- 1Institute for Food Quality and Food Safety, Foundation University of Veterinary Medicine Hannover, Hanover, Germany
- 2Department of Food Hygiene and Aquaculture, Faculty of Veterinary Medicine, Ferdowsi University of Mashhad, Mashhad, Iran
- 3Department Biological Safety, German Federal Institute for Risk Assessment, Berlin, Germany
- 4Institute for Veterinary Food Science, Justus-Liebig-University Giessen, Giessen, Germany
- 5Central Facility for Microscopy, Helmholtz Centre for Infection Research GmbH, Braunschweig, Germany
Campylobacter spp. are a major cause of bacterial foodborne diarrhea worldwide. While thermophilic Campylobacter species asymptomatically colonize the intestines of chickens, most human infections in industrial countries have been attributed to consumption of chicken meat or cross-contaminated products. Bacteriophages (phages) are natural predators of bacteria and their use at different stages of the food production chain has been shown to reduce the public health burden of human campylobacteriosis. However, regarding regulatory issues, the use of lytic phages in food is still under discussion and evaluation. This study aims to identify lytic phages suitable for reducing Campylobacter bacteria along the food production chain. Therefore, four of 19 recently recovered phages were further characterized in detail for their lytic efficacy against different Campylobacter field strains and their suitability under food production settings at different temperatures and pH values. Based on the results of this study, the phages vB_CjM-LmqsCP1-4 and vB_CjM-LmqsCP1-5 appear to be promising candidates for the reduction of Campylobacter jejuni in food production settings.
Introduction
Campylobacteriosis is the most frequently reported foodborne bacterial gastroenteritis in the European Union (EU) since 2005 (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2021), but represents also an important zoonosis worldwide (Skarp et al., 2016). In 2019, the European Food Safety Authority (EFSA) notified 220,682 campylobacteriosis cases in the EU. While 49.8% of the reported cases were acquired in the EU, 3% were considered to originate from other countries. However, for 47.2% of the human cases, further information on the travel status or the country of infection was unknown (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2021). In general, consumption of broiler meat is considered to be the most common cause for human infections (European Food Safety Authority [EFSA], 2010). Thermophilic Campylobacter spp. have been shown to be the most prevalent cause of campylobacteriosis. Campylobacter (C.) jejuni and C. coli are the most frequent species, while C. lari, C. fetus and C. upsaliensis have been detected less frequently in campylobacteriosis cases (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2021). Symptoms of Campylobacter enteritis include watery to hemorrhagic diarrhea and abdominal pain. Severe long-term sequelae like the Guillain-Barré syndrome, reactive arthritis, and erythema nodosum can occur in rare cases subsequent to the infection (Nachamkin, 2002; Moore et al., 2005; Skarp et al., 2016).
Campylobacter jejuni and C. coli can colonize the intestine of various wild, domestic, and farm animals (Mulder et al., 2020; European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2021). In broiler flocks, they can usually be detected after 2–3 weeks of housing and spread rapidly until almost all animals of the flock are colonized without showing clinical symptoms (Shreeve et al., 2000; Newell and Fearnley, 2003; Newell et al., 2011). During defeathering, evisceration, and further processing, the pathogens can be transferred to the carcasses (Lee and Newell, 2006; Sasaki et al., 2013), and by cross-contamination during food preparation, contamination of other foodstuffs can occur (Luber et al., 2006; Cardoso et al., 2021). Current estimations predict that a reduction in the Campylobacter load in chicken ceca by 2 log10 units can diminish the risk for human infection, arising from consumed broiler meat, by 42%. However, a 3 log10 unit reduction would lead to a 58% lower risk for human infections (European Food Safety Authority [EFSA] et al., 2020; European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2021).
Bacteriophages (phages) are viruses that specifically infect bacterial genera, species or strains. Currently, the International Committee on Taxonomy of Viruses (ICTV) recognizes nine Campylobacter viruses (ICTV, 2019) that all have an AT-rich double stranded DNA and belong to the family Myoviridae (Bradley’s morphotype A1) (Sails et al., 1998) in the order of Caudovirales. Phages in this family are non-enveloped viruses, with a head-tail structure encompassing a contractile tail. Campylobacter viruses which are part of the Siphoviridae family, have recently been described, but are not yet recognized by the ICTV (Nowaczek et al., 2019). The ICTV currently recognizes a scheme based on sequence analysis that subdivides the family into the two genera Firehammerviruses, including four recognized members [type species CP220, formerly named Cp220likevirus (2014) and Cp220virus (2015–2018)], and Fletcherviruses, including five recognized members [type species CP8, formerly named Cp8unalikevirus (2014) and Cp8virus (2015–2018)] (Javed et al., 2014; Adams et al., 2016; ICTV, 2019; Jackel et al., 2019). Moreover, there are currently two unclassified Firehammerviruses, as well as one classified and 21 unclassified Fletcherviruses listed by the NCBI taxonomy browser (Schoch et al., 2020). A former typing scheme divides Campylobacter phages into three groups based on their genome sizes (group I ∼320 kb, group II ∼184 kb, and group III ∼138 kb). These groups could also be predicted according to their prevailing host specificity (Sails et al., 1998; Javed et al., 2014). Fletcherviruses were assigned to phage group III and Firehammerviruses to phage group II before ICTV classification was established (Hammerl et al., 2011; Javed et al., 2014; Jackel et al., 2019; Ushanov et al., 2020).
The restricted host range of the Campylobacter phages described so far allows specific targeting of pathogenic species (Richards et al., 2019), but at the same time raises the need to combine various phages in cocktails for effective applications in food production settings, where different, unknown Campylobacter strains might be encountered (Chan et al., 2013; Kittler et al., 2013). Combining phages also offers the advantage of preventing bacterial resistance development that may occur as a result of phage application (Fischer et al., 2013; Azam and Tanji, 2019). Nonetheless, while some phage-resistant bacterial cells might occur even if cocktails were used (Fischer et al., 2013), phage-resistant bacteria were shown to exhibit a reduced colonization fitness (Kittler et al., 2014; Oechslin, 2018; Azam and Tanji, 2019). Phages can be used at multiple stages of the food production process without changing the organoleptic properties of the products, such as odor or taste. They can be applied during meat production pre- or post-harvest, and for bio sanitation (Connerton et al., 2011; Polaska and Sokolowska, 2019) as mandated by a farm to fork strategy (Nauta et al., 2007). However, a lack of data and approval procedures still impedes the implementation of phages as an efficient alternative for Campylobacter biocontrol (European Food Safety Authority [EFSA], 2009; European Food Safety Authority [EFSA] et al., 2020).
Application of Campylobacter phages in food production settings has been considered as a promising technology (European Food Safety Authority [EFSA] et al., 2020; Zbikowska et al., 2020). Previous studies highlighted the need for (i) a lytic phage collection directed against current Campyobacter field strains (Carrillo et al., 2005), (ii) more precise and faster methods to identify and select useful phages (Xie et al., 2018; Zbikowska et al., 2020), and (iii) a better understanding of bacteriophage kinetics to allow for advanced considerations concerning dosing and timing of phage applications (Cairns et al., 2009; Hammerl et al., 2014; Loessner et al., 2020). Furthermore, genome determination of Campylobacter phages was also shown to be laborious due to prevailing DNA modification and extensive repetitive sequences, including hypermutable polyGtracts, resulting in low amounts of extracted DNA and/or incomplete genomes (Hammerl et al., 2011; Carvalho et al., 2012; Jackel et al., 2015; Crippen et al., 2019; Sorensen et al., 2021). However, all Campylobacter phages described to date are closely related in their genetic composition and exhibit highly conserved nucleotide sequences. To the best of our knowledge, all Campylobacter phages are free of genes encoding antimicrobial and/or virulence factors (Hammerl et al., 2015). Questions regarding the stability of the phage genomes are hard to evaluate as Campylobacter phages comprise homing endonucleases, which might be involved in adaption of the genomes according to the prevailing selection pressures (i.e., host specificity etc.). This study represents an initiative to increase the number of carefully characterized phages against C. jejuni. Furthermore, we aimed to determine differences in the performance of phages with similar host ranges or origins in liquid cultures of Campylobacter field strains and analyzed their stability upon storage, varying pH values and temperatures (Carrillo et al., 2005; Jonczyk et al., 2011) for estimating their suitability for practical application under real life settings.
Materials and Methods
Bacteria Strains, Typing and Growth Conditions
Information on the bacterial strains used in this study are summarized in Supplementary Table 1. Briefly, the used bacterial panel consisted of two type strains (DSM 4688, DSM 4689) and two reference strains (NCTC 11168, ATCC BAA-2151) as well as 23 field isolates from chicken samples collected from commercial poultry farms in Lower Saxony, Germany, in 2017 (Supplementary Table 1). Field isolates were typed to preselect a host panel representing a wide range of current field isolates by flaA-typing in accordance with Zhang et al. (2018) followed by analysis of flaA sequences (Eurofins NDSC Food Testing GmbH, Hamburg, Germany). SmaI-PFGE macrorestriction analysis of the bacterial isolates was conducted as previously described (Nawaz et al., 2003) with minor modifications, using a CHEFIII System (BioRAD laboratories GmbH, Feldkircen, Germany).
Campylobacter spp. stock cultures were stored at –80°C. Bacteria were cultivated on Columbia Agar sheep Blood “Plus” plates (Thermo Fischer Scientific Oxoid Deutschland GmbH, Wesel, Germany) at 42°C under microaerobic conditions (5% O2, 10% CO2, 85 N2, >80% humidity). Liquid cultures were prepared with brain-heart infusion (Carl Roth GmbH & Co., KG, Karlsruhe, Germany, X916) supplemented with 1 mM calcium chloride (CBHI).
Bacteriophage Isolation and Propagation
The soft-agar overlay technique was used to detect Campylobacter phages from different matrices. All samples (ncecal = 136, nfecal = 111, nneck skin = 54) originated from poultry farms in Lower Saxony, Germany. For sample preparation, approximately 2 g of fecal or cecal samples were dispersed in 10 ml SM-buffer [100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl (pH 7.5)] using an Ultra-Turrax T10 homogenizer (IKA-Werke GmbH & Co. KG, Staufen, Germany). In contrast, skin samples were thoroughly rinsed by massaging them in a plastic bag containing 10 ml SM-buffer. After overnight shaking at 4°C, the samples were centrifuged twice (1st step at 3,488 × g for 20 min, 2nd step at 13,000 × g for 10 min), and the supernatant was filtered through a 0.2 μm polyethylensulfon membrane (PES) syringe filter (Carl Roth GmbH & Co., KG). For further purification, the resulting filtrate was cocultured with C. jejuni strain NCTC 12662 as previously described (Fischer et al., 2013) with minor modifications. Briefly, bacterial overnight cultures were cultivated on blood agar plates for 14–15 h, and a suspension in 10 mM magnesium sulfate was adjusted to a McFarland of 1.0 (McF) (DEN-1 densitometer, Grant Instruments Grant Bio, Thermo Fischer Scientific GmbH, Schwerte, Germany). One-milliliter of bacterial suspension was incubated in 9 ml culture medium for approximately 4 h. Afterward, 100 μl of the culture was mixed with the sample filtrate, and incubated for a further 10 min. Subsequently, the mixture was added to 5 ml molten 0.4% NZCYM-soft agar (Carl Roth GmbH & Co., KG). The soft-agar was poured onto NZCYM plates containing 1.5% agar. After 24 h incubation period, the resulting plaques were purified by a successive threefold picking and plating procedure of single plaques. Subsequent propagation of phages was carried out with 0.7% NZCYM-soft agar until suspensions reached a concentration of approximately 108 plaque forming units (PFU) per ml for host range determination. Concentrations were determined by using serial dilutions of the phage lysates and duplicate plating of 100 μl of each dilution on C. jejuni NCTC 12662.
Host Range Determination and Initial Characterization
The host range of the individual phages was determined in accordance with the plaque assay method of Korf et al. (2020), with some modifications, while the susceptibility of the bacteria was indicated by the efficiency of plaquing (EOP), as defined by Sorensen et al. (2012). In short, square NZCYM-agar plates were overlaid with 7.9 ml NZCYM-soft agar, which had been inoculated with a 2 h pre-incubated liquid culture of one of the 29 Campylobacter in CBHI (Supplementary Table 1), and 10 μl of 10-fold serial diluted phage suspensions from each of the 19 phages was applied onto the overlay. Plates were incubated for 20 ± 2 h. Phage/bacteria combinations that produced visible plaques 2–3 times were used to calculate the relative difference of plaque forming ability by dividing the measured concentration on the respective Campylobacter isolate or strain by the concentration measured on the host strain NCTC 12662. Production of visible plaques and opaque inhibition zones without plaques in only one of three tests was counted as a negative result.
Initial characterization by HhaI- and VspI-macrorestriction with subsequent pulsed-field gel electrophoresis (PFGE) was performed as previously described (Hansen et al., 2007).
Efficacy Testing of Phages Bacteria Reduction Capability in Liquid Culture
The growth of two C. jejuni field strains, Cj18 or LH86, with and without exposure to four different Campylobacter bacteriophages vB_CjM-LmqsCP1-4 (CP1-4), vB_CjM-LmqsCP1-5 (CP1-5), vB_CjM-LmqsCP74-2c1 (CP74-2c1), and vB_CjM-LmqsCP132-3c (CP132-3c) at five different multiplicities of infection (MOIinput of 10, 1, 0.1, 0.01, and 0.001) (Danis-Wlodarczyk et al., 2021) was examined in liquid cultures using a Tecan Spark Microplate Reader with O2 and CO2 control, similar to methods described by Xie et al. (2018); Rajnovic et al. (2019), and Zachary et al. (2020) with some modifications. Briefly, phage suspensions were adjusted to 108, 107, 106, 105, and 104 PFU/ml, which were further diluted 10-fold with CBHI to achieve final dilutions of 107, 106, 105, 104, and 103 PFU/ml. Bacterial overnight cultures were adjusted to 3.0 McF in 10 mM MgSO4 and used to inoculate 50 ml CBHI that was then incubated for 3 h with shaking (130 rpm). Afterward, the culture was adjusted to 0.5 McF, diluted 100-fold with CBHI and the wells of a 48-well microplate were filled with 250 μl of this suspension and 250 μl of phage suspensions. Plates were incubated with double orbital shaking at 42 °C under microaerobic conditions (5% O2, 10% CO2, 85% N2, 108 rpm). Optical density (OD600) was measured hourly for 26 h. There were two replication wells per plate and the experiment was performed in triplicate.
At the end of the experiment, 100 μl were taken from each well with an MOIinput 10 and 0.001, and plated on mCCDA Agar (Thermo Fischer Scientific Oxoid GmbH, Wesel). After 24 h, one colony per plate was picked and tested for phage resistance by spotting 10 μl of a 10-fold dilution series of phages on NZCYM-soft agar overlays containing the respective bacterial isolate. Bacteria were considered phage-resistant, if no plaques were observed. The remaining culture in the well was treated with 2–3 drops of chloroform and the total phage titer was determined as described above.
Stability Testing of Phages
Temperature stability was tested by incubating 300 μl of phage suspensions at a concentration of 108 PFU/ml for 15 or 60 min at –20 in a freezer and at 50, 60, 70, or 80°C in a block heater (Grant Instruments Ltd., Royston, United Kingdom). Samples were taken after 0, 15 and 60 min. Experiments were performed in triplicate.
For pH-stability tests, 100 μl of phage suspensions were diluted 10-fold with pH-adjusted phosphate buffer at pH values of 2–12 (Carl Roth GmbH & Co., KG) or tap water to reach a total volume of 1 ml. Suspensions were stored at two temperatures (in an incubator at 22.3 ± 0.4°C or at 41.7 ± 0.7°C) for 24 h. The concentration of each suspension was determined after 24 h. Additional samples were taken for pH 2, 3, and 7 at the start of the experiment (0 h), after 15 min for pH 2, and 120 min for pH 3. Experiments were performed in triplicate.
Stability of phages during storage was assessed by storing 6 ml of phage suspension for 7 months at 4.5 ± 0.5°C or 6 weeks at 23.5 ± 0.7°C. Phage concentrations were determined at regular intervals. The experiment was repeated four times.
Negative-Staining of Phages
Phages were negatively stained with 2% aqueous uranyl acetate after being adsorbed for 15–30 s onto carbon film according to Valentine et al. (1968). After washing with TE buffer (10 mM TRIS, 1 mM EDTA, pH 6.9) samples were blotted onto filter paper and air dried. Samples were examined in a Zeiss TEM 910 transmission electron microscope (Zeiss, Oberkochen) at an acceleration voltage of 80 kV and at calibrated magnifications with a line replica. Images were recorded digitally with a Slow-Scan CCD-Camera (ProScan, 1,024 × 1,024, Proscan Elektronische Systeme GmbH, Scheuring, Germany) with ITEM-Software (Olympus Soft Imaging Solutions GmbH, Münster, Germany).
Extraction of Phage DNA and Genome Analysis
Phage suspensions were prepared as described above and phage particles pelleted by centrifugation at 24,000 × g (Avanti J-26S XP, Beckmann Coulter Inc., Brea, CA, United States) for 2 h. The pellet was resuspended with a small amount of SM-buffer and purified by CsCl-gradient (Optima XPN-100, Beckmann Coulter Inc., Brea, United States, 165,100 × g, 4°C, 2 h). Cesium chloride was removed from the phages via dialysis with SM-buffer. The resulting phage suspensions were used for electron micrographs, PFGE macrorestriction analysis and DNA isolation. Phage DNA was extracted from virions purified by cesium chloride density gradient (Sambrook and Russell, 2001) with the Wizard DNA Clean-Up System (Promega, Madison, WI, United States) in accordance with the manufacturer’s instructions, followed by ethanol precipitation.
Phage DNA was subjected to short read whole-genome sequencing in-house at the German Federal Institute for Risk Assessment, Berlin, Germany (BfR). DNA-sequencing libraries were generated using the Nextera XT DNA Library Flex Preparation Kit (Illumina Inc., San Diego, CA, United States) in accordance with the recommendations of the manufacturer. Short read, paired-end sequencing was conducted on different Illumina devices (i.e., MiSeq, NextSeq) using the MiSeq Reagent v3 600-cycle Kit (Illumina). The generated raw reads were subjected to the Aquamis in house pipeline (Deneke et al., 2021).
Real-time PCR in accordance with a protocol by Jackel et al. (2017) was performed for phage differentiation. PCR primers targeted the tail tube gene, ORF186 of CP21 for group II phages (CPGII-probe: FAM-CCGGATTGACTGTAGAAACA-BHQ-1) and the gene for the base plate wedge, ORF008 of CP81, for group III phages (CPGII-probe: Cy5-TGTAACTGCCCTGTTTGCTG-BBQ-650).
Data Analysis
Data preparation, visualization, and statistical analysis (one tailed Student’s t-tests and Dunnett’s test) were performed using R software version 4.0.2 (R Core Team, 2020) and the DescTools package version 0.99.40. The ComplexHeatmap package (Gu et al., 2016) was used for visualization of the host range. Phage particle size parameters (head length, head diameter, tail length) were determined by size analysis of virions visible on negatively stained electron micrographs with ImageJ version 1.51q in the Fiji bundle in combination with the ObjectJ plugin. Data analysis of flaA typing was performed by PubMLST (Jolley et al., 2018) and SmaI-PFGE macrorestriction interpretation was conducted by GelAnalyzer 19.1 (www.gelanalyzer.com, Istvan Lazar Jr., Ph.D. and Istvan Lazar Sr., Ph.D., CSc.).
Results
Phage Isolation and Host Range Determination
In total, 301 samples from chicken sources (chicken feces n = 111, cecal content n = 136 and neck skin n = 54) were examined for the presence of bacteriophages using the soft-agar overlay technique with C. jejuni strain NCTC 12662. After threefold serial purification and propagation of plaques, 19 purified phages remained. Four phages were isolated from one fecal sample in 2015, while nine were isolated from cecal samples, and six from chicken neck skin samples in 2017. Information on recovered phages and sample origin are shown in Supplementary Table 2.
The host ranges of all purified phages were analyzed on 28 different Campylobacter isolates (20 C. jejuni and 8 C. coli) as displayed in Figure 1. All bacterial isolates included showed different flaA types (Supplementary Table 1) and/or could be distinguished by SmaI-PFGE macrorestriction analysis (personal communication).
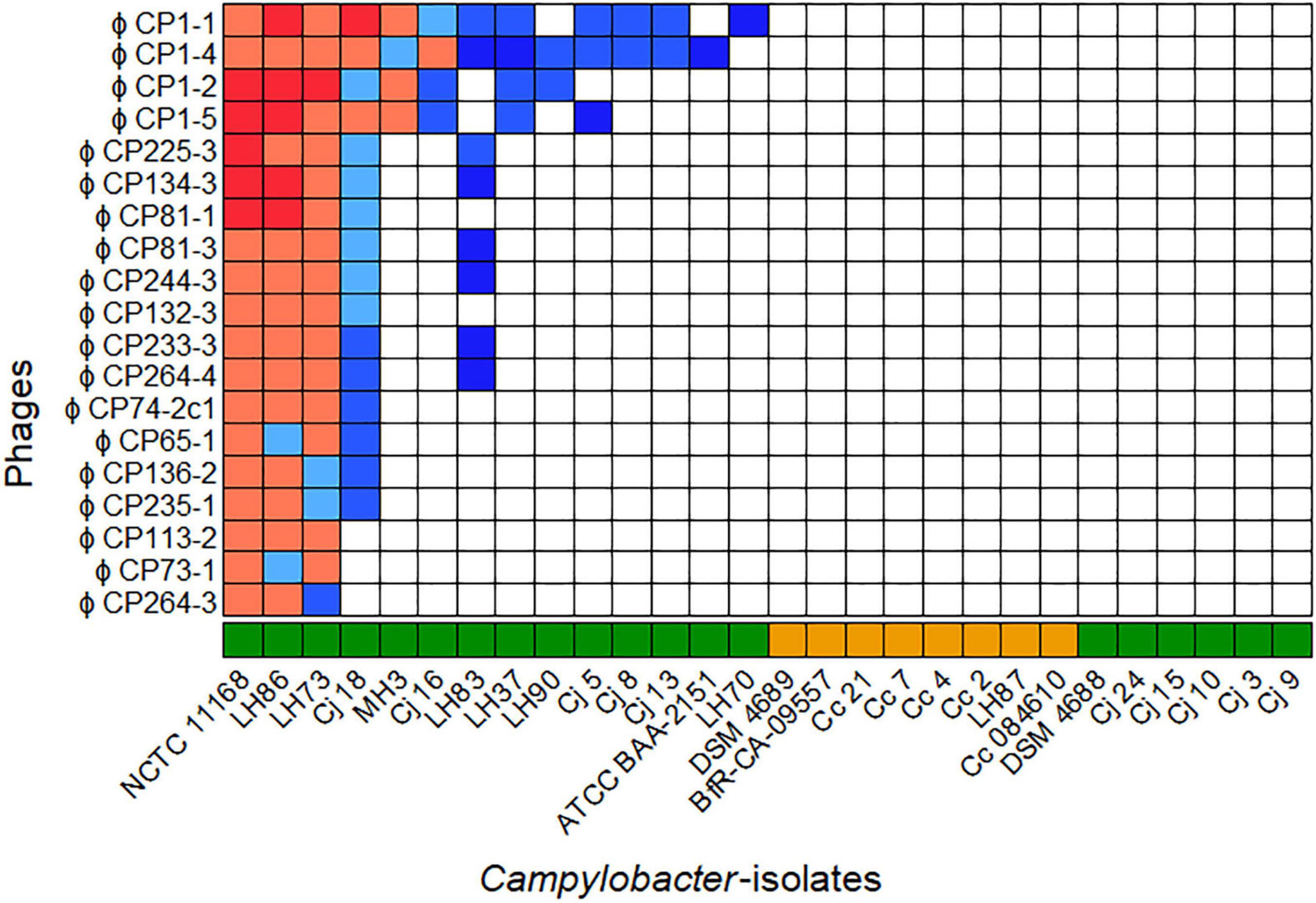
Figure 1. Host ranges of 19 phages isolated in 2015 and 2017 on 28 Campylobacter spp. Overall, seven host range patterns could be distinguished. The bacteriophages vB_CjM-LmqsCP1-4 (CP1-4), vB_CjM-LmqsCP1-5 (CP1-5), vB_CjM-LmqsCP74-2c1 (CP74-2c1), and vB_CjM-LmqsCP132-3c (CP132-3c) were selected for further investigations, showing three different host range patterns. Host range evaluation was performed by spotting serial dilutions of bacteriophages onto NZCYM-soft agar overlays inoculated with Campylobacter ( C. jejuni,
C. jejuni,  C. coli). Experiments were performed in triplicate. The concentration (plaque forming units/ml) on different hosts was calculated and divided by the concentration determined on C. jejuni NCTC 12662. Color coding was used to visualize the EOP values(
C. coli). Experiments were performed in triplicate. The concentration (plaque forming units/ml) on different hosts was calculated and divided by the concentration determined on C. jejuni NCTC 12662. Color coding was used to visualize the EOP values( x > 1,
x > 1,  1 ≤ x < 0.9,
1 ≤ x < 0.9,  0.9 ≤ x < 0.8,
0.9 ≤ x < 0.8,  0.8 ≤ x < 0.6,
0.8 ≤ x < 0.6,  0.6 ≤ x < 0, □ x = 0).
0.6 ≤ x < 0, □ x = 0).
Of the 28 Campylobacter isolates tested, 14 C. jejuni isolates were susceptible to at least one of the phages, while no plaque formation was observed on six C. jejuni and all C. coli isolates. The isolated phages displayed seven different lytic profiles. While four phages showed unique patterns, two groups of six and one group of three phages exhibited similar host ranges.
Four bacteriophages showing three of the seven lytic profiles and deriving from three isolation sources were selected for further characterization. The phages CP1-4 and CP1-5 were chosen according to their different host ranges, but they had the same sample origin, while the phages CP74-2c1 and CP132-3c had the same host range, but originated from different samples.
The four phages formed clear plaques on their host strain NCTC 12662 as shown in Figures 2A–D. Plaque diameters ranged from 0.75 to 1.86 mm (n = 30) after 24 h incubation on 0.7% NZCYM-soft agar (Supplementary Table 3). Negatively stained electron micrographs of all four phages showed that that they had isometric heads, contractile tails and tail fibers, leading to the conclusion that they belonged to the Myoviridae family (Figures 2E–H). The mean values for head length, head diameter, and tail length of the phages were very similar, except for CP74-2c1, which displayed a longer tail, and CP1-5, that showed an overall smaller head size than all other analyzed phages (Supplementary Table 3).
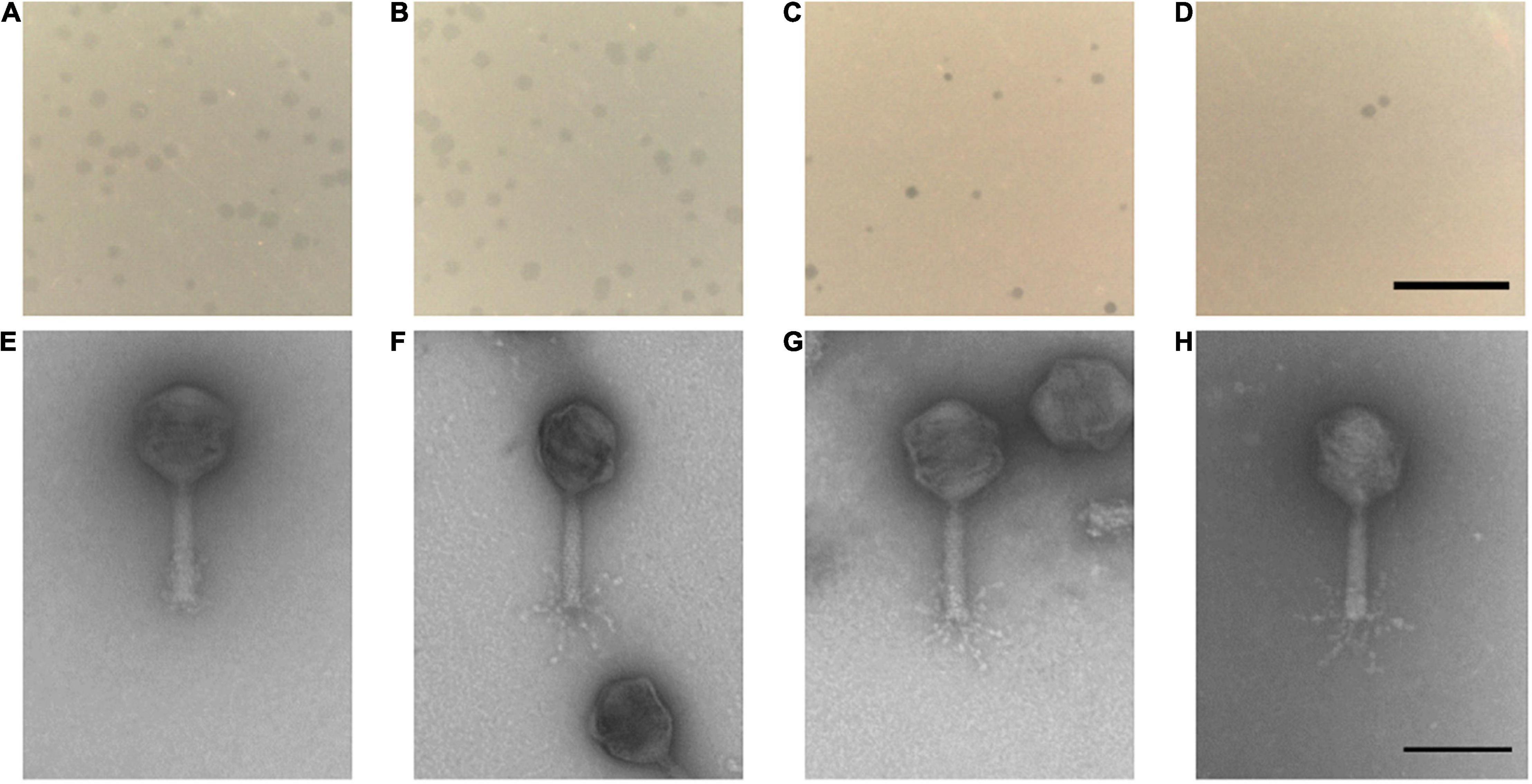
Figure 2. (A–D) Plaque morphology of the four examined myovirus phages (E–H) and micrographs showing the morphology of the virions. All four phages vB_CjM-LmqsCP132-3c (A,E), vB_CjM-LmqsCP1-5 (B,F), vB_CjM-LmqsCP74-2c1 (C,G), vB_CjM-LmqsCP1-4 (D,H) formed clear plaques on C. jejuni strain NCTC 12662; scale bar represents 2 mm. The virions consisted of an icosahedral head and a contractile tail structure with tail fibers; negatively stained phage particles with 2% uranylacetate, scale bar represents 100 nm.
Bacterial Reduction by Phages in Liquid Culture
Growth experiments were performed by measuring optical density (OD600) of C. jejuni field strains Cj18 and LH86 in liquid cultures with and without exposure to the four phages CP1-4, CP1-5, CP74-2c1, and CP132-3c at five different MOIinput (10, 1, 0.1, 0.01, and 0.001), using a Tecan Spark Multiplate reader over a time period of 26 h.
Both bacterial isolates were susceptible to all four phages according to their host ranges (Figure 1), and were chosen for testing the phage’s efficiency for reducing bacterial population growth and overall cell density. The EOP values of the four phages on the two Campylobacter isolates varied between 0.93 and 1.07, except for the combinations of Cj18 with CP74-2c1 or CP132-3c: Their EOP values were determined to be approximately 0.8.
Generally, bacterial growth was reduced in most liquid cultures that were exposed to bacteriophages compared to the untreated controls (Figure 3). Cultures containing Cj18 and CP74-2c1 or CP132-3c only showed reduced bacterial population growth if a high MOIinput of 10 was used in cultures containing the phages. Interestingly, in other experiments using an MOIinput of 10, most bacteria grew better than in experiments using a lower MOIinput of 0.001 after approx. 20 h (Figures 3C,D). Furthermore the data showed an initial rise and subsequent rapid decrease in optical density for LH86 in combination with CP74-2c1 or CP132-3c at an MOIinput of 0.01 and 0.001 (Figures 3G,H).
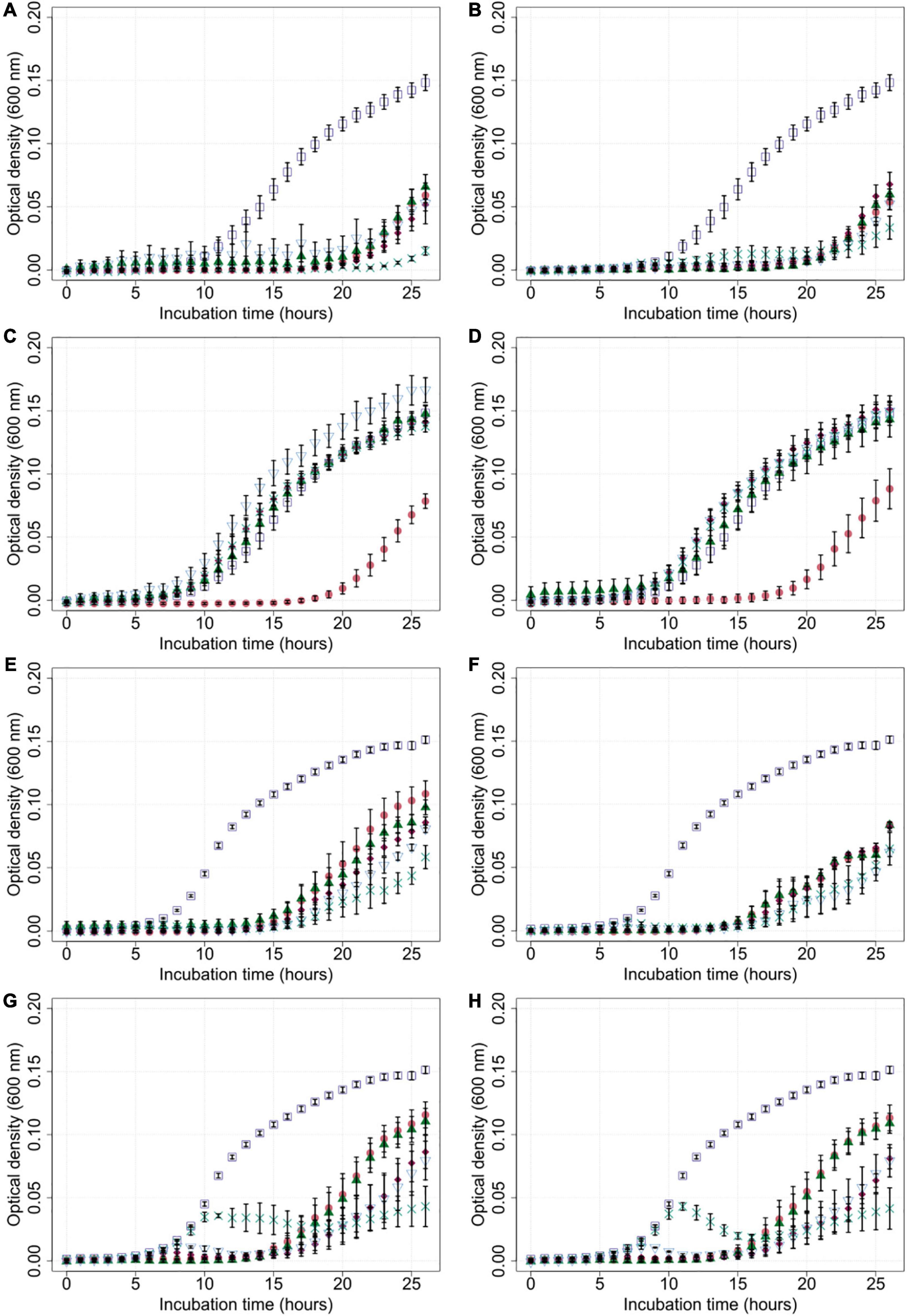
Figure 3. Growth of Campylobacter field isolates Cj18 and LH86 in presence of different phages. The growth of C. jejuni field strains Cj18 (A–D) and LH86 (E–H) was inhibited by all phages. Different multiplicities of infection (MOIinput) used, resulted in different reduction efficiencies. Experiments were performed in a Tecan Spark Microplate Reader under microaerobic conditions at 42°C. Optical density was measured every hour at 600 nm for 26 h. (A,E) vB_CjM-LmqsCP1-4, (B,F) vB_CjM-LmqsCP1-5, (C,G) vB_CjM-LmqsCP74-2c1 and (D,H) vB_CjM-LmqsCP132-3c were added to achieve different MOIinput:  no phages added,
no phages added,  MOIinput 10,
MOIinput 10,  MOIinput 1,
MOIinput 1,  MOIinput 0.1,
MOIinput 0.1,  MOIinput 0.01, and
MOIinput 0.01, and  MOIinput 0.001. Curves represent mean OD600 values of triplicate experiments with two replications per plate. Error bars indicate standard error of the mean.
MOIinput 0.001. Curves represent mean OD600 values of triplicate experiments with two replications per plate. Error bars indicate standard error of the mean.
Subsequently, the area under the curve (AUC) was calculated by spline fitting of the growth curves (Figure 4). The AUC values for most experiments with added phages were significantly reduced compared to the controls without phage exposure. Nevertheless, the AUC values equaled or exceeded those of experiments without phage exposure in case of experiments in which Cj18 was combined with CP74-2c1 or CP132-3c at an MOIinput of 1–0.001.
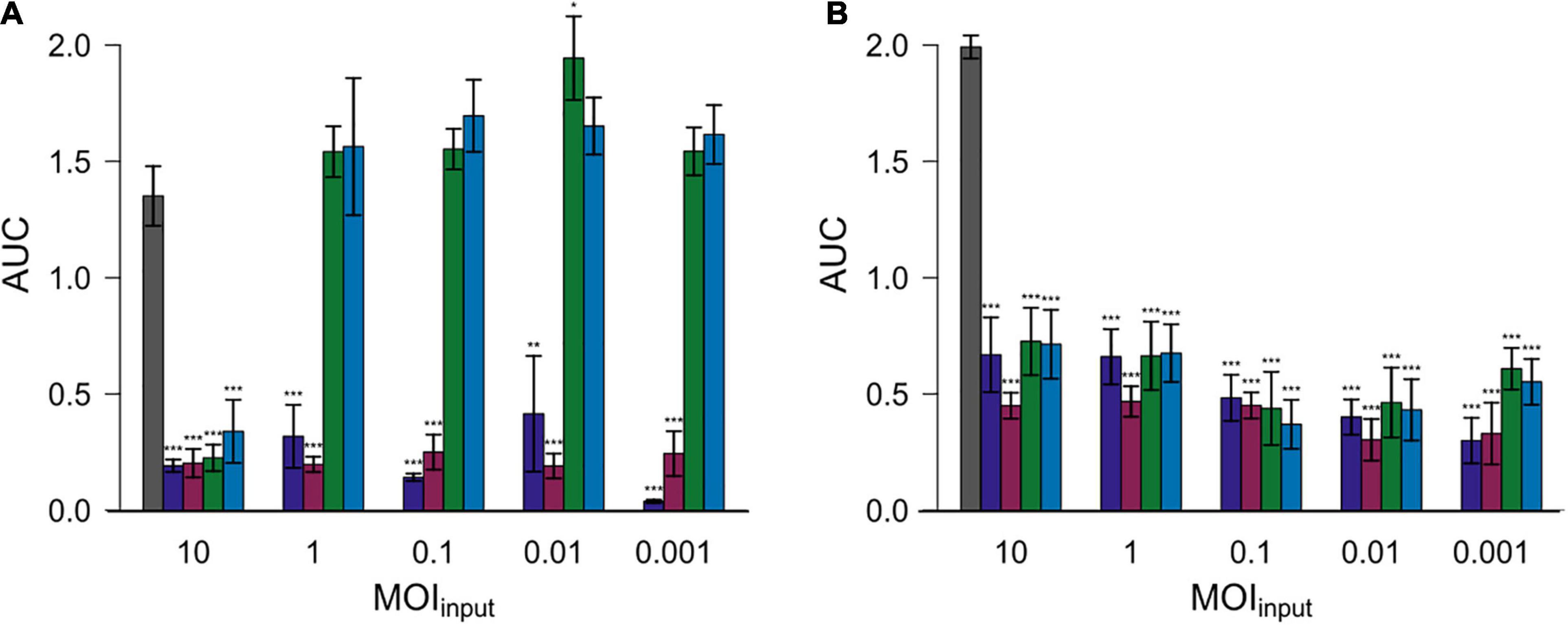
Figure 4. Areas under the curve (AUC) after 26 h for two different Campylobacter isolates. Campylobacter jejuni Cj18 ( A) and LH86 (
A) and LH86 ( B) were exposed to four different phages (
B) were exposed to four different phages ( vB_CjM-LmqsCP1-4,
vB_CjM-LmqsCP1-4,  vB_CjM-LmqsCP1-5,
vB_CjM-LmqsCP1-5,  vB_CjM-LmqsCP74-2c1 and
vB_CjM-LmqsCP74-2c1 and  vB_CjM-LmqsCP132-3c) at five different MOIinput (10, 1, 0.1, 0.01, and 0.001). The change of optical density over time was recorded. The AUC values were calculated by spline fitting (t = 0–26 h). Dunnett’s test with a 95% confidence level was used to compare the AUC values of the exposed samples to the untreated comparisons (significance codes indicate range of p-values: *0.01, **0.001, ***>0.001).
vB_CjM-LmqsCP132-3c) at five different MOIinput (10, 1, 0.1, 0.01, and 0.001). The change of optical density over time was recorded. The AUC values were calculated by spline fitting (t = 0–26 h). Dunnett’s test with a 95% confidence level was used to compare the AUC values of the exposed samples to the untreated comparisons (significance codes indicate range of p-values: *0.01, **0.001, ***>0.001).
After each experiment with an MOIinput of 0.001 or 10, bacterial isolates were recovered and tested for phage resistance with all four phages. Additionally, final phage concentrations in these wells were determined.
While these tests included only a limited number of bacterial isolates, Campylobacter isolates that showed resistance against at least one bacteriophage could be recovered from all experiments (Supplementary Table 4). Interestingly, one of the 27 recovered bacterial isolates was still susceptible to three bacteriophages after the experiment, while seven were susceptible to two phages under the same conditions. Furthermore, only three of the 27 isolates showed susceptibility toward the bacteriophages they were exposed to, after the experiments.
In wells with an MOIinput of 10, the starting concentrations were 107 PFU/ml. At the end, final phage concentrations ranged between 106 and 107.5 PFU/ml and were therefore very similar to the phage concentrations at the start of the experiment. However, final concentrations in the experiments using Cj18 with the phages CP74-2c1 or CP132-3c were lower and ranged between 104 and 105 PFU/ml. The final concentrations exceeded the starting concentrations of 103 PFU/ml noticeably by 103–104.5 PFU/ml at the end of the experiments with an MOIinput of 0.001. No phages could be recovered after the growth experiments of Cj18 at an MOIinput of 0.001 in combination with the phages CP74-2c1 and CP132-3c.
Bacteriophage Stability Under Different pH and Temperature Conditions
To determine the pH stability of the investigated phages, they were diluted 10-fold in pH adjusted phosphate buffered solutions, and exposed to temperatures of 22 and 42°C for 24 h (Figures 5A,B).
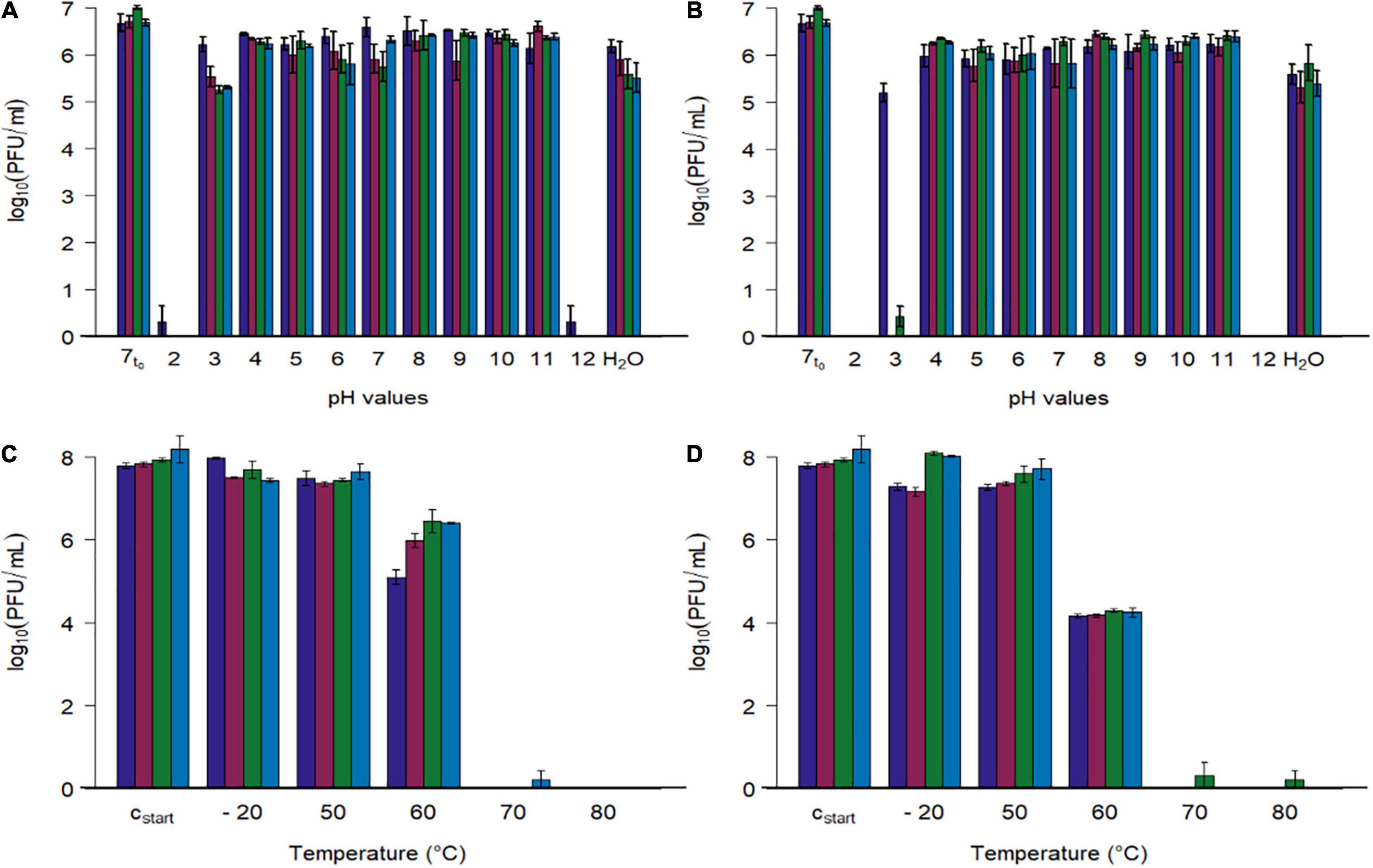
Figure 5. Stability tests of four phages at different pH values and different temperatures. (A,B) Phage concentration after exposure to different pH values (2–12) and tab water (H2O) at two temperatures (A: 22.3 ± 0.4°C, B: 41.7 ± 0.4). (C,D) Phage concentration after exposure to different temperatures (–20, 50, 60, 70, 80°C) at pH 7.4 (C: 15 min, D: 60 min). Initial concentration of SM-buffered phage solutions was 108 plaque forming units/ml (cstart), which were diluted tenfold with pH adjusted phosphate buffer for the first experiment (A,B, 7t0 = 1:10th dilution in pH 7 buffer at 0 h). Phage identity is represented as followes ( vB_CjM-LmqsCP1-4,
vB_CjM-LmqsCP1-4,  vB_CjM-LmqsCP1-5,
vB_CjM-LmqsCP1-5,  vB_CjM-LmqsCP74-2c1 and
vB_CjM-LmqsCP74-2c1 and  vB_CjM-LmqsCP132-3c). Columns represent the mean of the log10 transformed concentrations; experiments were performed in triplicate and error bars indicate the standard error of the mean.
vB_CjM-LmqsCP132-3c). Columns represent the mean of the log10 transformed concentrations; experiments were performed in triplicate and error bars indicate the standard error of the mean.
All four bacteriophages lost their lytic activity when exposed to pH values below 3 or above 11 at 22 or 42°C for 24 h (Figure 5), while they remained active after being exposed to pH values ranging from 4 to 11 or tap water under similar conditions. Results show that exposing the phages to buffer solutions with a pH value of 2 at 42°C immediately decreased their activity below detection limit, while after 24 h at pH 3 and 22°C, the analyzed phages retained their lytic activity. After 24 h at pH 3 and 22°C, the mean concentrations of the phages CP1-5, CP74-2c1, and CP132-3c were significantly reduced (CP1-5: 105.54 PFU/ml, p < 0.05; CP74-2c1: 105.27 PFU/ml, p < 0.05; CP132-3c: 105.32 PFU/ml, p < 0.05) in comparison to the controls (∼107 PFU/ml starting concentration at pH 7), while this was not the case for CP1-4, with a final concentration of 106.22 PFU/ml. However, after 24 h at pH 3 and 42°C, concentrations of CP1-5 and CP132-3c were reduced below detection limit, while CP1-4 was reduced to 105.21 PFU/ml and only single plaques were observed for CP74-2c.
To examine the stability at pH 3 in detail, phage concentrations of the more susceptible phages CP1-5, CP74-2c, and CP132-3c were determined after 120 min at pH 3 and 42°C. The concentrations of the three phages were significantly reduced (CP1-5: 105.30 PFU/ml, p < 0.05; CP74-2c1 103.57 PFU/ml, p < 0.05; CP132-3c: 104.69PFU/ml, p < 0.05) when compared to concentrations at the beginning of the experiment. The obtained data were used to calculate reduction rates by linear regression. CP1-5 showed the lowest reduction rate of 100.41 (PFU/ml)/1 h at pH 3 and 42°C, followed by 132_3c and CP74-2c1 with respective reduction rates of 100.78 (PFU/ml)/1 h and 101.30 (PFU/ml)/1 h.
Subsequently, phage stability was evaluated by exposing buffered phage suspensions with titers of 108 PFU/ml to –20, 50, 60, 70, and 80°C for 15 or 60 min (Figures 5C,D). Temperatures were chosen to simulate conditions during cooking or cold chain transportation. Concentrations of the four phages were not reduced during 15 min. storage at temperatures ranging from –20°C to 50°C, while their activity was completely lost at 70 and 80°C after 15 and 60 min. Concentrations of CP1-4 and CP1-5 were significantly reduced after 60 min at –20°C (CP1-4: 107.30, p < 0.05; CP1-5: 107.18, p < 0.05) and 50°C (CP1-4: 107.29, p < 0.05; CP1-5 107.30, p < 0.05) when compared to the control (∼108 PFU/ml). The other phages remained stable under these conditions. After 15 min at 60°C, significant reductions in concentrations of all four phages were observed compared to the starting concentrations (CP1-4: 105.10 PFU/ml, p < 0.05; CP1-5: 105.99, p < 0.05; CP74-2c1: 106.45 PFU/ml, p < 0.05; CP132-3c: 106.41, p < 0.05), and even greater reductions occurred after 60 min.
The storage stability of the investigated phages was tested at pH 7.5 for 7 months at 4.5 ± 0.5°C and 6 weeks at 23.5 ± 0.7°C. Results of these experiments were used to determine the average reduction rates by linear regression for 1 month. At 4.5°C, the average reduction rate was calculated to range from 1.02 to 1.20 (PFU/ml)/month, while at 23.5°C, the rates increased and ranged from 1.29 to 1.82 (PFU/ml)/month for the examined phages.
Bacteriophage DNA
For in-depth analysis, all extracted phage DNA (without an additional pre-amplification using phi29 polymerase; Kropinski et al., 2011; Crippen et al., 2019) was subjected to short-read whole-genome sequencing analysis on different Illumina Inc. devices. However, while all quality parameters of the prepared DNA and DNA sequencing libraries fulfilled the necessary requirements, no or only very low amounts of raw reads could be generated. Based on the available sequence information, no phage genome sequences could be derived.
Phage DNA was used for genome length estimations by PFGE. Estimations ranged from 144 kb for the smallest genome (CP1-4), to lengths between 148 and 152 kb for the genomes of the other three phages (CP1-5, CP74-2c1, and CP132-3c). DNA of all four phages was susceptible to cutting by the restriction enzyme HhaI (Supplementary Table 3), and real-time PCR identified them as group III phages (data not shown).
Discussion
Nineteen C. jejuni-specific bacteriophages from chicken samples were isolated in this study. The host range of all isolated phages was evaluated by testing them against a panel of well-characterized C. jejuni and C. coli isolates, comprising current field strains and using a highly reliable spot test format. Applied methods for host range analysis vary widely among currently published studies and no validation of reproducibility for these methods is currently available (Sails et al., 1998; Hansen et al., 2007; Hwang et al., 2009; Janez et al., 2014; Sorensen et al., 2015). While some host range panels include more isolates compared to this study a representation of the current epidemiological situation in broiler production (Carrillo et al., 2005) and further information on the used strains, such as origin and typing results, were considered to be more valuable compared to a broad but less defined panel.
From the newly isolated phages, CP1-4, CP1-5, CP74-2c1, and CP132-3c were selected for further characterization. The phages were chosen based on the assumption that the origin or host range of phages might influence their performance in liquid culture. Based on their morphology, the analyzed phages could be classified as members of the Myoviridae in the order Caudivirales (Sails et al., 1998). They were further classified as group III phages as their genome sizes were similar to 138 kb, their genomes were susceptible to HhaI digestion (Sails et al., 1998; Hansen et al., 2007; Javed et al., 2014), their head sizes close to 100 nm (Sails et al., 1998), and they exclusively infected C. jejuni strains (Jackel et al., 2019). Subsequent real-time PCR results confirmed this identification. Results from previous studies indicated that most group III phages use capsule polysaccharide receptors to attach to their hosts (Sorensen et al., 2015; Zampara et al., 2017). Sequenced members of this group belong to the genus Fletcherviruses (Hammerl et al., 2011; Javed et al., 2014; Jackel et al., 2019; Ushanov et al., 2020). They have been used for reducing Campylobacter on chicken meat in different studies and proved more efficient in binding to Campylobacter cells at low temperatures than the flagellotropic group II phages (Atterbury et al., 2003; Goode et al., 2003; Zampara et al., 2017). While some studies did not result in significant Campylobacter reduction on the treated products (Orquera et al., 2012), other studies reported a significant reduction by these phages (Thung et al., 2020). Experiments on reduction of Campylobacter in broiler chickens by using group III phages showed significant reductions in some of the settings and varying duration of the reducing effect (Carrillo et al., 2005; Wagenaar et al., 2005; Scott et al., 2007; Kittler et al., 2013). Additionally, first trials under commercial conditions were carried out, showing the general suitability of group III phages for use in food production settings (Kittler et al., 2013).
Whole-genome sequencing of phage genomes is mandatory for final evaluation of their suitability and approval in most countries. However, in accordance with results of our study, genome determination of Campylobacter phages has been described to be challenging due to (i) problems regarding preparation of necessary DNA amounts and (ii) the presence of DNA modification hampering DNA processing by using enzymatic procedures. Thus, the suitability of alternative sequencing procedures (i.e., use of different DNA library preparation procedures, alternative sequencing approaches) needs to be determined. It needs to be pointed out here that phage DNA pre-amplification with Phi29 polymerase has been shown as an appropriate possibility to omit enhancing effects for DNA sequencing and manipulation for some Campylobacter phages (Kropinski et al., 2011; Crippen et al., 2019). Further studies need to be conducted to check whether the phage genomes of this study can be also determined by conducting this additional amplification step. However, if possible further amplification steps (PCR) should be omitted to yield a low level of nucleotide alterations in the genomes caused by additional processing (i.e., PCR amplification).
To evaluate the efficiency of the selected phages in reducing Campylobacter, two C. jejuni field strains (Cj18 and LH86), which were susceptible to all four phages, were exposed to those phages at different MOIinput for 26 h [similar to the experiments by Carrillo et al. (2005) or Scott et al. (2007)]. All phages were able to reduce bacterial growth at an MOIinput of 10 (Figure 3), resulting in lower cell densities and reduced AUC values compared to the untreated controls (Figure 4). While significant reductions in AUC values were observed in most experiments using a lower MOIinput, the experiments using Cj18 with the phages CP74-2c1 and CP132-3c did not result in decreased AUC’s (Figure 4A). They showed a substantial decrease of the AUC in experiments using an MOIinput between 1 and 10, indicating lysis without replication (passive inundation), as was suggested for a similar case by El-Shibiny et al. (2009).
However, when comparing AUC values of experiments using different MOIinput (Figure 4), they do not indicate a dose dependence. Moreover, after ∼ 20 h, many OD600 values for MOIinput 10 were higher than those for MOIinput 0.001, indicating a potential negative correlation between MOIinput and OD600 values, while a positive correlation was expected as found in experiments by Rajnovic et al. (2019) for E. coli bacteriophages (Rajnovic et al., 2019). In 2009, El-Shibiny et al. (2009) reported similar results for in vivo tests with the group II phage CP220. In this study, the lowest phage dose resulted in the highest reduction in C. jejuni HPC5 cell numbers in chickens.
At the start of the growth experiment, phages were added at a concentration of ∼107 PFU/ml. Phage titers at the end of the experiment showed similar concentrations within 106–108 PFU/ml, while bacteria growth was indicated by high OD600 value (see above). The reasons for finding high phage and bacteria numbers at the same time at an MOIinput of 10 are unclear, but mathematical models (Levin et al., 1977; Cairns et al., 2009; Bull et al., 2014; Loessner et al., 2020) have predicted that phages and bacteria could coexist at high numbers. Therefore, further studies are necessary to elucidate the relationship of Campylobacter phages and bacteria in order to improve efficacy testing and practical phage application. In experiments using the Campylobacter isolate LH86 and the phages CP74-2c1 and CP132-3c at an MOIinput below 0.1, an initial rise and subsequent decline in OD600 values were observed (Figures 3G,H). Similar observations were made during experiments using E. coli as host bacteria in other studies (Bull et al., 2014; Tolen et al., 2018; Rajnovic et al., 2019). It was assumed that except for Cj18 with CP74-2c1 and CP132-3c, the other Campylobacter populations could show a similar growth but this was masked by the detection limit at low OD600 values. Further tests are needed for a better understanding of these observations and their relevance for practical application.
In experiments combining Cj18 with CP74-2c1 or CP132-3c, significantly reduced AUC values were observed at an MOIinput of 10 only (Figure 4A), while an MOIinput of 1 or lower resulted in AUC values equal to or in one instance even significantly higher than the control (Figure 4A). Results from host range analysis indicated phage amplification, but showed reduced EOP values for the two phages on this strain. Experiments with an MOIinput of 10 and 0.001 resulted in reduced final phage concentrations that were 2–3 log10 units lower compared to the starting concentrations. These findings indicate that the loss of virions was not compensated by virion production, potentially explaining the finding that bacterial reduction only occurred if phage concentrations exceeded bacterial concentrations. In this case, bacterial reduction would have relied on initial phage concentrations and not on phage replication. Similar results of non-lytic interactions impacting bacterial growth have been reported by a previous study (Carrillo et al., 2005).
Although the number of bacterial isolates tested for phage susceptibility was limited and carful interpretation is necessary, all tested combinations of bacteria and phages resulted in resistant bacterial colonies at the end of the experiment. Interestingly, Cj18 colonies that were resistant against CP74-2c1 and CP132-3c were not resistant against CP1-4 and CP1-5. This could indicate that these phages infected bacteria by using different recognition sites. However, a higher number of tested isolates and further tests would be necessary to confirm these results.
A broad host range and a high lytic efficiency are requirements for phage application under practical conditions, while stability against adverse effects in food production settings is a prerequisite for their utilization. Application of phages in primary production exposes them to room temperature at dosing and to the chicken’s body temperature of 42 °C after ingestion. Low pH values between 3 and 5 (Mabelebele et al., 2017) are encountered for 2–3 h (Svihus and Itani, 2019) when passing through the chicken’s crop, proventriculus, and gizzard. In this regard, temperature stability at 22 and 42°C in combination with different pH levels was tested as proposed by Carrillo et al. (2005). All four phage were stable in buffered solutions with pH values ranging from pH 4 to pH 11 at 22 and 42°C, while reduced stability was observed after exposure to pH 2 and 12. Thung et al. (2020) reported similar findings for the phage Cj01. Interestingly, just one bacteriophage (CP1-4), was able to retain stability at a pH of 3 at 42°C for 24 h, while the other three phages (CP1-5, CP74-2c1, and CP132-3c) proved only stable during exposure to pH 3 for at least 2 h at 42°C. At pH 3 and 22°C, the stability was less pronounced. Based on these results, the selected phages, CP1-4, CP1-5, CP74-2c1, and CP132-3c would be able to resist environmental stresses during application in primary production settings of commercial food production. CP1-4, however, showed even higher stability than Campylobacter phages from previous studies.
To guarantee a sufficient shelf life for storage under practical conditions and to evaluate stability on food products at retail level, the average reduction rate of phage activity per month was calculated. At 4.5°C the average reduction rate for all four phages ranged from 1.02 to 1.20 (PFU/ml)/month, which allowed to extrapolate that in 1 year of storage an average of not more than one log10 unit of phage activity would be lost. Similar stabilities of Campylobacter-specific bacteriophages were reported by Hammerl et al. (2014). At 23.5°C, the rates increased and ranged from 1.29 to 1.82 (PFU/ml)/month. These results show a sufficient stability of the selected phages for food production settings. As the phages proved stable at temperature levels found in household refrigerators, it became important to show that heat treatment, as it would occur during food preparation, was sufficient to inactivate the bacteriophages. An exposure to 70°C for 15 min was sufficient to deactivate all four bacteriophages, while 15 min at 60°C already caused a significant reduction in phage titers.
The reductions in concentrations of CP1-4 and CP1-5 after 60 min at –20 and 50°C are unexpected (Figure 5D), as this would indicate short-term stability at –20 and 50°C. However, results presented by Figures 5A,B contradict this conclusion, as they prove a high stability of these two phages at 22 and 42°C at pH 7 for 24 h. Interestingly, the increased stability of CP1-4 against acidic pH values did not correspond to an increased temperature stability. This combination of high pH-stability and easy heat inactivation would be a favorable combination in phages used in a farm to fork approach.
Concluding Remarks
Phage CP1-4 proved to be the most promising candidate for a wide variety of applications, as it provides a broad host range, high stability at low pH values, and heat inactivation at moderate temperatures. The phage CP1-5 originated from the same sample as CP1-4, however, its host range included five bacteria less than that of CP1-4. It appeared equally efficient in bacteria reduction based on growth experiments in liquid culture and displayed a significantly reduced pH stability, illustrating that phages with different characteristics can be isolated from the same sample.
On the other hand, the phages CP74-2c1 and CP132-3c have been rejected for commercial use based on missing efficiency at medium and lower phage doses in liquid cultures with one of the selected field strains. The obtained results illustrate the need for kinetic tests (optical density based or otherwise) on different Campylobacter to elucidate overall phage performance, as host range analysis with only laboratory strains does not allow for a rational phage choice. Problems during DNA extraction as well as the presence of extensive repetitive sequences on the genomes of Camplyobacter phages make the genetic evaluation laborious and force a search for better analysis protocols (Sorensen et al., 2021).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
CK and SK did funding acquisition. SK and SS did study design and planning. Bacteriophage isolation was performed by GS and SK, DNA isolation was done by SS and CJ. Bacteriophage genome sequencing was done by JH, electron micrographs were taken by MR, host range determination was jointly done by EP and SS, while the rest the experiments and data analysis were performed by SS. SS and SK conceptualized and wrote the draft manuscript. SS, GS, JH, CK, EP, MR, CJ, MP, and SK helped with writing and editing the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the German Federal Ministry of Education and Research (BMBF) through the zoonoses research consortium PAC-Campylobacter (project IP5/01KI1725E).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.761223/full#supplementary-material
References
Adams, M. J., Lefkowitz, E. J., King, A. M., Harrach, B., Harrison, R. L., Knowles, N. J., et al. (2016). Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch. Virol. 161, 2921–2949. doi: 10.1007/s00705-016-2977-6
Atterbury, R. J., Connerton, P. L., Dodd, C. E. R., Rees, C. E. D., and Connerton, I. F. (2003). Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl. Environ. Microb. 69, 6302–6306. doi: 10.1128/Aem.69.10.6302-6306.2003
Azam, A. H., and Tanji, Y. (2019). Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 103, 2121–2131. doi: 10.1007/s00253-019-09629-x
Bull, J. J., Vegge, C. S., Schmerer, M., Chaudhry, W. N., and Levin, B. R. (2014). Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS One 9:e94690. doi: 10.1371/journal.pone.0094690
Cairns, B. J., Timms, A. R., Jansen, V. A. A., Connerton, I. F., and Payne, R. J. H. (2009). Quantitative models of in vitro bacteriophage-host dynamics and their application to phage therapy. PLoS Pathog. 5:e1000253. doi: 10.1371/journal.ppat.1000253
Cardoso, M. J., Ferreira, V., Truninger, M., Maia, R., and Teixeira, P. (2021). Cross-contamination events of Campylobacter spp. in domestic kitchens associated with consumer handling practices of raw poultry. Int. J. Food Microbiol. 338:108984. doi: 10.1016/j.ijfoodmicro.2020.108984
Carrillo, C. L., Atterbury, R. J., El-Shibiny, A., Connerton, P. L., Dillon, E., Scott, A., et al. (2005). Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71, 6554–6563. doi: 10.1128/Aem.71.11.6554-6563.2005
Carvalho, C. M., Kropinski, A. M., Lingohr, E. J., Santos, S. B., King, J., and Azeredo, J. (2012). The genome and proteome of a Campylobacter coli bacteriophage vB_CcoM-IBB_35 reveal unusual features. Virol. J. 9:35. doi: 10.1186/1743-422X-9-35
Chan, B. K., Abedon, S. T., and Loc-Carrillo, C. (2013). Phage cocktails and the future of phage therapy. Future Microbiol. 8, 769–783. doi: 10.217/Fmb.13.74
Connerton, P. L., Timms, A. R., and Connerton, I. F. (2011). Campylobacter bacteriophages and bacteriophage therapy. J. Appl. Microbiol. 111, 255–265. doi: 10.1111/j.1365-2672.2011.05012.x
Crippen, C. S., Lee, Y. J., Hutinet, G., Shajahan, A., Sacher, J. C., Azadi, P., et al. (2019). Deoxyinosine and 7-deaza-2-deoxyguanosine as carriers of genetic information in the DNA of Campylobacter viruses. J. Virol. 93, e01111–19. doi: 10.1128/JVI.01111-19
Danis-Wlodarczyk, K., Dabrowska, K., and Abedon, S. T. (2021). Phage therapy: the pharmacology of antibacterial viruses. Curr. Issues Mol. Biol. 40, 81–163. doi: 10.21775/cimb.040.081
Deneke, C., Brendebach, H., Uelze, L., Borowiak, M., Malorny, B., and Tausch, S. H. (2021). Species-specific quality control, assembly and contamination detection in microbial isolate sequences with AQUAMIS. Genes 12:644. doi: 10.3390/genes12050644
European Food Safety Authority [EFSA] (2009). Scientific opinion of the panel on biological hazards on a request from European Commission on the use and mode of action of bacteriophages in food production. EFSA J. 7:1076. doi: 10.2903/j.efsa.2009.1076
European Food Safety Authority [EFSA] (2010). Scientific Opinion on Quantification of the risk posed by broiler meat to human campylobacteriosis in the EU. EFSA J. 8:1437. doi: 10.2903/j.efsa.2010.1437
European Food Safety Authority [EFSA], Koutsoumanis, K., Allende, A., Alvarez-Ordonez, A., Bolton, D., Bover-Cid, S., et al. (2020). Update and review of control options for Campylobacter in broilers at primary production. EFSA J. 18:e06090. doi: 10.2903/j.efsa.2020.6090
El-Shibiny, A., Scott, A., Timms, A., Metawea, Y., Connerton, P., and Connerton, I. (2009). Application of a group II Campylobacter bacteriophage to reduce strains of Campylobacter jejuni and Campylobacter coli colonizing broiler chickens. J. Food Prot. 72, 733–740. doi: 10.4315/0362-028x-72.4.733
European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC] (2021). The European Union One Health 2019 zoonoses report. EFSA J. 19:e06406. doi: 10.2903/j.efsa.2021.6406
Fischer, S., Kittler, S., Klein, G., and Glunder, G. (2013). Impact of a single phage and a phage cocktail application in broilers on reduction of Campylobacter jejuni and development of resistance. PLoS One 8:e78543. doi: 10.1371/journal.pone.0078543
Goode, D., Allen, V. M., and Barrow, P. A. (2003). Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microb. 69, 5032–5036. doi: 10.1128/Aem.69.8.5032-5036.2003
Gu, Z. G., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Hammerl, J. A., Jäckel, C., Alter, T., Janzcyk, P., Stingl, K., Knüver, M. T., et al. (2014). Reduction of Campylobacter jejuni in Broiler Chicken by Successive Application of Group II and Group III Phages. PLoS One 9:e114785. doi: 10.1371/journal.pone.0114785
Hammerl, J. A., Jackel, C., and Hertwig, S. (2015). Genetics of Campylobacter phages. Berl. Munch. Tierarztl. Wochenschr. 128, 148–154. doi: 10.2376/0005-9366-128-148
Hammerl, J. A., Jackel, C., Reetz, J., Beck, S., Alter, T., Lurz, R., et al. (2011). Campylobacter jejuni group III phage CP81 contains many T4-like genes without belonging to the T4-type phage group: implications for the evolution of T4 phages. J. Virol. 85, 8597–8605. doi: 10.1128/Jvi.00395-11
Hansen, V. M., Rosenquist, H., Baggesen, D. L., Brown, S., and Christensen, B. B. (2007). Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiol. 7:90. doi: 10.1186/1471-2180-7-90
Hwang, S., Yun, J., Kim, K. P., Heu, S., Lee, S., and Ryu, S. (2009). Isolation and characterization of bacteriophages specific for Campylobacter jejuni. Microbiol. Immunol. 53, 559–566. doi: 10.1111/j.1348-0421.2009.00163.x
ICTV (2019). ICTV Master Species List 2019.v1. Available Online at: https://talk.ictvonline.org/files/master-species-lists/ (accessed October 23, 2020).
Jackel, C., Hammerl, J. A., and Hertwig, S. (2019). Campylobacter phage isolation and characterization: what we have learned so far. Methods Protoc. 2:18. doi: 10.3390/mps2010018
Jackel, C., Hammerl, J. A., Rau, J., and Hertwig, S. (2017). A multiplex real-time PCR for the detection and differentiation of Campylobacter phages. PLoS One 12:e0190240. doi: 10.1371/journal.pone.0190240
Jackel, C., Hammerl, J. A., Reetz, J., Kropinski, A. M., and Hertwig, S. (2015). Campylobacter group II phage CP21 is the prototype of a new subgroup revealing a distinct modular genome organization and host specificity. BMC Genomics 16:629. doi: 10.1186/s12864-015-1837-1
Janez, N., Kokosin, A., Zaletel, E., Vranac, T., Kovac, J., Vuckovic, D., et al. (2014). Identification and characterisation of new Campylobacter group III phages of animal origin. FEMS Microbiol. Lett. 359, 64–71. doi: 10.1111/1574-6968.12556
Javed, M. A., Ackermann, H. W., Azeredo, J., Carvalho, C. M., Connerton, I., Evoy, S., et al. (2014). A suggested classification for two groups of Campylobacter myoviruses. Arch. Virol. 159, 181–190. doi: 10.1007/s00705-013-1788-2
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Jonczyk, E., Klak, M., Miedzybrodzki, R., and Gorski, A. (2011). The influence of external factors on bacteriophages-review. Folia Microbiol. 56, 191–200. doi: 10.1007/s12223-011-0039-8
Kittler, S., Fischer, S., Abdulmawjood, A., Glunder, G., and Klein, G. (2013). Effect of bacteriophage application on Campylobacter jejuni loads in commercial broiler flocks. Appl. Environ. Microbiol. 79, 7525–7533. doi: 10.1128/Aem.02703-13
Kittler, S., Fischer, S., Abdulmawjood, A., Glunder, G., and Klein, G. (2014). Colonisation of a phage susceptible Campylobacter jejuni population in two phage positive broiler flocks. PLoS One 9:e94782. doi: 10.1371/journal.pone.0094782
Korf, I. H. E., Kittler, S., Bierbrodt, A., Mengden, R., Rohde, C., Rohde, M., et al. (2020). In vitro evaluation of a phage cocktail controlling infections with Escherichia coli. Viruses 12:1470. doi: 10.3390/v12121470
Kropinski, A. M., Arutyunov, D., Foss, M., Cunningham, A., Ding, W., Singh, A., et al. (2011). Genome and Proteome of Campylobacter jejuni Bacteriophage NCTC 12673. Appl. Environ. Microbiol. 77, 8265–8271. doi: 10.1128/Aem.05562-11
Lee, M. D., and Newell, D. G. (2006). Campylobacter in poultry: filling an ecological niche. Avian Dis. 50, 1–9. doi: 10.1637/7474-111605R.1
Levin, B. R., Stewart, F. M., and Chao, L. (1977). Resource-limited growth, competition, and predation - a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111, 3–24. doi: 10.1086/283134
Loessner, H., Schlattmeier, I., Anders-Maurer, M., Bekeredjian-Ding, I., Rohde, C., Wittmann, J., et al. (2020). Kinetic fingerprinting links bacteria-phage interactions with emergent dynamics: rapid depletion of Klebsiella pneumoniae indicates phage synergy. Antibiotics 9:408. doi: 10.3390/antibiotics9070408
Luber, P., Brynestad, S., Topsch, D., Scherer, K., and Bartelt, E. (2006). Quantification of Campylobacter species cross-contamination during handling of contaminated fresh chicken parts in kitchens. Appl. Environ. Microbiol. 72, 66–70. doi: 10.1128/AEM.72.1.66-70.2006
Mabelebele, M., Norris, D., Brown, D., Ginindza, M. M., and Ngambi, J. W. (2017). Breed and sex differences in the gross anatomy, digesta pH and histomorphology of the gastrointestinal tract of Gallus Gallus Domesticus. Braz. J. Poult. Sci. 19, 339–346. doi: 10.1590/1806-9061-2016-0275
Moore, J. E., Corcoran, D., Dooley, J. S., Fanning, S., Lucey, B., Matsuda, M., et al. (2005). Campylobacter. Vet. Res. 36, 351–382. doi: 10.1051/vetres:2005012
Mulder, A. C., Franz, E., de Rijk, S., Versluis, M. A. J., Coipan, C., Buij, R., et al. (2020). Tracing the animal sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res. 187:116421. doi: 10.1016/j.watres.2020.116421
Nachamkin, I. (2002). Chronic effects of Campylobacter infection. Microbes Infect. 4, 399–403. doi: 10.1016/S1286-4579(02)01553-8
Nauta, M. J., Jacobs-Reitsma, W. F., and Havelaar, A. H. (2007). A risk assessment model for Campylobacter in broiler meat. Risk Anal. 27, 845–861. doi: 10.1111/j.1539-6924.2006.00834.x
Nawaz, M. S., Khan, S. A., Khan, A. A., Nayak, R., Steele, R., Paine, D., et al. (2003). Molecular characterization of fluoroquinolone-resistant Campylobacter spp. isolated from poultry. Poult. Sci. J. 82, 251–258. doi: 10.1093/ps/82.2.251
Newell, D. G., Elvers, K. T., Dopfer, D., Hansson, I., Jones, P., James, S., et al. (2011). Biosecurity-based interventions and strategies to reduce Campylobacter spp. on poultry farms. Appl. Environ. Microbiol. 77, 8605–8614. doi: 10.1128/Aem.01090-10
Newell, D. G., and Fearnley, C. (2003). Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69, 4343–4351. doi: 10.1128/aem.69.8.4343-4351.2003
Nowaczek, A., Urban-Chmiel, R., Dec, M., Puchalski, A., Stepien-Pysniak, D., Marek, A., et al. (2019). Campylobacter spp. and bacteriophages from broiler chickens: characterization of antibiotic susceptibility profiles and lytic bacteriophages. Microbiologyopen 8:e784. doi: 10.1002/mbo3.784
Oechslin, F. (2018). Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10:351. doi: 10.3390/v10070351
Orquera, S., Golz, G., Hertwig, S., Hammerl, J., Sparborth, D., Joldic, A., et al. (2012). Control of Campylobacter spp. and Yersinia enterocolitica by virulent bacteriophages. J. Mol. Genet. Med. 6, 273–278. doi: 10.4172/1747-0862.1000049
Polaska, M., and Sokolowska, B. (2019). Bacteriophages - a new hope or a huge problem in the food industry. AIMS Microbiol. 5, 324–346. doi: 10.3934/microbiol.2019.4.324
R Core Team (2020). R: A Language And Environment For Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
Rajnovic, D., Munoz-Berbel, X., and Mas, J. (2019). Fast phage detection and quantification: an optical density-based approach. PLoS One 14:e0216292. doi: 10.1371/journal.pone.0216292
Richards, P. J., Connerton, P. L., and Connerton, I. F. (2019). Phage biocontrol of Campylobacter jejuni in chickens does not produce collateral effects on the gut microbiota. Front. Microbiol. 10:476. doi: 10.3389/fmicb.2019.00476
Sails, A. D., Wareing, D. R. A., Bolton, F. J., Fox, A. J., and Curry, A. (1998). Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. J. Med. Microbiol. 47, 123–128. doi: 10.1099/00222615-47-2-123
Sambrook, J., and Russell, D. W. (2001). Molecular cloning: a laboratory manual (3 - volume set). New York: Cold Spring Harbor Laboratory Press.
Sasaki, Y., Maruyama, N., Zou, B., Haruna, M., Kusukawa, M., Murakami, M., et al. (2013). Campylobacter cross-contamination of chicken products at an abattoir. Zoonoses Public Health 60, 134–140. doi: 10.1111/j.1863-2378.2012.01509.x
Schoch, C. L., Ciufo, S., Domrachev, M., Hotton, C. L., Kannan, S., Khovanskaya, R., et al. (2020). NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database 2020:baaa062. doi: 10.1093/database/baaa062
Scott, A. E., Timms, A. R., Connerton, P. L., El-Shibiny, A., and Connerton, I. F. (2007). Bacteriophage influence Campylobacter jejuni types populating broiler chickens. Environ. Microbiol. 9, 2341–2353. doi: 10.1111/J.1462-2920.2007.01351.x
Shreeve, J. E., Toszeghy, M., Pattison, M., and Newell, D. G. (2000). Sequential spread of Campylobacter infection in a multipen broiler house. Avian Dis. 44, 983–988. doi: 10.2307/1593076
Skarp, C. P. A., Hanninen, M. L., and Rautelin, H. I. K. (2016). Campylobacteriosis: the role of poultry meat. Clin. Microbiol. Infect. 22, 103–109. doi: 10.1016/j.cmi.2015.11.019
Sorensen, M. C. H., Gencay, Y. E., Birk, T., Baldvinsson, S. B., Jackel, C., Hammerl, J. A., et al. (2015). Primary isolation strain determines both phage type and receptors recognised by Campylobacter jejuni bacteriophages. PLoS One 10:e0116287. doi: 10.1371/journal.pone.0116287
Sorensen, M. C. H., Van Alphen, L. B., Fodor, C., Crowley, S. M., Christensen, B. B., Szymanski, C. M., et al. (2012). Phase variable expression of capsular polysaccharide modifications allows Campylobacter jejuni to avoid bacteriophage infection in chickens. Front. Cell. Infect. Microbiol. 2:11. doi: 10.3389/fcimb.2012.00011
Sorensen, M. C. H., Vitt, A., Neve, H., Soverini, M., Ahern, S. J., Klumpp, J., et al. (2021). Campylobacter phages use hypermutable polyG tracts to create phenotypic diversity and evade bacterial resistance. Cell Rep. 35:109214. doi: 10.1016/j.celrep.2021.109214
Svihus, B., and Itani, K. (2019). Intestinal Passage and Its Relation to Digestive Processes. J. Appl. Poult. Res. 28, 546–555. doi: 10.3382/japr/pfy027
Thung, T. Y., Lee, E., Mahyudin, N. A., Radzi, C. W. J. W. M., Mazlan, N., Tan, C. W., et al. (2020). Partial characterization and in vitro evaluation of a lytic bacteriophage for biocontrol of Campylobacter jejuni in mutton and chicken meat. J. Food Saf. 40:e12770. doi: 10.1111/jfs.12770
Tolen, T. N., Xie, Y., Hairgrove, T. B., Gill, J. J., and Taylor, T. M. (2018). Evaluation of commercial prototype bacteriophage intervention designed for reducing O157 and non-O157 shiga-toxigenic Escherichia coli (STEC) on beef cattle hide. Foods 7:114. doi: 10.3390/foods7070114
Ushanov, L., Lasareishvili, B., Janashia, I., and Zautner, A. E. (2020). Application of Campylobacter jejuni phages: challenges and perspectives. Animals 10:279. doi: 10.3390/ani10020279
Valentine, R. C., Shapiro, B. M., and Stadtman, E. R. (1968). Regulation of glutamine synthetase. XII. electron microscopy of the enzyme from Escherichia coli. Biochemistry 7, 2143–2152. doi: 10.1021/bi00846a017
Wagenaar, J. A., Van Bergen, M. A. P., Mueller, M. A., Wassenaar, T. M., and Carlton, R. A. (2005). Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet. Microbiol. 109, 275–283. doi: 10.1016/j.vetmic.2005.06.002
Xie, Y. C., Wahab, L., and Gill, J. J. (2018). Development and validation of a microtiter plate-based assay for determination of bacteriophage host range and virulence. Viruses 10:189. doi: 10.3390/v10040189
Zachary, J. S., Matthew, R. T., Kevin, M., and Dominic, S. (2020). The Virulence Index: a Metric for Qualitative Analysis of Phage Virulence. Phage 1, 27–36. doi: 10.1089/phage.2019.0001
Zampara, A., Sørensen, M. C. H., Elsser-Gravesen, A., and Brøndsted, L. (2017). Significance of phage-host interactions for biocontrol of Campylobacter jejuni in food. Food Control 73, 1169–1175. doi: 10.1016/j.foodcont.2016.10.033
Zbikowska, K., Michalczuk, M., and Dolka, B. (2020). The use of bacteriophages in the poultry industry. Animals 10:872. doi: 10.3390/ani10050872
Keywords: Campylobacter virus, phage biocontrol, one health approach, phage therapy, novel antimicrobials, poultry meat, efficiency of plaquing, bacteriophages
Citation: Steffan SM, Shakeri G, Hammerl JA, Kehrenberg C, Peh E, Rohde M, Jackel C, Plotz M and Kittler S (2021) Isolation and Characterization of Group III Campylobacter jejuni–Specific Bacteriophages From Germany and Their Suitability for Use in Food Production. Front. Microbiol. 12:761223. doi: 10.3389/fmicb.2021.761223
Received: 19 August 2021; Accepted: 05 November 2021;
Published: 09 December 2021.
Edited by:
Greta Gölz, Freie Universität Berlin, GermanyReviewed by:
Martine Camilla Holst Sørensen, University of Copenhagen, DenmarkJessica C. Sacher, Phage Directory LLC., United States
Copyright © 2021 Steffan, Shakeri, Hammerl, Kehrenberg, Peh, Rohde, Jackel, Plotz and Kittler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie Kittler, c29waGllLmtpdHRsZXJAdGloby1oYW5ub3Zlci5kZQ==
 Severin Michael Steffan
Severin Michael Steffan Golshan Shakeri2
Golshan Shakeri2 Jens Andre Hammerl
Jens Andre Hammerl Corinna Kehrenberg
Corinna Kehrenberg Manfred Rohde
Manfred Rohde Claudia Jackel
Claudia Jackel Sophie Kittler
Sophie Kittler