- 1College of Bioscience and Biotechnology, Yangzhou University, Yangzhou, Jiangsu, China
- 2Jiangsu Key Laboratory of Zoonosis/Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, Jiangsu, China
- 3Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agri-food Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, Jiangsu, China
- 4Joint International Research Laboratory of Agriculture and Agri-Product Safety, Yangzhou University, Yangzhou, Jiangsu, China
Livestock-associated Staphylococcus aureus (LA-MRSA) has been of increasing concern due to its potential risk to humans. This study investigated the prevalence of MRSA in pig production in Eastern China and determined the genomic characteristics of pig-associated MRSA isolates by whole-genome sequencing (WGS). A total of 1,318 samples were collected from pig farms and pig slaughterhouses, and 150 S. aureus were identified, including 63 MRSA isolates and 87 MSSA isolates. MRSA was detected in all pig farms and pig slaughterhouses. The antimicrobial susceptibility test revealed that all MRSA isolates were multidrug-resistant. The WGS and MLST analysis demonstrated that 56 MRSA isolates belonged to clonal complex (CC) 398, and seven MRSA isolates belonged to CC9. All LA-MRSA isolates were absent of phiSa3 phage containing immune evasion cluster (IEC) and possessed an intact hlb gene. In addition, genes associated with Panton-Valentine leukocidin, typically indicative of human adaptation, were not detected. The analysis of antibiotic resistance genes (ARGs) demonstrated that all MRSA isolates contained multiple ARGs. All MRSA isolates had Plthe mecA gene and at least one tetracycline resistance gene. Both tetM and tetK were detected in all MRSA CC398 isolates, while tetL was detected in all MRSA CC9 isolates. The phenicol resistance gene fexA was detected in 51 MRSA isolates, while the linezolid resistance gene cfr was detected in 60 MRSA isolates. The emergence of LA-MRSA CC398 in four pig farms and one slaughterhouse in this study indicates the spread of this clonal complex in the pig production sector in Eastern China. Further investigations are required to understand the potential transmission routes of LA-MRSA CC398 within the pork production chain in China and to assess the potential risks to humans.
1 Introduction
Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) is an emerging animal-associated food safety problem in many parts of the world. Particular focus has been on pork production as pigs are universally known as the main reservoir for LA-MRSA. In Europe and North America, LA-MRSA clonal complex (CC) 398 is the predominant CC type, which is associated with the primary pig industry, and is also prevalent in poultry, cattle, sheep, and animal-origin meat products (Larsen et al., 2015; Sieber et al., 2018). Meanwhile, occupational exposure during livestock production has been regarded as the critical risk factor for LA-MRSA transmission in humans and the secondary spread to the community (Aires-De-Sousa, 2017). A European study demonstrated that 21.5% of LA-MRSA from clinical isolates belonged to CC398 in the Netherlands, while the ratio was around 9.7% in Belgium and Spain, 15.6% in Slovenia, and 16.7% in Demark (Kinross et al., 2017).
In China, CC9 is the predominant LA-MRSA CC type in pig production (Zou et al., 2022). However, LA-MRSA CC398 has already been sporadically reported in several surveillance studies in different provinces in China. A prevalence study has demonstrated that 38% of pig-associated S. aureus isolates belonged to LA-MRSA CC398, indicating the emergence of LA-MRSA CC398 in pig production (Cui et al., 2022). Besides, LA-MRSA CC398 has also been sporadically reported in pork and bulk milk in China (Monaco et al., 2013; Van De Sande-Bruinsma et al., 2015). This study primarily aimed to investigate the prevalence of MRSA in pig farms and slaughterhouses in Eastern China based on whole-genome sequencing (WGS) analysis. S. aureus isolates were collected from pig farms and slaughterhouses in Eastern China and subjected to WGS and MLST analyses. Since LA-MRSA CC398 has been widely associated with the pig industry and occupational infections in humans, we investigated the phylogenetic relationship of LA-MRSA CC398 isolates from this study to human-associated MRSA CC398 and LA-MRSA CC398 from both China and other countries. The phylogenetic analysis was performed to investigate the emergence and spread of LA-CC398 in the Chinese pig industry.
2 Materials and methods
2.1 Staphylococcus aureus isolation and identification
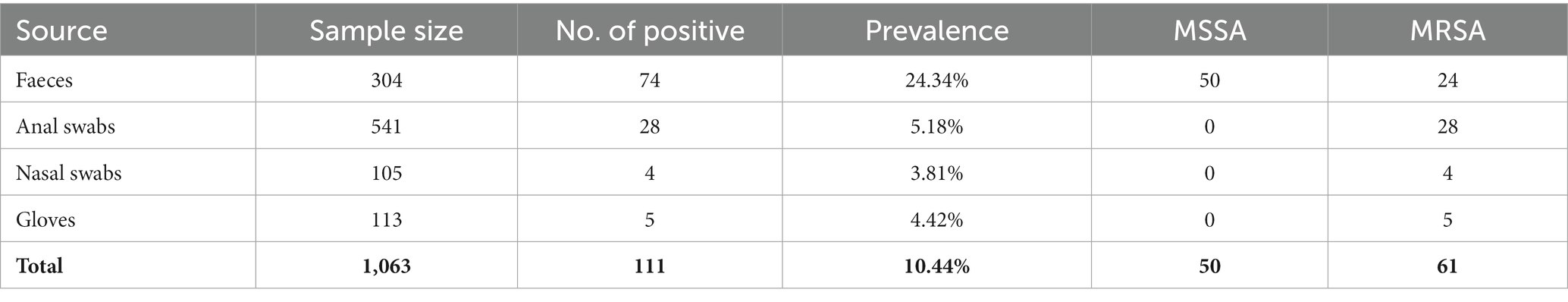
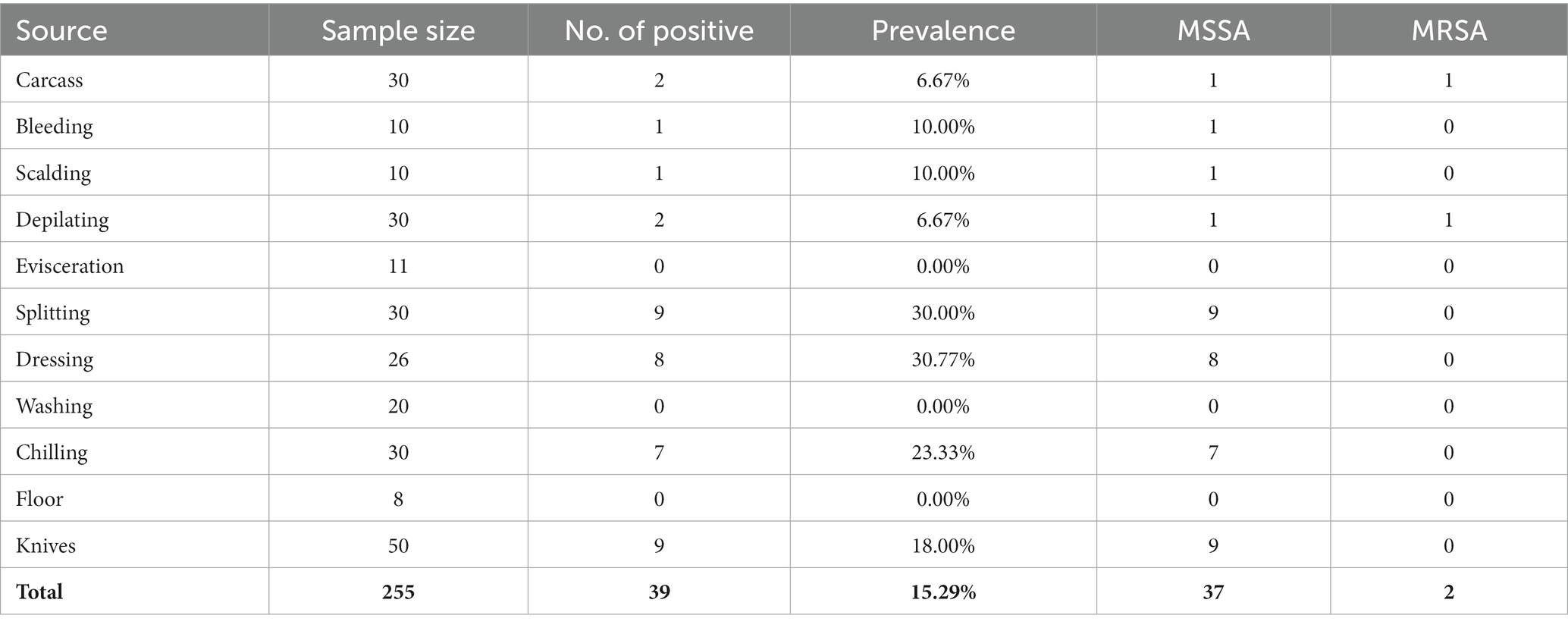
Samples were collected from 2018 to 2021 at four pig farms in Jiangsu province (farms A to D) and two pig slaughterhouses in Shanghai (slaughterhouses A and B), China. A total of 1,318 samples from pig farms were collected, including 304 fecal samples, 541 anal swab samples, 105 nasal swab samples, and 113 swab samples of gloves (Table 1). A total of 255 samples from two pig slaughterhouses were collected, including 30 samples from pig carcasses before slaughtering, 167 swab samples during the pig slaughtering process, 8 swab samples of the slaughtering floor, and 50 swab samples of knives during the slaughtering process (Table 2). All swab samples were stored in collection tubes containing Cary-Blair agar. Detailed sampling information is provided in Supplementary Table S1.
All samples were subjected to S. aureus isolation within 24 h, according to the literature previously described by Li et al. (2020). Briefly, each swab sample was enriched in 10 mL trypticase soy broth (TSB) with 6.5% NaCl and inoculated overnight at 37°C, 180 rpm. Ten μL enriched aliquots were inoculated on CHROMagar™ Staph aureus plates (CHROmagar, Paris, France) and incubated overnight at 37°C for the isolation of S. aureus. The presumptive S. aureus clones were identified by the presence of the spa gene, and MRSA was identified by the presence of the mecA or mecC gene (Tang et al., 2017).
2.2 Spa typing
All confirmed S. aureus isolates were analysed for spa typing (Supplementary Tables S2, S3). spa genes were detected with the primers spa-1113f and spa-1514f by the PCR program according to the previous description (Harmsen et al., 2003). The PCR product for each isolate was sequenced by Sanger sequencing (Genscript Biotech Corporation, Nanjing, China). The spa type was analysed by the Ridom Spa Server database (http://spaserver.ridom.de/, accessed on 4 February 2022).
2.3 Antimicrobial susceptibility test for MRSA
Antimicrobial susceptibility of all MRSA isolates was performed by disk diffusion according to the guidelines of the Clinical and Laboratory Standards Institute (2020). MRSA isolates from overnight TSA plates were resuspended in 0.9% saline to achieve the turbidity of 0.5 McFarland standard. The suspended cultures were evenly spread on the M.H. agar plate, and the antibiotic disk was tightly on the agar surface before being inoculated overnight at 37°C. The diameter of the antibiotic-resistant halo was measured, and results were interpreted according to the CLSI standard. A total of 10 antibiotics (Oxoid™, United Kingdom) were included: cefoxitin (FOX, 30 μg), chloramphenicol (C, 30 μg), ciprofloxacin (CIP, 5 μg), clindamycin (DA, 2 μg), erythromycin (E, 15 μg), gentamicin (C.N., 10 μg), linezolid (LZD, 10 μg), penicillin G (P, 10 units), tetracycline (T.E., 30 μg), and trimethoprim/sulfamethoxazole (SXT, 1:19, 25 μg). S. aureus ATCC25923 was included for quality control.
2.4 Whole genome sequencing of MRSA and comparative genomic analysis
All MRSA isolates were whole genome sequenced (WGS) by the Nova seq6000 (Illumina, the US). Raw reads were trimmed and filtered by the NGSQC toolkit and assembled with de novo assembly by SPAdes 3.15.5 (Bankevich et al., 2012). WGS data were submitted to the European Nucleotide Archive database (ENA) with the accession number PRJEB62143. The multi-locus sequence typing (MLST) was obtained by submitting the WGS data of each MRSA isolate to the database (https://pubmlst.org/saureus/, accessed on July 30, 2022). All MRSA isolates were tested for the core genome SNP analysis, and the genetic relationships of MRSA were revealed by the phylogenetic tree. To compare our MRSA CC398 isolates to the isolates from other countries, we downloaded the isolates data from Price et al. (2012). The phylogenetic tree of all MRSA isolates in this study and phylogeny was constructed by Parsnp v1.7.4 (https://harvest.readthedocs.io/en/latest/content/parsnp.html, accessed on August 3, 2022). Antimicrobial resistance genes of MRSA were detected by the ResFinder 4.0 database (Florensa et al., 2022). Virulence factors of all MRSA isolates were analysed by BLASTn against the Virulence Factors Database (VFDB) with >80% sequencing homology and coverage (Chen et al., 2005). Genes encoding virulent factors included scn encoding staphylococcal complement inhibitor; chp encoding chemotaxin inhibitory protein; sak encoding staphylokinase; sea encoding staphylococcal enterotoxin toxin A; hlb encoding β-hemolysin; lukF-PV and lukS-PV encoding Panton-Valentine leucocidin (PVL); tsst-1 encoding toxic shock syndrome toxin-1.
3 Results
3.1 Prevalence of Staphylococcus aureus in early-stage pig production
A total of 1,318 samples were collected from pig farms and pig slaughterhouses. In total, 11.38% of the samples were positive for S. aureus. MRSA was detected in pig samples from all farms and slaughterhouses, and no animal delivery occurred between farms and slaughterhouses (Supplementary Table S1). MSSA was detected in farm D, slaughterhouse A, and slaughterhouse B (Supplementary Table S1). In total, 63 MRSA isolates and 77 MSSA isolates were identified. The MLST analysis demonstrated that 56 MRSA isolates belonged to CC398, while 7 MRSA isolates belonged to CC9.
S. aureus was isolated from 10.44% of the farm samples, including 50 methicillin-susceptible S. aureus (MSSA) isolates and 61 MRSA isolates (Table 1). The distribution of S. aureus in different samples is diverse, including 24.34% from feces, 5.18% from anal swabs, 3.81% from nasal swabs, and 4.42% from gloves for workers (Table 1). At the slaughtering stage, S. aureus was isolated from 39 of 225 samples with a prevalence of 15.29%, including 27 MSSA isolates and 2 MRSA isolates (Table 2). Samples from splitting and dressing had the highest prevalence of S. aureus approximately 30%, followed by swab samples during washing, which had a prevalence of 26.67%. During the chilling step, the prevalence of S. aureus was 23.33% (Table 2). Other slaughtering processes with S. aureus positive included pig carcasses before slaughtering, bleeding, scalding, depilation, and chilling (Table 2). It should be noted that 18% of knives during the slaughtering process were S. aureus positive (Table 2).
3.2 Antibiotic resistance of MRSA isolates from pig production
Antibiotic susceptibility tests showed that all MRSA isolates were resistant to cefoxitin and penicillin. Most MRSA isolates were resistant to clindamycin (95.24%) and chloramphenicol (76.19%), while some were resistant to erythromycin (49.21%). Two MRSA isolates showed resistance to gentamicin (3.17%). One MRSA isolate YZU 4385 displayed resistance to linezolid (1.59%), while YZU4403 showed resistance to trimethoprim/sulfamethoxazole (1.59%) (Supplementary Table S4). A total of 20 multidrug-resistant (MDR) patterns were observed, with the predominant pattern of FOX-C-DA-P-TE, followed by FOX-C-DA-E-P-TE. MDR patterns of each MRSA isolate are illustrated in Supplementary Table S4.
3.3 Genomic characterisation of Staphylococcus aureus isolates from pig farms and pig slaughterhouses
In total, 13 spa types were detected in all S. aureus isolates (Figure 1; Supplementary Tables S2, S3). Most MRSA isolates (71.4%) belonged to spa type t034, followed by 11.5% belonging to t899, and 8.2% belonging to t011 (Supplementary Table S2). Eight spa types were detected in MSSA isolates, of which spa type t286 was predominant with a ratio of 32.2%, followed by spa type t571 (26.4%) and t899 (17.2%) (Supplementary Table S3).
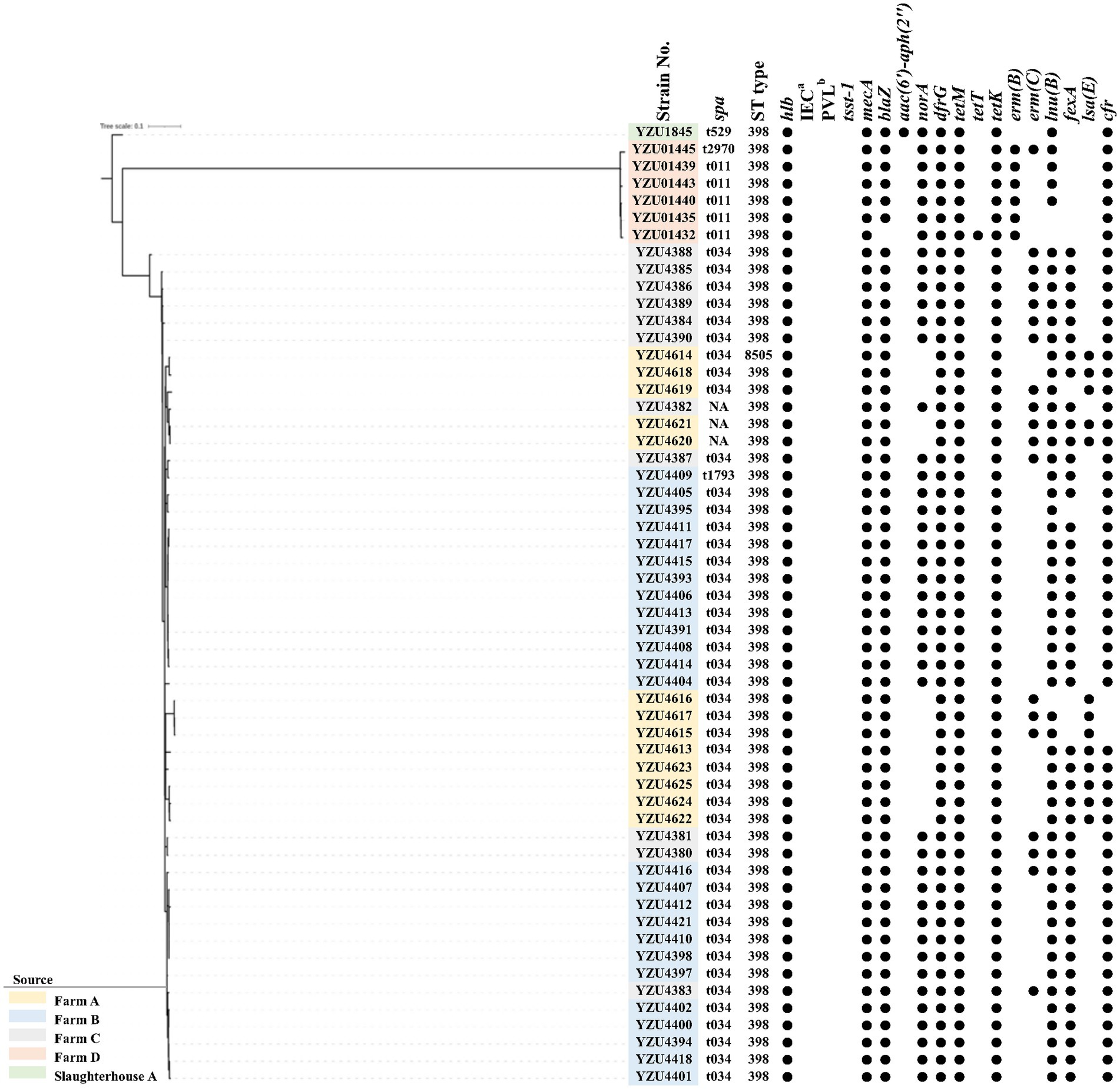
Figure 1. The phylogenetic tree of MRSA CC398 in terms of the core genome multi-locus sequence type (cgSNP). The filled circle (●) represents the presence of a gene. aimmune evasion cluster (IEC). bthe Panton-Valentine leucocidin (PVL).
MLST analysis showed that MRSA isolates were grouped into two CCs, of which 56 isolates belonged to CC398 and 7 isolates belonged to CC9. MRSA CC398 isolates contained two ST types, of which 55 MRSA isolates belonged to ST398, and one isolate was a new ST type ST8505 (Figure 1). The phylogenetic analysis divided the MRSA CC398 isolates into three clusters with a predominant cluster of 44 MRSA t034/ST398 isolates, 1 t1794/ST398 isolate, and 3 ST398 isolates without spa type. MRSA isolates t011/ST398 and t2970/ST398 were grouped into a cluster with all isolates from farm D, while MRSA t529/ST398 were from slaughterhouse A (Figure 1). The phylogenetic analysis divided MRSA CC9 into two clusters, of which one isolate with t899/ST1378 was detected from pig carcasses in slaughterhouse B, and the other cluster with MRSA t899/ST9 isolates was from farm B and farm D (Figure 2). Virulence factors were analysed in all MRSA isolates from pig farms and slaughterhouses.
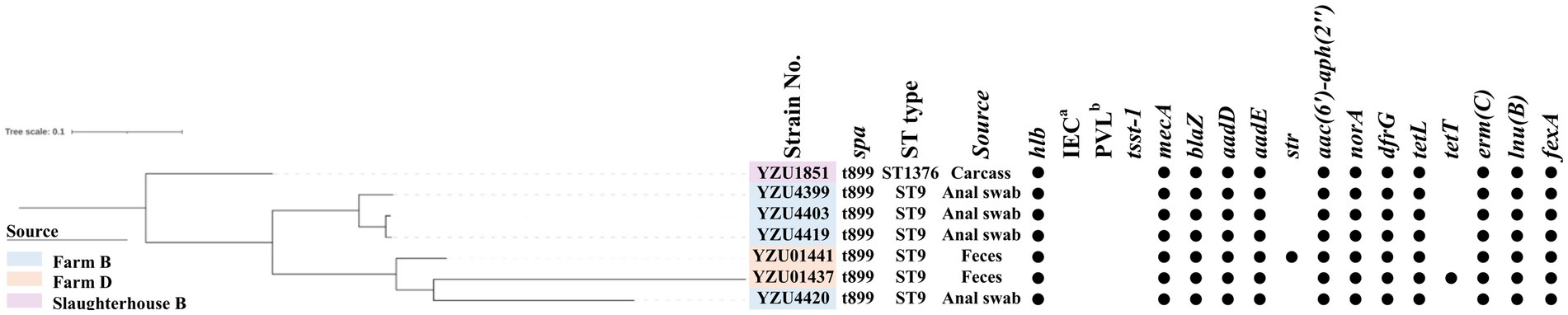
Figure 2. The phylogenetic tree of MRSA CC9 in terms of the core genome multi-locus sequence type (cgMLST). The filled circle (●) represents the presence of a gene. aimmune evasion cluster (IEC). b the Panton-Valentine leucocidin (PVL).
In total, 11 antibiotic resistance genes (ARGs) were detected from 56 MRSA CC398 isolates, while 18 ARGs were detected from 7 MRSA CC9 isolates. All MRSA isolates contain the mecA gene encoding the low-affinity penicillin-binding protein PBP 2a. Both blaZ encoding β-lactamase and dfrG for trimethoprim resistance were presented in 62 out of 63 MRSA isolates. Tetracycline-resistant genes were identified in all MRSA isolates, of which CC398 isolates harboured tetM, tetT, and tetK, while CC9 isolates harboured tetT and tetL. The emr(C) gene for the erythromycin resistance was present in 19 out of 55 MRSA CC398 isolates and all MRSA CC9 isolates, while emr(B) was only detected in six CC398 isolates. Aminoglycoside-resistant genes aadD and aadE were detected in CC9 isolates, while aac(6′)-aph(2″) was detected in all CC9 isolates and one CC398 isolate. The norA gene was detected in 43 out of 56 MRSA CC398 isolates and all MRSA CC9 isolates, which were associated with fluoroquinolone resistance in S. aureus. The lsa(B) gene encoding the ABC transporter for low-level clindamycin resistance was presented in 13 MRSA CC398 isolates. The macrolide-resistant gene lnu(B) was detected in 53 MRSA CC398 isolates and 6 MRSA CC9 isolates. Phenicol resistance genes fexA were detected in both CC398 and CC9 isolates, while the cfr gene was only detected in CC398 isolates.
To evaluate the transmission of MRSA CC398 among countries, we also conducted a core genome-based phylogenetic analysis of the 56 MRSA CC398 isolates and 67 MRSA CC398 isolates from a previous study (Price et al., 2012) (Supplementary Figure S1). The results showed that the MRSA CC398 isolates were grouped into two clusters with LA-MRSA CC398 isolates from Europe, which were genetically different from CC398 isolates from humans in China. All MRSA CC398 isolates were absent of phiSa3 carrying chp, scn, and sak for human-immune evasion cluster and contained an intact hlb gene in this study. Similar to MRSA from other countries, the tet(M) gene was present in all MRSA CC398 isolates from this study (Figures 1 and Supplementary Figure S1). Besides, lukF-PV and lukS-PV encoding PVL and tsst-1 encoding toxic shock syndrome toxin-1 were also absent in all MRSA isolates (Figure 1).
4 Discussion
This study investigated the distribution of MRSA and MSSA from pig farms and slaughterhouses in Eastern China. In total, 11.38% of pig-associated samples were S. aureus positive. MRSA was detected in all four pig farms and two pig slaughterhouses, with a prevalence of 10.44% in pig farms and 15.29% in pig slaughterhouses, respectively. Other surveillance studies showed that the occurrence of MRSA in pig production was diverse in different locations in China, which is 49% in Shandong, 45% in Shanghai, 19.4% in Qinghai, and 5.17% in Henan (Li et al., 2017; Sun et al., 2019; Cui et al., 2022). The prevalence of MRSA in pig production was diverse in Western countries, with 77% in the US, 80% in Denmark, 69.7% in Belgium, and 99.5% in the Netherlands (Crombé et al., 2012; Sun et al., 2015; Dierikx et al., 2016; Schulz et al., 2019). In Japan, MRSA was detected among pigs in slaughterhouses, diseased pigs on farms, imported breeding pigs, and farm dust, with a prevalence of 5.2, 3.4, 28.8, and 0.06%, respectively (Ozawa et al., 2022). This discrepancy in MRSA in pig production suggests the diversity of MRSA distribution across different geographical locations.
WGS analysis has demonstrated that 56 out of 63 MRSA isolates in this study belonged to CC398, whereas only seven of the MRSA isolates belonged to CC9. Remarkably, MRSA CC398 was detected in all pig farms as well as in slaughterhouse A, indicating the widespread dissemination of LA-MRSA CC398 in the pig production sector of Eastern China. A similar study conducted in Qinghai province, China, detected 26 out of 67 MRSA CC398 isolates with spa type t011 (Cui et al., 2022), whereas this study observed the predominant LA-MRSA CC398 with spa type t034. In addition to the above observations, it is worth noting that the dominant clone of pig-associated LA-MRSA in China belongs to LA-MRSA CC9. Li et al. (2017) conducted a pig-associated MRSA study in four provinces in China, and all 270 MRSA isolates were classified as CC9. Furthermore, a large-scale LA-MRSA study in Shandong, China, revealed that out of 92 MRSA isolates obtained from pig samples, 91 were LA-MRSA CC9, and one isolate was LA-MRSA CC398 through WGS analysis (Sun et al., 2019). We further analysed the phylogenetic relationship of MRSA CC398 isolates in this study genome with MRSA CC398 isolates from Price et al. (2012). The results showed that all MRSA CC398 isolates from this study were closely related to the MRSA CC398 isolates from pig samples all over the world (Supplementary Figure S1). This study observed that LA-MRSA CC398 was predominated in pig farms in Eastern China for the first time, and the close phylogenetic relationship of MRSA CC398 isolates in this study indicated an emergency of global MRSA ST398 in domestic pig production.
The multidrug-resistant phenotype of MRSA isolates in this study was similar to that of MRSA strains from livestock and humans (Li et al., 2017; Cui et al., 2020; Kruger-Haker et al., 2023). All isolates were resistant to cefoxitin and tetracycline with the presence of the mecA gene, tet(M), tet(K), tet(L), or tet(T), which are correlated to the LA-MRSA CC398 and CC9 antimicrobial characteristics (Kock et al., 2010). Most isolates were resistant to clindamycin (95.24%) and chloramphenicol (76.19%), which were well matched with the presence of lnu(B) and fexA. Thirteen LA-MRSA CC398 isolates harboured lnu(B) plus lsa(E) encoding lincomycin and clindamycin resistance, which was detected in a previous study in the UK (Kruger-Haker et al., 2023). The presence of the fexA gene in LA-MRSA CC398 was reported for the first time in China, while a previous study demonstrated a low prevalence of fexA in CC398 in the UK (Kruger-Haker et al., 2023). A previous study demonstrated that the prevalence of fexA in MRSA CC9 isolates was 69.1% (Yu et al., 2021). The phenicol-lincosamide-oxazolidinone-pleuromutilin-streptogramin resistance gene cfr was highly prevalent in LA-MRSA CC398 isolates, and one isolate showed resistance to linezolid. Although clindamycin and linezolid are not sanctioned in pig farming, the extensive usage of other antibiotics, such as lincomycin, florfenicol, and tiamulin in food-producing animals can exert selective pressure, thereby fostering the dissemination of the cfr gene (Schouls et al., 2022; Kruger-Haker et al., 2023). Eight MRSA isolates contained aminoglycoside-resistant gene aac(6′)-aph(2″), and two isolates showed resistance to gentamicin. The emergence and proliferation of these multidrug-resistant isolates in pigs consequently impose a substantial risk upon individuals with sustained contact with livestock.
5 Conclusion
This study investigated the prevalence of S. aureus in pig production in Eastern China. MRSA has been detected in all pig farms and slaughterhouses. The high prevalence of LA-MRSA CC398 in both pig farms and slaughterhouses has provided a significant indication of the emergence and spread of LA-MRSA CC398 in the Chinese pig production industry. The phylogenetic analysis demonstrated a close relationship between LA-MRSA CC398 isolates in this study and isolates from pig samples in other countries, indicating that international trade could be a potential for LA-MRSA CC398 transmission. Further studies are required to comprehensively understand LA-MRSA CC398 in the pig industry and its potential risks to livestock and humans in the context of pork production in China.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by the Research Ethics Committee of Yangzhou University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LZ: Data curation, Investigation, Methodology, Writing – original draft. ZJ: Data curation, Investigation, Methodology, Writing – original draft. ZW: Data curation, Formal analysis, Software, Writing – review & editing. YL: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing. XJ: Resources, Supervision, Writing – review & editing. QL: Resources, Validation, Writing – review & editing. YT: Conceptualization, Formal analysis, Funding acquisition, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Youth Foundation of Jiangsu Province (BK20190883), the University Natural Science Research Project of Jiangsu Province (19KJB230007), and the Foundation of Green Young and Golden Phenix of Yangzhou.
Acknowledgments
The authors thank Ruoyun Ji from the Faculty of Bioscience and Biotechnology, Yangzhou University, for the sample collection and technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1267885/full#supplementary-material
References
Aires-De-Sousa, M. (2017). Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin. Microbiol. Infect. 23, 373–380. doi: 10.1016/j.cmi.2016.11.002
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Clinical and Laboratory Standards Institute (2020). Performance standards for antimicrobial susceptibility testing : 30th (Wayne, PA).
Crombé, F., Willems, G., Dispas, M., Hallin, M., Denis, O., Suetens, C., et al. (2012). Prevalence and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus among pigs in Belgium. Microb. Drug Resist. 18, 125–131. doi: 10.1089/mdr.2011.0138
Cui, M., Ali, T., Li, J., Song, L., Shen, S., Li, T., et al. (2022). New clues about the global MRSA ST398: emergence of MRSA ST398 from pigs in Qinghai China. Int. J. Food Microbiol. 378:109820. doi: 10.1016/j.ijfoodmicro.2022.109820
Cui, M., Li, J., Ali, T., Kalim, K., Wang, H., Song, L., et al. (2020). Emergence of livestock-associated MRSA ST398 from bulk tank milk, China. J. Antimicrob. Chemother. 75, 3471–3474. doi: 10.1093/jac/dkaa367
Dierikx, C. M., Hengeveld, P. D., Veldman, K. T., de Haan, A., van der Voorde, S., Dop, P. Y., et al. (2016). Ten years later: still a high prevalence of MRSA in slaughter pigs despite a significant reduction in antimicrobial usage in pigs the Netherlands. J. Antimicrob. Chemother. 71, 2414–2418. doi: 10.1093/jac/dkw190
Florensa, A. F., Kaas, R. S., Clausen, P., Aytan-Aktug, D., and Aarestrup, F. M. (2022). ResFinder - an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 8:000748. doi: 10.1099/mgen.0.000748
Harmsen, D., Claus, H., Witte, W., Rothganger, J., Claus, H., Turnwald, D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003
Kinross, P., Petersen, A., Skov, R., Van Hauwermeiren, E., Pantosti, A., Laurent, F., et al. (2017). Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European economic area countries, 2013. Euro Surveill. 22, 16–00696. doi: 10.2807/1560-7917.ES.2017.22.44.16-00696
Kock, R., Becker, K., Cookson, B., Van Gemert-Pijnen, J. E., Harbarth, S., Kluytmans, J., et al. (2010). Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 15:19688. doi: 10.2807/ese.15.41.19688-en
Kruger-Haker, H., Ji, X., Hanke, D., Fiedler, S., Fessler, A. T., Jiang, N., et al. (2023). Genomic diversity of methicillin-resistant Staphylococcus aureus CC398 isolates collected from diseased swine in the German national resistance monitoring program GERM-vet from 2007 to 2019. Microbiol. Spectr. 11:e0077023. doi: 10.1128/spectrum.00770-23
Larsen, J., Petersen, A., Sørum, M., Stegger, M., van Alphen, L., Valentiner-Branth, P., et al. (2015). Meticillin-resistant Staphylococcus aureus CC398 is an increasing cause of disease in people with no livestock contact in Denmark, 1999 to 2011. Euro. Surveill. 20:10.2807/1560-7917.ES.2015.20.37.30021. doi: 10.2807/1560-7917.es.2015.20.37.30021
Li, J., Jiang, N., Ke, Y., Fessler, A. T., Wang, Y., Schwarz, S., et al. (2017). Characterization of pig-associated methicillin-resistant Staphylococcus aureus. Vet. Microbiol. 201, 183–187. doi: 10.1016/j.vetmic.2017.01.017
Li, Y., Tang, Y., Ren, J., Huang, J., Li, Q., Ingmer, H., et al. (2020). Identification and molecular characterization of Staphylococcus aureus and multi-drug resistant MRSA from monkey faeces in China. Transbound. Emerg. Dis. 67, 1382–1387. doi: 10.1111/tbed.13450
Monaco, M., Pedroni, P., Sanchini, A., Bonomini, A., Indelicato, A., and Pantosti, A. (2013). Livestock-associated methicillin-resistant Staphylococcus aureus responsible for human colonization and infection in an area of Italy with high density of pig farming. BMC Infect. Dis. 13:258. doi: 10.1186/1471-2334-13-258
Ozawa, M., Furuya, Y., Akama, R., Harada, S., Matsuda, M., Abo, H., et al. (2022). Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolated from pigs in Japan. Vet. Microbiol. 273:109523. doi: 10.1016/j.vetmic.2022.109523
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in Livestock. mBio 3:e00305-1. doi: 10.1128/mbio.00305-11
Schouls, L. M., Veldman, K., Brouwer, M. S. M., Dierikx, C., Witteveen, S., Van Santen-Verheuvel, M., et al. (2022). Cfr and fexA genes in methicillin-resistant Staphylococcus aureus from humans and livestock in the Netherlands. Commun. Med. 2:135. doi: 10.1038/s43856-022-00200-w
Schulz, J., Boklund, A., Toft, N., and Halasa, T. (2019). Effects of control measures on the spread of LA-MRSA among Danish pig herds between 2006 and 2015 – a simulation study. Sci. Rep. 9:691. doi: 10.1038/s41598-018-37075-8
Sieber, R. N., Skov, R. L., Nielsen, J., Schulz, J., Price, L. B., Aarestrup, F. M., et al. (2018). Drivers and dynamics of methicillin-resistant livestock-associated Staphylococcus aureus CC398 in pigs and humans in Denmark. mBio. 9. doi: 10.1128/mbio.02142-18
Sun, C. T., Chen, B. L., Hulth, A., Schwarz, S., Ji, X., Nilsson, L. E., et al. (2019). Genomic analysis of Staphylococcus aureus along a pork production chain and in the community, Shandong Province, China. Int. J. Antimicrob. Agents 54, 8–15. doi: 10.1016/j.ijantimicag.2019.03.022
Sun, J., Yang, M., Sreevatsan, S., and Davies, P. R. (2015). Prevalence and characterization of Staphylococcus aureus in growing pigs in the USA. PLoS ONE. 10:e0143670. doi: 10.1371/journal.pone.0143670
Tang, Y., Larsen, J., Kjeldgaard, J., Andersen, P. S., Skov, R., and Ingmer, H. (2017). Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 249, 72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001
Van De Sande-Bruinsma, N., Leverstein Van Hall, M. A., Janssen, M., Nagtzaam, N., Leenders, S., De Greeff, S. C., et al. (2015). Impact of livestock-associated MRSA in a hospital setting. Antimicrob. Resist. Infect. Control 4:11. doi: 10.1186/s13756-015-0053-8
Yu, F., Cienfuegos-Gallet, A. V., Cunningham, M. H., Jin, Y., Wang, B., Kreiswirth, B. N., et al. (2021). Molecular evolution and adaptation of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) sequence type 9. mSystems 6:e0049221. doi: 10.1128/mSystems.00492-21
Keywords: Staphylococcus aureus , LA-MRSA, pig production, clonal complex 398, antibiotic resistance
Citation: Zheng L, Jiang Z, Wang Z, Li Y, Jiao X, Li Q and Tang Y (2023) The prevalence of Staphylococcus aureus and the emergence of livestock-associated MRSA CC398 in pig production in eastern China. Front. Microbiol. 14:1267885. doi: 10.3389/fmicb.2023.1267885
Edited by:
Chunlei Shi, Shanghai Jiao Tong University, ChinaReviewed by:
Raphael Niklaus Sieber, State Serum Institute (SSI), DenmarkSilpak Biswas, Calcutta School of Tropical Medicine, India
Copyright © 2023 Zheng, Jiang, Wang, Li, Jiao, Li and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyue Tang, dGFuZ3l5QHl6dS5lZHUuY24=
†These authors have contributed equally to this work
 Lina Zheng
Lina Zheng Zhongyi Jiang2,3†
Zhongyi Jiang2,3† Xinan Jiao
Xinan Jiao Qiuchun Li
Qiuchun Li Yuanyue Tang
Yuanyue Tang