- 1Engineering and Research Center for Southwest Biopharmaceutical Resource of National Education Ministry of China, Guizhou University, Guiyang, Guizhou, China
- 2Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 3School of Science, Mae Fah Luang University, Chiang Rai, Thailand
- 4School of Food and Pharmaceutical Engineering, Guizhou Institute of Technology, Guiyang, Guizhou, China
- 5International Relations Unit, The Open University of Sri Lanka, Nawala, Nugegoda, Sri Lanka
Diaporthales is a species-rich order of fungi that includes endophytes, saprobes, and pathogens associated with forest plants and crops. They may also occur as parasites or secondary invaders of plant tissues injured or infected by other organisms or inhabit living animal and human tissues, as well as soil. Meanwhile, some severe pathogens wipe out large-scale cultivations of profitable crops, timber monocultures, and forests. Based on morphological and phylogenetic analyses of combined ITS, LSU, tef1-α, and rpb2 sequence data, generated using maximum likelihood (ML), maximum parsimony (MP), and MrBayes (BI), we introduce two new genera of Diaporthales found in Dipterocarpaceae in Thailand, namely Pulvinaticonidioma and Subellipsoidispora. Pulvinaticonidioma is characterized by solitary, subglobose, pycnidial, unilocular conidiomata with the internal layers convex and pulvinate at the base; hyaline, unbranched, septate conidiophores; hyaline, phialidic, cylindrical to ampulliform, determinate conidiogenous cells and hyaline, cylindrical, straight, unicellular, and aseptate conidia with obtuse ends. Subellipsoidispora has clavate to broadly fusoid, short pedicellate asci with an indistinct J- apical ring; biturbinate to subellipsoidal, hyaline to pale brown, smooth, guttulate ascospores that are 1-septate and slightly constricted at the septa. Detailed morphological and phylogenetic comparisons of these two new genera are provided in this study.
Introduction
Diaporthales is an order of ascomycetous fungi belonging to the subclass Diaporthomycetidae (Sordariomycetes) that dwell on terrestrial or aquatic taxa of plants, animals, and in soil (Senanayake et al., 2017, 2018; Wijayawardene et al., 2022). Senanayake et al. (2017, 2018) provided a recent treatment of the order and examined, described, and illustrated worldwide specimens and listed 27 families in Diaporthales. Many studies of this order have led to an explosion of species, including a total of 29 families (Crous et al., 2019; Guterres et al., 2019). Jiang et al. (2020) redefined the family Cryphonectriaceae and established two new families for the order, with a total of 31 families in Diaporthales. In the latest outline of the fungi and fungus-like taxa, Wijayawardene et al. (2022) accepted 32 families in the order.
Diaporthales contains both sexual and asexual morphs. The sexual morph is characterized by immersed stromata or substrata, brown or black perithecial ascomata with elongated beaks, sometimes with long papilla, deliquescent paraphyses at maturity, commonly unitunicate, thick-walled asci that are either evanescent with short stalks or intact, often floating free within the centrum at maturity, and have a refractive ring at the apex, containing 2–32 spores (Alexopoulus and Mims, 1978; Hawksworth et al., 1995; Castlebury et al., 2002; Rossman et al., 2007; Fan et al., 2018; Senanayake et al., 2018; Hyde et al., 2020b; Jiang et al., 2020). The asexual morph of Diaporthales is generally coelomycetous, rarely hyphomycetous, bearing their phialidic, rarely annellidic, conidiogenous cells, and conidia in acervuli or pycnidia with or without well-developed stromata. Since it has fewer distinguishing traits, proper identification at the genus and species levels is typically dependent on sequence data (Castlebury et al., 2002; Jiang et al., 2020).
In this study, we collected three interesting species from dead twigs and fruits of Dipterocarpaceae sp. from Thailand. The morphological characteristics indicated that these three taxa belong to the order Diaporthales. Furthermore, a phylogenetic analysis using a combination of ITS, LSU, tef1-α, and rpb2 sequence data confirmed them as distinct lineages within Diaporthales. Therefore, two new genera named Pulvinaticonidioma and Subellipsoidispora are described herein, with detailed descriptions and illustrations.
Materials and methods
Sample collection, isolation, and morphological studies
Fresh samples of decaying fruits and twigs from Dipterocarpaceae sp. were collected at the Mushroom Research Center, Chiang Mai, Thailand, in 2019. Samples were observed using a stereomicroscope (Motic SMZ-171). The detailed method of collection, observation of specimens, and isolation were carried out as references in the study by Senanayake et al. (2020) and Tang et al. (2022). The Tarosoft (R) Image Frame Work application (IFW 0.97 version) was used to take measurements, and the photoplates were made by Adobe Photoshop CS6 (Adobe Systems, USA). The type specimens were deposited in the Mae Fah Luang University Herbarium (MFLU), Chiang Rai, Thailand, and the ex-type cultures were deposited in the Culture Collection at Mae Fah Luang University (MFLUCC). Index Fungorum (2023) and Faces of Fungi numbers were acquired as detailed by Jayasiri et al. (2015). New species are established as recommended by Chethana et al. (2021a), and the records of new taxa in the Greater Mekong Subregion were uploaded to the GMS database (Chaiwan et al., 2021).
DNA extraction, PCR amplification, and sequencing
Fresh mycelia were prepared from the living culture that grew for 28 days and stored in the refrigerator at −20°C. DNA extraction, polymerase chain reaction (PCR) amplifications, sequencing, and phylogenetic analyses were carried out following the study by Tang et al. (2022). The manufacturer's instructions were followed while using the genomic DNA extraction kits [Sangon Biotech (Shanghai) Co., Ltd., China], in order to obtain DNA. The genes and primers used in this study were as follows: for internal transcribed spacer region (ITS), ITS5 and ITS4 (White et al., 1990); 28S large subunit rDNA region (LSU), LR0R, and LR5 (Vilgalys and Hester, 1990; Cubeta et al., 1991); translation elongation factor 1-alpha (tef1-α), EF1-728F, and EF2 (O'Donnell et al., 1998; Carbone and Kohn, 1999); and for RNA polymerase II second largest subunit (rpb2), frpb2-5f , and frpb2-7cr (Liu et al., 1999) genes. The PCR was carried out in a volume of 50 μl. The reagents that were used in the polymerase chain reaction were as follows: the DNA template (2 μl), forward primers (2 μl), reverse primers (2 μl), 2 ×Taq PCR Master Mix (25 μl), and 19 μl of ddH2O (double-distilled water). The annealing temperature was set to 52°C for 1 min and extension at 72°C for 90 s in LSU and ITS, followed by 35 cycles; 56°C for 1 min and extension at 72°C for 90 s in tef1-α, followed by 35 cycles; and 55°C for 1 min and extension at 72°C for 90 s in rpb2 followed by 35 cycles. The products of PCR were checked on 1% agarose gels and sent to Sangon Biotech (Shanghai) Co., Ltd., China for sequencing.
Phylogenetic analyses
The forward and reverse primers of the newly generated sequence were assembled by the Contig Express v3.0.0 application, and the most similar taxa were found by BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) in NCBI. A combination of sequence data (ITS, LSU, tef1-α, and rpb2) of Cryphonectriaceae and Coryneaceae in GenBank (Tables 1, 2) was downloaded for phylogenetic analyses. Sequence data of each region were aligned by the online version of MAFFT v. 7 (https://mafft.cbrc.jp/alignment/server/index.html) (Katoh et al., 2017), through the “auto” option. Multiple genes were combined by SequenceMatrix (Vaidya et al., 2011). The aligned sequences were trimmed by manually adjusting and using trimAl v 1.2, with the “-gt 0.6” option (Capella-Gutiérrez et al., 2009). The phylogenetic analyses in this study were based on the maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference (BI), by using a combined sequence dataset of ITS, LSU, tef1-α, and rpb2. The analysis of maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference (BI) was processed in the CIPRES web portal (Miller et al., 2010) using the “RAxML-HPC v.8 on XSEDE” tool, “PAUP on XSEDE” tool, and “MrBayes on XSEDE” tool, respectively (Huelsenbeck and Ronquist, 2001; Swofford, 2002; Stamatakis et al., 2008; Ronquist et al., 2012).
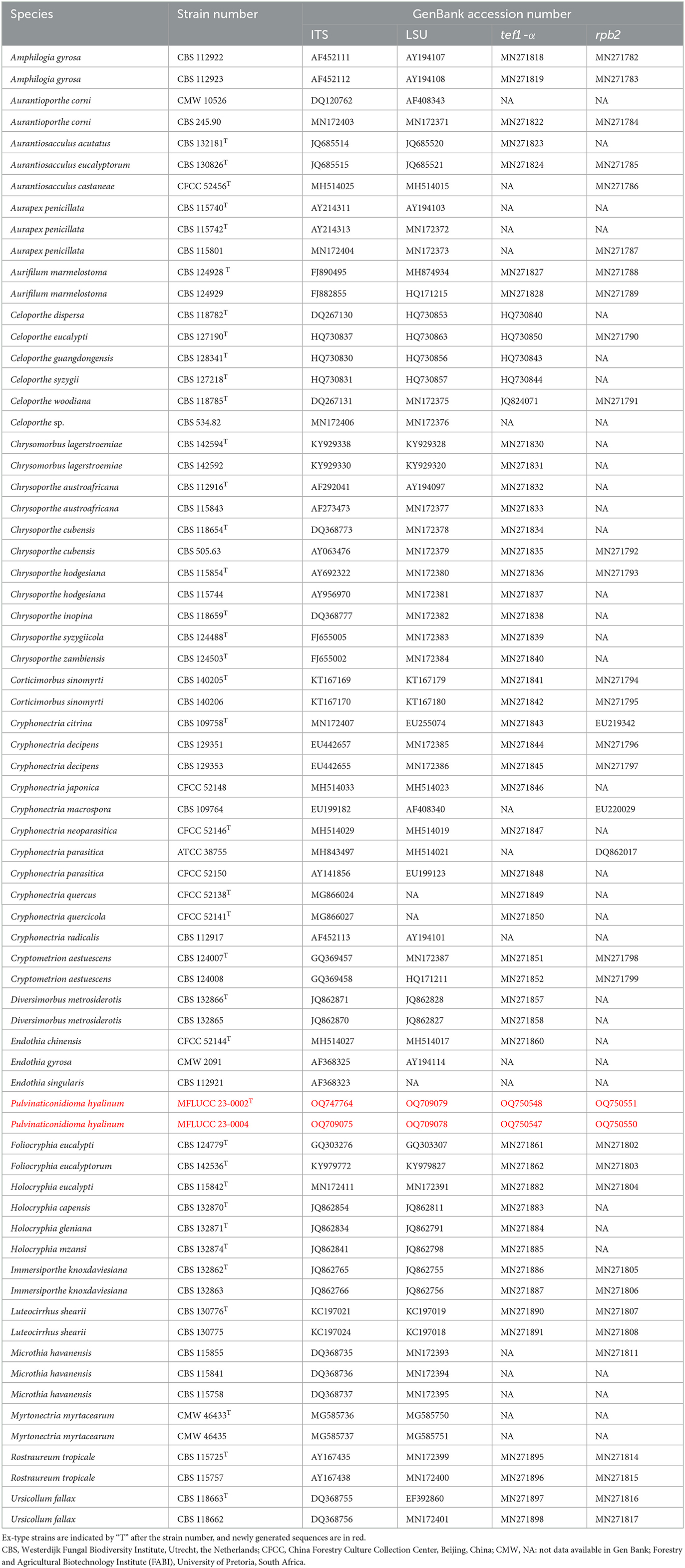
Table 1. Taxa used in this study for Cryphonectriaceae and their GenBank accession numbers for ITS, LSU, tef1-α, and rpb2 sequence data.
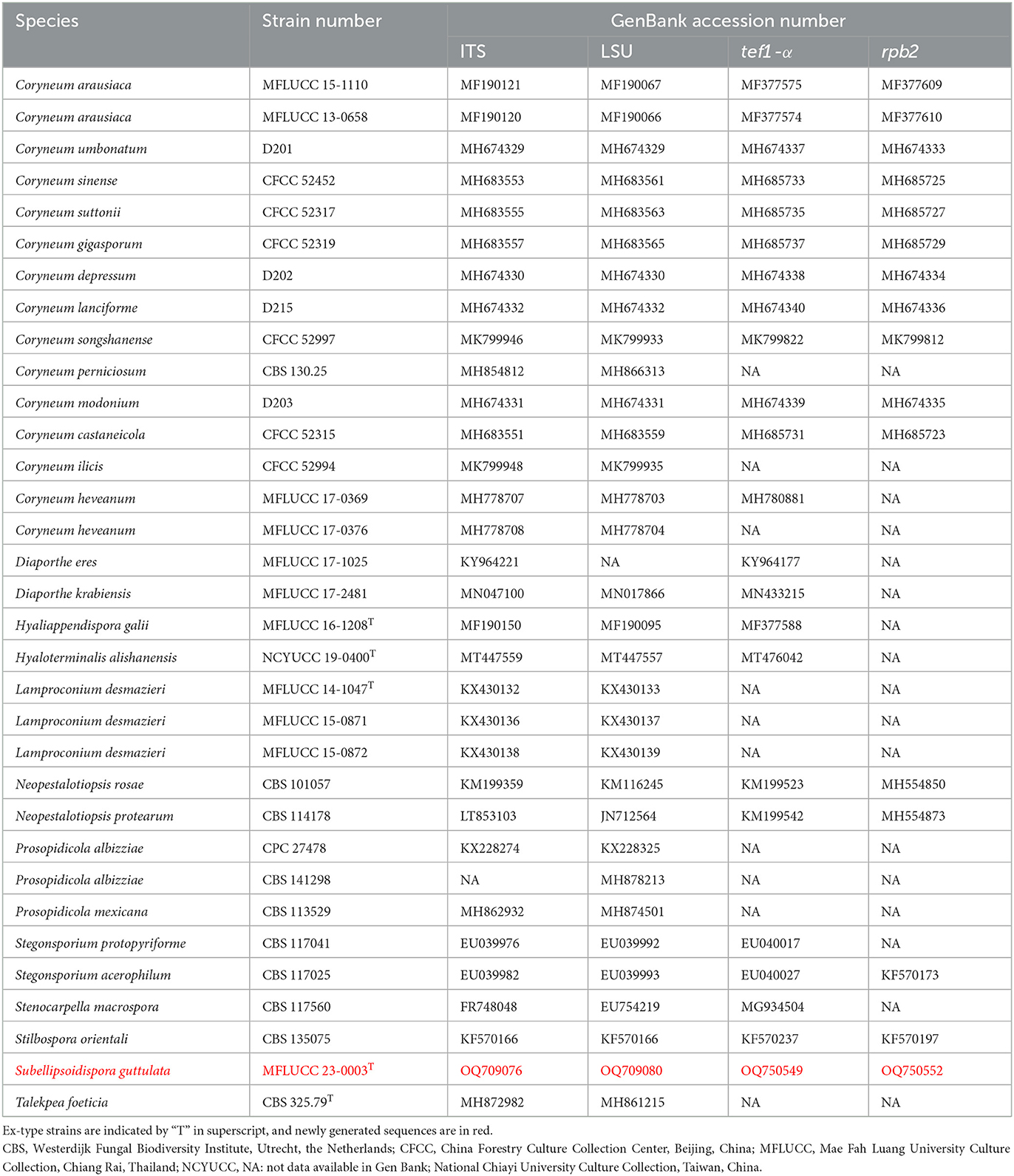
Table 2. Taxa used in this study for Coryneaceae and their GenBank accession numbers for ITS, LSU, tef1-α, and rpb2 sequence data.
For ML analysis, the GTRGAMMA+I-Invar model of nucleotide evolution was used, and RAxML rapid bootstrapping was set to 1,000 bootstrap replicates (Stamatakis et al., 2008).
For MP analysis, 1,000 random taxa addition was used to infer trees. With branches of zero length collapsed and all multiple parsimonious trees saved, the value of Maxtrees was set to 5,000. For trees produced using various optimal criteria, parsimony score values for tree length (TL), consistency index (CI), retention index (RI), and homoplasy index (HI) were determined. To evaluate the clade stability, 1,000 iterations of the Bootstrap (BT) method were utilized, each comprising 100 trials of random stepwise addition of taxa (Hillis and Bull, 1993).
For BI, MrModeltest v2 was used for the selection of the best-fit model for each gene region. The Markov chain Monte Carlo (MCMC) algorithm was launched with four chains running concurrently from a random tree topology. When the divided frequencies' average standard deviation dropped below 0.01, the procedure was immediately terminated. The burn-in factor was set at 25%, and the sampling interval for trees was set to every 1,000th generation. The posterior probabilities (PP) for the remaining trees were computed (Dissanayake et al., 2020). Adobe Illustrator version 51.1052.0.0 and FigTree version 1.4.0 were further used to view trees (Adobe Inc., San Jose, California, United States).
Results
Phylogenetic analyses
For the phylogenetic analyses, a combined dataset of ITS, LSU, tef1-α, and rpb2 sequences was used. The dataset of Cryphonectriaceae included 70 taxa, with Foliocryphia eucalypti (CBS 124779) and Foliocryphia eucalyptorum (CBS 142536) as outgroups. The data matrix comprised 2,860 total characteristics, including gaps (ITS: 1–481 bp, LSU: 482–1,290 bp, tef1-α: 1,291–1,858 bp, and rpb2: 1,859–2,564 bp). Phylogenetic reconstructions with broadly comparable topologies were produced by the combined dataset of ML, MP, and BI analyses. The top-scoring ML tree with a final ML optimization likelihood value of −16,383.140512 (ln) is shown in Figure 1. In the ML analysis, the GTRGAMMA + I-Invar model was used, and the results showed 1,022 unique alignment patterns and 27.97% of indeterminate characteristics or gaps. Base frequency estimates were as follows: A = 0.229377, C = 0.266423, G = 0.271764, and T = 0.232436; substitution rates were as follows: AC = 1.760988, AG = 4.032209, AT = 1.914644, CG = 1.261342, CT = 8.527324, and GT = 1.000000; gamma distribution shape parameter alpha = 0.176927; and the tree length was 1.784127. The findings of the MP analysis showed that 2,564 characteristics remained unchanged, 103 were changeable but parsimoniously uninformative, and 733 were parsimoniously informative. The following values were displayed by the most parsimonious tree: TL = 2693, CI = 0.494, RI = 0.779, RC = 0.385, and HI = 0.506. The best-fit models for the BI analysis were GTR + I + G for ITS, LSU, tef1-α, and rpb2. With a final average standard deviation of split frequencies of 0.009895, Bayesian posterior probabilities (BYPP) from MCMC were analyzed. A new taxon correlated with the Cryphonectriaceae clade and is sister to Chrysomorbus. It is distinct from all other Cryphonectriaceae genera sampled herein, although with no support (Figure 1).
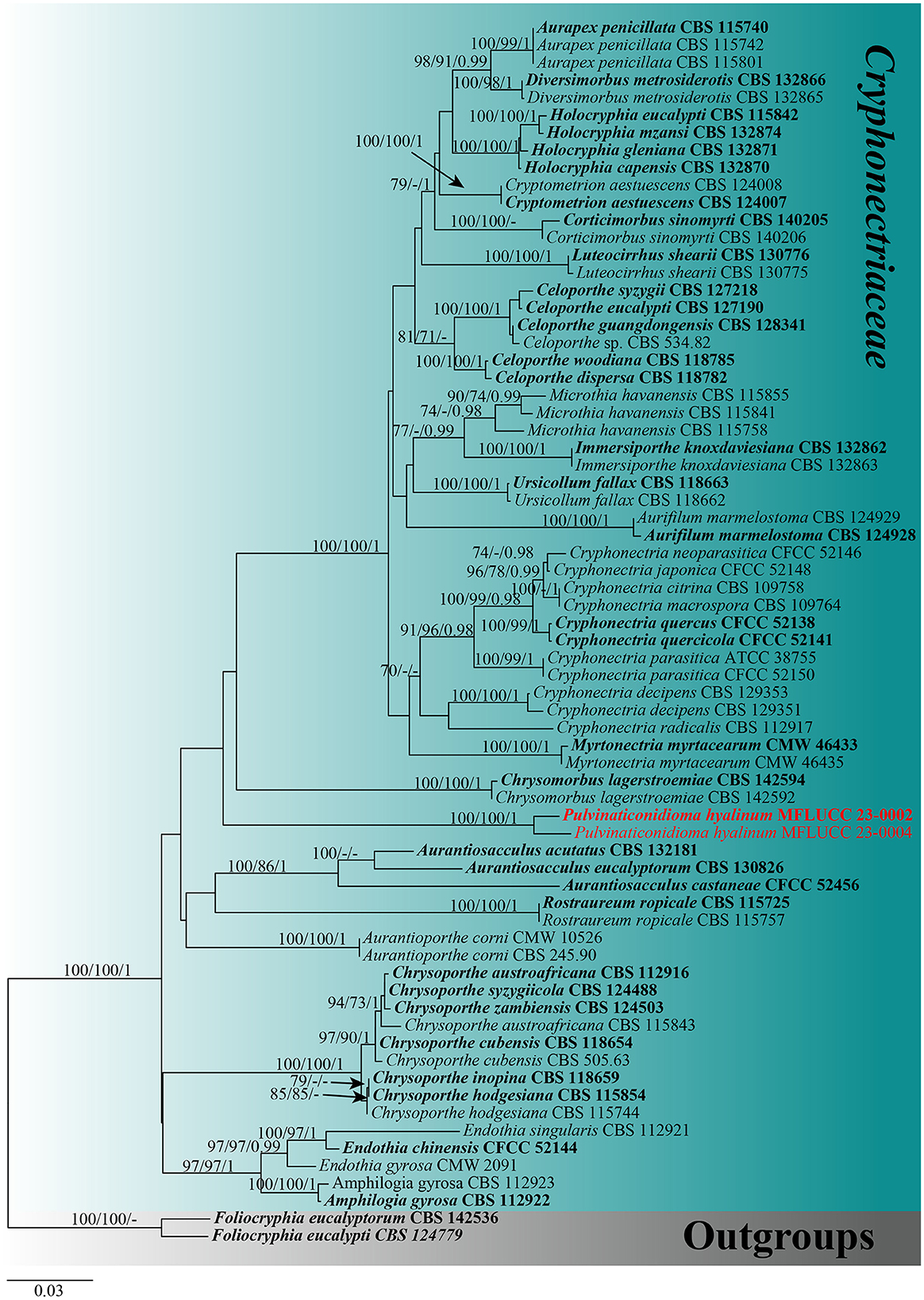
Figure 1. Maximum likelihood (RAxML) tree, based on the analysis of a combined dataset of ITS, LSU, tef1-α, and rpb2 sequence data. The tree is rooted with Foliocryphia eucalypti (CBS 124779) and Foliocryphia eucalyptorum (CBS 142536). Bootstrap support values for ML and MP ≥70% and Bayesian posterior probabilities (BYPP) ≥0.95 are given near the nodes, respectively. Ex-type strains are in bold, and the new isolates are in red.
For the tree of Coryneaceae, the combined sequence dataset of 33 taxa was used with Neopestalotiopsis protearum (CBS 114178) and Neopestalotiopsis rosae (CBS 101057) as the outgroups. The data matrix comprised 2,977 total characteristics, including gaps (ITS: 1–597 bp, LSU: 598–1,426 bp, tef1-α: 1,427–2,123 bp, and rpb2: 2,124–3,151 bp). Based on the results of phylogenetic analysis, the top-scoring RAxML tree with a final ML optimization likelihood value of −19,448.697623 (ln) is shown in Figure 2. The GTRGAMMA + I-Invar model was applied to the RAxML analysis, and the findings revealed 1,332 distinct alignment patterns and 33.88% of ambiguous characteristics or gaps. The following were the base frequency estimates: A = 0.237835, C = 0.267649, G = 0.278605, and T = 0.215911; the substitution rates: AC = 1.607401, AG = 1.967526, AT = 1.403753, CG = 1.150806, CT = 5.717313, and GT = 1.000000; the gamma distribution shape parameter alpha = 0.260733; and the tree length = 3.464265. The results of the MP analysis revealed that 3,151 characteristics remained constant, 271 were variable and parsimoniously uninformative, and 1,142 were parsimoniously informative. The most frugal tree resulted in TL = 3,542, CI = 0.636, RI = 0.684, RC = 0.435, and HI = 0.364 as its values. For the BI analysis, the best-fit models were GTR+G for ITS, tef1-α, and rpb2 and SYM + I + G for LSU. The BYPP from MCMC were examined with a final average standard deviation of split frequencies of 0.009847. Based on the results of phylogenetic analysis of the combined ITS, LSU, tef1-α, and rpb2 sequencing data, the new taxon is related to Coryneum, Hyaloterminalis, and Talekpea within Coryneaceae, with statistical support of 72% ML and 1 BYPP. It differs from any other Coryneaceae genus sampled here (Figure 2).
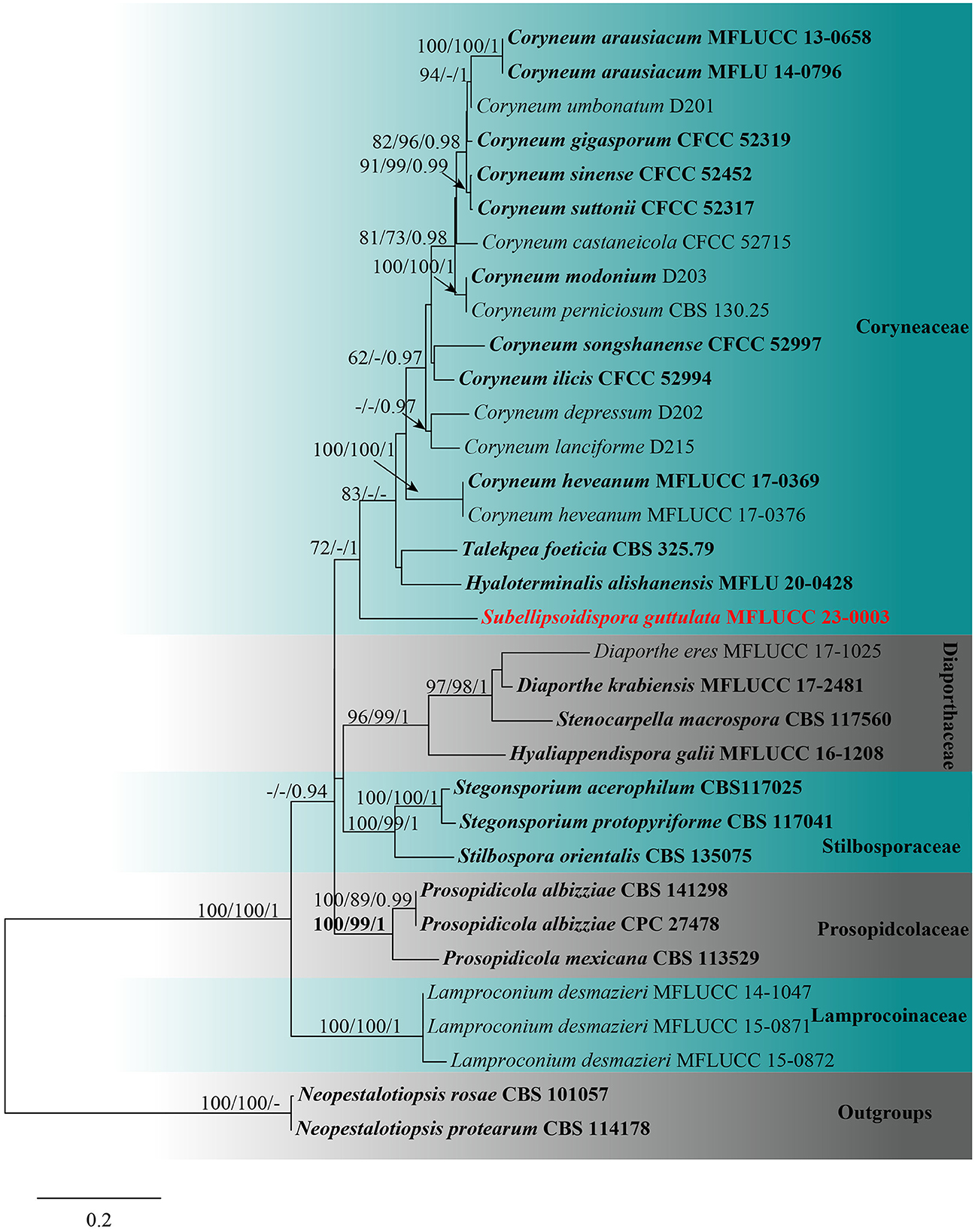
Figure 2. Maximum likelihood (RAxML) tree, based on the analysis of a combined dataset of ITS, LSU, tef1-α, and rpb2 sequence data. The tree is rooted with Neopestalotiopsis protearum (CBS 114178) and Neopestalotiopsis rosae (CBS 101057). Bootstrap support values for ML and MP ≥70% and Bayesian posterior probabilities (BYPP) ≥0.95 are given near the nodes, respectively. Ex-type strains are in bold, and the new isolates are in red.
Taxonomy
Cryphonectriaceae Gryzenh. & M.J. Wingf., Mycologia 98: 246. 2006.
Index Fungorum number: IF510585; Facesoffungi number: FoF03455.
Sexual morph see Jiang et al. (2020). Asexual morph Conidiomata semi-immersed to erumpent on the substrate, solitary, subglobose to pulvinate, pyriform, uni- to multiloculate, yellow, orange to fuscous black; necks absent or present with one to several attenuated necks. Conidiophores sometimes reduced to conidiogenous cells, cylindrical, hyaline, septate, or not. Conidiogenous cells hyaline, smooth, phialidic, ampulliform, inconspicuous, lining the inner cavity of conidiomata, with attenuate or truncate apices. Conidia hyaline, cylindrical, minute, seldom sigmoid, or slightly curved, aseptate (Jiang et al., 2020).
Notes: Cryphonectriaceae was described by Gryzenhout et al. (2006) to accommodate the Cryphonectria-Endothia complex based on LSU sequence data, and it mainly comprises plant pathogens (Vermeulen et al., 2011). Recently, Jiang et al. (2020) reevaluated this family based on morphology and combined ITS, LSU, tef1-α, and rpb2 multi-gene phylogenetic analysis. It now contains 22 genera and 56 species (Jiang et al., 2020; this study).
Type genus: Cryphonectria (Sacc.) Sacc. & D. Sacc.
Pulvinaticonidioma X. Tang, Jayaward, J.C. Kang & K.D. Hyde, gen. nov.
Index Fungorum number: IF900388; Faceoffungi number: FOF 13992
Etymology: The generic name refers to the pulvinate conidiomata.
Type species: Pulvinaticonidioma hyalinum X. Tang, Jayaward, J.C. Kang & K.D. Hyde.
Subclass classification: Sordariomycetes, Diaporthales, Cryphonectriaceae.
Saprobic on Dipterocarpaceae sp. Sexual morph not observed. Asexual morph Coelomycetous. Conidiomata immersed to semi-immersed in the substrate, solitary, glabrous or rough, pycnidial, subglobose, unilocular, thick-walled, ostiolate, brown to dark brown. Ostiole central, single with slightly protruding ostiolar papilla. Conidiomata wall composed of thick-walled, pale brown to dark brown cells of textura angularis at the exterior, and convex and pulvinate at the base. Conidiophores hyaline reduced to conidiogenous cells. Conidiogenous cells phialidic, cylindrical to ampulliform, determinate, smooth-walled, hyaline. Conidia hyaline, cylindrical, with obtuse ends, straight, unicellular, aseptate, thick- and smooth-walled.
Notes: Pulvinaticonidioma is characterized by solitary, subglobose, pycnidial conidiomata, phialidic, conidiogenous cells, and aseptate hyaline conidia. This matches with the morphological characteristics of Cryphonectriaceae (Jiang et al., 2020). Phylogenetically, Pulvinaticonidioma clusters with Chrysomorbus (Figures 3, 4). Both Pulvinaticonidioma and Chrysomorbus have a coelomycetous asexual morph (Chen et al., 2018). The former differs from the species in Chrysomorbus in having unilocular, glabrous or rough, thick-walled, ostiolate conidiomata with hyaline cells of textura angularis at the exterior, convex and pulvinate at the base; aseptate, straight, cylindrical, unicellular, and hyaline conidia with obtuse ends. After the comprehensive consideration based on the morphological and phylogenetic analysis, we, herein, introduce Pulvinaticonidioma as a new genus in Cryphonectriaceae, with Pulvinaticonidioma hyalinum as the type.
Pulvinaticonidioma hyalinum X. Tang, Jayaward, J.C. Kang & K.D. Hyde, sp. nov.
Index Fungorum number: IF900390; Faceoffungi number: FOF 13993
Etymology: The epithet refers to the hyaline conidia.
Holotype: MFLU 23-0052.

Figure 3. Pulvinaticonidioma hyalinum (MFLU 23-0052, Holotype). (a) Fallen pod of Dipterocarpaceae sp. (b) Conidiomata on Dipterocarpaceae sp. (c) Section of conidioma. (d) Conidioma wall (e) Ostiole. (f) Conidiogenous cells. (g–i) Conidia. (j) Germinated conidium. (k) Colonies on PDA. (l) Reverse of culture. Scale bars: (b) 500 μm, (c, d) 100 μm, (e, g–j) 20 μm, and (f) 10 μm.
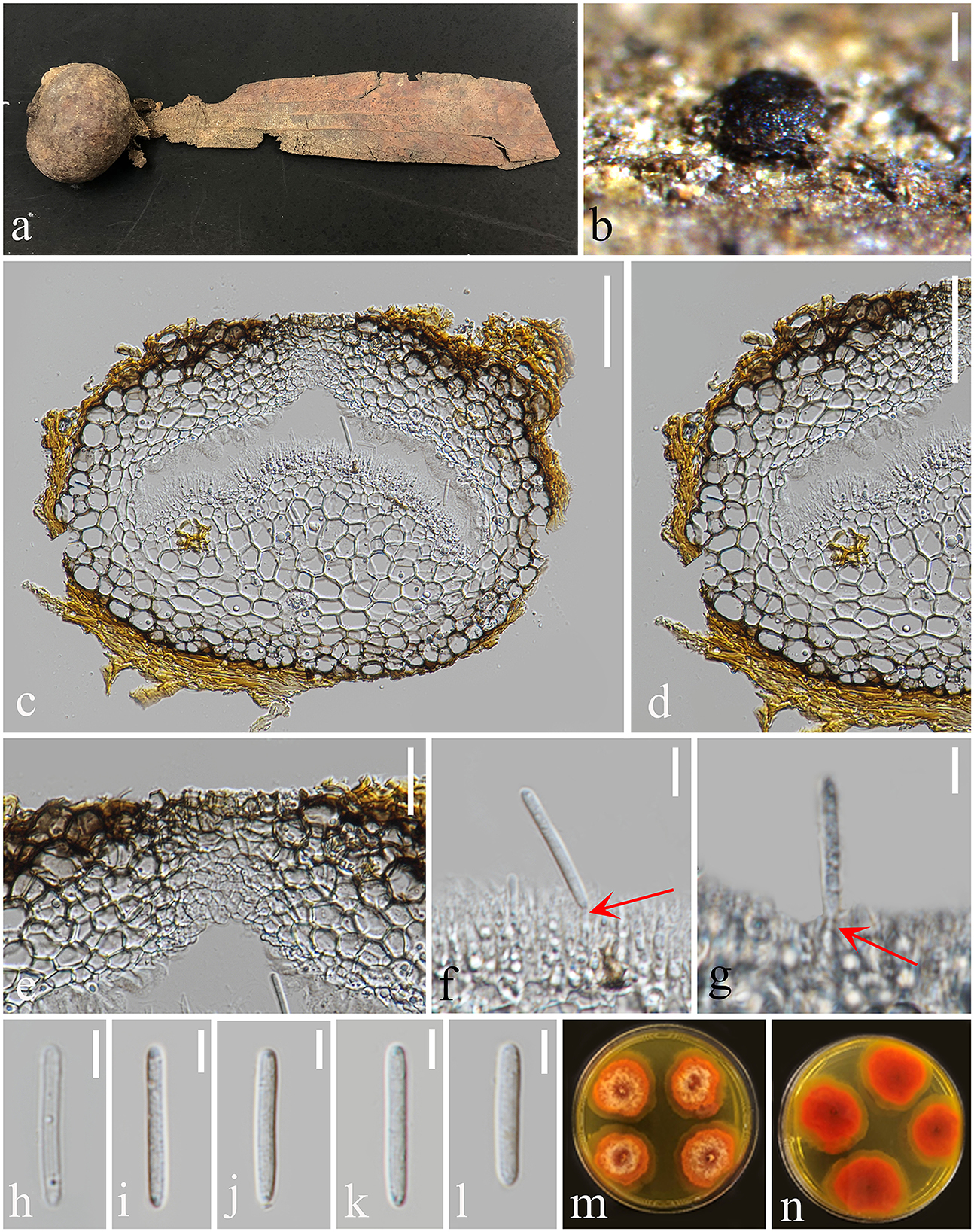
Figure 4. Pulvinaticonidioma hyalinum (MFLU 23-0053, Paratype). (a) Fallen pod of Dipterocarpaceae sp. (b) Conidiomata on Dipterocarpaceae sp. (c) Section of conidioma. (d) Conidioma wall. (e) Ostiole. (f, g) Conidiogenous cells. (h–l) Conidia. (m) Colonies from above. (n) The reverse of culture. Scale bars: (b) 200 μm, (c, d) 50 μm, (e) 20 μm, and (f–l) 5 μm.
Saprobic on Dipterocarpaceae sp. Sexual morph not observed. Asexual morph Coelomycetous. Conidiomata 297–473 × 211–316 μm (x̄ = 375 × 267 μm, n = 20), immersed to semi-immersed in substrate, solitary, glabrous or rough, pycnidial, subglobose, unilocular, thick-walled, ostiolate, brown to dark brown. Ostiole 51–65 × 34–48 μm (x̄ = 58 × 42 μm, n = 10), central, single with slightly protruding ostiolar papilla. Conidiomata wall 50–88 μm (x̄ = 70 μm, n = 20) wide, composed of thick-walled, pale brown to dark brown cells of textura angularis at the exterior, convex and pulvinate at the base 103–202 μm high (x̄ = 144 μm, n = 20). Conidiophores hyaline, reduced to conidiogenous cells. Conidiogenous cells 6–11.5 × 1.8–3.4 μm (x̄ = 7 × 2.5 μm, n = 20), phialidic, cylindrical to ampulliform, determinate, smooth-walled, hyaline. Conidia 15–20 × 2–3 μm (x̄ = 17 × 2.5 μm, n = 20) hyaline, cylindrical, with obtuse ends, straight, unicellular, aseptate, thick- and smooth-walled.
Culture characters: Conidia germinated on PDA within 24 h, and germ tubes produced from one end. The culture was incubated at room temperature. Colonies reached 45 mm diameter after 15 days, flat, spreading, fluffy colonies, circular with irregular lightly orange outer ring, cottony. The surface is lightly rough, with orange-red colonies, cream-colored hyphae attached to the center of the colony, with an irregular orange-yellow edge. The reverse orange-red, more orange-yellow at the margins, not pigmented.
Material examined: Thailand, Chiang Mai province, Mae Taeng District, on the fruits (pericarp and wings of the pod) of Dipterocarpaceae, 8 August 2019, Xia Tang, Dip17 (MFLU 23-0052, holotype; ex-type living culture, MFLUCC 23-0002), on the fruits of Dipterocarpaceae, 23 October 2020, Xia Tang, Dip41 (MFLU 23-0053, paratype; ex-paratype living culture, MFLUCC 23-0004).
Notes: The two Pulvinaticonidioma hyalinum collections, showing similar morphology clustered together with ML = 100, MP = 100, and BYPP = 1 support (Figure 1). The base pair differences between the two strains were as follows: ITS = 0.7% (4/557), LSU = 0% (0/811), tef1-α = 6.2% (38/613), and rpb2 = 1% (11/983), respectively, and we identified them as the same species following the guidelines for species delineation proposed by Chethana et al. (2021a). Pulvinaticonidioma hyalinum matches the characteristics of Cryphonectriaceae and is similar in having unilocular conidiomata without necks and conidiomata walls made of cells of textura globulosa (Jiang et al., 2020). However, P. hyalinum differs from the type species of Cryphonectriaceae, Chrysomorbus lagerstroemiae in their fruiting body, conidiomata wall, conidiophores, and conidia. Pulvinaticonidioma hyalinum has brown to dark brown conidiomata with slightly protruding ostiolar papilla, hyaline cells of textura angularis at the exterior, interior layers that are convex and pulvinate at the base, and unbranched conidiophores, while Ch. lagerstroemiae has uni- to multilocular, conidiomata lacking ostioles, with convoluted locules, and occasionally aseptate conidia with separating septa and branching conidiophores. The conidiogenous cells in P. hyalinum are phialidic, cylindrical to ampulliform with hyaline, straight, aseptate, unicellular, conidia with obtuse ends, while Ch. lagerstroemiae has flask-shaped conidiogenous cells with attenuated apices and minute, cylindrical conidia with obtuse ends, that are hyaline, fusoid to oval, aseptate, and exuded as orange droplets (Chen et al., 2018). The phylogenetic analysis of the combined ITS, LSU, tef1-α, and rpb2 sequence data showed that P. hyalinum belongs to Cryphonectriaceae and forms a separate lineage sister to Chrysomorbus. Although the bootstrap values are low, the phylogenetic analysis supports the placement of our new taxa in Cryphonectriaceae, as well as the possibility of other close relatives that have not yet been discovered; hence, their placement within the family is subjected to change. The base pair differences between P. hyalinum and the type species of Chrysomorbus, viz. Ch. lagerstroemiae were as follows: ITS = 5% (27/539), LSU = 1.4% (11/811), and tef1-α = 26.5% (151/569), respectively. Based on the phylogenetic analysis and morphological comparison of the nearest genus, we, herein, introduce Pulvinaticonidioma as a new genus to accommodate the new collection, P. hyalinum.
Coryneaceae Corda, Icon. fung. (Prague) 3: 36 (1839) amend.
Index Fungorum number: IF80650; Facesoffungi number: FoF06868;
Saprobes and pathogens exist on dead wood and living plants, respectively. Sexual morph: Stromata erumpent, solitary, comprising pseudoparenchymatous cells. Ectostromatic comprising small cells of textura prismatica, brown to black, disk well or poorly developed. Ascomata brown to black, ostiolate, aggregated, immersed, arranged in valsoid configuration, perithecial, coriaceous, globose to subglobose, papillate. Papilla central, upright, sometimes converging, broad, comprising brown cells of textura porrecta. Peridium thick-walled, comprising outer, brown cells of textura angularis and inner, thick-walled, hyaline, compressed cells of textura angularis. Paraphyses attached to the base, cellular, broad, septate, longer than asci. Asci ellipsoid to cylindrical, unitunicate, 8-spored, pedicellate, rounded at the apex with a J-, apical ring. Ascospores hyaline or initially hyaline, brown at maturity, overlapping uni- to biseriate, irregularly fasciculate, ellipsoid, 1–3-septate, fusoid or elongate, sometimes end-cells pointed, often distoseptate, pale brown or hyaline end-cells, straight or curved not constricted at the septa, guttulate, smooth-walled (added from Hyde et al., 2020b). Asexual morph: see Hyde et al. (2020b) and Rathnayaka et al. (2020).
Type genus: Coryneum Nees
Notes: Coryneaceae was described by Corda (1839) to accommodate Coryneum as the type genus. Rathnayaka et al. (2020) amended the description of this family to accommodate these genera based on their morphological characteristics and treated Talekpea and Hyaloterminalis in Coryneaceae. Until now, there are three genera included in Coryneaceae, viz. Coryneum (Nees von Esenbeck, 1816), Hyaloterminalis, and Talekpea (Rathnayaka et al., 2020; Wijayawardene et al., 2022).
Subellipsoidispora X. Tang, Jayaward, J.C. Kang & K.D. Hyde, gen. nov.
Index Fungorum number: IF900389; Faceoffungi number: FOF 13994
Etymology: The epithet refers to the subellipsoidal ascospores.
Type species: Subellipsoidispora guttulata X. Tang, Jayaward, J.C. Kang and K.D. Hyde
Subclass classification: Sordariomycetes, Diaporthales, and Coryneaceae.
Saprobic on Dipterocarpaceae sp. Asexual morph Not observed. Sexual morph Ascomata perithecial, erumpent, scattered, solitary, coriaceous, immersed, globose to subglobose, papillate, ostiolate, dark brown to black. The Ostiole canal narrowing toward the base, internally covered by hyaline periphyses, cells around the base small, thick-walled, and brown. Peridium comprising brown, compressed, cells of textura angularis. Hamathecium composed of cylindrical, unbranched, straight to flexible, smooth, hyaline, septate paraphyses slightly constricted at the septa, tapering toward to end, longer than asci. Asci 8-spored, unitunicate, clavate to broadly fusoid, short pedicellate, apex blunt, with an indistinct, J- apical ring, evanescent. Ascospores overlapping uniseriate to biseriate, biturbinate to subellipsoidal, 1-septate, slightly constricted at the septa, guttulate, smooth, hyaline to pale brown.
Notes: Subellipsoidispora share characteristics with Coryneaceae, such as perithecial, coriaceous, ostiolate, brown to black ascomata; with thick-walled peridium having outer and inner brown cells of textura angularis and hyaline, compressed cells of textura angularis, respectively; paraphyses are longer than asci; clavate to broadly fusoid, 8-spored asci with J- apical ring; guttulate and smooth, hyaline to pale brown and straight ascospores (Hyde et al., 2020b). Coryneaceae contains three genera, viz. Coryneum, Hyaloterminalis, and Talekpea (Rathnayaka et al., 2020). Both Subellipsoidispora and Coryneum have the ascomycetous sexual morph, while Talekpea and Hyaloterminalis have a hyphomycetous asexual morph (Senanayake et al., 2017, 2018). Subellipsoidispora differs from the species in Coryneum in having scattered, solitary ascomata; a thick-walled ostiolar canal narrowing toward the base, internally covered by hyaline periphyses, a peridium of brown-walled, compressed, cells of textura angularis, clavate to broadly fusoid, short pedicellate asci and biturbinate to subellipsoidal, 1-septate, guttulate ascospores, slightly constricted at the septa. In the phylogenetic analysis, Subellipsoidispora clusters in Coryneaceae and forms a separate lineage sister to Hyaloterminalis and Talekpea (Figure 2). Based on its unique morphology (Figure 5) and phylogenetic evidence (Figure 1), Subellipsoidispora is introduced as a new genus of Coryneaceae, and the sexual morph is described in this study, awaiting the discovery of its asexual morph.
Subellipsoidispora guttulata X. Tang, Jayaward, J.C. Kang & K.D. Hyde, sp. nov.
Index Fungorum number: IF900391; Faceoffungi number: FOF 13995
Etmology: Name referring to the hyaline ascospores.
Holotype: MFLU 23-0054.
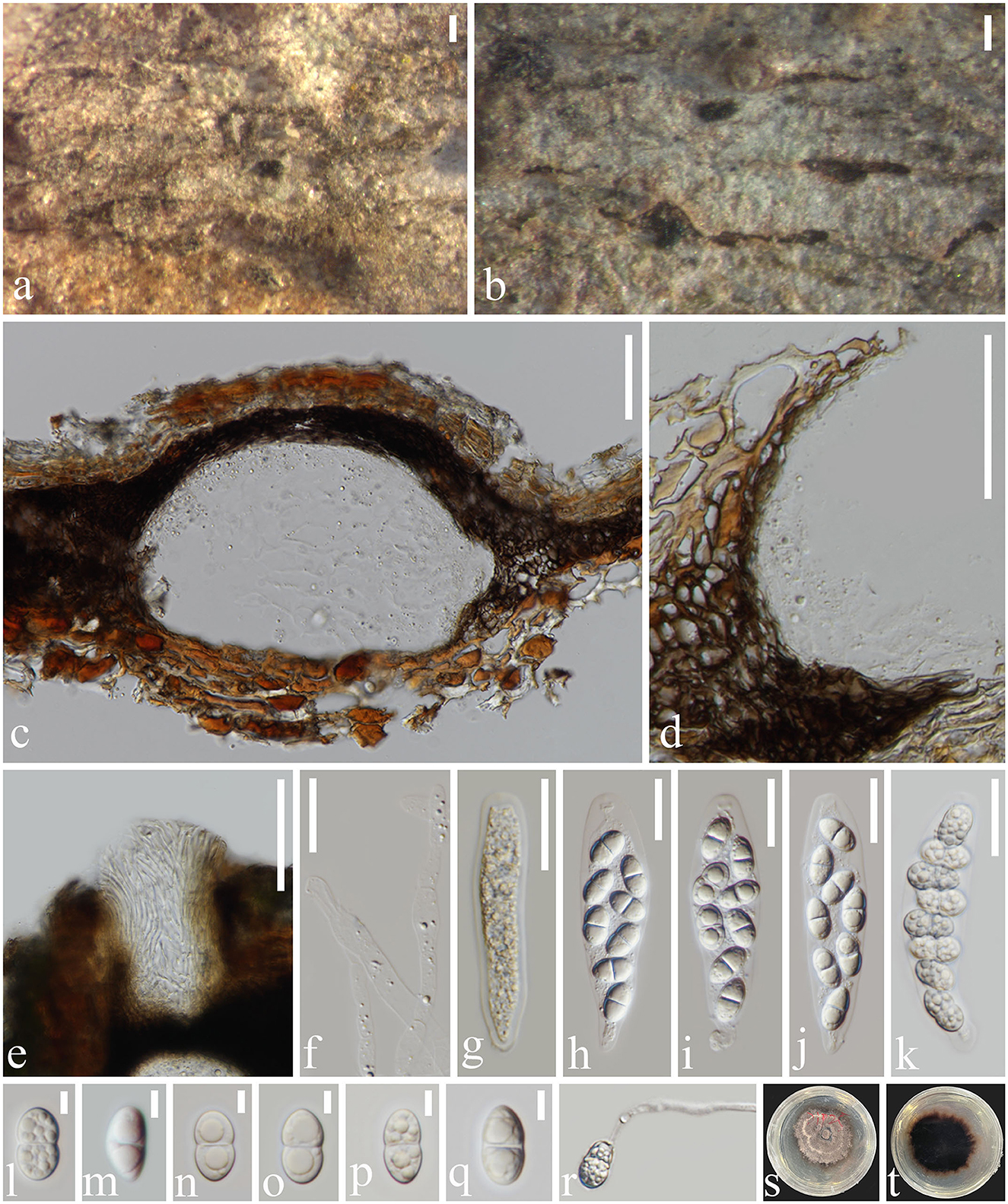
Figure 5. Subellipsoidispora guttulata (MFLU 23-0054, holotype) (a, b) Appearance of ascomata on host substrate. (c) Section of an ascoma. (d) Peridium. (e) Ostiole. (f) Paraphyses. (g–k) Asci from immature to mature. (l–q) Ascospores. (r) Germinated ascospore. (s) Colony on PDA. (t) The reverse of culture. Scale bars: (a, b) 200 μm, (c–e) 50 μm, (f–k) 20 μm, and (l–q) 5 μm.
Saprobic on dead barks of Dipterocarpaceae sp. Sexual morph Ascomata 117–270 × 71–155 μm (x̄ = 199 × 105 μm, n = 20), immersed, scattered, solitary, globose to subglobose, dark brown to black, coriaceous, ostiolate, papillate. The Ostiole canal narrowing toward the base, internally covered by hyaline periphyses, cells around the base small, thick-walled, and brown. Peridium 8–28 μm wide (x̄ = 18 μm, n = 20), comprising brown, compressed, cells of textura angularis. Paraphyses 3–6 μm wide (x̄ = 5.5 μm, n = 30), cylindrical, unbranched, straight to flexible, smooth, hyaline, septate, slightly constricted at the septa, tapering toward to end, longer than asci. Asci 67–90 × 13–24 μm (x̄ = 79 × 19 μm, n = 20), 8-spored, unitunicate, clavate to broadly fusoid, short pedicellate, apex blunt, with an indistinct, J- apical ring, evanescent. Ascospores 13–16 × 5–9 μm (x̄ = 14 × 7 μm, n = 20), overlapping uniseriate to biseriate, biturbinate to subellipsoidal, 1-septate, slightly constricted at the septa rounded at both ends, guttulate, smooth-walled, hyaline to pale brown. Asexual morph not observed.
Culture characters: Colonies grown on PDA and incubated at 25°C reached a diameter of 40 mm after 2 weeks, flat, spreading, fluffy, with a pale brown ring interlaced in the colonies. Surface lightly rough with brown mycelium, colonies somewhat raised in the middle, and with an irregular edge. The reverse side dark brown with an irregular, penniform, brown edge, and not pigmented.
Material examined: Thailand, Chiang Mai Province, Mae Taeng district, on dead bark of Dipterocarpaceae, 15 July 2020, Xia Tang, Dip25 (MFLU 23-0054, holotype; ex-type living culture, MFLUCC 23-0003).
Notes: Subellipsoidispora guttulata is similar to Coryneum umbonatum in having immersed, coriaceous, brown to black ascomata, and unitunicate asci with an indistinct J- apical ring. However, S. guttulata differs from C. umbonatum in having clavate to broadly fusoid, short pedicellate asci and subellipsoidal, 1-septate, guttulate, hyaline to pale brown ascospores, while C. umbonatum has ellipsoid to cylindrical, stalk pedicellate asci, and ellipsoid, fusoid or elongate, distoseptate, straight or curved spores that are brown at maturity (Senanayake et al., 2018). Phylogenetic analysis showed that S. guttulata belongs to Coryneaceae and forms a basal lineage sister to Coryneum, an ascomycetous genus, Hyaloterminalis and Talekpea, a hyphomycetous and monotypic genus. The base pair differences between S. guttulata and C. umbonatum were as follows: ITS = 7.7% (45/581), LSU = 3.2% (26/842), and rpb2 = 21.7% (223/1029), and the differences between S. guttulata and Talekpea foeticia were as follows: ITS = 12.5% (65/520) and LSU = 2% (17/843). Based on its phylogenetic and morphological analyses, we place S. guttulata as the type species of Subellipsoidispora in Coryneaceae.
Discussion
Diaporthales (Sordariomycetes) is an order that contains saprobic, endophytic, and pathogenic taxa with a wide distribution on a variety of hosts (Barr, 1978; Castlebury et al., 2002; Rossman et al., 2007; Senanayake et al., 2017, 2018; Fan et al., 2018; Jiang et al., 2020). The pathogenic members cause great economic losses, such as chestnut blight, caused by Cryphonectria parasitica (Cryphonectriaceae) (Gryzenhout et al., 2006; Rigling and Prospero, 2018; Gomdola et al., 2022), polar and willow canker on Populus and Salix, caused by Cytospora chrysosperma (Cytosporaceae) (Fan et al., 2014, 2020; Wang et al., 2015), and stem-end rot of citrus fruits infected by Diaporthe citri (Huang et al., 2013). Researchers have carried out their research on secondary metabolites in Diaporthaceae and Gnomoniaceae (Chepkirui and Stadler, 2017; Wu et al., 2019). As saprobes, they cause the degradation of wood, such as Apiosporopsis carpinea (Apiosporopsidaceae) on the overwintered leaves of Carpinus betulus (Senanayake et al., 2017) and Pseudoplagiostoma dipterocarpicola on the decaying wood of Dipterocarpaceae (Tang et al., 2022). As endophytes, they live in medicinal plants and are used for studies that investigate antimicrobial activities, e.g., Diaporthe spp., which were isolated from the hosts Copaifera langsdorffii and C. pubiflora (de Carvalho et al., 2021). Antibacterial activity has been demonstrated using extracts of two unidentified Diaporthe spp. and D. miriciae (Carvalho et al., 2018).
As more taxonomic studies of fungi are being conducted, the focus has steadily shifted from morphology to a combination of molecular phylogeny and morphology, serving as the foundation for the mainstream approach (Senanayake et al., 2017, 2018; Jiang et al., 2020; Chethana et al., 2021a; Maharachchikumbura et al., 2021). Initially, Castlebury et al. (2002) accepted Cytosporaceae, Diaporthaceae, Gnomoniaceae, and Melanconidaceae in Diaporthales by using LSU sequence data. Réblová et al. (2004) established a new family Togniniaceae to accommodate Togninia and its Phaeoacremonium anamorphs using LSU and SSU sequence data. Later, the family Togniniaceae was transferred into Togniniales from Diaporthales using LSU, SSU, tef1-α, and rpb2 sequence data (Gramaje et al., 2015; Maharachchikumbura et al., 2015, 2016). The use of multi-gene analysis for the identification of Diaporthales species was seen in subsequent studies, such as the combination of ITS-beta-tubulin (tub2) and ITS-LSU (Gryzenhout et al., 2006; Mostert et al., 2006; Cheewangkoon et al., 2010; Crous et al., 2012; Voglmayr et al., 2012, 2017; Suetrong et al., 2015; Réblová et al., 2016; Du et al., 2017; Yang et al., 2018; Maharachchikumbura et al., 2021). Voglmayr and Jaklitsch (2014) demonstrated through the evaluation of Stegonsporium and Stilbospora that LSU alone did not always contain sufficient phylogenetic resolution to identify consistently well-supported phylogenetic relationships at the generic level, and our research results matched this as well. Subsequently, Schizoparmaceae was revised using a combination of LSU, rpb2, ITS, and tef1-α (Alvarez et al., 2016). Combining DNA sequence data of ITS, LSU, tef1-α, and rpb2 is advised by Senanayake et al. (2017, 2018) and Fan et al. (2018) to evaluate the phylogenetic relationships of diaporthalean families. Jiang et al. (2020) used the combination of ITS, LSU, tef1-α, and rpb2 to redefine the family Cryphonectriaceae and to describe two new families, viz. Foliocryphiaceae and Mastigosporellaceae. With the increasing number of studies and knowledge on the diversity of lifestyles in Diaporthales, identifying its species has become difficult. The utilization of protein genes makes it possible to have a precise placement in Diaporthales, as proven in recent studies (Senanayake et al., 2017, 2018; Jiang et al., 2020). Thus, we suggest analyzing the families in Diaporthales via both morphological and molecular traits and the specific genes of each family for multigene phylogenetic analysis.
Members of the Dipterocarpaceae are economically significant trees generating lumber, camphor, and resin and are common in Southeast Asia (Maury-Lechon and Curtet, 1998). In this study, two new genera, namely Pulvinaticonidioma and Subellipsoidispora, were found on Dipterocarpaceae species in Thailand and were introduced. We introduce our collections as new genera based on unique features, such as the characteristics of the conidiomata, conidiogenous cells, and conidial appearance, as observed in the new taxon, Pulvinaticonidioma hyalinum when compared with other known genera in Cryphonectriacea. The results of the ML, MP, and MrBayes analyses also support that this is a new genus in Cryphonectriaceae (Figure 1). Similarly, the second collection Subellipsoidispora guttulata is morphologically distinct from other known genera in Coryneaceae in having unique characteristics in their asci and the shape of ascospores, and the phylogeny supports it as a new genus in Coryneaceae (Figure 2). To date, eight species of microfungi on Dipterocarpaceae have been described from Thailand, viz. Hermatomyces thailandica, Lauriomyces sakaeratensis, Lembosia xyliae, Pseudoplagiostoma dipterocarpi, P. dipterocarpicola, Pestalotiopsis shoreae, Pulvinaticonidioma hyalinum, and Subellipsoidispora guttulata (Suwannarach et al., 2016; Chethana et al., 2021b; Farr and Rossman, 2022; Tang et al., 2022; This study). Among these species, Pseudoplagiostoma dipterocarpi is an endophyte, while the rest are saprobes. It is remarkable that in this study, we found two new genera in a family that has been relatively well studied but on lesser studied hosts. This indicates that many more taxa will be discovered with further surveys on Dipterocarpaceae and other poorly studied hosts (Hyde et al., 2020a; Bhunjun et al., 2022).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
XT conducted the experiments, analyzed the data, and wrote the manuscript. Y-ZL, RJ, KH, and J-CK planned the experiments. XT and LD analyzed the data. XT and X-MC conducted the experiments. Y-ZL, RJ, KH, LD, IG, Y-PX, and J-CK revised the manuscript. Y-ZL, KH, and J-CK funded the experiments. All authors revised and agreed to the published version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (NSFC Grants Nos. 32170019, 31670027, and 31460011) and the Open Fund Program of Engineering Research Center of Southwest Bio-Pharmaceutical Resources, Ministry of Education, Guizhou University No. GZUKEY20160702 each provided funding for this project. The authors acknowledge the Thailand Research Fund grant entitled Impact of climate change on fungal diversity and biogeography in the Greater Mekong Sub-region (RDG6130001) and the National Research Council of Thailand (NRCT) grant, Total fungal diversity in a given forest area with implications toward species numbers, chemical diversity and biotechnology (grant no. N42A650547).
Acknowledgments
The authors would like to thank Dr. Shaun Pennycook for his input on the new fungus name (Pulvinaticonidioma hyalinum and Subellipsoidispora guttulate) and Dr. Wen-Jing Li for checking the description of the new taxa. The authors also acknowledge Mae Fah Luang University, Guizhou University, and Guizhou Institute of Technology for their support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alvarez, L. V., Groenewald, J. Z., and Crous, P. W. (2016). Revising the Schizoparmaceae: Coniella and its Q19 synonyms Pilidiella and Schizoparme. Stud. Mycol. 85, 1–34. doi: 10.1016/j.simyco.2016.09.001
Barr, M. E. (1978). The Diaporthales in North America: with emphasis on Gnomonia and its segregates. Mycologia Memoir. 7, 1–232.
Bhunjun, C. S., Niskanen, T., Suwannarach, N., Wannathes, N., Chen, Y. J., McKenzie, E. H. C., et al. (2022). The numbers of fungi: are the most speciose genera truly diverse? Fungal Divers. 114, 387–462. doi: 10.1007/s13225-022-00501-4
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Carvalho, C. D., Ferreira-D'Silva, A., Wedge, D. E., Cantrell, C. L., and Rosa, L. H. (2018). Antifungal activities of cytochalasins produced by Diaporthe miriciae, an endophytic fungus associated with tropical medicinal plants. Can. J. Microbiol. 64, 835–843. doi: 10.1139/cjm-2018-0131
Castlebury, L. A., Rossman, A. Y., Jaklitsch, W. J., and Vasilyeva, L. N. (2002). A preliminary overview of the Diaporthales based on large subunit nuclear ribosomal DNA sequences. Mycologia 94, 1017–1031. doi: 10.1080/15572536.2003.11833157
Chaiwan, N., Gomdola, D., Wang, S., Monkai, J., Tibpromma, S., Doilom, M., et al. (2021). https://gmsmicrofungi.org: an online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere 12, 1513–1526. doi: 10.5943/mycosphere/12/1/19
Cheewangkoon, R., Groenewald, J. Z., Verkley, G. J. M., Hyde, K. D., Wingfield, M. J., Gryzenhout, M., et al. (2010). Re-evaluation of Cryptosporiopsis eucalypti and Cryptosporiopsis-like species occurring on Eucalyptus leaves. Fungal Divers. 44, 89–105. doi: 10.1007/s13225-010-0041-5
Chen, S. F., Liu, Q. L., Li, G. Q., Wingfield, M. J., and Roux, J. (2018). A new genus of Cryphonectriaceae isolated from Lagerstroemia speciosa in southern China. Plant Pathol. 67, 107–123. doi: 10.1111/ppa.12723
Chepkirui, C., and Stadler, M. (2017). The genus Diaporthe: a rich source of diverse and bioactive metabolites. Mycol. Prog. 16, 477–494. doi: 10.1007/s11557-017-1288-y
Chethana, K. W. T., Manawasinghe, I. S., Hurdeal, V. G., Bhunjun, C. S., Appadoo, M. A., Gentekaki, E., et al. (2021a). What are fungal species and how to delineate them? Fungal Divers. 109, 1–25. doi: 10.1007/s13225-021-00483-9
Chethana, K. W. T., Niranjan, M., Dong, W., Samarakoon, M. C., Bao, D. F., Calabon, M. S., et al. (2021b). AJOM new records and collections of fungi: 101-150. Asian J. Mycol. 4, 113–260. doi: 10.5943/ajom/4/1/8
Crous, P. W., Schumacher, R. K., Akulov, A., Thangavel, R., Hernández-Restrepo, M., Carnegie, A. J., et al. (2019). New and interesting fungi. 2. Fungal Syst. Evol. 3, 57–134. doi: 10.3114/fuse.2019.03.06
Crous, P. W., Summerell, B. A., Alfenas, A. C., Edwards, J., Pascoe, I. G., Porter, I. J., et al. (2012). Genera of diaporthalean coelomycetes associated with leaf spots of tree hosts. Persoonia 28, 66–75. doi: 10.3767/003158512X642030
Cubeta, M. A., Echandi, E., Abernethy, T., and Vilgalys, R. (1991). Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 81, 1395–1400. doi: 10.1094/Phyto-81-1395
de Carvalho, C. R., Maia, M. Q., Sobral, M., Pereira, G. M. D., da Silva, K., Vital, M. J. S., et al. (2021). Diversity and antimicrobial activity of culturable endophytic fungi associated with the neotropical ethnomedicinal plants Copaifera langsdorffii and Copaifera pubiflora. S. Afr. J. Bot. 142, 305–315. doi: 10.1016/j.sajb.2021.06.021
Dissanayake, A. J., Bhunjun, C. S., Maharachchikumbura, S. S. N., and Liu, J. K. (2020). Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 11, 2652–2676. doi: 10.5943/mycosphere/11/1/18
Du, Z., Hyde, K. D., Yang, Q., Liang, Y. M., and Tian, C. M. (2017). Melansporellaceae: a novel family of Diaporthales (Ascomycota). Phytotaxa 305, 191–200. doi: 10.11646/phytotaxa.305.3.6
Fan, X. L., Bezerra, J. D., Tian, C. M., and Crous, P. W. (2018). Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 40, 119–134. doi: 10.3767/persoonia.2018.40.05
Fan, X. L., Bezerra, J. D., Tian, C. M., and Crous, P. W. (2020). Cytospora (Diaporthales) in China. Persoonia 45, 1–45. doi: 10.3767/persoonia.2020.45.01
Fan, X. L., Tian, C. M., Yang, Q., Liang, Y. M., You, C. J., Zhang, Y. B., et al. (2014). Cytospora from Salix in northern China. Mycotaxon 129, 303–315. doi: 10.5248/129.303
Farr, D. F., and Rossman, A. Y. (2022). Fungal Databases, U.S. National Fungus Collections, ARS, USDAA. Available online at: https://nt.ars-grin.gov/fungaldatabases/ (accessed January 2, 2023).
Gomdola, D., Bhunjun, C. S., Hyde, K. D., Jeewon, R., Pem, D., Jayawardena, R. S., et al. (2022). Ten important forest fungal pathogens: a review on their emergence and biology. Mycosphere 13, 612–671. doi: 10.5943/mycosphere/13/1/6
Gramaje, D., Mostert, L., Groenewald, J. Z., and Crous, P. W. (2015). Phaeoacremonium: from esca disease to phaeohyphomycosis. Fungal Biol. 119, 759–783. doi: 10.1016/j.funbio.2015.06.004
Gryzenhout, M., Myburg, H., and Wingfield, B. D. (2006). Cryphonectriaceae (Diaporthales), a new family including Cryphonectria, Chrysoporthe, Endothia and allied genera. Mycologia 98, 239–249. doi: 10.1080/15572536.2006.11832696
Guterres, D. C., Galvão-Elias, S., dos Santos, M. D. D. M., de Souza, B. C. P., de Almeida, C. P., Pinho, D. B., et al. (2019). Phylogenetic relationships of Phaeochorella parinarii and recognition of a new family, Phaeochorellaceae (Diaporthales). Mycologia 111, 660–675. doi: 10.1080/00275514.2019.1603025
Hawksworth, D. L., Kirk, P. M., Sutton, B. C., and Pegler, D. N. (1995). Ainsworth and Bisby's Dictionary of the Fungi, 8th ed. reprinted. Oxford: CAB International.
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Huang, F., Hou, X., Dewdney, M. M., Fu, Y., Chen, G., Hyde, K. D., et al. (2013). Diaporthe species occurring on citrus in China. Fungal Divers. 61, 237–250. doi: 10.1007/s13225-013-0245-6
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hyde, K. D., Jeewon, R., Chen, Y. J., Bhunjun, C. S., Calabon, M. S., Jiang, H.B., et al. (2020a). The numbers of fungi: is the descriptive curve flattening? Fungal Divers. 103, 219–271. doi: 10.1007/s13225-020-00458-2
Hyde, K. D., Norphanphoun, C., Maharachchikumbura, S. S. N., Bhat, D. J., Jones, E. B. G., Bundhun, D., et al. (2020b). Refined families of Sordariomycetes. Mycosphere 11, 305–1059. mycosphere/11/1/7
Index Fungorum (2023). Available online at: http://www.indexfungorum.org (accessed March 24, 2023).
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, J., Buyck, B., Cai, L., et al. (2015). The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jiang, N., Fan, X., Tian, C., and Crous, P. W. (2020). Reevaluating Cryphonectriaceae and allied families in Diaporthales. Mycologia 112, 267–292. doi: 10.1080/00275514.2019.1698925
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformatics 20, 1160–1166. doi: 10.1093/bib/bbx108
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Maharachchikumbura, S. S. N., Chen, Y., Ariyawansa, H. A., Hyde, K. D., Haelewaters, D., Perera, R. H., et al. (2021). Integrative approaches for species delimitation in Ascomycota. Fungal Divers. 109, 155–179. doi: 10.1007/s13225-021-00486-6
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Bhat, J. D., Dayarathne, M. D., et al. (2016). Families of Sordariomycetes. Fungal Divers. 79, 1–317. doi: 10.1007/s13225-016-0369-6
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. G. B., McKenzie, E. H. C., Huang, S. K., Abdel-Wahab, M. A., et al. (2015). Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 72, 199–301. doi: 10.1007/s13225-015-0331-z
Maury-Lechon, G., and Curtet, L. (1998). “Biogeography and evolutionary systematics of Dipterocarpaceae,” in A Review of Dipterocarps: Taxonomy, Ecology and Silviculture, eds S. Appanah and J. M. Turnbull (Indonesia: Center for International Forestry Research), 5–44.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in 2010 Gateway Computing Environments Workshop (GCE) (New Orleans, LA: Institute of Electrical and Electronics Engineers), 1–8. doi: 10.1109/GCE.2010.5676129
Mostert, L., Groenewald, J. Z., Summerbell, R. C., Gams, W., and Crous, P. W. (2006). Taxonomy and pathology of Togninia (Diaporthales) and its Phaeoacremonium anamorphs. Stud. Microbiol. 54, 1–115. doi: 10.3114/sim.54.1.1
Nees von Esenbeck, C. G. (1816). Das System der Pilze und Schwämme. Würzburg: Stahelsche Buchhandlung, 334. doi: 10.5962/bhl.title.110007
O'Donnell, K., Kistler, H. C. O., Cigelnik, E. L., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Nat. Acad. Sci. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Rathnayaka, A. R., Wanasinghe, D. N., Dayarathne, M. C., Chethana, K. T., Bhat, D. J., Kuo, C. H., et al. (2020). Hyaloterminalis, a novel genus of Coryneaceae in order Diaporthales. Phytotaxa 474, 132–144. doi: 10.11646/phytotaxa.474.2.3
Réblová, M., Miller, A. N., Rossman, A. Y., Seifert, K. A., Crous, P. W., Hawksworth, D. L., et al. (2016). Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7, 131–153. doi: 10.5598/imafungus.2016.07.01.08
Réblová, M., Mostert, L., Gams, W., and Crous, P. W. (2004). New genera in the Calosphaeriales: Togniniella and its anamorph Phaeocrella, and Calosphaeriophora as anamorph of Calosphaeria. Stud. Microbiol. 50, 533–550.
Rigling, D., and Prospero, S. (2018). Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol. Plant Pathol. 19, 7–20. doi: 10.1111/mpp.12542
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian Phylogenetic Inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rossman, A. Y., Farr, D. F., and Castlebury, L. A. (2007). A review of the phylogeny and biology of the Diaporthales. Mycoscience 48, 135–144. doi: 10.47371/mycosci.MYC48135
Senanayake, I. C, Crous, P. W., Groenewald, J. Z., Maharachchikumbura, S. S. N., Jeewon, R., Phillips, A. J. L., Bhat, J. D., et al. (2017). Families of Diaporthales based on morphological and Phylogenetic evidence. Stud. Microbiol. 86, 217–296. doi: 10.1016/j.simyco.2017.07.003
Senanayake, I. C., Jeewon, R., Chomnunti, P., Wanasinghe, D. N., Norphanphoun, C., Karunarathna, A., et al. (2018). Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Divers. 93, 241–443. doi: 10.1007/s13225-018-0410-z
Senanayake, I. C., Jeewon, R., Hyde, K. D., Bhat, J. D., and Cheewangkoon, R. (2020). Taxonomy and phylogeny of Leptosillia cordylinea sp. nov. from China. Phytotaxa 435, 213–226. doi: 10.11646/phytotaxa.435.3.1
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Suetrong, S., Klaysuban, A., Sakayaroj, J., Preedanon, S., Rung-areerate, P., Phongpaichit, S., et al. (2015). Tirisporellaceae, a new family in the order Diaporthales (Sordariomycetes, Ascomycota). Cryptogam. Mycol. 36, 319–330. doi: 10.7872/crym/v36.iss3.2015.319
Suwannarach, N., Kumla, J., and Lumyong, S. (2016). Pseudoplagiostoma dipterocarpi sp. nov., a new endophytic fungus from Thailand. Mycoscience 57, 118–122. doi: 10.1016/j.myc.2015.12.002
Swofford, D. L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony (and other methods), version 4.0 b10. Sunderland: Sinauer Associates.
Tang, X., Jayawardena, R. S., Stephenson, S. L., and Kang, J. C. (2022). A new species Pseudoplagiostoma dipterocarpicola (Pseudoplagiostomataceae, Diaporthales) found in northern Thailand on members of the Dipterocarpaceae. Phytotaxa 543, 233–243. doi: 10.11646/phytotaxa.543.4.3
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vermeulen, M., Gryzenhout, M., Wingfield, M. J., and Roux, J. (2011). New records of the Cryphonectriaceae from southern Africa including Latruncellus aurorae gen. sp. nov. Mycologia 103, 554–569. doi: 10.3852/10-283
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Voglmayr, H., Castlebury, L. A., and Jaklitsch, W. M. (2017). Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales). Persoonia 38, 136–155. doi: 10.3767/003158517X694768
Voglmayr, H., and Jaklitsch, W. M. (2014). Stilbosporaceae resurrected: generic reclassification and speciation. Pers.: Mol. Phylogeny Evol. Fungi 33, 61–82. doi: 10.3767/003158514X684212
Voglmayr, H., Rossman, A. Y., Castlebury, L. A., and Jaklitsch, W. M. (2012). Multigene phylogeny and taxonomy of the genus Melanconiella (Diaporthales). Fungal Divers. 57, 1–44. doi: 10.1007/s13225-012-0175-8
Wang, Y. L., Lu, Q., Decock, C., Li, Y. X., and Zhang, X. Y. (2015). Cytospora species from Populus and Salix in China with C. davidiana sp. nov. Fungal Biol. 119, 420–432. doi: 10.1016/j.funbio.2015.01.005
White, T. J., Bruns, T., Lee, S. J. W. T., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Sánchez-García, M., Goto, B. T., Saxena, R. K., et al. (2022). Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13, 53–453. doi: 10.5943/mycosphere/13/1/2
Wu, G., Schuelke, T. A., and Broders, K. (2019). The genome of the butternut canker pathogen, Ophiognomonia clavigignenti-juglandacearum shows an elevated number of genes associated with secondary metabolism and protection from host resistance responses in comparison with other members of the Diaporthales. bioRxiv. 820977. doi: 10.1101/820977
Keywords: 2 new taxa, morphology, multi-gene phylogeny, saprophytic fungi, Sordariomycetes, taxonomy
Citation: Tang X, Lu Y-Z, Dissanayake LS, Goonasekara ID, Jayawardena RS, Xiao Y-P, Hyde KD, Chen X-M and Kang J-C (2023) Two new fungal genera (Diaporthales) found on Dipterocarpaceae in Thailand. Front. Microbiol. 14:1169052. doi: 10.3389/fmicb.2023.1169052
Received: 18 February 2023; Accepted: 09 May 2023;
Published: 05 June 2023.
Edited by:
Anna Gała̧zka, Institute of Soil Science and Plant Cultivation, PolandReviewed by:
Samantha Chandranath Karunarathna, Qujing Normal University, ChinaRajesh Jeewon, University of Mauritius, Mauritius
Copyright © 2023 Tang, Lu, Dissanayake, Goonasekara, Jayawardena, Xiao, Hyde, Chen and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-Chuan Kang, amNrYW5nQGd6dS5lZHUuY24=
 Xia Tang
Xia Tang Yong-Zhong Lu
Yong-Zhong Lu Lakmali S. Dissanayake1
Lakmali S. Dissanayake1 Ishani D. Goonasekara
Ishani D. Goonasekara Yuan-Pin Xiao
Yuan-Pin Xiao Kevin D. Hyde
Kevin D. Hyde Ji-Chuan Kang
Ji-Chuan Kang