- 1Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan, China
- 2Engineering Research Center for Replacement Technology of Feed Antibiotics of Yunnan College, Kunming, Yunnan, China
Numerous different species of LAB are used in different fields due to their unique characteristics. However, Lacticaseibacillus chiayiensis, a newly established species in 2018, has limited microorganism resources, and lacks comprehensive evaluations of its properties. In this study, L. chiayiensis AACE3, isolated from fermented blueberry, was evaluated by genomic analysis and in vitro assays of the properties. The genome identified genes associated with biofilm formation (luxS, ccpA, brpA), resistance to oxidative stress (tpx, trxA, trxB, hslO), tolerance to acidic conditions (dltA, dltC), resistance to unfavorable osmotic pressure (opuBB, gbuA, gbuB, gbuC), and adhesion (luxS, dltA, dltC). The AACE3 showed 112 unique genes, relative to the other three L. chiayiensis strains. Among them, the presence of genes such as clpP, pepO, and feoA suggests a possible advantage of AACE3 over other L. chiayiensis in terms of environmental adaptation. In vitro evaluation of the properties revealed that AACE3 had robust antibacterial activity against eight common pathogens: Streptococcus agalactiae, Staphylococcus aureus, Escherichia coli, Salmonella enteritidis, Salmonella choleraesuis, Shigella flexneri, Pseudomonas aeruginosa, and Klebsiella pneumoniae. In addition, AACE3 showed more than 80% survival rate in all tests simulating gastrointestinal fluid, and it exhibited high antioxidant capacity. Interestingly, the cell culture supernatant was superior to intact organisms and ultrasonically crushed bacterial extracts in all tests of antioxidant capacity. These results suggested that the antioxidant capacity may originate from certain metabolites and extracellular enzymes produced by AACE3. Moreover, AACE3 was a moderate biofilm producer due to the self-agglomeration effect. Taken together, L. chiayiensis AACE3 appears to be a candidate strain for combating the growing incidence of pathogen infections and antioxidant production.
1. Introduction
Lactic acid bacteria (LAB) can produce a variety of extracellular polysaccharides, antimicrobial compounds and short-chain fatty acids, and have the advantage of being non-pathogenic, acid tolerant and bile tolerant (Mojgani et al., 2015; Vasconcelos et al., 2019). Several studies have confirmed that LAB have a variety of applications such as use in food fermentation to enhance food flavor, as food additives to promote host health, while LAB-produced bacteriocins are used in food preservation and antimicrobial therapy (Gerritsen et al., 2011; Kok and Hutkins, 2018; Xiang et al., 2022; Sharma et al., 2023). For example, the levels of organic acids, ethyl acetate and aromatic alcohols were increased and the organoleptic quality of the juice was improved by fermenting goji berry juice with Lactobacillus paracasei (Duan et al., 2023). During the livestock industry, the supplementation of diets with Lactobacillus acidophilus significantly reduced the abundance of potentially pathogenic Escherichia coli and increased the number of beneficial microorganisms in broilers intestine. The level of ileal amino acid digestion and humoral immunity were also increased (Zhang and Kim, 2014). In addition, Lactobacillus salivarius produced bacteriocin XJS01 showed excellent therapeutic and healing-promoting effects against Staphylococcus aureus infection in mouse skin wounds (Xiang et al., 2022). Thus, the application and exploitation of functional LAB strains in the food fermentation industry, livestock industry and other related industries has greatly increased over the years (De Melo Pereira et al., 2018). However, more studies have emerged that focus on the rational improvement of industrially useful LAB strains (Zhang et al., 2022). Therefore, more precise confirmation of the phenotypes of novel LAB strains will certainly be required before their adoption for better application purposes.
LAB must meet certain well-known criteria to fulfill their intended functions and specific applications. For example, they must be able to withstand harsh conditions such as acidity, bile salts, and proteases in the human and animal digestive tracts (Ticho et al., 2019). They should also be capable of colonizing and adhering to diverse environments, which typically depends on self-aggregation and hydrophobic properties, and is reflected in biofilm formation (Lo Curto et al., 2011). Additionally, they must be able to produce antibacterial substances like bacteriocins to combat pathogens, and should have precise taxonomy and be easily identifiable for accurate application (Vizoso Pinto et al., 2006; Sanders et al., 2010). Notably, functional genomics analysis of LAB can ensure the accurate classification of strains and provide information on genetic features, metabolic capacity, and the correlation between genotypes and phenotypes (Guinane et al., 2016; Li et al., 2017). In the post-genomic era, the selection of LAB has relied heavily on phenotypic characterization to meet the needs of applications in different fields. Therefore, technological advances in functional genomics can help to rapidly identify new isolates and evaluate their performance.
Lacticaseibacillus chiayiensis, a new lactobacillus species, was first isolated from cattle manure collected in Chiayi city (Taiwan, China) in 2018 (Huang et al., 2018). L. chiayiensis, which is closely related to L. casei and Lactobacillus zeae in terms of genomics (Kim et al., 2021). To date, only three L. chiayiensis strains have been reported with genomic information, only one L. chiayiensis strain has a complete reported genome, and few studies have integrated functional genomic data with the properties of L. chiayiensis (Kim et al., 2021). Therefore, it is critically important to build a comprehensive profile of the characteristics and genomic information of newly isolated L. chiayiensis strains.
In this study, whole-genome sequencing was conducted on L. chiayiensis AACE3 to evaluate the organism’s potential properties at the genomic level. In addition, the genomic variations between the AACE3 strain and other L. chiayiensis strains were examined by comparative genomic analysis. Finally, the in-vitro characteristics of L. chiayiensis AACE3 were experimentally evaluated, including its stress tolerance in a simulated gastrointestinal environment, antagonism profile against pathogenic bacteria, biofilm formation behavior and antioxidant capacity. These findings provide new perspectives for the comprehensive evaluation of newly isolated LAB.
2. Materials and methods
2.1. Bacterial strains and culture conditions
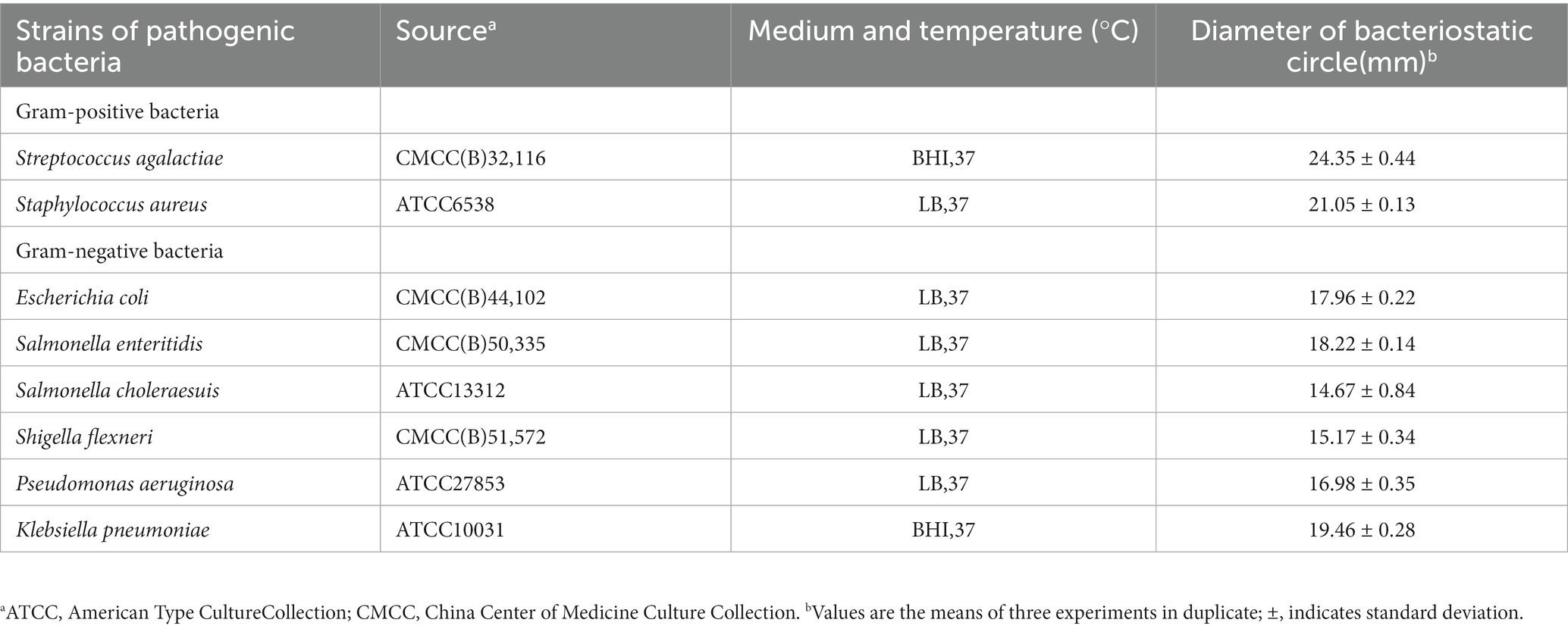
Lacticaseibacillus chiayiensis AACE3 was isolated from fermented blueberry. LAB were routinely grown in the study in MRS broth at 37°C for 24 h and reinoculated in fresh MRS broth at 37°C. The pathogens used in this study and their growth conditions are presented in Table 1. All the strains used in this study are stored at the Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming city, China.
2.2. Whole-genome sequencing, assembly, and annotation
Total genomic DNA was extracted by SDS extraction combined with column purification. DNA purity and quantity were confirmed spectrophotometrically at 260 nm using a NanoDrop One spectrophotometer (NanoDrop Technologies, Wilmington, DE). A combination of the Illumina platform (Illumina, TX, USA) and Oxford Nanopore platform (Oxford Nanopore Technologies, Oxford, UK) was used to sequence the genomes. After filtering low-quality and short-length reads from raw sequencing data, the reads were assembled using Unicycler V0.4.9 (Wick et al., 2017). Prokka V1.12 (Seemann, 2014) was utilized to predict protein-coding RNA for the assembled genome. The prediction of rRNA and tRNA sequences was performed by RNAmmer V1.2 (Lagesen et al., 2007) and Aragorn V1.2 (Laslett and Canback, 2004), respectively. Functional gene annotation was performed based on clusters of orthologous groups (COGs) (Galperin et al., 2015). The identification of secondary metabolite biosynthesis gene clusters was conducted with antiSMASH version 5.1.2 (Blin et al., 2019). The bacteriocin synthesis gene cluster of the strain was analyzed with BAGEL4 (van Heel et al., 2018).
2.3. Comparative genomic analysis
For comparative genomic and phylogenetic analyses, genomic data of L. chiayiensis and genetic information of other LAB strains were retrieved from the NCBI database,1 and the genomic information is shown in Supplementary Table S1. Average nucleotide identity (ANI) analysis was performed for the complete genome assembly using FastANI v2.0 (Jain et al., 2018). The 16S rRNA gene sequence was extracted from the whole-genome sequence, and a phylogenetic tree was constructed in MEGA-X using the neighbor-joining method. Initially, the ClustalW multiple sequence comparison method is selected for alignment, and then the Neighbor-Joining Tree is constructed. The Substitution model for calculating the genetic distance used the Maximum Composite Likelihood. The columns with more null positions in the multiple sequence comparison are processed by pairwise deletion. BPGA (Bacterial Pan Genome Analysis tool v. 1.3) was used to analyze the pangenome and core genome of the strains with default parameters (Chaudhari et al., 2016), and core gene evolutionary trees were constructed to infer the evolutionary relationships of the strains. The phylogenetic tree based on the core genes was constructed in BPGA (Bacterial Pan Genome Analysis tool v. 1.3) software using the neighbor joining method with default parameters. The “core genome” is typically defined as a set of genes shared by all strains (Lawrence and Hendrickson, 2005). COG functional classification assignment of core genomic and pangenomic genes of the AACE3 strain using USEARCH in the BPGA tool. Unique genes of AACE3 were annotated by using the KEGG database for pathway analysis.
2.4. Assessment of in vitro characteristics
2.4.1. Antibacterial activity
The antibacterial activity and antimicrobial spectrum of L. chiayiensis AACE3 against gram-positive (2) and gram-negative (6) indicators were determined by the modified Oxford cup double-layer plate method (Voulgari et al., 2010). Briefly, L. chiayiensis AACE3 was inoculated in MRS broth and cultured at 37°C for 24 h. The obtained fresh cultures were centrifuged at 8000 × g at 4°C for 15 min in a high-speed refrigerated centrifuge (Thermo Scientific, Waltham, MA, USA). The cell-free supernatant (CFS) was sterilized using a 0.22 μm filter membrane before use. Then, the indicated pathogens (100 μL, 1 × 107 CFU/mL) were mixed with 5 mL of semisolid LB agar medium and quickly poured onto the surface of solid LB agar medium. The Oxford cups were placed on the agar surface, 200 μL of CFS was added, and the plates were allowed to stand at 4°C until the CFS had fully diffused. Then, the plates were transferred to a constant-temperature incubator at 37°C for 24 h to observe the bacterial inhibitory activity.
2.4.2. Tolerance to simulated gastrointestinal conditions and bile salts
In vitro simulated gastrointestinal fluid and bile salt tolerance models were developed based on previous studies with modifications (Zhao et al., 2021). The artificial gastric juice was prepared by mixing 10 g/L pepsin (Solarbio, Beijing, China) with 16.4 mL of 0.1 mol/L HCl, and the pH was adjusted to 2.0, 3.0, and 4.0 with 1 mol/L NaOH. The prepared solutions were sterilized using a 0.22 μm filter membrane before use. For the artificial intestinal juice preparation, 10.0 g/L trypsin (Solarbio, Beijing, China) and 6.8 g KH2PO4 were mixed in 500 mL of sterile ddH2O, and the pH was adjusted to 6.8 with 1 mol/L NaOH; the solution was then filtered through a 0.22 μm membrane. The LAB suspension was inoculated in 5 mL of simulated intestinal juice and simulated gastric juice at pH 2.0, 3.0 and 4.0 and incubated at 37°C for 3 and 4 h. The LAB suspension was inoculated in MRS broth containing 0.3% bile salts and cultured at 37°C for 24 h. The survival rate was calculated using the plate count method with the following formula:
where N0 is the total number of viable bacteria in the untreated group, and N1 is the total number of viable bacteria after treatment with artificial gastrointestinal juice or bile salts.
2.4.3. Autoaggregation
The autoaggregation tests were performed according to a previously described method (Kim et al., 2021). Briefly, bacterial cultures were centrifuged and resuspended in PBS, and the absorbance at 600 nm (OD600, A0) was adjusted to 0.6. The adjusted mixture was shaken and vortexed for 20 s and then incubated at room temperature for 3 h and 24 h. The absorbance of the supernatant at 600 nm was measured at 3 h and 24 h (At). The autoaggregation was calculated with the following formula:
2.4.4. Cell surface hydrophobicity
The cell surface hydrophobicity was determined as previously described (Fadare et al., 2022). Freshly cultured LAB strains were centrifuged and resuspended in PBS, and the absorbance at 600 nm (OD0) was adjusted to 0.8 ± 0.2. Then, 1 mL of ethyl acetate or xylene was added to 5 mL of LAB suspension and vortexed for 2 min. After incubation at 37°C for 1 h, the aqueous phase was collected, and the absorbance at 600 nm was measured. The hydrophobicity value was calculated according to the following formula:
where OD0 and OD indicate the absorbance before and after mixing with ethyl acetate or xylene, respectively.
2.4.5. Determination of biofilm formation
The biofilm formation of L. chiayiensis AACE3 was quantitatively determined following previously reported methods with minor modifications (Benmouna et al., 2020). The bacterial cultures (grown for 24 h) were washed twice with PBS and resuspended to an OD600 of 1.0. Fifty microliters of bacterial suspension and 150 μL sterile MRS broth were added to a 96-well plate and incubated at 37°C for 24 h, while 200 μL of sterile MRS broth was used as a negative control. After 24 h, the medium in the wells was poured out, and sterile PBS was slowly added to the wells to wash the unattached bacteria; three repeat washes were performed. After drying at room temperature for 20 min, the biofilms were fixed with 10% methanol solution for 30 s. Then, 1% crystalline violet dye was used to stain the biofilms for 20 min, and the stained biofilms were washed three times with sterile water to remove excess stain. Finally, 200 μL of anhydrous ethanol was added to each well, and the optical density of the stained adnexal cells was measured at 595 nm. ODt and ODc represent the results of the experimental and control groups, respectively. The biofilm production capacity was measured as follows: ODt ≤ ODc indicated nonbiofilm producer (0), ODc < ODt ≤ 2ODc indicated a weak biofilm producer (+), 2ODc < ODt ≤ 4ODc indicated a moderate biofilm producer (++), and ODt > 4ODc indicated a robust biofilm producer (+++).
2.5. Antioxidant activity
2.5.1. Preparation of samples
The CFS of the strains, the washed and resuspended intact organism (IO), and the ultrasonically fragmented extract of the organism were used for the antioxidant viability test. The CFS was collected by centrifugation (8,000 × g, 15 min) of AACE3 (grown for 24 h) at 4°C. IOs were washed three times with PBS (pH 7.4) and resuspended in PBS at a concentration of 1.0 × 109 CFU/mL. Ultrasonically crushed bacterial extracts (UCBEs) were obtained by ultrasonic crushing in an ice bath for 15 min, and the supernatant was obtained by centrifugation (8,000 × g, 15 min) at 4°C. The CFS and UCBE were filtered with 0.22 μm filter membranes and used for further experiments.
2.5.2. Hydroxyl radical scavenging
The hydroxyl radical scavenging assay was performed as previously reported (Oyaizu, 1988). Then, 2.5 mmol/L 1,10-phenanthroline (1.0 mL), 2.5 mmol/L FeSO4 (1.0 mL), and PBS (1.0 mL, pH 7.4) were added to 1 mL of each sample. After thorough mixing, 20 mmol/L H2O2 (1 mL) was added, and the sample was placed in a constant-temperature water bath at 37°C for 90 min, and the absorbance at 517 nm was measured. The hydroxyl radical scavenging rate was calculated by the following formula:
The absorbance value of the samples measured in the experimental group was As. Distilled water was used instead of the experimental sample in a blank group to measure the absorbance value Ab, and distilled water was used instead of H2O2 to measure the absorbance value An.
2.5.3. DPPH radical scavenging
The measurement of DPPH radical scavenging capacity was based on a previous study with minor adjustments (Lin and Yen, 1999). One milliliter of preprepared sample was mixed with 1 mL of 0.2 mmol/L DPPH solution and placed in the dark for 30 min, and the absorbance at 517 nm was measured. The DPPH radical scavenging rate was calculated by the following formula:
The absorbance value of the samples measured in the experimental group was As. Distilled water was used instead of the experimental sample as a control group to measure the absorbance value Ac, and absolute alcohol was used instead of H2O2 to measure the absorbance value Ab.
2.5.4. Superoxide anion scavenging ability
The superoxide anion radical scavenging performance was evaluated according to a previous study (Li et al., 2012). One milliliter of preprepared sample and 3.0 mL of Tris–HCl solution (pH 8.2, 50 mM) were mixed and placed at room temperature for 20 min. Then, 0.4 mL of 25 mmol/L pyrogallol was added, and the sample was placed in the dark for 4 min. Then, 0.5 mL of 8 mmol/L HCl was added to end the reaction. The absorbance at 325 nm was recorded. The superoxide anion radical scavenging rate was calculated by the following formula:
where As represents the absorbance of each sample. The blank contained distilled water instead of the sample used in the reaction system.
2.6. Statistical analysis
All experimental data were obtained from at least three independent replicates and are expressed as the mean ± standard deviation (SD). The statistical significance between two groups was assessed by using the unpaired two-tailed t test by using GraphPad Prism 8.0.2 (GraphPad Software Inc., La Jolla, CA, USA). The statistically significant differences between multiple groups were analyzed by one-way ANOVA by using GraphPad Prism 8.0.2 (GraphPad Software Inc., La Jolla, CA, USA). The criterion for statistical significance was a p value <0.05.
3. Results
3.1. General genomic characteristics
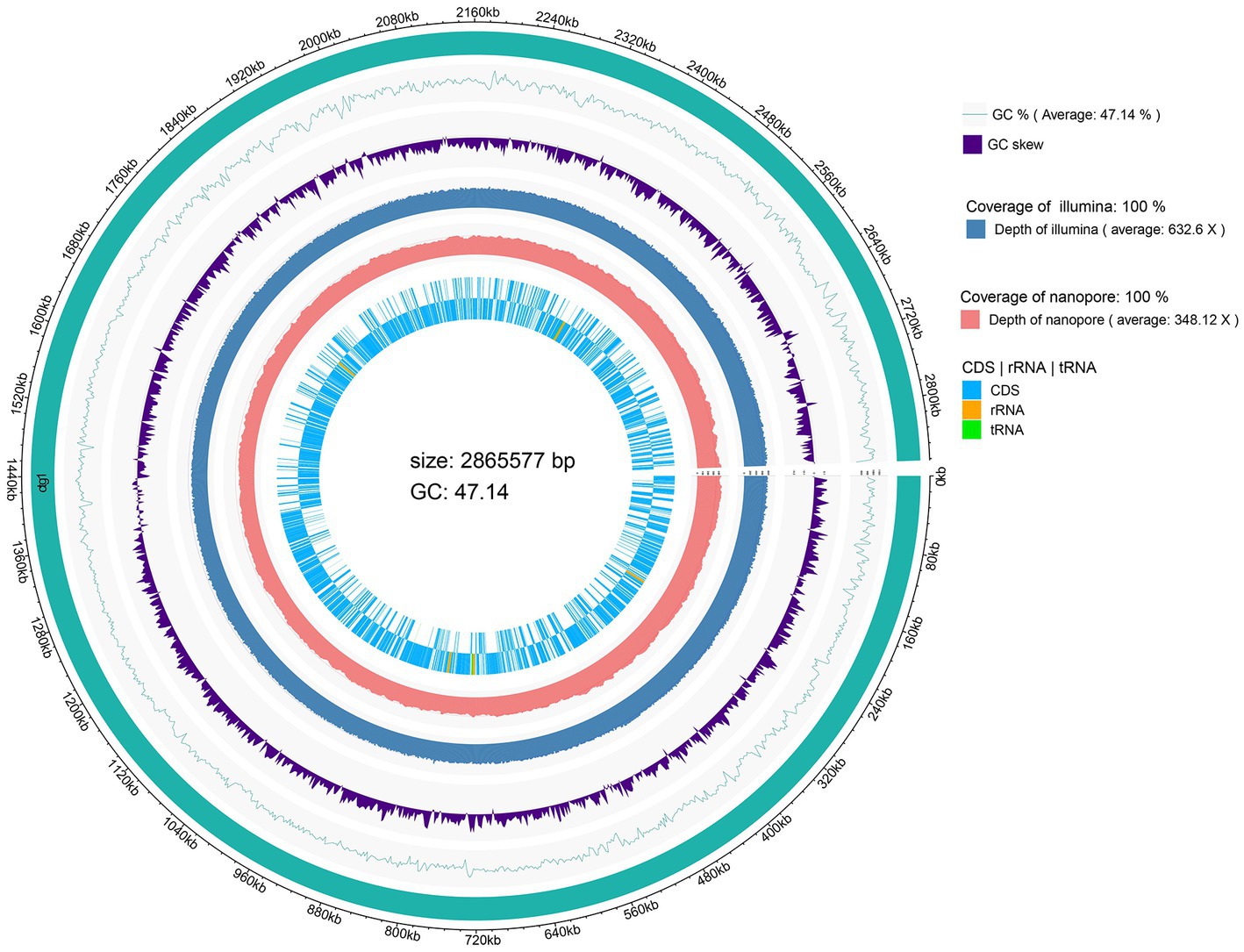
The genome size of L. chiayiensis AACE3 was 2,865,577 bp, with an average GC content of 47.14% (Figure 1). A total of 2,758 protein-coding sequences (CDSs) were identified among 2,870 predicted genes. In addition, 261 pseudogenes, 58 tRNAs, 15 rRNA manipulators (including 5 23S rRNAs, 5 16S rRNAs, 5 5S rRNAs) and 3 clustered regularly interspaced short palindromic repeat (CRISPR) sequences were predicted in the genome. All protein domains were classified into 21 functional categories according to the COG database, mainly carbohydrate transport and metabolism (G, 206), translation/ribosome structure and biogenesis (J, 145), amino acid transport and metabolism (E, 118), cell wall/membrane/envelope biogenesis (M, 80) and replication/recombination/repair (L, 79) (Supplementary Figure S1). The whole-genome sequence of L. chiayiensis has been submitted to GenBank with the accession number CP107523.1.
3.2. Phylogenetic analysis of Lacticaseibacillus chiayiensis
The phylogenetic relationships of L. chiayiensis AACE3 with other related species were analyzed. The phylogenetic tree was constructed based on 16S rRNA sequences, which showed that the L. chiayiensis AACE3 strain shared 99.86–100% identity with other L. chiayiensis isolates (Figure 2A). However, the identity with L. zeae, Lacticaseibacillus paracasei, and Lacticaseibacillus rhamnosus was more than 98%, exceeding the species classification threshold of 97% for 16S rRNA (Supplementary Table S2), indicating failure to effectively detect species-specific differences. The ANI assay between the genomes of L. chiayiensis AACE3 and other strains clearly distinguished L. chiayiensis from L. zeae, L. paracasei, and L. rhamnosus (Figure 2B). The genome of L. chiayiensis AACE3 was most similar to that of L. chiayiensis NCYUAS (99.81%) compared to those of other L. chiayiensis strains. In addition, in a core genome-based phylogenetic analysis, the evolutionary relationship between strain AACE3 and strain NCYUAS was shown to be the closest, with this result being similar to that obtained by ANI analysis (Figure 2C).
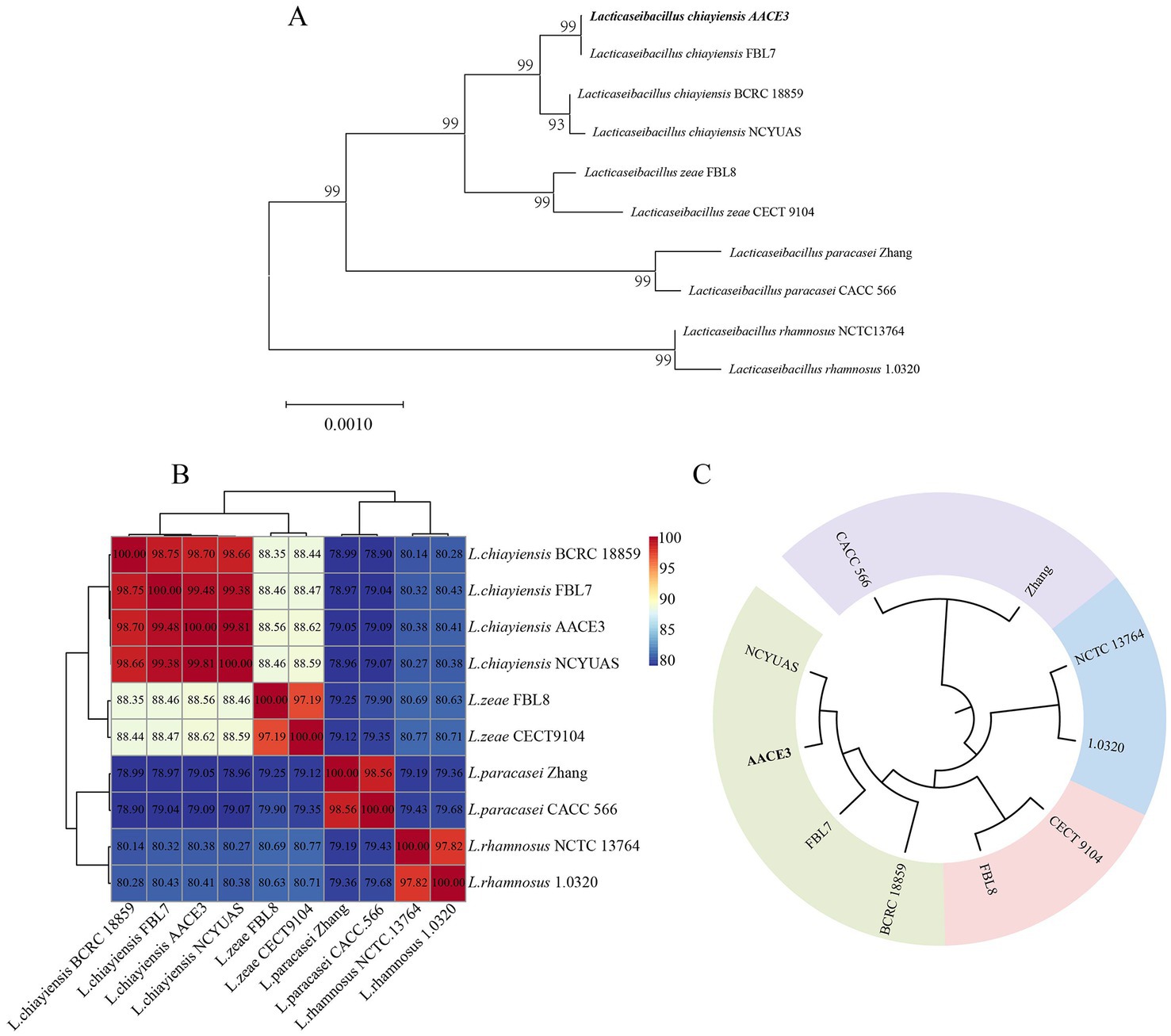
Figure 2. Phylogenetic analysis of L. chiayiensis AACE3 based on (A) 16S rRNA gene sequences and (B) average nucleotide identity (C) of the core genome.
3.3. Genomic synteny and pangenomic and core genome analyses
To explore the genetic differences between the L. chiayiensis strains, L. chiayiensis FBL7 and L. chiayiensis AACE3, which were selected as complete at the genomic level, were analyzed for genomic synteny. As shown in Figures 3A, a large homologous region between L. chiayiensis FBL7 and L. chiayiensis AACE3 was found, and the close relationship between the two strains was consistent.
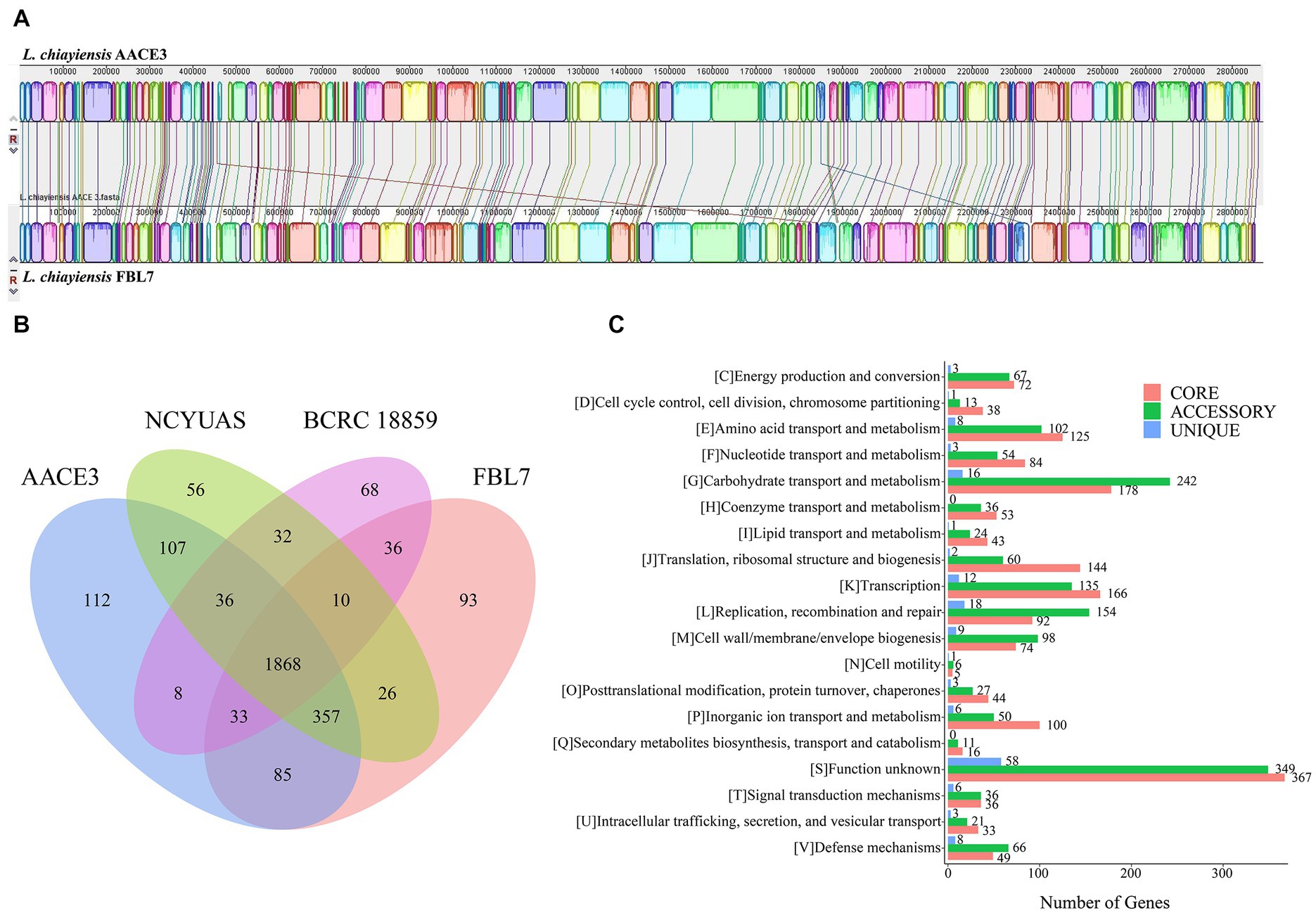
Figure 3. Comparative genomic analysis of L. chiayiensis AACE3 with three closely related Lacticaseibacillus species. (A) Genome-to-genome alignment of L. chiayiensis AACE3 and L. chiayiensis FBL7 using a progressive mauve software with a window of 1,000 nucleotides. Boxes with the same color indicate the syntenic regions. (B) Venn diagram showing the number of genes of orthologous CDSs shared and unique among the three strains L. chiayiensis AACE3, L. chiayiensis FBL7, L. chiayiensis NCYUAS and L. chiayiensis BCRC 18859. (C) The number of core, accessory, and unique genes among L. chiayiensis genomes assigned in cluster of orthologous group (COG) categories.
Four pangenomes of the L. chiayiensis strains consisted of 1868 core genes, 1896 accessory genes and 329 unique genes (Figure 3B). Among them, 1719 genes in the core genome were assigned to 19 COG categories. Most core genes were assigned to the functional categories of carbohydrate transport and metabolism (G, 178), transcription (K, 166) and translation, ribosomal structure and biogenesis (J, 144) (Figure 3C).
To further investigate the genetic characteristics and functional properties of L. chiayiensis AACE3, unique genes of L. chiayiensis AACE3 were classified using the COG and KEGG annotation databases. The 112 unique genes belonging to L. chiayiensis AACE3 encoded 106 hypothetical proteins and 6 known proteins, of which only 28 unique genes were annotated by the COG database and were classified into 10 COG categories (Supplementary Table S3). Two genes were assigned to the defense mechanisms category, containing genes encoding ABC-type bacteriocin lantibiotic exporters (locus_01810). Two genes were assigned to the posttranslational modification, protein turnover, and chaperones category, and one gene was assigned to the intracellular trafficking, secretion, and vesicular transport category, containing genes encoding the ATP-dependent clp protease proteolytic subunit (locus_00468). Two genes were assigned to the inorganic ion transport and metabolism category, including the feoA (locus_01741) gene. In addition, unique genes, including the lexA repressor (locus_00434), a key regulator in the bacterial SOS response, were detected.
3.4. Annotation of genes coded by Lacticaseibacillus chiayiensis AACE3 that are implicated in stress response
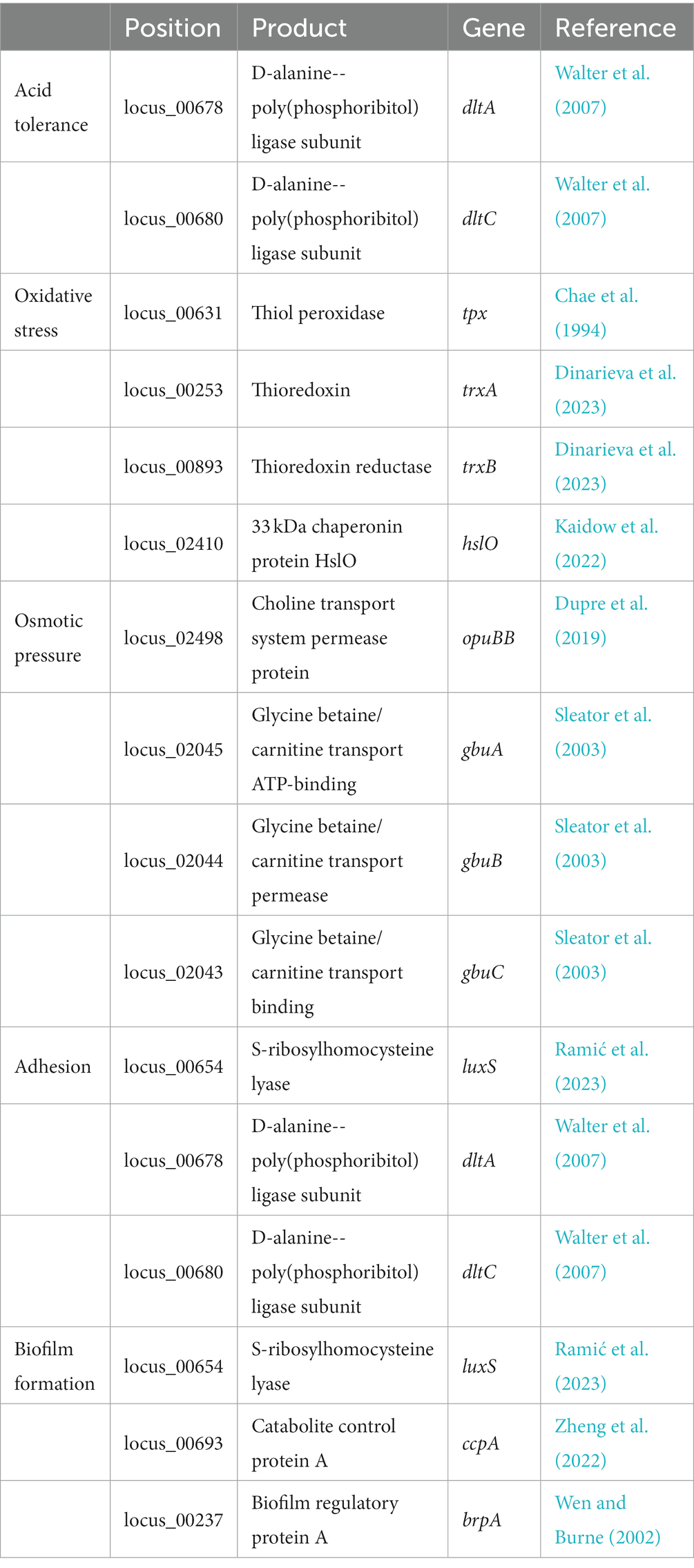
Several proteins encoded in the L. chiayiensis AACE3 genome that are associated with tolerance to stress factors, such as unfavorable osmotic pressure (i.e., anuBB, ghux, gbuB, gbuC), and acidic environments (i.e., dltA, dltC), have been identified. In addition, the presence of luxS, dltA, and dltC genes in the genome was shown to be associated with bacterial adhesion and colonization. As well as genes responsible for resistance to oxidative stress tpx, trxA, trxB and hslO and biofilm formation luxS, ccpA and brpA genes were also identified (Table 2).
3.5. Secondary metabolite prediction analysis
According to the antiSMASH database, the strain has five ribosomal synthesis and posttranslational modification peptide product (RiPP) clusters, all of which may have the ability to produce bacteriocins (Figure 4A). Annotation of each gene for the five RiPP clusters is shown in Supplementary Table S4. The first and second RiPP-like clusters are based on the sakacin-P bacteriocin-encoding gene as the core biosynthetic gene. The core biosynthetic genes in the third and fourth clusters are Blp family class II bacteriocin genes. The fifth cluster is based on the ABC transporter protein as the core gene. In addition, unlike in other L. chiayiensis strains, ABC-type bacteriocin lantibiotic exporter-related protein genes and some hypothetical protein genes are present in the second and fourth clusters of L. chiayiensis AACE3.
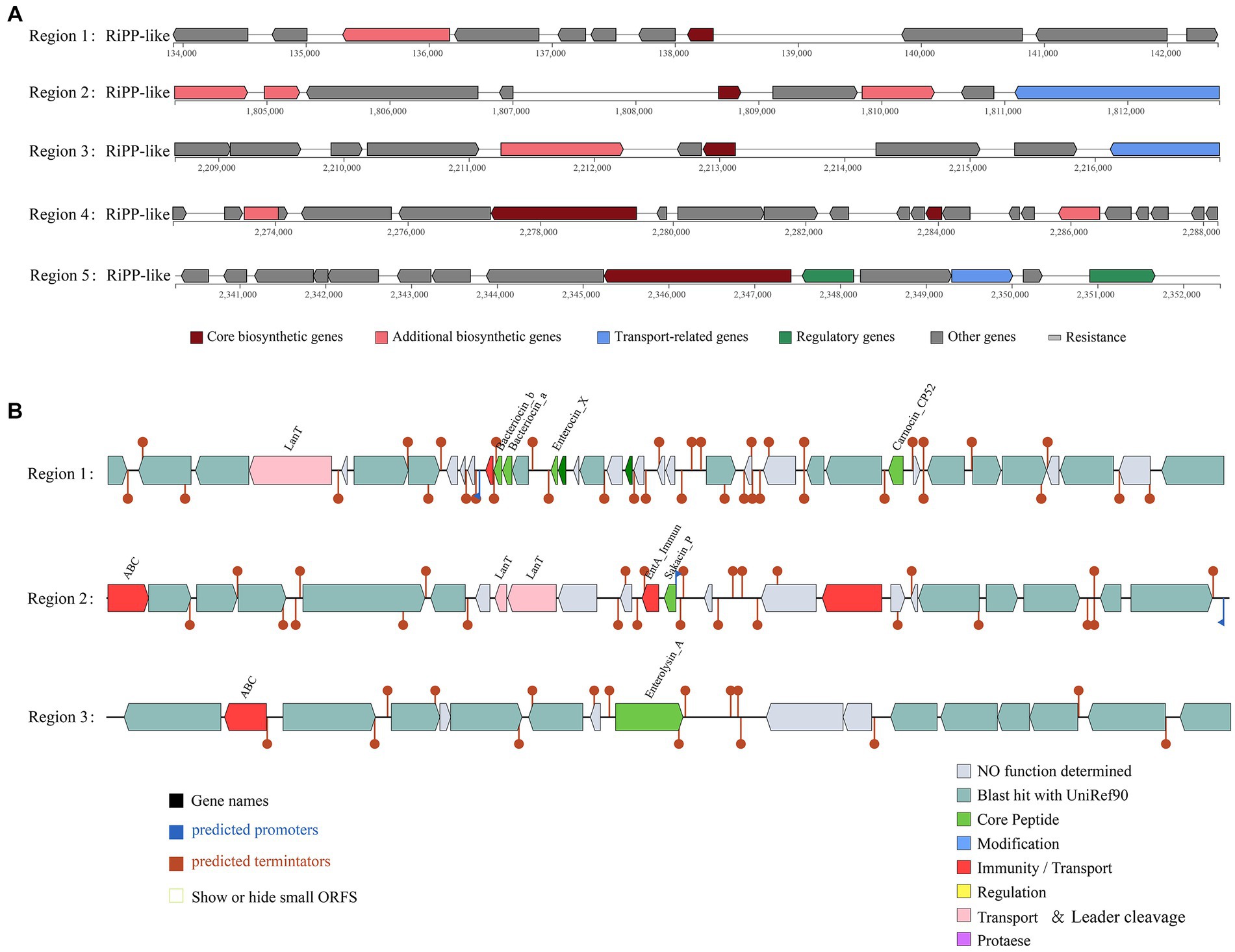
Figure 4. Prediction of antimicrobial activity-associated protein structures in the genome of L. chiayiensis AACE3. (A) Synthetic gene cluster of ribosomal synthesis and posttranslational modification peptide product (RiPP; based on antiSMASH database prediction). (B) Three bacteriocin (region_1: Enterocin_X, region_2: Sakacin_P and region_3: Enterolysin_A) synthesis gene clusters (predicted based on the BAGEL4 database).
The prediction of bacteriocin gene clusters from the BAGEL4 database showed that three bacteriocin synthesis gene clusters were present (Figure 4B). One cluster had enterocin_X, carnocin_CP52 and two antimicrobial peptides as core genes and included an immunity gene and a transport protein gene, and one had sakacin_P as a core gene and included three bacteriocin immunity/transport genes. The third cluster contained enterolysin_A as a core gene and an ABC transporter protein gene. In addition, several other genes related to bacteriocin secretion and processing and bacteriocin autoimmune genes such as lagD and mccF were also included in these three clusters (Supplementary Table S5).
3.6. Assessment of in vitro characteristics
3.6.1. Antimicrobial activity of Lacticaseibacillus chiayiensis AACE3
Results for the inhibition activity of L. chiayiensis AACE3 against eight pathogenic strains are shown in Table 1. The results indicate that AACE3 has broad-spectrum antibacterial activity against common gram-positive and gram-negative pathogens. L. chiayiensis AACE3 showed the best inhibition activity against Streptococcus agalactiae CMCC (B) 32116 and Staphylococcus aureus ATCC 6538, with inhibition zone diameters of 24.35 ± 0.44 mm and 21.05 ± 0.13 mm, respectively. Moreover, AACE3 showed robust antibacterial activity against gram-negative indicators such as Klebsiella pneumoniae ATCC 10031 (inhibitory zone diameter of 19.46 ± 0.28 mm), Salmonella enteritidis CMCC (B) 50335 (inhibitory zone diameter of 18.22 ± 0.14 mm) and Escherichia coli CMCC (B) 44102 (inhibitory zone diameter of 17.96 ± 0.22 mm).
3.6.2. Simulated gastrointestinal digestion and bile salt tolerance assay
Table 3 shows the survival rate of L. chiayiensis AACE3 under in vitro simulated gastrointestinal digestive juice conditions. AACE3 showed the highest tolerance to the intestinal juice, with a survival rate of 99.87%. The survival rates under gastric juice treatment under all three pH gradients were more than 80%, and the highest survival rate was 98.91% at pH 4.0. AACE3 had a survival rate of 84.32 ± 5.62% after 24 h of 0.3% bile salt treatment. In conclusion, L. chiayiensis AACE3 showed good tolerance to extreme environmental factors such as acidity, bile salts and proteases.
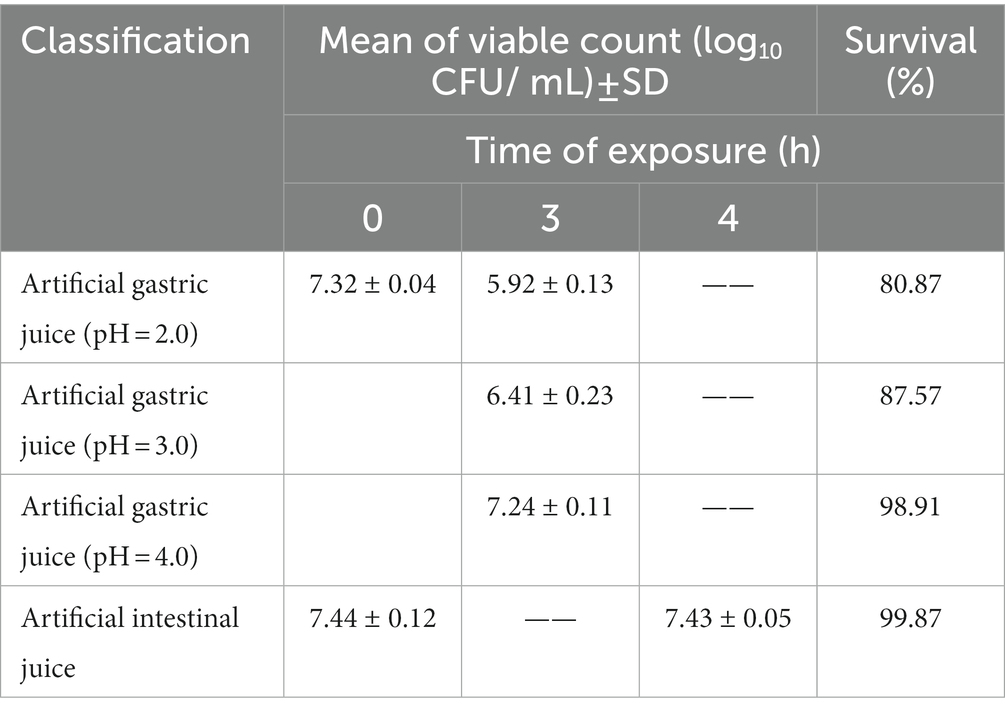
Table 3. Mean counts of L. chiayiensis AACE3 under exposure to simulated gastrointestinal fluid conditions.
3.6.3. Autoaggregation
The autoaggregation ability of strain AACE3 was examined, and the autoaggregation rate was 29.85 ± 2.56% at 3 h. After 24 h, the autoaggregation rate reached 61.25 ± 4.22%.
3.6.4. Cell surface hydrophobicity and biofilm formation capacity
AACE3 showed a decrease in optical density in the presence of ethyl acetate, with a hydrophobicity index of 32.56 ± 4.85%. The biofilm formation ability of L. chiayiensis AACE3 was quantitatively assessed by crystalline violet staining. The results showed that the biofilm formation ability of the experimental group was 3.39-fold higher than that of the negative control, which was a moderate biofilm producer (++).
3.7. Antioxidant properties
The hydroxyl radical scavenging ability, DPPH radical scavenging ability and superoxide anion scavenging ability were analyzed to demonstrate the antioxidant capacity of strain AACE3 (Figure 5). The results showed that strain AACE3 exhibited good antioxidant capacity, and the CFS retained the highest antioxidant capacity. The DPPH radical scavenging ability and superoxide anion scavenging ability of the CFS were 91.67 ± 5.00% and 92.15 ± 3.97%, respectively. No significant difference between the resuspended intact organisms and crushed cell extracts was observed. In addition, the hydroxyl radical scavenging abilities of the supernatant (62.03 ± 9.39%) and resuspended IOs (61.79 ± 2.70%) were not significantly different.
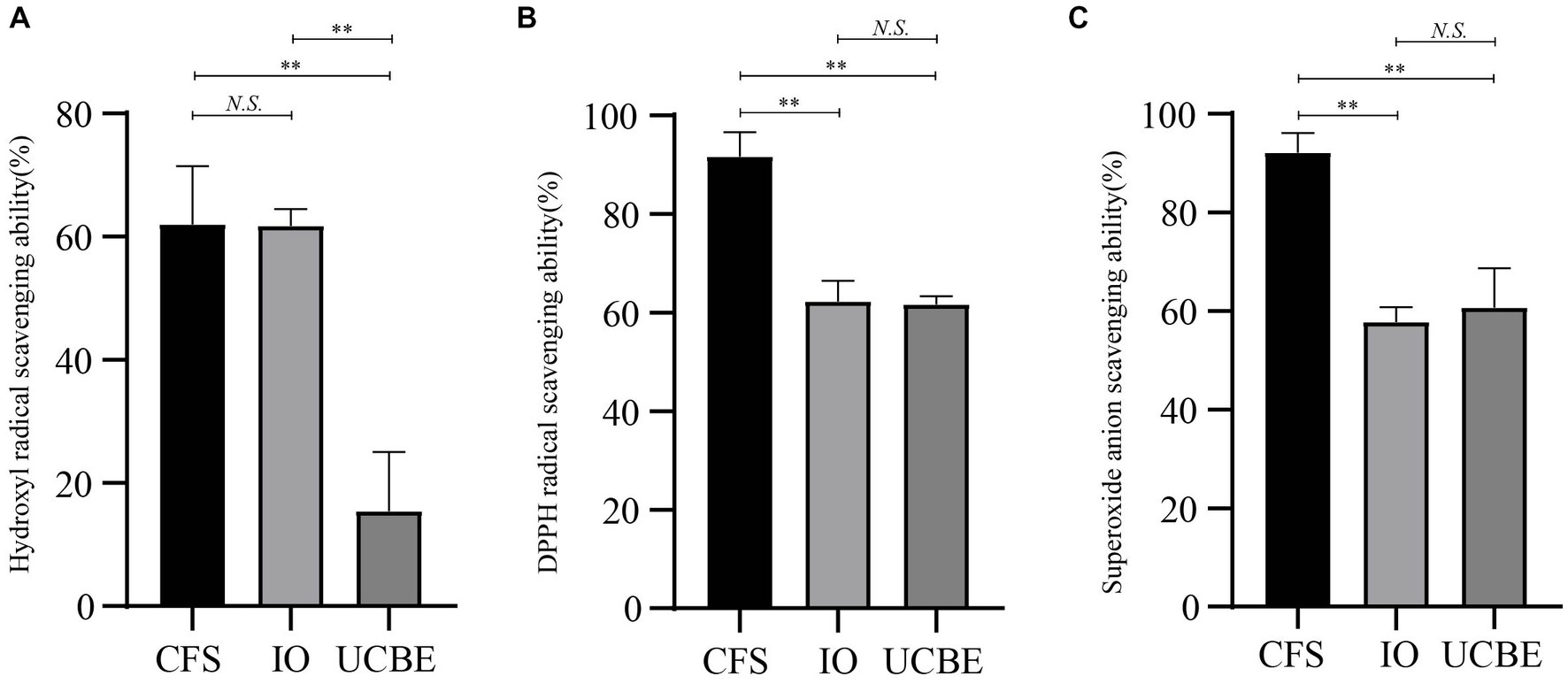
Figure 5. Antioxidant properties of L. chiayiensis AACE3. (A) Hydroxyl radical scavenging capacity, (B) DPPH radical scavenging capacity, (C) superoxide anion radical scavenging capacity. CFS, cell-free supernatant; IO, intact organisms; UCBE, ultrasonically crushed bacterial extract. **p < 0.01, N.S. denotes not significant.
4. Discussion
Discovery of new LAB and evaluation of their properties are crucial for meeting the needs for application in different fields. In this study, L. chiayiensis AACE3 isolated from fermented blueberry was comprehensively investigated by in vitro characterization and comparative genomic analysis. The genome size of strain AACE3 was 2,865,577 bp, and the GC content was 47.14%. In previous studies, it was reported that genome size and GC content can reflect the environmental adaptability of strains, with larger genome sizes indicating greater environmental adaptability (Bentkowski et al., 2015; Jia et al., 2022). Strains with large genomes are more likely to live in open environments than those with small genomes (Kant et al., 2017). In contrast, the strains living in fixed ecological niches (e.g., oral and intestinal tracts) are prone to loss and truncation of some useless gene fragments during evolution, resulting in smaller genome sizes (Duar et al., 2017). Compared with the genome sizes of common Lactobacillus strains, ranging from 1.3 to 3.3 Mb, the larger genome of AACE3 suggests that it may live in open environments, with the potential to adapt to a wide ecological niche.
Analysis of the 16S rRNA sequence of AACE3 revealed 99.86–100% identity to L. chiayiensis strains, but the similarities to L. zeae, L. paracasei, L. casei, and L. rhamnosus strains were all above 99% and > 97% of the interspecific classification threshold (Stackebrandt and Goebel, 1994). In 2008, the International Committee for Systematics of Prokaryotes (ICSP) declared that L. casei, L. paracasei (subsp. paracasei and subsp. tolerans) and L. rhamnosus are three closely related species in the L. casei group for which neither phenotypic nor 16S rRNA gene sequence-based identification methods provide sufficient resolution for accurate identification (Vásquez et al., 2005; Tindall, 2008; Colombo et al., 2014). The same problem has been noted for L. chiayiensis AACE3. Next, according to ANI identification, the identity between AACE3 and other L. chiayiensis genomes was 98.70–99.81%, which was greater than the 95% classification threshold (Konstantinidis and Tiedje, 2005). The genomic sequence identity with the L. zeae strain was 88.56–88.62%, with the L. paracasei strain was 79.05–79.09%, and with the L. rhamnosus strain was 80.38–80.41%. In addition, in the core genome-based phylogenetic analysis, four strains of L. chiayiensis with similar affinities were clustered into one group. The results confirmed that the 16S rRNA gene identity of all four species was above 97%, so it could not be used as an identification criterion. In contrast, whole-genome sequence-based analysis accurately distinguished the four species, suggesting that genome sequences can better reflect the evolutionary relationships of strains compared with 16S rRNA sequences.
Moreover, comparative genomic analysis revealed that AACE3 contains 112 unique genes that were not found in the other three L. chiayiensis strains. After KEGG annotation, genes encoding a casein lytic proteinase P (clpP, ko: K01358), an oligopeptidase (pepO, ko: K07386), and a ferrous iron transport protein A (feoA, ko: K04758) were identified. All these genes are closely related to bacterial survival and environmental adaptation. Casein lytic protease P is a central regulator of protein homeostasis in bacteria; clpP and its corresponding protease complex have the ability to remove or degrade improperly synthesized, damaged, denatured, aggregated or unwanted proteins from the bacterial cytosol, resulting in the maintenance of a dynamic balance of intracellular proteins under normal bacterial metabolism and stress stimulation (Sauer and Baker, 2011; Culp and Wright, 2017; Glynn, 2017). The oligopeptidase pepO hydrolyzes oligopeptides to free amino acids. While most LAB are amino acid auxotrophs, in environments lacking free amino acids (e.g., the milk fermentation process chain), LAB must activate certain gene pathways for hydrolyzing casein in milk to produce free amino acids for growth (Chavagnat et al., 2000). Moreover, the free amino acids produced enter the fermented product and can promote the synthesis of flavor compounds (Gomez et al., 1998). FeoA encodes a small basic cytoplasmic protein with 75–85 residues (Cartron et al., 2006), which enhances iron uptake in the presence of feoB (Kammler et al., 1993). BlpA (ko: K20344) and comA (ko: K12292) are related to bacteriocin production and biofilm formation. The blp locus is the key to bacteriocin production and immunity in Streptococcus pneumoniae, which was activated by blpA when protecting the bacteria from foreign bacteriocin invasion (Son et al., 2011). ComA was reported to function as a population-sensing regulator, enhancing the formation of bacterial biofilms and the production of bacteriostatic substances (Wang et al., 2016). In summary, L. chiayiensis AACE3 has evolved several genes in diverse environments compared with other L. chiayiensis strains. These genes may enhance the ability of AACE3 to compete with other microorganisms by producing bacteriocins and forming biofilms in a complex microbial environment. Moreover, the presence of genes such as clpP, pepO, and feoA indicated that AACE3 may have the ability to withstand harsh environmental conditions. These characteristics are directly related to the ecological niche in which AACE3 is found.
Regarding the functional properties during in vitro studies. L. chiayiensis AACE3 has broad-spectrum inhibitory activity against common gram-positive and gram-negative pathogens. Pathogen antagonism has been considered critical for balancing the normal intestinal microbiota and preventing microecological dysbiosis (Yang et al., 2015). Moreover, it is crucial for LAB application in food supplementation, farming, and other fields. The inhibition of pathogens occurs mainly due to the ability of LAB to produce certain inhibitory substances, such as bacteriocins, H2O2, lactic acid, and acetic acid (Tambekar et al., 2009). In this study, gene clusters related to bacteriocin production were also detected in the genome of the AACE3 strain. These gene clusters may have the ability to produce blp family class II bacteriocin, sakacin-P, enterolysin_A and enterocin_X. Blp family class II bacteriocin has broad-spectrum activity against Enterococcus faecalis and Streptococcus pyogenes (Renye et al., 2019). Sakacin-P inhibits the growth of a variety of gram-positive pathogens, especially Listeria monocytogenes, a common and persistent pathogen in the food industry (Katla et al., 2001). Enterolysin_A is a heat-resistant bacteriocin with broad-spectrum activity that degrades the cell wall of sensitive cells (Nilsen et al., 2003). Enterocin_X is a novel dual-peptide heat-resistant bacteriocin composed of two antimicrobial peptides (Xα and Xβ), the complementarity of which enhances its antibacterial activity (Hu et al., 2010).
Furthermore, notably, the resistance of bacteria to acid and protease environments and the ability of adhesion are considered critical in various applications, such as probiotic purposes, fermentation industry or food preservation (Monteagudo Mera et al., 2019). During adhesion, bacteria first form a biofilm barrier by self-aggregation (Saito et al., 2019). Cell surface hydrophobicity acts as a driving factor to promote adherence (Haddaji et al., 2015). In this study, L. chiayiensis AACE3 had a good survival rate in a simulated artificial gastrointestinal fluid environment and in the presence of 0.3% bile salts. And AACE3 showed good autoaggregation and biofilm formation ability and high cell hydrophobicity. In addition, the antioxidant capacity of LAB has been shown to reduce oxidation-mediated cellular damage and play an important role in the prevention of diabetes, heart disease and intestinal inflammation (Kaushik et al., 2009; Wang et al., 2017). L. chiayiensis AACE3 showed good performance in the DPPH radical scavenging test, superoxide anion scavenging test and hydroxyl radical scavenging test. In particular, the antioxidant capacity of CFS was the highest, which indicated that the antioxidant capacity of AACE3 could be attributed to multiple metabolites. The antioxidant capacity of peptides produced by LAB fermentation has been previously reported (Embiriekah et al., 2018). LAB produce a variety of extracellular polysaccharides demonstrated to be associated with free radical scavenging activity (Liu et al., 2011). In summary, the presence of bacteriocin gene clusters, comparative genomic analysis and in vitro demonstration of the properties indicate L. chiayiensis AACE3 may have strong environmental adaptability. It may have promising applications in antimicrobial therapy and production of antioxidant active substances in adverse environmental production.
5. Conclusion
In this study, the whole genome of the LAB strain AACE3 isolated from blueberry fermentation broth was sequenced and identified as L. chiayiensis. Genomic characterization and comparative genomic analysis suggested that AACE3 may have the potential to adapt to a wide range of ecological niches. In vitro evaluation of the properties indicated that AACE3 has broad-spectrum antibacterial activity and the ability to survive in unfavorable environments, such as in the presence of acid, bile salts, and proteases. In addition, antioxidant tests showed that L. chiayiensis AACE3 has good antioxidant capacity and the potential to reduce cellular damage caused by excessive oxidative stress in the host. Meanwhile, the presence of various genes in the genome provided genotypic validation of the results. Overall, this study provides a molecular basis for further investigation of the functional mechanism underlying the properties of this strain and a direction for subsequent application.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP107523.1.
Author contributions
X-DL conducted experiments, data sorting, and writing–preparation of original manuscript. Y-CL, R-SY, and XK conducted data analysis, reviewed, and edited the manuscript. W-GX assists in the analysis. FW, Q-LZ, and W-PZ reviewed and revised the manuscript. L-BL designed the study, provided reagents, materials, and revised manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Yunnan Major Scientific and Technological Projects (Grant No. 202202AG050008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1168378/full#supplementary-material
Footnotes
References
Benmouna, Z., Dalache, F., Zadi-Karam, H., Karam, N. E., and Vuotto, C. (2020). Ability of three lactic acid Bacteria to grow in sessile mode and to inhibit biofilm formation of pathogenic Bacteria. Adv. Exp. Med. Biol. 1282, 105–114. doi: 10.1007/5584_2020_495
Bentkowski, P., Van Oosterhout, C., and Mock, T. (2015). A model of genome size evolution for prokaryotes in stable and fluctuating environments. Genome Biol. Evol. 7, 2344–2351. doi: 10.1093/gbe/evv148
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Cartron, M. L., Maddocks, S., Gillingham, P., Craven, C. J., and Andrews, S. C. (2006). Feo–transport of ferrous iron into bacteria. Biometals 19, 143–157. doi: 10.1007/s10534-006-0003-2
Chae, H. Z., Chung, S. J., and Rhee, S. G. (1994). Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269, 27670–27678. doi: 10.1016/s0021-9258(18)47038-x
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 1–10. doi: 10.1038/srep24373
Chavagnat, F., Meyer, J., and Casey, M. G. (2000). Purification, characterisation, cloning and sequencing of the gene encoding oligopeptidase PepO from Streptococcus thermophilus a. FEMS Microbiol. Lett. 191, 79–85. doi: 10.1111/j.1574-6968.2000.tb09322.x
Colombo, M., de Oliveira, A. E. Z., de Carvalho, A. F., and Nero, L. A. (2014). Development of an alternative culture medium for the selective enumeration of Lactobacillus casei in fermented milk. Food Microbiol. 39, 89–95. doi: 10.1016/j.fm.2013.11.008
Culp, E., and Wright, G. D. (2017). Bacterial proteases, untapped antimicrobial drug targets. J. Antibiot. 70, 366–377. doi: 10.1038/ja.2016.138
de Melo Pereira, G. V., de Oliveira Coelho, B., Magalhães Júnior, A. I., Thomaz-Soccol, V., and Soccol, C. R. (2018). How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 36, 2060–2076. doi: 10.1016/j.biotechadv.2018.09.003
Dinarieva, T. Y., Klimko, A. I., Kahnt, J., Cherdyntseva, T. A., and Netrusov, A. I. (2023). Adaptation of Lacticaseibacillus rhamnosus CM MSU 529 to aerobic growth: a proteomic approach. Microorganisms 11:313. doi: 10.3390/microorganisms11020313
Duan, W., Guan, Q., Zhang, H. L., Wang, F. Z., Lu, R., Li, D. M., et al. (2023). Improving flavor, bioactivity, and changing metabolic profiles of goji juice by selected lactic acid bacteria fermentation. Food Chem. 408:135155. doi: 10.1016/j.foodchem.2022.135155
Duar, R. M., Lin, X. B., Zheng, J., Martino, M. E., Grenier, T., Pérez-Muñoz, M. E., et al. (2017). Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 41, S27–S48. doi: 10.1093/femsre/fux030
Dupre, J. M., Johnson, W. L., Ulanov, A. V., Li, Z., Wilkinson, B. J., and Gustafson, J. E. (2019). Transcriptional profiling and metabolomic analysis of Staphylococcus aureus grown on autoclaved chicken breast. Food Microbiol. 82, 46–52. doi: 10.1016/j.fm.2019.01.004
Embiriekah, S., Bulatović, M., Borić, M., Zarić, D., and Rakin, M. (2018). Antioxidant activity, functional properties and bioaccessibility of whey protein hydrolysates. Int. J. Dairy Technol. 71, 243–252. doi: 10.1111/1471-0307.12428
Fadare, O., Singh, V., Enabulele, O., Shittu, O., and Pradhan, D. (2022). In vitro evaluation of the synbiotic effect of probiotic Lactobacillus strains and garlic extract against Salmonella species. LWT 153:112439. doi: 10.1016/j.lwt.2021.112439
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Gerritsen, J., Smidt, H., Rijkers, G. T., and de Vos, W. M. (2011). Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 6, 209–240. doi: 10.1007/s12263-011-0229-7
Glynn, S. E. (2017). Multifunctional mitochondrial AAA proteases. Front. Mol. Biosci. 4:34. doi: 10.3389/fmolb.2017.00034
Gomez, M. J., Gaya, P., Nunez, M., and Médina, M. (1998). Streptococcus thermophilus as adjunct culture for a semi-hard cows' milk cheese. Lait 78, 501–511. doi: 10.1051/lait:1998547
Guinane, C. M., Crispie, F., and Cotter, P. D. (2016). Value of microbial genome sequencing for probiotic strain identification and characterization: promises and pitfalls. Gut Brain Axis, 45–60. doi: 10.1016/B978-0-12-802304-4.00004-9
Haddaji, N., Mahdhi, A. K., Krifi, B., Ismail, M. B., and Bakhrouf, A. (2015). Change in cell surface properties of Lactobacillus casei under heat shock treatment. FEMS Microbiol. Lett. 362:fnv047. doi: 10.1093/femsle/fnv047
Hu, C. B., Malaphan, W., Zendo, T., Nakayama, J., and Sonomoto, K. (2010). Enterocin X, a novel two-peptide Bacteriocin from Enterococcus faecium KU-B5, has an antibacterial Spectrum entirely different from those of its component peptides. Appl. Environ. Microbiol. 76, 4542–4545. doi: 10.1128/AEM.02264-09
Huang, C. H., Liou, J. S., Lee, A. Y., Tseng, M., Miyashita, M., Huang, L., et al. (2018). Polyphasic characterization of a novel species in the Lactobacillus casei group from cow manure of Taiwan: description of L. chiayiensis sp. nov. Syst. Appl. Microbiol. 41, 270–278. doi: 10.1016/j.syapm.2018.01.008
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jia, J., Liu, M., Feng, L., and Wang, Z. (2022). Comparative genomic analysis reveals the evolution and environmental adaptation of Acinetobacter johnsonii. Gene 808:145985. doi: 10.1016/j.gene.2021.145985
Kaidow, A., Ishii, N., Suzuki, S., Shiina, T., and Kasahara, H. (2022). Reactive oxygen species accumulation is synchronised with growth inhibition of temperature-sensitive recAts polA Escherichia coli. Arch. Microbiol. 204:396. doi: 10.1007/s00203-022-02957-z
Kammler, M., Schön, C., and Hantke, K. (1993). Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175, 6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993
Kant, R., Palva, A., and von Ossowski, I. (2017). An in silico pan-genomic probe for the molecular traits behind Lactobacillus ruminis gut autochthony. PLoS One 12:e0175541. doi: 10.1371/journal.pone.0175541
Katla, T., Møretrø, T., Aasen, I. M., Holck, A., Axelsson, L., and Naterstad, K. (2001). Inhibition of Listeria monocytogenes in cold smoked salmon by addition of sakacin P and/or live Lactobacillus sakei cultures. Food Microbiol. 18, 431–439. doi: 10.1006/fmic.2001.0420
Kaushik, J. K., Kumar, A., Duary, R. K., Mohanty, A. K., Grover, S., and Batish, V. K. (2009). Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS One 4:e8099. doi: 10.1371/journal.pone.0008099
Kim, E., Yang, S. M., Kim, D., and Kim, H. Y. (2021). Complete genome sequencing and comparative genomics of three potential probiotic strains, Lacticaseibacillus casei FBL6, Lacticaseibacillus chiayiensis FBL7, and Lacticaseibacillus zeae FBL8. Front. Microbiol. 12:794315. doi: 10.3389/fmicb.2021.794315
Kok, C. R., and Hutkins, R. (2018). Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr. Rev. 76, 4–15. doi: 10.1093/nutrit/nuy056
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Laslett, D., and Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. doi: 10.1093/nar/gkh152
Lawrence, J. G., and Hendrickson, H. (2005). Genome evolution in bacteria: order beneath chaos. Curr. Opin. Microbiol. 8, 572–578. doi: 10.1016/j.mib.2005.08.005
Li, B., Jin, D., Etareri Evivie, S., Li, N., Yan, F., Zhao, L., et al. (2017). Safety assessment of Lactobacillus helveticus KLDS1.8701 based on whole genome sequencing and Oral toxicity studies. Toxins 9:301. doi: 10.3390/toxins9100301
Li, S., Zhao, Y., Zhang, L., Zhang, X., Huang, L., Li, D., et al. (2012). Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 135, 1914–1919. doi: 10.1016/j.foodchem.2012.06.048
Lin, M. Y., and Yen, C. L. (1999). Antioxidative ability of lactic acid bacteria. J. Agric. Food Chem. 47, 1460–1466. doi: 10.1021/jf981149l
Liu, C. F., Tseng, K. C., Chiang, S. S., Lee, B. H., Hsu, W. H., and Pan, T. M. (2011). Immunomodulatory and antioxidant potential of Lactobacillus exopolysaccharides. J. Sci. Food Agric. 91, 2284–2291. doi: 10.1002/jsfa.4456
Lo Curto, A., Pitino, I., Mandalari, G., Dainty, J. R., Faulks, R. M., and John Wickham, M. S. (2011). Survival of probiotic lactobacilli in the upper gastrointestinal tract using an in vitro gastric model of digestion. Food Microbiol. 28, 1359–1366. doi: 10.1016/j.fm.2011.06.007
Mojgani, N., Hussaini, F., and Vaseji, N. (2015). Characterization of indigenous Lactobacillus strains for probiotic properties. Jundishapur J. Microbiol. 8:e59798. doi: 10.5812/jjm.17523
Monteagudo Mera, A., Rastall, R. A., Gibson, G. R., Charalampopoulos, D., and Chatzifragkou, A. (2019). Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 103, 6463–6472. doi: 10.1007/s00253-019-09978-7
Nilsen, T., Nes, I. F., and Holo, H. (2003). Enterolysin a, a Cell Wall-degrading Bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69, 2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003
Oyaizu, M. (1988). Antioxidative activities of browning products of glucosamine fractionated by organic solvent and thin-layer chromatography. Nippon Shokuhin Kogyo Gakkaishi 35, 771–775. doi: 10.3136/nskkk1962.35.11_771
Ramić, D., Jug, B., Šimunović, K., Tušek Žnidarič, M., Kunej, U., Toplak, N., et al. (2023). The role ofluxSin Campylobacter jejuni beyond intercellular signaling. Microbiol Spectr. 11, e02572–e02522. doi: 10.1128/spectrum.02572-22
Renye, J. A., Somkuti, G. A., and Steinberg, D. H. (2019). Thermophilin 109 is a naturally produced broad spectrum bacteriocin encoded within the blp gene cluster of Streptococcus thermophilus. Biotechnol. Lett. 41, 283–292. doi: 10.1007/s10529-018-02637-3
Saito, K., Tomita, S., and Nakamura, T. (2019). Aggregation of Lactobacillus brevis associated with decrease in pH by glucose fermentation. Biosci. Biotechnol. Biochem. 83, 1523–1529. doi: 10.1080/09168451.2019.1584522
Sanders, M. E., Akkermans, L. M. A., Haller, D., Hammerman, C., Heimbach, J. T., Hörmannsperger, G., et al. (2010). Safety assessment of probiotics for human use. Gut Microbes 1, 164–185. doi: 10.4161/gmic.1.3.12127
Sauer, R. T., and Baker, T. A. (2011). AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612. doi: 10.1146/annurev-biochem-060408-172623
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sharma, S., Sharma, N., and Kaushal, N. (2023). Utilization of novel bacteriocin synthesized silver nanoparticles (AgNPs) for their application in antimicrobial packaging for preservation of tomato fruit. Front. Sustain. Food Syst. 7:38. doi: 10.3389/fsufs.2023.1072738
Sleator, R. D., Gahan, C. G. M., and Hill, C. (2003). A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 69, 1–9. doi: 10.1128/AEM.69.1.1-9.2003
Son, M. R., Shchepetov, M., Adrian, P. V., Madhi, S. A., de Gouveia, L., von Gottberg, A., et al. (2011). Conserved mutations in the pneumococcal Bacteriocin transporter gene, blpA, result in a complex population consisting of producers and cheaters. MBio 2, e00179–e00111. doi: 10.1128/mBio.00179-11
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Tambekar, D., Bhutada, S., Choudhary, S., and Khond, M. (2009). Assessment of potential probiotic bacteria isolated from milk of domestic animals. J. Appl. Biosci. 15, 815–819.
Ticho, A. L., Malhotra, P., Dudeja, P. K., Gill, R. K., and Alrefai, W. A. (2019). Intestinal absorption of bile acids in health and disease. Compr. Physiol. 10, 21–56. doi: 10.1002/cphy.c190007
Tindall, B. (2008). The type strain of Lactobacillus casei is ATCC 393, ATCC 334 cannot serve as the type because it represents a different taxon, the name Lactobacillus paracasei and its subspecies names are not rejected and the revival of the name 'Lactobacillus zeae' contravenes rules 51b (1) and (2) of the international code of nomenclature of Bacteria Opinion 82. Int. J. Syst. Evol. Microbiol. 58, 1764–1765. doi: 10.1099/ijs.0.2008/005330-0
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/nar/gky383
Vasconcelos, F. M., Silva, H. L. A., Poso, S. M. V., Barroso, M. V., Lanzetti, M., Rocha, R. S., et al. (2019). Probiotic Prato cheese attenuates cigarette smoke-induced injuries in mice. Food Res. Int. 123, 697–703. doi: 10.1016/j.foodres.2019.06.001
Vásquez, A., Molin, G., Pettersson, B., Antonsson, M., and Ahrné, S. (2005). DNA-based classification and sequence heterogeneities in the 16S rRNA genes of Lactobacillus casei/paracasei and related species. Syst. Appl. Microbiol. 28, 430–441. doi: 10.1016/j.syapm.2005.02.011
Vizoso Pinto, M. G., Franz, C. M. A. P., Schillinger, U., and Holzapfel, W. H. (2006). Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int. J. Food Microbiol. 109, 205–214. doi: 10.1016/j.ijfoodmicro.2006.01.029
Voulgari, K., Hatzikamari, M., Delepoglou, A., Georgakopoulos, P., Litopoulou-Tzanetaki, E., and Tzanetakis, N. (2010). Antifungal activity of non-starter lactic acid bacteria isolates from dairy products. Food Control 21, 136–142. doi: 10.1016/j.foodcont.2009.04.007
Walter, J., Loach, D. M., Alqumber, M., Rockel, C., Hermann, C., Pfitzenmaier, M., et al. (2007). D-Alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 9, 1750–1760. doi: 10.1111/j.1462-2920.2007.01292.x
Wang, P., Guo, Q., Li, S., Lu, X., Zhang, X., and Ma, P. (2016). Functional analysis of quorum sensing regulator ComA on the production of antifungal active compounds and biofilm formation in Bacillus subtilis NCD-2. Acta Phytophylacica Sinica 43, 193–200. doi: 10.13802/j.cnki.zwbhxb.2016.02.003
Wang, Y., Wu, Y., Wang, Y., Fu, A., Gong, L., Li, W., et al. (2017). Bacillus amyloliquefaciens SC06 alleviates the oxidative stress of IPEC-1 via modulating Nrf2/Keap1 signaling pathway and decreasing ROS production. Appl. Microbiol. Biotechnol. 101, 3015–3026. doi: 10.1007/s00253-016-8032-4
Wen, Z. T., and Burne, R. A. (2002). Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68, 1196–1203. doi: 10.1128/AEM.68.3.1196-1203.2002
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xiang, Y. Z., Wu, G., Yang, L. Y., Yang, X. J., Zhang, Y. M., Lin, L. B., et al. (2022). Antibacterial effect of bacteriocin XJS01 and its application as antibiofilm agents to treat multidrug-resistant Staphylococcus aureus infection. Int. J. Biol. Macromol. 196, 13–22. doi: 10.1016/j.ijbiomac.2021.11.136
Yang, F., Hou, C., Zeng, X., and Qiao, S. (2015). The use of lactic acid Bacteria as a probiotic in swine diets. Pathogens 4, 34–45. doi: 10.3390/pathogens4010034
Zhang, Z. F., and Kim, I. H. (2014). Effects of multistrain probiotics on growth performance, apparent ileal nutrient digestibility, blood characteristics, cecal microbial shedding, and excreta odor contents in broilers. Poult. Sci. 93, 364–370. doi: 10.3382/ps.2013-03314
Zhang, W., Lai, S., Zhou, Z., Yang, J., Liu, H., Zhong, Z., et al. (2022). Screening and evaluation of lactic acid bacteria with probiotic potential from local Holstein raw milk. Front. Microbiol. 13:918774. doi: 10.3389/fmicb.2022.918774
Zhao, L., Wang, S., Dong, J., Shi, J., Guan, J., Liu, D., et al. (2021). Identification, characterization, and antioxidant potential of Bifidobacterium longum subsp. longum strains isolated from feces of healthy infants. Front. Microbiol. 12:756519. doi: 10.3389/fmicb.2021.756519
Keywords: Lacticaseibacillus chiayiensis, whole-genome sequencing, comparative genomics, antimicrobial activity, antioxidation
Citation: Li X-D, Lin Y-C, Yang R-S, Kang X, Xin W-G, Wang F, Zhang Q-L, Zhang W-P and Lin L-B (2023) Genomic and in-vitro characteristics of a novel strain Lacticaseibacillus chiayiensis AACE3 isolated from fermented blueberry. Front. Microbiol. 14:1168378. doi: 10.3389/fmicb.2023.1168378
Edited by:
Eric Altermann, Massey University, New ZealandReviewed by:
Kurt Selle, North Carolina State University, United StatesRenpeng Du, Heilongjiang University, China
Copyright © 2023 Li, Lin, Yang, Kang, Xin, Wang, Zhang, Zhang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lian-Bing Lin, bGlubGJAa3VzdC5lZHUuY24=
†These authors have contributed equally to this work
 Xin-Dong Li
Xin-Dong Li Yi-Cen Lin1,2†
Yi-Cen Lin1,2† Xin Kang
Xin Kang Wei-Gang Xin
Wei-Gang Xin Qi-Lin Zhang
Qi-Lin Zhang Wen-Ping Zhang
Wen-Ping Zhang