- 1Marine College, Shandong University, Weihai, China
- 2Key Laboratory of Mariculture, Ministry of Education, Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 3Department of Zoology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 4Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 5Department of Life Sciences, Natural History Museum, London, United Kingdom
During an investigation on freshwater peritrichs, a new colonial sessilid ciliate, Campanella sinica n. sp., was isolated from aquatic plants in an artificial freshwater pond in Qingdao, China. Specimen observations of this species were performed both in vivo and using silver staining. C. sinica n. sp. is characterized by the appearance of the mature colony, which is up to 2 cm high and contains more than 1,000 zooids, the asymmetric horn-shaped zooids, strongly everted and multi-layered peristomial lip, the slightly convex peristomial disc, and the well-developed haplokinety and polykinety, which make more than four circuits of the peristome before descending into the infundibulum. The small subunit ribosomal DNA (SSU rDNA), 5.8s rDNA and its flank internal transcribed spacers (ITS1-5.8s rDNA-ITS2), and large subunit ribosomal DNA (LSU rDNA) are sequenced and used for phylogenetic analyses which reveal that the family Epistylididae Kahl, 1933 is non-monophyletic whereas the genus Campanella is monophyletic and nests within the basal clade of the sessilids. The integrative results support the assertion that the genus Campanella represents a separate lineage from other epistylidids, suggesting a further revision of the family Epistylididae is needed. We revise Campanella including the transfer into this genus of a taxon formerly assigned to Epistylis, which we raise to species rank, i.e., Campanella ovata (Nenninger, 1948) n. grad. & n. comb. (original combination Epistylis purneri f. ovata Nenninger, 1948). In addition, we provide a key to the identification of the species of Campanella.
Introduction
Ciliated protists (ciliates) have a widespread distribution and are found almost everywhere on the earth’s surface where there is sufficient water for their survival (Lynn, 2008; Zhao et al., 2020; Huang et al., 2021; Liu et al., 2021). Members of the subclass Peritrichia Stein, 1859 are characterized by their well-developed oral ciliature and vestigial somatic ciliature and are divided into two orders, Sessilida Kahl, 1933 and Mobilida Kahl, 1933 (Song et al., 2009; Gao et al., 2016; Wang et al., 2017a, 2018, 2019, 2020; Hu et al., 2019; Lu et al., 2019; Chen et al., 2020b; Wu et al., 2020, 2021). Sessilids are widely distributed in a variety of aquatic environments where they play an important role as consumers of bacteria and other microorganisms (Lynn, 2008; Shen and Gu, 2016). At least 800 species of sessilids, representing about 110 genera, have been described, although Foissner et al. (2010) estimated the number of genera to be 105–140 (Lynn, 2008; Song et al., 2009; Lu et al., 2019, 2020; Wu et al., 2020). Knowledge and understanding of the diversity, taxonomy, systematics, and evolution of sessilids are rapidly advancing with the increased application of methods, such as differential interference contrast microscopy, silver staining, and molecular phylogenetic analysis (Gentekaki et al., 2017; Lu et al., 2019, 2020; Wu et al., 2020). However, only a small proportion of sessilid species have been investigated using such methods (Zhuang et al., 2018; Jiang et al., 2019; Wu et al., 2020).
Recent molecular phylogenetic analyses have cast doubts on the validity, monophyly, and/or membership for several peritrich genera and families, for example, the family Epistylididae Kahl, 1933, species of which are distributed among several separate clades in gene trees (Miao et al., 2001, 2004; Sun et al., 2016; Zhuang et al., 2018). According to Lynn (2008), the family Epistylididae comprises 11 genera and is mainly characterized by the sessile trophont that: (1) is attached to substrate via either a non-contractile stalk or the scopula; (2) has an everted peristomial lip and a slightly projecting peristomial disc; and (3) has an oral ciliature that makes more than one turn around the peristome before entering the infundibulum (Lynn, 2008; Irwin and Lynn, 2015; Lu et al., 2020). One of the most distinctive genera of epistylidids is Campanella Goldfuss, 1820. Campanella is colonial with a non-contractile stalk and large zooids that have a multi-layered peristomial lip, an oral ciliature that makes several circuits around the peristome before descending into the infundibulum and a reticulate silverline system (Nenninger, 1948; Yu et al., 1995; Lynn, 2008). Three valid species of Campanella have been described, namely Campanella umbellaria (Linnaeus, 1758) Goldfuss, 1820 (the type species), C. hanchuansensis Yu et al., 1995 and C. purneri (Nenninger, 1948) Yu et al., 1995, but only C. umbellaria has been studied using modern methods (Miao et al., 2004; Shi et al., 2004; Wang et al., 2011). Nevertheless, phylogenetic analyses based on small subunit ribosomal DNA (SSU rDNA) sequences have challenged the traditional classification of Campanella suggesting that this genus may not belong to the family Epistylididae (Miao et al., 2004; Utz et al., 2010; Sun et al., 2016).
In the present study, we describe a new species, C. sinica n. sp., isolated from a freshwater pond in Qingdao, China, based on observations of specimens both in vivo and following silver staining. The phylogeny of this species based on SSU rDNA, ITS1-5.8S rDNA-ITS2, and large subunit ribosomal DNA (LSU rDNA) sequences is also analyzed, offering new insights into the evolutionary relationships of sessilids. Moreover, we reevaluate the classification of epistylidid-like species based on the morphological and phylogenetic results. In addition, the taxonomy and nomenclature of the species of Campanella are reviewed and a key to their identification is supplied.
Materials and Methods
Sampling and Observation
Campanella sinica n. sp. was collected between 27 March and 27 July 2020 from aquatic plants growing in an artificial freshwater pond (N36°03ꞌ45ꞌꞌ; E120°20ꞌ10ꞌꞌ) in Ocean University of China, Qingdao, China (Figure 1, inset). The water and sediment in the pond came from the Lake Weishan wetland (N34°43ꞌ59ꞌꞌ; E117°9ꞌ22ꞌꞌ). Live cells were observed and measured under a Zeiss AXIO Imager D2 microscope (Zeiss, Germany). The ciliature and silverline system were revealed by the protargol staining method and dry silver nitrate staining method, respectively (Klein, 1958; Foissner, 2014). The protargol powder was synthesized following the method of Pan et al. (2013). All measurements were performed at 400–1,000× magnifications. Drawings of live cells, ciliature, and silverline system were based on direct observations and photomicrographs. Classification and terminology are mainly according to Warren (1986) and Lynn (2008), respectively.
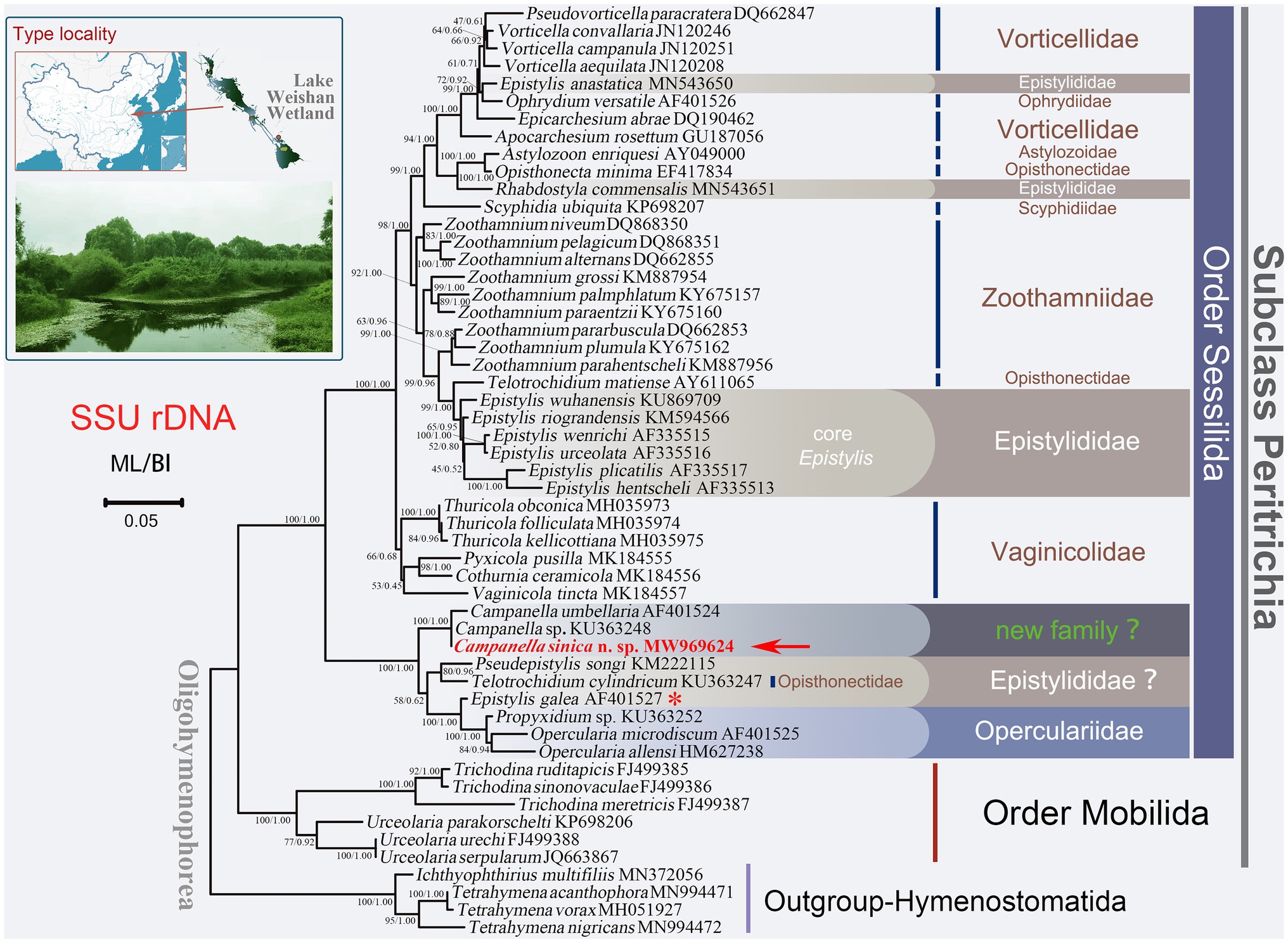
Figure 1. Maximum likelihood (ML) tree inferred from small subunit ribosomal DNA (SSU rDNA) sequence data. Sequence of Campanella sinica n. sp. is in red and indicated by an arrow. Numbers given at nodes of branches are the ultrafast bootstrap percent (UF) values for ML analysis and posterior probability (PP) values for BI analysis. The scale bar corresponds to five substitutions per 100 nucleotide positions. The asterisk represents the validity of this sequence needs to be verified.
DNA Extraction, PCR Amplification, and Sequencing
One to five zooids were isolated and washed several times in double distilled water using glass micropipettes under a dissecting microscope to remove contamination. Genomic DNA was extracted following the methods described by Zhang et al. (2020). The SSU rDNA, ITS1-5.8S rDNA-ITS2, and partial LSU rDNA sequences were amplified using primers 82F (5ꞌ-GAA ACT GCG AAT GGC TC-3ꞌ), 18SR (5ꞌ-TGA TCC TTC TGC AGG TTC ACC TAC-3ꞌ), 5.8SF (5ꞌ-GTA GGT GAA CCT GCG GAA GGA TC-3ꞌ), and R3 (5ꞌ- CAT TCG GCA GGT GAG TTG TTA CAC -3ꞌ), respectively (Medlin et al., 1988; Gong et al., 2007; Huang et al., 2018). Q5® Hot Start High-Fidelity DNA Polymerase (New England BioLabs, United States) was used for PCR to minimize the possibility of amplification errors. The fragments were sequenced biodirectionally by the Tsingke Biological Technology Company (Qingdao, China).
Phylogenetic Analyses
Phylogenetic analyses were performed both with single-gene datasets of SSU rDNA, ITS1-5.8S rDNA-ITS2, and LSU rDNA separately, and with a concatenated dataset of all three genes. In addition to the newly obtained sequences, other sequences downloaded from the GenBank database (for accession numbers, see Figures 1, 2), selected based on earlier studies (Sun et al., 2016; Zhuang et al., 2018), were used in the phylogenetic analyses. The boundary of the ITS1-5.8S rDNA-ITS2 was identified according to the methods described by Sun et al. (2010, 2013). A total of 53 SSU rDNA sequences including Campanella sinica n. sp., 48 peritrichs and four hymenostomatids as outgroup taxa were used to construct the SSU rDNA trees, while the analyses of ITS1-5.8S rDNA-ITS2, LSU rDNA, and concatenated data contained 28, 26, and 19 taxa, respectively (Figures 1, 2). Before constructing the trees, the selected sequences were aligned by MAFFT and further refined by Guidance 2 (https://guidance.tau.ac.il/ver2/; Katoh and Standley, 2013; Sela et al., 2015). The final alignments used for subsequent phylogenetic analyses comprised 1,605 sites for the SSU rDNA, 521 sites for the ITS1-5.8S rDNA-ITS2, 1,749 sites for the LSU rDNA, and 3,702 sites for the concatenated (SSU rDNA-ITS1-5.8S rDNA-ITS2-LSU rDNA) dataset.
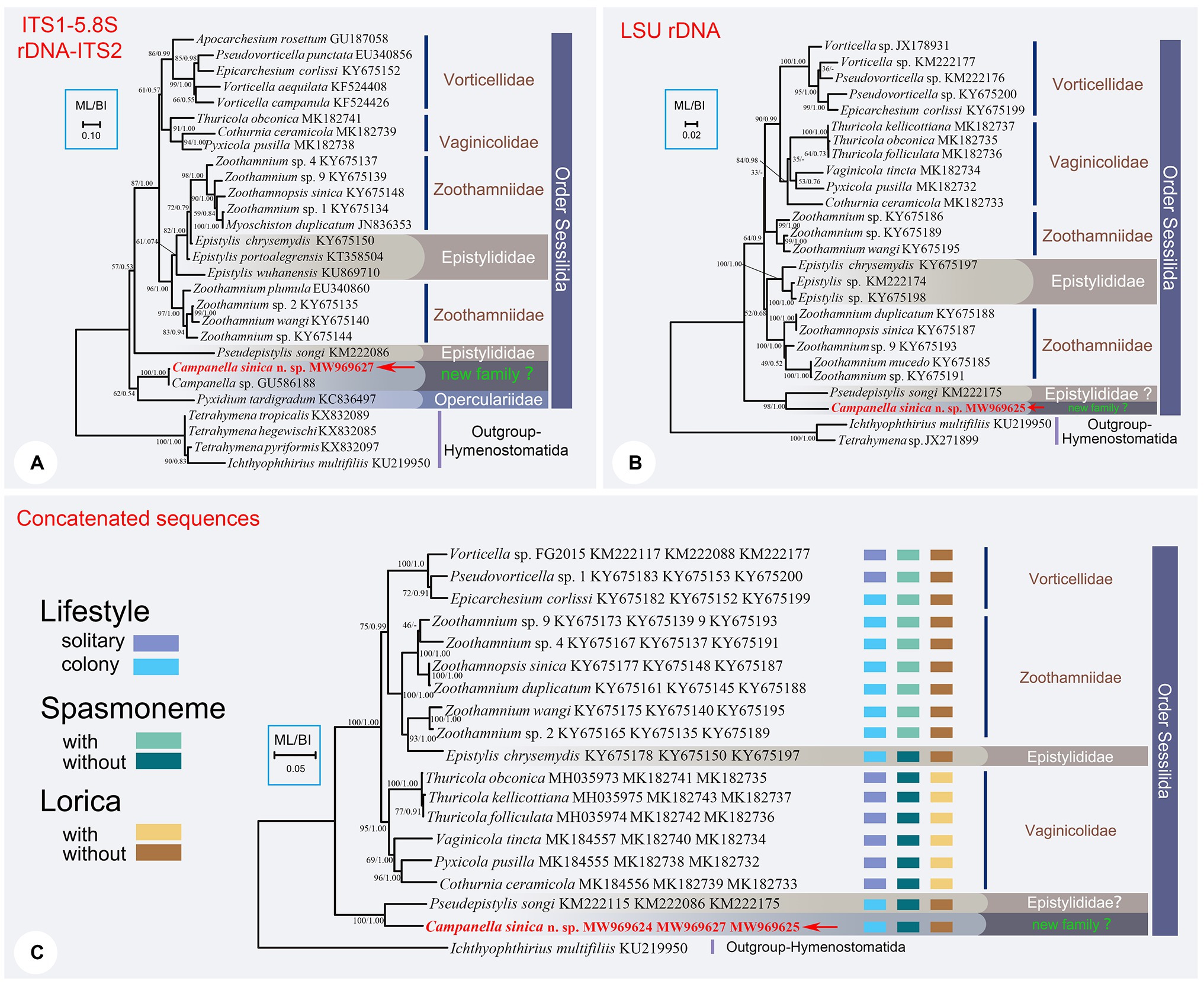
Figure 2. The ML trees inferred by ITS1-5.8S rDNA-ITS2 region (A), LSU rDNA sequence (B), and the concatenated data of SSU rDNA, ITS1-5.8S rDNA-ITS2, and LSU rDNA sequences (C). In each tree, Campanella sinica n. sp. is in red and indicated by an arrow. Numbers given at nodes of branches are the UF values for ML analysis and PP values for BI analysis. The scale bar corresponds to ten, two, and five substitutions per 100 nucleotide positions in (A-C), respectively.
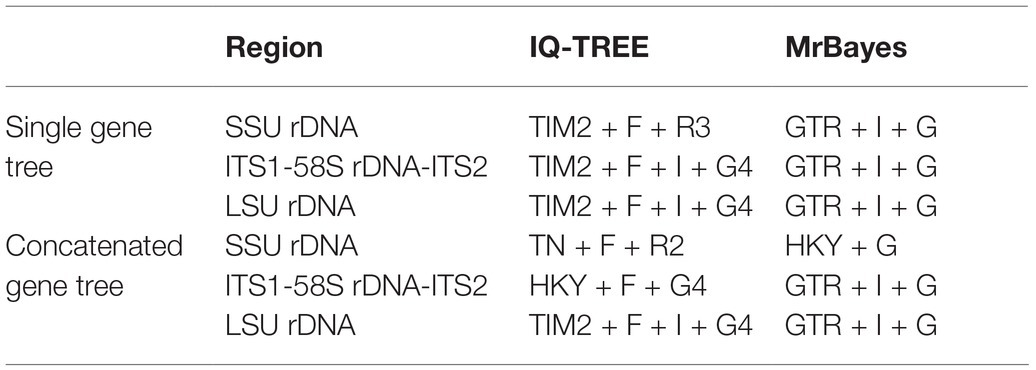
The best-fit models for maximum likelihood (ML) and Bayesian inference (BI) analyses were calculated by ModelFinder under Bayesian Information Criterion (Table 1; Kalyaanamoorthy et al., 2017). The concatenated sequences were treated with partitioned analyses to give the best-fit models for different regions.
Maximum likelihood analysis was carried out by IQ-TREE v2.0 with 10000 Ultrafast bootstrap replicates (Minh et al., 2020). BI analysis was carried out by MrBayes v3.2.7 (Ronquist et al., 2012). Markov chain Monte Carlo simulations were run for 1,000,000 generations with sampling every 100 generations. The first 25% of trees were discarded as burn-in. The run continued until the SD of split frequencies was below 0.01, and the effective sample size was > 200 (Rambaut et al., 2014). Phylogenetic trees were viewed by MEGA v.7 (Kumar et al., 2016) or Figtree.1
Sequence Comparisons Between Campanella sinica n. sp. and Its Closely Related Congeners
The SSU rDNA and ITS1-5.8S rDNA-ITS2 sequences of C. sinica n. sp. were separately aligned with closely related taxa by MAFFT v.7 with L-INS-I strategy (Katoh and Standley, 2013). The ends of alignments were refined by eye. The final comparisons were edited and viewed by Bioedit v.7 (Hall, 1999) and TBtools (Chen et al., 2020a). The nucleotide differences between each of the new sequences and those species with high sequence similarities are shown in Figure 3.
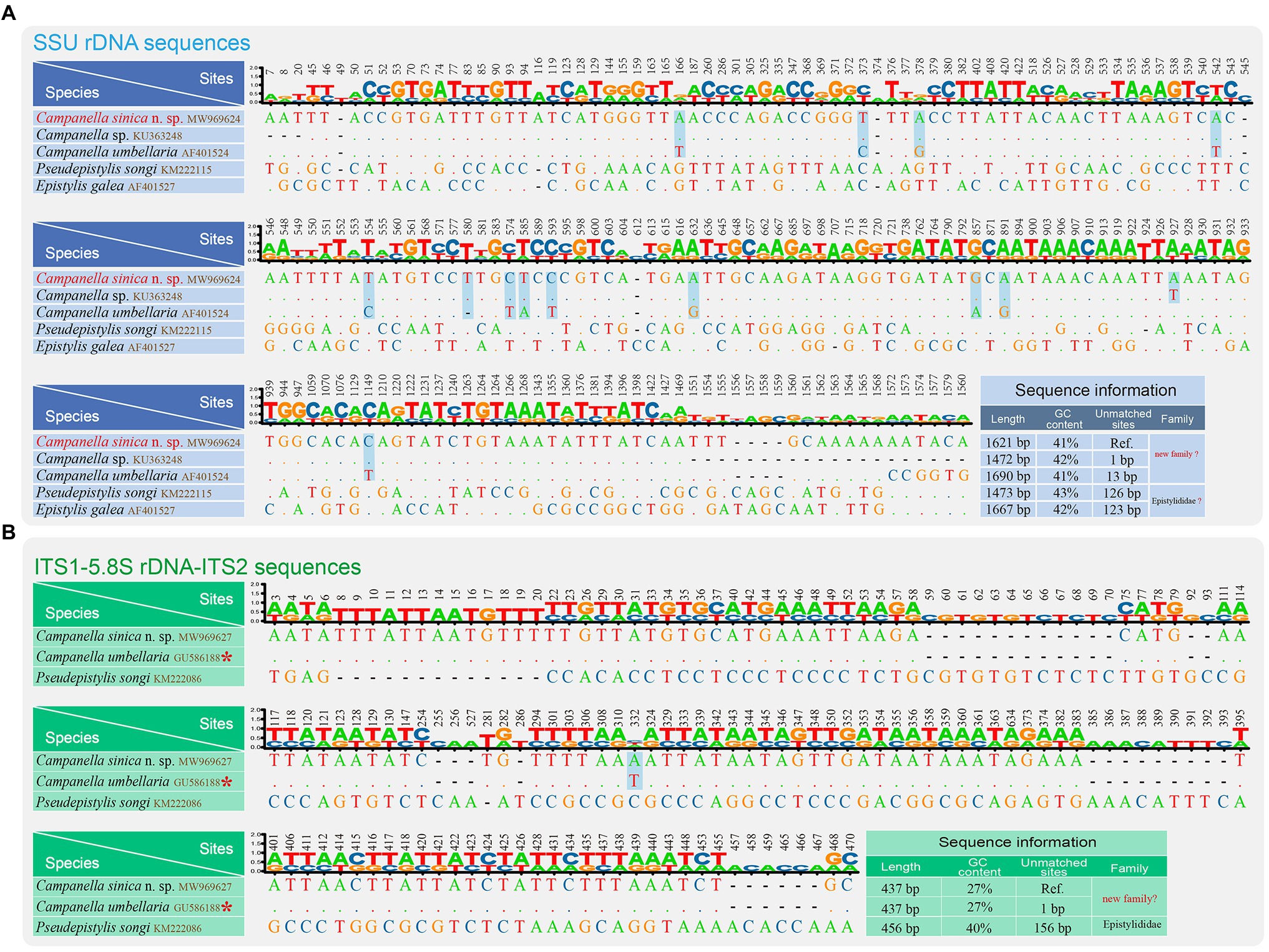
Figure 3. Nucleotide differences among Campanella sinica n. sp. and closely related taxa based on SSU rDNA sequences (A) and ITS1-5.8S rDNA-ITS2 sequences (B). The numbers in the header indicate the unmatched site positions. The sequences of Campanella sinica n. sp. in this study are shown in red. The variable sites of C. sinica n. sp. and the other Campanella species are shown in gray. Ref., reference sequence. The asterisk indicates the validity of this sequence needs to be verified.
Results
ZooBank Registration
Present work: urn:lsid:zoobank.org:pub:7D581595-D090-4750-B26B-27B1033356FF.
Campanella sinica n. sp.: urn:lsid:zoobank.org:act:D25C2BDA-EB29-457D-8DC0-B7CF4C62B72.
Subclass Peritrichia Stein, 1859
Order Sessilida Kahl, 1933
Family Epistylididae Kahl, 1933
Genus Campanella Goldfuss, 1820
Campanella sinica n. sp.
Diagnosis
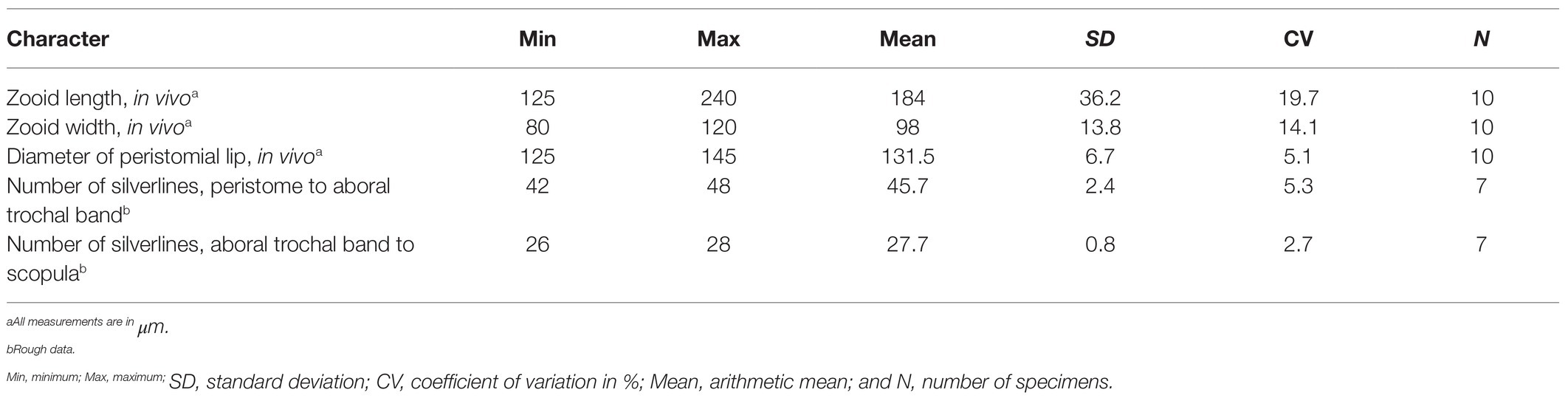
Mature colony up to 2 cm high with up to 1,000 zooids and dichotomously branched stalk. Zooids asymmetric horn-shaped, about 125–240 × 80–120 μm in vivo. Peristomial lip strongly everted. Haplokinety and polykinety makes 4.5–5 circuits around peristome before descending into infundibulum. Peristomial disc slightly convex. Contractile vacuole ventrally located beneath peristomial lip. Macronucleus C-shaped, transversely oriented. Infundibular polykineties 1 and 2 (P1, P2) each with three equal-length rows, infundibular kinety 3 (P3) with up to six rows. Abstomal end of P3 converges with P2. Transverse silverlines numbering about 42–48 from peristome to trochal band and about 26–28 from trochal band to scopula.
Type Locality and Ecological Features
Lake Weishan wetland (N34°43ꞌ59ꞌꞌ; E117°9ꞌ22ꞌꞌ), a freshwater wetland in Jining, Shandong Province, China. Water temperature of the artificial pond: 20°C.
Deposition of Slides
One protargol slide with the holotype specimen circled in ink (registration number: WT2020072201-01; Figure 4N), and two “dry” silver nitrate slides with paratype specimens (registration numbers: WZ2020052701-01, 02) were deposited in the Laboratory of Protozoology, Ocean University of China, Qingdao, China.
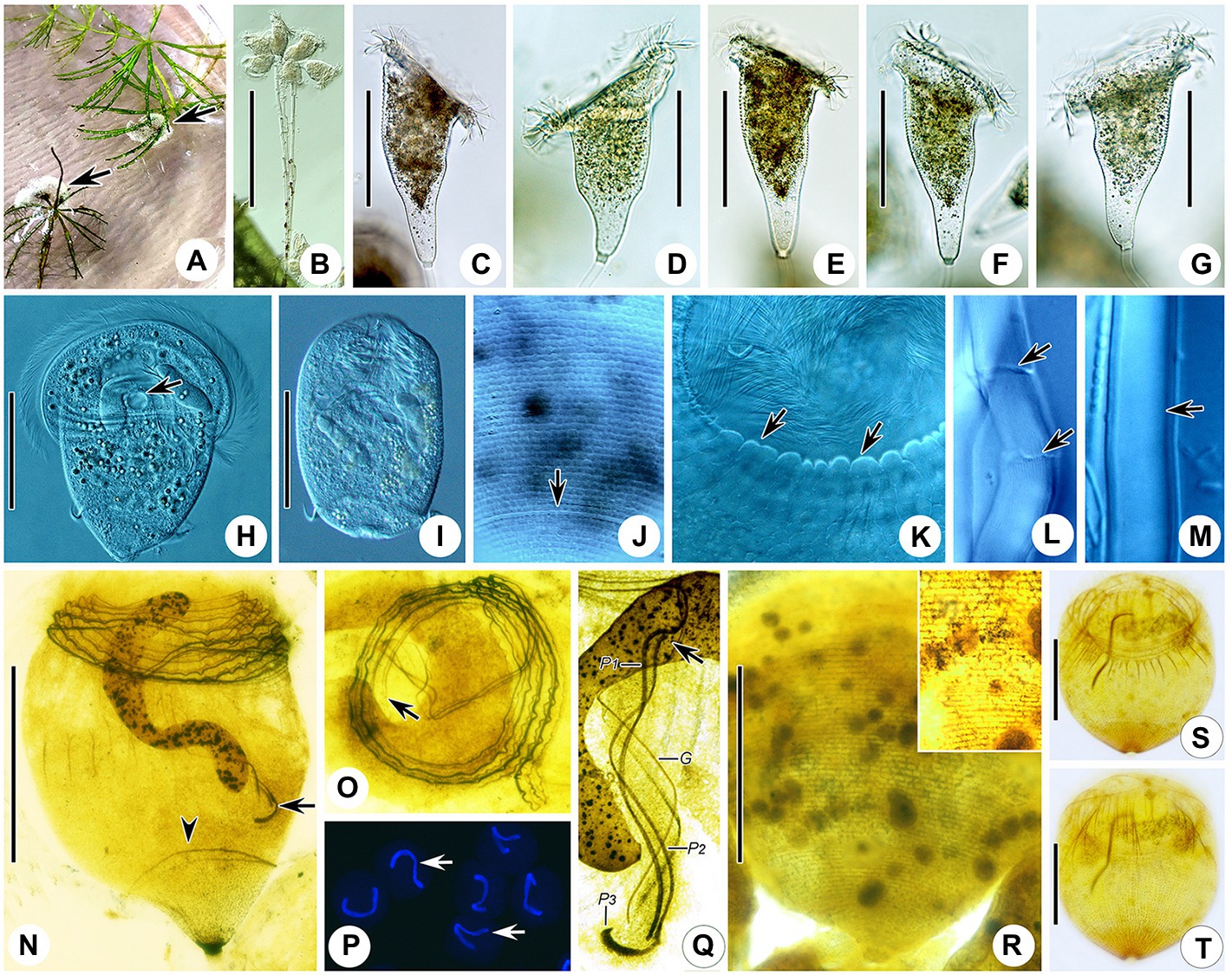
Figure 4. Photomicrographs of Campanella sinica n. sp. in vivo (A–M), after protargol staining (N,O,Q,S,T), after Hoechst-33342 staining (P), and after dry nitrate silver staining (R). (A) Colony (arrows) on aquatic plant. (B) Immature colony. (C–G) Different individuals, showing the variation of zooid shape. (H) A compressed zooid, arrow indicates the contractile vacuole. (I) Telotroch. (J) Pellicular striations, arrow indicates the aboral trochal band. (K) Detail of peristomial region of a contracted zooid, arrows indicate the peristomial lip. (L,M) Showing that the stalk is hollow (arrows). (N) Holotype specimen, showing the infraciliature and macronucleus, arrow indicates P3, arrowhead indicates the aboral trochal band. (O) Oral ciliature, apical view, arrow indicates the beginning of the oral ciliature. (P) Showing the macronucleus (arrows). (Q) Infundibular polykineties, arrow marks the anterior end of P2. (R) Silverline system. (S,T) Myoneme system. G, germinal kinety; P1–3, infundibular polykineties 1–3. Scale bars = 80 μm.
Etymology
The species-group name “sinica” refers to the country where the sample was collected.
Description
Zooids about 125–240 × 80–120 μm in vivo, asymmetrical horn-shaped, gradually tapering to scopula (Figures 4B–G, 5A; Table 2). Peristomial lip multi-layered, about 125–145 μm in diameter in vivo, relatively thick and strongly everted (Figures 4C–G, 5A). Peristomial disc slightly convex and elevated above peristome in fully extended zooids (Figures 4C–G, 5A). Oral cilia sturdy, about 12 μm long (Figures 4C–H, 5A). Telotroch cylindrical, with peristomial region fully contracted (Figures 4I, 5B).
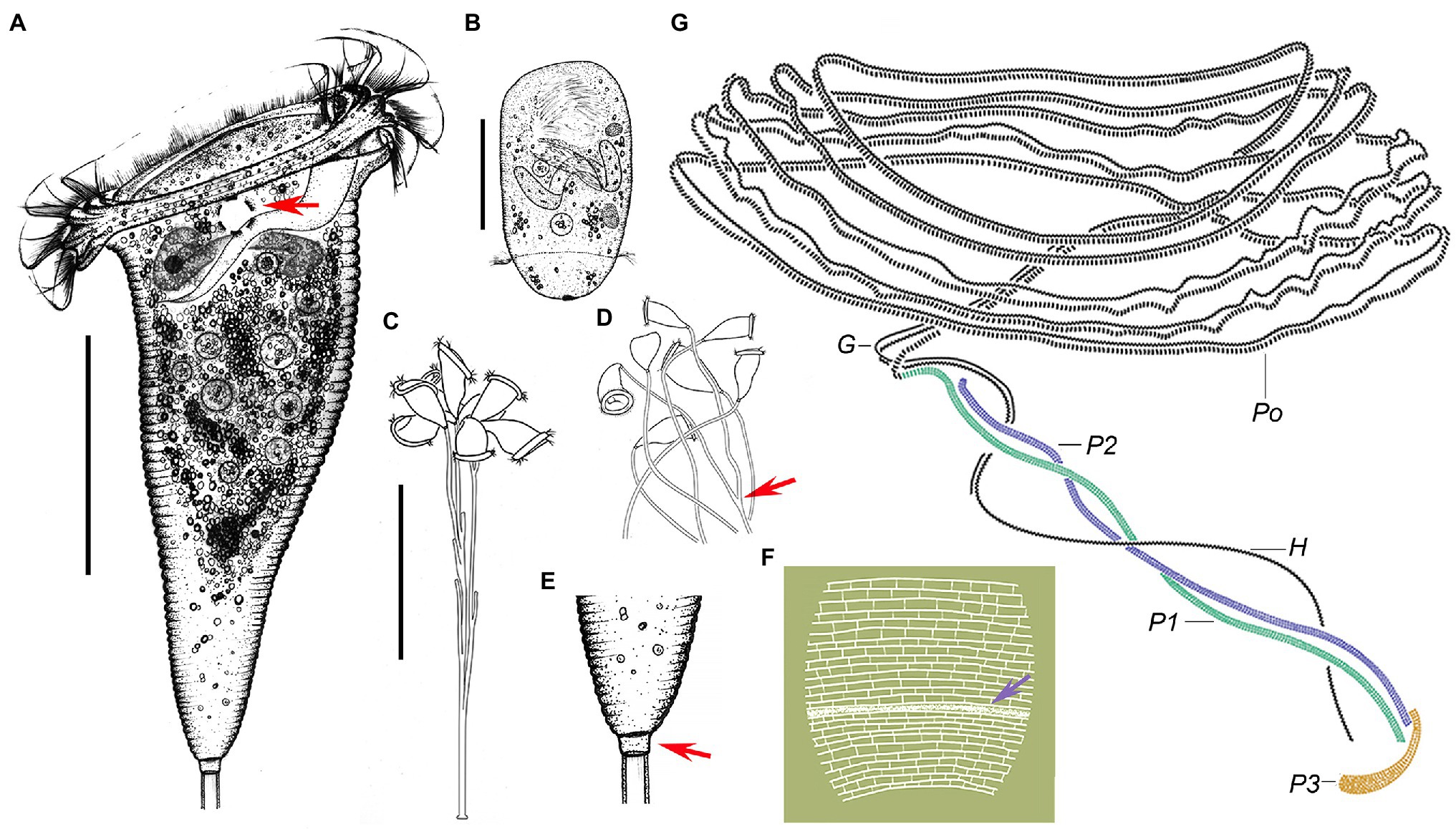
Figure 5. Campanella sincia n. sp. in vivo (A–E), after dry silver nitrate staining (F) and protargol (G) staining. (A) Typical zooid, arrow indicates the contractile vacuole. (B) Teletroch. (C,D) Immature colonies at low magnification, showing the branching pattern of the stalk, arrow in (D) shows an example of dichotomous branching. (E) Junction of stalk and zooid at high magnification, arrow shows the scopula. (F) Partial silverline system, arrow marks trochal band. (G) Oral ciliature. G, germinal kinety; H, haplokinety; Po, polykinety; and P1–3, infundibular polykineties 1–3. Scale bars: 50 μm in (A,B); 100 μm in (C).
Cytoplasm colorless, usually contains several grayish/darkish granules, 7 μm in diameter (Figures 4C, 5A). Contractile vacuole about 15 μm in diameter, located at ventral wall of infundibulum beneath peristomial lip (Figures 4H, 5A). Macronucleus relatively large, irregular C-shaped, transversely oriented in anterior half of zooid (Figures 4H,N,P, 5A). Pellicular striations reticulate, conspicuous when viewed at magnifications above 200× (Figures 4J,K,R, 5F). Trochal band dikinetid and located three-quarters of the way down length of zooid (Figures 4J, 5F). Scopula prominent, same width as stalk (Figures 4C–G, 5A,E).
Mature colony up to 2 cm high, contains more than 1,000 zooids (Figure 4A). Stalk straight, smooth, and hollow (Figures 4L,M, 5A,E), dichotomously branched with zooids regularly located in pairs (Figures 4B, 5C,D). Colony-founding zooid often sits on a relatively short stalk (Figure 5D).
Haplokinety and polykinety make approximately 4.5–5 circuits around peristome before entering infundibulum where they make a further circuit (Figures 4N,O, 5G). Polykinety transforms into infundibular polykinety 1 (P1). P1 accompanied by infundibular polykineties 2 and 3 (P2 and P3) within infundibulum. P1 and P2 composed of three equal-length rows of kinetosomes (Figures 4Q, 5G). P3 composed of about six rows (rough data) of kinetosomes (Figures 4Q, 5G). Germinal kinety (G) lies parallel to haplokinety (H) in abstomal region of infundibulum. Filamentous reticulum (FR) lies beneath wall of infundibulum parallel with haplokinety. Adstomally, P3 extends far beyond ends of P1 and P2, and sharply curves to make nearly 1/2 circuit around margin of cytostome (Figures 4Q, 5G).
Silverline system reticulate, with 42–48 (n = 7) transverse silverlines from peristome to aboral trochal band, and 26–28 (n = 7) transverse silverlines from aboral trochal band to scopula (Figures 4R, 5F). Transverse silverlines in anterior region of cell slightly more widely spaced than those in posterior region (Figures 4R, 5F). Myoneme system complex, composed of longitudinal fibers, medium-length longitudinal fibers, short longitudinal fibers, linking fibers, and support fibers (Figures 4S,T).
Phylogenetic Analyses
The GenBank accsssion numbers, lengths, and GC contents of the newly obtained rDNA sequences (SSU rDNA, ITS1-5.8S rDNA-ITS2, and LSU) are shown in Table 3.
In all the cases, the ML and BI trees have similar topologies, therefore, only the ML tree is presented for each gene or region. In the SSU rDNA tree, the order Sessilida is monophyletic, whereas the families Epistylididae, Opisthonectidae Foissner, 1976, Vorticellidae Ehrenberg, 1838, and Zoothamniidae Sommer, 1951 were non-monophyletic (Figure 1). Campanella species (C. umbellaria, C. sinica n. sp., and Campanella sp.) formed a fully supported clade (ML/BI, 100/1.00) that was sister to a clade comprising two epistylidids (Epistylis galea Ehrenberg, 1831 and Pseudepistylis songi Peng et al., 2007), three opercularids (Opercularia microdiscum Fauré-Fremiet, 1904, O. allensi Stokes, 1887, and Propyxidium sp.), and the opisthonectid Telotrochidium cylindricum (Figure 1). The above-mentioned species formed the basal clade within the Sessilida with maximal support (ML/BI, 100/1.00). The remaining sessilids formed a clade with maximal support (ML/BI, 100/1.00). Within this clade, loricate species of the family Vaginicolidae formed the basal branch, albeit with low support (ML/BI, 66/0.68). Species of the family Zoothamniidae were divided into separate clades. The majority of epistylidids (core Epistylis species) nested within Zoothamniidae (Figure 1), while E. anastatica (Linnaeus, 1767) Ehrenberg, 1830 and another epistylidid (Rhabostyla commensalis Möbius, 1888) nested within the Vorticellidae, and E. galea Ehrenberg, 1831 grouped with the operculariids, although the identity of this latter sequence awaits verification (Figure 1).
The topology of the ITS1–5.8S rDNA–ITS2 tree was broadly similar to that of the SSU rDNA tree except the position of family Vaginicolidae, which clustered with the Vorticellidae to form the crown group of sessilids rather than branching basally. Three colonial species, Campanella, C. sinica n. sp., and Propyxidium taradigradum Van der Land, 1964, formed the basal clade of the Sessilida. The remaining sessilids clustered into similar groups as those in the SSU rDNA tree. Each of the families Zoothamniidae, Epistylididae, Operculariidae, and Vorticellidae was non-monophyletic (Figure 2A).
In the LSU rDNA tree, Campanella sinica n. sp. and P. songi formed a strongly supported clade (ML/BI, 98/1.00) which was basal within the Sessilida (Figure 2B). The topology of the concatenated sequence tree was similar to that of the SSU rDNA tree, supporting the assertion that Campanella and Epistylis are distantly related. Epistylis chrysemydis Bishop and Jahn, 1941 nested within the Zoothamnidae (Figure 2C).
We also compared nucleotide differences of SSU rDNA and ITS1-5.8S rDNA-ITS2 sequences between C. sinica n. sp. and other closely related taxa (Figure 3). The SSU rDNA sequence most similar to C. sinica n. sp. was Campanella sp. (KU363248) with one variable site, followed by C. umbellaria (AF01524) with 13 variable sites. The GC contents of the SSU rDNA sequences of Campanella species, Epistylis gelea, and P. songi ranged from 41 to 43% (Figure 3A).
The ITS1-5.8S rDNA-ITS2 sequence most similar to C. sinica n. sp. was C. umbellaria (GU586188) with one variable site, followed by Pseudepistylis songi (KM222086) with 156 variable sites. The GC contents of the ITS1-5.8S rDNA-ITS2 sequences of Campanella species and P. songi were 27 and 40%, respectively (Figure 3B).
Discussion
Review of Campanella Species
Campanella umbellaria (Linnaeus, 1758) Goldfuss, 1820 is commonly found in eutrophic freshwaters (Linnaeus, 1758; Foissner et al., 1992). It is characterized by its symmetrical inverted bell-shaped zooid and horseshoe-shaped macronucleus (Foissner et al., 1992; Figures 6A,B). According to Sládeček and Sládečková (1974) and Foissner et al. (1992), the synonyms of this species are as follows: Hydra umbellaria Linnaeus, 1758, Epistylis flavicans Ehrenberg, 1838, Epistylis grandis Ehrenberg, 1838, Epistylis tincta Stokes, 1887, and Epistylis liebmanni Nenninger, 1948. It is noteworthy that no illustration of E. tincta was provided in the original description (Stokes, 1887). Kahl (1935) considered that E. tincta should belong to the genus Campanella and mentioned the difficulty in separating E. tincta from C. umbellaria. Yu et al. (1995) also accepted that E. tincta and E. liebmanni should belong to the genus Campanella but either overlooked or rejected the assertion that they are junior synonyms of C. umbellaria, referring to them as C. tincta and C. liebermanni (which is presumably a misspelling of “liebmanni”), respectively. Based on the descriptions provided by Stokes (1887) and Nenninger (1948), we agree with Sládeček and Sládečková (1974) and Foissner et al. (1992), who suggested that E. tincta and E. liebmanni (and as a consequence C. tincta and C. liebermanni) are junior synonyms of C. umbellaria. Nevertheless, variations are recognized in different populations of C. umbellaria, so it is possible that cryptic species might exist among these populations (Sládeček and Sládečková, 1974; Foissner et al., 1992). Further investigations including both morphological and molecular methods therefore need to be carried out to accurately characterize such populations.
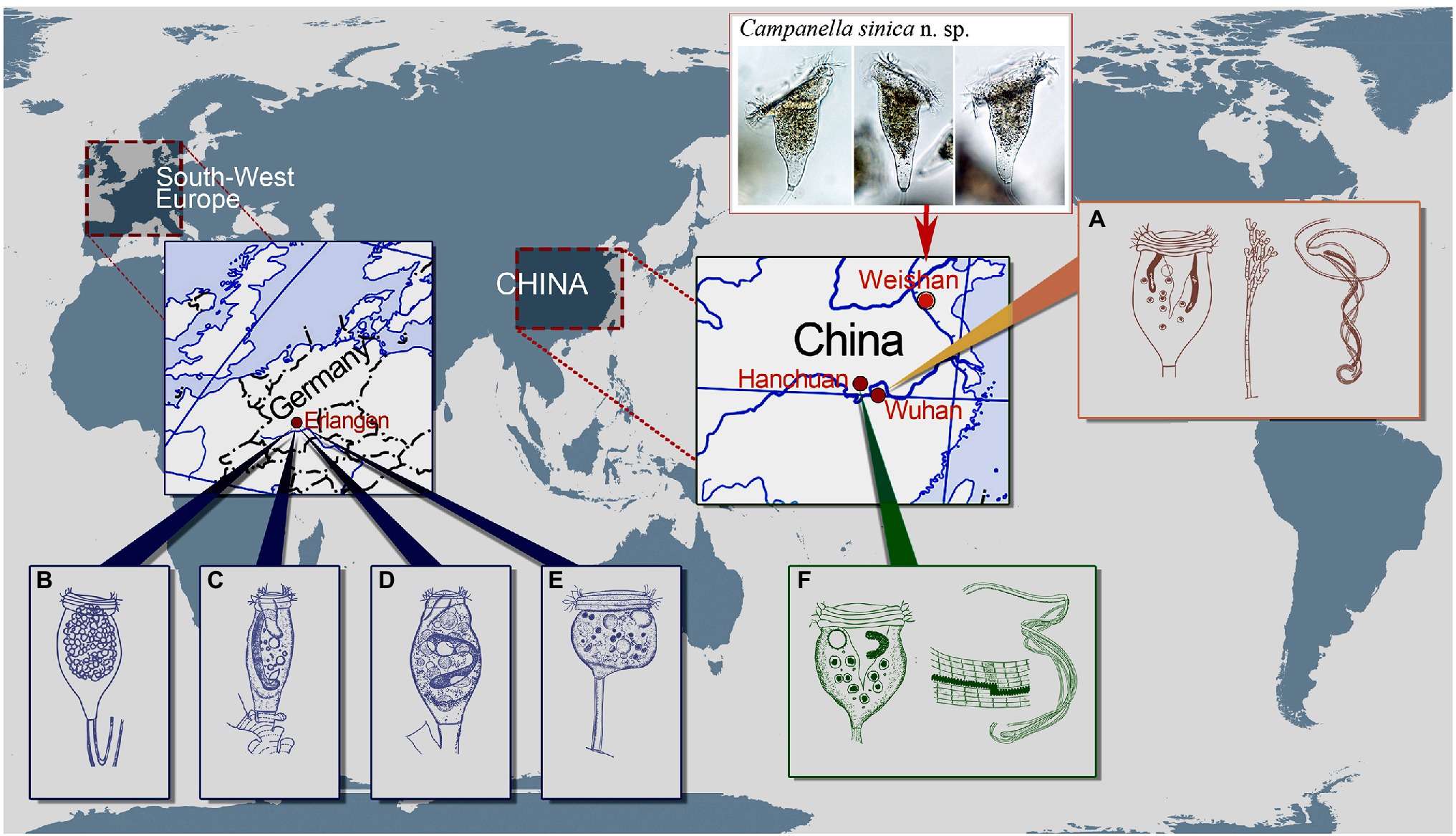
Figure 6. Schematic drawings of Campanella species showing some localities of C. umbellaria (A,B,E) and type localities of C. purneri (C), C. ovata n. grad. & n. comb. (D), and C. hanchuanensis (F). (A) After Shi et al. (2004) and Shen and Gu (2016). (B–E) After Nenninger (1948). (F) After Yu et al. (1995) and Shen and Gu (2016).
Besides E. liebmanni, Nenninger (1948) described other two taxa that she assigned to the genus Epistylis, i.e., E. purneri and E. purneri f. ovata, both of which should belong to the genus Campanella due to the presence in each of a multi-layered peristomial lip and an oral ciliature that makes more than four circuits around peristome before descending into the infundibulum (Figures 6B–D). Epistylis purneri and E. purneri f. ovata are similar but they differ from each other as follows: (1) the zooid shape of E. purneri is cylindrical, whereas that of E. purneri f. ovata is pyriform and (2) the stalk of E. purneri has several conspicuous transverse annular bulges, whereas that of E. purneri f. ovata has smooth surface (Figures 6C,D). Consequently, E. purneri f. ovata should be elevated to species rank. Yu et al. (1995) established the name C. purneri for E. purneri although without providing any justification. Here, we establish Campanella ovata (Nenninger, 1948) n. grad. & n. comb for E. purneri f. ovata (original combination Epistylis purneri f. ovata Nenninger, 1948). Nenninger (1948) also reported a new sessilid species, Opisthostyla tritora, which resembles colony-founding individuals of C. umbellaria (Figure 6E). Opisthostyla Stokes, 1886 is characterized by its solitary lifestyle and long, non-contractile stalk that is curved near the point of attachment to its substrate and acts in a spring-like manner throwing the organism backward when the zooid contracts (Stokes, 1886; Shen and Gu, 2016). However, the stalk of O. tritora has neither the structure nor the behavior that characterizes Opisthostyla. Therefore, O. tritora was probably misidentified by Nenninger (1948) and should belong to the genus Campanella. Horst (2018) listed O. tritora in the synonyms of C. umbellaria but did not provide any comments on this misidentification. Due to the lack of important morphological data for O. tritora, further studies are needed to determine the correct taxonomy of this species.
Yu et al. (1995) reported a new Campanella species, C. hanchuanensis, from a freshwater pond in Hubei Province, China (Figure 6F). In the original description, evidence for the presence of a reticulate (vs. transverse) silverline system is ambiguous (Yu et al., 1995). However, in a subsequent report, Shen and Gu (2016) confirmed that the silverline system of C. hanchuanensis is reticulate. Although C. hanchuanensis closely resembles C. umbellaria, they can be separated as follows: (1) the stalk of C. hanchuanensis is curved, whereas that of C. umbellaria is straight and (2) the trochal band of C. hanchuanensis consists of six rows of kinetosomes with four compact middle rows and two separated peripheral rows, whereas that of C. umbellaria comprises two compact rows only (Stiller, 1941; Yu et al., 1995; Shen and Gu, 2016).
Comparison of Campanella sinica n. sp. With Morphologically Similar Taxa
Campanella sinica n. sp. closely resembles C. umbellaria from which it can be separated as follows: (1) the zooid of C. sinica n. sp. is asymmetrical horn-shaped, while that of C. umbellaria is symmetrical and inverted bell-shaped; (2) the stalk of C. sinica n. sp. is smooth, whereas that of C. umbellaria has transverse annular bulges; and (3) the P3 of C. sinica n. sp. makes less than half a circuit within the infundibulum, whereas that of C. umbellaria makes 1–1.5 circuits. Campanella sinica n. sp. can be easily separated from C. purneri and C. ovata n. grad. & n. comb. by its asymmetrical horn-shaped (vs. elongated cylindrical) zooid, and convex, slightly elevated (vs. conspicuously elevated) peristomial disc. Compared with C. hanchuanensis, C. sinica n. sp. has a straight (vs. curved) stalk, asymmetrical horn-shaped zooids (vs. symmetrical inverted bell-shaped zooids), and the trochal band consists of several compact rows (vs. four compact middle rows and two separated peripheral rows). Based on these differences, C. sinica n. sp. can be easily distinguished from C. hanchuanensis.
Key to the Identification of Campanella Species
The genus Campanella contains five valid species, i.e., C. umbellaria, C. purneri, C. ovata n. grad. & n. comb., C. hanchuanensis, and C. sinica n. sp. We here supply a key to their identification.
1 Zooid cylindrical 2
- Zooid inverted bell-shaped or asymmetric 3
2 Stalk surface with several conspicuous transverse annular bulges C. purneri
- Stalk surface smooth C. ovata
3 Zooid inverted bell-shaped 4
- Zooid asymmetric C. sinica
4 Stalk straight C. umbellaria
- Stalk curved C. hanchuanensis
Phylogenetic Analyses of the Genus Campanella and Other Sessilids
Campanella has been reported from freshwater habitats worldwide (Nenninger, 1948; Sládeček and Sládečková, 1974; Shen and Gu, 2016). Species of Campanella are characterized by their colonial lifestyle, dichotomously branched non-contractile stalk, everted multi-layered peristomial lip, and well-developed haplokinety and polykiney that make more than four circuits around the peristome before descending into the infundibulum. Traditionally, this genus has been assigned to the family Epistylididae because of the non-contractile stalk and the well-defined, everted peristomial lip (Corliss, 1979; Lynn, 2008). However, phylogenetic analyses based on molecular data have challenged this classification. Miao et al. (2004), for example, suggested that the genus Campanella should be independent of the family Epistylididae and probably represents a separate lineage based on phylogenetic analyses of SSU rDNA sequences. Nonetheless, this study was solely based on single gene analyses and did not provide morphological data to support this conclusion.
In the present study, Campanella nests within the basal clade of the sessilids and is distinctly separated from other epistylidids in all phylogenetic trees, which is consistent with the findings of Miao et al. (2004). Based on the present phylogenetic analyses, Campanella species show a close relationship with species of the family Operculariidae (Figures 1, 2A). However, operculariids are characterized by the absence of a peristomial lip, separating them from Campanella which has a conspicuous peristomial lip. Thus, the genus Campanella should not be assigned to the family Operculariidae. In two of the present phylogenetic trees, i.e., LSU rDNA and concatenated data, the closest relative of C. sinica n. sp. is the epistylidid Pseudepistylis songi, while in the SSU rDNA tree, these two species, along with another epistylidid, namely Epistylis galea, nest within the basal clade of the sessilids. Nevertheless, Campanella can be clearly separated from both P. songi and E. galea by its multi-layered (vs. single-layered) peristomial lip and the haplokinety and ploykinety which make more than four turns around the peristome (vs. less than two turns) before descending into the infundibulum (Foissner et al., 1992; Peng et al., 2007). Compared with all families within the order Sessilida, Campanella is morphologically distinguishable by its multi-layered peristomial lip and the unique oral ciliature. Thus, a combination of the morphological and molecular phylogenetic data suggests that the genus Campanella should belong to a separate family within the order Sessilida as proposed by Miao et al. (2004). However, the present and previous studies show that most families in the order Sessilida are non-monophyletic (Miao et al., 2004; Sun et al., 2012, 2013, 2016; Zhuang et al., 2018). Furthermore, Epistylis galea, Pseudepistylis songi, and Rhabdostyla commensalis are also independent of core epistylidids in the present phylogenetic trees. Therefore, the establishment of new family-level taxa for genera such as Campanella is premature and should await a re-evaluation of the family Epistylididae based on detailed morphological and accurate molecular data.
Although at least 800 nominal species of sessilids have been reported from various aquatic environments worldwide, the taxonomy and classification of this group are confusing (Foissner et al., 2010; Sun et al., 2016; Wang et al., 2017b; Zhang et al., 2019; Zhou et al., 2019a,b). The order Sessilida is subdivided into 14 families based on phenotypic characters, such as lifestyle modes (solitary or colonial), stalk structures (with or without spasmoneme), lorica (presence or absence), and living habits (sessile or free-swimming) (Lynn, 2008). However, the validity of several of these families has been challenged in recent years following the application of molecular phylogenetic analyses, mainly based on SSU rDNA sequence data (Miao et al., 2001, 2004; Utz and Eizirik, 2007; Utz et al., 2010; Zhan et al., 2013; Jiang et al., 2019). Miao et al. (2001) provided the first analyses of phylogenetic relationships within the subclass Peritrichia based on SSU rDNA sequences. Subsequent molecular phylogenetic studies have suggested that family or genus assignments of many taxa should be reevaluated (Clamp and Williams, 2006; Utz and Eizirik, 2007; Williams and Clamp, 2007). Members of the basal clade of the Sessilida assemblage (e.g., Campanella umbellaria, Opercularia microdiscum, and Propyxidium sp.) are thought likely to possess the plesiomorphic characters of sessilids (Sun et al., 2016). The results of the present study support the findings of previous studies that the family Epistylididae is polyphyletic and should be divided into several groups and/or various of its members should be re-assigned to other families (Sun et al., 2012, 2013; Zhou et al., 2019b; Lu et al., 2020). The clustering pattern in the phylogenetic trees suggests that the major epistylidid group (core Epistylis) evolved from species of the family Zoothamniidae, which indicates that these underwent loss of the stalk spasmoneme. It is noteworthy that in the tree based on the concatenated dataset, E. chrysemydis is nested within the Zoothamnidae thus supporting this hypothesis (Figure 2C). Zhuang et al. (2018) reported a population of E. chrysemydis with a hollow stalk containing a central bundle of fibers that is similar to the spasmoneme of Zoothamnium. Species of Campanella have a hollow stalk suggesting that stalks with this structure may also represent an ancestral trait of the sessilids. Furthermore, based on their gene sequence similarities, Campanella spp. (represented by SSU rDNA sequence KU343248 and ITS1-5.8S rDNA-ITS2 sequence GU586188) and C. sinica n. sp. are probably conspecific, although it is not possible to verify this due to the lack of vouchered specimens or morphological data for the former taxa.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/ (MW969624, MW969627, and MW969625).
Author Contributions
ZW, LL, and CW conceived and designed the paper. ZW and TW carried out the live observations, protargol staining, and phylogenetic analyses. ZW, TW, BL, AW, YC, XZ, SA-F, LL, and CW wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of China (project numbers: 31772431, 32030015, and 32000300) and the Researchers Supporting Project (RSP-2021/7) of the King Saud University, Saudi Arabia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Mr. Changjun Mu (Weishan Special Aquaculture Base) and Mr. Ya Wang (Weishan Fishery Development Service Center) for the institutional support. We are also grateful to the editor and reviewers for their constructive suggestions.
Footnotes
References
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Wang, C. D., Pan, B., Lu, B. R., Li, C., Shen, Z., et al. (2020b). Single-cell genomic sequencing of three peritrichs (Protista, Ciliophora) reveals less biased stop codon usage and more prevalent programmed ribosomal frameshifting than in other ciliates. Front. Mar. Sci. 7:602323. doi: 10.3389/fmars.2020.602323
Clamp, J. C., and Williams, D. (2006). A molecular phylogenetic investigation of Zoothamnium (Ciliophora, Peritrichia, Sessilida). J. Eukaryot. Microbiol. 53, 494–498. doi: 10.1111/j.1550-7408.2006.00132.x
Corliss, J. O. (1979). The Ciliated Protozoa. Characterization, Classification and Guide to the Literature. Oxford, UK: Pergamon Press.
Foissner, W. (2014). An update of 'basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa'. Int. J. Syst. Evol. Microbiol. 64, 271–292. doi: 10.1099/ijs.0.057893-0
Foissner, W., Berger, H., and Kohmann, F. (1992). Taxonomische und Ökologische Revision Der Ciliaten Des Saprobiensystem, Band II: Peritrichia, Heterotrichida, Odontostomatida. Informationsberichte des Bayer. München: Bayerisches Landesamt für Wasserwirtschaft.
Foissner, W., Blake, N., Wolf, K., Breiner, H. W., and Stoeck, T. (2010). Morphological and molecular characterization of some peritrichs (Ciliophora Peritrichida) from tank bromeliads including two new genera: Orborhabdostyla and Vorticellides. Acta Protozool. 48, 291–319.
Gao, F., Warren, A., Zhang, Q., Gong, J., Miao, M., Sun, P., et al. (2016). The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep. 6:24874. doi: 10.1038/srep24874
Gentekaki, E., Kolisko, M., Gong, Y., and Lynn, D. (2017). Phylogenomics solves a long-standing evolutionary puzzle in the ciliate world: The subclass Peritrichia is monophyletic. Mol. Phylogenet. Evol. 106, 1–5. doi: 10.1016/j.ympev.2016.09.016
Gong, J., Kim, S.-J., Kim, S.-Y., Min, G.-S., Roberts, D. M., Warren, A., et al. (2007). Taxonomic redescriptions of two ciliates, Protogastrostyla pulchra n. g., n. comb. and Hemigastrostyla enigmatica (Ciliophora: Spirotrichea, Stichotrichia), with phylogenetic analyses based on 18S and 28S rRNA gene sequences. J. Eukaryot. Microbiol. 54, 468–478. doi: 10.1111/j.1550-7408.2007.00288.x
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids. Symp. Ser. 41, 95–98.
Horst, S. (2018). Kompendium der auf Wirbellosen epizoisch lebenden limnischen sessilen Peritrichia (Ciliophora) West-, Mittel- und Osteuropas. Denisia 41, 47–294.
Hu, X. Z., Lin, X. F., and Song, W. B. (2019). Ciliate Atlas: Species Found in the South China Sea. Beijing, China: Science Press.
Huang, H., Yang, J., Huang, S., Gu, B., Wang, Y., Wang, L., et al. (2021). Spatial distribution of planktonic ciliates in the western Pacific Ocean: along the transect from Shenzhen (China) to Pohnpei (Micronesia). Mar. Life Sci. Technol. 3, 103–115. doi: 10.1007/s42995-020-00075-7
Huang, J. B., Zhang, T., Zhang, Q., Li, Y., Warren, A., Pan, H., et al. (2018). Further insights into the highly derived haptorids (Ciliophora, Litostomatea): phylogeny based on multigene data. Zool. Scr. 47, 231–242. doi: 10.1111/zsc.12269
Irwin, N. A., and Lynn, D. H. (2015). Molecular phylogeny of mobilid and sessilid ciliates symbiotic in eastern Pacific limpets (Mollusca: Patellogastropoda). J. Eukaryot. Microbiol. 62, 543–552. doi: 10.1111/jeu.12208
Jiang, C. Q., Wang, G. Y., Xiong, J., Yang, W. T., Sun, Z. Y., Feng, J. M., et al. (2019). Insights into the origin and evolution of Peritrichia (Oligohymenophorea, Ciliophora) based on analyses of morphology and phylogenomics. Mol. Phylogenet. Evol. 132, 25–35. doi: 10.1016/j.ympev.2018.11.018
Kahl, A. (1935). Urtiere oder Protozoa. I. Wimpertiere oder Ciliata (Infusoria). 4. Peritricha und Chonotricha. Tierwelt Dtl. 30, 651–886.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14:587. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Klein, B. M. (1958). The “dry” silver method and its proper use. J. Protozool. 5, 99–103. doi: 10.1111/j.1550-7408.1958.tb02535.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, Q., Zhao, Q., McMinn, A., Yang, E. J., and Jiang, Y. (2021). Planktonic microbial eukaryotes in polar surface waters: recent advances in high-throughput sequencing. Mar. Life Sci. Technol. 3, 94–102. doi: 10.1007/s42995-020-00062-y
Lu, B. R., Li, L. F., Hu, X. Z., Ji, D. D., Al-Rasheid, K. A. S., and Song, W. B. (2019). Novel contributions to the peritrich family Vaginicolidae (Protista: Ciliophora), with morphological and phylogenetic analyses of poorly known species of Pyxicola, Cothurnia and Vaginicola. Zool. J. Linn. Soc. 187, 1–30. doi: 10.1093/zoolinnean/zlz009
Lu, B. R., Shen, Z., Zhang, Q., Hu, X., Warren, A., and Song, W. B. (2020). Morphology and molecular analyses of four epibiotic peritrichs on crustacean and polychaete hosts, including descriptions of two new species (Ciliophora, Peritrichia). Eur. J. Protistol. 73:125670. doi: 10.1016/j.ejop.2019.125670
Lynn, D. (2008). The ciliated Protozoa: Characterization, Classification, and Guide to the Literature. 3rd Edn. Dordrecht: Springer.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Miao, W., Fen, W., Yu, Y., Zhang, X., and Shen, Y. (2004). Phylogenetic relationships of the subclass Peritrichia (Oligohymenophorea, Ciliophora) inferred from small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 51, 180–186. doi: 10.1111/j.1550-7408.2004.tb00543.x
Miao, W., Yu, Y. H., and Shen, Y. F. (2001). Phylogenetic relationships of the subclass Peritrichia (Oligohymenophorea, Ciliophora) with emphasis on the genus Epistylis, inferred from small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 48, 583–587. doi: 10.1111/j.1550-7408.2001.tb00194.x
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Nenninger, U. (1948). Die Peritrichen der Umgebung von Erlangen mit besonderer Berücksichtigung der Wirtsspezifität. Zool. Jb. Syst. 77, 169–266.
Pan, X. M., Bourland, W. A., and Song, W. B. (2013). Protargol synthesis: an in-house protocol. J. Eukaryot. Microbiol. 60, 609–614. doi: 10.1111/jeu.12067
Peng, X., Shi, X., and Warren, A. (2007). Establishment of a new peritrich ciliate genus, Pseudepistylis n. gen. (Ciliophora: Peritrichia: Epistylididae), with a description of a new freshwater species, Pseudepistylis songi n. sp. from Wenzhou, China. Zootaxa 1524, 35–45. doi: 10.11646/zootaxa.1524.1.3
Rambaut, A., Drummond, A., and Suchard, M. (2014). Tracer v1. 6. Available at: http://beast.bio.ed.ac.uk.Tracer (Accessed June 12, 2017).
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Shen, Y. F., and Gu, M. R. (2016). Fauna Sinica: Invertebrata Ciliophora Oligohymenophorea Peritrichida. Beijing: Science Press.
Shi, X., Warren, A., Yu, Y., and Shen, Y. (2004). Infraciliature and myoneme system of Campanella umbellaria (Protozoa, Ciliophora, Peritrichida). J. Morphol. 261, 43–51. doi: 10.1002/jmor.10225
Sládeček, V., and Sládečková, A. (1974). Massenentwicklung von Campanella umbellaria L. in den Filteranlagen eines Dampfkraftwerkes. Acta Hydrochim. Hydrobiol. 2, 79–82. doi: 10.1002/aheh.19740020107
Song, W. B., Warren, A., and Hu, X. Z. (2009). Free-living Ciliates in the Bohai and Yellow Seas. China: Science Press.
Stiller, J. (1941). Einige Gewässer der Umgebung von Szeged und ihre Peritrichenfauna. Arch. Hydrobiol. 38, 313–435.
Sun, P., Clamp, J. C., and Xu, D. (2010). Analysis of the secondary structure of ITS transcripts in peritrich ciliates (Ciliophora, Oligohymenophorea): implications for structural evolution and phylogenetic reconstruction. Mol. Phylogenet. Evol. 56, 242–251. doi: 10.1016/j.ympev.2010.02.030
Sun, P., Clamp, J. C., Xu, D., Huang, B., and Shin, M. K. (2016). An integrative approach to phylogeny reveals patterns of environmental distribution and novel evolutionary relationships in a major group of ciliates. Sci. Rep. 6:21695. doi: 10.1038/srep21695
Sun, P., Clamp, J. C., Xu, D., Huang, B., Shin, M. K., and Turner, F. (2013). An ITS-based phylogenetic framework for the genus Vorticella: finding the molecular and morphological gaps in a taxonomically difficult group. Proc. Biol. Sci. 280:20131177. doi: 10.1098/rspb.2013.1177
Sun, P., Xu, D., Clamp, J. C., and Shin, M. K. (2012). Molecular and morphological characterization of a poorly known marine ciliate, Myoschiston duplicatum Precht 1935: implications for phylogenetic relationships between three morphologically similar genera – Zoothamnium, Myoschiston, and Zoothamnopsis (Ciliophora, Peritrichia, Zoothamniidae). J. Eukaryot. Microbiol. 59, 163–170. doi: 10.1111/j.1550-7408.2011.00609.x
Utz, L. R., and Eizirik, E. (2007). Molecular phylogenetics of subclass Peritrichia (Ciliophora: Oligohymenophorea) based on expanded analyses of 18S rRNA sequences. J. Eukaryot. Microbiol. 54, 303–305. doi: 10.1111/j.1550-7408.2007.00260.x
Utz, L. R., Simao, T. L., Safi, L. S., and Eizirik, E. (2010). Expanded phylogenetic representation of genera Opercularia and Epistylis sheds light on the evolution and higher-level taxonomy of peritrich ciliates (Ciliophora: Peritrichia). J. Eukaryot. Microbiol. 57, 415–420. doi: 10.1111/j.1550-7408.2010.00497.x
Wang, Z., Bourland, W. A., Zhou, T., Yang, H., Zhang, C., and Gu, Z. (2020). Morphological and molecular characterization of two Trichodina (Ciliophora, Peritrichia) species from freshwater fishes in China. Eur. J. Protistol. 72:125647. doi: 10.1016/j.ejop.2019.125647
Wang, Z., Deng, Q., Zhou, T., Yang, H., and Gu, Z. (2018). First record of two ectoparasitic ciliates of the genus Trichodina (Ciliophora: Trichodinidae) parasitizing gills of an invasive freshwater fish, Micropercops swinhonis, in Tibet. Parasitol. Res. 117, 2233–2242. doi: 10.1007/s00436-018-5910-y
Wang, H., Shi, X., Clamp, J. C., Liu, G., Wu, S., and Ma, L. (2011). A new interpretation of stomatogenesis in a peritrich ciliate: using Campanella umbellaria as a model system. J. Morphol. 272, 987–1006. doi: 10.1002/jmor.10965
Wang, Z., Zhou, T., and Gu, Z. (2017a). New data of two trichodinid ectoparasites (Ciliophora: Trichodinidae) from farmed freshwater fishes in Hubei, china. Eur. J. Protistol. 60, 50–59. doi: 10.1016/j.ejop.2017.04.002
Wang, Z., Zhou, T., Guo, Q., and Gu, Z. (2017b). Description of a new freshwater ciliate Epistylis wuhanensis n. sp. (Ciliophora, Peritrichia) from China, with a focus on phylogenetic relationships within family Epistylididae. J. Eukaryot. Microbiol. 64, 394–406. doi: 10.1111/jeu.12375
Wang, Z., Zhou, T., Yang, H., and Gu, Z. (2019). First diagnosis of ectoparasitic ciliates (Trichodina and Chilodonella) on farmed juvenile yellow catfish, Tachysurus fulvidraco in China. Aquac. Res. 50, 3275–3285. doi: 10.1111/are.14285
Warren, A. (1986). A revision of the genus Vorticella (Ciliophora: Peritrichida). Bull. Br. Mus. Nat. Hist. 50, 1–57. doi: 10.5962/p.18297
Williams, D., and Clamp, J. C. (2007). A molecular phylogenetic investigation of Opisthonecta and related genera (Ciliophora, Peritrichia, Sessilida). J. Eukaryot. Microbiol. 54, 317–323. doi: 10.1111/j.1550-7408.2007.00262.x
Wu, T., Li, Y., Lu, B. R., Shen, Z., Song, W. B., and Warren, A. (2020). Morphology, taxonomy and molecular phylogeny of three marine peritrich ciliates, including two new species: Zoothamnium apoarbuscula n. sp. and Z. apohentscheli n. sp. (Protozoa, Ciliophora, Peritrichia). Mar. Life Sci. Technol. 2, 334–348. doi: 10.1007/s42995-020-00046-y
Wu, T., Li, Y., Zhang, T., Hou, J., Mu, C., Warren, A., et al. (2021). Morphology and molecular phylogeny of three Epistylis species found in freshwater habitats in China, including the description of E. foissneri n. sp. (Ciliophora, Peritrichia). Eur. J. Protistol. 78:125767. doi: 10.1016/j.ejop.2021.125767
Yu, Y., Feng, W., and Shen, Y. (1995). Description of Campanella hanchuanensis sp. nov. (Ciliophora, Peritrichida): general morphology and isozyme pattern. Rep. Suwa Hydrobiol. 9, 87–94.
Zhan, Z., Xu, K., and Dunthorn, M. (2013). Evaluating molecular support for and against the monophyly of the Peritrichia and phylogenetic relationships within the Mobilida (Ciliophora, Oligohymenophorea). Zool. Scr. 42, 213–226. doi: 10.1111/j.1463-6409.2012.00568.x
Zhang, T., Dong, J., Cheng, T., Duan, L., and Shao, C. (2020). Reconsideration of the taxonomy of the marine ciliate Neobakuella aenigmatica moon et al., 2019 (Protozoa, Ciliophora, Hypotrichia). Mar. Life Sci. Technol. 2, 97–108. doi: 10.1007/s42995-020-00032-4
Zhang, Y., Shen, Z., Zhang, F., Yu, Y., Li, J., and Lin, X. F. (2019). Taxonomy and phylogeny of Pseudovorticella littoralis sp. n. and P. alani sp. n. (Ciliophora: Peritrichia) from coastal waters of southern China. Eur. J. Protistol. 71:125635. doi: 10.1016/j.ejop.2019.125635
Zhao, Y., Dong, Y., Li, H., Lin, S., Huang, L., Xiao, T., et al. (2020). Grazing by microzooplankton and copepods on the microbial food web in spring in the southern Yellow Sea, china. Mar. Life Sci. Technol. 2, 442–455. doi: 10.1007/s42995-020-00047-x
Zhou, T., Wang, Z., Yang, H., and Gu, Z. (2019a). Morphological and molecular identification of epibiontic sessilid Epistylis semiciculus n. sp. (Ciliophora, Peritrichia) from Procambarus clarkia (Crustacea, Decapoda) in China. Int. J. Parasitol. Parasites Wildl. 10, 289–298. doi: 10.1016/j.ijppaw.2019.09.006
Zhou, T., Wang, Z., Yang, H., and Gu, Z. (2019b). Two new colonial peritrich ciliates (Ciliophora, Peritrichia, Sessilida) from China: with a note on taxonomic distinction between Epistylididae and Operculariidae. Eur. J. Protistol. 70, 17–31. doi: 10.1016/j.ejop.2019.05.003
Keywords: rDNA sequences, morphology, peritrichs, sessilid ciliate, phylogenetic analyses
Citation: Wang Z, Wu T, Lu B, Chi Y, Zhang X, Al-Farraj SA, Song W, Warren A, Li L and Wang C (2021) Integrative Studies on a New Ciliate Campanella sinica n. sp. (Protista, Ciliophora, Peritrichia) Based on the Morphological and Molecular Data, With Notes on the Phylogeny and Systematics of the Family Epistylididae. Front. Microbiol. 12:718757. doi: 10.3389/fmicb.2021.718757
Edited by:
Weiwei Liu, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Igor Dovgal, A.O. Kovalevsky Institute of Marine Biological Research, RussiaLaura Utz, Pontifical Catholic University of Rio Grande do Sul, Brazil
Copyright © 2021 Wang, Wu, Lu, Chi, Zhang, Al-Farraj, Song, Warren, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lifang Li, cWRfbGlsaXlAc2luYS5jb20=; Chundi Wang, Y2h1bmRpd2FuZzEwMkBmb3htYWlsLmNvbQ==
†These authors have contributed equally to this work
 Zhe Wang
Zhe Wang Tong Wu
Tong Wu Borong Lu
Borong Lu Yong Chi
Yong Chi Xue Zhang
Xue Zhang Saleh A. Al-Farraj3
Saleh A. Al-Farraj3 Weibo Song
Weibo Song Lifang Li
Lifang Li Chundi Wang
Chundi Wang