- 1Shaanxi Key Laboratory of Earth Surface System and Environmental Carrying Capacity, College of Urban and Environmental Sciences, Northwest University, Xi’an, China
- 2Key Laboratory of Agro-Ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
- 3College of Resources and Environment, Hunan Agricultural University, Changsha, China
- 4College of Biodiversity, Conservation Southwest Forestry University, Kunming, China
Water and fertilizer managements are the most common practices to maximize crop yields, and their long-term impact on soil microbial communities has been extensively studied. However, the initial response of microbes to fertilization and soil moisture changes remains unclear. In this study, the immediate effects of nitrogen (N)-fertilizer application and moisture levels on microbial community of paddy soils were investigated through controlled incubation experiments. Amplicon sequencing results revealed that moisture had a stronger influence on the abundance and community composition of total soil bacteria, as well as ammonia oxidizing-archaea (AOA) and -bacteria (AOB). Conversely, fertilizer application noticeably reduced the connectivity and complexity of the total bacteria network, and increasing moisture slightly exacerbated these effects. NH4+-N content emerged as a significant driving force for changes in the structure of the total bacteria and AOB communities, while NO3−-N content played more important role in driving shifts in AOA composition. These findings indicate that the initial responses of microbial communities, including abundance and composition, and network differ under water and fertilizer managements. By providing a snapshot of microbial community structure following short-term N-fertilizer and water treatments, this study contributes to a better understanding of how soil microbes respond to long-term agriculture managements.
1. Introduction
Water and nutrients availability are two critical factors that limit global crop productivity. Chemical fertilizer application has been one of the most important practices of modern agriculture (Soman et al., 2017; Chen et al., 2019; Li G. C. et al., 2021). Fertilization enhances soil fertility by increasing nutrient levels. However, current estimates suggest that the efficiency of fertilizer use is relatively low, with only 40%–50% of the applied nitrogen (N)-fertilizer being utilized by crops (Bijay-Singh and Craswell, 2021). Overfertilization not only depletes resources but also gives rise to various environmental issues, including water pollution, soil degradation, greenhouse gas emissions, and loss of biodiversity (Francioli et al., 2016).
Microorganisms, as an integral component of the soil ecosystem, play a crucial role in maintaining soil quality and functions, and they are highly sensitive to changes in soil conditions (Li et al., 2016; Pan et al., 2016; Chen et al., 2017; Banerjee et al., 2018). Microorganisms drive the biogeochemical cycles of soil carbon and nitrogen, thus regulating the size of the soil carbon pool and the emissions of greenhouse gases like methane and N2O into the atmosphere (Chai et al., 2019). Different agriculture management practices influence soil microbial communities through various mechanisms (Liang et al., 2012). For instance, manure application improves soil texture and organic matter content, thereby stimulating the growth and activity of soil microorganisms (Gomiero et al., 2011; Tang et al., 2019). In addition to benefiting crop yield, fertilization also provides nutrients and substrates for soil microbes (Feng et al., 2015; Yu et al., 2019). Soil moisture directly or indirectly affects microbial activities by regulating oxygen concentration and nutrient availability (Drenovsky et al., 2004; Pan et al., 2016). Furthermore, the presence of free water connecting soil particles can facilitate the mobility of nutrient and microbial cell, thereby shaping the structure of the soil microbial community (Zhou et al., 2002; Yuan et al., 2016).
Both fertilizer application and water content play crucial roles in sustaining crop productivity. However, their impact on soil microbial community structure and associated ecosystem function is not always positive. For instance, the addition of fertilizer can stimulate microbial activity, leading to the decomposition of organic matter and denitrification, which can result in soil carbon loss and the emission of greenhouse gases (Canarini et al., 2016; Bastida et al., 2017; Tian et al., 2017).
Moisture levels and the availability of N-compounds are considered important factors in regulating the communities of nitrifying and denitrifying microbes (Qin et al., 2021; Liu et al., 2022), whose activities determine soil N2O emission fluxes (Wang et al., 2019; Jia et al., 2021). Interestingly, the highest N2O emissions are often observed at different moisture levels, suggesting that microbial communities in diverse environments respond differently to the same factor. This response is influenced by synergistic interactions with other elements such as soil type, vegetation and historical conditions (e.g., Liu et al., 2018; Qin et al., 2020).
Contrasting responses in nitrification rates and denitrification enzyme activities to moisture and N-fertilizer addition have been reported in grassland soils from China and Australia (Long et al., 2017). Additionally, a microcosm simulation study highlighted the legacy effects of soil moisture on microbial community structure and the transcription of genes encoding key enzymes involved in N-cycling (Banerjee et al., 2016). In the Three Gorges Reservoir environment, ammonia oxidizing archaea (AOA) have been suggested to be more adaptive than ammonia oxidation bacteria (AOB) to water level fluctuations, while AOB have shown greater competitiveness than AOA in riparian soils of the Miyun Reservoir in Beijing (Wang et al., 2016; Liu et al., 2018).
The effects of agricultural practices on soil microbial communities and their activities are intricate, often requiring long-term implementation to observe significant changes. For instance, changes in microbial diversity have been captured after decades of organic framing (Hartmann et al., 2015) or reduced tillage (Tyler, 2019). However, microbial community changes are dynamic and exhibit spatiotemporal heterogeneity. Therefore, snapshots taken after long-term treatments provide limited insight into how microbes initiate their responses and adapt to these conditions. Additionally, the initial response of microbial communities to specific soil conditions remains unclear.
In this study, we aimed to investigate the responses of soil microbial communities and key ammonia-oxidizing microbes to short-term changes in moisture levels, represented by different water filled pore space (WFPS) values, and nitrogen fertilization, through high-throughput 16S rRNA and amoA genes sequencing. The abundance, community structure and network connections of total bacteria and ammonia-oxidizing microbes were scrutinized to address two primary objectives: (1) understanding how the soil bacterial community responds to short-term N-fertilization and water treatments, and (2) examining the combined effects of moisture levels and fertilization on the indigenous microbial community.
2. Materials and methods
2.1. Soil sample preparation
Paddy soil samples were collected from the National Agroecological Research Station (111°26′ E, 28°55′ N, altitude: 92.2–125.3 m) in Taoyuan, Hunan Province, China. The region is characterized by a subtropical humid monsoon climate, with an average annual temperature of 16.5°C, an average annual precipitation of 1,448 mm, an average daily sunshine duration of 15 h and 13 min, and an annual frost-free period of 283 days. Soil samples (0–20 cm) were collected, sieved (<2 mm), and stored. The soil is silty clay, developed from red clay, comprising 31.1% clay (<0.002 mm), 53.0% silt (0.002–0.05 mm), and 15.9% sand (0.05–2.00 mm). The other main soil properties were as follows: pH, 5.06; Soil Organic Matter, 34.73 g kg−1; Total Phosphorus, 0.66 g kg−1; Total N, 2.22 g kg−1; Total Potassium, 11.76 g kg−1 (Qin et al., 2020).
2.2. Experimental design
In order to initiate the activity of microorganisms, the soil was pre-incubated for 2 days under dark conditions at 25°C, with 25% WFPS. Then microcosm culture experiments were set up in 1000 mL glass jars, each with 200 g (dry weight) preconditioned soil and was covered with a film to facilitate gas exchange. Two fertilization conditions, with nitrogen-fertilizer (NF) and without (CK) were prepared. For the NF treatment, NH4NO3 was applied at 720 μg N/g dry soil, which was equivalent to 200 kg N/ha on a surface area basis (200 g soils possess a surface area of 72 cm2). For each treatment, five soil moisture levels, namely, 25%, 50%, 75%, 100%, and 125% WFPS were maintained, as previously described (Qin et al., 2020). All microcosms were incubated at 25°C for 96 h.
2.3. DNA extraction
At the end of incubation, 0.5 g of soil samples were used for DNA extraction as previously described (Qin et al., 2020), DNA quality and concentration were measured using a spectrophotometer (NanoDrop ND-1000; ThermoFisher Scientific, Germany). For each incubation, three extractions were performed, and the DNA were pooled and stored at −80°C for further analysis.
2.4. PCR amplification
The AOA amoA gene, AOB amoA gene, and 16S rRNA gene sequences were amplified from soil DNA by PCR. A 25 μL PCR reaction contained 30 mM Tris-HCl (pH 8.3), 50 mM potassium chloride, 1.5 mM magnesium chloride, 10 μg bovine serum albumin, 200 μM of each deoxyribonucleoside triphosphate, 1.5 U of Taq DNA polymerase, 25 ng soil DNA and respective primers. Deionized water instead of DNA was used as a negative control. AOA, AOB amoA genes were amplified using Arch-amoA 23F (5′-ATGGTCTGGCTWAGACG-3′) and Arch-amoA 616R (5′-GGGGTTTCTACTGGTGGT-3′); amoA-1F (5′-GGGG TTTCTACTGGTGGT-3′) and amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′), respectively; and for 16S rRNA the primer pair 1369F (5′-CGGTGAATACGTTCYCGG-3′) and 1492R (5′-GGWTACCTTGTTACGACT-3′) was used (Sahan and Muyzer, 2008). The amplification conditions for amoA genes were as follows: 95°C for 5 min, followed by 40 cycles: 94°C for 45 s, 60°C for 1 min, 72°C for 1 min; and a final extension at 72°C for 10 min. The PCR procedure for 16S rRNA was the same, except the annealing temperature was 54°C.
2.5. qPCR
qPCR was performed using ABI7900HT (Applied Biosystems, Foster City, CA, United States). qPCR was performed by the SYBR Green method. 16S rRNA-1369F/1492R was used to quantify total bacteria abundance, using Arch-amoA 23F/A616R and amoA-1F/2R for AOA and AOB, respectively (Rotthauwe et al., 1997; Tourna et al., 2008). Plasmids containing the target fragments diluted in 10 x series were used to make qPCR standard curves. The reaction systems are all 10 μ L, including:5 μL 2× SYBR green mix II (TaKaRa Biotechnology Co. Ltd., Dalian, China), 1 μL (10 μM) of forward and reverse primers, 0.2 μL 50× Rox Reference Dye (TaKaRa Biotechnology Co. Ltd., Dalian, China), 5 ng DNA template and deionized water.
The 16S rRNA qPCR procedure was as follows: 95°C pre-denaturation for 30 s; followed by 40 cycles of 95°C denaturation for 5 s, 60°C annealing for 30 s, 72°C extension for 30 s; and then a final extension at 72°C for 1 min. The AOA and AOB amoA gene qPCR procedures were the same, except that the annealing temperature was 53°C and 55°C, respectively. Deionized water instead of soil DNA was used as a negative control to determine DNA contamination (Vestergaard et al., 2017). Three parallels were done for each sample. The results were required to have an amplification efficiency greater than 95%, R2 > 0.999, and a single peak for the melting curve.
2.6. AOA, AOB amoA gene and total 16S rRNA amplicon sequencing
The PCR processing, sequencing, and analysis of Illumina MiSeq sequencing data were performed as described in Qin et al. (2020). PCR mixtures contained 4 μL of 5× TransStart FastPfu buffer, 0.8 μL of forward primer (5 μM), 0.8 μL of reverse primer (5 μM), 2 μL of 2.5 mM dNTPs, 10 ng of template DNA and filled up to 20 μL with ddH2O (TransGen, Beijing, China). PCR products were purified using AxyPrep DNA purification kit (Axygen Bio, United States), and were pooled in equimolar. Paired-end sequencing was performed on the Illumina Miseq platform (Illumina, San Diego, United States) at Shanghai Majorbio BioPharm Technology Co., Ltd., Shanghai. Raw FASTQ files were demultiplexed and quality filtered using QIIME2-2021.4. Barcodes were trimmed, low quality and chimeric sequences were deleted. All samples were normalized to a similar sequencing depth using MOTHUR. Operational clustering of taxonomic units (OTUs) was performed using UPARSE v7.1, based on a cut-off of 97% similarity (Yang et al., 2018). Taxonomic classification was using RDP and FunGene-amoA database for 16S rRNA and amoA sequences, respectively. Representative sequences of each OTU were analyzed using NCBI BLAST to further validate their taxonomy.
2.7. Network analysis
Co-occurrence network analyses were performed to explore how nitrogen fertilization and moisture treatments affect the co-occurrence patterns of microbial communities. Interaction networks were constructed using CoNet v1.1.1 in Cytoscape v.3.6.1 based on the Pearson and Spearman correlation values, mutual information similarity, and Bray–Curtis and Kullback–Leibler dissimilarity measures. All networks were visualized using the Fruchterman–Reingold layout with 9,999 permutations and implemented in Gephi. Global network properties such as average path length, average clustering coefficient, and positive and negative correlation of links are calculated.
2.8. Statistical analysis
Analysis of variance (ANOVA) was run in SPSS v. 18.0 and used to test the significant effects of moisture and fertilization treatments on soil physiochemical properties, microbial gene abundance and diversity. The copy numbers of all functional genes were log-transformed, and the normality of all data was checked before ANOVA. Principal co-ordinates analysis (PCoA) was used to assess the similarities between the community composition of AOA, AOB and total bacteria. Redundancy analysis (RDA) was used to evaluate the effect of soil properties (exchangeable NH4+-N, NO3−-N, and DOC) on the community composition of AOA, AOB and total bacteria. ANOSIM (analysis of similarities) based on the Bray–Curtis distances of OTUs was used to measure the effects of moisture, fertilization, and their interactions on the community composition of AOA, AOB and total bacteria. PCoA, RDA and ANOSIM analysis were performed using R statistical software.
3. Results
3.1. Soil characteristics
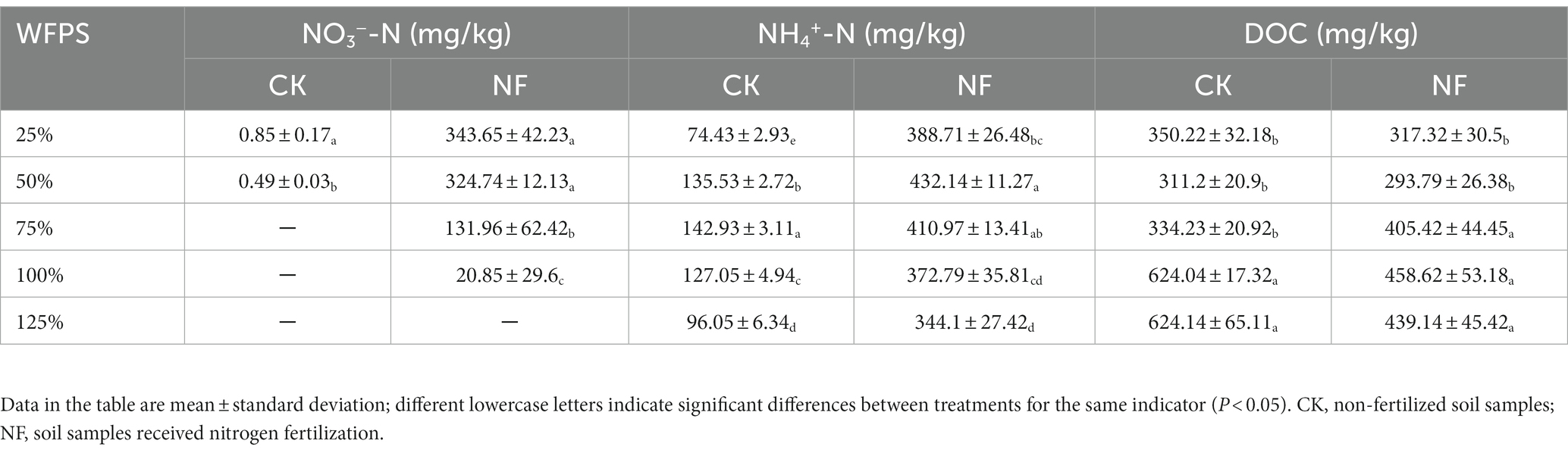
N-fertilizer application clearly increased inorganic N content in the NF treatments, in which nitrate concentrations significantly dropped with the increasing WFPS, and was undetectable in the 125% WFPS incubations, indicating a higher nitrate-consuming activity in higher moisture soils. Nitrate concentration was very low in all CK incubations, regardless the WFPS levels. Ammonium concentration peaked at moderate moisture level of 75% and 50% WFPS for the CK and NF incubations, respectively. However, if treating ammonium in the CK incubations as indigenous background and deducting it from corresponding NF incubations, the remaining ammonium (net NH4+) in NF incubations showed a decreasing trend along increasing WFPS levels, suggesting a possible active net ammonium consumption under higher moisture conditions (Table 1). DOC level in both treatments was similar under low moisture condition (25% and 50% WFPS), and it significantly increased when moisture was high (100% and 125% WFPS), especially in these of the CK treatment (Table 1).
3.2. Soil microbial abundance and diversity
Compared to diversity, the abundance of soil bacteria was more sensitive to water management, but not to N-fertilizer application (Table 2). For total bacteria, the abundance, Shannon index and PD index were affected by moisture levels. The total bacteria abundance was the lowest at 25% WFPS, while the Shannon and PD indexes were the lowest at 125% WFPS, irrespective of fertilization. For AOA and AOB communities, water management only affected their abundance, but not Shannon and PD index. AOB was always more abundant than AOA in all treatments (Figure 1). Fertilizer application had significant effect on total bacteria diversity, and decreased their Shannon index, especially under higher moisture conditions.
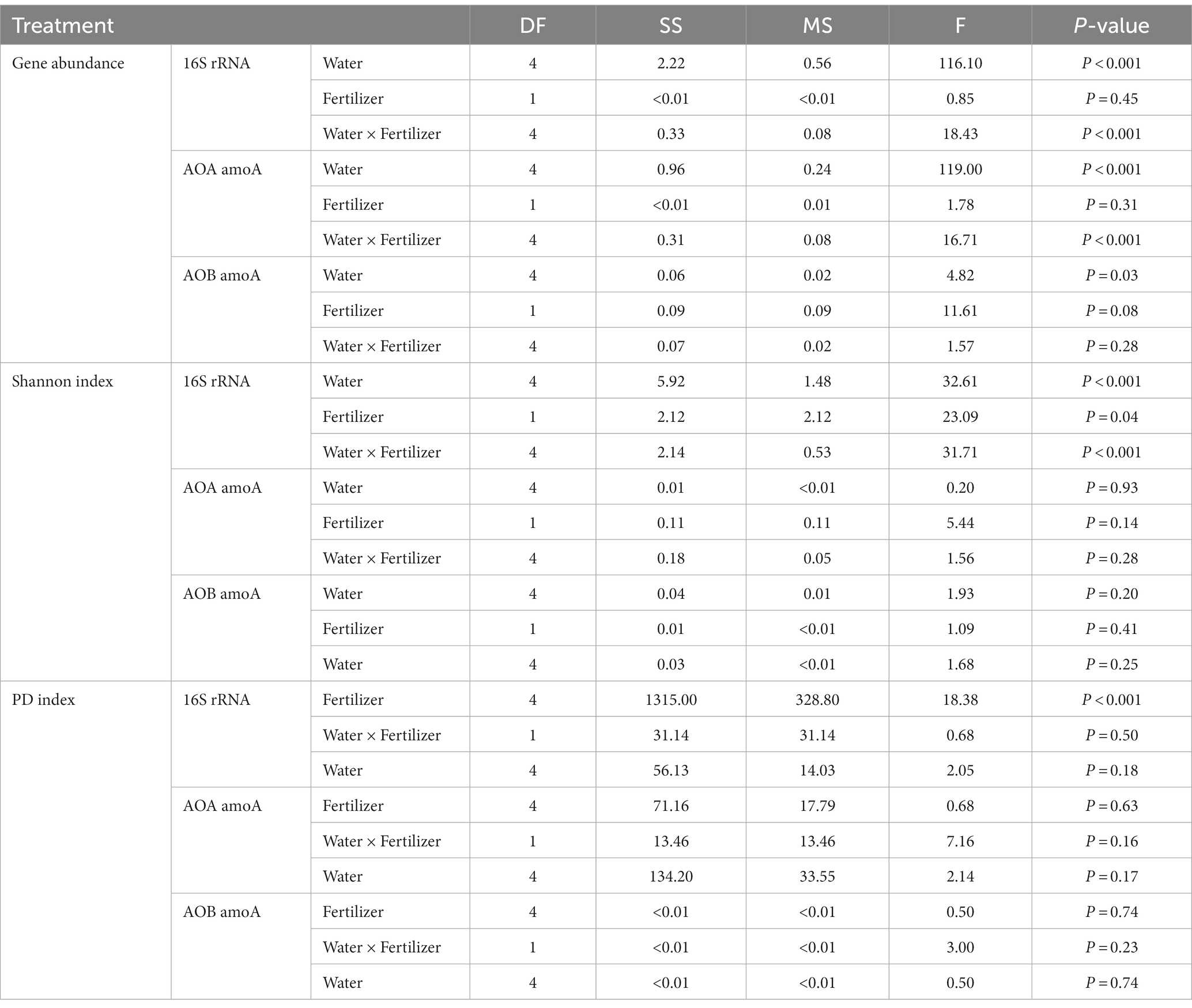
Table 2. Differences in the abundance and diversity indices of 16S rRNA, AOA and AOB affected by fertilization and water treatments.
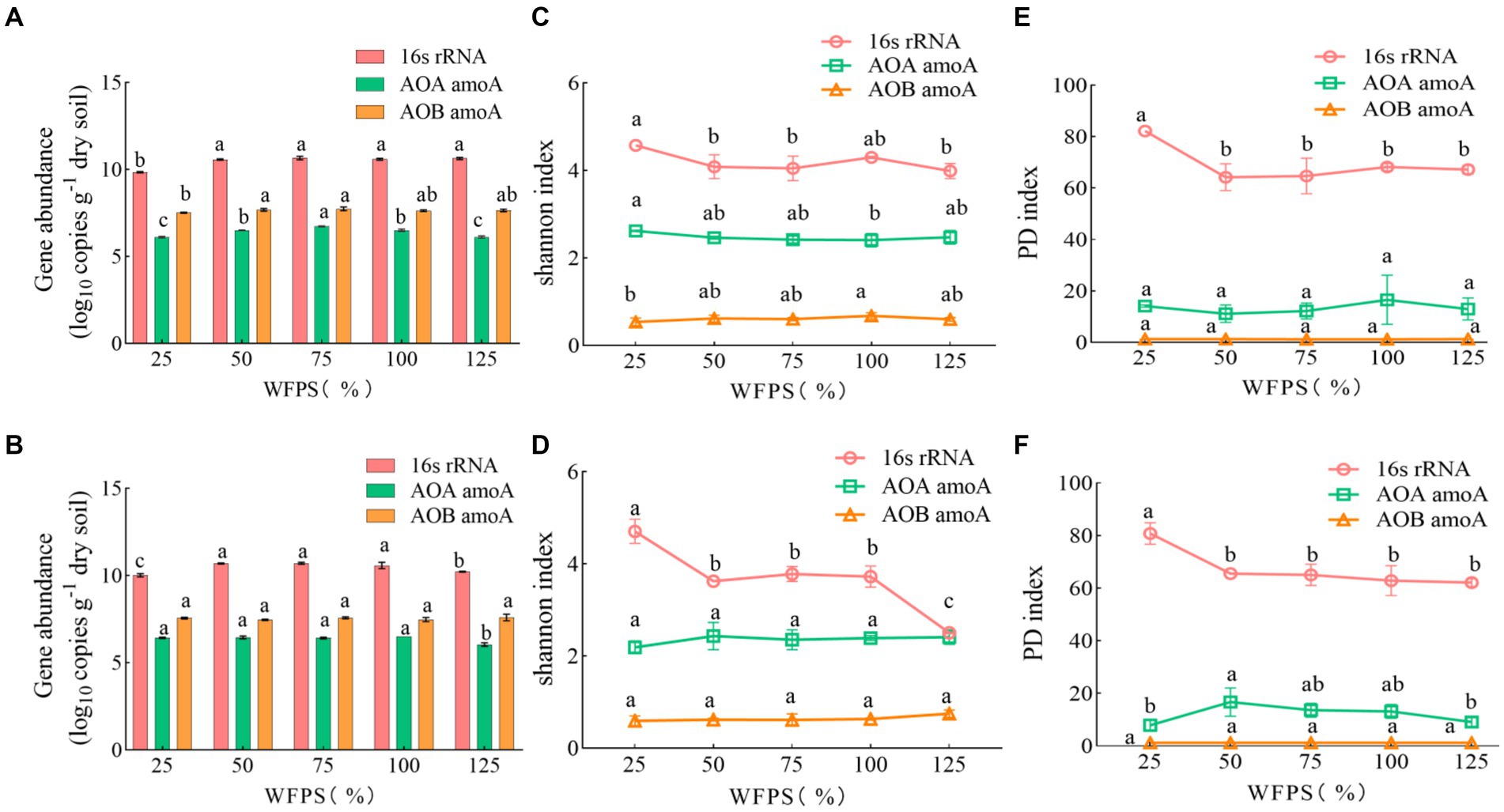
Figure 1. The abundance and diversity index of soil microbes under different WFPS and fertilization treatment conditions. Data are presented as mean and standard error (n = 3). Data labeled by different letters indicate significant differences (p < 0.05). (A,C,E) None-fertilized soil samples (CK); (B,D,F) Soil samples received nitrogen fertilization (NF).
3.3. Soil bacterial community composition
Under 25% and 125% WFPS, the respective microbial composition on phylum level remained very similar between the CK and NF treatments. While under other WFPS conditions, fertilization significantly enhanced the relative abundance of Firmicutes, mainly at the expense of Proteobacteria abundance loss, when compared to the non-fertilized incubations (Figure 2A). The most dominant bacteria phyla were Firmicutes, Proteobacteria, Actinobacteria, Chloroflexi, Planctomycetes, Acidobacteria, and Verrucomicrobia (Figure 2A). The shift of relative abundance and composition of ammonia-oxidizing microorganisms at the OTU level appeared to be more responsive to water content changes (Figures 2B,C).
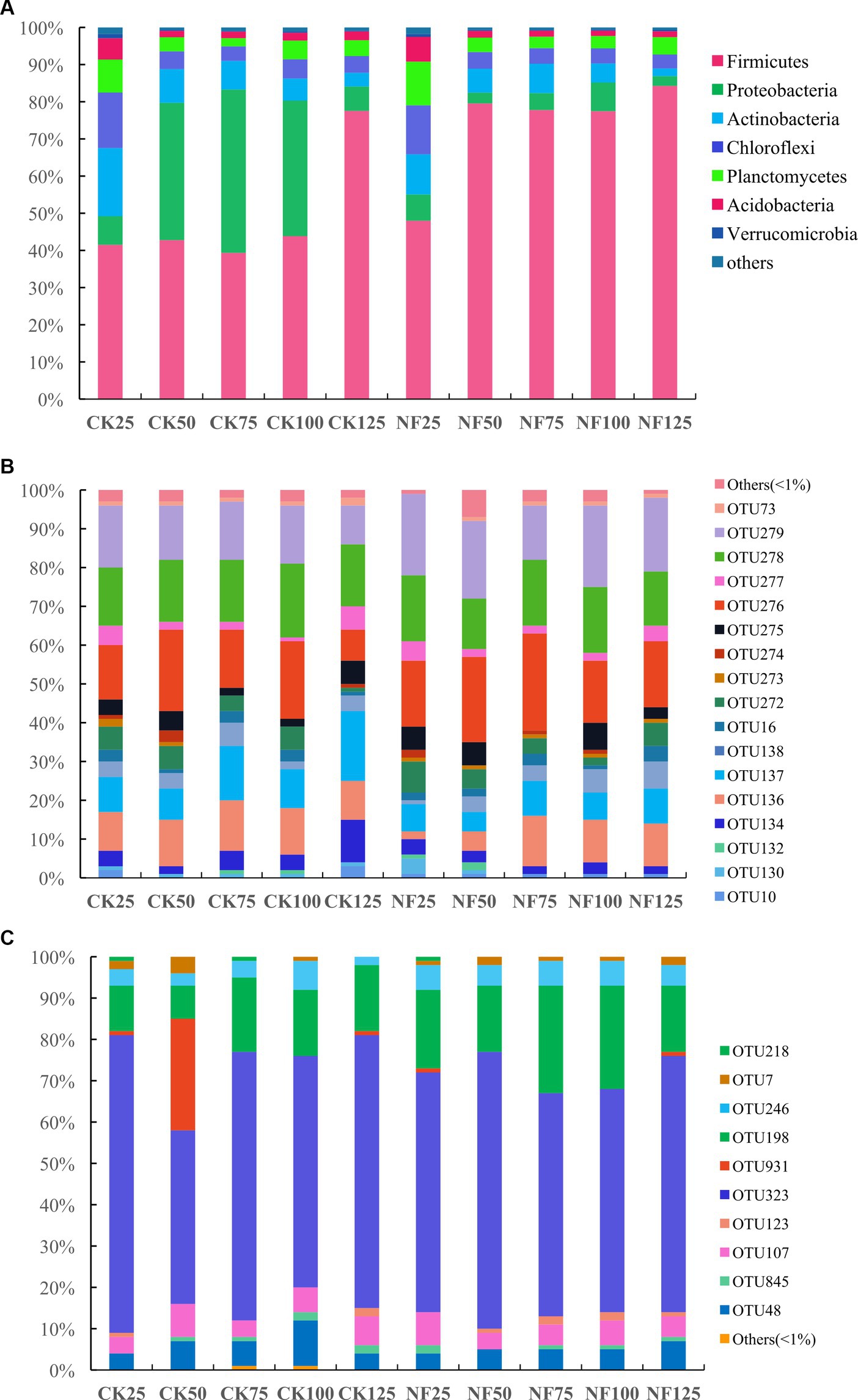
Figure 2. Relative abundance of soil microbes at the phylum (total bacteria) or OTU (AOA and AOB) level. CK, none-fertilized soil samples; NF, soil samples with nitrogen-fertilization; 25, 50, 75, 100, and 125 indicate the respective WFPS levels: 25%, 50%, 75%, 100%, and 125%. (A) total bacteria; (B) AOA; and (C) AOB.
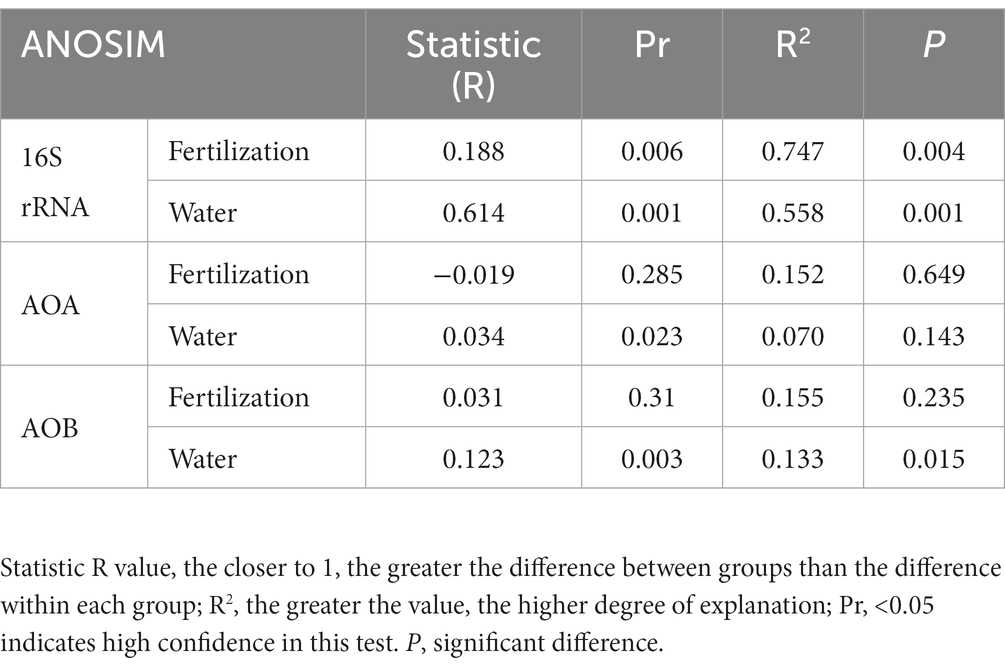
ANOSIM analysis indicated that both fertilization and water content had a significant effect on total bacteria community (p < 0.05), while their influence on AOA and AOB was less significant (Table 3). PCoA analysis showed that the total bacterial community of most incubations were separated between treatments of fertilization and non-fertilized controls (Figure 3A). The NF and CK treatments at 50%–100% WFPS were mostly grouped into respective clusters, albeit the microbial communities of both treatments at 25% and 125% WFPS were loosely distributed, indicating that “extreme” moisture conditions exerted additional influence on the bacterial community composition (Figure 3A). While, the communities of ammonia-oxidizing microorganisms could not be clearly divided between fertilization and non-fertilization treatments, nor among different WFPS levels, presumably due to the relatively slow growth rate of ammonia oxidizing microbes (Figures 3B,C). RDA analysis indicated that NO3−, NH4+ and DOC played an important role in shaping the community structure of total bacteria, AOA and AOB. Among them, the content of NO3−-N had a greater effect on AOA, and NH4+-N had the largest effect on total bacteria and AOB (Figure 4; Table 4).
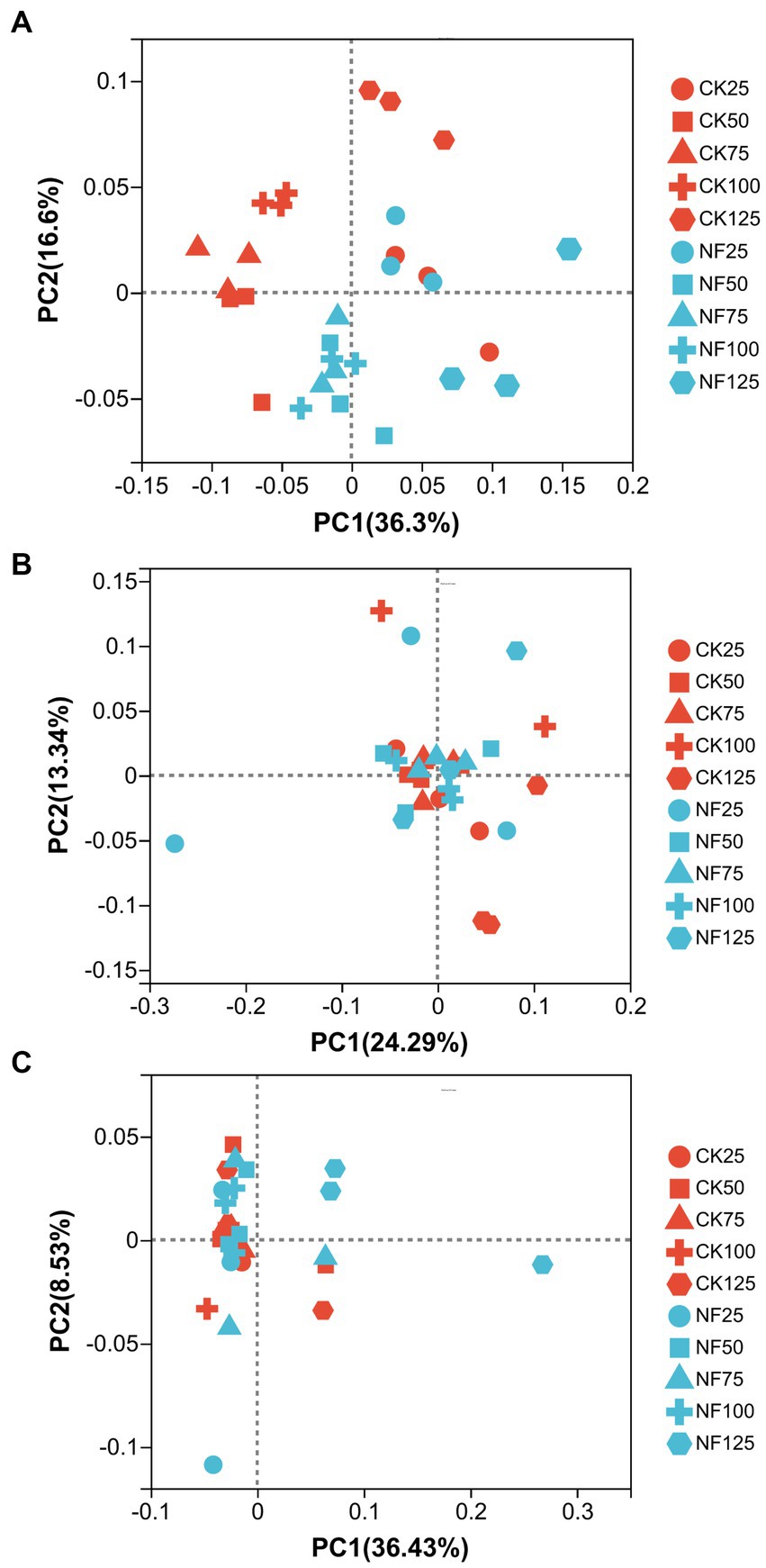
Figure 3. Principal coordinate analysis (PCoA) of the bacterial community structure. CK, soil samples with no N application; NF, soil samples with N-fertilization; 25, 50, 75, 100, and 125 represent WFPS level is 25%, 50%, 75%, 100%, 125%. (A) Total bacteria; (B) AOA; (C) AOB.
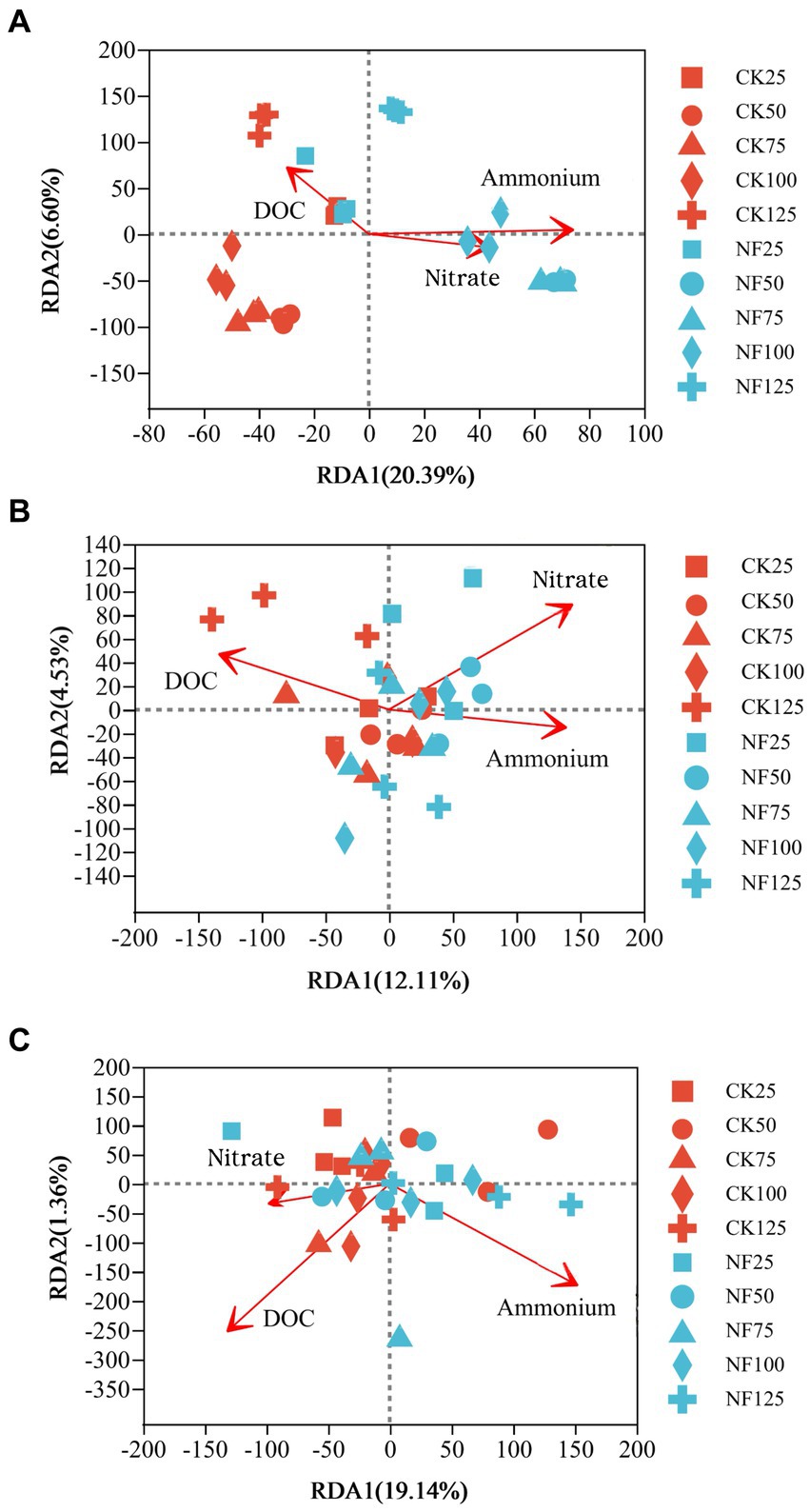
Figure 4. Redundancy analysis (RDA) of the effect of soil characteristics on the microbial community composition. CK, soil samples with no nitrogen application; NF, soil samples with nitrogen application; 25, 50, 75, 100, 125 represent WFPS level is 25%, 50%, 75%, 100%, 125%. (A) Total bacteria; (B) AOA; (C) AOB.
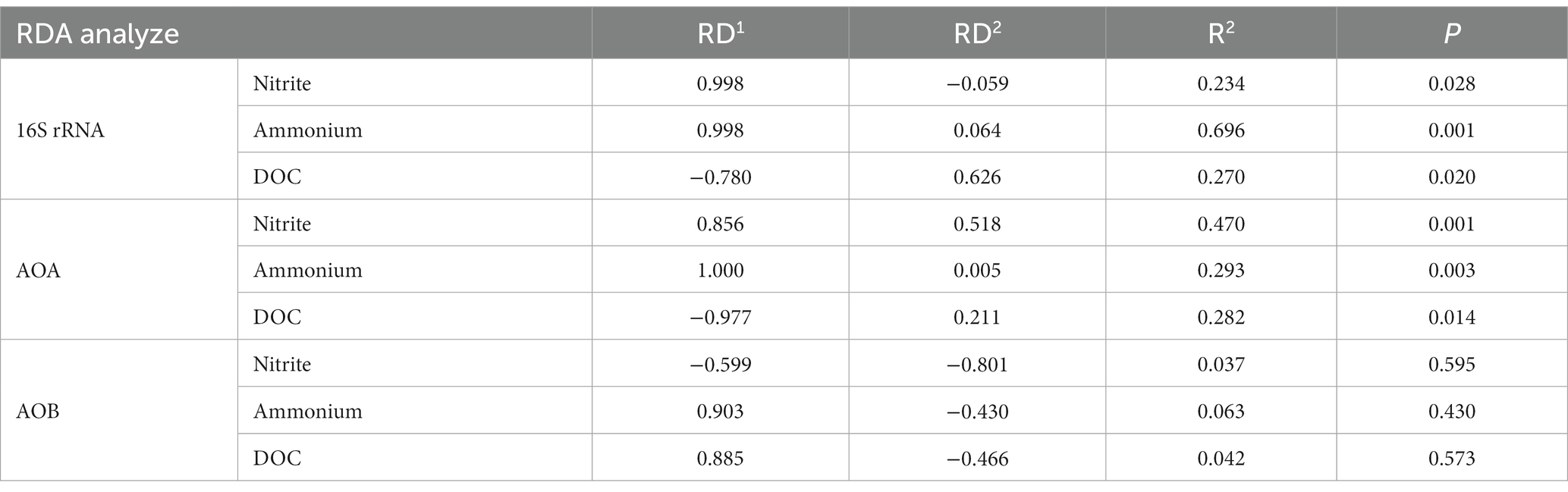
Table 4. Redundancy analysis (RDA) of the effect of different soil characteristics on microbial community composition.
3.4. Network analysis
Microbial network analyses showed that fertilizer application reduced the connectivity and complexity of total bacterial networks (Figure 5; Table 5). Compared to the CK treatments, the average number of nodes and edges decreased by 45.37% and 75.84%, respectively. Higher WFPS levels led to stronger decrease in the network density and clustering coefficients in NF than in CK treatment, suggesting that increasing moisture aggravated the effects of fertilization on the complexity of bacterial networks. The modularity pattern also changed with fertilization and high moisture. In the non-fertilized incubations with relatively low WFPS, two large modules were formed, each containing a number of OTUs with relatively high connectivity (Figure 5A). And all dominant phyla were represented by well-connected OTUs approximately according to their relative abundances, suggesting a rather stable microbial community was maintained. Increasing moisture or nitrogen fertilization diminished modularity, with fewer nodes forming modules. And Firmicutes, or Chloroflexi and Planctomycetes became the respective dominant taxa with well-connected OTUs (Figures 5B,C). Under simultaneous N-fertilization and high moisture, the microbial network did not form clear modules, and highly connected nodes were mostly Firmicutes.
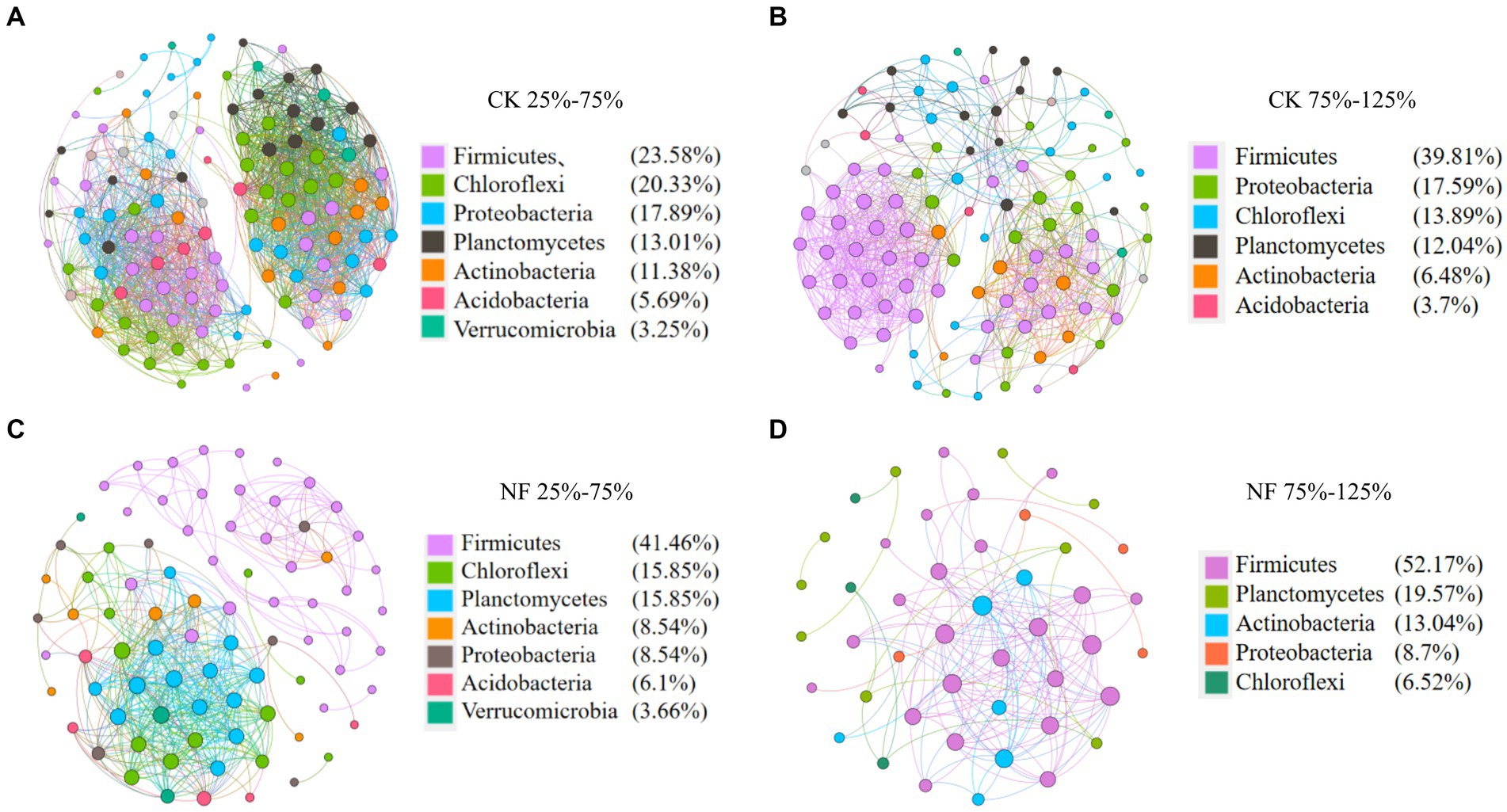
Figure 5. Co-occurrence analysis of total bacteria. Nodes are colored according to microbial taxonomy (phylum), edges indicate correlations among nodes. CK, none-fertilized soil samples; NF, soil samples received N-fertilization; 25, 50, 75, 100, 125 indicate respective WFPS level is 25%, 50%, 75%, 100%, 125%. (A) CK 25%–75%; (B) CK 75%–125%; (C) NF25%–75%; (D) NF 75%–125%.
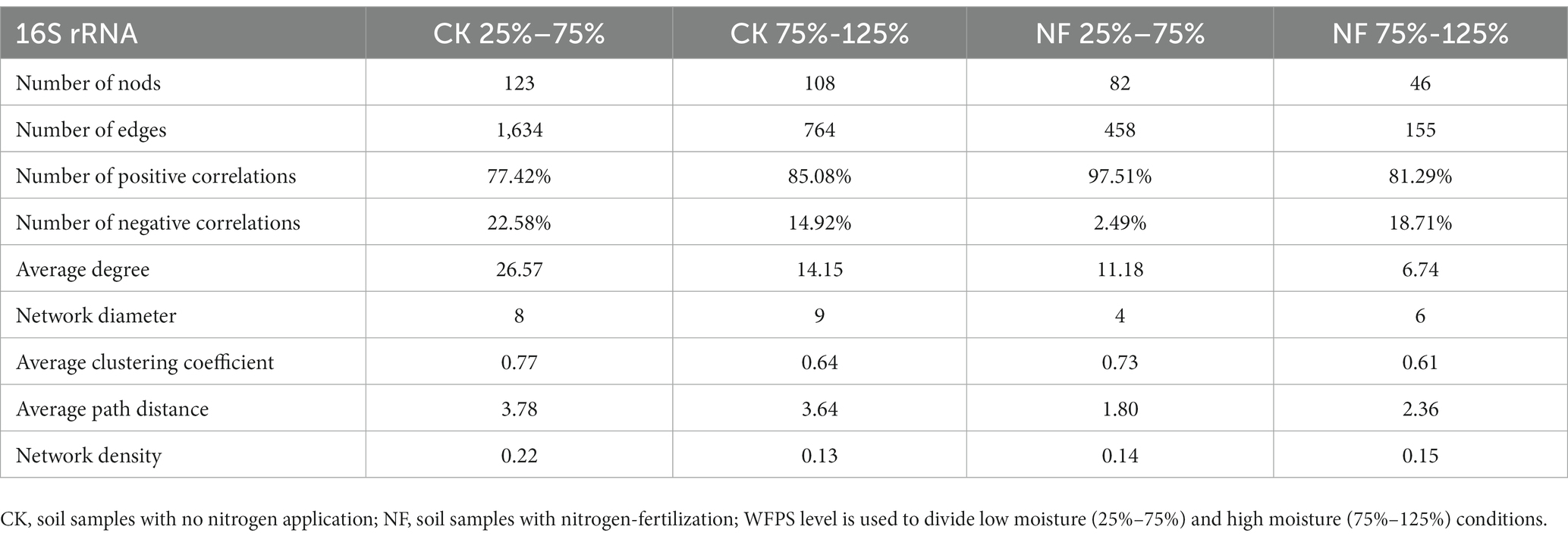
Table 5. Node-level topological features of the co-occurrence network of fertilization and non-fertilized treatments grouped by moisture levels.
4. Discussion
Soil microbial communities play an essential role in various ecological processes, including nutrient cycling, organic matter turnover, greenhouse gas emission, and soil fertility and structure maintenance. Nitrogen compounds and moisture are considered key factors shaping bacterial community in soils (Wang S. et al., 2018; Wang R. et al., 2018). However, common agricultural practices aimed at maximizing crop productivity, such as fertilization, can introduce excessive nitrogen into agroecosystems, and may have adverse ecological effects. Excessive nitrogen supply may reduce soil microbial biomass, alter microbial diversity, community structure and enzyme activity (Wang et al., 2023). Therefore, understanding how soil microbial communities respond to nitrogen fertilization and moisture fluctuations is crucial for developing sustainable agricultural practices. While several studies have examined the long-term impact of nitrogen addition on soil microbial diversity and richness in different environments, the results have been inconsistent (Fierer and Jackson, 2006; Jing et al., 2015). Investigating the immediate changes in microbial communities following short-term nitrogen fertilization and moisture variations can provide insights into the microbial response processes under long-term agricultural management.
In this study, NH4NO3 was provided as the nitrogen fertilizer, introducing both nitrate and ammonium to the NF treatments. However, after 96 h incubation, nitrate concentration in the NF incubations decreased with increasing WFPS and became undetectable under 125% WFPS. The “net NH4+” in the NF treatments also showed a decreasing trend with higher WFPS levels (Table 1), indicating a higher consumption rate of both nitrate and ammonium under increased moisture conditions. Rice paddies are well known nitrogen cycling hotspots, and denitrification and anammox are likely the main processes responsible for nitrate and ammonium consumption, especially under high WFPS levels (Zhu et al., 2011). Ammonium was available in significant concentrations, which likely supported a higher abundance of AOB compared to AOA in all incubations (Figure 1). AOA are more dominant in oligotrophic environments, while AOB tend to dominate in eutrophic habitats (Jung et al., 2022). Among the AOA OTUs, OTU276, OTU278 and OTU279 were consistently abundant in almost all incubations, while OTU323 was the most dominant AOB (Figure 2B). These OTUs had their closest cultured relatives as Nitrososphaera viennensis EN76 and Nitrosospira lacus (Tourna et al., 2011; Urakawa et al., 2015), with amoA sequence similarities below 80 and 86%, respectively. Hence, the AOA and AOB OTUs identified in this study were mostly uncultured, indicating a vast unexplored diversity of ammonia-oxidizing microbes in agriculture ecosystems. The abundance and composition of AOA and AOB shifted after the short-term incubation, but it remains unclear if these microbes directly contributed to the removal of ‘net NH4+’, particularly under higher WFPS conditions that were anoxic. Future studies should conduct a comprehensive analysis of the rates of different N-cycling processes, including the recently discovered oxygenic denitrification and nitrate-driven anaerobic methane oxidation (Zhu et al., 2020), to better understand their differential responses to N addition.
Soil moisture levels have been identified as a significant factor influencing soil microbial composition and activity (Shi et al., 2018). Adequate moisture levels can promote microbial growth and metabolic activities (Francioli et al., 2016; Yang et al., 2019). In our study, the microbial diversity and abundance responded differently to short-term fertilization and water management. Our results showed that total soil bacteria abundance was more responsive to water content than to fertilization (Table 2). This suggests that microbial growth was generally more strongly influenced by water content than nitrogen addition in the short-term incubation. This could be because the paddy soil was not N-limited, since there was sufficient residual ammonium present (Table 1). However, fertilization did increase bacteria diversity, likely due to the introduction of nitrate, which serves as a favorable electron acceptor for microbial respiration.
Firmicutes were the most dominant bacteria, especially in the NF treatments with higher WFPS (Figure 2). In the CK treatment with saturated moisture (125% WFPS), the relative abundance of Firmicutes (84.2%) was significantly higher than in other CK incubations (approximately 40%). This indicates a favorable response of Firmicutes to nitrogen addition and high moisture, consistent with previous findings (e.g., Supramaniam et al., 2016). Many Firmicutes are capable of forming spores, which confer high resistance to environmental stresses and enable quick response to substrate availability, making them one of the most common microbes in soils (Filippidou et al., 2016; Donhauser et al., 2021). Proteobacteria were abundant only in the CK treatments with moderate moisture levels (50%, 75%, and 100% WFPS). Interestingly, the relative abundance of Acidobacteria, Actinobacteria, Chloroflexi, and Planctomycetes was significant in both the CK and NF treatments with the lowest moisture (25% WFPS) but diminished in all other higher moisture incubations (Figure 2A). This indicates the influence of water management on soil microbial community structure. Acidobacteria and Chloroflexi members are often considered oligotrophs, and several studies have reported a decrease in their abundance with increasing concentrations of NO3−-N and NH4+-N (Zhou et al., 2015; Wang R. et al., 2018) and higher water content (Zhalnina et al., 2015; Zhou et al., 2015; Li H. et al., 2021). However, some long-term field studies have found had no significant effect of water content on soil bacterial communities (Zhang et al., 2021), possibly because there was sufficient time for the local microbial community to recover from and adapt to moisture changes (Azarbad et al., 2020). These inconsistent findings highlight the need to study the immediate responses of microbes to environmental changes.
In the present study, after a four-day incubation, it was observed that bacterial community structure, rather than total bacterial abundance, showed a stronger response to fertilization. Nitrogen fertilization has been demonstrated to be a significant factor in shaping soil microbial community (Yuan et al., 2015; Wu et al., 2021; Jin et al., 2022), probably by providing substrates and energy sources for indigenous microbes or due to nutrients imbalances resulting from a pulse input of N (Eo and Park, 2016). The bacterial abundance remained relatively stable after the 96-h incubation, which is consistent with the fact that soil bacteria generally have very slow growth rates, with a doubling time being over 100 days (Harris and Paul, 1994). Nevertheless, the shift in microbial community indicates a differential response among distinct groups of microbes. In long-term fertilizer treatments, both the structure and abundance of soil bacterial communities can also be influenced by indirect environmental changes, such as soil acidification resulting from nitrification (Zhou et al., 2015; Wang et al., 2016; Lin et al., 2020). However, some studies suggest that soil parent material plays a key role in shaping agricultural soil bacterial communities from a long-term perspective (Sheng et al., 2023).
The number of nodes, edges and average clustering coefficients decreased in the microbial networks of the NF treatment and higher moisture incubations, and the interaction pattern among soil microbes also shifted (Table 5). In the non-fertilized treatment, increasing moisture reduced the ratio of negative interactions, which represent microbial competition and are considered important for stabilizing microbial communities and their ecological services (van der Heijden et al., 2010; Hoek et al., 2016). These results indicated that both fertilization and higher water content rapidly reduced the complexity and stability of soil bacterial communities. Based on the degree of each node, key taxa identified in the microbial network mostly belonged to Firmicutes, Proteobacteria, Actinobacteria, Chloroflexi and Planctomycetes, indicating their crucial role in paddy soils under fertilization and water management. In recent years, the construction of microbial network has become increasingly popular for high-throughput sequencing data analysis and been helpful in uncovering interactions from complex datasets (Deng et al., 2012; Faust and Raes, 2012). However, it should be noted that the inference of biotic interactions from the cooccurrence or absence of sequences may not always reliable, and conclusions should be drawn with caution (Faust, 2021).
5. Conclusion
The composition and abundance of soil bacteria and ammonia-oxidizing microorganisms in paddy fields demonstrated varying degrees of response to fertilization and moisture, with a higher sensitivity to changes in water content levels. The impact of fertilization on the stability and complexity of bacteria networks was evident and slightly exacerbated by elevated moisture. Changes in the total bacteria and AOB community showed significant correlations with NH4+-N content, while NO3−-N content played a crucial role in driving changes in the AOA community structure. Studying the initial changes in microbial community composition and structure in response to water and fertilizer applications can provide valuable insights into how soil microbes adapt to long-term agricultural practices, thus aiding in the development of more sustainable agricultural approaches.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
LH, HQ, JW, DY, and LZ carried out the experiments. HQ, LH, BZ, and JG did sequencing and data analysis. LH, HQ, JG, and BZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by the National Key Research and Development Program of China (2021YFD1901203), the National Natural Science Foundation of China (project no. 42177104), and the National Ecosystem Science Data Center (no. NESDC20210204).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Azarbad, H., Tremblay, J., Giard-Laliberté, C., Bainard, L. D., and Yergeau, E. (2020). Four decades of soil water stress history together with host genotype constrain the response of the wheat microbiome to soil moisture. FEMS Microbiol. Ecol. 96:fiaa098. doi: 10.1093/femsec/fiaa098
Banerjee, S., Helgason, B., Wang, L., Winsley, T., Ferrari, B. C., and Siciliano, S. D. (2016). Legacy effects of soil moisture on microbial community structure and N2O emissions. Soil Biol. Biochem. 95, 40–50. doi: 10.1016/j.soilbio.2015.12.004
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Bastida, F., Torres, I. F., Hernández, T., and García, C. (2017). The impacts of organic amendments: do they confer stability against drought on the soil microbial community? Soil Biol. Biochem. 113, 173–183. doi: 10.1016/j.soilbio.2017.06.012
Bijay-Singh, C. E., and Craswell, E. (2021). Fertilizers and nitrate pollution of surface and ground water: an increasingly pervasive global problem. SN Appl. Sci. 3:518. doi: 10.1007/s42452-021-04521-8
Canarini, A., Carrillo, Y., Mariotte, P., Ingram, L., and Dijkstra, F. A. (2016). Soil microbial community resistance to drought and links to C stabilization in an Australian grassland. Soil Biol. Biochem. 103, 171–180. doi: 10.1016/j.soilbio.2016.08.024
Chai, R., Ye, X., Ma, C., Wang, Q., Tu, R., Zhang, L., et al. (2019). Greenhouse gas emissions from synthetic nitrogen manufacture and fertilization for main upland crops in China. Carbon Balance Manag. 14:20. doi: 10.1186/s13021-019-0133-9
Chen, J. H., Chen, D., Xu, Q. F., Fuhrmann, J. J., Li, L. Q., Pan, G. X., et al. (2019). Organic carbon quality, composition of main microbial groups, enzyme activities, and temperature sensitivity of soil respiration of an acid paddy soil treated with biochar. Biol. Fertil. Soils 55, 185–197. doi: 10.1007/s00374-018-1333-2
Chen, Z. M., Wang, H., Liu, X., Zhao, X., Lu, D., Zhou, J., et al. (2017). Changes in soil microbial community and organic carbon fractions under short-term straw return in a rice-wheat cropping system. Soil Tillage Res. 165, 121–127. doi: 10.1016/j.still.2016.07.018
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., Zhou, J., et al. (2012). Molecular ecological network analyses. Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113
Donhauser, J., Qi, W., Bergk-Pinto, B., and Frey, B. (2021). High temperatures enhance the microbial genetic potential to recycle C and N from necromass in high-mountain soils. Glob. Chang. Biol. 27, 1365–1386. doi: 10.1111/gcb.15492
Drenovsky, R. E., Vo, D., Graham, K. J., and Scow, K. M. (2004). Soil water content and organic carbon availability are major determinants of soil microbial community composition. Microb. Ecol. 48, 424–430. doi: 10.1007/s00248-003-1063-2
Eo, J., and Park, K. C. (2016). Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 231, 176–182. doi: 10.1016/j.agee.2016.06.039
Faust, K. (2021). Open challenges for microbial network construction and analysis. ISME J. 15, 3111–3118. doi: 10.1038/s41396-021-01027-4
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Feng, Y. Z., Chen, R., Hu, J., Zhao, F., Wang, J., Chu, H., et al. (2015). Bacillus asahii comes to the fore in organic manure fertilized alkaline soils. Soil Biol. Biochem. 81, 186–194. doi: 10.1016/j.soilbio.2014.11.021
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. PNAS 103, 626–631. doi: 10.1073/pnas.0507535103
Filippidou, S., Wunderlin, T., Junier, T., Jeanneret, N., Dorador, C., Molina, V., et al. (2016). Combination of extreme environmental conditions favor the prevalence of endospore-forming firmicutes. Front. Microbiol. 7:1707. doi: 10.3389/fmicb.2016.01707
Francioli, D., Schulz, E., Lentendu, G., Wubet, T., Buscot, F., and Reitz, T. (2016). Mineral vs. organic amendments: microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Front. Microbiol. 14:1446. doi: 10.3389/fmicb.2016.01446
Gomiero, T., Pimentel, D., and Paoletti, M. G. (2011). Environmental impact of different agricultural management practices: conventional vs. Organic agriculture. Crit. Rev. Plant Sci. 30, 95–124. doi: 10.1080/07352689.2011.554355
Harris, D., and Paul, E. A. (1994). Measurement of bacterial growth rates in soil. Appl. Soil Ecol. 1, 277–290. doi: 10.1016/0929-1393(94)90005-1
Hartmann, M., Frey, B., Mayer, J., Maeder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hoek, T. A., Axelrod, K., Biancalani, T., Yurtsev, E. A., Liu, J., and Gore, J. (2016). Resource availability modulates the cooperative and competitive nature of a microbial cross feeding mutualism. PLoS Biol. 14:e1002540. doi: 10.1371/journal.pbio.1002540
Jia, M., Gao, Z., Gu, H., Zhao, C., Liu, M., Liu, F., et al. (2021). Effects of precipitation change and nitrogen addition on the composition, diversity, and molecular ecological network of soil bacterial communities in a desert steppe[J]. PLoS One 16:e0248194. doi: 10.1371/journal.pone.0248194
Jin, Y., Chen, Z., He, Y., White, J. F., and Li, C. (2022). Effects of achnatherum inebrians ecotypes and endophyte status on plant growth, plant nutrient, soil fertility and soil microbial community. Soil Sci. Soc. Am. J. 86, 1028–1042. doi: 10.1002/saj2.20420
Jing, X., Sanders, N. J., Shi, Y., Chu, H., Classen, A. T., Zhao, K., et al. (2015). The links between ecosystem multifunctionality and above- and belowground biodiversity are mediated by climate. Nat. Commun. 6:8159. doi: 10.1038/ncomms9159
Jung, M. Y., Sedlacek, C. J., Kits, K. D., Mueller, A. J., Rhee, S. K., Hink, L., et al. (2022). Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 16, 272–283. doi: 10.1038/s41396-021-01064-z
Li, G. C., Niu, W., Sun, J., Zhang, W., Zhang, E., and Wang, J. (2021). Soil moisture and nitrogen content influence wheat yield through their effects on the root system and soil bacterial diversity under drip irrigation. Land Degrad. Dev. 32, 3062–3076. doi: 10.1002/ldr.3967
Li, H., Wang, H., Jia, B., Li, D., Fang, Q., and Li, R. (2021). Irrigation has a higher impact on soil bacterial abundance, diversity and composition than nitrogen fertilization. Sci. Rep. 11:6. doi: 10.1038/s41598-021-96234-6
Li, H., Xu, Z. W., Yang, S., Li, X. B., Top, E. M., Wang, R. Z., et al. (2016). Responses of soil bacterial communities to nitrogen deposition and precipitation increment are closely linked with above ground community variation. Microb. Ecol. 71, 974–989. doi: 10.1007/s00248-016-0730-z
Liang, Q., Chen, H., Gong, Y., Fan, M., Yang, H., Lal, R., et al. (2012). Effects of 15 years of manure and inorganic fertilizers on soil organic carbon fractions in a wheat-maize system in the North China plain. Nutr. Cycl. Agroecosyst. 92, 21–33. doi: 10.1007/s10705-011-9469-6
Lin, Y. X., Ye, G., Ding, W., Hu, H. W., Zheng, Y., Fan, J., et al. (2020). Niche differentiation of comammox Nitrospira and canonical ammonia oxidizers in soil aggregate fractions following 27-year fertilizations. Agric. Ecosyst. Environ. 304:107147. doi: 10.1016/j.agee.2020.107147
Liu, Y., Liu, J., Yao, P., Ge, T., Qiao, Y., Zhao, M., et al. (2018). Distribution patterns of ammonia-oxidizing archaea and bacteria in sediments of the eastern China marginal seas. Syst. Appl. Microbiol. 41, 658–668. doi: 10.1016/j.syapm.2018.08.008
Liu, H., Zheng, X., Li, Y., Yu, J., Ding, H., Sveen, T. R., et al. (2022). Soil moisture determines nitrous oxide emission and uptake. Sci. Total Environ. 822:153566. doi: 10.1016/j.scitotenv.2022.153566
Long, X., Shen, J., Wang, J., Zhang, L., Di, H., and He, J. (2017). Contrasting response of two grassland soils to N addition and moisture levels: N2O emission and functional gene abundance. J. Soil. Sediment. 17, 384–392. doi: 10.1007/s11368-016-1559-2
Pan, F. X., Li, Y. Y., Chapman, S. J., and Yao, H. (2016). Effect of rice straw application on microbial community and activity in paddy soil under different water status [J]. Environ. Sci. Pollut. Res. 23, 5941–5948. doi: 10.1007/s11356-015-5832-5
Qin, H., Wang, D., Xing, X., Tang, Y., Wei, X., Chen, X., et al. (2021). A few key nirK- and nosZ-denitrifier taxa play a dominant role in moisture-enhanced N2O emissions in acidic paddy soil. Geoderma 385:114917. doi: 10.1016/j.geoderma.2020.114917
Qin, H., Xing, X., Tang, Y., Zhu, B., Wei, X., Chen, X., et al. (2020). Soil moisture and activity of nitrite- and nitrous oxide-reducing microbes enhanced nitrous oxide emissions in fallow paddy soils. Biol. Fertil. Soils 56, 53–67. doi: 10.1007/s00374-019-01403-5
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997
Sahan, E., and Muyzer, G. (2008). Diversity and spatio-temporal distribution of ammonia-oxidizing archaea and bacteria in sediments of the Westerschelde estuary. FEMS Microbiol. Ecol. 64, 175–186. doi: 10.1111/j.1574-6941.2008.00462.x
Sheng, R., Xu, H., Xing, X., Zhang, W., Hou, H., Qin, H., et al. (2023). The role of inherited characteristics from parent materials in shaping bacterial communities in agricultural soils. Geoderma 433:116455. doi: 10.1016/j.geoderma.2023.116455
Shi, L., Zhang, H., Liu, T., Mao, P., Zhang, W., Shao, Y., et al. (2018). An increase in precipitation exacerbates negative effects of nitrogen deposition on soil cations and soil microbial communities in a temperate forest. Environ. Pollut. 235, 293–301. doi: 10.1016/j.envpol.2017.12.083
Soman, C., Li, D. F., Wander, M. M., and Kent, A. D. (2017). Long-term fertilizer and crop-rotation treatments differently affect soil bacterial community structure. Plant and Soil 413, 145–159. doi: 10.1007/s11104-016-3083-y
Supramaniam, Y., Chong, C. W., Silvaraj, S., and Tan, K. P. (2016). Effect of short term variation in temperature and water content on the bacterial community in a tropical soil. Appl. Soil Ecol. 107, 279–289. doi: 10.1016/j.apsoil.2016.07.003
Tang, H. M., Xiao, X. P., Li, C., Tang, W. G., Cheng, K. K., Wang, K., et al. (2019). Effects of rhizosphere and long-term fertilization practices on the activity and community structure of denitrifiers under double-cropping rice field. Commun. Soil Sci. Plant Anal. 50, 682–697. doi: 10.1080/00103624.2019.1589480
Tian, J. H., Wei, K., Condron, L. M., Chen, Z. H., Xu, Z. W., Feng, J., et al. (2017). Effects of elevated nitrogen and precipitation on soil organic nitrogen fractions and nitrogen-mineralizing enzymes in semi-arid steppe and abandoned cropland. Plant and Soil 417:6. doi: 10.1007/s11104-017-3253-6
Tourna, M., Freitag, T. E., Nicol, G. W., and Prosser, J. I. (2008). Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10, 1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x
Tourna, M., Stieglmeier, M., Spang, A., Koenneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. PNAS 108, 8420–8425. doi: 10.1073/pnas.1013488108
Tyler, H. L. (2019). Bacteria community composition under long-term reduced tillage and no till management. J. Appl. Microbiol. 126, 1797–1807. doi: 10.1111/jam.14267
Urakawa, H., Garcia, J. C., Nielsen, J., Le, V. Q., Kozlowski, J. A., Stein, L. Y., et al. (2015). Nitrosospira lacus sp. nov., a psychrotolerant, ammonia-oxidizing bacterium from sandy lake sediment. Int. J. Syst. Evol. Microbiol. 65, 242–250. doi: 10.1099/ijs.0.070789-0
van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2010). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Vestergaard, G., Schulz, S., Schöler, A., and Schloter, M. (2017). Making big data smart—how to use metagenomics to understand soil quality. Biol. Fertil. Soils 53, 479–484. doi: 10.1007/s00374-017-1191-3
Wang, X., Feng, J., Ao, G., Qin, W., Han, M., Shen, Y., et al. (2023). Globally nitrogen addition alters soil microbial community structure, but has minor effects on soil microbial diversity and richness. Soil Biol. Biochem. 179:108982. doi: 10.1016/j.soilbio.2023.108982
Wang, X., Hefting, M. M., Schwark, L., and Zhu, G. (2019). Anammox and denitrification separately dominate microbial N-loss in water saturated and unsaturated soils horizons of riparian zones. Water Res. 162, 139–150. doi: 10.1016/j.watres.2019.06.052
Wang, S., Wang, X., Han, X., and Deng, Y. (2018). Higher precipitation strengthens the microbial interactions in semiarid grassland soils. Glob. Ecol. Biogeogr. 27, 570–580. doi: 10.1111/geb.12718
Wang, C., Xiao, H., Liu, J., Zhou, J., and Du, D. (2016). Insights into the effects of simulated nitrogen deposition on leaf functional traits of Rhus Typhina. Pol. J. Environ. Stud. 25, 1279–1284. doi: 10.15244/pjoes/61788
Wang, R., Xiao, Y., Lv, F., Hu, L., Wei, L., Yuan, Z., et al. (2018). Bacterial community structure and functional potential of rhizosphere soils as influenced by nitrogen addition and bacterial wilt disease under continuous sesame cropping. Appl. Soil Ecol. 125, 117–127. doi: 10.1016/j.apsoil.2017.12.014
Wu, Y., Liu, J., Shaaban, M., and Hu, R. (2021). Dynamics of soil n2o emissions and functional gene abundance in response to biochar application in the presence of earthworms. Environ. Pollut. 268:115670. doi: 10.1016/j.envpol.2020.115670
Yang, J., Chen, W., Hou, H. J., Chen, C. L., Qin, H. L., and Lv, D. Q. (2019). Difference in response of soil bacterial to soil moisture and temperature in the hilly red soil region of subtropics of southern China. Chin J Trop Crops 40, 609–615. doi: 10.3969/j.issn.1000-2561.2019.03.028
Yang, Y. D., Hu, Y. G., Wang, Z. M., and Zeng, Z. H. (2018). Variations of the nirS-, nirK-, and nosZ-denitrifying bacterial communities in a northern Chinese soil as affected by different long-term irrigation regimes. Environ. Sci. Pollut. R. 25, 14057–14067. doi: 10.1007/s11356-018-1548-7
Yu, H. L., Ling, N., Wang, T. T., Zhu, C., Wang, Y., Wang, S. J., et al. (2019). Responses of soil biological traits and bacterial communities to nitrogen fertilization mediate maize yields across three soil types. Soil Tillage Res. 185, 61–69. doi: 10.1016/j.still.2018.08.017
Yuan, X., Knelman, J. E., Gasarch, E., Wang, D., Nemergut, D. R., and Seastedt, T. R. (2016). Plant community and soil chemistry responses to long-term nitrogen inputs drive changes in alpine bacterial communities. Ecology 97, 1543–1554. doi: 10.1890/15-1160.1
Yuan, H. C., Wu, H., Ge, T. D., Li, K. L., Wu, J. S., and Wang, J. R. (2015). Effects of long-term fertilization on bacterial and archaeal diversity and community structure within subtropical red paddy soils[J]. Chin. J. Appl. Ecol. 26, 1807–1813.
Zhalnina, K., Dias, R., de Quadros, P. D., Davis, R. A., Camargo, F. A., Clark, I. M., et al. (2015). Soil pH determines microbial diversity and composition in the park grass experiment. Microbiology. Ecology 69, 395–406. doi: 10.1007/s00248-014-0530-2
Zhang, R., Gu, J., and Wang, X. (2021). Responses of soil bacterial and fungi after 36 years fertilizer, straw cover, and irrigation management practices in Northwest China. Soil Use Manage. 37, 843–854. doi: 10.1111/sum.12671
Zhou, J., Guan, D., Zhou, B., Zhao, B., Ma, M., Qin, J., et al. (2015). Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in Northeast China. Soil Biol. Biochem. 90, 42–51. doi: 10.1016/j.soilbio.2015.07.005
Zhou, J., Xia, B., Treves, D. S., Wu, L. Y., Marsh, T. L., O’Neill, R. V., et al. (2002). Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68, 326–334. doi: 10.1128/AEM.68.1.326-334.2002
Zhu, B., Wang, Z., Kanaparthi, D., Kublik, S., Ge, T., Casper, P., et al. (2020). Long-read amplicon sequencing of nitric oxide dismutase (nod) genes reveal diverse oxygenic Denitrifiers in agricultural soils and Lake sediments. Microb. Ecol. 80, 243–247. doi: 10.1007/s00248-020-01482-0
Keywords: paddy soil, fertilization, moisture levels, microbial community, AOA, AOB, network analysis
Citation: Han L, Qin H, Wang J, Yao D, Zhang L, Guo J and Zhu B (2023) Immediate response of paddy soil microbial community and structure to moisture changes and nitrogen fertilizer application. Front. Microbiol. 14:1130298. doi: 10.3389/fmicb.2023.1130298
Edited by:
Huan Zhong, Nanjing University, ChinaReviewed by:
Yucheng Wu, Institute of Soil Science, Chinese Academy of Sciences (CAS), ChinaXi-En Long, Nantong University, China
Yongping Kou, Chengdu Institute of Biology, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Han, Qin, Wang, Yao, Zhang, Guo and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiahua Guo, amlhaHVhX2d1b0Bud3UuZWR1LmNu; Baoli Zhu, YmFvbGkuemh1QGlzYS5hYy5jbg==
 Linrong Han
Linrong Han Hongling Qin
Hongling Qin Jingyuan Wang2,3
Jingyuan Wang2,3 Jiahua Guo
Jiahua Guo Baoli Zhu
Baoli Zhu