- 1Safe and Sustainable Value Chain, World Vegetable Center, Shanhua, Taiwan
- 2Research Center of Information and Communication Technologies, University of A Coruña, A Coruña, Spain
- 3Bio-Cheminformatics Research Group, Universidad de Las Américas, Quito, Ecuador
- 4Seed and Seedling Management Section, Taiwan Seed Improvement and Propagation Station, Ministry of Agriculture, Taichung, Taiwan
- 5Facultad de Ingeniería y Ciencias Aplicadas, Universidad de Las Américas, Quito, Ecuador
The introduction of plant pathogens can quickly reshape disease dynamics in island agro-ecologies, representing a continuous challenge for local crop management strategies. Xanthomonas pathogens causing tomato bacterial spot were probably introduced in Taiwan several decades ago, creating a unique opportunity to study the genetic makeup and adaptive response of this alien population. We examined the phenotypic and genotypic identity of 669 pathogen entries collected across different regions of Taiwan in the last three decades. The analysis detected a major population shift, where X. euvesicatoria and X. vesicatoria races T1 and T2 were replaced by new races of X. perforans. After its introduction, race T4 quickly became dominant in all tomato-growing areas of the island. The genomic analysis of 317 global genomes indicates that the Xanthomonas population in Taiwan has a narrow genetic background, most likely resulting from a small number of colonization events. However, despite the apparent genetic uniformity, X. perforans race T4 shows multiple phenotypic responses in tomato lines. Additionally, an in-depth analysis of effector composition suggests diversification in response to local adaptation. These include unique mutations on avrXv3 which might allow the pathogen to overcome Xv3/Rx4 resistance gene. The findings underscore the dynamic evolution of a pathogen when introduced in a semi-isolated environment and provide insights into the potential management strategies for this important disease of tomato.
Introduction
Tomato (Solanum lycopersicum L.) is one of the most important vegetable crops worldwide. Yet, most tomato-growing areas are challenged by bacterial spot caused by Xanthomonas species. This disease induces necrotic spots on aboveground tissues, leading to defoliation, yield losses, and reduced fruit quality and marketability (Pohronezny and Volin, 1983). An increase in the incidence of bacterial spot has been reported in different regions of the world (Hamza et al., 2010; Horvath et al., 2012; Beran et al., 2014; Kebede et al., 2014; Abbasi et al., 2015; Potnis et al., 2015; Osdaghi et al., 2016; Araújo et al., 2017; Burlakoti et al., 2018). In the field, however, symptoms can be caused by different Xanthomonas species, including X. vesicatoria, X. gardneri, X. euvesicatoria, and, X. perforans (Jones et al., 2004). A more recent classification also suggests that X. euvesicatoria may include two distinct pathovars: euvesicatoira and perforans (Constantin et al., 2016). While historically, X. euvesicatoria and X. vesicatoria have been the predominant species infecting tomato, recent reports pointed out the emergence of X. perforans in the United States (Horvath et al., 2012; Ma, 2015), Ethiopia (Kebede et al., 2014), Canada (Abbasi et al., 2015), and Brazil (Araújo et al., 2017), among others. Xanthomonas pathogens causing bacterial spot can be classified into at least five different races (Asuta-Monge et al., 2000; Jones et al., 2004). Rapid changes in the pathogen population represent a significant challenge for disease management strategies because no tomato variates show resistance to all of these races. While the disease is well-studied, the emergence of new Xanthomonas strains and races poses a continuous challenge for disease management. Understanding the genetic makeup and adaptation pathways of these bacterial pathogens is crucial for developing effective control strategies.
To proliferate in the plant tissues, most bacteria pathogens rely on the translocation of type III effector proteins into the host cell. The Xanthomonas outer proteins (Xop) are a big group of effectors with diverse biochemical functions that target different subcellular compartments (Büttner and Bonas, 2010). Effectors interfere with plant processes to modulate plant defense response and facilitate access to host nutrients (Toruño et al., 2016). During host colonization, Xanthomonas are known to deliver between 23 and 37 different type III effectors, although individual strains might carry a variable repertoire, most likely due to redundancy in functions (Büttner and Bonas, 2010). Plants also evolved networks of protein sensors that recognize the effectors and elicit a response that restricts pathogen advance (Jones and Dangl, 2006). For instance, the tomato resistance protein Bs4 triggers a hypersensitive response (HR) when recognizing the cognate avirulence protein AvrBs4 from Xanthomonas T1 race (Ballvora et al., 2001). The same is true for the effector AvrXv3 (xopAF) from Xanthomonas T3 race, which is recognized by the nucleotide-binding leucine-rich repeat (NBS-LRR) protein Xv3/Rx4, present in S. pimpinellifolium (Zhang et al., 2021). Understanding the distribution of effector repertoires in pathogen populations is crucial for supporting disease management strategies and guiding breeding efforts in modern agriculture.
Plant pathogens often experience a process of selection and diversification as they adapt to new hosts or environments (Brasier, 1987). This is particularly true in semi-isolated areas, such as islands, where introducing new pathogens can drastically shape the existing population structure. The study of Xanthomonas species in Taiwan is particularly relevant since tomato cultivation is relatively new. While the Dutch introduced tomatoes as an ornamental plant in 1622 (Chen, 2005), high-scale cultivation only started when varieties like Ponderosa and Marglobe were brought in from the United States in the early 1930s. Since then, tomato production has occupied the central and southern areas of the island, where hot and humid environments are conducive to the disease. The first report of bacterial spot was in the early 1930s (Okabe, 1933), and recent reports suggest the presence of X. vesicatoria, X. euvesicatoria, and X. perforans (Lue et al., 2010; Burlakoti et al., 2018). This study aims to understand the genetic makeup of Xanthomonas pathogens causing bacterial spot in Taiwan and uncover the pathway of adaptation to the novel environment.
Materials and methods
Pathogen collection and molecular characterization
We analyzed the records of Xanthomonas strains causing bacterial spots in Taiwan over the past 34 years (Supplementary Table S1). Of these records, 293 were obtained from a previous study conducted by Burlakoti et al. (2018), which included collections from 1989 to 2016. We also included 376 bacterial strains isolated from tomato plants displaying disease symptoms between 2017 and 2023 (Supplementary Table S1). All bacterial strains were stored at −80°C in 30% glycerol at the World Vegetable Center. DNA was extracted through the boiled bacterial suspension (concentration: 1*108 CFU/mL) for each bacterial culture. The species identification was done using touchdown multiplex-PCR with species-specific primers following the reaction described in Lue et al. (2010). The primer set of species-specific primers for X. euvesicatoria (RST13/RST14), X. vesicatoria (Xv-1f/Xv-1r), X. perforans (HpaF-f/HpaF-r), X. gardneri (BS-XgF/BS-XgR). The PCR products were confirmed using 1.5% agarose gel electrophoresis. The gel was stained with HealthView Nucleic Acid Stain (Cat. GN-NAS-100) and visualized under ultraviolet (UV) light. X. euvesicatoria, X. vesicatoria, X. perforans, and X. gardneri amplified a product of 560-bp, 197-bp, 300-bp, and 154-bp, respectively.
Race identification, pathogenicity test, and resistance screening
All strains were characterized into Races using the hypersensitive reactions on 4 differential hosts (Stall et al., 2009), including S. lycopersicum Bonny best L305 (without resistance gene), Hawaii7998 (with resistance gene rx-1, rx-2, rx-3), Hawaii7981 (with resistance gene Xv3), and S. pennelii LA716 (with resistance gene Xv4). Strains were classified into T0, T1, T2, T3, T4, and T5 Races. Furthermore, 1 T5, 24 T4, and 5 T3 X. perforans strains were selected to characterize additional virulence patterns. The strains were inoculated on S. lycopersicum L305 (susceptible line) and H7981 (with resistance gene Xv3), S. pimpinellifolium L3707 (resistant to race T3 and T4), and S. lycopersicum var. cerasiforme PI114490 (high-level resistance) using the dip-inoculation method. Seedlings at the 4–6 true leaves stage were inverted and immersed in bacterial suspension (1*108 CFU/mL) containing 0.03% surfactant (Silwett L−77) for 30 s. The inoculated plants were kept in a growth room with warm and high humidity conditions (first 48 h at 90 ± 5% RH, then transfer to 80 ± 5% RH and 28–35°C), suitable for disease progress. The experiment was laid out in a randomized complete block design (RCBD) with 3 replications, each replication containing 5 plants/strain/host genotypes. Disease severity was recorded according to Horsfall-Barratt’s rating scale (0–11) at 14 days post-inoculation, where 0 = 0%, 1 = 0–3%, 2 = 3–6%, 3 = 6–12%, 4 = 12–25%, 5 = 25–50%, 6 = 50–75%, 7 = 75–88%, 8 = 88–94%, 9 = 94–97%, 10 = 97–100%, 11 = 100% diseased tissue (Horsfall and Barratt, 1945). The disease reaction of tomato genotypes was categorized as referred by Hutton et al. (2010), where resistance (R) = mean scale ≤4; moderate resistance (M) = 4 < mean scale ≤5; susceptible (S) = mean scale >5. These patterns allowed the selection of a representative strain from the main virulence group for resistance screening. A total of 86 S. pimpinellifolium and 14 S. lycopersicum var. cerasiforme accessions were screened with a representative strain to identify new sources of resistance. The resistance screening method was the same as described above.
Genome assembly, core genome determination, and spanning tree reconstruction
The genomes were sequenced at BGI Genomics in China, using a DNBseq platform with a short-insert library to obtain 150 bp paired end reads. The Velvet algorithm was utilized for assembling the Xanthomonas genomes (Zerbino and Birney, 2008). To determine the optimal K-mer size and coverage cutoff for each specific genome, the VelvetOptimiser script was employed (Gladman et al., 2017). For detailed statistics of the genome sequencing and assembly, refer to Supplementary Table S3. Subsequently, the contigs originating from X. euvesicatoria and X. perforans strains were mapped using the genomic reference of X. euvesicatoria pv. vesicatoria str. 85–10 (NC_007508) and X. perforans strain GEV872 (NZ_CP116305) respectively. Correspondingly, the contigs derived from X. vesicatoria were aligned against the genomic reference of X. vesicatoria strain LMG911 (NZ_CP018725.1) using the Geneious plugin minimap2 (Li, 2018). This approach yielded 95.5% (on average) of the reference genome for both Taiwanese X. vesicatoria and X. perforans genomes and 94.4% in average for X. euvesicatoria genomes. Genome annotation was done within Geneious version 2023.2.1.1 We also annotated the genomes contained within the scaffolds generated by Velvet. The core genome of X. euvesicatoria, X. vesicatoria, and X. perforans was leveraged with all the available annotated genomes in GenBank (as of December 22, 2022), along with additional strains from Taiwan. The get_homologues software (Contreras-Moreira et al., 2017) was applied to discern the core genomic elements. Subsequently, the identified core genome was input for selecting phylogenomic markers using the get_phylomarkers software (Vinuesa et al., 2018). The phylogenetic tree was built with the 62 robust markers as ascertained by the get_phylomarkers approach. Spanning trees were constructed using a dataset of 296 genomes available in the NCBI database for X. perforans, X. euvesicatoria, and X. vesicatoria in conjunction with the 21 Taiwan assembled genomes. Thirty informative phylomarkers were selected and concatenated to form a representative sequence for each genome. Software PHYLOViZ (Francisco et al., 2012) was used to reconstruct the spanning tree.
Effectors annotation and analysis
To establish a dataset of Xanthomonas effectors, we used the database resource contained in the Euro Xanth webpage.2 This resource compiles 80 effectors, out of which 70 possess reference sequences in the NCBI Nucleotide database. The corresponding effector genes were obtained in GenBank format. With the objective of discerning effectors present in the Taiwanese genomes, the Geneious annotation tool was employed. Annotation of all the 70 reference sequences was conducted using a 70% similarity threshold. As a result, 48 effectors were identified among the Taiwanese genomes. These effectors include avrBs1, avrBs2, avrXccA1, HpaA, xopA, xopAA, xopAD, xopAE, xopAF, xopAG1, xopAI, xopAJ, xopAK, xopAM, xopAP, xopAQ, xopAR, xopAU, xopAV1, xopAW, xopAX, xopAY, xopAZ, xopB, xopBA, xopC, xopD, xopE, xopE2, xopE3, xopF1, xopF2, xopG, xopI, xopJ1, xopJ2, xopJ3, xopJ4, xopK, xopM, xopN, xopO, xopP, xopQ, xopS, xopV, xopX, and xopZ1. The sequences of the identified effectors in each genome were subsequently extracted, translated, and grouped. A BLASTp analysis was conducted using the GenBank database, focusing on the species X. perforans, X. euvesicatoria, and X. vesicatoria. BLASTp results that match with the effector annotation were retained. We identified unique alleles by performing protein alignments and all the BLAST hits found in the databases.
Results and discussion
Xanthomonas populations in Taiwan experienced a major shift
To investigate the historical dynamics of Xanthomonas populations, we mined a dataset of 669 strains causing bacterial spot in Taiwan (Supplementary Table S1). The strains were isolated across 82 tomato-growing areas in 13 major districts in the last 34 years (1989–2023). Along with previous samples collected between 1989 and 2016 (293), we also obtained 376 Xanthomonas strains during the 2017 to 2023 surveys. Overall, we identified three main Xanthomonas species in our collection using DNA markers: X. vesicatoria, X. euvesicatoria, and X. perforans (Figure 1; Supplementary Table S1). We were unable to find any samples corresponding to X. gardneri. We plotted the time distribution of all Xanthomonas strains and found a significant shift in Xanthomonas species, a trend that began in the early 2000s, as previously described in Burlakoti et al. (2018), and persists to the present day. The data consistently show the replacement of X. euvesicatoria and X. vesicatoria by X. perforans as the dominant population from the early 2000s until 2023 (Figure 1A).
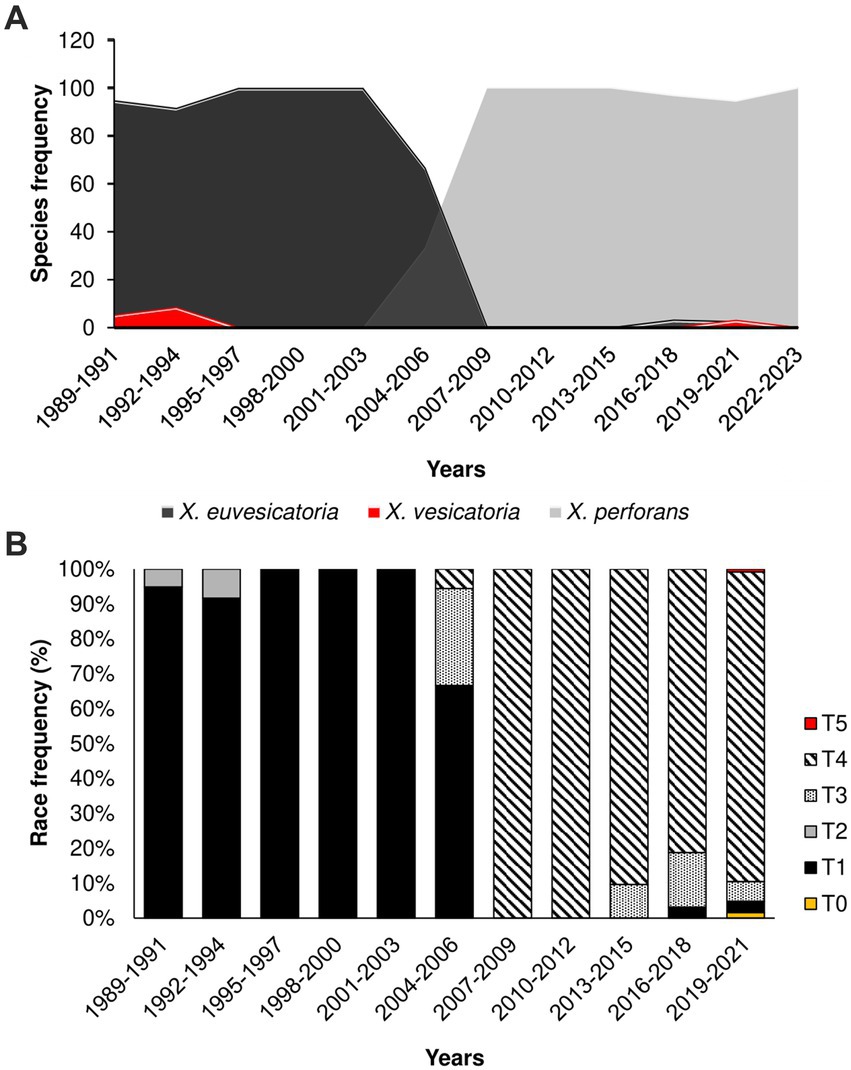
Figure 1. The population shift of Xanthomonas species causing Bacterial Spot in Taiwan started in 2004–2006; (A) Relative frequency of Xanthomonas species in the last 34 years (1989–2023); (B) Relative frequency of race groups among Xanthomonas species causing bacteria spot collected in Taiwan.
Changes in Xanthomonas species prevalence have also been observed in different regions over the past two decades (Kebede et al., 2014; Timilsina et al., 2015; Araújo et al., 2017). For instance, Timilsina et al. (2019) reported a shift from X. euvesicatoria to X. perforans in Florida’s tomato-growing areas. Other reports describe a similar phenomenon in North Carolina, Indiana, Ohio, and other regions in the Midwest United States (Ma et al., 2011; Horvath et al., 2012; Egel et al., 2018; Adhikari et al., 2019; Bernal et al., 2022). Rapid shifts in pathogen species of Xanthomonas are likely the result of fitness advantage. For instance, resistance to copper-based compounds, commonly used in Taiwan, might explain how resistance populations might become dominant in time. However, Burlakoti et al. (2018) find no significant differences in copper resistance phenotypes among Xanthomonas species. It has been suggested that the increase in X. perforans may be due to the production of antimicrobial peptides, also known as bacteriocins (Marutani-Hert et al., 2020). However, this factor does not fully explain why pepper does not experience a similar surge in X. perforans infections in Taiwan (Burlakoti et al., 2018). It is more plausible that other factors, such as pathogenicity and resistance components, play a pivotal role in shaping the observed trends of bacterial spot in tomato and pepper (Adhikari et al., 2020). Further investigation into these factors is essential to comprehensively understand the dynamics of Xanthomonas species shifts in different crops and regions.
Xanthomonas perforans T4 Race is now dominant in Taiwan
To investigate the change in pathogenicity of the Xanthomonas population in Taiwan, we inoculated 156 strains from the late collection onto four tomato differentials. All the races (T0-T5) were identified. The integrated data on pathogenicity unveiled a shift from T1 and T2 races, collected before the 2000s, toward T3 and T4 races collected after this period (Figure 1B). Race T4 showed remarkable prevalence, with more than 95% of the samples collected after 2015 (Figure 1B), suggesting a fitness advantage of this pathogenicity group over T3. Race designation as T3 or T4 is based only on the presence or absence of hypersensitive response (HR) caused by the recognition of avrXv3 by the cognate Xv3 (Robbins et al., 2009). Further genetic analysis found no differences between T3 and T4 genomes, except for crucial mutations that disrupt the avrXv3 effector in T4 strains (details below). In Taiwan, there is no report on the frequency of Xv3, but a draft estimate suggests that 20% of tomato hybrids might have Xv3 (Chou, personal communication). As a result, one possible explanation for the increase in T4 races is that the host is driving selection due to the presence of Xv3. However, a recent population swift in the Midwestern United States involving an X. perforans T4 population occurred in the absence of the resistance Xv3 locus (Bernal et al., 2022), suggesting that other factors might also be involved. This new T4 population (2017–2020) appears to be genetically identical, probably due to recent migration events (Bernal et al., 2022).
We further identified additional virulence patterns within T3, T4, and T5 populations by inoculating 1 T5, 24 T4, and 5 T3 strains on a different tomato panel. Based on the phenotypic response, which was classified as resistance (R), moderate resistance (M), and susceptible (S), we found eight different pattern combinations among the small sample (Figure 2A; Supplementary Figure S1; Supplementary Table S1). Race T4 strains were classified into 8 patterns, suggesting that the X. perforans population in Taiwan harbors diverse phenotypes and might be going through a process of local diversification. Phenotypic heterogeneity in plant pathogens in response to the environment has been described even for clonal populations (Delgado et al., 2013; Mariz-Ponte et al., 2022). Finally, we look for new sources of resistance against predominant virulent disease by screening the highly virulent T4 strain (Xant314) against an S. pimpinellifolium-diversity panel. We found only one accession (VI007032) with high resistance levels (Figure 2B; Supplementary Table S2). VI007032, which showed a slight symptom of bacterial spot, will be further tested as a potential resistance donor. The data suggest a fitness advantage of race T4 but also a process of phenotypic diversification.
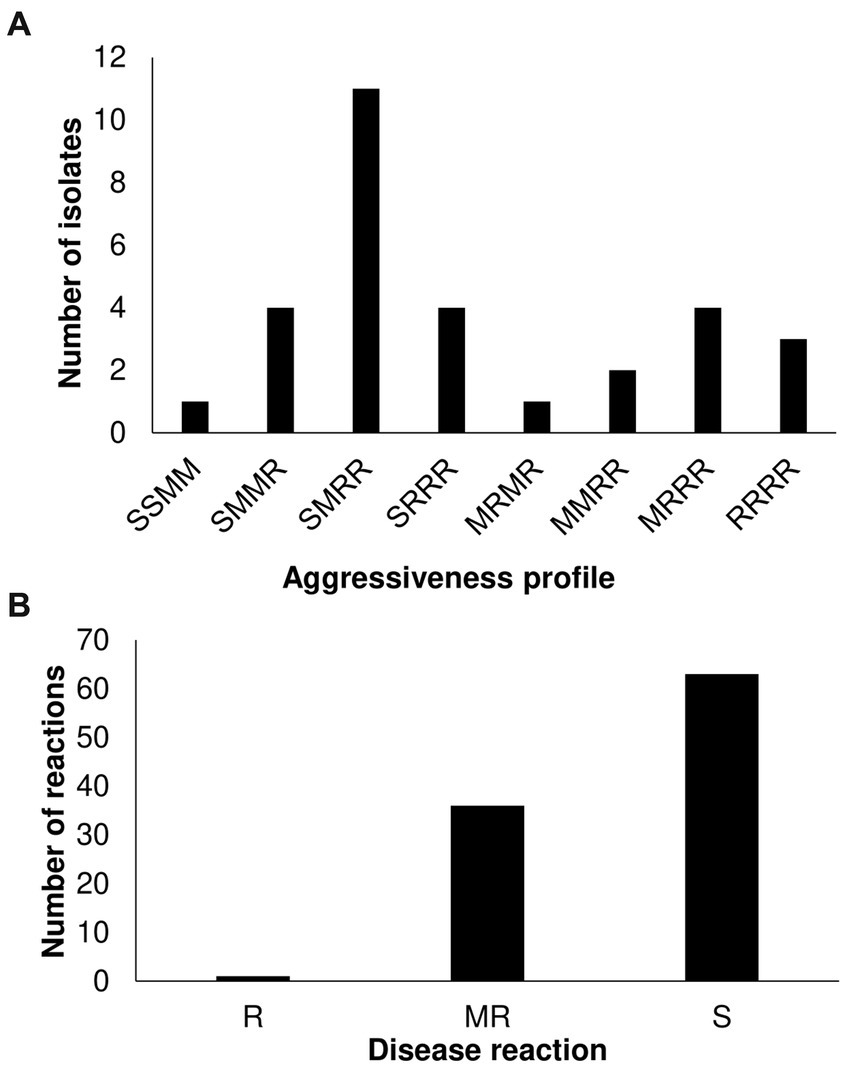
Figure 2. Characterization of Xanthomonas perforans race T3 and T4 strains on Solanaceae germplasm. (A) Virulence patterns of 30 X. perforans race T3, T4, and T5 against four reference tomato accessions. The letters in order represent the disease reaction of tomato variety (L305, H7981, L3707, PI114490) classified as resistance (R): mean scale <=4, moderate resistance (M): 4 < mean scale <=5, and susceptible (S): mean scale >5. (B) Screening of 86 S. pimpinellifolium and 14 S. lycopersicum var. cerasiforme accessions with strain Xant314.
Xanthomonas in Taiwan come from a narrow genetic background
To investigate the genetic variation of Xanthomonas causing bacterial spot in Taiwan, we obtained complete genome sequences for 21 representative strains collected in highland and lowland tomato-growing areas over 30 years (Supplementary Table S2). The assembled genomes (235-245X coverage range) represent X. perforans (11), X. euvesicatoria (6), X. vesicatoria (4), and include T1 to T5 races (Supplementary Table S3). Overall, we found indications of a narrow genetic background within Xanthomonas species from Taiwan. Using phylogenetic reconstruction from 317 genomes, we clearly distinguish between Xanthomonas strains clustering at the species level and forming single clusters across the tree (Supplementary Figure S2). The spanning tree constructed from concatenated phylomarkers linked the Taiwanese X. perforans strains to a single North American genotype, particularly represented in the United States and Canada (Figure 3). We identified two clades that derived from the same X. perforans genotypes. The first clade included strains collected in 2005, and the second clade included strains from 2016 (Figure 3).
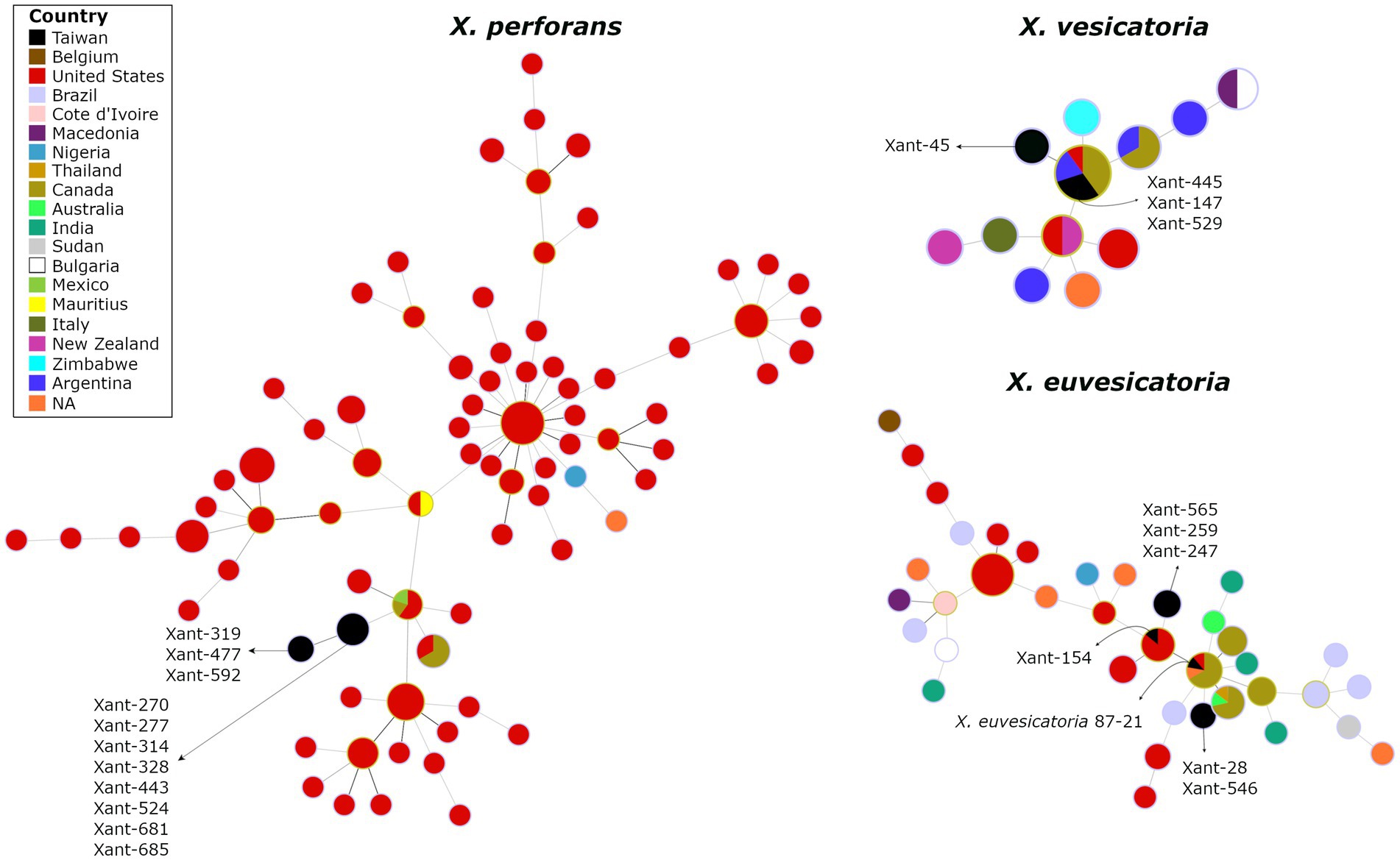
Figure 3. The spanning tree of Xanthomonas species causing Bacterial Spot in Taiwan depicts the potential linkage of X. perforans, X, euvesicatoria, and X. vesicatoria to North American populations. The circles represent genetic clusters that are linked by lines. The colors inside the circles represent the countries where those clusters have been found. The genomes from Taiwan are depicted in black.
Interestingly, we found no genetic substructure that clusters T3, T4, or T5 groups, suggesting that these might be derived. In addition, the Split decomposition tree found no evidence of recombination among Xanthomonas in Taiwan (data not shown). These preliminary results challenge the hypothesis of multiple colonization events on the island and suggest that X. perforans in Taiwan result from a single introduction (one or few genotypes). It is worth noting that the abundance of genomes from North America in our databases may introduce some bias into the results. The lack of recombination, the absence of structure among races (Supplementary Table S1), and the virulence pattern on reference tomato accessions (Figure 2) also suggest that the original X. perforans diversified locally in the last two decades. A similar scenario has been described recently where a clonal population of X. perforans that dominate extensive areas shows the accumulation of genetic variation (Bernal et al., 2022). The case of X. vesicatoria appears to follow the trend, as it is characterized by a single genotype. A single introduction, probably during the late 80s, was likely responsible for bringing a race T1 to Taiwan (Figure 3). The spanning tree of X. euvesicatoria detected two main clusters, suggesting more than one colonization event. Both colonization events of race T2 are likely to come from North America (Figure 3). One cluster was associated with Taiwanese strain 87–21, collected in the late 80s (Bouzar et al., 1994), and the other was associated with strain Xant154, collected in 1994 (Burlakoti et al., 2018). The genetic data of representative Xanthomonas genomes from Taiwan points to North America as the main source. Discrete genotypes were likely introduced and then diversified locally. The pathogen could have been introduced during the importation of tomato varieties, but other explanations cannot be excluded.
The effector composition of Xanthomonas populations reveals diversification and local adaptation
To characterize the effector composition of the Taiwanese population, we compared the Xanthomonas outer proteins (Xop) repertoires of 317 genomes and found evidence of diversification and local adaptation in Taiwan. Out of 70 Xops reported in the global database,3 only 48 were present in the target genomes (Supplementary Table S3). Similar to previous studies, we found a variable number of effectors in each strain (Schwartz et al., 2015; Roach et al., 2019), where X. perforans displayed 23 to 31 Xops while X. euvesicatoria displayed 30 to 31 Xops. We only found 11 Xop effectors in all the X. vesicatoria strains (Supplementary Table S3). Based on the presence/absence of effectors, the local X. perforans, X. euvesicatoria, and X. vesicatoria strains showed a unique composition compared to the global population (Figure 4A; Supplementary Figure S2). We identified at least 13 effector profiles in a sample of 21 genomes, with most of the unique profiles occurring in X. perforans genomes (8) compared to X. euvesicatoria genomes (4). The observed pattern is unlikely to result from an assembly or annotation issue since all the X. vesicatoria genomes assembled the same Xop genes and formed a single profile. The diversity in effector composition observed in X. perforans (Figure 4) aligns with the occurrence of multiple virulence patterns within T4 and T3 strains (Figure 2). Diversification of virulence factors has been reported in other pathogens and is likely the result of local adaptation (Quibod et al., 2016; Richards et al., 2019).
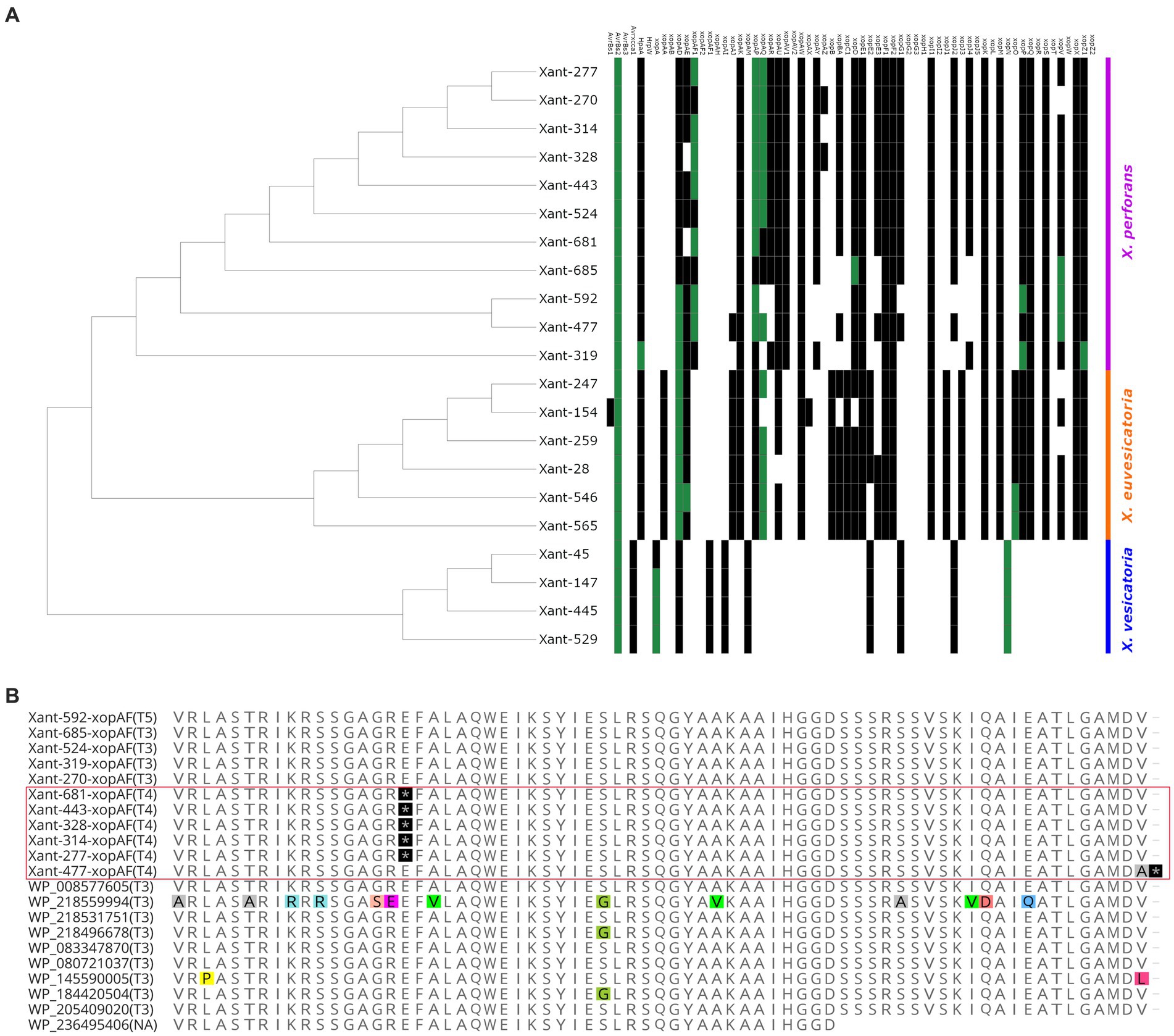
Figure 4. Evidence of distinctive effector composition in Xanthomonas from Taiwan. (A) The heat map tree illustrates the presence or absence of Xanthomonas outer proteins (Xop) present in each strain. Green squares indicate the presence of a unique allele found in a Xanthomonas genome. (B) Partial sequence alignment of XopAF showing exclusive nonsense mutations in the X. perforans T4 race from Taiwan (highlighted by the red square). Nonsense mutations are depicted in black. Polymorphic residues are depicted with colors. The race of each strain is identified within parentheses.
The comparative analysis of the Xops dataset also identified 13 effectors with rare alleles only present in Taiwan (Supplementary Table S4). These variations are characterized by one or more nonsynonymous mutations (Supplementary Table S4), which are most likely the result of adaptation to the local environment. Interestingly, we found most of the rare alleles in X. perforans genomes (Figure 4A). For instance, the strain Xant319 showed a unique amino acid substitution (S741G) in the highly conserved region of the protein xopZ1. We found derived alleles of xopD and xopV in X. perforans strains associated with recent collections (Supplementary Table S4), which might suggest more recent mutations. All of these genes have been reported to contribute to Xanthomonas colonization in various host systems (Furutani et al., 2009; Medina et al., 2018; Deb et al., 2020), and therefore, the observed mutations might have a fitness role in the local environment. In addition, strains Xant314 and Xant328 contain the effector xopAZ, which is absent from any of the genomes analyzed but was first reported in X. arboricola and X. cynarae (Kara et al., 2017). A few effectors also showed Indels and frameshift mutations that potentially disrupt their function (Supplementary Table S5). One clear example represents mutations in xopAF/avrXv3 associated with race T4. Loss of avirulence function in this gene has been associated with a range of changes in the protein sequence (Timilsina et al., 2016; Bernal et al., 2022). We found two nonsense mutations in xopAF alleles from X. perforans T4 strains from Taiwan (Figure 4B). These mutations appear to be the only difference between xopAF alleles from race T3 and T4, suggesting that T4 strains emerged from T3 genetic backgrounds by random mutation (Bernal et al., 2022).
Interestingly, the effector repertoires of X. vesicatoria strains consist of only 11 genes (Figure 4A; Supplementary Table S3), which is shorter than any of the global X. vesicatoria genomes reported so far. The pathogenicity capabilities of these X. vesicatoria strains with a minimal number of effectors suggest a high plasticity of the pathogen associated with the redundancy of effector functions (Castaneda et al., 2005; Long et al., 2018). The overall data indicates that the genetic background of the Xanthomonas population from Taiwan appears to be narrow and most likely the result of a few colonization events that caused profound changes in population structure. However, the phenotypic pattern and the effector composition suggest that such a genetically uniform population is diversifying due to local adaptation.
Conclusion
The introduction of Xanthomonas pathogens into Taiwan has led to significant shifts in disease dynamics across tomato-growing areas, particularly evident in the rapid dominance of X. perforans race T4. Despite that only a few pathogen genotypes managed to colonize the island, the population quickly diversified in response to local adaptation. These findings emphasize the complexity of managing introduced plant pathogens in semi-isolated environments and underscore the importance of ongoing research for effective disease responses.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
J-RC: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing – review & editing. KA-C: Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Validation, Writing – review & editing. D-YX: Data curation, Resources, Validation, Writing – review & editing. H-CC: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – review & editing. LA-M: Conceptualization, Data curation, Formal analysis, Validation, Writing – review & editing. Y-PL: Formal analysis, Software, Writing – review & editing. VA-J: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. RO: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Scientists at the World Vegetable Center are partially funded by The Ministry of Foreign Affairs and The Ministry of Agriculture in Taiwan (107AS-4.5.1-ST-a2, 108AS-1.1.2-ST-a4, 109AS-1.1.2-ST-a3, 110AS-1.2.5-ST-a1). Researchers at Universidad de Las Américas receive funding from internal sources, as part of the program PRG.BIO.23.14.01. This work received financial support from the Consortium of International Agricultural Research Centers/Consultative Group on International Agricultural Research (CGIAR).
Acknowledgments
The authors would like to thank Chuang, M. H., Lin, C. H., and Wang, J. F. for their technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1408885/full#supplementary-material
Footnotes
References
Abbasi, P. A., Khabbaz, S. E., Weselowski, B., and Zhang, L. (2015). Occurrence of copper-resistant strains and a shift in Xanthomonas spp. causing tomato bacterial spot in Ontario. Can. J. Microbiol. 61, 753–761. doi: 10.1139/cjm-2015-0228
Adhikari, P., Adhikari, T. B., Louws, F. J., and Panthee, D. R. (2020). Advances and challenges in bacterial spot resistance breeding in tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 21:1734. doi: 10.3390/ijms21051734
Adhikari, P., Adhikari, T. B., Timilsina, S., Meadows, I., Jones, J. B., Panthee, D. R., et al. (2019). Phenotypic and genetic diversity of Xanthomonas perforans populations from tomato in North Carolina. Phytopathology 109, 1533–1543. doi: 10.1094/PHYTO-01-19-0019-R
Araújo, E. R., Costa, J. R., Ferreira, M. A. S. V., and Quezado-Duval, A. M. (2017). Widespread distribution of Xanthomonas perforans and limited presence of X. gardneri in Brazil. Plant Pathol. 66, 159–168. doi: 10.1111/ppa.12543
Asuta-Monge, G., Minsavage, G. V., Stall, R. E., Vallejos, C. E., Davis, M. J., and Jones, J. B. (2000). Xv4-vrx4: a new gene-for-gene interaction identified between Xanthomonas campestris pv. vesicatoria race T3 and wild tomato relative Lycopersicon pennellii. Mol. Plant Microbe Interact. 13, 1346–1355. doi: 10.1094/MPMI.2000.13.12.1346
Ballvora, A., Pierre, M., van den Ackerveken, G., Schornack, S., Rossier, O., Ganal, M., et al. (2001). Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol. Plant-Microbe Interact. 14, 629–638. doi: 10.1094/MPMI.2001.14.5.629
Beran, P., Mraz, I., Kokoskova, B., and Bohata, A. (2014). Monitoring the occurrence of bacterial spot of tomato and pepper in the Czech Republic and development of new PCR primers for detection of Xanthomonas vesicatoria. Eur. J. Plant Pathol. 141, 617–621. doi: 10.1007/s10658-014-0554-y
Bernal, E., Rotondo, F., Roman-Reyna, V., Klass, T., Timilsina, S., Minsavage, G. V., et al. (2022). Migration drives the replacement of Xanthomonas perforans races in the absence of widely deployed resistance. Front. Microbiol. 13:826386. doi: 10.3389/fmicb.2022.826386
Bouzar, H., Jones, J. B., Stall, R. E., Hodge, N. C., Minsavage, G. V., Benedict, A. A., et al. (1994). Physiological, chemical, serological, and pathogenic analyses of a worldwide collection of Xanthomonas campestris pv. vesicatoria strains. Phytopathology 84, 663–671. doi: 10.1094/Phyto-84-663
Brasier, C. M. (1987). “The dynamics of fungal speciation” in Evolutionary biology of the Fungi. eds. A. D. M. Rayner, C. M. Brasier, and D. Moore (Cambridge, UK: Cambridge University Press), 231–260.
Burlakoti, R. R., Hsu, C. F., Chen, J. R., and Wang, J. F. (2018). Population dynamics of Xanthomonads associated with bacterial spot of tomato and pepper during 27 years across Taiwan. Plant Dis. 102, 1348–1356. doi: 10.1094/PDIS-04-17-0465-RE
Büttner, D., and Bonas, U. (2010). Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. doi: 10.1111/j.1574-6976.2009.00192.x
Castaneda, A., Reddy, J. D., El-Yacoubi, B., and Gabriel, D. W. (2005). Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant-Microbe Interact. 18, 1306–1317. doi: 10.1094/MPMI-18-1306
Chen, Z. C. (2005). Tomato. In: M. H. Huang, Yang, L. C. The guidance of agriculture in Taiwan. Taipei: Harvest Farm Magazine, pp. 517–532.
Constantin, E. C., Cleenwerck, I., Maes, M., Baeyen, S., Van Malderghem, C., De Vos, P., et al. (2016). Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 65, 792–806. doi: 10.1111/ppa.12461
Contreras-Moreira, B., Cantalapiedra, C. P., García-Pereira, M. J., Gordon, S. P., Vogel, J. P., Igartua, E., et al. (2017). Analysis of plant pan-genomes and transcriptomes with GET_HOMOLOGUES-EST, a clustering solution for sequences of the same species. Front. Plant Sci. 8:184. doi: 10.3389/fpls.2017.00184
Deb, S., Ghosh, P., Patel, H. K., and Sonti, R. V. (2020). Interaction of the Xanthomonas effectors XopQ and XopX results in induction of rice immune responses. Plant J. 104, 332–350. doi: 10.1111/tpj.14924
Delgado, R. A., Monteros-Altamirano, A. R., Li, Y., Visser, R. G. F., van der Lee, T. A. J., and Vosman, B. (2013). Large subclonal variation in Phytophthora infestans populations associated with Ecuadorian potato landraces. Plant Pathol. 62, 1081–1088. doi: 10.1111/ppa.12039
Egel, D. S., Jones, J. B., Minsavage, G. V., Creswell, T., Ruhl, G., Maynard, E., et al. (2018). Distribution and characterization of Xanthomonas strains causing bacterial spot of tomato in Indiana. Plant Health Prog. 19, 319–321. doi: 10.1094/PHP-07-18-0041-BR
Francisco, A. P., Vaz, C., Monteiro, P. T., Melo-Cristino, J., Ramirez, M., and Carriço, J. A. (2012). PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. doi: 10.1186/1471-2105-13-87
Furutani, A., Takaoka, M., Sanada, H., Noguchi, Y., Oku, T., Tsuno, K., et al. (2009). Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 22, 96–106. doi: 10.1094/mpmi-22-1-0096
Gladman, S., Page, A., and Seemann, T. (2017). Velvet optimiser [computer software]. Available at: https://github.com/tseemann/VelvetOptimiser/tree/master.
Hamza, A., Robene-Soustrade, I., Jouen, E., Gagnevin, L., Lefeuvre, P., Chiroleu, F., et al. (2010). Genetic and pathological diversity among Xanthomonas strains responsible for bacterial spot on tomato and pepper in the Southwest Indian Ocean region. Plant Dis. 94, 993–999. doi: 10.1094/PDIS-94-8-0993
Horsfall, J. G., and Barratt, R. W. (1945). An improved grading system for measuring plant disease. Phytopathology 35:655,
Horvath, D. M., Stall, R. E., Jones, J. B., Pauly, M. H., Vallad, G. E., Dahlbeck, D., et al. (2012). Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS One 7:e42036. doi: 10.1371/journal.pone.0042036
Hutton, S. F., Scott, J. W., and Yang, W. (2010). Identification of QTL associated with resistance of bacterial spot race T4 in tomato. Theor. Appl. Genet. 121, 1275–1287. doi: 10.1007/s00122-010-1387-5
Jones, J., and Dangl, J. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, J. B., Lacy, G. H., Bouzar, H., Minsavage, G. V., Stall, R. E., and Schaad, N. W. (2004). Bacterial spot-worldwide distribution, importance, and review. Acta. Hort. 695, 27–33. doi: 10.17660/ActaHortic.2005.695.1
Kara, S., Timilsina, S., Jacques, M. A., Potnis, N., Vallad, G., Fischer-Le Saux, M., et al. (2017). A novel type III Xop effector in Xanthomonas cynarae associated with rapid cell death. Phytopathology 107, S5.1–S5.142. doi: 10.1094/PHYTO-107-12-S5.1
Kebede, M., Timilsina, S., Ayalew, A., Admassu, B., Potnis, N., Minsavage, G. V., et al. (2014). Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. Eur. J. Plant Pathol. 140, 677–688. doi: 10.1007/s10658-014-0497-3
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Long, J., Song, C., Yan, F., Zhou, J., Zhou, H., and Yang, B. (2018). Non-TAL effectors from Xanthomonas oryzae pv. oryzae suppress peptidoglycan-triggered MAPK activation in rice. Front. Plant Sci. 9:1857. doi: 10.3389/fpls.2018.01857
Lue, Y. S., Deng, W. L., Wu, Y. F., Cheng, A. S., Hsu, S. T., and Tzeng, K. C. (2010). Characterization of Xanthomoans associated with bacterial spot of tomato and pepper in Taiwan. Plant Path. Bull. 19, 181–190,
Ma, X. (2015). Characterization and management of bacterial leaf spot of processing tomato in Ohio. [Ph. D. thesis]. [Department of Plant Pathology]. Columbus, OH: University of Ohio State.
Ma, X., Lewis Ivey, M. L., and Miller, S. A. (2011). First report of Xanthomonas gardneri causing bacterial spot of tomato in Ohio and Michigan. Plant Dis. 95:1584. doi: 10.1094/PDIS-05-11-0448
Mariz-Ponte, N., Gimranov, E., Rego, R., Moura, L., Santos, C., and Tavares, F. (2022). Distinct phenotypic behaviours within a clonal population of Pseudomonas syringae pv. actinidiae. PLoS One 17:e0269343. doi: 10.1371/journal.pone.0269343
Marutani-Hert, M., Hert, A. P., Tudor-Nelson, S. M., Preston, J. F., Minsavage, G. V., Stall, R. E., et al. (2020). Characterization of three novel genetic loci encoding bacteriocins associated with Xanthomonas perforans. PLoS One 15:e0233301. doi: 10.1371/journal.pone.0233301
Medina, C. A., Reyes, P. A., Trujillo, C. A., Gonzalez, J. L., Bejarano, D. A., Montenegro, N. A., et al. (2018). The role of type III effectors from Xanthomonas axonopodis pv. manihotis in virulence and suppression of plant immunity. Mol. Plant Pathol. 19, 593–606. doi: 10.1111/mpp.12545
Okabe, N. (1933). Bacterial diseases of plants occurring in Formosa II: bacterial leaf spot of tomato. J. Soci. Trop. Agric. 5, 26–36,
Osdaghi, E., Taghavi, S. M., Hamzehzarghani, H., and Lamichhane, J. R. (2016). Occurrence and characterization of the bacterial spot pathogen Xanthomonas euvesicatoria on pepper in Iran. J. Phytopathol. 164, 722–734. doi: 10.1111/jph.12493
Pohronezny, K., and Volin, R. B. (1983). The effect of bacterial spot on yield and quality of fresh market tomatoes. HortScience 18, 69–70. doi: 10.21273/HORTSCI.18.1.69
Potnis, N., Timilsina, S., Strayer, A., Shantharaj, D., Barak, J. D., Paret, M. L., et al. (2015). Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 16, 907–920. doi: 10.1111/mpp.12244
Quibod, I., Perez-Quintero, A., Booher, N., Dossa, G. S., Grande, G., Szurek, B., et al. (2016). Effector diversification contributes to Xanthomonas oryzae pv. oryzae phenotypic adaptation in a semi-isolated environment. Sci. Rep. 6:34137. doi: 10.1038/srep34137
Richards, J. K., Stukenbrock, E. H., Carpenter, J., Liu, Z., Cowger, C., Faris, J. D., et al. (2019). Local adaptation drives the diversification of effectors in the fungal wheat pathogen Parastagonospora nodorum in the United States. PLoS Genet. 15:e1008223. doi: 10.1371/journal.pgen.1008223
Robbins, M. D., Darrigues, A., Sim, S. C., Masud, M. A., and Francis, M. D. (2009). Characterization of hypersensitive resistance to bacterial spot race T3 (Xanthomonas perforans) from tomato accession PI 128216. Phytopathology 99, 1037–1044.
Roach, R., Mann, R., Gambley, C. G., Chapman, T., Shivas, R. G., and Rodoni, B. (2019). Genomic sequence analysis reveals diversity of Australian Xanthomonas species associated with bacterial leaf spot of tomato, capsicum, and chilli. BMC Genomics 20:310. doi: 10.1186/s12864-019-5600-x
Schwartz, A. R., Potnis, N., Milsina, S., Wilson, M., Patane, J., Martins, J., et al. (2015). Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front. Microbiol. 6:535. doi: 10.3389/fmicb.2015.00535
Stall, R. E., Jones, J. B., and Minsavage, G. V. (2009). Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu. Rev. Phytopathol. 47, 265–284. doi: 10.1146/annurev-phyto-080508-081752
Timilsina, S., Abrahamian, P., Potnis, N., Minsavage, G. V., White, F. F., Staskawicz, B. J., et al. (2016). Analysis of sequenced genomes of Xanthomonas perforans identifies candidate targets for resistance breeding in tomato. Phytopathology 106, 1097–1104. doi: 10.1094/PHYTO-03-16-0119-FI
Timilsina, S., Jibrin, M. O., Potnis, N., Minsavage, G. V., Kebede, M., Schwartz, A., et al. (2015). Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Appl. Env. Microb. 81, 1520–1529. doi: 10.1128/AEM.03000-14
Timilsina, S., Pereira-Martin, J. A., Minsavage, G. V., Iruegas-Bocardo, F., Abrahamian, P., Potnis, N., et al. (2019). Multiple recombination events drive the current genetic structure of Xanthomonas perforans in Florida. Front. Microb. 10:448. doi: 10.3389/fmicb.2019.00448
Toruño, T. Y., Stergiopoulos, I., and Coaker, G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. doi: 10.1146/annurev-phyto-080615-100204
Vinuesa, P., Ochoa-Sánchez, L. E., and Contreras-Moreira, B. (2018). GET_PHYLOMARKERS, a software package to select optimal orthologous clusters for phylogenomics and inferring pan-genome phylogenies, used for a critical geno-taxonomic revision of the genus stenotrophomonas. Front. Microb. 9:771. doi: 10.3389/fmicb.2018.00771
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Keywords: bacterial spot, local adpatation, diversification, pathogen introduction, effectors
Citation: Chen J-R, Aguirre-Carvajal K, Xue D-Y, Chang H-C, Arone-Maxwell L, Lin Y-P, Armijos-Jaramillo V and Oliva R (2024) Exploring the genetic makeup of Xanthomonas species causing bacterial spot in Taiwan: evidence of population shift and local adaptation. Front. Microbiol. 15:1408885. doi: 10.3389/fmicb.2024.1408885
Edited by:
Md. Motaher Hossain, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Mihaela Niculae, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaDoron Teper, Agricultural Research Organization (ARO), Israel
Sarah Refi Hind, University of Illinois at Urbana-Champaign, United States
Copyright © 2024 Chen, Aguirre-Carvajal, Xue, Chang, Arone-Maxwell, Lin, Armijos-Jaramillo and Oliva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vinicio Armijos-Jaramillo, dmluaWNpby5hcm1pam9zQHVkbGEuZWR1LmVj; Ricardo Oliva, cmljYXJkby5vbGl2YUB3b3JsZHZlZy5vcmc=
 Jaw-Rong Chen
Jaw-Rong Chen Kevin Aguirre-Carvajal
Kevin Aguirre-Carvajal Dao-Yuan Xue4
Dao-Yuan Xue4 Ya-Ping Lin
Ya-Ping Lin Vinicio Armijos-Jaramillo
Vinicio Armijos-Jaramillo Ricardo Oliva
Ricardo Oliva