- 1College of Veterinary Medicine, Northwest A&F University, Yangling, China
- 2School Hospital, Northwest A&F University, Yangling, China
Type I interferons (IFN), a family of cytokines widely expressed in various tissues, play important roles in anti-infection immunity. Nevertheless, it is not known whether Brucella spp. could interfere with IFN-I production induced by other pathogens. This study investigated the regulatory roles of Brucella outer membrane protein (Omp)25 on the IFN-I signaling pathway and found that Omp25 inhibited the production of IFN-β and its downstream IFN-stimulated genes induced by various DNA viruses or IFN-stimulatory DNA in human, murine, porcine, bovine, and ovine monocyte/macrophages or peripheral blood mononuclear cells. Brucella Omp25 suppressed the phosphorylation of stimulator of IFN genes (STINGs) and IFN regulatory factor 3 and nuclear translocation of phosphorylated IFN regulatory factor 3 in pseudorabies virus- or herpes simplex virus-1-infected murine, human, or porcine macrophages. Furthermore, we found that Brucella Omp25 promoted cyclic guanosine monophosphate–adenosine monophosphate synthase (cGAS) degradation via the proteasome-dependent pathway, resulting in a decreased cyclic guanosine monophosphate–adenosine monophosphate production and downstream signaling activation upon DNA virus infection or IFN-stimulatory DNA stimulation. Mapping the predominant function domain of Omp25 showed that the amino acids 161 to 184 of Omp25 were required for Omp25-induced cGAS degradation, among which five amino acid residues (R176, Y179, R180, Y181, and Y184) were required for the inhibitory effect of Omp25 on IFN-β induction. Altogether, our results demonstrated that Brucella Omp25 inhibits cGAS STING signaling pathway-induced IFN-β via facilitating the ubiquitin–proteasome-dependent degradation of cGAS in various mammalian monocyte/macrophages.
Introduction
Brucella spp. is a gram-negative facultative intracellular bacterium that causes brucellosis with clinical manifestations of arthritis, endocarditis, meningitis, abortion, and infertility (particularly in cattle) in humans and animals, resulting in significant economic losses to the livestock industry (Baldi and Giambartolomei, 2013a, b; de Figueiredo et al., 2015). Brucella has the propensity to localize inside macrophages, where it inhibits innate immune responses (Campos et al., 2014). The major outer membrane proteins (Omps) of Brucella spp. have been identified as the predominant virulence factors in promoting Brucella spp. infection and pathogenicity and the coinfection with other pathogens (Machelart et al., 2018; Pasquevich et al., 2019). Previous studies have shown that Omp25 plays a critical role in suppressing tumor necrosis factor-alpha (TNF-α) and interleukin (IL)-12 expression induced by other pathogens (Jubier-Maurin et al., 2001; Cui et al., 2017; Luo et al., 2017). However, it is unknown if Brucella infection might increase other pathogen infections by interfering with the induction of type I interferon (IFN) and the function of major Omps.
According to their apparent molecular mass, the Omps were classified into 36- to 38-kDa OMPs or group 2 porin proteins, 25- to 27-kDa, and 31- to 34-kDa OMPs, which belong to group 3 proteins (Cloeckaert et al., 2002). Omp10 is a kind of outer membrane lipoprotein belonging to 31- to 34-kDa Omps, which is necessary for Brucella spp. 544 virulence (Tibor et al., 2002). Omp2a is one of the porin proteins belonging to 36- to 38-kDa Omps (Cloeckaert et al., 2002). Omp25, an outer membrane protein of Brucella, belonging to the group 3 proteins (Guzman-Verri et al., 2002; Vizcaino et al., 2004), is a Brucella key virulence factor (Glowacka et al., 2018) and is involved in the negative regulation of TNF-α production during infection of human macrophages (Jubier-Maurin et al., 2001). In addition, Omp25 is often regulated by environmental signals and plays an important role in bacterial pathogenesis by enhancing the adaptability to various environments (Caro-Hernandez et al., 2007). Our previous studies have demonstrated that Brucella suis Omp25 downregulates the production of TNF-α via regulation of different microRNAs to promote intracellular survival in porcine and murine macrophages (Luo et al., 2017) and negatively regulates IL-12 production at both transcriptional and post-transcriptional levels through regulation of programmed cell death protein 1 signaling by inducing miR-155, -23b, and -21-5p (Cui et al., 2017). However, the role of Omp25 in regulating the production of IFN-β has not been defined.
The present study investigated the roles of Omp25 in the regulation of DNA viruses or IFN-stimulatory DNA (ISD)-induced IFN-β expression. We found that Omp25 effectively inhibited the cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) synthase (cGAS)–stimulator of IFN genes (STINGs)-mediated induction of IFN-β and downstream IFN-stimulated genes (ISGs) upon DNA viruses or ISD stimulation by promoting the degradation of cGAS. Furthermore, we determined the specific domains of Omp25 responsible for cGAS degradation by reverse transcription-quantitative polymerase chain reaction (qRT-PCR) or Western blotting for quantitative measurement of messenger RNA (mRNA) and protein levels and its regulation of the IFN-I signaling pathway.
Materials and Methods
Sample Collections
Blood samples were collected from 105 bovines from China’s northwest district, including three provinces (Shanxi, Ningxia, and Xinjiang), and most samples were collected from bovines with obvious clinical symptoms (such as abortion, conjunctivitis, and rhinotracheitis). Some samples were collected from clinically healthy animals by conventional detection. Whole blood samples were collected into ethylenediaminetetraacetic acid (EDTA)-coated tubes and stored at −20°C until DNA isolation. Blood samples were collected into tubes without an anticoagulant for separation of serum, allowing to clot at room temperature, and centrifuged at 2,000 rpm/min for 10 min. Serum was collected and kept at −80°C. Other clinically common viral infections (such as bovine viral diarrhea virus, parainfluenza virus, and bovine papillomavirus) were excluded from Brucella-positive samples by enzyme-linked immunosorbent assay (ELISA). According to the manufacturer’s instructions, commercial ELISA kits were used to detect the antibodies against Brucella spp. (F751174-A, Fankewei, Shanghai, China), bovine herpesvirus (BoHV)-1 (F75165-A, Fankewei, Shanghai, China), BoHV-4 (F75169-A, Fankewei, Shanghai, China), bovine viral diarrhea virus (F6732-A, Fankewei, Shanghai, China), parainfluenza virus (F75170-A, Fankewei, Shanghai, China), and bovine papillomavirus (F75022-A, Fankewei, Shanghai, China). All operations were carried out at the biosafety level 2 laboratory.
Cell Culture
Porcine alveolar macrophages (3D4/21 cells), mouse macrophage cell line (RAW264.7 cells), human monocytic cell line THP-1, Vero cells, Madin–Darby bovine kidney cell line (MDBK), and human embryonic kidney 293 T cell line (HEK-293T cells) were all purchased from American Type Tissue Culture (Manassas, VA, United States) and stored in our laboratory. RAW264.7 cells, Vero cells, MDBK cells, and HEK-293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Hyclone, Thermo Scientific HyClone, Beijing, China). 3D4/21 and THP-1 cells were maintained in Roswell Park Memorial Institute-1640 medium (Invitrogen, Carlsbad, CA, United States) with 10% heat-inactivated FBS, sodium pyruvate, non-essential amino acids. All cells were cultured in a fully humidified atmosphere containing 5% carbon dioxide at 37°C.
Viruses and Bacteria Strains
Porcine parvovirus (PPV China-XY) (Genbank: MK993540) was isolated and stored in our laboratory. Pseudorabies virus (PRV GD0304) (Genbank: MH582511.1), herpes simplex virus-1 (HSV-1) (Genbank: KC140233.1), BoHV-1 (NM2_2016) (Genbank: MT179818.1), and BoHV-4 J4034 (Genbank: MN735177.1) were stored in our laboratory.
Brucella abortus (Genbank: X79284.1), B. suis (Genbank: U39397.1), Brucella melitensis (Genbank: JF918759.1), Brucella canis (Genbank: U39358.1), Brucella ovis (Genbank: U33004.1), and Brucella neotomae (Genbank: U39359.1) were also stored in our laboratory.
Cell Isolation and Culture
Peripheral blood mononuclear cells (PBMCs) were prepared from fresh-heparinized venous blood of bovine and ovine by centrifugation over Ficoll-Histopaque (Sigma, St. Louis, MO, United States) as described (Huang et al., 2011). M/MΦs cells were enriched from PBMCs using Ficoll-Percoll gradients (GE Healthcare) and further purified by anti-CD14 magnetic beads with column purification according to the manufacturer’s instructions (Miltenyi Biotec; purity of cells >95%). The enriched cells were cultured in Roswell Park Memorial Institute-1640 medium supplemented with 10% FBS.
Lentivirus Production
The full-length omp25 (Genbank: X79284.1), omp10 (Genbank: L27995.1), and omp2a (Genbank: AY008719.1) were synthesized and cloned to T vector by Sangon Biotech. The omp25 (Δ161-184) and omp25-5M were amplified by overlap PCR, all sequences were confirmed by sequencing analysis (Sangon Biotech, Shanghai, China), and then they were all cloned into PCDH-CMV-MCS-EF1-copGFP (Addgene, Cambridge, MA, United States) vector according to the manufacturer’s instructions in our laboratory, which were then co-transfected with psPAX2 and PMD2-VSV (Addgene, Cambridge, MA, United States) plasmids into HEK-293T cells for 48 h to generate individual lentivirus particles. Supernatants containing lentiviral particles were harvested and concentrated, and viral titer was determined through infecting HEK-293T cells (Sastry et al., 2002).
Enzyme-Linked Immunosorbent Assay
3D4/21, THP-1, and RAW264.7 cells that adhered to six-well plates were infected with 100 multiplicity of infections (MOIs) of lentivirus infection for 48 h and then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 24 h. According to the manufacturer’s instructions, supernatants were harvested to measure IFN-β secretion by ELISA kit (R&D, Minneapolis, MN, United States).
RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction
3D4/21, THP-1, and RAW264.7 cells that adhered to six-well plates were infected with 100 MOIs of lentivirus infection for 48 h and then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 6 h. Bovine PBMCs and ovine PBMCs that adhered to six-well plates were infected with 100 MOIs of lentivirus infection for 48 h, then infected with BoHV-1 or transfected with ISD (2 μg/ml) for another 6 h. IFN-β, ISG56, and C-X-C motif chemokine ligand 10 (IP-10) mRNAs were quantified by qRT-PCR. The primer sequences for real-time PCR are shown in Table 1. According to the manufacturer’s protocol, the total cellular RNA was isolated by TRIZOL. RNA concentration and purity were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States), and equal amounts of RNA (1 μg) were used for qRT-PCR analysis. Relative mRNA levels were detected by an Applied Biosystems QuantStudio 6&7 (Applied Biosystems, Grand Island, NY, United States) and calculated using β-actin as endogenous control, respectively, following the 2–Δ Δ Ct method.
Western Blotting
The cells cultured in a 100-mm-diameter dish (Thermo Fisher Scientific, Waltham, MA, United States) were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 36 or 48 h, then treated with dimethyl sulfoxide, chloroquine (CQ; 20 μM), or MG132 (5 μM) in the presence of cycloheximide (CHX; 100 μg/ml) for another 12 h or further infected by HSV-1 (10 MOI) or PRV (1 MOI) for 6 h; cells were suspended in radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Waltham, MA, United States) supplemented with protease inhibitor (Sigma Aldrich, St. Louis, MO, United States) on ice for 30 min; cytosol and nuclear fractions were isolated according to manufacturer’s instruction (Thermo Fisher Scientific, Waltham, MA, United States). In detail, infected cells were briefly treated with trypsin-EDTA and gently resuspended in DMEM, then centrifuged at 3,000 × g/min for 5 min and washed with PBS. After completely aspirating the PBS, 200 μl of nuclear isolation buffer [1.28-M sucrose, 40-mM Tris–hydrochloride (pH 7.5), 20-mM magnesium chloride, and 4% Triton X-100], 200-μl PBS, and 600-μl water were added to each sample, and the pellets were gently resuspended and placed on ice for 20 min. Samples were centrifuged at 2,500/min for 15 min. Supernatants (cytoplasmic fractions) were collected, and then the pellets (nuclear fraction) were gently resuspended in 500-μl 0.1-M PBS. Finally, the protein was eluted by boiling for 10 min in a 2 × loading buffer. Forty-microgram proteins in each lane were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinyl difluoride membranes (Millipore Corp., Atlanta, GA, United States) for Western blotting. After blocking the membrane with 5% non-fat dry milk for 2 h, we incubated it with primary antibodies at 4°C overnight. The cells were treated with ISD (Invivogen tlrl-ISDN, Cayla, France), protein synthesis inhibitor (CHX; Sigma, St. Louis, MO, United States), lysosome inhibitor (CQ; Sigma, St. Louis, MO, United States), and proteasome inhibitors (MG132, Beyotime, Shanghai, China). Primary antibodies included mouse anti-β-actin (A00702; Genscript, Nanjing, China), mouse anti-tubulin (A01410-100; Genscript, Nanjing, China), rabbit anti-cGAS (31659; CST, Danvers, MA, United States), rabbit anti-phospho-STING (72971; 50907; CST, Danvers, MA, United States), rabbit anti-STING (ab181125; Abcam, Cambridge, MA, United Kingdom), rabbit anti-STING (19851-1-AP; Proteintech, Chicago, IL, United States), anti-phospho-IFN regulatory factor 3 (IRF3) (4947; CST, Danvers, MA, United States), rabbit anti-IRF3 (4302; CST, Danvers, MA, United States), mouse anti-HA antibody (H3663; Sigma, St. Louis, MO, United States), rabbit anti-Flag antibody (701629, Invitrogen, Carlsbad, CA), and rabbit anti-histone H3 (4499; CST, Danvers, MA, United States). The anti-cGAS polyclonal antibody was obtained from a mouse immunized with purified full-length cGAS protein expressed by the pET32a vector in Escherichia coli. Horseradish peroxidase-conjugated anti-mouse immunoglobulin G (BM2002; Wuhan Boster Biotech, Wuhan, China) or anti-rabbit immunoglobulin G (BA1058; Wuhan Boster Biotech, Wuhan, China) was incubated at room temperature for 1 h. According to the manufacturer’s instructions, enhanced chemiluminescence (Bio-Rad, Hercules, CA, United States) was used for detection.
Cyclic Guanosine Monophosphate–Adenosine Monophosphate Activity Assay
Infected or transfected cells were trypsinized and washed with PBS. Pelleted cells were resuspended in the indicated volume of PBS andlysed by freezing at −80°C. Cell extracts were nuclease and heat-treated as previously described (Gao et al., 2013) with modifications. Briefly, cell extracts were incubated with ∼1 U/μl Benzonase (Sigma-Aldrich, St. Louis, MO, United States) for 30 min at 37°C. Cell extracts were then heated at 95°C for 5 min and centrifuged for 5 min at maximum speed (∼16,000 × g) in an Eppendorf microcentrifuge. RAW264.7, THP-1, and 3D4/21 cells were used as a reporter cell line to measure cGAMP production. Cells were permeabilized as described previously (Woodward et al., 2010) with modifications. Briefly, media was aspirated from RAW264.7, THP-1 or 3D4/21 cells, and digitonin permeabilization solution [50-mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.0), 100-mM potassium chloride, 85-mM sucrose, 3-mM magnesium chloride, 0.2% bovine serum albumin, 1-mM adenosine triphosphate, 0.1-mM dithiothreitol, and 10-μg/ml digitonin] was added with 25 μl per well-treated cell extracts or 2′-3′-cGAMP (Invivogen tlrl-cGAS, Cayla, France) as a positive control. RAW264.7, THP-1, and 3D4/21 reporter cells were incubated with extracts for 30 min at 37°C and then replaced with supplemented media. RNA was harvested 6 h after the initial addition of extracts, and qRT-PCR was performed, as described earlier.
Immunoprecipitation Assay
According to the manufacturer’s instructions, cells were cultured in a 100-mm diameter dish and co-transfected with indicated plasmids using the Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, United States); 24 h later, the cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP. At 36-h post-infection, cells were treated with MG132 (5 μM) for another 12 h, then the cells lysed with lysis buffer [150-mM sodium chloride, 50-mM Tris–hydrochloride (pH 7.4), 1% Nonidet P-40, 0.5% Triton X-100, 1-mM EDTA, 0.1% sodium deoxycholate, 1-mM dithiothreitol, 0.2-mM phenylmethylsulfonyl fluoride, and protease inhibitor protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, United States)] on ice for 30 min. The cell lysate supernatant was collected by centrifuging for 15 min at 13,000 × g and incubated with protein A-agarose (Santa Cruz, California, CA, United States) for pre-cleaning of non-specific proteins for 1 h at 4°C. The samples were centrifuged, and the supernatant was incubated with the indicated antibodies overnight at 4°C. The complex was incubated with protein A-agarose again for 30 min at room temperature and then centrifuged at 2,000 × g for 10 s and washed three times with PBS. Finally, the bound proteins were eluted by boiling for 10 min in 2 × loading buffer and analyzed by Western blotting using the indicated antibodies.
Herpes Simplex Virus 1 and Pseudorabies Virus Plaque Assay
Vero cells were seeded into six-well plates at a density of 2 × 106 viable cells per well. On the following day, cells were infected with 100 MOI per cell of Lv-Omp25 or Lv-GFP. At 48 h after infection, cells were incubated with HSV-1 (10 MOI) or PRV (1 MOI) for 2 h at room temperature on a rocking apparatus, then aspirated the inoculum and washed with PBS three times, overlayed the cells with 2 ml of 2 × DMEM supplemented with 2% methylcellulose, 2% FBS per well. Plates were incubated at 37°C and 5% carbon dioxide for 3 days. Cells were stained with 1-ml 1% crystal violet hydrate solution containing 0.6% sodium chloride and 2% formaldehyde per well for 30 min to visualize plaques.
Ethics Statement
This research was approved by the Institutional Animal Care and Use Committee of Northwest A&F University and was performed according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China. No other specific permissions were required for these activities. This study did not involve endangered or protected species.
Statistical Analysis
The results are representative of three independent experiments. The data are presented as mean ± standard error of the mean (standard deviation). Comparisons between the two groups were performed by unpaired Student t-test, whereas multiple group data were analyzed by analysis of variance, followed by Bonferroni post hoc test. Statistically significant and very significant results were defined as P < 0.05 and P < 0.01.
Results
Brucella spp.-Positive Cattle Are More Susceptible to Bovine Herpes Viruses-1 and -4
Firstly, we detected the infection rate and morbidity rate of BoHV-1 and BoHV-4 in Brucella spp.-positive and -negative samples. The results showed that the infection and morbidity rates were higher in Brucella spp.-positive cattle than Brucella spp.-negative cattle (Figures 1A,B). Then, we tested the levels of IFN-β in Brucella spp.-positive and -negative cattle that co-infected with BoHV-1 or BoHV-4, and we found that IFN-β levels were lower in Brucella spp.-positive cattle than that in Brucella spp.-negative cattle (Figure 1C). To further confirm the relationships between IFN-β level and BoHV-1 and BoHV-4 viral load, ELISA was applied to detect the production of IFN-β in different cattle infected with BoHV-1 or BoHV-4; qRT-PCR was applied to detect the copies of BoHV-1 or BoHV-4 in these cattle. The result showed that the level of IFN-β was inversely correlated with BoHV-1 or BoHV-4 copies in these cattle that infected with or without Brucella spp. (Figures 1D,E). Then, we detected replication of BoHV-1 and BoHV-4 in MDBK cells with or without infected Lv-Omp25. MDBK cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 48 h and then infected with BoHV-1 (1 MOI) or BoHV-4 (1 MOI) for another 6 h; we found that Brucella spp. Omp25 inhibited BoHV-1 or BoHV-4 copies in MDBK cells (Figures 1F,G). These data suggested that Brucella spp. infection could inhibit IFN-β to promote BoHV-1 or BoHV-4 infection.
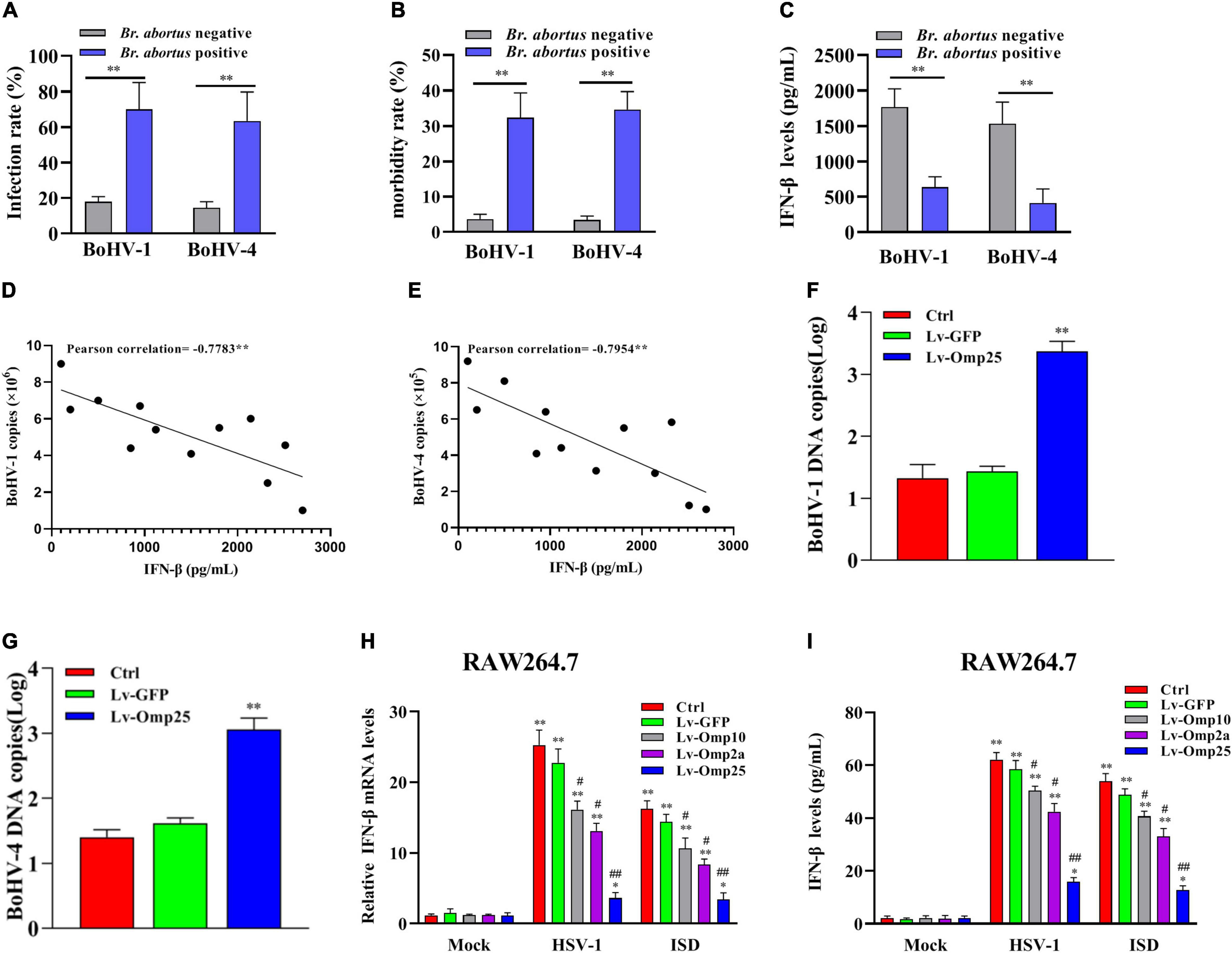
Figure 1. Brucella spp.-positive cattle are more susceptible to BoHV-1 and BoHV-4. (A,B) Serum were collected from Brucella spp.-positive or -negative cattle, and all samples were detected with specific ELISA kits. **P < 0.01 versus Brucella spp.-negative group. (C) IFN-β levels were lower in Brucella spp.-positive cattle than that in Brucella spp.-negative cattle. Serum was collected from Brucella spp.-positive or -negative cattle coinfection BoHV-1 or BoHV-4; level of IFN-β was detected with ELISA kit. **P < 0.01 versus Brucella spp.-negative group. (D,E) Level of IFN-β is inversely correlated with either BoHV-1 or BoHV-4 copies in these cattle that infected with or without Brucella spp. ELISA was applied to detect production of IFN-β in different cattle infected with BoHV-1 or BoHV-4 virus; qRT-PCR was applied to detect copies of BoHV-1 or BoHV-4 in these cattle. **P < 0.01 demonstrated that level of IFN-β is inversely correlated with BoHV-1 or BoHV-4 copies. (F,G) Brucella spp. Omp25 inhibits replication of BoHV-1 and BoHV-4 in MDBK cells. MDBK cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 48 h and then infected with BoHV-1 (1 MOI) or BoHV-4 (1 MOI) for another 6 h, qRT-PCR was applied to detect copies of BoHV-1 or BoHV-4. (H,I) Brucella spp. Omp25 is key protein to inhibit expression of IFN-β induced by HSV-1 or ISD in RAW264.7. RAW264.7s were infected with 100 multiplicity of infection (MOI) of Lv-Omp25, Lv-Omp10, Lv-Omp2a, or Lv-GFP for 48 h and then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 6 or 24 h. Cells were harvested and subjected to qRT-PCR analysis (H) or ELISA analysis (I). *P < 0.05, **P < 0.01 versus mock group, #P < 0.05, ##P < 0.01 versus control or Lv-GFP-infected cells in same secondary or infection stimulation groups. Results are means ± SEMs of three independent experiments.
Next, we evaluated the effects of three outer membrane proteins of Brucella spp. (Omp25, Omp10, and Omp2a) on IFN-β production. RAW264.7 were infected with 100 MOI of Lv-Omp25, Lv-Omp10, Lv-Omp2a, or Lv-GFP for 48 h and then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 6 or 24 h followed by qRT-PCR detection of IFN-β mRNA levels and ELISA detection of IFN-β secretion levels. We found that Omp25, Omp10, and Omp2a could inhibit IFN-β production, but Omp25 showed a most significant inhibitory effect (Figures 1H,I). Thus, we explored how Omp25 inhibits IFN-β production induced by the DNA virus in this study.
Omp25 Inhibits DNA Virus and Interferon-Stimulatory DNA-Induced Interferon-β Production in Various Mammalian Monocyte/Macrophages
To investigate the direct effects of Omp25 on IFN-β production in monocyte cells or macrophages, we examined and compared the IFN-β mRNA and protein levels between various Lv-Omp25-infected and Lv-GFP-infected mammalian monocyte/macrophage cells. Upon DNA viruses [HSV-1 (10 MOI), PPV (1 MOI), PRV (1 MOI), and BoHV-1 (1 MOI)] infection or ISD (2 μg/ml) stimulation, IFN-βs were significantly induced in both mRNA and protein levels in THP-1, 3D4/21 cells, bovine PBMCs, and ovine PBMCs (Figures 2A–F), but we found that the levels of IFN-β mRNA in Lv-Omp25-infected cells were lower than that in Lv-GFP-infected various mammalian cells and control cells regardless of DNA viruses infection or ISD stimulation (Figures 2A,C,E,F). We also observed that the production of IFN-β induced by DNA viruses or ISD (2 μg/ml) was also lower in Lv-Omp25-infected cells than in Lv-GFP-infected and control cells (Figures 2B,D). Correspondingly, the replication of PRV and HSV-1 were increased in Lv-Omp25-infected cells relative to either Lv-GFP-infected or control cells in the plaque assay (Figures 2G,H). These results indicated that Brucella Omp25 could directly inhibit the production of DNA viruses and ISD-induced IFN-β expression at both protein and mRNA levels in various mammalian monocyte/macrophages.
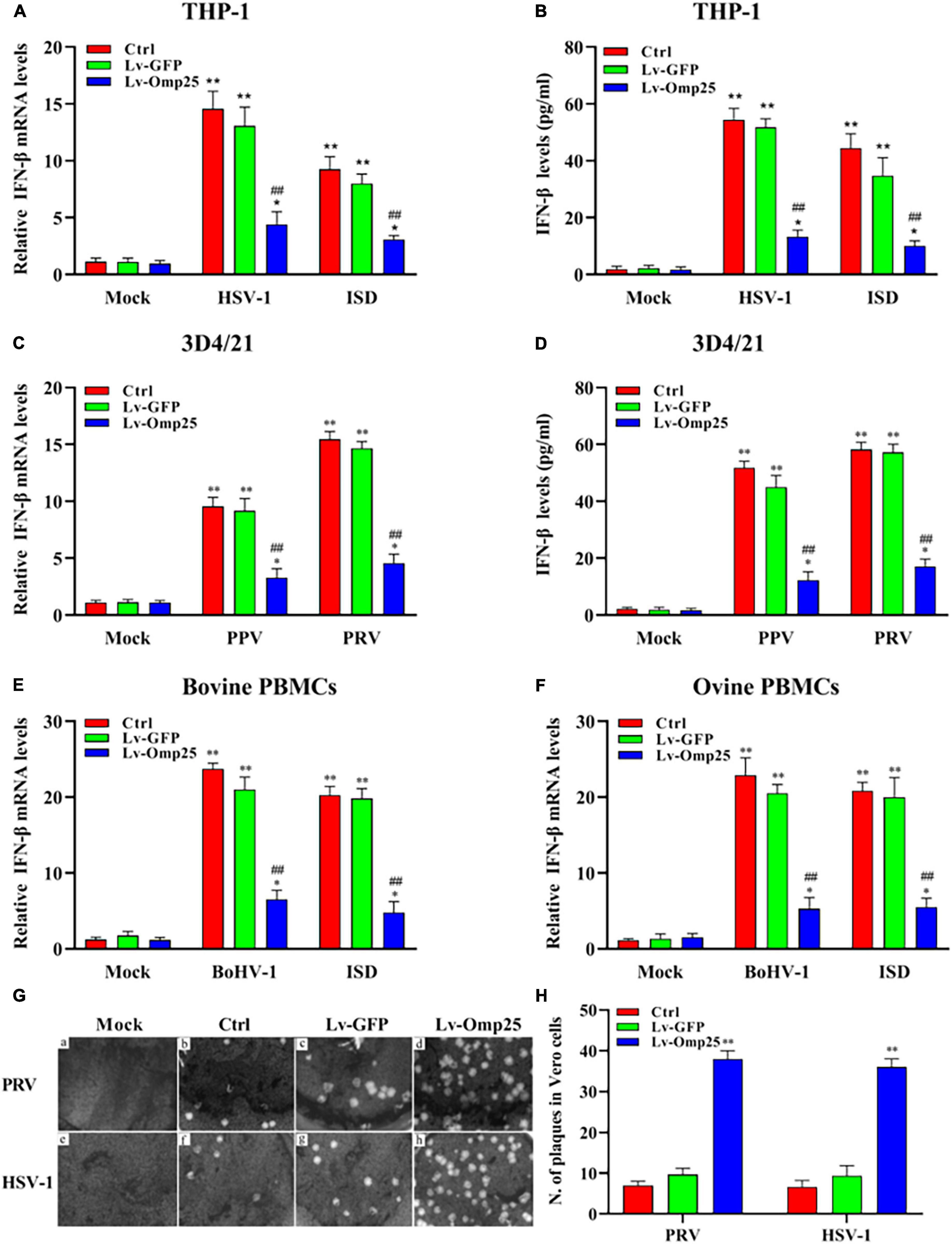
Figure 2. Omp25 inhibits DNA virus and ISD-induced IFN-β expression in various mammalian monocyte/macrophages. (A) Omp25 inhibits IFN-β expression in THP-1 cells. THP-1 cells were infected with 100 multiplicity of infection (MOI) of Lv-Omp25 or Lv-GFP for 48 h and then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 6 h. Cells were harvested and subjected to qRT-PCR analysis. (B) Omp25 inhibits IFN-β production in THP-1 cells. THP-1 cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 48 h and then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 24 h. Cells were harvested and subjected to ELISA analysis. (C,D) Omp25 inhibits PPV- or PRV-induced IFN-β transcription and expression in 3D4/21 cells. Cells were infected and treated as discussed earlier and then infected with PPV (1 MOI) or PRV (1 MOI) for another 6 or 24 h. Cells were harvested and subjected to qRT-PCR analysis or ELISA analysis. (E,F) Omp25 inhibits BoHV-1-induced IFN-β transcription in bovine and ovine PBMCs. Cells were infected and treated as in discussed earlier and then infected with BoHV-1 (1 MOI) or stimulated with ISD (2 μg/ml) for another 6 h. Cells were harvested and subjected to qRT-PCR analysis. (G,H) Omp25 inhibits PRV and HSV-1 replication. Vero cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 48 h and then infected with HSV-1 (10 MOI) or PRV (1 MOI) for 1 h and overlayed with methylcellulose for 3 days, and then, cells were stained with 5% crystal violet hydrate solution for 30 min (G). Number of plaque by taking average number of viruses in per well (H). Results are means ± SEMs of three independent experiments. *P < 0.05, **P < 0.01 versus mock group, #P < 0.05, ##P < 0.01 versus control or Lv-GFP-infected cells in same secondary infection or stimulation groups.
Outer Membrane Protein 25 Inhibits DNA Viruses and Interferon-Stimulatory DNA-Induced Transcription of Interferon-Stimulated Gene
To investigate the effects of Omp25 on transcription of IFN-β-stimulated genes, we analyzed ISG56 and C-X-C motif chemokine ligand 10 (IP-10) expressions by qRT-PCR in various mammalian monocyte/macrophages. Upon DNA virus infection or ISD (2 μg/ml) stimulation, the transcription of ISG56 in cells expressing Omp25 was markedly lower than in Lv-GFP-infected or control cells (Figures 3A,C,E,G,I). Similarly, the transcriptional levels of IP-10 were also lower in Omp25-expressing cells than in Lv-GFP-infected or control cells (Figures 3B,D,F,H,J). Meanwhile, the ISG56 and IP-10 mRNA levels were higher in DNA virus-infected cells or ISD (2 μg/ml)-stimulated cells than in mock treatment (Figures 3A–J). These results demonstrated that Omp25 inhibited DNA viruses and ISD-induced transcription of ISG.
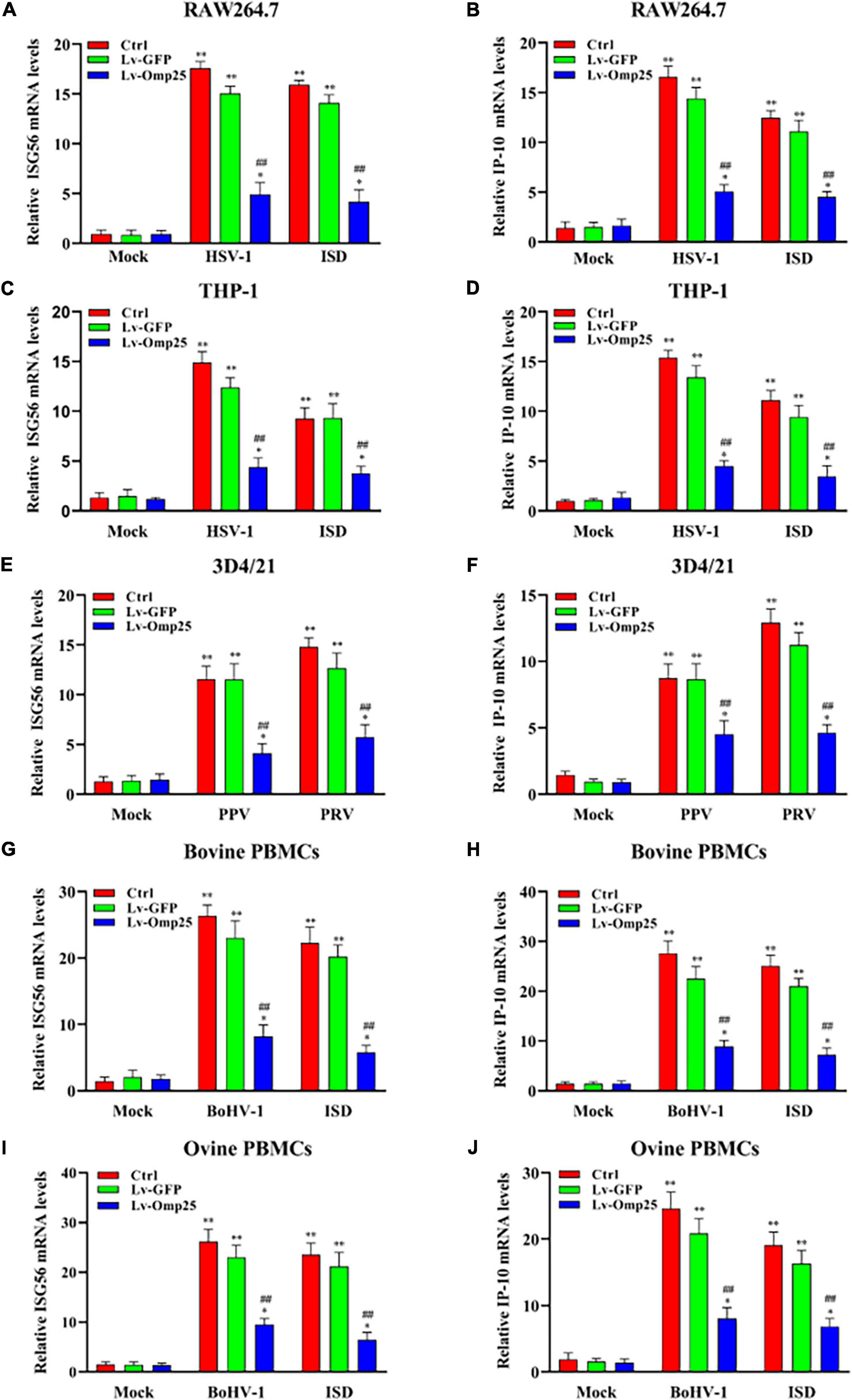
Figure 3. Omp25 inhibits DNA virus and ISD-induced ISG56 and IP-10 expression in various mammalian monocyte/macrophages. Cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 48 h and then infected with HSV-1 (10 MOI), PPV (1 MOI), PRV (1 MOI), or BoHV-1 (1 MOI) or transfected with ISD (2 μg/ml) for another 6 h. Cells were harvested and subjected to qRT-PCR analysis to measure ISG56 mRNA levels (A,C,E,G,I) or IP-10 mRNA levels (B,D,F,H,J). *P < 0.05, **P < 0.01 versus mock group, #P < 0.05, ##P < 0.01 versus Lv-GFP-infected cells in same groups.
Outer Membrane Protein 25 Negatively Regulates DNA Viruses-Activated Stimulator of Interferon Genes and Interferon Regulatory Factor 3 Phosphorylation
To further investigate how Omp25 inhibited IFN-I production, we tested phosphorylation of STING and IRF3 in RAW264.7, THP-1, and 3D4/21 cells. The results showed that following HSV-1 or PRV infection, the phosphorylation of STING and IRF3 significantly decreased in cells overexpressing Omp25 compared with that in Lv-GFP-infected or control cells (Figures 4A–C), and also less nuclear p-IRF3 were detected in Lv-Omp25-infected cells compared to that in Lv-GFP-infected or control cells (Figures 4D–F). Thus, these data suggested that Omp25 negatively regulated DNA viruses-activated STING and IRF3 phosphorylation.
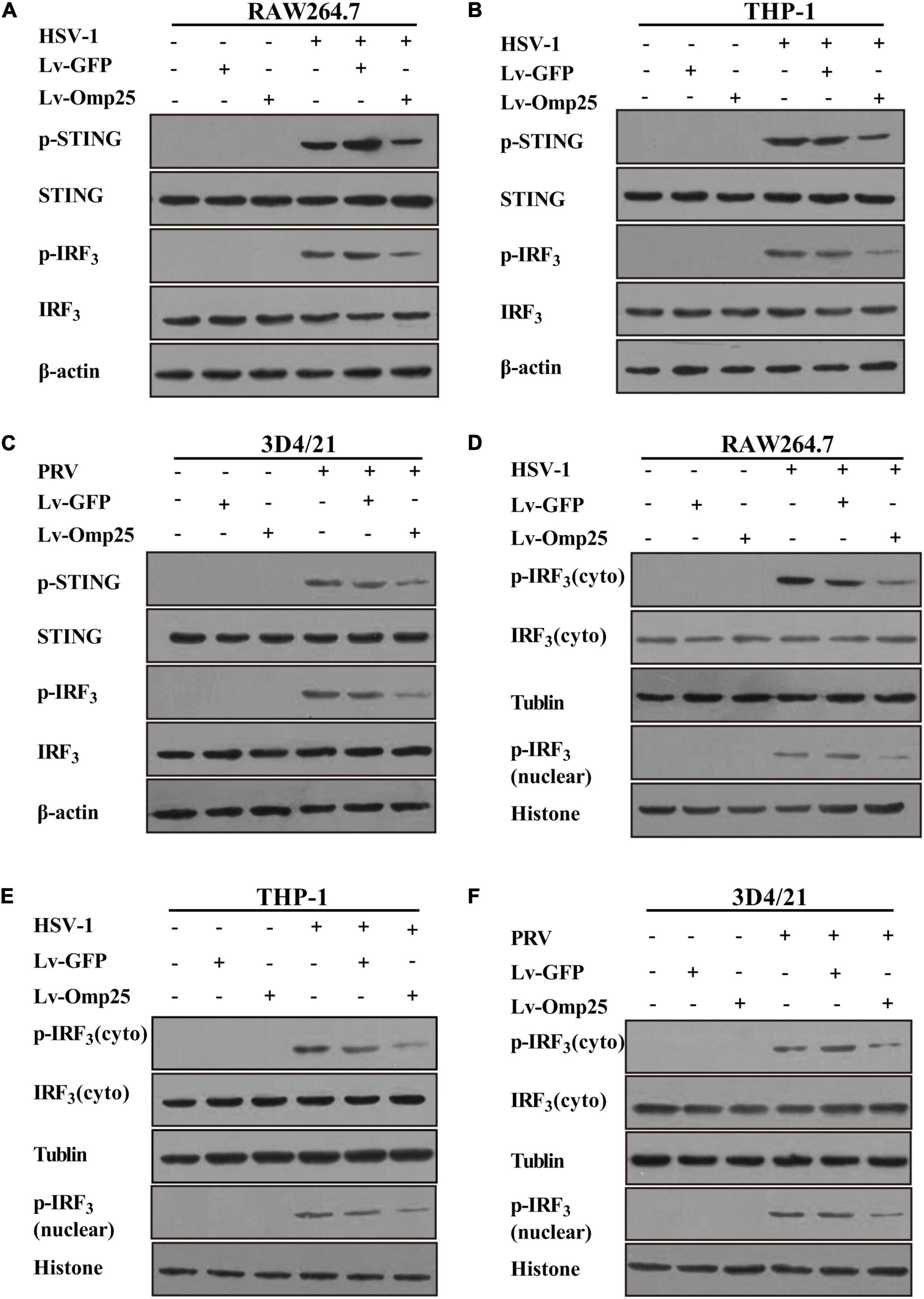
Figure 4. Omp25 inhibits activation of cGAS/STING pathway. (A–C) Omp25 inhibits phosphorylation of STING and IRF3 (p-STING, p-IRF3) in murine, human, and porcine macrophages. Cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 48 h and then further infected by HSV-1 (10 MOI) or PRV (1 MOI) for 6 h. Cells were harvested and subjected to detect level of p-STING and p-IRF3 by Western blotting. (D–F) Omp25 affects distribution of cytoplasmic and nuclear p-IRF3. Cells were infected and treated as in panels (A–C), cytoplasm and nuclear were separated, and level of p-IRF3 in cytoplasm and nuclear were detected by Western blotting.
Outer Membrane Protein 25 Dampens Interferon-β Induction by Reducing the Activity of Cyclic Guanosine Monophosphate–Adenosine Monophosphate Synthase
To further define the potential mechanisms of Omp25 in regulating the cGAS-STING signaling pathway, we examined the production of cGAMP in RAW264.7, THP-1, and 3D4/21 cells infected with Lv-GFP or Lv-Omp25 under the DNA virus infection and ISD (2 μg/ml) stimulation. cGAMP activity was measured in cell extracts using a modified bioassay by assessing the induction of IFN-β transcripts in a permeabilized secondary reporter cells (RAW264.7, THP-1, and 3D4/21 cells). As shown in Figures 5A–C, cGAMP activity was significantly decreased in the cells infected with Lv-Omp25 than in the cells infected with Lv-GFP or control. These results indicated that Omp25 dampened IFN-β production by reducing the activity of cGAS.
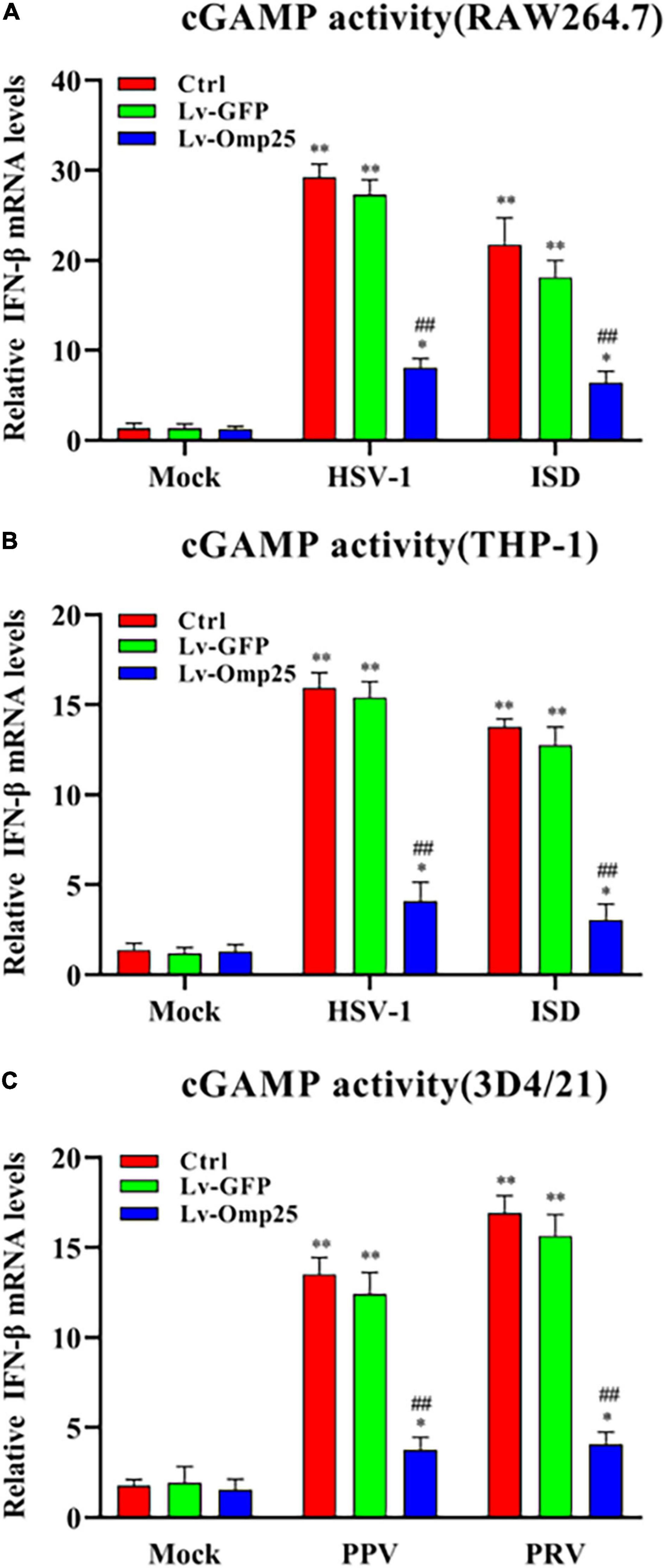
Figure 5. Omp25 inhibits cGAS activity in human, murine, and porcine macrophages. (A,B) Omp25 inhibits cGAS activity in RAW264.7 or THP-1 cells. Cells were infected with Lv-Omp25 or Lv-GFP at an MOI of 100 for 48 h, then infected with HSV-1 (10 MOI) or stimulated by ISD (2 μg/ml) another 6 h. Extracts from infected cells were prepared, DNase and heat-treated, and incubated with permeabilized RAW264.7 cells or THP-1 cells for 6 h. IFN-β RNA induction was analyzed by qRT-PCR and normalized to that of β-actin to assess levels of cGAMP in cell extracts. (C) Omp25 inhibits cGAS activity in porcine 3D4/21 cells. Cells were infected and treated as in panels (A,B). Extracts from infected cells were prepared and incubated with permeabilized 3D4/21 cells for 6 h. IFN-β RNA induction was analyzed by qRT-PCR. *P < 0.05, **P < 0.01 versus mock group, #P < 0.05, ##P < 0.01 versus Lv-GFP-infected cells in same groups.
Outer Membrane Protein 25 Induces the Ubiquitin–Proteasome-Dependent Cyclic Guanosine Monophosphate–Adenosine Monophosphate Synthase Degradation
Because Omp25 inhibited cGAS-induced IFN-I production by reducing cGAMP production, we further detected the effects of Omp25 on cGAS protein levels. Firstly, we detected the interaction between Omp25 and cGAS. HEK293T cells were co-transfected with the vector expressing HA-cGAS and Flag-Omp25 for 24 h. The results showed that cGAS interacted with Omp25 (Figures 6A,B). Then, we tested the effect of Omp25 on cGAS in RAW264.7 and 3D4/21 cells. Cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP. At 36-h post-infection, cells were treated with or without protein synthesis inhibitor (CHX, 100 μg/ml), then the level of cGAS protein was determined by Western blotting. The results showed that the protein level of cGAS was significantly reduced in Lv-Omp25-infected cells compared with Lv-GFP-infected or control cells whenever these cells were treated with or without CHX (Figures 6C,D), suggesting that Omp25 can promote cGAS degradation.
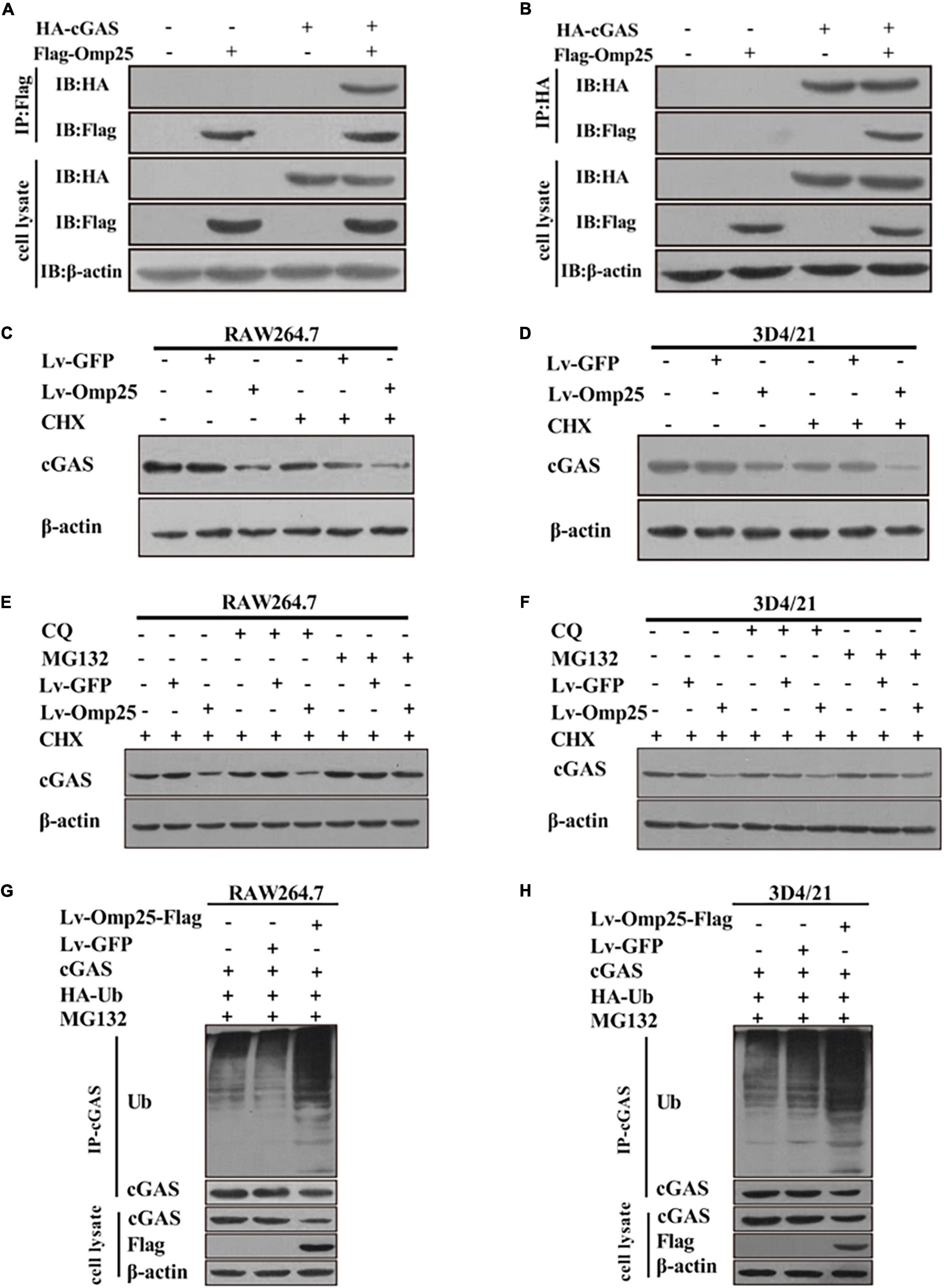
Figure 6. Omp25 reduces level of cGAS protein and induces proteasome-dependent degradation of cGAS. (A,B) Omp25 interacted with cGAS. Cells were co-expressed with Flag-Omp25 and HA-cGAS for 24 h, and cell lysates were subjected to a co-IP assay using anti-Flag antibody, anti-HA antibody, and anti-β-actin antibody. (C,D) Omp25 degraded cGAS in murine and porcine macrophages. RAW264.7 or 3D4/21 cells were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 36 h, then treated with or without CHX (100 μg/ml) for another 12 h. Level of cGAS was detected by Western blotting. (E,F) Omp25 promoted degradation of cGAS via proteasome-dependent pathway. RAW264.7 or 3D4/21 were infected with 100 MOI of Lv-Omp25 or Lv-GFP for 36 h and treated with dimethyl sulfoxide, CQ (20 μM), or MG132 (5 μM) in presence of CHX for another 12 h, followed by Western blotting to detect levels of cGAS. (G,H) Omp25 induced cGAS degradation through ubiquitin–proteasome pathway in RAW264.7 and 3D4/21 cells. Cells were transfected with vector expressing HA-Ub by Lipofectamine 3000 for 24 h, then infected with 100 MOI of Lv-Omp25 or Lv-GFP. At 36-h post-infection, cells were treated with MG132 (5 μM) for another 12 h; ubiquitination level of cGAS protein was determined by Western blotting.
Therefore, to clarify which pathway was responsible for the cGAS degradation induced by Omp25 in the proteasome pathway and lysosome pathway, we compared the cGAS degradation in the Lv-Omp25-infected cells treated with either proteasome inhibitor (MG132, 5 μM) or lysosome inhibitor (CQ, 20 μM). Results showed that MG132 treatment could restore the level of cGAS protein in Lv-Omp25-infected cells, similar to the level as that in Lv-GFP-infected or control cells in the presence of CHX, whereas CQ treatment could not (Figures 6E,F). Then, we detected the effect of Omp25 on cGAS ubiquitination in RAW264.7 and 3D4/21 cells. Cells were transfected with the vector expressing HA-Ub for 24 h, then infected with 100 MOI of Lv-Omp25 or Lv-GFP. At 36-h post-infection, cells were treated with MG132 (5 μM) for another 12 h before collecting, then the ubiquitination level of cGAS protein was determined by Western blotting. The results showed that the level of cGAS ubiquitination was markedly increased in Lv-Omp25-infected cells compared with Lv-GFP-infected or control cells (Figures 6G,6H). Altogether, these results suggested that Omp25 induced cGAS degradation through the ubiquitin–proteasome pathway.
161 to 184 Amino Acids of Outer Membrane Protein 25 Are Critical for Induction of Cyclic Guanosine Monophosphate–Adenosine Monophosphate Synthase Degradation
To identify the key functional motifs and amino acid residues of Omp25 required to induce cGAS degradation, we constructed 10 deletion mutants of Omp25 (Figure 7A). We examined and compared the inhibitory effects of these Omp25 mutants on the HSV-1 (10 MOI)- or ISD (2 μg/ml)-induced IFN-β expression in mRNA and protein levels. Results showed that Omp25 fragments 51–213, 108–213, 134–213, 161–213, and 1–184 significantly reduced cGAS-induced IFN-β production and full-length Omp25, whereas Omp25 fragments 1–50, 1–107, 1–133, 1–160, and 185–213 failed to exhibit a significant inhibitory effect on cGAS-induced IFN-β production (Figures 7B,C). These results suggested that the fragment (amino acid 161–184) of Omp25 was required to induce cGAS degradation. To further confirm this domain was critical for induction of cGAS degradation, we constructed an Omp25 (161–184 aa) deletion mutant Omp25Δ161-184 and detected the ability of this mutant to induce cGAS degradation. Results showed that Omp25Δ161-184 deletion mutant appeared a significantly attenuated inhibitory ability in transcription and expression of IFN-β induced by HSV-1 infection and ISD stimulation compared with full-length Omp25 but still exhibited a certain inhibitory effect on IFN-β induction relative to Lv-GFP or control (Figures 7D,E). Meanwhile, upon HSV-1 (10 MOI) or ISD (2 μg/ml) stimulation, Omp25Δ161-184 showed a weaken inhibitory effect on the levels of cGAS and the production of cGAMP relative to full-length Omp25 but did decrease cGAS and cGAMP production relative to Lv-GFP infected or control cells (Figures 7F,G). Correspondingly, the phosphorylation of STING and IRF3 was higher in Lv-Omp25Δ161-184-infected cells than that in Lv-Omp25-infected cells but still lower than that in Lv-GFP-infected or control cells (Figure 7H). Taken together, the 161 to 184 amino acids of Omp25 were critical for induction of cGAS degradation and inhibition of IFN-β induction.
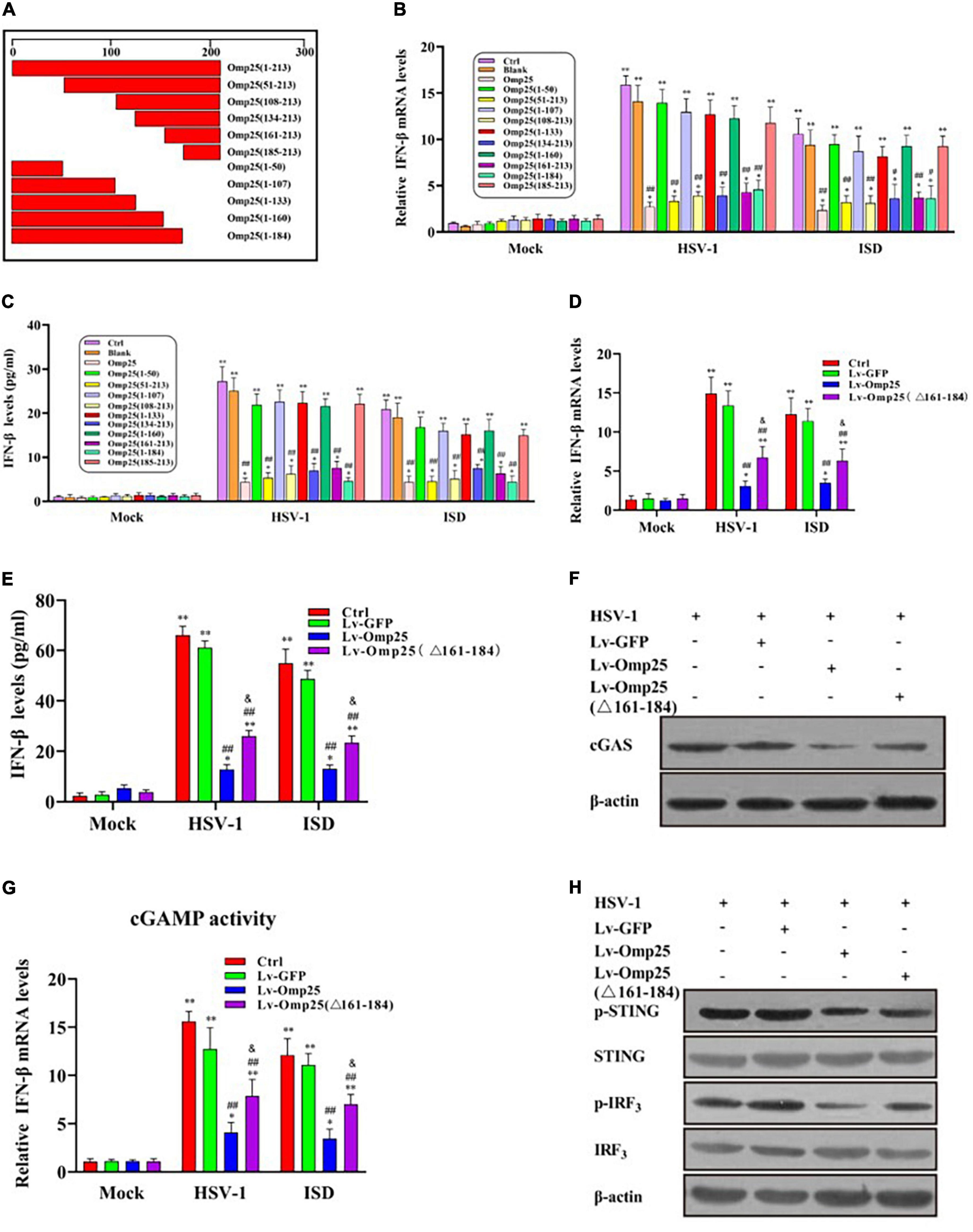
Figure 7. Effect of Omp25 deletion mutants on cGAS in cells. (A) Schematic diagram of Omp25 deletion mutants. (B,C) Mapping regions of Omp25 that decrease IFN-β mRNA levels or protein levels. RAW264.7 cells were infected with different deletion mutants for 48 h, then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 6 or 24 h. Cells were harvested and subjected to qRT-PCR analysis or ELISA analysis to examine expression of IFN-β. (D,E) Lv-Omp25Δ161–184 showed an attenuated inhibitory effect in transcription and expression of IFN-β. RAW264.7 cells were infected with Lv-Omp25, Lv-Omp25Δ161–184, or Lv-GFP for 48 h, then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 6 or 24 h. Cells were harvested and subjected to qRT-PCR analysis or ELISA analysis to examine expression of IFN-β. (F,G) Omp25Δ161–184 showed a weak inhibitory effect on levels of cGAS and production of cGAMP. Cells were treated as in panels (D,E); protein levels of cGAS and levels of cGAMP in cell extracts were measured by Western blotting or qRT-PCR as described in section “Materials and Methods.” (H) Lv-Omp25Δ161–184 showed a weakened inhibitory effect on phosphorylation of STING and IRF3. Cells were treated as in panels (D,E), and phosphorylation of STING and IRF3 were measured by Western blotting. *P < 0.05, **P < 0.01 versus mock group, #P < 0.05, ##P < 0.01 versus Lv-GFP- infected cells in same groups, &P < 0.05 versus Lv-Omp25-infected cells in same groups.
Amino Acid Residues (R176, Y179, R180, Y181, and Y184) Are Critical for Outer Membrane Protein 25 to Inhibit Interferon β Induction
Sequence alignment of Brucella spp. Omp25 protein revealed that the amino acid residues 158–198 were highly conserved in different Brucella species (Figure 8A). Therefore, to identify the key amino acid residues in Omp25 (161–184) that played a critical role in Omp25 inhibition of IFN-β expression, we screened out five amino acid residues (R176, Y179, R180, Y181, and Y184) from the region (aa 158 to 198) (Figure 8B). When these five amino acid residues were replaced by alanine, we observed that the corresponding mutant, Omp25-5M, showed an attenuated inhibitory effect on HSV-1- or ISD-induced IFN-β but remained a certain inhibitory effect on IFN-β induction compared with Lv-GFP-infected or control cells (Figures 8C,D). Furthermore, upon HSV-1 (10 MOI) infection or ISD (2 μg/ml) stimulation, the protein level and activity of cGAS were higher in mutant Omp25-5M-expressing cells than that in wild-type Omp25-expressing cells, although the protein level and activity of cGAS in both Omp25-5M-expressing cells and Omp25-expressing cells were still lower than that in Lv-GFP-infected or control cells (Figures 8E,F). At the same time, the phosphorylation of STING and IRF3 was increased in Omp25-5M-expressing cells compared with Omp25-expressing cells but still lower than that in Lv-GFP-infected or control cells (Figure 8G). Altogether, these results demonstrated that amino acid residues (R176, Y179, R180, Y181, and Y184) were critical for Omp25 to inhibit IFN-β induction.
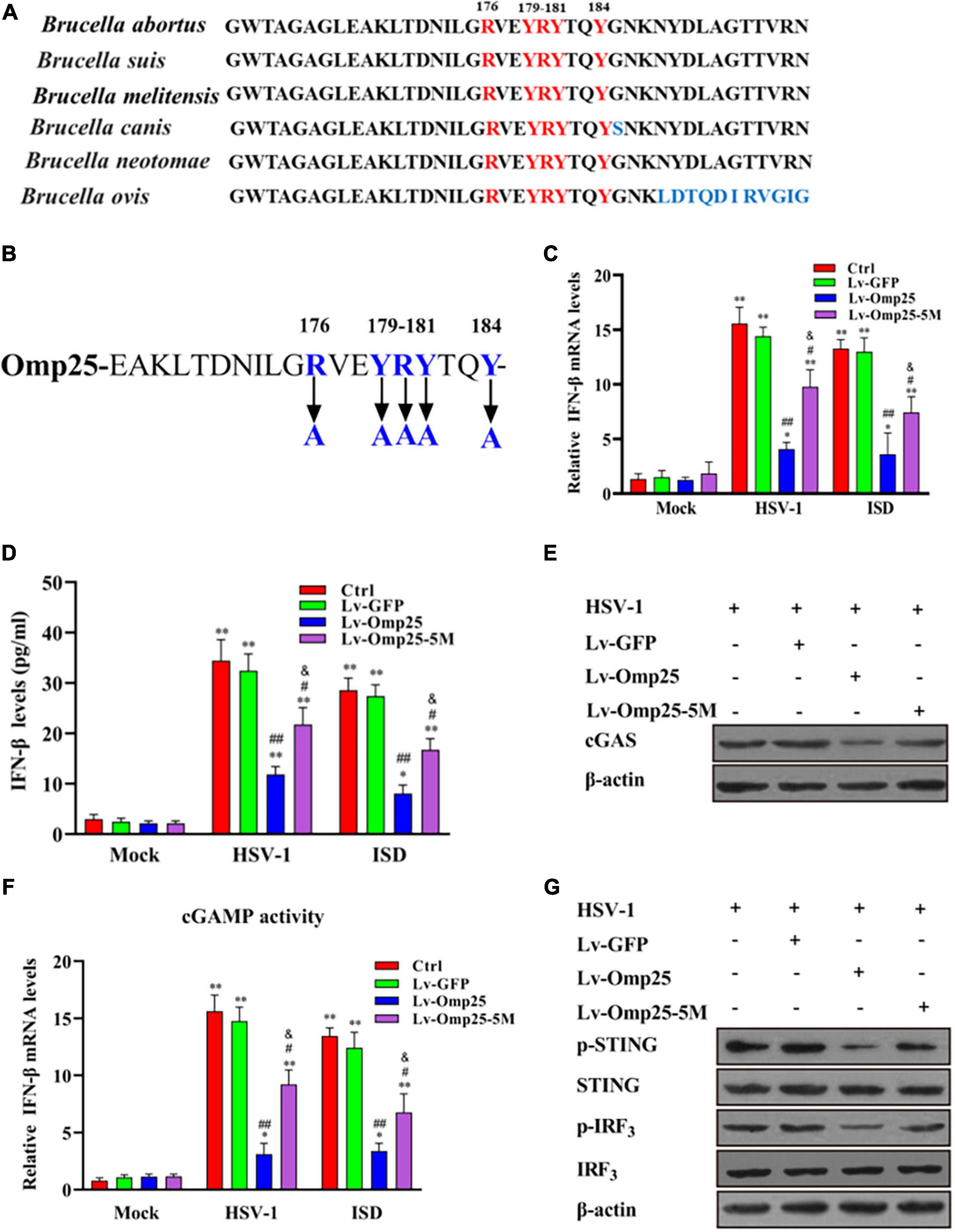
Figure 8. Determination of key amino acid residue in Omp25 plays a critical role in Omp25 inhibition of IFN-β expression. (A) Alignment of Omp25 high conserved amino acid motif in different Brucella species. (B) Schematic diagram of Omp25 mutation in R176, Y179, R180, Y181, and Y184. (C–G) RAW264.7 cells were infected with Lv-Omp25, Lv-Omp25-5M, or Lv-GFP for 48 h, then infected with HSV-1 (10 MOI) or transfected with ISD (2 μg/ml) for another 6 or 24 h. Cells were harvested and subjected to qRT-PCR analysis or ELISA analysis to examine expression of IFN-β (C,D); Western blotting was performed to detect levels of cGAS (E); qRT-PCR was used to detect levels of cGAMP in cell extracts as described in section “Materials and Methods” (F), and phosphorylation levels of STING and IRF3 were measured by Western blotting (G). *P < 0.05, **P < 0.01 versus mock group, #P < 0.05, ##P < 0.01 versus Lv-GFP-infected cells in same groups. &P < 0.05 versus Lv-Omp25 infected cells in same groups.
Discussion
It is well known that IFN-Is, such as IFN-α and IFN-β, are playing important roles in antiviral and antibacterial infections (Borden et al., 2007; Monroe et al., 2010; McNab et al., 2015). Cheng et al. (2018), Huang B. et al. (2018), and Marinho et al. (2018) have reported that both viral and bacterial intracellular pathogens can induce IFN-I by the cGAS/STING signal pathway. It has been reported that induction of IFN-β by Mycobacterium tuberculosis and Listeria depends on cytoplasmic cGAS sensing (Hansen et al., 2014; Collins et al., 2015). However, the microbes have developed mechanisms to manipulate this innate immune sensing pathway. For example, some proteins expressed by viruses increase infectivity by interfering with cGAS activation (Wu et al., 2015; Aguirre et al., 2017; Huang Z. F. et al., 2018). In this study, we demonstrated that Omp25 inhibited the expression of IFN-β induced by DNA viruses or ISD via degradation of cGAS depending on the ubiquitin–proteasome pathway in mammalian monocyte/macrophages.
Brucellosis, caused by Brucella, is a worldwide zoonosis affecting animal and human health, which often causes immunosuppression (Renoux and Renoux, 1977). Forestier et al. (2000) reported that the membrane ingredients of Brucella spp. (such as lipopolysaccharide) might inhibit T-cell activation from causing immunosuppression. Brucella spp. prevent immune activation of macrophages by inducing CD4+CD25+ T cells to produce the anti-inflammatory cytokine IL-10 during early infection (Xavier et al., 2013). In addition, B. melitensis suppresses gamma IFN production and delays memory responses through exhausting CD8+T cells (Durward-Diioia et al., 2015). We found that the cattle infected with Brucella spp. were susceptible to BoHV-1 and BoHV-4 during our epidemiological investigation. This phenomenon might associate with some component of Brucella spp. that could promote the replication of PRV and HSV-1 via suppression of the expression of IFN-I. Thus, we investigated the roles of Omp25 in this process, as Omp25 is involved in inhibiting the activity of T cells and macrophages in our previous studies (Cui et al., 2017; Luo et al., 2017).
Cyclic guanosine monophosphate–adenosine monophosphate synthase, a DNA sensor, is essential for IFN-β expression during viral and bacterial infections (Zhang et al., 2014; Collins et al., 2015; Huang Z. F. et al., 2018). Viruses and bacteria have evolved various strategies to prevent activation of the cGAS/STING pathway, resulting in the evasion of host innate immunity. For example, the dengue virus protein NS2B promoting cGAS degradation in autophagy–lysosome-dependent manner results in inhibition of IFN-I production during infection (Aguirre et al., 2017). HSV-1 UL41 was demonstrated to evade the cGAS/STING-mediated DNA-sensing pathway by degrading cGAS mRNA via its RNase activity, and HSV-1 VP22 was shown to interact with cGAS and inhibited the enzymatic activity of cGAS (Su and Zheng, 2017; Huang J. et al., 2018; Zheng, 2018). In this study, our results showed that Omp25 that promoted cGAS degradation via a ubiquitin–proteasome-dependent pathway led to a decreased cGAMP production in 3D4/21 cells and RAW264.7 cells. Ivashkiv and Donlin (2014) have reported that IRF3 is a master transcriptional factor regulating IFN-I gene induction and innate immune defenses. Some viral proteins can modulate the activation of IRF3 through different processes, including IRF3 dimerization, nuclear localization, and binding to target gene promoters (Schneider et al., 2014). Several HSV-1 proteins, including US3, VP24, VP16, and VZV ORF61, were shown to target IRF3 and inhibit IFN-I production (Zhu et al., 2011; Wang et al., 2013; Xing et al., 2013; Zhang et al., 2016), and PRV UL13 inhibits the IFN-β signaling pathway by targeting IRF3 for ubiquitination and degradation (Lv et al., 2020). This study detected the phosphorylated IRF3 in the cytoplasm and nucleus. The results showed that phosphorylated IRF3 in the nucleus was significantly decreased in Omp25-expressing cells than the cells without Omp25 expression. Thus, we speculate that Omp25 interferes with the translocation of IRF3 to the nucleus, thereby resulting in inhibition for IFN-β transcription.
In summary, the data presented in this work demonstrated that Omp25 promoted the degradation of cGAS by ubiquitin–proteasome to interfere with the activation of the cGAS/STING signaling pathway, to inhibit the production of IFN-β, resulting in the cells with Omp25 expression more easily infected by other pathogens. To the best of our knowledge, this was the first description of Omp25 inhibiting the expression of IFN-β via targeting cGAS for degradation. Meanwhile, we cleared the key functional domain of Omp25 that was critical for suppressing the expression of IFN-β. Altogether, our findings presented a novel role of Omp25 in regulating IFN-β production, which might provide novel insights for brucellosis prevention.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
RL, WL, YH, and DT conceived and designed the experiments and wrote and revised the manuscript. RL, WL, XY, FZ, ZW, XW, and XZ performed the experiments and analyzed the data. RL, WL, and QD revised the manuscript. All the authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grants 31872447 and 31972686 to DT and YH). This work was also supported by the Key R&D Program of Shaanxi Province (grants 2020NY-010 and 2018ZDCXL-NY-02-07), by the Project of Xi’an University of Science and Technology (grant 20NYYF0032) and by the Fundamental Research Funds for the Central Universities (grant 2452017023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Life Science Research Core Services of Northwest A&F University for some key technical support assistance.
References
Aguirre, S., Luthra, P., Sanchez-Aparicio, M. T., Maestre, A. M., Patel, J., Lamothe, F., et al. (2017). Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2:17037.
Baldi, P. C., and Giambartolomei, G. H. (2013a). Immunopathology of Brucella infection. Recent Pat. Antiinfect. Drug Discov. 8, 18–26. doi: 10.2174/1574891x11308010005
Baldi, P. C., and Giambartolomei, G. H. (2013b). Pathogenesis and pathobiology of zoonotic brucellosis in humans. Rev. Sci. Tech. 32, 117–125. doi: 10.20506/rst.32.1.2192
Borden, E. C., Sen, G. C., Uze, G., Silverman, R. H., Ransohoff, R. M., Foster, G. R., et al. (2007). Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6, 975–990. doi: 10.1038/nrd2422
Campos, P. C., Gomes, M. T., Guimaraes, G., Costa Franco, M. M., Marim, F. M., and Oliveira, S. C. (2014). Brucella abortus DNA is a major bacterial agonist to activate the host innate immune system. Microbes Infect. 16, 979–984. doi: 10.1016/j.micinf.2014.08.010
Caro-Hernandez, P., Fernandez-Lago, L., de Miguel, M. J., Martin-Martin, A. I., Cloeckaert, A., Grillo, M. J., et al. (2007). Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect. Immun. 75, 4050–4061. doi: 10.1128/iai.00486-07
Cheng, W. Y., He, X. B., Jia, H. J., Chen, G. H., Jin, Q. W., Long, Z. L., et al. (2018). The cGas-sting signaling pathway is required for the innate immune response against ectromelia virus. Front. Immunol. 9:1297.
Cloeckaert, A., Vizcaıìno, N., Paquet, J.-Y., Bowden, R. A., and Elzer, P. H. (2002). Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol. 90, 229–247. doi: 10.1016/s0378-1135(02)00211-0
Collins, A. C., Cai, H., Li, T., Franco, L. H., Li, X. D., Nair, V. R., et al. (2015). Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host Microbe 17, 820–828. doi: 10.1016/j.chom.2015.05.005
Cui, B., Liu, W., Wang, X., Chen, Y., Du, Q., Zhao, X., et al. (2017). Brucella Omp25 Upregulates miR-155, miR-21-5p, and miR-23b to Inhibit Interleukin-12 production via modulation of programmed death-1 signaling in human monocyte/macrophages. Front. Immunol. 8:708.
de Figueiredo, P., Ficht, T. A., Rice-Ficht, A., Rossetti, C. A., and Adams, L. G. (2015). Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am. J. Pathol. 185, 1505–1517.
Durward-Diioia, M., Harms, J., Khan, M., Hall, C., Smith, J. A., and Splitter, G. A. (2015). CD8+ T cell exhaustion, suppressed gamma interferon production, and delayed memory response induced by chronic Brucella melitensis infection. Infect. Immun. 83, 4759–4771. doi: 10.1128/iai.01184-15
Forestier, C., Deleuil, F., Lapaque, N., Moreno, E., and Gorvel, J. P. (2000). Brucella abortus lipopolysaccharide in murine peritoneal macrophages acts as a down-regulator of T cell activation. J. Immunol. 165, 5202–5210. doi: 10.4049/jimmunol.165.9.5202
Gao, D., Wu, J., Wu, Y. T., Du, F., Aroh, C., Yan, N., et al. (2013). Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906. doi: 10.1126/science.1240933
Glowacka, P., Zakowska, D., Naylor, K., Niemcewicz, M., and Bielawska-Drozd, A. (2018). Brucella - virulence factors, pathogenesis and treatment. Pol. J. Microbiol. 67, 151–161. doi: 10.21307/pjm-2018-029
Guzman-Verri, C., Manterola, L., Sola-Landa, A., Parra, A., Cloeckaert, A., Garin, J., et al. (2002). The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U. S. A. 99, 12375–12380. doi: 10.1073/pnas.192439399
Hansen, K., Prabakaran, T., Laustsen, A., Jorgensen, S. E., Rahbaek, S. H., Jensen, S. B., et al. (2014). Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33, 1654–1666. doi: 10.15252/embj.201488029
Huang, B., Zhang, L., Lu, M., Li, J., and Lv, Y. (2018). PCV2 infection activates the cGAS/STING signaling pathway to promote IFN-beta production and viral replication in PK-15 cells. Vet. Microbiol. 227, 34–40. doi: 10.1016/j.vetmic.2018.10.027
Huang, J., You, H., Su, C., Li, Y., Chen, S., and Zheng, C. (2018). Herpes simplex virus 1 tegument Protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J. Virol. 92:JVI.00841-18.
Huang, Y., Lei, Y., Zhang, H., Zhang, M., and Dayton, A. (2011). Interleukin-12 treatment down-regulates STAT4 and induces apoptosis with increasing ROS production in human natural killer cells. J. Leukoc. Biol. 90, 87–97. doi: 10.1189/jlb.1210674
Huang, Z. F., Zou, H. M., Liao, B. W., Zhang, H. Y., Yang, Y., Fu, Y. Z., et al. (2018). Human cytomegalovirus protein UL31 inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microbe 24, 69–80 e64.
Ivashkiv, L. B., and Donlin, L. T. (2014). Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49. doi: 10.1038/nri3581
Jubier-Maurin, V., Boigegrain, R. A., Cloeckaert, A., Gross, A., Alvarez-Martinez, M. T., Terraza, A., et al. (2001). Major outer membrane protein Omp25 of Brucella suis is involved in inhibition of tumor necrosis factor alpha production during infection of human macrophages. Infect. Immun. 69, 4823–4830. doi: 10.1128/iai.69.8.4823-4830.2001
Luo, X., Zhang, X., Wu, X., Yang, X., Han, C., Wang, Z., et al. (2017). Brucella downregulates tumor necrosis factor-alpha to promote intracellular survival via Omp25 regulation of different MicroRNAs in porcine and murine macrophages. Front. Immunol. 8:2013.
Lv, L., Cao, M., Bai, J., Jin, L., Wang, X., Gao, Y., et al. (2020). PRV-encoded UL13 protein kinase acts as an antagonist of innate immunity by targeting IRF3-signaling pathways. Vet. Microbiol. 250:108860. doi: 10.1016/j.vetmic.2020.108860
Machelart, A., Potemberg, G., Van Maele, L., Demars, A., Lagneaux, M., De Trez, C., et al. (2018). Allergic asthma favors brucella growth in the lungs of infected mice. Front. Immunol. 9:1856.
Marinho, F. V., Benmerzoug, S., Rose, S., Campos, P. C., Marques, J. T., Bafica, A., et al. (2018). The cGAS/STING pathway is important for dendritic cell activation but is not essential to induce protective immunity against mycobacterium tuberculosis infection. J. Innate Immun. 10, 239–252. doi: 10.1159/000488952
McNab, F., Mayer-Barber, K., Sher, A., Wack, A., and O’Garra, A. (2015). Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103. doi: 10.1038/nri3787
Monroe, K. M., McWhirter, S. M., and Vance, R. E. (2010). Induction of type I interferons by bacteria. Cell Microbiol. 12, 881–890. doi: 10.1111/j.1462-5822.2010.01478.x
Pasquevich, K. A., Carabajal, M. V., Guaimas, F. F., Bruno, L., Roset, M. S., Coria, L. M., et al. (2019). Omp19 enables Brucella abortus to evade the antimicrobial activity from host’s proteolytic defense system. Front. Immunol. 10:1436.
Renoux, M., and Renoux, G. (1977). Brucellosis, immunodepression, and levamisole. Lancet 1:372. doi: 10.1016/s0140-6736(77)91181-3
Sastry, L., Johnson, T., Hobson, M. J., Smucker, B., and Cornetta, K. (2002). Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 9, 1155–1162. doi: 10.1038/sj.gt.3301731
Schneider, W. M., Chevillotte, M. D., and Rice, C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Su, C., and Zheng, C. (2017). Herpes simplex Virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff Protein, UL41. J. Virol. 91:e02414-16.
Tibor, A., Wansard, V., Bielartz, V., Delrue, R. M., Danese, I., Michel, P., et al. (2002). Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 70, 5540–5546. doi: 10.1128/iai.70.10.5540-5546.2002
Vizcaino, N., Caro-Hernandez, P., Cloeckaert, A., and Fernandez-Lago, L. (2004). DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6, 821–834. doi: 10.1016/j.micinf.2004.04.009
Wang, S., Wang, K., Lin, R., and Zheng, C. (2013). Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J. Virol. 87, 12814–12827. doi: 10.1128/jvi.02355-13
Woodward, J. J., Iavarone, A. T., and Portnoy, D. A. (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. doi: 10.1126/science.1189801
Wu, J. J., Li, W., Shao, Y., Avey, D., Fu, B., Gillen, J., et al. (2015). Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18, 333–344. doi: 10.1016/j.chom.2015.07.015
Xavier, M. N., Winter, M. G., Spees, A. M., Nguyen, K., Atluri, V. L., Silva, T. M., et al. (2013). CD4+ T cell-derived IL-10 promotes Brucella abortus persistence via modulation of macrophage function. PLoS Pathog. 9:e1003454. doi: 10.1371/journal.ppat.1003454
Xing, J., Ni, L., Wang, S., Wang, K., Lin, R., and Zheng, C. (2013). Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J. Virol. 87, 9788–9801. doi: 10.1128/jvi.01440-13
Zhang, D., Su, C., and Zheng, C. (2016). Herpes simplex Virus 1 serine Protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory Factor 3. J. Virol. 90, 5824–5829. doi: 10.1128/jvi.00186-16
Zhang, Y., Yeruva, L., Marinov, A., Prantner, D., Wyrick, P. B., Lupashin, V., et al. (2014). The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J. Immunol. 193, 2394–2404. doi: 10.4049/jimmunol.1302718
Zheng, C. (2018). Evasion of cytosolic DNA-stimulated innate immune responses by herpes simplex Virus 1. J. Virol. 92:e00099-17.
Keywords: Brucella spp., Omp25, macrophage, cyclic guanosine monophosphate–adenosine monophosphate synthase, interferon β
Citation: Li R, Liu W, Yin X, Zheng F, Wang Z, Wu X, Zhang X, Du Q, Huang Y and Tong D (2021) Brucella spp. Omp25 Promotes Proteasome-Mediated cGAS Degradation to Attenuate IFN-β Production. Front. Microbiol. 12:702881. doi: 10.3389/fmicb.2021.702881
Received: 30 April 2021; Accepted: 22 June 2021;
Published: 29 July 2021.
Edited by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranReviewed by:
Chunfu Zheng, University of Calgary, CanadaKo Fujimori, Osaka University of Pharmaceutical Sciences, Japan
Yongtao Li, Henan Agricultural University, China
Copyright © 2021 Li, Liu, Yin, Zheng, Wang, Wu, Zhang, Du, Huang and Tong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Huang, eW9uZ2h1YW5nQG53c3VhZi5lZHUuY24=; Dewen Tong, ZHd0b25nQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Ruizhen Li1†
Ruizhen Li1† Wenli Liu
Wenli Liu Xiangrui Yin
Xiangrui Yin Zhenyu Wang
Zhenyu Wang Yong Huang
Yong Huang Dewen Tong
Dewen Tong