- Department of Microbiology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China
Introduction: Probiotic Lactobacillus strains had been investigated for the potential to protect against infection caused by the major fungal pathogen of human, Candida albicans. Besides antifungal activity, lactobacilli demonstrated a promising inhibitory effect on biofilm formation and filamentation of C. albicans. On the other hand, two commonly isolated non-albicans Candida species, C. tropicalis and C. parapsilosis, have similar characteristics in filamentation and biofilm formation with C. albicans. However, there is scant information of the effect of lactobacilli on the two species.
Methods: In this study, biofilm inhibitory effects of L. rhamnosus ATCC 53103, L. plantarum ATCC 8014, and L. acidophilus ATCC 4356 were tested on the reference strain C. albicans SC5314 and six bloodstream isolated clinical strains, two each of C. albicans, C. tropicalis, and C. parapsilosis.
Results and Discussion: Cell-free culture supernatants (CFSs) of L. rhamnosus and L. plantarum significantly inhibited in vitro biofilm growth of C. albicans and C. tropicalis. L. acidophilus, conversely, had little effect on C. albicans and C. tropicalis but was more effective on inhibiting C. parapsilosis biofilms. Neutralized L. rhamnosus CFS at pH 7 retained the inhibitory effect, suggesting that exometabolites other than lactic acid produced by the Lactobacillus strain might be accounted for the effect. Furthermore, we evaluated the inhibitory effects of L. rhamnosus and L. plantarum CFSs on the filamentation of C. albicans and C. tropicalis strains. Significantly less Candida filaments were observed after co-incubating with CFSs under hyphae-inducing conditions. Expressions of six biofilm-related genes (ALS1, ALS3, BCR1, EFG1, TEC1, and UME6 in C. albicans and corresponding orthologs in C. tropicalis) in biofilms co-incubated with CFSs were analyzed using quantitative real-time PCR. When compared to untreated control, the expressions of ALS1, ALS3, EFG1, and TEC1 genes were downregulated in C. albicans biofilm. In C. tropicalis biofilms, ALS3 and UME6 were downregulated while TEC1 was upregulated. Taken together, the L. rhamnosus and L. plantarum strains demonstrated an inhibitory effect, which is likely mediated by the metabolites secreted into culture medium, on filamentation and biofilm formation of C. albicans and C. tropicalis. Our finding suggested an alternative to antifungals for controlling Candida biofilm.
1. Introduction
Candida species are commensal fungi that colonize healthy human skin, gastrointestinal, and genital tracts. They are also opportunistic pathogens, leading to mucosal and systemic infections (Kim and Sudbery, 2011). Candida are one of the most important agents of fungal infection worldwide and the most frequently isolated fungal species in healthcare-associated infections (Bongomin et al., 2017). Over 90% of invasive Candida infections were caused by five species, Candida albicans, C. glabrata, C. tropicalis, C. parapsilosis, and C. krusei, with C. albicans accounted for 47–66% of cases (Pfaller and Diekema, 2007; Pfaller et al., 2019). Being the predominant cause of fungal infection, C. albicans has become the best-studied Candida species (Kabir et al., 2012). Although C. albicans remains as the most common isolated single species, a progressive shift to increased prevalence of non-albicans Candida species has been observed over the past decades (Pfaller et al., 2019). In the global SENTRY Antifungal Surveillance Program, the frequency of C. albicans was reported to gradually decrease from 57.4% of invasive Candida isolates in 1997–2001 to 46.4% in 2015–2016 (Pfaller et al., 2019).
Compared to C. albicans, the emerging non-albicans Candida species exhibit reduced antifungal susceptibility. For example, C. krusei is considered as intrinsically resistant to fluconazole. C. glabrata has reduced susceptibility to azoles, while C. parapsilosis shows elevated minimum inhibitory concentration to echinocandins (Clinical and Laboratory Standards Institute, 2008). Among the isolates collected in 2006–2016 for the SENTRY Program, the fluconazole resistance rate of C. albicans was 0.3%, while that of C. glabrata, C. parapsilosis, and C. tropicalis was 8.1, 3.9, and 3.2%, respectively (Pfaller et al., 2019). For echinocandins, the resistance rates were low in C. albicans (0–0.1%) and C. parapsilosis (0–0.1%) but were higher in C. tropicalis (0.5–0.7%), C. krusei (0–1.7%), and C. glabrata (1.7–3.5%).
Among the four common isolated non-albicans Candida species, C. tropicalis and C. parapsilosis are phylogenetically closely related to C. albicans (Butler et al., 2009). Similar to C. albicans, C. tropicalis, and C. parapsilosis can undergo yeast-filament transition, which is an important virulence factor of C. albicans for invading host epithelial cells and causing tissue damage (Sudbery, 2011). C. albicans and C. tropicalis produce true hyphae and pseudohyphae, while C. parapsilosis produces only pseudohyphae (Lackey et al., 2013). These three species are also capable of forming biofilm composed of both yeast cells and filaments.
Formation of Candida biofilm on abiotic surfaces of indwelling medical devices, such as dentures, endotracheal tubes, intragastric balloons, urinary catheters, and intravascular catheters, can lead to device failure and Candida infections (Costa et al., 2022). A meta-analysis of 31 studies revealed that the rate of biofilm formation among Candida strains isolated from blood culture of hospitalized patients varied from 16 to 100% with a pooled rate of 80.0% (Atiencia-Carrera et al., 2022). The pooled attributable mortality of Candida-related bloodstream infection was 37.9%, while the mortality rate with biofilm-forming Candida strains was 70.0%. Moreover, Candida biofilm has shown significantly decreased susceptibility to some commonly used antifungal agents, such as fluconazole and amphotericin B, when compared to the planktonic cells (Silva et al., 2017). Biofilm protects Candida cells from antifungal agents, leading to persistent infection that is difficult to eliminate without removal of the infected implant (Lohse et al., 2018).
In view of the emergence of antifungal resistance, the use of probiotic Lactobacillus species has been suggested as an alternative therapeutic option against Candida infection (Matsubara et al., 2016a). Several clinical trials reported that the administration of Lactobacillus improved the outcome of vaginal candidiasis and reduced colonization of Candida in oral cavity of elderly people and in gastrointestinal tract of preterm neonates (Roy et al., 2014; Kovachev and Vatcheva-Dobrevska, 2015; Kraft-Bodi et al., 2015). In vitro studies also demonstrated that lactobacilli possess anticandidal ability, inhibitory effect on adhesion to host cells, and inhibitory effect on biofilm formation and filamentation of C. albicans (Köhler et al., 2012; Coman et al., 2015; Vilela et al., 2015; Matsubara et al., 2016b; Song and Lee, 2017). Interestingly, as demonstrated with cell-free supernatants and in agar overlay assays, direct interaction of lactobacilli with C. albicans cells is unnecessary for the inhibitory effect, suggesting that the effect is exhibited by the metabolites produced by lactobacilli (Matsubara et al., 2016b; Liao et al., 2019).
Although the effect of lactobacilli on C. albicans has been extensively studied, there is a relative lack of studies on non-albicans Candida species. The aim of this study was to investigate the inhibitory effect of L. rhamnosus, L. plantarum, and L. acidophilus on biofilm formation of C. albicans and closely related non-albicans Candida species, namely C. tropicalis and C. parapsilosis. In addition, we evaluated the change in filamentation and the expression of filamentation- and biofilm-related genes of C. albicans and C. tropicalis when incubated with cell-free supernatants of L. rhamnosus and L. plantarum.
2. Materials and methods
2.1. Strains and culture conditions
A laboratory reference strain C. albicans SC5314 and six Candida clinical strains (C. albicans A14 and A69, C. tropicalis T18S and T38R, and C. parapsilosis P149 and P152) were used in the study. The six clinical strains were initially isolated from blood culture samples, retrieved from our archival laboratory collection and selected because of their strong biofilm-forming ability. All strains were identified with matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using a Bruker Biotyper (Bruker, United States) and the manufacturer’s database (MBT Compass Library DB-7311) with a score of ≥1.7 (Vlek et al., 2014). Prior to experiment, Candida cells were first subcultured from glycerol stock culture onto Sabouraud dextrose agar (SDA) and incubated at 35°C for at least 1 day to assess the purity and viability of the culture.
Type strains L. rhamnosus GG ATCC 53103 (LGG), L. plantarum ATCC 8014 (LP8014), and L. acidophilus ATCC 4356 (LA4356) were used. Stock cultures were prepared by resuspending bacterial cells in de Man, Rogosa and Sharpe (MRS) broth with 10% glycerol and stored at −80°C until use. Before experiments, the bacterial cells were first streaked from stock culture onto MRS agar and incubated at 37°C under microaerophilic conditions (5% O2, provided by CampyGen, Thermo Fisher Scientific, United States) for 2 days.
2.2. Biofilm inhibition
Candida strains were grown in yeast extract peptone dextrose (YPD) broth at 37°C with shaking (200 r.p.m.) for 18 h. The cells were harvested by centrifugation, washed with phosphate-buffered saline (PBS), and resuspended to 106–107 colony-forming unit (CFU)/mL in RPMI 1640 medium (with L-glutamine, buffered with MOPS; Thermo Fisher Scientific, United States). For Lactobacillus, Lactobacillus colonies were collected from MRS plates and inoculated into MRS broth. After incubated microaerophilically at 37°C with shaking (200 r.p.m.) for 18 h, the liquid cultures were centrifuged to separate spent media and bacterial cells. Cell-free supernatant (CFS) was prepared by filtering the spent media through a membrane filter with a 0.22 μm pore size (Merck Millipore, United States). MColorpHast pH indicator strip (Millipore, United States) was used to measure the pH of CFS. The harvested Lactobacillus cells were washed twice with PBS and resuspended in MRS broth to 1 × 107 CFU/ml.
The incubation of biofilm was divided into two parts, adhesion phase and formation phase (Kuhn et al., 2002). Briefly, in adhesion phase, 100 μl of Candida inoculum was seeded into the wells of flat bottom 96-well plates. Plates were prepared in duplicate and then incubated statically at 37°C for 90 min. After incubation, spent medium was aspirated from the well. The wells were washed with PBS to remove non-adherent cells. In the formation phase, 100 μl of RPMI medium and 50 μl of Lactobacillus CFS, living cell suspension, or MRS broth as control were added per wells. There were five replicate wells for each condition per plate. The plates were incubated statically at 37°C for an additional 24 h. After incubation, a MColorpHast pH strip was used to check the pH of spent medium. The biofilms were quantified by two methods, XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] reduction assay and crystal violet staining.
For XTT reduction assay, 150 μl of XTT (0.5 mg/ml in PBS, Sigma-Aldrich, United States) with 1 μM of menadione (Sigma-Aldrich, United States) was added per well (Pierce et al., 2008). The plate was incubated in dark at 37°C for 2.5 h. The resulting-colored supernatant (120 μl) was transferred to a round bottom 96-well plate and its absorbance was measured at 492 nm using a LED microplate reader (Ledetect 96, Labexim Products, Austria). For crystal violet staining, the biofilm in the well was first fixed with 150 μl of 99% methanol, then stained with 150 μl of 0.0625% w/v crystal violet (CV) solution (Peeters et al., 2008). To solubilize the stain, 225 μl of 33% acetic acid was added per well. The solution (150 μl) was transferred to a round bottom plate and its absorbance was measured at 570 nm (Silva et al., 2009).
To examine whether a lower pH contributed to the inhibitory effect of CFS, the assay was repeated using MRS broth acidified to pH 4.0 with lactic acid, and LGG CFS neutralized to pH 7.0 with sodium hydroxide.
2.3. Filamentation inhibition
The C. albicans and C. tropicalis strains were tested against the CFSs of lactobacilli LGG or LP8014, which were prepared from 18 h-old culture as previously described and stored at −20°C until use. MRS broth was used as control. The assay was performed following the method described by Wang et al. (2017) with some modifications. In a 1.5 ml Eppendorf tube, 144 μl of Candida suspension in RPMI medium at a concentration of 1 × 107 CFU/ml was mixed with 300 μl of Lactobacillus CFS or MRS broth and then topped up to a total volume of 900 μl with RPMI. Additional 10% v/v of fetal bovine serum were added for C. tropicalis strains to induce filamentation. After incubating statically at 37°C for 4 h, the mixture was vortexed for 20 s and 20 μl of it was loaded into a Fuchs-Rosenthal counting chamber (Marienfeld, Germany). Microphotographs of ten squares with an area of 0.0625 mm2 each in the chamber were captured using an AmScope MD500 digital eyepiece camera (AmScope, United States) under a light microscope with a 10× objective lens. The number of filaments within the ten squares was counted from the photographic images.
2.4. Gene expression in biofilm
To determine the effects of Lactobacillus CFS on the transcription of biofilm-related genes in C. albicans and C. tropicalis, the gene expression levels of ACT1, ALS1, ALS3, BCR1, EFG1, TEC1, RIP1, and UME6 of C. albicans and the corresponding orthologs in C. tropicalis were evaluated using quantitative real-time polymerase chain reaction (qPCR). Except primers for RIP1, which were adopted from previous literature, primer for other genes was designed using the NCBI/Primer-BLAST tool1 based on the gene sequences available in GenBank (Nailis et al., 2006). The specificity of amplification was evaluated in silico by performing Basic Local Alignment Search Tool (BLAST) analysis against the NCBI GenBank nucleotide collection (nr) and experimentally by agarose gel electrophoresis of the PCR products. The sequences of primers are listed in Tables 1, 2.
To extract RNA, biofilms of C. albicans and C. tropicalis were formed in a flat bottom, cell culture-treated 48-well plate (Thermo Fisher Scientific, United States) using the two-phase procedures described for biofilm inhibition, with the volumes of inoculum and CFS (or MRS broth) increased to 200 and 100 μl, respectively. After incubated statically at 37°C for 24 h, RNA was extracted from biofilms using the TRIzol™ Plus RNA Purification Kit (Thermo Fisher Scientific, United States) with homogenization done on a Fastprep-24 5G instrument (MP Biomedicals, United States) with lysing matrix Y 2 ml tubes at a speed of 6.0 m/s for 30 s twice, with a cooling interval on ice for 3 min. The RNA was eluted with 30 μl of RNase-free water preheated to 60°C. RNA samples were DNase treated using the TURBO DNA-free Kit (Thermo Fisher Scientific, United States) according to the manufacturer’s instruction. Complementary DNA (cDNA) was reverse transcribed from 500 ng of RNA by the High-Capacity cDNA Reverse Transcription Kit with RNase inhibitor (Thermo Fisher Scientific, United States). cDNA samples were stored at −80°C until use.
qPCR was performed in 10 μl reaction volume using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, United States) on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, United States). Within a single qPCR run (96-well plate), amplification of the eight targeted genes was performed using cDNA samples prepared from the biofilms of a same Candida strain grown under three experimental conditions [MRS broth (control), LGG CFS, or LP8014 CFS] in triplicate wells. No-template controls were included for each gene and each qPCR run. The reactions started from a primary denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Specificity of amplification was confirmed by the presence of a single peak in melting curve analysis. ACT1 and RIP1 genes were used as reference genes (Nailis et al., 2006; Silva et al., 2011; Lackey et al., 2013). Expression levels of the genes of interest were normalized to the reference genes using the following formula: , where E represents the amplification efficiency and ΔCt is the difference in cycle threshold (Ct) values of the treatment and control group (Hellemans et al., 2007).
2.5. Statistical analysis
Experiments were repeated in triplicate at different occasions. Results of experiment groups in the biofilm inhibition assay were expressed as a percentage relative to the control (MRS) and compared to the control using Student’s t-test. Results of filamentation assay were compared to control using one-way analysis of variance (ANOVA) followed by Dunnett’s test. Expression levels of genes in biofilm were expressed as NRQ as described. Student’s t-test was used to compare the log2-transformed NRQ values of experimental group to control group of the same Candida strain. A value of p < 0.05 was considered significant in all tests. Statistical analyses were performed using Prism (Version 9, GraphPad Software, United States) and Microsoft Excel (Office 365, Microsoft, United States).
3. Results
3.1. Biofilm inhibition
The LGG CFS significantly reduced the biomass of the biofilms produced by five Candida strains and the metabolic activity of three strains (Figure 1B). The inhibition was most pronounced (>50% in biomass, p < 0.01) in C. albicans, followed by C. tropicalis. The LP8014 CFS also shown inhibitory effects to some extent and significantly reduced the biomass and metabolic activity of biofilm formed by C. albicans SC5314 (Figure 1D, P < 0.05). However, living LGG and LP8014 cell suspensions had little to no effect (Figures 1A,C). On the other hand, LA4356’s cells significantly reduced both biomass and metabolic activity of the two C. parapsilosis strains while its CFS also had similar effect (Figures 1E,F).
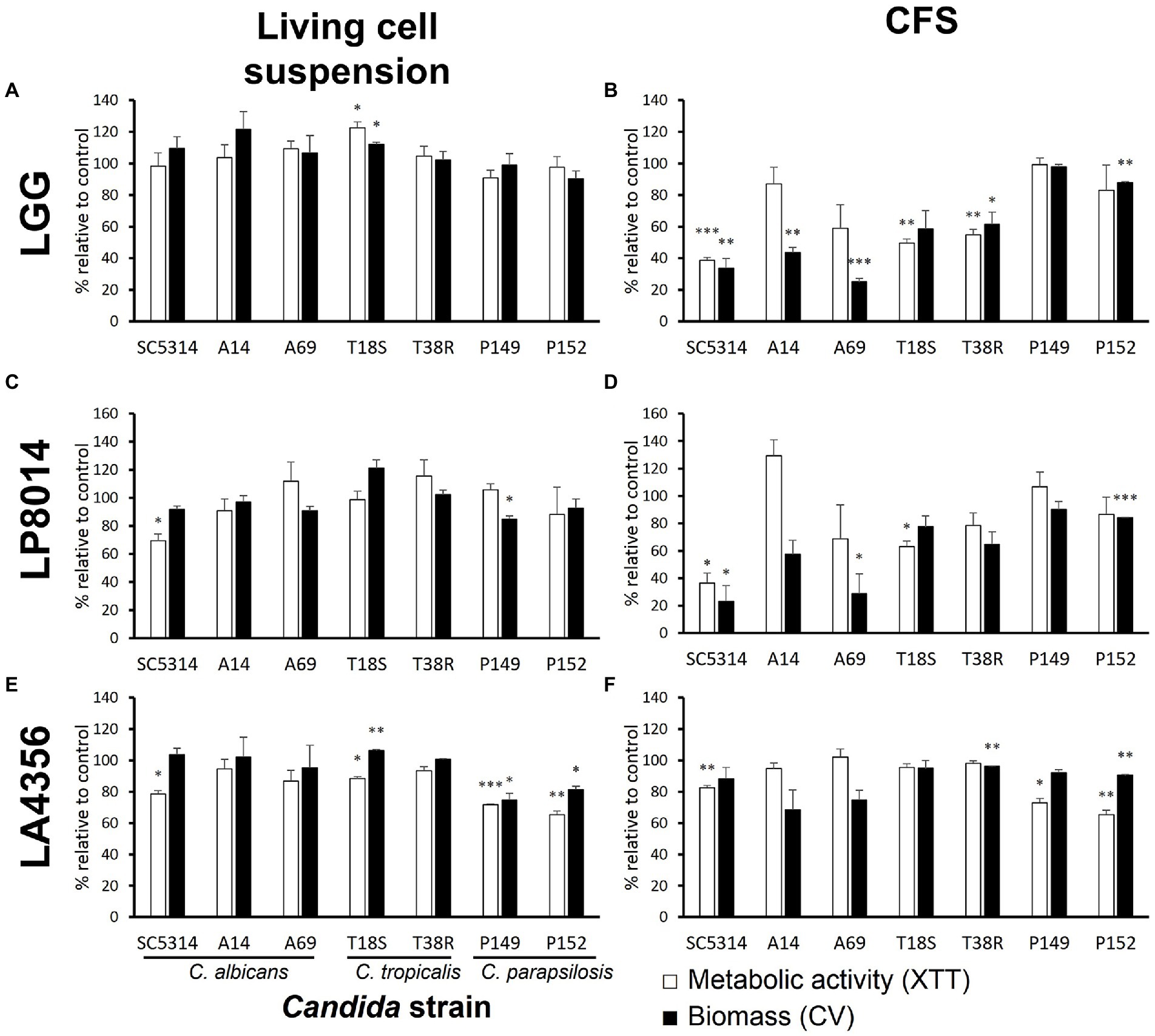
Figure 1. Effects of living cell suspension and CFS of three Lactobacillus strains on biofilm formation of seven Candida strains. (A,B) L. rhamnosus GG, (C,D) L. plantarum ATCC 8014, (E,F) L. acidophilus ATCC 4356. Data are shown as percentage relative to paired control (MRS broth) and the mean ± SEM of triplicate experiments. Comparison with control was performed with Student’s t-test. *p < 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Both LGG CFS and LP8014 CFS had a median pH of 4.0, but LA4356 CFS was less acidic (median pH 5). When used in inhibitory assay, the CFSs lowered the median pH of the spent medium after 24-h incubation from 7.5 (control, MRS broth) to 6.0 (LGG), 6.5 (LP8014), and 7.0 (LA4356), respectively. The medium pH of spent medium was 7.0 in the living cell groups of the three Lactobacillus strains. However, when incubated with RPMI medium without Candida inoculum, all three Lactobacillus strains were able to reduce the medium pH to 4.5 after 24 h.
To examine whether a lower pH contributed to the inhibitory effect of CFS, the assay was repeated with MRS broth acidified to pH 4.0, and LGG CFS neutralized to pH 7.0. As shown in Figure 2, significant inhibition by acidified MRS was only found in biomass of C. albicans A14 (35.3% of reduction) and metabolic activity of T18S (12.9%) when compared to MRS control. Neutralized LGG CFS only shown significant reduced inhibitory effect on metabolic activity of C. albicans A69 and on biomass of A14 and A69 when compared to untreated LGG CFS.
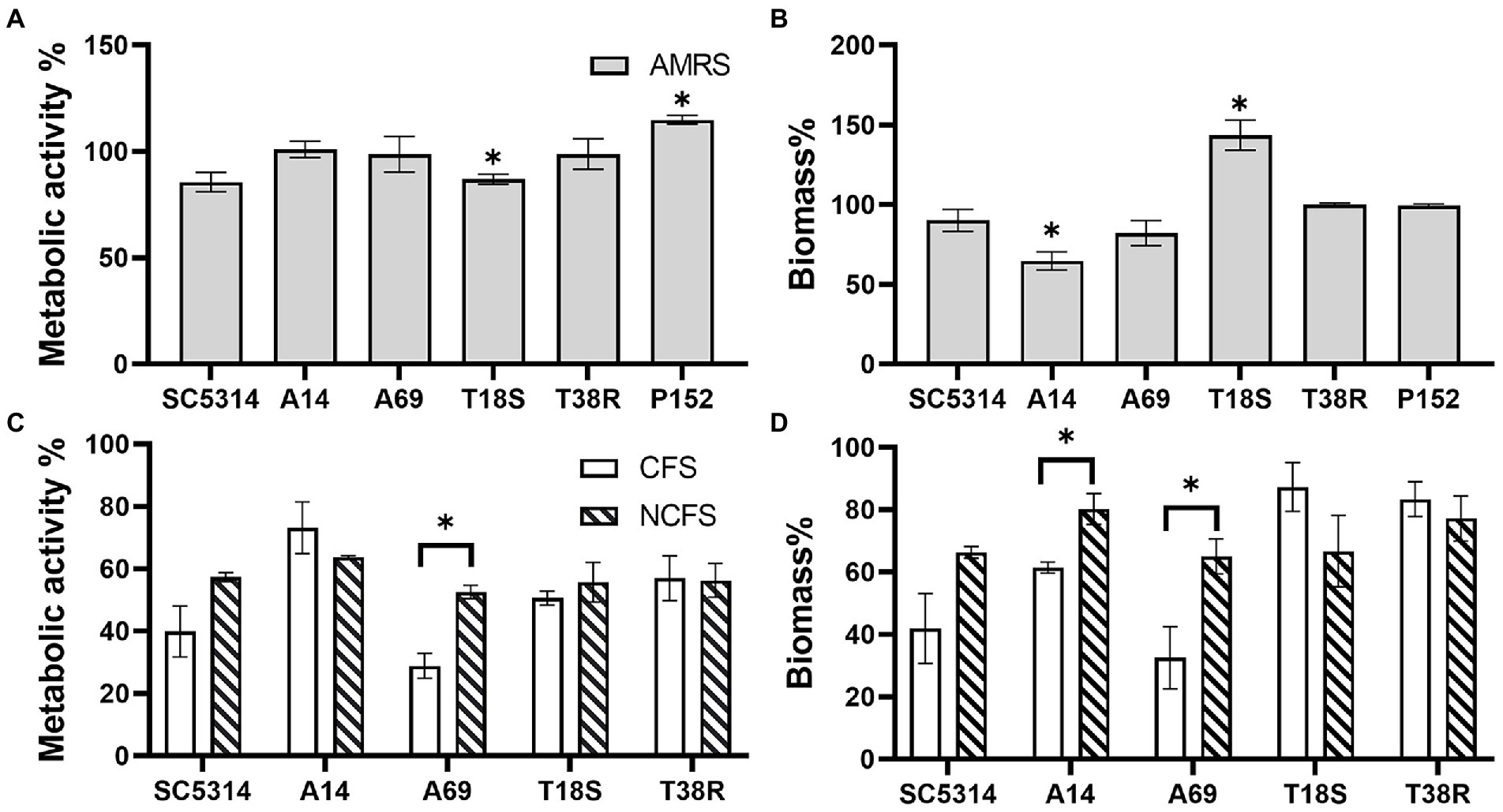
Figure 2. Effects of acidified MRS broth (AMRS) and neutralized CFS (NCFS) on Candida biofilm. Data are shown as percentage relative to a paired control (untreated MRS) and as mean ± SEM of triplicate experiments. (A,B) Effects of AMRS on the metabolic activity and biomass of biofilms produced by three C. albicans (SC5314, A14, and A69), two C. tropicalis (T18S and T38R), and one C. parapsilosis (P152) strains. Asterisks indicate data that are significantly different from control (MRS) at p < 0.05 as determined by one-sample t-test. (C,D) Effects of untreated CFS (pH 4.0) and NCFS (pH 7.0) of LGG on the C. albicans and C. tropicalis strains. Comparison between CFS, NCFS, and control (MRS) was done with one-way ANOVA with Tukey’s test. Asterisks indicate significant difference between CFS and NCFS results at p < 0.05.
3.2. Filamentation inhibition
As the CFSs of LGG and LP8014 showed pronounced inhibitory effects on biofilms of C. albicans and C. tropicalis, their effects on the filamentation of C. albicans and C. tropicalis were further investigated. In the filamentation assay, a large number of Candida cells developed into filaments after incubating in RPMI medium mixed with MRS broth (control). However, many Candida cells remained in yeast form in the presence of Lactobacillus CFSs. Figure 3 shows the examples of cell morphology of each tested Candida strains under different conditions. Filamentations of C. albicans SC5314 and C. tropicalis T18S and T38R (Figures 4A,D,E) were statistically significantly inhibited by both LGG CFS and LP8014 CFS (p < 0.05). The number of filaments of C. albicans A14 was significantly reduced by LGG CFS (p < 0.05) but not LP8014 CFS when compared to control (Figure 4B). The filamentation of C. albicans A69 when incubated with CFSs was inhibited but not significantly different from the control (Figure 4C).
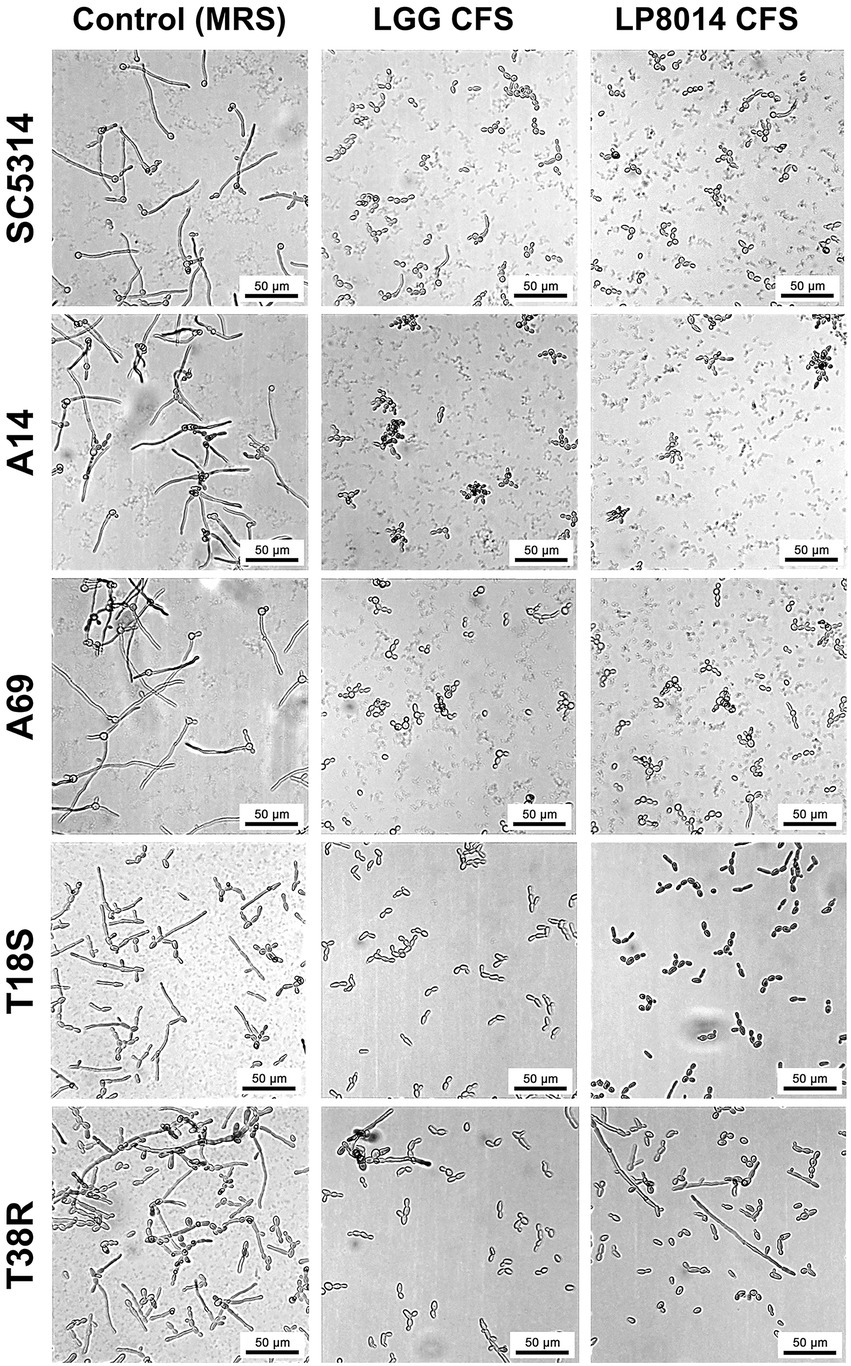
Figure 3. Light micrographs of C. albicans and C. tropicalis cells in the filamentation assay. In each row, images of one Candida strain under three conditions: Control (left), LGG CFS (middle), and LP8014 CFS (right) are shown. Scale-bar: 50 μm.
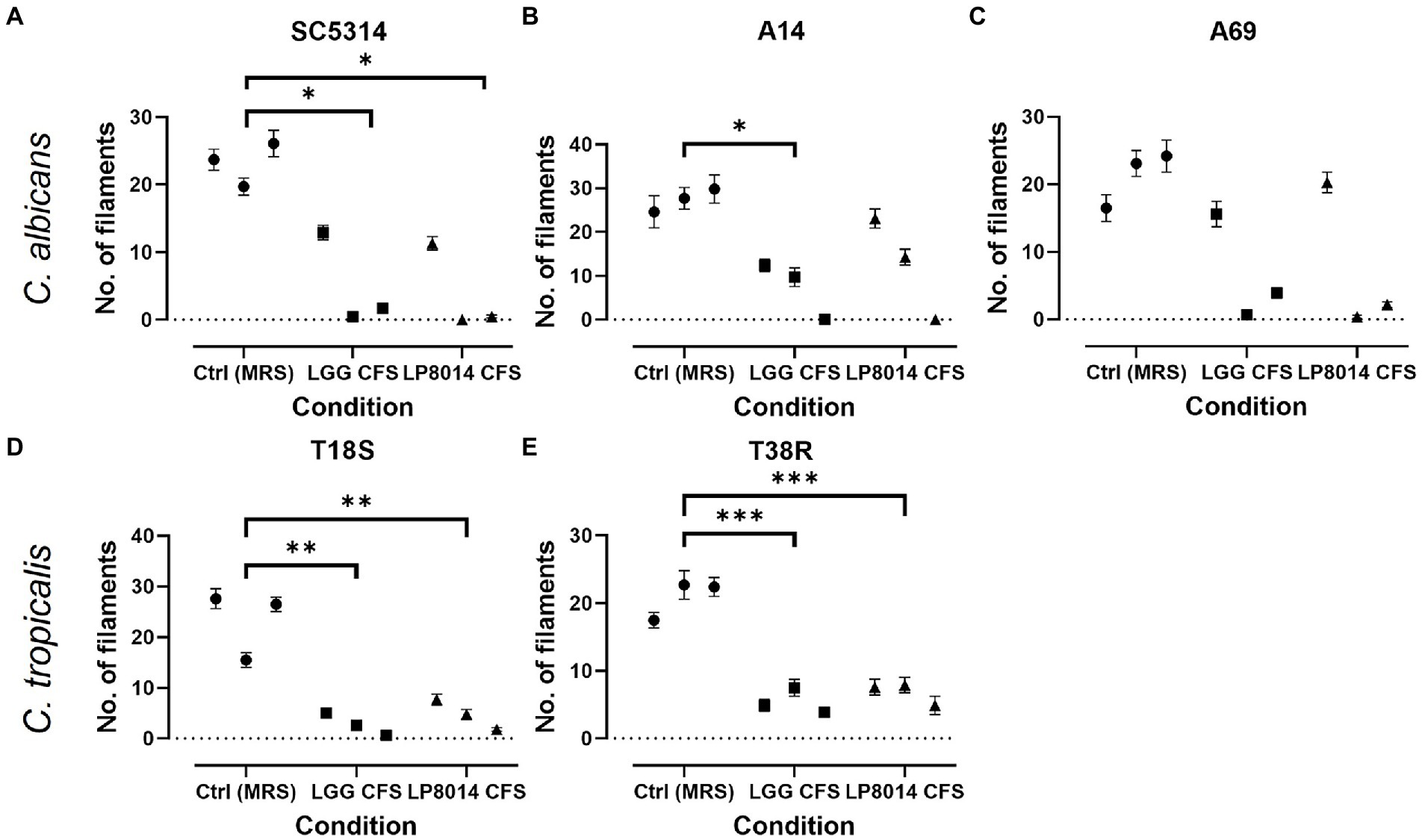
Figure 4. Quantification of filament in the filamentation inhibition assays. (A) C. albicans SC5314, (B) C. albicans A14, (C) C. albicans A69, (D) C. tropicalis T18S, and (E) C. tropicalis T38R. Each data point represents the mean ± SEM of the number of filaments in one experiment. Comparisons with the control group were performed with results of triplicate experiments and Dunnett’s test. *p < 0.05; **p ≤ 0.01; ***p ≤ 0.001.
3.3. Gene expression in biofilm
Expression of ALS1, ALS3, BCR1, EFG1, TEC1, and UME6 genes in biofilms of C. albicans and C. tropicalis strains incubated with LGG or LP8014 CFSs was quantified by qPCR. Although being significant only in some of the strains, there were consistent downregulations in ALS1, EFG1, and TEC1 genes caused by LGG CFS, and in ALS3 and EFG1 genes by LP8014 CFS in the biofilm of the C. albicans strains SC5314, A14, and A69 (Figure 5). On the other hand, ALS3 and UME6 genes were downregulated and TEC1 gene was upregulated by both CFSs in C. tropicalis T18S and T38R. Interestingly, the expression levels of ALS1 in T38R were significantly increased to 1.648- and 1.770-fold by both CFSs (Table 3). The expression level of UME6 gene was downregulated by LP8014 CFS in SC5314, T18S, and T38R and by LGG CFS in T18S, but the differences were not statistically significant.
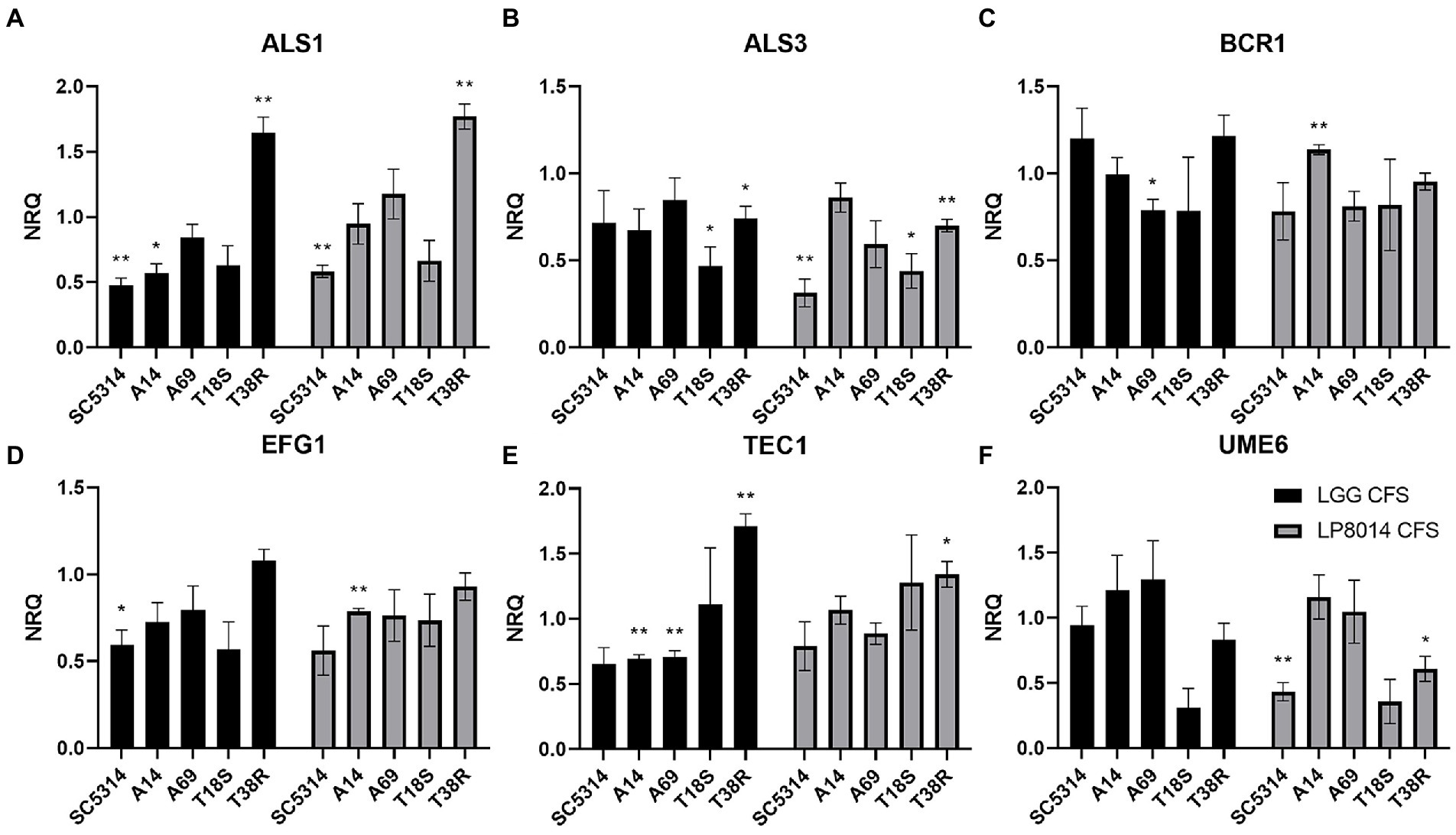
Figure 5. Expression levels filamentation and biofilm-related genes in biofilms of C. albicans and C. tropicalis incubated with LGG or LP8014 CFSs. (A) ALS1, (B) ALS3, (C) BCR1, (D) EFG1, (E) TEC1, and (F) UME6. Expression levels were expressed as the normalized relative quantity (NRQ). NRQ of the control group (MRS) is constantly 1.0 and not shown on the figures. Values represent the mean ± SEM of triplicate experiments. Comparison with control group was performed with the binary logarithm of NRQ by Student’s t-test. *p < 0.05; **p ≤ 0.01.
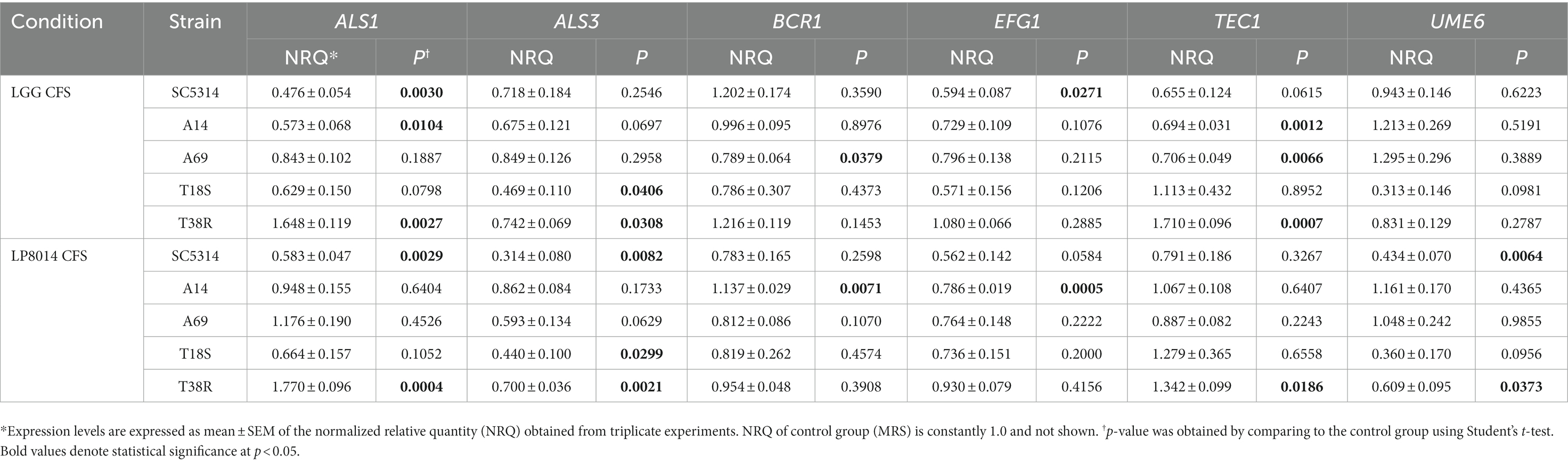
Table 3. Expression levels filamentation and biofilm-related genes in biofilms of Candida albicans and Candida tropicalis incubated with LGG or LP8014 CFSs.
4. Discussion
Lactobacillus species have been reported to exhibit an inhibitory effect on biofilm formation and filamentation of C. albicans; however, few studies have described their effect on the non-albicans Candida species. In the present study, we studied the anti-biofilm effect of cell suspensions and CFSs of three Lactobacillus strains on C. albicans, C. tropicalis, and C. parapsilosis. All three of the Lactobacillus strains are laboratory reference strains and were shown to have inhibitory effect on C. albicans by previous research (Kheradmand et al., 2014; Vilela et al., 2015; Song and Lee, 2017). LGG CFS shown the strongest anti-biofilm activity against C. albicans and C. tropicalis strains, followed by LP8014 CFS. The C. parapsilosis were more susceptible to LA4356 than LGG and LP8014. Previous studies have reported the anti-biofilm activity of Lactobacillus cell suspensions on C. albicans, however, we detected only a modest effect of cell suspension on Candida biofilm in our study (Vilela et al., 2015; Matsubara et al., 2016b; Ribeiro et al., 2017). It is noteworthy that different methods of preparing the cell suspensions were used. In our study, Lactobacillus cells were harvested and resuspended in fresh media, while cell suspensions used in other studies were directly diluted from a pre-incubated liquid culture to the desired concentration. Vilela et al. (2015) have tested the anti-biofilm activity of L. acidophilus culture at different growth phases (4, 6, 18, and 24 h) on C. albicans biofilms and found that the reduction of C. albicans viable cells in biofilm compared to control was only statistically significant for the 24-h L. acidophilus culture, suggesting that inhibitory activity of Lactobacillus is possibly associated with the growth stage. In the present study, C. parapsilosis was found to be less susceptible to CFSs of LGG and LP8014. Similar Candida species-specific manner of the Lactobacillus inhibitory activity was reported in previous research. Tan et al. (2018) observed that biofilm formation of C. parapsilosis was less suppressed by the supernatant of L. gasseri and L. rhamnosus than C. tropicalis and C. krusei. Parolin et al. (2015) investigated the antifungal activities of CFS of Lactobacillus strains isolated from vaginal swabs on C. albicans and non-albicans Candida species but found that no strains were effective against C. krusei and C. parapsilosis, even when being effective on other Candida species.
Our study showed an inhibitory effect of L. rhamnosus (LGG) and L. plantarum (LP8014) CFSs on Candida biofilm. This finding corroborates the results of numerous studies using other Lactobacillus species and strains (Vilela et al., 2015; James et al., 2016; Matsubara et al., 2016b; Ribeiro et al., 2017; Matsuda et al., 2018; Tan et al., 2018). The indirect interaction between Lactobacillus cells and Candida biofilm suggests that soluble exometabolites secreted into the culture medium by Lactobacillus are possibly responsible for the inhibitory effect. Production of lactic acid and the resulted low pH (4.5) environment has been accounted for candicidal activity of L. rhamnosus and Lactobacillus reuteri (Köhler et al., 2012). However, in our biofilm assay, the lowest pH of medium measured was 6.0 and caused by the addition of LGG CFS. Moreover, acidified MRS broth showed little inhibitory effect on biofilm, and the inhibitory effect was not diminished in neutralized LGG CFS when compared with MRS control (p < 0.05, except for biomass of C. tropicalis T18S and T38R) and only reduced when compared with untreated LGG CFS in C. albicans A14 and A69. These results suggested that there are other factors, rather than pH, contributed to the inhibitory effect.
Apart from lactic acid, Lactobacillus have been described to produce other exometabolites with antimicrobial and anti-biofilm effects, such as small chain fatty acids, hydrogen peroxide, antimicrobial peptides, and biosurfactants (Gudiña et al., 2010; Nguyen et al., 2011; Rushdy and Gomaa, 2013; Ceresa et al., 2015). Sodium butyrate, a small chain fatty acid produced by Lactobacillus species, is a known histone deacetylase inhibitor (Garnaud et al., 2016). Nguyen et al. (2011) demonstrated that sodium butyrate can significantly inhibit biofilm formation of C. albicans and C. parapsilosis, by 80 and 65%, respectively, at a concentration of 10 mM. Song and Lee investigated the antifungal activity of spent culture medium of L. rhamnosus and L. casei and found that the growth of C. albicans in both yeast- and hyphal-predominant conditions were significantly inhibited with 50% v/v spent culture medium (Song and Lee, 2017). However, proteinase K-treated spent culture medium of both species lost the inhibitory effect, suggesting that the effect was due to antifungal peptides. A biosurfactant produced by a Lactobacillus brevis strain CV8LAC was shown to inhibit C. albicans biofilm formation by co-incubation and precoating the substrate while demonstrated no inhibition on the fungal growth in planktonic and sessile form (Ceresa et al., 2015). Similarly, liposomes and hyalurosomes containing the biosurfactant isolated from a vaginal strain Lactobacillus crispatus BC1 were shown to inhibit biofilm development in C. albicans and non-albicans Candida species (Abruzzo et al., 2021). It is possible that these metabolites and similar compounds also contribute to the inhibitory effect of CFS demonstrated in our study, but further investigation is needed to identify the exact active compounds and their mechanisms.
Filaments are key structural component of mature biofilm of C. albicans (Kuhn et al., 2002). Mutants of hyphal-related genes showed defects in forming hyphae and biofilm formation as well (Ramage et al., 2002; Nobile and Mitchell, 2005). Therefore, we investigated the effect of CFSs of LGG and LP8014 on the filamentation of the C. albicans and C. tropicalis strains. The filamentation of C. albicans and C. tropicalis cells was greatly reduced when co-incubated with the CFSs. These results are consistent with other studies that demonstrated inhibitory effects of Lactobacillus cell suspensions and CFSs on C. albicans in filamentation assay of similar designs (Vilela et al., 2015; Ribeiro et al., 2017; Matsuda et al., 2018; Rossoni et al., 2018b). Matsubara et al. (2016b) observed a significant reduction of hyphal elements in C. albicans biofilm treated with L. rhamnosus supernatant by confocal laser scanning microscopy. In a monolayer model of oral epithelial cells, preincubating the epithelial cells with LGG suspension for 12 h before C. albicans infection was shown to significantly reduce hyphal length and invasion of C. albicans (Mailänder-Sánchez et al., 2017). In contrast, Lactobacillus iners, the abundance of which in the vaginal microbiome was associated with vaginal dysbiosis and bacterial vaginosis, was demonstrated to promote filamentation and biofilm formation, and upregulate the expression of hyphal-specific genes HWP1 and ECE1 in C. albicans strains with weak to moderate biofilm-forming ability (Sabbatini et al., 2021; Zheng et al., 2021).
Environmental pH is known to induce morphological differentiation in C. albicans. Growth in yeast from is favored in acidic conditions, whereas neutral and alkaline conditions prompt hyphal growth (Davis, 2003). We found that the addition of LGG and LP8014 CFSs reduced the pH of the medium from 7.5 of the control group (MRS broth) to 6 and 7. As Nadeem et al. (2013) have observed that germ tube formation in C. albicans at pH 6.4 was moderately lower than at pH 7.4 and further reduced at pH 5.4, we could not exclude the effect of the slightly decreased pH in the inhibition of filamentation observed in our study, but other metabolites of Lactobacillus are suggested to contribute. For example, in a recent study, 1-acetyl-β-carboline (1-ABC), a small molecule isolated from the culture supernatant of Lactobacillus species, was reported to block C. albicans filamentation in a concentration-dependent manner (MacAlpine et al., 2021). It was proposed that 1-ABC inhibits C. albicans Yak1 kinase, which is a member of the dual-specificity tyrosine-regulated kinase (DYRK) family and necessary for upregulation of hypha-induced genes in C. albicans (Goyard et al., 2008). Purified 1-ABC was also shown to inhibit filamentation of C. dubliniensis and C. tropicalis, in which Yak1 is conserved (MacAlpine et al., 2021). The authors also demonstrated that acidic pH alone was insufficient to achieve the same degree of filamentation inhibition caused by Lactobacillus culture supernatant.
To elucidate the mechanisms of the anti-biofilm activity of Lactobacillus CFS, we measured the expression of six biofilm-related genes, namely ALS1, ALS3, BCR1, EFG1, TEC1, and UME6, in the biofilm produced by C. albicans or C. tropicalis co-incubated with LGG and LP8014 CFSs. ALS1 and ALS3, belonging to the agglutinin-like sequence (ALS) gene family, encode cell wall glycoproteins Als1 and Als3, respectively (Hoyer, 2001). Expression of ALS1 is detectable in both yeast or hyphae form, while ALS3 transcribed exclusively in germ tubes and hyphae (Green et al., 2005). In C. tropicalis, ALS1-, ALS2-, and ALS3- like genes were more upregulated in sessile cells, suggesting their roles in biofilm formation (Galán-Ladero et al., 2019). BCR1, encoding a transcription factor Bcr1, is upregulated in C. albicans hyphae but not required for normal hyphal development (Nobile and Mitchell, 2005). Bcr1 is required for the full expression of several cell wall proteins, including Als1 and the hyphal-specific Als3, Hwp1, and Hyr1. EFG1 encodes the transcription factor Efg1, which is essential for yeast-hyphal transition and biofilm formation of C. albicans (Stoldt et al., 1997; Ramage et al., 2002). Efg1 is activated by upstream cAMP pathway and regulates expression of hyphal-specific genes and downstream transcription factors such as Tec1 and Eed1 (Sudbery, 2011). Mancera et al. (2015) have reported that EFG1 ortholog in C. tropicalis has conserved function on filamentation and biofilm formation. TEC1 encodes transcription factor Tec1, deletion of which results in defect of hyphal formation and biofilm formation, and suppression of expression of secreted aspartyl protease (SAP) family proteins Sap4-6 in C. albicans (Schweizer et al., 2000; Nobile and Mitchell, 2005). In C. tropicalis, deletion mutants of BCR1, BRG1, TEC1, EFG1, or NDT80 homologs produced significantly less biofilm biomass, suggesting the conserved roles of these genes in biofilm formation (Tseng et al., 2020). UME6, a common downstream target of regulators Efg1, Chp1, and Ras1, strictly or partially controls the expression of hyphae-specific genes in C. albicans (Zeidler et al., 2009). Constitutive high-level expression of UME6 orthologs in C. tropicalis and C. parapsilosis also enhances filamentation and biofilm formation (Lackey et al., 2013).
In our study, we observed a downregulation in EFG1 and TEC1 genes by CFS in the three C. albicans strains, which is in agreement with the findings of previous studies on C. albicans biofilm (James et al., 2016; Matsuda et al., 2018; Rossoni et al., 2018a). These studies, however, also reported significant downregulation of ALS3 and BCR1 genes in Lactobacillus-inhibited biofilm, which was not the case in our results. Interestingly, in our study, the expressions of ALS1, BCR1, and TEC1 genes were upregulated in C. tropicalis T38R by LGG CFS although LGG CFS was shown to suppress biofilm formation and filamentation of T38R, suggesting a strain-dependent gene response. The upregulation of ALS1 in T38R could possibly be a compensatory upregulation due to the suppression of ALS3 (Liu and Filler, 2011). Such compensatory upregulations had been demonstrated by the upregulation of ALS2 in als4/als4 mutant and vice versa, and the upregulation of SAP5 in sap1-3 mutant and SAP2 in sap 4–6 mutant of C. albicans (Zhao et al., 2005; Naglik et al., 2008). The expression of UME6 in C. albicans SC5314, C. tropicalis T18S and T38R was downregulated by 0.434-, 0.360- (p = 0.0956), and 0.609-fold, respectively, suggesting that the mechanism of action of LP8014 CFS involves the repression of UME6.
There are a number of limitations to the current study need to be taken into account. First, we used two semiquantitative methods, CV staining and XTT reduction assay, to measure Candida biofilm formation. However, neither of the assays provide information about effect of Lactobacillus on the three-dimensional architecture of the Candida biofilm. Second, the dosage of inhibitory agents was not optimized. Ideally, the experiment should be repeated with a range of concentrations to examine the presence of dose-dependent effect and determine the most effective concentration. Third, the study lacked a characterization of the metabolites in LGG and LP8014 CFSs, which showed strong inhibitory effect. Analyses such as thermal stability, protease stability, activity of hydrogen peroxide, and metabolite profiling by liquid chromatography-mass spectrometry would provide information to identify the active components and elucidate mechanisms of action if performed. Another limitation is that we selected few target genes for the gene expression analysis from the biofilm regulatory network of Candida, which involves multiple signaling pathways and more than 50 genes (Finkel and Mitchell, 2011). Other well-known biofilm-related genes, such as HWP1, which encodes a cell wall adhesin required in biofilm formation, and NRG1, a negative regulator of filamentation, were not included (Braun et al., 2001; Nobile et al., 2006). To unravel the elusive mechanisms of the effect of Lactobacillus CFS, genomic-wide transcriptional profiling by technologies such as RNA-seq is a better approach (Chong et al., 2018).
5. Conclusion
The present study evaluated the inhibitory effect of three lactobacilli strains, LGG, LP8014, and LA4356, on C. albicans, C. tropicalis, and C. parapsilosis. Significant inhibitory effects were demonstrated by the CFSs of LGG and LP8014 on filamentation and biofilm formation of both C. albicans and C. tropicalis. Hypha-related gene expressions in biofilm were also changed in response to the addition of CFSs. However, C. parapsilosis biofilms were not susceptible to the inhibitory effect of these two lactobacilli strains. The pathogen-specific inhibition suggests a common mechanism of action, presumably mediated by exometabolites of the lactobacilli, on C. albicans and C. tropicalis. The active components and the potential clinical application of LGG and LP8014 CFSs remain to be investigated.
Author’s note
The content of this manuscript has previously been presented in part as a poster at the 29th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), 13–16 April 2019, Amsterdam, Netherlands, and published as part of the doctoral thesis of YP (Poon, 2022). A preliminary version of this manuscript has been released as a preprint at BioRxiv (Poon and Hui, 2022).
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YP performed the experiments and wrote the manuscript. MH reviewed and revised the manuscript. All authors read and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Abruzzo, A., Giordani, B., Parolin, C., De Gregorio, P. R., Foschi, C., Cerchiara, T., et al. (2021). Lactobacillus crispatus BC1 biosurfactant delivered by Hyalurosomes: an advanced strategy to counteract Candida biofilm. Antibiotics 10:33. doi: 10.3390/antibiotics10010033
Atiencia-Carrera, M. B., Cabezas-Mera, F. S., Tejera, E., and Machado, A. (2022). Prevalence of biofilms in Candida spp. bloodstream infections: a meta-analysis. PLoS One 17:e0263522. doi: 10.1371/journal.pone.0263522
Bongomin, F., Gago, S., Oladele, R. O., and Denning, D. W. (2017). Global and multi-National Prevalence of fungal diseases—estimate precision. J. Fungi 3:57. doi: 10.3390/jof3040057
Braun, B. R., Kadosh, D., and Johnson, A. D. (2001). NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20, 4753–4761. doi: 10.1093/emboj/20.17.4753
Butler, G., Rasmussen, M. D., Lin, M. F., Santos, M. A. S., Sakthikumar, S., Munro, C. A., et al. (2009). Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662. doi: 10.1038/nature08064
Ceresa, C., Tessarolo, F., Caola, I., Nollo, G., Cavallo, M., Rinaldi, M., et al. (2015). Inhibition of Candida albicans adhesion on medical-grade silicone by a Lactobacillus-derived biosurfactant. J. Appl. Microbiol. 118, 1116–1125. doi: 10.1111/jam.12760
Chong, P. P., Chin, V. K., Wong, W. F., Madhavan, P., Yong, V. C., and Looi, C. Y. (2018). Transcriptomic and genomic approaches for unravelling Candida albicans biofilm formation and drug resistance—an update. Genes 9:540. doi: 10.3390/genes9110540
Clinical and Laboratory Standards Institute (2008). Reference method for broth dilution antifungal susceptibility testing of yeasts-third edition: Approved standard M27-A3. CLSI document M27-A3 and supplement S (Wayne, PA, USA).
Coman, M. M., Verdenelli, M. C., Cecchini, C., Silvi, S., Orpianesi, C., Caspani, M., et al. (2015). In vitro evaluation on HeLa cells of protective mechanisms of probiotic lactobacilli against Candida clinical isolates. J. Appl. Microbiol. 119, 1383–1390. doi: 10.1111/jam.12947
Costa, P. D. S., Prado, A., Bagon, N. P., Negri, M., and Svidzinski, T. I. E. (2022). Mixed fungal biofilms: from Mycobiota to devices, a new challenge on clinical practice. Microorganisms 10:1721. doi: 10.3390/microorganisms10091721
Davis, D. (2003). Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44, 1–7. doi: 10.1007/s00294-003-0415-2
Finkel, J. S., and Mitchell, A. P. (2011). Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9, 109–118. doi: 10.1038/nrmicro2475
Galán-Ladero, M. Á., Blanco-Blanco, M. T., Fernández-Calderón, M. C., Lucio, L., Gutiérrez-Martín, Y., Blanco, M. T., et al. (2019). Candida tropicalis biofilm formation and expression levels of the CTRG ALS-like genes in sessile cells. Yeast 36, 107–115. doi: 10.1002/yea.3370
Garnaud, C., Champleboux, M., Maubon, D., Cornet, M., and Govin, J. (2016). Histone deacetylases and their inhibition in Candida species. Front. Microbiol. 7:1238. doi: 10.3389/fmicb.2016.01238
Goyard, S., Knechtle, P., Chauvel, M., Mallet, A., Prévost, M.-C., Proux, C., et al. (2008). The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albicans. Mol. Biol. Cell 19, 2251–2266. doi: 10.1091/mbc.e07-09-0960
Green, C. B., Zhao, X., Yeater, K. M., and Hoyer, L. L. (2005). Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology 151, 1051–1060. doi: 10.1099/mic.0.27696-0
Gudiña, E. J., Teixeira, J. A., and Rodrigues, L. R. (2010). Isolation and functional characterization of a biosurfactant produced by Lactobacillus paracasei. Colloids Surf. B Biointerfaces 76, 298–304. doi: 10.1016/j.colsurfb.2009.11.008
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. doi: 10.1186/gb-2007-8-2-r19
Hoyer, L. L. (2001). The ALS gene family of Candida albicans. Trends Microbiol. 9, 176–180. doi: 10.1016/S0966-842X(01)01984-9
James, K. M., MacDonald, K. W., Chanyi, R. M., Cadieux, P. A., and Burton, J. P. (2016). Inhibition of Candida albicans biofilm formation and modulation of gene expression by probiotic cells and supernatant. J. Med. Microbiol. 65, 328–336. doi: 10.1099/jmm.0.000226
Kabir, M. A., Hussain, M. A., and Ahmad, Z. (2012). Candida albicans: a model organism for studying fungal pathogens. ISRN Microbiol. 2012, 538615–538694. doi: 10.5402/2012/538694
Kheradmand, E., Rafii, F., Yazdi, M. H., Sepahi, A. A., Shahverdi, A. R., and Oveisi, M. R. (2014). The antimicrobial effects of selenium nanoparticle-enriched probiotics and their fermented broth against Candida albicans. DARU J. Pharm. Sci. 22:48. doi: 10.1186/2008-2231-22-48
Kim, J., and Sudbery, P. (2011). Candida albicans, a major human fungal pathogen. J. Microbiol. 49, 171–177. doi: 10.1007/s12275-011-1064-7
Köhler, G. A., Assefa, S., and Reid, G. (2012). Probiotic interference of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 with the opportunistic fungal pathogen Candida albicans. Infect. Dis. Obstet. Gynecol. 2012:636474. doi: 10.1155/2012/636474
Kovachev, S. M., and Vatcheva-Dobrevska, R. S. (2015). Local probiotic therapy for vaginal Candida albicans infections. Probiot. Antimicrob. Proteins 7, 38–44. doi: 10.1007/s12602-014-9176-0
Kraft-Bodi, E., Jørgensen, M. R., Keller, M. K., Kragelund, C., and Twetman, S. (2015). Effect of probiotic bacteria on oral Candida in frail elderly. J. Dent. Res. 94, 181s–186s. doi: 10.1177/0022034515595950
Kuhn, D. M., Chandra, J., Mukherjee, P. K., and Ghannoum, M. A. (2002). Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect. Immun. 70, 878–888. doi: 10.1128/IAI.70.2.878-888.2002
Lackey, E., Vipulanandan, G., Childers, D. S., and Kadosh, D. (2013). Comparative evolution of morphological regulatory functions in Candida species. Eukaryot. Cell 12, 1356–1368. doi: 10.1128/EC.00164-13
Liao, H., Liu, S., Wang, H., Su, H., and Liu, Z. (2019). Enhanced antifungal activity of bovine lactoferrin-producing probiotic Lactobacillus casei in the murine model of vulvovaginal candidiasis. BMC Microbiol. 19:7. doi: 10.1186/s12866-018-1370-x
Liu, Y., and Filler, S. G. (2011). Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 10, 168–173. doi: 10.1128/EC.00279-10
Lohse, M. B., Gulati, M., Johnson, A. D., and Nobile, C. J. (2018). Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 16, 19–31. doi: 10.1038/nrmicro.2017.107
MacAlpine, J., Daniel-Ivad, M., Liu, Z., Yano, J., Revie, N. M., Todd, R. T., et al. (2021). A small molecule produced by lactobacillus species blocks Candida albicans filamentation by inhibiting a DYRK1-family kinase. Nat. Commun. 12:6151. doi: 10.1038/s41467-021-26390-w
Mailänder-Sánchez, D., Braunsdorf, C., Grumaz, C., Müller, C., Lorenz, S., Stevens, P., et al. (2017). Antifungal defense of probiotic lactobacillus rhamnosus GG is mediated by blocking adhesion and nutrient depletion. PLoS One 12, –e0184438. doi: 10.1371/journal.pone.0184438
Mancera, E., Porman, A. M., Cuomo, C. A., Bennett, R. J., and Johnson, A. D. (2015). Finding a missing gene: EFG1 regulates morphogenesis in Candida tropicalis. G3 5, 849–856. doi: 10.1534/g3.115.017566
Matsubara, V. H., Bandara, H. M. H. N., Mayer, M. P. A., and Samaranayake, L. P. (2016a). Probiotics as antifungals in mucosal candidiasis. Clin. Infect. Dis. 62, 1143–1153. doi: 10.1093/cid/ciw038
Matsubara, V. H., Wang, Y., Bandara, H., Mayer, M. P. A., and Samaranayake, L. P. (2016b). Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 100, 6415–6426. doi: 10.1007/s00253-016-7527-3
Matsuda, Y., Cho, O., Sugita, T., Ogishima, D., and Takeda, S. (2018). Culture supernatants of Lactobacillus gasseri and L. crispatus inhibit Candida albicans biofilm formation and adhesion to HeLa cells. Mycopathologia 183, 691–700. doi: 10.1007/s11046-018-0259-4
Nadeem, S. G., Shafiq, A., Hakim, S. T., Anjum, Y., and Kazm, S. U. (2013). Effect of growth media, pH and temperature on yeast to hyphal transition in Candida albicans. Open J. Med. Microbiol. 3, 185–192. doi: 10.4236/ojmm.2013.33028
Naglik, J. R., Moyes, D., Makwana, J., Kanzaria, P., Tsichlaki, E., Weindl, G., et al. (2008). Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology 154, 3266–3280. doi: 10.1099/mic.0.2008/022293-0
Nailis, H., Coenye, T., Van Nieuwerburgh, F., Deforce, D., and Nelis, H. J. (2006). Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 7:25. doi: 10.1186/1471-2199-7-25
Nguyen, L. N., Lopes, L. C. L., Cordero, R. J. B., and Nosanchuk, J. D. (2011). Sodium butyrate inhibits pathogenic yeast growth and enhances the functions of macrophages. J. Antimicrob. Chemother. 66, 2573–2580. doi: 10.1093/jac/dkr358
Nobile, C. J., and Mitchell, A. P. (2005). Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15, 1150–1155. doi: 10.1016/j.cub.2005.05.047
Nobile, C. J., Nett, J. E., Andes, D. R., and Mitchell, A. P. (2006). Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5, 1604–1610. doi: 10.1128/ec.00194-06
Parolin, C., Marangoni, A., Laghi, L., Foschi, C., Ñahui Palomino, R. A., Calonghi, N., et al. (2015). Isolation of vaginal lactobacilli and characterization of anti-Candida activity. PLoS One 10, –e0131220. doi: 10.1371/journal.pone.0131220
Peeters, E., Nelis, H. J., and Coenye, T. (2008). Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72, 157–165. doi: 10.1016/j.mimet.2007.11.010
Pfaller, M. A., and Diekema, D. J. (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163. doi: 10.1128/CMR.00029-06
Pfaller, M. A., Diekema, D. J., Turnidge, J. D., Castanheira, M., and Jones, R. N. (2019). Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997–2016. Open forum. Infect. Dis. 6, S79–S94. doi: 10.1093/ofid/ofy358
Pierce, C. G., Uppuluri, P., Tristan, A. R., Wormley, F. L., Mowat, E., Ramage, G., et al. (2008). A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 3, 1494–1500. doi: 10.1038/nprot.2008.141
Poon, Y. (2022). Inhibitory effect on biofilm formation and filamentation of C. albicans, C. tropicalis, and C. parapsilosis by probiotic lactobacillus strains. Doctoral thesis. Hong Kong SAR, China: The Chinese University of Hong Kong.
Poon, Y., and Hui, M. (2022). Inhibitory effect of lactobacilli supernatants on biofilm and filamentation of C. albicans, C. tropicalis, and C. parapsilosis. bioRxiv. doi: 10.1101/2022.11.26.518025
Ramage, G., VandeWalle, K., López-Ribot, J. L., and Wickes, B. L. (2002). The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 214, 95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x
Ribeiro, F. C., de Barros, P. P., Rossoni, R. D., Junqueira, J. C., and Jorge, A. O. C. (2017). Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 122, 201–211. doi: 10.1111/jam.13324
Rossoni, R. D., de Barros, P. P., de Alvarenga, J. A., Ribeiro, F. D. C., Velloso, M. D. S., Fuchs, B. B., et al. (2018a). Antifungal activity of clinical Lactobacillus strains against Candida albicans biofilms: identification of potential probiotic candidates to prevent oral candidiasis. Biofouling 34, 212–225. doi: 10.1080/08927014.2018.1425402
Rossoni, R. D., dos Santos Velloso, M., Figueiredo, L. M. A., Martins, C. P., Jorge, A. O. C., and Junqueira, J. C. (2018b). Clinical strains of lactobacillus reduce the filamentation of Candida albicans and protect Galleria mellonella against experimental candidiasis. Folia Microbiol. 63, 307–314. doi: 10.1007/s12223-017-0569-9
Roy, A., Chaudhuri, J., Sarkar, D., Ghosh, P., and Chakraborty, S. (2014). Role of enteric supplementation of probiotics on late-onset sepsis by Candida species in preterm low birth weight neonates: a randomized, double blind, placebo-controlled trial. N. Am. J. Med. Sci. 6, 50–57. doi: 10.4103/1947-2714.125870
Rushdy, A. A., and Gomaa, E. Z. (2013). Antimicrobial compounds produced by probiotic Lactobacillus brevis isolated from dairy products. Ann. Microbiol. 63, 81–90. doi: 10.1007/s13213-012-0447-2
Sabbatini, S., Visconti, S., Gentili, M., Lusenti, E., Nunzi, E., Ronchetti, S., et al. (2021). Lactobacillus iners cell-free supernatant enhances biofilm formation and hyphal/Pseudohyphal growth by Candida albicans vaginal isolates. Microorganisms 9:2577. doi: 10.3390/microorganisms9122577
Schweizer, A., Rupp, S., Taylor, B. N., Röllinghoff, M., and Schröppel, K. (2000). The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38, 435–445. doi: 10.1046/j.1365-2958.2000.02132.x
Silva, S., Henriques, M., Martins, A., Oliveira, R., Williams, D., and Azeredo, J. (2009). Biofilms of non-Candida albicans Candida species: quantification, structure and matrix composition. Med. Mycol. 47, 681–689. doi: 10.3109/13693780802549594
Silva, S., Hooper, S. J., Henriques, M., Oliveira, R., Azeredo, J., and Williams, D. W. (2011). The role of secreted aspartyl proteinases in Candida tropicalis invasion and damage of oral mucosa. Clin. Microbiol. Infect. 17, 264–272. doi: 10.1111/j.1469-0691.2010.03248.x
Silva, S., Rodrigues, C. F., Araújo, D., Rodrigues, M. E., and Henriques, M. (2017). Candida species biofilms’ antifungal resistance. J. Fungi 3:8. doi: 10.3390/jof3010008
Song, Y.-G., and Lee, S.-H. (2017). Inhibitory effects of Lactobacillus rhamnosus and Lactobacillus casei on Candida biofilm of denture surface. Arch. Oral Biol. 76, 1–6. doi: 10.1016/j.archoralbio.2016.12.014
Stoldt, V. R., Sonneborn, A., Leuker, C. E., and Ernst, J. F. (1997). Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16, 1982–1991. doi: 10.1093/emboj/16.8.1982
Sudbery, P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9, 737–748. doi: 10.1038/nrmicro2636
Tan, Y., Leonhard, M., Moser, D., Ma, S., and Schneider-Stickler, B. (2018). Inhibitory effect of probiotic lactobacilli supernatants on single and mixed non-albicans Candida species biofilm. Arch. Oral Biol. 85, 40–45. doi: 10.1016/j.archoralbio.2017.10.002
Tseng, Y.-K., Chen, Y.-C., Hou, C.-J., Deng, F.-S., Liang, S.-H., Hoo, S. Y., et al. (2020). Evaluation of biofilm formation in Candida tropicalis using a silicone-based platform with synthetic urine medium. Microorganisms 8:660. doi: 10.3390/microorganisms8050660
Vilela, S. F. G., Barbosa, J. O., Rossoni, R. D., Santos, J. D., Prata, M. C. A., Anbinder, A. L., et al. (2015). Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella. Virulence 6, 29–39. doi: 10.4161/21505594.2014.981486
Vlek, A., Kolecka, A., Khayhan, K., Theelen, B., Groenewald, M., Boel, E., et al. (2014). Interlaboratory comparison of sample preparation methods, database expansions, and cutoff values for identification of yeasts by matrix-assisted laser desorption ionization–time of flight mass spectrometry using a yeast test panel. J. Clin. Microbiol. 52, 3023–3029. doi: 10.1128/JCM.00563-14
Wang, S., Wang, Q., Yang, E., Yan, L., Li, T., and Zhuang, H. (2017). Antimicrobial compounds produced by vaginal Lactobacillus crispatus are able to strongly inhibit Candida albicans growth, hyphal formation and regulate virulence-related gene expressions. Front. Microbiol. 8:564. doi: 10.3389/fmicb.2017.00564
Zeidler, U., Lettner, T., Lassnig, C., Müller, M., Lajko, R., Hintner, H., et al. (2009). UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9, 126–142. doi: 10.1111/j.1567-1364.2008.00459.x
Zhao, X., Oh, S. H., Yeater, K. M., and Hoyer, L. L. (2005). Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 151, 1619–1630. doi: 10.1099/mic.0.27763-0
Keywords: Candida, Lactobacillus, biofilm, filamentation, gene expression, non-albicans Candida species
Citation: Poon Y and Hui M (2023) Inhibitory effect of lactobacilli supernatants on biofilm and filamentation of Candida albicans, Candida tropicalis, and Candida parapsilosis. Front. Microbiol. 14:1105949. doi: 10.3389/fmicb.2023.1105949
Edited by:
Claudia Monari, University of Perugia, ItalyReviewed by:
Terenzio Cosio, University of Rome Tor Vergata, ItalyAntónio Machado, Universidad San Francisco de Quito, Ecuador
Copyright © 2023 Poon and Hui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yeuklan Poon, eWV1a2xhbnBvb25AbGluay5jdWhrLmVkdS5oaw==
†ORCID: Yeuklan Poon https://orcid.org/0000-0001-8706-1890
Mamie Hui https://orcid.org/0000-0001-6598-8054
 Yeuklan Poon
Yeuklan Poon Mamie Hui
Mamie Hui