- 1Key Laboratory of Animal Immunology of the Ministry of Agriculture, Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Zhengzhou, China
- 2School of Physical Education and Health Administration, Henan Finance University, Zhengzhou, China
Introduction
Zoonotic spillover is one of the most serious threats to public health. Outbreaks of zoonotic viruses, such as avian influenza virus (Miranda et al., 2022) and monkeypox virus (Bragazzi et al., 2022), as well as the ongoing epidemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (da Silva Torres et al., 2022) highlight their hazards to the global public health and economy.
Arteriviruses are enveloped positive-strand RNA viruses in the Nidovirale order along with coronaviruses (Snijder et al., 2013). These viruses have been previously shown to infect equids, mice, swine, and non-human primates (Brinton and Snijder, 2008). Of note, simian hemorrhagic fever virus (SHFV), an arterivirus endangering non-human primates, has been recently reported to be poised for spillover to human via an intracellular receptor CD163, which raises an alert on a potential cross-species transmission (Warren et al., 2022). Under One Health, we assessed the potential cross-species transmission of arteriviruses and further evaluated the interspecies receptor usage by arteriviruses to express our concern over the emerging risk of arteriviruses to breach the interspecies boundaries among animals and human.
Cross-species transmission of arteriviruses
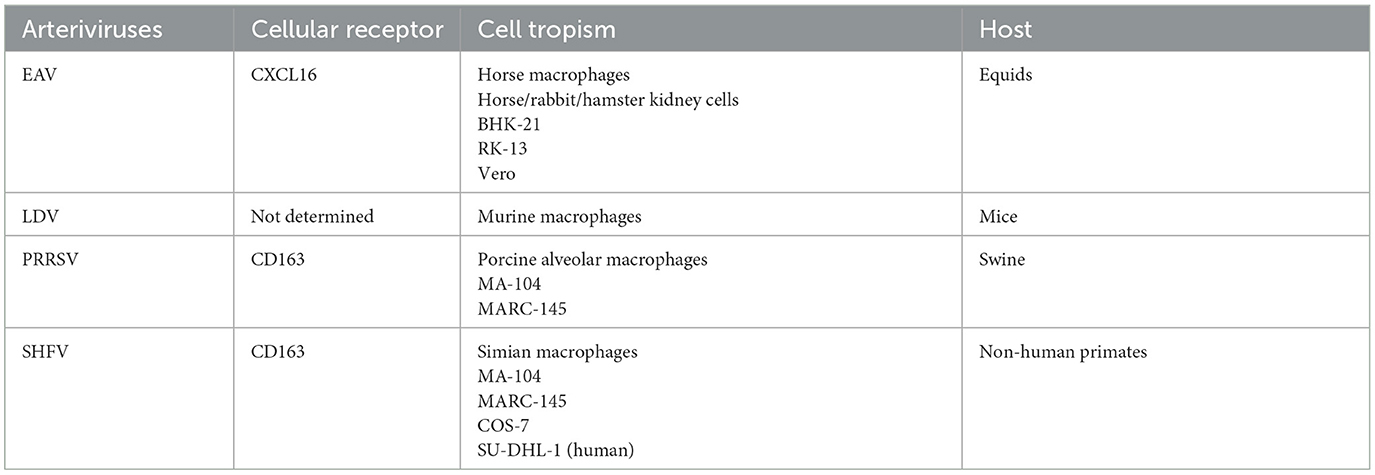
As shown in Figure 1A, arteriviruses include equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), porcine reproductive and respiratory syndrome virus (PRRSV), and SHFV (Snijder et al., 2013). These viruses establish natural persistent infections in equids (EAV), mice (LDV), swine (PRRSV), and non-human primates (SHFV), and propagate primarily in their respective host macrophages (Brinton and Snijder, 2008). However, certain arteriviruses replicate efficiently in a wide variety of cells without species specificity (Table 1).
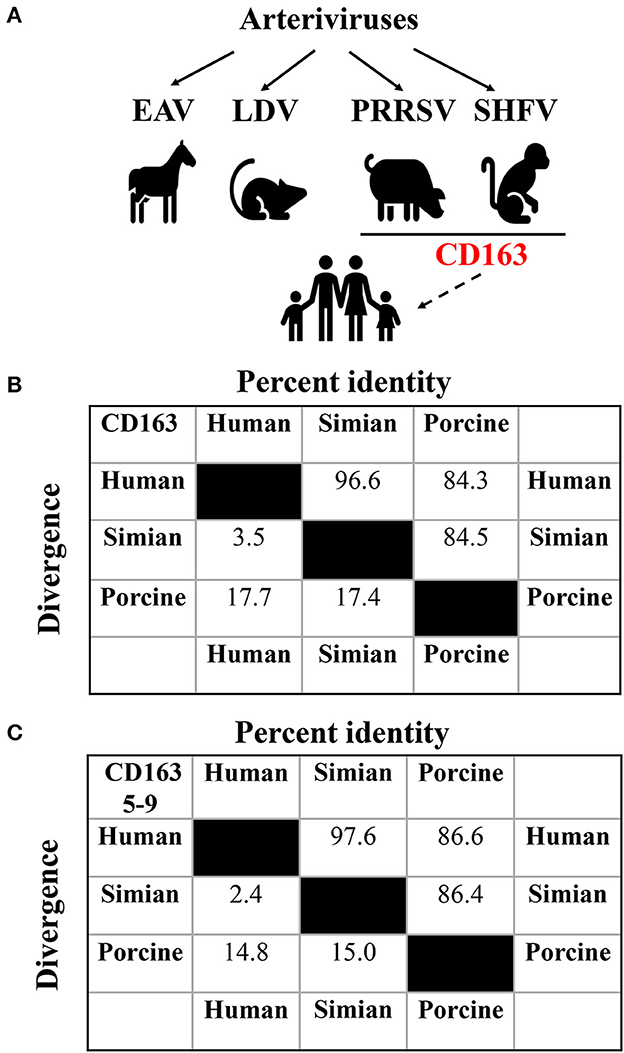
Figure 1. Assessment of potential arteriviral spillover to human. (A) Potential cross-species transmission of arteriviruses. The solid lines indicate documented transmission, while the dashed one indicates potential transmission. The putative generic receptor CD163 for SHFV and PRRSV spillover to human is highlighted in red. (B) Analysis of the sequence identity among human (UniProt entry Q86VB7), simian (UniProt entry Q2VLG4), and porcine (UniProt entry Q2VL90) CD163 using CLUSTAL W method of DNASTAR Lasergene software (Madison, USA). (C) Analysis of the sequence identity among human, simian, and porcine CD163 SRCR5-9 using CLUSTAL W method of DNASTAR Lasergene software as described in (B).
For example, EAV is able to proliferate in primary horse, rabbit, and hamster kidney cells as well as various cell lines, such as baby hamster kidney (BHK)-21, rabbit kidney (RK)-13, and African green monkey kidney Vero cells (Payne, 2017). PRRSV is capable of infecting African green monkey kidney epithelial MA-104 and its derivative MARC-145 cells (Kim et al., 1993). In addition to simian macrophages, SHFV has been shown to multiply in MA-104 and MARC-145 cells, whereas it is recently demonstrated to enter into and replicate in human histiocytic SU-DHL-1 cells as described above (Warren et al., 2022). Therefore, arteriviruses possess an inherent potential for dissemination to other cell species.
Interspecies receptor usage by arteriviruses
As a host cellular receptor usually determines viral cell tropism and even host range, we further evaluated the interspecies receptor usage by arteriviruses based on analyzing the amino acid sequence identities of arteriviral receptors amongst different species.
CD163 is a class I scavenger receptor (SR) containing nine SR cysteine-rich (SRCR) domains (SRCR1-9), and functions in multiple physiological and pathological processes (Van Gorp et al., 2010). Simian CD163 is a crucial cell factor for SHFV entry into MA-104 and MARC-145 cells (Caı̀ et al., 2015), and human CD163 is exploited by SHFV as an intracellular receptor to infect SU-DHL-1 cells (Warren et al., 2022). Therefore, we firstly determined the amino acid sequence identity between human and simian CD163 for SHFV. In Figure 1B, human and simian CD163 share almost identical amino acid sequences (96.6% identity). Since CD163 SRCR5-9 is supposed to be the putative arteriviral interaction domain (Warren et al., 2022), we further conducted a comparative analysis of the amino acid sequences between human and simian CD163 SRCR5-9, and it shows a higher sequence identity (97.6% identity, Figure 1C). These high sequence identities may partly explain why SHFV can utilize human CD163 for transmission to human cells. In the meantime, these analyses remind us of another potential arteriviral spillover by PRRSV, which utilizes porcine CD163 as an indispensable receptor (Whitworth et al., 2016). As mentioned above, PRRSV infects simian CD163-expressed MA-104 and MARC-145 cells as SHFV does (Kim et al., 1993). More importantly, expression of CD163 from different species (including porcine, simian, and human CD163) renders permissiveness to PRRSV infection in numerous refractory cells (Calvert et al., 2007). Interestingly, porcine CD163 only shows 84.5% and 84.3% in sequence identity compared to simian and human versions, respectively (Figure 1B), whereas porcine CD163 SRCR5-9 shows 86.4 and 86.6% in sequence identity compared to simian and human versions, respectively (Figure 1C). These analyses suggest that CD163 can function as a generic receptor for PRRSV to cross cell species despite sequence divergence within a certain range.
Equine CXCL16 has been identified as an entry receptor for EAV in equine cells. Additionally, non-permissive human embryonic kidney (HEK)-293T cells become susceptible to EAV infection after stably expressing equine CXCL16 (Sarkar et al., 2016). However, CXCL16 from different species share a relatively low sequence identity (data not shown). As a consequence, whether CXCL16 is a determinant for EAV broad cell tropism remains to be demonstrated, and there may exist other genuine generic receptors for EAV.
Discussion
Indeed, the cell tropism of arteriviruses may not be applicable to their in vivo infections. Fortunately, infection by arteriviruses hasn't yet documented in human until now. However, three bat-originated coronaviruses, severe acute respiratory syndrome coronavirus, Middle East respiratory syndrome coronavirus, and SARS-CoV-2 have broken the original and intermediate host barriers to infect human upon recognizing human versions of receptors (Cui et al., 2019; Li et al., 2020; Frutos et al., 2021). It is acknowledged that coronavirus evolution via genetic mutation contributes to viral adaption to human receptors and cross-species transmission (Cui et al., 2019; Zhai et al., 2020). Considering this fact, we cannot preclude the possibility that SHFV and PRRSV may also evolve to gain a capability of infecting human via efficient usage of human CD163. As a consequence, comprehensive investigation on the interaction between SHFV/PRRSV and CD163 from different species is imperative to evaluate their potential spillover to human from the receptor perspective. Structural studies on SHFV/PRRSV proteins in complex with human, simian, or porcine CD163 will be beneficial for understanding receptor hotspots for viral binding and interspecies receptor usage.
In addition to receptors, host cellular restriction factors and immune responses, especially innate immune responses as the first line of defense, have been identified to determine viral cross-species transmission. Therefore, it is required to elucidate whether there exist certain host cellular restriction factors or different host innate immune response levels to prevent arteriviral replication in human.
SHFV causes a fatal hemorrhagic fever in macaque colonies worldwide and primate facilities in several countries. PRRSV is a pathogen of veterinary importance and leads to significant economic losses in the global swine industry. They actually pose an emerging threat to public health for close contact exists among human and arteriviral hosts, exemplified by non-human primate facility workers and infected non-human primates, or pig farmers and sick pigs. As the analyses described above suggest a potential interspecies transmission of arteriviruses, we propose a systemic surveillance in arteriviral hosts and people with close contact to prevent potential outbreaks through serological and virological tests along with other One Health approaches.
Author contributions
RL and GZ conceived the study. RL and HM conducted the analyses. RL wrote the manuscript. RL, HM, SQ, and GZ revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32272993), the Fund for Distinguished Young Scholars from Henan Academy of Agricultural Sciences (2021JQ01), the Earmarked Fund for CARS (CARS-35), and the Pig Industry Technology System Innovation Team Project of Henan Province (HARS-22-12-S).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bragazzi, N. L., Woldegerima, W. A., Iyaniwura, S. A., Han, Q., Wang, X., Shausan, A., et al. (2022). Knowing the unknown: the underestimation of monkeypox cases. Insights and implications from an integrative review of the literature. Front. Microbiol. 13, 1011049. doi: 10.3389/fmicb.2022.1011049
Brinton, M. A., and Snijder, E. J. (2008). “Arteriviruses” in Encyclopedia of Virology, 3rd Edn., eds B. W. J. Mahy and M. H. V. Van Regenmortel (Oxford: Academic Press), 176–186. doi: 10.1016/B978-012374410-4.00537-9
Caì, Y., Postnikova, E. N., Bernbaum, J. G., Yú, S. Q., Mazur, S., Deiuliis, N. M., et al. (2015). Simian hemorrhagic fever virus cell entry is dependent on CD163 and uses a clathrin-mediated endocytosis-like pathway. J. Virol. 89, 844–856. doi: 10.1128/JVI.02697-14
Calvert, J. G., Slade, D. E., Shields, S. L., Jolie, R., Mannan, R. M., Ankenbauer, R. G., et al. (2007). CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81, 7371–7379. doi: 10.1128/JVI.00513-07
Cui, J., Li, F., and Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192. doi: 10.1038/s41579-018-0118-9
da Silva Torres, M. K., Bichara, C. D. A., de Almeida, M., Vallinoto, M. C., Queiroz, M. A. F., Vallinoto, I., et al. (2022). The complexity of SARS-CoV-2 infection and the COVID-19 pandemic. Front. Microbiol. 13, 789882. doi: 10.3389/fmicb.2022.789882
Frutos, R., Serra-Cobo, J., Pinault, L., Lopez Roig, M., and Devaux, C. A. (2021). Emergence of bat-related betacoronaviruses: hazard and risks. Front. Microbiol. 12, 591535. doi: 10.3389/fmicb.2021.591535
Kim, H. S., Kwang, J., Yoon, I. J., Joo, H. S., and Frey, M. L. (1993). Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133, 477–483. doi: 10.1007/BF01313785
Li, R., Qiao, S., and Zhang, G. (2020). Analysis of angiotensin-converting enzyme 2 (ACE2) from different species sheds some light on cross-species receptor usage of a novel coronavirus 2019-nCoV. J. Infect. 80, 469–496. doi: 10.1016/j.jinf.2020.02.013
Miranda, M. N. S., Pingarilho, M., Pimentel, V., Torneri, A., Seabra, S. G., Libin, P. J. K., et al. (2022). A tale of three recent pandemics: influenza, HIV and SARS-CoV-2. Front. Microbiol. 13, 889643. doi: 10.3389/fmicb.2022.889643
Payne, S. (2017). “Chapter 18 - family arteriviridae” in Viruses, ed S. Payne (Oxford, UK: Academic Press), 159–163. doi: 10.1016/B978-0-12-803109-4.00018-0
Sarkar, S., Chelvarajan, L., Go, Y. Y., Cook, F., Artiushin, S., Mondal, S., et al. (2016). Equine arteritis virus uses equine CXCL16 as an entry receptor. J. Virol. 90, 3366–3384. doi: 10.1128/JVI.02455-15
Snijder, E. J., Kikkert, M., and Fang, Y. (2013). Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 94(Pt 10), 2141–2163. doi: 10.1099/vir.0.056341-0
Van Gorp, H., Delputte, P. L., and Nauwynck, H. J. (2010). Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol. Immunol. 47, 1650–1660. doi: 10.1016/j.molimm.2010.02.008
Warren, C. J., Yu, S., Peters, D. K., Barbachano-Guerrero, A., Yang, Q., Burris, B. L., et al. (2022). Primate hemorrhagic fever-causing arteriviruses are poised for spillover to humans. Cell 185, 3980–3991.e18. doi: 10.1016/j.cell.2022.09.022
Whitworth, K. M., Rowland, R. R., Ewen, C. L., Trible, B. R., Kerrigan, M. A., Cino-Ozuna, A. G., et al. (2016). Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22. doi: 10.1038/nbt.3434
Keywords: zoonotic spillover, cross-species transmission, arterivirus, SHFV, CD163
Citation: Li R, Ma H, Qiao S and Zhang G (2023) Potential arteriviral spillover: An emerging threat to public health? Front. Microbiol. 14:1156327. doi: 10.3389/fmicb.2023.1156327
Received: 01 February 2023; Accepted: 17 February 2023;
Published: 02 March 2023.
Edited by:
Qin Zhao, Northwest A&F University, ChinaReviewed by:
Biyun Xue, The University of Iowa, United StatesGaopeng Hou, Washington University in St. Louis, United States
Copyright © 2023 Li, Ma, Qiao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Li, bGlydWk4NjA2MjBAc2luYS5jb20=; Gaiping Zhang, emhhbmdnYWlwQDEyNi5jb20=
†These authors have contributed equally to this work
 Rui Li
Rui Li Hongfang Ma2†
Hongfang Ma2† Gaiping Zhang
Gaiping Zhang