- 1Natural Science Research Institute, Gangneung-Wonju National University, Gangneung, South Korea
- 2Department of Zoology, Al Azhar University, Assiut, Egypt
- 3Department of Biology, Gangneung-Wonju National University, Gangneung, South Korea
- 4Protist Research Team, Nakdonggang National Institute of Biological Resources, Sangju, South Korea
Tunicothrix halophila n. sp. was discovered in a hypersaline marine sample from Jeju Island, Korea. It is characterized by the highly reduced number of dorsal bristles. In addition, the main character of the genus Tunicothrix (e.g., alveolar layer) is absent/indistinct. To figure out its identity and phylogenetic relationship, we examined the new species based on modern morphological methods and molecular phylogenetic analyses. Since the parabirojimids are of basal position to core hypotrichs and a smaller data set could show incorrect phylogenetic relationships among the hypotrichs, we used a huge data set composed of 1,460 DNA sequences to infer the phylogenetic tree. The reduction of dorsal bristles is very likely a secondarily evolved character in hypotrichs, resulting in the independent phenotypic adaptation in the hypersaline ecosystems as shown in other hypersaline hypotrichs. Furthermore, the so-called right marginal row 1 in other congeners is found to produce a pretransverse and transverse cirrus and thus we recommend using the term frontoventral row. Based on our data, we can justify Tunicothrix halophila n. sp. as a new species; however, despite the phenotypic distinctiveness, we refrain to establish a new genus because of the missing data and the non-monophyly of Tunicothrix.
Introduction
The hypersaline habitats harbor a high number of undescribed ciliates (Ruinen, 1938; Borror, 1972; Wilbert and Kahan, 1981, 1986; Wilbert, 1986, 1995; Foissner, 1987, 1999, 2012; Foissner et al., 2002; Berger, 2006, 2011; Li et al., 2019). However, only few hypotrichous genera have been discovered so far from such extreme environments, for instance Afrothrix Foissner, 1999; Apourosomoida Foissner et al., 2002; Cladotricha Gaievska a, 1925; Erniella Foissner, 1987; Etoschothrix Foissner et al., 2002; Schmidingerothrix Foissner, 2012; and Schmidtiella Li et al., 2019. In terms of morphology of the halophile hypotrichs, most of them have lost most or all dorsal bristles/kineties, which are rather equidistant and mostly bipolar in other hypotrich ciliates (Berger, 1999, 2006, 2011; Foissner, 2012). The reduced ciliary pattern of these taxa is suggested to be a secondary oligomerization that might be a common ancestor of hypotrichs, but it differs from the ancestor hypothesized by Berger (2008, 2011) and is very likely evolved independently among distinct taxa (Foissner et al., 2014; Li et al., 2019). This is supported by the discovery of Schmidingerothrix extraordinaria Foissner, 2012, an extraordinary hypotrich from African hypersaline soils, which lost some of the most stable features of hypotrichs such as all dorsal bristles, paroral, and the middle ciliary row of adoral membranelles. On the other hand, Li et al. (2019) discovered Schmidtiella ultrahalophila from a solar saltern. This species has few dorsal bristles and shows a basal position to other hypotrichs in the phylogenetic trees. However, the simplified character states and the fewer genetic data available in public database hamper researchers from inferring the common ancestor.
From a hypersaline marine sample, we discovered a new Tunicothrix ciliate with a highly reduced number of dorsal bristles that differs from other members of the family Parabirojimidae Dai and Xu, 2011, and is also very rare within hypotrichs (Berger, 1999, 2006, 2008, 2011; Dai and Xu, 2011; Foissner, 2012). Further, the cortical alveolar layer (tunica), which is one of the most prominent characters of the genus Tunicothrix Xu et al., 2006 (Lin and Song, 2004; Xu et al., 2006; Dai and Xu, 2011), is absent or significantly reduced and can be observed only at high magnification. Possibly, this is another adaptation to hypersaline habitat.
To figure out the identity and phylogenetic relationship of the new species, we examined the population based on modern morphological methods (live observations, protargol impregnation, and SEM) and molecular phylogenetic analyses. To examine the phylogenetic position of the new species, we used a huge data set composed of 1,460 DNA sequences because the parabirojimids are of basal positions to core hypotrichs and a smaller data set (commonly used in ciliate taxonomy/phylogeny) could show incorrect phylogenetic relationships among the hypotrichs. As we know, the hypotrichs are full of homoplasies/plesiomorphies and scattered in 18S rRNA gene trees. Based on our data, and despite the phylogenetic distinctiveness of Tunicothrix halophila n. sp., we refrain from establishing a new genus because of missing data and the non-monophyly of the genus Tunicothrix.
Materials and Methods
Sample Collection and Identification
Tunicothrix halophila n. sp. was discovered in a coastal water sample (34.9‰, 2.1°C at the time of sampling) from Jeju Island, Korea. The sample was collected in February 2020 and transferred to the laboratory within a week. It was maintained in a Petri dish (13.5 cm in diameter) for a year at room temperature (15–20°C). Sterilized wheat grains were supplied to enrich bacteria as a food source. After 10 months, the salinity became 71‰ due to evaporation. It should be noted that the new species was the only hypotrich found in the culture at that time.
The new species was examined under a stereomicroscope (SZ11; Olympus, Tokyo, Japan) and light microscopes (BX53, IX73; Olympus) using bright field and differential interference contrast (DIC) at magnifications of 50–1,000×. About 10 ml of the culture water was fixed using saturated HgCl2, and the cells were washed three to five times with tap water to remove the fixative by centrifugation (3,000–5,000 rpm, 30–60 s). The protargol impregnation was conducted using a synthesized protargol and acetone developer (Foissner, 2014; Kim and Jung, 2017). It should be noted that a new batch of the synthesized protargol powder showed an optimum impregnation condition at pH 8.5–8.7, which is distinctly higher than previous batches synthesized in our laboratory (e.g., below pH 8.0). Attempts to induce formation of resting cysts were unsuccessful. General terminology follows Lynn (2008), and specific terms of parabirojimids follow Dai and Xu (2011) except for the term “right marginal row 1” (for details, see “Discussion” section). For assigning/describing the buccal lips, we follow Foissner and Al-Rasheid (2006).
DNA Extraction, PCR Amplification, and Sequencing
Five cells of Tunicothrix halophila n. sp. were isolated from the raw culture and washed with the culture water filtered by a 0.2-μm syringe filter (Minisart® Syringe Filter; Sartorius, G ttingen, Germany) more than five times. Each cell was then transferred to a 1.5-ml tube using a micro-capillary with minimum volume of the water. Genomic DNA of each cell was extracted using a REDExtract-N-Amp Tissue PCR Kit (Sigma, St. Louis, MO, United States). The conditions for PCR were as follows: initial denaturation at 94°C for 1 min 30 s, followed by 40 cycles of denaturation at 98°C for 10 s, annealing at 58.5°C for 30 s, and extension at 72°C for 3 min, and a final extension step at 72°C for 7 min. The 18S rRNA gene was amplified using two slightly modified primers (New Euk A and LSU rev4) from Sonnenberg et al. (2007). After the amplification, the amplicons were purified using a MG PCR Purification Kit (MGmed, Seoul, Korea). We completed direct sequencing and assembled sequence fragments using one of the five cells because the five sequences determined by the New Euk A were completely identical. The DNA sequencing was performed using an ABI 3700 sequencer (Applied Biosystems, Foster City, CA, United States), and the following internal primers were used: 18SF790v2, 18SF1470, and 18SR300. Sequence fragments were assembled using Geneious Prime 2019.2.3 (Kearse et al., 2012).
Scanning Electron Microscopy
Cells isolated from the raw culture were prepared for scanning electron microscopy following the protocol of Foissner (2014). Briefly, they were fixed for 30 min using an equal mix of 2% aqueous osmium tetroxide and 3% glutaraldehyde. The cells were attached on a coverslip using poly-L-lysine, dehydrated using an ethanol series (from 30% to 100%), and dried using a critical point dryer (EM CPD300, Leica, Vienna, Austria). Subsequently, they were coated with platinum using a sputter coater (EM SCD005, Leica) and JSM-IT500 (JEOL Ltd., Tokyo, Japan) was used for the observation.
Phylogenetic Analyses
To infer the phylogenetic relationship of Tunicothrix halophila n. sp., we retrieved 1,459 18S rRNA gene sequences belonging to the class Spirotrichea from GenBank. As an outgroup, four species/sequences belonging to the class Protocruziea were selected. These sequences were aligned using ClustalW (Thompson et al., 1994) implemented into Geneious Prime 2019.2.3 (Kearse et al., 2012). Overhangs from both ends of the alignment were trimmed to construct blunt ends using Geneious, resulting in a final matrix of 1,955 columns. The best-fit substitution model for phylogenetic analysis, TVM + I (0.1260) + G (0.6620) based on the Akaike information criterion (AIC), was selected using jModelTest 2.1.10 (Guindon and Gascuel, 2003; Darriba et al., 2012). IQ-TREE 1.6.12 was used to infer the maximum likelihood (ML) trees, with 1,000 bootstrap replicates (Nguyen et al., 2015). The trees were visualized using FigTree 1.4.41.
Results
ZooBank Registration
ZooBank registration number of present work: dXJuOmxzaWQ6em9vYmFuay5vcmc=:pub:7277C82F-18FF-483A-9747-BF22265B0518.
ZooBank registration number of Tunicothrix halophila n. sp.: dXJuOmxzaWQ6em9vYmFuay5vcmc=:act:D64923EA-0756-4347-9051-D1B1ADAF53C8.
Taxonomy and Morphological Description of Tunicothrix halophila n. sp.
Diagnosis
Body size 45–70 × 12–18 μm in vivo, body outline elliptical to slightly slender with slightly narrow posterior end; cortex flexible, but not contractile. 2 macronuclear nodules. Arc-shaped structures underneath pellicle, 3.0–4.0 μm long in vivo. 3 frontal, 1 parabuccal, and 1 buccal cirrus on frontoventral area; midventral complex composed of 1 cirral pair and 1 cirral row with 3–5 cirri; 1 frontoventral row with 9–12 cirri; 1 pretransverse, 3 transverse cirri; 1 left and 1 right marginal row, left marginal row distinctly shortened posteriorly. 3 dorsal kineties with sparse cilia; caudal cirri lacking. Bipartite adoral zone of membranelles composed of invariably 4 frontal and 9 ventral membranelles; undulating membranelles slightly curved leftward and optically crossing.
Type Locality
Hypersaline water collected from a puddle on a harbor in Jeju Island, South Korea (33°31′9″N, 126°31′44″E). This temporary puddle is right next to the seaside (<1 m) and formed by the waves crashing into the harbor and/or the water from fishermen unloading fishery products. As mentioned in the material and methods section, the salinity was 34.9‰ at the sampling site and the new species was absent at the beginning of the culture. By the evaporation for months in the laboratory, the salinity became high and the species appeared. Tunicothrix halophila n. sp. is very likely a hypersaline species well adapted to temporary habitats in the coastal area because there is no stable hypersaline ecosystem (e.g., solar saltern) near the type locality.
Type Material
The slide containing the holotype (NNIBRPR17559) and three paratype slides (NNIBRPR17560–17562) with protargol-impregnated specimens have been deposited in the Nakdonggang National Institute of Biological Resources, South Korea. Another paratype slide (GUC004219) has been deposited in the Jung-lab (J.-H. Jung) in Gangneung-Wonju National University.
Etymology
The species-group name halophila (feminine) is a composite of the Greek words halós (salt) and philos (preferring), referring to the hypersaline habitat where the species occurs.
Description
Body size in vivo 45–70 × 12–18 μm (n = 12), on average 36.7 × 10.9 μm after protargol impregnation (Figures 1A,B, 2A,B, 3A–F, 4A–C). Body outline elliptical to slightly slender, posterior end narrower than anterior, dorsoventrally flattened (Figure 2F); body flexible but not contractile; cortex smooth except of ventral and ventrolateral grooves along frontoventral and marginal rows; beak-like protrusion between frontal and ventral adoral membranelles (Figures 2A–E,I); alveolar layer-like structure indistinct in vivo, not observed at magnifications of 50–400× (n = 50 cells), observed in some stained cells (3 out of 19 cells examined; Figure 5). Cells grayish to slightly yellowish at low magnification because of ingested algae (Figure 2C). Nuclear apparatus composed of two globular to ellipsoidal macronuclear nodules and one or two micronuclei slightly left of body midline (Figures 1A,D,E, 3A,C,E,H). Typical cortical granules lacking, but colorless arc-shaped structures underneath pellicle, 3.0–4.0 μm long in vivo, not observed in protargol-impregnated specimens (Figure 2D).
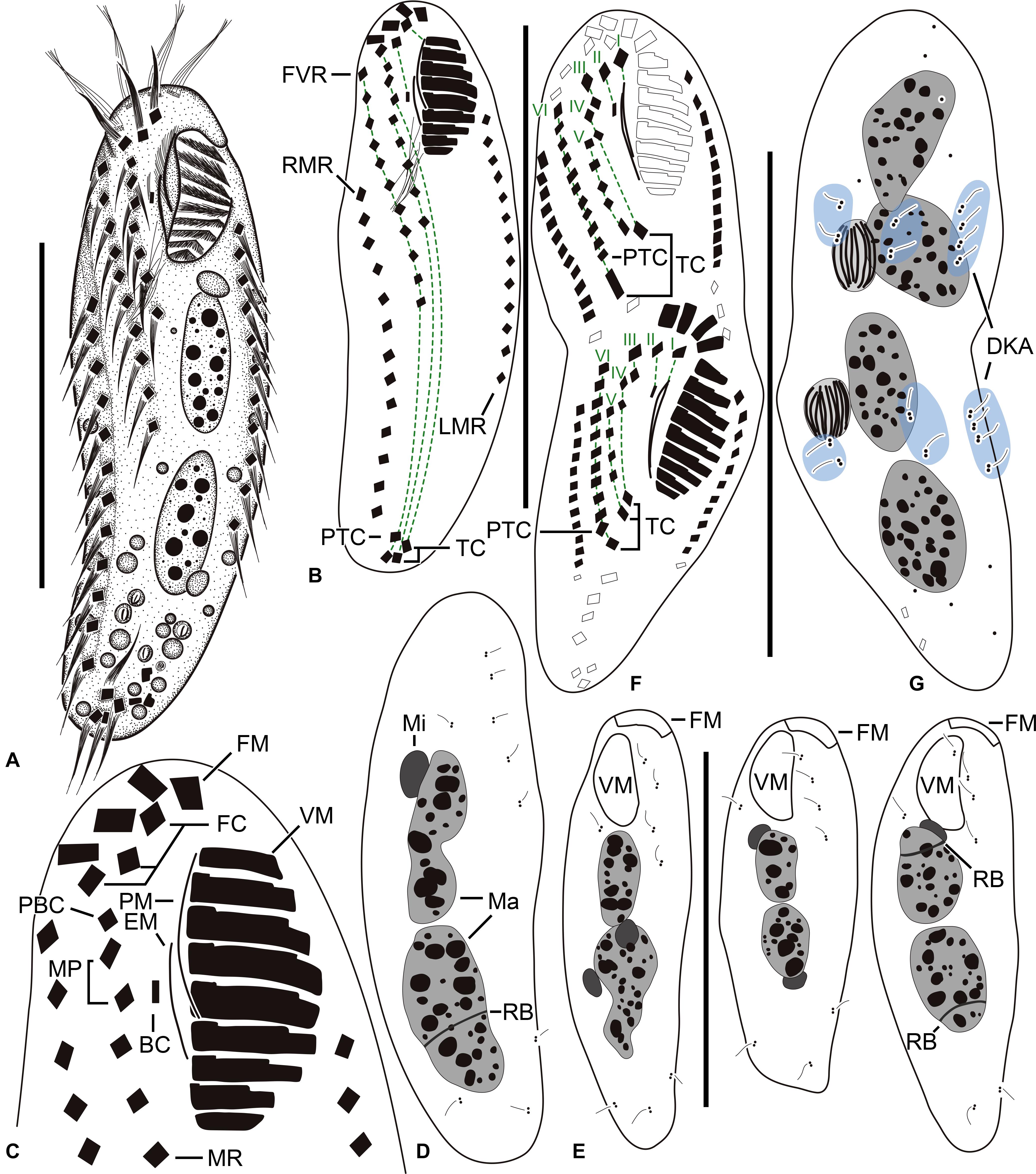
Figure 1. Tunicothrix halophila n. sp. in vivo (A) and after protargol impregnation (B–G). (A) Ventral view of a representative specimen. (B–D) Ventral (B,C) and dorsal (D) views of the holotype specimen. (E) Dorsal views showing dorsal bristles and nuclear apparatus. (F,G) Ventral (F), and dorsal (G) views of a late divider. BC, buccal cirrus; DKA, dorsal kinety anlagen; EM, endoral membrane; FC, frontal cirrus; FM, frontal adoral membranelles; I–VI, frontal-ventral transverse anlagen; FVR, frontoventral row; LMR, left marginal row; Ma, macronuclear nodules; Mi, micronuclei; MP, midventral pair; MR, midventral row; PBC, parabuccal cirrus; PTC, pretransverse cirri; RB, replication band; RMR, right marginal row; TC, transverse cirri; VM, ventral adoral membranelles. Scale bars 30 μm.
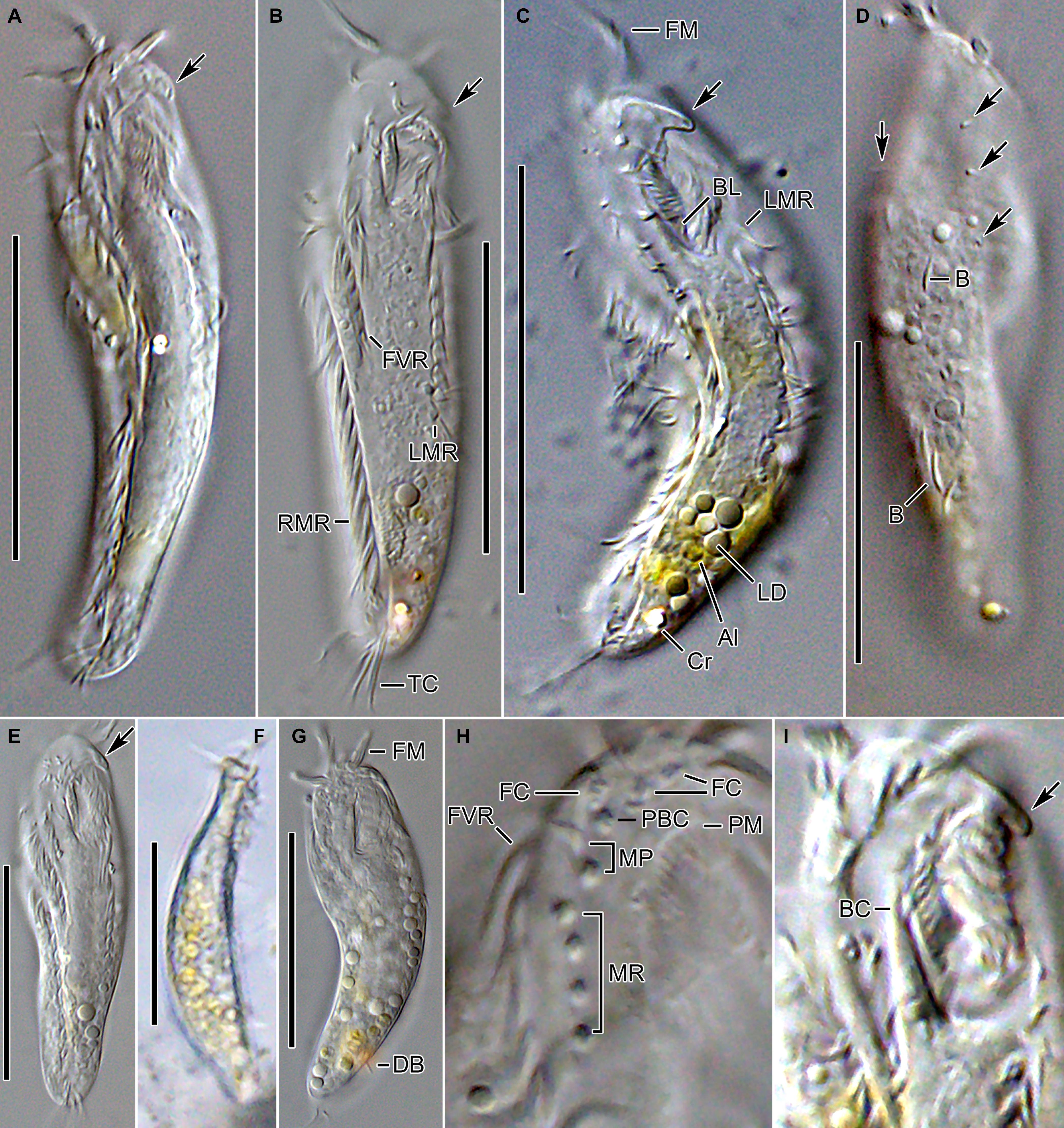
Figure 2. Tunicothrix halophila n. sp. in vivo. (A–C,E) Ventral views showing morphology. Arrows denote beak-like protrusion. (D) Dorsal view showing dorsal bristles (arrows) and arc-shaped cortical structures. (F) Lateral view. (G) Ordinary size of dorsal bristles. (H) Cirri on right frontoventral area. (I) Oral apparatus with beak-like protrusion (arrow). Al, algae; B, arc-shaped structures; BC, buccal cirrus; BL, left wall of buccal lip; Cr, cytoplasmic crystals; DB, dorsal bristles; FC, frontal cirri; FM, frontal adoral membranelles; FVR, frontoventral row; LD, lipid droplets; LMR, left marginal row; MP, midventral pair; MR, midventral row; PBC, parabuccal cirrus; PM, paroral membrane; RMR, right marginal row; TC, transverse cirri. Scale bars 30 μm.
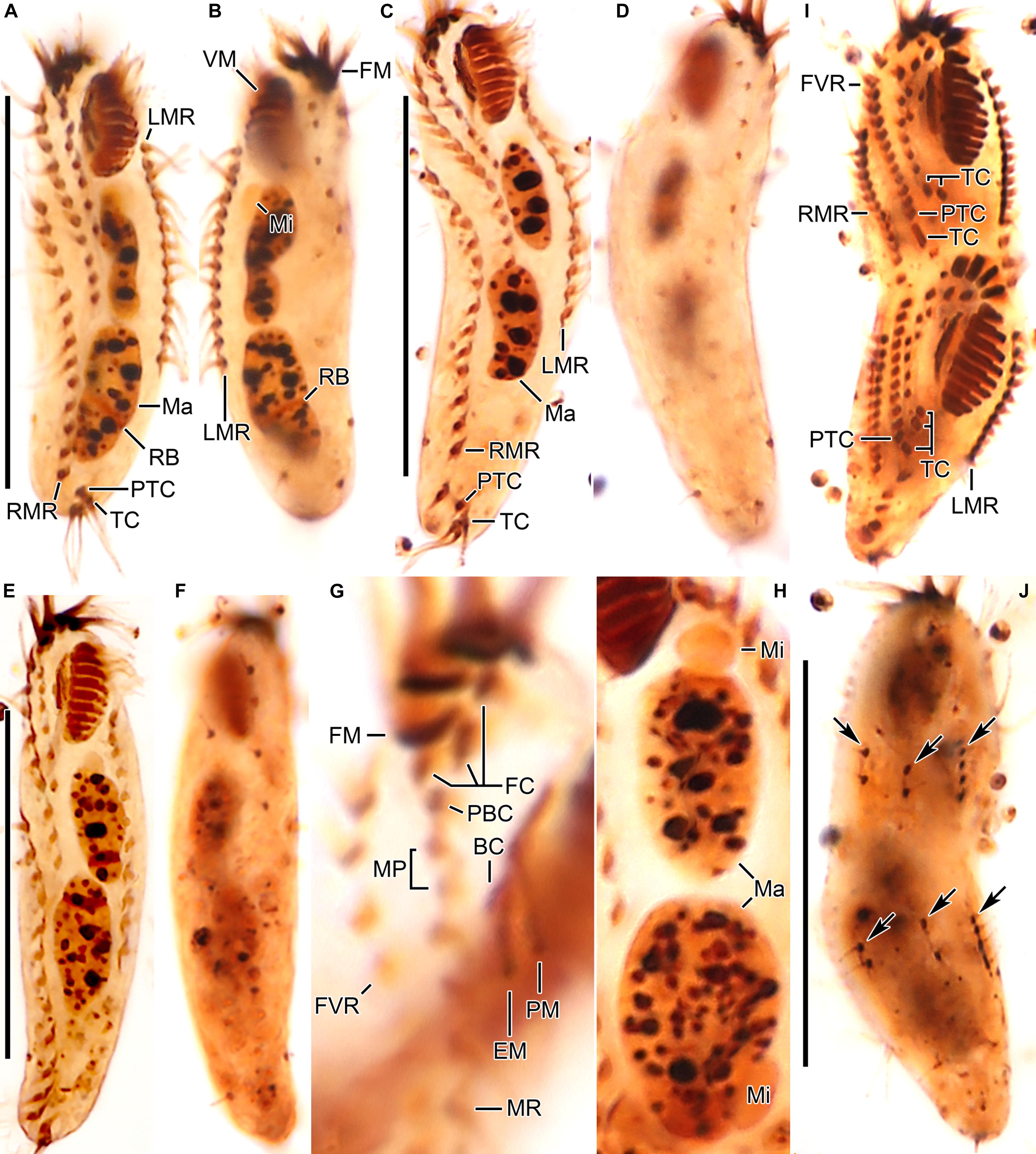
Figure 3. Tunicothrix halophila n. sp. after protargol impregnation. (A,B) Ventral (A) and dorsal (B) views of the holotype specimen. (C–F) Ventral (C,E) and dorsal (D,F) views. (G) Ventral view showing cirri on right frontoventral area. (H) Nuclear apparatus. (I,J) Ventral (I), and dorsal (J) view of a late divider. Arrows denote dorsal kinety anlagen. BC, buccal cirrus; DB, dorsal bristles; EM, endoral membrane; FC, frontal cirri; FM, frontal adoral membranelles; FVR, frontoventral row; LMR, left marginal row; Ma, macronuclear nodules; Mi, micronuclei; MP, midventral pair; MR, midventral row; PBC, parabuccal cirrus; PM, paroral membrane; PTC, pretransverse cirri; RB, replication band; RMR, right marginal row; TC, transverse cirri; VM, ventral adoral membranelles. Scale bars 30 μm.
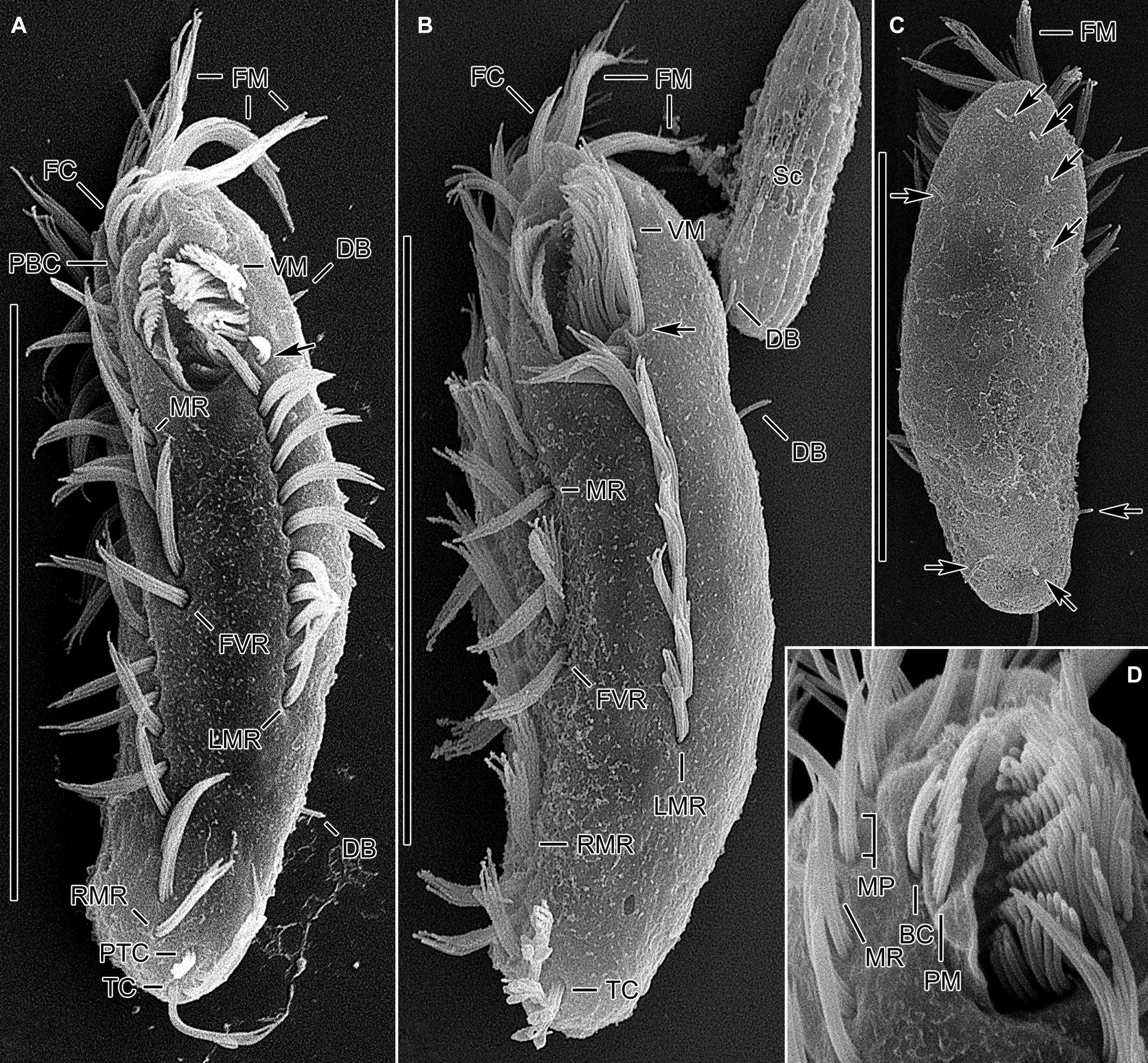
Figure 4. Tunicothrix halophila n. sp. in the scanning electron microscope. (A,B) Ventral and lateral view showing typical morphology. Arrows denote anteriormost left marginal cirrus curved leftward. (C) Dorsal view showing highly reduced dorsal bristles (arrows). (D) Oral apparatus. BC, buccal cirrus; DB, dorsal bristles; FC, frontal cirri; FM, frontal adoral membranelles; FVR, frontoventral row; LMR, left marginal row; MP, midventral pair; MR, midventral row; PBC, parabuccal cirrus; PM, paroral membrane; PTC, pretransverse cirri; RMR, right marginal row; Sc, unidentified scuticociliate; TC, transverse cirri; VM, ventral adoral membranelles. Scale bars 30 μm.
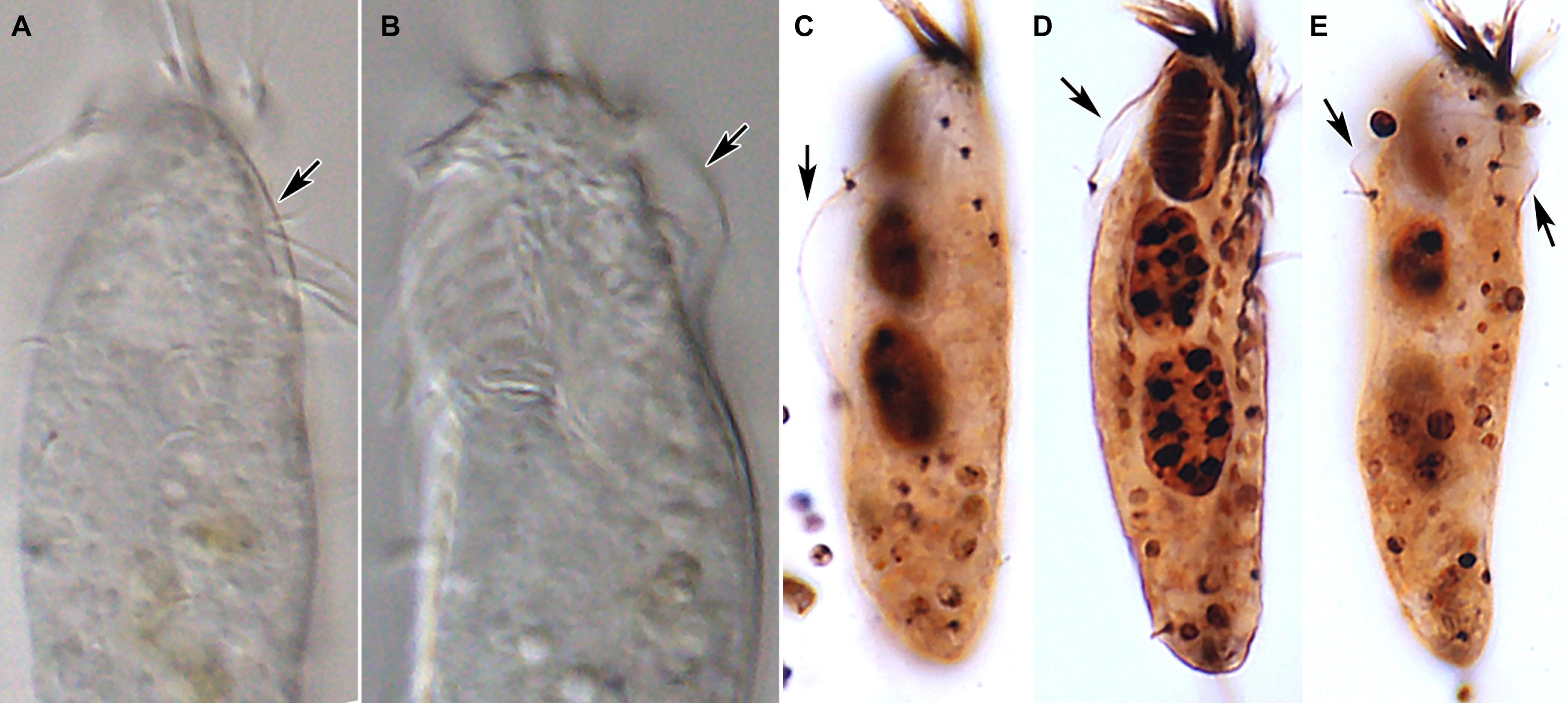
Figure 5. Tunicothrix halophila n. sp. in vivo (A,B) and after protargol impregnation (C–E). Arrows denote the alveolar layer-like structure observed in only few cells.
Cirri, except for buccal cirrus, 6–10 μm long in vivo and composed of four or six cilia (e.g., 2 × 2 or 2 × 3 arrangement) in SEM preparations, of them transverse cirri longest; buccal cirrus composed of three or four cilia in a row, 2–3 μm long in SEM preparations (Figures 2A–C,E, 4). Three frontal cirri, about 6–7 μm long; right frontal cirrus (III/1) below level of distal end of adoral zone of membranelles. Invariably one buccal cirrus right next to endoral membrane in vivo and 1 parabuccal (III/2) cirrus posterior to right frontal cirrus (Figures 1B,C, 2H,I, 3A,C,E,G, 4D). Midventral complex composed of one cirral pair and one cirral row; midventral pair terminates at level of buccal cirrus; midventral row composed of 3–5 cirri, extends to about 30% of body length. Frontoventral row (= right marginal row 1 in Tunicothrix rostrata) composed of 9–12 cirri, commences near level of parabuccal cirrus and terminates near mid-body. One pretransverse cirrus anterior and similar in size to transverse cirri; three transverse cirri arranged in J-shaped pattern. One left and one right marginal row; right row composed of 10–13 cirri, distinctly shortened anteriorly; left row, composed of 11–14 cirri, distinctly shortened posteriorly, anteriormost cirrus always slightly left to level of second cirrus.
Dorsal kineties very likely composed of three rows; sparsely ciliated (e.g., one or two bristles in row 1, one or two bristles in row 2, four to seven bristles in row 3; however, it should be noted that the “sparse” arrangement of bristles hampers accurate counting of the number of dorsal kineties); usually one bristle at anterior and/or posterior end of dorsal kineties 1 and 2; in dorsal kinety 3, usually four bristles anterior to mid-body and invariably one bristle at posterior body end; bristles about 2 μm long in vivo; caudal cirri lacking (Figures 1D,E,G, 2G, 3B,D,F,G, 4C).
Bipartite adoral zone (e.g., frontal and ventral membranelles) extends to about 25% of body length; DE value 0.29 on average; frontal and ventral zone invariably composed of 4 and 9 membranelles, respectively; cilia from frontal membranelles 7–12 μm long in vivo while those from ventral membranelles distinctly shorter (Figures 1B,C,E, 2A,C,E,G,I, 4A–C). Largest ventral membranelle composed of four ciliary rows and 3.3 μm wide in protargol preparations. Buccal cavity narrow and shallow. Undulating membranes slightly curved leftward and optically crossing at anterior region of endoral membrane; paroral membrane composed of one or two (slightly zigzaging) combined rows of basal bodies, cilia 2–3 μm long. Buccal lip flat-type; left wall of buccal lip distinct and 2–3 μm wide. Cytopharynx obliquely extending to right body margin in protargol preparations.
Late Divider
We found a late divider from protargol preparations (Figures 1F,G, 3I,J). The specimen shows six frontal-ventral-transverse cirral anlagen, of which the posteriormost cirri of anlagen IV–VI become pretransverse and transverse cirri. The midventral complex originates from anlagen IV (for midventral pair) and V (for midventral row). Three dorsal kinety anlagen for each daughter cell were developed. Considering the interphasic specimens (Table 1), new cilia very likely replace all parental ones; however, we cannot confirm the replacement of parental membranelles.
18S rDNA Phylogeny
The SSU rDNA sequence of Tunicothrix halophila n. sp. is 1,462 base pairs long and has a GC content of 46.7% (MZ147003). The pairwise similarities among the two congeners range from 96.66% (T. brachysticha GU574811) to 98.15% (T. wilberti GU437210). The family Parabirojimidae is monophyletic, and the sequence of T. halophila n. sp. shows a sister relationship to the clade composed of T. wilberti and Parabirojimia with full supporting value (Figure 6). However, the genus Tunicothrix is non-monophyletic because Parabirojimia sequences are nested within it. In addition, the clade of P. multinucleata and P. similis shows low supporting value (43) even though they cluster together. In other trees (data not shown) with small data sets, the genus Parabirojimia is paraphyletic because the clade composed of T. halophila n. sp. and T. wilberti are nested within it.
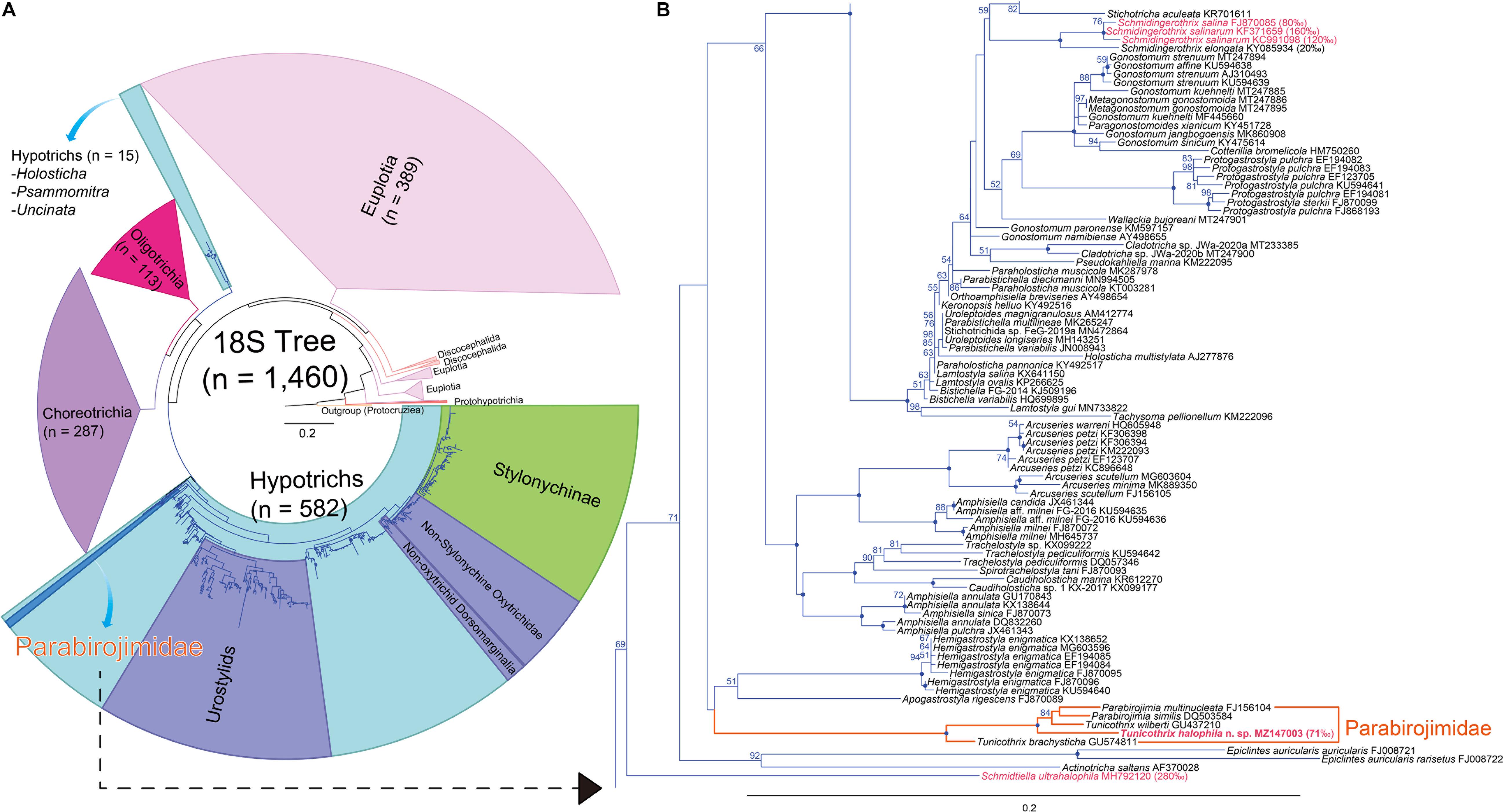
Figure 6. Maximum likelihood (ML) phylogenetic tree inferred from the 18S rRNA gene sequences (A,B). (A) Complete tree comprising 1,460 sequences. (B) The clades containing the family Parabirojimidae and other halophile hypotrichs. Bootstrap values are shown near nodes, values ≤ 50% were not shown, dots at nodes represent 100 bootstrap value. Newly obtained sequence is in bold. Species from hypersaline habitats are in pink color. The scale bar represents 20 nucleotide substitutions per 100 nt.
Discussion
Synonyms of “Right Marginal Row 1”
Many hypotrich ciliates (such as the gonostomatids, oxytrichids, and urostylids) are characterized by the presence of a cirral row which comprises few to many cirri on the right side of the buccal field and commences left and usually slightly ahead of the anterior end of the right marginal row. The terminology of this row depends on the number of cirri, its length, and its origin.
In the family Parabirojimidae, this row of cirri is long (usually as long as the right marginal row) and called “right ventral row” in T. wilberti (Lin and Song, 2004) and “right marginal row 1” in the other three species of Tunicothrix and Parabirojimia species (Hu et al., 2002; Xu et al., 2006; Chen et al., 2010; Dai and Xu, 2011). A similar row was called “frontoventral cirral row” in Schmidingerothrix (Lu et al., 2018). This row is represented by a few to many cirri in the gonostomatids and the core urostylids, while there are only two cirri in the oxytrichids called “frontoterminal cirri” (Berger, 1999, 2006, 2011; Foissner, 2016). On the other hand, the “amphisiellid median cirral row” definitely differs from that row because it is a mixed row composed of more than one true row. This organization is recognizable only during the ontogenetic process (Eigner and Foissner, 1994; Berger, 2008), and in his monograph, Berger (2008) used the neutral term “frontoventral row.”
Considering the ontogenesis, it seems that the origin of the hypotrich’s inner row (right marginal row 1) does not depend on its length. For instance, the long row of some hypotrichs originates intrakinetally (e.g., parabirojimids, schmidingerotrichids, and some gonostomatids) (Hu et al., 2002; Lin and Song, 2004; Wang et al., 2011, 2017; Foissner et al., 2014; Lu et al., 2018; present study) while in some other gonostomatids, it originates de novo or from oral primordium (e.g., Gonostomum strenuum (Song, 1990). However, this row could be considered a second (inner) marginal row if it originates intrakinetally and does not produce transverse cirri at the posterior end as in Cladotricha, Parabistichella, Paragonostomoides, and Schmidingerothrix (Berger, 2011; Foissner et al., 2014; Wang et al., 2017; Lu et al., 2018; Dong et al., 2020). On the other hand, it is considered a frontal-ventral-transverse row if it originates de novo (e.g., Eschaneustyla), from oral primordium (e.g., Gonostomum strenuum) or produces transverse cirrus despite its origin as in Metagonostomum and the parabirojimids (Hemberger, 1982, 1985; Song, 1990; Berger, 1999, 2006, 2011; Deng et al., 2018). Thus, we conclusively recommend using the neutral term “frontoventral row” for such row in the genus Tunicothrix.
Comparison of T. halophila n. sp. With Closely Related Species
The genus Tunicothrix consists of four species as follows: T. brachysticha Dai and Xu, 2011; T. multinucleata Dai and Xu, 2011; T. rostrata Xu et al., 2006 (type species); and T. wilberti (Lin and Song, 2004) Xu et al., 2006. Considering the body size, the alveolar layer, and the numbers of cirri and membranelles, T. halophila n. sp. can be easily distinguished from these congeners by having the smallest body size and the least numbers of cirri and membranelles (Lin and Song, 2004; Xu et al., 2006; Dai and Xu, 2011).
Of the four congeners, T. brachysticha resembles T. halophila n. sp. However, as mentioned above, T. brachysticha differs from the new species by the body length (55.0–77.0 μm vs. 30.2–47.7 μm after protargol impregnation), the alveolar layer (distinct vs. absent/indistinct), and the numbers of frontal (6–9 vs. 4) and ventral (14–17 vs. 9) adoral membranelles, cirri in midventral row (8–13 vs. 3–5), left marginal cirri (17–30 vs. 10–13), right marginal cirri (16–22 vs. 11–14), and cirri in frontoventral row (16–22 vs. 9–12). In addition, they differ in the number (ordinary, i.e., equidistantly arranged in bipolar rows vs. reduced in T. halophila n. sp.) of dorsal bristles (Dai and Xu, 2011).
Establishing a New Genus for T. halophila n. sp.
Tunicothrix halophila n. sp. differs from all other congeners by the absence/indistinctness (vs. presence) of the alveolar layer, which is a character state for the diagnosis/etymology of Tunicothrix (Xu et al., 2006). In addition, the highly reduced number of dorsal bristles (vs. ordinary in the type species T. rostrata) and the hypersaline habitat might be enough to establish a new genus. To justify the establishment of a new genus, however, further studies are needed on the following issues.
First, the genus Tunicothrix is non-monophyletic in the 18S rRNA gene tree (Zhang et al., 2018; Moon et al., 2020; the present study), i.e., T. brachysticha shows a basal position to the clade containing Parabirojimia and other Tunicothrix species. Second, the gene sequence of T. brachysticha (GU574811) was not obtained from the type population (Dai and Xu, 2011) that results in emphasizing a redescription of this species in terms of the morphology and DNA sequence simultaneously obtained from the same population. Third, we do not know the arrangement of dorsal bristles for T. brachysticha and T. multinucleata (Dai and Xu, 2011). All congeners including T. halophila n. sp. have three dorsal kineties, but Dai and Xu (2011) mentioned “dorsal kineties with sparse cilia” for the type population of T. multinucleata. Fourth, a detailed morphology of a Chinese population of T. wilberti (Huang et al., 2010; GenBank Acc. No. GU437210), which is necessary to clarify the non-monophyly of Tunicothrix in the gene tree (Figure 6), is unavailable. In addition, one (Wang et al., 2011) of the three populations of T. wilberti described from China (Lin and Song, 2004; Huang et al., 2010; Wang et al., 2011) differs from the type population by the widely (vs. ordinarily) spaced cilia (see Figure 4E in Wang et al., 2011). Further, the micrographs Figure 5 of Huang et al. (2010) and Figure 4F of Wang et al. (2011) likely belong to the same specimen. Thus, we cannot exclude that the populations of Huang et al. (2010) and Wang et al. (2011) are misidentifications.
In terms of the hypersaline habitat, T. halophila occurred at 71‰ while the other congeners are as follows: 31‰ for the type species/population T. rostrata (Xu et al., 2006); 18.5‰ for the type population of T. wilberti (Lin and Song, 2004), 20‰ for T. wilberti sensu Wang et al. (2011), salinity unavailable for T. wilberti sensu Huang et al. (2010); 29‰ for the type population of T. brachysticha (Dai and Xu, 2011); and 29‰ for the type population of T. multinucleata (Dai and Xu, 2011). The hypersaline habitat is very likely related with the reduction of dorsal bristles as in the genera Apourosomoida Foissner et al., 2002, Erniella Foissner, 1987, Etoschothrix Foissner et al., 2002, and Schmidtiella Li et al., 2019, or even the total loss of dorsal bristles as in Schmidingerothrix Foissner, 2012.
In conclusion, the reduction of dorsal ciliature in T. halophila n. sp. represents a diagnostic character state enough to establish a new genus. However, we refrain from establishing a new genus until the dorsal kinety pattern of other Tunicothrix species and populations is documented.
Secondarily Oligomerized Dorsal Bristles
According to Berger (2008, 2011), the last common ancestor of the hypotrichs very likely had 18 frontal-ventral-transverse cirri originated from six cirral anlagen and three bipolar dorsal kineties each bearing a caudal cirrus. From our gene tree (Figure 6) and a previous report (Li et al., 2019), Schmidtiella ultrahalophila showed a basal position to most hypotrichs, and it has six cirral anlagen and one dorsal kinety. In addition, other basal taxa such as Epiclintes and Actinotricha saltans have more cirral anlagen and more dorsal kineties, respectively (Song et al., 1991; Hu et al., 2009). It is rather difficult to infer the last common ancestor of hypotrichs from the 18S rRNA gene because the supporting values of their clades are low, and they are non-monophyletic (see Holosticha in Figure 6A). Very likely, this is caused by undersampling, for instance, based on the recent redefinition on the Hypotrichia (Da Silva Paiva, 2020), the genus Holosticha forms a clade within hypotrichs, but with the increase of taxon sampling, it showed a sister relationship to the clade composed of Oligotrichia and Choreotrichia (Gao et al., 2016; Sheng et al., 2018).
Considering the reduced number of dorsal bristles, we can find some hypersaline hypotrichs at basal positions of the 18S rRNA gene tree (Foissner, 2012; Foissner et al., 2014; Lu et al., 2018; Li et al., 2019). These hypotrichs have very few or no dorsal bristles at all. According to Foissner (2012) and Remmert (1992), the reduction of the dorsal ciliature is very likely a modification caused by the hypersaline habitat. As shown in Figure 6, these hypersaline hypotrichs are definitely non-monophyletic, supporting the secondary oligomerization hypothesis. However, the interior branches on the basal taxa of the gene tree show lower supporting values even with the huge increase of taxon sampling, so further studies are necessary for basal hypotrichs or those living in hypersaline habitats to clarify their evolutionary relationship/history.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Wm9vQmFuay5vcmc=; dXJuOmxzaWQ6em9vYmFuay5vcmc=:pub:7277C82F-18FF-483A-9747-BF22265B 0518; dXJuOmxzaWQ6em9vYmFuay5vcmc=:act:D64923EA-0756-4347-9051-D1B1ADAF53C8
Author Contributions
AO carried out all laboratory work (preparations, analyses, micrographs, etc.). JHM collected the water sample and maintained the raw culture. AO, SWN, and J-HJ wrote the manuscript. All authors revised the manuscript.
Funding
This work was supported by grants from the National Research Foundation of Korea (NRF) grand funded by the Korea Government (MSIT) (No. 2020R1C1C1011199), and from the Nakdonggang National Institute of Biological Resources (NNIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NNIBR202101102).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Center for Research Facilities at Gangneung-Wonju National University for their assistance in the analysis of the cell structure (CPD, SEM, etc.).
Footnotes
References
Berger, H. (1999). Monograph of the Oxytrichidae (Ciliophora, Hypotrichia). Monographiae Biologicae, Vol. 78. Dordrecht: Springer, 938–1080.
Berger, H. (2006). Monograph of the Urostyloidea (Ciliophora, Hypotricha). Monographiae Biologicae, Vol. 85. Dordrecht: Springer, 1304.
Berger, H. (2008). Monograph of the Amphisiellidae and Trachelostylidae (Ciliophora, Hypotricha). Monographiae Biologicae, Vol. 88. Dordrecht: Springer, 683–737.
Berger, H. (2011). Monograph of the Gonostomatidae and Kahliellidae (Ciliophora, Hypotricha). Monographiae Biologicae, Vol. 90. Dordrecht: Springer, 697–741.
Borror, A. C. (1972). Revision of the order Hypotrichida (Ciliophora. Protozoa). J. Protozool. 19, 1–23. doi: 10.1111/j.1550-7408.1972.tb03407.x
Chen, X., Gao, S., Song, W., Al-Rasheid, K. A. S., Warren, A., Gong, J., et al. (2010). Parabirojimia multinucleata spec. nov. and Anteholosticha scutellum (Cohn, 1866) Berger, 2003, marine ciliates (Ciliophora, Hypotrichida) from tropical waters in southern China, with notes on their small-subunit rRNA gene sequences. Int. J. Syst. Evol. Microbiol. 60, 234–243. doi: 10.1099/ijs.0.008037-0
Da Silva Paiva, T. (2020). Systematic redefinition of the Hypotricha (Alveolata, Ciliophora) based on combined analyses of morphological and molecular characters. Protist 171:125755. doi: 10.1016/j.protis.2020.125755
Dai, R., and Xu, K. (2011). Taxonomy and phylogeny of Tunicothrix (Ciliophora, Stichotrichia), with the description of two novel species, Tunicothrix brachysticha n. sp. and Tunicothrix multinucleata n. sp., and the establishment of Parabirojimidae n. fam. Int. J. Syst. Evol. Microbiol. 61, 1487–1496. doi: 10.1099/ijs.0.024463-0
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Deng, Y., Lu, X., Li, J., Ba, S., and Paiva, T. D. S. (2018). Morphology of two Eschaneustyla species (Ciliophora, Urostylida), with notes on morphogenesis of Eschaneustyla lugeri. Acta Protozool. 57, 69–78.
Dong, J. Y., Li, L. F., Fan, X. P., Ma, H. G., and Warren, A. (2020). Two Urosoma species (Ciliophora, Hypotrichia): a multidisciplinary approach provides new insights into their ultrastructure and systematics. Eur. J. Protistol. 72:125991.
Eigner, P., and Foissner, W. (1994). Divisional morphogenesis in Amphisiellides illuvialis n. sp., Paramphisiella caudata (Hemberger) and Hemiamphisiella terricola Foissner, and redefinition of the Amphisiellidae (Ciliophora, Hypotrichida). J. Eukaryot. Microbiol. 41, 243–261. doi: 10.1111/j.1550-7408.1994.tb01505.x
Foissner, W. (1987). Neue und wenig bekannte hypotriche und colpodide Ciliaten (Protozoa: Ciliophora) aus Böden und Moosen. Zool. Beitr. 31, 187–282.
Foissner, W. (1999). Notes on the soil ciliate biota (Protozoa, Ciliophora) from the Shimba Hills in Kenya (Africa): diversity and description of three new genera and ten new species. Biodivers. Conserv. 8, 319–389.
Foissner, W. (2012). Schmidingerothrix extraordinaria nov. gen., nov. spec., a secondarily oligomerized hypotrich (Ciliophora, Hypotricha, Schmidingerotrichidae nov. fam.) from hypersaline soils of Africa. Eur. J. Protistol. 48, 237–251. doi: 10.1016/j.ejop.2011.11.003
Foissner, W. (2014). An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int. J. Syst. Evol. Microbiol. 64, 271–292. doi: 10.1099/ijs.0.057893-0
Foissner, W. (2016). Terrestrial and Semiterrestrial Ciliates (Protozoa, Ciliophora) from Venezuela and Galápagos. Denisia, Vol. 35. Linz: Biologiezentrum/Oberösterreichisches Landesmuseum 912.
Foissner, W., Agatha, S., and Berger, H. (2002). Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib desert. Part I: text and line drawings. Denisia 5, 1–1063. doi: 10.1111/j.1469-7998.1989.tb02561.x
Foissner, W., and Al-Rasheid, K. (2006). A unified organization of the stichotrichine oral apparatus, including a description of the buccal seal (Ciliophora: Spirotrichea). Acta Protozool. 45, 1–16. doi: 10.11646/zootaxa.504.1.1
Foissner, W., Filker, S., and Stoeck, T. (2014). Schmidingerothrix salinarum nov. spec. is the molecular sister of the large oxytrichid clade (Ciliophora, Hypotricha). J. Eukaryot. Microbiol. 61, 61–74. doi: 10.1111/jeu.12087
Gaievska a, N. (1925). Sur deux nouveaux infusoires des mares sal es–Cladotricha koltzowii nov. gen. nov. sp. et Palmarium salinum nov. gen. nov. sp. Russk. Arkh. Protist. 4, 255–288.
Gao, F., Warren, A., Zhang, Q., Gong, J., Miao, M., Sun, P., et al. (2016). The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep. 6:24874.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hemberger, H. (1982). Revision der Ordnung Hypotrichida Stein (Ciliophora, Protozoa) an Hand von Protargolpräparaten und Morphogenesedarstellungen. Ph.D. dissertation. Bonn: Universität Bonn.
Hemberger, H. (1985). Neue gattungen und arten hypotricher ciliaten. Arch. Protistenk. 130, 397–417. doi: 10.1016/s0003-9365(85)80051-8
Hu, X., Fan, X., Lin, X., Gong, J., and Song, W. (2009). The morphology and morphogenesis of a marine ciliate, Epiclintes auricularis rarisetus nov. sspec. (Ciliophora, Epiclintidae), from the Yellow Sea. Eur. J. Protistol. 45, 281–291. doi: 10.1016/j.ejop.2009.01.005
Hu, X., Song, W., and Warren, A. (2002). Observations on the morphology and morphogenesis of a new marine urostylid ciliate, Parabirojimia similis nov. gen., nov. spec. (Protozoa, Ciliophora, Hypotrichida). Eur. J. Protistol. 38, 351–364. doi: 10.1078/0932-4739-00880
Huang, J., Yi, Z., Al-Farraj, S. A., and Song, W. (2010). Phylogenetic positions and taxonomic assignments of the systematically controversial genera, Spirotrachelostyla, Uroleptopsis and Tunicothrix (Protozoa, Ciliophora, Stichotrichia) based on small subunit rRNA gene sequences. Syst. Biodivers. 8, 409–416. doi: 10.1080/14772000.2010.508502
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, J. H., and Jung, J.-H. (2017). Cytological staining of protozoa: a case study on the impregnation of hypotrichs (Ciliophora: Spirotrichea) using laboratory-synthesized protargol. Anim. Cells Syst. 21, 412–418. doi: 10.1080/19768354.2017.1376707
Li, F., Qu, Z., Luo, D., Filker, S., Hu, X., and Stoeck, T. (2019). Morphology, morphogenesis and molecular phylogeny of a new obligate halophile ciliate, Schmidtiella ultrahalophila gen. nov., spec. nov. (Ciliophora, Hypotrichia) isolated from a volcanic crater on Sal (Cape Verde Islands). J. Eukaryot. Microbiol. 66, 694–706. doi: 10.1111/jeu.12714
Lin, X., and Song, W. (2004). A new marine ciliate, Erniella wilberti sp. n. (Ciliophora: Hypotrichida), from shrimp culturing waters in north China. Acta Protozool. 43, 55–60.
Lu, X., Huang, J., Shao, C., and Berger, H. (2018). Morphology, cell-division, and phylogeny of Schmidingerothrix elongata spec. nov. (Ciliophora, Hypotricha), and brief guide to hypotrichs with Gonostomum-like oral apparatus. Eur. J. Protistol. 62, 24–42. doi: 10.1016/j.ejop.2017.11.001
Lynn, D. H. (2008). The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature. New York: Springer.
Moon, J. H., Kim, J. H., Quintela-Alonso, P., and Jung, J.-H. (2020). Morphology, morphogenesis, and molecular phylogeny of Neobakuella aenigmatica n. sp. (Ciliophora, Spirotrichea, Bakuellidae). J. Eukaryot. Microbiol. 67, 54–65. doi: 10.1111/jeu.12753
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Ruinen, J. (1938). Notizen über ciliaten aus konzentrierten salzgewässern. Zool. Meded. 20, 243–256.
Sheng, Y., He, M., Zhao, F., Shao, C., and Miao, M. (2018). Phylogenetic relationship analyses of complicated class Spirotrichea based on transcriptomes from three diverse microbial eukaryotes: Uroleptopsis citrina, Euplotes vannus and Protocruzia tuzeti. Mol. Phylogenet. Evol. 129, 338–345. doi: 10.1016/j.ympev.2018.06.025
Song, W. (1990). A comparative analysis of the morphology and morphogenesis of Gonostomum strenua (Engelmann, 1862) (Ciliophora, Hypotrichida) and related species. J. Protozool. 37, 249–257. doi: 10.1111/j.1550-7408.1990.tb01136.x
Song, W., Shin, M. K., and Kim, W. (1991). Morphology and infraciliature of the marine ciliate Oxytricha saltans (Cohn, 1866) Kahl, 1932 (Protozoa, Ciliophora, Hypotrichida). Kor. J. Syst. Zool. 7, 233–240.
Sonnenberg, R., Nolte, A. W., and Tautz, D. (2007). An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front. Zool. 4:6. doi: 10.1186/1742-9994-4-6
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Wang, J., Ma, J., Qi, S., and Shao, C. (2017). Morphology, morphogenesis and molecular phylogeny of a new soil ciliate Paragonostomoides xianicum n. sp. (Ciliophora, Hypotrichia, Gonostomatidae). European Journal of Protistology 61, 233–243. doi: 10.1016/j.ejop.2017.07.001
Wang, Y., Hu, X., Huang, J., Al-Rasheid, K. A. S., and Warren, A. (2011). Characterization of two urostylid ciliates, Metaurostylopsis flavicana spec. nov. and Tunicothrix wilberti (Lin & Song, 2004) Xu et al., 2006 (Ciliophora, Stichotrichia), from a mangrove nature protection area in China. Int. J. Syst. Evol. Microbiol. 61, 1740–1750. doi: 10.1099/ijs.0.024935-0
Wilbert, N., and Kahan, D. (1981). Ciliates of solar lake on the red sea shore. Arch. Protistenk. 124, 70–95. doi: 10.1016/s0003-9365(81)80004-8
Wilbert, N., and Kahan, D. (1986). Semiplatyophrya foissneri nov. gen., nov. spec. und Perisincirra pori nov. spec., Ciliaten aus einem Salzboden des Sinai. Arch. Protistenk. 131, 129–138. doi: 10.1016/s0003-9365(86)80070-7
Xu, K., Lei, Y., and Choi, J. K. (2006). Tunicothrix rostrata n. g., n. sp., a new urostylid ciliate (Ciliophora, Stichotrichia) from the Yellow Sea. J. Eukaryot. Microbiol. 53, 485–493. doi: 10.1111/j.1550-7408.2006.00131.x
Zhang, T., Qi, H., Zhang, T., Sheng, Y., Warren, A., and Shao, C. (2018). Morphology, morphogenesis and molecular phylogeny of a new brackish water subspecies, Neourostylopsis flava paraflava nov. subsp. (Ciliophora, Hypotrichia, Urostylidae), with redefinition of the genus Neourostylopsis. Eur. J. Protistol. 66, 48–62. doi: 10.1016/j.ejop.2018.07.004
Keywords: ciliate, extreme habitat, ontogenesis, phylogeny, taxonomy
Citation: Omar A, Moon JH, Nam SW and Jung J-H (2021) Tunicothrix halophila n. sp., a Secondarily Oligomerized Parabirojimid Hypotrich (Ciliophora, Spirotrichea) From Hypersaline Costal Water in Korea. Front. Microbiol. 12:691361. doi: 10.3389/fmicb.2021.691361
Received: 06 April 2021; Accepted: 25 May 2021;
Published: 05 July 2021.
Edited by:
Per Juel Hansen, University of Copenhagen, DenmarkReviewed by:
Gabriela Küppers, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaXiaozhong Hu, Ocean University of China, China
Copyright © 2021 Omar, Moon, Nam and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-Ho Jung, amhqdW5nQGd3bnUuYWMua3I=
 Atef Omar
Atef Omar Ji Hye Moon
Ji Hye Moon Seung Won Nam
Seung Won Nam Jae-Ho Jung
Jae-Ho Jung