- 1College of Biological Sciences and Technology, Beijing Forestry University, Beijing, China
- 2National Engineering Research Center of Tree Breeding and Ecological Restoration, Beijing Forestry University, Beijing, China
- 3College of Biological and Chemical Engineering, Qilu Institute of Technology, Jinan, China
Introduction: Penicillium species exhibit a broad distribution in nature and play a crucial role in human and ecological environments.
Methods: Two Penicillium species isolated from the ancient Great Wall loess in the Mentougou District of Beijing, China, were identified and described as new species, namely, Penicillium acidogenicum and P. floccosum, based on morphological characteristics and phylogenetic analyses of multiple genes including ITS, BenA, CaM, and RPB2 genes.
Results: Phylogenetic analyses showed that both novel species formed a distinctive lineage and that they were most closely related to P. chrzaszczii and P. osmophilum, respectively.
Discussion: Penicillium acidogenicum is characterized by biverticillate conidiophores that produce globose conidia and is distinguished from similar species by its capacity to grow on CYA at 30°C. Penicillium floccosum is typically recognized by its restricted growth and floccose colony texture. The description of these two new species provided additional knowledge and new insights into the ecology and distribution of Penicillium.
1 Introduction
Penicillium is widely distributed in various habitats, including soil, plants, air, and indoor settings, and various types of foods (Frisvad and Samson, 2004; Houbraken and Samson, 2011; Visagie et al., 2014a). Penicillium fungi, such as P. rubens for penicillin production, P. citrinum for synthesizing cholesterol-lowering drug mevastatin, P. camemberti and P. roqueforti for cheese production, and P. oxalicum with biocontrol potential, have significant economic value in antibiotic production, pharmaceutical synthesis, biocontrol, food processing, and food safety (Giraud et al., 2010; Houbraken et al., 2011a; Tsang et al., 2018; Steenwyk et al., 2019; Dumas et al., 2020; Yang et al., 2022). In addition, Penicillium can have some negative effects in some cases, such as producing a variety of mycotoxins that can cause food contamination and even threaten human health (Frisvad et al., 2004; Perrone and Susca, 2017; Stefanello et al., 2022).
Penicillium, established by Link (1809), derives its name from the Latin word penicillus, meaning small brush or paintbrush. The infrageneric classification system of Penicillium was mainly based on morphological characteristics in the past 100 years, whereas this phenotype-based sectional classification has been replaced by a system based on a multigene phylogeny in recent decades (Visagie et al., 2014a; Houbraken et al., 2016, 2020). Subgenera, sections, and series are usually transformed from well-supported clades based on DNA sequence analyses. Next-generation sequencing technology has made it possible to obtain a growing number of complete or nearly complete fungal genomes. Phylogenetic analysis based on whole genome sequences to determine the taxonomic position of Penicillium and its subordinate members is becoming an important trend in the future (Yang et al., 2016). Currently, 558 species of Penicillium were accepted (Wang et al., 2023) and were grouped into two subgenera, namely, Aspergilloides and Penicillium, 32 sections and 89 series (Houbraken et al., 2020). Species classified in the same section or series share many common features. For example, the series Canescentia and Atroveneta are closely related in phylogeny, but they can be distinguished by different extrolite profiles and colony textures. Therefore, defining a new species into a section or series could be highly predicted for their functional characteristics (Houbraken et al., 2020).
Penicillium section Citrina comprises a diverse range of species that exhibit a broad distribution and usually occur in soil habitats. Members of this group are distinguished by their symmetrically biverticillate conidiophores and relatively small, globose to subglobose conidia (Houbraken et al., 2011b; Visagie et al., 2014b; Visagie and Yilmaz, 2023). Furthermore, members of this section have a high potential to produce the mycotoxins citrinin (Houbraken et al., 2011b; Dutra-Silva et al., 2021; Yin et al., 2021). Currently, this section includes 47 species (Houbraken et al., 2020; Andrade et al., 2021; Nguyen et al., 2021; Ashtekar et al., 2022; Tan and Shivas, 2022; Visagie and Yilmaz, 2023). Penicillium sect. Osmophila was introduced by Houbraken et al. (2016) for species producing bi-, ter-, and quarter-verticillate conidiophores and demonstrating comparable growth rates on CYA when they were incubated at 15 and 25°C. This section currently only contains two species (Houbraken et al., 2020). Members of this section are isolated from soil, and no specific metabolites have been found (Houbraken et al., 2016). Due to the difficulty of delimiting the species within these sections solely based on phenotypic characteristics, a polyphasic approach incorporating morphological, extrolite, genetic data, and multigene phylogenetic analysis has been extensively employed for species identification (Visagie et al., 2014a).
During a survey of Penicillium diversity in China, two strains isolated from the ancient Great Wall loess at Qingshui Town, Mentougou District, Beijing, were identified as two new species by multiphase classification. In this study, we provided the morphology of these new species and conducted the phylogenetic analyses using the internal transcribed spacer rDNA area (ITS), partial β-tubulin (BenA), calmodulin (CaM), and the RNA polymerase II second largest subunit (RPB2) genes and compared them with closely related species. The description of these two novel species is expected to enrich our comprehension of Penicillium ecology and distribution.
2 Materials and methods
2.1 Sampling and isolation
Soil samples were collected from loess at the base of the ancient Great Wall (Hongshui Kou section) (39°59′16”N, 115°28′54″E) in Mentougou District, Beijing, China. Cultures were isolated from the soil using the dilution plate method. Initially, 10 g of soil sample was thoroughly mixed with 90 mL of sterile water to prepare a soil suspension. This suspension was then serially diluted to 10−2, 10−3, and 10−4 concentrations. Subsequently, 100 μL of each diluted suspension was plated on potato glucose agar (PDA) with penicillin (50 ppm) and streptomycin (50 ppm) (Lin, 2010). All plates were incubated at 25°C. Type specimens (dry cultures) were deposited in the Fungarium (HMAS), Institute of Microbiology, Chinese Academy of Sciences. Ex-type strains (living cultures) were deposited in the China General Microbiological Culture Collection Centre (CGMCC).
2.2 Morphology
Colony characters were observed for strains grown on Czapek yeast autolysate agar (CYA), malt extract agar (MEA), yeast extract sucrose agar (YES), dichloran 18% glycerol agar (DG18), and creatine sucrose agar (CREA). The cultures were incubated at 25°C for 7 days, with extra CYA plates incubated at 30 and 37°C, which are useful for species distinction. Culture media preparation, inoculation technique, and incubation conditions followed the methods described by Visagie et al. (2014a). Color names and codes referred to the Color standards and color nomenclature (Ridgway, 1912). For microscopic observations, slides were made from colonies that have been growing on MEA for 7 days, using phenol glycerin solution as mounting fluid or staining with cotton blue. The isolates were tested for indole metabolite production using the Ehrlich reagent and a filter paper method (Lund, 1995). A violet ring observed within 10 min was considered a positive reaction, while any other color was defined as a negative response (Houbraken et al., 2016).
2.3 Observation of scanning electron microscope
Strains were grown for 5–7 days on MEA or PDA, and the conidiophores were mature. Agar blocks (4 × 4 mm) with conidial structures were cut with a blade before transferring to sterile Petri dishes. They were initially fixed with 2.5% glutaraldehyde at room temperature for 2 h, followed by an overnight incubation at 4°C. Subsequently, a gradient dehydration process involved varying ethanol concentrations (30, 50, 70, 95, and 100%), before a transition to tert-butanol (Zhang et al., 2016; Mukherjee et al., 2022). Finally, the samples were freeze-dried, sprayed with gold, and observed using FESEM (Hitachi SU8010, Japan).
2.4 DNA extraction, sequencing, and phylogenetic analysis
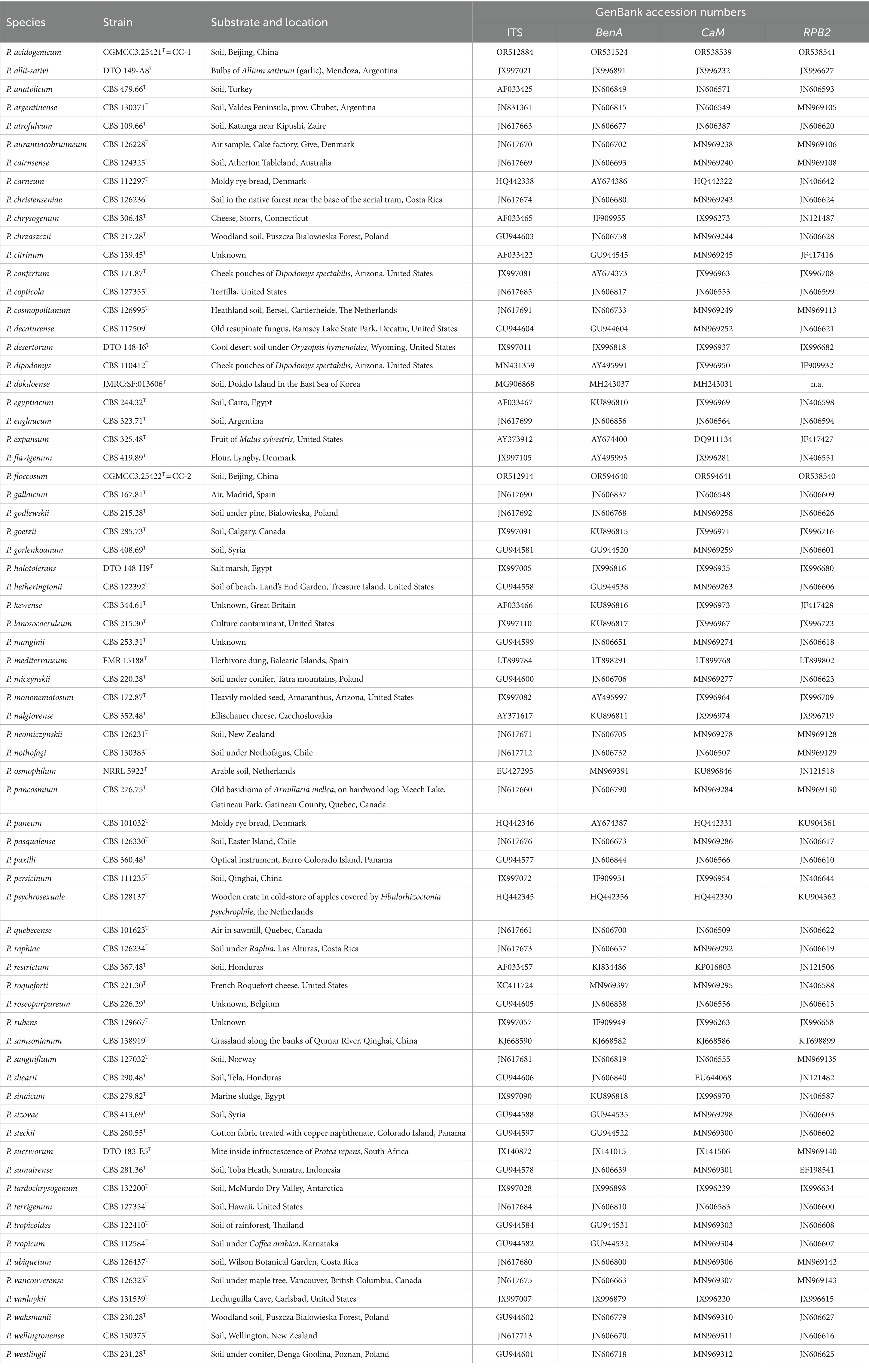
Strains were grown on PDA for 7 days and DNA was extracted using the E.Z.N.A.® Fungal DNA Mini Kit (Omega Bio-Tek, Inc., United States), involving fungal tissue disruption and lysis, isopropanol precipitation of DNA, precipitation of proteins, and DNA elution. Primers, PCR amplification, and DNA sequencing methods used for the ITS, BenA, CaM, and RPB2 genes were based on the description of Visagie et al. (2014a). The newly generated sequences were submitted to GenBank.1
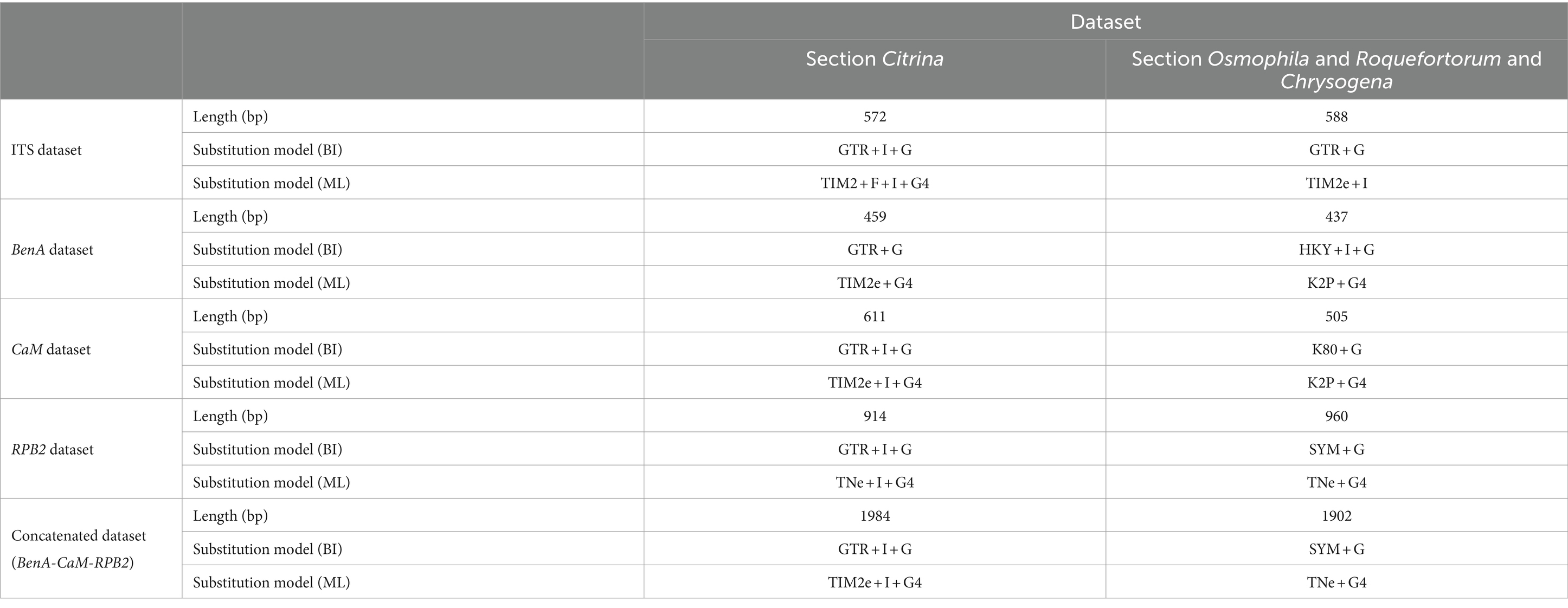
Sequence datasets were established using newly generated sequences and reference-type sequences retrieved from GenBank. All datasets were aligned using MEGA 11 implementing the Align by ClustalW option (Tamura et al., 2021). Datasets were analyzed using maximum likelihood (ML) and Bayesian inference (BI). ML analyses were performed within IQtree v. 1.6.12 (Nguyen et al., 2015) and tested by standard non-parametric bootstrap analyses for 1,000 replications (Visagie et al., 2021). The best model for ML was determined using ModelFinder (Kalyaanamoorthy et al., 2017), a fast model-selection method implemented in IQtree. Bayesian inference (BI) analyses were conducted using MrBayes v. 3.2.7 (Ronquist et al., 2012), with a sampling frequency of 100 and the exclusion of the initial 25% of trees as burn-in. The sequences used for phylogenetic analyses in this study are listed in Table 1. Gene sequence alignment datasets were stored in TreeBASE2 with the submission number 30834.
3 Results
3.1 Isolates and identification
Isolations resulted in six fungal isolates obtained from the ancient Great Wall loess, with two suspect new species CC-1(CGMCC 3.25421T) and CC-2(CGMCC 3.25422T). Through sequencing of ITS, BenA, CaM, and RPB2 genes, CC-1(CGMCC 3.25421T) generated gene fragments with sizes of 549 bp, 459 bp, 534 bp, and 1,070 bp, and CC-2(CGMCC 3.25422T) with sizes of 541 bp, 453 bp, 520 bp, and 1,053 bp, respectively. The blast results of ITS, BenA, CaM, and RPB2 genes showed that CC-1 was closely related to P. chrzaszczii (Identities: ITS: 99.62%, BenA: 95.62%, CaM: 96.21%, RPB2: 97.27%), while CC-2 was closely related to P. osmophilum (Identities: ITS: 98.34%, BenA: 96.22%, CaM: 95.31%, RPB2: 96.97%). Preliminary identification based on blast results of sufficient gene and morphological characteristics, strain CC-1 was designated as a member of Penicillium section Citrina and strain CC-2 was a member of section Osmophila, but neither strain could be identified as any known species, so further phylogenetic analyses were performed.
3.2 Phylogenetic analyses
Phylogenetic analyses of the Penicillium sections Citrina, Chrysogena, Osmophila, and Roquefortorum were conducted using the ITS, BenA, CaM, and RPB2 genes, along with a concatenation of the latter three genes, to determine the phylogenetic position of the new species (CGMCC 3.25421T and CGMCC 3.25422T). A total of 43 ex-(neo) type strains were involved in the analyses of the individual and combined datasets of section Citrina, while 26 ex-(neo) type strains were involved in the analyses of the sections Chrysogena, Osmophila, and Roquefortorum. A summary of the length and substitution models for each dataset is provided in Table 2.
3.2.1 Section Citrina
Phylogenetic analyses based on concatenated datasets (BenA-CaM-RPB2) divided section Citrina into nine clades (Figure 1), which is consistent with the study by Houbraken et al. (2020). Penicillium acidogenicum (CC-1 = CGMCC3.25421T) was introduced as a new species, comprising a well-supported distinct clade related to species P. chrzaszczii in series Westlingiorum (95% bs, 1.00 pp) (Figure 1). The phylogenetic analyses of single-gene revealed that the CaM phylogeny better resolved the relationship between these branches in section Citrina compared to the ITS, BenA, and RPB2 phylogenies. ITS has poor discriminatory ability in this section. BenA, CaM, and RPB2 can easily distinguish the new species, but BenA cannot reliably distinguish among P. decaturense, P. pancosmium, and P. ubiquetum (Figure 2).
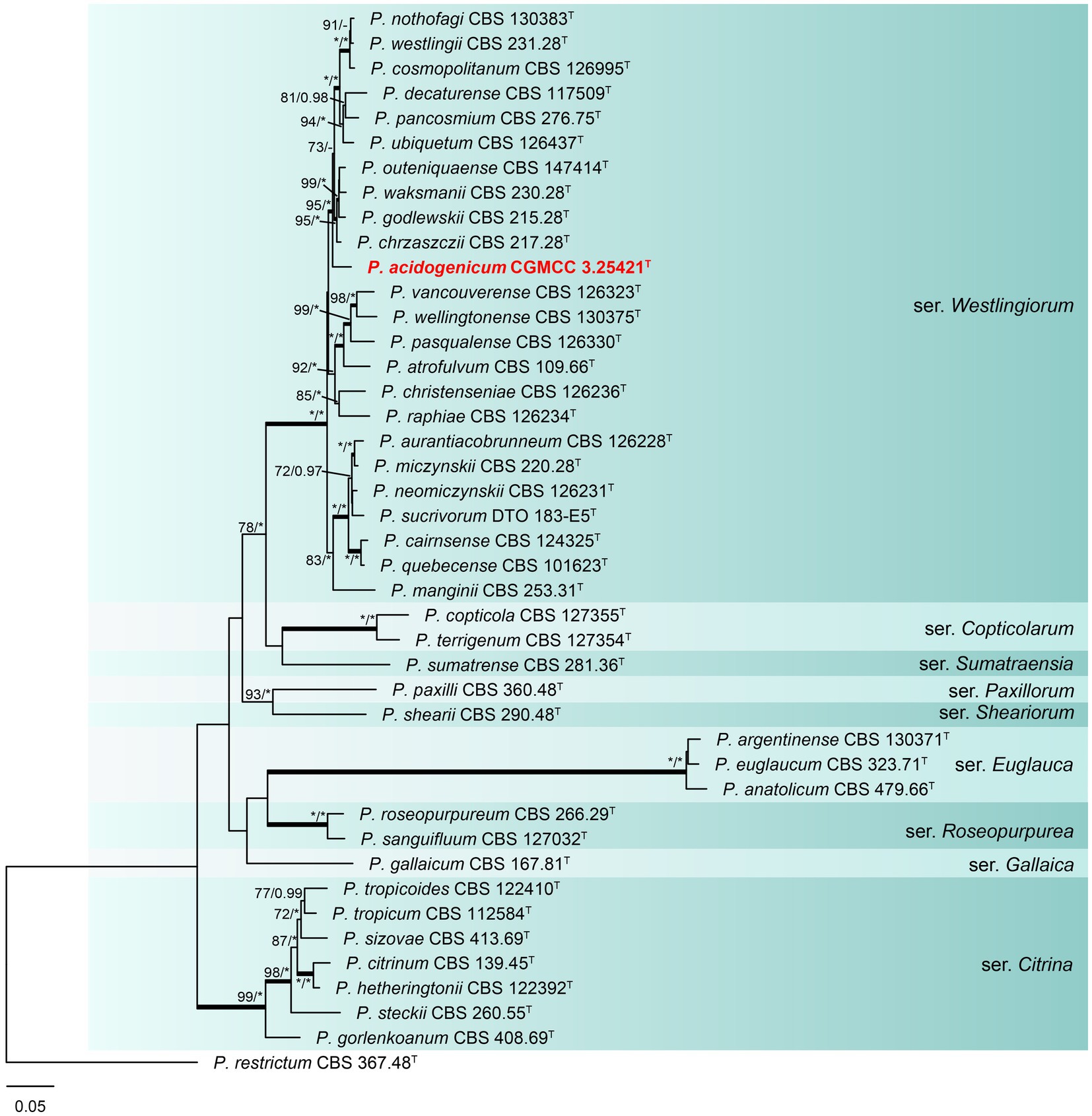
Figure 1. ML phylogenetic tree for concatenated datasets (BenA-CaM-RPB2) of Penicillium acidogenicum and Penicillium section Citrina. Penicillium restrictum was chosen as the outgroup. Bootstrap values (bs) above 70% or posterior probability (pp) above 0.95 are shown at nodes. The branches with more than 95% bs and 1.00 pp values are thickened. Names in red text indicate strains that belong to the new species in this study. *Indicates bs = 100% or pp = 1.00, T = ex-type strain.
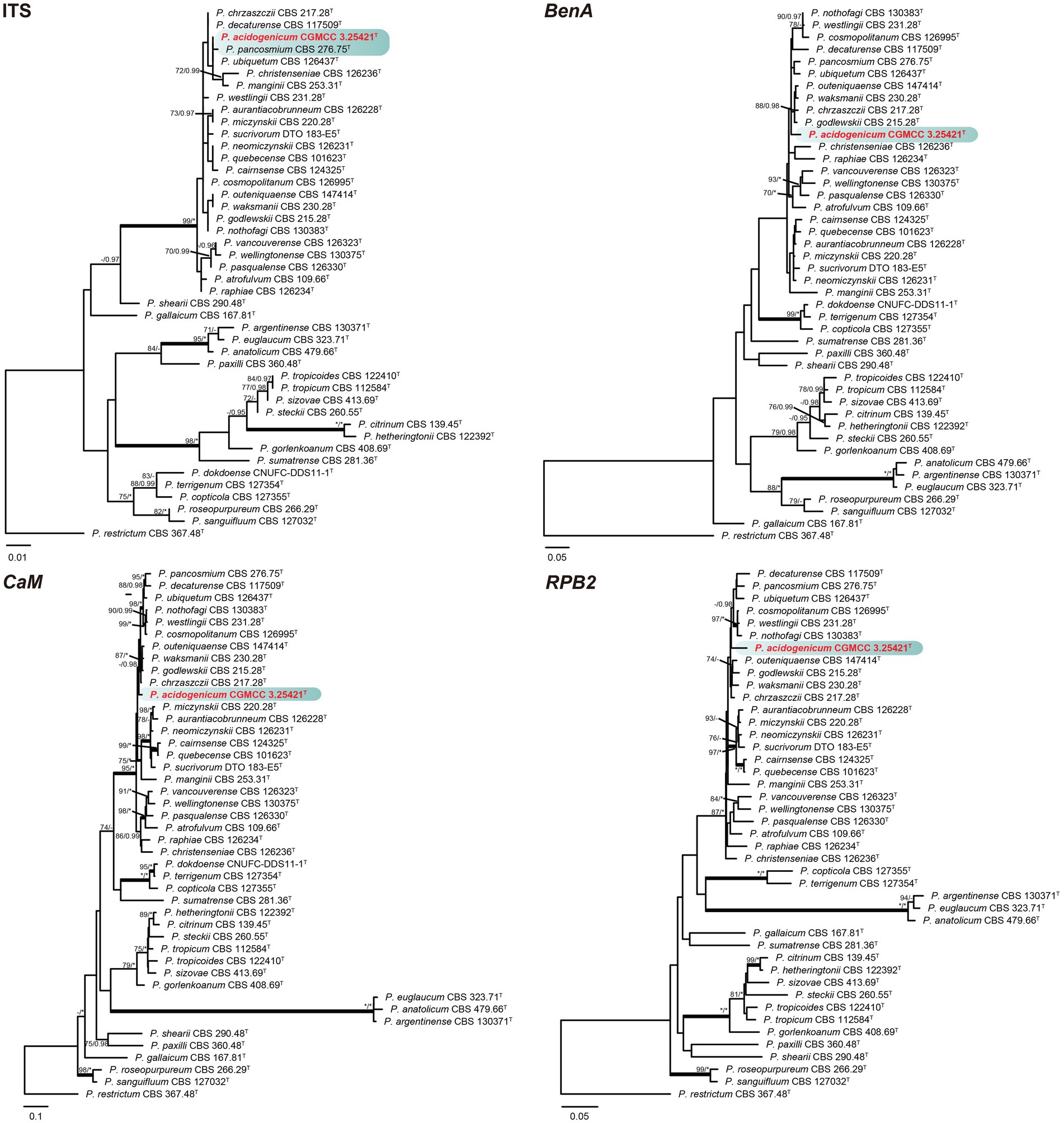
Figure 2. ML phylogenetic tree for individual gene dataset of ITS, BenA, CaM, and RPB2 of Penicillium acidogenicum and Penicillium section Citrina. Penicillium restrictum was chosen as the outgroup. Bootstrap values (bs) above 70% or posterior probability (pp) above 0.95 are shown at nodes. The branches with more than 95% bs and 1.00 pp values are thickened. Names in red text indicate strains that belong to the new species in this study. *Indicates bs = 100% or pp = 1.00, T = ex-type strain.
3.2.2 Section Chrysogena, Osmophila, and Roquefortorum
Phylogenetic analyses revealed a new species in section Osmophila and described it as Penicillium floccosum (CC-2 = CGMCC3.25422T). This species was grouped in a clade as P. osmophilum with robust support (100% bs, 1.00 pp) (Figure 3). In the single-gene phylogenies, P. floccosum and P. osmophilum formed a clade with a high degree of support, except for ITS (Figure 4). Compared with ITS, BenA, CaM, and RPB2 can easily distinguish all species of these sections.
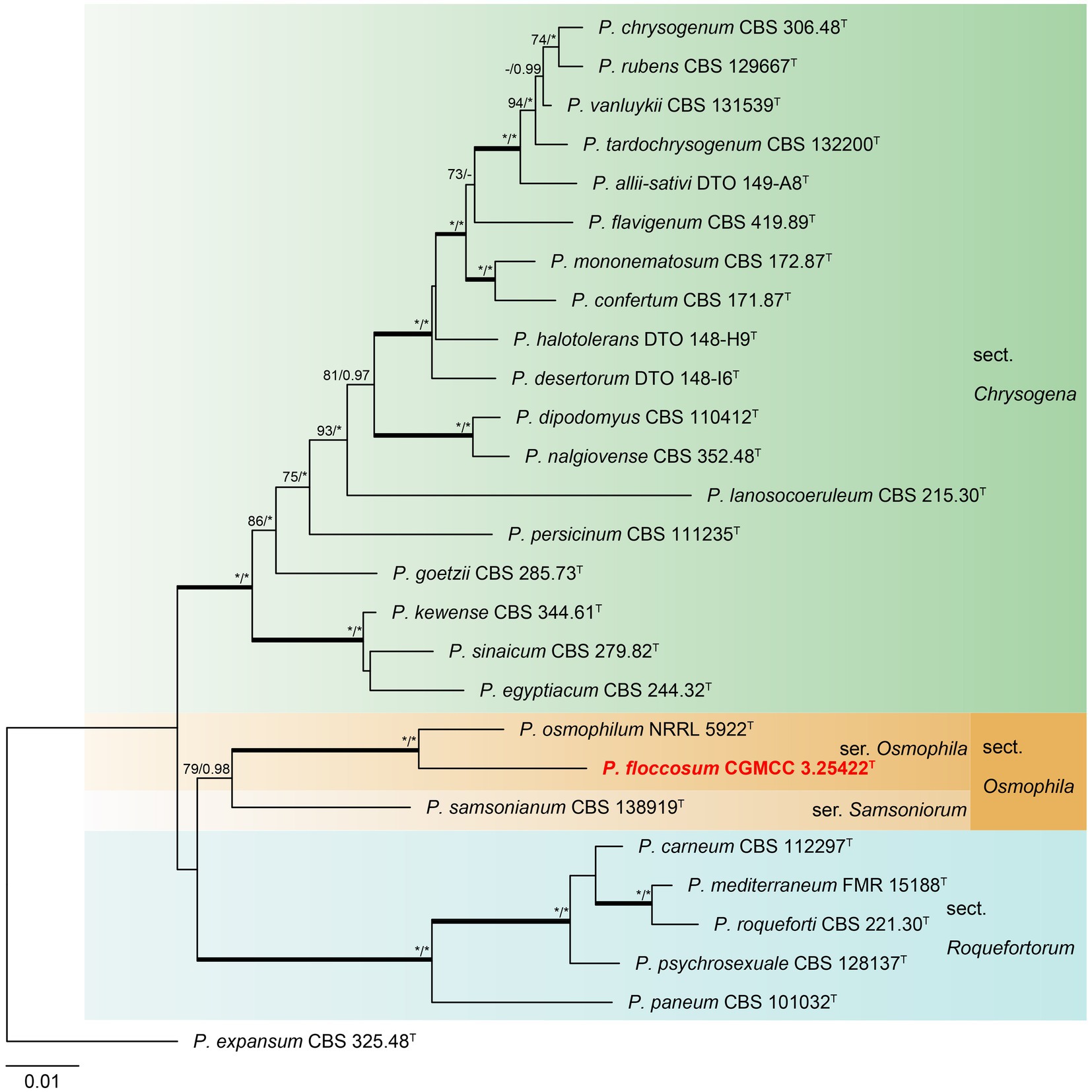
Figure 3. ML phylogenetic tree for concatenated datasets (BenA-CaM-RPB2) of Penicillium floccosum and Penicillium sections Chrysogena, Osmophila, and Roquefortorum. Penicillium expansum was chosen as the outgroup. Bootstrap values (bs) above 70% or posterior probability (pp) above 0.95 are shown at nodes. The branches with more than 95% bs and 1.00 pp values are thickened. Names in red text indicate strains that belong to the new species in this study. *Indicates bs = 100% or pp = 1.00, T = ex-type strain.
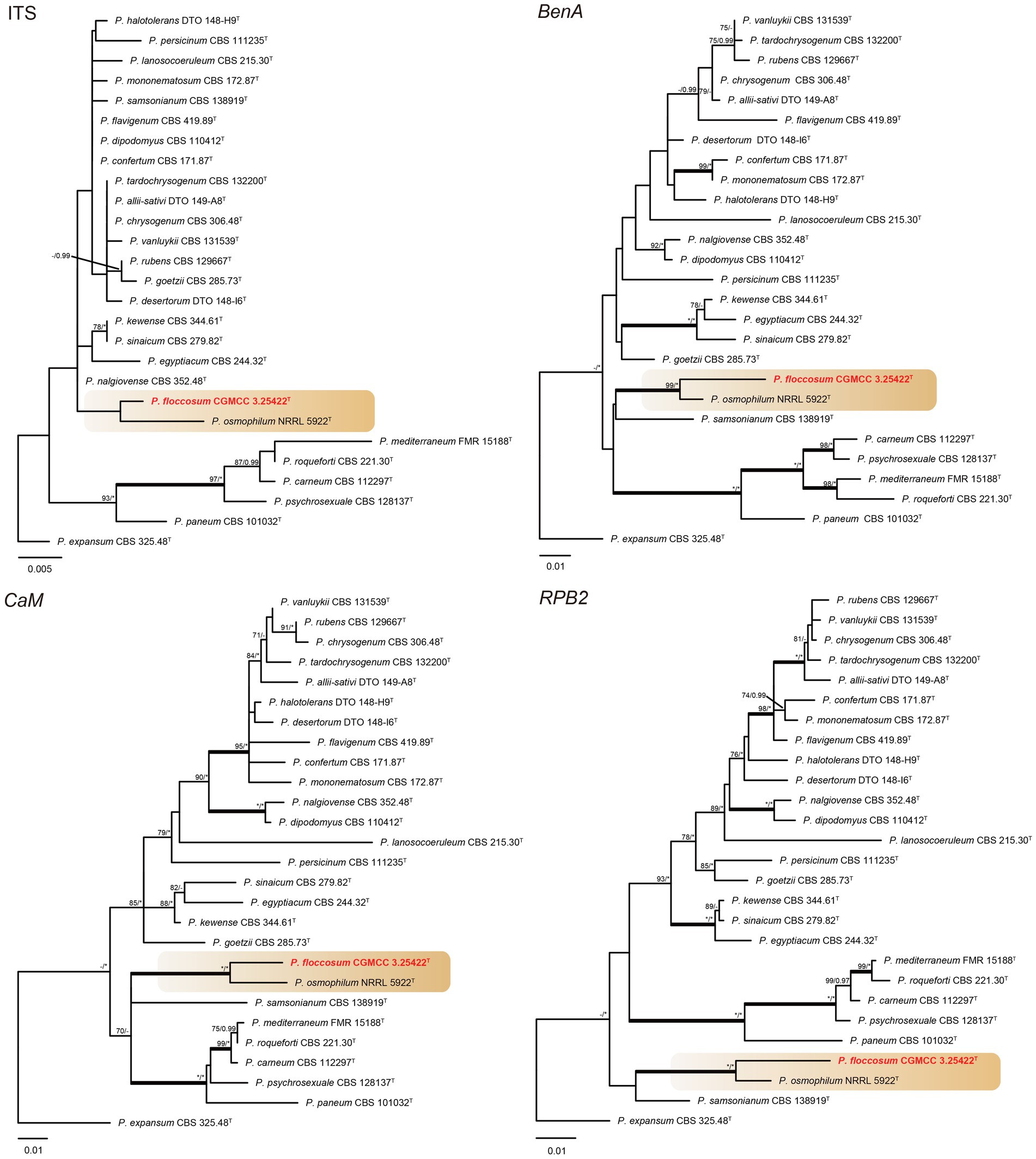
Figure 4. ML phylogenetic tree for individual gene dataset of ITS, BenA, CaM, and RPB2 of Penicillium floccosum and Penicillium sections Chrysogena, Osmophila, and Roquefortorum. Penicillium expansum was chosen as the outgroup. Bootstrap values (bs) above 70% or posterior probability (pp) above 0.95 are shown at nodes. The branches with more than 95% bs and 1.00 pp values are thickened. Names in red text indicate strains that belong to the new species in this study. *Indicates bs = 100% or pp = 1.00, T = ex-type strain.
3.3 Taxonomy
3.3.1 Penicillium acidogenicum R. N. Liang and G. Z. Zhao, sp. nov.
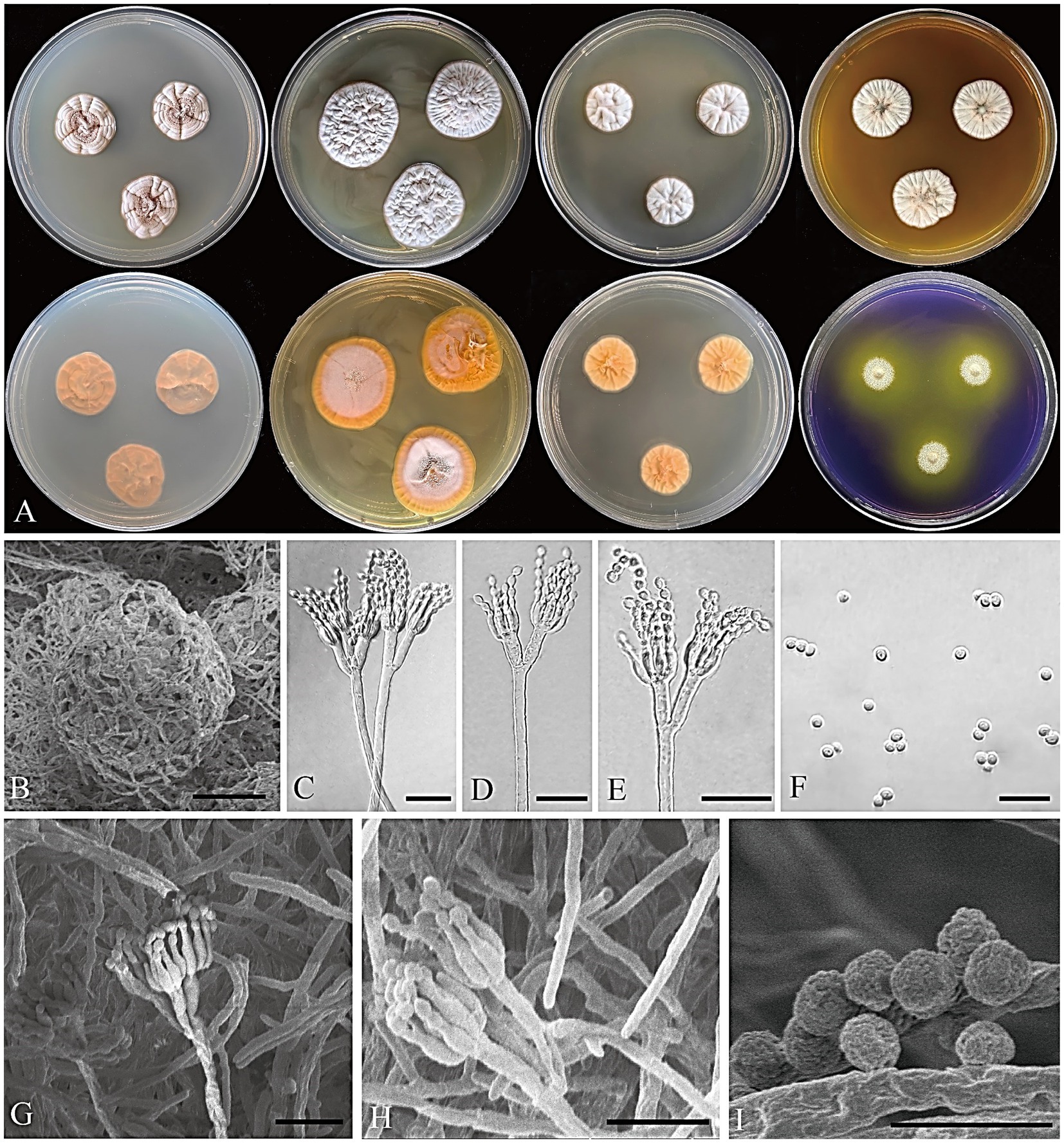
Figure 5. Penicillium acidogenicum CGMCC 3.25421. (A) Colonies on medium for 7 days (left to right, first row: CYA, YES, DG18, and MEA obverse; second row: CYA, YES, DG18 reverse, and CREA obverse); (B) SEM micrograph of cleistothecia; (C–E) Conidiophores; (F) Conidia; (G,H) SEM micrograph of conidiophores; (I) SEM micrograph of conidia. Scale bars: B = 50 μm, C–H = 10 μm, I = 5 μm.
MycoBank number: 850530.
Infrageneric classification: subgenus Aspergilloides, section Citrina, series Westlingiorum.
Etymology: “acidogenicum” refers to the acid-producing characteristics of colonies grown on CREA.
Type: CHINA. Beijing, Mentougou District, Qingshui Town, from the ancient Great Wall loess, 27 August 2022, collected by G. Z. Zhao, CC-1 (holotype HMAS 352643, dried culture; culture ex-type CGMCC 3.25421).
Colony diameter after 7 days (mm): CYA 18–21; MEA 20–23; YES 24–29; DG18 16–19; CREA: 10–13; CYA 30°C 14–16; CYA 37°C no growth.
Colony characteristics (7 days): CYA at 25°C: Colonies moderately deep, sulcate, slightly elevated at the center; margins entire; mycelium white; texture floccose and funicolose; sporulation moderate, conidia light grayish vinaceous (R. Pl. VIII); exudate clear; reverse light orange-yellow (R. Pl. LXXI); soluble pigment absent. CYA at 30°C: Colonies moderately deep, sulcate, slightly elevated at the center; margins entire; mycelium white; texture floccose and funicolose; sporulation moderate, conidia light grayish vinaceous (R. Pl. XXXII) to gray; exudate clear; reverse orange-pink (R. Pl. LII); soluble pigment absent. MEA at 25°C: Colonies moderately deep, radially sulcate; margins entire; mycelium white; texture floccose; sporulation sparse, conidia livid pink (R. Pl. IV); exudate clear; reverse cadmium orange (R. Pl. XLVIII); soluble pigment absent. YES at 25°C: Colonies moderately deep, sulcate, elevated at the center; margins entire; mycelium white; texture floccose; sporulation sparse to absent; exudate absent; reverse orange (R. Pl. L); soluble pigment absent. DG18 at 25°C: Colonies moderately deep, sulcate, raised at the center; margins entire; mycelium white; texture floccose; sporulation sparse to absent; exudate absent; reverse pale orange-yellow (R. Pl. LXX); soluble pigment absent. CREA at 25°C: weak growth, moderate acid production. Ehrlich reaction negative.
Micromorphology: Cleistothecia produced on CYA and MEA, 80–130 μm diam; conidiophores biverticillate; stipes smooth-walled, 80–235 × 2.5–3.5 μm; metulae divergent, 2–4 per stipe, 8–13 × 1.5–3 μm; phialides ampulliform, 4–8 per metula, 5.5–7.5 × 2–2.5 μm; conidia globose, finely rough, 2–3 μm diam.
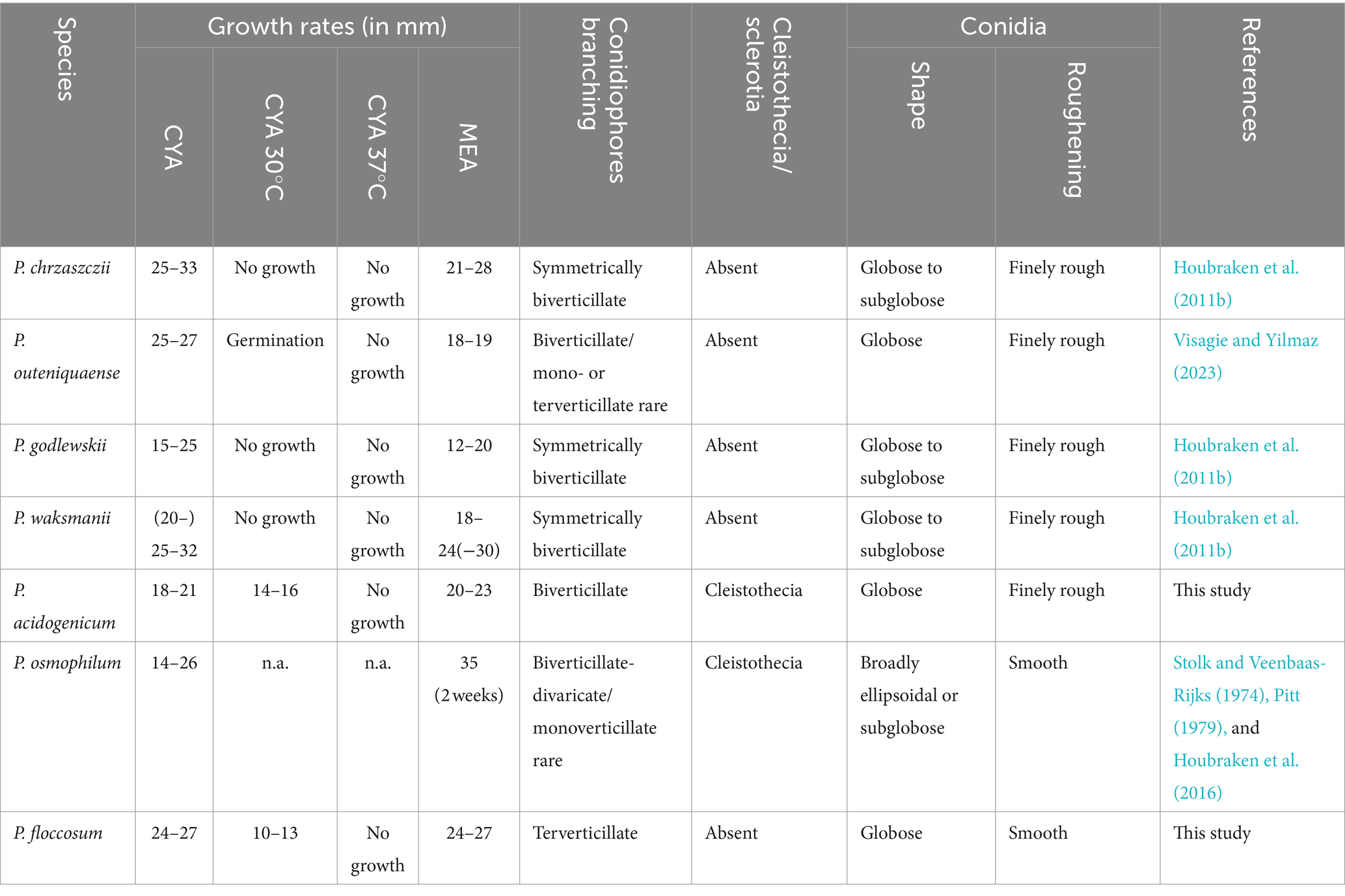
Notes: Phylogenetic analyses clustered Penicillium acidogenicum within a sister clade alongside 10 species, including P. chrzaszczii, P. godlewskii, P. waksmanii, P. outeniquaense, P. ubiquetum, P. pancosmium, P. decaturense, P. cosmopolitanum, P. westlingii, and P. nothofagi. The new species P. acidogenicum is phylogenetically most closely related to P. chrzaszczii, P. outeniquaense, P. waksmanii, and P. godlewskii. However, the former (colony 14–16 mm diam) can grow on CYA at 30°C, and the latter four cannot grow at 30°C. Penicillium acidogenicum can produce acid on CREA which is easily distinguished from the non-acid-producing character of closely related species (Table 3). The growth of P. acidogenicum is more restricted on DG18 (colony 16–19 mm) than P. chrzaszczii (20–27 mm), P. outeniquaense (20–21 mm), and P. waksmanii (16–27 mm) (Houbraken et al., 2011b; Visagie and Yilmaz, 2023).
3.3.2 Penicillium floccosum R. N. Liang and G. Z. Zhao, sp. nov.
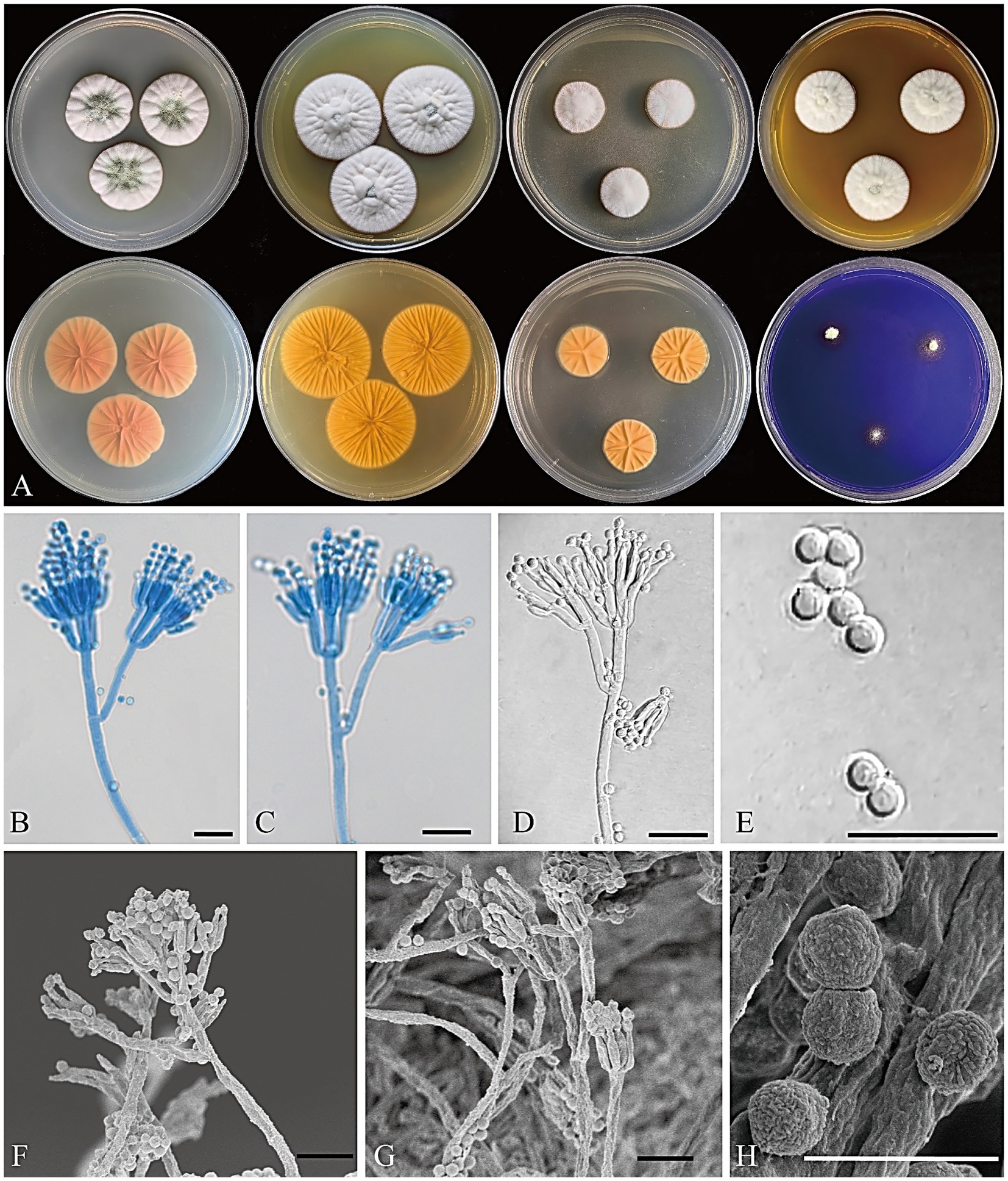
Figure 6. Penicillium floccosum CGMCC 3.25422. (A) Colonies on medium for 7 days (left to right, first row: CYA, YES, DG18, and MEA obverse; second row: CYA, YES, DG18 reverse, and CREA obverse); (B–D) Conidiophores; (E) Conidia; (F,G) SEM micrograph of conidiophores; (H) SEM micrograph of conidia. Scale bars: B–G = 10 μm, H = 5 μm.
MycoBank number: 850534.
Infrageneric classification: subgenus Penicillium, section Osmophila, series Osmophila.
Etymology: “floccosum” refers to its floccose colony texture.
Type: CHINA. Beijing, Mentougou District, Qingshui Town, from the ancient Great Wall loess, 27 August 2022, collected by G. Z. Zhao, CC-2 (holotype HMAS 352644, dried culture; culture ex-type CGMCC 3.25422).
Colony diameter after 7 days (mm): CYA 24–27; MEA 24–27; YES 31–34; DG18 18–19; CREA: 5–8; CYA 30°C 10–13; CYA 37°C no growth.
Colony characteristics (7 days): CYA at 25°C: Colonies moderately deep, radially sulcate, elevated at the margin; margins entire; mycelium white; texture floccose; sporulation moderate, conidia dark greenish glaucous (R. Pl. CXXXV); exudate clear; reverse peach red (R. Pl. XXXVII); soluble pigment absent. CYA at 30°C: Colonies moderately deep, radially sulcate, elevated at the center; margins entire; mycelium white; texture floccose; sporulation absent; exudate absent; reverse peach red (R. Pl. XXXVII); soluble pigment absent. MEA at 25°C: Colonies moderately deep, radially sulcate, elevated at the center; margins entire; mycelium white to pale yellow; texture floccose; sporulation sparse, conidia tea green (R. Pl. CXXII); exudate absent; reverse orange to cadmium orange (R. Pl. L); soluble pigment absent. YES at 25°C: Colonies moderately deep, radially sulcate, elevated at center; margins entire; mycelium white; texture floccose; sporulation sparse, conidia cadet gray (R. Pl. CLXXXV) to calamine blue (R. Pl. CLXXI); exudate absent; reverse cadmium yellow (R. Pl. LXVIII) to cadmium orange (R. Pl. L); soluble pigment absent. DG18 at 25°C: Colonies moderately deep, radially sulcate, raised at the center; margins entire; mycelium white; texture floccose; sporulation absent; exudate absent; reverse light orange–yellow (R. Pl. LXXI); soluble pigment absent. CREA at 25°C: weak growth, no acid production. Ehrlich reaction negative.
Micromorphology: Conidiophores terverticillate; stipes smooth to nearly smooth-walled, 70–300 × 2–4 μm; rami 11–21 × 1.5–3 μm; metulae divergent, 2–4 per branch/ramus, 8–12 × 1.5–2.5 μm; phialides ampulliform to cylindrical, 2–6 per metula, 6–7.5 × 1.5–2.5 μm; conidia globose, smooth, 2–3.5 μm diam.
Notes: Penicillium floccosum is classified in the section Osmophila and phylogenetically closely related to P. osmophilum. However, P. osmophilum produces ascomata, a feature lacking in the new species. Penicillium floccosum produces globose conidia and is distinguished from P. osmophilum which produces pear-shaped to ellipsoidal, occasionally subglobose conidia (Table 3; Stolk and Veenbaas-Rijks, 1974).
4 Discussion
The delimitation of Penicillium species currently relies on a polyphasic approach, typically including morphological characteristics, extrolites data, and multigene phylogenetic analyses. DNA sequence markers used for identification and phylogeny include ITS, BenA, CaM, and RPB2 genes (Houbraken et al., 2020). The ITS region is widely recognized as a universal barcode for fungi (Schoch et al., 2012). Nevertheless, in Penicillium, ITS is inadequate to distinguish between all closely related species, and secondary markers including BenA, CaM, and RPB2 genes are often needed to identify isolates to species accurately (Visagie et al., 2014a). BenA offers accurate identification of Penicillium species as do CaM and RPB2 (Visagie et al., 2016). Furthermore, RPB2 contains almost no introns, making it robust and easy to align for phylogenies (Vetrovsky et al., 2016). However, the RPB2 gene is usually difficult to amplify, probably because the RPB2 gene sequence varies significantly among different fungal species, and thus, the universal primers contain some degenerate bases that reduce the specificity of PCR amplifications (Liu et al., 1999; Diao et al., 2019). To enhance the all-inclusiveness of the RPB2 database, it is possible to design primers with higher specificity targeting Penicillium and reduce the generation of non-specific amplification products. On the other hand, cloning of the amplification products and sequencing of the recombinant plasmids can be performed to obtain the target gene sequence (Zhou and Gomez-Sanchez, 2000).
In this study, we introduced two new species Penicillium acidogenicum and P. floccosum, belonging to sections Citrina and Osmophila, respectively, based on a polyphasic approach. The morphological characteristics of the new species and their closely related species are summarized in Table 3. Section Citrina members are frequently found in soil and have also been recovered from indoor environments and food products (Samson et al., 2010; Houbraken et al., 2011b). This shows that members of the group have a relatively wide range of habitats. Accurate species identification is crucial for section Citrina strains. Some members of this group produce mycotoxins citrinin, which is widely recognized as a harmful contaminant in food and feed (Houbraken et al., 2010; Gao et al., 2013). For example, Schmidt-Heydt et al. (2019) performed whole genome sequencing on P. citrinum DSM 1997 and revealed the biosynthesis gene cluster for citrinin. Section Osmophila currently includes three species, P. osmophilum, P. samsonianum, and the new species P. floccosum identified in this study. Species in this group have restricted colony growth with smooth-walled conidiophores and conidia (Stolk and Veenbaas-Rijks, 1974; Houbraken et al., 2016) and are mostly isolated from soil, while P. osmophilum is also isolated from the roots of the plant Colobanthus quitensis (Hereme et al., 2020).
Soil fungi, distinguished by their abundance and diversity, play a fundamental role in the ecosystem. A few Ascomycota taxa including Penicillium species dominate the soil fungal communities (Egidi et al., 2019). The discovery of two new species from the ancient Great Wall loess in Beijing predicts that there may still be a large number of undescribed species in special soil habitats. Therefore, using a polyphasic approach to study Penicillium from the soil will enrich the species diversity of the genus and provide more ideas and insights for us to understand the function of fungi in the ecosystem.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
RL: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. QY: Investigation, Visualization, Writing – original draft. YL: Investigation, Visualization, Writing – original draft. GZ: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Validation, Writing – review & editing. GY: Conceptualization, Methodology, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Survey project on alien invasive species and grassland pests in the Mentougou District (2022HXFWSWXY038), the Fundamental Research Funds for the Central Universities at Beijing Forestry University (2021ZY61), and the University–Industry Collaborative Education Program (202102083002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Andrade, K. C. R., Fernandes, R. A., Pinho, D. B., De Freitas, M. M., Filho, E. X. F., Pessoa, A., et al. (2021). Sequencing and characterization of an L-asparaginase gene from a new species of Penicillium section Citrina isolated from Cerrado. Sci. Rep. 11:17861. doi: 10.1038/s41598-021-97316-1
Ashtekar, N., Rajeshkumar, K. C., Yilmaz, N., and Visagie, C. M. (2022). A new Penicillium section Citrina species and series from India. Mycol. Prog. 21:42. doi: 10.1007/s11557-022-01802-3
Diao, Y. Z., Chen, Q., Jiang, X. Z., Houbraken, J., Barbosa, R. N., Cai, L., et al. (2019). Penicillium section Lanata-divaricata from acidic soil. Cladistics 35, 514–549. doi: 10.1111/cla.12365
Dumas, E., Feurtey, A., De La Vega, R. C. R., Le Prieur, S., Snirc, A., Coton, M., et al. (2020). Independent domestication events in the blue-cheese fungus Penicillium roqueforti. Mol. Ecol. 29, 2639–2660. doi: 10.1111/mec.15359
Dutra-Silva, L., Pereira, G. E., Batista, L. R., and Matteoli, F. P. (2021). Fungal diversity and occurrence of mycotoxin producing fungi in tropical vineyards. World J. Microbiol. Biotechnol. 37:112. doi: 10.1007/s11274-021-03081-8
Egidi, E., Delgado-Baquerizo, M., Plett, J. M., Wang, J., Eldridge, D. J., Bardgett, R. D., et al. (2019). A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 10:2369. doi: 10.1038/s41467-019-10373-z
Frisvad, J., and Samson, R. (2004). Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and airborne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 49, 1–173.
Frisvad, J. C., Smedsgaard, J., Larsen, T. O., and Samson, R. A. J. (2004). Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 49, 201–241.
Gao, J. M., Yang, S. X., and Qin, J. C. (2013). Azaphilones: chemistry and biology. Chem. Rev. 113, 4755–4811. doi: 10.1021/cr300402y
Giraud, F., Giraud, T., Aguileta, G., Fournier, E., Samson, R., Cruaud, C., et al. (2010). Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. Int. J. Food Microbiol. 137, 204–213. doi: 10.1016/j.ijfoodmicro.2009.11.014
Hereme, R., Morales-Navarro, S., Ballesteros, G., Barrera, A., Ramos, P., Gundel, P. E., et al. (2020). Fungal endophytes exert positive effects on Colobanthus quitensis under water stress but neutral under a projected climate change scenario in Antarctica. Front. Microbiol. 11:264. doi: 10.3389/fmicb.2020.00264
Houbraken, J. A. M., Frisvad, J. C., and Samson, R. A. (2010). Taxonomy of Penicillium citrinum and related species. Fungal Divers. 44, 117–133. doi: 10.1007/s13225-010-0047-z
Houbraken, J., Frisvad, J. C., and Samson, R. A. (2011a). Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2, 87–95. doi: 10.5598/imafungus.2011.02.01.12
Houbraken, J., Frisvad, J. C., and Samson, R. A. (2011b). Taxonomy of Penicillium section Citrina. Stud. Mycol. 70, 53–138. doi: 10.3114/sim.2011.70.02
Houbraken, J., Kocsube, S., Visagie, C. M., Yilmaz, N., Wang, X. C., Meijer, M., et al. (2020). Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 95, 5–169. doi: 10.1016/j.simyco.2020.05.002
Houbraken, J., and Samson, R. A. (2011). Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 70, 1–51. doi: 10.3114/sim.2011.70.01
Houbraken, J., Wang, L., Lee, H. B., and Frisvad, J. C. (2016). New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia 36, 299–314. doi: 10.3767/003158516X692040
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). Model finder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Lin, X. G. (2010). Principles and methods of soil microbiology research. Beijing, China: Higher Education Press.
Link, J. H. F. (1809). Observationes in ordines plantarum naturales. Dissertatio 1ma. Magazin der Gesellschaft Naturforschenden Freunde Berlin, No. 3. pp. 3–42.
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lund, F. (1995). Differentiating Penicillium species by detection of indole metabolites using a filter paper method. Lett. Appl. Microbiol. 20, 228–231. doi: 10.1111/j.1472-765X.1995.tb00434.x
Mukherjee, D., Pramanik, K., Mandal, S., and Mandal, N. C. (2022). Augmented growth of cd-stressed rice seedlings with the application of phytostimulating, root-colonizing, cd-tolerant, leaf-endophytic fungi Colletotrichum spp. isolated from Eupatorium triplinerve. J. Hazard. Mater. 438:129508. doi: 10.1016/j.jhazmat.2022.129508
Nguyen, T. T. T., Kwan Noh, K. J., and Lee, H. B. (2021). New species and eight undescribed species belonging to the families Aspergillaceae and Trichocomaceae in Korea. Mycobiology 49, 534–550. doi: 10.1080/12298093.2021.1997461
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Perrone, G., and Susca, A. (2017). Penicillium species and their associated mycotoxins. Methods Mol. Biol. 1542, 107–119. doi: 10.1007/978-1-4939-6707-0_5
Pitt, J. I. (1979). The genus Penicillium and its Teleomorphic states Eupenicillium and Talaromyces. London, UK: Academic Press Inc., Ltd.
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Samson, R. A., Houbraken, J., Thrane, U., Frisvad, J. C., and Andersen, B. (2010). Food and indoor Fungi. Utrecht: CBS-KNAW Fungal Biodiversity Centre.
Schmidt-Heydt, M., Stoll, D., and Geisen, R. (2019). Whole-genome sequencing of the fungus Penicillium citrinum reveals the biosynthesis gene cluster for the mycotoxin citrinin. Microbiol. Resour. Announc. 8, e01419–e01418. doi: 10.1128/mra.01419-18
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 109, 6241–6246. doi: 10.1073/pnas.1117018109
Steenwyk, J. L., Shen, X. X., Lind, A. L., Goldman, G. H., and Rokas, A. (2019). A robust phylogenomic time tree for biotechnologically and medically important fungi in the genera aspergillus and Penicillium. MBio 10, e00925–e00919. doi: 10.1128/mBio.00925-19
Stefanello, A., Gasperini, A. M., and Copetti, M. V. (2022). Ecophysiology of OTA-producing fungi and its relevance in cured meat products. Curr. Opin. Food Sci. 45:100838. doi: 10.1016/j.cofs.2022.100838
Stolk, A. C., and Veenbaas-Rijks, J. W. (1974). Eupenicillium osmophilum sp. nov. Antonie Van Leeuwenhoek 40, 1–5. doi: 10.1007/BF00394547
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tan, Y. P., and Shivas, R. G. (2022). Nomenclatural novelties. Index of Australian Fungi, No. 3, pp. 1–21.
Tsang, C. C., Tang, J. Y. M., Lau, S. K. P., and Woo, P. C. Y. (2018). Taxonomy and evolution of aspergillus, Penicillium and Talaromyces in the omics era - past, present and future. Comput. Struct. Biotechnol. J. 16, 197–210. doi: 10.1016/j.csbj.2018.05.003
Vetrovsky, T., Kolarik, M., Zifcakova, L., Zelenka, T., and Baldrian, P. (2016). The rpb2 gene represents a viable alternative molecular marker for the analysis of environmental fungal communities. Mol. Ecol. Resour. 16, 388–401. doi: 10.1111/1755-0998.12456
Visagie, C. M., Frisvad, J. C., Houbraken, J., Visagie, A., Samson, R. A., and Jacobs, K. (2021). A re-evaluation of Penicillium section Canescentia, including the description of five new species. Persoonia 46, 163–187. doi: 10.3767/persoonia.2021.46.06
Visagie, C. M., Houbraken, J., Frisvad, J. C., Hong, S. B., Klaassen, C. H., Perrone, G., et al. (2014a). Identification and nomenclature of the genus Penicillium. Stud. Mycol. 78, 343–371. doi: 10.1016/j.simyco.2014.09.001
Visagie, C. M., Seifert, K. A., Houbraken, J., Samson, R. A., and Jacobs, K. (2014b). Diversity of Penicillium section Citrina within the fynbos biome of South Africa, including a new species from a Protea repens infructescence. Mycologia 106, 537–552. doi: 10.3852/13-256
Visagie, C. M., Seifert, K. A., Houbraken, J., Samson, R. A., and Jacobs, K. (2016). A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 7, 75–117. doi: 10.5598/imafungus.2016.07.01.06
Visagie, C. M., and Yilmaz, N. (2023). Along the footpath of Penicillium discovery: six new species from the Woodville big tree Forest Trail. Mycologia 115, 87–106. doi: 10.1080/00275514.2022.2135915
Wang, X. C., Zhang, Z. K., and Zhuang, W. Y. (2023). Species diversity of Penicillium in Southwest China with discovery of forty-three new species. J. Fungi 9:1150. doi: 10.3390/jof9121150
Yang, Y., Chen, M., Li, Z. W., Al-Haimi, A. M. S., De Hoog, S., Pan, W. H., et al. (2016). Genome sequencing and comparative genomics analysis revealed pathogenic potential in Penicillium capsulatum as a novel fungal pathogen belonging to Eurotiales. Front. Microbiol. 7:1541. doi: 10.3389/fmicb.2016.01541
Yang, Y. C., Li, K., Liu, C. X., Cheng, F., Liu, C., Quan, W. J., et al. (2022). Sanxiapeptin, a linear pentapeptide from Penicillium oxalicum, inhibited the growth of citrus green mold. Food Chem. 366:130541. doi: 10.1016/j.foodchem.2021.130541
Yin, G., Zhao, H., Pennerman, K. K., Jruick, W. M. III, Fu, M., Bu, L., et al. (2021). Genomic analyses of Penicillium species have revealed patulin and citrinin gene clusters and novel loci involved in oxylipin production. J. Fungi 7:743. doi: 10.3390/jof7090743
Zhang, Y., Liu, X., Wang, Y., Jiang, P., and Quek, S. (2016). Antibacterial activity and mechanism of cinnamon essential oil against Escherichia coli and Staphylococcus aureus. Food Control 59, 282–289. doi: 10.1016/j.foodcont.2015.05.032
Keywords: Aspergillaceae, DNA markers, phylogeny, taxonomy, new taxa
Citation: Liang R, Yang Q, Li Y, Yin G and Zhao G (2024) Morphological and phylogenetic analyses reveal two new Penicillium species isolated from the ancient Great Wall loess in Beijing, China. Front. Microbiol. 15:1329299. doi: 10.3389/fmicb.2024.1329299
Edited by:
Bing-Da Sun, Chinese Academy of Sciences, ChinaReviewed by:
Xin-Cun Wang, Chinese Academy of Sciences, ChinaRegina Sharmila Dass, Pondicherry University, India
Rungtiwa Phookamsak, Shenzhen University, China
Copyright © 2024 Liang, Yang, Li, Yin and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guozhu Zhao, emhhb2d6QGJqZnUuZWR1LmNu
 Ruina Liang
Ruina Liang Qiqi Yang1
Qiqi Yang1 Guozhu Zhao
Guozhu Zhao