- 1College of Traditional Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 2Department of Cardiology, Second Affiliated Hospital of Tianjin University of Traditional Chinese Medicine, Tianjin, China
Cardiometabolic diseases (CMD) are a cluster of complex syndromes characterized by cardiovascular damage resulting from metabolic dysregulation; however, their underlying mechanisms remain unclear. Recently, CMD research has paid considerable attention to the gut microbiota, though the emphasis has been on bacterial communities, and the gut mycobiome’s role is still not well understood. Hence, this review consolidates information on the correlation between the gut mycobiome and CMD and examines how the gut mycobiome may play a role in CMD progression. Accumulating evidence indicates that gut mycobiome dysbiosis, particularly the aberrant expansion of specific fungal genera such as Candida and Saccharomyces, is closely associated with the development and progression of cardiometabolic diseases. This association is primarily mediated through multiple mechanisms. For instance, fungal metabolites (enzymatic derivatives, alcohol) enhance intestinal lipid absorption, accelerate hepatic steatosis, and trigger systemic insulin resistance. Meanwhile, dysregulated cross-kingdom interactions disrupt intestinal barrier function, leading to endotoxemia and vascular inflammation, thereby promoting atherosclerosis. Additionally, fungal pathogen-associated molecular patterns activate the immune-metabolic axis, resulting in adipose tissue inflammation and glucose dysregulation. These pathways interact synergistically, collectively exacerbating metabolic dysfunction and cardiovascular damage. We also outline strategies targeting the gut mycobiome as a potential therapeutic approach for cardiometabolic diseases. By integrating current state-of-the-art insights, this review provides a critical reference for the development of novel mycobiome-based interventions in cardiometabolic disease management.
1 Introduction
Vertical transmission from the mother during the perinatal period initiates colonization of the human body by microorganisms (Wang et al., 2020). These microorganisms continue to adapt and evolve as the host matures, leading to the formation of a more stable adult microbiota (Kumbhare et al., 2019). Every person has a diverse and interlinked collection of microbial communities (Costello et al., 2009). Recent studies have confirmed that the partnership between microbiota and humans is vital for developing and healing diseases (Aggarwal et al., 2023). These microscopic entities are as numerous as the human cells they inhabit (Sender et al., 2016), primarily residing in the distal section of the gastrointestinal tract.
The gut microbiota comprises various microorganisms, including bacteria, archaea, fungi, and viruses, with bacteria being the predominant group (Sender et al., 2016). These gut microorganisms contribute to the host’s energy metabolism through their metabolites and byproducts and are essential for developing the host’s immune system (Carmody and Bisanz, 2023; Arnaiz-Villena et al., 2024), which supports physiological functions. Beyond these roles, the gut microbiota also exerts endocrine regulatory functions, modulating hormones such as leptin, ghrelin, and cortisol, thereby influencing host energy balance. It plays a crucial role in maintaining intestinal barrier integrity and produces diverse metabolites—including short-chain fatty acids (SCFAs), branched-chain fatty acids, and phenolic compounds—that can enter systemic circulation and affect cardiovascular physiology via anti-inflammatory effects, barrier enhancement, and signaling regulation (Datta et al., 2024; Zhang et al., 2025). Researchers have observed that disturbances in gut microbial communities are associated with various diseases, including neurodegenerative, cardiovascular, metabolic, and gastrointestinal disorders (Chen et al., 2021; Wei et al., 2025). Traditionally, investigations into how gut microbial populations relate to human health or illness have primarily centered on bacteria. However, the emergence of metagenomic high-throughput sequencing technologies has led to a greater appreciation of the gut mycobiome’s role in affecting the human host.
Although the gut mycobiome comprises only about 0.1% of the overall intestinal microbiota in healthy adults, it is a critically important component (Nash et al., 2017). The most dominant fungal taxa include Candida, Saccharomyces, Penicillium, and Malassezia (Bhunjun et al., 2024). As key intestinal components, they dynamically interact with bacterial communities and the host gut mucosal immune system, playing essential roles in regulating microbial ecosystem homeostasis (Kombrink et al., 2019; García et al., 2017). However, factors such as diet, antimicrobial use, geographical location, and age can contribute to gut fungal dysbiosis (Wu et al., 2021). The overproliferation of these opportunistic fungi directly disrupts intestinal mucosal homeostasis, leading to persistent and excessive immune activation (Huang et al., 2024), which promotes the progression of intestinal diseases. For instance, Beauveria bassiana-mediated upregulation of macrophage glycolytic pathways and IL-7 expression induces IL-22 secretion by type 3 innate lymphoid cells (ILC3s) through the AhR and STAT3 pathways, thereby exacerbating colorectal cancer (CRC) progression (Zhu et al., 2021). Moreover, the excessive proliferation of these opportunistic fungi not only affects the gut but can also trigger dysregulation in distal organs. For example, chronic alcohol consumption increases gut fungal abundance and promotes the translocation of fungal β-glucan into the portal circulation, which subsequently activates Kupffer cells via the C-type lectin-like receptor (CLEC7A) and upregulates IL-1β expression, ultimately leading to hepatocyte injury and hepatic inflammatory response (Thomas, 2017). The gut mycobiome’s association with multiple diseases has been the central theme of recent studies, particularly inflammatory bowel disease (Hsu et al., 2023), hepatic disorders (Zeng and Schnabl, 2024), autoimmune diseases (Jensen et al., 2024), neuropsychiatric conditions (Hadrich et al., 2025), and malignancies (Guglietta et al., 2025). The investigation into how the gut mycobiome might be related to cardiometabolic disease (CMD) is gaining more attention; however, this field currently remains deficient in systematic reviews and comprehensive syntheses. CMD is a clinical syndrome in which metabolic dysregulation triggers cardiovascular damage, characterized by a causal relationship between metabolic abnormalities and structural or functional cardiovascular impairments (Barteková et al., 2021; Sattar et al., 2020). This syndrome encompasses complex interactions between cardiovascular disorders (CVD), including hypertension, atherosclerosis (AS), and heart failure (HF), and metabolic disturbances, including insulin resistance, diabetes, and obesity, affecting 33–35% of adults and demonstrating significant associations with adverse cardiovascular events and all-cause mortality (Taha et al., 2022; Mechanick et al., 2020). According to the Global Burden of Disease (GBD) study and WHO reports, cardiometabolic diseases and related cardiovascular conditions account for more than one-third of global deaths each year, corresponding to approximately 17.9 million deaths annually, and remain the leading cause of morbidity and mortality worldwide (Dong et al., 2025; Saglietto et al., 2021). Epidemiological data indicate a continued global rise in cardiovascular disease incidence, with most patients exhibiting cardiometabolic risk factors, including obesity and dyslipidemia (Chen et al., 2025; Wang et al., 2024). China faces severe challenges, including persistently elevated cardiovascular mortality and rapidly increasing prevalence of CMD-related comorbidities (concurrent ≥2 metabolic-cardiovascular conditions), substantially worsening patient prognosis (Roth et al., 2020; In China TWCOTROCHAD and Hu, 2023). These risk factors are multifactorial, extending beyond genetic predisposition; lifestyle elements, particularly physical inactivity, smoking, and a suboptimal diet, constitute major reversible determinants (Chowdhury et al., 2023; Freisling et al., 2020). Amid accelerating population aging, the economic burden imposed by these diseases continues to escalate, presenting substantial challenges to public health systems globally (Han et al., 2021; Kivimäki et al., 2019). Despite the implementation of comprehensive prevention strategies, the global burden of CVD persists, with particularly pronounced impacts in Asia (Arena et al., 2015; Chew et al., 2023). Global life expectancy growth is projected to slow between 2016 and 2040, partially attributable to stagnating therapeutic advances in CVD management (Goh et al., 2024). Recent research has shown a strong connection between gut microbiota and the development of CMD (Yuan et al., 2024). Although most research has focused on bacteria, emerging evidence suggests that fungi play an important role. This review systematically examines current knowledge on gut mycobiome-CMD interactions, providing novel insights into future preventive and therapeutic strategies.
2 Methods: literature search and synthesis
This study employed a systematic literature review methodology, with comprehensive searches conducted in established databases, including PubMed, Web of Science, and the China National Knowledge Infrastructure. The core search strategy was exemplified by PubMed queries: (“gut mycobiome” OR “intestinal fungi” OR “fungal microbiome”) AND (“cardiometabolic diseases” OR “CMD” OR “atherosclerosis” OR “hypertension” OR “heart failure” OR “obesity” OR “diabetes” OR “fatty liver disease” OR “Metabolic syndrome”). During screening, priority was given to experimental or clinical studies examining gut mycobiome-CMD relationships or exploring the putative role of the mycobiome in CMD pathogenesis, which formed the basis of the inclusion criteria. Studies investigating acute infections caused by exogenous fungi leading to heart failure, as well as literature on free-living fungal organisms unrelated to the human microbiota (such as mushrooms), were excluded from this research. Subsequently, the included studies underwent rigorous evaluation to decipher how the gut mycobiome modulates CMD initiation and progression through intestinal microecological regulation.
3 Association between CMD and gut mycobiome
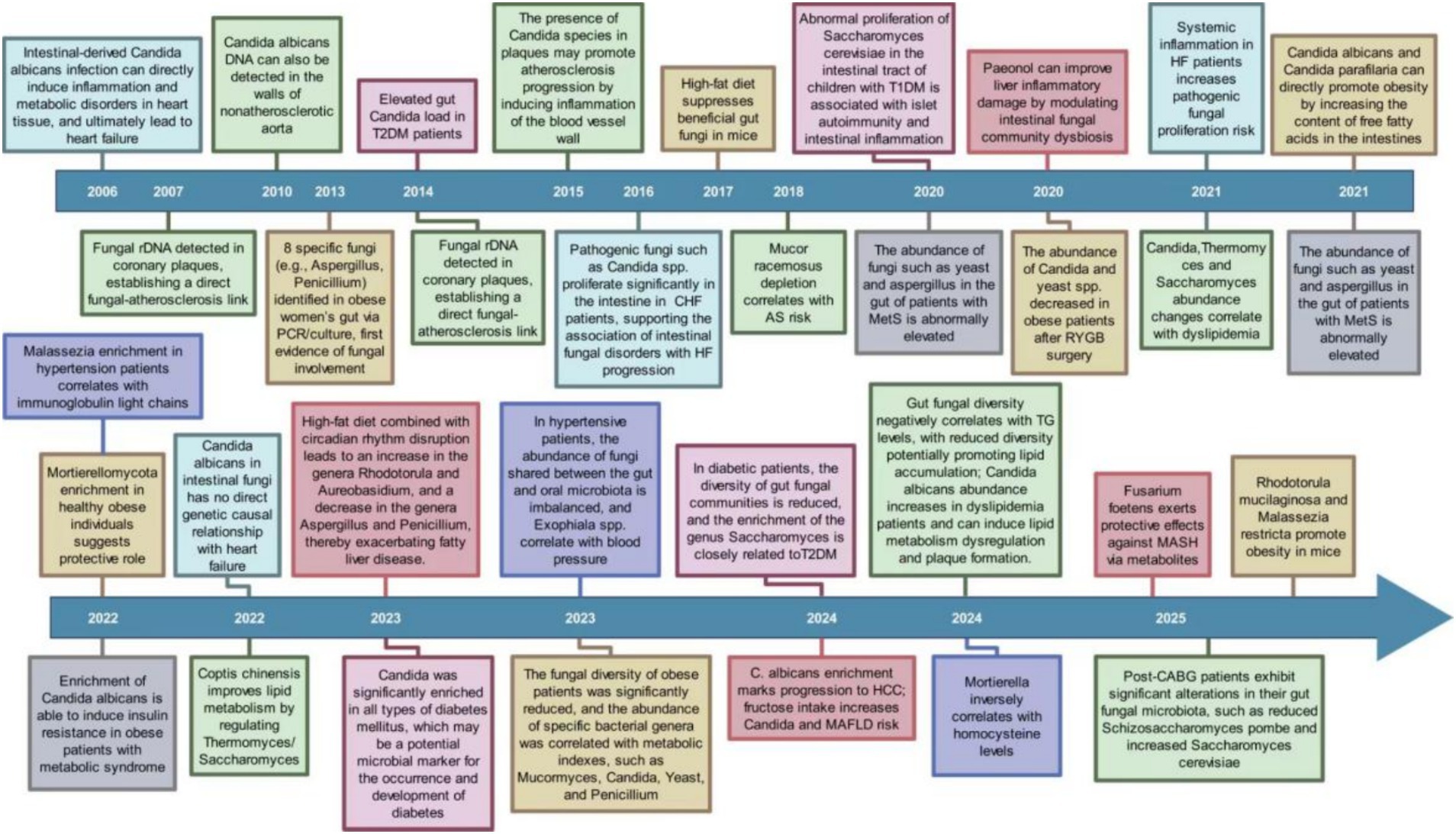
Research into how the gut mycobiome is connected to CMD was initiated in 2006. In the last twenty years, studies have progressively revealed links between the gut mycobiome and diseases such as heart failure, hypertension, atherosclerosis, diabetes, and obesity (Figure 1). These researches has evolved from detecting fungi and observing changes in their abundance to investigating their impact on disease onset and progression, while also encompassing factors like diet and intervention strategies. As the years advance, both the scope and depth of these studies continue to expand. The gut mycobiome is underscored as a crucial factor in the initiation and progression of CMD by these findings.
3.1 Association between obesity and gut mycobiome
Amid socioeconomic development, lifestyle transformations are driving an increased prevalence of overweight or obesity and associated health risks globally (Zhou et al., 2024). This constitutes a pressing health challenge, irrespective of the nation’s development status (Wong et al., 2020). Obesity pathogenesis involves multifaceted mechanisms, with research indicating that gut microbiota dysbiosis-induced metabolic dysregulation and functional decline may be key contributors (Geng et al., 2022). The scientific importance of the gut mycobiome (Zhan et al., 2024) is attracting increasing attention.
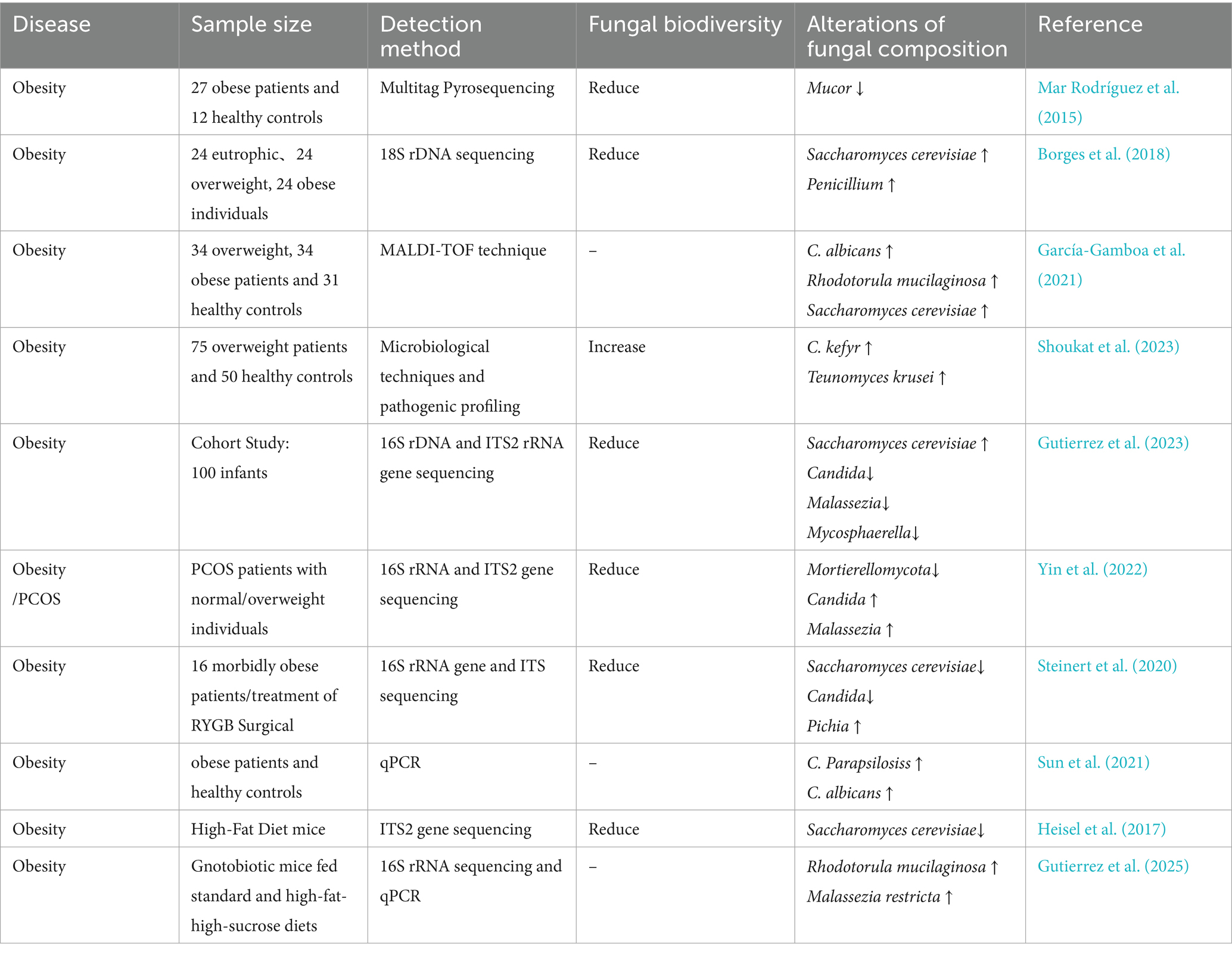
An early study (Gouba et al., 2013) identified eight fungal genera, including Aspergillus, Penicillium, and Malassezia, in the gut of obese women using polymerase chain reaction amplification and fungal culture, suggesting a potential role for fungi in host metabolism. Subsequent studies have established the structural features of the gut mycobiome in obese individuals, revealing that fungal diversity is significantly reduced (Szóstak et al., 2023) and that alterations in specific genera are correlated with metabolic parameters. Specifically, Mucor abundance was decreased in obese individuals (Mar Rodríguez et al., 2015), while levels of Candida, Saccharomyces cerevisiae, and Penicillium correlated positively with body weight, body mass index (BMI), and waist circumference (Salamati et al., 2015; Borges et al., 2018; García-Gamboa et al., 2021; Shoukat et al., 2023). In an infant cohort study, Saccharomyces demonstrated potential relevance, with its abundance positively correlating with BMI and potentially predicting childhood obesity risk (Gutierrez et al., 2023). Another study found a significantly elevated phylum-level abundance of Mortierellomycota in metabolically healthy obese individuals, suggesting a potentially protective role (Yin et al., 2022). Roux-en-Y gastric bypass, known for its effectiveness in treating obesity, can help in reestablishing mycobiome balance. Post-surgery reductions in the abundance of Candida and Saccharomyces have been observed in obese individuals (Steinert et al., 2020). Candida species, such as C. albicans and C. parapsilosis, contribute to obesity induced by a high-fat diet by elevating intestinal free fatty acid levels (García-Gamboa et al., 2021; Sun et al., 2021).
Animal models play a vital role in examining how the gut mycobiome is causally related to obesity. A high-fat diet impacts the gut bacterial community in mice and significantly suppresses the abundance of beneficial fungi (Heisel et al., 2017). Furthermore, antifungal treatment that disrupts the mycobiome exacerbates metabolic disorders in these models (Abdallah et al., 2020). Moreover, early colonization by specific fungi exerts a long-term impact on metabolic outcomes. Studies demonstrate that Rhodotorula mucilaginosa and Malassezia restricta promote obesity and the progression of metabolic disorders in mice (Gutierrez et al., 2025). Conversely, several interventions, including a low-carbohydrate diet (Yu et al., 2022), Litchi chinensis seed (Xiang et al., 2022), and avenanthramide B (Liu et al., 2025), can reverse obesity progression. These interventions promote weight loss by rebalancing the ratio of beneficial to pathogenic fungi.
In summary, elevated abundance of Candida spp., Saccharomyces cerevisiae, Penicillium spp., Rhodotorula mucilaginosa, and Malassezia restricta may be involved in the development and onset of obesity. Conversely, Mortierellomycota and Mucor spp. may confer potential protective roles against obesity (Table 1).
3.2 Association between diabetes and gut mycobiome
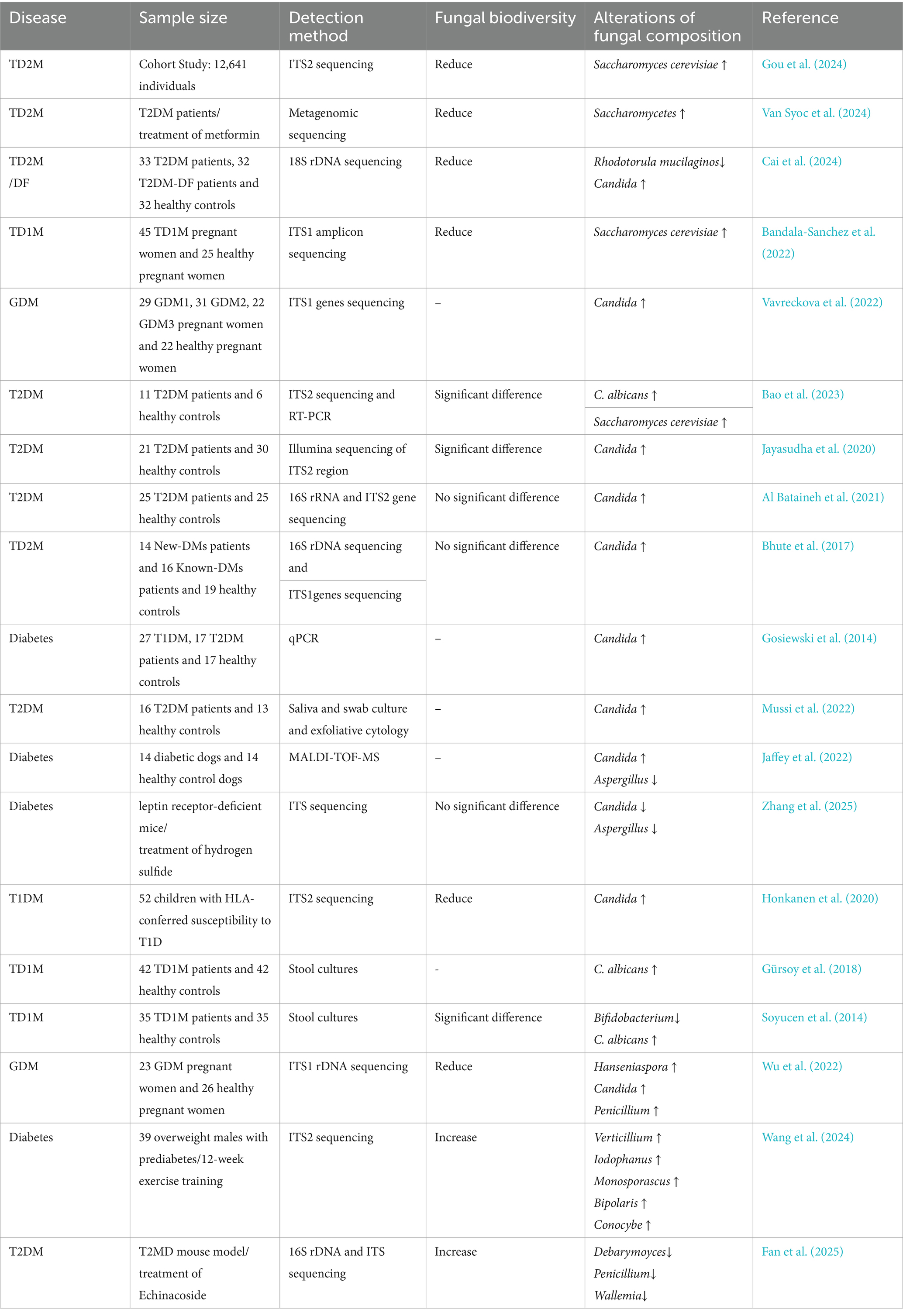
Diabetes mellitus is identified as a metabolic disorder with either an absolute or relative shortage of insulin and/or resistance to insulin in the organs it affects (American Diabetes Association, 2014). It can lead to various vascular and peripheral neuropathic complications, severely impairing the quality of life of affected individuals worldwide (Ahmad et al., 2022). Established risk factors, including genetics, family history, and metabolic dysregulation, have been demonstrated to play substantial roles in the development and progression of diabetes (Yu et al., 2024). Recent research has elucidated a close association between gut microbiota and diabetes (Gurung et al., 2020; Zhou et al., 2022). A landmark study published in Science demonstrated that early-life exposure to C. dubliniensis during infancy confers protection against diabetes development (Hill et al., 2025). This study utilized human infant fecal samples to colonize murine models, revealing that mice colonized with samples from infants aged 7–12 months exhibited significantly elevated insulin expression in pancreatic islets and increased circulating serum insulin levels. Subsequent mechanistic investigations demonstrated that cell wall components derived from fungi such as Aspergillus dublinensis induce infiltration of islet-resident macrophages (IRMs) and promote β-cell regeneration in diabetic murine models, thereby attenuating diabetes incidence (Hill et al., 2025). These results further scientific insights into the involvement of the gut mycobiome in diabetes development.
Previous studies have consistently documented reduced diversity in gut fungal communities among individuals with type 2 diabetes mellitus (T2DM) (Yan et al., 2024; Gou et al., 2024; Van Syoc et al., 2024). This dysbiotic pattern is markedly exacerbated in patients with diabetic complications such as diabetic foot ulcers (Cai et al., 2024). A parallel reduction in mycobiome diversity has been observed in the cohorts of type 1 diabetes (T1DM) and gestational diabetes mellitus (GDM) (Bandala-Sanchez et al., 2022; Vavreckova et al., 2022). This evidence indicates that gut mycobiome dysbiosis, characterized by diminished diversity, is a common pathophysiological feature across subtypes of diabetes mellitus. Investigations into signature fungal taxa have revealed significant enrichment of the genus Candida across all major forms of diabetes mellitus. Studies have demonstrated significantly elevated intestinal Candida burden in patients with T2DM compared to healthy controls (Bao et al., 2023; Saleem et al., 2022; Jayasudha et al., 2020; Al Bataineh et al., 2021; Bhute et al., 2017; Rodrigues et al., 2019; Gosiewski et al., 2014). Further reinforcing this observation, research indicates that microbial translocation occurs between oral and gut microbiomes in diabetic populations (Pacheco-Yanes et al., 2023), correspondingly increasing Candida abundance in the oral cavity (Mussi et al., 2022). Animal model studies have further demonstrated increased intestinal Candida colonization in diabetic dogs (Jaffey et al., 2022) and mice (Zhang et al., 2025). Moreover, Candida colonization rates are significantly higher in patients with T1DM (Honkanen et al., 2020; Gürsoy et al., 2018; Soyucen et al., 2014) and gestational diabetes (Vavreckova et al., 2022) than in healthy subjects. These findings suggest that aberrant Candida proliferation may represent a potential microbial marker for the pathogenesis of diabetes mellitus. Yeasts have also demonstrated significant potential in diabetic populations. The intestinal abundance of Rhodotorula mucilaginosa is closely associated with disease progression in T2DM studies (Cai et al., 2024), while alterations in Saccharomycetes class abundance correlate with the response to metformin treatment in patients with T2DM (Van Syoc et al., 2024). Cohort studies have revealed that Saccharomyces enrichment strengthens the association between conventional risk factors (obesity and insulin resistance) and T2DM, suggesting its potential as a predictive biomarker (Gou et al., 2024). In GDM research, changes in the abundance of taxa, such as Hanseniaspora, directly correlate with dysglycemia (Wu et al., 2022). Conversely, late-pregnancy intestinal enrichment of Saccharomyces cerevisiae in T1DM mothers frequently coincides with elevated inflammatory markers (Bandala-Sanchez et al., 2022). Similarly, aberrant Saccharomyces proliferation in children with T1DM is associated with pancreatic autoimmunity and intestinal inflammation. These findings further implicate Saccharomycetes in the pathogenesis of diabetes (Honkanen et al., 2020). Furthermore, elevated intestinal levels of Fusarium and Malassezia furfur in patients with T2DM underscore the disease-specific signatures within the mycobiome. Regarding interventions, 12-week exercise training ameliorated insulin resistance by enriching beneficial fungi such as Verticillium (Wang et al., 2024). Additionally, Echinacoside suppresses hepatic gluconeogenesis by remodeling bacterial-fungal interaction networks (Fan et al., 2025), while exogenous H2S improves metabolism by reducing the abundance of Candida and Aspergillus (Liu et al., 2022). These approaches provide novel therapeutic avenues for precise interventions.
Reduced gut mycobiome diversity and increased abundance of genera, including Candida, Saccharomyces, and Hanseniaspora, may contribute to diabetes progression, whereas exercise-induced enrichment of Verticillium demonstrates potentially protective roles in diabetes amelioration (Table 2).
3.3 Association between AS and gut mycobiome
AS is a prevalent condition that poses a substantial threat to human health, with coronary artery disease as a primary manifestation. A hallmark feature of AS lesions is the subendothelial accumulation of lipids within specific arterial regions, accompanied by smooth muscle cell proliferation and deposition of fibrous matrix components, resulting in atherosclerotic plaque formation (Shao et al., 2024). When AS affects coronary arteries, complete vascular occlusion can occur, potentially triggering myocardial infarction. Similarly, obstructions in cerebral vessels can cause cerebral infarction, commonly termed as a stroke. This pathological process constitutes the fundamental pathological basis of ischemic cardiocerebrovascular diseases (Lu and Daugherty, 2015). AS pathogenesis involves multifactorial complexity. Previous studies indicate that relevant theories primarily involve inflammation (Wolf and Ley, 2019), lipid accumulation (Bäck et al., 2019), oxidative stress (Batty et al., 2022), and endothelial injury; however, no single theory fully accounts for the entire pathogenesis of AS. Beyond the established risk factors, hypertension, diabetes, hypercholesterolemia, smoking, stressors, and environmental determinants significantly accelerate lesion progression (Libby, 2021). Epidemiological data have confirmed that AS is a principal contributor to global cardiovascular mortality and morbidity. In Western populations, AS-related deaths constitute approximately 50% of all cardiovascular fatalities (Ramji and Davies, 2015). Consequently, elucidating AS pathogenesis and developing targeted therapeutic strategies have substantial clinical and scientific implications.
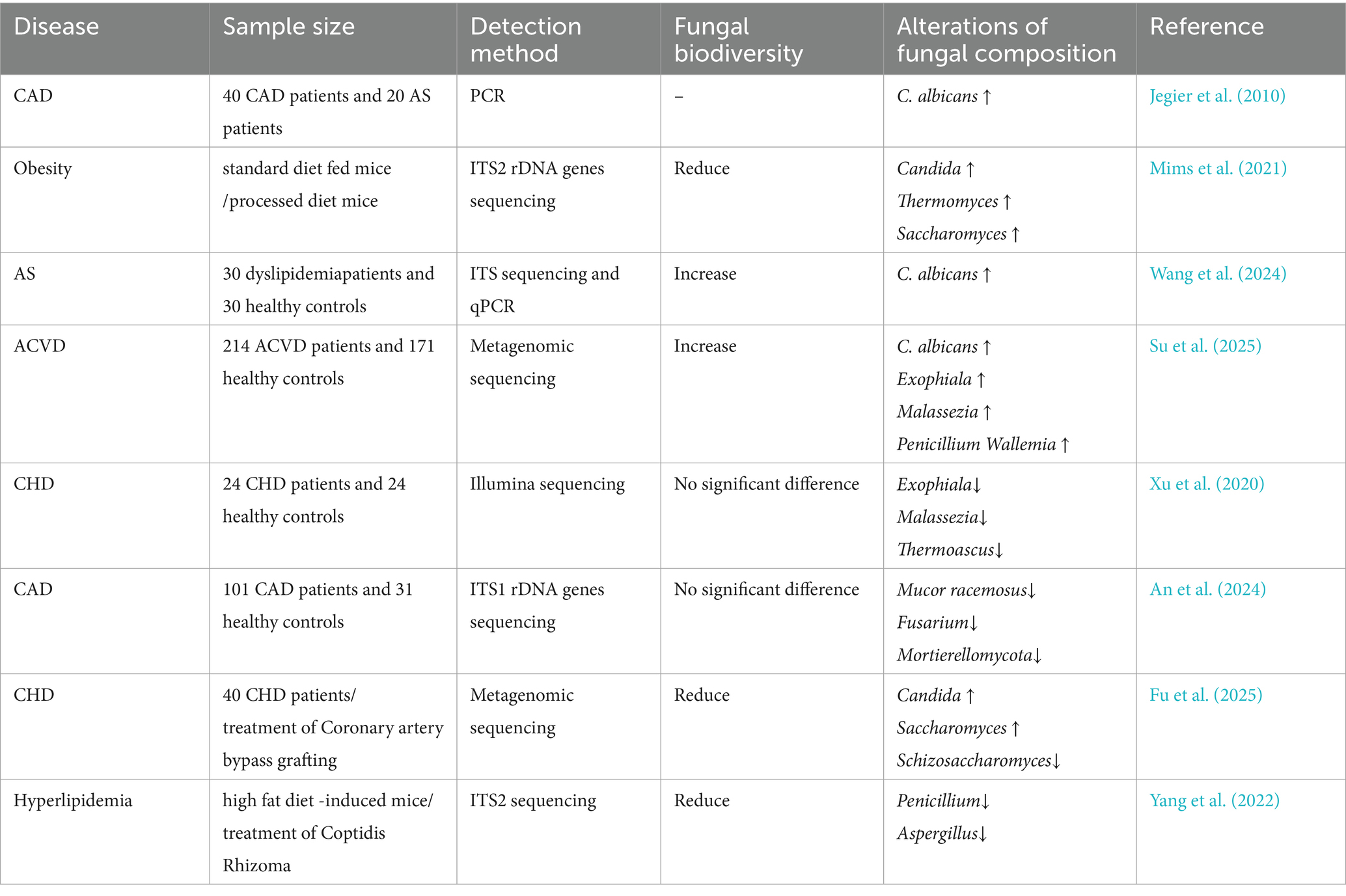
Early investigations have established a direct link between fungi and AS. In 2007, a study employing fungal rDNA detection within coronary plaques initially identified mycotic components in atherosclerotic lesions (Ott et al., 2007). Subsequent research further identified detectable C. albicans DNA even in non-atherosclerotic aortic walls, with a significant correlation with vascular inflammation marker expression (Jegier et al., 2010). Masoumi and Nurgeldiyeva et al. substantiated the presence of Candida spp. within plaques and proposed their potential role in promoting atherogenesis by inducing vascular inflammation (Masoumi et al., 2015; Nurgeldiyeva et al., 2014). These findings provide the initial evidence of fungal involvement in the pathological progression of AS.
Recent research has increasingly focused on the interplay between the gut mycobiome and host lipid metabolism dysregulation, as well as its impact on atherogenesis. Empirical evidence reveals an inverse correlation between gut fungal diversity indices and triglyceride (TG) levels, suggesting that diminished mycobiotic diversity may contribute to pathological lipid accumulation (Estrada et al., 2024). Alterations in the abundance of intestinal genera, including Candida, Thermomyces, and Saccharomyces, correlated significantly with lipid metabolism dysregulation (Mims et al., 2021). C. albicans has emerged as a primary pathogenic driver, exhibiting an elevated abundance in patients with dyslipidemia. Research has demonstrated that its metabolic byproduct, formyl-methionine, provokes aberrant lipid metabolism and promotes plaque development (Wang et al., 2024). Furthermore, patients with atherosclerotic CVD exhibited significantly increased abundances of the genera Exophiala and Malassezia (Su et al., 2025). However, contradictory findings have emerged in patients with coronary heart disease, where a decreased abundance of these fungal taxa has been observed (Xu et al., 2020). This suggests that a structural imbalance in the gut mycobiome may potentiate AS progression, concurrently underscoring the importance of further clinical validation. Abundances of Mucor racemosus (Chacón et al., 2018) and Fusarium spp. (An et al., 2024) reduced significantly, demonstrating negative associations with AS risk, thereby suggesting their therapeutic potential as protective fungal taxa. In clinical intervention studies, patients undergoing coronary artery bypass grafting altered significantly gut mycobiome, characterized by decreased Schizosaccharomyces pombe and increased Saccharomyces cerevisiae abundance. These shifts suggest that surgical intervention may modulate fungal communities and influence postoperative prognosis (Fu et al., 2025). Regarding pharmacological interventions, Coptis chinensis ameliorates lipid metabolism disorders by modulating Thermomyces spp. and Saccharomyces spp., revealing its dual-targeting potential toward gut bacteria and fungi (Yang et al., 2022). This provides preliminary evidence for future fungus-targeted therapeutics.
Intestinal Candida spp., Thermomyces spp., and Saccharomyces spp. may constitute key drivers of AS progression, whereas Mucor racemosus and Fusarium spp. may represent protective fungal taxa (Table 3).
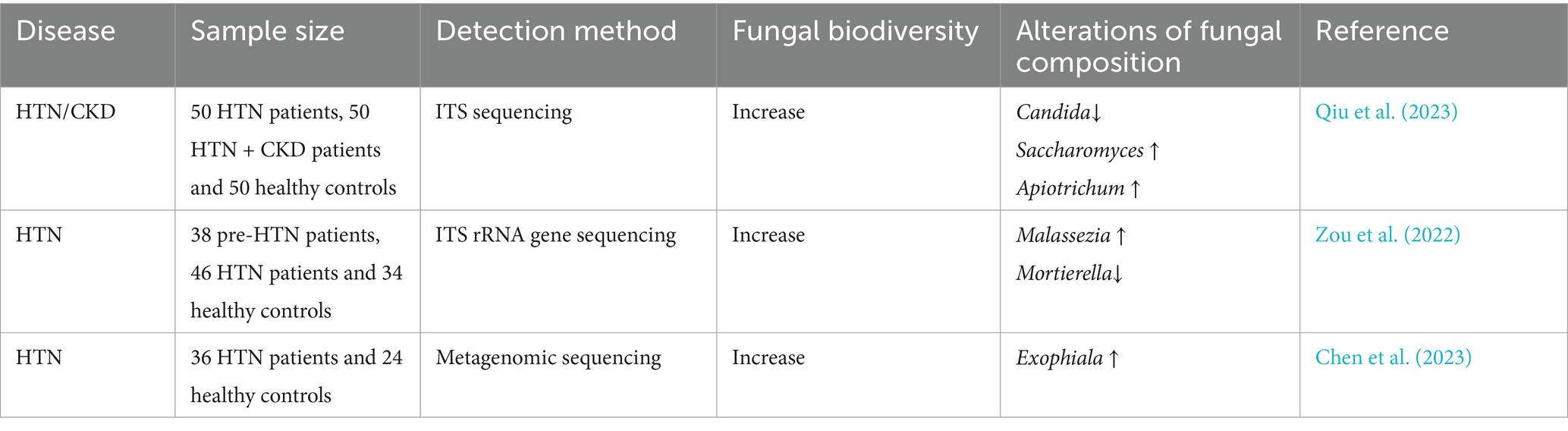
3.4 Association between hypertension and gut mycobiome
Hypertension is a cardiovascular syndrome characterized by sustained elevation of systemic arterial pressure. This condition poses significant risks for a spectrum of cardiocerebrovascular diseases, including stroke, coronary heart disease, and HF. According to the first Global Report on Hypertension released by the World Health Organization in 2023, data from 2019 revealed that 33% of the global population aged 30–79 years had hypertension. Approximately 10 million deaths annually are attributable to elevated systolic pressure (Kario et al., 2024). Studies have confirmed that overweight, obesity, dyslipidemia, and insulin resistance are established risk factors for hypertension (Tang et al., 2022; Guo et al., 2025), with metabolic disorders contributing to over half of hypertension cases while directly promoting vascular dysfunction and blood pressure elevation (Zhu et al., 2016).
Emerging research has revealed that the gut mycobiome mediates metabolic dysregulation and elevates blood pressure. Despite the absence of statistically significant differences in gut mycobiome diversity between hypertensive patients and healthy controls (Qiu et al., 2023; Zou et al., 2022), specific alterations in fungal abundance are significantly associated with blood pressure regulation. Malassezia demonstrated significant enrichment in hypertensive cohorts, and its abundance positively correlated with serum immunoglobulin light chain (κ/λ) concentrations. This implicates its pathogenic role in hypertension via immune activation (Zou et al., 2022). Conversely, Mortierella prevails predominantly in normotensive populations (Zou et al., 2022), presenting an inverse association with homocysteine, an established hypertension biomarker, suggesting a protective function through metabolic modulation (An et al., 2024). Several species of Exophiala (E. xenobiotica and E. mesophila) demonstrated direct positive correlations with systolic and diastolic blood pressure levels. Concurrently, patients with hypertension exhibit dysbiosis in shared fungal communities across gut-oral axes (Chen et al., 2023). Furthermore, the genera Nakaseomyces and Saccharomyces are specifically associated with diastolic pressure (Qiu et al., 2023).
The genera Malassezia, Exophiala, Nakaseomyces, and Saccharomyces demonstrate pathophysiological contributions to blood pressure elevation, whereas Mortierella potentially confers protective cardiometabolic benefits (Table 4).
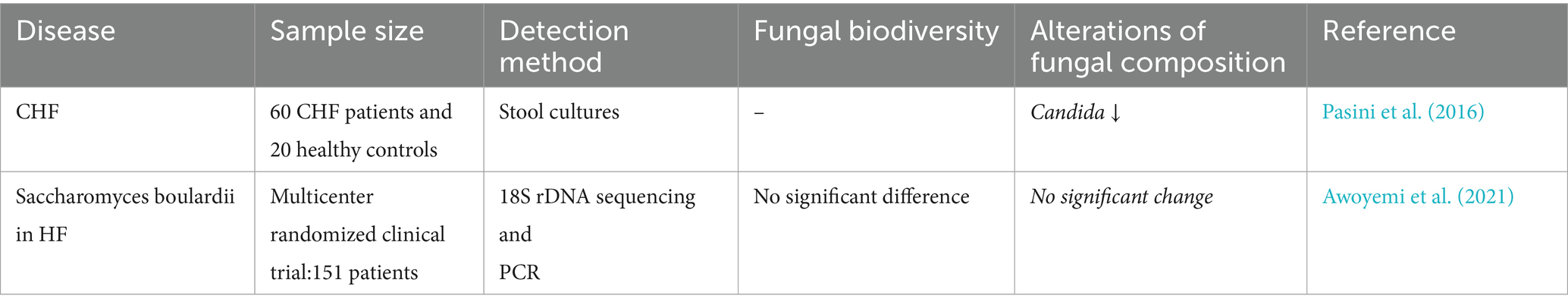
3.5 Association between HF and gut mycobiome
HF is a clinical syndrome primarily driven by structural and/or functional cardiac abnormalities. This pathophysiological cascade elevates the intracardiac pressures at rest or during exertion and/or compromises cardiac output, manifesting as diverse clinical symptoms (McDonagh et al., 2021). HF is an end-stage manifestation of CVD, including hypertension, and is intrinsically linked to cardiac decompensation. Despite prolonged investigations into the association between HF progression and gut microbiota alterations (Sandek et al., 2007), a definitive causal link remains unestablished (Tang et al., 2019).
A prior Mendelian randomization analysis revealed no genetic causal relationship between C. albicans gut colonization and HF pathogenesis (Luo et al., 2022), whereas discordant conclusions have emerged from independent cohort studies. Animal studies have demonstrated that in C5-deficient mice, gut-derived C. albicans infection directly triggers cardiac inflammatory damage and metabolic dysregulation, ultimately culminating in heart failure (Mullick et al., 2006). Clinical investigations have revealed a significant proliferation of pathogenic fungi, including Candida spp., in the gut mycobiome of patients with chronic heart failure (CHF), concomitant with elevated intestinal permeability and systemic inflammation. These alterations are positively correlated with disease severity, further supporting the association between gut mycobiome dysbiosis and HF progression (Pasini et al., 2016). Moreover, heightened inflammatory cytokine levels in CHF may compromise immune function and increase susceptibility to pathogenic fungal overgrowth (García-Torre et al., 2021). This provides evidence of a complex, mutually reinforcing relationship between HF and gut mycobiome perturbations involving immune competence, medication exposure, and inflammatory status. Therapeutically, targeted probiotic supplementation (Saccharomyces boulardii) failed to improve cardiac function or microbial diversity in the GutHeart trial (Awoyemi et al., 2021), indicating complexity in microbiota modulation strategies.
Gut mycobiota, particularly Candida spp., could be crucial in the beginning and advancement of HF (Table 5). However, current evidence remains limited, warranting further in-depth investigations.
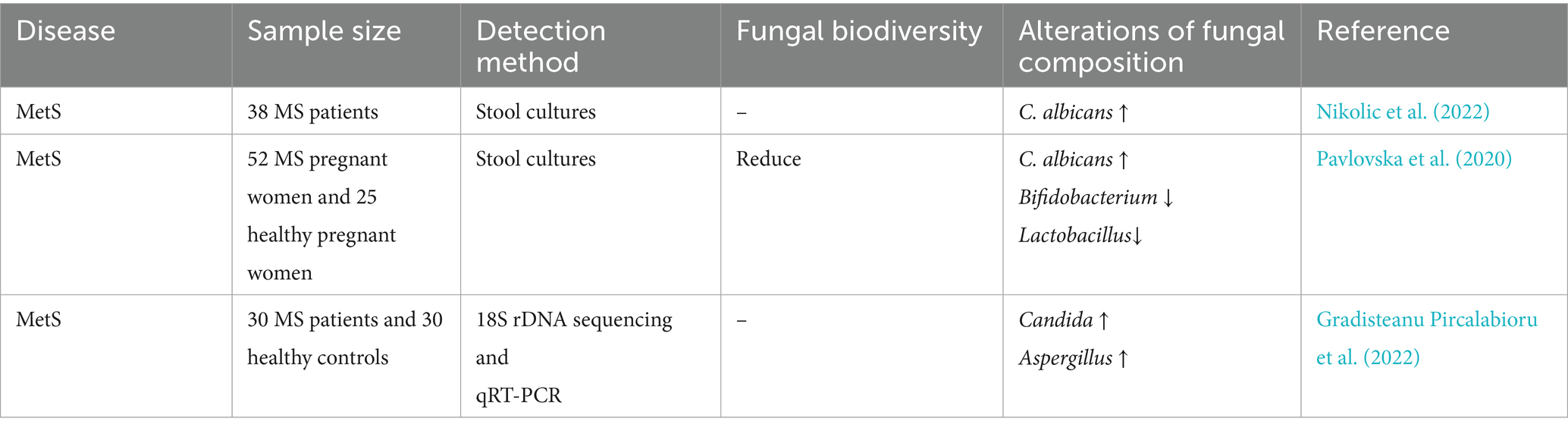
3.6 Association between metabolic syndrome (MetS) and gut mycobiome
MetS is a pathological state characterized by dysregulated metabolism of carbohydrates, lipids, and proteins. Its four core features are obesity, dyslipidemia, hepatic steatosis, and insulin resistance (Ndumele et al., 2023). Globally, MetS affects approximately 25% of the population and contributes to two-thirds of noncommunicable disease-related mortality, and its prevalence continues to rise worldwide (Saklayen, 2018). Evidence suggests an association between MetS and gut mycobiome dysbiosis.
Previous clinical investigations have demonstrated that the enrichment of C. albicans in the gut induces insulin resistance in obese patients with MetS, potentially serving as a key mediator linking metabolic dysregulation and immune imbalance (Nikolic et al., 2022). Further studies revealed that vitamin D3 deficiency may drive opportunistic fungal overgrowth, including Candida spp., thereby increasing the susceptibility of patients with psoriasis to MetS (Marchlewicz et al., 2024). This association is more pronounced in pregnant women, whose MetS correlates with gut microbial dysbiosis characterized by overgrowth of pathogenic fungi (C. albicans) and reduced abundance of Bifidobacterium and Lactobacillus (Pavlovska et al., 2020). Additionally, elevated gut fungal burdens of Saccharomyces and Aspergillus spp. have been documented in patients with MetS (Gradisteanu Pircalabioru et al., 2022). In parallel, dysregulation of adipokines constitutes another central feature of MetS. Pro-inflammatory adipokines such as TNF-α, IL-6, leptin, and resistin are upregulated, whereas anti-inflammatory adipokines including adiponectin and omentin are downregulated. This imbalance drives chronic low-grade inflammation, insulin resistance, and vascular dysfunction. Emerging evidence suggests that gut fungal dysbiosis, particularly Candida overgrowth, may exacerbate this adipokine imbalance by promoting systemic inflammation and thereby worsening metabolic and cardiovascular complications (Datta et al., 2025). Regarding probiotic fungal interventions, the administration of Saccharomyces boulardii improves insulin sensitivity and ameliorates lipid metabolic disorders in patients with MetS by suppressing pathogenic fungal overgrowth (Egea et al., 2023).
The evidence suggests that C. albicans might play a vital role in the pathogenesis of MetS; however, the implications of Saccharomyces and Aspergillus spp. require further mechanistic substantiation (Table 6).
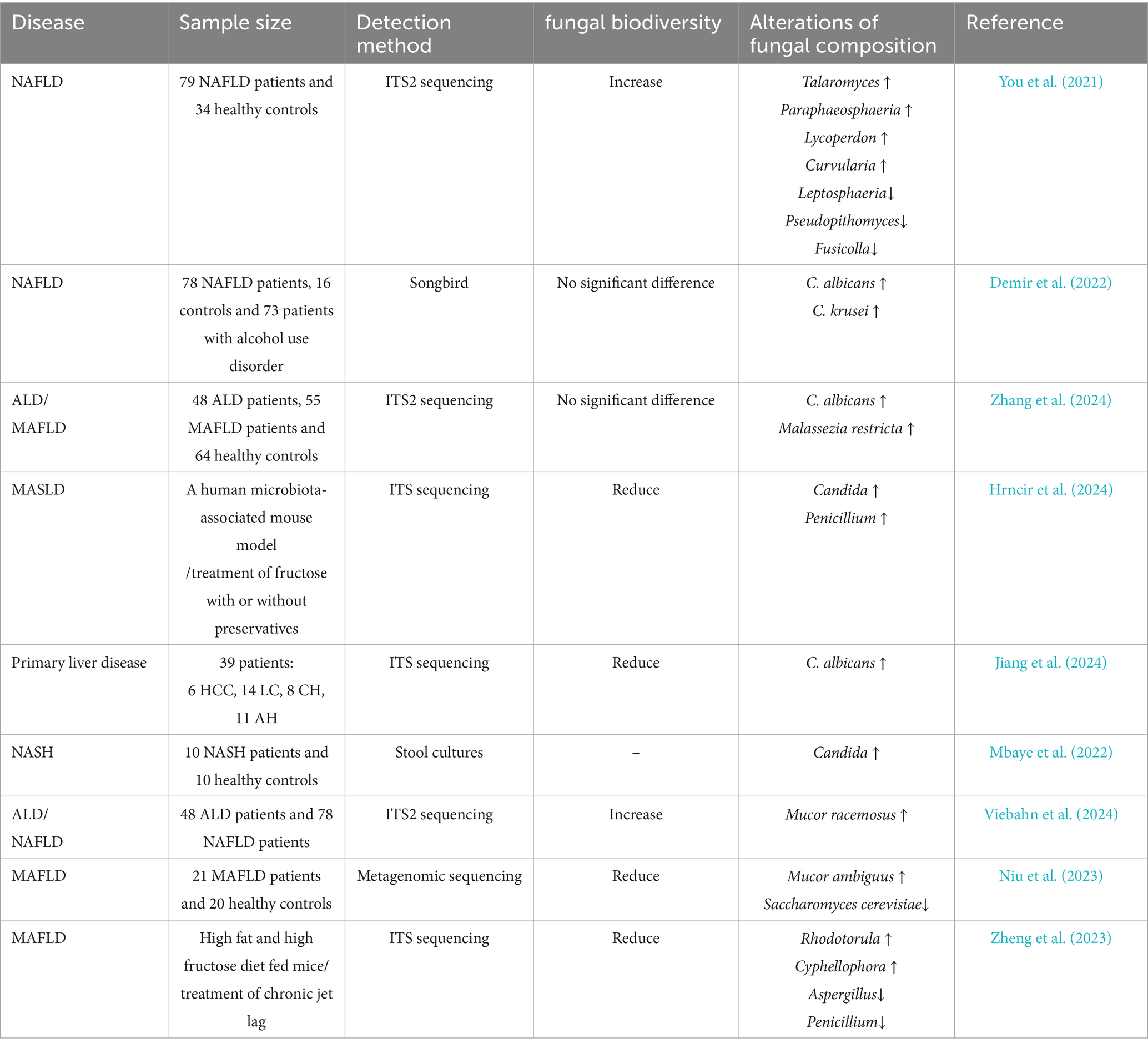
3.7 Association between metabolic dysfunction-associated fatty liver disease (MAFLD) and gut mycobiome
MAFLD is a chronic metabolic stress-related liver injury in genetically predisposed individuals, primarily driven by overnutrition and insulin resistance (Targher et al., 2024). Currently, the most prevalent chronic liver disorder has historically been termed non-alcoholic fatty liver disease (NAFLD) (Eslam et al., 2020). It has been redefined to emphasize its pathogenic links to T2DM dysregulation, involving multifaceted factors such as obesity and T2DM (Jurek et al., 2025). The growing exploration of gut microbial ecosystems further implicates mycobiome dysbiosis in MAFLD pathogenesis.
Accumulating evidence suggests that intestinal mycobiome dysbiosis occurs in patients with MAFLD. Fungal compositional analyses demonstrated an elevated abundance of multiple fungal taxa in NAFLD-affected guts, alterations that may drive hepatic injury, dysregulated lipid metabolism, and disease progression (You et al., 2021). Dysbiosis within the genus Candida is particularly prominent, with C. albicans and C. krusei significantly increased in MAFLD cohorts (Demir et al., 2022; Zhang et al., 2024; Hrncir et al., 2024). The increased abundance of C. albicans further serves as a biomarker for progression from fatty liver or cirrhosis to early-stage hepatocellular carcinoma (Jiang et al., 2024). Intestinal Pichia kudriavzevii and Candida spp. have been demonstrated to endogenously generate ethanol and TG via fructose-dependent pathways, directly contributing to non-alcoholic steatohepatitis pathogenesis (Mbaye et al., 2022). Concurrently, the abundance of genus Mucor is correlated with hepatic inflammation and fibrosis, with the subspecies Mucor ambiguus demonstrating significant associations with obesity, dyslipidemia, and liver injury biomarkers (Demir et al., 2022; Viebahn et al., 2024; Niu et al., 2023). Environmental and dietary risk factors further exacerbate mycobiome dysbiosis. For instance, a high-fat diet combined with chronodisruption (chronic jet lag) disrupts microbial circadian oscillations, elevates Rhodotorula and Cyphellophora spp., and reduces Aspergillus and Penicillium spp., thereby aggravating fatty liver pathology (Zheng et al., 2023). Moreover, high-fructose intake amplifies the abundance of Candida spp. by remodeling bacterial-fungal cross-kingdom networks, mediating host metabolic dysfunction, and elevating the MAFLD risk (Hrncir et al., 2024). Therapeutically, the intestinal fungus Fusarium foetens exerts hepatoprotective effects against metabolic dysfunction-associated steatohepatitis by secreting bioactive secondary metabolites (Zhou et al., 2025). Concurrently, the phytochemical paeonol demonstrates therapeutic potential by ameliorating histopathological injury by remolding mycobiome dysbiosis (Wu et al., 2020; Ge et al., 2021).
In summary, Candida spp., Mucor spp., Pichia kudriavzevii, and other Candida yeasts correlated positively with MAFLD progression, whereas Fusarium foetens exhibited protective potential (Table 7).
4 Gut mycobiome-mediated mechanisms in CMD progression
The gut mycobiome participates in the onset and progression of cardiometabolic diseases through multiple mechanisms. Existing evidence suggests that it may contribute to disease progression via mechanisms such as the release of metabolites, cross-kingdom interactions, and the immune-metabolic axis (Figure 2).
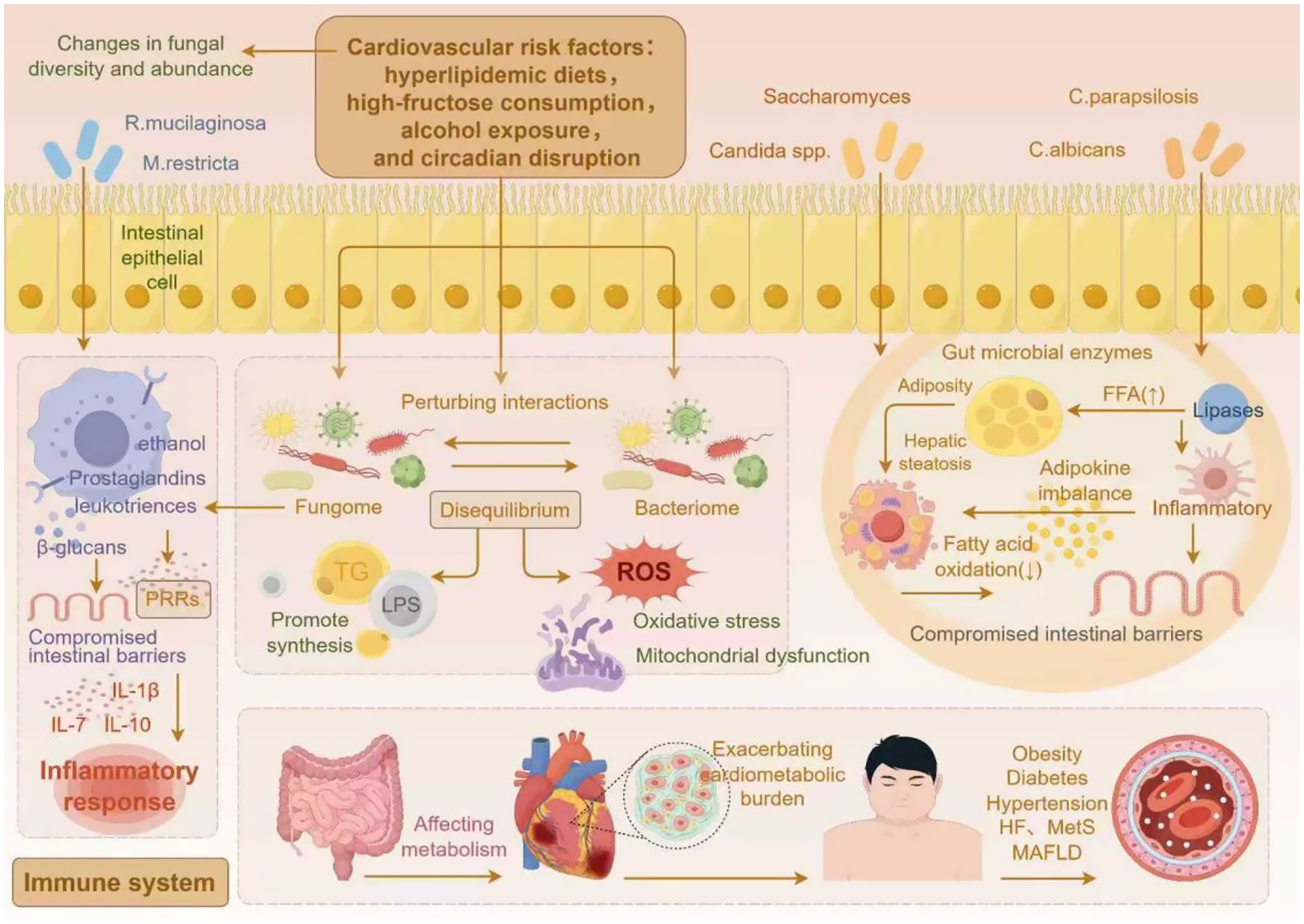
Figure 2. The gut mycobiome modulates cardiometabolic disease progression via associated mechanisms (by Figdraw).
4.1 Gut mycobiome-derived metabolites promote lipid absorption and accumulation
The gut mycobiome directly participates in host metabolism and modulates physiological processes through its derived metabolites. Gut microbial enzymes represent an emerging research focus, as they catalyze metabolic reactions that enhance the breakdown and absorption of nutrients (Jiang et al., 2024). Recent evidence has indicated that the gut microbiota and their enzymatic derivatives significantly drive the progression of T2DM (Wang et al., 2023) and MAFLD (Luo et al., 2025). Candida parapsilosis secretes lipases that hydrolyze dietary fats, thereby elevating circulating FFA concentrations (Sun et al., 2021). FFA accumulation promotes obesity through multiple pathways, primarily by inducing inflammation that disrupts intestinal barrier integrity, secondarily by enhancing energy absorption and surplus in the gut, and ultimately by activating fat-storage pathways while inhibiting fatty acid oxidation, thereby promoting adiposity and hepatic steatosis, collectively exacerbating cardiometabolic burden (Sun et al., 2021). Furthermore, excessive fructose intake has been definitively linked to an elevated risk of CMD (Zhang et al., 2020; Soleimani et al., 2023; Drożdż et al., 2021). High-fructose diets alter bacterial-fungal cross-kingdom interactions, significantly enriching Candida spp. and Saccharomyces (Hrncir et al., 2024). These proliferating gut fungi metabolize fructose to ethanol while driving de novo TG synthesis. Both endogenously generated ethanol and TG accelerate MASLD development (Mbaye et al., 2022).
4.2 Gut mycobiome-bacterial community cross-kingdom interaction dysbiosis and its mechanistic implications
Research has established that the intestinal fungome and bacteriome maintain homeodynamic equilibrium through reciprocal modulation, collectively preserving gut ecosystem stability (Underhill and Iliev, 2014; Elnagar, 2024). This interaction mainly manifests as competition for nutrients, co-metabolism of dietary components, or modulation of the host immune system (Santus et al., 2021). Moreover, gut fungi can inhibit bacterial colonization. For example, the presence of Candida albicans reduces the efficacy of fecal microbiota transplantation (FMT) in treating Clostridioides difficile infection, and further studies have shown that Candida albicans suppresses bacterial engraftment (Zuo et al., 2018). Conversely, gut bacteria can also inhibit fungal proliferation; for instance, colonization by Lactobacillus has been shown to suppress the expansion of Candida (Strus et al., 2005). Moreover, gut fungi and bacteria not only antagonize each other, but also cooperate to promote pathogenesis. For example, Escherichia coli enhances the invasion of Candida albicans into intestinal epithelium in an in vitro model (Yang et al., 2016); another study showed that monocolonization with gut fungi alone does not induce physiological changes in germ-free mice, whereas co-colonization with fungi and bacteria significantly exacerbates colonic inflammation. These findings further highlight that the interaction between fungi and bacteria is fundamental to the onset and progression of disease (Zhang et al., 2022; Krüger et al., 2019). It is well known that diet and lifestyle, as major modifiable factors of CMD, are among the most effective regulators of microbial community interactions (Guiducci et al., 2023). Studies show that a high-fat diet is associated with increased abundance of Candida and decreased abundance of Prevotella (Heisel et al., 2017; Peroumal et al., 2022). High-carbohydrate diets show a positive correlation with elevated abundances of Candida and Methanobrevibacter (Hoffmann et al., 2013). Furthermore, high-fat, high-fructose diets (HFFD) increase the abundance of pathogenic fungi (Aspergillus, Penicillium, and Candida), while reducing the abundance of bacterial taxa including Lachnospiraceae_NK4A136_group and Harryflintia (Zhao et al., 2024; Zhao et al., 2025). Moreover, many of these bacteria with reduced abundance are probiotics capable of producing short-chain fatty acids (SCFAs), and the association between decreased SCFA levels and CMD has been extensively studied and well established (Neves et al., 2015; Wang et al., 2025). These findings confirm that diet-induced alterations in fungal and bacterial communities are closely associated with host metabolic dysregulation (glucose and lipid metabolism) and vascular inflammatory phenotypes (Zhao et al., 2024; Zhao et al., 2025). Furthermore, circadian disruption, an emerging cardiometabolic risk factor, worsens hepatic inflammation and lipid metabolism dysregulation by perturbing gut mycobiota-bacteriome interactions during chronic jetlag (Zheng et al., 2023). These findings indicate that dysregulation of the fungal-bacterial interaction network impairs microbial regulation of host energy harvest and immune homeostasis, thereby triggering inflammation due to intestinal barrier defects, as well as abnormalities in short-chain fatty acid and bile acid metabolism (Wang et al., 2020), ultimately driving the pathological progression of CMD. Further fungal transplantation experiments have reinforced this understanding. For instance, single gavage administration of Candida albicans increased the relative abundance of Staphylococcus and Mucispirillum, while decreasing the abundance of Akkermansia. Staphylococcus may produce LPS, which can enter the bloodstream through a compromised intestinal mucosal barrier, thereby triggering chronic low-grade inflammation—a key mediator in the development of insulin resistance, diabetes, obesity, and atherosclerosis (Bao et al., 2023). Furthermore, multiple studies have confirmed that supplementation with Akkermansia can ameliorate cardiometabolic diseases (Gofron et al., 2024; Niu et al., 2024), while a reduction in Akkermansia abundance further exacerbates disease pathogenesis. Although existing evidence supports the synergistic role of opportunistic fungal proliferation alongside harmful bacterial overgrowth and beneficial bacterial suppression in the pathogenesis of cardiometabolic diseases, it remains unclear whether gut fungi and bacteria also modulate CMD progression through antagonistic interactions—a question that represents a promising avenue for future research.
4.3 Gut mycobiome-immune-metabolic axis crosstalk mechanisms
Gut mycobiome drives metabolic dysregulation by modulating the immune system. First, gut fungal-derived metabolites (including candidalysin, prostaglandins, formyl-methionine [Wang et al., 2024), leukotrienes, and ethanol (Santelmann and Howard, 2005; Noverr et al., 2002; Tan et al., 2019)] can induce inflammatory responses. For example, candidalysin is a fungal peptide toxin secreted by Candida albicans that directly induces IL-1α, IL-1β, IL-8, IL-36, and the NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3) inflammasome (Zhou et al., 2024). These inflammatory mediators are known to play critical roles in the development of insulin resistance, T2DM, obesity, vascular endothelial injury, atherosclerosis, and NAFLD (Li et al., 2025). Prostaglandins (PGs) are a class of eicosanoids, comprising homologs from series 1, 2, and 3. Studies indicate that gut fungi such as Cryptococcus neoformans and Candida albicans can secrete prostaglandins via de novo synthesis or through the conversion of exogenous arachidonic acid (Noverr et al., 2001). Among these, PGE₂ has been most extensively studied and participates in regulating diverse physiological and pathological processes, associated with diseases such as diabetes, hypertension, obesity, non-alcoholic fatty liver disease, and cardiovascular diseases (Wang et al., 2021). On one hand, PGE₂ binds to the EP3 receptor to activate extracellular signal-regulated kinase 1/2 (ERK1/2), thereby inducing serine phosphorylation of insulin receptor substrate 1 (IRS1) and ultimately exacerbating insulin resistance (Henkel et al., 2009). On the other hand, PGE₂ induces β-cell dysfunction via the IL-1β-mediated NF-κB activation pathway and promotes hepatic lipid accumulation through NF-κB signaling (Chung et al., 2015). In summary, PGE₂ effectively mediates NF-κB pathway activation and dysregulation of glucose and lipid metabolism, triggering a cascade of pathological alterations in vascular endothelium (Xu et al., 2020), liver, and adipose tissue, thereby driving the progression of cardiometabolic diseases. Formyl-methionine is a metabolite that can be produced by Candida albicans (Wang et al., 2024). Studies show that the concentration of formyl-methionine in circulation is closely associated with the risk of various cardiovascular diseases (Cai et al., 2021). This substance can activate the HIF-2α signaling pathway in the gut, thereby promoting ceramide synthesis and ultimately accelerating the progression of atherosclerosis (Wang et al., 2024). Previous studies have demonstrated that ceramide, an emerging compound, is closely associated with CMD (Wilkerson et al., 2024). This bioactive lipid can induce insulin resistance, oxidative stress, and atherosclerotic plaque progression (Spaggiari et al., 2024). Secondly, gut fungi can also induce inflammatory responses through cell wall components. Upon recognition by host pattern recognition receptors (PRRs), their cell wall components (chitin, mannans, and β-glucans) trigger inflammatory cascades that exacerbate metabolic disorders (Cheng et al., 2011; Gringhuis et al., 2012). For instance, Rhodotorula mucilaginosa and Malassezia restricta, when recognized by host PRRs, drive pathological lipid accumulation by inducing intestinal low-grade inflammation and enhancing nutrient uptake, thereby promoting high-fat diet-induced obesity (Gutierrez et al., 2025). Fungal β-glucans translocate across damaged intestinal barriers, bind to the CLEC7A receptor on Kupffer cells, and trigger IL-1β-dependent hepatic inflammation and fibrosis (Yang et al., 2017), subsequently accelerating the progression of steatohepatitis. Similarly, in mouse models, Candida albicans-derived β-glucans released via the dectin-1 receptor activate inflammatory cascades to induce insulin resistance; conversely, eliminating C. albicans or blocking this pathway significantly alleviates insulin resistance and halts the progression of metabolic disorders (Bao et al., 2023). The aforementioned mechanistic studies reinforce the causal link between gut fungi and CMD. Gut fungal derivatives promote the pathogenesis and progression of metabolic cardiovascular disorders via immunoinflammatory mechanisms, as well as through pathways inducing insulin resistance and lipid metabolic disorders. Furthermore, the roles of gut bacterial derivatives—such as bile acids, LPS, and trimethylamine N-oxide (TMAO)—in CMD have been well-established (Kondapalli et al., 2025; Du et al., 2025). However, whether metabolites derived from bacteria and fungi exert synergistic or antagonistic effects in CMD remains unclear, and their interactive mechanisms warrant further elucidation in future studies.
5 Therapeutic strategies targeting the gut mycobiome
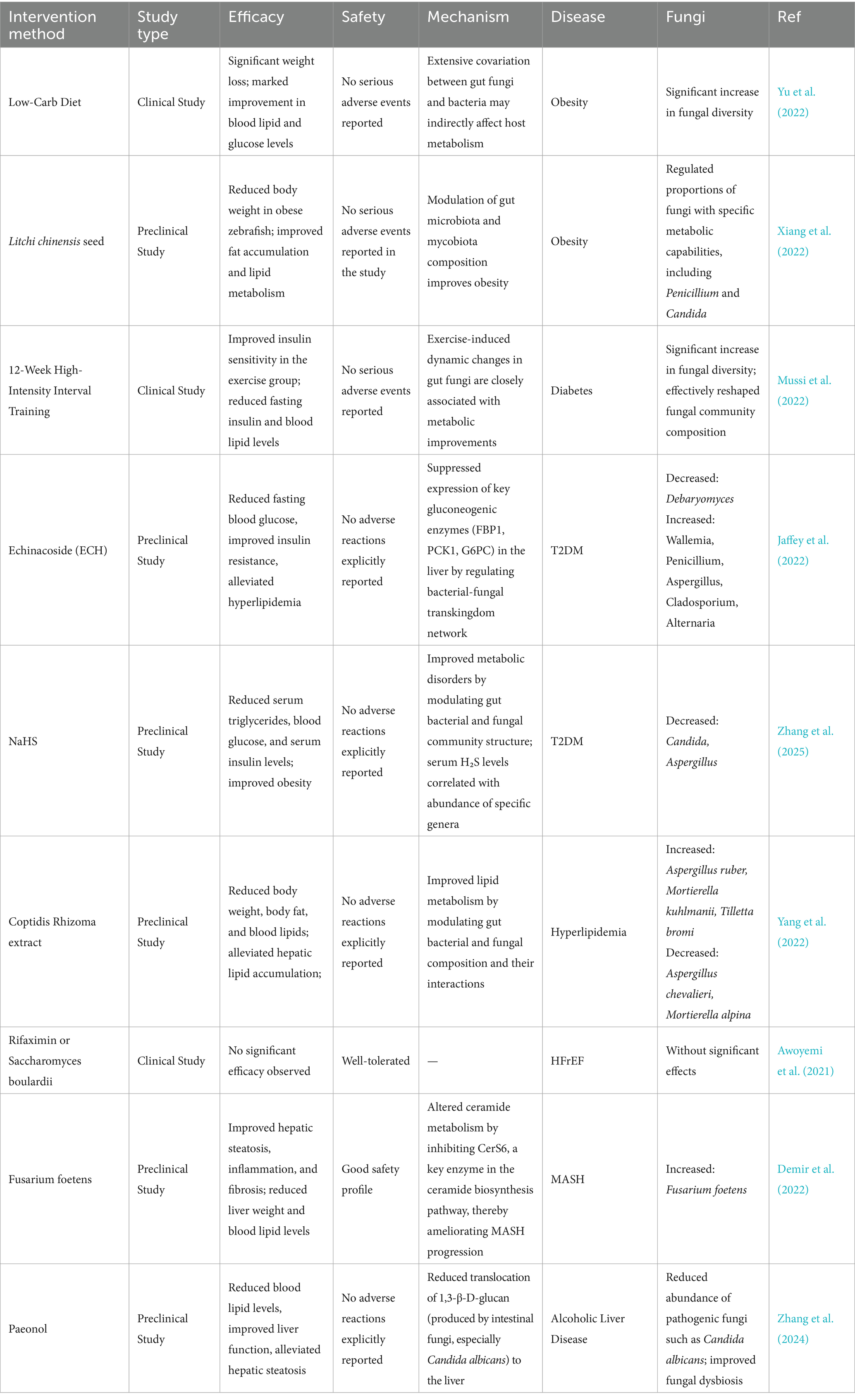
The gut mycobiome is closely associated with the onset and progression of cardiometabolic diseases. Recent evidence suggests that multiple potential drugs or therapies can modulate the progression of CMD by regulating the gut mycobiota (Table 8). Therefore, exploring intervention strategies based on the gut mycobiome may offer novel approaches for treating cardiometabolic disorders. Current methods for modulating the gut mycobiome can be broadly categorized into two main types: targeted therapies against gut fungi and their derivatives, and comprehensive modulation of the gut mycobiome-immune-metabolism axis. Specific interventions include: antifungal agents, microbial enzyme-specific inhibitors, clearance of endogenous metabolites, probiotic fungal supplementation, Dectin-1 antagonists, dietary interventions (Mediterranean diet, low-carbohydrate diet, high-fiber diet), natural compounds, and physical exercise (Figure 3).
5.1 Therapeutic approaches targeting gut fungi and their metabolites
Targeted modulation of gut fungi and their derivatives holds therapeutic promise for the integrative management of cardio-metabolic diseases. First, previous studies have demonstrated that pharmacological ablation of C. albicans using conventional antifungals (amphotericin B or fluconazole) significantly ameliorates insulin resistance and metabolic derangements (Wang et al., 2024). Fluconazole monotherapy also exhibits promising efficacy in attenuating AS progression by suppressing C. albicans proliferation, which concurrently curbs systemic inflammation and ceramide accumulation, thereby reducing cardiometabolic risk (Bao et al., 2023). Given that amphotericin B and fluconazole are broad-spectrum antifungal agents whose long-term use may incur significant side effects, whether antifungal therapy should be implemented as a potential clinical intervention for cardiometabolic diseases thus requires further clinical investigation and evidence-based validation. Second, targeted inhibition of microbial enzymatic activity is a promising therapeutic strategy. The groundbreaking discovery of microbial DPP4 isozymes has unveiled a novel mechanism (Wu et al., 2023). Consequently, developing specific inhibitors against fungal enzymes, such as C. parapsilosis lipase, can attenuate excessive FFA production, thereby ameliorating oxidative lipotoxicity and hepatic steatosis (Sun et al., 2021); this has significant therapeutic implications for CMD amelioration. Furthermore, the depletion of fungal-derived metabolites (endogenous ethanol or ceramides) confers distinct physiological benefits. Notably, ceramides, designated as the ‘putative second cholesterol’ (Zhang et al., 2025), render pharmacological modulation of their biosynthetic pathways, such as serine palmitoyltransferase (SPT), a novel therapeutic paradigm for CMD management. Consequently, Myriocin (thermozymocidin), a potent SPT inhibitor, demonstrates substantial translational promise for mitigating CMDs (Gengatharan et al., 2025).
5.2 Integrated modulation of gut mycobiome-immune-metabolic axis
As previously indicated, fungal β-glucan, a prototypical pathogen-associated molecular pattern, serves as a master regulator of host immunity. Its binding to Dectin-1 receptors on immune cells primarily instigates inflammatory cascades (Liu et al., 2025). Pioneering studies have suggested β-glucan-specific nanobodies hold therapeutic promise owing to their target-blocking capacity (Liu et al., 2023). Developing Dectin-1 antagonists is a viable therapeutic alternative (Li et al., 2024) to mitigate inflammation-driven metabolic derangements. Moreover, dysbiosis within the gut mycobiome-bacteriome interactome fuels cardiometabolic pathogenesis via hub-mediated immuno-metabolic axes. Accumulating clinical evidence supports the therapeutic efficacy of fungal-targeted transplantation or supplementation with probiotic fungi. Nevertheless, cutting-edge findings suggest that dietary modulation is the linchpin for reconfiguring gut microbiota, surpassing conventional microbiota transfer (Kennedy et al., 2025). Established paradigms, including Mediterranean, low-carbohydrate, and high-fiber diets, demonstrate a documented potential to correct metabolic derangements (Gao et al., 2024). Additionally, natural compounds constitute an indispensable therapeutic arsenal against CMD. Previous studies have demonstrated that Litchi chinensis seed (Xiang et al., 2022), echinacoside (Fan et al., 2025), Coptidis Rhizoma (Yang et al., 2022), and paeonol (Wu et al., 2020) exhibit dual lipid-glycemic modulatory efficacy, recalibrating fungal-bacterial interactomes to remodel gut ecological architectures. Furthermore, the indisputably beneficial role of exercise serves as a vital adjunct strategy for weight management (Jung et al., 2015), mitigation of metabolic dysregulation (Wewege et al., 2018), and cardiovascular health enhancement (Yue et al., 2022; Ramos et al., 2015). Contemporary research reveals that compared to sedentary counterparts, a 12-week regimen of high-intensity interval training significantly augmented the abundance of gut mycobiota including Verticillium, Ceratocystis, and Sarocladium. This elevation in fungal populations demonstrated significant correlation with exercise-induced improvements in key metabolic parameters—namely blood glucose levels, blood pressure, and lipid profiles (Wang et al., 2024).
6 Limitations
Despite the growing body of evidence linking the gut mycobiome to CMD, several important limitations must be acknowledged. First, the majority of existing studies are cross-sectional or conducted with small cohorts, which limits the generalizability and reproducibility of the findings. Large-scale, longitudinal, and multi-center studies are still lacking, making it difficult to draw firm conclusions about temporal dynamics and causal relationships. Second, most current evidence remains correlative. Although associations between specific fungal taxa (e.g., Candida and Saccharomyces) and CMD have been observed, direct mechanistic and causal links are insufficiently established. Experimental validation, particularly in vivo models and interventional trials, is urgently needed to confirm the pathogenic or protective roles of fungi in metabolic and cardiovascular pathways. Third, methodological and technical constraints remain a significant barrier. Current high-throughput sequencing techniques often lack sufficient depth and taxonomic resolution for fungi compared to bacteria. For example, 18S rRNA gene sequencing is widely employed for eukaryotic classification, yet it demonstrates limited resolution at the fungal species level, particularly in samples with highly diverse microbial communities (e.g., gut microbiota) (Hibbett et al., 2007; Jansen et al., 2024). In contrast, Internal Transcribed Spacer (ITS) sequencing—especially targeting the ITS1 and ITS2 regions—has become the predominant approach for fungal identification and diversity analysis in the gut (Diaz and Dongari-Bagtzoglou, 2021). However, the ITS region exhibits substantial length variation, which can introduce PCR amplification bias. Furthermore, its analytical outcomes are heavily dependent on reference sequence databases and cannot distinguish between viable and non-viable organisms. These factors may compromise identification accuracy; therefore, primer selection requires particular caution (Schoch et al., 2012; Nilsson et al., 2009). Secondly, in fine-scale species-level classification, shotgun metagenomic sequencing theoretically offers superior accuracy compared to ITS sequencing, as it provides access to more comprehensive genomic information, including functional genes. Despite its potential in mycobiome research, shotgun metagenomics faces significant challenges in gut fungal analysis: fungi typically constitute a minor fraction of the gut microbiota, with their extremely low DNA abundance often overshadowed by overwhelming bacterial DNA (Huseyin et al., 2017; Thomas et al., 2012; Xie et al., 2023). Furthermore, while emerging technologies such as MALDI-TOF and DART mass spectrometry have been explored for fungal strain classification, their utility and robustness in holistically profiling complex gut mycobiota—particularly regarding comprehensive taxonomic resolution and functional characterization—require rigorous validation and optimization (Neurauter et al., 2025). Furthermore, fungal cultivation is challenging due to slow growth rates and complex nutritional requirements, which hinders species-level characterization and functional analyses. Improvements in sequencing coverage, primer design, and fungal-specific bioinformatics pipelines are required to advance the field. Finally, translation into clinical practice remains limited. While preliminary evidence suggests that antifungal therapies, probiotic fungal supplementation, and dietary interventions may hold therapeutic potential, clinical studies validating these approaches are scarce. The safety, efficacy, and long-term effects of gut mycobiome-targeted therapies require rigorous evaluation before clinical application can be realized. Taken together, these limitations underscore that our current understanding of gut mycobiome–CMD interactions remains in its infancy. Addressing these challenges will be crucial for transforming correlative insights into mechanistic understanding and developing clinically applicable diagnostic and therapeutic strategies.
7 Conclusion and perspectives
This is the first systematic review to explore the link between the gut mycobiome and CMD. By incorporating mycobiota evidence into CMD pathogenesis models, we provide a comprehensive overview of the latest progress in this field. In contrast to previous reviews that primarily focused on bacterial microbiota-CMD correlations, this systematic demonstration reveals the impact of intestinal fungi on disease progression via multiple pathways, including the secretion of metabolites and the regulation of the immune-metabolic axis. This study formulates a new research framework by integrating advanced findings from metagenomic and clinical cohort studies, moving beyond traditional bacteria-centric strategies. This holistic perspective highlights previously undervalued therapeutic targets within the fungal microbiota and creates opportunities for novel CMD intervention strategies. Therefore, dietary modulation, supplementation with probiotic fungi, natural compounds, and targeted therapies against the immune-metabolic axis, when synergistically combined with conventional pharmacotherapies, may represent key strategies for the effective management of cardiometabolic diseases in the future.
In future studies, clinical validation of fungal biomarkers in CMD remains pivotal for translational applications. Accumulating evidence suggests elevated abundances of the genera Candida and Saccharomyces in CMD, encompassing diabetes, obesity, and atherosclerosis (AS). However, large-scale cohort validation is urgently required. Future research should deploy multicenter, prospective cohorts to authenticate fungal signatures and integrate mycobiome biomarkers with key clinical indices (BMI, glycemia, lipidemia, and hypertension) to establish precision diagnostic matrices for CMD. These efforts will catalyze the development of mechanism-based antifungal therapeutic pipelines. Moreover, cardiometabolic-protective fungal consortia, exemplified by Mortierella spp. and Mucor racemosus, which demonstrate significant inverse associations with CMD pathogenesis, warrant mechanistic dissection. Emerging evidence posits that therapeutic modulation of the gut mycobiota equilibrium may constitute the definitive frontier for next-generation metabolic therapeutics.
To address this gap, advancing from association to causality requires deepening mechanistic understanding of gut mycobiome-CMD crosstalk. This research nexus remains nascent, yet existing studies delineate Candida -dominated fungal consortia that orchestrate metabolic reprogramming through effector metabolites and cell wall components. The precise pathological cascades by which fungal-derived molecules perturb distal organ homeostasis via spatiotemporal circulatory trafficking demand systematic deconvolution. Integrated metabolomic, proteomic, and single-cell topological mapping will be essential to unravel these mechanisms, bridging the gap between observational data and causal inference. Multi-omics technologies, as a recently emerged integrative biomedical tool, provide a novel framework for elucidating the complexity of the gut microbiome (Chetty and Blekhman, 2024). Metagenomics enables precise classification of gut fungal strains and assessment of the potential functions of the fungal community through DNA profiling; metaproteomics identifies key proteins and their contributions to host biochemical pathways via protein expression profiling of gut fungi and the host; metatranscriptomics reveals which genes are actively expressed under specific conditions by RNA expression profiling, thereby elucidating the actual functional state of gut fungi; and metabolomics detects small-molecule metabolites generated from the interaction between gut fungi and the host (Duan et al., 2025). When analyzing multi-omics characteristics of the gut mycobiome, host multi-omics data and phenotypic data (e.g., diet, lifestyle habits, clinical indicators) must be concurrently collected. For instance, the FoCus cohort study integrated multi-omics data across phenomics, microbiome, metabolome, genome, metagenome, and nutritional profiling levels to systematically investigate the stepwise progression of CMD from healthy status to overt metabolic dysregulation and ultimately to cardiovascular disease (Geisler et al., 2022). The multi-omics testing generates vast amounts of data, and effectively analyzing and interpreting these complex biological insights has become the central challenge in current research (Rozera et al., 2025). Although a consensus on optimal integration strategies has not yet been reached, existing approaches—such as dimensionality reduction and clustering analysis, correlation analysis, Bayesian networks, machine learning, and AI model development—can be leveraged for application (Chetty and Blekhman, 2024; Wu et al., 2024). For example, a multi-omics study investigating the mechanism by which exercise ameliorates type 2 diabetes effectively integrated gut mycobiome, metabolome, and proteome data through correlation analysis, revealing gut mycobiome-mediated metabolic changes (Wang et al., 2024). Another study performed integrated analysis of multi-omics data for NAFLD using Bayesian networks (Chella Krishnan et al., 2018). Additionally, researchers can leverage newly developed software packages to integrate multi-omics data; for instance, in the cardiometabolic disease cohort (MetaCardis), the tool successfully revealed associations between the microbiome and metabolome (Muller et al., 2024). Furthermore, by integrating Mendelian randomization (MR) analysis, multi-omics data—including the gut mycobiome—can be systematically assessed for potential causal relationships with CMD (He et al., 2024). Comprehensive application of these strategies enables effective integration of multi-omics data, thereby facilitating a deeper understanding of the complex role of the gut mycobiome in CMD.
Moreover, disease prevention during critical early-life windows presents a promising avenue for long-term CMD risk reduction and should be a focus of future research and public health strategies. During the birthing process, infants acquire maternal fungal communities through contact with the vaginal canal and maternal skin (Willis et al., 2019), as well as through postnatal skin-to-skin contact (Heisel et al., 2019; Schei et al., 2017) or breastfeeding (Schei et al., 2017). Research indicates vertical transmission of these fungal communities from mother to infant: when specific fungal DNA is present in the mother’s gut, the probability of detecting corresponding fungal DNA in the infant’s gut significantly increases (Boutin et al., 2021; Fujimura et al., 2016). The diversity of the infant gastrointestinal fungal community markedly expands during the first year of life, with dominant genera undergoing stage-specific shifts (Boutin et al., 2021; Fujimura et al., 2016). Specifically, Candida predominates at birth; by 6 months of age, Malassezia and Cystofilobasidium gradually become dominant; and by 18 months, Trichosporon emerges as the primary genus (Zeng et al., 2025). The early establishment of the gut mycobiota is influenced by multiple factors. Maternal factors—including geography, environment, diet, mode of delivery, and antibiotic use—affect the composition of breast milk fungal communities. Infant factors—such as feeding practices, antibiotic exposure, and age—also shape the gut fungal composition (Rodriguez et al., 2024). Among these, mode of delivery (cesarean section) is the most extensively studied. For instance, infants or mice born via cesarean section exhibit higher risks of developing obesity and diabetes in adulthood compared to those delivered vaginally (Cox et al., 2014; Jensen et al., 2022; Keag et al., 2018). This underscores the critical importance of microbial community establishment during early-life critical windows, where the presence of specific microbial taxa contributes to optimal immune development and healthy metabolic homeostasis (Zeng et al., 2025). Landmark research identifies gut-adapted Candida dubliniensis colonization during the neonatal window as a master regulator of pancreatic β-cell ontogeny, establishing a groundbreaking conceptual framework for understanding diabetes pathogenesis. This discovery necessitates expanding investigations into the establishment trajectories of early-life gut mycobiota and their causal entanglement with CMD. Here, gut-fungal reprogramming emerges as a viable substrate for primordial interceptive therapeutics, offering a novel paradigm for disease prevention.
In addition to mechanistic and clinical priorities, overcoming persistent technical challenges is critical to accelerating progress in the field. Overcoming persistent technical challenges is critical to accelerating progress in the field. Notwithstanding significant advancements in gut mycobiome research, technical limitations such as inadequate sequencing coverage, limited taxonomic discrimination, challenging cultivation requirements, and low strain isolation efficiency remain unresolved. Future efforts must prioritize the development of fungal-specific primers and advanced metagenomic assembly algorithms to achieve species-level classification accuracy. Such technological evolution will expedite the translation of fundamental mycobiome discoveries into precision diagnostic and therapeutic strategies for CMD, ultimately realizing the bench-to-bedside paradigm.
Author contributions
XW: Conceptualization, Writing – original draft. ZG: Conceptualization, Writing – original draft. JW: Data curation, Investigation, Visualization, Writing – review & editing. DG: Data curation, Funding acquisition, Investigation, Visualization, Writing – review & editing. QX: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. SH: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China [82374195] and the Tianjin Municipal Education Commission General Project of Scientific Research Plan [2023KJ168].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdallah, Q. A., Mims, T., Watts, S., Al Abdallah, Q., Pingili, A., Bajwa, A., et al. (2020). Intestinal fungi as regulators of obesity and glucose tolerance. FASEB J. 34:1. doi: 10.1096/fasebj.2020.34.s1.09898
Aggarwal, N., Kitano, S., Puah, G. R. Y., Kittelmann, S., Hwang, I. Y., and Chang, M. W. (2023). Microbiome and human health: current understanding, engineering, and enabling technologies. Chem. Rev. 123, 31–72. doi: 10.1021/acs.chemrev.2c00431
Ahmad, E., Lim, S., Lamptey, R., Webb, D. R., and Davies, M. J. (2022). Type 2 diabetes. Lancet 400, 1803–1820. doi: 10.1016/S0140-6736(22)01655-5
Al Bataineh, M. T., Dash, N. R., Bel Lassen, P., Banimfreg, B. H., Nada, A. M., Belda, E., et al. (2021). Author correction: revealing links between gut microbiome and its fungal community in type 2 diabetes mellitus among Emirati subjects: a pilot study. Sci. Rep. 11:15102. doi: 10.1038/s41598-021-94439-3
American Diabetes Association (2014). Diagnosis and classification of diabetes mellitus. Diabetes Care 37, S81–S90. doi: 10.2337/dc14-S081
An, K., Jia, Y., Xie, B., Gao, J., Chen, Y., Yuan, W., et al. (2024). Alterations in the gut mycobiome with coronary artery disease severity. EBioMedicine 103:105137. doi: 10.1016/j.ebiom.2024.105137
Arena, R., Guazzi, M., Lianov, L., Whitsel, L., Berra, K., Lavie, C. J., et al. (2015). Healthy lifestyle interventions to combat noncommunicable disease-a novel nonhierarchical connectivity model for key stakeholders: a policy statement from the American Heart Association, European Society of Cardiology, European Association for Cardiovascular Prevention and Rehabilitation, and American College of Preventive Medicine. Eur. Heart J. 36, 2097–2109. doi: 10.1093/eurheartj/ehv207
Arnaiz-Villena, A., Juarez, I., Vaquero-Yuste, C., Lledo, T., Martin-Villa, J. M., and Suarez-Trujillo, F. (2024). Complex interactions between the human major histocompatibility complex (MHC) and microbiota: their roles in disease pathogenesis and immune system regulation. Biomedicine 12:1928. doi: 10.3390/biomedicines12081928
Awoyemi, A., Mayerhofer, C., Felix, A. S., Hov, J. R., Moscavitch, S. D., Lappegård, K. T., et al. (2021). Rifaximin or Saccharomyces boulardii in heart failure with reduced ejection fraction: results from the randomized GutHeart trial. EBioMedicine 70:103511. doi: 10.1016/j.ebiom.2021.103511
Bäck, M., Yurdagul, A. Jr., Tabas, I., Öörni, K., and Kovanen, P. T. (2019). Inflammation and its resolution in atherosclerosis: mediators and therapeutic opportunities. Nat. Rev. Cardiol. 16, 389–406. doi: 10.1038/s41569-019-0169-2
Bandala-Sanchez, E., Roth-Schulze, A. J., Oakey, H., Penno, M. A. S., Bediaga, N. G., Naselli, G., et al. (2022). Women with type 1 diabetes exhibit a progressive increase in gut Saccharomyces cerevisiae in pregnancy associated with evidence of gut inflammation. Diabetes Res. Clin. Pract. 184:109189. doi: 10.1016/j.diabres.2022.109189
Bao, L., Zhang, Y., Zhang, G., Jiang, D., and Yan, D. (2023). Abnormal proliferation of gut mycobiota contributes to the aggravation of type 2 diabetes. Commun. Biol. 6:226. doi: 10.1038/s42003-023-04591-x
Barteková, M., Adameová, A., Görbe, A., Ferenczyová, K., Pecháňová, O., Lazou, A., et al. (2021). Natural and synthetic antioxidants targeting cardiac oxidative stress and redox signaling in cardiometabolic diseases. Free Radic. Biol. Med. 169, 446–477. doi: 10.1016/j.freeradbiomed.2021.03.045
Batty, M., Bennett, M. R., and Yu, E. (2022). The role of oxidative stress in atherosclerosis. Cells 11:3843. doi: 10.3390/cells11233843
Bhunjun, C. S., Chen, Y. J., Phukhamsakda, C., Boekhout, T., Groenewald, J. Z., McKenzie, E. H. C., et al. (2024). What are the 100 most cited fungal genera? Stud. Mycol. 108, 1–411. doi: 10.3114/sim.2024.108.01
Bhute, S. S., Suryavanshi, M. V., Joshi, S. M., Yajnik, C. S., Shouche, Y. S., and Ghaskadbi, S. S. (2017). Gut microbial diversity assessment of Indian Type-2-diabetics reveals alterations in eubacteria, Archaea, and eukaryotes. Front. Microbiol. 8:214. doi: 10.3389/fmicb.2017.00214
Borges, F. M., de Paula, T. O., Sarmiento, M. R. A., de Oliveira, M. G., Pereira, M. L. M., Toledo, I. V., et al. (2018). Fungal diversity of human gut microbiota among eutrophic, overweight, and obese individuals based on aerobic culture-dependent approach. Curr. Microbiol. 75, 726–735. doi: 10.1007/s00284-018-1438-8
Boutin, R. C. T., Sbihi, H., McLaughlin, R. J., Hahn, A. S., Konwar, K. M., Loo, R. S., et al. (2021). Composition and associations of the infant gut fungal microbiota with environmental factors and childhood allergic outcomes. mBio 12:e0339620. doi: 10.1128/mBio.03396-20
Cai, N., Gomez-Duran, A., Yonova-Doing, E., Kundu, K., Burgess, A. I., Golder, Z. J., et al. (2021). Mitochondrial DNA variants modulate N-formylmethionine, proteostasis and risk of late-onset human diseases. Nat. Med. 27, 1564–1575. doi: 10.1038/s41591-021-01441-3
Cai, Y., Li, Y., Xiong, Y., Geng, X., Kang, Y., and Yang, Y. (2024). Diabetic foot exacerbates gut mycobiome dysbiosis in adult patients with type 2 diabetes mellitus: revealing diagnostic markers. Nutr. Diabetes 14:71. doi: 10.1038/s41387-024-00328-9
Carmody, R. N., and Bisanz, J. E. (2023). Roles of the gut microbiome in weight management. Nat. Rev. Microbiol. 21, 535–550. doi: 10.1038/s41579-023-00888-0
Chacón, M. R., Lozano-Bartolomé, J., Portero-Otín, M., Rodríguez, M. M., Xifra, G., Puig, J., et al. (2018). The gut mycobiome composition is linked to carotid atherosclerosis. Benef Microbes. 9, 185–198. doi: 10.3920/BM2017.0029
Chella Krishnan, K., Kurt, Z., Barrere-Cain, R., Sabir, S., Das, A., Floyd, R., et al. (2018). Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Syst. 6, 103–115.e7. doi: 10.1016/j.cels.2017.12.006
Chen, Q., Huang, S., Wang, X., Peng, J., Wang, P., Luo, R., et al. (2025). The burden of diseases attributable to high body mass index in Asia from 1990 - 2019: results from the global burden of disease study 2019. Ann. Med. 57:2483977. doi: 10.1080/07853890.2025.2483977
Chen, B. Y., Lin, W. Z., Li, Y. L., Bi, C., Du, L. J., Liu, Y., et al. (2023). Characteristics and correlations of the Oral and gut fungal microbiome with hypertension. Microbiol Spectr. 11:e0195622. doi: 10.1128/spectrum.01956-22
Chen, Y., Zhou, J., and Wang, L. (2021). Role and mechanism of gut microbiota in human disease. Front. Cell. Infect. Microbiol. 11:625913. doi: 10.3389/fcimb.2021.625913
Cheng, S. C., van de Veerdonk, F. L., Lenardon, M., Stoffels, M., Plantinga, T., Smeekens, S., et al. (2011). The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J. Leukoc. Biol. 90, 357–366. doi: 10.1189/jlb.1210702
Chetty, A., and Blekhman, R. (2024). Multi-omic approaches for host-microbiome data integration. Gut Microbes 16:2297860. doi: 10.1080/19490976.2023.2297860
Chew, N. W. S., Chong, B., Kuo, S. M., Jayabaskaran, J., Cai, M., Zheng, H., et al. (2023). Trends and predictions of metabolic risk factors for acute myocardial infarction: findings from a multiethnic nationwide cohort. Lancet Reg. Health. 37:100803. doi: 10.1016/j.lanwpc.2023.100803
Chowdhury, S. R., Chandra Das, D., Sunna, T. C., Beyene, J., and Hossain, A. (2023). Global and regional prevalence of multimorbidity in the adult population in community settings: a systematic review and meta-analysis. EClinicalMedicine. 57:101860. doi: 10.1016/j.eclinm.2023.101860
Chung, M. Y., Mah, E., Masterjohn, C., Noh, S. K., Park, H. J., Clark, R. M., et al. (2015). Green tea lowers hepatic COX-2 and prostaglandin E2 in rats with dietary fat-induced nonalcoholic steatohepatitis. J. Med. Food 18, 648–655. doi: 10.1089/jmf.2014.0048
Costello, E. K., Lauber, C. L., Hamady, M., Fierer, N., Gordon, J. I., and Knight, R. (2009). Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697. doi: 10.1126/science.1177486
Cox, L. M., Yamanishi, S., Sohn, J., Alekseyenko, A. V., Leung, J. M., Cho, I., et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721. doi: 10.1016/j.cell.2014.05.052
Datta, S., Koka, S., and Boini, K. M. (2025). Understanding the role of Adipokines in Cardiometabolic dysfunction: a review of current knowledge. Biomolecules. 15:612. doi: 10.3390/biom15050612
Datta, S., Pasham, S., Inavolu, S., Boini, K. M., and Koka, S. (2024). Role of gut microbial metabolites in cardiovascular diseases-current insights and the road ahead. Int. J. Mol. Sci. 25:10208. doi: 10.3390/ijms251810208
Demir, M., Lang, S., Hartmann, P., Duan, Y., Martin, A., Miyamoto, Y., et al. (2022). The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 76, 788–799. doi: 10.1016/j.jhep.2021.11.029
Diaz, P. I., and Dongari-Bagtzoglou, A. (2021). Critically appraising the significance of the Oral Mycobiome. J. Dent. Res. 100, 133–140. doi: 10.1177/0022034520956975
Dong, G., Ryan, T. E., and Esser, K. A. (2025). Animal models of exercise and Cardiometabolic disease. Circ. Res. 137, 139–162. doi: 10.1161/CIRCRESAHA.124.325704
Drożdż, K., Nabrdalik, K., Hajzler, W., Kwiendacz, H., Gumprecht, J., and Lip, G. Y. H. (2021). Metabolic-associated fatty liver disease (MAFLD), diabetes, and cardiovascular disease: associations with fructose metabolism and gut microbiota. Nutrients 14:103. doi: 10.3390/nu14010103
Du, W., Zou, Z. P., Ye, B. C., and Zhou, Y. (2025). Gut microbiota and associated metabolites: key players in high-fat diet-induced chronic diseases. Gut Microbes 17:2494703. doi: 10.1080/19490976.2025.2494703
Duan, D., Wang, M., Han, J., Li, M., Wang, Z., Zhou, S., et al. (2025). Advances in multi-omics integrated analysis methods based on the gut microbiome and their applications. Front. Microbiol. 15:1509117. doi: 10.3389/fmicb.2024.1509117
Egea, M. B., Oliveira Filho, J. G., and Lemes, A. C. (2023). Investigating the efficacy of Saccharomyces boulardii in metabolic syndrome treatment: a narrative review of what is known so far. Int. J. Mol. Sci. 24:12015. doi: 10.3390/ijms241512015
Elnagar, R. M. (2024). Cross interaction between bacterial and fungal microbiota and their relevance to human health and disease: mechanistic pathways and prospective therapy. Biosci. Microbiota Food Health. 43, 309–320. doi: 10.12938/bmfh.2024-031
Eslam, M., Sanyal, A. J., and George, J.International Consensus Panel (2020). MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology 158, 1999–2014. doi: 10.1053/j.gastro.2019.11.312
Estrada, R., Romero, Y., Quilcate, C., Dipaz, D., Alejos-Asencio, C. S., Leon, S., et al. (2024). Age-dependent changes in Protist and fungal microbiota in a Peruvian cattle genetic nucleus. Life (Basel). 14:1010. doi: 10.3390/life14081010
Fan, L., Liu, J., Li, L., Yang, X., Zhao, Q., and Zhao, L. (2025). Echinacoside alleviates type 2 diabetes mellitus through inhibiting hepatic gluconeogenesis via gut bacterial-fungal trans-kingdom network reconstruction. Phytomedicine 142:156802. doi: 10.1016/j.phymed.2025.156802
Freisling, H., Viallon, V., Lennon, H., Bagnardi, V., Ricci, C., Butterworth, A. S., et al. (2020). Lifestyle factors and risk of multimorbidity of cancer and cardiometabolic diseases: a multinational cohort study. BMC Med. 18:5. doi: 10.1186/s12916-019-1474-7
Fu, Z., Jia, Y., Zhao, J., Guo, Y., Xie, B., An, K., et al. (2025). Perioperative multi-kingdom gut microbiota alters in coronary artery bypass grafting. Biomedicine 13:475. doi: 10.3390/biomedicines13020475
Fujimura, K. E., Sitarik, A. R., Havstad, S., Lin, D. L., Levan, S., Fadrosh, D., et al. (2016). Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191. doi: 10.1038/nm.4176
Gao, Y., Li, W., Huang, X., Lyu, Y., and Yue, C. (2024). Advances in gut microbiota-targeted therapeutics for metabolic syndrome. Microorganisms. 12:851. doi: 10.3390/microorganisms12050851
García, C., Tebbji, F., Daigneault, M., Liu, N. N., Köhler, J. R., Allen-Vercoe, E., et al. (2017). The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere. 2, e00555–e00517. doi: 10.1128/mSphere.00555-17
García-Gamboa, R., Kirchmayr, M. R., Gradilla-Hernández, M. S., Pérez-Brocal, V., Moya, A., and González-Avila, M. (2021). The intestinal mycobiota and its relationship with overweight, obesity and nutritional aspects. J. Hum. Nutr. Diet. 34, 645–655. doi: 10.1111/jhn.12864
García-Torre, A., Bueno-García, E., López-Martínez, R., Rioseras, B., Díaz-Molina, B., Lambert, J. L., et al. (2021). CMV infection is directly related to the inflammatory status in chronic heart failure patients. Front. Immunol. 12:687582. doi: 10.3389/fimmu.2021.687582
Ge, Y., Pan, M., Zhang, C., Wang, C., Ma, K., Yan, G., et al. (2021). Paeonol alleviates dextran sodium sulfate induced colitis involving Candida albicans-associated dysbiosis. Med. Mycol. 59, 335–344. doi: 10.1093/mmy/myaa053
Geisler, C., Schlicht, K., Knappe, C., Rohmann, N., Hartmann, K., Türk, K., et al. (2022). Cohort profile: the food chain plus (FoCus) cohort. Eur. J. Epidemiol. 37, 1087–1105. doi: 10.1007/s10654-022-00924-y
Geng, J., Ni, Q., Sun, W., Li, L., and Feng, X. (2022). The links between gut microbiota and obesity and obesity related diseases. Biomed. Pharmacother. 147:112678. doi: 10.1016/j.biopha.2022.112678
Gengatharan, J. M., Handzlik, M. K., Chih, Z. Y., Ruchhoeft, M. L., Secrest, P., Ashley, E. L., et al. (2025). Altered sphingolipid biosynthetic flux and lipoprotein trafficking contribute to trans-fat-induced atherosclerosis. Cell Metab. 37, 274–290.e9. doi: 10.1016/j.cmet.2024.10.016
Gofron, K., Berezowski, A., Gofron, M., Borówka, M., Dziedzic, M., Kazimierczak, W., et al. (2024). Akkermansia muciniphila - impact on the cardiovascular risk, the intestine inflammation and obesity. Acta Biochim. Pol. 71:13550. doi: 10.3389/abp.2024.13550
Goh, R. S. J., Chong, B., Jayabaskaran, J., Jauhari, S. M., Chan, S. P., Kueh, M. T. W., et al. (2024). The burden of cardiovascular disease in Asia from 2025 to 2050: a forecast analysis for East Asia, South Asia, South-East Asia, Central Asia, and high-income Asia Pacific regions. Lancet Reg Health West Pac. 49:101138. doi: 10.1016/j.lanwpc.2024.101138
Gosiewski, T., Salamon, D., Szopa, M., Sroka, A., Malecki, M. T., and Bulanda, M. (2014). Quantitative evaluation of fungi of the genus Candida in the feces of adult patients with type 1 and 2 diabetes - a pilot study. Gut Pathog. 6:43. doi: 10.1186/s13099-014-0043-z
Gou, W., Wang, H., Su, C., Fu, Y., Wang, X., Gao, C., et al. (2024). The temporal dynamics of the gut mycobiome and its association with cardiometabolic health in a nationwide cohort of 12,641 Chinese adults. Cell Rep Med. 5:101775. doi: 10.1016/j.xcrm.2024.101775
Gouba, N., Raoult, D., and Drancourt, M. (2013). Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PLoS One 8:e59474. doi: 10.1371/journal.pone.0059474
Gradisteanu Pircalabioru, G., Ilie, I., Oprea, L., Picu, A., Petcu, L. M., Burlibasa, L., et al. (2022). Microbiome, Mycobiome and related metabolites alterations in patients with metabolic syndrome-a pilot study. Meta 12:218. doi: 10.3390/metabo12030218
Gringhuis, S. I., Kaptein, T. M., Wevers, B. A., Theelen, B., van der Vlist, M., Boekhout, T., et al. (2012). Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat. Immunol. 13, 246–254. doi: 10.1038/ni.2222
Guglietta, S., Li, X., and Saxena, D. (2025). Role of Fungi in tumorigenesis: promises and challenges. Annu. Rev. Pathol. 20, 459–482. doi: 10.1146/annurev-pathmechdis-111523-023524
Guiducci, L., Nicolini, G., and Forini, F. (2023). Dietary patterns, gut microbiota remodeling, and Cardiometabolic disease. Meta 13:760. doi: 10.3390/metabo13060760
Guo, Z., Guo, X., Xu, H., Chu, H., Tian, Y., Wang, S., et al. (2025). Association between metabolic score for insulin resistance (METS-IR) and hypertension: a cross-sectional study based on NHANES 2007-2018. Lipids Health Dis. 24:64. doi: 10.1186/s12944-025-02492-y
Gürsoy, S., Koçkar, T., Atik, S. U., Önal, Z., Önal, H., and Adal, E. (2018). Autoimmunity and intestinal colonization by Candida albicans in patients with type 1 diabetes at the time of the diagnosis. Korean J. Pediatr. 61, 217–220. doi: 10.3345/kjp.2018.61.7.217
Gurung, M., Li, Z., You, H., Rodrigues, R., Jump, D. B., Morgun, A., et al. (2020). Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 51:102590. doi: 10.1016/j.ebiom.2019.11.051
Gutierrez, M. W., Mercer, E. M., Moossavi, S., Laforest-Lapointe, I., Reyna, M. E., Becker, A. B., et al. (2023). Maturational patterns of the infant gut mycobiome are associated with early-life body mass index. Cell Rep Med. 4:100928. doi: 10.1016/j.xcrm.2023.100928
Gutierrez, M. W., van Tilburg Bernardes, E., Ren, E., Kalbfleisch, K. N., Day, M., Lameu, E. L., et al. (2025). Early-life gut mycobiome core species modulate metabolic health in mice. Nat. Commun. 16:1467. doi: 10.1038/s41467-025-56743-8
Hadrich, I., Turki, M., Chaari, I., Abdelmoula, B., Gargouri, R., Khemakhem, N., et al. (2025). Gut mycobiome and neuropsychiatric disorders: insights and therapeutic potential. Front. Cell. Neurosci. 18:1495224. doi: 10.3389/fncel.2024.1495224
Han, Y., Hu, Y., Yu, C., Guo, Y., Pei, P., Yang, L., et al. (2021). Lifestyle, cardiometabolic disease, and multimorbidity in a prospective Chinese study. Eur. Heart J. 42, 3374–3384. doi: 10.1093/eurheartj/ehab413
He, J. L., Zhao, Y. W., Yang, J. L., Ju, J. M., Ye, B. Q., Huang, J. Y., et al. (2024). Enhanced interactions among gut mycobiomes with the deterioration of glycemic control. Med. 5, 909–925.e7. doi: 10.1016/j.medj.2024.03.023
Heisel, T., Montassier, E., Johnson, A., Al-Ghalith, G., Lin, Y. W., Wei, L. N., et al. (2017). High-fat diet changes fungal microbiomes and Interkingdom relationships in the murine gut. mSphere. 2, e00351–e00317. doi: 10.1128/mSphere.00351-17
Heisel, T., Nyaribo, L., Sadowsky, M. J., and Gale, C. A. (2019). Breastmilk and NICU surfaces are potential sources of fungi for infant mycobiomes. Fungal Genet. Biol. 128, 29–35. doi: 10.1016/j.fgb.2019.03.008
Henkel, J., Neuschäfer-Rube, F., Pathe-Neuschäfer-Rube, A., and Püschel, G. P. (2009). Aggravation by prostaglandin E2 of interleukin-6-dependent insulin resistance in hepatocytes. Hepatology 50, 781–790. doi: 10.1002/hep.23064
Hibbett, D. S., Binder, M., Bischoff, J. F., Blackwell, M., Cannon, P. F., Eriksson, O. E., et al. (2007). A higher-level phylogenetic classification of the fungi. Mycol. Res. 111, 509–547. doi: 10.1016/j.mycres.2007.03.004
Hill, J. H., Bell, R., Barrios, L., Baird, H., Ost, K., Greenewood, M., et al. (2025). Neonatal fungi promote lifelong metabolic health through macrophage-dependent β cell development. Science 387:eadn0953. doi: 10.1126/science.adn0953
Hoffmann, C., Dollive, S., Grunberg, S., Chen, J., Li, H., Wu, G. D., et al. (2013). Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. doi: 10.1371/journal.pone.0066019
Honkanen, J., Vuorela, A., Muthas, D., Orivuori, L., Luopajärvi, K., Tejesvi, M. V. G., et al. (2020). Fungal Dysbiosis and intestinal inflammation in children with Beta-cell autoimmunity. Front. Immunol. 11:468. doi: 10.3389/fimmu.2020.00468
Hrncir, T., Trckova, E., and Hrncirova, L. (2024). Synergistic effects of fructose and food preservatives on metabolic dysfunction-associated Steatotic liver disease (MASLD): from gut microbiome alterations to hepatic gene expression. Nutrients 16:3722. doi: 10.3390/nu16213722
Hsu, C., Ghannoum, M., Cominelli, F., and Martino, L. D. (2023). Mycobiome and inflammatory bowel disease: role in disease pathogenesis, current approaches and novel nutritional-based therapies. Inflamm. Bowel Dis. 29, 470–479. doi: 10.1093/ibd/izac156
Huang, H., Wang, Q., Yang, Y., Zhong, W., He, F., and Li, J. (2024). The mycobiome as integral part of the gut microbiome: crucial role of symbiotic fungi in health and disease. Gut Microbes 16:2440111. doi: 10.1080/19490976.2024.2440111
Huseyin, C. E., Rubio, R. C., O'Sullivan, O., Cotter, P. D., and Scanlan, P. D. (2017). The fungal frontier: a comparative analysis of methods used in the study of the human gut mycobiome. Front. Microbiol. 8:1432. doi: 10.3389/fmicb.2017.01432
In China TWCOTROCHADHu, S. S. (2023). Report on cardiovascular health and diseases in China 2021: an updated summary. J. Geriatr. Cardiol. 20, 399–430. doi: 10.26599/1671-5411.2023.06.001
Jaffey, J. A., Okwumabua, O., Graves, T. K., Al-Nakkash, L., Monasky, R., Wilson, A., et al. (2022). Evaluation of Candida spp. and other fungi in feces from dogs with naturally occurring diabetes mellitus. Vet. Sci. 9:567. doi: 10.3390/vetsci9100567
Jansen, G. J., Schouten, G. P., and Wiersma, M. (2024). Advancements in analytical methods for studying the human gut microbiome. J. Biol. Methods 12:e99010038. doi: 10.14440/jbm.2024.0050
Jayasudha, R., Das, T., Kalyana Chakravarthy, S., Sai Prashanthi, G., Bhargava, A., Tyagi, M., et al. (2020). Gut mycobiomes are altered in people with type 2 diabetes mellitus and diabetic retinopathy. PLoS One 15:e0243077. doi: 10.1371/journal.pone.0243077
Jegier, B., Jaszewski, R., Kurnatowski, P., Kuba, K., and Lelonek, M. (2010). Mycotic DNA in non-atherosclerotic aortic wall of coronary patients is associated with sICAM-1 expression. Circ. J. 74, 749–753. doi: 10.1253/circj.cj-09-0632
Jensen, E. T., Bertoni, A. G., Crago, O. L., Rotter, J. I., Chen, Y. I., Wood, A., et al. (2022). Cesarean delivery and insulin sensitivity in the older adult: the microbiome and insulin longitudinal evaluation study. J. Endocr. Soc. 6:bvac072. doi: 10.1210/jendso/bvac072
Jensen, O., Trujillo, E., Hanson, L., and Ost, K. S. (2024). Controlling Candida: immune regulation of commensal fungi in the gut. Infect. Immun. 92:e0051623. doi: 10.1128/iai.00516-23
Jiang, Z., Mei, L., Li, Y., Guo, Y., Yang, B., Huang, Z., et al. (2024). Enzymatic regulation of the gut microbiota: mechanisms and implications for host health. Biomolecules. 14:1638. doi: 10.3390/biom14121638
Jiang, S., Xu, L., Chen, Y., Shu, Z., Lv, L., Zhao, Y., et al. (2024). Longitudinal gut fungal alterations and potential fungal biomarkers for the progression of primary liver disease. Sci. China Life Sci. 67, 1183–1198. doi: 10.1007/s11427-023-2458-1
Jung, M. E., Bourne, J. E., Beauchamp, M. R., Robinson, E., and Little, J. P. (2015). High-intensity interval training as an efficacious alternative to moderate-intensity continuous training for adults with prediabetes. J. Diabetes Res. 2015:191595. doi: 10.1155/2015/191595
Jurek, J. M., Zablocka-Sowinska, K., Clavero Mestres, H., Reyes Gutiérrez, L., Camaron, J., and Auguet, T. (2025). The impact of dietary interventions on metabolic outcomes in metabolic dysfunction-associated Steatotic liver disease (MASLD) and comorbid conditions, including obesity and type 2 diabetes. Nutrients 17:1257. doi: 10.3390/nu17071257
Kario, K., Okura, A., Hoshide, S., and Mogi, M. (2024). The WHO global report 2023 on hypertension warning the emerging hypertension burden in globe and its treatment strategy. Hypertens. Res. 47, 1099–1102. doi: 10.1038/s41440-024-01622-w
Keag, O. E., Norman, J. E., and Stock, S. J. (2018). Long-term risks and benefits associated with cesarean delivery for mother, baby, and subsequent pregnancies: systematic review and meta-analysis. PLoS Med. 15:e1002494. doi: 10.1371/journal.pmed.1002494
Kennedy, M. S., Freiburger, A., Cooper, M., Beilsmith, K., St George, M. L., Kalski, M., et al. (2025). Diet outperforms microbial transplant to drive microbiome recovery in mice. Nature 642, 747–755. doi: 10.1038/s41586-025-08937-9
Kivimäki, M., Singh-Manoux, A., Pentti, J., Sabia, S., Nyberg, S. T., Alfredsson, L., et al. (2019). Physical inactivity, cardiometabolic disease, and risk of dementia: an individual-participant meta-analysis. BMJ 365:l1495. doi: 10.1136/bmj.l1495
Kombrink, A., Tayyrov, A., Essig, A., Stöckli, M., Micheller, S., Hintze, J., et al. (2019). Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 13, 588–602. doi: 10.1038/s41396-018-0293-8
Kondapalli, N., Katari, V., Dalal, K. K., Paruchuri, S., and Thodeti, C. K. (2025). Microbiota in gut-heart Axis: metabolites and mechanisms in cardiovascular disease. Compr. Physiol. 15:e70024. doi: 10.1002/cph4.70024
Krüger, W., Vielreicher, S., Kapitan, M., Jacobsen, I. D., and Niemiec, M. J. (2019). Fungal-bacterial interactions in health and disease. Pathogens 8:70. doi: 10.3390/pathogens8020070
Kumbhare, S. V., Patangia, D. V. V., Patil, R. H., Shouche, Y. S., and Patil, N. P. (2019). Factors influencing the gut microbiome in children: from infancy to childhood. J. Biosci. 44:49. doi: 10.1007/s12038-019-9860-z
Li, Y., Huang, M., Cardinale, S., Su, Y., Peters, D. E., Slusher, B. S., et al. (2024). Dectin-1 as a therapeutic target for inflammatory bowel disease. Adv. Pharmacol. 101, 237–264. doi: 10.1016/bs.apha.2024.10.002
Li, J., Zhao, S., Zhang, X., Fan, M., Wan, J., Lin, R., et al. (2025). Association of Systemic Inflammatory Response Index with the cardiometabolic multimorbidity among US adults: a population-based study. Brain Circ. 11, 39–47. doi: 10.4103/bc.bc_32_24
Libby, P. (2021). The changing landscape of atherosclerosis. Nature 592, 524–533. doi: 10.1038/s41586-021-03392-8
Liu, Y., Huang, K., Guan, X., Li, S., Song, H., Zhang, Y., et al. (2025). Oat avenanthramide B alleviates high-fat diet-induced obesity via regulating fatty acid metabolism involved in gut bacteria and fungi remodeling. Food Sci. Human Wellness 14:9250124. doi: 10.26599/FSHW.2024.9250124
Liu, X., Sui, J., Li, C., Wang, Q., Peng, X., Meng, F., et al. (2023). The preparation and therapeutic effects of β-glucan-specific nanobodies and nanobody-natamycin conjugates in fungal keratitis. Acta Biomater. 169, 398–409. doi: 10.1016/j.actbio.2023.08.019
Liu, C., Yang, L., Wang, Z., Zhu, H., Luo, Q., Wu, D., et al. (2025). Qi-Huang decoction alleviates DSS-induced colitis with Candida albicans dysbiosis by enhancing innate immune response through Dectin-1-associated signaling. Phytomedicine 140:156613. doi: 10.1016/j.phymed.2025.156613
Liu, J., Zhao, W., Gao, Z. W., Liu, N., Zhang, W. H., and Ling, H. (2022). Effects of exogenous hydrogen sulfide on diabetic metabolic disorders in db/db mice are associated with gut bacterial and fungal microbiota. Front. Cell. Infect. Microbiol. 12:801331. doi: 10.3389/fcimb.2022.801331
Lu, H., and Daugherty, A. (2015). Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 35, 485–491. doi: 10.1161/ATVBAHA.115.305380
Luo, Q., Hu, Y., Chen, X., Luo, Y., Chen, J., and Wang, H. (2022). Effects of gut microbiota and metabolites on heart failure and its risk factors: a two-sample Mendelian randomization study. Front. Nutr. 9:899746. doi: 10.3389/fnut.2022.899746
Luo, X., Wang, K., and Jiang, C. (2025). Gut microbial enzymes and metabolic dysfunction-associated steatohepatitis: function, mechanism, and therapeutic prospects. Cell Host Microbe 33, 836–853. doi: 10.1016/j.chom.2025.04.020
Mar Rodríguez, M., Pérez, D., Javier Chaves, F., Esteve, E., Marin-Garcia, P., Xifra, G., et al. (2015). Obesity changes the human gut mycobiome. Sci. Rep. 5:14600. doi: 10.1038/srep14600
Marchlewicz, M., Sagan, P., Grabowska, M., Kiedrowicz, M., Kruk, J., Gill, K., et al. (2024). The role of vitamin D3 deficiency and colonization of the oral mucosa by Candida yeast-like fungi in the pathomechanism of psoriasis. J. Clin. Med. 13:6874. doi: 10.3390/jcm13226874
Masoumi, O., Shahzadi, M., Kordbacheh, P., Zaini, F., Mahmoudi, S., Mahmoudi, M., et al. (2015). Detection of fungal elements in atherosclerotic plaques using mycological, pathological and molecular methods. Iran. J. Public Health 44, 1121–1125.
Mbaye, B., Borentain, P., Magdy Wasfy, R., Alou, M. T., Armstrong, N., Mottola, G., et al. (2022). Endogenous ethanol and triglyceride production by gut Pichia kudriavzevii, Candida albicans and Candida glabrata yeasts in non-alcoholic steatohepatitis. Cells 11:3390. doi: 10.3390/cells11213390
McDonagh, T. A., Metra, M., Adamo, M., Gardner, R. S., Baumbach, A., Böhm, M., et al. (2021). 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 42, 3599–3726. doi: 10.1093/eurheartj/ehab368
Mechanick, J. I., Farkouh, M. E., Newman, J. D., and Garvey, W. T. (2020). Cardiometabolic-based chronic disease, adiposity and Dysglycemia drivers: JACC state-of-the-art review. J. Am. Coll. Cardiol. 75, 525–538. doi: 10.1016/j.jacc.2019.11.044
Mims, T. S., Abdallah, Q. A., Stewart, J. D., Watts, S. P., White, C. T., Rousselle, T. V., et al. (2021). The gut mycobiome of healthy mice is shaped by the environment and correlates with metabolic outcomes in response to diet. Commun Biol. 4:281. doi: 10.1038/s42003-021-01820-z
Muller, E., Shiryan, I., and Borenstein, E. (2024). Multi-omic integration of microbiome data for identifying disease-associated modules. Nat. Commun. 15:2621. doi: 10.1038/s41467-024-46888-3
Mullick, A., Leon, Z., Min-Oo, G., Berghout, J., Lo, R., Daniels, E., et al. (2006). Cardiac failure in C5-deficient a/J mice after Candida albicans infection. Infect. Immun. 74, 4439–4451. doi: 10.1128/IAI.00159-06
Mussi, M. C. M., Fernandes, K. S., and Gallottini, M. H. C. (2022). A call for further research on the relation between type 2 diabetes and oral candidiasis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 134, 206–212. doi: 10.1016/j.oooo.2022.02.009
Nash, A. K., Auchtung, T. A., Wong, M. C., Smith, D. P., Gesell, J. R., Ross, M. C., et al. (2017). The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 5:153. doi: 10.1186/s40168-017-0373-4
Ndumele, C. E., Rangaswami, J., Chow, S. L., Neeland, I. J., Tuttle, K. R., Khan, S. S., et al. (2023). Cardiovascular-kidney-metabolic health: a presidential advisory from the American Heart Association. Circulation 148, 1606–1635. doi: 10.1161/CIR.0000000000001184
Neurauter, M., Vinzelj, J. M., Strobl, S. F. A., Kappacher, C., Schlappack, T., Badzoka, J., et al. (2025). Application of MALDI TOF and DART mass spectrometry as novel tools for classification of anaerobic gut fungi strains. Anal. Bioanal. Chem. 417, 3245–3256. doi: 10.1007/s00216-025-05846-8
Neves, A. L., Chilloux, J., Sarafian, M. H., Rahim, M. B., Boulangé, C. L., and Dumas, M. E. (2015). The microbiome and its pharmacological targets: therapeutic avenues in cardiometabolic diseases. Curr. Opin. Pharmacol. 25, 36–44. doi: 10.1016/j.coph.2015.09.013
Nikolic, D. M., Dimitrijevic-Sreckovic, V., Ranin, L. T., Stojanovic, M. M., Ilic, I. D., Gostiljac, D. M., et al. (2022). Homeostatic microbiome disruption as a cause of insulin secretion disorders. Candida albicans, A new factor in pathogenesis of diabetes: a STROBE compliant cross-sectional study. Medicine (Baltimore) 101:e31291. doi: 10.1097/MD.0000000000031291
Nilsson, R. H., Ryberg, M., Abarenkov, K., Sjökvist, E., and Kristiansson, E. (2009). The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol. Lett. 296, 97–101. doi: 10.1111/j.1574-6968.2009.01618.x
Niu, C., Tu, Y., Jin, Q., Chen, Z., Yuan, K., Wang, M., et al. (2023). Mapping the human oral and gut fungal microbiota in patients with metabolic dysfunction-associated fatty liver disease. Front. Cell. Infect. Microbiol. 13:1157368. doi: 10.3389/fcimb.2023.1157368
Niu, H., Zhou, M., Zogona, D., Xing, Z., Wu, T., Chen, R., et al. (2024). Akkermansia muciniphila: a potential candidate for ameliorating metabolic diseases. Front. Immunol. 15:1370658. doi: 10.3389/fimmu.2024.1370658
Noverr, M. C., Phare, S. M., Toews, G. B., Coffey, M. J., and Huffnagle, G. B. (2001). Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun. 69, 2957–2963. doi: 10.1128/IAI.69.5.2957-2963.2001
Noverr, M. C., Toews, G. B., and Huffnagle, G. B. (2002). Production of prostaglandins and leukotrienes by pathogenic fungi. Infect. Immun. 70, 400–402. doi: 10.1128/IAI.70.1.400-402.2002
Nurgeldiyeva, M. J., Hojakuliyev, B. G., and Muhammedov, M. B. (2014). Correlation of atherogenesis with an infection of Candida albicans. Int. J. Clin. Exp. Med. 7, 2137–2143
Ott, S. J., El Mokhtari, N. E., Rehman, A., Rosenstiel, P., Hellmig, S., Kühbacher, T., et al. (2007). Fungal rDNA signatures in coronary atherosclerotic plaques. Environ. Microbiol. 9, 3035–3045. doi: 10.1111/j.1462-2920.2007.01415.x
Pacheco-Yanes, J., Reynolds, E., Li, J., and Mariño, E. (2023). Microbiome-targeted interventions for the control of oral-gut dysbiosis and chronic systemic inflammation. Trends Mol. Med. 29, 912–925. doi: 10.1016/j.molmed.2023.08.006
Pasini, E., Aquilani, R., Testa, C., Baiardi, P., Angioletti, S., Boschi, F., et al. (2016). Pathogenic gut Flora in patients with chronic heart failure. JACC Heart Fail. 4, 220–227. doi: 10.1016/j.jchf.2015.10.009
Pavlovska, O. M., Pavlovska, K. M., Heryak, S. M., Khmil, S. V., and Gorban, N. Y. (2020). Intestinal dysbiosis as a possible predictor of very early preterm labor in pregnant women with metabolic syndrome. J. Med. Life 13, 200–205. doi: 10.25122/jml-2020-0027
Peroumal, D., Sahu, S. R., Kumari, P., Utkalaja, B. G., and Acharya, N. (2022). Commensal fungus Candida albicans maintains a long-term mutualistic relationship with the host to modulate gut microbiota and metabolism. Microbiol Spectr. 10:e0246222. doi: 10.1128/spectrum.02462-22
Qiu, J., Zhao, L., Cheng, Y., Chen, Q., Xu, Y., Lu, Y., et al. (2023). Exploring the gut mycobiome: differential composition and clinical associations in hypertension, chronic kidney disease, and their comorbidity. Front. Immunol. 14:1317809. doi: 10.3389/fimmu.2023.1317809
Ramji, D. P., and Davies, T. S. (2015). Cytokines in atherosclerosis: key players in all stages of disease and promising therapeutic targets. Cytokine Growth Factor Rev. 26, 673–685. doi: 10.1016/j.cytogfr.2015.04.003
Ramos, J. S., Dalleck, L. C., Tjonna, A. E., Beetham, K. S., and Coombes, J. S. (2015). The impact of high-intensity interval training versus moderate-intensity continuous training on vascular function: a systematic review and meta-analysis. Sports Med. 45, 679–692. doi: 10.1007/s40279-015-0321-z
Rodrigues, C. F., Rodrigues, M. E., and Henriques, M. (2019). Candida sp. infections in patients with diabetes mellitus. J. Clin. Med. 8:76. doi: 10.3390/jcm8010076
Rodriguez, K. A., Gurung, M., Talatala, R., Rearick, J. R., Ruebel, M. L., Stephens, K. E., et al. (2024). The role of early life gut Mycobiome on child health. Adv. Nutr. 15:100185. doi: 10.1016/j.advnut.2024.100185
Roth, G. A., Mensah, G. A., Johnson, C. O., Addolorato, G., Ammirati, E., Baddour, L. M., et al. (2020). Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J. Am. Coll. Cardiol. 76, 2982–3021. doi: 10.1016/j.jacc.2020.11.010
Rozera, T., Pasolli, E., Segata, N., and Ianiro, G. (2025). Machine learning and artificial intelligence in the multi-omics approach to gut microbiota. Gastroenterology 169, 487–501. doi: 10.1053/j.gastro.2025.02.035
Saglietto, A., Manfredi, R., Elia, E., D'Ascenzo, F., DE Ferrari, G. M., Biondi-Zoccai, G., et al. (2021). Cardiovascular disease burden: Italian and global perspectives. Minerva Cardiol. Angiol. 69, 231–240. doi: 10.23736/S2724-5683.21.05538-9
Saklayen, M. G. (2018). The global epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 20:12. doi: 10.1007/s11906-018-0812-z
Salamati, S., Martins, C., and Kulseng, B. (2015). Baker's yeast (Saccharomyces cerevisiae) antigen in obese and normal weight subjects. Clin Obes. 5, 42–47. doi: 10.1111/cob.12079
Saleem, A., Ikram, A., Dikareva, E., Lahtinen, E., Matharu, D., Pajari, A. M., et al. (2022). Unique Pakistani gut microbiota highlights population-specific microbiota signatures of type 2 diabetes mellitus. Gut Microbes 14:2142009. doi: 10.1080/19490976.2022.2142009
Sandek, A., Bauditz, J., Swidsinski, A., Buhner, S., Weber-Eibel, J., von Haehling, S., et al. (2007). Altered intestinal function in patients with chronic heart failure. J. Am. Coll. Cardiol. 50, 1561–1569. doi: 10.1016/j.jacc.2007.07.016
Santelmann, H., and Howard, J. M. (2005). Yeast metabolic products, yeast antigens and yeasts as possible triggers for irritable bowel syndrome. Eur. J. Gastroenterol. Hepatol. 17, 21–26. doi: 10.1097/00042737-200501000-00005
Santus, W., Devlin, J. R., and Behnsen, J. (2021). Crossing kingdoms: how the Mycobiota and fungal-bacterial interactions impact host health and disease. Infect. Immun. 89:e00648-20. doi: 10.1128/IAI.00648-20
Sattar, N., Gill, J. M. R., and Alazawi, W. (2020). Improving prevention strategies for cardiometabolic disease. Nat. Med. 26, 320–325. doi: 10.1038/s41591-020-0786-7
Schei, K., Avershina, E., Øien, T., Rudi, K., Follestad, T., Salamati, S., et al. (2017). Early gut mycobiota and mother-offspring transfer. Microbiome 5:107. doi: 10.1186/s40168-017-0319-x
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 109, 6241–6246. doi: 10.1073/pnas.1117018109
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and Bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Shao, R., Chen, R., Zheng, Q., Yao, M., Li, K., Cao, Y., et al. (2024). Oxidative stress disrupts vascular microenvironmental homeostasis affecting the development of atherosclerosis. Cell Biol. Int. 48, 1781–1801. doi: 10.1002/cbin.12239
Shoukat, M., Ullah, F., Tariq, M. N., Din, G., Khadija, B., and Faryal, R. (2023). Profiling of potential pathogenic candida species in obesity. Microb. Pathog. 174:105894. doi: 10.1016/j.micpath.2022.105894
Soleimani, M., Barone, S., Luo, H., and Zahedi, K. (2023). Pathogenesis of hypertension in metabolic syndrome: the role of fructose and salt. Int. J. Mol. Sci. 24:4294. doi: 10.3390/ijms24054294
Soyucen, E., Gulcan, A., Aktuglu-Zeybek, A. C., Onal, H., Kiykim, E., and Aydin, A. (2014). Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatr. Int. 56, 336–343. doi: 10.1111/ped.12243
Spaggiari, R., Angelini, S., Di Vincenzo, A., Scaglione, G., Morrone, S., Finello, V., et al. (2024). Ceramides as emerging players in cardiovascular disease: focus on their Pathogenetic effects and regulation by diet. Adv. Nutr. 15:100252. doi: 10.1016/j.advnut.2024.100252
Steinert, R. E., Rehman, A., Souto Lima, E. J., Agamennone, V., Schuren, F. H. J., Gero, D., et al. (2020). Roux-en-Y gastric bypass surgery changes fungal and bacterial microbiota in morbidly obese patients-a pilot study. PLoS One 15:e0236936. doi: 10.1371/journal.pone.0236936
Strus, M., Kucharska, A., Kukla, G., Brzychczy-Włoch, M., Maresz, K., and Heczko, P. B. (2005). The in vitro activity of vaginal Lactobacillus with probiotic properties against Candida. Infect. Dis. Obstet. Gynecol. 13, 69–75. doi: 10.1080/10647440400028136
Su, G., Huang, P., Liu, D., Xing, G., Guo, R., Li, S., et al. (2025). Gut mycobiome alterations and network interactions with the bacteriome in patients with atherosclerotic cardiovascular disease. Microbiol Spectr. 13:e0218224. doi: 10.1128/spectrum.02182-24
Sun, S., Sun, L., Wang, K., Qiao, S., Zhao, X., Hu, X., et al. (2021). The gut commensal fungus, Candida parapsilosis, promotes high fat-diet induced obesity in mice. Commun Biol. 4:1220. doi: 10.1038/s42003-021-02753-3
Szóstak, N., Handschuh, L., Samelak-Czajka, A., Tomela, K., Schmidt, M., Pruss, Ł., et al. (2023). Host factors associated with gut Mycobiome structure. mSystems. 8:e0098622. doi: 10.1128/msystems.00986-22
Taha, M. B., Rao, N., Vaduganathan, M., Cainzos-Achirica, M., Nasir, K., and Patel, K. V. (2022). Implementation of Cardiometabolic centers and training programs. Curr. Diab. Rep. 22, 203–212. doi: 10.1007/s11892-022-01459-y
Tan, T. G., Lim, Y. S., Tan, A., Leong, R., and Pavelka, N. (2019). Fungal symbionts produce prostaglandin E2 to promote their intestinal colonization. Front. Cell. Infect. Microbiol. 9:359. doi: 10.3389/fcimb.2019.00359
Tang, W. H. W., Li, D. Y., and Hazen, S. L. (2019). Dietary metabolism, the gut microbiome, and heart failure. Nat. Rev. Cardiol. 16, 137–154. doi: 10.1038/s41569-018-0108-7
Tang, N., Ma, J., Tao, R., Chen, Z., Yang, Y., He, Q., et al. (2022). The effects of the interaction between BMI and dyslipidemia on hypertension in adults. Sci. Rep. 12:927. doi: 10.1038/s41598-022-04968-8
Targher, G., Byrne, C. D., and Tilg, H. (2024). MASLD: a systemic metabolic disorder with cardiovascular and malignant complications. Gut 73, gutjnl-2023-330595–gutjnl-2023-330702. doi: 10.1136/gutjnl-2023-330595
Thomas, H. (2017). Gut microbiota: intestinal fungi fuel the inflammatory fire in alcoholic liver disease. Nat. Rev. Gastroenterol. Hepatol. 14:385. doi: 10.1038/nrgastro.2017.78
Thomas, T., Gilbert, J., and Meyer, F. (2012). Metagenomics - a guide from sampling to data analysis. Microb Inform Exp. 2:3. doi: 10.1186/2042-5783-2-3
Underhill, D. M., and Iliev, I. D. (2014). The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 14, 405–416. doi: 10.1038/nri3684
Van Syoc, E., Nixon, M. P., Silverman, J. D., Luo, Y., Gonzalez, F. J., Elbere, I., et al. (2024). Changes in the type 2 diabetes gut mycobiome associate with metformin treatment across populations. MBio 15:e0016924. doi: 10.1128/mbio.00169-24
Vavreckova, M., Galanova, N., Kostovcik, M., Krystynik, O., Ivanovova, E., Roubalova, R., et al. (2022). Specific gut bacterial and fungal microbiota pattern in the first half of pregnancy is linked to the development of gestational diabetes mellitus in the cohort including obese women. Front. Endocrinol. (Lausanne). 13:970825. doi: 10.3389/fendo.2022.970825
Viebahn, G., Hartmann, P., Lang, S., Demir, M., Zhang, X., Fouts, D. E., et al. (2024). Fungal signature differentiates alcohol-associated liver disease from nonalcoholic fatty liver disease. Gut Microbes 16:2307586. doi: 10.1080/19490976.2024.2307586
Wang, Y., Chen, J., Ni, Y., Liu, Y., Gao, X., Tse, M. A., et al. (2024). Exercise-changed gut mycobiome as a potential contributor to metabolic benefits in diabetes prevention: an integrative multi-omics study. Gut Microbes 16:2416928. doi: 10.1080/19490976.2024.2416928
Wang, P. X., Deng, X. R., Zhang, C. H., and Yuan, H. J. (2020). Gut microbiota and metabolic syndrome. Chin. Med. J. 133, 808–816. doi: 10.1097/CM9.0000000000000696
Wang, C., Liu, Z., Zhou, T., Wu, J., Feng, F., Wang, S., et al. (2025). Gut microbiota-derived butyric acid regulates calcific aortic valve disease pathogenesis by modulating GAPDH lactylation and butyrylation. iMeta 4:e70048. doi: 10.1002/imt2.70048
Wang, S., Ryan, C. A., Boyaval, P., Dempsey, E. M., Ross, R. P., and Stanton, C. (2020). Maternal vertical transmission affecting early-life microbiota development. Trends Microbiol. 28, 28–45. doi: 10.1016/j.tim.2019.07.010
Wang, Y., Wang, X., Wang, C., and Zhou, J. (2024). Global, regional, and National Burden of cardiovascular disease, 1990-2021: results from the 2021 global burden of disease study. Cureus. 16:e74333. doi: 10.7759/cureus.74333
Wang, K., Zhang, Z., Hang, J., Liu, J., Guo, F., Ding, Y., et al. (2023). Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target. Science 381:eadd5787. doi: 10.1126/science.add5787
Wang, W., Zhong, X., and Guo, J. (2021). Role of 2-series prostaglandins in the pathogenesis of type 2 diabetes mellitus and non-alcoholic fatty liver disease (review). Int. J. Mol. Med. 47:114. doi: 10.3892/ijmm.2021.4947
Wang, X., Zhou, S., Hu, X., Ye, C., Nie, Q., Wang, K., et al. (2024). Candida albicans accelerates atherosclerosis by activating intestinal hypoxia-inducible factor2αsignaling. Cell Host Microbe 32:e7, 964–979. doi: 10.1016/j.chom.2024.04.017
Wei, X., Zhang, X., Cui, M., Huang, L., Gao, D., and Hua, S. (2025). An exploration of the relationship between gut Virome and cardiovascular disease: a comprehensive review. Rev. Cardiovasc. Med. 26:36386. doi: 10.31083/RCM36386
Wewege, M. A., Thom, J. M., Rye, K. A., and Parmenter, B. J. (2018). Aerobic, resistance or combined training: a systematic review and meta-analysis of exercise to reduce cardiovascular risk in adults with metabolic syndrome. Atherosclerosis 274, 162–171. doi: 10.1016/j.atherosclerosis.2018.05.002
Wilkerson, J. L., Tatum, S. M., Holland, W. L., and Summers, S. A. (2024). Ceramides are fuel gauges on the drive to cardiometabolic disease. Physiol. Rev. 104, 1061–1119. doi: 10.1152/physrev.00008.2023
Willis, K. A., Purvis, J. H., Myers, E. D., Aziz, M. M., Karabayir, I., Gomes, C. K., et al. (2019). Fungi form interkingdom microbial communities in the primordial human gut that develop with gestational age. FASEB J. 33, 12825–12837. doi: 10.1096/fj.201901436RR
Wolf, D., and Ley, K. (2019). Immunity and inflammation in atherosclerosis. Circ. Res. 124, 315–327. doi: 10.1161/CIRCRESAHA.118.313591
Wong, M. C. S., Huang, J., Wang, J., Chan, P. S. F., Lok, V., Chen, X., et al. (2020). Global, regional and time-trend prevalence of central obesity: a systematic review and meta-analysis of 13.2 million subjects. Eur. J. Epidemiol. 35, 673–683. doi: 10.1007/s10654-020-00650-3
Wu, N., Mo, H., Mu, Q., Liu, P., Liu, G., and Yu, W. (2022). The gut Mycobiome characterization of gestational diabetes mellitus and its association with dietary intervention. Front. Microbiol. 13:892859. doi: 10.3389/fmicb.2022.892859
Wu, J., Singleton, S. S., Bhuiyan, U., Krammer, L., and Mazumder, R. (2024). Multi-omics approaches to studying gastrointestinal microbiome in the context of precision medicine and machine learning. Front. Mol. Biosci. 10:1337373. doi: 10.3389/fmolb.2023.1337373
Wu, J., Wu, D., Ma, K., Wang, T., Shi, G., Shao, J., et al. (2020). Paeonol ameliorates murine alcohol liver disease via mycobiota-mediated Dectin-1/IL-1β signaling pathway. J. Leukoc. Biol. 108, 199–214. doi: 10.1002/JLB.3MA0120-325RR
Wu, X., Xia, Y., He, F., Zhu, C., and Ren, W. (2021). Intestinal mycobiota in health and diseases: from a disrupted equilibrium to clinical opportunities. Microbiome. 9:60. doi: 10.1186/s40168-021-01024-x
Wu, G., Zhao, N., and Zhao, L. (2023). Microbial-host isozyme: a novel target in "drug the bug" strategies for diabetes. Cell Metab. 35, 1677–1679. doi: 10.1016/j.cmet.2023.09.008
Xiang, J. Y., Chi, Y. Y., Han, J. X., Kong, P., Liang, Z., Wang, D., et al. (2022). Litchi chinensis seed prevents obesity and modulates the gut microbiota and mycobiota compositions in high-fat diet-induced obese zebrafish. Food Funct. 13, 2832–2845. doi: 10.1039/d1fo03991a
Xie, Z., Canalda-Baltrons, A., d'Enfert, C., and Manichanh, C. (2023). Shotgun metagenomics reveals interkingdom association between intestinal bacteria and fungi involving competition for nutrients. Microbiome 11:275. doi: 10.1186/s40168-023-01693-w
Xu, A., Deng, F., Chen, Y., Kong, Y., Pan, L., Liao, Q., et al. (2020). NF-κB pathway activation during endothelial-to-mesenchymal transition in a rat model of doxorubicin-induced cardiotoxicity. Biomed. Pharmacother. 130:110525. doi: 10.1016/j.biopha.2020.110525
Xu, J., Zhang, Y., Wang, X., Ren, X., and Liu, Y. (2020). Changes and roles of intestinal fungal microbiota in coronary heart disease complicated with nonalcoholic fatty liver disease. Am. J. Transl. Res. 12, 3445–3460.
Yan, Q., Li, S., Yan, Q., Huo, X., Wang, C., Wang, X., et al. (2024). A genomic compendium of cultivated human gut fungi characterizes the gut mycobiome and its relevance to common diseases. Cell 187, 2969–2989.e24. doi: 10.1016/j.cell.2024.04.043
Yang, Y., Cao, S., Xu, W., Zang, C., Zhang, F., Xie, Y., et al. (2022). Dual modulation of gut bacteria and fungi manifests the gut-based anti-hyperlipidemic effect of Coptidis Rhizoma. Biomed. Pharmacother. 153:113542. doi: 10.1016/j.biopha.2022.113542
Yang, A. M., Inamine, T., Hochrath, K., Chen, P., Wang, L., Llorente, C., et al. (2017). Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest. 127, 2829–2841. doi: 10.1172/JCI90562
Yang, W., Zhou, Y., Wu, C., and Tang, J. (2016). Enterohemorrhagic Escherichia coli promotes the invasion and tissue damage of enterocytes infected with Candida albicans in vitro. Sci. Rep. 6:37485. doi: 10.1038/srep37485
Yin, G., Chen, F., Chen, G., Yang, X., Huang, Q., Chen, L., et al. (2022). Alterations of bacteriome, mycobiome and metabolome characteristics in PCOS patients with normal/overweight individuals. J. Ovarian Res. 15:117. doi: 10.1186/s13048-022-01051-8
You, N., Xu, J., Wang, L., Zhuo, L., Zhou, J., Song, Y., et al. (2021). Fecal Fungi Dysbiosis in nonalcoholic fatty liver disease. Obesity (Silver Spring) 29, 350–358. doi: 10.1002/oby.23073
Yu, M. G., Gordin, D., Fu, J., Park, K., Li, Q., and King, G. L. (2024). Protective factors and the pathogenesis of complications in diabetes. Endocr. Rev. 45, 227–252. doi: 10.1210/endrev/bnad030
Yu, D., Xie, L., Chen, W., Qin, J., Zhang, J., Lei, M., et al. (2022). Dynamics of the gut Bacteria and Fungi accompanying Low-carbohydrate diet-induced weight loss in overweight and obese adults. Front. Nutr. 9:846378. doi: 10.3389/fnut.2022.846378
Yuan, L., Li, Y., Chen, M., Xue, L., Wang, J., Ding, Y., et al. (2024). Therapeutic applications of gut microbes in cardiometabolic diseases: current state and perspectives. Appl. Microbiol. Biotechnol. 108:156. doi: 10.1007/s00253-024-13007-7
Yue, T., Wang, Y., Liu, H., Kong, Z., and Qi, F. (2022). Effects of high-intensity interval vs. moderate-intensity continuous training on cardiac rehabilitation in patients with cardiovascular disease: a systematic review and Meta-analysis. Front Cardiovasc Med. 9:845225. doi: 10.3389/fcvm.2022.845225
Zeng, S., and Schnabl, B. (2024). Gut mycobiome alterations and implications for liver diseases. PLoS Pathog. 20:e1012377. doi: 10.1371/journal.ppat.1012377
Zeng, S., Zhou, M., Mu, D., and Wang, S. (2025). Clinical implications of maternal multikingdom transmissions and early-life microbiota. Lancet Microbe. 6:101042. doi: 10.1016/j.lanmic.2024.101042
Zhan, H., Wan, Y., Sun, Y., Xu, Z., Zhang, F., Yang, K., et al. (2024). Gut mycobiome alterations in obesity in geographically different regions. Gut Microbes 16:2367297. doi: 10.1080/19490976.2024.2367297
Zhang, F., Aschenbrenner, D., Yoo, J. Y., and Zuo, T. (2022). The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe. 3, e969–e983. doi: 10.1016/S2666-5247(22)00203-8
Zhang, Y., Ji, X., Chang, K., Yin, H., Zhao, M., and Zhao, L. (2025). The regulatory effect of chitooligosaccharides on islet inflammation in T2D individuals after islet cell transplantation: the mechanism behind Candida albicans abundance and macrophage polarization. Gut Microbes 17:2442051. doi: 10.1080/19490976.2024.2442051
Zhang, C., Li, L., Zhang, Y., and Zeng, C. (2020). Recent advances in fructose intake and risk of hyperuricemia. Biomed. Pharmacother. 131:110795. doi: 10.1016/j.biopha.2020.110795
Zhang, S., Lin, H., Wang, J., Rui, J., Wang, T., Cai, Z., et al. (2025). Sensing ceramides by CYSLTR2 and P2RY6 to aggravate atherosclerosis. Nature 641, 476–485. doi: 10.1038/s41586-025-08792-8
Zhang, D., Wang, Q., Li, D., Chen, C., Lv, Y., Huang, S., et al. (2024). Different fungal signatures in ALD and MAFLD. Front. Microbiol. 15:1510507. doi: 10.3389/fmicb.2024.1510507
Zhang, Y., Wu, H., Jin, M., Feng, G., and Wang, S. (2025). The gut-heart axis: unveiling the roles of gut microbiota in cardiovascular diseases. Front Cardiovasc Med. 12:1572948. doi: 10.3389/fcvm.2025.1572948
Zhao, Z., Zhong, L., Wu, J., Zeng, G., Liu, S., Deng, Y., et al. (2025). Modulation of gut Mycobiome and serum metabolome by a MUFA-Rich diet in Sprague Dawley rats fed a high-fructose, high-fat diet. Foods. 14:506. doi: 10.3390/foods14030506
Zhao, Z., Zhong, L., Zhou, P., Deng, Y., Liu, G., Li, P., et al. (2024). Impact of dietary fatty acid composition on the intestinal microbiota and fecal metabolism of rats fed a high-fructose/high-fat diet. Nutrients 16:3774. doi: 10.3390/nu16213774
Zheng, R., Xiang, X., Shi, Y., Qiu, A., Luo, X., Xie, J., et al. (2023). Chronic jet lag alters gut microbiome and mycobiome and promotes the progression of MAFLD in HFHFD-fed mice. Front. Microbiol. 14:1295869. doi: 10.3389/fmicb.2023.1295869
Zhou, X. D., Chen, Q. F., Yang, W., Zuluaga, M., Targher, G., Byrne, C. D., et al. (2024). Burden of disease attributable to high body mass index: an analysis of data from the global burden of disease study 2021. EClinicalMedicine 76:102848. doi: 10.1016/j.eclinm.2024.102848
Zhou, S., Li, M., Wang, P., Guo, C., Zhang, J., Luo, X., et al. (2025). A symbiotic filamentous gut fungus ameliorates MASH via a secondary metabolite-CerS6-ceramide axis. Science 388:adp5540. doi: 10.1126/science.adp5540
Zhou, Z., Sun, B., Yu, D., and Zhu, C. (2022). Gut microbiota: An important player in type 2 diabetes mellitus. Front. Cell. Infect. Microbiol. 12:834485. doi: 10.3389/fcimb.2022.834485
Zhou, X., Zhang, X., and Yu, J. (2024). Gut mycobiome in metabolic diseases: mechanisms and clinical implication. Biom. J. 47:100625. doi: 10.1016/j.bj.2023.100625
Zhu, Y., Shi, T., Lu, X., Xu, Z., Qu, J., Zhang, Z., et al. (2021). Fungal-induced glycolysis in macrophages promotes colon cancer by enhancing innate lymphoid cell secretion of IL-22. EMBO J. 40:e105320. doi: 10.15252/embj.2020105320
Zhu, Z., Xiong, S., and Liu, D. (2016). The gastrointestinal tract: an initial organ of metabolic hypertension? Cell. Physiol. Biochem. 38, 1681–1694. doi: 10.1159/000443107
Zou, Y., Ge, A., Lydia, B., Huang, C., Wang, Q., and Yu, Y. (2022). Gut mycobiome dysbiosis contributes to the development of hypertension and its response to immunoglobulin light chains. Front. Immunol. 13:1089295. doi: 10.3389/fimmu.2022.1089295
Keywords: gut mycobiome, fungi, gut microbiota dysbiosis, cardiometabolic diseases, fungal metabolites
Citation: Wei X, Guo Z, Wang J, Gao D, Xu Q and Hua S (2025) Gut mycobiome in cardiometabolic disease progression: current evidence and future directions. Front. Microbiol. 16:1659654. doi: 10.3389/fmicb.2025.1659654
Edited by:
Trina Ekawati Tallei, Sam Ratulangi University, IndonesiaReviewed by:
Muhammad Afzaal, Government College University Faisalabad, PakistanSayantap Datta, University of Houston, United States
Copyright © 2025 Wei, Guo, Wang, Gao, Xu and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Xu, NDA3Mjc5MjQ2QHFxLmNvbQ==; Shengyu Hua, dGcxMjEyMUAxNjMuY29t
†These authors have contributed equally to this work
 Xiaoyu Wei
Xiaoyu Wei Zixin Guo
Zixin Guo Jingyang Wang2
Jingyang Wang2