- 1School of Science, Mae Fah Luang University, Chiang Rai, Thailand
- 2Center of Chemical Innovation for Sustainability (CIS), Mae Fah Luang University, Chiang Rai, Thailand
Polyester urethanes (PUR) are widely used in industries and have led to a worldwide plastic waste problem. Thus, novel solutions for PUR degradation are required to reduce environmental pollution. This work investigates the PUR biodegradation efficiency of 33 fungal species using a polyester-polyurethane colloid branded Impranil DLN (Impranil) compared to Aspergillus niger, which served as the positive control. The biodegradation is evaluated based on its ability to clear Impranil in media. Eleven fungi can clear Impranil in both solid- and liquid-medium assays. The highest degradation was attributed to Embarria clematidis cultured with Impranil as a carbon source. The degradation was confirmed by the Sturm test, Fourier-transform infrared (FTIR) spectroscopy, and gas chromatography-mass spectrometry (GC-MS). From the Sturm test, CO2 at a concentration of 0.85 g/L was found in E. clematidis cultured with 150 mL of Impranil solution after a 2-week incubation period while the CO2 at a concentration of 0.53 g/L was detected from A. niger in the same conditions. The biodegradation was further confirmed by evaluating the clearance percentage of supernatant of E. clematidis and A. niger culturing with Impranil from the Sturm test. The clearance percentage of E. clematidis and A. niger supernatant was 88.84 and 48.97%, respectively. Moreover, the degradation of soft segment and breakdown of ester linkages were observed, as evidenced by the decrease of the carbonyl (1,715 cm–1) and N-H stretching (1,340 cm–1 and 1,020 cm–1) FTIR spectral peaks, respectively. GC-MS detected 3Z-heptenol, 5Z-octenol, 2E,4E-hexadienol acetate, and 3E,6Z-nonadienol as degradation products from the E. clematidis culture supernatant. This fungus was screened for its ability to produce extracellular esterase, protease, and urease enzymes. Extracellular esterase, very low urease, and no protease activities were detected in the culture supernatant of E. clematidis in the presence of Impranil. E. clematidis can degrade Impranil partially via hydrolysis of ester linkages by cell-bound esterases at a considerable rate without any prior treatment. This fungus not only degraded Impranil but also mineralized them into CO2 and H2O. E. clematidis can be applied in the process of biochemical depolymerization of PUR for the pure monomers recycling.
Introduction
Plastic has tremendously used due to its excellent physical properties such as versatility, malleability, chemical, and physical resistance, and ease of production compared to other materials. Polyester polyurethane (PUR) is a major plastic material that is widely used in many industrial applications to produce foams, elastomers, coatings, and adhesives (Howard, 2002). The estimated production of PUR is approximately 8 million tons annually (Matsumura et al., 2006). It is mainly produced via the nucleophilic reaction from two monomers (a diol and a highly toxic and reactive diisocyanate) and is derived from the toxic substance phosgene in the presence of some additives. Reprocessing of PUR generates toxic residues, such as aldehydes, ammonia, cyanide, isocyanates, nitrogen oxides, and vinyl chloride, which are hazardous to humans, disrupt ecological processes, and contribute to environmental pollution (Cregut et al., 2013). PUR is also physically stable, making it resistant to biological degradation (Yang et al., 2001). Moreover, the large amount of produced PUR materials has resulted in a massive increase in plastic waste (Miskolczi et al., 2004). Hence, effective methods for the degradation of plastic wastes are needed to solve the problems posed by PUR waste accumulation.
Recently, various bacteria and fungi have been demonstrated to degrade PUR materials. Fungi in some genera have been reported to degrade PUR, including Alternaria, Aspergillus, Phoma, Penicillium, Plectosphaerella, Geomyces, Nectria, and Neonectria (Cosgrove et al., 2007; Schmidt et al., 2017). In addition, Aspergillus niger, Aspergillus flavus, Aspergillus vesicolor, Penicillium funicutosum, Aureobasidium pullulaus, some species of Trichoderma sp., and Chyaetomium glubosum also can degrade PUR reported by Darby and Kaplan (1968). Some bacterial species, including a few species of Corynebacterium genus, Enterobacter agglomerans, and Bacillus subtilis have also been reported to efficiently degrade PUR samples (Rowe and Howard, 2002; Mahajan and Gupta, 2015; Schmidt et al., 2017). However, few organisms have been demonstrated to degrade PUR as the sole carbon source (Shah et al., 2008; Russell et al., 2011).
Several reports have indicated that the active enzymes involved in microbial degradation of PUR are esterases, lipases, proteases, and ureases, suggesting the degradation of the PUR substrate by the cleavage of the ester bond (Tokiwa and Calabia, 2004; Shah et al., 2008; Kale et al., 2015; Danso et al., 2019; Ru et al., 2020). These enzymes were isolated from various microorganisms such as Pseudomonas fluorescens, Comamonas acidovorans, Pseudomonas chlororaphis, Pseusomonas aeruginosa, Acinetobacter gerneri, Bacillus subtilis MZA-75, Comamonas acidovorans TB-35, Aspergillus terreus, and Chaetomium globosum (Allen et al., 1999; Yang et al., 2001; Shah et al., 2008). Although both fungi and bacteria have been reported to degrade PUR, the actual mechanism of the biodegradation process remains unknown. The ability of these microorganisms to degrade PUR offers the possibility of being able to degrade other complex polymers as well (Russell et al., 2011). A massive number of microorganisms could have the degradation ability to colonize the surface of PUR (Glaser, 2019). Therefore, investigating other PUR-degrading microorganisms and their enzymatic activities is essential for efficient PUR waste management (Liu et al., 2021).
This study presents the Impranil biodegradation potential by various fungi. The study also demonstrates their ability to degrade Impranil on a culture plate, in a liquid medium, and with Impranil as the sole carbon source. Analysis of its CO2 production, enzymatic activities, and degradation products was also performed.
Materials and Methods
Fungal Strains
Thirty-four fungal species were selected for this study (Table 1). The fungus A. niger was obtained from the Thailand Institute of Scientific and Technological Research, Bangkok, Thailand while the other strains were obtained from the culture collection of the Institute of Excellence in Fungal Research, Mae Fah Luang University, Thailand.
Screening of Impranil-Degrading Activity
All fungi were screened for their Impranil-degrading activity on solid-medium assay (Russell et al., 2011). Impranil, an anionic aliphatic aqueous PUR dispersion (Bayer MaterialScience, New Jersey, United States), was used to prepare the polymer solution. A solid medium was prepared following a protocol by Russell et al. (2011), which involves mixing 19 mM NaH2PO4, 33.5 mM K2HPO4, 7.6 mM (NH4)2SO4, 2.5 mM Na citrate, 250 μM MgSO4, 19 μM thiamine, 0.05% Casamino acids, 147 μM FeCl3⋅6H2O, 14 μM ZnCl2⋅4H2O, 12 μM CoCl2⋅6H2O, 12 μM Na2MoO4⋅2H2O, 10 μM CaCl2⋅2H2O, 11 μM CuCl2, 12 μM MnCl2, 12 μM H3BO3, 1.8 mM HCl, and 15 g of agar in 1,000 mL deionized water. Impranil (10 mL) was added to the medium after autoclaving to prevent deformation. A plug of fungi (5 mm diameter), which was cultured on potato dextrose agar (PDA) for 1 week, was placed at the center of solid medium plates. All plates were further kept at room temperature for 2 weeks. The plates containing the agar plug of A. niger were used as the positive control. The Impranil-degrading activity of the fungi was evaluated by inspecting for halos of clearance and colony diameter after 2 weeks of incubation.
Analysis of Clearance
Solid-Medium Assay
The fungi that showed Impranil-degrading activity were further assayed for their clearance via the solid-medium screening assay, according to the method of Russell et al. (2011). The solid medium (20 mL) was placed in sterile tubes. A plug of fungi (5 mm diameter) cultured on PDA for 1 week was placed on top of each test tube before 2 weeks of incubation at room temperature. The fungus A. niger was used as a positive control for Impranil degradation. After 2 weeks of incubation, Impranil degradation was evaluated via the clearance of the medium, which changed from opaque to translucent. The clearance depth was measured from the top of the medium to the lowest point of the visible clearance.
Liquid-Medium Assay
The fungi that showed Impranil-degrading activity was also assayed for their clearance via the liquid-medium screening assay, according to a modified method described by Russell et al. (2011). The liquid medium was prepared using the same composition found in the solid medium but without agar. The liquid medium (150 mL) was added to a sterile culture flask and inoculated with five plugs of fungus grown on PDA for 1 week. All culture flasks were further incubated at room temperature for 2 weeks. At the end of incubation, the fungal mycelium was separated from the culture supernatant using vacuum filtration. The obtained liquid supernatant was homogenized by vigorous shaking and the absorbance of the supernatant liquid was measured using a UV-visible PerkinElmer spectrophotometer at a wavelength of 600 nm. The sterile deionized water was used as a blank. Series dilutions of liquid medium prepared in sterile water were used to construct a standard curve for converting absorbance to percent clearance. The fungus A. niger was used as a positive control.
Sole Carbon Source Assay
The fungi that showed Impranil-degrading activity were tested for their ability to use Impranil as the sole carbon source, following a modified method described by Russell et al. (2011). To ensure that there was no other carbon source, Impranil was only used as the sole carbon source for metabolism and fungal growth. The fungus A. niger was tested as a positive control. Each fungus was cultured in a potato dextrose broth (PDB) for 1 week. Stock cultures were homogenized by vigorous shaking and further centrifuged at 3,000 rpm for 5 min. The supernatant was removed and the fungal pellets were washed three times with sterile distilled water to confirm the complete removal of residual PDB. The fungal pellet was then added to 150 mL of liquid medium, which was prepared similar to the liquid-medium assay without agar and salts. The visual clearance of the opaque culture supernatant was determined every 2 days for 2 weeks by measuring the optical density at 600 nm.
CO2 Production Assay
The amount of CO2 released by E. clematidis in the presence and absence of Impranil as the sole carbon source after a 2-week inoculation was investigated. The modified Sturm test was used in this experiment. Sterile was first flowed through 1 M KOH solution to remove atmospheric CO2 before analysis. The amount of dissolved CO2 in the medium broth was determined every 2 days for 2 weeks by titration method (Ameen et al., 2016). The medium broth was filtered and 25 mL of filtrate was mixed with 0.05 mL of 0.1 N Na2S2O3 and 2 drops of methyl orange. The solution was titrated with 0.02 M NaOH solution until the solution turned yellow. After that, two drops of phenolphthalein were added to the solution, and titration was continued until the solution turned pink. The volume of the used titrant was recorded. The CO2 amount was calculated using the following equation: [A × B × 50 × 1,000]/V, where A = volume of NaOH in mL, B = normality of NaOH, and V = volume of sample in mL. The fungus A. niger was used as a positive control.
Fourier-Transform Infrared Spectroscopy Analysis
Degradation by E. clematidis in the presence of Impranil as the sole carbon source in a liquid medium was also characterized by FTIR (Álvarez-Barragán et al., 2016). After the 2-week incubation, the mycelium was removed by filtration filter paper and 1 mL of each filtrate was placed in sample cell before its spectra was collected using FTIR (Lumos, Bruker) with 64 scans at a resolution of 4 cm–1. All spectra were presented within the wavenumber range of 500–4,000 cm–1. Deionized water spectrum was used for the background spectrum. The results were compared to the spectra obtained from the A. niger culture in the presence of Impranil as the sole carbon source after 2-week inoculation and the liquid medium without fungal culture.
GC-MS Analysis
After 2-week incubation, 20 mL of filtered culture supernatants of E. clematidis and A. niger in the presence of Impranil as the sole carbon source were partitioned with 20 mL of dichloromethane (Sigma Chemical Co., St. Louis) using a separatory funnel. The volatile organic compounds was analyzed using the GC-MS system (Agilent Technologies, Santa Clara, CA, United States). The capillary column is used as an HP-5 ms column (30 m × 0.25 mm i.d., 0.25 μm film thickness) (Agilent Technologies, Santa Clara, CA, United States). The oven temperature was initially held at 60°C and then increased at a rate of 3°C/min to a final temperature of 220°C. The injector temperature was set to 250°C. Helium was used as a carrier gas with a flow rate of 1 mL/min. Electron impact mass spectra were collected at 70 eV over the range of m/z 30–300. The ion source and analyzer temperatures were set to 250 and 230°C, respectively. Identification of compounds was done by comparing their mass spectra with those found in the NIST14 Mass Spectral Library.
Enzymatic Activity Assay
The enzymatic activity of esterase, lipase, and urease was measured using spectrophotometry. These enzymes were measured every 2 days from cultures of E. clematidis in the presence of Impranil as the sole carbon source following the protocol of Álvarez-Barragán et al. (2016). The filtrates were centrifuged at 10,000 × g for 10 min and the cell pellet was removed from the supernatant. The obtained supernatants were further filtered through 0.45 nm-pore size Millipore membranes, obtaining a solution with a total volume of 3 mL. Next, three dialysis rounds were sequentially carried out against 50 mM potassium phosphate buffer (pH 7.0) for 2 h (in 1,000 mL of buffer), 24 h (in 2,000 mL of buffer), and 2 h (in 1,000 mL of buffer). For 0–7 days of incubation, 1 μg of protein was used, whereas, for 8–14 days of incubation, 4 μg of protein was used for the enzymatic assays. Esterase activity was determined by the hydrolysis of p-nitrophenyl acetate (Sigma-Aldrich, United Kingdom). The reaction was prepared by mixing 50 mM potassium phosphate (pH 7.0), 100 mL of enzyme extract, and 5 mM p-NPA in a final volume of 1 mL, following a protocol introduced by Oceguera-Cervantes et al. (2007). Protease activity was determined by casein hydrolysis, following the modified method of Oceguera-Cervantes et al. (2007). The reaction solution contained 100 mM potassium phosphate (pH 7.0), 50 mL of enzyme extract, and 0.5% casein in a final volume of 1 before incubation at 37°C for 24 h. Urease activity was determined based on the amount of ammonia from a phenol hypochlorite assay, following a protocol by Oceguera-Cervantes et al. (2007). The reaction solution contained 15 mM potassium phosphate (pH 7.0), 100 mL of enzyme extract, and 3.8 mM urea in a final volume of 1.3 mL. The solutions were determined spectrophotometrically at 405, 280, and 636 nm for esterase, lipase, and urease activity assay, respectively. The results were compared to those obtained from the liquid medium without fungal culture and the A. niger in the presence and absence of Impranil.
Statistical Analysis
All experiments were performed in triplicates. Data from each experiment were presented in terms of the mean ± standard deviation. Data were subjected to Analysis of Variance (ANOVA), followed by post hoc multiple pairwise comparisons using Duncan’s multiple range tests. The statistical analyses were conducted using the SPSS 20.0 software (IBM Corp.; 2011, NY, United States).
Results
Screening of Impranil-Degrading Fungi
Thirty-three fungal species were tested on their ability to degrade Impranil as compared to A. niger. After 2 weeks of incubation, the visible halo of clearance around the colony was evaluated and recorded. It was found that halos were detected in 12 fungal species, including P. rubi, O. tanaceti, M. clematidis, R. scabrispora, E. clematidis, S. lathyri, and P. bromicola, N. multiseptata, V. chlamydospore, H. obiones, C. fructicola, and A. niger. The colony diameter of these active fungi was also measured (Figure 1). E. clematidis exhibited the biggest colony diameter (7.3 cm). Some fungi, including A. niger, V. chlamydospore, H. obiones, C. fructicola, N. multiseptata, S. lathyri, P. bromicola, and R. scabrispora, exhibited a moderate colony diameter (3.4–4.7 cm), while M. clematidis, P. rubi, and O. tanaceti exhibited a small colony diameter of less than 3.0 cm.
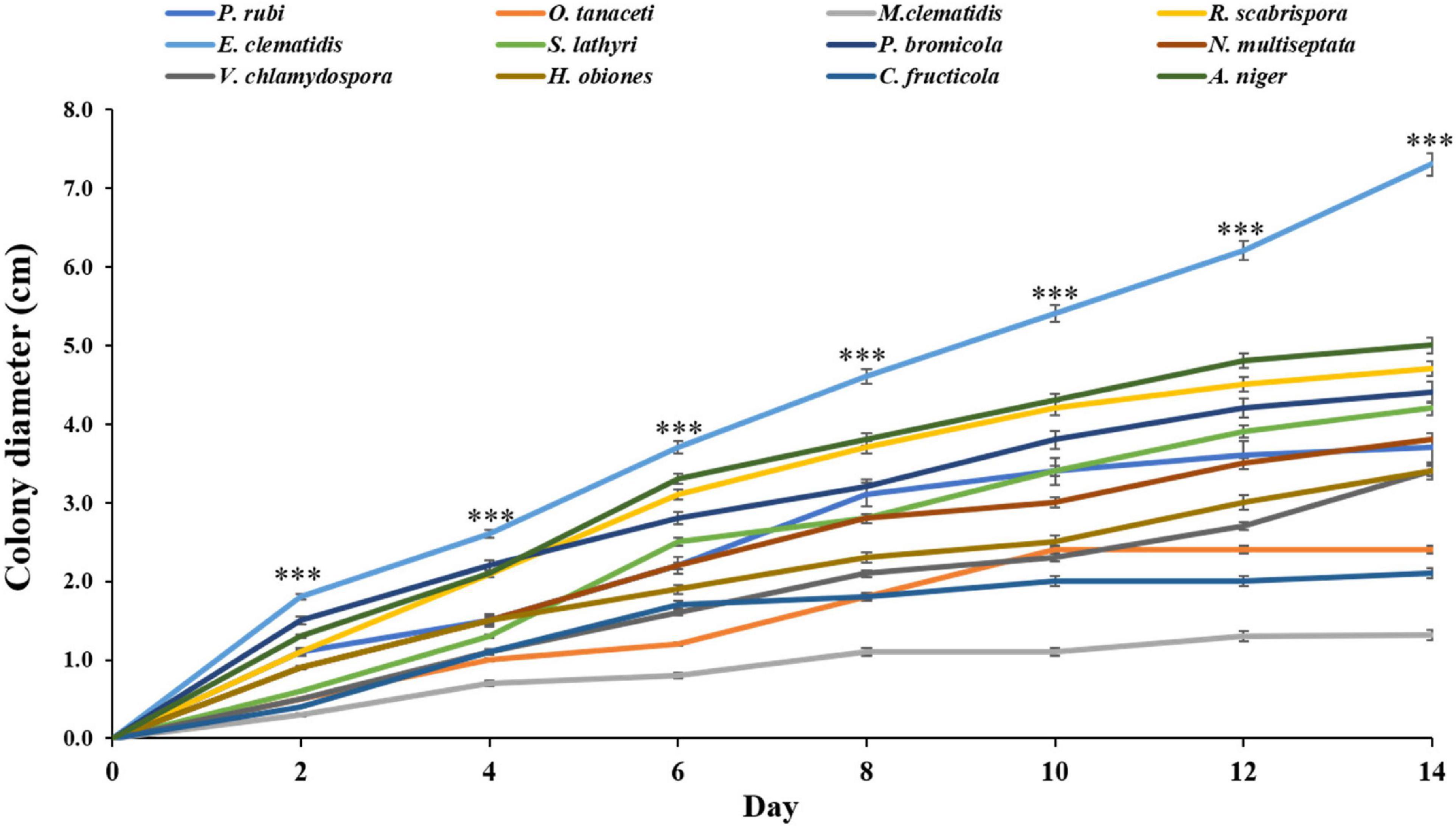
Figure 1. Colony diameter of active fungi on solid medium plates after 2-week inoculation. Sign *** above the points indicates a significant difference (p < 0.001) (ANOVA, followed by Dunnett’s test).
Analysis of Impranil Clearance
The active fungi were tested in Impranil clearance assays using solid- and liquid-medium. In the solid-medium assay, each fungal plug was inoculated in test tubes with the solid medium and the clearance depth was measured after 2 weeks of incubation (Figure 2). All of the 12 active fungi visibly cleared the solid medium. Four fungi (S. lathyri, C. fructicola, R. scabrispora, and E. clematidis) cleared the medium more efficiently than the positive control fungus A. niger (2.24 cm). The most efficient fungi for Impranil degradation was E. clematidis with an average clearance depth of 3.28 cm. The Impranil clearance percentage of the active fungi was also measured in a liquid-medium assay (Figure 3). The liquid cultures were cleared to the point of visual transparency at the top of the liquid culture, extending down the length of the flask. The relative order of the liquid clearance assay was similar to those observed for the solid-medium assay. The E. clematidis was the most active fungus in the liquid-medium assay, followed by R. scabrispora, C. fructicola, and S. lathyri with a clearance rate of 88.84, 64.68, 53.19, and 50.05%, respectively. These fungi cleared the liquid culture better than A. niger, the positive fungal control, after 2 weeks of incubation.
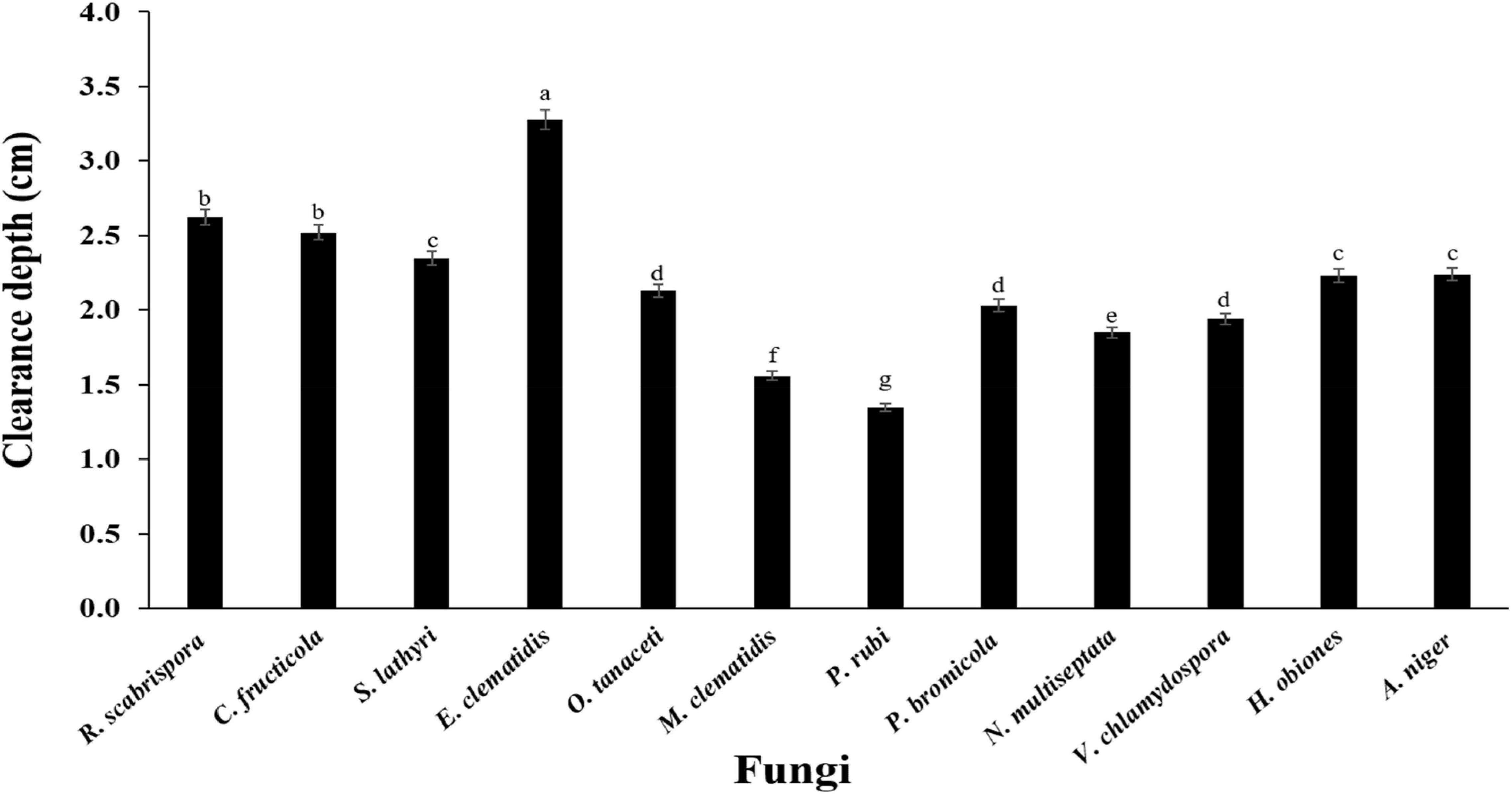
Figure 2. Clearance depth of solid medium of active fungi after 2-week inoculation. Different letters indicate significant differences (p < 0.05) (ANOVA, followed by Duncan’s multiple range test).
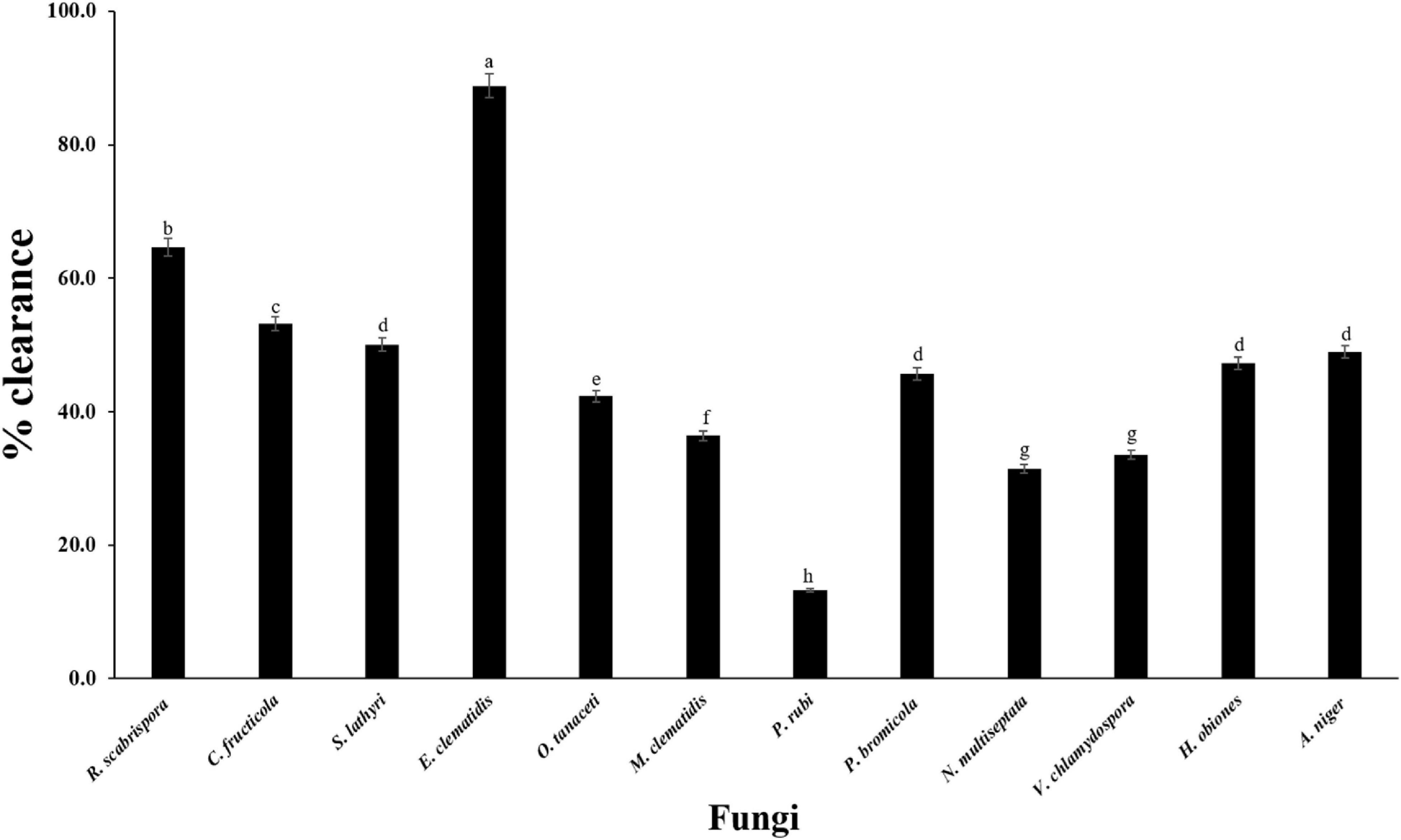
Figure 3. Clearance percentage of culture supernantant of active fungi after 2-week inoculation. Different letters indicate significant differences (p < 0.05) (ANOVA, followed by Duncan’s multiple range test).
Sole Carbon Source Assay
All active fungi were tested for their ability to degrade Impranil in liquid culture using Impranil as the sole carbon source. If fungus grows in the liquid medium, it means that Impranil can be used as the sole carbon source for its metabolism and growth. The tested fungi from the solid- and liquid-medium assays demonstrated Impranil degradation activity even under this minimal medium condition. To compare the degradation rates of Impranil, the active fungi were grown for 2 weeks. The optical density of the culture supernatant of active fungi after a 2-week inoculation period is depicted in Figure 4. The cultures were visually transparent by the end of the 2 weeks. E. clematidis, with an optical density value of 0.21, has the highest clearance rate among the active fungi including the control A. niger, which showed an optical density value of 0.35. The lowest clearance rate was found in C. fructicola with an optical density value of 0.71.
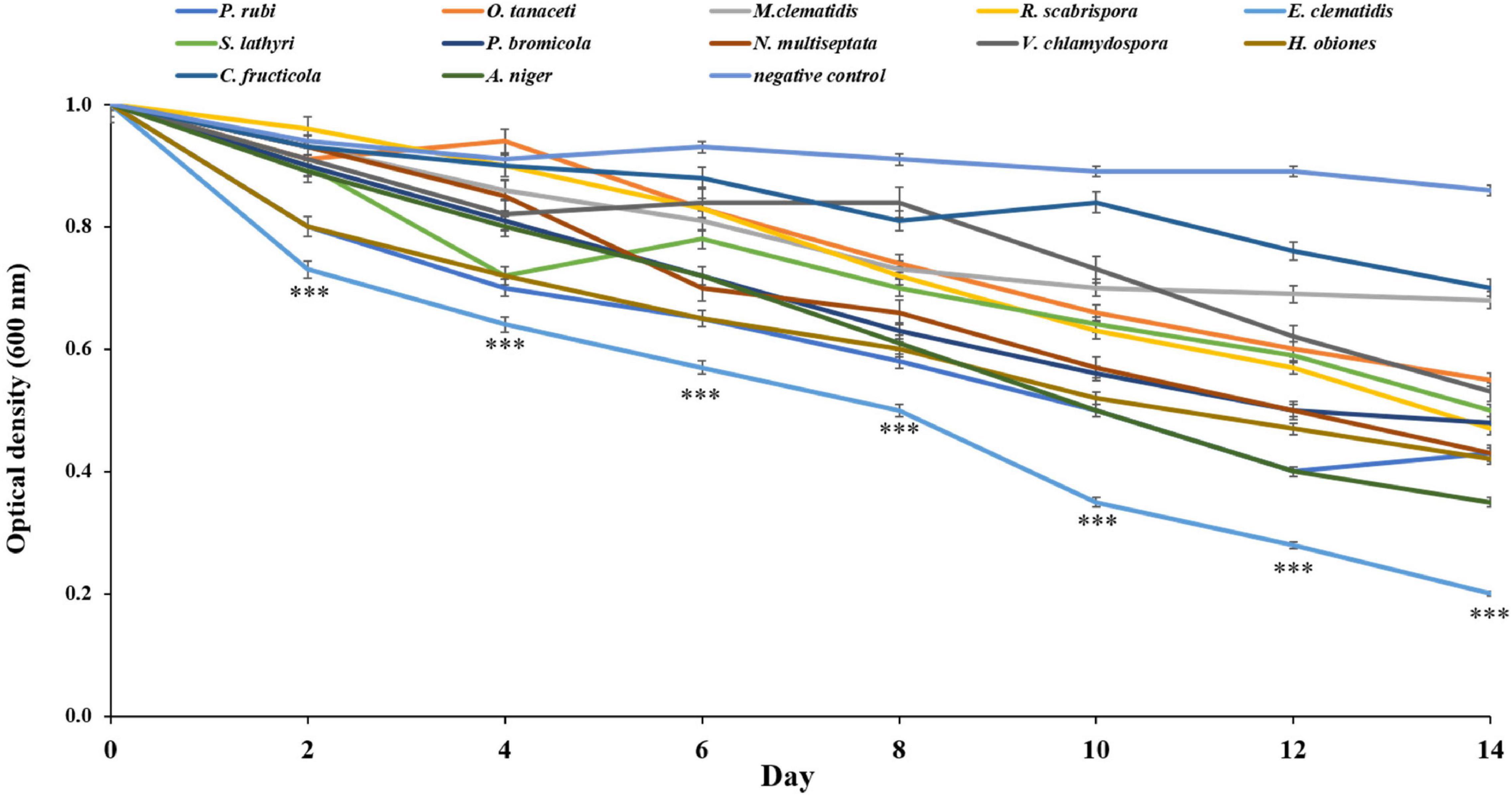
Figure 4. Optical density of culture supernatant of active fungi after 2-week inoculation. ***Above the points indicates a significant difference (p < 0.001) (ANOVA, followed by Dunnett’s test).
CO2 Production
The amounts of CO2 released by E. clematidis and A. niger in the presence and absence of Impranil after a 2-week incubation are shown in Figure 5. E. clematidis in the presence of Impranil demonstrated the highest CO2 amount among the tested treatments, with the highest amount detected on day 8 of the incubation. The CO2 concentration from the other treatments ranged from 0.00 to 0.85 g/L. A. niger in the presence of Impranil also demonstrated a high amount of CO2, ranging from 0.00 to 0.53 g/L, with the highest CO2 amount detected on day 8 as well. Similar CO2 amounts were detected in E. clematidis and A. niger in the absence of Impranil, ranging from 0.00 to 0.27 g/L and 0.00 to 0.24 g/L, respectively, after a 2-week incubation.
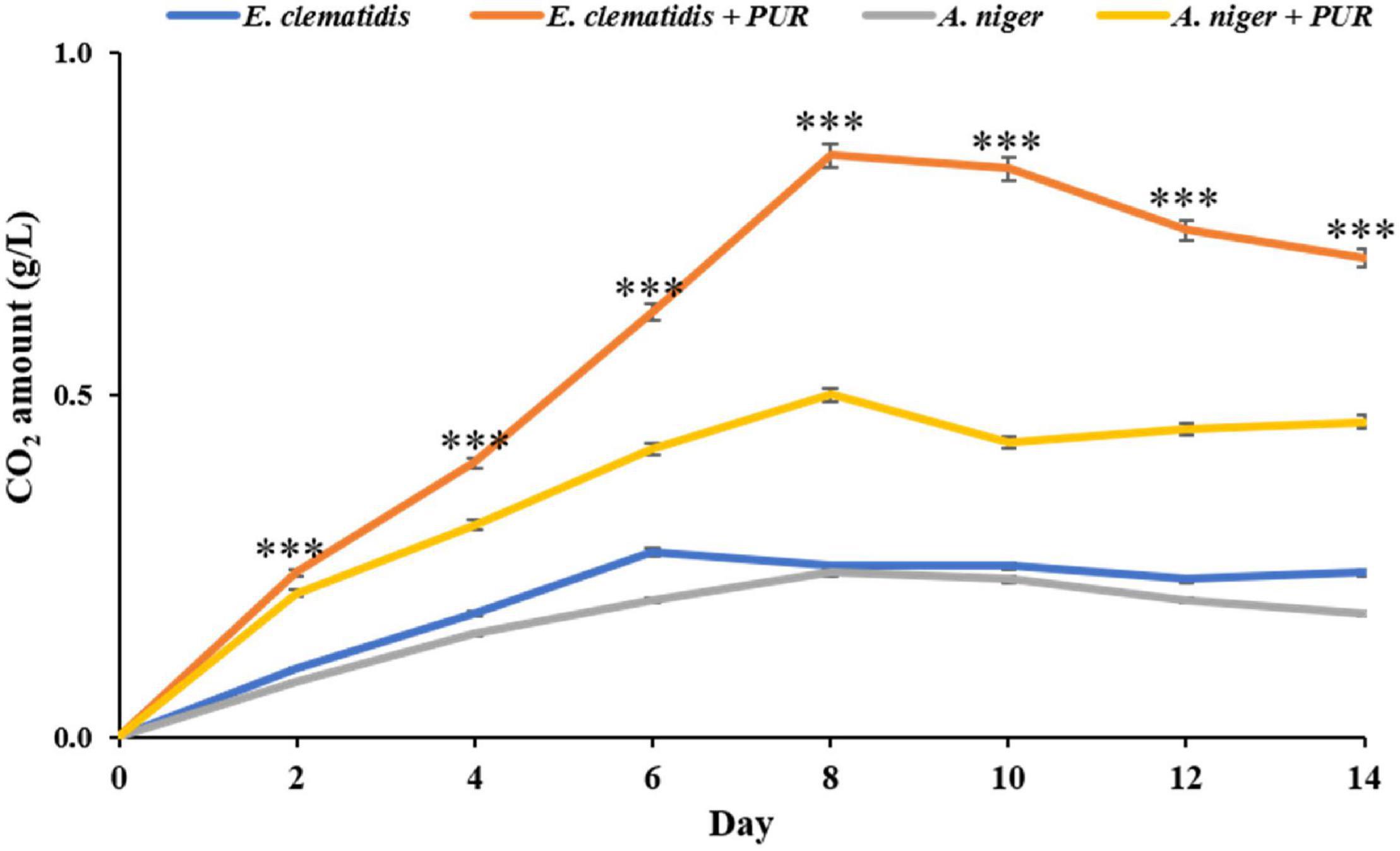
Figure 5. CO2 amount released by E. clematidis and A. niger in the presence and absence of Impranil as the sole carbon source after 2-week inoculation. ***Above the points indicates a significant difference (p < 0.001) (ANOVA, followed by Dunnett’s test).
Fourier-Transform Infrared Spectroscopy Analysis
The chemical structure of culture broth during the Impranil degradation by E. clematidis was investigated using FTIR spectroscopy (Figure 6). The liquid medium without fungi displayed a large absorption peak at 1,715 cm–1, corresponding to the carbonyl (C=O) stretch of the ester linkage in the Impranil (Figure 6A). In addition, the low intensities of the C-N-H bending (1,340 cm–1) and stretching (1,020 cm–1) modes of the urethane group were also observed in the liquid medium without fungi. However, a significant decrease in the relative intensities of the peaks at 1,715 cm–1, 1,340 cm–1, and 1,020 cm–1 was detected in the visually transparent culture broths of E. clematidis (Figure 6C) and A. niger (Figure 6B) in the presence of Impranil as sole carbon sources. In addition, a strong peak broadening was observed at 3,370–3,135 cm–1 for the liquid medium without fungi, while a medium peak broadening at the same spectral window was observed for E. clematidis and A. niger in the presence of Impranil.
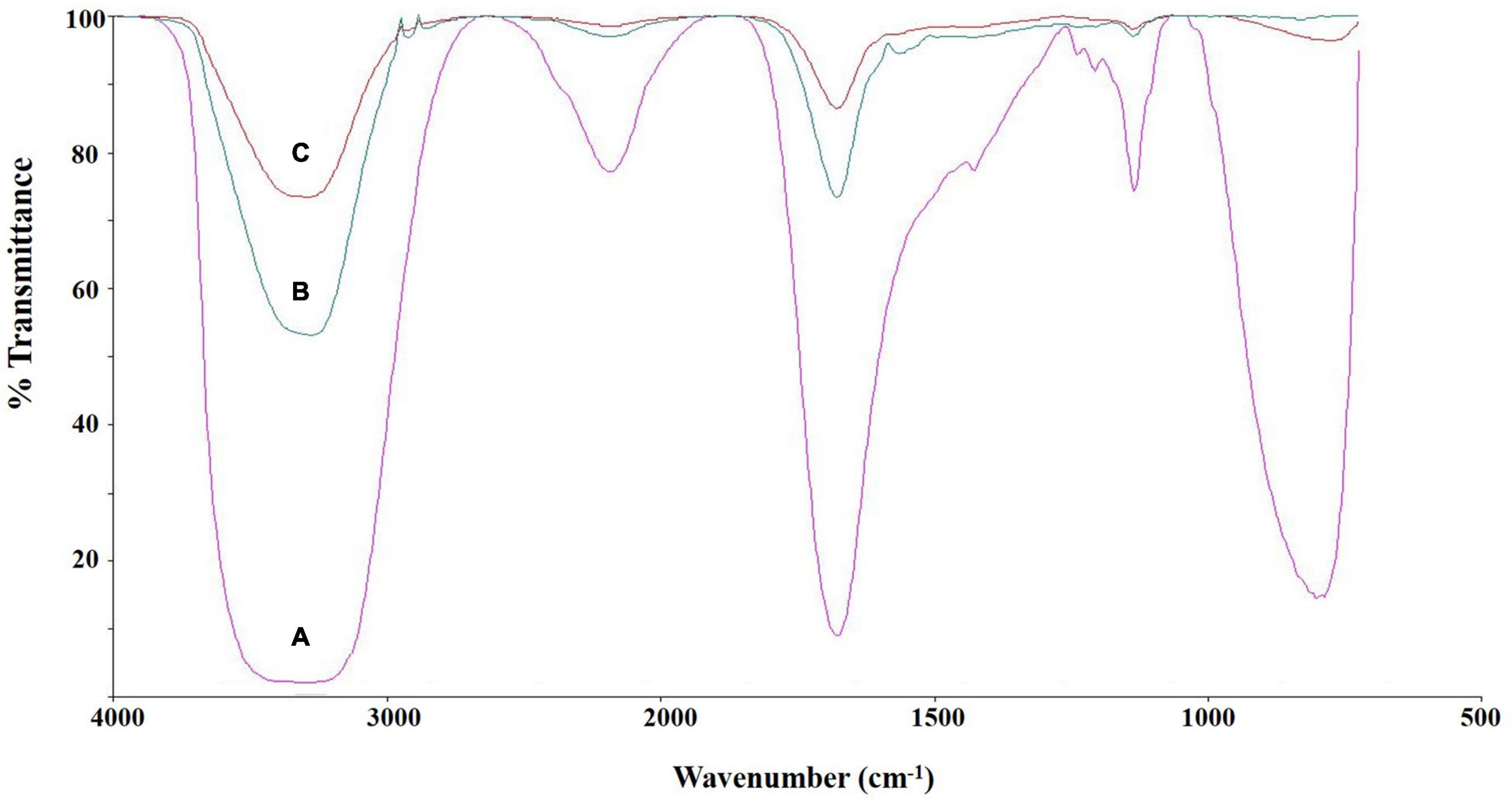
Figure 6. FTIR spectra of culture supernatant of E. clematidis (C), and A. niger (B) in the presence of Impranil as sole carbon source, and Impranil liquid medium without culturing of fungi (A) after 2-week inoculation.
GC-MS Analysis
The volatile organic compounds of the culture supernatant of E. clematidis and A. niger in the presence of Impranil after a 2-week incubation period is shown in Figure 7 and Table 2, respectively. Eight chemical compounds were detected among both fungal cultures in presence of Impranil after 2-week incubation (Table 2). The major compounds in the E. clematidis culture supernatant in the presence of Impranil were 3Z-heptenol, 5Z-octenol, 2E,4E-hexadienol acetate, and 3E,6Z-nonadienol. Meanwhile, the compounds 5Z-octenol, 2E,4E-hexadienol acetate, 3Z-hexenyl isobutanoate, and phenyl-tert-butanol were detected as the major compounds in the A. niger culture supernatant in the presence of Impranil.
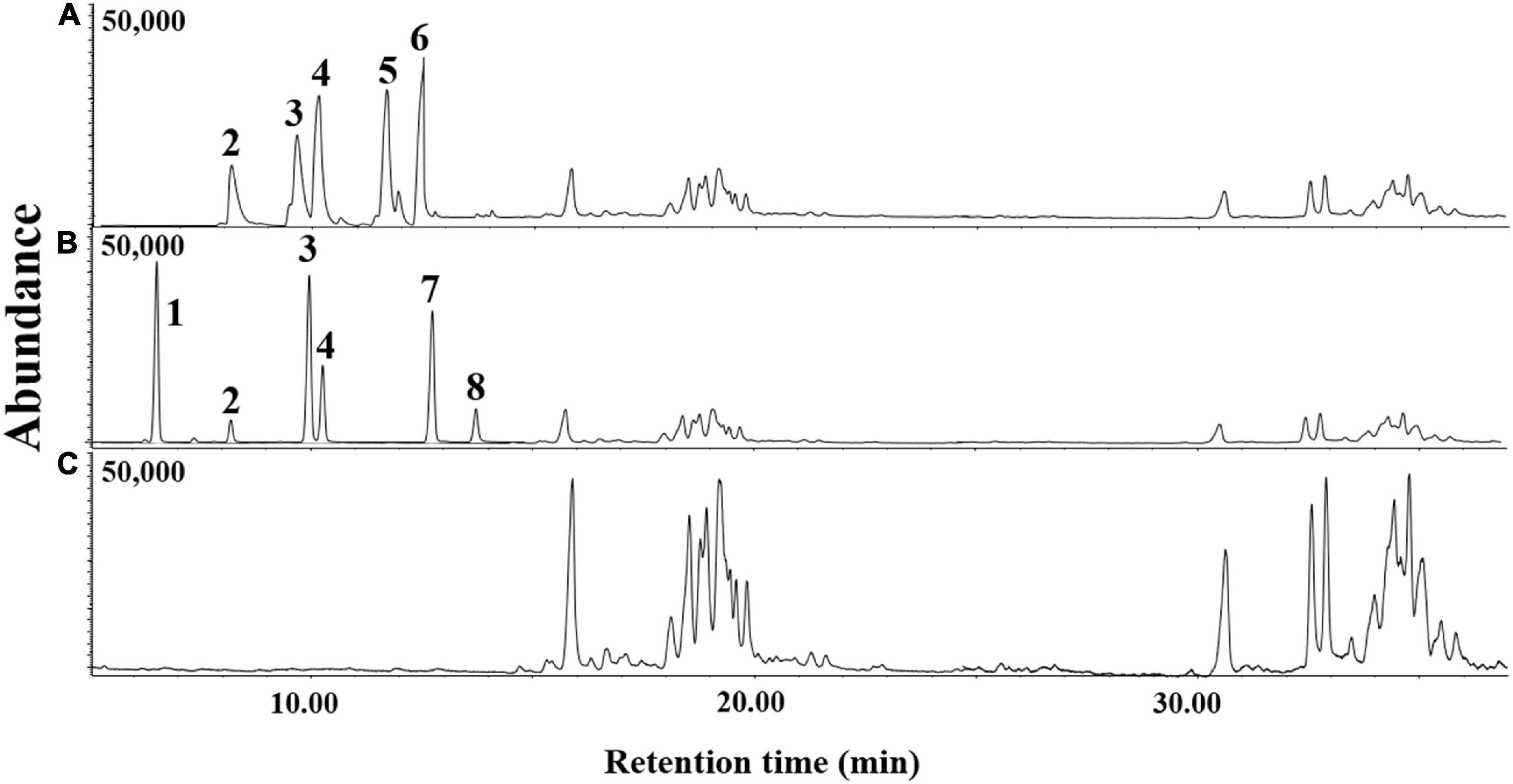
Figure 7. GC-MS chromatograms of dichloromethane extracts of A. niger (A) and E. clematidis (B) culture supernatant in the presence of Impranil as the sole carbon source, and Impranil liquid medium without culturing of fungi (C) after 2-week inoculation.

Table 2. Volatile organic compounds of dichloromethane extracts E. clematidis and A. niger culture supernatants in the presence of Impranil as sole carbon source after 2-week inoculation.
Enzymatic Activity Assay
Protease activity was not found in supernatant extracts of E. clematidis and A. niger in the presence of Impranil after a 2-week incubation, and very low urease activity was determined among all treatments, based on the amount of NH3, ranging from 0.14 to 0.21 μM/min/μg. However, a high amount of esterase enzyme was detected in the supernatant extracts of E. clematidis and A. niger, as shown in Figure 8. Low esterase activity (< 1.0 mM/min/mg) was detected in cell supernatants of A. niger in the absence of Impranil after a 2-week incubation. The highest enzymatic activity was detected in the supernatant of the E. clematidis fungal culture broth in the presence of Impranil. A gradual increase in enzymatic activity in the E. clematidis cell in the presence of Impranil was found on days 0–8 (0.00–2.45 mM/min/mg), which then gradually decreased to 1.81 mM/min/mg after 2 weeks of incubation. The enzymatic activity of E. clematidis cell in the absence of Impranil increased from 0.52 to 1.31 mM/min/mg from day 0 to day 10 but then decreased to 1.14 mM/min/mg after day 14. In addition, the highest esterase activity of A. niger in the presence of Impranil was found on day 12 (0.88 mM/min/mg), followed by 0.72 mM/min/mg on day 14.
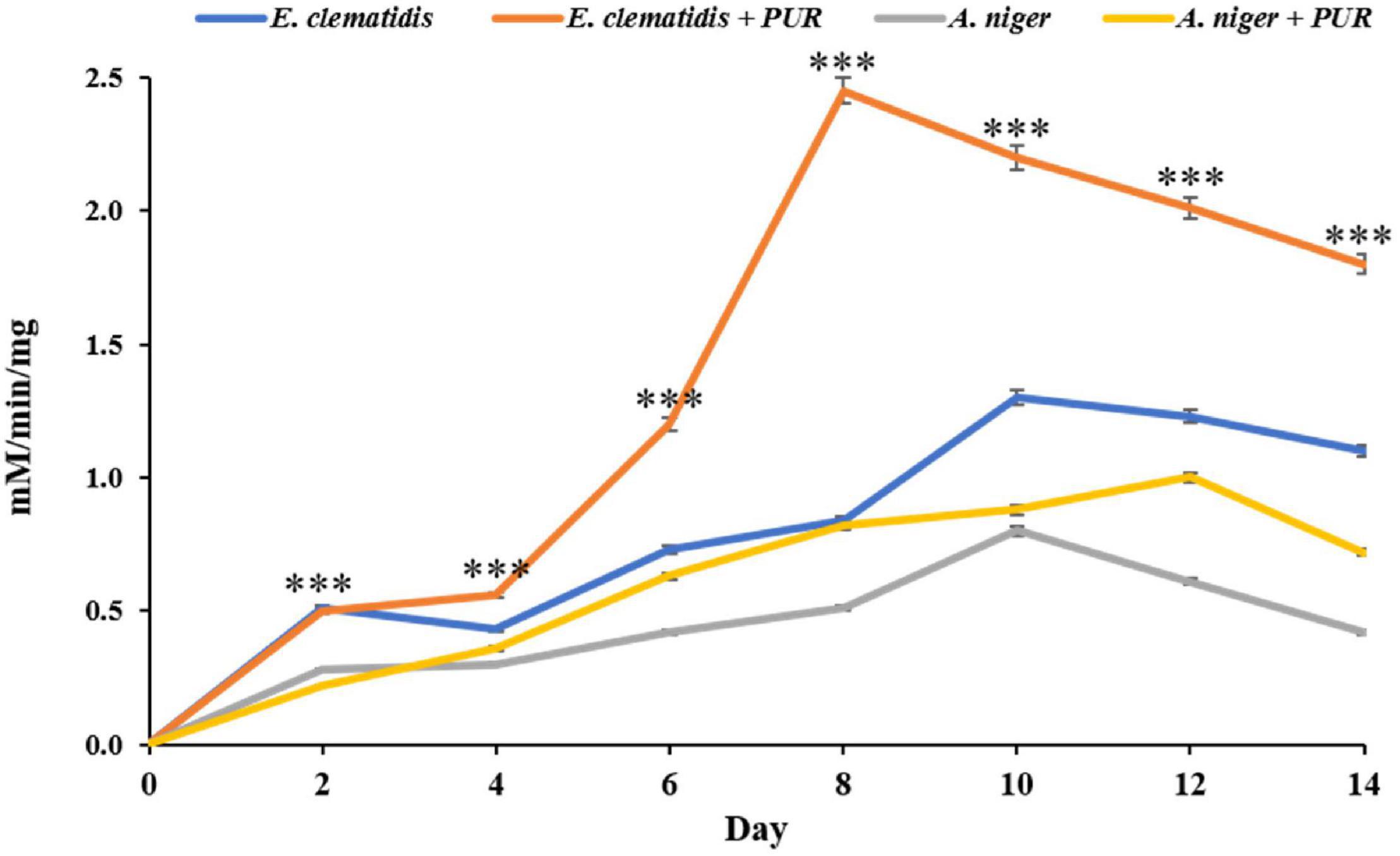
Figure 8. Esterase activity of E. clematidis and A. niger in the presence and absence of Impranil as the sole carbon source after 2-week inoculation. ***Above the points indicates a significant difference (p < 0.001) (ANOVA, followed by Dunnett’s test).
Discussion
All Impranil-degrading fungi belong to the phylum Ascomycota. Several fungal species in this phylum have been reported to have PUR degradation ability, such as Pestalotiopsis microspore (Russell et al., 2011), Cladosporium herbarum (Álvarez-Barragán et al., 2016), Cladosporium cladosporioides (Álvarez-Barragán et al., 2016), Penicillium griseofulvum (Brunner et al., 2018), Alicycliphilus spp. (Oceguera-Cervantes et al., 2007), and Xepiculopsis graminea (Álvarez-Barragán et al., 2016). Although many fungi have already been reported as PUR degraders, most of the species that are described in this work, as well as their ability to degrade Impranil, have been rarely reported. Impranil was found to employ as a representative of PUR in searching for PUR-degrading microorganisms (Russell et al., 2011; Nakkabi et al., 2015; Hung et al., 2016; Magnin et al., 2019) due to their various useful attributes resulting it ideal for biodegradation assays such as its commercial availability, application as a coating material, stability to pH 4–8, tolerance to high temperature up to 80°C, and rapid rate of dissolution compared to other polyurethanes. However, the formulation of Impranil is proprietary and its colloidal state has ultimately made Impranil a precarious choice for polyurethane-degrading microorganisms (Biffinger et al., 2015). The active fungi showed different Impranil degradation abilities, probably due to differences in the physiology, biochemistry, and genetics among the fungal species (Brunner et al., 2018). The fungus E. clematidis showed the highest Impranil degradation ability among all the active fungi that were tested, based on the formation of a halo in medium plates. It is classified as part of the genus Allophaeosphaeria of the Phaeosphaeriaceae family. However, we found no references related to the degradation of E. clematidis. Thus, this is the first report on the ability of E. clematidis to degrade Impranil. Recently, Phaeosphaeriaceae was evaluated as a big family, consisting of more than 300 species of economically important plant pathogens, endophytes, or saprobes on plants (Phookamsak et al., 2014). The degradation activity suggests that the fungi in this family might be a promising source of biodiversity for testing important bioremediation activities.
We found that E. clematidis in solid and liquid screening assays (with Impranil as the sole carbon source) had the highest rate of Impranil degradation compared to other fungi in this study. This observation indicates that E. clematidis growth in the medium and its metabolism in the medium could be harnessed for Impranil degradation. The growth of E. clematidis was investigated through a modified Sturm test, which is based on the amount of CO2 released by the fungal culture in the presence and absence of Impranil. The growth of the fungus E. clematidis in the presence of ester hydrolysis products revealed that it utilized the products as a carbon source and mineralized to CO2 and H2O. Various studies used CO2 evolution tests to evaluate the degradation ability of microorganisms and found that several microbes released high amounts of CO2 during culturing, with polymer as a carbon source (Nakkabi et al., 2015; Bano et al., 2017; Hung et al., 2019). In the present study, the quantity of CO2 released by E. clematidis in the presence of Impranil as a carbon source was significantly higher than when Impranil is absent, suggesting that Impranil biodegradation was enhanced by fungal activity, as reflected by the corroborative levels of enzyme production and CO2 liberation.
Chemical composition analysis of the E. clematidis culture was performed using FTIR spectroscopy and GC-MS. Both techniques provide evidence to predict the mechanism of how the fungus attacks the polymer during biodegradation (Mahalakshmi et al., 2012; Shah et al., 2013; Muhonja et al., 2018). As far as the mechanism of degradation of Impranil is concerned, the fungus E. clematidis easily degraded the aliphatic segment to form various alcohol compounds detected by the GC-MS. However, the formulation of Impranil is proprietary (Biffinger et al., 2015). These compounds could be derived from both the polyester and polyisocyanate portions of PUR (Shah et al., 2013). FTIR analysis revealed that E. clematidis degraded the polyester portion of the Impranil. The dramatic variation of the carbonyl signal (1,715 cm–1) in the FTIR spectra were related to the attack of the ester bonds that are present in the polyol fractions, as well as the attack of the urethane groups (Mahalakshmi et al., 2012; Shah et al., 2013; Álvarez-Barragán et al., 2016; Muhonja et al., 2018). Russell et al. (2011) also reported that a decrease in the peak intensity at 1,715 cm–1 is consistent with the hydrolysis of the ester bond in the urethane linkage of PUR. Moreover, the decrease in the C-N-H bond peaks at 1,340 cm–1, and 1,020 cm–1 indicates that the urethane groups were eliminated. The reduction of the C=O and C-N-H signals confirms the degradation of Impranil by the fungus E. clematidis. The FTIR spectrum measured from the liquid medium without fungi showed a strong peak broadening at 3,270–3,335 cm–1, indicating the overlap of N-H and O-H- related peaks (Oprea, 2010). FTIR spectroscopy has been applied to monitor the variation in characteristic peaks related to polymer biodegradation.
Evidence of Impranil biodegradation by the fungus E. clematidis was also obtained by GC-MS analysis. New chemical compounds were detected after a 2-week incubation of E. clematidis and A. niger in the medium. The reduction of compounds containing ester bonds at the 15:00–38:00 min-mark increased alcohol compounds, probably due to the hydrolysis of the ester bonds in the polymer chain of PUR (Howard, 2002; Russell et al., 2011; Álvarez-Barragán et al., 2016). Ester compounds, 2E,4E-hexadienol acetate, and 3Z-hexenyl isobutanoate, may be synthesized by the combination of PUR precursors that are degraded by the fungi (Russell et al., 2011; Álvarez-Barragán et al., 2016). This work suggests that the use of fungi to degrade PUR waste presents an alternative, sustainable way of recovering PUR precursors for recycling. However, the compounds generated during the degradation of PUR by fungi might be related to oxidative reactions (Oprea, 2010; Yarahmadi et al., 2017).
FTIR spectroscopy and GC-MS were successfully applied for the analysis of chemical structure changes due to biodegradation by microorganisms. For example, Álvarez-Barragán et al. (2016) studied the biodegradative activities of selected environmental fungi on PUR. FTIR and GC-MS analyses revealed structural changes in PUR. A decrease in carbonyl groups (1,729 cm–1) and N-H bonds (1,540 and 1,261 cm–1) was measured by FTIR spectroscopy whereas a decrease in ester compounds and an increase in alcohols and hexane diisocyanate were shown by GC-MS analysis. The results obtained by both techniques indicated the hydrolysis of ester and urethane bonds. Shah et al. (2013) confirmed the degradation of PUR by Pseudomonas aeruginosa strain MZA-85 using FTIR and GC-MS. IR spectra indicated that the peak at 1,725 cm–1, which corresponds to ester linkage, decreased in the treated PUR samples, showing ester hydrolysis. The peak at 1,685 cm–1, obtained from the treated PUR sample, represents the overlap of carboxylic acid and amide carboxyl peaks, while the broadened N-H peak at 3,325 cm–1 represents the presence of free hydroxyl groups in the treated sample. The peak at 1,135 cm–1, which is attributed to the ester group was absent in the treated sample, confirming ester hydrolysis and the production of free hydroxyl groups after incubation with P. aeruginosa strain MZA-85. In addition, GC-MS analysis demonstrated two compounds, including 1,4-butanediol and adipic acid, which were detected in the treated sample only.
Many studies reported the esterase activities of microorganisms in the PUR degradation (Akutsu et al., 1998; Rowe and Howard, 2002; Mohanan et al., 2020). The enzymatic degradation of PUR occurs in two stages including adsorption of enzymes on the polymer surface in the first stage, followed by hydro-peroxidation/hydrolysis of the bonds in the second stage (Akutsu et al., 1998; Mohanan et al., 2020). Sato (1981) reported the protease, urease, and esterase activities produced by A. niger while culturing with PUR. Similarly, the structural changes due to Impranil degradation by the fungus E. clematidis are also correlated with the same enzymes. However, the rate of polymer biodegradation varied depending on several factors including chemical structures, and molecular weights (Mohanan et al., 2020). Of these three enzymes, esterase appears to be the most responsible for Impranil degradation. It is noted that the enzyme responsible for PUR degradation is extracellular, secreted, and diffusible (Oceguera-Cervantes et al., 2007; Loredo-Treviño et al., 2012; Hung et al., 2016). Extracellular enzymes secreted by microorganisms have played an important role in polymer degradation via depolymerization reaction by breaking down polymers into smaller molecules (Howard, 2002). The polymer chain is degraded by extracellular enzymes and becomes a degradation product. It can then be assimilated into microbial cells and used as carbon sources for producing carboxylic acid and alcohol that ultimately undergo respiration to produce energy (Kay et al., 1991; Nakajima-Kambe et al., 1995; Hung et al., 2016). In this study, extracellular enzymes esterase, lipase, and urease were investigated. E. clematidis was able to produce esterase enzymes which are believed to play a role in Impranil degradation. This result was in agreement with the study of Biffinger et al. (2015) showing the partial degradation of Impranil was significantly achieved by enzyme esterase. They reported that the enzymes (a Pseudomonas sp. esterase showed significant esterase activities and partially cleared Impranil by partial hydrolysis. Esterase enzymes have been isolated and reported by several studies to degrade PUR. Noor et al. (2020) found esterase in the outer membrane of P. aeruginosa, demonstrating a particular affinity to long-chain acyl esters. Purified esterase was also isolated from the outer membrane of PUR-degrading bacteria C. acidovorans strain TB35 and P. aeruginosa PAO1 (Akutsu et al., 1998; Wilhelm et al., 1999). Mukherjee et al. (2011) also reported the ability of esterase enzymes from P. aeruginosa to degrade polyurethane diol. Moreover, esterase was purified from a PUR-degrading fungus, Curvularia senegalensis (Loredo-Treviño et al., 2012). The esterase enzyme attacked the ester bond cleavage in the soft segments of PUR. Yu et al. (2010) reported the role of esterase in the catalysis of the cleavage of ester bonds in the polymer chain in PUR. In addition, 6-hydroxyhexanoate was generated from the hydrolysis of the ester linkages in polycaprolactone polyol-based PUR by an esterase enzyme (Magnin et al., 2019). Shah et al. (2013) also described that esterase that was purified from the strain P. aeruginosa MZA-85 specifically damaged the ester bonds in the aliphatic or soft segment, leading to the depolymerization of the polymer chain to degradation products. These products could be further metabolized through different metabolic pathways (Brzostowicz et al., 2002; Yoshida et al., 2016). However, there is limited information about the mechanism of degradation of Impranil by esterase secreted from E. clematidis. Therefore, the mechanism of how esterase is secreted by E. clematidis should be further studied in detail. Optimal conditions for the fungal activity must also be improved before applying it on a larger scale for commercial use.
Conclusion
This work establishes that various fungi are useful biodiversity sources with potential application in Impranil degradation. It is interesting to note that E. clematidis had the highest capacity for Impranil degrading. FTIR spectroscopy and GC-MS analysis indicate that Impranil was successfully degraded by E. clematidis, with Impranil as the sole carbon source. The fungus E. clematidis degraded the polymer chain of Impranil, constituting the soft segment, and produced degradation products such as 5Z-octenol, and 2E,4E-hexadienol acetate. The ester and urethane groups in the polymer chain of Impranil were attacked via the esterase enzyme. Further studies, which explain the mechanism behind the esterase enzyme production of E. clematidis and its role in PUR degradation, are required for its potential application in polyurethane waste management.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SK and TS performed experiments on solid- and liquid-medium assays. SK performed FTIR, GC-MS analysis, Sturm test, enzyme activity assay, analyzed data, and wrote the first draft of the manuscript. PP edited, reviewed the manuscript, and supervised the work. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the Mae Fah Luang University and the Royal Golden Jubilee Ph.D. Program through (grant no. PHD/0193/2560).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Center of Excellence in Fungal Research, Mae Fah Luang University for providing all fungi in this study. We are grateful to Dr. Parinya Panuwet from the Gangarosa Department of Environmental Health, Rollins School of Public Health at Emory University, for providing his assistance in laboratory result interpretation.
References
Akutsu, Y., Nakajima-Kambe, T., Nomura, N., and Nakahara, T. (1998). Purification and properties of a polyester polyurethane-degrading enzyme from Comamonas acidovorans TB-35. Appl. Environ. Microbiol. 64, 62–67. doi: 10.1128/AEM.64.1.62-67.1998
Allen, A. B., Hilliard, N. P., and Howard, G. T. (1999). Purification and characterization of a soluble polyurethane degrading enzyme from Comamonas acidovorans. Int. Biodeterior. Biodegrad. 43, 37–41. doi: 10.1016/S0964-8305(98)00066-3
Álvarez-Barragán, J., Domínguez-Malfavón, L., Vargas-Suárez, M., González-Hernández, R., Aguilar-Osorio, G., and Loza-Tavera, H. (2016). Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 82, 5225–5235. doi: 10.1128/AEM.01344-16
Ameen, F., Moslem, M., Hadi, S., and Al-Sabri, A. E. (2016). Biodegradation of diesel fuel hydrocarbons by mangrove fungi from red sea coast of Saudi Arabia. Saudi J. Biol. Sci. 23, 211–218. doi: 10.1016/j.sjbs.2015.04.005
Bano, K., Kuddus, M., Zaheer, M. R., Zia, Q., Khan, M. F., Gupta, A., et al. (2017). Microbial enzymatic degradation of biodegradable plastics. Curr. Pharm. Biotechnol. 18, 429–440. doi: 10.2174/1389201018666170523165742
Biffinger, J. C., Barlow, D. E., Cockrell, A. L., Cusick, K. D., Hervey, W. J., Fitzgerald, L. A., et al. (2015). The applicability of Impranil® DLN for gauging the biodegradation of polyurethanes. Polym. Degrad. Stab. 120, 178–185. doi: 10.1016/j.polymdegradstab.2015.06.020
Brunner, I., Fischer, M., Rüthi, J., Stierli, B., and Frey, B. (2018). Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PLoS One 13:0202047. doi: 10.1371/journal.pone.0202047
Brzostowicz, P., Blasko, M., and Rouvière, P. (2002). Identification of two gene clusters involved in cyclohexanone oxidation in Brevibacterium epidermidis strain HCU. Appl. Microbiol. Biotechnol. 58, 781–789. doi: 10.1007/s00253-002-0968-x
Cosgrove, L., McGeechan, P. L., Robson, G. D., and Handley, P. S. (2007). Fungal communities associated with degradation of polyester polyurethane in soil. Appl. Environ. Microbiol. 73, 5817–5824. doi: 10.1128/AEM.01083-07
Cregut, M., Bedas, M., Durand, M. J., and Thouand, G. (2013). New insights into polyurethane biodegradation and realistic prospects for the development of a sustainable waste recycling process. Biotechnol. Adv. 31, 1634–1647. doi: 10.1016/j.biotechadv.2013.08.011
Danso, D., Chow, J., and Streit, W. R. (2019). Plastics: environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 85:e1095-19. doi: 10.1128/AEM.01095-19
Darby, R. T., and Kaplan, A. M. (1968). Fungal susceptibility of polyurethanes. J. Appl. Microbiol. 16, 900–905. doi: 10.1128/am.16.6.900-905.1968
Glaser, J. A. (2019). Biological Degradation of Polymers in the Environment, Vol. 1. London: IntechOpen, 13.
Howard, G. T. (2002). Biodegradation of polyurethane: a review. Int. Biodeterior. Biodegrad. 49, 245–252. doi: 10.1016/S0964-8305(02)00051-3
Hung, C. S., Barlow, D. E., Varaljay, V. A., Drake, C. A., Crouch, A. L., and Russell, J. N. Jr., et al. (2019). The biodegradation of polyester and polyester polyurethane coatings using Papiliotrema laurentii. Int. Biodeterior. Biodegrad. 139, 34–43. doi: 10.1016/j.ibiod.2019.02.002
Hung, C. S., Zingarelli, S., Nadeau, L. J., Biffinger, J. C., Drake, C. A., Crouch, A. L., et al. (2016). Carbon catabolite repression and Impranil polyurethane degradation in Pseudomonas protegens strain Pf-5. Appl. Environ. Microbiol. 82, 6080–6090. doi: 10.1128/AEM.01448-16
Kale, S. K., Deshmukh, A. G., Dudhare, M. S., and Patil, V. B. (2015). Microbial degradation of plastic: a review. J. Biochem. Technol. 6, 952–961.
Kay, M. J., Morton, L. H. G., and Prince, E. L. (1991). Bacterial degradation of polyester polyurethane. Int. Biodeter. 27, 205–222. doi: 10.1016/0265-3036(91)90012-G
Liu, J., He, J., Xue, R., Xu, B., Qian, X., Xin, F., et al. (2021). Biodegradation and up-cycling of polyurethanes: progress, challenges, and prospects. Biotechnol. Adv. 48:107730. doi: 10.1016/j.biotechadv.2021.107730
Loredo-Treviño, A., Gutiérrez-Sánchez, G., Rodríguez-Herrera, R., and Aguilar, C. N. (2012). Microbial enzymes involved in polyurethane biodegradation: a review. J. Polym. Environ. 20, 258–265. doi: 10.1007/s10924-011-0390-5
Magnin, A., Pollet, E., Perrin, R., Ullmann, C., Persillon, C., Phalip, V., et al. (2019). Enzymatic recycling of thermoplastic polyurethanes: synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manage. 85, 141–150. doi: 10.1016/j.wasman.2018.12.024
Mahajan, N., and Gupta, P. (2015). New insights into the microbial degradation of polyurethanes. RSC Adv. 5, 41839–41854. doi: 10.1039/C5RA04589D
Mahalakshmi, V., Siddiq, A., and Andrew, S. N. (2012). Analysis of polyethylene degrading potentials of microorganisms isolated from compost soil. Int. J. Pharm. Biol. Arch. 3, 1190–1196.
Matsumura, S., Soeda, Y., and Toshima, K. (2006). Perspectives for synthesis and production of polyurethanes and related polymers by enzymes directed toward green and sustainable chemistry. Appl. Microbiol. Biotechnol. 70, 12–20. doi: 10.1007/s00253-005-0269-2
Miskolczi, N., Bartha, L., Deak, G., and Jover, B. (2004). Thermal degradation of municipal plastic waste for production of fuel-like hydrocarbons. Polym. Degrad. Stab. 86, 357–366. doi: 10.1016/j.polymdegradstab.2004.04.025
Mohanan, N., Montazer, Z., Sharma, P. K., and Levin, D. B. (2020). Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 11:580709. doi: 10.3389/fmicb.2020.580709
Muhonja, C. N., Makonde, H., Magoma, G., and Imbuga, M. (2018). Biodegradability of polyethylene by bacteria and fungi from Dandora dumpsite Nairobi-Kenya. PLoS One 13:e0198446. doi: 10.1371/journal.pone.0198446
Mukherjee, K., Tribedi, P., Chowdhury, A., Ray, T., Joardar, A., Giri, S., et al. (2011). Isolation of a Pseudomonas aeruginosa strain from soil that can degrade polyurethane diol. Biodegradation 22, 377–388. doi: 10.1007/s10532-010-9409-1
Nakajima-Kambe, T., Onuma, F., Kimpara, N., and Nakahara, T. (1995). Isolation and characterization of a bacterium which utilizes polyester polyurethane as a sole carbon and nitrogen source. FEMS Microbiol. Lett. 129, 39–42. doi: 10.1111/j.1574-6968.1995.tb07554.x
Nakkabi, A., Sadiki, M., Fahim, M., Ittobane, N., IbnsoudaKoraichi, S., Barkai, H., et al. (2015). Biodegradation of poly (ester urethane)s by Bacillus subtilis. Int. J. Environ. Res. 9, 157–162. doi: 10.22059/IJER.2015.885
Noor, H., Satti, S. M., Din, S. U., Farman, M., Hasan, F., Khan, S., et al. (2020). Insight on esterase from Pseudomonas aeruginosa strain S3 that depolymerize poly (lactic acid) (PLA) at ambient temperature. Polym. Degrad. Stab. 174:109096. doi: 10.1016/j.polymdegradstab.2020.109096
Oceguera-Cervantes, A., Carrillo-García, A., López, N., Bolaños-Nuñez, S., Cruz-Gómez, M. J., Wacher, C., et al. (2007). Characterization of the polyurethanolytic activity of two Alicycliphilus sp. strains able to degrade polyurethane and N-methylpyrrolidone. Appl. Environ. Microbiol. 73, 6214–6223. doi: 10.1128/AEM.01230-07
Oprea, S. (2010). The effect of chain extenders structure on properties of new polyurethane elastomers. Polym. Bull. 65, 753–766. doi: 10.1007/s00289-009-0242-9
Phookamsak, R., Liu, J. K., McKenzie, E. H., Manamgoda, D. S., Ariyawansa, H., Thambugala, K. M., et al. (2014). Revision of phaeosphaeriaceae. Fungal Divers. 68, 159–238. doi: 10.1007/s13225-014-0308-3
Rowe, L., and Howard, G. T. (2002). Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int. Biodeterior. Biodegrad. 50, 33–40. doi: 10.1016/S0964-8305(02)00047-1
Ru, J., Huo, Y., and Yang, Y. (2020). Microbial degradation and valorization of plastic wastes. Front. Microbiol. 11:442. doi: 10.3389/fmicb.2020.00442
Russell, J. R., Huang, J., Anand, P., Kucera, K., Sandoval, A. G., Dantzler, K. W., et al. (2011). Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 77, 6076–6084. doi: 10.1128/AEM.00521-11
Sato, M. (1981). Deterioration of polyurethane filaments with fungal enzymes Aspergillus niger FERM J-1 and Cladosporium cladosporioides FERM J-8. Seni Gakkaishi 37, T290–T300.
Schmidt, J., Wei, R., Oeser, T., Dedavid e Silva, L. A., Breite, D., Schulze, A., et al. (2017). Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers 9:65. doi: 10.3390/polym9020065
Shah, A. A., Hasan, F., Akhter, J. I., Hameed, A., and Ahmed, S. (2008). Degradation of polyurethane by novel bacterial consortium isolated from soil. Ann. Microbiol. 58, 381–386. doi: 10.1007/BF03175532
Shah, Z., Hasan, F., Krumholz, L., Aktas, D. F., and Shah, A. A. (2013). Degradation of polyester polyurethane by newly isolated Pseudomonas aeruginosa strain MZA-85 and analysis of degradation products by GC-MS. Int. Biodeterior. Biodegrad. 77, 114–122. doi: 10.1016/j.ibiod.2012.11.009
Tokiwa, Y., and Calabia, B. P. (2004). Review degradation of microbial polyesters. Biotechnol. Lett. 26, 1181–1189.
Wilhelm, S., Tommassen, J., and Jaeger, K. E. (1999). A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 181, 6977–6986. doi: 10.1128/JB.181.22.6977-6986.1999
Yang, X. F., Vang, C., Tallman, D. E., Bierwagen, G. P., Croll, S. G., and Rohlik, S. (2001). Weathering degradation of a polyurethane coating. Polym. Degrad. Stab. 74, 341–351. doi: 10.1016/S0141-3910(01)00166-5
Yarahmadi, N., Vega, A., and Jakubowicz, I. (2017). Accelerated ageing and degradation characteristics of rigid polyurethane foam. Polym. Degrad. Stab. 138, 192–200. doi: 10.1016/j.polymdegradstab.2017.03.012
Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., et al. (2016). A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 351, 1196–1199. doi: 10.1126/science.aaf8305
Keywords: polyester polyurethane, biodegradation, Embarria clematidis, Fourier-transform infrared spectroscopy, gas chromatography-mass spectrometry, esterase
Citation: Khruengsai S, Sripahco T and Pripdeevech P (2022) Biodegradation of Polyester Polyurethane by Embarria clematidis. Front. Microbiol. 13:874842. doi: 10.3389/fmicb.2022.874842
Received: 13 February 2022; Accepted: 16 May 2022;
Published: 14 June 2022.
Edited by:
Tahar Mechichi, University of Sfax, TunisiaReviewed by:
Sikandar I. Mulla, REVA University, IndiaSamantha Chandranath Karunarathna, Qujing Normal University, China
Ahmad Ali Pourbabaee, University of Tehran, Iran
Subhankar Chatterjee, Pondicherry University, India
Copyright © 2022 Khruengsai, Sripahco and Pripdeevech. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patcharee Pripdeevech, cGF0Y2hhcmVlLnByaUBtZnUuYWMudGg=
 Sarunpron Khruengsai1
Sarunpron Khruengsai1 Patcharee Pripdeevech
Patcharee Pripdeevech