Crous et al., 2018, 2020; Wanasinghe et al., 2018; Zhang et al., 2018; Cordeiro et al., 2020; Lima et al., 2020; de Lima et al., 2021; Zhao et al., 2021), and nine species have been recorded in China (Zhang et al., 2018; Zheng and Liu, 2018; Zhao et al., 2021).
Recently, seven strains of Absidia were collected from China but could not be assigned to any described species. Herein, morphological, physiological, and molecular phylogenetics [internal transcribed spacer (ITS) and D1–D2 domains of LSU rDNA] are presented to support them to four new species in Absidia sensu stricto, and consequently, a revised synoptic key to all the 13 known species of Absidia in China is provided.
Materials and Methods
Isolation and Strains
Strains were isolated from the soil collected in Hubei province, Shanxi province, Xinjiang province, and Yunnan province, China. Soil samples (1 g) were suspended in 100 mL sterilized water and shaken vigorously. Then, a 100 μL of the suspension was added onto a potato dextrose agar (PDA; Benny, 2008) plate with antibiotics streptomycin sulfate (100 mg/mL) and ampicillin (100 mg/mL). The plate was incubated at 27°C and examined daily with a stereo microscope (SMZ1500, Nikon Corporation, Japan). Upon the presence of colonies, a single colony was picked and transferred to new PDA plates. Living cultures were deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC). Dried cultures were deposited in the Herbarium Mycologicum Academiae Sinicae, Beijing, China (HMAS).
Morphology and Growth Experiments
Pure cultures were established in triplicate, respectively, with malt extract agar (MEA; Benny, 2008), modified synthetic mucor agar (SMA; Zheng and Chen, 2001), and PDA plates. For morphological observation, they were incubated at 27°C for 4–7 days and examined daily under a microscope (Axio Imager A2, Carl Zeiss Microscopy, Germany). For determining maximum growth temperatures, pure cultures were initially incubated at 32°C for 4 days, and then the incubation temperature was adjusted until the colonies stopped growing. The color of colonies was designated according to Ridgway (1912).
DNA Extraction, Polymerase Chain Reaction Amplification, and Sequencing
Mycelia were grown at 27°C for 5 days on PDA plates, and then cell DNAs were extracted using a kit (GO-GPLF-400, GeneOnBio Corporation, Changchun, China). The ITS and D1–D2 domain of LSU rDNA were amplified with primer pairs NS5M and LR5M (Wang et al., 2014). The polymerase chain reaction (PCR) procedure was as follows: an initial temperature at 95°C for 5 min; then 30 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 60 s, and extension at 72°C for 60 s; and finally an extra extension at 72°C for 10 min. PCR products were purified and then sequenced with primers ITS5 (White et al., 1990) and LR5M at BGI Tech Solutions Beijing Liuhe Co., Limited, Beijing, China. All newly generated sequences were deposited in GenBank and National Microbiology Data Center (NMDC,2 Table 1).
Phylogenetic Analyses
The software platform Geneious 8.13 was used to assemble and proofread DNA sequences. All the sequences were realigned using AliView version 3.0 (Larsson, 2014). The sequence alignments and phylogenetic trees were deposited at TreeBase (submission ID 27734). Sequences of Cunninghamella elegans and Cunninghamella blakesleeana retrieved from GenBank were used as outgroups in the ITS and LSU analyses following Hoffmann et al. (2007).
Phylogenetic analyses were carried out using maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI). MP phylogenetic analyses followed Zhao and Wu (2017), and the tree construction was performed in PAUP∗ version 4.0b10 (Swofford, 2002). All characters were equally weighted, and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1,000 random sequence additions. Max-trees were set to 5,000; branches of zero length were collapsed, and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap analysis with 1,000 replicates (Felsenstein, 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled CI (RC), and homoplasy index (HI) were calculated for each maximum parsimonious tree generated.
Maximum likelihood phylogenetic analyses were conducted with raxmlGUI 2.0 beta (Edler et al., 2020). A general time reversible model was used with a gamma-distributed rate variation (GTR + G) and 1,000 bootstrap replicates.
Bayesian inference phylogenetic analyses was calculated with MrBayes 3.2.7a by a general time-reversible model with an estimate of the proportion of invariant sites and a gamma distribution for variable rates across sites (GTR + I + G; Ronquist et al., 2012). Four Markov chains were run simultaneously from random starting trees, for 2,400,000 generations (ITS) or 300,000 generations (LSU). Trees were sampled every 100 generations. The chains stopped once the average standard deviation of split frequencies decreased lower than 0.01. The first one-fourth generations were discarded as burn-in. A majority rule consensus tree of all remaining trees was calculated. Branches were considered as significantly supported if they received ML bootstrap > 75%, MP bootstrap > 75%, or Bayesian posterior probabilities > 0.95.
Results
Phylogenetic Analyses
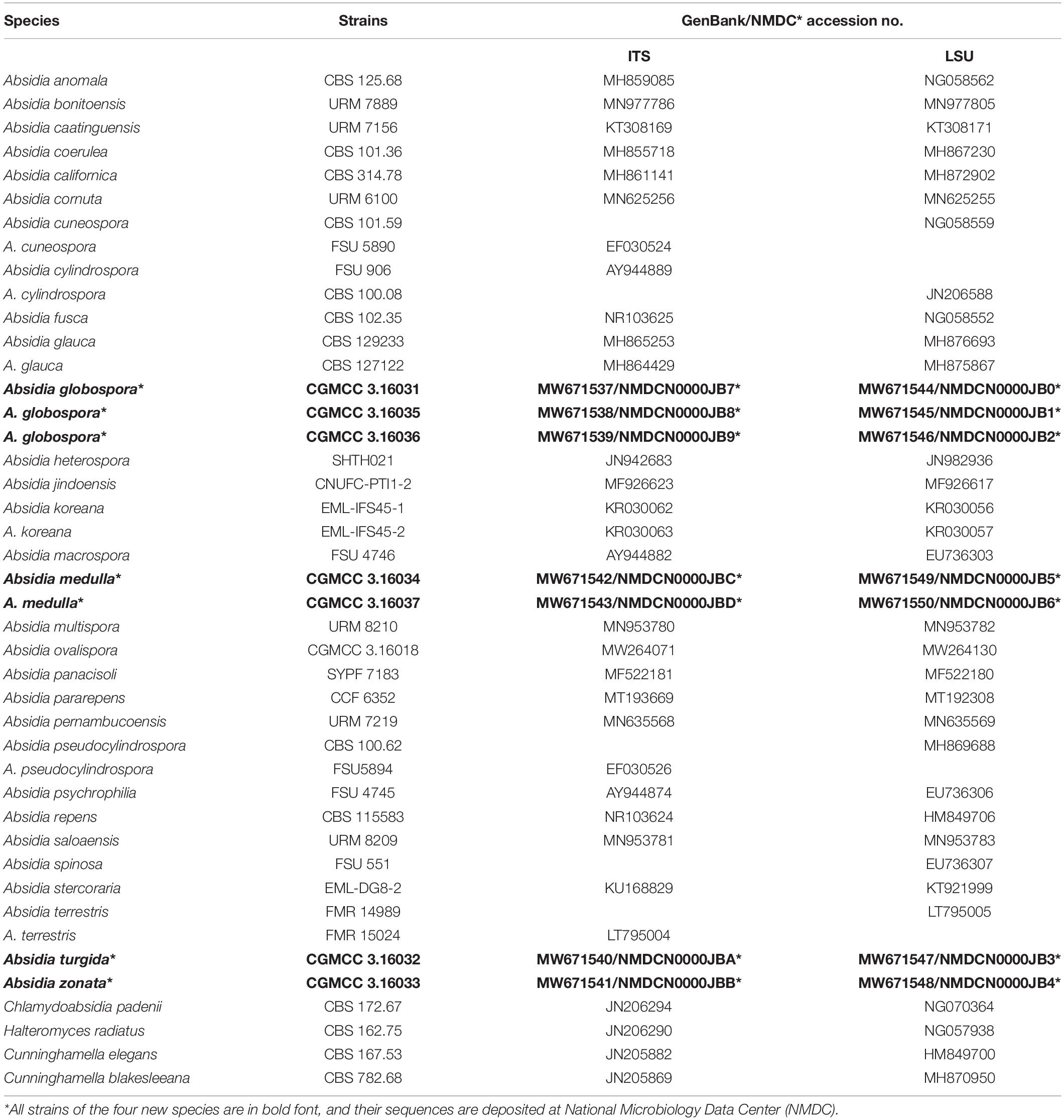
The ITS dataset included sequences from 38 strains representing 33 species of Absidia and related genera. The dataset had an aligned length of 903 characters, of which 248 characters were constant, 136 were variable and parsimony-uninformative, and 519 were parsimony-informative. MP analyses yielded two equally parsimonious trees (TL = 4163, CI = 0.3394, HI = 0.6606, RI = 0.3446, RC = 0.1170). At the end of the inference, the average standard deviation of split frequencies was 0.009990. All BI, ML, and MP phylogenetic trees resulted in similar topologies. The phylogram (Figure 1) consists of three clades, although with relatively low support values: (1) except the A. pararepens and A. bonitoensis, all members in the cylindrospora clade produce cylindric sporangiospores; (2) all members in the globospora clade produce globose sporangiospores; and (3) the Absidia cuneospora G.F. Orr & Plunkett separately groups as a cuneospora clade, forming conical sporangiospores (Orr and Plunkett, 1959).
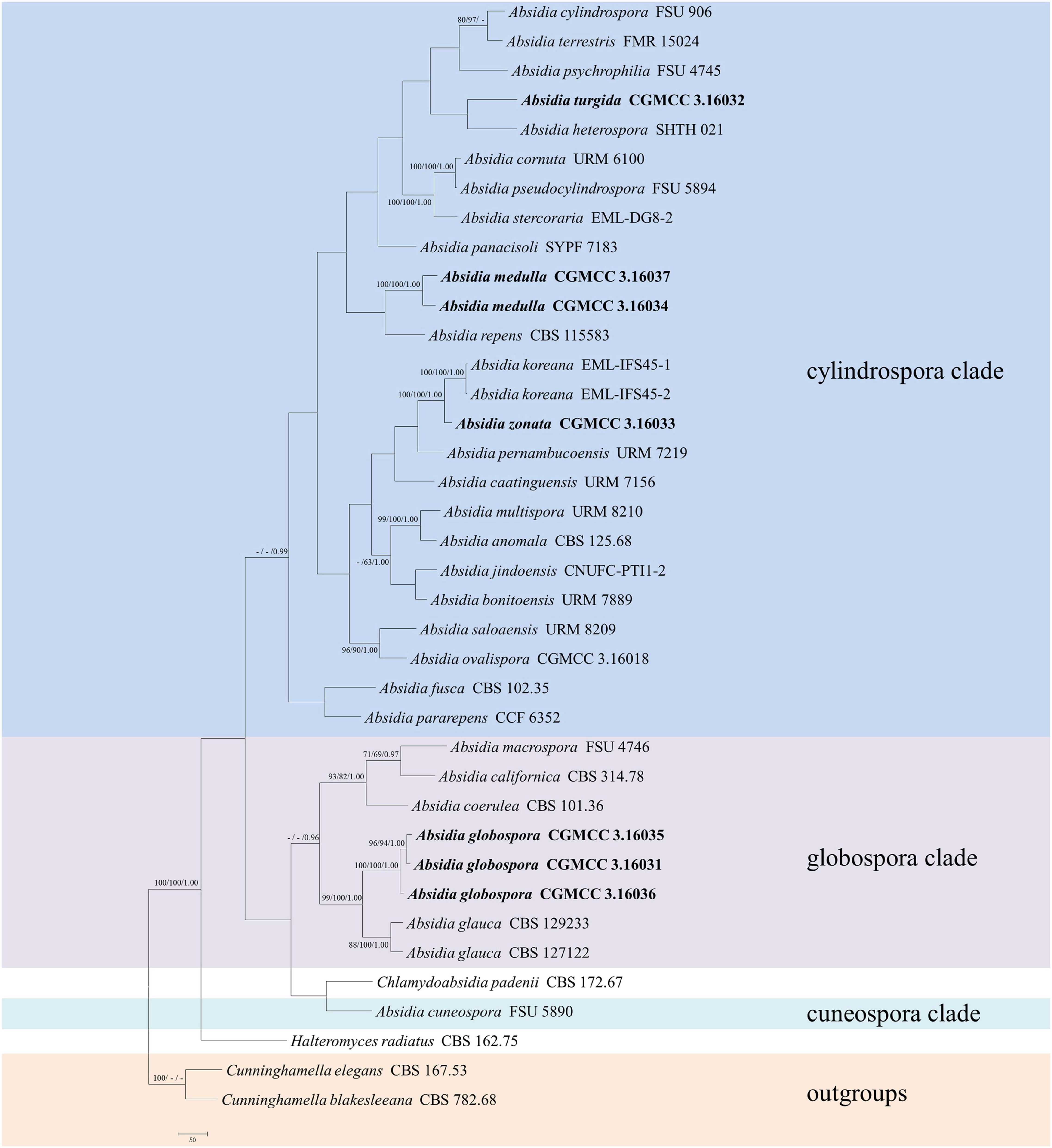
Figure 1. The maximum parsimony strict consensus tree illustrating the phylogeny of four new species of Absidia and related species in Cunninghamellaceae based on ITS sequences. Cunninghamella elegans and Cunninghamella blakesleeana serve as outgroups. Branches are labeled with maximum likelihood bootstrap values higher than 70%, maximum parsimony bootstrap values higher than 50%, and Bayesian posterior probabilities more than 0.95. The lower left scale represents steps.
The LSU dataset included sequences from 39 strains representing 34 species within Absidia. The dataset had an aligned length of 967 characters, of which 562 characters were constant, 117 were variable and parsimony-uninformative, and 288 were parsimony-informative. MP analyses yielded 20 equally parsimonious trees (TL = 1422, CI = 0.4339, HI = 0.5661, RI = 0.6443, RC = 0.2795). At the end of the inference, the average standard deviation of split frequencies was 0.009767. All BI, ML, and MP phylogenetic trees resulted in similar topologies. The phylogram (Figure 2) consists of three clades, similar to the ITS phylogram (Figure 1) but with relatively high support values, in detail, clade cylindrospora, globospora, and cuneospora with a support of 80/-/0.99, 98/98/1.00, and 100/100/1.00, respectively.
Figure 2. The maximum parsimony strict consensus tree illustrating the phylogeny of four new species of Absidia and related species in Absidia based on LSU sequences. Cunninghamella elegans and Cunninghamella blakesleeana serve as outgroups. Branches are labeled with maximum likelihood bootstrap values higher than 70%, maximum parsimony bootstrap values higher than 50%, and Bayesian posterior probabilities more than 0.95. The lower left scale represents steps.
Taxonomic Treatments
Absidia globospora T.K. Zong & X.Y. Liu, sp. nov.
Fungal names: FN570833 (Figures 3, 4).

Figure 3. Morphologies of Absidia globospora CGMCC 3.16031. (A) Sporangium; (B,C) columellae; (D) sporangiospores; (E,F) rhizoids; (G) swelling on sporangiospores; (H) sympodial sporangiophores. Scale bars: (A–C,E–G) 20 μm; (D) 5 μm; (H) 100 μm.
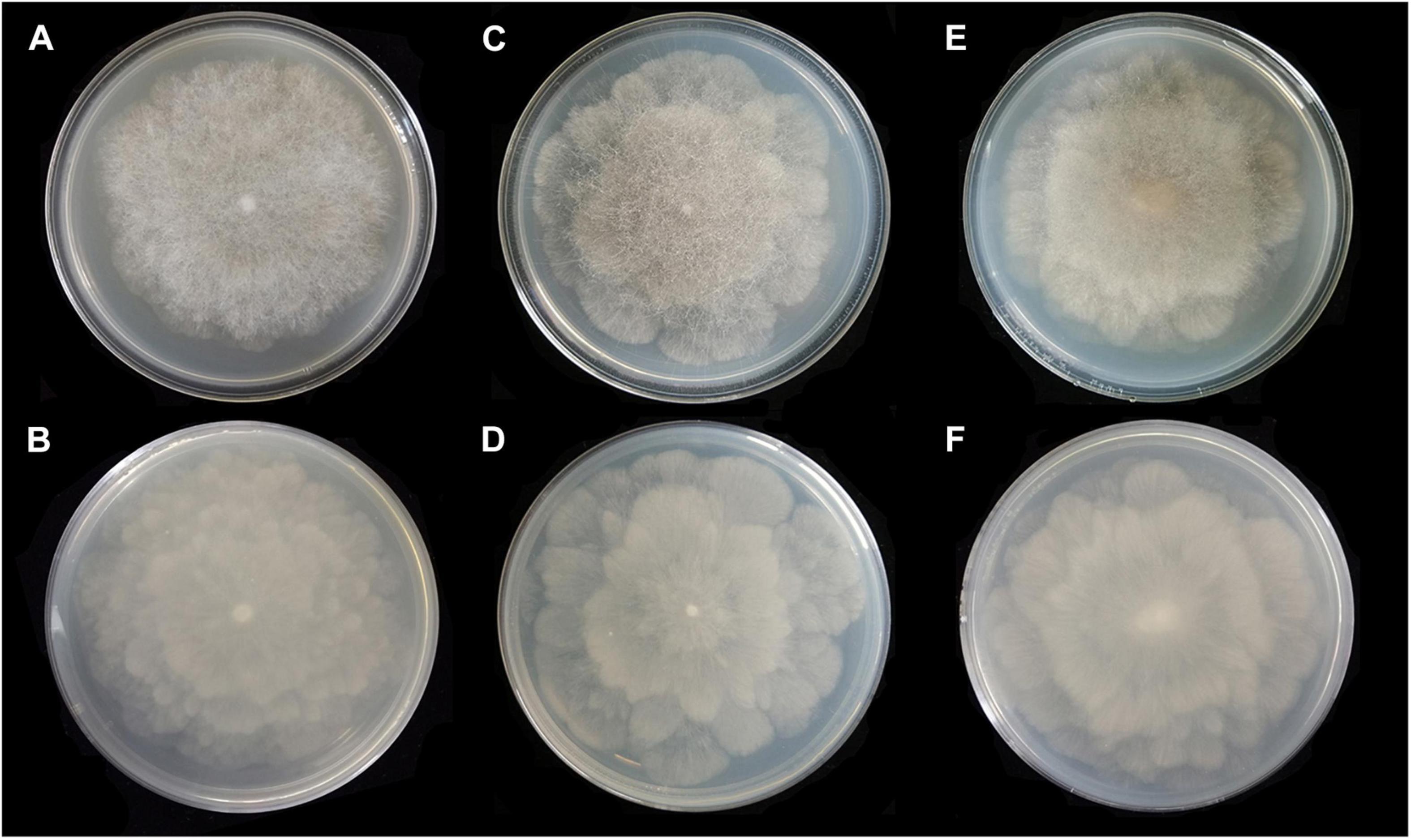
Figure 4. Colonies of Absidia globospora CGMCC 3.16031 at 27°C after 6 days on MEA (A) obverse, (B) reverse; on SMA (C) obverse, (D) reverse; on PDA (E) obverse, and (F) reverse.
Holotype: China. Hubei Province, Shennongjia Forestry District, from soil sample, 20 August 1984, Chen Guiqing (HMAS 249881, living culture CGMCC 3.16031. GenBank: ITS = MW671537, LSU = MW671544. NMDC: ITS = NMDCN0000JB7, LSU = NMDCN0000JB0).
Paratype: China. Shanxi Province, Baoji, Tangyu County, Taibaishan National Forest Park, from soil sample, 11 October 2002, Wang Xuewei (CGMCC 3.16035. GenBank: ITS = MW671538, LSU = MW671545. NMDC: ITS = NMDCN0000JB8, LSU = NMDCN0000JB1); Hubei Province, Shennongjia District, Hubei Shennongjia Forest Ecosystem National Field Scientific Observation and Research Station, from an unknown substrate, 16 October 2002, Wang Xuewei (CGMCC 3.16036. GenBank: ITS = MW671539, LSU = MW671546. NMDC: ITS = NMDCN0000JB9, LSU = NMDCN0000JB2).
Etymology: globospora (Lat.) referring to the shape of sporangiospores.
Description: Colonies on MEA, irregularly zonate, attaining 73-mm diameter after 6 days at 27°C, white at first and then elm green (R17) to dark cress green (R31). Hyphae hyaline at first, becoming brown when mature (6.0–)8.0–13.5(–14.5)-μm diameter. Stolons branched, hyaline to brown, smooth, with few septa near the base of sporangiophores (6.0–)7.0–10.5-μm diameter. Rhizoids root-like, branched mostly twice and rarely repeatedly, with a septum at the base. Sporangiophores erect or slightly bent, 1–5 in whorls, unbranched, simple, monopodial or sympodial, hyaline, or brown, with a septum (9.5–)11.0–21.5 μm below apophyses, sometimes a swelling beneath sporangia (45.0–)65.0–350.0(–440.0) × 5.0–8.5(–9.5) μm. Apophyses distinct, slightly pigmented (3.0–)4.0–14.0(–20.0) μm high, 4.0–11.0(–13.5) μm wide at the base, and 11.0–24.0(–26.5) μm wide at the top. Sporangia globose, multispored, deliquescent-walled (20.5–)23.0–50.0(–55.5) × (17.0–)23.0–40.0(–57.0) μm. Columellae hemispherical, hyaline, smooth, sometimes with a 1–3.5 μm papillary projection at the apex, 12.5–33.5(–48.5) × (8.5–)10.0–31.5(–46.5) μm. Collars present or absent, but indistinct if present. Sporangiospores globose, hyaline, smooth, 3.0–4.0(–4.5) × 2.5–3.5(–4.0) μm. Chlamydospores absent. Zygospores not observed.
Media and temperatures: Colonies on SMA, flower-shaped, attaining 74-mm diameter after 6 days at 27°C, white at first and then olive-citrine (R16) to Kronberg’s green (R31). Colonies on PDA, flower-shaped, attaining 73-mm diameter after 6 days at 27°C, white at first and then cossack green (R6) to cerro green (R5). No growth at 29°C.
Absidia medulla T.K. Zong & X.Y. Liu, sp. nov.
Fungal names: FN570836 (Figures 5–7).
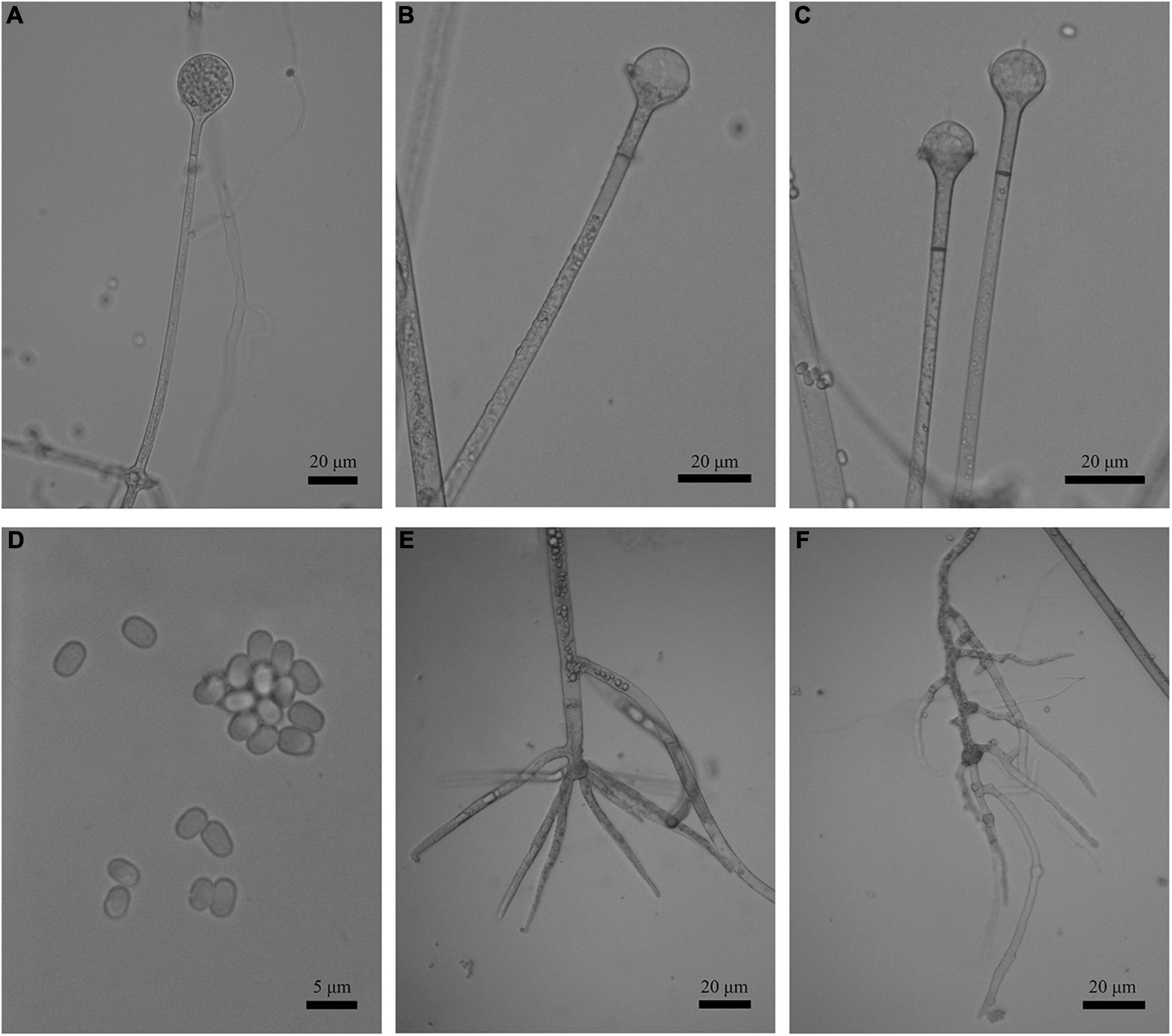
Figure 5. Morphologies of Absidia medulla CGMCC 3.16034. (A) Sporangium; (B,C) columellae; (D) sporangiospores; (E,F) rhizoids. Scale bars: (A–C,E,F) 20 μm; (D) 5 μm.
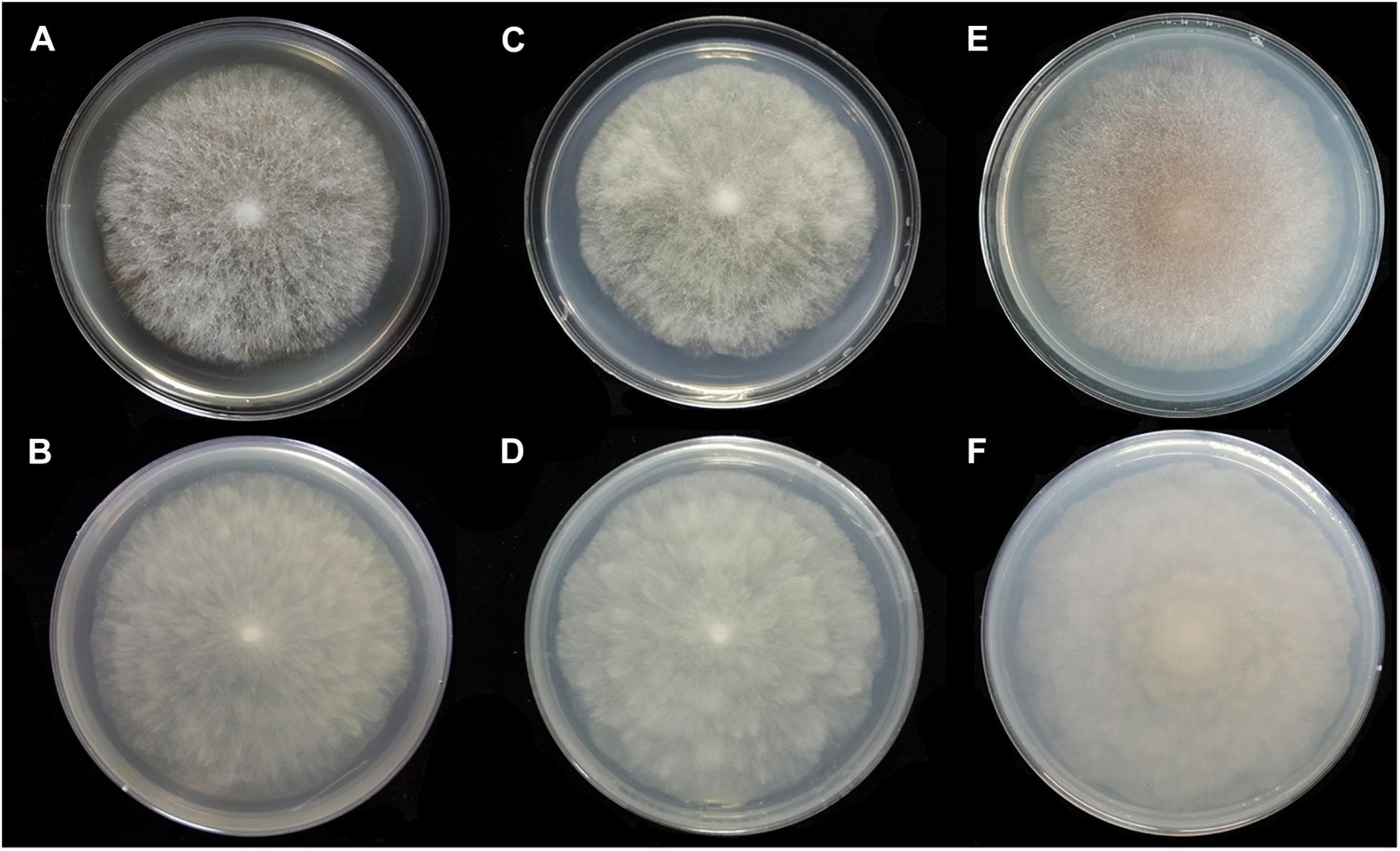
Figure 6. Colonies of Absidia medulla CGMCC 3.16034 at 27°C after 5 days on MEA (A) obverse, (B) reverse; on SMA (C) obverse, (D) reverse; after 7 days on PDA (E) obverse, and (F) reverse.
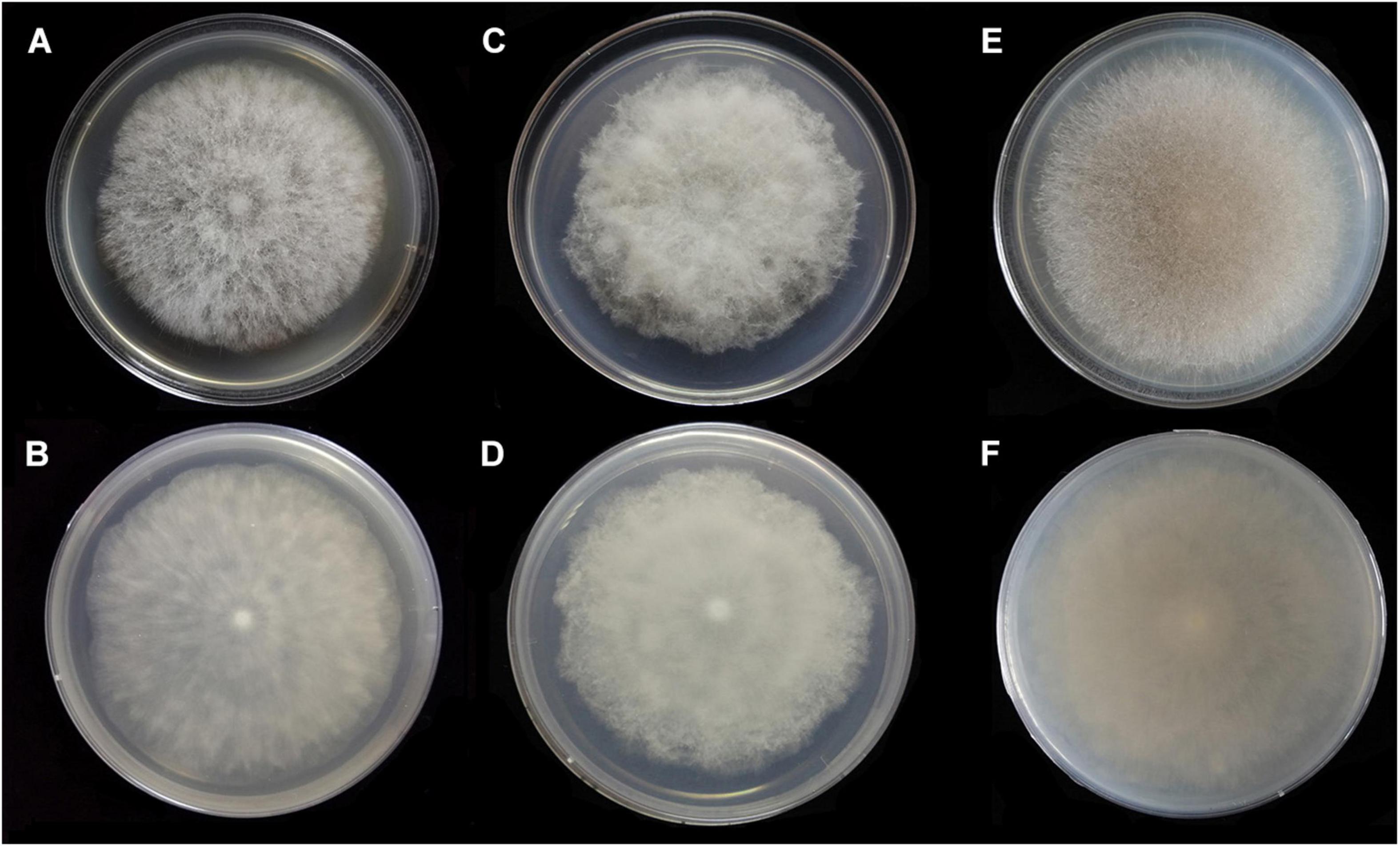
Figure 7. Colonies of Absidia medulla CGMCC 3.16037 at 27°C after 5 days on MEA (A) obverse, (B) reverse; after 6 days on SMA (C) obverse, (D) reverse; after 5 days on PDA (E) obverse, and (F) reverse.
Holotype: China. Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Xishuangbanna, from soil sample, 16 June 1992, Hu Fumei (HMAS 249884, living culture CGMCC 3.16034. GenBank: ITS = MW671542, LSU = MW671549. NMDC: ITS = NMDCN0000JBC, LSU = NMDCN0000JB5).
Paratype: China. Yunnan Province, Kunming, Yunnan Nationalities Village, from soil sample, 25 August 1995, Guo Yinglan (CGMCC 3.16037. GenBank: ITS = MW671543, LSU = MW671550. NMDC: ITS = NMDCN0000JBD, LSU = NMDCN0000JB6).
Etymology: medulla (Lat.) referring to the spine-like shape of rhizoids.
Description: Colonies on MEA, regularly zonate, attaining 74-mm diameter after 5 days at 27°C, white at first and then smoke gray (R46), sparse, but abundantly sporulated. Hyphae hyaline at first, becoming brown when mature, septate in age (5.0–)7.0–15.5-μm diameter. Stolons branched, smooth, with few septa near the base of sporangiophores, 3.5- to 6.5-μm diameter. Rhizoids root-like or spine-like, singly to multiply branched, with a septum at the base. Sporangiophores erect or slightly bent, 1–6 in whorls, unbranched, simple or monopodial, rarely sympodial, hyaline, with a septum 12.5–20.5(–27.5) μm below apophyses (50.0–)75.0–200.0(–220.0) × (2.5–)3.0–6.0(–7.5) μm. Apophyses slightly pigmented, 3.0–8.0(–8.5) μm high, 3.0–5.5(–6.5) μm wide at the base, and 7.5–16.5(–17.5) μm wide at the top. Sporangia globose to pyriform, multispored, deliquescent-walled (12.0–)16.0–30.5(–41.0) × (11.5–)15.0–30.0(–32.5) μm. Columellae hemispherical, hyaline, smooth, generally with a single 1.0- to 4.5-μm-long projection, 8.5–20.5 × 7.0–17.5 μm. Collars present or absent, distinct if present. Sporangiospores cylindrical to oval, hyaline, smooth, 3.0–4.5 × 2.0–3.0(–3.5) μm. Chlamydospores absent. Zygospores not observed.
Media and temperatures: Colonies on SMA, cottony, regularly zonate, attaining 70-mm diameter after 5 days at 27°C, white at first and then pale olive-gray (R51). Colonies on PDA, regularly zonate, attaining 74-mm diameter after 5 days at 27°C, white at first and then snuff brown (R29) to deep olive (R40) in center. No growth at 33°C.
Absidia turgida T.K. Zong & X.Y. Liu, sp. nov.
Fungal names: FN570834 (Figures 8, 9).
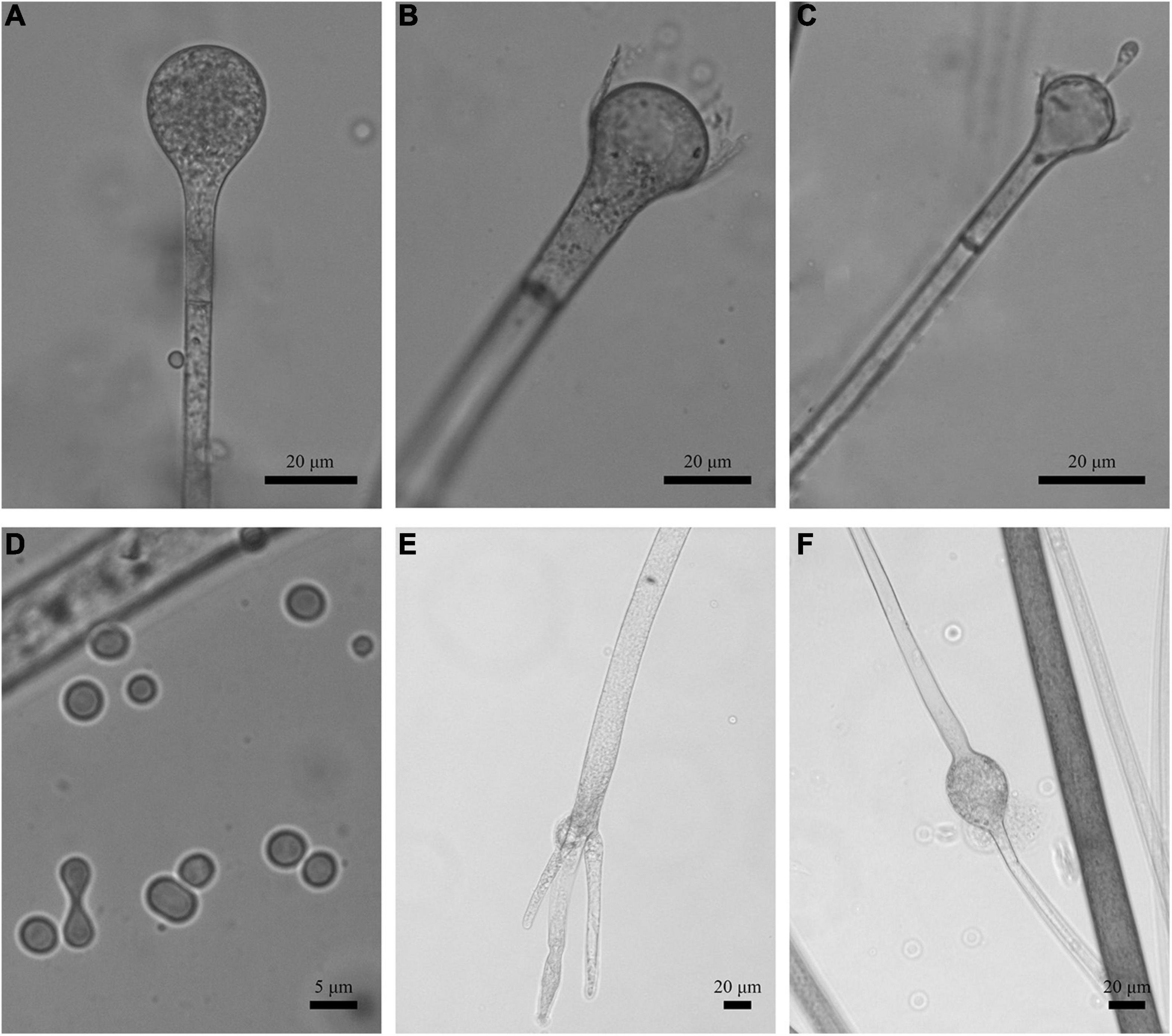
Figure 8. Morphologies of Absidia turgida CGMCC 3.16032. (A) Sporangium; (B,C) columellae; (D) sporangiospores; (E) rhizoids; (F) swollen on hyphae. Scale bars: (A–C,E,F) 20 μm; (D) 5 μm.
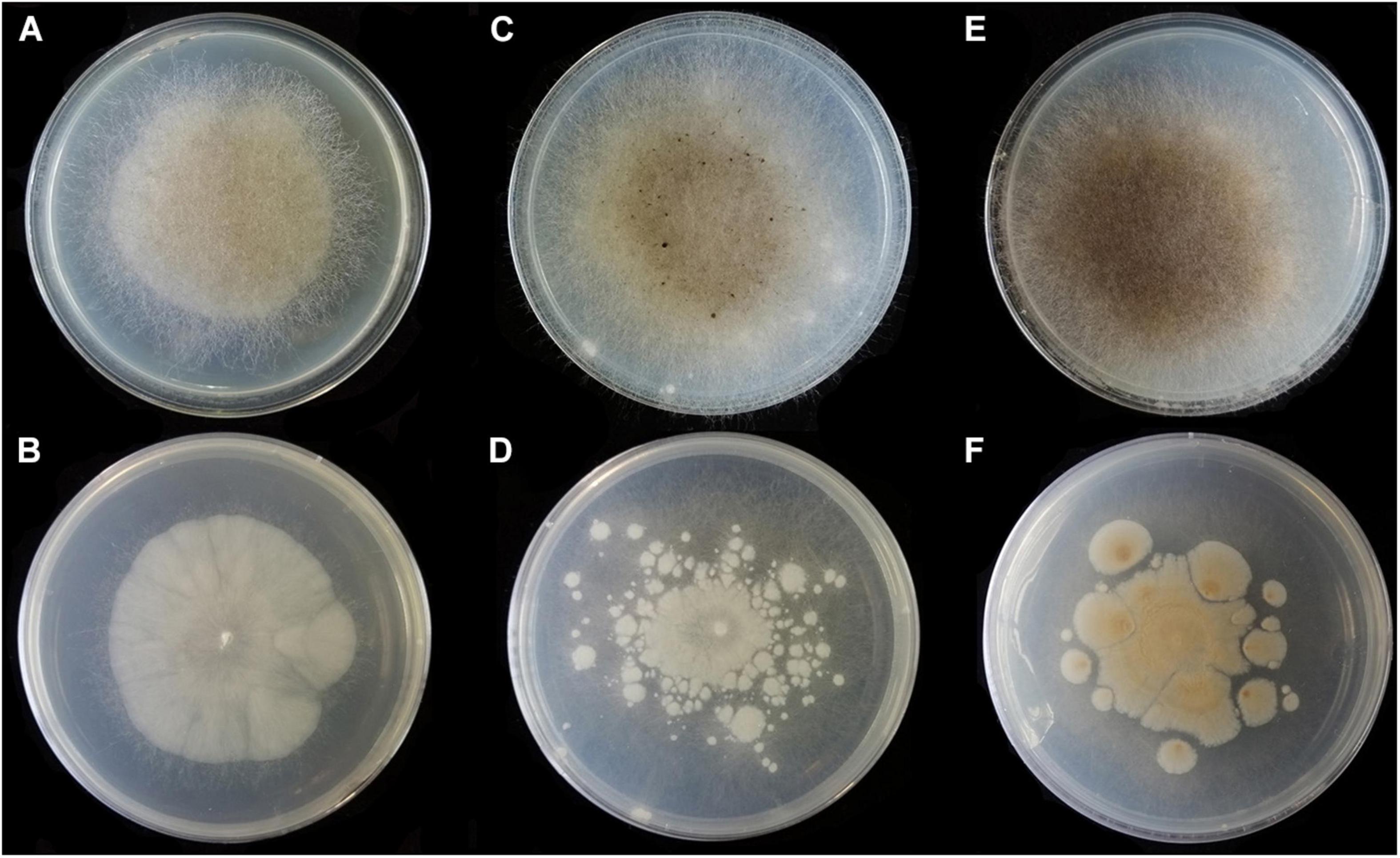
Figure 9. Colonies of Absidia turgida CGMCC 3.16032 at 27°C after 12 days on MEA (A) obverse, (B) reverse; on SMA (C) obverse, (D) reverse; on PDA (E) obverse, and (F) reverse.
Holotype: China. Xinjiang Uygur Autonomous Region, Urumqi, Urumqi County, Xiejiagou Natural Scenic Resort, from soil sample, 7 June 2002, Wang Xuewei (HMAS 249882, living culture CGMCC 3.16032. GenBank: ITS = MW671540, LSU = MW671547. NMDC: ITS = NMDCN0000JBA, LSU = NMDCN0000JB3).
Etymology: turgida (Lat.) referring to the swollen hyphae and the inflate projection on columellae.
Description: Colonies on MEA, irregularly radially gaped, attaining 23-mm diameter after 3 days, 35-mm diameter after 7 days, 50-mm diameter after 12 days at 27°C, white at first and then drab gray to drab (R45), sparse, but abundantly sporulated. Hyphae hyaline at first, becoming brown when mature, occasionally swollen, 9.0- to 23.0-μm diameter. Stolons branched, smooth, with few septa near the base of sporangiophores, 8.5- to 16.0-μm diameter. Rhizoids root-like, thick, short or comparatively long, simple or 2–3 branched, with a septum at the base. Sporangiophores erect or slight bent, 1–4 in whorls, unbranched or sometimes simple, hyaline, with a septum (17.0–)21.0–39.5(–43.5) μm below apophyses, 125.0–350.0(–370.0) × (3.5–)4.5–10.0(–11.0) μm. Apophyses distinct, unpigmented (4.5–)5.0–13.5(–16.5) μm high, 3.5–10.0 μm wide at the base, and (10.0–)11.0–22.0(–23.5) μm wide at the top. Sporangia globose to pyriform, multispored, deliquescent-walled, 20.5–42.5 × 20.0–41.5(–46.0) μm. Columellae mostly hemispherical, sometimes conical, hyaline, smooth, with a single clavate projection, up to 9.5 μm in length, with a bulbous swelling at top (13.0–)14.5–25.0(–26.5) × (10.0–)11.5–21.5 μm. Collars present or absent, distinct if present. Sporangiospores variable, globose, cylindrical or irregular, hyaline, smooth, 4.0–5.0(–6.5) × 3.0–4.0 μm when cylindrical, 3.5–4.5 × 3.0–4.0 μm or 2.0- to 2.5-μm diameter when globose. Chlamydospores absent. Zygospores not observed.
Media and temperatures: Colonies on SMA, sporadic, nebula-shaped, attaining 22-mm diameter after 3 days, 32-mm diameter after 7 days, 47-mm diameter after 12 days at 27°C, white at first and then pale drab-gray to light cinnamon-drab (R45). Colonies on PDA, irregularly tree ring-shaped, attaining 21-mm diameter after 3 days, 32-mm diameter after 7 days, 44-mm diameter after 12 days at 27°C, growing slowly when aerial hyphae reaching the lid of the petri dish, white at first and then drab to hair brown (R45). No growth at 33°C.
Absidia zonata T.K. Zong & X.Y. Liu, sp. nov.
Fungal names: FN570835 (Figures 10, 11).
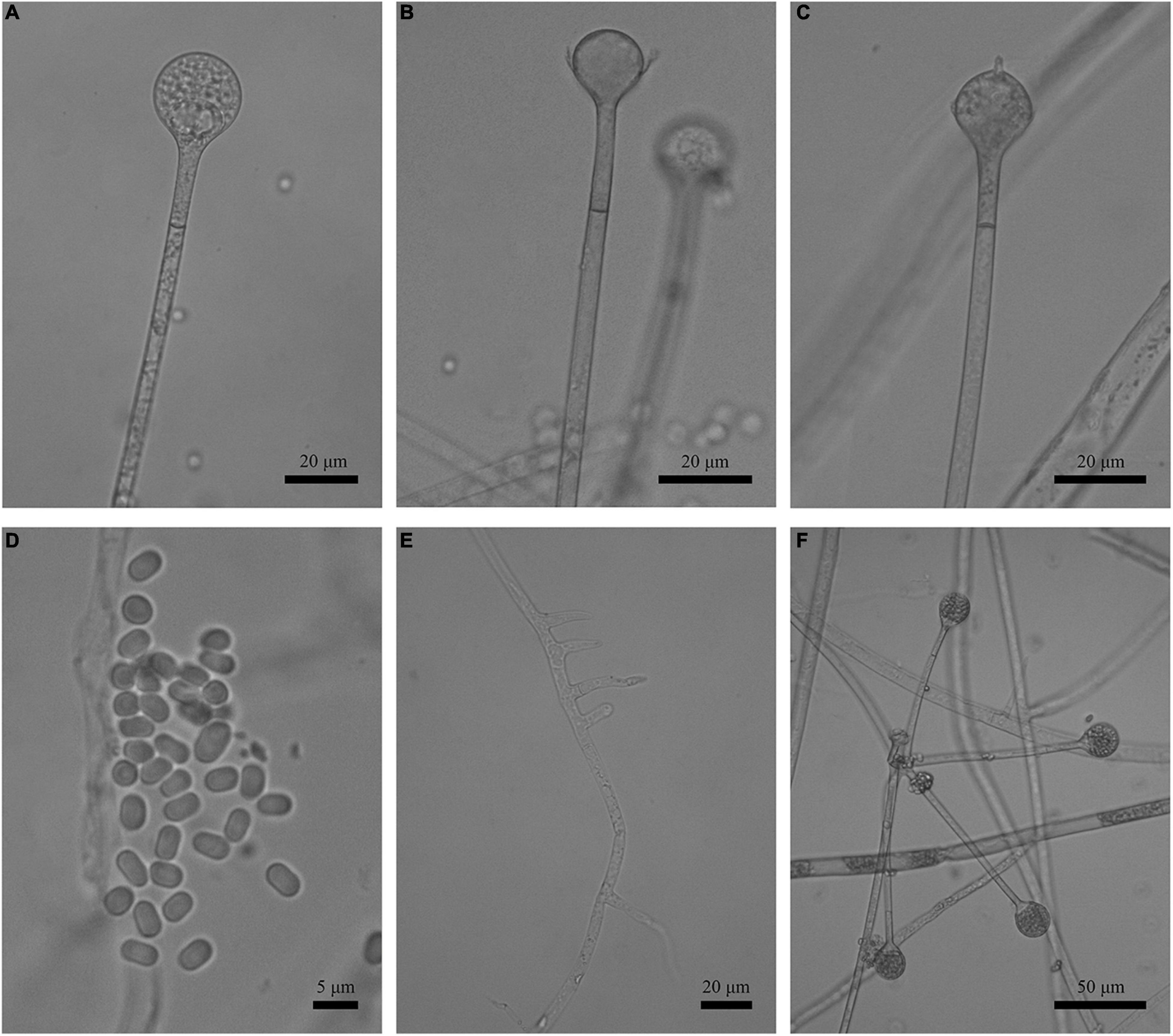
Figure 10. Morphologies of Absidia zonata CGMCC 3.16033. (A) Sporangium; (B,C) columellae; (D) sporangiospores; (E) rhizoids; (F) verticillately branched sporangiophores. Scale bars: (A–C,E) 20 μm; (D) 5 μm; (F) 50 μm.
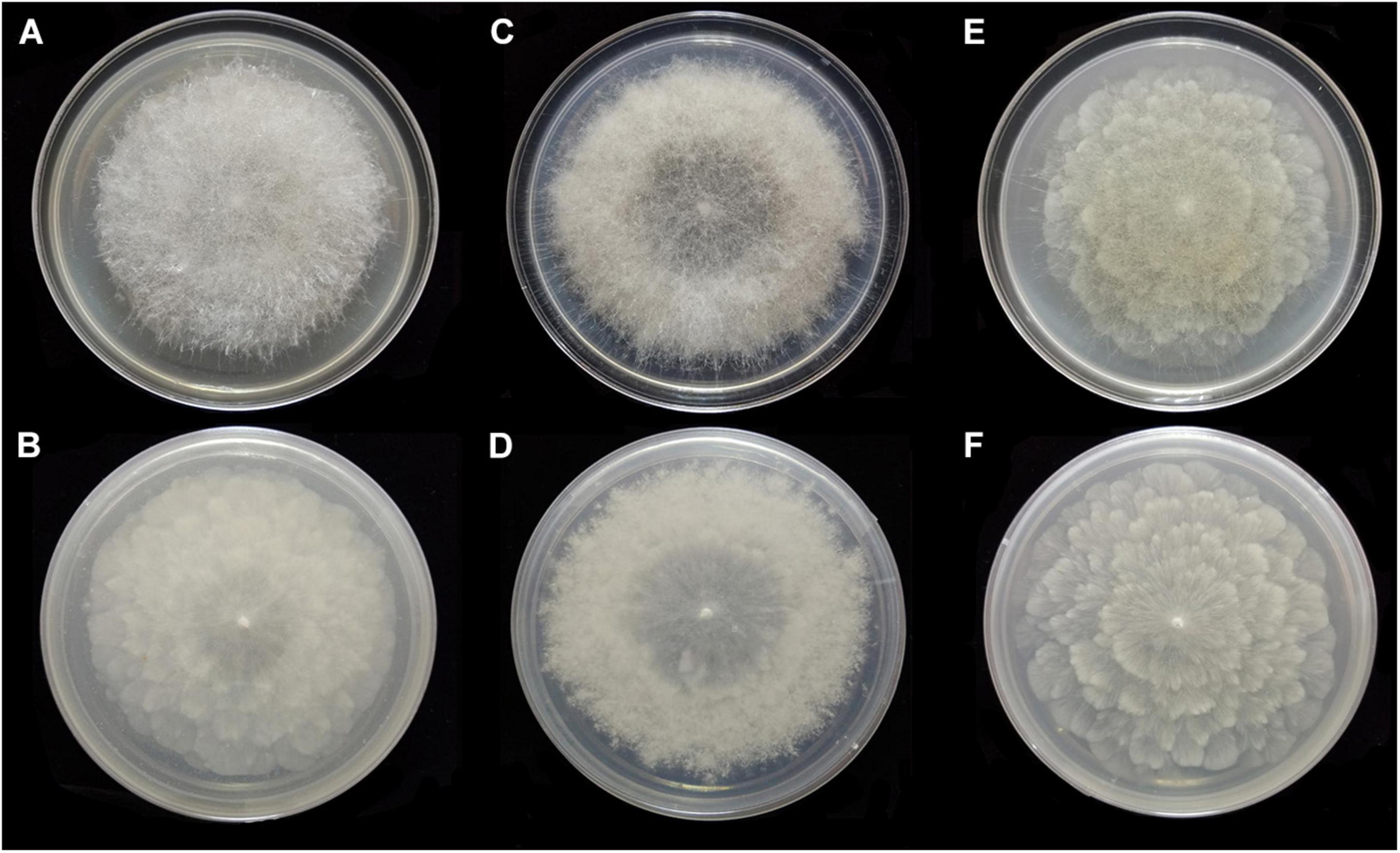
Figure 11. Colonies of Absidia zonata CGMCC 3.16033 at 27°C after 8 days on MEA (A) obverse, (B) reverse; on SMA (C) obverse, (D) reverse; after 7 days on PDA (E) obverse, and (F) reverse.
Holotype: China. Beijing (39°57′58′′N, 116°11′43′′E), from soil sample, 31 December 2019, Liu Xiaoyong (HMAS 249883, living culture CGMCC 3.16033. GenBank: ITS = MW671541, LSU = MW671548. NMDC: ITS = NMDCN0000JBB, LSU = NMDCN0000JB4).
Etymology: zonata (Lat.) referring to the zonate colony.
Description: Colonies on MEA, regularly concentric ring zonate, attaining 69-mm diameter after 8 days at 27°C, white at first and then smoke gray (R46). Hyphae hyaline at first, becoming brown when mature, 5.0- to 10.5-μm diameter. Stolons branched, smooth, with few septa near the base of sporangiophores, 4.0- to 8.0-μm diameter. Rhizoids root-like or tentaculiform, simple or 2–3 branched, with a septum at the base. Sporangiophores erect or slightly bent, 1–5(–8) in whorls, unbranched, sometimes simple, rarely monopodial, hyaline, with a septum 16.0–26.5 μm below apophyses (44.0–)55.0–180.0(–280.0) × 2.5–5.5(–6.0) μm. Apophyses distinct, slightly pigmented, 3.0–8.0(–8.5) μm high, 3.0–5.5(–6.5) μm wide at the base, and 7.5–16.5(–17.5) μm wide at the top. Sporangia globose to pyriform, multispored, deliquescent-walled, 14.0–27.0 × 12.5–26.5 μm. Columellae hemispherical, hyaline, smooth, generally presenting a single spinous projection, up to 2.0–3.5 μm in length, 9.5–19.0 × (6.0–)7.5–14.5(–16.5) μm. Collars present or absent, distinct if present. Sporangiospores mostly cylindrical, sometimes oval, hyaline, smooth, 3.5–4.5(–6.0) × 2.0–3.0(–3.5) μm. Chlamydospores absent. Zygospores not observed.
Media and temperatures: Colonies on SMA, rough around the edges, concentric ring-shaped, attaining 69-mm diameter after 8 days at 27°C, white. Colonies on PDA, regularly wavy zonate, attaining 72-mm diameter after 7 days at 27°C, white at first and then lime green (R31). No growth at 38°C.
Key to the known species of Absidia in China
1. Sporangiospores typically globose; Colonies greenish ……………………………2
1. Sporangiospores typically cylindrical, oval, or other shaped; Colonies not greenish………………………3
2. Maximum temperatures below 30°C; Sporangiophores not reaching 10 μm in width; Sporangia rarely reaching 55-μm diameter.………………….………A. globospora
2. Maximum temperatures above 30°C; Sporangiophores reaching 12 μm in width; Sporangia mostly 50- to 60-μm diameter.……………………………A. glauca
3. Columellae without distinct apical projections ……………………………………A. heterospora
3. Columellae with apical projections ……………………………4
4. Sporangiospores variable, sometimes irregular in shape and size…………………5
4. Sporangiospores invariable, always regular …………………………6
5. Hyphae without swelling, <9-μm diameter; sporangiophores sometimes simple, more often monopodial or verticillate; columellae sometimes with a short projection…………………………………Absidia idahoensis
5. Hyphae occasionally swelling, >9-μm diameter; Sporangiophores unbranched or sometimes simple; columellae always with a projection up to 9 μm in length ………………………………………A. turgida
6. Abundant secondary sporangia in older cultures ………………………A. repens
6. No abundant secondary sporangia in older cultures ………………………………7
7. Sporangiophores never in pairs or in whorls ……………………………A. panacisoli
7. Sporangiophores in pairs or in whorls ………………………………………………8
8. Sporangiophores no more than 6 in whorls ………………………………………9
8. Sporangiophores as many as 7–11 in whorls ……………………………………11
9. Maximum temperatures below 35°C. ……………………………………A. medulla
9. Maximum temperatures above 35°C ……………………………………………10
10. Sporangiophores up to 6 in whorls, occasionally swollen below the sporangia; Collar absent; Sporangiospores ovoid to ellipsoid….…………………A. ovalispora
10. Sporangiophores up to 4 in whorls, no swollen; Collar always present; Sporangiospores cylindrical………………………………………A. cylindrospora
11. Rhizoids typically aseptate…………………………… A. spinosa
11. Rhizoids generally or rarely septate ……………………………………………12
12. The projections on columellae < 5 μm in length, taper at the end…………………………………………A. zonata
12. The projections on columellae > 5 μm in length, rounded at the top……………………A. pseudocylindrospora
Discussion
Phylogenetically, the ITS (Figure 1) and LSU (Figure 2) trees show that four new species cluster in different clades of Absidia. The A. globospora (100/100/1.00 for both ITS and LSU) is located in the globospora clade and most closely related to A. glauca Hagem (88/100/1.00 for ITS). Their sibling relationship is completely supported by ITS and LSU, with 99/100/1.00 and 100/100/1.00 support values, respectively. Physiologically, A. globospora is similar to A. glauca in heterothallism but differs in maximum growth temperature (37 vs. 29°C). Morphologically, A. globospora is similar to A. glauca in forming green colonies and globose sporangiospores. However, A. glauca differs in its wider sporangiophores (up to 12 μm), glaucous stolons, and larger sporangia (mostly 50- to 60-μm diameter, Ellis and Hesseltine, 1965).
The other three new species are placed in the cylindrospora clade where A. zonata is most closely related to A. koreana (100/100/1.00 for ITS, 99/95/1.00 for LSU), which is strongly supported with a high value of 100/100/1.00 or 98/99/0.99. Both A. zonata and A. koreana are physiologically similar in maximum growth temperatures but morphologically differentiated by characteristics on SMA media (Table 2; Ariyawansa et al., 2015). The width of sporangiospores in A. koreana are more narrow and uniform. In its protologue, A. koreana did not form projections, but the figure in the original article illustrated projections. A. turgida is basal to A. heterospora in ITS tree (Figure 1) or next to A. repens Tiegh. and A. pararepens in the LSU tree (Figure 2). A. medulla is closely related to A. repens in ITS tree (Figure 1) or A. saloaensis and A. ovalispora in LSU tree (Figure 2).

Table 2. Comparisons of morphological characteristics of Absidia zonata and Absidia koreana on SMA media at 25°C.
The two strains, CGMCC 3.16034 and CGMCC 3.16037, of A. medulla are similar in maximum growth temperature, micromorphology, and even colonies on MEA and PDA media, but slightly different in colonies on SMA media. The ex-paratype CGMCC 3.16037 is more floccose and thicker and grows more slowly than the ex-holotype CGMCC 3.16034 when they are incubated on SMA at 27°C, and it lacks white concentric rings from the reverse side of the colony (Figure 7).
The species Chlamydoabsidia padenii Hesselt. & J.J. Ellis and Halteromyces radiatus Shipton & Schipper are obviously nested within Absidia in LSU tree (Figure 2). However, morphologically unique multiseptate, easily pigmented aerial chlamydospores were developed in C. padenii, whereas dumbbell-shaped sporangia were formed in H. radiatus (Hesseltine and Ellis, 1966; Shipton and Schipper, 1975).
The genus Absidia was proposed to be divided into several groups distinguishable by their sporangiospores (Kwaśna et al., 2006; Hoffmann et al., 2007, 2009b; Hoffmann and Voigt, 2008; Hoffmann, 2010), which is confirmed in the present study with three well-supported clades, i.e., cylindrospora clade, globospora clade, and cuneospora clade (Figures 1, 2). Two exceptions are worth noting, specifically, both A. pararepens and A. bonitoensis have sub-globose to globose sporangiospores, even though they are in the cylindrospora clade (Crous et al., 2020).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
T-KZ, C-LZ, and X-YL contributed to conception and design of the study. T-KZ wrote the draft of the manuscript. C-LZ and X-YL improved the manuscript. T-KZ, HZ, C-LZ, and X-YL observed and described the morphology. X-LL and L-YR collected the molecular data. All authors contributed to manuscript revision, proofread, and approved the submitted version.
Funding
The study is supported by the National Natural Science Foundation of China (Grant No. 31970009) and the Yunnan Fundamental Research Project (Grant No. 202001AS070043).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ariyawansa, H. A., Hyde, K. D., Jayasiri, S. C., Buyck, B., Chethana, K. T., Dai, D. Q., et al. (2015). Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 75, 27–274. doi: 10.1007/s13225-015-0346-5
Benny, G. L. (2008). Methods used by Dr. RK Benjamin, and other mycologists, to isolate zygomycetes. Aliso 26, 37–61. doi: 10.5642/aliso.20082601.08
Chen, J., Fan, F., Qu, G., Tang, J., Xi, Y., Bi, C., et al. (2020). Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone. Metab. Eng. 57, 31–42. doi: 10.1016/j.ymben.2019.10.006
Cordeiro, T. R. L., Nguyen, T. T. T., Lima, D. X., Silva, S. B. G. D., Lima, C. F. D., Leitão, J. D., et al. (2020). Two new species of the industrially relevant genus Absidia (Mucorales) from soil of the Brazilian Atlantic Forest. Acta Bot. Bras. 34, 549–558. doi: 10.1590/0102-33062020abb0040
Crous, P. W., Luangsa-Ard, J. J., Wingfield, M. J., Carnegie, A. J., Hernández-Restrepo, M., Lombard, L., et al. (2018). Fungal Planet description sheets: 785–867. Persoonia 41, 238–417. doi: 10.3767/persoonia.2018.41.12
Crous, P. W., Wingfield, M. J., Chooi, Y. H., Gilchrist, C. L., Lacey, E., Pitt, J. I., et al. (2020). Fungal Planet description sheets: 1042–1111. Persoonia 44, 301–459. doi: 10.3767/persoonia.2020.44.11
de Lima, C. L. F., Lima, D. X., Cordeiro, T. R. L., Lee, H. B., Nguyen, T. T. T., Gurgel, L. M. S., et al. (2021). Absidia bonitoensis (Mucorales, Mucoromycota), a new species isolated from the soil of an upland Atlantic forest in Northeastern Brazil. Nova Hedwigia 112, 241–251. doi: 10.1127/nova_hedwigia/2021/0614
Edler, D., Klein, J., Antonelli, A., and Silvestro, D. (2020). raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. [Preprint]. doi: 10.1101/800912
Ellis, J. J., and Hesseltine, C. W. (1965). The genus Absidia: globose-spored species. Mycologia 57, 222–235. doi: 10.1080/00275514.1965.12018205
Ellis, J. J., and Hesseltine, C. W. (1966). Species of Absidia with ovoid sporangiospores. II. Sabouraudia 5, 59–77. doi: 10.1080/00362176785190111
Felsenstein, J. (1985). Confidence limits on phylogenetics: an approach using bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Hawksworth, D. L., Kirk, P. M., Sutton, B. C., and Pegler, D. N. (1995). Ainsworth & Bisby’s Dictionary of the Fungi, 8th Edn. UK: CABI Publishing.
Hesseltine, C. W., and Ellis, J. J. (1961). Notes on Mucorales, especially Absidia. Mycologia 53, 406–426. doi: 10.1080/00275514.1961.12017970
Hesseltine, C. W., and Ellis, J. J. (1964). The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia 56, 568–601. doi: 10.1080/00275514.1964.12018145
Hesseltine, C. W., and Ellis, J. J. (1966). Species of Absidia with ovoid sporangiospores. I. Mycologia 58, 761–785. doi: 10.1080/00275514.1966.12018369
Hoffmann, K. (2010). “Identification of the genus Absidia (Mucorales, Zygomycetes): a comprehensive taxonomic revision,” in Molecular Identification of Fungi, eds Y. Gherbawy and K. Voigt (Berlin: Springer).
Hoffmann, K., and Voigt, K. (2008). Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biol. 11, 537–554. doi: 10.1111/j.1438-8677.2008.00145.x
Hoffmann, K., Discher, S., and Voigt, K. (2007). Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol. Res. 111, 1169–1183. doi: 10.1016/j.mycres.2007.07.002
Hoffmann, K., Telle, S., Walther, G., Eckart, M., Kirchmair, M., Prillinger, H., et al. (2009b). “Diversity, genotypic identification, ultrastructural and phylogenetic characterization of zygomycetes from different ecological habitats and climatic regions: limitations and utility of nuclear ribosomal DNA barcode markers,” in Current Advances in Molecular Mycology, eds Y. Gherbawy, R. Mach, and M. Rai (New York, NY: Nova Science Publishers, Inc), 263–312.
Hoffmann, K., Walther, G., and Voigt, K. (2009a). Mycocladus vs. Lichtheimia: a correction (Lichtheimiaceae fam. nov., Mucorales, Mucoromycotina). Mycol. Res. 113, 275–278.
Kaczmarek, M. B., Struszczyk-Swita, K., Li, X., Szczêsna-Antczak, M., and Daroch, M. (2019). Enzymatic modifications of chitin, chitosan, and chitooligosaccharides. Front. Bioeng. Biotech. 7:243. doi: 10.3389/fbioe.2019.00243
Kwaśna, H., Elaine, W., and Bateman, G. L. (2006). Phylogenetic relationships among Zygomycetes from soil based on ITS1/2 rDNA sequences. Mycol. Res. 110, 501–510. doi: 10.1016/j.mycres.2006.02.004
Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278. doi: 10.1093/bioinformatics/btu531
Li, G. J., Hyde, K. D., Zhao, R. L., Hongsanan, S., Abdel-Aziz, F. A., Abdel-Wahab, M. A., et al. (2016). Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78, 1–237. doi: 10.1007/s13225-016-0366-9
Lima, D. X., Cordeiro, T. R., De Souza, C. A., De Oliveira, R. J., Lee, H. B., Souza-Motta, C. M., et al. (2020). Morphological and molecular evidence for two new species of Absidia from Neotropic soil. Phytotaxa 446, 61–71. doi: 10.11646/phytotaxa.446.1.8
Orr, G. F., and Plunkett, O. A. (1959). A new species of Absidia from California. Mycologia 51, 203–209. doi: 10.1080/00275514.1959.12024813
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Boil. 61, 539–542. doi: 10.1093/sysbio/sys029
Shipton, W. A., and Schipper, M. A. (1975). Halteromyces, a new genus in the Mucorales. Antonie van Leeuwenhoek 41, 337–342. doi: 10.1007/BF02565068
Swofford, D. L. (2002). PAUP∗: Phylogenetic Analysis Using Parsimony (∗and Other Methods). Version 4.0b10. Sunderland, MA: Sinauer Associates.
van Tieghem, P. (1876). Troisième mémoire sur les Mucorinées. Annales des Siences Naturelles Botanique 4, 312–399.
Wanasinghe, D. N., Phukhamsakda, C., Hyde, K. D., Jeewon, R., Lee, H. B., Jones, E. G., et al. (2018). Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 89, 1–236. doi: 10.1007/s13225-018-0395-7
Wang, Y. N., Liu, X. Y., and Zheng, R. Y. (2014). Umbelopsis changbaiensis sp. nov. from China and the typification of Mortierella vinacea. Mycol. Prog. 13, 657–669. doi: 10.1007/s11557-013-0948-9
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Zhang, T. Y., Yu, Y., Zhu, H., Yang, S. Z., Yang, T. M., Zhang, M. Y., et al. (2018). Absidia panacisoli sp. nov., isolated from rhizosphere of Panax notoginseng. Int. J. Syst. Evol. Microbiol. 68, 2468–2472. doi: 10.1099/ijsem.0.002857
Zhao, C. L., and Wu, Z. Q. (2017). Ceriporiopsis kunmingensis sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Mycol. Prog. 16, 93–100. doi: 10.1007/s11557-016-1259-8
Zhao, H., Zhu, J., Zong, T. K., Liu, X. L., Ren, L. Y., Lin, Q., et al. (2021). Two New Species in the Family Cunninghamellaceae from China. Mycobiology 49, 142–150. doi: 10.1080/12298093.2021.1904555
Zheng, R. Y., and Chen, G. Q. (2001). A monograph of Cunninghamella. Mycotaxon 80, 1–75. doi: 10.1007/978-3-319-23534-9_1
Keywords: morphology, molecular phylogeny, taxonomy, Mucoromycetes, Mucoromycota
Citation: Zong T-K, Zhao H, Liu X-L, Ren L-Y, Zhao C-L and Liu X-Y (2021) Taxonomy and Phylogeny of Four New Species in Absidia (Cunninghamellaceae, Mucorales) From China. Front. Microbiol. 12:677836. doi: 10.3389/fmicb.2021.677836
Received: 10 March 2021; Accepted: 12 July 2021;
Published: 04 August 2021.
Edited by:
Hyang Burm Lee, Chonnam National University, South KoreaReviewed by:
Kerstin Voigt, Friedrich Schiller University Jena, GermanyThuong Nguyen, Chonnam National University, South Korea
Copyright © 2021 Zong, Zhao, Liu, Ren, Zhao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Yong Liu, bGl1eGlhb3lvbmdAaW0uYWMuY24=; Chang-Lin Zhao, ZnVuZ2ljaGFuZ2xpbnpAMTYzLmNvbQ==
 Tong-Kai Zong
Tong-Kai Zong Heng Zhao
Heng Zhao Xiao-Ling Liu
Xiao-Ling Liu Li-Ying Ren
Li-Ying Ren Chang-Lin Zhao
Chang-Lin Zhao Xiao-Yong Liu
Xiao-Yong Liu