- 1NHC Key Laboratory of Food Safety Risk Assessment, China National Center for Food Safety Risk Assessment, Beijing, China
- 2Qingdao Municipal Center for Disease Control and Prevention, Qingdao Institute of Preventive Medicine, Qingdao, China
- 3Pharmaceutical Department, Qingdao Traditional Chinese Medicine Hospital (Qingdao Hiser Hospital) Qingdao Hiser Hospital Affiliated of Qingdao University, Qingdao, China
- 4Key Laboratory of Alkene-carbon Fibres-based Technology and Application for Detection of Major Infectious Diseases, MOE Key Laboratory of Geriatric Diseases and Immunology, Pasteurien College, Suzhou Medical College, Soochow University, Suzhou, China
- 5Center for Food Safety, University of Georgia, Griffin, GA, United States
- 6Guangdong University of Technology, Guangzhou, China
Salmonella, especially antimicrobial resistant strains, remains one of the leading causes of foodborne bacterial disease. Retail chicken is a major source of human salmonellosis. Here, we investigated the prevalence, antimicrobial resistance (AMR), and genomic characteristics of Salmonella in 88 out of 360 (24.4%) chilled chicken carcasses, together with 86 Salmonella from humans with diarrhea in Qingdao, China in 2020. The most common serotypes were Enteritidis and Typhimurium (including the serotype I 4,[5],12:i:-) among Salmonella from both chicken and humans. The sequence types were consistent with serotypes, with ST11, ST34 and ST19 the most dominantly identified. Resistance to nalidixic acid, ampicillin, tetracycline and chloramphenicol were the top four detected in Salmonella from both chicken and human sources. High multi-drug resistance (MDR) and resistance to third-generation cephalosporins resistance were found in Salmonella from chicken (53.4%) and humans (75.6%). In total, 149 of 174 (85.6%) Salmonella isolates could be categorized into 60 known SNP clusters, with 8 SNP clusters detected in both sources. Furthermore, high prevalence of plasmid replicons and prophages were observed among the studied isolates. A total of 79 antimicrobial resistant genes (ARGs) were found, with aac(6′)-Iaa, blaTEM-1B, tet(A), aph(6)-Id, aph(3″)-Ib, sul2, floR and qnrS1 being the dominant ARGs. Moreover, nine CTX-M-type ESBL genes and the genes blaNMD-1, mcr-1.1, and mcr-9.1 were detected. The high incidence of MDR Salmonella, especially possessing lots of mobile genetic elements (MGEs) in this study posed a severe risk to food safety and public health, highlighting the importance of improving food hygiene measures to reduce the contamination and transmission of this bacterium. Overall, it is essential to continue monitoring the Salmonella serotypes, implement the necessary prevention and strategic control plans, and conduct an epidemiological surveillance system based on whole-genome sequencing.
1 Introduction
Salmonella is one of the common pathogens causing sporadic cases or outbreaks of gastroenteritis (Jain et al., 2020). In 2010, there were an estimated 153 million cases of non-typhoidal Salmonella enteric infections worldwide, of which about 50% were foodborne (Kirk et al., 2015; WHO, 2022). In the United States, foodborne salmonellosis causes an estimated 212,500 infections and 90 deaths annually (CDC, 2019). In China, Salmonella is the second most common bacteria causing foodborne outbreaks (Sun et al., 2021). In fact, consuming contaminated foods, especially poultry meat, directly threatens human health, which appears to be one of the major sources of human infection (Heredia and García, 2018; Pan et al., 2019; Yang et al., 2020). Poultry meat in the food production supply chain have been frequently associated with human salmonellosis cases and are an important cause of Salmonella transmission between poultry farms and humans (Antunes et al., 2016; Rincón-Gamboa et al., 2021). Furthermore, Salmonella is also a significant repository of antimicrobial resistance genes, posing substantial challenges to public health and security (Jajere, 2019).
Over recent years, resistance to cephalosporins have been increasingly reported in humans and poultry industry and the contribution of poultry products to the dissemination of extended-spectrum β-lactamase (ESBL) producing Salmonella strains, and the associated dangers for human health, is well documented in certain countries (Antunes et al., 2016). Of concern is the increased incidence of infections caused by ESBL-producing organisms, including Salmonella, because they are resistant not only to most of the β-lactam antimicrobials but also to other antimicrobial classes, leaving few treatment options and the potential for clinical outcomes worse than those of infections caused by non-ESBL-producing strains (Wei et al., 2019; Wang et al., 2020a). Meanwhile, carbapenems and colistin are often used as the last-line treatment for infections caused by multidrug-resistant (MDR), especially ESBL-producing gram-negative bacteria. However, selective pressure from the overuse or misuse of both antimicrobials has resulted in the emergence of carbapenem- and colistin-resistant Enterobacteriaceae (CRE) (Nang et al., 2019; Octavia et al., 2020). Regarding Salmonella isolates, recent studies have demonstrated their resistance to these critical antimicrobial drugs (Wang et al., 2017; Fan et al., 2022; Zhang et al., 2022b).
The New Delhi metallo (NDM)-β-lactamases that can hydrolyze almost all β-lactam antibiotics, have become one of the most commonly reported carbapenemase resistance mechanisms worldwide (Poirel et al., 2010). According to the Comprehensive Antibiotic Resistance Database (CARD1), 43 variants of NDM genes have been identified in 2022, with NDM-1 and NDM-5 being particularly widespread among Enterobacteriaceae including Salmonella (Iovleva and Doi, 2017). Colistin resistance in bacteria has become another significant threat to food safety and public health, and its development was mainly attributed to the plasmid-mediated mcr genes. Currently, a variety of mcr genes, including mcr-1 to -10 have been reported with mcr-9.1 being prevalent in Salmonella (Ling et al., 2020; Hussein et al., 2021). Hence, the increasing antimicrobial resistance in Salmonella needs to be monitored.
Our previous investigation has described the emergence of the blaNDM-1 gene in Salmonella recovered from chicken carcass in slaughterhouse, while the mcr genes were also detected from living chicken in Qingdao (Wang et al., 2017; Zou et al., 2021). However, there is limited available information on the surveillance and genomics studies of Salmonella in retail chicken in Qingdao. In the current study, we therefore investigated the prevalence, AMR, and genomic characteristics of Salmonella from chilled chicken carcasses and humans with diarrhea in Qingdao. Specially, we identified four Salmonella isolates carrying the plasmid-borne mcr-1.1, mcr-9.1, and NDM-1 genes in this study.
2 Materials and methods
2.1 Bacterial isolation
A total of 360 chilled chicken carcasses were collected from local markets in Qingdao, China in 2020. All samples were subjected to qualitative analysis for Salmonella using an enrichment method described by the National Food Safety Standard of China-Food microbiological examination, Salmonella (GB 4789.4–2016). Finally, presumptive Salmonella was selected for biochemical confirmation using API 20E test identification test strips (bioMérieux, Marcy l′ Etoile, France), as well as for molecular identification using PCR assay targeting the invA gene (Ed-Dra et al., 2018). In addition, 86 Salmonella isolates from humans with diarrhea collected from enteric clinic settings in the study year in Qingdao, were also included in the study (Supplementary Table S1). The isolates were serotyped using O- and H-antigens by slide agglutination with hyperimmune sera, and the serotypes of Salmonella samples were identified following the Kauffmann-White scheme (Grimont and Weill, 2007). All confirmed Salmonella isolates were stored in brain heart infusion broth with 40% glycerol (Land Bridge, Beijing, China) at −80°C. One isolate was retained from each sample. All the procedures performed in studies involving human participants were in accordance with the ethical standards of the Research Ethics Committee of China National Center of Food Safety Risk Assessment, Beijing, China (approval no. 2014003).
2.2 Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) of the Salmonella isolates was evaluated using the broth dilution method by the Biofosun Gram-negative panels (Shanghai Biofosun Biotech, China) by the manufacturer’s instructions. The MICs of 13 antimicrobial agents, including ampicillin (AMP), ceftazidime (CAZ), cefotaxime (CTX), cefoxitin (CFX), imipenem (IPM), meropenem (MEM), trimethoprim-sulfamethoxazole (SXT), gentamicin (GEN), tetracycline (TET), ciprofloxacin (CIP), nalidixic acid (NAL), chloramphenicol (CHL), and polymyxin E (CT) were determined, and the results were interpreted using Clinical and Laboratory Standards Institute guidelines (CLSI, 2017). Escherichia coli ATCC 25922 was used as quality control. Isolates showed resistance to all tested antimicrobials in this study were defined as pan-resistance, while isolates showed resistance to at least three classes of antimicrobials were defined as MDR.
2.3 Whole-genome sequencing and in silico analysis
Whole-genome sequencing of the Salmonella isolates from both chicken and humans was carried out using the Illumina NovaSeq PE150 at the Beijing Novogene Bioinformatics Technology Co., Ltd. Trimmomatic (Bolger et al., 2014), FastQC,2 SPAdes v3.14 (Bankevich et al., 2012), and Prokka v1.14.5 (Seemann, 2014) were used for reads quality control, assembly, and annotation. MLST v2.19.03 was used for identifying the sequence type (ST). SeqSero2 v1.2.14 was used for identifying the Salmonella serotypes (Zhang et al., 2019). Antimicrobial resistance genes (ARGs) were identified using ResFinder v4.0 (Bortolaia et al., 2020). Virulence genes were identified by ABRicate v1.015 using the VFDB database (Liu et al., 2022) with 80% identity and 80% query coverage cutoffs. Plasmid replicon types were identified using PlasmidFinder v2.1 (Carattoli and Hasman, 2020). Prophage predictions were carried out using PHASTER to explore intact prophages (Arndt et al., 2016). Pan-genome analysis was performed using Roary v3.13.0 (Page et al., 2015). Core genome phylogeny was built using PhyML v3.3.20200621 (Guindon et al., 2010). The gene presence/absence of ARGs, virulence genes, plasmid replication types, and prophage types, along with the phylogenetic tree were visualized through R script (v3.6.2) with the package ggtree (Yu, 2020). The genomes were submitted to EnteroBase for cgMLST profiling (Zhou et al., 2020) and submitted to NCBI Pathogen Detection portal for SNP clustering (Timme et al., 2020).
2.4 Statistical analysis
The Chi-square test was performed to analyze differences between groups. Data analysis was performed using SPSS 20.0 (SPSS, Chicago, United States). All statistical tests were two-sided; p < 0.05 were considered statistically significant.
2.5 Data availability statement
The sequences obtained in this study have been deposited in the NGDC Genome Sequence Archive6 under accession number CRA012442.
3 Results
3.1 Distribution of Salmonella serotypes and sequence types in retail chilled chicken carcasses and humans with diarrhea
A total of 88 (24.4%) Salmonella isolates were recovered from 360 retail chilled chicken carcasses in the local markets in Qingdao, China in 2020. Using both traditional and in silico serotyping methods, which yielded identical results, we determined that the 88 isolates represented 18 different serotypes (Figure 1, Supplementary Table S1). The serotypes of Enteritidis, Indiana, and Typhimurium were frequently detected (each was 17.0%, 15/88) among Salmonella from retail chilled chicken carcasses, followed by Derby (12.5%, 11/88), Agona (6.8%, 6/88), and Thompson (5.7%, 5/88) (Supplementary Table S2). The results also demonstrated that the 86 isolates collected from humans with diarrhea belonged to 15 different serotypes, with Enteritidis (23.3%, 20/86), Typhimurium (17.4%, 15/86), and I 4,[5],12:i:- (17.4%, 15/86) frequently detected, followed by Agona (8.1%, 7/86), London (7.0%, 6/86), and Derby (4.7%, 4/86) (Supplementary Table S2). There were six and three serotypes were isolated only in retail chilled chicken carcasses or humans with diarrhea, respectively.
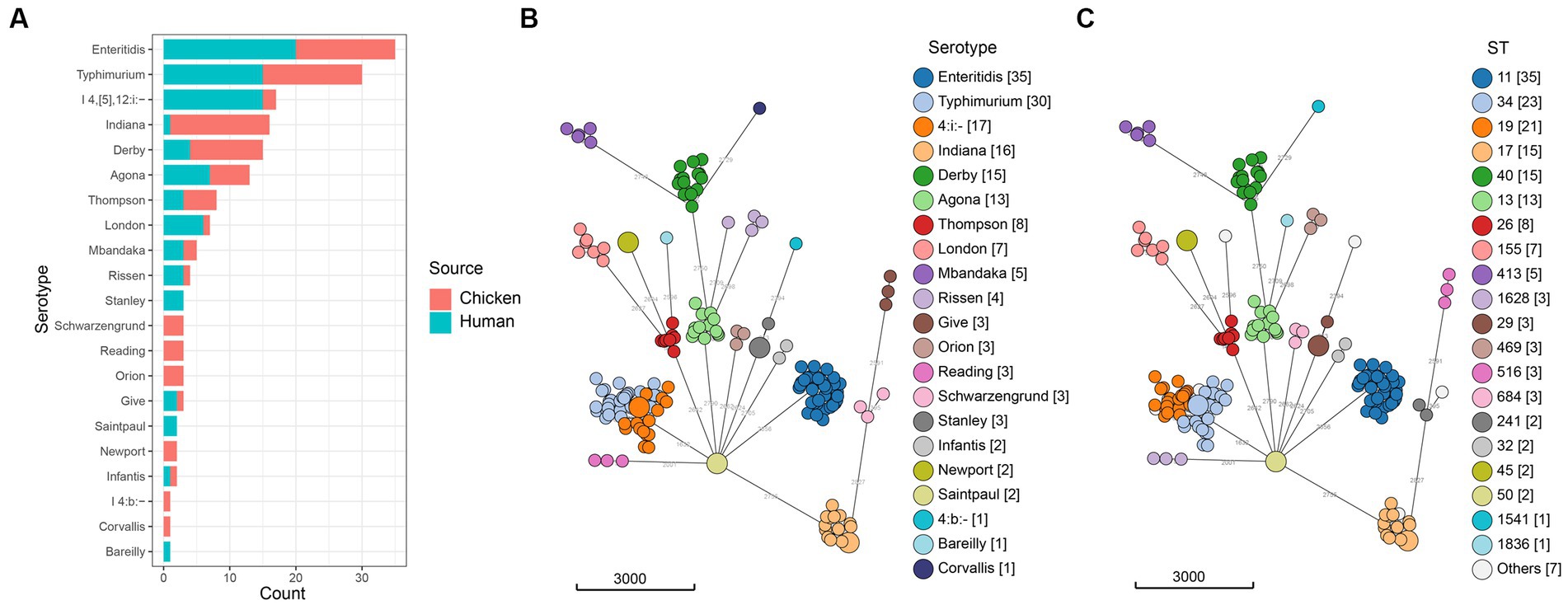
Figure 1. Distribution of Salmonella isolates. (A) Number of isolates from retail chilled chicken carcasses and humans with diarrhea. (B) Minimum spanning tree of the 174 Salmonella genomes colored by serotype. (C) Minimum spanning tree of the 174 Salmonella genomes colored by ST. The minimum spanning tree was built using cgMLST allelic profiles with the MSTreeV2 algorithm.
The sequence types of all 174 studied Salmonella isolates showed strong clustering by serotypes (Figure 1, Supplementary Table S3). The most prevalent sequence type was ST11 (20.1%, 35/174, S. Enteritidis), followed by ST34 (13.2%, 23/174, S. Typhimurium/S. I 4,[5],12:i:-), ST19 (12.1%, 21/174, S. Typhimurium). The ST17 (8.6%, 15/174), ST40 (8.6%, 15/174), and ST13 (7.5%, 13/174) clones were detected in S. Indiana, S. Derby, and S. Agona, respectively. We also noticed that each serotype was associated with one ST, except for S. Typhimurium (ST19, ST34, ST36, ST99, and ST3557), S. Indiana (ST17 and ST3558), S. Rissen (ST469 and ST1836), and S. Schwarzengrund (ST96 and ST241).
3.2 Antimicrobial resistance phenotype
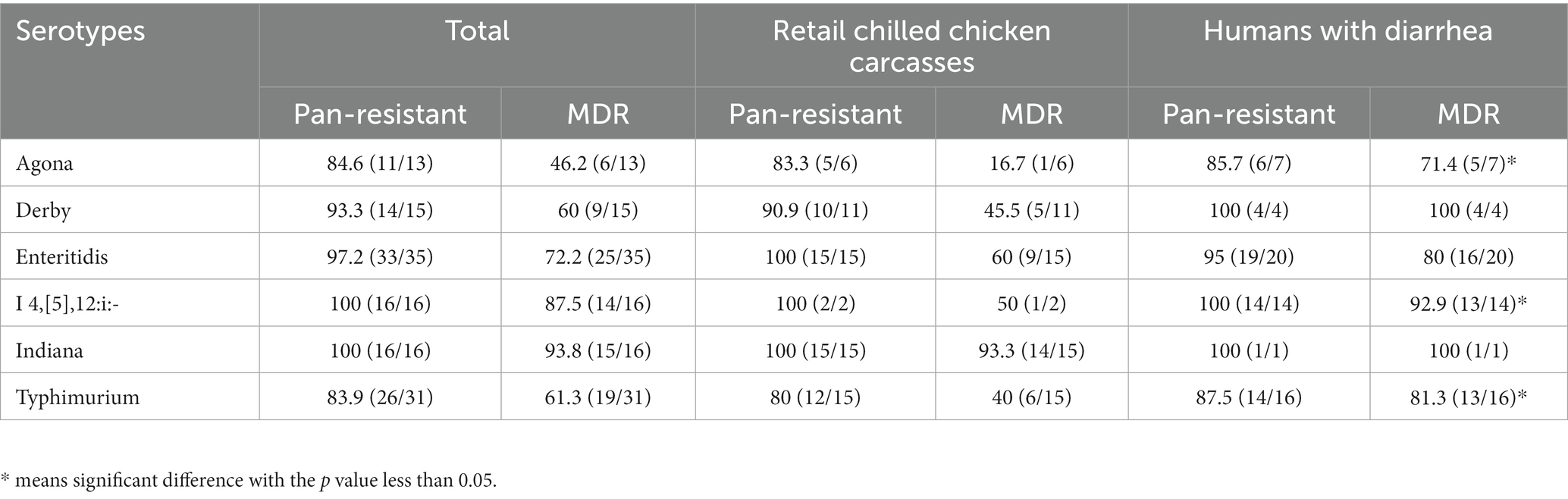
The results showed that 85.2% (75/88) of the Salmonella from retail chilled chicken carcasses were resistant to at least one antimicrobial agent, whereas 13 isolates (14.8%) were susceptible to all tested antimicrobials (Supplementary Table S1). In detail, the top four resistant antimicrobials were nalidixic acid (60.2%, 53/88), ampicillin (51.1%, 45/88), tetracycline (47.7%, 42/88) and chloramphenicol (40.9%, 36/88) (Figure 2A). Meanwhile, 47 isolates (53.4%) were MDR (i.e., could resist ≥3 antimicrobial classes) (Figure 2B). A total of 23 isolates (26.1%) were resistant to at least one of the third-generation cephalosporins (ceftazidime and cefotaxime), including 13 S. Indiana (86.7%, 13/15), 5 S. Thompson (5/5), 2 S. Typhimurium (13.3%, 2/15), 1 S. Agona (1/6), 1 S. Rissen (1/1), and 1 S. Enteritidis (6.7%, 1/15) (Supplementary Table S1). Most of these 23 isolates showed concurrent resistance to ciprofloxacin (n = 17). Notably, Polymyxin E resistance was observed in 16 isolates, including 8 S. Enteritidis, 2 S. Derby, and one each of S. Agona, S. I 4,[5],12:i:-, S. Indiana, S. Reading, S. Schwarzengrund, and S. Typhimurium.
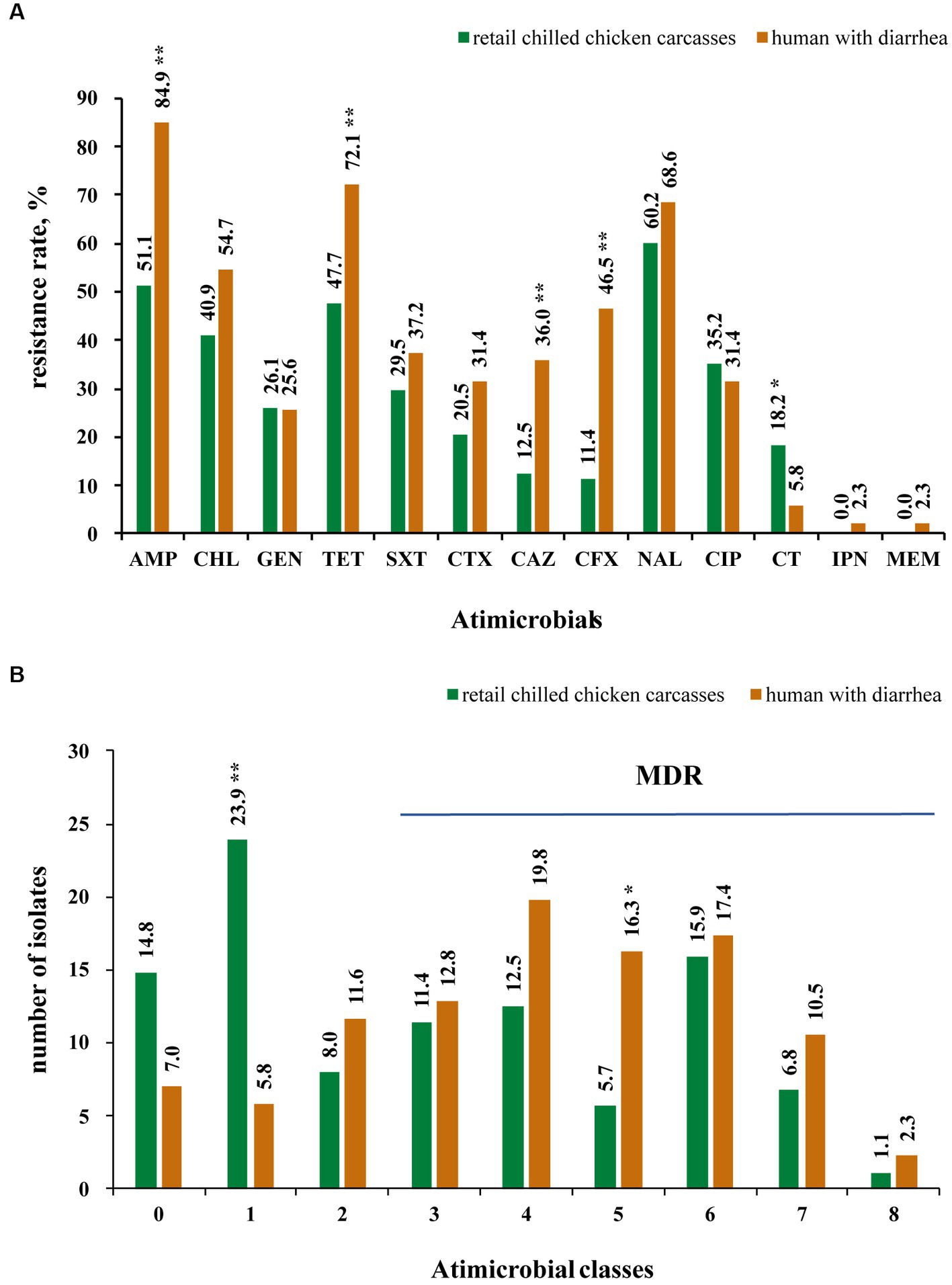
Figure 2. Antimicrobial resistance of 174 Salmonella isolates collected from retail chilled chicken carcasses and humans with diarrhea in this study. (A) Antimicrobial resistance rate of 174 Salmonella isolates to 13 antimicrobials. (B) Number of Salmonella isolates showing resistance to studied antimicrobials. * and ** mean significant difference with the p value less than 0.05 and 0.01, respectively.
Among the Salmonella isolates from humans with diarrhea, 93.0% (80/86) displayed resistance to at least one antimicrobial agent, while the remaining 6 isolates (7.0%) were susceptible to all tested antimicrobials (Supplementary Table S1). In detail, the top four resistant antimicrobials were also found to be ampicillin (84.9%, 73/86), tetracycline (72.1%, 62/86), nalidixic acid (68.6%, 59/86), and chloramphenicol (54.7%, 47/86), while 65 isolates (75.6%) were MDR (Figures 2A,B). A total of 34 isolates (39.5%) were resistant to at least one of the third-generation cephalosporins (ceftazidime and cefotaxime), including 9 S. Enteritidis (45%, 9/20), 8 S. Typhimurium (50%, 8/16), 7 S. I 4,[5],12:i:- (50%, 7/14), 2 S. Derby (2/4), 2 S. Saintpaul (2/2), 2 S. Stanley (2/3), one of each S. Agona (1/7), S. Bareilly (1/1), S. Infantis (1/1), S. Mbandaka (1/3), and S. Thompson (1/3), while 13 of these 34 isolates showed concurrent resistance to ciprofloxacin (Supplementary Table S1). Five isolates including 4 S. Enteritidis and 1 S. Agona showed resistant to polymyxin E. Moreover, two isolates (one of each S. Enteritidis and S. Infantis) were found to be resistant to carbapenems (imipenem and meropenem).
Furthermore, the Salmonella isolates from humans with diarrhea displayed higher resistance rates to several antimicrobials, including ampicillin, tetracycline, ceftazidime, and cefotaxime (p < 0.01), than the isolates from retail chilled chicken carcasses (Figure 2A). In addition, the human-associated Salmonella isolates showed higher MDR (p < 0.05) than those from retail chilled chicken carcasses (Figure 2B).
The AMR of the mainly detected Salmonella serotypes (n > 10) from retail chilled chicken carcasses and humans with diarrhea were estimated that the S. Indiana isolates exhibited the most pan-resistance (100%) and MDR (93.8%), followed by S. I 4,[5],12:i:- (pan-resistance, 100%; MDR, 87.5%), and S. Enteritidis (pan-resistance, 97.1%; MDR, 71.4%) (Table 1). Of each serotype, the human-associated isolates S. Agona (71.4%), S. I 4,[5],12:i:- (92.9%), and S. Typhimurium (81.3%) showed higher MDR than those of chicken-associated isolates (16.7, 50, and 40%, respectively).
3.3 Phylogenetic analysis revealed genomic diversity of Salmonella
All of the 174 isolates including 88 chicken-associated and 86 human-associated Salmonella isolates were subjected to whole genome sequencing (WGS). Based on data from the NCBI Pathogen Detection, 149 out of the 174 isolates (85.6%) could be categorized into known SNP clusters (n = 60). The distribution of SNP clusters differed between retail chicken and humans, with 27 unique SNP clusters found in chicken source and 25 unique cluster found in human source (Figure 3). Eight SNP clusters (PDS000078427.2, PDS000043691.33, PDS000096798.86, PDS000106143.144, PDS000144678.1, PDS000004748.67, PDS000101103.13, and PDS000026869.266) were detected in both sources, which belonged to six serotypes (S. London, S. Indiana, S. I 4,[5],12:i:-, S. Agona, S. Typhimurium, and S. Enteritidis) (Supplementary Table S4), revealing diverse origins of Salmonella isolated in this study.
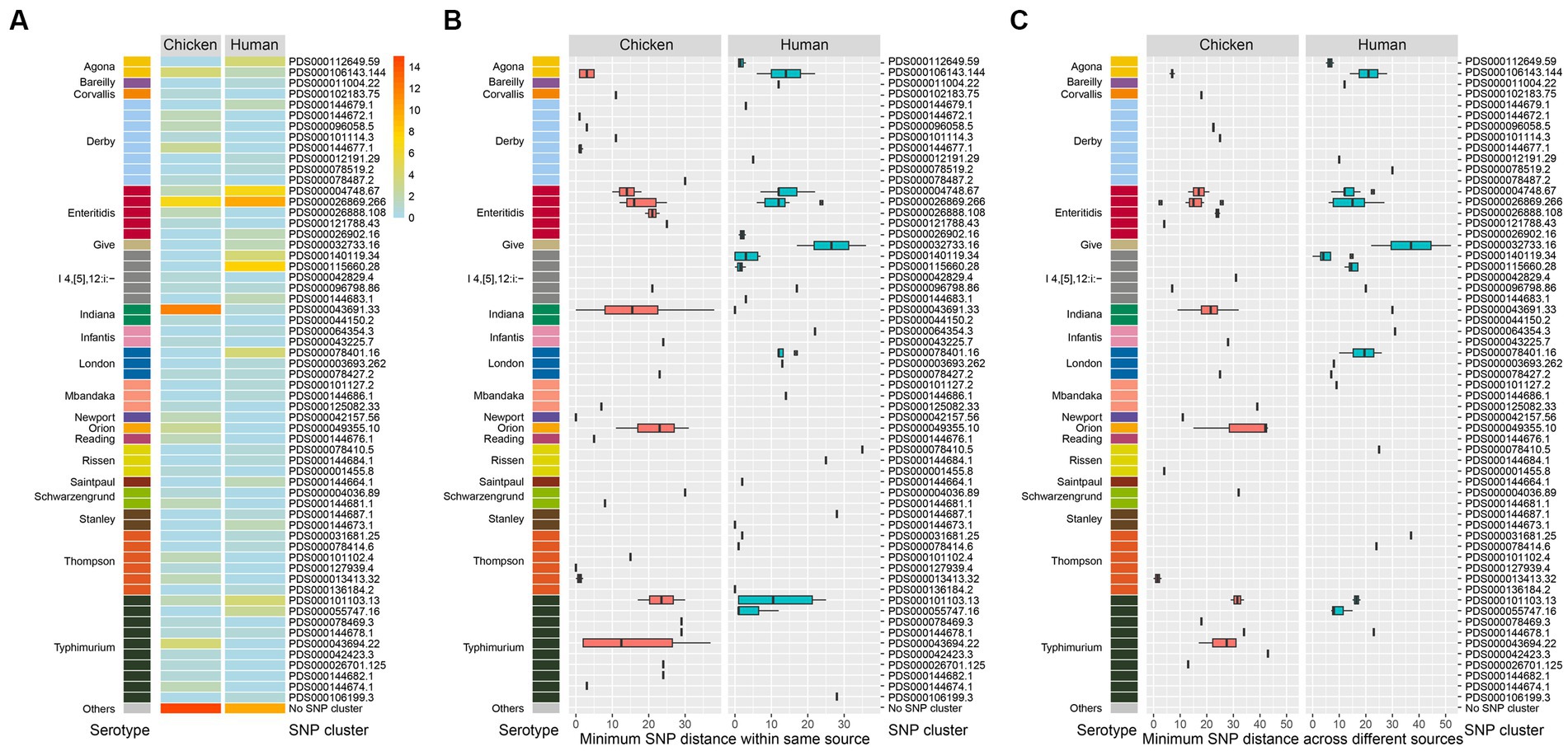
Figure 3. Genomic diversity of Salmonella isolates. (A) Heat map of serotype distribution and the designated SNP clusters from different sources. Color indicates the number of isolates. (B) Box plot of minimum SNP distance within same source of the designated SNP clusters of different serotypes. (C) Box plot of minimum SNP distance across different sources of the designated SNP clusters of different serotypes.
Notably, the top six prevalent SNP clusters among the 174 isolates included PDS000026869.266, PDS000043691.33, PDS000004748.67, PDS000115660.28, PDS000101103.13, and PDS000106143.144. All of these, except PDS000115660.28, were detected in both chicken and humans. PDS000115660.28 only contained isolates from humans. The most prevalent SNP cluster PDS000026869.266 contained 10 isolates from humans and 7 isolates from retail chicken.
S. Typhimurium/S. I 4,[5],12:i:- displayed the highest diversity with 15 detected SNP clusters, followed by S. Derby with 8, S. Thompson with 6, and S. Enteritidis with 5 (Figure 3A). Among the designated SNP clusters (n = 35) of the chicken isolates, 21 of them had a minimum SNP distance within environmental source below 21 (Figure 3B), while 14 of them had a minimum SNP distance within human source below 21 (Figure 3C). In contrast, 24 of the designated SNP clusters (n = 33) of the human-associated isolates had a minimum SNP distance within human source below 21 (Figure 3B), while 15 clusters had a minimum SNP distance within environmental source below 21 (Figure 3C). These findings demonstrated that a large number of the designated SNP clusters contained isolates circulating in both human and environmental sources (Pightling et al., 2018). For the 25 isolates that did not fit into designated SNP clusters, 15 were from retail chicken and 10 were from humans. The most common serotypes among these were S. Typhimurium, S. Enteritidis, and S. Agona.
3.4 MGE profiles
Plasmid replicons were detected in a majority of the bacterial isolates from this study, with only 9.8% (17/174) showing no evidence of plasmids in their genomes. Ten of the human-associated isolates and seven of the chicken-associated isolates had no detected plasmid replicons. A total of 38 plasmid replicons were identified among the 174 isolates, with 5 isolates showing the highest number of 7 plasmid replicons (Figure 4A). The most frequently identified plasmid replicons were Col(pHAD28), IncFIB(S), and IncFII(S) (Figure 4A). Conversely, less common types such as Col(MG828), ColpVC, FII(Cf), IncFIB(pHCM2), IncFIB(pNDM-Mar), IncFII(p96A), and pSL483 were present in certain single isolate, respectively. Additionally, the prevalence of plasmid replicons exhibited variations over isolates from retail chicken and humans. For instance, strains isolated in chicken showed a higher prevalence of IncHI2A, IncHI2 and pKPC-CAV1321, while those isolated in humans showed a higher prevalence of IncI1-I(Alpha) and Col440II. When considering different serotypes, S. Typhimurium and S. Indiana exhibited the largest number of plasmid types (16 in total, respectively), followed by S. I 4,[5],12:i:- (15 in total), and S. Enteritidis (14 in total) (Figure 4A). Notably, isolates from S. Enteritidis were more likely to carry plasmid replicons of IncFIB(S), IncFII(S), and IncX1, isolates from S. Typhimurium were more likely to carry Col(pHAD28), IncFIB(S), and IncFII(S), and isolates from S. Indiana were more likely to carry IncHI2A, IncHI2, and pKPC-CAV1321 (Figure 4A). Among the designated SNP clusters, PDS000043691.33 (S. Indiana), PDS000115660.28 (S. I 4,[5],12:i:-), PDS000042829.4 (S. I 4,[5],12:i:-), and PDS000144683.1 (S. I 4,[5],12:i:-) each harbored the highest number of plasmid replicon types, with seven types each (Figure 4B). Chicken-associated SNP clusters had a higher median number of plasmid replicons (3) compared to human-associated SNP clusters (2) (Figure 4B).
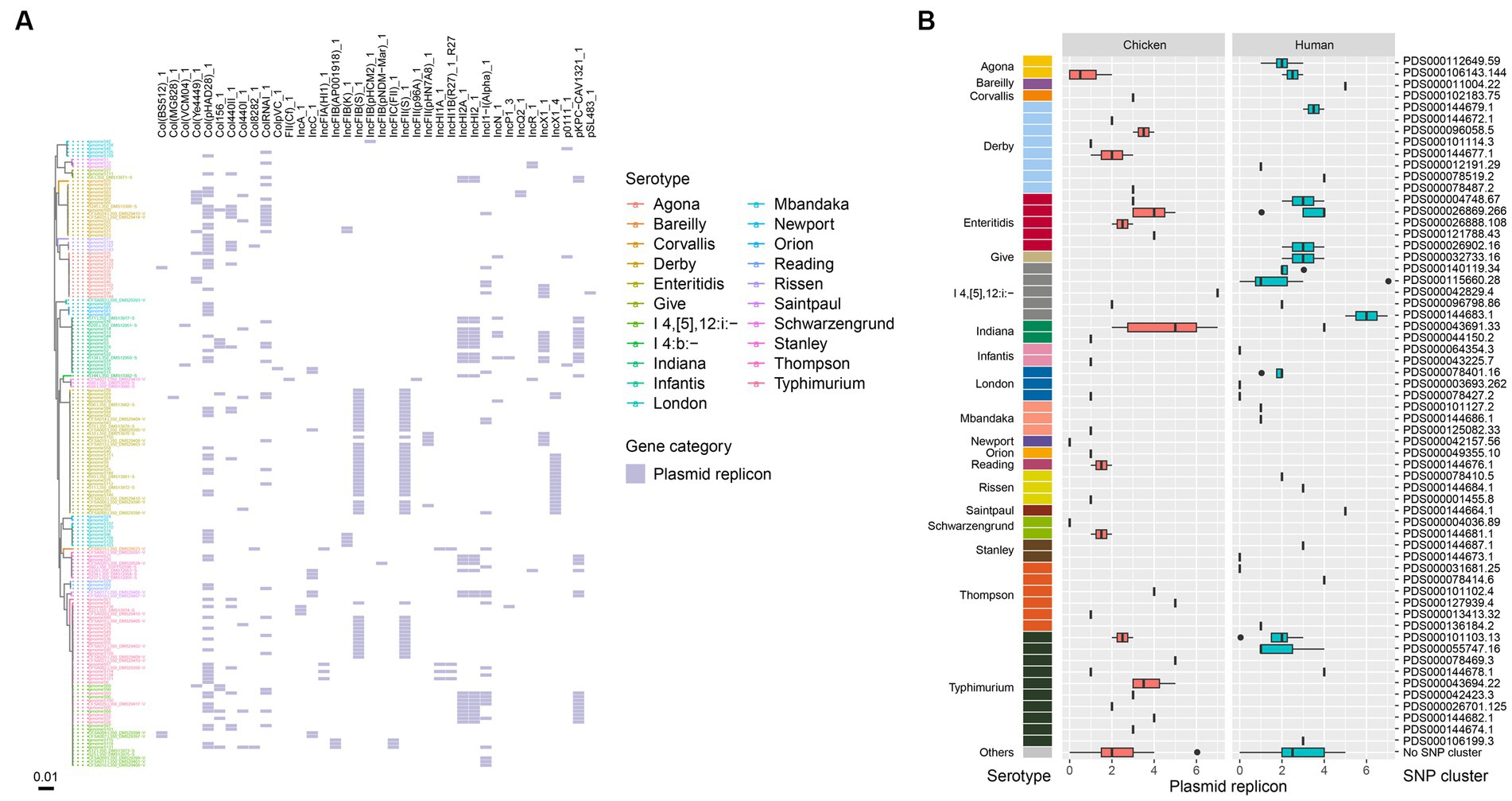
Figure 4. Plasmid replicons in the Salmonella genomes. (A) Core genome phylogeny of the Salmonella genomes and distribution of plasmid replicons. Color of the heatmap indicates the presence of plasmid replicons. (B) Box plot of plasmid replicon counts of the designated SNP clusters of different serotypes.
We also observed a high prevalence of intact prophages among the isolates, with only 17 lacking any. A total of 45 intact prophages were identified among the 174 isolates, with 31 isolates harboring only one prophage type (Figure 5A). Most isolates (72.4%, 126/174) contained at least two intact prophages. Seven isolates harbored more than 5 intact prophages, which belonged to serotypes such as Mbandaka, Typhimurium, Schwarzengrund, and Reading. Notably, 64 isolates, representing various serotypes, contained both Gifsy (Gifsy-1/Gifsy-2) and Salmon 118,970 sal3 prophages (Figure 5A). Among the various prophages identified, Gifsy-2 was more commonly present in isolates from humans (41/86) than retail chicken (26/88). When examining different serotypes, the Gifsy and Salmon 118,970 sal3 prophages were more likely to be carried by S. Enteritidis, S. Typhimurium, and S. I 4,[5],12:i:-, and were less frequently found in other serotypes. All the S. Enteritidis isolates carried Gifsy-2 and Salmon 118,970 sal3 prophages (Figure 5A). It is worth noting that all the isolates of S. I 4,[5],12:i:- carried Gifsy-2 other than Gifsy-1 prophage, while isolates of S. Typhimurium carried both types of Gifsy-1 and Gifsy-2 prophages. Most S. Indiana isolates (9/16) did not carry any intact prophages, while a small portion of S. Indiana isolates (6/16) carried only one intact prophage. The two S. Newport isolates were found to be free of intact prophages. In addition, SNP clusters that were associated a high number of intact prophage types (more than 5) contained PDS000144674.1 of S. Typhimurium (2/30), PDS000144676.1 of S. Reading (2/3), PDS000004036.89 of S. Schwarzengrund (1/3), and PDS000125082.33 of S. Mbandaka (1/5) (Figure 5B).
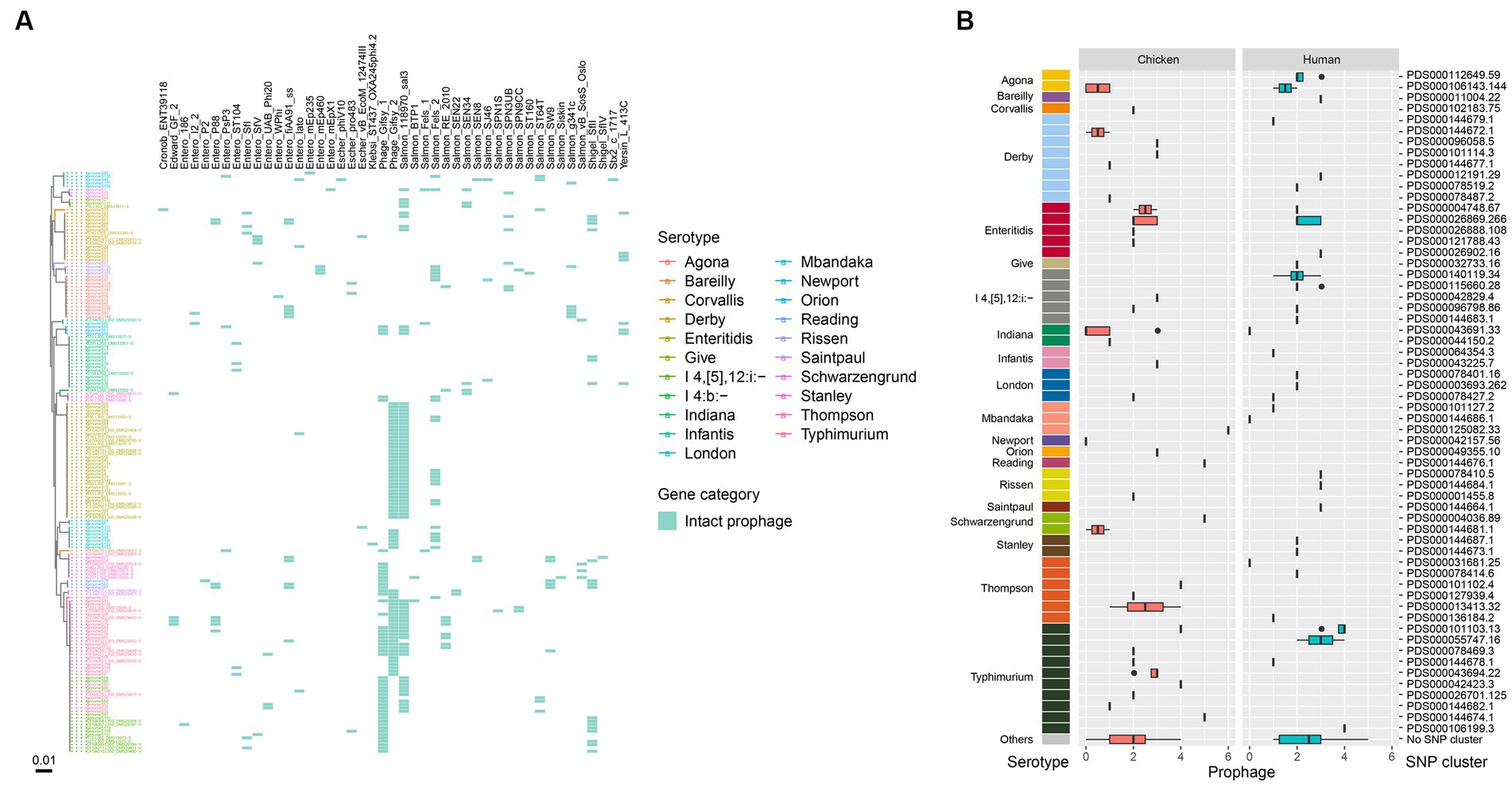
Figure 5. Prophages in the Salmonella genomes. (A) Core genome phylogeny of the Salmonella genomes and distribution of intact prophages. Color of the heatmap indicates the presence of prophages. (B) Box plot of prophage counts of the designated SNP clusters of different serotypes.
3.5 ARG profiles
To determine the presence of resistance genes, all 174 genome sequences were screened against the ResFinder database. A total of 79 different antibiotic resistance genes (ARGs) were identified in the isolates (Figure 6A). The majority of these ARGs belonged to beta-lactam resistance (n = 19) and aminoglycoside resistance (n = 19) categories. Among the CTX-M-type beta-lactamases, 9 different blaCTX-M genes were detected, with blaCTX-M-55 (n = 11), blaCTX-M-65 (n = 10), and blaCTX-M-14 (n = 6) being the most prevalent (Figure 6A).
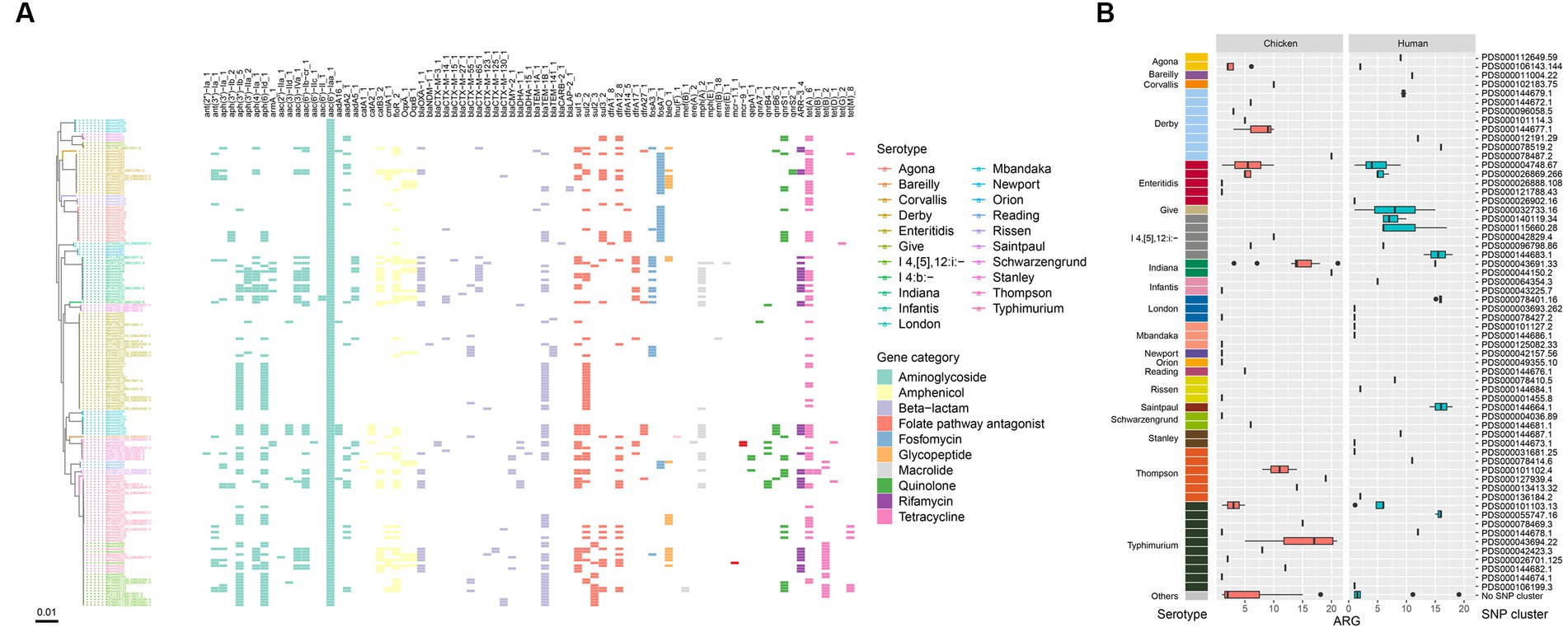
Figure 6. ARGs in the Salmonella genomes. (A) Core genome phylogeny of the Salmonella genomes and distribution of ARGs. Color of the heatmap indicates the presence of ARGs. (B) Box plot of ARG counts of the designated SNP clusters of different serotypes.
Notably, all isolates were positive for the aminoglycoside resistance gene aac(6′)-Iaa, and a large number of them carried the beta-lactam resistance gene blaTEM-1B (n = 67), the tetracycline resistance gene tet(A) (n = 64), aminoglycoside resistance genes aph(6)-Id (n = 65) and aph(3″)-Ib (n = 60), the sulfonamide resistance gene sul2 (n = 50), and the amphenicol resistance gene floR (n = 50) (Figure 6A). Six types of quinolone resistance genes were found, including qnrS1 (n = 23), qnrB4 (n = 7), qnrB6 (n = 6), qepA1 (n = 3), qnrS2 (n = 2), and qnrA7 (n = 1). Additionally, one S. Typhimurium isolate was positive for the colistin resistance gene mcr-1.1, and two S. Thompson isolates carried mcr-9.1 (Figure 6A). All these mcr-positive isolates were from chicken. One S. Enteritidis isolate was positive for the blaNDM-1 gene. Furthermore, the mcr-1.1 gene was detected on an IncHI2 plasmid replicon, while the blaNDM-1 gene was carried on an IncC plasmid. The plasmid carrying the mcr-9.1 gene could not be identified as the gene was located on small contigs with a length of ~2,500 bp (data not shown).
Due to the high prevalence of genes related to aminoglycoside and beta-lactam resistance, all isolates contained at least one ARG in their genomes (Figure 6A). Three isolates, genomeS31, genomeS44, and genomeS52, harbored the largest number of ARGs (n = 21), which belonged to two serotypes, S. Indiana and S. Typhimurium. When considering different serotypes, S. Indiana exhibited the largest number of ARGs (14.5 in median), followed by S. Thompson (12.5 in median), S. I 4,[5],12:i:- (6 in median), and S. Typhimurium (5.5 in median). SNP clusters that were associated with higher number of ARGs included PDS000043691.33 (14 in median, S. Indiana), PDS000044150.2 (20, S. Indiana), PDS000055747.16 (16 in median, S. Typhimurium), PDS000043694.22 (17 in median, S. Typhimurium), PDS000144683.1 (15.5 in median, S. I 4,[5],12:i:-), PDS000127939.4 (19, S. Thompson), PDS000078401.16 (16 in median, S. London), PDS000144664.1 (16 in median, S. Saintpaul), and PDS000078519.2 (16, S. Derby) (Figure 6B).
It is worth noting that most of the ARGs were detected on plasmid sequences (Supplementary Figure S1A). The aminoglycoside resistance gene aac(6′)-Iaa presented in all the isolates was found located on chromosome sequences (Supplementary Figure S1B). Additionally, a total of 40 isolates were found carrying no ARGs other than the aac(6′)-Iaa gene, which belonged to a variety of serotypes such as S. Typhimurium (n = 10), S. Enteritidis (n = 9), S. Mbandaka (n = 5), etc.
4 Discussion
Salmonella is one of the most widespread pathogenic foodborne bacteria in Chinese food products and retail chicken have been recognized as one of the major sources of human salmonellosis (Paudyal et al., 2018). Moreover, there has been an increase in reports of the dissemination of resistant Salmonella from chicken to humans through the chicken production chain (Kongsanan et al., 2021; Manzari et al., 2022). In this study, we surveyed the prevalence, serovar distribution, MDR, and genomic characteristics of Salmonella in chilled chicken carcass and humans with diarrhea sources in Qingdao, China in 2020.
Overall, 24.4% of the retail chilled chicken carcass samples purchased in 2020 in local markets contained Salmonella. This prevalence is lower than what was found in other provinces (average, 39.3%) in China (Wang et al., 2020a), Brazil (31.5%) (Perin et al., 2020), but higher than those in Korea (14.1%) (La et al., 2021), the USA (17.9%) (Peng et al., 2016), and European Union (7.1%) (Gonçalves-Tenório et al., 2018). Thus, chicken meat in retail may be an important source of human salmonellosis in China. Besides, data reported that Salmonella prevalence in chilled poultry meat was statistically higher than that of frozen poultry meat and ambient poultry meat in China (Sun et al., 2021). In China, immersion chilling is employed more frequently and Chinese consumers have a preference for chilled meat (25% market share) over frozen meat (15% market share) (Liu et al., 2017). However, once a sample is contaminated with Salmonella during the immersion process, the contamination may spread among the whole batch of carcasses, leading to an increase in the prevalence of pathogens on finished products (Marmion et al., 2021). Therefore, in response to potential public health pressures, more effective intervention strategies during immersion chilling should be implemented to control the quality and safety of chicken products.
Serotype results suggested a wide range of Salmonella serotypes present in retail chilled chicken carcasses and humans with diarrhea. The dominance of S. Enteritidis and S. Typhimurium among the studied Salmonella isolates, is consistent with the data obtained from chicken and human previously in China and other regions worldwide (EFSA and ECDC, 2018; Balasubramanian et al., 2019; Wang et al., 2020a). Besides, S. Indiana is another frequently detected serotype among chicken samples and is also detected from humans with diarrhea in this study. Accordingly, since 2009, the presence of S. Indiana has seen a remarkable increase, which has become one of the top three common serotypes in China (Zhang et al., 2022a). S. Indiana has been isolated from a wide and diverse variety of sample sources in 18 provinces throughout China and was most frequently recovered from food, followed by animals, the environment and then people, especially children under 6 years old with low immunity (Gong et al., 2017). What is more, Salmonella serovars Enteritidis, Typhimurium, and Indiana are also reported as the most common serotypes associated with human infections and outbreaks (Kuang et al., 2018b; Wu et al., 2018). Thus, the high prevalence of these Salmonella serotypes in chicken carcasses and humans with diarrhea indicates a significant risk to public health. The monitoring of the emergence and prevalence of these Salmonella serotypes is essential for the better control of salmonellosis.
The combination of MLST and serotype detection can facilitate research on the hereditary and evolutionary relationships of Salmonella (Xu et al., 2022). The most common Salmonella sequence types were found to be ST11, ST34, and ST19 in this study, where the corresponding serotypes were S. Enteritidis and S. Typhimurium (including S. I 4,[5],12:i:-). This is consistent with findings from previous reports in China (Wang et al., 2020b; Xu et al., 2022). Accordingly, among S. Enteritidis worldwide, ST11 is the predominant sequence type, accounting for 89% of the sequence types in the EnteroBase database (Ashton et al., 2016). However, a recent report from Malaysia showed that ST1925 was dominantly detected among chicken-associated S. Enteritidis strains (Zakaria et al., 2022). In America, the major prevalent STs are ST34, ST33, and ST11 (Pethplerdprao et al., 2017), which shows that STs may be associated with regions. Furthermore, the other mainly identified sequence types, ST13 (S. Agona), ST17 (S. Indiana), and ST40 (S. Derby), were also previously reported to be associated with salmonellosis in human (Luo et al., 2022; Sun et al., 2022; De Jesus Bertani et al., 2023). Observations from the present study therefore provided important evidence and confirmed further that these types of Salmonella can serve an important role in human diarrhea in Qingdao, China.
Herein, we found that 53.4 and 75.6% of retail chilled chicken carcass- and humans with diarrhea-associated Salmonella isolates were MDR, respectively. High percentages of MDR strains were also observed in Salmonella isolated from poultry (Li et al., 2022), food animals (Tang et al., 2023) and humans (Chen et al., 2022) in various regions of China. The surge in antimicrobial-resistant Salmonella isolates is recognized as a crucial public health issue (Eng et al., 2015). The major of Salmonella isolates among both sources were resistant to four antimicrobials of nalidixic acid, ampicillin, tetracycline and chloramphenicol, which are the first-line drugs used against bacterial infection in animal farms worldwide (Nhung et al., 2016; Lekagul et al., 2019). This finding is consistent with the literature from different countries, including China (Tamang et al., 2011; Cai et al., 2016; EFSA and ECDC, 2018). Our results also suggested that Salmonella isolates in this study showed high level resistance to third generation cephalosporins (ceftazidime and cefotaxime) and quinolone (ciprofloxacin), which are the first-line drugs used against bacterial infection in clinics (Tack et al., 2020). These findings are regarded as significant threats to public health, leading to limited drugs of choice for salmonellosis infection treatment in animals and humans. The high AMR rate of S. Enteritidis, S. Typhimurium (including S. I 4,[5],12:i:-) and S. Indiana, with a widespread AMR spectrum is of concern. Moreover, several Salmonella isolates showed resistance to colistin, which is considered as one of the last-resort therapeutic options for the defense of multidrug-resistant Enterobacteriaceae (Elbediwi et al., 2019). This finding of colistin resistance agreed with results reported by other recent studies in Salmonella isolates from poultry and humans (Fortini et al., 2022; Tang et al., 2022). Of note, two carbapenems (imipenem and meropenem) resistant Salmonella isolates were also found in this study. Therefore, the spread of MDR Salmonella isolates, especially resistant to these clinically important antimicrobials, including fluoroquinolone, third-generation cephalosporins, colistin and carbapenems, will result in challenges with public health.
Phylogenetic analysis revealed a high level of genomic diversity of the Salmonella isolates collected from Qingdao region in China. Eight SNP clusters containing both chicken and human isolated collected in this study were identified through NCBI Pathogen Detection. Six of these clusters (PDS000043691.33, PDS000096798.86, PDS000106143.144, PDS000004748.67, PDS000101103.13, and PDS000026869.266) were featured by involving a large number of strains from different countries, indicating these isolates had been widely spread around the world. In contrast, the other two clusters (PDS000078427.2 and PDS000144678.1) formed distinct, locally restricted clades circulating in a limited number of cities in China. Most of the chicken and human isolates shared no common SNP clusters, indicating multiple origins of these isolates. Interestingly, the major SNP clusters found in the present study comprised numerous strains from clinical infections in China and other countries. This highlights the importance of conducting additional research on the dissemination of strains associated with these SNP clusters.
Mobile genetic elements (MGEs), such as bacteriophages and plasmids, are DNA segments that carry genes involving with DNA movement within genomes (Frost et al., 2005). The presence of intact Salmon 118,970 sal3, Gifsy-2, and Gifsy-1 was found at a higher frequency in the genomes of S. Typhimurium, S. I 4,[5],12:i:-, and S. Enteritidis, whereas such prophage types were seldom observed in other prevalent serotypes such as S. Indiana, S. Agona, and S. Derby. These findings indicate the presence of prophage types is associated with serotypes and may be related to the evolutionary history of different serotypes. Recently, a study of a large-scale investigation on S. Typhimurium has reported the common presence of the above-mentioned prophages in S. Typhimurium isolated in China (Wang et al., 2023). Furthermore, the specific phage, Salmon 118,970 sal3 was not only found in Salmonella, but also found in E. coli (Li et al., 2020) and Morganella morganii (Minnullina et al., 2019), suggesting that this phage has a relatively wide host range. The presence of Gifsy-1 and Gifsy-2 and their contribution to virulence in S. Typhimurium has been widely reported (Ho and Slauch, 2001).
Plasmids play a crucial role as the primary vehicles for horizontal gene transfer (HGT) in bacteria (De La Cruz and Davies, 2000). Among the isolates, the Col(pHAD28) plasmid replicon was the most common, and the IncF plasmid replicon was the second most common. Interestingly, the IncF is also one of the most prevalent plasmid replicons in Salmonella isolated from food animals in the United States (Mcmillan et al., 2019). The presence of Col(pHAD28) plasmid replicon was observed in a total of 14 serotypes identified in this study, indicating its high occurrence in Salmonella isolated from Qingdao region in China. Continuous monitoring of the spread of this plasmid is essential, as several studies have reported the presence of PMQR genes within this plasmid (Li et al., 2021; Hurtado et al., 2022).
Genomic analysis showed that the Salmonella isolates harbored different antimicrobial resistant genes. All of the studied Salmonella isolates harbored the aac(6′)-Iaa gene. However, aac(6′)-Iaa gene and similar genes usually are transcriptionally silent and rarely become transcriptionally active. AAC(6′)-Iaa is a typical member of the [A] family in that it acetylates tobramycin, kanamycin, and amikacin effectively but acetylates gentamicin ineffectively (Salipante and Hall, 2003). The mere presence of this gene does not confer aminoglycoside resistance in Salmonella (Magnet et al., 1999; Neuert et al., 2018). Besides, 81.4% (96/118) of the isolates resistant to the aminoglycoside class harbored different aminoglycoside resistance genes in addition to the aac(6′)-Iaa gene. Among diverse mechanisms of aminoglycoside resistance, enzymatic modification is the most prevalent mechanism in pathogenic bacteria, including Salmonella (Ramirez and Tolmasky, 2010; Biswas et al., 2019). Moreover, our study revealed 52/57 (91.2%) of the isolates resistant to the third-generation cephalosporins harbored different beta-lactam resistance genes, which were common detected in S. Enteritidis, S. Typhimurium, S. I 4,[5],12:i:-, and S. Indiana. The bla genes control the resistance to beta-lactam antimicrobials by hydrolyzing the beta-lactam ring, leading to antibiotic inactivation (Jacoby, 2009; Eguale et al., 2017). In this study, the frequently detected blaTEM-1B gene (37.9%, 67/174) conferring resistance to ampicillin, is the dominant beta-lactam in most Salmonella serotypes worldwide (Eguale et al., 2017). Meanwhile, nine different CTX-M-type ESBL-producing were found in this study. During the last decade, the most frequently encountered (particularly in areas of Europe and Asia) ESBL genes were those encoding the CTX-M enzyme family, primarily carried by transferable plasmids and transposons (Cantón et al., 2012). The emergence of CTX-M-type ESBL-producing Salmonella has been reported in clinical cases, animals, and food samples worldwide, including China (Brown et al., 2018; Wang et al., 2020a). Five different tet resistance genes were found in 88/174 (50.6%) of the studied isolates, 84 of which were resistant to tetracyclines. Additionally, six plasmid-mediated quinolone resistance (PMQR) genes, with qnrS1 prevalently detected, were identified in among the examined isolates belonging to different serotypes. These critical genes in Salmonella isolates from both chicken and human related samples present a tremendous public health concern. It is essential that the existence of the acquired antimicrobial resistant genes in bacterial genomes does not inevitably confer phenotypic resistance and vice versa.
Through NCBI’s Pathogen Detection platform, a total of 60 SNP clusters was categorized using isolates collected in this study (Supplementary Table S4). A SNP cluster is a cluster of isolates where each isolate is less than or equal to 50 SNPs distant from others, which offers higher resolution than serotype and MLST for outbreak surveillance purpose. In the designated SNP clusters, we identified several SNP clusters carrying PMQR genes. Some of these SNP clusters contained a large number of isolates from various isolation locations harboring specific PMQR genes, for example, qnrS1 in PDS000112649.59 of S. Agona, qnrB6 in PDS000102183.75 of S. Corvallis, and qnrS1 in PDS000115660.28 of S. 4,[5],12:i:-; qnr-carrying strains with the same sequence types have been reported to be circulating globally with a high rate of quinolone resistance (Fernández et al., 2018; Zhang et al., 2020; Cadel-Six et al., 2021; Lee et al., 2021; Chen et al., 2023).
On the other hand, there are qnr-carrying isolates collected uniquely or mostly from China in SNP clusters with small numbers of matched isolates such as PDS000078519.2 of S. Derby, PDS000013413.32 of S. Thompson, and PDS000055747.16 of S. Typhimurium, indicating that these strains were circulating locally in specific regions in China and tend to be emerging threat to public health. Consistently, in a recent study investigating ciprofloxacin resistance in Salmonella from humans, food and animals collected in Shanghai, China, qnr-positive isolates were mostly S. Thompson, S. Derby and S. Typhimurium (Kuang et al., 2018a).
Apart from qnr-carrying isolates, this study also detected three mcr-carrying isolates. The isolate harboring mcr-1.1 was found clustered in the SNP cluster PDS000042829.4 of S. I 4,[5],12:i:-, which contained a total of four mcr-1.1-carrying isolates uniquely from China, with one isolated from a diarrheal patient (Sun et al., 2023) and another one isolated from ready-to-eat pork (Wang et al., 2018). The two mcr-9.1-carrying isolates were designated to the SNP cluster PDS000101102.4 of S. Thompson. This SNP cluster had a total of 14 isolates collected from China, of which 11 were mcr-9.1 positive and 9 were qnrB4 positive. Interestingly, there are 8 isolates carrying both mcr-9.1 and qnrB4. A recent study also reported a S. I 4,[5],12:i:- isolate carrying both mcr-9.1 and qnrA1 and it was found that these clinically relevant resistance genes were driven by IncHI2-ST1 plasmids (Vázquez et al., 2023).
Additionally, the blaNDM-1-carrying S. Enteritidis isolate was associated with the SNP cluster PDS000004748.67, which had a total of 258 matched isolates, but no other isolates within this cluster were blaNDM-1 positive, suggesting the blaNDM-1 positive isolate has recently acquired plasmid carrying blaNDM-1 gene. It should be paid great attention to as the NDM-1-harboring plasmids confer resistance to all β-lactams (Huang et al., 2017).
In summary, the frequently detection of Salmonella from retail chilled chicken carcasses in Qingdao, China, highlighted the importance of improving food hygiene measures to reduce the contamination and transmission of this bacterium. We found a considerable diversity of Salmonella serotypes and sequence types among both chicken and human, with MDR, posing a severe risk to food safety and public health. The finding of eight worldwide spread SNP clusters in this study emphasized additional research on the dissemination of strains associated with these SNP clusters. The MGE results showed high prevalence of plasmid replicons and prophages among Salmonella from both sources, and the presence of these MGEs associated with specific serotypes indicated different evolutionary history of these serotypes. Given the high prevalence of CTX-M type ESBL-producing and PMQR genes identified in this study, we strongly recommend that both clinical and veterinary sectors routinely test for AMR when resistance to these antimicrobials is detected in Salmonella. This will enhance monitoring and guide the selection of effective treatments. Overall, it is essential to continue monitoring the Salmonella serotypes, implement the necessary prevention and strategic control plans, and conduct an epidemiological surveillance system based on whole-genome sequencing.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
WW: Writing – original draft, Writing – review & editing. JC: Writing – original draft, Writing – review & editing. FLiu: Writing – review & editing. YH: Writing – review & editing. FLi: Writing – review & editing. XD: Writing – review & editing. YD: Writing – review & editing. SL: Writing – original draft, Writing – review & editing. JX: Writing – review & editing. ZZ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Comparative analysis of domestic and foreign detection method performance index evaluation system and special research on database construction (2018001001), and the China Food Safety Talent Competency Development Initiative: CFSA 523 Program funded by the China National Center for Food Safety Risk Assessment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1295769/full#supplementary-material
Footnotes
1. ^ https://card.mcmaster.ca/home
2. ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc
3. ^ https://github.com/tseemann/mlst
4. ^http://www.denglab.info/SeqSero2
References
Antunes, P., Mourão, J., Campos, J., and Peixe, L. (2016). Salmonellosis: the role of poultry meat. Clin. Microbiol. Infect. 22, 110–121. doi: 10.1016/j.cmi.2015.12.004
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Ashton, P. M., Nair, S., Peters, T. M., Bale, J. A., Powell, D. G., Painset, A., et al. (2016). Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4:e1752. doi: 10.7717/peerj.1752
Balasubramanian, R., Im, J., Lee, J. S., Jeon, H. J., Mogeni, O. D., Kim, J. H., et al. (2019). The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccin. Immunother. 15, 1421–1426. doi: 10.1080/21645515.2018.1504717
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Biswas, S., Li, Y., Elbediwi, M., and Yue, M. (2019). Emergence and dissemination of mcr-carrying clinically relevant Salmonella Typhimurium monophasic clone ST34. Microorganisms 7:298. doi: 10.3390/microorganisms7090298
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Brown, A. C., Chen, J. C., Watkins, L. K. F., Campbell, D., Folster, J. P., Tate, H., et al. (2018). Ctx-m-65 extended-spectrum β-lactamase-producing Salmonella enterica serotype Infantis, United States. Emerg. Infect. Dis. 24, 2284–2291. doi: 10.3201/eid2412.180500
Cadel-Six, S., Cherchame, E., Douarre, P.-E., Tang, Y., Felten, A., Barbet, P., et al. (2021). The spatiotemporal dynamics and microevolution events that favored the success of the highly clonal multidrug-resistant monophasic Salmonella Typhimurium circulating in Europe. Front. Microbiol. 12:651124. doi: 10.3389/fmicb.2021.651124
Cai, Y., Tao, J., Jiao, Y., Fei, X., Zhou, L., Wang, Y., et al. (2016). Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 222, 56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli, A., and Hasman, H. (2020). Plasmidfinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
CDC (2019). Antibiotic resistance threats in the United States. Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. (Accessed October 2022).
Chen, J., Ed-Dra, A., Zhou, H., Wu, B., Zhang, Y., and Yue, M. (2022). Antimicrobial resistance and genomic investigation of non-typhoidal Salmonella isolated from outpatients in Shaoxing city, China. Front. Public Health 10:988317. doi: 10.3389/fpubh.2022.988317
Chen, K., Xie, M., Wang, H., Chan, E. W.-C., and Chen, S. (2023). Intercontinental spread and clonal expansion of ColRNA1 plasmid-bearing Salmonella Corvallis ST1541 strains: a genomic epidemiological study. One Health Adv. 1, 1–15. doi: 10.1186/s44280-023-00017-9
CLSI (2017). Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA.
De Jesus Bertani, A. M., Vieira, T., Reis, A. D., Dos Santos, C. A., De Almeida, E. A., Camargo, C. H., et al. (2023). Whole genome sequence analysis of the first reported isolate of Salmonella Agona carrying blaCTX-M-55 gene in Brazil. Sci. Rep. 13:2299. doi: 10.1038/s41598-023-29599-5
De La Cruz, F., and Davies, J. (2000). Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8, 128–133. doi: 10.1016/S0966-842X(00)01703-0
Ed-Dra, A., Karraouan, B., Allaoui, A. E., Khayatti, M., Ossmani, H. E., Filali, F. R., et al. (2018). Antimicrobial resistance and genetic diversity of Salmonella Infantis isolated from foods and human samples in Morocco. J. Glob. Antimicrob. Resist. 14, 297–301. doi: 10.1016/j.jgar.2018.05.019
EFSA and ECDC (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 16:e05500. doi: 10.2903/j.efsa.2018.5500
Eguale, T., Birungi, J., Asrat, D., Njahira, M. N., Njuguna, J., Gebreyes, W. A., et al. (2017). Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in Central Ethiopia. Antimicrob. Resist. Infect. Control 6:13. doi: 10.1186/s13756-017-0171-6
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018). Microorganisms 7:461. doi: 10.3390/microorganisms7100461
Eng, S.-K., Pusparajah, P., Ab Mutalib, N.-S., Ser, H.-L., Chan, K.-G., and Lee, L.-H. (2015). Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 8, 284–293. doi: 10.1080/21553769.2015.1051243
Fan, J., Cai, H., Fang, Y., He, J., Zhang, L., Xu, Q., et al. (2022). Molecular genetic characteristics of plasmid-borne mcr-9 in Salmonella enterica serotype typhimurium and Thompson in Zhejiang, China. Front. Microbiol. 13:852434. doi: 10.3389/fmicb.2022.852434
Fernández, J., Guerra, B., and Rodicio, M. R. (2018). Resistance to carbapenems in non-typhoidal Salmonella enterica serovars from humans, animals and food. Vet. Sci. 5:40. doi: 10.3390/vetsci5020040
Fortini, D., Owczarek, S., Dionisi, A. M., Lucarelli, C., Arena, S., Carattoli, A., et al. (2022). Colistin resistance mechanisms in human Salmonella enterica strains isolated by the National Surveillance Enter-net Italia (2016-2018). Antibiotics (Basel) 11:102. doi: 10.3390/antibiotics11010102
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Gonçalves-Tenório, A., Silva, B. N., Rodrigues, V., Cadavez, V., and Gonzales-Barron, U. (2018). Prevalence of pathogens in poultry meat: a meta-analysis of European published surveys. Foods 7:69. doi: 10.3390/foods7050069
Gong, J., Kelly, P., and Wang, C. (2017). Prevalence and antimicrobial resistance of Salmonella enterica serovar Indiana in China (1984-2016). Zoonoses Public Health 64, 239–251. doi: 10.1111/zph.12328
Grimont, P. A. D., and Weill, F.-X. (2007). “Antigenic formulae of the Salmonella serovars” in WHO collaborating centre for reference and research on Salmonella, vol. 9, 1–166.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Heredia, N., and García, S. (2018). Animals as sources of food-borne pathogens: a review. Anim. Nutr. 4, 250–255. doi: 10.1016/j.aninu.2018.04.006
Ho, T. D., and Slauch, J. M. (2001). OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183, 1495–1498. doi: 10.1128/JB.183.4.1495-1498.2001
Huang, J., Deng, S., Ren, J., Tu, J., Ye, M., and Wang, M. (2017). Characterization of a blaNDM-1-harboring plasmid from a Salmonella enterica clinical isolate in China. Mol. Med. Rep. 16, 1087–1092. doi: 10.3892/mmr.2017.6733
Hurtado, R., Barh, D., Weimer, B. C., Viana, M. V. C., Profeta, R., Sousa, T. J., et al. (2022). WGS-based lineage and antimicrobial resistance pattern of Salmonella Typhimurium isolated during 2000-2017 in Peru. Antibiotics (Basel) 11:1170. doi: 10.3390/antibiotics11091170
Hussein, N. H., Al-Kadmy, I. M. S., Taha, B. M., and Hussein, J. D. (2021). Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol. Biol. Rep. 48, 2897–2907. doi: 10.1007/s11033-021-06307-y
Iovleva, A., and Doi, Y. (2017). Carbapenem-resistant Enterobacteriaceae. Clin. Lab. Med. 37, 303–315. doi: 10.1016/j.cll.2017.01.005
Jacoby, G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jain, P., Chowdhury, G., Samajpati, S., Basak, S., Ganai, A., Samanta, S., et al. (2020). Characterization of non-typhoidal Salmonella isolates from children with acute gastroenteritis, Kolkata, India, during 2000-2016. Braz. J. Microbiol. 51, 613–627. doi: 10.1007/s42770-019-00213-z
Jajere, S. M. (2019). A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 12, 504–521. doi: 10.14202/vetworld.2019.504-521
Kirk, M. D., Pires, S. M., Black, R. E., Caipo, M., Crump, J. A., Devleesschauwer, B., et al. (2015). World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 12:e1001921. doi: 10.1371/journal.pmed.1001921
Kongsanan, P., Angkititrakul, S., Kiddee, A., and Tribuddharat, C. (2021). Spread of antimicrobial-resistant Salmonella from poultry to humans in Thailand. Jpn. J. Infect. Dis. 74, 220–227. doi: 10.7883/yoken.JJID.2020.548
Kuang, D., Zhang, J., Xu, X., Shi, W., Chen, S., Yang, X., et al. (2018a). Emerging high-level ciprofloxacin resistance and molecular basis of resistance in Salmonella enterica from humans, food and animals. Int. J. Food Microbiol. 280, 1–9. doi: 10.1016/j.ijfoodmicro.2018.05.001
Kuang, D., Zhang, J., Xu, X., Shi, W., Yang, X., Su, X., et al. (2018b). Increase in ceftriaxone resistance and widespread extended-spectrum β-lactamases genes among Salmonella enterica from human and nonhuman sources. Foodborne Pathog. Dis. 15, 770–775. doi: 10.1089/fpd.2018.2468
La, T. M., Kim, T., Lee, H. J., Lee, J. B., Park, S. Y., Choi, I. S., et al. (2021). Whole-genome analysis of multidrug-resistant Salmonella Enteritidis strains isolated from poultry sources in Korea. Pathogens 10:1615. doi: 10.3390/pathogens10121615
Lee, S., Park, N., Yun, S., Hur, E., Song, J., Lee, H., et al. (2021). Presence of plasmid-mediated quinolone resistance (PMQR) genes in non-typhoidal Salmonella strains with reduced susceptibility to fluoroquinolones isolated from human salmonellosis in Gyeonggi-do, South Korea from 2016 to 2019. Gut Pathog. 13:35. doi: 10.1186/s13099-021-00431-7
Lekagul, A., Tangcharoensathien, V., and Yeung, S. (2019). Patterns of antibiotic use in global pig production: a systematic review. Vet. Anim. Sci. 7:100058. doi: 10.1016/j.vas.2019.100058
Li, Y., Kang, X., Ed-Dra, A., Zhou, X., Jia, C., Müller, A., et al. (2022). Genome-based assessment of antimicrobial resistance and virulence potential of isolates of non-Pullorum/Gallinarum Salmonella serovars recovered from dead poultry in China. Microbiol. Spectr. 10:e0096522. doi: 10.1128/spectrum.00965-22
Li, Y., Ma, X., Li, C., Dai, X., and Zhang, L. (2020). Occurrence and genomic characterization of ESBL-producing Escherichia coli ST29 strains from swine with abundant virulence genes. Microb. Pathog. 148:104483. doi: 10.1016/j.micpath.2020.104483
Li, L., Olsen, R. H., Wang, C., Song, A., Xiao, J., Meng, H., et al. (2021). First report of two foodborne Salmonella enterica subsp. enterica serovar Bovismorbificans isolates carrying a novel mega-plasmid harboring Bla(DHA-1) and qnrB4 genes. Int. J. Food Microbiol. 360:109439. doi: 10.1016/j.ijfoodmicro.2021.109439
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., and Walsh, T. R. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, R., Xing, L., Zhou, G., and Zhang, W. (2017). What is meat in China? Anim. Front. 7, 53–56. doi: 10.2527/af.2017.0445
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–d917. doi: 10.1093/nar/gkab1107
Luo, M., She, Y., Jiang, Y., Xie, L., Yang, C., Qiu, Y., et al. (2022). Population dynamics and antimicrobial resistance of Salmonella derby ST40 from Shenzhen, China. Front. Microbiol. 13:1065672. doi: 10.3389/fmicb.2022.1065672
Magnet, S., Courvalin, P., and Lambert, T. (1999). Activation of the cryptic aac(6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J. Bacteriol. 181, 6650–6655. doi: 10.1128/JB.181.21.6650-6655.1999
Manzari, M., Fani, F., Alebouyeh, M., Moaddeli, A., Rahnamaye Farzami, M., Amin Shahidi, M., et al. (2022). Multidrug-resistant Salmonella strains from food animals as a potential source for human infection in Iran. Comp. Immunol. Microbiol. Infect. Dis. 90-91:101898. doi: 10.1016/j.cimid.2022.101898
Marmion, M., Ferone, M. T., Whyte, P., and Scannell, A. G. M. (2021). The changing microbiome of poultry meat; from farm to fridge. Food Microbiol. 99:103823. doi: 10.1016/j.fm.2021.103823
Mcmillan, E. A., Gupta, S. K., Williams, L. E., Jové, T., Hiott, L. M., Woodley, T. A., et al. (2019). Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 10:832. doi: 10.3389/fmicb.2019.00832
Minnullina, L., Pudova, D., Shagimardanova, E., Shigapova, L., Sharipova, M., and Mardanova, A. (2019). Comparative genome analysis of uropathogenic Morganella morganii strains. Front. Cell. Infect. Microbiol. 9:167. doi: 10.3389/fcimb.2019.00167
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841X.2018.1492902
Neuert, S., Nair, S., Day, M. R., Doumith, M., Ashton, P. M., Mellor, K. C., et al. (2018). Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front. Microbiol. 9:592. doi: 10.3389/fmicb.2018.00592
Nhung, N. T., Cuong, N. V., Thwaites, G., and Carrique-Mas, J. (2016). Antimicrobial usage and antimicrobial resistance in animal production in Southeast Asia: a review. Antibiotics (Basel) 5:37. doi: 10.3390/antibiotics5040037
Octavia, S., Chew, K. L., Chew, K. L., Lin, R. T. P., and Teo, J. W. P. (2020). Multidrug-resistant Salmonella enterica serovar London carrying Bla(NDM-1) encoding plasmid from Singapore. Clin. Microbiol. Infect. 26, 963–966. doi: 10.1016/j.cmi.2020.01.033
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Pan, H., Zhou, X., Chai, W., Paudyal, N., Li, S., Zhou, X., et al. (2019). Diversified sources for human infections by Salmonella enterica serovar Newport. Transbound. Emerg. Dis. 66, 1044–1048. doi: 10.1111/tbed.13099
Paudyal, N., Pan, H., Liao, X., Zhang, X., Li, X., Fang, W., et al. (2018). A meta-analysis of major foodborne pathogens in Chinese food commodities between 2006 and 2016. Foodborne Pathog. Dis. 15, 187–197. doi: 10.1089/fpd.2017.2417
Peng, M., Salaheen, S., Almario, J. A., Tesfaye, B., Buchanan, R., and Biswas, D. (2016). Prevalence and antibiotic resistance pattern of Salmonella serovars in integrated crop-livestock farms and their products sold in local markets. Environ. Microbiol. 18, 1654–1665. doi: 10.1111/1462-2920.13265
Perin, A. P., Martins, B. T. F., Barreiros, M. A. B., Yamatogi, R. S., Nero, L. A., and dos Santos Bersot, L. (2020). Occurrence, quantification, pulse types, and antimicrobial susceptibility of Salmonella sp. isolated from chicken meat in the state of Paraná, Brazil. Braz. J. Microbiol. 51, 335–345. doi: 10.1007/s42770-019-00188-x
Pethplerdprao, P., Supa-Amornkul, S., Panvisavas, N., and Chaturongakul, S. (2017). Salmonella enterica multilocus sequence typing and its correlation with serotypes. Food Biotechnol. 31, 73–79. doi: 10.1080/08905436.2017.1301820
Pightling, A. W., Pettengill, J. B., Luo, Y., Baugher, J. D., Rand, H., and Strain, E. (2018). Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front. Microbiol. 9:1482. doi: 10.3389/fmicb.2018.01482
Poirel, L., Lagrutta, E., Taylor, P., Pham, J., and Nordmann, P. (2010). Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54, 4914–4916. doi: 10.1128/AAC.00878-10
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Rincón-Gamboa, S. M., Poutou-Piñales, R. A., and Carrascal-Camacho, A. K. (2021). Analysis of the assessment of antimicrobial susceptibility. Non-typhoid Salmonella in meat and meat products as model (systematic review). BMC Microbiol. 21:223. doi: 10.1186/s12866-021-02268-1
Salipante, S. J., and Hall, B. G. (2003). Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol. Biol. Evol. 20, 653–659. doi: 10.1093/molbev/msg074
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sun, R. Y., Fang, L. X., Ke, B. X., Sun, J., Wu, Z. W., Feng, Y. J., et al. (2023). Carriage and transmission ofmcr-1in Salmonella typhimurium and its monophasic 1,4,[5],12:i:- variants from diarrheal outpatients: a 10-year genomic epidemiology in Guangdong, southern China. Microbiol. Spectr. 11:e0311922. doi: 10.1128/spectrum.03119-22
Sun, R. Y., Guo, W. Y., Zhang, J. X., Wang, M. G., Wang, L. L., Lian, X. L., et al. (2022). Phylogenomic analysis of Salmonella Indiana ST17, an emerging MDR clonal group in China. J. Antimicrob. Chemother. 77, 2937–2945. doi: 10.1093/jac/dkac243
Sun, T., Liu, Y., Qin, X., Aspridou, Z., Zheng, J., Wang, X., et al. (2021). The prevalence and epidemiology of salmonella in retail raw poultry meat in China: a systematic review and meta-analysis. Foods 10:2757. doi: 10.3390/foods10112757
Sun, L., Zhang, H., Chen, J., Chen, L., Qi, X., and Zhang, R. (2021). Epidemiology of foodborne disease outbreaks caused by nontyphoidal Salmonella in Zhejiang province, China, 2010–2019. Foodborne Pathogens and Disease, 18, 880–886. doi: 10.1089/fpd.2021.0006
Tack, B., Vanaenrode, J., Verbakel, J. Y., Toelen, J., and Jacobs, J. (2020). Invasive non-typhoidal Salmonella infections in sub-Saharan Africa: a systematic review on antimicrobial resistance and treatment. BMC Med. 18:212. doi: 10.1186/s12916-020-01652-4
Tamang, M. D., Nam, H. M., Kim, T. S., Jang, G. C., Jung, S. C., and Lim, S. K. (2011). Emergence of extended-spectrum beta-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J. Clin. Microbiol. 49, 2671–2675. doi: 10.1128/JCM.00754-11
Tang, B., Elbediwi, M., Nambiar, R. B., Yang, H., Lin, J., and Yue, M. (2022). Genomic characterization of antimicrobial-resistant Salmonella enterica in duck, chicken, and pig farms and retail markets in eastern China. Microbiol. Spectr. 10:e0125722. doi: 10.1128/spectrum.01257-22
Tang, B., Siddique, A., Jia, C., Ed-Dra, A., Wu, J., Lin, H., et al. (2023). Genome-based risk assessment for foodborne Salmonella enterica from food animals in China: a one health perspective. Int. J. Food Microbiol. 390:110120. doi: 10.1016/j.ijfoodmicro.2023.110120
Timme, R. E., Wolfgang, W. J., Balkey, M., Venkata, S. L. G., Randolph, R., Allard, M., et al. (2020). Optimizing open data to support one health: best practices to ensure interoperability of genomic data from bacterial pathogens. One Health Outlook 2, 1–11. doi: 10.1186/s42522-020-00026-3
Vázquez, X., Fernández, J., Alkorta, M., De Toro, M., Rodicio, M. R., and Rodicio, R. (2023). Spread of Bla(ctx-m-9) and other clinically relevant resistance genes, such as mcr-9 and qnra1, driven by inchi2-st1 plasmids in clinical isolates of monophasic Salmonella enterica serovar typhimurium ST34. Antibiotics (Basel) 12:547. doi: 10.3390/antibiotics12030547
Wang, W., Baloch, Z., Zou, M., Dong, Y., Peng, Z., Hu, Y., et al. (2018). Complete genomic analysis of a Salmonella enterica serovar typhimurium isolate cultured from ready-to-eat pork in China carrying one large plasmid containing mcr-1. Front. Microbiol. 9:616. doi: 10.3389/fmicb.2018.00616
Wang, Y., Liu, Y., Lyu, N., Li, Z., Ma, S., Cao, D., et al. (2023). The temporal dynamics of antimicrobial-resistant Salmonella enterica and predominant serovars in China. Natl. Sci. Rev. 10:nwac269. doi: 10.1093/nsr/nwac269
Wang, W., Peng, Z., Baloch, Z., Hu, Y., Xu, J., Zhang, W., et al. (2017). Genomic characterization of an extensively-drug resistance Salmonella enterica serotype Indiana strain harboring Bla(NDM-1) gene isolated from a chicken carcass in China. Microbiol. Res. 204, 48–54. doi: 10.1016/j.micres.2017.07.006
Wang, X., Wang, H., Li, T., Liu, F., Cheng, Y., Guo, X., et al. (2020b). Characterization of Salmonella spp. isolated from chickens in Central China. BMC Vet. Res. 16:299. doi: 10.1186/s12917-020-02513-1
Wang, W., Zhao, L., Hu, Y., Dottorini, T., Fanning, S., Xu, J., et al. (2020a). Epidemiological study on prevalence, serovar diversity, multidrug resistance, and ctx-m-type extended-spectrum β-lactamases of Salmonella spp. from patients with diarrhea, food of animal origin, and pets in several provinces of China. Antimicrob. Agents Chemother. 64:aac, 00092–00020. doi: 10.1128/AAC.00092-20
Wei, Z., Xu, X., Yan, M., Chang, H., Li, Y., Kan, B., et al. (2019). Salmonella typhimurium and Salmonella Enteritidis infections in sporadic diarrhea in children: source tracing and resistance to third-generation cephalosporins and ciprofloxacin. Foodborne Pathog. Dis. 16, 244–255. doi: 10.1089/fpd.2018.2557
WHO (2022). Food safety. Available at: https://www.who.int/en/news-room/fact-sheets/detail/food-safety (Accessed October 2022).
Wu, C., Yan, M., Liu, L., Lai, J., Chan, E. W., and Chen, S. (2018). Comparative characterization of nontyphoidal Salmonella isolated from humans and food animals in China, 2003-2011. Heliyon 4:e00613. doi: 10.1016/j.heliyon.2018.e00613
Xu, L., He, Q., Tang, Y., Wen, W., Chen, L., Li, Y., et al. (2022). Multi-locus sequence and drug resistance analysis of Salmonella infection in children with diarrhea in Guangdong to identify the dominant ST and cause of antibiotic-resistance. Exp. Ther. Med. 24:678. doi: 10.3892/etm.2022.11614
Yang, X., Huang, J., Zhang, Y., Liu, S., Chen, L., Xiao, C., et al. (2020). Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 713:136385. doi: 10.1016/j.scitotenv.2019.136385
Yu, G. (2020). Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinformatics 69:e96. doi: 10.1002/cpbi.96
Zakaria, Z., Hassan, L., Sharif, Z., Ahmad, N., Mohd Ali, R., Amir Husin, S., et al. (2022). Virulence gene profile, antimicrobial resistance and multilocus sequence typing of Salmonella enterica subsp. enterica serovar Enteritidis from chickens and chicken products. Animals (Basel) 12:97. doi: 10.3390/ani12010097
Zhang, S., Den Bakker, H. C., Li, S., Chen, J., Dinsmore, B. A., Lane, C., et al. (2019). SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl. Environ. Microbiol. 85, e01746–e01719. doi: 10.1128/AEM.01746-19
Zhang, Z., He, S., Xu, X., Chang, J., Zhan, Z., Cui, Y., et al. (2022a). Antimicrobial susceptibility and molecular characterization of Salmonella enterica serovar Indiana from foods, patients, and environments in China during 2007–2016. Food Control 131:108427. doi: 10.1016/j.foodcont.2021.108427
Zhang, Z., Tian, X., and Shi, C. (2022b). Global spread of mcr-producing Salmonella enterica isolates. Antibiotics (Basel) 11:998. doi: 10.3390/antibiotics11080998
Zhang, C. Z., Zhang, Y., Ding, X. M., Lin, X. L., Lian, X. L., Trampari, E., et al. (2020). Emergence of ciprofloxacin heteroresistance in foodborne Salmonella enterica serovar Agona. J. Antimicrob. Chemother. 75, 2773–2779. doi: 10.1093/jac/dkaa288
Zhou, Z., Alikhan, N. F., Mohamed, K., Fan, Y., and Achtman, M. (2020). The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 30, 138–152. doi: 10.1101/gr.251678.119
Keywords: Salmonella, retail chicken, genome sequencing, humans with diarrhea, antimicrobial resistance
Citation: Wang W, Cui J, Liu F, Hu Y, Li F, Zhou Z, Deng X, Dong Y, Li S and Xiao J (2023) Genomic characterization of Salmonella isolated from retail chicken and humans with diarrhea in Qingdao, China. Front. Microbiol. 14:1295769. doi: 10.3389/fmicb.2023.1295769
Edited by:
Atte Johannes Von Wright, University of Eastern Finland, FinlandReviewed by:
Sunghyun Yoon, National Center for Toxicological Research (FDA), United StatesAbdelaziz Ed-Dra, Université Sultan Moulay Slimane, Morocco
Copyright © 2023 Wang, Cui, Liu, Hu, Li, Zhou, Deng, Dong, Li and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinping Dong, ZG9uZ3lpbnBpbmdAY2ZzYS5uZXQuY24=; Shaoting Li, c2hhb3RpbmcubGlAZ2R1dC5lZHUuY24=; Jing Xiao, eGlhb2ppbmdAY2ZzYS5uZXQuY24=
†These authors have contributed equally to this work
 Wei Wang
Wei Wang Jing Cui2†
Jing Cui2† Yujie Hu
Yujie Hu Fengqin Li
Fengqin Li Xiangyu Deng
Xiangyu Deng Yinping Dong
Yinping Dong Shaoting Li
Shaoting Li