- 1School of Life Sciences, Arizona State University, Tempe, AZ, United States
- 2Biodesign Center for Fundamental and Applied Microbiomics, Arizona State University, Tempe, AZ, United States
- 3Center for Biological Physics, Arizona State University, Tempe, AZ, United States
- 4Department of Engineering Science, The University of Electro-Communications, Tokyo, Japan
The Fo ATP synthase, the bacterial flagellar motor, and the bacterial type 9 secretion system (T9SS) are the three known proton motive force driven biological rotary motors. In this review, we summarize the current information on the nuts and bolts of T9SS. Torque generation by T9SS, its role in gliding motility of bacteria, and the mechanism via which a T9SS-driven swarm shapes the microbiota are discussed. The knowledge gaps in our current understanding of the T9SS machinery are outlined.
The Diversity of Bacterial Protein Secretion Systems
The hydrophobic lipid bilayer of the bacterial cell membrane stops proteins from diffusing outside the cell. However, physiological processes such as the degradation and uptake of complex extracellular nutrition sources, adhesion to biotic and abiotic surfaces, colonization, motility, virulence, and interbacterial antagonism require transport of proteins across membranes. Protein secretion systems serve this purpose and form a pore that provides a gateway to a select group of proteins. Thus far, eleven bacterial protein secretion systems have been discovered (Figure 1). Additionally, the Sec and Tat transporters (Kudva et al., 2013), Chaperon-usher pathway (Waksman and Hultgren, 2009), the YidC insertase (Hennon et al., 2015), Sortases (Spirig et al., 2011), and a TonB-dependent machinery (Gómez-Santos et al., 2019) are also involved in bacterial protein export. Majority of this review is focused on the bacterial Type 9 Secretion System (T9SS), its mechanism of rotation, gliding motility, and collective behavior. However, without an appreciation of the diversity of bacterial secretion machineries, the beauty of T9SS can be lost on the reader. Hence, we initiate this review with a brief and up-to date overview of bacterial protein secretion machineries.
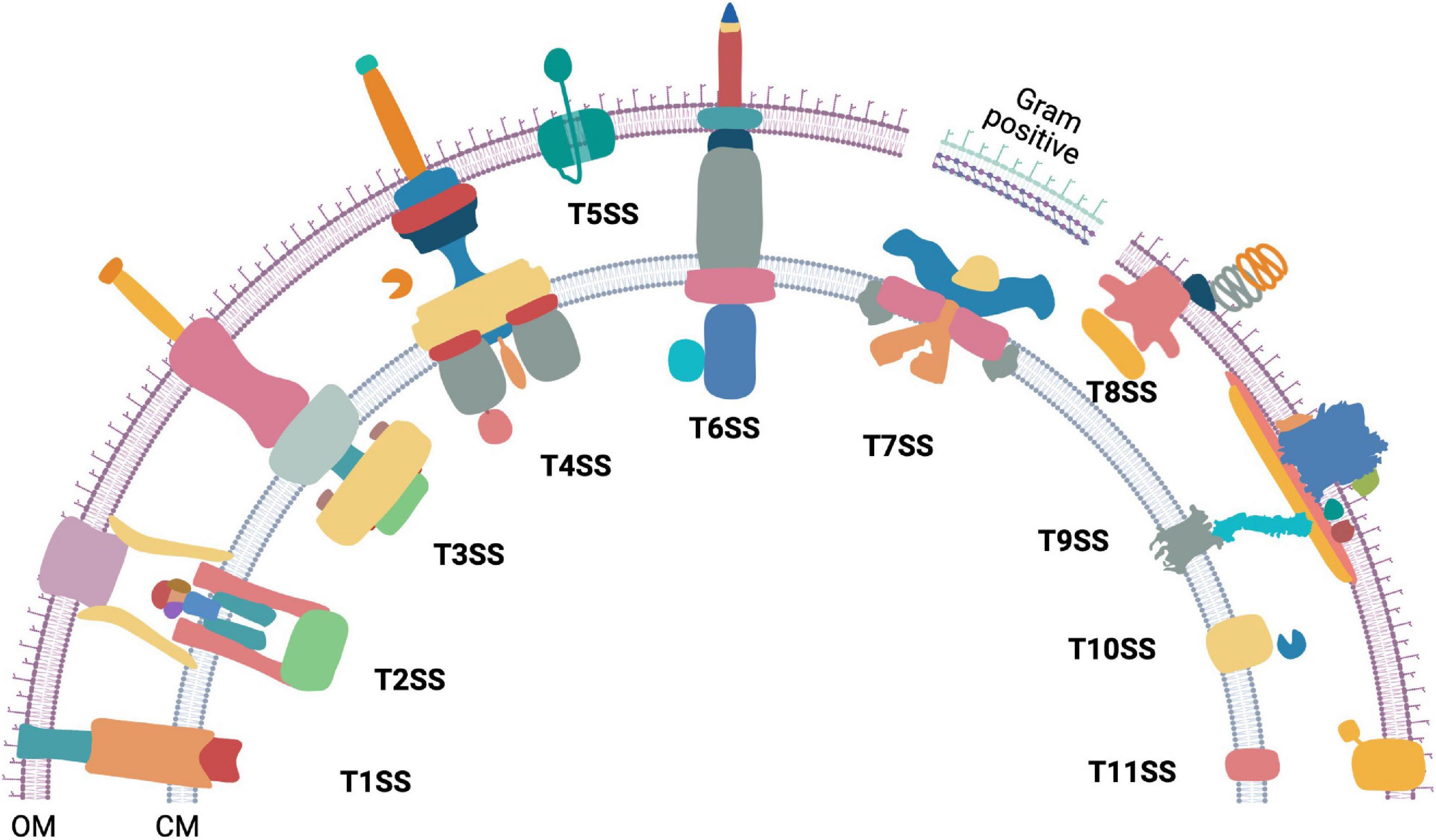
Figure 1. The diversity of bacterial protein secretion systems. Graphical representation of the currently known bacterial protein secretion systems ranging from Type 1 to Type 11 and their localization in the outer (OM) and cytoplasmic membrane (CM). Except T7SS (Gram positive), all the above shown secretion systems are found in Gram negative bacteria. Occurrence of T7SS mediated protein secretion is found within some Gram positive members of the phylum Actinobacteria that have an outer lipid layer.
Type 1 to Type 11 bacterial secretion systems are known to facilitate extracellular transport of proteins. Some bacterial secretion systems work in conjunction with either the Sec or the Tat transporters, whereas others work independently. The bacterial Sec protein export pathway allows transport of unfolded proteins through the cytoplasmic membrane (Papanikou et al., 2007; Kusters and Driessen, 2011; Denks et al., 2014) while the Twin Arginine translocation (Tat) system allows transport of folded proteins across the cytoplasmic membrane (Palmer and Berks, 2012). Interestingly, some protein secretion systems are associated with machineries that power bacterial motility. Examples include the Type 2 Secretion System (T2SS) associated type IV pilus (Craig et al., 2019), the Type 3 Secretion System (T3SS) associated with the flagellar motor (Abrusci et al., 2013), and the T9SS that is integral to gliding motility of Bacteroidetes (Sato et al., 2010; Shrivastava et al., 2013; Zhu and McBride, 2014; Kita et al., 2016; Lauber et al., 2018; Shrivastava and Berg, 2020; Hennell James et al., 2021).
The Type 1 Secretion System (T1SS) is comprised of an ABC transporter-like protein complex spanning the cytoplasmic membrane. It is coupled with a viral fusion protein and a β-barrel channel that spans the outer membrane. T1SS enables secretion of unfolded protein substrates from the cytoplasm to the extracellular milieu (Spitz et al., 2019). The T2SS secretes proteins that are already exported to the periplasm via Sec or Tat pathway. The multiprotein piston-like pseudopilus component of the T2SS pushes its folded protein substrates out of the outer membrane secretin in a process powered by ATP hydrolysis (Korotkov and Sandkvist, 2019; Naskar et al., 2021). The T3SS forms an injectisome and transports proteins from the cytosol either to the extracellular milieu or across the plasma membrane of a eukaryotic host cell (Portaliou et al., 2016). The core of both the bacterial flagellar apparatus and the injectisome have a T3SS. While there is some debate on the evolution of injectisome and the flagellum, it has been proposed that the flagellum is the ancestor of the injectisome-T3SS complex (Diepold and Armitage, 2015). The Type 4 Secretion System (T4SS) is found in both eubacteria and archaea. It exports both proteins and DNA from the cytosol of T4SS containing bacteria to either a prokaryotic or a eukaryotic host cell (Li et al., 2019). Additionally, a class of T4SS is also employed by bacteria to take up extracellular DNA (Hofreuter et al., 2001; Li et al., 2019). The Type 5 Secretion System (T5SS), also known as autotransporter, contains a single protein that spans the Gram negative bacterial outer membrane. The translocator domain of the T5SS protein is a β-barrel. In most cases, its secretory substrate (passenger domain) is a polypeptide chain contiguous to the translocator domain (Fan et al., 2016; Meuskens et al., 2019). The Type 6 Secretion System (T6SS) is formed by the association of homologs of T4SS components and bacteriophage contractile tails. It is widely employed to inject effector proteins from the cytoplasm directly across other bacterial or eukaryotic host membranes (Cianfanelli et al., 2016; Coulthurst, 2019). The Type 7 Secretion System (T7SS) is found in Gram positive bacteria of the genus Mycobacterium and other members of the phylum Actinobacteria. Five paralogous esx loci are found in Mycobacteria and a recent high resolution Cryo-EM structure of an Esx-5 paralog revealed the formation of an inner membrane pore (Bunduc et al., 2020; Beckham et al., 2021; Rivera-Calzada et al., 2021). The Type 8 Secretion System (T8SS), widely known as the curli biogenesis pathway spans the outer membrane of Gram-negative bacteria. It secretes monomeric curli particles and enables their extracellular assembly and nucleation (Van Gerven et al., 2015; Bhoite et al., 2019). The Type 9 Secretion System (T9SS) spans both membranes and is found in the Gram negative Fibrobacteres-Chlorobi-Bacteroidetes superphylum. The substrates are delivered to the periplasm by Sec transport pathway and T9SS secretes them across the outer membrane. The T9SS contains a motor that drives secretion. This is one of the only three known biological rotary motors driven by a proton motive force (pmf) and is the focus of this review. The components of Type 10 Secretion System (T10SS) comprise of holins and peptidoglycan modifying enzymes. T10SS secretes hydrolytic enzymes and toxins (Palmer et al., 2021). Recently, the presence of a Type 11 Secretion System (T11SS) was proposed. It comprises of DUF560 family proteins and it transports periplasmic cargo across the outer membrane (Grossman et al., 2021).
A Brief History of Type 9 Secretion System
Proteins that are now known to form the core of the bacterial type 9 secretion system (T9SS) were discovered around similar timelines in two model organisms Flavobacterium johnsoniae and Porphyromonas gingivalis. Transposon mutagenesis led to the discovery that F. johnsoniae GldK, GldL, GldM, and GldN are required for motility and chitin utilization (Braun et al., 2005). Similarly, it was identified that P. gingivalis PorT (Sato et al., 2005) and SprA/Sov are required for the transport of gingipain proteases (Saiki and Konishi, 2010). Later, it was shown that PorT of F. johnsoniae is also required for the secretion of chitinase and that GldK, GldL, GldM, and GldN form a multi-protein complex. Hence, it was inferred that these set of proteins form a secretion system that was initially named as the Por secretion system (PorSS) (Sato et al., 2010). It was later found that these proteins are prevalent in the Bacteroidetes phylum (McBride and Zhu, 2013; Shrivastava, 2013) and they are required for secretion of multiple classes of proteins. Because the components of the T9SS are not similar to those of other secretion systems, it was renamed as T9SS (Shrivastava et al., 2013). In recent years, the T9SS field has moved forward by leaps and bounds. We now know that T9SS forms a rotary machinery that enables gliding motility (Shrivastava et al., 2015) and we know the identity and structure of the ion channel that powers rotation (Shrivastava et al., 2015; Hennell James et al., 2021). Additionally, structures of several T9SS proteins have recently been resolved (Gorasia et al., 2016; Lauber et al., 2018; Leone et al., 2018; Trinh et al., 2020; Hennell James et al., 2021). T9SS secretes many enzymes, adhesins, and virulence factors. The signatures of T9SS secreted proteins have recently been characterized (Sato et al., 2013; de Diego et al., 2016; Kulkarni et al., 2017, 2019).
Diversity of Substrates Secreted by Type 9 Secretion System
Type 9 secretion system driven protein export occurs in conjunction with the Sec transport machinery. T9SS substrates have a N-terminal Sec signal peptide and a C-terminal T9SS signal sequence (CTD). Via the Sec system, T9SS substrates reach the periplasm and the N-terminal Sec signal peptide is cleaved. T9SS CTD containing periplasmic proteins are exported either to the extracellular milieu or they attach to the outer membrane (OM) (Slakeski et al., 2011; Kulkarni et al., 2017, 2019). T9SS secreted extracellular proteins lose their CTD (Sato et al., 2013) and it is unclear whether CTD cleavage occurs during or after transport. The T9SS CTD is divided into two protein domain families namely TIGR04183 (Type A CTD) and TIGR04131 (Type B CTD). Type A CTD have seven β strands and lg-like folds which are thought to interact with the structural components of T9SS (de Diego et al., 2016; Lasica et al., 2016). Super folder GFP (sfGFP) fused with T9SS CTD and N-terminal Sec signal peptide is secreted by T9SS (Kulkarni et al., 2017, 2019) which further confirms that substrate recognition by T9SS only occurs due to the CTD. Additionally, a T9SS secreted chitinase possesses a CTD different from both Type A and Type B CTD (Kharade and McBride, 2014). We found that on InterPro (Blum et al., 2021) 179,000 different bacterial proteins are annotated to have TIGR04183 domain and 28,000 different proteins are annotated to have TIGR04131 domain. Many of T9SS encoding proteomes belong to bacteria of the phylum Bacteroidetes. The environmental microbe F. johnsoniae UW101 encodes 40 Type A and 12 Type B CTD containing proteins. These include the motility adhesins SprB (Nelson et al., 2008) and RemA (Shrivastava et al., 2012). Additionally, multiple enzymes including a chitinase are secreted by the T9SS of F. johnsoniae (Kharade and McBride, 2014, 2015). SprB is one of the largest known adhesin protein (669 kDa) and cells lacking SprB are severely deficient for gliding motility (Nelson et al., 2008). SprB moves spirally on the bacterial cell-surface (Nakane et al., 2013; Shrivastava et al., 2016) and is driven by pmf (Nakane et al., 2013). Adhesion of SprB to an external substratum enables screw-like motion of the rod-shaped F. johnsoniae (Shrivastava et al., 2016). The human oral microbe P. gingivalis W83 encodes 17 Type A CTD-containing proteins (including the virulent gingipain proteases) and one Type B CTD-containing protein. In contrast, another prominent human oral microbe Capnocytophaga ochracea ATCC 27872 encodes 2 Type A CTD containing proteins and 8 Type B CTD containing proteins.
Many of the shared T9SS components identified in F. johnsoniae and P. gingivalis were given different names (e.g., Gld vs. Por). These nuts and bolts of T9SS enable both gliding motility and protein secretion (Figure 2A). Below, we compile information on T9SS proteins from both model organisms F. johnsoniae and P. gingivalis.
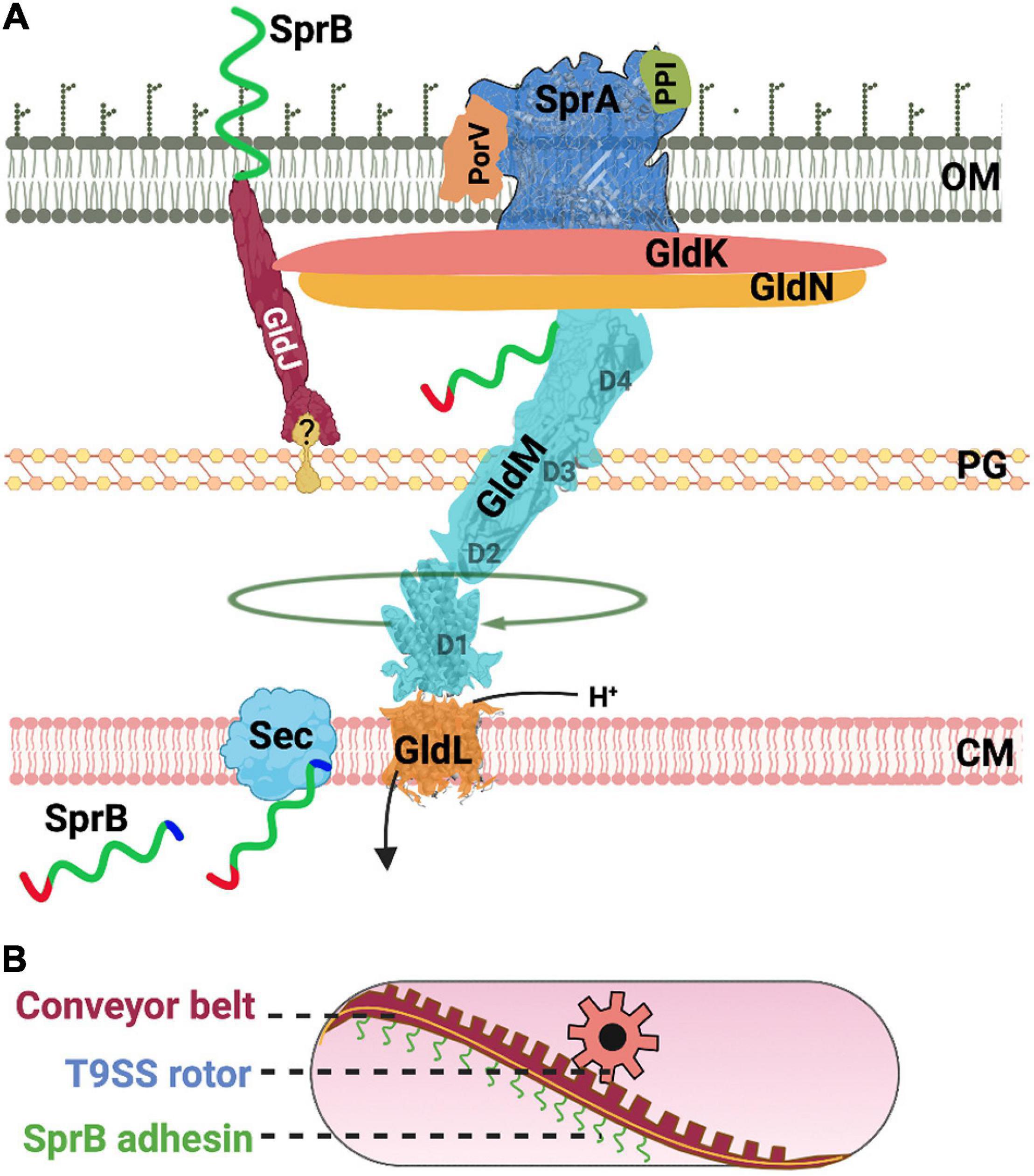
Figure 2. Structure and function of the T9SS. (A) A cartoon of the nuts and bolts of the T9SS motor that drives protein secretion and gliding motility. T9SS substrates (SprB shown as an example) are transported to the periplasm via the Sec transport pathway. The CTD of T9SS substrates is cleaved during transport. A recent model suggests that the proton channel GldL powers the rotation of T9SS. (B) A cartoon of the molecular rack and pinion machinery that drives gliding motility. A model based on recent data suggests that the rotary T9SS pinion drives a cell-surface conveyor belt (rack). Cell-surface adhesins such as SprB are secreted by T9SS and are loaded onto the conveyor belt. Interaction of SprB with an external substratum results in screw-like gliding motility of the bacterial cell.
The Nuts and Bolts That Form the Core of Rotary Type 9 Secretion System
GldK/PorK (Fjoh_1853/PGN_1676) is attached to the outer membrane via a lipid anchor and is localized in the periplasmic space (Shrivastava et al., 2013; Vincent et al., 2017). GldK/PorK of P. gingivalis forms a ring that has an outer diameter of 50 nm and a pore of 35 nm. This ring comprises 32–36 GldK/PorK monomers (Gorasia et al., 2016, Gorasia D. et al., 2020).
GldN/PorN (Fjoh_1856/PGN_1673) forms a ring and is associated with GldK/PorK in a 1:1 stoichiometry (Gorasia et al., 2016, Gorasia D. et al., 2020). GldN/PorN is localized in the periplasm. Flavobacterium johnsoniae has a paralog of GldN/PorN called as GldO. The two proteins are partially redundant. In the genomic context, gldO of F. johnsoniae is located next to gldN (Rhodes et al., 2010).
GldL/PorL (Fjoh_1854/PGN_1675) is a cytoplasmic membrane protein (Shrivastava, 2013) and is found near the axis of rotation of a tethered cell (Shrivastava and Berg, 2020). Flavobacterium johnsoniae cells containing amino acid substitution of protonatable residues of GldL/PorL (E49Q and E49D) are completely deficient for secretion (Hennell James et al., 2021). Additionally, secretion is reduced for strains that contain GldL/PorL Y13A and K27A substitutions. This suggests that GldL/PorL is an ion channel that generates pmf driven power stroke for the rotation of T9SS (Hennell James et al., 2021). Aspartate and glutamate residues of ion channels are known to play important roles in the generation of pmf (Zhou et al., 1998; Vorburger et al., 2009). GldL/PorL has two glutamates, one is in the transmembrane helix 2 (TMH2) (E49) and the other (E59) is located between TMH2 and cytoplasmic domain. Cells with GldL/PorL E49A substitution are unable to secrete SprB and fail to attach to a glass surface. Cells with GldL/PorL E59A substitution secrete SprB to the surface yet they are non-motile (Vincent et al., 2021).
GldM/PorM (Fjoh_1855/PGN_1674) interacts with GldL/PorL to form an asymmetric complex with two subunits of GldL/PorL surrounded by five subunits of GldM/PorM. It has one transmembrane domain that contains a protonable residue. Y17A substitution in the transmembrane domain of GldM/PorM resulted in reduced secretion. GldM/PorM has four periplasmic domains D1–D4. X-ray crystallography of GldM/PorM showed a hinge between D2 and D3 (Leone et al., 2018) whereas cryo-EM structural determination of truncated GldM/PorM in complex with GldL/PorL showed a bend between D1 and D2 (Hennell James et al., 2021).
The Mechanism of Rotation and Comparison of Type 9 Secretion System With the Bacterial Flagellar Motor
Similar to the flagellar FlgH-FlgI LP ring that forms a molecular bushing, GldK/PorK-N rings of T9SS localize in the outer membrane and periplasmic region. Like GldK/PorK, FlgH is a lipoprotein and like GldN/PorN, FlgI is a periplasmic protein. The LP ring of the flagellar motor interacts with the distal end of the driveshaft (rod), supports stable rotation of the rod, and reduces friction (Yamaguchi et al., 2021). The GldK/PorK-N ring may play a similar role in T9SS rotation. If it does, this implies that T9SS might have a currently undiscovered driveshaft. Alternatively, the mechanism of torque generation could be unique and the rotary component (Gld/PorK-N ring) could be placed on the periplasmic face of the outer membrane of T9SS containing bacteria.
Based on structural data, a model was recently presented where due to asymmetry, GldL/PorL enables rotation of GldM/PorM (Hennell James et al., 2021). The only experimental evidence for rotation of T9SS is from tethered cell analysis (Shrivastava et al., 2015) and it depicts rotation of an outer membrane protein. How does rotary GldM/PorM cross the peptidoglycan barrier and transmit rotation to an outer membrane protein? In one scenario, it might be possible that GldM/PorM might pass through a pore in the peptidoglycan. This pore could be formed by a protein that has a structure like the P ring bushing of the flagellar motor. Given the current structural data, the ring shaped GldN/PorN appears to be a candidate for the bushing. GldM/PorM might be the driveshaft that passes the peptidoglycan via the GldN/PorN bushing, and it can induce rotation of the outer membrane associated GldK/PorK ring. However, one caveat to this model is that GldN/PorN ring is shown to directly interact with the GldK/PorK ring (Gorasia et al., 2016). It is possible that the linkage between GldK/PorK and GldN/PorN is weak and transient. Hence, GldM/PorM driven GldK/PorK might rotate relative to the static GldN/PorN bushing. There is also a possibility that GldN/PorN might not form the bushing and it rotates along with GldK/PorK ring. This scenario implies that another protein might from the pore that allows passage of GldM/PorM through the peptidoglycan. Future structural and biochemical data will help fill the current knowledge gaps regarding the mechanism of rotation of T9SS.
PorE (Multiple homologs in F. johnsoniae/PGN_1296) is a periplasmic protein that is anchored to inner leaflet of outer membrane (Heath et al., 2016). Sequence analysis and modeling suggest that PGN_1296 has four domains (i) a tetratricopeptide repeat domain (TPR, residues 25–149); (ii) a β–propeller domain (WD40, residues 167–435); (iii) a carboxypeptidase regulatory domain-like fold (CRD, residues 441–527) and (iv) an OmpA_C-like putative peptidoglycan-binding domain (PBD) (residues 534–668). The OmpA_C (PBD) structure of PorE consists of a three-stranded β-sheet (β1–3) and five α-helices (α1–5) (Trinh et al., 2020).
The MotA–MotB stator units of the flagellar motor are attached to the cell wall by a peptidoglycan binding domain of MotA. Neither GldL/PorL nor GldM/PorM contain a detectable peptidoglycan binding domain. For the Gld/PorLM complex to function as a stator unit, attachment to a rigid part of the cell is necessary. This might be achieved by the PBD of PorE. However, there is no direct evidence for interaction between PorE and Gld/PorLM and whether PorE enables T9SS anchoring remains to be seen. P. gingivalis cells lacking PorE have reduced T9SS activity (Heath et al., 2016). The Role of PorE homolog of F. johnsoniae is unknown. There are multiple homologs of PorE in F. johnsoniae. Future experimental evidence might clear our understanding of anchoring of GldL/PorL. In an alternate scenario, it is possible that PorE might not interact with core T9SS proteins and GldL/PorL might be anchored via a cytoskeleton protein.
SprF/PorP (Multiple homologs in F. johnsoniae/PGN_1677) of P. gingivalis has been reported to interact individually with PorE, Gld/PorK and GldM/PorM (Gorasia D. et al., 2020) and could be the connection between T9SS and the peptidoglycan bound PorE. However, if this scenario is correct, neither GldK/PorK ring or GldM/PorM can rotate. It might imply that like LP rings of the flagellar motor, both GldK/PorK and GldN/PorN act as bushings. Alternatively, as discussed below, the interaction of SprF/PorP with PorE might not have any role in motility and it might only occur during the process of protein secretion.
SprF/PorP is required for secretion of TIGR04131 domain containing proteins. In the genomic context, a cell often has multiple copies of SprF/PorP and genes encoding SprF/PorP are often localized next to genes encoding TIGR04131 domain containing proteins (Kulkarni et al., 2019). As an example, one F. johnsoniae SprF encoding gene (Fjoh_0978) is localized downstream of the mobile cell-surface adhesin SprB encoding gene (Fjoh_0979). SprF/PorP is required for secretion of SprB, and it might be possible that SprF interacts with SprB during transport. If correct, this might imply that the interaction of SprF/PorP with GldK/PorK and GldM/PorM happens solely during transport and that it might help the T9SS pore recognize TIGR04131 containing substrates.
SprA/Sov (Fjoh_1653/PGN_0832) is required for secretion SprB, RemA, chitinase, and gingipains (Saiki and Konishi, 2007; Shrivastava et al., 2013). It forms the largest single polypeptide outer membrane ß-barrel (36 strand) known and acts as the protein translocon of the T9SS. SprA is found in two states one with PorV and the other with a plug protein. In both the states, SprA is bound to a lipoprotein Peptidyl-prolyl cis-trans isomerase (PPI). SprA forms a transmembrane β-barrel structure which rises about 20 Å above the membrane on the cell-surface. Two folded inserts are present between β-barrel strand 7–8 and 11–12. They form a 50 Å high cap structure that seals the extracellular end of the barrel. The cap insert consist of two domains with a β-sheet structure and the first domain surrounding the second domain from both the sides (Lauber et al., 2018).
PorV (Fjoh_1555/PGN_0023) is bound to the outer membrane pore of SprA and acts as a gate. There are highly conserved packing interactions between PorV and the lateral opening of SprA. When in complex with SprA, the PorV barrel is tilted by 25°. While in a free state, it is proposed that PorV adopts a more vertical position in the membrane bilayer (Lauber et al., 2018). F. johnsoniae cells lacking PorV are deficient in secretion of the motility adhesin RemA, the chitinase ChiA, and many other proteins. However, cells lacking PorV have the ability to secrete SprB hence they form spreading colonies on agar (Kharade and McBride, 2015).
Peptidyl-prolyl cis-trans isomerase (PPI: Fjoh_4997/PGN_0742/PGN_0744) binds to the lateral face of SprA opposite to PorV. PPIases are chaperones which catalyze the cis/trans isomerization of proline and act as a regulatory switch during protein folding (Dunyak and Gestwicki, 2016). Deletion of this PPI has no notable effect on T9SS function or gliding motility (Lauber et al., 2018). However, another predicted PPI, GldI (discussed later), is essential for motility.
Plug (Fjoh_1759/putative PGN_0144) can be bound to SprA ß-barrel when SprA is not in a complex with PorV. The plug protein mutant neither has gliding defect nor is defective in T9SS secretion. When PorV is not bound to SprA, the plug prevents leakage of small molecules through the SprA pore (Lauber et al., 2018).
SprT/PorT (Fjoh_1466/PGN_0778) is an outer membrane protein (Nguyen et al., 2009). Weak activity of gingipains Kgp and RgpA are observed in lysates and culture supernatants of P. gingivalis cells that lack PorT (Sato et al., 2005). F. johnsoniae cells lacking SprT form non-spreading colonies on agar. They fail to secrete SprB to the cell surface, and are deficient in chitin utilization (Sato et al., 2010).
SprE/PorW (Fjoh_1051/PGN_1877) is required for the secretion of an array of T9SS proteins (Gorasia D. G. et al., 2020). F. johnsoniae cells lacking SprE exhibit very little motility and are deficient in chitin utilization (Rhodes et al., 2011). It is reported that PorW of P. gingivalis acts as a bridge between Sov/SprA translocon and PorK/N rings (Gorasia et al., 2022).
Additional Proteins Required for Gliding Motility
It is proposed that a molecular rack and pinion assembly enables gliding motility of Bacteroidetes (Shrivastava and Berg, 2020). Here, T9SS rotary motor is the pinion that pushes a conveyor belt on the cell-surface (Figure 2B). As described above, the identity of proteins that form T9SS is now well-known, however, the nature of proteins that form the rack or the conveyor belt are mysterious. Comparative genomic analysis has shown that non-motile Bacteroidetes lack several accessory Gld proteins. It is possible that the additional Gld proteins help in the formation of the rack or the conveyor belt (rack) which is only found in motile Bacteroidetes. The localization and function of the additional Gld proteins are described below.
GldJ (Fjoh_1557) is a lipoprotein that appears to form helical tracks on the cell-surface (Braun and McBride, 2005) and it might be one of the central components of the gliding machinery. GldJ is required for the stability of GldK (Johnston et al., 2018). However, F. johnsoniae cells lacking GldK/PorK, GldL/PorL, GldM/PorM, and GldN/PorN have wild-type like levels of GldJ (Braun et al., 2005). Cells lacking either 8 or 13 C-terminal amino acids (AA) of GldJ have functional T9SS, however, cells lacking C-terminal 13 AA of GldJ are completely non-motile and cells lacking C-terminal 8 AA of GldJ exhibit very little motility. This is the first instance where the motility phenotype is completely separated from T9SS secretion (Johnston et al., 2018). GldJ has 30% sequence similarity with F. johnsoniae GldK/PorK. It is a homolog of a sulfatase activating enzyme (SUMF1) but it lacks the active site of SUMF1 (Braun and McBride, 2005).
GldA (Fjoh_1516) belongs to the ATP binding cassette family of transport proteins (ABC transporter proteins) essential for gliding motility. GldA has Walker “A” and “B” motifs, which are characteristics of an ATP hydrolyzing domain of an ABC transporter (Agarwal et al., 1997). Furthermore, GldA point mutations in ABC transporter putative active site G40R result in reduction of cellular levels of GldJ and GldK/PorK. Similarly, a point mutation in ABC transporter signature sequence LSKGYRQR (site of point mutation underlined) shows that a functional GldA is required for the stability of both GldJ and GldK/PorK (Braun et al., 2005; Johnston et al., 2018).
GldB (Fjoh_1793) is a lipoprotein essential for gliding motility and cells lacking GldB have a defective T9SS (Hunnicutt and McBride, 2000). GldB is required for the stability of both GldJ and GldK/PorK (Braun and McBride, 2005; Johnston et al., 2018).
GldD (Fjoh_1540) is a lipoprotein essential for gliding motility (Hunnicutt and McBride, 2001; McBride et al., 2003). Furthermore, F. johnsoniae cells lacking GldD do not propel latex beads. GldD is required for the stability of both GldJ and GldK/PorK (Braun and McBride, 2005; Johnston et al., 2018).
GldF (Fjoh_2722) is essential for gliding motility (Hunnicutt et al., 2002). Both GldF and GldG are required for membrane localization of GldA (Agarwal et al., 1997; Hunnicutt et al., 2002). GldF is required for the stability of GldK/PorK (Johnston et al., 2018).
GldG (Fjoh_2721). Cells lacking GldG are non-motile (Hunnicutt et al., 2002). GldG is predicted to be a transmembrane protein that interacts with GldA to form an ABC transporter (Agarwal et al., 1997; Hunnicutt et al., 2002).
GldH (Fjoh_0890) is a lipoprotein essential for gliding motility. Cell lacking GldH do not propel latex beads and are unable to utilize chitin (McBride et al., 2003). GldH is required for the stability of GldJ and GldK/PorK (Braun and McBride, 2005; Johnston et al., 2018).
GldI (Fjoh_2369) mutants form non-spreading colonies on agar. GldI has a PPI domain (McBride and Braun, 2004). Cells lacking GldI do not propel latex beads and are unable to utilize chitin. GldI is required for the stability of GldJ and GldK/PorK (Braun and McBride, 2005; Johnston et al., 2018).
Accessory Proteins Not Required for Gliding Motility
Evolutionary forces can drive modifications in components of biological machineries from different model organisms. As examples, the shape of bacterial flagellar motor varies amongst organisms (Chen et al., 2011; Beeby et al., 2016) and the Fo ATP synthase varies across taxa (Nirody et al., 2020). The Fo rotor c-ring stoichiometry ranges from eight subunits in mammalian mitochondrion (Zhou et al., 2015) to 17 in the bacterium Burkholderia pseudomallei (Schulz et al., 2017). Following this trend of organism-based modifications of molecular machines, three proteins described below interact with the T9SS of P. gingivalis but they are not essential for the functioning of the F. johnsoniae T9SS.
PorU (Fjoh_1556/PGN_0022) PorU of P. gingivalis is a part of the attachment complex that also involves PorZ, PorV, and PorQ. This complex modifies the T9SS substrates with an anionic lipopolysaccharide (A-LPS) and attaches them to the cell surface (Gorasia et al., 2015; Glew et al., 2017). PorU is a sortase which cleaves the CTD signal of the T9SS substrate and attaches the newly generated CTD of T9SS substrate to the cell surface via A-LPS (Glew et al., 2012, 2017). P. gingivalis cells lacking PorU have a partial T9SS defect and gingipain secretion is reduced (Glew et al., 2012). However, the F. johnsoniae PorU is not required for secretion of ChiA, RemA, or SprB, indicating that it does not play an essential role in the secretion of proteins by T9SS of F. johnsoniae (Kharade and McBride, 2015).
PorZ (Fjoh_0707/PGN_0509) and PorQ (Fjoh_2755/PGN_0645) PorZ of P. gingivalis interacts with A-LPS and PorU. Hence, it may play a role in providing A-LPS substrate to PorU (Madej et al., 2021). P. gingivalis cells lacking PorZ are defective in gingipain secretion (Lasica et al., 2016). PorQ is required for the association of PorZ with the outer membrane (Lasica et al., 2016). F. johnsoniae does not need PorQ (Fjoh_2755) for either gliding motility or chitin utilization (Shrivastava, 2013). No porZ mutant of F. johnsoniae has been reported thus far.
Type 9 Secretion System Driven Swarm Behavior of Flavobacteria
It has long been known that F. johnsoniae forms spreading colonies on agar plates (Wolkin and Pate, 1984; Gorski et al., 1993). A recent study showed that the cells at high density are connected with one another and that in a starved environment they move in counterclockwise trajectories (Nakane et al., 2021). A F. johnsoniae swarm exhibits a vortex pattern that spontaneously appears as a lattice that is integrated into a large-scale circular plate (Figure 3A). Notably, the rotational direction of the circular plate is counterclockwise (Figure 3B). This two-dimensional pattern possibly appears due to the collision of cells and it further induces a nematic alignment of dense cells (Sumino et al., 2012). It is hypothesized that swarm behavior of T9SS driven Bacteroidetes may be involved in survival strategies and the efficient search of nutrients (Figure 3C; Nakane et al., 2021). A three-dimensional spherical microcolony was recently studied in a microfluidic device and it was reported that this biofilm-like microcolony self-assembles by the gliding motility (Li et al., 2021). It has also been reported that gliding motility contributes to the robustness of a F. johnsoniae biofilm (Eckroat et al., 2021). The colony of Flavobacterium strain Iridescent 1 (IR1) displays a bright brilliant green structural coloration, which is caused by the 2D close-packed cell arrangement aided by gliding motility (Johansen et al., 2018; Hamidjaja et al., 2020). This may provide photoprotection to either bacteria or host. Additionally, this structure might lead to the optimum cellular organization for the degradation of biological polymers (Johansen et al., 2018). The role of the structural coloration of the colony in the natural environment needs to be studied further. It is evident that our understanding of biofilm formation of F. johnsoniae and its dependence on swarm behavior is currently in its nascent stage. Future investigations in this area might reveal details about bacterial cooperation and behavior.
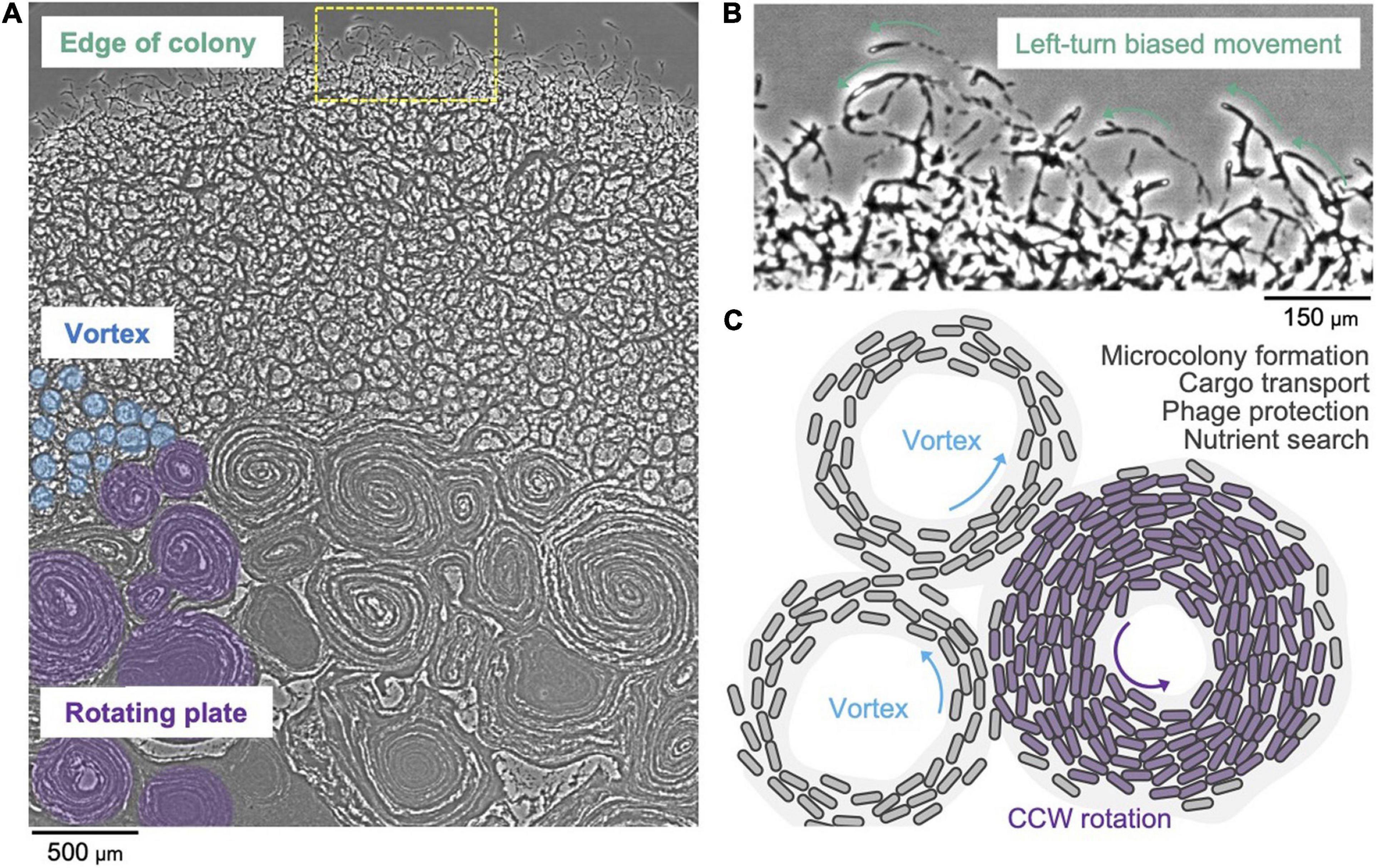
Figure 3. Collective motion of gliding F. johnsoniae. (A) Colony spreading pattern in a starved environment. (B) Left-turn biased cell movement at the edge of the colony. (C) Schematic of the vortex pattern formation and its possible roles.
Benefits of Swarm Behavior to the Microbiota
Type 9 secretion systems are found in bacterial genera that are abundant in the human oral and gut microbiota. Examples include the genera Prevotella, Bacteroides, Parabacteroides, Capnocytophaga, Tanerella, and Porphyromonas. Capnocytophaga sp. exhibit robust gliding motility (Holt et al., 1979; Leadbetter et al., 1979; Shrivastava et al., 2018) and are found in abundance in the oral microbiota. They are prominent members of human supra-gingival and sub-gingival biofilms (Mark Welch et al., 2016). In contrast with the wild-type strains isolated from the human oral microbiota, cells of C. ochracea that lack T9SS are non-motile and are deficient for biofilm formation (Kita et al., 2016).
Capnocytophaga gingivalis swarm in a multilayered counter clockwise vortex pattern (Shrivastava et al., 2018). C. gingivalis cells transport non-motile bacteria as cargo and shape the spatial organization of a polymicrobial community (Shrivastava et al., 2018). A C. gingivalis swarm can also transport bacteriophages as cargo (Ratheesh et al., 2021) and can increase the kinetics of death of a colony of prey bacteria. Via a combination of vortexing, layering, transportation of other bacteria, and phage transportation, T9SS-driven microbes play a crucial role in shaping a polymicrobial community. This amazing molecular machinery holds many secrets, which once unraveled, can significantly enrich our understanding of microbiology.
Author Contributions
AT and AS contributed all sections besides the ones described below and created Figure 2. JG and AS contributed the section on diversity of protein secretion systems and created Figure 1. DN contributed the section on swarm behavior of Flavobacteria and created Figure 3. All authors contributed to the article and approved the submitted version.
Funding
AS was supported by NIH grant DE026826.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrusci, P., Vergara-Irigaray, M., Johnson, S., Beeby, M. D., Hendrixson, D. R., Roversi, P., et al. (2013). Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104. doi: 10.1038/nsmb.2452
Agarwal, S., Hunnicutt, D. W., and McBride, M. J. (1997). Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. U.S.A. 94, 12139–12144. doi: 10.1073/pnas.94.22.12139
Beckham, K. S. H., Ritter, C., Chojnowski, G., Ziemianowicz, D. S., Mullapudi, E., Rettel, M., et al. (2021). Structure of the mycobacterial ESX-5 type VII secretion system pore complex. Sci. Adv. 7:eabg9923. doi: 10.1126/sciadv.abg9923
Beeby, M., Ribardo, D. A., Brennan, C. A., Ruby, E. G., Jensen, G. J., and Hendrixson, D. R. (2016). Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc. Natl. Acad. Sci. U.S.A. 113, E1917–E1926. doi: 10.1073/pnas.1518952113
Bhoite, S., van Gerven, N., Chapman, M. R., and Remaut, H. (2019). Curli biogenesis: bacterial amyloid assembly by the type VIII secretion pathway. Ecosal Plus 8, ecosalplus.ESP-0037-2018. doi: 10.1128/ecosalplus.ESP-0037-2018
Blum, M., Chang, H.-Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. doi: 10.1093/nar/gkaa977
Braun, T. F., and McBride, M. J. (2005). Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J. Bacteriol. 187, 2628–2637. doi: 10.1128/JB.187.8.2628-2637.2005
Braun, T. F., Khubbar, M. K., Saffarini, D. A., and McBride, M. J. (2005). Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J. Bacteriol. 187, 6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005
Bunduc, C. M., Bitter, W., and Houben, E. N. G. (2020). Structure and function of the mycobacterial type VII secretion systems. Annu. Rev. Microbiol. 74, 315–335. doi: 10.1146/annurev-micro-012420-081657
Chen, S., Beeby, M., Murphy, G. E., Leadbetter, J. R., Hendrixson, D. R., Briegel, A., et al. (2011). Structural diversity of bacterial flagellar motors: structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981. doi: 10.1038/emboj.2011.186
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016). Aim, load, fire: the type vi secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Coulthurst, S. (2019). The type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. doi: 10.1099/mic.0.000789
Craig, L., Forest, K. T., and Maier, B. (2019). Type IV pili: dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440. doi: 10.1038/s41579-019-0195-4
de Diego, I., Ksiazek, M., Mizgalska, D., Koneru, L., Golik, P., Szmigielski, B., et al. (2016). The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal β-sandwich domain. Sci. Rep. 6:23123. doi: 10.1038/srep23123
Denks, K., Vogt, A., Sachelaru, I., Petriman, N.-A., Kudva, R., and Koch, H.-G. (2014). The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31, 58–84. doi: 10.3109/09687688.2014.907455
Diepold, A., and Armitage, J. P. (2015). Type III secretion systems: the bacterial flagellum and the injectisome. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370:20150020. doi: 10.1098/rstb.2015.0020
Dunyak, B. M., and Gestwicki, J. E. (2016). Peptidyl-Proline Isomerases (PPIases): targets for natural products and natural product-inspired compounds. J. Med. Chem. 59, 9622–9644. doi: 10.1021/acs.jmedchem.6b00411
Eckroat, T. J., Greguske, C., and Hunnicutt, D. W. (2021). The type 9 secretion system is required for Flavobacterium johnsoniae biofilm formation. Front. Microbiol. 12:660887. doi: 10.3389/fmicb.2021.660887
Fan, E., Chauhan, N., Udatha, D. B. R. K. G., Leo, J. C., and Linke, D. (2016). Type V secretion systems in bacteria. Microbiol. Spectr. 4. doi: 10.1128/microbiolspec.VMBF-0009-2015
Glew, M. D., Veith, P. D., Chen, D., Gorasia, D. G., Peng, B., and Reynolds, E. C. (2017). PorV is an outer membrane shuttle protein for the type IX secretion system. Sci. Rep. 7:8790. doi: 10.1038/s41598-017-09412-w
Glew, M. D., Veith, P. D., Peng, B., Chen, Y.-Y., Gorasia, D. G., Yang, Q., et al. (2012). PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J. Biol. Chem. 287, 24605–24617. doi: 10.1074/jbc.M112.369223
Gómez-Santos, N., Glatter, T., Koebnik, R., Świątek-Połatyńska, M. A., and Søgaard-Andersen, L. (2019). A TonB-dependent transporter is required for secretion of protease PopC across the bacterial outer membrane. Nat. Commun. 10:1360. doi: 10.1038/s41467-019-09366-9
Gorasia, D. G., Glew, M. D., Veith, P. D., and Reynolds, E. C. (2020). Quantitative proteomic analysis of the type IX secretion system mutants in Porphyromonas gingivalis. Mol. Oral Microbiol. 35, 78–84. doi: 10.1111/omi.12283
Gorasia, D. G., Lunar Silva, I., Butler, C. A., Chabalier, M., Doan, T., Cascales, E., et al. (2022). Protein interactome analysis of the type IX secretion system identifies PorW as the missing link between the PorK/N ring complex and the Sov Translocon. Microbiol. Spectr. 10:e0160221. doi: 10.1128/spectrum.01602-21
Gorasia, D. G., Veith, P. D., Chen, D., Seers, C. A., Mitchell, H. A., Chen, Y.-Y., et al. (2015). Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog. 11:e1005152. doi: 10.1371/journal.ppat.1005152
Gorasia, D. G., Veith, P. D., Hanssen, E. G., Glew, M. D., Sato, K., Yukitake, H., et al. (2016). Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog. 12:e1005820. doi: 10.1371/journal.ppat.1005820
Gorasia, D., Chreifi, G., Seers, C., Butler, C., Heath, J., Glew, M., et al. (2020). In situ structure and organisation of the type IX secretion system. bioRxiv [Preprint]. doi: 10.1101/2020.05.13.094771
Gorski, L., Godchaux, W., and Leadbetter, E. R. (1993). Structural specificity of sugars that inhibit gliding motility of Cytophaga johnsonae. Arch. Microbiol. 160, 121–125. doi: 10.1007/BF00288713
Grossman, A. S., Mauer, T. J., Forest, K. T., and Goodrich-Blair, H. (2021). A widespread bacterial secretion system with diverse substrates. mBio 12:e0195621. doi: 10.1128/mBio.01956-21
Hamidjaja, R., Capoulade, J., Catón, L., and Ingham, C. J. (2020). The cell organization underlying structural colour is involved in Flavobacterium IR1 predation. ISME J. 14, 2890–2900. doi: 10.1038/s41396-020-00760-6
Heath, J. E., Seers, C. A., Veith, P. D., Butler, C. A., Nor Muhammad, N. A., Chen, Y.-Y., et al. (2016). PG1058 is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One 11:e0164313. doi: 10.1371/journal.pone.0164313
Hennell James, R., Deme, J. C., Kjær, A., Alcock, F., Silale, A., Lauber, F., et al. (2021). Structure and mechanism of the proton-driven motor that powers type 9 secretion and gliding motility. Nat. Microbiol. 6, 221–233. doi: 10.1038/s41564-020-00823-6
Hennon, S. W., Soman, R., Zhu, L., and Dalbey, R. E. (2015). YidC/Alb3/Oxa1 family of insertases. J. Biol. Chem. 290, 14866–14874. doi: 10.1074/jbc.R115.638171
Hofreuter, D., Odenbreit, S., and Haas, R. (2001). Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system: H. pylori natural transformation competence. Mol. Microbiol. 41, 379–391. doi: 10.1046/j.1365-2958.2001.02502.x
Holt, S. C., Leadbetter, E. R., and Socransky, S. S. (1979). Capnocytophaga: new genus of gram-negative gliding bacteria. II. Morphology and ultrastructure. Arch. Microbiol. 122, 17–27. doi: 10.1007/BF00408041
Hunnicutt, D. W., and McBride, M. J. (2000). Cloning and characterization of the Flavobacterium johnsoniae gliding-motility genes gldB and gldC. J. Bacteriol. 182, 911–918. doi: 10.1128/JB.182.4.911-918.2000
Hunnicutt, D. W., and McBride, M. J. (2001). Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J. Bacteriol. 183, 4167–4175. doi: 10.1128/JB.183.14.4167-4175.2001
Hunnicutt, D. W., Kempf, M. J., and McBride, M. J. (2002). Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 184, 2370–2378. doi: 10.1128/JB.184.9.2370-2378.2002
Johansen, V. E., Catón, L., Hamidjaja, R., Oosterink, E., Wilts, B. D., Rasmussen, T. S., et al. (2018). Genetic manipulation of structural color in bacterial colonies. Proc. Natl. Acad. Sci. U.S.A. 115, 2652–2657. doi: 10.1073/pnas.1716214115
Johnston, J. J., Shrivastava, A., and McBride, M. J. (2018). Untangling Flavobacterium johnsoniae gliding motility and protein secretion. J. Bacteriol. 200, e00362-17. doi: 10.1128/JB.00362-17
Kharade, S. S., and McBride, M. J. (2014). Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J. Bacteriol. 196, 961–970. doi: 10.1128/JB.01170-13
Kharade, S. S., and McBride, M. J. (2015). Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J. Bacteriol. 197, 147–158. doi: 10.1128/JB.02085-14
Kita, D., Shibata, S., Kikuchi, Y., Kokubu, E., Nakayama, K., Saito, A., et al. (2016). Involvement of the Type IX secretion system in Capnocytophaga ochracea gliding motility and biofilm formation. Appl. Environ. Microbiol. 82, 1756–1766. doi: 10.1128/AEM.03452-15
Korotkov, K. V., and Sandkvist, M. (2019). Architecture, function, and substrates of the Type II secretion system. EcoSal Plus 8, ecosallus.ES–0034–2018. doi: 10.1128/ecosalplus.ESP-0034-2018
Kudva, R., Denks, K., Kuhn, P., Vogt, A., Müller, M., and Koch, H.-G. (2013). Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534. doi: 10.1016/j.resmic.2013.03.016
Kulkarni, S. S., Johnston, J. J., Zhu, Y., Hying, Z. T., and McBride, M. J. (2019). The carboxy-terminal region of Flavobacterium johnsoniae SprB facilitates its secretion by the type IX secretion system and propulsion by the gliding motility machinery. J. Bacteriol. 201, e218–e219. doi: 10.1128/JB.00218-19
Kulkarni, S. S., Zhu, Y., Brendel, C. J., and McBride, M. J. (2017). Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J. Bacteriol. 199, e00884–16. doi: 10.1128/JB.00884-16
Kusters, I., and Driessen, A. J. M. (2011). SecA, a remarkable nanomachine. Cell. Mol. Life Sci. 68, 2053–2066. doi: 10.1007/s00018-011-0681-y
Lasica, A. M., Goulas, T., Mizgalska, D., Zhou, X., de Diego, I., Ksiazek, M., et al. (2016). Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci. Rep. 6:37708. doi: 10.1038/srep37708
Lauber, F., Deme, J. C., Lea, S. M., and Berks, B. C. (2018). Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564, 77–82. doi: 10.1038/s41586-018-0693-y
Leadbetter, E. R., Holt, S. C., and Socransky, S. S. (1979). Capnocytophaga: new genus of gram-negative gliding bacteria. I. General characteristics, taxonomic considerations and significance. Arch. Microbiol. 122, 9–16. doi: 10.1007/BF00408040
Leone, P., Roche, J., Vincent, M. S., Tran, Q. H., Desmyter, A., Cascales, E., et al. (2018). Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space. Nat. Commun. 9:429. doi: 10.1038/s41467-017-02784-7
Li, C., Hurley, A., Hu, W., Warrick, J. W., Lozano, G. L., Ayuso, J. M., et al. (2021). Social motility of biofilm-like microcolonies in a gliding bacterium. Nat. Commun. 12:5700. doi: 10.1038/s41467-021-25408-7
Li, Y. G., Hu, B., and Christie, P. J. (2019). Biological and structural diversity of type IV secretion systems. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.PSIB-0012-2018
Madej, M., Nowakowska, Z., Ksiazek, M., Lasica, A. M., Mizgalska, D., Nowak, M., et al. (2021). PorZ, an essential component of the type IX secretion system of Porphyromonas gingivalis, delivers anionic lipopolysaccharide to the PorU sortase for transpeptidase processing of T9SS cargo proteins. mBio 12, e02262–20. doi: 10.1128/mBio.02262-20
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E., and Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800. doi: 10.1073/pnas.1522149113
McBride, M. J., and Braun, T. F. (2004). GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 186, 2295–2302. doi: 10.1128/JB.186.8.2295-2302.2004
McBride, M. J., and Zhu, Y. (2013). Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J. Bacteriol. 195, 270–278. doi: 10.1128/JB.01962-12
McBride, M. J., Braun, T. F., and Brust, J. L. (2003). Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 185, 6648–6657. doi: 10.1128/JB.185.22.6648-6657.2003
Meuskens, I., Saragliadis, A., Leo, J. C., and Linke, D. (2019). Type V secretion systems: an overview of passenger domain functions. Front. Microbiol. 10:1163. doi: 10.3389/fmicb.2019.01163
Nakane, D., Odaka, S., Suzuki, K., and Nishizaka, T. (2021). Large-Scale vortices with dynamic rotation emerged from monolayer collective motion of gliding Flavobacteria. J. Bacteriol. 203:e0007321. doi: 10.1128/JB.00073-21
Nakane, D., Sato, K., Wada, H., McBride, M. J., and Nakayama, K. (2013). Helical flow of surface protein required for bacterial gliding motility. Proc. Natl. Acad. Sci. U.S.A. 110, 11145–11150. doi: 10.1073/pnas.1219753110
Naskar, S., Hohl, M., Tassinari, M., and Low, H. H. (2021). The structure and mechanism of the bacterial type II secretion system. Mol. Microbiol. 115, 412–424. doi: 10.1111/mmi.14664
Nelson, S. S., Bollampalli, S., and McBride, M. J. (2008). SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 190, 2851–2857. doi: 10.1128/JB.01904-07
Nguyen, K.-A., Żylicz, J., Szczesny, P., Sroka, A., Hunter, N., and Potempa, J. (2009). Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiol. Read. Engl. 155, 328–337. doi: 10.1099/mic.0.024323-0
Nirody, J. A., Budin, I., and Rangamani, P. (2020). ATP synthase: evolution, energetics, and membrane interactions. J. Gen. Physiol. 152:e201912475. doi: 10.1085/jgp.201912475
Palmer, T., and Berks, B. C. (2012). The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10, 483–496. doi: 10.1038/nrmicro2814
Palmer, T., Finney, A. J., Saha, C. K., Atkinson, G. C., and Sargent, F. (2021). A holin/peptidoglycan hydrolase-dependent protein secretion system. Mol. Microbiol. 115, 345–355. doi: 10.1111/mmi.14599
Papanikou, E., Karamanou, S., and Economou, A. (2007). Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5, 839–851. doi: 10.1038/nrmicro1771
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Zorzini, V., and Economou, A. (2016). Type III secretion: building and operating a remarkable nanomachine. Trends Biochem. Sci. 41, 175–189. doi: 10.1016/j.tibs.2015.09.005
Ratheesh, N. K., Calderon, C. A., Zdimal, A. M., and Shrivastava, A. (2021). Bacterial swarm-mediated phage transportation disrupts a biofilm inherently protected from phage penetration. bioRxiv [Preprint]. doi: 10.1101/2021.06.25.449910
Rhodes, R. G., Samarasam, M. N., Shrivastava, A., van Baaren, J. M., Pochiraju, S., Bollampalli, S., et al. (2010). Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J. Bacteriol. 192, 1201–1211. doi: 10.1128/JB.01495-09
Rhodes, R. G., Samarasam, M. N., Van Groll, E. J., and McBride, M. J. (2011). Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J. Bacteriol. 193, 5322–5327. doi: 10.1128/JB.05480-11
Rivera-Calzada, A., Famelis, N., Llorca, O., and Geibel, S. (2021). Type VII secretion systems: structure, functions and transport models. Nat. Rev. Microbiol. 19, 567–584. doi: 10.1038/s41579-021-00560-5
Saiki, K., and Konishi, K. (2007). Identification of a Porphyromonas gingivalis novel protein sov required for the secretion of gingipains. Microbiol. Immunol. 51, 483–491. doi: 10.1111/j.1348-0421.2007.tb03936.x
Saiki, K., and Konishi, K. (2010). The role of Sov protein in the secretion of gingipain protease virulence factors of Porphyromonas gingivalis. FEMS Microbiol. Lett. 302, 166–174. doi: 10.1111/j.1574-6968.2009.01848.x
Sato, K., Naito, M., Yukitake, H., Hirakawa, H., Shoji, M., McBride, M. J., et al. (2010). A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 276–281. doi: 10.1073/pnas.0912010107
Sato, K., Sakai, E., Veith, P. D., Shoji, M., Kikuchi, Y., Yukitake, H., et al. (2005). Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 280, 8668–8677. doi: 10.1074/jbc.M413544200
Sato, K., Yukitake, H., Narita, Y., Shoji, M., Naito, M., and Nakayama, K. (2013). Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol. Lett. 338, 68–76. doi: 10.1111/1574-6968.12028
Schulz, S., Wilkes, M., Mills, D. J., Kühlbrandt, W., and Meier, T. (2017). Molecular architecture of the N-type ATPase rotor ring from Burkholderia pseudomallei. EMBO Rep. 18, 526–535. doi: 10.15252/embr.201643374
Shrivastava, A. K. (2013). Cell Surface Adhesins, Exopolysaccharides and the Por (Type IX) Secretion System of Flavobacterium johnsoniae. Theses and Dissertations. Milwaukee, WI: University of Wisconsin-Milwaukee.
Shrivastava, A., and Berg, H. C. (2020). A molecular rack and pinion actuates a cell-surface adhesin and enables bacterial gliding motility. Sci. Adv. 6:eaay6616. doi: 10.1126/sciadv.aay6616
Shrivastava, A., Johnston, J. J., van Baaren, J. M., and McBride, M. J. (2013). Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J. Bacteriol. 195, 3201–3212. doi: 10.1128/JB.00333-13
Shrivastava, A., Lele, P. P., and Berg, H. C. (2015). A rotary motor drives Flavobacterium gliding. Curr. Biol. 25, 338–341. doi: 10.1016/j.cub.2014.11.045
Shrivastava, A., Patel, V. K., Tang, Y., Yost, S. C., Dewhirst, F. E., and Berg, H. C. (2018). Cargo transport shapes the spatial organization of a microbial community. Proc. Natl. Acad. Sci. U.S.A. 115, 8633–8638. doi: 10.1073/pnas.1808966115
Shrivastava, A., Rhodes, R. G., Pochiraju, S., Nakane, D., and McBride, M. J. (2012). Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J. Bacteriol. 194, 3678–3688. doi: 10.1128/JB.00588-12
Shrivastava, A., Roland, T., and Berg, H. C. (2016). The screw-like movement of a gliding bacterium is powered by spiral motion of cell-surface adhesins. Biophys. J. 111, 1008–1013. doi: 10.1016/j.bpj.2016.07.043
Slakeski, N., Seers, C. A., Ng, K., Moore, C., Cleal, S. M., Veith, P. D., et al. (2011). C-Terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis. J. Bacteriol. 193, 132–142. doi: 10.1128/JB.00773-10
Spirig, T., Weiner, E. M., and Clubb, R. T. (2011). Sortase enzymes in Gram-positive bacteria: sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 82, 1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x
Spitz, O., Erenburg, I. N., Beer, T., Kanonenberg, K., Holland, I. B., and Schmitt, L. (2019). Type I secretion systems—one mechanism for all? Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.PSIB-0003-2018
Sumino, Y., Nagai, K. H., Shitaka, Y., Tanaka, D., Yoshikawa, K., Chaté, H., et al. (2012). Large-scale vortex lattice emerging from collectively moving microtubules. Nature 483, 448–452. doi: 10.1038/nature10874
Trinh, N. T. T., Tran, H. Q., Van Dong, Q., Cambillau, C., Roussel, A., and Leone, P. (2020). Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment. Sci. Rep. 10:7384. doi: 10.1038/s41598-020-64115-z
Van Gerven, N., Klein, R. D., Hultgren, S. J., and Remaut, H. (2015). Bacterial amyloid formation: structural insights into curli biogensis. Trends Microbiol. 23, 693–706. doi: 10.1016/j.tim.2015.07.010
Vincent, M. S., Canestrari, M. J., Leone, P., Stathopulos, J., Ize, B., Zoued, A., et al. (2017). Characterization of the Porphyromonas gingivalis type IX secretion trans-envelope PorKLMNP core complex. J. Biol. Chem. 292, 3252–3261. doi: 10.1074/jbc.M116.765081
Vincent, M. S., Hervada, C. C., Sebban-Kreuzer, C., Guenno, H. L., Chabalier, M., Kosta, A., et al. (2021). Dynamic proton-dependent motors power type IX secretion and gliding adhesin movement in Flavobacterium. bioRxiv [Preprint]. doi: 10.1101/2021.10.19.464928
Vorburger, T., Stein, A., Ziegler, U., Kaim, G., and Steuber, J. (2009). Functional role of a conserved aspartic acid residue in the motor of the Na(+)-driven flagellum from Vibrio cholerae. Biochim. Biophys. Acta 1787, 1198–1204. doi: 10.1016/j.bbabio.2009.05.015
Waksman, G., and Hultgren, S. J. (2009). Structural biology of the chaperone–usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7, 765–774. doi: 10.1038/nrmicro2220
Wolkin, R. H., and Pate, J. L. (1984). Translocation of motile cells of the gliding bacterium Cytophaga johnsonae depends on a surface component that may be modified by sugars. Microbiology 130, 2651–2669. doi: 10.1099/00221287-130-10-2651
Yamaguchi, T., Makino, F., Miyata, T., Minamino, T., Kato, T., and Namba, K. (2021). Structure of the molecular bushing of the bacterial flagellar motor. Nat. Commun. 12:4469. doi: 10.1038/s41467-021-24715-3
Zhou, A., Rohou, A., Schep, D. G., Bason, J. V., Montgomery, M. G., Walker, J. E., et al. (2015). Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife 4:e10180. doi: 10.7554/eLife.10180
Zhou, J., Sharp, L. L., Tang, H. L., Lloyd, S. A., Billings, S., Braun, T. F., et al. (1998). Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180, 2729–2735. doi: 10.1128/JB.180.10.2729-2735.1998
Keywords: bacterial motility, T9SS, gliding motility, bacterial swarming, Flavobacteria
Citation: Trivedi A, Gosai J, Nakane D and Shrivastava A (2022) Design Principles of the Rotary Type 9 Secretion System. Front. Microbiol. 13:845563. doi: 10.3389/fmicb.2022.845563
Received: 29 December 2021; Accepted: 17 January 2022;
Published: 10 May 2022.
Edited by:
Beiyan Nan, Texas A&M University, United StatesReviewed by:
Mark J. McBride, University of Wisconsin–Milwaukee, United StatesBen Berks, University of Oxford, United Kingdom
Copyright © 2022 Trivedi, Gosai, Nakane and Shrivastava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abhishek Shrivastava, YXNocml2YXN0YXZhQGFzdS5lZHU=
 Abhishek Trivedi
Abhishek Trivedi Jitendrapuri Gosai
Jitendrapuri Gosai Daisuke Nakane
Daisuke Nakane Abhishek Shrivastava
Abhishek Shrivastava