- 1Division of Biosciences, College of Health, Medicine and Life Sciences, Brunel University London, Uxbridge, United Kingdom
- 2Department of Biology, University of York, York, United Kingdom
Faithful transmission of the genome from one generation to the next is key to life in all cellular organisms. In the majority of bacteria, the genome is comprised of a single circular chromosome that is normally replicated from a single origin, though additional genetic information may be encoded within much smaller extrachromosomal elements called plasmids. By contrast, the genome of a eukaryote is distributed across multiple linear chromosomes, each of which is replicated from multiple origins. The genomes of archaeal species are circular, but are predominantly replicated from multiple origins. In all three cases, replication is bidirectional and terminates when converging replication fork complexes merge and ‘fuse’ as replication of the chromosomal DNA is completed. While the mechanics of replication initiation are quite well understood, exactly what happens during termination is far from clear, although studies in bacterial and eukaryotic models over recent years have started to provide some insight. Bacterial models with a circular chromosome and a single bidirectional origin offer the distinct advantage that there is normally just one fusion event between two replication fork complexes as synthesis terminates. Moreover, whereas termination of replication appears to happen in many bacteria wherever forks happen to meet, termination in some bacterial species, including the well-studied bacteria Escherichia coli and Bacillus subtilis, is more restrictive and confined to a ‘replication fork trap’ region, making termination even more tractable. This region is defined by multiple genomic terminator (ter) sites, which, if bound by specific terminator proteins, form unidirectional fork barriers. In this review we discuss a range of experimental results highlighting how the fork fusion process can trigger significant pathologies that interfere with the successful conclusion of DNA replication, how these pathologies might be resolved in bacteria without a fork trap system and how the acquisition of a fork trap might have provided an alternative and cleaner solution, thus explaining why in bacterial species that have acquired a fork trap system, this system is remarkably well maintained. Finally, we consider how eukaryotic cells can cope with a much-increased number of termination events.
Introduction
Bacteria show a considerable degree of variability in terms of genome size and organization. Genomes can be as small as 0.159 Mbp, such as in the symbiotic bacterium Carsonella ruddii, which carries only 182 genes (Nakabachi et al., 2006), and as large as 13 Mbp in the myxobacterium Sorangium cellulosum, with 9,376 predicted open reading frames (Schneiker et al., 2007). The majority of bacteria carry a single chromosome, in the form of a covalently closed circle. About 5% carry multiple chromosomes (Touchon and Rocha, 2016). These species include Vibrio cholerae and close relatives, with two (Touchon and Rocha, 2016) and Paracoccus denitrificans, with three (Winterstein and Ludwig, 1998). A few species in the Actinomycetales, including the genus Streptomyces (Kirby, 2011), have linear chromosomes or a mixed state. For instance, Agrobacterium tumefaciens has one circular and one linear chromosome, along with two mega-plasmids (Nester, 2015).
In all bacteria, replication initiation is tightly regulated (Jameson and Wilkinson, 2017; Katayama et al., 2017; Hansen and Atlung, 2018) and restricted to a single origin (oriC) per chromosome (Gao and Zhang, 2008; Gao, 2015). Two replisomes are recruited at oriC by the main initiator protein, DnaA, forming two replication fork complexes that bidirectionally duplicate the genome. Bidirectional synthesis continues until the replication fork complexes fuse in an area opposite oriC where replication terminates (Masters and Broda, 1971; Prescott and Kuempel, 1972; Wake, 1972, 1973; Gyurasits and Wake, 1973). For bacteria with linear chromosomes replication ceases when fork complexes reach the chromosome ends (Cui et al., 2007; Rudolph et al., 2013). In both cases, the chromosome is divided into two replichores, each replicated with a defined directionality (Figure 1A).
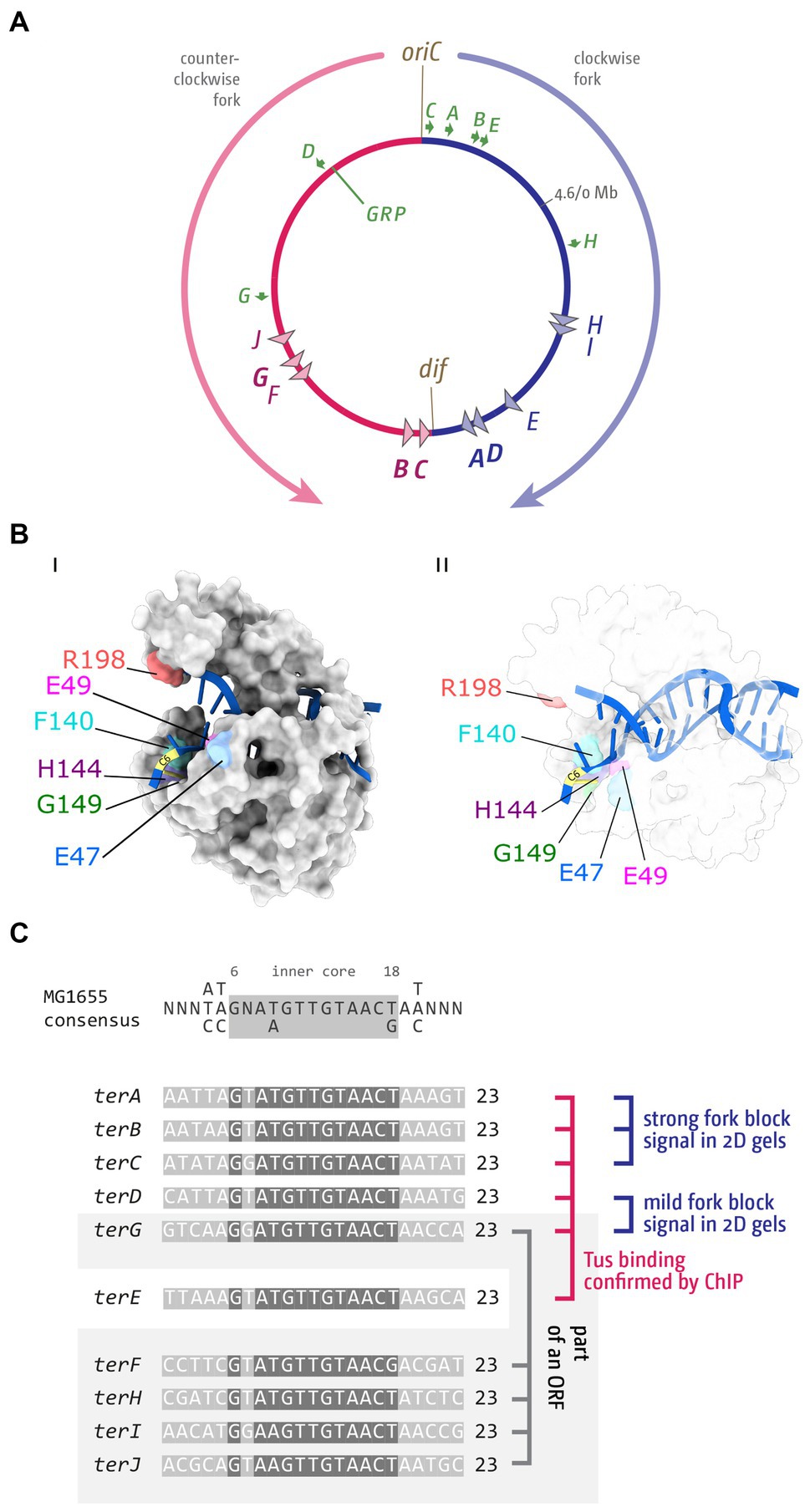
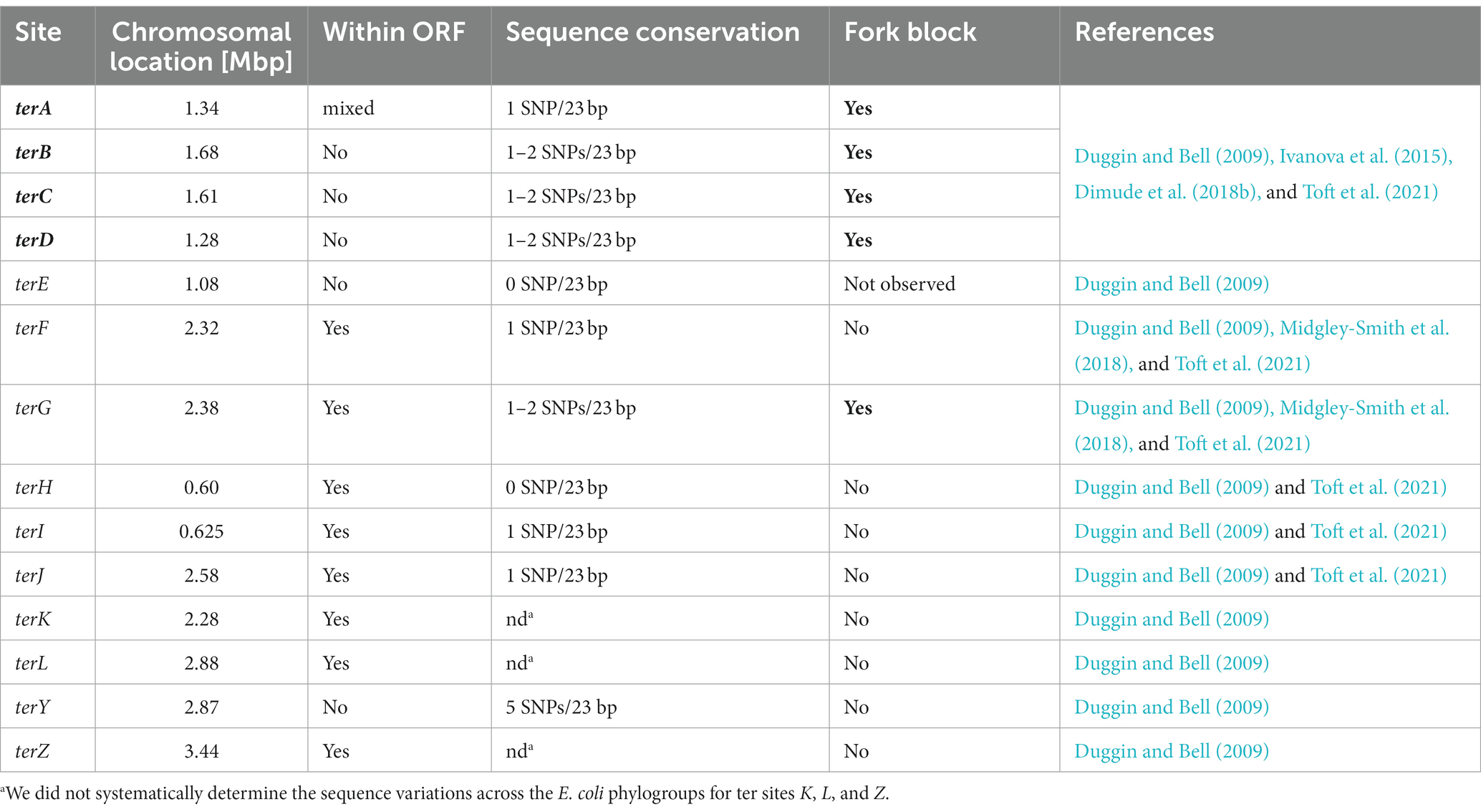
Figure 1. Chromosome structure and Tus-ter trap in Escherichia coli. (A) Schematic representation of the E. coli chromosome. Two replication forks are initiated at a single origin termed oriC and move in opposite directions along the DNA until they approach one another and fuse within the terminus region opposite oriC. A replication fork trap is formed in the terminus region via terminator sequences (terA–J) which are arranged as two opposed groups, with the pink terminators oriented to block movement of the clockwise replication fork and the blue terminators oriented to block the counter-clockwise fork. The locations of the dif chromosome dimer resolution site is marked. Locations of the rrn operons, which are particularly highly transcribed under fast growth conditions, are shown by green arrows, with the arrow pointing in the direction in which transcribing RNA polymerase molecules travel. ‘GRP’ indicates the location of a cluster of genes encoding ribosomal proteins, almost all of which are transcribed co-directionally with replication. (B) Structure of Tus-ter (PDB ID: 2I06) (Singleton et al., 2001). (Bi) Illustration of the “locked” conformation formed by DNA unwinding at the nonpermissive face. The cytosine base at position 6 of ter (C6), which flips into a specific binding site on the nonpermissive face of Tus to form the ‘lock’, is indicated, and important amino acid residues contributing to the locked conformation highlighted. (Bii) As above, but a transparent view is shown to allow easier visualisation of the DNA in general and the flipped C6 in particular. (C) Sequences of ter sites A–J from E. coli MG1655. The ter consensus sequence is shown at the top. Base pairs 1–5, 7, and 19–23 show a higher degree of variability and are shaded in a lighter grey. The highly conserved ter core (6, 8–18) is highlighted by a darker grey. Tus binding (Toft et al., 2021), in vivo fork blocking observed by 2D gel electrophoresis (Duggin and Bell, 2009) and analysis whether a ter site is part of an open reading frame (Duggin and Bell, 2009; Goodall et al., 2021) are indicated (see text for further details).
The fact that replication is restricted to two replication fork complexes dictates that, for bacteria with a circular chromosome, each round of chromosome duplication is concluded by exactly one termination event in an area opposite oriC. For termination two main mechanisms were initially considered in the literature: the free fusion of moving forks, or site-specific termination. Forks may proceed around the chromosome freely and fuse wherever they happen to meet. Alternatively, there might be distinct locations where fork progression is blocked, forcing fork fusion to take place in a distinct location (Masters and Broda, 1971; Bird et al., 1972; Hendrickson and Lawrence, 2007; Kono et al., 2012).
Replication fork complexes will encounter a wide variety of potential blocks on their way to the terminus area, such as stable protein-DNA complexes, G4 quadruplex DNA or DNA lesions (Lang and Merrikh, 2018; Marians, 2018; Bianco, 2020; Linke et al., 2021). Many of these obstacles are stochastic in nature and can occur at different chromosomal locations for each round of genome duplication. However, there are also mechanisms that have specifically evolved to arrest replication complexes (Hyrien, 2000; Rothstein et al., 2000; Labib and Hodgson, 2007; Hizume and Araki, 2019). The first examples of distinct blocking points for replication complexes were identified in plasmid systems. Replication of the conjugative R plasmid R6K was reported to be either unidirectional or bidirectional (de Graaff et al., 1978), based on a complex mechanism of replication initiation (Abhyankar et al., 2003, 2004; Rakowski and Filutowicz, 2013), but progression of both the clockwise and counter-clockwise forks is blocked by a specific terminus (Crosa et al., 1976). In the R1 plasmid, a distinct termination system is in use to specifically enforce unidirectional replication. Between the minimal origin, oriR, and the start site for leading strand replication, two ter sites are located, terRL and terRR (Hill et al., 1988b). terRL blocks the counter-clockwise fork almost immediately. Thus, the plasmid is replicated unidirectionally via the clockwise fork until it is blocked at terRR (Krabbe et al., 1997).
The first chromosomal fork block was identified in E. coli (Kuempel et al., 1977; Louarn et al., 1977), and shortly afterwards in B. subtilis (Weiss et al., 1981). In E. coli the integration of an ectopic replication origin into the chromosome was used to show that progression of replication forks was blocked in the termination area (Kuempel et al., 1977; Louarn et al., 1977, 1979). By using a temperature-sensitive version of the main replication initiator protein DnaA to inactivate oriC, and inserting an ectopic P2 prophage origin to drive replication initiation at a site 1 Mbp away from oriC, the authors showed that termination still occurred diametrically opposite oriC, revealing that progression of replication must be impeded in an area opposite oriC (Kuempel et al., 1977; Louarn et al., 1977, 1979). Prompted by these results, the use of sophisticated marker frequency analysis revealed specific termination sites, initially called T1 and T2 and later changed to ter, in line with plasmid-based systems such as R6K and R1, with the Louarn group identifying the innermost ter sites terA and terC, and the Kuempel lab identifying terA and terB (Figure 1A; de Massy et al., 1987; Hill et al., 1987, 1988a; Hill and Marians, 1990). In addition, the Kuempel lab identified a region close to terB that was needed for ter sites to act as a block, and they named the terminus function termination utilization substance (tus) (Hill et al., 1988a). Further analysis revealed the sequence of the tus gene (Hill et al., 1989), and in vitro work by Thomas Hill and Ken Marians showed how the binding of Tus protein to a ter sequence can arrest progression of DNA replication (Hill and Marians, 1990; Figure 1B).
By developing ter consensus sequences, the use of radioactively labeled probes allowed identification of additional ter sites D, E, and F (François et al., 1989; Hidaka et al., 1991; Sharma and Hill, 1992). Once the whole genome sequence of E. coli was available (Blattner et al., 1997), bioinformatics analysis resulted in the identification of terG–terJ (Neidhardt et al., 1996). Thus, the E. coli chromosome is divided into two replichores, one replicated clockwise and the other counter-clockwise, with 5 ter sites being oriented to block progression of the clockwise replication fork, while the other 5 ter sites block the counter-clockwise moving replication fork complex (Figure 1A). Finally, bioinformatics analysis also allowed the identification of the putative and weak ter sites K, L, Y, and Z (Duggin and Bell, 2009).
The specific termination site identified in the Gram-positive Bacillus subtilis (Weiss et al., 1981) was later characterized to consist of a cluster of ter sites blocking progression of the clockwise replication fork complex, while a second cluster blocked progression of the counter-clockwise replication fork complex (Wake, 1997; Neylon et al., 2005; Rudolph et al., 2019). While acting in a similar fashion, the ter sequences show very little similarity to the E. coli sequences. Moreover, the B. subtilis terminator protein, RTP, has little structural similarity to Tus. Importantly, these differences indicate that the two termination systems developed via convergent evolution (Franks et al., 1995; Neylon et al., 2005).
In this review we will discuss how the architecture of bacterial chromosomes limits pathological consequences that can arise when replication forks fuse and explore why a fork trap in the termination area might provide an evolutionary benefit by limiting the local and global impacts caused by fork fusions. Based on the pathological consequences that can arise as a result of fork fusions, we suggest that the strict use of only a single origin per chromosome observed in all bacteria helps to reduce the number of termination events to exactly one, thereby limiting pathologies triggered by fork fusions.
Mechanism of fork arrest by Tus-ter in Escherichia coli
How can a single Tus-ter complex be such an efficient block to the progression of replication fork complexes? Functional ter sites bound by the monomeric terminator protein Tus block replication in a polar manner. Key to this polar activity is that Tus binding is asymmetric, a feature that has been extensively studied (Neylon et al., 2005; Elshenawy et al., 2015; Pandey et al., 2015; Berghuis et al., 2018). Arrest of an approaching replication fork complex is triggered when the double-stranded DNA immediately adjacent to one face of Tus, called the non-permissive face, is unwound as the fork approaches (Mulcair et al., 2006). The structural change in the opened double helix induces specific contacts between the DNA and Tus that result in the formation of ‘locked complex’, which will block progression of the oncoming replisome with surprising efficiency, a mechanism referred to as a ‘mousetrap’ (Mulcair et al., 2006). Specifically, a base on the leading strand template (C in position 6) flips into a binding site on Tus, guided by nearby residues (Figure 1B and Table 1). This mechanism of ‘clicking into place’ essentially blocks any movement along the DNA by a ratchet-type mechanism, and also slows the dissociation of Tus (Mulcair et al., 2006; Berghuis et al., 2015; Elshenawy et al., 2015; Pandey et al., 2015). This ‘locked’ Tus-ter complex can be generated by unwinding of the double-stranded DNA alone; DnaB helicase is not essential for its formation (Bastia et al., 2008; Berghuis et al., 2015; Elshenawy et al., 2015; Toft et al., 2022). Some stabilization by specific protein–protein interaction between Tus and DnaB has been reported (Mulugu et al., 2001), but to what extent they contribute towards the efficient arrest is not clear. Multiple labs have highlighted key residues which have major roles in the formation of a successful Tus-ter lock (Table 1). Mutants in these residues alter the C6 binding, ssDNA guiding, and slowing the momentum of the approaching replisome.
Table 1. Key amino acid residues of E. coli Tus protein and their role in blocking progression of replication fork complexes.
If individual Tus-ter complexes block replication progression so effectively, why are so many ter sites found in the E. coli chromosome? It has become increasingly clear that not all sequences identified as ter sites block forks. Also, even a ter site with a strong blocking activity is not an absolute block. In a study using 2-dimensional neutral-neutral gel electrophoresis analysis, Duggin and Bell were able to show that in wild type cells the vast majority of blocked forks occur at the innermost ter sites of the fork trap system. Highest levels were detected at terC and terB, followed by terA. In this study rather mild levels of signal were found for terD and terG (Figure 1C), and the lowest levels were identified for terH and I, while ter sites E, F and J showed no signal in wild type cells (Duggin and Bell, 2009). In a recent study by the Schaeffer lab, ChiP-Seq was used to identify which ter sites are actually occupied by Tus protein. The authors identified ter sites A–E and G (Figure 1C), whereas no Tus occupancy was observed at any of the other ter sites (Toft et al., 2021).
These observations fit well with our own data. The replichore arrangement can be significantly skewed by introducing additional ectopic copies of the oriC sequence into the chromosome. In a strain with two active replication origins (Wang et al., 2011; Ivanova et al., 2015; Dimude et al., 2018b), one fork reaches Tus-ter complexes much earlier than the other. Depending on whether the ectopic origin is located in the left-hand or right-hand replichore, forks get arrested at terC/B or terA/D (Figure 2A), which can be visualized by using high-resolution marker frequency analysis (MFA) from whole genome sequencing (WGS) (Ivanova et al., 2015; Dimude et al., 2018b; Syeda et al., 2020). These replication profiles, generated by WGS, reveal initiation sites of synthesis, such as oriC, as distinct peaks, whereas termination sites form a v-shaped valley, with a replication gradient between initiation and termination sites (Skovgaard et al., 2011; Figure 2Bi). Our MFA analyses confirm that the four innermost ter sites A–D form a strong block. All four are low points in the replication profile (Figures 2Bii,iii), in line with the idea that individual Tus-ter complexes can be overcome occasionally, but the strong ‘step’ in the replication profile confirms the considerable block that is formed, especially by the innermost ter sites A and C/B.
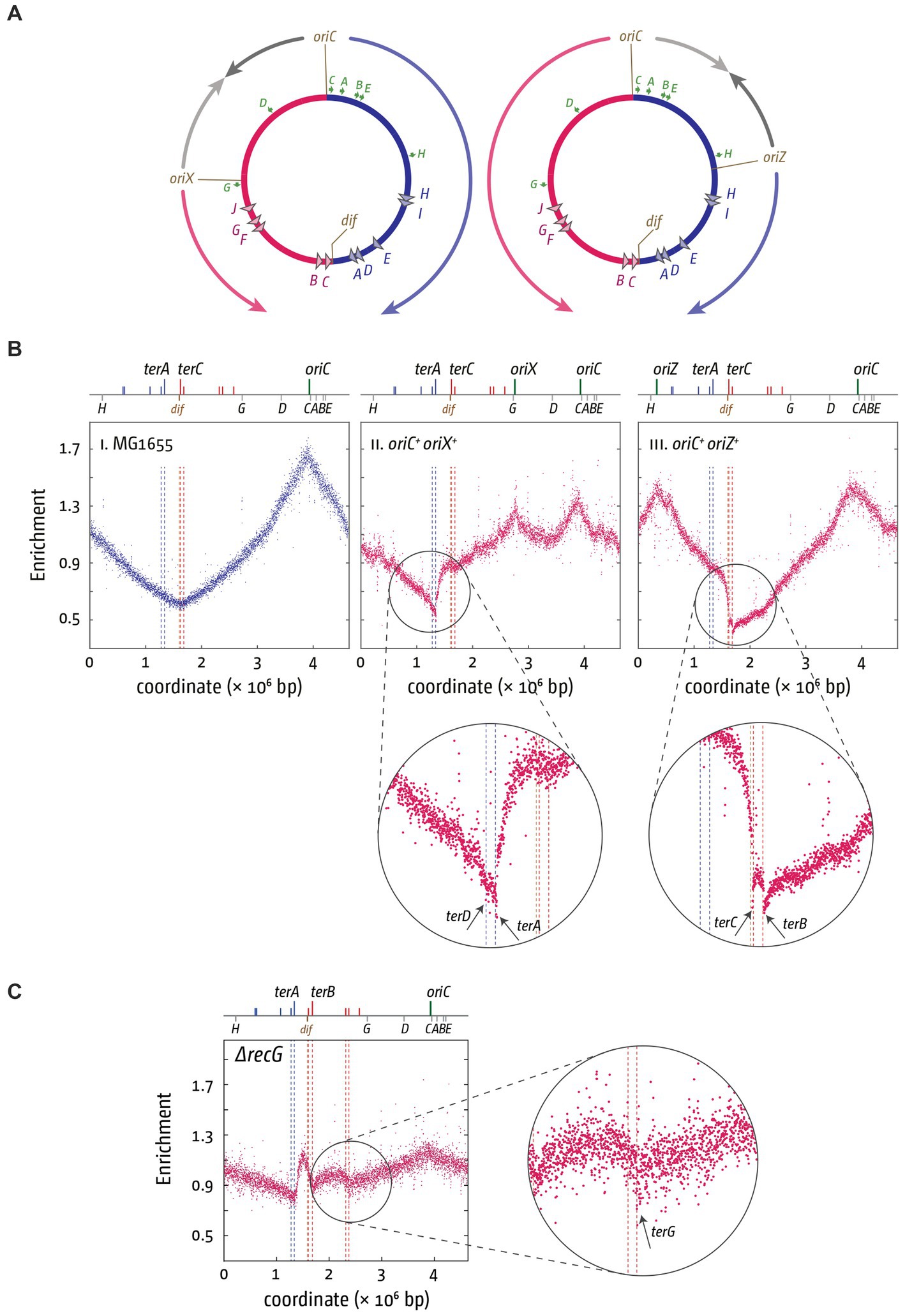
Figure 2. Altered replichore structure in E. coli cells with additional ectopic replication origins. (A) Schematic representation of E. coli chromosomes with additional ectopic replication origins oriX (left) and oriZ (right), respectively. Positions of oriC, ter sites as well as the dif site and rrn operons A–E, G, and H are shown. (Bi–iii) Marker frequency analysis (MFA) in MG1655, oriC+ oriX+ and oriC+ oriZ+ cells. The number of reads (normalized against reads for a stationary phase wild type control) is plotted against the chromosomal location. A schematic representation of the E. coli chromosome showing positions of oriC, oriX and oriZ (green lines) and ter sites (all above) as well as dif and rrn operons A–E, G, and H (all below) is shown above the plotted data. The MFA raw data were taken from Ivanova et al. (2015) and Dimude et al. (2018b) and re-plotted to allow axis scale changes, if necessary. The areas where forks are blocked by Tus-ter complexes are highlighted by circles and details magnified below to highlight the fork blocking ability of ter sites A, B, C, and D. (C) Over-replication in the termination area of ΔrecG cells growing in M9 minimal salts with glucose. The area where forks escaping the innermost ter sites are blocked by Tus-ter complexes are highlighted by a circle, and details magnified to the side to highlight the fork blocking ability of ter site G. The MFA raw data were taken from Midgley-Smith et al. (2018) and re-plotted.
Additional information comes from the replication profiles of strains that show over-replication of the termination area. Over-replication of the chromosome can be triggered by the absence of certain proteins, such as Tus, RecG or 3′ exonucleases, resulting in a peak of synthesis between ter sites A and B/C (Markovitz, 2005; Duggin et al., 2008; Rudolph et al., 2009, 2013; Wendel et al., 2014; Midgley-Smith et al., 2019). Synthesis can proceed beyond ter sites C and B under certain conditions in cells lacking RecG helicase (Figure 2C; Midgley-Smith et al., 2018). The precise molecular mechanism for this regular progression of synthesis beyond ter sites C and B is not clear. However, the ChIP-Seq data from the Schaeffer lab would predict that escape synthesis should proceed past terF before getting blocked by terG (Toft et al., 2021). This is indeed precisely what we observed, supporting the idea that terF is not blocking the progression of forks in normal cells, whereas terG can block ongoing synthesis in wild type cells (Figure 2C). Thus, while the majority of the work of blocking forks is done by three of the innermost ter sites A and B/C, with terD clearly being active (Figure 2B) but contributing relatively little, as observed before (Duggin and Bell, 2009), some progressing synthesis can also be blocked by more distal ter sites, such as terG.
Given that the innermost Tus-ter complexes in particular very efficiently block progression of ongoing synthesis, the fusion of replication fork complexes will occur almost exclusively within the innermost termination area. However, this does not answer the question of whether Tus-ter complexes are a regular component of termination, or if they are only involved when progression of the two replication fork complexes becomes asynchronous, for example if one of the two complexes is arrested by a DNA lesion, a secondary structure or a protein-DNA complex for any length of time.
Early DNA labeling experiments demonstrated that fork fusion events typically occur close to the numerical midpoint of the chromosome and away from Tus-ter complexes (Bouché et al., 1982). This finding is supported by our own MFA-based replication profiles. MFA-based replication profiles use 1 kb windows per individual data point, which means resolution is relatively coarse (Skovgaard et al., 2011; Rudolph et al., 2013). However, within the limits of this resolution, our data demonstrate that the fork fusion point, as determined by LOESS regression, is in precisely the same location in the presence and absence of Tus protein (Rudolph et al., 2013; Dimude et al., 2016). The only change observed is a mild distortion of the marker frequencies in the vicinity of the fusion point (see Dimude et al. (2016) and Goodall et al. (2021) for a direct comparison of the replication profiles in the presence and absence of Tus terminator proteins). In contrast, in cells in which the replichore arrangement is artificially skewed by an additional ectopic origin, the fork fusion point is significantly shifted if the tus gene is deleted, as expected (Ivanova et al., 2015; Dimude et al., 2018b; Syeda et al., 2020). These findings suggest that a significant proportion of fork fusion reactions take place in a relatively short stretch between the dif chromosome dimer resolution site and terB, regardless of whether Tus is present or absent (Rudolph et al., 2013; Ivanova et al., 2015; Dimude et al., 2016). However, the fact that the termination area changes shape in the replication profile of cells lacking Tus demonstrates that Tus-ter complexes do contribute to termination on a regular basis (Rudolph et al., 2013; Dimude et al., 2015). Indeed, in a modelling study by Kono and colleagues it was demonstrated that a mixture of Tus-ter based and free fusion reactions was shown to result in GC-skew profiles that represented the experimental data most closely (Kono et al., 2012). Additionally, the authors did observe significant differences of the GC-skew profiles in bacteria which do not have a Tus or RTP-based fork trap system (Kono et al., 2012). Taken together, the data suggest that replication often terminates via the free fusion of forks, but if a fork trap system is present this also contributes to the termination process on a regular basis.
Fork trap systems in other bacterial species
Galli and co-workers recently analyzed the bacterial species that have a Tus-ter fork trap system. They showed that the presence of a replication fork trap in the termination area of the chromosome is a specialized feature of a relatively small group of bacteria (Galli et al., 2019). This suggests that perhaps the majority of bacterial species do not have a fork trap mechanism. Proteins with homology to Tus can be found in most Enterobacteriales, the Pseudoalteromonas and most Aeromonadales (Galli et al., 2019). As already highlighted, a fork trap system can be found in B. subtilis, but distribution amongst Bacillus species is even more restricted and the dissimilarity to Tus-ter strongly suggests that its origin is different and has arisen as a result of convergent evolution, highlighting the importance of a fork trap (Franks et al., 1995; Neylon et al., 2005; Galli et al., 2019). Indeed, the binding mechanisms of the terminator proteins to their ter sites are different, with Tus being a polar monomer and the B. subtilis terminator protein, RTP, being a dimer that requires a conformation change to arrest forks (Eli Bussiere et al., 1995; Franks et al., 1995; Pai et al., 1996; Griffiths et al., 1998; Wilce et al., 2001). In the B. subtilis chromosome, terminator sites are formed via two ‘half-sites’, the A and the B site, each bound by an RTP dimer (Lewis et al., 1990). Thus, in total four RTP monomers bind to each ter sequence (Lewis et al., 1990). However, binding of an RTP dimer to the A site is much weaker than binding of the second dimer to the B site. Consequently, forks arriving at the B site first will be blocked, while forks arriving at the A site can proceed (Smith and Wake, 1992), which is fundamentally different to the E. coli Tus-ter system. Interestingly, the transcriptional regulation of the genes encoding for RTP and Tus are achieved through auto-regulation, with a ter site being located in the promotor area of the respective genes (Natarajan et al., 1991; Ahn et al., 1993).
The phylogenetic analysis by Galli and co-workers resulted in the interesting hypothesis that the E. coli fork trap system might have been domesticated from plasmid-based precursor systems such as in R1 (Figure 3A). The authors highlight that in the Pseudoalteromonadacae, tus is always located directly adjacent to the origin of the secondary chromosome, which is a mega plasmid (Galli et al., 2019; Figure 3B). Indeed, the GC skew for the secondary chromosome in Pseudoalteromonas haloplanktis fits a unidirectional replication mode (Médigue et al., 2005; Xie et al., 2021). A fork trap in close proximity to the origin, together with the unidirectional mode of replication, is indeed very reminiscent of the system found in the R1-type plasmids (Figure 3A; Krabbe et al., 1997).
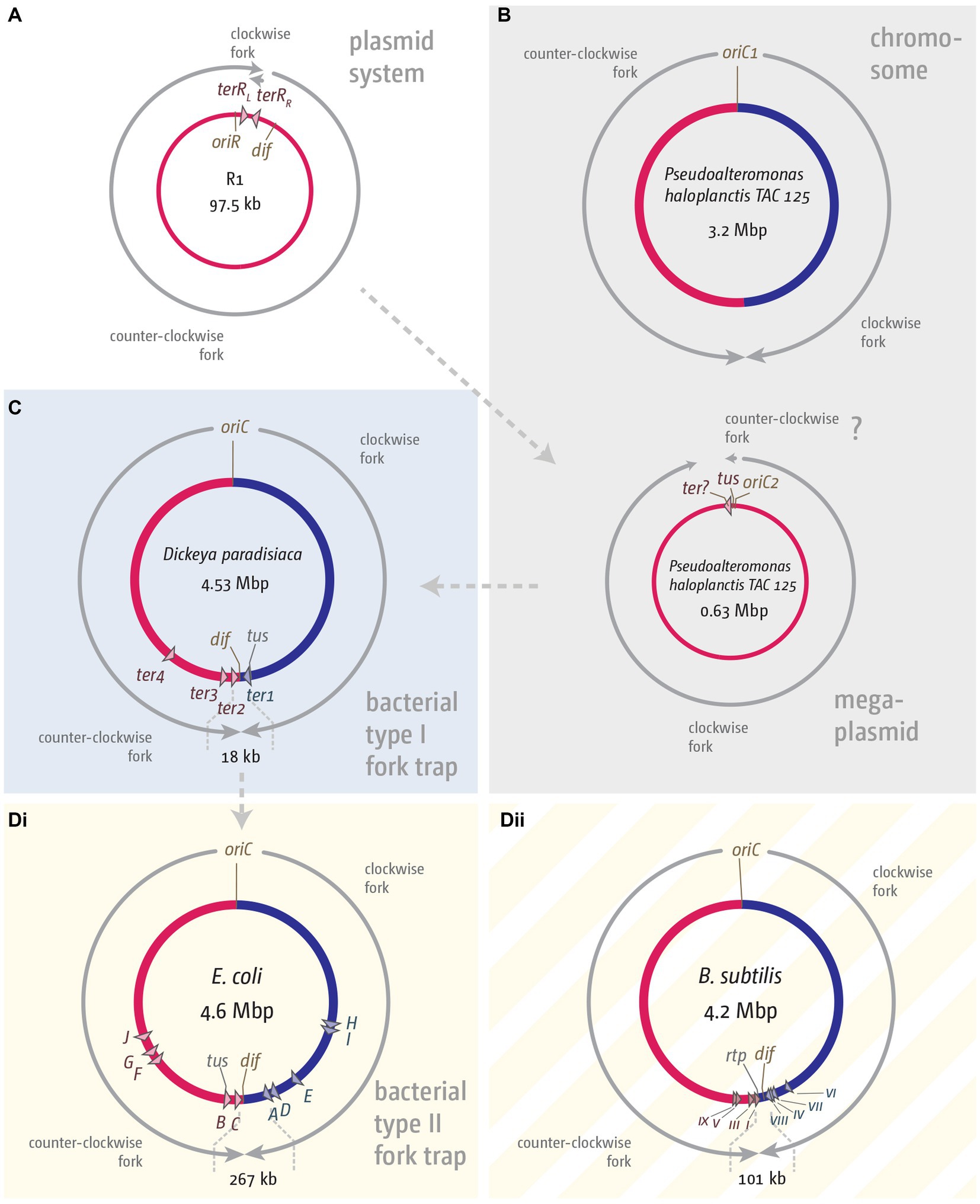
Figure 3. Comparison of fork trap systems in various plasmids and bacterial chromosomes. (A) Replication dynamics and fork trap system in the plasmid R1. (B) Replication and fork trap features of the primary and secondary chromosome in Pseudoalteromonas haloplanctis TAC 125. The secondary chromosome contains a tus gene, which is located next to the origin of replication, oriC2. The presence of a ter site is only implicated and therefore marked with a question mark. (C) Type I fork trap system in Dickeya paradisiaca, as described in Toft et al. (2022). (D) Type II fork trap system in E. coli (Di) and the fork trap system in Bacillus subtilis (Dii). See main text for further details.
If the fork trap system was domesticated in this fashion, it is easy to imagine that the unidirectional mode of replication will become rate-limiting for the size of chromosomes, a problem easily solved by a ‘remodelling’ of the chromosome that resulted in the integration of sequences between the origin and the Tus-ter complexes (Figure 3C). A prediction from this hypothesis would be that there might be chromosomes with a single ter site limiting progression of one of the replichores, and where the tus gene is associated with this innermost ter site. In a recent study by Toft and colleagues it was indeed highlighted that two distinct fork trap architectures exist (Toft et al., 2021). The fork trap system in E. coli is classed as a type II system, because the ter site regulating transcription of the tus gene is terB, which is a secondary rather than the innermost ter site (Toft et al., 2021). Outside of the Enterobacteriaceae, another system exists which the authors classified as a type I system. In organisms such as Edwardsiella tarda or Dickeya paradisiaca a single ter site is found to block progression of one replichore, and also acts as a transcriptional repressor of the tus gene (Toft et al., 2021, 2022), exactly as predicted. In organisms such as Edwardsiella tarda and Cedecea neteri the second replichore is also restricted by only a single ter site, whereas in organisms such as Dickeya paradisiaca two functional ter sites are used (Toft et al., 2021, 2022). The authors speculate that the type I system is the ancestral system, whereas the type II system is more divergent (Toft et al., 2021, 2022), which seems overall to fit well with the notion of a plasmid-derived system (Figure 3; Galli et al., 2019).
However, while the classification into a type I and II system is helpful to some degree, the systems are likely to be more similar than they appear upon casual inspection. There is no doubt that the number of innermost ter sites differ across species, together with the association and relative location of the tus genes. But the outer ter sites F–J in E. coli are all part of open reading frames, which means that, regardless of whether it ever was a functioning ter site or whether the sequence similarity is coincidence, the sequence will be conserved as part of the maintenance of the genes they are in. Both arguments are not mutually exclusive, but the ChIP-Seq data from the Schaeffer lab highlight that the occupancy of ter sites F and H–J is low (Toft et al., 2021). Indeed, when we analyzed the sequence conservation of all ter sites across all phylogenetic groups in E. coli, we found a revealing trend: for the innermost ter sites A–D, which clearly have a role in blocking forks (Duggin and Bell, 2009; Toft et al., 2021), strict conservation was observed for all bases that make contact with Tus. However, especially at bases 1–5 and 7 numerous variations were found, which are unlikely to affect Tus binding (Goodall et al., 2021). Interestingly, the same pattern was observed for terG (Goodall et al., 2021), the one ter site which is not part of the inner fork trap, but for which Tus binding and significant fork blocking activity was demonstrated (Duggin and Bell, 2009; Midgley-Smith et al., 2018; Toft et al., 2021). For ter sites E, F and H–J far fewer sequence variations were observed, a trend that extends beyond the 23 bp ter sequence (Goodall et al., 2021), in line with the idea that the sequence conservation might be driven to a large extent by the ORF they are in Table 2.
In this analysis terE stands out, because it is not part of an ORF (Table 2), but is still highly conserved (Goodall et al., 2021). terE is located within the efeUOB operon, which, in enterohaemorrhagic E. coli O157:H7, encodes an iron transport system (Cao et al., 2007). terE is located between efeU, a pseudo-gene in E. coli K12 (Grosse et al., 2006), and efeO. In our sequence analysis across all E. coli phylogroups we did not find any variations of terE, nor of the wider region around the actual terE sequence (Goodall et al., 2021). This conservation makes it tempting to speculate that it is located in a sequence environment with an important regulatory function, even though we do not know what this function is. In contrast, when we analyzed terY, a pseudo ter site that neither acts as a part of the fork trap nor is it located within an ORF (Table 2), we observed significant sequence variations across the phylogenetic groups, as well as changes in its relative location, in line with the idea that this sequence snippet is subjected to unselective evolutionary drift (Goodall et al., 2021).
Taken together, the data suggest that we can simplify the fork trap system in E. coli and related bacteria by focusing on the innermost ter sites A–D as those being active in termination. The main difference between the type I and type II systems remains whether the border of the fork trap is defined by a single ter site associated with the tus gene (type I), or whether a cluster of ter sites is used, with tus associated with one of the secondary ter sites (type II) (Toft et al., 2022). All of the other ter sites in E. coli are likely to be still present because of selection pressures unrelated to termination issues.
Fork trap systems can pose problems for DNA replication
The defined architecture of the bacterial chromosome, where a single origin results in each replichore being replicated by a single replication fork complex, dictates that any restriction to fork movement will become problematic if a fork is held up unexpectedly, for example at a small DNA lesion or a protein-DNA complex (Lang and Merrikh, 2018; Marians, 2018; Bianco, 2020; Linke et al., 2021). Any stalled fork has to be reactivated to complete chromosome replication, as the only other fork available is blocked by Tus-ter complexes, a clear disparity to the concept of dormant origins in eukaryotic cells (Blow et al., 2011; Shima and Pederson, 2017). If fork reactivation cannot be achieved and the other fork is arrested by the multiple Tus-ter complexes in its way, the chromosome will remain only partially replicated, with potentially lethal consequences for the cell.
The difficulties caused by this scenario can be shown experimentally in cells in which the replichore arrangement is distorted. This was done in B. subtilis in which a ter site was integrated in blocking orientation into an ectopic location of the chromosome, on the edge of the replication fork trap system. Forks were blocked for a significant period of time, causing significant morphological changes of the affected cells (Franks and Wake, 1996). Indeed, similar experiments were done in E. coli by the Michel lab, showing that arrested forks only can be reactivated if a second round of replication reaches the arrested primary fork, resulting in the generation of linear DNA molecules that engage in homologous recombination and allow reactivation of forks. If recombination is inactivated, the presence of an ectopic ter site proved lethal (Bidnenko et al., 2002, 2006).
In our own work we used strains where, instead of using ectopic ter sequences, the origin was moved 1 Mbp into an ectopic position. In this scenario one of the two forks initiated will duplicate ¼ of the chromosome before being arrested at Tus-ter complexes, while the other fork has to duplicate the remaining ¾ of the chromosome. In addition, this fork has to replicate ¼ of the chromosome in an orientation opposite to normal. The doubling time of such a construct was approximately 40 min, twice that of the 20 min normally observed for wild type cells (Ivanova et al., 2015). Inactivation of the replication fork trap by the deletion of tus reduced the doubling time to ~30 min (Ivanova et al., 2015). Together, the findings in these experimental systems strongly highlight the impact the replication fork trap has to impede the movement of replisomes (Franks and Wake, 1996; Bidnenko et al., 2002; Ivanova et al., 2015).
However, even if one fork is blocked at an obstacle only for a limited period of time, the replication fork trap still poses a threat to the completion of DNA synthesis. At fork speeds of 700–1,000 nt/s (Baker and Bell, 1998; Pham et al., 2013) even a short delay of one of the forks will cause the second to get arrested at a Tus-ter complex. This means that, upon reactivation, the temporarily arrested fork will eventually reach a replication fork complex that is stably arrested at a Tus-ter complex. While the arrest of a single replisome by Tus-ter complexes has received considerable attention, and a substantial number of studies have looked at the effects of Tus-ter on the progression of reconstituted replication fork complexes in vitro (Khatri et al., 1989; Lee et al., 1989; Hill and Marians, 1990; MacAllister et al., 1990; Gottlieb et al., 1992; Hiasa and Marians, 1994; Neylon et al., 2000), the fusion reaction of a freely moving fork with another stably arrested at a Tus-ter complex has not really been studied, even though this is likely to be physiologically very relevant. We were recently able to reconstitute this specific reaction in vitro. We used a plasmid template with an oriC sequence which, upon adding all the purified E. coli replisome components, together with the DnaA and DnaC initiator proteins, allows the formation of fully functional replication fork complexes (Jameson et al., 2021). The plasmid substrate contains terB, which, upon addition of Tus, will block progression of one of the two established forks (Figure 4A). The second fork moving in the opposite direction can be blocked at an array of 22 copies of the lacO sequence if the lac repressor protein LacI is added. The block is temporary, because addition of IPTG allows release of the block and continuation of synthesis until the second fork reaches the Tus-ter complex from the permissive side (Figure 4A). This precisely mimics the scenario of a moving fork running into another stably arrested at a Tus-ter complex (Jameson et al., 2021).
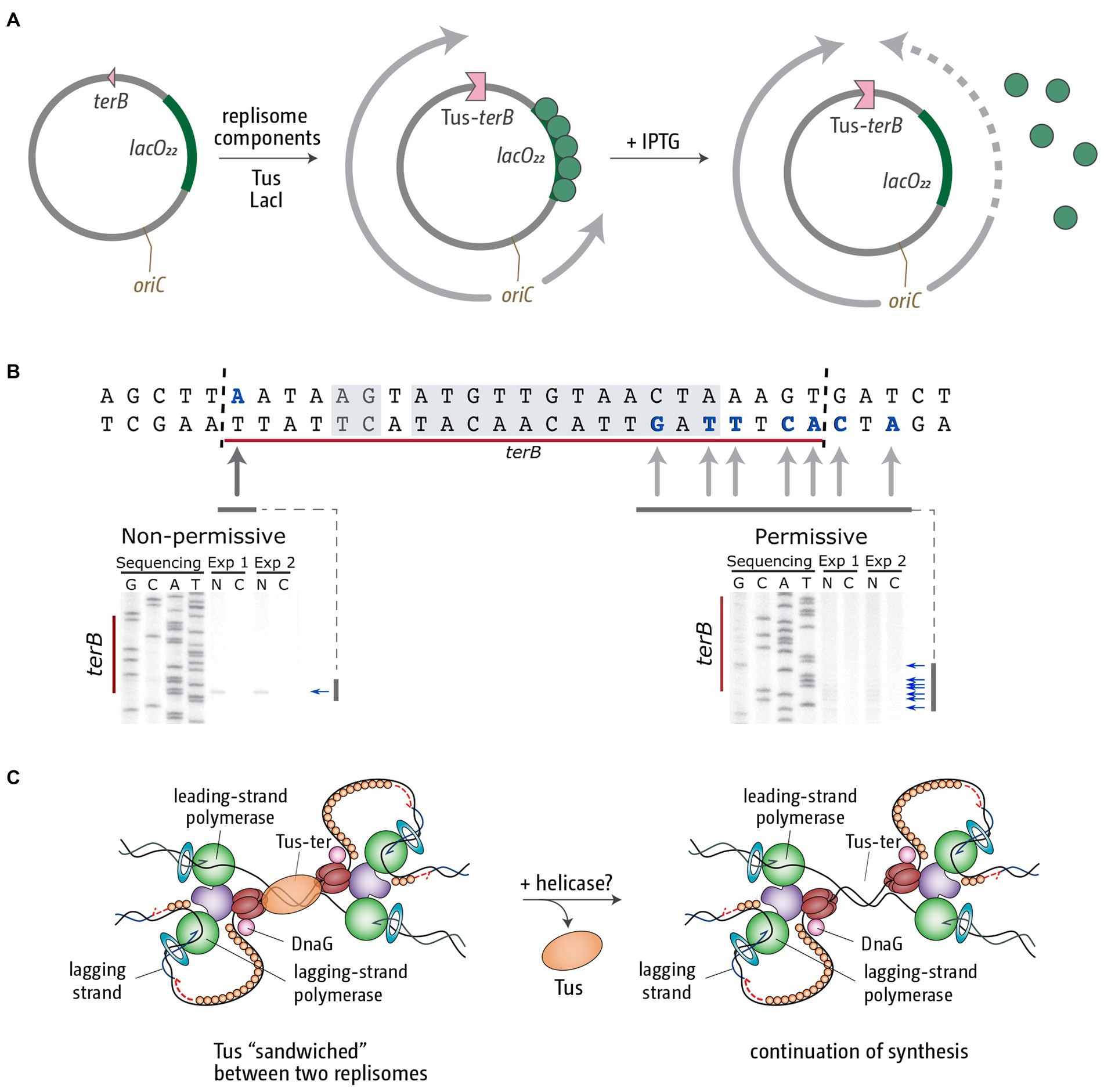
Figure 4. Replication fork fusion can be controlled in vitro to occur at Tus-terB. (A) pKJ1 replication assay template, indicating the location of replication initiation at oriC, the terB site and the lacO22 array. Replisome movement is shown pre (middle) and post (right) LacI-lacO block removal. (B) terB sequence indicating leading strand stop locations (bold and blue) and the Tus-binding site (shaded area). Mapping analysis of the leading strand products approaching Tus-terB in the non-permissive and permissive direction is shown below. Nicked products (stop sites) are indicated by arrows. Mapping data were reproduced from Jameson et al. (2021). (C) Schematic representation of the termination intermediates generated by a fork fusion event taking place at Tus-terB. We believe the most likely candidate for the removal of Tus from this super-complex in vivo is an as yet unidentified helicase.
Termination mapping analysis revealed that in this scenario the template also remains under-replicated. We observed a gap of between 15 and 24 bp, which matches the footprint of Tus (Figure 4B; Jameson et al., 2021). Thus, it is very tempting to speculate that, as a result of this particular type of fusion reaction, Tus remains stably bound, sandwiched between two replication fork complexes (Figure 4C; Jameson et al., 2021). We currently do not know how this complex is resolved in vivo. It seems likely that a helicase needs to be involved to remove the bound Tus protein and facilitate progression of at least one of the two forks to facilitate completion of DNA synthesis (Figure 4C). However, we do not currently know which helicase might be involved in this reaction (Jameson et al., 2021).
Nevertheless, both scenarios highlight that, within the defined architecture of the E. coli chromosome, a fork trap poses a risk that can prove fatal. We therefore wanted to investigate whether we could find a fork trap system that was inactivated by point mutations in specific E. coli strains. E. coli can be found in the gastrointestinal tract of a wide variety of warm-blooded animals (Stoppe et al., 2017). These populations are shaped by factors specific to the host as well as the environment, resulting in a high degree of diversity. This diversity is reflected in a core genome that contains approximately 2,200 genes, while the pan-genome contains more than 13,000 genes (Rasko et al., 2008). However, despite this variability, our sequence analysis did not reveal even a single example of a phylogenetic group where the replication fork trap appears to be non-functional (Goodall et al., 2021). Thus, while a fork trap clearly is not essential, and a wide variety of bacterial species appear not to utilize a fork trap (Galli et al., 2019), it appears that, once acquired, such a fork trap system is strictly maintained (Neylon et al., 2005; Goodall et al., 2021; Toft et al., 2022). Together with the fork trap system in B. subtilis, which has a similar functionality but has likely arisen by convergent evolution (Franks et al., 1995; Neylon et al., 2005), these findings suggest that a fork trap system is involved in mitigating a problem important enough to selectively maintain the fork trap system.
Are replication-transcription conflicts responsible for the evolution of fork traps?
One proposed role for the fork trap is maintaining the co-directionality of transcription and replication (Brewer, 1988). Head-on collisions of replication and transcription complexes pose a challenge to cells (Rudolph et al., 2007; Kim and Jinks-Robertson, 2012; McGlynn et al., 2012; Merrikh et al., 2012; Lang and Merrikh, 2018), a challenge that becomes especially significant if collisions occur at highly-transcribed genes such as the operons for ribosomal RNA (Wang et al., 2007; Boubakri et al., 2010; Srivatsan et al., 2010; De Septenville et al., 2012; Ivanova et al., 2015; Dimude et al., 2018a; Hawkins et al., 2019). Especially highly transcribed genes, such as rrn operons and genes encoding ribosomal proteins, are transcribed co-directionally with the direction of replication (Brewer, 1988; McLean et al., 1998). If a fork would proceed beyond the termination area, perhaps because the other is held up at an obstacle, the proceeding fork will now encounter these genomic areas in the dangerous head-on orientation, which is normally prevented by a fork trap.
Is this likely to be the driver behind the strict maintenance of a fork trap area? In E. coli all rrn operons and the majority of genes encoding ribosomal proteins are located in the origin-proximal half of the chromosome (McLean et al., 1998; Dimude et al., 2016). Forks proceeding from the termination area in the wrong orientation would have to duplicate 1 Mbp and more before they would even reach these highly transcribed areas. At least under laboratory conditions, and within the resolution limit of the MFA used, the fork fusion point is surprisingly consistent (Rudolph et al., 2013; Dimude et al., 2016; Goodall et al., 2021), which is perfectly in line with the relatively small percentages of stalled forks detected by 2D gel electrophoresis (Duggin and Bell, 2009). Together, this suggests that long delays of a single fork are a rare event.
For the rest of the E. coli chromosome co-directionality is approximately 55% (McLean et al., 1998). However, this does not necessarily mean that directionality is random. Data from Merrikh and colleagues have shown that head-on transcription-replication conflicts increase the incidence of mutagenesis that can result in adaptive mutations, which actually increase cell survival (Merrikh et al., 2012, 2016; Merrikh, 2017; Merrikh and Merrikh, 2018). This arises from the fact that many stress-induced genes are encoded on the lagging strand, thereby triggering head-on collisions with transcription machinery and, as a result, location-specific mutagenesis (Schroeder et al., 2016, 2020). Thus, while co-directionality reduces conflicts and for this reason is favored for highly transcribed genes, head-on collisions might be advantageous during times of stress, as they are useful for adaptive mutagenesis (Lang and Merrikh, 2018; Schroeder et al., 2020). Still, these effects are likely to be more subtle than the consequences of head-on collisions at highly-transcribed locations.
To gain at least some insight into whether forks escaping the termination area are causing any systematic problems before reaching the highly-transcribed rrn operons we used cells which contain ectopic replication origins in addition to oriC, as described above, and in which the fork trap is inactivated by the deletion of the tus gene. In Δtus cells carrying an ectopic replication origin called oriZ roughly half way into the clockwise replichore, the fork fusion point is shifted significantly into the counter-clockwise replichore, with one fork escaping the termination area and duplicating roughly 500 kb before fusing with the opposing fork (Ivanova et al., 2015). The mirrored situation takes place in Δtus cells carrying an ectopic replication origin called oriX roughly half way into the counter-clockwise replichore (Dimude et al., 2018b). Our marker frequency analysis did not reveal any systematic delays of the forks escaping the termination area, very much in contrast to the situation observed at rrn operons (Ivanova et al., 2015; Dimude et al., 2018a,b; Hawkins et al., 2019). Thus, we currently do not have any data supporting the idea that avoiding the negative consequences of head-on replication-transcription clashes in the origins-distal half of the chromosome is contributing in a major way to the evolutionary maintenance of the replication fork trap, even though control of directionality imposed by the fork trap almost certainly will be an added bonus. It would be interesting to analyze mutation rates and spectrums in the area replicated opposite to normal to also investigate the potentially beneficial effects of replication-transcription conflicts in this area of the chromosome.
Making sense of the replication fork trap
A role in preventing over-replication
If we assume that the fork trap system was derived from an ancestral system that came from plasmids, it is important to consider what is known about the physiological role of such plasmid-born fork trap systems. As described before, for R1 plasmid replication the fork trap mechanism enforces unidirectional DNA replication. Two ter sites, terRL and terRR, are located between the minimal origin sequence and the start site for leading strand synthesis (Hill et al., 1988b). DNA replication proceeding counter-clockwise is almost immediately blocked by terRL, whereas the clockwise fork duplicates most of the plasmid DNA until it is blocked at terRR (Krabbe et al., 1997). Interestingly, for R1 replication the absence of Tus protein was shown to result in the accumulation of complex branched DNA structures, plasmid multimers and rolling circle replication intermediates, which interfered with successful plasmid segregation (Krabbe et al., 1997). These findings strongly suggest that the replication fork complex, upon encountering the already replicated part of the plasmid, continues to unwind the already generated nascent section if it is not arrested by a Tus-ter complex. The resulting intermediates then serve as substrates for the continuation of replication in various ways (Krabbe et al., 1997). This idea is supported by biochemical studies showing that Tus-ter complexes forming a fork trap can prevent over-replication of a circular plasmid substrate, both by a traditional in vitro study (Hiasa and Marians, 1994) and the recently developed elegant “replication chain reaction” (Hasebe et al., 2018), lending support to a function of the bacterial replication fork trap in preventing over-replication and, as a result, segregation difficulties, which would pose a strong selection pressure.
Is there any indication that the fork trap might have a similar function for the E. coli chromosome? It was indeed reported that the deletion of tus resulted in low levels of chromosomal over-replication, an effect significantly exacerbated by specific point mutants in the polA gene, which encodes for DNA polymerase I (Markovitz, 2005), a polymerase involved in maintenance and repair processes, such as Okazaki fragment maturation and various DNA repair pathways (Kurth and O’Donnell, 2009). We were able to confirm over-replication of the termination area in a strain with a polA2 point mutation (Midgley-Smith et al., 2018). These data are in line with the idea that the fork trap, together with specific proteins such as polymerase I, is involved in bringing DNA replication to a successful conclusion (Markovitz, 2005; Duggin et al., 2008; Midgley-Smith et al., 2018; Rudolph et al., 2019). Similarly, absence of the terminator protein RTP in B. subtilis results in an increased formation of chromosomal dimers, which could arise from a related mechanism (Lemon et al., 2001).
What might be the molecular structures that arise as part of termination? Systematic analysis of a number of mutants that show over-replication of the termination area have given important insights. Significant levels of over-replication of the termination area were observed in cells lacking RecG helicase (Rudolph et al., 2009, 2013; Wendel et al., 2014; Dimude et al., 2015), an effect that is completely suppressed by point mutations inactivating the helicase activity of the main replication restart protein PriA (Rudolph et al., 2013; Dimude et al., 2015). Even more specifically, the over-replication is suppressed in a PriA point mutation called srgA1, encoding PriA[L557P], a PriA protein with a very specific change of the substrate activity. It unwinds a replication fork with both a leading and a lagging strand at the branch point as efficiently as wild type PriA, but it has lost the ability to unwind a fork in which the leading strand is missing (Gregg et al., 2002), which is, effectively, a 3′ flap structure. This result strongly suggests that a 3′ flap-like structure is involved in triggering the observed over-replication, and we hypothesized that it arises if, as part of a fork fusion reaction (Figures 5i,ii), the helicase of one replication fork complex displaces the leading strand polymerase of the opposing fork (Figure 5iii). Both template and primer are held in the active center of the replicative polymerase under tension (Joyce and Steitz, 1995; Baker and Bell, 1998; Xu and Dixon, 2018), and a ‘nudge’ of the leading strand polymerase by the DnaB helicase of the opposing fork might be enough to temporarily melt primer and template.
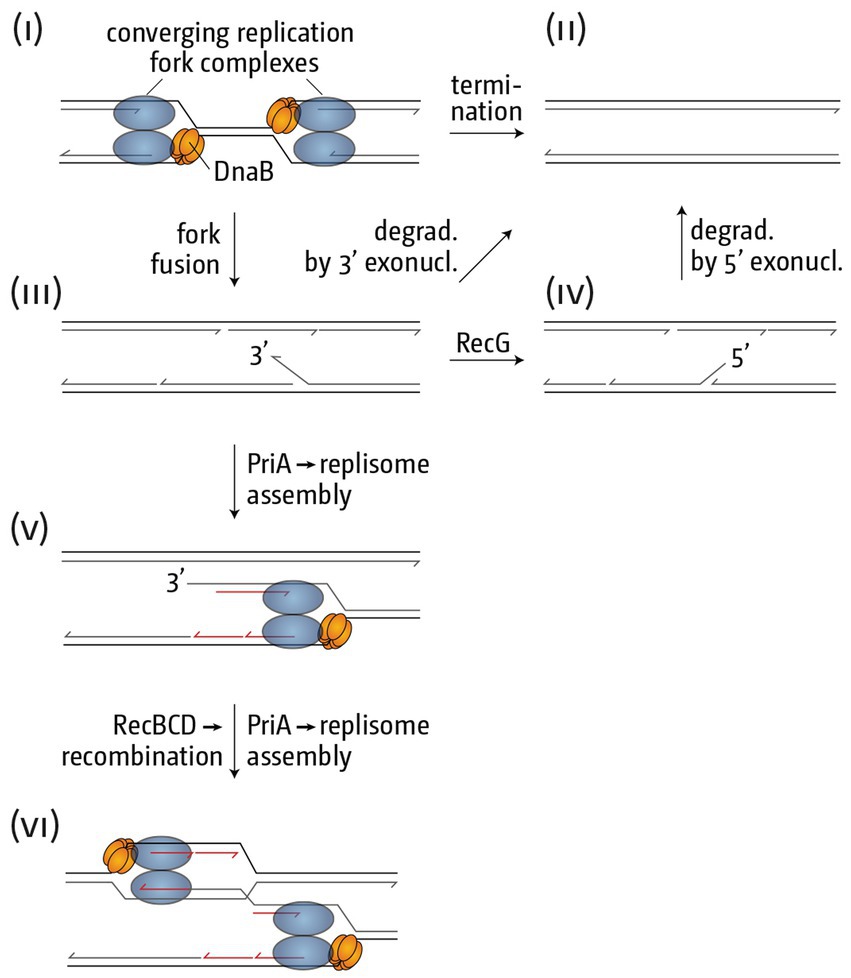
Figure 5. Schematic illustrating how replication fork fusions might trigger over-replication in the termination area. (i) Two merging replication fork complexes. (ii) Successful termination event where the two replisomes are disassembled, synthesis of all strands is completed and all nascent strands are successfully sealed using DNA ligase. (iii) As part of the fork fusion reaction the helicase of one replication fork complex might displace the leading strand polymerase of the opposing fork, resulting in the formation of a 3’ flap structure, which might be degraded by proteins such as 3’ exonucleases, resulting in successful termination. Note that the formation of a 3’ flap can occur at both forks. However, for simplicity the schematic shows only one such reaction. (iv) A 3’ flap is one of the best substrates for RecG helicase, which would convert it into a 5’ flap. Upon degradation by 5’ exonucleases successful termination can be achieved. (v) If 3’ flaps persist they are also a substrate for the restart protein PriA, which will establish a replisome, thereby not only over-replicating an already fully replicated area of the genome, but also generating a double-stranded DNA end. (vi) The dsDNA end can engage in homologous recombination, resulting in the formation of a displacement or D-loops, which PriA will use to set up yet another replication fork complex, thereby exacerbating the problem of over-replication.
RecG helicase shows a particularly high affinity for 3′ flaps and processes these by converting them into a 5′ flap structure (Figure 5iv; McGlynn and Lloyd, 2001; Tanaka and Masai, 2006). In addition, 3′ flaps will also be degraded by 3′ single-stranded exonucleases, and we have shown that the absence of multiple 3′ exonucleases in particular results in high levels of over-replication in the termination area (Rudolph et al., 2010, 2013; Midgley-Smith et al., 2019). Thus, we hypothesize that the absence of either RecG or 3′ exonucleases results in the persistence of 3′ flap-like structures, a substrate for PriA, even though the affinity of PriA for a 3′ flap is lower than that of RecG (Tanaka and Masai, 2006). PriA processing will result in the assembly of a new replication fork, the progression of which will not only result in chromosomal over-replication, but also the formation of a double-stranded DNA end (Figure 5v). dsDNA ends will be rapidly processed by the homologous recombination proteins RecBCD and RecA, leading to the formation of a displacement loop (D-loop) (Rudolph et al., 2009, 2013, 2019; Dimude et al., 2016; Midgley-Smith et al., 2019). D-loops are yet another substrate for PriA, which means an opportunity for the formation of more replication forks, thus setting up a positive feedback loop of over-replication (Figure 5vi).
What data support the idea that over-replication is triggered at termination inter mediates generated by freely-fusing forks, and not at structures such as replisomes blocked by Tus-ter complexes? The latter cannot be the case, because over-replication was initially found in cells lacking Tus protein (Markovitz, 2005) and several recent studies have confirmed the presence of over-replication in Δtus cells (Rudolph et al., 2013; Dimude et al., 2015; Midgley-Smith et al., 2018, 2019). Thus, the synthesis observed is triggered independently of the fork trap system, very likely directly at fusing replication fork complexes.
In addition, in a recent analysis we were able to show that over-replication can be triggered in the absence of RecG in defined ectopic chromosomal areas if forks are artificially forced to fuse in these areas (Midgley-Smith et al., 2018). Again, we used a strain that harbors an ectopic replication origin, oriZ, half-way into the right-hand replichore (Wang et al., 2011; Ivanova et al., 2015). Forks coming from oriC and oriZ fuse approximately at position 4.5 Mbp of the chromosome (Ivanova et al., 2015), and we artificially engineered an ectopic fork trap in this location, with ter sequences inserted at positions 4.4 and 4.6 Mbp (Figure 6A). No over-replication was observed in this location in oriC+ oriZ+ cells in the presence of RecG, but in its absence a clear peak of over-replication was observed (Figure 6B; Midgley-Smith et al., 2018).
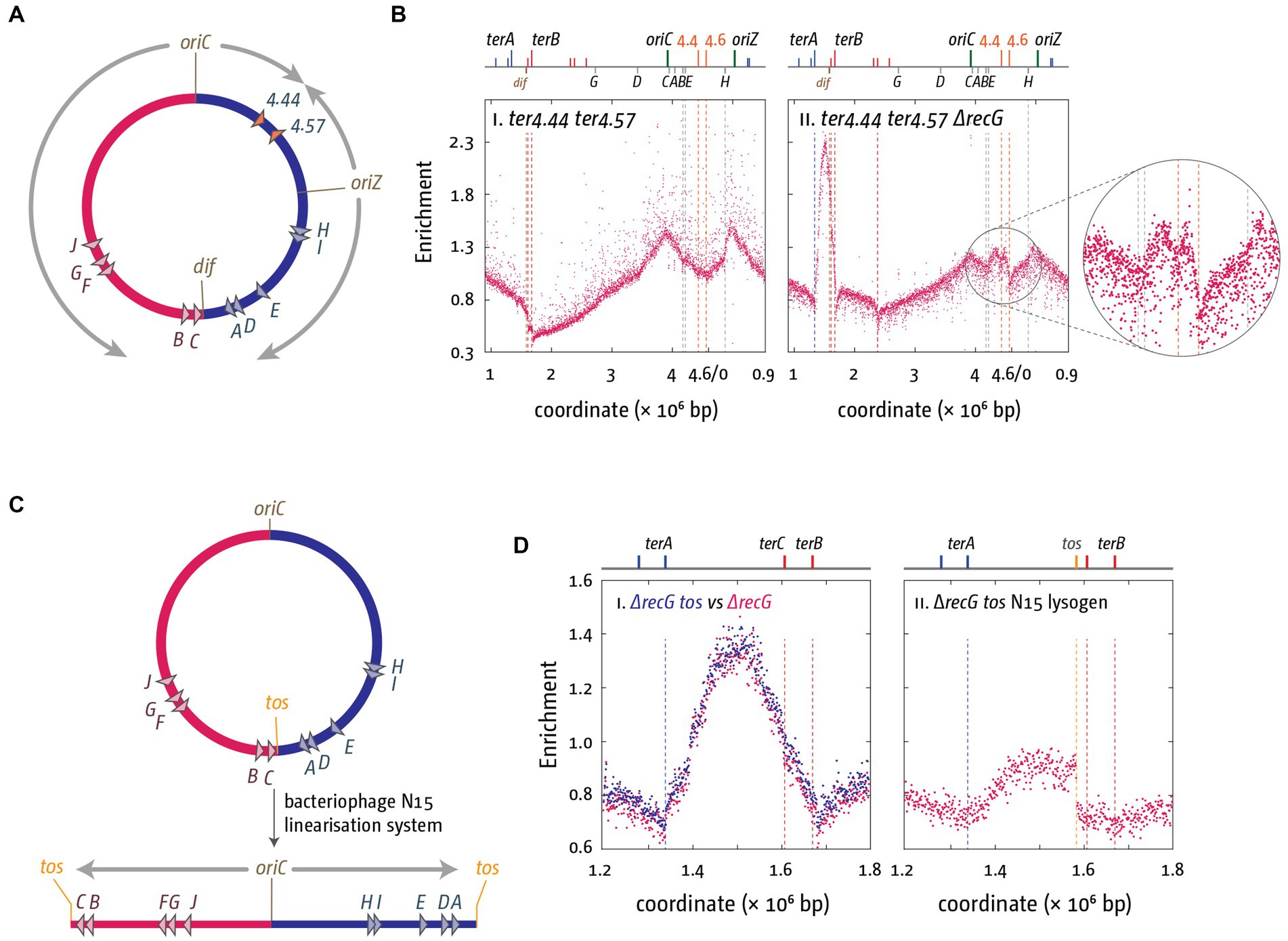
Figure 6. Over-replication at fork fusion sites can be modulated in E. coli if termination sites are artificially altered. (A) Schematic representation of the chromosome of oriC+ oriZ+ cells with additional ter sites integrated either side of the ectopic fork fusion area. (B) Chromosomal marker frequency analysis (MFA) of oriC+ oriZ+ and oriC+ oriZ+ ΔrecG cells with an ectopic replication fork trap in the presence and absence of RecG helicase. The numbers of reads (normalized against reads for a stationary phase wild type control) are plotted against the chromosomal location. A schematic representation of the E. coli chromosome showing positions of oriC, oriZ, native and ectopic ter sites (above) as well as dif and rrn operons A–E, G, and H (below) is shown above the plotted data. The data were re-plotted from Midgley-Smith et al. (2018). (C) Schematic representation of the linearization of the E. coli chromosome, as described in Cui et al. (2007) and Rudolph et al. (2013). The integrated bacteriophage N15 linearization site tos is highlighted. Processing by the N15 telomerase results in the formation of a linear chromosome with covalently-closed hairpin ends, which prevents forks meeting each other. (D) Effect of chromosome linearization on origin-independent synthesis in ΔrecG cells. Shown is the number of reads (normalized against the reads for a stationary-phase wild-type control) plotted against the chromosomal location. In panel (Di) data sets for recG and recG tos are plotted together for direct comparison, while panel (Dii) shows the data for the linearised construct. Data replotted from Rudolph et al. (2013).
We also investigated what happens if replication forks are prevented from fusing. The E. coli chromosome can be stably linearized using the bacteriophage N15 linearization system. Cui and colleagues successfully integrated the tos linearization sequence into the termination area of the chromosome (Cui et al., 2007). Expression of the bacteriophage N15 telomerase resulted in the processing of the tos linearization sequence into two hairpin ends, which will prevent forks from meeting each other (Figure 6C; Cui et al., 2007; Rudolph et al., 2013). If fork fusion reactions are responsible for the observed over-replication, chromosome linearization should result in a significant reduction of the over-replication. This was indeed what we observed. Over-replication was significantly reduced and entirely depended on forks coming from oriC (Figure 6D; Rudolph et al., 2013; Dimude et al., 2015). Taken together both findings strongly support the idea that the over-replication observed in the termination area is triggered by the fusion of replication fork complexes. They are of particular importance in the light of a recent elegant study by Raghunathan and co-workers showing that in cells lacking the DNA adenine methylase, Dam, a peak of over-replication is observed in the termination area, and cells are capable of growing in the absence of a functional origin (Raghunathan et al., 2019). The authors postulate that an increased number of recombination events in all chromosomal areas triggers significant levels of recombination-dependent replication, which also results in over-replication of the termination area and, because of the fork trap, results in a peak in the marker frequency analysis (Raghunathan et al., 2019). Indeed, a role for RecG in controlling recombination-dependent replication was reported before (Azeroglu et al., 2016; Azeroglu and Leach, 2017), which might explain the small peak of synthesis remaining in ΔrecG cells in which the chromosome is linearized (Figure 6D). However, as recombination-dependent synthesis is triggered independently of fork fusion events, the reduction of over-replication in ΔrecG cells with a linear chromosome strongly suggests that it is not responsible for the significant levels of synthesis observed in the termination area in cells lacking RecG helicase.
Thus, research by our own and other labs shows that resolving the observed over-replication requires a surprising number of proteins, including RecG helicase, DNA polymerase I, the exonucleases ExoI, ExoVII, SbcCD and RecJ, the homologous recombination proteins RecBC, RecD and RuvABC and others (Markovitz, 2005; Rudolph et al., 2013; Wendel et al., 2014, 2018; Dimude et al., 2015, 2018a; Sinha et al., 2017, 2018; Midgley-Smith et al., 2018, 2019). In addition, our recent in vitro results suggest that more proteins are yet to be discovered (Jameson et al., 2021). As many of these intermediates arise as a direct result of fork fusions, it is tempting to speculate that they can arise generally as part of termination in bacterial DNA replication, and indeed many of the above proteins are well conserved in prokaryotes (and some beyond). Thus, the necessary proteins exist to bring termination to a successful conclusion, regardless of the presence of absence of a fork trap mechanism. However, a fork trap, once acquired, seems to add an additional layer of protection.
What are the particular benefits of a fork trap system? Bacterial cells go to great lengths to ensure that exactly one complete copy of the chromosome is generated per cell cycle (Boye et al., 2000; Skarstad and Katayama, 2013). However, if the over-replication triggered at termination intermediates is allowed to proceed, it results in uncontrolled over-replication of the chromosome, which can undermine the regulatory steps employed to control genome duplication. In cells lacking both RecG and Tus the over-replication that is triggered in the termination area can proceed into both replichores, eventually leading to dangerous head-on collisions with transcribing RNA polymerase complexes at highly transcribed genes such as rrn operons, as described above. Such collisions can be alleviated by so-called rpo* point mutations, point mutations in genes for some of the RNA polymerase subunits which destabilize ternary RNA polymerase complexes (Trautinger et al., 2005; Rudolph et al., 2007). If an rpo* mutation is introduced in cells lacking RecG and Tus, the synthesis in these cells is robust enough to allow cell growth and division even if the entire native origin is deleted (Rudolph et al., 2013; Dimude et al., 2015). Inhibition of oriC firing in ΔrecG Δtus rpo* cells revealed a replication profile that was inverted, with highest levels of synthesis in the termination area where forks normally fuse, while lowest levels were observed in the area around where oriC normally is located (Rudolph et al., 2013; Dimude et al., 2015). In contrast, in cells with a fork trap such over-replication is blocked by Tus-ter complexes (Rudolph et al., 2013; Dimude et al., 2015).
One danger of replication fork complexes escaping the termination area is that these forks will fuse somewhere in the replichore with forks coming from oriC, which poses the risk of repeating the problem (Figure 7i). In contrast, based on our genetics data we have postulated before that fork fusion events at a Tus-ter complex might be less prone to triggering over-replication (Midgley-Smith et al., 2019), even though additional proteins might be required to resolve replisome-Tus-ter-replisome super-complexes (Figure 4; Jameson et al., 2021). Thus, if replication fork complexes are stably arrested at Tus-ter complexes until the next round of synthesis arrives from oriC, the resolution of these complexes might result in successful termination (Figure 7ii). In addition, there is evidence of processing by several nucleases in the termination area, including RecBCD and SbcCD (Rudolph et al., 2013; Courcelle et al., 2015; Azeroglu et al., 2016; Sinha et al., 2017, 2018; Dimude et al., 2018a). The molecular details are complex and will depend on the precise intermediates that are being formed, but if over-replication is blocked relatively quickly, degradation could contribute to simply resolving that situation and achieve successful termination (Figure 7ii), a situation that is much harder to achieve if over-replication is allowed to proceed into the replichore in a direction opposite to normal.
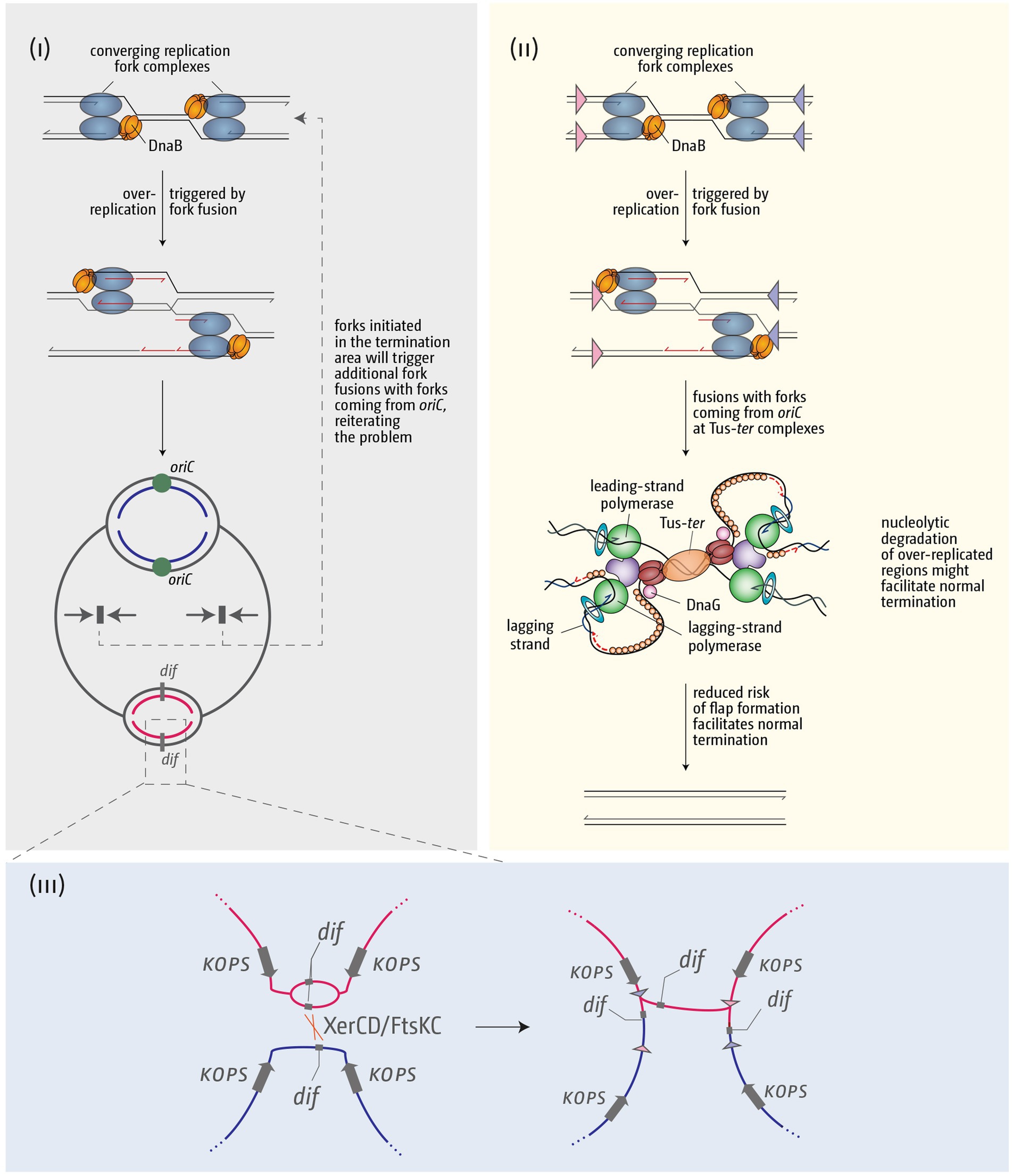
Figure 7. Schematic illustrating how over-replication triggered by replication fork fusions might trigger problems for chromosome duplication and segregation, and how the presence of a fork trap area might help in resolving these issues. (i) Over-replication triggered in the termination area by fork fusion reactions will result in additional collisions with forks coming from oriC, thereby reiterating and exacerbating the problem. (ii) A fork fusion reaction by a freely moving fork encountering a second stably arrested at a Tus-ter complex is less likely to result in formation of a 3’ flap and subsequent over-replication, thereby facilitating successful termination. (iii) If termination induces over-replication and the chromosome dimer resolution site dif is duplicated, this can lead to problems of the chromosome dimer resolution mechanism. See text for further details.
Fork trap systems have multiple benefits
An additional benefit of a fork trap system was recently observed by Hamilton and colleagues. They measured DNA synthesis induced by χ sites, which switch the functionality of RecBCD from dsDNA degradation to the formation of a 3′ ssDNA overhang that will be bound by RecA and which can engage in recombination-dependent replication. By using a plasmid-based system the authors were able to show that the addition of ter sites successfully limited the amount of replication induced by χ sites, which helped to improve plasmid stability (Hamilton et al., 2023).
Furthermore, any process that involves processive translocation of large molecular machines along DNA will trigger superhelical stress (Chedin and Benham, 2020). This will be particularly problematic if the overall number of replication forks is increased by over-replication forks escaping the termination area. Superhelical stress can result in increased R-loop formation, as R-loops can help to absorb negative superhelicity (Chedin and Benham, 2020). However, increased levels of R-loops can also threaten genomic integrity (Crossley et al., 2019).
In addition, processing of fork fusion intermediates by homologous recombination proteins will increase the frequency of recombination events in the termination area, and elevated levels of recombination were indeed reported for this area in E. coli (Horiuchi et al., 1994). Aspects such as over-replication of already replicated DNA, increased levels of R-loops and increased recombination frequencies have all been found to contribute to genomic instability (Finkel et al., 2007; Blow and Gillespie, 2008; Alexander and Orr-Weaver, 2016; Tomasetti et al., 2017; Crossley et al., 2019), highlighting the benefits of a replication fork trap system and adding to the explanation of why fork trap systems, once acquired, are so consistently maintained.
Another aspect where the fork trap area might play an important role is the chromosome dimer resolution site dif. Chromosomal dimers can form as a consequence of an odd number of recombination events. FtsK translocase, directed by FtsK-Orienting Polar Sequences (KOPS) towards the termination area and dif, controls a site-specific recombination system that resolves chromosomal dimers, which involves the site-specific recombinase XerCD and the dif site (Bigot, 2005; Levy et al., 2005; Barre, 2007; Sherratt et al., 2010). In our analysis of the different E. coli phylogroups we found that dif is consistently located between the innermost ter sites A and C (Figure 8). This is also the case for a variety of other bacteria with a fork trap, including Salmonella enterica, Klebsiella variicola and Klebsiella pneumoniae, Edwardsiella tarda, Cedecea neteri and Dickeya paradisiaca (Goodall et al., 2021; Toft et al., 2022). It is likely that this defined location of the dif site reflects biological importance. For the successful resolution of chromosomal dimers exactly two dif sites of a dimer have to be brought together, followed by XerCD processing (Castillo et al., 2017). If more than two dif sites are present, for example because one stretch containing a dif site is duplicated via termination-induced over-replication, this will be problematic, because a recombination event between the wrong dif sites will not resolve the chromosomal dimer (Figure 7iii; Goodall et al., 2021). Similarly, if two dif sites are available for processing before replication is complete, which might be the case if the fork fusion point is located in a different area of the chromosome to dif, this will be problematic, as it could trigger unwanted processing events. Indeed, we showed recently that the doubling time of a strain in which the dif site is moved approximately 100 kb out of the inner termination area is significantly longer (27 vs. 22 min) (Goodall et al., 2021). The importance of limiting the number of available dif sites is also highlighted by the fact that in Vibrio cholerae, which carries two circular chromosome, the dif sites of the different chromosomes have particular sequence changes, thereby avoiding inter-chromosomal attempts of processing (Castillo et al., 2017).
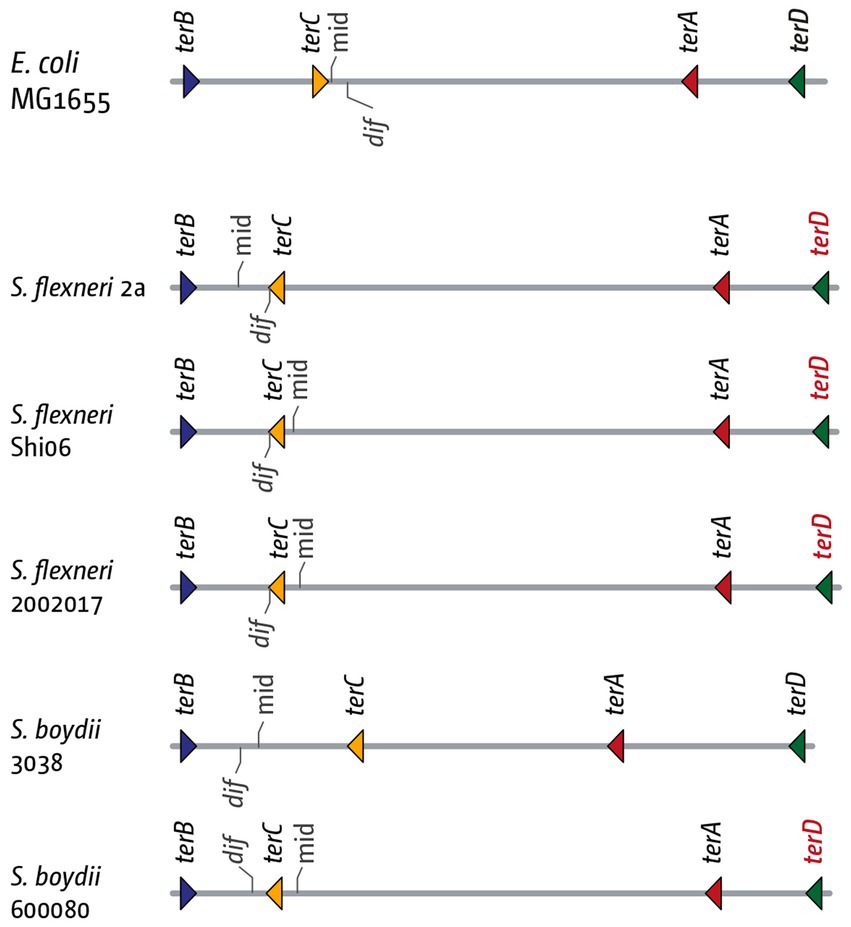
Figure 8. Details of the inner replication fork trap architecture in genomes from three Shigella flexneri and two Shigella boydii genomes. The inner ter sites and the chromosome dimer resolution site dif are marked. The orientation of the ter sites are indicated by the direction of the triangle (forks encountering the tip of the triangle would get blocked). The orientation of the dif chromosome dimer resolution site is indicated by the marker pointing upwards (indicating the (+)-strand) or downwards (indicating the (−)-strand). The terD sites highlighted in red indicate a change of the G6 residue, which makes this ter site much less efficient at blocking a progressing replication fork.
In this context we stumbled across an intriguing detail in Shigella. When analyzing the termination area in Shigella flexneri and Shigella boydii strains, we found that, in the majority of genomes investigated, the dif site was located between ter sites B and C, rather than A and C (Goodall et al., 2021). This change was accompanied with a consistent inversion of terC, as well as a mutation of the critical C6 base in terD (Figure 7). In other words, in these strains the innermost termination area is flanked by terB on one side, and ter sites C, A and D on the other, with D often inactivated, as the critical C6 is mutated (Kamada et al., 1996; Neylon et al., 2005). Thus, it seems that dif moves with the location of the inner termination area, or vice versa (Goodall et al., 2021). In contrast, in the absence of a replication fork trap the delay of one fork will result in progression of the opposite fork, leading to the duplication of the dif site before chromosome duplication is complete, generating intermediates that are not as efficiently processed at the late stages of the duplication process. Thus, by restricting the continuation of synthesis, a fork trap system is likely to alleviate all of these problems.
Conclusion
The chromosomal architecture in bacteria is quite remarkable: the strict limitation to a single replication origin (Gao and Zhang, 2008; Gao, 2015) dictates that the speed of replication directly influences growth. Faster growth will require faster replication fork speeds, and under fast growth conditions DNA replication in E. coli is indeed almost 15 × faster than DNA synthesis in eukaryotic cells (Pham et al., 2013). To speed up genome duplication, bacteria such as E. coli use overlapping rounds of replication, while the seemingly simple solution of multiple origins is not used. Both in archaea and eukaryotic cells the use of multiple origins is the norm (Early et al., 2004; Ausiannikava and Allers, 2017). What difference allows the use of multiple origins? Eukaryotic cells have much larger genomes, the duplication of which is facilitated by an increased number of replication forks (Leonard and Méchali, 2013), but this is not a satisfactory explanation, as archaeal chromosomes and also the chromosomes of, e.g., Saccharomyces cerevisiae are of similar size or even smaller than the E. coli chromosome (Belda et al., 2019) and still several origins are present.
Multiple factors will have contributed to the development of these distinct chromosomal architectures. However, as highlighted above, termination of DNA replication can trigger the formation of intermediates that cause a variety of downstream problems. These problems include chromosomal over-replication, threatening the control that restricts genome duplication to once per cell cycle, localized increases in recombination which will destabilize the genome, topological problems, which may result in secondary effects, such as increased levels of R-loops, and problems for the processes associated with the late stages of genome duplication, such as decatenation and chromosome dimer resolution. Given the large number of possible problems, it is perhaps not surprising that a large number of conserved protein activities are reported to be involved in the rapid and effective processing of fork fusion intermediates. However, it is tempting to speculate that one easy and efficient way to restrict the number of fork fusion events is by restricting the number of origins to its minimum – a single event. We believe that a fork trap system, while not essential, adds an additional mechanistic layer to the coordination of fork movement. It enables intermediates that arise in a defined area of the chromosome to be trapped, which facilitates their processing, as well as the coordination of termination with concatenation and chromosome dimer resolution (Figure 7), explaining why, once acquired, it was so consistently retained.
But why is the increased number of origins less of a problem in archaea and eukaryotic cells? Our data strongly suggest that 3′ flaps are an important termination intermediate as replication fork complexes fuse in bacteria (Rudolph et al., 2010, 2013; Dimude et al., 2015; Midgley-Smith et al., 2019). 3′ flap formation is driven by the replicative helicase DnaB encircling the lagging strand template (LeBowitz and McMacken, 1986). In contrast, the archaeal and eukaryotic replisomes use CMG helicase to unwind the parental duplex, which will not generate a 3′ flap structure, as it is not encircling the lagging strand template (Abid Ali and Costa, 2016), which makes it likely that the precise intermediates that are generated as forks fuse differ significantly from the situation in bacteria.
However, this does not mean that the termination stage of DNA replication is unproblematic in eukaryotic cells. On the contrary, recent work by numerous labs has highlighted that the fusion of two forks needs to be carefully orchestrated as well. Similar to bacterial helicases such as RecG, the helicases Pif1 and Rrm3 play an important role in facilitating fork fusions and bringing the replication process to a successful conclusion (Steinacher et al., 2012; Deegan et al., 2019), even though replisomes appear not to slow as they move towards each other (Dewar et al., 2015; Dewar and Walter, 2017). Furthermore, upon completion of synthesis the disassembly of the replisome components needs to be carefully coordinated (Maric et al., 2014; Moreno et al., 2014; Moreno and Gambus, 2015; Sonneville et al., 2017; Zhou et al., 2021).
We are only beginning to understand the molecular mechanisms that are at play during this fascinating stage of the replication process. Future mechanistic studies in bacteria, archaea and eukaryotic cells will without doubt provide additional important mechanistic details of how replication is concluded efficiently and with high accuracy.
Author contributions
DG and CR wrote the initial draft of the article. DG, DW, MH, and CR were involved in extensive edits of the entire text. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by Research Grant BB/N014995/1 from the Biotechnology and Biological Sciences Research Council to CR and MH, and Research Grant BB/W000393/1 from the Biotechnology and Biological Sciences Research Council to CR.
Acknowledgments
The authors would like to thank Stelinda Peros and the four reviewers for their constructive critical comments. The authors would especially like to thank Robert Lloyd for his insightful comments and corrections on this review, and for his support and the many interesting discussions on the topic over the years.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abhyankar, M. M., Reddy, J. M., Sharma, R., Büllesbach, E., and Bastia, D. (2004). Biochemical investigations of control of replication initiation of plasmid R6K. J. Biol. Chem. 279, 6711–6719. doi: 10.1074/jbc.M312052200
Abhyankar, M. M., Zzaman, S., and Bastia, D. (2003). Reconstitution of R6K DNA replication in vitro using 22 purified proteins. J. Biol. Chem. 278, 45476–45484. doi: 10.1074/jbc.M308516200
Abid Ali, F., and Costa, A. (2016). The MCM helicase motor of the eukaryotic replisome. J. Mol. Biol. 428, 1822–1832. doi: 10.1016/j.jmb.2016.01.024
Ahn, K. S., Malo, M. S., Smith, M. T., and Wake, R. G. (1993). Autoregulation of the gene encoding the replication terminator protein of Bacillus subtilis. Gene 132, 7–13. doi: 10.1016/0378-1119(93)90508-Z
Alexander, J. L., and Orr-Weaver, T. L. (2016). Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 30, 2241–2252. doi: 10.1101/gad.288142.116
Ausiannikava, D., and Allers, T. (2017). Diversity of DNA replication in the Archaea. Genes 8:E56. doi: 10.3390/genes8020056
Azeroglu, B., and Leach, D. R. F. (2017). RecG controls DNA amplification at double-strand breaks and arrested replication forks. FEBS Lett. 591, 1101–1113. doi: 10.1002/1873-3468.12583
Azeroglu, B., Mawer, J. S. P., Cockram, C. A., White, M. A., Hasan, A. M. M., Filatenkova, M., et al. (2016). RecG directs DNA synthesis during double-Strand break repair. PLoS Genet. 12:e1005799. doi: 10.1371/journal.pgen.1005799
Baker, T. A., and Bell, S. P. (1998). Polymerases and the replisome: machines within machines. Cells 92, 295–305. doi: 10.1016/S0092-8674(00)80923-X
Barre, F.-X. (2007). FtsK and SpoIIIE: the tale of the conserved tails. Mol. Microbiol. 66, 1051–1055. doi: 10.1111/j.1365-2958.2007.05981.x
Bastia, D., Zzaman, S., Krings, G., Saxena, M., Peng, X., and Greenberg, M. M. (2008). Replication termination mechanism as revealed by Tus-mediated polar arrest of a sliding helicase. Proc. Natl. Acad. Sci. U. S. A. 105, 12831–12836. doi: 10.1073/pnas.0805898105
Belda, I., Ruiz, J., Santos, A., Wyk, N. V., and Pretorius, I. S. (2019). Saccharomyces cerevisiae. Trends Genet. 35, 956–957. doi: 10.1016/j.tig.2019.08.009
Berghuis, B. A., Dulin, D., Xu, Z.-Q., van Laar, T., Cross, B., Janissen, R., et al. (2015). Strand separation establishes a sustained lock at the Tus-Ter replication fork barrier. Nat. Chem. Biol. 11, 579–585. doi: 10.1038/nchembio.1857
Berghuis, B. A., Raducanu, V.-S., Elshenawy, M. M., Jergic, S., Depken, M., Dixon, N. E., et al. (2018). What is all this fuss about Tus? Comparison of recent findings from biophysical and biochemical experiments. Crit. Rev. Biochem. Mol. Biol. 53, 49–63. doi: 10.1080/10409238.2017.1394264
Bianco, P. R. (2020). DNA helicase-SSB interactions critical to the regression and restart of stalled DNA replication forks in Escherichia coli. Genes 11:471. doi: 10.3390/genes11050471
Bidnenko, V., Ehrlich, S. D., and Michel, B. (2002). Replication fork collapse at replication terminator sequences. EMBO J. 21, 3898–3907. doi: 10.1093/emboj/cdf369
Bidnenko, V., Lestini, R., and Michel, B. (2006). The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol. Microbiol. 62, 382–396. doi: 10.1111/j.1365-2958.2006.05382.x
Bigot (2005). KOPS: DNA motifs that control E-coli chromosome segregation by orienting the FtsK translocase. EMBO J. 24, 3770–3780. doi: 10.1038/sj.emboj.7600835
Bird, R. E., Louarn, J., Martuscelli, J., and Caro, L. (1972). Origin and sequence of chromosome replication in Escherichia coli. J. Mol. Biol. 70, 549–566. doi: 10.1016/0022-2836(72)90559-1
Blattner, F. R., Plunkett, G., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi: 10.1126/science.277.5331.1453
Blow, J. J., Ge, X. Q., and Jackson, D. A. (2011). How dormant origins promote complete genome replication. Trends Biochem. Sci. 36, 405–414. doi: 10.1016/j.tibs.2011.05.002
Blow, J. J., and Gillespie, P. J. (2008). Replication licensing and cancer--a fatal entanglement? Nat. Rev. Cancer 8, 799–806. doi: 10.1038/nrc2500
Boubakri, H., de Septenville, A. L., Viguera, E., and Michel, B. (2010). The helicases DinG, rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 29, 145–157. doi: 10.1038/emboj.2009.308
Bouché, J. P., Gélugne, J. P., Louarn, J., Louarn, J. M., and Kaiser, K. (1982). Relationships between the physical and genetic maps of a 470 x 10(3) base-pair region around the terminus of Escherichia coli K12 DNA replication. J. Mol. Biol. 154, 21–32.
Boye, E., Løbner-Olesen, A., and Skarstad, K. (2000). Limiting DNA replication to once and only once. EMBO Rep. 1, 479–483. doi: 10.1093/embo-reports/kvd116
Brewer, B. J. (1988). When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cells 53, 679–686.
Cao, J., Woodhall, M. R., Alvarez, J., Cartron, M. L., and Andrews, S. C. (2007). EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65, 857–875. doi: 10.1111/j.1365-2958.2007.05802.x
Castillo, F., Benmohamed, A., and Szatmari, G. (2017). Xer site specific recombination: double and single recombinase systems. Front. Microbiol. 8:453. doi: 10.3389/fmicb.2017.00453
Chedin, F., and Benham, C. J. (2020). Emerging roles for R-loop structures in the management of topological stress. J. Biol. Chem. 295, 4684–4695. doi: 10.1074/jbc.REV119.006364
Courcelle, J., Wendel, B. M., Livingstone, D. D., and Courcelle, C. T. (2015). RecBCD is required to complete chromosomal replication: implications for double-strand break frequencies and repair mechanisms. DNA Repair 32, 86–95. doi: 10.1016/j.dnarep.2015.04.018
Crosa, J. H., Luttropp, L. K., and Falkow, S. (1976). Mode of replication of the conjugative R-plasmid RSF1040 in Escherichia coli. J. Bacteriol. 126, 454–466.
Crossley, M. P., Bocek, M., and Cimprich, K. A. (2019). R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411. doi: 10.1016/j.molcel.2019.01.024
Cui, T., Moro-oka, N., Ohsumi, K., Kodama, K., Ohshima, T., Ogasawara, N., et al. (2007). Escherichia coli with a linear genome. EMBO Rep. 8, 181–187. doi: 10.1038/sj.embor.7400880
de Graaff, J., Crosa, J. H., Heffron, F., and Falkow, S. (1978). Replication of the nonconjugative plasmid RSF1010 in Escherichia coli K-12. J. Bacteriol. 134, 1117–1122. doi: 10.1128/jb.134.3.1117-1122.1978
de Massy, B., Béjar, S., Louarn, J., Louarn, J. M., and Bouché, J. P. (1987). Inhibition of replication forks exiting the terminus region of the Escherichia coli chromosome occurs at two loci separated by 5 min. Proc. Natl. Acad. Sci. U. S. A. 84, 1759–1763.
De Septenville, A. L., Duigou, S., Boubakri, H., and Michel, B. (2012). Replication fork reversal after replication-transcription collision. PLoS Genet. 8:e1002622. doi: 10.1371/journal.pgen.1002622
Deegan, T. D., Baxter, J., Ortiz Bazán, M. Á., Yeeles, J. T. P., and Labib, K. P. M. (2019). Pif1-family helicases support fork convergence during DNA replication termination in eukaryotes. Mol. Cell 74, 231–244.e9. doi: 10.1016/j.molcel.2019.01.040
Dewar, J. M., Budzowska, M., and Walter, J. C. (2015). The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350. doi: 10.1038/nature14887
Dewar, J. M., and Walter, J. C. (2017). Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 18, 507–516. doi: 10.1038/nrm.2017.42
Dimude, J. U., Midgley-Smith, S. L., and Rudolph, C. J. (2018a). Replication-transcription conflicts trigger extensive DNA degradation in Escherichia coli cells lacking RecBCD. DNA Repair 70, 37–48. doi: 10.1016/j.dnarep.2018.08.002
Dimude, J. U., Midgley-Smith, S. L., Stein, M., and Rudolph, C. J. (2016). Replication termination: containing fork fusion-mediated pathologies in Escherichia coli. Genes 7:40. doi: 10.3390/genes7080040
Dimude, J. U., Stein, M., Andrzejewska, E. E., Khalifa, M. S., Gajdosova, A., Retkute, R., et al. (2018b). Origins left, right, and Centre: increasing the number of initiation sites in the Escherichia coli chromosome. Genes 9:376. doi: 10.3390/genes9080376
Dimude, J. U., Stockum, A., Midgley-Smith, S. L., Upton, A. L., Foster, H. A., Khan, A., et al. (2015). The consequences of replicating in the wrong orientation: bacterial chromosome duplication without an active replication origin. mBio :6. doi: 10.1128/mBio.01294-15
Duggin, I. G., and Bell, S. D. (2009). Termination structures in the Escherichia coli chromosome replication fork trap. J. Mol. Biol. 387, 532–539. doi: 10.1016/j.jmb.2009.02.027
Duggin, I. G., Wake, R. G., Bell, S. D., and Hill, T. M. (2008). The replication fork trap and termination of chromosome replication. Mol. Microbiol. 70, 1323–1333. doi: 10.1111/j.1365-2958.2008.06500.x
Early, A., Drury, L., and Diffley, J. F. X. (2004). Mechanisms involved in regulating DNA replication origins during the cell cycle and in response to DNA damage. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 359, 31–38. doi: 10.1098/rstb.2003.1362
Eli Bussiere, D., Bastia, D., and Stephen White, W. (1995). Crystal structure of the replication terminator protein from B. subtilis at 2.6 Å. Cells 80, 651–660. doi: 10.1016/0092-8674(95)90519-7
Elshenawy, M. M., Jergic, S., Xu, Z.-Q., Sobhy, M. A., Takahashi, M., Oakley, A. J., et al. (2015). Replisome speed determines the efficiency of the Tus-Ter replication termination barrier. Nature 525, 394–398. doi: 10.1038/nature14866
Finkel, T., Serrano, M., and Blasco, M. A. (2007). The common biology of cancer and ageing. Nature 448, 767–774. doi: 10.1038/nature05985
François, V., Louam, J., and Louarn, J.-M. (1989). The terminus of the Escherichia coli chromosome is flanked by several polar polar replication pause sites. Mol. Microbiol. 3, 995–1002. doi: 10.1111/j.1365-2958.1989.tb00250.x
Franks, A., Griffiths, A. A., and Wake, R. G. (1995). Identification and characterization of new DNA replication terminators in Bacillus subtilis. Mol. Microbiol. 17, 13–23. doi: 10.1111/j.1365-2958.1995.mmi_17010013.x
Franks, A. H., and Wake, R. G. (1996). Replication fork arrest at relocated replication terminators on the Bacillus subtilis chromosome. J. Bacteriol. 178, 4258–4265. doi: 10.1128/jb.178.14.4258-4265.1996
Galli, E., Ferat, J.-L., Desfontaines, J.-M., Val, M.-E., Skovgaard, O., Barre, F.-X., et al. (2019). Replication termination without a replication fork trap. Sci. Rep. 9:8315. doi: 10.1038/s41598-019-43795-2
Gao, F. (2015). Bacteria may have multiple replication origins. Front. Microbiol. 6:324. doi: 10.3389/fmicb.2015.00324
Gao, F., and Zhang, C.-T. (2008). Ori-finder: A web-based system for finding oriC s in unannotated bacterial genomes. BMC Bioinformatics 9:79. doi: 10.1186/1471-2105-9-79
Goodall, D. J., Jameson, K. H., Hawkins, M., and Rudolph, C. J. (2021). A fork trap in the chromosomal termination area is highly conserved across all Escherichia coli phylogenetic groups. Int. J. Mol. Sci. 22:7928. doi: 10.3390/ijms22157928
Gottlieb, P. A., Wu, S., Zhang, X., Tecklenburg, M., Kuempel, P., and Hill, T. M. (1992). Equilibrium, kinetic, and footprinting studies of the Tus-Ter protein-DNA interaction. J. Biol. Chem. 267, 7434–7443.
Gregg, A. V., McGlynn, P., Jaktaji, R. P., and Lloyd, R. G. (2002). Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 9, 241–251. doi: 10.1016/s1097-2765(02)00455-0
Griffiths, A. A., Andersen, P. A., and Wake, R. G. (1998). Replication terminator protein-based replication fork-arrest systems in various Bacillus species. J. Bacteriol. 180, 3360–3367. doi: 10.1128/JB.180.13.3360-3367.1998
Grosse, C., Scherer, J., Koch, D., Otto, M., Taudte, N., and Grass, G. (2006). A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol. Microbiol. 62, 120–131. doi: 10.1111/j.1365-2958.2006.05326.x
Gyurasits, E. B., and Wake, R. G. (1973). Bidirectional chromosome replication in Bacillus subtilis. J. Mol. Biol. 73, 55–63. doi: 10.1016/0022-2836(73)90158-7
Hamilton, N. A., Jehru, A. E., Samples, W. N., Wendel, B. M., Mokhtari, P. D., Courcelle, C. T., et al. (2023). Chi sequences switch the RecBCD helicase-nuclease complex from degradative to replicative modes during the completion of DNA replication. J. Biol. Chem. 299:103013. doi: 10.1016/j.jbc.2023.103013
Hansen, F. G., and Atlung, T. (2018). The DnaA tale. Front. Microbiol. 9:319. doi: 10.3389/fmicb.2018.00319
Hasebe, T., Narita, K., Hidaka, S., and Su’etsugu, M. (2018). Efficient arrangement of the replication fork trap for in vitro propagation of monomeric circular DNA in the chromosome-replication cycle reaction. Life Basel Switz. 8:43. doi: 10.3390/life8040043
Hawkins, M., Dimude, J. U., Howard, J. A. L., Smith, A. J., Dillingham, M. S., Savery, N. J., et al. (2019). Direct removal of RNA polymerase barriers to replication by accessory replicative helicases. Nucleic Acids Res. 47, 5100–5113. doi: 10.1093/nar/gkz170
Hendrickson, H., and Lawrence, J. G. (2007). Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites. Mol. Microbiol. 64, 42–56. doi: 10.1111/j.1365-2958.2007.05596.x
Hiasa, H., and Marians, K. J. (1994). Tus prevents overreplication of oriC plasmid DNA. J. Biol. Chem. 269, 26959–26968.
Hidaka, M., Kobayashi, T., and Horiuchi, T. (1991). A newly identified DNA replication terminus site, TerE, on the Escherichia coli chromosome. J. Bacteriol. 173, 391–393.
Hill, T. M., Henson, J. M., and Kuempel, P. L. (1987). The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc. Natl. Acad. Sci. U. S. A. 84, 1754–1758. doi: 10.1073/pnas.84.7.1754
Hill, T. M., Kopp, B. J., and Kuempel, P. L. (1988a). Termination of DNA replication in Escherichia coli requires a trans-acting factor. J. Bacteriol. 170, 662–668.
Hill, T. M., and Marians, K. J. (1990). Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc. Natl. Acad. Sci. U. S. A. 87, 2481–2485.
Hill, T. M., Pelletier, A. J., Tecklenburg, M. L., and Kuempel, P. L. (1988b). Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cells 55, 459–466.
Hill, T. M., Tecklenburg, M. L., Pelletier, A. J., and Kuempel, P. L. (1989). Tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 86, 1593–1597. doi: 10.1073/pnas.86.5.1593
Hizume, K., and Araki, H. (2019). Replication fork pausing at protein barriers on chromosomes. FEBS Lett. 593, 1449–1458. doi: 10.1002/1873-3468.13481
Horiuchi, T., Fujimura, Y., Nishitani, H., Kobayashi, T., and Hidaka, M. (1994). The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J. Bacteriol. 176, 4656–4663.
Ivanova, D., Taylor, T., Smith, S. L., Dimude, J. U., Upton, A. L., Mehrjouy, M. M., et al. (2015). Shaping the landscape of the Escherichia coli chromosome: replication-transcription encounters in cells with an ectopic replication origin. Nucleic Acids Res. 43, 7865–7877. doi: 10.1093/nar/gkv704
Jameson, K. H., Rudolph, C. J., and Hawkins, M. (2021). Termination of DNA replication at Tus-ter barriers results in under-replication of template DNA. J. Biol. Chem. 297:101409. doi: 10.1016/j.jbc.2021.101409
Jameson, K. H., and Wilkinson, A. J. (2017). Control of initiation of DNA replication in Bacillus subtilis and Escherichia coli. Genes 8:22. doi: 10.3390/genes8010022
Joyce, C. M., and Steitz, T. A. (1995). Polymerase structures and function: variations on a theme? J. Bacteriol. 177, 6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995
Kamada, K., Horiuchi, T., Ohsumi, K., Shimamoto, N., and Morikawa, K. (1996). Structure of a replication-terminator protein complexed with DNA. Nature 383, 598–603. doi: 10.1038/383598a0
Katayama, T., Kasho, K., and Kawakami, H. (2017). The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 8:2496. doi: 10.3389/fmicb.2017.02496
Khatri, G. S., MacAllister, T., Sista, P. R., and Bastia, D. (1989). The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cells 59, 667–674. doi: 10.1016/0092-8674(89)90012-3
Kim, N., and Jinks-Robertson, S. (2012). Transcription as a source of genome instability. Nat. Rev. Genet. 13, 204–214. doi: 10.1038/nrg3152
Kirby, R. (2011). Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol. Lett. 319, 1–10. doi: 10.1111/j.1574-6968.2011.02242.x
Kono, N., Arakawa, K., and Tomita, M. (2012). Validation of bacterial replication termination models using simulation of genomic mutations. PLoS One 7:e34526. doi: 10.1371/journal.pone.0034526
Krabbe, M., Zabielski, J., Bernander, R., and Nordström, K. (1997). Inactivation of the replication-termination system affects the replication mode and causes unstable maintenance of plasmid R1. Mol. Microbiol. 24, 723–735.
Kuempel, P. L., Duerr, S. A., and Seeley, N. R. (1977). Terminus region of the chromosome in Escherichia coli inhibits replication forks. Proc. Natl. Acad. Sci. U. S. A. 74, 3927–3931.
Kurth, I., and O’Donnell, M. (2009). Replisome dynamics during chromosome duplication. EcoSal Plus 3:10.1128/ecosalplus.4.4.2. doi: 10.1128/ecosalplus.4.4.2
Labib, K., and Hodgson, B. (2007). Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 8, 346–353. doi: 10.1038/sj.embor.7400940
Lang, K. S., and Merrikh, H. (2018). The clash of macromolecular titans: replication-transcription conflicts in Bacteria. Annu. Rev. Microbiol. 72, 71–88. doi: 10.1146/annurev-micro-090817-062514
LeBowitz, J. H., and McMacken, R. (1986). The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 261, 4738–4748. doi: 10.1016/S0021-9258(17)38564-2
Lee, E. H., Kornberg, A., Hidaka, M., Kobayashi, T., and Horiuchi, T. (1989). Escherichia coli replication termination protein impedes the action of helicases. Proc. Natl. Acad. Sci. U. S. A. 86, 9104–9108. doi: 10.1073/pnas.86.23.9104
Lemon, K. P., Kurtser, I., and Grossman, A. D. (2001). Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98, 212–217. doi: 10.1073/pnas.011506098
Leonard, A. C., and Méchali, M. (2013). DNA replication origins. Cold Spring Harb. Perspect. Biol. 5:a010116. doi: 10.1101/cshperspect.a010116
Levy, O., Ptacin, J. L., Pease, P. J., Gore, J., Eisen, M. B., Bustamante, C., et al. (2005). Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc. Natl. Acad. Sci. U. S. A. 102, 17618–17623. doi: 10.1073/pnas.0508932102
Lewis, P. J., Ralston, G. B., Christopherson, R. I., and Wake, R. G. (1990). Identification of the replication terminator protein binding sites in the terminus region of the Bacillus subtilis chromosome and stoichiometry of the binding. J. Mol. Biol. 214, 73–84. doi: 10.1016/0022-2836(90)90147-E
Linke, R., Limmer, M., Juranek, S. A., Heine, A., and Paeschke, K. (2021). The relevance of G-quadruplexes for DNA repair. Int. J. Mol. Sci. 22:12599. doi: 10.3390/ijms222212599
Louarn, J., Patte, J., and Louarn, J. M. (1977). Evidence for a fixed termination site of chromosome replication in Escherichia coli K12. J. Mol. Biol. 115, 295–314. doi: 10.1016/0022-2836(77)90156-5
Louarn, J., Patte, J., and Louarn, J. M. (1979). Map position of the replication terminus on the Escherichia coli chromosome. Mol. Gen. Genet. MGG 172, 7–11. doi: 10.1007/BF00276208
MacAllister, T., Khatri, G. S., and Bastia, D. (1990). Sequence-specific and polarized replication termination in vitro: complementation of extracts of tus- Escherichia coli by purified Ter protein and analysis of termination intermediates. Proc. Natl. Acad. Sci. U. S. A. 87, 2828–2832. doi: 10.1073/pnas.87.7.2828
Marians, K. J. (2018). Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 87, 217–238. doi: 10.1146/annurev-biochem-062917-011921
Maric, M., Maculins, T., De Piccoli, G., and Labib, K. (2014). Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 346:1253596. doi: 10.1126/science.1253596
Markovitz, A. (2005). A new in vivo termination function for DNA polymerase I of Escherichia coli K12. Mol. Microbiol. 55, 1867–1882. doi: 10.1111/j.1365-2958.2005.04513.x
Masters, M., and Broda, P. (1971). Evidence for the bidirectional replications of the Escherichia coli chromosome. Nature. New Biol. 232, 137–140. doi: 10.1038/newbio232137a0
McGlynn, P., and Lloyd, R. G. (2001). Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. 98, 8227–8234. doi: 10.1073/pnas.111008698
McGlynn, P., Savery, N. J., and Dillingham, M. S. (2012). The conflict between DNA replication and transcription. Mol. Microbiol. 85, 12–20. doi: 10.1111/j.1365-2958.2012.08102.x
McLean, M. J., Wolfe, K. H., and Devine, K. M. (1998). Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 47, 691–696.
Médigue, C., Krin, E., Pascal, G., Barbe, V., Bernsel, A., Bertin, P. N., et al. (2005). Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15, 1325–1335. doi: 10.1101/gr.4126905
Merrikh, H. (2017). Spatial and temporal control of evolution through replication-transcription conflicts. Trends Microbiol. 25, 515–521. doi: 10.1016/j.tim.2017.01.008
Merrikh, C. N., and Merrikh, H. (2018). Gene inversion potentiates bacterial evolvability and virulence. Nat. Commun. 9:4662. doi: 10.1038/s41467-018-07110-3
Merrikh, C. N., Weiss, E., and Merrikh, H. (2016). The accelerated evolution of lagging Strand genes is independent of sequence context. Genome Biol. Evol. 8, 3696–3702. doi: 10.1093/gbe/evw274
Merrikh, H., Zhang, Y., Grossman, A. D., and Wang, J. D. (2012). Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 10, 449–458. doi: 10.1038/nrmicro2800
Midgley-Smith, S. L., Dimude, J. U., and Rudolph, C. J. (2019). A role for 3′ exonucleases at the final stages of chromosome duplication in Escherichia coli. Nucleic Acids Res. 47, 1847–1860. doi: 10.1093/nar/gky1253
Midgley-Smith, S. L., Dimude, J. U., Taylor, T., Forrester, N. M., Upton, A. L., Lloyd, R. G., et al. (2018). Chromosomal over-replication in Escherichia coli recG cells is triggered by replication fork fusion and amplified if replichore symmetry is disturbed. Nucleic Acids Res. 46, 7701–7715. doi: 10.1093/nar/gky566
Moreno, S. P., Bailey, R., Campion, N., Herron, S., and Gambus, A. (2014). Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 346, 477–481. doi: 10.1126/science.1253585
Moreno, S. P., and Gambus, A. (2015). Regulation of unperturbed DNA replication by Ubiquitylation. Genes 6, 451–468. doi: 10.3390/genes6030451
Mulcair, M. D., Schaeffer, P. M., Oakley, A. J., Cross, H. F., Neylon, C., Hill, T. M., et al. (2006). A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cells 125, 1309–1319. doi: 10.1016/j.cell.2006.04.040
Mulugu, S., Potnis, A., Shamsuzzaman, T., Alexander, K., and Bastia, D. (2001). Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc. Natl. Acad. Sci. U. S. A. 98, 9569–9574. doi: 10.1073/pnas.171065898
Nakabachi, A., Yamashita, A., Toh, H., Ishikawa, H., Dunbar, H. E., Moran, N. A., et al. (2006). The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. doi: 10.1126/science.1134196
Natarajan, S., Kelley, W. L., and Bastia, D. (1991). Replication terminator protein of Escherichia coli is a transcriptional repressor of its own synthesis. Proc. Natl. Acad. Sci. U. S. A. 88, 3867–3871.
Neidhardt, F. C., Curtiss, R., Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., et al. (eds). (1996). Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. Washington, D.C.: ASM Press.
Nester, E. W. (2015). Agrobacterium: nature’s genetic engineer. Front. Plant Sci. 5:730. doi: 10.3389/fpls.2014.00730
Neylon, C., Brown, S. E., Kralicek, A. V., Miles, C. S., Love, C. A., and Dixon, N. E. (2000). Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: a model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 39, 11989–11999. doi: 10.1021/bi001174w
Neylon, C., Kralicek, A. V., Hill, T. M., and Dixon, N. E. (2005). Replication termination in Escherichia coli: structure and antihelicase activity of the Tus-Ter complex. Microbiol. Mol. Biol. Rev. MMBR 69, 501–526. doi: 10.1128/MMBR.69.3.501-526.2005
Pai, K. S., Bussiere, D. E., Wang, F., Hutchison, C. A. 3rd, White, S. W., and Bastia, D. (1996). The structure and function of the replication terminator protein of Bacillus subtilis: identification of the ‘winged helix’ DNA-binding domain. EMBO J. 15, 3164–3173. doi: 10.1002/j.1460-2075.1996.tb00679.x
Pandey, M., Elshenawy, M. M., Jergic, S., Takahashi, M., Dixon, N. E., Hamdan, S. M., et al. (2015). Two mechanisms coordinate replication termination by the Escherichia coli Tus-Ter complex. Nucleic Acids Res. 43, 5924–5935. doi: 10.1093/nar/gkv527
Pham, T. M., Tan, K. W., Sakumura, Y., Okumura, K., Maki, H., and Akiyama, M. T. (2013). A single-molecule approach to DNA replication in Escherichia coli cells demonstrated that DNA polymerase III is a major determinant of fork speed. Mol. Microbiol. 90, 584–596. doi: 10.1111/mmi.12386
Prescott, D. M., and Kuempel, P. L. (1972). Bidirectional replication of the chromosome in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 69, 2842–2845. doi: 10.1073/pnas.69.10.2842
Raghunathan, N., Goswami, S., Leela, J. K., Pandiyan, A., and Gowrishankar, J. (2019). A new role for Escherichia coli dam DNA methylase in prevention of aberrant chromosomal replication. Nucleic Acids Res. 47, 5698–5711. doi: 10.1093/nar/gkz242
Rakowski, S. A., and Filutowicz, M. (2013). Plasmid R6K replication control. Plasmid 69, 231–242. doi: 10.1016/j.plasmid.2013.02.003
Rasko, D. A., Rosovitz, M. J., Myers, G. S. A., Mongodin, E. F., Fricke, W. F., Gajer, P., et al. (2008). The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190, 6881–6893. doi: 10.1128/JB.00619-08
Rothstein, R., Michel, B., and Gangloff, S. (2000). Replication fork pausing and recombination or “gimme a break”. Genes Dev. 14, 1–10. doi: 10.1101/gad.14.1.1
Rudolph, C. J., Corocher, T.-A., Grainge, I., and Duggin, I. G. (2019). Termination of DNA replication in prokaryotes, Chichester: John Wiley & Sons, Ltd, 1–15
Rudolph, C. J., Dhillon, P., Moore, T., and Lloyd, R. G. (2007). Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 6, 981–993. doi: 10.1016/j.dnarep.2007.02.017
Rudolph, C. J., Mahdi, A. A., Upton, A. L., and Lloyd, R. G. (2010). RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics 186, 473–492. doi: 10.1534/genetics.110.120691
Rudolph, C. J., Upton, A. L., and Lloyd, R. G. (2009). Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol. Microbiol. 74, 940–955. doi: 10.1111/j.1365-2958.2009.06909.x
Rudolph, C. J., Upton, A. L., Stockum, A., Nieduszynski, C. A., and Lloyd, R. G. (2013). Avoiding chromosome pathology when replication forks collide. Nature 500, 608–611. doi: 10.1038/nature12312
Schneiker, S., Perlova, O., Kaiser, O., Gerth, K., Alici, A., Altmeyer, M. O., et al. (2007). Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25, 1281–1289. doi: 10.1038/nbt1354
Schroeder, J. W., Hirst, W. G., Szewczyk, G. A., and Simmons, L. A. (2016). The effect of local sequence context on mutational bias of genes encoded on the leading and lagging strands. Curr. Biol. CB 26, 692–697. doi: 10.1016/j.cub.2016.01.016
Schroeder, J. W., Sankar, T. S., Wang, J. D., and Simmons, L. A. (2020). The roles of replication-transcription conflict in mutagenesis and evolution of genome organization. PLoS Genet. 16:e1008987. doi: 10.1371/journal.pgen.1008987
Sharma, B., and Hill, T. M. (1992). TerF, the sixth identified replication arrest site in Escherichia coli, is located within the rcsC gene. J. Bacteriol. 174, 7854–7858.
Sherratt, D. J., Arciszewska, L. K., Crozat, E., Graham, J. E., and Grainge, I. (2010). The Escherichia coli DNA translocase FtsK. Biochem. Soc. Trans. 38, 395–398. doi: 10.1042/BST0380395
Shima, N., and Pederson, K. D. (2017). Dormant origins as a built-in safeguard in eukaryotic DNA replication against genome instability and disease development. DNA Repair 56, 166–173. doi: 10.1016/j.dnarep.2017.06.019
Singleton, M. R., Scaife, S., and Wigley, D. B. (2001). Structural analysis of DNA replication fork reversal by RecG. Cells 107, 79–89. doi: 10.1016/s0092-8674(01)00501-3
Sinha, A. K., Durand, A., Desfontaines, J.-M., Iurchenko, I., Auger, H., Leach, D. R. F., et al. (2017). Division-induced DNA double strand breaks in the chromosome terminus region of Escherichia coli lacking RecBCD DNA repair enzyme. PLoS Genet. 13:e1006895. doi: 10.1371/journal.pgen.1006895
Sinha, A. K., Possoz, C., Durand, A., Desfontaines, J.-M., Barre, F.-X., Leach, D. R. F., et al. (2018). Broken replication forks trigger heritable DNA breaks in the terminus of a circular chromosome. PLoS Genet. 14:e1007256. doi: 10.1371/journal.pgen.1007256
Skarstad, K., and Katayama, T. (2013). Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5:a012922. doi: 10.1101/cshperspect.a012922
Skovgaard, O., Bak, M., Løbner-Olesen, A., and Tommerup, N. (2011). Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 21, 1388–1393. doi: 10.1101/gr.117416.110
Smith, M. T., and Wake, R. G. (1992). Definition and polarity of action of DNA replication terminators in Bacillus subtilis. J. Mol. Biol. 227, 648–657. doi: 10.1016/0022-2836(92)90214-5
Sonneville, R., Moreno, S. P., Knebel, A., Johnson, C., Hastie, C. J., Gartner, A., et al. (2017). CUL-2LRR-1 and UBXN-3 drive replisome disassembly during DNA replication termination and mitosis. Nat. Cell Biol. 19, 468–479. doi: 10.1038/ncb3500
Srivatsan, A., Tehranchi, A., MacAlpine, D. M., and Wang, J. D. (2010). Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 6:e1000810. doi: 10.1371/journal.pgen.1000810
Steinacher, R., Osman, F., Dalgaard, J. Z., Lorenz, A., and Whitby, M. C. (2012). The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev. 26, 594–602. doi: 10.1101/gad.184663.111
Stoppe, N. C., Silva, J. S., Carlos, C., Sato, M. I. Z., Saraiva, A. M., Ottoboni, L. M. M., et al. (2017). Worldwide phylogenetic group patterns of Escherichia coli from commensal human and wastewater treatment plant isolates. Front. Microbiol. 8:2512. doi: 10.3389/fmicb.2017.02512
Syeda, A. H., Dimude, J. U., Skovgaard, O., and Rudolph, C. J. (2020). Too much of a good thing: how ectopic DNA replication affects bacterial replication dynamics. Front. Microbiol. 11:534. doi: 10.3389/fmicb.2020.00534
Tanaka, T., and Masai, H. (2006). Stabilization of a stalled replication fork by concerted actions of two helicases. J. Biol. Chem. 281, 3484–3493. doi: 10.1074/jbc.M510979200
Toft, C. J., Moreau, M. J. J., Perutka, J., Mandapati, S., Enyeart, P., Sorenson, A. E., et al. (2021). Delineation of the ancestral Tus-dependent replication fork trap. Int. J. Mol. Sci. 22:13533. doi: 10.3390/ijms222413533
Toft, C. J., Sorenson, A. E., and Schaeffer, P. M. (2022). A soft Tus-Ter interaction is hiding a fail-safe lock in the replication fork trap of Dickeya paradisiaca. Microbiol. Res. 263:127147. doi: 10.1016/j.micres.2022.127147
Tomasetti, C., Li, L., and Vogelstein, B. (2017). Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334. doi: 10.1126/science.aaf9011
Touchon, M., and Rocha, E. P. C. (2016). Coevolution of the organization and structure of prokaryotic genomes. Cold Spring Harb. Perspect. Biol. 8:a018168. doi: 10.1101/cshperspect.a018168
Trautinger, B. W., Jaktaji, R. P., Rusakova, E., and Lloyd, R. G. (2005). RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell 19, 247–258. doi: 10.1016/j.molcel.2005.06.004
Wake, R. G. (1972). Visualization of reinitiated chromosomes in Bacillus subtilis. J. Mol. Biol. 68, 501–509. doi: 10.1016/0022-2836(72)90102-7
Wake, R. G. (1973). Circularity of the Bacillus subtilis chromosome and further studies on its bidirectional replication. J. Mol. Biol. 77, 569–575. doi: 10.1016/0022-2836(73)90223-4
Wake, R. G. (1997). Replication fork arrest and termination of chromosome replication in Bacillus subtilis. FEMS Microbiol. Lett. 153, 247–254. doi: 10.1111/j.1574-6968.1997.tb12581.x
Wang, J. D., Berkmen, M. B., and Grossman, A. D. (2007). Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 104, 5608–5613. doi: 10.1073/pnas.0608999104
Wang, X., Lesterlin, C., Reyes-Lamothe, R., Ball, G., and Sherratt, D. J. (2011). Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc. Natl. Acad. Sci. U. S. A. 108, E243–E250. doi: 10.1073/pnas.1100874108
Weiss, A. S., Hariharan, I. K., and Wake, R. G. (1981). Analysis of the terminus region of the Bacillus subtilis chromosome. Nature 293, 673–675. doi: 10.1038/293673a0
Wendel, B. M., Cole, J. M., Courcelle, C. T., and Courcelle, J. (2018). SbcC-SbcD and ExoI process convergent forks to complete chromosome replication. Proc. Natl. Acad. Sci. U. S. A. 115, 349–354. doi: 10.1073/pnas.1715960114
Wendel, B. M., Courcelle, C. T., and Courcelle, J. (2014). Completion of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 111, 16454–16459. doi: 10.1073/pnas.1415025111
Wilce, J. A., Vivian, J. P., Hastings, A. F., Otting, G., Folmer, R. H. A., Duggin, I. G., et al. (2001). Structure of the RTP–DNA complex and the mechanism of polar replication fork arrest. Nat. Struct. Biol. 8, 206–210. doi: 10.1038/84934
Winterstein, C., and Ludwig, B. (1998). Genes coding for respiratory complexes map on all three chromosomes of the Paracoccus denitrificans genome. Arch. Microbiol. 169, 275–281. doi: 10.1007/s002030050572
Xie, B.-B., Rong, J.-C., Tang, B.-L., Wang, S., Liu, G., Qin, Q.-L., et al. (2021). Evolutionary trajectory of the replication mode of bacterial replicons. mBio 12:e02745-20. doi: 10.1128/mBio.02745-20
Xu, Z.-Q., and Dixon, N. E. (2018). Bacterial replisomes. Curr. Opin. Struct. Biol. 53, 159–168. doi: 10.1016/j.sbi.2018.09.006
Keywords: termination of DNA replication, DNA replication, DNA segregation, RecG helicase, Tus-ter complexes, bacterial chromosome dynamics, chromosomal architecture
Citation: Goodall DJ, Warecka D, Hawkins M and Rudolph CJ (2023) Interplay between chromosomal architecture and termination of DNA replication in bacteria. Front. Microbiol. 14:1180848. doi: 10.3389/fmicb.2023.1180848
Edited by:
Ian Grainge, The University of Newcastle, AustraliaReviewed by:
Julia Grimwade, Florida Institute of Technology, United StatesIain G. Duggin, University of Technology Sydney, Australia
Hua Xiang, Chinese Academy of Sciences (CAS), China
Gregory Marczynski, McGill University, Canada
Copyright © 2023 Goodall, Warecka, Hawkins and Rudolph. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian J. Rudolph, Y2hyaXN0aWFuLnJ1ZG9scGhAYnJ1bmVsLmFjLnVr
 Daniel J. Goodall
Daniel J. Goodall Dominika Warecka
Dominika Warecka Michelle Hawkins
Michelle Hawkins Christian J. Rudolph
Christian J. Rudolph