- 1Department of Dental Emergency, School and Hospital of Stomatology, Wenzhou Medical University, Wenzhou, China
- 2School and Hospital of Stomatology, Wenzhou Medical University, Wenzhou, China
- 3Department of Endodontics, School and Hospital of Stomatology, Wenzhou Medical University, Wenzhou, China
- 4Department of Pediatric Dentistry, School and Hospital of Stomatology, Wenzhou Medical University, Wenzhou, China
Streptococcus mutans (S. mutans), a prime conditionally cariogenic organism, produces membrane vesicles (MVs) containing proteins, nucleic acids, and lipids, including cariogenic virulence factors. Factors including culture conditions, peptide signals, bacterial strains, and genes affect the size and contents of MVs. Based on the composition of their contents, MVs play a wide range of roles in self-regulation, microbial interspecies communication, and microbe–host interactions, which have important potential applications in the fields of vaccine research and disease treatment. In this study, we summarize recent developments in the biogenesis, influencing factors, composition, and functions of S. mutans MVs to lay a theoretical foundation for their potential clinical application and future research.
1 Introduction
Streptococcus mutans (S. mutans) is an important cariogenic bacterium in the oral cavity that produces various biological factors, such as adhesin and glucosyltransferases (Gtfs), which promote the adhesion and aggregation of other bacteria, ultimately resulting in the formation of a thick biofilm (Banas, 2004). It can produce three types of glucosyltransferases (Gtfs) that utilize sucrose to produce extracellular polysaccharides, which are the main components of the three-dimensional extracellular matrix of plaque biofilms (Ren et al., 2016). In addition, S. mutans dynamically releases extracellular deoxyribonucleic acid (eDNA), which strongly interacts with extracellular polysaccharides, synergistically reinforcing microbial adherence and promoting biofilm formation (Klein et al., 2015). In addition, S. mutans can effectively assist the biofilm formation and the maintenance of other oral cariogenic microbial species, such as Candida albicans (C. albicans) and Lactobacillus spp., to colonize the tooth surface (Wen et al., 2017; Guo et al., 2021). Taken together, S. mutans possesses various mechanisms for forming thick biofilms, which not only benefit its own biofilm formation itself but also promote other microbial biofilms. Combined with its ability to produce and tolerate acid, it eventually promotes a cariogenic environment.
Membrane vesicles (MVs) were first discovered to originate from Vibrio cholerae and were considered to be the products of normal physiological processes during bacterial development. Their components were not thoroughly analyzed. This process was initially thought to be related to the excretion of products containing cholera toxins (Chatterjee and Das, 1967). After this initial detection, an increasing number of MVs have been identified in different Gram-negative bacteria, such as Escherichia coli, Burkholderia thailandensis (B. thailandensis), and the periodontal pathogen Porphyromonas gingivalis (McBroom et al., 2006; Toyofuku et al., 2019; Sartorio et al., 2021; Wang et al., 2021). Later, researchers investigated whether Gram-positive bacteria could also produce MVs, and several studies found spherical lipid bilayer structures in their supernatants, including Staphylococcus aureus (S. aureus), Bacillus anthracis, Enterococcus faecalis, and the opportunistic cariogenic bacterium S. mutans (Lee et al., 2009; Barnes et al., 2012; Brown et al., 2015). The MVs, all of which were nanostructures with diameters of 20–400 nm, were enclosed by a coated lipid bilayer membrane. The reported components included lipid molecules, nucleotides [such as DNA and ribonucleic acid (RNA)], proteins (such as enzymes and toxins), and immunogenic peptidoglycan (Klimentová and Stulík, 2015; Kroniger et al., 2018; Dell'Annunziata et al., 2021). MVs play vital roles in bacterial growth, proliferation, pathogenicity, bacterial interactions, and microbe–host interactions (Brown et al., 2015; Rainey et al., 2019; Briaud and Carroll, 2020; Juodeikis and Carding, 2022). For instance, MVs derived from B. thailandensis display anti-biofilm effects on S. mutans, whereas MVs from S. aureus have been shown to enhance the development of airway hypersensitivity to inhaled allergens (Bitto et al., 2020; Wang et al., 2021).
The first successful extraction of MVs from a supernatant culture solution of S. mutans was reported in 2014 (Liao et al., 2014). In this study, classical vesicular structures of MVs were identified in cell-free supernatants using uranyl acetate staining (Liao et al., 2014). This study also revealed that MVs from S. mutans actively released DNA to assist in autologous biofilm formation (Liao et al., 2014). Subsequently, they have been reported in an increasing number of studies. Recent studies have demonstrated that S. mutans MVs harbor nucleic acids, proteins, and lipids, including multiple cariogenic virulence factors that may be involved in self-regulation, microbial interspecies communication, and microbe–host interactions (Iwabuchi et al., 2021; Rainey et al., 2019). This review focuses on the biogenesis, composition, and functions of S. mutans MVs. We aim to provide a theoretical basis for future research on S. mutans MVs through this mini-review.
2 Streptococcus mutans MV biogenesis
All Gram-positive bacteria have a 20–40 nm thick cell wall, which aids in resisting0 extreme conditions such as strong osmotic pressure changes, DNA-damaging agents, antibiotics, and some toxic chemical reagents (Liu et al., 2009; Bose et al., 2020). Peptidoglycan, a major component of the cell wall, in addition to polysaccharides and proteins, acts as a barrier that blocks the release of MVs. Current research indicates that MV biogenesis within Gram-positive bacteria occurs through either autolysin-dependent or endolysin-dependent pathways (Toyofuku et al., 2017; Abe et al., 2021).
In the autolysin-dependent process, the extent of peptidoglycan cross-linking and autolysin activity regulate MV production by altering the permeability of the cell wall in Gram-positive bacteria (Abe et al., 2021). As peptidoglycan hydrolases, autolysins facilitate the release of MVs by increasing the porosity of the thick Gram-positive cell wall. They always localize to the septum, where they exhibit peptidoglycan hydrolase activity, leading to the isolation of MVs from bacteria (Abe et al., 2021). For example, S. aureus can promote the fluidity of its cytoplasmic membrane using modulins, followed by the breakdown of peptidoglycan through autolysins, which can be encoded by sle1 and atl, leading to the release of MVs (Wang and Lee, 2024). Recent research has indicated that S. mutans can release MVs through an autolysin-dependent mechanism, although the details of this process remain largely unclear (Figure 1). Specifically, S. mutans MVs are released via a cell-to-cell communication system mediated by peptide signals called the Com system (Nagasawa et al., 2025). It can regulate the expression of the autolysin (LytF)-encoding gene lytF to further control the release of autolysins, which further modulate the production of MVs (Nagasawa et al., 2025). Electron microscopy images indicated that MV release was accompanied by cell death in a subpopulation of cells, which benefited the remaining cells (Nagasawa et al., 2025). Moreover, it is worth noting that the autolysin AtlA (encoded by altA) with peptidoglycan-degrading activity is likely a major contributor to the biogenesis of S. mutans MVs and is readily detectable within these vesicles (Morales-Aparicio et al., 2020). However, further studies are required to identify their specific roles during the process of S. mutans MV biogenesis.
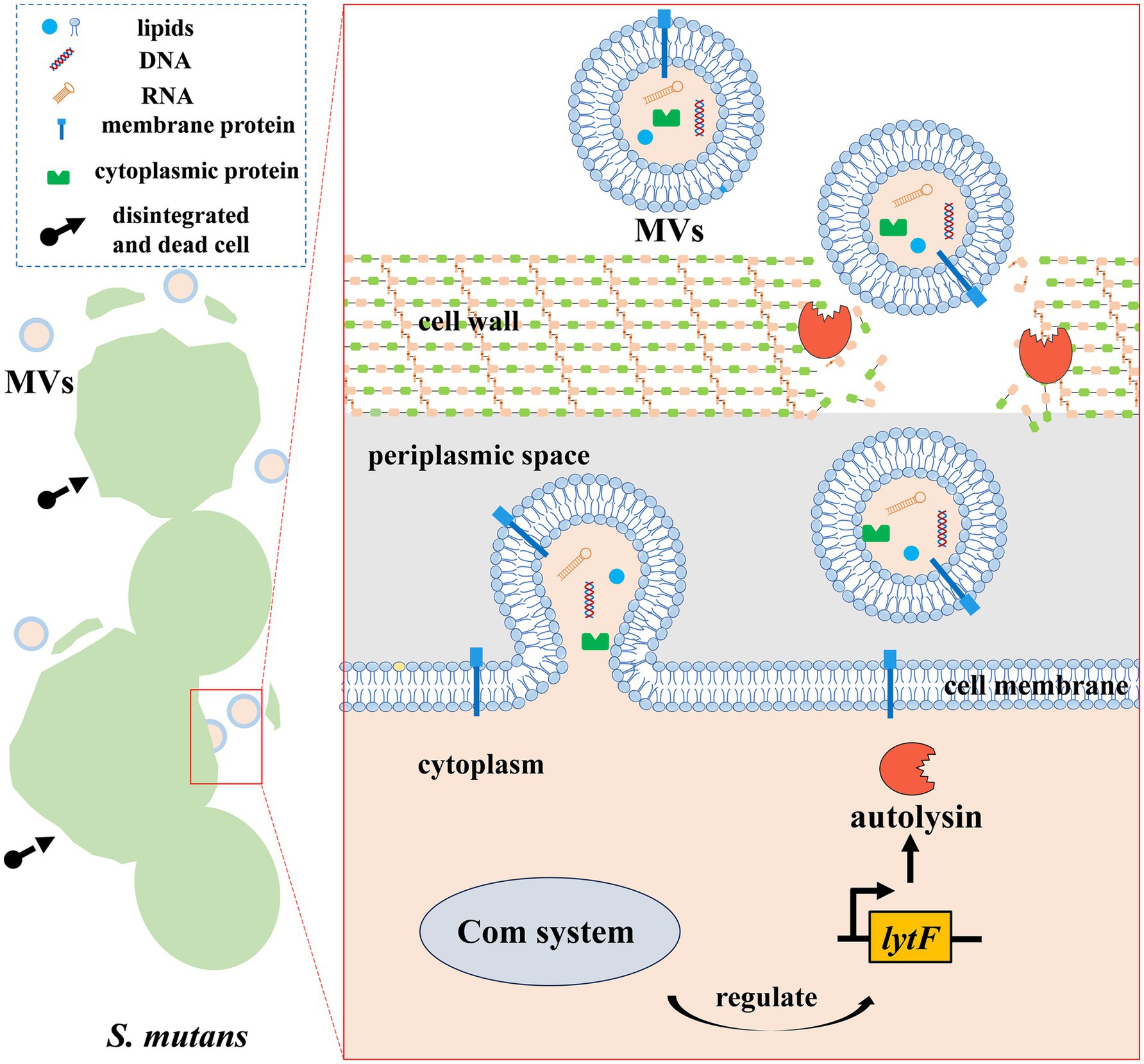
Figure 1. S. mutans MV biogenesis. MV biogenesis in S. mutans appears to occur in an autolysin-dependent manner. The lytF-encoding autolysin can be regulated by the Com system of S. mutans. Autolysin, a peptidoglycan hydrolase, facilitates the release of MVs by increasing the porosity of the cell wall, finally triggering cells’ disintegration and death.
Recent studies have revealed that, in addition to autolysins, phage-derived endolysins can induce MVs in Gram-positive bacteria. The expression of endolysin, which is encoded by a defective prophage, triggers vesicle formation and release in Gram-positive bacteria (Toyofuku et al., 2017). Similar to the explosive cell lysis observed in Gram-negative bacteria, the enzymatic action of endolysins weakens peptidoglycan, causing bacterial contents protrude outward and be released as MVs in certain Gram-positive bacteria. Another group of bacteria undergoes a process called “bubbling cell death,” which results from a loss of cell integrity, and this also leads to the release of MVs (Toyofuku et al., 2023). More research is required to further explore whether S. mutans can produce MVs through this mechanism, although no associated genes have been found in S. mutans to date (Nagasawa et al., 2025).
3 Factors affecting Streptococcus mutans MVs
MV biogenesis is a highly regulated and active process (Brown et al., 2014; Lee et al., 2018). Several factors have been implicated in affecting S. mutans MVs, including culture conditions, peptide signals, bacterial strains, and gene regulation.
3.1 Culture conditions
pH and culture medium have been reported as two culture conditions that affect S. mutans MVs. First, the properties of S. mutans MVs are regulated by pH (Cao et al., 2020; Wen et al., 2021; Iwabuchi et al., 2021). In particular, the initial pH of the culture environment appears to play an important role in MV biogenesis. MVs prepared from S. mutans under different initial pH conditions exhibited different sizes. Although particles of 0–200 nm2 dominated in MVs extracted from both pH 6.0 and pH 8.0 culture media, a higher proportion of MVs exceeding 1,000 nm2 was found under alkaline conditions at pH 8.0 (Iwabuchi et al., 2021). Another study reported similar trends, where the diameter of S. mutans MVs at pH 7.5 was significantly larger than that at pH 5.5 (Cao et al., 2020). In addition, biofilm formation triggered by the treatment with different MVs from S. mutans under various initial pH conditions exhibited different results; these different MVs affected the structure and characteristics of the S. mutans biofilm (Iwabuchi et al., 2021). Although the specific mechanism may require further investigation, it is clear that MVs under different pH conditions are significantly different, not only in size but also in content. S. mutans produces larger MVs under neutral conditions (pH 7.5), despite harboring approximately 10-fold less protein content (standardized by bacterial colony-forming units) compared to acidic conditions (pH 5.5) (Cao et al., 2020).
In addition to pH, the culture medium also influences the characteristics of S. mutans MVs (Nagasawa et al., 2025). Brain heart infusion (BHI) is a complex medium that is commonly used for oral bacterial cultures, including S. mutans. Interestingly, the response of S. mutans to autologous SigX (alternative sigma factor)-inducing peptide (XIP) is restricted by this type of medium. In contrast, chemically defined medium (CDM) is a peptide-free culture medium that supports S. mutans growth and limits the function of self-generated competence-stimulating peptides (CSPs) (Son et al., 2012). Proteins within S. mutans MVs from the BHI medium and CDM were found to be quite different (Nagasawa et al., 2025). Moreover, S. mutans wild-type MVs isolated from BHI could induce the biofilm formation of S. mutans ΔgtfBC, a strain that lacks the corresponding coding products, glucosyltransferase B (GtfB) and glucosyltransferase C (GtfC), and could barely form biofilms (Nagasawa et al., 2025). In contrast, S. mutans wild-type MVs isolated from CDM have limited effects on the biofilm formation of S. mutans ΔgtfBC (Nagasawa et al., 2025).
3.2 Peptide signals
In the autolysin-dependent process of S. mutans MV biogenesis, MV release occurs via a cell-to-cell communication system mediated by peptide signals, called the Com system (Nagasawa et al., 2025). The Com system consists of an upstream CSP-mediated ComDE pathway and a downstream XIP-involved ComRS pathway, which can regulate sigX to further control the autolysin-encoding gene lytF, the product of which is responsible for S. mutans MV release by targeting peptidoglycans (Khan et al., 2016; Nagasawa et al., 2025). Therefore, CSPs with 18 amino acids or XIP with seven amino acids may contribute to S. mutans MV biogenesis. As expected, the exogenous addition of CSP or XIP to BHI or CDM promoted S. mutans MV formation compared to the corresponding medium without peptide signals (Nagasawa et al., 2025). In addition, both CSP and XIP altered the protein contents of S. mutans MVs isolated from the corresponding medium (Nagasawa et al., 2025).
3.3 Bacterial strains
Different strains of S. mutans can produce different numbers of MVs (Wen et al., 2021). For instance, according to a quantitative analysis, S. mutans 27–3, a clinical strain isolated from a patient with active caries, can produce approximately 8-fold more MVs than S. mutans UA159 under the same conditions (Wen et al., 2021). Moreover, the above results are in line with qualitative observations from transmission electron microscopy (TEM), which revealed many more vesicular structures surrounding the cells of S. mutans 27–3 than S. mutans UA159 (Wen et al., 2021). The whole genome sequencing of S. mutans 27–3 revealed significant differences compared to S. mutans UA159, including the addition of 192 genes and the deletion of 275 genes. This may be related to the increase in MV yields (Wen et al., 2021). Evidence suggests that these genes are implicated in S. mutans MV biogenesis.
3.4 Genes
In addition to culture conditions, peptide signals, and bacterial strains, the properties of S. mutans MVs, including size, quantity, and content, are regulated by specific genes.
The lytF-encoding product, LytF, has been reported to induce cell death in a subpopulation of S. mutans and to promote eDNA production (Nagasawa et al., 2020). Interestingly, lytF-expressing S. mutans cells were abundant near the base of the biofilm, while all cells within the biofilm received the CSP signal, which could induce the expression of lytF (Nagasawa et al., 2020). S. mutans MV biogenesis has been reported to occur in an autolysin-dependent manner; the autolysin (LytF)-encoding gene lytF can undoubtedly affect S. mutans MVs. The S. mutans ΔlytF strain produced fewer MVs than its wild-type strain under CSP or XIP treatment. Moreover, the defect of the S. mutans ΔlytF strain in producing MVs was restored in a lytF-complemented strain. These results confirm the involvement of lytF in S. mutans MV biogenesis (Nagasawa et al., 2025).
GtfB and GtfC are encoded by gtfB and gtfC, respectively, and they are two of the most important glycosyltransferases involved in insoluble glucan synthesis in S. mutans. These two important cariogenic virulence factors are present in S. mutans MVs, as revealed by anti-GTF antiserum (Senpuku et al., 2019). These two GTF-encoding genes influence S. mutans MV biogenesis through multiple pathways. First, MVs isolated from strains deficient in gtfB and gtfC display markedly different effects on autologous and other oral microbial biofilm formation compared to those from the wild-type strain. The effects of S. mutans MVs were restricted significantly under S. mutans ΔgtfBC MV. For example, the remarkable enhancement effects of MVs from the wild-type strain on S. mutans UA 159 biofilm formation were not found in MVs from the S. mutans ΔgtfBC strain (Senpuku et al., 2019). The significant repression effects of MVs from wild-type S. mutans on the biofilm formation of Streptococcus gordonii (S. gordonii) and Streptococcus sanguinis (S. sanguinis) were lost when the MVs were replaced with the ones from the S. mutans ΔgtfBC strain (Cui et al., 2022). The protein content of MVs was decreased in S. mutans ΔgtfC and ΔgtfBC strains compared to MVs from the wild-type strain. In contrast, MVs from S. mutans ΔgtfB had a similar protein concentration compared to the wild-type strain. In addition, S. mutans and its ΔgtfB strain had larger MVs than ΔgtfC and ΔgtfBC strains (Nakamura et al., 2020). These results prove that GtfC, but not GtfB, influences the protein content and size of S. mutans MVs (Nakamura et al., 2020).
SMU_833, a putative glycosyltransferase encoded by smu_833, is recognized as an important virulence factor in S. mutans. The deficiency of smu_833 resulted in no changes in the overall biofilm biomass, but it caused changes in biofilm architecture, decreased acidogenesis in vitro, and reduced virulence in a rat caries model (Rainey et al., 2019). In addition, it can alter the interactions between eDNA and glucan, the two primary biofilm matrix constituents (Jakubovics and Burgess, 2015). The deficiency of smu_833 led to a reduction in glucan levels, which resulted from a decrease in Gtfs (GtfB and GtfC) and enhanced eDNA generation. Notably, the increase in eDNA was accompanied by improved release of MVs. The increase in eDNA and MVs as a result of the smu_833 deletion appears to compensate for the defects in Gtfs, making up for any biofilm biomass changes to some extent (Rainey et al., 2019).
Furthermore, SrtA, encoded by srtA, is a transpeptidase that covalently combines several surface-associated proteins with peptidoglycans within the cell wall and has been reported to play a role in MV biogenesis in S. mutans (Liao et al., 2014). The lack of srtA in S. mutans impairs the membrane localization and activity of the multifunctional adhesin P1 and other proteins, which subsequently affects bacterial adhesion and weakens biofilm formation. Therefore, SrtA is a significant protein that plays a role in biofilm formation (Liao et al., 2014). However, subsequent experiments showed that srtA deficiency did not disrupt the production of MVs significantly, as supported by transmission electron microscope observations (Liao et al., 2014). Quantitative analysis from another study suggested that the srtA deficiency strain had a higher MV particle concentration than the wild type (Morales-Aparicio et al., 2020). The protein profile of MVs was significantly altered by srtA deficiency (Liao et al., 2014; Morales-Aparicio et al., 2020). Detailed analysis using Western blotting revealed that MVs extracted from the S. mutans ΔsrtA strain produced lower levels of adhesin P1, glucan-binding proteins B (GbpB) and C (GbpC), and Gtfs compared to MVs released by the wild-type strain (Liao et al., 2014). In addition to differences in content, physical properties of ΔsrtA MVs, analyzed by nanoparticle tracking analysis, displayed a larger mean diameter than the wild-type MVs (Morales-Aparicio et al., 2020). Overall, srtA in S. mutans not only affects MV quantity but also the protein component and size (Liao et al., 2014; Morales-Aparicio et al., 2020).
Similar to SrtA, the 4′-phosphopantetheinyl transferase Sfp has been reported to affect MV biogenesis. Sfp deficiency by sfp mutation in other Gram-positive bacteria impairs the production of MVs and results in defects in biofilm formation (Brown et al., 2014). In S. mutans, the sfp homolog mubP (smu_1334c) is located within a prevalent large genomic island called TnSmu2 and affects MV biogenesis (Wu et al., 2010). In contrast to srtA, sfp deficiency results in lower MV particle concentration compared to its wild type (Morales-Aparicio et al., 2020). In addition, proteomic analyses have shown that sfp mutation also affects the protein composition of MVs (Morales-Aparicio et al., 2020; Wen et al., 2021). This indicates that protein transport from bacteria to MVs is selective and active, and multiple factors may affect this process during different delivery phases (Wen et al., 2021). In addition, the diameter of Δsfp MVs differed from that of wild-type strain MVs (Morales-Aparicio et al., 2020; Wen et al., 2021).
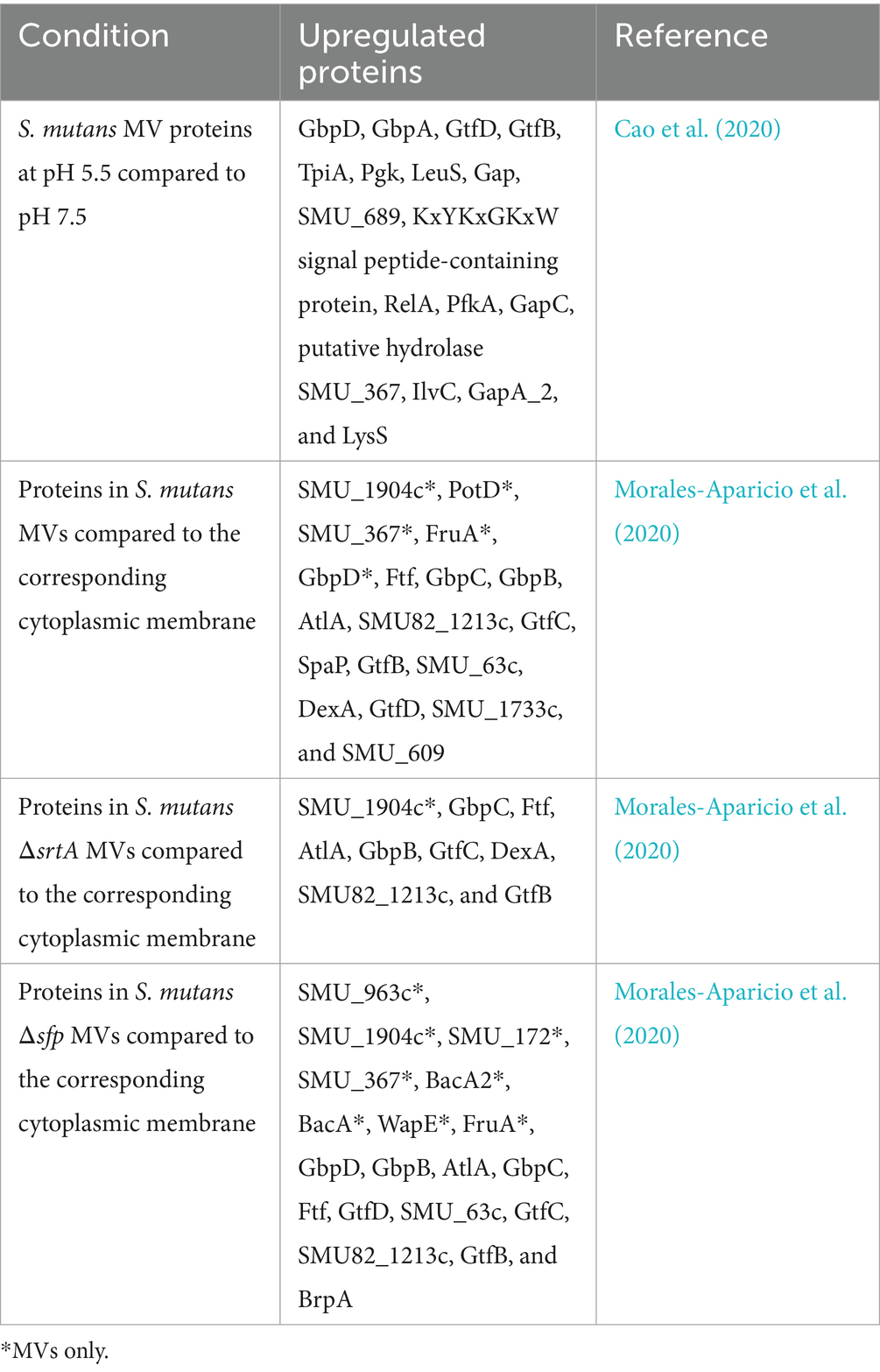
The OpuB transporter, encoded by opuB, was shown to play a critical role in the biogenesis of MVs and affected the composition of S. mutans MVs. For biogenesis, the opuB-deficient (Δ opuB) strain produced smaller and more MVs than S. mutans UA159 at pH 7.5 (Wang C. et al., 2025; Wang W. et al., 2025). However, there was no significant difference in MV quantity or size when the opuB-deficient strain was compared to the wild type at acidic pH 5.5 (Wang C. et al., 2025; Wang W. et al., 2025). When S. mutans MV composition was examined, the knockout of opuB impacted the lipid concentration and composition of MVs (Wang C. et al., 2025; Wang W. et al., 2025). In addition, 108 and 279 proteins in MVs were altered by more than 2-fold in the opuB-deficient strain under pH 7.5 and pH 5.5 conditions, respectively (Wang C. et al., 2025; Wang W. et al., 2025). Genes currently reported to affect S. mutans MVs are listed in Table 1.
In addition to the factors discussed above, other elements involved in S. mutans MV biogenesis need to be investigated. It is clear that components of S. mutans MV-related genes and their regulation play a role in S. mutans MV biosynthesis. Therefore, composition analysis and identification of S. mutans MVs may contribute to the control of MV biogenesis.
4 Composition of Streptococcus mutans MVs
Recently, increasing research attention has been devoted to the content of S. mutans MVs, mainly focusing on proteins, lipids, and nucleic acids.
4.1 Proteins
S. mutans MVs contain many proteins, and the MV protein content has been adopted as a measurement standard to quantify MVs (Cao et al., 2020). Proteomic analyses have identified proteins within MVs that are associated with several biological processes (Cao et al., 2020). A total of 509 proteins were detected in S. mutans MVs, comprising 351 proteins at pH 5.5 and 495 proteins at pH 7.5 (Cao et al., 2020). Although MVs with smaller sizes had significantly higher protein content (normalized by bacterial colony-forming units) under acidic conditions (pH 5.5) compared to pH 7.5, 344 proteins were detected at both pH 5.5 and pH 7.5. They included metabolic enzymes, membrane transporters, secretory proteins, signal peptidase, proteases, structural components of the ribosome, cell wall-associated hydrolases, and lysozymes (Cao et al., 2020). Notably, many virulence factors of S. mutans, such as Gtfs, surface protein antigen P1 (SpaP), glucan-binding proteins (Gbps), lactate dehydrogenase (LDH), and dextranase (DexA), have been identified in MVs using proteomic analysis (Cao et al., 2020).
In addition to the regulatory effects on the protein content due to pH changes, the deletion of some genes, such as srtA and spf, undoubtedly changes the protein composition and quantity of MVs, as mentioned above (Cao et al., 2020; Morales-Aparicio et al., 2020). It was reported that MVs from the S. mutans Δsfp strain shared 61.16% protein similarity with its wild-type strain MVs (Morales-Aparicio et al., 2020). The similarity in MV protein composition was 28.10% when the S. mutans ΔsrtA strain was compared to its wild-type strain (Morales-Aparicio et al., 2020). Comparatively, these two strains with gene mutations shared 28.51% similarity in MV protein composition (Morales-Aparicio et al., 2020). These results illustrate that the transport of proteins to S. mutans MVs is a selective process that is substantially influenced by the presence of SrtA, and to a lesser extent, by Sfp (Cao et al., 2020; Morales-Aparicio et al., 2020). All proteins reported under different conditions are listed in Table 2.
4.2 Lipids
Lipids are vital structural constituents of bacterial cell membranes, including S. mutans, through which MVs are secreted (Morales-Aparicio et al., 2020). Hence, it is clear that outer membrane phospholipids are components of S. mutans MVs. Nevertheless, some lipids are found exclusively in MVs and not in the outer membrane of S. mutans (Morales-Aparicio et al., 2020). Lipids in MVs from S. mutans were analyzed using liquid chromatography-mass spectrometry, and approximately 30 individual lipids were identified in S. mutans MVs (Morales-Aparicio et al., 2020). The relative proportion of each lipid category varies between the cytomembrane and MVs. The analysis showed that cardiolipins and flavonoids are present in higher proportions in MVs compared to the cytomembrane (Morales-Aparicio et al., 2020). The lipid architecture plays an important role in environmental adaptation (Fozo and Quivey, 2004). The richness of monounsaturated long-chain fatty acids helps S. mutans improve its tolerance to the acidic environment generated during the fermentation of carbohydrates into organic acid end products (Morales-Aparicio et al., 2020). Interestingly, in MVs from S. mutans ΔsrtA and Δsfp strains, the level of monounsaturated long-chain fatty acids was significantly increased; this transformation may assist these strains to tolerate the acidic environment (Morales-Aparicio et al., 2020).
4.3 Nucleic acids
MVs from Gram-positive bacteria have been reported to harbor nucleic acids such as DNA and RNA, which can be delivered to other bacteria and facilitate horizontal gene transfer (HGT) (Schooling et al., 2009; Díaz-Garrido et al., 2021).
To identify whether S. mutans MVs can act as carriers for the release of eDNA, hydrolyzed MVs were used to detect eDNA existence. Unsurprisingly, the experiments confirmed the presence of eDNA in MVs (Liao et al., 2014). eDNA plays a crucial role in biofilm formation, including that of its own and several other bacteria (Senpuku et al., 2019; Wu et al., 2020). Several studies have also identified RNA in MVs, which are important for some physiological processes (Munhoz da Rocha et al., 2020). These RNAs include messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and long non-coding RNA (lncRNA) (Munhoz da Rocha et al., 2020). A recent study identified tRNA in S. mutans MVs that could facilitate cell proliferation together with the migration of the oral mucosa, support focal adhesion complex formation within organoids, and aid wound healing in a mouse model (Oh et al., 2025). RNA sequencing analysis identified “microRNA-like” molecules in S. mutans, suggesting that these RNAs may contribute to bacteria that are analogous to microRNAs within eukaryotes (Lee and Hong, 2012; Munhoz da Rocha et al., 2020).
In addition to the studies on MV components discussed above, further research on MV composition and its influencing factors is needed to further understand MVs and explore their potential functions and applications.
5 Functions of Streptococcus mutans MVs
Owing to the different compositions of MVs, it is possible that they perform different functions. However, many of these potential functions, based on their contents, have not yet been verified. Currently, major research advances have focused on self-regulation, microbial communication, and microbe–host interactions (Figure 2).
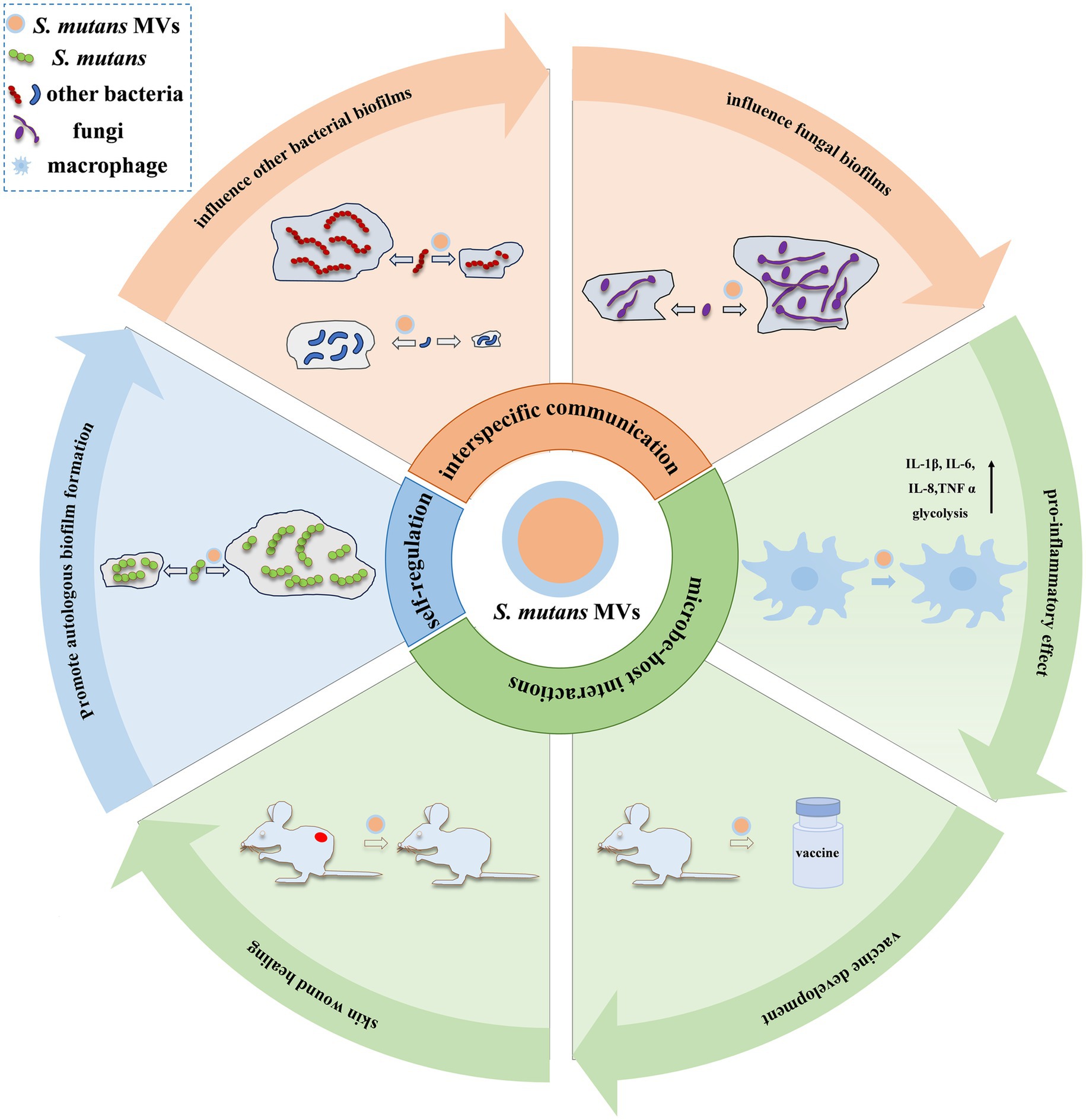
Figure 2. Functions of S. mutans MVs. Currently, major research advances in S. mutans MV functions have focused on self-regulation, microbial communication, and microbe–host interactions. The self-regulatory effect of S. mutans MVs has been mainly observed in autologous biofilm formation (Nakamura et al., 2020). In interspecies communication, S. mutans MVs display suppression (such as S. gordonii and S. sanguinis) (Cui et al., 2022) or enhancement (such as S. sanguinis, S. mitis, S. oralis, A. naeslundii, S. gordonii, and A. oris) (Senpuku et al., 2019) effects on bacterial biofilms and augment fungal biofilm development, such as C. albicans (Wu et al., 2020). In microbe–host interactions, S. mutans MVs have been reported to elevate the release of inflammatory cytokines (such as IL-1β, IL-6, IL-8, and TNF-α) in macrophages and induce cellular glycolysis (Song et al., 2024); stimulate an immune response to produce anti-Gtfs antibodies, which make the development of vaccines feasible (Nakamura et al., 2020); and promote skin wound healing in mice (Oh et al., 2025).
5.1 Self-regulation
The self-regulatory effect of S. mutans MVs has been mainly observed in autologous biofilm formation. A previous study showed that MVs derived from S. mutans contribute to autologous biofilm formation (Senpuku et al., 2019). The extracellular biofilm matrix of S. mutans mainly consists of glucan polysaccharides, eDNA, and lipoteichoic acid (Klein et al., 2015). A significant part of this process is the glucan matrix, which facilitates S. mutans adherence to the tooth surface, maintains mechanical stability, protects microorganisms from environmental assaults, reserves energy sources, limits the diffusion of substances into and out of the biofilm, and helps concentrate metal ions and other physiological nutrients (Schilling and Bowen, 1992; Koo et al., 2009). The glucan matrix of S. mutans is synthesized and organized using extracellular Gtfs. Surprisingly, one of the most important components in MVs secreted from S. mutans is Gtfs, which is a key enzyme in dental caries development (Nakamura et al., 2020). Several studies have suggested that S. mutans secretes MVs harboring Gtfs that can augment sucrose metabolism and promote autologous biofilm formation. Compared to MVs from the S. mutans ΔgtfBC strain, the MVs from the wild-type strain significantly enhanced biofilm formation (Senpuku et al., 2019). Interestingly, GtfB largely adheres to the MV surface (Nakamura et al., 2020). In contrast, GtfC is primarily present within MVs and regulates MV size and aggregation and S. mutans biofilm formation (Nakamura et al., 2020). Another factor that plays an important role in the formation of S. mutans biofilms on tissues within the oral cavity is eDNA, which contributes to adhesion and the accumulation of S. mutans, as well as the architecture and stability of autologous biofilms (Castillo Pedraza et al., 2017; Kim et al., 2018). To investigate the function of eDNA and MVs in S. mutans biofilm formation, Senpuku et al. reported the extraction of a purified complex consisting of DNA and MVs with Gtfs from S. mutans and incubation with the S. mutans ΔgtfBC strain. Interestingly, the results suggested that this complex induced Gtfs-dependent S. mutans ΔgtfBC biofilm formation (Senpuku et al., 2019). Moreover, short DNA fragments associated with S. mutans MVs can significantly promote autologous biofilm formation (Senpuku et al., 2019). MVs that had eDNA removed showed a different effect on S. mutans ΔgtfBC biofilm formation compared to MVs at a relatively low concentration (Senpuku et al., 2019). Therefore, it is inferred that MVs can assist in autologous biofilm formation, further enhancing the cariogenicity of S. mutans and promoting dental caries.
5.2 Interspecies communication
MVs are natural carriers of molecules that are protected by them, allowing long-distance delivery of these biological molecules and avoiding direct intercellular contact to safely reach their final destination (Gill et al., 2019). This characteristic endows S. mutans MVs with the function of interspecies communication by influencing biofilm formation in other species.
A recent study showed that MVs not only contribute to the biofilm formation of S. mutans but also influence the formation of other bacterial biofilms. S. sanguinis and S. gordonii are the initial colonizers of tooth surfaces. They compete with S. mutans for hydrogen peroxide (H2O2) and are countered by S. mutans through mutacin (Zhang et al., 2025). It has been reported that biofilm formation by S. gordonii and S. sanguinis is inhibited by S. mutans MVs, where the Gtfs in MVs play a role (Cui et al., 2022). When co-cultured with MVs from S. mutans, S. gordonii, and S. sanguinis, biofilm formation was significantly suppressed (Cui et al., 2022). In contrast, S. mutans ΔgtfBC MVs had no significant effect on the biofilm formation of these two species (Cui et al., 2022). In addition, S. mutans MVs suppressed the expression of their virulence genes, including GtfG (encoding glucosyltransferase in S. gordonii), GtfP (encoding glucosyltransferase in S. sanguinis), and SpxB (encoding pyruvate oxidase to produce H2O2) (Cui et al., 2022). Another study reported that biofilm formation by S. sanguinis, Streptococcus mitis, Streptococcus oralis, Actinomyces naeslundii, S. gordonii, and Actinomyces oris can be facilitated by S. mutans MVs (Senpuku et al., 2019). Further studies have shown that this facilitative action is GtfB- and GtfC-dependent, except in A. naeslundii, where MVs from S. mutans ΔgtfBC still display promotional effects (Senpuku et al., 2019). The differing effects of S. mutans MVs on S. gordonii and S. sanguinis biofilms observed in the two separate studies may have resulted from differences in culture conditions, bacterial strains, and experimental methods.
In addition to being involved in the communication between bacteria, S. mutans MVs also affect fungi, such as C. albicans, one of the most common colonizers within the oral cavity. MVs derived from S. mutans can augment the biofilm development of C. albicans and are Gtf-dependent (Wu et al., 2020). In addition, S. mutans MVs enhance the pathogenicity and carbohydrate metabolism of C. albicans. The enhanced pathogenicity of fungal biofilms was revealed in a bovine dentin demineralization experiment, where S. mutans MV-containing groups showed greater hardness loss, more exposure, and increased damage to dentin tubules (Wu et al., 2022). Promoted carbohydrate metabolism is mainly revealed by the increase in related metabolites and protein expression (Wu et al., 2022). In addition, when co-cultured with MVs, C. albicans biofilms have a three-dimensional structure with an abundant extracellular matrix, and C. albicans forms hyphal cells under biofilm-forming conditions (Wang C. et al., 2025; Wang W. et al., 2025).
Other potential functions of S. mutans MVs involved in interspecies communication include providing substrates for horizontal gene transfer (HGT) and regulating gene expression and protein translation, which are based on nucleic acid loading in MVs. HGT has recently been identified as an effective mechanism for microbiomes to interact, helping bacteria acquire new genetic traits in addition to plasmids (Arnold et al., 2022). The distribution of antimicrobial resistance genes is an example of this interaction and is considered a type of HGT (Yaron et al., 2000). S. mutans has been reported to release eDNA via MVs into developing biofilms; therefore, it is reasonable to infer that MVs from S. mutans also offer other competent bacteria an important source of transformation through this novel mechanism (Roberts and Kreth, 2014; Campoccia et al., 2021). In addition to eDNA, RNA is another type of nucleic acid found in MVs. As mentioned previously, multiple types of RNA can be delivered by S. mutans MVs (Munhoz da Rocha et al., 2020). It may contribute to bacterial communication by regulating gene expression via non-coding RNAs and protein translation via messenger RNAs (Munhoz da Rocha et al., 2020). The effects of RNA within S. mutans MVs have been demonstrated in microbe–host interactions (Tsatsaronis et al., 2018; Oh et al., 2025). However, whether nucleic acids within S. mutans MVs are involved in HGT, microbial interspecies regulation of gene expression, and protein translation requires further investigation.
5.3 Microbe–host interactions
The role of S. mutans MVs in microbe–host interactions is mainly reflected in their immunity. On the one hand, S. mutans MVs can trigger an immune response and induce a pro-inflammatory effect. On the other hand, S. mutans MVs have emerged as promising tools for the development of vaccines and immunotherapeutic strategies against infectious and non-infectious diseases (Nakao et al., 2011; Long et al., 2022).
A recent study showed that S. mutans MVs could notably elevate the release of inflammatory cytokines and induce macrophage glycolysis. When cultured with S. mutans MVs, the expression of macrophage pro-inflammatory cytokines, such as interleukin-1β (IL-1β), interleukin-6, interleukin-8, and tumor necrosis factor α, was significantly increased (Song et al., 2024). Among these highly expressed cytokines, IL-1β was particularly prominent, and its increased production induced by S. mutans MVs could occur through the activation of the nucleotide-binding oligomerization domain-like receptor protein 3 (NLRP3), absent in melanoma 2 (AIM2), apoptosis-associated speck-like protein containing CARD (ASC), and nucleotide-binding oligomerization domain-like receptor C4 (NLRC4) inflammasomes (Song et al., 2024). In addition, potassium ion efflux and adenosine triphosphate generation were involved in IL-1β production induced by S. mutans MVs (Song et al., 2024). Macrophage glycolysis is a crucial part of this pro-inflammatory process for classical activation (Song et al., 2024). In addition to these two main findings, S. mutans MVs promoted S. mutans colonization of oral epithelial cells and suppressed macrophage phagocytosis against S. mutans (Song et al., 2024).
In addition to their pro-inflammatory effects, S. mutans MVs can induce an immune response in the oral environment, making vaccine development feasible (Nakao et al., 2011; Long et al., 2022). GtfB and GtfC, which are closely associated with S. mutans MVs, are key cariogenic virulence factors that contribute to biofilm formation by themselves and through other microorganisms (Liao et al., 2014; Senpuku et al., 2019). Based on this evidence, S. mutans MVs are regarded as crucial virulence factors and targets for biofilm-associated disease control. It was reported that MVs from S. mutans wild-type, ΔgtfB, ΔgtfC, and ΔgtfBC strains produced anti-MV IgA and IgG antibodies after intranasal immunization of mice (Nakamura et al., 2020). Further investigation revealed that it is the antibodies induced by MVs from S. mutans wild-type and S. mutans ΔgtfB strains, rather than S. mutans ΔgtfC and S. mutans ΔgtfBC strains, that react with MV Gtfs (Nakamura et al., 2020). It is clear that S. mutans MVs harboring GtfC are operative mucosal immunogens that induce anti-Gtf antibody production (Nakamura et al., 2020). Using S. mutans MVs as antigens stimulated IgA and IgG antibody generation against to Gtfs successfully, which may be useful for future vaccine development. However, considering the extremely complex composition of S. mutans MVs, the potential side effects of this process require further investigation.
In addition to the two proven effects discussed above, S. mutans MVs have been shown to play a role in skin wound healing in mice (Oh et al., 2025). Specifically, S. mutans MVs not only promoted the proliferation of human oral organoids, assisted in the migration of oral epithelial cells, and enhanced the formation of focal adhesion complexes but also facilitated wound healing in the dorsal skin of mice (Oh et al., 2025). Further research has revealed that tRNA variants, the most abundant RNAs within S. mutans MVs, play a vital role in this process (Oh et al., 2025). Surprisingly, the tRNA mentioned above could take effect even when electroporated into Escherichia coli MVs, in addition to being packaged within S. mutans MVs (Oh et al., 2025). Further research revealed that the promotion of skin wound healing occurred through a Toll-like receptor 3-dependent mechanism (Oh et al., 2025). This study demonstrated that the use of S. mutans MVs and RNA cargo is a promising therapeutic strategy for skin wound rehabilitation (Oh et al., 2025).
6 Conclusion and future perspectives
In summary, S. mutans MVs containing multiple molecules, including virulence factors, have been recognized as powerful tools for S. mutans to survive and compete. As demonstrated above, they have diverse capabilities, including self-regulation, microbial interspecies communication, and microbe—host interactions. Further systematic and comprehensive clarification of MV biogenesis, composition, and function, not limited to S. mutans, will help us better understand the potential of MVs. Although the characteristics of MVs make the management of biofilm-associated diseases even more challenging, they provide a potential target for the control of these diseases. In addition, the MV-based development of vaccines or therapeutics is an important direction for future research. MVs can provide a protected environment for carrying this cargo, thereby demonstrating great potential as a tool for drug delivery.
Author contributions
LQ: Visualization, Writing – original draft. QC: Writing – original draft, Visualization. GZ: Visualization, Writing – original draft. HD: Writing – original draft. MX: Writing – original draft. JZ: Writing – original draft. LZ: Writing – original draft. YS: Writing – original draft. MW: Writing – original draft. YaP: Writing – original draft. JY: Conceptualization, Supervision, Writing – review & editing. YiP: Funding acquisition, Writing – review & editing, Supervision, Conceptualization. KZ: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work was supported by the Zhejiang Provincial Natural Science Foundation of China (grant number ZCLTGY24H1403), National Natural Science Foundation of China (grant number 82470971), Provincial and ministerial joint project (grant number WKJ-ZJ-2214), and Wenzhou Technology Bureau Project (grant number ZY2024030).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, K., Toyofuku, M., Nomura, N., and Obana, N. (2021). Autolysis-mediated membrane vesicle formation in Bacillus subtilis. Environ. Microbiol. 23, 2632–2647. doi: 10.1111/1462-2920.15502
Arnold, B. J., Huang, I. T., and Hanage, W. P. (2022). Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 20, 206–218. doi: 10.1038/s41579-021-00650-4
Banas, J. A. (2004). Virulence properties of Streptococcus mutans. Front. Biosci. 9, 1267–1277. doi: 10.2741/1305
Barnes, A. M., Ballering, K. S., Leibman, R. S., Wells, C. L., and Dunny, G. M. (2012). Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. MBio 3:e00193-00112. doi: 10.1128/mBio.00193-12
Bitto, N. J., Cheng, L., Johnston, E. L., Pathirana, R., Phan, T. K., Poon, I. K. H., et al. (2020). Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. J. Extracell. Vesicles 10:e12080. doi: 10.1002/jev2.12080
Bose, S., Aggarwal, S., Singh, D. V., and Acharya, N. (2020). Extracellular vesicles: an emerging platform in gram-positive bacteria. Microb Cell 7, 312–322. doi: 10.15698/mic2020.12.737
Briaud, P., and Carroll, R. K. (2020). Extracellular vesicle biogenesis and functions in gram-positive bacteria. Infect. Immun. 88:e00433-20. doi: 10.1128/IAI.00433-20
Brown, L., Kessler, A., Cabezas-Sanchez, P., Luque-Garcia, J. L., and Casadevall, A. (2014). Extracellular vesicles produced by the gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol. Microbiol. 93, 183–198. doi: 10.1111/mmi.12650
Brown, L., Wolf, J. M., Prados-Rosales, R., and Casadevall, A. (2015). Through the wall: extracellular vesicles in gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13, 620–630. doi: 10.1038/nrmicro3480
Campoccia, D., Montanaro, L., and Arciola, C. R. (2021). Tracing the origins of extracellular DNA in bacterial biofilms: story of death and predation to community benefit. Biofouling 37, 1022–1039. doi: 10.1080/08927014.2021.2002987
Cao, Y., Zhou, Y., Chen, D., Wu, R., Guo, L., and Lin, H. (2020). Proteomic and metabolic characterization of membrane vesicles derived from Streptococcus mutans at different pH values. Appl. Microbiol. Biotechnol. 104, 9733–9748. doi: 10.1007/s00253-020-10563-6
Castillo Pedraza, M. C., Novais, T. F., Faustoferri, R. C., Quivey, R. G., Terekhov, A., Hamaker, B. R., et al. (2017). Extracellular DNA and lipoteichoic acids interact with exopolysaccharides in the extracellular matrix of Streptococcus mutans biofilms. Biofouling 33, 722–740. doi: 10.1080/08927014.2017.1361412
Chatterjee, S. N., and Das, J. (1967). Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J. Gen. Microbiol. 49, 1–11. doi: 10.1099/00221287-49-1-1
Cui, G., Li, P., Wu, R., and Lin, H. (2022). Streptococcus mutans membrane vesicles inhibit the biofilm formation of Streptococcus gordonii and Streptococcus sanguinis. AMB Express 12:154. doi: 10.1186/s13568-022-01499-3
Dell'Annunziata, F., Folliero, V., Giugliano, R., De Filippis, A., Santarcangelo, C., Izzo, V., et al. (2021). Gene transfer potential of outer membrane vesicles of gram-negative bacteria. Int. J. Mol. Sci. 22:5985. doi: 10.3390/ijms22115985
Díaz-Garrido, N., Badia, J., and Baldomà, L. (2021). Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J Extracell Vesicles 10:e12161. doi: 10.1002/jev2.12161
Fozo, E. M., and Quivey, R. G. (2004). Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70, 929–936. doi: 10.1128/AEM.70.2.929-936.2004
Gill, S., Catchpole, R., and Forterre, P. (2019). Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 43, 273–303. doi: 10.1093/femsre/fuy042
Guo, H., Chen, Y., Guo, W., and Chen, J. (2021). Effects of extracellular DNA on dual-species biofilm formed by Streptococcus mutans and Candida albicans. Microb. Pathog. 154:104838. doi: 10.1016/j.micpath.2021.104838
Iwabuchi, Y., Nakamura, T., Kusumoto, Y., Nakao, R., Iwamoto, T., Shinozuka, O., et al. (2021). Effects of pH on the properties of membrane vesicles including glucosyltransferase in Streptococcus mutans. Microorganisms 9:2308. doi: 10.3390/microorganisms9112308
Jakubovics, N. S., and Burgess, J. G. (2015). Extracellular DNA in oral microbial biofilms. Microbes Infect. 17, 531–537. doi: 10.1016/j.micinf.2015.03.015
Juodeikis, R., and Carding, S. R. (2022). Outer membrane vesicles: biogenesis, functions, and issues. Microbiol. Mol. Biol. Rev. 86:e0003222. doi: 10.1128/mmbr.00032-22
Khan, R., Rukke, H. V., Høvik, H., Åmdal, H. A., Chen, T., Morrison, D. A., et al. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1, e00038–e00015. doi: 10.1128/mSystems.00038-15
Kim, M., Jeon, J., and Kim, J. (2018). Streptococcus mutans extracellular DNA levels depend on the number of bacteria in a biofilm. Sci. Rep. 8:13313. doi: 10.1038/s41598-018-31275-y
Klein, M. I., Hwang, G., Santos, P. H., Campanella, O. H., and Koo, H. (2015). Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front. Cell. Infect. Microbiol. 5:10. doi: 10.3389/fcimb.2015.00010
Klimentová, J., and Stulík, J. (2015). Methods of isolation and purification of outer membrane vesicles from gram-negative bacteria. Microbiol. Res. 170, 1–9. doi: 10.1016/j.micres.2014.09.006
Koo, H., Xiao, J., and Klein, M. I. (2009). Extracellular polysaccharides matrix--an often forgotten virulence factor in oral biofilm research. Int. J. Oral Sci. 1, 229–234. doi: 10.4248/IJOS.09086
Kroniger, T., Otto, A., and Becher, D. (2018). Proteomic analysis of bacterial (outer) membrane vesicles: progress and clinical potential. Expert Rev. Proteomics 15, 623–626. doi: 10.1080/14789450.2018.1505509
Lee, E. Y., Choi, D. Y., Kim, D. K., Kim, J. W., Park, J. O., Kim, S., et al. (2009). Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9, 5425–5436. doi: 10.1002/pmic.200900338
Lee, H.-J., and Hong, S.-H. (2012). Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol. Lett. 326, 131–136. doi: 10.1111/j.1574-6968.2011.02441.x
Lee, T., Jun, S. H., Choi, C. W., Kim, S. I., Lee, J. C., and Shin, J. H. (2018). Salt stress affects global protein expression profiles of extracellular membrane-derived vesicles of Listeria monocytogenes. Microb. Pathog. 115, 272–279. doi: 10.1016/j.micpath.2017.12.071
Liao, S., Klein, M. I., Heim, K. P., Fan, Y., Bitoun, J. P., Ahn, S.-J., et al. (2014). Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J. Bacteriol. 196, 2355–2366. doi: 10.1128/JB.01493-14
Liu, X. D., Duan, J., and Guo, L. H. (2009). Role of phosphoglucosamine mutase on virulence properties of Streptococcus mutans. Oral Microbiol. Immunol. 24, 272–277. doi: 10.1111/j.1399-302X.2009.00503.x
Long, Q., Zheng, P., Zheng, X., Li, W., Hua, L., Yang, Z., et al. (2022). Engineered bacterial membrane vesicles are promising carriers for vaccine design and tumor immunotherapy. Adv. Drug Deliv. Rev. 186:114321. doi: 10.1016/j.addr.2022.114321
McBroom, A. J., Johnson, A. P., Vemulapalli, S., and Kuehn, M. J. (2006). Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188, 5385–5392. doi: 10.1128/JB.00498-06
Morales-Aparicio, J. C., Lara Vasquez, P., Mishra, S., Barrán-Berdón, A. L., Kamat, M., Basso, K. B., et al. (2020). The impacts of Sortase a and the 4′-phosphopantetheinyl transferase homolog Sfp on Streptococcus mutans extracellular membrane vesicle biogenesis. Front. Microbiol. 11:570219. doi: 10.3389/fmicb.2020.570219
Munhoz da Rocha, I. F., Amatuzzi, R. F., Lucena, A. C. R., Faoro, H., and Alves, L. R. (2020). Cross-kingdom extracellular vesicles EV-RNA communication as a mechanism for host–pathogen interaction. Front. Cell. Infect. Microbiol. 10:593160. doi: 10.3389/fcimb.2020.593160
Nagasawa, R., Ito, T., Yamamoto, C., Unoki, M., Obana, N., Nomura, N., et al. (2025). Membrane vesicle production via cell-to-cell communication-induced autolysis in Streptococcus mutans. Microbiol Spectr 13:e0033425. doi: 10.1128/spectrum.00334-25
Nagasawa, R., Yamamoto, T., Utada, A. S., Nomura, N., and Obana, N. (2020). Competence-stimulating-peptide-dependent localized cell death and extracellular DNA production in Streptococcus mutans biofilms. Appl. Environ. Microbiol. 86:e02080-20. doi: 10.1128/AEM.02080-20
Nakamura, T., Iwabuchi, Y., Hirayama, S., Narisawa, N., Takenaga, F., Nakao, R., et al. (2020). Roles of membrane vesicles from Streptococcus mutans for the induction of antibodies to glucosyltransferase in mucosal immunity. Microb. Pathog. 149:104260. doi: 10.1016/j.micpath.2020.104260
Nakao, R., Hasegawa, H., Ochiai, K., Takashiba, S., Ainai, A., Ohnishi, M., et al. (2011). Outer membrane vesicles of Porphyromonas gingivalis elicit a mucosal immune response. PLoS One 6:e26163. doi: 10.1371/journal.pone.0026163
Oh, S. Y., Kim, D. Y., Lee, K. Y., Ha, D.-L., Kim, T.-L., Kwon, T.-G., et al. (2025). Streptococcus mutans-derived extracellular vesicles promote skin wound healing via tRNA cargo. J Nanobiotechnology 23:322. doi: 10.1186/s12951-025-03410-1
Rainey, K., Michalek, S. M., Wen, Z. T., Wu, H., and Nojiri, H. (2019). Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl. Environ. Microbiol. 85, e02247–e02218. doi: 10.1128/AEM.02247-18
Ren, Z., Chen, L., Li, J., and Li, Y. (2016). Inhibition of Streptococcus mutans polysaccharide synthesis by molecules targeting glycosyltransferase activity. J. Oral Microbiol. 8:31095. doi: 10.3402/jom.v8.31095
Roberts, A. P., and Kreth, J. (2014). The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front. Cell. Infect. Microbiol. 4:124. doi: 10.3389/fcimb.2014.00124
Sartorio, M. G., Pardue, E. J., Feldman, M. F., and Haurat, M. F. (2021). Bacterial outer membrane vesicles: from discovery to applications. Ann. Rev. Microbiol. 75, 609–630. doi: 10.1146/annurev-micro-052821-031444
Schilling, K. M., and Bowen, W. H. (1992). Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect. Immun. 60, 284–295. doi: 10.1128/iai.60.1.284-295.1992
Schooling, S. R., Hubley, A., and Beveridge, T. J. (2009). Interactions of DNA with biofilm-derived membrane vesicles. J. Bacteriol. 191, 4097–4102. doi: 10.1128/JB.00717-08
Senpuku, H., Nakamura, T., Iwabuchi, Y., Hirayama, S., Nakao, R., and Ohnishi, M. (2019). Effects of complex DNA and MVs with GTF extracted from Streptococcus mutans on the oral biofilm. Molecules 24:3131. doi: 10.3390/molecules24173131
Son, M., Ahn, S. J., Guo, Q., Burne, R. A., and Hagen, S. J. (2012). Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol. Microbiol. 86, 258–272. doi: 10.1111/j.1365-2958.2012.08187.x
Song, G., Li, M., Zhou, B., Qi, H., and Guo, J. (2024). Streptococcus mutans outer membrane vesicles affect inflammasome activation and the glycolysis of macrophages. Microb. Pathog. 196:106994. doi: 10.1016/j.micpath.2024.106994
Toyofuku, M., Cárcamo-Oyarce, G., Yamamoto, T., Eisenstein, F., Hsiao, C. C., Kurosawa, M., et al. (2017). Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 8:481. doi: 10.1038/s41467-017-00492-w
Toyofuku, M., Nomura, N., and Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24. doi: 10.1038/s41579-018-0112-2
Toyofuku, M., Schild, S., Kaparakis-Liaskos, M., and Eberl, L. (2023). Composition and functions of bacterial membrane vesicles. Nat. Rev. Microbiol. 21, 415–430. doi: 10.1038/s41579-023-00875-5
Tsatsaronis, J. A., Franch-Arroyo, S., Resch, U., and Charpentier, E. (2018). Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol. 26, 401–410. doi: 10.1016/j.tim.2018.02.009
Wang, W., Huang, Y., Lin, H., and Cao, Y. (2025). Role of opuB in modulating membrane vesicle composition and function in Streptococcus mutans under neutral and acidic conditions. Microorganisms 13:884. doi: 10.3390/microorganisms13040884
Wang, Y., Hoffmann, J. P., Baker, S. M., zu Bentrup, K. H., Wimley, W. C., Fuselier, J. A., et al. (2021). Inhibition of Streptococcus mutans biofilms with bacterial-derived outer membrane vesicles. BMC Microbiol. 21:34. doi: 10.1186/s12866-021-02296-x
Wang, X., and Lee, J. C. (2024). Staphylococcus aureus membrane vesicles: an evolving story. Trends Microbiol. 32, 1096–1105. doi: 10.1016/j.tim.2024.04.003
Wang, C., Zhang, Y., Gu, Z., Li, Z., Wu, Q., Xu, X., et al. (2025). Streptococcus mutans regulates ubiquitin modification of Candida albicans in the bacterial-fungal interaction. PLoS Pathog. 21:e1012887. doi: 10.1371/journal.ppat.1012887
Wen, Z. T., Jorgensen, A. N., Huang, X., Ellepola, K., Chapman, L., Wu, H., et al. (2021). Multiple factors are involved in regulation of extracellular membrane vesicle biogenesis in Streptococcus mutans. Mol Oral Microbiol 36, 12–24. doi: 10.1111/omi.12318
Wen, Z. T., Liao, S., Bitoun, J. P., De, A., Jorgensen, A., Feng, S., et al. (2017). Streptococcus mutans displays altered stress responses while enhancing biofilm formation by Lactobacillus casei in mixed-species consortium. Front. Cell. Infect. Microbiol. 7:524. doi: 10.3389/fcimb.2017.00524
Wu, C., Cichewicz, R., Li, Y., Liu, J., Roe, B., Ferretti, J., et al. (2010). Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl. Environ. Microbiol. 76, 5815–5826. doi: 10.1128/AEM.03079-09
Wu, R., Cui, G., Cao, Y., Zhao, W., and Lin, H. (2022). Streptococcus mutans membrane vesicles enhance Candida albicans pathogenicity and carbohydrate metabolism. Front. Cell. Infect. Microbiol. 12:940602. doi: 10.3389/fcimb.2022.940602
Wu, R., Tao, Y., Cao, Y., Zhou, Y., and Lin, H. (2020). Streptococcus mutans membrane vesicles harboring glucosyltransferases augment Candida albicans biofilm development. Front. Microbiol. 11:581184. doi: 10.3389/fmicb.2020.581184
Yaron, S., Kolling, G. L., Simon, L., and Matthews, K. R. (2000). Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66, 4414–4420. doi: 10.1128/AEM.66.10.4414-4420.2000
Keywords: Streptococcus mutans , membrane vesicles, biogenesis, composition, functions
Citation: Qiu L, Chen Q, Zheng G, Dong H, Xu M, Zhou J, Zhang L, Sun Y, Wang M, Pan Y, Yu J, Pan Y and Zhang K (2025) A brief review of membrane vesicles from Streptococcus mutans. Front. Microbiol. 16:1656926. doi: 10.3389/fmicb.2025.1656926
Edited by:
Shi Huang, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Shanshan Liu, Bengbu Medical College, ChinaL. Jeannine Brady, University of Florida, United States
Copyright © 2025 Qiu, Chen, Zheng, Dong, Xu, Zhou, Zhang, Sun, Wang, Pan, Yu, Pan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keke Zhang, emhhbmdra0B3bXUuZWR1LmNu; Yihuai Pan, eWlodWFpcGFuQHdtdS5lZHUuY24=; Jie Yu, NzM1Mzg1NDQzQHFxLmNvbQ==; Yan Sun, c3VueWFuMjI0NkB3bXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Lili Qiu1†
Lili Qiu1† Yan Sun
Yan Sun Yangyang Pan
Yangyang Pan Yihuai Pan
Yihuai Pan Keke Zhang
Keke Zhang