- 1Georgia Institute of Technology, School of Biological Sciences, Atlanta, GA, United States
- 2School of Materials, Chemistry and Chemical Engineering, Osaka Prefecture University, Sakai, Japan
- 3Savannah River National Laboratory, Environmental Sciences Section, Aiken, SC, United States
Microbial iodate (IO3–) reduction is a major component of the iodine biogeochemical reaction network in anaerobic marine basins and radioactive iodine-contaminated subsurface environments. Alternative iodine remediation technologies include microbial reduction of IO3– to iodide (I–) and microbial methylation of I– to volatile gases. The metal reduction pathway is required for anaerobic IO3– respiration by the gammaproteobacterium Shewanella oneidensis. However, the terminal IO3– reductase and additional enzymes involved in the S. oneidensis IO3– electron transport chain have not yet been identified. In this study, gene deletion mutants deficient in four extracellular electron conduits (EECs; ΔmtrA, ΔmtrA-ΔmtrDEF, ΔmtrA-ΔdmsEF, ΔmtrA-ΔSO4360) and DMSO reductase (ΔdmsB) of S. oneidensis were constructed and examined for anaerobic IO3– reduction activity with either 20 mM lactate or formate as an electron donor. IO3– reduction rate experiments were conducted under anaerobic conditions in defined minimal medium amended with 250 μM IO3– as anaerobic electron acceptor. Only the ΔmtrA mutant displayed a severe deficiency in IO3– reduction activity with lactate as the electron donor, which suggested that the EEC-associated decaheme cytochrome was required for lactate-dependent IO3– reduction. The ΔmtrA-ΔdmsEF triple mutant displayed a severe deficiency in IO3– reduction activity with formate as the electron donor, whereas ΔmtrA-ΔmtrDEF and ΔmtrA-ΔSO4360 retained moderate IO3– reduction activity, which suggested that the EEC-associated dimethylsulfoxide (DMSO) reductase membrane-spanning protein DmsE, but not MtrA, was required for formate-dependent IO3– reduction. Furthermore, gene deletion mutant ΔdmsB (deficient in the extracellular terminal DMSO reductase protein DmsB) and wild-type cells grown with tungsten replacing molybdenum (a required co-factor for DmsA catalytic activity) in defined growth medium were unable to reduce IO3– with either lactate or formate as the electron donor, which indicated that the DmsAB complex functions as an extracellular IO3– terminal reductase for both electron donors. Results of this study provide complementary genetic and phenotypic evidence that the extracellular DMSO reductase complex DmsAB of S. oneidensis displays broad substrate specificity and reduces IO3– as an alternate terminal electron acceptor.
Introduction
Iodine is commonly found in the environment in the forms of iodide (I–; -1 oxidation state), iodate (IO3–;+5 oxidation state), and organic iodine compounds (Whitehead, 1984; Amachi, 2013; Kaplan et al., 2014; Fuge and Johnson, 2015). An unstable radioisotope of iodine, 129I, is a nuclear waste product produced during uranium and plutonium fission reactions and displays a long half-life of 16 million years (Timar et al., 2014). 129I is found in contaminated groundwater at the U.S. Department of Energy Savannah River and Hanford Superfund sites from a long history of nuclear weapons testing (Emerson et al., 2014; Timar et al., 2014; Bagwell et al., 2019). Following the 2011 Fukushima nuclear reactor catastrophe, westerly winds deposited a large portion of 129I in the Pacific Ocean, where radioactive IO3– and I– are the predominant 129I forms (Hou et al., 2007, 2013; Bluhm et al., 2011; Kenyon et al., 2020). IO3– is more thermodynamically stable in seawater than I–; however, significant quantities of I– are detected in anaerobic environments such as anaerobic basins and oxygen minimum zones in marine environments, which potentially indicates that microbial IO3– reduction is a major component of the iodine biogeochemical reaction network (Whitehead, 1984; Councell et al., 1997; Farrenkopf et al., 1997; Wong et al., 2002; Chance et al., 2007; Bluhm et al., 2010; Amachi, 2013; Kaplan et al., 2014; Fuge and Johnson, 2015; Guido-Garcia et al., 2015). Microbial IO3– reduction has also received recent attention as a component of alternative strategies for the remediation of waters and sediments contaminated with radioactive iodine inadvertently released to the environment (Amachi, 2013; Kaplan et al., 2014; Riley et al., 2016; Mok et al., 2018; Toporek et al., 2019). The presence of environmental 129I presents a significant risk of bioaccumulation in the human thyroid gland, as iodine is a biologically active element for humans and vertebrate animals as a constituent of the thyroid hormones, thyroxine, and triiodothyronine (Whitehead, 1984; Fuge and Johnson, 2015). Despite the human health concerns that surround the fate and transport of radioactive iodine in the environment, the molecular mechanism of microbial IO3– reduction remains poorly understood in the iodine biogeochemical cycle (Amachi, 2013; Gong and Zhang, 2013; Kaplan et al., 2014; Fuge and Johnson, 2015; Yeager et al., 2017).
Several IO3–-reducing microorganisms have been reported, which include Shewanella putrefaciens, Shewanella oneidensis, Desulfovibrio desulfuricans, Pseudomonas sp. strain SCT, and Rhizobiaceae bacterium strain DVZ35 (Councell et al., 1997; Farrenkopf et al., 1997; Amachi et al., 2007; Mok et al., 2018; Toporek et al., 2019). In particular, the facultative anaerobe S. oneidensis reduces a wide range of terminal electron acceptors, which includes oxidized forms of iron, manganese, nitrogen, sulfur, uranium, plutonium, technetium, and iodine (Farrenkopf et al., 1997; Venkateswaran et al., 1999; Neu et al., 2005; Newsome et al., 2014; Mok et al., 2018; Toporek et al., 2019). S. oneidensis transfers electrons to a variety of extracellular electron acceptors, which include Mn(III) and Fe(III) and Mn(IV) oxides (Cooper et al., 2016; White et al., 2016). To transfer the electrons to external metal oxides, S. oneidensis employs a variety of novel respiratory strategies, which include (i) direct enzymatic reduction via decaheme c-type cytochromes associated with extracellular electron conduits (EECs) located on the surface or surface extensions of the S. oneidensis outer membrane (Myers and Myers, 1992; DiChristina et al., 2002; Gorby et al., 2006), (ii) extracellular electron transfer via endogenous or exogenous electron shuttling compounds (Taillefert et al., 2007; Fennessey et al., 2010; Jones et al., 2010), and (iii) non-reductive Fe(III) solubilization by organic ligands to produce more readily reducible soluble organic Fe(III) complexes (Hernandez et al., 2004; Marsili et al., 2008; Roden et al., 2010).
The previous studies of other IO3–-reducing microorganisms indicated that nitrate (NO3–) reductase may catalyze the reduction of IO3– as an alternative electron acceptor (Tsunogai and Sase, 1969; Wong and Hung, 2001; Lee et al., 2018). However, results from the later studies indicated that neither assimilatory nor dissimilatory NO3– reductases are required for IO3– reduction by S. oneidensis (Mok et al., 2018). Recently, a putative periplasmic molybdopterin-dependent iodate reductase (Idr) system composed of four proteins (IdrA, IdrB, IdrP1, and IdrP2) was identified in Pseudomonas sp. strain SCT. The catalytic subunits IdrA and IdrB displayed amino acid sequence homology with the catalytic and electron transfer subunits of respiratory arsenite oxidase (Aio), and IdrA represented a novel clade within the dimethylsulfoxide (DMSO) reductase family (Yamazaki et al., 2020). Another estuarine bacterium, Denitromonas sp. IR-12, was also recently reported to utilize a molybdenum (Mo)-dependent IrdA for dissimilatory IO3– reduction (Reyes-Umana et al., 2022).
The IO3– reduction pathway of S. oneidensis shares electron transport components with EEC systems that reduce alternate electron acceptors such as metals, NO3–, sulfur compounds, DMSO, and trimethylamine N-oxide (Myers and Myers, 1992; DiChristina et al., 2002; Hernandez et al., 2004; Gorby et al., 2006; Taillefert et al., 2007; Marsili et al., 2008; Fennessey et al., 2010; Jones et al., 2010; Roden et al., 2010; Cooper et al., 2016; White et al., 2016). The electron transport pathways of S. oneidensis consist of upstream dehydrogenases linked via the menaquinone pool and the inner membrane-tethered c-type cytochrome CymA to downstream terminal reductase complexes associated with the metal-reducing EEC (Marsili et al., 2008; Roden et al., 2010; Gong and Zhang, 2013). The metal-reducing EEC of S. oneidensis is comprised of outer membrane β-barrel protein MtrB (and essential cysteine residue C42) (Wee et al., 2014) and decaheme c-type cytochromes, MtrA and MtrC (Shi et al., 2006, 2012; Coursolle and Gralnick, 2010; Richardson et al., 2012; Richter et al., 2012; Szeinbaum et al., 2014). MtrC is translocated to the outside face of the outer membrane through GspD, the outer membrane secretin of the type II protein secretion system (Cianciotto, 2005; McLaughlin et al., 2012; Cooper et al., 2016). Other proteins that are essential for electron transport to external metal oxides include the c-type cytochrome maturation permease CcmB (Dale et al., 2007) and the cAMP receptor protein CRP required for anaerobic respiratory gene expression by S. oneidensis (Saffarini et al., 2003). On the other hand, S. oneidensis EEC-associated DMSO reductase DmsAB is comprised of DmsA, which requires the Mo-binding co-factor molybdopterin, and the ferredoxin subunit DmsB, which contains [4Fe-4S] clusters as co-factors (Figure 1; May et al., 1988; Gralnick et al., 2006).
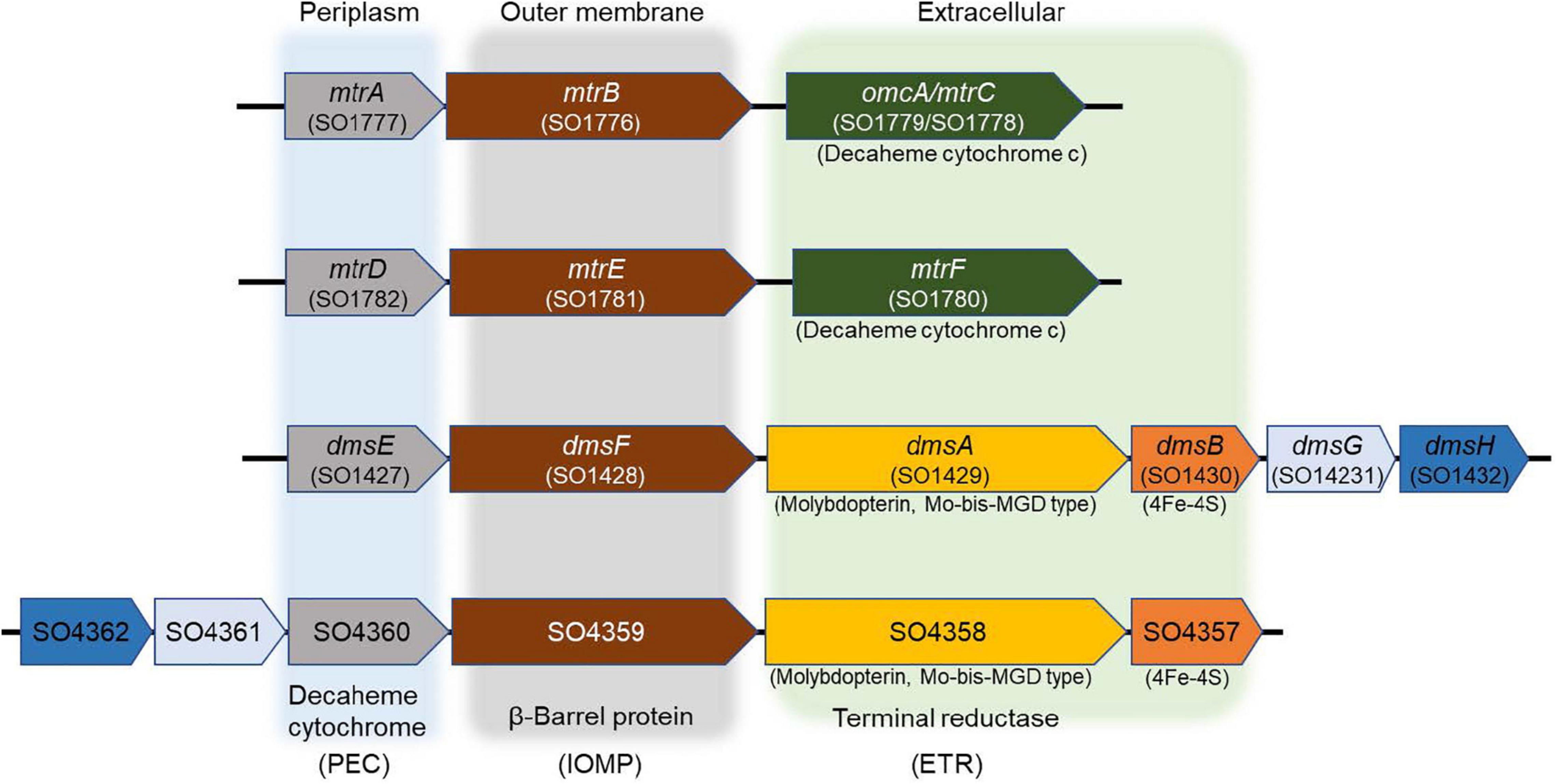
Figure 1. Mtr paralogs of extracellular electron conduits (EEC) system responsible for anaerobic respiration in S. oneidensis. PEC, periplasmic electron carrier; IOMP, integral outer membrane β-barrel protein; ETR, extracellular terminal reductase.
The previous work suggests that outer membrane (type II) protein secretion and metal reduction genes encoding the outer membrane MtrAB module of the EEC MtrCAB are required for IO3– reduction by S. oneidensis with lactate, but not formate, as the electron donor (Toporek et al., 2019). However, the metal-reducing c-type cytochrome MtrC associated with the EEC MtrAB was not required for IO3– reduction by S. oneidensis with any electron donor tested. These findings indicate that the IO3– electron transport pathway is modular, electron donor-dependent, and terminates with an as yet unidentified IO3– reductase that associates with an outer membrane EEC to deliver electrons extracellularly to IO3– (Toporek et al., 2019).
In addition to MtrCAB, the S. oneidensis genome harbors three additional gene clusters that encode the EECs MtrDEF, DmsEFAB, and SO4357-4360 (Bucking et al., 2010; Coursolle and Gralnick, 2010). MtrCAB and DmsEFAB are required for anaerobic reduction of Fe(III) and DMSO, respectively (Bucking et al., 2010; Coursolle and Gralnick, 2010; Schicklberger et al., 2013). Furthermore, several Mtr and Dms paralogs are functionally interchangeable (Coursolle and Gralnick, 2010, 2012; Schicklberger et al., 2013). For example, MtrD and DmsE may functionally replace MtrA (Coursolle and Gralnick, 2010), while MtrF and to a partial extent OmcA may functionally replace MtrC (Coursolle and Gralnick, 2010). MtrDEF reduces Fe(III) citrate at approximately half the rate of MtrCAB in ΔMtr mutants (Coursolle and Gralnick, 2012). While the expression of SO4359 and SO4360 alone was sufficient to complement an mtrB mutant under Fe(III) citrate-reducing conditions (Schicklberger et al., 2013). These findings led us to hypothesize that S. oneidensis reduces IO3– with separate lactate (MtrAB)- and formate-dependent EEC paralogs that deliver electrons extracellularly to IO3– (i.e., function as electron donor-dependent IO3– terminal reductases).
The main objective of this study was to identify the IO3– reductase of S. oneidensis. The experimental strategy consisted of the following steps: (i) construction of three EEC paralog mutants via the deletion of mtrDEF, dmsEF, and SO4360 in a ΔmtrA mutant host strain, and subsequent testing for IO3– reduction activity; (ii) replacement of Mo with tungsten (W) in defined growth medium and tests for IO3– reduction activity; and (iii) construction of DMSO reductase mutant ΔdmsB (deficient in DMSO reductase protein DmsB) and tests of ΔdmsB for IO3– reduction activity with formate and lactate as the electron donor.
Materials and Methods
Growth and Cultivation Conditions
Shewanella oneidensis strains were routinely cultured aerobically at 30°C in lysogeny broth (LB) (10 g/L–1 of NaCl, 10 g/L–1 of tryptone, and 5 g/L–1 of yeast extract). IO3– reduction rate experiments were conducted under anaerobic conditions in M1 minimal medium (Myers and Nealson, 1988; Supplementary Material) amended with 20 mM sodium formate or lactate as the electron donor and 250 μM IO3– as the anaerobic electron acceptor. Anaerobic growth of MR-1 on iodate is minimal under the incubation conditions used in this study. The toxicity threshold of MR-1 to iodate concentrations was previously determined in defined medium under aerobic and anaerobic incubation conditions (Toporek et al., 2019). When required for selection, gentamicin (20 μg ml–1) or chloramphenicol (25 μg ml–1) was amended to the appropriate growth medium for the selection of deletion mutant or the maintenance of recombinant plasmid vector pBBR1MCS (Kovach et al., 1995).
In-Frame Gene Deletion Mutagenesis
The genes mtrDEF, dmsEF, SO4360, and dmsB were deleted in frame from the S. oneidensis genome by following the previously described procedures (Supplementary Table 1; Burns and DiChristina, 2009). A dmsA deletion mutant was attempted but it was unsuccessful. Regions corresponding to 750 bp upstream and downstream of mtrDEF, dmsEF, SO4360, and dmsB were PCR amplified with Q5 High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA, United States) [primers D1/D2 and D3/D4 (Supplementary Table 2)] and subsequently joined using overlap extension PCR [primers D1/D4 (Supplementary Table 2)]. To construct ΔmtrA-ΔmtrDEF, ΔmtrA-ΔdmsEF, and ΔmtrA-ΔSO4360 mutants, the resulting fragments of mtrDEF, dmsEF, and SO4360 were cloned into suicide vector pKO2.0 (which does not replicate in S. oneidensis) and mobilized into S. oneidensis ΔmtrA (Szeinbaum et al., 2014) via conjugation with E. coli donor strain β2155 λ pir (Chung et al., 1989). In addition, the resulting fragment of dmsB was also cloned into suicide vector pKO2.0 and mobilized into wild-type S. oneidensis via conjugation with E. coli donor strain β2155 λ pir (Chung et al., 1989) to construct mutant ΔdmsB. S. oneidensis strains with the integrated plasmid were selected on LB agar containing gentamicin (20 μg ml–1). Single crossover integrations were verified using PCR with primers flanking the recombination region (TF/TR) and were resolved from the genomes by plating on LB agar lacking NaCl and containing sucrose [10% (wt/vol)]. The in-frame deletion strains (ΔmtrA-ΔmtrDEF, ΔmtrA-ΔdmsEF, ΔmtrA-ΔSO4360, and ΔdmsB) were verified by PCR with primers TF/TR (Supplementary Table 2). Genetic complementation analysis of the ΔdmsB strain was carried out by cloning the wild-type gene (after amplification from the S. oneidensis genome using primer set dB-F and dB-R; Supplementary Table 2) into broad-host-range cloning vector pBBR1MCS (Kovach et al., 1995) and conjugally transferring the recombinant vector into the respective mutant strains via biparental mating procedures (Chung et al., 1989).
IO3– and Dimethylsulfoxide Reduction Activity Assays
Mutant strains were initially inoculated in the liquid LB growth medium and incubated at 30°C for 24 h. About 10 ml of subcultures at an initial optical density at 600 nm (OD600) of 0.02 was incubated at 30°C for 24 h. Subcultures were centrifuged at 4,000 rpm for 30 min, resuspended in 10 ml of M1 growth medium amended with 10 mM formate and incubated aerobically at room temperature for 8 h. The preconditioned cells were inoculated in the 30-ml serum bottles at an initial OD600 of 0.1 in M1 growth medium amended with either 40 mM DMSO or 250 μM IO3– and 10 mM formate and incubated anaerobically via continuous sparging with 100% high-purity (hydrated) N2 gas. Cultures were incubated at room temperature with gentle stirring under anaerobic conditions maintained by continuous sparging with high-purity hydrated N2 gas. At preselected time points, OD600 was measured and IO3– concentrations were determined using the IO3–-triiodide formation method (Afkhami et al., 2001; Mok et al., 2018; Toporek et al., 2019) described below. Cells corresponding to OD = 0.1 contain 50 mg protein as measured by the Bradford assay (Bradford, 1976). DMSO reduction was monitored by measuring anaerobic growth at OD600. For substitution of molybdenum Mo with tungsten W in anaerobic IO3– reduction activity assays, Mo was replaced with equal molar concentration of W in M1 medium, and IO3– reduction activity was compared to IO3–-reduction activity in normal M1 medium containing Mo. For the cultivation of recombinant strains carrying pBBR1MCS or pBBR1MCS-dmsB, 25 μg ml–1 chloramphenicol and 0.1 mM IPTG were amended to the medium to maintain the plasmid and induce cloned dmsB gene expression, respectively.
Determination of IO3– Concentrations via IO3–-Triiodide Formation With I– at Acidic pH
The extent of IO3– reduction was determined using the IO3–-triiodide method (Afkhami et al., 2001; Mok et al., 2018; Toporek et al., 2019). Culture samples were added to the 96-well 500-μl microtiter plates. Sodium citrate buffer (0.1 M; pH 3.3) and potassium iodide solution (75 mM) were added to each well to initiate triiodide formation (IO3– + 5I– + 6H+ Δ 3H2O + 3I2). Absorbance at 352 nm was measured with a UV spectrophotometer (Multiskan Go; Thermo Scientific) within the first 3 min of reaction time. IO3– concentrations were determined from a previously generated calibration curve.
Results
IO3– Reduction Activity of Extracellular Electron Conduit Mutant Strains
To test the hypothesis that S. oneidensis employs periplasmic and outer membrane proteins other than MtrAB to deliver electrons to IO3–, three additional EEC gene mutant strains (ΔmtrDEF, ΔdmsEF, and ΔSO4360) were constructed in a ΔmtrA mutant host strain. A ΔmtrA gene deletion background was selected since ΔmtrA retained wild-type IO3– reduction activity with formate as electron donor (Toporek et al., 2019). Mtr proteins and their paralogs (e.g., DMS operon) are modular and can provide partial compensation for each other in the absence of a primary component (Coursolle and Gralnick, 2010). To avoid the possibility of MtrA compensating for the lack of DmsE, we constructed all dms mutants with a ΔmtrA background. The IO3– reduction activities of the three additional EEC mutant strains (ΔmtrAΔmtrDEF,ΔmtrAΔdmsEF, and ΔmtrAΔSO4360) were determined with either lactate or formate as the electron donor.
All mutant strains cultured with lactate displayed severely impaired (between 0 and 50% of wild-type activity) IO3– reduction activities when compared to wild-type rates (ΔmtrA, 7 and 7% of the wild-type rate and extent of reaction, respectively; ΔmtrAΔmtrDEF, 38 and 24% of the wild-type rate and extent of reaction, respectively; ΔmtrAΔdmsEF, 17 and 15% of the wild-type rate and extent of reaction, respectively; and ΔmtrAΔSO4360, 35 and 28% of the wild-type rate and extent of reaction, respectively; Table 1 and Figure 2). These results further confirm that EEC component MtrA is required for IO3– reduction with lactate as the electron donor (Toporek et al., 2019; Figure 1).
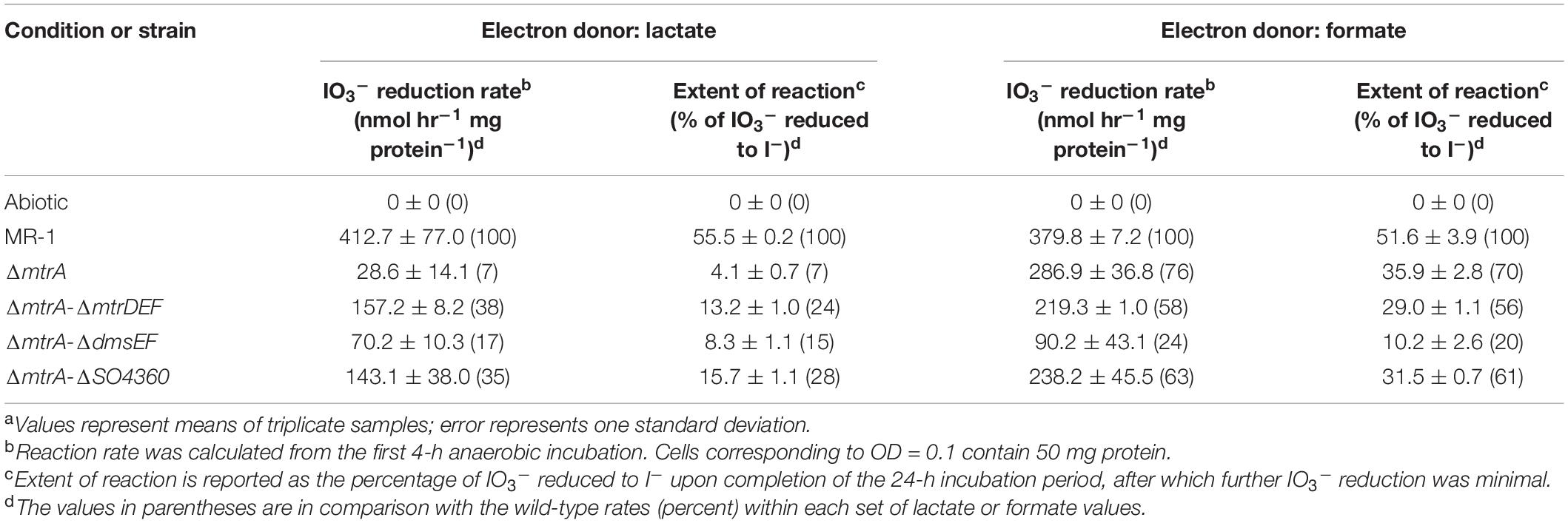
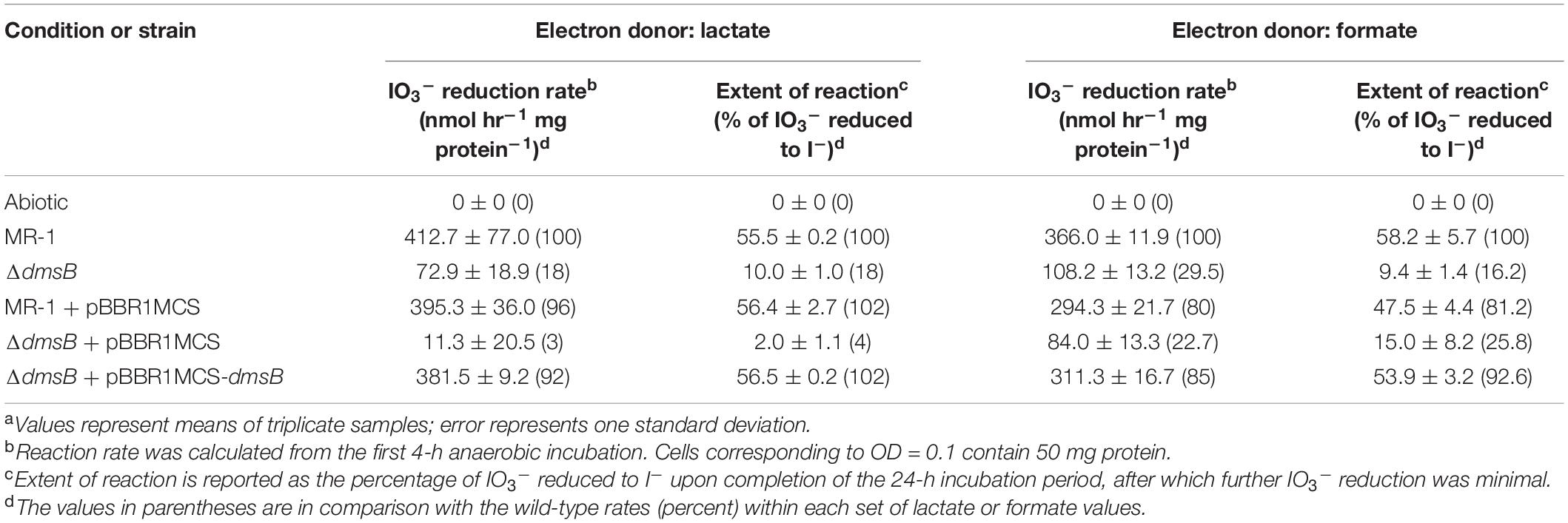
Table 1. IO3– reduction activities of wild-type and EEC paralog mutant strains of Shewanella oneidensis with lactate and formate as electron donors.a
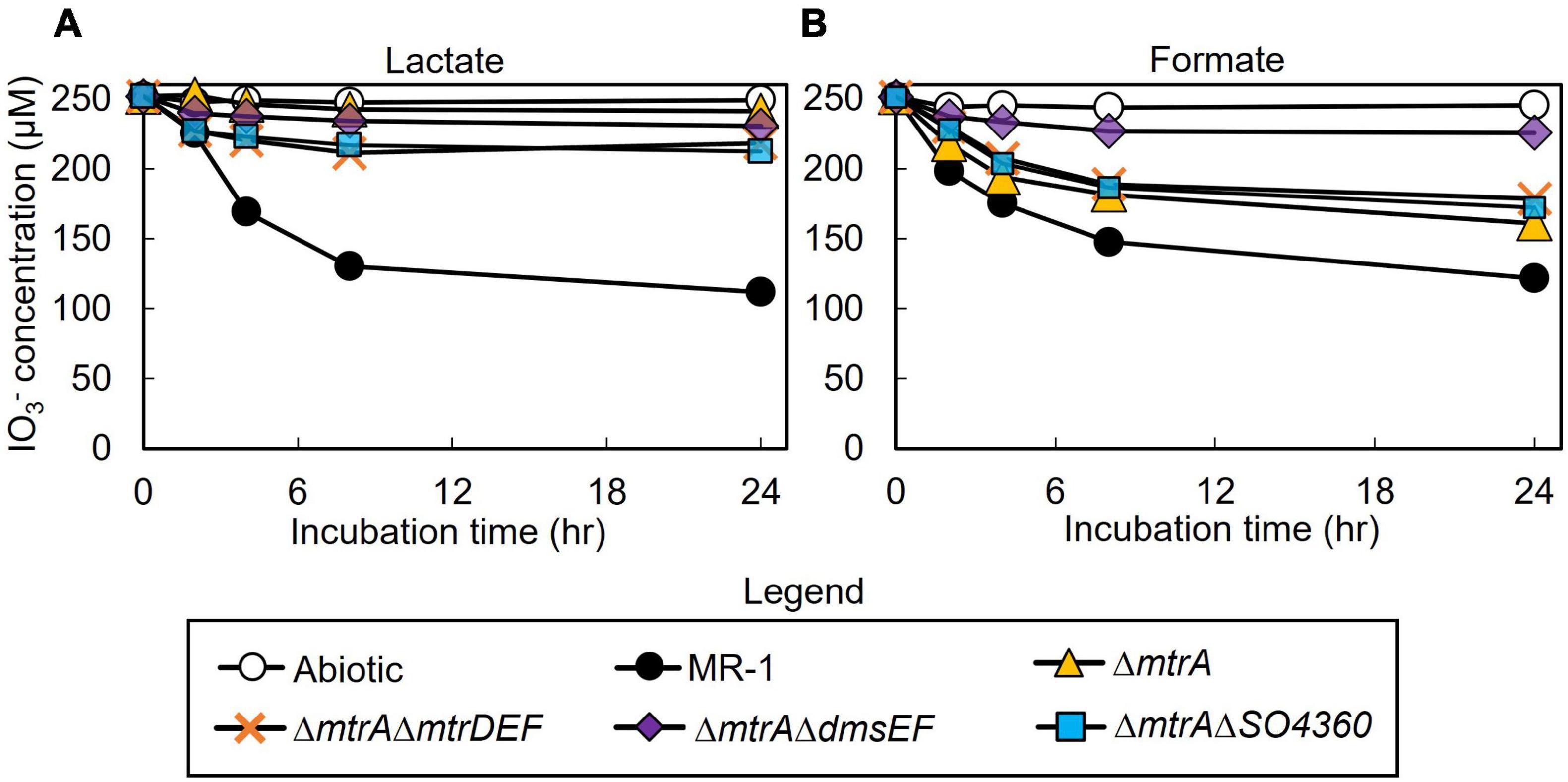
Figure 2. IO3– reduction activity of Shewanella oneidensis wild-type (MR-1) and ΔmtrA, ΔmtrAΔmtrDEF,ΔmtrAΔdmsEF, and ΔmtrAΔSO4360 mutants with IO3– as the electron acceptor and lactate (A) or formate (B) as the electron donor and their IO3– reduction rate. Values are means of triplicate samples from anaerobic incubations. Error bars represent one standard deviation.
The ΔmtrAΔmtrDEF and ΔmtrAΔSO4360 mutant strains provided with formate as the electron donor displayed moderately impaired (between 51 and 70% of wild-type activity) IO3– reduction activities when compared to wild-type rates (58 and 63% of the wild-type rate and 56 and 61% of the wild-type extents of reaction, respectively; Table 1 and Figure 2). The ΔmtrA mutant displayed a similar profile (between 71 and 102% of wild-type activity) with the wild-type strain (76% of the wild-type rate and 70% of the wild-type extent of reaction, respectively; Table 1 and Figure 2), while the ΔmtrAΔdmsEF mutant strain was severely impaired in IO3– reduction activity with formate as the electron donor (24% of the wild-type rate and 20% of the wild-type extent of reaction, respectively; Table 1 and Figure 2). These results indicate that EEC component DmsE, but not MtrA, and OMP component DmsF, but not MtrB, are required for IO3– reduction with formate as the electron donor (Figure 1).
Replacement of Mo With W in Defined Minimal Growth Medium and the Effect on IO3– Reduction Activity of Shewanella oneidensis
The S. oneidensis DMSO reductase DmsAB is composed of the molybdopterin-binding subunit DmsA and the ferredoxin subunit DmsB, which contains Mo and [4Fe-4S] clusters as co-factors, respectively (Figure 1; Gralnick et al., 2006). To test the hypothesis that S. oneidensis employs DmsAB as the IO3– terminal reductase we attempted to generate a ΔdmsA deletion mutant, as DmsA is the active component of DMSO reductase, but were unsuccessful. A previous study reported the similar inability to produce a ΔdmsA deletion mutant, which indicates that the dmsA deletion may be lethal (Gralnick et al., 2006). Mo is the critical catalytic element of the molybdopterin-binding DMSO reductase family, which includes DMSO reductase, nitrate reductase, and formate dehydrogenase (May et al., 1988; Hanzelmann and Mayer, 1998; McEwan et al., 2002; Waite and Trucesdale, 2003). W readily replaces Mo in molybdopterin-binding enzymes, yet equimolar Mo substitution with W results in loss of enzymatic activity of DMSO reductase family enzymes (May et al., 1988; Hanzelmann and Mayer, 1998; Hille, 2002; Waite and Trucesdale, 2003). The substitution of Mo with W did not affect cell fitness (Supplementary Figure 1). Depleting the wild-type strain MR-1 of Mo caused catalytic inactivation of the DmsA subunit and effectively generated a mutant strain deficient in DmsA catalytic activity. After growth in W-containing defined minimal medium, wild-type S. oneidensis reduced IO3– at severely impaired rates when incubated with either lactate or formate as the electron donor (62 and 42 nmol hr–1 mg protein–1, respectively), corresponding to only 17 and 11% of the rates measured after growth in Mo-containing defined minimal medium (lactate, 371 nmol hr–1 mg protein–1; formate, 366 nmol hr–1 mg protein–1, respectively) (Figure 3 and Table 2). These findings indicate that Mo is required for IO3– reduction with either lactate or formate as electron donor, potentially as the critical element of the molybdopterin-binding co-factor of DMSO reductase.
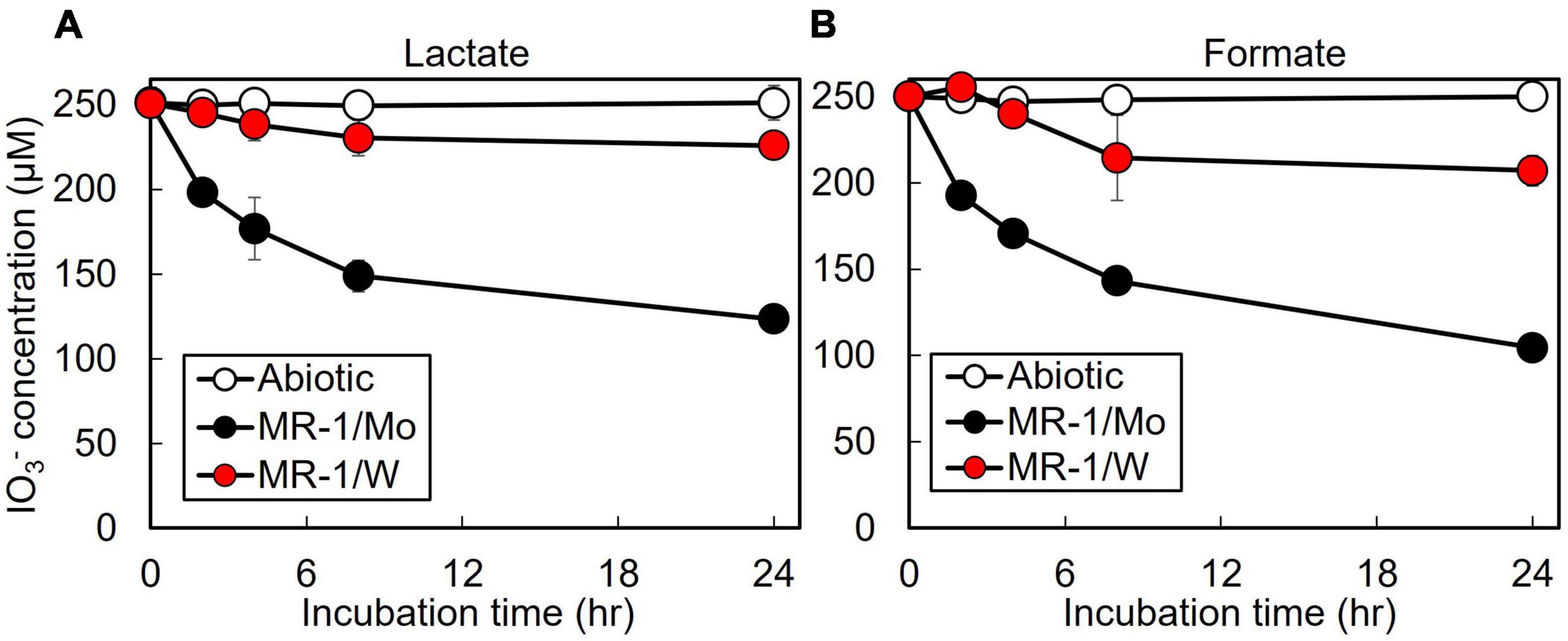
Figure 3. Effect of molybdenum (Mo) substitution with tungsten (W) on IO3– reduction activity of Shewanella oneidensis with IO3– as the electron acceptor and (A) lactate or (B) formate as the electron donor. Values are means of triplicate samples from anaerobic incubations. Error bars represent one standard deviation.
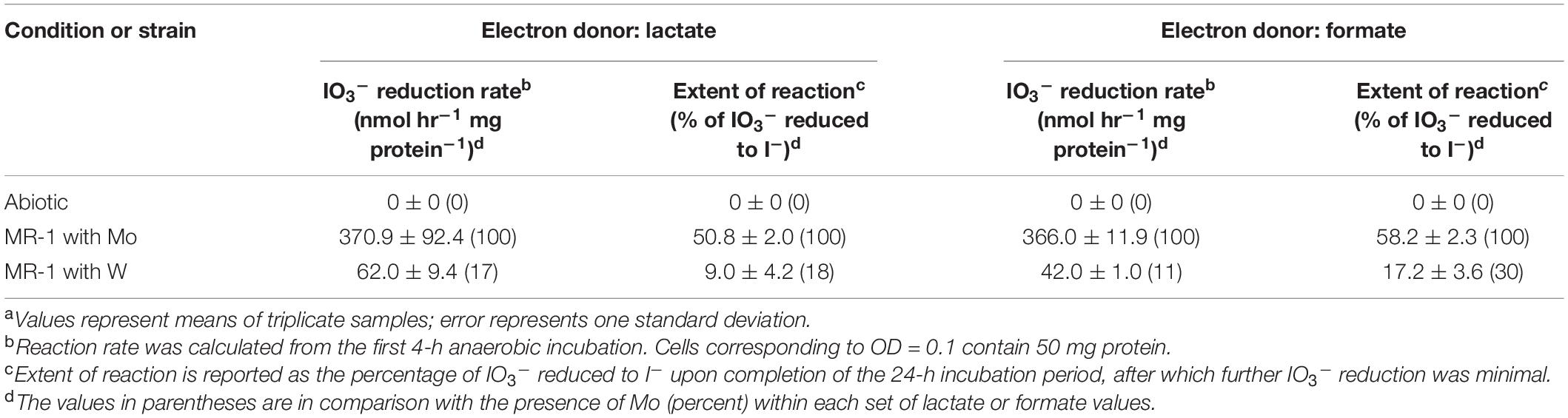
Table 2. Effect of replacement of molybdenum (Mo) with tungsten (W) on IO3– reduction activities of Shewanella oneidensis.a
IO3– Reduction Activity of ΔdmsB Mutant With Lactate or Formate as Electron Donor
To test the hypothesis that S. oneidensis employs DmsAB as the IO3– terminal reductase, we generated a ΔdmsB gene deletion mutant. In a previous study, mutant strain ΔdmsB was unable to grow anaerobically with DMSO as a terminal election acceptor (Gralnick et al., 2006). Fumarate reduction was not impaired by ΔdmsB deletion, which indicates that overall fitness is unaffected (Gralnick et al., 2006). In this study, ΔdmsB was also unable to grow with DMSO as electron acceptor and formate as the electron donor, while a ΔdmsB transconjugant strain provided with a wild-type copy of dmsB (ΔdmsB + pBBR1MCS-dmsB) grew at wild-type rates with DMSO as electron acceptor (Supplementary Figure 2). ΔdmsB was also severely impaired in IO3– reduction activity with lactate or formate as the electron donor (18 and 30% of the wild-type rate and 18 and 16% of the wild-type extents of reaction with lactate or formate, respectively; Figure 4 and Table 3), while the ΔdmsB + pBBR1MCS-dmsB transconjugant strain reduced IO3– at wild-type rates and extents of reaction (92 and 85% of the wild-type rate and 102 and 93% of the wild-type extents of reaction with lactate or formate, respectively; Figure 5 and Table 3). As expected, control MR-1 + pBBR1MCS (96 and 80% of the wild-type rate and 102 and 81% of the wild-type extents of reaction with lactate or formate, respectively; Figure 5 and Table 3) reduced IO3– at near-wild-type rates, and control ΔdmsB + pBBR1MCS (3 and 23% of the wild-type rate and 4 and 26% of the wild-type extents of reaction with lactate or formate, respectively; Figure 5 and Table 3) was severely affected. These results further indicate that DmsAB displays broad substrate specificity and reduces IO3– as an alternate terminal electron acceptor.
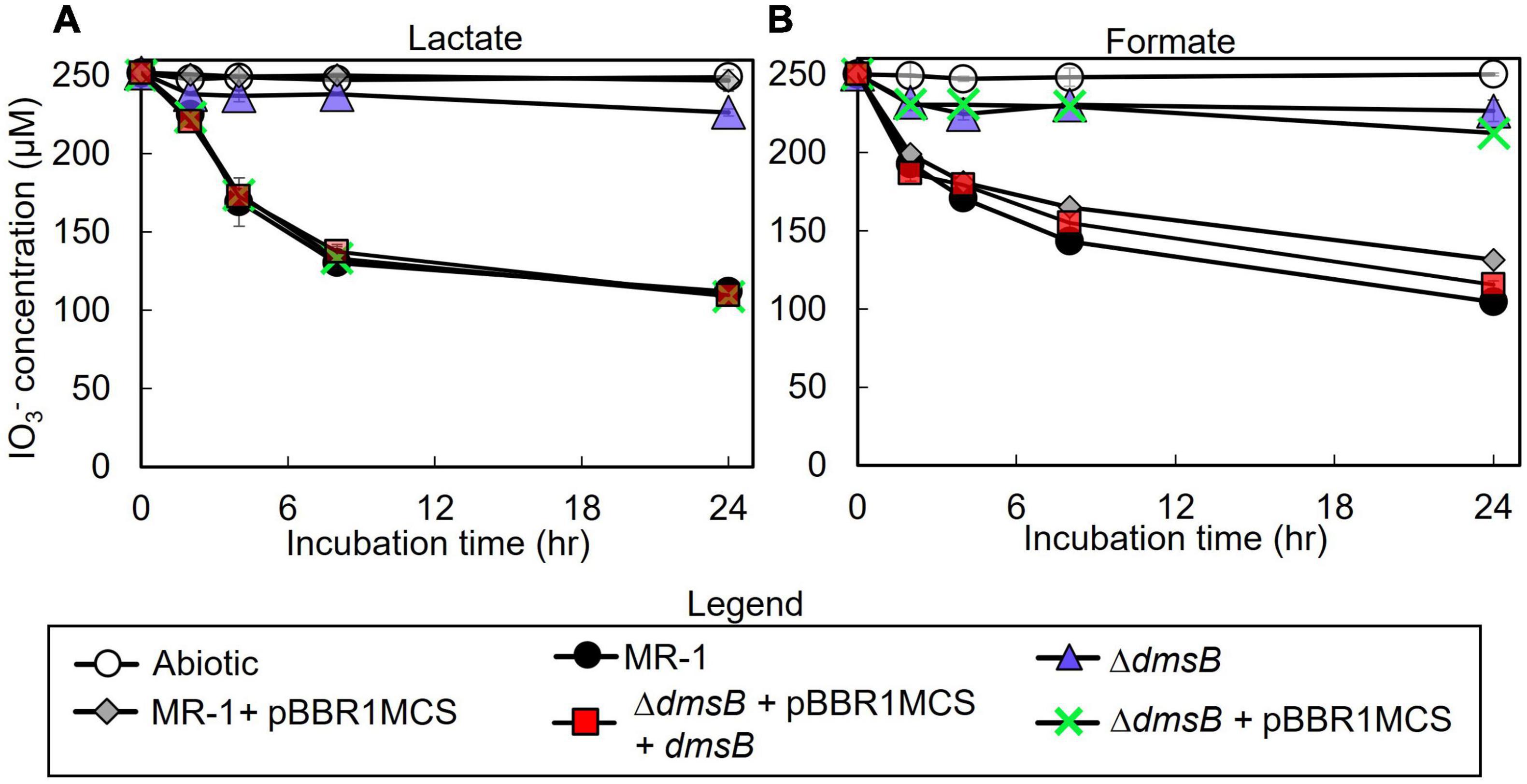
Figure 4. IO3– reduction activity of Shewanella oneidensis wild-type (MR-1) and ΔdmsB, ΔdmsB + pBBRdmsB,ΔdmsB + pBBR1MCS, MR-1 + pBBR1MCS strains with IO3– as the electron acceptor and (A) lactate or (B) formate as the electron donor and their IO3– reduction rate. Values are means of triplicate samples from anaerobic incubations. Error bars represent one standard deviation.
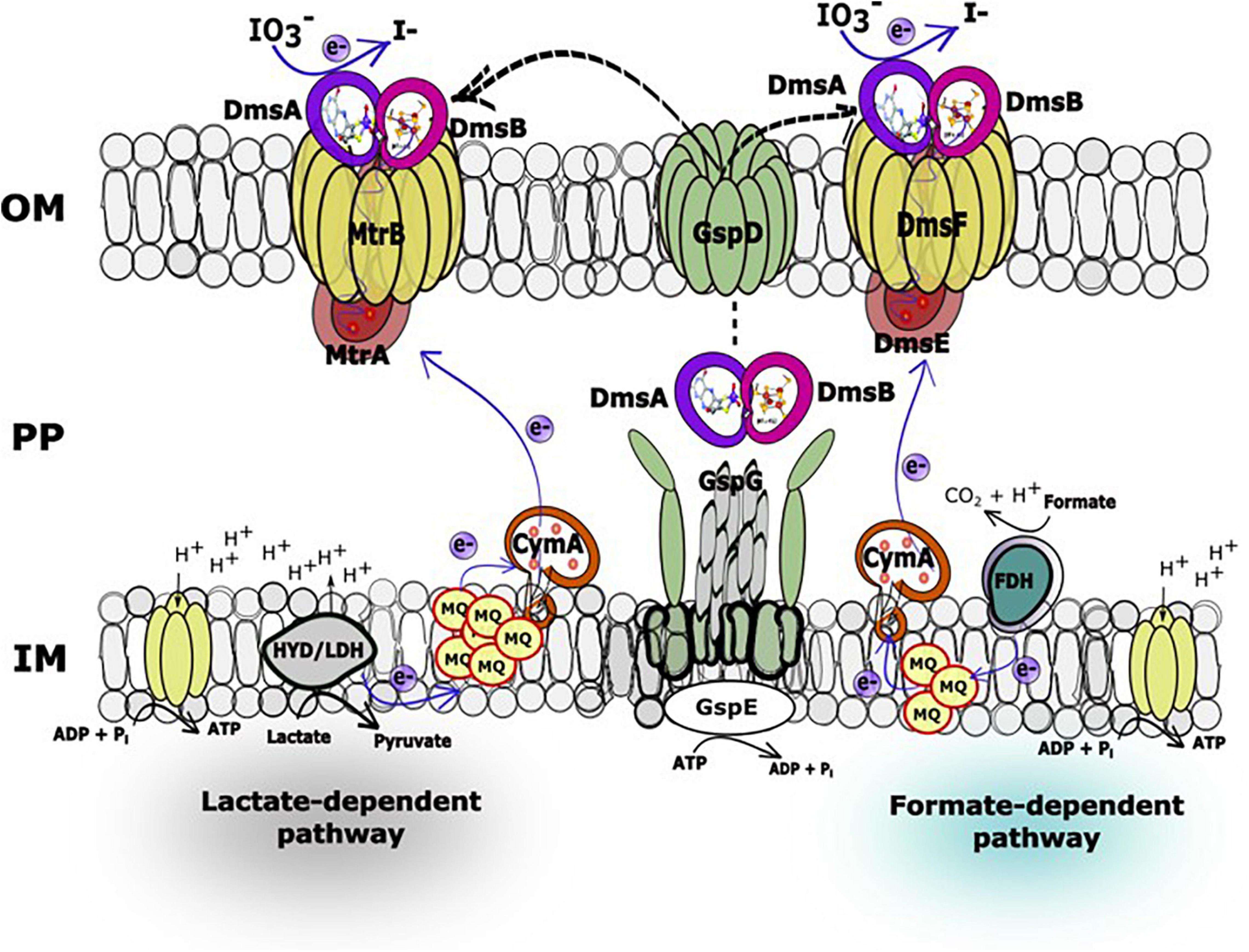
Figure 5. Hypothetical working model of the lactate (MtrAB)- and formate (DmsEF)-dependent IO3– reduction electron transport pathways in Shewanella oneidensis. In the IO3– reduction pathways, electrons originating from lactate dehydrogenase or formate dehydrogenase located at the head end of the electron transport chain are transferred to the inner membrane-localized menaquinone pool and subsequently to CymA, which facilitates electron transfer across the periplasmic space to decaheme c-type cytochromes MtrA or DmsE, respectively. At this location in the electron transport chain, the IO3– reduction pathways diverge to MtrAB (lactate as electron donor) or DmsEF (formate as electron donor) and terminate with DmsA and DmsB, both of which associate with MtrA (or DmsE) and β-barrel protein MtrB (or DmsF). DmsA and DmsB are both secreted extracellularly by the type II protein secretion system to form a ternary complex with the MtrAB (or DmsEF) EEC modules on the outside face of the outer membrane (Gralnick et al., 2006).
Discussion
The S. oneidensis genome encodes four EECs, each composed of three major components: periplasmic electron carrier (PEC), outer membrane β-barrel protein (OMP), and extracellular terminal reductase (ETR) (Figure 1). S. oneidensis EECs are involved in the reduction of Fe(III), Mn(IV), Mn(III), flavins, and DMSO (Bucking et al., 2010; Coursolle and Gralnick, 2010; Szeinbaum et al., 2014). Several EEC components are interchangeable and can functionally replace the corresponding paralog proteins (Coursolle and Gralnick, 2010, 2012; Schicklberger et al., 2013). S. oneidensis PECs include the decaheme c-type cytochromes MtrA, MtrD, DmsE, and SO4360 (Bucking et al., 2010; Coursolle and Gralnick, 2010; Schicklberger et al., 2013). MtrA is the primary PEC of the S. oneidensis Fe(III), Mn(IV), Mn(III), and flavin reduction systems. The overexpression of MtrD restores wild-type Fe(III)-citrate reduction rates to an ΔmtrA mutant (Bucking et al., 2010; Coursolle and Gralnick, 2010; Schicklberger et al., 2013). DmsE is primarily devoted to DMSO reduction (Gralnick et al., 2006; Bucking et al., 2010), but partially restores Fe(III)-citrate reduction to mtrA Mutants (Bucking et al., 2010; Coursolle and Gralnick, 2010; Schicklberger et al., 2013). SO4360 displays high amino acid sequence homology to other S. oneidensis PECs, but has not yet been assigned a respiratory function (Bucking et al., 2010; Coursolle and Gralnick, 2010; Schicklberger et al., 2013).
The four S. oneidensis ETRs are involved in Fe(III) or DMSO reduction and include the decaheme c-type cytochromes MtrC, MtrF, and OmcA. MtrF and OmcA functionally replace MtrC (Coursolle and Gralnick, 2010), while MtrDE only partially replaces MtrAB in the Fe(III)-citrate reduction pathways (Coursolle and Gralnick, 2012). Although DmsEF are the paralogs of MtrAB, DmsEF does not functionally replace MtrAB in the Fe(III)-citrate reduction pathway (Coursolle and Gralnick, 2012). The third and fourth ETRs (DmsAB and SO4358/SO4357) each contain both molybdopterin and 4Fe-4S clusters as co-factors, respectively (Gralnick et al., 2006; Bucking et al., 2010; Coursolle and Gralnick, 2010; Schicklberger et al., 2013). OMPs form a pore-like structure through the outer membrane that directs electron transfer between the PEC and ETR components (Beliaev and Saffarini, 1998; Hartshorne et al., 2009). The S. oneidensis genome harbors four OMP paralogs (MtrB, MtrE, DmsF, and SO4359) (Bucking et al., 2010; Coursolle and Gralnick, 2010; Schicklberger et al., 2013). SO4359, a paralog of DmsE, functionally replaces MtrB under Fe(III) citrate-reducing conditions (Schicklberger et al., 2013). The MtrAB module of MtrCAB is required for IO3– reduction by S. oneidensis with lactate (but not formate) as the electron donor (Toporek et al., 2019). Similar electron donor-dependent respiratory phenotypes of S. oneidensis were also previously reported with technetium [Tc(VII)] as the terminal electron acceptor (Payne and DiChristina, 2006). Based on these previous findings, we hypothesized that S. oneidensis employed an ETR other than MtrC to deliver electrons to IO3–.
In our previous report, a lactate (MtrAB)-dependent S. oneidensis IO3– reduction system was proposed (Toporek et al., 2019). In this working model, electrons originating from lactate dehydrogenase were transported via the menaquinone pool, CymA, and MtrAB to the unknown terminal IO3– reductase that was translocated to the outside face of the outer membrane via type II protein secretion, while IO3– reduction with formate as the electron donor was MtrABC-independent (Toporek et al., 2019). In the expanded working model with formate as the electron donor, electrons originating from formate dehydrogenase located at the head end of the electron transport chain are transferred to the inner membrane-localized menaquinone pool and subsequently to CymA (Toporek et al., 2019), which facilitates electron transfer across the periplasmic space to DmsE, a decaheme c-type cytochrome partially embedded in the outer membrane and encased in the β-barrel protein DmsF (Figure 5). DmsA and DmsB are secreted extracellularly by the type II protein secretion system and form a ternary complex with the DmsEF PEC module on the outside face of the outer membrane (Gralnick et al., 2006). The extracellular DMSO reductase DmsAB of S. oneidensis reduces IO3– as an alternate terminal electron acceptor. The SO4358/4357 complex, a paralog of DmsAB (Figure 1), is unlikely to be the preferred or alternate IO3– terminal reductase with lactate or formate as the electron donors.
The lactate-dependent S. oneidensis IO3– reduction system utilizes MtrAB and does not require DmsEF, but does require DmsAB as the IO3– terminal reductase. DmsAB is evolutionarily unrelated to MtrC, MtrF, or OmcA. Although the Mtr respiratory pathway is modular, MtrAB has not been reported to transfer electrons to extracellular reductases apart from MtrC, MtrF, or OmcA. However, previous work which tested all possible combinations of Mtr paralogs (displayed in Figure 1) were tested solely with Fe(III)-citrate as an electron acceptor (Schicklberger et al., 2013), and have not been tested with most other electron acceptors including IO3–. Compellingly, DmsE functionally replaces MtrA (Coursolle and Gralnick, 2012), and the DmsF paralog SO4359 functionally replaces MtrB under Fe(III) citrate-reducing conditions (Schicklberger et al., 2013). Furthermore, MtrAB forms a stable complex in the outer membrane without MtrC, and redox properties of MtrA are modulated upon the formation of an MtrCAB complex (Hartshorne et al., 2009). DmsAB may hypothetically localize to the outer face of the outer membrane through the type II secretion porin GspD and pair with MtrAB to reduce extracellular IO3– with lactate as the electron donor.
The extracellular IO3– reductase system of S. oneidensis differs from the periplasmic IO3– reductase (Idr) system of Pseudomonas sp. strain SCT and Denitromonas sp. IR-12 homologs that display amino acid sequence homology to respiratory arsenite oxidase; however, both systems require molybdopterin coordinating Mo (Yamazaki et al., 2020; Reyes-Umana et al., 2022). Results of this study provide new insights into the molecular mechanism of microbial IO3– reduction, yield details important to the biogeochemical cycling of iodine in marine systems, and provide information crucial to the development of alternative bioremediation technologies for the treatment of radioactive iodine-contaminated subsurface environments.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
H-DS, YT, JM, and RM performed the experiments. H-DS, YT, and TD wrote the manuscript. All authors had given approval to the final version of the manuscript.
Funding
The funding for this work was provided by the US Department of Energy Office of Environmental Management and the Richland Operations Office through a subcontract from the Pacific Northwest National Laboratory.
Author Disclaimer
Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the US Department of Energy Office of Environmental Management.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.852942/full#supplementary-material
References
Afkhami, A., Madrakian, T., and Zarei, A. R. (2001). “Spectrophotometric determination of periodate, iodate and bromate mixtures based on their reaction with iodide.”. Anal. Sci. 17, 1199–1202. doi: 10.2116/analsci.17.1199
Amachi, S. (2013). Iodine geochemistry and microbes: bacterial volatilization, accumulation, oxidation, reduction, sorption and dehalogenation of iodine. Chikyukagaku 47, 209–219.
Amachi, S., Kawaguchi, N., Muramatsu, Y., Tsuchiya, S., Watanabe, Y., Shinoyama, H., et al. (2007). Dissimilatory iodate reduction by marine Pseudomonas sp. strain SCT. Appl. Environ. Microbiol. 73, 5725–5730. doi: 10.1128/AEM.00241-07
Bagwell, C., Zhong, L., Wells, J., Mitroshkov, A., and Qafoku, N. P. (2019). Microbial methylation of iodide in unconfined aquifer sediments at the Hanford Site. USA. Front. Microbiol. 10, 2460. doi: 10.3389/fmicb.2019.02460
Beliaev, A. S., and Saffarini, D. A. (1998). Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180, 6292–6297. doi: 10.1128/JB.180.23.6292-6297.1998
Bluhm, K., Croot, P., Wuttig, K., and Lochte, K. (2010). Transformation of iodate to iodide in marine phytoplankton driven by cell senescence. Aquat. Biol. 11, 1–15. doi: 10.3354/ab00284
Bluhm, K., Croot, P. L., Huhn, O., Rohardt, G., and Lochte, K. (2011). Distribution of iodide and iodate in the Atlantic sector of the Southern Ocean during austral summer. Deep Sea Res. 58, 2733–2748. doi: 10.1016/j.dsr2.2011.02.002
Bradford, M. (1976). A rapid and sensitive method for quantification of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Bucking, C., Popp, F., Kerzenmacher, S., and Gescher, J. (2010). Involvement and specificity of Shewanella oneidensis outermembrane cytochromes in the reduction of soluble and solid-phase terminal electron acceptors. FEMS Microbiol. Lett. 306, 144–151. doi: 10.1111/j.1574-6968.2010.01949.x
Burns, J. L., and DiChristina, T. J. (2009). Anaerobic respiration of elemental sulfur and thiosulfate by Shewanella oneidensis MR-1 requires psrA, a homolog of the phsA gene of Salmonella enterica serovar Typhimurium LT2. Appl. Environ. Microbiol. 75, 5209–5217. doi: 10.1128/AEM.00888-09
Chance, R., Malin, G., Jickells, T., and Baker, A. R. (2007). Reduction of iodate to iodide by cold water diatom cultures. Mar. Chem. 105, 169–180. doi: 10.1016/j.marchem.2006.06.008
Chung, C. T., Niemela, S. L., and Miller, R. H. (1989). One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U S A 86, 2172–2175. doi: 10.1073/pnas.86.7.2172
Cianciotto, N. P. (2005). Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13, 581–588. doi: 10.1016/j.tim.2005.09.005
Cooper, R., Goff, J., Reed, B., Sekar, R., and DiChristina, T. J. (2016). “Breathing iron: molecular mechanism of microbial iron reduction by Shewanella oneidensis,” in Manual of Environmental Microbiology, 4th Edn, eds M. V. Yates, C. H. Nakatsu, R. V. Miller, and S. D. Pillai (Washington, DC: American Society for Microbiology).
Councell, T. B., Landa, E. R., and Lovley, D. R. (1997). Microbial reduction of iodate. Water Air. Soil. Pollut. 100, 99–106.
Coursolle, D., and Gralnick, J. A. (2010). Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 77, 995–1008. doi: 10.1111/j.1365-2958.2010.07266.x
Coursolle, D., and Gralnick, J. A. (2012). Reconstruction of extracellular respiratory pathways for iron (III) reduction in Shewanella oneidensis strain MR-1. Front. Microbiol. 3:56. doi: 10.3389/fmicb.2012.00056
Dale, J. R., Wade, R., and DiChristina, T. J. (2007). A conserved histidine in cytochrome c maturation permease CcmB of Shewanella putrefaciens is required for anaerobic growth below a threshold standard redox potential. J. Bacteriol. 189, 1036–1043. doi: 10.1128/JB.01249-06
DiChristina, T. J., Moore, C. M., and Haller, C. A. (2002). Dissimilatory Fe(III) and Mn(IV) reduction by Shewanella putrefaciens requires ferE, a homolog of the pulE (gspE) type II protein secretion gene. J. Bacteriol. 184, 142–151. doi: 10.1128/JB.184.1.142-151.2002
Emerson, H. P., Xu, C., Ho, Y. F., Zhang, S., Schwehr, K. A., Lilley, M., et al. (2014). Geochemical controls of iodine uptake and transport in Savannah River Site subsurface sediments. Appl. Geochem. 45, 105–113. doi: 10.1016/j.apgeochem.2014.03.002
Farrenkopf, A. M., Dollhopf, M. E., Chadhain, S. N., Luther, G. W., and Nealson, K. H. (1997). Reduction of iodate in seawater during Arabian Sea shipboard incubations and in laboratory cultures of the marine bacterium Shewanella putrefaciens strain MR-4. Mar. Chem. 57, 347–354. doi: 10.1016/s0304-4203(97)00039-x
Fennessey, C. M., Jones, M. E., Taillefert, M., and DiChristina, T. J. (2010). Siderophores are not involved in Fe(III) solubilization during anaerobic Fe(III) respiration by Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 76, 2425–2432. doi: 10.1128/AEM.03066-09
Fuge, R., and Johnson, C. C. (2015). Iodine and human health, the role of environmental geochemistry and diet, a review. Appl. Geochem. 63, 282–302. doi: 10.1016/j.apgeochem.2015.09.013
Gong, T. T., and Zhang, X. R. (2013). Determination of iodide, iodate and organo-iodine in waters with a new total organic iodine measurement approach. Water Res. 47, 6660–6669. doi: 10.1016/j.watres.2013.08.039
Gorby, Y. A., Yanina, S., McLean, J. S., Rosso, K. M., Moyles, D., Dohnalkova, A., et al. (2006). Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U.S.A. 103, 11358–11363. doi: 10.1073/pnas.0604517103
Gralnick, J. A., Vali, H., Lies, D. P., and Newman, D. K. (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U S A 103, 4669–4674. doi: 10.1073/pnas.0505959103
Guido-Garcia, F., Law, G. T. W., Lloyd, J. R., Lythgoe, P., and Morris, K. (2015). Bioreduction of iodate in sediment microcosms. Mineral. Mag. 79, 1343–1351. doi: 10.1180/minmag.2015.079.6.10
Hanzelmann, P., and Mayer, O. (1998). Effect of molybdenum and tungsten on biosynthesis of CO dehydrogenase and the molybdopterin cytosine-dinucleotide-type of molybdenum cofactor in Hydrogenophaga pseudoflava. Eur. J. Biochem. 255, 755–765. doi: 10.1046/j.1432-1327.1998.2550755.x
Hartshorne, R. S., Reardon, C. L., Ross, D., Nuester, J., Clarke, T. A., Gates, A. J., et al. (2009). Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. U.S.A. 106, 22169–22174. doi: 10.1073/pnas.0900086106
Hernandez, M. E., Kappler, A., and Newman, D. K. (2004). Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70, 921–928. doi: 10.1128/AEM.70.2.921-928.2004
Hou, X., Aldahan, A., Nielsen, S. P., Possnert, G., Nies, H., and Hedfors, J. (2007). Speciation of I-129 and I-127 in seawater and implications for sources and transport pathways in the North Sea. Environ. Sci. Technol. 41, 5993–5999. doi: 10.1021/es070575x
Hou, X. L., Povinec, P. P., Zhang, L. Y., Shi, K. L., Biddulph, D., Chang, C. C., et al. (2013). Iodine-129 in seawater offshore Fukushima: distribution, inorganic speciation, sources, and budget. Environ. Sci. Technol. 47, 3091–3098. doi: 10.1021/es304460k
Jones, M. E., Fennessey, C. M., DiChristina, T. J., and Taillefert, M. (2010). Shewanella oneidensis MR-1 mutants selected for their inability to produce soluble organic-Fe(III) complexes are unable to respire Fe(III) as anaerobic electron acceptor. Environ. Microbiol. 12, 938–950. doi: 10.1111/j.1462-2920.2009.02137.x
Kaplan, D. I., Denham, M. E., Zhang, S., Yeager, C., Xu, C., Schwehr, K. A., et al. (2014). Radioiodine biogeochemistry and prevalence in groundwater. Crit. Rev. Environ. Sci. Technol. 44, 2287–2335. doi: 10.1080/10643389.2013.828273
Kenyon, J. A., Buesseler, K. O., Casacuberta, N., Castrillejo, M., Otosaka, S., Masqueì, P., et al. (2020). Distribution and Evolution of Fukushima Dai-ichi derived 137Cs, 90Sr, and 129l in Surface Seawater off the Coast of Japan. Environ. Sci. Technol. 54, 15066–15075. doi: 10.1021/acs.est.0c05321
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. I. I., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Lee, B. D., Ellis, J. T., Dodwell, A., Eisenhauer, E. E. R., Saunders, D. L., and Lee, M. H. (2018). Iodate and nitrate transformation by Agrobacterium/Rhizobium related strain DVZ35 isolated from contaminated Hanford groundwater. J. Hazard. Mater. 350, 19–26. doi: 10.1016/j.jhazmat.2018.02.006
Marsili, E., Baron, D. B., Shikhare, I. D., Coursolle, D., Gralnick, J. A., and Bond, D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973. doi: 10.1073/pnas.0710525105
May, H. D., Patel, P. S., and Ferry, J. G. (1988). Effect of molybdenum and tungsten on synthesis and composition of formate dehydrogenase in Methanobacterium formicicum. J. Bacteriol. 170, 3384–3389. doi: 10.1128/jb.170.8.3384-3389.1988
McEwan, A. G., Ridge, J. P., McDevitt, C. A., and Hugenholtz, P. (2002). The DMSO reductase family of microbial molybdenum enzymes; molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 19, 3–21. doi: 10.1080/014904502317246138
McLaughlin, L. S., Haft, R. J. F., and Forest, K. T. (2012). Structural insights into the type II secretion nanomachine. Curr. Opin. Struct. Biol. 22, 208–216. doi: 10.1016/j.sbi.2012.02.005
Mok, J. K., Toporek, Y. J., Shin, H. D., Lee, B. D., Lee, M. H., and DiChristina, T. J. (2018). Iodate reduction by Shewanella oneidensis does not involve nitrate reductase. Geomicrobiol. J. 35, 570–579. doi: 10.1080/01490451.2018.1430189
Myers, C. R., and Myers, J. M. (1992). Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174, 3429–3438. doi: 10.1128/jb.174.11.3429-3438.1992
Myers, C. R., and Nealson, K. H. (1988). Bacterial manganese reduction and growth with manganese oxide as the sole electron-acceptor. Science 240, 1319–1321. doi: 10.1126/science.240.4857.1319
Neu, M. P., Icopini, G. A., and Boukhalfa, H. (2005). Plutonium speciation affected by environmental bacteria. Radiochim. Act. 93, 705–714. doi: 10.1016/j.jenvrad.2015.08.019
Newsome, L., Morris, K., and Lloyd, J. R. (2014). The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem. Geol. 363, 164–184. doi: 10.1016/j.chemgeo.2013.10.034
Payne, A. N., and DiChristina, T. J. (2006). A rapid mutant screening technique for detection of technetium [Tc (VII)] reduction-deficient mutants of Shewanella oneidensis MR-1. FEMS Microbiol. Lett. 259, 282–287. doi: 10.1111/j.1574-6968.2006.00278.x
Reyes-Umana, V., Henning, Z., Lee, K., Barnum, T. P., and Coates, J. D. (2022). Genetic and phylogenetic analysis of dissimilatory iodate-reducing bacteria identifies potential niches across the world’s oceans. ISME J. 16, 38–49. doi: 10.1038/s41396-021-01034-5
Richardson, D. J., Butt, J. N., Fredrickson, J. K., Zachara, J. M., Shi, L., Edwards, M. J., et al. (2012). The ‘porincytochrome’ model for microbe-to-mineral electron transfer. Mol. Microbiol. 85, 201–212. doi: 10.1111/j.1365-2958.2012.08088.x
Richter, K., Schicklberger, M., and Gescher, J. (2012). Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl. Environ. Microbiol. 78, 913–921. doi: 10.1128/AEM.06803-11
Riley, B. J., Vienna, J. D., Strachan, D. M., McCloy, J. S., and Jerden, Jr (2016). Materials and processes for the effective capture and immobilization of radioiodine: a review. J. Nucl. Mater. 470, 307–326. doi: 10.1016/j.jnucmat.2015.11.038
Roden, E. E., Kappler, A., Bauer, I., Jiang, J., Paul, A., Stoesser, R., et al. (2010). Extracellular electron transfer through microbial reduction of solid-phase humic substances. Nature Geosci. 3, 417–421. doi: 10.1016/j.scitotenv.2019.134683
Saffarini, D. A., Schultz, R., and Beliaev, A. (2003). Involvement of cyclic AMP (CAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 185, 3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003
Schicklberger, M., Sturm, G., and Gescher, J. (2013). Genomic plasticity enables a secondary electron transport pathway in Shewanella oneidensis. Appl. Environ. Microbiol. 79, 1150–1159. doi: 10.1128/AEM.03556-12
Shi, L., Chen, B. W., Wang, Z. M., Elias, D. A., Mayer, M. U., Gorby, Y. A., et al. (2006). Isolation of a high-affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 188, 4705–4714. doi: 10.1128/JB.01966-05
Shi, L., Rosso, K. M., Zachara, J. M., and Fredrickson, J. K. (2012). Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: a genomic perspective. Biochem. Soc. Trans. 40, 1261–1267. doi: 10.1042/BST20120098
Szeinbaum, N., Burns, J. L., and DiChristina, T. J. (2014). Electron transport and protein secretion pathways involved in Mn(III) reduction by Shewanella oneidensis. Environ. Microbiol. Rep. 6, 490–500. doi: 10.1111/1758-2229.12173
Taillefert, M., Beckler, J. S., Carey, E., Burns, J. L., Fennessey, C. M., and DiChristina, T. J. (2007). Shewanella putrefaciens produces an Fe(III)-solubilizing organic ligand during anaerobic respiration on insoluble Fe(III) oxides. J. Inorg. Biochem. 101, 1760–1767. doi: 10.1016/j.jinorgbio.2007.07.020
Timar, J., Elekes, Z., and Singh, B. (2014). Nuclear data sheets for A=129. Nucl. Data Sheets 121, 143–394. doi: 10.1016/j.nds.2014.09.002
Toporek, Y. J., Mok, J. K., Shin, H. D., Lee, B. D., Lee, M. H., and DiChristina, T. J. (2019). Metal reduction and protein secretion genes required for iodate reduction by Shewanella oneidensis. Appl. Environ. Microbiol. 85, e2115–e2118. doi: 10.1128/AEM.02115-18
Tsunogai, S., and Sase, T. (1969). Formation of iodide-iodine in the ocean. Deep Sea Res. Part 1 16, 489–496. doi: 10.1016/0011-7471(69)90037-0
Venkateswaran, K., Moser, D. P., Dollhopf, M. E., Lies, D. P., Saffarini, D. A., MacGregor, B. J., et al. (1999). Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49, 705–724. doi: 10.1099/00207713-49-2-705
Waite, T. J., and Trucesdale, V. W. (2003). Iodate reduction by isochrysis galbana is relatively insensitive to de-activated of nitrate reductase activity. Marine Chem. 81, 137–148. doi: 10.1016/s0304-4203(03)00013-6
Wee, S. K., Burns, J. L., and DiChristina, T. J. (2014). Identification of a molecular signature unique to metal-reducing gammaproteobacteria. FEMS Microbiol. Lett. 350, 90–99. doi: 10.1111/1574-6968.12304
White, G. F., Edwards, M. J., Gomez-Perez, L., Richardson, D. J., Butt, J. N., and Clarke, T. A. (2016). Mechanisms of bacterial extracellular electron exchange. Adv. Microb. Physiol. 68, 87–138. doi: 10.1016/bs.ampbs.2016.02.002
Whitehead, D. C. (1984). The distribution and transformations of iodine in the environment. Environ. Int. 10, 321–339. doi: 10.1016/0160-4120(84)90139-9
Wong, G. T. F., and Hung, C. C. (2001). Speciation of dissolved iodine: integrating nitrate uptake over time in the oceans. Cont. Shelf Res. 21, 113–128. doi: 10.1016/s0278-4343(00)00086-8
Wong, G. T. F., Piumsomboon, A. U., and Dunstan, W. M. (2002). The transformation of iodate to iodide in marine phytoplankton cultures. Mar. Ecol. Prog. Ser. 237, 27–39. doi: 10.3354/meps237027
Yamazaki, C., Kashiwa, S., Horiuchi, A., Kasahara, Y., Yamamura, S., and Amachi, S. A. (2020). novel dimethylsulfoxide reductase family of molybdenum enzyme. Environ. Microbiol. 22, 2196–2212. doi: 10.1111/1462-2920.14988
Yeager, C. M., Amachi, S., Grandbois, R., Kaplanm, D. I., Xu, C., Schwehr, K. A., et al. (2017). “Microbial transformation of iodine: from radioisotopes to iodine deficiency,” in Advances in Applied Microbiology, Vol. 101, eds S. Sariaslani and G. M. Gadd (Amsterdam: Elsevier Inc), 83–136. doi: 10.1016/bs.aambs.2017.07.002
Keywords: iodate reduction, radioactive iodine, bioremediation, DMSO reductase, Shewanella oneidensis, anaerobic respiration, formate metabolism, molybdopterin
Citation: Shin H-D, Toporek Y, Mok JK, Maekawa R, Lee BD, Howard MH and DiChristina TJ (2022) Iodate Reduction by Shewanella oneidensis Requires Genes Encoding an Extracellular Dimethylsulfoxide Reductase. Front. Microbiol. 13:852942. doi: 10.3389/fmicb.2022.852942
Received: 11 January 2022; Accepted: 07 March 2022;
Published: 14 April 2022.
Edited by:
Eric D. van Hullebusch, Université de Paris, FranceReviewed by:
Daniel R. Bond, University of Minnesota Twin Cities, United StatesYamini Jangir, California Institute of Technology, United States
Hor-Gil Hur, Gwangju Institute of Science and Technology, South Korea
Copyright © 2022 Shin, Toporek, Mok, Maekawa, Lee, Howard and DiChristina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas J. DiChristina, dGhvbWFzLmRpY2hyaXN0aW5hQGJpb2xvZ3kuZ2F0ZWNoLmVkdQ==
†These authors have contributed equally to this work and share first authorship
 Hyun-Dong Shin
Hyun-Dong Shin Yael Toporek1†
Yael Toporek1† Ruri Maekawa
Ruri Maekawa Brady D. Lee
Brady D. Lee Thomas J. DiChristina
Thomas J. DiChristina