- 1Department of Animal Science, University of California, Davis, Davis, CA, United States
- 2Evonik Operations GmbH, Hanau, Germany
The objective of this study was to investigate the effects of dietary supplementation of Bacillus (B.) amyloliquefaciens on growth performance, diarrhea, systemic immunity, and intestinal microbiota of weaned pigs experimentally infected with F18 enterotoxigenic Escherichia coli (ETEC). A total of 50 weaned pigs (7.41 ± 1.35 kg BW) were individually housed and randomly allotted to one of the following five treatments: sham control (CON-), sham B. amyloliquefaciens (BAM-), challenged control (CON+), challenged B. amyloliquefaciens (BAM+), and challenged carbadox (AGP+). The experiment lasted 28 days, with 7 days of adaptation and 21 days after the first ETEC inoculation. ETEC challenge reduced (P < 0.05) average daily gain (ADG) of pigs. Compared with CON+, AGP+ enhanced (P < 0.05) ADG, while B. amyloliquefaciens supplementation tended (P < 0.10) to increase ADG in pigs from days 0 to 21 post-inoculation (PI). The ETEC challenge increased (P < 0.05) white blood cell (WBC) count on days 7 and 21 PI, while BAM+ pigs tended (P < 0.10) to have low WBC on day 7 PI and had lower (P < 0.05) WBC on day 21 PI compared with CON+. In comparison to AGP+ fecal microbiota, BAM+ had a lower (P < 0.05) relative abundance of Lachnospiraceae on day 0 and Clostridiaceae on day 21 PI, but a higher (P < 0.05) relative abundance of Enterobacyeriaceae on day 0. In ileal digesta, the Shannon index was higher (P < 0.05) in BAM+ than in AGP+. Bray-Curtis PCoA displayed a difference in bacterial community composition in ileal digesta collected from sham pigs vs. ETEC-infected pigs on day 21 PI. Pigs in BAM+ had a greater (P < 0.05) relative abundance of Firmicutes, but a lower (P < 0.05) relative abundance of Actinomycetota and Bacteroidota in ileal digesta than pigs in AGP+. Ileal digesta from AGP+ had a greater (P < 0.05) abundance of Clostridium sensu stricto 1 but lower (P < 0.05) Bifidobacterium than pigs in BAM+. In conclusion, supplementation of B. amyloliquefaciens tended to increase ADG and had limited effects on the diarrhea of ETEC-infected pigs. However, pigs fed with B. amyloliquefaciens exhibit milder systemic inflammation than controls. B. amyloliquefaciens differently modified the intestinal microbiota of weaned pigs compared with carbadox.
Introduction
Newly weaned pigs experience a period of high stress from sudden environmental changes in housing and dietary changes from sow milk to a solid diet, which increases the risk of pigs experiencing post-weaning diarrhea induced by enterotoxigenic Escherichia coli (ETEC) (Fairbrother et al., 2005). Weaned pigs can experience watery diarrhea from ETEC disrupting the osmotic pressure in the intestines, leading pigs to undergo dehydration and reduced feed efficiency (Lee et al., 2016). Reduced feed intake equilibrates to reduced energy intake, which results in less growth and lower immunity in weaned pigs (Amezcua et al., 2002). Untreated pigs with post-weaning diarrhea can eventually lead to death, and the increasing mortality rate can negatively impact the economy of the swine industry. Therefore, ETEC pathogenicity must be suppressed within early contact.
In the swine industry, in-feed antibiotics were administered to treat post-weaning diarrhea and to prevent the spread of ETEC in weaned pigs. Some antibiotics, including carbadox, can be administered in-feed at a low dose to additionally promote pig growth and are commonly referred to as antibiotic growth promoters (AGP) (Lekagul et al., 2019). The continuous use of AGP increases the risk of antibiotic resistance, which can be transmitted zoonotically from pigs to humans, leading to increasing concerns for public health (Aarestrup, 2005). Thus, regulations and legislation have been applied in countries such as the European Union to ban or reduce the use of antibiotics in animal production, and the World Health Organization has developed a global action plan to increase awareness of using antibiotics for human health and livestock purposes (Casewell et al., 2003; World Health Organization, 2015). Although approximately 30 countries have restricted or banned the use of AGP, many other countries are still administering AGP in swine diets to prevent diarrhea and promote growth (Liao and Nyachoti, 2017). Hence, the swine industry is currently challenged to maintain health while improving the growth of newly weaned pigs without the use of AGP.
Categorized as direct-fed microbials, Bacillus spp. have shown to secrete secondary metabolites that may contribute to antimicrobial factors and can be easily isolated from soil (Sansinenea and Ortiz, 2011). In our previous study, B. subtilis DSM 25841 supplementation has been shown to reduce diarrhea and improve the growth performance of weaned pigs that were experimentally infected with ETEC (He et al., 2020b). Bacillus spp. also modify the intestinal microbiota when supplemented to pigs (Fouhse et al., 2016; Lee et al., 2019). Comparing the whole genomes, B. subtilis and Bacillus amyloliquefaciens have similar antibacterial synthetases, which lead B. amyloliquefaciens to also be categorized as direct-fed microbials (Koumoutsi et al., 2004). Salazar et al. (2017) have also identified bacteriocin-like substances in B. amyloliquefaciens that could inhibit the growth of pathogenic bacteria, and it has shown high-temperature resistance and pH stability, which may imply high survivability in the swine gastrointestinal tract. Findings in these previous studies suggest B. amyloliquefaciens as direct-fed microbials may have the potential to alleviate post-weaning diarrhea and enhance the growth performance of weaned pigs. However, limited research was reported on utilizing dietary B. amyloliquefaciens as in-feed supplementation on pig performance and intestinal health. Therefore, the present study aimed to investigate the effects of supplementing B. amyloliquefaciens on growth performance and systemic immunity of newly weaned pigs with or without the ETEC challenge and to compare the effects of B. amyloliquefaciens with carbadox on the fecal and ileal microbiota of weaned pigs.
Materials and methods
Animals and study design
The protocol for this experiment was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC# 20809) at the University of California, Davis (UC Davis). A total of 50 weaned pigs [21 days old, 7.41 ±1.35 kg body weight (BW)] with an equal number of barrows and gilts were obtained from the Swine Teaching and Research Center, and the experiment was conducted at the Cole facility at UC Davis. All pigs and their sows did not receive E. coli vaccines, antibiotic injections, or antibiotics in feed prior to the experiment. After weaning, pigs were individually housed (pen size: 0.61 × 1.22 m) and assigned into one of 5 treatment groups with 10 replicate pigs per treatment using a randomized complete block design with sex normalized by BW and litter as blocks and pig as the experimental unit. There were four treatments in a 2 × 2 factorial arrangement with two diets [control (CON) vs. 0.10% inclusion rate with 109 CFU/kg B. amyloliquefaciens (BAM) in the complete feed] and two challenges [sham (-) vs. ETEC (+)]. The fifth was an antibiotic (50 mg/kg of carbadox in the complete feed) treatment with an ETEC challenge (AGP+). Prior to weaning, tail samples were collected from all piglets to assess their susceptibility to ETEC F18 using the genotyping analysis described by Kreuzer et al. (2013). All pigs used in the present study were susceptible to ETEC F18.
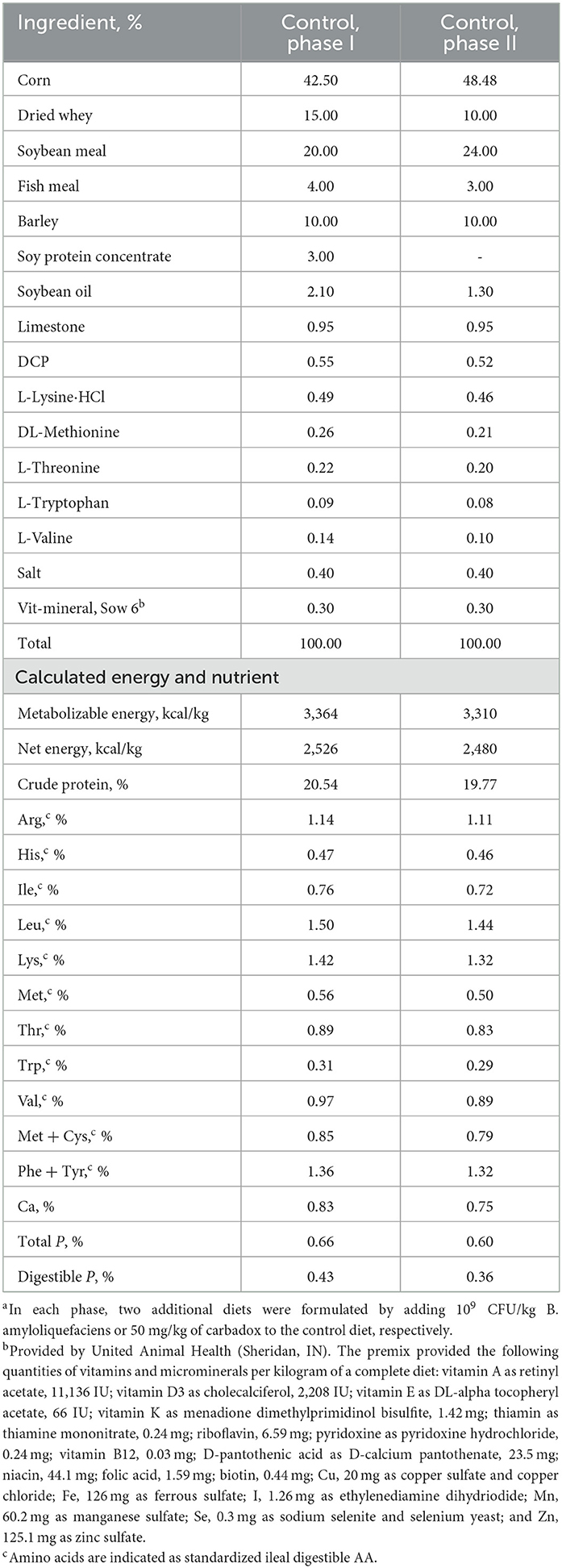
The experiment included a 7-day habituation period and 21 days after the first ETEC F18 inoculation (day 0). A 2-phase feeding program was used, with weeks 1 and 2 as Phase I and weeks 3 and 4 as Phase II. Hence, six diets were prepared. Spray-dried plasma, antibiotics, and high levels of zinc oxide were not included in the diets. All diets were formulated to meet pig nutritional requirements (NRC, 2012; Table 1).
Pigs in the ETEC challenge groups received 3 oral doses of ETEC F18 at 1010 CFU per dose. The F18 ETEC was cultured in Dr. Xunde Li's lab at the Western Institute for Food Safety and Security at UC Davis. The bacterial strain was originally isolated from a field disease outbreak by the University of Illinois Veterinary Diagnostic Lab (isolate number: U.IL-VDL # 05-27242), and the strain expresses heat-labile toxin, heat-stable toxin b, and Shiga-like toxins. On the terminating day of the experiment, all pigs were anesthetized by intramuscularly injecting 1 ml of a mixture of telazol (100 mg), ketamine (50 mg), and xylazine (50 mg) prior to an intracardiac injection of 78 mg sodium pentobarbital (Vortech Pharmaceuticals, Ltd., Dearborn, MI) per 1 kg of BW for euthanasia.
Clinical observations and sample collection
Fecal and alertness scores were recorded two times daily from days 0 to 21 post-inoculation (PI). The fecal score was measured by two independent evaluators, with scores ranging from 1 to 5 (1, normal feces; 2, moist feces; 3, mild diarrhea; 4, severe diarrhea; and 5, watery diarrhea). The alertness score of each pig was also assessed visually, with the score ranging from 1 to 3 (1, normal; 2, slightly depressed or listless; and 3, severely depressed or recumbent). The frequency of diarrhea was calculated by quantifying the number of pigs and days with a fecal score ≥ 3 or ≥ 4, respectively.
Fecal samples were collected from the rectum of each pig at the beginning of the experiment (day −7), on day 0 before ETEC inoculation, and on days 2, 7, 14, and 21 PI to perform a fecal culture. Whole blood samples were collected from the jugular vein of all pigs on days −7 and 0, and days 7, 14, and 21 PI. Fresh blood samples were submitted to the Comparative Pathology Laboratory at the University of California, Davis, to measure total and differential blood cell counts. A multiparameter, automated, and programmed hematology analyzer (Drew/ERBA Scientific 950 FS Hematological Analyzer, Drew Scientific Inc., Miami, FL) was used for the assay to optimally differentiate porcine blood. Feeder weights, feed allowances, and pig BWs were recorded weekly to calculate average daily gain (ADG), average daily feed intake (ADFI), and gain-to-feed ratio (G:F) from days −7 to 0, days 0 to 7 PI, days 7 to 14 PI, and days 14 to 21 PI. Additional batches of fecal samples collected from days −7 and 0 before ETEC inoculation and on days 7, 14, and 21 PI and ileal digesta collected on day 21 PI were immediately frozen in liquid nitrogen and stored at −80°C until microbiota analysis.
Fecal culture
Enterotoxigenic E. coli used in this experiment elaborates β-hemolysis and lactose fermentation, thus Columbia blood agar with 5% sheep blood and MacConkey agar were used to identify the percentage of β-hemolytic coliforms in feces. Fecal samples collected from the rectum of all pigs using cotton swabs on days 2, 7, 14, and 21 PI were used to perform fecal cultures. Briefly, fecal swabs were plated on Columbia blood agar and MacConkey agar using the quadrant streak plate method, and all plates were cultured in an air incubator at 37°C for 24 h. Total coliforms from both agars and β-hemolytic coliforms from blood agar were assessed visually using a scoring system ranging from 0 to 8 (0 = no bacterial growth, 8 = very heavy bacterial growth). The percentage of β-hemolytic coliforms in feces was calculated as the score ratio of β-hemolytic coliforms to total coliforms (Liu et al., 2013; He et al., 2020b).
Microbiota analysis
Bacterial DNA was extracted from fecal samples and ileal digesta using the Quick-DNA Fecal/Soil Microbe Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. DNA samples were amplified in duplicates by PCR in the V4 region of the 16S rRNA gene using primers 515F (5′-XXXXXXXXGTGTGCCAGCMGCCGCGGTAA-3′), which included an 8-bp barcode (X) unique to each sample followed by a 2 nt Illumina adapter (bold), and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Caporaso et al., 2012). The PCR reaction for each PCR well was composed of 2 μl of template DNA, 9.5 μl of nuclease-free water, 12.5 μl of GoTaq 2 × Master Mix (Promega, Madison, WI, USA), 0.5 μl of V4 reverse primer (10 μM), and 0.5 μl of barcoded forward primer (10 μM). Amplification was carried out in a thermocycler with the following settings: 94°C for 3 min for initializing denaturation; followed by 35 cycles of 94°C for 45 s, 50°C for 1 min, and 72°C for 1.5 min; and 72°C for 10 min for final elongation. The amplicon size for each sample was verified using agarose gel electrophoresis, and amplified samples were then pooled together, with the amount of sample added being quantified subjectively based on the band brightness in the agarose gel. The pooled sample was then purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and submitted to the UC Davis Genome Center DNA Technologies Core for 250 bp paired-end sequencing on the Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA).
Raw fastq files were first demultiplexed, and 8-bp barcodes were removed using saber (https://github.com/najoshi/sabre). Demultiplexed sequences were then imported into Quantitative Insights Into Microbial Ecology 2 (QIIME2; version 2020.8) to use the DADA2 plugin, which removes primers and lower-quality reads (Callahan et al., 2016; Bolyen et al., 2019, 2). Paired-end reads were denoised and merged, and chimeras were then removed to construct amplicon sequence variants (ASVs). Representative sequences for each ASV were aligned using MAFFT, and masked alignments were used to generate phylogenetic trees using FastTree2 (Price et al., 2010; Katoh and Standley, 2013). Python library scikit-learn was used to assign taxonomies based on representative sequences against Silva (version 138), which was pretrained in QIIME2 to be clipped in only the V4 hypervariable region and clustered at 99% sequence identity (Pedregosa et al., 2011; Quast et al., 2012; Bokulich et al., 2018).
Statistical analyses
All data excluding microbiota were analyzed using SAS (SAS Inst. Inc., Cary, NC). The normality of all data was verified using the UNIVARIATE procedure. Values that deviated from the treatment mean by more than 3 times the interquartile range were assumed to be outliers and removed. Measurements were analyzed by ANOVA using the PROC MIXED of SAS in a randomized complete block design with pigs as experimental units. The model included treatment as the main effect and blocks as random effects. The LSMEANS statement and the PDIFF option of PROC MIXED were used to separate treatment means. Chi-square was used to find significance in the frequency of diarrhea. Statistical significance and tendency were assessed as α = 0.05 and α = 0.10, respectively.
Shannon and Chao1 indices were measured for alpha diversity by using the estimate_richness function in phyloseq (McMurdie and Holmes, 2013). Bray-Curtis matrix was used to compare community composition among treatments and day for feces and only treatment for ileal digesta. The relative abundance of each taxon in each sample was calculated by dividing the number of taxa by the total number of filtered reads in each sample. Files were exported from QIIME2 and imported into R 4.1.0 for data visualization and statistical analysis (R Core Team, 2021). All microbiota analyses were performed using the phyloseq package, and data were visualized using the ggplot2 package (Wickham, 2011). The normality and homoscedasticity were tested using the Shapiro-Wilks test and the Bartlett test, respectively. For fecal microbiota, the linear mixed-effect model was fitted using the lme4 package, with treatment, site, day, and interaction as fixed effects and pigs as random effects (Bates et al., 2014, p. 4). The significance of each term in the model was determined using the F-test as a type 3 analysis of variance using the ANOVA function in the car package, followed by a group comparison using the cld function in the emmeans package (Fox and Weisberg, 2018; Lenth, 2021). When normality or homoscedasticity was not observed, a nonparametric test was performed using the Kruskal-Wallis sum-rank test using the agricolae package (de Mendiburu and de Mendiburu, 2019). Bray-Curtis dissimilarity was first tested for homoscedasticity using the betadisper function and confirmed with P > 0.05. The statistical significance for beta diversity was then tested using PERMANOVA and the vegan package (Oksanen et al., 2013). Statistical significance was assessed as α = 0.05 and statistical tendency as α = 0.10. The P-values were adjusted for multiple comparisons using the false discovery rate (FDR).
Results
Growth performance, diarrhea, and white blood cell profile
No difference was observed in pig BW among treatments on days −7 and 0, day 7 PI, and day 14 PI (Table 2). On day 21 PI, pigs in BAM- had the heaviest BW, and pigs in CON+ had the lowest BW among all treatments (P < 0.05). Pig's final BW was greater (P < 0.05) in CON- than CON+, and final BW was greater (P < 0.05) in AGP+ than in CON+. No difference in ADG, ADFI, and gain:feed was observed in pigs between CON- and BAM- throughout the experiment. ETEC inoculation reduced (P < 0.05) ADG from days 0 to 21 PI and gain:feed from days 14 to 21 PI when CON+ was compared with CON-. Supplementation of AGP enhanced (P < 0.05) ADG from days 0 to 21 PI compared with CON+. Compared with CON+, pigs fed the BAM+ diet tended (P < 0.10) to increase ADFI and ADG of weaned pigs from days 0 to 21 PI and final BW at day 21 PI.
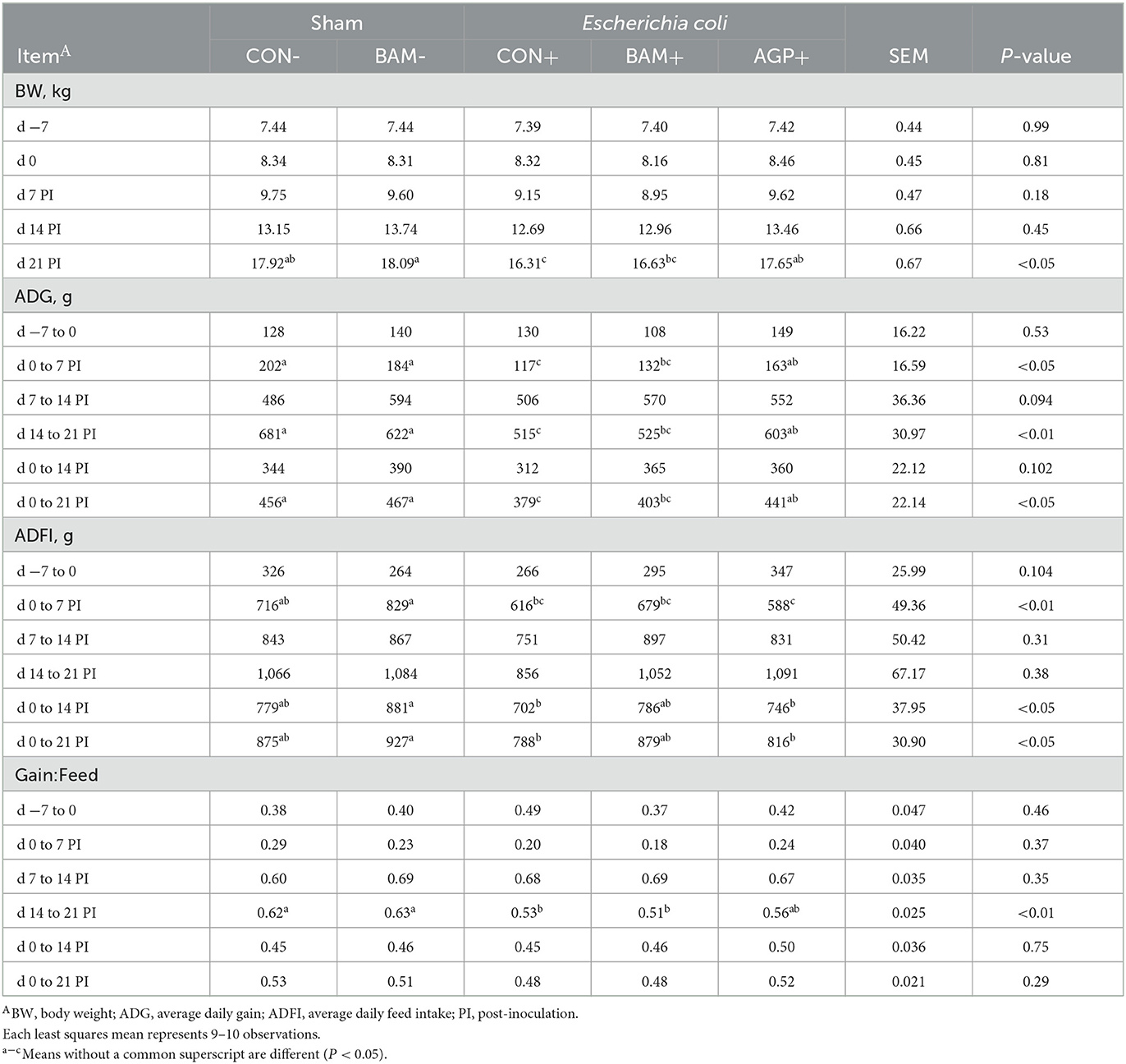
Table 2. Growth performance of weaned pigs fed a control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP).
Pigs in the sham groups (CON- and BAM-) had the lowest fecal score throughout the experiment (Figure 1). Pigs in BAM+ and CON+ had a greater (P < 0.05) fecal score from days 1 to 8 PI than pigs in CON-, while pigs in AGP+ were intermediate. While for all treatments the fecal score decreased as of day 9 to a level below diarrhea, between days 11 and 14 PI, pigs in BAM+ had the highest (P < 0.05) fecal score among treatments. After ETEC inoculation, the frequency of diarrhea (diarrhea score ≥ 3) was 31.36% in CON+ pigs, while the diarrhea frequency was 8.18% in CON- pigs (Figure 2). Pigs in CON+, BAM+, and AGP+ had higher (P < 0.05) frequency of diarrhea than pigs in CON- and BAM-, regardless of incidence (diarrhea score ≥ 3) and severity (diarrhea score ≥ 4). Supplementation of either BAM or AGP did not affect the frequency of diarrhea throughout the experiment, although pigs in AGP+ had a numerically lower frequency of diarrhea than pigs in CON+ and BAM+.
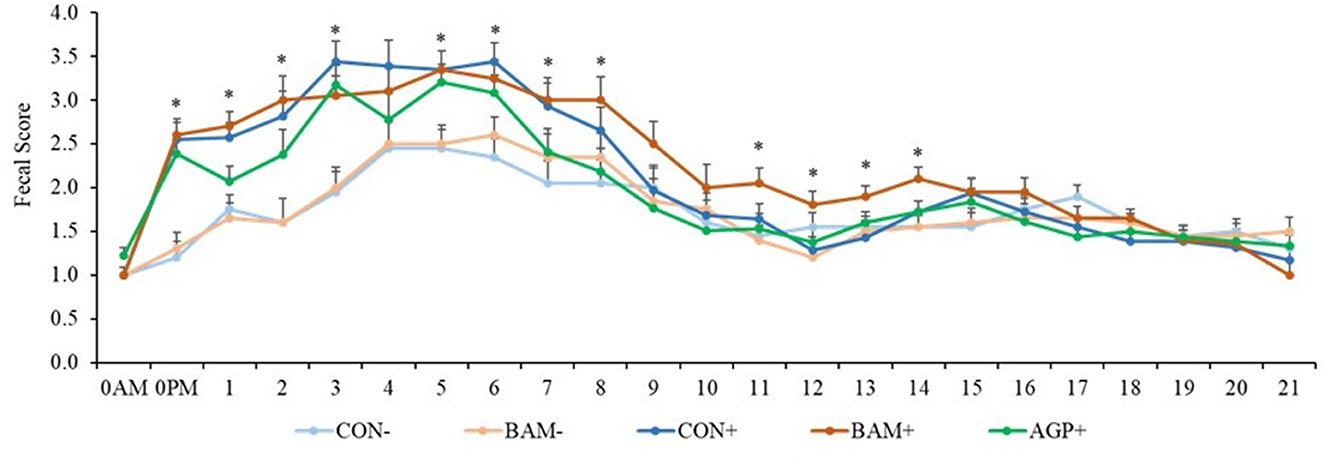
Figure 1. The daily fecal score of weaned pigs fed diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP) with or without enterotoxigenic Escherichia coli challenge. Fecal score = 1, normal feces; 2, moist feces; 3, mild diarrhea; 4, severe diarrhea; 5, watery diarrhea. PI, post-inoculation. *P < 0.05, indicating diarrhea scores were different among treatments. Each least squares mean represents 9–10 observations.
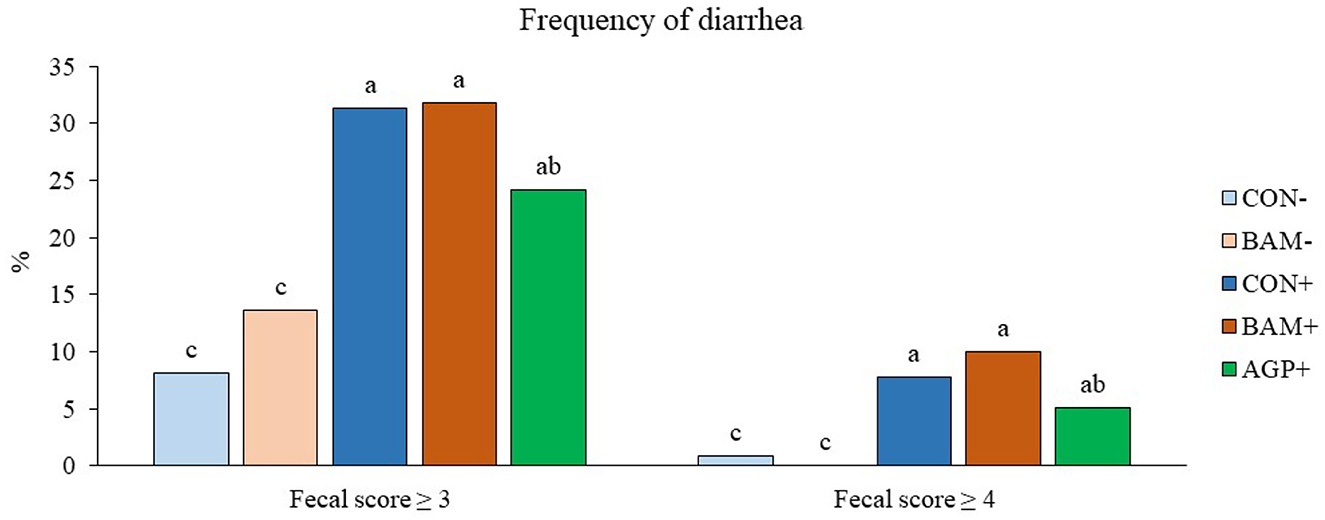
Figure 2. Frequency of diarrhea in weaned pigs fed diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP). The frequency of diarrhea was calculated as the percentage of pig days with a fecal score ≥ 3 or ≥ 4 in the total number of pig days. Each least squares mean represents 9–10 observations. a−cMeans without a common superscript are different (P < 0.05).
No β-hemolytic coliforms were detected in fecal samples of pigs in CON- and BAM- throughout the experiment, and no β-hemolytic coliforms were observed in all pigs on days −7 and 0, day 14 PI, and day 21 PI. On day 2 PI, AGP+ had a lower (P < 0.05) percentage of β-hemolytic coliforms proliferated on the blood agar plates than CON+ (Figure 3). On day 7 PI, no difference was observed among all three treatments under the ETEC challenge.
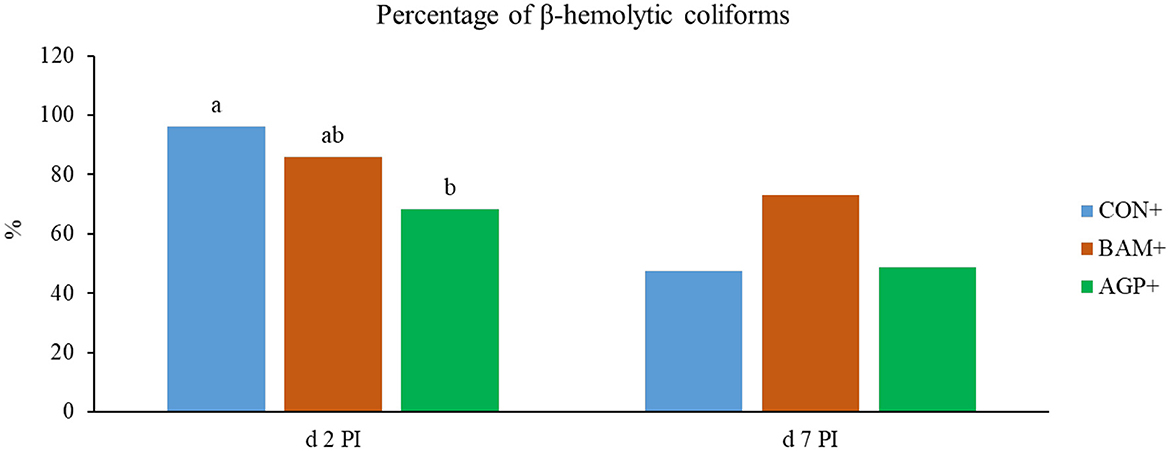
Figure 3. The percentage (%) of β-hemolytic total coliforms in fecal samples of Escherichia coli challenged weaned pigs fed a control diet (CON+) or diets supplemented with Bacillus amyloliquefaciens (BAM+) or antibiotics (AGP+). No β-hemolytic coliforms were observed in the fecal samples of pigs in the sham groups. No β-hemolytic coliforms were observed in the fecal samples of pigs before E. coli (ETEC) challenge and on days 14 and 21 post-inoculation (PI). Each least squares mean represents 9–10 observations. a, bMeans without a common superscript are different (P < 0.05).
No difference was observed in the white blood cell profile among the five treatments on day −7 (Table 3). On day 0, lymphocyte count was greater (P < 0.05) in AGP+ than in other treatments, except for BAM+. However, lymphocyte percentage was greater (P < 0.05) in BAM+ than in CON- and CON+. After ETEC inoculation, greater (P < 0.05) white blood cell and lymphocyte counts were observed in CON+ on days 7 and 21 PI, and a greater (P < 0.05) neutrophil count was observed in CON+ on day 14 PI, compared with CON-. Pigs in BAM+ had lower (P < 0.05) neutrophils on day 14 PI and lower (P < 0.05) white blood cells, neutrophils, and lymphocytes on day 21 PI, than pigs in CON+. Supplementation of AGP reduced (P < 0.05) neutrophil count on days 14 and 21 PI and increased (P < 0.05) lymphocyte percentage and monocyte percentage on day 21 PI compared with CON+. No difference was observed in the white blood cell profile of pigs between BAM+ and AGP+, with the exception that pigs in AGP+ had a greater (P < 0.05) monocyte percentage on days 7 and 21 PI but lower (P < 0.05) neutrophil percentage on day 21 PI than pigs in BAM+. No difference was observed in the white blood cell profile of pigs between BAM- and CON-. No difference was observed in the red blood cell profile of pigs among all treatments (Data were not shown).
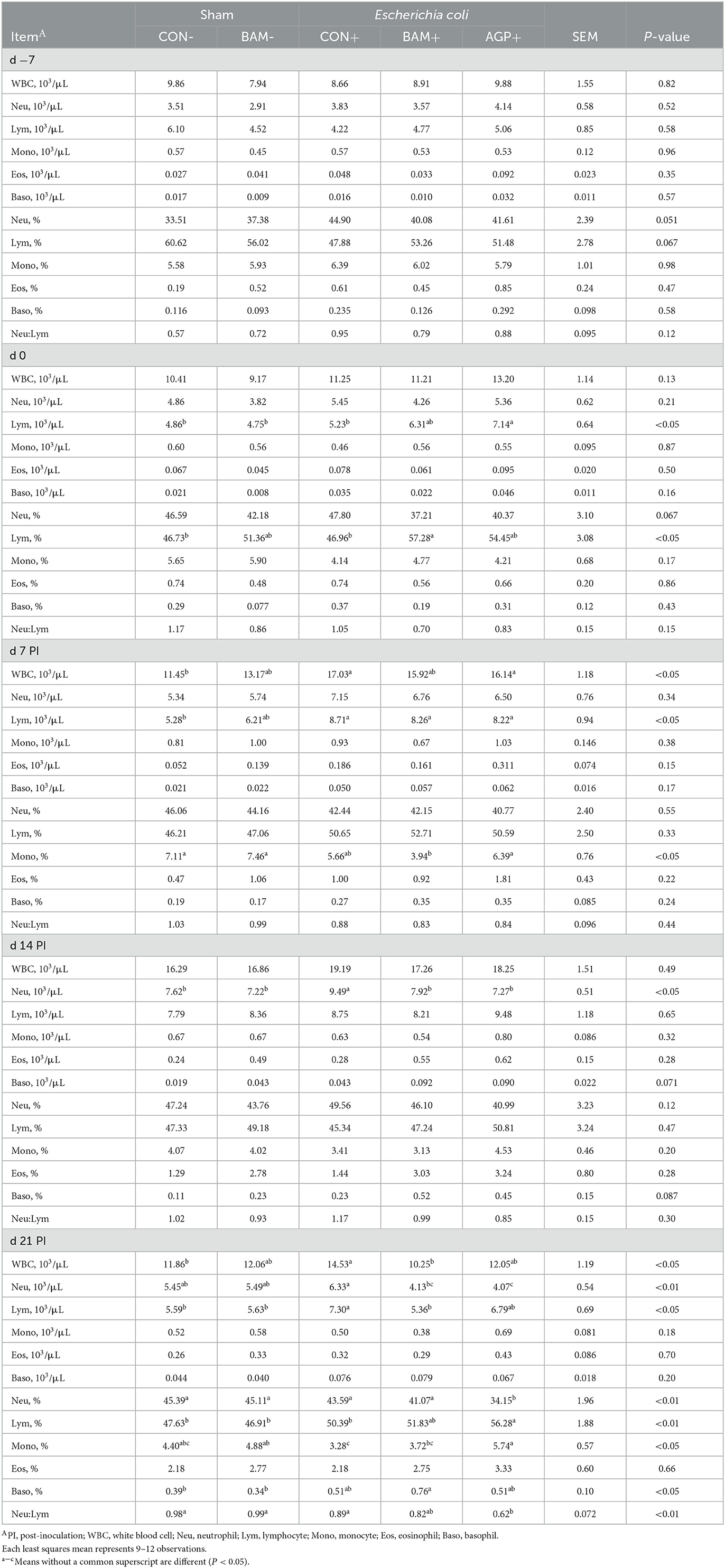
Table 3. Total and differential white blood cells in weaned pigs a fed control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP).
Fecal microbiota
The mean number of reads was 15,368, and the total number of taxa identified was 4,410 in the sequence data. An increase (P < 0.05) in Shannon and Chao1 indices was observed in feces when the age of the pig increased from day −7 to day 21 PI. However, no significant differences in Shannon and Chao1 indices were observed in feces among treatments throughout the experiment (Figure 4). In beta diversity, fecal samples collected on day −7 were clustered and separated from fecal samples collected on days 0, 7, 14, and 21 PI (Figure 5A). Fecal samples collected on day 0 were clustered away from fecal samples collected on day 21 PI. Fecal samples from all treatments were clustered together on day −7 (Figure 5B). On day 7 PI, fecal samples from AGP+ were moderately clustered away from CON- and CON+. AGP+ was clustered farther away from BAM+ and CON+ on day 21 PI.
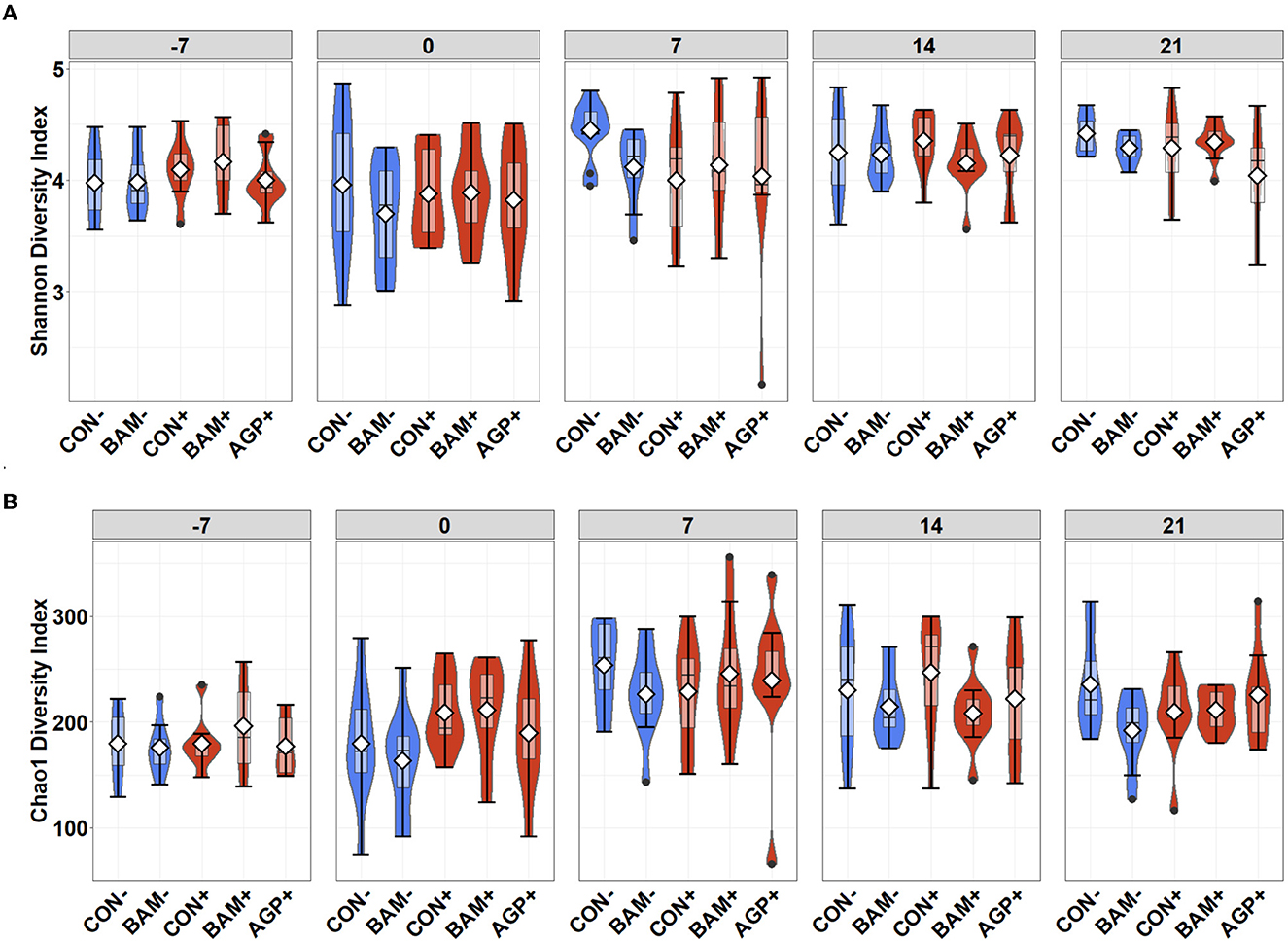
Figure 4. Alpha diversity as indicated by Shannon (A) and Chao1 (B) indices in feces of weaned pigs fed with control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM), or antibiotics (AGP) at the beginning of the experiment (day−7), the first day of E. coli (ETEC) inoculation (day 0), and days 7, 14, and 21 post-inoculation. No difference was observed in Shannon (A) and Chao1 (B) indices among treatments. Violin plots are colored whether not infected (blue) or infected with E. coli (red). Data are expressed as mean (diamond) ± SEM.
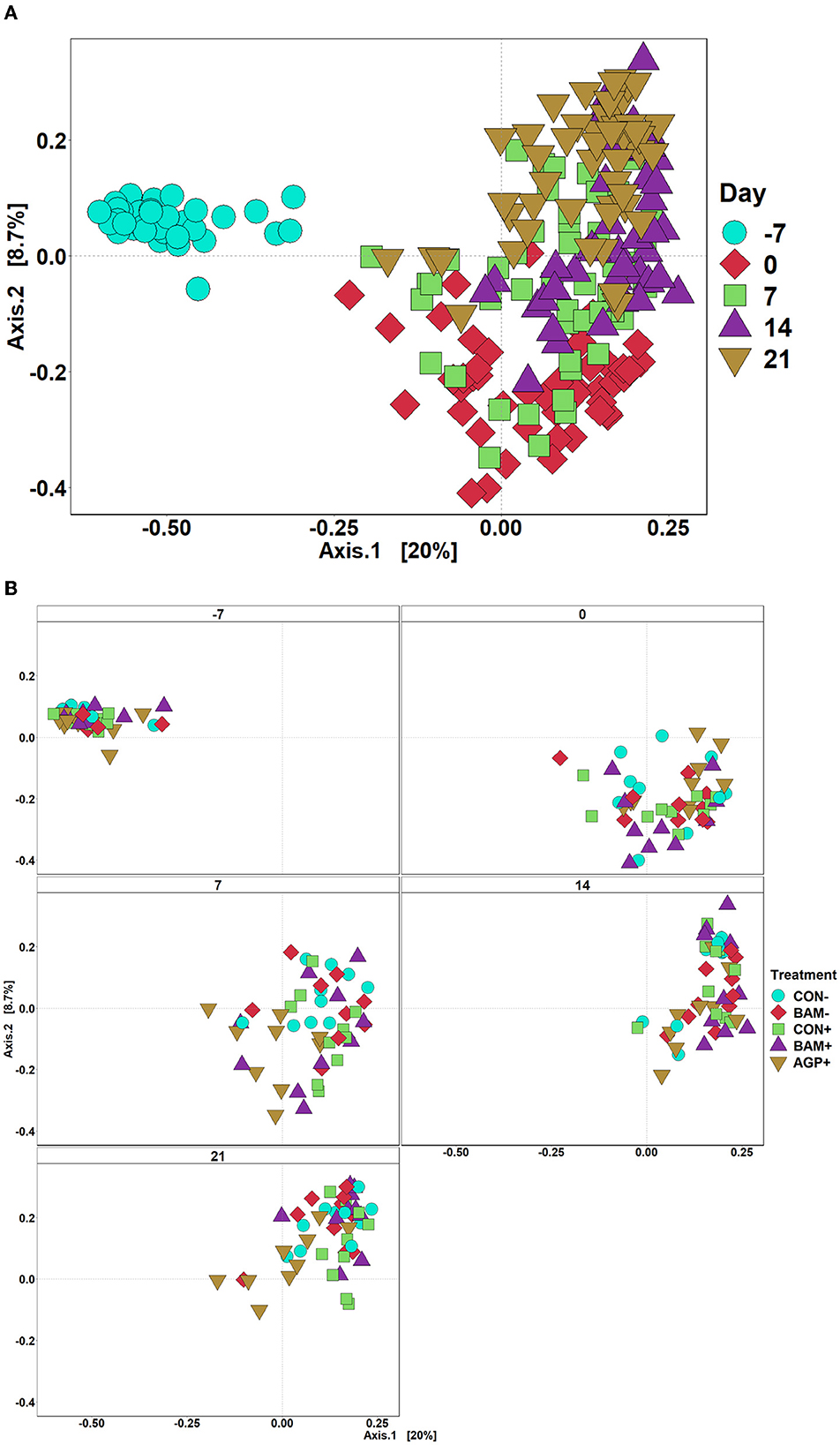
Figure 5. Principal coordinate analysis (PCoA) based on Bray-Curtis distance for beta diversity of fecal samples of weaned pigs fed with a control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP). Different symbols and shapes represent day fecal samples collected on days−7 and 0 before E. coli (ETEC) inoculation and days 7, 14, and 21 post-inoculation (A). Different symbols and shapes represent treatments (B). Each treatment has 9–10 observations.
The three most abundant phyla (most to least abundant) were Firmicutes, Bacteroidota, and Proteobacteria in the fecal samples of pigs from all treatments throughout the experiment. No difference was observed in the relative abundance of phyla in fecal samples on day −7 (Table 4). The relative abundance of Firmicutes was the highest on day 14 PI compared with other time points. The relative abundance of Proteobacteria was observed to be the highest (P < 0.05) on day 7 PI and the relative abundance of Bacteroidota was the highest (P < 0.05) on day 21 PI among all fecal collection days. At the family level, the relative abundance of Bacteroidaceae and Clostridiaceae was decreased (P < 0.05), while the relative abundance of Prevotellaceae, Lachnospiraceae, and Lactobacillaceae was increased (P < 0.05) in feces as the age of pig increased.
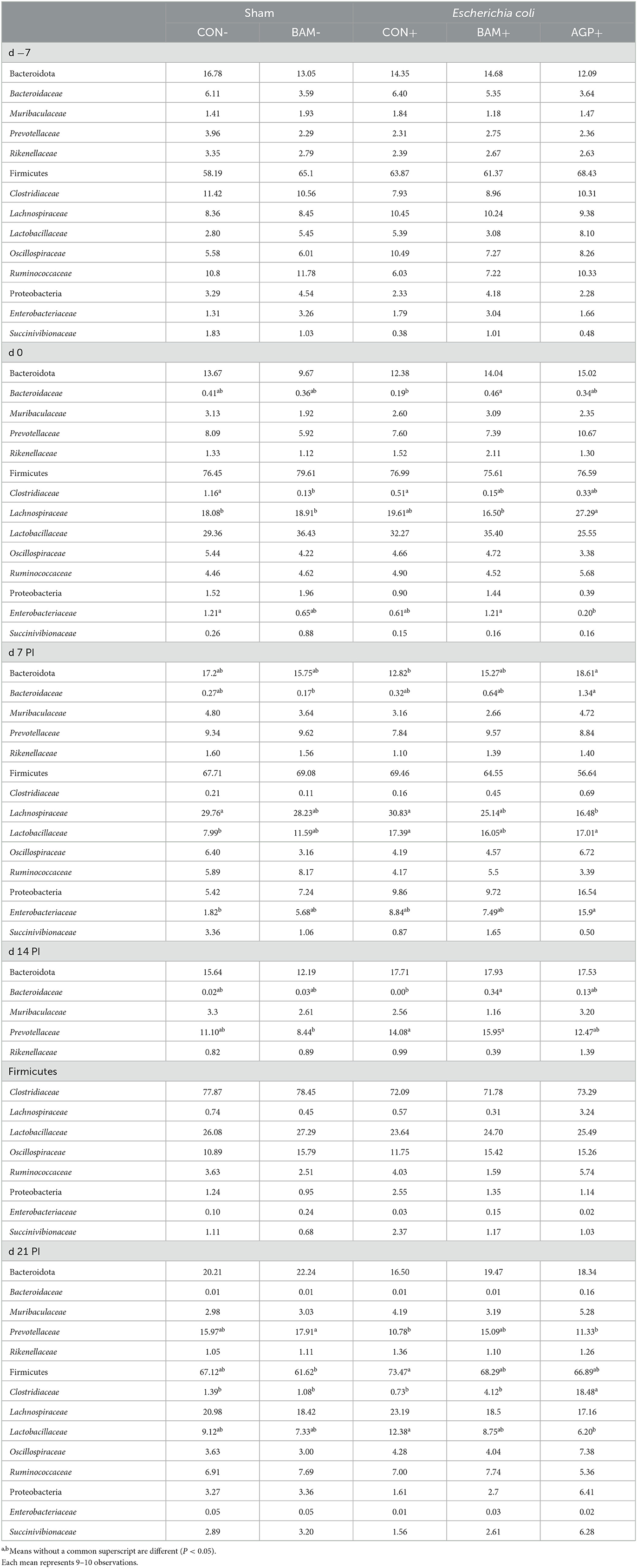
Table 4. Relative abundance (%) of Bacteroidota, Firmicutes, and Proteobacteria and their top families in feces of weaned pigs a fed control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP).
Dietary treatments had limited effects on the relative abundance of Firmicutes, Bacteroidota, and Proteobacteria in fecal samples collected on day 0, except that pigs in AGP+ had the greatest (P < 0.05) Lachnospiraceae but the lowest (P < 0.05) Enterobacteriaceae among all treatments. Pigs in BAM- had a relatively lower (P < 0.05) abundance of Clostridiaceae than pigs in CON-, while pigs in BAM+ had a relatively higher (P < 0.05) abundance of Bacteroidaceae than pigs in CON- on day 0. ETEC inoculation increased (P < 0.05) the relative abundance of Lactobacillaceae in feces on day 7 PI when CON+ was compared with CON-. Supplementation of AGP enhanced (P < 0.05) the relative abundance of Bacteroidota on day 7 PI and Clostridiaceae on day 21 PI, but AGP decreased (P < 0.05) the relative abundance of Lachnospiraceae on day 7 PI and Lactobacillaceae on day 21 PI compared with CON+. Pigs in AGP+ also had a greater (P < 0.05) relative abundance of Clostridiaceae in feces than pigs in BAM+ on day 21 PI.
Within the 12 most abundant genera, nine genera were classified under Firmicutes, two under Bacteroidota, and one under Proteobacteria (Table 5). The three most abundant genera in all fecal samples throughout the experiment were Lactobacillus, Blautia, and Prevotella. The relative abundance of these three genera was increased from days −7 to 0. However, the relative abundance of Lactobacillus in fecal samples from all pigs decreased (P < 0.05) from days 0 to 7 PI. No difference was observed in the most abundant genera in fecal samples of pigs between CON- and BAM- throughout the experiment. On day 0, pigs supplemented with AGP had a greater (P < 0.05) relative abundance of Prevotella than pigs in BAM+ and BAM- and had a greater (P < 0.05) relative abundance of Blautia than pigs in all other treatments. Compared CON+ with CON-, ETEC infection enhanced (P < 0.05) the relative abundance of Lactobacillus on day 7 PI and Megasphaera on day 21 PI but reduced (P < 0.05) the relative abundance of Coprococcus on day 14 PI and Streptococcus on day 21 PI. Supplementation of AGP reduced (P < 0.05) the relative abundance of Agathobacter on day 7 PI, the relative abundance of Dorea and Streptococcus on day 14 PI, and the relative abundance of Blautia, Dorea, Lactobacillus, and Megasphaera on day 21 PI compared with CON+. Compared with AGP+, pigs in BAM+ had a relatively higher (P < 0.05) abundance of Streptococcus on day 14 PI, and Prevotella, Megasphaera, and Streptococcus on day 21 PI in feces.
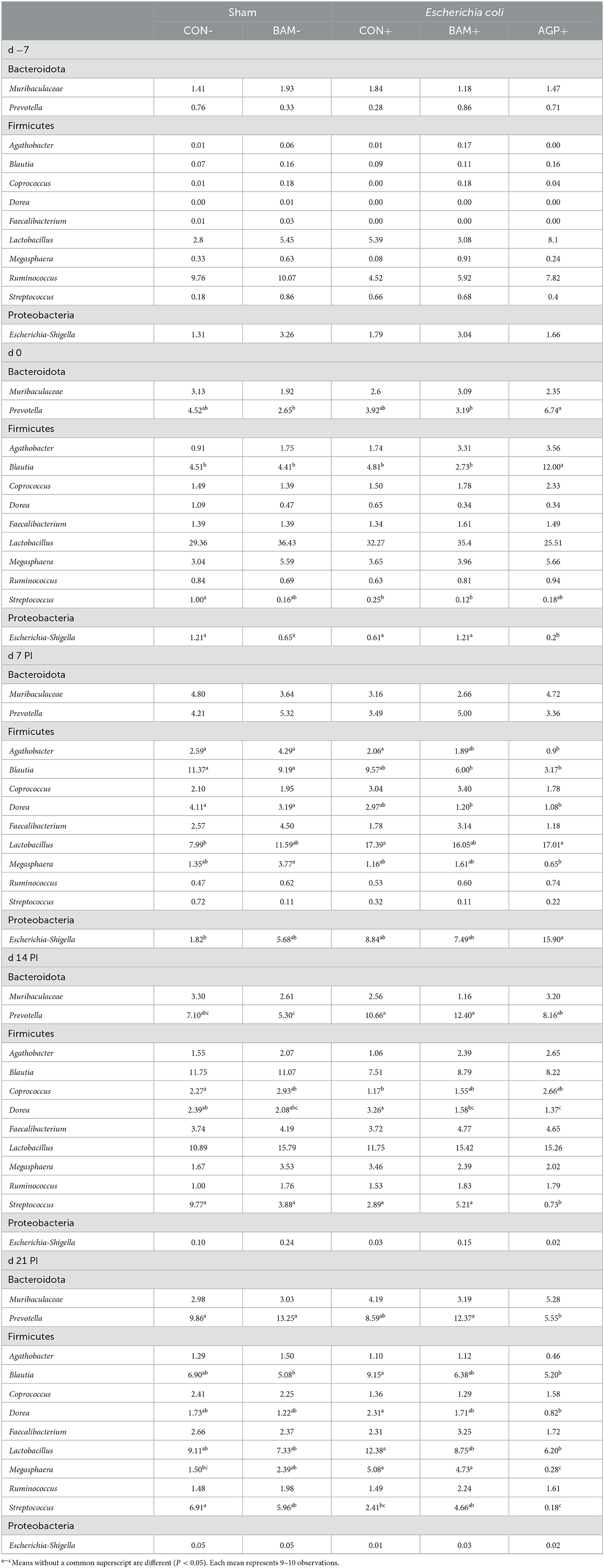
Table 5. Relative abundance (%) of most abundant genera from Bacteroidota, Firmicutes, and Proteobacteria in feces of weaned pigs fed control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP).
Ileal digesta microbiota
In ileal digesta, BAM+ had a greater (P < 0.05) Shannon index than AGP+ (Figure 6A). Pigs in CON+ had a greater (P < 0.05) Chao1 diversity and a tendency (P < 0.10) to be greater than pigs in BAM- and CON-, respectively, in terms of comparing Chao1 diversity (Figure 6B). In beta diversity, CON- and BAM- clusters were overlapping each other and separated from ETEC-infected groups, while the ETEC-infected groups had ileal digesta samples that were more dispersed from each other (Figure 7). Clusters for BAM+ and CON+ were overlapping each other, and the AGP+ cluster was partially isolated from other treatment clusters.
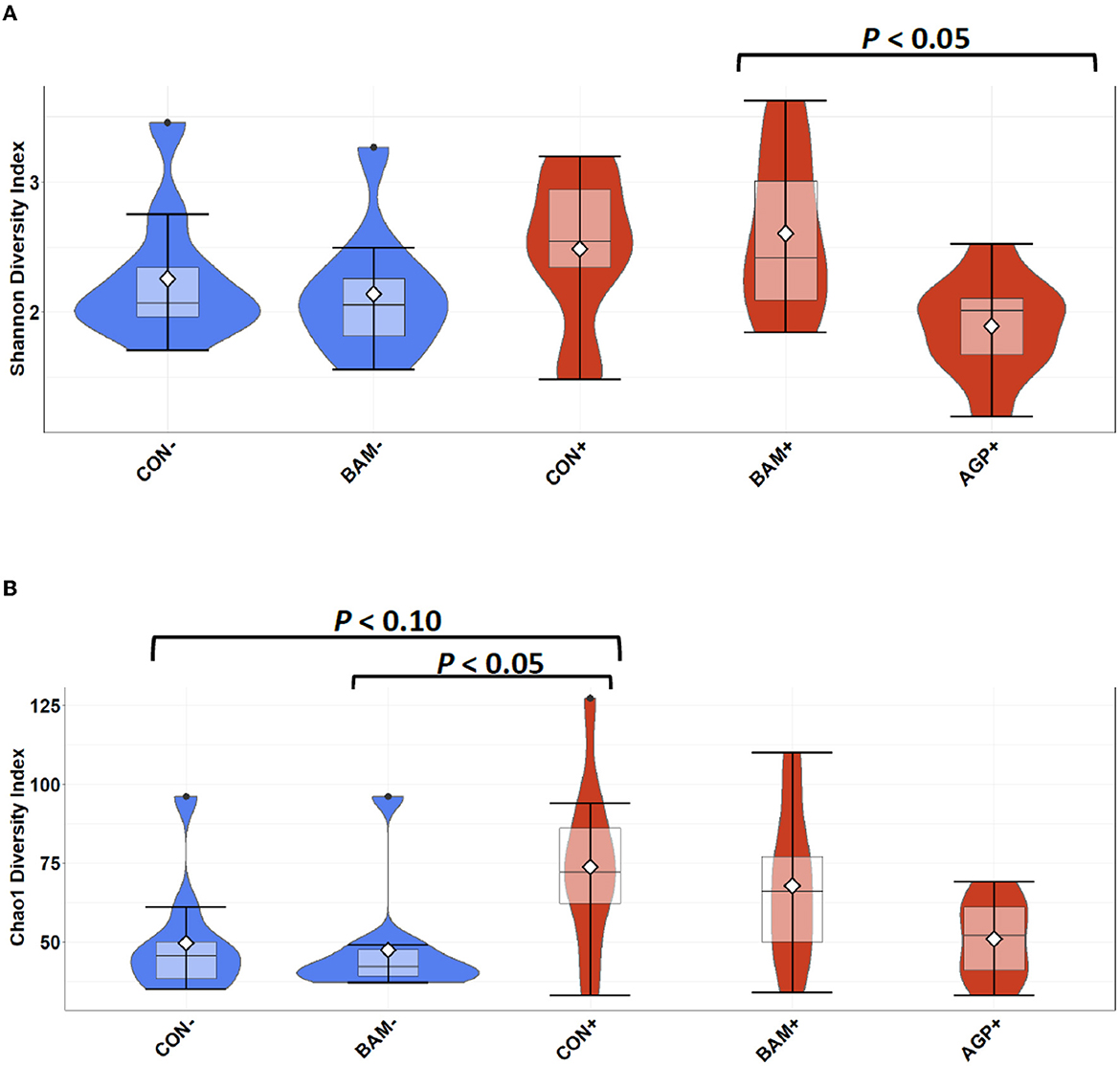
Figure 6. Alpha diversity as indicated by Shannon (A) and Chao1 (B) indices in ileal digesta collected from weaned pigs fed with a control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP) 21 days after enterotoxigenic E. coli (ETEC) inoculation. Violin plots are colored by ETEC infected or not. Data are expressed as mean (diamond) ± SEM.
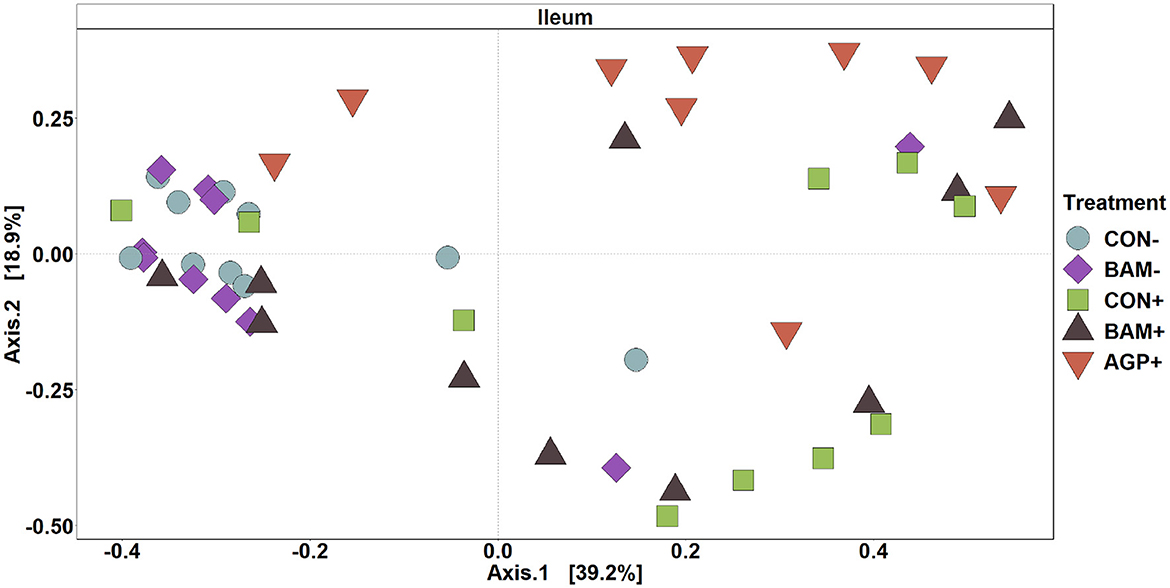
Figure 7. Principal coordinate analysis (PCoA) based on Bray-Curtis distance for beta diversity of ileal digesta of weaned pigs fed with a control (CON) diet or diets supplemented with Bacillus amyliloquefaciens (BAM) and antibiotics (AGP). Different symbols and shapes represent treatments. Each treatment has 9–10 observations.
The top four abundant phyla in ileal digesta collected on day 21 PI were Firmicutes, Proteobacteria, Actinomycetota, and Bacteroidota, from most to least abundant (Table 6). Pigs in CON+ had a greater (P < 0.05) relative abundance of Bacteroidota and Proteobacteria but a lower (P < 0.05) relative abundance of Firmicutes than pigs in CON-. At the family level, pigs in CON+ had a greater (P < 0.05) relative abundance of Atopobiaceae, Clostridiaceae, and Pasteurellaceae but a lower (P < 0.05) relative abundance of Lactobacillaceae than pigs in CON-. No difference was observed (P > 0.05) in ileal digesta microbiota between CON and BAM regardless of the ETEC challenge. Under the ETEC challenge, AGP enhanced (P < 0.05) the relative abundance of Firmicutes and Clostridiaceae but reduced (P < 0.05) the relative abundance of Bacteroidota and Atopobiaceae in ileal digesta compared with CON+. Compared with AGP+, BAM+ increased (P < 0.05) the relative abundance of Actinomycetota, Bacteroidota, Atopobiaceae, Bifidobacteriaceae, and Prevotellaceae but reduced (P < 0.05) the relative abundance of Firmicutes and Clostridiaceae in ileal digesta.
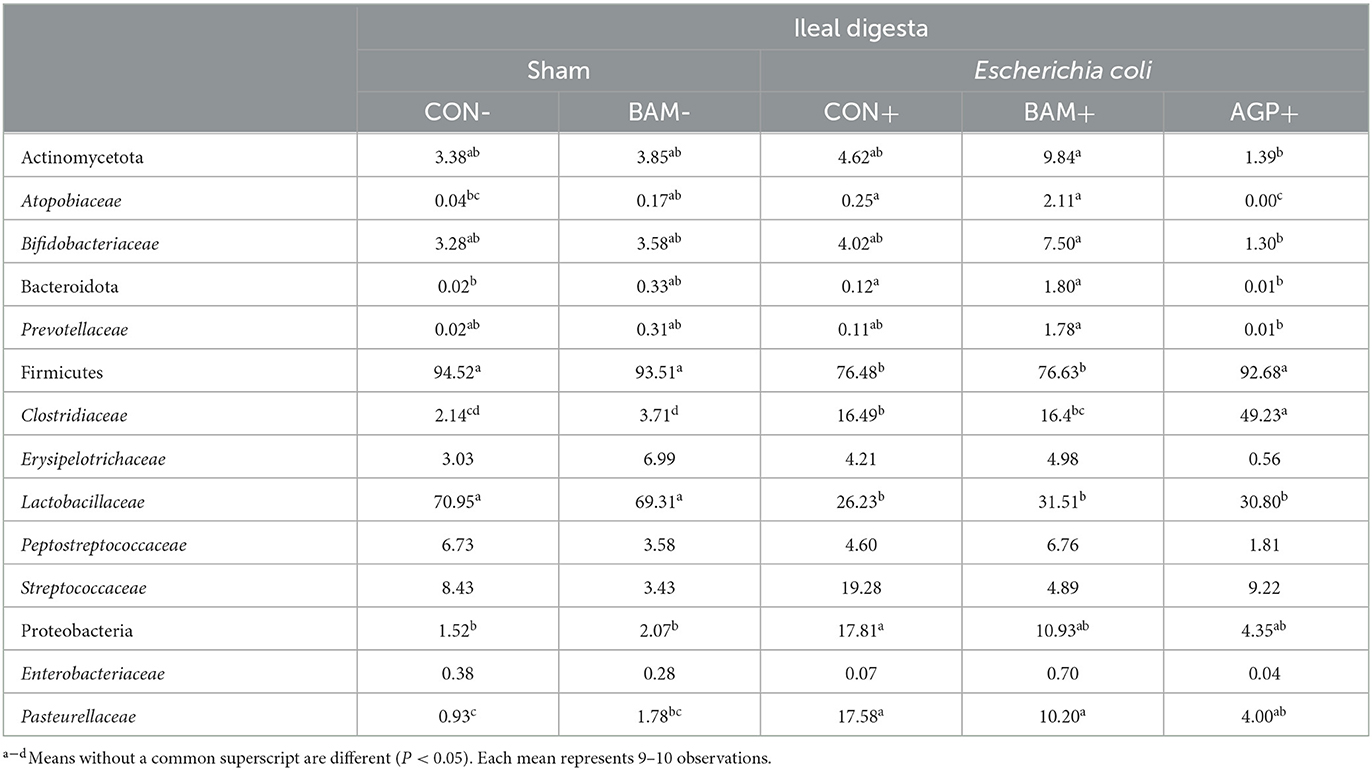
Table 6. Relative abundance (%) of Actinomycetota, Bacteroidota, Firmicutes, and Proteobacteria and their top families in ileal digesta of weaned pigs a fed control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP).
Within the eight most abundant genera, one was under Actinomycetota, six were under Firmicutes, and one was under Proteobacteria (Table 7). ETEC infection increased (P < 0.05) the relative abundance of Clostridium sensu stricto 1 and Actinobacillus but reduced (P < 0.05) the relative abundance of Lactobacillus when comparing pigs in CON+ vs. CON-. AGP+ had a greater (P < 0.05) relative abundance of Clostridium sensu stricto 1 and less (P < 0.05) relative abundance in Megasphaera than CON+. In addition, the relative abundance of Megasphaera and Bifidobacterium was greater (P < 0.05) in BAM+ than in AGP+, while AGP+ had a greater (P < 0.05) relative abundance of Clostridium sensu stricto 1 than BAM+.
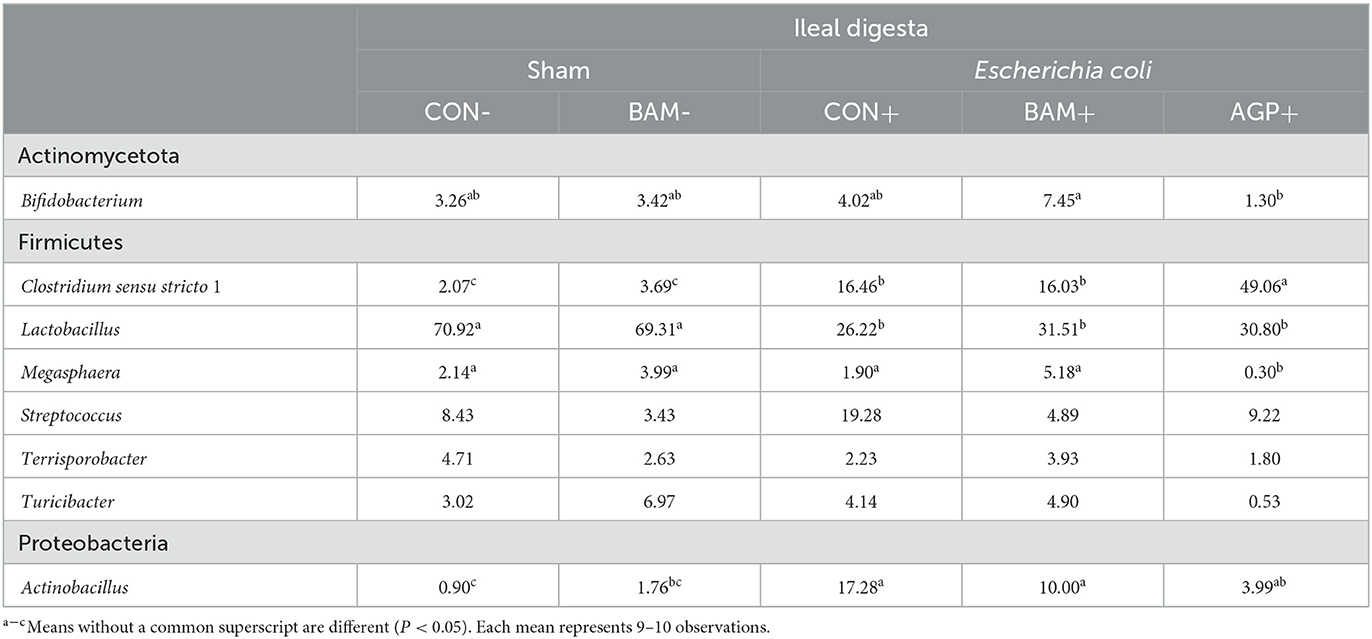
Table 7. Relative abundance (%) of most abundant genera from Firmicutes and Proteobacteria in ileal digesta of weaned pigs fed a control (CON) diet or diets supplemented with Bacillus amyloliquefaciens (BAM) or antibiotics (AGP).
Discussion
Antibiotics have been shown to induce prophages in the fecal samples of pigs over time, imposing a risk of developing antibiotic-resistant pathogens that could spread to humans (Allen et al., 2011). Complete eradication of antibiotic use at the post-weaning stage is desirable but currently not feasible; thus, developing alternative practices to treat post-weaning diarrhea and enhance the feed efficiency of weaned pigs is necessary (Angulo et al., 2005). Direct-fed microbials are looked upon to alleviate the intestinal damage caused by ETEC and to reduce the mortality rate of weaned pigs (Buntyn et al., 2016). Although B. amyloliquefaciens supplementations have been tested on broilers, limited studies have investigated the effects of B. amyloliquefaciens in weaned pigs. The present study observed that B. amyloliquefaciens supplementation tended to enhance growth performance and reduce systemic inflammation in weaned pigs challenged with ETEC F18. In addition, carbadox supplementation in the present study improved feed efficiency and alleviated diarrhea in ETEC-challenged weaned pigs. The gut microbiota were influenced differently between carbadox and B. amyloliquefaciens.
Consistent with our previous research, the increased frequency of diarrhea in pigs confirmed that the ETEC F18 strain inoculated into the pigs has successfully induced pathogenicity in the present study (Kim et al., 2019; He et al., 2020b). ETEC-infected pigs had reduced feed intake and weight gain and experienced severe diarrhea for approximately 6 days after the first ETEC inoculation, which falls under the average number of days when weaning pigs show diarrheal symptoms (Cox et al., 2012). No difference was observed in the growth performance of weaned pigs between the control and B. amyloliquefaciens in the sham group. Supplementation of B. amyloliquefaciens tended to enhance body weight, average daily gain, and feed intake of ETEC-infected pigs compared with controls. In consistency with previous research, supplementation of mixed strains of B. amyloliquefaciens enhanced feed efficiency of ETEC-infected pigs, while supplementation of a single strain of B. amyloliquefaciens reduced feed conversion ratio in broilers under necrotic enteritis challenge (Jerzsele et al., 2012; Becker et al., 2020). As expected, the supplementation of carbadox in the current study reduced the frequency of diarrhea and enhanced the growth rate of ETEC-challenged pigs. This observation was consistent with the results of Hung et al. (2020) and was supported by our fecal culture results, in which pigs fed with carbadox had fewer β-hemolytic coliforms in feces right after ETEC inoculation.
A complete blood count is crucial for evaluating the systemic severity of inflammation induced by bacterial infections, including ETEC. An increase in the number of white blood cells is commonly used to indicate the presence of systemic inflammation (Gordon-Smith, 2013). Our previous research also reported that the ETEC F18 challenge significantly increased the number of white blood cells, neutrophils, lymphocytes, and monocytes in weaned pigs at different time points (Liu et al., 2013; He et al., 2020a). In the present study, we also observed that ETEC inoculation increased total white blood cell counts and lymphocytes within 7 days post-inoculation. Reduced counts of lymphocytes and white blood cells in pigs supplemented with B. amyloliquefaciens or carbadox on days 14 and 21 PI suggest that both supplements may alleviate systemic inflammation caused by ETEC. These findings were also analogous to a study where B. amyloliquefaciens supplementation decreased white blood cell counts when broilers were challenged with lipopolysaccharides (Li et al., 2015). Without the ETEC challenge, B. amyloliquefaciens supplementation did not impact blood counts compared with negative controls. Similar results were also observed by Tang et al. (2018), in which no difference was observed in white and red blood cell counts and lymphocyte percentages when healthy laying hens were supplemented with B. amyloliquefaciens. The results from the current and previous studies suggest that B. amyloliquefaciens may have limited impacts on systemic immunity in animals when they are healthy.
The gut microbiota play an important role in reducing intestinal inflammation to promote a mutual relationship with the host (Lawley and Walker, 2013). Watery diarrhea induced by ETEC infection can disturb the gut microbiota, leading to difficulty suppressing inflammation (Bin et al., 2018). The alpha diversity in the present study showed an increase in microbial diversity and richness in fecal samples of ETEC-challenged pigs compared to sham pigs. This observation was contradicted with the findings by Pollock et al. (2018), in which fecal microbial diversity decreased over time when pigs were challenged with ETEC. The decreased microbial diversity observed by Pollock et al. (2018) may have occurred due to pigs being inoculated with ETEC at five different time points throughout the experiment, whereas pigs in the present study were inoculated with ETEC for 3 consecutive days after a 7-day adaptation period. However, the alpha diversity results in the present study agree with another study by Pollock et al. (2019), in which microbial diversity and richness in feces were not affected by the ETEC challenge. The increase in microbial diversity and richness in fecal samples over time and the overlapping samples in beta diversity between days 14 and 21 PI can be an indicator of maturity and stability in microbial diversity in pigs over time (Chen et al., 2017).
Firmicutes and Bacteroidota were the most abundant phyla in fecal microbiota throughout the experiment, which was expected in weaned piglets (Pajarillo et al., 2014; Yue et al., 2020; Luise et al., 2021). As expected, ETEC infection altered the microbial composition in the feces of pigs. The relative abundance of Escherichia–Shigella peaked 7 days after the first ETEC inoculation and then decreased on day 14 PI. This observation was supported by a gradual decrease in the diarrhea severity of ETEC-infected pigs in the present study, and it agrees with the results of Kim et al. (2022) that pigs underwent recovery from ETEC infection around 11 days after inoculation. Carbadox has been shown to decrease the relative abundance of taxa that are noted to be highly abundant in the fecal microbiota of pigs, which includes Lachnospiraceae, Blautia, and Lactobacillus. Carbadox is known to be a bactericidal primarily active against gram-positive bacteria; however, the mechanism behind this is yet to be known (Constable et al., 2017). In agreement with Lourenco et al. (2021), carbadox supplementation in the current study significantly decreased the relative abundance of Agathobacter on day 7 PI, Dorea and Streptococcus on day 14 PI, and Blautia and Dorea on day 21 PI, which are all gram-positive bacteria. Although some taxa, including Enterobacteriaceae and Clostridiaceae, are still abundant after carbadox exposure, the changes in gut bacterial biomass are unknown due to the limits of 16S rRNA sequencing. Moreover, the decrease in these taxa was observed on days 7 and 21 PI, indicating that carbadox may induce short- and long-term shifts in the fecal microbiota of weaned pigs by reducing microbial diversity (Holman et al., 2017). Supplementation of B. amyloliquefaciens had limited effects on fecal microbiota in pigs in the sham group. However, supplementation of B. amyloliquefaciens moderately affected fecal microbiota composition compared with control or carbadox under the ETEC challenge. On day 14 PI, pigs fed with B. amyloliquefaciens had less abundant Dorea than pigs in the positive control but had more abundant Streptococcus than pigs fed with carbadox. Supplementation of B. amyloliquefaciens also increased the relative abundance of Prevotella, Megasphaera, and Streptococcus compared with pigs fed with carbadox on day 21 PI.
Microbiota changes in the ileal digesta on day 21 PI were expected when pigs were challenged with ETEC or fed different diets. Unlike the proximate site of the digestive system, which has more bacterial barrier including stomach acid and bile salts, the ileum provides an optimal environment for ETEC proliferation in pigs (Gonzales et al., 2013; Roussel et al., 2020). Similar to the alpha diversity results in fecal microbiota, ETEC infection tended to increase microbial diversity and richness in the ileum. These findings were also concurrent with the findings by Pollock et al. (2019). The ETEC challenge in the current study decreased the relative abundance of Firmicutes but increased the relative abundance of Bacteroidota and Proteobacteria phyla in the ileum. The ratio of Firmicutes to Bacteroidota is widely accepted to play an important role in maintaining normal intestinal homeostasis, and a decrease in the Firmicutes:Bacteroidota ratio is usually observed in inflammatory bowel disease (Shen et al., 2018). However, this ratio can also be affected by an increase in other phyla during dysbiosis, such as the change in Proteobacteria. Growing evidence suggests that Proteobacteria is the most variable phylum, which contributes to microbial perturbation and may lead to increased disease risks (Morgan et al., 2012; Shin et al., 2015). The changes in ileal phyla are also explained by the changes in microbiota composition at family and genera levels. ETEC infection reduced the relative abundance of Lactobacillus (26.22% in positive control vs. 70.92% in negative control) but increased the relative abundance of Clostridium sensu stricto 1 and Actinobacillus in ileal digesta. Various Lactobacillus species have shown beneficial impacts on overall intestinal ecology; thus, the species are commonly being investigated as probiotic candidates in humans and pigs (de Vries et al., 2006; Suo et al., 2012; Sayan et al., 2018). It was also reported that commensal Lactobacillus can activate the host immunity to promote the overall health of mice (Holman et al., 2017; Nakamoto et al., 2017; Qi et al., 2019). Actinobacillus are gram-negative bacteria with most of the species characterized as commensals, but some species are considered pathogens in animal disease and the abundance was reported to increase in human disease as well (Rycroft and Garside, 2000; Denoth et al., 2021). Clostridium sensu stricto 1 is characterized as an opportunistic pathogen, which was reported to induce intestinal inflammation and reduce short-chain fatty acid production in pigs and poultry (Yang et al., 2019; Li et al., 2020; Hu et al., 2021). Thus, a reduction in Lactobacillus abundance and an increase in Actinobacillus and Clostridium sensu stricto 1 in the ileal digesta of ETEC-infected pigs confirm that ETEC infection remarkably disturbs the intestinal microbiota community of weaned pigs by potentially competing for colonization sites or nutrients with favorable bacteria. The present results also suggest that ETEC can cause a long-term perturbation in ileal microbiota throughout the weaning phase of pigs. Perturbation in the ileal microbiota may lead to unfavorable consequences in pig health, including immunosuppression and disrupted integrity in intestinal structure (Xia et al., 2022). In addition, more differences were observed in ileal microbiota than in fecal microbiota, which is likely due to the ileum being the major site of ETEC colonization (Martín-Rodríguez et al., 2022).
Under the ETEC challenge, pigs supplemented with B. amyloliquefaciens had an increased microbial diversity and relative abundance of Actinomycetota, particularly Bifidobacterium in the ileal digesta than pigs fed with carbadox. Actinomycetota plays an important role in maintaining gut homeostasis, especially their genus Bifidobacterium is also commonly investigated as potential probiotics, as they support the host immune system by stimulating the release of immunoglobulins in the intestinal mucosa (Holman et al., 2017; Binda et al., 2018; Sun et al., 2020). The presence of B. amyloliquefaciens may aid the growth of other microbes in the gut to outperform ETEC colonization (Dubreuil et al., 2016). Moreover, B. amyloliquefaciens supplementation modified the ileal microbiota differently compared with carbadox. Compared with B. amyloliquefaciens, carbadox further increased the abundance of Clostridium sensu stricto 1 and reduced the abundance of Prevotellaceae in ileal microbiota, which was also observed in a pig study with antibiotic growth promoter tylosin and a mouse study with enrofloxacin (Kim et al., 2016; Sun et al., 2019). The increased abundance of Firmicutes under carbadox supplementation may build on existing evidence of Firmicutes developing antimicrobial resistance genes due to consistent exposure to antibiotics (Anthony et al., 2022). Current results also suggest that carbadox supplementation may have long-term impacts on the ileal microbiota of weaned pigs, which may not be able to reestablish similarly to that of healthy weaned pigs (Yue et al., 2020). Our findings highlight taxa influenced by ETEC infection and/or dietary supplements; however, future studies should consider evaluating the functional genomes from the gut microbiota and assess the relationship between growth performance and immunity of the host to their microbiota.
Conclusion
As the use of antibiotic growth promoters becomes less desirable globally, alternative practices are imperative for enhancing growth and reducing post-weaning diarrhea in weaned pigs. The present study ultimately observed that supplementing B. amyloliquefaciens tended to increase feed intake and weight gain but had limited impacts on the diarrhea of weaned pigs infected with ETEC. However, pigs fed with B. amyloliquefaciens had relatively milder systemic inflammation than controls under disease-challenging conditions. In addition, pigs supplemented with B. amyloliquefaciens had a relatively higher abundance of Bifidobacterium but lower Clostridium sensu stricto 1 than carbadox when pigs were challenged with ETEC. The modulatory effects of B. amyloliquefaciens on immunity and ileal microbiota in pigs warrant further investigation. Taken altogether, supplementation of B. amyloliquefaciens solely may not provide growth enhancement and acute diarrheal alleviation of weaned pigs as similar to the addition of carbadox. Although various Bacillus spp. have shown the potential for promoting animal health and performance, findings in the present study suggest that there are differences between Bacillus spp. and their effects on different animal species. Nevertheless, manipulating the gut microbiota to overall improve pig health and eradicate ETEC pathogenicity is currently a captivating interest in the swine industry. To further assess the importance of gut microbiota to alleviate post-weaning diarrhea, future studies are suggested to employ metagenomics and to investigate correlations among gut microbiota, immunity, and growth performance of pigs undergoing diarrhea in a larger scale study.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA887417.
Ethics statement
The protocol for this experiment was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC# 20809) at the University of California, Davis (UC Davis).
Author contributions
YL designed and supervised the entire experiment. CJ conducted the animal experiment and performed sample analysis and data analysis. CJ, MK, and YL contributed to the result interpretation. BW and XL provided assistance for animal experiments. CJ and YL wrote the manuscript. BW, MK, and XL revised the manuscript. The final version was approved by all authors.
Conflict of interest
MK and JH were employed by Evonik Operations GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aarestrup, F. M. (2005). Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin. Pharmacol. Toxicol. 96, 271–281. doi: 10.1111/j.1742-7843.2005.pto960401.x
Allen, H. K., Looft, T., Bayles, D. O., Humphrey, S., Levine, U. Y., Alt, D., et al. (2011). Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2, e00260–e00211. doi: 10.1128/mBio.00260-11
Amezcua, R., Friendship, R. M., Dewey, C. E., Gyles, C., and Fairbrother, J. M. (2002). Presentation of postweaning Escherichia coli diarrhea in southern Ontario, prevalence of hemolytic E. coli serogroups involved, and their antimicrobial resistance patterns. Can. J. Vet. Res. 66, 73–78.
Angulo, F. J., Collignon, P., Wegener, H. C., Braam, P., and Butler, C. D. (2005). The routine use of antibiotics to promote animal growth does little to benefit protein undernutrition in the developing world. Clin. Infect. Dis. 41, 1007–1013. doi: 10.1086/433191
Anthony, W. E., Wang, B., Sukhum, K. V., D'Souza, A. W., Hink, T., Cass, C., et al. (2022). Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults. Cell Rep. 39, 110649. doi: 10.1016/j.celrep.2022.110649
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2014). Fitting linear mixed-effects models using lme4. arXiv. doi: 10.18637/jss.v067.i01
Becker, S. L., Li, Q., Burrough, E. R., Kenne, D., Sahin, O., Gould, S. A., et al. (2020). Effects of an F18 enterotoxigenic Escherichia coli challenge on growth performance, immunological status, and gastrointestinal structure of weaned pigs and the potential protective effect of direct-fed microbial blends. J. Anim. Sci. 98, skaa113. doi: 10.1093/jas/skaa113
Bin, P., Tang, Z., Liu, S., Chen, S., Xia, Y., Liu, J., et al. (2018). Intestinal microbiota mediates Enterotoxigenic Escherichia coli-induced diarrhea in piglets. BMC Vet. Res. 14, 385. doi: 10.1186/s12917-018-1704-9
Binda, C., Lopetuso, L. R., Rizzatti, G., Gibiino, G., Cennamo, V., and Gasbarrini, A. (2018). Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Digest. Liver Dis. 50, 421–428. doi: 10.1016/j.dld.2018.02.012
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome. 6, 90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Buntyn, J. O., Schmidt, T., Nisbet, D. J., and Callaway, T. R. (2016). The role of direct-fed microbials in conventional livestock production. 22, 335–55. doi: 10.1146/annurev-animal-022114-111123
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Casewell, M., Friis, C., Marco, E., McMullin, P., and Phillips, I. (2003). The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 52, 159–161. doi: 10.1093/jac/dkg313
Chen, L., Xu, Y., Chen, X., Fang, C., Zhao, L., and Chen, F. (2017). The maturing development of gut microbiota in commercial piglets during the weaning transition. Front. Microbiol. 8, 1688. doi: 10.3389/fmicb.2017.01688
Constable, P. D., Hinchcliff, K. W., Done, S. H., and Grünberg, W. (2017). “6 - Practical Antimicrobial Therapeutics,” in Veterinary Medicine (Eleventh Edition) Saunders, W. B. p. 153–174.
Cox, E., Loos, M., Coddens, A., Devriendt, B., Vesna, M., Vanrompay, D., et al. (2012). Post-Weaning E. coli Infections in Pigs and Importance of the Immune System. Maisons-Alfort: Actes des Journées AFMVP.
de Vries, M. C., Vaughan, E. E., Kleerebezem, M., and de Vos, W. M. (2006). Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16, 1018–1028. doi: 10.1016/j.idairyj.2005.09.003
Denoth, L., Juillerat, P., Kremer, A. E., Rogler, G., Scharl, M., Yilmaz, B., et al. (2021). Modulation of the mucosa-associated microbiome linked to the PTPN2 risk gene in patients with primary sclerosing cholangitis and ulcerative colitis. Microorganisms. 9, 1752. doi: 10.3390/microorganisms9081752
Dubreuil, J. D., Isaacson, R. E., and Schifferli, D. M. (2016). Animal enterotoxigenic Escherichia coli. EcoSal Plus. 7. doi: 10.1128/ecosalplus.ESP-0006-2016
Fairbrother, J. M., Nadeau, É., and Gyles, C. L. (2005). Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 6, 17–39. doi: 10.1079/AHR2005105
Fouhse, J. M., Zijlstra, R. T., and Willing, B. P. (2016). The role of gut microbiota in the health and disease of pigs. Anim. Fron. 6, 30–36. doi: 10.2527/af.2016-0031
Fox, J., and Weisberg, S. (2018). An R Companion to Applied Regression. Thousand Oaks, CA: Sage publications.
Gonzales, L., Ali, Z. B., Nygren, E., Wang, Z., Karlsson, S., Zhu, B., et al. (2013). Alkaline pH is a signal for optimal production and secretion of the heat labile toxin, LT in enterotoxigenic Escherichia coli (ETEC). PLoS ONE. 8, e74069. doi: 10.1371/journal.pone.0074069
Gordon-Smith, T. (2013). Structure and function of red and white blood cells. Medicine. 41, 193–199. doi: 10.1016/j.mpmed.2013.01.023
He, Y., Jinno, C., Kim, K., Wu, Z., Tan, B., Li, X., et al. (2020a). Dietary Bacillus spp. enhanced growth and disease resistance of weaned pigs by modulating intestinal microbiota and systemic immunity. J. Anim. Sci. Biotechnol. 11, 101. doi: 10.1186/s40104-020-00498-3
He, Y., Kim, K., Kovanda, L., Jinno, C., Song, M., Chase, J., et al. (2020b). Bacillus subtilis: a potential growth promoter in weaned pigs in comparison to carbadox. J. Anim. Sci. 98, skaa290. doi: 10.1093/jas/skaa290
Holman, D. B., Brunelle, B. W., Trachsel, J., and Allen, H. K. (2017). Meta-analysis to define a core microbiota in the swine gut. mSystems 2, e00004–17. doi: 10.1128/mSystems.00004-17
Hu, C., Niu, X., Chen, S., Wen, J., Bao, M., Mohyuddin, S. G., et al. (2021). A comprehensive analysis of the colonic flora diversity, short chain fatty acid metabolism, transcripts, and biochemical indexes in heat-stressed pigs. Front. Immunol. 12, 717723. doi: 10.3389/fimmu.2021.717723
Hung, Y.-T., Hu, Q., Faris, R. J., Guo, J., Urriola, P. E., Shurson, G. C., et al. (2020). Analysis of gastrointestinal responses revealed both shared and specific targets of zinc oxide and carbadox in weaned pigs. Antibiotics. 9, 463. doi: 10.3390/antibiotics9080463
Jerzsele, A., Szeker, K., Csizinszky, R., Gere, E., Jakab, C., Mallo, J. J., et al. (2012). Efficacy of protected sodium butyrate, a protected blend of essential oils, their combination, and Bacillus amyloliquefaciens spore suspension against artificially induced necrotic enteritis in broilers. Poult. Sci. 91, 837–843. doi: 10.3382/ps.2011-01853
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, J., Guevarra, R. B., Nguyen, S. G., Lee, J.-H., Jeong, D. K., and Unno, T. (2016). Effects of the antibiotics growth promoter tylosin on swine gut microbiota. J. Microbiol. Biotechnol. 26, 876–882. doi: 10.4014/jmb.1512.12004
Kim, K., He, Y., Jinno, C., Kovanda, L., Li, X., Bravo, D., et al. (2022). Supplementation of oligosaccharide-based polymer enhanced growth and disease resistance of weaned pigs by modulating intestinal integrity and systemic immunity. J. Anim. Sci. Biotechnol. 13, 10. doi: 10.1186/s40104-021-00655-2
Kim, K., He, Y., Xiong, X., Ehrlich, A., Li, X., Raybould, H., et al. (2019). Dietary supplementation of Bacillus subtilis influenced intestinal health of weaned pigs experimentally infected with a pathogenic E. coli. J. Animal Sci. Biotechnol. 10, 52. doi: 10.1186/s40104-019-0364-3
Koumoutsi, A., Chen, X.-H., Henne, A., Liesegang, H., Hitzeroth, G., Franke, P., et al. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186, 1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004
Kreuzer, S., Reissmann, M., and Brockmann, G. A. (2013). New fast and cost-effective gene test to get the ETEC F18 receptor status in pigs. Vet. Microbiol. 163, 392–394. doi: 10.1016/j.vetmic.2012.12.040
Lawley, T. D., and Walker, A. W. (2013). Intestinal colonization resistance. Immunology. 138, 1–11. doi: 10.1111/j.1365-2567.2012.03616.x
Lee, I. K., Kye, Y. C., Kim, G., Kim, H. W., Gu, M. J., Umboh, J., et al. (2016). Stress, nutrition, and intestinal immune responses in pigs—a review. Asian-Australas. J. Anim. Sci. 29, 1075–1082. doi: 10.5713/ajas.16.0118
Lee, N.-K., Kim, W.-S., and Paik, H.-D. (2019). Bacillus strains as human probiotics: characterization, safety, microbiome, and probiotic carrier. Food Sci. Biotechnol. 28, 1297–1305. doi: 10.1007/s10068-019-00691-9
Lekagul, A., Tangcharoensathien, V., and Yeung, S. (2019). Patterns of antibiotic use in global pig production: a systematic review. Vet. Animal Sci. 7, 100058. doi: 10.1016/j.vas.2019.100058
Lenth, R. V. (2021). Emmeans: Estimated Marginal Means, AKA Least-Squares Means; R Package Version 1. 6. Vienna: CRAN (the Comprehensive R Archive Network).
Li, Y., Zhang, H., Chen, Y. P., Yang, M. X., Zhang, L. L., Lu, Z. X., et al. (2015). Bacillus amyloliquefaciens supplementation alleviates immunological stress in lipopolysaccharide-challenged broilers at early age. Poult. Sci. 94, 1504–1511. doi: 10.3382/ps/pev124
Li, Y.-J., Li, J., and Dai, C. (2020). The role of intestinal microbiota and mast cell in a rat model of visceral hypersensitivity. J. Neurogastroenterol. Motil. 26, 529–538. doi: 10.5056/jnm20004
Liao, S. F., and Nyachoti, M. (2017). Using probiotics to improve swine gut health and nutrient utilization. Anim. Nutrit. 3, 331–343. doi: 10.1016/j.aninu.2017.06.007
Liu, Y., Song, M., Che, T. M., Almeida, J. A. S., Lee, J. J., Bravo, D., et al. (2013). Dietary plant extracts alleviate diarrhea and alter immune responses of weaned pigs experimentally infected with a pathogenic Escherichia coli. J. Anim. Sci. 91,5294–5306. doi: 10.2527/jas.2012-6194
Lourenco, J. M., Hampton, R. S., Johnson, H. M., Callaway, T. R., Rothrock, M. J. Jr., and Azain, M. J. (2021). The effects of feeding antibiotic on the intestinal microbiota of weanling pigs. Front. Vet. Sci. 8, 601394. doi: 10.3389/fvets.2021.601394
Luise, D., Sciellour, M. L., Buchet, A., Resmond, R., Clement, C., Rossignol, M.-N., et al. (2021). The fecal microbiota of piglets during weaning transition and its association with piglet growth across various farm environments. PLoS ONE. 16, e0250655. doi: 10.1371/journal.pone.0250655
Martín-Rodríguez, A. J., Joffr,é, E., and Sjöling, Å. (2022). “Bacterial gastrointestinal infections,” in Encyclopedia of Infection and Immunity, Rezaei, N. (Oxford: Elsevier) p. 72–81. doi: 10.1016/B978-0-12-818731-9.00105-1
McMurdie, P. J., and Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 8, e61217. doi: 10.1371/journal.pone.0061217
Morgan, X. C., Tickle, T. L., Sokol, H., Gevers, D., Devaney, K. L., Ward, D. V., et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79. doi: 10.1186/gb-2012-13-9-r79
Nakamoto, N., Amiya, T., Aoki, R., Taniki, N., Koda, Y., Miyamoto, K., et al. (2017). Commensal Lactobacillus controls immune tolerance during acute liver injury in mice. Cell Rep. 21, 1215–1226. doi: 10.1016/j.celrep.2017.10.022
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'hara, R. B., et al. (2013). Package ‘Vegan', Community Ecology Package, Version, Vol. 2. Vienna: CRAN (the Comprehensive R Archive Network).
Pajarillo, E. A. B., Chae, J.-P., Balolong, M. P., Kim, H. B., and Kang, D.-K. (2014). Assessment of fecal bacterial diversity among healthy piglets during the weaning transition. J. Gen. Appl. Microbiol. 60, 140–146. doi: 10.2323/jgam.60.140
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2011). Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830.
Pollock, J., Gally, D. L., Glendinning, L., Tiwari, R., Hutchings, M. R., and Houdijk, J. G. M. (2018). Analysis of temporal fecal microbiota dynamics in weaner pigs with and without exposure to enterotoxigenic Escherichia coli1, 2. J. Anim. Sci. 96, 3777–3790. doi: 10.1093/jas/sky260
Pollock, J., Hutchings, M. R., Hutchings, K. E. K., Gally, D. L., and Houdijk, J. G. M. (2019). Changes in the ileal, but not fecal, microbiome in response to increased dietary protein level and enterotoxigenic Escherichia coli exposure in pigs. Appl. Environ. Microbiol. 85, e01252–e01219. doi: 10.1128/AEM.01252-19
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 5, e9490. doi: 10.1371/journal.pone.0009490
Qi, H., Li, Y., Yun, H., Zhang, T., Huang, Y., Zhou, J., et al. (2019). Lactobacillus maintains healthy gut mucosa by producing L-Ornithine. Commun. Biol. 2, 1–14. doi: 10.1038/s42003-019-0424-4
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team. (2021). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Roussel, C., De Paepe, K., Galia, W., De Bodt, J., Chalancon, S., Leriche, F., et al. (2020). Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol. 18, 141. doi: 10.1186/s12915-020-00860-x
Rycroft, A. N., and Garside, L. H. (2000). Actinobacillus species and their role in animal disease. Vet. J 159, 18–36. doi: 10.1053/tvjl.1999.0403
Salazar, F., Ortiz, A., and Sansinenea, E. (2017). Characterisation of two novel bacteriocin-like substances produced by Bacillus amyloliquefaciens ELI149 with broad-spectrum antimicrobial activity. J. Glob. Antimicrob. Resist. 11, 177–182. doi: 10.1016/j.jgar.2017.08.008
Sansinenea, E., and Ortiz, A. (2011). Secondary metabolites of soil Bacillus spp. Biotechnol. Lett. 33, 1523–1538. doi: 10.1007/s10529-011-0617-5
Sayan, H., Assavacheep, P., Angkanaporn, K., and Assavacheep, A. (2018). Effect of Lactobacillus salivarius on growth performance, diarrhea incidence, fecal bacterial population and intestinal morphology of suckling pigs challenged with F4+ enterotoxigenic Escherichia coli. Asian-australas. J. Anim. Sci. 31, 1308–1314. doi: 10.5713/ajas.17.0746
Shen, Z. H., Zhu, C. X., Quan, Y. S., Yang, Z. Y., Wu, S., Luo, W. W., et al. (2018). Relationship between intestinal microbiota and ulcerative colitis: Mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 24, 5–14. doi: 10.3748/wjg.v24.i1.5
Shin, N.-R., Whon, T. W., and Bae, J.-W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Sun, L., Zhang, X., Zhang, Y., Zheng, K., Xiang, Q., Chen, N., et al. (2019). Antibiotic-induced disruption of gut microbiota alters local metabolomes and immune responses. Front. Cell. Infect. 9, 99. doi: 10.3389/fcimb.2019.00099
Sun, S., Luo, L., Liang, W., Yin, Q., Guo, J., Rush, A. M., et al. (2020). Bifidobacterium alters the gut microbiota and modulates the functional metabolism of T regulatory cells in the context of immune checkpoint blockade. Proc. Nat. Acad. Sci. 117, 27509–27515. doi: 10.1073/pnas.1921223117
Suo, C., Yin, Y., Wang, X., Lou, X., Song, D., Wang, X., et al. (2012). Effects of lactobacillus plantarumZJ316 on pig growth and pork quality. BMC Vet. Res. 8, 89. doi: 10.1186/1746-6148-8-89
Tang, R. Y., Wu, Z. L., Wang, G. Z., and Liu, W. C. (2018). The effect of Bacillus amyloliquefaciens on productive performance of laying hens. Ital. J. Anim. Sci. 17, 436–441. doi: 10.1080/1828051X.2017.1394169
Wickham, H. (2011). ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185. doi: 10.1002/wics.147
World Health Organization (2015). Antimicrobial Resistance–SEA/RC68/R3. Geneva, Switzerland: World Health Organization.
Xia, B., Wu, W., Fang, W., Wen, X., Xie, J., and Zhang, H. (2022). Heat stress-induced mucosal barrier dysfunction is potentially associated with gut microbiota dysbiosis in pigs. Animal Nutrit. 8, 289–299. doi: 10.1016/j.aninu.2021.05.012
Yang, W.-Y., Lee, Y., Lu, H., Chou, C.-H., and Wang, C. (2019). Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS ONE. 14, e0205784. doi: 10.1371/journal.pone.0205784
Keywords: Bacillus amyloliquefaciens, Escherichia coli challenge, microbiome, performance, systemic immunity, weaned pigs
Citation: Jinno C, Wong B, Klünemann M, Htoo J, Li X and Liu Y (2023) Effects of supplementation of Bacillus amyloliquefaciens on performance, systemic immunity, and intestinal microbiota of weaned pigs experimentally infected with a pathogenic enterotoxigenic E. coli F18. Front. Microbiol. 14:1101457. doi: 10.3389/fmicb.2023.1101457
Received: 17 November 2022; Accepted: 20 February 2023;
Published: 15 March 2023.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Bing Xia, Beijing University of Agriculture, ChinaHyeun Bum Kim, Dankook University, Republic of Korea
Copyright © 2023 Jinno, Wong, Klünemann, Htoo, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhong Liu, eWFobGl1QHVjZGF2aXMuZWR1
 Cynthia Jinno
Cynthia Jinno Braden Wong1
Braden Wong1 Xunde Li
Xunde Li Yanhong Liu
Yanhong Liu