- 1College of Agriculture, Guangxi University, Nanning, China
- 2Key Laboratory of Green Prevention and Control on Fruits and Vegetables in South China Ministry of Agriculture and Rural Affairs, Guangxi Key Laboratory of Biology for Crop Diseases and Insect Pests, Plant Protection Research Institute, Guangxi Academy of Agricultural Science, Nanning, Guangxi, China
- 3Baise Agricultural Scientific Research Institute, Baise, China
- 4College of Agricultural Engineering, Guangxi Vocational University of Agriculture, Nanning, China
Pitaya canker, caused by Neoscytalidium dimidiatum, is one of the most important fungal diseases that cause significant losses in production. To replace chemical pesticides, the use of biocontrol strains to manage plant diseases has been the focus of research. In this study, the bacterial strain AF01, identified as Paenibacillus polymyxa, exhibited significant antifungal effects against N. dimidiatum and four other pitaya fungal pathogens. The strain P. polymyxa AF01 produces 13 fusaricidins, which directly inhibit mycelial growth, spore germination and germ tube elongation by causing the membrane integrity and cell ultrastructure to incur irreversible damage. Pot experiment and yield test confirmed that AF01 provided preservative effects by reducing the disease index. In comparison to the untreated control groups, RNA-seq data showed that P. polymyxa AF01 selectively blocked some transcription and translation processes and inhibited RNA and DNA structural dynamics, energy production and conversion, and signal transduction, particularly cell wall biosynthesis, changes in membrane permeability, and impairment of protein biosynthesis. Thus, P. polymyxa AF01 could be potentially useful as a suitable biocontrol agent for pitaya canker.
1. Introduction
Pitaya (Hylocereus spp.), also known as dragon fruit, is a tropical and subtropical fruit native to Latin America, including Mexico, Central America, and tropical South America. Currently, pitaya cultivation is concentrated in Australia, China, Southeast Asia, Malaysia, Vietnam, Thailand, and the Philippines (Adnan et al., 2011). Pitaya contains numerous nutrients such as vitamins, minerals, fiber, protein, flavonoids, betanin, and polyphenols. Additionally, the beneficial effects of red pitaya on several human diseases have been reported (Ramli et al., 2016). Recently, canker caused by Neoscytalidium dimidiatum has become one of the most serious diseases affecting pitaya (Hong et al., 2020). The disease progression in pitaya plants begins with tiny dimples on the infected cladode; then, chlorotic spots develop and gradually turn yellow, even brown, and form flat, raised, hard brown scabs. As the disease progresses, the lesions develop a typical light-gray coloration with numerous dark spots. Under high-humidity conditions, lesions can cause softening and dissolution of tissues and structures, leading to the formation of cavities and collapse (Chuang et al., 2012). The N. dimidiatum fungus produces two types of conidia: one occurring in arthric chains in aerial mycelium or in ostiolate pycnidia embedded on the surface of mature lesions, and the other type is ellipsoid to nearly fusiform, hyaline even dark brown (Crous et al., 2006; Phillips et al., 2013). Conidia are difficult to eradicate and remain viable for several years. When diseased cladodes exist in orchard soil or drains, the lesions can continue to produce spores and infect young growth (Fullerton et al., 2018). If not controlled effectively, pitaya cankers can destroy the canopy, disfigure the fruit, and render orchards uneconomical. Current management strategies for pitaya canker, include the use of disease-resistant cultivars and chemical fungicides. However, there are several limitations to developing new disease-resistant cultivars to control pitaya canker, such as variations in cultivar resistance and differences in isolate virulence (Harsonowati et al., 2020). Therefore, farmers were forced to apply excessive doses and costly fungicide treatments throughout the infection season as susceptible cultivars grew. However, the frequent use of chemical pesticides results in high levels of pathogen resistance, pesticide residues, and environmental pollution, thereby becoming a human health concern (De Miccolis Angelini et al., 2014).
Considering these problems, new measures, such as the use of microbial fungicides, are desirable to control the disease and reduce the adverse effects of chemical fungicides. Biological control is one of the most promising and safe measures to suppress diseases in plants (Choudhary and Johri, 2009; Wu et al., 2022). For example, the adaptable metabolism, rapid growth, and high mobility of fluorescent pseudomonads make them valuable in plant disease control (Bakker et al., 2007; Du et al., 2017). Similarly, metabolites from Bacillales species activate plant immune regulators (Ryu et al., 2004). In general, antifungal substances produced by antagonistic bacteria have been reported to be the most important mechanism of inhibition of pathogen growth as it affects the lipid metabolism, amino acid metabolism, carbohydrate metabolism, membrane transport, and energy metabolism (Zuo et al., 2017). These antifungal substances had been identified as antagonistic proteins, enzymes and bacteriocins synthesized by the ribosomal pathway. Other antifungal substances include lipopeptides, polyketides, peptides, and volatile antifungal substances. With a broad range of antibiotic activity and low toxicity, cyclic lipopeptides (CLPs) synthesized by Bacillus spp. (iturins, surfactin, and fengycins, etc.), Pseudomonas spp. (massetolide A and putisolvin, etc.) and Paenibacillus spp. (fusaricidins, polymyxins) have been the focus of research in the control of plant diseases in recent years (Mülner et al., 2020; Soni et al., 2021). In nature products discovery, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), have become in crucial for the identification of microorganisms and chemotyping of secondary metabolites (Mülner et al., 2021). Compounds were detected from culture supernatants or surface extracts of colonies without the need for further purification prior to mass spectrometric analysis. This technique affords simple preparation and efficient investigation of complex families of natural compounds, such as the fusaricidins, iturins with high sensitivity and accuracy in minimum time (Costa et al., 2022; Tsai et al., 2022).
Recently, there have been examples of the biological control of diseases caused by N. dimidiatum, mainly with biocontrol fungi and bacteria. For example, Trichoderma harzianum and T. atroviride reduced pathogen growth. Biocontrol bacteria such as B. amyloliquefaciens, Penicillium rolfsii and B. subtilis have also shown strong in vitro antifungal effects on N. dimidiatum (Ratanaprom et al., 2021; Wang et al., 2021). However, the inhibitory effect of biocontrol agents against N. dimidiatum has not been well elucidated to date, including the effects on the viability of conidia and ultrastructural modifications, or the ability to locate potential targets of conidia and mycelia. Therefore, in this study, we aimed to investigate the effectiveness of biocontrol microbes in suppressing pitaya canker. The objectives of this study were (1) to screen and identify effective biocontrol bacteria against pitaya canker; (2) to determine the biological control mechanisms of a biocontrol bacterium of P. polymyxa; (3) to clarify the potential capability of the bacterium for reduction of pitaya canker; and (4) to explore the molecular mechanism to identify the major target sites using RNA-seq. The results obtained in the present study provide promising information on bacterial biocontrol agents for controlling pitaya canker.
2. Materials and methods
2.1. Isolation of microorganisms
Soil samples (35) were collected from the 5–10 cm deep rhizospheres of seven pitaya orchards in Guangxi Province, China. To study the microorganisms present in these samples, 5 g of soil was added to autoclaved distilled water in a shaker for 30 min at 200 rpm at 28°C. Serial dilutions ranging from 10−1 to 10−3 were created from the supernatant, which was then spread over PDA plates (potato, 200 g; glucose, 20 g; agar, 20 g; ddH2O, 1,000 mL). After a 72 h incubation at 28°C, single morphological colonies were selected and purified on PDA plates by repeated streaking.
2.2. Screening of antagonistic bacteria against Neoscytalidium dimidiatum
To determine the potential of these isolates as antagonists, preliminary screening was conducted using the dual culture method (Ferreira et al., 1991). Phytopathogenic isolate BH5 was used in this test. BH5 was known to cause pitaya canker and was identified as N. dimidiatum by an internal transcribed spacer gene sequencing, with the NCBI GenBank database accession number OP393909.1. Analysis using BLAST (Basic Local Alignment Search Tool) revealed 99% sequence similarities of the strains with those of N. dimidiatum (Genbak accession number MZ047291.1 and OL455801.1). After demonstrating noticeable antagonistic activity against N. dimidiatum, isolates with inhibitory capacity were stored in 20% (v/v) glycerol at −80°C, and the strain with the strongest inhibition ability (labeled AF01) was selected for further studies.
2.3. Antifungal spectrum tests
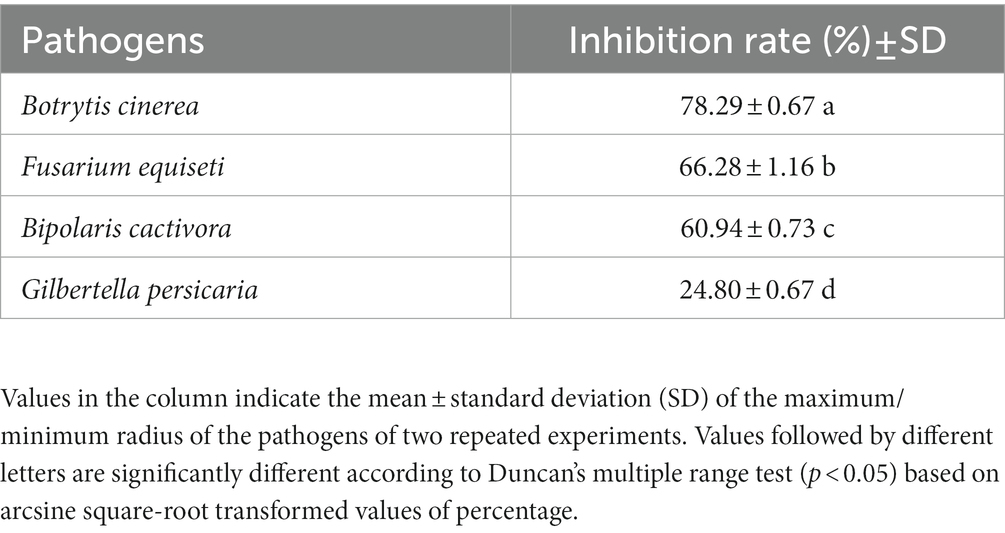
A collection of pitaya fungal pathogens of Fusarium equiseti (C), Botrytis cinerea (D), Bipolaris cactivora (E) and Gilbertella persicaria (F) were tested (Lin et al., 2018). Inhibition of the pathogens was assessed as described: 1 μL of AF01 bacterial culture was inoculated 2.5 cm away from the edge of the plate twice, with three replicate plates. The antimicrobial activity was calculated by the percentage of mycelium growth inhibition compared to the control according to the formula: (R1 − R2)/R1 × 100, where R1 and R2 were the radii of the pathogen colony in the control and the antagonist colony, respectively.
2.4. Identification of strain AF01
Morphological characteristics of AF01 were observed on nutrient agar (NA). To determine the 16 s rDNA gene sequence of strain AF01, genomic DNA was extracted using the TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China). The 16 s rDNA gene was amplified by PCR using the primers 27F:5′-AGAGTTTGATCCTGGCTCAG-3′; 1492R 5′-GGTTACCTTGTTACGACTT-3′. The 50 μL PCR mixture contained 25 μL 2 × Es Taq MasterMix (Dye), 2 μL 10 μM primers each, 2 μL genomic DNA, and 19 μL double distilled water. The PCR program was as follows: denaturation at 94°C for 2 min, 30 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, and final extension at 72°C for 2 min. The amplified PCR product was sequenced by Tsingke Biotechnology (Guangzhou, China), and the resulting sequences were subjected to BLASTN searches using the GenBank database. A phylogenetic tree was constructed using Phylosuit 1.2.2, based on the neighbor-joining method. The biochemical properties of strains AF01 was characterized using a GEN III MicroStation (BIOLOG, Hayward, CA, United States). A 150 μL bacterial suspension (108 CFU/mL) was added to each well of a GEN III MicroPlate containing carbon source utilization assays and 23 chemical sensitivity assays. The MicroPlates were incubated for 14 h to form phenotypic fingerprint through the reduction of the tetrazolium redox dye caused by increased respiration. The phenotypic pattern in the GEN III MicroPlate was analyzed using read on the BIOLOG automated microbial analysis system software (GEN_ III_v2.8.0.15G).
2.5. Effects of cultural conditions on biomass and inhibitory activities of strain AF01 against Neoscytalidium dimidiatum
To test the ability to produce antimicrobial lipopeptides on different media, strain AF01 was grown in 100 mL PDB (the same components except agar), Landy (glucose, 20 g; glutamic, acid 5 g; yeast extract, 1 g; K2HPO4, 1 g; MgSO4, 0.5 g; KCl, 0.5 g; CuSO4, 1.6 mg; Fe2(SO4)3, 1.2 mg; MnSO4, 0.4 mg; ddH2O, 1,000 mL), NA (beef extract, 3 g; peptone, 5 g; glucose, 2.5 g; ddH2O, 1,000 mL), LB (tryptone, 10 g; yeast extract, 5 g; NaCl, 10 g; ddH2O, 1,000 mL), GSC (glucose, 20 g; starch, 20 g; (NH4)2SO4, 20 g; yeast extract, 10 g; K2HPO4, 2.6 g; FeSO4· 7H2O, 0.1 g; MgSO4· 7H2O, 0.5 g; NaCl, 0.25 g; CaCO3, 9 g; ddH2O, 1,000 mL), BPYDB (beef extract, 3 g; peptone 5 g; yeast extract, 1 g; glucose, 10 g; ddH2O 1,000 mL). After shaking at 28°C for 72 h, each culture was centrifuged at 10,000 rpm for 10 min, and supernatants were filtered through a 0.22 μm filter into Oxford cups. The Oxford cups were placed equidistantly in molten PDA medium (20 mL/dish, 9 cm flat dish). After solidification, the oxford cups were removed and the plates were evenly coated with 200 μL of N. dimidiatum spore suspensions (107 CFU/mL). Then, each cell-free culture was added to the wells, and the diameter of the inhibition circle was measured by the crossover method after incubation for 72 h. Experiments in conical flasks cultivations were carried out in triplicate; mean value and standard deviation were calculated.
Strain AF01 was incubated in the optimal screening medium (1%, v:v) on a rotary shaker (180 rpm) at 28°C for different time courses ranged from 0 to 96 h. The OD600 values of the resultant cell suspensions were separately measured at each incubation time using the spectrophotometer. The inhibitory activities of the cell-free suspensions against N. dimidiatum at each incubation time were diluted into 1:10 with melted PDA. A N. dimidiatum plug (0.5 cm diameter) was placed at the center of a PDA plate and incubated at 28°C for 72 h and examined the inhibition rate, separately.
2.6. Effect of AF01 culture filtrate on hyphal growth of Neoscytalidium dimidiatum
Paenibacillus polymyxa AF01 was grown in a PDB liquid medium and incubated at 28°C, 180 rpm for 48 h, then the suspension was filtered through a 0.22 μm bacterial filter. Filtrate of AF01 was added to PDA at final concentrations of 2.5%, 5%, and 10%, and three plates were used for each treatment to examine the inhibition rate as 2.5. PDA plates without the AF01 culture filtrate were used as controls. A N. dimidiatum plug (0.5 cm diameter) was placed at the center of a PDA plate and incubated at 28°C for 72 h. The morphology of N. dimidiatum mycelia treated with AF01 culture filtrate was compared to that of mycelia untreated with AF01 under an optical microscope of OLYMPUS BX53 (OLYMPUS, Tokyo, Japan).
2.7. Effect of AF01 culture filtrate on spore germination of Neoscytalidium dimidiatum
Four days after inoculation, conidia were rinsed from N. dimidiatum dishes with 2.5%, 5%, and 10% AF01 culture filtrates to collect spores and adjusted to 107 cells/mL by a hemocytometer. Control was washed with PDB only. Each treatment had three replicates. Conidia suspensions were incubated at 28°C, and spore germination was counted when the control germination rate was over 90%. Approximately a hundred N. dimidiatum conidia per replicate were examined microscopically for germination. To measure the bud lengths, line tool of cellSens (Ver.2.2) was used to draw a straight line from the germ neck to the germ tip. The measure of the length of this line was taken as the approximate length of the germ tube. The inhibitions of germination and germ tube elongation were then calculated using the formula: Inhibitions (%) of conidia germination = (average germinated conidia in control − average germinated conidia in treatment)/average germinated conidia in control × 100. Inhibitions (%) of germ tube elongation = (average germ tube length in control − average germ tube length in treatment)/average germ tube length in control × 100.
2.8. Assay of cell contents leakage
Conidia suspensions (~106 cells/mL) of N. dimidiatum were grown in PDB media at 180 rpm for 3 days at 25°C. The mycelia and conidia were collected and washed three times with sterile distilled water, then resuspended in sterile distilled water containing 10% AF01 culture filtrate and shaken at 28°C for 0–6 h. The control was treated with 10% PDB. The supernatant was collected centrifugation at 6,000 rpm for 5 min. Then, the OD260 and OD280 values were detected to evaluate the leakage of nucleic acid and proteins from N. dimidiatum by NanoDrop2000 (Thermo, MA, United States). Electrolyte leakage was detected in a conductometer (LEICI, Shanghai, China).
2.9. MALDI-TOF MS analysis
The potential antifungal components in AF01 culture were detected and identified by MALDI-TOF MS as previously reported (Vater et al., 2015) with slightly modified. The culture of AF01 was centrifuged at 12,000 rpm, 10 min, and the supernatant was further filtered through a 0.22 μm bacterial filter to get the cell-free extraction of AF01, which was 10 times diluted with acetonitrile. Then 5 μL diluted cell-free extraction of AF01 was mixed with the same volume of matrix solution (a saturated solution of α-hydroxy-cinnamic acid in 50% aqueous acetonitrile containing 0.1% trifluoroacetic acid). The mixture (2.5 μL) was spotted on the target, air-dried, and measured. Mass spectra were obtained in positive ion and line modes with a QuanTOF MALDI-TOF mass spectrometer (IntelliBio, Shandong, China) equipped with a 337 nm nitrogen laser. For recording mass spectra in the range of 0.8 to 1.4 kDa, the focus mass was 1 kDa, the detector voltages were set at −0.44 kV, the laser pulse energy was set at 3.0 μJ, and each spectrum was recorded 800 times. The resulting mass/charge ratios (m/z) were used for component identification by comparing with the known fusaricidins.
2.10. Assembly screening for differentially expressed genes, gene ontology annotation, and Kyoto encyclopedia of genes and genomes enrichment
Strain N. dimidiatum BH5 was inoculated on a PDA plate with pre-placed cellophane and cultured for 36 h at 28°C. For the treatment, cell-free AF01 culture medium was diluted 1:10 with PDB, and 15 mL was added to plates co-cultured with N. dimidiatum. The control was treated with PDB alone. After 3 h, the AF01 culture filtrate was removed, and N. dimidiatum was washed thrice with sterile water. Treated and control N. dimidiatum mycelia were collected and subjected to intertranscriptomic analysis.
The TRIzol® Reagent (Plant RNA Purification Reagent) was used to extract total RNA from treated and control N. dimidiatum mycelia according to the manufacturer’s instructions (Invitrogen). DNase I (TaKaRa) was used to remove genomic DNA. The quality of RNA was verified by examining RNA degradation and contamination on 1% agarose gel. Then RNA quality was assessed using the 2,100 Bioanalyser (Agilent Technologies) and quantified using the NanoDrop2000. The sequencing library was only prepared from high-quality RNA samples with an OD260/ OD 280 = 1.8–2.2, OD260/OD230 ≥ 2.0, RIN ≥ 8.0, 28S:18S ≥ 1.0, and > 1 μg. The Illumina HiSeq X Ten platform (Illumina, San Diego, CA, United States) was used to sequence the cDNA libraries prepared by Shanghai Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China. Alignment of reads was done to the reference genome of N. dimidiatum BH5 (unpublished). The raw reads were subjected to quality control (QC; Q30 > 99.9%). Raw data were filtered to obtain high-quality sequencing data (clean). Significantly differentially expressed genes (DEGs) were identified based on a threshold of |log2 (foldchange)| ≥ 1 and P-adjust ≤ 0.05 using DESeq2 software tools. To further understand the biological functions of the DEGs, functional enrichment analysis was performed using Gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The significance of the enriched GO terms and metabolic pathways were determined at adjusted p ≤ 0.05 compared to the whole-transcriptome background. GO functional enrichment and KEGG pathway enrichment analyses were performed using the software tools Goatools1 and KOBAS,2 respectively.
2.11. Pot experiment
Disease-susceptible pitaya (Hylocereus polyrhizus) “Jindu No. 1” was used in this experiment. The seedlings of pitaya were transplanted into pots, and after 3 weeks, the plants with 2–3 cladodes in a core, with similar growth status and no mechanical injury, were selected. Plants were disinfected with 75% ethyl alcohol for 1 min, followed by washing with sterile distilled water three times then air dried. Fermented bacterial suspension of AF01 (~5 × 107 CFU/mL) was sprayed on the pitayas. Sterile water was used as the control. After 24 h, spore suspensions (~107 cells/mL) of the pathogens were sprayed onto the cladodes. The experiment used a randomized block design with three replicates of two pots each; 73 twigs were tested. All plants were maintained in light growth chambers and then were incubated at 28°C and 80% humidity. Disease severity was rated after 14 and 21 days post infection (dpi) by using an index of 0–9 (0 = healthy; 1 = lesion area less than 5%; 3 = lesion area 6%–10%; 5 = lesion area 11%–25%; 7 = lesion area 26%–50%; and 9 = lesion area 51%–100%). The disease index (DI) was then calculated using the formula: Disease index = [Σ (rating × number of plants rated)/(total number of plants × highest rating)] × 100.
2.12. Field efficacy assessment of the AF01 against pitaya canker
To confirm the preventive effects of pitaya canker under field conditions, experiments were conducted in a field naturally infested with N. dimidiatum in September of 2021. The field tests were conducted at the Guangxi Academy of Agricultural Sciences experimental station in Nanning, China (108.063786°E, 23.252453°N, soil pH: 5.64, soil type: loam, organic matter: 29.2 g/kg) cultivated with “Jindu No. 1” pitaya plants. Pitaya cankers occur regularly every year at this station. The field trial was started on September 4 and finished on September 25 in 2021. At the start, a batch of very young fruit (approximately 1 week after flowering) appeared. Pitaya were grown in 2.5 × 0.5 m (row spacing × plant space) plots. The experimental design was a complete randomized block, and each treatment area was 15 m2, consisting of 30 plants per replicate with three replicates. During the trial, the climatic conditions recorded by an automatic weather station (Campbell, Logan, Utah, United States): rainfall 190.5 mm; rainy days: 8; temperature 23.13°C–38.54°C, daily average temperature: 25.30–31.23°C, and RH 40.16%–99.9%. Treatments of strain AF01 and 250 g/L pyraclostrobin EC (BASF) were set up in the trial. Strain AF01 was grown in PDB under agitation (180 rpm) at 28°C for 48 h, and liquid culture were diluted into 10 × and 20 × (1 × 108 and 5 × 107 CFU/mL, respectively) with H2O. Pyraclostrobin (166.7 mg kg−1, active ingredient) was separately sprayed on the plants to control pitaya canker. Clean water was sprayed on the control blocks. All treatments were carried out with battery-pressurized backpack sprayer with a hollow cone nozzle delivering at 0.2 KPa. Each plot was sprayed with 2.5 L of liquid per plot, equivalent to 1666.7 L per hectare. The application was performed three times at 7-day intervals. All pitaya fruits with no symptoms at the first treatment were signed in each repeating area to investigate disease incidence. The DI investigation was performed twice: 7 days after 2nd application time, when occurrence of pitaya canker was significantly at CK plots. And the final investigation was performed 7 days after the last application. Disease grading for pitaya canker was based on a pot experiment. The formation for calculating the disease index as 2.10 and control efficiency was as follows: Control efficiency = [(disease index of control area − disease index of treatment area)/disease index of control area] × 100.
2.13. Statistical analysis
Statistical analyses were performed in SPSS software version 26 (SPSS, Chicago, IL, United States), significant differences (p < 0.05) were analyzed by one-way ANOVA with Duncan’s multiple range test.
3. Results
3.1. Isolation of biocontrol bacteria
A total of 113 bacterial isolates were collected from different sites, and five strains showed antagonistic activity against N. dimidiatum in vitro. The strain isolated from soil collected in Qinzhou, Guangxi Province, China, labeled AF01, showed the strongest antagonistic activity against N. dimidiatum with an inhibition rate of 76.80% (Figure 1B).
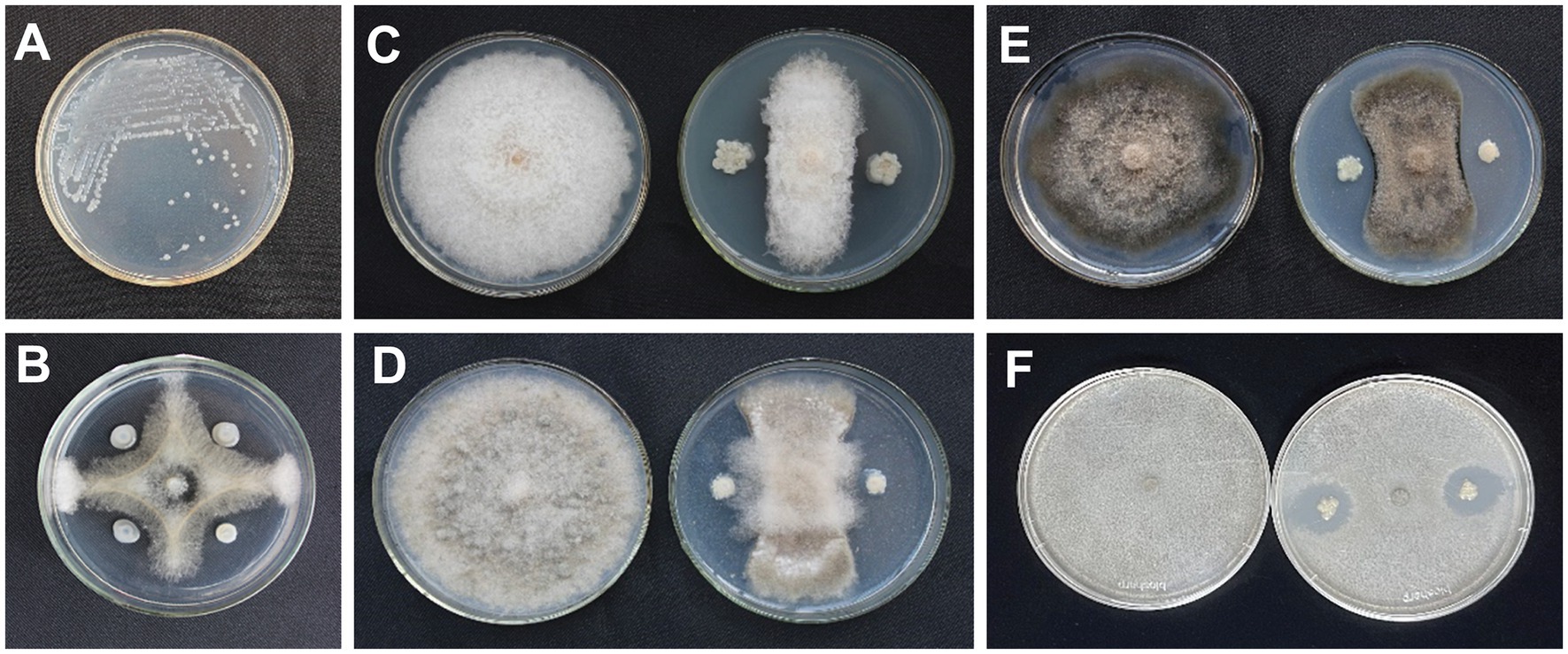
Figure 1. Colony morphology of Paenibacillus polymyxa AF01 on NA plate (A). Inhibition effect of P. polymyxa AF01 on mycelial growth of Neoscytalidium dimidiatum (B), Fusarium equiseti (C), Botrytis cinerea (D), Bipolaris cactivora (E), and Gilbertella persicaria (F) based on a dual culture method.
3.2. Inhibitory spectrum of strain AF01
Strain AF01 revealed strong inhibitory abilities against all the tested pathogens F. equiseti, B. cinerea, and B. cactivora. The highest percentage inhibition of radial growth (PIRG) value of 78.29% was observed for AF01 against B. cinerea (Figure 1D), and the lowest recorded, 24.80%, was observed against G. persicaria (Figure 1F). AF01 had a relatively weak inhibitory effect on B. cactivora (Figure 1E) and equiseti (Figure 1C), with 60.94% and 66.28% inhibition rates, respectively (Table 1).
3.3. Identification of bacterial isolates
AF01 is a rod-shaped Gram-negative bacterium that appears as a bulged surface of milky white, opaque colonies with irregular margins on an NA plate (Figure 1A). The beeswarm figure presented strains AF01 similar in the use of most carbon source substrates and assayed chemical compounds compared to the BIOLOG databases (Figure 2). Thereby, the strain AF01 was confirmed to be Paenibacillus polymyxa (SIM value:0.724, PROB value:0.910, and DIST value:2.878). In addition, BLAST analysis of the 16 s rDNA genes showed that the AF01 (GenBank database accession number: OQ601576) had an identity of 99% for Paenibacillus polymyxa (CP009909 and CP025957), suggesting that the strain was classified as Paenibacillus or Paenibacillus polymyxa. Phylogenetic analysis revealed the close clustering of strain AF01 with the sequences of Paenibacillus spp. Furthermore, AF01 and P. polymyxa were clustered together (Figure 3).
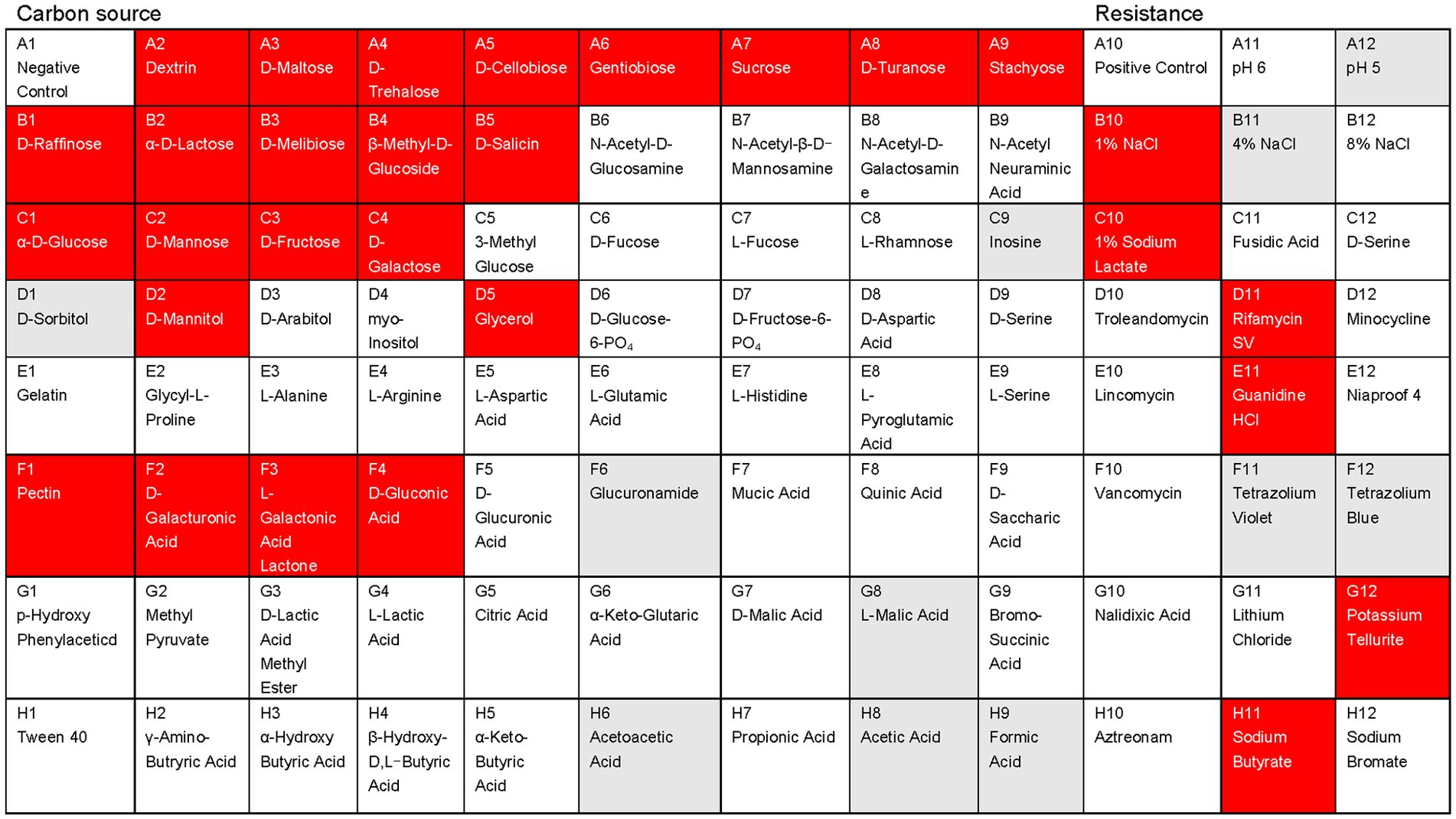
Figure 2. Physiological and biochemical characteristics analysis of the Paenibacillus polymyxa AF01using the Biolog GEN III MicroPlates system. Different patterns of 71 carbon sources and 23 susceptibility factors were observed in the studied microorganisms: red, positive; white, negative; gray, weak.
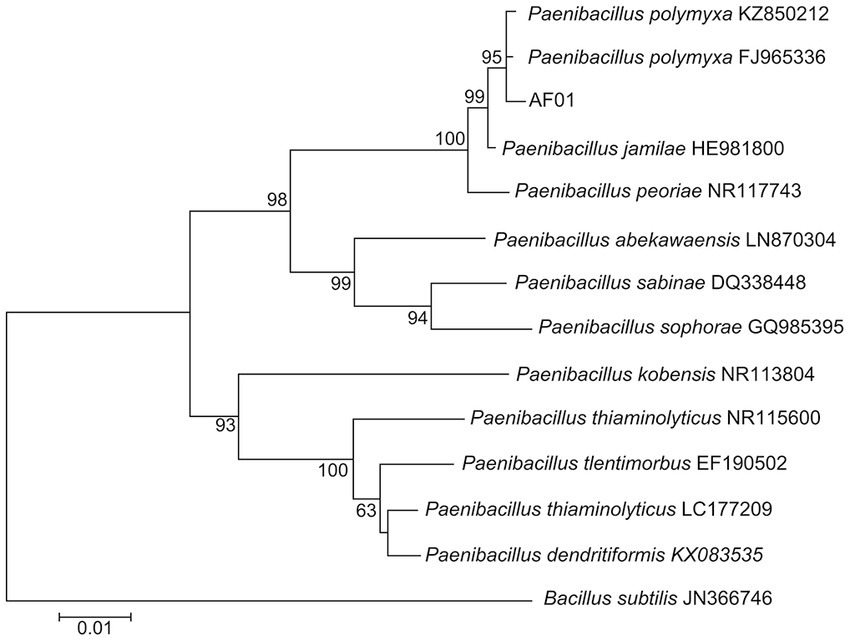
Figure 3. Neighbor joining phylogenetic tree showing the position of Paenibacillus polymyxa AF01 isolate with other species of Paenibacillus spp. and related taxa based on 16S rDNA gene sequences. Bootstrap values (expressed as percentages of 1,000 replications) are indicated at tree branch points.
3.4. Effects of cultural conditions on biomass and inhibitory activities of strain AF01 against Neoscytalidium dimidiatum
The inhibitory activity of strain AF01 against N. dimidiatum was detected in three of all media. The inhibition zone with the largest diameter (27.5 mm) were observed in PDB. The inhibit activities of strain AF01in GSC was higher than BPYDB and Landy. On the other hand, no inhibition zone was observed in LB and NA (Figure 4A). The bacterial biomass expressed by OD600 values and the inhibition rates of strain AF01 against N. dimidiatum obviously increased before 48 h of incubation and subsequently maintained at a stable level. The highest biomass (OD600 = 3.78) and peak inhibition rate (66.88%) occurred at the 56 and 48 h of incubation, respectively (Figure 4B).
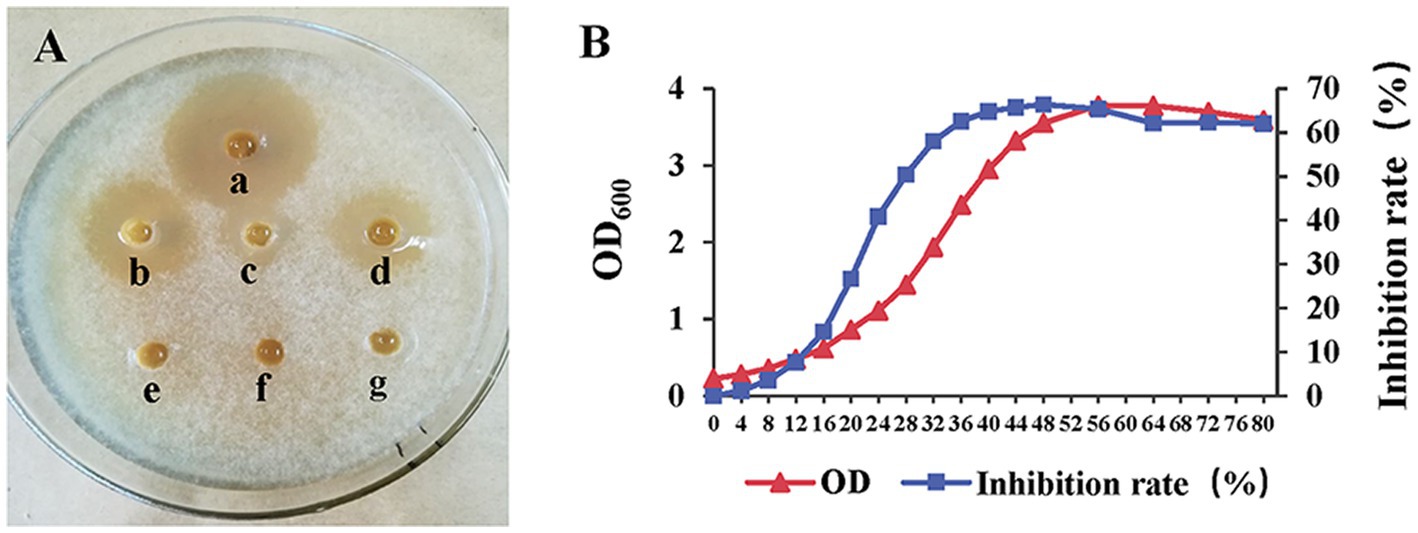
Figure 4. Agar well diffusion assay illustrating the growth inhibition of Neoscytalidium dimidiatum by cell-free supernatants extracted from different media (A). (a) PDB (inhibition zone 27.5 mm); (b) GSC (inhibition zone 2.24 mm); (c) Landy (inhibition zone 1.08 mm); (d) PBYDB (inhibition zone 1.86 mm); (e) NA (no inhibition zone); (f) LB (no inhibition zone); (g) ddH2O. Variation curves of cultural time on biomass (OD600) and inhibitory activity (inhibition rate) of strain AF01 against N. dimidiatum (B). The OD600 values and inhibition rates were separately measured at each incubation time.
3.5. Determination of antifungal activity of AF01
Under an optical microscope, two types of spores were observed in a ratio of about 1:1. There were significant differences in the antifungal abilities among the different application doses of the AF01 cultural filtrate in inhibiting N. dimidiatum. The AF01 cultural filtrate inhibited mycelial growth and spore germination by 9.47%–81.93% (Figure 5A) and 9.71%–62.22% (Figure 5B), respectively. Moreover, the inhibitions of germ tube elongation were 8.09%–63.97% (Figure 5C) at different treatments.
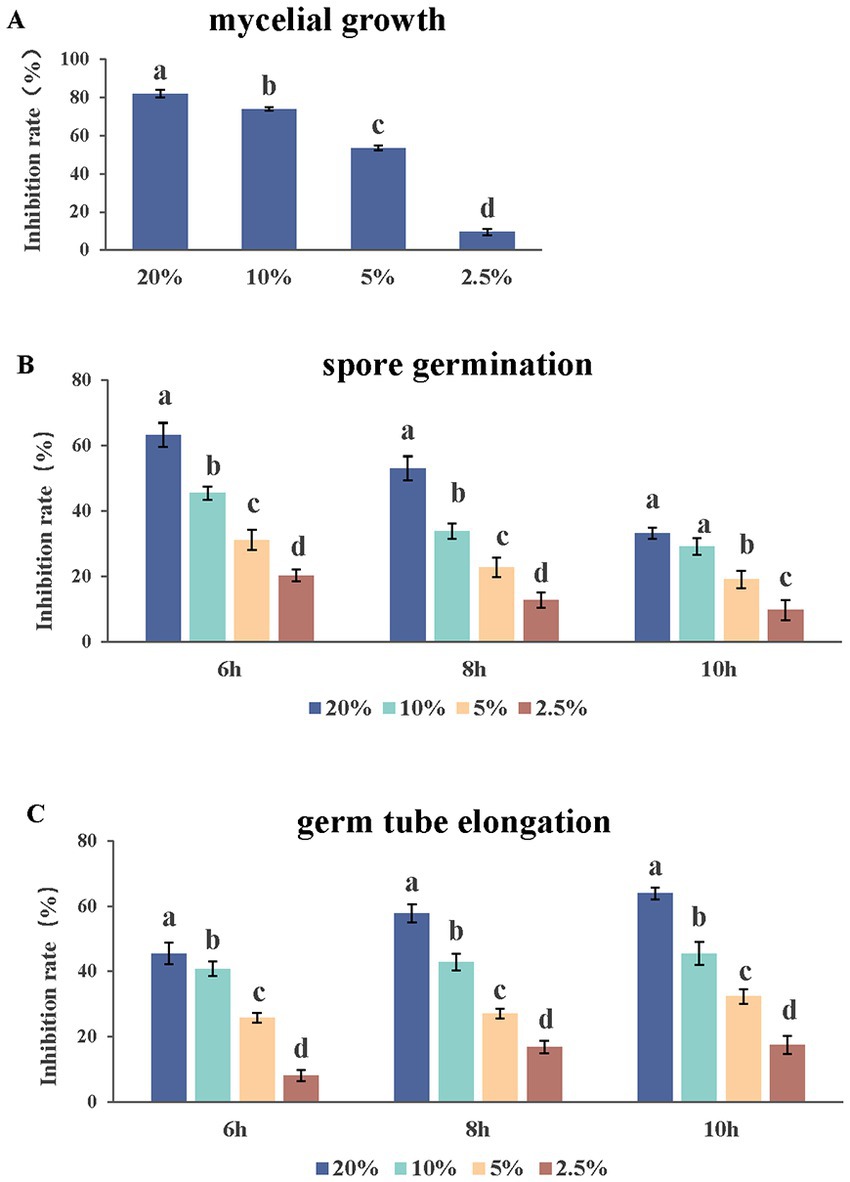
Figure 5. Effect of Paenibacillus polymyxa AF01 culture filtrate on three group of mycelial growth (A), spore germination (B) and germ tube elongation (C) of Neoscytalidium dimidiatum. AF01 filtrate was applied at 20%, 10%, 5%, and 2.5%. Error bars indicate standard deviation. Different letters above the bars indicate significant difference within each group according to Duncan’s multiple range test (p = 0.05) based on arcsine square-root transformed values of percentage.
Morphological changes in N. dimidiatum caused by AF01 culture filtrate were visualized using an OLYMPUS BX53 biological microscope at 600× magnification. The mycelia and germ tubes without AF01 cultural filtrate were normal and smooth but displayed marked structural changes when exposed to the AF01 cultural filtrate. Hyphae were severely deformed, with massive conglobation, uneven surfaces, expanded widths, whereas the germ tubes appeared shortened and swollen, and vacuolation also observed in conidia (Figure 6).
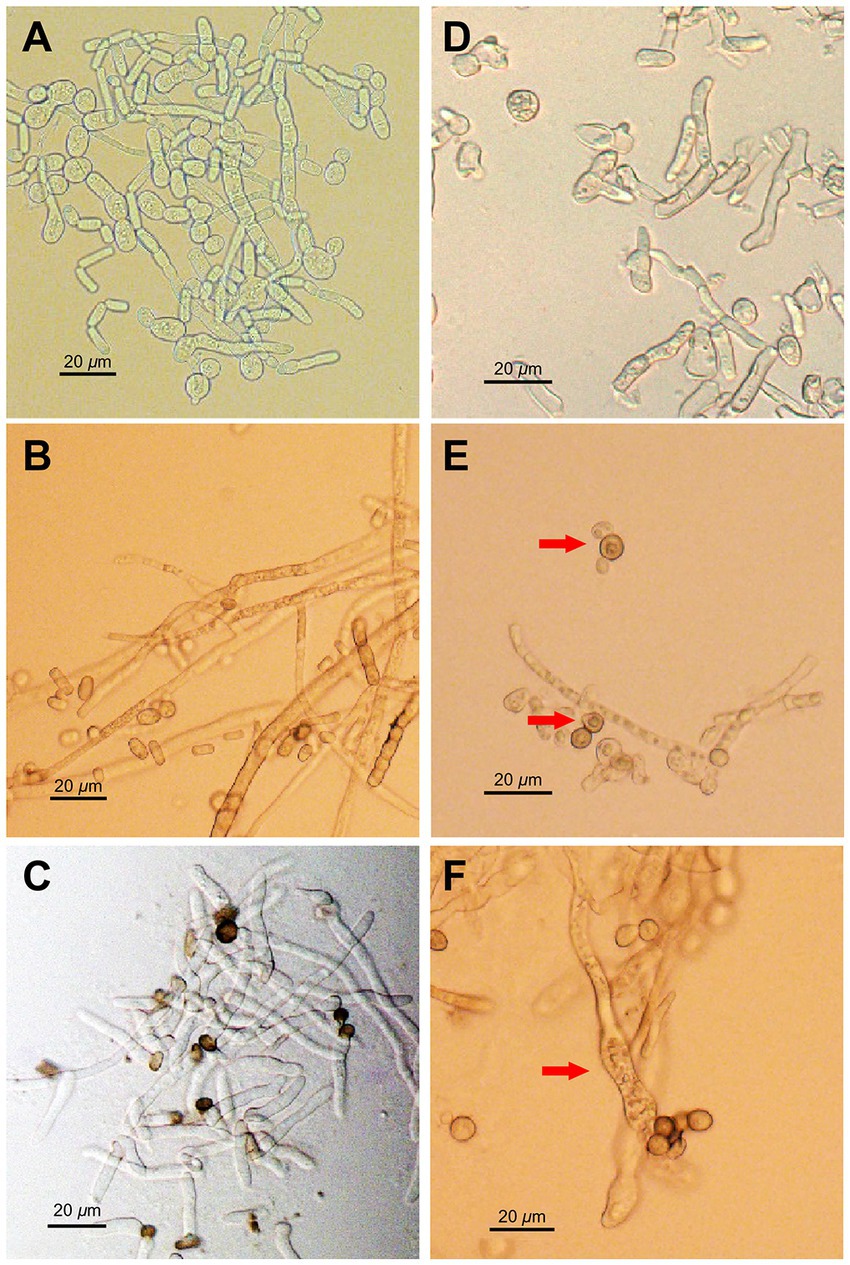
Figure 6. Inhibition effect of Paenibacillus polymyxa AF01 on phytopathogenic morphologies of Neoscytalidium dimidiatum observed via light microscopy. (A–C) The normal morphology in the control groups, and (D–F) the corresponding effect of P. polymyxa. Arrows indicate hyphal and conidia alterations with deformation (D), vacuolar (E), and swelling (F) of structures caused by P. polymyxa AF01.
3.6. Assay of cell contents leakage
Figure 7 shows the protein leakage assay used to determine cellular protein leakage. Once the bacterial cell membrane is irreversibly disrupted, cellular contents such as DNA, RNA, proteins and electrolyte leak out. For AF01-treated samples, the values of OD260 (Figure 7A), OD280 (Figure 7B) and electrical conductivity (Figure 7C) of the extracellular fluids increased rapidly, especially after 3 h, indicating cell membrane damage and the cell contents leakage.
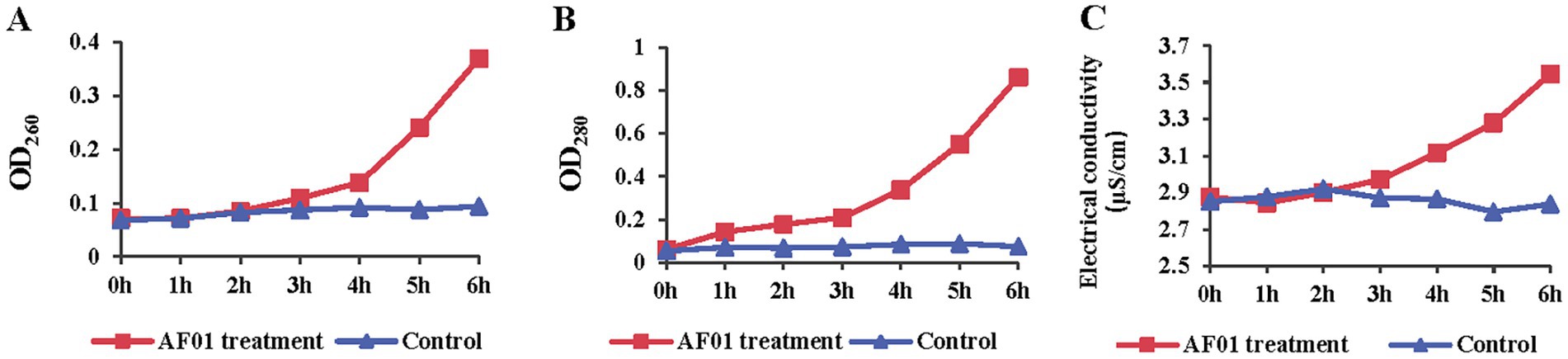
Figure 7. Membrane integrity change of Neoscytalidium dimidiatum in the presence of Paenibacillus polymyxa AF01 was evaluated by DNA leakage (A), protein (B) and electrolytes (C). N. dimidiatum was cultured with AF01 cultural filtrate. Supernatant were kept after centrifugation and quantified by absorption spectroscopy at 280 nm and 260 nm, respectively. Electrolyte leakage was detected with a conductometer.
3.7. Identification of antibiosis metabolites produced by strain AF01
With simple sample preparation and high sensitivity, MALDI-TOF-MS has become a useful method for rapid in situ detection of complex families of natural compounds, such as fusaricidins and polymyxins. In present study, this technique was employed to identify the main ingredients of the AF0F culture. As shown in Figure 8, MALDI-TOF-MS spectrum of AF01 culture obviously exhibited the mass peaks of fusaricidins (m/z 860–1,000) and polymyxins (m/z 1,150–1,250). By comparing with the mass data of known fusaricidins the obtained mass peaks was identified as fusaricidins A-D (Table 2; Vater et al., 2015). According to available data, fusaricidins were found at m/z = 869.6 which could be derived from fusaricidin A by substitution of Thr in positions 4 with Ser. Two others yet unknown fusaricidins with molecular masses of m/z = 954.9 and 969.0 were related to fusaricidins A and B with a mass difference of 71 Da, indicating modification by attachment of an alanine residue (Mülner et al., 2021; Tsai et al., 2022). In addition to these identified mass peaks, there remain many peaks in the spectrum to be further identified, e.g., m/z 915.7, 921.0, 935.8, 986.8, 1006.9, 1033.1, and 1095.1.
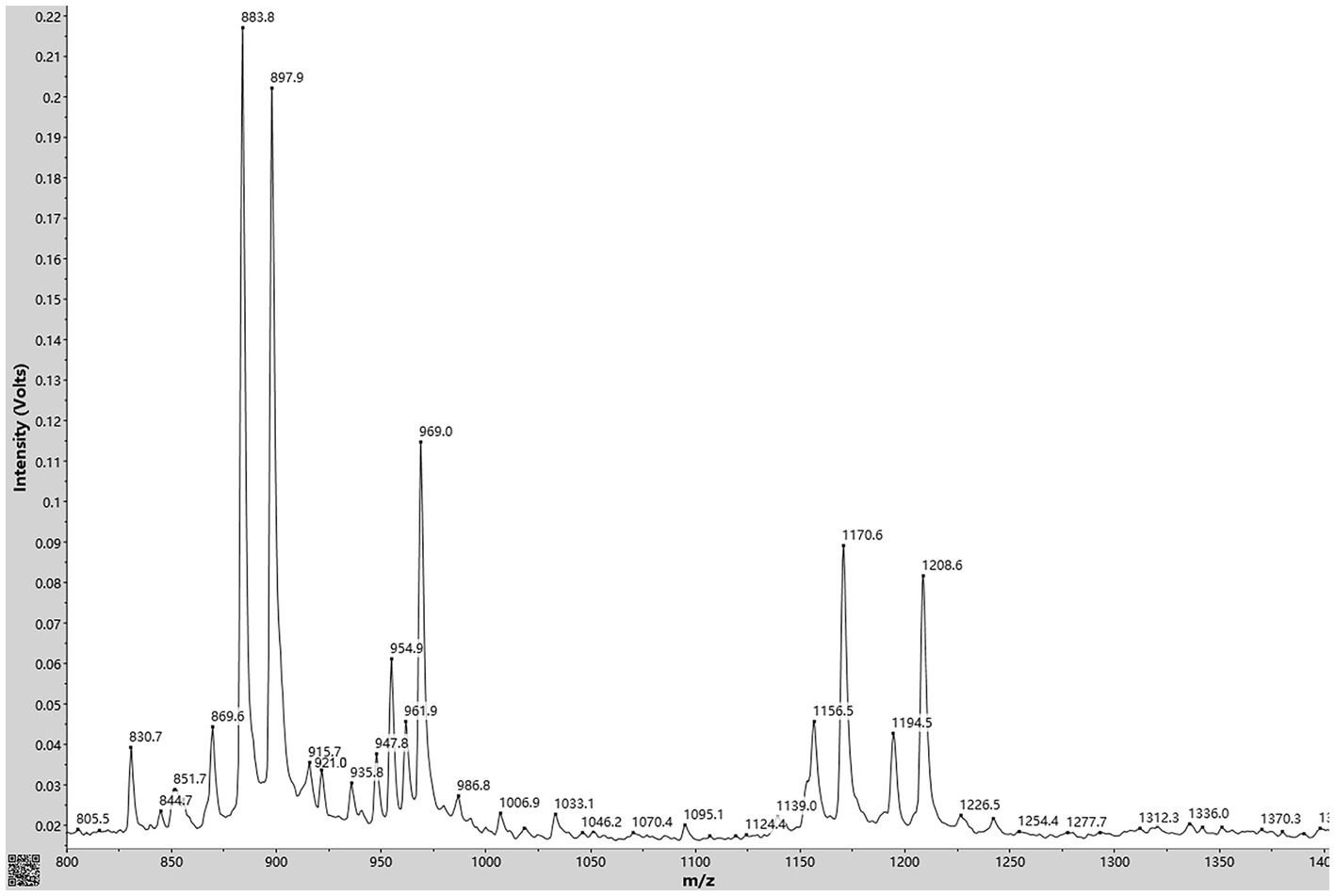
Figure 8. MALDI-TOF MS analysis of antimicrobial compounds produced by Paenibacillus polymyxa AF01 cultivated in PDB liquid medium. Mass peaks of fusaricidins were detected in the culture supernatant extracted with acetonitrile.
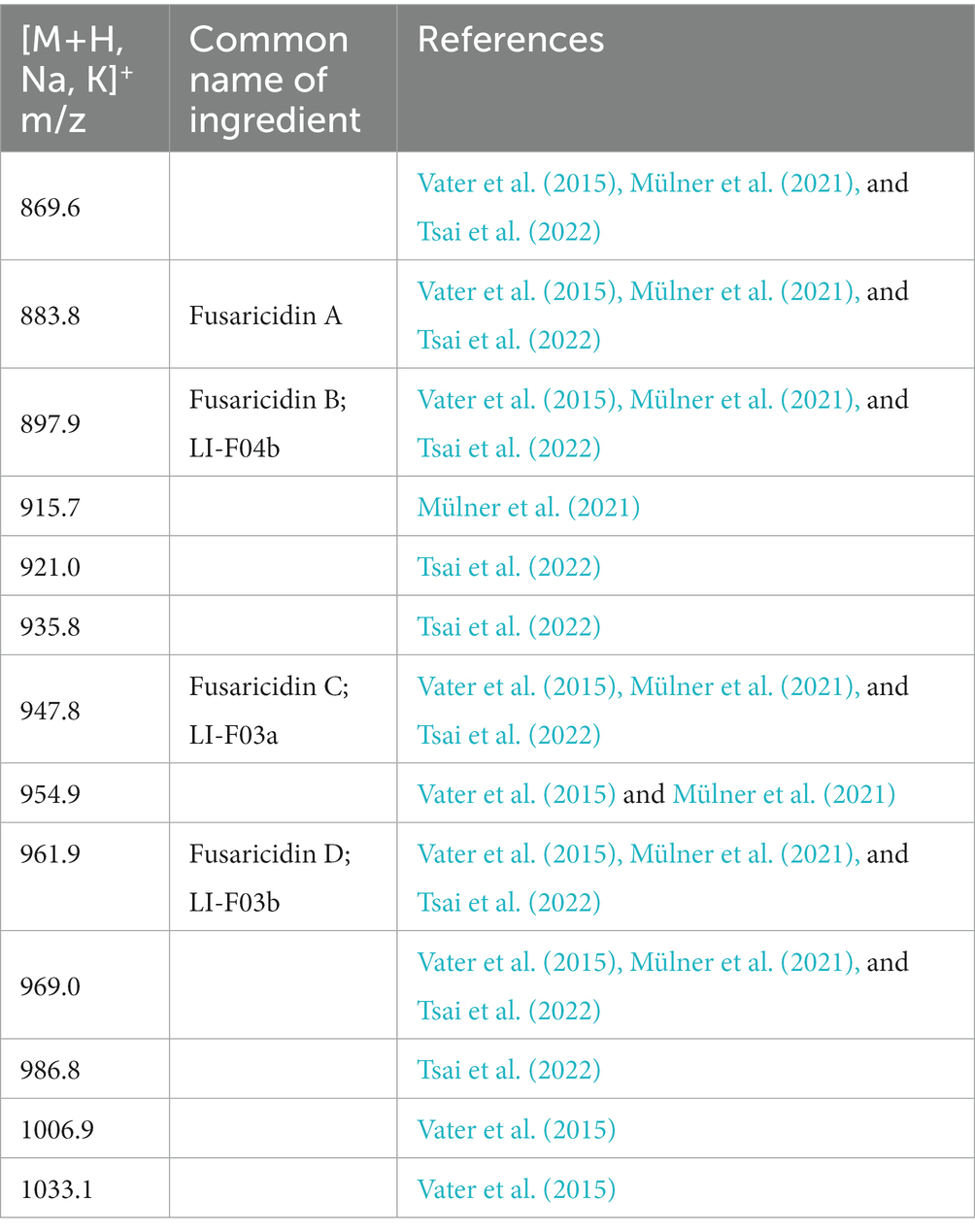
Table 2. Fusaricidins detected in culture filtrates of Paenibacillus polymyxa AF01 by MALDI-TOF mass spectrometry.
3.8. GO functions and KEGG pathway enrichment analysis of DEGs
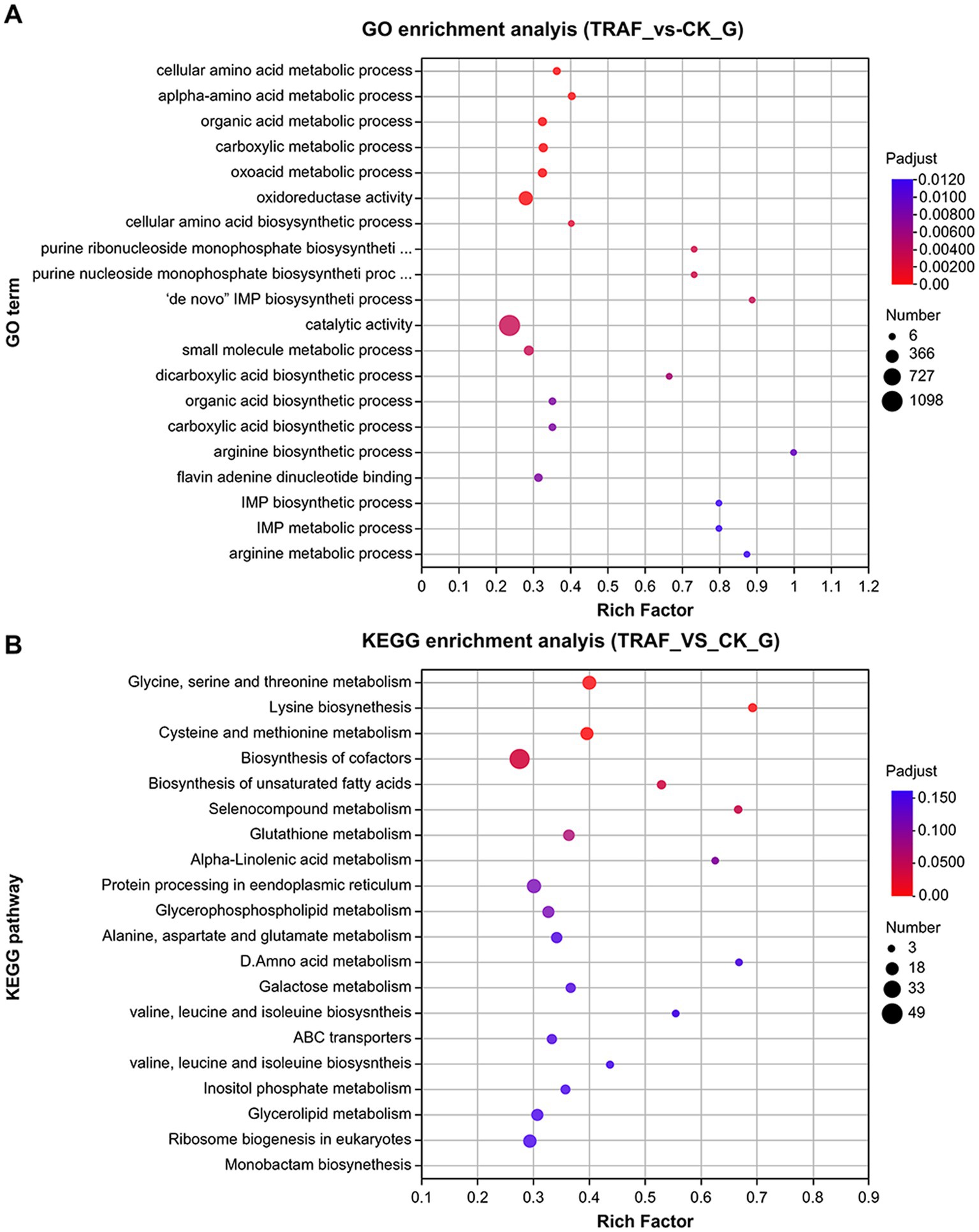
RNA-seq was used to analyze the transcriptomic response of N. dimidiatum BH5 after exposure to P. polymyxa AF01 for 3 h. Raw data were normalized and transformed into log2 values, and DESeq2 was used to analyze the differential gene expression of the samples. Subsequently, under co-cultivated conditions, strain BH5 exhibited significant differential gene expression with 2,248 genes, comprising 1,358 upregulated genes and 890 downregulated genes (|log2 FoldChange| ≥ 1). To understand the biological functions of the differentially expressed genes during co-culture, the annotated pathways of these genes were analyzed using the GO and KEGG databases. As a result, a total of 168 and 115 terms in the treatment group were enriched in the GO and KEGG, among which the top 20 terms of the GO and KEGG pathway are listed in Figures 9A,B (p < 0.05).
Compared to the control group, the genes involved in the biosynthesis of UDP-glucose, D-galacturonate, chitin, major structural components of fungal cell walls, and those related to pentose and glucuronate interconversions (ko00040, ko00520) were suppressed in response to P. polymyxa AF01 treatment for 3 h, particularly PLY (gene05693 and gene01489), which was downregulated by 3.92 and 3.56-fold, respectively (Table 3). Furthermore, the expression of genes associated with lipid metabolism (ko01040, ko00564, and ko00590) was prominently reduced following exposure to AF01, and some genes showed lower expression levels, such as GDE1 (gene08147), EPHX2 (gene03244), glpQ and ugpQ (gene09638), by more than 6-fold. These genes have been described as encoding hypothetical proteins, glycerophosphoryl diester phosphodiesterases, and alpha/beta hydrolase folds (Table 3). During protein processing in the endoplasmic reticulum, 25 genes exhibited differential expression. Of these, 23 were downregulated and two were upregulated. The downregulated factors play important roles in protein recognition, glucosylation, protein transport, glucosyl transfer, and receptor binding, as shown in Table 3.
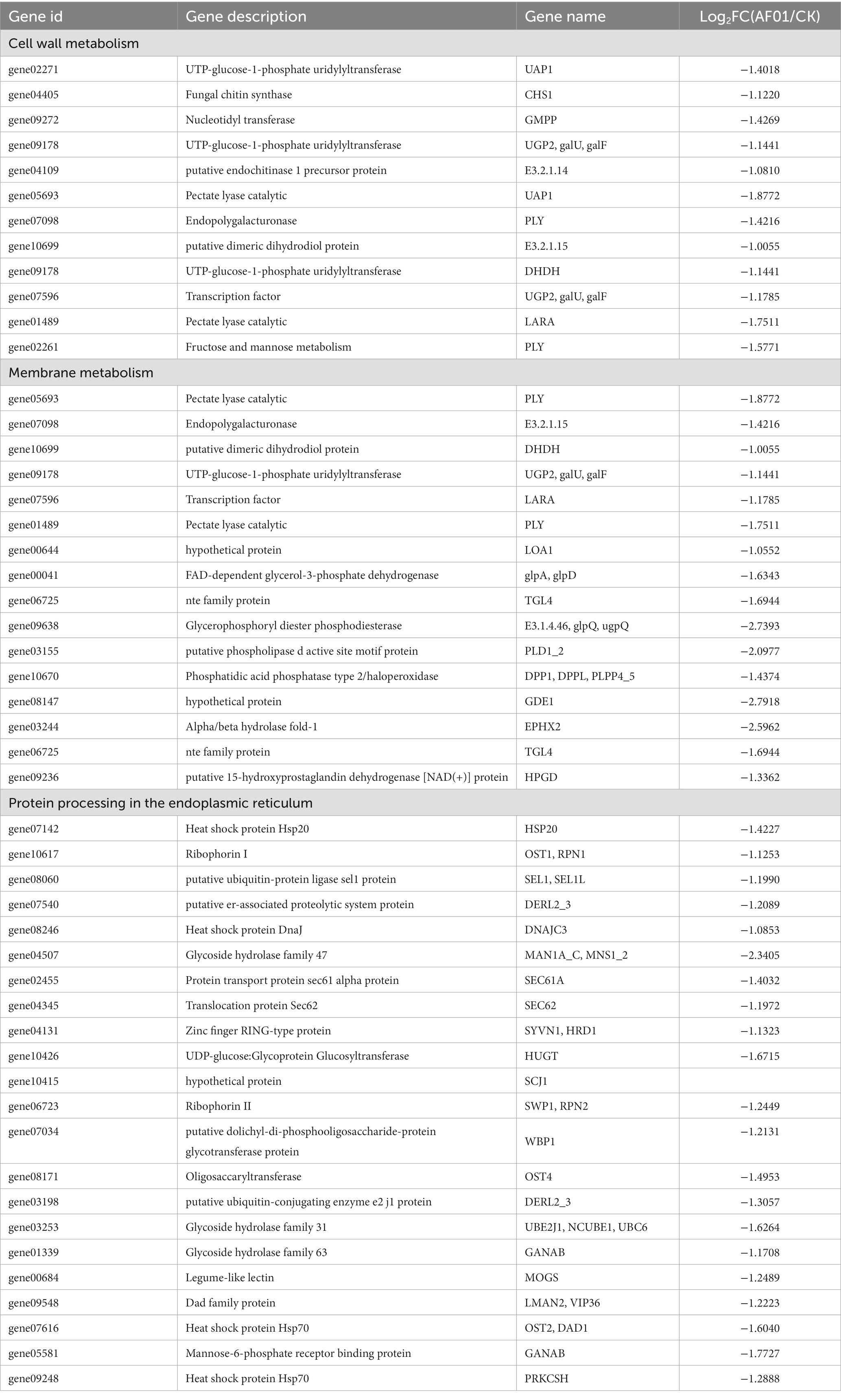
Table 3. The expression of genes in cell wall metabolism, membrane metabolism, and protein processing in the endoplasmic reticulum in Neoscytalidium dimidiatum BH5.
In addition, other genes downregulated in the treatment group were enriched in KEGG terms, including biosynthesis of cofactors, meiosis, chromatin structure and dynamics, peroxisomes, and the MAPK signaling pathway.
3.9. Efficacy of AF01 in controlling pitaya canker in pots
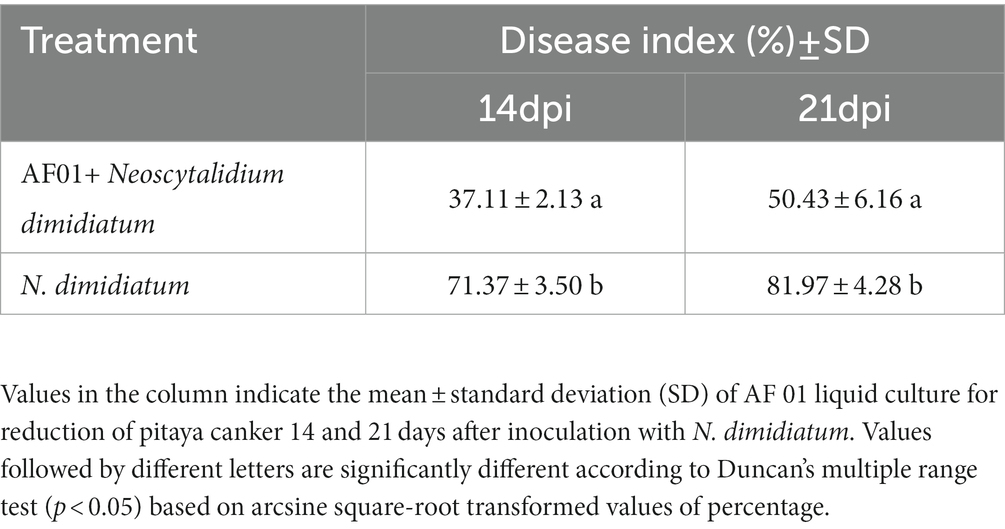
Disease suppression in pitaya was investigated to test the capacity of AF01 as a biocontrol agent. Typical symptoms of chlorotic spots appeared 10 days after inoculation with N. dimidiatum. The DI at 14 dpi reached 71.37 with control. By comparison, the P. polymyxa AF01 and pathogen treatment was 37.11, significantly lower than the foliage with only N. dimidiatum (Figures 10A,B). With age, severe canker symptoms, including necrosis, soft rot, and defoliation gradually appeared in the plants. At 21 dpi of inoculation, the DI of plants inoculated with N. dimidiatum was 81.97; however, in plants inoculated with only P. polymyxa AF01 and N. dimidiatum, was 50.43 (Figures 10C,D), suggesting a positive effect in controlling pitaya canker in this experiment (Table 4).

Figure 10. Symptom development in different groups of Neoscytalidium dimidiatum inoculation. Treatment: plants challenged with Paenibacillus polymyxa AF01 for 24 h and then with N. dimidiatum; control: plants challenged with N. dimidiatum. The pictures are representative of two independent experiments: treatment on the 14th day after N. dimidiatum inoculation (A); treatment on the 21th day after N. dimidiatum inoculation (C); control on the 14th and 21th day after N. dimidiatum inoculation (B,D).
3.10. Field efficacy assessment of the AF01 against pitaya canker
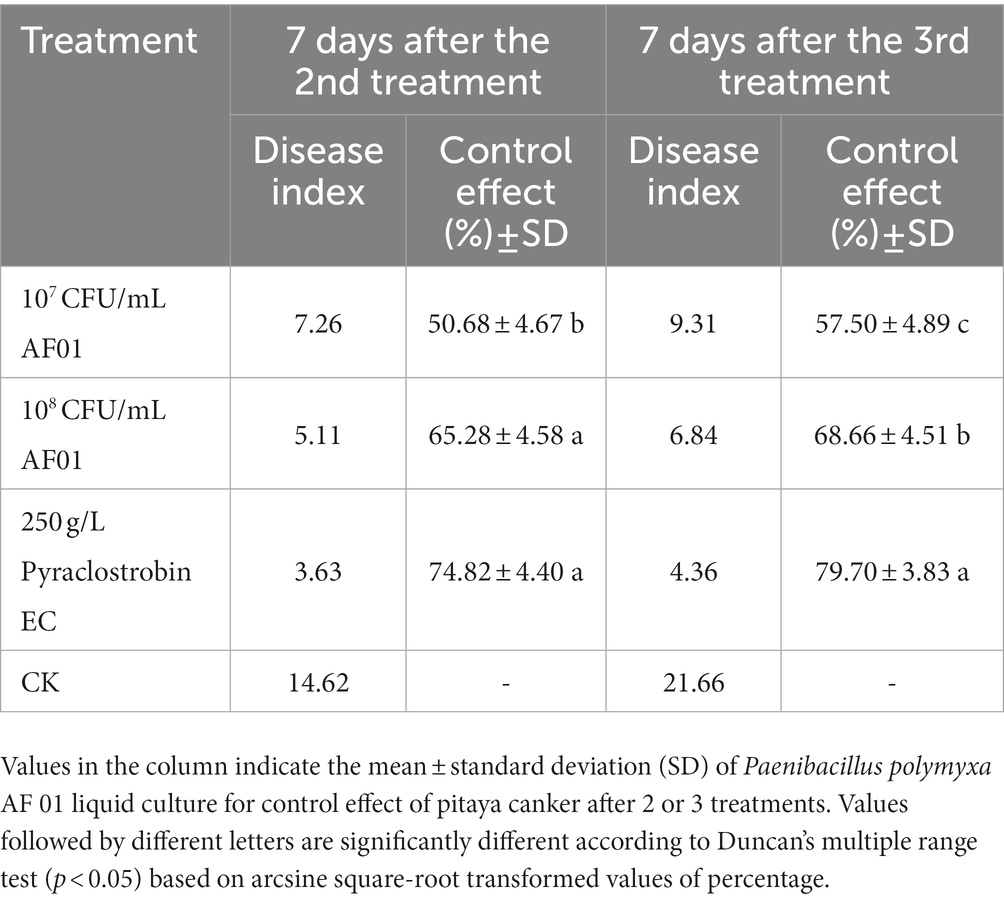
Paenibacillus polymyxa AF01 fermentation broth reduced pitaya canker caused by N. dimidiatum in field experiments. The fungicide treatment had the greatest effect of 74.82%. The efficacies of different concentrations (5 × 107 CFU/mL and 1 × 108 CFU/mL) of AF01 suspension were evaluated and shown to be 50.68 and 65.28%, respectively. The control efficacy increased as the application frequency increased; after three treatments, 107 CFU/mL, 108 CFU/mL AF01, and pyraclostrobine suppressed pitaya canker with efficacies of 57.50%, 68.66%, and 79.70%, respectively (Table 5). The results showed that AF01 fermentation broth provided preservative effects by reducing chlorotic spots, suberization, and pitaya decay.
4. Discussion
Pitaya is highly susceptible to microbial diseases, particularly those caused by N. dimidiatum (Pan et al., 2021). Therefore, developing an effective prevention and control strategy to control this disease during production is crucial and necessary. As humans become increasingly concerned about food safety and environmental pollution, biocontrol has become increasingly important in sustainable agriculture and industrial biotechnology over time (Gao et al., 2019; Xu et al., 2020). Paenibacillaceae are found in the rhizosphere of different crops and found in marine sediments, forest plants, and insect larvae. In the context of the current information, Paenibacillus spp. can benefit agriculture through antimicrobial activity, nitrogen fixation, and phosphate solubilization and strongly affected plant-parasitic nematodes, oomycetes, pathogenic bacteria, and fungal phytopathogens, including Fusarium oxysporum, B. cinerea, and Verticillium dahlia (Grady et al., 2016). Multiple anti-fungal activity mechanisms are influenced by antibiosis, competition for nutrients and biological niches, heavy parasitism, and the induction of plant resistance (Zhai et al., 2021). In our study, among the 113 bacterial strains isolated from rhizosphere samples, strain AF01 from Qinzhou, Guangxi Province, China, showed the strongest antagonistic activity against N. dimidiatum, F. equiseti, B. cinerea, B. cactivora, and G. persicaria. Combined with the 16 s rRNA result, AF01 has a 99% identity with that of P. polymyxa (CP009909 and CP025957). According to current studies, Paenibacillus is one of the eight genera in the family Paenibacillaceae and currently comprises ~200 species (Grady et al., 2016). Comparative 16S rRNA sequence analysis could not demonstrate that the genus Paenibacillus consisted of P. polymyxa and close relatives. The identification of microbial taxonomy requires a combination of physiological, biochemical and molecular techniques (von der Weid et al., 2000). In this study, the strain AF01 was identified as P. polymyxa based on the results obtained from GEN III MicroPlate tests, where most biochemical characteristics of strain AF01 were consistent with those of P. polymyxa. Based on biochemical characters and the phylogenetic tree, strain AF01 was classified as P. polymyxa.
In vitro effects of P. polymyxa AF01 on the mycelial growth, spore germination and germ tube elongation provided the first evidence related to their potential toxicity and efficacy against the pathogenic N. dimidiatum. Compared with spore germination and germ tube elongation, hyphae growth phase was more sensitive to the effect of AF01 culture filtrate, while germ tube growth was less affected, which is in agreement with other findings on the antifungal activity of P. polymyxa and other Paenibacillus spp. against fungal species such as Cylindrocarpon destructant, Rhizoctonia solani, Fusarium sp., Sclerotinia sclerotiorum, Pythium ultimum (Bae et al., 2004), Colletotrichum gloeosporioides (Ran et al., 2021), Pseudoperonospora cubensis, B. cinerea, Phytophthora capsica (Kim et al., 2009) and Magnaporthe oryzae (Ali et al., 2021). Microscopic observations revealed that P. polymyxa AF01 hampered the fungus spore causing mycelial tip enlargement, protoplast cleavage and abnormal morphological alternations of the spores. A marked enhancement of fungal cell permeability was reflected by the abnormal and massive spillage of nucleic acid, proteins and electrolyte from macromolecular cytoplasmic components owing to cell damage by P. polymyxa AF01. Dispersal of conidia are the origin of many secondary infections on pitaya. The antifungal effects of AF01 on fungal spore germination may play a role in disease control, as the pathogen life cycle may be disrupted.
Fusaricidins produced by P. polymyxa strains play an important strategy for controlling plant pathogenic. These compounds comprised a guanidinylated β-hydroxy fatty acid and a cyclic hexapeptide. Paenibacillus stains produced various types of fusaricidins (Gil et al., 2021), where enormous diversity exists even within those found in the same bacterial. MALDI-TOF MS is a practical technique for detecting secondary metabolites without additional purification, allowing the identification of active compounds in crude extracts via mass spectrometry. More than 20 variants of these compounds have been detected and characterized in detail in P. polymyxa-M1, including fusaricidins A–D, LI-F03, LI-F04, LI-F05, LI-F06, LI-F07, and LI-F08 and other novel fusaricidin derivatives (Vater et al., 2015). Using a similar method, we demonstrated that AF01 produced 13 forms of fusaricidins, showing a much higher complexity than expected. Among which, fusaricidin A and fusaricidin D were the main fusaricidin found. Inhibitions of mycelial growth, spore germination, germ tube elongation caused by AF01 culture filtrate were observed in the study. Furthermore, this inhibiting action maybe due to fusaricidins, which cause cytoplasm leakage such as intracellular nucleic acid, proteins and electrolyte. The finding was consistent with reports that the main mode of fusaricidin against bacteria is due to the disruption of membrane integrity.
In this study, N. dimidiatum BH5 mycelia and spores were damaged and had a rough, swollen, and large area of cavitation that even appeared in the center of the cytoplasm after treatment with P. polymyxa AF01, which was probably due to damage to cell function. Based on previous studies that used ultrastructural observations and proteomic analyses, the synthesis of secondary metabolites involves multiple pathways as well as the inhibition of lipid, carbohydrate, and amino acid metabolism, thiamine biosynthesis, energy metabolism, and reduced reactive oxygen species (ROS) scavenging capacity (Han et al., 2017; Mokhtar et al., 2020). Our results suggest that the efficacy of P. polymyxa against N. dimidiatum BH5 is mediated by its ability to selectively block critical cellular processes including the transcription and translation mechanisms as well as RNA and DNA structural dynamics, energy production and conversion, and signal transduction pathways. Additionally, it appears to affect cell walls, membrane permeability, and protein biosynthesis.
Fungal cells possess dynamic cell walls that protect them against environmental stress and changes in osmotic pressure, making them critical to the species biology and ecology (Gow et al., 2017). Cell membranes serve a similar function, controlling the movement of substances in and out of the cell, making them functionally important in biological systems (Kraft, 2013). However, changes in these structures can cause cell wall and plasma membrane dysfunction, resulting in the leakage of protoplasm from conidia and mycelia and the formation of vacuoles in spores and mycelia. Therefore, cell walls and plasma membranes are major targets of antifungal drugs and volatile compounds because of their crucial roles in fungal cell survival (Bakkali et al., 2008). Similar to the effect of iturin A on Phytophthora infestans and fusaricidin on Bacillus subtilis and Fusarium moniliforme (Han et al., 2017), RNA-seq results indicated that the expression levels of CHS1, galU, galF, PLY, glpA, glpD, glpQ, ugpQ, GDE1, MVK genes were significantly altered by P. polymyxa AF01 treatment. This might adversely affect the biosynthesis of Glu, Rha, Gal, glycerone-P, sterol precursor, 1,2-diacyl-glycerol-3p, arachidonic acid significantly and thus suppressed the growth of the N. dimidiatum.
The endoplasmic reticulum (ER) is an organelle responsible for metabolism, signal transduction, and the growth and development of organisms. P. polymyxa AF01 treatment inhibited the expression of MAN1A-C, MNS1-2, HUGT, GANAB, and PRKCSH, resulting in significant disruptions in the accurate folding, post-translational modification, and final assembly of the cellular proteome of N. dimidiatum BH5. It may also reduce the secretion of sufficient quantities of cell-wall-degrading enzymes, proteases, pectinases, and other enzymes required to penetrate plant surfaces and cause necrosis (Joubert et al., 2011).
ROS are highly reactive, can damage cellular components such as proteins, lipids, and nucleic acids, and harm various biomolecules within biological systems (Scott and Eaton, 2008; Yang et al., 2022). Accordingly, peroxiredoxin and catalase T are significantly induced to detoxify ROS, maintain the redox balance, and resist oxidative damage to the cells (Hossain et al., 2015). This study examined the effects of P. polymyxa AF01 on N. dimidiatum BH5 cells. Treatment with P. polymyxa AF01 to pitaya resulted in the upregulation of ACOX1, ACOX3, DAO, and AAO, whereas MPV17 was downregulated, suggesting that the treated cells accumulated ROS. These alterations result in changes in the permeability of the fungal membrane, impaired membrane transport and respiration, and the disintegration of the fungal cell wall.
Results of the pot experiment strongly supported that P. polymyxa AF01 significantly reduces the severity of pitaya canker. However, the actual agricultural field is far more complex than the pot experiment under laboratory conditions. Further field studies were carried out to verify the control effect as the disease control efficacy of strain AF01 reached 68.66%. Despite being less potent than chemically synthesized chemical fungicides, P. polymyxa AF01 has the benefit of being more environmentally friendly and conforming to the idea of sustainable development. To improve its biological control effect, optimal application rate, timing, and frequency of AF01 are also important issues that must be considered in field production.
Although P. polymyxa can improve plant defense abilities to reduce disease occurrence; our previous study showed that irrigating with AF01 did not significantly reduce pitaya canker. it is not yet clear whether AF01 can regulate defensive response to N. dimidiatum infection. Therefore, studies in this field should be conducted in further work.
5. Conclusion
Pitaya canker is a fungal disease threatening the pitaya fruit industry. P. polymyxa AF01, a bacterium exhibiting potent antifungal activity against the pathogen N. dimidiatum, was identified using a combinate analyses. The main antifungal products of P. polymyxa AF01 are fusaricidins, which possess a broad antibacterial spectrum. Based on our existing data and previous studies, a hypothesis was proposed regarding the mode of fusaricidin produced by P. polymyxa in suppressing pitaya canker: it was postulated that fusaricidins disturbed cell wall biosynthesis, changes in membrane permeability direct inhibitory effects on spore germination and hyphal growth of N. dimidiatum. Considering its high potential as a biocontrol agent, additional studies are warranted to explore the development and practical applications of P. polymyxa AF01.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA943517.
Author contributions
SL: conducting the experiments and writing the manuscript. XC: participating in experiments and revisions. LX: supervision. YZ: data and bioinformatics analyses. FZ: data curation. YL, LR, and XQ: participating in the design and experiments. JW: funding acquisition and writing-reviewed and edited. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Guangxi Natural Science Foundation Youth Fund (grant number: 2017GXNSFBA19823).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Adnan, L., Osman, A., and Abdul Hamid, A. (2011). Antioxidant activity of different extracts of red pitaya (Hylocereus polyrhizus) seed. Int. J. Food Prop. 14, 1171–1181. doi: 10.1080/10942911003592787
Ali, M. A., Lou, Y., Hafeez, R., Li, X., Hossain, A., Xie, T., et al. (2021). Functional analysis and genome mining reveal high potential of biocontrol and plant growth promotion in nodule-inhabiting bacteria within Paenibacillus polymyxa complex. Front. Microbiol. 11:618601. doi: 10.3389/fmicb.2020.618601
Bae, Y. S., Park, K., and Kim, C. H. (2004). Bacillus spp. as biocontrol agents of root rot and phytophthora blight on ginseng. Plant Pathol. J. 20, 63–66. doi: 10.5423/PPJ.2004.20.1.063
Bakkali, F., Averbeck, S., Averbeck, D., and Idaomar, M. (2008). Biological effects of essential oils–a review. Food Chem. Toxicol. 46, 446–475. doi: 10.1016/j.fct.2007.09.106
Bakker, P. A., Pieterse, C. M., and Van Loon, L. (2007). Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 97, 239–243. doi: 10.1094/PHYTO-97-2-0239
Choudhary, D. K., and Johri, B. N. (2009). Interactions of Bacillus spp. and plants–with special reference to induced systemic resistance (ISR). Microbiol. Res. 164, 493–513. doi: 10.1016/j.micres.2008.08.007
Chuang, M., Ni, H., Yang, H., Shu, S., Lai, S., and Jiang, Y. (2012). First report of stem canker disease of pitaya (Hylocereus undatus and H. polyrhizus) caused by Neoscytalidium dimidiatum in Taiwan. Plant Dis. 96:906. doi: 10.1094/PDIS-08-11-0689-PDN
Costa, A., Corallo, B., Amarelle, V., Stewart, S., Pan, D., Tiscornia, S., et al. (2022). Paenibacillus sp. strain UY79, isolated from a root nodule of Arachis villosa, displays a broad spectrum of antifungal activity. Appl. Environ. Microbiol. 88, e0164521–e0101621. doi: 10.1128/AEM.01645-21
Crous, P. W., Slippers, B., Wingfield, M. J., Rheeder, J., Marasas, W. F., Philips, A. J., et al. (2006). Phylogenetic lineages in the Botryosphaeriaceae. Stud. Mycol. 55, 235–253.
De Miccolis Angelini, R. M., Rotolo, C., Masiello, M., Gerin, D., Pollastro, S., and Faretra, F. (2014). Occurrence of fungicide resistance in populations of Botryotinia fuckeliana (Botrytis cinerea) on table grape and strawberry in southern Italy. Pest Manag. 70, 1785–1796. doi: 10.1002/ps.3711
Du, N., Shi, L., Yuan, Y., Sun, J., Shu, S., and Guo, S. (2017). Isolation of a potential biocontrol agent Paenibacillus polymyxa NSY50 from vinegar waste compost and its induction of host defense responses against Fusarium wilt of cucumber. Microbiol. Res. 202, 1–10. doi: 10.1016/j.micres.2017.04.01
Ferreira, J., Matthee, F., and Thomas, A. (1991). Biological control of Eutypa lata on grapevine by an antagonistic strain of Bacillus subtilis. Phytopathology 81, 283–287. doi: 10.1094/phyto-81-283
Fullerton, R., Sutherland, P., Hieu, R. R. N. T., Thu, N. N. A., Linh, D. T., Thanh, N. T. K., et al. (2018). The life cycle of dragon fruit canker caused by Neoscytalidium dimidiatum and implications for control. In: Dragon Fruit Regional Network Initiation Workshop 9, 71–80.
Gao, D., Wang, D., Chen, K., Huang, M., Xie, X., and Li, X. (2019). Activation of biochemical factors in CMV-infected tobacco by ningnanmycin. Pest. Biochem. Physiol. 156, 116–122. doi: 10.1016/j.pestbp.2019.02.012
Gil, J., Pastar, I., Houghten, R. A., Padhee, S., Higa, A., Solis, M., et al. (2021). Novel cyclic Lipopeptides Fusaricidin analogs for treating wound infections. Front. Microbiol. 12:708904. doi: 10.3389/fmicb.2021.708904
Gow, N. A., Latge, J. P., and Munro, C. A. (2017). The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spec. 5:5.3. 01. doi: 10.1128/microbiolspec.FUNK-0035-2016
Grady, E. N., Macdonald, J., Liu, L., Richman, A., and Yuan, Z. C. (2016). Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Fact. 15, 203–218. doi: 10.1186/s12934-016-0603-7
Han, J., Wang, F., Gao, P., Ma, Z., Zhao, S., Lu, Z., et al. (2017). Mechanism of action of AMP-jsa9, a LI-F-type antimicrobial peptide produced by Paenibacillus polymyxa JSa-9, against Fusarium moniliforme. Fungal Genet. Biol. 104, 45–55. doi: 10.1016/j.fgb.2017.05.002
Harsonowati, W., Marian, M., and Narisawa, K. (2020). The effectiveness of a dark septate endophytic fungus, Cladophialophora chaetospira SK51, to mitigate strawberry Fusarium wilt disease and with growth promotion activities. Front. Microbiol. 11:585. doi: 10.3389/fmicb.2020.00585
Hong, C. F., Gazis, R., Crane, J. H., and Zhang, S. (2020). Prevalence and epidemics of Neoscytalidium stem and fruit canker on pitahaya (Hylocereus spp.) in South Florida. Plant Dis. 104, 1433–1438. doi: 10.1094/PDIS-10-19-2158-RE
Hossain, M. A., Bhattacharjee, S., Armin, S. M., Qian, P., Xin, W., Li, H. Y., et al. (2015). Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 6:420. doi: 10.3389/fpls.2015.00420
Joubert, A., Simoneau, P., Campion, C., Bataillé Simoneau, N., Iacomi Vasilescu, B., Poupard, P., et al. (2011). Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol. Microbiol. 79, 1305–1324. doi: 10.1111/j.1365-2958.2010.07522.x
Kim, S. G., Khan, Z., Jeon, Y. H., and Kim, Y. H. (2009). Inhibitory effect of Paenibacillus polymyxa GBR-462 on Phytophthora capsici causing phytophthora blight in chili pepper. J. Phytopathol. 157, 329–337. doi: 10.1111/j.1439-0434.2008.01490.x
Kraft, M. L. (2013). Plasma membrane organization and function: moving past lipid rafts. Mol. Biol. Cell 24, 2765–2768. doi: 10.1091/mbc.e13-03-0165
Lin, S. Y., Xian, X. Y., Wei, X. M., Wei, J. G., and Zhu, G. N. (2018). Isolation and identification of pathogens for the postharvest diseases in Pitaya fruits. S. china Fruit. 47, 6–12. doi: 10.13938/j.issn.1007-1431.20170469
Mokhtar, N. F. K., Hashim, A. M., Hanish, I., Zulkarnain, A., Raja Nhari, R. M. H., Abdul Sani, A. A., et al. (2020). The discovery of new antilisterial proteins from Paenibacillus polymyxa Kp10 via genome mining and mass spectrometry. Front. Microbiol. 11:960. doi: 10.3389/fmicb.2020.00960
Mülner, P., Schwarz, E., Dietel, K., Herfort, S., Jähne, J., Lasch, P., et al. (2021). Fusaricidins, polymyxins and volatiles produced by Paenibacillus polymyxa strains DSM 32871 and M1. Pathogens 10:1485. doi: 10.3390/pathogens10111485
Mülner, P., Schwarz, E., Dietel, K., Junge, H., Herfort, S., Weydmann, M., et al. (2020). Profiling for bioactive peptides and volatiles of plant growth promoting strains of the Bacillus subtilis complex of industrial relevance. Front. Microbiol. 11:1432. doi: 10.3389/fmicb.2020.01432
Pan, L., Zhang, R., Lu, F., Liu, C., and Fu, J. (2021). Comparative proteomic analysis of the fungal pathogen Neoscytalidium dimidiatum infection in the pitaya. Environ. Biotechnol. 62, 649–659. doi: 10.1007/s13580-021-00341-2
Phillips, A., Alves, A., Abdollahzadeh, J., Slippers, B., Wingfield, M. J., Groenewald, J., et al. (2013). The Botryosphaeriaceae: genera and species known from culture. Stud. Mycol. 76, 51–167. doi: 10.3114/sim0021
Ramli, N. S., Ismail, P., and Rahmat, A. (2016). Red pitaya juice supplementation ameliorates energy balance homeostasis by modulating obesity-related genes in high-carbohydrate, high-fat diet-induced metabolic syndrome rats. BMC Complement. Altern. Med. 16, 243–213. doi: 10.1186/s12906-016-1200-3
Ran, J., Jiao, L., Zhao, R., Zhu, M., Shi, J., Xu, B., et al. (2021). Characterization of a novel antifungal protein produced by Paenibacillus polymyxa isolated from the wheat rhizosphere. J. Sci. Food Agric. 101, 1901–1909. doi: 10.1002/jsfa.10805
Ratanaprom, S., Nakkanong, K., Nualsri, C., Jiwanit, P., Rongsawat, T., and Woraathakorn, N. (2021). Overcoming encouragement of dragon fruit plant (Hylocereus undatus) against stem brown spot disease caused by Neoscytalidium dimidiatum using Bacillus subtilis combined with codium bicarbonate. Plant Pathol. J. 37, 205–214. doi: 10.5423/PPJ.OA.01.2021.0007
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Kloepper, J. W., and Paré, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583
Scott, B., and Eaton, C. J. (2008). Role of reactive oxygen species in fungal cellular differentiations. Curr. Opin. Microbiol. 11, 488–493. doi: 10.1016/j.mib.2008.10.008
Soni, R., Rawal, K., and Keharia, H. (2021). Genomics assisted functional characterization of Paenibacillus polymyxa HK4 as a biocontrol and plant growth promoting bacterium. Microbiol. Res. 248:126734. doi: 10.1016/j.micres.2021.126734
Tsai, S. H., Chen, Y. T., Yang, Y. L., Lee, B. Y., Huang, C. J., and Chen, C. Y. (2022). The potential biocontrol agent Paenibacillus polymyxa TP3 produces Fusaricidin-type compounds involved in the antagonism against gray Mold pathogen Botrytis cinerea. Phytopathology 112, 775–783. doi: 10.1094/PHYTO-04-21-0178-R
Vater, J., Niu, B., Dietel, K., and Borriss, R. (2015). Characterization of novel fusaricidins produced by Paenibacillus polymyxa-M1 using MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 26, 1548–1558. doi: 10.1007/s13361-015-1130-1
Von Der Weid, I., Paiva, E., Nóbrega, A., Van Elsas, J. D., and Seldin, L. (2000). Diversity of Paenibacillus polymyxa strains isolated from the rhizosphere of maize planted in Cerrado soil. Res. Microbiol. 151, 369–381. doi: 10.1016/S0923-2508(00)00160-1
Wang, F., Zhang, R., Yuan, Z., and Chen, P. (2021). Biological prevention and control of pitaya fruit canker disease using endophytic fungi isolated from papaya. Arch. Microbiol. 203, 4033–4040. doi: 10.1007/s00203-021-02378-4
Wu, Z., Ma, G., Zhu, H., Chen, M., Huang, M., Xie, X., et al. (2022). Plant viral coat proteins as biochemical targets for antiviral compounds. J. Agric. Food Chem. 70, 8892–8900. doi: 10.1021/acs.jafc.2c02888
Xu, P., Xie, S., Liu, W., Jin, P., Wei, D., Yaseen, D. G., et al. (2020). Comparative genomics analysis provides new strategies for bacteriostatic ability of Bacillus velezensis HAB-2. Front. Microbiol. 11:594079. doi: 10.3389/fmicb.2020.594079
Yang, Q., Yang, J., Wang, Y., Du, J., Zhang, J., Luisi, B. F., et al. (2022). Broad-spectrum chemicals block ROS detoxification to prevent plant fungal invasion. Curr. Biol. 32:e3886, 3886–3897.e6. doi: 10.1016/j.cub.2022.07.022
Zhai, Y., Zhu, J. X., Tan, T. M., Xu, J. P., Shen, A. R., Yang, X. B., et al. (2021). Isolation and characterization of antagonistic Paenibacillus polymyxa HX-140 and its biocontrol potential against Fusarium wilt of cucumber seedlings. BMC Microbiol. 21, 75–12. doi: 10.1186/s12866-021-02131-3
Zuo, C., Zhang, W., Chen, Z., Chen, B., and Huang, Y. (2017). RNA sequencing reveals that endoplasmic reticulum stress and disruption of membrane integrity underlie dimethyl trisulfide toxicity against Fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol. 8:1365. doi: 10.3389/fmicb.2017.01365
Keywords: Paenibacillus polymyxa, Neoscytalidium dimidiatum, pitaya canker, fusaricidins, RNA-Seq
Citation: Lin S, Chen X, Xie L, Zhang Y, Zeng F, Long Y, Ren L, Qi X and Wei J (2023) Biocontrol potential of lipopeptides produced by Paenibacillus polymyxa AF01 against Neoscytalidium dimidiatum in pitaya. Front. Microbiol. 14:1188722. doi: 10.3389/fmicb.2023.1188722
Edited by:
Wensheng Fang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Xin Xie, Guizhou University, ChinaOrlando Borras-Hidalgo, Qilu University of Technology, China
Copyright © 2023 Lin, Chen, Xie, Zhang, Zeng, Long, Ren, Qi and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiguang Wei, MTk4NjAwMzhAZ3h1LmVkdS5jbg==
 Shanyu Lin
Shanyu Lin Xiaohang Chen
Xiaohang Chen Ling Xie2
Ling Xie2 Yan Zhang
Yan Zhang Liyun Ren
Liyun Ren