- 1Stomatological Hospital, School of Stomatology, Southern Medical University, Guangzhou, China
- 2Department of Oral Mucosal Diseases, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou, China
Cancer remains a significant global challenge, with an estimated 47% increase in cancer patients from 2020 to 2040. Increasing research has identified microorganism as a risk factor for cancer development. The oral cavity, second only to the colon, harbors more than 700 bacterial species and serves as a crucial microbial habitat. Although numerous epidemiological studies have reported associations between oral microorganisms and major systemic tumors, the relationship between oral microorganisms and cancers remains largely unclear. Current research primarily focuses on respiratory and digestive system tumors due to their anatomical proximity to the oral cavity. The relevant mechanism research mainly involves 47% dominant oral microbial population that can be cultured in vitro. However, further exploration is necessary to elucidate the mechanisms underlying the association between oral microbiota and tumors. This review systematically summarizes the reported correlations between oral microbiota and common cancers while also outlining potential mechanisms that may guide biological tumor treatment.
1. Introduction
Cancer is the leading cause of mortality worldwide, with an estimated global population of 24.8 million cancer patients projected for 2040 (Sung et al., 2021). Female breast cancer has emerged as the most prevalent malignancy globally, followed by lung, liver, stomach and colorectal cancer (CRC) (Cao et al., 2021). During tumor malignant transformation, certain characteristics that contribute to tumor progression may be acquired, such as indefinite replicative potential and incorporation of polymorphic microbiomes (Hanahan, 2022). The understanding of tumors is expanding alongside advancements in Shotgun metagenomic sequencing technique which have facilitated investigations into diverse microbial communities within tumors (Meslier et al., 2022). Over 3 × 103 species of microorganisms are found on the mucosal surface, with more than 90% residing in the colon (Xiao et al., 2020; Sepich-Poore et al., 2021). Currently, only 11 species have been identified as carcinogenic microorganisms: seven viruses, three parasites and a single bacterium known as Helicobacter pylori (H. pylori) (Cullin et al., 2021).
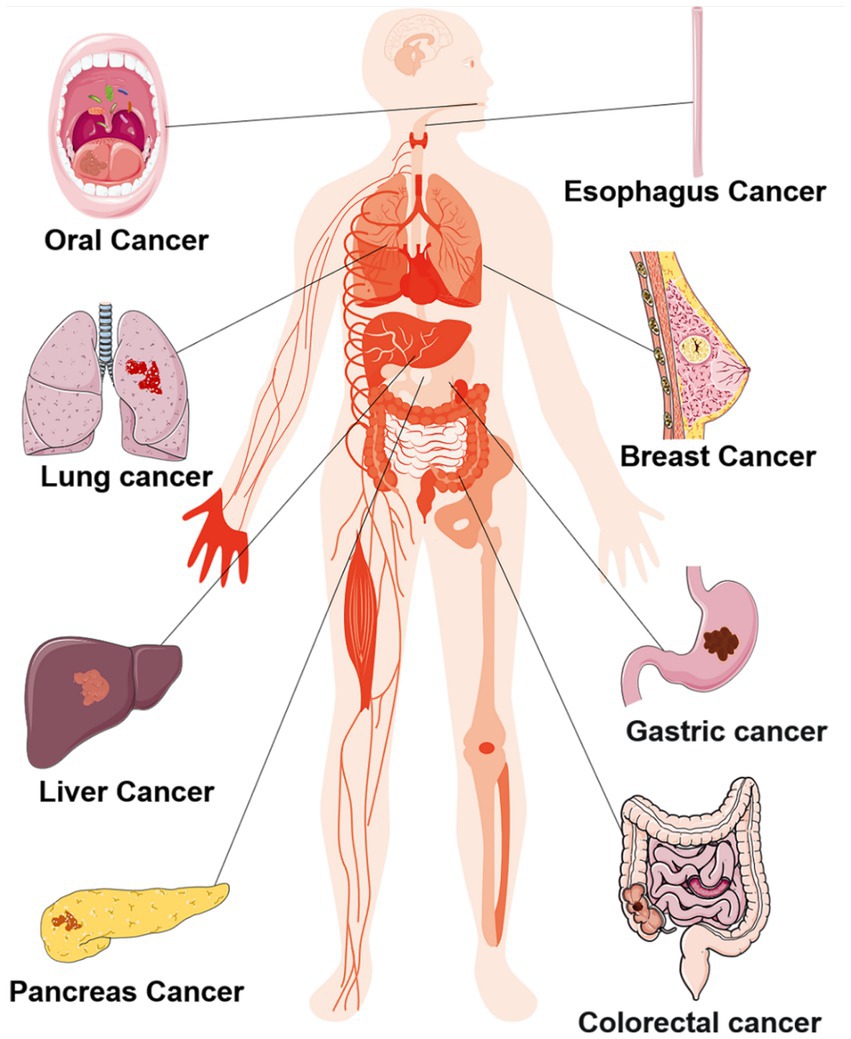
The oral cavity maintains an optimal temperature of 37°C and a pH range of 6.5–7.5, creating an exceptionally favorable environment for the survival of oral microorganisms (Koliarakis et al., 2019). According to the human oral microbiome database, more than 700 bacterial species inhabiting various ecological niches within the oral cavity (Gao et al., 2018; Koliarakis et al., 2019; Mark Welch et al., 2020). These oral microorganisms mutually benefit each other and collectively maintain the homeostasis of the oral ecosystem, however, this delicate balance is disrupted under pathological conditions (Li X. et al., 2022). Dysbiosis typically manifests as alterations in microbial composition characterized by a decrease in beneficial bacteria and an increase in potentially pathogenic bacteria (DeGruttola et al., 2016). Furthermore, dysbiosis not only give rise to local lesions such as caries and periodontal disease but also exerts systemic effects on distant organs, contributing to systemic diseases including cancers shown as Figure 1 (Sedghi et al., 2021). The main mechanism facilitating translocation of oral microorganisms include: (i) Interconnected anatomical structure between the oral cavity, respiratory tract and digestive tract allow for entry of oral microorganisms into these systems through saliva, air inhalation and food ingestion; (ii) Hematogenous and lymphogenous transmission: traumatic events like tooth extraction can result in invasion of blood circulation by oral microorganisms leading to distant metastasis (Mo et al., 2022).
According to the existing researches, the mechanisms by which oral microbes promote tumor occurrence and development are as follows: (i) Induce and aggravate chronic inflammation and promote the occurrence and development of tumors by the components of themselves (Gagliani et al., 2014; Niklander, 2021). (ii) Regulate cells proliferation and apoptosis by disrupting cell cycle and tumor signal transduction (Meurman, 2010; Deo and Deshmukh, 2020; Bakhti and Latifi-Navid, 2021). (iii) Indirectly metabolize substances, including sulfides, nitrosamines, hydroxyl radical, acetaldehyde, deoxycholic acid and toxins, which interfere with tumor occurrence, metastasis and recurrence (Doherty and Cleveland, 2013; Belcheva et al., 2014; Teles et al., 2020). (iv) Regulate host immune response (Figure 2). Common oral microorganisms, such as Fusobacterium nucleatum (F. nucleatum) and Porphyromonas gingivalis (P. gingivalis), can promote the infiltration of immunosuppressive cells and interfere with the function of immune killer cells, thereby protecting tumor cells from immune system surveillance and clearance (Steffen et al., 2000; Colucci, 2015; Wen et al., 2020). In summary, the objective of this review is to provide a comprehensive analysis of the interface between oral microbiota and systemic cancer, while elucidating the underlying mechanisms involved.
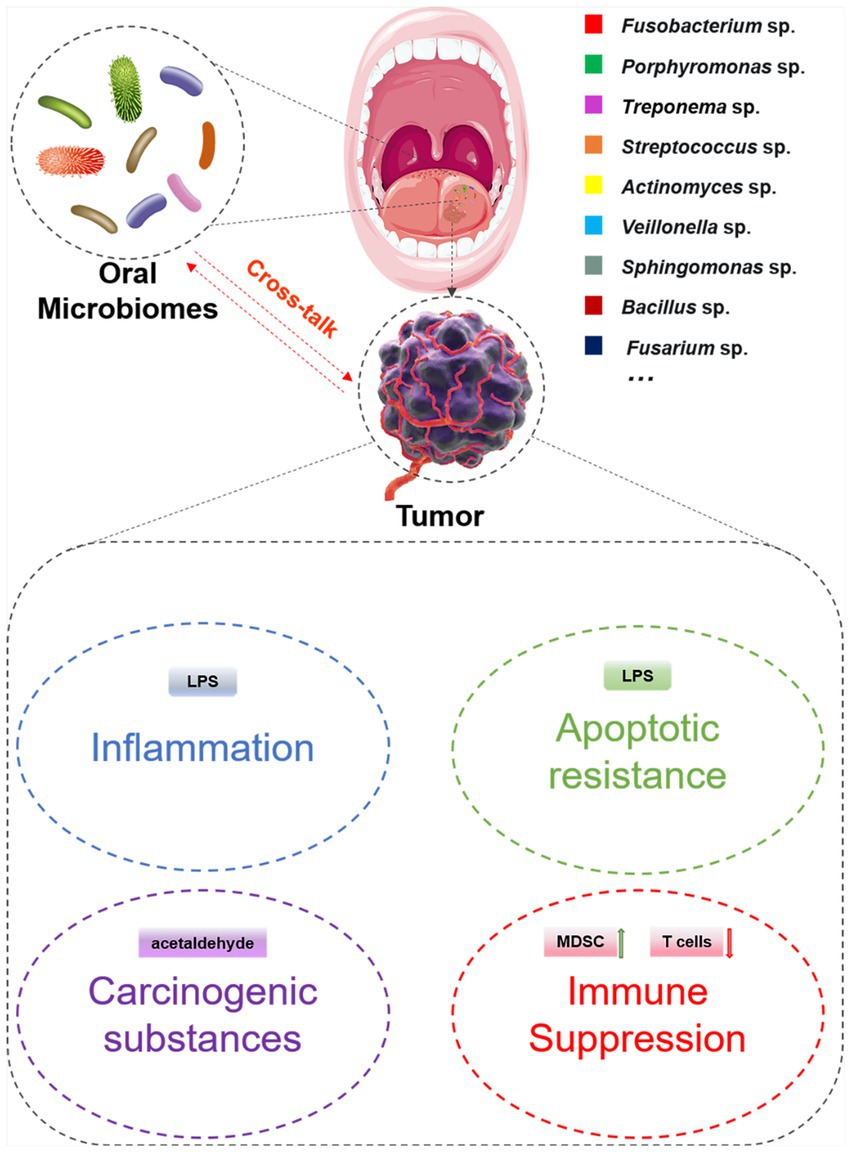
Figure 2. The potential mechanism associated with the cross-talk between oral microbiomes and tumors.
2. Oral microbiome and cancer
2.1. Lung carcinoma
Lung carcinoma has the highest mortality rate at around 18%, equivalent to 1,796,144 cases (Sung et al., 2021). Smoking is the primary etiological factor for lung cancer, while other contributing factors include air pollution and infection (Liu and Dong, 2021). Numerous studies have provided evidence supporting a close association between lung cancer and carcinogenic viruses such as HPV and HIV, which is significantly elevate the risk of developing lung cancer (Zhai et al., 2015; Cribbs et al., 2020). Although HPV and HIV can also be detected in the oral cavity, they do not constitute the dominant microbiota. Prevailing bacteria species in the oral cavity encompass P. gingivalis, F. nucleatum, Treponema dentinosum and Streptococcus (Zhang et al., 2019). However, the relationship between lung cancer and major oral microorganisms has mostly been constrained to observational studies. Shi et al. (2021) discovered a strong correlation between poor oral hygiene resulting from periodontal disease and an increased susceptibility of lung cancer. Utilizing sequencing technology in cohort studies has revealed higher alpha diversity within oral microbes is associated with a reduced risk of developing lung cancer, and they also identified distinct representative microbial genera as potential indicators for monitoring lung cancer (Hosgood et al., 2021; Vogtmann et al., 2022; Zhou et al., 2022). It is evident that oral microorganisms and their microbial derivatives, including proteins, endotoxins, and other metabolites, can be transmitted to the respiratory tract or even through direct inhalation or hematogenous dissemination, thereby influencing the occurrence and progression of lung cancer (Ma et al., 2023). Furthermore, certain oral microbiota can directly induce chronic inflammation, immune and activation of carcinogenic signaling pathways in order to promote the development of lung cancer. Tsay conducted an analysis of airway rinse samples of 39 lung cancer patients and 36 non-cancer patients, revealing a higher abundance of Streptococcus and Veillonella in the lower airways of lung cancer patients. Furthermore, through transcriptome analysis of airway epithelial cells and in vitro experiments, they revealed that Streptococcus and Veillonella were related to the up-regulation of ERK and PI3K signaling pathways during carcinogenesis (Tsay et al., 2018). Similarly, Yang et al. (2018) identified an enrichment of Sphingomonas and Bacillus in the saliva of non-smoking female lung cancer patients, suggested that dysbiosis in oral saliva microbiota may regulate the apoptosis of lung cancer cells through the p53 pathway. But the limitation of these studies is the absence of in vivo experiments. It is noteworthy that Helicobacter pylori (H. pylori), the bacterium responsible for stomach cancer, can also be detected in the oral cavity (Zhang et al., 2022). Oral H. pylori has the potential to induce significant and persistent inflammation in the lung lining through direct inhalation or hematogenous transfer to bronchus or lung tissue, thereby promoting malignant transformation and tumor growth (GonzÁlez et al., 2018). The presence of H. pylori infection can potentially impact the efficacy of immunotherapy in patients with non-small cell lung cancer, and mechanistic investigations have elucidated that H. pylori has the ability to modulate dendritic cell cross-presentation activity, suppress CD8+ T cell response against tumors, and influence both innate and adaptive immune responses in the host (Oster et al., 2022). It is undeniable that the mechanism of oral microbiota and lung cancer is still in its infancy, and more exploration is needed in the future.
2.2. Colorectal cancer
CRC ranks the second in the world with a mortality rate of 9.4% (Sung et al., 2021). Colorectal mucosa harbors a diverse microbial community, and emerging evidence suggests a close association between CRC and gastrointestinal (GI) microbes (Tilg et al., 2018). However, the colonization of oral microbiota in the gut is hindered by the presence of an oral-gut barrier (Tugizov, 2016; Moutsopoulos and Konkel, 2020). Successful translocation of oral microorganisms to the gut requires overcoming two major challenges: (i) Traversing the upper digestive tract chemical barrier composed of acid and bile and (ii) surpassing intestinal colonization resistance mediated by commensal bacteria (Buffie and Pamer, 2013; Mo et al., 2022). Once this barrier is breached, ectopic colonization of oral microorganisms in the colorectal site can disrupt microbial ecology and lead to the occurrence of colorectal inflammation, thus creating an environment conducive to tumor progression (Reitano et al., 2021). The latest mechanistic investigations have revealed that oral microorganisms exert additional detrimental effects on the colon through their hydrolysis of glycoproteins present on the surface of colonic epithelial cells, degradation of mucin and extracellular matrix components, as well as synthesis of carcinogenic metabolites, reactive oxygen species, and polyamines (Cueva et al., 2020). Currently, numerous literatures have reported that intestinal dysbiosis is directly or indirectly related to the CRC (Gao et al., 2015; Wirbel et al., 2019). Moreover, oral and pharyngeal swabs, saliva, fecal samples and tumor tissue samples were used to reveal significant differences in oral microbial composition between CRC patients and healthy controls, which provides biological markers for the diagnosis of CRC (Kostic et al., 2013; Gao et al., 2015; Guven et al., 2019; Zhang et al., 2020; Wang Y. et al., 2021; Yu et al., 2022). Although the utility of throat swabs in assessing microbial composition has been subject to scrutiny, their sensitivity significantly increased from 53 to 76% with the inclusion of fecal samples (Flemer et al., 2018), but its universality needs to be further studied.
Cumulative evidences demonstrated that oral microorganisms, including but are not limited to P. gingivalis, F. nucleatum and Streptococcus, exhibit significantly increased abundance in tumors and feces of patients with CRC (Ahn et al., 2013; Kostic et al., 2013; McCoy et al., 2013; Drewes et al., 2017; Uchino et al., 2021). Two studies have similarly found that P. gingivalis can not only mediate the occurrence of enteritis by activating the NLRP3 inflammasome, but also activate the MAPK/ERK signaling pathway (Mu et al., 2020; Wang X. et al., 2021). In addition, it has been reported that tissue-infiltrating P. gingivalis can colonize dendritic cells and macrophages to escape clearance of immune system, resulting in systemic dissemination (Carrion et al., 2012). Approximately 40% infiltrated F. nucleatum in CRC originates from the oral cavity (Komiya et al., 2019). F. nucleatum has emerged as a major driver of CRC due to its anaerobic nature, high invasiveness, FAP2-dependent colorectal adhesion, and glucose free metabolism (Drewes et al., 2017; Osman et al., 2021). Moreover, its high abundance within CRC is associated with tumor metastasis, recurrence, chemotherapy resistance and reduced efficacy of radiotherapy (Yu et al., 2017; Dong et al., 2021; Chen S. et al., 2022; Ou et al., 2022). The potential mechanisms underlying these effects include: (i) Directly influencing tumor cells by regulating tumor metabolism and enhancing the stemness of cancer stem cells; (ii) Modulating T cell-mediated immunity and recruiting myeloid-derived suppressor cells to mediate tumor immune suppression; (iii) Producing inflammatory factors that create a pro-inflammatory microenvironment to promote CRC progression; (iv)Targeting TLR4/MyD88 signaling pathway to induce autophagy to promote chemotherapy resistance of CRC (Kostic et al., 2013; Yu et al., 2017; Ou et al., 2022). Interestingly, F. nucleatum has been found to activate the STING signaling pathway and up-regulate the expression of PD-L1, thus exhibiting a remarkable sensitivity toward PD-L1 immune blockade therapy (Gao Y. et al., 2021). Additionally, Streptococcus has been implicated in mediating inflammation and induce an immunosuppressive tumor microenvironment (TME) dominated by myeloid-derived immunosuppressive cells (MDSC) and tumor-associated macrophages, thereby promoting CRC progression (Long et al., 2019; Taylor et al., 2021).
2.3. Hepatocellular cancer
Hepatocellular cancer (HCC) ranks third in terms of mortality rate, with 830,180deaths (4.7%) reported (Sung et al., 2021). Well-established risk factors for high incidence of HCC include alcohol consumption and infection (Liu and Dong, 2021). However, limited knowledge exists regarding the association between oral microbiota and HCC. Currently, Streptococcus, Porphyromonas, Actinomyces, Fusarium and Fusobacterium are among the oral microorganisms that have be suggested to be potentially linked to HCC progression (Lu et al., 2016; Li et al., 2020). Regrettably, the mechanism of its role in HCC remains to be further studied. Nevertheless, research has demonstrated a close association between oral microorganisms, specifically P. gingivalis, and the development of hepatitis and alcoholic liver disease (Gao et al., 2023). P. gingivalis is capable of inducing intestinal microbial dysbiosis and impairing the integrity of the intestinal mucosal barrier, thereby facilitating the migration of enterobacteriaceae to the liver (Nakajima et al., 2015). Additionally, P. gingivalis can disrupt the balance between Th17/Treg cells in the intestinal tract, leading to hepatitis and promoting ferroptosis in hepatocytes (Yao et al., 2023). Hence, it is plausible to postulate that the oral microbiota might be implicated in the pathogenesis of liver cancer; however, further empirical evidence is warranted.
2.4. Gastric cancer
H. pylori infection is a well-established risk factor for gastric cancer, which ranks as the fourth most common cancer globally, with the new incidence of gastric cancer in China was 44% in 2020 (Liu and Dong, 2021). An analysis of the microbiome of gastric cancer in Mexico and China revealed that the dominant bacteria was H. pylori, followed by oral microorganisms---Proteobacteria and Firmicutes (Yu et al., 2017). However, only 1–3% of H. pylori infected patients will develop into gastric cancer, so it is reasonable to speculate that other biological factors may be involved in the occurrence and development of gastric cancer (Chen et al., 2019). Study have exhibited elevated abundance of Peptostreptococcus stomatis, Streptococcus anginosus, Parvimonas micra, Slackia exigua and Dialister pneumosintes in gastric cancer (Coker et al., 2018). A recent study indicated that more than half of gastric cancer patients tested positive for Fusobacterium, Clostridium and Lactobacillus, suggesting their potential as biomarkers for early diagnosis of gastric cancer (Hsieh et al., 2018). P. gingivalis has also been reported to be associated with an increased risk of gastric cancer in Asians (Yang et al., 2022). However, the precise underlying mechanism remains unclear.
2.5. Breast cancer
Breast cancer is the fifth most commonly diagnosed tumor worldwide (Sung et al., 2021). Its risk factors primarily include overweight, family genetic history and unhealthy lifestyle such as smoking and excessive alcohol consumption (Liu and Dong, 2021). Previous studies have reported the presence of oral microorganisms in breast milk and speculated on potential transmission routes including: (i) Penetration through the skin and nipple; (ii) Colonization by translocation from the digestive tract and genital tract; (iii) Invasion through blood and lymphatic circulation system, leading to the location in breast lobules and ducts (Urbaniak et al., 2012). However, some scholars suggested that Proteobacteria and Firmicutes are the predominant microbial groups found in the breast tissue, which exhibit tolerence toward fatty acids infiltration. Therefore, it can be considered that there are unique and diverse microbial groups in the breast (Urbaniak et al., 2014).
For a considerable duration, it has been postulated that alterations in the intestinal microbiota within breast tissue can exert influence on hormone levels and contribute to the pathogenesis of breast cancer (Laborda-Illanes et al., 2020). Meanwhile, limited attention has been given to the oral microbiota in the mammary gland within existing literature. A meta-analysis has revealed a significant association between periodontal disease and oral microbial infection with breast cancer, thereby suggesting that periodontal disease may serves as a potential risk factor for the development of breast cancer (Shao et al., 2018). Another study compared oral rinse samples from 50 breast cancer patients and 20 healthy controls, finding no significant difference in microbial communities (Wang et al., 2017). This result may be constrained by the limited sample size of the included studies. Recently, a sequencing study involving 369 breast cancer cases, 93 non-malignant cases and 419 healthy controls revealed a decrease relative abundance of representative oral microorganisms, namely P. gingivalis and F. nucleatum, in breast cancer patients (Wu et al., 2022). However, the study did not directly examine the microbiome in breast tumor. Therefore, the association between oral microbiota and breast cancer is somewhat unreliable. Parhi et al. (2020) verified that F. nucleatum could spread through blood and colonize breast cancer sites in a FAP2-dependent manner, thereby impairing anti-tumor immunity and promoting the progression of breast cancer, while the corresponding metronidazole antibacterial treatment could delay the progression of tumor. In addition, F. nucleatum colonized in breast cancer can also activate NF-κB through TLR4/MyD88 pathway, creating an immunosuppressive TME by recruiting immunosuppressive cells, and promoting the immune escape of breast cancer cells by MYC-dependent up-regulation of PD-L1 and CD47 in breast cancer (Van der Merwe et al., 2021). In conclusion, oral microbiota is involved in the progression of breast cancer, and targeted elimination may be beneficial for the treatment of breast cancer.
2.6. Other cancer
Due to the anatomical relationship between oral cavity and digestive tract, there is a higher likelihood of cross-talk between oral microorganisms and tumors in digestive system (Tuominen and Rautava, 2021). In addition to the above-mentioned tumors, oral cancer, esophageal cancer and pancreatic cancer have also been reported to be closely related to oral microbiome (Binder Gallimidi et al., 2015; Fan et al., 2018; Wang et al., 2019; Yamamura et al., 2019). The oral cavity is the main habitat for oral microorganisms, and almost all oral microorganisms have been implicated in the progression of oral cancer.
Oral microorganisms that have been clearly associated with the progression of oral cancer include but are not limited to Enterococcus faecalis, F. nucleatum and P. gingivalis (Zhao et al., 2017; Metsäniitty et al., 2021). Enterococcus faecalis can promote the proliferation of oral cancer cells through the H2O2-mediated EGFR signaling pathway (Boonanantanasarn et al., 2012). F. nucleatum promotes the progression of oral cancer by regulating cell cycle, inducing oral inflammation, regulating anti-tumor immune response and promoting epithelial-mesenchymal transition of oral cancer cells (McIlvanna et al., 2021; Shao et al., 2021). The carcinogenic mechanisms of P. gingivalis and F. nucleatum share similarities (Lafuente Ibáñez de Mendoza et al., 2020; Wen et al., 2020; Yao et al., 2021). Interestingly, an in vitro transcriptomic study involving three oral squamous cell carcinoma cell lines, CAL27, SCC4, and SCC25, and three oral commensals, Streptococcus, Neisseria aureus, and Haemophilus parainfluis, with P. gingivalis as a positive control, found that only Streptococcus exhibited significant antitumor properties alone. The other two commensal bacteria showed the coexistence of pro-tumor and anti-tumor effects, while P. gingivalis only had pro-tumor effects on SCC4 cell line (Baraniya et al., 2022). In addition, it has also been reported that P. gingivalis can mediate autophagy to induce G1 phase arrest in oral squamous cell carcinoma cells (Cho et al., 2014). Therefore, the exploration of oral microorganisms with anti-tumor effects may provide a new strategy for cancer treatment.
Six studies used 16S rRNA sequencing to analyze saliva and oral swab samples collected from patients with esophageal cancer, revealing significant alterations of oral microorganisms. Although the dominant groups were different, they all suggested that oral microorganisms may be potential screening markers for esophageal cancer (Chen et al., 2015; Wang et al., 2019; Li et al., 2021; Chen X. et al., 2022; Jiang et al., 2022; Nomburg et al., 2022). Currently, P. gingivalis and F. nucleatum are the primary oral microorganisms reported to be associated with esophageal cancer (Wang et al., 2019; Gao S. et al., 2021; Liu et al., 2021; Lamont et al., 2022). The definite mechanism by which P. gingivalis promotes esophageal cancer involves the activation of NF-κB (Meng et al., 2019), PTEN/Akt (Liang et al., 2020), EMT (Chen et al., 2021) and apoptosis resistance signaling pathway (Gao S. et al., 2021), which can regulate proliferation, induce inflammation, create immunosuppressive TME and mediate chemotherapy resistance. F. nucleatum can also activate NOD1/RIPK2/NF-κB signaling pathway (Nomoto et al., 2022), AHR/CYP1A1 (Yin et al., 2023), NLRP3 inflammasome (Liang et al., 2022) and autophagy-related signaling pathway (Liu et al., 2021). It also mediates the infiltration of MDSC and regulatory T cells to regulate tumor immunity (Zhang et al., 2021; Liang et al., 2022).
Furthermore, recent studies have highlighted the potential of P. gingivalis and F. nucleatum as important biomarkers for distinguishing pancreatic cancer patients (Fan et al., 2018; Wei et al., 2020). Notably, the oral microorganism F. nucleatum has been found to stimulate the secretion of cytokines, including GM-CSF and CXCL1, thus promoting tumor cell proliferation and migration (Udayasuryan et al., 2022). However, the relationship between other oral microbiota and pancreatic cancer remains elusive.
3. Discussion
With the development of technology and interdisciplinary science, the research on microorganisms and cancer has become a hot topic. Due to the anatomical connection, the current reports mainly focus on digestive system tumors, and mainly investigate the common dominant bacteria in the oral microbiome, such as P. gingivalis and F. nucleatum. This review provides a systematic overview of the association between oral microbiota and the world’s most common malignancies, and summarizes the possible mechanisms reported in the literature. At present, it is believed that oral microorganisms are widely associated with the malignant transformation and progression of tumors. Changes in the abundance of oral microorganisms may be used as potential biomarkers for predicting tumorigenesis, and targeted elimination of related harmful oral microorganisms is expected to become a new strategy for cancer treatment. However, it has also been reported that oral microorganisms play a dual role in tumor biology, which either promotes or inhibits tumor depends on the interaction between microorganisms, host and TME. Oral microorganisms have the ability to colonize extra-oral organs, such as the lungs, colorectum, and stomach (Park et al., 2021). They employ their own virulence factors and metabolites to disrupt the epithelial barrier and extracellular matrix, induce an inflammatory microenvironment and immunosuppressive tumor microenvironment, thereby influencing both local and distant tumors (Li S. et al., 2022; Ma et al., 2023). The anaerobic conditions within the tumor microenvironment further facilitate the accumulation of anaerobic oral microorganisms in tumors (Wu et al., 2021). Intratumoral microorganisms have been found to enhance anti-tumor immunity through activation of the STING signaling pathway, stimulation of T cells and NK cells, formation of intratumoral tertiary lymphoid structures (TLS), as well as microbial-derived antigen presentation mechanisms; Conversely, they can also dampen anti-tumor immune responses by upregulating ROS levels, promoting an anti-inflammatory environment, impairing T cell function, and inducing immunosuppression (Yang et al., 2023). Moreover, spatial transcriptomics analysis revealed a decrease of cytotoxic T cells within regions characterized by higher bacterial accumulation in tumors, accompanied by more potent immunosuppressive effects (Galeano Niño et al., 2022). Studies on oral microbiota also present a dual perspective: The Fap2 protein, expressed by F. nucleatum, interacts with human inhibitory receptors TIGIT to facilitate the evasion of tumor cells from NK cells and T cells (Gur et al., 2015). Additionally, it has been demonstrated that F. nucleatum can activate the STING signaling pathway and enhance the expression of PD-L1, thereby exhibiting heightened susceptibility toward PD-L1 immune blockade therapy (Gao Y. et al., 2021). Therefore, the conflicting conclusions need to be further verified, and the beneficial microorganisms against cancer need to be further explored to broaden the treatment options for cancer.
Author contributions
ZL, W-JL, Y-YZ, and G-TY: conceptualization. ZL and W-JL: formal analysis and writing the original draft. ZL, WJ-L, HCu, K-LZ, HCh, Y-YZ, and G-TY: writing review and editing. Y-YZ and G-TY: supervision and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China [82103404], Guangdong Basic and Applied Basic Research Foundation [2020A1515110719], Southern Medical University Excellent Youth Scholars Training Program [2020YQPY008], Guangzhou Basic and Applied Basic Research Foundation [202102020687], Stomatological Hospital of Southern Medical University Startup Funds [No. PY2020001, PY2019026], Medical Research Fund of Guangdong Province [No. B2020081].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahn, J., Sinha, R., Pei, Z., Dominianni, C., Wu, J., Shi, J., et al. (2013). Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 105, 1907–1911. doi: 10.1093/jnci/djt300
Bakhti, S. Z., and Latifi-Navid, S. (2021). Oral microbiota and Helicobacter pylori in gastric carcinogenesis: what do we know and where next? BMC Microbiol. 21:71. doi: 10.1186/s12866-021-02130-4
Baraniya, D., Chitrala, K. N., and Al-Hebshi, N. N. (2022). Global transcriptional response of oral squamous cell carcinoma cell lines to health-associated oral bacteria-an in vitro study. J. Oral Microbiol. 14:2073866. doi: 10.1080/20002297.2022.2073866
Belcheva, A., Irrazabal, T., Robertson, S. J., Streutker, C., Maughan, H., Rubino, S., et al. (2014). Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cells 158, 288–299. doi: 10.1016/j.cell.2014.04.051
Binder Gallimidi, A., Fischman, S., Revach, B., Bulvik, R., Maliutina, A., Rubinstein, A. M., et al. (2015). Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 6, 22613–22623. doi: 10.18632/oncotarget.4209
Boonanantanasarn, K., Gill, A. L., Yap, Y., Jayaprakash, V., Sullivan, M. A., and Gill, S. R. (2012). Enterococcus faecalis enhances cell proliferation through hydrogen peroxide-mediated epidermal growth factor receptor activation. Infect. Immun. 80, 3545–3558. doi: 10.1128/IAI.00479-12
Buffie, C. G., and Pamer, E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. doi: 10.1038/nri3535
Cao, W., Chen, H. D., Yu, Y. W., Li, N., and Chen, W. Q. (2021). Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin. Med. J. 134, 783–791. doi: 10.1097/CM9.0000000000001474
Carrion, J., Scisci, E., Miles, B., Sabino, G. J., Zeituni, A. E., Gu, Y., et al. (2012). Microbial carriage state of peripheral blood dendritic cells (DCs) in chronic periodontitis influences DC differentiation, atherogenic potential. J. Immunol. 189, 3178–3187. doi: 10.4049/jimmunol.1201053
Chen, M. F., Lu, M. S., Hsieh, C. C., and Chen, W. C. (2021). Porphyromonas gingivalis promotes tumor progression in esophageal squamous cell carcinoma. Cell. Oncol. (Dordr.) 44, 373–384. doi: 10.1007/s13402-020-00573-x
Chen, S., Zhang, L., Li, M., Zhang, Y., Sun, M., Wang, L., et al. (2022). Fusobacterium nucleatum reduces METTL3-mediated m(6)a modification and contributes to colorectal cancer metastasis. Nat. Commun. 13:1248. doi: 10.1038/s41467-022-28913-5
Chen, X., Winckler, B., Lu, M., Cheng, H., Yuan, Z., Yang, Y., et al. (2015). Oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS One 10:e0143603. doi: 10.1371/journal.pone.0143603
Chen, X., Xian, B., Wei, J., Chen, Y., Yang, D., Lai, X., et al. (2022). Predictive value of the presence of Prevotella and the ratio of Porphyromonas gingivalis to Prevotella in saliva for esophageal squamous cell carcinoma. Front. Cell. Infect. Microbiol. 12:997333. doi: 10.3389/fcimb.2022.997333
Chen, X. H., Wang, A., Chu, A. N., Gong, Y. H., and Yuan, Y. (2019). Mucosa-associated microbiota in gastric Cancer tissues compared with non-cancer tissues. Front. Microbiol. 10:1261. doi: 10.3389/fmicb.2019.01261
Cho, T. J., Wee, S. W., Woo, V. H., Choi, J. I., Kim, S. J., Shin, H. I., et al. (2014). Porphyromonas gingivalis-induced autophagy suppresses cell proliferation through G1 arrest in oral cancer cells. Arch. Oral Biol. 59, 370–378. doi: 10.1016/j.archoralbio.2014.01.001
Coker, O. O., Dai, Z., Nie, Y., Zhao, G., Cao, L., Nakatsu, G., et al. (2018). Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67, 1024–1032. doi: 10.1136/gutjnl-2017-314281
Colucci, F. (2015). An oral commensal associates with disease: chicken, egg, or red herring? Immunity 42, 208–210. doi: 10.1016/j.immuni.2015.01.024
Cribbs, S. K., Crothers, K., and Morris, A. (2020). Pathogenesis of HIV-related lung disease: immunity, infection, and inflammation. Physiol. Rev. 100, 603–632. doi: 10.1152/physrev.00039.2018
Cueva, C., Silva, M., Pinillos, I., Bartolomé, B., and Moreno-Arribas, M. V. (2020). Interplay between dietary polyphenols and Oral and gut microbiota in the development of colorectal Cancer. Nutrients 12:625. doi: 10.3390/nu12030625
Cullin, N., Azevedo Antunes, C., Straussman, R., Stein-Thoeringer, C. K., and Elinav, E. (2021). Microbiome and cancer. Cancer Cell 39, 1317–1341. doi: 10.1016/j.ccell.2021.08.006
DeGruttola, A. K., Low, D., Mizoguchi, A., and Mizoguchi, E. (2016). Current understanding of Dysbiosis in disease in human and animal models. Inflamm. Bowel Dis. 22, 1137–1150. doi: 10.1097/MIB.0000000000000750
Deo, P. N., and Deshmukh, R. (2020). Oral microbiome and oral cancer–the probable nexus. J Oral Maxillofac Pathol. 24, 361–367. doi: 10.4103/jomfp.JOMFP_20_20
Doherty, J. R., and Cleveland, J. L. (2013). Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123, 3685–3692. doi: 10.1172/JCI69741
Dong, J., Li, Y., Xiao, H., Zhang, S., Wang, B., Wang, H., et al. (2021). Oral microbiota affects the efficacy and prognosis of radiotherapy for colorectal cancer in mouse models. Cell Rep. 37:109886. doi: 10.1016/j.celrep.2021.109886
Drewes, J. L., White, J. R., Dejea, C. M., Fathi, P., Iyadorai, T., Vadivelu, J., et al. (2017). High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 3:34. doi: 10.1038/s41522-017-0040-3
Fan, X., Alekseyenko, A. V., Wu, J., Peters, B. A., Jacobs, E. J., Gapstur, S. M., et al. (2018). Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 67, 120–127. doi: 10.1136/gutjnl-2016-312580
Flemer, B., Warren, R. D., Barrett, M. P., Cisek, K., Das, A., Jeffery, I. B., et al. (2018). The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67, 1454–1463. doi: 10.1136/gutjnl-2017-314814
Gagliani, N., Hu, B., Huber, S., Elinav, E., and Flavell, R. A. (2014). The fire within: microbes inflame tumors. Cells 157, 776–783. doi: 10.1016/j.cell.2014.03.006
Galeano Niño, J. L., Wu, H., LaCourse, K. D., Kempchinsky, A. G., Baryiames, A., Barber, B., et al. (2022). Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817. doi: 10.1038/s41586-022-05435-0
Gao, L., Xu, T., Huang, G., Jiang, S., Gu, Y., and Chen, F. (2018). Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell 9, 488–500. doi: 10.1007/s13238-018-0548-1
Gao, S., Liu, Y., Duan, X., Liu, K., Mohammed, M., Gu, Z., et al. (2021). Porphyromonas gingivalis infection exacerbates oesophageal cancer and promotes resistance to neoadjuvant chemotherapy. Br. J. Cancer 125, 433–444. doi: 10.1038/s41416-021-01419-5
Gao, Y., Bi, D., Xie, R., Li, M., Guo, J., Liu, H., et al. (2021). Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct. Target. Ther. 6:398. doi: 10.1038/s41392-021-00795-x
Gao, Y., Zhang, P., Wei, Y., Ye, C., Mao, D., Xia, D., et al. (2023). Porphyromonas gingivalis exacerbates alcoholic liver disease by altering gut microbiota composition and host immune response in mice. J. Clin. Periodontol. 50, 1253–1263. doi: 10.1111/jcpe.13833
Gao, Z., Guo, B., Gao, R., Zhu, Q., and Qin, H. (2015). Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 6:20. doi: 10.3389/fmicb.2015.00020
GonzÁlez, I., Araya, P., and Rojas, A. (2018). Helicobacter pylori infection and lung Cancer: new insights and future challenges. Zhongguo Fei Ai Za Zhi 21, 658–662. doi: 10.3779/j.issn.1009-3419.2018.09.03
Gur, C., Ibrahim, Y., Isaacson, B., Yamin, R., Abed, J., Gamliel, M., et al. (2015). Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42, 344–355. doi: 10.1016/j.immuni.2015.01.010
Guven, D. C., Dizdar, O., Alp, A., Akdoğan Kittana, F. N., Karakoc, D., Hamaloglu, E., et al. (2019). Analysis of Fusobacterium nucleatum and Streptococcus gallolyticus in saliva of colorectal cancer patients. Biomark. Med 13, 725–735. doi: 10.2217/bmm-2019-0020
Hanahan, D. (2022). Hallmarks of Cancer: new dimensions. Cancer Discov. 12, 31–46. doi: 10.1158/2159-8290.CD-21-1059
Hosgood, H. D., Cai, Q., Hua, X., Long, J., Shi, J., Wan, Y., et al. (2021). Variation in oral microbiome is associated with future risk of lung cancer among never-smokers. Thorax 76, 256–263. doi: 10.1136/thoraxjnl-2020-215542
Hsieh, Y. Y., Tung, S. Y., Pan, H. Y., Yen, C. W., Xu, H. W., Lin, Y. J., et al. (2018). Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric Cancer in Taiwan. Sci. Rep. 8:158. doi: 10.1038/s41598-017-18596-0
Jiang, Z., Wang, J., Qian, X., Zhang, Z., and Wang, S. (2022). Oral microbiota may predict the presence of esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. doi: 10.1007/s00432-022-04393-4
Koliarakis, I., Messaritakis, I., Nikolouzakis, T. K., Hamilos, G., Souglakos, J., and Tsiaoussis, J. (2019). Oral Bacteria and intestinal Dysbiosis in colorectal Cancer. Int. J. Mol. Sci. 20:4146. doi: 10.3390/ijms20174146
Komiya, Y., Shimomura, Y., Higurashi, T., Sugi, Y., Arimoto, J., Umezawa, S., et al. (2019). Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut 68, 1335–1337. doi: 10.1136/gutjnl-2018-316661
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Laborda-Illanes, A., Sanchez-Alcoholado, L., Dominguez-Recio, M. E., Jimenez-Rodriguez, B., Lavado, R., Comino-Méndez, I., et al. (2020). Breast and gut microbiota action mechanisms in breast Cancer pathogenesis and treatment. Cancers (Basel). 12:2465. doi: 10.3390/cancers12092465
Lafuente Ibáñez de Mendoza, I., De Mendoza, I., Maritxalar Mendia, X., García de la Fuente, A. M., Quindós Andrés, G., and Aguirre Urizar, J. M. (2020). Role of Porphyromonas gingivalis in oral squamous cell carcinoma development: a systematic review. J. Periodontal Res. 55, 13–22. doi: 10.1111/jre.12691
Lamont, R. J., Fitzsimonds, Z. R., Wang, H., and Gao, S. (2022). Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma. Periodontol. 89, 154–165. doi: 10.1111/prd.12425
Li, D., Xi, W., Zhang, Z., Ren, L., Deng, C., Chen, J., et al. (2020). Oral microbial community analysis of the patients in the progression of liver cancer. Microb. Pathog. 149:104479. doi: 10.1016/j.micpath.2020.104479
Li, S., He, M., Lei, Y., Liu, Y., Li, X., Xiang, X., et al. (2022). Oral microbiota and tumor-a new perspective of tumor pathogenesis. Microorganisms 10:2206. doi: 10.3390/microorganisms10112206
Li, X., Liu, Y., Yang, X., Li, C., and Song, Z. (2022). The Oral microbiota: community composition, influencing factors, pathogenesis, and interventions. Front. Microbiol. 13:895537. doi: 10.3389/fmicb.2022.895537
Li, Z., Dou, L., Zhang, Y., He, S., Zhao, D., Hao, C., et al. (2021). Characterization of the Oral and esophageal microbiota in esophageal precancerous lesions and squamous cell carcinoma. Front. Cell. Infect. Microbiol. 11:714162. doi: 10.3389/fcimb.2021.714162
Liang, G., Wang, H., Shi, H., Zhu, M., An, J., Qi, Y., et al. (2020). Porphyromonas gingivalis promotes the proliferation and migration of esophageal squamous cell carcinoma through the miR-194/GRHL3/PTEN/Akt Axis. ACS Infect. Dis. 6, 871–881. doi: 10.1021/acsinfecdis.0c00007
Liang, M., Liu, Y., Zhang, Z., Yang, H., Dai, N., Zhang, N., et al. (2022). Fusobacterium nucleatum induces MDSCs enrichment via activation the NLRP3 inflammosome in ESCC cells, leading to cisplatin resistance. Ann. Med. 54, 989–1003. doi: 10.1080/07853890.2022.2061045
Liu, H., and Dong, Z. (2021). Cancer etiology and prevention principle: "1 + X". Cancer Res. 81, 5377–5395. doi: 10.1158/0008-5472.CAN-21-1862
Liu, Y., Baba, Y., Ishimoto, T., Tsutsuki, H., Zhang, T., Nomoto, D., et al. (2021). Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br. J. Cancer 124, 963–974. doi: 10.1038/s41416-020-01198-5
Long, X., Wong, C. C., Tong, L., Chu, E. S. H., Ho Szeto, C., Go, M. Y. Y., et al. (2019). Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol. 4, 2319–2330. doi: 10.1038/s41564-019-0541-3
Lu, H., Ren, Z., Li, A., Zhang, H., Jiang, J., Xu, S., et al. (2016). Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci. Rep. 6:33142. doi: 10.1038/srep33142
Ma, Q., Li, X., Jiang, H., Fu, X., You, L., You, F., et al. (2023). Mechanisms underlying the effects, and clinical applications, of oral microbiota in lung cancer: current challenges and prospects. Crit. Rev. Microbiol. 11, 1–22. doi: 10.1080/1040841X.2023.2247493
Mark Welch, J. L., Ramírez-Puebla, S. T., and Borisy, G. G. (2020). Oral microbiome geography: Micron-scale habitat and niche. Cell Host Microbe 28, 160–168. doi: 10.1016/j.chom.2020.07.009
McCoy, A. N., Araújo-Pérez, F., Azcárate-Peril, A., Yeh, J. J., and Sandler, R. S., Keku TO (2013). Fusobacterium is associated with colorectal adenomas. PLoS One 8:e53653. doi: 10.1371/journal.pone.0053653
McIlvanna, E., Linden, G. J., Craig, S. G., Lundy, F. T., and James, J. A. (2021). Fusobacterium nucleatum and oral cancer: a critical review. BMC Cancer 21:1212. doi: 10.1186/s12885-021-08903-4
Meng, F., Li, R., Ma, L., Liu, L., Lai, X., Yang, D., et al. (2019). Porphyromonas gingivalis promotes the motility of esophageal squamous cell carcinoma by activating NF-κB signaling pathway. Microbes Infect. 21, 296–304. doi: 10.1016/j.micinf.2019.01.005
Meslier, V., Quinquis, B., Da Silva, K., Plaza Oñate, F., Pons, N., Roume, H., et al. (2022). Benchmarking second and third-generation sequencing platforms for microbial metagenomics. Sci Data. 9:694. doi: 10.1038/s41597-022-01762-z
Metsäniitty, M., Hasnat, S., Salo, T., and Salem, A. (2021). Oral microbiota-a new frontier in the pathogenesis and Management of Head and Neck Cancers. Cancers (Basel) 14:46. doi: 10.3390/cancers14010046
Meurman, J. H. (2010). Infectious and dietary risk factors of oral cancer. Oral Oncol. 46, 411–413. doi: 10.1016/j.oraloncology.2010.03.003
Mo, S., Ru, H., Huang, M., Cheng, L., Mo, X., and Yan, L. (2022). Oral-intestinal microbiota in colorectal cancer: inflammation and immunosuppression. J. Inflamm. Res. 15, 747–759. doi: 10.2147/JIR.S344321
Moutsopoulos, N. M., and Konkel, J. E. (2020). Healthy mouth, healthy gut: a dysbiotic oral microbiome exacerbates colitis. Mucosal Immunol. 13, 852–854. doi: 10.1038/s41385-020-00341-y
Mu, W., Jia, Y., Chen, X., Li, H., Wang, Z., and Cheng, B. (2020). Intracellular Porphyromonas gingivalis promotes the proliferation of colorectal Cancer cells via the MAPK/ERK signaling pathway. Front. Cell. Infect. Microbiol. 10:584798. doi: 10.3389/fcimb.2020.584798
Nakajima, M., Arimatsu, K., Kato, T., Matsuda, Y., Minagawa, T., Takahashi, N., et al. (2015). Oral administration of P. gingivalis induces Dysbiosis of gut microbiota and impaired barrier function leading to dissemination of Enterobacteria to the liver. PLoS One 10:e0134234. doi: 10.1371/journal.pone.0134234
Niklander, S. E. (2021). Inflammatory mediators in Oral Cancer: pathogenic mechanisms and diagnostic potential. Front. Oral Health 2:642238. doi: 10.3389/froh.2021.642238
Nomburg, J., Bullman, S., Nasrollahzadeh, D., Collisson, E. A., Abedi-Ardekani, B., Akoko, L. O., et al. (2022). An international report on bacterial communities in esophageal squamous cell carcinoma. Int. J. Cancer 151, 1947–1959. doi: 10.1002/ijc.34212
Nomoto, D., Baba, Y., Liu, Y., Tsutsuki, H., Okadome, K., Harada, K., et al. (2022). Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression via the NOD1/RIPK2/NF-κB pathway. Cancer Lett. 530, 59–67. doi: 10.1016/j.canlet.2022.01.014
Osman, M. A., Neoh, H. M., Ab Mutalib, N. S., Chin, S. F., Mazlan, L., Raja Ali, R. A., et al. (2021). Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci. Rep. 11:2925. doi: 10.1038/s41598-021-82465-0
Oster, P., Vaillant, L., Riva, E., McMillan, B., Begka, C., Truntzer, C., et al. (2022). Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut 71, 457–466. doi: 10.1136/gutjnl-2020-323392
Ou, S., Wang, H., Tao, Y., Luo, K., Ye, J., Ran, S., et al. (2022). Fusobacterium nucleatum and colorectal cancer: from phenomenon to mechanism. Front. Cell. Infect. Microbiol. 12:1020583. doi: 10.3389/fcimb.2022.1020583
Parhi, L., Alon-Maimon, T., Sol, A., Nejman, D., Shhadeh, A., Fainsod-Levi, T., et al. (2020). Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 11:3259. doi: 10.1038/s41467-020-16967-2
Park, S. Y., Hwang, B. O., Lim, M., Ok, S. H., Lee, S. K., Chun, K. S., et al. (2021). Oral-gut microbiome Axis in gastrointestinal disease and Cancer. Cancers (Basel). 13:2124. doi: 10.3390/cancers13092124
Reitano, E., De’Angelis, N., Gavriilidis, P., Gaiani, F., Memeo, R., Inchingolo, R., et al. (2021). Oral bacterial microbiota in digestive Cancer patients: a systematic review. Microorganisms. 9:2585. doi: 10.3390/microorganisms9122585
Sedghi, L., DiMassa, V., Harrington, A., Lynch, S. V., and Kapila, Y. L. (2021). The oral microbiome: role of key organisms and complex networks in oral health and disease. Periodontol. 87, 107–131. doi: 10.1111/prd.12393
Sepich-Poore, G. D., Zitvogel, L., Straussman, R., Hasty, J., Wargo, J. A., and Knight, R. (2021). The microbiome and human cancer. Science 371:eabc4552. doi: 10.1126/science.abc4552
Shao, J., Wu, L., Leng, W. D., Fang, C., Zhu, Y. J., Jin, Y. H., et al. (2018). Periodontal disease and breast Cancer: a Meta-analysis of 1,73,162 participants. Front. Oncol. 8:601. doi: 10.3389/fonc.2018.00601
Shao, W., Fujiwara, N., Mouri, Y., Kisoda, S., Yoshida, K., Yoshida, K., et al. (2021). Conversion from epithelial to partial-EMT phenotype by Fusobacterium nucleatum infection promotes invasion of oral cancer cells. Sci. Rep. 11:14943. doi: 10.1038/s41598-021-94384-1
Shi, J., Yang, Y., Xie, H., Wang, X., Wu, J., Long, J., et al. (2021). Association of oral microbiota with lung cancer risk in a low-income population in the southeastern USA. Cancer Causes Control 32, 1423–1432. doi: 10.1007/s10552-021-01490-6
Steffen, M. J., Holt, S. C., and Ebersole, J. L. (2000). Porphyromonas gingivalis induction of mediator and cytokine secretion by human gingival fibroblasts. Oral Microbiol. Immunol. 15, 172–180. doi: 10.1034/j.1399-302x.2000.150305.x
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi: 10.3322/caac.21660
Taylor, J. C., Gao, X., Xu, J., Holder, M., Petrosino, J., Kumar, R., et al. (2021). A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PLoS Pathog. 17:e1009182. doi: 10.1371/journal.ppat.1009182
Teles, F. R. F., Alawi, F., Castilho, R. M., and Wang, Y. (2020). Association or causation? Exploring the Oral microbiome and Cancer links. J. Dent. Res. 99, 1411–1424. doi: 10.1177/0022034520945242
Tilg, H., Adolph, T. E., Gerner, R. R., and Moschen, A. R. (2018). The intestinal microbiota in colorectal Cancer. Cancer Cell 33, 954–964. doi: 10.1016/j.ccell.2018.03.004
Tsay, J. J., Wu, B. G., Badri, M. H., Clemente, J. C., Shen, N., Meyn, P., et al. (2018). Airway microbiota is associated with upregulation of the PI3K pathway in lung Cancer. Am. J. Respir. Crit. Care Med. 198, 1188–1198. doi: 10.1164/rccm.201710-2118OC
Tugizov, S. (2016). Human immunodeficiency virus-associated disruption of mucosal barriers and its role in HIV transmission and pathogenesis of HIV/AIDS disease. Tissue Barriers 4:e1159276. doi: 10.1080/21688370.2016.1159276
Tuominen, H., and Rautava, J. (2021). Oral microbiota and Cancer development. Pathobiology 88, 116–126. doi: 10.1159/000510979
Uchino, Y., Goto, Y., Konishi, Y., Tanabe, K., Toda, H., Wada, M., et al. (2021). Colorectal Cancer patients have four specific bacterial species in Oral and gut microbiota in common-a metagenomic comparison with healthy subjects. Cancers (Basel). 13:3332. doi: 10.3390/cancers13133332
Udayasuryan, B., Ahmad, R. N., Nguyen, T. T. D., Umaña, A., Monét Roberts, L., Sobol, P., et al. (2022). Fusobacterium nucleatum induces proliferation and migration in pancreatic cancer cells through host autocrine and paracrine signaling. Sci. Signal. 15(756):eabn4948 15:eabn4948. doi: 10.1126/scisignal.abn4948
Urbaniak, C., Burton, J. P., and Reid, G. (2012). Breast, milk and microbes: a complex relationship that does not end with lactation. Womens Health (Lond) 8, 385–398. doi: 10.2217/WHE.12.23
Urbaniak, C., Cummins, J., Brackstone, M., Macklaim, J. M., Gloor, G. B., Baban, C. K., et al. (2014). Microbiota of human breast tissue. Appl. Environ. Microbiol. 80, 3007–3014. doi: 10.1128/AEM.00242-14
Van der Merwe, M., Van Niekerk, G., Botha, A., and Engelbrecht, A. M. (2021). The onco-immunological implications of Fusobacterium nucleatum in breast cancer. Immunol. Lett. 232, 60–66. doi: 10.1016/j.imlet.2021.02.007
Vogtmann, E., Hua, X., Yu, G., Purandare, V., Hullings, A. G., Shao, D., et al. (2022). The oral microbiome and lung cancer risk: An analysis of 3 prospective cohort studies. J. Natl. Cancer Inst. 114, 1501–1510. doi: 10.1093/jnci/djac149
Wang, H., Altemus, J., Niazi, F., Green, H., Calhoun, B. C., Sturgis, C., et al. (2017). Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 8, 88122–88138. doi: 10.18632/oncotarget.21490
Wang, Q., Rao, Y., Guo, X., Liu, N., Liu, S., Wen, P., et al. (2019). Oral microbiome in patients with Oesophageal squamous cell carcinoma. Sci. Rep. 9:19055. doi: 10.1038/s41598-019-55667-w
Wang, X., Jia, Y., Wen, L., Mu, W., Wu, X., Liu, T., et al. (2021). Porphyromonas gingivalis promotes colorectal carcinoma by activating the hematopoietic NLRP3 Inflammasome. Cancer Res. 81, 2745–2759. doi: 10.1158/0008-5472.CAN-20-3827
Wang, Y., Zhang, Y., Qian, Y., Xie, Y. H., Jiang, S. S., Kang, Z. R., et al. (2021). Alterations in the oral and gut microbiome of colorectal cancer patients and association with host clinical factors. Int. J. Cancer 149, 925–935. doi: 10.1002/ijc.33596
Wei, A. L., Li, M., Li, G. Q., Wang, X., Hu, W. M., Li, Z. L., et al. (2020). Oral microbiome and pancreatic cancer. World J. Gastroenterol. 26, 7679–7692. doi: 10.3748/wjg.v26.i48.7679
Wen, L., Mu, W., Lu, H., Wang, X., Fang, J., Jia, Y., et al. (2020). Porphyromonas gingivalis promotes Oral squamous cell carcinoma progression in an immune microenvironment. J. Dent. Res. 99, 666–675. doi: 10.1177/0022034520909312
Wirbel, J., Pyl, P. T., Kartal, E., Zych, K., Kashani, A., Milanese, A., et al. (2019). Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689. doi: 10.1038/s41591-019-0406-6
Wu, H., Zhong, D., Zhang, Z., Wu, Y., Li, Y., Mao, H., et al. (2021). A Bacteria-inspired morphology genetic biomedical material: self-propelled artificial microbots for metastatic triple negative breast Cancer treatment. ACS Nano 15, 4845–4860. doi: 10.1021/acsnano.0c09594
Wu, Z., Byrd, D. A., Wan, Y., Ansong, D., Clegg-Lamptey, J. N., Wiafe-Addai, B., et al. (2022). The oral microbiome and breast cancer and nonmalignant breast disease, and its relationship with the fecal microbiome in the Ghana breast health study. Int. J. Cancer 151, 1248–1260. doi: 10.1002/ijc.34145
Xiao, J., Fiscella, K. A., and Gill, S. R. (2020). Oral microbiome: possible harbinger for children's health. Int. J. Oral Sci. 12:12. doi: 10.1038/s41368-020-0082-x
Yamamura, K., Izumi, D., Kandimalla, R., Sonohara, F., Baba, Y., Yoshida, N., et al. (2019). Intratumoral Fusobacterium nucleatum levels predict therapeutic response to neoadjuvant chemotherapy in esophageal squamous cell carcinoma. Clin. Cancer Res. 25, 6170–6179. doi: 10.1158/1078-0432.CCR-19-0318
Yang, J., Mu, X., Wang, Y., Zhu, D., Zhang, J., Liang, C., et al. (2018). Dysbiosis of the salivary microbiome is associated with non-smoking female lung Cancer and correlated with immunocytochemistry markers. Front. Oncol. 8:520. doi: 10.3389/fonc.2018.00520
Yang, L., Li, A., Wang, Y., and Zhang, Y. (2023). Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct. Target. Ther. 8:35. doi: 10.1038/s41392-022-01304-4
Yang, Y., Long, J., Wang, C., Blot, W. J., Pei, Z., Shu, X., et al. (2022). Prospective study of oral microbiome and gastric cancer risk among Asian, African American and European American populations. Int. J. Cancer 150, 916–927. doi: 10.1002/ijc.33847
Yao, C., Lan, D., Li, X., Wang, Y., Qi, S., and Liu, Y. (2023). Porphyromonas gingivalis is a risk factor for the development of nonalcoholic fatty liver disease via ferroptosis. Microbes Infect. 25:105040. doi: 10.1016/j.micinf.2022.105040
Yao, Y., Shen, X., Zhou, M., and Tang, B. (2021). Periodontal pathogens promote Oral squamous cell carcinoma by regulating ATR and NLRP3 Inflammasome. Front. Oncol. 11:722797. doi: 10.3389/fonc.2021.722797
Yin, H., Zhang, J., Zhang, H., Li, Q., Qiu, H., Hong, K., et al. (2023). Fusobacterium nucleatum promotes proliferation in oesophageal squamous cell carcinoma via AHR/CYP1A1 signalling. FEBS J. 290, 837–854. doi: 10.1111/febs.16619
Yu, G., Torres, J., Hu, N., Medrano-Guzman, R., Herrera-Goepfert, R., Humphrys, M. S., et al. (2017). Molecular characterization of the human stomach microbiota in gastric Cancer patients. Front. Cell. Infect. Microbiol. 7:302. doi: 10.3389/fcimb.2017.00302
Yu, L., Zhao, G., Wang, L., Zhou, X., Sun, J., Li, X., et al. (2022). A systematic review of microbial markers for risk prediction of colorectal neoplasia. Br. J. Cancer 126, 1318–1328. doi: 10.1038/s41416-022-01740-7
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes Chemoresistance to colorectal Cancer by modulating autophagy. Cells 170, 548–563.e516. doi: 10.1016/j.cell.2017.07.008
Zhai, K., Ding, J., and Shi, H. Z. (2015). HPV and lung cancer risk: a meta-analysis. J. Clin. Virol. 63, 84–90. doi: 10.1016/j.jcv.2014.09.014
Zhang, L., Chen, X., Ren, B., Zhou, X., and Cheng, L. (2022). Helicobacter pylori in the Oral cavity: current evidence and potential survival strategies. Int. J. Mol. Sci. 23:13646. doi: 10.3390/ijms232113646
Zhang, N., Liu, Y., Yang, H., Liang, M., Wang, X., Wang, M., et al. (2021). Clinical significance of Fusobacterium nucleatum infection and regulatory T cell enrichment in esophageal squamous cell carcinoma. Pathol. Oncol. Res. 27:1609846. doi: 10.3389/pore.2021.1609846
Zhang, S., Kong, C., Yang, Y., Cai, S., Li, X., Cai, G., et al. (2020). Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics. 10, 11595–11606. doi: 10.7150/thno.49515
Zhang, W. L., Wang, S. S., Wang, H. F., Tang, Y. J., Tang, Y. L., and Liang, X. H. (2019). Who is who in oral cancer? Exp. Cell Res. 384:111634. doi: 10.1016/j.yexcr.2019.111634
Zhao, H., Chu, M., Huang, Z., Yang, X., Ran, S., Hu, B., et al. (2017). Variations in oral microbiota associated with oral cancer. Sci. Rep. 7:11773. doi: 10.1038/s41598-017-11779-9
Keywords: oral microbiota, cancer, tumor, inflammation, tumor microenvironment
Citation: Lan Z, Liu W-J, Cui H, Zou K-L, Chen H, Zhao Y-Y and Yu G-T (2023) The role of oral microbiota in cancer. Front. Microbiol. 14:1253025. doi: 10.3389/fmicb.2023.1253025
Edited by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandReviewed by:
Sunil Banskar, University of Arizona, United StatesArmando Rojas, Catholic University of the Maule, Chile
Copyright © 2023 Lan, Liu, Cui, Zou, Chen, Zhao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guang-Tao Yu, Z3Vhbmd0YW8xOTg2QHNtdS5lZHUuY24=; Yu-Yue Zhao, emhhb3l1eXVlQHdodS5lZHUuY24=
†These authors have contributed equally to this work
 Zhou Lan
Zhou Lan Wei-Jia Liu2†
Wei-Jia Liu2† Guang-Tao Yu
Guang-Tao Yu