- 1Hunan Agricultural University, Changsha, China
- 2Changsha Tobacco Company of Hunan Province, Changsha, China
- 3Chenzhou Tobacco Company of Hunan Province, Chenzhou, China
- 4Wenshan Tobacco Company of Yunnan Province, Wenshan, China
- 5CAS Key Laboratory for Environmental Biotechnology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China
- 6Institute for Marine Science and Technology, Shandong University, Qingdao, China
- 7Tobacco Company of Hunan Province, Changsha, China
- 8College of Resources and Environment, University of Chinese Academy of Sciences, Beijing, China
In the plant rhizosphere and endosphere, some fungal and bacterial species regularly co-exist, however, our knowledge about their co-existence patterns is quite limited, especially during invasion by bacterial wilt pathogens. In this study, the fungal communities from soil to endophytic compartments were surveyed during an outbreak of tobacco wilt disease caused by Ralstonia solanacearum. It was found that the stem endophytic fungal community was significantly altered by pathogen invasion in terms of community diversity, structure, and composition. The associations among fungal species in the rhizosphere and endosphere infected by R. solanacearum showed more complex network structures than those of healthy plants. By integrating the bacterial dataset, associations between fungi and bacteria were inferred by Inter-Domain Ecological Network (IDEN) approach. It also revealed that infected samples, including both the rhizosphere and endosphere, had more complex interdomain networks than the corresponding healthy samples. Additionally, the bacterial wilt pathogenic Ralstonia members were identified as the keystone genus within the IDENs of both root and stem endophytic compartments. Ralstonia members was negatively correlated with the fungal genera Phoma, Gibberella, and Alternaria in infected roots, as well as Phoma, Gibberella, and Diaporthe in infected stems. This suggested that those endophytic fungi may play an important role in resisting the invasion of R. solanacearum.
Introduction
Ralstonia solanacearum, the causative agent of soil-borne bacterial wilt disease in plants, is often found in agricultural land used for tobacco cultivation. Once the pathogen invades the plant root system, it rapidly spreads to the stem, causing an internal system imbalance of the entire tobacco plant, accelerating senescence and death (Li et al., 2017; Wei et al., 2018). As important members of the plant microecosystem (Nilsson et al., 2019), fungi play a vital role in promoting the material cycle of agro-ecosystem and affecting plant growth and health (Lilleskov et al., 2011; Zhang et al., 2021). Therefore, dynamic changes in the structure and composition of the plant soil fungal community can indicate the alterations of soil micro-ecological environment (Shi et al., 2020). Plant endophytic fungi exist within the host plant and interact closely with other microorganisms to promote plant growth, resist the invasion of plant pathogens, and improve the disease resistance of host plant through their own metabolites or induction of the host’s metabolites (Arnold, 2007; Mousa et al., 2016; Bastias et al., 2017). However, our knowledge about how the soil and endophytic fungal communities change under the invasion of R. solanacearum is quite limited.
With the surge of research on microbial communities in various ecological environments, the interdomain relationships between different types of microbial communities has attracted great attention (Kapitan et al., 2019; Wang et al., 2020). The association of plant bacterial and fungal communities is critical to overall microbial community structure and plant health (Hassani et al., 2018), and intrigues numerous botanists and microbiologists. Plant microbial communities live and colonize several zones including the bulk soil, rhizosphere soil, phyllosphere, and endosphere (Bulgarelli et al., 2013; Kumar et al., 2017). They play a vital role in the acquisition of nutrients by plants, mutual defense, and co-evolution (Martin et al., 2017; Fitzpatrick et al., 2018). Understanding the relationship between related microorganisms in natural ecosystems may help us better explore and learn about the assembly, diversity, and stability of plant-related communities (Haq et al., 2014; van der Putten, 2017). Recently, researchers have attempted to identify plant-related microbial communities from bacteria to fungi, as well as their associations, by means of microbiome annotation database or molecular-based experimental methods, broadening our basic knowledge of these types of microbial communities (Jackrel et al., 2017; Bamisile et al., 2018; Levy et al., 2018). Endophytic fungi are often considered to be beneficial to their host plants (Qian et al., 2019), because they may play various ecological roles such as promoting plant growth, enhancing the absorption of nutrient, resistance against various plant pathogens, as well as tolerance against various biotic and abiotic stresses (Waqas et al., 2012; Jia et al., 2016; Terhonen et al., 2016; Ripa et al., 2019). Current studies on tobacco wilt, a bacterial disease, mainly focus on the diversity and structural composition of the tobacco soil microbial community and its correlation with soil physicochemical properties, as well as the associations between the pathogen and bacterial species (Jiang et al., 2017; Li et al., 2017). However, the associations between the closely related bacterial and fungal communities in various zones of the plant-soil microecosystem under the invasion of R. solanacearum and the roles of endophytic fungi remains unclear.
The purposes of the current study are to: (i) Illuminate the characteristics of fungal communities in the bulk and rhizosphere soils and the root and stem endophytic compartments of healthy tobacco plants and those infected by R. solanacearum; (ii) Reveal the associations among species of fungal communities from various zones of the plant-soil microecosystem via molecular ecological network analysis; (iii) Explore the associations between fungi and bacteria through interdomain ecological network (IDEN) analysis; (iv) Study the associations between pathogenic Ralstonia members and fungi through sub-network analysis, and to explore fungal biocontrol resources that may antagonize the bacterial wilt pathogen. From the obtained results, we will provide a new strategy and theoretical support for enriching the study of tobacco endophytic fungal resources and for exploring the antagonistic fungal resources targeting R. solanacearum.
Materials and Methods
Sample Collection and Processing
The general collecting locations and methods of collecting and processing for soil and endophytic samples have been described previously in detail (Hu Q.L. et al., 2020), and will only be summarized here. Detailed location and other information were list in Supplementary Table 1. Eighty samples of bulk soil, rhizosphere soil, roots, and stems for tobacco cultivar Yunyan 87, including healthy and severely infected by R. solanacearum (grade 5–9 infection) (Chinese Standards, 2008), were collected from 5 different tobacco field sites located in the Chenzhou tobacco-growing region of Hunan province of China on June 2016 (mature stage of tobacco). The samples included 10 bulk soils samples of healthy tobacco (HBS), 10 bulk soil samples of wilt-infected tobacco (IBS), 10 rhizosphere samples of healthy tobacco (HRS), 10 rhizosphere samples of wilt-infected tobacco (IRS), 10 root samples of healthy tobacco (HR), 10 root samples of wilt-infected tobacco (IR), 10 stem samples of healthy tobacco (HS), 10 stem samples of wilt-infected tobacco (IS). Bulk soil samples were collected by shaking soil off tobacco roots. After shaking off bulk soils, the rhizosphere soils adhering to plant roots were collected in 50 mL tubes containing PBS (0.1% Tween 80) with a brush. After stirring for 5 min, the resulting suspension was then poured into a sterile centrifuge tube, this process was repeated a further two times. The suspensions were mixed and centrifuged for 5 min at 2,000 × g. The resulting sediment pellets were collected as the rhizosphere soils, which were stored at –80°C until DNA extraction. The roots and stems were processed immediately by washing consecutively with 75% ethanol, 2.5% sodium hypochlorite, and sterile water. They were then cut into small pieces and homogenized using a mortar and pestle with PBS, then transferred into a centrifuge tube for further treatment. Ultimately, the resulting sediment pellets from soil samples and resulting cell pellet from endophytic samples were stored in a freezer at –80°C until DNA extraction.
DNA Extraction and High-Throughput Sequencing
Total DNA was extracted in duplicate using the FastDNATM SPIN kit (MP Biomedicals) following the manufacturer’s instructions. Methods of PCR amplification and high-throughput sequencing for 16S rRNA genes followed those of Zhang et al. (2017). The V5-V6 region of 16S rRNA gene was amplied by the 799F (5′-AACMGGATTAGATACC CKG-3′)/1115R (5′-AGGGTTGCGCTCGTTG-3′) primers, and ITS2 fragment was amplified using the primer pair 5.8F (5′-AACTTTYRRCAAYGGAT CWCT-3′)/4R (5′-AGCCTCCGCTTATTGATATGCTTAART-3′) (Taylor et al., 2016). PCR amplification was performed in a 50 μL reaction system including 5 μL DNA template, 2.5 U of Taq DNA Polymerase (TaKaRa), 1 × Taq buffer, 75 μM dNTP and 0.3 μM of each primer. The thermal cycle operations for ITS2 fragment was performed as follows: 94°C for 1 min; following 35 cycles of 94°C for 20 s, 57°C for 25 s, and 68°C for 45 s, with a final elongation step at 68°C for 10 min, and finally stored at 4°C. The recovered products were qualified and quantified by a NanoDrop Spectrophotometer (Nano-100, Aosheng Instrument Co., Ltd.). Subsequently, the purified amplicons were pooled together and sequenced on Miseq sequencing machine (Illumina) at Central South University, Changsha, China.
Sequence Processing and Analysis
Preprocessing of raw reads of 16S rRNA genes and ITS2 fragment were submitted to an in-house pipeline1 integrated with various bioinformatics tools (Feng et al., 2017). All reads were assigned to individual sample according to their barcodes, allowing for a single mismatch. After trimming off the barcode and primer sequences, the pair-ended sequences for 16S rRNA genes were merged and their quality was checked by Flash program (Kong, 2011). The pair-ended sequences for ITS2 fragment with forward and reverse primers combinations were trimmed off. Subsequently, the sequences were passed through the ITSx program to remove the ITS flanking regions and non-fungal sequences (Bengtsson-Palme et al., 2013). Next, sequences were clustered into operational taxonomic units (OTUs) using UPARSE (SRP101823 for 16S rRNA gene sequences, and SRP123067 for ITS sequences) with a 97% sequence similarity threshold (Edgar, 2013). Ultimately, an OTU table was created and the total read counts were resampled before use in downstream analyses.
Statistical Analysis
The statistical significance of differences between two groups were tested by Wilcoxon test. Two measurements of alpha-diversity, Richness and Chao1, were calculated to assess the diversity of fungal communities. Richness was obtained by counting the number of species displayed in the OTU table. The Chao1 value was calculated using Mothur software (Chao, 1984; Schloss et al., 2009). Principal coordinate analysis (PCoA) was used to analyze the β-diversity of fungal communities in the bulk soil, rhizosphere soil, and root and stem endophytic compartments for both healthy and infected plants. Dissimilarity tests for soil and endophytic microbial community structures between healthy and infected samples were performed by using PERMANOVA based on Jaccard distance. Differences in soil and plant endophytic community compositions from healthy and infected samples were determined using an analysis of variance (ANOVA).
Random Matrix Theory Based Molecular Ecology Networks
To reveal the associations among fungal species in soil and endophytic fungal communities, from both healthy and infected samples, we constructed phylogenetic molecular ecological networks (MEN) via a Random Matrix Theory (RMT)-based approach (Deng et al., 2012) in molecular ecological network analysis pipeline (MENA)2 (Deng et al., 2016).
Interdomain Ecological Network Construction
The topology of ecological networks can represent the assembly process of microbial communities (Layeghifard et al., 2017), and the connections between interacting species can be used to predict ecosystem stability (Thebault and Fontaine, 2010). Recently, Feng et al. (2019) set up a workflow to construct IDEN, to find the association between two taxonomic groups (i.e., aboveground plants and underground bacteria) in ecological surveys. This method provided technical support for our analysis of the interdomain microbial associations between fungal and bacterial communities of soil and endophytes.
To elucidate associations between fungi and bacteria in soil and endophytic communities, by integrating the bacterial dataset (NCBI SRA database, accession PRJNA540089), interdomain ecological networks via SparCC approaches based on the inter-domain ecological network analysis pipeline3 workflow (Feng et al., 2019) were constructed. The threshold value for generating regional IDEN was 0.30, with 0.05 significance, to filter the non-correlated associations. The obtained adjacent matrix associated with the bipartite graph consisted of 1 or 0, showing presence/absence of corresponding fungi-bacteria association. The topological properties (connectance, links per species, specialization asymmetry, and web asymmetry) were calculated to explore alterations in associations between fungi and bacteria in the soil and endophytic communities under the invasion of R. solanacearum. The SparCC method with default parameters (Feng et al., 2019) was used for correlation analysis of specific associations between the pathogen and fugal members. The constructed networks were visualized using Gephi 0.9.2 software (Bastian et al., 2009). The keystone microorganisms were identified by the Zi-Pi plot based on the nodes’ roles within their own network (Deng et al., 2012).
Results
Effects of Wilt Pathogen Invasion on the Fungal Community Diversity and Structure of Soil and Tobacco Endophyte
Using high-throughput sequencing, a total of 8, 317 operational taxonomic units (OTUs) were obtained from 80 samples, 7, 216 OTUs among the soil samples and 2, 786 OTUs among the endophytic samples. Both observed Chao1 (Figure 1A) and estimated richness (Figure 1B) demonstrated no significant difference in fungal community diversity between healthy and infected tobacco plants regarding the bulk soil, rhizosphere soil, and root endophytes. However, the fungal richness of infected stems was significantly higher than that of healthy counterparts (Wilcoxon test, P < 0.05). As shown in Figure 1C, the endophytic fungal communities for healthy and infected plants were clearly separated in the PCoA plot, while the bulk, and rhizosphere soil communities of healthy and infected plants partially overlapped, indicating significant differences in fungal community structure between the healthy and infected endophytic compartments (PERMANOVA, P < 0.05).
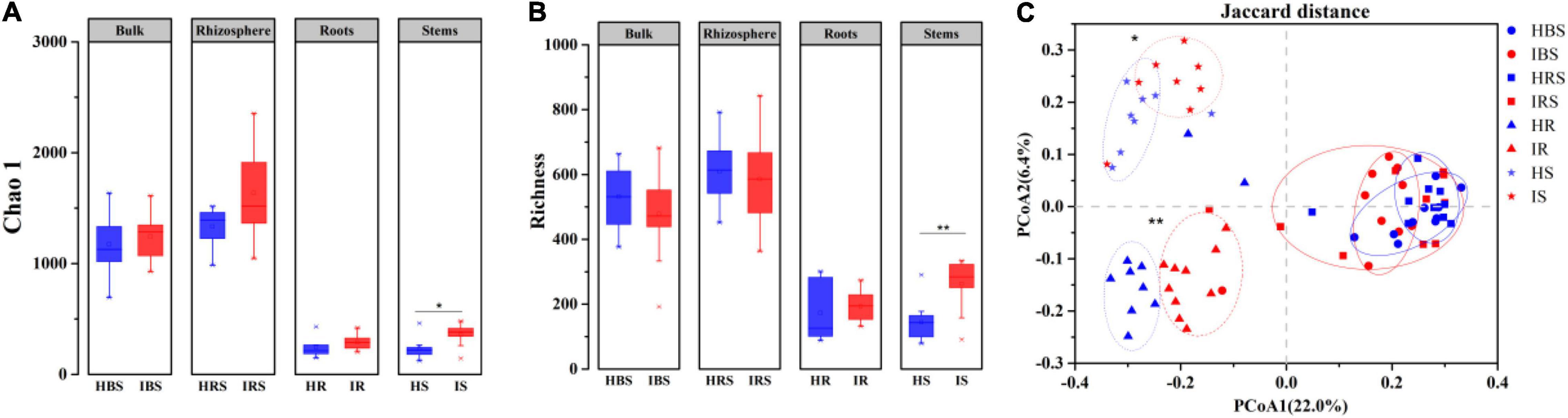
Figure 1. Diversity and structure of fungal communities in the healthy and infected bulk soil, rhizosphere soil, and root and stem endophytic compartments based on internal transcribed spacer (ITS) sequencing. (A) Richness. (B) Chao 1 index. (C) PCoA plot of fungal communities in healthy and infected plants. Note: blue and red indicate healthy and infected samples, respectively. HBS: bulk soils samples of healthy tobacco, IBS: bulk soil samples of wilt-infected tobacco, HRS: rhizosphere samples of healthy tobacco, IRS: rhizosphere samples of wilt-infected tobacco, HR: root samples of healthy tobacco, IR: root samples of wilt-infected tobacco, HS: stem samples of healthy tobacco, IS: stem samples of wilt-infected tobacco. Wilcoxon test and PERMANOVA were used to examine the statistical significance of differences for alpha and beta diversity, respectively. Difference was significant at *, 0.05; **, 0.01.
Effects of Wilt Pathogen Invasion on the Fungal Community Composition of Soil and Tobacco Endophyte
The composition of tobacco soils and endophytic fungal communities at the phylum level is shown in Figure 2A (with relative abundance > 1%). In the soil communities, the most abundant fungal phyla were Ascomycota, Basidiomycota, and Zygomycota, followed by Chytridiomycota and Glomeromycota. In the tobacco endophytic fungal communities, the most abundant fungal phyla were Ascomycota and Basidiomycota, followed by Zygomycota, Chytridiomycota, and Glomeromycota. The composition of tobacco soil and endophytic fungal communities at the genus level is shown in Figure 2B (with relative abundance > 1%). In the soil communities, the fungal genera with higher relative abundance included Mortierella, Aleuria, Cyberlindnera, Cryptococcus, Plectosphaerella, Gibberella, Mucor, Debaryomyces, Podospora, Entoloma, Paraphoma, Rhodotorula, Fusarium, Conocybe, and Guehomyces. In the tobacco endophytic fungal communities, the fungal genera with higher relative abundance included Plectosphaerella, Paraphoma, Gibberella, Rhodotorula, Alternaria, Ceratobasidium, Nectria, Davidiella, Haematonectria, Bionectria, and Thanatephorus. The genera whose abundance increased significantly in the infected soil samples as compared to healthy soil samples were Gibberella, Cryptococcus, Mucor, Nectria, Debaryomyces, and Haematonectria, and the genera that displayed significantly deceased abundance were Rhodotorula, Ceratobasidium, Cyberlindnera, Podospora, Conocybe, Monoblepharis, Paraconiothyrium, and Phoma. The genera whose abundance increased significantly in the endophytic communities of infected samples compared to those of healthy samples were Haematonectria, Gibberella, Ceratobasidium, Nectria, Bionectria, and Didymella, and the genera with significantly deceased abundance were Cryptococcus, Didymella, Mortierella, Paraphoma, Davidiealla, Phoma, and Mucor. In summary, the fungal community compositions and relative abundance were different between the healthy and infected soil communities, between the healthy and infected endophytic communities, and between the soil and endophytic communities (Supplementary Table 2, P < 0.05).
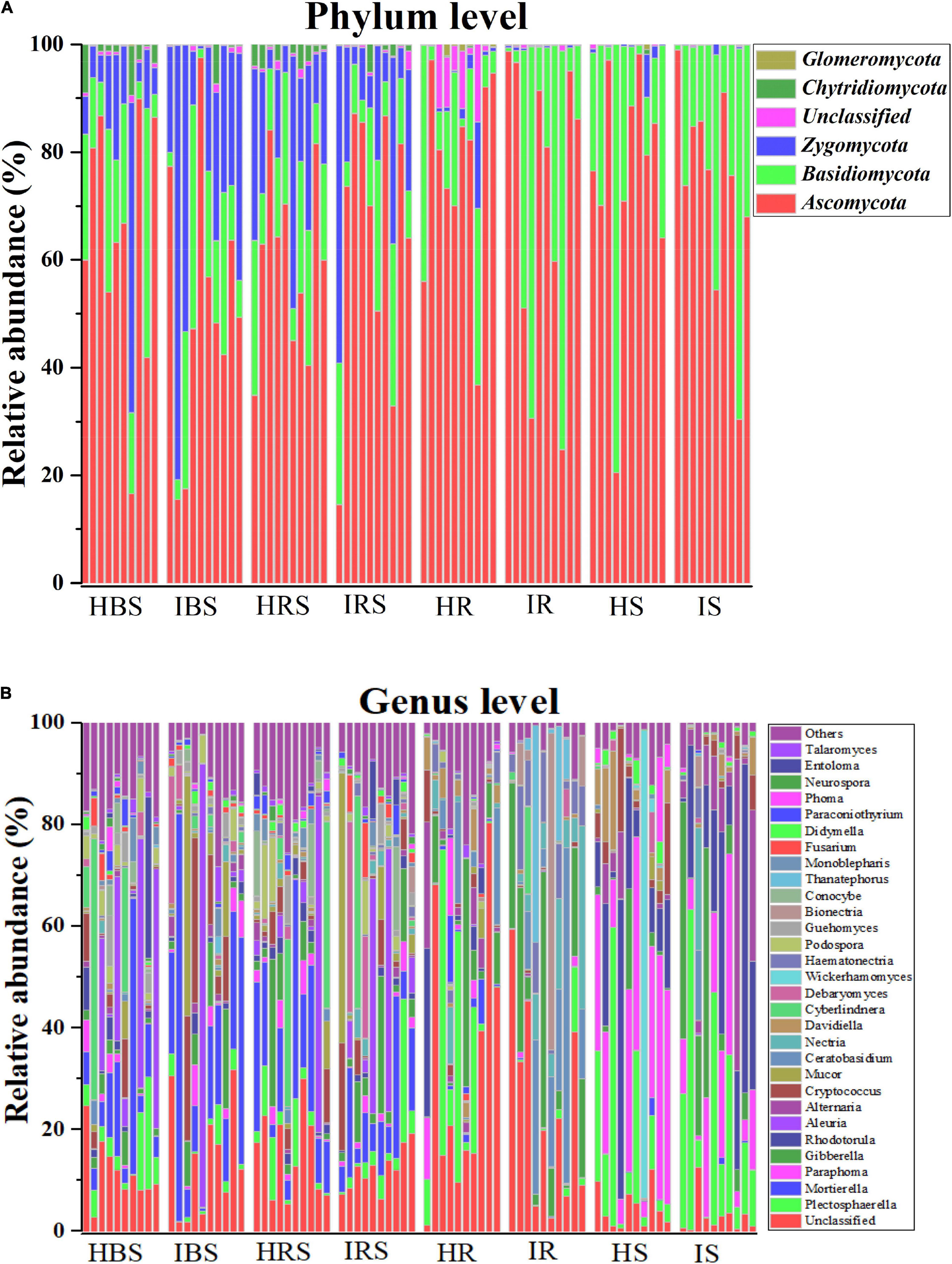
Figure 2. Comparison of the community compositions of soil and plant endophytic fungi from healthy and infected samples. (A) Comparison of the community compositions of the healthy and infected bulk soil, rhizosphere soil, and root and stem endophytic compartments with relative abundance > 1% OTUs at the phylum level. (B) Comparison of the community compositions of the healthy and infected bulk soil, rhizosphere soil, and root and stem endophytic compartments with relative abundance > 1% OTUs at the genus level.
The Species Associations Among Fungal Communities
In the network structures of rhizosphere soil, root endophytes, and stem endophytes, the fungal network structures of the infected samples showed a higher complexity and more links than those of healthy samples (Table 1). For example, the number of network nodes and links of the infected rhizosphere soil were 187 and 518, respectively, and those of the healthy rhizosphere soil were 179 and 367, respectively. In addition, the average clustering coefficients (avgCCs) of the empirical networks of all tested samples (0.02–0.67) were higher than those of the corresponding random networks (0.01–0.228), suggesting that the eight constructed networks all had typical small-world network characteristics (Watts and Strogatz, 1998). The modularity (M) values of all empirical networks (0.534–0.91) were significantly higher than the M-values of the corresponding random networks (0.339–0.860), indicating modular topological features of constructed networks (Newman, 2016). The visualized networks were constructed to intuitively display the associations among microorganisms in the fungal communities of healthy and infected samples, as shown in Figure 3. In general, there were obvious differences in the network topological structure between infected and healthy samples from all four of the investigated microecosystem zones.
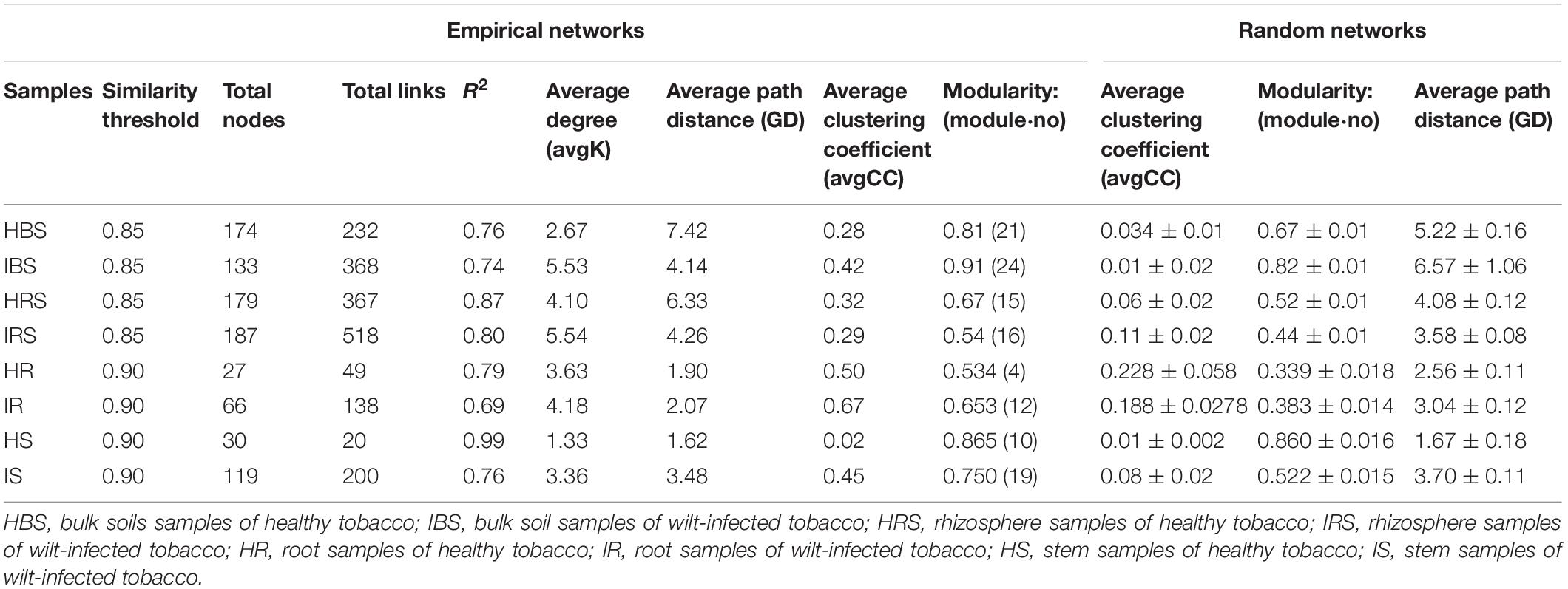
Table 1. Topological features of the soil and endophytic fungal community networks in healthy and infected samples.
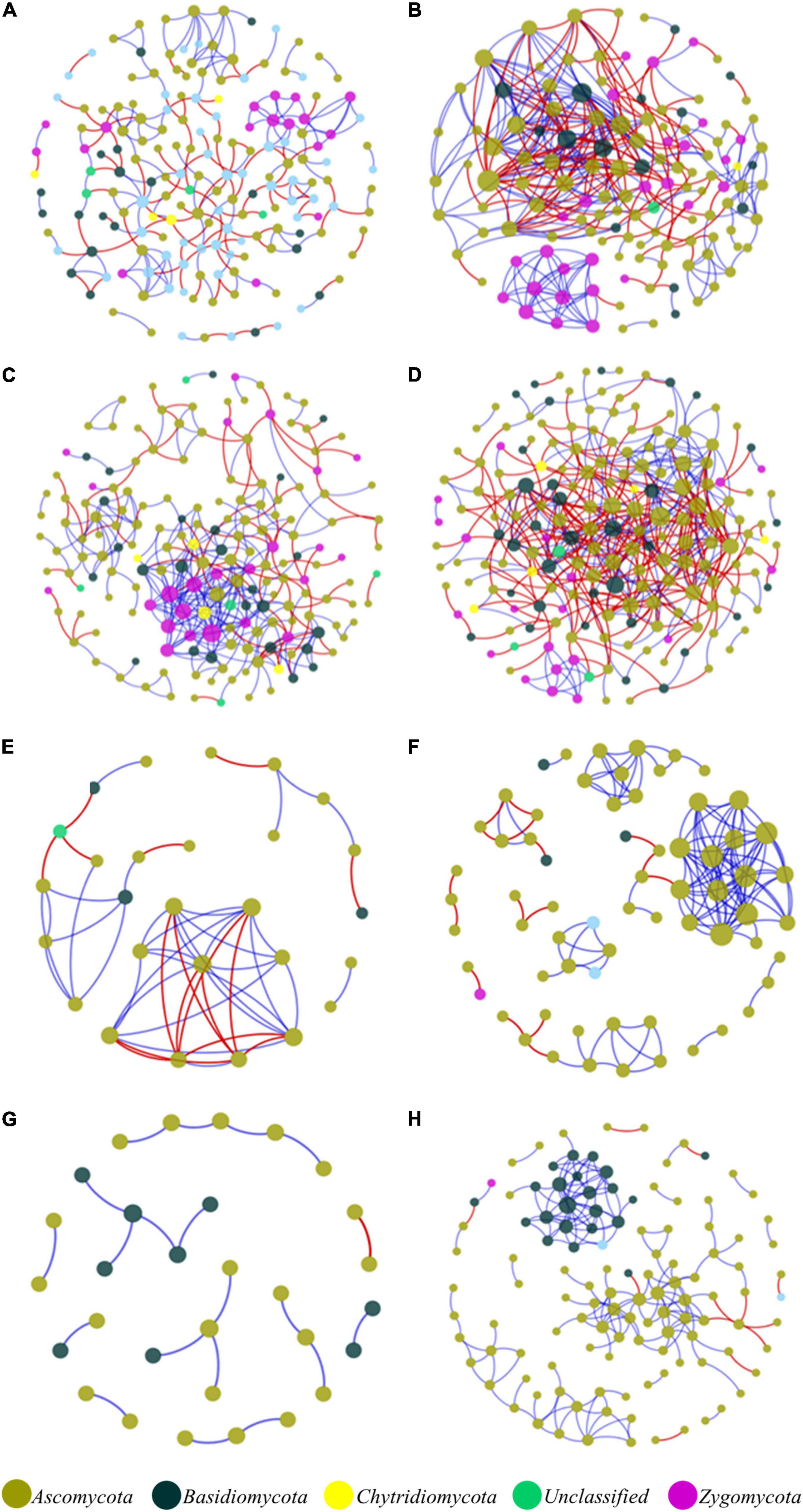
Figure 3. Molecular ecological networks of microorganism associations in the fungal communities of healthy and infected samples. (A) Healthy bulk soil. (B) Infected bulk soil. (C) Healthy rhizosphere soil. (D) Infected rhizosphere soil. (E) Healthy root endophytic compartment. (F) Infected root endophytic compartment. (G) Healthy stem endophytic compartment. (H) Infected stem endophytic compartment. Node color represents phylum-level taxonomy. Blue links indicate positive correlations between nodes, and red links indicate negative correlations.
IDEN Between Fungal and Bacterial Communities
The eight constructed networks of fungi-bacteria associations in healthy and infected samples from the surrounding bulk soil, rhizosphere soil, and root and stem endophytic compartments showed some basic bipartite topological structures, e.g., the modular structure and high web asymmetry of two species groups (Table 2), and all showed significant topological differences (Figure 4). According to the topological indices of the networks (Table 2), the total numbers of network nodes and links of the infected samples were all higher than the corresponding healthy samples, indicating more complex and compact bacterial-fungal associations in the infected samples. The higher value of web asymmetry for networks of infected root and stems endophytes showed that more fungi were involved in the interdomain associations. Instead of higher positive and negative associations for soil samples, the IDENs contained large proportions of negative associations in infected root and stem endophytic compartments compared to healthy samples, with more fungal nodes involved (1,298 vs. 266 for roots, and 493 vs. 131 for stems), suggesting resistance relationships between bacteria and fungi. Module detection analysis further demonstrated smaller modularity and fewer modules for IDENs of the endophytic compartments, implying closer fungi-bacteria associations. In addition, we found the greatest number of network links and the highest number of links per species in the IDEN of infected root endophytic compartment, indicating larger proportions of bacterial-fungal associations among the infected root endophytes.
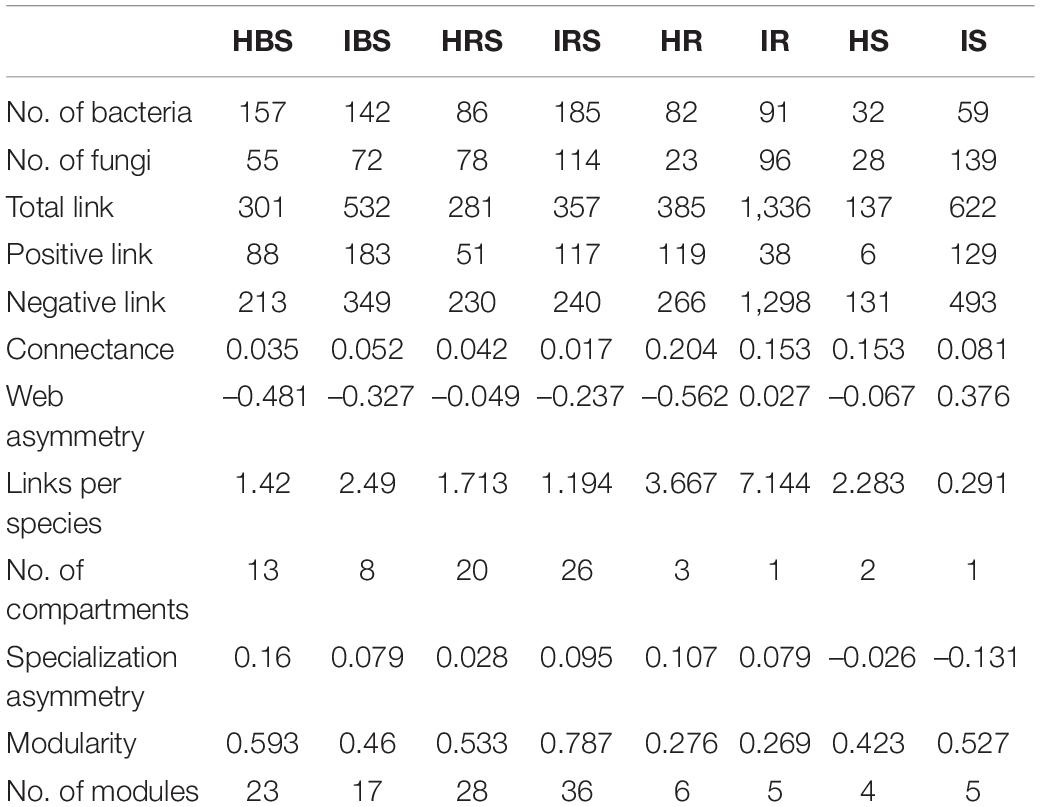
Table 2. Interdomain network topology features of healthy and infected soil and endophytic microbial communities.
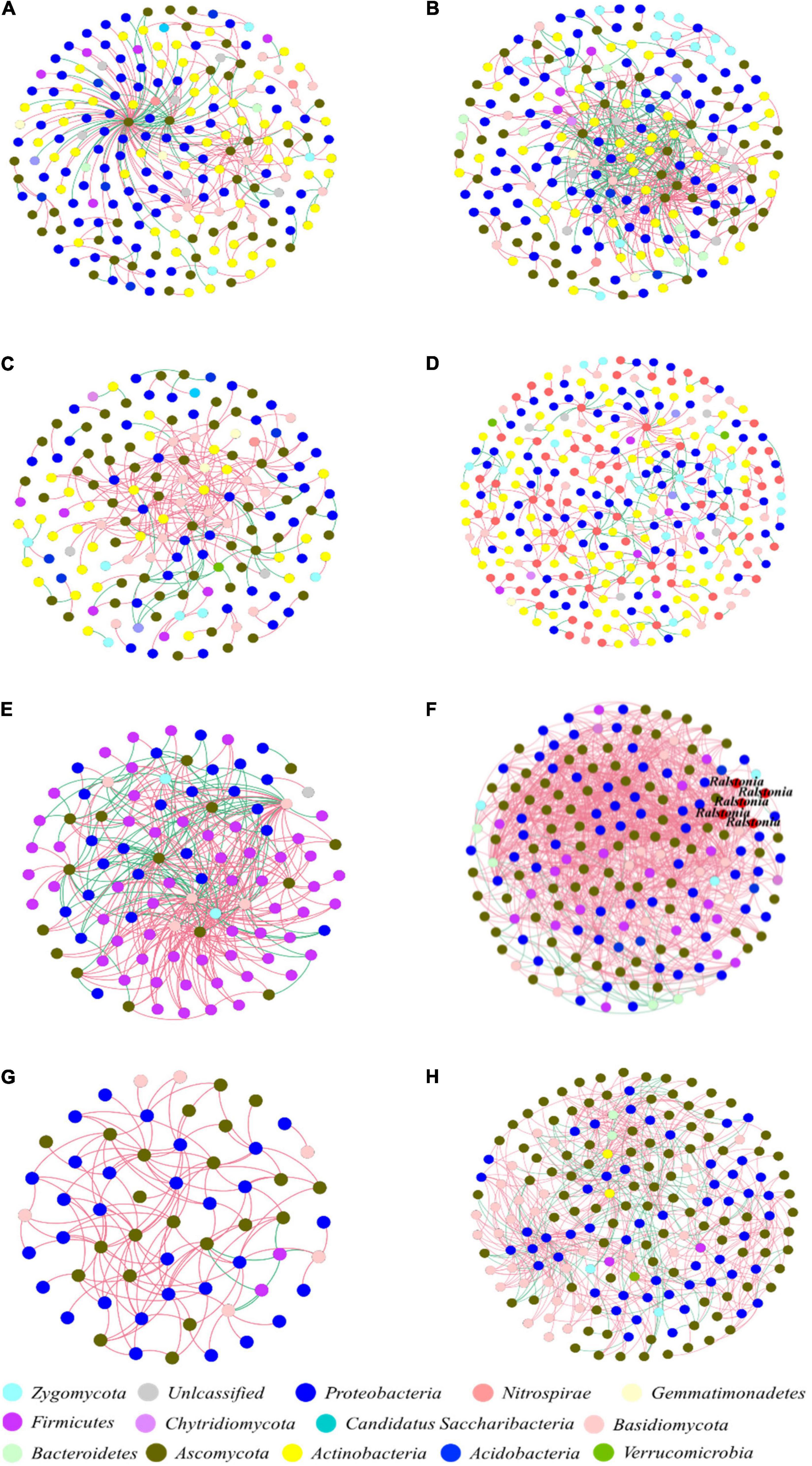
Figure 4. Interdomain ecological networks of the bacterial-fungal associations of healthy and infected samples. (A) Healthy bulk soil. (B) Infected bulk soil. (C) Healthy rhizosphere soil. (D) Infected rhizosphere soil. (E) Healthy root endophytic compartment. (F) Infected root endophytic compartment. (G) Healthy stem endophytic compartment. (H) Infected stem endophytic compartment. Node color indicates phylum-level taxonomy, and the pathogen Ralstonia was labeled at the genus level. Blue links indicate positive correlations between nodes, and red links indicate negative correlations.
Mining of Keystone Fungal Genus With Biocontrol Potential Against Bacterial Wilt Pathogen
The Zi-Pi plots were drawn to exhibit the distribution of network nodes OTUs based on the modular topology (Figure 5). The topology of each node (genus) could be measured by its within-module connectivity (Zi = 2.5) and among-module connectivity (Pi = 0.62). According to the simplified categorization, all nodes in the networks were distributed among four subcategories: Peripherals, Connectors, Module hubs and Network hubs. Nodes that belonged to the latter three subcategories were considered to be keystone microbial microorganisms playing a critical role in the network (Zhou et al., 2011; Jiang et al., 2015) and were marked in the corresponding networks. Interestingly, the bacterial wilt pathogenic Ralstonia members were found to be the keystone genus in the IDENs of the infected root and stem endophytic compartments.
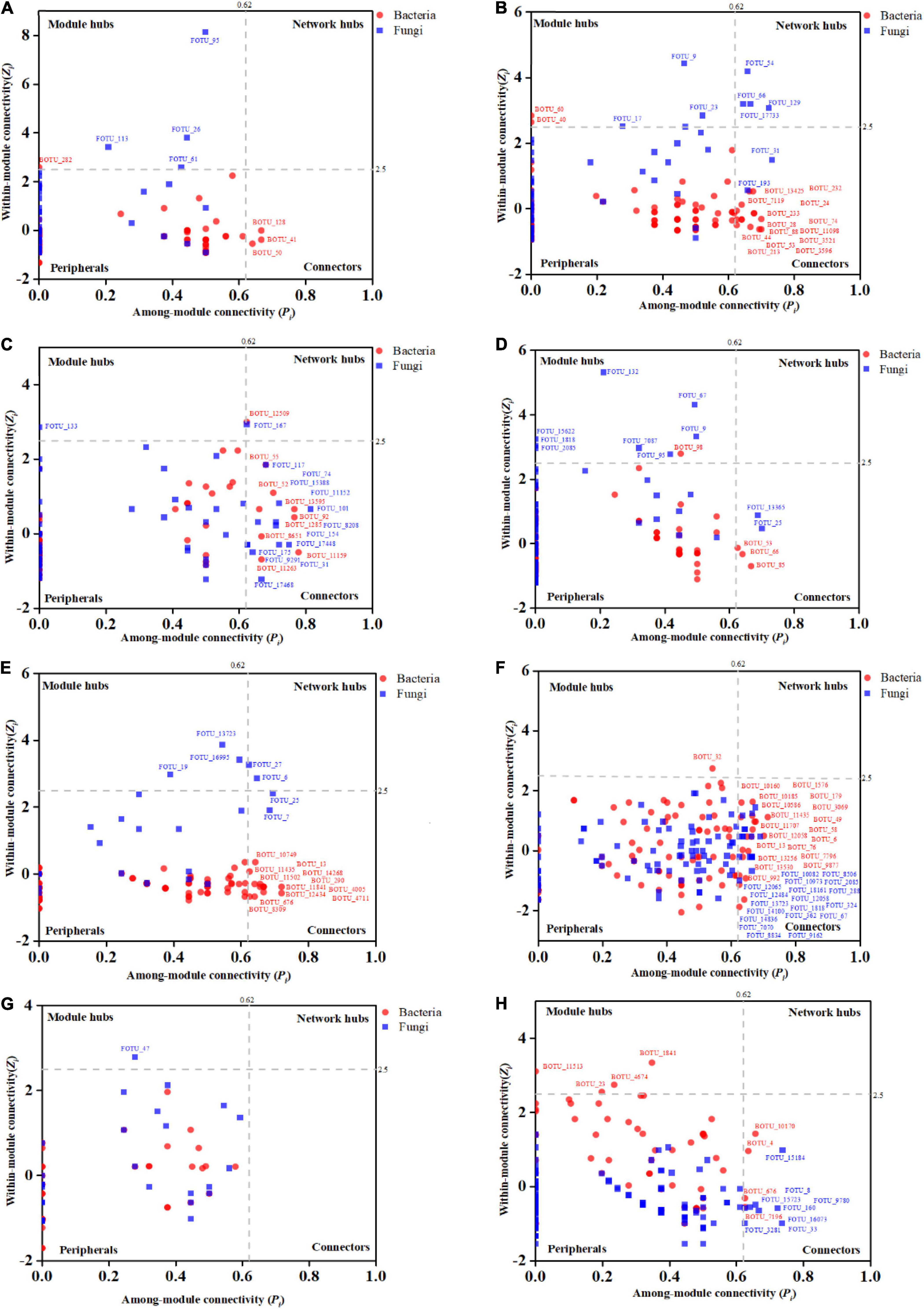
Figure 5. The Zi-Pi plots exhibit the distributions of OTUs based on their topology. The symbols represent OTUs in the bacterial (red dots) and fungal (blue squares) networks. The threshold values of Zi and Pi for categorizing OTUs are 2.5 and 0.62, respectively. (A) Healthy bulk soil. (B) Infected bulk soil. (C) Healthy rhizosphere soil. (D) Infected rhizosphere soil. (E) Healthy root endophytic compartment. (F) Infected root endophytic compartment. (G) Healthy stem endophytic compartment; (H) Infected stem endophytic compartment.
To find the fungal genus closely associated with Ralstonia members, we further analyzed the associations between Ralstonia members and fungal genus in IDENs for the infected root and stem endophytic compartments. The results showed that the fungal genera Phoma, Gibberella, Alternaria, Haematonectria, Cryptococcus, Podospora, Spodiobolus, Malassezia, Aleuria, Dioszgia, Davidiealla, and unclassified genera were negatively correlated with the pathogenic Ralstonia members in the root endophytic communities (Figure 6), and the fungal genera Phoma, Gibberella, Diaporthe, Didymella, and unclassified genera were negatively correlated with Ralstonia members in the stem endophytic communities (Figure 6). No positive correlations between fungal genus and the pathogen were found in the root and stem endophytic communities.
Figure 6. Entangled webs of fungi-bacteria and fungi-pathogen associations in the microbial communities of the surrounding soil, rhizosphere, and root and stem endophytic compartments. The healthy plant (left) successfully suppressed invasion of pathogenic Ralstonia. The infected (right) plant was successfully invaded by the pathogen. The Interdomain ecological networks of the associations between pathogenic bacteria (red) and other fungal species in the root and stem endophytic compartments of the infected samples. Each node is marked at the genus level. Red links indicate negative correlations.
Discussion
The occurrence of plant diseases is closely related to the microbial diversity in the soil and endophytic compartments (Shi et al., 2019; Ulloa-Munoz et al., 2020). Our previous studies have shown that the diversity of endophytic bacterial communities in the roots and stems of plants with bacterial wilt infected was significantly higher than that of healthy samples (Hu Q.L. et al., 2020). In this study, there was no significant difference in the diversity of the root endophytic fungal community between infected and healthy samples. Endophytic fungal community diversity of the infected stem samples was significantly higher than that of the healthy samples. This may be explained by that the bacterial wilt pathogen invasion success within root endophyte could result in the destruction of plant’s defense system. The bacterial wilt pathogen and fungal members interacted closely in the roots, inducing more fungal communities to migrate into the stem, and therefore resulted in an increased diversity of the stem endophytic fungal community compared to the healthy samples (Tan and Zou, 2001; Thebault and Fontaine, 2010; Kefi et al., 2012). Moreover, high diversity and close associations among various microorganism are beneficial to the stability of microbial communities, thereby boosting the microbial community’s resistance to pathogen invasion (McCann, 2000; Wehner et al., 2010; van Elsas et al., 2012; Mallon et al., 2015). The increased diversity of fungal communities in stem endophytes may be a middle-late-stage immune response of the plant to the bacterial wilt pathogen invasion.
The invasion of bacterial wilt pathogen may cause changes in the fungal community composition of the various zones of the tobacco microecosystem. From the perspective of relative abundance, fungal composition displayed significant changes at the genus level between infected and healthy samples in the bulk soil, rhizosphere soil, and root and stem endophytic compartments. In the infected bulk soil and rhizosphere soil, the relative abundances of Rhodotorula, Ceratobasidium, Cyberlindnera, Podospora, Conocybe, Monoblepharis, Paraconiothyrium, and Phoma were significantly decreased, whereas the relative abundances of Gibberella, Cryptococcus, Mucor, Nectria, Debaryomyces, and Haematonectria were significantly increased, compared to the corresponding healthy samples. Such changes in composition might be because the invasion of bacterial wilt disease made pathogenic Ralstonia members the dominant species in soil and thus altered the composition of the soil fungal community. In the infected endophytic samples, the genera that significantly declined were Cryptococcus, Didymella, Mortierella, Paraphoma, Davidiealla, Phoma, and Mucor. Many of the secondary metabolites produced by these endophytic fungi have been reported to have inhibiting or antibacterial abilities (Melo et al., 2014; Xia et al., 2015; Li et al., 2018). These results indicated that the beneficial endophytes were either actively repelled by the host immune system or defeated by the more dominant migrating microbial community (Lundberg et al., 2012; Velásquez et al., 2017). The relative abundances of Haematonectria, Gibberella, Ceratobasidium, Nectria, Bionectria, and Didymella were significantly enhanced in the infected endophytic samples, indicating that they may benefit during the pathogen invasion process. It is possible they are opportunists that took advantage of the potential niche opened by pathogen invasion and entered the plant endophytic compartment (Lundberg et al., 2012). The compositional changes of these fungal communities may be caused by changes in root exudates or complex changes in the plant immune system during pathogen invasion (Martinoia and Baetz, 2014; dos Santos et al., 2020), and this promoted the differential recruitment and/or differential rejection of microorganisms to resist the invasion of bacterial wilt pathogen in plant roots and stems (Kwak et al., 2018).
Microbe-microbe associations are essential for the function of microecosystems in soil and endophytic compartments (Barberán et al., 2012). Molecular ecological network (MEN) analysis has been increasingly employed to explore potential microbial associations in various ecosystems (de Menezes et al., 2015). However, there are few reports on microbial associations in the fungal community of plants under invasion by bacterial wilt pathogen. In this study, we applied network analysis to quantify and visualize the associations among microorganisms of the fungal community under the invasion of R. solanacearum. The results showed that the fungal networks of infected samples had higher complexity and more links than the healthy samples in the rhizosphere soil and root and stem endophytic compartments. Furthermore, the corresponding topological structures demonstrated significant differences as well. Together this indicated that the invasion of bacterial wilt pathogen changed the composition of the soil fungal community and further strengthened the associations among species in the fungal community. The highly connected and modularized fungal community associations were conducive to regulating the stability of the community (Eisenhauer et al., 2013; Downing et al., 2014; Tardy et al., 2014), thereby controlling the propagation and colonization of the pathogen. Hence, it is necessary to study the associations of microorganisms in soil and endophytic fungal communities for more effective prevention and control of diseases.
Soil is one of the main habitats for bacteria and fungi (Effmert et al., 2012). Endophytic microorganisms, including fungi and bacteria, live in the intercellular or intracellular spaces of plant tissues. The associations between fungi and bacteria are part of the communication network maintaining the balance of this microhabitat (Bamisile et al., 2018). We adopted IDENs to analyze the association network between fungi and bacteria in each zone of the plant-soil microecosystem, and found more complex and tighter fungal-bacterial associations in the infected samples than the corresponding healthy samples for all tested zones. In addition, the IDEN of the infected root endophytic compartment presented the most network links and the highest number of links per microorganism, suggesting a closer bacterial-fungal associations in this network. Interestingly, the Zi-Pi plots demonstrated that the pathogenic Ralstonia members were the keystone genus in the root and stem endophytic bacterial-fungal association networks. The reason for these results may be that with the invasion of the bacterial wilt pathogen, more soil fungi and bacteria developed a mutually beneficial relationship and entered the plant root endophytic community together, resulting in more complex associations among microorganisms (Hu Q.L. et al., 2020). It may also be that the competition of nutrient resources or niche space caused more diversified associations between fungi and bacterial microorganisms (Ghoul and Mitri, 2016). A third possibility is that because the microecological balance was broken by the pathogen invasion, leading to more intense antagonistic relationships between fungal, other bacterial members, and the pathogen (García-Bayona and Comstock, 2018; Hu J. et al., 2020). These phenomena were more prominent in infected root and stem endophytic compartments.
To further clarify which fungi interacted closely with the pathogenic Ralstonia members in the endophytic roots and stems, we built sub-networks centered on the pathogen, Ralstonia members, and included its associations with fungi. The results showed that the root endophytic fungal genera Phoma, Gibberella, Alternaria, Haematonectria, Cryptococcus, Podospora, Spodiobolus, Malassezia, Aleuria, Dioszgia, and Davidiealla, and the stem endophytic fungal genera Phoma, Gibberella, Diaporthe, and Didymella were all negatively correlated with Ralstonia members. Plant-associated endophytic fungi are rich sources of novel bioactive and structurally diverse secondary metabolites and other natural products, which were generally considered to protect their host plants by blocking or inhibiting the appropriate pathogenic microorganisms (Rustamova et al., 2020). According to previous research, the active compound named as barceloneic acid C isolated and purified from the secondary metabolites of the endophytic fungus Phoma sp. JS752, isolated from Phragmites communis Trinius, demonstrated an antibacterial activity against pathogenic gram-positive bacteria Listeria monocytogenes and Staphylococcus pseuditermedius, and gram-negative bacteria such as Escherichia coli and Salmonella typhimurium (Xia et al., 2015). The purified secondary metabolites [(3S)-3,6,7-trihydroxy-α-tetralone, Cercosporamide, β-Sitosterol and trichodermin] of Phoma sp. ZJWCF006, which was screened and isolated from the Arisaema erubescens endophytes, showed remarkable antibacterial activity against four plant fungal pathogens (Fusarium oxysporium, Rhizoctonia solani, Colletotrichum gloeosporioides, and Magnaporthe oryzae) and two plant bacterial pathogens (Xanthomonas campestris and Xanthomonas oryzae) (Wang et al., 2012). The secondary metabolity compound ergosterol peroxide from the endophytic fungus Gibberella moniliformis JS1055, isolated from a halophyte Vitex rotundifolia (Kim et al., 2018), exhibited moderate inhibitory activity against bacteria Staphylococcus aureus and Escherichia coli (Zhu et al., 2017). The endophytic fungus Alternaria alternata AE1, isolated from Azadirachta indica A. Juss, could produce highly effective bioactive metabolites that showed a strong inhibitory effect on pathogenic bacteria Listeria monocytogenes and Escherichia coli (Chatterjee et al., 2019). The antibacterial activities of compounds phomosines A and C produced by endophytic fungus Diaporthe sp. F2934, isolated from the tropical plant Aegle marmelos, showed an antibacterial activity against a variety of gram-negative and gram-positive bacteria, and its inhibitory zone diameter (IZD) against Staphylococcus aureus was 20% larger than the standard antibiotic vancomycin (Sousa et al., 2016). It can be seen that these fungi revealed by our study and their secondary metabolites have been reported with antibacterial ability or activity against some bacteria, and they may have potential resistance to bacterial wilt pathogen invasion.
Conclusion
The bacteria wilt pathogen Ralstonia members and the infected root endophytic fungal genera Phoma, Gibberella, Alternaria, Haematonectria, Cryptococcus, Podospora, Spodiobolus, Malassezia, Aleuria, Dioszgia, and Davidiealla, as well as the infected stem endophytic fungal genera Phoma, Gibberella, Didymella, and Diaporthe, were negatively correlated, and these fungi may be potential biocontrol resources in dealing with tobacco bacterial wilt disease. At present, there are few reports on the exploration and application of tobacco endophytic fungal resources. This study will provide potential ideas and theoretical support for enriching the study of tobacco endophytic fungal resources and controlling tobacco bacterial wilt disease. Further experiment with species isolation and verification is needed to confirm these findings.
Data Availability Statement
16S rRNA and ITS gene sequencing data of all samples were submitted to the NCBI SRA database (https://www.ncbi.nlm.nih.gov/) under accession numbers PRJNA540089 and PRJNA735450, respectively.
Author Contributions
LT, WZ, YX, and SG performed the main experiments and analyzed data. LT, YD, and QH planned and designed the research, wrote the manuscript with substantial input from PL, SW, ZZ, and KF. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Foundation for Tobacco Science of Changsha Tobacco Company of Hunan Province (Nos 19-21A01 and 19-21A02) and the Wenshan Tobacco Company of Yunnan Province of China (No. 2021530000241033).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank James Walter Voordeckers for carefully editing the grammar of the manuscript and for some valuable suggestions for this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.722626/full#supplementary-material
Footnotes
References
Arnold, A. E. (2007). Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol. Rev. 21, 51–66. doi: 10.1016/j.fbr.2007.05.003
Bamisile, B. S., Dash, C. K., Akutse, K. S., Keppanan, R., and Wang, L. (2018). Fungal endophytes: beyond herbivore management. Front. Microbiol. 9:544. doi: 10.3389/fmicb.2018.00544
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Proceedings of the International AAAI Conference on Web and Social Media, Palo Alto, CA.
Bastias, D. A., Martínez- Ghersa, M. A., Ballaré, C. L., and Gundel, P. E. (2017). Epichloë fungal endophytes and plant defenses: not just alkaloids. Trends Plant Sci. 22, 939–948. doi: 10.1016/j.fbr.2020.06.001
Bengtsson-Palme, J., Ryberg, M., Hartmann, M., Branco, S., Wang, Z., Godhe, A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for use in environmental sequencing. Methods Ecol. Evol. 4, 914–919. doi: 10.1111/2041-210X.12073
Bulgarelli, D., Schlaeppi, K., Spaepen, S., van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Chatterjee, S., Ghosh, R., and Mandal, N. C. (2019). Production of bioactive compounds with bactericidal and antioxidant potential by endophytic fungus Alternaria alternata AE1 isolated from Azadirachta indica A. Juss. PLoS One 14:e0214744. doi: 10.1371/journal.pone.0214744
Chinese Standards (2008). Grade and Investigation Method of Tobacco Diseases and Insect Pests, GB/T 23222–22008. Washington, DC: Chinese Standards.
de Menezes, A. B., Prendergast-Miller, M. T., Richardson, A. E., Toscas, P., Farrell, M., Macdonald, L. M., et al. (2015). Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ. Microbiol. 17, 2677–2689. doi: 10.1111/1462-2920.12559
Deng, Y., Jiang, Y. H., Yang, Y., He, Z. L., Luo, F., and Zhou, J. Z. (2012). Molecular ecological network analyses. BMC Bioinform. 13:e113. doi: 10.1186/1471-2105-13-113
Deng, Y., Zhang, P., Qin, Y. J., Tu, Q. C., Yang, Y. F., He, Z. L., et al. (2016). Network succession reveals the importance of competition in response to emulsified vegetable oil amendment for uranium bioremediation. Environ. Microbiol. 18, 205–218. doi: 10.1111/1462-2920.12981
dos Santos, T. C., Leandro, M. R., Maia, C. Y., Rangel, P., Soares, F. S., Reis, R., et al. (2020). Arabidopsis thaliana exudates induce growth and proteomic changes in Gluconacetobacter diazotrophicus. Peerj 8:e9600. doi: 10.7717/peerj.9600
Downing, A. L., Brown, B. L., and Leibold, M. A. (2014). Multiple diversity-stability mechanisms enhance population and community stability in aquatic food webs. Ecology 95, 173–184. doi: 10.1890/12-1406.1
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/NMETH.2604
Effmert, U., Kalderas, J., Warnke, R., and Piechulla, B. (2012). Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38, 665–703. doi: 10.1007/s10886-012-0135-5
Eisenhauer, N., Schulz, W., Scheu, S., and Jousset, A. (2013). Niche dimensionality links biodiversity and invasibility of microbial communities. Funct. Ecol. 27, 282–288. doi: 10.1111/j.1365-2435.2012.02060.x
Feng, K., Zhang, Y. G., He, Z. L., Ning, D. L., and Deng, Y. (2019). Interdomain ecological networks between plants and microbes. Mol. Ecol. Resour. 19, 1565–1577. doi: 10.1111/1755-0998.13081
Feng, K., Zhang, Z. J., Cai, W. W., Liu, W. Z., Xu, M. Y., Yin, H. Q., et al. (2017). Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol. Ecol. 26, 6170–6182. doi: 10.1111/mec.14356
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
García-Bayona, L., and Comstock, L. E. (2018). Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456
Ghoul, M., and Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
Haq, I. U., Zhang, M. Z., Yang, P., and van Elsas, J. D. (2014). The interactions of bacteria with fungi in soil: emerging concepts. Adv. Appl. Microbiol. 89, 185–215. doi: 10.1016/B978-0-12-800259-9.00005-6
Hassani, M. A., Duran, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:e58. doi: 10.1186/s40168-018-0445-0
Hu, J., Wei, Z., Kowalchuk, G. A., Xu, Y. C., Shen, Q. R., and Jousset, A. (2020). Rhizosphere microbiome functional diversity and pathogen invasion resistance build up during plant development. Environ. Microbiol. 22, 5005–5018. doi: 10.1111/1462-2920.15097
Hu, Q. L., Tan, L., Gu, S. S., Xiao, Y. S., Xiong, X. Y., Zeng, W. A., et al. (2020). Network analysis infers the wilt pathogen invasion associated with non-detrimental bacteria. NPJ Biofilms Microbiol. 6:e8. doi: 10.1038/s41522-020-0117-2
Jackrel, S. L., Owens, S. M., Gilbert, J. A., and Pfister, C. A. (2017). Identifying the plant-associated microbiome across aquatic and terrestrial environments: the effects of amplification method on taxa discovery. Mol. Ecol. Resour. 17, 931–942. doi: 10.1111/1755-0998.12645
Jia, M., Chen, L., Xin, H. L., Zheng, C. J., Rahman, K., and Qin, L. P. (2016). A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front. Microbiol. 7:e906. doi: 10.3389/fmicb.2016.00906
Jiang, G., Wei, Z., Xu, J., Chen, H. L., Zhang, Y., She, X. M., et al. (2017). Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 8:1549. doi: 10.3389/fpls.2017.01549
Jiang, Y. J., Sun, B., Li, H. X., Liu, M. Q., Chen, L. J., and Zhou, S. (2015). Aggregate-related changes in network patterns of nematodes and ammonia oxidizers in an acidic soil. Soil Biol. Biochem. 88, 101–109. doi: 10.1016/j.soilbio.2015.05.013
Kapitan, M., Niemiec, M. J., Steimle, A., Frick, J. S., and Jacobsen, I. D. (2019). Fungi as part of the microbiota and interactions with intestinal bacteria. Curr. Top. Microbiol. Immunol. 422, 265–301. doi: 10.1007/82_2018_117
Kefi, S., Berlow, E. L., Wieters, E. A., Navarrete, S. A., Petchey, O. L., Wood, S. A., et al. (2012). More than a meal. Integrating non-feeding interactions into food webs. Ecol. Lett. 15, 291–300. doi: 10.1111/j.1461-0248.2011.01732.x
Kim, J. W., Ryu, J., and Shim, S. H. (2018). Chemical investigation on an endophytic fungus Gibberella moniliformis JS1055 derived from a halophyte Vitex rotundifolia. Nat. Prod. Sci. 24, 189–193. doi: 10.20307/nps.2018.24.3.189
Kong, Y. (2011). Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98, 152–153. doi: 10.1016/j.ygeno.2011.05.009
Kumar, J., Singh, D., Ghosh, P., and Kumar, A. (2017). “Endophytic and epiphytic modes of microbial interactions and benefits,” in Plant-Microbe Interactions in Agro-Ecological Perspectives, eds D. Pratap Singh, H. Bahadur Singh, and R. Prabha (Berlin: Springer), 227–253.
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Layeghifard, M., Hwang, D. M., and Guttman, D. S. (2017). Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 25, 217–228. doi: 10.1016/j.tim.2016.11.008
Levy, A., Gonzalez, I. S., Mittelviefhaus, M., Clingenpeel, S., Paredes, S. H., Miao, J. M., et al. (2018). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Li, C. S., Hu, Z. Q., Liu, Q. S., Wu, X. H., and Cao, S. G. (2018). Two new tricycloalternarenes from Hawaiian endophytic fungus Didymella sp FT433. Tetrahedron. Lett. 9, 3381–3383. doi: 10.1016/j.tetlet.2018.07.061
Li, X., Liu, Y., Cai, L., Zhang, H., Shi, J., and Yuan, Y. (2017). Factors affecting the virulence of Ralstonia solanacearum and its colonization on tobacco roots. Plant Pathol. 66, 1345–1356. doi: 10.1111/ppa.12675
Lilleskov, E. A., Hobbie, E. A., and Horton, T. R. (2011). Conservation of ectomycorrhizal fungi: exploring the linkages between functional and taxonomic responses to anthropogenic N deposition. Fungal Ecol. 4, 174–183. doi: 10.1016/j.funeco.2010.09.008
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Mallon, C. A., Elsas, J. D. V., and Salles, J. F. (2015). Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23, 719–729. doi: 10.1016/j.tim.2015.07.013
Martin, F. M., Uroz, S., and Barker, D. G. (2017). Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356:eaad4501. doi: 10.1126/science.aad4501
Martinoia, E., and Baetz, U. (2014). Root exudates: the hidden part of plant defense. Trends Plant Sci. 19, 90–98. doi: 10.1016/j.tplants.2013.11.006
Melo, I. S., Santos, S. N., Rosa, L. H., Parma, M. M., Silva, L. J., Queiroz, S. C. N., et al. (2014). Isolation and biological activities of an endophytic Mortierella alpina strain from the Antarctic moss Schistidium antarctici. Extremophiles 18, 15–23. doi: 10.1007/s00792-013-0588-7
Mousa, W. K., Shearer, C. R., Limay-Rios, V., Ettinger, C. L., Eisen, J. A., and Raizada, M. N. (2016). Root-hair endophyte stacking in finger millet creates a physicochemical barrier to trap the fungal pathogen Fusarium graminearum. Nat. Microbiol. 1:e16167. doi: 10.1038/NMICROBIOL.2016.167
Newman, M. E. J. (2016). Modularity and community structure in networks. Proc. Natl. Acad. Sci. U.S.A. 103, 8577–8582. doi: 10.1073/pnas.0601602103
Nilsson, R. H., Anslan, S., Bahram, M., Wurzbacher, C., Baldrian, P., and Tedersoo, L. (2019). Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17, 95–109. doi: 10.1038/s41579-018-0116-y
Qian, X., Li, H. Z., Wang, Y. L., Wu, B. W., Wu, M. S., Chen, L., et al. (2019). Leaf and root endospheres harbor lower fungal diversity and less complex fungal co-occurrence patterns than rhizosphere. Front. Microbiol. 10:e1015. doi: 10.3389/fmicb.2019.01015
Ripa, F. A., Cao, W., Tong, S., and Sun, J. (2019). Assessment of plant growth promoting and abiotic stress tolerance properties of wheat endophytic fungi. BioMed Res. Int. 2019, 1–12. doi: 10.1155/2019/6105865
Rustamova, N., Bozorov, K., Efferth, T., and Yili, A. (2020). Novel secondary metabolites from endophytic fungi: synthesis and biological properties. Phytochem. Rev. 19, 425–448. doi: 10.1007/s11101-020-09672-x
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shi, W., Li, M., Wei, G., Tian, R. M., Li, C. P., Wang, B., et al. (2019). The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 7:e14. doi: 10.1186/s40168-019-0629-2
Shi, Y., Delgado-Baquerizo, M., Li, Y., Yang, Y., Zhu, Y. G., Peñuelas, J., et al. (2020). Abundance of kinless hubs within soil microbial networks are associated with high functional potential in agricultural ecosystems. Environ. Int. 142:e105869. doi: 10.1016/j.envint.2020.105869
Sousa, J. P. B., Aguilar-Perez, M. M., Arnold, A. E., Rios, N., Coley, P. D., Kursar, T. A., et al. (2016). Chemical constituents and their antibacterial activity from the tropical endophytic fungus Diaporthe sp F2934. J. Appl. Microbiol. 120, 1501–1508. doi: 10.1111/jam.13132
Tan, R. X., and Zou, W. X. (2001). Endophytes: a rich source of functional metabolites. Nat. Prod. Rep. 18, 448–459. doi: 10.1039/b100918o
Tardy, V., Mathieu, O., Lévêque, J., Terrat, S., Chabbi, A., Lemanceau, P., et al. (2014). Stability of soil microbial structure and activity depends on microbial diversity. Environ. Microbiol. Rep. 6, 173–183. doi: 10.1111/1758-2229.12126
Taylor, D. L., Walters, W. A., Lennon, N. J., Bochicchio, J., Krohn, A., Caporaso, J. G., et al. (2016). Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for illumina amplicon sequencing. Appl. Environ. Microbiol. 82, 7217–7226. doi: 10.1128/AEM.02576-16
Terhonen, E., Sipari, N., and Asiegbu, F. O. (2016). Inhibition of phytopathogens by fungal root endophytes of Norway spruce. Biol. Control 99, 53–63. doi: 10.1016/j.biocontrol.2016.04.006
Thebault, E., and Fontaine, C. (2010). Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853–856. doi: 10.1126/science.1188321
Ulloa-Munoz, R., Olivera-Gonzales, P., Castaneda-Barreto, A., Villena, G. K., and Tamariz-Angeles, C. (2020). Diversity of endophytic plant-growth microorganisms from Gentianella weberbaueri and Valeriana pycnantha, highland Peruvian medicinal plants. Microbiol. Res. 223:e126413. doi: 10.1016/j.micres.2020.126413
van der Putten, W. H. (2017). Belowground drivers of plant diversity. Science 355, 134–135. doi: 10.1126/science.aal4549
van Elsas, J. D., Chiurazzi, M., Mallon, C. A., Elhottova, D., Kristufek, V., and Salles, J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. doi: 10.1073/pnas.1109326109
Velásquez, A. C., Oney, M., Huot, B., Xu, S., and He, S. Y. (2017). Diverse mechanisms of resistance to Pseudomonas syringae in a thousand natural accessions of Arabidopsis thaliana. New Phytol. 214, 1673–1687. doi: 10.1111/nph.14517
Wang, F., Xin, C. Y., Liu, J. X., Ran, Z. N., Zhao, C. L., and Song, Z. Y. (2020). Interactions between invasive fungi and symbiotic bacteria. World J. Microbiol. Biotechnol. 36:e137. doi: 10.1007/s11274-020-02913-3
Wang, L. W., Xu, B. G., Wang, J. Y., Su, Z. Z., Lin, F. C., Zhang, C. L., et al. (2012). Bioactive metabolites from Phoma species, an endophytic fungus from the Chinese medicinal plant Arisaema erubescens. Appl. Microbiol. Biot. 93, 1231–1239. doi: 10.1007/s00253-011-3472-3
Waqas, M., Khan, A. L., Kamran, M., Hamayun, M., Kang, S. M., Kim, Y. H., et al. (2012). Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules 17, 10754–10773. doi: 10.3390/molecules170910754
Watts, D. J., and Strogatz, S. H. (1998). Collective dynamics of ‘small-world’ networks. Nature 393, 440–442. doi: 10.1038/30918
Wehner, J., Antunes, P. M., Powel, J. R., Mazukatow, J., and Rillig, M. C. (2010). Plant pathogen protection by arbuscular mycorrhizas: a role for fungal diversity? Pedobiologia 53, 197–201. doi: 10.1016/j.pedobi.2009.10.002
Wei, Z., Hu, J., Gu, Y., Yin, S. X., Xu, Y. C., Jousset, A., et al. (2018). Ralstonia solanacearum pathogen disrupts bacterial rhizosphere microbiome during an invasion. Soil Biol. Bioch. 118, 8–17. doi: 10.1016/j.soilbio.2017.11.012
Xia, X., Kim, S., Bang, S., Lee, H. J., Liu, C., Park, C. I., et al. (2015). Barceloneic acid C, a new polyketide from an endophytic fungus Phoma sp JS752 and its antibacterial activities. J. Antibiot. 68, 139–141. doi: 10.1038/ja.2014.116
Zhang, X. X., Zhang, R. J., Gao, J. S., Wang, X. C., Fan, F. L., Ma, X. T., et al. (2017). Thirty-one years of rice-rice-green manure rotations shape the rhizosphere microbial community and enrich beneficial bacteria. Soil Biol. Biochem. 104, 208–217. doi: 10.1016/j.soilbio.2016.10.023
Zhang, Z. Q., Zhang, J., and Jiao, S. (2021). Fungi show broader environmental thresholds in wet than dry agricultural soils with distinct biogeographic patterns. Sci. Total Environ. 750:e141761. doi: 10.1016/j.scitotenv.2020.141761
Zhou, J. Z., Deng, Y., Luo, F., He, Z. L., and Yang, Y. F. (2011). Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio 2:e00122-11. doi: 10.1128/mBio.00122-11
Keywords: bacterial wilt invasion, soil microbiota, endophytic microbiota, molecular ecological network, biocontrol fungal resources
Citation: Tan L, Zeng W, Xiao Y, Li P, Gu S, Wu S, Zhai Z, Feng K, Deng Y and Hu Q (2021) Fungi-Bacteria Associations in Wilt Diseased Rhizosphere and Endosphere by Interdomain Ecological Network Analysis. Front. Microbiol. 12:722626. doi: 10.3389/fmicb.2021.722626
Received: 09 June 2021; Accepted: 11 August 2021;
Published: 06 September 2021.
Edited by:
Hai-Ming Zhao, Jinan University, ChinaReviewed by:
Tengxiang Lian, South China Agricultural University, ChinaBing Yang, Sichuan Academy of Giant Panda, Chengdu, China
Biying Zhao, Jiangsu University, China
Juntao Wang, Western Sydney University, Australia
Wensheng Qin, Lakehead University, Canada
Copyright © 2021 Tan, Zeng, Xiao, Li, Gu, Wu, Zhai, Feng, Deng and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ye Deng, eWVkZW5nQHJjZWVzLmFjLmNu; Qiulong Hu, aHVxaXVsb25nbmV0QDEyNi5jb20=
 Lin Tan1
Lin Tan1 Kai Feng
Kai Feng Ye Deng
Ye Deng Qiulong Hu
Qiulong Hu